- 1The First Affiliated Hospital, Guangdong Pharmaceutical University, Guangzhou, China
- 2School of Basic Medicine, Hubei University of Arts and Science, Xiangyang, China
Obesity is increasing globally and is closely associated with a range of metabolic disorders, including metabolic associated fatty liver disease, diabetes, and cardiovascular diseases. An effective strategy to combat obesity involves stimulating brown and beige adipocyte thermogenesis, which significantly enhances energy expenditure. Recent research has underscored the vital role of PRDM16 in the development and functionality of thermogenic adipocytes. Consequently, PRDM16 has been identified as a potential therapeutic target for obesity and its related metabolic disorders. This review comprehensively examines various studies that focus on combating obesity by directly targeting PRDM16 in adipose tissue.
1 Introduction
Obesity is becoming a global epidemic and is closely associated with various chronic diseases, such as type 2 diabetes, cardiovascular disorders, and hypertension (1). In recent years, non-shivering thermogenesis in adipose tissues has emerged as a promising therapeutic target for obesity and its related comorbidities. Adipocyte thermogenesis occurs when white adipocytes or beige precursor cells transform into beige adipocytes in white adipose tissue (WAT), and brown adipocytes are activated in brown adipose tissue (BAT). This process involves increased expression of thermogenic genes and proteins, such as uncoupling protein 1 (UCP1), which is highly expressed in beige and brown adipocytes (thermogenic adipocytes) and can produce heat by dissipating the proton motive force without generating adenosine triphosphate (ATP). UCP1 is the most extensively studied mediator of thermogenesis. However, a variety of additional mechanisms have also been identified to promote thermogenic energy expenditure in adipocytes. These include fatty acid oxidation, creatine phosphorylation, and calcium cycling (2). These changes enhance the adipocytes’ capacity to produce heat, thereby increasing energy expenditure and promoting weight loss. Numerous studies have demonstrated that enhancing adipocyte thermogenesis can reduce fat mass and improve metabolic health (3–6). It is noted that human brown adipose tissue is not only present in the interscapular region of infants but is also identified in various anatomical parts of adults (7, 8). Cold exposure or adrenergic receptor agonists can activate brown adipocytes in humans (9–12). The adrenergic stress response to prolonged cold exposure has been observed to induce browning of subcutaneous white adipose tissue (sWAT) in rodents. In contrast, in humans, ten days of cold exposure, which resulted in the BAT activation, but did not lead to the browning of sWAT (13). Interestingly, burn injury can result in prolonged elevations of circulating norepinephrine levels, which in turn leads to an increase in UCP1-positive beige adipocytes within the sWAT. This observation has been linked to the severe adrenergic stress response and the browning of sWAT in humans (14). Furthermore, BAT secretes a multitude of cytokines that facilitate communication with other organ systems, potentially contributing to the systemic metabolic effects (15).
The mechanisms underlying adipocyte thermogenesis involve transcriptional regulation, environmental cues, and signaling pathways. The key transcription factor PRDM16 plays a vital role in adipocyte thermogenesis. The PRDM protein family consists of 17 members that structurally contain a conserved N-terminal PR (PRDI-BF1 and RIZ1 homology) domain, which is similar to the SET (suppressor of variegation, enhancer of zeste, and trithorax) domain found in many histone lysine methyltransferases (HMTs). The PRDM proteins also contain a variable number of zinc finger domains, which directly bind with DNA. PRDM proteins are involved in various cellular processes, which include cell fate decision (16, 17). The PRDM16 gene, which was also named MDS1/EVI1-like gene 1 (MEL1) by Mochizuki et al., was first discovered in patients with acute myeloid leukemia and the myelodysplastic syndrome (18). The human PRDM16 gene (located on chromosome 11p36.32) and the mouse Prdm16 gene (located on chromosome 4qE2) contain 17 exons and encode a protein with a positive regulatory (PR) domain, an inhibitory domain (RD), two zinc finger DNA binding domains (ZF1 and ZF2), and an acidic domain (AD) (19, 20).
Recent studies have provided comprehensive information about the role of PRDM16 in adipose tissue. This review summarizes the function and regulation of PRDM16 in adipose tissue and proposes the application of this transcription factor as a potential target for treating obesity and its related metabolic disorders. Additionally, the review includes a summary of references that report proteins directly interacting with PRDM16, as well as factors influencing the expression of Prdm16 mRNA and protein. These studies have also elucidated the physiological roles of these genes in the obesity and its related metabolic diseases.
2 Function of PRDM16 in adipose tissue
The physiological role of PRDM16 in adipose tissue, along with its mechanism of action, a topic of significant interest in the field of endocrinology and metabolism, is summarized below:
2.1 Direct function in adipose tissue
2.1.1 Inducing brown adipocyte characteristics in white adipocytes
PRDM16 is significantly enriched in brown adipocytes as compared to white adipocytes. When Prdm16 is overexpressed in white adipocytes, a robust brown fat phenotype is activated, leading to a remarkable increase in uncoupled respiration. Conversely, when Prdm16 is knocked down using shRNA in brown adipocytes, the brown characteristics are significantly reduced. Mechanistically, PRDM16 directly binds to the promoters of Ucp1 and PPAR gamma coactivator 1 alpha (Pgc1α), key regulators of brown adipocyte thermogenic function. Additionally, PRDM16 interacts with the PGC1α transcriptional coactivator to modulate brown fat determination and function (21).
White fat depots include subcutaneous adipose tissue and several intra-abdominal adipose tissues, including mesenteric adipose tissue (22). Adiposity is a major risk factor for metabolic diseases in both humans and rodents (23–25). In contrast, the expansion of subcutaneous adipose tissue has been suggested to promote insulin sensitivity in both rodents and humans (26–28). The expression levels of Prdm16 are higher in subcutaneous white adipocytes than in other abdominal white adipocytes in mice. Transgenic expression of Prdm16 in white fat, under the -5kb aP2 promoter/enhancer, strongly promoted the development of beige adipocytes in subcutaneous adipose tissues but not in epididymal adipose tissues (29). When mice with overexpressed Prdm16 were fed on high-fat diets, increased energy expenditure, reduced body weight gain, and improved glucose tolerance were observed (29). In adipose-specific Prdm16 knockout mice (Adipo-Prdm16 KO), which were generated by Prdm16lox/lox and Adiponectin-cre mice, minimal effects on classical brown fat were noted. However, beige adipocyte function was markedly inhibited in subcutaneous white adipose tissue. When fed on high-fat diets, Adipo-Prdm16 KO developed obesity, severe insulin resistance, and hepatic steatosis. The subcutaneous adipose tissue in Adipo-Prdm16 KO mice acquired many key properties of visceral fat, including decreased thermogenic gene expression, increased inflammatory gene expression, and macrophage accumulation (30). Furthermore, adipocyte progenitor cells derived from human white adipose tissue demonstrate beige adipocyte characteristics upon differentiation into white adipocytes, as revealed through single-cell RNA sequencing analysis. These cells exhibit a pronounced enrichment of markers traditionally associated with beige adipocytes, including PGC1α, PRDM16, and UCP1 (31). These findings indicate that PRDM16 is essential for the formation of beige adipocytes and enhances the health effects of subcutaneous adipose tissue.
2.1.2 Controlling the switch of myoblasts into brown adipocytes
Although white and brown adipocytes both contain lipid droplets and display some morphological similarities, their developmental origins differ markedly. Brown adipocytes share the Myf5-positive precursor with myoblasts. PRDM16 is the key effector that controls the bidirectional cell fate switch between skeletal myoblasts and brown fat cells. Loss of Prdm16 in brown preadipocytes blocks brown adipogenesis and promotes muscle differentiation. Conversely, ectopic expression of Prdm16 in myoblasts induces their differentiation into brown adipocytes. Studies have shown that the adipogenic conversion of myoblasts to brown adipocytes by PRDM16 depends on the presence of rosiglitazone, a specific agonist for peroxisome proliferator-activated receptor gamma (PPARγ). PPARγ is the master regulator of adipogenic differentiation (32, 33). PPARγ alone can convert myogenic cells into adipocytes, but not into brown adipocytes; the latter requires PRDM16 expression. PRDM16 binds to PPARγ, thereby activating its transcriptional function and driving the differentiation of myoblasts into brown adipocytes (34). In a single-cell RNA sequencing analysis of human perivascular adipose tissue (PVAT), a brown-like fat depot, Angueira et al. identified that preadipocytes express the adipogenic gene PPARγ, while mature adipocytes express thermogenic genes UCP1 and PRDM16 (35).
The global homozygous Prdm16-deficient mouse died at birth. The putative BAT from the Prdm16 knockout (KO) mice at late-stage embryos (E17) exhibited substantially larger lipid droplets, accompanied by lower brown adipocyte-selective gene expression and higher skeletal myogenic gene expression. Interestingly, embryonic BAT development in mice that selectively lacked Prdm16 in the Myf5 lineage was normal, with no observable discrepancies when compared with control mice. In contrast to juvenile mice, Myf5-Prdm16 KO mice that were more than 6 months old exhibited profound morphological whitening in BAT. However, tissue temperature dramatically dropped or oxygen consumption was marginally raised in Myf5-Prdm16 KO mice upon cold exposure or norepinephrine (NE) stimulation. Prdm3, closely related to Prdm16 based on sequence and structure, compensates for the ablation of Prdm16 in embryonic brown fat development. Prdm3 not only regulates white adipocyte differentiation (36), but also could induce the expression of Ucp1 and Pgc1α. The double KO of Prdm3 and Prdm16 in the Myf5 lineage caused a more obvious white fat phenotype than in WT and Prdm16 KO mice (37). The above studies suggest that PRDM16 is crucial for determining thermogenic adipocyte fate by interacting with PGC1α and PPARγ together.
2.2 Indirect function in adipose tissue
PRDM16 not only directly regulates the formation and function of brown and beige fat but also indirectly modulates the expression of other genes to control obesity and its related metabolic diseases (Figure 1).
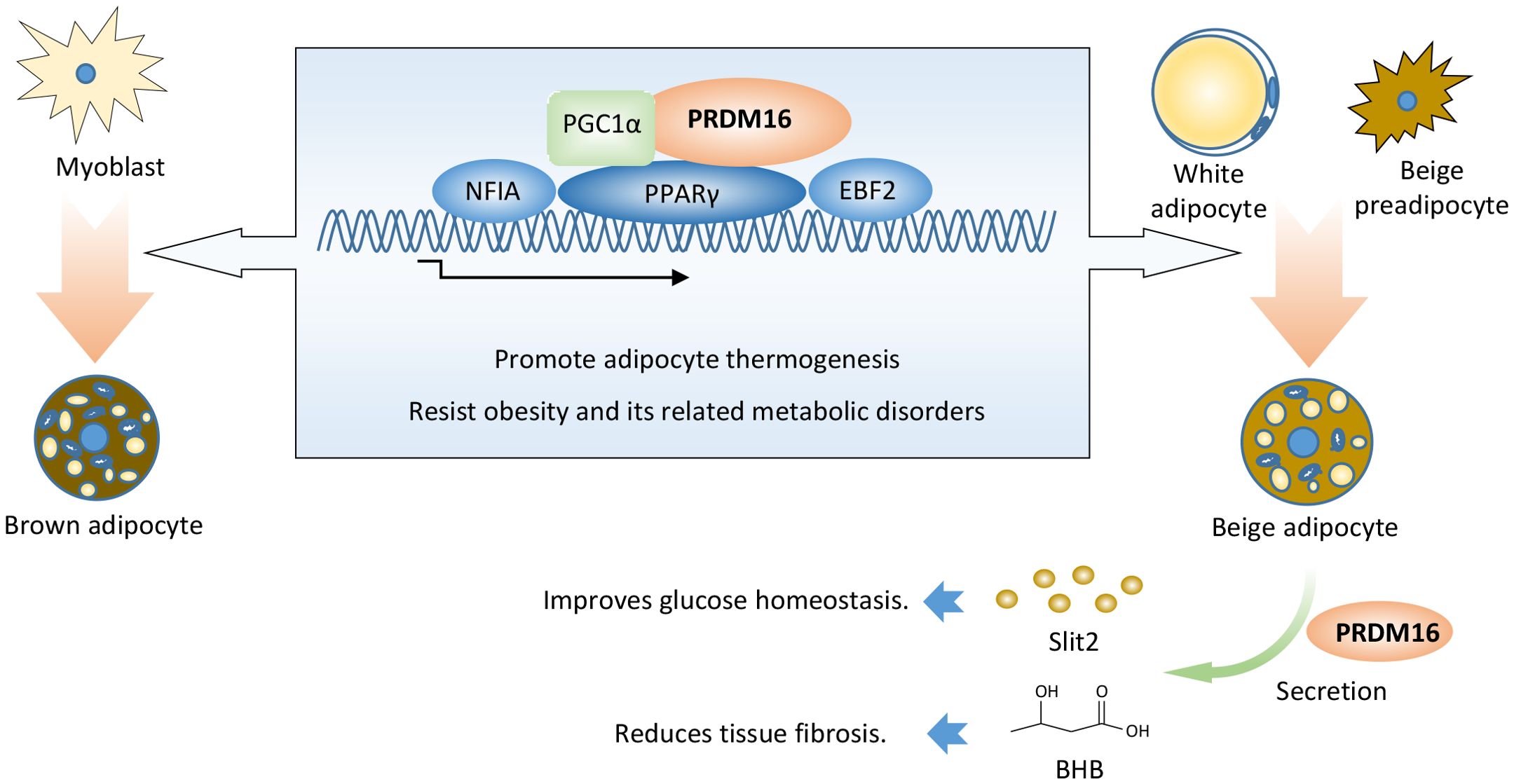
Figure 1. PRDM16 promotes thermogenic adipocyte function and formation. PRDM16 coordinates with PPARγ, PGC1α, EBF2, and NFIA to determine thermogenic adipocyte fate. PRDM16 regulates the expression of other genes, thereby indirectly modulating adipose tissue function. PRDM16 promotes the secretion of SLIT2 protein in beige adipocytes, thereby inducing adipocyte thermogenesis and improving insulin sensitivity. PRDM16 induces BHB secretion, which aids the differentiation of beige adipocytes and represses adipose fibrosis. BHB: β-hydroxybutyrate.
