
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Energy Res. , 29 May 2020
Sec. Bioenergy and Biofuels
Volume 8 - 2020 | https://doi.org/10.3389/fenrg.2020.00093
This article is part of the Research Topic Advancements in Bio-Based Products from Renewable Feedstocks: Drop-In Replacement and Beyond View all 7 articles
 Austin D. Comer1
Austin D. Comer1 Joshua P. Abraham1
Joshua P. Abraham1 Alexander J. Steiner1
Alexander J. Steiner1 Travis C. Korosh1
Travis C. Korosh1 Andrew L. Markley1
Andrew L. Markley1 Brian F. Pfleger1,2*
Brian F. Pfleger1,2*Current sources of fermentation feedstocks, i.e., corn, sugar cane, or plant biomass, fall short of demand for liquid transportation fuels and commodity chemicals in the United States. Aquatic phototrophs including cyanobacteria have the potential to supplement the supply of current fermentable feedstocks. In this strategy, cells are engineered to accumulate storage molecules including glycogen, cellulose, and/or lipid oils that can be extracted from harvested biomass and fed to heterotrophic organisms engineered to produce desired chemical products. In this manuscript, we examine the production of glycogen in the model cyanobacteria, Synechococcus sp. strain PCC 7002, and subsequent conversion of cyanobacterial biomass by an engineered Escherichia coli to octanoic acid as a model product. In effort to maximize glycogen production, we explored the deletion of catabolic enzymes and overexpression of GlgC, an enzyme that catalyzes the first committed step toward glycogen synthesis. We found that deletion of glgP increased final glycogen titers when cells were grown in diurnal light. Overexpression of GlgC led to a temporal increase in glycogen content but not in an overall increase in final titer or content. The best strains were grown, harvested, and used to formulate media for growth of E. coli. The cyanobacterial media was able to support the growth of an engineered E. coli and produce octanoic acid at the same titer as common laboratory media.
Current sources of fermentation feedstocks, such as corn, sugar cane, or plant biomass, fall short of demand for liquid transportation fuels and commodity chemicals in the United States. For example, an optimistic estimate suggests that ~1.5 billion tons of biomass, e.g., lignocelluosic wastes and energy crops, is available in the United States (Langholtz et al., 2016), equivalent to the energy in ~4.4 billion barrels of oil. If all this energy could be converted to fuels, it would represent ~75% of the petroleum imported and produced domestically in the country (~5.9 billion barrels of oil). While terrestrial biomass represents a large feedstock opportunity, additional sustainable sources of fermentation feedstocks will be needed to meet the demand of a growing biomanufacturing economy. Cyanobacteria and eukaryotic algae can address this limitation and provide an additional sustainable feedstock for the bioeconomy. Cyanobacteria produce many storage compounds, i.e., glycogen, cyanophycin, polyphosphate, polyhydroxyalkanoates, during photosynthetic growth in order to provide energy and nutrients for continued survival in the dark. These nutrient-rich cells can be recovered and used to formulate media (Figure 1A) to feed heterotrophic organisms (Hoover et al., 2011; Aikawa et al., 2014; Möllers et al., 2014), including those engineered to synthesize biofuels and other chemical building blocks. The rapid growth and CO2 fixation potential of cyanobacteria provides a significant advantage when comparing the productivity from a given land area; algae and cyanobacteria yields are upward of 40 g/m2/days (Clippinger et al., 2019) approaching current Brazilian sugarcane yields (FAO, 2020). Many species of cyanobacteria can grow in brackish or salt waters and utilize wastewater as a source of nitrogen and phosphate (Korosh et al., 2018). In order to leverage cyanobacteria biomass as a potential fermentation feedstock, strains must be engineered to maximize flux to desired storage compounds.
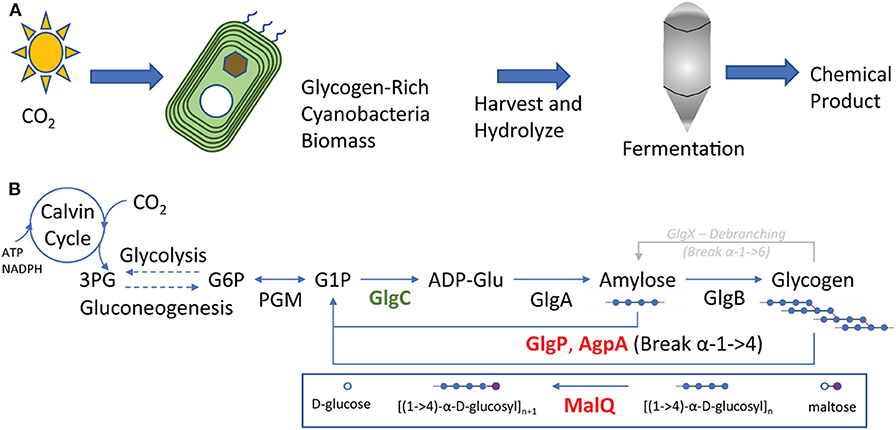
Figure 1. (A) Cultivation of aquatic phototrophs as a feedstock for bioprocessing. Photosynthesis enables the fixation of carbon dioxide into reduced metabolites including energy storage molecules. Cells rich in storage molecules can be harvested, hydrolyzed, and fed to heterotrophs for production of commodity chemicals. The process is analogous to production of terrestrial plant biomass and processing to biofuels and chemicals. (B) Glycogen is produced from the products of the Calvin cycle, producing glucose 1-phosphate though (G1P) gluconeogenesis and phosphoglucomutase (PGM). G1P is converted to glycogen through GlgC and GlgA. Glycogen structure is altered by branching and debranching enzymes, GlgB and GlgX. Finally, glycogen is degraded by linear depolymerases GlgP and AgpA as well as the branched chain pathway including MalQ. Break α-1->4 nomenclature refers to breaking the α-1->4 glycosidic bond.
