
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Energy Res., 08 April 2022
Sec. Bioenergy and Biofuels
Volume 10 - 2022 | https://doi.org/10.3389/fenrg.2022.842284
 Franziska Steger1*
Franziska Steger1* İpek Ergal2
İpek Ergal2 Armin Daubek3
Armin Daubek3 Nadine Loibl1
Nadine Loibl1 Lydia Rachbauer3,4
Lydia Rachbauer3,4 Werner Fuchs1
Werner Fuchs1 Simon K.-M. R. Rittmann2
Simon K.-M. R. Rittmann2 Günther Bochmann1
Günther Bochmann1Acetic acid is an essential industrial building block and can be produced by acetogenic bacteria from molecular hydrogen (H2) and carbon dioxide (CO2). When gasses are supplied as substrates, bioreactor design plays an important role for their availability. Trickle-bed bioreactors (TBs) have an enhanced gas-to-liquid mass transfer and cells remain in the system by forming a biofilm on the carriers. So far, TBs have been investigated extensively for bio-methanation processes, whereas studies for their use in acetic acid production are rare. In this study, we evaluated the reproducibility of two parallel TBs for acetic acid production from H2:CO2 (= 70:30) by a mixed culture with a gas flow rate of 3.8 mL min−1 and a medium flow rate of 10 mL min−1. Additionally, the effect of glucose addition during the starting phase on the resulting products and microbial composition was investigated by setting up a third TB2. Partial medium exchanges to decrease the internal acetic acid concentration (AAC) combined with recycling of withdrawn cells had a positive impact on acetic acid production rates with maxima of around 1 g L−1 d−1 even at high AACs of 19–25 g L−1. Initial glucose addition resulted in the accumulation of unwanted butyric acid up to concentrations of 2.60 ± 0.64 g L−1. The maximum AAC of 40.84 g L−1 was obtained without initial glucose addition. The main families identified in the acetogenic TBs were Peptococcaceae, Ruminococcaceae, Planococcaceae, Enterobacteriaceae, Clostridiaceae, Lachnospiraceae, Dysgonomonadaceae and Tannerellaceae. We conclude that a TB is a viable solution for conversion of H2/CO2 to acetate using an anaerobic enrichment culture.
Several renewable energies, like wind and solar power, are characterized by high fluctuations, as a result of the prevailing conditions, and therefore demand storage solutions (Jafari et al., 2022; Polleux et al., 2022). As such, technologies like the power-to-gas concept have been developed, contributing to a flexible energy storage system (Mehrjerdi et al., 2022). The power-to-gas concept entails producing electricity that is utilized to generate hydrogen gas (H2) through the electrolysis of water (Götz et al., 2016). Technologies for the storage of H2, in turn, must be implemented, but have several limitations since H2 is highly flammable and may not be injected at high amounts in the already existing natural gas grid (Alamri and Alamri, 2009; Hadjipaschalis et al., 2009; Romeo et al., 2022). A promising solution is the conversion of H2 into liquid hydrocarbons, the so-called power-to-liquid concept (Herz et al., 2021). Thermochemical Fischer-Tropsch (FT) synthesis is a well-established process for producing liquid hydrocarbons from H2 and CO/CO2. However, the required elevated temperatures and pressures (200–350°C and 0.5–3 MPa) involve high power input (Gupta et al., 2021). An additional major drawback is the strong dependence on a suitable feed gas composition and purity (Griffin and Schultz, 2012; Montoya Sánchez et al., 2022).
Microbial gas fermentation is considerably less dependent on gas purity and gas composition, and, additionally, is significantly less energy-intensive than FT synthesis (Munasinghe and Khanal, 2010; Griffin and Schultz, 2012). Currently, there are various methods to capture CO2 from flue gases (Bond et al., 2001; Kanniche et al., 2010; Zhang et al., 2020) and the subsequent fixation of CO2 by microbial gas fermentation is beneficial with respect to environmental issues. Some anaerobic bacteria catalyze the conversion of gaseous H2 and CO2 into liquid products like alcohols and acids such as formic acid and acetic acid (Abubackar et al., 2018; Cheng et al., 2018; Müller, 2019; Xu et al., 2015). Being an essential industrial building block that today is mainly produced from fossil oil, the bioformation of acetic acid is a cornerstone in the production of green chemicals and fuels (Wagner, 2002; Cheung et al., 2012; Merli et al., 2021).
Acetic acid producing bacteria, so-called acetogens, are a diverse group of anaerobic, autotrophic bacteria that utilize the Wood-Ljungdahl pathway for energy conservation. This diverse group has different morphologies, habitats, and physiological properties to produce acetic acid from H2 and CO2 (Schuchmann and Müller, 2014). Up until 2013, acetogens appeared in 21 genera, with Clostridium and Acetobacterium comprising the highest number of acetogenic species (Drake et al., 2013). Many studies have applied acetogenic pure cultures to produce acetic acid (Hu et al., 2013; Kantzow et al., 2015; Groher and Weuster-botz, 2016; Abubackar et al., 2018; Cheng et al., 2018). However, the requirement of sterilization and the limited metabolic capacity of pure cultures constrain the production of acetic acid. The application of acetogenic mixed cultures, on the other hand, is more attractive, due to the lack of a sterilization requirement and the adaptive capacity resulting from the microbial diversity (Kleerebezem and van Loosdrecht, 2007; Zhang et al., 2013; Xu et al., 2015).
Several studies described that trickle-bed bioreactors (TBs) have an enhanced gas-to-liquid mass transfer combined with low power consumption (Klasson et al., 1992; Bengelsdorf et al., 2018). In TBs, cells remain in the system by forming a biofilm on the carriers. The medium trickles downwards on the carriers while gas flows counter- or co-currently (Bredwell et al., 1999). TBs have been investigated for bio-methanation processes quite extensively (Rachbauer et al., 2016; Ullrich et al., 2018; Burkhardt et al., 2019), whereas studies for their use in acetic acid production are rare (Klasson et al., 1990; Devarapalli et al., 2016; Devarapalli et al., 2017). This study is the first to use acetogenic mixed cultures in TBs. A question that comes up for large-scale implementation of gas fermentation is the start-up phase of TBs. Initial glucose addition can be a tool to deplete the remaining molecular oxygen (O2) in the system due to its consumption by aerobic and facultative anaerobic organisms. The depletion of O2 not only enables the growth of obligate anaerobic organisms, but also minimizes the risk of O2 and H2 forming potentially explosive mixtures. Additional to their autotrophic growth, most acetogens are capable of growing heterotrophically (Schuchmann and Müller, 2016). Hence, another advantage of an initial glucose addition is that specific growth rates of the desired acetogens are increased via their heterotrophic metabolism. In the model acetogen Acetobacterium woodii, only 0.3 mol of ATP per mole of acetic acid are generated during autotrophic growth (Schuchmann and Müller, 2014), whereas 4.3 mol of ATP are generated during its heterotrophic growth, converting 1 mol of glucose into 3 mol of acetic acid (Schuchmann and Müller, 2016).
In this study, we evaluated the reproducibility of 2 TBs for acetic acid production from H2 and CO2 utilizing mixed cultures. Initial glucose addition (pretreatment) and partial medium exchange(s) (MEs) were applied over the course of the fermentations. The influence of glucose addition on the resulting products and microbial composition was investigated by setting up a third TB without pretreatment. Acetic acid production rates (AAPRs) were determined at different ranges of acetic acid concentration (AACs) to provide a knowledge base for future continuous processes. Recycling of cells which were suspended in the withdrawn liquid phase during MEs was investigated, as well as the AAPR potential of solely the immobilized biofilm-forming cells.
The ingredients (Supplementary Table S1) were dissolved one by one in RO water while stirring. Fermentation medium (henceforth referred to simply as medium) was used for all TBs and contained variable amounts of yeast extract (YE) and 8 g L−1 2-bromethane-sulfonic acid (BES) to suppress methanogens. For the vitamin solution, vitamin K1 and lipoic acid were dissolved in 1 mL 96% ethanol, Niacin in 1 mL RO-water, and Hemin in 1 mL 1 mol L−1 NaOH before adding them into the purchased Kao & Michayluk vitamin solution (PhytoTech Labs, USA). A whole-cell conversion (WCC) medium without essential nutrients for cell growth was applied for biofilm investigation. Ready-made media were purged with gaseous nitrogen (N2) for at least 30 min before introducing them into the bioreactors to ensure anoxic conditions.
