- 1Institute of Reproductive & Stem Cell Engineering, Center of Reproductive Health, School of Basic Medical Science, Central South University, Changsha, Hunan, China
- 2Center of Genetics, Changsha Jiangwan Maternity Hospital, Changsha, Hunan, China
Background: Previous study suggested evidence for coexistence and similarities between endometriosis and polycystic ovary syndrome (PCOS), but it is unclear regarding the shared genetic architecture and causality underlying the phenotypic similarities observed for endometriosis and PCOS.
Methods: By leveraging summary statistics from public genome-wide association studies regarding endometriosis (European-based: N=470,866) and PCOS (European-based: N=210,870), we explored the genetic correlation that shared between endometriosis and PCOS using linkage disequilibrium score regression. Shared risk SNPs were derived using PLACO (Pleiotropic analysis under composite null hypothesis) and FUMA (Functional Mapping and Annotation of Genetic Associations). The potential causal association between endometriosis and PCOS was investigated using two-sample Mendelian randomization (MR). Linkage disequilibrium score for the specific expression of genes analysis (LDSC-SEG) were performed for tissue enrichment analysis. The expression profiles of the risk gene in tissues were further examined.
Results: A positive genetic association was observed between endometriosis and PCOS. 12 significant pleiotropic loci shared between endometriosis and PCOS were identified. Genetic associations between endometriosis and PCOS were particularly enriched in uterus, endometrium and fallopian tube. Two-sample MR analysis further indicated a potential causative effect of endometriosis on PCOS, and vice versa. Microarray and RNA-seq verified the expressions of SYNE1 and DNM3 were significantly altered in the endometrium of patients with endometriosis or PCOS compared to those of control subjects.
Conclusion: Our study indicates the genetic correlation and shared risk genes between PCOS and endometriosis. These findings provide insights into the potential mechanisms behind their comorbidity and the future development of therapeutics.
1 Introduction
Endometriosis is characterized by abnormal presence of endometrial glands and stroma outside the uterus (1, 2), of which symptoms include dysmenorrhea, pelvic pain and infertility. There are approximately 10% of reproductive-aged women diagnosed with this condition. It is a benign disease but with many malignant characteristics, such as estrogen dependence, recurrence and invasiveness (3). The economic burden exceeds US$22 billion in the USA alone (4). Polycystic ovary syndrome (PCOS), with a prevalence rate of approximately 10% (5), is also a disorder affecting reproductive-aged women, which is characterized by enlarged and dysfunctional ovaries, resistance to insulin, excess androgen levels (6, 7). During pregnancy in PCOS patients, their hospitalization rate is twice higher than that of general population, because of pregnancy complications such as diabetes and pre-eclampsia.
The relationship between endometriosis and PCOS is controversial. Some studies suggested evidence for coexistence and similarities between endometriosis and PCOS (8, 9). For example, endometriosis and PCOS have similar symptoms, both of which can lead to infertility and miscarriage (10). Holoch et al. reported high rates of endometriosis in women with PCOS (>70%) (11). In endometrium, endometriosis patients and PCOS patients have similar defects (12, 13). The receptivity of the endometrium to embryos decreases, and the expression of genes related to supporting blastocyst attachment (MUC1, L-Selectin) in the endometrium is abnormal. The genes that affect the interaction between mother and fetus also express abnormally (14). Adipokines, secreted by white or brown adipose tissues, plays a central role in energy metabolism. The expression level of some adipokines, such as leptin, chemerin, adiponectin, apelin, have similar changes in both endometriosis and PCOS (15). In ovary, CXC chemokines, chemotactic and secreted cytokines, were dysregulated in both PCOS and endometriosis, which may promote the progression of both disease (16). Endometriosis and PCOS also have similar hormone dysregulation. Endometriosis is an estrogen dependent disease accompanied by progesterone resistance. This hormonal imbalance can lead to exacerbation of inflammation, increase of pelvic pain, and reduction of the endometrium receptivity to embryo implantation (17). In PCOS, estrogen activity is also increased and accompanied by progesterone resistance, which promotes endometrial growth (18). Dysregulation of the gut microbiota composition can lead to several diseases in reproductive endocrine system (19). There are similar changes in gut microbiota in patients with endometriosis and PCOS (19). Some studies claimed endometriosis and PCOS are diametric disorders (11). In the ovaries of patients with endometriosis, the rate of follicle recruitment and degeneration accelerates, leading to accelerated depletion of ovarian reserves. In the ovaries of PCOS patients, the speed of follicle transition from primordial reserve to dynamic reserve slows down, and the number of antral follicles increased (11). They supposed that endometriosis and PCOS represent diametric outcomes of variation in hypothalamic-pituitary-gonadal axis development. Endometriosis is mediated by low prenatal and postnatal testosterone, while PCOS is mediated by high prenatal testosterone. Therefore, the association between endometriosis and PCOS needs further exploration.
Both endometriosis and PCOS are heritable disease. It was estimated that about 51% of the variation in endometriosis risk is heritable according to a study including 3,096 female twins in Australian women (20). Genome-wide association studies (GWASs) have identified several independent single-nucleotide polymorphisms (SNPs) for endometriosis. In a Japanese ancestry, rs10965235 in CDKN2BAS on chromosome 9p21.3 was identified (21); In a US GWAS from European-ancestry women, rs1519761 on 2q23.3 were identified (22); In an European-ancestry GWAS and from a meta-analysis of European and Japanese ancestry GWAS data, rs7521902 near WNT4 on 1p36.12, rs13391619 in GREB1 on 2p25.1, rs4141819 on 2p14, rs7739264 near ID4 on 6p22.3, rs12700667 on 7p15.2, rs1537377 near CDKN2B-AS1 (independent of rs10965235) on 9p21.3 and rs10859871 near VEZT on 12q22 were identified (23, 24); In an Icelandic GWAS, rs17773813 near KDR on 4q12 and rs519664 in TTC39B on 9p22 were identified (25). Sapkota et al. identified five novel genetic loci (ESR1, CYP19A1, HSD17B1, VEGF and GnRH) which are associated with genes involved in sex steroid regulation and function (26) based on GWAS. These genetic loci may be beneficial for providing diagnostic markers for endometriosis. Based on GWAS, genetic association between endometriosis and other diseases, such as depression, anxiety, and ovarian cancer was also identified. These results implicated endometriosis share a common genetic basis with multiple diseases. Genetic factors has a role in the development of PCOS. It was estimated that the heritability of PCOS is 70% (27). Using linkage and association studies within the population or families, almost 100 susceptibility genes related to PCOS were identified (28). Through GWAS, 11 susceptibility loci mapping to DENND1A, THADA, LHCGR, FSHR, INSR, TOX3, YAP1, RAB5B, c9orf3, HMGA2, and SUMO1P1/ZNF217 have been identified in Han Chinese populations, and polymorphism in CYP11A, CYP17, CYP19, CYP21, β-HSD also result in the phenotypic expression of PCOS (29). However, the shared genetic architecture and causality between endometriosis and PCOS remains largely unknown, and no shared risk loci been reported previously.
