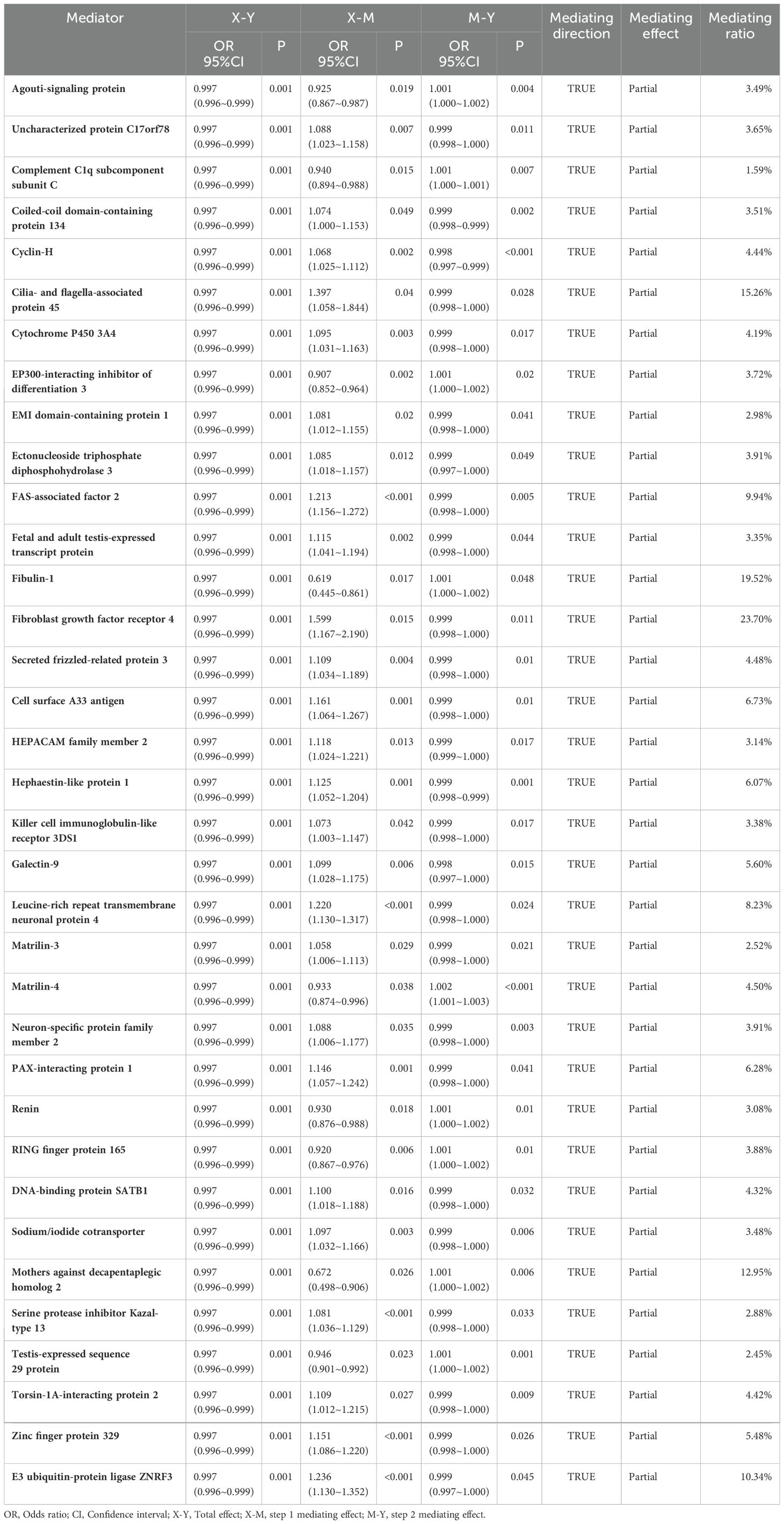
- 1Department of Nephrology, The First Hospital of Jilin University, Changchun, China
- 2Department of Hepatology, The First Hospital of Jilin University, Changchun, China
Background: The relationship between selenium and renal function has always attracted widespread attention. Increased selenium level has been found to cause impaired renal function in our previous study, but the mechanism is not clear. In this study, we evaluate the potential mediating effects of plasma proteome in the association of selenium level and renal function to understand the mechanisms of selenium’s effect on renal function.
Methods: Utilizing two-sample two-step mediating mendelian randomization (MR) methodology to investigate the genetically causal relationship between selenium level and renal function as well as the role of the plasma proteome in mediating them. Additionally, the mediating proteins were enriched and analyzed through bioinformatics to understand the potential mechanisms of selenium effects on renal function.
Results: In the MR analysis, an increase in selenium level was found to decrease estimated glomerular filtration rate (eGFR). Specifically, for each standard deviation (SD) increase in selenium levels, eGFR levels are reduced by 0.003 SD [Beta (95% CI): -0.003 (-0.004 ~ -0.001), P=0.001, with no observed heterogeneity and pleiotropy]. Through mediation analysis, 35 proteins have been determined mediating the genetically causal effects of selenium on the levels of eGFR, including Fibroblast growth factor receptor 4 (FGFR4), Fibulin-1, Cilia- and flagella-associated protein 45, Mothers against decapentaplegic homolog 2 (SMAD2), and E3 ubiquitin-protein ligase ZNRF3, and the mediation effect rates of these proteins ranged from 1.59% to 23.70%. In the enrichment analysis, 13 signal transduction pathways, including FGFR4 mutant receptor activation and Defective SLC5A5 causing thyroid dyshormonogenesis 1, were involved in the effect of selenium on eGFR levels.
Conclusion: Our finding has revealed the underlying mechanism by which increased selenium level lead to deterioration of renal function, effectively guiding the prevention of chronic kidney disease and paving the way for future studies.
1 Introduction
Chronic kidney disease (CKD) is defined as an abnormality in the structure or function of kidney, lasting more than 3 months, with specific implications for health (1). It could progress to end-stage renal disease (ESRD), which is fatal without renal replacement therapy (kidney transplantation or dialysis). Moreover, because of its effect on cardiovascular risk, CKD could directly affect the global burden of death caused by cardiovascular disease (2). Over 10% of the world’s population carries CKD and in the United States, more than 30 million people have CKD (3). In addition, with the rapid increase in the prevalence of risk factors such as hypertension, diabetes and obesity, the burden of CKD will become even heavier in the future. Therefore, CKD as a major public health problem has attracted more and more attention.
Selenium is an essential microelement for mammals and is widely distributed in all tissues in the body (4). Elemental selenium is biologically inactive by itself. It is involved in the synthesis of 25 selenoproteins in the form of selenocysteine and selenomethionine to perform its biological functions (5). Selenoproteins play important functions in the body such as combating oxidative stress, regulating immune system, regulating thyroxine metabolism and fertility (6). However, the biological activities of about 50% of selenoproteins are unknown and their roles in biology remain to be elucidated (7). Kidney is the organ with the highest selenium content (8). Many observational studies have found a strong association between selenium and CKD (9–11). Selenium levels are usually lower in CKD patients than normal people (12, 13), but whether selenium supplementation can benefit CKD patients is still controversial (14, 15).