Three parallel trickle-bed bioreactors (TB1-3, Figure 1) consisted of glass columns, with an empty column volume of 387 mL each, a diameter of 3.3 cm and a total height of 45 cm. The packing height of the carriers (Bioflow nine PE black, RTV Process Equipment GmbH, Germany) was 33.5 cm. The gas volume of each packed column was 316 mL. The ends of the glass columns were closed with screw caps (GL45, DURAN Group, Germany) which contained a lid-element with three quick connectors attached to tubing (PUN-H-6x1, FESTO, Germany) for the equal distribution of gas entry and gas exit as well as medium circulation.
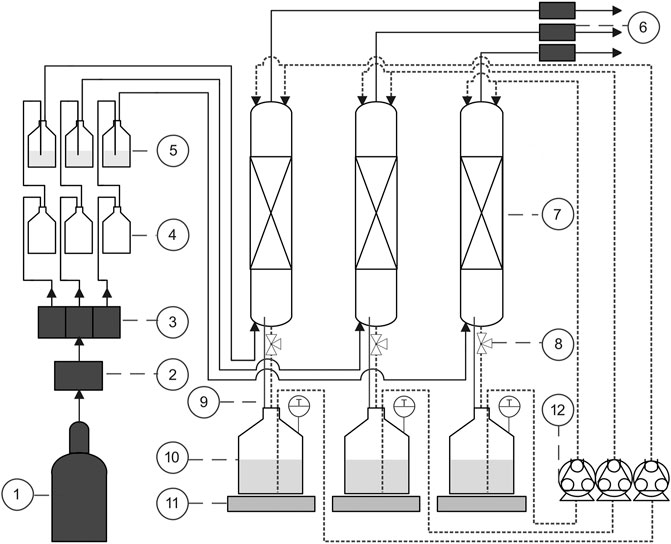
FIGURE 1. Schematic drawing of three parallel TBs with gas cylinder (1), MFC (2), gas distributer (3), liquid traps (4), humidification flasks (5), MGCs (6), three columns with heating jackets (7), sampling ports (8), pressure balancing tubes (9), three medium collection flasks (10) which were located on magnetic stirrers (11) and contained magnetic stir bars, and peristaltic pumps (12).
After trickling through the columns, the medium (400 mL) was drained and collected in closed 1 L medium collection flasks. From there, the medium was continuously recirculated to the column at a rate of 9.8 mL min−1 using a peristaltic pump (PD 5201, Heidolph, Germany). Three tubes were connected to each medium collection flask: a medium drain tube, a circulation tube, and a pressure balancing tube. The medium circulation tubes split up in two before entering on the tops of the columns to dispense the medium over the carriers more homogeneously. The drain tube contained a three-way valve for liquid sampling (sampling port). The pressure balancing tube between the bottom of the column and the medium collection flask ensured pressure equalization and facilitated drainage. Heating jackets were wrapped around the columns and medium collection flasks to regulate the temperature to 30 ± 1°C. The temperature was monitored online in the medium collection flasks. The pH was adjusted to 7 ± 0.3 once per day with 4 mol L−1 NaOH.
The gas flow rate from the gas cylinder (H2:CO2 = 70:30, Linde, Ireland) was controlled via a mass flow controller (MFC, type 8,711, Bürkert, Germany). A self-constructed gas distributor distributed the gas flow equally over the three columns. The gas passed liquid traps to protect the gas distributor and MFC in case unwanted overpressure should occur in the following humidification flasks where the gas was saturated by sparging through RO-water. The gas entered the columns at the bottom end and exited on the top before passing milligascounters (type: MGC-1 V3.4 PMMA, Ritter, Germany) for gas consumption measurement. The gas flow rate through each column was set to 3.8 ± 0.4 mL min−1. The gas flow rate was determined prior to the experiments via the MGCs downstream of the columns when the TBs were filled with water.
Two trickle-bed bioreactors (TB1 and TB2) were filled with 400 mL of medium containing 2 g L−1 YE and 1 g L−1 glucose to enhance O2 depletion and specific growth rates of acetogens. After flushing the bioreactors for 1 h with substrate gas at 9.2 ± 0.4 mL min−1 per TB, 10 mL of mixed culture inoculum 1, were introduced through the sampling port. The gas flow rate was reduced to 3.8 ± 0.4 mL min−1 per TB. After 2 days, an additional 1 g L−1 glucose was added to each bioreactor. After 5 days, the medium was completely exchanged with 400 mL of medium containing 2 g L−1 YE. To start the main acetic acid production phase, 10 mL of mixed culture inoculum 2 were introduced to each bioreactor. Inoculum 1 varied from inoculum 2 (Supplementary Table S2), as it was supplemented with sewage sludge from the aeration tank of the municipal sewage plant in Tulln, Austria. During the main acetic acid production phase, partial medium exchanges (MEs) were conducted (described in Medium Exchanges). For microbial composition analysis, 10 mL of liquid sample was withdrawn from each TB. Samples of days 28 and 36 (MCA1) were pooled together as well as samples of days 57 and 64 (MCA2) to gain an insight on acetic acid production and respective microbial composition over a time range of 1 week, instead of the snapshots.
To investigate the effect of initial glucose addition on the fermentation, the third bioreactor (TB3) was filled with 400 mL medium containing 0.5 g L−1 YE and inoculated with 10 mL of inoculum 2 without the preceding addition of glucose and inoculum 1. Before inoculation, the bioreactor was gassed with substrate gas at 28 ± 0.1 mL min−1 for 3 h to exchange the gas volume of the medium flask and column. The fermentation was started at the same time as ME1 was conducted in TB1 and TB2 (Table 1). In TB3, only ME2 and ME3 (explained in Medium Exchanges) were conducted. For microbial composition analysis, 10 mL liquid samples were withdrawn. Samples of days 16 and 24 were pooled together as well as days 45 and 52, similar to the samples from pretreatment TBs.
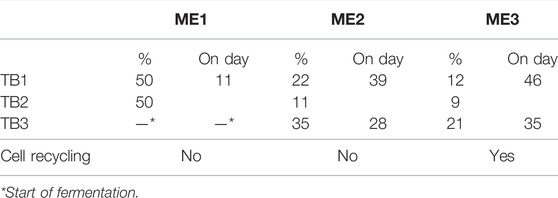
TABLE 1. Percentages of medium that were exchanged at different time points during fermentations of TB1-3.
MEs are listed in Table 1. For ME1-3, the medium with 0.5 g L−1 YE was introduced. During ME1, half of the medium was exchanged to reduce the YE concentration in TB1 and TB2. In contrast to ME1 and ME2, cells from the withdrawn liquid phase were recycled during ME3 by centrifuging. The pellet was resuspended in the fresh medium.
To investigate the AAPR of solely the biofilm that built-up on the carriers, the medium was completely exchanged in TB1 and TB2 on day 67 and in TB3 on day 56. The fresh medium lacked essential nutrients (WCC medium, Supplementary Table S1). The cells in the withdrawn medium were not recycled back into the bioreactors. The biofilm remained untouched on the carriers of the bioreactors. The investigation was conducted for 1 week. At the end of the investigation, the bioreactors were opened and samples of biofilm were gathered from the carriers for microbial composition analysis.
Optical density (OD600) was measured at 600 nm in 1 mL cuvettes (VWR, United States) using a spectrophotometer (DR 2800, Hach-Lange GmbH, Germany). The pH was measured offline with a pH-meter (FiveEasy/pH/mV, Mettler Toledo, Switzerland). Volatile fatty acids including acetic acid, butyric acid, and propionic acid as well as ethanol were quantified using high pressure liquid chromatography (Agilent 1,100 Series HPLC System with G1362A refractive index detector, Agilent Technology, CA USA) as previously described by Rachbauer et al. (2016).
All samples were frozen in liquid nitrogen and stored at −80°C. The samples were shipped on dry ice to the University of Vienna for microbial composition analysis. The microbial composition of the samples was obtained through amplicon sequencing of the V3-V4 region of the prokaryotic ribosomal small subunit (16S) gene. Primer design for amplicon sequencing is described elsewhere (Caporaso et al., 2010). The primer sequences containing adaptors are represented in Supplementary Table S3.
