- 1Department of Animal Science and the Reproductive and Developmental Sciences Program, Michigan State University, East Lansing, MI, United States
- 2Department of Obstetrics, Gynecology, and Reproductive Sciences and Center for Reproductive Science and Medicine, University of California, San Diego, La Jolla, CA, United States
- 3Department of Radiology and Precision Health Program, Michigan State University, East Lansing, MI, United States
- 4Department of Psychology, University of California, San Diego, La Jolla, CA, United States
- 5Center for Circadian Biology, University of California, San Diego, La Jolla, CA, United States
Background: The suprachiasmatic nucleus (SCN) within the hypothalamus is a key brain structure required to relay light information to the body and synchronize cell and tissue level rhythms and hormone release. Specific subpopulations of SCN neurons, defined by their peptide expression, regulate defined SCN output. Here we focus on the vasoactive intestinal peptide (VIP) expressing neurons of the SCN. SCN VIP neurons are known to regulate circadian rhythms and reproductive function.
Methods: To specifically study SCN VIP neurons, we generated a novel knock out mouse line by conditionally deleting the SCN enriched transcription factor, Ventral Anterior Homeobox 1 (Vax1), in VIP neurons (Vax1Vip; Vax1fl/fl:VipCre).
Results: We found that Vax1Vip females presented with lengthened estrous cycles, reduced circulating estrogen, and increased depressive-like behavior. Further, Vax1Vip males and females presented with a shortened circadian period in locomotor activity and ex vivo SCN circadian period. On a molecular level, the shortening of the SCN period was driven, at least partially, by a direct regulatory role of VAX1 on the circadian clock genes Bmal1 and Per2. Interestingly, Vax1Vip females presented with increased expression of arginine vasopressin (Avp) in the paraventricular nucleus, which resulted in increased circulating corticosterone. SCN VIP and AVP neurons regulate the reproductive gonadotropin-releasing hormone (GnRH) and kisspeptin neurons. To determine how the reproductive neuroendocrine network was impacted in Vax1Vip mice, we assessed GnRH sensitivity to a kisspeptin challenge in vivo. We found that GnRH neurons in Vax1Vip females, but not males, had an increased sensitivity to kisspeptin, leading to increased luteinizing hormone release. Interestingly, Vax1Vip males showed a small, but significant increase in total sperm and a modest delay in pubertal onset. Both male and female Vax1Vip mice were fertile and generated litters comparable in size and frequency to controls.
Conclusion: Together, these data identify VAX1 in SCN VIP neurons as a neurological overlap between circadian timekeeping, female reproduction, and depressive-like symptoms in mice, and provide novel insight into the role of SCN VIP neurons.
Introduction
The circadian system is responsible for coordinating circadian and time-of-day signals throughout the body. On a cellular level, circadian rhythms are produced by the molecular clock, a transcription/translation feedback loop that generates 24-hour cell autonomous rhythms (1–3). Alterations in molecular clock function can lead to changes in circadian behaviors and fertility, as shown in transgenic mouse models with a loss of Bmal1, a gene required for molecular circadian rhythm generation (4–8). Along with proper molecular clock function, the suprachiasmatic nucleus (SCN), located in the ventral hypothalamus, is the central pacemaker of the brain and serves to coordinate external timing signals and physiological processes throughout the body. Importantly, the SCN requires a finely balanced neuropeptide expression to maintain circadian rhythms (9). Correct levels of vasoactive intestinal peptide (VIP), along with other neuropeptides such as arginine vasopressin (AVP), somatostatin, and gastric releasing peptide, are required for SCN output controlling synchrony both within and downstream of the SCN, including behavioral and tissue level circadian rhythms. Both the molecular clock and neuropeptide expression must function correctly to maintain strong and synchronized circadian rhythms (10–13). SCN VIP-expressing neurons play an important role in aligning SCN function to the time of day by relaying photic information from the optic nerve to generate synchrony among SCN neurons and SCN output (14, 15).
One of the well-established downstream effects of SCN VIP neurons is the regulation of neuroendocrine function, including the regulation of gonadal sex steroids needed for female fertility (6, 16–19). In males, the role of VIP neurons with regard to reproductive function is less clear. It has been suggested that VIP may be involved in testicular health (20, 21), but no reproductive phenotype has been reported in any Vip knock-out mouse line to our knowledge. Additional studies have demonstrated that impaired SCN function or disrupted circadian rhythms in the SCN or periphery do not, or only modestly, disrupt male fertility, supporting the idea that male fertility is more robust than female fertility and often resilient to circadian disruption (22–26). In contrast, female fertility relies on precise synchronization between hormone release and peripheral tissue function (24, 27, 28). To regulate the reproductive axis, VIP neurons project directly and indirectly through SCN AVP neurons and kisspeptin neurons (29–32) to gonadotropin-releasing hormone (GnRH) neurons. GnRH is released through both pulse and surge modes into the median eminence, promoting the pituitary to release luteinizing hormone (LH) and follicle-stimulating hormone (FSH). The timing of the LH surge is particularly important in females at both the level of the hypothalamus and the ovary for optimal ovulation (33, 34). This surge requires coordinated input to GnRH neurons from both the SCN and kisspeptin neurons. In the ovary, the molecular clock is required in ovarian theca cells to increase LH receptor expression around the time of day of the LH surge, facilitating ovulation (35).