In current study, using large-scale GWAS summary statistics (Figure 1), we aimed to investigate the genetic correlation, causal association, and shared risk loci with potential functions between endometriosis and PCOS, and to provide insights into their comorbidity.
2 Methods
2.1 Datasets
2.1.1 GWAS summary statistics
GWAS summary results for endometriosis were obtained from the GWAS catalog database (https://www.ebi.ac.uk/gwas/, GCST90269970) comprising 21,779 cases and 449,087 controls of European ancestry (30). GWAS summary results for PCOS (consortium definition) were obtained from FinnGen R9 release (https://r9.finngen.fi/) compromising 31,548 cases and 179,322 controls of European ancestry.
2.1.2 GTEx data and Franke lab data
GTEx data is a public data resource of gene expression in 53 nondiseased human primary tissues (31). First of all, we downloaded the GTEx dataset (32), and then chose the lite version of the GTEx V8 expression quantitative trait locus (eQTL) summary data (P<1×10−5). The Franke lab data set is an aggregation of publicly available microarray gene expression data sets comprising 152 tissues in human, mouse, and rat (33). We downloaded the Franke lab publicly available gene expression data from the DEPICT website (https://data.broadinstitute.org/mpg/depict/depict_download/tissue_expression).
2.1.3 Microarray dataset and bulk-tissue RNA sequencing gene expression data
Two datasets, including GSE7305 and GSE226146, were extracted from Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). GSE7305 contained ectopic endometrium of eight endometriosis patients and endometrium of eight healthy controls, and the dataset was based on GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array) (34). GSE226146 contained endometrium of three PCOS patients and endometrium of three healthy controls, and the dataset was based on GPL16791 (Illumina HiSeq 2500) (35). All samples were collected during the secretory phase of the menstrual cycle.
2.2 Statistical analyses
2.2.1 Global genetic correlation analysis
We used stratified linkage disequilibrium scoring regression (S-LDSC) together with the baseline-LD model to estimate single trait SNP heritabilities (h2SNP) for endometriosis and PCOS. As an extension of S-LDSC, the baseline-LD modeling approach is based on continuous SNP heritability partitioning rather than binary annotation sets. Using the 1000 genomes dataset as a reference panel for the underlying LD structure and limited to HapMap3 SNP (36).
We used bivariate LDSC to estimate the genetic correlation (rg) between endometriosis and PCOS. Under the multigene model, the slope of the z-score regression from the LD scores was used to estimate rg:
where and are the z-scores of SNP j from trait 1 and trait 2, and N2 are the sample sizes of trait 1 and trait 2, ρg is the genetic covariance, lj is the LD score, M is the number of SNPs, Ns is the number of overlapping samples, ρ is the phenotypic correlation in the overlapping samples, and and are the heritability of SNPs in trait 1 and trait 2 (37). To assess possible sample overlap between the traits in the pooled GWAS data, we performed LDSC correlation analyses of unconstrained genetic covariate intercepts. LDSC with constrained intercepts was likewise performed as a sensitivity analysis.
2.2.2 Identification of pleiotropic loci
Pleiotropic analysis under composite null hypothesis (PLACO) has been employed to investigate pleiotropic loci associated with complex traits using GWAS association statistics (38). During the process, the squared Z score was calculated for each variant, and SNPs with very high Z2 (> 80) values were removed. In addition, given the potential correlation between the two diseases, the correlation matrix of Z was estimated, and its matrix was included in the analysis. Finally, the hypothesis of no pleiotropy was tested using the level-α cross-over-unit test (IUT) method, and the final pleiotropy p-value was determined. Significant pleiotropic variants were defined as single-nucleotide variants with P-values less than 5 × 10−8. The Functional Mapping and Annotation of Genetic Associations (FUMA) tool was used to delineate potential pleiotropic loci. In the FUMA, SNPs can be annotated based on their physical location to genes, we used FUMA SNP2GENE workflow to define the genomic risk loci and annotate the candidate SNPs. These SNPs were then mapped to genes using ANNOVAR software.
2.2.3 Functional analysis for pleiotropic genes
To gain biological insights for the pleiotropic SNPs, based on PLACO results, we performed multi-marker analysis of genomic annotation (MAGMA) analysis (39) on the genes located in or overlapping with the pleiotropic loci to identify candidate pleiotropic genes. We used the MAGMA analysis in the FUMA platform (40).
The Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology-Based Annotation System (version 3.0) (41) was used to conduct pathway enrichment analysis and Gene Ontology (GO) was used to better understand the biological mechanisms associated with endometriosis and PCOS.
2.2.4 Mendelian randomization analysis
We applied two-sample bidirectional Mendelian randomization analysis to identify potential causal relationships between endometriosis and PCOS (42). In total, we employed five methods. The main method applied in this study is the inverse variance weighting (IVW). The method pooled the Wald ratio estimates for each SNP, which were obtained by dividing the SNP outcome estimate by the SNP exposure estimate, using the random effects inverse variance method, which weights each ratio according to its standard error and obtains the average chance effect estimate between the two features (43). To increase the reliability and validity of our results, we performed additional analyses using the MR-Egger method, the weighted median method, simple mode, and the weighted mode. MR-Egger regression can be used to detect and correct bias due to horizontal pleiotropy (44). These four methods are not as effective as IVW in detecting true causal effects and are therefore used to complement the findings of IVW.
Valid instrumental variables (IVs) were selected based on three main assumptions: that IVs (1) are associated with exposure; (2) do not depend on confounders; and (3) do not have a direct effect on the outcome (44). Utilizing PLINK, the MR approaches identified independent instrumental SNPs by LD clumping (LD r2< 0.001, within 1000-kb windows). For the MR analysis in this study, we selected SNPs with genome-wide significance (P ≤ 5 × 10−8) or SNPs with locus-wide significance level (1 × 10−6) of the exposure trait as IVs, based on the results of heterogeneity analysis (45). F statistics for each instrument were estimated by F = β2/SE2 (46). As sensitivity analysis, the heterogeneity Q statistics were calculated. An MR pleiotropy residual sum and outlier (MR-PRESSO) (47) test were performed to detect and correct for horizontal pleiotropy, detecting pleiotropic outliers, and providing outlier-corrected estimates. We then conducted five MR analyses to estimate the causal effect after excluding outlier SNPs.