DNA was extracted using a modified version of the protocol by Griffiths et al. (2000). Details can be found in Supplementary Chapter 1.3.2. Nucleic acid quantification was performed with NanoDrop (NanoDrop Technologies, USA) and DNA from the samples was stored at −20°C until analysis. The PCR parameters and reagents were optimized according to the S7 Fusion DNA polymerase (Biozym, Oldendorf, Germany). Details are shown in Supplementary Chapter 1.3.3. Each sample was amplified in triplicate and after the amplification, they were checked with electrophoresis. The triplicates were combined, and purified using the QIAquick PCR purification kit (Qiagen, Netherlands) according to the manufacturer’s instructions. Cleaned DNA for sequencing was quantified using a Qubit™2.0 Fluorometer (Invitrogen) using the dsDNA HS Assay Kit. Equimolar ratios of DNA from the individual samples were prepared and 16S rRNA gene amplicon sequencing was performed on an Ilumina MiSeq (paired-end 300-bp reads) at the Vienna Biocenter Core Facilities (VBCF).
After the raw data was retrieved from the sequencing facility, the qualities of the16S amplicon sequences from each sample were checked using FastQC (Andrew, 2020). The sequence reads (https://www.ncbi.nlm.nih.gov/sra/PRJNA791644) were processed with the QIIME2 (v.2018.11) (Bolyen et al., 2019) and the variants were predicted using the DADA2 (Callahan et al., 2016) pipeline within QIIME2. For taxonomic annotations the VSEARCH (Rognes et al., 2016) tool with the SILVA (v. 132) (Quast et al., 2013) database was used. The alpha diversity and the phylogeny-based measures of the samples were calculated in QIIME2. Relative abundance was calculated as the number of sequences affiliated with that taxonomic group divided by the total number of sequences per sample and visualized using SigmaPlot version 13.0. Statistical analyses were performed using SigmaPlot version 13.0.
Before inoculation, glucose was added to TB1 and TB2 at a concentration of 1 g L−1 to enhance O2 depletion by aerobic and facultative anaerobic microorganisms. This procedure was repeated on day 2. Glucose was completely consumed within 1 day. Until day 5, AAC continuously increased to 2.65 g L−1 and 2.50 g L−1 in TB1 and TB2, respectively (Supplementary Figure S1B). Propionic acid accumulated until day 5 to 0.18 g L−1 and 0.62 g L−1 in TB1 and TB2, respectively (Supplementary Figure S1C). Lactic acid production and its subsequent consumption was observed within 1 day in both bioreactors (Supplementary Figure S1D).
For the main acetic acid production phase, the medium was exchanged completely for the medium with 2 g L−1 YE without glucose, and inoculum 2 was added to the bioreactors. Within 1 week, OD600 reached 0.44 and 0.49 in TB1 and TB2, respectively. Subsequently, the OD600 (Supplementary Figure S2) decreased to 0.05 and 0.15 in TB1 and TB2 by the end of the fermentation while acetic acid (Figure 2A) was continuously produced. This indicates that the cells were successfully immobilized on the carriers rather than suspended in the liquid medium. Due to the acidification and readjustment, the pH fluctuated between 5.5 and 7.0. This sort of pH regulation is an effective tool to reduce product inhibition due to decreased levels of undissociated acetic acid at pH-values above 6 (Wang and Wang, 1984; Sakai et al., 2005).
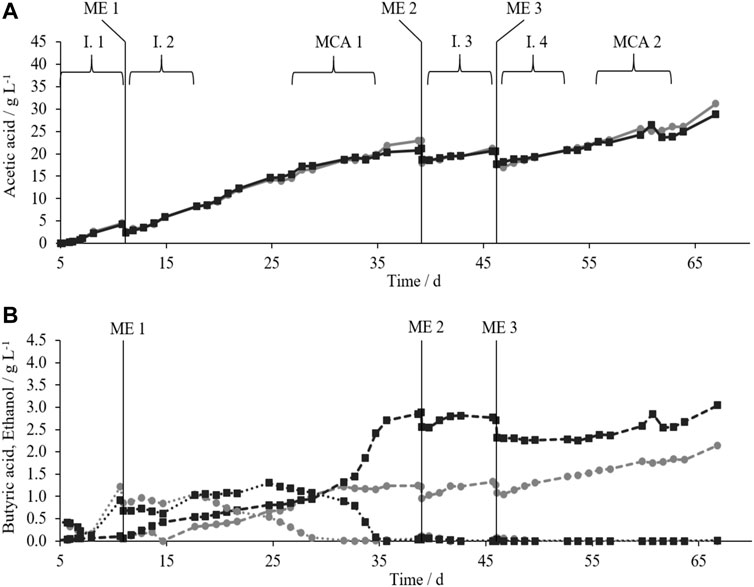
FIGURE 2. (A) AAC (solid lines) and (B) BAC (dashed lines) and ethanol concentration (dotted lines) during acetic acid production in TB1 (grey circles) and TB2 (black squares) with MEs 1–3 (vertical lines); time intervals (I.) for calculation of AAPR after MEs; time intervals for microbial composition analysis (MCA).
Initially, AAC progressed similarly in TB1 and TB2. In the 1st week of the main acetic acid production phase until ME1 (where medium was 50% exchanged with medium containing 0.5 g L−1 YE), TB1 and TB2 produced 4.53 g L−1 and 4.27 g L−1 of acetic acid, respectively, while ethanol accumulated to about 1 g L−1 in both bioreactors (Figure 2B). By the end of the experiment, AAC increased to maxima of 31.19 g L−1 in TB1 and 28.79 g L−1 in TB2. Gas was consumed throughout the fermentations (Supplementary Figure S3), except in TB1 in the week after ME3, where the output gas flow was up to 20% higher than the input gas flow. The reason could be gas production by microorganisms or the release of a preceding unintended overpressure build-up directly after ME3. The latter could have added to the increased AAPR in TB1 compared to TB2 during interval 4 (I.4) as seen in Table 2 (Demler and Weuster-Botz, 2011).
Analysis of the C-balances between days 5 and 67 (Supplementary Tables S4, 5) showed that minima of 69% and 66% of C contained in acetic acid must be derived from CO2 rather than from liquid components in TB1 and TB2, respectively. According to literature, the C content of YE was estimated by 0.4 g g−1 (Holwerda et al., 2012; Thompson et al., 2017). The total input of C contained in the YE was 32 and 31 mmol while the total output of C contained in acetic acid was 429 mmol and 368 mmol in TB1 and TB2, respectively, between day 5 and 67.
Between ME1 and ME2, ethanol concentrations in both bioreactors initially stayed at high levels. Ethanol peaks of 1.22 g L−1 on day 11 in TB1 and 1.30 g L−1 on day 25 in TB2 were followed by a complete consumption of ethanol in both bioreactors. Meanwhile, butyric acid concentrations (BAC) increased to 2.15 g L−1 and 3.05 g L−1 in TB1 and TB2, respectively, until the end of the experiment. To summarize, TB2 produced slightly less acetic acid but more butyric acid than TB1. Butyric acid could be produced from acetic acid with ethanol as electron donor (Steinbusch et al., 2011).
The goal for ME2 and ME3 was to readjust the AAC to 15–20 g L−1. AAPRs were calculated for time intervals (I.s) after each ME at varying AACs (Table 2). The highest AAPRs (0.88 g L−1d−1 in TB1 and 0.92 g L−1d−1 in TB2) were reached after ME1 when AAC was between 3–8 g L−1. At higher AAC, AAPRs decreased. This is in line with reported product inhibition by Kantzow et al. (2015), where inhibition of the acetogen Acetobacterium woodi started at AACs of 8–12 g L−1. For product purification, a higher AAC in the withdrawn medium is favored (Uribe Santos et al., 2020). In a study by Sakai et al. (2005), cell recycling combined with product removal was a suitable strategy to compensate for product inhibition in repeated batch cultures with Moorella thermoacetica. Therefore, recycling of cells from the withdrawn liquid phase was conducted during ME3 to evaluate if AAPR could be increased at higher AACs. Cell recycling during ME3 had a positive impact on AAPR versus without cell recycling during ME2. At AACs of 17–21 g L−1, AAPRs increased from 0.44 to 0.64 g L−1d−1 in TB1 and from 0.32 to 0.42 g L−1d−1 in TB2 during I.3 and I.4, respectively.
Microbial composition analysis was performed to confirm the presence of acetogens in TB1 and TB2. AAPR during time intervals between pooled samples (MCA1 and MCA2) were calculated (Table 2). Similar to the results presented in Main Acetic Acid Production Phase, TB1 showed 27 and 40% higher AAPRs than TB2 during MCA1 and MCA2, respectively. While in total 32% more butyric acid accumulated in TB2 compared to TB1, despite the equal starting conditions for both TBs. Microbial composition analysis was conducted to elucidate the findings.
