- 1Centre for Bioinnovation, University of the Sunshine Coast, Maroochydore, QLD, Australia
- 2School of Science, Technology and Engineering, University of the Sunshine Coast, Maroochydore, QLD, Australia
- 3Centre for AgriBioscience, Agriculture Victoria, Bundoora, VIC, Australia
- 4Institute for Marine and Antarctic Studies (IMAS), University of Tasmania, Hobart, TAS, Australia
- 5Department of Biology, Colorado State University, Fort Collins, CO, United States
- 6University of California-Davis Bodega Marine Laboratory, Bodega Bay, CA, United States
G protein-coupled receptors (GPCRs) are an ancient family of signal transducers that are both abundant and consequential in metazoan endocrinology. The evolutionary history and function of the GPCRs of the decapod superfamilies of gonadotropin-releasing hormone (GnRH) are yet to be fully elucidated. As part of which, the use of traditional phylogenetics and the recycling of a diminutive set of mis-annotated databases has proven insufficient. To address this, we have collated and revised eight existing and three novel GPCR repertoires for GnRH of decapod species. We developed a novel bioinformatic workflow that included clustering analysis to capture likely GnRH receptor-like proteins, followed by phylogenetic analysis of the seven transmembrane-spanning domains. A high degree of conservation of the sequences and topology of the domains and motifs allowed the identification of species-specific variation (up to ~70%, especially in the extracellular loops) that is thought to be influential to ligand-binding and function. Given the key functional role of the DRY motif across GPCRs, the classification of receptors based on the variation of this motif can be universally applied to resolve cryptic GPCR families, as was achieved in this work. Our results contribute to the resolution of the evolutionary history of invertebrate GnRH receptors and inform the design of bioassays in their deorphanization and functional annotation.
1 Introduction
The decapods are an order of crustaceans that constitute an important global source of dietary protein for humans and are being harvested in increasingly large numbers using aquaculture facilities (1). While finite in number, the members of the decapod order are hyper-diverse in biology (for example, freshwater and salt water crabs, shrimps, lobsters and crayfish) (2), which represents a real challenge in understanding decapod biology. Moreover, a better understanding of the endocrinology that controls the decapod growth and fecundity is a major goal of crustacean aquaculture biologists (3, 4). To date, neuropeptides of the decapod gonadotropin-releasing hormone (GnRH)-like superfamily have been implicated in the reproduction-related maturation of ovaries and proliferation of oocytes (5), raising the likelihood that the characterization of the GnRH-like receptors and their role in signal transduction promises the development of important future aquaculture technologies.
G protein-coupled receptors (GPCRs) are a family of ancient cell membrane spanning signal transducers that are found across the eukaryotic tree of life (including crustaceans and humans), making them the key target for medical drugs (6, 7). Despite their ancestral origin, GPCRs display remarkably conserved topological domains that are defined by their relationship to the hydrophobic phospholipid bi-layered cell membrane. Canonically, these include the extracellular N-terminus, intracellular C-terminus, seven transmembrane spans (7TMs), three extracellular loops (ECLs) and three intracellular loops (ICLs) (8). While the N-terminus and ECLs compose the extracellular ligand binding sites, the C-terminus and ICLs compose the intracellular binding site(s) of the titular associated G protein (9). Members of decapod GnRH-like receptors are GPCRs (10).
The identification of the GnRH decapeptide in the hypothalamo-pituitary regulation of reproduction was a major step in both basic and clinical reproductive endocrinology, winning Roger Guillemin and Andrew Schally the 1977 Nobel Prize in Physiology and/or Medicine. This decapeptide was found to date back 400 million of years in evolution, raising the question of what this peptide function was in the absence of a hypothalamus or pituitary {Kochman, 2012 #2894}.
The ancestral history of the receptors of the superfamily of invertebrate GnRH-like neuropeptides is unresolved (5), undergoing proposed nomenclature reassignment (11), and requires further evidence that contributes to its resolution. By interpolation, the same holds true for the crustacean counterparts. The functional annotation of orphan GPCRs is resource-intensive and not trivial. In fact, few neuropeptide receptors have been deorphanized in crustaceans (12–14), lagging behind functional neuropeptide GPCR deorphanization in insects (15). New databases which were recently developed including CrustyBase (16) and CrusTome (17) provide ample opportunity to bridge this gap by exploring expression patterns and phylogenetic analyses of genes across multiple pancrustacean species. Therefore, any measure that appreciably narrows the focus by reducing the relevant search space is of potential worth. To date, the bioinformatic annotation of crustacean GPCRs using Basic Local Alignment Search Tool (BLAST) databases and traditional phylogenetic analysis has proven insufficient in the description of GnRH-like GPCRs. Understanding any species-specific sequence variation and the congruence of conserved, function-related GPCR domains and motifs, serves to better define the phylogeny of GnRH-like receptors and contextualize their functional annotation studies.
Within the context of the conserved topological domains, GPCRs have been observed to possess several highly conserved and function-related aa motifs including ‘DRY’, ‘CWxP’ and ‘NPxxY’, which are located at the union of transmembrane domain 3 (TM3) and ICL2, within TM6, and with TM7, respectively. The DRY motif is thought to create a network of interactions (including ‘salt bridges’ or ‘ionic locks’) that keep the receptors in the inactive conformation, ultimately playing a crucial role in the activation of the receptor upon ligand binding (18). The binding of the G protein to the GPCR has also been observed to occur in a binding pocket that is topologically and functionally related to the DRY motif locus (19). Similarly, conserved CWxP and NPxxY motifs have been implicated in GPCR activation, and ligand and G protein binding (20).
In this study, we describe well-resolved evolutionary histories of decapod GnRH-like receptors. We annotate clades of receptors of GnRH (GnRHR), corazonin (CrzR), red pigment concentrating hormone (RPCHR), adipokinetic hormone/corazonin-related peptide (ACPR1 and ACPR2), within the context of the closely-related receptors of vasopressin (VR) and crustacean cardioactive peptide (CCAPR).
2 Methods
2.1 Standardization of GPCR-encoding transcripts
The putative GPCR-encoding transcripts of eleven decapod species (including crab, shrimp, crayfish and lobster from both freshwater and seawater) were compiled into a single catalogue using the contemporary methods described below. Previously published GPCRomes (i.e., all proteins identified as putative GPCRs of an organism) that have been incorporated into this study include: Procambarus clarkii (21), Sagmariasus verreauxi (10), Nephrops norvegicus (22), Carcinus maenas (23), Gecarcinus lateralis (24) and Penaeus monodon (25). Publicly available transcriptomic reads accessed from NCBI Transcriptome Sequence Assembly (TSA) database include: Homarus americanus, Cherax quadricarinatus and Eriochier sinensis. Novel GPCRomes were generated for Panulirus ornatus, C. quadricarinatus and E. sinensis and are presented as part of this study. A summary of the species, tissues, assembly technology, and origin of transcriptomic reads is summarized in Table 1.
2.2 Contemporary methods employed in the identification of the putative GPCR-encoding transcripts of Panulirus ornatus, Eriocheir sinensis and Cherax quadricarinatus
The sampling of tissue, RNA extraction, RNA quality assurance and RNA read sequencing for P. ornatus have been outlined previously (26–28). The resulting read sequences were subjected to the de novo assembly algorithm of CLC Genomics Workbench v9.5 (https://www.qiagenbioinformatics.com/) and cDNA was translated into the longest open reading frames (ORFs) using CLC. All ORFs were then screened against the Pfam database for predicted GPCR structural domains using the CLC platform with the Pfam module (v29). Search results displaying maximum E-values of 10-3 were extracted and iteratively used as a reference list. Additionally, a search was conducted on UniprotKB database using the terms “family: G protein coupled receptor 1 family and taxonomy: Arthropoda [6656]” for Rhodopsin receptors, and “family: G protein coupled receptor 2 family and taxonomy: Arthropoda [6656]” for Secretin receptors. Rhodopsin and Secretin reference BLAST databases were generated from these lists and a local BLASTp search was conducted to discriminate the class A (Rhodopsin-like) and class B (Secretin-like) decapod GPCRs. The N-terminus and C-terminus of all predicted GPCR-encoding transcripts were trimmed to also yield lists of 7TM domains (including ECLs and ICLs).
