- College of Medicine, Alfaisal University, Riyadh, Saudi Arabia
Type 2 diabetes mellitus (T2DM) is a rapidly escalating global health concern, with its prevalence projected to increase significantly in the near future. This review delves into the intricate role of epigenetic modifications - including DNA methylation, histone acetylation, and micro-ribonucleic acid (miRNA) expression - in the pathogenesis and progression of T2DM. We critically examine how these epigenetic changes contribute to the onset and exacerbation of T2DM by influencing key pathogenic processes such as obesity, insulin resistance, β-cell dysfunction, cellular senescence, and mitochondrial dysfunction. Furthermore, we explore the involvement of epigenetic dysregulation in T2DM-associated complications, including diabetic retinopathy, atherosclerosis, neuropathy, and cardiomyopathy. This review highlights recent studies that underscore the diagnostic and therapeutic potential of targeting epigenetic modifications in T2DM. We also provide an overview of the impact of lifestyle factors such as exercise and diet on the epigenetic landscape of T2DM, underscoring their relevance in disease management. Our synthesis of the current literature aims to illuminate the complex epigenetic underpinnings of T2DM, offering insights into novel preventative and therapeutic strategies that could revolutionize its management.
1 Introduction
Type 2 diabetes mellitus (T2DM) is a persistent metabolic condition associated with increasing concentrations of glucose in the blood characterized by impaired insulin action and/or inadequate insulin secretion. The global prevalence of T2DM has been rising quickly. A study by Saeedi et al. has projected the global incidence of T2DM to reach 8% by 2030 and 8.6% by 2045 (1). Therefore, there is a critical need for preventative measures to reduce the global health burden of the disease.
Epigenetics is a field that describes functional changes to the genome without changes to the nucleotide sequences of the deoxyribonucleic acid (DNA) (2). Several epigenetic modifications have been identified. The most commonly studied mechanism is DNA methylation, a crucial epigenetic modification, primarily functions to regulate gene expression by adding methyl groups to cytosine residues in DNA, often leading to the suppression of gene activity and affecting cellular differentiation, development, and disease pathogenesis (3). This process almost always occurs in CpG regions, where 70% of methylation events occur (4). Epigenetic modifications encompass a diverse range of mechanisms that regulate gene expression at the chromatin level. One significant type of epigenetic modification is histone acetylation. This process involves the addition of an acetyl group to the lysine residues of histone proteins, an action catalyzed by enzymes known as lysine acetyltransferases (5). Histone acetylation plays a crucial role in modulating the accessibility of chromatin and thereby influencing gene transcription. Beyond acetylation, the chromatin landscape is shaped by various other epigenetic modifications, including methylation, phosphorylation, ubiquitination, and sumoylation, each contributing uniquely to the regulation of gene expression.
Additionally, non-coding ribonucleic acids (ncRNAs) represent another essential facet of epigenetics (3). These RNA molecules, which are transcribed but not translated into proteins, are involved in genetic silencing. Beyond this, emerging evidence highlights the role of ncRNAs in orchestrating epigenetic modifications, further underscoring their significance in gene regulation (3, 6).
In addition to these mechanisms, the epitranscriptome represents a burgeoning area of interest in epigenetics, particularly in its relation to T2DM (7). The epitranscriptome involves modifications to RNA molecules, such as methylation, that do not alter their primary sequence but can significantly impact RNA stability, localization, and translation (8). These RNA modifications are akin to epigenetic changes as they regulate gene expression post-transcriptionally and are dynamically reversible (8). Such epitranscriptomic modifications offer another layer of genetic regulation and have been increasingly implicated in the pathogenesis of metabolic disorders like T2DM (8, 9).
Data have suggested that these epigenetic modifications are essential to the development of several diseases (10). Among these diseases, recent epigenetic studies have highlighted the role of epigenetic modifications in the pathogenesis of T2DM (11–13). Additionally, these mechanisms could be used to predict the risk of T2DM and, potentially, as therapeutic targets (14–16). The association between epigenetic modifications and T2DM is subject to ongoing debate. This uncertainty largely stems from the limited scope of existing literature and research in this area. As a result, establishing a definitive causal relationship between epigenetic changes and the development of T2DM remains a complex and unresolved issue (17, 18). However, a recent study by Juvinao-Quintero et al. identified a causal effect of the 24-dehydrocholesterol reductase gene DHCR24 (a gene linked to lipid metabolism) methylation in T2DM risk (11). Therefore, studying and targeting the epigenetics associated with T2DM could open the door for preventative and therapeutic measures.
In this article, we synthesize the latest literature describing the role of epigenetics in various aspects of T2DM pathogenicity, including obesity, insulin resistance (IR), β-cell dysfunction, cellular senescence, and mitochondrial dysfunction. Then, we describe the epigenetics mechanisms through which exercise and dietary choices influence the risk of T2DM. Finally, we also describe the role of epigenetics in T2DM-related complications, including atherosclerosis, diabetic retinopathy, neuropathy, and cardiomyopathy.
2 Epigenetics in obesity
Obesity has been steadily increasing in the past decades, reaching a global prevalence rate of 14% in 2019 (19). Obesity is associated with several pathologies, including ischemic heart disease, non-alcoholic fatty liver disease, chronic kidney disease, several cancers, and T2DM (19–22). The mechanisms linking obesity and T2DM are well-established and have been reviewed extensively (23–25).
Epigenetic modifications have been shown to play a significant role in the development of obesity. Xu et al. conducted a genome-wide methylation on blood samples from 48 obese and 48 lean individuals aged 14-20 years (26). The authors identified more than 20,000 differentially methylated (DMC) and differentially variable (DVC) CpG sites associated with obesity status. Both DMC and DVC sites were able to predict obesity case-control status in the cohort (26). Lastly, genes containing DMCs and DVCs showed significant enrichment of genes associated with obesity-related disorders, including hypertension, dyslipidemia, and T2DM (26). Dagaard et al. uncovered a pivotal epigenetic mechanism that triggers obesity (27). They identified a Trim28-dependent network, which operates in a non-Mendelian, “on/off” manner, fundamentally influencing body weight distribution. This network is characterized by the decreased expression of key imprinted genes, including Nnat, Peg3, Cdkn1c, and Plagl1. Intriguingly, manipulating these genes can replicate the observed bi-stable, stochastic obesity phenotype (27). Their findings not only shed light on the enigmatic heritability of obesity but also highlight the potential for discrete polyphenism in obesity’s genetic underpinnings, both in mice and humans. Dick et al. identified five CpG cites associated with increased body mass index (BMI); among which, three are located within HIF3A (28). HIF3A is a transcription factor for hypoxia inducible factor-3 alpha responsible for regulating the body’s response to hypoxia (2). More recently, Janjanam et al. found that DMC sites on NANOS1 and SOX14 in newborns can modulate the risk of increased body weight at later stages of life (29). Methylation of specific CpG sites is not only associated with increased risk of obesity but can result in decreased risk of obesity as well. For example, Huang et al. found that a 1% increase in methylation in TAOK3 decreased the odds of child obesity by 0.91 (30). The TAOK3 gene encodes the STE20-type protein kinase TOAK3, which exacerbates lipid storage and oxidative/endoplasmic stress in hepatocytes (31).
Several studies have also highlighted the role of histone acetylation in obesity. Hypertrophied adipocytes, the main cause of obesity, exhibit increased acetylation of histone H3 at lysine 9 and global hypomethylation of DNA (32). A study by Pessoa Rodrigues et al. studied the role of the histone H4 lysine 16 acetyltransferase MOF in metabolic regulation (33). Although Mof depletion resulted in resistance to weight gain, these modifications led to an insulin-glucose imbalance, resulting in T2D predisposition. MOF was also found to promote the proliferator-activated receptor-γ (PPARγ) and downstream transcriptional signals. Additionally, treatment with the PPARγ agonist thiazolidinedione restored glucose uptake in Mof-deficient mice (33). Collectively, these studies indicate that epigenetics play a key role in the development of obesity and obesity-related diseases (Figure 1).
