- 1School of Biological Science and Engineering, North Minzu University, Yinchuan, China
- 2Ningxia Grape and Wine Technology Innovation Center, North Minzu University, Yinchuan, China
- 3Bioenergy Research Centre, Department of Bioinformatics and Biotechnology, Government College University Faisalabad, Faisalabad, Pakistan
- 4School of Food and Wine, Ningxia University, Yinchuan, China
- 5National Institute for Biotechnology and Genetic Engineering College, Pakistan Institute of Engineering and Applied Sciences (NIBGE-C, PIEAS), Faisalabad, Pakistan
Efficient biotransformation of lignocellulosic biomass to second-generation (2G) bioethanol requires promising strains harboring built-in resistance against limitations imposed by pretreated lignocellulose-derived compounds. Ethanol fermentation and stress tolerance of yeast cells are almost simultaneously exposed to sequence variations and multiple inhibitory factors during the phases of proliferation, metabolism, and productivity. Several studies have extensively concentrated on identification or characterization of genes which confer resistance to various stresses and yeast tolerance enhancement through genetic breeding. However, the investigation of individual genes is inadequate to explain the global molecular mechanism. Herewith, “OMICs-approaches,” including genomics, transcriptomics, proteomics, and metabolomics, which are comprehensively aimed at comparative, functional profiling of the whole metabolic network, have elucidated complex cellular reactions under stressful conditions. This review briefly discusses the research progress in the field of multi-OMICs with a special focus on stress-responsive factors in frequently used S. cerevisiae. It also highlights how to promote metabolic-engineered strains for increased tolerance and higher production yield, which should be deeply exploited to achieve robustness during the lignocellulose-to-ethanol conversion process.
1 Introduction
Biofuels have emerged as alternatives to fossil-based liquid fuels due to their renewable and carbon-neutral nature. Generally, bioethanol is produced from food-crops including sugarcane and maize, which causes grain shortage. Meanwhile, crops as a sustainable source would take up a huge amount of arable land; for instance, 300% of the total US area would be needed to cultivate maize to meet the energy demands of the US alone (Usmani et al., 2021). Lignocellulosic biomass from agricultural practices, forests, and marginal lands is believed to be a potential resource for future fuel supply (Mehmood et al., 2017a). Unfortunately, biological conversion of lignocellulose faces difficulties mainly due to the stubbornness alleviated by lignin and hemicellulose, often requiring physicochemical pretreatments to break the recalcitrance of the biomass (Pant et al., 2022). Various inhibitory compounds derived from the pretreatment belong to four different groups, namely, organic acids (e.g., acetic acid), phenols (e.g., phenol), aldehydes (e.g., furfural), and ketones; these chemicals and their toxicity cannot be decomposed during enzymatic hydrolysis and the fermentation stage (Liu, 2011; Sjulander and Kikas, 2020). Unlike simple substrate (glucose) and product inhibition (ethanol) in VHG (very high gravity) fermentation, multiple inhibitory effects of the lignocellulosic hydrolysate pose remarkable problems to the ethanol production efficiency of brewing yeast, which also cannot be alleviated by using traditional methods of bioreaction engineering (Melendez et al., 2022).
Apart from toxic byproducts, higher ethanol titers and elevated temperatures also exert stress on the Saccharomyces cerevisiae, that is, the workhorse to produce cellulosic bioethanol, which significantly compromises productivity and yield. If achieved, high concentration of ethanol does not only reduce the energy consumption for ethanol recovery by distillation but also reduces stillage discharge (Liu et al., 2019). However, high-concentration ethanol inhibits yeast cells, resulting in paused fermentation (Stanley et al., 2010; Vamvakas and Kapolos, 2020). On the other hand, elevated temperature facilitates the control of contamination, but S. cerevisiae cannot usually tolerate more than 34°C (Cripwell et al., 2020). Consequently, a chilled water system is needed for almost all fuel ethanol plants, since regular cooling of water from the cooling tower with a temperature only 2–3°C lower than the environmental temperature cannot cool down the fermenters in summer when the temperature reaches as high as 35°C, which always increases the cost of production. Apparently, it is believed that thermo-tolerant strains can address this issue.
The adaptation-based response of S. cerevisiae to the above stresses that adversely influence growth and metabolism can be sorted into ethanol-, inhibitor-, oxidative-/thermal-tolerance, and intriguing joint cellular protections including membrane integrity, organelle modifications, and so forth (Li et al., 2022). Rapid advancements in “OMICs” and metabolic engineering techniques have paved a way to elucidate the underlying mechanisms with characteristic phenotypes under the stressful conditions followed by targeted pathway engineering to develop stress-tolerant strains for biofuel production (Jayakody and Jin, 2021; Mehmood et al., 2021). Hence, in this review, the recent progress on efficient ethanol production made through OMICs-based studies including comparative genomics or multi-OMICs to identify the molecular mechanism of stress tolerance is discussed. Second, the up/down regulation of key genes from bioethanol strains depending on specific-conditioned inhibitions is overviewed. Moreover, some fine-tuning approaches for tolerance improvement of recombinants, as well as inevitable challenges or striving directions of lignocellulosic bioethanol production, are subsequently pointed out.
2 Role of Comparative Genomics in Elucidating Yeast Tolerance Mechanisms
The 2G (de novo or re-sequencing) and third-generation (single molecule sequencing) DNA sequencing have been applied to reveal genetic traits of yeasts. The whole genome of model S. cerevisiae S288c was sequenced (Engel et al., 2013). Since then, comprehensive investigation of comparative genomics was performed to study numerous structural variations, genomic loci, and nucleotide mutations for assigning diversity of S. cerevisiae strains. Compared with S288c, SNPs (single nucleotide polymorphisms), Indels (insertions/deletions), CNV (copy number variation), and unique ORFs (open reading frames) from genomics of special niche isolated strains were identified, for instance, the wild strain YJM789, containing 60,000 SNPs, 6,000 indels, and several unique ORFs that might have been acquired through gene transfer (Wei et al., 2007) as well as functional gene conversions from meiotic recombination (Sun et al., 2019). Another example is Kyokai No. 7 (K7) in Japanese sake brewery, a closely related strain to the S288c, but sub-telomeric polymorphisms and large inversions were found. Besides, it was pinpointed that two DNA segments located on the chromosome numbers I and VII of S288c were replaced by the Ty elements of K7 (Akao et al., 2011); whereafter, chromosomal recombination defect of haploids of K7 was examined (Shimoi et al., 2019).
Comparison of genome-scale variations among species is being used in confirming the crucial independent domestication or artificial selection that took place during the evolutionary history. The sequenced-genome of interspecific hybrid and wine-making strain VIN7 showed several SNPs and chromosomal rearrangements between the diploid S. cerevisiae and the haploid S. kudriavzevii. A distance tree, based on SNP variations, was constructed among VIN7 and 24 other S. cerevisiae strains which demonstrated that it had a European origin (Borneman et al., 2012). Genome assemblies of six commercial S. cerevisiae strains containing four wine (Lalvin QA23, AWRI796, Vin13, and VL3) and two brewing strains (Fosters O and Fosters B) with S. cerevisiae strains (S288c, YJM789, JAY291, RM11-1a, and EC1118) have revealed clear signature sequences which implied that each industrial strain has signature genetic variations comprising SNPs, large-scale Indels, and putative novel genes between them (Borneman et al., 2011). The genome variability of 16 yeast strains, of both laboratory and commercial origins, where sub-telomeric instability, differences of Ty element insertion regions, copy number changes, or depletion were listed, was studied (Carreto et al., 2008). Likewise, a survey of a collection of 63 diverse strains from diverse ecological niches manifested nearly two million SNPs and the occurrence of 3,985 deletions of a length >200 bp (Schacherer et al., 2009). An investigation of 83 strains confirmed interspecific hybridization and pervasive CNV in wild and industrial strains (Dunn et al., 2012). The genomes of 93 strains, collected from different geographical sites, indicated that most of the genetic or phenotypic variations were quantitative (Strope et al., 2015). These studies pointed to the promising prospect of studying genome-scale change to elucidate the origin of novel genetic elements.
Phenotypic impact of genomic variations provides the evidence for enabling strains to endure different stresses, so the resistance potential of the strains can be harnessed through inferring the genetic basis of tolerance. Genomic features of the yeast PE-2 derived diploid (JAY270) and haploid (JAY291) strains uncovered higher zygosity and polymorphism (∼2 SNPs/kb) in the JAY270 between homologous chromosomes, and chromosomal rearrangements were limited to the peripheral sites of chromosomes, which may illustrate adaptation of two strains to ethanol, thermal, and oxidative stresses (Argueso et al., 2009). The CNVs of important industrial strains, BG-1, CAT-1, PE-2, SA-1, and VR-1, which are being used to produce fuel ethanol from sugarcane in Brazil, were characterized, and substantial augmentations of the SNO and SNZ genes were discovered, which exhibited the adaptive competence to biomass utilization in an industrial environment (Stambuk et al., 2009). A similar event was also discovered in the Brazilian strain JAY291 derived mutant which could tolerate 16–17% ethanol. It was observed that three closely located genes, namely, MKT1, SWS2, and APJ1, were responsible for affecting ethanol tolerance. Mkt1 was recruited to form a complex that may mediate posttranscriptional regulation of HO endonuclease gene. Sws2 is a mitochondrial ribosomal protein that is essential for respiratory growth. Apj1 is the chaperone of Hsp40 with a role in SUMO-mediated protein degradation (Swinnen et al., 2012). The genome of S. cerevisiae CAT-1 strain contained ∼36,000 homozygous and ∼30,000 heterozygous SNPs, several duplications, and deletions, particularly at the telomeric regions, whereas some genes like IRA1 and IRA2, encoding GTPase-activating proteins, were associated with bioethanol production (Babrzadeh et al., 2012). Alterations in gene expression involved central carbon metabolism, antioxidation activities, and membrane conformations that might be responsible for multiple stress tolerance of bioethanol-producing and tolerant strains YJS329 and ZK2. ELO1 (encoding a fatty acid elongase) overexpression improved acetic acid tolerance of both strains, and introduction of ZNF1 (Zinc cluster transcription factor) into ZK2 increased ethanol, H2O2, and acetic acid tolerance (Zheng et al., 2013). An isolate (S3-110) capable of tolerating higher ethanol titer under the VHG condition was obtained through whole genome shuffling of an industrial strain ZTW1. The strain could tolerate 55°C and oxidative and ethanol stresses, which was attributed to the CNVs of the stress-responsive genes (Zheng et al., 2014). In particular, the ZTW1 was confirmed as an aneuploidy strain; its changed DNA dosage, such as CNVs in chromosomal segments or CNVs of functional genes, might have improved its fermentation rate and tolerance to various stresses (Zhang et al., 2016).
A set of stress-responsive genes in different metabolic pathways were selected by genome-wide scale analysis of yeast in the laboratory and industry that annotated novel functions for regulatory and protein-coding genes. For BY4743 strain, 331 genes were found to confer hypersensitivity to oxidative stress (3 mM H2O2). Among these genes, 71 genes were essential, which were associated with nucleolus, rRNA, and tRNA synthesis (Okada et al., 2014). For BY4741 strain, 650 genes were speculated to be involved in acetic acid stress (70–110 mM, pH 4.5), which mediated cellular processes including carbohydrate metabolism, pH homeostasis, cell wall assembly, biogenesis of vacuoles, and mitochondria. More than 75% of those genes were first certified for acetic acid tolerance (Mira et al., 2010a). Pereira et al. (2011) reported 8 genes linked with concurrent resistance to stress imposed by higher concentrations of ethanol, glucose, and acetic acid; meanwhile, 11 genes were associated with lignocellulosic-derived stress tolerance. In addition, BUD31 and HPR1 were found to have a key role in ethanol yield and fermentation rate. PHO85, VRP1, and YGL024w were indispensable for maximal ethanol production. In a subsequent analysis, genes responsible for vitamin metabolism, peroxisomal and mitochondrial functions, and biogenesis of ribosomes and microtubules were identified in wheat straw hydrolysate resistance (Pereira et al., 2011; Pereira et al., 2014). Besides, genome sequencing of the natural yeast strain MUCL28177 with quantitative trait loci (QTL) mapping led to the discovery that several new QTLs are responsible for heat stress and MKT1, PRP42, and SMD2 as causative genes suggests RNA processing is crucial in thermotolerance (Yang et al., 2013). PRP42 and SMD2 encoding U1 snRNP protein and subunit of U2-type pre-spliceosome complex, respectively.
Due to a mixture of lignocellulose-derived hydrolysates, for existing multiple stresses, five genes, namely, ERG2 (encoding C-8 sterol isomerase in ergosterol biosynthesis), PRS3 (encoding pyrophosphate synthetase), RAV1 (encoding subunit of RAVE complex), RPB4 (encoding RNA polymerase II subunit), and VMA8 (encoding subunit of the domain of V-ATPase), which were demonstrated as the tolerant genes, contributed to maintaining the cell viability and maximal fermentation rate during fermentation of wheat straw hydrolysates (Pereira et al., 2014). Ubp7, Art5, Nrg1 (stress response transcriptional repressors), and Gdh1 (glutamate dehydrogenase) of strain R57 evolved via genome shuffling were selected as biological determinants for tolerance of spent sulfite liquor, which provided certain targets for strain breeding (Pinel et al., 2015). The industrial strain ISO12, which is tolerant to inhibitors at 37°C, unfolded several SNPs and Indels in its parental strain ethanol Red. Besides, CYC3 (encoding cytochrome C heme lyase), MTL1 (encoding putative plasma membrane sensor), and FLO genes (encoding flocculating proteins) have explained positive selection in thermal and inhibitor stresses (Wallace-Salinas et al., 2015). Therefore, composited stress-related genes must be screened for hydrolysate fermentation to achieve desired properties that are highly resistant to compound inhibitors.
Furthermore, S. cerevisiae NCIM3186 (Goud and Ulaganathan, 2015), NCIM3107 (Ulaganathan et al., 2015), IR-2 (Sahara et al., 2014), BG-1 (Coutouné et al., 2017), and BT0510 (Costa et al., 2021) are being utilized in production of bioethanol from various feedstocks, such as sweet sorghum, sugarcane, or the wild-type FMY097 (Nagamatsu et al., 2019) and Pf-1 (Kanamasa et al., 2019) that isolated at special habitats. These strains have been genome-sequenced, which will conceivably yield fresh genetic insights into genomics diversity and evolutionary adaptation.
As a prerequisite, there is an urgent demand for screening and sequencing of newly separated and industrial strains, which could resolve significative variations in genetics and correlated mutations on evolution through comparative genome analysis. Fundamental work on data mining of genomics and function validation on stress tolerance still needs to be done and evaluated.
3 Multi-OMICs Analysis for Tolerant Phenotypes Under Characteristic Stresses
Multi-OMICs other than genomics, including transcriptomics, proteomics, and metabolomics, have been implemented to explore mechanisms of stress tolerance (Amer and Baidoo, 2021). Similarly, microarray analysis and gene deletion libraries, at a genome-wide, transcriptional, and translational level, can interpret the functional elements responding to characteristic stresses. Here, representative OMICs studies of S. cerevisiae tolerance in the past decades are summarized in Table 1.
