
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 18 January 2024
Sec. Neuroendocrine Science
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.1292024
This article is part of the Research Topic Steroids and the Brain - Volume II View all 6 articles
Glucocorticoids are key executors of the physiological response to stress. Previous studies in mice showed that the androgen receptor (AR) influenced the transcriptional outcome of glucocorticoid treatment in white and brown adipocytes and in the liver. In the brain, we observed that chronic hypercorticism induced changes in gene expression that tended to be more pronounced in male mice. In the present study, we investigated if glucocorticoid signaling in the brain could be modulated by androgen. After chronic treatment with corticosterone, dihydrotestosterone, a combination of both, and corticosterone in combination with the AR antagonist enzalutamide, we compared the expression of glucocorticoid receptor (NR3C1, also abbreviated GR) target genes in brain regions where AR and GR are co-expressed, namely: prefrontal cortex, hypothalamus, hippocampus, ventral tegmental area and substantia nigra. We observed that androgen affected glucocorticoid signaling only in the prefrontal cortex and the substantia nigra. Dihydrotestosterone and corticosterone independently and inversely regulated expression of Sgk1 and Tsc22d3 in prefrontal cortex. AR antagonism with enzalutamide attenuated corticosterone-induced expression of Fkbp5 in the prefrontal cortex and of Fkbp5 and Sgk1 in the substantia nigra. Additionally, in the substantia nigra, AR antagonism increased expression of Th and Slc18a1, two genes coding for key components of the dopaminergic system. Our data indicate that androgen influence over glucocorticoid stimulation in the brain is not a dominant phenomenon in the context of high corticosterone levels, but can occur in the prefrontal cortex and substantia nigra.
Glucocorticoids (GCs) regulate many physiological processes, including energy homeostasis, peripheral circadian signaling, and adaptation to stress (1). In humans the predominant form of endogenous glucocorticoid is cortisol, whereas in mice it is corticosterone. In the brain, glucocorticoids influence diverse processes, such as neurogenesis, neuronal activity, emotional regulation and cognition (2). Glucocorticoids target both neurons and glial cells (oligodendrocytes, astrocytes and microglia), which we have previously described to undergo extensive changes in mice with chronically elevated corticosterone levels (3, 4).
Corticosterone binds to the glucocorticoid receptor (NR3C1, hereafter GR) and the mineralocorticoid receptor (MR). GR and MR belong to the superfamily of steroid nuclear receptors, which also includes the androgen receptor (AR) (5–9). Upon ligand binding, nuclear receptors homo- or heterodimerize and translocate to the cell nucleus in order to activate or repress gene transcription (10). The DNA binding domains of AR and GR are very similar, and both receptors can bind to common response elements, possibly as heterodimers (11). Modulation of glucocorticoid signaling by androgen has been demonstrated in peripheral organs, via several potential mechanisms. In liver, brown and white adipose tissues, GR-driven gene expression is potentiated when AR is activated, and such potentiation is abolished when an AR antagonist is administered (12, 13). AR is expressed in different regions of the brain, and it has been demonstrated that AR agonism in the brain influences hypothalamic pituitary adrenal (HPA) axis activity (14–16).
Previous studies addressed sex differences in the response to stressors, of which glucocorticoids are an integral part (17). However, less is known of modulation of glucocorticoid activity by sex hormones in specific brain regions or in transcriptional/translational profiles specific to particular brain cell types. Such relationships can be explored via animal models of chronic hormonal imbalances. Recently, we observed sex-dependent differences in the brain in the AdKO2.0 mouse (a mouse model with a deletion of the regulatory subunit of the PKA specific to the adrenal gland, which provokes adrenal enlargement and chronic hypercorticosteronemia) in comparison to wild types, including changes in brain volume, as well as in the expression of diverse mRNAs and proteins (3, 18). Notably, male mice presented larger differences than females in myelin basic protein, hippocampal aquaporin 4 and allograft inflammatory factor1 mRNA expression in cortex and hippocampus (4). We hypothesized that these findings could be explained by modulation of GC-signaling by androgens, given the sexual differences in circulating androgens and AR expression in the brain (19).
In order to explore the influence of androgen over glucocorticoid signaling in the brain, we measured the expression of a selection of genes which are responsive to GR activation by glucocorticoids. For this we selected brain areas that are known to co-express AR and GR (20). We utilized tissue from mice in which manipulations affect GR signaling in peripheral tissues (12). First, we compared mice chronically exposed to high levels of corticosterone, dihydrotestosterone, or a combination of both, to evaluate the possible potentiation effects of AR agonism on GR transcriptional activity. In addition, we investigated further AR influence on GR-driven gene expression by treating mice with corticosterone in combination with the AR antagonist enzalutamide.
We previously published data from these mouse cohorts, with respect to liver transcriptome (12, 21). Male C57BL6/J mice (Jackson Laboratory) were housed in conventional cages (3-4 mice per cage) under controlled temperature and 12:12 light-dark cycle, with access to regular chow diet and water ad libitum. At 8 weeks of age, a slow-release pellet was subcutaneously implanted, superior to the cervical vertebrae. In experiment 1, pellets contained one of the following treatments: 100 mg cholesterol (vehicle - VEH), 20 mg corticosterone + 80 mg cholesterol (CORT), 5 mg dihydrotestosterone + 20 mg corticosterone + 75 mg cholesterol (DHT+CORT), or 5 mg dihydrotestosterone + 95 mg cholesterol (DHT). In experiment 2, pellets contained one of the following treatments: 100 mg cholesterol (VEH) or 20 mg corticosterone (Sigma-Aldrich) + 80 mg cholesterol (CORT). AR antagonist Enzalutamide (ENZ; MedChemExpress) was administered via diet supplementation (357 mg drug per kg diet; resulting in an estimated dose of 40 mg/kg/day) to a subgroup of mice implanted with CORT pellets (CORT+ENZ). After 14 days of treatment, mice were euthanized and perfused intracardially with ice cold PBS for 5 minutes. After perfusion, brains were extracted from skulls, placed on aluminum foil and snap frozen on dry ice, and stored at -80°C until further processing.
60 µm coronal sections were obtained in a cryotome (Thermo Scientific) and micro-dissected with a coring tool (Fine Science Tools), 0.8 or 1.0 mm of diameter. Five brain regions were collected using stereotactic coordinates from Paxinos and Franklin Mouse brain atlas, namely: prefrontal cortex (PFC) (bregma 1.10 to 0.62), ventromedial hypothalamus (VMH) (bregma -1.34 to -1.94), dorsal hippocampus (HIP) (bregma -1.34 to -2.06), ventral tegmental area (VTA) and substantia nigra (SN) (bregma -2.92 to -3.80). The complete dorsal hippocampus was collected by double-punching, using the 1.0 mm needle. Samples were kept frozen until further processing. The areas were selected based on prominent coexpression of AR and GR mRNA in the mouse brain as determined by in situ hybridization (22).
We determined that we could test androgen-glucocorticoid crosstalk in the brain by measuring the expression of GR responsive genes under chronic GR activation in contexts of AR activation or inactivation by means of androgen or androgen antagonist chronic treatment. GR responsive genes were selected irrespective of their biological function, using procedure described by Viho et al. (2022) (23). Briefly, selection was made by comparing previous rodent brain and neuron studies reporting transcriptomic responses upon glucocorticoid treatment (24) to hippocampal RNA-seq data from mice previously injected with corticosterone (23) and to data from two ChIP-seq studies addressing rat brain response to corticosterone injections (25, 26), and applying the following predefined filtering criteria: GC regulation, directional consistency and genomic association to binding sites for GR, MR or both. The genes we include in the present study were validated for response to chronic corticosterone treatment as used in the present experiment.
Total RNA was extracted from micropunches using TriPure™ Isolation Reagent (Merck), following the manufacturer’s protocol. First strand cDNA was synthesized with M-MLV RT system (Promega), to a final concentration of 1 ng/µL cDNA. qPCR reactions were run with iQ™ SYBR® Green Supermix (Bio-Rad) in real-time PCR detection system (Bio-Rad). Expression levels were normalized to reference gene Rn18s. QuantiTect® Primer Assays were purveyed by Qiagen. All information about primers used is contained in Supplementary Table 1. All primer pairs were exon spanning and had amplification efficacies of > 90%.