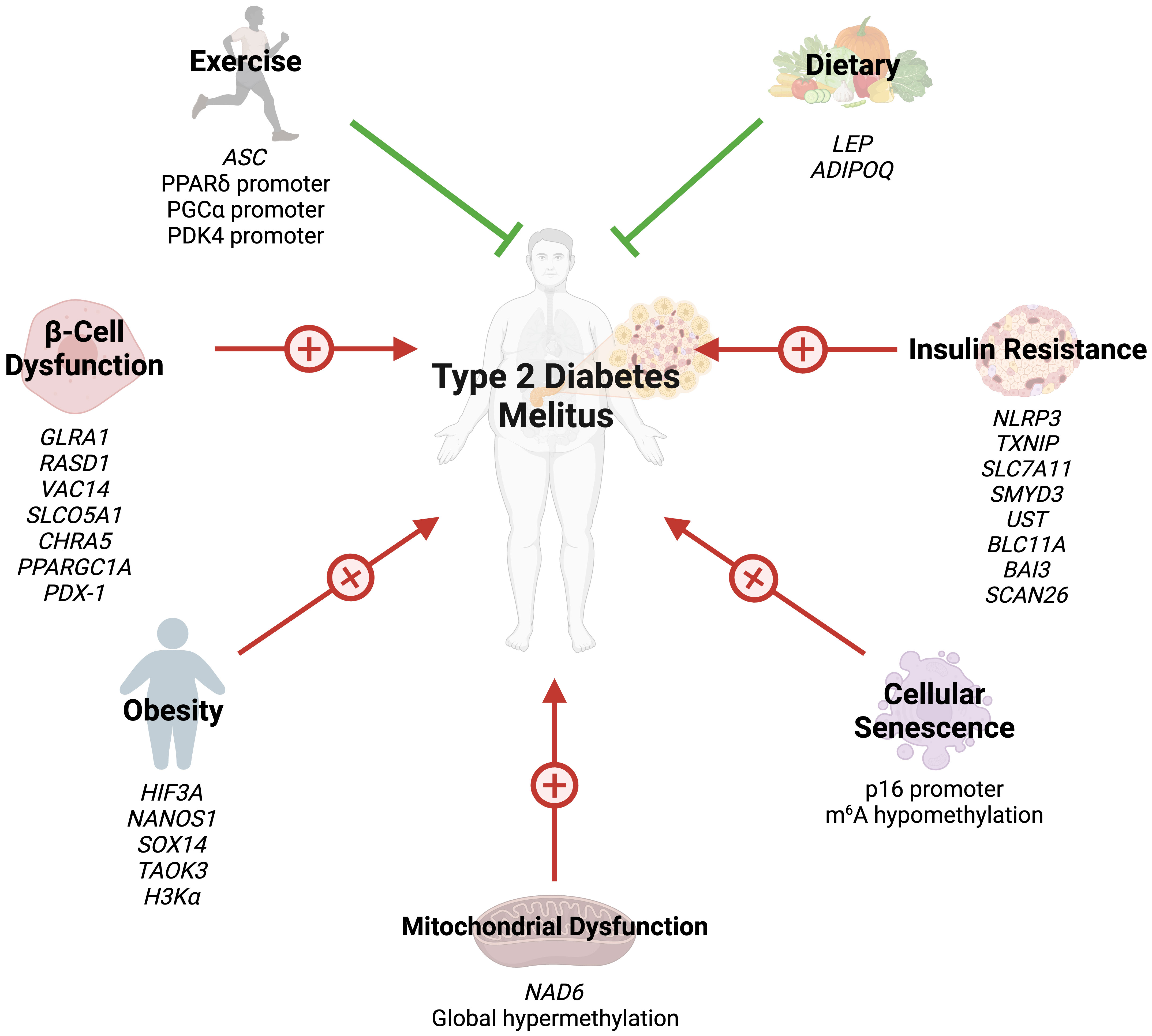
Figure 1 Several epigenetic modifications can modulate the risk of T2DM through obesity, insulin resistance, beta-cell dysfunction, cellular senescence, mitochondrial dysfunction, exercise, and dietary choices.
3 Epigenetics in insulin resistance and β-cell dysfunction
The cornerstones of T2DM pathogenesis are IR and β-cell dysfunction, resulting in reduced insulin secretion (34). Understanding the various pathways that contribute to beta cell homeostasis and failure is essential for determining the pathophysiology of T2DM. β-cells of the pancreas synthesize insulin and store insulin. Through the action of the glucose transporter GLUT2, glucose enters β-cells where it is generates adenosine triphosphate (ATP) through the process of glycolysis. When the ATP/adenosine diphosphate ratio rises in hyperglycemic states, ATP-operated potassium channels are closed, depolarizing the cell membrane and opening voltage-gated calcium channels. Calcium influx triggers insulin release from secretory granules, which in turn regulates glucose levels in physiologic conditions (35).
IR is often the first trigger of T2DM and is initially offset by marked insulin release (36). However, increased insulin demands eventually lead to β-cell failure and T2DM progression. The role of epigenetics in both IR and β-cell failure has been described in the literature. Global DNA methylation in lymphocytes and visceral adipose tissue positively correlates with IR (37). On the other hand, a cross-ancestry epigenome-wide association study identified five CpG sites inversely associated with IR: three in TXNIP, one in SLC7A11, and another in ZSCAN26 (38). TXNIP encodes for thioredoxin-interacting protein (TXNIP) which promotes β-cell apoptosis in hyperglycemic states (39). TXNIP depletion has been found to promote Akt/Bcl-xL pathways and β-cell mass, hence protecting against T2DM in mice (40). Zhang et al. compared DNA methylation in insulin sensitive and IR obese women (41). The authors identified four significant DMC sites: SMYD3, UST, BCL11A, and BAI3, which are involved in glycosaminoglycan and chondroitin/dermatan sulfate synthetic processes.
β-cell mass is reduced by up to 60% in T2DM (42), and loss of cellular function further progresses the condition (43). As previously described, several pathways regulating β-cell mass can be modified by epigenetics (38). The insulin promoter gene is methylated in human pancreatic islet cells of T2DM patients (44). Glycated hemoglobin levels correlate both positively and inversely with methylation of several CpG sites in the same patients (44). Additionally, GLRA1, RASD1, VAC14, SLCO5A1, and CHRNA5 genes in human islet cells exhibit DNA methylation changes after exposure to high glucose levels (45). Similarly, DNA methylation of PPARγ PPARGC1A gene promoter in islet cells is near double in T2DM patients compared to their non-diabetic counterparts (46).
The transcription factor pancreatic duodenal homeobox-1 (PDX-1) plays a key role in pancreatic development and regulates mature islet cell physiology (47). Knockout of the PDX-1 gene results in failure of pancreatic bud formation and mice death from hyperglycemia (47). Epigenetic modifications of PDX-1 in pancreatic cells have been extensively studied in T2DM. PDX-1 methylation is significantly increases after high glucose exposure in pancreatic islet cells (45). Additionally, PDX-1 methylation is significantly greater in T2DM pancreas cells (48).
β-cell malfunction and apoptosis are strongly linked to inflammation, especially low-grade chronic inflammation (49). Oxidative stress, which is characterized by an imbalance between the production of reactive oxygen species (ROS) and antioxidant defense systems, has a significant impact on β-cell failure. In addition to causing β-cell death and disrupting signaling pathways, ROS can reduces insulin secretion and predisposes patients to T2DM (50). The epigenetic link between inflammation and β-cell is understudied; however, emerging evidence has highlighted the role of epigenetics in this relationship. For example, IL-1β induces abnormal DNA methylation of islet cells, and treatment with methyltransferase inhibitors partially reverses IL-1β-induced β-cell dysfunction (51, 52).
MicroRNAs (miRNAs) are pivotal in the epigenetic interplay between inflammation and β-cell function, a critical facet in the pathogenesis of T2DM. The deregulation of specific miRNAs, such as miR-126 and miR-146a, which are notably downregulated in T2DM patients, exemplifies this connection (12). The alteration in the expression levels of these miRNAs can be attributed to epigenetic modifications, such as DNA methylation and histone modifications, which affect the transcription and stability of miRNA genes. For instance, miR-126, which has demonstrated high diagnostic accuracy for T2DM with a sensitivity of 91% and specificity of 97%, is subject to such epigenetic regulation (12).
Moreover, miR-375, predominantly expressed in the human pancreas, is regulated epigenetically through promoter methylation and interactions with circular RNAs (53). This miRNA, a key regulator of inflammation, has been implicated in various diseases including Grave’s disease, type 1 diabetes mellitus, and inflammatory bowel disease, and its levels are associated with insulin resistance markers in obese children (53, 54).