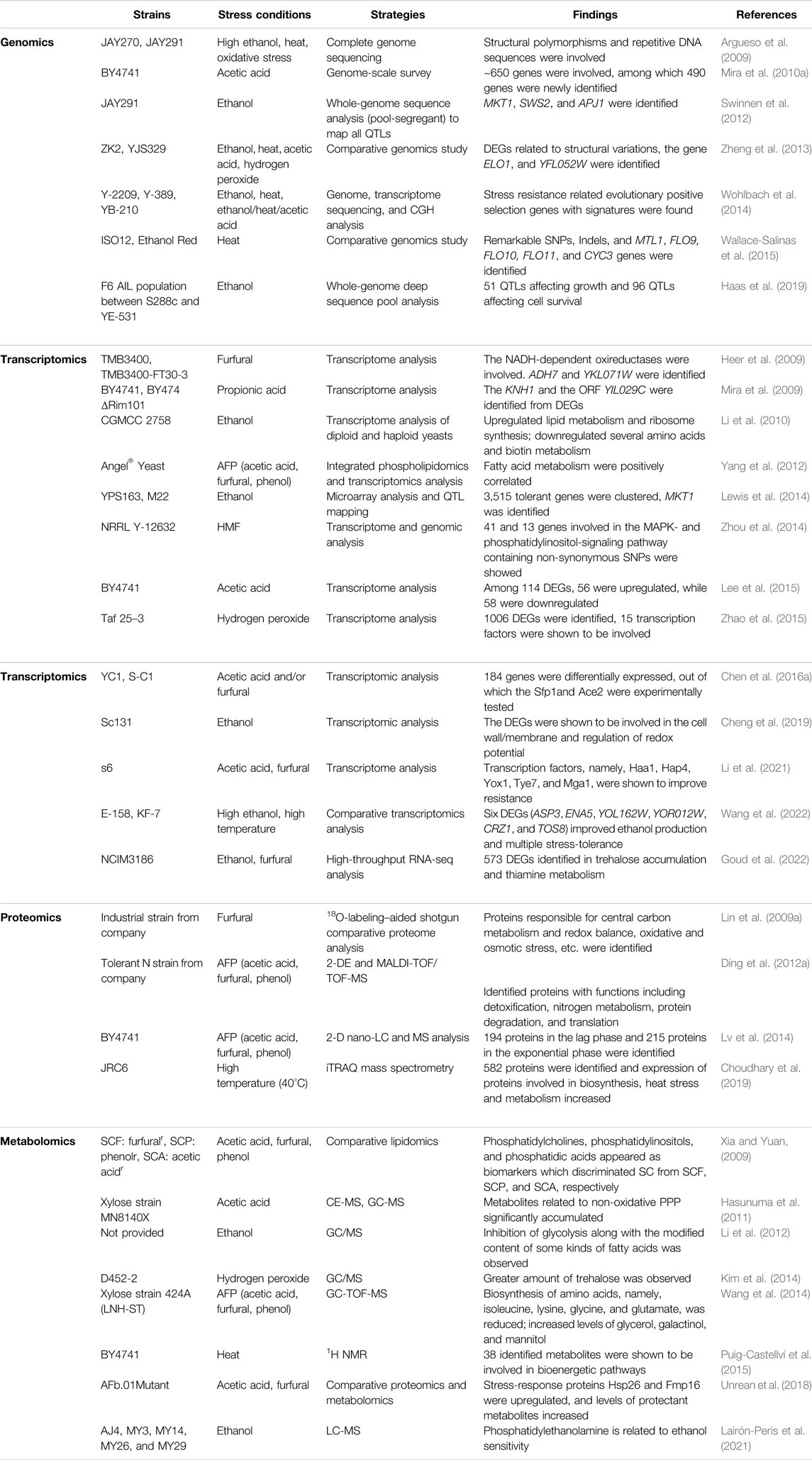
TABLE 1. Representative examples of “OMICs” study of S. cerevisiae stress tolerance for fuel bioethanol.
Keeping in view its applications, transcriptomics or RNA sequencing unveils the relationship of phenotypes at variable transcriptional levels and detects hot spots of gene engineering that help develop strains for industry (Negi et al., 2022). Although market price of protein-seq is more expensive than that of RNA-seq, mass spectrometry technologies have highlighted the great potential to precisely profile the proteome, to map protein–protein interactions, and to distinguish the protein modifications (Lenz and Dihazi, 2016). Based on optimal experimental design, metabolomics has been used in bioprocessing of ethanol production to account for differential metabolite profiles caused by various feedstocks (Abdelnur et al., 2014) and inhibitors (Chen Z. et al., 2016). There are two catalogs of metabolome: 1) untargeted metabolomics, which represents the characterization of simultaneous qualitative and quantitative analysis of a large number of metabolites, and 2) targeted metabolomics, which includes quantitative data on a pre-defined set of compounds, like lipidome (Sévin et al., 2015). Yeast lipidome is critical in balancing fatty acid metabolism, maintaining the plasma membrane integrity, and adapting to ambient surroundings, including sphingolipids, glycerophospholipids, and so on, which, thus, immediately provides insights into explicating adjustments of metabolic reaction for both the intra- and extra-cellular stimuli (Yuan et al., 2011; Klose et al., 2012). Taking lipidomics analyses of a furfural-, a phenol-, and an acetic acid–tolerant strain as an example, their metabolomes were discriminated by phosphatidylcholines, phosphatidylinositols, and phosphatidic acids, individually, when compared to their initial strain (Xia and Yuan, 2009).
It has been demonstrated over the last decades that considerable data can be quickly collected by multi-OMICs sequencing, illustrative of the tolerant phenotypes under characteristic stresses that seem to become much easier, but it is critical to center on a set of key data and then analyze the real reason underlying the intrinsic mechanism. The complexity of OMICs-derived interpretation lies not only in the regulations of tolerance such as synergy and antagonism but in the superimposed effects of stimulation or suppression from inhibitors generated by different materials and pretreatment methods on yeast cells. Primarily, faced with toxicities of ethanol and hydrolysates and co-existing pressures of thermal and oxidative stress, the molecular evidence reflects a paradox that the tolerating performance of strains engineered for multi-substrate conversion that is directly affected before ethanol production; these events should be concerned, as shown in Figure 1.
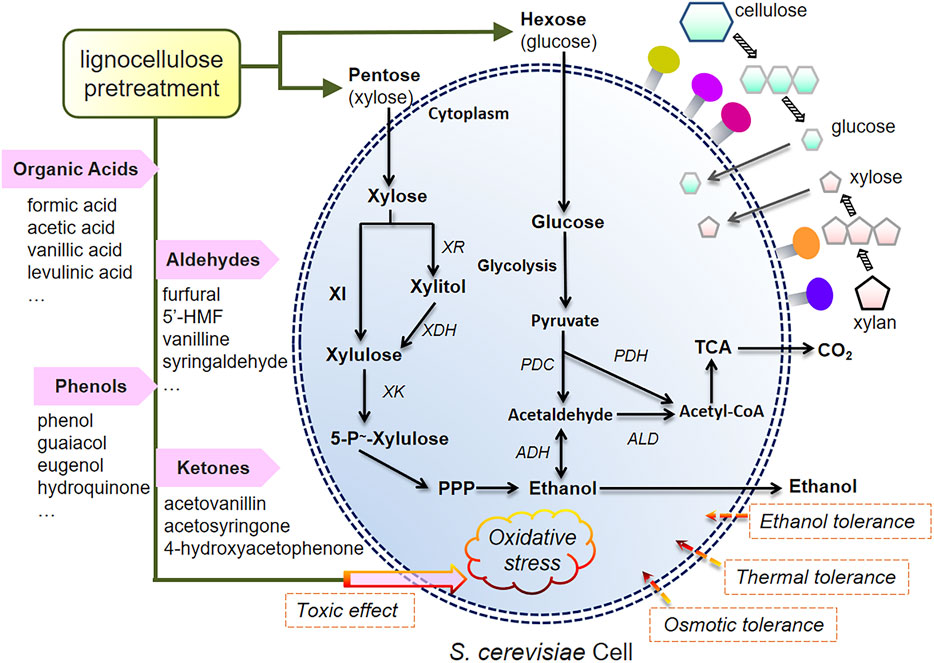
FIGURE 1. Stress tolerance in S. cerevisiae against pretreated hydrolysate inhibitors and metabolic pathways of xylose-fermenting or CBP strains during lignocellulosic ethanol fermentation.
3.1 Ethanol Tolerance
S. cerevisiae is an excellent cell factory for producing ethanol, yet it is also sensitive to a critical threshold level of ethanol concentration. Ethanol stress is one of the earliest tolerances to be focused during fermentation; many studies uncovered the genetic response for strains in ethanol tolerance (Lairón-Peris et al., 2021). Gene expression and pathway clustering mainly associated with the cell wall, membrane organization, nucleus, mitochondrion, cellular enzymes activities, lipid composition, and ethanol stress-responsive factors, along with the interplay of in-depth integrated gene networks, like Msn2/4 regulators, and implicating the cAMP-protein kinase pathway in signal transduction regulation, could assist to establish general stress-tolerance profiles in yeast (Ma and Liu, 2010c; Vamvakas and Kapolos, 2020). Nevertheless, in lignocellulose-ethanol fermentation, because of multiple inhibitors, the yield and ethanol production is sharply reduced, so the cell tolerance is often intrinsically fragile.
Under 5–8% (v/v) ethanol stress, the tryptophan biosynthesis of brewing strain IFO2347 was upregulated, and overexpressing the tryptophan permease gene TAT2 and tryptophan biosynthesis related TRP genes conferred ethanol stress tolerance (Hirasawa et al., 2007). The genes encoding ATP synthesis in mitochondria and ribosomal proteins were highly upregulated in A1 strain, derived from S. cerevisiae FY834 strain, in the presence of 10% (v/v) ethanol (Dinh et al., 2009). Gene expression profiling of the diploid strain CGMCC 2758 and two homologous haploid strains under 3 and 7% (v/v) ethanol concentration revealed that genes responsible for lipid synthesis (e.g. FAA1) and biosynthesis of ribosomes were upregulated, while genes associated with amino acid synthesis (TRP2/3), metabolism of biotin (BIO3/4/5), and folic acid were downregulated (Li et al., 2010). When a tolerant strain, NRRL Y-50316, was exposed to 8% (v/v) ethanol stress, expressed genes engaged in glycolysis, trehalose metabolism, heat shock, pentose phosphate pathway (PPP), and fatty/amino acid metabolism were identified. Among these, 82 candidate genes were confirmed to share binding motifs of transcription factors of Hsf1, Msn4/2, Pdr1/3, and Yap1 (Ma M. and Liu L. Z., 2010).
In addition, the metabolic analysis of diploid (α/a) and haploid (α,a) strains proved that the diploid strains have increased glycolytic intermediates and amino acids when exposed to 3 and 7% (v/v) ethanol stress. So, a higher change in these metabolites, such as precursors of phospholipids and unsaturated fatty acids, in the haploid strains indicated the vulnerability of the haploid strains to ethanol stress (Ding et al., 2010). In another study, 29 different metabolites were found from metabolomic profiling of a stress-exposed strain. With concomitant inhibition of glycolysis, the levels of fatty acids including hexadecanoic, octadecanoic, and palmitelaidic acids were increased to maintain the integrity of the plasma membrane, and amino acids were accumulated (Li et al., 2012).
Similarly, the YDR307W and YHL042W were verified to be linked with improved tolerance to ethanol in BY4743 strain derived from a cross between BY4742 and BY4741, which are generated from S288c (Kasavi et al., 2014). A total of 3515 genes were differentially expressed in 5% (v/v) ethanol stress response for the S288c, M22, and YPS163 strains. Moreover, MKT1 has been certified to be associated with ethanol tolerance (Lewis et al., 2014). The transcriptional levels of two ethanol tolerant mutants of S. cerevisiae, namely, PMT7Δ and YHL042WΔ, were compared with the reference strain under 8% (v/v) ethanol stress. A total of 60 genes were ethanol-responsive, including transcription factors like Yap1, Phd1, Msn2, and Gcn4 (Kasavi et al., 2016). In terms of improving ethanol tolerance of S. cerevisiae, 36 compounds were included in the metabolomics of 14 mutants of S. cerevisiae BY4742, and further evidence of valine and inositol in upregulation of ethanol tolerance was confirmed by deleting LEU4 and LEU9 genes and INM1 and INM2 genes, resulting in the reduction of valine accumulation and inositol concentration (Ohta et al., 2016). 108 intracellular stress-responsive metabolites were identified in ethanol-adapted strain iETS3 compared with wild-type BY4741 under 5% (v/v) ethanol stress, which were mostly illustrated to be the intermediates of central carbon metabolism, and changed metabolites involved in cell membranes, trehalose biosynthesis, and glutamate metabolism (Kim et al., 2016).
Recently, further analyses were conducted in five representative strains (AJ4, MY3, MY14, MY26, and MY29) of S. cerevisiae for a range of varying tolerance with 6 and 10% (v/v) ethanol treatment indicating lipidomic differences, or permeable membrane compositions associated with possible involvement in ethanol sensitivity (Lairón-Peris et al., 2021). From phenomics perspectives, the functional divergences among ethanol tolerant strains were introduced, jointly integrating OMICs data and network analysis of regulatory expression, suggesting a cluster of long non-coding RNAs (lncRNAs) that bind or interact with DNA or proteins, which play a highlighted role in RNA/protein storage and degradation that promotes tolerance (Wolf et al., 2021). The reconstruction strategy of ethanol tolerance on energy metabolism and pathways involved in cell cycle progression was uncovered via quantitative proteomics and genomic changes. Under 6% (v/v) ethanol stress, energy of resistant strains was mainly produced from glycolysis and ethanol fermentation, other than the responses of the ancestral S288c, which switched to cell respiration and utilizing the electron transport chain in the mitochondria (Sostaric et al., 2021).
Up to now, there are some inconsistencies that exist in practice between integrated OMICs data, and confirmatory experiments are needed for a rigorous verification, though the ethanol tolerance mechanism has been generally studied. It is reported that exogenous addition of arginine and tryptophan in the medium or increased level of valine could enhance ethanol tolerance and accumulation of content of cell protectant proline, aiding against various stresses (Cheng et al., 2016; Auesukaree, 2017); however, expression of genes within amino acid biosynthesis pathways does not always keep upregulating, which may just represent anabolic metabolism in cell growth regulation. The association of amino acid metabolism with stress tolerance largely depends on fluctuating stressful conditions; if the stress condition becomes more acutely severe, it is hard to speculate whether reduced metabolism of amino acid may or may not help with the cell tolerance and survival, for which the mechanism must be pursued for an overall convinced proof instead of the selective confirmations.
3.2 Hydrolysate Inhibitor Tolerance
3.2.1 Acetic Acid, Furfural, and Hydroxymethylfurfural
Acetic acid is one of the organic weak acids, produced directly from the hydrolysis of acetyl groups in hemicelluloses during pretreatment of biomass, and it serves as a byproduct of glucose metabolism, which stimulates stress response in S. cerevisiae (Mira and Teixeira, 2013; Jönsson and Martín, 2016). Mechanisms of acetic acid tolerance can be categorized into several aspects in brief: 1) activated transporters: Fps1 aquaglyceroporin channel, Pma1, a ATPase that pumps cellular protons, and Ade2 acetate transmembrane transporter; 2) Haa1 was proved very critical to regulating 80% of acetic acid resistance, to enable moderate effects of two important regulators, Msn2 and Nrg1; 3) proteins involved in the cell wall and membrane, phospholipid synthesis, amino acid synthesis, ion homeostasis, and carbohydrate metabolism are susceptible to varied levels of acetic acid; and 4) lethal concentrations of acetic acid induce protein misfolding, organelle dysfunction, and cell death (Giannattasio et al., 2013; Ndukwe et al., 2020). Besides, modulating the purine biosynthesis has also shown to confer tolerance against acetic acid stress (Zhang et al., 2016). The transcriptomics of BY4741 released 56 and 58 genes, whose expression was modified after 12 h of exposure to 0.6% acetic acid (pH 4.5), respectively. The genes DBP2, ASC1, and GND1 were involved in acetic acid tolerance (Lee et al., 2015). The problem still exists, in that differentially expressed genes (DEGs) from elucidation of mechanisms cannot guarantee to accurately guide the strain engineering for apparent phenotypes in acid tolerance, owing to post-transcriptional and post-translational modifications and a complicated regulation network of associated proteins. On the contrary, increased acetic acid tolerance does not necessarily and determinately lead to change in fermentation efficiency (Mira et al., 2010b; Jönsson and Martín, 2016).