Slit2, a 180 kDa member of the Slit extracellular protein family, was identified as a secreted factor of beige adipocytes from aP2-Prdm16 transgenic mice. Full-length Slit2 is cleaved to generate several smaller fragments. The C-terminal fragment of Slit2 (Slit2-C) is an active thermogenic moiety. Slit2-C induces the thermogenesis of brown and beige adipocytes by activating the PKA signaling pathway. It promotes adipose thermogenesis, augments energy expenditure, and improves glucose homeostasis in vivo. However, the protease that generates Slit2-C from full-length Slit2, along with the effective surface receptor of Slit2-C, remains unknown (38).
Aging impairs beige adipogenesis and promotes fibrogenesis (39, 40). Genetic loss of Prdm16 mimics the effects of aging, whereas increasing PRDM16 in aged mice reduces fibrosis and restores beige adipogenic potential. LC-MS analysis revealed that PRDM16-driven fatty acid oxidation produces a paracrine factor, beta-hydroxybutyrate (BHB). BHB locally or selectively accumulates in the inguinal WAT (iWAT) of young animals, thereby suppressing myofibrogenesis and stimulating beige adipogenic competency in precursor cells. BHB catabolism is mediated by BDH1 and is required for beige fat differentiation. BHB supplementation in aged animals reduced adipose fibrosis and promoted beige fat formation. However, it is still unclear whether BHB would have antifibrotic or pro-beige adipogenic effects in human adipose tissue (41).
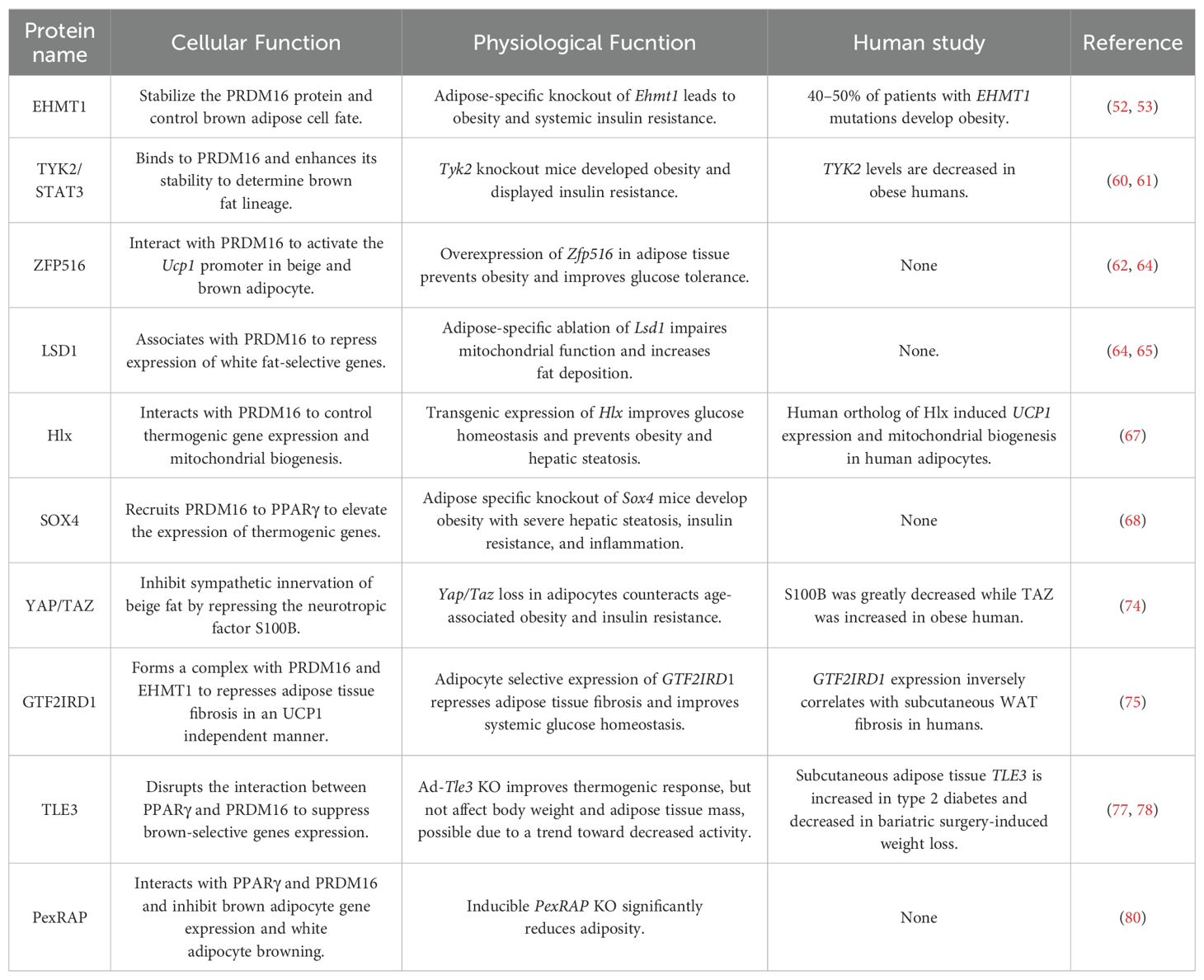
3 Proteins resist obesity and its related metabolic disorders by directly interacting with PRDM16
Different types of proteins have been reported to regulate adipocyte thermogenesis or fibrosis to combat adiposity in adipose tissue by directly interacting with the PRDM16 protein (Figure 2; Table 1).
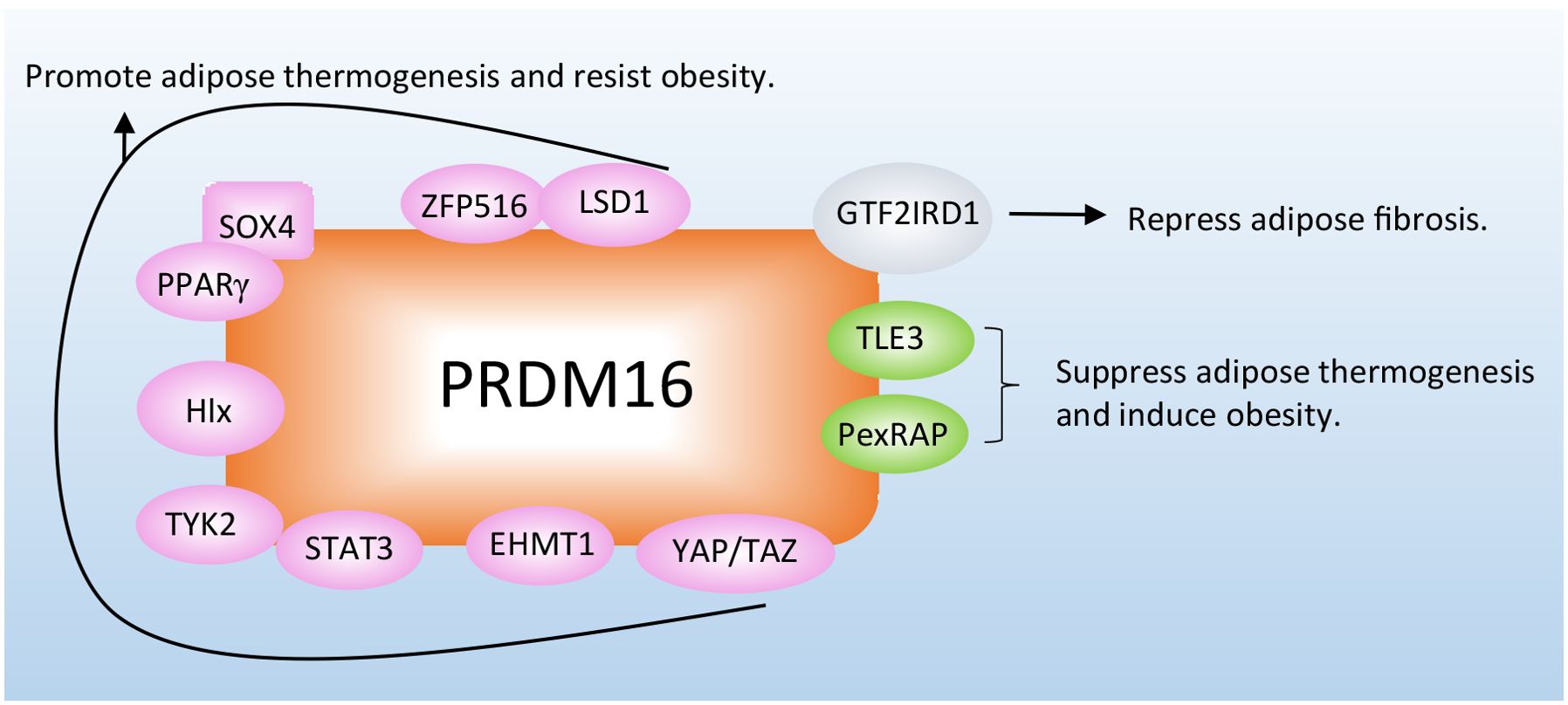
Figure 2. Proteins combat obesity and its related metabolic disorders by directly interacting with PRDM16. The proteins interact with PRDM16 to regulate adipocyte thermogenesis, depending on different mechanisms. More detailed information about these proteins is listed in Table 1.
3.1 Sequence-specific transcriptional factors to determine cell fate of thermogenic adipocytes
Although PRDM16 contains a DNA-binding domain, this domain is dispensable for PRDM16’s role in determining the fate of thermogenic adipocytes. The R998Q mutant allele of PRDM16 completely loses its DNA-binding ability but still retains the capability to interact with PPARγ to activate the brown fat gene program (21). PRDM16 indirectly binds to the genome via transcription factors that recognize specific motifs and directly binds to DNA, including PPARγ. Although PPARγ can interact with PRDM16 to convert myoblasts into brown adipocytes, its expression levels are quite low in primary and immortalized myoblasts. The Bruce M. Spiegelman group analyzed proteomic transcriptional complexes formed with wild-type (WT) PRDM16 or different mutant alleles of PRDM16 and found that C/EBP-β, prior to PPARγ, cooperates with PRDM16 to initiate the conversion of myoblasts to brown adipocytes. Forced expression of PRDM16 and C/EBP-β is sufficient to induce a fully functional brown fat program in naive fibroblastic cells, including in murine and human skin fibroblasts (42).
