- 1South West Sydney Limb Preservation and Wound Research, Ingham Institute for Applied Medical Research, Sydney, NSW, Australia
- 2South West Clinical School, Faculty of Medicine, University of New South Wales, Sydney, NSW, Australia
- 3Department of Endocrinology, Royal Prince Alfred Hospital, Sydney, NSW, Australia
- 4Garvan Institute of Research, Sydney, NSW, Australia
- 5Animal Health Laboratory, Department of Natural Resources and Environment Tasmania, Tasmania, TAS, Australia
- 6Infectious Disease and Microbiology, Ingham Institute for Applied Medical Research, Sydney, NSW, Australia
- 7School of Medicine Antibiotic Resistance and Mobile Elements Groups, Ingham Institute for Applied Medical Research, Sydney, NSW, Australia
- 8Liverpool Diabetes Collaboration, Ingham Institute of Applied Medical Research, Sydney, NSW, Australia
Aims/hypothesis: The gut microbiota play crucial roles in the digestion and degradation of nutrients, synthesis of biological agents, development of the immune system, and maintenance of gastrointestinal integrity. Gut dysbiosis is thought to be associated with type 2 diabetes mellitus (T2DM), one of the world’s fastest growing diseases. The aim of this systematic review is to identify differences in the composition and diversity of the gut microbiota in individuals with T2DM.
Methods: A systematic search was conducted to identify studies reporting on the difference in gut microbiota composition between individuals with T2DM and healthy controls. Relevant studies were evaluated, and their characteristics and results were extracted using a standardized data extraction form. The studies were assessed for risk of bias and their findings were reported narratively.
Results: 58 observational studies published between 2010 and 2024 were included. Beta diversity was commonly reported to be different between individuals with T2DM and healthy individuals. Genera Lactobacillus, Escherichia-Shigella, Enterococcus, Subdoligranulum and Fusobacteria were found to be positively associated; while Akkermansia, Bifidobacterium, Bacteroides, Roseburia, Faecalibacteirum and Prevotella were found to be negatively associated with T2DM.
Conclusions: This systematic review demonstrates a strong association between T2DM and gut dysbiosis, as evidenced by differential microbial abundances and altered diversity indices. Among these taxa, Escherichia-Shigella is consistently associated with T2DM, whereas Faecalibacterium prausnitzii appears to offer a protective effect against T2DM. However, the heterogeneity and observational nature of these studies preclude the establishment of causative relationships. Future research should incorporate age, diet and medication-matched controls, and include functional analysis of these gut microbes.
Systematic review registration: https://www.crd.york.ac.uk/prospero/, identifier CRD42023459937.
1 Introduction
The human body hosts a vast population of microorganisms, including archaebacteria, viruses, fungi and eubacteria (also referred to as bacteria), collectively referred to as microbiota. The period of initial gut colonization in humans remains a contentious topic, with some studies suggesting such colonization occurs in utero, while others refute this suggestion. Regardless, it is widely accepted that in humans, the infant gut microbiota is rapidly populated near the time of birth, typically achieving stability between the ages of 2 and 5 (1).
Due to factors such as peristalsis, pH, oxygen and biological products, the microbiota varies throughout different parts of the gastrointestinal tract. The small intestine contains fewer microorganisms due to a faster transit time, acidic environment, and the presence of bile and pancreatic secretions. In contrast, the large intestine hosts billions of microorganisms, mainly dominated by anaerobic bacteria, including Firmicutes, Bacteroides, Actinobacteria, Proteobacteria and Verrucomicrobia (2). This is the primary site where the microbiota interact with the human host (3).
Gut microbiota are involved in core human bodily functions including digestion and nutrient degradation, synthesis of biological agents, immune system development and maintenance of gut integrity (4). Significant factors that influence the microbiotia gut composition include age, gender, geographical location and diet. Additionally, prebiotics and probiotics have been used to change the composition of gut microbiota and induce beneficial effects. It has also been suggested that early microbial transfer during the formation and development of the gut microbiota may play a role in the inheritability of human conditions such as neurological diseases and obesity (5).
The gut bacterial microbiome has been associated with the pathophysiology of multiple chronic diseases, one of which is Type 2 diabetes mellitus (T2DM) (6–8). Type 2 diabetes mellitus (T2DM) is characterized by chronic hyperglycemia due to decreased insulin secretion by pancreatic beta cells and increased insulin resistance. Rapid urbanization, nutrition transition and sedentary lifestyles have led to a drastic rise in cases (9). In 2018 there were over 500 million cases of T2DM globally (172). In Australia, the number of patients with T2DM increased to 1 million accounting for 2.3% ($2.7 billion AUD) of total disease expenditure in 2015-2016.
Increasing evidence shows that alterations in gut bacterial microbiota plays a crucial role in the development of T2DM. Gut bacterial dysbiosis in individuals with T2DM is thought to cause systemic inflammation and altered metabolism, leading to increased peripheral insulin resistance (4). Over time, this can lead to the development of complications such as diabetes related foot complications. Hence, it is crucial to identify bacteria contributing to the development and exacerbation of this disease, as well as those that play a protective role in preventing it.
2 Aim of systematic review
Several studies have established that the composition and function of gut bacterial microbiota in individuals with T2DM are different from healthy individuals. Despite this, the specific microbial changes remain largely unknown. This systematic review aims to provide an updated review on whether the gut bacterial microbiota profile of individuals with T2DM differ from healthy individuals. Mechanisms contributing to the pathophysiology of T2DM will also be discussed.
3 Methods
3.1 Search strategy
We performed a detailed systematic review of published data according to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta Analyses) guidelines. The methodological approach was registered in PROSPERO (International prospective register of systematic reviews) database under protocol number CRD42023459937.
Embase and PubMed literature search was performed on articles between Jan 1st 2010 and April 15th 2024. The search strategy combined MESH (Medline) and free terms using the boolean operators “AND” and “OR”. “Diabetes Mellitus”, “gut microbiome”, “intestinal flora” and “gastrointestinal microbiome” were terms used in the search. A complementary search was carried out in the references of studies included. The search protocol is shown below:
((“Diabetes Mellitus”[Majr: NoExp] OR “Diabetes Mellitus, Type 2”[Majr: NoExp] OR T2D[Text Word] OR type 2 diabetes[Text Word] OR “type 2 diabetes mellitus”[Title/Abstract:~2]) AND (“Gastrointestinal Microbiome”[Majr] OR gut micro*[Text Word] OR intestine flora[Text Word] OR intestinal flora[Text Word] OR gut flora[Text Word] OR intestine micro*[Text Word] OR intestinal micro*[Text Word] OR Gastrointestinal micro*[Text Word])) NOT (animals[Mesh] NOT humans[Mesh])
3.2 Eligibility criteria
All original peer reviewed research publications were considered. Eligible studies included observational human studies specifically examining gut microbiota in T2DM patients compared with control groups.
Exclusions: studies on type 1 diabetes mellitus or gestational diabetes; those without control groups; longitudinal studies; studies on children or adolescents aged <18 years or in the elderly aged >80 years; non-English studies; studies with only abstracts available; and studies with high risk of bias.
Microbial taxa were defined as positively or negatively associated with T2DM if p value <0.05 when comparing taxa abundance between individuals with T2DM and healthy controls. For linear discriminant analysis (LDA), a score of >4 indicated a positively association, while <4 indicated a negative association. For prospective studies with interventions, the baseline result was used. For studies with more than one population group, results were only reported to be positively or negatively associated if both groups demonstrated the result. Microbial taxa without reported p values, p values >0.05 or LDA values <4 were classified as non-significant and into an increased, decreased or equivocal (equal abundance or not reported) trend.
The titles and abstracts of all identified studies were reviewed by two independent authors. Studies were assessed using the Newcastle–Ottawa Quality Assessment Scale. This instrument included three domains: selection, comparability, and outcomes. High risk of bias was determined when some of the domains did not receive a point, in which case that study was excluded. Ambiguities in selection criteria were resolved by discussions between at least 3 researchers.
3.3 Data extraction
The data extracted from the studies included in this systematic review are summarized in Supplementary Table 1 with the following information: author and year of publication, country and period of study/seasons (if available), sample size and characterization of the study population, method used to evaluate the gut microbiota and bacteria analyzed (if applicable), and outcomes.
4 Results and discussion
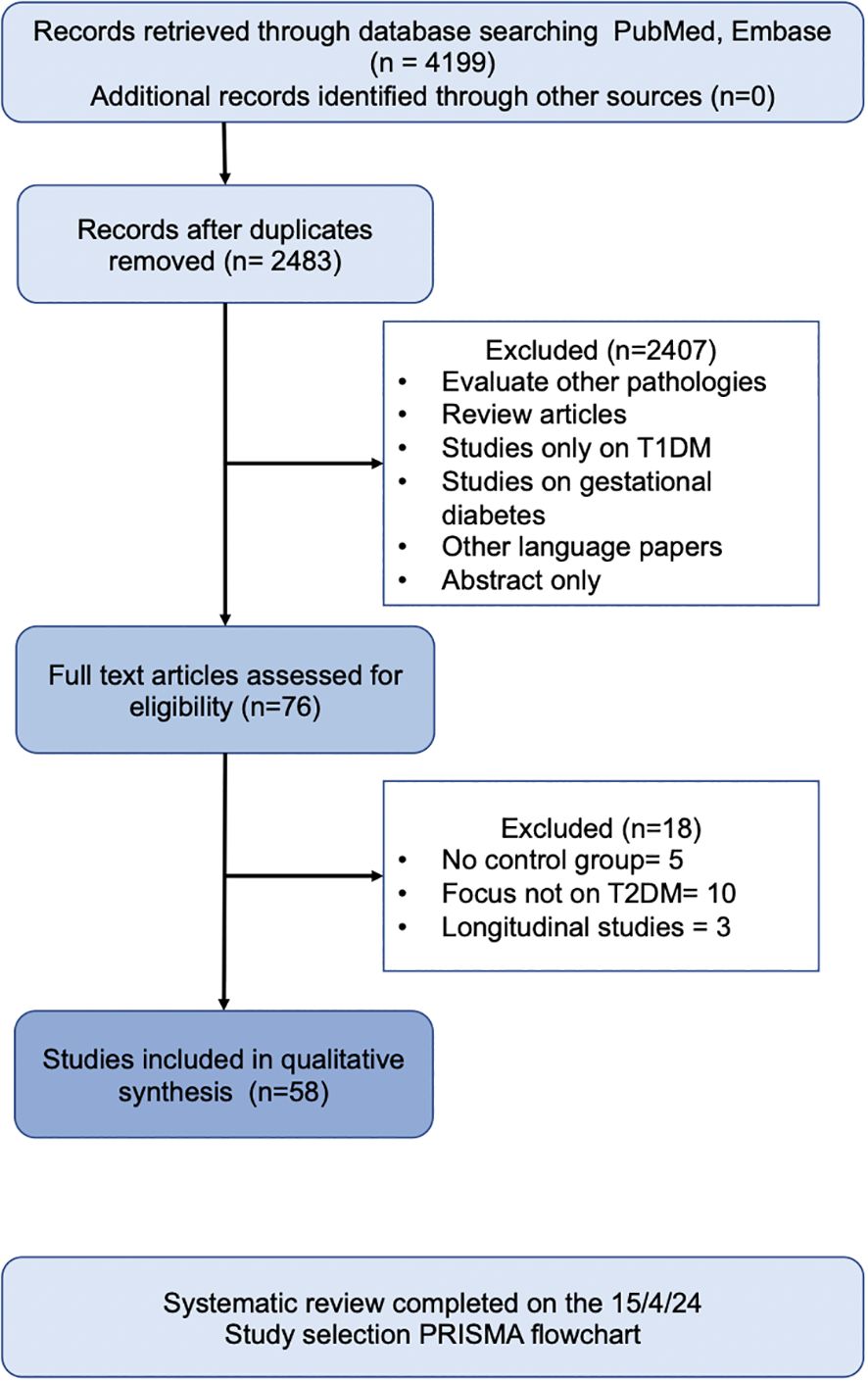
In total, 58 human observational studies were included in this review (Figure 1). The majority of these studies reported associations between specific taxa and the development and exacerbation of T2DM. However, no taxa were universally agreed upon to be positively or negatively associated with T2DM.
4.1 Alpha and beta diversity
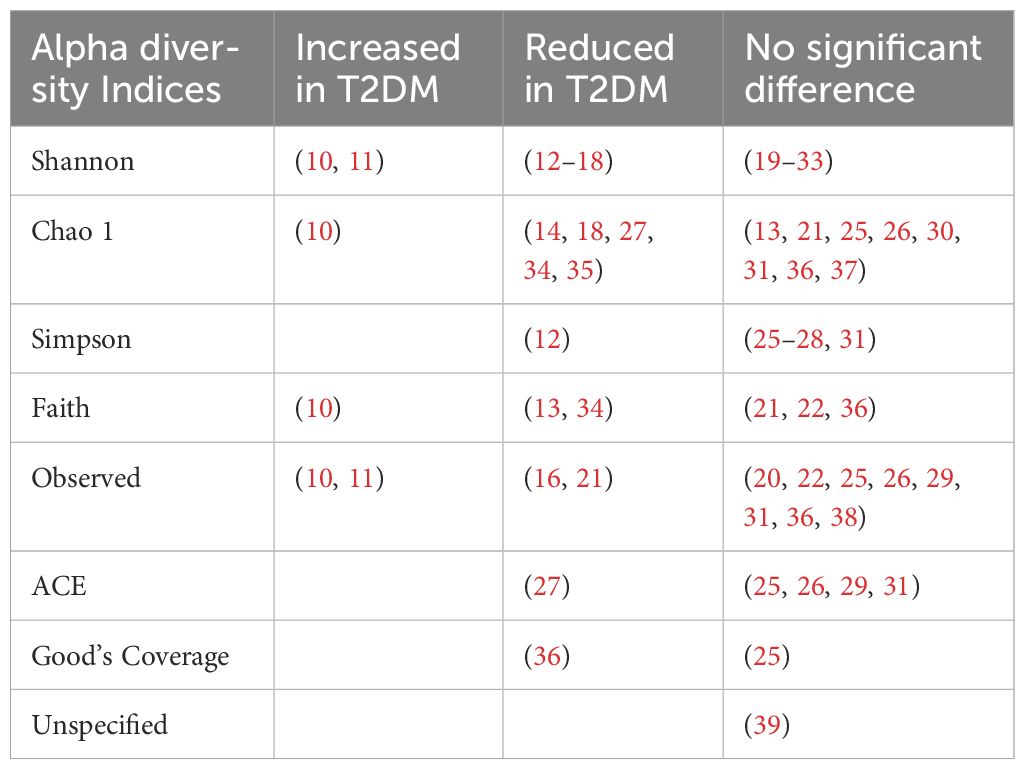
4.1.1 Alpha diversity
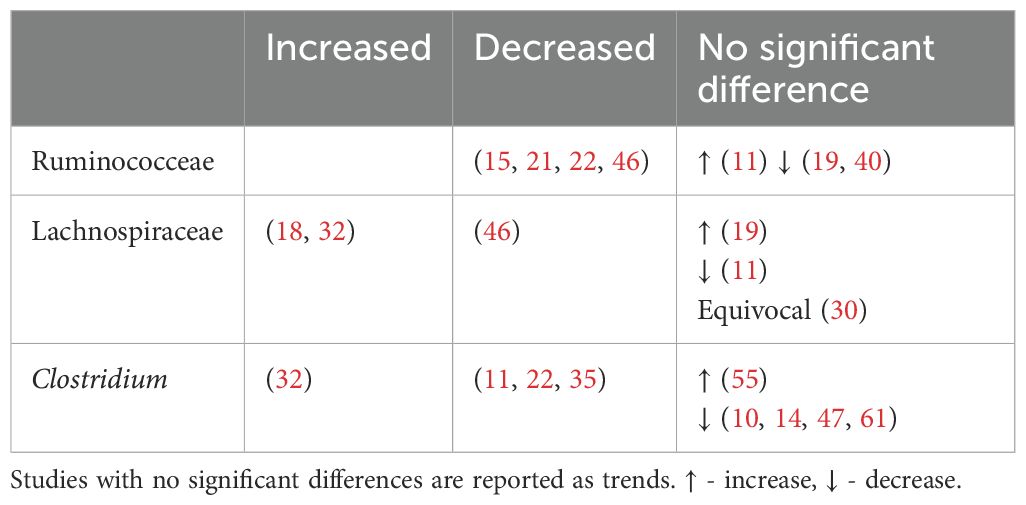
Alpha diversity refers to the microbial species diversity (richness) within a functional community. Reported indices included the Shannon index, Chao1 index, Simpson index, Faith index, Observed index, Abundance-based Coverage Estimator (ACE) index and Good’s Coverage. The Shannon index was the most commonly reported metric. A p value of <0.05 was deemed statistically significant. Most analyses reported no difference in alpha diversity between T2DM individuals and healthy controls (Table 1). Alpha diversity metrics varied by ethnicity, oral antihyperglycemic agents and other environmental factors (20, 31, 40). Higher diversity was observed in treatment naïve T2DM individuals compared to those receiving treatment (41).
