- Department of Biology, Debre Berhan University, Debre Berhan, Ethiopia
Besides organic compounds such as lactose and proteins, cheese whey is rich in other nutrients. Damping of these valuable compounds to the environment, first, harms the environment, and second, it wastes valuable resources. Therefore, this review aims to find out the current progress on the valorization of cheese whey for ethanol production. Efficient ethanol-producing yeasts like Saccharomyces cerevisiae have no pathway to utilize lactose and, therefore, they can be co-cultured with microbes that can produce β-galactosidase. In addition, chemical, biological, and physical hydrolysis of lactose can be used to produce ethanol from cheese whey. Ethanol production from unsterilized or unpasteurized whey is very promising and this reduces the production cost significantly. This suggests that the ethanol-producing yeasts are competent against the lactic acid bacteria that are commonly found in cheese whey. Despite the presence of central metabolic genes associated with ethanol production from different sugars in some yeasts, these yeasts can’t ferment the different sugars and this is basically due to a lack of the different sugar transport systems in the yeasts. Therefore, additions of different sugars to whey to increase the sugar content for economical ethanol production are impaired by catabolite repressions. However, catabolite repression can be significantly reduced by metabolic engineering by targeting sugar transporter proteins like the major facilitator superfamily (MFS), particularly LAC, CEL2, HGT, RAG, and KHT. Therefore, this enhances ethanol production from cheese whey supplemented with a variety of sugars. Currently, nanoparticles and metal-organic frameworks coated immobilization of S. cerevisiae produced higher ethanol from lignocellulosic substrates than the classical carries such as alginates; however, studies of such immobilizing materials on Kluveromyces spp for ethanol production are very limited, and open for research. Electro-fermentation, an emerging bioprocess to control microbial fermentative metabolism, boosts ethanol production, enables the production of 14% (v/v) ethanol, and shortens the fermentation time of high sugar-containing whey. Generally, utilizing efficient yeast (possibly by adaptive evolution and genetic engineering) at optimal fermenting conditions enabled to production of economical ethanol from cheese whey that contains higher sugars (greater than 15%) at the large-scale cheese processing industries.
1 Introduction
Increasing energy imports, high energy prices, different challenges on petroleum supplies, and greater recognition of the environmental consequences of fossil fuels have driven interest in searching for and utilizing biofuels. Biofuels are any kind of fuel obtained from biological materials. The biofuel includes wood, biogas and biomethane, bioethanol, biohydrogen vegetable oil, biodiesel, bioethers, and bioelectricity. Among these, bioethanol is the dominantly used biofuel for the transport sector. Bioethanol is produced from food-based starch, lignocellulosic feedstock, and different wastes of food processing industries such as beer spent grain, corn steep liquor, molasses, and cheese whey. Bioethanol production from cheese whey is getting attention for two reasons. First, it is rich in nutrients for microbial fermentations. Second, disposing of cheese whey to the environment brings serious pollution since the whey has high biological oxygen demand (BOD) and chemical oxygen demand (COD), exceeding the standard limits set by the national and international regulatory bodies (Azzouni et al., 2019; Sebastián-Nicolás et al., 2020; Sáenz-Hidalgo et al., 2021). More than 90% of the COD and BOD of chees whey is contributed by lactose and higher than 50% of cheese whey is released into the environment without treatment (Asunis et al., 2020). Treating cheese whey is a very expensive process and it needs to valorize the cheese whey with a value-adding fermentation process that produces different biological products such as biofuels, lactic acids, biopolymers, bioelectrochemicals, volatile fatty acids, distilled whey-based sprit (whey vodka), and whey vinegar (Díez-Antolínez et al., 2018; Asunis et al., 2020; Zotta et al., 2020; Sáenz-Hidalgo et al., 2021).
Worldwide production of cheese whey is estimated to be 190 billion kg per year (Asunis et al., 2020), showing a 1%-2% annual growth rate (Sharma et al., 2018). In 2015, Dairy Farmers of America (USA) and Anchor ethanol Ltd. (New Zealand) are producing eight million gallons per year of ethanol from whey fermentation (Ryan and Walsh, 2016). According to the 2023 report of the Renewable Fuels Association (RFA), the global fuel ethanol production reaches 28 Billion gallons in 2022, of which a major share was contributed by the United States of America (55%, 15.4 Billion gallons) and Brazil (26%, 7.42 Billion gallons), the remaining 19% was contributed by the rest of the world (Europe, Asia, Canada, Latin America, and others) (RFA, 2023). The African contribution to the world ethanol market is insignificant. RFA (2023) also reported that in 2022, many ethanol producers make progress toward net-zero carbon emissions by promoting ethanol production and utilization by 2050 or sooner. In the United States of America, more than 94% of ethanol was produced from corn starch whereas Brazil produces it from sugar cane and this brings competition for food since the world, particularly Africa and Asia, is suffering from food insecurity. Utilizing wastes like cheese whey and molasses rich in sugar for ethanol production contributes its part to mitigating these drawbacks.
Microorganisms are the heart of fermentations to produce all these valuable products from cheese whey and microorganisms that ferment lactose are economically important to valorize cheese whey. Among different microorganisms including lactic acid bacteria, fungi, and yeast that produce ethanol from cheese, yeasts are superior to bacteria and fungi to produce ethanol at the industry level (Tesfaw and Assefa, 2014; Valdez Castillo et al., 2021). The ability to grow in lactose as a sole carbon is the main feature required to select yeasts for cheese fermentation targeting ethanol production. The most studied microorganisms of ethanol-producing yeasts from lactose include Kluyveromyces lactis, Kluyveromyces marxianus, and Candida tropicals (de Carvalho et al., 2020; Sampaio et al., 2020; Tesfaw et al., 2021b; Valdez Castillo et al., 2021). Debaryomyces hansenii, Lachancea thermotolerans, Kluyveromyces waltii, C. pseudotropicalis, and K. fragilis are also known to ferment the lactose present in the whey (Dragone et al., 2011; Farkas et al., 2019; Valdez Castillo et al., 2021). However, the lactose fermenting yeasts are less tolerant to ethanol and they are highly sensitive to oxygen since they are Crabtree negative, and genetic manipulations and adaptive evolution are commonly used to increase ethanol tolerance capability and reduce oxygen sensitivity (Mo et al., 2019; Karim et al., 2020; Dekker et al., 2021).
The lactose content of cheese whey (maximum of 5% w/v) is not sufficient to produce economical ethanol so concentrating the lactose of cheese via a vacuum evaporator or distillation or/and adding external cheap carbon sources are used to solve such kind of limitations (Sampaio et al., 2020; Tesfaw et al., 2021b; Leonel et al., 2021; Zou and Chang, 2022). However, the addition of external non-lactose or non-glucose substrates results in catabolite repression although metabolic engineering was employed to allow the entrance of mixed sugars without being impaired by the presence of glucose (Kwon et al., 2020; Murata et al., 2022).
In this paper, the microbial processes useful for the production of economical ethanol from cheese whey are discussed. The whey stabilization methods, the ethanol tolerance of Kluyveromyces spp, the effect of nutrient supplementations on cheese whey, and yeast growth variables were considered in this review. The potential of recently introduced techniques such as nanoparticle and metal-organic-frame coated immobilization, electro-fermentation, and electro-activated whey utilization for enhancing ethanol production is also discussed.
2 Cheese whey
Cheese whey is the watery portion formed during the coagulation of casein in cheese making; it is a byproduct of the dairy industry. Whey is often considered a waste because it has a high organic load (Pescuma et al., 2015). Utilizing whey to produce ethanol is biologically important to treat the wastes besides the ethanol produced.
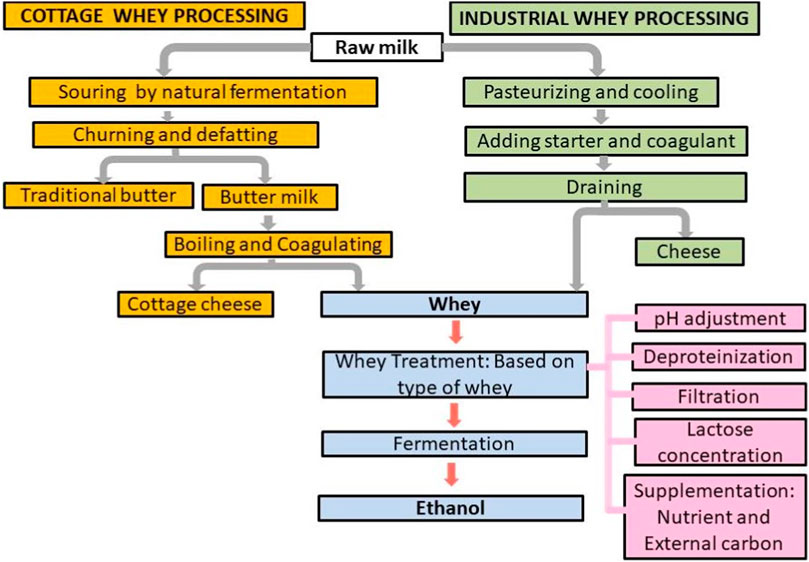
The types and properties of cheese whey depend on the type of milk and milk sources (sheep, goat, cow, or buffalo) used for cheese production, the method employed for casein coagulation, and the fermentation conditions of cheese production (Figure 1). Traditionally, the milk is fermented by the natural undefined microorganisms existing in the milk which are introduced into the milk during milk processing, and cow handling and the natural microorganisms acidify the milk for cheese making. However, industrially, defined starter cultures are added to bring the required cheese properties and aroma (Figure 1). As a result, the type of whey drained off varies. In addition, the amount of water, detergents, and sanitizing agents used to clean the container of the milk or cheese whey also affects the composition of cheese whey (Asunis et al., 2020). The technology used to produce the whey significantly affects the chemical composition of the whey (Liu et al., 2016). The composition of the whey varies with the composition of the milk, the type of cheese made, and the cheese-making process employed (Koushki et al., 2012). In addition, the composition of cheese whey depends on milk quality, animal feed, and animal breed (Pasotti et al., 2017). Broadly speaking, the two types of cheese whey are made. First, acidic whey (pH 4.5–5) is produced by fermentation (direct acidification of milk through the activity of lactobacilli) or/and the addition of organic acids or minerals; second, sweet whey (pH 5-6) is obtained by the adding proteolytic enzymes (enzymatic coagulation process with rennet containing the protease chymosin) (Liu et al., 2016; Sáenz-Hidalgo et al., 2021).
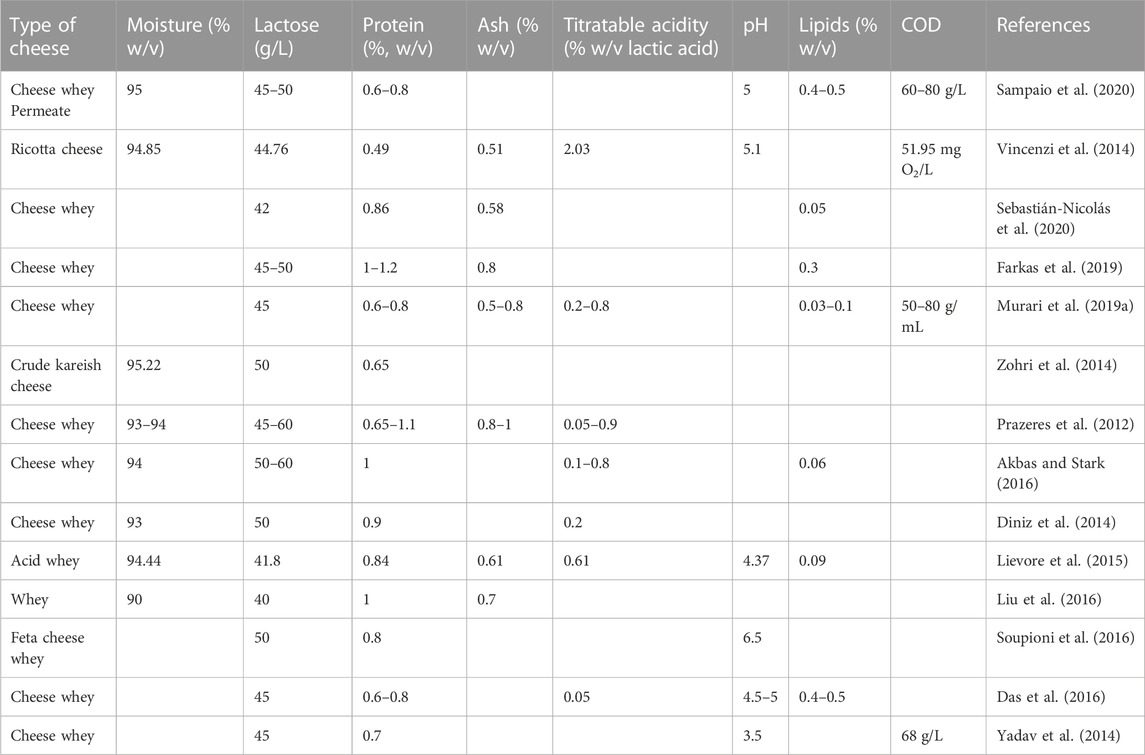
In general, 55% of milk nutrients such as lactose, vitamins, and minerals (Sáenz-Hidalgo et al., 2021) and 20% of milk proteins (Asunis et al., 2020) are retained in cheese whey. Table 1 shows the composition of various cheeses used for ethanol production and the lactose content ranges from 4.18% to 6% depending on the type of cheese. The lactose content of whey is between 4.5%–5.0% (Ariyanti and Hadiyanto, 2013; Pescuma et al., 2015), and 5%-6% (Christensen et al., 2011). The lactose content in acidic whey is smaller than in sweet whey since lactose is fermented into lactic acid in acidic whey (Pescuma et al., 2015). It is lactose that is the rate-limiting step in ethanol production from whey and the presence of lactose in whey (the sole carbon source for ethanol production) limits the growth of other microorganisms (Ariyanti and Hadiyanto, 2013). The whey was to be used directly for ethanol production soon after it was made. The cold preservation of whey can elongate not more than 2 weeks. The lactic acid bacteria and other microbes in the whey significantly reduce the lactose content of the whey when stored for more than 2 weeks (Tesfaw et al., 2021b). They also reported that 20%–25% of lactose was reduced within 2 weeks even if it was preserved in the refrigerator at 4°C and reducing lactose reduced ethanol concentration by 50%.
In most whey, the protein content ranges from 0.8% to 1.0% (Christensen et al., 2011; Pescuma et al., 2015). In Table 1, the protein content of the cheese whey was found to be between 0.49% and 1.1%; most of the researchers reported a protein content range of 0.8%–1%. Acid whey contains lower levels of proteins than sweet whey (Lievore et al., 2015). The fat present in whey varies significantly (Akbas and Stark, 2016; Farkas et al., 2019). Christensen et al. (2011) reported 0.06% of fats in whey whereas Koushki et al. (2012) found 3% fat. In the other study, Farkas et al. (2019) reported 0.3% (w/v) lipids in the 50% solids retained in the whey. The lactic acid present in sweet and acid whey was 0.00% up to 0.8% respectively (Pescuma et al., 2015). In addition, the whey also contained citric acids, minerals, B group, and vitamins non-protein nitrogen sources in small quantities (Pescuma et al., 2015). The ash content of the whey is found to be between 0.51 and 1.0 (%, w/v) (Table 1). Acid whey contains a higher ash content than sweet whey (Lievore et al., 2015).