Nuclear factor I-A (NFIA) is regarded as a transcriptional regulator of brown adipocytes, as identified through genome-wide open chromatin analysis. NFIA and PPARγ co-localize at brown adipocyte-specific enhancers to facilitate PPARγ binding to these enhancers and drive active transcription. Overexpression of Nfia in myoblasts promotes brown adipocyte differentiation while inhibiting myogenic differentiation. Conversely, Nfia KO impairs the expression of brown fat-specific genes and elevates the expression of muscle genes. Interestingly, NFIA does not physically bind to PRDM16, and PRDM16 is dispensable for NFIA’s effects. NFIA and PRDM16 operate in parallel to each other (43). Additionally, NFIA in adipocytes can down-regulate pro-inflammatory cytokine genes to ameliorate adipose tissue inflammation by binding to the regulatory region of the Ccl2 gene. CCL2 expression is negatively correlated with NFIA expression in human adipose tissue (44).
Early B-cell factor 2 (Ebf2) is also a marker gene of brown and beige adipogenic precursor cells. Ebf2-expressing precursor cells from brown adipose tissue and white adipose tissue differentiate into brown and beige adipocytes, respectively (45). PPARγ genome-wide Chromatin immunoprecipitation (ChIP)-Seq revealed that the EBF DNA binding motif was highly enriched in brown adipose-specific PPARγ binding sites. EBF2 increases PPARγ binding at brown-specific sites. Conversely, it significantly reduces PPARγ binding at white-specific sites. It is important to note that EBF2 is recruited to the Prdm16 locus before PPARγ. When overexpressed in myoblasts or white adipose stromal cells, EBF2 recruits PPARγ to its brown-selective binding sites to drive a brown fat-specific differentiation program. Ebf2-deficient mice typically die soon after birth. An analysis of BAT in late-stage mouse embryos showed that Ebf2 KO resulted in the loss of brown-specific characteristics and thermogenic capacity in brown adipocytes and tissue (46). Consistently, EBF2 physically interacts with the chromatin remodeler BRG1 and the BAF chromatin remodeling complex component DPF3 to activate brown fat identity genes (47). Furthermore, transgenic expression of Ebf2 in adipose tissue under the aP2 promoter/enhancer powerfully stimulates beige fat development and protects animals against obesity (48). In human white adipose tissue, a subtype of adipocytes with potential thermogenic properties has been identified, characterized by the expression of thermogenic markers PPARGC1A and EBF2 through single-cell RNA sequencing analysis (49). While a direct interaction between PRDM16 and EBF2 has not been reported, multiple independent studies have confirmed that these two factors co-localize at the brown adipocyte-specific enhancers, suggesting that PRDM16 could potentially directly interact with EBF2 to form a substantial transcriptional complex.
3.2 Co-factors and histone modification enzymes to determine cell fate of thermogenic adipocytes
High-resolution liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) showed that EHMT1, which is the only methyltransferase with enzymatic activity on H3K9 mono- or di-methylation, was in the PRDM16 complex (50). The PR-domain of PRDM16 shares high homology with methyltransferase SET domains (51). The PRDM16 complex purified from brown adipocytes expressing the PR mutant exhibited significant methyltransferase activity on H3, due to the presence of EHMT1. Consistent with this, both WT PRDM16 and the PR mutant could convert myoblasts into brown adipocytes. Loss of Ehmt1 in brown adipocytes reduces brown fat characteristics. Conversely, overexpression of Ehmt1 positively promotes the expression of BAT-selective genes by stabilizing the PRDM16 protein. Adipose-specific ablation of Ehmt1 leads to a significant reduction in BAT-mediated adaptive thermogenesis and the development of severe obesity and systemic insulin resistance. Notably, patients with EHMT1 mutations develop obesity, possibly due to impaired adaptive thermogenesis. Moreover, activation of classical brown adipocytes in the adult human perirenal depot required PRDM16-EHMT1 complex expression (52, 53).
The Mediator complex, an evolutionarily conserved multiprotein entity, is integral to transcriptional regulation. It facilitates this process by directly interacting with RNA Polymerase II and coordinating the activities of various co-activators and co-repressors at the chromatin level (54). In adipocytes, the MED1 subunit of the Mediator complex modulates the function of key transcription factors, including PPARγ, PGC-1α, and C/EBPβ, which also interact with PRDM16 (55, 56).
PRDM16 is recruited to the chromatin in brown adipose tissue via its DNA-binding partners, including C/EBPβ and PPARγ. Notably, PRDM16 deficiency does not compromise the DNA-binding capacity of these partners to their respective target sites; yet the presence of MED1 at these locations is reduced. Moreover, silencing Med1 in mature brown adipocytes results in a marked decrease in the transcription of genes specific to brown fat. Through its zinc finger domains, PRDM16 directly engages with MED1, positioning it as a super-enhancer for these genes. Furthermore, this interaction is critical for the formation of complex chromatin structures at key gene loci specific to brown adipose tissue (57, 58).
Tyrosine kinase 2 (Tyk2) is a member of the Jak kinase family. Numerous cytokines and growth factors activate the Jak/STAT pathway to stimulate tyrosine phosphorylation of the STAT transcription factors, thereby inducing the expression of early response genes (59). Mice lacking Tyk2 become progressively obese and exhibit abnormal BAT morphology and function. Constitutively active Stat3 (CAStat3) and PRDM16 restore the differentiation of Tyk2 KO brown adipocytes. Furthermore, altered BAT morphology and function were restored, and obesity was reversed in Tyk2 KO mice expressing CAStat3 in BAT. Interestingly, a mutated form of Tyk2, which lacks tyrosine kinase activity (Tyk2KD), also rescues the differentiation of Tyk2 KO brown preadipocytes in vitro. It is important to note that Tyk2 KO mice expressing a Tyk2KD transgene in BAT display normal brown adipocyte differentiation and resist the obese phenotype of Tyk KO mice. Mechanistically, STAT3 binds to PRDM16 and enhances the stability of the PRDM16 protein, thereby maintaining brown adipocyte differentiation. Both Tyk2WT and Tyk2KD can also interact with PRDM16 and PGC1α to regulate brown fat differentiation (60, 61).
3.3 Co-factors to positively regulate functions of thermogenic adipocytes
High-throughput screening using the Ucp1 promoter revealed that ZFP516 is a transcriptional activator of Ucp1. ZFP516 directly binds to the proximal region of the Ucp1 promoter to activate Ucp1 expression by interacting with PRDM16. Zfp516 is induced by cold and sympathetic stimulation through the cAMP-CREB/ATF2 pathway. Ectopic expression of Zfp516 represses the myogenic program and enhances brown adipocyte differentiation. Although ablation of Zfp516 is embryonically lethal, BAT mass in KO embryos is still drastically reduced. Overexpression of Zfp516 in adipose tissue (aP2-Zfp516) promotes the browning of iWAT, increases body temperature and energy expenditure, and prevents diet-induced obesity (62).
LSD1, also known as KDM1A, demethylates the monomethylated or dimethylated histone H3 Lys4 (H3K4me1 or H3K4me2), which are histone markers closely associated with active transcription (63). Quantitative mass spectrometry is used to identify LSD1 from nuclear extracts of primary brown adipocytes with a PRDM16-specific antibody. The brown adipocytes from Adipo-Lsd1-KO mice exhibited increased expression of white-selective genes, a blunted response to norepinephrine, and reduced flavin adenine dinucleotide (FAD) levels. It is important to note that Adipo-Lsd1-KO mice have more fat mass and lower energy expenditure levels. In line with this result, Ucp1-Lsd1-KO mice also gained more body weight on a high-fat diet than control littermates. LSD1 regulates the metabolism of brown adipocytes by cooperating with PRDM16 and ZFP516 (64–66).
Transcription factor Hlx is highly expressed in BAT and iWAT. It is translationally upregulated by β3-adrenergic signaling in iWAT. The knockdown of Hlx in BAT and iWAT adipocytes had no effect on adipogenesis but decreased Ucp1 expression and strongly suppressed oxygen consumption. Hlx homozygous null mice are embryonically lethal. Hlx heterozygous KO mice exhibit defects in beige adipocyte formation in iWAT and develop glucose intolerance and high-fat-induced hepatic steatosis. Conversely, transgenic expression of Hlx promotes the browning of white adipocytes, improves glucose homeostasis, and prevents obesity and hepatic steatosis. Hlx directly interacts with PRDM16 to control BAT-selective gene expression and mitochondrial biogenesis (67).
Whole transcriptome deep sequencing (RNA-seq) analysis of iWAT after cold stimulation identified transcription factor SOX4 as a new regulator of white adipocyte browning. Mice with either Adiponectin-Cre or Ucp1-Cre deletion of Sox4 exhibited significant cold intolerance, decreased energy expenditure, and impaired beige adipocyte formation. These mice also developed obesity, severe hepatic steatosis, insulin resistance, and inflammation after being fed on a high-fat diet. ShRNA knockdown of Sox4 in beige adipocytes reduced energy metabolism and the expression of thermogenic genes. SOX4 recruits PRDM16 to PPARγ, thereby forming a transcriptional complex and elevating the expression of thermogenic genes (68).
YAP and TAZ are key molecular effectors of the Hippo pathway in mammals (69). YAP/TAZ acts as a PPARγ co-repressor that regulates adipocyte differentiation. They are required for adipocyte survival in diet-induced obesity and promote obesity-related adipose tissue fibrosis (70, 71). Adipocyte-specific KO of Yap/Taz (Yapfl/fl; Tazfl/fl with AdipoqCre mice) induces S100b expression, thereby stimulating sympathetic innervation and biogenesis of functional beige fat in both iWAT and visceral WAT. S100b is a neurotrophic factor that stimulates the sympathetic innervation of thermogenic fat (72). Calsyntenin 3β was initially suggested to enhance the secretion of S100b in adipocytes, as indicated in reference (72). However, contrasting findings were reported in a subsequent study, which demonstrated that Calsyntenin 3β does not regulate S100b secretion and innervation, as detailed in reference (73). YAP/TAZ competes with C/EBPβ for binding sites on the ZF-2 domain of PRDM16 to inhibit S100b transcription. YAP/TAZ disrupts the formation of the PRDM16-C/EBPβ complex, thereby acting as a brake on beige fat innervation (74).
GTF2IRD1, a cold-inducible and BAT-enriched transcriptional factor, interacts with both PRDM16 and EHMT1 to form a transcriptional complex that represses adipose tissue fibrosis in an UCP1-independent manner. Repressed adipose tissue fibrosis was observed in transgenic mice, where variant 5 of Gtf2ird1 is driven by the aP2 promoter/enhancer in adipose tissues. This repression was accompanied by improved systemic glucose homeostasis, independent of body weight loss. Deleting Gtf2ird1 promoted fibrosis in a cell-autonomous manner. GTF2IRD1 represses the transcription of transforming growth factor β-dependent pro-fibrosis genes by recruiting PRDM16 and EHMT1 to their promoter/enhancer regions. Additionally, GTF2IRD1 expression inversely correlates with human subcutaneous WAT fibrosis. The EHMT1-PRDM16-GTF2IRD1 complex plays a crucial role in repressing obesity-associated adipose tissue fibrosis and enhancing systemic glucose homeostasis (75).
3.4 Co-factors to negatively regulate functions of thermogenic adipocytes
Transducin-like enhancer protein 3 (TLE3) is an adipogenic coregulator that functions synergistically with PPARγ to stimulate adipogenesis (76). TLE3 counteracts the brown fat program by competing with PRDM16 for interaction with PPARγ. TLE3 and PRDM16 reciprocally regulate BAT- and WAT-selective gene expression. In aP2-Tle3 transgenic mice, impaired fatty acid oxidation and adipocyte thermogenesis were observed compared to control mice. Conversely, mice lacking Tle3 in adipose tissue (Adipo-Tle3 KO) exhibited enhanced thermogenesis in inguinal white adipose depots and were protected from age-dependent deterioration of brown adipose tissue function (77). In humans, TLE3 expression in subcutaneous adipose tissue is increased in type 2 diabetes and decreased following bariatric surgery-induced weight loss (78).
PexRAP synthesizes ether-linked phospholipids that are potential partial agonists for PPARγ and also promotes white adipogenesis (79). Homozygous KO of PexRAP in mice resulted in embryonic lethality. To overcome this, Rosa-CreER mice were used to generate tamoxifen-inducible global PexRAP KO (PexRAP-iKO) animals. In PexRAP-iKO mice, enhanced adipocyte browning, increased energy expenditure, and decreased adiposity were observed. Similar to TLE3, PexRAP disrupts the formation of the PRDM16-PPARγ complex by interacting with PPARγ, thereby inhibiting white adipocyte browning (80).