In this manuscript, we describe efforts to maximize glycogen accumulation in Synechococcus sp. strain PCC 7002, previously known as Agmenellum quadruplicatum. PCC 7002 was originally isolated from coastal ocean waters (Van Baalen, 1962; Stevens and van Baalen, 1973) and has been shown capable of rapid autotrophic growth (~2–3 h doubling time) in many water sources (e.g., fresh, saline, wastewater); growing best in brackish water (Aikawa et al., 2014). PCC 7002 is naturally transformable and genetically tractable with a wide array of synthetic biology tools (Begemann et al., 2013; Markley et al., 2015; Zess et al., 2015; Gordon et al., 2016; Ruffing et al., 2016; Santos-Merino et al., 2019; Vavitsas et al., 2019; Xia et al., 2019). Further, models of metabolism, regulation, carbon fixation, and process systems have been developed for the strain (Vu et al., 2013; Clark et al., 2014; Cameron et al., 2015; Hendry et al., 2016; McClure et al., 2016; Yenkie et al., 2016). These advantageous features provide an opportunity to use PCC 7002 to generate a tailorable biomass feedstock that does not compete with current food production or potable water supplies.
Glycogen is a polymer of glucose molecules linked with alpha-1,4 bonds for linear chains and by alpha-1,6 bonds at branch points (Cifuente et al., 2019). It is easier to break down than lignocellulosic biomass and produces fewer toxins during hydrolysis. This makes glycogen accumulating organisms attractive as a feedstock for value-added fuel and chemical production by heterotrophic organisms (Aikawa et al., 2014; Möllers et al., 2014). In many cyanobacteria, glycogen is the main source of energy storage by which cells can regulate energy availability between periods of light and dark, as well as periods of high and low nutrients (Luan et al., 2019). As shown in Figure 1B, Glycogen molecules are synthesized by polymerizing activated glucose units made from the products of photosynthesis (Cifuente et al., 2019). Glucose-6-phosphate, made via gluconeogenesis, is isomerized to glucose-1-phosphate by phosphoglucomutase, and converted to ADP-glucose by GlgC (Zilliges, 2014). Finally, ADP-glucose is polymerized by GlgA into glycogen (Cifuente et al., 2019). Glycogen branching is controlled by the branching and debranching enzymes GlgB and GlgX, respectively (Suzuki et al., 2007; Zilliges, 2014; Cifuente et al., 2019). These enzymes can adjust how compact the glycogen granules are based on the amount of branching, which can impact the volume efficiency of energy storage.
In order to access the energy stored in glycogen, cyanobacteria must extract sugar units from the polymer and send them through central metabolism, producing ATP and waste metabolites (e.g., CO2, ethanol, etc.). Glycogen can be degraded linearly by GlgP and AgpA (Figure 1B). Once linear degradation to a branch point is complete, the last four glucose units must be cleaved by the debranching enzyme GlgX and are degraded by a number of enzymes including MalQ, which is responsible for rearranging glucose monomers and dimers (maltose) between glucose polymers, including breaking maltotetraose and maltotriose units into smaller components (Zilliges, 2014). These pathways allow cellular access to energy and carbon through glycolysis and central metabolism from stored glycogen.
Historically, glycogen production has been probed at a few points within this pathway as well as through environmental changes (Ducat et al., 2012; Xu et al., 2012; Guerra et al., 2013; Joseph et al., 2014; McNeely et al., 2014; Qian et al., 2018). For PCC 7002, glycogen production is induced by different forms of nutrient starvation including, most commonly, nitrogen starvation (Aikawa et al., 2014). Theglobal nitrogen control factor, ntcA, controls this response and can be triggered by other forms of nutrient or light limitation (Liu and Yang, 2014; Forchhammer and Selim, 2019). Historically, the glgC node has seen the most manipulation in PCC 7002. Groups have worked to knock out this node to reduce glycogen accumulation within the cell to increase the available energy for chemical production (van der Woude et al., 2014). A similar analysis and result was achieved for a ΔglgA strain (Xu et al., 2012). In this study, we explored genetic changes that would lead to enhanced glycogen productivity. We explored the overexpression of GlgC to enhance synthesis rates and deletion of catabolic pathways to prevent consumption of the desired storage molecules. We found that deletion of glgP increased final glycogen titers when cells were grown in diurnal light. Overexpression of GlgC led to a temporal increase in glycogen content but not in an overall increase in glycogen titer or content at the end of production. The best strain (ΔglgPΔagpA) was grown, harvested, and used to formulate media for growth of an engineered E. coli NHL17 in order to produce octanoic acid (Hernández Lozada et al., 2018). The cyanobacterial media was able to support growth and the E. coli produced octanoic acid at the same titer as in common laboratory media.
Biomass titers were estimated in one of two ways (1) optical density at 730 nm (OD730) and (2) dry cell weight. OD730 measurements include both glycogen and other biomass components. Dry cell weight measurements were performed by filling a pre-weighed conical tube with a recorded volume of cell culture. The sample was centrifuged at 5,000 × g for 20 min. The supernatant was aspirated, and the resulting pellet was washed with 1 volume of water before being flash frozen in liquid nitrogen or frozen overnight in a −80°C freezer. Once frozen, the samples were lyophilized, and final weights were measured.
Glycogen content was measured by quantifying glucose released by hydrolysis of lyophilized biomass samples. Desiccated biomass (5–20 mg) was added to a 5 mL glass reactor and resuspended in 1 mL of 4% (v/v) sulfuric acid. The reactor was sealed with a cap containing a second septa to prevent vapor loss during hydrolysis. The reactors were placed in a silicon oil bath at 121°C for 1 h and cooled in an ice water bath for 20 min. Samples were transferred into 1.7 mL microfuge tubes, centrifuged for 10 min at 20,000 × g, filtered through a 0.22 μm filter, and stored at −20°C until analysis. The protocol was based on the biomass hydrolysis process reported by the National Renewable Energy Lab (NREL) (Ruiz and Date, 1996). Glucose concentration in the hydrolyzed sample or E. coli media was measured by HPLC. A 5 mM sulfuric acid mobile phase was used to separate metabolites in the hydrolysate over a Rezek ROA-Organic Acid column. Samples quantified using a refractive index detector relative to a prepared standard curve.