The whey and whey permeate physicochemical properties are being improved by the electro-activation technology (Karim and Aïder, 2020; Aidarbekova and Aider, 2021; Aider-Kaci et al., 2023) and it is a method of treating the whey with an external electric field inside anion–cation exchange membranes modulated reactor. Single-cell protein, organic acids, aroma volatiles, lactulose, and other metabolites have been produced from electro-activated whey and whey permeates by K. marxianus and bacteria including Lacticaseibacillus, Lactobacillus, Bifidobacterium, and Streptococcus. However, bioethanol production from electro-activated whey is at an infant stage (Kareb et al., 2018). Karim and Aider (2022) studied the potentials of electro-activated cheese whey for ethanol production and the highest ethanol was obtained at non-electro-activated whey suggesting further research on the ethanol production capability of electro-activated whey.
3 Ethanol-producing microorganisms from lactose
Although the yeasts that aerobically assimilate lactose are widespread, few ferment it (Tavares and Malcata, 2016). Kluyveromyces species have been used extensively for bioethanol production from cheese whey. These include K. fragilis (Dragone et al., 2011; Akbas and Stark, 2016), K. marxianus (Zhou et al., 2019; Zotta et al., 2020; Tesfaw et al., 2021b; Zou and Chang, 2022), and K. lactis (Ariyanti and Hadiyanto, 2013; Das et al., 2016). Besides Kluyveromyces, some strains of Candida species like C. pseudotropicalis (Ariyanti and Hadiyanto, 2013; Akbas and Stark, 2016; Das et al., 2016), C. krusei (Ikegami et al., 2009), C. inconspicua, and C. xylopsoci (Azzouni et al., 2019; Farkas et al., 2019) and C. kefyr (Koushki et al., 2012; Akbas and Stark, 2016) are also known for their ability to produce ethanol from lactose. Hoshida and Akada (2017) isolated Pichia kudriavzevii from kefir, and the yeast was found to ferment lactose; however, the yeast is a slow grower in lactose-containing media.
K. marxianus and K. lactis are most commonly employed for ethanol production since they are efficient and tolerant to lower pH of whey (Roohina et al., 2016). Despite their phylogenetic closeness, K. marxianus is involved in better fermentative metabolism than the respiring K. lactis, especially at higher temperatures and K. marxianus is superior to K. lactis for ethanol production (Zotta et al., 2020). K. marxianus is thermotolerant yeast with high growth yield and β-galactosidase activity. It is a good candidate to produce ethanol from whey even at high temperatures. It was a suitable microorganism for producing ethanol from lactose fermentation, showing a maximum alcohol production efficiency of 96.5% (Pescuma et al., 2015). K. marxianus consumes all lactose present in whey within 16 h (Ariyanti and Hadiyanto, 2013), indicating its efficiency in ethanol production. In the other study, the yeast K. lactis NRRL Y-8279 was found to be more efficient than K. marxianus ATCC 36907 for the co-production of β-galactosidase and ethanol using “coalho” cheese whey as substrate (de Carvalho et al., 2020).
Besides Kluyveromyces species, Candida species (C. kefir, C. inconspicua, and C. xylopsoci) are promising in lactose hydrolysis and ethanol production although the conversion capacity of the sugars into ethanol is lower in Candida species (Azzouni et al., 2019; Farkas et al., 2019) and Candida species can utilize higher levels of lactose compared to Kluyveromyces species (Das et al., 2017; Sáenz-Hidalgo et al., 2021). In addition, many nutritional requirements of Candida species (which is fastidious) make ethanol production by Candida less competitive. Furthermore, most of the Candida species are very sensitive to ethanol concentrations, i.e., inhibited by low ethanol concentrations with a low conversion yield (40%) (Tavares and Malcata, 2016).
Kluyveromyces species are inhibited by moderate sugar and salt concentrations in whey and show low ethanol tolerance compared to S. cerevisiae (Akbas and Stark, 2016). Utilizing S. cerevisiae to produce ethanol from whey containing lactose makes the production process economical. Nevertheless, S. cerevisiae lacks the assimilatory pathway to convert lactose in whey directly into ethanol. However, ethanol can be produced by S. cerevisiae after the whey is treated with β-galactosidase to convert lactose into glucose and galactose (Tomaszewska and Białończyk, 2016). In addition to pre-hydrolysis via chemical or biological methods (Tomaszewska and Białończyk, 2016; Zhou et al., 2019; Schmidt et al., 2020), protoplast fusions of S. cerevisiae and Kluyveromyces spp, exogenous expression of the lactose hydrolase gene in S. cerevisiae (genes from E. coli, A. niger, and Kluyveromyces spp) are strategies to confer lactose utilization to S. cerevisiae (Zou and Chang, 2022). In addition, Lactococcus lactis has been engineered to produce ethanol from lactose present in whey; however, many nutritional requirements (being fastidious nature) are the main drawbacks of utilizing L. lactis for economically competitive ethanol production (Liu et al., 2016).
Besides yeasts and genetically modified bacteria and yeast, some filamentous fungal species such as Aspergillus oryzae, Neurospora intermedia, and Neolentinus lepideus can naturally utilize lactose and produce ethanol using cheese whey as a carbon source although ethanol concentration was lower than that of the yeasts (Sar et al., 2022). N. lepideus could produce ethanol not only from glucose, galactose, and lactose but also from the five-carbon sugar xylose (Okamoto et al., 2019). Furthermore, Aspergillus niger was able to produce optimal ethanol from lactose-dominating lactoserum at a pH of 5.9 and a temperature of 30°C hence it can produce ethanol from sweet whey (Azzouni et al., 2019). In addition to its ability to produce ethanol from whey, the fungus can also be used to produce ethanol from chemically as well as biologically hydrolyzed lignocellulose. The advantage of ethanol production by N. lepideus is its ability to ferment the lactose in the presence of high levels of calcium, the mineral commonly found in whey (Okamoto et al., 2019). They also reported that N. lepideus can also ferment milk-rich fats to produce ethanol.
4 Ethanol fermentation
Different yeasts produce ethanol under different conditions (Table 2). Ethanol can be produced from non-deproteinized, non-diluted, and non-sterilized whey (Zafar and Owais, 2006; Tesfaw et al., 2021b). K. marxianus was capable of maintaining high productivity at low pH in non-sterilized whey. It was found that K. marxianus is a very robust microorganism capable of producing ethanol at high temperatures and low pH in whey by taking over lactic acid bacteria present in the whey (Christensen et al., 2011). Continuous fermentation results in higher ethanol production and reduces fermentation time than batch processes (Gabardo et al., 2016; Asunis et al., 2020).
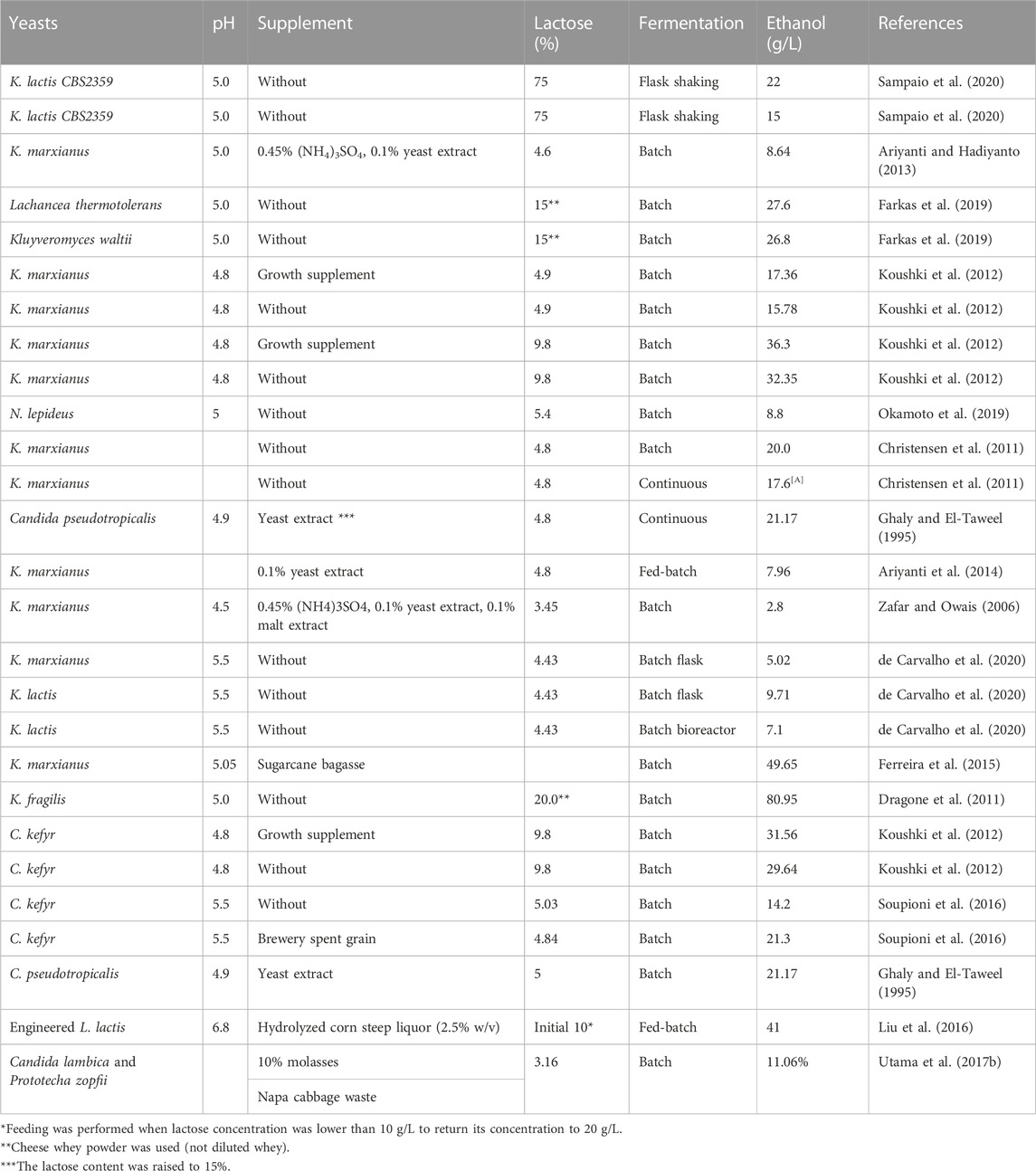
TABLE 2. Bioethanol production from different types of cheese whey with varying pH, supplements, lactose concentration, and fermentation conditions.
5 Pasteurization versus unpasteurized whey
Pasteurization or sterilization of cheese whey increases industrial ethanol production time and cost. Various researches have been conducted to determine the effects of unpasteurized cheese whey on ethanol production. The possible microbes that predominantly flourish the cheese whey are lactose-consuming bacteria, particularly lactic acid bacteria. In Christensen et al. (2011) study, K. marxianus was able to produce 22 g/L ethanol from unpasteurized cheese whey containing 46 g/L lactose via batch fermentation. This suggests that K. marxianus can take over the lactic acid bacteria added during cheese production. However, if the lactic acid bacteria dominate the cheese whey before ethanol fermentation, the lactose content of the cheese whey drops significantly reduces and with it the ethanol production (Tesfaw et al., 2021b).
6 Growth conditions for ethanol-producing yeasts from lactose
Ethanol production is highly dependent on the growth conditions of the yeast of interest. Temperature, pH, incubation time, salinity, solid load, and carbon source concentration are the main factors responsible for bioethanol production from whey. Diniz et al. (2014) prioritized the growth factors and concluded that temperature was the most important factor affecting ethanol production from chees whey using K. marxianus UFV-3, followed by pH, inoculum size, and lactose concentration.
6.1 Temperature
Reduction of cooling costs, decreasing the risk of contamination, nonstop switching from fermentation to distillation, and simultaneous saccharification and fermentation are the benefits of high-temperature fermentation to produce ethanol (Karim et al., 2020). The ability of K. marxianus to grow and ferment optimally at higher temperatures has received attention to this day, as this facilitates cooling during large-scale fermentations where heat transfer has been shown to be a limiting factor.
K. marxianus was able to produce a maximum of ethanol (6-7 g/L) at 30–35°C in fed-batch culture; however, ethanol production declined significantly at 40°C due to the inactivity of enzymes (Ariyanti et al., 2014). Similarly, a temperature range of 32.5°C–35.0°C was found to be optimal for the production of ethanol from cheese whey by K. marxianus URM 7404 (Murari et al., 2019b). Ferreira et al. (2015) reported that optimal ethanol products (49.65 g/L) were obtained from K. marxianus on ricotta whey containing hydrolysates from sugar cane bagasse at pH 5.05, agitation of 65 rpm, and a temperature of 40°C. However, the decline in ethanol production by K. marxianus occurred significantly at 40°C, possibly due to the reduction of activity or/and inactivity of enzymes (Ariyanti et al., 2014). In the other study, Zou and Chang (2022) reported that some strains of K. marxianus are highly thermotolerant, able to grow and produce ethanol at 43°C with lactose and/or whey permeate as the sole carbon source. Similarly, Leonel et al. (2021) reported that temperature ranges of 44 and 52°C are the distinctive feature of K. marxianus. A few other studies reported different but thermophilic optimal temperatures for optimal ethanol production; for example, K. marxianus was capable to produce higher ethanol at a temperature of 52°C (Mo et al., 2019), 45–52°C (Li et al., 2018), and 47°C (da Silva et al., 2018).
The benefits of high-temperature fermentation generally include reduced cooling cost, less contamination, more viscous fermentation broth, and an optimal condition for most enzymes (Hoshida and Akada, 2017). As the study by Hoshida and Akada (2017) shows, the ability of K. marxianus ETP87 to produce ethanol from whey at 45°C is promising to apply the yeast for ethanol production from warm whey before it is dominated by lactic acid bacteria (Tesfaw et al., 2021b). However, the higher temperature intensifies the inhibitory effect of other factors such as ethanol and salt.
Among Candida species, C. inconspicua and C. xylopsoci efficiently produced ethanol from lactoserum at temperatures of 38°C and 42°C. The optimal temperature and pH for C. kefyr are 30°C and 4.8 for ethanol production from cheese whey (Koushki et al., 2012). Kluyveromyces species are more thermotolerant than Candida species.
6.2 pH
The deviation of the external pH from its optimal state strongly affects the energy production process, enzymatic activity, other protein structures, cell membrane permeability, and metabolite transport in yeast cells (Orij et al., 2011; Peña et al., 2015). However, yeast cells developed different mechanisms including pH compartmentalization, control of cytosolic pH by proton translocating ATPases, control of cytosolic pH by cation transporters, and regulation of organellar pH to adapt to the deviation of the pH (Orij et al., 2011).
The pH of cheese whey ranges from 3.5 to 6.5 (Table 1); the ethanol-producing yeasts from cheese whey have to tolerate this pH. Even though K. marxianus tolerate a wide range of pH 2.5–9 (Ha-Tran et al., 2021), optimal ethanol is produced at pH 5-6 (Table 2). The ethanol yield from whey of K. marxianus URM 7404 is comparable with theoretical values at pH ranging from 4.8 to 5.3 (Murari et al., 2019b). C. inconspicua and C. xylopsoci were able to produce optimal ethanol from lactoserum at pH 4.5 and 4 (Azzouni et al., 2019). The lower pH optimality is remarkable for the produce ethanol from cheese whey at the industrial level since it is not necessary to adjust the most acidic pH of cheese whey. In the other studies, the cheese whey is mostly acidic and ethanol can be efficiently produced without controlling the pH of the cheese whey (Christensen et al., 2011; Tesfaw et al., 2021b). Similarly, an ethanol concentration of 7.96 g/L was produced by K. marxianus from non-pH-adjusted acidic whey supplemented with 0.1% yeast extract using a fed-batch fermentation type (Ariyanti et al., 2014). When ethanol is produced from non-pH adjusted whey (particularly when the pH is below 4.0), it takes a longer fermentation time since some time is elapsed for the lower pH adaptation, a longer lag phase (Mohd Azhar et al., 2017).