2.3 Novel bioinformatic workflow for the annotation of GPCRs
Predicted class A (Rhodopsin-like) decapod GPCR-encoding transcripts were then subjected to a bioinformatic workflow which was designed to assist in the classification and functional annotation of GPCRs by reducing the relevant search spaces. Key steps in the workflow included clustering analysis (incorporating deorphanized and predicted orthologues), and the determination of the topological GPCR domains and conserved motifs (that is DRY, CWxP, NPxxY). The relationships between the sequences of the function-related DRY motif locus and the phylogeny of the 7TM domains was then analyzed for predicted receptors of the GnRH-like superfamily of neuropeptides. The congruence and compartmentalization of species, conserved motifs and phylogenetic clades was then qualified collectively, allowing the annotation of receptors into clades.
Firstly, all available class A decapod GPCR ORFs were downloaded from the Interpro database using the search terms ‘Rhodopsin-like’ and ‘7TM’. A combined list of class A decapod GPCRs was compiled that incorporated the transcripts from Interpro, our eleven decapod species and selectively ‘seeded’ decapod GnRH-like receptor transcripts that have previously been functionally or bioinformatically annotated. Clustering analysis was then performed on the combined list using the CLANS2 algorithm (29) with a P-value of 1e-50 for 10,000 rounds in three dimensions. The resulting clusters were then collapsed to two dimensions for visualization. In brief, CLANS2 performs all-against-all BLAST searches in the three-dimensional space where clustering is performed using attractive forces proportional to the negative logarithm of the BLAST P-values, and a uniform repulsive force.
A list of the available functionally annotated receptors of the decapod superfamily of GnRH-like neuropeptides (that is, GnRH, corazonin, RPCH, ACP, CCAP and vasopressin) was compiled as seeds. The seeded transcripts included the functionally annotated CCAPR of Scylla paramamosain (Sp_CCAPR) (30), and corazonin receptor (Cm_CrzR: accession number MF974386) and RPCH receptor (Cm_RPCHR: accession number MF974387) of Cm (31), as well as the bioinformatically annotated vasopressin receptors (VRs) of Portunus pelagicus (Pp_VR: accession number MZ147830.1) and of Litopenaeus vannamei (Lv_VR: accession number ROT66533.1), and the ACP receptor of Macrobrachium rosenbergii (Mr_ACPR) (32). Transcripts that clustered with these seeds were extracted and considered putative GnRHR-like transcripts.
The candidate GnRHR-like transcripts were then subjected to the online machine-learning DeepTMHMM algorithm in the prediction of GPCR topological domains (including N-terminus, C-terminus, ECLs, ICLs, and 7TM) (33). Identification of these domains enabled the subsequent prediction of the highly conserved DRY, CWxP, and NPxxY motifs. The aa sequences of both the 7TM (including ECLs and ICLs) and the transcript ORFs of the GnRHR-like receptors were aligned using the MUSCLE algorithm (34) implemented in MEGA-X (35). Then neighbor joining phylogenetic trees were constructed (bootstrap values of 1,000) using Mega-X. The resulting phylogenetic clades that were constructed using both the 7TMs (including ECLs and ICLs) and the ORFs were compared and contrasted to assess the propriety of using either method in this study.
Snake plots were generated using Protter (wlab.ethz.ch/protter) (36) for the functionally annotated Cm_CrzR, Cm_RPCHR and Sp_CCAPR, and three transcripts from this catalogue (Gl_GPCR_A7, Po_GPCR_A146 and Cm_GPCR_A117) that respectively shared a high degree of sequence identity. Snake plots of the ACPR1 and ACPR2 clades were generated to compare the intra-clade and inter-clade similarities and differences, and especially the ECLs. All snake plots were augmented with the species, conserved GPCR motifs, and a color-coded annotation.
The aa sequences of the 7TM (including ECLs and ICLs) of the GnRHR-like transcripts were aligned and Neighbor Joining phylogenetic trees were constructed, as described above. The phylogenetic trees were also labeled with species and associated DRY motif sequence. The compartmentalization of labels and the strength of phylogenetic clades were analyzed and visually represented. In considering the two ACPR clades (grey and orange), that lacked a functionally annotated constituent, we identified the sequences of the conserved motifs of the two functionally annotated ACPR-like receptor variants, BNGR-A28 (accession number: NP_001127726.1) and BNGR-A29 (accession number: NP_001127745.1) of silkworm moth Bombyx mori (37). The sequences of the conserved DRY, CWxP, NPxxY motifs of all receptor transcripts were tabulated and sorted. The congruence and compartmentalization of species and conserved motifs was analyzed, and color-coded annotations were then ascribed.
A representative of each of the color-coded GnRHR-like clades was subjected to CrustyBase expression pattern analysis. That is, a tBLASTN search of the aa was conducted for the following four molt-related transcriptomes: multiple tissues, 11 stages of embryo development, 12 stages of larval development of the ornate spiny lobster (P. ornatus), and Y-organ across five molt stages of the blackback land crab (G. lateralis). The contigs with E-values less than approximately 1e-50 were all downloaded. The GPCR topological domains (predicted using DeepTMHMM) and conserved GPCR motifs (DRY, CWxP and NPxxY) were sequenced for these contigs to determine which clade they actually represented. This is, as opposed to just returning the lowest E-value in the tBLASTN search. The expression patterns of the legitimized contigs were then compiled for each of the transcriptome contexts and are presented in the results.
3 Results
3.1 Catalogue of GPCRome sequences
The class A and class B GPCR-encoding transcripts of P. clarkii, H. americanus, S. verreauxi, Cancer borealis, N. norvegicus, C. maenas, G. lateralis, and P. monodon, whose aa sequences have been predicted previously by the authors and others, were subjected to contemporary workflows and compiled in one convenient spreadsheet (retaining original names, as applicable). We also present the novel GPCR-encoding transcripts of P. ornatus, C. quadricarinatus and E. sinensis (see Supplementary Material S1).
3.2 Clustering analysis and determination of GPCR topological domains
Transcript ORFs that clustered strongly with the seeded transcripts of predicted receptors of the GnRH superfamily of neuropeptides (that is, displaying E-values less than 1e-50) were identified and extracted (see Supplementary Material S1). Figure 1 depicts the clustering of the class A receptor transcripts (Figure 1A) and the constituent members of the GnRHR-like superfamily (Figure 1B). The topological GPCR domains of the GnRHR-like predicted ORFs, including the N-terminus and C-terminus, ICLs and ECLs, and the canonical 7TM domains have been predicted and are presented in Supplementary Material S1. DRY, CWxP, and NPxxY motif loci were sequenced for the GnRHR-like transcripts and have also been reported (see Supplementary Material S1).