Data from experiment 1 were compared by means of 2-way ANOVA, with corticosterone and DHT as factors. Data from experiment 2 were compared by 1-way ANOVA. The level of significance was set at α < 0.05. For pairwise comparison post hoc Tukey test was used.
To facilitate comparison between the complementary experiments 1 and 2, we present data from both experiments in the same figures.
As a strategy to identify effects of androgens on glucocorticoid signaling, we evaluated gene expression in brain areas that co-express AR and GR. In experiment 1, we determined the levels of mRNA expression of AR and GR, as regulation of the receptors may be a mechanism for (mutual) regulation of responsiveness. In the PFC, corticosterone increased AR mRNA expression (p=0.02, Figure 1A). In the ventral tegmental area and the substantia nigra, dihydrotestosterone led to a decrease in AR mRNA levels (main effect for DHT: p=0.01 and p=0.02, respectively; Figures 1D, E). Gr in the substantia nigra was significantly downregulated by corticosterone (p=0.02) and dihydrotestosterone (p=0.05) (Figure 1J). Posthoc analysis showed lower GR mRNA levels in DHT+CORT group compared to all the others.
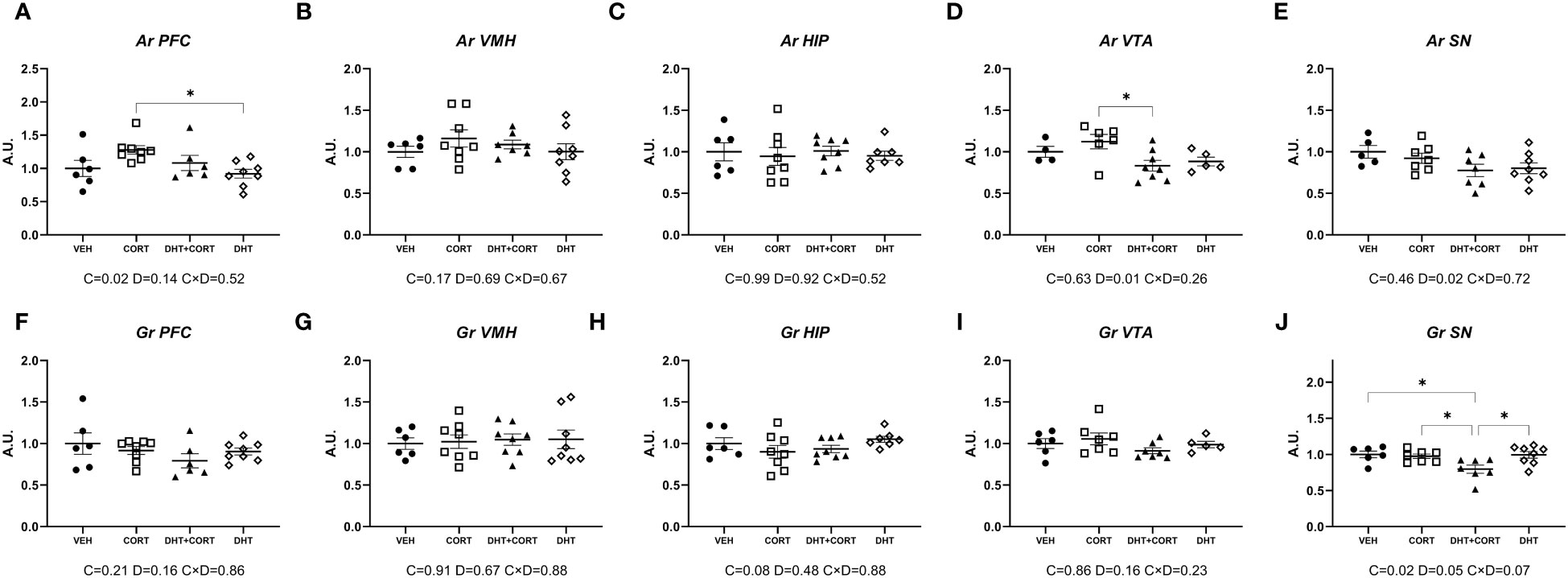
Figure 1 AR and Gr expression. Expression of Androgen Receptor (Ar, A-E) and Glucocorticoid Receptor (Gr, F-J) mRNA in selected brain areas, obtained from experiment 1. 2-way ANOVA main effects (p<0.05) are depicted below the x axis as: C, Corticosterone; D, Dihydrotestosterone; CxD, Interaction. *p<0.05.
Overall, AR and GR regulation was only observed in the PFC and in dopaminergic areas. In the substantia nigra corticosterone and dihydrotestosterone cooperated in the downregulation of GR.
The availability of a mouse hippocampal scRNA-seq dataset allowed us to determine co-expression of AR and GR in different types of cells, and this would a priori define potential direct effect of androgens on GC signaling. Figure 2A shows that AR and GR are co-expressed in neuronal cells mainly. In contrast, most of the corticosterone target genes that we selected are co-expressed with AR and GR in both neurons and non-neuronal cells. In experiment 1, corticosterone significantly increased hippocampal expression of Fkbp5, Pla2g3 and Mfsd2a (Figures 2B, E, G; p<0.01 in all cases). Although no significant interaction effects with dihydrotestosterone were found, posthoc analysis showed a significant induction of Fkbp5 and Mfsd2a relative to VEH only in the DHT+CORT group. Pla2g3 was clearly induced by corticosterone, independent of dihydrotestosterone. Dihydrotestosterone, but not corticosterone, significantly increased Sgk1 expression (p=0.01, Figure 2C). In experiment 2, 1-way ANOVA analyses reached statistical significance for Fkbp5, Sgk1, Pla2g3, Tsc22d3 and Mfsd2a (p<0.05 in all cases, Figures 2H, I, K–M). Posthoc analyses showed that in all cases expression increased in the CORT group compared to VEH group. Tsc22d3 and Mfsd2a expression increased also in the CORT +ENZ group compared to VEH group. These data indicate that gene expression in the hippocampus is responsive to GCs and DHT and that they might cooperate in the regulation of specific genes.