Given their importance for both circadian timekeeping and reproductive health, the role of VIP neurons in the SCN is an important area of investigation in the context of female reproduction. Previous work has shown that changes in VIP alter circadian rhythms in mice (36–38), with full-body VIP knock-out female mice having disruptions in both the circadian and reproductive systems, demonstrating the importance of VIP in both processes (19). Such changes in VIP can also negatively impact reproductive hormone release through dysregulation of the required neurocircuitry for LH and FSH release (39–41), which can, in turn, lead to dysregulation of female reproductive sex steroids (42), an imbalance that can have negative effects on both reproductive function and mood (43, 44). Evidence supports that disrupted circadian rhythms and changes in estrogen and progesterone might be contributing factors to depressive symptoms in humans (41, 45). Together this evidence indicates a potential shared origin between circadian deregulation, reproductive deficits, and mood changes at the level of VIP neurons.
In this study, our goal was to determine if abnormal SCN VIP neuron function causes disruption in circadian timekeeping, fertility, and mood. We deleted the SCN-enriched transcription factor, Ventral anterior homeobox 1 (VAX1) within the VIP neurons of mice. Our previous work has shown that VAX1 is required for SCN development (46, 47) and maintains a function in late development of the SCN and VIP neurons, where it is required for VIP expression, SCN output and female fertility (25). Due to the specific overlap between VAX1 and VIP in the SCN, shown here, the use of Vax1flox/flox:VipCre mice provides a model to specifically investigate how weakened, but not ablated, SCN VIP neuron function regulates reproductive function, SCN circadian output, and mood. Identifying the shared genetic underpinnings of the association of reproductive and mood disorders is a required first step towards future development of efficient strategies to improve hormone related mood disorders. We hypothesize that VAX1 in postnatal VIP neurons is required for VIP neuron function, where loss of VAX1 in VIP neurons causes weakened SCN output, leading to female, but not male, subfertility and increased depressive-like symptoms.
Materials and methods
Mouse breeding
All animal procedures were performed according to protocols approved by the University of California, San Diego Institutional Animal Care and Use Committee and the Institutional Animal Care and Use Committee of Michigan State University and conducted in accordance with the Guide for the Care and Use of Laboratory Animals (48). Mice were maintained on a light/dark cycle of LD [12 h light, 12 h dark, average light intensity ~150-350 lux within the cage]. Based on the newly proposed light reporting method by (49, 50), we determined the relative perception of light by mice using the mouse α-opics equivalent daylight illuminance (EDI, Supplementary Figure 1). We calculated that the α-opics for our experimental mice on LD to be melanopsin = 43.2 ± 16.9 lux, rod = 51.6 ± 19.8 lux, s-cone = 0.03 ± 0.01 lux, and m-cone = 57.1 ± 21.7 lux. Lights ON = ZT0 (6:00), lights OFF = ZT12 (18:00), Zeitgeber time (ZT). Vax1flox/flox (Vax1tm1c(KOMP)Mbp, MGI: 5796178) (51), were crossed with mice heterozygous for the VipCre/wt allele (JAX #010908). Period2::Luciferase (PER2::LUC) mice were purchased from JAX (strain B6.129S6-Per2tm1Jt/J, JAX #006852) and crossed with the offspring of Vax1flox/flox and VipCre/wt mice. We did not detect any significant differences in mice heterozygous for the VipCre/wt allele as compared to Vax1flox/flox mice, thus we pooled the Vax1flox/flox and VipCre/wt control groups into one control group, referred to as Ctrl. Genotyping primer sequences were as follows: Vax1-wtF: CCAGTAAGAGCCCCTTTGGG, Vax1-floxF: GCCGGAACCGAAGTTCCTA; Vax1-R: CGGATAGACCCCTTGGCATC; CreF: GCATTACCGGTCGTAGCAACGAGTG, CreR: GAACGCTAGAGCCTGTTTTGCACGTTC. Mice were kept on a C57BL/6J background and were screened for germline recombination. Mice with germline recombination were excluded from the studies. VIP-tdTomato mice were generated by crossing VipCre/wt mice with Rosa-tdTomato reporter mice (JAX# 007914). Mice were euthanized by cervical dislocation followed by decapitation.