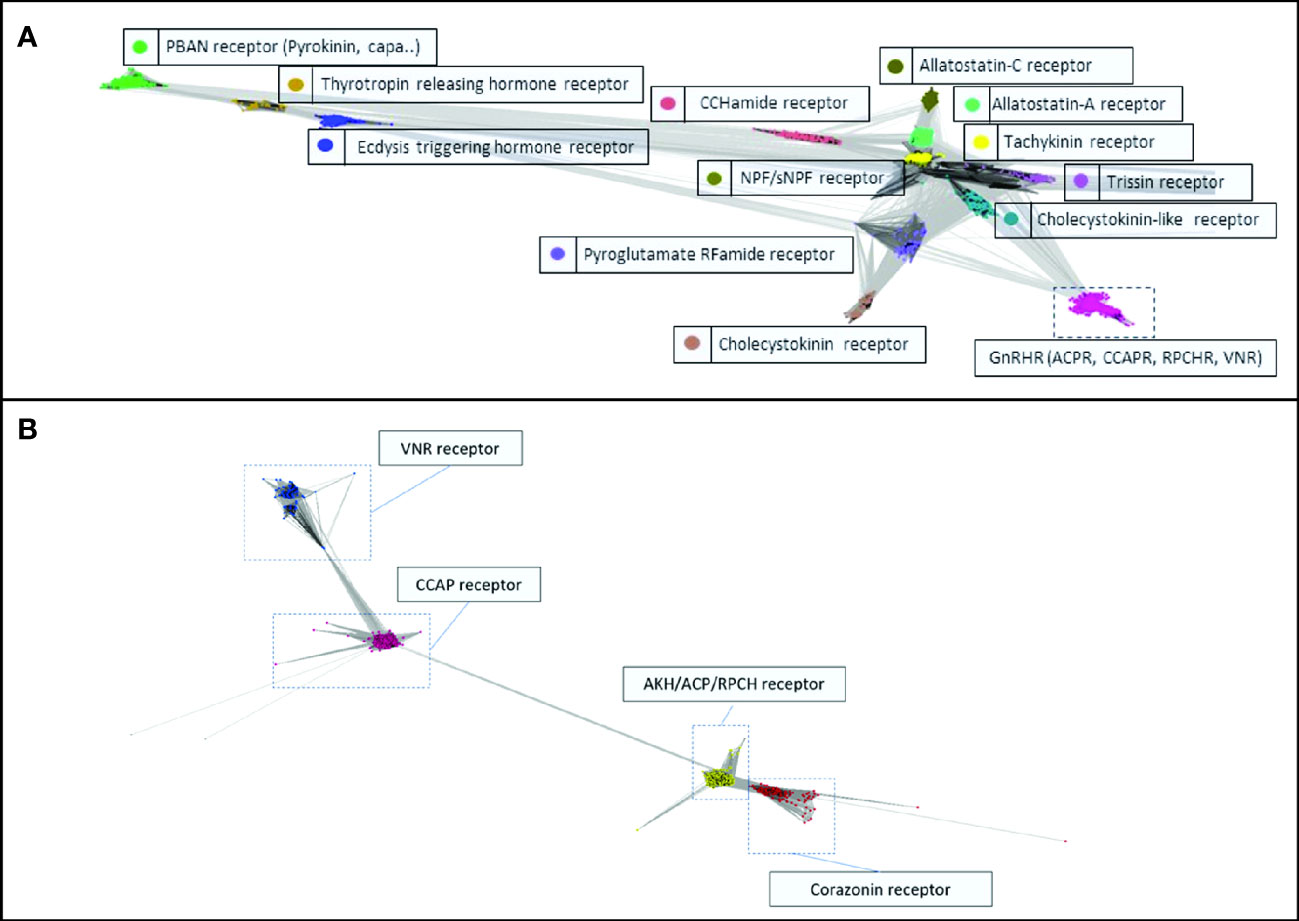
Figure 1 Cluster analysis of (A) the class A GPCR-encoding transcripts, and (B) the Gonadotropin-Releasing Hormone (GnRH) superfamily of receptor-encoding transcripts with predicted ligands. The images were generated using the CLANS2 algorithm with E-values less than 1e-50.
3.3 Diversity of DRY motif locus sequences
Table 2 summarizes the diversity observed in the aa sequences of the function-related DRY motif loci of this dataset of decapod GPCRs and, for comparison, a dataset of human GPCRs analyzed by others (8). Noting the high degree of conservation of the DRY motif, and the similarity between datasets.
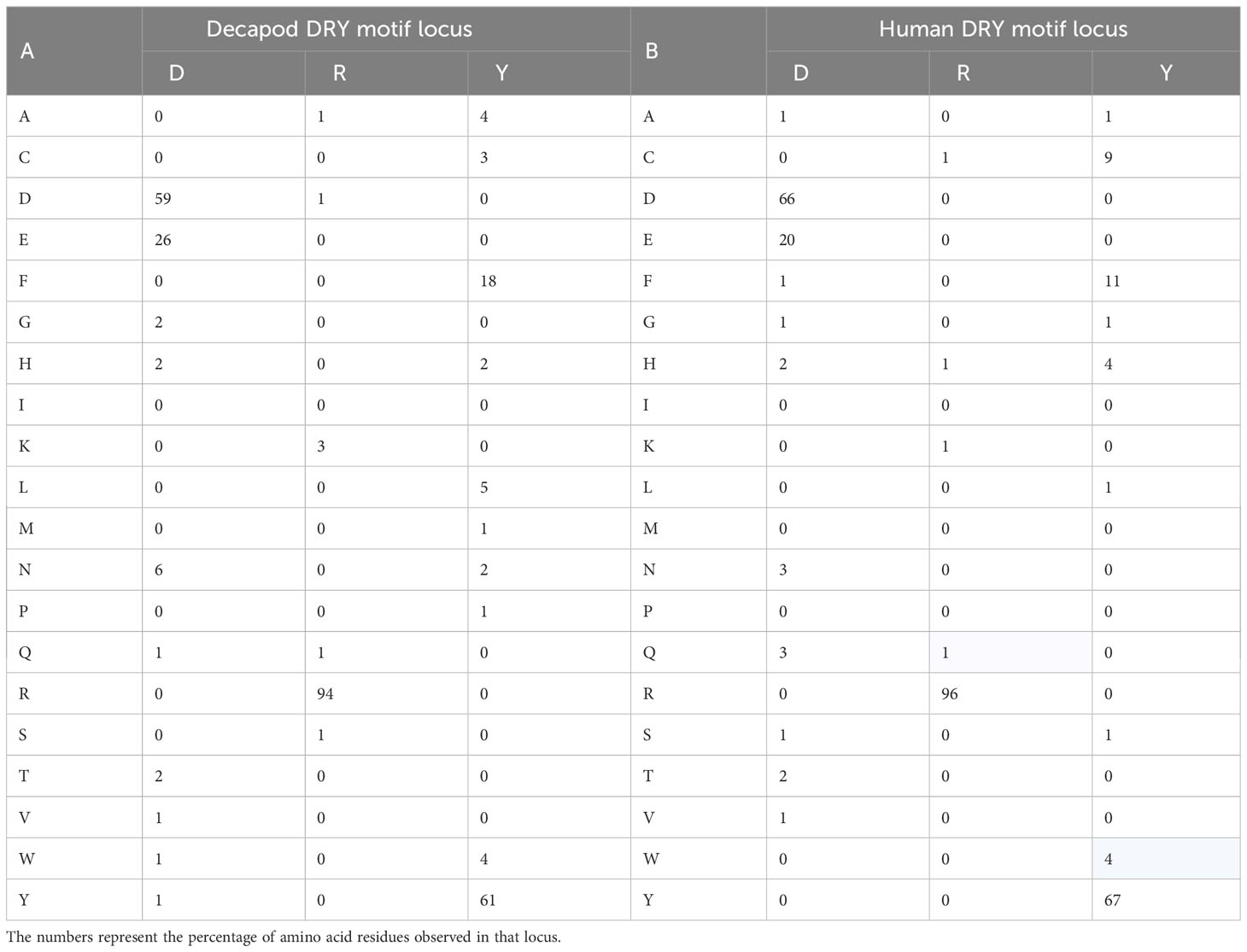
Table 2 Heatmaps depicting the diversity of the amino acid sequences of GPCR-encoding transcript DRY motif loci of A) the eleven decapod species reported in this study (n=1289), and B) of a dataset of human GPCRs that was reported by others (n=270) (8).
Supplementary Material S2: Table 1 depicts the diversity of aa residues observed at the DRY motif locus for the 1289 decapod GPCR-encoding transcripts of this study. The conserved DRY motif was observed in 40.8% of the transcripts. Whereas ERY (10.6%), DRF (8.8%), ERF (6.8%), GRF (1.8%), DKY (1.7%), DSY (0.9%), DRC (0.7%), ERC (0.6%), GRY (0.5%), and VRY (0.3%) collectively were observed in 32.7% of the transcripts. Meaning that the variation observed in the DRY motif loci of 73.5% of the transcripts is explained by a point mutation in a single nucleotide of an aa codon. DRY locus motifs containing R as the second aa residue were observed in 93.8% of the transcripts (refer to Table 2 and the left-hand column of Supplementary Material S2: Table 1). Interestingly, in the third aa or ‘Y’ locus of the DRY motif, an aromatic aa (that is, W = tryptophan, Y = tyrosine, F= phenylalanine and H= histidine) was observed in 84.5% of the transcripts. Moreover, DRM and GRF motifs were only observed in 0.9% and 1.8% of the GPCR-encoding transcripts, respectively. The clusters of motifs that deviated from the DRY sequence represented in Supplementary Material S2: Table 1 (that is, away from the upper left corner) are noted as being biologically less probable, as is detailed in the accompanying notes (Supplementary Material S2). These deviations raise the possibility of function-related consequence if they are not bioinformatic anomalies (for example GRF sequence in the DRY locus).