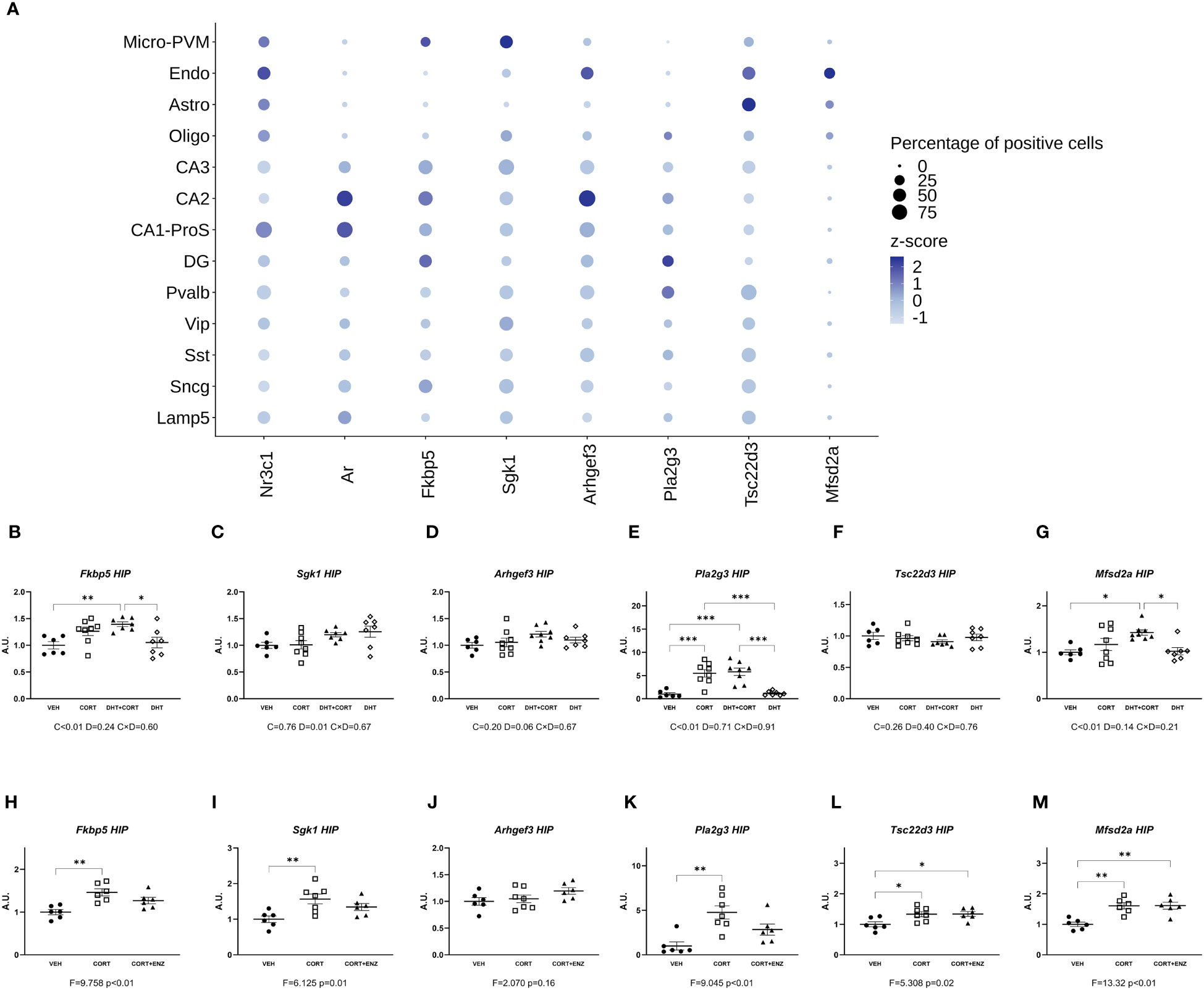
Figure 2 Gene expression in Hippocampus. (A) Normalized expression of GR, MR, AR and GR responsive genes in hippocampal cells under basal conditions. The dots represent gene centered log-normalized average expression (z-score) as color and the percentage of positive cells as dot size. Micro-PVM, microglia-perivascular macrophages; Endo, endothelial cells; Astro, astrocytes; Oligo, oligodendrocytes; CA3, CA2, CA1-ProS and DG: glutamatergic neurons from Cornu Ammonis areas 3, 2, 1 and dentate gyrus, respectively; Pvalb, Vip. Sst, Sncg and Lamp5: GABAergic neurons expressing Parvalbumin, Vasoactive Intestinal Peptide, Somatostatin, gamma-synuclein, or Lysosome-associated membrane glycoprotein 5, respectively. (B-G) Expression of GR responsive genes in hippocampus, obtained from experiment 1. 2-way ANOVA main effects are depicted below the x axis as: C, Corticosterone; D, Dihydrotestosterone; CxD, Interaction. (H-M) expression of GR responsive genes in hippocampus, obtained from experiment 2. 1-way ANOVA values of F-statistic and p are depicted below the x axis. VEH, vehicle; CORT, Corticosterone; CORT+ENZ, Corticosterone in combination with Enzalutamide. *p<0.05, **p<0.01, ***p<0.001.
In the prefrontal cortex, in experiment 1, corticosterone significantly increased mRNA expression for all genes measured (main effects p ≤ 0.01, Figures 3A-F). Additionally, dihydrotestosterone significantly decreased Sgk1 and Tsc22d3 expression (p<0.05 in both cases, Figures 3B, E). No interaction effects were found. Posthoc comparisons showed that for all genes the expression was significantly increased in the CORT group compared to the DHT group. Fkbp5 and Pla2g3 expression was also significantly higher in the CORT group compared to VEH and DHT groups. Sgk1 expression in the DHT+CORT group was significantly lower than in the CORT group but higher than in the DHT group, and expression in the DHT group was significantly lower than in the VEH group. Overall, we found independent effects of corticosterone and dihydrotestosterone on gene expression in PFC, and these had opposite directions for those genes that were affected by both treatments.
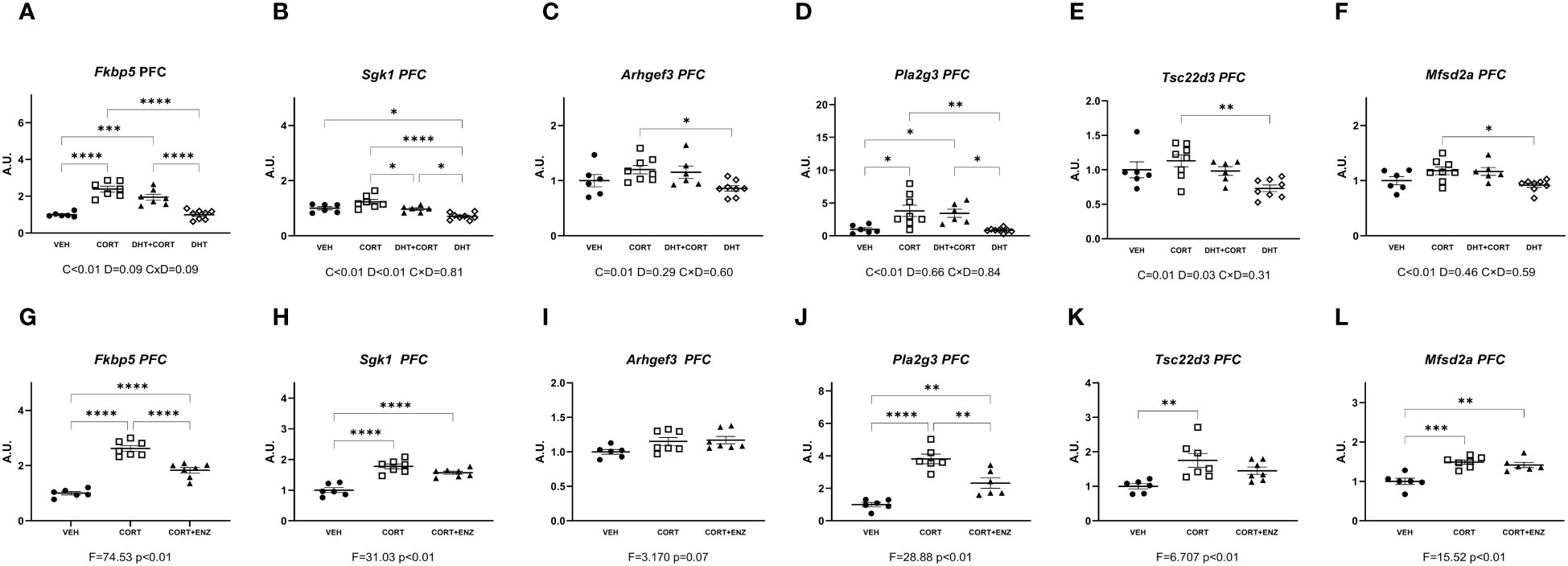
Figure 3 Gene expression in prefrontal cortex. (A-F) Expression of GR responsive genes in prefrontal cortex (PFC), obtained from experiment 1. 2-way ANOVA main effects are depicted below the x axis as: C, Corticosterone; D, Dihydrotestosterone; CxD, Interaction. (G-L) expression of GR responsive genes in prefrontal cortex (PFC), obtained from experiment 2. 1-way ANOVA values of F-statistic and p are depicted below the x axis. VEH, vehicle; CORT, Corticosterone; CORT+ENZ, Corticosterone in combination with Enzalutamide. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
In experiment 2, 1-way ANOVA analyses indicated statistical significance for all genes except Arghef1 (Figures 3G-L). Posthoc analysis indicated that these genes were all induced by corticosterone. Fkbp5 and Pla2g3 expression in the CORT+ENZ group was intermediate to that in the VEH and CORT groups. Sgk1 and Mfsd2a expression was increased both in CORT and CORT+ENZ groups compared to VEH group, and Tsc22d3 expression was increased in the CORT group compared to the VEH group. These results indicate that PFC is particularly responsive to GCs, and that enzalutamide attenuated the corticosterone effects for some genes. This indicates that androgens may affect GC signaling in this brain region. However, in terms of possible mechanism, it should be noted that the AR agonist and antagonist both tended to downregulate/attenuate gene expression. Thus, there may be different mechanism in action in the two different settings that we evaluated.
In the ventromedial hypothalamus, in experiment 1, only Pla2g3 expression was induced by corticosterone (main effect p<0.01, Figure 4D). Dihydrotestosterone significantly increased Arhgef3 and Mfsd2a expression (p=0.05 and p=0.01, respectively; Figures 4C, F). In experiment 2, 1-way ANOVA analyses indicated statistical significance for Fkbp5, Sgk1, Pla2g3, Tsc22d3 and Mfsd2a (p ≤ 0.01 in all cases, Figures 4G, H, J–L). Posthoc analyses showed that Fkbp5, Sgk1, Tsc22d3 and Mfsd2a expression was increased in CORT and CORT+ENZ groups compared to VEH group, and in Pla2g3 expression was increased only in the CORT group compared to VEH group. These results suggest that in ventromedial hypothalamus most of the genes we selected are responsive to GCs. Some of these genes also are induced by dihydrotestosterone, and the effect of enzalutamide was very limited.