Furfural and HMF are aldehyde inhibitors and major chemicals of hydrolysate from the dehydration of lignocellulose-derived pentose and hexose, individually. 1) A furfural-resistant strain proved that NADH-dependent oxidoreductases are the foremost resistance mechanism at lower concentrations of furfural. Proteins linked to the tricarboxylic acid (TCA) cycle were being overexpressed, and proteins responsible for glycerol synthesis were less expressed in the industrial S. cerevisiae under 17 g/L furfural stress. Furfural stress-responsive proteins were distributed on the pathways including protein unfolding, oxidative stress, osmotic stress, DNA damage, nutrient starvation, central carbon metabolism, alcohol dehydrogenation, and the redox balance (Lin et al., 2009a). For the same situation, under 8 g/L furfural stress, a panel of 24 proteins related to diverse cellular reactions including oxidative stress (Ahp1, Gre2, Grx4, and Tsa1), protein unfolding (Egd2, Hsc82, Hsp31, Kar2, and Ssa1), osmotic and salt stress (Act1, Gpd1, Gre2, and Rhr2), DNA damage (Stm1 and Rps3), and pH stress (Gre2) were differentially expressed (Lin et al., 2009b). 2) Compared with S288c, an industrial strain NRRL Y-12632 displayed 32,000 SNPs, and 54 genes containing non-synonymous SNPs were shown to confer HMF adaptation (Zhou et al., 2014). Meanwhile, gene regulatory networks were reconstructed, and the transcription factors Yap1 and Pdr3 were identified for HMF tolerance in S. cerevisiae NRRL Y-12632 strain (Song et al., 2009). Almost 365 candidate genes of BY4742 strain were involved in HMF (30 mM) tolerance. Genes encoding functional enzymes, such as ADH6, ARI1, and OYE3, were regulated by YAP1 and PDR genes; genes associated with cellular transporters, such as TPO1, RSB1, YOR1, and SNQ2; genes involved in protein degradation or modifications like SHP1 and SSA4 were regulated by RPN4 and HSF1 (Ma and Liu, 2010b).
However, for the double tolerant yeast, the in situ detoxification and conversion pathways can reduce the contents of HMF and furfural coupled with NADH and/or NADPH-dependent redox balance and catalyzed by aldehyde reductases (Liu et al., 2008). Hundreds of DEGs have been speculated to be associated in furfural treatment, HMF treatment, and their combined treatment. Relevant mechanisms like Yap1 regulator, Ald4 mitochondrial aldehyde dehydrogenase, and genes of the PDR family, ABC transporters, and glucose metabolic pathways displayed close relation of combating with toxic threats of furfural and HMF (Almeida et al., 2007; Liu, 2011). Under the impaired toxicity of furfural or HMF, cell viability and gene expression were often repressed severely because these inhibitors exert the most toxic effect compared to any other class of inhibitors (Jayakody and Jin, 2021).
3.2.2 Mixture of Hydrolysate Inhibitors
Acetic acid, furfural, HMF, or even phenol are representative inhibitors from pretreated lignocellulose, yeast stress tolerance encountering a mixture of inhibitors that could be assessed; therefore, OMICs-based studies exploited the detailed correlation of various proportions of combined inhibitors and the corresponding gene expression (Doğan et al., 2014; Jönsson and Martín, 2016; Sardi et al., 2016). Under sensitive stress conditions, metabolites with specific concentrations have been analyzed and the differential flux distribution has been generated; however, our scant understanding of the complicated mechanism that occurred upon interactions of multi-inhibitors required expansion.
In reference to furfural stress, transcriptional and translational control genes were decreased, but stress-responsive genes were increased. In response to acetic acid, genes responsible for metabolizing carbohydrates, encoding mitochondrial ribosomal proteins, and carbohydrate metabolism were decreased, while genes related to amino acid metabolism were increased (Li and Yuan, 2010). Under the inhibitors acetic acid, furfural, and their mixture, the comparative transcriptomic analysis was applied on the S. cerevisiae YC1 strain and 184 consensus genes were determined to be regulated differentially. Among these, two RNA polymerase II transcription factors, Sfp1 and Ace2, were experimentally validated (Chen et al., 2016a).
Membrane permeability and fluidity were correlated with the upregulation of fatty acid metabolic genes in S. cerevisiae cells which were obtained from phospholipidomics and transcriptomics when exposed to the stress imposed by a mixture of phenol, acetic acid, and furfural (Yang et al., 2012). The S. cerevisiae T2 strain was cultured in diluted hardwood spent sulfite liquor containing 0.55% acetic acid, 0.1% furfural, and 0.3% HMF, and it was discovered that acetic acid mainly affected energy generation, uptake systems, and metabolism of the yeast, while furfural and HMF altered gene expression of the redox balance (Bajwa et al., 2013).
Effects of 5.3 g/L acetic acid (A), 1.3 g/L furfural (F), 0.5 g/L phenol (p), and their mixture (AFP) on the metabolite profiling of Angel® tolerant yeast were investigated. The metabolism of amino acids and central carbon was inhibited by acetic acid, while the phenol and furfural stresses both perturbed the membrane integrity. Nonetheless, AFP inhibitors exerted antagonistic impact on the synthesis of tryptophan, cadaverine, inositol, and threonine (Ding et al., 2011). Subsequently, after multiple rounds of random mutagenesis, the metabolic profile of the tolerant strain in the presence of AFP was modified. The concentrations of most amino acids, inositol, and phenethylamine were remarkably increased, whereas those of the ones involved in carbon metabolism (glycolysis and TCA), protein degradation, and pyrimidine ribonucleotide synthesis (uracil and cytosine) were declined (Ding et al., 2012b). The metabolome of a xylose-fermenting S. cerevisiae 424A was compared with its parental strain 4124 in response to AFP inhibition, which indicated variations in the content of trehalose, cadaverine, glutamate, and g-aminobutyric acid (GABA). Moreover, reduced levels of glutamate, glycine, lysine, and isoleucine and the accumulation of glycerol, galactinol, and mannitol were detected in strain 424A (LNH-ST) during xylose fermentation. It is worth noting that xylose utilization of strain 424A (LNH-ST) was more disturbed by inhibitors than glucose utilization (Wang et al., 2014). Proteins responsible for detoxification (Ahp1 and Hsp26), nitrogen metabolism (Gdh1 and Met1), protein folding, degradation, and translation (Ssc1, Ubp14, and Efb1), oxidative stress and unfolded protein response (Grx1, Gre2, and Asc1), and oxidative stress–related activities (Ahp1 and Grx1) were being overexpressed, under culturing of combined acetic acid (5.3 g/L), furfural (1.3 g/L), and phenol (0.5 g/L) (Ding et al., 2012a). Multiple inhibitors including acetic acid (5.3 g/L), furfural (1.3 g/L), and phenol (0.5 g/L) showed that yeast tolerance depends on the cultivation period. The BY4741 strain was exposed to the AFP in batch fermentation, followed by a proteome analysis of the cells collected at different stages. There were 194 and 215 unique proteins expressed differentially during the lag phase and exponential phase, respectively (Lv et al., 2014). Recently, the thousands of S. cerevisiae mutants developed through overexpression or deletion of unique genes have proved altered stress tolerance to inhibitors (acetic acid, furans, and phenolic compounds) or inhibitor combinations, which can guide successful attempts to engineer strain for bioconversion of lignocellulose biomass to ethanol (Cámara et al., 2022). Furthermore, in order to describe or understand combined inhibition exerted by mixed inhibitors, for one thing, the 50% inhibition (IC50) or possible dosage effect of individual inhibitors, as well as the complete inhibitory concentration of hydrolysate, must be re-estimated because there could be synergistic effects on inhibition of cell growth and ethanol production in the mixture. Thus, it follows that global transcriptional regulators that compatibly respond to multiple inhibitors might be a comprehensive direction for strain modification.
3.3 Oxidation and Thermal Tolerance
3.3.1 Oxidative Stress
Yeast cells are vulnerable to the oxidation and thermal stress in ethanol-producing plants; their regulation mechanism has some overlaps and an interaction effect; accordingly, OMICs has been employed as a tailor-made approach for multiple-effect or changed gene profiling (Morano et al., 2012; Bleoanca et al., 2014; Mejía-Barajas et al., 2017). In general, accumulation of ROS (reactive oxygen species) happens during normal metabolic activities, but their accumulation can also be triggered by environmental stimuli, such as ethanol, thermal stress, hydrolysate inhibitors (e.g., furfural and phenolic compounds), and oxidants (Morano et al., 2012). When incremental ROS inevitably leads to oxidative stress, imbalanced oxidation–reduction reactions could be broadly adjusted originally by activities of antioxidants, such as peroxidases, catalase, superoxide dismutase, and glutathione-related enzymes, but excessive formation of ROS or failure of antioxidant defenses to impede severe oxygen toxicity compromises the cell viability (Morano et al., 2012; Poljsak et al., 2013). It has been reported that oxidative stress causes lipid peroxidation, degradation of proteins, DNA damage and subsequent disruption of cell homeostasis, as well as autophagy and programmed cell death (Farrugia and Balzan, 2012; Zhao et al., 2015).
An oxidation stress exerted by 2 mM H2O2 induced the differential expression of mutant strain taf 25–3, where 15 transcription factors related to fatty acid degradation, amino acid synthesis, carbon metabolism, and peroxisomal functions were found to be responsive. Moreover, PKA (protein kinase A) and MAPK (Mitogen-activated protein kinases) were also coordinated (Zhao et al., 2015). Cellular oxidative damage may be induced on S. cerevisiae BY4741 and BY4741ΔRIM101; DEGs involved in protein catabolism, pH homeostasis, and vacuolar function, in particular KNH1, associated with cell wall β-1, 6-glucan synthesis, and the uncharacterized ORF_YIL029C, were highlighted (Mira et al., 2009). For a global analysis of yeast protein expression of a stress-tolerant KNU5377 strain, several proteins (Sod1, Sod2, Tsa1, Ahp1, Abp1, Sam1, Sax2, and Wcx2) were involved in oxidative stress; meanwhile, hypothetic proteins (YNL281w, YGR279c, YPL273w, YKL133c, and YKR074w) were detected, which changed their roles in stress-responsive proteins (Kim et al., 2007). Herein, for efficient scavenging of ROS and its harmful effects, molecular mechanisms of oxidation and anti-oxidation should be fully elucidated.
3.3.2 Thermal Tolerance
Thermotolerance of S. cerevisiae can generate a fall in production costs of cooling systems and also bring an improvement on fermenting efficiency of lignocellulose-ethanol, so thermotolerant mutants obtained through directed adaptive evolution should survive at higher temperatures than the wild-type strain (Abdel-Banat et al., 2010). Meanwhile, it has still been hard to break the limitation of the upper temperature tolerance (42°C) during fermentation. The investigations of the mechanism emphasize the importance of the trehalose accumulation and metabolism, stability of the entire proteome, and heat shock factor (HSF) that is highly conserved (Verghese et al., 2012). In the heat-shock response, HSF-centered regulation is intricate and special, in which targets associated with aspects of cellular function to protect against heat stress, such as components of the cell wall, plasma membrane, cytoskeleton, transporters, chaperones, bioprocessing of ubiquitination and proteolysis, carbon metabolism, energy generation, and signal transduction, attach to the defense of oxidative stress (Morano et al., 2012; Caspeta and Nielsen, 2015).
To identify the genes of S. cerevisiae M206111 at 40°C conferring the innate and/or acquired thermotolerance, at the late stage of fermentation, one-third of the genes were picked out, among which proteins for glycolysis, trehalose biosynthesis, ethanol production, and heat-shock tolerance were transcription-regulated; in particular, the HSP26 exhibited the highest upregulation at 6 h (Chen et al., 2013). A pool of genome-scale S. cerevisiae deletion strains combined deep-sequencing and obtained an assessment of genes influencing metabolism, cellular signaling, and chromatin that are functionally crucial for thermotolerance (Gibney et al., 2013). Upon exposure to 41°C, the deletion of DFG5, a putative glycosidase/glycosyltransferase, could increase thermal tolerance and decrease concentration of ROS in BY4741. Comparative transcriptome analysis revealed DEGs (38 upregulated and 23 downregulated) in the mutant Dfg5Δ, and deletion of 11 genes among the downregulated genes was verified to enhance thermal tolerance (Nasution et al., 2015). Proteomics of an industrial strain, ScY01, were remodeled in transient heat shock and prolonged thermal stress situations followed by experiments, and it was discovered that transcription factors Mig1 and Srb2 can be targeted for enhancement of thermal tolerance (Xiao et al., 2018). The investigation of metabolomics of the BY4741ΔSSK2 mutant at 42°C indicated that 91 intracellular metabolites were emphasized, and pathways of fatty acids, amino acid metabolism, the cell wall, and the membrane were considered as pivotal signatures in thermo-tolerance due to SSK2 gene deficiency in the MAPK cascade (Kim et al., 2021). Herein, we see that studies lead to disclosing mechanisms of cellular reactions on heat stress and to discriminating between associated hot spots that determine the variability of thermal phenotypes, such as DFG5, HSP26, and SSK2.
Additionally, quite a few of the S. cerevisiae strains characterized by resistance to high temperature were isolated and reported; thermotolerant IR2-9a (Edgardo et al., 2008), PTCC5269 M3 and Areni M7 (Mehdikhani et al., 2011), mbc 2 (Cha et al., 2015), DBKKU Y-53 (Nuanpeng et al., 2016), KKU-VN8 (Techaparin et al., 2017), DMKU 3-S087 (Pattanakittivorakul et al., 2019), and TC-5 (Boonchuay et al., 2021) exhibited remarkable advantages for ethanol production, particularly from sweet sorghum, molasses, and corncob residue, at temperatures that are all equal to or greater than 40°C; further research efforts with OMICs sequencing and analysis of the above mutants are required for strain improvement toward industrial application.
4 Strategies to Develop Robust Strains for Efficient Ethanol Production
The S. cerevisiae possesses a prominent merit: ethanol fermentation is almost simultaneously coupled with its powerful cell growth. Starch-based fermentation brings about high ethanol yield, often accompanied by alcohol tolerance of cells; when compared to the fermentation with lignocellulosic hydrolysates, the first dilemma is posed by typical inhibitors that give rise to gravely suppressed cell growth. Therefore, taking aim at efficient ethanol production and effective resistance to inhibitory factors, the preferable strategies of strain improvement should be balanced and controlled. Arguably, identified genes participating in the glycolysis might be assigned to take charge of tolerance enhancement, as shown in Figure 2 (information obtained from Saccharomyces Genome Database; Hasunuma et al., 2014b), if it is possible to be accomplished expectantly by genetic modification. Nevertheless, other strategies on strain engineering are still required to widen substrate utilization, to achieve higher ethanol titer, and to confer tolerance to inhibitors by inverse metabolic engineering. With the advent of technology and the assist of OMICs tools, it should be emphasized that attention to tolerance should not be minimized so that promoted specific growth rate and cell reproductive capacity and improved fermentation performance will be subsequently achieved.