Wheel-running behavior
Female and male mice aged 8-12 weeks at the start of the experiment were single-housed in cages containing metal running wheels and wheel revolutions were monitored using magnetic sensors. All cages were contained in a light-tight cabinet with programmable lighting conditions and rooms were monitored for temperature and humidity. Sylvania T8 32-Watt 4100K fluorescent bulbs (F032/841/ECO) were used to provide light to the cabinets. Food and water were available ad libitum during the entire experiment. After 1-week acclimation to the polypropylene cages (17.8 × 25.4 × 15.2 cm or 33.2 × 15 × 13.2 cm) containing a metal running wheel (11.4 cm diameter or 11 cm diameter, respectively), locomotor activity rhythms were monitored with a VitalView data collection system (Version 4.2, Minimitter, Bend OR) that integrated into 6 minute bins the number of magnetic switch closures triggered by half wheel rotations or full wheel rotations, respectively. Running wheel activity was initially monitored for 2 weeks in a standard 12 h light/12 h dark cycle (LD). Subsequently, mice were monitored for 4 weeks in constant darkness (DD), with wheel running data analyzed from weeks 2-4 (14 days) in DD. Cage changes were scheduled at 3-week intervals. The light intensity varied between 268-369 lux inside the mouse cages. Wheel running activity was analyzed using ClockLab Analysis (ActiMetrics) by an experimenter blind to the experimental group. The circadian period was analyzed by constructing a least-squares regression line through a minimum of 13 daily activity onsets. Daily onset and offset of activity, defined as a period of 5 h of activity following 5 h of inactivity (onset) or a period of 5 h of inactivity following 5 h of activity (offset), were used to calculate the length of the active phase (alpha). Chi2 periodograms were generated for periods from 0 to 36 h, with significance set at 0.001. Activity profiles were generated for weeks 2-4 in DD using the estimated chi2 periodogram tau for the same time period. Total daily counts for mice on wheels with 2 sensors were calculated over 24 h, during both LD and DD.
Porsolt forced swim test
Female mice aged 9-13 weeks were single-housed for two weeks on LD, then placed in a transparent 7 by 24 inch cylinder filled 2/3 full with 20-21°C water. Swim tests were completed during the mice active phase and started at ZT13. Mice were recorded in dim red light (5 lux, α-opics: melanopsin = 0.03, rod = 0.20, s-cone = 0, m-cone = 0.54, Supplementary Figures 1C, F) for 6 minutes before they were removed from the water, dried with a cloth, and returned to their cages for one hour before euthanasia and blood collection from the abdominal aorta. Videos were scored by an observer blinded to the experimental group using the sampling method where the mouse is determined to be either floating or swimming every 30 seconds for 5 minutes, with the first minute of each video going unscored and serving as an adjustment period. The percent of intervals where the mouse was observed to be floating is reported (52).
Estrous cyclicity, sperm count, and fertility assessment
For the fertility assessment, virgin 8- to 12-week-old male and female Ctrl, and Vax1Vip mice were housed with opposite-sex Ctrl mice (53, 54). The number of litters and the number of pups per litter were recorded over a period of 4 months, as described previously (54). Estrous cyclicity was monitored by vaginal lavage with 20 µl H2O daily between ZT3 and ZT5 for 16-18 days. The lavage solution was dried on a slide and stained with 0.5% methylene blue. Cytology was visually examined and scored. Ovary and uterus weights were collected after euthanasia in diestrus. Following euthanasia in males, testes and epididymis were collected and weighed. Sperm was collected from the epididymis of male mice in M2 media (Sigma #M7167). The epididymis was cut in half and sperm were expelled by gently pressing down on the epididymis and then left in M2 media at room temperature for 15 min. The numbers of total and motile sperm were counted from a 1:10 dilution of the M2 media containing sperm by using a hemocytometer. The second epididymis was cut into small pieces and left 15 minutes at room temperature in M2 media. The solution was homogenized frequently to help liberate the sperm. The solution was filtered using a cell streamer (70 µm, Falcon #352350) and sperm were diluted 1:10 with MQ before counting total number of sperm heads.
Pubertal onset
Pubertal onset was established by visual inspection of preputial separation (PPS) in males and vaginal opening (VO) in females, as described previously (54). Body weight was recorded daily until pubertal onset was observed.
Immunohistochemistry staining
Tissues were collected between ZT3 and ZT5 from adult male and proestrus female mice on LD light cycle and fixed overnight at 4°C in 60% ethanol, 10% formaldehyde, and 10% glacial acetic acid. Tissues were washed in 70% ethanol and embedded in paraffin. Single immunohistochemistry on 10 µm coronal brain sections embedded in paraffin was performed as previously described (24). The primary antibody was rabbit anti-VIP (Immunostar #20077, 1:1000, RRID : AB_572270). Sections were incubated in 1:300 secondary anti-rabbit IgG (Vector Laboratories, #BA-1000). Secondary antibodies were purchased from Vector labs, and colorimetric VIP (purple staining) and DAB (brown staining) assays (Vector laboratories) revealed the primary antibodies.
VipCre/wt:Rosa-tdTomato+/- mice (n=4; 2 female and 2 male) were sacrificed between ZT4-6 at 6 weeks of age and brains immersed in 4% PFA overnight at 4°C. Brains were transferred to 30% sucrose until sectioned 40 μm thick with a cryostat. Sections were stored in cryoprotectant at -80°C. Prior to staining, sections were washed overnight in PBS at 4°C. Sections underwent antigen retrieval for 20 minutes in citrate buffer, followed by a wash and blocking for 30 minutes at room temperature using an Avidin/Biotin blocking kit (Vector Labs) with 5% normal goat serum. Sections were briefly washed and then stained following the protocol for the Mouse on Mouse Basic Immunodetection kit (Vector Labs) using a mouse anti-VAX1 (1:100; Origene RRID : AB_2941013). Slices underwent Vectastain ABC kit (Vector Labs) followed by TSA treatment (Akoya Biosciences) for 10 minutes and finally streptavidin-conjugated secondary (1:200) for 30 minutes. Slices were mounted, air-dried, and coverslipped with Prolong Gold with DAPI (ThermoFisher). Slices were imaged on a Nikon Eclipse Ti2-E using a Lumencor SpectraX LED and acquired using a DS-Qi2 CMOS camera. One SCN section per animal was analyzed using QuPath (55). All image manipulations were applied homogenously to the entire image.
