- 1Genetics and Bioinformatics Department, Dasman Diabetes Institute, Kuwait City, Kuwait
- 2Department of Medical Laboratories, Ahmadi Hospital, Kuwait Oil Company (KOC), Ahmadi, Kuwait
- 3Saad Al-Abdullah Academy for Security Sciences, Ministry of Interior, Shuwaikh, Kuwait
Background: Numerous studies have linked mitochondrial dysfunction to the development of type 2 diabetes (T2D) by affecting glucose-stimulated insulin secretion in pancreatic beta cells and reducing oxidative phosphorylation in insulin-responsive tissues. Given the strong genetic underpinnings of T2D, research has explored the connection between mitochondrial DNA haplogroups, specific variants, and the risk and comorbidities of T2D. For example, haplogroups F, D, M9, and N9a have been linked to an elevated risk of T2D across various populations. Additionally, specific mitochondrial DNA variants, such as the rare mtDNA 3243 A>G and the more prevalent mtDNA 16189 T>C, have also been implicated in heightened T2D risk. Notably, these associations vary among different populations. Given the high incidence of T2D in the Gulf Cooperation Council countries, this study investigates the correlation between T2D and mitochondrial haplogroups and variants in Arab populations from the Gulf region.
Methods: This analysis involved mitochondrial haplogroup and variant testing in a cohort of 1,112 native Kuwaiti and Qatari individuals, comprising 685 T2D patients and 427 controls. Complete mitochondrial genomes were derived from whole exome sequencing data to examine the associations between T2D and haplogroups and mitochondrial DNA variants.
Results: The analysis revealed a significant protective effect of haplogroup H against T2D (odds ratio [OR] = 0.65; P = 0.022). This protective association persisted when adjusted for age, sex, body mass index (BMI) and population group, with an OR of 0.607 (P = 0.021). Furthermore, specific mitochondrial variants showed significant associations with T2D risk after adjustment for relevant covariates, and some variants were exclusively found in T2D patients.
Conclusion: Our findings confirm that the maternal haplogroup H, previously identified as protective against obesity in Kuwaiti Arabs, also serves as a protective factor against T2D in Arabs from the Gulf region. The study also identifies mitochondrial DNA variants that either increase or decrease the risk of T2D, underscoring their role in cellular energy metabolism.
Introduction
Type 2 diabetes (T2D) is a chronic adult-onset metabolic disorder characterized by high blood sugar due to insulin resistance or inadequate insulin production by pancreatic beta cells. Major complications of T2D include retinopathy, nephropathy, and neuropathy, which can lead to blindness, renal failure, impotence, and foot amputation, respectively (1). The International Diabetes Federation (IDF) has reported that 73 million people have diabetes in the Middle East and North Africa region, with this number projected to increase to 95 million by 2030. Kuwait has a high prevalence of 25.5%, and Qatar has a prevalence of 16.4% among the total adult population (2).
T2D is known to have a heritable component, yet the interactions between environmental and genetic factors complicate their identification. The currently known susceptibility genes for T2D, identified through linkage analysis, candidate gene approaches, and genome-wide association studies on the nuclear genome, contribute only a small percentage to the high estimated genetic heritability of T2D (3–5), suggesting the presence of missing heritability (6).
Mitochondria play a critical role in generating cellular energy, primarily through the process of oxidative phosphorylation (OXPHOS), where adenosine triphosphate (ATP) is synthesized within the inner mitochondrial membrane. Beyond energy production, mitochondria are key players in fatty acid metabolism and the production of reactive oxygen species (ROS), which are essential for oxidative stress responses and the regulation of apoptosis (7). Mitochondrial dysfunction, including reduced oxidative phosphorylation capacity, has been linked to insulin resistance and T2D in classic target organs such as the liver, fat, and muscle (8–11).
The mitochondrial genome (mtDNA) is a maternally inherited, circular, double-stranded molecule that is independent of nuclear DNA. Each cell contains several thousand mitochondria, each housing multiple copies of mtDNA (12, 13). A notable feature of the mtDNA is its proneness to high rates of genetic alteration compared to nuclear DNA especially the D-loop region which is a major non-coding region that oversees mitochondrial transcription and replication (14). Haplogroups, which group individuals based on similar mtDNA variants, provide insight into genetic diversity and evolutionary history across different populations. These variations, including single-nucleotide polymorphisms (SNPs) and insertions or deletions (INDELs), are increasingly recognized as potential markers for diagnosing and understanding complex common disorders (15, 16). Some mtDNA mutations are heteroplasmic, leading to variability in phenotypic expression because the proportion of mtDNA copies with the pathogenic variant is higher than that of wild-type mtDNA copies (17).
There are mitochondrial DNA mutations associated with T2D exhibiting population-wide variation in allele frequencies, for instance, a rare mutation, mtDNA 3243 A>G in the tRNA(Leu)(UUR) gene, causes maternally inherited diabetes and deafness (MIDD); however, its frequency varies across different ethnicities (18, 19). Another example is a common variant, mtDNA 16189 T>C which is associated with an increased risk of T2D in populations with a high frequency of this variant, such as Asian populations (20), but not in European (21) and North African populations (22), where the frequency is low (23). Additionally, the mtDNA haplogroup is proposed to be involved in risk prediction of T2D. Haplogroups F, D, and M9 are associated with an increased risk of T2D (21, 24, 25). However, these haplogroup associations are not consistent across populations; for example, haplogroup N9a is associated with an increased risk of T2D in southern Chinese populations (26), whereas haplogroup N9a is associated with protection against T2D in Japanese and Korean populations (24).
Despite extensive research on mitochondrial dysfunction and diabetes in other regions, there is a significant gap in studies focused on the Arab Gulf region. Studying mitochondrial variants and haplogroups in this region is crucial due to the high prevalence of type 2 diabetes and the unique genetic makeup of these populations. The primary objective of this study is to fill this gap by analysing mtDNA variants and haplogroups in Kuwait and Qatar to understand their impact on diabetes susceptibility. By way of utilizing whole mitochondrial genome sequences extracted from whole exome data of 1,112 individuals from Kuwait and Qatar, this study aims to clarify the relationship of mtDNA haplogroups/variants and type 2 diabetes, potentially enhancing precision medicine by improving the identification of individuals at risk based on their genetic profiles.
Materials and methods
Ethics declaration
This study received approval from the Institutional Ethical Review Committee at Dasman Diabetes Institute, following the ethical principles outlined in the Declaration of Helsinki. While the Qatari whole-exome sequence data utilized in this research were previously published in a separate study by Fakhro et al. (27) and O’Beirne et al. (28), the study adhered to ethical standards, and written informed consent was obtained from all participants. The ethical approval for the study, including the use of the previously collected samples, was granted under the project reference number RA HM 2019-025.
Whole exome data
This secondary analysis involved a cohort of 1,112 unrelated participants from Kuwait and Qatar, collected through national genome sequencing programs designed to investigate genomic variants at the population level. DNA from whole blood of 348 Kuwaiti individuals was sequenced using the Illumina HiSeq platform with TruSeq and Nextera capture kits, as previously described by John et al. (29) (SRA accession: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1162699). DNA from whole blood of 764 Qatari participants with available clinical data (Supplementary Table S1) was sequenced using the Illumina HiSeq platform with the SureSelect Agilent V5 kit, which captures the entire mitochondrial genome (27, 28) (SRA accession: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA290484).