4.1.2 Beta diversity
Beta diversity describes the amount of differentiation and dissimilarities between gut bacterial microbiota communities. The most common beta diversity metric used was the unweighted Unifrac distance. A p value of < 0.05 was deemed significant. The majority of studies reported a significant difference in beta diversity in individuals with T2DM compared to healthy controls (Table 2).
4.2 Phylum analysis - prevalence of firmicutes, bacteroidetes and the firmicutes/bacteroidetes ratios
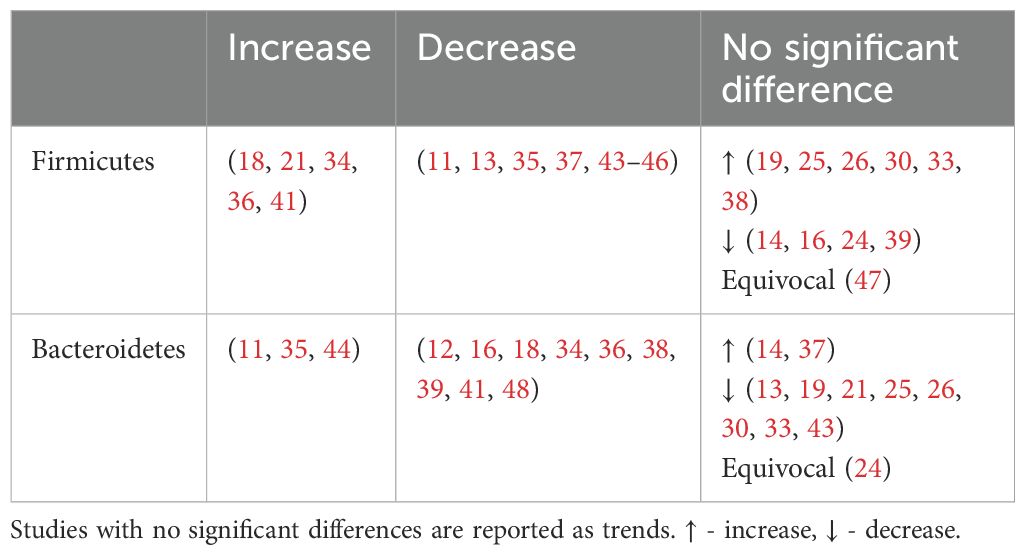
This review focuses on the phylum and genus levels of gut bacteria. The human gut bacterial microbiota consists mainly of Firmicutes and Bacteroidetes, which make up over 90% of the community. The remaining 10% includes phyla like Proteobacteria, Actinobacteria and Verrucomicrobia. In individuals with T2DM, the most commonly altered phyla are Firmicutes and Bacteroidetes (Figure 2, Table 3).
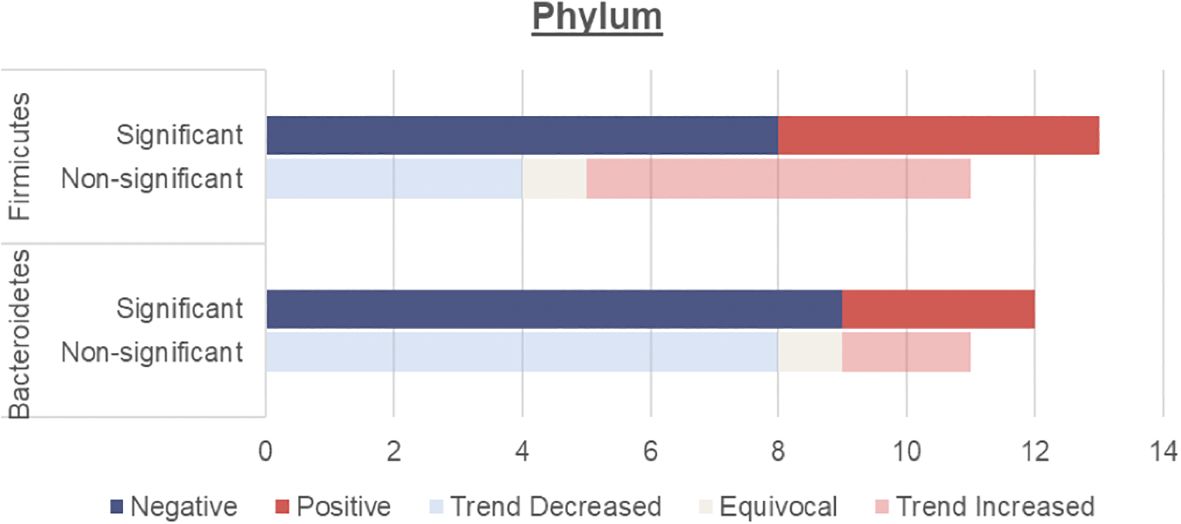
Figure 2. Number of human studies reporting Firmicutes and Bacteroidetes abundance and their association with T2DM. Studies were classified as having a significant association with T2DM (either positive or negative) if the p values were <0.05. Studies were classified as having a non-significant association with T2DM if they did not report on p values, had p values >0.05 or an LDA value <4 or >-4. These studies were then further classified into a non-significant association but trend increased, equivocal (equal abundance or not reported), or trend decreased.
4.2.1 Firmicutes and bacteroidetes
Overall, an unchanged Firmicutes and reduced Bacteroidetes abundance were observed among individuals with T2DM.
An unchanged Firmicutes abundance may be due to a simultaneous increase in opportunistic Firmicutes pathogens such as Enterococcus (Table 4), Eisenbergiella (16) Acidaminococcus (29, 41) and a decrease in beneficial Firmicutes microbes including Faecalibacterium and Roseburia (Table 5)
Meanwhile, Bacteroidetes are thought to be beneficial to human health with several genera including Bacteroides and Prevotella considered an untapped resource for next-generation prebiotics. Both these taxa, proposed to mitigate metabolic endotoxaemia and inflammation, were reduced among individuals with T2DM (Table 5). Bacteroidetes have negative correlation with fasting blood glucose levels (27, 36), corresponding with their reduced levels in T2DM.
4.2.2 The firmicutes/bacteroidetes ratio
The Firmicutes/Bacteroidetes (F/B) ratio (Table 6) represents the relationship between two dominant phyla and is commonly used as a marker of gut dysbiosis.
The F/B ratio was not consistently associated with clinical parameters. Larsen et al. found a positive correlation between the Bacteroidetes to Firmicutes ratio and plasma glucose (37) while Wang et al. reported a positive correlation between the F/B ratio and body mass index (BMI), fasting blood glucose levels and HBA1c (27). Other studies found no correlation with fasting, postprandial blood glucose levels (30), age, HBA1c or lipid profile (39). This suggests that while the F/B ratio indicates dysbiosis, it does not specifically predict metabolic outcomes.
4.3 Genera analysis - bacteria involved in type 2 diabetes
4.3.1 Genera of bacteria found to be increased in individuals with type 2 diabetes
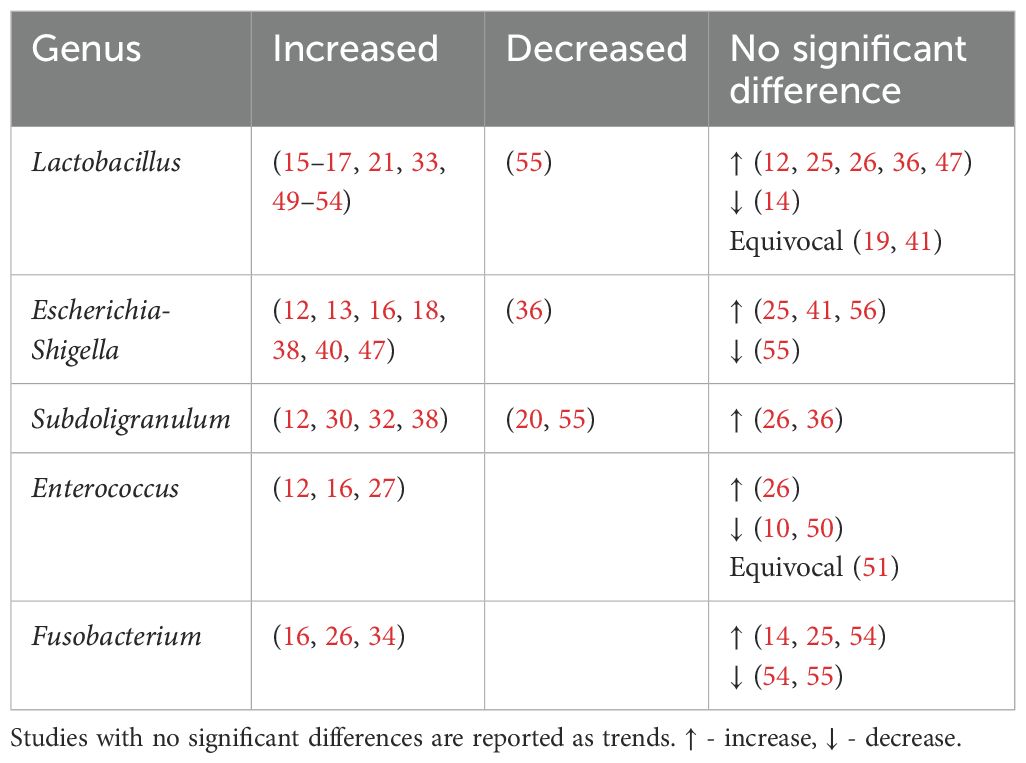
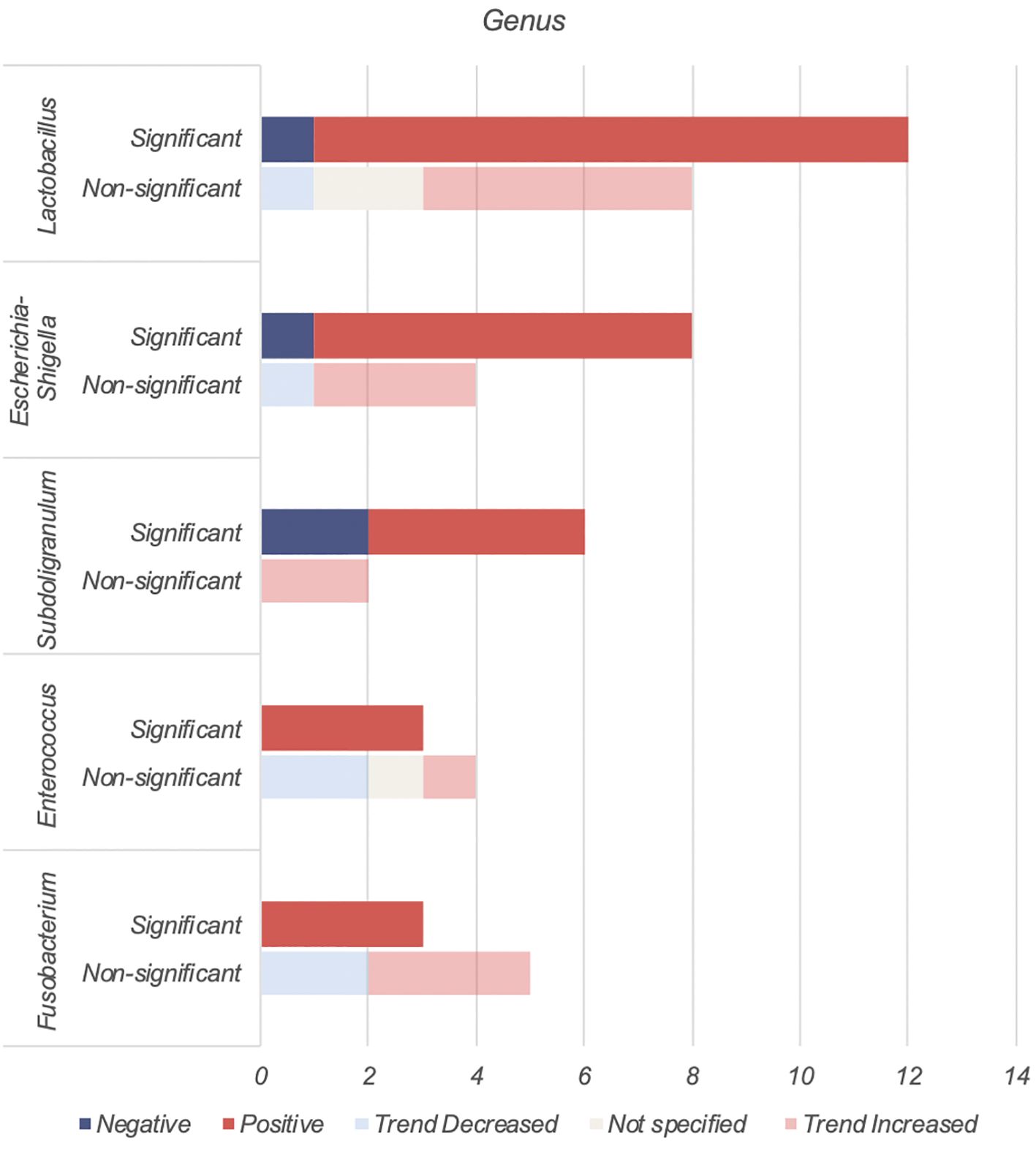
Lactobacillus, Escherichia-Shigella, Enterococcus, Subdoligranulum and Fusobacteria were found to be positively associated with T2DM (Table 4, Figure 3).