2.2.5 LDSC-SEG analysis
We performed linkage disequilibrium score for the specific expression of genes analysis (LDSC-SEG) to investigate whether SNP heritability for endometriosis and PCOS was significant for trait tissue correlation inference (48). 53 tissue types are available for GTEx (v.8), and 152 tissues types are available for Franke lab. To further understand the (dis)similarity across traits, we partitioned heritability using stratified LDSC-SEG leveraging genome-wide genetic variants of endometriosis and PCOS. LDSC-SEG was conducted to evaluate the degree of SNP-heritability enrichment of endometriosis and PCOS in specific tissues. And P value< 0.05 indicated suggestive significance.
2.2.6 Validation of expressions of the risk gene
The expression profiles of risk genes, which are genes shared SNPs identified by PLACO, were evaluated for the risk gene by the datasets GSE7305 and GSE226146. These two raw datasets were analyzed by GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/), which is an online tool employed to compare two or more samples in different datasets. P-value<0.05 and |logFC| >0.5 were defined as differentially expressed genes (DEGs). In GSE7305, we compared DEGs between ectopic endometrium of endometriosis patients and endometrium of healthy controls. In GSE226146, we compared DEGs between endometrium of PCOS patients and endometrium of healthy controls.
3 Results
3.1 Global genetic correlation between endometriosis and PCOS
We first applied stratified linkage disequilibrium (LD) score regression (S-LDSC) with the baseline-LD model to estimate the SNP-based heritability of endometriosis and PCOS. The observed-scale SNP heritability (unconstrained intercept) was 1.58% for endometriosis, and 2.97% for PCOS. We then used the bivariate LDSC to estimate the genetic correlation between endometriosis and PCOS, genetic correlation (without constrained intercept) between endometriosis and PCOS were identified (rg = 0.56, P = 1.81×10-21). The calculated values of the intercept of the genetic covariance indicated slight sample overlap (Supplementary Table S1). We used a constrained intercept for LDSC without assuming an overall stratification with slightly higher genetic correlations, and the results were still significant (Supplementary Table S1).
3.2 Identification of shared risk SNPs between endometriosis and PCOS
Based on evidence for significant genetic correlations between endometriosis and PCOS, we performed PLACO to identify risk SNPs underlying the joint phenotypes endometriosis-PCOS. We identified a total of 3,805 single nucleotide variants (SNVs) that exhibited potential polymorphic variants associated with both traits by PLACO (Supplementary Table S2). MAGMA analysis yielded 40 significant pleiotropic genes (Supplementary Tables S3, S4). FUMA further identified 12 independent genomic risk loci that exhibited pleiotropic effects (Supplementary Table S5).
3.3 Enrichment analysis for identified pleiotropic genes
GO enrichment analysis indicated that the 40 pleiotropic genes were significantly enriched in filopodium assembly (P = 1.64 × 10-4), focal adhesion (P = 8.06 × 10-4), and GTPase activity (P = 4.11 × 10-4). The results of the KEGG enrichment analysis demonstrated significant enrichment of genes involved in N-Glycan biosynthesis (P = 4.65×10-3), endocrine and other factor-regulated calcium reabsorption (P = 5.21×10-3), and VEGF signaling pathway (P = 6.42×10-3). Figure 2 and Supplementary Table S6 present the GO and KEGG pathways with the top 10 minimum P-values.
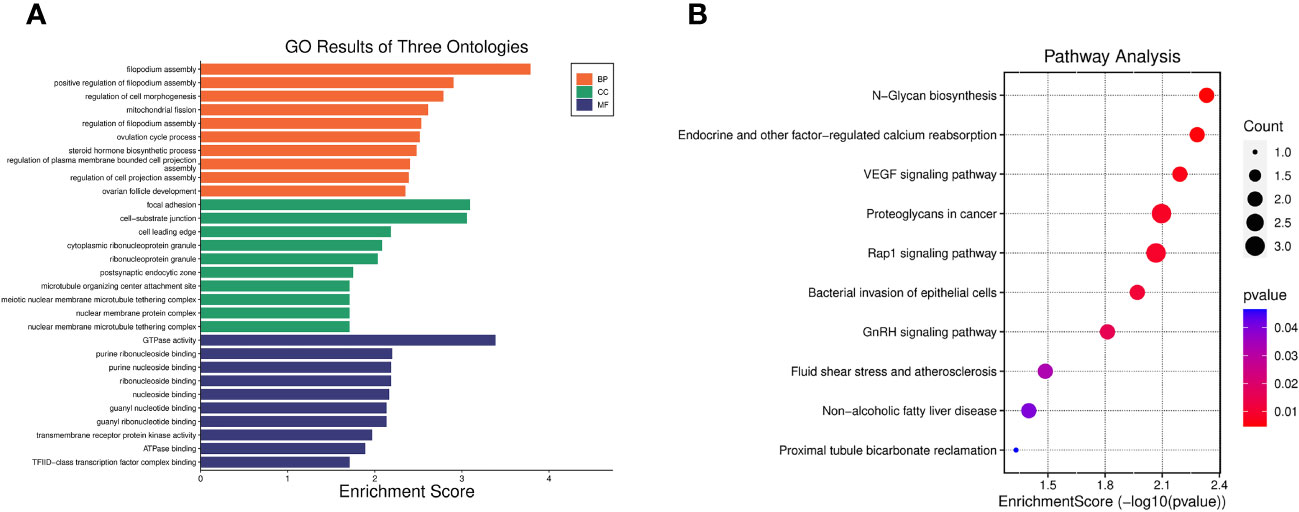
Figure 2 Enrichment analysis of 40 pleiotropic genes. (A) Top 10 significant GO results of three ontologies. BP, biological process; CC, cellular component; MF, molecular function. (B) Top 10 significant pathways in terms of the KEGG pathway enrichment analyses.