6.3 Incubation time
Solid particles present in whey adversely affect the ethanol fermentation process, particularly the fermentation time. The rate of lactose utilization and ethanol formation increased linearly with increasing whey solids content (Koushki et al., 2012). If the whey is not concentrated or an external carbon source is added, all lactose present in the whey can be consumed by Kluyveromyces within 12 h (de Carvalho et al., 2020) and 16 h (Ariyanti and Hadiyanto, 2013) for ethanol production and thus waiting for more incubation time reduces the ethanol titers since the Kluyveromyces spp utilize ethanol as a carbon source during carbon source deficiency. Another study showed that lactose utilization began within 24 h and was completed after 72 h (Christensen et al., 2011). Generally, the incubation time will be shorter when other growth variables are in optimal conditions. The existence of different sugars (Beniwal et al., 2017) and the lower pH (mainly when it is less than 4.0) (Mohd Azhar et al., 2017) elongate the incubation time.
6.4 Oxygen concentration
The fermentation performance of whey lactose by Kluyveromyces is potentially affected by oxygen availability. Oxygen is one of the factors responsible for sugar consumption preference. For example, only under high aeration lactose consumed first, followed by glucose and fructose when K. lactis was grown on a medium containing carobs sugars mixed with whey (Rodrigues et al., 2016). Similarly, sufficient ethanol is produced from high lactose media when the oxygen concentration increases; no ethanol was produced from high lactose under anaerobic conditions (Diniz et al., 2014). Generally, the oxygen concentration preference of Kluyveromyces spp is wide since Kluyveromyces spp yeasts can perform fermentation and respiratory processes simultaneously (de Carvalho et al., 2020).
However, a few strains of K. marxianus NCYC3396 and UFV-3 displayed good growth under anaerobic conditions (Madeira-Jr and Gombert, 2018). The ability of yeast to grow in anaerobic conditions increases when other growing conditions become optimal. Besides being the final electron acceptor, oxygen is also involved in the synthesis of fatty acids, heme proteins, nicotinic acids, ergosterol, and deoxyribonucleic acids (Madeira-Jr and Gombert, 2018).
The low oxygen concentration initiated a decrease in growth rate and an increase in ethanol concentration compared to anaerobic fermentation (Leonel et al., 2021). Ethanol was already detected at oxygen saturation level around 80%, an expected behavior, that may have been attributed to some catabolite repression at glucose concentrations between 10 and 20 g/L (Fonseca et al., 2013). The oxygen demand increases when the lactose concentration in the media increases (Guimarães et al., 2010). Generally, a high concentration of ethanol is produced under anaerobic conditions, although a low concentration of oxygen is required for the production of for sterol and other metabolites (Murari et al., 2019a).
6.5 Crabtree effect
The Crabtree effect, also known as aerobic fermentation, allows the yeast to grow and ferment higher rates with high sugar and oxygen availability. Nevertheless, the lactose fermenting yeasts such as K. marxianus and K. lactis are Crabtree-negative yeasts (Karim et al., 2020), suggesting the inability to efficiently ferment sugars to ethanol, resulting in lower ethanol. The inability of K. marxianus to ferment in the absence of oxygen is associated with the absence of a functional sterol uptake mechanism as a key factor in oxygen demand (Radecka et al., 2015). Genetic manipulation enabled K. marxianus to grow in the complete absence of oxygen. Heterologous expression of a squalene-tetrahymanol cyclase allowed the oxygen-independent synthesis of the sterol surrogate tetrahymanol in K. marxianus (Dekker et al., 2021) and ethanol production from lactose could be maximized since the aerobic respiratory that utilizes the lactose is declined. Generally, enabling K. marxianus to grow anaerobically represents an important step towards the application of this thermotolerant yeast in large-scale anaerobic bioprocesses.
Under aerobic conditions, K. marxianus boosts acetyl-CoA biosynthesis from pyruvate (evidenced by an increase in the transcript abundances of the PDA1 and LAT1 genes contributing to the rise of acetyl-CoA synthesis under aerobic conditions) and utilizes the oxidative TCA cycle; a decrease in glucose uptake rate (evidenced by decreased hexokinase activity) has also been reported; these effects results in strong suppression of ethanol production (Sakihama et al., 2019). Furthermore, aeration increased the mRNA expression levels of K. marxianus ALD2, ADH2, and ACS1 responsible the for the assimilation of ethanol to acetyl-CoA (Sakihama et al., 2019).
Deleting genes that favor aerobic respiration enhances ethanol production. For example, deleting KlNDI1, a gene encoding the single internal NADH: ubiquinone Oxidoreductase, in the mitochondria of K. lactis (KlNdi1p), shifted the respiratory to fermentative metabolism and this enhanced the performance of the Δklndi1 strain (mutant) for bioethanol production from lactose present in cheese whey (González-Siso et al., 2015).
6.6 Salt concentration
Salting is a common practice in cheese-making for various reasons. Adding salt helps to enhance flavor, dry the curd, develop a good rind, and control bacteria and other harmful microbes in the cheese. Sodium chloride (NaCl) followed by calcium chloride (CaCl2) is the most common salt employed in cheese processing (Lukjanenko et al., 2014). Depending on its origin and processing type, whey contains different concentrations of salts (mainly NaCl and CaCl2). Sweet whey contains lower salt than acidic whey. Blaschek et al. (2007) reported an average salt range of 3.1%–4.8% in whey; high salt content and increased processing and disposal costs resulted in the underutilization of salty whey in the dairy industry. The effect of salt concentration (2, 4, 6, 8, and 10% NaCl) on the growth and bioethanol production by K. marxianus strains was studied by Ertuğrul Karatay et al. (2019) and optimal ethanol was reported to be 4% NaCl. Further increment of the salt concentration decreased the bioethanol concentration. The decrement of ethanol productivity at higher salt concentrations could be associated with osmotic effects. Salt decreased the process efficiency of ethanol production (Tomaszewska and Białończyk, 2016). Similarly, the addition of NaCl and Na2SO4 to cheese whey harmed growth and consequently reduced the ethanol titer by more than 70% (Sayed et al., 2018).
The yeasts exhibited different sensitization with varying salts. For example, K. marxianus is more sensitive to sodium than to potassium (Illarionov et al., 2021). The negative correlation between the salt concentration and the ethanol production rate was found in the concentration range of 10–40 g/L of both CaCl2 and NaCl (Lukjanenko et al., 2014). There was no significant difference in ethanol production from cheese whey containing lower salts (less than 2 g/L) compared to chees whey without external salts, suggesting that the cheese whey contained sufficient NaCl and CaCl2 required for the growth of K. marxianus. The effects of salts on ethanol synthesis were more pronounced at higher lactose concentration (Lukjanenko et al., 2014) or other non-optimal growth conditions.
7 Cheese whey as a nitrogen source during bioethanol production from different carbon sources
Demiray et al. (2016) studied whey, (NH4)2SO4, and soy wheat as nitrogen sources to produce ethanol from dilute acid (1.5%) hydrolyzed carrot pomace using Pichia stipitis and they found higher ethanol content from cheese whey supplemented carrot pomace hydrolysate than soy wheat. However, using (NH4)2SO4 as a nitrogen source yielded more ethanol than cheese whey even if (NH4)2SO4 is more expensive than its organic counterparts like cheese whey and soy wheat. In Ferreira et al. (2015) study, the mixture of ricotta cheese and sugar cane bagasse was fermented by thermotolerant K. marxianus CCT 7735 to produce bioethanol. The results demonstrated that a mixture of the cellulosic residue of sugarcane bagasse and ricotta whey is promising for ethanol production because the ethanol yield in the mixture was higher than that in the single substrate of sugarcane bagasse.
8 Supplements to whey
Distillation cost during dawn-stream processing makes ethanol production less economically competitive. At least 4% (w/w) ethanol should be produced to reduce distillation costs (Liu et al., 2016; Díez-Antolínez et al., 2018). On the one hand, ultrafiltration or/and reverse osmosis could be used to concentrate the lactose; however, this still increases production costs. On the other hand, since the lactose content of most whey is not higher than 5% (w/v) (Tables 1, 2), different cheap carbon sources supplementation to whey is highly appreciated to increase the carbon source concentration. In addition, growth factors and mineral fortifications to whey also improve ethanol productivity since they improve the health of ethanol-producing microbes during ethanol fermentation. Generally, different supplements were added to cheese whey with varying effects on ethanol titer (Table 2), and the choice of supplements depends on the type of cheese whey deficient in specific nutrients. Nutritional supplementation (other than lactose) becomes more significant to cheese permeate than cheese whey for good-performance ethanol production because some nutrients might be lost or not available during cheese permeate-making (Murari et al., 2019a). Adding growth factors to whey could also enhance ethanol production (Table 1). Even if the addition of extraneous sugars from different sources raises the ethanol content significantly, the supplementation of yeast extract, malt extract, and ammonium sulfate does not significantly increase ethanol productivity significantly (Koushki et al., 2012).
8.1 Molasses
Despite the abundant availability and low cost of cheese whey, ethanol production from whey is economically less competitive due to the low lactose content compared to other wastes such as molasses or/and corn (Díez-Antolínez et al., 2018). Supplementation of different carbon sources, mainly molasses, and lactose is commonly recommended to boost the ethanol titer. Ethanol production from whey can be enhanced by adding supplemental sugar sources. For example, the adding of extraneous lactose and sucrose to whey increases ethanol yield (Tomaszewska and Białończyk, 2016).
Ertuğrul Karatay et al. (2019) studied the effect of the addition of 6, 8, 10, 10, 12, and 14% molasses to whey and found that the maximum ethanol (7.92 g/L) at 10% molasses than a lower and higher percentage of molasses and whey alone. Ethanol productivity decreased when more than 10% molasses was supplemented with whey (Ertuğrul Karatay et al., 2019) and this might be due to substrate inhibition. However, the impacts of high sugar concentration can be solved by using a fed-batch fermentation strategy (Liu et al., 2016). However, Tesfaw et al. (2021b) reported optimal ethanol production (17.28 g/L) from fortification with 25% molasses; however, the 25% molasses was diluted five-fold. Balia et al. (2018) studied ethanol production from the combination of Mozzarella cheese whey and sugar cane molasses using indigenous yeasts and high ethanol content (8.49%) was obtained when 15% molasses was added to Mozzarella cheese whey and fermented by indigenous mixed culture of C. tropicalis and Blastoschizomyces capitatus at an equal ratio. The existence of different kinds of sugars in whey creates lag phases after the consumption of each kind of sugar, further which increases elongates the fermentation time of ethanol (Beniwal et al., 2017).
8.2 Lignocellulosic feedstock
Lignocellulosic hydrolysates are the cheapest and most abundant sources of carbon. Blending of cheese whey with lignocellulosic hydrolysates can be used to increase the carbon content of the whey. Some strains of K marxianus utilize and ferment the pentose sugars, dominantly found in sugar in lignocellulosic hydrolysates (Tesfaw et al., 2021a; Leonel et al., 2021), making the yeast an ideal candidate for the production of ethanol from whey mixed with lignocellulosic feedstock. In addition, some strains of K. marxianus produce ethanol from xylose under aerobic conditions. However, mixing the chemically hydrolyzed lignocellulose with whey brings the inhibitors and reduces ethanol productivity (Tesfaw et al., 2021a; Leonel et al., 2021). The whey mixed with hydrolysates of sugar cane bagasse, spent breweries grain, and corn cob resulted in high ethanol titer (Zou and Chang, 2022).
8.3 External lactose
The challenge of adding molasses and sugars other than lactose or glucose to cheese whey is that the diauxic growth occurs when different carbon sources are present in the cheese whey (Sar et al., 2022). Supplementing lactose as a carbon source solves such a challenge, although adding pure lactose increases production costs. The concentration of lactose has a significant impact on the production of ethanol. Roohina et al. (2016) studied the effect of supplementation of lactose concentration ranging from 42 to 150 g/L to chees whey and found that a lactose addition of 70 g/L lactose to whey enhanced ethanol production compared to the raw cheese whey (42 g/L lactose). However, raising the lactose concentration higher than 70 g/L significantly decreased ethanol production due to possible substrate limitations. The high lactose concentration results in high osmotic pressure and this exerts an inhibitory effect on the less tolerant K. marxianus. In the other study, optimal ethanol production was reported in cheese whey containing 61.0–65 g/L lactose (Murari et al., 2019b). Similarly utilizing concentrated whey powder is another alternative to solve the existence of diluted lactose in whey but this requires yeasts with high a tolerance to lactose and ethanol (Zou and Chang, 2022).
8.4 Fatty acids
The fatty acid composition of the membrane of the yeast has a great role in stress tolerance. Even if most strains of K. marxianus are thermotolerant, the addition of fatty acids such as linoleic acid, oleic acid, araquidic acid, and linolenic acid enhanced the thermotolerant activity of K. marxianus SLP1. This is probably due to changing the fluidity of the membrane to the required level. Similarly, supplementation of exogenous lipids rich in sterols and unsaturated fatty acids enhances fermentative performance and ethanol tolerance of yeasts (Guimarães et al., 2010).
8.5 Yeast extract and peptone
The addition of yeast extracts promoted the biomass growth of the yeasts and consequently increased bioethanol production (Murari et al., 2019b) since yeast extracts contain amino acids (cysteine, methionine, glutamic acid, glycine, aspartic acid, and alanine) and B complex vitamins (pyridoxine, thiamine, niacin, pantothenic acid, folic acid, and biotin) (Vieira et al., 2016). Similarly, higher ethanol (15.92 g/L) was produced by K. marxianus when the yeast fermented the cheese whey supplemented with peptone and yeast extracts than unsupplemented whey (10.65 g/L ethanol) (Tesfaw et al., 2021b). In the Murari et al. (2019b) study, the addition of yeast extract alone to whey increased the ethanol production but the ethanol concentration was lower when yeast extract and minerals [(NH4)2SO4; K3PO4, MgSO4; ZnSO4] together were added to whey.
8.6 Minerals
The ethanol produced from cheese whey supplemented with minerals such as (NH4)2SO4, K3PO4, MgSO4, and ZnSO4 was not satisfactory compared with unsupplemented cheese whey (Murari et al., 2019b). Besides osmotic pressure generated by added minerals, this might be due to the richness of cheese whey with minerals including KCl, NaCl, and calcium salts (predominantly phosphate) (Dragone et al., 2009). In another study, the addition of (NH4)2SO4; K3PO4, MgSO4; ZnSO4 together with the yeast to cheese whey reduced the ethanol concentration (Murari et al., 2019b). However, fermentation of crude cheese whey containing external mono ammonium phosphate (NH4H2PO4, 0.25%) and ammonium hydroxide (NH4OH, 0.07%) by K. marxianus increased ethanol concentrations (by 13%) (Koushki et al., 2012).
9 Sugar transport
Most of the inability to ferment different sugars and glucose repression is linked to the kinetics of sugar transport and sugar transport is an important part of ethanol production from different substrates. The Major Facilitator Superfamily (MFS) are cell membrane-located proteins expressed in all life forms for import or export of target substrates. Although initially believed to transport sugars, MFS is responsible for the import and export of metabolites, amino acids, drugs, oligosaccharides, and other compounds (Carvalho-Silva and Spencer-Martins, 1990; Yan, 2013; Västermark and Saier, 2014). The MFS that transport sugars contain different transporter proteins such as HGT1, HGT2, RAG1, LAC12, KHT1, and KHT2, (Varela et al., 2019a).
K. lactis uses Rag1p for glucose, Hgt1 for glucose and galactose, and Lac12 for lactose (Varela et al., 2019a). In yeasts, most monosaccharides are transported via the HXT classes of transporters (Donzella et al., 2021). Even if the HXT protein efficiently transports hexoses, it has low affinity for xylose and/or arabinose, and pentose transport is hindered by the presence of glucose. The protein encoded by HGT gene clusters in K. marxianus can transport arabinose and xylose (Knoshaug et al., 2015; Donzella et al., 2021). Donzella et al. (2021) also investigated that K. marxianus HGTs encoded pentose transporters such as KMAR 10531 have high affinity for arabinose and xylose while HXT-like proteins such as KMAR 10529 and KMAR 50027 are low-affinity arabinose/xylose transporters.