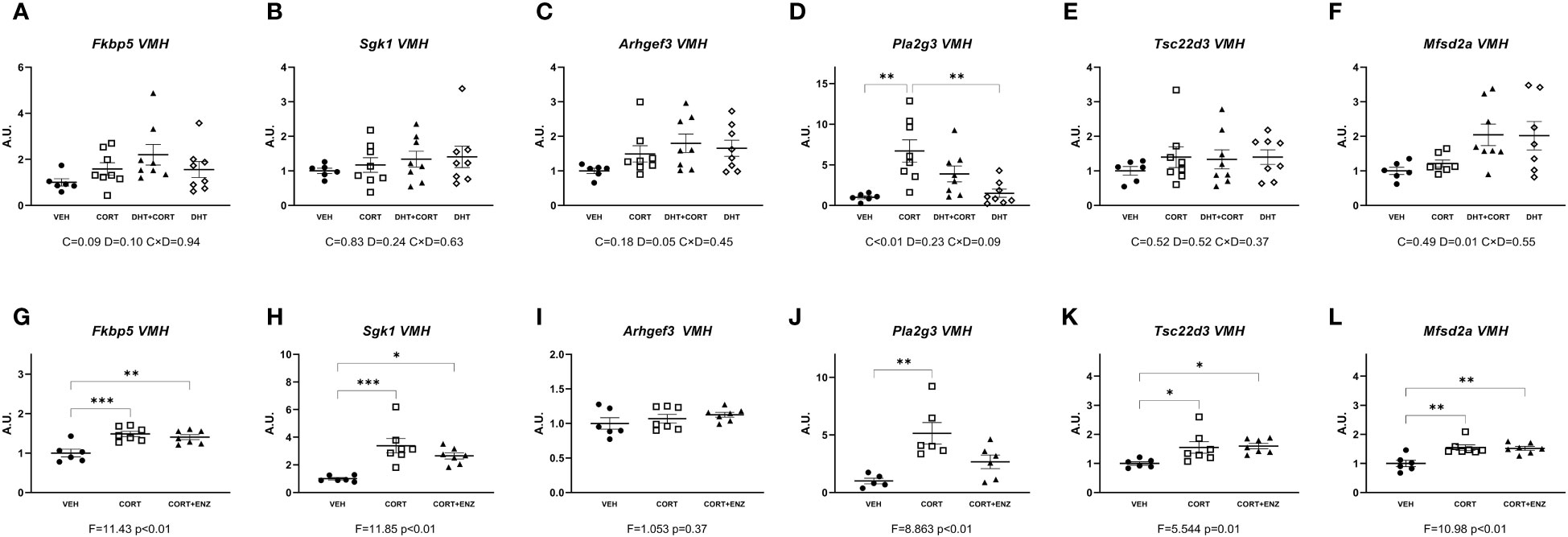
Figure 4 Gene expression in ventromedial hypothalamus. (A-F) Expression of GR responsive genes in ventromedial hypothalamus (VMH), obtained from experiment 1. 2-way ANOVA main effects are depicted below the x axis as: C, Corticosterone; D, Dihydrotestosterone; CxD, Interaction. (G-L) Expression of GR responsive genes in ventromedial hypothalamus (VMH), obtained from experiment 2. 1-way ANOVA values of F-statistic and p are depicted below the x axis. VEH, vehicle; CORT, Corticosterone; CORT+ENZ, Corticosterone in combination with Enzalutamide. *p<0.05, **p<0.01, ***p<0.001.
In the ventral tegmental area, in experiment 1, corticosterone significantly increased Fkbp5, Sgk1, Pla2g3 and Mfsd2a expression (main effect p ≤ 0.01 in all cases; Figures 5A, B, D, F). Dihydrotestosterone significantly decreased Arhgef3 and Tsc22d3 expression (p=0.02 and p<0.01, Figures 5C, E). Posthoc analysis for Fkbp5 and Pla2g3 showed that expression was increased in the CORT and DHT+CORT groups compared to both VEH and DHT groups, while in Arhgef3 expression was increased in the CORT group compared to the DHT group, and Mfsd2a expression was increased in the DHT+CORT group compared to both VEH and DHT groups, and in the CORT group compared to the DHT group. In experiment 2, 1-way ANOVA analyses reached statistical significance for Fkbp5, Sgk1 and Pla2g3 and Mfsd2a (p<0.05 in all cases, Figures 5G, H, J, L). Posthoc analyses showed increased expression of Fkbp5 in the CORT group compared to the VEH group, increased expression of Sgk1 and Pla2g3 in CORT and CORT+ENZ groups compared to VEH group, and increased expression of Mfsd2a in CORT+ENZ group compared to VEH group.
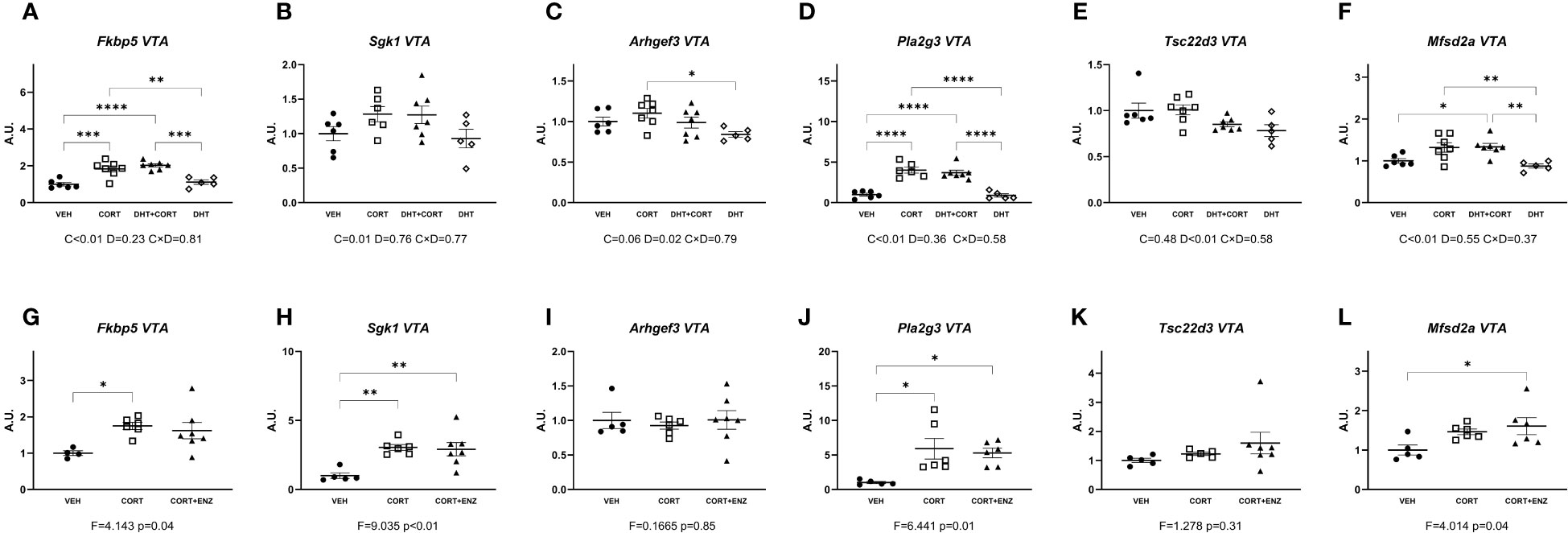
Figure 5 Gene expression in ventral tegmental area. (A-F) Expression of GR responsive genes in ventral tegmental area (VTA), obtained from experiment 1. 2-way ANOVA main effects are depicted below the x axis as: C, Corticosterone; D, Dihydrotestosterone; CxD, Interaction. (G-L) Expression of GR responsive genes in ventral tegmental area (VTA), obtained from experiment 2. 1-way ANOVA values of F-statistic and p are depicted below the x axis. VEH, vehicle; CORT, Corticosterone; CORT+ENZ, Corticosterone in combination with Enzalutamide. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
These data show that both corticosterone and DHT are able to regulate gene expression. In experiment 1 the target genes did not overlap, and directionality of the effects was opposite. The independent effects of the hormones was confirmed in experiment 2, where enzalutamide had no effects on corticosterone-induced gene expression.