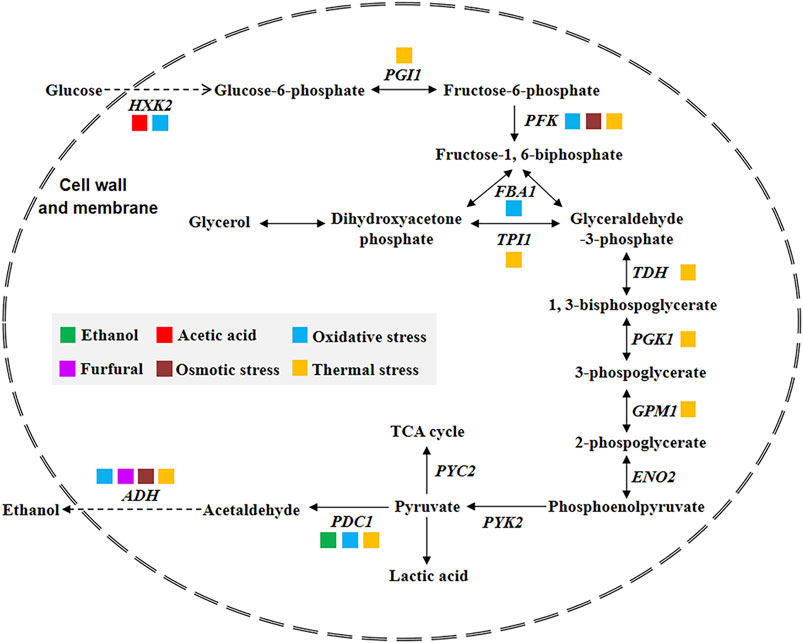
FIGURE 2. Schematics of stress-associated genes involved in the glycolysis pathway for ethanol production of S. cerevisiae. Colorful solid-squares represent identified stress tolerances of genes (HXK, hexokinase; PGI, phosphoglucoisomerase; PFK, phosphofructokinase; FBA, fructose 1,6-bisphosphate aldolase; TPI, triose phosphate isomerase; TDH, glyceraldehydes-3-phosphate dehydrogenase; PGK, 3-phosphoglycerate kinase; GPM, phosphoglyceromutase; ENO, enolase; PYK, pyruvate kinase; PYC, pyruvate carboxylase; PDC, pyruvate decarboxylase; ADH, alcohol dehydrogenase).
4.1 Inverse Metabolic Engineering
The regularly intrinsic metabolism pathways can be optimized or directedly altered via inverse metabolic engineering (IME) for efficiently producing desired phenotypes, which a systematic process including 1) choosing the target genes, 2) construction of engineered strain, and 3) high-throughput screening and fine-tuning on strain breeding (Mehmood et al., 2017b; Pereira et al., 2021). Combined with multi-OMICs data analyses, genetic manipulation has been successfully applied in overexpressing decisive genes affecting metabolic flow, gene deletion, or mutation for blocking and alleviating the competing pathway, even heterologous expression (Liu et al., 2013; Chen and Dou, 2016; Roy et al., 2020); additionally, some approaches such as global transcription machinery engineering (gTME) and multiplex automated genome engineering (MAGE) could be selectively utilized to support the IME for increased alcohol fermentation and stress tolerance (Liu and Jiang, 2015; Adebami et al., 2021). Promoted alcohol tolerance is elicited by overexpressing indigenous genes INO1 (encoding an inositol-3-phosphate synthase), HAL1 (encoding a cytoplasmic protein), and DOG1 (encoding 2-deoxyglucose-6-phosphate phosphatase involved in glucose metabolism) of S. cerevisiae or a truncated form of MSN2 in CEN. PK2-1D strain (Hong et al., 2010). Gene MSN2, a stress-responsive transcriptional activator, often confirmed that regulating of transcription would float along with a wide variety of stress conditions. In parallel, overexpression of WHI2 gene that encodes a cytoplasmic globular scaffold protein rendered the SR8 strain boosting tolerance and fermentation under acetic acid (3.5 g/L, pH 4.0) stress, but its deletion-mutant was expressively susceptible to acetic acid, which is explored using the IME method compared to its wild-type. Furthermore, co-expression of WHI2 and PSR1, a phosphatase encoding gene, had a synergistic impact on acetic acid resistance (Chen et al., 2016b). Inversely, overexpressing the ADH7 gene and the ORF YKL071W increased furfural resistance (Heer et al., 2009). Disruption of OAZ1 (encoding an ornithine decarboxylase) and TPO1 (encoding a polyamine transporter), which enable the D452-2 strain to tolerate furan derivatives (e.g., furfural, HMF, and acetic acid) or phenols, its dominant recombinant produced by co-disruption above coupled with SPE3 overexpression, which demonstrated 33 and 60% shortening of the lag phase under the stress of 4 g/L acetic acid, 2 g/L HMF, and furfural, respectively (Almeida et al., 2007; Kim et al., 2015). By co-analysis of genome and transcriptome sequencing, reverse engineering of Ras2 (a GTP-binding protein in the RAS-cAMP-PKA signaling pathway), Hsf1 (heat shock transcription factor), and Sum1 (one regulator of replication initiation) confirmed their special relevance to acetic acid and thermal tolerance in ethanol production (Salas-Navarrete et al., 2022). The cases are examples of the applications of directed genetic technology in strain improvement; therefore, an increase in efforts needs to be maintained for the greater tolerance.
4.2 Xylose Pathway Engineering
The S. cerevisiae prioritizes glucose as a first choice among extracellular carbon sources and simultaneously lacks the xylose utilization pathway, while xylose is one of the main dissolved saccharides present in the hydrolysate or had reached up to two-fifths of the total biomass in agricultural and wood residues (Cunha et al., 2020). Introduction of heterologous expression of xylose reductase (XR), xylitol dehydrogenase (XDH), and xylulokinase (XK) or xylose isomerase (XI) into S. cerevisiae that is capable of converting xylose to xylulose-5-phosphate could artificially link up C5 and C6 sugar metabolism; nevertheless, imbalanced cofactor, substrate affinity, or extra metabolic burden are considered to be the bottlenecks in genetically engineered mutants for the failure of smoothly co-fermenting xylose and glucose, apart from unignored cell resistance to inhibitors of hydrolysate, which is also more significant, and which are, subsequently, ameliorated by evolutionary engineering (Nogué and Karhumaa, 2015; Lee et al., 2021). Overexpression of WHI2 conferred promoted fermentation of glucose/xylose under acetic acid stress which improved ethanol productivity by 5-folds (Chen et al., 2016b). Disruption of PHO13 that encoded a conserved phosphatase in xylose fermenting BY4741 strain promoted a conversion from xylose to ethanol under organic acids (e.g., acetic acid) and furfural stresses, as well as an increased bioethanol production from cellulose supplemented with rice straw hydrolysate (Fujitomi et al., 2012). Synergistic effect of PHO13 deletion and overexpression of TAL1, a gene encoding a transaldolase of the non-oxidative pentose phosphate pathway, were released on xylose-utilizing strain NAPX37 during xylose fermentation along with weak acid (formic, acetic, or levulinic acids) inhibition; meanwhile, enhanced ethanol production and reduced xylitol accumulation were measured when the weak acid is present or not (Li et al., 2014). Intracellular ADH isozymes (alcohol dehydrogenases) could lessen furan derivatives, and overexpression of mutated ADH1 gene in the xylose-metabolizing YPH499XU strain raised xylose utilization in the presence of 5-HMF (Ishii et al., 2013). In another study, co-expressing TAL1 and ADH1 in the MT8-1X strain were well performed on increased ethanol production not only from xylose medium supplemented with furfural but also from undetoxified hydrolysate with toxic compounds (Hasunuma et al., 2014b). Accelerated cell biomass growth and xylose fermentation were achieved by overexpressing Haa1, an acetic acid–responsive transcriptional activator and a possible regulator of cell wall integrity (Sakihama et al., 2015). Then, enhanced xylose metabolism and weak acid tolerance were observed through overexpressing RTC3 and ANB1; RTC3 gene with unknown function is involved in RNA metabolism, and Anb1 still served as the translation elongation factor eIF-5A (Hasunuma et al., 2016). In xylose-consuming mutants initiated from the industrial PE-2 strain, Haa1 and/or Prs3 (a phosphoribosyl pyrophosphate synthetase) over-expression facilitates xylose fermentation and acetic acid tolerance, further boosting the traits of the new strain toward efficient production in the plant (Cunha et al., 2018). It is therefore worth studying how the influence of xylose fermentation broth used for incubation and pre-culture medium supplemented with weak acids on yeast cells results in xylose pathway–engineered S. cerevisiae acclimatized to acid stress and xylose fermentation, owing to their unique properties.
4.3 Engineered S. cerevisiae for Efficient Consolidated Bioprocessing
Recently, the compact-designed strategy of pretreatment biomass-to-bioethanol conversion called consolidated bioprocessing (CBP), which offers advantages over separate hydrolysis and fermentation, has become popular. The CBP utilizes a platform of comprehensively engineered strains producing enzymes on-site, followed by hydrolysis and fermentation as a continuous workflow (Drosos et al., 2021). In most cases, CBP-based strains encounter a variety of toxic compounds that hamper link-coupled growth and ethanol production or harsh environments such as high temperature or metal ions. Owing to those difficulties, the tolerance mechanism of engineered S. cerevisiae as predicted possesses a promising potential to be exploited for ligno-bioethanol refinery (Rani Singhania et al., 2022). Crucially, the optimal temperature of S. cerevisiae for saccharification (50°C) and fermentation (28–37°C) is entirely different; hence, establishment of thermotolerant mutants suitable for elevated heat stresses can undoubtedly reduce inconsistency in temperature of the CBP (Hasunuma and Kondo, 2012). Some examples focused on the thermal endurance of CBP strains are worth noting that transformed genes are selected and amplified from thermotolerant species, such as Kluyveromyces marxianus, is an efficacious approach (van Zyl et al., 2013). Overexpression of genes encoding β-glucosidase from Aspergillus aculeatus, endoxylanase from Trichoderma reesei, xylosidase from Aspergillus oryzae, xylose reductase and xylitol dehydrogenase from Scheffersomyces stipitis, and Adh1/Fdh1/Tal1/Xks1 of S. cerevisiae in industrial strain Sun049 results in a high-quality recombinant that is hemicellulolytic, xylose-utilizing, and inhibitor-stress tolerant (Hasunuma et al., 2014a). On the other hand, through protein engineering, improved tolerance to ethanol and osmotic stress can be realized by introducing a glycosylable protein with the hydrophobic property when designed proteins have been incorporated at the N-terminal agglutinin (Aga2) and displayed on the cell surface (Perpiñá et al., 2015). Improved osmotic and heat stress and heterogeneously secreted enzyme activity were gained by overexpressing either YHB1 or SET5 genes (Lamour et al., 2019). Although referred genes encoding heat shock proteins, chaperones, and regulators within the heat sensitive mechanism were uncovered (Lu et al., 2012; Jarolim et al., 2013), acquiring thermal tolerance is still a problem to be further resolved by the IME for CBP strains to meet the industrial goals (Table 2).
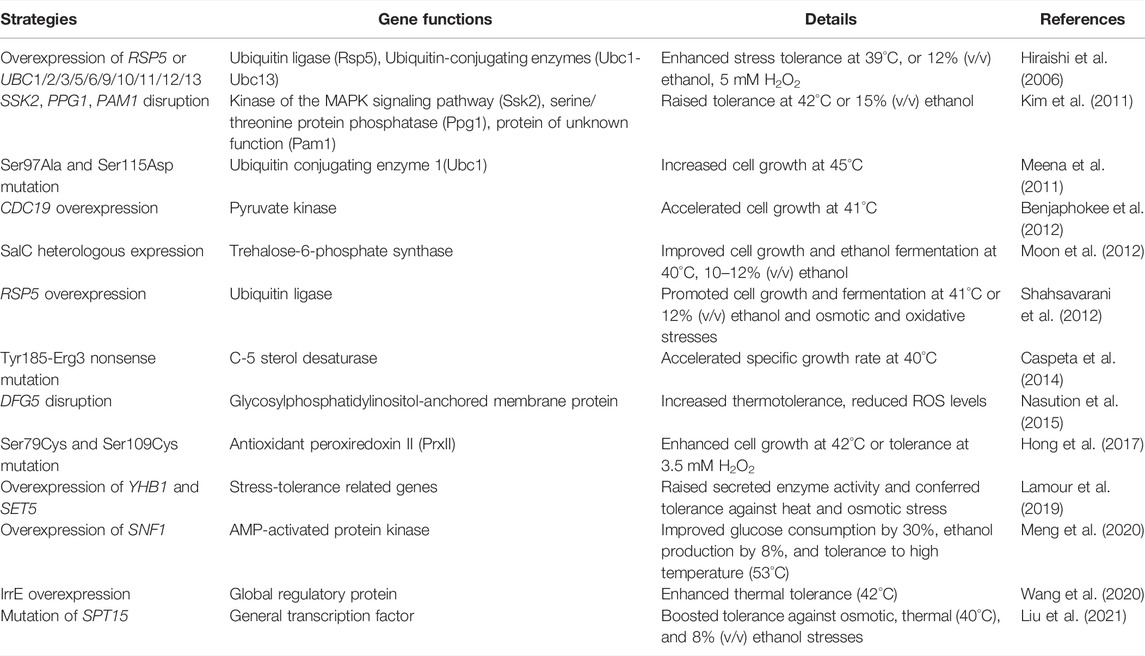
TABLE 2. Representative examples of improved thermal tolerance of S. cerevisiae by genetic engineering.
5 Current Challenges and Recommendations for Future Research
For a feasible production of lignocellulose ethanol, technical challenges lie in various inhibitors derived from physiochemial pretreatments of agricultural and forestry wastes, followed by detoxification processes that also unavoidably remove the dissolved sugars, thereby decreasing the sugar yield and content. The mixture of fermentable and non-fermentable saccharides enables the fermenting strain to be less efficient in substrate utilization and ethanol yield; nevertheless, the robustness of the metabolically engineered strain is not simply realized.
Moreover, S. cerevisiae cannot tolerate multiple stress factors, some of which are inescapable due to the harsh production process and lead to the adverse impact on the specific growth rate and ethanol productivity. Accordingly, mutual enhancement of tolerance and fermentation performance is a not-so-trivial pursuit, which severely affects the fermentation efficiency on an industrial scale and brings discouraged economic costs.
The challenges mentioned are plotted and shown in Figures 1, 3; future research directions are recommended in S. cerevisiae strains for highly efficient and sustainable 2G fuel-ethanol production and beyond as follows:
1) Multi-omics sequencing and data mining should be widely applied, and deep-level investigations are needed for potentially important yeasts in commercial use. It is demonstrated that joint study of static genomics and another dynamic-omics indeed contributes to satisfying the demand of identification or modulation from genotype to phenotype and vice versa, but owing to complexity, the related work is rarely reported in the ethanol fermentation industry and is still an exquisite subject that attracts research interest.