4.3.1.1 Lactobacillus
The Lactobacillus genus comprises of over 200 physiologically diverse gram-positive, non-spore forming lactic acid bacteria. Despite its positive association with T2DM, Lactobacillus species such as Lactobacillus paracasei (63), Lactobacillus fermentum (64) Lactobacillus acidophilus and Lactobacillus rhamnosus (65, 66) have demonstrated anti-inflammatory properties or benefits on host metabolism as a combination probiotic with Bifidobacterium lactic (65, 66).
The positive association of Lactobacillus with T2DM may therefore be driven by Metformin. Metformin, a first-line antihyperglycemic agent for treatment of T2DM, may alter bacterial abundances depending on the taxon’s resistance or sensitivity to the drug. In 2015, using 784 human gut metagnomes, Forslund et al. confirmed this positive association between metformin and Lactobacillus (55).
Among the eleven studies that reported an increase in Lactobacillus abundance (15–17, 21, 33, 49–54), only three studies (21, 50, 52) accounted for metformin use. Among these, one study found higher Lactobacillus levels regardless of metformin use (50), one found higher levels only in participants on unspecified oral antihyperglycemic agents (21), while the last study found no difference when accounting for metformin (52). More studies on treatment naïve T2DM or controlled for Metformin use are warranted.
4.3.1.2 Escherichia-Shigella
The Escherichia-Shigella genus, part of the family Enterobacteriaceae, includes multiple opportunistic pathogens (67). These gram-negative bacteria produce proinflammatory components such as lipopolysaccharide (LPS) and peptidoglycans, leading to intestinal and systemic inflammation (12). This systemic inflammation and consequent insulin resistance are key drivers for T2DM.
Unsurprisingly, Escherichia-Shigella abundance correlates with variables related to diabetes and obesity, including insulin resistance, diminished beta cell function (56), fasting glucose (41), HBA1c and BMI (47). This genus has been implicated in T2DM complications such as peripheral neuropathy (68), autonomic neuropathy (69), retinopathy (70), diabetic nephropathy (71) and chronic diabetic foot infections (72). Escherichia-Shigella has also been associated with an increasing abundance from healthy controls, pre-diabetes to T2DM (56). An increase in Escherichia-Shigella has also been associated with metformin use (13, 73). The outlier study that reported decreased Escherichia-Shigella abundance may be due to dietary or environmental differences (36).
4.3.1.3 Subdoligranulum
Subdoligranulum are anaerobic, spore-free gram-negative bacteria (12). This genera remains relatively underexplored and has only two known species - Subdoligranulum variabile and Subdoligranulum didolesgii. Four studies (12, 30, 32, 38) found Subdoligranulum more common in T2DM (Table 4) while two studies reported a negative association between T2DM and Subdoligranulum variabile (46, 74). These discrepancies may be related to species-specific properties.
Subdoligranulum has been linked to both promotion (75) and reduction of chronic inflammation (74). Subdoligranulum didolesgii has been associated with rheumatoid arthritis by triggering synovitis, while Subdoligranulum variabile has anti-inflammatory properties through short chain fatty acid (SCFA) production. Decreased levels of Subdoligranulum variabile in T2DM individuals may be suggestive of an overall state of inflammation (46).
Subdoligranulum’s positive association with T2DM may be influenced by metformin use (55). Of four studies reporting increased Subdoligranulum, two did not report metformin use (12, 32), one excluded metformin users (30), and one found an increase regardless of metformin use (38).
4.3.1.4 Enterococcus
Enterococcus are gram-positive facultative anaerobic cocci found in intestinal microbiota and on the skin. Some species are opportunistic pathogens causing severe infections such as bacterial endocarditis and spontaneous bacterial peritonitis, while others (Enterococcus durans) produce anti-inflammatory SCFAs (76).
Enterococcus may contribute to the development of T2DM through two mechanisms. Firstly, Enterococcus faecalis secretes matrix metalloprotease gelatinase causing chronic intestinal inflammation and impaired gut barrier integrity (77), leading to systemic inflammation. Secondly, Enterococcus has been linked to impaired glucose homeostasis. Associations include higher HBA1c (16, 27), fasting (27) and post prandial (16) glucose levels, and impaired beta cell function (27). Mechanistically this may relate to overgrowth of enterococcus leading to proportional decreases in beneficial anti-inflammatory bacteria (50).
4.3.1.5 Fusobacterium
Fusobacterium are anaerobic gram-negative rod bacteria. Similar to Enterococcus, this genus is part of the regular colorectal microbiota. Fusobacterium, in particular Fusobacterium nucleatum, has been associated with increased production of inflammatory cytokines such as IL-6, IL-8, TNF-α and COX-2 (78). This may contribute to the chronic inflammatory state seen in T2DM. Fusobacterium has also been associated with diabetic nephropathy (79) and its species found increased among individuals with T2DM (23, 44).
4.3.2 Genera of bacteria found to be reduced in individuals with type 2 diabetes
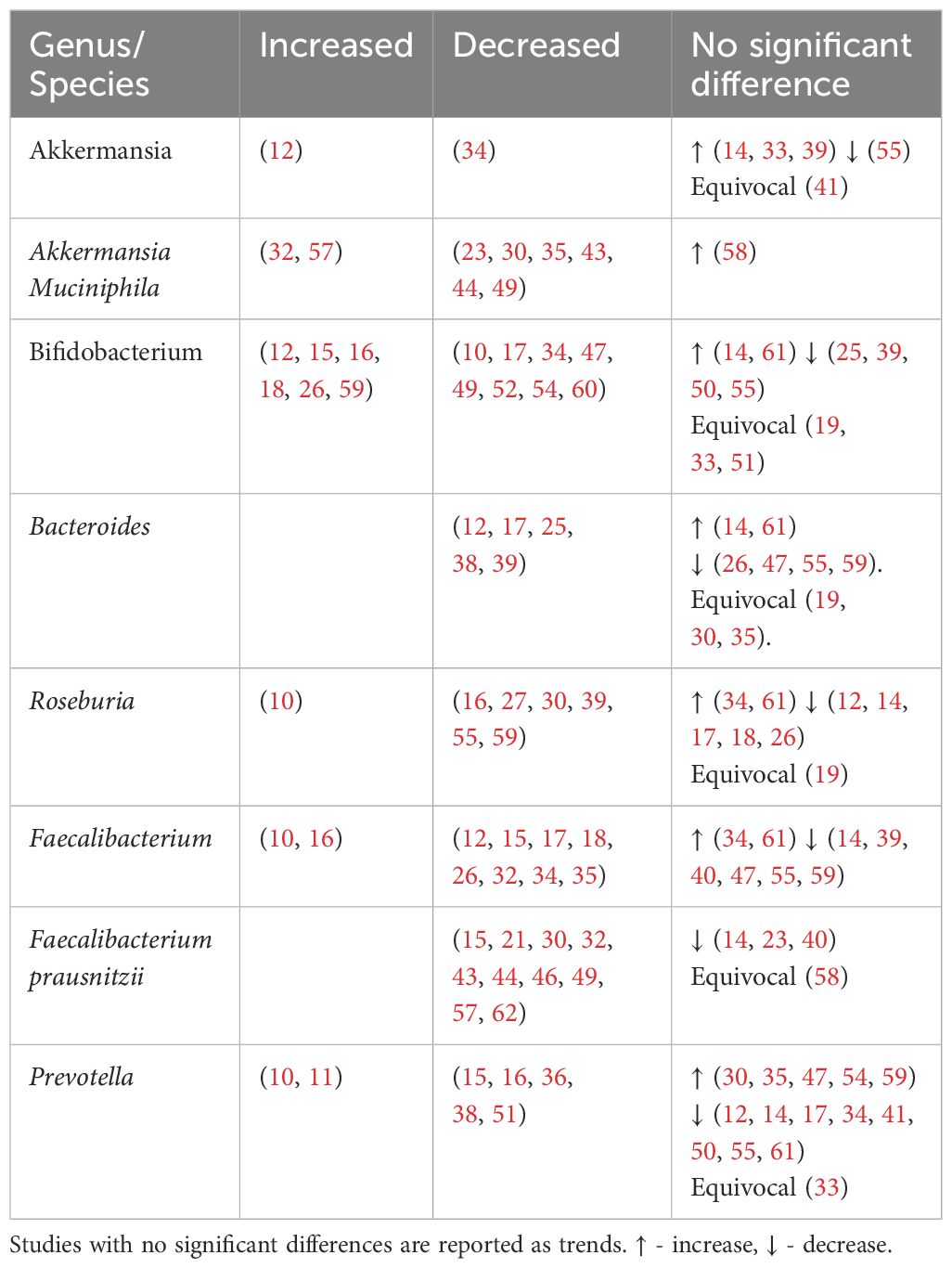
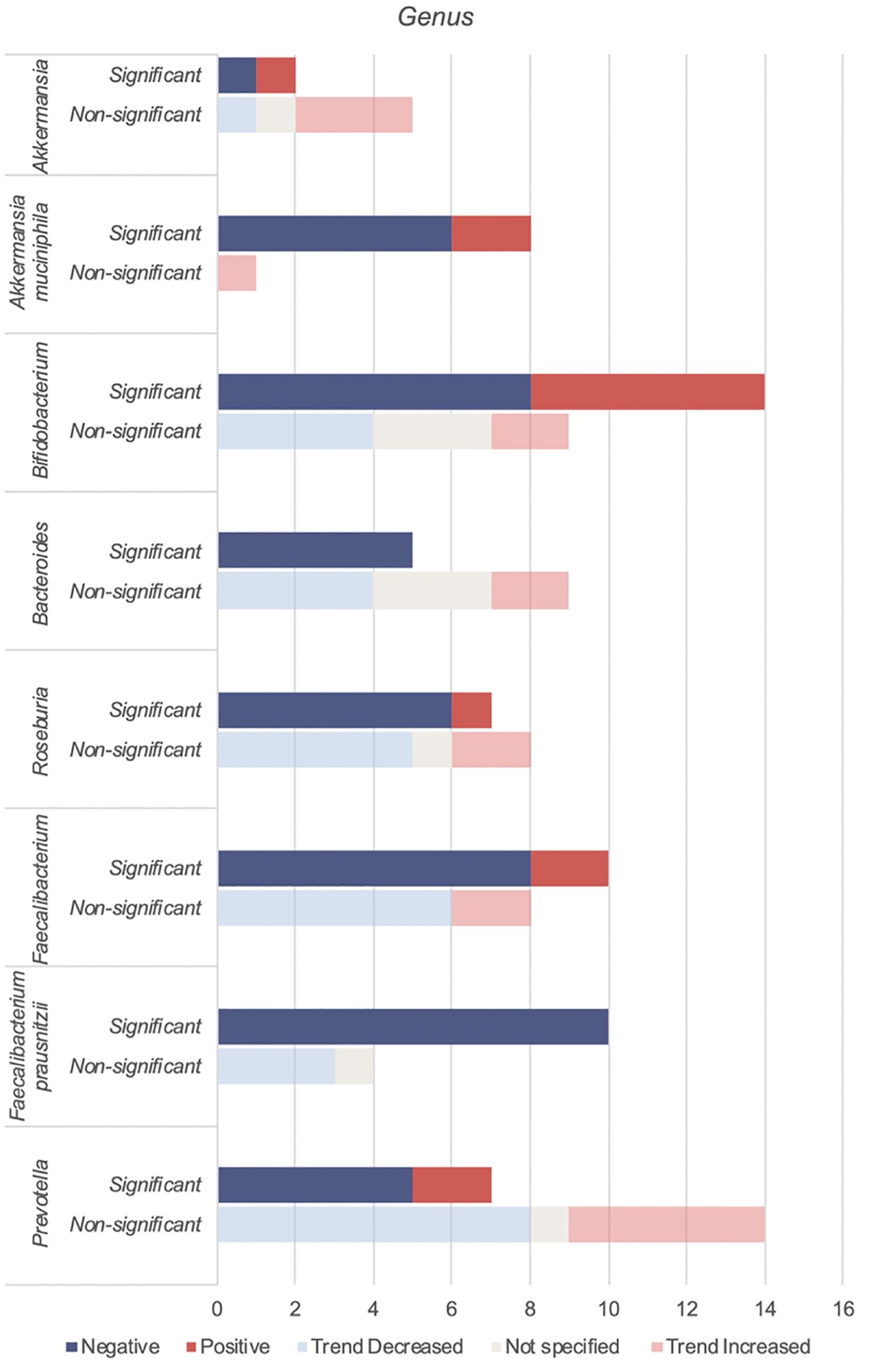
Akkermansia, Bifidobacterium, Bacteroides, Roseburia, Faecalibacteirum and Prevotella were found to be negatively associated with T2DM (Table 5, Figure 4). Species abundance of Bifidobacterium, Bacteroides, Roseburia and Prevotella can be found in Supplementary Table 2.
4.3.2.1 Akkermansia
Akkermansia is gram-negative bacterium belonging to the Verrucomicrobia phylum. Akkermansia mucinphilia, a symbiont microbe colonizing the human intestinal mucosal barrier, is a promising next generation probiotic. It plays a critical role in the maintenance of intestinal barrier, production of anti-inflammatory cytokines and SCFA benefiting host metabolism. In diabetic rat models, administration of live attenuated Akkermansia reduced oxidative stress, lipotoxicity, LPS and inflammation (80). In individuals with T2DM, combined probiotics containing Akkermansia muciniphila reduced HBA1c and postprandial glucose control (81).
Reduced levels of Akkermansia mucinphilia are associated with T2DM. Akkermansia is inversely correlated with HBA1c and fasting glucose and positively with anti-oxidants (41).
4.3.2.2 Bifidobacterium
Bifidobacterium is a dominant non-spore-forming, gram-positive taxa that help maintain balances between the various intestinal floras (82). Key Bifidobacterium species include Bifidobacteroim bifidum, Bifidobacterium adolescentis and Bifidobacterium longum. These species have been used as probiotics in humans (65, 66, 83) and administered in animal studies (84, 85) leading to reduced cytokine production and improved metabolic parameters such as glucose and HBA1c (66, 84).
