
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 26 February 2024
Sec. Obesity
Volume 15 - 2024 | https://doi.org/10.3389/fendo.2024.1334342
This article is part of the Research Topic Effects and Mechanisms of Bariatric Surgery in Relieving Obesity and Its Complications View all 12 articles
Early-onset obesity is a rising health concern influenced by heredity. However, many monogenic obesity variants (MOVs) remain to be discovered due to differences in ethnicity and culture. Additionally, patients with known MOVs have shown limited weight loss after bariatric surgery, suggesting it can be used as a screening tool for new candidates. In this study, we performed whole-exome sequencing (WES) combined with postoperative data to detect candidate MOVs in a cohort of 62 early-onset obesity and 9 late-onset obesity patients. Our findings demonstrated that patients with early-onset obesity preferred a higher BMI and waist circumference (WC). We confirmed the efficacy of the method by identifying a mutation in known monogenic obesity gene, PCSK1, which resulted in less weight loss after surgery. 5 genes were selected for further verification, and a frameshift variant in CAMKK2 gene: NM_001270486.1, c.1614dup, (p. Gly539Argfs*3) was identified as a novel candidate MOV. This mutation influenced the improvement of metabolism after bariatric surgery. In conclusion, our data confirm the efficacy of WES combined with postoperative data in detecting novel candidate MOVs and c.1614dup (CAMKK2) might be a promising MOV, which needs further confirmation. This study enriches the human monogenic obesity mutation database and provides a scientific basis for clinically accurate diagnosis and treatment.
The increase in obesity presents a significant health issue as it has become a major contributor to the global occurrence of chronic diseases, including type 2 diabetes, cardiovascular diseases and some types of cancer (1, 2). Of particular concern is early-onset obesity, which refers to obesity that develops during childhood or adolescence. The prevalence of early-onset obesity is on the rise in low-income and middle-income countries, as well as many high-income countries (3). The role of genetics in the transmission of early-onset obesity and its associated health conditions is undeniable. Through genome-wide association studies (GWAS) and whole-exome sequencing (WES), numerous genetic loci linked to obesity have been identified (4, 5). However, a significant portion of patients with obesity remains unexplained by the currently identified individual genes. This implies that numerous unknown genes are yet to be discovered, and this genetic diversity is especially enhanced by differences in ethnicity and culture.
Bariatric surgery has emerged as an effective treatment for severe obesity, leading to sustained weight loss over a 10-year period and improvements in comorbidities and quality of life (6). However, it has been reported that patients carrying known MOVs experience lower weight loss after undergoing bariatric surgery (7), suggesting long-term negative effects of MOVs on patients following bariatric surgery. We therefore wondered whether this could be used to screening novel candidate MOVs.
In the current study, we employed WES in combination with weight data obtained after bariatric surgery to identify candidate MOVs that are enriched in patients with early-onset obesity. We hypothesized that candidate MOVs would be selectively enriched in the cases and filtered out in the control group. Our study successfully identified PCSK1, a known monogenic obesity gene, in 4 patients who experienced less weight loss after surgery, thereby confirming the validity of our method. Additionally, our study provides evidence for another candidate MOV found in the coding sequence of the CAMKK2 gene.
These findings highlight the potential of using WES combined with post-surgical weight data to identify and characterize candidate MOVs. By expanding our understanding of the genetic factors contributing to obesity, we can gain insights into the underlying mechanisms and potentially develop more personalized and targeted approaches for the management and treatment of obesity.
Obese patients (BMI >30kg/m2) who underwent bariatric surgery at the Gastrointestinal Minimally Invasive Center of the Third People’s Hospital of Chengdu during years of 2021 to 2022 were selected. The early-onset obesity group included subjects who were obese before 10 years old while the control group were after, according to the criteria of the Working Group for Obesity in China (WGOC) (8). Finally, a total of 62 early-onset obesity and 9 controls were included. Patient blood samples were collected, and preoperative clinical data and postoperative follow-up information were recorded. All procedures involving human subjects were approved by the Ethics Committee of The Third People’s Hospital of Chengdu. The clinical trial registration number is ChiCTR2300073353. The volunteers were provided with full information about the study, and their informed consent was obtained.
Genomic DNA was isolated from peripheral blood using the Blood Genome DNA Extraction Kit (TIANGEN Biotech, Beijing, China) according to manufacturer’s protocols. Whole-exome sequencing analysis was performed using the Illumina HiSeq2500 platform (Illumina San Diego, CA, USA) with 150 bp paired-end reads. All reads were mapped against the hg38 reference genome using the Burrow–Wheeler aligner (BWA). The identification of variants was performed using a custom pipeline that utilizes the GATK Best Practices workflow (9).