2) The molecular mechanisms of resistance to a variety of inhibitors should be elucidated. Also, it is essential to functionally validate novel genes or ORFs in stress tolerance. The comprehensive illustration has incorporated intricate networks of gene regulation associated with stress-regulating metabolism of great importance, which should be gradually tackled, and has remained partially solved, or even unsolved. The synergistic effects of several stresses from hydrolysate, such as oxidative, osmotic stress, and even toxic heavy metals on yeast cells must be studied in detail.
3) Enhancement of conversion efficiency of bioethanol by innovative approaches is still insufficient. There is a hope that some research teams focused on actual utilization of raw materials of lignocellulosic residues persisted in the construction of recombinant CBP S. cerevisiae, despite the fragile performance of fermentation. Undoubtedly, a decline in growth and tolerance is pivotal, mainly caused by metabolic burden, cofactor imbalance, etc. So, tolerance improvement is desirable for engineered strains harboring heterogeneous pathways that could metabolize xylose and arabinose or secrete enzymes for hydrolysis. Thus, many efforts clearly remain to be made for overcoming the barriers.
4) From the perspective of synthetic biology, reconstruction or synthesis of yeast chromosomes suitable for 2G bioethanol fermentation should probably be considered, and as a systematic design tool, emerging technologies, for example, CRISPR-Cas9, have been used in an attempt to achieve precise gene editing or DNA rearrangement from the level of genomes (Sambasivam et al., 2020; Sandhya et al., 2020). Toward the ambitious dual goals of excellent stress tolerance and ethanol fermentation, it will certainly take a longer time to be explored and fulfilled.
FIGURE 3. Research strategy of stress tolerance improvement using “OMICs”-based technologies for the 2G fuel ethanol with challenges and applications.
6 Conclusion
As a vigorous cell factory, S. cerevisiae may support the reliable supply of transportation fuel, which has exhibited a credible bioconversion efficiency of mixed sugars- and grain-substrates to cost-effective production of ethanol. Combined with obvious advantages of lignocellulose utilization, lignocellulosic feedstock is environment-friendly, abundantly available, and low-cost for green bio-refinery. Despite this, bottlenecks remained; the limitations are not merely profitable ones encountered by the pilot-scale and large-scale fermentation; the application prospect of transformation of hemicelluloses of lignocellulose to ethanol is worthy of more attention.
During past decades, a lot of progress has been made on tolerance research of S. cerevisiae (Mitsui and Yamada, 2021). Especially, the multi-omics analysis of transcriptome, proteome, and metabolome, linked with comparative yeast genomics that focusing on ethanol tolerance, inhibitions of hydrolysates, oxidative, and thermal stresses, was concluded. These studies revealed the microbial genetic background or adaption mechanisms and provided targeted DNA modifications; meanwhile, profiling characteristics of physiological reaction and cell metabolism in response to condition-specific environments offered the beneficial experience for continuing research in this field. S. cerevisiae strains aimed at broadening the substrate spectrum and effectively integrating conversion processes have been developed.
Author Contributions
J-RX wrote the original draft and provided funding for the manuscript. MM helped in writing and improving the language. LW and NA revised the manuscript. H-JM helped review the manuscript.
Funding
This work was supported by the Scientific Research Start-up Fund of North Minzu University (2020KYQD03) and the Ningxia Key R&D Project (Special Project for Introduced Talents) (2020BEB04013).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors greatly acknowledge the IMBE lab in Shanghai Jiaotong University and Haijun Ma for his assistance in North Minzu University (Ningxia Natural Science Foundation 2021AAC03182).
References
Abdel-Banat, B. M. A., Hoshida, H., Ano, A., Nonklang, S., and Akada, R. (2010). High-temperature Fermentation: How Can Processes for Ethanol Production at High Temperatures Become Superior to the Traditional Process Using Mesophilic Yeast? Appl. Microbiol. Biotechnol. 85, 861–867. doi:10.1007/s00253-009-2248-5
Abdelnur, P. V., Caldana, C., and Martins, M. C. M. (2014). Metabolomics Applied in Bioenergy. Chem. Biol. Technol. Agric. 1, 1–9. doi:10.1186/s40538-014-0022-0
Adebami, G. E., Kuila, A., Ajunwa, O. M., Fasiku, S. A., and Asemoloye, M. D. (2021). Genetics and Metabolic Engineering of Yeast Strains for Efficient Ethanol Production. J. Food Process Eng. e13798, 1–30. doi:10.1111/jfpe.13798
Akao, T., Yashiro, I., Hosoyama, A., Kitagaki, H., Horikawa, H., and Watanabe, D. (2011). Whole-genome Sequencing of Sake Yeast Saccharomyces cerevisiae Kyokai No. 7. DNA Res. 18, 423–434. doi:10.1093/dnares/dsr029
Almeida, J. R., Modig, T., Petersson, A., Hähn‐Hägerdal, B., Lidén, G., and Gorwa‐Grauslund, M. F. (2007). Increased Tolerance and Conversion of Inhibitors in Lignocellulosic Hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 82, 340–349. doi:10.1002/jctb.1676
Amer, B., and Baidoo, E. E. K. (2021). Omics-driven Biotechnology for Industrial Applications. Front. Bioeng. Biotechnol. 9, 613307. doi:10.3389/fbioe.2021.613307
Argueso, J. L., Carazzolle, M. F., Mieczkowski, P. A., Duarte, F. M., Netto, O. V. C., Missawa, S. K., et al. (2009). Genome Structure of a Saccharomyces cerevisiae Strain Widely Used in Bioethanol Production. Genome Res. 19, 2258–2270. doi:10.1101/gr.091777.109
Auesukaree, C. (2017). Molecular Mechanisms of the Yeast Adaptive Response and Tolerance to Stresses Encountered during Ethanol Fermentation. J. Biosci. Bioeng. 124, 133–142. doi:10.1016/j.jbiosc.2017.03.009
Babrzadeh, F., Jalili, R., Wang, C. L., Shokralla, S., Pierce, S., Robinson-Mosher, A., et al. (2012). Whole-genome Sequencing of the Efficient Industrial Fuel-Ethanol Fermentative Saccharomyces cerevisiae Strain CAT-1. Mol. Genet. Genomics 287, 485–494. doi:10.1007/s00438-012-0695-7
Bajwa, P. K., Ho, C. Y., Chan, C. K., Martin, V. J. J., Trevors, J. T., and Lee, H. (2013). Transcriptional Profiling of Saccharomyces cerevisiae T2 Cells upon Exposure to Hardwood Spent Sulphite Liquor: Comparison to Acetic Acid, Furfural and Hydroxymethylfurfural. Antonie Leeuwenhoek 103, 1281–1295. doi:10.1007/s10482-013-9909-1
Benjaphokee, S., Koedrith, P., Auesukaree, C., Asvarak, T., Sugiyama, M., Kaneko, Y., et al. (2012). CDC19 Encoding Pyruvate Kinase Is Important for High-Temperature Tolerance in Saccharomyces cerevisiae. New Biotechnol. 29, 166–176. doi:10.1016/j.nbt.2011.03.007
Bleoanca, I., Silva, A. R. C., Pimentel, C., Rodrigues-Pousada, C., and Menezes, R. D. (2014). Relationship between Ethanol and Oxidative Stress in Laboratory and Brewing Yeast Strains. J. Biosci. Bioeng. 116, 697–705. doi:10.1016/j.jbiosc.2013.05.037
Boonchuay, P., Techapun, C., Leksawasdi, N., Seesuriyachan, P., Hanmoungjai, P., Watanabe, M., et al. (2021). Bioethanol Production from Cellulose-Rich Corncob Residue by the Thermotolerant Saccharomyces cerevisiae TC-5. J. Fungi 7, 547. doi:10.3390/jof7070547
Borneman, A. R., Desany, B. A., Riches, D., Affourtit, J. P., Forgan, A. H., Pretorius, I. S., et al. (2012). The Genome Sequence of the Wine Yeast VIN7 Reveals an Allotriploid Hybrid Genome with Saccharomyces cerevisiae and Saccharomyces Kudriavzevii Origins. FEMS Yeast Res. 12, 88–96. doi:10.1111/j.1567-1364.2011.00773.x
Borneman, A. R., Desany, B. A., Riches, D., Affourtit, J. P., Forgan, A. H., Pretorius, I. S., et al. (2011). Whole-genome Comparison Reveals Novel Genetic Elements that Characterize the Genome of Industrial Strains of Saccharomyces cerevisiae. PLoS Genet. 7, e1001287. doi:10.1371/journal.pgen.1001287
Cámara, E., Olsson, L., Zrimec, J., Zelezniak, A., Geijer, C., and Nygård, Y. (2022). Data Mining of Saccharomyces cerevisiae Mutants Engineered for Increased Tolerance towards Inhibitors in Lignocellulosic Hydrolysates. Biotechnol. Adv. 57, 107947. doi:10.1016/j.biotechadv.2022.107947
Carreto, L., Eiriz, M. F., Gomes, A. C., Pereira, P. M., Schuller, D., Santos, M. A. S., et al. (2008). Comparative Genomics of Wild Type Yeast Strains Unveils Important Genome Diversity. BMC Genomics 9, 524. doi:10.1186/1471-2164-9-524
Caspeta, L., Chen, Y., Ghiaci, P., Feizi, A., Buskov, S., Hallstrom, B. M., et al. (2014). Biofuels. Altered Sterol Composition Renders Yeast Thermotolerant. Science 346, 75–78. doi:10.1126/science.1258137
Caspeta, L., and Nielsen, J. (2015). Thermotolerant Yeast Strains Adapted by Laboratory Evolution Show Trade-Off at Ancestral Temperatures and Preadaptation to Other Stresses. mBio 6, e00431. doi:10.1128/mBio.00431-15
Cha, Y. L., An, G. H., Yang, J., Moon, Y. H., Yu, G. D., and Ahn, J. W. (2015). Bioethanol Production from Miscanthus Using Thermotolerant Saccharomyces cerevisiae Mbc 2 Isolated from the Respiration-Deficient Mutants. Renew. Energy 80, 259–265. doi:10.1016/j.renene.2015.02.016
Chen, Q., Fang, Y., Zhao, H., Zhang, G. H., and Jin, Y. L. (2013). Transcriptional Analysis of Saccharomyces cerevisiae, during High-Temperature Fermentation. Ann. Microbiol. 63, 1433–1440. doi:10.1007/s13213-013-0606-0
Chen, R., and Dou, J. (2016). Biofuels and Bio-Based Chemicals from Lignocellulose: Metabolic Engineering Strategies in Strain Development. Biotechnol. Lett. 38, 213–221. doi:10.1007/s10529-015-1976-0
Chen, Y. Y., Sheng, J. Y., Jiang, T., Stevens, J., Feng, X. Y., and Wei, N. (2016a). Transcriptional Profiling Reveals Molecular Basis and Novel Genetic Targets for Improved Resistance to Multiple Fermentation Inhibitors in Saccharomyces cerevisiae. Biotechnol. Biofuels 9, 9. doi:10.1186/s13068-015-0418-5
Chen, Y. Y., Stabryla, L., and Wei, N. (2016b). Improved Acetic Acid Resistance in Saccharomyces cerevisiae by Overexpression of the WHI2 Gene Identified through Inverse Metabolic Engineering. Appl. Environ. Microbiol. 82, 2156–2166. doi:10.1128/AEM.03718-15
Chen, Z., Zheng, Z., Yi, C. F., Wang, F. L., Niu, Y. P., and Li, H. (2016c). Intracellular Metabolic Changes in Saccharomyces cerevisiae and Promotion to Ethanol Tolerance during the Bioethanol Fermentation Process. Rsc Adv. 6, 105046–105055. doi:10.1039/c6ra19254h
Cheng, L., Zhang, X., Zheng, X. J., Wu, Z. F., and Weng, P. F. (2019). RNA-seq Transcriptomic Analysis of Green Tea Polyphenols Regulation of Differently Expressed Genes in Saccharomyces cerevisiae under Ethanol Stress. World J. Microbiol. Biotechnol. 35, 59. doi:10.1007/s11274-019-2639-4
Cheng, Y., Du, Z., Zhu, H., Guo, X., and He, X. (2016). Protective Effects of Arginine on Saccharomyces cerevisiae against Ethanol Stress. Sci. Rep. 6, 31311. doi:10.1038/srep31311
Choudhary, J., Singh, S., Tiwari, R., Goel, R., and Nain, L. (2019). An iTRAQ Based Comparative Proteomic Profiling of Thermotolerant Saccharomyces cerevisiae JRC6 in Response to High Temperature Fermentation. Curr. Proteomics 16, 289–296. doi:10.2174/1570164616666190131145217
Costa, A. C. T., Hornick, J., Antunes, T. F. S., Santos, A. M. C., Fernandes, A. A. R., Broach, J. R., et al. (2021). Complete Genome Sequence and Analysis of a Saccharomyces cerevisiae Strain Used for Sugarcane Spirit Production. Braz. J. Microbiol. 52, 1087–1095. doi:10.1007/s42770-021-00444-z
Coutouné, N., Mulato, A. T. N., Riaño-pachón, D. M., and Oliveira, J. V. D. (2017). Draft Genome Sequence of Saccharomyces cerevisiae Barra Grande (BG-1), a Brazilian Industrial Bioethanol-Producing Strain. Microbiol. Resour. Announc. 5, e00111–17. doi:10.1128/genomeA.00111-17
Cripwell, R. A., Favaro, L., Viljoen-Bloom, M., and van Zyl, W. H. (2020). Consolidated Bioprocessing of Raw Starch to Ethanol by Saccharomyces cerevisiae: Achievements and Challenges. Biotechnol. Adv. 42, 107579. doi:10.1016/j.biotechadv.2020.107579
Cunha, J. T., Costa, C. E., Ferraz, L., Romani, A., Johansson, B., Sa-Correia, I., et al. (2018). HAA1, and PRS3 Overexpression Boosts Yeast Tolerance towards Acetic Acid Improving Xylose or Glucose Consumption: Unravelling the Underlying Mechanisms. Appl. Microbiol. Biotechnol. 102, 4589–4600. doi:10.1007/s00253-018-8955-z