Cyanobacterial cultures were grown in 60 mL of Media A in a 250 mL baffled flask unless otherwise specified (Ludwig and Bryant, 2011). Typically, cultures were grown under continuous light provided by LED strips mounted at the top of a shaking incubator (illumination from above), held at 37°C, and had a constant carbon dioxide concentration of 1%. Cultures were supplemented with water to adjust for volume loss due to evaporation. Continuous light experiments were run at 100% of maximum incubator light intensity. Diurnal cultures were provided light in varying intensity with a cycle programmed into an 8-cycle controller. The controller was programmed by setting the length of time and light intensity for each of the 8 segments of the controller scheme (Figure 3A). Light intensity was measured as a percentage of maximum intensity, which was equal to ~180 μmol photons m−2 s−1.
Genetic alterations were made using the natural homologous recombination ability and competency of PCC 7002 (Stevens and Porter, 1980), antibiotic resistance, and/or a counterselection protocol dependent on the acsA gene—encoding acetyl-coenzyme A synthetase (Begemann et al., 2013). Donor DNA was provided in the form of plasmid (E. coli shuttle vectors) containing ~500 base pair homology regions to the target integration site. The homology regions flanked an expression cassette for the native acsA gene, foreign aadA (specR/strepR), foreign APH(3')-Ia (kanR) and/or foreign aacC1 (gentR) gene. DNA was transformed by adding 1–1.5 μg of purified plasmid to 1 mL of an overnight culture of cells grown to an OD730 of 1. The cultures were then placed at 37°C under illumination for 16 h. The cells were plated on 50 μM acrylic acid (acsA locus) 100 μg/mL streptomycin, 100 μg/ml kanamycin, or 100 μg/ml gentamycin (glpK locus) to select for recombinants. The GlgC+ and ΔglgC strains were derived from wild-type PCC 7002 and the depolymerase strains (ADC001, ADC002, ADC003, and ADC004) were made from BPSyn_014, a strain with a barcode deletion in the acsA locus (Begemann et al., 2013). Annotated plasmid maps used in strain construction are provided as Supplementary Material in GenBank format and a complete list of strains is provided in Table 1.
The PCC 7002 ΔglpPΔagpA strain was inoculated from overnight cultures to an OD730 of 0.05 in 250 mL shake flasks to a final volume of 60 mL of Media A. Cultures were grown at 27°C and 150 RPM with 1% CO2 and 200 μmol photons m−2 s−1 provided by light emitting diode strips. Prior to measuring OD730, we adjusted for evaporative losses by replacing lost mass with sterile water.
After 8 days, we harvested cultures by centrifugation at 10,000 x g for 10 min. Samples were resuspended in water and vacuum aspirated twice to remove salts, flash frozen, and lyophilized. Once dehydrated, ~100 mg of desiccated biomass was suspended in 2 mL of 4% (v/v) H2SO4 inside a glass reactor with a second septa to prevent evaporation. Acid hydrolysis was performed for 2 h at 121°C in a silicone oil bath. Samples were then titrated to a pH of 6.75 to match Luria-Bertani (LB) broth with NaOH and passed through a 0.22 μm nylon filter to remove particulate matter and sterilize. After measuring glucose content of the hydrolysate, we diluted the media to 10 g/L glucose with sterile water. An appropriate amount of glucose was added to M9 minimal media to match. Additionally, we prepared a 50%/50% mixture of M9 minimal media (without glucose) and hydrolysate. M9 Hydrolysate media was formulated by a 1:1 mixture of M9 minimal media and hydrolysate, leading to total salt concentrations of 0.1 mM MgSO4, 0.05 mM CaCl2, 23.9 mM Na2HPO4, 11 mM KH2PO4, 4.28 mM NaCl, and 9.35 mM NH4Cl.
E. coli NHL17 (Hernández Lozada et al., 2018) was inoculated from an overnight culture to an OD600 of 0.05 into 3 mL of respective liquid medium in test tubes and grown at 37°C. Cultures were grown until an OD600 of 0.3 then induced with 50 μM IPTG and moved to 30°C with vigorous shaking at 250 RPM.
After 24 h post-induction, 500 μL of culture was combined with 2.5 μL of 100 mg/mL heptanoic acid, 100 mg/L non-anoic acid, and 100 mg/L pentadecanoic acid in a methanol solution. Samples were prepared with 50 μL of 12 M HCl and 500 μL of a 116:34 solution of methanol and pyridine then vortexed for 5 min. Fatty acids were derivatized into fatty acid methyl esters using two volumes of 50 μL methylchloroformate with 30 s of vortexing after each addition. Reactions were quenched with 800 μL of chloroform and 800 μL of 100 g/L NaHCO3. The chloroform layer was collected and analyzed using a GC-FID (Shimadzu GC-2010) with an AOC-20i auto-injector and a 30 m, 0.25 mm DB FATWAX-UI column. The GC temperature protocol began at 120°C for 2 min, ramped to 140°C (at 5°C/min) and held for 3 min, then ramped to 250°C (at 20°C/min) and held for 8 min.
When engineering production of small molecules in cyanobacteria, glycogen synthesis is often blocked by disruption of glgC to redirect carbon flux to desired products, for example lactic acid (van der Woude et al., 2014). The ΔglgC deletion prevents the activation glucose-1-P to ADP-glucose, a committed step toward glycogen synthesis (Zilliges, 2014). We hypothesized that, conversely, GlgC overexpression would increase flux to glycogen by pulling flux from the sugar-phosphate pool. We overexpressed glgC by adding a second copy to the genome in the glpK locus under the control of the strong IPTG-inducible promoter, PcLac143. To test the impact of GlgC overexpression (GlgC+), cultures of the modified strain, wild-type, and a ΔglgC mutant were grown under continuous (Figure 2) and diurnal light (Figure 3) in the presence or absence of 0.1mM IPTG. Total biomass, OD730, and glycogen titer were quantified as described in the methods section.