To identify the microbial composition at a high accuracy, a targeted amplicon study was conducted. The V3-V4 region was selected for microbial composition analysis. This region is considered the most effective for paired-end sequencing with increasing coverage for long sequences and one of the most frequently targeted regions of the 16S rRNA gene (Mizrahi-Man et al., 2013; Franzén et al., 2015). Hence, Illumina MiSeq sequencing technology (paired-end 300-bp reads) based on the 16S rRNA gene (V3-V4 region) was applied to assess the changes in the consortium, and identify the differences between the MCAs in TB1-2 and TB3.
The main families identified in the microbial composition analysis of TBs were Enterobacteriaceae, Clostridiaceae, Ruminococcaceae, Lachnospiraceae, Peptococcaceae, Planococcaceae, Dysgonomonadaceae, Tannerellaceae (Figure 3A). The overview of relative abundances can be found in Supplementary Table S6. Methanogens were not detected due to the fact that BES was added to the fermentations (Supplementary Table S1). As expected, due to equal starting conditions, the presence of these organisms was similar in TB1 and TB2, and altered (either decreased or increased) comparably from MCA1 to MCA2. However, even though the trend was the same, the degree of alteration of microbial composition varied.
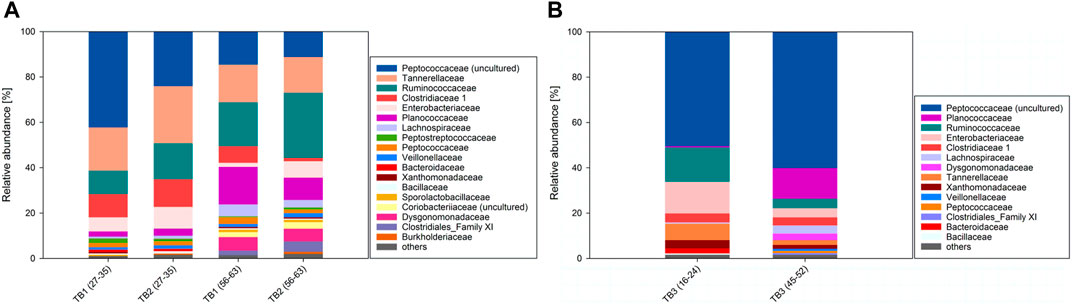
FIGURE 3. (A) Microbial composition of TB1 and TB2 with pretreatment during intervals MCA1 and MCA2. (B) Microbial composition of TB3 without pretreatment during intervals MCA1 and MCA2.
In TB1, families Enterobacteriaceae, Tannerellaceae and Clostridiaceae 1, decreased by 4.4, 2.5 and 2.9% from MCA1 to MCA2, while in TB2 these ratios were 2.3, 9.5 and 10.7% from MCA1 to MCA2, respectively. Enterobacteriaceae and Tannerellaceae are known to produce acetic acid from glucose (Sakamoto et al., 2007; Octavia and Lan, 2013). Initial glucose addition might have stimulated the early growth of Enterobacteriaceae and Tannerellaceae, while their presence started to decreased once glucose was completely depleted after day 5 in TB1 and TB2. A drastic decrease in the Clostridiaceae 1 family, which contains acetogens, resulted in an 8% reduction of maximum AAC in TB2 compared to TB1. There was no obvious reason why this drastic decrease occurred in TB2 but not in TB1. Potentially, Clostridiaceae 1 were outrun by other organisms in TB2. Ruminococcaceae, for example, increased by 12.9% from MCA1 to MCA2 in TB2, but only by 9.0% in TB1. Ruminococcaceae contain butyrate producing-bacteria (Esquivel-Elizondo et al., 2017), reasonably explaining the 32% higher butyric acid amount in TB2 compared to TB1.
A significant decrease of Peptococcaceae and Peptostreptococcaceae by 27.6 and 11.9% was observed in TB1 and TB2, respectively. Some species of Peptococcaceae and Peptostreptococcaceae are known to metabolize amino acids, YE and/or glucose to H2, CO2 and short chain fatty acids including acetic acid and butyric acid (Rogosa, 1971; Slobodkin, 2014). Their decrease is consistent with the depletion of initially added glucose and the YE reduction during the MEs.
Other Firmicutes (Planococcaceae, Lachnospiraceae) and Bacteroidetes (Dysgonomonadaceae) increased. Planococcaceae perform incomplete oxidation of organic compounds to acetic acid (Robertson et al., 2001; Spring and Rosenzweig, 2006; Villemur et al., 2006). Their increase can be explained by the addition of 2 g YE L−1 during ME one and addition of 0.5 g YE L−1 during MEs 2–3. It has been shown that acetate is predominantly produced by the phylum Bacteroidetes, whereas butyrate is the main product of the phylum Firmicutes (Ismail et al., 2011). Most acetogens, which produce acetic acid from H2/CO2, belong to the class of Clostridia of the phylum Firmicutes, including the families Lachnopsiraceae and Clostridiaceae (Drake et al., 2008), explaining the increase of Lachnospiraceae in both TBs. However, a higher increase of Lachnospiraceae by 4.5% was observed in TB1 compared to 2.0% in TB2. Higher percentages of Clostridiaceae 1 and Lachnospiraceae in TB1 (7.4 and 5.3%, respectively) compared to TB2 (1.5 and 3.2%, respectively) explain the higher AAPR of 0.47 g L−1 d−1 in TB1 compared to 0.28 g L−1 d−1 TB2 during MCA2.
In TB3, the main acetic acid production phase started directly with inoculum 2 and proceeded without initial glucose addition with 0.5 g L−1 YE. The OD600 increased to a maximum of 0.48 after 14 days and afterwards gradually decreased to 0.10 by the end of the fermentation (Supplementary Figure S4) while acetic acid was continuously produced (Figure 4A), indicating that the cells were immobilized in the column.
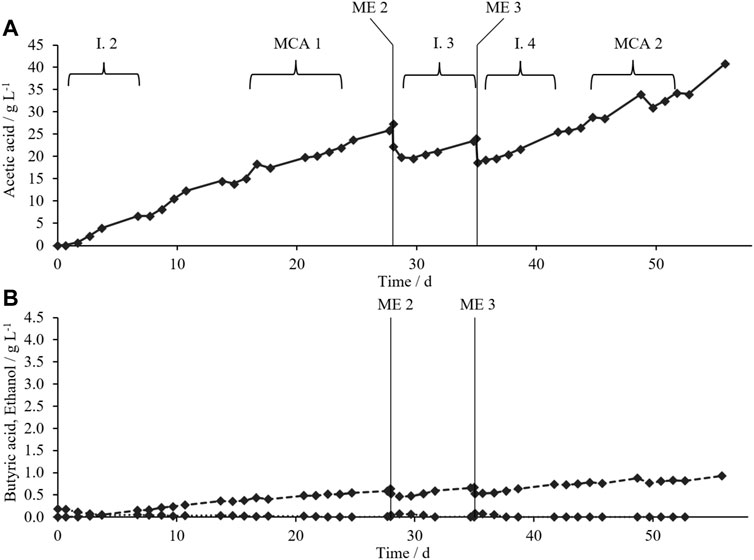
FIGURE 4. (A) AAC (solid lines) and (B) BAC (dashed lines) and ethanol concentration (dotted lines) during acetic acid production in TB2 (black diamonds) with MEs 2–3 (vertical lines); time intervals (I.) for calculation of AAPR after MEs; time intervals for microbial composition analysis (MCA).
After a short lag phase of 2 days, AAC accumulated to 27.36 g L−1 until ME2. Butyric acid accumulated to 0.93 g L−1, whereas ethanol was not detected (Figure 4B). TB3 reached a high final AAC of 40.84 g L−1 with a still-rising trend. Xu et al. (2015) investigated acetogenic mixed cultures in 1 L serum bottles and reached a maximum AAC of 57 g L−1 with daily pH adjustment. The highest maximum AAC reported for acetogenic pure culture was 59 g L−1 with A. woodii (Kantzow et al., 2015). We hypothesize that similar high AACs could have been reached in TB3 with a prolonged process time. Wang et al. (2017) reported a maximum AAC of 42.4 g L−1 with a thermophilic (55°C) mixed culture in hollow-fiber membrane (HFM) bioreactors. Similar to our study, butyric acid was the other byproduct.