K. lactis and K. marxianus have evolved a unique mechanism to utilize glucose and galactose simultaneously (Liu et al., 2022). However, there are significant differences between K. marxianus in terms of lactose utilization and the variation is due to the Lac12 gene that encodes lactose permease, a protein that transports lactose (Varela et al., 2019a). Although the existence of four LAC12 copies might indicate that the difference in lactose utilization might be attributed to multiple genes, Varela et al. (2017) found that a single gene is responsible for the differences in lactose consumption by K marxianus. Similarly, despite the presence of four genes encoding Lac12 proteins, only one of the Lac12 proteins can transport lactose (Varela et al., 2019a). Other copies of LAC12 may not be functional or have alternative substrates. Once the lactose has entered the cells, β-galactosidase, a protein encoded by the Lac4 gene, hydrolyses it to glucose and galactose for utilization via glycolytic and Leloir pathways, respectively.
Besides lactose, LAC12 in K. lactis also transports cellobiose, a hydrolytic product of cellulose used for ethanol production (Varela et al., 2019b; Murata et al., 2022) and cellobiose uptake and hydrolysis are associated with the LAC12-CEL2 cluster in K. marxianus, K. lactis var. Lactis, K. dobzhanskii, K. aestuarii, K. nonfermentans, and K. wickerhamii although a variation of lactose and cellobiose exists among them (Varela et al., 2019b). Like LAC12, CEL2 (a gene responsible for cellobiose utilization and eventually fermentation) is negatively regulated by glucose repression in K. marxianus and glucose repression promotes cellobiose utilization and ethanol production. Murata et al. (2022) suggested that cellobiose is first rearranged and changed to glucose by the CEL2 protein and then ethanol production continues.
The Lac12 protein works via a proton-symport mechanism and is saturable at high substrate concentrations (da Silveira et al., 2019). Varela et al. (2017) discussed that the Lac12p protein transports galactose and the transporter has high affinity for lactose and low-affinity for galactose in K. lactis. The strains of K. marxianus that lack proton-symport cannot ferment lactose (Carvalho-Silva and Spencer-Martins, 1990).
In K. marxianus, the regulation of LAC4 is weekly repressed by glucose and the LAC4 promotor showed relatively low leakage expression of LAC4 in glucose medium and strong induction by lactose (Rajkumar et al., 2019; Liu et al., 2022). In K. lactis, the transient repression of glucose during initial induction of LAC4 is not mediated by the repressor Mig1 in the LAC4 promotor; however, Mig1 turns off the expression of Gal1 (encoding galactokinase, which mediates the catabolism of galactose) in the presence of glucose and Lac9-mediated transcription activation of LAC4 is prevented by low-level Gal1 (Liu et al., 2022). Finally, the ability to uptake sugar is an important genetic engineering target to optimize ethanol production by yeasts.
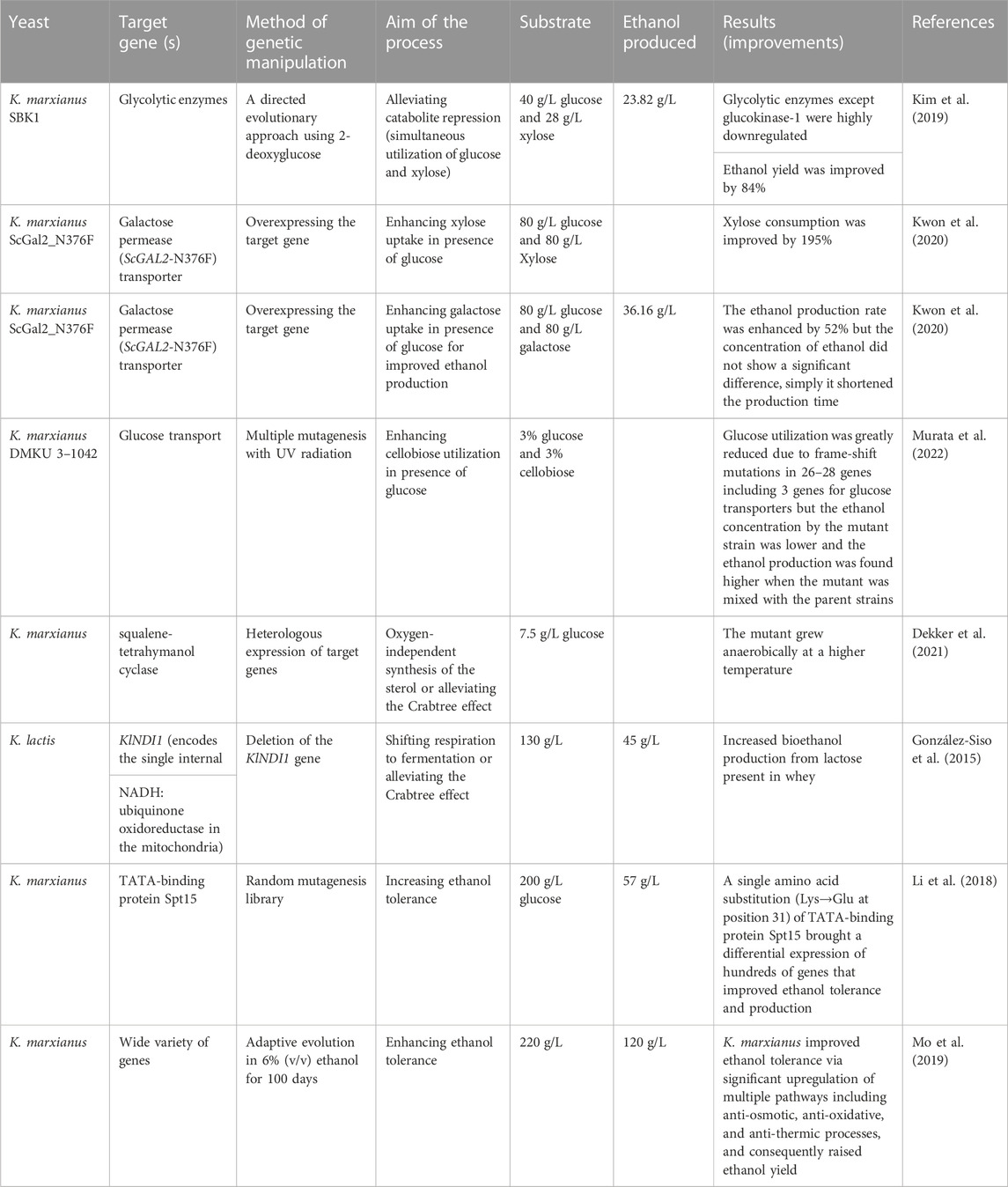
10 Catabolite repression
Various studies supplementing cheese whey with external carbon sources has been conducted to increase ethanol productivity of K. marxianus (Díez-Antolínez et al., 2018; Leonel et al., 2021; Sar et al., 2022). However, glucose represses the usage of other sugars, leading to diauxic growth and a reduction in ethanol yield (Tesfaw et al., 2021b). In recent years, however, genetic engineering of sugar-specific transporters alleviates such a challenge and glucose-other sugar mixtures can be co-fermented simultaneously since sugar-specific transporters cannot be affected by the presence of glucose (Table 3). For example, a genetically engineered K. marxianus overexpressed xylose-specific transporter (GAL2-N376F) enabling for the production of 50.10 g/L from the co-fermentation of glucose and xylose (Kim et al., 2015). K. marxianus SBK, a mutant developed through a directed evolutionary approach using 2-deoxyglucoses, was capable of co-fermenting glucose and xylose simultaneously by alleviating catabolite repression and the mutant produced 23.82 g/L ethanol at 40°C from a mixture of 40 g/L glucose and 28 g/L xylose (Kim et al., 2019). Similarly, overexpressing the mutant sugar transporter ScGAL2-N376F (derived from S. cerevisiae) in K. marxianus KCTC 17555ΔURA3 strain improved xylose utilization by 195% compared to the parental strains (Kwon et al., 2020). The ScGAL2-N376F mutant also enhanced sugar consumption (by 48%) and ethanol production (by 52%) rates when a mixture of glucose and galactose was used as the carbon source. Hua et al. (2019) reported that overexpression of xylose reductase and xylose-specific transporter genes enhanced the xylose utilization rate from the mixture of xylose and glucose. These methods enable to supplement of lignocellulosic feedstock to whey since pentose sugars are a substantial portion of lignocellulosic hydrolysates for ethanol production.
Besides overexpressing genes associated with sugar transport and metabolism to enhance non-glucose sugars fermentation, removing glucose repression is another strategy to alleviate their catabolite repression. Mig1 (transcriptional factor involved in glucose repression) represses the expression of several genes such as GAL83, SUC2, MAL62, LAC4, and LAC12 (genes associated with the transport and utilization of galactose, sucrose maltose, and lactose) when glucose is present, mainly at higher concentrations (Nurcholis et al., 2019). In K. marxianus, disruption of KmMIG1, the gene of the multicopy inhibitor of GAL gene expression, alleviated the glucose repression on some non-glucose sugars such as galactose, sucrose, and raffinose (Hua et al., 2019). In addition, Murata et al. (2022) showed that UV mutagenesis at the LAC12-CEL2 cluster of K. marxianus removes the repression of glucose impacts on cellobiose sugars transport.
Therefore, disrupting Mig1 using different techniques reduces the repression of glucose on other sugar transport and metabolism. Nurcholis et al. (2019) investigated disrupted mutants of genes for Mig1 of K. marxianus and found that a kanMX4-inserted mutant was able to utilize sucrose in the presence of glucose. However, the central metabolic pathways particularly the glycolytic pathways might be affected by disrupting Mig1. Generally, overexpressing genes associated with specific non-glucose sugar transporters and removing glucose repression by disrupting the regulator-associated genes enhance non-glucose sugar transportation, utilization, and fermentation.
11 Ethanol tolerance of lactose fermenting yeasts
The naturally occurring and biologically important metabolite, ethanol, can have a wide effect on ethanol-producing yeasts, and ethanol can affect nutrient metabolism, protein biosynthesis, protein function, membrane structure, and small molecule transport (Vanegas Juan et al., 2012). Ethanol-producing yeasts evolve a wide range of mechanisms to tolerate the ethanol they produce as well as the extraneous ethanol. Among these, modification of plasma membrane composition by increasing unsaturated lipids and ergosterol is widely used by the yeasts as a survival factor for high ethanol tolerance (de Moura Ferreira et al., 2022).
In contrast to S. cerevisiae, Kluyveromyces spp has low ethanol tolerance (Akbas and Stark, 2016). Tesfaw et al. (2021c) studied the survival percentage of K. marxianus at 10, 15, and 20% extraneous ethanol and found that the yeast survival percentage was 62.1% and 17.7% for exposure of 10% and 17% ethanol (v/v), respectively. The yeast viability at 10% external ethanol doesn’t guarantee the ability to produce ethanol at this ethanol concentration (Tesfaw et al., 2021c). The growth of K. marxianus was strongly inhibited and its central metabolic pathway was hindered by an ethanol concentration of 6% (v/v) (Diniz et al., 2017). Similarly, ethanol-producing Candida spp. cannot grow when the extraneous ethanol concentration was 6% (v/v) (Azzouni et al., 2019). In K. marxianus, dawnregulation of genes encoding unsaturated fatty acid biosynthesis was reported at high ethanol concentrations and the unsaturated fatty acid did not increase at high ethanol concentrations (Diniz et al., 2017; Ha-Tran et al., 2020). Like Ha-Tran et al. (2020) review, K. lactis exhibits low ethanol tolerance, and the degree of fatty acid unsaturation is reduced in the presence of external ethanol (Diniz et al., 2017). Besides high ergosterol presence in the cell membrane, ethanol tolerance is also linked to many factors such as heat shock proteins, amino acids, and trehalose (Saini et al., 2017).
11.1 Genetic manipulation to enhance ethanol tolerance
Different techniques have been used to enhance the ethanol tolerance of Kluyveromyces. Besides the classical methods like immobilization of the yeast, genetic manipulations have increased significantly (Table 3). Adaptive evolution of a haploid wild-type K. marxianus FIM1 for 100-days (about 450 generations) using 6% (v/v) external ethanol at a pH of 5.5 and enabled to tolerate ethanol up to 10% (v/v) (Leonel et al., 2021). The improvement in ethanol tolerance and production with K. marxianus was engineered through a single amino acid substitution (K31E) of the TATA-binding protein Spt15 and this resulted in a differential expression of hundreds of genes that enable the yeast to develop ethanol tolerance (Li et al., 2018). Similarly, Mo et al. (2019) investigated to get K. marxianus mutant that tolerates higher ethanol concentration via adaptive evolution in 6% (v/v) ethanol and they found that K. marxianus improved ethanol tolerance through a significant upregulation of multiple pathways including anti-osmotic, anti-thermic processes, and anti-oxidative, and this enabled to raise ethanol yield in high-ethanol circumstance and industrial high-temperature.
12 Immobilization of yeasts
Ethanol production from whey is retarded by the ethanol (inhibitors) produced by the yeasts themselves during batch fermentation. The most commonly employed ethanol-producing yeasts from whey are categorized under Kluyveromyces, which is less ethanol tolerant than the industrially efficient S. cerevisiae. The inhibitors including ethanol retard yeast growth and slow ethanol fermentation and finally leading to cell death.
12.1 Immobilization of yeasts with classical carriers
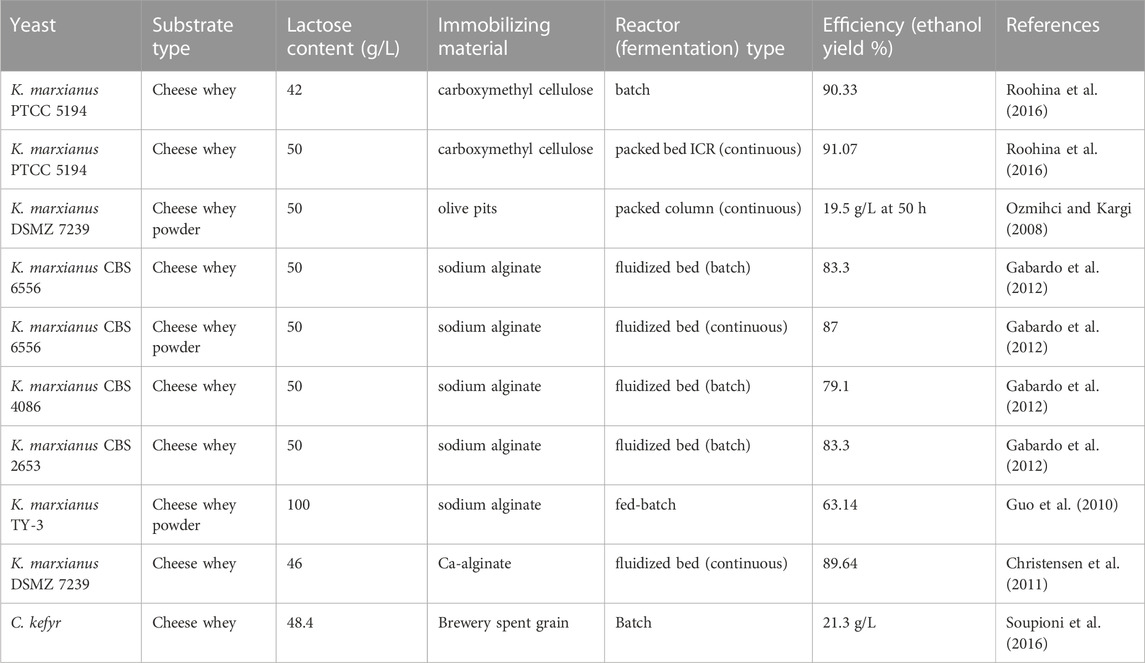
Immobilization of the yeast cells with the solid matrix could be used to solve such challenges. Carboxyl methyl cellulose, olive pits, sodium alginate, and calcium alginate are commonly used as immobilizing agents during ethanol production from whey (Table 2). Cell immobilization increases substrate utilization, enhances productivity, and improves ethanol yield (Roohina et al., 2016). In addition, it lowers the cost of production, retains stability of yeast activity, and makes yeast biomass separation from the bulk easy during dawn processing (Tesfaw and Assefa, 2014). For example, immobilization of K marxianus with calcium alginate enhances the ethanol production from cheese whey by 23%; however, the use of immobilized yeasts for ethanol production also has limitations such as limited nutrient diffusion, chemical and physical instability of the gel, the high cell densities in the gel beads, metabolites due to the gel matrix, non-regenerability of the beads, and immobilization increases costs (Christensen et al., 2011).