Additionally, miR-200 plays a proinflammatory role in vascular smooth cells of diabetic mice and is involved in β-cell damage and enhanced insulin resistance, with its effects being mediated through epigenetic modulation of signaling pathways (55–57). Furthermore, the knockdown of miRNAs such as -24, -26, -182, and -148 leads to a decrease in insulin promoter activity and insulin mRNA expression, further illustrating the epigenetic influence on miRNA-mediated regulation of β-cell function (58).
Hence, miRNAs serve not only as key components in the inflammatory response associated with T2DM but also as integral players in the epigenetic regulation of genes crucial for insulin secretion and β-cell function. Their potential utility as diagnostic and therapeutic markers in T2DM underscores the importance of understanding their epigenetic regulation mechanisms. The role of epigenetics in IR and β-cell failure is summarized in (Figure 1).
4 Epigenetics and cellular senescence in T2DM
Cellular senescence is a cellular hallmark of aging and is defined by irreversible cell cycle arrest due to telomere attrition (21). Cell senescence results in rapid development of age-related comorbidities and is associated with several conditions, including obesity, chronic kidney disease, and cancer (21, 59). Cellular senescence is mediated by upregulation of several cell cycle inhibitors, including p16INK4a, p15INK4b, p19ARF, and p21CIP (60). Markers of cellular senescence are significantly increased in T2DM β-cells (61). Additionally, glycated hemoglobin, a marker of long-term diabetic control, is negatively correlated with telomere length in T2DM patients (62). Global DNA methylation of β-cells increases with age (63). Specifically, promoters of β-cell proliferation become methylated with increasing age (63).
Studies assessing the role of epigenetic modifications of cellular senescence markers in T2DM are rare. N6-methyladenosine (m6A) refers to methylation of adenosine at the nitrogen-6 position and is the most common internal messenger RNA modification in humans (64). Alterations in m6A promote ROS, telomere shortening, DNA damage, and pro-inflammatory functions, eventually leading to cellular senescence (65). T2DM results in m6A hypomethylation of genes involved in cell cycle progression, insulin release, and several insulin signaling pathways (66). Reduced m6A in β-cells results in cell cycle arrest and impairs insulin release via decreased AKT phosphorylation and PDX1 protein levels (66). This modification also enhances insulin messenger RNA translation, and depletion has been associated with T2DM-like states (67). Dietary changes can also hinder cellular senescence through epigenetic changes. Li et al. demonstrated that glucose restriction decreases p16 expression by exerting changes in histone acetylation and p16 promoter methylation (68). Collectively, these findings reveal that epigenetics regulate cellular aging in T2DM (Figure 1). However, further studies are needed to identify additional epigenetic changes in T2DM-associated cellular senescence.
5 Epigenetics and mitochondrial dysfunction in T2DM
Due to their key role in metabolic functions, it is unsurprising that mitochondrial dysfunction is a common characteristic among T2DM. T2DM and obesity patients have less mitochondria compared to their lean, non-diabetic counterparts (69). T2DM patients also exhibit significant reductions in electron transport chain activity (69). Furthermore, T2DM mitochondria demonstrate no response to insulin stimulus in several studies (70, 71). Mitochondria are the primary sources of ROS, which retards GLUT4 translocation and insulin signaling pathways (72, 73).
Recently, studies have shed light on the role of epigenetic modifications of mitochondrial DNA (mtDNA) in the context of T2DM. mtDNA is hypermethylated by hyperglycemic conditions (74). NADH-dehydrogenase-6 (NAD6), the only protein encoded by mtDNA L-strand, is greatly reduced in obese and T2DM populations, as a result of hypermethylation of NAD6 (75). Hypermethylation is promoted by free fatty acids which induce DNA methyltransferase-1 mitochondrial translocation via AMPK activation (75). Inhibiting NAD6 results in systemic IR, and DNA methyltransferase-1 inhibition exerts opposite effects and improves insulin sensitivity in mice (75). mtDNA methylation results in capillary cell apoptosis and treatment with a DNA methyltransferase-1 inhibitor blunts these effects in diabetic retinopathy (74). These findings demonstrate that mtDNA epigenetic modifications can play a crucial role in the development of IR (Figure 1).
6 Epigenetic modifications related to exercise and diet
Emerging evidence has highlighted the effects of exercise on epigenetic modifications, specifically DNA methylation (Figure 1). Although several cross-sectional studies have identified epigenetic modifications related to exercise, their findings are greatly varying and contradictory (76). However, interventional studies have provided more clarity. Barrès et al. studied the effects of acute exercise on DNA methylation in human skeletal muscles (76). Acute exercise induced hypomethylation of PGCα, PDK4, and PPARδ promoters, resulting in increased expression of the proteins in a dose-dependent manner. Similar results can be seen in chronic exercise, which promotes PDK4 expression in normal patients, but not diabetics (77). Chronic exercise also enhances ASC gene methylation, which is responsible for IL-1β and -18 production (78).
Dietary choices can also exert their impact on the epigenetics of T2DM. Nilsson et al. revealed DNA hypomethylation with concurrently reduced folate levels in T2DM levels (79). The authors hypothesized that reduced folate levels could lead to these epigenetic changes. Fasting has been shown to alter the epigenetic profile of humans. Hjort et al. found that 36 hour fasting increases methylation of LEP (leptin) and ADIPOQ (adiponectin) in the subcutaneous adipose tissue of patients with normal birth weight (80). The choice of dietary fat type also plays a great role in epigenetic modifications. Alexander et al. conducted a randomized, controlled trial where patients were either given excessive saturated fatty acids or polyunsaturated fatty acids (81). The methylation of 4875 CpG sites was affected differently between the two arms, and the mean methylation of 1444 genes in adipose tissue was altered by overeating. Finally, the baseline methylation of 12 CpG sites (9 genes) was associated with the degree of weight gain during the trial (81). Collectively, these findings demonstrate that lifestyle factors modify the epigenetic profiles of humans, possibly preventing or promoting the development of T2DM.
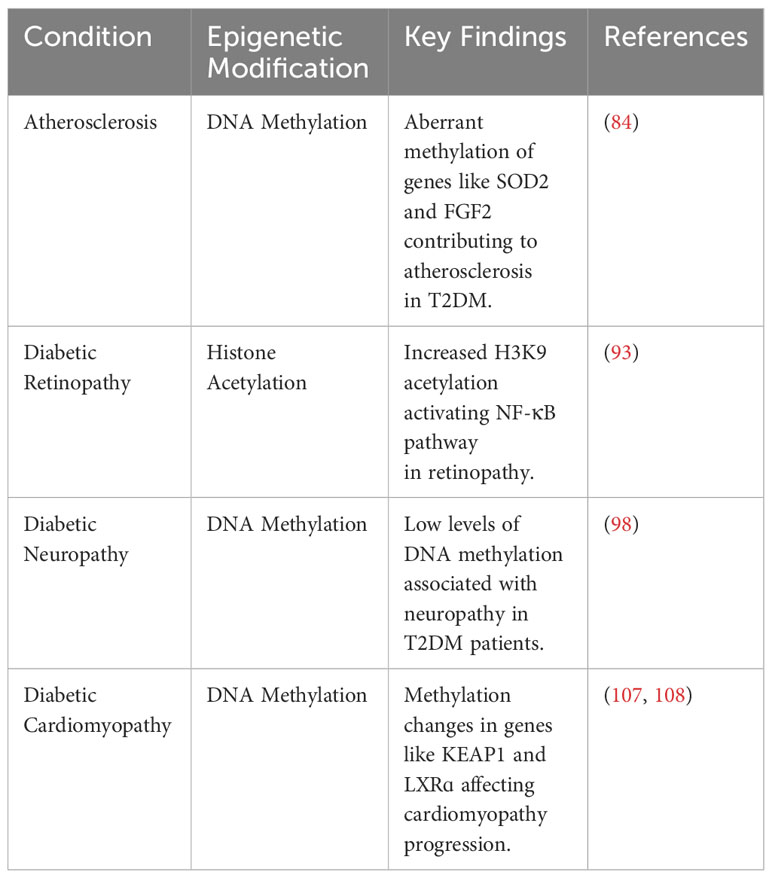
7 Epigenetic modifications in the complications of T2DM
T2DM is associated with a wide spectrum of complications, including retinopathy, nephropathy, neuropathy, atherosclerosis, and cardiomyopathy (82). In the following section, we summarize how epigenetic mechanisms promote and modify T2DM-related complications (Figure 2).