Variants were filtered according to minor allele frequency (MAF) ≥ 5% in the 1000 Genomes and Exome Aggregation Consortium (ExAC) databases. Variants showed in the controls and synonymous variants, assumed to be benign or likely benign, were also excluded. Frameshift, stop gain and stoploss were considered damaging. The nonsynonymous variants were predicted to be damaging by annotation in dbNSFP database. In detail, different prediction software (SIFT, Polyphen2_HDIV, Polyphen2_HVAR, LRT, MutationTaster, MutationAssessor, FATHMM, PROVEAN, MetaSVM, MetaLR, M-CAP, CADD, fathmm-MKL) were used to assign scores for the nonsynonymous variants. Considering that some software may lack annotations for certain mutations, for those with available scores, if half or more of the software predicts damaging, the variant is considered deleterious. Then, Criteria of the American College of Medical Genetics and Genomics (ACMG) were used to determine the pathogenicity of variants, building on the automatic classification provided by VarSome (https://varsome.com/). Finally, variants for further verification were selected based on mutation category and literature data.
The early-onset subjects were divided into groups of non-carriers and carriers depending on whether they carried the particular variant in each comparison, e.g., non-carriers of PCSK1 mutation vs carriers of PCSK1 mutation. In the postoperative period of bariatric surgery, patients’ weight was followed up at 1 month, 3 months, 6 months, and 12 months, and the percent of weight loss was calculated. Independent t tests were conducted to compare the percentage of weight loss between the two groups at each time point, and the P values were adjusted via the FDR. Paired t tests were used to compare HDL, LDL, TC and TG between baseline and 12 months after surgery. To compare the percentage changes in HDL, LDL, TC and TG from baseline to 12 months post-surgery between CAMKK2 non-carriers and carriers, independent t tests were also performed. A value of 0.05 was considered to be statistically significant. Statistical analyses were carried out with the use of IBM SPSS Statistics, GraphPad Prism 8 and R software.
All individuals included in the whole exome sequencing (WES) and assessments were classified as obese (BMI >30 kg/m2). Control subjects were selected from individuals who developed obesity after the age of 10 (n=9), while the early-onset obesity group consisted of individuals who developed obesity before the age of 10 (n=62). The clinical characteristics of both groups are summarized in Table 1.
The results showed a significant difference in BMI and waist circumference (WC) between the control and early-onset obesity groups (P< 0.05), with the latter exhibiting higher BMI and WC, suggesting that early-onset obesity has a more pronounced impact on overall weight gain compared to individuals who develop obesity later in life. However, there is no significant change in waist-hip ratio (WHR), visceral fat level (VFL), and percent body fat (PBF) between them, indicating that early-onset obesity has a greater impact on weight rather than on fat distribution, potentially influenced by genetic factors that influence appetite and energy homeostasis.
WES was performed for 62 early-onset obesity and 9 late-onset obesity and an average sequencing depth of 100× was obtained. Variants were selected according to mutation damaging prediction software, the American College of Medical Genetics (ACMG) guidelines (10) and literature data for further analysis. Candidate MOVs were verified by the weight loss data after bariatric surgery, as previous study reported that MOVs lead to less weight loss after bariatric surgery (7).
A total of 567 variants (variant frequency ≥5%) predicted as deleterious were identified in patients with early-onset obesity (including nonsynonymous SNVs and frameshift Indels). 11 were predicted pathogenic (P) or likely pathogenic (LP) and 60 were considered variants of uncertain significance (VUS) according to ACMG guidelines (Supplementary Table 1). We found 1 missense variant in the PCSK1 gene (c.242G>A) (Table 2) that was already known to be associated with monogenic early-onset obesity (11, 12), which mutation led to less weight loss after bariatric surgery (7). We also found 5 genes (2 frameshift insertion, 2 frameshift deletion and 1 stop-loss) reported associating with obesity, adipogenesis or previously linked to obesity-associated traits by genome-wide association studies: SLC25A5 (13), AIM2 (14), SNX16 (15), CAMKK2 (16–18), PDE11A (19) (Table 2) and they were selected for further verification.