Due to higher ethanol production and reduced fermentation time, continuous fermentation is potentially superior to batch processes (Gabardo et al., 2012; Gabardo et al., 2016; Asunis et al., 2020). Fluidized bed, packed bed ICR, and packed column are the most common bioreactor type to produce ethanol via continuous fermentation (Table 4).
12.2 Nanoparticle and metal-organic framework (MOF) based immobilization
Immobilization protects the ethanol-producing yeasts and the enzymes from the inhibitory effects of intermediate metabolites such as alcohols and organic acids and external inhibitors. It allows cells and enzymes to withstand harsher environmental conditions than free cells and enzymes. Particularly, the immobilization of the β-galactosidase from Kluyveromyces species with nanoparticles increases the reusing cycles of the enzyme with a higher percentage of the ability to retain the original activity. Beniwal et al. (2018) studied β-galactosidase immobilized on silicon dioxide nanoparticles for the hydrolysis of whey and co-immobilized cultures of K. marxianus and S. cerevisiae in a single-stage batch process resulting in a high bioethanol yield (63.9 g/L). They also reported that immobilizing β-galactosidase with nanoparticles increased the reuse of the enzyme up to 15 times without major loss of catalytic activity. Liu et al. (2012) studied the immobilization of β-galactosidase from K. fragilis with the magnetic poly (glycidyl methacrylate–ethylene glycol dimethacrylate–hydroxyethyl methacrylate) nanobeads with reactive epoxy groups; the activity of novel magnetic nanobeads immobilized β-galactosidase was improved 2.6-folds compared with commercial epoxy support; and the magnetic nanobeads immobilized biocatalyst retained 81.5% of its original activity after 10 reaction cycles. Similarly, encapsulating β-D-galactosidase from K. lactis by silicon dioxide nanoparticles enabled to retain the enzyme activities (more than 50%) after 11 cycles, and the nanoparticle-based immobilization shortened the hydrolysis time (Verma et al., 2012). Half of the initial activity of β-galactosidase was retained after reusing it 20 times when the enzyme was immobilized by a polysiloxane–polyvinyl alcohol magnetic (mPOS–PVA) composite (Neri et al., 2008).
Besides the immobilization of β-galactosidase enzyme, research on yeast cells immobilized by nanoparticles to improve ethanol production from whey is limited. Immobilized cells of S. cerevisiae using magnetic nanoparticles were capable to produce higher bioethanol production with a productivity of 264 g/L/h (Sekoai et al., 2019). Similarly, a reduction in fermentation time, higher ethanol titer, and productivity were demonstrated by S. cerevisiae immobilizing with dark brown chitosan-coated manganese ferrite nanoparticles (Núñez Caraballo et al., 2021). In Firoozi et al. (2022) investigation, S. cerevisiae was immobilized with L-lysine-coated magnetite nanoparticles (MNPs), produced via a one-pot system, and the immobilized yeasts were evaluated for ethanol production from molasses as a carbon source. Their results also showed that the sugar consumption rate of the immobilized yeasts was higher than that of the free yeast cells and the nanoparticle coating immobilization had no negative effects on S. cerevisiae metabolism or ethanol production. Besides easy magnetic separation, the benefits of applying amino acids like L-lysine in the structures of iron oxide nanoparticles include decreased toxicity, increased high surface activity, and biocompatibility. However, the roles of nanoparticles to encapsulate Kluyveromyces species for optimal ethanol production from cheese whey are open for research. Most of the studies on nanoparticle application in biofuel production are related to ethanol production from lignocellulosic biomass, particularly during biomass pretreatment, hydrolysis, and fermentation (Sanusi et al., 2021; Verma et al., 2022).
Metal-organic frameworks (MOFs) consist of positively charged metals and organic ligands (pores) surrounding the metals; and due to hollow (porous) structures, MOFs have a large internal surface area to hold compounds. Since MOFs are highly inert hosts, selective, stable, tunable, easily separable, and vastly reusable, they are getting attractive attention as encapsulating agents (Wei et al., 2019; Silva et al., 2022). MOFs are commonly used to immobilize enzymes. Wei et al. (2019) studied the encapsulation of β-galactosidase within MOFs such as zeolitic imidazolate framework-8 (ZIF-8), Zn-MOF-74, or UiO-66-NH2 via a ball milling process, a solid-state mechanochemical strategy that is rapid and minimizes the use of organic solvents and strong acids and they reported that the encapsulated enzyme exhibits increased resistance to proteases, even in the presence of acids.
The production of an exoskeleton for S. cerevisiae cells was reported by Liang et al. (2016) and during their experimentation the yeast cells were first coated with exogenous β-galactosidase, followed by the crystallization of MOF (particularly ZIF-8), where external β-galactosidase had two services: first nucleation agent for MOF crystallization and second, the enzyme breaks down lactose to glucose and galactose that serves as energy sources during starvation. Furthermore, the homogenous ZIF-8 coating can protect yeast cells from UV radiation, and toxic agents particularly large cytotoxic proteins and anti-fungal agents (Liang et al., 2016; An et al., 2019). The protective shell also enabled the yeasts to survive the extreme oligotrophic environments for more than a week (Liang et al., 2017).
13 Two steps ethanol production from cheese whey
S. cerevisiae produces more ethanol and is more tolerant to ethanol stress than Kluyveromyces spp. The good fermentation capacity and ethanol tolerance [allowing the production of ethanol up to 20% (v/v)], its ability to grow under anaerobic conditions, its GRAS (generally regarded as safe), and extensive industrial and scientific knowledge that has been developed although a long time enabled the industrial microbiologist to choose S. cerevisiae in first plane for industrial alcoholic fermentation (Guimarães et al., 2010). However, S. cerevisiae cannot utilize the lactose in cheese whey to produce bioethanol since it lacks the enzymes β-galactosidase and lactose permease (Gabardo et al., 2016; Díez-Antolínez et al., 2018). In addition, there are still certain biotechnological aspects in which Kluyveromyces cannot compete with Saccharomyces industrially (Guo et al., 2010). Coculture and two steps ethanol production processes are commonly employed to produce ethanol from whey using S. cerevisiae.
Chemical or enzymatic hydrolysis of lactose in cheese whey is first needed to produce ethanol by S. cerevisiae. Lactose in whey can be degraded to glucose and galactose by inorganic acids (sulfuric and hydrochloric acids) and carbonic acid (Coté et al., 2004). However, acid hydrolysis of lactose has caused sugar loss and inhibitors like hydroxymethyl furfural by caramelization reactions since the hydrolysis is commonly performed at elevated temperatures (180–250°C). In addition, acid hydrolysis of lactose can result in adding a demineralization process, protein denaturation, and the formation of undesirable products (Zhou et al., 2019). Enzymatic hydrolysis (by β-galactosidase) of lactose in whey is preferred over acid hydrolysis. β-galactosidase which can be produced by bacteria, yeast, and filamentous fungi, hydrolyze lactose into monomeric sugars such as glucose and galactose (Schmidt et al., 2020).
The challenge of two-step ethanol production is the presence of diauxic growth of the yeasts in the presence of a mixture of glucose and galactose, which reduces the efficiency of ethanol production (Sar et al., 2022). Glucose represses the absorption and utilization of galactose by the yeasts when present in the mixture and later decreases the utilization of lactose (Jin et al., 2016; Zou and Chang, 2022). Furthermore, the conversion of galactose into glycolytic intermediates requires additional steps and energy since glycolytic enzymes are not galactose specific and the galactose is converted to glucose-6-phosphate via the Leloir pathway that involves different steps and reduced to ethanol (Díez-Antolínez et al., 2018).
14 Coculture method for ethanol production from whey
The step-by-step approach discussed above has at least three drawbacks. First, the chemical hydrolysis of lactose release inhibitors such as hydroxymethyl furfural, which significantly reduce yeast growth. Second, the enzymatic process requires β-galactosidase and others; the enzymes increase production costs. Third, enzymatic hydrolysis can be affected by catabolite repression (Guimarães et al., 2010). The mixed culture approach of ethanol-producing S. cerevisiae with β-galactosidase-producing microbes is highly valuable to tackle such drawbacks. In addition, the recombinant S. cerevisiae expressing β-galactosidase is another alternative solution to produce ethanol directly from whey lactose using S. cerevisiae. However, the genetically modified S. cerevisiae and other strains are often discarded for the production of food-grade ethanol (Zotta et al., 2020). In addition, the genetically engineered S. cerevisiae strains have shown undesirable traits such as genetic instability, low growth, and reduced ethanol production (Díez-Antolínez et al., 2018).
Guo et al. (2010) studied cheese whey powder containing high concentration of lactose for co-cultivation of K. marxianus and S. cerevisiae to produce ethanol and the mixed cells produced higher ethanol than the monoculture of S. cerevisiae. Besides enabling hydrolyzing of the lactose in the whey, S. cerevisiae was able to produce more ethanol indicating that K. marxianus had other contributions favoring bioethanol production. Similarly, batch fermentation of cheese whey with the mixed culture of S. cerevisiae and K. marxianus resulted in higher lactose consumption and ethanol production than yeast monocultures (Farkas et al., 2019). The impact of coculture becomes more pronounced when the lactose in whey is higher by external supplementation of lactose or different cheese whey concentrating methods. Farkas et al. (2019) found a higher ethanol content (80–82 g/L) in a mixed culture of S. cerevisiae and K. marxianus when the lactose concentration was raised to 150 g/L. The Crabtree-negative K. marxianus ethanol yield is lower than that of S. cerevisiae {Zou, 2022 #114} (Zou and Chang, 2022), and coculturing K. marxianus with S. cerevisiae enables us to solve such a challenge. In the mixed culture, it was found that the amount of ethanol increased when the mixed cells were immobilized with alginate.
The use of a mixed culture in the whey fermentation reduced the substrate inhibitions and then increased the ethanol concentration, which is why 57.6 g/L ethanol through the mixed culture of S. cerevisiae and K. marxianus in a ratio of 1:1 from cheese whey powder (containing 152 g/L) lactose (Farkas et al., 2019). The mixed culture resulted in higher lactose consumption and higher ethanol production than yeast monocultures (Rodrigues et al., 2016). Similarly, more ethanol was produced by mixed culture of K. marxianus and Candida krusei using whey as the fermentation media. During co-cultivation, it was found that co-immobilization of both galactose-producing microbes and ethanol-producing yeast for ethanol production is more effective than suspension cultures, especially for high-temperature ethanol production (Zou and Chang, 2022). Co-cultivation between Kluyveromyces spp and S. cerevisiae enhances ethanol tolerance (Zou and Chang, 2022) and higher ethanol titer could be produced in co-culture than in mono-culture.
15 Electro-fermentation for bioethanol production from whey
Electro-fermentation is an emerging bioprocess that electrochemically controls microbial fermentative metabolism using electrodes. Besides modifying the media by changing the redox balances, imbalanced fermentations might be generated since the electrodes can act as either electron sinks or sources (Moscoviz et al., 2016; Sriram et al., 2022). It is the oxidation-reduction potential of the growing media that controls the metabolism of the microorganisms and any factor affecting the oxidation-reduction potential such as the changing electrochemistry via applying electricity or other means, can have a positive or negative impact have on the on the fermentation of microbes. However, the capability of electro-fermentation to regulate redox instability boosts the overall metabolites activities of the microorganisms and hence improved metabolic products such as ethanol, methane, hydrogen gas, and biopolymers could be produced (Chandrasekhar et al., 2021).
Mathew et al. (2015) studied the enhancement of ethanol production by S. cerevisiae via electrostatically accelerated fermentation of glucose and a voltage was applied to the fermentation mixture without causing a current (a purely electrostatic version). Their investigation showed that electro-fermentation enabled the production of 14% (v/v) ethanol by the active S. cerevisiae cells in 20 h compared to 7%–8% (v/v) ethanol concentration in 50–70 h via the traditional fermentation and the method does not consume external energy since it is electrostatic (without external electricity). In Joshi et al. (2019) study, S. cerevisiae and W. anomalous were grown in 5% glucose and lignocellulosic hydrolysates to enhance ethanol production in an electrochemical cell. The ethanol production by S. cerevisiae (non-xylose fermenting yeast) and W. anomalous (xylose-fermenting yeast) from 5% glucose was enhanced by 19.8% and 23.7%, respectively, in an electrochemical cell when externally supplied with 4 V, as compared to the control cultures (no external voltage). Cultivation of S. cerevisiae and W. anomalous using glucose as a substrate in the platinum nanoparticle-coated platinum anode and the neutral red-coated graphite cathode, respectively, potentially increased ethanol production (52.8%). The increase in ethanol production reached 61.5% when glucose was replaced with lignocellulosic biomass hydrolysate. Faster cell growth resulting in the production of 53 g/L ethanol was reported when external −1.5 V is applied to the electrochemical bioreactor containing 120 g/L glucose and S. cerevisiae (Kumar et al., 2018). Dhungana et al. (2022) studied ethanol production from lignocellulosic materials by a fungal consortium composed of a cellulolytic A. niger and lignocellulolytic G. sessile together with S. cerevisiae via electro-fermentation, which resulted in electro-fermentation increasing ethanol production by 12% and 18% with a mixed culture of G. sessile—S. cerevisiae and G. sessile- A. niger - S. cerevisiae, respectively.
Generally, the benefits of electro-fermentation are being selective, efficient sugar (carbon) utilization minimized use of additives for pH control or redox balance, better cell growth, better product purity, and in some cases improved product recovery (Schievano et al., 2016; Joshi et al., 2019). Most studies on electro-fermentation have focused on biogas and biohydrogen production using bacteria (Sriram et al., 2022) and ethanol production by bacteria via electro-fermentation is also known (Kumar et al., 2018; Chandrasekhar et al., 2021). Ethanol production by yeasts, particularly S. cerevisiae, involving electro-fermentation, has also been documented; however, there is no study on ethanol production from whey through the new technology. Hence, it is a new area of research as the bioelectrochemical system is affected by the chemical composition of the media (Yamada et al., 2022) and the whey composition is completely different from a substrate utilized by S. cerevisiae to produce ethanol.
16 Economics of ethanol production from cheese
Due to the expensive technology, the small and medium-sized dairy industries cannot produce value-added products from their residues; some cheese factories process approximately 50% of their cheese whey into powder cheese whey and condensed cheese whey instead of producing valuable products such as ethanol, organic acids (Osorio-González et al., 2022) and the remaining 50% of the cheese whey of small and medium-sized industries is damped off to water bodies (Tesfaw et al., 2021b). Asides from the expensive technology needed to commercialize of cheese whey, few studies have shown the economic feasibility of producing ethanol from cheese whey; however, the economic feasibilities of cellulosic, molasses, and starch-based ethanol is dealt with in detail.