Analysis of the C-balance (Supplementary Table S7) showed that a minimum of 85% of C contained in acetic acid must have derived from CO2 rather than from liquid components. The total input of C contained in YE was 11 mmol while the total output of C contained in acetic acid was 651 mmol in TB3, from day 1 to day 56. Gas consumption was noticed throughout the fermentation (Supplementary Figure S5).
As for TB1 and TB2, the goal for ME2 and ME3 was to reset the AAC to 15–20 g L−1. AAPRs were calculated for time intervals after MEs (Table 3) as in experiments with pretreatment (Main Acetic Acid Production Phase). The highest AAPR (1.12 g L−1 d−1) was observed during the 1st week I.2. AAPRs decreased with elevated AACs without cell recycling. Cell recycling from the withdrawn liquid phase during ME3 must be highlighted here as it enabled similarly high AAPR (1.08 g L−1 d−1) at a significantly elevated AAC of 19–25 g L−1. The AAPR during I.4 increased by 61% compared to I.3 without cell recycling.
The microbial composition of TB3 contained the same main families as TB1 and TB2 (Microbial Composition Analysis of Trickle-Bed Bioreactors With Pretreatment). However, community structure altered differently in TB3, which we attribute to the lack of pretreatment (initial glucose addition) in TB3, in direct contrast to TB1 and TB2. The overview of relative abundances can be found in Supplementary Table S8. In contrast to TB1 and TB2 with pretreatment, where butyric acid producing Ruminococcaceae increased (Microbial Composition Analysis of Trickle-Bed Bioreactors With Pretreatment), Ruminococcaceae decreased by 11.1% from MCA1 to MCA2 in TB3 (Figure 3B). Hence, in total 57 and 70% less butyric acid accumulated in TB3 compared to TB1 and TB2, respectively. Also, percentages of families Enterobacteriaceae and Tannerellaceae, which produce acids from glucose (Octavia and Lan, 2013; Duysburgh et al., 2019), decreased by 9.9 % and 5.2%, respectively, from MCA1 to MCA2 in TB3, likely due to the lack of pretreatment.
The families Dysgonomonadaceae, Peptococcaceae and Planococcaceae, on the other hand, increased by 2.9, 9.7 and 13.0% from MCA1 to MCA2. Dysgonomonadaceae belongs to the phylum Bacteroidetes which is known for acetic acid production (Ismail et al., 2011). Additionally, the Peptococcaceae population, that oxidizes propionate to acetate, formate, and H2, might be dominant in TB3 (Müller et al., 2010). As said in Microbial Composition Analysis of Trickle-Bed Bioreactors With Pretreatment, some of the members of the Planococcaceae perform incomplete oxidation of organic compounds to acetate (Robertson et al., 2001; Spring and Rosenzweig, 2006; Villemur et al., 2006). Hence, the increase of Dysgonomonadaceae, Peptococcaceae and Planococcaceae could be an indication for the higher AAPR of 0.76 g L−1 d−1 in TB3 (Table 3) compared to 0.47 g L−1 d−1 and 0.28 g L−1 d−1 in TB1 and TB2 (Table 2) during MCA2.
In contrast to TB1 and TB2, the percentage of Clostridiaceae 1 did not decrease but stayed at 3.6%, while Lachnospiraceae increased by 3.0% from MCA1 to MCA2 in TB3. Many members of the Clostridiaceae 1 and Lachnospiraceae are prominent acetogens, producing acetic acid from H2/CO2 (Drake et al., 2008). Their presence explains the higher maximum AAC in TB3 (31 and 42% compared to TB1 and TB2) as observed in our results in main acetic acid production phases of Trickle-bed bioreactors with and without pretreatment.
Additionally, the Peptococcaceae population that oxidized propionate to acetate, formate, and H2, might be dominant in TB3 (Müller et al., 2010). As said in Microbial Composition Analysis of Trickle-Bed Bioreactors With Pretreatment, some of the members of Planococcaceae perform incomplete oxidation of organic compounds to acetate (Robertson et al., 2001; Spring and Rosenzweig, 2006; Villemur et al., 2006). This could be another indication for higher AAC, although less YE was added to TB3 than to TB1 and TB2.
In contrast to TB3, TB1 and TB2 received pretreatment in the form of the initial addition of glucose and inoculation with inoculum 1 which was supplemented with sewage sludge. Aerobic and facultative anaerobic organisms were supposed to deplete O2 and subsequently decrease under the anaerobic and almost autotrophic conditions of the main acetic acid production phase. On day 5, the medium was completely exchanged to a medium with 2 g L−1 YE, to enhance the formation of an acetogenic biofilm, and inoculation with inoculum 2 was conducted. At this point, TB3 was started with 0.5 g L−1 YE and inoculated with inoculum 2. Subsequently, the YE concertation was only lowered in TB1 and TB2 during partial medium exchanges with a medium containing 0.5 g L−1 YE. Hence, the TB1 and TB2 contained more non-CO2 C-sources, especially in the starting phase.
TB1 and TB2 developed similarly regarding the accumulation of acetic acid and butyric acid (Main Acetic Acid Production Phase of Trickle-Bed Bioreactors With Pretreatment) as well as regarding the microbial composition (Microbial Composition Analysis of Trickle-Bed Bioreactors With Pretreatment). Whereas TB3 developed differently (Main Acetic Acid Production Phase and Microbial Composition Analysis of Trickle-Bed Bioreactor Without Pretreatment). C-balance analysis (Supplementary Sections 2.1.4 and 2.2.3) showed that acetic acid was successfully produced in all TBs from H2/CO2. In TBs with pretreatment, C contained in acetic acid constituted 90 ± 3% of total C contained in produced volatile fatty acids, while C contained in butyric acid was 10 ± 3%. Relatively fewer side products and more acetic acid accumulated in TB3 without pretreatment, where C contained in acetic acid constituted 97% of total C contained in the produced volatile fatty acids, while C contained in butyric acid was only 3%. Wang et al. (2017) also observed the side-production of butyric acid during conversion of H2/CO2 to acetic acid with thermophilic mixed culture. However, acetic acid still accounted for ca. 99% of total volatile fatty acids, while their medium did not contain YE and glucose at all. In summary, the enhanced formation of side product (butyric acid) at the expense of acetic acid production seems to overshadow the beneficial effect of initial glucose addition for O2 depletion as well as of YE addition for enhancing the acetogenic biofilm formation.
AAC lag phases of about 2 days occurred in both scenarios (with and without pretreatment). Hence, the pretreatment and higher amounts (2 g L−1) of YE did not promote early AAPR. Direct inoculation to 0.5 g L−1 YE without glucose addition resulted in the highest AAPR, of 1.12 g L−1 d−1, during the main acetic acid production phases (achieved in TB3). Similar observations were made in a previous study, where the decrease of YE concentration from 0.5 to 0.25 g L−1 during acetic acid production with A. woodii did not affect the AAC lag phase (Steger et al., 2017).
Cell recycling from withdrawn culture broth had an accelerating effect on AAPR in all TBs during ME3, indicating that cells in the liquid phase contributed to AAPRs, additionally to immobilized cells in the columns. Thereby, initially high AAPR (1.12 g L−1 d−1) was fully regained at significantly higher AAC (19–25 g L−1) in TB3. In TB1 and TB2, initially high AAPRs (0.90 ± 0.03 g L−1 d−1) were only partly regained after ME 3 with 0.53 ± 0.16 g L−1 d−1 at higher AAC (18–21 g L−1). The positive effect of cell recycling or retention combined with product removal was already shown by other studies (Suzuki et al., 1993; Sakai et al., 2005; Kantzow et al., 2015; Steger et al., 2017). Kantzow et al. (2015) used a CSTR with a submerged HFM module to increase the biomass of A. woodii and to reach one of the highest AAPRs reported so far during continuous fermentation (146.8 g L−1 d−1). However, membrane blockage might be a challenge when HFM is applied for cell retention. Suzuki et al. (1993) increased the mean AAPR from 0.60 g L−1d−1 in batch mode to 0.76 g L−1 d−1 in repeated batch mode with flocculated cells of A. woodii. Steger et al. (2017) reached a constant AAPR of 1.2 g L−1 d−1 when A. woodii was immobilized on linen in a continuous CSTR fermentation.