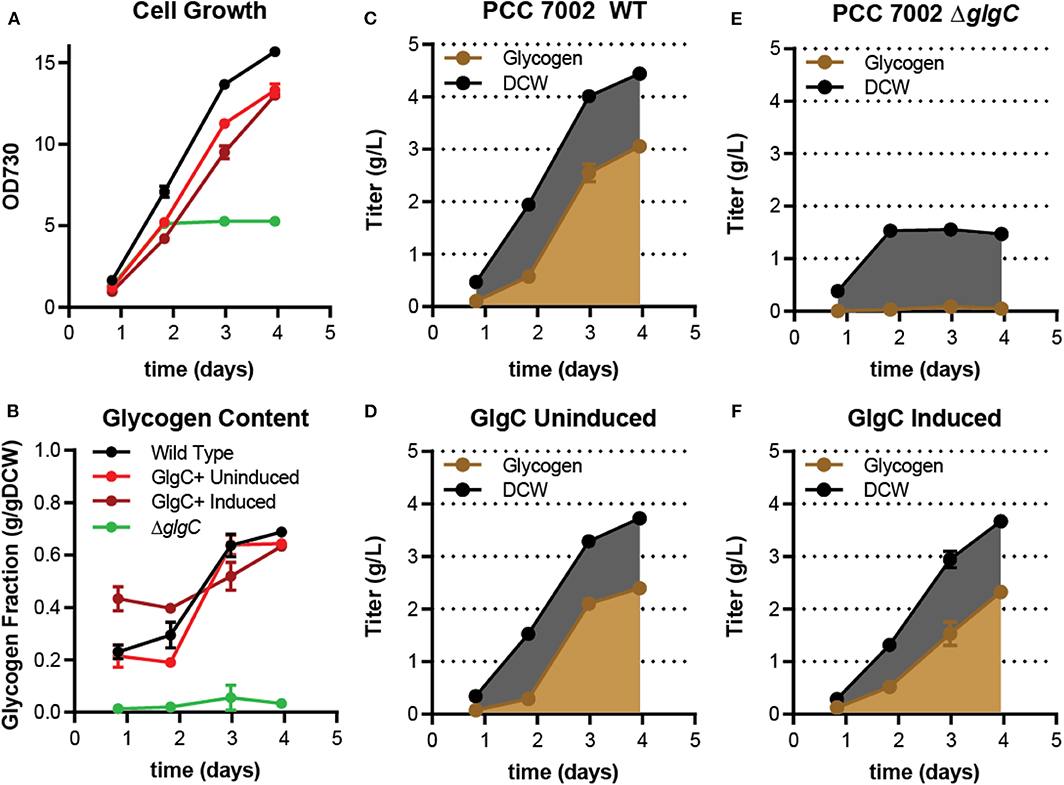
Figure 2. Biomass and glycogen production by PCC 7002 strains under continuous light conditions in media A. (A) Shows the biomass production measured by optical density at 730 nm. (B) Reports the fraction of the total dry cell weight that was glycogen over time. (C–F) Show the titer of glycogen (brown) and total DCW (black) over time for each of the four listed cultures. The difference between the DCW and glycogen content represents the productive biomass. Each culture was performed in triplicate. Error bars represent standard deviation.
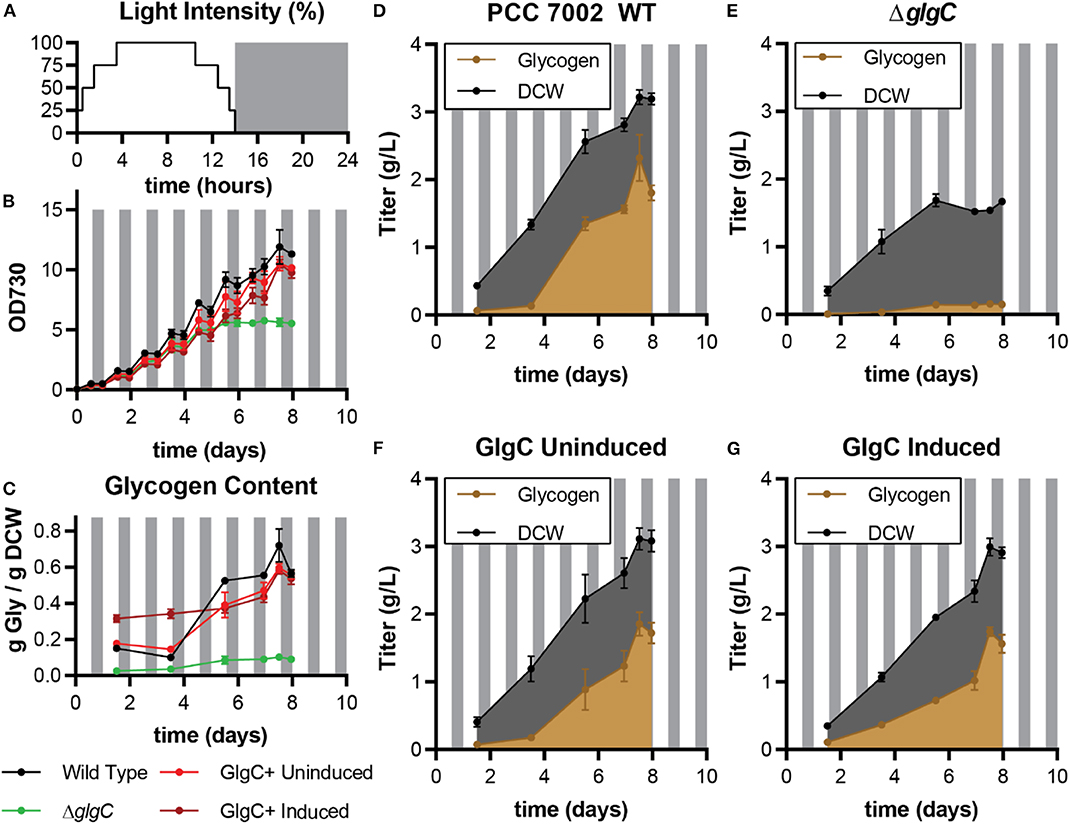
Figure 3. Biomass and glycogen production by PCC 7002 strains under diurnal light conditions in media A. (A) Diurnal light intensity was programmed as an 8-step variation with each step varying the fraction of maximum light output. (B) Shows the biomass production measured optical density at 730 nm. (C) Reports the fraction of the total dry cell weight that was glycogen over time. (D–G) Show the titer of glycogen (brown) and total DCW (black) over time for each of the four listed cultures. Each culture was performed in triplicate. Error bars represent standard deviation.