Multiplex in situ hybridization assay
To examine Vip, Avp, Nms, and Vax1 mRNA when adult male and female hormones are most comparable, brains were collected at ZT4-8 in young mice, adult males, and diestrus females. To examine Avp, Vip, and Bmal1 mRNA around the time of the LH surge in Ctrl and Vax1Vip mice, brains were collected at ZT13-16 in proestrus females. Multiplex in situ hybridization detection of mouse (Mus musculus) mRNAs was performed with RNAscope® LS Multiplex Fluorescent Reagent Kit (Advanced Cell Diagnostics, cat no. 322800) for 3-plex assay in addition to RNAscope® LS 4-Plex Ancillary Kit (Advanced Cell Diagnostics, cat no. 322830) for 4-plex assay following vendor’s standard protocol for FFPE tissue sections with minor modifications. RNAscope® assays were performed on a Leica Bond autostainer as described (56) with the following probes: RNAscope® 2.5 LS Probe – Mm-Arntl (also known as Bmal1) [aryl hydrocarbon receptor nuclear translocator-like (Arntl) transcript variant 1 mRNA, cat no. 438748-C1] or RNAscope® 2.5 LS Probe - Mm-Vax1 mRNA – [musculus ventral anterior homeobox 1 (Vax1), cat no. 805108-C1]; RNAscope® 2.5 LS Probe – Mm-Avp-C2 [arginine vasopressin (Avp) mRNA, cat no. 401398-C2]; RNAscope® 2.5 LS Probe – Mm-Vip-C3 [vasoactive intestinal polypeptide (Vip) mRNA, cat no. 415968-C3]; and RNAscope® 2.5 LS Probe – Mm-Nms-C4 [neuromedin S (Nms) transcript variant 1, cat no. 472338-C4]. Stock Mm-Nms-C4 probe was diluted at 1:50 in a pre-diluted C1 probe as recommended by the vendor, whereas stock Mm-Avp-C2 and Mm-Vip-C3 were further diluted to 1:100 in appropriate pre-diluted C1 probe due to saturating signal in the pilot experiment. Tissue slides were counterstained with DAPI and scanned with an Aperio Versa imaging system with 20X objective with customized narrow-width band excitation and emission filter cubes as described (56). The Aperio Cellular IF Algorithm (Leica Biosystems, No: 23CIFWL) was used for automated cell enumeration and segmentation based on nuclear DAPI staining. Cells were classified based on the expression levels of one or more mRNAs. In images taken at P2, Vax1 staining was oversaturated, so they were re-imaged for proper mRNA visualization. Representative images at P2 were displayed with increased contrast, applied to all channels to compare with P10 and P60 in Figure 1.
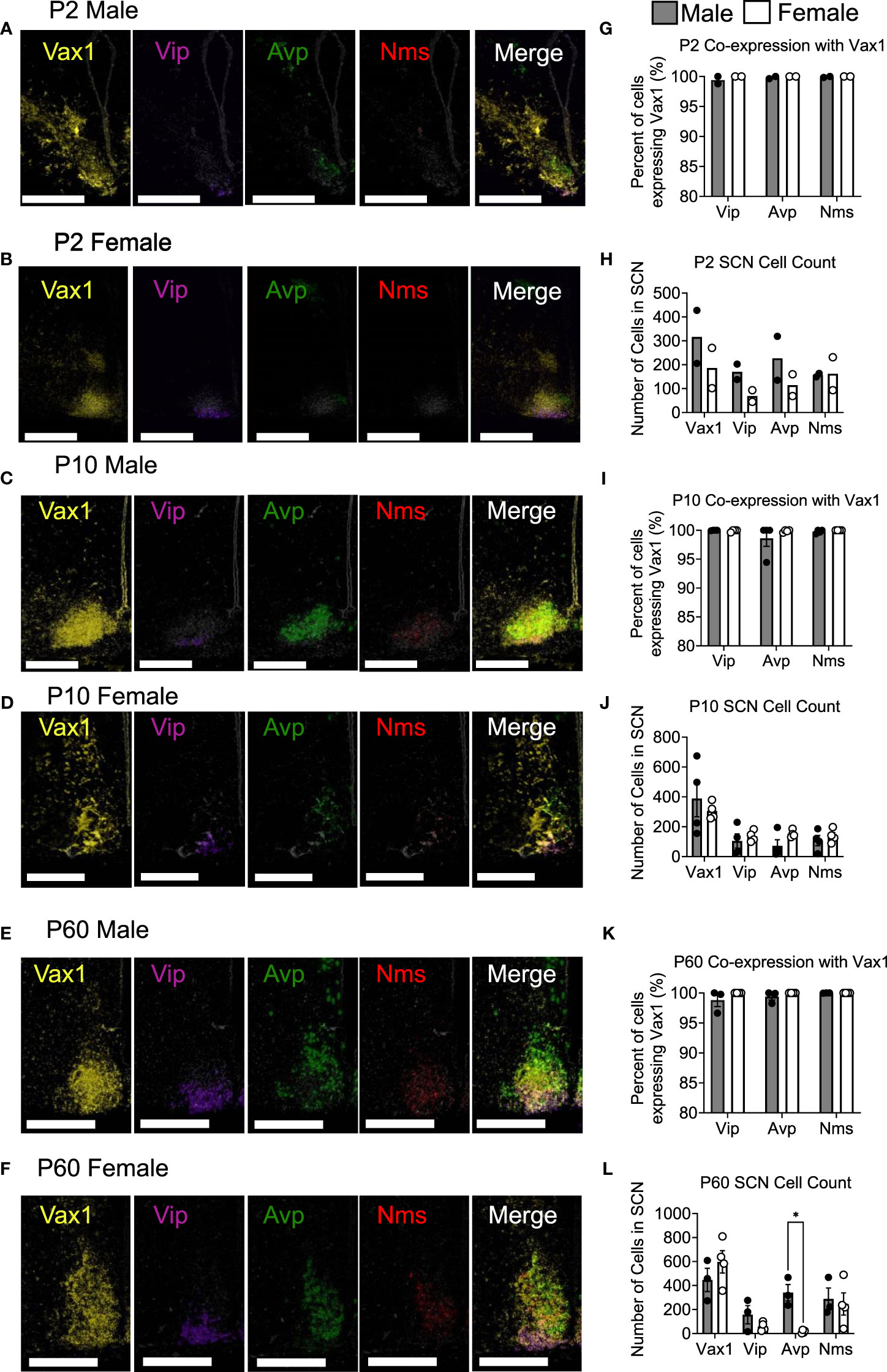
Figure 1 Vax1 is expressed in SCN Vip neurons from the early postnatal period through adulthood in both males and females. RNAscope® ISH representative images (A–F). Images at P2 are displayed with increased contrast, applied to all channels, to compare with P10 and P60. Percent co-expression between Vax1 and Vip, Avp, and Nms-expressing neurons (G, I, K) and cell counts (H, J, L) in P2, P10 and P60 males and females at ZT5. Scale bar is 300 µm. Student’s t-test *, p <0.05.
GnRH and Kisspeptin Challenges and Hormonal Assays