At the end of the fermentations, AAC in TB3 was about 10 g L−1 higher than in TB1 and TB2, with 40.84 g L−1. In the literature, similarly high AACs were reached with mixed culture by Xu et al. (2015) with 57 g L−1 and Wang et al. (2017) with 42.4 g L−1, both without the addition of YE and glucose, confirming the observation made during the current study, that YE addition counteracts the accumulation of acetic acid with mixed culture in TB systems as well.
The medium was completely exchanged in all three bioreactors and substituted with WCC medium without essential nutrients for cell growth to investigate the AAPR potential of solely the biofilms on the carriers for 1 week. In all 3 TBs, OD600 increased from about 0.1 to 0.22, 0.36 and 0.16 in TB1, TB2, and TB3, respectively (Supplementary Figure S6), indicating that cells detached from the biofilm. However, residues of the old medium might have permitted little growth of microorganisms as MEs were conducted without washing steps.
Final BACs were low with 0.42 g L−1, 0.51 g L−1, and 0.26 g L−1 in TB1, TB2, and TB3, respectively (Supplementary Figures S7B, D). Ethanol formation was not detected in all TBs. Acetic acid was produced in all 3 TBs indicating that acetogens were successfully retained in the biofilms in the columns. AACs increased after a lag phase of about 2 days in TB1-3 (Supplementary Figures S7A, C), as already seen during the main acetic acid production phases of TB1-3 (Figure 2A and4a). TB2 showed the highest increase of OD600 together with the highest AAPR of 1.15 g L−1 d−1 during this study (Table 4). AAPR seemed to correlate with OD600 as TB3 showed the lowest AAPR of 0.82 g L−1 d−1 together with the lowest OD600. This result was surprising because, initially, TB3 produced more acetic acid than TB2 (Figures 2, 4). One possible explanation is that acetic acid was also formed by acetogens that were suspended in the liquid phase of the TBs, which would explain the positive effect of cell recycling during MEs. Microbial composition of the biofilms was similar for the 3 TBs (Supplementary Figure S8). The main families were Peptococcaceae, Tannerellaceae, Clostridicaea 1, Ruminococcaceae, Planococcaceae and Enterobacteriaceae. The overview of relative abundances can be found in Supplementary Table S9. Potential roles of these families were already discussed in microbial composition analysis of Trickle-bed bioreactors with and without pretreatment. Especially Clostridiaceae 1, which are well-known for their acetic acid production (Drake et al., 2008), were found in higher percentages in the biofilms (11.6, 5.1 and 7.4% in TB1, TB2 and TB3) than in the liquid phases of the TBs during the MCA2 (7.4, 1.5 and 3.6% in TB1, TB2 and TB3).
The current study proves that TBs are convenient systems for the production of acetic acid from H2 and CO2 achieving high AACs (at least 40.84 g L−1) and AAPRs (up to 1.15 g L−1 d−1). Higher AAPRs were reached when AACs were below 8 g L−1. However, recycling of cells from withdrawn liquid phase during medium exchanges could compensate for product inhibition regarding AAPR. Acetogens were successfully retained in the biofilms in the columns, whereas, the cells in the liquid phase added a significant amount to AAPR. Thus, we recommend the recycling of withdrawn cells during continuous or semi-continuous mode in the starting phase of the fermentation.
The microbial composition of the liquid phases developed comparably in 2 TBs with pretreatment (glucose addition). On the contrary, microbial composition altered differently in the third TB without pretreatment. Inoculation to almost autotrophic conditions (0.5 g L−1 YE) without pretreatment was advantageous over initial glucose addition and higher amounts of YE (2 g L−1). Glucose and YE addition did not, as hypothesized, shorten the lag-phase of acetogens, but increased the growth of non-acetic acid producers and therefore the formation of side products (butyric acid and ethanol).
YE was not contained in the added medium during the investigation of the AAPR potential of the biofilms. However, acetic acid was produced at AAPRs of up to 1.15 g L−1 d−1 within 1 week. This is an important result as YE is an expensive additive and therefore should be omitted for industrial applications.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/PRJNA791644.
FS, LR and GB planned the experiments. FS, AD and NL performed the experiments. İE and SK-MRR performed the microbial composition analysis. FS and İE wrote the manuscript. WF, SK-MRR and GB supervised the work. All authors discussed the collected data and corrected the manuscript before submission.
The presented study was developed in the course of a research project. We kindly thank the Austrian Research Promotion Agency (FFG) for funding of PTLiquid (FFG project no. 865091).
Authors AD and LR were employed by BEST—Bioenergy and Sustainable Technologies GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2022.842284/full#supplementary-material
We acknowledge Angus S. Hilts, MSc for assistance in proofreading the manuscript.
Abubackar, H. N., Veiga, M. C., and Kennes, C. (2018). Production of Acids and Alcohols from Syngas in a Two-Stage Continuous Fermentation Process. Bioresour. Technol. 253, 227–234. doi:10.1016/j.biortech.2018.01.026
Alamri, B. R., and Alamri, A. R. (2009). “Technical Review of Energy Storage Technologies when Integrated with Intermittent Renewable Energy,” in 1st International Conference on Sustainable Power Generation and Supply, Nanjing, China, 6-7 April 2009. SUPERGEN ’09, art. no. 5348055. doi:10.1109/SUPERGEN.2009.5348055
Andrew, S. (2020). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (Accessed September 9, 2020).
Bengelsdorf, F. R., Beck, M. H., Erz, C., Hoffmeister, S., Karl, M. M., Riegler, P., et al. (2018). Bacterial Anaerobic Synthesis Gas (Syngas) and CO2+H2 Fermentation. Adv. Appl. Microbiol. 103, 143–221. doi:10.1016/bs.aambs.2018.01.002
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi:10.1038/s41587-019-0209-9
Bond, G. M., Stringer, J., Brandvold, D. K., Simsek, F. A., Medina, M.-G., and Egeland, G. (2001). Development of Integrated System for Biomimetic CO2 Sequestration Using the Enzyme Carbonic Anhydrase. Energy Fuels 15 (2), 309–316. doi:10.1021/ef000246p
Bredwell, M. D., Srivastava, P., and Worden, R. M. (1999). Reactor Design Issues for Synthesis-Gas Fermentations. Biotechnol. Prog. 15 (5), 834–844. doi:10.1021/bp990108m
Burkhardt, M., Jordan, I., Heinrich, S., Behrens, J., Ziesche, A., and Busch, G. (2019). Long Term and Demand-Oriented Biocatalytic Synthesis of Highly Concentrated Methane in a Trickle Bed Reactor. Appl. Energ. 240, 818–826. doi:10.1016/j.apenergy.2019.02.076
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 13 (7), 581–583. doi:10.1038/nmeth.3869
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 7 (5), 335–336. doi:10.1038/nmeth.f.303
Cheng, H.-H., Syu, J.-C., Tien, S.-Y., and Whang, L.-M. (2018). Biological Acetate Production from Carbon Dioxide by Acetobacterium Woodii and Clostridium Ljungdahlii: The Effect of Cell Immobilization. Bioresour. Technol. 262, 229–234. doi:10.1016/j.biortech.2018.04.069