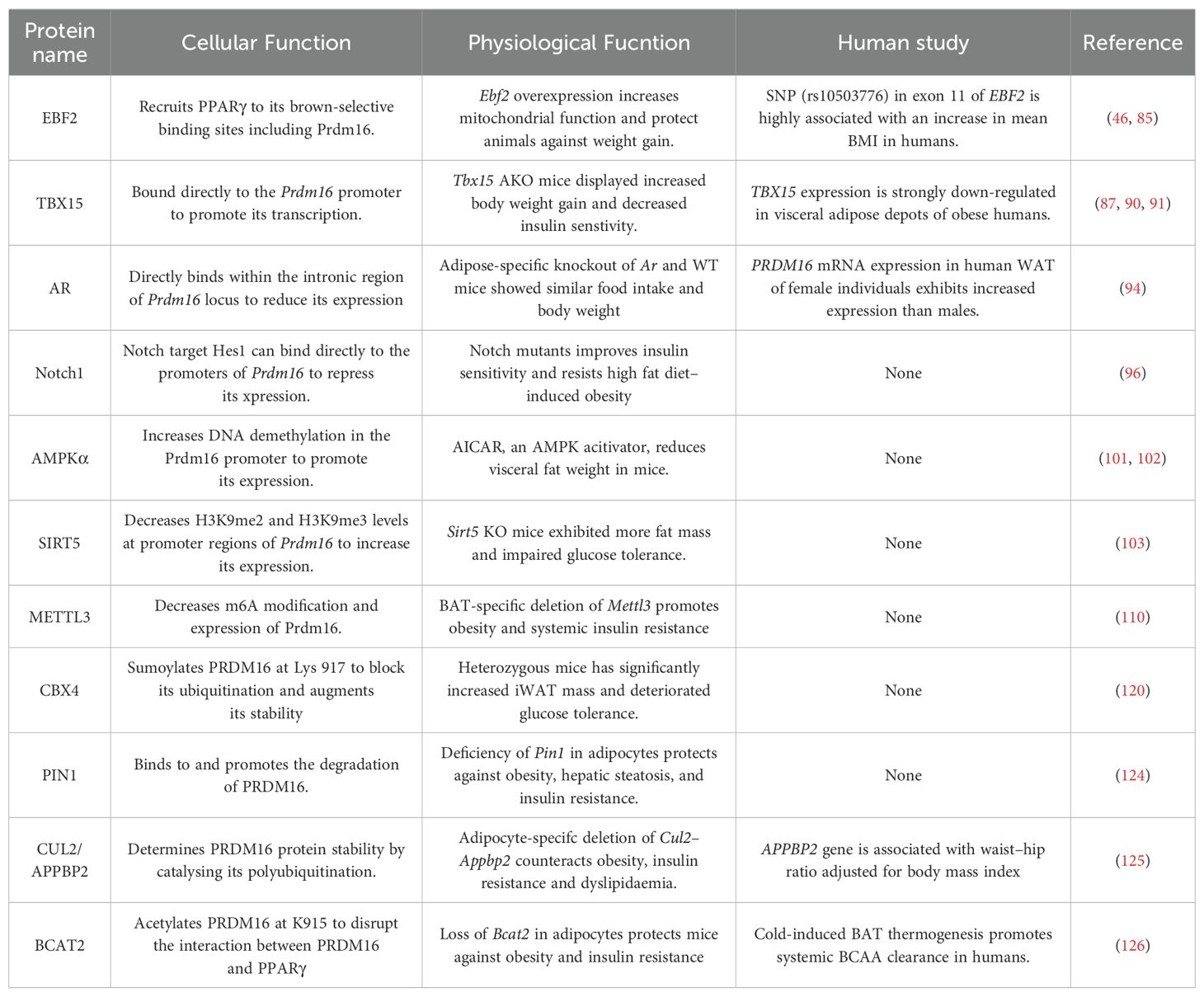
4 Proteins combat obesity by directly regulating Prdm16 expression in adipose tissue
The amounts and stabilization of PRDM16 protein are critical in controlling the formation and function of thermogenic adipocytes and in combating obesity. The upstream regulation of PRDM16 has been extensively investigated at both transcriptional and post-transcriptional levels (Figure 3; Table 2).
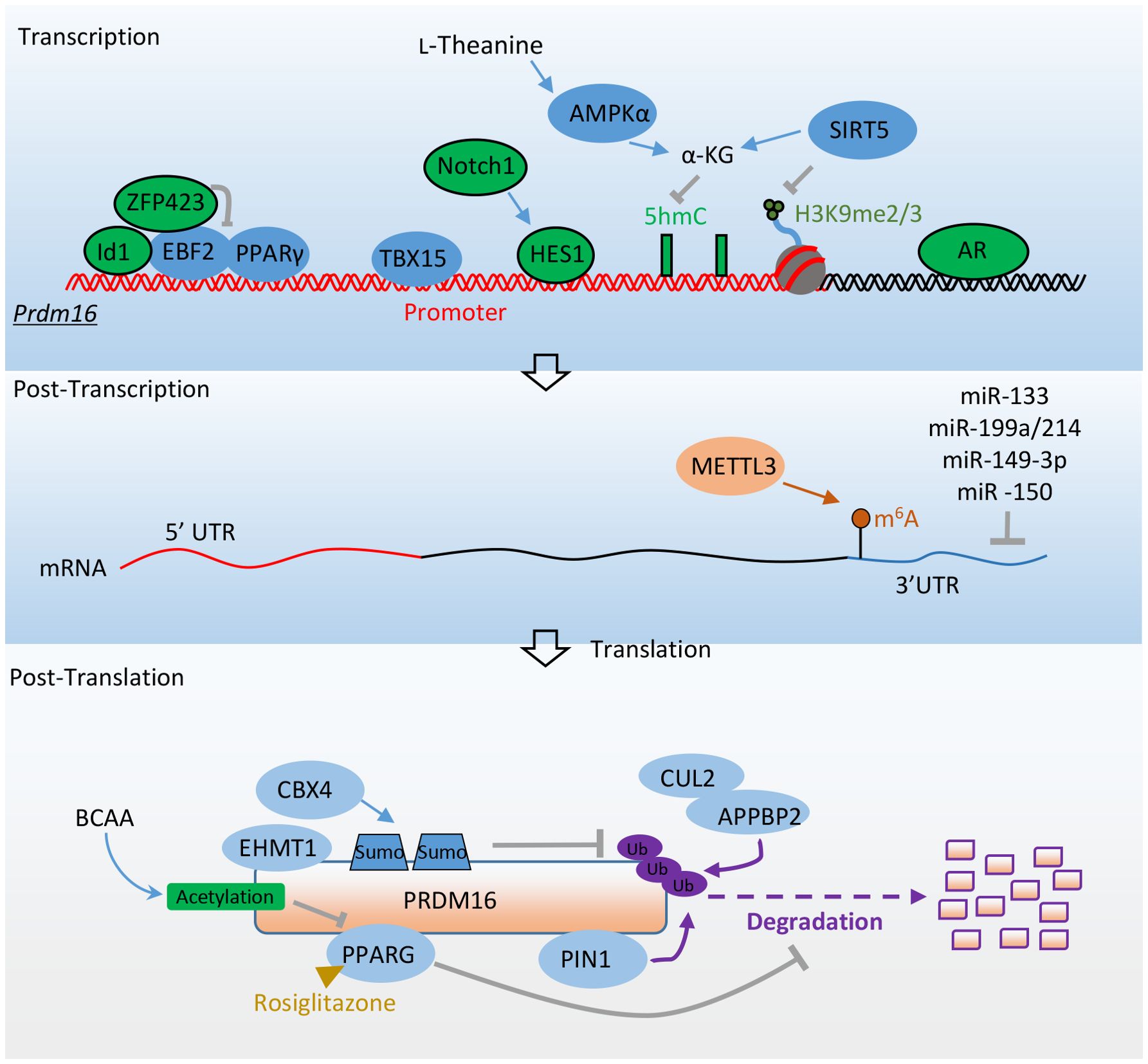
Figure 3. Factors promote adipocyte thermogenesis and resist obesity by directly regulating Prdm16’s expression. Transcriptional factors EBF2, PPARγ, and TBX15 promote the transcription of Prdm16 mRNA. ZFP423 and ID1 inhibit Prdm16 mRNA transcription by binding with EBF2, while HES1 and AR repress its transcription. AMPKα and SIRT5 enhance Prdm16 mRNA transcription through epigenetic regulation. At the post-transcriptional level, METTL3 stabilizes Prdm16 mRNA through m6A modification. Various microRNAs regulate the stability of Prdm16 mRNA by targeting its 3’ untranslated region (3’UTR). At the post-translational level, the interaction between PPARγ and EHMT1 with PRDM16 enhances its stability. PIN1 promotes the degradation of PRDM16 via the ubiquitin-proteasome system. CBX4 sumoylates PRDM16, preventing its degradation. The CUL2–APPBP2 complex reduces PRDM16 protein stability by catalyzing its polyubiquitination. BCAA promotes the acetylation of PRDM16, disrupting its interaction with PPARγ. More detailed information about these factors is provided in Table 2.
4.1 Transcriptional level regulation
Inhibitor of differentiation 1 (Id1) is a helix-loop-helix transcription factor with a significant role in cell proliferation and differentiation (81). In aP2 promoter-driven Id1 transgenic mice, there was a propensity for high-fat diet-induced obesity, along with reduced BAT thermogenesis and energy expenditure. Id1 suppresses BAT thermogenesis by binding to PGC1α and repressing its transcriptional activity. The absence of Id1 enhances the expression of beige genes in WAT. Additionally, the differentiation of thermogenic genes is increased in embryonic fibroblasts of Id1-deficient mice. At the molecular level, Id1 directly binds to EBF2 and suppresses its transcriptional activity, leading to the down-regulation of Prdm16 expression (82).
The zinc finger transcriptional coregulator 423 (ZFP423) is a transcriptional factor crucial for preadipocyte determination (83). It suppresses the fate of thermogenic adipocytes, with Zfp423 mRNA being significantly more abundant in white adipocytes compared to brown adipocytes. Inducible deletion of Zfp423 in the adipocytes of adult mice leads to the accumulation of beige adipocytes in the iWAT depot. This loss of Zfp423 is associated with increased energy expenditure and relatively lower diet-induced weight gain. Conversely, overexpression of Zfp423 converts BAT to a more unilocular, white adipocyte-like phenotype, suppressing the expression of thermogenic genes. Zfp423-deficient adipocytes can be converted to beige adipocytes in a cell-autonomous manner. ZFP423 recruits the NuRD corepressor complex to EBF2-bound thermogenic gene enhancers, including Prdm16. Conversely, deletion of Zfp423 induces a coregulator switch from the NuRD complex to the BAF complex on EBF2, resulting in a shift in PPARγ occupancy to thermogenic gene enhancers (84, 85).
T-box 15 (Tbx15) belongs to a phylogenetically conserved family, which has a similar characteristic sequence in the DNA binding domain and is involved in various developmental processes (86). Tbx15 is highly expressed in brown adipose tissue and iWAT, minimally expressed in visceral white adipose tissue. In mice, siRNA knockdown of Tbx15 in brown and inguinal adipose stromal cells impaired their differentiation into mature adipocytes (87). Paradoxically, overexpression of Tbx15 in 3T3-L1 preadipocytes blunted their differentiation into adipocytes (88). In vivo, the heterozygous deletion of Tbx15 resulted in glucose intolerance and obesity when the mice were fed on high-fat diets (89). Furthermore, Adipo-Tbx15 KO mice displayed increased body weight gain and decreased whole-body energy expenditure in response to high-fat diets. In humans, TBX15 expression is strongly downregulated in visceral adipose depots of overweight and obese individuals (90). TBX15 could directly bind to the Prdm16 promoter to enhance its transcription and participate in adipocyte browning induced by the adrenergic signaling pathway (91).
Androgens play a crucial role in regulating body fat distribution and function. Their excess is significantly correlated with body mass index, abdominal obesity, and insulin resistance (92, 93). Using the Prdm16 luciferase knock-in reporter mouse model, it has been observed that the androgen receptor (AR) shows the strongest negative correlation with Prdm16. Activation of androgen-AR signaling suppresses Prdm16 expression and reduces the beiging process in beige adipocytes. Adipocyte-selective ablation of Ar promotes beige adipogenesis, whereas its adipocyte-specific overexpression inhibits the browning of white adipocytes. A sex dimorphism in Prdm16 mRNA expression is also evident in human WAT, with female individuals exhibiting higher expression levels than males. Additionally, an analysis of a recent single-nuclei RNA sequencing dataset from human visceral adipose tissue indicates a trend toward increased PRDM16 expression in female adipocytes compared to male adipocytes (49). ChIP analysis reveals that AR directly binds to the intronic region of the Prdm16 locus, negatively regulating its expression (94).