To test the validity of the post-operative data to identify candidate MOVs, analysis was first performed on PCSK1 mutation carriers, and the results showed that the PCSK1 mutation resulted in less weight loss with a significant difference (FDR<0.05) (Figure 1), indicating that the method is effective. Therefore, to validate whether other gene variants are candidates, analysis were performed between non-carriers and carriers of each variant, including c.450del (SLC25A5), c.1029dup (AIM2), c.1033T>C (SNX16), c.1614dup (CAMKK2), c.20_21del (PDE11A). Compared to the non-carriers, CAMKK2 variant carriers exhibited poorer weight loss with a significant difference (FDR<0.001) (Figure 2), with no overlapping individuals with PCSK1 variant carriers, indicating that the effect of the CAMKK2 variant on weight loss is independent. AIM2 variant carriers also showed less weight loss from 1 month until 12 months after surgery, but similar with other three gene variants (c.450del (SLC25A5), c.1033T>C (SNX16), c.20_21del (PDE11A) carriers, the difference was not statistically significant compared to the non-carriers group (Supplementary Figure 1). In order to rule out other genetic mutations carried by CAMKK2 mutation carriers resulting in less weight loss, we took the intersection of all the mutations carried by the 8 CAMKK2 mutation carriers, and none of the mutations were screened. Therefore, we consider c.1614dup (CAMKK2) might be a promising MOV but needs further confirmation.
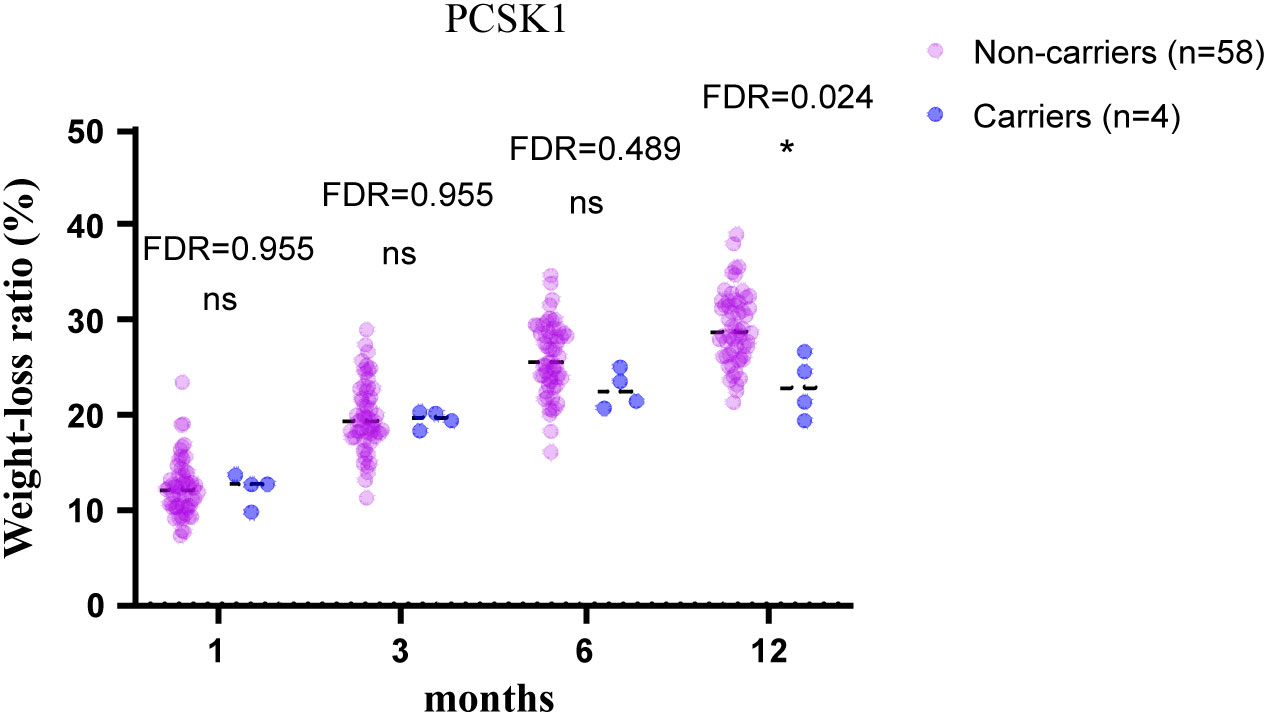
Figure 1 12-month weight-loss ratio for obese individuals with/without the PCSK1 mutation after bariatric surgery. PCSK1 variant led to less weight loss after bariatric surgery compared with PCSK1 non-carriers. Independent t tests were conducted at 1, 3, 6, 12 months, respectively, and the P values were adjusted via the FDR. *FDR<0.05, ns, no significance.