Apart from SCFA production, in vivo and in vitro studies show that Bifidobacterium administration markedly decreased intestinal permeability by increasing tight junction expression and reducing inflammatory cytokines such as IL-6 and TNF-α (86). This reduces metabolic endotoxaemia, systemic inflammation and may explain its overall negative association with T2DM (Table 5). An increase in Bifidobacterium has been attributed to antihyperglycemic agents (16) or a U shaped association with T2DM (26, 59).
4.3.2.3 Bacteroides
Bacteroides is a gram-negative obligate anaerobic taxa constituting approximately 25% of the intestinal gut microbiota. As commensals, these taxa generally maintain a beneficial relationship with the human gut. Overall, Bacteroides species including Bacteroides fragilis, Bacteroides thetaiotamicron, Bacteroides vulgutas or Bacteroides dorei have been associated with a protective effect against T2DM through anti-inflammatory properties (87) and an improved gut barrier integrity from mucus (88) and SCFA production (89). Bacteroides species also have a structurally different LPS that is less pro-inflammatory than classical enterobacterial LPS (90). Discrepancies in Bacteroides abundance (Table 5) may be due to the bacteriostatic and bactericidal effect of metformin (55) or potential pathogenic Bacteroides species that can contribute to chronic inflammation (39).
4.3.2.4 Roseburia
Roseburia is a gram-positive, SCFA-producing member of the Firmicutes phylum that inhabits the human colon. Roseburia has been identified as a pathognomonic bacteria in T2DM (91) with significant lower levels in participants. Reduced species include Roseburia hominis (23, 46), Roseburia intestinalis and Roseburia inulinivorans (32, 53, 55). Roseburia improves glucose homeostasis and intestinal permeability through SCFA production and anti-inflammatory properties (92). Gut microbiota transplantations from lean donors to recipients with metabolic syndrome led to increased fecal Roseburia and butyrate levels, correlating with improved insulin sensitivity (93).
4.3.2.5 Faecalibacterium
Faecalibacterium are human gut colonizers and well-known SCFA producers. Faecalibacterium and Faecalibacterium prausnitzii were consistently reduced in T2DM (Table 5), with the later being highly discriminant (91). In mice, Faecalibacterium prausnitzii administration was associated with improved glucose levels and HBA1c, making it a promising orally administered probiotic (94). Faecalibacterium is negatively associated with HBA1c (39).
4.3.2.6 Prevotella
Prevotella has been linked to both pathogenic effects including systemic inflammation and insulin resistance (95) and beneficial effects like SCFA production (96) and reduced gut permeability via increased production of tight junction proteins (97). Prevotella is negatively correlated with HBA1c (16, 41, 98), but positively with blood glucose (10, 41). The discrepancies within the Prevotella genus may be due to diet (24) and genetic diversity within its species (99).
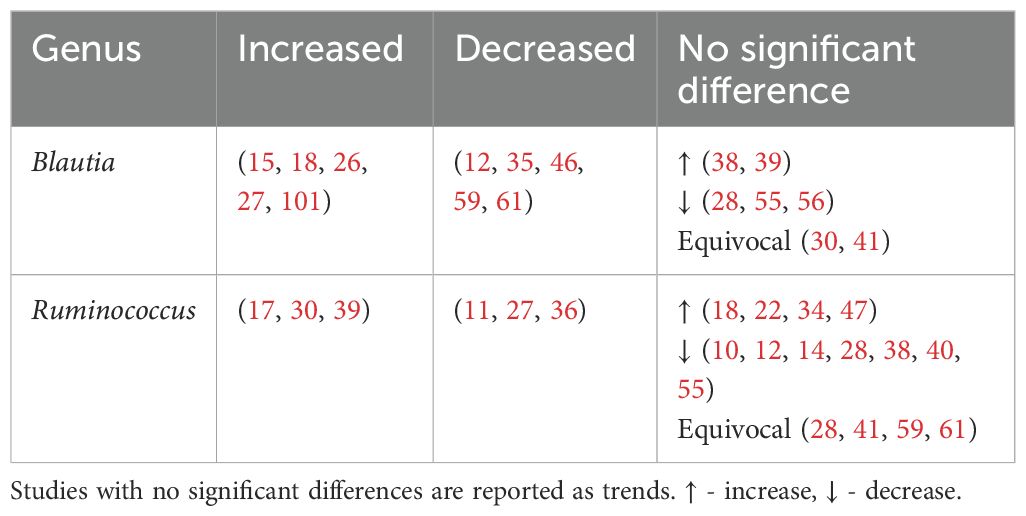
4.3.3 Genera of bacteria found to have mixed findings in type 2 diabetes
Unlike previous reviews (100), Blautia and Ruminococcus were found to have mixed associations (Table 7).
4.4 Microbiota effects on metabolism in type 2 diabetes individuals
In T2DM, gut dysbiosis leads to increased systemic inflammation and an unfavorable host metabolism (Figure 5). This is due to an increase in pro-inflammatory cytokine and LPS production, increased gut permeability enabling bacterial endotoxin translocation, and reduced beneficial gut metabolites. Ultimately, systemic inflammation induces insulin resistance and contributes to chronic hyperglycemia and development of complications.
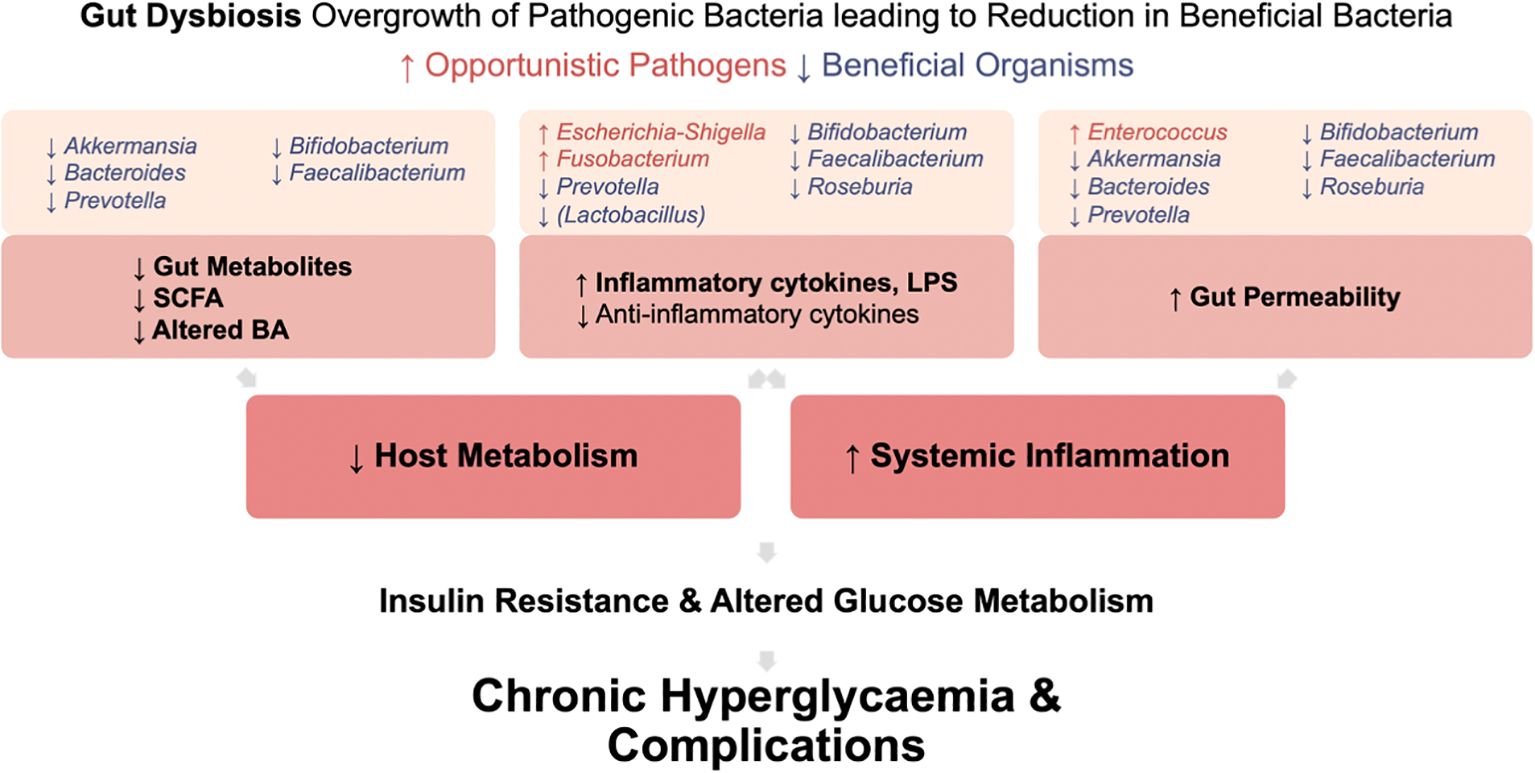
Figure 5. Mechanisms by which gut dysbiosis contributes to the development and progression of T2DM. Gut dysbiosis in T2DM leads to increased systemic inflammation and an unfavorable host metabolism. This occurs due to increased production of pro-inflammatory cytokine and LPS, increased gut permeability enabling bacterial endotoxin translocation, and reduced production of beneficial gut metabolites. Ultimately, this systemic inflammation induces insulin resistance. Coupled with altered glucose metabolism in T2DM, these factors contribute to chronic hyperglycemia and the development of complications.
4.4.1 Increased gut permeability
Patients with T2DM have increased intestinal permeability compared to age, sex and BMI matched controls (102). This results in translocation of gut microbes and their products into the bloodstream, in turn causing metabolic endotoxaemia and increased systemic inflammation. This is supported by elevated blood levels of bacterial cell wall products and circulating intestinal bacteria in individuals with pre-diabetes (103) and T2DM (51).
Gut bacterial dysbiosis increases gut permeability via three mechanisms: alterations in expression, canalization and distribution of tight junction proteins; overactivation of the endocannabinoid system; and altered production of beneficial gut metabolites including SCFA and bile acids.
4.4.1.1 Alterations in tight junction proteins
The intestinal lining composed of epithelial cells assisted by tight junctions (TJ), acts as a physical barrier against microorganisms and antigens. TJ controls intestinal permeability (104). In T2DM, reduction in beneficial microbes Bacteroides, Bifidobacterium, Faecalibacterium, Roseburia and Akkermansia, leads to decreased gene expression and therefore reduced localization, production, and distribution of TJ proteins. This results in increased gut permeability.
Mouse studies show that pre-treatment with Bifidobacterium (86), Bacteroides vulgatus, Bacteroides dorei (105) or Prevotella histicola (97), upregulates TJ genes leading to reduced intestinal permeability and inflammation. Bacteroides fragilis (106–108), Bacteroides facies (109), Bifidobacterium bifidum (110), Bifidobacterium adolescentis (111) and Bifidobacterium longum (112) have also been found to increase TJ proteins.
Faecalibacterium prausnitzii and Roseburia intestinalis reduce gut permeability by production of butyrate and upregulation of TJ proteins (89, 109). Butyrate is essential for colonic epithelial cells, offering anti-inflammatory properties and protecting against pathogens (30). In db/db mice, Faecalibacterium prausnitzii also produces microbial anti-inflammatory molecule, increasing TJ expression and restoring the damaged intestinal barrier (113).
Akkermansia muciniphila decreases gut permeability by promoting TJ protein expression via its outer membrane protein Amuc_1100. Additionally, it improves intestinal TJ via AMPK activation in the epithelium (114) and modulation of the endocannabinoid system (115).
Less understood are the bacteria Rumincoccaeceae and Blautia which may be associated with increased gut permeability (116). Further studies are needed to confirm these findings and understand their mechanisms.
4.4.1.2 Endocannabinoid system
There is growing evidence that the endocannabinoid system regulates intestinal inflammation and mucosal barrier permeability, thus influencing T2DM pathophysiology.
The endocannabinoid system, historically associated with cognitive and emotional processes, also regulates intestinal inflammation. The two main endocannabinoids are anadamide (AEA) and 2 arachidonylglycerol (2-AG). They act primarily through cannabinoid receptors CB1R and CB2R. CB1R is expressed in gastrointestinal epithelial cells and myenteric and submucosal plexuses while CB2R may be found on enteric neurons (117).
Overactivation of CB1R via AEA and 2-AG leads to increased gut permeability (117). In T2DM mice models, CB1R antagonists were shown to decrease gut permeability by reducing inflammation and alterations in TJ proteins (118). Akkermansia muciniphila antagonises CB1R through its outer membrane protein Amuc_1100, reducing gut permeability, LPS levels and systemic inflammation (115). Bacteroides fragilis also affects epithelial barrier permeability through the endocannabinoid system (119).
Oxidative stress, inflammation, and insulin secretion contribute to T2DM and its complications. Although unrelated to gut permeability, CB2R activation decreases inflammation and oxidative stress and promotes pancreatic insulin secretion via calcium signal regulation (120). This suggests potential benefits of CB2R agonists in T2DM management.
4.4.2 Alteration to the gut metabolites
The gut microbiota acts as a metabolic organ and facilitates nutrient and energy harvesting from food. It produces metabolites that regulate host metabolism including SCFA and bile acids which maintain the intestinal barrier (4). Alterations in the gut microbiota is thus associated with alteration to the gut metabolites which in turn contributes to T2DM and its complications.
4.4.2.1 Alteration to short chain fatty acids
SCFAs are produced by gut microbiota from non-digestible carbohydrates. They provide energy to colonocytes, reduce inflammation and regulate satiety (121). The most common SCFAs are acetate, propionate and butyrate, and are predominantly produced by anaerobic Bacteroidetes and Firmicutes phyla.
SCFAs have multiple beneficial effects such as maintaining gut permeability, modulating host metabolism and anti-inflammatory effects. Reduced levels of SCFA-producing bacteria including Bacteroides, Bifidobacterium, Faecalibacterium, Prevotella and Akkermansia. are associated with T2DM. This is reflected by the reduced acetate (38), propinionate (38, 98), butyrate (38, 98) and other SCFA (38, 51) concentrations in T2DM fecal samples. Functional analysis of gut microbiota showed reduced SCFA-producing pathways in T2DM compared to controls (61).
Individuals with T2DM related complications had lower SCFA fecal concentrations than those without complications (38). Increased dysbiosis severity and reduced production of SCFA may contribute to the development and progression of T2DM complications.
4.4.2.1.1 Alteration to SCFA resulting in decreased gut barrier integrity
SCFA help to maintain gut barrier integrity through a number of mechanisms. This includes promoting epithelial growth and innate responses to microbes, providing energy to intestinal epithelial cells via beta-oxidation in the mitochondrial tricarboxylic acid cycle and maintaining an anaerobic gut environment hostile to opportunistic aerobic pathogens (122). SCFA also stabilize transcription factors that protect the barrier and activate genes for TJ proteins thus preventing bacterial and LPS translocation and systemic inflammation (89). Lower SCFA concentrations in T2DM may therefore to altered microbiota diversity and increased intestinal permeability, predisposing to insulin resistance through metabolic endotoxaemia.