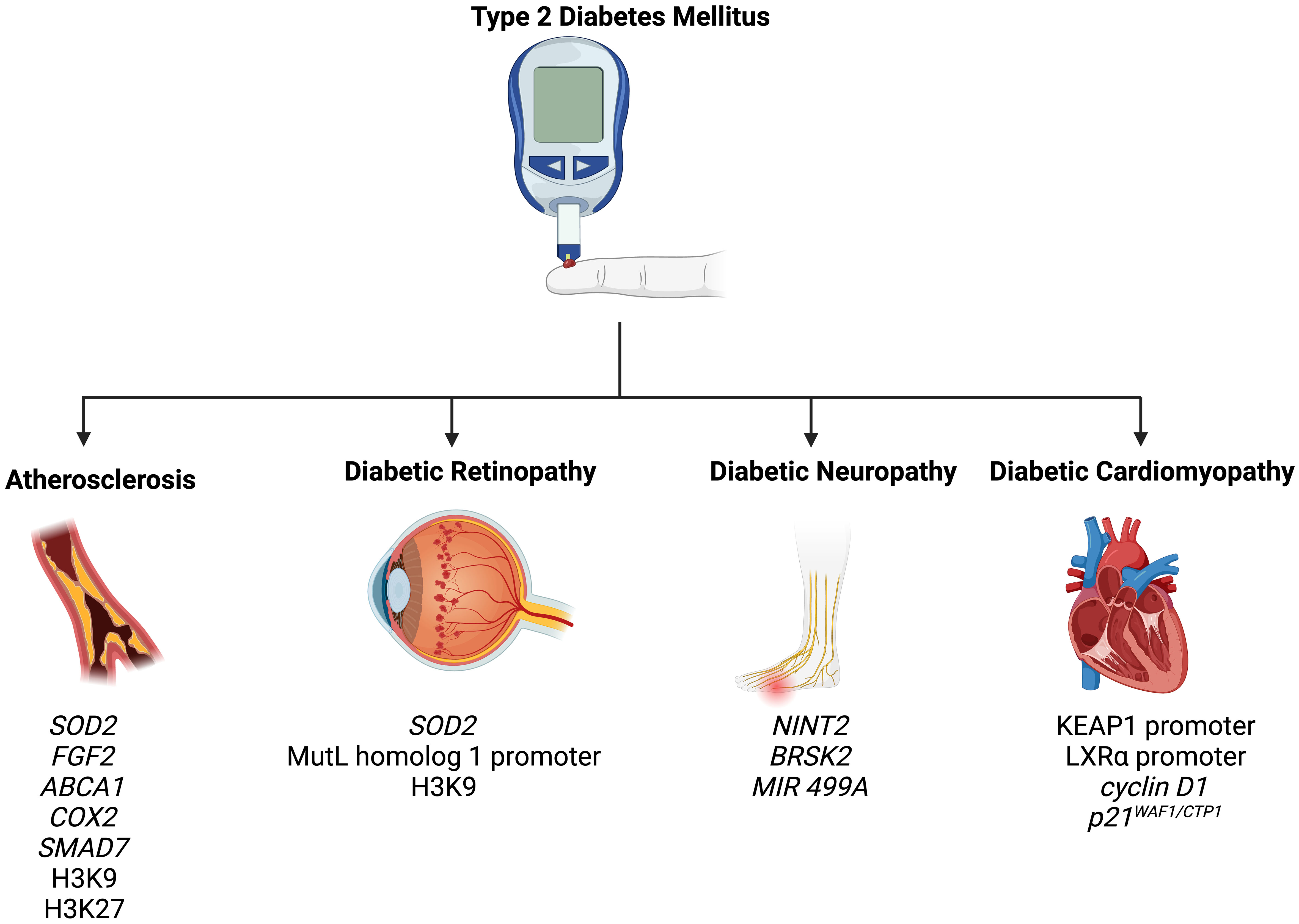
Figure 2 Epigenetic modifications play a role in the pathogenesis of T2DM-related complications, including atherosclerosis, diabetic retinopathy, diabetic neuropathy, and diabetic cardiomyopathy.
7.1 Atherosclerosis
Atherosclerosis is an extremely common complication of T2DM, presenting in nearly 30% of patients (83). Dyslipidemia, hyperglycemia, and chronic inflammation collectively play major roles in atherosclerotic plaque formation in T2DM (84). Aberrant methylation of several genes, including SOD2, FGF2, ABCA1, COX2, and SMAD7, have been identified as key progressor of T2DM-related atherosclerosis (84). Increased acetylation of histone 3 lysine 9 and histone 3 lysine 27 is seen in atherosclerotic plaques (85). Histone 3 lysine 9 acetylation in smooth muscles and macrophages is associated with greater severity of atherosclerosis (85). On the other hand, reduced methylation of the same histones is noted in such plaques.
Several miRNAs can be used as biomarkers of coronary artery disease (CAD). miR -126, -208a, -210, and -370 have shown promising diagnostic accuracy in CAD diagnosis among T2DM patients (86–88). Similarly, miR -195-5p and -451a have been used to detect transient and acute ischemic strokes in diabetic patients (89). miR -32-3p, -106-5p, -1246, and -532-5p have also been utilized as markers of acute ischemic strokes in the general population (90). These studies highlight the promising role of epigenetics as diagnostic markers for atherosclerosis in T2DM patients.
7.2 Diabetic retinopathy
Retinopathy is a significant diabetic consequence that can impair vision and lead to blindness. Diabetic retinopathy is associated with global DNA methylation, which is predictive of for the disease (odds ratio: 1.53) (91). Methylation of the promoter for MutL homolog 1, an enzyme responsible for repairing base mismatches in mtDNA, is increased in diabetic retinopathy, preventing the enzyme from correcting mtDNA damage (92). These effects, however, are reversed by administration of DNA methyltransferase-1 inhibitors. Increased histone 3 lysine 9 acetylation promotes diabetic retinopathy by activating the proinflammatory NF-κB pathway (93). Additionally, methylation of histone 4 lysine 20 at SOD2 (superoxide dismutase-2) downregulates the antioxidant, exacerbating diabetic retinopathy and reinforcing metabolic memory of retinal cells (93). Several miRNAs have also been identified as potential biomarkers in the sera and plasma of diabetic retinopathy patients, such as miR -21, -27b, -126, and -190-5p, further solidifying the role of epigenetics in diabetic retinopathy (94).
7.3 Diabetic neuropathy
Diabetic neuropathy is the most common and debilitating complication of T2DM, present in more than one-third of diabetics (95–97). Low levels of DNA methylation is associated with diabetic neuropathy in T2DM patients (98). Recent studies have highlighted that diabetes mellitus effects genes crucial for nerve regeneration and functionality. For example, Gastoł et al. demonstrated that type 1 diabetics with neuropathy have greater methylation of NINJ2 (ninjurin-2, responsible for nerve regeneration) and lower methylation of BRSK2 (BR serine/threonine kinase-2, responsible for nerve functions) than diabetics without neuropathy (99). Reduced mtDNA copies has also been linked to diabetic neuropathy and is associated with the homozygous variant genotype for the polymorphism rs3746444 of MIR499A (100). Genetic variations in miR -146a, -128a, and -27 also increase susceptibility to diabetic neuropathy among T2DM patients (101).
Several miRNAs have fallen under the spotlight in patients with diabetic neuropathy. miR -33 and -380 are expressed in nociceptive neurons and are key mediators of diabetic pain (102). Circulating miRNAs, such as miR -199a-3p and -499A, could potentially be utilized to detect diabetic retinopathy and have been implicated in the disease (103). However, further studies are needed to determine their diagnostic accuracy in T2DM patients.