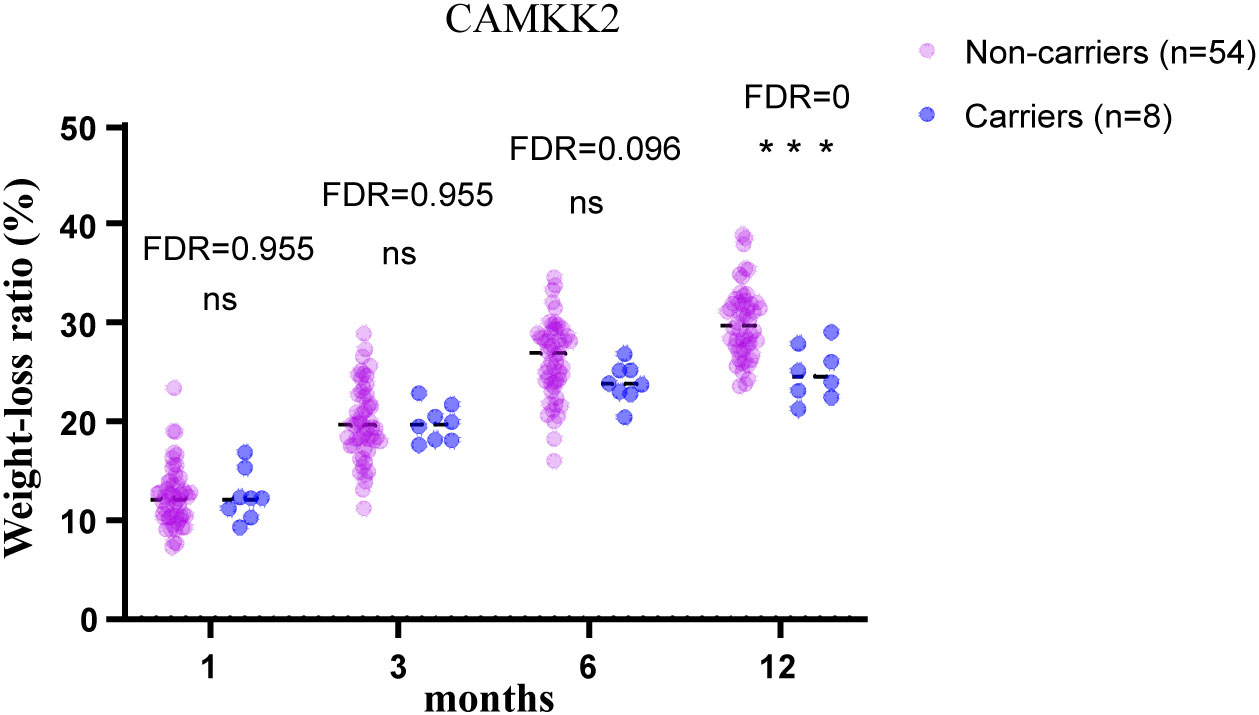
Figure 2 12-month postoperative weight-loss ratio in obese patients with/without CAMKK2 mutation. CAMKK2 variant carriers exhibited poorer weight loss after bariatric surgery compared with CAMKK2 non-carriers. Independent t tests were performed at each time point and the P values were adjusted via the FDR. ***FDR<0.001, ns, no significance.
The calcium/calmodulin dependent protein kinase kinase 2 (CAMKK2) is a member of the serine/threonine protein kinase family and Ca(2+)/calmodulin-dependent protein kinase subfamily requiring the presence of calcium and calmodulin to be active. The c.1614dup (CAMKK2) variant involves the insertion of an A base at position 1615, which is identical to position 1614. This results in the mutation of glycine to arginine at position 539 of the CAMKK2 protein and a premature truncation from 556 amino acids to 540 amino acids. It is important to note that this mutation is predicted to cause nonsense-mediated mRNA decay (NMD), a process by which mRNAs that contain premature termination codons are degraded, indicating that even though it only reduces 16 amino acids, it is critical for the stability of this protein.