Hormonal challenges were done using kisspeptin and GnRH intraperitoneal (i.p.) injections at ZT3-4. Kiss-10 (catalog #42-431, Batch 7A, Fisher Science, 30 nmoles/mouse) was injected into males or diestrus females and blood was collected from the mouse from the tail vein before (time 0) and after i.p. injection at time points 5, 10, 15, 30, and 45 minutes. Tail blood was collected before (time 0) and 10 minutes after i.p. injection of GnRH (Millipore Sigma, catalog #L7134, 1 μg/kg dose). For all other serum hormone analyses the mice were killed by cervical dislocation, and blood was collected from the abdominal aorta between ZT3 and ZT6. Blood was allowed to clot for 1 hour at room temperature, then centrifuged (room temperature, 15 minutes, 2,600× g).
Serum was collected and stored at -80°C before analysis for estradiol at the Center for Research in Reproduction, Ligand Assay, and Analysis Core, University of Virginia (Charlottesville), by Luminex analysis for LH and FSH on MILLIPLEX MAP Mouse Pituitary Magnetic Bead Panel (Millipore Sigma #MPTMAG-49k) or a competitive enzyme-linked immunosorbent assay (ELISA) kit (EIACORT, ThermoFisher) for corticosterone. Coefficients of variance (CVs) were based on the variance of samples in the standard curve run in duplicate. Reportable range: estradiol: 3–300 pg/ml, CV = <20%; LH: lower detection limit: 5.6 pg/ml, CV < 15%; FSH: lower detection limit: 25.3 pg/ml, CV < 15%; corticosterone: lower detection limit: 0.87 μg/dl, CV = <20%. Samples were run in singlets.
Ex vivo tissue recordings of PER2::LUC expression
For circadian rhythm organotypic explant studies, tissues from mice expressing the PER2::LUC circadian reporter were collected and analyzed as previously described (57). Male and proestrus and diestrus PER2::LUC females were placed on LD and euthanized at ZT3-4 via isoflurane inhalation and cervical dislocation. The brain was removed immediately and placed in an ice-cold, CO2-saturated Hank’s Balanced Salt Solution (HBSS) for approximately 1 hour. Using a Vibratome (Leica), coronal brain sections of 300 μm were collected and the SCN was dissected from the slices in ~2x2 mm squares and placed on a 30 mm Millicell membrane (Millipore-Sigma) in a 35 mm cell culture plate containing 1 mL Neurobasal-A Medium (Gibco) with 1% Glutamax (Gibco), B27 supplement (2%; 12349-015, Gibco), and 1 mM luciferin (BD Biosciences). The lid was sealed to the plate using vacuum grease to ensure an air-tight seal. Plated tissues were loaded into a LumiCycle luminometer (Actimetrics) inside a 35˚C non-humidified incubator at ZT6-6.5, and recordings were started. The bioluminescence was counted for 70 seconds every 10 minutes for 6 days (day 1 – day 7 of recording time). PER2::LUC rhythm data were analyzed using LumiCycle Analysis software (Actimetrics) by an experimenter blind to the experimental group. Data were detrended by subtraction of the 24 h running average, smoothed with a 2 h running average, and fitted to a damped sine wave (LM Fit, damped). The period was defined as the time in hours between the peaks of the fitted curve. Amplitude was defined as the value of the second peak and phase was defined as the time of the first peak. Data from proestrus and diestrus female SCN recordings were pooled as no significant differences in PER2::LUC period or amplitude were found.