Mendelian randomization (MR) is a research approach that utilizes genetic variation as an instrumental variables to examine whether exposure factor has a causal effect on health outcome (16). Compared to conventional methods, MR could avoid reverse causation bias and attenuates the interference of confounding factors, so it is becoming increasingly popular in epidemiologic studies (17). Using MR method, our team has previously found that increased selenium levels in the body could lead to a decrease of glomerular filtration rate and an increase in blood urea nitrogen levels, suggesting that selenium supplementation in CKD patients should be taken with great caution (18). However, the mechanism by which increased selenium levels in the body lead to the deterioration of renal function remains unclear. Genome-wide association studies (GWAS) have detected genetic variants associated with plasma proteome levels (19, 20), which provide an opportunity to utilize MR to reveal the underlying mechanisms behind the genetically causal relationship between exposure and outcome, as many circulating proteins always act as the principal regulators of molecular pathways (21).
In the current study, we further selected 3282 circulating plasma proteome levels to explore their potential mediating effect in the association of selenium levels and renal function. By evaluating these mediation effects, we can gain insights into the mechanistic pathways through which the increased selenium levels might influence the risk of renal failure, paving the way for potential therapeutic interventions.
2 Materials and methods
2.1 Overall study design
This study utilized two-sample two-step mediating MR methodology to investigate the potential mediating effect of the plasma proteome in the genetically causal relationship between selenium levels and renal function (22), which was conducted in three stages (Figure 1). The first stage determined the genetically causal effects of selenium levels on renal function. The second stage focused on investigating the genetically causal effects of selenium levels on the plasma proteome and the genetically causal effects of the plasma proteome on renal function. The third stage aimed to investigate and quantify the potential mediating role of the plasma proteome in the genetically causal relationship between selenium levels and renal function, and perform enrichment analysis to the mediating proteins through bioinformatics for understanding the potential mechanisms involved in selenium’s influence on renal function. We reviewed our analytical process in accordance with the STROBE-MR Checklist to ensure the reliability of the results, with the checklist information available in Supplementary Table 1.
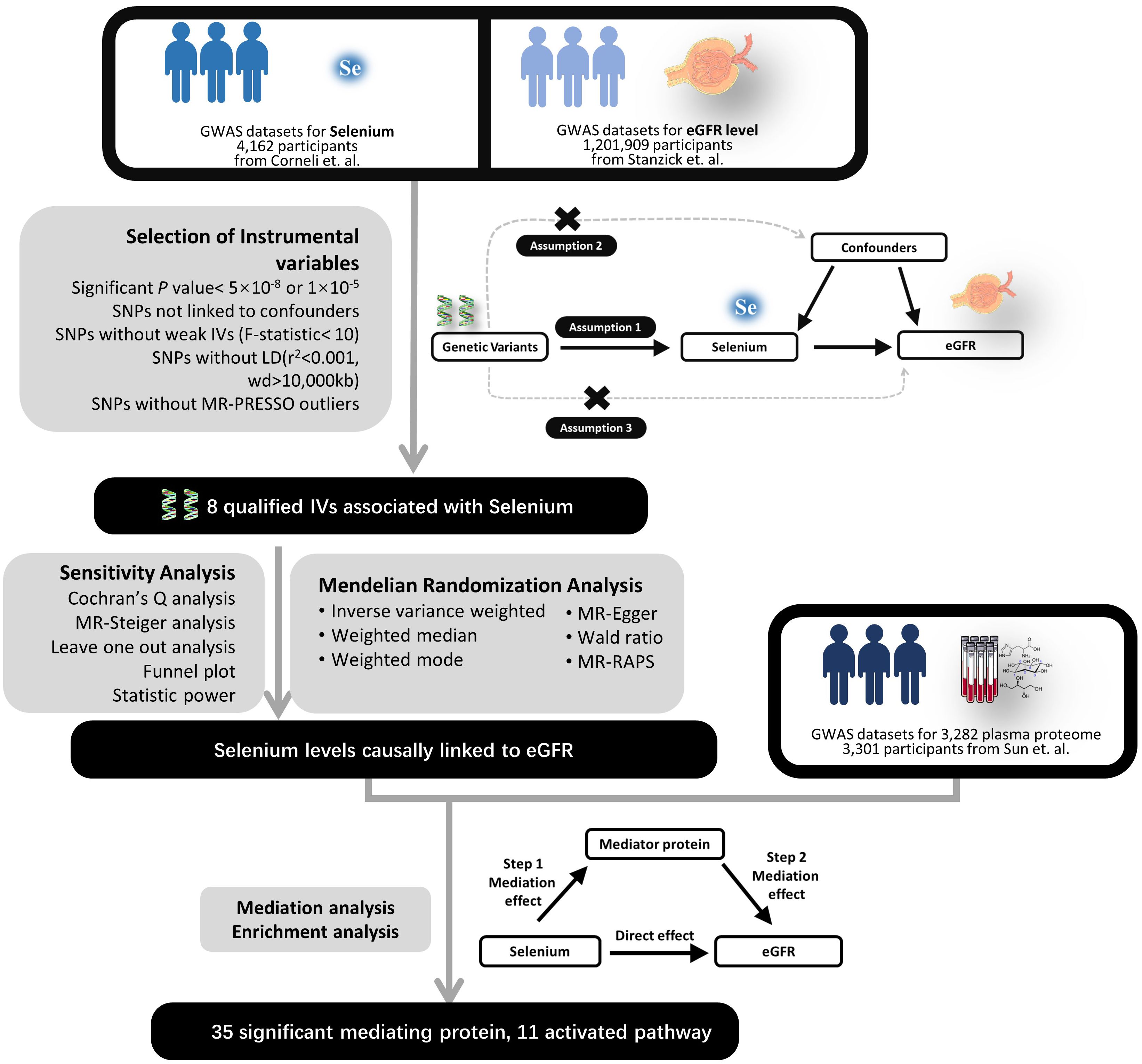
Figure 1. Experimental design flowchart. LD, Linkage Disequilibrium; SNP, Single Nucleotide Polymorphism; IV, Instrumental Variable; eGFR, Estimated Glomerular Filtration Rate; MR, Mendelian Randomization; RAPS, Robust Adjusted Profile Score.
2.2 GWAS data sources
The instrumental variables closely associated with selenium levels were obtained from a large GWAS meta-analysis of toenail and blood selenium (23). Toenail selenium concentrations were collected from 4,162 individuals of European descent from four cohorts in the United States (adjusted for genetic associations based on sex, age, smoking status and study-specific covariates) (24–27). Blood selenium concentrations were collected from 2603 Australian twin pairs and their families and 2874 pregnant women from England (adjusted for genetic associations based on sex, age and within- family relatedness) (28). GWAS summary data for plasma proteome levels were gathered from the genomic atlas of the human plasma proteome published by Sun et al. in 2018 (19), which genome-wide tested 3282 plasma proteins of 10.6 million putative autosomal variants in 3301 healthy participants from 25 centers in England. GWAS summary data for kidney function (eGFR) were derived from the largest GWAS meta-analysis for kidney function to date (29), which included the data from UK Biobank (n = 436561) and Chronic Kidney Disease Genetics (CKDGen) Consortium (n = 765348). In UKB, GFR was estimated based on the CKD-EPI formula, whereas in CKDGen, GFR was estimated based on the CKD-EPI formula (for individuals > 18 years) and the Schwartz formula (for individuals ≤ 18 years). All participants have provided informed consent. Table 1 details the characteristics of the involved GWAS datasets, and information on all GWAS datasets included in the study is presented in Supplementary Table 2. Based on the information from the data set sources, there is no sample overlap in the MR analysis of this study.