3.4 Mendelian randomization
We performed a two-sample bidirectional mendelian randomization analysis to explore whether there is a potential causal relationship between endometriosis and PCOS. The valid IVs were evaluated and screened based on previously mentioned hypothesized criteria. And all SNPs used in the MR analysis were strong instruments (F-statistics >10), indicating a low probability of weak instrumental bias. Various bidirectional mendelian randomization analysis methods were conducted for a more stable results. We first selected SNPs with genome-wide significance (P ≤ 5 × 10−8) as instrumental variables (IVs). Using IVW, genetic liability to endometriosis (β= 0.18, P = 3.09×10-6) was significantly associated with an increased risk of PCOS, with no significant evidence of heterogeneity (Supplementary Table S7). The estimates remained directionally consistent using weighted median approach (Figure 3; Supplementary Table S7). MR-PRESSO analysis revealed no outliers in the results. In the reverse MR analysis, using IVW, genetic liability to PCOS (β= 1.16, P = 3.71×10-3) was significantly associated with an increased risk of endometriosis. However, heterogeneity test showed that Q_pval is less than 0.05 (Supplementary Table S7). Therefore, we selected SNPs with locus-wide significance level (1 × 10−6) as IVs. Using IVW, genetic liability to PCOS (β=0.44, P = 1.52×10-3) was significantly associated with an increased risk of endometriosis, with no significant evidence of heterogeneity (Supplementary Table S7). The estimates remained directionally consistent using weighted median and weighted model (Supplementary Table S7). MR-PRESSO analysis revealed no outliers in the results. Reciprocal causation indicated shared pathogenesis between endometriosis and PCOS.
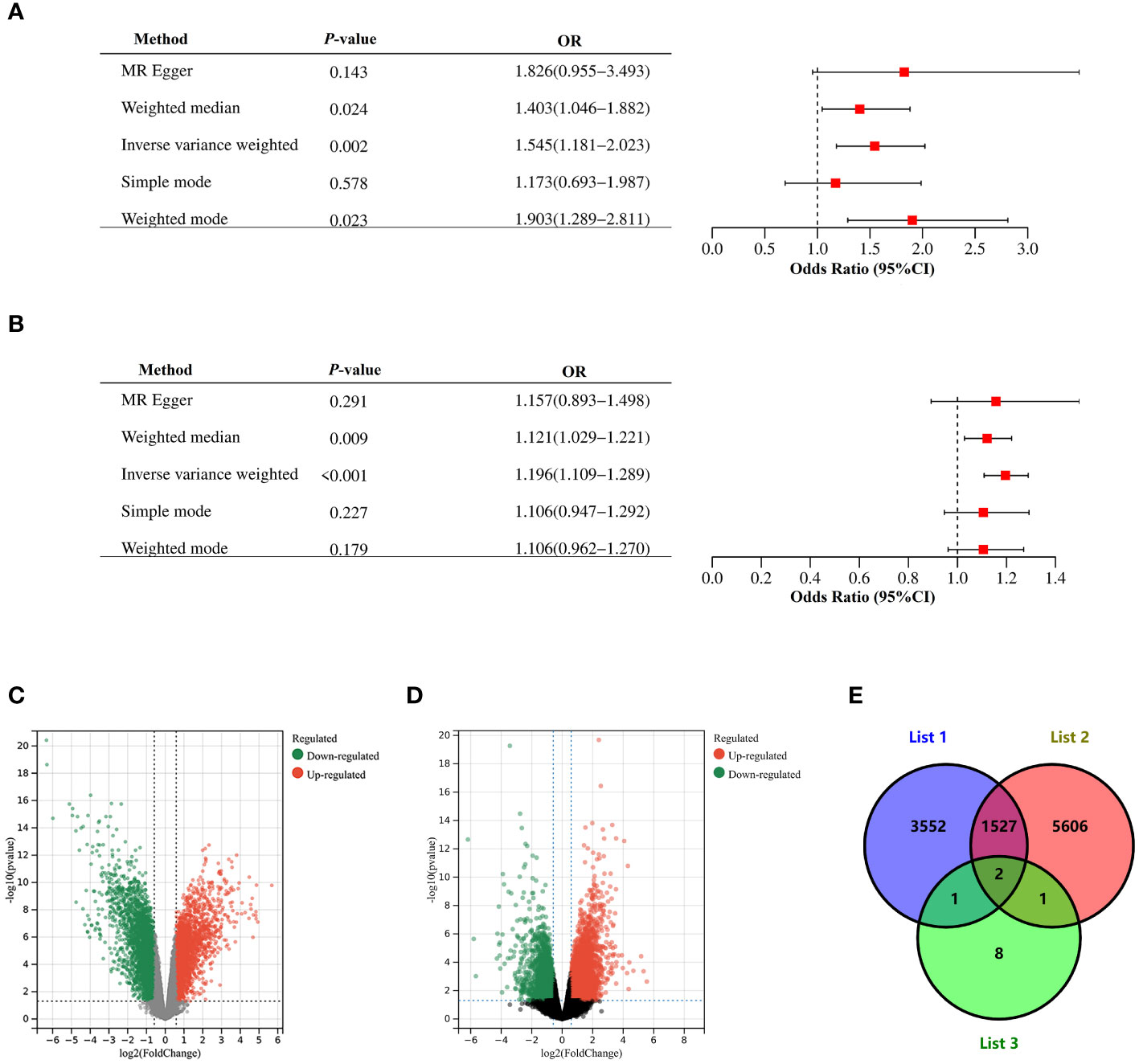
Figure 3 Mendelian randomization and Validation of expressions of the risk gene. (A) Causal effect of PCOS on endometriosis. (B) Causal effect of endometriosis on PCOS. Estimates and 95% CI were represented with square points and error bars, respectively. In the x-axis, the odds ratio (OR) is the effect of one standard deviation increase in exposure on outcome, at OR = 1, the vertical dashed line represents the reference. (C) Volcano plots of DEGs between endometrium of endometriosis patients and endometrium of healthy controls in GSE7305. (D) Volcano plots of DEGs between endometrium of PCOS patients and endometrium of healthy controls in GSE226146. Red dots represent significantly up-regulated DEGs, green dots represent significantly down-regulated DEGs, gray dots indicate no significant difference. Genes satisfying the criteria adjust P< 0.05 and |logFC| >0.5 were considered significant. Panel (E) is the Venn diagram showing common DEGs among List 1, List 2 and List 3. List 1 indicated DEGs in GSE7305, List 2 indicated DEGs in GSE226146, and List 3 indicated 12 pleiotropic genes.
3.5 Tissue-level SNP heritability enrichment
We used LDSC-SEG to assess tissue-level enrichment of SNP heritability for endometriosis and PCOS using genotypic tissue expression (GTEx) and Franke lab data. We identified FDR significant (P< 0.05) SNP heritability enrichment for endometriosis across ten tissues (Supplementary Table S8). For PCOS, six tissue were significantly enriched (P< 0.05) (Supplementary Table S9). These results indicated shared genetic basis for endometriosis and PCOS may mainly enriched in uterus, endometrium and fallopian tubes. Therefore, in following validation of expressions of the risk genes, we selected expression profile analysis of endometrium in endometriosis and PCOS.