7.4 Diabetic cardiomyopathy
Diabetic cardiomyopathy is defined as myocardial disease in diabetic patients which cannot be attributed to any common cardiovascular etiologies or risk factors (104). The condition can be seen in 12% of diabetics and its prevalence is expected to rise exponentially in the coming years (105). The role of epigenetics in diabetic cardiomyopathy has been demonstrated by several studies. Tao et al. found that DNA methyltransferase-1 triggers cardiac fibroblast autophagy in diabetic cardiac fibrosis models by inhibiting the androgen receptor axis (106). Inhibiting the enzyme restores increases androgen receptor expression in these models. Promoter DNA demethylation of KEAP1 is seen in the myocardium of diabetic patients (107). Kelch-like ECH-associated protein-1 (KEAP1) binds to NF-E2-related factor 2 (Nrf2), resulting in its proteasomal degradation (107). Nrf2 activates several antioxidant enzymes; hence, its degradation is key to diabetic cardiomyopathy progression. Expression of the protein liver X receptor-ɑ (LXRɑ) gradually increases with diabetic cardiomyopathy progression as a result of promoter demethylation (108). Chronic activation of LXRɑ induces β-cell apoptosis and, accordingly, predisposes to T2DM (109). Interestingly, LXRɑ improves myocardial glucose tolerance and blunts cardiac hypertrophy in T2DM mice (110). It could be possible that LXRɑ mediates protective effects in acute settings and eventually becomes pathogenic in more chronic settings. Further studies are required to demonstrate the role of LXRɑ and associated epigenetic modifications in diabetic cardiomyopathy. Epigenetics also influence cardiac cell cycle arrest in diabetic cardiomyopathy. Complete methylation of the cyclin D1 and complete demethylation of p21WAF1/CIP1 gene have been noted in early in the development of the condition (111).
In conclusion, epigenetic modifications are crucial mediators of T2DM-related comorbidities. Furthermore, a wide spectrum of circulating miRNAs can be used to predict and diagnose these conditions in this patient population. A comprehensive summary of the various epigenetic modifications involved in the pathogenesis of T2DM is presented in Table 1.
8 Conclusion
Epigenetic research offers the potential to enhance T2DM prevention and treatment by unearthing novel molecular mechanisms responsible for the progression of the disease and its associated complications. Dysregulation of epigenetic modifications induce T2DM by promoting several cornerstones of the disease, such as obesity, IR, β-cell dysfunction, mitochondrial dysfunction, and cellular senescence. Additionally, lifestyle choices, including diet and exercise, also promote and reduce the risk of T2DM through epigenetic modifications. Preclinical studies have identified a wide range of therapeutic targets, such as DNA methyltransferase-1, with promising results. Clinical studies are needed to determine whether these studies can be translated to the human population. Additionally, further studies are needed to solidify the role of epigenetics in cellular senescence and mitochondrial dysfunction among T2DM patients.
Author contributions
CJ: Writing – original draft. TA: Writing – original draft, Writing – review & editing. ZA: Writing – original draft. BS: Visualization, Writing – original draft, Writing – review & editing. AA: Writing – original draft. KA: Writing – review & editing. AY: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The figures in this manuscript were made using BioRender.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the international diabetes federation diabetes Atlas, 9(th) edition. Diabetes Res Clin Pract (2019) 157:107843. doi: 10.1016/j.diabres.2019.107843
2. Ling C, Rönn T. Epigenetics in human obesity and type 2 diabetes. Cell Metab (2019) 29(5):1028–44. doi: 10.1016/j.cmet.2019.03.009
3. Klimczak S, Śliwińska A. Epigenetic regulation of inflammation in insulin resistance. Semin Cell Dev Biol (2022) 154(Pt C):185–92. doi: 10.1016/j.semcdb.2022.09.004
4. Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res (1982) 10(8):2709–21. doi: 10.1093/nar/10.8.2709
5. Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol (2022) 23(5):329–49. doi: 10.1038/s41580-021-00441-y
6. Klimczak S, Śliwińska A. Epigenetic regulation of inflammation in insulin resistance. Semin Cell Dev Biol (2024) 154:185–92. doi: 10.1016/j.semcdb.2022.09.004
7. Geng X, Li Z, Yang Y. Emerging role of epitranscriptomics in diabetes mellitus and its complications. Front Endocrinol (2022) 13. doi: 10.3389/fendo.2022.907060
8. Cosentino C, Cnop M, Igoillo-Esteve M. The tRNA Epitranscriptome and Diabetes: Emergence of tRNA Hypomodifications as a Cause of Pancreatic β-Cell Failure. Endocrinology. (2019) 160(5):1262–74. doi: 10.1210/en.2019-00098
9. Yin L, Zhu X, Novák P, Zhou L, Gao L, Yang M, et al. The epitranscriptome of long noncoding RNAs in metabolic diseases. Clinica Chimica Acta (2021) 515:80–9. doi: 10.1016/j.cca.2021.01.001
11. Juvinao-Quintero DL, Sharp GC, Sanderson ECM, Relton CL, Elliott HR. Investigating causality in the association between DNA methylation and type 2 diabetes using bidirectional two-sample Mendelian randomisation. Diabetologia. (2023) 66(7):1247–59. doi: 10.1007/s00125-023-05914-7
12. Benko J, Sarlinova M, Mikusova V, Bolek T, Pec MJ, Halasova E, et al. MiR-126 and miR-146a as markers of type 2 diabetes mellitus: a pilot study. Bratislava Med J (2023) 124(07):527–33. doi: 10.4149/BLL_2023_081
13. Sultan S, Almalki S. Analysis of global DNA methylation and epigenetic modifiers (DNMTs and HDACs) in human foetal endothelium exposed to gestational and type 2 diabetes. Epigenetics. (2023) 18(1):2201714. doi: 10.1080/15592294.2023.2201714
14. Dhawan S, Natarajan R. Epigenetics and type 2 diabetes risk. Curr Diabetes Rep (2019) 19(8):47. doi: 10.1007/s11892-019-1168-8
15. Ling C, Bacos K, Rönn T. Epigenetics of type 2 diabetes mellitus and weight change — a tool for precision medicine? Nat Rev Endocrinol (2022) 18(7):433–48. doi: 10.1038/s41574-022-00671-w
16. Singh R, Chandel S, Dey D, Ghosh A, Roy S, Ravichandiran V, et al. Epigenetic modification and therapeutic targets of diabetes mellitus. Bioscience Rep (2020) 40(9):BSR20202160. doi: 10.1042/BSR20202160
17. Vaiserman A, Lushchak O. Developmental origins of type 2 diabetes: Focus on epigenetics. Ageing Res Rev (2019) 55:100957. doi: 10.1016/j.arr.2019.100957
18. Vaag A, Brøns C, Gillberg L, Hansen NS, Hjort L, Arora GP, et al. Genetic, nongenetic and epigenetic risk determinants in developmental programming of type 2 diabetes. Acta Obstetricia Gynecologica Scandinavica. (2014) 93(11):1099–108. doi: 10.1111/aogs.12494
19. Boutari C, Mantzoros CS. A 2022 update on the epidemiology of obesity and a call to action: as its twin COVID-19 pandemic appears to be receding, the obesity and dysmetabolism pandemic continues to rage on. Metabolism. (2022) 133:155217. doi: 10.1016/j.metabol.2022.155217