2.3 Selection of instrumental variables and mendelian randomization analysis
To ensure the reliability of MR analyses, three central assumptions must be met (30): (1) the instrumental variables are closely associated with exposure, (2) the instrumental variables independent of any confounders, (3) the instrumental variables affect the outcome only through exposure rather than any other causal pathway. Based on the above guidelines, we used the following steps to select instrumental variables: single nucleotide polymorphisms (SNPs) associated with exposure were selected at the genome-wide significant level (p-value < 5 × 10-8) or Genome-wide potential significance level (p-value < 1 × 10-5). SNPs exhibiting linkage disequilibrium, ambiguity, or being palindromic were excluded, and weak instruments were eliminated based on an F-statistic < 10 (31). The MR-PRESSO method was employed to detect and remove pleiotropic SNPs. The MR-Steiger test was utilized to identify and exclude SNPs with incorrect directionality of association. The web tool PhenoScanner and the R package “gwasrapidd” are utilized respectively to examine each included variation for its association with other traits, and SNPs associated with confounding factors are excluded. MR analyses were performed using inverse-variance weighting (IVW) (32), MR-Egger regression (33), weighted median (WM) (34), and MR-Robust Adjusted Profile Scores (MR-RAPS) (35).
2.4 Sensitivity analyses
Sensitivity analyses mainly include horizontal pleiotropy, heterogeneity tests and leave-one-out analysis. Horizontal pleiotropy was assessed using the MR-Egger method. When horizontal pleiotropy is detected, MR-Egger regression is preferred. Heterogeneity was assessed using Cochran’s Q test. According to MR operational guidelines, the random-effects IVW model is employed as the primary analysis method regardless of the presence of heterogeneity. We performed the leave-one-out analyses and plotted the funnel plots. In addition, MR-Steiger test was also performed to determine the overall causal direction was correct (36). Finally we calculated the statistical power to clarify the reliability of the negative results (18).
2.5 Mediation analyses of plasma proteome and enrichment analyses of pathways
In the mediation analyses, MR analysis of selenium levels on plasma proteome was performed in the first step, and MR analysis of plasma proteome on renal function was performed in the second step. Proteins that were significant in both steps of the MR analyses had a partial mediation effect. The mediation effect is calculated by multiplying the effect of first step and second step, while the mediation effect rate is the proportion of the mediation effect to the total effect. Then pathway enrichment of the discovered mediator proteins was performed in Reactome. Reactome is a peer-reviewed pathway database that provides intuitive bioinformatics tools for visualizing, interpreting and analyzing pathway information to support genome analyses (37).
2.6 Statistical analysis and graphing
For visualization, scatter plots were drawn for each SNP in this study, showing the relationship between exposure factors and outcome effects. Funnel plots were utilized to assess possible directional effects and pleiotropy. Forest plots were drawn for final causal estimation, presenting the results for each SNP and the results of the overall MR analysis. The full Mendelian randomization figure results can be found in Supplementary Figures 1-4. Statistical calculations and results visualization were conducted using R software (version 4.3.1) and R software packages “TwoSample MR”, “MR-PRESSO”, “mr.raps”.
3 Results
3.1 Dataset characteristics and screening for instrumental variables
In the MR analysis, we initially screened 12 SNPs related to selenium levels, which primarily originate from chromosomes 5 and 21 (Figure 2A), and no weak instrumental variables were found; the MR-PRESSO test detected no SNPs with pleiotropy; the MR-Steiger test identified no SNPs with reverse causality. No SNPs associated with potential confounders (e.g., daily diet, occupational exposures, smoking, etc.) were found, but four SNPs potentially related to the outcomes were excluded. Ultimately, eight qualified SNPs were included in the study. Specifically, among these instrumental variables, there were three SNPs that upregulated selenium levels (rs3797535, rs11951068, and rs1789953), and five SNPs that downregulated selenium levels (rs234709, rs558133, rs567754, rs6859667 and rs705415). Details of these SNPs leading to changes in selenium body levels in are shown in Figure 2B. In the mediation MR analysis, we initially screened 249,286 SNPs related to the exposure, finding no weak instrumental variables. A total of 49,690 SNPs were absent in the outcome database and thus excluded; 23,526 ambiguous SNPs and palindromic SNPs were deleted when merging datasets. After Bonferroni correction, 3,203 SNPs directly related to the outcome were excluded. Ultimately, 172,870 qualified SNPs were included in the study. SNPs closely associated with eGFR levels are primarily distributed across multiple chromosomes (Figure 3A), with the top SNP being rs1617634. These SNPs mutations all contribute to the variations of eGFR (Figure 3B).
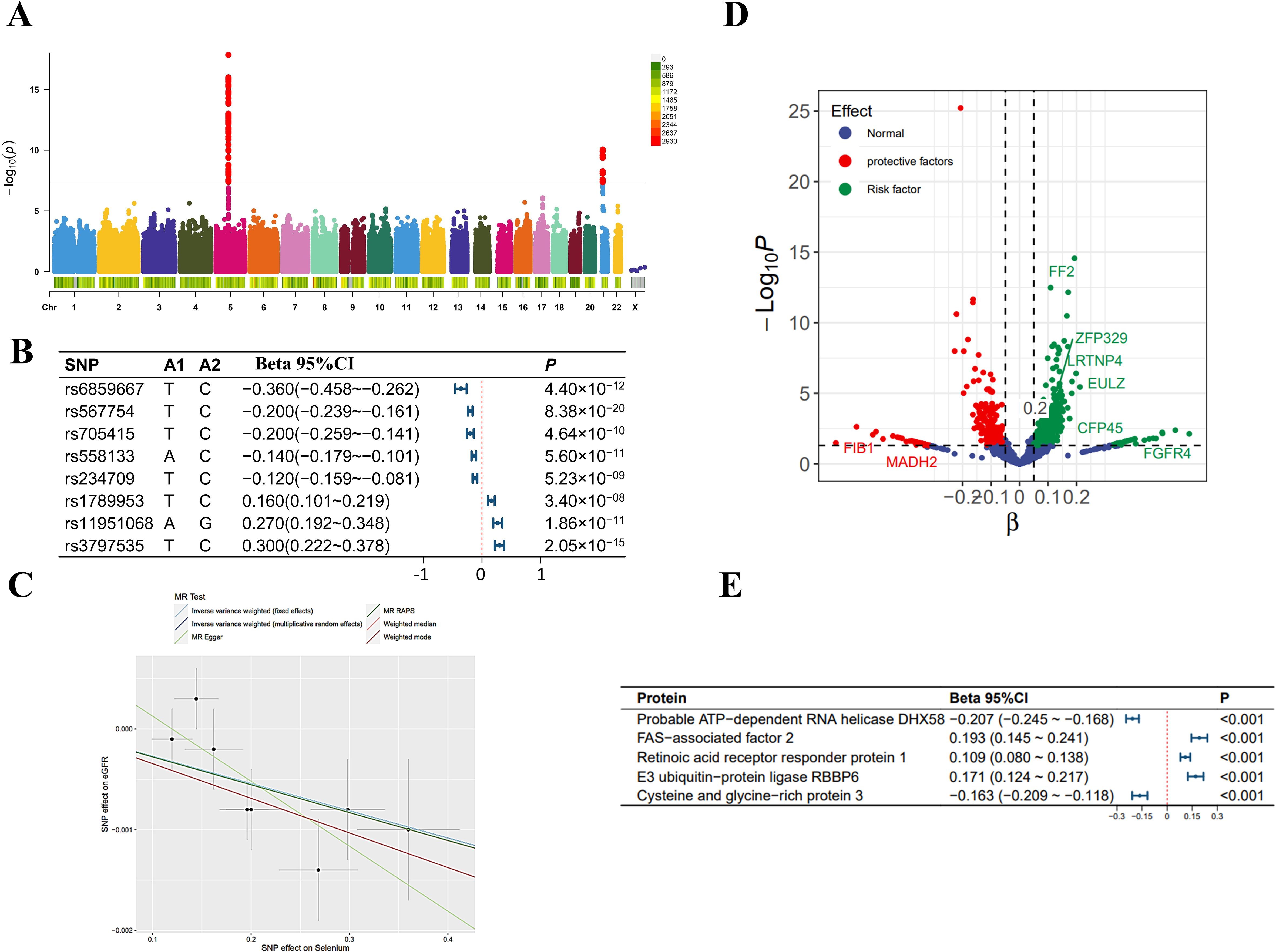
Figure 2. Selenium-related analysis diagram. (A) Manhattan plot of genome-wide association study summary data on selenium levels; (B) The Single Nucleotide Polymorphisms (SNPs) significantly associated with selenium levels involved as instrumental variables; (C) Scatter plot and regression curve of Mendelian Randomization exploring the association between selenium levels and eGFR; (D) Scatter plot of the effect-significance level of selenium levels on plasma proteins, with named proteins indicating mediating effects; (E) The top 5 plasma proteins most significantly influenced by selenium levels. Effects are scaled as “per 1 standard deviation”.