Direct fermentation of raw cheese to ethanol is not economically feasible due to low lactose content resulting in 2%-3% (v/v) ethanol (Zou and Chang, 2022). Utama et al. (2017b) studied the economic feasibility of bioconversion of cheese-making wastes to bioethanol using a joint cost allocation with the market price method approach and the break-even point and the result showed that bioethanol production has financial benefit of up to US$ 3816.96 per month (ethanol price per liter at US$3.02 with a hypothetical market price of US$2.21/L and a cost per unit after split-off of US$0.81/L), reducing the variable cost of main product by14.73%, saved disposal cost, and attained the breakeven point in 3.53 months, and they invested US$17197.28 to treat 24,000 L of cheese whey per month. Utilizing distillery waste for bio-fertilizer production enhances the economic feasibility of ethanol production (Utama et al., 2017a).
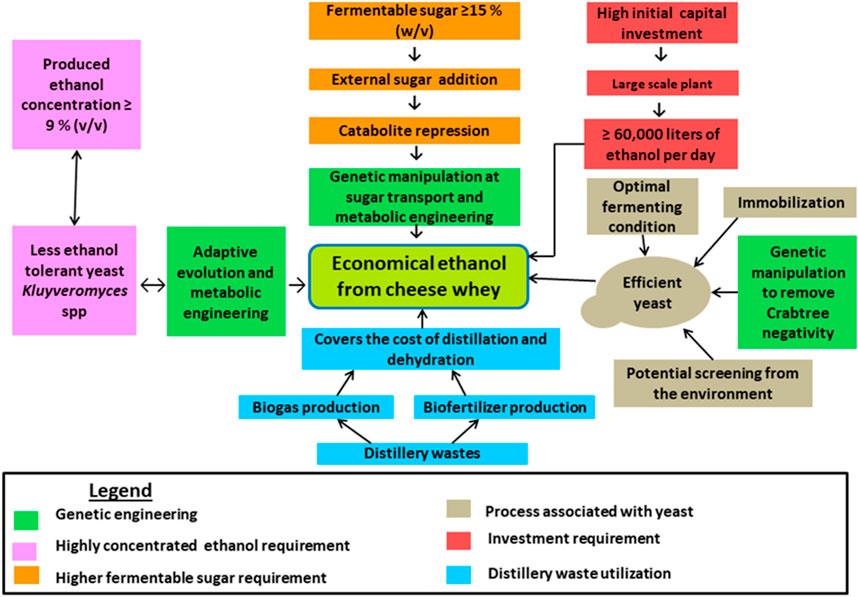
The key factor hampering economical ethanol production from cheese whey is the low lactose present in whey and economical ethanol could be produced when the lactose concentration is raised to 200 g/L (Valta et al., 2017), despite the presence of 40–50 g/L lactose in whey (Table 2). On the other hand, to be economically feasible, biogas from effluent treatment should be used to recover the alcohol (dehydration and distillation), the plant has to produce 60,000 L of ethanol per day (5 million gallons a year), the fermentable sugars should be 15% (w/v) to produce 9%-10% (v/v) ethanol, and the recovery cost has to be reduced below $0.2 per liter (Ling, 2008). These enforce to add the external lactose making the production cost higher, and this necessity for the fortification of the whey with cheap sugar sources like molasses and other industrial wastes. However, the mixing of different sugars results in catabolite repression that might be solved by genetic manipulations, particularly, in the sugar transport systems and metabolic engineering. Furthermore, increasing the sugar content of cheese whey requires high tolerance to ethanol and osmotic pressure (Zou and Chang, 2022). Generally, optimizing different processing conditions related to ethanol production such as increasing fermentable sugars, creating optimal growth conditions for the yeast, producing efficient ethanol-producing yeast via metabolic engineering and classical methods, improving ethanol titer (more than 9% v/v), utilizing distillery wastes for income generation to reduce the production cost, and increasing the investment cost enable to produce economical ethanol from whey (Figure 2).
17 Conclusion and feature directions
Cheese whey is the main and voluminous waste generated during dairy product processing. It has high BOD and COD hence it harms the environment if it is released into the environment without treatment. Valorization of cheese whey for various biological products is the best choice but this paper focused on the current trends of bioethanol production from whey via the classical fermentation technologies and recently evolved ethanol optimization methods such as electro-fermentation and immobilizing the microbes with nanoparticle and MOF-coated supporting material. The ability of K. marxianus to ferment lactose at a higher temperature makes it the best strain choice among Kluyveromyces and Candida species since it reduces cooling costs and prevents contamination by lactic acid bacteria and other microbes. The K. marxianus is competent enough against lactic acid bacteria to produce ethanol without pasteurizing or sterilizing the whey. The efficient ethanol-producing yeasts (like S. cerevisiae) that lack β-galactosidase can be employed to produce ethanol from whey by coculturing with microbes that have β-galactosidase or by hydrolyzing lactose using chemical, biological, or/and physical methods. In addition, most lactose fermenting yeasts such as K. marxianus, K. lactis, and Candida kefyr are not tolerant to ethanol. Researches on ethanol tolerance by genetic modification are common but few kinds of research are reported on adaptive evolution and other non-genetic modification methods hence it is still open for research. The main challenges to producing economical ethanol from cheese whey are the low concentration of lactose in the whey and as a result lower ethanol titer. This increases the distillation cost at the industrial level hence this problem can be solved by adding cheap external carbon sources to the whey; however, this results in catabolite repression which needs to be solved by genetic engineering tools. Economical ethanol production is feasible if different production processes are optimized and investment cost is increased.
Author contributions
Since it is a sole authorship, the entire manuscript is written by AT. The author confirms being the sole contributor of this work and has approved it for publication. All authors contribution to the article and approved the submitted version.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aidarbekova, S., and Aider, M. (2021). Use of electro-activated whey as ingredient in fermented milk production: Proof of the concept of the technological feasibility. ACS Food Sci. Technol. 1, 1349–1359. doi:10.1021/acsfoodscitech.1c00230
Aider-Kaci, F. A., Aidarbekova, S., and Aider, M. (2023). Impact of electro-activated whey on growth, acid and bile resistance of Lacticaseibacillus rhamnosus GG and Lactobacillus acidophilus ATCC 4356. Heliyon 9, e13154. doi:10.1016/j.heliyon.2023.e13154
Akbas, M. Y., and Stark, B. C. (2016). Recent trends in bioethanol production from food processing byproducts. J. Industrial Microbiol. Biotechnol. 43, 1593–1609. doi:10.1007/s10295-016-1821-z
An, H., Li, M., Gao, J., Zhang, Z., Ma, S., and Chen, Y. (2019). Incorporation of biomolecules in Metal-Organic Frameworks for advanced applications. Chem. Rev. 384, 90–106. doi:10.1016/j.ccr.2019.01.001
Ariyanti, D., Aini, A. P., and Pinundi, D. S. (2014). Optimization of ethanol production from whey through fed-batch fermentation using Kluyveromyces marxianus Energy. Procedia 47, 108–112.
Ariyanti, D., and Hadiyanto, H. (2013). Ethanol production from whey by Kluyveromyces marxianus in batch fermentation system: Kinetics parameters estimation. Bull. Chem. React. Eng. Catal. 7, 179–184. doi:10.9767/bcrec.7.3.4044.179-184
Asunis, F., De Gioannis, G., Dessì, P., Isipato, M., Lens, P. N., Muntoni, A., et al. (2020). The dairy biorefinery: Integrating treatment processes for cheese whey valorisation. J. Environ. Manag. 276, 111240. doi:10.1016/j.jenvman.2020.111240
Azzouni, D., Lahkimi, A., Louaste, B., Taleb, M., Chaouch, M., and Eloutassi, N. (2019). “Performance of Aspergillus Niger and Kluyveromyces marxianus for optimized bioethanol production from dairy waste,” in Advanced intelligent systems for sustainable development (AI2SD’2018) (Cham: Springer International Publishing), 162–175.
Balia, R. L., Kurnani, T. B. A., and Utama, G. L. (2018). The combination of Mozzarella cheese whey and sugarcane molasses in the production of bioethanol with the inoculation of indigenous yeasts. J. Jpn. Inst. Energy 97, 266–269. doi:10.3775/jie.97.266
Beniwal, A., Saini, P., Kokkiligadda, A., and Vij, S. (2017). Physiological growth and galactose utilization by dairy yeast Kluyveromyces marxianus in mixed sugars and whey during fermentation. Dur. Ferment. 3 Biotech. 7, 349. doi:10.1007/s13205-017-0985-1
Beniwal, A., Saini, P., Kokkiligadda, A., and Vij, S. (2018). Use of silicon dioxide nanoparticles for β-galactosidase immobilization and modulated ethanol production by co-immobilized K marxianus and S cerevisiae in deproteinized cheese whey. cerevisiae deproteinized cheese whey LWT 87, 553–561. doi:10.1016/j.lwt.2017.09.028
Blaschek, K. M., Wendorff, W. L., and Rankin, S. A. (2007). Survey of salty and sweet whey composition from various cheese plants in Wisconsin. Wis. J. Dairy Sci. 90, 2029–2034. doi:10.3168/jds.2006-770
Carvalho-Silva, M., and Spencer-Martins, I. (1990). Modes of lactose uptake in the yeast species Kluyveromyces marxianus. Kluyveromyces Marx. Antonie Leeuwenhoek 57, 77–81. doi:10.1007/BF00403158
Chandrasekhar, K., Naresh Kumar, A., Kumar, G., Kim, D.-H., Song, Y.-C., and Kim, S.-H. (2021). Electro-fermentation for biofuels and biochemicals production: Current status and future directions. Curr. status future Dir. Bioresour. Technol. 323, 124598. doi:10.1016/j.biortech.2020.124598
Christensen, A. D., Kádár, Z., Oleskowicz-Popiel, P., and Thomsen, M. H. (2011). Production of bioethanol from organic whey using Kluyveromyces marxianus. J. industrial Microbiol. Biotechnol. 38, 283–289. doi:10.1007/s10295-010-0771-0
Coté, A., Brown, W. A., Cameron, D., and van Walsum, G. P. (2004). Hydrolysis of lactose in whey permeate for subsequent fermentation to ethanol. J. Dairy Sci. 87, 1608–1620. doi:10.3168/jds.S0022-0302(04)73315-9
da Silva, F. L., de Oliveira Campos, A., dos Santos, D. A., Magalhães, E. R. B., de Macedo, G. R., and dos Santos, E. S. (2018). Valorization of an agroextractive residue—carnauba straw—For the production of bioethanol by simultaneous saccharification and fermentation (SSF). Renew. Energy 127, 661–669. doi:10.1016/j.renene.2018.05.025
da Silveira, F. A., Diniz, R. H. S., Sampaio, G., Brandão, R. L., da Silveira, W. B., and Castro, I. M. (2019). Sugar transport systems in Kluyveromyces marxianus CCT 7735. Antonie Leeuwenhoek 112, 211–223. doi:10.1007/s10482-018-1143-4
Das, B., Das, M., Bhattacharjee, S., and Bhattacharjee, C. (2017). Ethanol production from deproteinized cheese whey powder in a batch fermentation process: Optimization of process and kinetic modelling. Desalination Water Treat. 64, 198–206. doi:10.5004/dwt.2017.20238
Das, M., Raychaudhuri, A., and Ghosh, S. K. (2016). Supply chain of bioethanol production from whey: A review. A Rev. Procedia Environ. Sci. 35, 833–846. doi:10.1016/j.proenv.2016.07.100
de Carvalho, C. T., de Oliveira Júnior, S. D., de Brito Lima, W. B., de Medeiros, F. G. M., de Sá Leitão, A. L. O., dos Santos, E. S., et al. (2020). Potential of “coalho” cheese whey as lactose source for β-galactosidase and ethanol co-production by Kluyveromyces spp. yeasts. yeasts Prep. Biochem. Biotechnol. 50, 925–934. doi:10.1080/10826068.2020.1771731
de Moura Ferreira, M. A., da Silveira, F. A., and da Silveira, W. B. (2022). Ethanol stress responses in Kluyveromyces marxianus: Current knowledge and perspectives. Appl. Microbiol. Biotechnol. 106, 1341–1353. doi:10.1007/s00253-022-11799-0
Dekker, W. J. C., Ortiz-Merino, R. A., Kaljouw, A., Battjes, J., Wiering, F. W., Mooiman, C., et al. (2021). Engineering the thermotolerant industrial yeast Kluyveromyces marxianus for anaerobic growth. Metab. Eng. 67, 347–364. doi:10.1016/j.ymben.2021.07.006
Demiray, E., Karatay, S. E., Dönmez, S., and Dönmez, G. (2016). The usage of carrot pomace for bioethanol production. J. Chil. Chem. Soc. 61, 2996–2998.