Cheung, H., Tanke, R. S., and Torrence, G. P. (2012). “Acetic Acid,” in Encyclopedia of Industrial Chemistry (Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA), 1, 209–237. doi:10.1002/14356007.a01_045.pub2
Demler, M., and Weuster-Botz, D. (2011). Reaction Engineering Analysis of Hydrogenotrophic Production of Acetic Acid by Acetobacterium Woodii. Biotechnol. Bioeng. 108 (2), 470–474. doi:10.1002/bit.22935
Devarapalli, M., Atiyeh, H. K., Phillips, J. R., Lewis, R. S., and Huhnke, R. L. (2016). Ethanol Production during Semi-Continuous Syngas Fermentation in a Trickle Bed Reactor Using Clostridium Ragsdalei. Bioresour. Technol. 209, 56–65. doi:10.1016/j.biortech.2016.02.086
Devarapalli, M., Lewis, R., and Atiyeh, H. (2017). Continuous Ethanol Production from Synthesis Gas by Clostridium Ragsdalei in a Trickle-Bed Reactor. Fermentation 3 (2), 23. doi:10.3390/fermentation3020023
Drake, H. L., Gößner, A. S., and Daniel, S. L. (2008). Old Acetogens, New Light. Ann. N Y Acad. Sci. 1125, 100–128. doi:10.1196/annals.1419.016
Drake, H. L., Küsel, K., and Matthies, C. (2013). “Acetogenic Prokaryotes,” in The Prokaryotes: Prokaryotic Physiology and Biochemistry. Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer-Verlag Berlin Heidelberg), 3–60. doi:10.1007/978-3-642-30141-4_61
Duysburgh, C., Van den Abbeele, P., Krishnan, K., Bayne, T. F., and Marzorati, M. (2019). A Synbiotic Concept Containing Spore-Forming Bacillus Strains and a Prebiotic Fiber Blend Consistently Enhanced Metabolic Activity by Modulation of the Gut Microbiome In Vitro. Int. J. Pharmaceutics: X 1, 100021. doi:10.1016/j.ijpx.2019.100021
Esquivel-Elizondo, S., Ilhan, Z. E., Garcia-Peña, E. I., and Krajmalnik-Brown, R. (2017). Insights into Butyrate Production in a Controlled Fermentation System via Gene Predictions. MSystems 2 (4), e00051–17. doi:10.1128/msystems.00051-17
Franzén, O., Hu, J., Bao, X., Itzkowitz, S. H., Peter, I., and Bashir, A. (2015). Improved OTU-Picking Using Long-Read 16S rRNA Gene Amplicon Sequencing and Generic Hierarchical Clustering. Microbiome 3, 43. doi:10.1186/s40168-015-0105-6
Götz, M., Lefebvre, J., Mörs, F., McDaniel Koch, A., Graf, F., Bajohr, S., et al. (2016). Renewable Power-To-Gas: A Technological and Economic Review. Renew. Energ. 85, 1371–1390. doi:10.1016/j.renene.2015.07.066
Griffin, D. W., and Schultz, M. A. (2012). Fuel and Chemical Products from Biomass Syngas: A Comparison of Gas Fermentation to Thermochemical Conversion Routes. Environ. Prog. Sustain. Energ. 31 (2), 219–224. doi:10.1002/ep.11613
Griffiths, R. I., Whiteley, A. S., O'Donnell, A. G., and Bailey, M. J. (2000). Rapid Method for Coextraction of DNA and RNA from Natural Environments for Analysis of Ribosomal DNA- and rRNA-Based Microbial Community Composition. Appl. Environ. Microbiol. 66 (12), 5488–5491. doi:10.1128/AEM.66.12.5488-5491.2000
Groher, A., and Weuster-botz, D. (2016). Comparative Reaction Engineering Analysis of Different Acetogenic Bacteria for Gas Fermentation. J. Biotechnol. 228, 82–94. doi:10.1016/j.jbiotec.2016.04.032
Gupta, P. K., Kumar, V., and Maity, S. (2021). Renewable Fuels from Different Carbonaceous Feedstocks: A Sustainable Route through Fischer-Tropsch Synthesis. J. Chem. Technol. Biotechnol. 96 (4), 853–868. doi:10.1002/jctb.6644
Hadjipaschalis, I., Poullikkas, A., and Efthimiou, V. (2009). Overview of Current and Future Energy Storage Technologies for Electric Power Applications. Renew. Sustain. Energ. Rev. 13 (6–7), 1513–1522. doi:10.1016/j.rser.2008.09.028
Herz, G., Rix, C., Jacobasch, E., Müller, N., Reichelt, E., Jahn, M., et al. (2021). Economic Assessment of Power-To-Liquid Processes - Influence of Electrolysis Technology and Operating Conditions. Appl. Energ. 292, 116655. doi:10.1016/j.apenergy.2021.116655
Holwerda, E. K., Hirst, K. D., and Lynd, L. R. (2012). A Defined Growth Medium with Very Low Background Carbon for culturingClostridium Thermocellum. J. Ind. Microbiol. Biotechnol. 39 (6), 943–947. doi:10.1007/s10295-012-1091-3
Hu, P., Rismani-Yazdi, H., and Stephanopoulos, G. (2013). Anaerobic CO2 fixation by the Acetogenic bacteriumMoorella Thermoacetica. Aiche J. 59 (9), 3176–3183. doi:10.1002/aic.14127
Ismail, N. A., Ragab, S. H., ElBaky, A. A., Shoeib, A. R. S., Alhosary, Y., and Fekry, D. (2011). Frequency of Firmicutes and Bacteroidetes in Gut Microbiota in Obese and normal Weight Egyptian Children and Adults. Arch. Med. Sci. 7 (3), 501–507. doi:10.5114/aoms.2011.23418
Jafari, M., Botterud, A., and Sakti, A. (2022). Decarbonizing Power Systems: A Critical Review of the Role of Energy Storage. Renew. Sustain. Energ. Rev. 158, 112077. doi:10.1016/j.rser.2022.112077
Kanniche, M., Gros-Bonnivard, R., Jaud, P., Valle-Marcos, J., Amann, J.-M., and Bouallou, C. (2010). Pre-Combustion, Post-Combustion and Oxy-Combustion in Thermal Power Plant for CO2 Capture. Appl. Therm. Eng. 30 (1), 53–62. doi:10.1016/j.applthermaleng.2009.05.005
Kantzow, C., Mayer, A., and Weuster-Botz, D. (2015). Continuous Gas Fermentation by Acetobacterium Woodii in a Submerged Membrane Reactor with Full Cell Retention. J. Biotechnol. 212, 11–18. doi:10.1016/j.jbiotec.2015.07.020
Klasson, K. T., Ackerson, M. D., Clausen, E. C., and Gaddy, J. L. (1992). Bioconversion of Synthesis Gas into Liquid or Gaseous Fuels. Enzyme Microb. Technol. 14 (8), 602–608. doi:10.1016/0141-0229(92)90033-K
Klasson, K. T., Elmore, B. B., Vega, J. L., Ackerson, M. D., Clausen, E. C., and Gaddy, J. L. (1990). Biological Production of Liquid and Gaseous Fuels from Synthesis Gas. Appl. Biochem. Biotechnol. 24-25 (1), 857–873. doi:10.1007/BF02920300
Kleerebezem, R., and van Loosdrecht, M. C. (2007). Mixed Culture Biotechnology for Bioenergy Production. Curr. Opin. Biotechnol. 18 (3), 207–212. doi:10.1016/j.copbio.2007.05.001
Mehrjerdi, H., Saboori, H., and Jadid, S. (2022). Power-to-Gas Utilization in Optimal Sizing of Hybrid Power, Water, and Hydrogen Microgrids with Energy and Gas Storage. J. Energ. Storage 45, 103745. doi:10.1016/j.est.2021.103745
Merli, G., Becci, A., Amato, A., and Beolchini, F. (2021). Acetic Acid Bioproduction: The Technological Innovation Change. Sci. Total Environ. 798, 149292. doi:10.1016/j.scitotenv.2021.149292
Mizrahi-Man, O., Davenport, E. R., and Gilad, Y. (2013). Taxonomic Classification of Bacterial 16S rRNA Genes Using Short Sequencing Reads: Evaluation of Effective Study Designs. PLoS ONE 8 (1), e53608. doi:10.1371/journal.pone.0053608
Montoya Sánchez, N., Link, F., Chauhan, G., Halmenschlager, C., El‐Sayed, H. E. M., Sehdev, R., et al. (2022). Conversion of Waste to Sustainable Aviation Fuel via Fischer-Tropsch Synthesis: Front‐end Design Decisions. Energ. Sci. Eng., 1–27. doi:10.1002/ese3.1072
Müller, N., Worm, P., Schink, B., Stams, A. J. M., and Plugge, C. M. (2010). Syntrophic Butyrate and Propionate Oxidation Processes: From Genomes to Reaction Mechanisms. Environ. Microbiol. Rep. 2 (4), 489–499. doi:10.1111/j.1758-2229.2010.00147.x
Müller, V. (2019). New Horizons in Acetogenic Conversion of One-Carbon Substrates and Biological Hydrogen Storage. Trends Biotechnol. 37 (12), 1344–1354. doi:10.1016/j.tibtech.2019.05.008