3.4 Conserved motifs of the receptors of the Gonadotropin-Releasing Hormone (GnRH)-like neuropeptides
The snake plots in Figure 2 depict the topological domains of three pairs of GnRHR-like proteins. The receptors on the left-hand side are the functionally annotated receptors of corazonin, RPCH and CCAP. The three receptors on the right-hand side (Gl_GPCR_A7, Po_GPCR_A146 and Cm_GPCR_A117) are transcripts from this dataset that share aa sequence identity with the annotated receptors. Note the high degree of similarity of the N-termini, ECLs, ICLs, C-termini, conserved N-glycosylation sites and conserved motifs (that is, DRY, CWxP and NPxxY). Observed differences in the lengths of N-terminus and C-terminus may be a sequencing artefact.
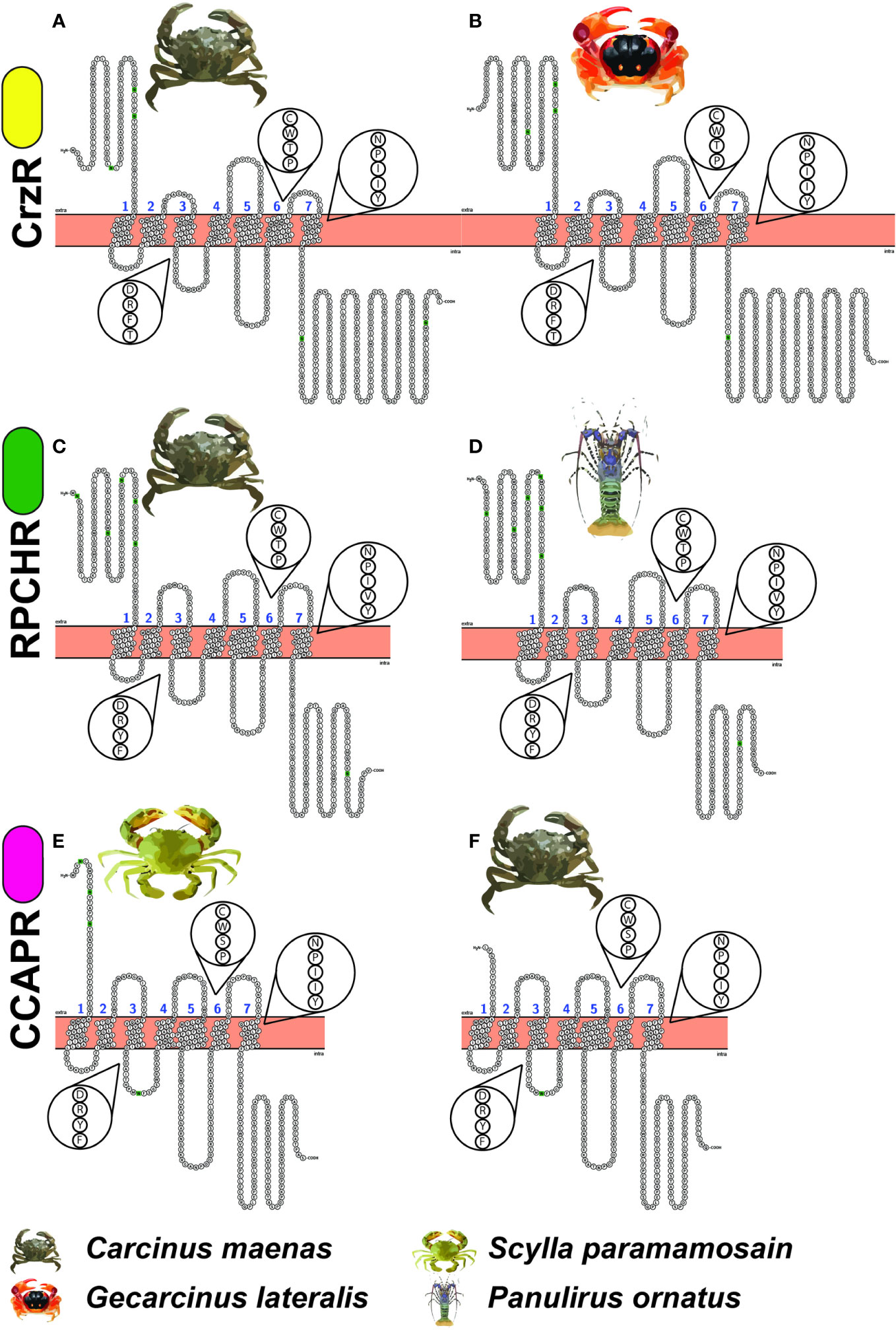
Figure 2 Snake plots of the functionally annotated receptors of (A) Corazonin (Cm_CrzR), (C) Red Pigment Concentrating Hormone (Cm_RPCHR) and (E) Crustacean Cardioactive Peptide (Sp_CCAPR), compared with GPCR-encoding transcripts (B) Gl_GPCR_A7 (D) Po_GPCR_A146 and (F) Cm_GPCR_A117 of this dataset. The exploded views depict the conserved function-related DRY, CWxP and NPxxY motifs. Noting the similarity of the GPCR topological domains that are represented including the N-terminus (H2N), C-terminus (COOH), three intracellular loops, three extracellular loops, canonical seven transmembrane domains, N-glycosylation sites (depicted by green) and conserved motifs. Yellow, green and pink ovals mean next to the receptor names correspond to the DRY motifs listed in Table 3.
3.5 Relationships between the DRY motifs and the seven transmembrane (7TM) domains of the Gonadotropin-Releasing Hormone (GnRH) superfamily of receptors
The relationship between the phylogeny of the 7TMs (including ICLs and ECLs) of the GnRHR-like protein transcripts, and the associated sequence of the DRYx motif locus and species is summarized in Figure 3. Noting the high degree of compartmentalization of the labels (that is, species and DRY motif sequence), and very high bootstrap values within color-coded clades. Moreover, the receptor pairs depicted in the snake plots of Figure 2 cluster together.