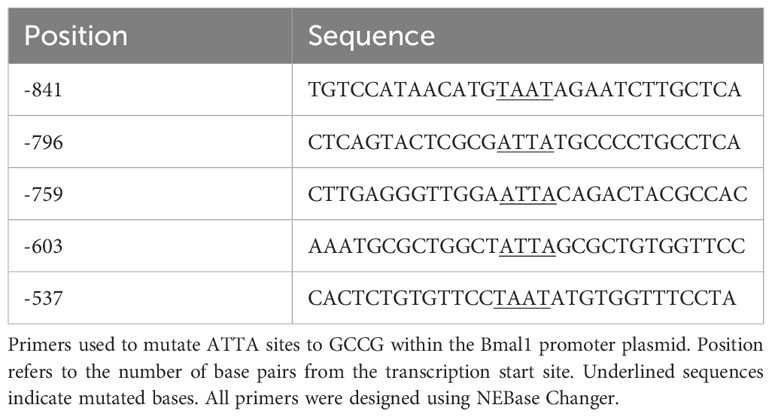
Cell culture and transient transfections
NIH3T3 (American Type Culture Collection) and COS-1 (American Type Culture Collection) cells were cultured in DMEM (Mediatech), containing 10% fetal bovine serum (Gemini Bio), and 1x penicillin-streptomycin (Life Technologies/Invitrogen) in a humidified 5% CO2 incubator at 37˚C. For luciferase assays, NIH3T3 cells were seeded into 24-well plates (Nunc) at 30,000 cells per well. For electrophoretic mobility shift assays (EMSA) COS-1 cells were plated at 1.5 million cells/10 cm dish. Transient transfections for luciferase assays were performed using PolyJet™ (SignaGen Laboratories, Rockville, MD), whereas Fugene was used for plasmid overexpression for EMSA, following the manufacturer’s recommendations. Transfection of cells was performed 48 h after the cells were plated. COS-1 cells were transfected with Vax1/DKK-Flag or CMV6/DKK-Flag overexpression plasmids (20 ng/well, Origene Technologies, Rockville, MD) and harvested at sub-confluency 48–56 h after transient transfections in 10 cm dishes (Nunc). Transient transfections for luciferase assays were done following the manufacturer’s recommendations. NIH3T3 cells for luciferase assays were co-transfected with 150 ng/well of Bmal1-luciferase or Per2-luciferase reporters, 100 ng/well thymidine kinase-β-galactosidase reporter plasmid, which served as an internal control (54), as well as mouse Vax1/pCMV6 overexpression plasmid (20 ng/well, Origene Technologies, Rockville, MD), or its empty vector control (pCMV6). To generate the Bmal1-luciferase plasmid the Bmal1 sequence between -966 bp to +140 bp from the Bmal1 transcriptional start site was excised from the pABpuro-BluF plasmid (Addgene, Plasmid #46824) with PCR primers (F: gggctacaacagaacaactaac, R: taaacaggcacctccgt). The PCR product was inserted into the pGL3-basic backbone between the Mlu-HF and XhoI sites using the Quick Ligation Kit (New England Biolabs). Site directed mutagenesis of the homeodomain binding sites (ATTA and ATTA-like) in the mouse Bmal1-luciferase plasmid was performed using the NEB Q5 Site-Directed Mutagenesis Protocol (New England Biolabs Inc.), following manufacturer’s instructions. Primers for NEB Q5 site-directed mutagenesis were designed using the NEB Base Changer (Table 1). To equalize the amount of DNA transfected into cells, we systematically equalized plasmid concentrations by adding the corresponding inactive plasmid backbone. Cells were harvested 24 h after transfection in lysis buffer [100 mM potassium phosphate (pH 7.8) and 0.2% Triton X-100]. Luciferase values were normalized to β-galactosidase values to control for transfection efficiency. Values were further normalized by expression as fold change compared to the pGL3 control plasmid, as indicated in the figure legends. Data represent the mean ± SEM of at least three independent experiments done in duplicate and triplicate.
Cytoplasmic and nuclear extracts and Electrophoretic Mobility Shift Assay (EMSA)
COS-1 cells were scraped in hypotonic buffer (20 mM Tris-HCl, pH 7.4, 10 mM NaCl, 1 mM MgCl2, 10 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1x protease inhibitor cocktail; Sigma-Aldrich) and left on ice to swell. Cells were lysed and nuclei were collected by centrifugation (4°C, 1700 g, 4 minutes). Nuclear proteins were extracted on ice for 30 minutes in hypertonic buffer [20 mM HEPES, pH 7.9, 20% glycerol, 420 mM KCl, 2 mM MgCl2, 10 mM NaF, 0.1 mM EDTA, 0.1 mM EGTA, 1x protease inhibitor cocktail (Sigma-Aldrich), and 1 mM phenylmethylsulfonylfluoride]. Debris was eliminated by centrifugation (4°C, 20,000 g, 10 min), and the supernatant was snap-frozen and stored at -80°C.