Notch receptors (Notch1-Notch4) are activated by binding to Dll or Jag family ligands, followed by cleavage to release the Notch intracellular domain (NICD). NICD then binds with the Rbpj transcriptional complex to activate the transcription of downstream targets, including Hes (95). Adipose-specific inactivation of Notch1 triggers the browning of white adipocytes and elevates the expression of thermogenic genes. In contrast, adipose-specific activation of Notch1 (N1ICD) inhibits the browning of white adipocytes and is associated with higher expression levels of Hes1 and Hey1 compared to WT mice. Overexpression of the transcriptional repressor Hes1 in adipose stromal cells of WT mice results in lower levels of Prdm16 and Pgc1α. At the molecular level, the Notch target HES1 can directly bind to the promoters of Prdm16 and Pgc1α, repressing their expression and thereby inhibiting thermogenic gene programs (96).
AMP-activated protein kinase (AMPK) indirectly regulates BAT thermogenic function by influencing the hypothalamus and the adrenergic nervous system (97). It directly regulates brown adipogenesis by elevating α-ketoglutarate (αKG) production and promoting DNA demethylation in the Prdm16 promoter of progenitor cells. Prdm16 is enriched with CpG sites surrounding its transcription start site (TSS) (98). Active DNA demethylation, essential for initiating its expression, is mediated by the ten-eleven translocation hydroxylases (TETs) (99). The TET catalytic reaction requires αKG, a key metabolite of the Krebs cycle, thereby linking metabolism to epigenetic modifications (100). Ablation of AMPKα1 reduces the activity of isocitrate dehydrogenase 2 and cellular αKG levels, leading to an increase in 5-methylcytosine (5mC) and a decrease in 5-hydroxymethylcytosine (5hmC) in the Prdm16 promoter. Consequently, AMPKα1 knockout suppresses brown adipogenesis, while its activation promotes brown adipogenesis (101). L-theanine, a non-proteinogenic amino acid, enhances AMPKα phosphorylation, to increase αKG levels and to activate DNA demethylation on the Prdm16 promoter. When L-theanine was administered to high-fat diet-fed mice, it resulted in increased energy expenditure, reduced obesity, and improved glucose tolerance and insulin sensitivity (102).
SIRT5, a member of the sirtuin family, is essential for brown adipocyte differentiation and the expression of brown adipogenic genes. Knockdown of Sirt5 reduces intracellular αKG concentrations while elevating H3K9me2 and H3K9me3 levels at the promoter regions of Pparγ and Prdm16 loci. This inhibits the expression of Pparγ and Prdm16, thereby impairing adipocyte thermogenesis. Sirt5 deficiency in mice reduces the browning capacity in iWAT and leads to insulin resistance and obesity (103). Other sirtuins contribute to adipocyte function by modulating other genes. For example, SIRT1 facilitates the browning of white adipose tissue by deacetylating PPARγ (104). SIRT2 impedes adipocyte differentiation by deacetylating FOXO1 (105). SIRT3 is known to regulate mitochondrial function and thermogenesis in brown adipocytes, enhancing the expression of PGC1α (106).
4.2 Posttranscriptional level regulation
Methyltransferase-like 3 (METTL3), a key RNA methyltransferase, is highly expressed in BAT and plays an essential role in postnatal BAT development and maturation. RNA m6A modification is one of the most prevalent mRNA modifications in eukaryotes. It can be catalyzed by m6A writer proteins such as METTL3, recognized by m6A reader proteins, and removed by m6A eraser proteins (107). RNA m6A modification regulates most RNA processing steps, including mRNA splicing, stability, and translation efficiency, which further modulate other biological processes (108, 109). Deletion of Mettl3 driven by the Ucp1 promoter severely impairs the maturation of BAT by reducing m6A peaks and the expression of Prdm16, Pparγ, and Ucp1. This significantly reduces BAT-mediated adaptive thermogenesis and promotes high-fat diet-induced obesity and systemic insulin resistance (110).
Non-coding strands of RNA, including microRNAs, regulate gene expression at the transcriptional level (111). Prdm16 mRNA expression is controlled by several miRNAs, including miR-133 and miR-199a/214, which directly target the 3’ untranslated region (3’ UTR) of Prdm16 and negatively regulate its expression. These microRNAs are potential therapeutic targets for inducing brown or beige adipocyte lineage differentiation, thereby increasing energy expenditure and reducing fat mass in obese individuals. Inhibition of miR-133 leads to the differentiation of precursors in BAT and skeletal muscle stem cells into mature brown adipocytes, increasing whole-body energy expenditure, improving glucose tolerance, and impeding the development of diet-induced obesity (112, 113). Conversely, overexpression of the miR-199a/214 cluster suppresses the differentiation of brown adipocytes, while knockdown of this cluster increases thermogenic gene expression in beige adipocytes (114). Overexpression of miR-149-3p promotes a visceral-like switch during inguinal preadipocyte differentiation (115, 116). The KH-type splicing regulatory protein (KSRP), an RNA-binding protein, enhances the expression of miR-150 in iWAT, attenuating the expression of brown fat genes (117). Taken together, Prdm16 mRNA expression is regulated by various microRNAs to facilitate the expression of brown-selective genes and resist obesity.
4.3 Post-translational level regulation
PRDM16 protein is short-lived and rapidly degraded, making the increase of its lifetime in adipose tissue a promising strategy for combating obesity. Several studies have shed light on the regulation of PRDM16 protein stabilization in adipose tissue.
The full agonist of PPARγ, rosiglitazone, induces the expression of brown fat genes such as Ucp1 in primary adipocytes differentiated from the stromal-vascular fraction (SVF) of iWAT, but not in those from epididymal WAT. Prdm16 mRNA expression is significantly higher in iWAT than in epididymal WAT. ShRNA knockdown of Prdm16 blunts the effects of the PPARγ agonist-induced browning of inguinal white adipocytes. Conversely, transgenic expression of Prdm16, together with rosiglitazone, synergistically induces the expression of thermogenic genes and the emergence of UCP1-positive multilocular adipocytes in vivo. The white-to-brown fat conversion induced by rosiglitazone is partly regulated through the enhanced stability of PRDM16 protein and inhibition of the ubiquitin-proteasome pathway (118).
CBX4, a Polycomb group protein, acts as a SUMO E3 ligase for PRDM16 and mediates its sumoylation. The expression of Cbx4 is enriched in brown adipose tissue and is induced by acute cold exposure. CBX4 sumoylates PRDM16 at lysine 917, preventing its ubiquitination-mediated degradation and enhancing its stability mediated by EHMT1. Homozygous Cbx4 knockout mice die almost immediately after birth (119). Heterozygous Cbx4 knockout reduces PRDM16 protein levels, impairs white adipocyte browning, significantly increases iWAT mass, and deteriorates glucose tolerance and cold tolerance. Furthermore, fat-specific Cbx4 knockdown or overexpression through adenovirus injection remodels iWAT, decreasing or increasing adipocyte thermogenesis by down-regulating or up-regulating PRDM16 protein levels, respectively (120).
Pin1 is a unique enzyme associated with the phospho-serine or phosphor-threonine-proline-containing motif of numerous substrates where it controls their functions and stability (121). Pin1 expression in adipose tissue is markedly increased by obesity. As a result, Pin1 KO mice are highly resistant to diet-induced obesity (122, 123). Adipocyte-specific Pin1 KO (adipo-Pin1 KO) promotes thermogenesis, resists high-fat diet-induced obesity, and improves glucose tolerance. At the molecular level, PIN1 binds to PRDM16 and promotes its degradation through the ubiquitin-proteasome system, suppressing non-shivering thermogenesis (124).
Cullin–RING member 2 (CUL2) functions as a scaffold protein by interacting with an E2 enzyme, elongin B (ELOB), elongin C (ELOC), and a substrate-specific receptor (amyloid precursor protein-binding protein (APPBP2)). APPBP2 directly interacts with PRDM16. CUL2–APPBP2 is an ubiquitin E3 ligase that determines PRDM16 protein stability by catalyzing its polyubiquitination, without altering its mRNA expression. Inhibiting CUL2–APPBP2 sufficiently extended the half-life of the PRDM16 protein and promoted the biogenesis of beige adipocytes. Adipocyte-specific deletion of Cul2 or Appbp2 counteracted diet-induced obesity, glucose intolerance, and insulin resistance while promoting adipocyte thermogenesis (125).
Branched-chain amino acids (BCAAs), including leucine, isoleucine, and valine, are converted to branched-chain keto acids (BCKAs) by BCAT1 (BCATc, cytosolic branched-chain aminotransferase) or BCAT2 (BCATm, mitochondrial branched-chain aminotransferase). Adipose tissue Bcat2 KO mice exhibit increased iWAT browning and thermogenesis, and have an increased ability to resist high-fat diet-induced obesity. BCKA supplementation rescues Bcat2 KO mice from high-fat diet-induced obesity. Moreover, telmisartan, an anti-hypertension drug and direct inhibitor of BCAT2, enhances WAT browning and reduces adiposity. Mechanistically, acetyl-CoA derived from BCKA promotes acetylation of PRDM16 at K915, thereby disrupting its interaction with PPARγ. This prevents iWAT browning and BAT thermogenesis (126, 127). Furthermore, BAT actively utilizes BCAA in the mitochondria for thermogenesis and enhances systemic BCAA clearance in both mice and humans upon cold exposure. A BAT-specific defect in BCAA catabolism can attenuate systemic BCAA clearance and thermogenesis, which lead to diet-induced obesity and glucose intolerance (126). Additionally, BAT catabolize BCAA in the mitochondria, serving as nitrogen donors for the biosynthesis of non-essential amino acids and glutathione. This process which helps to reduce oxidative stress and increase hepatic insulin signaling is independent of BAT thermogenesis (128).
5 Discussion and prospectives
The activity and expression of PRDM16 in adipose tissue are closely associated with adipocyte thermogenesis and resistance to obesity, as evidenced by these distinguished studies. Consequently, directly targeting PRDM16 proteins in thermogenic adipose tissue emerges as a potential strategy for combating obesity and related metabolic disorders. Interventions that increase the expression of PRDM16, reduce its degradation, or stabilize the PRDM16 protein could enhance the formation and function of brown and beige adipocytes. Numerous studies have reported that various drugs can alleviate obesity and diabetes by modulating the expression and function of PRDM16. For example, resveratrol is known to activate adipocyte thermogenesis and counteract obesity induced by a high-fat diet by upregulating the expression of PRDM16 (129). Similarly, rutaecarpine has been shown to promote adipocyte thermogenesis via the AMPK-PRDM16 signaling pathway (130). A comprehensive summary of studies focusing on drugs targeting PRDM16 is available in another review (131).
Humans with active BAT exhibit a healthier body fat distribution, characterized by reduced visceral adipose tissue and increased subcutaneous adipose tissue. Additionally, these individuals demonstrate improved metabolic profiles, including lower levels of blood glucose and lipids, as well as decreased liver fat accumulation. A significant association between the presence of BAT and enhanced cardiometabolic health has also been observed in overweight or obese individuals (132, 133). Exposure to mild cold has been shown to activate adrenergic receptor signaling pathways, which in turn enhance glucose and fatty acid uptake, as well as oxidative metabolism within human BAT (134, 135). Oral administration of mirabegron, a β3 adrenergic receptor agonist approved for the treatment of overactive bladder, has the potential to activate human BAT and augment whole-body energy expenditure (136, 137). Despite this, significant weight loss in obese patients following mirabegron treatment has not yet been conclusively demonstrated (138). In human subjects, the most efficacious stimulation of adipose tissue has been observed with higher doses of mirabegron (139). However, potential cardiovascular side effects may restrict the clinical use of such dosages (140).