4.4.2.1.2 Alteration to SCFA resulting in altered glucose and lipid metabolism
SCFA influence glucose and appetite regulation. In human in vivo studies, rectal infusions of SCFA mixtures led to a rise in plasma peptides YY (123–125) and glucagon peptide-1 (GLP-1) (123). This resulted in appetite control, increased insulin sensitivity and increased pancreatic beta cell concentrations (4, 126). SCFA also modulate glucose and lipid metabolism. Propionate suppresses hepatic gluconeogenesis, while acetate and butyrate reduce lipogenesis and increase leptin secretion (122). In mouse models, SCFA increase food intake via parasympathetic activity and support glucose stimulated insulin secretion (127). Reduced levels of SCFA may therefore lead to poor appetite control, hyperglycemia, hyperlipidemia and insulin resistance.
4.4.2.1.3 Alteration to SCFA results in increased inflammation
SCFA exhibit anti-inflammatory properties. Butyrate inhibits NF-kB activation, reducing pro-inflammatory cytokines like TNF-α, IL-6, IL-2, IL-8 and promotes IL-10 production via GPR109A, maintaining a balance between pro and anti-inflammatory T cells (128). Lower SCFA levels may contribute to chronic inflammatory state and insulin resistance in T2DM.
4.4.2.1.4 Alteration to SCFA negatively disrupting the gut environment
Butyrate producing bacteria compete with gram-negative bacteria, maintaining microflora balance and inhibit pathogenic strains. They also maintain an anaerobic environment by enhancing coloncyte oxygen consumption and stabilizing hypoxia inducible factor (122). Depletion of butyrate producing bacteria can lead an increase in opportunistic pathogens like Fusobacterium, which releases harmful by-products perpetuating the inflammatory cycle (129).
4.4.2.2 Alteration to bile acids
Bile acids, known for their role in digestion of dietary fats, have recently gained attention due to their possible influence on metabolic processes, particularly in the context of T2DM. Primary bile acids (PBAs), cholic acid (CA) and chenodeoxycholic acid (CDCA) are synthesized from cholesterol in hepatocytes and released into the duodenum. They are then uncoupled by bile saline hydrolysase before being converted into more hydrophobic secondary bile acids (SBAs) through bile acid deconjugation and the rate limiting 7α-dehydroxylase enzyme. Bacteroides and Enterococcus are involved in the initial deconjugation, while Bifidobacterium, Lactobacillus and Enterococcus utilize bile saline hydrolase. Meanwhile, selected bacteria from the Lachnospiraceae and Ruminococcaceae family perform the subsequent 7α-dehydroxylase conversion of CA and CDCA to generate the SBAs deoxycholic acid (DCA) and lithocholic acid (LCA) respectively (130). The abundance of these bacteria are described in Table 8.
Interestingly, the profiles of bile acids in patients with T2DM vary across different studies. Some studies indicate higher levels of total bile acids, PBA and SBA, among individuals with T2DM (131, 132). In contrast, other studies have found no significant differences in total serum bile acid levels between T2DM patients and controls (133). Nonetheless, the majority of these studies do suggest a relationship between increased insulin resistance and higher total bile acids (132, 133), highlighting the therapeutic potential of targeting bile acids in T2DM. Alterations in bile acids have been associated with complications of T2DM including cardiovascular disease (134) and diabetic kidney disease (135).
4.4.2.2.1 Alteration of bile acids resulting in altered glucose metabolism
Bile acids regulate glucose homeostasis through the Farnesoid X receptor (FXR) and Takeda-G-protein-receptor 5 (TGR5) (136). PBAs preferentially activate FXR, while SBAs favor TGR5. Activation of TGR5 appears to have a beneficial effect on glucose metabolism by stimulating release of GLP-1 from enteroendocrine cells, which enhances insulin secretion, slows gastric emptying and reduces appetite (137). Interestingly, both deactivation and activation of FXR have been linked to positive effects on glycemic regulation. For example, intestinal FXR activation has been associated with reduced hepatic gluconeogenesis (138, 139) and contribute to glucagon fasting-induced hepatic gluconeogenesis (140). FXR deficiency has been linked to increased GLP-1 plasma concentrations (138, 141). Nonetheless, hepatic FXR deficiency in mice has been shown to increase gluconeogenesis, worsening glucose intolerance and insulin resistance (142). This FXR paradox highlights the complexity of FXR signaling, and suggests that the role of FXR in metabolic dysfunction may differ between the liver and intestine (143).
The systematic effects of various secondary bile acids on glycemic control have been demonstrated in both humans and animal models. For example, administration of ursodeoxycholic acid (UDCA) has been shown to improve post-prandial glucose levels and GLP-1 secretion (144), reduce metabolic syndrome (145) and increase the survival rate of pancreatic beta cells (146, 147). Additionally, intrajejunal and rectal taurocholic acid led to decreased blood glucose levels and the release of satiety hormones GLP-1 and Peptide YY (148, 149). Meanwhile, metformin, a drug commonly prescribed for T2DM, has been suggested to modulate primary and secondary bile acid levels and alter the expression of their receptors, thereby enhancing insulin sensitivity (150).
Specifically, among the taxa that differ significantly in individuals with T2DM, Lactobacillus and Bifidobacterium have been suggested to play a role in modulating bile acids and improving glycemic control. In a recent randomized control trial, a probiotic product containing Lactobacillus casei, Lactobacillus plantarum, Lactobacillus rhamnosus, Bifidobacterium animalis subsp. lactis M8 and Bifidobacterium animalis subsp. lactis V9. led to reductions in HbA1c and fasting blood glucose levels, along with increased insulin secretion. Faecal metabolite analysis demonstrated an increase in both CDCA and hyodeoxycholic, a component of hyoholic acid shown to upregulate GLP-1 secretion via TGR5 (139). The study suggested that specific bile acids may activate various receptors, which in turn promotes GLP-1 secretion, thereby reducing blood glucose levels (151). Collectively, these findings highlight the potential therapeutic value of bile acids in T2DM.
4.4.2.2.2 Alteration to bile acids affecting gut barrier integrity
Alterations in bile acid profiles affect intestinal permeability through regulation of TJ proteins. In murine models, DCA reduces TJ protein Zona-Occludens-1, thereby increasing gut permeability (152). Primary biliary acids CDCA and CA, and secondary biliary acids DCA, increase epithelial permeability through phosphorylation of occludin in intestinal Caco cells (153). At high concentrations DCA is cytotoxic to intestinal stem cells and goblet cells, thereby impairing gut permeability (154). Conversely, LCA reduces intestinal permeability by ameliorating TNF-α induced disruption of TJ proteins (155). In murine models, an increase in LCA and DCA was associated with increased colon expression of TGR5 and TJ proteins, thereby improving gut-barrier integrity (156). Human studies demonstrate that elevated levels of LCA and DCA have anti-inflammatory properties within the colon (157). Bile acids have both beneficial and detrimental effects on intestinal permeability, and further studies are required to understand their specific impacts.
4.4.2.2.3 Alteration in bile acids resulting in systemic inflammation
Bile acids have been shown to inhibit the induction of pro-inflammatory genes and the production of inflammatory cytokines by macrophages via FXR and TGFR-5 receptors (158). In mice models, the production of secondary bile acids, such as LCA and UDCA, ameliorated colitis and reduced the production of proinflammatory cytokines TNF- α, IL-17A and IL-6 (156). Alteration in bile acids can thus lead to decreased anti-inflammatory effects and contribute as well as exacerbate the chronic low-grade inflammatory state in T2DM.
In summary, bile acids play a role in modulating intestinal permeability, systemic inflammation, and glucose homeostasis, thereby contributing to the pathogenesis of T2DM. While bile acids represent a promising therapeutic target, the precise abundance of various bile acids in T2DM and their effects on different receptors, particularly FXR, remain unclear. Further studies are needed to confirm these alterations and clarify the specific interactions involved.
4.4.2.3 Increased systemic inflammation
T2DM is associated with chronic low-grade systemic inflammation caused by metabolic endotoxaemia and cytokine stimulation by microbes leading to oxidative stress, macrophage activity and insulin resistance. Insulin resistance occurs due to activation of the inflammatory cascade, subsequent activation of serine kinases, insulin receptor substrate serine phosphorylation and consequent insulin signaling inhibition causing cellular insulin resistance (159).
4.4.2.3.1 Metabolic endotoxaemia
In T2DM, metabolic endoxaemia occurs due to increased production of toxic bacterial components and increased gut permeability enabling translocation of these products into the systemic circulation.
Gram-negative bacteria, such as Fusobacterium and Escherichia-Shigella, produce LPS an endotoxin that activates immune responses by binding to pattern recognition receptors such as toll-like receptor 4 (TLR4), NLRP3 inflammasome and NOD-like receptors which are expressed on the surfaces of antigen presenting cells. This leads to release of pro-inflammatory cytokines IL-1, IL-7, TNF-α release (121) and insulin resistance via inhibition of insulin signaling (159). Gut dysbiosis in T2DM increases LPS synthesis (46, 57) with higher plasma levels of LPS (15, 51) and TLR4 receptor activation (15) observed.
IAP is an enzyme which mitigates intestinal inflammation through detoxification of pathogen toxins and regulation of gut microbes (160). It de-phosphorylates LPS, reducing its toxicity and lowering systemic inflammation (161). In mice, IAP was shown to reverse metabolic endotoxaemia (162). Very low levels of fecal IAP have been reported in T2DM patients (163). Bifidobacterium species, Faecalibacterium prausnitzii, Roseburia species and other butyrate producing bacteria modulate IAP activity (164). A decrease in these anti-inflammatory, butyrate producing bacteria may contribute to chronic systemic inflammation in T2DM.
4.4.2.3.2 Cytokine modulation
T2DM is associated with elevated pro-inflammatory cytokines. Bacterial taxa such as Escherichia-Shigella and Fusobacterium are increased in T2DM and correlate with higher levels of pro-inflammatory cytokines like IL-17, TNF-α and IL-6 (165).
Conversely, beneficial microbes Roseburia intestinalis (166), Prevotella histicola(97), Faecalibacterium prausnitzii (167), Bifidobacterium longum (167), Bacteroides fragilis (87, 168), Akkermansia muciniphila(169), Lactobacillus paracasei (63) and Lactobacillus fermentum (64) promote anti-inflammatory cytokine IL-10 production and suppress pro-inflammatory cytokines (87, 92, 166, 167, 170, 171). Butyrate producing bacteria such as Roseburia, Faecalibacterium and Subdoligranulum also decreases pro-inflammatory cytokine production by inhibiting NF-kB, a major transcription factor essential for inflammatory responses (128).
4.4.2.4 Preferential growth of pathogenic microbiota
Pathogenic bacteria including Enterococcus and Escherichia-Shigella may outcompete beneficial bacteria, such as Faecalibacterium, Roseburia and Bifidobacterium, perpetuating negative effects on gut health and inflammation.
5 Limitations
This systematic review has several limitations. The significant variation in methodology across various human observational studies made it difficult to draw definitive conclusions. Differences in inclusion and exclusion criteria, and varied methods for controlling factors such as age, BMI, diet and medication, affected bacterial abundances and hindered efforts for consistent comparisons. Furthermore, few studies provided raw data on bacterial abundances or reported non-significant bacterial abundances, complicating quantitative data pooling for any specific bacteria.
Most studies did not account for the effects of metformin and other oral anti-hyperglycemic agents, which are known to alter certain bacterial abundances. This review could not control for their use, highlighting the need for future large-scale studies to at least account for, if not control, the effects of these diabetes medications.
Majority of the studies utilized 16s RNA gene sequencing, with few studies utilizing metagenomic sequencing. This meant that it was rare to identify microbes at species or strain levels and may account for some discrepancies at the genus level. Moreover, few studies examined functional alterations in T2DM and correlated it to individual bacterial taxa. Therefore, only associations but not causations between taxa and T2DM could be determined. Future research should assess the functional potential of the gut microbiome in individuals with T2DM.
Finally, the pathogenesis, perpetuation and management of T2DM is multifactorial and various clinical factors including genetics, other comorbidities, adherence to therapies and presence of complications all play a critical role. Future studies should measure these factors, and consider their interplay with gut microbiota in T2DM.
6 Conclusion
This systematic review demonstrates that T2DM is strongly associated with gut dysbiosis, as evidenced by differential microbial abundances, altered F/B ratio and changed diversity indices. Through increased gut permeability, decreased SCFA production and modulation of inflammatory cytokines, gut dysbiosis leads to increased systemic inflammation and disrupted glucose homeostasis.