3.6 Validation of expressions of the risk genes
In dataset GSE7305, which contained ectopic endometrium of endometriosis patients and endometrium of healthy controls, 5,082 DEGs were sifted out in total (Figure 3C). In dataset GSE226146, which contained endometrium of PCOS patients and endometrium of healthy controls, 7,136 DEGs were sifted out in total (Figure 3D). In addition, 12 independent genomic risk loci were identified through PLACO and FUMA. Only two genes, including SYNE1 and DNM3, existed in these three datasets (Figure 3E). The expression of SYNE1 and DNM3 were significantly decreased in endometrium of both endometriosis and PCOS compared to control subjects. These results indicated SYNE1 and DNM3 are shared genes in endometriosis and PCOS, promoting changes of endometrium in both endometriosis and PCOS.
4 Discussion
By leveraging large GWAS datasets and tissue specific expression data, our study provided evidence of causality and shared genetic architecture between endometriosis and PCOS.
We found a significant positive genetic correlation between endometriosis and PCOS, which supported the hypothesis that genetic factors play an important role in the comorbidity of these two diseases.
Based on the PLACO, 40 significant pleiotropic genes were identified. GO and KEGG enrichment analysis for these 40 pleiotropic genes indicated biological mechanisms associated with endometriosis and PCOS, and results showed that there are lots of similar pathways involved in endometriosis and PCOS, which may explain the similarities between these two diseases. For example, the receptivity of the endometrium to embryos in both endometriosis and PCOS decreases, and “Regulation of cell morphogenesis” (49–51), “focal adhesion” , “regulation of cell projection assembly” (52) are associated with the development of endometrial receptivity for blastocyst implantation. In addition, in endometriosis and PCOS, estrogen activity is increased and accompanied by progesterone resistance, and “steroid hormone biosynthetic process” (53) are associated with hormone imbalance. There are also differences between these two diseases. For example, “mitochondrial fission” (53) and “GTPase activity” (54, 55) plays opposite roles in two diseases, which may explain the differences between these two diseases, such as the speed of follicle recruitment and degeneration.
In addition to the IVW methods, different statistical methods for Mendelian randomization analysis were used to evaluate the robustness of our results. Since several methods for the causal estimates of endometriosis on PCOS, or estimates of PCOS on endometriosis, were statistically significant, the effect was considered robust in this study. Reciprocal causation indicated shared pathogenesis between endometriosis and PCOS. Our result is also consistent with studies proposed by Yang et al, which indicated that pathogenesis of both endometriosis and PCOS included oxidative stress, inflammation, immune dysregulation (38). Inflammation, resulting from immune dysregulation, is one of the main mechanisms that triggers cell metastasis and invasion, which are critical for development of endometriosis (56). Oxidative stress markers are elevated in endometriosis in comparison to control groups (57). High levels of oxidative stress promote inflammation, angiogenesis and cell proliferation, which may promote the development of endometriosis (57). Oxidative stress and inflammatory markers are significantly correlated with increasing androgen levels by upgrading the activity of enzymes involved in steroidogenesis, which may explain their role in PCOS (58–60).
Functional enrichment for gene expression in multiple tissues and cells was also investigated using the GTEx datasets and Franke lab. We identified ten tissues, with significant SNP heritability enrichment for the endometriosis trait. The enrichment results of the PCOS trait were mainly reflected in six tissues. Additionally, we identified heritability enrichment for endometriosis and PCOS in the uterus, endometrium and fallopian tubes. Consistent with studies proposed by Palomba et al. and Liu et al., there are lots of similarities in endometrium between endometriosis and PCOS (61, 62). For example, compared with healthy controls, pro-inflammatory pathways were enhanced, estrogen receptors were upregulated while progesterone receptors were downregulated, markers of epithelial cells were decreased, in endometrium of both endometriosis and PCOS. However, the similarity between endometriosis and PCOS in the fallopian tubes is currently unclear and needs further research.
Based on the PLACO and FUMA, 12 independent genomic risk loci for endometriosis and PCOS were also identified. In addition, microarray dataset and RNA sequencing dataset indicated the mRNA expressions of SYNE1 and DNM3 were downregulated in endometrium of patients with endometriosis or PCOS compared with control groups. SYNE1 encodes nesprin-1, a nuclear envelope protein, which is critical for binding between cytoskeleton, nuclear envelope and other subcellular compartments (63, 64). Nesprin-1 deficiency leads to abnormal nuclear morphology, cell mobility, and cytoskeleton organization (63, 65). In muscle, SYNE1 is involved in anchoring specialized myonuclei underneath the neuromuscular junctions, and SYNE1 mutations may cause ataxia (66). SYNE1 is also downregulated in numerous malignancies, and had notable alterations in 10% of gynecologic malignancies (67). One particular mechanism of SYNE1 in malignancies is based on the fact that nuclei are the stiffest organelles in most cells (68). Since nesprin proteins regulate nuclear morphology and cell mobility, SYNE1 downregulation in cancer may increase nuclear malleability so that cells can migrate through narrow tissue spaces to invade neighboring tissues. In endometriosis, based on GWAS, meta-analysis, and whole-exome sequencing (WES) analysis on a cohort of 80 endometriosis patients, SYNE1 is identified as a candidate gene in endometriosis (69). The relationship between SYNE1 and PCOS, and the mechanisms of SYNE1 in endometrium of endometriosis or PCOS is currently unclear. Endometriosis is a benign disease but exhibit many malignant features, including invasive, progressive and estrogen-dependent growth (70). PCOS patients may exhibit endometrial hyperplasia and an increased risk of endometrial cancer (71). Therefore, we speculate that downregulation of SYNE1 may promote migration and invasion of endometrial cells in endometriosis or PCOS by regulation nuclear malleability and morphology. In addition, Sur et al. proposed SYNE1 is involved in sex steroid hormone pathways (26). Endometriosis is a estrogen-dependent and progesterone resistance disease, and the endometrium of women with PCOS often exhibits elevated estrogen activity and progesterone resistance (18). Downregulation of SYNE1 may also promote development of endometriosis or PCOS by regulating sex steroid hormone, such as estrogen. DNM3 is a tumor suppressor factor. For example, in hepatocellular carcinoma, the downregulation of DNM3 promoted cell proliferation by increasing cell cycle-associated proteins, including cyclin D1, and the upregulation of DNM3 induced cell apoptosis and inhibited tumor growth (72). The relationship between DNM3 and endometriosis or PCOS is currently unknown. Endometriosis exhibit many malignant features, such as invasive and progressive. PCOS patients may exhibit endometrial hyperplasia. Therefore, we suppose that DNM3 may also promote endometrial cells proliferation and invasion through modulating cell cycle-associated or apoptosis-associated proteins. However, additional research is needed to confirm these possible associations and to elucidate the mechanisms involved.