In continuous light, the GlgC+ strain, regardless of induction state, experienced a slight growth defect relative to the wild type (Figure 2A). Induction of the GlgC+ slightly enhanced the growth defect. After 4 days, the GlgC+ strain, both uninduced or when induced, achieved about 80% of the total dry cell weight and OD730 achieved by the wild type (OD730 ~15; 4.5 g/L DCW; Figures 2B,C,E,F). The ΔglgC mutant achieved lower final OD730 (~5 under both conditions) and DCW (1.5 g/L) than any of the other strains (Figure 2D). This is primarily due to the lack of glycogen in this strain. In each of the strains, productive biomass, i.e., the non-glycogen DCW, reached a maximum of 1.5 g/L by day 2 or 3 (the induced GlgC+ strain took an extra day) and remained unchanged thereafter. After this point the increase in cell mass can be attributed solely to glycogen accumulation.
These dynamics are likely due to a shift from replete media to nitrogen-limited conditions. The final biomass titer and the duration of productive biomass generation was proportional to the amount of nitrate fed to cultures (Figure S1). Correspondingly the amount of nitrogen in the media correlated to the point at which cells reached a maximum glycogen content (~60–80% of DCW); cultures with minimal nitrate stopped growing early and filled with glycogen earlier. Cultures replete with nitrate took longer to reach starvation, ultimately generating higher biomass and glycogen titers. The profile of the GlgC+ strain, which showed rapid glycogen accumulation and higher glycogen content at early times, is similar to strains that were starved for nitrogen with the exception that biomass and glycogen titers ultimately were no better than those of the wild type.
Any large-scale cultivation of cyanobacteria for sugar production will depend on natural light, which varies in intensity throughout each day. As such, we studied the impact of each genetic variation under diurnal light conditions. Variable glycogen content during a diurnal cycle is caused by the production and consumption cycles of glycogen that occur to regulate cell growth across light/dark cycles (Pattanayak et al., 2014). Therefore, we simulated a natural day-night cycle in our incubator shaker using a series of stepped light intensities to yield a 10-hour dark cycle with a 14-h variable light cycle that more closely represents outdoor light delivery than simplistic on/off systems (Figure 3A). The effect of diurnal growth can be seen in the “saw-tooth” pattern (OD730 dropped slightly during the dark phase—Figure 3B) as well as the final three glycogen content measurements (Figure 3C). In these diurnal conditions, the growth rates of each strain were indistinguishable; both glycogen producing strains yielded ~3 g/L DCW by day 8, while the ΔglgC strain topped out at 1.5 g/L DCW after ~5 days (Figures 3D–G). The productive biomass ceased to increase after day ~3 at a value of ~1.5 g/L. The induced GlgC+ again demonstrated a higher glycogen content early in the culture but was matched by the wild type after additional days in culture. As before, the delayed glycogen accumulation tracked with the amount of nitrogen in the media, here titers and content were delayed relative to continuous light conditions. From these experiments, we concluded that GlgC overexpression alone was not sufficient to improve the volumetric titer of glycogen relative to wild type but could alter the dynamics of specific glycogen content and volumetric productivity. These enhancements could be exploited in cultures harvested at low cell densities.
Glycogen is used as a form of energy storage for use at night and cyanobacteria access the energy by depolymerizing glycogen and processing the sugar units through central metabolism. To maximize glycogen accumulation, we hypothesized that cells lacking key enzymes in the glycogen breakdown pathways would result in faster production and/or higher titers of glycogen. We considered three gene knockouts at two key nodes in glycogen metabolism: malQ, glgP, and agpA (Figure 1B). We created knockouts at these key nodes and measured the resulting changes in glycogen content and titer. The ΔmalQ strain (ADC001) showed a consistent growth defect compared to an isogenic wild type strain carrying a resistance marker (ALM220) as shown by the decreased biomass titer (Figure S2). Both strains, when grown in Media A, had higher glycogen content than the same cells grown in the presence of extra nitrate (+N). The glycogen titer and content were not improved by the ΔmalQ mutation under either media condition. Given the growth defect, further work with the ΔmalQ strain was not pursued. The ΔagpA mutation did not impact glycogen production, but deletion of glgP had a significant impact on final glycogen titer (Figure 4F). Where the wild type produced 3.0 ± 0.3 g/L of glycogen, the ΔglgP and ΔglgPΔagpA strains produced 3.7 ± 0.1 g/L and 3.6 ± 0.1 g/L, respectively (P < 0.05). In each case, the amount of productive biomass stopped increasing by day 5 with the remaining increase in DCW attributed to glycogen accumulation (Figures 4A–E). Knocking out glycogen depolymerases under diurnal light conditions had a stronger effect than in continuous light conditions (data not shown). From our data, we concluded that the ΔglgP mutation increased glycogen accumulation compared to diurnally grown wild type cells.
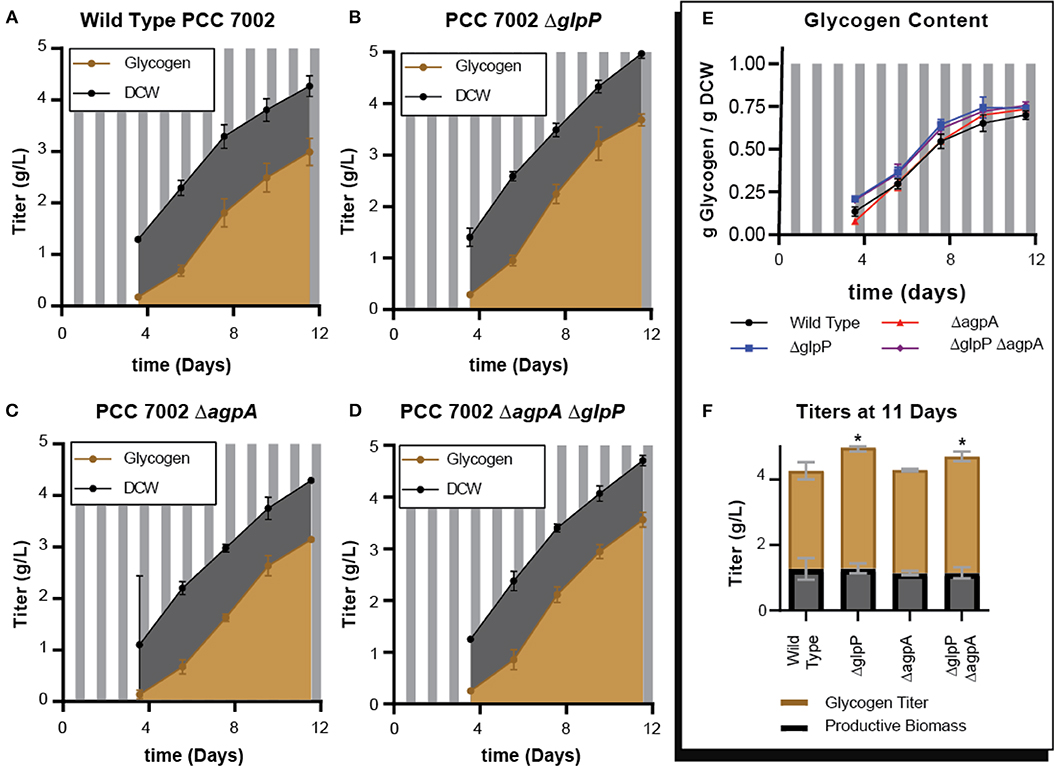
Figure 4. Biomass and glycogen production by PCC 7002 strains grown under diurnal light conditions (see Figure 3A) in media A. This experiment investigates the value of knocking out linear glycogen phosphorylases in the glycogen catabolism pathway under more restrictive diurnal conditions. (A–D) Show the accumulation of glycogen and productive biomass over time. Gray areas represent incubation without light. (E) Shows the fractional glycogen content. In each strain the content approached 75% by the end of the culture. (F) Shows the breakdown of total DCW as glycogen (brown) and productive biomass (black). The ΔglgP and ΔglgPΔagpA strains produced more glycogen than the corresponding wild type (t-test with P < 0.05). Error bars represent standard deviation.