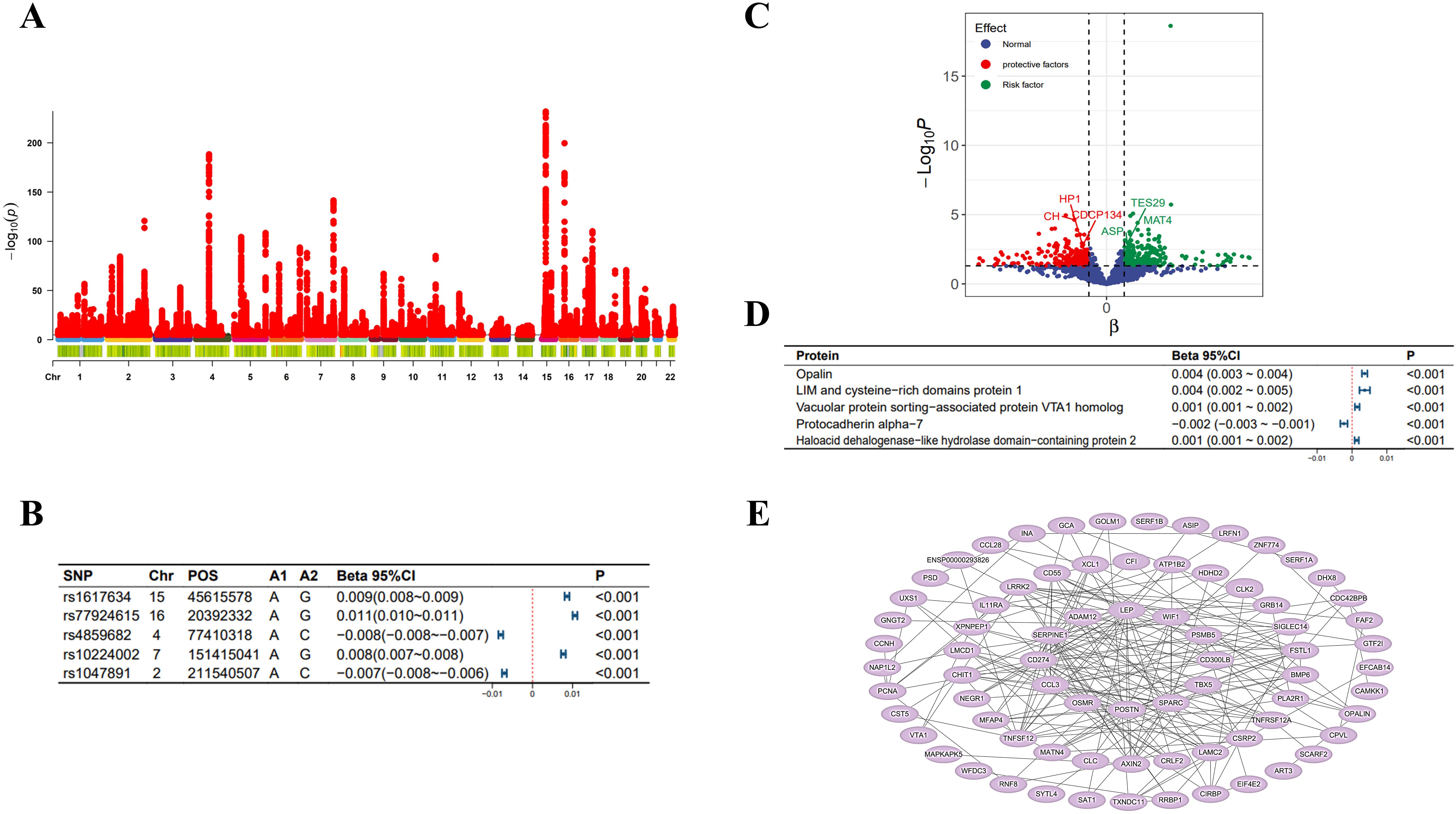
Figure 3. eGFR-related analysis diagram. (A) Manhattan plot of genome-wide association study summary data on eGFR levels; (B) The top 5 SNPs most significantly associated with eGFR levels; (C) Scatter plot of the effect-significance level of plasma proteins on eGFR levels, with named proteins indicating mediating effects; (D) The top 5 plasma proteins most significantly affecting eGFR levels; (E) Schematic diagram of plasma protein interactions affecting eGFR levels. Effects are scaled as “per 1 standard deviation”.
3.2 Mendelian randomization analysis
As shown in Figure 2C, in the MR analysis, for each standard deviation (SD) increase in selenium levels, eGFR levels are reduced by 0.003 SD [Beta (95% CI): -0.003 (-0.004 ~ -0.001), P=0.001, with no observed heterogeneity and pleiotropy]. Significant plasma protein effects mediated by selenium were shown in Figure 2D, among which the top 5 plasma proteins most significantly influenced by selenium levels were Probable ATP-dependent RNA helicase DHX58 (its level decreased by 0.207 SD for 1 SD increase in selenium level), FAS-associated factor 2 (its level increased by 0.193 SD for 1 SD increase in selenium level), Retinoic acid receptor responder protein 1 (its level increased by 0.109 SD for 1 SD increase in selenium level), E3 ubiqui-tin-protein ligase RBBP6 (its level increased by 0.171 SD for 1 SD increase in selenium level) and Cysteine and glycine-rich protein 3 (its level decreased by 0.163 SD for 1 SD increase in selenium level). Details of the effects of changes in selenium levels on them are shown in Figure 2E. The effect-significance level of plasma proteins on eGFR levels were shown in Figure 3C, among which the top 5 plasma proteins most significantly affecting eGFR levels were Opalin, LIM and cysteine-rich domains protein 1, Vacuolar protein sorting-associated protein VTA1 homolog, Protocadherin alpha-7 and Haloacid dehalogenase-like hydro-lase domain-containing protein 2 (Figure 3D). In addition, protein interaction analyses were performed and found sufficient interactions between the significant plasma proteins mediating eGFR alteration (Figure 3E). In the proteomic mediator analysis, 35 plasma proteins with mediating effects were identified, with mediation effect rates ranging from 1.59% to 23.70%. The full Mendelian randomization results and sensitivity analyses can be found in Supplementary Tables 3-8. Specifically, the top five proteins with the highest mediation effect rates were Fibroblast growth factor receptor 4, Fibu-lin-1, Cilia and flagella-associated protein 45, Mothers against decapentaplegic homolog 2, and E3 ubiquitin-protein ligase ZNRF3 (Table 2, Figures 4A, B).