20. Lauby-Secretan B, Scoccianti C, Loomis D, Grosse Y, Bianchini F, Straif K. Body fatness and cancer — Viewpoint of the IARC working group. New Engl J Med (2016) 375(8):794–8. doi: 10.1056/NEJMsr1606602
21. Arabi T, Shafqat A, Sabbah BN, Fawzy NA, Shah H, Abdulkader H, et al. Obesity-related kidney disease: Beyond hypertension and insulin-resistance. Front Endocrinol (2023) 13. doi: 10.3389/fendo.2022.1095211
22. Emerging Risk Factors Collaboration, Wormser D, Kaptoge S, Di Angelantonio E, Wood AM, Pennells L. The Emerging Risk Factors C. Separate and combined associations of body-mass index and abdominal adiposity with cardiovascular disease: Collaborative analysis of 58 prospective studies. Lancet (2011) 377(9771):1085–95. doi: 10.1016/S0140-6736(11)60105-0
23. Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. (2006) 444(7121):840–6. doi: 10.1038/nature05482
24. Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest (2017) 127(1):1–4. doi: 10.1172/JCI92035
25. Cătoi AF, Pârvu A, Mureşan A, Busetto L. Metabolic mechanisms in obesity and type 2 diabetes: Insights from bariatric/metabolic surgery. Obes Facts. (2015) 8(6):350–63. doi: 10.1159/000441259
26. Xu X, Su S, Barnes VA, De Miguel C, Pollock J, Ownby D, et al. A genome-wide methylation study on obesity. Epigenetics. (2013) 8(5):522–33. doi: 10.4161/epi.24506
27. Dalgaard K, Landgraf K, Heyne S, Lempradl A, Longinotto J, Gossens K, et al. Trim28 haploinsufficiency triggers bi-stable epigenetic obesity. Cell. (2016) 164(3):353–64. doi: 10.1016/j.cell.2015.12.025
28. Dick KJ, Nelson CP, Tsaprouni L, Sandling JK, Aïssi D, Wahl S, et al. DNA methylation and body-mass index: a genome-wide analysis. Lancet (2014) 383(9933):1990–8. doi: 10.1016/S0140-6736(13)62674-4
29. Janjanam VD, Ewart S, Zhang H, Jiang Y, Arshad H, Ziyab AH, et al. Offspring epigenetic markers at birth related to gestational BMI predict offspring BMI-trajectories from infancy to 26 years. Obes Sci Practice. (2023) 9(4):424–34. doi: 10.1002/osp4.660
30. Huang RC, Garratt ES, Pan H, Wu Y, Davis EA, Barton SJ, et al. Genome-wide methylation analysis identifies differentially methylated CpG loci associated with severe obesity in childhood. Epigenetics. (2015) 10(11):995–1005. doi: 10.1080/15592294.2015.1080411
31. Xia Y, Caputo M, Cansby E, Anand SK, Sütt S, Henricsson M, et al. STE20-type kinase TAOK3 regulates hepatic lipid partitioning. Mol Metab (2021) 54:101353. doi: 10.1016/j.molmet.2021.101353
32. Baldini F, Zeaiter L, Diab F, Zbeeb H, Cuneo L, Pagano A, et al. Nuclear and chromatin rearrangement associate to epigenome and gene expression changes in a model of in vitro adipogenesis and hypertrophy. Biochim Biophys Acta Mol Cell Biol Lipids. (2023) 1868(10):159368. doi: 10.1016/j.bbalip.2023.159368
33. Pessoa Rodrigues C, Chatterjee A, Wiese M, Stehle T, Szymanski W, Shvedunova M, et al. Histone H4 lysine 16 acetylation controls central carbon metabolism and diet-induced obesity in mice. Nat Commun (2021) 12(1):6212. doi: 10.1038/s41467-021-26277-w
34. Galicia-Garcia U, Benito-Vicente A, Jebari S, Larrea-Sebal A, Siddiqi H, Uribe KB, et al. Pathophysiology of type 2 diabetes mellitus. Int J Mol Sci (2020) 21(17):6275. doi: 10.3390/ijms21176275
35. Buteau J, Foisy S, Rhodes CJ, Carpenter L, Biden TJ, Prentki M. Protein kinase Czeta activation mediates glucagon-like peptide-1-induced pancreatic beta-cell proliferation. Diabetes. (2001) 50(10):2237–43. doi: 10.2337/diabetes.50.10.2237
36. Groop LC. Insulin resistance: the fundamental trigger of type 2 diabetes. Diabetes Obes Metab (1999) 1 Suppl 1:S1–7. doi: 10.1046/j.1463-1326.1999.0010s1001.x
37. Małodobra-Mazur M, Alama A, Bednarska-Chabowska D, Pawelka D, Myszczyszyn A, Dobosz T. Obesity-induced insulin resistance via changes in the DNA methylation profile of insulin pathway genes. Adv Clin Exp Med (2019) 28(12):1599–607. doi: 10.17219/acem/110321
38. Fragoso-Bargas N, Elliott HR, Lee-Ødegård S, Opsahl JO, Sletner L, Jenum AK, et al. Cross-Ancestry DNA Methylation marks of insulin resistance in pregnancy: An integrative Epigenome-Wide association study. Diabetes. (2022) 72(3):415–26. doi: 10.2337/db22-0504
39. Cha-Molstad H, Saxena G, Chen J, Shalev A. Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. J Biol Chem (2009) 284(25):16898–905. doi: 10.1074/jbc.M109.010504
40. Chen J, Hui ST, Couto FM, Mungrue IN, Davis DB, Attie AD, et al. Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta-cell mass and protects against diabetes. FASEB J (2008) 22(10):3581–94. doi: 10.1096/fj.08-111690
41. Zhang S-J, Wang Y, Yang Y-L, Zheng H. Aberrant DNA methylation involved in obese women with systemic insulin resistance. Open Life Sci (2018) 13(1):201–7. doi: 10.1515/biol-2018-0024
42. Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. (2003) 52(1):102–10. doi: 10.2337/diabetes.52.1.102
43. Clark A, Jones LC, de Koning E, Hansen BC, Matthews DR. Decreased insulin secretion in type 2 diabetes: A problem of cellular mass or function? Diabetes (2001) 50 Suppl 1:S169–71. doi: 10.2337/diabetes.50.2007.S169
44. Yang BT, Dayeh TA, Kirkpatrick CL, Taneera J, Kumar R, Groop L, et al. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA1c levels in human pancreatic islets. Diabetologia. (2011) 54(2):360–7. doi: 10.1007/s00125-010-1967-6
45. Hall E, Dekker Nitert M, Volkov P, Malmgren S, Mulder H, Bacos K, et al. The effects of high glucose exposure on global gene expression and DNA methylation in human pancreatic islets. Mol Cell Endocrinology. (2018) 472:57–67. doi: 10.1016/j.mce.2017.11.019
46. Ling C, Del Guerra S, Lupi R, Rönn T, Granhall C, Luthman H, et al. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. (2008) 51(4):615–22. doi: 10.1007/s00125-007-0916-5
47. Ashizawa S, Brunicardi FC, Wang X-P. PDX-1 and the pancreas. Pancreas. (2004) 28(2):109–20. doi: 10.1097/00006676-200403000-00001
48. Yang BT, Dayeh TA, Volkov PA, Kirkpatrick CL, Malmgren S, Jing X, et al. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol Endocrinology. (2012) 26(7):1203–12. doi: 10.1210/me.2012-1004
49. Desgraz R, Bonal C, Herrera PL. β-Cell regeneration: the pancreatic intrinsic faculty. Trends Endocrinol Metab (2011) 22(1):34–43. doi: 10.1016/j.tem.2010.09.004
50. Newsholme P, Haber EP, Hirabara SM, Rebelato EL, Procopio J, Morgan D, et al. Diabetes associated cell stress and dysfunction: role of mitochondrial and non-mitochondrial ROS production and activity. J Physiol (2007) 583(Pt 1):9–24. doi: 10.1113/jphysiol.2007.135871
51. Wang TY, Liu XJ, Xie JY, Yuan QZ, Wang Y. Cask methylation involved in the injury of insulin secretion function caused by interleukin1-β. J Cell Mol Med (2020) 24(24):14247–56. doi: 10.1111/jcmm.16041
52. Roshanzamir N, Hassan-Zadeh V. Methylation of specific CpG sites in IL-1β and IL1R1 genes is affected by Hyperglycaemia in Type 2 diabetic patients. Immunol Investigations. (2020) 49(3):287–98. doi: 10.1080/08820139.2019.1656227
53. Liu Y, Wang Q, Wen J, Wu Y, Man C. MiR-375: A novel multifunctional regulator. Life Sci (2021) 275:119323. doi: 10.1016/j.lfs.2021.119323