Obesity arises from an energy imbalance, characterized by excessive energy intake or insufficient energy expenditure. To combat obesity, one approach is to enhance energy expenditure, including the promotion of adipocyte thermogenesis. Another strategy involves reducing energy intake. Glucagon-like peptide-1 (GLP-1) receptor agonists, such as liraglutide and semaglutide, have proven effective in resisting obesity and improving hyperglycemia, insulin sensitivity, and blood pressure by inhibiting food intake, as demonstrated in both preclinical and clinical studies. Interestingly, in preclinical experiments, liraglutide and semaglutide have also shown the ability to induce adipocyte thermogenesis, evidenced by elevated expression of UCP1 and PRDM16 proteins and improvements in insulin sensitivity (141, 142). However, the administration of GLP-1 receptor agonists in humans does not appear to significantly contribute to weight loss through increasing adipocyte thermogenesis. Clinical trials have shown that once-daily liraglutide administration in individuals with obesity and type 2 diabetes reduces body weight and decreases resting energy expenditure, but does not alter the proportion of fat within brown adipose tissue (143). Similarly, once-weekly semaglutide administration in individuals with obesity reduced food intake and fat mass without changing resting energy expenditure (144). Therefore, while GLP-1 receptor agonists may promote adipocyte thermogenesis in rodents, their role in this mechanism in humans remains to be clearly defined (145).
To date, no clinically approved drugs effectively increase adipocyte thermogenesis for the treatment of obesity. Compared to β3 adrenergic receptor agonist and GLP-1 receptor agonist, PRDM16 protein could represent a novel and promising therapeutic approach for combating obesity and its associated metabolic disorders. However, unlike these agonists, PRDM16 is not a direct drug target. Therefore, alternative pathways must be modulated to regulate the expression and function of PRDM16 in adipose tissue. Furthermore, Prdm16 is not exclusively expressed in thermogenic adipocytes like Ucp1; it is also expressed in various other tissues including skeletal and cardiac muscle. Prdm16 has been implicated in the regulation of hematopoietic and neural stem cell maintenance, cardiac development, and other pathological conditions (146–149). In the absence of PRDM16, there is an increase in reactive oxygen species and apoptosis within hematopoietic stem cells. Global mutations in PRDM16 are associated with cleft palate, altered craniofacial development, and impaired cardiac development. In humans, PRDM16 mutations have been identified as a cause of cardiomyopathy. Brown adipocytes share a common precursor with myocytes, and PRDM16 is a pivotal effector in the conversion of myoblasts into brown adipocytes. Therefore, further studies are warranted to examine the potential effects of drugs targeted at PRDM16 on muscle development and function in vivo. In summary, it is still a long way to combat obesity and its related metabolic disorders by targeting PRDM16 protein in thermogenic adipose tissue.
Author contributions
LM: Writing – original draft, Writing – review & editing. JL: Writing – original draft. YH: Writing – original draft. TN: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported, in part, by the Natural Science Joint Foundation of Hubei Province (2023AFD038), the Natural Science Foundation of Guangdong Province (2022A1515010283), the National Natural Science Foundation of China (81970729), and the Initial Funding of Hubei University of Arts and Science (2059200).
Acknowledgments
The authors express their gratitude to all the reviewers for their valuable contributions and to EditSprings for their linguistic assistance in preparing this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ng M, Fleming T, Robinson M, Thomson B, Graetz N, Margono C, et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. (2014) 384:766–81. doi: 10.1016/S0140-6736(14)60460-8
2. Roesler A, Kazak L. UCP1-independent thermogenesis. Biochem J. (2020) 477:709–25. doi: 10.1042/BCJ20190463
3. Carpentier AC, Blondin DP, Haman F, Richard D. Brown adipose tissue-A translational perspective. Endocrine Rev. (2023) 44:143–92. doi: 10.1210/endrev/bnac015
4. Wang W, Seale P. Control of brown and beige fat development. Nat Rev Mol Cell Biol. (2016) 17:691–702. doi: 10.1038/nrm.2016.96
5. Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev. (2013) 27:234–50. doi: 10.1101/gad.211649.112
6. Stanford KI, Middelbeek RJ, Townsend KL, An D, Nygaard EB, Hitchcox KM, et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J Clin Invest. (2013) 123:215–23. doi: 10.1172/JCI62308
7. Cypess AM, White AP, Vernochet C, Schulz TJ, Xue R, Sass CA, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med. (2013) 19:635–9. doi: 10.1038/nm.3112
8. Zwick RK, Guerrero-Juarez CF, Horsley V, Plikus MV. Anatomical, physiological, and functional diversity of adipose tissue. Cell Metab. (2018) 27:68–83. doi: 10.1016/j.cmet.2017.12.002
9. van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. (2009) 360:1500–8. doi: 10.1056/NEJMoa0808718
10. Yoneshiro T, Aita S, Matsushita M, Okamatsu-Ogura Y, Kameya T, Kawai Y, et al. Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obes (Silver Spring). (2011) 19:1755–60. doi: 10.1038/oby.2011.125
11. Yoneshiro T, Aita S, Matsushita M, Kayahara T, Kameya T, Kawai Y, et al. Recruited brown adipose tissue as an antiobesity agent in humans. J Clin Invest. (2013) 123:3404–8. doi: 10.1172/JCI67803
12. Cypess AM, Weiner LS, Roberts-Toler C, Franquet Elía E, Kessler SH, Kahn PA, et al. Activation of human brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metab. (2015) 21:33–8. doi: 10.1016/j.cmet.2014.12.009
13. van der Lans AA, Hoeks J, Brans B, Vijgen GH, Visser MG, Vosselman MJ, et al. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. J Clin Invest. (2013) 123:3395–403. doi: 10.1172/JCI68993
14. Sidossis LS, Porter C, Saraf MK, Børsheim E, Radhakrishnan RS, Chao T, et al. Browning of subcutaneous white adipose tissue in humans after severe adrenergic stress. Cell Metab. (2015) 22:219–27. doi: 10.1016/j.cmet.2015.06.022
15. Villarroya F, Cereijo R, Villarroya J, Giralt M. Brown adipose tissue as a secretory organ. Nat Rev Endocrinol. (2017) 13:26–35. doi: 10.1038/nrendo.2016.136
16. Fog CK, Galli GG, Lund AH. PRDM proteins: important players in differentiation and disease. Bioessays. (2012) 34:50–60. doi: 10.1002/bies.201100107
17. Hohenauer T, Moore AW. The Prdm family: expanding roles in stem cells and development. Development. (2012) 139:2267–82. doi: 10.1242/dev.070110
18. Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J, et al. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. (2000) 96:3209–14. doi: 10.1182/blood.V96.9.3209
19. Nishikata I, Sasaki H, Iga M, Tateno Y, Imayoshi S, Asou N, et al. A novel EVI1 gene family, MEL1, lacking a PR domain (MEL1S) is expressed mainly in t(1;3)(p36;q21)-positive AML and blocks G-CSF-induced myeloid differentiation. Blood. (2003) 102:3323–32. doi: 10.1182/blood-2002-12-3944
20. Chi J, Cohen P. The multifaceted roles of PRDM16: adipose biology and beyond. Trends Endocrinol Metab. (2016) 27:11–23. doi: 10.1016/j.tem.2015.11.005
21. Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. (2007) 6:38–54. doi: 10.1016/j.cmet.2007.06.001
22. Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. (2007) 131:242–56. doi: 10.1016/j.cell.2007.10.004
23. Manolopoulos KN, Karpe F, Frayn KN. Gluteofemoral body fat as a determinant of metabolic health. Int J Obes (Lond). (2010) 34:949–59. doi: 10.1038/ijo.2009.286
24. Porter SA, Massaro JM, Hoffmann U, Vasan RS, O’Donnel CJ, Fox CS. Abdominal subcutaneous adipose tissue: a protective fat depot? Diabetes Care. (2009) 32:1068–75. doi: 10.2337/dc08-2280
25. Ross R, Fortier L, Hudson R. Separate associations between visceral and subcutaneous adipose tissue distribution, insulin and glucose levels in obese women. Diabetes Care. (1996) 19:1404–11. doi: 10.2337/diacare.19.12.1404
26. Snijder MB, Visser M, Dekker JM, Goodpaster BH, Harris TB, Kritchevsky SB, et al. Low subcutaneous thigh fat is a risk factor for unfavourable glucose and lipid levels, independently of high abdominal fat. The Health ABC Study. Diabetologia. (2005) 48:301–8. doi: 10.1007/s00125-004-1637-7
27. Kim JY, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest. (2007) 117:2621–37. doi: 10.1172/JCI31021
28. Fox CS, Massaro JM, Hoffmann U, Pou KM, Maurovich-Horvat P, Liu CY, et al. Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation. (2007) 116:39–48. doi: 10.1161/CIRCULATIONAHA.106.675355
29. Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. (2011) 121:96–105. doi: 10.1172/JCI44271
30. Cohen P, Levy Julia D, Zhang Y, Frontini A, Kolodin Dmitriy P, Svensson Katrin J, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. (2014) 156:304–16. doi: 10.1016/j.cell.2013.12.021
31. Raajendiran A, Ooi G, Bayliss J, O’Brien PE, Schittenhelm RB, Clark AK, et al. Identification of metabolically distinct adipocyte progenitor cells in human adipose tissues. Cell Rep. (2019) 27:1528–40.e7. doi: 10.1016/j.celrep.2019.04.010
32. Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. (1994) 79:1147–56. doi: 10.1016/0092-8674(94)90006-X
33. Rosen ED, Sarraf P, Troy AE, Bradwin G, Moore K, Milstone DS, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. (1999) 4:611–7. doi: 10.1016/S1097-2765(00)80211-7
34. Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. (2008) 454:961–7. doi: 10.1038/nature07182
35. Angueira AR, Sakers AP, Holman CD, Cheng L, Arbocco MN, Shamsi F, et al. Defining the lineage of thermogenic perivascular adipose tissue. Nat Metab. (2021) 3:469–84. doi: 10.1038/s42255-021-00380-0
36. Ishibashi J, Firtina Z, Rajakumari S, Wood KH, Conroe HM, Steger DJ, et al. An Evi1-C/EBPβ complex controls peroxisome proliferator-activated receptor γ2 gene expression to initiate white fat cell differentiation. Mol Cell Biol. (2012) 32:2289–99. doi: 10.1128/MCB.06529-11
37. Harms MJ, Ishibashi J, Wang W, Lim HW, Goyama S, Sato T, et al. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab. (2014) 19:593–604. doi: 10.1016/j.cmet.2014.03.007
38. Svensson KJ, Long JZ, Jedrychowski MP, Cohen P, Lo JC, Serag S, et al. A secreted slit2 fragment regulates adipose tissue thermogenesis and metabolic function. Cell Metab. (2016) 23:454–66. doi: 10.1016/j.cmet.2016.01.008
39. Ouellet V, Routhier-Labadie A, Bellemare W, Lakhal-Chaieb L, Turcotte E, Carpentier AC, et al. Outdoor temperature, age, sex, body mass index, and diabetic status determine the prevalence, mass, and glucose-uptake activity of 18F-FDG-detected BAT in humans. J Clin Endocrinol Metab. (2011) 96:192–9. doi: 10.1210/jc.2010-0989
40. Pfannenberg C, Werner MK, Ripkens S, Stef I, Deckert A, Schmadl M, et al. Impact of age on the relationships of brown adipose tissue with sex and adiposity in humans. Diabetes. (2010) 59:1789–93. doi: 10.2337/db10-0004