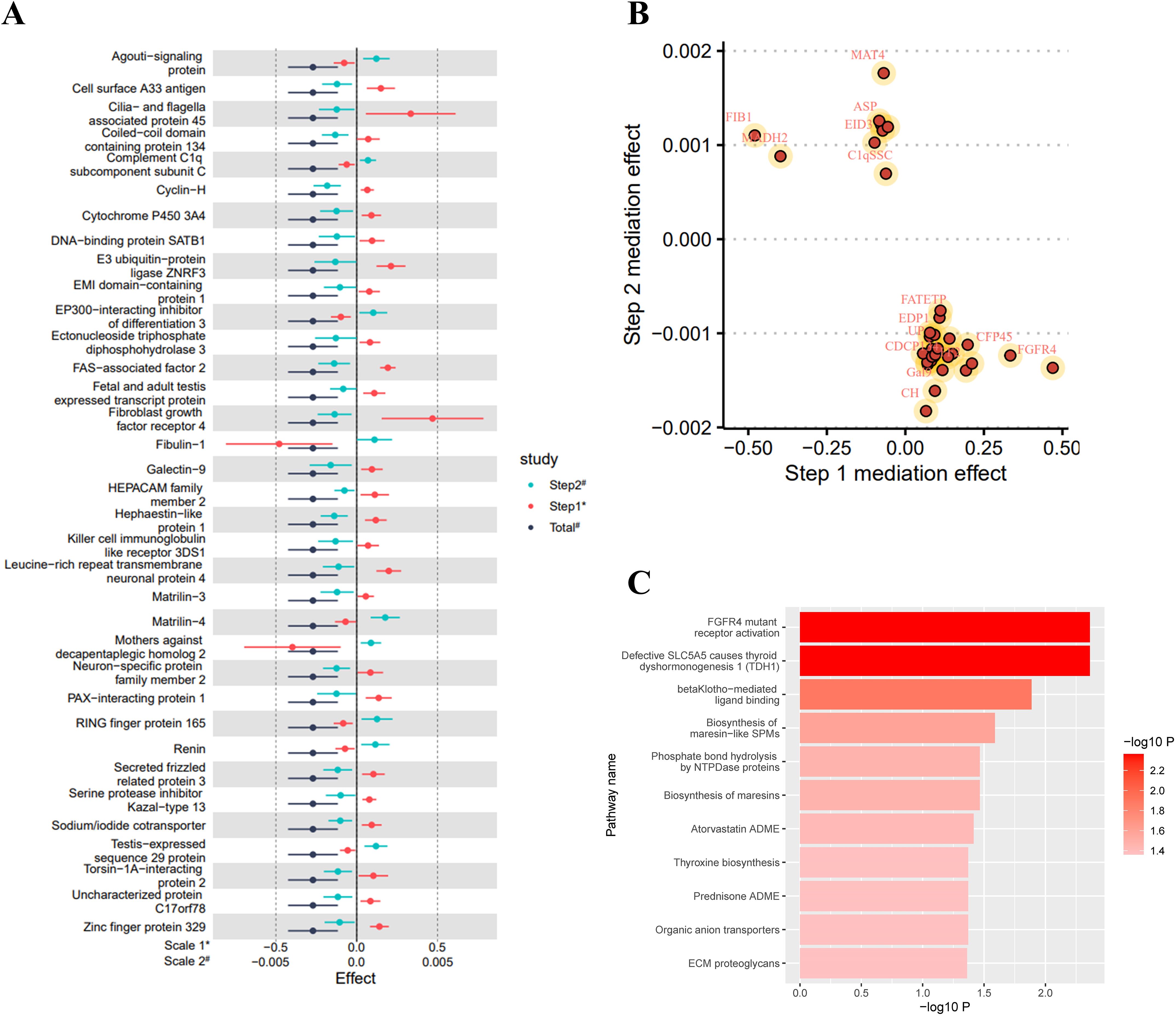
Figure 4. Mediation analysis and enrichment analysis results. (A) Forest plot of the total effect and two-step mediation effect; (B) Two-step effect scatter plot of mediating effect proteins; (C) Metabolic or signal transduction pathways mediating the effect of selenium levels on eGFR levels. Effects are scaled as “per 1 standard deviation”.
3.3 Activation of metabolic and signaling pathways
In the enrichment analysis, 11 mediating pathways were identified that are implicated in the decrease of eGFR levels mediated by an increase in selenium levels. The five most significant pathways were FGFR4 mutant receptor activation (Reactions 5/5, Entities 1/1, pValue 0.004), Defective SLC5A5 causes thyroid dyshormonogenesis 1 (Reactions 1/1, Entities 1/1, pValue 0.004), betaKlotho-mediated ligand binding (Reactions 2/2, Entities 3/, pValue 0.013), Biosynthesis of maresin-like SPMs (Reactions 1/3, Entities 1/6, pValue 0.026), and Biosynthesis of maresins (Reactions 1/5, Entities 1/8, pValue 0.034) (Figure 4C; Supplementary Table 9).
4 Discussion
As a trace element, selenium is essential to the human body, and due to its important role in combating oxidative stress, regulating immune system, regulating thyroxine metabolism and fertility (6), there has been an increasing focus on selenium supplementation. As selenium levels in CKD patients are usually lower than normal (12, 13), they are more likely to favor selenium supplementation without professional guidance. Notably, it is important to note that the safe intake range for selenium is very narrow, and the optimal daily intake of selenium for the human body is only 30-400 μg day−1 adult−1 (38), below which can lead to selenium deficiency, and above which can produce symptoms of toxicity, such as hair loss, thyroid dysfunction, and neurological damage (39). Therefore, selenium intake needs to be finely regulated. Moreover, selenium is not biologically active by itself, and it plays its biological roles mainly via selenoproteins (5). Currently, most of the functionally known selenoproteins of eukaryotes are oxidoreductases with antioxidant properties (40, 41). However, it is worth noting that taking GPX1 as an example, while it has anti-inflammatory and antioxidant effects, it has also been found to be associated with insulin resistance and type 2 diabetes (42), which fully illustrates the complexity of selenoproteins’ biological functions. Beneficial or harmful depending on the levels of selenoproteins in the body and other factors, and the biological effects of most selenoproteins, especially on kidney, remain unclear. Furthermore, the benefits and harms of selenium are dependent on its dose and form (43). With increasing selenium intake, selenoprotein expression tends to saturate and the excess selenium bind nonspecifically to human proteins through the substitution of methionine by selenomethionine (SeMet) resulting the production of non-specific selenium-containing proteins, and the non-specific selenium-containing proteins could cause damage to health (44, 45). Hence, lack or excess of selenium is a relative concept. CKD patients are characterized by decreased dietary intake, reduced intestinal resorption, increased urinary protein excretion, and reduced ability in synthesizing selenoproteins (46). Therefore, when supplemented with selenium, they are more susceptible to produce non-specific selenium-containing proteins and thus to toxic effects of selenium. Results of this study found that increased selenium levels could lead to decreased renal function, so supplementation of selenium must be taken with extreme caution, especially for patients with CKD.