Dhungana, P., Prajapati, B., Bhatt, P., Regmi, D., Yadav, M., and Maharjan, S. (2022). Production of bioethanol from Saccharum spontaneum by simultaneous saccharification and electro-fermentation using mixed culture of microbes. Biofuels 14, 191–199. doi:10.1080/17597269.2022.2126083
Díez-Antolínez, R., Hijosa-Valsero, M., Paniagua-García, A. I., Garita-Cambronero, J., and Gómez, X. (2018). Yeast screening and cell immobilization on inert supports for ethanol production from cheese whey permeate with high lactose loads. PloS one 13, e0210002. doi:10.1371/journal.pone.0210002
Diniz, R. H., Rodrigues, M. Q., Fietto, L. G., Passos, F. M., and Silveira, W. B. (2014). Optimizing and validating the production of ethanol from cheese whey permeate by Kluyveromyces marxianus UFV-3. UFV-3 Biocatal. Agric. Biotechnol. 3, 111–117. doi:10.1016/j.bcab.2013.09.002
Diniz, R. H. S., Villada, J. C., Alvim, M. C. T., Vidigal, P. M. P., Vieira, N. M., Lamas-Maceiras, M., et al. (2017). Transcriptome analysis of the thermotolerant yeast Kluyveromyces marxianus CCT 7735 under ethanol stress. Appl. Microbiol. Biotechnol. 101, 6969–6980. doi:10.1007/s00253-017-8432-0
Donzella, L., Varela, J. A., Sousa, M. J., and Morrissey, J. P. (2021). Identification of novel pentose transporters in Kluyveromyces marxianus using a new screening platform. Research 21, foab026. doi:10.1093/femsyr/foab026
Dragone, G., Mussatto, S. I., e Silva, J. B. A., and Teixeira, J. A. (2011). Optimal fermentation conditions for maximizing the ethanol production by Kluyveromyces fragilis from cheese whey powder. Biomass Bioenergy 35, 1977–1982. doi:10.1016/j.biombioe.2011.01.045
Dragone, G., Mussatto, S. I., Oliveira, J. M., and Teixeira, J. A. (2009). Characterisation of volatile compounds in an alcoholic beverage produced by whey fermentation. Food Chem. 112, 929–935. doi:10.1016/j.foodchem.2008.07.005
Ertuğrul Karatay, S., Demiray, E., and Dönmez, G. (2019). Bioethanol production by newly isolated halotolerant Kluyveromyces marxianus strains. Environ. Prog. Sustain. Energy 38, 542–547. doi:10.1002/ep.12935
Farkas, C., Rezessy-Szabó, J. M., Gupta, V. K., Bujna, E., Pham, T. M., Pásztor-Huszár, K., et al. (2019). Batch and fed-batch ethanol fermentation of cheese-whey powder with mixed cultures of different yeasts. Energies 12, 4495. doi:10.3390/en12234495
Ferreira, P. G., da Silveira, F. A., dos Santos, R. C. V., Genier, H. L. A., Diniz, R. H. S., Ribeiro, J. I., et al. (2015). Optimizing ethanol production by thermotolerant Kluyveromyces marxianus CCT 7735 in a mixture of sugarcane bagasse and ricotta whey. Food Sci. Biotechnol. 24, 1421–1427. doi:10.1007/s10068-015-0182-0
Firoozi, F. R., Raee, M. J., Lal, N., Ebrahiminezhad, A., Teshnizi, S. H., Berenjian, A., et al. (2022). Application of magnetic immboilization for ethanol biosynthesis using Saccharomyces cerevisiae. Sep. Sci. Technol. 57, 777–787. doi:10.1080/01496395.2021.1939376
Fonseca, G. G., de Carvalho, N. M. B., and Gombert, A. K. (2013). Growth of the yeast Kluyveromyces marxianus CBS 6556 on different sugar combinations as sole carbon and energy source. Appl. Microbiol. Biotechnol. 97, 5055–5067. doi:10.1007/s00253-013-4748-6
Gabardo, S., Pereira, G. F., Klein, M. P., Rech, R., Hertz, P. F., and Ayub, M. A. Z. (2016). Dynamics of yeast immobilized-cell fluidized-bed bioreactors systems in ethanol fermentation from lactose-hydrolyzed whey and whey permeate. Bioprocess Biosyst. Eng. 39, 141–150. doi:10.1007/s00449-015-1498-0
Gabardo, S., Rech, R., and Ayub, M. A. Z. (2012). Performance of different immobilized-cell systems to efficiently produce ethanol from whey: Fluidized batch, packed-bed and fluidized continuous bioreactors. J. Chem. Technol. Biotechnol. 87, 1194–1201. doi:10.1002/jctb.3749
Ghaly, A. E., and El-Taweel, A. A. (1995). Effect of nutrient supplements addition on ethanol production from cheese whey usingCandida pseudotropicalis under batch condition. Appl. Biochem. Biotechnol. 53, 107–131. doi:10.1007/BF02788602
González-Siso, M. I., Touriño, A., Vizoso, Á., Pereira-Rodríguez, Á., Rodríguez-Belmonte, E., Becerra, M., et al. (2015). Improved bioethanol production in an engineered K luyveromyces lactisstrain shifted from respiratory to fermentative metabolism by deletion of NDI1. Microb. Biotechnol. 8, 319–330. doi:10.1111/1751-7915.12160
Guimarães, P. M. R., Teixeira, J. A., and Domingues, L. (2010). Fermentation of lactose to bio-ethanol by yeasts as part of integrated solutions for the valorisation of cheese whey. Advances 28, 375–384. doi:10.1016/j.biotechadv.2010.02.002
Guo, X., Zhou, J., and Xiao, D. (2010). Improved ethanol production by mixed immobilized cells of Kluyveromyces marxianus and Saccharomyces cerevisiae from cheese whey powder solution fermentation. Appl. Biochem. Biotechnol. 160, 532–538. doi:10.1007/s12010-008-8412-z
Ha-Tran, D. M., Lai, R.-Y., Nguyen, T. T. M., Huang, E., Lo, S.-C., and Huang, C.-C. (2021). Construction of engineered RuBisCO Kluyveromyces marxianus for a dual microbial bioethanol production system. PLOS ONE 16, e0247135. doi:10.1371/journal.pone.0247135
Ha-Tran, D. M., Nguyen, T. T. M., and Huang, C.-C. (2020). Kluyveromyces marxianus: Current state of omics studies, strain improvement strategy and potential industrial implementation. Ind. Implement. Ferment. 6, 124. doi:10.3390/fermentation6040124
Hoshida, H., and Akada, R. (2017). “High-temperature bioethanol fermentation by conventional and nonconventional yeasts,” in Biotechnology of yeasts and filamentous fungi. Editor A. A. Sibirny (Cham: Springer International Publishing), 39–61. doi:10.1007/978-3-319-58829-2_2
Hua, Y., Wang, J., Zhu, Y., Zhang, B., Kong, X., Li, W., et al. (2019). Release of glucose repression on xylose utilization in Kluyveromyces marxianus to enhance glucose-xylose co-utilization and xylitol production from corncob hydrolysate. Microb. Cell Factories 18, 24. doi:10.1186/s12934-019-1068-2
Ikegami, T., Morita, T., Nakayama, S., Negishi, H., Kitamoto, D., Sakaki, K., et al. (2009). Processing of ethanol fermentation broths by Candida krusei to separate bioethanol by pervaporation using silicone rubber-coated silicalite membranes. J. Chem. Technol. Biotechnol. 84, 1172–1177. doi:10.1002/jctb.2151
Illarionov, A., Lahtvee, P.-J., Kumar, R., and Zhou, N.-Y. (2021). Potassium and sodium salt stress characterization in the yeasts Saccharomyces cerevisiae, Kluyveromyces marxianus, and Rhodotorula toruloides. Kluyveromyces marxianus, Rhodotorula toruloides Appl. Environ. Microbiol. 87, 03100. doi:10.1128/AEM.03100-20
Jin, Y., Parashar, A., Mason, B., and Bressler, D. C. (2016). Simultaneous hydrolysis and co-fermentation of whey lactose with wheat for ethanol production. Bioresour. Technol. 221, 616–624. doi:10.1016/j.biortech.2016.09.063
Joshi, J., Dhungana, P., Prajapati, B., Maharjan, R., Poudyal, P., Yadav, M., et al. (2019). Enhancement of ethanol production in electrochemical cell by Saccharomyces cerevisiae (CDBT2) and Wickerhamomyces anomalus (CDBT7). Front. Energy Res. 7, 70. doi:10.3389/fenrg.2019.00070
Kareb, O., Champagne, C. P., Jean, J., Gomaa, A., and Aïder, M. (2018). Effect of electro-activated sweet whey on growth of Bifidobacterium, Lactobacillus, and Streptococcus strains under model growth conditions. Food Res. Int. 103, 316–325. doi:10.1016/j.foodres.2017.10.060
Karim, A., and Aider, M. (2022). Comprehensive utilisation of electro-activated whey-based media in cell growth, metabolite production and aroma compounds synthesis using a starter culture originated from kefir grains. Int. Dairy J. 126, 105276. doi:10.1016/j.idairyj.2021.105276
Karim, A., and Aïder, M. (2020). Contribution to the process development for lactulose production through complete valorization of whey permeate by using electro-activation technology versus a chemical isomerization process. ACS Omega 5, 28831–28843. doi:10.1021/acsomega.0c04178
Karim, A., Gerliani, N., and Aïder, M. (2020). Kluyveromyces marxianus: An emerging yeast cell factory for applications in food and biotechnology. Int. J. Food Microbiol. 333, 108818. doi:10.1016/j.ijfoodmicro.2020.108818
Kim, J.-S., Park, J.-B., Jang, S.-W., and Ha, S.-J. (2015). Enhanced xylitol production by mutant Kluyveromyces marxianus 36907-FMEL1 due to improved xylose reductase activity. Appl. Biochem. Biotechnol. 176, 1975–1984. doi:10.1007/s12010-015-1694-z
Kim, S.-B., Kwon, D.-H., Park, J.-B., and Ha, S.-J. (2019). Alleviation of catabolite repression in Kluyveromyces marxianus: The thermotolerant SBK1 mutant simultaneously coferments glucose and xylose. Biotechnol. Biofuels 12, 90. doi:10.1186/s13068-019-1431-x
Knoshaug, E. P., Vidgren, V., Magalhães, F., Jarvis, E. E., Franden, M. A., Zhang, M., et al. (2015). Novel transporters from Kluyveromyces marxianus and Pichia guilliermondii expressed in Saccharomyces cerevisiae enable growth on l -arabinose and d -xylose. L-arabinose D-xylose Yeast 32, 615–628. doi:10.1002/yea.3084
Koushki, M., Jafari, M., and Azizi, M. (2012). Comparison of ethanol production from cheese whey permeate by two yeast strains. J. food Sci. Technol. 49, 614–619. doi:10.1007/s13197-011-0309-0
Kumar, P., Chandrasekhar, K., Kumari, A., Sathiyamoorthi, E., and Kim, B. S. (2018). Electro-fermentation in aid of Bioenergy and biopolymers. Biopolym. Energies 11, 343. doi:10.3390/en11020343
Kwon, D.-H., Kim, S.-B., Park, J.-B., and Ha, S.-J. (2020). Overexpression of mutant galactose permease (ScGal2_N376F) effective for utilization of glucose/xylose or glucose/galactose mixture by engineered Kluyveromyces marxianus. J. Microbiol. Biotechnol. 30, 1944–1949. doi:10.4014/jmb.2008.08035
Leonel, L. V., Arruda, P. V., Chandel, A. K., Felipe, M. G. A., and Sene, L. (2021). Kluyveromyces marxianus: A potential biocatalyst of renewable chemicals and lignocellulosic ethanol production. Crit. Rev. Biotechnol. 41, 1131–1152. doi:10.1080/07388551.2021.1917505
Li, P., Fu, X., Li, S., and Zhang, L. (2018). Engineering TATA-binding protein Spt15 to improve ethanol tolerance and production in Kluyveromyces marxianus. Kluyveromyces marxianus Biotechnol. Biofuels 11, 207. doi:10.1186/s13068-018-1206-9
Liang, K., Richardson, J. J., Cui, J., Caruso, F., Doonan, C. J., and Falcaro, P. (2016). Metal–organic framework coatings as cytoprotective exoskeletons for living cells. Adv. Mater. 28, 7910–7914. doi:10.1002/adma.201602335
Liang, K., Richardson, J. J., Doonan, C. J., Mulet, X., Ju, Y., Cui, J., et al. (2017). An enzyme-coated metal–organic framework shell for synthetically adaptive cell survival. Angew. Chem. Int. Ed. 56, 8510–8515. doi:10.1002/anie.201704120
Lievore, P., Simões, D. R. S., Silva, K. M., Drunkler, N. L., Barana, A. C., Nogueira, A., et al. (2015). Chemical characterisation and application of acid whey in fermented milk. J. Food Sci. Technol. 52, 2083–2092. doi:10.1007/s13197-013-1244-z
Ling, K. C. (2008). Whey to ethanol: A biofuel role for dairy cooperatives? Washington DC: University of Minnesota.
Liu, B., Wu, P., Zhou, J., Yin, A., Yu, Y., and Lu, H. (2022). Characterization and optimization of the LAC4 upstream region for low-leakage expression in Kluyveromyces marxianus. Yeast 39, 283–296. doi:10.1002/yea.3682
Liu, H., Liu, J., Tan, B., Zhou, F., Qin, Y., and Yang, R. (2012). Covalent immobilization of Kluyveromyces fragilis β-galactosidase on magnetic nanosized epoxy support for synthesis of galacto-oligosaccharide. Bioprocess Biosyst. Eng. 35, 1287–1295. doi:10.1007/s00449-012-0716-2
Liu, J., Dantoft, S. H., Würtz, A., Jensen, P. R., and Solem, C. (2016). A novel cell factory for efficient production of ethanol from dairy waste. dairy waste Biotechnol. Biofuels 9, 33. doi:10.1186/s13068-016-0448-7
Lukjanenko, J., Kovtuna, K., Scherbaka, R., and Vigants, A. (2014). Bioethanol and biomass production by Kluyveromyces marxianus during lactose fermentation at different salts and substrate concentrations. J. Biotechnol. 185, S122. doi:10.1016/j.jbiotec.2014.07.417
Madeira-, J. V., and Gombert, A. K. (2018). Towards high-temperature fuel ethanol production using Kluyveromyces marxianus: On the search for plug-in strains for the Brazilian sugarcane-based biorefinery. sugarcane-based biorefinery Biomass bioenergy 119, 217–228. doi:10.1016/j.biombioe.2018.09.010
Mathew, A. S., Wang, J., Luo, J., and Yau, S. T. (2015). Enhanced ethanol production via electrostatically accelerated fermentation of glucose using Saccharomyces cerevisiae. Sci. Rep. 5, 15713. doi:10.1038/srep15713
Mo, W., Wang, M., Zhan, R., Yu, Y., He, Y., and Lu, H. (2019). Kluyveromyces marxianus developing ethanol tolerance during adaptive evolution with significant improvements of multiple pathways. Biotechnol. Biofuels 12, 63. doi:10.1186/s13068-019-1393-z
Mohd Azhar, S. H., Abdulla, R., Jambo, S. A., Marbawi, H., Gansau, J. A., Mohd Faik, A. A., et al. (2017). Yeasts in sustainable bioethanol production: A review. Biochem. Biophysics Rep. 10, 52–61. doi:10.1016/j.bbrep.2017.03.003
Moscoviz, R., Toledo-Alarcón, J., Trably, E., and Bernet, N. (2016). Electro-fermentation: How to drive fermentation using electrochemical systems. Trends Biotechnol. 34, 856–865. doi:10.1016/j.tibtech.2016.04.009
Murari, C. S., da Silva, D. C. M. N., Schuina, G. L., Mosinahti, E. F., and Del Bianchi, V. L. (2019a). Bioethanol production from dairy industrial coproducts. Bioethanol Prod. Dairy Industrial Coproducts BioEnergy Res. 12, 112–122. doi:10.1007/s12155-018-9949-5
Murari, C. S., Machado, W. R. C., Schuina, G. L., and Del Bianchi, V. L. (2019b). Optimization of bioethanol production from cheese whey using Kluyveromyces marxianus URM 7404. URM 7404 Biocatal. Agric. Biotechnol. 20, 101182. doi:10.1016/j.bcab.2019.101182
Murata, M., Pattanakittivorakul, S., Manabe, T., Limtong, S., and Yamada, M. (2022). Mutants with enhanced cellobiose-fermenting ability from thermotolerant Kluyveromyces marxianus DMKU 3-1042, which are beneficial for fermentation with cellulosic biomass. Fuels 3, 232–244. doi:10.3390/fuels3020015
Neri, D. F. M., Balcão, V. M., Carneiro-da-Cunha, M. G., Carvalho, L. B., and Teixeira, J. A. (2008). Immobilization of β-galactosidase from Kluyveromyces lactis onto a polysiloxane–polyvinyl alcohol magnetic (mPOS–PVA) composite for lactose hydrolysis. Catal. Commun. 9, 2334–2339. doi:10.1016/j.catcom.2008.05.022
Núñez Caraballo, A., Iliná, A., Ramos González, R., Aguilar, C. N., Michelena Álvarez, G., Flores Gallegos, A. C., et al. (2021). Sustainable ethanol production from sugarcane molasses by Saccharomyces cerevisiae immobilized on chitosan-coated manganese ferrite. Front. Sustain. Food Syst. 5. doi:10.3389/fsufs.2021.683170
Nurcholis, M., Nitiyon, S., Suprayogi, , Rodrussamee, N., Lertwattanasakul, N., Limtong, S., et al. (2019). Functional analysis of Mig1 and Rag5 as expressional regulators in thermotolerant yeast Kluyveromyces marxianus. Kluyveromyces marxianus Appl. Microbiol. Biotechnol. 103, 395–410. doi:10.1007/s00253-018-9462-y
Okamoto, K., Nakagawa, S., Kanawaku, R., and Kawamura, S. (2019). Ethanol production from cheese whey and expired milk by the Brown rot fungus Neolentinus lepideus. Fermentation 5, 49. doi:10.3390/fermentation5020049
Orij, R., Brul, S., and Smits, G. J. (2011). Intracellular pH is a tightly controlled signal in yeast. Biochimica Biophysica Acta (BBA) - General Subj. 1810, 933–944. doi:10.1016/j.bbagen.2011.03.011
Osorio-González, C. S., Gómez-Falcon, N., Brar, S. K., and Ramírez, A. A. (2022). Cheese whey as a potential feedstock for producing renewable biofuels: A review. A Rev. Energies 15, 6828. doi:10.3390/en15186828
Ozmihci, S., and Kargi, F. (2008). Ethanol production from cheese whey powder solution in a packed column bioreactor at different hydraulic residence times. Biochem. Eng. J. 42, 180–185. doi:10.1016/j.bej.2008.06.017
Pasotti, L., Zucca, S., Casanova, M., Micoli, G., Cusella De Angelis, M. G., and Magni, P. (2017). Fermentation of lactose to ethanol in cheese whey permeate and concentrated permeate by engineered Escherichia coli. BMC Biotechnol. 17, 48. doi:10.1186/s12896-017-0369-y
Peña, A., Sánchez, N. S., Álvarez, H., Calahorra, M., and Ramírez, J. (2015). Effects of high medium pH on growth, metabolism and transport in Saccharomyces cerevisiae. FEMS Yeast Res. 15, fou005. doi:10.1093/femsyr/fou005
Pescuma, M., de Valdez, G. F., and Mozzi, F. (2015). Whey-derived valuable products obtained by microbial fermentation. Appl. Microbiol. Biotechnol. 99, 6183–6196. doi:10.1007/s00253-015-6766-z
Prazeres, A. R., Carvalho, F., and Rivas, J. (2012). Cheese whey management: A review. J. Environ. Manag. 110, 48–68. doi:10.1016/j.jenvman.2012.05.018
Radecka, D., Mukherjee, V., Mateo, R. Q., Stojiljkovic, M., Foulquié-Moreno, M. R., and Thevelein, J. M. (2015). Looking beyond Saccharomyces: The potential of non-conventional yeast species for desirable traits in bioethanol fermentation. FEMS Yeast Res. 15, fov053. doi:10.1093/femsyr/fov053
Rajkumar, A. S., Varela, J. A., Juergens, H., Daran, J.-M. G., and Morrissey, J. P. (2019). Biological parts for Kluyveromyces marxianus synthetic biology. Front. Bioeng. Biotechnol. 7, 97. doi:10.3389/fbioe.2019.00097
Rodrigues, B., Lima-Costa, M., Constantino, A., Raposo, S., Felizardo, C., Gonçalves, D., et al. (2016). Growth kinetics and physiological behavior of co-cultures of Saccharomyces cerevisiae and Kluyveromyces lactis, fermenting carob sugars extracted with whey. fermenting carob sugars Extr. whey Enzyme Microb. Technol. 92, 41–48. doi:10.1016/j.enzmictec.2016.06.012
Roohina, F., Mohammadi, M., and Najafpour, G. D. (2016). Immobilized Kluyveromyces marxianus cells in carboxymethyl cellulose for production of ethanol from cheese whey: Experimental and kinetic studies. Bioprocess Biosyst. Eng. 39, 1341–1349. doi:10.1007/s00449-016-1610-0
Ryan, M. P., and Walsh, G. (2016). The biotechnological potential of whey. Bio/Technology 15, 479–498. doi:10.1007/s11157-016-9402-1
Sáenz-Hidalgo, H. K., Guevara-Aguilar, A., Buenrostro-Figueroa, J. J., Baeza-Jiménez, R., Flores-Gallegos, A. C., and Alvarado-González, M. (2021). “Biotechnological valorization of whey: A by-product from the dairy industry,” in Bioprocessing of agri-food residues for production of bioproducts (New Jersey and Canada: Apple Academic Press), 159–200.