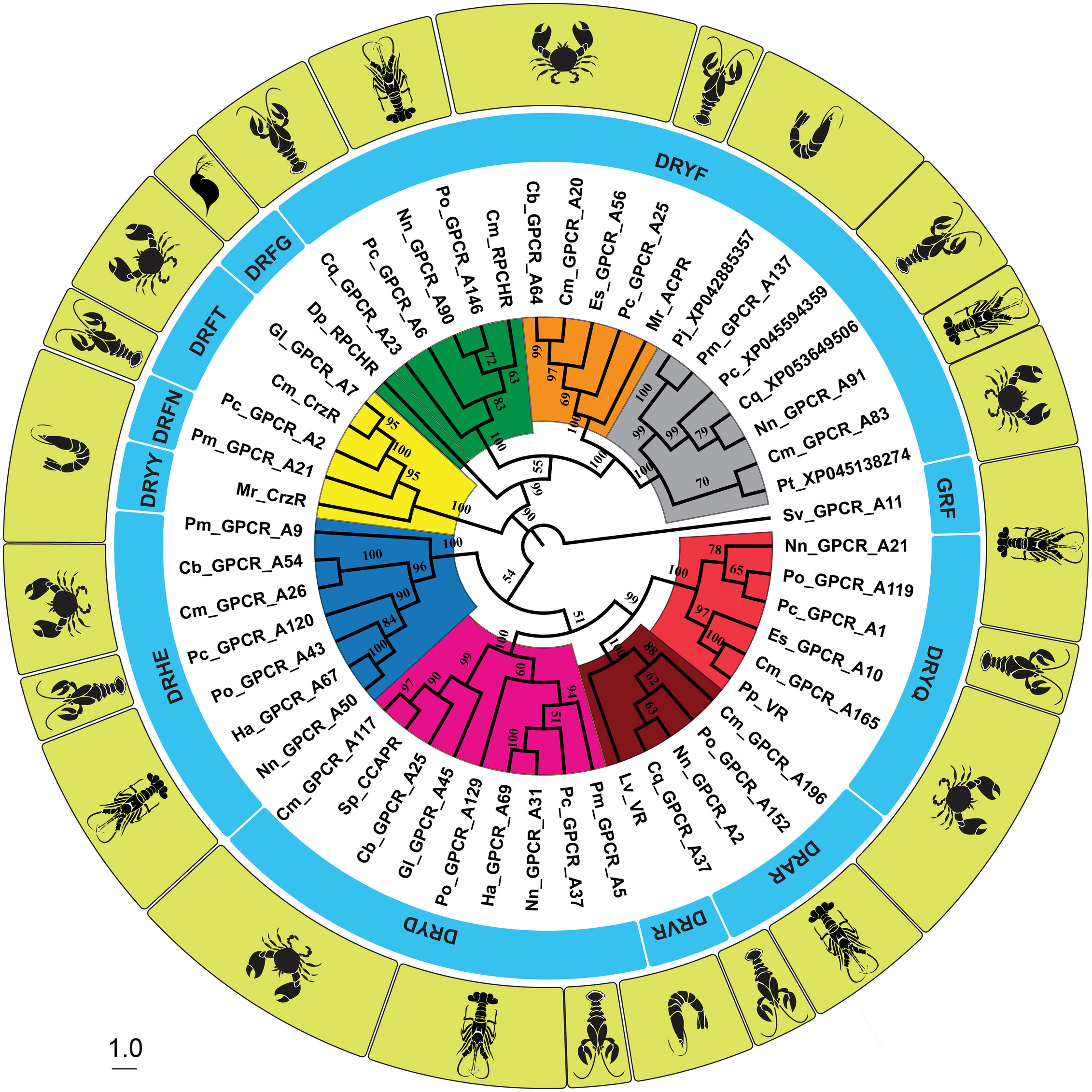
Figure 3 Neighbor-joining cladogram using the seven transmembrane domains, and intracellular and extracellular loops of the Gonadotropin-Releasing Hormone (GnRH) superfamily of receptor transcripts. Labeling of the tree includes the name of the receptor (including the decapod species), the sequence of the functional-relevant ‘DRYx’ motif locus, and an image of the species. The Muscle algorithm was employed in the sequence alignment and bootstrap values of 1000 were used. Legend: Pc, Procambarus clarkii Louisiana crawfish; Ha, Homarus americanus American lobster; Sv, Sagmariasus verreauxi eastern rock lobster; Cb, Cancer borealis Jonah crab; Nn, Nephrops norvegicus Norwegian lobster; Cm, Carcinus maenas Green shore crab; Gl, Gecarcinus lateralis blackback land crab; Pm, Penaeus monodon giant tiger prawn; Po, Panulirus ornatus ornate rock lobster; Cq, Cherax quadricarinatus Australian red claw crayfish; Es, Eriocheir sinensis Chinese mitten crab; Sp, Scylla paramamosain green mud crab; Pp, Portunus pelagicusblue swimmer crab; Lv, Litopenaeus vannameiwhite leg shrimp; Mr, Macrobrachium rosenbergii giant freshwater prawn; Pj, Penaeus japonicus kuruma shrimp and Dp, Daphnia pulex water flea. Distinct clades are color-labeled.
Referring again to the snake plots (Figure 2) and the DRYx motif (Figure 3), we present this as evidence that Gl_GPCR_A7, Po_GPCR_A146, and Cm_GPCR_A117 are orthologues of Cm_CrzR, Cm_RPCHR and Sp_CCAPR, respectively. Thereby representing candidates for functional annotation as receptors of corazonin, RPCH, and CCAP, respectively.
3.6 Annotation of the decapod GnRHR-like superfamily of receptors
The Neighbor Joining phylogenetic trees of both the 7TMs (including ECLs and ICLs) and of the ORFs of the GnRHR-like receptors (seen in Figure 3 and Supplementary Material S2: Figure 1, respectively) are presented for comparison. The eight discernible and color-coded clades were composed of the same constituent receptors and had similar bootstrap values. The yellow, green and magenta clades contained the functionally annotated receptors of corazonin, RPCH and CCAP, respectively. The maroon and light red clades contained the two bioinformatically annotated variants of the VR. The blue clade contains Ha_GPCR_A67 of which GNRH2R (accession number: XP_042226332.1) (38) is a truncated version. Notably, the GnRHRs (blue) were more closely related to the VR/CCAPR clade than the CrzR/ACPR1/ACPR2/RPCHR clade, respectively (albeit with the low bootstrap values of 54 and 34). Snake plots of the ACPR1 (orange clade) and ACPR2 (grey clade) receptors are included in Supplementary Material S2: Figures 2, 3, respectively. Comparison and contrast of the topological domains depicted in these figures reveals a high degree of intra-clade similarity, and inter-clade similarity but with a discernible degree of inter-clade difference. For example, the length of ECL3, which may have implications for function and/or ligand binding. The orange clade contained the bioinformatically annotated ACPR1 receptor, Mr_ACPR.
Table 3 summarizes the canonical conserved motifs (DRY, NPxxY, and CWxP) observed in GnRHR-like transcripts of our dataset and in the aa sequences of GPCRs annotated by others (functionally and bioinformatically). Noting the high degree of conservation, congruence and compartmentalization of sequence motif variants within clades of the same predicted ligand, and by extension, the same predicted function. Members of the GnRHR clade (blue) were observed to possess a distinctive DRHEAV sequence in the DRYxxx motif, and lack any discernible CWxP motif, which was exceptional in the dataset tested. Further we noted that tyrosine (Y) is aromatic, hydrophobic and non-charged, whereas histidine (H) is aromatic, hydrophobic and positively charged, which likely has implications for functionality. Altogether, suggesting that the GnRHR receptors (blue) represent a putative novel GPCR model. Moreover, as new models they will likely need novel bioassays for functional annotation. Considering the ACPR clades (orange and grey), the silkworm moth B. mori ACPR-like BNGR-A28 and BNGR-A29 had conserved DRYxxx, CWxP and NPxxY motifs of DRFFAI, CWFP and DPLVY, and DRFFAV, CWLP and NPLVY, respectively. Noting a, not unexpected, lack of total congruence between the conserved motifs of the two ACP receptors of B. mori (Taxonomical order: Lepidoptera) and the decapods of this study (Taxonomical order: Crustacea) (see Table 3). Considering all of the above, this suggests that the yellow, green, magenta, maroon, light red, blue, orange and grey clades represent CrzR, RPCHR, CCAPR, VR1, VR2, and the newly annotated GnRHR, ACPR1 and ACPR2, respectively.