In this study, we found that increased levels of selenium in body would lead to an increase of plasma protein levels such as FAS-associated factor 2 (FAF2), Retinoic acid receptor responder protein 1 (RARRES1). It may be related to the pro-oxidant effect of selenium compounds. As selenium levels increase, the selenium compounds, such as selenite, also increase, and selenite could lead to an excess of reactive oxygen species (ROS) in the cell, triggering oxidative stress (47). RARRES1 protein is involved in the regulation of signaling pathways in response to oxidative damage, so their up-regulation may reflect an adaptive reaction of the body to maintain cellular homeostasis facing oxidative challenges (48). The pro-oxidative effects of selenium compounds can also induce the activation of apoptosis and protein degradation pathways. FAF2, an apoptosis-associated protein involved in ubiquitination and protein degradation, may increase its expression in response to oxidative stress-induced protein damage (49). This reflects a complex mechanism of cellular regulation of oxidative damage and provides insight into the potential mechanisms of selenium toxicity.
Many circulating proteins are key regulators of molecular pathways, so exploring their potential mediating effect in the association of selenium levels and renal function could help to understand the underlying mechanisms of selenium’s influence on renal function. In this study, a total of 35 plasma proteins mediating between selenium levels and renal function were identified, and the three with the highest mediating effects were fibroblast growth factor receptor 4 (FGFR4), fibulin-1 (FBLN1), and cilia-and flagella-associated protein 45 (CFAP45). CKD patients have elevated serum levels of fibroblast growth factor (FGF), of which FGF23 has a strong dose-dependent association with cardiovascular morbidity, chronic inflammation and progression of kidney disease (50). FGFR4 is one of the receptors for FGF23. Activation of hepatic FGFR4 causes the production of inflammatory cytokines in the liver, thereby exacerbating the chronic inflammation in vivo and participating in the progression of CKD (51). In addition, it has been shown that FGF23 could participate in renal fibrosis by amplifying the activation of local renal fibroblast in injury and perpetuating pro-fibrotic signaling through FGFR4 (52). FBLN1 is an extracellular matrix (ECM) protein, which interacts with many ECM molecules (53). As a secreted plasma protein, FBLN1 has been found to be a good potential biomarker for renal impairment, significantly elevated in the plasma of CKD patients (54). The mechanism underlying the elevation of FBLN1 in patients with impaired renal function remains unknown. In the present study, we found that decreased plasma FBLN1 level could lead to the impairment of renal function, which is counterintuitive and suggests that, like BNP in heart failure, increased FBLN1 may be a compensatory result when renal function is impaired. However, the specific mechanisms behind this need to be studied further. CFAP45 protein is a static structural component of the ciliary axoneme and critical for motor cilia stability (55). Cilia is widely distributed in the proximal and distal tubules as well as the collecting ducts of kidney. Normal structure and function of cilia are essential for renal organogenesis and maintenance of epithelial cell differentiation and proliferation, and persistent ciliary dysfunction contributes to both early and progressive renal disease (56). However, the study on the relationship between CFAP45 and the kidney is still lacking, and its effect on renal cilia and renal function needs to be further explored in the future.
Eleven pathways, including thyroxine biosynthesis and defective SLC5A5 causes thyroid dyshormonogenesis were found to mediate the effects of selenium levels on renal function in the enrichment analysis. Selenium is important for thyroxine biosynthesis. The thyroid gland expresses selenoproteins such as glutathione peroxidase, selenoprotein P and thioredoxin reductase, which together form the antioxidant system to mitigate the damage to thyroid cells caused by reactive oxygen species generated during thyroxine biosynthesis, and thus maintains the normal function of thyroid tissue (57). In addition, iodothyronine deiodinases is also a selenoprotein. It converts the inactive precursor thyroxine to the active triiodothyronine. Abnormal selenium levels in the body affect the expression and activity of iodothyronine deiodinases, which can lead to disruption of thyroxine metabolism (58). The kidney is an important target organ for thyroxine action. Thyroxine affects protein synthesis and cell growth and therefore has an important role in the growth and development of kidney (59). Thyroxine regulates the activity of tubular ion transporters (Na+, K+, 2Cl− co-transport protein, etc.), which modulates tubuloglomerular feedback and affects glomerular filtration rate (60). It also regulates the expression of glomerular vasodilatory factors, such as vascular endothelial growth factor, and the synthesis and secretion of various components of the renin-angiotensin-aldosterone system, to modulate renal perfusion (61). Additionally, it has been found that abnormal increases or decreases of thyroxine are strongly associated with the development and progression of renal fibrosis (62, 63). These evidences support our finding that thyroxine disruption may be a potential mechanism by which selenium affects renal function.
Our two-sample MR analysis uses stringent criteria and provides compelling insights into the intricate relationship between the levels of selenium in vivo and renal function, suggesting that increased selenium levels are associated with decreased renal function. Furthermore, our mediation analysis revealed the important role of plasma proteome as mediators, depicting the causal trajectories from increased selenium levels to decreased renal function, which has profound significance in constructing a blueprint from the levels of micronutrients in vivo to the pathogenesis of CKD, thereby effectively guiding the prevention of CKD and delaying its progression.
However, it is crucial to acknowledge certain limitations in our study. First, the genetic analyses in this study were performed on individuals of European ancestry, so generalization of the findings to other ethnic populations is limited. Second, selenium levels in populations are closely related to geographic variation, thus also limiting the generalization of the findings to different geographic populations. Third, our data were derived from publicly available aggregated statistics and the raw clinical outcome data for each individual were not available, thus preventing further population stratification analyses. Fourth, this study used the relative scale of “per 1 SD” to determine the association of selenium with eGFR, and did not use an absolute scale due to limitations of the original data. Lastly, we did not perform experimental validations for the identified mediating proteins and potential mechanisms by which selenium affects renal function, and the role of environmental factors and the influence of genetic factors interacting with the environment on this causal effect should be further explored in future studies.
5 Conclusions
Our comprehensive MR analysis has revealed the complex relationship between selenium levels and renal function, and found that increased selenium level is one genetically causative factor for kidney function impairment, suggesting that supplementation of selenium must be taken with extreme caution, especially for patients with CKD. More importantly, we reveal the underlying mechanism by which increased selenium levels lead to deterioration of renal function, effectively guiding the prevention of CKD and paving the way for future studies.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SF: Formal analysis, Methodology, Writing – original draft. MQ: Formal analysis, Investigation, Writing – original draft. ZY: Data curation, Formal analysis, Investigation, Writing – original draft. SS: Software, Visualization, Writing – review & editing. FM: Project administration, Writing – review & editing. FL: Resources, Software, Visualization, Writing – original draft. ZX: Data curation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Grant No. 81974094) and Natural Science Foundation of Jilin Province (Grant No. 20210101259JC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2024.1410463/full#supplementary-material
Supplementary Figure 1 | Forest plots of all the mendelian randomization analyses in this study.
Supplementary Figure 2 | Scatter plots of all the mendelian randomization analyses in this study.
Supplementary Figure 3 | Leave-one-out plots of all the mendelian randomization analyses in this study.
Supplementary Figure 4 | Funnel plots of all the mendelian randomization analyses in this study.