Participants were diagnosed with Type 2 Diabetes based on the American Diabetes Association (ADA) criteria, including fasting blood glucose ≥126 mg/dL and/or HbA1C ≥6.5%. The study included individuals of Arab ethnicity with confirmed ancestry up to three generations. For detailed inclusion criteria, please refer to Hebbar et al. (30) for the Kuwaiti cohort and O’Beirne et al. (28) for the Qatari cohort.
mtDNA sequences, variant calling, and annotation
The FASTQ paired-end Illumina reads were aligned to the human reference assembly GRCh37 with the default setting of Burrows-Wheeler Aligner (BWA-MEM) version v07-17 (31). After alignment, duplicate reads were removed, and the mtDNA sequence (NC_012920.1) was extracted utilizing Picard tool version 2.20.2 (http://broadinstitute.github.io/picard) and SAMtools version 0.1.19 (32). Subsequently, the mtDNA coverage and Genomic Variant Call Format (GVCF) files were generated for each sample with the Genome Analysis Tool Kit (GATK) version v3.8-1-0 (33). Using GATK HaploCaller, the combined 1,112 GVCF files were genotyped, producing a Variant Calling Format (VCF) file that contained the identified mtDNA variants. Finally, mtDNA annotations were performed using the Ensembl Variant Effect Predictor (VEP) (34), the Mitomap (https://www.mitomap.org/) database (35), and Varsome website (https://varsome.com).
mtDNA haplogroup assignment
To determine the mitochondrial haplogroup profiles for all 1,112 individuals, the mtDNA VCF files were analysed using HaploGrep 2 (36) with the phylotree build PT17-FU1 (accessed on February 2, 2024).
Statistical analyses
Statistical analyses were performed using R software version 3.6.2 (https://www.R-project.org/). Descriptive statistics were derived for demographic and anthropometric data, including age, BMI, and T2D status. Continuous variables were summarized as mean ± standard deviation (SD) or as median and interquartile range (IQR). The Chi-square test was used to assess the statistical significance of associations between categorical variables (sex and population) and T2D status. The Mann-Whitney U test was used to examine the associations between age, BMI, and T2D status. A p-value of less than 0.05 was considered statistically significant.
To identify hidden relationships among the samples due to potential covariates, principal component analysis (PCA) was conducted on the complete set of mtDNA variants. PCA was carried out using the PCAtools package in R, and the results were visualized on a biplot, highlighting the principal components (PCs) that captured the most variation in the data.
Fisher’s exact test was employed to test for nominal associations between T2D and mtDNA haplogroups. The odds ratio (OR) and 95% confidence intervals (CI) were calculated for each haplogroup, with a p-value <0.05 set as the threshold for statistical significance. Additionally, logistic regression was implemented using IBM® SPSS® Statistics Version 25 software to adjust for covariates, including sex, age, BMI, and population. Finally, PLINK version 1.9 (71) was used to identify mtDNA variants associated with T2D, with a two-tailed p-value <0.05. In addition, PLINK tool was used for conditional analysis on the top leading variant with the lowest p-value. Independence of associated variants was assessed using LD pruning with the parameters (–indep-pairwise 50 5 0).
Results
Study demographic and clinical profile
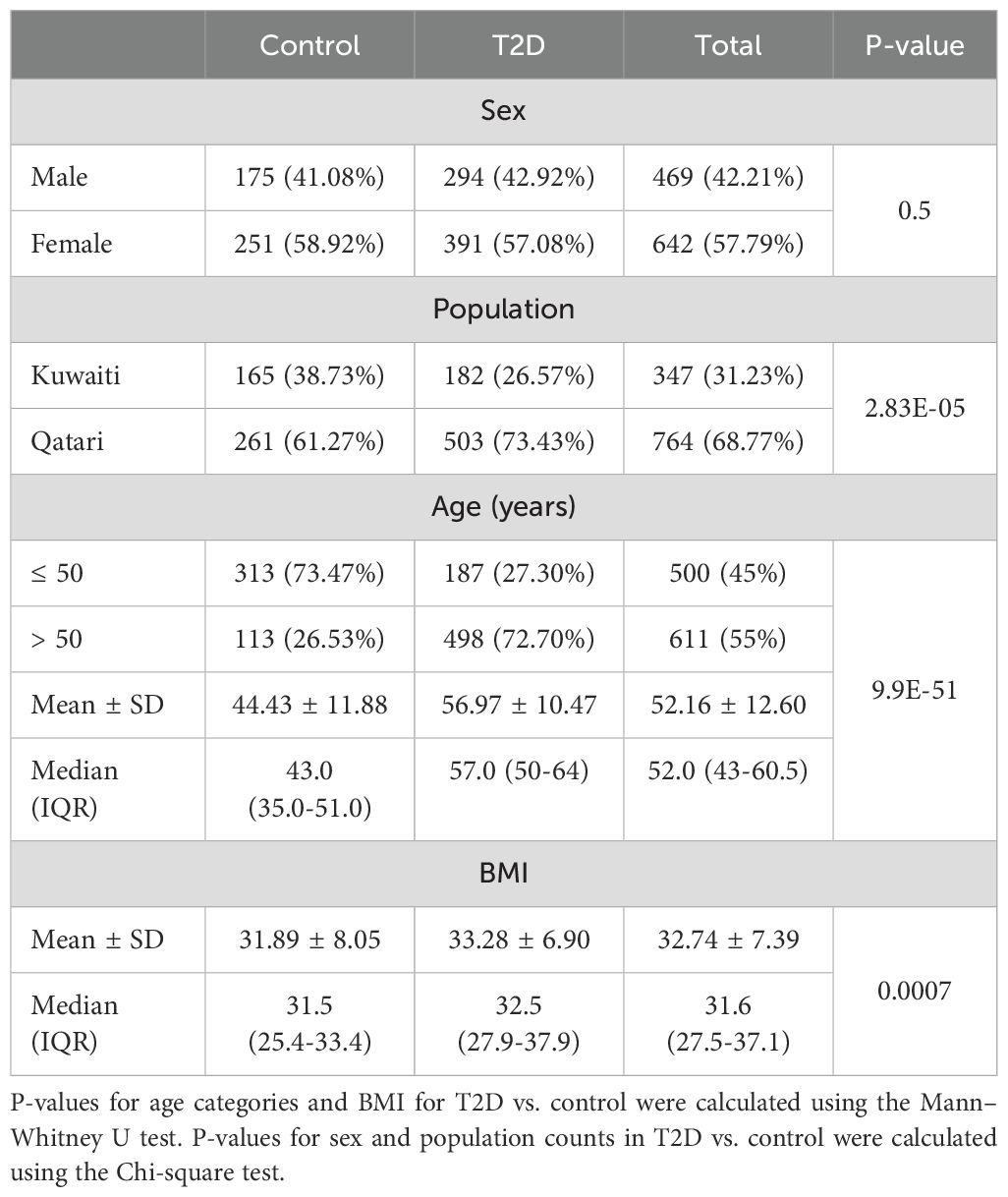
The samples in our study, including those referenced from previous research (27, 29), underwent relatedness assessments based on nuclear DNA to ensure unrelated samples. Table 1 presents descriptive statistics for the dataset containing 1,112 Kuwaiti and Qatari individuals. The Mann-Whitney U test displayed significant differences between T2D and control individuals with regards to age categories and BMI scores. Additionally, the Chi-square test showed a significant result for the distribution of T2D and control individuals with regards to population (Kuwaiti and Qatari). However, no significant difference was found for sex distribution between the two groups.
mtDNA variants
A total of 1,850 high-quality variants were identified after passing filters and quality control steps. These identified mtDNA variants include SNPs and INDELs, with an average coverage of 75X across the entire mtDNA. The genomic-control inflation factor (λ) was 1 in tests that accounted for all the covariates. Since this value is close to 1.0, it indicates that there is no need to adjust the association statistics for genomic-control inflation.
mtDNA haplogroups
The average haplogroup quality score was above 92%, obtained using HaploGrep 2.4.0, which identified 15 mitochondrial haplogroups (B, E, H, HV, I, J, K, L, M, N, R, T, U, W, X). PCA was performed on the mitochondrial DNA variants from both cohorts to explore potential hidden relationships among the samples. The PCA results revealed that the 1,112 samples clustered primarily according to their mitochondrial haplogroups (Figure 1A), reflecting the rich ancestral information encoded in the mtDNA. Notably, the samples did not cluster based on population groups (Kuwaiti or Qatari), indicating that the primary genetic variation is driven by haplogroup differences rather than geographical origin. In Figure 1B, the PCA plot shows no distinct clustering based on gender or diabetes status, further emphasizing that mitochondrial haplogroups are the dominant factor influencing the genetic structure observed in this analysis.