Saini, P., Beniwal, A., Kokkiligadda, A., and Vij, S. (2017). Evolutionary adaptation of Kluyveromyces marxianus strain for efficient conversion of whey lactose to bioethanol. Biochemistry 62, 69–79. doi:10.1016/j.procbio.2017.07.013
Sakihama, Y., Hidese, R., Hasunuma, T., and Kondo, A. (2019). Increased flux in acetyl-CoA synthetic pathway and TCA cycle of Kluyveromyces marxianus under respiratory conditions. Sci. Rep. 9, 5319. doi:10.1038/s41598-019-41863-1
Sampaio, F. C., de Faria, J. T., da Silva, M. F., de Souza Oliveira, R. P., and Converti, A. (2020). Cheese whey permeate fermentation by Kluyveromyces lactis: A combined approach to wastewater treatment and bioethanol production. Environ. Technol. 41, 3210–3218. doi:10.1080/09593330.2019.1604813
Sanusi, I. A., Suinyuy, T. N., and Kana, G. E. B. (2021). Impact of nanoparticle inclusion on bioethanol production process kinetic and inhibitor profile. Biotechnol. Rep. 29, e00585. doi:10.1016/j.btre.2021.e00585
Sar, T., Harirchi, S., Ramezani, M., Bulkan, G., Akbas, M. Y., Pandey, A., et al. (2022). Potential utilization of dairy industries by-products and wastes through microbial processes: A critical review. Sci. Total Environ. 810, 152253. doi:10.1016/j.scitotenv.2021.152253
Sayed, W., Cabrol, A., Abdallah, R., Taha, S., Amrane, A., and Djelal, H. (2018). Enhancement of ethanol production from synthetic medium model of hydrolysate of macroalgae. Renew. Energy 124, 3–10. doi:10.1016/j.renene.2017.10.094
Schievano, A., Sciarria, T. P., Vanbroekhoven, K., De Wever, H., Puig, S., Andersen, S. J., et al. (2016). Electro-fermentation–merging electrochemistry with fermentation in industrial applications. Trends Biotechnol. 34, 866–878. doi:10.1016/j.tibtech.2016.04.007
Schmidt, C. M., Nedele, A.-K., and Hinrichs, J. (2020). Enzymatic generation of lactulose in sweet and acid whey: Feasibility study for the scale up towards robust processing. Food Bioprod. Process. 119, 329–336. doi:10.1016/j.fbp.2019.11.015
Sebastián-Nicolás, J. L., González-Olivares, L. G., Vázquez-Rodríguez, G. A., Lucho-Constatino Carlos, A., Castañeda-Ovando, A., and Cruz-Guerrero, A. E. (2020). Valorization of whey using a biorefinery. Bioprod. Biorefining 14, 1010–1027. doi:10.1002/bbb.2100
Sekoai, P. T., Ouma, C. N. M., du Preez, S. P., Modisha, P., Engelbrecht, N., Bessarabov, D. G., et al. (2019). Application of nanoparticles in biofuels: An overview. Overv. Fuel 237, 380–397. doi:10.1016/j.fuel.2018.10.030
Sharma, D., Manzoor, M., Yadav, P., Sohal, J. S., Aseri, G. K., and Khare, N. (2018). “Bio-valorization of dairy whey for bioethanol by stress-tolerant yeast,” in Fungi and their role in sustainable development: Current perspectives. Editors P. Gehlot, and J. Singh (Singapore: Springer Singapore), 349–366. doi:10.1007/978-981-13-0393-7_20
Silva, A. R., Alexandre, J. Y., Souza, J. E., Neto, J. G. L., de Sousa Júnior, P. G., Rocha, M. V., et al. (2022). The chemistry and applications of metal–organic frameworks (MOFs) as industrial enzyme immobilization systems. Molecules 27, 4529. doi:10.3390/molecules27144529
Soupioni, M., Vlachou, K., Psarologou, A., and Bekatorou, A. (2016). Abating whey organic load through ethanol and lactic acid production by kefir. Toxicol. Environ. Chem. 98, 1191–1199. doi:10.1080/02772248.2016.1220559
Sriram, S., Wong, J. W. C., and Pradhan, N. (2022). Recent advances in electro-fermentation technology: A novel approach towards balanced fermentation. Bioresour. Technol. 360, 127637. doi:10.1016/j.biortech.2022.127637
Tavares, T., and Malcata, F. X. (2016). “Whey and whey powders: Fermentation of whey,” in Encyclopedia of food and health (Oxford: Academic Press), 486–492. doi:10.1016/B978-0-12-384947-2.00749-2
Tesfaw, A., and Assefa, F. (2014). Current trends in bioethanol production by Saccharomyces cerevisiae: Substrate, inhibitor reduction, growth variables, coculture, and immobilization international. Sch. Res. Notices 2014, 532852. doi:10.1155/2014/532852
Tesfaw, A., Kosebent, D., Oner, E. T., and Assefa, F. (2021a). Bioethanol production from grass pea and wild oat hydrolysates using S. cerevisiae ETP53, K. marxianus ETP87, and P. Ferment. ETP22 Discov. Energy 1, 1–12.
Tesfaw, A., Oner, E. T., and Assefa, F. (2021b). Evaluating crude whey for bioethanol production using non-Saccharomyces yeast, Kluyveromyces marxianus. Kluyveromyces Marx. SN Appl. Sci. 3, 42. doi:10.1007/s42452-020-03996-1
Tesfaw, A., Oner, E. T., and Assefa, F. (2021c). Optimization of ethanol production using newly isolated ethanologenic yeasts. Biochem. Biophysics Rep. 25, 100886. doi:10.1016/j.bbrep.2020.100886
Tomaszewska, M., and Białończyk, L. (2016). Ethanol production from whey in a bioreactor coupled with direct contact membrane distillation. Today 268, 156–163. doi:10.1016/j.cattod.2016.01.059
Utama, G. L., Kurnani, T. B. A., and Balia, R. L. (2017a). Reducing cheese-making by-product disposal through ethanol fermentation and the utilization of distillery waste for fertilizer. Geomate J. 13, 103–107.
Utama, G. L., Kurnani, T. B. A., Sunardi, S., Cahyandito, M. F., and Balia, R. L. (2017b). Joint cost allocation of cheese-making wastes bioconversions into ethanol and organic liquid fertilizer Bulgarian. J. Agric. Sci. 23, 1016–1020.
Valdez Castillo, M., Tahmasbi, H., Pachapur, V. L., Brar, S. K., Vuckovic, D., Sitnikov, D., et al. (2021). Production of aroma and flavor-rich fusel alcohols by cheese whey fermentation using the Kluyveromyces marxianus and Debaryomyces hansenii yeasts in monoculture and co-culture modes. J. Chem. Technol. Biotechnol. 96, 6763–2367. doi:10.1002/jctb.6763
Valta, K., Damala, P., Angeli, E., Antonopoulou, G., Malamis, D., and Haralambous, K. J. (2017). Current treatment technologies of cheese whey and wastewater by Greek cheese manufacturing units and potential valorisation opportunities. Valoris. Oppor. Waste Biomass Valorization 8, 1649–1663. doi:10.1007/s12649-017-9862-8
Vanegas Juan, M., Contreras Maria, F., Faller, R., and Longo Marjorie, L. (2012). Role of unsaturated lipid and ergosterol in ethanol tolerance of model yeast biomembranes. Biomembr. Biophysical J. 102, 507–516. doi:10.1016/j.bpj.2011.12.038
Varela, J. A., Montini, N., Scully, D., Van der Ploeg, R., Oreb, M., Boles, E., et al. (2017). Polymorphisms in the LAC12 gene explain lactose utilisation variability in Kluyveromyces marxianus strains. FEMS Yeast Res. 17. doi:10.1093/femsyr/fox021
Varela, J. A., Puricelli, M., Montini, N., and Morrissey, J. P. (2019a). Expansion and diversification of MFS transporters in Kluyveromyces marxianus. Front. Microbiol. 9, 3330. doi:10.3389/fmicb.2018.03330
Varela, J. A., Puricelli, M., Ortiz-Merino, R. A., Giacomobono, R., Braun-Galleani, S., Wolfe, K. H., et al. (2019b). Origin of lactose fermentation in Kluyveromyces lactis by interspecies transfer of a neo-functionalized gene cluster during domestication. Curr. Biol. 29, 4284–4290. doi:10.1016/j.cub.2019.10.044
Västermark, Å., and Saier, M. H. (2014). Major Facilitator Superfamily (MFS) evolved without 3-transmembrane segment unit rearrangements. Proc. Natl. Acad. Sci. 111, E1162–E1163. doi:10.1073/pnas.1400016111
Verma, D., Paul, J. S., Tiwari, S., and Jadhav, S. K. (2022). A review on role of nanomaterials in bioconversion of sustainable fuel bioethanol. Waste Biomass Valorization 13, 4651–4667. doi:10.1007/s12649-022-01843-5
Verma, M. L., Barrow, C. J., Kennedy, J. F., and Puri, M. (2012). Immobilization of β-d-galactosidase from Kluyveromyces lactis on functionalized silicon dioxide nanoparticles: Characterization and lactose hydrolysis. Int. J. Biol. Macromol. 50, 432–437. doi:10.1016/j.ijbiomac.2011.12.029
Vieira, E. F., Carvalho, J., Pinto, E., Cunha, S., Almeida, A. A., and Ferreira, I. M. (2016). Nutritive value, antioxidant activity and phenolic compounds profile of brewer’s spent yeast extract. J. Food Compos. Analysis 52, 44–51. doi:10.1016/j.jfca.2016.07.006
Vincenzi, A., Maciel, M. J., Burlani, E., Oliveira, E., Volpato, G., Lehn, D. N., et al. (2014). Ethanol bio-production from ricotta cheese whey by several strains of the yeast Kluyveromyces. Am. J. Food Technol. 9, 281–291. doi:10.3923/ajft.2014.281.291
Wei, T.-H., Wu, S. H., Huang, Y. D., Lo, W. S., Williams, B. P., Chen, S. Y., et al. (2019). Rapid mechanochemical encapsulation of biocatalysts into robust metal–organic frameworks. Nat. Commun. 10, 5002. doi:10.1038/s41467-019-12966-0
Yadav, J. S. S., Bezawada, J., Elharche, S., Yan, S., Tyagi, R. D., and Surampalli, R. Y. (2014). Simultaneous single-cell protein production and COD removal with characterization of residual protein and intermediate metabolites during whey fermentation by K. marxianus. marxianus Bioprocess Biosyst. Eng. 37, 1017–1029. doi:10.1007/s00449-013-1072-6
Yamada, S., Takamatsu, Y., Ikeda, S., Kouzuma, A., and Watanabe, K. (2022). Towards application of electro-fermentation for the production of value-added chemicals from biomass feedstocks. Front. Chem. 9, 805597. doi:10.3389/fchem.2021.805597
Yan, N. (2013). Structural advances for the major facilitator superfamily (MFS) transporters. MFS) Transp. Trends Biochem. Sci. 38, 151–159. doi:10.1016/j.tibs.2013.01.003
Zafar, S., and Owais, M. (2006). Ethanol production from crude whey by Kluyveromyces marxianus. Kluyveromyces marxianus Biochem. Eng. J. 27, 295–298. doi:10.1016/j.bej.2005.05.009
Zhou, X., Hua, X., Huang, L., and Xu, Y. (2019). Bio-utilization of cheese manufacturing wastes (cheese whey powder) for bioethanol and specific product (galactonic acid) production via a two-step bioprocess. Bioresour. Technol. 272, 70–76. doi:10.1016/j.biortech.2018.10.001
Zohri, A.-N. A., Gomah, N. H., and Ali, M. A. (2014). Utilization of cheese whey for bio-ethanol production. Univers. J. Microbiol. Res. 2, 57–73. doi:10.13189/ujmr.2014.020401
Zotta, T., Solieri, L., Iacumin, L., Picozzi, C., and Gullo, M. (2020). Valorization of cheese whey using microbial fermentations. Appl. Microbiol. Biotechnol. 104, 2749–2764. doi:10.1007/s00253-020-10408-2
Keywords: bioethanol, cheese whey, kluyveromyces, valorization, immobilization
Citation: Tesfaw A (2023) The current trends of bioethanol production from cheese whey using yeasts: biological and economical perspectives. Front. Energy Res. 11:1183035. doi: 10.3389/fenrg.2023.1183035
Received: 09 March 2023; Accepted: 04 May 2023;
Published: 15 May 2023.
Edited by:
Halil Durak, Yüzüncü Yıl University, TürkiyeReviewed by:
Rahmiye Zerrin Yarbay, Bilecik Şeyh Edebali University, TürkiyeRicha Arora, Punjab Agricultural University, India
Copyright © 2023 Tesfaw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asmamaw Tesfaw, YXNtYW1hd3Rlc2Zhd0BkYnUuZWR1LmV0
 Asmamaw Tesfaw
Asmamaw Tesfaw