54. Iacomino G, Lauria F, Russo P, Venezia A, Iannaccone N, Marena P, et al. The association of circulating miR-191 and miR-375 expression levels with markers of insulin resistance in overweight children: An exploratory analysis of the I.Family Study. Genes Nutr (2021) 16(1):10. doi: 10.1186/s12263-021-00689-1
55. Magenta A, Ciarapica R, Capogrossi MC. The emerging role of miR-200 family in cardiovascular diseases. Circ Res (2017) 120(9):1399–402. doi: 10.1161/CIRCRESAHA.116.310274
56. Reddy MA, Jin W, Villeneuve L, Wang M, Lanting L, Todorov I, et al. Pro-Inflammatory role of MicroRNA-200 in vascular smooth muscle cells from diabetic mice. Arteriosclerosis Thrombosis Vasc Biol (2012) 32(3):721–9. doi: 10.1161/ATVBAHA.111.241109
57. Belgardt B-F, Ahmed K, Spranger M, Latreille M, Denzler R, Kondratiuk N, et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med (2015) 21(6):619–27. doi: 10.1038/nm.3862
58. Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N, et al. miRNAs control insulin content in pancreatic β-cells via downregulation of transcriptional repressors. EMBO J (2011) 30(5):835–45. doi: 10.1038/emboj.2010.361
59. Shafqat S, Arana Chicas E, Shafqat A, Hashmi SK. The Achilles’ heel of cancer survivors: Fundamentals of accelerated cellular senescence. J Clin Invest (2022) 132(13):e158452. doi: 10.1172/JCI158452
60. Narasimhan A, Flores RR, Robbins PD, Niedernhofer LJ. Role of cellular senescence in type II diabetes. Endocrinology. (2021) 162(10):1–12. doi: 10.1210/endocr/bqab136
61. Sone H, Kagawa Y. Pancreatic beta cell senescence contributes to the pathogenesis of type 2 diabetes in high-fat diet-induced diabetic mice. Diabetologia. (2005) 48(1):58–67. doi: 10.1007/s00125-004-1605-2
62. Tamura Y, Izumiyama-Shimomura N, Kimbara Y, Nakamura K, Ishikawa N, Aida J, et al. β-cell telomere attrition in diabetes: Inverse correlation between HbA1c and telomere length. J Clin Endocrinol Metab (2014) 99(8):2771–7. doi: 10.1210/jc.2014-1222
63. Avrahami D, Li C, Zhang J, Schug J, Avrahami R, Rao S, et al. Aging-Dependent demethylation of regulatory elements correlates with chromatin state and improved β cell function. Cell Metab (2015) 22(4):619–32. doi: 10.1016/j.cmet.2015.07.025
64. Frye M, Harada BT, Behm M, He C. RNA modifications modulate gene expression during development. Science. (2018) 361(6409):1346–9. doi: 10.1126/science.aau1646
65. Pan G, Feng Y, Jin P, Kai Y, Xiao Z. m6A methylation in cellular senescence of age-associated diseases. Acta Biochim Biophys Sin. 55(8):1168–83. doi: 10.3724/abbs.2023107
66. De Jesus DF, Zhang Z, Kahraman S, Brown NK, Chen M, Hu J, et al. m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nat Metab (2019) 1(8):765–74. doi: 10.1038/s42255-019-0089-9
67. Wilinski D, Dus M. N(6)-adenosine methylation controls the translation of insulin mRNA. Nat Struct Mol Biol (2023) 30(9):1260–4. doi: 10.1038/s41594-023-01048-x
68. Li Y, Tollefsbol TO. p16INK4a suppression by glucose restriction contributes to human cellular lifespan extension through SIRT1-Mediated epigenetic and genetic mechanisms. PloS One (2011) 6(2):e17421. doi: 10.1371/journal.pone.0017421
69. Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes. (2005) 54(1):8–14. doi: 10.2337/diabetes.54.1.8
70. Asmann YW, Stump CS, Short KR, Coenen-Schimke JM, Guo Z, Bigelow ML, et al. Skeletal muscle mitochondrial functions, mitochondrial DNA copy numbers, and gene transcript profiles in type 2 diabetic and nondiabetic subjects at equal levels of low or high insulin and euglycemia. Diabetes. (2006) 55(12):3309–19. doi: 10.2337/db05-1230
71. Stump CS, Short KR, Bigelow ML, Schimke JM, Nair KS. Effect of insulin on human skeletal muscle mitochondrial ATP production, protein synthesis, and mRNA transcripts. Proc Natl Acad Sci (2003) 100(13):7996–8001. doi: 10.1073/pnas.1332551100
72. Tirosh A, Potashnik R, Bashan N, Rudich A. Oxidative stress disrupts insulin-induced cellular redistribution of insulin receptor Substrate-1 and Phosphatidylinositol 3-Kinase in 3T3-L1 Adipocytes. J Biol Chem (1999) 274(15):10595–602. doi: 10.1074/jbc.274.15.10595
73. Rudich A, Tirosh A, Potashnik R, Hemi R, Kanety H, Bashan N. Prolonged oxidative stress impairs insulin-induced GLUT4 translocation in 3T3-L1 adipocytes. Diabetes. (1998) 47(10):1562–9. doi: 10.2337/diabetes.47.10.1562
74. Mishra M, Kowluru RA. Epigenetic modification of mitochondrial DNA in the development of diabetic retinopathy. Invest Opthalmology Visual Science. (2015) 56(9):5133. doi: 10.1167/iovs.15-16937
75. Cao K, Lv W, Wang X, Dong S, Liu X, Yang T, et al. Hypermethylation of hepatic mitochondrial ND6 provokes systemic insulin resistance. Advanced Sci (2021) 8(11):2004507. doi: 10.1002/advs.202004507
76. Voisin S, Eynon N, Yan X, Bishop DJ. Exercise training and DNA methylation in humans. Acta Physiologica. (2015) 213(1):39–59. doi: 10.1111/apha.12414
77. Kulkarni SS, Salehzadeh F, Fritz T, Zierath JR, Krook A, Osler ME. Mitochondrial regulators of fatty acid metabolism reflect metabolic dysfunction in type 2 diabetes mellitus. Metabolism. (2012) 61(2):175–85. doi: 10.1016/j.metabol.2011.06.014
78. Nakajima K, Takeoka M, Mori M, Hashimoto S, Sakurai A, Nose H, et al. Exercise effects on methylation of ASC gene. Int J Sports Med (2010) 31(9):671–5. doi: 10.1055/s-0029-1246140
79. Nilsson E, Matte A, Perfilyev A, de Mello VD, Käkelä P, Pihlajamäki J, et al. Epigenetic alterations in human liver from subjects with type 2 diabetes in parallel with reduced folate levels. J Clin Endocrinol Metab (2015) 100(11):E1491–E501. doi: 10.1210/jc.2015-3204
80. Hjort L, Jørgensen SW, Gillberg L, Hall E, Brøns C, Frystyk J, et al. 36 h fasting of young men influences adipose tissue DNA methylation of LEP and ADIPOQ in a birth weight-dependent manner. Clin Epigenetics. (2017) 9(1):40. doi: 10.1186/s13148-017-0340-8
81. Perfilyev A, Dahlman I, Gillberg L, Rosqvist F, Iggman D, Volkov P, et al. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial1, 2, 3. Am J Clin Nutr (2017) 105(4):991–1000. doi: 10.3945/ajcn.116.143164
82. Forbes JM, Cooper ME. Mechanisms of diabetic complications. Physiol Rev (2013) 93(1):137–88. doi: 10.1152/physrev.00045.2011
83. Einarson TR, Acs A, Ludwig C, Panton UH. Prevalence of cardiovascular disease in type 2 diabetes: A systematic literature review of scientific evidence from across the world in 2007–2017. Cardiovasc Diabetology. (2018) 17(1):83. doi: 10.1186/s12933-018-0728-6
84. Poznyak A, Grechko AV, Poggio P, Myasoedova VA, Alfieri V, Orekhov AN. The diabetes Mellitus–Atherosclerosis connection: The Role of lipid and glucose metabolism and chronic inflammation. Int J Mol Sci (2020) 21(5):1835. doi: 10.3390/ijms21051835
85. Greißel A, Culmes M, Burgkart R, Zimmermann A, Eckstein HH, Zernecke A, et al. Histone acetylation and methylation significantly change with severity of atherosclerosis in human carotid plaques. Cardiovasc Pathol (2016) 25(2):79–86. doi: 10.1016/j.carpath.2015.11.001