Figure 1. PCA of 1,112 Samples of Gulf Region Arabs Based on Their mtDNA Variants. The PCA was conducted on all mitochondrial DNA variants identified in the two cohorts. PC1 (Principal Component 1) and PC2 (Principal Component 2) on the x- and y-axes represent the first two principal components, which capture the largest proportion of genetic variance in the dataset. The percentage of variance explained by each component is indicated on the axes. (A) The samples are colour-coded based on their mitochondrial haplogroups, reflecting the rich ancestral information contained in the mtDNA, and different shapes describe the population groups. (B) The same PCA plot is overlaid with gender represented by different colours and diabetes status represented by different shapes.
mtDNA haplogroups associated with T2D
Figure 2 presents the frequencies of mitochondrial haplogroups in the Gulf region Arabs of Kuwaiti and Qatari population based on diabetes phenotype. The frequency of the H haplogroup was higher in the control groups compared to the diabetes groups in both the Kuwaiti (17.5% vs. 11%) and the Qatari (12.3% vs. 9.3%) cohorts (Figure 2). The most common haplogroups were J followed by U haplogroup. Haplogroups with frequencies below 3% were categorized as “others”.
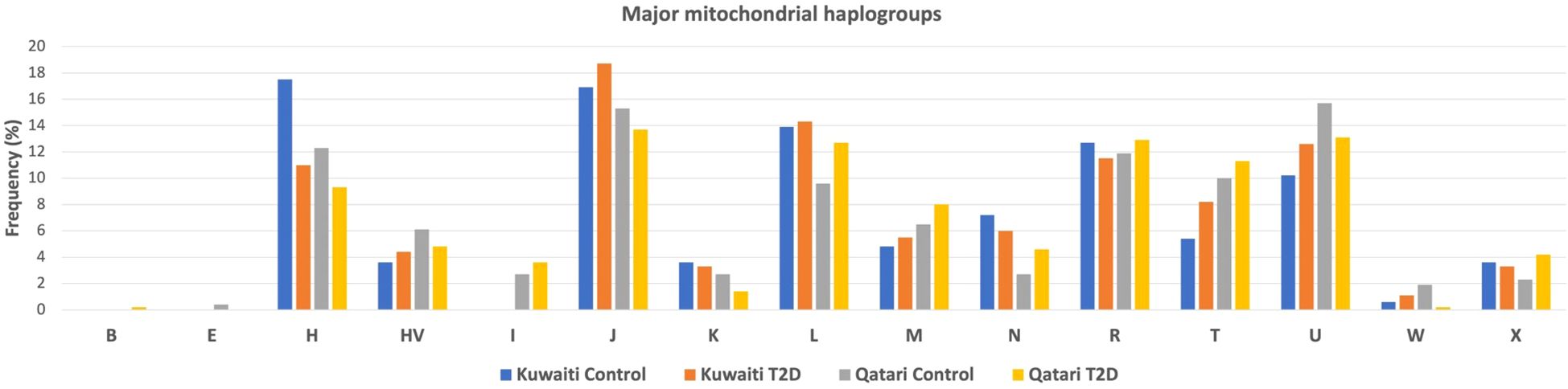
Figure 2. Distribution of mtDNA Haplogroups of gulf region arabs based on their diabetes status. Allocation of major mtDNA haplogroup prevalence in T2D and control groups within the Kuwaiti and Qatari populations.