To evaluate the effect of c.1614dup (CAMKK2) on metabolism after bariatric surgery, we compared lipid levels between baseline and 12 months post-surgery between carriers and non-carriers of the CAMKK2 mutation (Figure 3A), and compared their percentage change from baseline to 12 months after surgery (Figure 3B). Final data were collected from 43 non-carriers and 7 carriers of the mutation due to loss of follow-up in some patients. Both groups showed a statistically significant increase in HDL cholesterol levels (mean HDL levels increased from 1.15mmol/L to 1.44mmol/L in CAMKK2 mutation non-carriers, FDR = 0, and from 1.17mmol/L to 1.30mmol/L in CAMKK2 carriers, FDR = 0.038) (Figure 3A), and there was a significant difference in the percentage change between the two groups (CAMKK2 non-mutation carriers 27.3%, CAMKK2 mutation carriers 11.5%, FDR = 0.046) (Figure 3B). LDL cholesterol was down-regulated in both groups. However, the changes between baseline and 12 months after surgery were statistically significant in CAMKK2 mutation non-carriers but not in carriers (Figure 3A), likely due to the limitation of the sample size. Furthermore, there was no significant difference in the percentage change between the two groups (Figure 3B). Meanwhile, both groups exhibited a statistically significant decrease in triglyceride (TG) levels (Figure 3A) and the percentage decrease was greater in individuals without the CAMKK2 mutation (29.5%) compared to those with the mutation (14%) (FDR = 0.029) (Figure 3B). However, no significant changes in total cholesterol (TC) levels were observed in either group (Figure 3A) or in percentage change between the two groups (Figure 3B). The results indicate that patients, regardless of whether they carry the CAMKK2 mutation or not, experience improved metabolism after undergoing bariatric surgery. However, the degree of improvement varies, with non-carriers of the CAMKK2 mutation showing significant changes in elevated levels of HDL and reduced levels of TG compared to carriers of the CAMKK2 mutation.
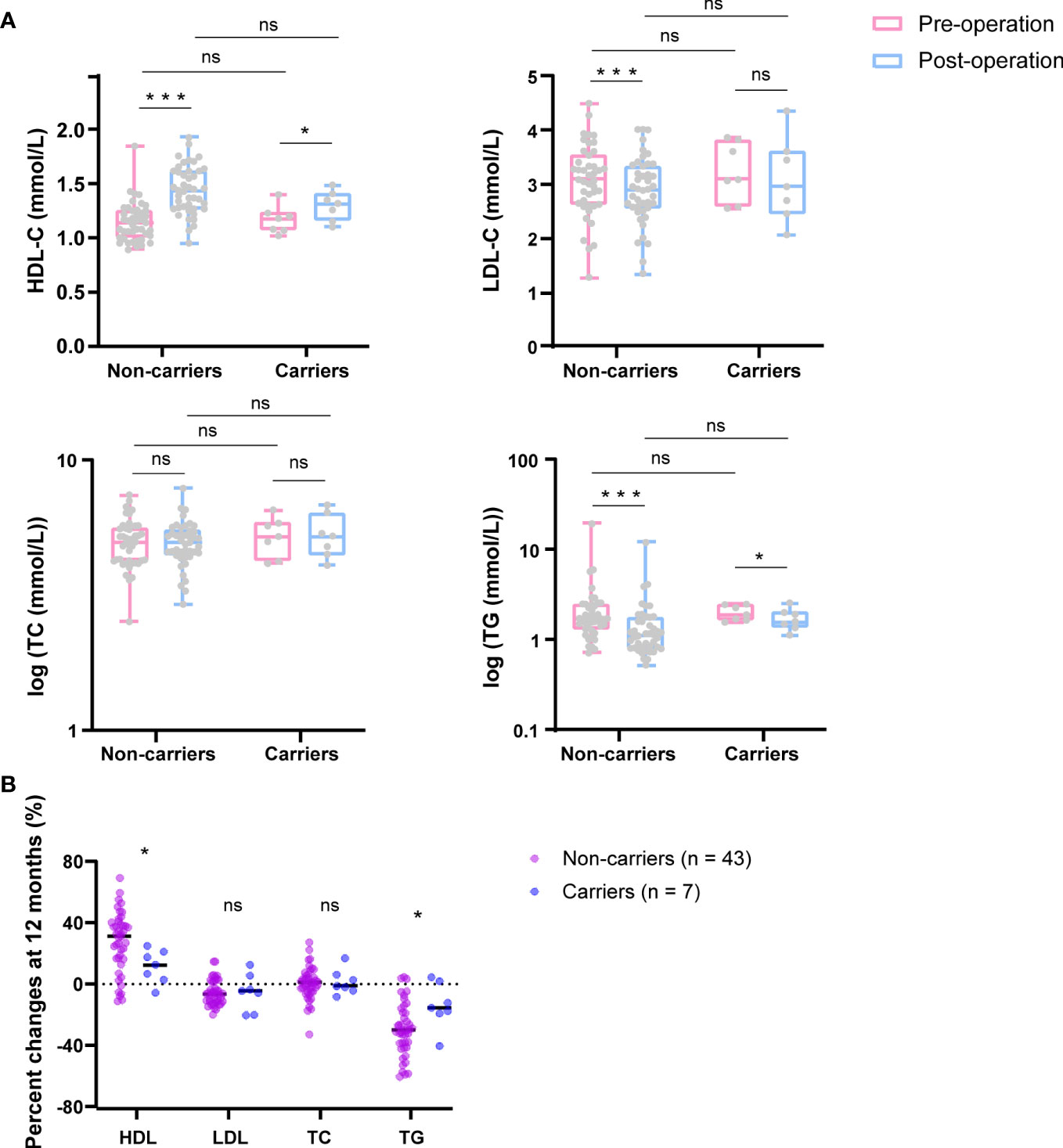
Figure 3 CAMKK2 variant influences metabolism improvement after bariatric surgery. (A) Changes in HDL, LDL, TC, and TG after bariatric surgery (12months) (blue) compared with baseline (pink) between CAMKK2 mutation non-carriers (n = 43) and carriers (n = 7). (B) Percentage changes from baseline to 12 months post-surgery of HDL, LDL, TC and TG. TC and TG were logarithmically transformed using GraphPad Prism 8. Compared t tests and independent t tests were performed, and the P values were adjusted via the FDR. *FDR<0.05, ***FDR<0.001, ns, no significance.