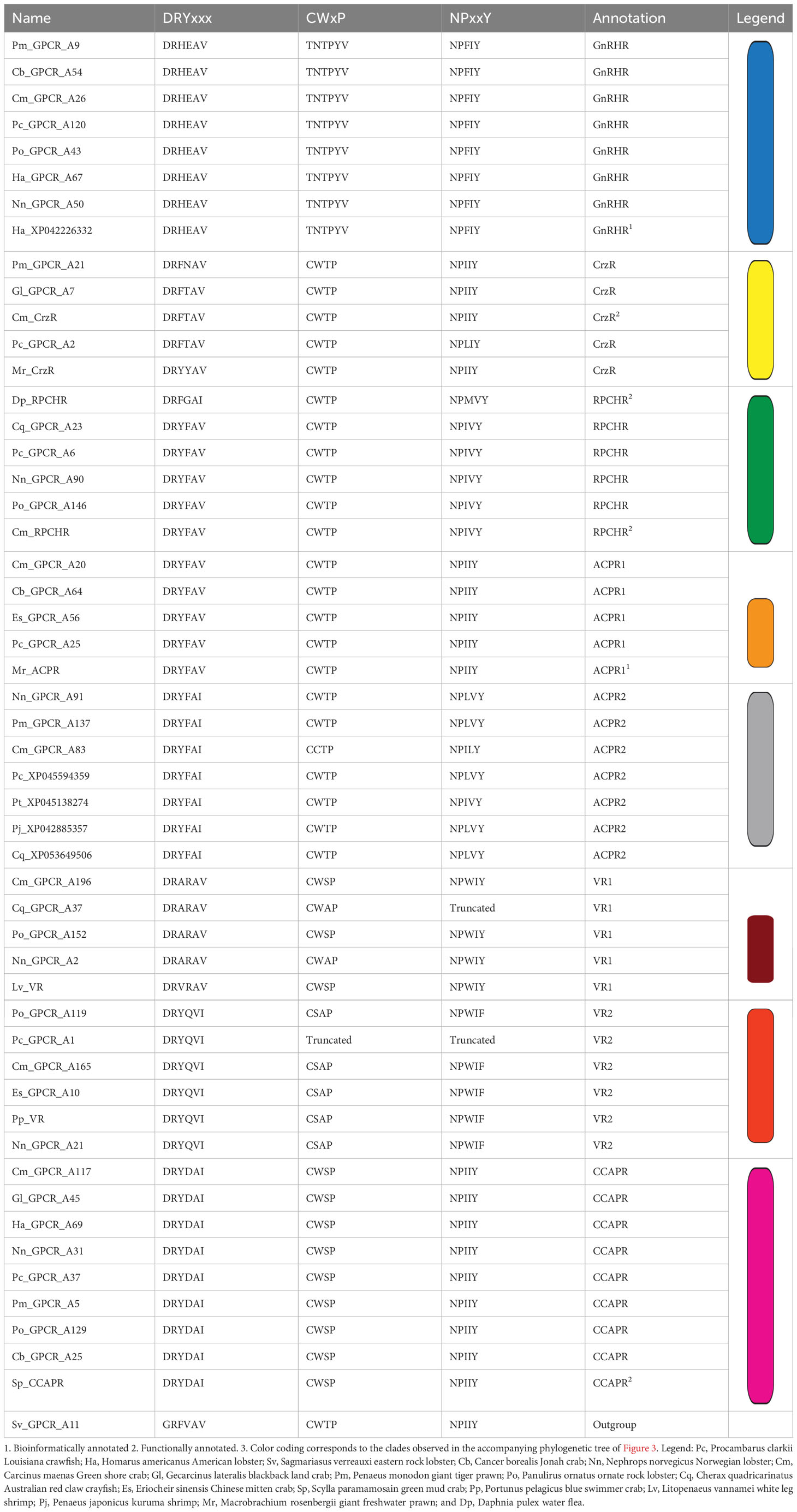
Table 3 Conserved GPCR motifs (that is, DRYxxx, CWxP and NPxxY) observed in transcripts of the GnRHR-like receptors of this dataset and in protein sequences published on NCBI.
Figure 4 summarizes the findings of the CrustyBase expression pattern analysis of the GnRHR-like transcripts. In general, across tissues of P. ornatus, gene expression levels were low, however, the highest expression was in neural tissues (eyestalks, brain, thoracic ganglia), antennal gland, and the adult testis. Higher expression of GnRHR-like and CCAPR-like were observed throughout Po embryo and larval developmental stages, respectively. Notably the expression of CrzR-like was elevated over the molt stages of G. lateralis. ACPR2 (grey) and VR2 (light red) were observed to be expressed in the adult testis, suggesting putative reproduction-related function.
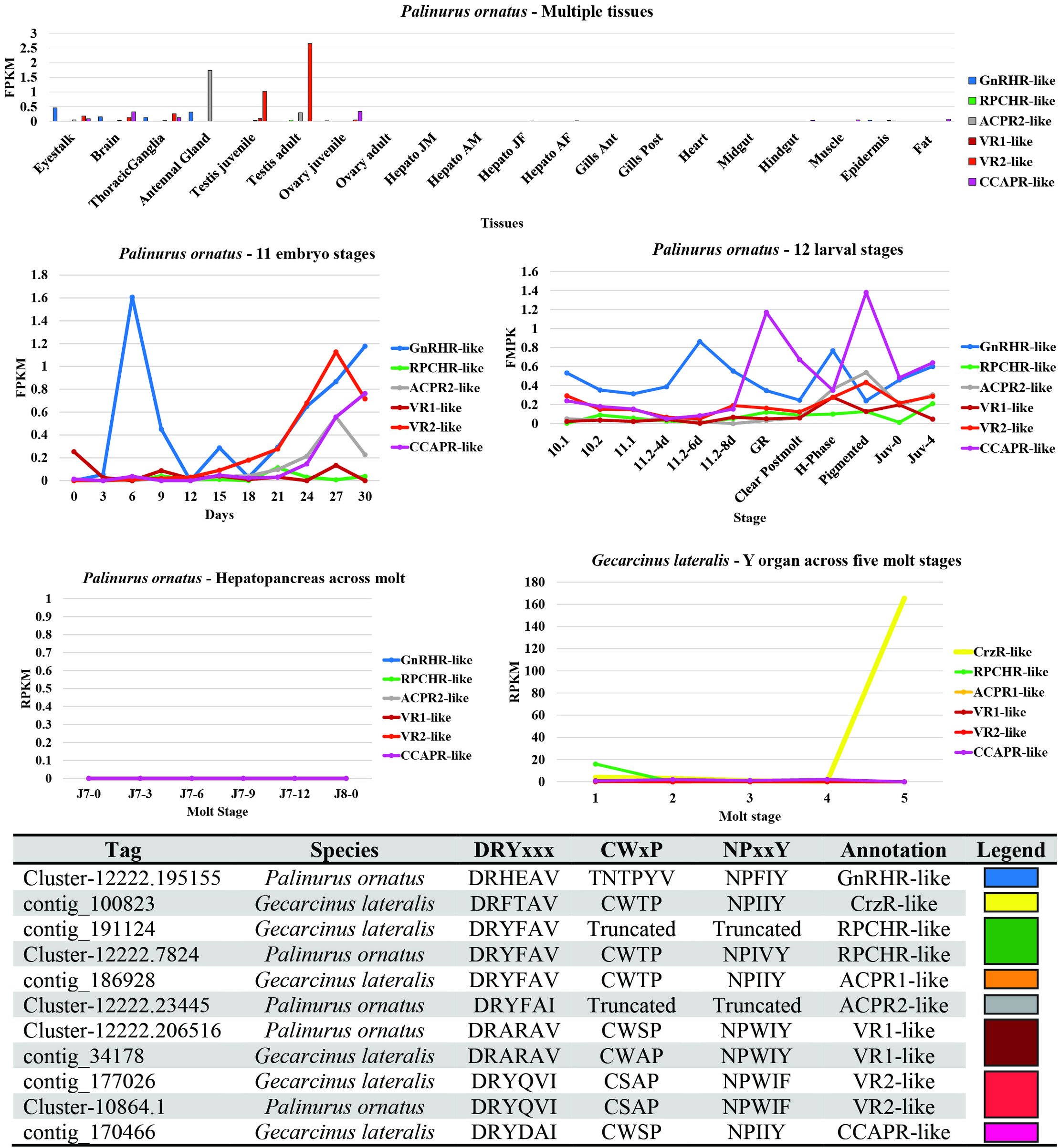
Figure 4 Transcriptomic-related expression patterns of decapod GnRHR-like receptors using the CrustyBase database (39). Tag numbers represent CrustyBase designation.
4 Discussion
We present several key findings in our study of the GPCRs of decapod species. Foundationally, the GPCRomes of eleven decapod species have been compiled into one convenient source, which also includes the novel GPCR repertoires of three species. We have also developed a novel bioinformatic workflow to augment traditional methods of annotating GPCRs, whose application extends beyond the decapod order. The catalogue of GPCRomes established in this study is a significant achievement in the field of decapod biology for several reasons. Firstly, it is the largest collection of its kind to have been published thus far, comprising a large-scale dataset of GPCR aa sequences. Despite its size, the catalogue is organized in a way that makes it both manageable and easily augmentable (Supplementary File S1), ensuring that it remains a valuable resource for researchers over time. Additionally, the inclusion of transcripts derived from a diversity of both species and transcriptomic resources provides valuable insights into the species-specificity of GPCR domains and motifs, rendering it an indispensable tool for the analysis of GPCR amino acid sequences in decapod species.