To demonstrate the utility of cyanobacteria biomass as a fermentation feedstock, we cultured PCC 7002 ΔglgPΔagpA and harvested the resulting biomass. The cell pellet was washed, lyophilized, and hydrolyzed in acid to release fermentable molecules. The hydrolysate was neutralized and supplemented with M9 salts to formulate media for E. coli. This approach is appropriate in a lab setting for high accuracy measurement, but would be infeasible within an industrial setting; an alternate methodology would be need to be developed. We decided to compare the growth and product titer in hydrolysate and standard M9 glucose lab media containing the same level of glucose (~10 g/L). Although this normalizes for glucose content, there is additional protein content and other molecules present in cyanobacterial hydrolysate that could promote bacterial growth. The test was performed by cultivating NHL17, an E. coli engineered to express a highly active C8-specific thioesterase in a β-oxidation null background. The final OD600 was recorded as a measure of cell growth and the titer of glucose and octanoic acid was quantified to determine the product titer and yield (Figure 5). Cells grown in pure hydrolysate produced small quantities of octanoic acid whereas minimal media and the hydrolysate formulated with M9 salts produced equal amounts (~300 mg/L) at similar yields (0.045–0.06 g FFA/ g glucose consumed). The lower performance in pure hydrolysate, where cells grew to about half of the final biomass (1.1 vs. 2.1 OD600 at 24 h), is likely due to the combination of octanoic acid stress and lack of pH buffering. The equivalent final titers indicate that cyanobacteria biomass could be used to formulate fermentation media for industrial applications. Further fine tuning is likely needed as all cultures failed to consume the full 10 g/L of glucose in 24 h.
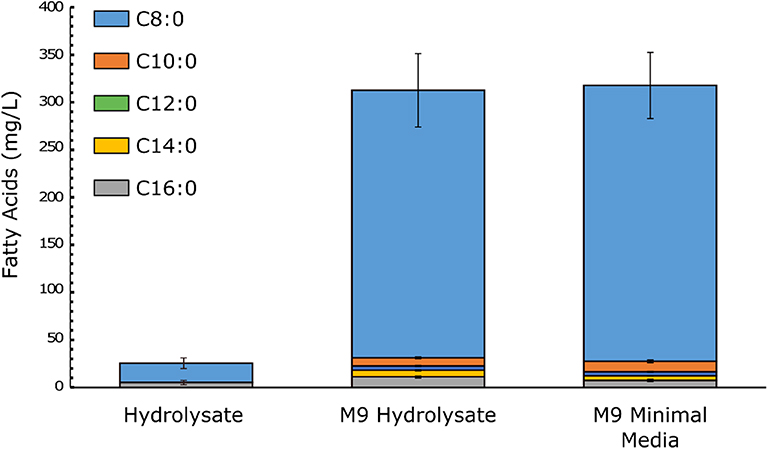
Figure 5. Titers of octanoic acid and other free fatty acids produced by E. coli NHL17. Cells were grown in media containing ~10 g/L glucose. M9 Hydrolysate media was formulated by a 1:1 mixture of M9 minimal media and hydrolysate, leading to total salt concentrations of 0.1 mM MgSO4, 0.05 mM CaCl2, 23.9 mM Na2HPO4, 11 mM KH2PO4, 4.28 mM NaCl, and 9.35 mM NH4Cl. The optical density at 24 h was 1.1, 2.4, and 1.5, respectively. Error bars represent standard deviation.
Based on the data above, we conclude that nitrogen starvation has a stronger impact on the timing and production rate of glycogen than genetic manipulations in the pathway. While GlgC overexpression led to a short temporal increase in glycogen content, wild type cells could achieve the same glycogen titer later in the cell culture. Glycogen degradation was not a substantial sink because deletion of depolymerase increased final titers by ~20% under diurnal conditions. It was interesting to note that the increase in biomass titer (DCW) could be split into two phases. In the first, productive cell mass accumulated and glycogen content was low (<25%). In the second, presumably after nitrogen was depleted from the media, any increase in total DCW could be attributed to increased glycogen titer. This experiment shows that strong nitrogen starvation can be used as a control for glycogen accumulation or if cells could be harvested efficiently at low densities, GlgC overexpression could reduce the culture times required to maximize glycogen content. The cyanobacterial media was able to support the growth of an engineered E. coli and the production of octanoic acid at the same titer as common laboratory media.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
AC, AM, and BP designed the study. AC, AM, AS, JA, and TK constructed strains and performed experiments. AC, AM, and BP wrote the manuscript. All authors contributed to data analysis and manuscript editing.