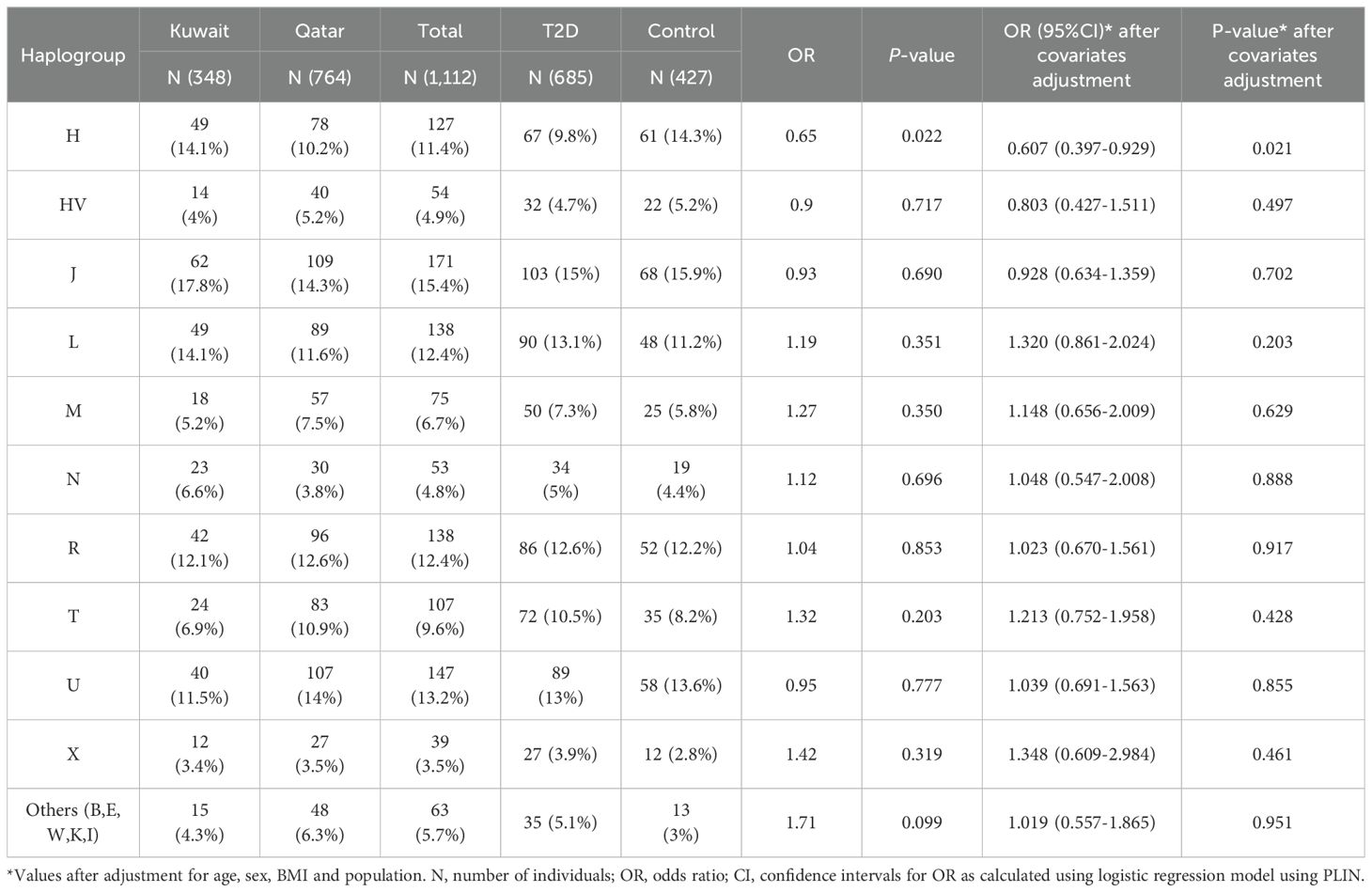
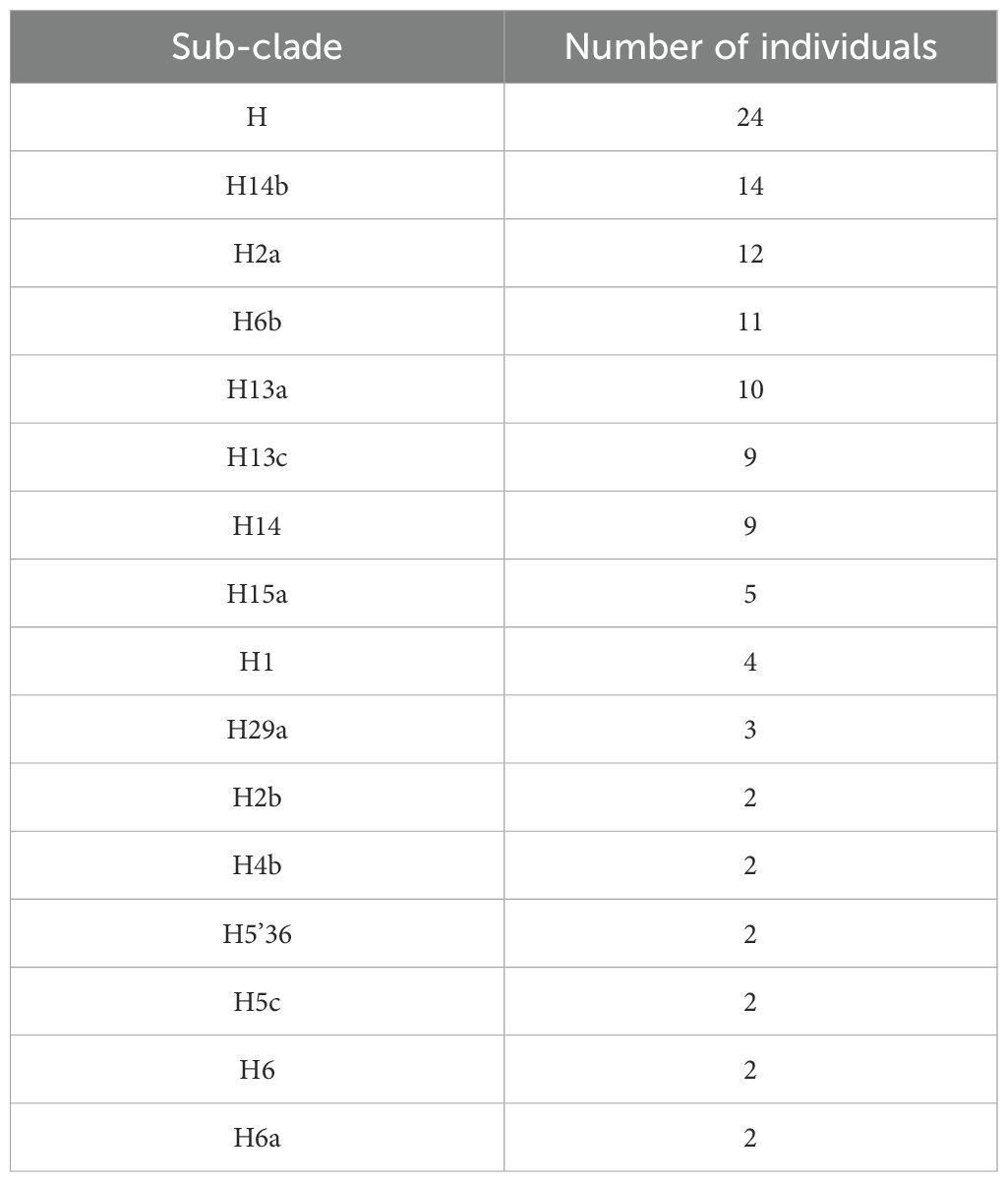
When conducting separate mitochondrial haplogroup analysis using the Fisher exact test on the Kuwaiti population (Supplementary Table S2) and the Qatari population (Supplementary Table S3), we did not find significant associations. However, when we combined the two populations as Arabs in the Gulf region, thereby increasing the number of samples, we observed a significant association using the multivariate logistic regression analysis (Table 2). Specifically, the H haplogroup had a protective effect against obesity (odds ratio [OR] = 0.65; p = 0.022). This effect remained significant after adjusting for age, sex, BMI, and population (OR/95% CI = 0.607/0.397-0.929; p = 0.021). Further analysis showed that most individuals with the H haplogroup belonged to the H14b and H2a subclades (Table 3).
mtDNA variants associated with T2D
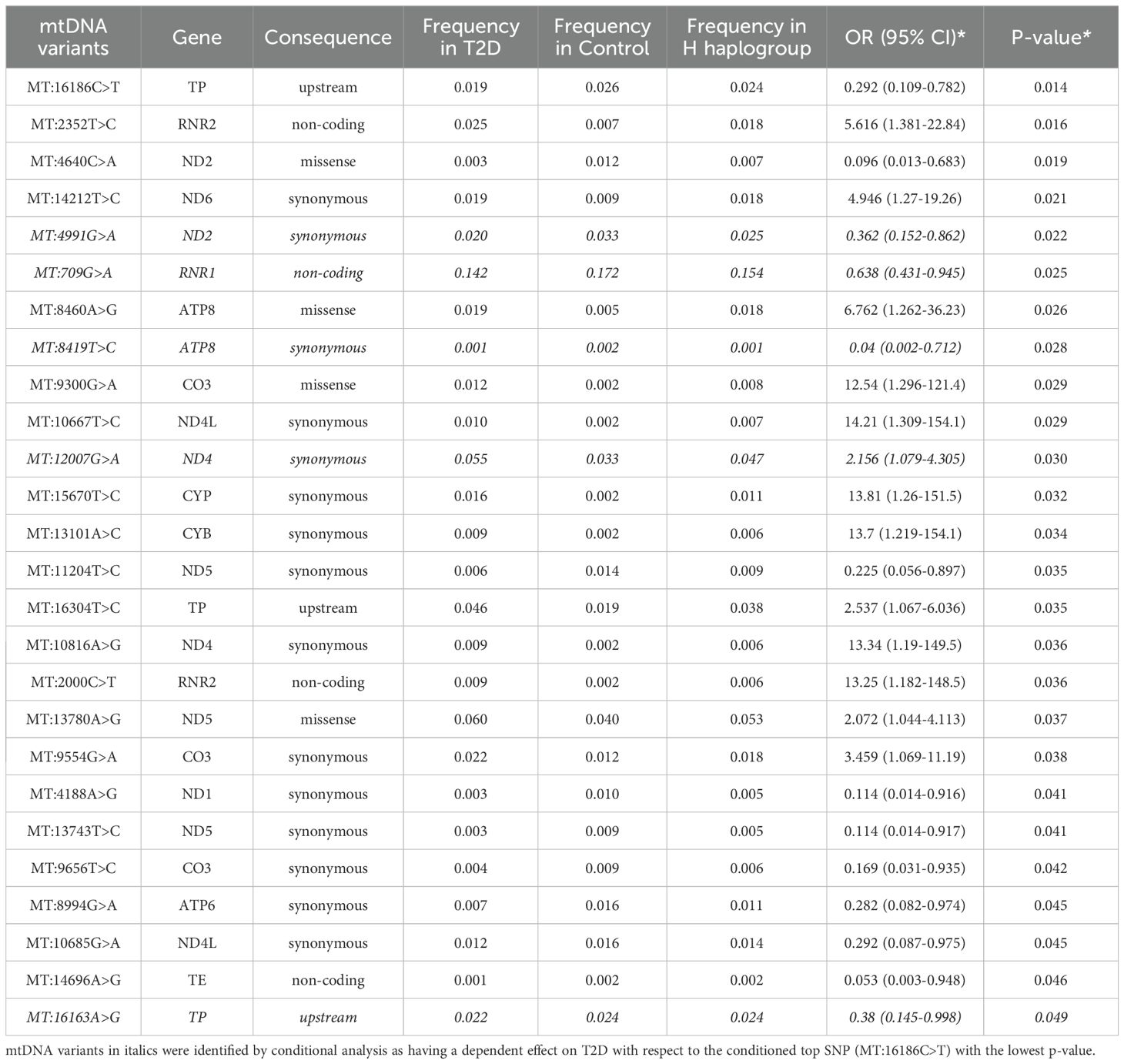
The analysis identified 26 mtDNA variants that were nominally associated with T2D in the Arab Gulf region cohort, as presented in Table 4. These associations were determined after adjusting for key covariates including age, sex, mtDNA haplogroup, and population group. Of these 26 variants, 13 were found to be positively correlated with an increased risk of T2D, while the remaining 13 exhibited a negative correlation, suggesting a potential protective effect.