Oligonucleotide probes are listed in Table 1. All synthetic oligonucleotides were made by IDT (San Diego, CA). Annealed double-stranded oligonucleotides (1 pmol/µl) were end-labeled with T4 Polynucleotide Kinase (New England Biolabs, Ipswich, MA) and [γ32P]ATP (7000 Ci/mmol; MP Biomedicals, Solon, OH). Probes were purified using Micro Bio-Spin 6 Chromatography Columns (Bio-Rad). Binding reactions contained 2 µg nuclear protein and 1 fmol of labeled probe in 10 mM HEPES (pH 7.9), 25 mM KCl, 2.5 mM MgCl2, 1% glycerol, 0.1% Nonidet P-40, 0.25 mM EDTA, 0.25% BSA, 1 mM dithiothreitol, and 350 ng poly(dI-dC). For super-shift experiments, 2 µg mouse anti-DKK (Flag antibody, Origene #TA50011) or 2 µg of normal mouse IgG (Santa Cruz Biotechnology, #sc2025) were added to the reaction. Samples were incubated for 20 minutes at room temperature before loading on a 5% non-denaturing polyacrylamide gel in 0.25x Tris-borate EDTA buffer. Gels were run for 2 h at 200 V, dried under vacuum, and exposed to film for 2-5 d at room temperature.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 8, using Student’s t-test, one-way ANOVA, or two-way ANOVA, followed by post hoc analysis by Tukey or Bonferroni as indicated in figure legends, with p < 0.05 to indicate significance. All data were analyzed as independent measures except for wheel-running activity, which was analyzed via a two-way repeated-measures ANOVA. PER2::LUC timing of first peak phase relationships was analyzed in R via a Circular Analysis of Variance High Concentration F-Test, with a corrected confidence level of p < 0.01667 to account for family-wise error.
Results
Characterization of Vax1 expression in Vip, Avp, and Nms neurons in the male and female SCN
Vax1 is highly expressed in the developing mouse SCN and becomes refined to the hypothalamus, primarily in the SCN in the early postnatal period (25, 58). Although conditional deletion of Vax1 in late neuronal development using the SynapsinCre allele reduced VIP expression in the adult SCN (25), it remains unknown if all VIP expressing neurons co-express VAX1 and how postnatal deletion of Vax1 in VIP neurons impacts VIP expression. Because VAX1 is highly expressed in the developing SCN, we first asked how VAX1 expression changed after birth and into adulthood in males and females. To answer this, we performed multiplex RNAscope® assay at postnatal day 2 (P2), P10, and P60 (adult) at ZT5 in males and females. We found that all SCN Vip-expressing cells at P2 co-express Vax1 [Figures 1A, B, G; male (n = 2) 99.4 ± 0.6%, female (n = 2) 100% ± 0%], a pattern maintained at P10 [Figures 1C, D, I; male (n = 4) 99.98 ± 0.021%, female (n = 4) 99.91 ± 0.09%], and P60 [Figures 1E, F, K; male (n = 3) 98.80 ± 1.08%, female (n = 4) 100 ± 0%]. In addition to the high co-expression with Vip, Vax1 is highly expressed throughout the SCN at P2, P10, and P60 (Figures 1A–F), where both Avp and Nms-expressing cells also exhibited full overlap with Vax1 in both sexes (Figures 1G, I, K). Interestingly, we found a sex difference in the number of cells expressing Avp at P60, where females had fewer Avp+ cells compared to males [Figure 1L; n = 7, t(5) = 5.671, p = 0.0024], a difference that was not present prior to puberty (P10). Although we did not see a significant sex difference in the number of Vip neurons at any age, the concentration of Vip, as evaluated through Vip probe signal intensity, was significantly lower in females than males at P10 [t(3) = 6.01, p = 0.037, not shown] and trended lower at P60 [t(5) = 2.47, p = 0.16, not shown]. To determine if this modest sex difference in Vip concentration translated to a sex difference in peptide levels, we performed IHC in adult male and female brains. Adult female mice had a significant reduction in the intensity of VIP peptide in the SCN [Figures 2A, B, t(11) = 3.874, p = 0.0026] as compared to males.