References
1. Adamczak M, Surma S. Metabolic acidosis in patients with CKD: epidemiology, pathogenesis, and treatment. Kidney Dis (Basel). (2021) 7:452–67. doi: 10.1159/000516371
2. Akchurin OM. Chronic kidney disease and dietary measures to improve outcomes. Pediatr Clin North Am. (2019) 66:247–67. doi: 10.1016/j.pcl.2018.09.007
3. Luo S, Grams ME. Epidemiology research to foster improvement in chronic kidney disease care. Kidney Int. (2020) 97:477–86. doi: 10.1016/j.kint.2019.11.010
4. Rayman MP. Selenium and human health. Lancet. (2012) 379:1256–68. doi: 10.1016/s0140-6736(11)61452-9
5. Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev. (2014) 94:739–77. doi: 10.1152/physrev.00039.2013
6. Huang Z, Rose AH, Hoffmann PR. The role of selenium in inflammation and immunity: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. (2012) 16:705–43. doi: 10.1089/ars.2011.4145
7. Mojadadi A, Au A, Salah W, Witting P, Ahmad G. Role for selenium in metabolic homeostasis and human reproduction. Nutrients. (2021) 13:3256. doi: 10.3390/nu13093256
8. Zachara BA, Pawluk H, Bloch-Boguslawska E, Sliwka KM, Korenkiewicz J, Skok Z, et al. Tissue level, distribution, and total body selenium content in healthy and diseased humans in Poland. Arch Environ Health. (2001) 56:461–6. doi: 10.1080/00039890109604483
9. Alehagen U, Aaseth J, Alexander J, Brismar K, Larsson A. Selenium and coenzyme Q10 supplementation improves renal function in elderly deficient in selenium: observational results and results from a subgroup analysis of a prospective randomized double-blind placebo-controlled trial. Nutrients. (2020) 12:3780. doi: 10.3390/nu12123780
10. Zitouni K, Steyn M, Lyka E, Kelly FJ, Cook P, Ster IC, et al. Derepression of glomerular filtration, renal blood flow and antioxidant defense in patients with type 2 diabetes at high-risk of cardiorenal disease. Free Radic Biol Med. (2020) 161:283–9. doi: 10.1016/j.freeradbiomed.2020.10.005
11. Kiss I. Importance of selenium homeostasis in chronic and end-stage kidney diseases. Orv Hetil. (2013) 154:1641–7. doi: 10.1556/oh.2013.29733
12. Sabé R, Rubio R, García-Beltrán L. Reference values of selenium in plasma in population from Barcelona. Comparison with several pathologies. J Trace Elem Med Biol. (2002) 16:231–7. doi: 10.1016/s0946-672x(02)80050-5
13. Wu CY, Wong CS, Chung CJ, Wu MY, Huang YL, Ao PL, et al. The association between plasma selenium and chronic kidney disease related to lead, cadmium and arsenic exposure in a Taiwanese population. J Hazard Mater. (2019) 375:224–32. doi: 10.1016/j.jhazmat.2019.04.082
14. Shen Y, Yin Z, Lv Y, Luo J, Shi W, Fang J, et al. Plasma element levels and risk of chronic kidney disease in elderly populations (≥ 90 Years old). Chemosphere. (2020) 254:126809. doi: 10.1016/j.chemosphere.2020.126809
15. Omrani H, Golmohamadi S, Pasdar Y, Jasemi K, Almasi A. Effect of selenium supplementation on lipid profile in hemodialysis patients. J Renal Inj Prev. (2016) 5:179–82. doi: 10.15171/jrip.2016.38
16. Birney E. Mendelian randomization. Cold Spring Harb Perspect Med. (2022) 12: a041302. doi: 10.1101/cshperspect.a041302
17. Yuan S, Xu F, Li X, Chen J, Zheng J, Mantzoros CS, et al. Plasma proteins and onset of type 2 diabetes and diabetic complications: Proteome-wide Mendelian randomization and colocalization analyses. Cell Rep Med. (2023) 4:101174. doi: 10.1016/j.xcrm.2023.101174
18. Fu S, Zhang L, Ma F, Xue S, Sun T, Xu Z. Effects of selenium on chronic kidney disease: A mendelian randomization study. Nutrients. (2022) 14:4458. doi: 10.3390/nu14214458
19. Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. Genomic atlas of the human plasma proteome. Nature. (2018) 558:73–9. doi: 10.1038/s41586-018-0175-2
20. Ferkingstad E, Sulem P, Atlason BA, Sveinbjornsson G, Magnusson MI, Styrmisdottir EL, et al. Large-scale integration of the plasma proteome with genetics and disease. Nat Genet. (2021) 53:1712–21. doi: 10.1038/s41588-021-00978-w
21. Finan C, Gaulton A, Kruger FA, Lumbers RT, Shah T, Engmann J, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. (2017) 9:eaag1166. doi: 10.1126/scitranslmed.aag1166
22. Burgess S, Daniel RM, Butterworth AS, Thompson SG. Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int J Epidemiol. (2015) 44:484–95. doi: 10.1093/ije/dyu176
23. Cornelis MC, Fornage M, Foy M, Xun P, Gladyshev VN, Morris S, et al. Genome-wide association study of selenium concentrations. Hum Mol Genet. (2015) 24:1469–77. doi: 10.1093/hmg/ddu546
24. Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR Jr., et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. (1988) 41:1105–16. doi: 10.1016/0895-4356(88)90080-7
25. Jordan JM, Helmick CG, Renner JB, Luta G, Dragomir AD, Woodard J, et al. Prevalence of knee symptoms and radiographic and symptomatic knee osteoarthritis in African Americans and Caucasians: the Johnston County Osteoarthritis Project. J Rheumatol. (2007) 34:172–80.