In the substantia nigra, in experiment 1, corticosterone significantly increased the expression of evaluated genes except for Tsc22d3 (Figures 6A-F). Posthoc analyses showed that Fkbp5 expression in the CORT group was higher than in both VEH and DHT groups. Sgk1 expression was higher in the CORT group compared to the VEH group, and expression of Pla2g3 and Mfsd2a was increased in both CORT and DHT+CORT groups compared to both VEH and DHT groups. In experiment 2, 1-way ANOVA analyses indicated statistical significance for the same genes as in experiment 1(Figures 6G-L). Posthoc analyses showed that Fkbp5 and Sgk1 expression in the CORT+ENZ group was intermediate to CORT and VEH groups. The expression of Arhgef3 and Mfsd2a was increased in CORT and CORT+ENZ groups compared to the VEH group, and Pla2g3 expression was increased in CORT group compared to VEH group.

Figure 6 Gene expression in Substantia Nigra. (A-F) Expression of GR responsive genes in substantia nigra (SN), obtained from experiment 1. 2-way ANOVA main effects are depicted below the x axis as: C, Corticosterone; D, Dihydrotestosterone; CxD, Interaction. (G-L) Expression of GR responsive genes in substantia nigra (SN), obtained from experiment 2. 1-way ANOVA values of F-statistic and p are depicted below the x axis. VEH, vehicle; CORT, Corticosterone; CORT+ENZ, Corticosterone in combination with Enzalutamide. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
These data show that corticosterone but not dihydrotestosterone had intrinsic effects on the selected target genes. Enzalutamide attenuated the induction of expression by corticosterone for several genes.
Given the availability of the tissue and the relatively direct link to physiological function, we also probed regulation of genes related to dopamine signaling in the ventral tegmental area and substantia nigra. In experiment 1, dihydrotestosterone significantly decreased expression of Ddc and Slc18a1 in the ventral tegmental area (main effect p<0.01 in both cases; Figures 7B, J). Posthoc analysis showed that Ddc expression was decreased in the DHT+CORT group compared to both VEH and CORT groups, and that Slc18a1 expression was decreased in the DHT+CORT group compared to CORT group. In experiment 2, 1-way ANOVA analyses reached statistical significance for Slc18a1 expression in the ventral tegmental area (p=0.02, Figure 7N) and for the expression of Th and Slc18a1 in the substantia nigra (p<0.05 in both cases, Figures 7 G, P). Posthoc analyses showed that Slc18a1 expression in the ventral tegmental area was increased in the CORT+ENZ group compared to the VEH group, and that in the substantia nigra Th expression was increased in the CORT+ENZ group compared to the CORT group, while Slc18a1 expression was increased in the CORT+ENZ group compared to the VEH group. Of note, the markers for dopamine signaling were the only instance in which dihydrotestosterone and enzalutamide showed opposite effects, with down-regulation by dihydrotestosterone and the occasional upregulation after enzalutamide (although the specific genes/areas were less consistent).
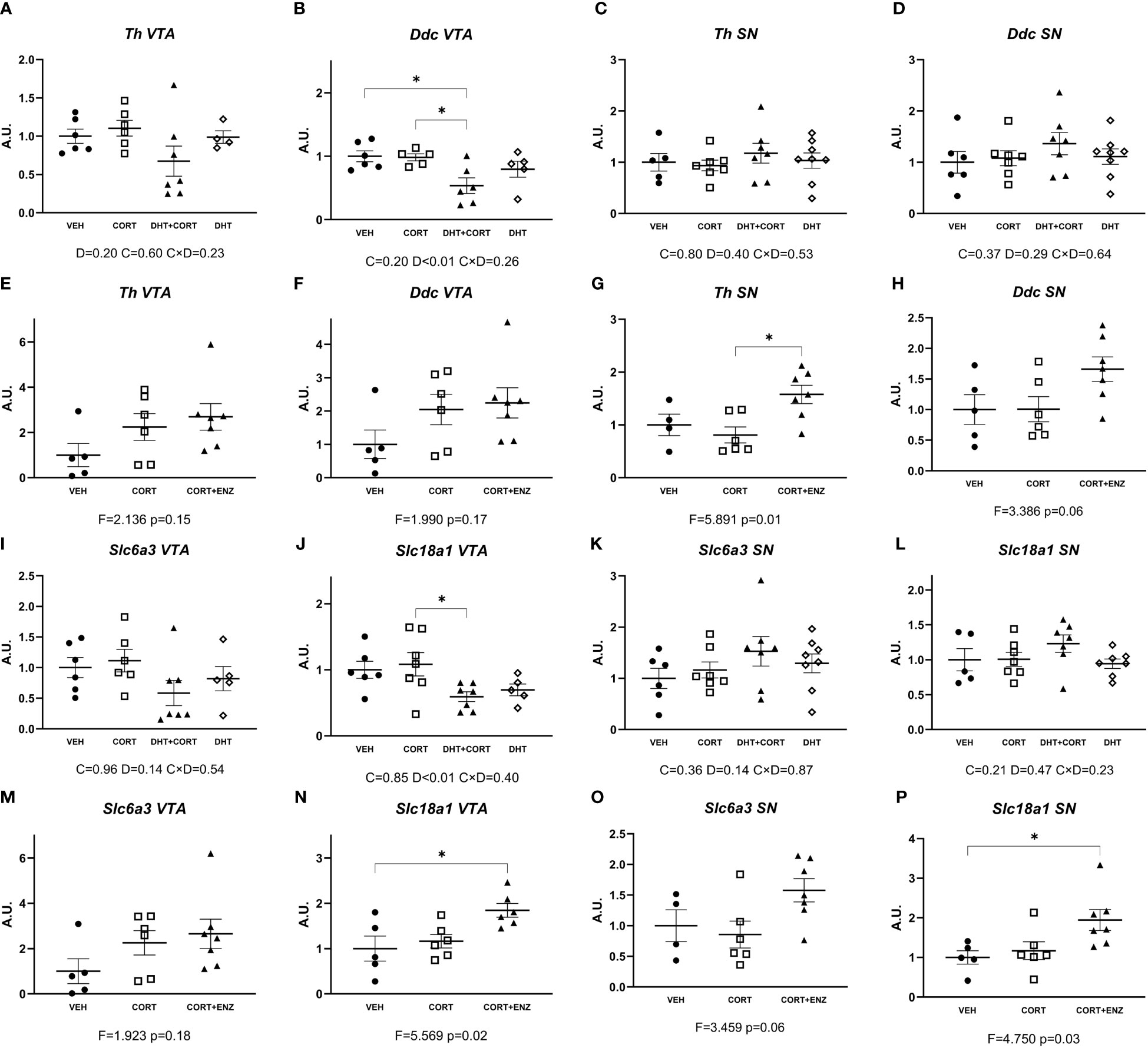
Figure 7 Dopamine synthesis and transport genes expression in VTA and SN. (A–D) Expression of dopamine synthesis genes in ventral tegmental area (VTA) and substantia nigra (SN), obtained from experiment 1. (E–H) Expression of dopamine synthesis genes in VTA and SN, obtained from experiment 2. (I–L) Expression of dopamine transport genes in VTA and SN, obtained from experiment 1. (M–P) Expression of dopamine transport genes in VTA and SN, obtained from experiment 2. In (A–D) and (I–L), 2-way ANOVA main effects are depicted below the x axis as: C, Corticosterone; D, Dihydrotestosterone; CxD, Interaction. In (E–H) and (M–P), 1-way ANOVA values of F-statistic and p are depicted below the x axis. VEH: vehicle, CORT, Corticosterone; CORT+ENZ, Corticosterone in combination with Enzalutamide. *p<0.05.
In this work we aimed to test the hypothesis that androgen signaling could influence glucocorticoid signaling in the brain, which could explain our previous observation that chronic hypercorticosteronemia had more pronounced effects in male mice (3, 4) as compared to female mice. We and others had also previously observed that androgens influence GR activity in liver and adipose tissues (12, 13). Peripheral AR modulation of GR transcriptional activity was found only in regimes of chronic exposure to GCs and might be due to AR upregulation (21). Since AR-GR heterodimerization has been shown in vitro (11), we addressed the possibility that androgen exerts an effect over glucocorticoid signaling in brain areas where AR and GR are co-expressed (22, 27, 28).