Cunha, J. T., Soares, P. O., Baptista, S. L., Costa, C. E., and Domingues, L. (2020). Engineered Saccharomyces cerevisiae for Lignocellulosic Valorization: a Review and Perspectives on Bioethanol Production. Bioengineered 11, 883–903. doi:10.1080/21655979.2020.1801178
Ding, M. Z., Li, B. Z., Cheng, J. S., and Yuan, Y. J. (2010). Metabolome Analysis of Differential Responses of Diploid and Haploid Yeast to Ethanol Stress. OMICS A J. Integr. Biol. 14, 553–561. doi:10.1089/omi.2010.0015
Ding, M. Z., Wang, X., Liu, W., Cheng, J. S., Yang, Y., and Yuan, Y. J. (2012a). Proteomic Research Reveals the Stress Response and Detoxification of Yeast to Combined Inhibitors. PLoS One 7, e43474. doi:10.1371/journal.pone.0043474
Ding, M. Z., Wang, X., Yang, Y., and Yuan, Y. J. (2012b). Comparative Metabolic Profiling of Parental and Inhibitors-Tolerant Yeasts during Lignocellulosic Ethanol Fermentation. Metabolomics 8, 232–243. doi:10.1007/s11306-011-0303-6
Ding, M. Z., Wang, X., Yang, Y., and Yuan, Y. J. (2011). Metabolomic Study of Interactive Effects of Phenol, Furfural, and Acetic Acid on Saccharomyces cerevisiae. OMICS A J. Integr. Biol. 15, 647–653. doi:10.1089/omi.2011.0003
Dinh, T. N., Nagahisa, K., Yoshikawa, K., Hirasawa, T., Furusawa, C., and Shimizu, H. (2009). Analysis of Adaptation to High Ethanol Concentration in Saccharomyces cerevisiae, Using DNA Microarray. Bioprocess Biosyst. Eng. 32, 681–688. doi:10.1007/s00449-008-0292-7
Doğan, A., Demirci, S., Aytekin, A. Ö., and Şahin, F. (2014). Improvements of Tolerance to Stress Conditions by Genetic Engineering in Saccharomyces cerevisiae during Ethanol Production. Appl. Biochem. Biotechnol. 174, 28–42. doi:10.1007/s12010-014-1006-z
Drosos, A., Boura, K., Dima, A., Soupioni, M., Nigam, P. S., Kanellaki, M., et al. (2021). A Cell-Factory Model of Saccharomyces cerevisiae Based on Bacterial Cellulose without GMO for Consolidated Bioprocessing of Starch. Food Bioprod. Process. 128, 202–214. doi:10.1016/j.fbp.2021.05.006
Dunn, B., Richter, C., Kvitek, D. J., Pugh, T., and Sherlock, G. (2012). Analysis of the Saccharomyces cerevisiae Pan-Genome Reveals a Pool of Copy Number Variants Distributed in Diverse Yeast Strains from Differing Industrial Environments. Genome Res. 22, 908–924. doi:10.1101/gr.130310.111
Edgardo, A., Carolina, P., Manuel, R., Juanita, F., and Jaime, B. (2008). Selection of Thermotolerant Yeast Strains Saccharomyces cerevisiae for Bioethanol Production. Enzyme Microb. Technol. 43, 120–123. doi:10.1016/j.enzmictec.2008.02.007
Engel, S. R., Dietrich, F. S., Fisk, D. G., Binkley, G., Balakrishnan, R., Costanzo, M. C., et al. (2013). The Reference Genome Sequence of saccharomyces Cerevisiae: Then and Now. G3 Genes Genomes Genet. 4, 389–398. doi:10.1534/g3.113.008995
Farrugia, G., and Balzan, R. (2012). Oxidative Stress and Programmed Cell Death in Yeast. Front. Oncol. 2, 64. doi:10.3389/fonc.2012.00064
Fujitomi, K., Sanda, T., Hasunuma, T., and Kondo, A. (2012). Deletion of the PHO13 Gene in Saccharomyces cerevisiae Improves Ethanol Production from Lignocellulosic Hydrolysate in the Presence of Acetic and Formic Acids, and Furfural. Bioresour. Technol. 111, 161–166. doi:10.1016/j.biortech.2012.01.161
Giannattasio, S., Guaragnella, N., Ždralević, M., and Marra, E. (2013). Molecular Mechanisms of Saccharomyces cerevisiae Stress Adaptation and Programmed Cell Death in Response to Acetic Acid. Front. Microbiol. 4, 33. doi:10.3389/fmicb.2013.00033
Gibney, P. A., Lu, C., Caudy, A. A., Hess, D. C., and Botstein, D. (2013). Yeast Metabolic and Signaling Genes Are Required for Heat-Shock Survival and Have Little Overlap with the Heat-Induced Genes. Proc. Natl. Acad. Sci. U. S. A. 110, 4393–4402. doi:10.1073/pnas.1318100110
Goud, B. S., Kim, J. H., and Ulaganathan, K. (2022). Identification of Genes Associated with Stress Tolerance of High Ethanol-Producing Saccharomyces cerevisiae Strain, NCIM3186, by Differential Gene Expression Analysis. BioEnergy Res., 1–13. doi:10.1007/s12155-021-10389-8
Goud, B. S., and Ulaganathan, K. (2015). Draft Genome Sequence of Saccharomyces cerevisiae NCIM3186 Strain Used in the Production of Bioethanol from Sweet Sorghum. Microbiol. Resour. Announc. 3, e00813–15. doi:10.1128/genomeA.00813-15
Haas, R., Horev, G., Lipkin, E., Kesten, I., Portnoy, M., Buhnik-Rosenblau, K., et al. (2019). Mapping Ethanol Tolerance in Budding Yeast Reveals High Genetic Variation in a Wild Isolate. Front. Genet. 10, 998. doi:10.3389/fgene.2019.00998
Hasunuma, T., Hori, Y., Sakamoto, T., Ochiai, M., Hatanaka, H., and Kondo, A. (2014a). Development of a GIN11/FRT-Based Multiple-Gene Integration Technique Affording Inhibitor-Tolerant, Hemicellulolytic, Xylose-Utilizing Abilities to Industrial Saccharomyces cerevisiae Strains for Ethanol Production from Undetoxified Lignocellulosic Hemicelluloses. Microb. Cell Factories 13, 145. doi:10.1186/s12934-014-0145-9
Hasunuma, T., Ismail, K. S. K., Nambu, Y., and Kondo, A. (2014b). Co-expression of TAL1 and ADH1 in Recombinant Xylose-Fermenting Saccharomyces cerevisiae Improves Ethanol Production from Lignocellulosic Hydrolysates in the Presence of Furfural. J. Biosci. Bioeng. 117, 165–169. doi:10.1016/j.jbiosc.2013.07.007
Hasunuma, T., and Kondo, A. (2012). Development of Yeast Cell Factories for Consolidated Bioprocessing of Lignocellulose to Bioethanol through Cell Surface Engineering. Biotechnol. Adv. 30, 1207–1218. doi:10.1016/j.biotechadv.2011.10.011
Hasunuma, T., Sakamoto, T., and Kondo, A. (2016). Inverse Metabolic Engineering Based on Transient Acclimation of Yeast Improves Acid-Containing Xylose Fermentation and Tolerance to Formic and Acetic Acids. Appl. Microbiol. Biotechnol. 100, 1027–1038. doi:10.1007/s00253-015-7094-z
Hasunuma, T., Sanda, T., Yamada, R., Yoshimura, K., Ishii, J., and Kondo, A. (2011). Metabolic Pathway Engineering Based on Metabolomics Confers Acetic and Formic Acid Tolerance to a Recombinant Xylose-Fermenting Strain of Saccharomyces cerevisiae. Microb. Cell Factories 10, 2. doi:10.1186/1475-2859-10-2
Heer, D., Heine, D., and Sauer, U. (2009). Resistance of Saccharomyces cerevisiae to High Concentrations of Furfural Is Based on NADPH-dependent Reduction by at Least Two Oxireductases. Appl. Environ. Microbiol. 75, 7631–7638. doi:10.1128/AEM.01649-09
Hiraishi, H., Mochizuki, M., and Takagi, H. (2006). Enhancement of Stress Tolerance in Saccharomyces cerevisiae by Overexpression of Ubiquitin Ligase Rsp5 and Ubiquitin-Conjugating Enzymes. Biosci. Biotechnol. Biochem. 70, 2762–2765. doi:10.1271/bbb.60250
Hirasawa, T., Yoshikawa, K., Nakakura, Y., Nagahisa, K., Furusawa, C., Katakura, Y., et al. (2007). Identification of Target Genes Conferring Ethanol Stress Tolerance to Saccharomyces cerevisiae Based on DNA Microarray Data Analysis. J. Biotechnol. 131, 34–44. doi:10.1016/j.jbiotec.2007.05.010
Hong, M. E., Lee, K. S., Yu, B. J., Sung, Y. J., Park, S. M., Koo, H. M., et al. (2010). Identification of Gene Targets Eliciting Improved Alcohol Tolerance in Saccharomyces cerevisiae through Inverse Metabolic Engineering. J. Biotechnol. 149, 52–59. doi:10.1016/j.jbiotec.2010.06.006
Hong, S. H., Lee, S. S., Chung, J. M., Jung, H. S., Singh, S., Mondal, S., et al. (2017). Site-specific Mutagenesis of Yeast 2-Cys Peroxiredoxin Improves Heat or Oxidative Stress Tolerance by Enhancing its Chaperone or Peroxidase Function. Protoplasma 254, 327–334. doi:10.1007/s00709-016-0948-0
Ishii, J., Yoshimura, K., Hasunuma, T., and Kondo, A. (2013). Reduction of Furan Derivatives by Overexpressing NADH-dependent Adh1 Improves Ethanol Fermentation Using Xylose as Sole Carbon Source with Saccharomyces cerevisiae Harboring XR-XDH Pathway. Appl. Microbiol. Biotechnol. 97, 2597–2607. doi:10.1007/s00253-012-4376-6
Jarolim, S., Ayer, A., Pillay, B., Gee, A. C., Phrakaysone, A., Perrone, G. G., et al. (2013). Saccharomyces cerevisiae Genes Involved in Survival of Heat Shock. G3 (Bethesda, Md) 3, 2321–2333. doi:10.1534/g3.113.007971
Jayakody, L. N., and Jin, Y. S. (2021). In-depth Understanding of Molecular Mechanisms of Aldehyde Toxicity to Engineer Robust Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 105, 2675–2682. doi:10.1007/s00253-021-11213-1
Jönsson, L. J., and Martín, C. (2016). Pretreatment of Lignocellulose: Formation of Inhibitory By-Products and Strategies for Minimizing Their Effects. Bioresour. Technol. 199, 103–112. doi:10.1016/j.biortech.2015.10.009
Kanamasa, S., Yamaguchi, D., Machida, C., Fujimoto, T., Takahashi, A., Murase, M., et al. (2019). Draft Genome Sequence of Saccharomyces cerevisiae Strain Pf-1, Isolated from Prunus Mume. Microbiol. Resour. Announc. 8, e01169–19. doi:10.1128/MRA.01169-19
Kasavi, C., Eraslan, S., Arga, K. Y., Oner, E. T., and Kirdar, B. (2014). A System Based Network Approach to Ethanol Tolerance in Saccharomyces cerevisiae. BMC Syst. Biol. 8, 90. doi:10.1186/s12918-014-0090-6
Kasavi, C., Eraslan, S., Oner, E. T., and Kirdar, B. (2016). An Integrative Analysis of Transcriptomic Response of Ethanol Tolerant Strains to Ethanol in Saccharomyces cerevisiae. Mol. Biosyst. 12, 464–476. doi:10.1039/c5mb00622h
Kim, H. S., Kim, N. R., Yang, J., and Choi, W. (2011). Identification of Novel Genes Responsible for Ethanol And/or Thermotolerance by Transposon Mutagenesis in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 91, 1159–1172. doi:10.1007/s00253-011-3298-z
Kim, I., Yun, H., and Jin, I. (2007). Comparative Proteomic Analyses of the Yeast Saccharomyces cerevisiae KNU5377 Strain against Menadione-Induced Oxidative Stress. J. Microbiol. Biotechnol. 17, 207–217. doi:10.1113/jphysiol.1991.sp018819
Kim, S., Kim, J., Song, J. H., Jung, Y. H., Choi, I. S., Choi, W., et al. (2016). Elucidation of Ethanol Tolerance Mechanisms in Saccharomyces cerevisiae by Global Metabolite Profiling. Biotechnol. J. 11, 1221–1229. doi:10.1002/biot.201500613
Kim, S. K., Jin, Y. S., Choi, I. G., Park, Y. C., and Seo, J. H. (2015). Enhanced Tolerance of Saccharomyces cerevisiae to Multiple Lignocellulose-Derived Inhibitors through Modulation of Spermidine Contents. Metab. Eng. 29, 46–55. doi:10.1016/j.ymben.2015.02.004
Kim, S., Shin, M., Choi, W., and Kim, K. H. (2021). Comparative Metabolite Profiling of Wild Type and Thermo-Tolerant Mutant of Saccharomyces cerevisiae. Process Biochem. 111, 62–68. doi:10.1016/j.procbio.2021.10.006
Kim, T. Y., Oh, E. J., Jin, Y. S., and Oh, M. K. (2014). Improved Resistance against Oxidative Stress of Engineered Cellobiose-Fermenting Saccharomyces cerevisiae Revealed by Metabolite Profiling. Biotechnol. Bioprocess Eng. 19, 951–957. doi:10.1007/s12257-014-0301-4
Klose, C., Surma, M. A., Gerl, M. J., Meyenhofer, F., Shevchenko, A., and Simons, K. (2012). Flexibility of a Eukaryotic Lipidome-Iinsights from Yeast Lipidomics. PLoS One 7, e35063. doi:10.1371/journal.pone.0035063
Lairón-Peris, M., Routledge, S. J., Linney, J. A., Alonso-del-Real, J., Spickett, C. M., Pitt, A. R., et al. (2021). Analysis of Lipid Composition Reveals Mechanisms of Ethanol Tolerance in the Model Yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 87, e00440–21. doi:10.1128/AEM.00440-21
Lamour, J., Wan, C., Zhang, M. M., Zhao, X. Q., and Den Haan, R. (2019). Overexpression of Endogenous Stress-Tolerance Related Genes in Saccharomyces cerevisiae Improved Strain Robustness and Production of Heterologous Cellobiohydrolase. FEMS Yeast Res. 19, foz035. doi:10.1093/femsyr/foz035
Lee, J. W., Yook, S., Koh, H., Rao, C. V., and Jin, Y. S. (2021). Engineering Xylose Metabolism in Yeasts to Produce Biofuels and Chemicals. Curr. Opin. Biotechnol. 67, 15–25. doi:10.1016/j.copbio.2020.10.012
Lee, Y., Nasution, O., Choi, E., Choi, I. G., Kim, W., and Choi, W. (2015). Transcriptome Analysis of Acetic-Acid-Treated Yeast Cells Identifies a Large Set of Genes Whose Overexpression or Deletion Enhances Acetic Acid Tolerance. Appl. Microbiol. Biotechnol. 99, 6391–6403. doi:10.1007/s00253-015-6706-y
Lenz, C., and Dihazi, H. (2016). Introduction to Proteomics Technologies. Methods Mol. Biol. 1362, 3–27. doi:10.1007/978-1-4939-3106-4_1
Lewis, J. A., Broman, A. T., Will, J., and Gasch, A. P. (2014). Genetic Architecture of Ethanol-Responsive Transcriptome Variation in Saccharomyces cerevisiae Strains. Genetics 198, 369–382. doi:10.1534/genetics.114.167429