Our novel bioinformatic workflow is summarized in the following steps. A non-redundant and exhaustive dataset of decapod class A GPCR transcripts was compiled. Clustering analysis was performed using the aa sequences of annotated GPCRs as seeds. Strongly clustering transcripts were extracted, curated for bioinformatic integrity, subjected to a GPCR domain predicting algorithm, and conserved motifs were sequenced. The phylogeny of the 7TM-spanning domains was constructed and labeled with species and conserved DRY motif. Congruence and compartmentalization of the species and conserved motifs was tabulated. All of which represents a superior method of annotating GPCRs than a traditional phylogenetic and BLAST analysis. More specifically, we communicate an absence of the highly conserved CWxP motif in the clade of GnRHR receptors, which was unique among GnRHR-like receptors.
At a time when then the description of the evolutionary history of invertebrate GnRHR-like proteins is in need of novel methods and tools (5), we present our contribution that is consistent with the GnRHR nomenclature proposed by Zandawala et al. (11). We have constructed phylogenies for the GnRHR-like transcripts based on both the 7TM-spanning domains and the entire ORFs. From which we observed congruence between the clades (and relative bootstrap values) that were formed using both methods, suggesting that either method has utility in the analysis of the evolutionary history of decapod GPCRs. Consequently, we have chosen to use the 7TM-spanning domains with their higher degree of sequence conservation when constructing our phylogenies. However, we advocate that the consideration of the sequences of the N-terminus and C-terminus are crucial to the understanding of the function (that is, ligand binding and G protein interaction, respectively) of GPCRs. Our communicated phylogenies contribute to the resolution of decapod and invertebrate GnRHR-like receptors. Further research will address the G proteins involved in GnRH-like receptor signaling.
Several of the factors that influence the transcriptomic analysis described here include the species, spatiotemporal circumstance, life cycle stage and environmental conditions of the subject. Our dataset is large-scale and diverse, however, larger datasets of greater diversity only stand to strengthen future evaluation of the presence and absence of GPCR-encoding transcripts which is crucial in the solidification of our understanding of species-specific variations. Furthermore, the classical nomenclature for annotating GPCRs is heavily biased toward the binding of a single ligand neuropeptide to its single cognate GPCR receptor with unmistakably high affinity. This has proven adequate for the deorphanisation of a multitude of GPCRs, but proves challenging, in the annotation of the GnRHR-like superfamily of receptors. This is because the neuropeptides of this arthropod superfamily are known to display promiscuous binding with the consequence of pleiotropic and overlapping biological effects (37, 40). With this in mind, we have used color-coding of the clades as an intermediate nomenclature that facilitates seamless communication between this and future datasets while the interpretation of the non-singular relationships of the GnRH-like neuropeptides and GPCRs is strengthened. We contend that this study has produced a readily extensible dataset and future-compatible workflow that constitute a tractable prototyping tool in the future functional annotation of decapod GPCRs.
5 Conclusions
In this study we have compiled the most comprehensive publicly available catalogue of decapod GPCR-encoding transcripts, to date. We have developed a novel workflow that includes clustering analysis to determine superfamilies of GPCRs (for example, GnRHR-like receptors), sequencing of the membrane-related domains and conserved function-related motifs, phylogenetic analysis of the seven transmembrane-spanning domains with species and conserved motif labels, and visual representation of the conservation of (that is, congruence and compartmentalization) of the domains and motifs. We have constructed well-resolved phylogenies and noted a high degree of conservation of the sequences and topology of the domains and motifs. This conservation has allowed the identification of species-specific variation (especially in the extracellular loops) that is thought to be influential to ligand binding and function. Our results contribute to the resolution of the evolutionary history of invertebrate GnRHRs, and inform the design of bioassays in the deorphanisation and functional annotation of GPCRs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
SB: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing – original draft. TVN: Conceptualization, Data curation, Investigation, Software, Visualization, Writing – review & editing. SC: Writing – review & editing. AE: Writing – review & editing. QF: Funding acquisition, Writing – review & editing. GS: Funding acquisition, Writing – review & editing. DM: Funding acquisition, Writing – review & editing. TV: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Australian Government with funding from the Australian Research Council (http://www.arc.gov.au/, accessed on 4 May 2023) Industrial Transformation Research Hub (project number IH190100014) and the National Science Foundation (IOS-1257732 and IOS-1922701).
Acknowledgments
The authors would like to thank Dr. Susan Glendinning for her valuable suggestions while reading the manuscript. We would like to thank Dr. Minh Nhut Tran for his assistance in retrieving the GPCR databases used for this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2024.1348465/full#supplementary-material
Supplementary Material S1 | GPCR-encoding transcripts for eleven decapod species, and GnRHR-like and CHHR-like topological domains.
Supplementary Material S2 | Analysis of DRY locus sequence diversity, GnRHR-like ORF based phylogeny, Snake plots of ACPR1 and ACPR2.
References
1. Metian M, Troell M, Christensen V, Steenbeek J, Pouil S. Mapping diversity of species in global aquaculture. Rev Aquaculture (2020) 12:1090–100. doi: 10.1111/raq.12374
2. Wolfe JM, Breinholt JW, Crandall KA, Lemmon AR, Lemmon EM, Timm LE, et al. A phylogenomic framework, evolutionary timeline and genomic resources for comparative studies of decapod crustaceans. Proc R Soc B (2019) 286:20190079. doi: 10.1098/rspb.2019.0079
3. Raviv S, Parnes S, Sagi A. Coordination of reproduction and molt in decapods. In: Reproductive biology of Crustaceans. Enfield, New Hampshire, USA: CRC Press (2008). p. 365–90.
4. Mykles DL. Signaling pathways that regulate the crustacean molting gland. Front Endocrinol (2021) 12:674711. doi: 10.3389/fendo.2021.674711
5. Dufour S, Quérat B, Tostivint H, Pasqualini C, Vaudry H, Rousseau K. Origin and evolution of the neuroendocrine control of reproduction in vertebrates, with special focus on genome and gene duplications. Physiol Rev (2020) 100:869–943. doi: 10.1152/physrev.00009.2019
6. Anantakrishnan S, Naganathan AN. Thermodynamic architecture and conformational plasticity of GPCRs. Nat Commun (2023) 14:128. doi: 10.1038/s41467-023-35790-z
7. Schöneberg T, Hofreiter M, Schulz A, Römpler H. Learning from the past: evolution of GPCR functions. Trends Pharmacol Sci (2007) 28:117–21. doi: 10.1016/j.tips.2007.01.001
8. Mirzadegan T, Benkö G, Filipek S, Palczewski K. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry (2003) 42:2759–67. doi: 10.1021/bi027224+
9. Wheatley M, Wootten D, Conner MT, Simms J, Kendrick R, Logan RT, et al. Lifting the lid on GPCRs: the role of extracellular loops. Br J Pharmacol (2012) 165:1688–703. doi: 10.1111/j.1476-5381.2011.01629.x