In our experiments, the effects of androgen signaling on brain glucocorticoid-induced gene expression were modest. A priori, there are several ways by which steroid hormones may affect each other. We cannot exclude that the picture would be different in other experimental settings. For one thing, the used corticosterone treatment was earlier shown to not affect basal testosterone levels (12). However, the treatment with 20 mg pellets of corticosterone led to constant and very high levels of circulating hormone. This situation may mimic Cushing’s disease, or regimes in which high doses of synthetic glucocorticoids are used. We would expect that at these levels GRs would be likely maximally occupied, and at such saturated conditions potential modulatory effects of androgens may have been masked. And so, while our experimental design can reveal some forms of GR modulation via androgen receptors, future experiments should include more physiological hormone levels. Other limitations of our data is that we quantified bulk mRNA levels, which may have masked cell-type specific effects that may be present. Also, we did not measure protein levels, which could had helped us to infer the basal availability of AR and GR in each brain region.
Overall, we found that almost all the GR-responsive genes that we selected were upregulated by corticosterone treatment in most brain regions, while dihydrotestosterone had more limited effects. Although we observed significant effects of AR activation by dihydrotestosterone, we did not observe any statistically significant interaction effects between GC and androgens. We propose that in cases such as Fkbp5 and Pla2g3 in the prefrontal cortex, and Fkbp5 and Sgk1 in the substantia nigra, a tonic activation of AR by endogenous androgens would be enough to positively affect GR signaling, which we could unveil after blocking AR activity by means of enzalutamide. This scenario has been demonstrated previously in the study of neurobiology of sexual and aggressive behaviors (29). In our results, in the presence of enzalutamide, corticosterone treatment was still effective in upregulating Fkbp5, Sgk1 and Pla2g3 expression, yet the effect was not as pronounced when AR was blocked (Figures 3G, J, 6G, H).
The fact that dihydrotestosterone and enzalutamide did not have opposite effects may also point to off-target effects of enzalutamide. These in all likelihood would not involve MR, GR or progesterone receptor (PR), given the high selectivity of enzalutamide for AR (30). Enzalutamide can induce pregnane X receptor (PXR) activity, but with an EC50 of more than 1µM (31). We propose that any AR independent effect of enzalutamide should therefore involve an unknown mediator. Here our conclusion is limited by the fact that we did not measure protein levels of AR and GR, as well as that we did not assess the levels of AR and GR mRNA in the experiment 2, so we cannot exclude the possibility that enzalutamide might have influenced AR and GR levels. To our knowledge this has been addressed only in prostate and bladder cancer models, with inconsistent results (32, 33). Alternative ways to further assess this would include castrating animals or the use of AR-knockout animals.
Corticosterone significantly increased the expression of Fkbp5 in the prefrontal cortex and substantia nigra (Figures 3A, 6A), and Pla2g3 in the prefrontal cortex (Figure 3D) in comparison to vehicle. These increases were attenuated when corticosterone was administered in combination with enzalutamide (Figures 3G, J, 6G). We had observed previously that in liver and in brown adipose tissue enzalutamide ablated the upregulation of GR target genes caused by corticosterone (12). Our data showed that in the prefrontal cortex, corticosterone treatment increased AR mRNA expression, as we have previously shown in the mouse liver (21), which suggests that AR is contributing to Fkbp5 and Pla2g3 upregulation upon glucocorticoid treatment. The upregulation of AR expression is the only mechanism for an effect of androgens on glucocorticoid target genes for which there is evidence in our data.
Colocalization of AR and GR in different brain areas (20, 34) would be a prerequisite for an effect of androgens on GR-mediated transcription that would depend on direct protein-protein interactions between the receptors (11). With data derived from hippocampal scRNA-seq (23, 24), we tested potential dual regulation via AR and GR for genes that would be co-expressed in particular hippocampal cell types. We found that basal co-expression of AR and GR does not per se predict influence of AR over GR regulation of downstream target genes. However, for hippocampal Sgk1, the basal expression patterns predicted gene regulation. Sgk1 showed significant upregulation by dihydrotestosterone. This gene showed appreciable basal expression in the CA1-3 pyramidal cells, which also expressed AR (Figure 2A).
Outside of the hippocampus we found significant dihydrotestosterone induced upregulation of Arhgef3 and Mfsd2a in the ventromedial hypothalamus (Figures 4C, F), and significant independent corticosterone and dihydrotestosterone effects in Sgk1 and Tsc22d3 in prefrontal cortex (Figures 3B, E). The ventromedial hypothalamus Arhgef3 increase is probably an exclusive AR effect since corticosterone did not influence it in either experiment (Figures 4C, I). In the ventromedial hypothalamus increased Mfsd2a expression is more difficult to interpret since in experiment 1 the increase in expression is a dihydrotestosterone effect, but in experiment 2 both corticosterone and corticosterone in combination with enzalutamide increase expression (Figures 4F, L). This could indicate that other transcription factors interact with both GR and AR independently. In the case of the prefrontal cortex, we propose that the significant independent corticosterone and dihydrotestosterone effects in Sgk1 and Tsc22d3 (Figures 3B, E) reflect opposite transcriptional regulatory actions of AR and GR. These effects are consistent with basal expression of the receptors, and could be explained further with use of scRNA-seq data in these brain areas. Datasets recently obtained and published by Bhattacherjee et al. (35) and Hook et al. (36) could be used as a starting point for future experiments addressing possible cell-type specific functional interactions between GR and AR, be it via direct protein-protein interaction, or in more indirect ways.
Our data showed an upregulatory effect by corticosterone and a downregulatory effect by dihydrotestosterone in the prefrontal cortex on Sgk1 and Tsc22d3 expression. These genes are well known as shared target genes for MR, GR, and AR via established glucocorticoid response elements (GREs) in different tissues (37–39). Sgk1 has been established as a gene upregulated by AR activation in both healthy and prostate cancer cells (40, 41). However, recently it was shown that dihydrotestosterone abolished Sgk1 increases caused by dexamethasone stimulation in triple-negative breast cancer cells (42). This discrepancy may be related to differences in coregulator presence per cell type or at the particular chromatin locus, considering that coregulator expression varies throughout brain regions (43), and that specific configurations of coregulator expression can modify expression of nuclear receptor target genes. For example, enhancing the expression of SRC1a abolishes a dexamethasone-induced increase in Crh expression in the amygdala, but has no effect in Fkbp5 (44). Given the functional diversity of the different brain regions and areas we can speculate that Sgk1 bidirectional regulation by GR and AR is due to molecular make-up of the constituent cells of the prefrontal cortex and hippocampus.
In a previous study on steroid receptor colocalization in the mouse brain, the co-expression of GR, AR and the nuclear receptor coregulator Pak6 in the midbrain dopaminergic regions stood out (20). Here we extended our ‘pure GR target gene’ approach with markers for dopaminergic signaling: two factors pertaining to the dopamine synthesis pathway and two dopamine transporters, in both the ventral tegmental area and the substantia nigra. Our data indicate that midbrain dopaminergic systems are regulated by androgens. We found that in ventral tegmental area, dihydrotestosterone lowered the expression of Ddc and Slc18a1. In the substantia nigra we observed that enzalutamide caused significant increases in the expression of Th and Slc18a1. Our results suggest that AR activation attenuates dopamine synthesis and presynaptic loading in the ventral tegmental area, and that its inactivation potentiates synthesis and vesicular loading. This is in accordance to previous results in which it has been observed that androgens decrease tyrosine hydroxylase (TH) expression in ventral tegmental area and substantia nigra (45) in a mechanism related to apoptosis (46, 47), as well as expression of dopamine transporter (DAT) (the translational product of Slc6a3) in prefrontal cortex (48). In contrast, in distinct experimental setups androgens might also exert upregulatory activities on dopaminergic system. Activated AR can increase Th promoter activity in cell lines (49). In several studies in adolescent male rats androgens where shown to stimulate TH and Slc6a3 mRNA or protein expression (50–53). The discrepancies between these observations and the results obtained by us and others could be influenced by either the age of the animals or possibly by a species difference between mice and rats.