Among the positively associated variants, MT:2352T>C in the RNR2 gene stands out as a significant non-coding variant, with an odds ratio (OR) of 5.616 (95% CI = 1.381-22.84; p = 0.016), indicating a strong association with T2D. Similarly, the MT:8460A>G variant, a missense mutation in the ATP8 gene, was associated with T2D (OR = 6.762; 95% CI = 1.262-36.23; p = 0.026). This variant affects a key component of the mitochondrial ATP synthase complex, potentially leading to impaired ATP production and subsequent disruptions in insulin secretion and glucose metabolism.
Conversely, several variants were identified as having a protective association with T2D. Five of these variants, including MT:16186C>T, MT:4991G>A, MT:4188A>G, MT:709G>A, and MT:11204T>C, showed a significant negative correlation with T2D, indicating a protective effect. For example, MT:16186C>T, located in the non-coding D-loop region, exhibited a protective effect against T2D (OR = 0.292; 95% CI = 0.109-0.782; p = 0.014). This variant is likely involved in the regulation of mitochondrial DNA replication and transcription, possibly reducing the risk of T2D by improving mitochondrial function. Similarly, MT:4991G>A in the ND2 gene and MT:4188A>G in the ND1 gene, which are both involved in encoding subunits of NADH dehydrogenase, also demonstrated protective effects, suggesting that these variants might enhance oxidative phosphorylation efficiency, thereby mitigating T2D risk. MT:709G>A in the RNR1 gene and MT:11204T>C in the ND5 gene were additionally found to have protective associations, further emphasizing the role of mitochondrial variants in influencing metabolic health and disease resistance.
Further conditional analysis, focusing on the top leading variant MT:16186C>T, identified 20 additional variants that remained significantly associated with T2D, suggesting that these variants exert an independent effect on the risk of the disease. However, five variants from the initial 26 lost their significance after controlling for MT:16186C>T, indicating that their association with T2D was likely dependent on this leading variant. This dependency highlights the complex genetic interactions among mtDNA variants that contribute to T2D susceptibility.
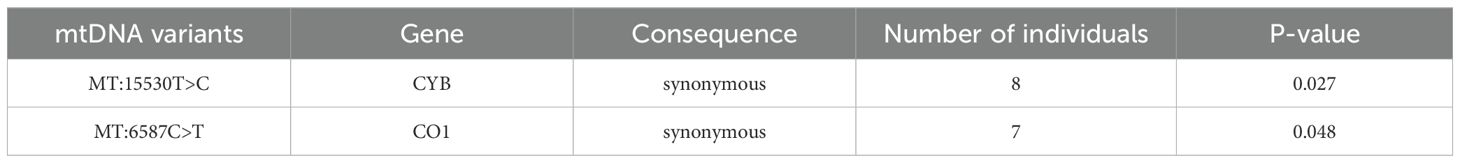
In total, 514 mtDNA variants were identified exclusively in individuals with T2D, as detailed in Supplementary Table S4. Table 5 highlights several significant variants that are uniquely present in the T2D group, including MT:15530T>C in the CYB gene and MT:6587C>T in the CO1 gene. These variants have been implicated in previous studies of metabolic disorders, further supporting their relevance in T2D within the Arab Gulf population.
Discussion
In this study, we explored the association between mitochondrial haplogroups and T2D by combining data from Kuwaiti and Qatari cohorts, which individually might not have shown significant associations, possibly due to limited sample sizes. Given the environmental and genetic similarities of these populations (42), we combined the cohorts to increase the statistical power and offer a comprehensive view of the genetic landscape of Arabs in the Gulf region. The PCA analysis of mitochondrial haplogroups confirmed the ethnic homogeneity between the two populations. This integrated approach enhanced the reliability of our findings, enabling us to identify genetic markers and risk factors with greater confidence, thus providing valuable insights into the genetic factors associated with T2D.
In the current study, the mitochondrial H haplogroup was found to have a protective effect against obesity in the combined populations (OR/95% CI = 0.607/0.397-0.929; p = 0.021) after adjusting for age, sex, BMI, and population. Furthermore, the prevalence of individuals with the H haplogroup was higher in the control groups than in the T2D groups in both populations, although this difference was not statistically significant. Additional analysis revealed that most individuals with the H haplogroup belonged to the H14b and H2a subclades. Haplogroup H, although most prevalent in Europe, has a notable presence in the Arabian Peninsula. Studies have shown mean frequencies of around 9.4% in the region, with variations across different countries and regions (43). This distribution supports the relevance of our findings within the Gulf Arab populations. Furthermore, this haplogroup has been previously associated with a protective effect against obesity in Arabs living in Kuwait (44). Given that obesity is a significant risk factor for T2D, this protective effect might contribute to a reduced risk of T2D in individuals with haplogroup H. Furthermore, the H2a maternal lineage, common in the Saudi Arabian population, is associated with lean individuals in the Arab population living in Kuwait (44), likely contributing to the reduced T2D risk observed in the H2a subclade.
However, studies on Chinese Uyghur (45) and Bangladeshi populations (46) reported that haplogroup H was associated with an increased risk of T2D. This inconsistency with our results may be due to several factors. First, genetic background effects can differ significantly across populations, leading to inconsistent haplogroup associations. The different frequencies of H subclades across populations further complicate this, as our study identified specific subclades of haplogroup H, whereas these studies did not. Second, the Bangladeshi study only sequenced the D-loop hypervariable region I in their haplogroup classification (46). We previously observed a 71% concordance in mitochondrial haplogroup profiling between variants from the full mitochondrial genome extracted from whole-exome data and the D-loop region data from conventional Sanger sequencing. This suggests that using only the D-loop region could lead to haplogroup misclassification (47). Additionally, the small sample sizes in these studies may have contributed to the differing results. Another explanation involves observations of tightly coupled oxidative phosphorylation (OXPHOS) haplogroups, such as haplogroup H, which use fewer calories to produce more ATP. Another tightly coupled haplogroup has been associated with both elite athletes (48) and obesity (49, 50) in the Japanese population. Researchers hypothesize that the “thrifty genotype” could explain this phenomenon: while high ATP production efficiency benefits athletes, it predisposes individuals to obesity if they become sedentary later in life (48).
The prevalence of individuals with mitochondrial haplogroup T is higher in the T2D group compared to the control group in the Kuwaiti population, the Qatari population, and the combined population representing Arabs in the Gulf region. However, the difference in frequency was not significant. However, a significant association was observed for haplogroup T with an increased risk of T2D in the white Brazilian population (51). The T haplogroup was also associated with obesity in the southern Italian population (52) and in Austrian juveniles and adults (53).
mtDNA variants have been implicated as possible genetic causes of T2D. The T-to-C change at nucleotide position 16189, which creates a homopolymeric polycytosine (poly-C) tract between positions 16184 and 16193 within the D-loop region, has been linked to T2D in specific ethnic groups (20, 54, 55). This variant’s diabetogenic effect is thought to be connected to mtDNA replication issues and influenced (56) by factors such as increased body weight (57) and oxidative stress (58). However, the association has been inconsistent across different populations (23).
In our study involving Arabs from the Gulf region, we identified a significant protective variant: 16186 C>T (OR/95% CI = 0.292/0.109-0.782) after adjusting for covariates. This variant was the most significant in our study. Located within the same poly-C tract in the D-loop region, it replaces a cytosine with thymine, possibly impacting replication or regulation processes differently from the 16189 T>C variant, which was not prioritized in our study.
We found that the variant MT:2352T>C (OR/95% CI = 5.616/1.381-22.84) is positively correlated with T2D in our study. This variant was previously associated with decreased mitochondrial inner membrane potential during oxidative phosphorylation. The minor allele of this SNP is almost absent in individuals of Caucasian ancestry but is prevalent in those of African American ancestry, where it has been linked to an increased risk of fibromyalgia (59). Given its prevalence in the Arab population, its association with decreased cellular energy metabolism suggests a potential mechanism for its link to T2D through mitochondrial dysfunction and impaired insulin sensitivity (60).