Among these microbes, Escherichia-Shigella is consistently associated with T2DM, while Faecalibacterium, in particular Faecalibacterium prausnitzii appears to offer a protective effect against T2DM. However, the heterogenicity and observational nature of these studies hinder establishment of causative relationships. Future research should control for factors such as age, diet and medication use, and incorporate functional analysis of these gut microbes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SC: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Writing – original draft, Writing – review & editing. ML: Methodology, Writing – review & editing. DC: Methodology, Writing – review & editing. SJ: Conceptualization, Formal analysis, Project administration, Supervision, Writing – review & editing. NL: Conceptualization, Formal analysis, Investigation, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We would like to acknowledge A/Prof Matthew Malone who was involved in the initial process of the systematic review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2024.1486793/full#supplementary-material
References
1. Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas M-E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. (2016) 8:42. doi: 10.1186/s13073-016-0303-2
2. De Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. (2022) 71:1020–32. doi: 10.1136/gutjnl-2021-326789
4. Arora A, Behl T, Sehgal A, Singh S, Sharma N, Bhatia S, et al. Unravelling the involvement of gut microbiota in type 2 diabetes mellitus. Life Sci. (2021) 273:119311. doi: 10.1016/j.lfs.2021.119311
5. Ussar S, Fujisaka S, Kahn CR. Interactions between host genetics and gut microbiome in diabetes and metabolic syndrome. Mol Metab. (2016) 5:795–803. doi: 10.1016/j.molmet.2016.07.004
6. Wu X, He B, Liu J, Feng H, Ma Y, Li D, et al. Molecular insight into gut microbiota and rheumatoid arthritis. Int J Of Mol Sci. (2016) 17:431–1. doi: 10.3390/ijms17030431
7. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. (2015) 6. doi: 10.1038/ncomms7528
8. Karlsson FH, Fåk F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. (2012) 3. doi: 10.1038/ncomms2266
9. Waly M, Mustafa ME, Ali A, Al-Shuhaibi YM, Al-Farsi Y. The global burden of type 2 diabetes: A review. Int J Of Biol Med Res. (2010) 1:326–9. doi: 10.2991/jegh.k.191028.001
10. Yang HT, Liu JK, Xiu WJ, Tian TT, Yang Y, Hou XG, et al. Gut microbiome-based diagnostic model to predict diabetes mellitus. Bioengineered. (2021) 12:12521–34. doi: 10.1080/21655979.2021.2009752
11. Doumatey AP, Adeyemo A, Zhou J, Lei L, Adebamowo SN, Adebamowo C, et al. Gut microbiome profiles are associated with type 2 diabetes in urban africans. Front In Cell And Infection Microbiol. (2020) 10. doi: 10.3389/fcimb.2020.00063
12. Zhao X, Zhang Y, Guo R, Yu W, Zhang F, Wu F, et al. The alteration in composition and function of gut microbiome in patients with type 2 diabetes. J Of Diabetes Res. (2020) 2020. doi: 10.1155/2020/8842651
13. Maskarinec G, Raquinio P, Kristal BS, Setiawan VW, Wilkens LR, Franke AA, et al. The gut microbiome and type 2 diabetes status in the multiethnic cohort. PloS One. (2021) 16:E0250855. doi: 10.1371/journal.pone.0250855
14. Talukdar R, Sarkar P, Jakkampudi A, Sarkar S, Aslam M, Jandhyala M, et al. The gut microbiome in pancreatogenic diabetes differs from that of type 1 and type 2 diabetes. Sci Rep. (2021) 11:10978. doi: 10.1038/s41598-021-90024-w
15. Saleem A, Ikram A, Dikareva E, Lahtinen E, Matharu D, Pajari AM, et al. Unique Pakistani gut microbiota highlights population-specific microbiota signatures of type 2 diabetes mellitus. Gut Microbes. (2022) 14. doi: 10.1080/19490976.2022.2142009
16. Wang J, Li W, Wang C, Wang L, He T, Hu H, et al. Enterotype bacteroides is associated with A high risk in patients with diabetes: A pilot study. J Of Diabetes Res. (2020) 2020. doi: 10.1155/2020/6047145
17. Candela M, Biagi E, Soverini M, Consolandi C, Quercia S, Severgnini M, et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic ma-pi 2 diet. Br J Of Nutr. (2016) 116:80–93. doi: 10.1017/S0007114516001045
18. Guo XJ, Dai SX, Lou JD, Ma XX, Hu XJ, Tu LP, et al. Distribution characteristics of oral microbiota and its relationship with intestinal microbiota in patients with type 2 diabetes mellitus. Front In Endocrinol. (2023) 14:1119201. doi: 10.3389/fendo.2023.1119201
19. Kitten AK, Ryan L, Lee GC, Flores BE, Reveles KR. Gut microbiome differences among mexican americans with and without type 2 diabetes mellitus. PloS One. (2021) 16. doi: 10.1371/journal.pone.0251245
20. Alvarez-Silva C, Kashani A, Hansen TH, Pinna NK, Anjana RM, Dutta A, et al. Trans-ethnic gut microbiota signatures of type 2 diabetes in Denmark and India. Genome Med. (2021) 13. doi: 10.1186/s13073-021-00856-4
21. Bhute SS, Suryavanshi MV, Joshi SM, Yajnik CS, Shouche YS, Ghaskadbi SS. Gut microbial diversity assessment of Indian type-2-diabetics reveals alterations in eubacteria, archaea, and eukaryotes. Front In Microbiol. (2017) 8. doi: 10.3389/fmicb.2017.00214
22. Afolayan AO, Adebusoye LA, Cadmus EO, Ayeni FA. Insights into the gut microbiota of Nigerian elderly with type 2 diabetes and non-diabetic elderly persons. Heliyon. (2020) 6:E03971. doi: 10.1016/j.heliyon.2020.e03971
23. Zhong H, Ren H, Lu Y, Fang C, Hou G, Yang Z, et al. Distinct gut metagenomics and metaproteomics signatures in prediabetics and treatment-naive type 2 diabetics. Ebiomedicine. (2019) 47:373–83. doi: 10.1016/j.ebiom.2019.08.048
24. Lambeth SM, Carson T, Lowe J, Ramaraj T, Leff JW, Luo L, et al. Composition, diversity and abundance of gut microbiome in prediabetes and type 2 diabetes. Diabetes Obes And Metab. (2015) 2:1–7. doi: 10.15436/2376-0949.15.031
25. Zhang Z, Tian T, Chen Z, Liu L, Luo T, Dai J. Characteristics of the gut microbiome in patients with prediabetes and type 2 diabetes. Peerj. (2021) 9. doi: 10.7717/peerj.10952
26. Almugadam BS, Liu Y, Chen SM, Wang CH, Shao CY, Ren BW, et al. Alterations of gut microbiota in type 2 diabetes individuals and the confounding effect of antidiabetic agents. J Of Diabetes Res. (2020) 2020:7253978. doi: 10.1155/2020/7253978
27. Wang G, Lyu Q, Yang T, Cui S, Niu K, Gu R, et al. Association of intestinal microbiota markers and dietary pattern in chinese patients with type 2 diabetes: the henan rural cohort study. Front In Public Health. (2022) 10:1046333. doi: 10.3389/fpubh.2022.1046333
28. Tang LT, Feng L, Cao HY, Shi R, Luo BB, Zhang YB, et al. Comparative study of type 2 diabetes mellitus-associated gut microbiota between the dai and han populations. World Jorunal Of Diabetes. (2023) 14:1766–83. doi: 10.4239/wjd.v14.i12.1766
29. Al Bataineh MT, Dash NR, Lassen PB, Banimfreg BH, Nada AM, Belda E, et al. Revealing links between gut microbiome and its fungal community in type 2 diabetes mellitus among emirati subjects: A pilot study. Sci Rep. (2020) 10. doi: 10.1038/s41598-020-66598-2
30. Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PloS One. (2013) 8:A497. doi: 10.1371/journal.pone.0071108
31. Wang Y, Luo X, Mao X, Tao Y, Ran X, Zhao H, et al. Gut microbiome analysis of type 2 diabetic patients from the chinese minority ethnic groups the uygurs and kazaks. PloS One. (2017) 12. doi: 10.1371/journal.pone.0172774
32. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. (2012) 490:55–60. doi: 10.1038/nature11450
33. Remely M, Dworzak S, Hippe B, Zwielehner J, Aumüller E, Brath H, et al. Abundance and diversity of microbiota in type 2 diabetes and obesity. J Of Diabetes Metab. (2013) 4:2.
34. Li Q, Chang Y, Zhang K, Chen H, Tao S, Zhang Z. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci Rep. (2020) 10. doi: 10.1038/s41598-020-62224-3
35. Fassatoui M, Saffarian A, Mulet C, Jamoussi H, Gamoudi A, Ben Halima Y, et al. Gut microbiota profile and the influence of nutritional status on bacterial distribution in diabetic and healthy Tunisian subjects. Bioscience Rep. (2023) 43. doi: 10.1042/BSR20220803
36. Ahmad A, Yang W, Chen G, Shafiq M, Javed S, Ali Zaidi SS, et al. Analysis of gut microbiota of obese individuals with type 2 diabetes and healthy individuals. PloS One. (2019) 14:E0226372. doi: 10.1371/journal.pone.0226372
37. Larsen N, Vogensen FK, Van Den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS One. (2010) 5:E9085. doi: 10.1371/journal.pone.0009085
38. Zhao L, Lou H, Peng Y, Chen S, Zhang Y, Li X. Comprehensive relationships between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications. Endocrine. (2019) 66:526–37. doi: 10.1007/s12020-019-02103-8
39. Salamon D, Sroka-Oleksiak A, Kapusta P, Szopa M, Mrozińska S, Ludwig-Słomczyńska AH, et al. Characteristics of gut microbiota in adult patients with type 1 and type 2 diabetes based on next−Generation sequencing of the 16s rrna gene fragment. Polish Arch Of Internal Med. (2018) 128:336–43. doi: 10.20452/pamw.4246
40. Kwan SY, Sabotta CM, Joon A, Wei P, Petty LE, Below JE, et al. Gut microbiome alterations associated with diabetes in mexican americans in south texas. Msystems. (2022) 7. doi: 10.1128/msystems.00033-22
41. Gaike AH, Paul D, Bhute S, Dhotre DP, Pande P, Upadhyaya S, et al. The gut microbial diversity of newly diagnosed diabetics but not of prediabetics is significantly different from that of healthy nondiabetics. Msystems. (2020) 5. doi: 10.1128/mSystems.00578-19
42. Gravdal K, Kirste KH, Grzelak K, Kirubakaran GT, Leissner P, Saliou A, et al. Exploring the gut microbiota in patients with pre-diabetes and treatment naïve diabetes type 2 - A pilot study. BMC Endocrine Disord. (2023) 23:179. doi: 10.1186/s12902-023-01432-0
43. Fassatoui M, Lopez-Siles M, Diaz-Rizzolo DA, Jmel H, Naouali C, Abdessalem G, et al. Gut microbiota imbalances in Tunisian participants with type 1 and type 2 diabetes mellitus. Bioscience Rep. (2019) 39. doi: 10.1042/BSR20182348
44. Gradisteanu Pircalabioru G, Liaw J, Gundogdu O, Corcionivoschi N, Ilie I, Oprea L, et al. Effects of the lipid profile, type 2 diabetes and medication on the metabolic syndrome-associated gut microbiome. Int J Of Mol Sci. (2022) 23. doi: 10.3390/ijms23147509
45. Hoang HT, Le DH, Le TTH, Nguyen TTN, Chu HH, Nguyen NT. Metagenomic 16s rdna amplicon data of microbial diversity of guts in Vietnamese humans with type 2 diabetes and nondiabetic adults. Data Brief. (2021) 34:106690. doi: 10.1016/j.dib.2020.106690
46. Wang TY, Zhang XQ, Chen AL, Zhang J, Lv BH, Ma MH, et al. A comparative study of microbial community and functions of type 2 diabetes mellitus patients with obesity and healthy people. Appl Microbiol And Biotechnol. (2020) 104:7143–53. doi: 10.1007/s00253-020-10689-7
47. Pushpanathan P, Srikanth P, Seshadri KG, Selvarajan S, Pitani RS, Kumar TD, et al. Gut microbiota in type 2 diabetes individuals and correlation with monocyte chemoattractant protein1 and interferon gamma from patients attending A tertiary care centre in chennai, India. Indian J Of Endocrinol And Metab. (2016) 20:523–30. doi: 10.4103/2230-8210.183474
48. Li L, Li C, Lv M, Hu Q, Guo L, Xiong D. Correlation between alterations of gut microbiota and mir-122-5p expression in patients with type 2 diabetes mellitus. Ann Of Trans Med. (2020) 8. doi: 10.21037/atm-20-6717
49. Ghaemi F, Fateh A, Sepahy AA, Zangeneh M, Ghanei M, Siadat SD. Intestinal microbiota composition in Iranian diabetic, pre-diabetic and healthy individuals. J Of Diabetes And Metab Disord. (2020) 19:1199–203. doi: 10.1007/s40200-020-00625-x
50. Chen PC, Chien YW, Yang SC. The alteration of gut microbiota in newly diagnosed type 2 diabetic patients. Nutrition. (2019) 63-64:51–6. doi: 10.1016/j.nut.2018.11.019
51. Sato J, Kanazawa A, Ikeda F, Yoshihara T, Goto H, Abe H, et al. Gut dysbiosis and detection of “Live gut bacteria” In blood of Japanese patients with type 2 diabetes. Diabetes Care. (2014) 37:2343–50. doi: 10.2337/dc13-2817
52. Lê KA, Li Y, Xu X, Yang W, Liu T, Zhao X, et al. Alterations in fecal lactobacillus and bifidobacterium species in type 2 diabetic patients in southern China population. Front In Physiol. (2012) 3:496. doi: 10.3389/fphys.2012.00496
53. Wang X, Xu X, Xia Y. Further analysis reveals new gut microbiome markers of type 2 diabetes mellitus. Antonie Van Leeuwenhoek. (2017) 110:445–53. doi: 10.1007/s10482-016-0805-3
54. Sedighi M, Razavi S, Navab-Moghadam F, Khamseh ME, Alaei-Shahmiri F, Mehrtash A, et al. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microbial Pathogenesis. (2017) 111:362–9. doi: 10.1016/j.micpath.2017.08.038
55. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. (2015) 528:262–6. doi: 10.1038/nature15766
56. Diener C, Reyes-Escogido MDL, Jimenez-Ceja LM, Matus M, Gomez-Navarro CM, Chu ND, et al. Progressive shifts in the gut microbiome reflect prediabetes and diabetes development in A treatment-naive mexican cohort. Front In Endocrinol. (2021) 11. doi: 10.3389/fendo.2020.602326
57. Demirci M, Taner Z, Keskin FE, Ozyazar M, Kiraz N, Kocazeybek BS, et al. Similar bacterial signatures in the gut microbiota of type 1 and type 2 diabetes patients and its association with G protein-coupled receptor 41 and 43 gene expression. J Of Diabetes And Metab Disord. (2022) 21:1359–68. doi: 10.1007/s40200-022-01068-2
58. Remely M, Hippe B, Zanner J, Aumueller E, Brath H, Haslberger AG. Gut microbiota of obese, type 2 diabetic individuals is enriched in faecalibacterium prausnitzii, akkermansia muciniphila and peptostreptococcus anaerobius after weight loss. Endocrine Metab And Immune Disord - Drug Targets. (2016) 16:99–106. doi: 10.2174/1871530316666160831093813