In conclusion, in our data, interplay between the glucocorticoid and androgen signaling axes in the male mouse brain was restricted to the prefrontal cortex, the substantia nigra and perhaps the hippocampus. An upregulation of AR expression by glucocorticoids is the mechanism for which there is evidence in our data, analogous to earlier observations in the liver of male mice (21). GR-driven transcription is largely resilient to AR influence, at least at the relatively high dose of corticosterone that we used. In the case of prefrontal cortex, AR activation has a downregulating effect on GR transcription of Sgk1 and Tsc22d3 only. Also, in both prefrontal cortex and substantia nigra, an intact AR signal appears to contribute to GR transcription of Fkbp5 and Sgk1. Lastly, blockade of AR increased the expression of the dopamine-related genes Th and Slc18a1. Future works will help to clarify the influence of AR in brain cell processes and signaling routes that can explain sexual differences in health and disease.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://osf.io/j5wtd/.
The animal study was approved by the ethical committee of Leiden University Medical Center. The study was conducted in accordance with the local legislation and institutional requirements.
JA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Validation, Writing – original draft, Writing – review & editing. HS: Investigation, Project administration, Resources, Writing – review & editing. EV: Data curation, Writing – review & editing. JK: Conceptualization, Funding acquisition, Writing – review & editing. OM: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. JA was funded by CONACyT (grant number 440584). JK and OCM receive research funding from Corcept Therapeutics. The funder was not involved in the study design, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
OM and JK collaborate with and receive funding from Corcept Therapeutics who develop GR antagonists for a variety of indications related to hypercorticism.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1292024/full#supplementary-material
1. Swarbrick M, Zhou H, Seibel M. MECHANISMS IN ENDOCRINOLOGY: Local and systemic effects of glucocorticoids on metabolism: new lessons from animal models. Eur J Endocrinol (2021) 185(5):R113–R29. doi: 10.1530/EJE-21-0553
2. Hodges TE, Puri TA, Blankers SA, Qiu W, Galea LAM. Steroid hormones and hippocampal neurogenesis in the adult mammalian brain. Vitam Horm (2022) 118:129–70. doi: 10.1016/bs.vh.2021.11.003
3. Amaya JM, Suidgeest E, Sahut-Barnola I, Dumontet T, Montanier N, Pages G, et al. Effects of long-term endogenous corticosteroid exposure on brain volume and glial cells in the adKO mouse. Front Neurosci (2021) 15:604103. doi: 10.3389/fnins.2021.604103
4. Amaya JM, Viho EMG, Sips HCM, Lalai RA, Sahut-Barnola I, Dumontet T, et al. Gene expression changes in the brain of a Cushing's syndrome mouse model. J Neuroendocrinol (2022) 34(4):e13125. doi: 10.1111/jne.13125
5. Cain DW, Cidlowski JA. Specificity and sensitivity of glucocorticoid signaling in health and disease. Best Pract Res Clin Endocrinol Metab (2015) 29(4):545–56. doi: 10.1016/j.beem.2015.04.007
6. Vandewalle J, Luypaert A, De Bosscher K, Libert C. Therapeutic mechanisms of glucocorticoids. Trends Endocrinol Metab (2018) 29(1):42–54. doi: 10.1016/j.tem.2017.10.010
7. Fuller PJ, Yang J, Young MJ. Mechanisms of mineralocorticoid receptor signaling. Vitam Horm (2019) 109:37–68. doi: 10.1016/bs.vh.2018.09.004
8. Mifsud KR, Reul J. Mineralocorticoid and glucocorticoid receptor-mediated control of genomic responses to stress in the brain. Stress (2018) 21(5):389–402. doi: 10.1080/10253890.2018.1456526
9. Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol (2010) 72:247–72. doi: 10.1146/annurev-physiol-021909-135917
10. De Bosscher K, Desmet SJ, Clarisse D, Estebanez-Perpina E, Brunsveld L. Nuclear receptor crosstalk - defining the mechanisms for therapeutic innovation. Nat Rev Endocrinol (2020) 16(7):363–77. doi: 10.1038/s41574-020-0349-5
11. Chen S, Wang J, Yu G, Liu W, Pearce D. Androgen and glucocorticoid receptor heterodimer formation. A possible mechanism for mutual inhibition of transcriptional activity. J Biol Chem (1997) 272(22):14087–92. doi: 10.1074/jbc.272.22.14087
12. Spaanderman DCE, Nixon M, Buurstede JC, Sips HC, Schilperoort M, Kuipers EN, et al. Androgens modulate glucocorticoid receptor activity in adipose tissue and liver. J Endocrinol (2018) 240(1):51–63. doi: 10.1530/JOE-18-0503
13. Gasparini SJ, Swarbrick MM, Kim S, Thai LJ, Henneicke H, Cavanagh LL, et al. Androgens sensitise mice to glucocorticoid-induced insulin resistance and fat accumulation. Diabetologia (2019) 62(8):1463–77. doi: 10.1007/s00125-019-4887-0
14. Green MR, Zeidan M, Hodges TE, McCormick CM. Age-dependent regulation by androgens of gene expression in the anterior hypothalamus and stress-induced release of adrenal hormones in adolescent and adult male rats. J Neuroendocrinol (2019) 31(6):e12714. doi: 10.1111/jne.12714
15. Rybka KA, Sturm KL, De Guzman RM, Bah S, Jacobskind JS, Rosinger ZJ, et al. Androgen regulation of corticotropin releasing factor receptor 1 in the mouse brain. Neuroscience (2022) 491:185–99. doi: 10.1016/j.neuroscience.2022.04.005
16. Marrocco J, McEwen BS. Sex in the brain: hormones and sex differences. Dialogues Clin Neurosci (2016) 18(4):373–83. doi: 10.31887/DCNS.2016.18.4/jmarrocco
17. Zuloaga DG, Heck AL, De Guzman RM, Handa RJ. Roles for androgens in mediating the sex differences of neuroendocrine and behavioral stress responses. Biol Sex Differ (2020) 11(1):44. doi: 10.1186/s13293-020-00319-2
18. Sahut-Barnola I, de Joussineau C, Val P, Lambert-Langlais S, Damon C, Lefrançois-Martinez A-M, et al. Cushing's syndrome and fetal features resurgence in adrenal cortex–specific prkar1a knockout mice. PloS Genet (2010) 6(6):e1000980. doi: 10.1371/journal.pgen.1000980
19. Lu SF, McKenna SE, Cologer-Clifford A, Nau EA, Simon NG. Androgen receptor in mouse brain: sex differences and similarities in autoregulation. Endocrinology (1998) 139(4):1594–601. doi: 10.1210/endo.139.4.5863
20. Mahfouz A, Lelieveldt BP, Grefhorst A, van Weert LT, Mol IM, Sips HC, et al. Genome-wide coexpression of steroid receptors in the mouse brain: Identifying signaling pathways and functionally coordinated regions. Proc Natl Acad Sci U S A (2016) 113(10):2738–43. doi: 10.1073/pnas.1520376113
21. Buurstede JC, Paul SN, De Bosscher K, Meijer OC, Kroon J. Hepatic glucocorticoid-induced transcriptional regulation is androgen-dependent after chronic but not acute glucocorticoid exposure. FASEB J (2022) 36(4):e22251. doi: 10.1096/fj.202101313R
22. Dugger BN, Morris JA, Jordan CL, Breedlove SM. Androgen receptors are required for full masculinization of the ventromedial hypothalamus (VMH) in rats. Horm Behav (2007) 51(2):195–201. doi: 10.1016/j.yhbeh.2006.10.001
23. Viho EMG, Buurstede JC, Berkhout JB, Mahfouz A, Meijer OC. Cell type specificity of glucocorticoid signaling in the adult mouse hippocampus. J Neuroendocrinol (2022) 34(2):e13072. doi: 10.1111/jne.13072
24. Juszczak GR, Stankiewicz AM. Glucocorticoids, genes and brain function. Prog Neuropsychopharmacol Biol Psychiatry (2018) 82:136–68. doi: 10.1016/j.pnpbp.2017.11.020