This material is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program under Award Number DE-SC0019404 and by the US National Science Foundation (EFRI-1240268, GEO-1215871). TK was recipient of a National Institutes of Health (NIH) Biotechnology Training Fellowship (NIGMS-5 T32 GM08349) and a fellowship from the University of Wisconsin— Madison (UW—Madison) College of Engineering's Graduate Engineering Research Scholars (GERS) program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenrg.2020.00093/full#supplementary-material
Aikawa, S., Nishida, A., Ho, S.-H., Chang, J.-S., Hasunuma, T., and Kondo, A. (2014). Glycogen production for biofuels by the euryhaline cyanobacteria Synechococcus sp. strain PCC 7002 from an oceanic environment. Biotechnol. Biofuels 7:88. doi: 10.1186/1754-6834-7-88
Begemann, M. B., Zess, E. K., Walters, E. M., Schmitt, E. F., Markley, A. L., and Pfleger, B. F. (2013). An organic acid based counter selection system for cyanobacteria. PLoS ONE 8:e76594. doi: 10.1371/journal.pone.0076594
Cameron, J. C., Gordon, G. C., and Pfleger, B. F. (2015). Genetic and genomic analysis of RNases in model cyanobacteria. Photosynth. Res. 126, 171–183. doi: 10.1007/s11120-015-0076-2
Cifuente, J. O., Comino, N., Trastoy, B., D'Angelo, C., and Guerin, M. E. (2019). Structural basis of glycogen metabolism in bacteria. Biochem. J. 476, 2059–2092. doi: 10.1042/BCJ20170558
Clark, R. R. L., Cameron, J. J. C., Root, T. T. W. T., and Pfleger, B. B. F. (2014). Insights into the industrial growth of cyanobacteria from a model of the carbon-concentrating mechanism. AIChE J. 60, 1269–1277. doi: 10.1002/aic.14310
Clippinger, J., Davis, R., Clippinger, J., and Davis, R. (2019). Techno-Economic Analysis for the Production of Algal Biomass via Closed Photobioreactors : Future Cost Potential Evaluated Across a Range of Cultivation System Designs Techno-Economic Analysis for the Production of Algal Biomass via Closed Photobioreactor (Golden, CO: National Renewable Energy Lab). doi: 10.2172/1566806
Ducat, D. C., Avelar-Rivas, J. A., Way, J. C., Silver, P. A., and Silvera, P., a. (2012). Rerouting carbon flux to enhance photosynthetic productivity. Appl. Environ. Microbiol. 78, 2660–2668. doi: 10.1128/AEM.07901-11
FAO (2020). Food and Agriculture Organization of the United Nations [WWW Document]. Avaliable online at: http://www.fao.org/faostat/en/#data/QC
Forchhammer, K., and Selim, K. A. (2019). Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 44, 33–53.doi: 10.1093/femsre/fuz025
Gordon, G. C., Korosh, T. C., Cameron, J. C., Markley, A. L., Begemann, M. B., and Pfleger, B. F. (2016). CRISPR interference as a titratable, trans-acting regulatory tool for metabolic engineering in the cyanobacterium Synechococcus sp. strain PCC 7002. Metab. Eng. 38, 170–179. doi: 10.1016/j.ymben.2016.07.007
Guerra, L. T., Xu, Y., Bennette, N., McNeely, K., Bryant, D. A., and Dismukes, G. C. (2013). Natural osmolytes are much less effective substrates than glycogen for catabolic energy production in the marine cyanobacterium Synechococcus sp. strain PCC 7002. J. Biotechnol. 166, 65–75. doi: 10.1016/j.jbiotec.2013.04.005
Hendry, J. I., Prasannan, C. B., Joshi, A., Dasgupta, S., and Wangikar, P. P. (2016). Metabolic model of Synechococcus sp. PCC 7002: prediction of flux distribution and network modification for enhanced biofuel production. Bioresour. Technol. 213, 190–197. doi: 10.1016/j.biortech.2016.02.128
Hernández Lozada, N. J., Lai, R.-Y. Y., Simmons, T. R., Thomas, K. A., Chowdhury, R., Maranas, C. D., et al. (2018). Highly active C8-Acyl-ACP thioesterase variant isolated by a synthetic selection strategy. ACS Synth. Biol. 7, 2205–2215. doi: 10.1021/acssynbio.8b00215
Hoover, S. W., Marner, W. D., Brownson, A. K., Lennen, R. M., Wittkopp, T. M., Yoshitani, J., et al. (2011). Bacterial production of free fatty acids from freshwater macroalgal cellulose. Appl. Microbiol. Biotechnol. 91, 435–446. doi: 10.1007/s00253-011-3344-x
Joseph, A., Aikawa, S., Sasaki, K., Matsuda, F., Hasunuma, T., and Kondo, A. (2014). Increased biomass production and glycogen accumulation in apcE gene deleted Synechocystis sp. pcc 6803. AMB Express 4:17. doi: 10.1186/s13568-014-0017-z
Korosh, T. C., Dutcher, A., Pfleger, B. F., and McMahon, K. D. (2018). Inhibition of Cyanobacterial growth on a municipal wastewater sidestream is impacted by temperature. mSphere 3:00538-17. doi: 10.1128/mSphere.00538-17
Langholtz, M. H., Stokes, B. J., and Eaton, L. M. (2016). 2016 Billion-Ton Report: Advancing Domestic Resources For A Thriving Bioeconomy. doi: 10.2172/1271651
Liu, D., and Yang, C. (2014). The Nitrogen-regulated response regulator NrrA controls cyanophycin synthesis and glycogen catabolism in the cyanobacterium synechocystis sp. pcc 6803. J. Biol. Chem. 289, 2055–2071. doi: 10.1074/jbc.M113.515270
Luan, G., Zhang, S., Wang, M., and Lu, X. (2019). Progress and perspective on cyanobacterial glycogen metabolism engineering. Biotechnol. Adv. 37, 771–786. doi: 10.1016/j.biotechadv.2019.04.005
Ludwig, M., and Bryant, D. A. (2011). Transcription profiling of the model cyanobacterium Synechococcus sp. strain pcc 7002 by next-gen (SOLiDTM) sequencing of cDNA. Front. Microbiol. 2:41. doi: 10.3389/fmicb.2011.00041