Li, B., Liu, N., and Zhao, X. (2022). Response Mechanisms of Saccharomyces cerevisiae to the Stress Factors Present in Lignocellulose Hydrolysate and Strategies for Constructing Robust Strains. Biotechnol. Biofuels Bioprod. 15, 28. doi:10.1186/s13068-022-02127-9
Li, B., Wang, L., Wu, Y. J., Xia, Z. Y., Yang, B. X., and Tang, Y. Q. (2021). Improving Acetic Acid and Furfural Resistance of Xylose-Fermenting Saccharomyces cerevisiae Strains by Regulating Novel Transcription Factors Revealed via Comparative Transcriptomic Analysis. Appl. Environ. Microbiol. 87, e00158–21. doi:10.1128/AEM.00158-21
Li, B. Z., Cheng, J. S., Ding, M. Z., and Yuan, Y. J. (2010). Transcriptome Analysis of Differential Responses of Diploid and Haploid Yeast to Ethanol Stress. J. Biotechnol. 148, 194–203. doi:10.1016/j.jbiotec.2010.06.013
Li, B. Z., and Yuan, Y. J. (2010). Transcriptome Shifts in Response to Furfural and Acetic Acid in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 86, 1915–1924. doi:10.1007/s00253-010-2518-2
Li, H., Ma, M. L., Luo, S., Zhang, R. M., Han, P., and Hu, W. (2012). Metabolic Responses to Ethanol in Saccharomyces cerevisiae Using a Gas Chromatography Tandem Mass Spectrometry-Based Metabolomics Approach. Int. J. Biochem. Cell Biol. 44, 1087–1096. doi:10.1016/j.biocel.2012.03.017
Li, Y. C., Gou, Z. X., Liu, Z. S., Tang, Y. Q., Akamatsu, T., and Kida, K. (2014). Synergistic Effects of TAL1 Over-expression and PHO13 Deletion on the Weak Acid Inhibition of Xylose Fermentation by Industrial Saccharomyces cerevisiae Strain. Biotechnol. Lett. 36, 2011–2021. doi:10.1007/s10529-014-1581-7
Lin, F. M., Qiao, B., and Yuan, Y. J. (2009a). Comparative Proteomic Analysis of Tolerance and Adaptation of Ethanologenic Saccharomyces cerevisiae to Furfural, a Lignocellulosic Inhibitory Compound. Appl. Environ. Microbiol. 75, 3765–3776. doi:10.1128/AEM.02594-08
Lin, F. M., Tan, Y., and Yuan, Y. J. (2009b). Temporal Quantitative Proteomics of Saccharomyces cerevisiae in Response to a Nonlethal Concentration of Furfural. Proteomics 9, 5471–5483. doi:10.1002/pmic.200900100
Liu, C. G., Xiao, Y., Xia, X. X., Zhao, X. Q., Peng, L., Srinophakun, P., et al. (2019). Cellulosic Ethanol Production: Progress, Challenges and Strategies for Solutions. Biotechnol. Adv. 37, 491–504. doi:10.1016/j.biotechadv.2019.03.002
Liu, L. Q., Redden, H., and Alper, H. S. (2013). Frontiers of Yeast Metabolic Engineering: Diversifying beyond Ethanol and Saccharomyces. Curr. Opin. Biotechnol. 24, 1023–1030. doi:10.1016/j.copbio.2013.03.005
Liu, W. S., and Jiang, R. R. (2015). Combinatorial and High-Throughput Screening Approaches for Strain Engineering. Appl. Microbiol. Biotechnol. 99, 2093–2104. doi:10.1007/s00253-015-6400-0
Liu, Y. F., Lin, Y. P., Guo, Y. F., Wu, F. L., Zhang, Y. Y., Qi, X. N., et al. (2021). Stress Tolerance Enhancement via SPT15 Base Editing in Saccharomyces cerevisiae. Biotechnol. Biofuels 14, 155. doi:10.1186/s13068-021-02005-w
Liu, Z. L. (2011). Molecular Mechanisms of Yeast Tolerance and In Situ Detoxification of Lignocellulose Hydrolysates. Appl. Microbiol. Biotechnol. 90, 809–825. doi:10.1007/s00253-011-3167-9
Liu, Z. L., Moon, J., Andersh, B. J., Slininger, P. J., and Weber, S. (2008). Multiple Gene-Mediated NAD(P)H-dependent Aldehyde Reduction Is a Mechanism of In Situ Detoxification of Furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 81, 743–753. doi:10.1007/s00253-008-1702-0
Lu, Y., Cheng, Y. F., He, X. P., Guo, X. N., and Zhang, B. R. (2012). Improvement of Robustness and Ethanol Production of Ethanologenic Saccharomyces cerevisiae, under Co-stress of Heat and Inhibitors. J. Industrial Microbiol. Biotechnol. 39, 73–80. doi:10.1007/s10295-011-1001-0
Lv, Y. J., Wang, X., Ma, Q., Bai, X., Li, B. Z., Zhang, W. W., et al. (2014). Proteomic Analysis Reveals Complex Metabolic Regulation in Saccharomyces cerevisiae Cells against Multiple Inhibitors Stress. Appl. Microbiol. Biotechnol. 98, 2207–2221. doi:10.1007/s00253-014-5519-8
Ma, M., and Liu, L. Z. (2010a). Quantitative Transcription Dynamic Analysis Reveals Candidate Genes and Key Regulators for Ethanol Tolerance in Saccharomyces cerevisiae. BMC Microbiol. 10, 169. doi:10.1186/1471-2180-10-169
Ma, M., and Liu, Z. L. (2010b). Comparative Transcriptome Profiling Analyses during the Lag Phase Uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as Key Regulatory Genes in Genomic Adaptation to the Lignocellulose Derived Inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11, 660. doi:10.1186/1471-2164-11-660
Ma, M., and Liu, Z. L. (2010c). Mechanisms of Ethanol Tolerance in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 87, 829–845. doi:10.1007/s00253-010-2594-3
Meena, R. C., Thakur, S., Nath, S., and Chakrabarti, A. (2011). Tolerance to Thermal and Reductive Stress in Saccharomyces cerevisiae Is Amenable to Regulation by Phosphorylation-Dephosphorylation of Ubiquitin Conjugating Enzyme 1 (Ubc1) S97 and S115. Yeast 28, 783–793. doi:10.1002/yea.1904
Mehdikhani, P., Bari, M. R., and Hovsepyan, H. (2011). Screening of Saccharomyces cerevisiae for High Tolerance of Ethanol Concentration and Temperature. Afr. J. Microbiol. Res. 5, 2654–2660.
Mehmood, M. A., Shahid, A., Malik, S., Wang, N., Javed, M. R., Haider, M. N., et al. (2021). Advances in Developing Metabolically Engineered Microbial Platforms to Produce Fourth-Generation Biofuels and High-Value Biochemicals. Bioresour. Technol. 337, 125510. doi:10.1016/j.biortech.2021.125510
Mehmood, M. A., Ibrahim, M., Rashid, U., Nawaz, M., Ali, S., Hussain, A., et al. (2017a). Biomass Production for Bioenergy Using Marginal Lands. Sustain. Prod. Consum. 9, 3–21. doi:10.1016/j.spc.2016.08.003
Mehmood, M. A., Shahid, A., Xiong, L., Ahmad, N., Liu, C. G., Bai, F. W., et al. (2017b). Chapter Five-Development of Synthetic Microbial Platforms to Convert Lignocellulosic Biomass to Biofuels. Adv. Bioenergy 2, 233–278. doi:10.1016/bs.aibe.2016.12.001
Mejía-Barajas, J. A., Montoya-Pérez, R., Salgado-Garciglia, R., Aguilera-Aguirre, L., Cortés-Rojo, C., Mejía-Zepeda, R., et al. (2017). Oxidative Stress and Antioxidant Response in a Thermotolerant Yeast. Braz. J. Microbiol. 48, 326–332. doi:10.1016/j.bjm.2016.11.005
Melendez, J. R., Mátyás, B., Hena, S., Lowy, D. A., and Salous, A. E. (2022). Perspectives in the Production of Bioethanol: a Review of Sustainable Methods, Technologies, and Bioprocesses. Renew. Sustain. Energy Rev. 160, 112260. doi:10.1016/j.rser.2022.112260
Meng, L., Liu, H. L., Lin, X., Hu, X. P., Teng, K. R., and Liu, S. X. (2020). Enhanced Multi-Stress Tolerance and Glucose Utilization of Saccharomyces cerevisiae by Overexpression of the SNF1 Gene and Varied Beta Isoform of Snf1 Dominates in Stresses. Microb. Cell Fact. 19, 134. doi:10.1186/s12934-020-01391-4
Mira, N. P., Lourenço, A. B., Fernandes, A. R., Becker, J. D., and Sa-Correia, I. (2009). The RIM101 Pathway Has a Role in Saccharomyces cerevisiae Adaptive Response and Resistance to Propionic Acid and Other Weak Acids. FEMS Yeast Res. 9, 202–216. doi:10.1111/j.1567-1364.2008.00473.x
Mira, N. P., Palma, M., Guerreiro, J. F., and Sa-Correia, I. (2010a). Genome-wide Identification of Saccharomyces cerevisiae Genes Required for Tolerance to Acetic Acid. Microb. Cell Factories 9, 79. doi:10.1186/1475-2859-9-79
Mira, N. P., and Teixeira, M. C. (2013). Microbial Mechanisms of Tolerance to Weak Acid Stress. Front. Microbiol. 4, 416. doi:10.3389/fmicb.2013.00416
Mira, N. P., Teixeira, M. C., and Sá-Correia, I. (2010b). Adaptive Response and Tolerance to Weak Acids in Saccharomyces cerevisiae: a Genome-wide View. OMICS A J. Integr. Biol. 14, 525–540. doi:10.1089/omi.2010.0072
Mitsui, R., and Yamada, R. (2021). Chapter 16 - Saccharomyces cerevisiae as a Microbial Cell Factory. Microb. Cell Factories Eng. Prod. Biomol. 319–333. doi:10.1016/B978-0-12-821477-0.00004-0
Moon, M. H., Ryu, J., Choeng, Y. H., Hong, S. K., Kang, H. A., and Chang, Y. K. (2012). Enhancement of Stress Tolerance and Ethanol Production in Saccharomyces cerevisiae by Heterologous Expression of a Trehalose Biosynthetic Gene from. Streptomyces albus. Biotechnol. Bioprocess Eng. 17, 986–996. doi:10.1007/s12257-012-0148-5
Morano, K. A., Grant, C. M., and Moye-Rowley, W. S. (2012). The Response to Heat Shock and Oxidative Stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195. doi:10.1534/genetics.111.128033
Nagamatsu, S. T., Teixeira, G. S., de Mello, F. D. B., Tizei, P. A. G., Nakagawa, B. T. G., de Carvalho, L. M., et al. (2019). Genome Assembly of a Highly Aldehyde-Resistant Saccharomyces cerevisiae SA1-Derived Industrial Strain. Microbiol. Resour. Announc. 8, e00071–19. doi:10.1128/MRA.00071-19
Nasution, O., Lee, J., Srinivasa, K., Choi, I. G., Lee, Y. M., Kim, E., et al. (2015). Loss of Dfg5 Glycosylphosphatidylinositol-Anchored Membrane Protein Confers Enhanced Heat Tolerance in Saccharomyces cerevisiae. Environ. Microbiol. 17, 2721–2734. doi:10.1111/1462-2920.12649
Ndukwe, J. K., Aliyu, G. O., Onwosi, C. O., Chukwu, K. O., and Ezugworie, F. N. (2020). Mechanisms of Weak Acid-Induced Stress Tolerance in Yeasts: Prospects for Improved Bioethanol Production from Lignocellulosic Biomass. Process Biochem. 90, 118–130. doi:10.1016/j.procbio.2019.11.009
Negi, A., Shukla, A., Jaiswar, A., Shrinet, J., and Jasrotia, R. S. (2022). Chapter 6 - Applications and Challenges of Microarray and RNA-Sequencing. Bioinformatics 91–103. doi:10.1016/B978-0-323-89775-4.00016-X
Nogué, V. S., and Karhumaa, K. (2015). Xylose Fermentation as a Challenge for Commercialization of Lignocellulosic Fuels and Chemicals. Biotechnol. Lett. 37, 761–772. doi:10.1007/s10529-014-1756-2
Nuanpeng, S., Thanonkeo, S., Yamada, M., and Thanonkeo, P. (2016). Ethanol Production from Sweet Sorghum Juice at High Temperatures Using a Newly Isolated Thermotolerant Yeast Saccharomyces cerevisiae DBKKU Y-53. Energies 9, 253. doi:10.3390/en9040253
Ohta, E., Nakayama, Y., Mukai, Y., Bamba, T., and Fukusaki, E. (2016). Metabolomic Approach for Improving Ethanol Stress Tolerance in Saccharomyces cerevisiae. J. Biosci. Bioeng. 121, 399–405. doi:10.1016/j.jbiosc.2015.08.006
Okada, N., Ogawa, J., and Shima, J. (2014). Comprehensive Analysis of Genes Involved in the Oxidative Stress Tolerance Using Yeast Heterozygous Deletion Collection. FEMS Yeast Res. 14, 425–434. doi:10.1111/1567-1364.12136
Pant, S., and Kuila, A.Ritika (2022). “Chapter 8 - Pretreatment of Lignocellulosic Biomass for Bioethanol Production,” in Advanced Biofuel Technologies. Amsterdam: Elsevier Inc., 177–194. doi:10.1016/B978-0-323-88427-3.00008-8
Pattanakittivorakul, S., Lertwattanasakul, N., Yamada, M., and Limtong, S. (2019). Selection of Thermotolerant Saccharomyces cerevisiae for High Temperature Ethanol Production from Molasses and Increasing Ethanol Production by Strain Improvement. Antonie Leeuwenhoek 112, 975–990. doi:10.1007/s10482-019-01230-6
Pereira, F. B., Guimarães, P. M. R., Gomes, D. G., Mira, N. P., Teixeira, M. C., Sa-Correia, I., et al. (2011). Identification of Candidate Genes for Yeast Engineering to Improve Bioethanol Production in Very High Gravity and Lignocellulosic Biomass Industrial Fermentations. Biotechnol. Biofuels 4, 57. doi:10.1186/1754-6834-4-57
Pereira, F. B., Teixeira, M. C., Mira, N. P., Sa-Correia, I., and Domingues, L. (2014). Genome-wide Screening of Saccharomyces cerevisiae Genes Required to Foster Tolerance towards Industrial Wheat Straw Hydrolysates. J. Industrial Microbiol. Biotechnol. 41, 1753–1761. doi:10.1007/s10295-014-1519-z
Pereira, R., Ishchuk, O. P., Li, X., Liu, Q., Liu, Y., Otto, M., et al. (2021). “Metabolic Engineering of Yeast,” in Metabolic Engineering. Hoboken: WILEY-VCH GmbH, 689–733. doi:10.1002/9783527823468.ch18
Perpiñá, C., Vinaixa, J., Andreu, C., and del Olmo, M. (2015). Development of New Tolerant Strains to Hydrophilic and Hydrophobic Organic Solvents by the Yeast Surface Display Methodology. Appl. Microbiol. Biotechnol. 99, 775–789. doi:10.1007/s00253-014-6048-1
Pinel, D., Colatriano, D., Jiang, H., Lee, H., and Martin, V. J. J. (2015). Deconstructing the Genetic Basis of Spent Sulphite Liquor Tolerance Using Deep Sequencing of Genome Shuffled Yeast. Biotechnol. Biofuels 8, 53. doi:10.1186/s13068-015-0241-z
Poljsak, B., Šuput, D., and Milisav, I. (2013). Achieving the Balance between ROS and Antioxidants: when to Use the Synthetic Antioxidants. Oxidative Med. Cell. Longev. 2013, 956792. doi:10.1155/2013/956792