41. Wang W, Ishibashi J, Trefely S, Shao M, Cowan AJ, Sakers A, et al. A PRDM16-driven metabolic signal from adipocytes regulates precursor cell fate. Cell Metab. (2019) 30:174–89 e5. doi: 10.1016/j.cmet.2019.05.005
42. Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. (2009) 460:1154–8. doi: 10.1038/nature08262
43. Hiraike Y, Waki H, Yu J, Nakamura M, Miyake K, Nagano G, et al. NFIA co-localizes with PPARγ and transcriptionally controls the brown fat gene program. Nat Cell Biol. (2017) 19:1081–92. doi: 10.1038/ncb3590
44. Hiraike Y, Saito K, Oguchi M, Wada T, Toda G, Tsutsumi S, et al. NFIA in adipocytes reciprocally regulates mitochondrial and inflammatory gene program to improve glucose homeostasis. Proc Natl Acad Sci. (2023) 120:e2308750120. doi: 10.1073/pnas.2308750120
45. Wang W, Kissig M, Rajakumari S, Huang L, H-w L, Won K-J, et al. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc Natl Acad Sci. (2014) 111:14466–71. doi: 10.1073/pnas.1412685111
46. Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. (2013) 17:562–74. doi: 10.1016/j.cmet.2013.01.015
47. Shapira SN, Lim H-W, Rajakumari S, Sakers AP, Ishibashi J, Harms MJ, et al. EBF2 transcriptionally regulates brown adipogenesis via the histone reader DPF3 and the BAF chromatin remodeling complex. Genes Dev. (2017) 31:660–73. doi: 10.1101/gad.294405.116
48. Stine RR, Shapira SN, Lim H-W, Ishibashi J, Harms M, Won K-J, et al. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Mol Metab. (2016) 5:57–65. doi: 10.1016/j.molmet.2015.11.001
49. Emont MP, Jacobs C, Essene AL, Pant D, Tenen D, Colleluori G, et al. A single-cell atlas of human and mouse white adipose tissue. Nature. (2022) 603:926–33. doi: 10.1038/s41586-022-04518-2
50. Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. (2005) 19:815–26. doi: 10.1101/gad.1284005
51. Pinheiro I, Margueron R, Shukeir N, Eisold M, Fritzsch C, Richter FM, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. (2012) 150:948–60. doi: 10.1016/j.cell.2012.06.048
52. Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. (2013) 504:163–7. doi: 10.1038/nature12652
53. Nagano G, Ohno H, Oki K, Kobuke K, Shiwa T, Yoneda M, et al. Activation of classical brown adipocytes in the adult human perirenal depot is highly correlated with PRDM16-EHMT1 complex expression. PloS One. (2015) 10:e0122584. doi: 10.1371/journal.pone.0122584
54. Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. (2003) 12:1137–49. doi: 10.1016/S1097-2765(03)00391-5
55. Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell Dev Biol. (2011) 22:759–68. doi: 10.1016/j.semcdb.2011.07.022
56. Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. (2010) 11:761–72. doi: 10.1038/nrg2901
57. Harms MJ, Lim HW, Ho Y, Shapira SN, Ishibashi J, Rajakumari S, et al. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. (2015) 29:298–307. doi: 10.1101/gad.252734.114
58. Iida S, Chen W, Nakadai T, Ohkuma Y, Roeder RG. PRDM16 enhances nuclear receptor-dependent transcription of the brown fat-specific Ucp1 gene through interactions with Mediator subunit MED1. Genes Dev. (2015) 29:308–21. doi: 10.1101/gad.252809.114
59. Strobl B, Stoiber D, Sexl V, Mueller M. Tyrosine kinase 2 (TYK2) in cytokine signalling and host immunity. Front Bioscience. (2011) 16:3214–32. doi: 10.2741/3908
60. Derecka M, Gornicka A, Koralov SB, Szczepanek K, Morgan M, Raje V, et al. Tyk2 and Stat3 regulate brown adipose tissue differentiation and obesity. Cell Metab. (2012) 16:814–24. doi: 10.1016/j.cmet.2012.11.005
61. Raje V, Derecka M, Cantwell M, Meier J, Szczepanek K, Sisler JD, et al. Kinase inactive tyrosine kinase (Tyk2) supports differentiation of brown fat cells. Endocrinology. (2017) 158:148–57. doi: 10.1210/en.2015-2048
62. Dempersmier J, Sambeat A, Gulyaeva O, Paul SM, Hudak CS, Raposo HF, et al. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol Cell. (2015) 57:235–46. doi: 10.1016/j.molcel.2014.12.005
63. Seckl JR, Walker BR. Minireview: 11beta-hydroxysteroid dehydrogenase type 1- a tissue-specific amplifier of glucocorticoid action. Endocrinology. (2001) 142:1371–6. doi: 10.1210/endo.142.4.8114
64. Sambeat A, Gulyaeva O, Dempersmier J, Tharp Kevin M, Stahl A, Paul Sarah M, et al. LSD1 interacts with zfp516 to promote UCP1 transcription and brown fat program. Cell Rep. (2016) 15:2536–49. doi: 10.1016/j.celrep.2016.05.019
65. Zeng X, Jedrychowski MP, Chen Y, Serag S, Lavery GG, Gygi SP, et al. Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation. Genes Dev. (2016) 30:1822–36. doi: 10.1101/gad.285312.116
66. Lin JZ, Farmer SR. LSD1-a pivotal epigenetic regulator of brown and beige fat differentiation and homeostasis. Genes Dev. (2016) 30:1793–5. doi: 10.1101/gad.288720.116
67. Huang L, Pan D, Chen Q, Zhu LJ, Ou J, Wabitsch M, et al. Transcription factor Hlx controls a systematic switch from white to brown fat through Prdm16-mediated co-activation. Nat Commun. (2017) 8:68. doi: 10.1038/s41467-017-00098-2
68. Shen H, He T, Wang S, Hou L, Wei Y, Liu Y, et al. SOX4 promotes beige adipocyte-mediated adaptive thermogenesis by facilitating PRDM16-PPARγ complex. Theranostics. (2022) 12:7699–716. doi: 10.7150/thno.77102
69. Koo JH, Guan KL. Interplay between YAP/TAZ and metabolism. Cell Metab. (2018) 28:196–206. doi: 10.1016/j.cmet.2018.07.010
70. Wang L, Wang S, Shi Y, Li R, Günther S, Ong YT, et al. YAP and TAZ protect against white adipocyte cell death during obesity. Nat Commun. (2020) 11:5455. doi: 10.1038/s41467-020-19229-3
71. El Ouarrat D, Isaac R, Lee YS, Oh DY, Wollam J, Lackey D, et al. TAZ is a negative regulator of PPARγ activity in adipocytes and TAZ deletion improves insulin sensitivity and glucose tolerance. Cell Metab. (2020) 31:162–73.e5. doi: 10.1016/j.cmet.2019.10.003
72. Zeng X, Ye M, Resch JM, Jedrychowski MP, Hu B, Lowell BB, et al. Innervation of thermogenic adipose tissue via a calsyntenin 3β-S100b axis. Nature. (2019) 569:229–35. doi: 10.1038/s41586-019-1156-9
73. Qian K, Tol MJ, Wu J, Uchiyama LF, Xiao X, Cui L, et al. CLSTN3β enforces adipocyte multilocularity to facilitate lipid utilization. Nature. (2023) 613:160–8. doi: 10.1038/s41586-022-05507-1
74. Huang X, Li X, Shen H, Zhao Y, Zhou Z, Wang Y, et al. Transcriptional repression of beige fat innervation via a YAP/TAZ-S100B axis. Nat Commun. (2023) 14:7102. doi: 10.1038/s41467-023-43021-8
75. Hasegawa Y, Ikeda K, Chen Y, Alba DL, Stifler D, Shinoda K, et al. Repression of adipose tissue fibrosis through a PRDM16-GTF2IRD1 complex improves systemic glucose homeostasis. Cell Metab. (2018) 27:180–94 e6. doi: 10.1016/j.cmet.2017.12.005
76. Villanueva CJ, Waki H, Godio C, Nielsen R, Chou WL, Vargas L, et al. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. (2011) 13:413–27. doi: 10.1016/j.cmet.2011.02.014
77. Villanueva CJ, Vergnes L, Wang J, Drew BG, Hong C, Tu Y, et al. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARγ specifies lipid storage versus thermogenic gene programs. Cell Metab. (2013) 17:423–35. doi: 10.1016/j.cmet.2013.01.016
78. Ortega FJ, Serrano M, Rodriguez-Cuenca S, Moreno-Navarrete JM, Gómez-Serrano M, Sabater M, et al. Transducin-like enhancer of split 3 (TLE3) in adipose tissue is increased in situations characterized by decreased PPARγ gene expression. J Mol Med. (2014) 93:83–92. doi: 10.1007/s00109-014-1207-5
79. Lodhi IJ, Yin L, Jensen-Urstad AP, Funai K, Coleman T, Baird JH, et al. Inhibiting adipose tissue lipogenesis reprograms thermogenesis and PPARγ activation to decrease diet-induced obesity. Cell Metab. (2012) 16:189–201. doi: 10.1016/j.cmet.2012.06.013
80. Lodhi IJ, Dean JM, He A, Park H, Tan M, Feng C, et al. PexRAP inhibits PRDM16-mediated thermogenic gene expression. Cell Rep. (2017) 20:2766–74. doi: 10.1016/j.celrep.2017.08.077
81. Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. (2000) 113:3897–905. doi: 10.1242/jcs.113.22.3897
82. Patil M, Sharma BK, Elattar S, Chang J, Kapil S, Yuan J, et al. Id1 promotes obesity by suppressing brown adipose thermogenesis and white adipose browning. Diabetes. (2017) 66:1611–25. doi: 10.2337/db16-1079
83. Gupta RK, Arany Z, Seale P, Mepani RJ, Ye L, Conroe HM, et al. Transcriptional control of preadipocyte determination by Zfp423. Nature. (2010) 464:619–23. doi: 10.1038/nature08816
84. Shao M, Ishibashi J, Kusminski CM, Wang QA, Hepler C, Vishvanath L, et al. Zfp423 maintains white adipocyte identity through suppression of the beige cell thermogenic gene program. Cell Metab. (2016) 23:1167–84. doi: 10.1016/j.cmet.2016.04.023
85. Shao M, Zhang Q, Truong A, Shan B, Vishvanath L, Li L, et al. ZFP423 controls EBF2 coactivator recruitment and PPARγ occupancy to determine the thermogenic plasticity of adipocytes. Genes Dev. (2021) 35:1461–74. doi: 10.1101/gad.348780.121
86. Sheeba CJ, Logan MP. The roles of T-box genes in vertebrate limb development. Curr Topics Dev Biol. (2017) 122:355–81. doi: 10.1016/bs.ctdb.2016.08.009
87. Gburcik V, Cawthorn WP, Nedergaard J, Timmons JA, Cannon B. An essential role for Tbx15 in the differentiation of brown and “brite” but not white adipocytes. Am J Physiol Endocrinol Metab. (2012) 303:E1053–60. doi: 10.1152/ajpendo.00104.2012
88. Gesta S, Bezy O, Mori MA, Macotela Y, Lee KY, Kahn CR. Mesodermal developmental gene Tbx15 impairs adipocyte differentiation and mitochondrial respiration. Proc Natl Acad Sci USA. (2011) 108:2771–6. doi: 10.1073/pnas.1019704108
89. Lee KY, Singh MK, Ussar S, Wetzel P, Hirshman MF, Goodyear LJ, et al. Tbx15 controls skeletal muscle fibre-type determination and muscle metabolism. Nat Commun. (2015) 6:8054. doi: 10.1038/ncomms9054
90. Gesta S, Blüher M, Yamamoto Y, Norris AW, Berndt J, Kralisch S, et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci USA. (2006) 103:6676–81. doi: 10.1073/pnas.0601752103
91. Sun W, Zhao X, Wang Z, Chu Y, Mao L, Lin S, et al. Tbx15 is required for adipocyte browning induced by adrenergic signaling pathway. Mol Metab. (2019) 28:48–57. doi: 10.1016/j.molmet.2019.07.004
92. Reinehr T, de Sousa G, Roth CL, Andler W. Androgens before and after weight loss in obese children. J Clin Endocrinol Metab. (2005) 90:5588–95. doi: 10.1210/jc.2005-0438
93. Yanase T, Fan W, Kyoya K, Min L, Takayanagi R, Kato S, et al. Androgens and metabolic syndrome: lessons from androgen receptor knock out (ARKO) mice. J Steroid Biochem Mol Biol. (2008) 109:254–7. doi: 10.1016/j.jsbmb.2008.03.017
94. Zhao S, Nie T, Li L, Long Q, Gu P, Zhang Y, et al. Androgen receptor is a negative regulator of PRDM16 in beige adipocyte. Advanced Sci. (2023) 10:e2300070. doi: 10.1002/advs.202300070
95. Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. (1998) 393:382–6. doi: 10.1038/30756
96. Bi P, Shan T, Liu W, Yue F, Yang X, Liang XR, et al. Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nat Med. (2014) 20:911–8. doi: 10.1038/nm.3615
97. Martínez de Morentin PB, González-García I, Martins L, Lage R, Fernández-Mallo D, Martínez-Sánchez N, et al. Estradiol regulates brown adipose tissue thermogenesis via hypothalamic AMPK. Cell Metab. (2014) 20:41–53. doi: 10.1016/j.cmet.2014.03.031
98. Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. (2008) 454:766–70. doi: 10.1038/nature07107
99. Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. (2011) 473:398–402. doi: 10.1038/nature10008
100. Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. (2014) 156:45–68. doi: 10.1016/j.cell.2013.12.019
101. Yang Q, Liang X, Sun X, Zhang L, Fu X, Rogers CJ, et al. AMPK/α-ketoglutarate axis dynamically mediates DNA demethylation in the prdm16 promoter and brown adipogenesis. Cell Metab. (2016) 24:542–54. doi: 10.1016/j.cmet.2016.08.010
102. Peng WQ, Xiao G, Li BY, Guo YY, Guo L, Tang QQ. l-theanine activates the browning of white adipose tissue through the AMPK/α-ketoglutarate/prdm16 axis and ameliorates diet-induced obesity in mice. Diabetes. (2021) 70:1458–72. doi: 10.2337/db20-1210
103. Shuai L, Zhang LN, Li BH, Tang CL, Wu LY, Li J, et al. SIRT5 regulates brown adipocyte differentiation and browning of subcutaneous white adipose tissue. Diabetes. (2019) 68:1449–61. doi: 10.2337/db18-1103
104. Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. (2012) 150:620–32. doi: 10.1016/j.cell.2012.06.027
105. Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol Biol Cell. (2009) 20:801–8. doi: 10.1091/mbc.e08-06-0647
106. Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. (2005) 280:13560–7. doi: 10.1074/jbc.M414670200
107. Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. (2015) 29:1343–55. doi: 10.1101/gad.262766.115
108. Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. (2018) 561:556–60. doi: 10.1038/s41586-018-0538-8
109. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. (2016) 537:369–73. doi: 10.1038/nature19342
110. Wang Y, Gao M, Zhu F, Li X, Yang Y, Yan Q, et al. METTL3 is essential for postnatal development of brown adipose tissue and energy expenditure in mice. Nat Commun. (2020) 11:1648. doi: 10.1038/s41467-020-15488-2
111. Zhou JY, Li L. MicroRNAs are key regulators of brown adipogenesis. Biochim Biophys Acta. (2014) 1841:1590–5. doi: 10.1016/j.bbalip.2014.08.009
112. Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol. (2012) 14:1330–5. doi: 10.1038/ncb2612
113. Yin H, Pasut A, Soleimani VD, Bentzinger CF, Antoun G, Thorn S, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. (2013) 17:210–24. doi: 10.1016/j.cmet.2013.01.004
114. He L, Tang M, Xiao T, Liu H, Liu W, Li G, et al. Obesity-Associated miR-199a/214 Cluster Inhibits Adipose Browning via PRDM16-PGC-1α Transcriptional Network. Diabetes. (2018) 67:2585–600. doi: 10.2337/db18-0626
115. Ding H, Zheng S, Garcia-Ruiz D, Hou D, Wei Z, Liao Z, et al. Fasting induces a subcutaneous-to-visceral fat switch mediated by microRNA-149-3p and suppression of PRDM16. Nat Commun. (2016) 7:11533. doi: 10.1038/ncomms11533
116. Zheng S, Guo S, Sun G, Shi Y, Wei Z, Tang Y, et al. Gain of metabolic benefit with ablation of miR-149-3p from subcutaneous adipose tissue in diet-induced obese mice. Mol Ther Nucleic Acids. (2019) 18:194–203. doi: 10.1016/j.omtn.2019.07.024
117. Chou CF, Lin YY, Wang HK, Zhu X, Giovarelli M, Briata P, et al. KSRP ablation enhances brown fat gene program in white adipose tissue through reduced miR-150 expression. Diabetes. (2014) 63:2949–61. doi: 10.2337/db13-1901
118. Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. (2012) 15:395–404. doi: 10.1016/j.cmet.2012.01.019
119. Liu B, Liu YF, Du YR, Mardaryev AN, Yang W, Chen H, et al. Cbx4 regulates the proliferation of thymic epithelial cells and thymus function. Development. (2013) 140:780–8. doi: 10.1242/dev.085035
120. Chen Q, Huang L, Pan D, Zhu LJ, Wang YX. Cbx4 sumoylates prdm16 to regulate adipose tissue thermogenesis. Cell Rep. (2018) 22:2860–72. doi: 10.1016/j.celrep.2018.02.057
121. Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. (2007) 8:904–16. doi: 10.1038/nrm2261
122. Uchida T, Furumai K, Fukuda T, Akiyama H, Takezawa M, Asano T, et al. Prolyl isomerase Pin1 regulates mouse embryonic fibroblast differentiation into adipose cells. PloS One. (2012) 7:e31823. doi: 10.1371/journal.pone.0031823
123. Nakatsu Y, Sakoda H, Kushiyama A, Zhang J, Ono H, Fujishiro M, et al. Peptidyl-prolyl cis/trans isomerase NIMA-interacting 1 associates with insulin receptor substrate-1 and enhances insulin actions and adipogenesis. J Biol Chem. (2011) 286:20812–22. doi: 10.1074/jbc.M110.206904
124. Nakatsu Y, Matsunaga Y, Yamamotoya T, Ueda K, Inoue MK, Mizuno Y, et al. Prolyl isomerase pin1 suppresses thermogenic programs in adipocytes by promoting degradation of transcriptional co-activator PRDM16. Cell Rep. (2019) 26:3221–30.e3. doi: 10.1016/j.celrep.2019.02.066
125. Wang Q, Li H, Tajima K, Verkerke ARP, Taxin ZH, Hou Z, et al. Post-translational control of beige fat biogenesis by PRDM16 stabilization. Nature. (2022) 609:151–8. doi: 10.1038/s41586-022-05067-4
126. Yoneshiro T, Wang Q, Tajima K, Matsushita M, Maki H, Igarashi K, et al. BCAA catabolism in brown fat controls energy homeostasis through SLC25A44. Nature. (2019) 572:614–9. doi: 10.1038/s41586-019-1503-x
127. Ma QX, Zhu WY, Lu XC, Jiang D, Xu F, Li JT, et al. BCAA-BCKA axis regulates WAT browning through acetylation of PRDM16. Nat Metab. (2022) 4:106–22. doi: 10.1038/s42255-021-00520-6
128. Verkerke ARP, Wang D, Yoshida N, Taxin ZH, Shi X, Zheng S, et al. BCAA-nitrogen flux in brown fat controls metabolic health independent of thermogenesis. Cell. (2024) 187:2359–74.e18. doi: 10.1016/j.cell.2024.03.030
129. Pan MH, Koh YC, Lee TL, Wang B, Chen WK, Nagabhushanam K, et al. Resveratrol and oxyresveratrol activate thermogenesis via different transcriptional coactivators in high-fat diet-induced obese mice. J Agric Food Chem. (2019) 67:13605–16. doi: 10.1021/acs.jafc.9b05963
130. Liu X, Zhang Y, Chu Y, Zhao X, Mao L, Zhao S, et al. The natural compound rutaecarpine promotes white adipocyte browning through activation of the AMPK-PRDM16 axis. Biochem Biophys Res Commun. (2021) 545:189–94. doi: 10.1016/j.bbrc.2021.01.080
131. Jiang N, Yang M, Han Y, Zhao H, Sun L. PRDM16 regulating adipocyte transformation and thermogenesis: A promising therapeutic target for obesity and diabetes. Front Pharmacol. (2022) 13. doi: 10.3389/fphar.2022.870250
132. Wibmer AG, Becher T, Eljalby M, Crane A, Andrieu PC, Jiang CS, et al. Brown adipose tissue is associated with healthier body fat distribution and metabolic benefits independent of regional adiposity. Cell Rep Med. (2021) 2:100332. doi: 10.1016/j.xcrm.2021.100332
133. Becher T, Palanisamy S, Kramer DJ, Eljalby M, Marx SJ, Wibmer AG, et al. Brown adipose tissue is associated with cardiometabolic health. Nat Med. (2021) 27:58–65. doi: 10.1038/s41591-020-1126-7
134. Ouellet V, Labbe SM, Blondin DP, Phoenix S, Guerin B, Haman F, et al. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J Clin Invest. (2012) 122:545–52. doi: 10.1172/JCI60433
135. UD M, Raiko J, Saari T, Kudomi N, Tolvanen T, Oikonen V, et al. Human brown adipose tissue [(15)O]O2 PET imaging in the presence and absence of cold stimulus. Eur J Nucl Med Mol Imaging. (2016) 43:1878–86. doi: 10.1007/s00259-016-3364-y
136. Baskin AS, Linderman JD, Brychta RJ, McGehee S, Anflick-Chames E, Cero C, et al. Regulation of human adipose tissue activation, gallbladder size, and bile acid metabolism by a beta3-adrenergic receptor agonist. Diabetes. (2018) 67:2113–25. doi: 10.2337/db18-0462
137. O’Mara AE, Johnson JW, Linderman JD, Brychta RJ, McGehee S, Fletcher LA, et al. Chronic mirabegron treatment increases human brown fat, HDL cholesterol, and insulin sensitivity. J Clin Invest. (2020) 130:2209–19. doi: 10.1172/JCI131126
138. Finlin BS, Memetimin H, Zhu B, Confides AL, Vekaria HJ, El Khouli RH, et al. The β3-adrenergic receptor agonist mirabegron improves glucose homeostasis in obese humans. J Clin Invest. (2020) 130:2319–31. doi: 10.1172/JCI134892
139. Loh RKC, Formosa MF, La Gerche A, Reutens AT, Kingwell BA, Carey AL. Acute metabolic and cardiovascular effects of mirabegron in healthy individuals. Diabetes Obes Metab. (2019) 21:276–84. doi: 10.1111/dom.13516
140. Dąbrowska AM, Dudka J. Mirabegron, a selective β3-adrenergic receptor agonist, as a potential anti-obesity drug. J Clin Med. (2023) 12:6897. doi: 10.3390/jcm12216897
141. Zhang N, Zhou H, Xu Y, Zhang Y, Yu F, Gui L, et al. Liraglutide promotes UCP1 expression and lipolysis of adipocytes by promoting the secretion of irisin from skeletal muscle cells. Mol Cell Endocrinol. (2024) 588:112225. doi: 10.1016/j.mce.2024.112225
142. Martins FF, Marinho TS, Cardoso LEM, Barbosa-da-Silva S, Souza-Mello V, Aguila MB, et al. Semaglutide (GLP-1 receptor agonist) stimulates browning on subcutaneous fat adipocytes and mitigates inflammation and endoplasmic reticulum stress in visceral fat adipocytes of obese mice. Cell Biochem Funct. (2022) 40:903–13. doi: 10.1002/cbf.3751
143. van Eyk HJ, Paiman EHM, Bizino MB, SL IJ, Kleiburg F, Boers TGW, et al. Liraglutide decreases energy expenditure and does not affect the fat fraction of supraclavicular brown adipose tissue in patients with type 2 diabetes. Nutrition Metabolism Cardiovasc Diseases: NMCD. (2020) 30:616–24. doi: 10.1016/j.numecd.2019.12.005
144. Blundell J, Finlayson G, Axelsen M, Flint A, Gibbons C, Kvist T, et al. Effects of once-weekly semaglutide on appetite, energy intake, control of eating, food preference and body weight in subjects with obesity. Diabetes Obes Metab. (2017) 19:1242–51. doi: 10.1111/dom.12932
145. Drucker DJ. GLP-1 physiology informs the pharmacotherapy of obesity. Mol Metab. (2022) 57:101351. doi: 10.1016/j.molmet.2021.101351
146. Aguilo F, Avagyan S, Labar A, Sevilla A, Lee DF, Kumar P, et al. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood. (2011) 117:5057–66. doi: 10.1182/blood-2010-08-300145
147. Chuikov S, Levi BP, Smith ML, Morrison SJ. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat Cell Biol. (2010) 12:999–1006. doi: 10.1038/ncb2101
148. Cibi DM, Bi-Lin KW, Shekeran SG, Sandireddy R, Tee N, Singh A, et al. Prdm16 deficiency leads to age-dependent cardiac hypertrophy, adverse remodeling, mitochondrial dysfunction, and heart failure. Cell Rep. (2020) 33:108288. doi: 10.1016/j.celrep.2020.108288
Keywords: obesity, brown adipocyte, beige adipocyte, PRDM16, UCP1
Citation: Mao L, Lu J, Hou Y and Nie T (2024) Directly targeting PRDM16 in thermogenic adipose tissue to treat obesity and its related metabolic diseases. Front. Endocrinol. 15:1458848. doi: 10.3389/fendo.2024.1458848
Received: 03 July 2024; Accepted: 28 August 2024;
Published: 16 September 2024.
Edited by:
Claire Joanne Stocker, Aston University, United KingdomReviewed by:
Endre Károly Kristóf, University of Debrecen, HungaryYuta Hiraike, The University of Tokyo, Japan
Manu Sudhakar, Amrita Vishwa Vidyapeetham University, India
Copyright © 2024 Mao, Lu, Hou and Nie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Nie, bmllX3Rhb0BoYnVhcy5lZHUuY24=
 Liufeng Mao
Liufeng Mao Jinli Lu1
Jinli Lu1 Tao Nie
Tao Nie