Among the positively associated variants in our study, we identified a significant missense variant, MT:8460A>G, within the ATP8 gene. This gene encodes a subunit of ATP synthase in complex V, which utilizes the proton electrochemical gradient across the inner membrane during oxidative phosphorylation to synthesize ATP from adenosine diphosphate (61). Variants within the ATP8 gene have been shown to impact β-cell function and ROS generation in animal models (62) and have been associated with T2D in genetic studies (45).
Additionally, we identified a missense (MT:9300G>A) and a synonymous (MT:9554G>A) as significant variants in the cytochrome c oxidase subunit III (CO3) gene. A different variant in the CO3 gene has been linked to MIDD (63).
Variants in the Cytochrome B gene (CYB) have also been linked to MIDD (64) and T2D (39). In our study, we identified a significant synonymous variant (MT:13101A>C) in the CYB gene.
We found several significant variants positively correlated with T2D located in the nicotinamide adenine dinucleotide (NADH) dehydrogenase subunit genes (ND4, ND4L, and ND6) of respiratory complex I. NADH dehydrogenase is essential for energy generation; thus, variants within its encoding genes could result in metabolic disorders (65). Variants in these genes have been associated with MIDD (66) and T2D (67, 68). Similarly, we also found a significant synonymous variant in the RNR2 gene (MT:2000C>T), which encodes Humanin, a peptide shown to improve insulin sensitivity in animal models of diabetes mellitus (69). We also identified two significant synonymous variants that were found exclusively in the T2D groups: MT:15530T>C in the CYB gene and MT:6587C>T in the cytochrome c oxidase subunit I (COI) gene. A variant in the COI gene has been linked to MIDD (63).
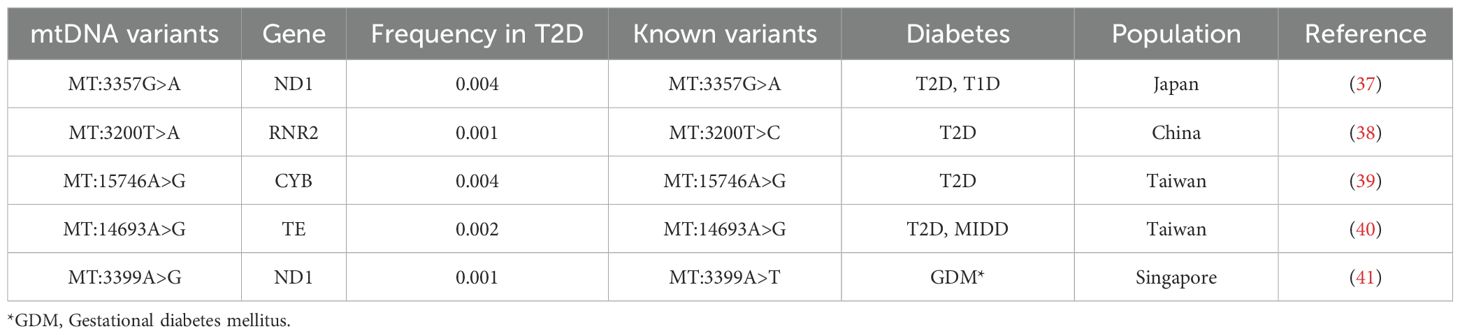
Finally, we examined known mtDNA positions associated with T2D exclusively in individuals with T2D (70), and we identified five variants. Three of these are exact known variants from ND1 (37), CYB (39), and mitochondrially encoded tRNA glutamic acid (TE) (40) genes, all of which are associated with T2D. The other two variants share the same positions with known variants in literature but have different nucleotide changes. One is located within RNR2 at position MT:3200 (38) with a T>A change, and the other is within ND1 at position MT:3399 (41) with an A>G change (Table 6).
This study acknowledges several limitations. First, the associations between mitochondrial haplogroups and T2D, as well as mitochondrial variants and T2D, did not reach statistical significance after applying Bonferroni correction, might indicate that the sample size is not large enough to accommodate multiple testing’s. Furthermore, diabetes is a multifactorial disorder influenced by a variety of factors. Including adjustments for lifestyle variables, dietary habits, and meal frequency could provide a more comprehensive analysis. Lastly, our results have not been validated in a larger, independent cohort of Arabs from the Gulf region, which is necessary to confirm these findings.
Conclusion
In conclusion, this study provides important evidence that mitochondrial genetics play a significant role in modulating T2D risk in Arab populations of the Gulf region. The protective effect of haplogroup H, as well as the identification of specific disease-associated variants, highlight the value of investigating mitochondrial markers as part of a precision medicine approach to understanding and managing T2D in this high-risk population. Further research is warranted to elucidate the functional mechanisms underlying these genetic associations and to translate these findings into improved clinical screening and prevention strategies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The studies involving humans were approved by Ethical Review Committee at Dasman Diabetes Institute. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
MD: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. NA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. HA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. SJ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. RN: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft. TT: Writing – original draft, Writing – review & editing. FA-M: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The Kuwait Foundation for the Advancement of Sciences (KFAS).
Acknowledgments
We extend our sincere thanks to the Kuwait Foundation for the Advancement of Sciences (KFAS) for their institutional funding support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2024.1443737/full#supplementary-material
References
1. World Health Organization. The WHO global diabetes compact. Geneva (2023). Available at: https://www.who.int/initiatives/the-who-global-diabetes-compact.
2. International Diabetes Federation. IDF diabetes atlas. 10th edn. Brussels, Belgium (2021). Available at: https://www.diabetesatlas.org.
3. Medici F, Hawa M, Ianari A, Pyke DA, Leslie RD. Concordance rate for type II diabetes mellitus in monozygotic twins: actuarial analysis. Diabetologia. (1999) 42:146–50. doi: 10.1007/s001250051132
4. Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance–a population-based twin study. Diabetologia. (1999) 42:139–45. doi: 10.1007/s001250051131
5. Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. (2000) 49:2201–7. doi: 10.2337/diabetes.49.12.2201
6. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. (2009) 461:747–53. doi: 10.1038/nature08494
7. Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. The role of mitochondria in metabolism and cell death. Biochem Biophys Res Commun. (2017) 482:426–31. doi: 10.1016/j.bbrc.2016.11.088
8. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. (2003) 34:267–73. doi: 10.1038/ng1180
9. Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U.S.A. (2003) 100:8466–71. doi: 10.1073/pnas.1032913100
10. Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circ Res. (2008) 102:401–14. doi: 10.1161/CIRCRESAHA.107.165472
11. Softic S, Meyer JG, Wang GX, Gupta MK, Batista TM, Lauritzen HPMM, et al. Dietary sugars alter hepatic fatty acid oxidation via transcriptional and post-translational modifications of mitochondrial proteins. Cell Metab. (2019) 30:735–753.e734. doi: 10.1016/j.cmet.2019.09.003
12. Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet. (1997) 13:450–5. doi: 10.1016/s0168-9525(97)01266-3
13. Naue J, Hörer S, Sänger T, Strobl C, Hatzer-Grubwieser P, Parson W, et al. Evidence for frequent and tissue-specific sequence heteroplasmy in human mitochondrial DNA. Mitochondrion. (2015) 20:82–94. doi: 10.1016/j.mito.2014.12.002
14. Fernández-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol. (2003) 88:41–56. doi: 10.1113/eph8802514
15. Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. (2010) 51:440–50. doi: 10.1002/em.20586
16. Wallace DC. A mitochondrial bioenergetic etiology of disease. J Clin Invest. (2013) 123:1405–12. doi: 10.1172/JCI61398
17. Stewart JB, Chinnery PF. The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet. (2015) 16:530–42. doi: 10.1038/nrg3966
18. Tsukuda K, Suzuki Y, Kameoka K, Osawa N, Goto Y, Katagiri H, et al. Screening of patients with maternally transmitted diabetes for mitochondrial gene mutations in the tRNA[Leu(UUR)] region. Diabetes Med. (1997) 14:1032–7. doi: 10.1002/(SICI)1096-9136(199712)14:12<1032::AID-DIA504>3.0.CO;2-Y
19. Saker PJ, Hattersley AT, Barrow B, Hammersley MS, Horton V, Gillmer MD, et al. UKPDS 21: low prevalence of the mitochondrial transfer RNA gene (tRNA(Leu(UUR))) mutation at position 3243bp in UK Caucasian type 2 diabetic patients. Diabetes Med. (1997) 14:42–5. doi: 10.1002/(SICI)1096-9136(199701)14:1<42::AID-DIA295>3.0.CO;2-T
20. Park KS, Chan JC, Chuang LM, Suzuki S, Araki E, Nanjo K, et al. A mitochondrial DNA variant at position 16189 is associated with type 2 diabetes mellitus in Asians. Diabetologia. (2008) 51:602–8. doi: 10.1007/s00125-008-0933-z
21. Wang PW, Lin TK, Weng SW, Liou CW. Mitochondrial DNA variants in the pathogenesis of type 2 diabetes - relevance of asian population studies. Rev Diabetes Stud. (2009) 6:237–46. doi: 10.1900/RDS.2009.6.237
22. Hsouna S, Ben Halim N, Lasram K, Arfa I, Jamoussi H, Bahri S, et al. Association study of mitochondrial DNA polymorphisms with type 2 diabetes in Tunisian population. Mitochondrial DNA. (2015) 26:367–72. doi: 10.3109/19401736.2013.836508
23. Chinnery PF, Elliott HR, Patel S, Lambert C, Keers SM, Durham SE, et al. Role of the mitochondrial DNA 16184-16193 poly-C tract in type 2 diabetes. Lancet. (2005) 366:1650–1. doi: 10.1016/S0140-6736(05)67492-2
24. Fuku N, Park KS, Yamada Y, Nishigaki Y, Cho YM, Matsuo H, et al. Mitochondrial haplogroup N9a confers resistance against type 2 diabetes in Asians. Am J Hum Genet. (2007) 80:407–15. doi: 10.1086/512202
25. Liao WQ, Pang Y, Yu CA, Wen JY, Zhang YG, Li XH. Novel mutations of mitochondrial DNA associated with type 2 diabetes in Chinese Han population. Tohoku J Exp Med. (2008) 215:377–84. doi: 10.1620/tjem.215.377
26. Fang H, Hu N, Zhao Q, Wang B, Zhou H, Fu Q, et al. mtDNA haplogroup N9a increases the risk of type 2 diabetes by altering mitochondrial function and intracellular mitochondrial signals. Diabetes. (2018) 67:1441–53. doi: 10.2337/db17-0974
27. Fakhro KA, Staudt MR, Ramstetter MD, Robay A, Malek JA, Badii R, et al. The Qatar genome: a population-specific tool for precision medicine in the Middle East. Hum Genome Var. (2016) 3:16016. doi: 10.1038/hgv.2016.16
28. O’Beirne SL, Salit J, Rodriguez-Flores JL, Staudt MR, Abi Khalil C, Fakhro KA, et al. Exome sequencing-based identification of novel type 2 diabetes risk allele loci in the Qatari population. PloS One. (2018) 13:e0199837. doi: 10.1371/journal.pone.0199837
29. John SE, Antony D, Eaaswarkhanth M, Hebbar P, Channanath AM, Thomas D, et al. Assessment of coding region variants in Kuwaiti population: implications for medical genetics and population genomics. Sci Rep. (2018) 8:16583. doi: 10.1038/s41598-018-34815-8
30. Hebbar P, Nizam R, Melhem M, Alkayal F, Elkum N, John SE, et al. Genome-wide association study identifies novel recessive genetic variants for high TGs in an Arab population. J Lipid Res. (2018) 59:1951–66. doi: 10.1194/jlr.P080218
31. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv. (2013), 1303. doi: 10.48550/arXiv.1303.3997
32. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. (2009) 25:2078–9. doi: 10.1093/bioinformatics/btp352
33. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. (2010) 20:1297–303. doi: 10.1101/gr.107524.110
34. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. (2016) 17:122. doi: 10.1186/s13059-016-0974-4
35. Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, et al. mtDNA variation and analysis using mitomap and mitomaster. Curr Protoc Bioinf. (2013) 44(123):1.23.1–1.23.26. doi: 10.1002/0471250953.bi0123s44
36. Weissensteiner H, Pacher D, Kloss-Brandstätter A, Forer L, Specht G, Bandelt HJ, et al. HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic Acids Res. (2016) 44:W58–63. doi: 10.1093/nar/gkw233
37. Ohkubo K, Yamano A, Nagashima M, Mori Y, Anzai K, Akehi Y, et al. Mitochondrial gene mutations in the tRNA(Leu(UUR)) region and diabetes: prevalence and clinical phenotypes in Japan. Clin Chem. (2001) 47:1641–8. doi: 10.1093/clinchem/47.9.1641
38. Yang T, Lam CW, Tsang MW, Tong SF, Kam GY, Chan LY, et al. Novel mitochondrial 16S rRNA mutation 3200T–>C, associated with adult-onset type 2 diabetes. Chin Med J (Engl). (2002) 115:753–8.
39. Loo JH, Trejaut JA, Yen JC, Chen ZS, Ng WM, Huang CY, et al. Mitochondrial DNA association study of type 2 diabetes with or without ischemic stroke in Taiwan. BMC Res Notes. (2014) 7:223. doi: 10.1186/1756-0500-7-223
40. Tzen CY, Thajeb P, Wu TY, Chen SC. Melas with point mutations involving tRNALeu (A3243G) and tRNAGlu(A14693g). Muscle Nerve. (2003) 28:575–81. doi: 10.1002/mus.10473
41. Chen Y, Liao WX, Roy AC, Loganath A, Ng SC. Mitochondrial gene mutations in gestational diabetes mellitus. Diabetes Res Clin Pract. (2000) 48:29–35. doi: 10.1016/s0168-8227(99)00138-2
42. Dashti M, Ateyah K, Alroughani R, Al-Temaimi R. Replication analysis of variants associated with multiple sclerosis risk. Sci Rep. (2020) 10:7327. doi: 10.1038/s41598-020-64432-3
43. Abu-Amero KK, Larruga JM, Cabrera VM, González AM. Mitochondrial DNA structure in the arabian peninsula. BMC Evol Biol. (2008) 8:45. doi: 10.1186/1471-2148-8-45
44. Eaaswarkhanth M, Melhem M, Sharma P, Nizam R, Al Madhoun A, Chaubey G, et al. Mitochondrial DNA D-loop sequencing reveals obesity variants in an Arab population. Appl Clin Genet. (2019) 12:63–70. doi: 10.2147/TACG.S198593
45. Jiang W, Li R, Zhang Y, Wang P, Wu T, Lin J, et al. Mitochondrial DNA mutations associated with type 2 diabetes mellitus in chinese uyghur population. Sci Rep. (2017) 7:16989. doi: 10.1038/s41598-017-17086-7
46. Saha SK, Saba AA, Hasib M, Rimon RA, Hasan I, Alam MS, et al. Evaluation of D-loop hypervariable region I variations, haplogroups and copy number of mitochondrial DNA in Bangladeshi population with type 2 diabetes. Heliyon. (2021) 7:e07573. doi: 10.1016/j.heliyon.2021.e07573
47. Dashti M, Alsaleh H, Eaaswarkhanth M, John SE, Nizam R, Melhem M, et al. Delineation of mitochondrial DNA variants from exome sequencing data and association of haplogroups with obesity in Kuwait. Front Genet. (2021) 12:626260. doi: 10.3389/fgene.2021.626260
48. Mikami E, Fuku N, Takahashi H, Ohiwa N, Scott RA, Pitsiladis YP, et al. Mitochondrial haplogroups associated with elite Japanese athlete status. Br J Sports Med. (2011) 45:1179–83. doi: 10.1136/bjsm.2010.072371