59. Tamura A, Murabayashi M, Nishiya Y, Mizushiri S, Hamaura K, Ito R, et al. Interrelations between gut microbiota composition, nutrient intake and diabetes status in an adult Japanese population. J Of Clin Med. (2022) 11. doi: 10.3390/jcm11113216
60. Wu X, Ma C, Han L, Nawaz M, Gao F, Zhang X, et al. Molecular characterisation of the faecal microbiota in patients with type ii diabetes. Curr Microbiol. (2010) 61:69–78. doi: 10.1007/s00284-010-9582-9
61. Inoue R, Ohue-Kitano R, Tsukahara T, Tanaka M, Masuda S, Inoue T, et al. Prediction of functional profiles of gut microbiota from 16s rrna metagenomic data provides A more robust evaluation of gut dysbiosis occurring in Japanese type 2 diabetic patients. J Of Clin Biochem And Nutr. (2017) 61:217–21. doi: 10.3164/jcbn.17-44
62. Navab-Moghadam F, Sedighi M, Khamseh ME, Alaei-Shahmiri F, Talebi M, Razavi S, et al. The association of type ii diabetes with gut microbiota composition. Microbial Pathogenesis. (2017) 110:630–6. doi: 10.1016/j.micpath.2017.07.034
63. Zeng Z, Guo X, Zhang J, Yuan Q, Chen S. Lactobacillus paracasei modulates the gut microbiota and improves inflammation in type 2 diabetic rats. Food Funct. (2021) 12:6809–20. doi: 10.1039/D1FO00515D
64. Jang YJ, Kim WK, Han DH, Lee K, Ko G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes. (2019) 10:696–711. doi: 10.1080/19490976.2019.1589281
65. Ejtahed HS, Mohtadi-Nia J, Homayouni-Rad A, Niafar M, Asghari-Jafarabadi M, Mofid V. Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition. (2012) 28:539–43. doi: 10.1016/j.nut.2011.08.013
66. Mohamadshahi M, Veissi M, Haidari F, Shahbazian H, Kaydani GA, Mohammadi F. Effects of probiotic yogurt consumption on inflammatory biomarkers in patients with type 2 diabetes. Bioimpacts. (2014) 4:83–8. doi: 10.5681/bi.2014.007
67. Devanga Ragupathi NK, Muthuirulandi Sethuvel DP, Inbanathan FY, Veeraraghavan B. Accurate differentiation of escherichia coli and shigella serogroups: challenges and strategies. New Microbes And New Infections. (2018) 21:58–62. doi: 10.1016/j.nmni.2017.09.003
68. Wang Y, Ye X, Ding D, Lu Y. Characteristics of the intestinal flora in patients with peripheral neuropathy associated with type 2 diabetes. J Of Int Med Res. (2020) 48:300060520936806. doi: 10.1177/0300060520936806
69. Du Y, Neng Q, Li Y, Kang Y, Guo L, Huang X, et al. Gastrointestinal autonomic neuropathy exacerbates gut microbiota dysbiosis in adult patients with type 2 diabetes mellitus. Front In Cell And Infection Microbiol. (2021) 11:804733. doi: 10.3389/fcimb.2021.804733
70. Das T, Jayasudha R, Chakravarthy S, Prashanthi GS, Bhargava A, Tyagi M, et al. Alterations in the gut bacterial microbiome in people with type 2 diabetes mellitus and diabetic retinopathy. Sci Rep. (2021) 11. doi: 10.1038/s41598-021-82538-0
71. Tao S, Li L, Li L, Liu Y, Ren Q, Shi M, et al. Understanding the gut-kidney axis among biopsy-proven diabetic nephropathy, type 2 diabetes mellitus and healthy controls: an analysis of the gut microbiota composition. Acta Diabetologica. (2019) 56:581–92. doi: 10.1007/s00592-019-01316-7
72. Radzieta M, Malone M, Ahmad M, Dickson HG, Schwarzer S, Jensen SO, et al. Metatranscriptome sequencing identifies escherichia are major contributors to pathogenic functions and biofilm formation in diabetes related foot osteomyelitis. Front In Microbiol. (2022) 13:956332. doi: 10.3389/fmicb.2022.956332
73. Cao TTB, Wu KC, Hsu JL, Chang CS, Chou C, Lin CY, et al. Effects of non-insulin anti-hyperglycemic agents on gut microbiota: A systematic review on human and animal studies. Frontiners In Endocrinol. (2020) 11:573891. doi: 10.3389/fendo.2020.573891
74. Van Hul M, Le Roy T, Prifti E, Dao MC, Paquot A, Zucker J-D, et al. From correlation to causality: the case of subdoligranulum. Gut Microbes. (2020) 12:1849998. doi: 10.1080/19490976.2020.1849998
75. Chriswell ME, Lefferts AR, Clay MR, Hsu AR, Seifert J, Feser ML, et al. Clonal iga and igg autoantibodies from individuals at risk for rheumatoid arthritis identify an arthritogenic strain of subdoligranulum. Sci Translation Med. (2022) 14:Eabn5166. doi: 10.1126/scitranslmed.abn5166
76. Carasi P, Racedo SM, Jacquot C, Elie AM, Serradell ML, Urdaci MC. Enterococcus durans ep1 A promising anti-inflammatory probiotic able to stimulate siga and to increase faecalibacterium prausnitzii abundance. Front In Immunol. (2017) 8:88. doi: 10.3389/fimmu.2017.00088
77. Steck N, Hoffmann M, Sava IG, Kim SC, Hahne H, Tonkonogy SL, et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology. (2011) 141:959–71. doi: 10.1053/j.gastro.2011.05.035
78. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its fada adhesin. Cell Host Microbe. (2013) 14:195–206. doi: 10.1016/j.chom.2013.07.012
79. Zhang L, Lu Q-Y, Wu H, Cheng Y-L, Kang J, Xu Z-G. The intestinal microbiota composition in early and late stages of diabetic kidney disease. Microbiol Spectr. (2023) 11:E00382–23. doi: 10.1128/spectrum.00382-23
80. Zhang L, Qin Q, Liu M, Zhang X, He F, Wang G. Akkermansia muciniphila can reduce the damage of gluco/lipotoxicity, oxidative stress and inflammation, and normalize intestine microbiota in streptozotocin-induced diabetic rats. Pathog And Dis. (2018) 76. doi: 10.1093/femspd/fty028
81. Perraudeau F, Mcmurdie P, Bullard J, Cheng A, Cutcliffe C, Deo A, et al. Improvements to postprandial glucose control in subjects with type 2 diabetes: A multicenter, double blind, randomized placebo-controlled trial of A novel probiotic formulation. BMJ Open Diabetes Res &Amp; Care. (2020) 8:E001319. doi: 10.1136/bmjdrc-2020-001319
82. Ghoddusi HB, Tamime AY. Microflora of the intestine | Biology of bifidobacteria. In: Batt CA, Tortorello MLeditors. Encyclopedia of food microbiology, 2nd ed. Academic Press, Oxford (2014).
83. Moroti C, Souza Magri LF, De Rezende Costa M, Cavallini DC, Sivieri K. Effect of the consumption of A new symbiotic shake on glycemia and cholesterol levels in elderly people with type 2 diabetes mellitus. Lipids In Health And Dis. (2012) 11:29. doi: 10.1186/1476-511X-11-29
84. Le TK, Hosaka T, Nguyen TT, Kassu A, Dang TO, Tran HB, et al. Bifidobacterium species lower serum glucose, increase expressions of insulin signaling proteins, and improve adipokine profile in diabetic mice. Biomed Res. (2015) 36:63–70. doi: 10.2220/biomedres.36.63
85. Moya-Pérez A, Neef A, Sanz Y. Bifidobacterium pseudocatenulatum cect 7765 reduces obesity-associated inflammation by restoring the lymphocyte-macrophage balance and gut microbiota structure in high-fat diet-fed mice. PloS One. (2015) 10:E0126976. doi: 10.1371/journal.pone.0126976
86. Ling X, Linglong P, Weixia D, Hong W. Protective effects of bifidobacterium on intestinal barrier function in lps-induced enterocyte barrier injury of caco-2 monolayers and in A rat nec model. PloS One. (2016) 11:E0161635. doi: 10.1371/journal.pone.0161635
87. Chang YC, Ching YH, Chiu CC, Liu JY, Hung SW, Huang WC, et al. Tlr2 and interleukin-10 are involved in bacteroides fragilis-mediated prevention of dss-induced colitis in gnotobiotic mice. PloS One. (2017) 12:E0180025. doi: 10.1371/journal.pone.0180025
88. Wrzosek L, Miquel S, Noordine M-L, Bouet S, Chevalier-Curt MJ, Robert V, et al. Bacteroides thetaiotaomicron and faecalibacterium prausnitziiinfluence the production of mucus glycans and the development of goblet cells in the colonic epithelium of A gnotobiotic model rodent. BMC Biol. (2013) 11:61. doi: 10.1186/1741-7007-11-61
89. Wang H-B, Wang P-Y, Wang X, Wan Y-L, Liu Y-C. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein claudin-1 transcription. Digestive Dis And Sci. (2012) 57:3126–35. doi: 10.1007/s10620-012-2259-4
90. Jacobson AN, Choudhury BP, Fischbach MA. The biosynthesis of lipooligosaccharide from bacteroides thetaiotaomicron. Mbio. (2018) 9. doi: 10.1128/mBio.02289-17
91. Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in european women with normal, impaired and diabetic glucose control. Nature. (2013) 498:99–103. doi: 10.1038/nature12198
92. Zhu C, Song K, Shen Z, Quan Y, Tan B, Luo W, et al. Roseburia Intestinalis inhibits interleukin−17 excretion and promotes regulatory T cells differentiation in colitis. Mol Med Rep. (2018) 17:7567–74. doi: 10.3892/mmr.2018.8833
93. Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. (2012) 143:913–6.E7. doi: 10.1053/j.gastro.2012.06.031
94. Kallassy J, Gagnon E, Rosenberg D, Silbart LK, Mcmanus SA. Strains of Faecalibacterium prausnitzii and its extracts reduce blood glucose levels, percent HbA1c, and improve glucose tolerance without causing hypoglycemic side effects in diabetic and prediabetic mice. BMJ Open Diabetes Res Care. (2023) 11:E003101. doi: 10.1136/bmjdrc-2022-003101
95. Larsen JM. The immune response to prevotella bacteria in chronic inflammatory disease. Immunology. (2017) 151:363–74. doi: 10.1111/imm.2017.151.issue-4
96. Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella. Cell Metab. (2015) 22:971–82. doi: 10.1016/j.cmet.2015.10.001
97. Marietta EV, Murray JA, Luckey DH, Jeraldo PR, Lamba A, Patel R, et al. Suppression of inflammatory arthritis by human gut-derived prevotella histicola in humanized mice. Arthritis Rheumatol. (2016) 68:2878–88. doi: 10.1002/art.v68.12
98. Adachi K, Sugiyama T, Yamaguchi Y, Tamura Y, Izawa S, Hijikata Y, et al. Gut microbiota disorders cause type 2 diabetes mellitus and homeostatic disturbances in gut-related metabolism in Japanese subjects. J Of Clin Biochem And Nutr. (2019) 64:231–8. doi: 10.3164/jcbn.18-101
99. Guerreiro CS, Calado Â, Sousa J, Fonseca JE. Diet, microbiota, and gut permeability-the unknown triad in rheumatoid arthritis. Front In Med. (2018) 5:349. doi: 10.3389/fmed.2018.00349
100. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. Ebiomedicine. (2020) 51. doi: 10.1016/j.ebiom.2019.11.051
101. Egshatyan L, Kashtanova D, Popenko A, Tkacheva O, Tyakht A, Alexeev D, et al. Gut microbiota and diet in patients with different glucose tolerance. Endocrine Connections. (2016) 5:1–9. doi: 10.1530/EC-15-0094
102. Cox AJ, Zhang P, Bowden DW, Devereaux B, Davoren PM, Cripps AW, et al. Increased intestinal permeability as A risk factor for type 2 diabetes. Diabetes Metab. (2017) 43:163–6. doi: 10.1016/j.diabet.2016.09.004
103. Camargo A, Jimenez-Lucena R, Alcala-Diaz JF, Rangel-Zuñiga OA, Garcia-Carpintero S, Lopez-Moreno J, et al. Postprandial endotoxemia may influence the development of type 2 diabetes mellitus: from the cordioprev study. Clin Nutr. (2019) 38:529–38. doi: 10.1016/j.clnu.2018.03.016
104. Lee B, Moon KM, Kim CY. Tight junction in the intestinal epithelium: its association with diseases and regulation by phytochemicals. J Of Immunol Res. (2018) 2018:2645465. doi: 10.1155/2018/2645465
105. Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T, et al. Bacteroides vulgatus and bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation. (2018) 138:2486–98. doi: 10.1161/CIRCULATIONAHA.118.033714
106. Sofi MH, Wu Y, Ticer T, Schutt S, Bastian D, Choi HJ, et al. A single strain of bacteroides fragilis protects gut integrity and reduces gvhd. JCI Insight. (2021) 6. doi: 10.1172/jci.insight.136841
107. Zhou Q, Shen B, Huang R, Liu H, Zhang W, Song M, et al. Bacteroides fragilis strain zy-312 promotes intestinal barrier integrity via upregulating the stat3 pathway in A radiation-induced intestinal injury mouse Model. Front In Nutr. (2022) 9:1063699. doi: 10.3389/fnut.2022.1063699
108. He Q, Niu M, Bi J, Du N, Liu S, Yang K, et al. Protective effects of A new generation of probiotic bacteroides fragilis against colitis in vivo and in vitro. Sci Rep. (2023) 13:15842. doi: 10.1038/s41598-023-42481-8
109. Mohebali N, Ekat K, Kreikemeyer B, Breitrück A. Barrier protection and recovery effects of gut commensal bacteria on differentiated intestinal epithelial cells in vitro. Nutrients. (2020) 12. doi: 10.3390/nu12082251
110. Hsieh CY, Osaka T, Moriyama E, Date Y, Kikuchi J, Tsuneda S. Strengthening of the intestinal epithelial tight junction by bifidobacterium bifidum. Physiol Rep. (2015) 3. doi: 10.14814/phy2.12327
111. Roberts JL, Liu G, Darby TM, Fernandes LM, Diaz-Hernandez ME, Jones RM, et al. Bifidobacterium adolescentis supplementation attenuates fracture-induced systemic sequelae. Biomedicine Pharmacotherapy. (2020) 132:110831. doi: 10.1016/j.biopha.2020.110831
112. Caviglia GP, Tucci A, Pellicano R, Fagoonee S, Rosso C, Abate ML, et al. Clinical response and changes of cytokines and zonulin levels in patients with diarrhoea-predominant irritable bowel syndrome treated with bifidobacterium longum es1 for 8 or 12 weeks: A preliminary report. J Of Clin Med. (2020) 9. doi: 10.3390/jcm9082353
113. Xu J, Liang R, Zhang W, Tian K, Li J, Chen X, et al. Faecalibacterium prausnitzii-derived microbial Anti-inflammatory molecule regulates intestinal integrity in diabetes mellitus mice via modulating tight junction protein expression. J Of Diabetes. (2020) 12:224–36. doi: 10.1111/1753-0407.12986
114. Chelakkot C, Choi Y, Kim DK, Park HT, Ghim J, Kwon Y, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp And Mol Med. (2018) 50:E450. doi: 10.1038/emm.2017.282
115. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. (2017) 23:107–13. doi: 10.1038/nm.4236