Markley, A. L., Begemann, M. B., Clarke, R. E., Gordon, G. C., and Pfleger, B. F. (2015). Synthetic biology toolbox for controlling gene expression in the cyanobacterium Synechococcus sp. strain pcc 7002. ACS Synth. Biol. 4, 595–603. doi: 10.1021/sb500260k
McClure, R. S., Overall, C. C., McDermott, J. E., Hill, E. A., Markillie, L. M., McCue, L. A., et al. (2016). Network analysis of transcriptomics expands regulatory landscapes in Synechococcus sp. pcc7002. Nucleic Acids Res. 44, 8810–8825. doi: 10.1093/nar/gkw737
McNeely, K., Kumaraswamy, G. K., Guerra, T., Bennette, N., Ananyev, G., and Dismukes, G. C. (2014). Metabolic switching of central carbon metabolism in response to nitrate: application to autofermentative hydrogen production in cyanobacteria. J. Biotechnol. 182–183, 83–91. doi: 10.1016/j.jbiotec.2014.04.004
Möllers, K., Cannella, D., Jørgensen, H., and Frigaard, N. U. (2014). Cyanobacterial biomass as carbohydrate and nutrient feedstock for bioethanol production by yeast fermentation. Biotechnol. Biofuels 7:64. doi: 10.1186/1754-6834-7-64
Pattanayak, G. K., Phong, C., and Rust, M. J. (2014). Rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Curr. Biol. 24, 1934–1938. doi: 10.1016/j.cub.2014.07.022
Qian, X., Zhang, Y., Lun, D. S., and Dismukes, G. C. (2018). Rerouting of metabolism into desired cellular products by nutrient stress: fluxes reveal the selected pathways in cyanobacterial photosynthesis. ACS Synth. Biol. 7, 1465–1476. doi: 10.1021/acssynbio.8b00116
Ruffing, A. M., Jensen, T. J., and Strickland, L. M. (2016). Genetic tools for advancement of Synechococcus sp. pcc 7002 as a cyanobacterial chassis. Microb. Cell Fact. 15, 1–14. doi: 10.1186/s12934-016-0584-6
Ruiz, R., and Date, T. E. (1996). “Determination of carbohydrates in biomass by high performance liquid chromatography,” in Laboratory Analytical Procedure No. 002 (Golden, CO: National Renewable Research Laboratory).
Santos-Merino, M., Singh, A. K., and Ducat, D. C. (2019). New applications of synthetic biology tools for cyanobacterial metabolic engineering. Front. Bioeng. Biotechnol. 7:33. doi: 10.3389/fbioe.2019.00033
Stevens, S. E., and Porter, R. D. (1980). Transformation in Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. U.S.A. 77, 6052–6056. doi: 10.1073/pnas.77.10.6052
Stevens, S. E., and van Baalen, C. (1973). Characteristics of nitrate reduction in a mutant of the blue-green alga agmenellum quadruplicatum. Plant Physiol. 51, 350–356. doi: 10.1104/pp.51.2.350
Suzuki, E., Umeda, K., Nihei, S., Moriya, K., Ohkawa, H., Fujiwara, S., et al. (2007). Role of the GlgX protein in glycogen metabolism of the cyanobacterium, Synechococcus elongatus PCC 7942. Biochim. Biophys. Acta Gen. Subj. 1770, 763–773. doi: 10.1016/j.bbagen.2007.01.006
Van Baalen, C. (1962). Studies on marine blue-green algae. Bot. Mar. 4. doi: 10.1515/botm.1962.4.1-2.129
van der Woude, A. D., Angermayr, S. A., Puthan Veetil, V., Osnato, A., and Hellingwerf, K. J. (2014). Carbon sink removal: Increased photosynthetic production of lactic acid by Synechocystis sp. pcc 803 in a glycogen storage mutant. J. Biotechnol. 184, 100–102. doi: 10.1016/j.jbiotec.2014.04.029
Vavitsas, K., Crozet, P., Vinde, M. H., Davies, F., Lemaire, S. D., and Vickers, C. E. (2019). The synthetic biology toolkit for photosynthetic microorganisms. Plant Physiol. 181, 14–27. doi: 10.1104/pp.19.00345
Vu, T. T., Hill, E. a., Kucek, L. a., Konopka, A. E., et al. (2013). Computational evaluation of Synechococcus sp. pcc 7002 metabolism for chemical production. Biotechnol. J. 8, 619–630. doi: 10.1002/biot.201200315
Xia, P., Ling, H., Foo, J. L., and Chang, M. W. (2019). Synthetic biology toolkits for metabolic engineering of cyanobacteria. Biotechnol. J. 14:1800496. doi: 10.1002/biot.201800496
Xu, Y., Tiago Guerra, L., Li, Z., Ludwig, M., Charles Dismukes, G., and Bryant, D. A. (2012). Altered carbohydrate metabolism in glycogen synthase mutants of Synechococcus sp. strain pcc 7002: Cell factories for soluble sugars. Metab. Eng. 16, 56–67. doi: 10.1016/j.ymben.2012.12.002
Yenkie, K. M., Wu, W. Z., Clark, R. L., Pfleger, B. F., Root, T. W., and Maravelias, C. T. (2016). A roadmap for the synthesis of separation networks for the recovery of bio-based chemicals: Matching biological and process feasibility. Biotechnol. Adv. 34, 1362–1383. doi: 10.1016/j.biotechadv.2016.10.003
Keywords: cyanobacteria, glycogen, metabolic engineenng, biofuels, feedstock
Citation: Comer AD, Abraham JP, Steiner AJ, Korosh TC, Markley AL and Pfleger BF (2020) Enhancing Photosynthetic Production of Glycogen-Rich Biomass for Use as a Fermentation Feedstock. Front. Energy Res. 8:93. doi: 10.3389/fenrg.2020.00093
Received: 01 March 2020; Accepted: 30 April 2020;
Published: 29 May 2020.
Edited by:
Shihui Yang, Hubei University, ChinaReviewed by:
Shuyi Zhang, Massachusetts Institute of Technology, United StatesCopyright © 2020 Comer, Abraham, Steiner, Korosh, Markley and Pfleger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian F. Pfleger, cGZsZWdlckBlbmdyLndpc2MuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.