Identifying the underlying causes of early-onset obesity is a challenging task due to the complex interplay of genetic and environmental factors. WES has emerged as an effective tool for detecting novel candidate genes associated with these disorders. In a study conducted by Li et al., the researchers investigated whether MOVs influence the effectiveness of bariatric surgery (7). Similarly, the long-term outcomes of bariatric surgery were evaluated in patients with bi-allelic mutations in known monogenic obesity genes such as POMC, LEPR, and MC4R (20). The findings from both studies suggested that carriers of monogenic obesity gene mutations exhibit less weight loss after bariatric surgery, indicating that bariatric surgery could potentially be used as a screening method for detecting MOVs following WES analysis. In the present study, WES was performed on a cohort of 62 individuals with early-onset obesity and 9 individuals with late-onset obesity. This analysis led to the identification of a known monogenic obesity gene, PCSK, with a mutation that was associated with less weight loss in our study. This finding further validated the effectiveness of using WES combined with postoperative data as a method for detecting candidate MOVs. Then, we selected 5 variants in genes previously reported to be associated with obesity, adipogenesis or linked to obesity-associated traits by genome-wide association studies (SLC25A5, AIM2, SNX16, CAMKK2 and PDE11A) for further analysis to identify candidate MOVs. In combination with weight data after bariatric surgery, we finally identified c.1614dup (CAMKK2) as a novel candidate MOV.
CAMKK2 is expressed both in hypothalamus (17) and preadipocytes (16). It plays a key role in the calcium/calmodulin-dependent (CaM) kinase cascade through the phosphorylation of the downstream kinases, including CaMKI, CaMKIV and AMP-activated protein kinase (AMPK) (17, 21–23). Among these, AMPK is a key regulator of cellular energy balance, which is involved in the leptin-melanocortin signaling pathway (24). CAMKK2 null mice developed obesity, insulin resistance, and less glucose tolerance with standard chow whereas displayed a considerably smaller increase in adiposity and adipocyte size with high-fat diet compared with WT mice by regulating NPY and therefore affecting appetite (17). NPY is a known appetite regulator and its mutation led to obesity and metabolic syndrome (25). In vitro, inhibition or deletion of CAMKK2 in preadipocytes promoted their differentiation into mature adipocytes, which could be rescued by AMPK activation (16). These studies suggest that the CAMKK2 variant carriers identified in this study may have followed a normal diet structure instead of a high-fat diet, which surprisingly lead to weight gain. After bariatric surgery, all patients, including CAMKK2 variant carriers, are instructed to follow a specific dietary structure, which is more closely aligned with a standard diet, rather than a high-fat diet, leading to suboptimal weight loss.
Among the other genes variants that lacked statistical significance, AIM2 necessitates further validation. AIM2, a member of the IFN-inducible HIN200 protein family (26), is a cytoplasmic double-stranded DNA sensor and a tumor suppressor (27, 28). AIM2 knockout mice exhibited increased both subcutaneous and visceral adiposity compared to WT, with no differences in food intake. For the reason, AIM2 null mice showed a reduction in energy expenditure and impaired function of their brown adipose tissue. Furthermore, AIM2 knockout led to insulin resistance and glucose intolerant (14). AIM2 variants have not been reported to cause obesity yet, and in our study, a frameshift variant of AIM2 was found in 4 patients who had a family history of obesity. Although their weight loss after bariatric surgery did not reach statistical significance, it is important to note that the sample size was limited, suggesting additional studies with larger sample sizes or different populations may help to clarify whether AIM2 is a candidate monogenic obesity gene.