116. Pedersen C, Ijaz UZ, Gallagher E, Horton F, Ellis RJ, Jaiyeola E, et al. Fecal enterobacteriales enrichment is associated with increased in vivo intestinal permeability in humans. Physiolology Rep. (2018) 6:E13649. doi: 10.14814/phy2.13649
117. Pesce M, D’alessandro A, Borrelli O, Gigli S, Seguella L, Cuomo R, et al. Endocannabinoid-related compounds in gastrointestinal diseases. J Of Cell And Mol Med. (2018) 22:706–15. doi: 10.1111/jcmm.2018.22.issue-2
118. Muccioli GG, Naslain D, Bäckhed F, Reigstad CS, Lambert DM, Delzenne NM, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol. (2010) 6:392. doi: 10.1038/msb.2010.46
119. Malek A, Ahmadi Badi S, Karimi G, Bizouarn T, Irian S, Siadat SD. The effect of bacteroides fragilis and its postbiotics on the expression of genes involved in the endocannabinoid system and intestinal epithelial integrity in caco-2 cells. J Of Diabetes Metab Disord. (2023) 22:1417–24. doi: 10.1007/s40200-023-01264-8
120. Kumawat VS, Kaur G. Therapeutic potential of cannabinoid receptor 2 in the treatment of diabetes mellitus and its complications. Eur J Of Pharmacol. (2019) 862:172628. doi: 10.1016/j.ejphar.2019.172628
121. Vallianou NG, Stratigou T, Tsagarakis S. Microbiome and diabetes: where are we now? Diabetes Res And Clin Pract. (2018) 146:111–8. doi: 10.1016/j.diabres.2018.10.008
122. Fusco W, Lorenzo MB, Cintoni M, Porcari S, Rinninella E, Kaitsas F, et al. Short-chain fatty-acid-producing bacteria: key components of the human gut microbiota. Nutrients. (2023) 15. doi: 10.3390/nu15092211
123. Freeland KR, Wolever TM. Acute effects of intravenous and rectal acetate on glucagon-like peptide-1, peptide yy, ghrelin, adiponectin and tumour necrosis factor-alpha. Br J Of Nutr. (2010) 103:460–6. doi: 10.1017/S0007114509991863
124. Canfora EE, van der Beek CM, Jocken JWE, Goossens GH, Holst JJ, Olde Damink SWM, et al. Colonic infusions of short-chain fatty acid mixtures promote energy metabolism in overweight/obese men: A randomized crossover trial. Sci Rep. (2017) 7. doi: 10.1038/s41598-017-02546-x
125. Van Der Beek CM, Canfora EE, Lenaerts K, Troost FJ, Damink S, Holst JJ, et al. Distal, not proximal, colonic acetate infusions promote fat oxidation and improve metabolic markers in overweight/obese men. Clin Sci. (2016) 130:2073–82. doi: 10.1042/CS20160263
126. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. (2008) 57:1470–81. doi: 10.2337/db07-1403
127. Tokarz VL, Macdonald PE, Klip A. The cell biology of systemic insulin function. J Of Cell Biol. (2018) 217:2273–89. doi: 10.1083/jcb.201802095
128. Lee C, Kim BG, Kim JH, Chun J, Im JP, Kim JS. Sodium butyrate inhibits the nf-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an il-10 independent manner. Int Immunopharmacol. (2017) 51:47–56. doi: 10.1016/j.intimp.2017.07.023
129. Deleu S, Machiels K, Raes J, Verbeke K, Vermeire S. Short chain fatty acids and its producing organisms: an overlooked therapy for ibd? Ebiomedicine. (2021) 66. doi: 10.1016/j.ebiom.2021.103293
130. Zheng Y, Yue C, Zhang H, Chen H, Liu Y, Li J. Deoxycholic acid and lithocholic acid alleviate liver injury and inflammation in mice with klebsiella pneumoniae-induced liver abscess and bacteremia. J Of Inflammation Res. (2021) 14:777–89. doi: 10.2147/JIR.S298495
131. Mantovani A, Dalbeni A, Peserico D, Cattazzo F, Bevilacqua M, Salvagno GL, et al. Plasma bile acid profile in patients with and without type 2 diabetes. Metabolites. (2021) 11. doi: 10.3390/metabo11070453
132. Zhu W, Wang S, Dai H, Xuan L, Deng C, Wang T, et al. Serum total bile acids associate with risk of incident type 2 diabetes and longitudinal changes in glucose-related metabolic traits. J Diabetes. (2020) 12:616–25. doi: 10.1111/1753-0407.13040
133. Lee S-G, Lee Y-H, Choi E, Cho Y, Kim J-H. Fasting serum bile acids concentration is associated with insulin resistance independently of diabetes status. Clin Chem And Lab Med (Cclm). (2019) 57:1218–28. doi: 10.1515/cclm-2018-0741
134. Lu Q, Chen J, Jiang L, Geng T, Tian S, Liao Y, et al. Gut microbiota-derived secondary bile acids, bile acids receptor polymorphisms, and risk of cardiovascular disease in individuals with newly diagnosed type 2 diabetes: A cohort study. Am J Of Clin Nutr. (2024) 119:324–32. doi: 10.1016/j.ajcnut.2023.08.023
135. Xu J, Wang N, Yang L, Zhong J, Chen M. Intestinal flora and bile acid interactions impact the progression of diabetic kidney disease. Frontiners In Endocrinol. (2024) 15:1441415. doi: 10.3389/fendo.2024.1441415
136. Wahlström A, Sayin SI, Marschall H-U, Bäckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. (2016) 24:41–50. doi: 10.1016/j.cmet.2016.05.005
137. Drucker DJ. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab. (2018) 27:740–56. doi: 10.1016/j.cmet.2018.03.001
138. Xie C, Jiang C, Shi J, Gao X, Sun D, Sun L, et al. An intestinal farnesoid X receptor-ceramide signaling axis modulates hepatic gluconeogenesis in mice. Diabetes. (2017) 66:613–26. doi: 10.2337/db16-0663
139. Zheng X, Chen T, Jiang R, Zhao A, Wu Q, Kuang J, et al. Hyocholic acid species improve glucose homeostasis through A distinct tgr5 and fxr signaling mechanism. Cell Metab. (2021) 33:791–803.E7. doi: 10.1016/j.cmet.2020.11.017
140. Ploton M, Mazuy C, Gheeraert C, Dubois V, Berthier A, Dubois-Chevalier J, et al. The nuclear bile acid receptor fxr is A pka- and foxa2-sensitive activator of fasting hepatic gluconeogenesis. J Hepatol. (2018) 69:1099–109. doi: 10.1016/j.jhep.2018.06.022
141. Trabelsi M-S, Daoudi M, Prawitt J, Ducastel S, Touche V, Sayin SI, et al. Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine L cells. Nat Commun. (2015) 6. doi: 10.1038/ncomms8629
142. Cariou B, Van Harmelen K, Duran-Sandoval D, Van Dijk TH, Grefhorst A, Abdelkarim M, et al. The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J Of Biol Chem. (2006) 281:11039–49. doi: 10.1074/jbc.M510258200
143. Sonne DP. Mechanisms in endocrinology: fxr signalling: A novel target in metabolic diseases. Eur J Of Endocrinol. (2021) 184:R193–205. doi: 10.1530/EJE-20-1410
144. Murakami M, Une N, Nishizawa M, Suzuki S, Ito H, Horiuchi T. Incretin secretion stimulated by ursodeoxycholic acid in healthy subjects. Springer Plus. (2013) 2:20. doi: 10.1186/2193-1801-2-20
145. Mahmoud AA, Elshazly SM. Ursodeoxycholic acid ameliorates fructose-induced metabolic syndrome in rats. PloS One. (2014) 9:E106993. doi: 10.1371/journal.pone.0106993
146. Mooranian A, Negrulj R, Al-Salami H, Morahan G, Jamieson E. Designing anti-diabetic β-cells microcapsules using polystyrenic sulfonate, polyallylamine, and A tertiary bile acid: morphology, bioenergetics, and cytokine analysis. Biotechnol Prog. (2016) 32:501–9. doi: 10.1002/btpr.2223
147. Cariou B, Staels B. Fxr: A promising target for the metabolic syndrome? Trends In Pharmacol Sci. (2007) 28:236–43. doi: 10.1016/j.tips.2007.03.002
148. Wu T, Bound MJ, Standfield SD, Jones KL, Horowitz M, Rayner CK. Effects of taurocholic acid on glycemic, glucagon-like peptide-1, and insulin responses to small intestinal glucose infusion in healthy humans. J Of Clin Endocrinol Metab. (2013) 98:E718–22. doi: 10.1210/jc.2012-3961
149. Adrian TE, Gariballa S, Parekh KA, Thomas SA, Saadi H, Al Kaabi J, et al. Rectal taurocholate increases L cell and insulin secretion, and decreases blood glucose and food intake in obese type 2 diabetic volunteers. Diabetologia. (2012) 55:2343–7. doi: 10.1007/s00125-012-2593-2
150. Hou Y, Zhai X, Wang X, Wu Y, Wang H, Qin Y, et al. Research progress on the relationship between bile acid metabolism and type 2 diabetes mellitus. Diabetol Metab Syndrome. (2023) 15:235. doi: 10.1186/s13098-023-01207-6
151. Chen Y, Shen X, Ma T, Yu X, Kwok L-Y, Li Y, et al. Adjunctive probio-X treatment enhances the therapeutic effect of A conventional drug in managing type 2 diabetes mellitus by promoting short-chain fatty acid-producing bacteria and bile acid pathways. Am Soc For Microbiol. (2023) 8:E01300–22. doi: 10.1128/msystems.01300-22
152. Liu L, Dong W, Wang S, Zhang Y, Liu T, Xie R, et al. Deoxycholic acid disrupts the intestinal mucosal barrier and promotes intestinal tumorigenesis. Food Funct. (2018) 9:5588–97. doi: 10.1039/C8FO01143E
153. Agus A, Clément K, Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. (2021) 70:1174–82. doi: 10.1136/gutjnl-2020-323071
154. Huang D, Xiong M, Xu X, Wu X, Xu J, Cai X, et al. Bile acids elevated by high-fat feeding induce endoplasmic reticulum stress in intestinal stem cells and contribute to mucosal barrier damage. Biochem And Biophys Res Commun. (2020) 529:289–95. doi: 10.1016/j.bbrc.2020.05.226
155. Yao B, He J, Yin X, Shi Y, Wan J, Tian Z. The protective effect of lithocholic acid on the intestinal epithelial barrier is mediated by the vitamin D receptor via A sirt1/nrf2 and nf-κb dependent mechanism in caco-2 cells. Toxicol Lett. (2019) 316:109–18. doi: 10.1016/j.toxlet.2019.08.024
156. Zhou C, Wang Y, Li C, Xie Z, Dai L. Amelioration of colitis by A gut bacterial consortium producing anti-inflammatory secondary bile acids. Microbiol Spectr. (2023) 11:E03330–22. doi: 10.1128/spectrum.03330-22
157. Ward JBJ, Lajczak NK, Kelly OB, O’dwyer AM, Giddam AK, Franco P, et al. Ursodeoxycholic acid and lithocholic acid exert anti-inflammatory actions in the colon. Am J Of Physiol Gastrointestinal And Liver Physiol. (2017) 312:G550–8. doi: 10.1152/ajpgi.00256.2016
158. Gao R, Meng X, Xue Y, Mao M, Liu Y, Tian X, et al. Bile acids-gut microbiota crosstalk contributes to the improvement of type 2 diabetes mellitus. Front In Pharmacol. (2022) 13:1027212. doi: 10.3389/fphar.2022.1027212
159. Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends In Immunol. (2004) 25:4–7. doi: 10.1016/j.it.2003.10.013
160. Malo MS, Moaven O, Muhammad N, Biswas B, Alam SN, Economopoulos KP, et al. Intestinal alkaline phosphatase promotes gut bacterial growth by reducing the concentration of luminal nucleotide triphosphates. Am J Of Physiolology | Gastrointestinal And Liver Physiol. (2014) 306:G826–38. doi: 10.1152/ajpgi.00357.2013
161. Fawley J, Gourlay DM. Intestinal alkaline phosphatase: A summary of its role in clinical disease. J Of Surg Res. (2016) 202:225–34. doi: 10.1016/j.jss.2015.12.008
162. Everard A, Geurts L, Van Roye M, Delzenne NM, Cani PD. Tetrahydro iso-alpha acids from hops improve glucose homeostasis and reduce body weight gain and metabolic endotoxemia in high-fat diet-fed mice. PloS One. (2012) 7:E33858. doi: 10.1371/journal.pone.0033858
163. Malo MS. A high level of intestinal alkaline phosphatase is protective against type 2 diabetes mellitus irrespective of obesity. Ebiomedicine. (2015) 2:2016–23. doi: 10.1016/j.ebiom.2015.11.027
164. Santos GM, Ismael S, Morais J, Araújo JR, Faria A, Calhau C, et al. Intestinal alkaline phosphatase: A review Of This Enzyme role in the intestinal barrier function. Microorganisms. (2022) 10. doi: 10.3390/microorganisms10040746
165. Dong Y, Liao W, Tang J, Fei T, Gai Z, Han M. Bifidobacterium bla80 mitigates colitis by altering gut microbiota and alleviating inflammation. Amb Express. (2022) 12:67. doi: 10.1186/s13568-022-01411-z
166. Shen Z, Zhu C, Quan Y, Yang J, Yuan W, Yang Z, et al. Insights into roseburia intestinalis which alleviates experimental colitis pathology by inducing anti-inflammatory responses. J Of Gastroenterol And Hepatol. (2018) 33:1751–60. doi: 10.1111/jgh.2018.33.issue-10
167. Qiu X, Zhang M, Yang X, Hong N, Yu C. Faecalibacterium prausnitzii upregulates regulatory T cells and anti-inflammatory cytokines in treating tnbs-induced colitis. J Of Crohn’s And Colitis. (2013) 7:E558–68. doi: 10.1016/j.crohns.2013.04.002
168. Cohen-Poradosu R, Mcloughlin RM, Lee JC, Kasper DL. Bacteroides fragilis-stimulated interleukin-10 contains expanding disease. J Of Infect Dis. (2011) 204:363–71. doi: 10.1093/infdis/jir277
169. Seregin SS, Golovchenko N, Schaf B, Chen J, Pudlo NA, Mitchell J, et al. Nlrp6 protects il10(-/-) mice from colitis by limiting colonization of akkermansia muciniphila. Cell Rep. (2017) 19:733–45. doi: 10.1016/j.celrep.2017.03.080
170. Hoffmann TW, Pham HP, Bridonneau C, Aubry C, Lamas B, Martin-Gallausiaux C, et al. Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. Isme J. (2016) 10:460–77. doi: 10.1038/ismej.2015.127
171. Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of crohn disease patients. Proc Of Natl Acad Of Sci Of United States Of America. (2008) 105:16731–6. doi: 10.1073/pnas.0804812105
Keywords: gut microbiota, diabetes, gut dysbiosis, diabetes mellitus, systematic review
Citation: Chong S, Lin M, Chong D, Jensen S and Lau NS (2025) A systematic review on gut microbiota in type 2 diabetes mellitus. Front. Endocrinol. 15:1486793. doi: 10.3389/fendo.2024.1486793
Received: 26 August 2024; Accepted: 18 December 2024;
Published: 17 January 2025.
Edited by:
Nigel Irwin, Ulster University, United KingdomReviewed by:
Jerzy Beltowski, Medical University of Lublin, PolandWendong Huang, City of Hope, United States
Copyright © 2025 Chong, Lin, Chong, Jensen and Lau. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Serena Chong, c2VyZW5hLmNob25nMUBoZWFsdGgubnN3Lmdvdi5hdQ==
†ORCID: Serena Chong, orcid.org/0000-0003-1829-5540
Mike Lin, orcid.org/0000-0002-0388-4193
Slade Jensen, orcid.org/0000-0003-4489-589X
Namson S. Lau, orcid.org/0000-0002-9400-8470
 Serena Chong
Serena Chong Mike Lin3,4†
Mike Lin3,4†