25. van Weert L, Buurstede JC, Mahfouz A, Braakhuis PSM, Polman JAE, Sips HCM, et al. NeuroD factors discriminate mineralocorticoid from glucocorticoid receptor DNA binding in the male rat brain. Endocrinology (2017) 158(5):1511–22. doi: 10.1210/en.2016-1422
26. Buurstede JC, van Weert L, Colucci P, Gentenaar M, Viho EMG, Koorneef LL, et al. Hippocampal glucocorticoid target genes associated with enhancement of memory consolidation. Eur J Neurosci (2022) 55(9-10):2666–83. doi: 10.1111/ejn.15226
27. Commins D, Yahr P. Autoradiographic localization of estrogen and androgen receptors in the sexually dimorphic area and other regions of the gerbil brain. J Comp Neurol (1985) 231(4):473–89. doi: 10.1002/cne.902310406
28. Low KL, Tomm RJ, Ma C, Tobiansky DJ, Floresco SB, Soma KK. Effects of aging on testosterone and androgen receptors in the mesocorticolimbic system of male rats. Horm Behav (2020) 120:104689. doi: 10.1016/j.yhbeh.2020.104689
29. Cunningham RL, Lumia AR, McGinnis MY. Androgen receptors, sex behavior, and aggression. Neuroendocrinology (2012) 96(2):131–40. doi: 10.1159/000337663
30. Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science (2009) 324(5928):787–90. doi: 10.1126/science.1168175
31. Dellal H, Boulahtouf A, Alaterre E, Cuenant A, Grimaldi M, Bourguet W, et al. High content screening using new U2OS reporter cell models identifies harmol hydrochloride as a selective and competitive antagonist of the androgen receptor. Cells (2020) 9(6):1469. doi: 10.3390/cells9061469
32. Abazid A, Martin B, Choinowski A, McNeill RV, Brandenburg LO, Ziegler P, et al. The androgen receptor antagonist enzalutamide induces apoptosis, dysregulates the heat shock protein system, and diminishes the androgen receptor and estrogen receptor beta1 expression in prostate cancer cells. J Cell Biochem (2019) 120(10):16711–22. doi: 10.1002/jcb.28929
33. Kawahara T, Ide H, Kashiwagi E, El-Shishtawy KA, Li Y, Reis LO, et al. Enzalutamide inhibits androgen receptor-positive bladder cancer cell growth. Urol Oncol (2016) 34(10):432.e15–23. doi: 10.1016/j.urolonc.2016.05.016
34. Kerr JE, Beck SG, Handa RJ. Androgens modulate glucocorticoid receptor mRNA, but not mineralocorticoid receptor mRNA levels, in the rat hippocampus. J Neuroendocrinol (1996) 8(6):439–47. doi: 10.1046/j.1365-2826.1996.04735.x
35. Bhattacherjee A, Djekidel MN, Chen R, Chen W, Tuesta LM, Zhang Y. Cell type-specific transcriptional programs in mouse prefrontal cortex during adolescence and addiction. Nat Commun (2019) 10(1):4169. doi: 10.1038/s41467-019-12054-3
36. Hook PW, McClymont SA, Cannon GH, Law WD, Morton AJ, Goff LA, et al. Single-cell RNA-seq of mouse dopaminergic neurons informs candidate gene selection for sporadic parkinson disease. Am J Hum Genet (2018) 102(3):427–46. doi: 10.1016/j.ajhg.2018.02.001
37. Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev (2007) 21(16):2005–17. doi: 10.1101/gad.1564207
38. Jin HJ, Kim J, Yu J. Androgen receptor genomic regulation. Transl Androl Urol (2013) 2(3):157–77. doi: 10.3978/j.issn.2223-4683.2013.09.01
39. Munkley J, Maia TM, Ibarluzea N, Livermore KE, Vodak D, Ehrmann I, et al. Androgen-dependent alternative mRNA isoform expression in prostate cancer cells. F1000Res (2018) 7:1189. doi: 10.12688/f1000research.15604.1
40. Shanmugam I, Cheng G, Terranova PF, Thrasher JB, Thomas CP, Li B. Serum/glucocorticoid-induced protein kinase-1 facilitates androgen receptor-dependent cell survival. Cell Death Differ (2007) 14(12):2085–94. doi: 10.1038/sj.cdd.4402227
41. Dhiman VK, Attwood K, Campbell MJ, Smiraglia DJ. Hormone stimulation of androgen receptor mediates dynamic changes in DNA methylation patterns at regulatory elements. Oncotarget (2015) 6(40):42575–89. doi: 10.18632/oncotarget.6471
42. Kanai A, McNamara KM, Iwabuchi E, Miki Y, Onodera Y, Guestini F, et al. Significance of glucocorticoid signaling in triple-negative breast cancer patients: a newly revealed interaction with androgen signaling. Breast Cancer Res Treat (2020) 180(1):97–110. doi: 10.1007/s10549-020-05523-7
43. Meijer OC, Steenbergen PJ, De Kloet ER. Differential expression and regional distribution of steroid receptor coactivators SRC-1 and SRC-2 in brain and pituitary. Endocrinology (2000) 141(6):2192–9. doi: 10.1210/endo.141.6.7489
44. Zalachoras I, Verhoeve SL, Toonen LJ, van Weert LT, van Vlodrop AM, Mol IM, et al. Isoform switching of steroid receptor co-activator-1 attenuates glucocorticoid-induced anxiogenic amygdala CRH expression. Mol Psychiatry (2016) 21(12):1733–9. doi: 10.1038/mp.2016.16
45. Johnson ML, Day AE, Ho CC, Walker QD, Francis R, Kuhn CM. Androgen decreases dopamine neurone survival in rat midbrain. J Neuroendocrinol (2010) 22(4):238–47. doi: 10.1111/j.1365-2826.2010.01965.x
46. Cunningham RL, Giuffrida A, Roberts JL. Androgens induce dopaminergic neurotoxicity via caspase-3-dependent activation of protein kinase Cdelta. Endocrinology (2009) 150(12):5539–48. doi: 10.1210/en.2009-0640
47. Seo MH, Jin BR, Kim HJ, An HJ, Yeo S. Reduction of tyrosine hydroxylase expression and increase of alpha-synuclein in the substantia nigra in a rat model of benign prostatic hyperplasia. Neurosci Lett (2022) 769:136386. doi: 10.1016/j.neulet.2021.136386
48. Du X, McCarthny CR, Notaras M, van den Buuse M, Hill RA. Effect of adolescent androgen manipulation on psychosis-like behaviour in adulthood in BDNF heterozygous and control mice. Horm Behav (2019) 112:32–41. doi: 10.1016/j.yhbeh.2019.03.005
49. Jeong H, Kim MS, Kwon J, Kim KS, Seol W. Regulation of the transcriptional activity of the tyrosine hydroxylase gene by androgen receptor. Neurosci Lett (2006) 396(1):57–61. doi: 10.1016/j.neulet.2005.11.011
50. Li L, Kang YX, Ji XM, Li YK, Li SC, Zhang XJ, et al. Finasteride inhibited brain dopaminergic system and open-field behaviors in adolescent male rats. CNS Neurosci Ther (2018) 24(2):115–25. doi: 10.1111/cns.12781
51. Tomm RJ, Seib DR, Kachkovski GV, Schweitzer HR, Tobiansky DJ, Floresco SB, et al. (2022) Androgen synthesis inhibition increases behavioural flexibility and mPFC tyrosine hydroxylase in gonadectomized male rats. J Neuroendocrinol (2022) 34(6):e13128. doi: 10.1111/jne.13128
52. Purves-Tyson TD, Owens SJ, Double KL, Desai R, Handelsman DJ, Weickert CS. Testosterone induces molecular changes in dopamine signaling pathway molecules in the adolescent male rat nigrostriatal pathway. PloS One (2014) 9(3):e91151. doi: 10.1371/journal.pone.0091151
Keywords: androgen receptor, glucocorticoid receptor, brain, dopamine, prefrontal cortex, substantia nigra
Citation: Amaya JM, Sips HCM, Viho EMG, Kroon J and Meijer OC (2024) Restricted effects of androgens on glucocorticoid signaling in the mouse prefrontal cortex and midbrain. Front. Endocrinol. 14:1292024. doi: 10.3389/fendo.2023.1292024
Received: 10 September 2023; Accepted: 21 December 2023;
Published: 18 January 2024.
Edited by:
Takayoshi Ubuka, International Cancer Laboratory Co., Ltd., JapanReviewed by:
Joanna Spencer-Segal, University of Michigan, United StatesCopyright © 2024 Amaya, Sips, Viho, Kroon and Meijer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jorge Miguel Amaya, Si5NLkEuQW1heWFfRmVybmFuZGV6QGx1bWMubmw=
†Present address: Eva MG. Viho, Department of Genes and Environment, Max Planck Institute of Psychiatry, Munich, Germany
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.