Munasinghe, P. C., and Khanal, S. K. (2010). Biomass-Derived Syngas Fermentation into Biofuels: Opportunities and Challenges. Bioresour. Technol. 101 (13), 5013–5022. doi:10.1016/j.biortech.2009.12.098
Octavia, S., and Lan, R. (2013). “The Family Enterobacteriaceae,” in The Prokaryotes: Gammaproteobacteria. Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin Heidelberg: Springer), 225–286. doi:10.1007/978-3-642-38922-1
Polleux, L., Guerassimoff, G., Marmorat, J.-P., Sandoval-moreno, J., and Schuhler, T. (2022). An Overview of the Challenges of Solar Power Integration in Isolated Industrial Microgrids with Reliability Constraints. Renew. Sustain. Energ. Rev. 155, 111955. doi:10.1016/j.rser.2021.111955
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 41 (D1), D590–D596. doi:10.1093/nar/gks1219
Rachbauer, L., Voitl, G., Bochmann, G., and Fuchs, W. (2016). Biological Biogas Upgrading Capacity of a Hydrogenotrophic Community in a Trickle-Bed Reactor. Appl. Energ. 180, 483–490. doi:10.1016/j.apenergy.2016.07.109
Robertson, W. J., Bowman, J. P., Franzmann, P. D., and Mee, B. J. (2001). Desulfosporosinus Meridiei Sp. nov., a Spore-Forming Sulfate-Reducing Bacterium Isolated from Gasolene-Contaminated Groundwater. Int. J. Syst. Evol. Microbiol. 51 (1), 133–140. doi:10.1099/00207713-51-1-133
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 4 (10), e2584. doi:10.7717/peerj.2584
Rogosa, M. (1971). Peptococcaceae, a New Family to Include the Gram-Positive, Anaerobic Cocci of the Genera Peptococcus, Peptostreptococcus, and Ruminococcus. Int. J. Syst. Bacteriol. 21 (3), 234–237. doi:10.1099/00207713-21-3-234
Romeo, L. M., Cavana, M., Bailera, M., Leone, P., Peña, B., and Lisbona, P. (2022). Non-Stoichiometric Methanation as Strategy to Overcome the Limitations of Green Hydrogen Injection into the Natural Gas Grid. Appl. Energ. 309, 118462. doi:10.1016/j.apenergy.2021.118462
Sakai, S., Nakashimada, Y., Inokuma, K., Kita, M., Okada, H., and Nishio, N. (2005). Acetate and Ethanol Production from H2 and CO2 by Moorella Sp. Using a Repeated Batch Culture. J. Biosci. Bioeng. 99 (3), 252–258. doi:10.1263/jbb.99.252
Sakamoto, M., Kitahara, M., and Benno, Y. (2007). Parabacteroides Johnsonii Sp. nov., Isolated from Human Faeces. Int. J. Syst. Evol. Microbiol. 57 (2), 293–296. doi:10.1099/ijs.0.64588-0
Schuchmann, K., and Müller, V. (2014). Autotrophy at the Thermodynamic Limit of Life: A Model for Energy Conservation in Acetogenic Bacteria. Nat. Rev. Microbiol. 12 (12), 809–821. doi:10.1038/nrmicro3365
Schuchmann, K., and Müller, V. (2016). Energetics and Application of Heterotrophy in Acetogenic Bacteria. Appl. Environ. Microbiol. 82 (14), 4056–4069. doi:10.1128/AEM.00882-164056-4069
Slobodkin, A. (2014). “The Family Peptostreptococcaceae,” in The Prokaryotes: Firmicutes and Tenericutes. Editors E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin Heidelberg: Springer), 291–302. doi:10.1007/978-3-642-30120-9_217
Spring, S., and Rosenzweig, F. (2006). “The Genera Desulfitobacterium and Desulfosporosinus: Taxonomy,” in The Prokaryotes: A Handbook on the Biology of Bacteria. Editors M. Dworkin, S. Falkow, E. Rosenberg, and E. Stackebrandt. 3rd ed. (Berlin: Springer Nature), 771–786. doi:10.1007/0-387-30744-3_24
Steger, F., Rachbauer, L., Windhagauer, M., Montgomery, L. F. R., and Bochmann, G. (2017). Optimisation of Continuous Gas Fermentation by Immobilisation of Acetate-Producing Acetobacterium Woodii. Anaerobe 46, 96–103. doi:10.1016/j.anaerobe.2017.06.010
Steinbusch, K. J. J., Hamelers, H. V. M., Plugge, C. M., and Buisman, C. J. N. (2011). Biological Formation of Caproate and Caprylate from Acetate: Fuel and Chemical Production from Low Grade Biomass. Energy Environ. Sci. 4 (1), 216–224. doi:10.1039/c0ee00282h
Suzuki, T., Matsuo, T., Ohtaguchi, K., and Koide, K. (1993). Continuous Production of Acetic Acid from CO2 in Repeated-Batch Cultures Using Flocculated Cells of Acetobacterium Woodii. J. Chem. Eng. Jpn. 26 (5), 459–462. doi:10.1252/jcej.26.459
Thompson, K. A., Summers, R. S., and Cook, S. M. (2017). Development and Experimental Validation of the Composition and Treatability of a New Synthetic Bathroom Greywater (SynGrey). Environ. Sci. Water Res. Technol. 3 (6), 1120–1131. doi:10.1039/c7ew00304h
Ullrich, T., Lindner, J., Bär, K., Mörs, F., Graf, F., and Lemmer, A. (2018). Influence of Operating Pressure on the Biological Hydrogen Methanation in Trickle-Bed Reactors. Bioresour. Technol. 247, 7–13. doi:10.1016/j.biortech.2017.09.069
Uribe Santos, D. L., Delgado Dobladez, J. A., Águeda Maté, V. I., Álvarez Torrellas, S., and Larriba Martínez, M. (2020). Recovery and Purification of Acetic Acid from Aqueous Mixtures by Simulated Moving Bed Adsorption with Methanol and Water as Desorbents. Separat. Purif. Technol. 237, 116368. doi:10.1016/j.seppur.2019.116368
Villemur, R., Lanthier, M., Beaudet, R., and Lépine, F. (2006). The Desulfitobacterium Genus. FEMS Microbiol. Rev. 30 (5), 706–733. doi:10.1111/j.1574-6976.2006.00029.x
Wagner, F. S. (2002). “Acetic Acid,” in Kirk‐Othmer Encyclopedia of Chemical Technology (Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA), 1, 115–136. doi:10.1002/0471238961.0103052023010714.a01.pub2
Wang, G., and Wang, D. I. C. (1984). Elucidation of Growth Inhibition and Acetic Acid Production by Clostridium Thermoaceticum. Appl. Environ. Microbiol. 47 (2), 294–298. Available at: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC239662/. (Accessed October 02, 2021). doi:10.1128/aem.47.2.294-298.1984
Wang, Y.-Q., Yu, S.-J., Zhang, F., Xia, X.-Y., and Zeng, R. J. (2017). Enhancement of Acetate Productivity in a Thermophilic (55 °C) Hollow-Fiber Membrane Biofilm Reactor with Mixed Culture Syngas (H2/CO2) Fermentation. Appl. Microbiol. Biotechnol. 101 (6), 2619–2627. doi:10.1007/s00253-017-8124-9
Xu, S., Fu, B., Zhang, L., and Liu, H. (2015). Bioconversion of H2/CO2 by Acetogen Enriched Cultures for Acetate and Ethanol Production: the Impact of pH. World J. Microbiol. Biotechnol. 31 (6), 941–950. doi:10.1007/s11274-015-1848-8
Zhang, F., Ding, J., Zhang, Y., Chen, M., Ding, Z.-W., van Loosdrecht, M. C. M., et al. (2013). Fatty Acids Production from Hydrogen and Carbon Dioxide by Mixed Culture in the Membrane Biofilm Reactor. Water Res. 47 (16), 6122–6129. doi:10.1016/j.watres.2013.07.033
Keywords: acetic acid, H2/CO2 conversion, gas fermentation, trickle-bed (bio) reactors, mixed culture, microbial composition analysis
Citation: Steger F, Ergal İ, Daubek A, Loibl N, Rachbauer L, Fuchs W, Rittmann SK-MR and Bochmann G (2022) Trickle-Bed Bioreactors for Acetogenic H2/CO2 Conversion. Front. Energy Res. 10:842284. doi: 10.3389/fenrg.2022.842284
Received: 23 December 2021; Accepted: 03 March 2022;
Published: 08 April 2022.
Edited by:
Claire Dumas, INRAE Occitanie Toulouse, FranceReviewed by:
Obulisamy Parthiba Karthikeyan, South Dakota School of Mines and Technology, United StatesCopyright © 2022 Steger, Ergal, Daubek, Loibl, Rachbauer, Fuchs, Rittmann and Bochmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Franziska Steger, RnJhbnppc2thLnN0ZWdlckBib2t1LmFjLmF0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.