49. Fuku N, Oshida Y, Takeyasu T, Guo LJ, Kurata M, Yamada Y, et al. Mitochondrial ATPase subunit 6 and cytochrome B gene polymorphisms in young obese adults. Biochem Biophys Res Commun. (2002) 290:1199–205. doi: 10.1006/bbrc.2002.6330
50. Okura T, Koda M, Ando F, Niino N, Tanaka M, Shimokata H. Association of the mitochondrial DNA 15497G/A polymorphism with obesity in a middle-aged and elderly Japanese population. Hum Genet. (2003) 113:432–6. doi: 10.1007/s00439-003-0983-8
51. Crispim D, Canani LH, Gross JL, Tschiedel B, Souto KE, Roisenberg I. The European-specific mitochondrial cluster J/T could confer an increased risk of insulin-resistance and type 2 diabetes: an analysis of the m.4216T > C and m.4917A > G variants. Ann Hum Genet. (2006) 70:488–95. doi: 10.1111/j.1469-1809.2005.00249.x
52. Nardelli C, Labruna G, Liguori R, Mazzaccara C, Ferrigno M, Capobianco V, et al. Haplogroup T is an obesity risk factor: mitochondrial DNA haplotyping in a morbid obese population from southern Italy. BioMed Res Int. (2013) 2013:631082. doi: 10.1155/2013/631082
53. Ebner S, Mangge H, Langhof H, Halle M, Siegrist M, Aigner E, et al. Mitochondrial haplogroup T is associated with obesity in Austrian juveniles and adults. PloS One. (2015) 10:e0135622. doi: 10.1371/journal.pone.0135622
54. Kim JH, Park KS, Cho YM, Kang BS, Kim SK, Jeon HJ, et al. The prevalence of the mitochondrial DNA 16189 variant in non-diabetic Korean adults and its association with higher fasting glucose and body mass index. Diabetes Med. (2002) 19:681–4. doi: 10.1046/j.1464-5491.2002.00747.x
55. Weng SW, Liou CW, Lin TK, Wei YH, Lee CF, Eng HL, et al. Association of mitochondrial deoxyribonucleic acid 16189 variant (T->C transition) with metabolic syndrome in Chinese adults. J Clin Endocrinol Metab. (2005) 90:5037–40. doi: 10.1210/jc.2005-0227
56. Liou CW, Chen JB, Tiao MM, Weng SW, Huang TL, Chuang JH, et al. Mitochondrial DNA coding and control region variants as genetic risk factors for type 2 diabetes. Diabetes. (2012) 61:2642–51. doi: 10.2337/db11-1369
57. Liou CW, Lin TK, Huei Weng H, Lee CF, Chen TL, Wei YH, et al. A common mitochondrial DNA variant and increased body mass index as associated factors for development of type 2 diabetes: Additive effects of genetic and environmental factors. J Clin Endocrinol Metab. (2007) 92:235–9. doi: 10.1210/jc.2006-0653
58. Lin TK, Chen SD, Wang PW, Wei YH, Lee CF, Chen TL, et al. Increased oxidative damage with altered antioxidative status in type 2 diabetic patients harboring the 16189 T to C variant of mitochondrial DNA. Ann N Y Acad Sci. (2005) 1042:64–9. doi: 10.1196/annals.1338.007
59. van Tilburg MAL, Parisien M, Boles RG, Drury GL, Smith-Voudouris J, Verma V, et al. A genetic polymorphism that is associated with mitochondrial energy metabolism increases risk of fibromyalgia. Pain. (2020) 161:2860–71. doi: 10.1097/j.pain.0000000000001996
60. Blagov A, Nedosugova L, Kirichenko T, Sukhorukov V, Melnichenko A, Orekhov A. Mitochondrial dysfunction as a factor of energy metabolism disorders in type 2 diabetes mellitus. Front Biosci (Schol Ed). (2024) 16:5. doi: 10.31083/j.fbs1601005
61. Jonckheere AI, Smeitink JA, Rodenburg RJ. Mitochondrial ATP synthase: architecture, function and pathology. J Inherit Metab Dis. (2012) 35:211–25. doi: 10.1007/s10545-011-9382-9
62. Weiss H, Wester-Rosenloef L, Koch C, Koch F, Baltrusch S, Tiedge M, et al. The mitochondrial Atp8 mutation induces mitochondrial ROS generation, secretory dysfunction, and β-cell mass adaptation in conplastic B6-mtFVB mice. Endocrinology. (2012) 153:4666–76. doi: 10.1210/en.2012-1296
63. Tabebi M, Mkaouar-Rebai E, Mnif M, Kallabi F, Ben Mahmoud A, Ben Saad W, et al. A novel mutation MT-COIII m.9267G>C and MT-COI m.5913G>A mutation in mitochondrial genes in a Tunisian family with maternally inherited diabetes and deafness (MIDD) associated with severe nephropathy. Biochem Biophys Res Commun. (2015) 459:353–60. doi: 10.1016/j.bbrc.2015.01.151
64. Li W, Zhang W, Li F, Wang C. Mitochondrial genetic analysis in a Chinese family suffering from both mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes and diabetes. Int J Clin Exp Pathol. (2015) 8:7022–7.
65. Flaquer A, Baumbach C, Kriebel J, Meitinger T, Peters A, Waldenberger M, et al. Mitochondrial genetic variants identified to be associated with BMI in adults. PloS One. (2014) 9:e105116. doi: 10.1371/journal.pone.0105116
66. Crispim D, Estivalet AA, Roisenberg I, Gross JL, Canani LH. Prevalence of 15 mitochondrial DNA mutations among type 2 diabetic patients with or without clinical characteristics of maternally inherited diabetes and deafness. Arq Bras Endocrinol Metabol. (2008) 52:1228–35. doi: 10.1590/s0004-27302008000800005
67. Tawata M, Ohtaka M, Iwase E, Ikegishi Y, Aida K, Onaya T. New mitochondrial DNA homoplasmic mutations associated with Japanese patients with type 2 diabetes. Diabetes. (1998) 47:276–7. doi: 10.2337/diab.47.2.276
68. Tawata M, Hayashi JI, Isobe K, Ohkubo E, Ohtaka M, Chen J, et al. A new mitochondrial DNA mutation at 14577 T/C is probably a major pathogenic mutation for maternally inherited type 2 diabetes. Diabetes. (2000) 49:1269–72. doi: 10.2337/diabetes.49.7.1269
69. Voigt A, Jelinek HF. Humanin: a mitochondrial signaling peptide as a biomarker for impaired fasting glucose-related oxidative stress. Physiol Rep. (2016) 4(9):e12796. doi: 10.14814/phy2.12796
70. Dabravolski SA, Orekhova VA, Baig MS, Bezsonov EE, Starodubova AV, Popkova TV, et al. he role of mitochondrial mutations and chronic inflammation in diabetes. Int J Mol Sci. (2021) 22(13):6733. doi: 10.3390/ijms22136733
Keywords: mitochondria, type 2 diabetes, haplogroups, mtDNA variants, Arab
Citation: Dashti M, Ali NM, Alsaleh H, John SE, Nizam R, Thanaraj TA and Al-Mulla F (2024) Association of mitochondrial haplogroup H with reduced risk of type 2 Diabetes among Gulf Region Arabs. Front. Endocrinol. 15:1443737. doi: 10.3389/fendo.2024.1443737
Received: 04 June 2024; Accepted: 01 November 2024;
Published: 26 November 2024.
Edited by:
Leandro Ceotto Freitas Lima, The University of Texas Health Science Center at San Antonio, United StatesReviewed by:
Kátia Michelle Freitas, Minas Gerais State University, BrazilRaúl Cuauhtémoc Baptista Rosas, University of Guadalajara, Mexico
Copyright © 2024 Dashti, Ali, Alsaleh, John, Nizam, Thanaraj and Al-Mulla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thangavel Alphonse Thanaraj, YWxwaG9uc2UudGhhbmdhdmVsQGRhc21hbmluc3RpdHV0ZS5vcmc=; Fahd Al-Mulla, ZmFoZC5hbG11bGxhQGRhc21hbmluc3RpdHV0ZS5vcmc=
 Mohammed Dashti
Mohammed Dashti Naser M. Ali
Naser M. Ali Hussain Alsaleh
Hussain Alsaleh Sumi Elsa John
Sumi Elsa John Rasheeba Nizam
Rasheeba Nizam Thangavel Alphonse Thanaraj
Thangavel Alphonse Thanaraj Fahd Al-Mulla
Fahd Al-Mulla