Our research also has several limitations. First, the vast majority of participants in our study were of European ancestry and not extrapolating to other ancestries. Second, we included only data from autosomes in our study (except MR analyses), because the analysis software does not apply to sex chromosomes. Third, we assessed tissue-specific heritability enrichment based on the top 10% of most specific genes overlooking the effects of other genes. Fourth, our inferred causality is presumptive because it is generated based on GWAS summary statistics. Larger and more robust endometriosis and PCOS GWAS are needed to clarify potential causal relationships. Last, some studies proposed that endometrium and PCOS are diametric disorders, which seems contradictory to our research. We speculated that this may be due to the fact that the two diseases are partially positively correlated, and partially negatively correlated. Future longitudinal studies and experimental work are also necessary to investigate the biological mechanisms behind the observed genetic relationships.
In conclusion, we found significant genetic correlations between endometriosis and PCOS and identified common risk SNPs. The causal relationship between endometriosis and PCOS is reciprocal. SYNE1 and DNM3 were potential shared genes between endometriosis and PCOS. These findings may provide insights into the common genetic architecture between endometriosis and PCOS, and contribute to a better understanding of their pathogenesis as well as future therapeutic strategies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HT: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. PL: Writing – review & editing, Writing – original draft, Validation, Supervision, Software, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. HX: Writing – review & editing, Writing – original draft, Software, Project administration, Investigation, Data curation, Conceptualization.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We appreciate the researchers who published the GWAS summary data and the participants who participated in these investigations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2024.1359236/full#supplementary-material
References
1. Giudice LC. Clinical practice. Endometriosis. N Engl J Med. (2010) 362:2389–98. doi: 10.1056/NEJMcp1000274
2. Clement PB. The pathology of endometriosis: a survey of the many faces of a common disease emphasizing diagnostic pitfalls and unusual and newly appreciated aspects. Adv Anat Pathol. (2007) 14:241–60. doi: 10.1097/PAP.0b013e3180ca7d7b
3. Wang D, Luo Y, Wang G, Yang Q. CircATRNL1 promotes epithelial-mesenchymal transition in endometriosis by upregulating Yes-associated protein 1 in vitro. Cell Death Dis. (2020) 11:594. doi: 10.1038/s41419-020-02784-4
4. Simoens S, Dunselman G, Dirksen C, Hummelshoj L, Bokor A, Brandes I, et al. The burden of endometriosis: costs and quality of life of women with endometriosis and treated in referral centres. Hum Reprod. (2012) 27:1292–9. doi: 10.1093/humrep/des073
5. Liu D, Gao X, Pan XF, Zhou T, Zhu C, Li F, et al. The hepato-ovarian axis: genetic evidence for a causal association between non-alcoholic fatty liver disease and polycystic ovary syndrome. BMC Med. (2023) 21:62. doi: 10.1186/s12916-023-02775-0
6. Rothenberg SS, Beverley R, Barnard E, Baradaran-Shoraka M, Sanfilippo JS. Polycystic ovary syndrome in adolescents. Best Pract Res Clin Obstet Gynaecol. (2018) 48:103–14. doi: 10.1016/j.bpobgyn.2017.08.008
7. Witchel SF, Oberfield SE, Pena AS. Polycystic ovary syndrome: pathophysiology, presentation, and treatment with emphasis on adolescent girls. J Endocr Soc. (2019) 3:1545–73. doi: 10.1210/js.2019-00078
8. Kujanpaa L, Arffman RK, Pesonen P, Korhonen E, Karjula S, Järvelin MR, et al. Women with polycystic ovary syndrome are burdened with multimorbidity and medication use independent of body mass index at late fertile age: A population-based cohort study. Acta Obstet Gynecol Scand. (2022) 101:728–36. doi: 10.1111/aogs.14382
9. Singh KB, Patel YC, Wortsman J. Coexistence of polycystic ovary syndrome and pelvic endometriosis. Obstet Gynecol. (1989) 74:650–2.
10. Lessey BA. Assessment of endometrial receptivity. Fertil Steril. (2011) 96:522–9. doi: 10.1016/j.fertnstert.2011.07.1095
11. Dinsdale NL, Crespi BJ. Endometriosis and polycystic ovary syndrome are diametric disorders. Evol Appl. (2021) 14:1693–715. doi: 10.1111/eva.13244
12. Margarit L, Taylor A, Roberts MH, Hopkins L, Davies C, Brenton AG, et al. MUC1 as a discriminator between endometrium from fertile and infertile patients with PCOS and endometriosis. J Clin Endocrinol Metab. (2010) 95:5320–9. doi: 10.1210/jc.2010-0603
13. Margarit L, Gonzalez D, Lewis PD, Hopkins L, Davies C, Conlan RS, et al. L-selectin ligands in human endometrium: comparison of fertile and infertile subjects. Hum Reprod. (2009) 24:2767–77. doi: 10.1093/humrep/dep247
14. Khatun M, Arffman RK, Lavogina D, Kangasniemi M, Laru J, Ahtikoski A, et al. Women with polycystic ovary syndrome present with altered endometrial expression of stanniocalcin-1dagger. Biol Reprod. (2020) 102:306–15. doi: 10.1093/biolre/ioz180
15. Schuler-Toprak S, Ortmann O, Buechler C, Treeck O. The complex roles of adipokines in polycystic ovary syndrome and endometriosis. Biomedicines. (2022) 10. doi: 10.3390/biomedicines10102503
16. Ullah A, Wang MJ, Wang YX, Shen B. CXC chemokines influence immune surveillance in immunological disorders: Polycystic ovary syndrome and endometriosis. Biochim Biophys Acta Mol Basis Dis. (2023) 1869:166704. doi: 10.1016/j.bbadis.2023.166704
17. Marquardt RM, Kim TH, Shin JH, Jeong JW. Progesterone and estrogen signaling in the endometrium: what goes wrong in endometriosis? Int J Mol Sci. (2019) 20. doi: 10.3390/ijms20153822
18. Savaris RF, Groll JM, Young SL, DeMayo FJ, Jeong JW, Hamilton AE, et al. Progesterone resistance in PCOS endometrium: a microarray analysis in clomiphene citrate-treated and artificial menstrual cycles. J Clin Endocrinol Metab. (2011) 96:1737–46. doi: 10.1210/jc.2010-2600
19. Qi X, Yun C, Pang Y, Qiao J. The impact of the gut microbiota on the reproductive and metabolic endocrine system. Gut Microbes. (2021) 13:1–21. doi: 10.1080/19490976.2021.1894070