26. Colditz GA, Hankinson SE. The Nurses’ Health Study: lifestyle and health among women. Nat Rev Cancer. (2005) 5:388–96. doi: 10.1038/nrc1608
27. Chu NF, Spiegelman D, Yu J, Rifai N, Hotamisligil GS, Rimm EB. Plasma leptin concentrations and four-year weight gain among US men. Int J Obes Relat Metab Disord. (2001) 25:346–53. doi: 10.1038/sj.ijo.0801549
28. Evans DM, Zhu G, Dy V, Heath AC, Madden PA, Kemp JP, et al. Genome-wide association study identifies loci affecting blood copper, selenium and zinc. Hum Mol Genet. (2013) 22:3998–4006. doi: 10.1093/hmg/ddt239
29. Stanzick KJ, Li Y, Schlosser P, Gorski M, Wuttke M, Thomas LF, et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nat Commun. (2021) 12:4350. doi: 10.1038/s41467-021-24491-0
30. Davey Smith G, Holmes MV, Davies NM, Ebrahim S. Mendel’s laws, Mendelian randomization and causal inference in observational data: substantive and nomenclatural issues. Eur J Epidemiol. (2020) 35:99–111. doi: 10.1007/s10654-020-00622-7
31. Li F, Liu Y, Wang Z, Zhao Q, Li Y, Tang T. A mendelian randomization study with populations of European ancestry rules out a causal relationship between inflammatory bowel disease and colorectal cancer. Front Genet. (2022) 13:949325. doi: 10.3389/fgene.2022.949325
32. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. (2013) 37:658–65. doi: 10.1002/gepi.21758
33. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
34. Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res. (2019) 4:186. doi: 10.12688/wellcomeopenres.15555.3
35. Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. J Ann Stat. (2020) 3:1742–1769,28. doi: 10.1214/19-AOS1866
36. Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PloS Genet. (2017) 13:e1007081. doi: 10.1371/journal.pgen.1007081
37. Rothfels K, Milacic M, Matthews L, Haw R, Sevilla C, Gillespie M, et al. Using the reactome database. Curr Protoc. (2023) 3:e722. doi: 10.1002/cpz1.722
38. Gao X, Ye C, Ma H, Zhang Z, Wang J, Zhang ZH, et al. Research advances in preparation, stability, application, and possible risks of nanoselenium: focus on food and food-related fields. J Agric Food Chem. (2023) 71:8731–45. doi: 10.1021/acs.jafc.3c02714
39. Vinceti M, Filippini T, Wise LA. Environmental selenium and human health: an update. Curr Environ Health Rep. (2018) 5:464–85. doi: 10.1007/s40572-018-0213-0
40. Mostert V. Selenoprotein P: properties, functions, and regulation. Arch Biochem Biophys. (2000) 376:433–8. doi: 10.1006/abbi.2000.1735
41. Zachara BA. Selenium and selenium-dependent antioxidants in chronic kidney disease. Adv Clin Chem. (2015) 68:131–51. doi: 10.1016/bs.acc.2014.11.006
42. Zhang L, Zeng H, Cheng WH. Beneficial and paradoxical roles of selenium at nutritional levels of intake in healthspan and longevity. Free Radic Biol Med. (2018) 127:3–13. doi: 10.1016/j.freeradbiomed.2018.05.067
43. Rayman MP. Selenium intake, status, and health: a complex relationship. Hormones (Athens). (2020) 19:9–14. doi: 10.1007/s42000-019-00125-5
44. Rayman MP. The use of high-selenium yeast to raise selenium status: how does it measure up? Br J Nutr. (2004) 92:557–73. doi: 10.1079/bjn20041251
46. Ortaç E, Ozkaya O, Saraymen R, Yildiz N, Bedir A, Buyan N, et al. Low hair selenium and plasma glutathione peroxidase in children with chronic renal failure. Pediatr Nephrol. (2006) 21:1739–45. doi: 10.1007/s00467-006-0245-9
47. Brigelius-Flohé R, Flohé L. Selenium and redox signaling. Arch Biochem Biophys. (2017) 617:48–59. doi: 10.1016/j.abb.2016.08.003
48. Roy A, Ramalinga M, Kim OJ, Chijioke J, Lynch S, Byers S, et al. Multiple roles of RARRES1 in prostate cancer: Autophagy induction and angiogenesis inhibition. PloS One. (2017) 12:e0180344. doi: 10.1371/journal.pone.0180344
49. Imai Y, Nakada A, Hashida R, Sugita Y, Tanaka T, Tsujimoto G, et al. Cloning and characterization of the highly expressed ETEA gene from blood cells of atopic dermatitis patients. Biochem Biophys Res Commun. (2002) 297:1282–90. doi: 10.1016/s0006-291x(02)02380-x
50. Fliser D, Kollerits B, Neyer U, Ankerst DP, Lhotta K, Lingenhel A, et al. Fibroblast growth factor 23 (FGF23) predicts progression of chronic kidney disease: the Mild to Moderate Kidney Disease (MMKD) Study. J Am Soc Nephrol. (2007) 18:2600–8. doi: 10.1681/asn.2006080936
51. Singh S, Grabner A, Yanucil C, Schramm K, Czaya B, Krick S, et al. Fibroblast growth factor 23 directly targets hepatocytes to promote inflammation in chronic kidney disease. Kidney Int. (2016) 90:985–96. doi: 10.1016/j.kint.2016.05.019
52. Smith ER, Holt SG, Hewitson TD. FGF23 activates injury-primed renal fibroblasts via FGFR4-dependent signaling and enhancement of TGF-β autoinduction. Int J Biochem Cell Biol. (2017) 92:63–78. doi: 10.1016/j.biocel.2017.09.009
53. Cangemi C, Skov V, Poulsen MK, Funder J, Twal WO, Gall MA, et al. Fibulin-1 is a marker for arterial extracellular matrix alterations in type 2 diabetes. Clin Chem. (2011) 57:1556–65. doi: 10.1373/clinchem.2011.162966
54. Neiman M, Hedberg JJ, Dönnes PR, Schuppe-Koistinen I, Hanschke S, Schindler R, et al. Plasma profiling reveals human fibulin-1 as candidate marker for renal impairment. J Proteome Res. (2011) 10:4925–34. doi: 10.1021/pr200286c
55. Deniz E, Pasha M, Guerra ME, Viviano S, Ji W, Konstantino M, et al. CFAP45, a heterotaxy and congenital heart disease gene, affects cilia stability. Dev Biol. (2023) 499:75–88. doi: 10.1016/j.ydbio.2023.04.006
56. Bai Y, Wei C, Li P, Sun X, Cai G, Chen X, et al. Primary cilium in kidney development, function and disease. Front Endocrinol (Lausanne). (2022) 13:952055. doi: 10.3389/fendo.2022.952055
57. Rotondo Dottore G, Leo M, Casini G, Latrofa F, Cestari L, Sellari-Franceschini S, et al. Antioxidant actions of selenium in orbital fibroblasts: A basis for the effects of selenium in graves’ Orbitopathy. Thyroid. (2017) 27:271–8. doi: 10.1089/thy.2016.0397
58. Wang F, Li C, Li S, Cui L, Zhao J, Liao L. Selenium and thyroid diseases. Front Endocrinol (Lausanne). (2023) 14:1133000. doi: 10.3389/fendo.2023.1133000
59. Vargas F, Moreno JM, Rodríguez-Gómez I, Wangensteen R, Osuna A, Alvarez-Guerra M, et al. Vascular and renal function in experimental thyroid disorders. Eur J Endocrinol. (2006) 154:197–212. doi: 10.1530/eje.1.02093
60. Katz AI, Lindheimer MD. Renal sodium- and potassium-activated adenosine triphosphatase and sodium reabsorption in the hypothyroid rat. J Clin Invest. (1973) 52:796–804. doi: 10.1172/jci107243
61. Michel B, Grima M, Coquard C, Welsch C, Barthelmebs M, Imbs JL. Effects of triiodothyronine and dexamethasone on plasma and tissue angiotensin converting enzyme in the rat. Fundam Clin Pharmacol. (1994) 8:366–72. doi: 10.1111/j.1472-8206.1994.tb00814.x
62. Lu X, Chen Z, Liang H, Li Z, Zou X, Luo H, et al. Thyroid hormone inhibits TGFβ1 induced renal tubular epithelial to mesenchymal transition by increasing miR34a expression. Cell Signal. (2013) 25:1949–54. doi: 10.1016/j.cellsig.2013.06.005
Keywords: selenium, renal function, plasma proteins, Mendelian randomization analysis, mediation analysis
Citation: Fu S, Qian M, Yuan Z, Su S, Ma F, Li F and Xu Z (2024) A new perspective on selenium’s impact on renal function: European population-based analysis of plasma proteome-mediated Mendelian randomization study. Front. Endocrinol. 15:1410463. doi: 10.3389/fendo.2024.1410463
Received: 01 April 2024; Accepted: 28 August 2024;
Published: 12 September 2024.
Edited by:
Xianli Gao, Jiangsu University, ChinaReviewed by:
Marco Vinceti, University of Modena and Reggio Emilia, ItalyTao Hou, Huazhong Agricultural University, China
Copyright © 2024 Fu, Qian, Yuan, Su, Ma, Li and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonggao Xu, emhvbmdnYW9Aamx1LmVkdS5jbg==
 Shaojie Fu
Shaojie Fu Man Qian
Man Qian Zishu Yuan1
Zishu Yuan1 Sensen Su
Sensen Su Fuzhe Ma
Fuzhe Ma Fan Li
Fan Li Zhonggao Xu
Zhonggao Xu