86. Amr KS, Abdelmawgoud H, Ali ZY, Shehata S, Raslan HM. Potential value of circulating microRNA-126 and microRNA-210 as biomarkers for type 2 diabetes with coronary artery disease. Br J BioMed Sci (2018) 75(2):82–7. doi: 10.1080/09674845.2017.1402404
87. Al-Kafaji G, Al-Mahroos G, Abdulla Al-Muhtaresh H, Sabry MA, Abdul Razzak R, Salem AH. Circulating endothelium-enriched microRNA-126 as a potential biomarker for coronary artery disease in type 2 diabetes mellitus patients. Biomarkers (2017) 22(3-4):268–78. doi: 10.1080/1354750X.2016.1204004
88. Liu H, Yang N, Fei Z, Qiu J, Ma D, Liu X, et al. Analysis of plasma miR-208a and miR-370 expression levels for early diagnosis of coronary artery disease. BioMed Rep (2016) 5(3):332–6. doi: 10.3892/br.2016.726
89. Giordano M, Trotta MC, Ciarambino T, D'Amico M, Galdiero M, Schettini F, et al. Circulating MiRNA-195-5p and -451a in diabetic patients with transient and acute ischemic stroke in the emergency department. Int J Mol Sci (2020) 21(20):7615. doi: 10.3390/ijms21207615
90. Li P, Teng F, Gao F, Zhang M, Wu J, Zhang C. Identification of circulating MicroRNAs as potential biomarkers for detecting acute ischemic stroke. Cell Mol Neurobiology. (2015) 35(3):433–47. doi: 10.1007/s10571-014-0139-5
91. Maghbooli Z, Hossein-nezhad A, Larijani B, Amini M, Keshtkar A. Global DNA methylation as a possible biomarker for diabetic retinopathy. Diabetes Metab Res Rev (2015) 31(2):183–9. doi: 10.1002/dmrr.2584
92. Mohammad G, Radhakrishnan R, Kowluru RA. Epigenetic modifications compromise mitochondrial DNA quality control in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci (2019) 60(12):3943–51. doi: 10.1167/iovs.19-27602
93. Zhong Q, Kowluru RA. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes. (2011) 60(4):1304–13. doi: 10.2337/db10-0133
94. Zhao X, Ling F, Gw Z, Yu N, Yang J, Xin Xy. The correlation between MicroRNAs and diabetic retinopathy. Front Immunol (2022) 13. doi: 10.3389/fimmu.2022.941982
95. Maser RE, Steenkiste AR, Dorman JS, Nielsen VK, Bass EB, Manjoo Q, et al. Epidemiological correlates of diabetic neuropathy. Report from Pittsburgh epidemiology of diabetes complications study. Diabetes. (1989) 38(11):1456–61. doi: 10.2337/diab.38.11.1456
96. Callaghan BC, Cheng HT, Stables CL, Smith AL, Feldman EL. Diabetic neuropathy: Clinical manifestations and current treatments. Lancet Neurology. (2012) 11(6):521–34. doi: 10.1016/S1474-4422(12)70065-0
97. Jankovic M, Novakovic I, Nikolic D, Mitrovic Maksic J, Brankovic S, Petronic I, et al. Genetic and epigenomic modifiers of diabetic neuropathy. Int J Mol Sci (2021) 22(9):4887. doi: 10.3390/ijms22094887
98. Zhang H-H, Han X, Wang M, Hu Q, Li S, Wang M, et al. The association between genomic DNA methylation and diabetic peripheral neuropathy in patients with type 2 diabetes mellitus. J Diabetes Res (2019) 2019:2494057. doi: 10.1155/2019/2494057
99. Gastoł J, Kapusta P, Polus A, Pitera E, Biela M, Wołkow P, et al. Epigenetic mechanism in search for the pathomechanism of diabetic neuropathy development in diabetes mellitus type 1 (T1DM). Endocrine. (2020) 68(1):235–40. doi: 10.1007/s12020-019-02172-9
100. Latini A, Borgiani P, De Benedittis G, D'Amato C, Greco C, Lauro D, et al. Mitochondrial DNA copy number in peripheral blood is reduced in type 2 diabetes patients with polyneuropathy and associated with a MIR499A Gene Polymorphism. DNA Cell Biol (2020) 39(8):1467–72. doi: 10.1089/dna.2019.5326
101. Ciccacci C, Morganti R, Di Fusco D, D'Amato C, Cacciotti L, Greco C, et al. Common polymorphisms in MIR146a, MIR128a and MIR27a genes contribute to neuropathy susceptibility in type 2 diabetes. Acta Diabetol (2014) 51(4):663–71. doi: 10.1007/s00592-014-0582-2
102. Bali KK, Gandla J, Rangel DR, Castaldi L, Mouritzen P, Agarwal N, et al. A genome-wide screen reveals microRNAs in peripheral sensory neurons driving painful diabetic neuropathy. Pain. (2021) 162(5):1334–51. doi: 10.1097/j.pain.0000000000002159
103. Xourgia E, Papazafiropoulou A, Melidonis A. Circulating microRNAs as biomarkers for diabetic neuropathy: A novel approach. World J Exp Med (2018) 8(3):18–23. doi: 10.5493/wjem.v8.i3.18
104. Hao J, Liu Y. Epigenetics of methylation modifications in diabetic cardiomyopathy. Front Endocrinol (2023) 14. doi: 10.3389/fendo.2023.1119765
105. Dannenberg L, Weske S, Kelm M, Levkau B, Polzin A. Cellular mechanisms and recommended drug-based therapeutic options in diabetic cardiomyopathy. Pharmacol Ther (2021) 228:107920. doi: 10.1016/j.pharmthera.2021.107920
106. Tao H, Shi P, Xuan H-Y, Ding X-S. DNA methyltransferase-1 inactivation of androgen receptor axis triggers homocysteine induced cardiac fibroblast autophagy in diabetic cardiac fibrosis. Arch Biochem Biophysics. (2020) 692:108521. doi: 10.1016/j.abb.2020.108521
107. Liu ZZ, Zhao XZ, Zhang XS, Zhang M. Promoter DNA demethylation of Keap1 gene in diabetic cardiomyopathy. Int J Clin Exp Pathol (2014) 7(12):8756–62.
108. Cheng Y, Liu G, Pan Q, Guo S, Yang X. Elevated expression of liver X receptor alpha (LXRα) in myocardium of Streptozotocin-Induced diabetic rats. Inflammation. (2011) 34(6):698–706. doi: 10.1007/s10753-010-9281-5
109. Choe SS, Choi AH, Lee JW, Kim KH, Chung JJ, Park J, et al. Chronic activation of liver X receptor induces beta-cell apoptosis through hyperactivation of lipogenesis: Liver X receptor-mediated lipotoxicity in pancreatic beta-cells. Diabetes. (2007) 56(6):1534–43. doi: 10.2337/db06-1059
110. Cannon MV, Silljé HHW, Sijbesma JWA, Khan MAF, Steffensen KR, Van Gilst WH, et al. LXRα improves myocardial glucose tolerance and reduces cardiac hypertrophy in a mouse model of obesity-induced type 2 diabetes. Diabetologia. (2016) 59(3):634–43. doi: 10.1007/s00125-015-3827-x
Keywords: epigenetics, diabetes mellitus, type 2 diabetes complications, obesity, insulin resistance, beta-cell dysfunction, cellular senescence, mitochondrial dysfunction
Citation: Jazieh C, Arabi TZ, Asim Z, Sabbah BN, Alsaud AW, Alkattan K and Yaqinuddin A (2024) Unraveling the epigenetic fabric of type 2 diabetes mellitus: pathogenic mechanisms and therapeutic implications. Front. Endocrinol. 15:1295967. doi: 10.3389/fendo.2024.1295967
Received: 19 September 2023; Accepted: 04 January 2024;
Published: 22 January 2024.
Edited by:
Jean-François Tanti, INSERM U1065 Centre Meíditerraneíen de Meídecine Moleículaire, FranceReviewed by:
Valerie Grandjean, INSERM U1065 Centre Meíditerraneíen de Meídecine Moleículaire, FranceSathishkumar Chandrakumar, University of California, Los Angeles, United States
Shafeeq Ahmed Mohammed, University Hospital Zürich, Switzerland
Copyright © 2024 Jazieh, Arabi, Asim, Sabbah, Alsaud, Alkattan and Yaqinuddin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed Yaqinuddin, YXlhcWludWRkaW5AYWxmYWlzYWwuZWR1
†These authors have contributed equally to this work and share first authorship
 Cham Jazieh
Cham Jazieh Tarek Ziad Arabi
Tarek Ziad Arabi Zohaib Asim
Zohaib Asim Belal Nedal Sabbah
Belal Nedal Sabbah Khaled Alkattan
Khaled Alkattan Ahmed Yaqinuddin
Ahmed Yaqinuddin