20. Mehedintu C, Plotogea MN, Ionescu S, Antonovici M. Endometriosis still a challenge. J Med Life. (2014) 7:349–57.
21. Uno S, Zembutsu H, Hirasawa A, Takahashi A, Kubo M, Akahane T, et al. A genome-wide association study identifies genetic variants in the CDKN2BAS locus associated with endometriosis in Japanese. Nat Genet. (2010) 42:707–10. doi: 10.1038/ng.612
22. Albertsen HM, Chettier R, Farrington P, Ward K. Genome-wide association study link novel loci to endometriosis. PloS One. (2013) 8:e58257. doi: 10.1371/journal.pone.0058257
23. Painter JN, Anderson CA, Nyholt DR, Macgregor S, Lin J, Lee SH, et al. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. (2011) 43:51–4. doi: 10.1038/ng.731
24. Nyholt DR, Low SK, Anderson CA, Painter JN, Uno S, Morris AP, et al. Genome-wide association meta-analysis identifies new endometriosis risk loci. Nat Genet. (2012) 44:1355–9. doi: 10.1038/ng.2445
25. Steinthorsdottir V, Thorleifsson G, Aradottir K, Feenstra B, Sigurdsson A, Stefansdottir L, et al. Common variants upstream of KDR encoding VEGFR2 and in TTC39B associate with endometriosis. Nat Commun. (2016) 7:12350. doi: 10.1038/ncomms12350
26. Sapkota Y, Steinthorsdottir V, Morris AP, Fassbender A, Rahmioglu N, De Vivo I, et al. Meta-analysis identifies five novel loci associated with endometriosis highlighting key genes involved in hormone metabolism. Nat Commun. (2017) 8:15539. doi: 10.1038/ncomms15539
27. Stener-Victorin E, Deng Q. Epigenetic inheritance of polycystic ovary syndrome - challenges and opportunities for treatment. Nat Rev Endocrinol. (2021) 17:521–33. doi: 10.1038/s41574-021-00517-x
28. Chaudhary H, Patel J, Jain NK, Joshi R. The role of polymorphism in various potential genes on polycystic ovary syndrome susceptibility and pathogenesis. J Ovarian Res. (2021) 14:125. doi: 10.1186/s13048-021-00879-w
29. Watson HJ, Yilmaz Z, Thornton LM, Hübel C, Coleman JRI, Gaspar HA, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. (2012) 44:1020–5. doi: 10.1038/ng.2384
30. Rahmioglu N, Mortlock S, Ghiasi M, Møller PL, Stefansdottir L, Galarneau G, et al. The genetic basis of endometriosis and comorbidity with other pain and inflammatory conditions. Nat Genet. (2023) 55:423–36. doi: 10.1038/s41588-023-01323-z
31. Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature. (2017) 550:204–13. doi: 10.1038/nature24277
32. Finucane HK, Reshef YA, Anttila V, Slowikowski K, Gusev A, Byrnes A, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat Genet. (2018) 50:621–9. doi: 10.1038/s41588-018-0081-4
33. Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. (2015) 6:5890. doi: 10.1038/ncomms6890
34. Hever A, Roth RB, Hevezi P, Marin ME, Acosta JA, Acosta H, et al. Human endometriosis is associated with plasma cells and overexpression of B lymphocyte stimulator. Proc Natl Acad Sci U S A. (2007) 104:12451–6. doi: 10.1073/pnas.0703451104
35. Xu X, Yang A, Tian P, Zhang K, Liu Y, Wang Y, et al. Expression profile analysis of LncRNAs and mRNAs in pre-receptive endometrium of women with polycystic ovary syndrome undergoing in vitro fertilization-embryo transfer. BMC Med Genomics. (2024) 17:26. doi: 10.1186/s12920-024-01806-w
36. Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. (2015) 47:291–5. doi: 10.1038/ng.3211
37. Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. (2015) 47:1236–41. doi: 10.1038/ng.3406
38. Yang HL, Zhou WJ, Gu CJ, Meng YH, Shao J, Li DJ, et al. Pleiotropic roles of melatonin in endometriosis, recurrent spontaneous abortion, and polycystic ovary syndrome. Am J Reprod Immunol. (2018) 80:e12839. doi: 10.1111/aji.12839
39. de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PloS Comput Biol. (2015) 11:e1004219. doi: 10.1371/journal.pcbi.1004219
40. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. (2017) 8:1826. doi: 10.1038/s41467-017-01261-5
41. Bu D, Luo H, Huo P, Wang Z, Zhang S, He Z, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. (2021) 49:W317–25. doi: 10.1093/nar/gkab447
42. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. (2017) 46:1734–9. doi: 10.1093/ije/dyx034
43. Bull CJ, Bell JA, Murphy N, Sanderson E, Davey SG, Timpson NJ, et al. Adiposity, metabolites, and colorectal cancer risk: Mendelian randomization study. BMC Med. (2020) 18:396. doi: 10.1186/s12916-020-01855-9
44. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
45. Long Y, Tang L, Zhou Y, Zhao S, Zhu H. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med. (2023) 21:66. doi: 10.1186/s12916-023-02761-6
46. Chen J, Zhang X. D-MANOVA: fast distance-based multivariate analysis of variance for large-scale microbiome association studies. Bioinformatics. (2021) 38:286–8. doi: 10.1093/bioinformatics/btab498
47. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. (2018) 50:693–8. doi: 10.1038/s41588-018-0099-7
48. Bryois J, Skene NG, Hansen TF, Kogelman L, Watson HJ, Liu Z, et al. Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson’s disease. Nat Genet. (2020) 52:482–93. doi: 10.1038/s41588-020-0610-9
49. Liang J, Li K, Chen K, Liang J, Qin T, He J, et al. Regulation of ARHGAP19 in the endometrial epithelium: a possible role in the establishment of uterine receptivity. Reprod Biol Endocrinol. (2021) 19:2. doi: 10.1186/s12958-020-00689-7
50. Murphy CR. Uterine receptivity and the plasma membrane transformation. Cell Res. (2004) 14:259–67. doi: 10.1038/sj.cr.7290227