10. Buckley SJ, Fitzgibbon QP, Smith GG, Ventura T. In silico prediction of the G-protein coupled receptors expressed during the metamorphic molt of Sagmariasus verreauxi (Crustacea: Decapoda) by mining transcriptomic data: RNA-seq to repertoire. Gen Comp Endocrinol (2016) 228:111–27. doi: 10.1016/j.ygcen.2016.02.001
11. Zandawala M, Tian S, Elphick MR. The evolution and nomenclature of GnRH-type and corazonin-type neuropeptide signaling systems. Gen Comp Endocrinol (2018) 264:64–77. doi: 10.1016/j.ygcen.2017.06.007
12. Alexander JL, Oliphant A, Wilcockson DC, Audsley N, Down RE, Lafont R, et al. Functional characterization and signaling systems of corazonin and red pigment concentrating hormone in the green shore crab, Carcinus maenas. Front Neurosci (2018) 11:752–2. doi: 10.3389/fnins.2017.00752
13. Marco HG, Verlinden H, Vanden Broeck J, Gäde G. Characterisation and pharmacological analysis of a crustacean G protein-coupled receptor: The red pigment-concentrating hormone receptor of Daphnia pulex. Sci Rep (2017) 7:6851–12. doi: 10.1038/s41598-017-06805-9
14. Alexander JL, Oliphant A, Wilcockson DC, Brendler-Spaeth T, Dircksen H, Webster SG. Pigment dispersing factors and their cognate receptors in a crustacean model, with new insights into distinct neurons and their functions. Front Neurosci (2020) 14. doi: 10.3389/fnins.2020.595648
15. Caers J, Verlinden H, Zels S, Vandersmissen HP, Vuerinckx K, Schoofs L. More than two decades of research on insect neuropeptide GPCRs: an overview. Front Endocrinol (2012) 3:151. doi: 10.3389/fendo.2012.00151
16. Hyde CJ, Fitzgibbon QP, Elizur A, Smith GG, Ventura T. CrustyBase: an interactive online database for crustacean transcriptomes. BMC Genomics (2020) 21:637. doi: 10.1186/s12864-020-07063-2
17. Perez-Moreno JL, Kozma MT, DeLeo DM, Bracken-Grissom HD, Durica DS, Mykles DL. CrusTome: a transcriptome database resource for large-scale analyses across Crustacea. G3 (Bethesda) (2023) 13. doi: 10.1093/g3journal/jkad098
18. Rovati GE, Capra V, Shaw VS, Malik RU, Sivaramakrishnan S, Neubig RR. The DRY motif and the four corners of the cubic ternary complex model. Cell Signalling (2017) 35:16–23. doi: 10.1016/j.cellsig.2017.03.020
19. Weis WI, Kobilka BK. The molecular basis of G protein–coupled receptor activation. Annu Rev Biochem (2018) 87:897–919. doi: 10.1146/annurev-biochem-060614-033910
20. Seo MJ, Heo J, Kim K, Chung KY, Yu W. Coevolution underlies GPCR-G protein selectivity and functionality. Sci Rep (2021) 11:7858. doi: 10.1038/s41598-021-87251-6
21. Veenstra JA. The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen Comp Endocrinol (2015) 224:84–95. doi: 10.1016/j.ygcen.2015.06.013
22. Nguyen TV, Rotllant GE, Cummins SF, Elizur A, Ventura T. Insights into sexual maturation and reproduction in the Norway lobster (Nephrops norvegicus) via in silico prediction and characterization of neuropeptides and G protein-coupled receptors. Front Endocrinol (2018) 9:430. doi: 10.3389/fendo.2018.00430
23. Oliphant A, Alexander JL, Swain MT, Webster SG, Wilcockson DC. Transcriptomic analysis of crustacean neuropeptide signaling during the moult cycle in the green shore crab, Carcinus maenas. BMC Genomics (2018) 19:1–26. doi: 10.1186/s12864-018-5057-3
24. Tran NM, Mykles DL, Elizur A, Ventura T. Characterization of G-protein coupled receptors from the blackback land crab Gecarcinus lateralis Y organ transcriptome over the molt cycle. BMC Genomics (2019) 20:1–20. doi: 10.1186/s12864-018-5363-9
25. Nguyen TV, Ryan LW, Nocillado J, Le Groumellec M, Elizur A, Ventura T. Transcriptomic changes across vitellogenesis in the black tiger prawn (Penaeus monodon), neuropeptides and G protein-coupled receptors repertoire curation. Gen Comp Endocrinol (2020) 298:113585. doi: 10.1016/j.ygcen.2020.113585
26. Hyde CJ, Fitzgibbon QP, Elizur A, Smith GG, Ventura T. Transcriptional profiling of spiny lobster metamorphosis reveals three new additions to the nuclear receptor superfamily. BMC Genomics (2019) 20:1–14. doi: 10.1186/s12864-019-5925-5
27. Lewis CL, Fitzgibbon QP, Smith GG, Elizur A, Ventura T. Transcriptomic analysis and time to hatch visual prediction of embryo development in the ornate spiny lobster (Panulirus ornatus). Front Mar Sci (2022) 9:1009. doi: 10.3389/fmars.2022.889317
28. Ventura T, Chandler JC, Nguyen TV, Hyde CJ, Elizur A, Fitzgibbon QP, et al. Multi-tissue transcriptome analysis identifies key sexual development-related genes of the ornate spiny lobster (Panulirus ornatus). Genes (2020) 11:1150. doi: 10.3390/genes11101150
29. Frickey T, Lupas A. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics (2004) 20:3702–4. doi: 10.1093/bioinformatics/bth444
30. Bao C, Yang Y, Zeng C, Huang H, Ye H. Identifying neuropeptide GPCRs in the mud crab, Scylla paramamosain, by combinatorial bioinformatics analysis. Gen Comp Endocrinol (2018) 269:122–30. doi: 10.1016/j.ygcen.2018.09.002
31. Alexander JL, Oliphant A, Wilcockson DC, Audsley N, Down RE, Lafont R, et al. Functional characterization and signaling systems of corazonin and red pigment concentrating hormone in the green shore crab, Carcinus maenas. Front Neurosci (2018) 11:752. doi: 10.3389/fnins.2017.00752
32. Suwansa-Ard S, Zhao M, Thongbuakaew T, Chansela P, Ventura T, Cummins SF, et al. Gonadotropin-releasing hormone and adipokinetic hormone/corazonin-related peptide in the female prawn. Gen Comp Endocrinol (2016) 236:70–82. doi: 10.1016/j.ygcen.2016.07.008
33. Hallgren J, Tsirigos KD, Pedersen MD, Almagro Armenteros JJ, Marcatili P, Nielsen H, et al. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. BioRxiv (2022). doi: 10.1101/2022.04.08.487609
34. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res (2004) 32:1792–7. doi: 10.1093/nar/gkh340
35. Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X. molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol (2018) 35:1547. doi: 10.1093/molbev/msy096
36. Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics (2014) 30:884–6. doi: 10.1093/bioinformatics/btt607
37. Sehadova H, Takasu Y, Zaloudikova A, Lin Y-H, Sauman I, Sezutsu H, et al. Functional analysis of adipokinetic hormone signaling in Bombyx mori. Cells (2020) 9:2667. doi: 10.3390/cells9122667
38. Polinski JM, Zimin AV, Clark KF, Kohn AB, Sadowski N, Timp W, et al. The American lobster genome reveals insights on longevity, neural, and immune adaptations. Sci Adv (2021) 7:eabe8290. doi: 10.1126/sciadv.abe8290
39. Hyde CJ, Fitzgibbon QP, Elizur A, Smith GG, Ventura T. CrustyBase: an interactive online database for crustacean transcriptomes. BMC Genomics (2020) 21:1–10. doi: 10.1186/s12864-020-07063-2
Keywords: GPCR, evolutionary history, GnRHR, conserved motifs, decapoda
Citation: Buckley SJ, Nguyen TV, Cummins SF, Elizur A, Fitzgibbon QP, Smith GS, Mykles DL and Ventura T (2024) Evaluating conserved domains and motifs of decapod gonadotropin-releasing hormone G protein-coupled receptor superfamily. Front. Endocrinol. 15:1348465. doi: 10.3389/fendo.2024.1348465
Received: 02 December 2023; Accepted: 18 January 2024;
Published: 20 February 2024.
Edited by:
Jeff M. P. Holly, University of Bristol, United KingdomReviewed by:
Yves Combarnous, Centre National de la Recherche Scientifique (CNRS), FrancePierre De Meyts, Université catholique de Louvain, Belgium
Copyright © 2024 Buckley, Nguyen, Cummins, Elizur, Fitzgibbon, Smith, Mykles and Ventura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sean J. Buckley, c2Vhbi5idWNrbGV5QG1haWwuY29t; Tomer Ventura, dHZlbnR1cmFAdXNjLmVkdS5hdQ==
 Sean J. Buckley1,2*
Sean J. Buckley1,2* Tuan Viet Nguyen
Tuan Viet Nguyen Scott F. Cummins
Scott F. Cummins Abigail Elizur
Abigail Elizur Quinn P. Fitzgibbon
Quinn P. Fitzgibbon Gregory S. Smith
Gregory S. Smith Donald L. Mykles
Donald L. Mykles Tomer Ventura
Tomer Ventura