Furthermore, in 70 genes predicted pathology, likely pathology and variants of uncertain significance, MPP2 is predominantly expressed in the brain (Genotype-Tissue Expression), which is an established key region in the regulation of energy homeostasis and food intake. However, its role in obesity or metabolic disorders has not been reported and need to be further explored in the future.
In summary, the present investigation indicates that individuals with early-onset obesity have significantly higher BMI and waist circumstance (WC) compared to subjects with late-onset obesity. This data verified the efficiency of WES combined with postoperative data to identify candidate monogenic obesity genes and also identified c.1614dup (CAMKK2) as a promising MOV in a cohort of patients experiencing early-onset obesity. Further research with larger sample sizes is needed to confirm these findings and to determine the long-term effects of the c.1614dup (CAMKK2) variant on metabolism.
WES combined with weight data following bariatric surgery is an effective strategy for identifying novel candidate MOVs and c.1614dup (CAMKK2) might be a new MOV.
The datasets presented in this study can be found in online repositories. The accession number is PRJNA1041755 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1041755).
The studies involving humans were approved by The Ethics Committee of The Third People’s Hospital of Chengdu. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
YW: Investigation, Validation, Writing – original draft, Visualization, Writing – review & editing. CY: Data curation, Validation, Writing – review & editing. JW: Resources, Writing – review & editing. LJ: Investigation, Writing – review & editing. ZR: Resources, Writing – review & editing. TZ: Funding acquisition, Supervision, Writing – review & editing, Project administration. YL: Funding acquisition, Supervision, Writing – review & editing, Project administration.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by The National Natural Science Foundation of China (82370884, 82170887).
The authors thank all patients involved in this project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2024.1334342/full#supplementary-material
1. Piché ME, Tchernof A, Després JP. Obesity phenotypes, diabetes, and cardiovascular diseases. Circ Res (2020) 126(11):1477–500. doi: 10.1161/CIRCRESAHA.120.316101
2. Lega IC, Lipscombe LL. Review: diabetes, obesity, and cancer-pathophysiology and clinical implications. Endocrine Rev (2020) 41(1):bnz014. doi: 10.1210/endrev/bnz014
3. Jebeile H, Kelly AS, O'Malley G, Baur LA. Obesity in children and adolescents: epidemiology, causes, assessment, and management. Lancet Diabetes endocrinol (2022) 10(5):351–65. doi: 10.1016/S2213-8587(22)00047-X
4. Loos RJF, Yeo GSH. The genetics of obesity: from discovery to biology. Nat Rev Genet (2022) 23(2):120–33. doi: 10.1038/s41576-021-00414-z
5. Littleton SH, Berkowitz RI, Grant SFA. Genetic determinants of childhood obesity. Mol diagnosis Ther (2020) 24(6):653–63. doi: 10.1007/s40291-020-00496-1
6. Nguyen NT, Kim E, Vu S, Phelan M. Ten-year outcomes of a prospective randomized trial of laparoscopic gastric bypass versus laparoscopic gastric banding. Ann surgery (2018) 268(1):106–13. doi: 10.1097/SLA.0000000000002348
7. Li Y, Zhang H, Tu Y, Wang C, Di J, Yu H, et al. Monogenic obesity mutations lead to less weight loss after bariatric surgery: a 6-year follow-up study. Obes Surg (2019) 29(4):1169–73. doi: 10.1007/s11695-018-03623-4
8. Group of China Obesity Task Force. Body mass index reference norm for screening overweight and obesity in Chinese children and adolescents. Chin J Epidemiol (2004) 25:97–102.
9. Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinf (2010) 11:548. doi: 10.1186/1471-2105-11-548
10. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med (2015) 17(5):405–24. doi: 10.1038/gim.2015.30
11. Folon L, Baron M, Toussaint B, Vaillant E, Boissel M, Scherrer V, et al. Contribution of heterozygous PCSK1 variants to obesity and implications for precision medicine: a case-control study. Lancet Diabetes endocrinol (2023) 11(3):182–90. doi: 10.1016/S2213-8587(22)00392-8
12. Ramos-Molina B, Martin MG, Lindberg I. PCSK1 variants and human obesity. Prog Mol Biol Trans science (2016) 140:47–74. doi: 10.1016/bs.pmbts.2015.12.001
13. Zhu S, Wang W, Zhang J, Ji S, Jing Z, Chen YQ. Slc25a5 regulates adipogenesis by modulating ERK signaling in OP9 cells. Cell Mol Biol letters (2022) 27(1):11. doi: 10.1186/s11658-022-00314-y
14. Gong Z, Zhang X, Su K, Jiang R, Sun Z, Chen W, et al. Deficiency in AIM2 induces inflammation and adipogenesis in white adipose tissue leading to obesity and insulin resistance. Diabetologia (2019) 62(12):2325–39. doi: 10.1007/s00125-019-04983-x
15. Wu Y, Wang W, Jiang W, Yao J, Zhang D. An investigation of obesity susceptibility genes in Northern Han Chinese by targeted resequencing. Medicine (2017) 96(7):e6117. doi: 10.1097/MD.0000000000006117
16. Lin F, Ribar TJ, Means AR. The Ca2+/calmodulin-dependent protein kinase kinase, CaMKK2, inhibits preadipocyte differentiation. Endocrinology (2011) 152(10):3668–79. doi: 10.1210/en.2011-1107
17. Anderson KA, Ribar TJ, Lin F, Noeldner PK, Green MF, Muehlbauer MJ, et al. Hypothalamic CaMKK2 contributes to the regulation of energy balance. Cell Metab (2008) 7(5):377–88. doi: 10.1016/j.cmet.2008.02.011
18. Najar MA, Rex DAB, Modi PK, Agarwal N, Dagamajalu S, Karthikkeyan G, et al. A complete map of the Calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) signaling pathway. J Cell communication Signaling (2021) 15(2):283–90. doi: 10.1007/s12079-020-00592-1
19. Ohlsson T, Lindgren A, Engström G, Jern C, Melander O. A stop-codon of the phosphodiesterase 11A gene is associated with elevated blood pressure and measures of obesity. J hypertension (2016) 34(3):445–51; discussion 51. doi: 10.1097/HJH.0000000000000821
20. Poitou C, Puder L, Dubern B, Krabusch P, Genser L, Wiegand S, et al. Long-term outcomes of bariatric surgery in patients with bi-allelic mutations in the POMC, LEPR, and MC4R genes. Surg Obes related Dis (2021) 17(8):1449–56. doi: 10.1016/j.soard.2021.04.020
21. Ye C, Zhang D, Zhao L, Li Y, Yao X, Wang H, et al. CaMKK2 suppresses muscle regeneration through the inhibition of myoblast proliferation and differentiation. Int J Mol Sci (2016) 17(10):1695. doi: 10.3390/ijms17101695
22. Matsushita M, Nairn AC. Characterization of the mechanism of regulation of Ca2+/ calmodulin-dependent protein kinase I by calmodulin and by Ca2+/calmodulin-dependent protein kinase kinase. J Biol Chem (1998) 273(34):21473–81. doi: 10.1074/jbc.273.34.21473
23. Marcelo KL, Means AR, York B. The ca(2+)/calmodulin/caMKK2 axis: nature's metabolic camshaft. Trends Endocrinol metabolism: TEM (2016) 27(10):706–18. doi: 10.1016/j.tem.2016.06.001
24. Pollard AE, Martins L, Muckett PJ, Khadayate S, Bornot A, Clausen M, et al. AMPK activation protects against diet induced obesity through Ucp1-independent thermogenesis in subcutaneous white adipose tissue. Nat Metab (2019) 1(3):340–9. doi: 10.1038/s42255-019-0036-9
25. Olza J, Gil-Campos M, Leis R, Rupérez AI, Tojo R, Cañete R, et al. Influence of variants in the NPY gene on obesity and metabolic syndrome features in Spanish children. Peptides (2013) 45:22–7. doi: 10.1016/j.peptides.2013.04.007
26. Cresswell KS, Clarke CJ, Jackson JT, Darcy PK, Trapani JA, Johnstone RW. Biochemical and growth regulatory activities of the HIN-200 family member and putative tumor suppressor protein, AIM2. Biochem Biophys Res Commun (2005) 326(2):417–24. doi: 10.1016/j.bbrc.2004.11.048
27. Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature (2009) 458(7237):514–8. doi: 10.1038/nature07725
Keywords: monogenic obesity, WES, bariatric surgery, CAMKK2, weight loss
Citation: Wang Y, Yang C, Wen J, Ju L, Ren Z, Zhang T and Liu Y (2024) Whole-exome sequencing combined with postoperative data identify c.1614dup (CAMKK2) as a novel candidate monogenic obesity variant. Front. Endocrinol. 15:1334342. doi: 10.3389/fendo.2024.1334342
Received: 07 November 2023; Accepted: 09 January 2024;
Published: 26 February 2024.
Edited by:
Yayun Wang, Air Force Medical University, ChinaReviewed by:
Søren Madsen, The University of Sydney, AustraliaCopyright © 2024 Wang, Yang, Wen, Ju, Ren, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tongtong Zhang, MTYzenR0b25nQDE2My5jb20=; Yanjun Liu, bGl1eWFuanVuQHN3anR1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.