51. Tu Z, Wang Q, Cui T, Wang J, Ran H, Bao H, et al. Uterine RAC1 via Pak1-ERM signaling directs normal luminal epithelial integrity conducive to on-time embryo implantation in mice. Cell Death Differ. (2016) 23:169–81. doi: 10.1038/cdd.2015.98
52. Nikas G. Cell-surface morphological events relevant to human implantation. Hum Reprod. (1999) 14 Suppl 2:37–44. doi: 10.1093/humrep/14.suppl_2.37
53. Yilmaz BD, Bulun SE. Endometriosis and nuclear receptors. Hum Reprod Update. (2019) 25:473–85. doi: 10.1093/humupd/dmz005
54. Yarahmadi G, Dehghanian M, Sandoghsaz RS, Savaee M, Shamsi F, Vahidi MM. Evaluation of NF1 and RASA1 gene expression in endometriosis. Eur J Obstet Gynecol Reprod Biol X. (2022) 15:100152. doi: 10.1016/j.eurox.2022.100152
55. Xu A, Fan Y, Liu S, Sheng L, Sun Y, Yang H. GIMAP7 induces oxidative stress and apoptosis of ovarian granulosa cells in polycystic ovary syndrome by inhibiting sonic hedgehog signalling pathway. J Ovarian Res. (2022) 15:141. doi: 10.1186/s13048-022-01092-z
56. Cacciottola L, Donnez J, Dolmans MM. Can endometriosis-related oxidative stress pave the way for new treatment targets? Int J Mol Sci. (2021) 22. doi: 10.3390/ijms22137138
57. Ito F, Yamada Y, Shigemitsu A, Akinishi M, Kaniwa H, Miyake R, et al. Role of oxidative stress in epigenetic modification in endometriosis. Reprod Sci. (2017) 24:1493–502. doi: 10.1177/1933719117704909
58. Gonzalez F, Rote NS, Minium J, Kirwan JP. Reactive oxygen species-induced oxidative stress in the development of insulin resistance and hyperandrogenism in polycystic ovary syndrome. J Clin Endocrinol Metab. (2006) 91:336–40. doi: 10.1210/jc.2005-1696
59. Yilmaz M, Bukan N, Ayvaz G, Karakoc A, Toruner F, Cakir N, et al. The effects of rosiglitazone and metformin on oxidative stress and homocysteine levels in lean patients with polycystic ovary syndrome. Hum Reprod. (2005) 20:3333–40. doi: 10.1093/humrep/dei258
60. Siddiqui S, Mateen S, Ahmad R, Moin S. A brief insight into the etiology, genetics, and immunology of polycystic ovarian syndrome (PCOS). J Assist Reprod Genet. (2022) 39:2439–73. doi: 10.1007/s10815-022-02625-7
61. Palomba S, Piltonen TT, Giudice LC. Endometrial function in women with polycystic ovary syndrome: a comprehensive review. Hum Reprod Update. (2021) 27:584–618. doi: 10.1093/humupd/dmaa051
62. Liu H, Lang JH. Is abnormal eutopic endometrium the cause of endometriosis? The role of eutopic endometrium in pathogenesis of endometriosis. Med Sci Monit. (2011) 17:RA92–9. doi: 10.12659/msm.881707
63. Sur I, Neumann S, Noegel AA. Nesprin-1 role in DNA damage response. Nucleus-Phila. (2014) 5:173–91. doi: 10.4161/nucl.29023
64. Indelicato E, Nachbauer W, Fauth C, Krabichler B, Schossig A, Eigentler A, et al. SYNE1-ataxia: Novel genotypic and phenotypic findings. Parkinsonism Relat Disord. (2019) 62:210–4. doi: 10.1016/j.parkreldis.2018.12.007
65. Cartwright S, Karakesisoglou I. Nesprins in health and disease. Semin Cell Dev Biol. (2014) 29:169–79. doi: 10.1016/j.semcdb.2013.12.010
66. Noreau A, Bourassa CV, Szuto A, Levert A, Dobrzeniecka S, Gauthier J, et al. SYNE1 mutations in autosomal recessive cerebellar ataxia. JAMA Neurol. (2013) 70:1296–31. doi: 10.1001/jamaneurol.2013.3268
67. Harbin LM, Lin N, Ueland FR, Kolesar JM. SYNE1 mutation is associated with increased tumor mutation burden and immune cell infiltration in ovarian cancer. Int J Mol Sci. (2023) 24. doi: 10.3390/ijms241814212
68. Zwerger M, Ho CY, Lammerding J. Nuclear mechanics in disease. Annu Rev BioMed Eng. (2011) 13:397–428. doi: 10.1146/annurev-bioeng-071910-124736
69. Santin A, Spedicati B, Morgan A, Lenarduzzi S, Tesolin P, Nardone GG, et al. Puzzling out the genetic architecture of endometriosis: whole-exome sequencing and novel candidate gene identification in a deeply clinically characterised cohort. Biomedicines. (2023) 11. doi: 10.3390/biomedicines11082122
70. Flores I, Rivera E, Ruiz LA, Santiago OI, Vernon MW, Appleyard CB. Molecular profiling of experimental endometriosis identified gene expression patterns in common with human disease. Fertil Steril. (2007) 87:1180–99. doi: 10.1016/j.fertnstert.2006.07.1550
71. Hu M, Zhang Y, Li X, Cui P, Sferruzzi-Perri AN, Brannstrom M, et al. TLR4-associated IRF-7 and NFkappaB signaling act as a molecular link between androgen and metformin activities and cytokine synthesis in the PCOS endometrium. J Clin Endocrinol Metab. (2021) 106:1022–40. doi: 10.1210/clinem/dgaa951
Keywords: endometriosis, polycystic ovary syndrome, genome-wide association study, shared genetic architecture, genetics
Citation: Tan H, Long P and Xiao H (2024) Dissecting the shared genetic architecture between endometriosis and polycystic ovary syndrome. Front. Endocrinol. 15:1359236. doi: 10.3389/fendo.2024.1359236
Received: 21 December 2023; Accepted: 15 April 2024;
Published: 29 April 2024.
Edited by:
Hannu Kullervo Martikainen, University of Oulu, FinlandReviewed by:
Sally Mortlock, The University of Queensland, AustraliaEdgar Ricardo Vázquez-Martínez, National Autonomous University of Mexico, Mexico
Copyright © 2024 Tan, Long and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongmei Xiao, aG14aWFvQGNzdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Hangjing Tan1†
Hangjing Tan1† Panpan Long
Panpan Long Hongmei Xiao
Hongmei Xiao