Puig-Castellví, F., Alfonso, I., Piña, B., and Tauler, R. (2015). A Quantitative 1H NMR Approach for Evaluating the Metabolic Response of Saccharomyces cerevisiae, to Mild Heat Stress. Metabolomics 11, 1612–1625. doi:10.1007/s11306-015-0812-9
Rani Singhania, R., Kumar Patel, A., Singh, A., Haldar, D., Soam, S., Chen, C. W., et al. (2022). Consolidated Bioprocessing of Lignocellulosic Biomass: Technological Advances and Challenges. Bioresour. Technol. 354, 127153. doi:10.1016/j.biortech.2022.127153
Roy, S., Radivojevic, T., Forrer, Mark., Martí, J. M., Jonnalagadda, V., Backman, T., et al. (2020). Multiomics Data Collection, Visualization, and Utilization for Guiding Metabolic Engineering. Front. Bioeng. Biotechnol. 9, 612893. doi:10.1101/2020.10.15.341909
Sahara, T., Fujimori, K. E., Nezuo, M., Tsukahara, M., Tochigi, Y., Ohgiya, S., et al. (2014). Draft Genome Sequence of Saccharomyces cerevisiae IR-2, a Useful Industrial Strain for Highly Efficient Production of Bioethanol. Genome Announc. 2, e01160–13. doi:10.1128/genomeA.01160-13
Sakihama, Y., Hasunuma, T., and Kondo, A. (2015). Improved Ethanol Production from Xylose in the Presence of Acetic Acid by the Overexpression of the HAA1 Gene in Saccharomyces cerevisiae. J. Biosci. Bioeng. 119, 297–302. doi:10.1016/j.jbiosc.2014.09.004
Salas-Navarrete, P. C., de Oca Miranda, A. I. M., Martínez, A., and Caspeta, L. (2022). Evolutionary and Reverse Engineering to Increase Saccharomyces cerevisiae Tolerance to Acetic Acid, Acidic pH, and High Temperature. Appl. Microbiol. Biotechnol. 106, 383–399. doi:10.1007/s00253-021-11730-z
Sambasivam, V., Rao, D. N., and Chandrasegaran, S. (2020). Rewriting the Genome of the Model Eukaryote Saccharomyces cerevisiae. Resonance 25, 801–816. doi:10.1007/s12045-020-0997-8
Sandhya, D., Jogam, P., Narnoliya, L. K., Srivastava, A., and Jadaun, J. S. (2020). Sustainable Production of Biofuels through Synthetic Biology Approach. Hoboken: John Wiley & Sons. doi:10.1002/9781119593065.ch14
Sardi, M., Rovinskiy, N., Zhang, Y., and Gasch, A. P. (2016). Leveraging Genetic-Background Effects in Saccharomyces cerevisiae to Improve Lignocellulosic Hydrolysate Tolerance. Appl. Environ. Microbiol. 82, 5838–5849. doi:10.1128/AEM.01603-16
Schacherer, J., Shapiro, J. A., Ruderfer, D. M., and Kruglyak, L. (2009). Comprehensive Polymorphism Survey Elucidates Population Structure of Saccharomyces cerevisiae. Nature 458, 342–345. doi:10.1038/nature07670
Sévin, D. C., Kuehne, A., Zamboni, N., and Sauer, U. (2015). Biological Insights through Nontargeted Metabolomics. Curr. Opin. Biotechnol. 34, 1–8. doi:10.1016/j.copbio.2014.10.001
Shahsavarani, H., Sugiyama, M., Kaneko, Y., Chuenchit, B., and Harashima, S. (2012). Superior Thermotolerance of Saccharomyces cerevisiae for Efficient Bioethanol Fermentation Can Be Achieved by Overexpression of RSP5 Ubiquitin Ligase. Biotechnol. Adv. 30, 1289–1300. doi:10.1016/j.biotechadv.2011.09.002
Shimoi, H., Hanazumi, Y., Kawamura, N., Yamada, M., Shimizu, S., Suzuki, T., et al. (2019). Meiotic Chromosomal Recombination Defect in Sake Yeasts. J. Biosci. Bioeng. 127, 190–196. doi:10.1016/j.jbiosc.2018.07.027
Sjulander, N., and Kikas, T. (2020). Origin, Impact and Control of Lignocellulosic Inhibitors in Bioethanol Production - a Review. Energies 13, 4751. doi:10.3390/en13184751
Song, M., Ouyang, Z., and Liu, Z. L. (2009). Discrete Dynamical System Modelling for Gene Regulatory Networks of 5-hydroxymethylfurfural Tolerance for Ethanologenic Yeast. Iet Syst. Biol. 3, 203–218. doi:10.1049/iet-syb.2008.0089
Sostaric, N., Arslan, A., Carvalho, B., Plech, M., Voordeckers, K., Verstrepen, K. J., et al. (2021). Integrated Multi-Omics Analysis of Mechanisms Underlying Yeast Ethanol Tolerance. J. Proteome Res. 20, 3840–3852. doi:10.1021/acs.jproteome.1c00139
Stambuk, B. U., Dunn, B., Alves, S. L. J., Duval, E. H., and Sherlock, G. (2009). Industrial Fuel Ethanol Yeasts Contain Adaptive Copy Number Changes in Genes Involved in Vitamin B1 and B6. Genome Res. 19, 2271–2278. doi:10.1101/gr.094276.109
Stanley, D., Bandara, A., Fraser, S., Chambers, P. J., and Stanley, G. A. (2010). The Ethanol Stress Response and Ethanol Tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 109, 13–24. doi:10.1111/j.1365-2672.2009.04657.x
Strope, P. K., Skelly, D. A., Kozmin, S. G., Mahadevan, G., Stone, E. A., Magwene, P. M., et al. (2015). The 100-genomes Strains, an S. cerevisiae Resource that Illuminates its Natural Phenotypic and Genotypic Variation and Emergence as an Opportunistic Pathogen. Genome Res. 25, 762–774. doi:10.1101/gr.185538.114
Sun, X., Ni, L., Wang, G., and Zhang, X. (2019). Signatures of Directional Selection in a Hybrid Yeast Population. Genome 62, 749–760. doi:10.1139/gen-2019-0075
Swinnen, S., Schaerlaekens, K., Pais, T., Claesen, J., Hubmann, G., Yang, Y. D., et al. (2012). Identification of Novel Causative Genes Determining the Complex Trait of High Ethanol Tolerance in Yeast Using Pooled-Segregant Whole-Genome Sequence Analysis. Genome Res. 22, 975–984. doi:10.1101/gr.131698.111
Techaparin, A., Thanonkeo, P., and Klanrit, P. (2017). High-temperature Ethanol Production Using Thermotolerant Yeast Newly Isolated from Greater Mekong Subregion. Braz. J. Microbiol. 48, 461–475. doi:10.1016/j.bjm.2017.01.006
Ulaganathan, K., Goud, B. S., Reddy, M. M., Kumar, V. P., Radhakrishna, S., and Balsingh, J. (2015). Genome Sequence of Saccharomyces cerevisiae NCIM3107, Used in Bioethanol Production. Microbiol. Resour. Announc. 3, e01557–14. doi:10.1128/genomeA.01557-14
Unrean, P., Gätgens, J., Klein, B., Noack, Stephan., and Champreda, V. (2018). Elucidating Cellular Mechanisms of Saccharomyces cerevisiae Tolerant to Combined Lignocellulosic-Derived Inhibitors Using High-Throughput Phenotyping and Multiomics Analyses. FEMS Yeast Res. 18, foy106. doi:10.1093/femsyr/foy106
Usmani, Z., Sharma, M., Awasthi, A., Lukk, T., Tuohy, M., Gong, L., et al. (2021). Lignocellulosic Biorefineries: the Current State of Challenges and Strategies for Efficient Commercialization. Renew. Sustain. Energy Rev. 148, 111258. doi:10.1016/j.rser.2021.111258
Vamvakas, S. S., and Kapolos, J. (2020). Factors Affecting Yeast Ethanol Tolerance and Fermentation Efficiency. World J. Microbiol. Biotechnol. 36, 114. doi:10.1007/s11274-020-02881-8
van Zyl, W. H., den Haan, R., and la Grange, D. C. (2013). “Developing Cellulolytic Organisms for Consolidated Bioprocessing of Lignocellulosics,” in Biofuel Technologies (Berlin, Heidelberg: Springer), 189–220. doi:10.1007/978-3-642-34519-7_8
Verghese, J., Abrams, J., Wang, Y. Y., and Morano, K. A. (2012). Biology of the Heat Shock Response and Protein Chaperones: Budding Yeast (Saccharomyces cerevisiae) as a Model System. Microbiol. Mol. Biol. Rev. 76, 115–158. doi:10.1128/MMBR.05018-11
Wallace-Salinas, V., Brink, D. P., Ahrén, D., and Gorwa-Grauslund, M. F. (2015). Cell Periphery-Related Proteins as Major Genomic Targets behind the Adaptive Evolution of an Industrial Saccharomyces cerevisiae Strain to Combined Heat and Hydrolysate Stress. BMC Genomics 16, 514. doi:10.1186/s12864-015-1737-4
Wang, L., Li, B., Su, R. R., Wang, S. P., Xia, Z. Y., Xie, C. Y., et al. (2022). Screening Novel Genes by a Comprehensive Strategy to Construct Multiple Stress-Tolerant Industrial Saccharomyces cerevisiae with Prominent Bioethanol Production. Biotechnol. Biofuels 15, 1–19. doi:10.1186/s13068-022-02109-x
Wang, L., Wang, X., He, Z. Q., Zhou, S. J., Xu, L., Tan, X. Y., et al. (2020). Engineering Prokaryotic Regulator IrrE to Enhance Stress Tolerance in Budding Yeast. Biotechnol. Biofuels 13, 193. doi:10.1186/s13068-020-01833-6
Wang, X., Jin, M. J., Balan, V., Jones, A. D., Li, X., Li, B. Z., et al. (2014). Comparative Metabolic Profiling Revealed Limitations in Xylose-Fermenting Yeast during Co-fermentation of Glucose and Xylose in the Presence of Inhibitors. Biotechnol. Bioeng. 111, 152–164. doi:10.1002/bit.24992
Wei, W., McCusker, J. H., Hyman, R. W., Jones, T., Ning, Y., Cao, Z., et al. (2007). Genome Sequencing and Comparative Analysis of Saccharomyces cerevisiae Strain YJM789. Proc. Natl. Acad. Sci. U. S. A. 104, 12825–12830. doi:10.1073/pnas.0701291104
Wohlbach, D. J., Rovinskiy, N., Lewis, J. A., Sardi, M., Schackwitz, W. S., Martin, J. A., et al. (2014). Comparative Genomics of Saccharomyces cerevisiae Natural Isolates for Bioenergy Production. Genome Biol. Evol. 6, 2557–2566. doi:10.1093/gbe/evu199
Wolf, I. R., Marques, L. F., Almeida, L. F. D., Lázari, L. C., Moraes, L. N. D., Cardoso, L. H., et al. (2021). The Ethanol Tolerance in Saccharomyces cerevisiae under a Phenomics Perspective. bioRxiv. doi:10.1101/2021.08.04.455136
Xia, J. M., and Yuan, Y. J. (2009). Comparative Lipidomics of Four Strains of Saccharomyces cerevisiae Reveals Different Responses to Furfural, Phenol, and Acetic Acid. J. Agric. Food Chem. 57, 99–108. doi:10.1021/jf802720t
Xiao, W. D., Duan, X. X., Lin, Y. P., Cao, Q. C., Li, S. S., Guo, Y. F., et al. (2018). Distinct Proteome Remodeling of Industrial Saccharomyces cerevisiae in Response to Prolonged Thermal Stress or Transient Heat Shock. J. Proteome Res. 17, 1812–1825. doi:10.1021/acs.jproteome.7b00842
Yang, J., Ding, M. Z., Li, B. Z., Liu, Z. L., Wang, X., and Yuan, Y. J. (2012). Integrated Phospholipidomics and Transcriptomics Analysis of Saccharomyces cerevisiae with Enhanced Tolerance to a Mixture of Acetic Acid, Furfural, and Phenol. OMICS A J. Integr. Biol. 16, 374–386. doi:10.1089/omi.2011.0127
Yang, Y. D., Foulquié-Moreno, M. R., Clement, L., Erdei, É., An, T., Schaerlaekens, K., et al. (2013). QTL Analysis of High Thermotolerance with Superior and Downgraded Parental Yeast Strains Reveals New Minor QTLs and Converges on Novel Causative Alleles Involved in RNA Processing. PLoS Genet. 9, e1003693. doi:10.1371/journal.pgen.1003693
Yuan, Y. J., Ding, M. Z., Xia, J. M., and Cheng, J. S. (2011). “Metabolomics for Ethanologenic Yeast,” in Microbial Stress Tolerance for Biofuels. Editor Z. Liu (Berlin, Heidelberg: Springer), 22, 239–258. Microbiology Monographs. doi:10.1007/978-3-642-21467-7_10
Zhang, K., Zhang, L. J., Fang, Y. H., Jin, X. N., Qi, L., Wu, X. C., et al. (2016). Genomic Structural Variation Contributes to Phenotypic Change of Industrial Bioethanol Yeast Saccharomyces cerevisiae. FEMS Yeast Res. 16, fov118. doi:10.1093/femsyr/fov118
Zhao, H. W., Chen, J. Y., Liu, J. J., and Han, B. Z. (2015). Transcriptome Analysis Reveals the Oxidative Stress Response in Saccharomyces cerevisiae. Rsc Adv. 5, 22923–22934. doi:10.1039/c4ra14600j
Zheng, D. Q., Chen, J., Zhang, K., Gao, K. H., Li, O., Wang, P. M., et al. (2014). Genomic Structural Variations Contribute to Trait Improvement during Whole-Genome Shuffling of Yeast. Appl. Microbiol. Biotechnol. 98, 3059–3070. doi:10.1007/s00253-013-5423-7
Zheng, D. Q., Liu, T. Z., Chen, J., Zhang, K., Li, O., Zhu, L., et al. (2013). Comparative Functional Genomics to Reveal the Molecular Basis of Phenotypic Diversities and Guide the Genetic Breeding of Industrial Yeast Strains. Appl. Microbiol. Biotechnol. 97, 2067–2076. doi:10.1007/s00253-013-4698-z
Zhou, Q., Liu, Z. L., Ning, K., Wang, A. H., Zeng, X. W., and Xu, J. (2014). Genomic and Transcriptome Analyses Reveal that MAPK- and Phosphatidylinositol-Signaling Pathways Mediate Tolerance to 5-Hydroxymethyl-2-Furaldehyde for Industrial Yeast Saccharomyces cerevisiae. Sci. Rep. 4, 6556. doi:10.1038/srep06556
Keywords: Saccharomyces cerevisiae, stress tolerance, omics, strain improvement, fuel ethanol production
Citation: Xu J-R, Mehmood MA, Wang L, Ahmad N and Ma H-J (2022) OMICs-Based Strategies to Explore Stress Tolerance Mechanisms of Saccharomyces cerevisiae for Efficient Fuel Ethanol Production. Front. Energy Res. 10:884582. doi: 10.3389/fenrg.2022.884582
Received: 26 February 2022; Accepted: 29 April 2022;
Published: 06 June 2022.
Edited by:
Md. Asraful Alam, Zhengzhou University, ChinaReviewed by:
Zhaojun Wei, Hefei University of Technology, ChinaMuhammad Zohaib Nawaz, University of Agriculture Faisalabad, Pakistan
Copyright © 2022 Xu, Mehmood, Wang, Ahmad and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian-Ren Xu, eHVqaWFucmVuQG5tdS5lZHUuY24=; Muhammad Aamer Mehmood, ZHJhYW1lckBnY3VmLmVkdS5waw==
 Jian-Ren Xu
Jian-Ren Xu Muhammad Aamer Mehmood
Muhammad Aamer Mehmood Lan Wang4
Lan Wang4 Niaz Ahmad
Niaz Ahmad