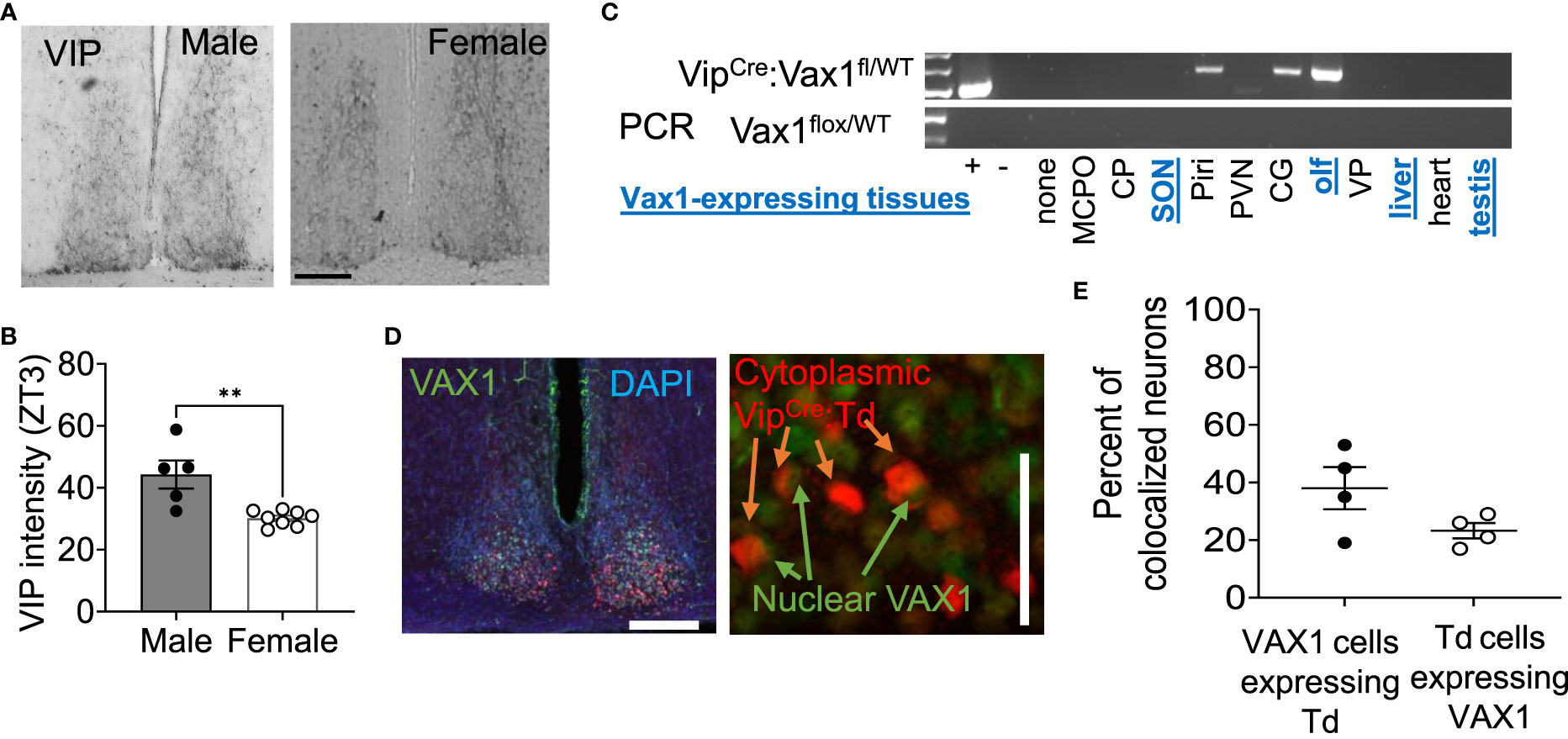
Figure 2 VIP peptide is sexually dimorphic in the SCN. (A) Example image and (B) quantification of SCN IHC for VIP in adult males and females at ZT3. Scale bar 100µm. Student’s t-test, **, p < 0.01 (C) RT-PCR shows VipCre recombination of the Vax1flox allele in indicated tissues. Abbreviations stand for olfactory bulb (olf), magnocellular preoptic area (MCPO), caudoputamen (CP), supraoptic nucleus (SON), piraform area (Piri), paraventricular nucleus (PVN), cingulate gyrus (CG), and ventral palidum (VP). Blue, underlined text indicates tissues that are known to express Vax1. (D) Example images and quantification (E) of dual IHC for VipCre:Td (red, cytoplasm) and VAX1 (green, nuclear) expressing cells in the adult SCN (DAPI, nuclei). Scale 50 µm.
As Vax1 is ~100% co-expressed with Vip from P2 until P60 in males and females (Figure 1), we next generated a conditional knockout mouse to determine how the loss of VAX1 in VIP neurons would impact SCN function. Using our RNAscope® ISH data from Figure 1, we first visually inspected all stained sections for potential Vax1 expression in non-SCN Vip cells. The olfactory bulb is the only additional brain area that expresses Vax1 which is also targeted by the VipCre allele (Figure 2C). In the scenario, the VipCre allele does target some Vax1 expressing cells in the olfactory bulb, a change in reproductive behavior could impact fertility data because olfaction is required for normal male reproductive behavior. To validate that the VipCre allele targeted VAX1 expressing neurons of the SCN, we generated VipCre:RosaTdTomato (VipCre:Td) mice allowing the identification of all neurons that are targeted by the VipCre allele (Figures 2D, E). Using dual IHC, we quantified the colocalization of VIP- and VAX1-expressing neurons in the SCN. Using tdTomato as a marker for VIP neurons in sections from VipCre:Td animals, we found 38 ± 7% of SCN tdTomato+ neurons colocalized with VAX1 (n=4, 2 per sex, 36-101 neurons per animal). An average of 23 ± 5% of VAX1 neurons colocalized with tdTomato (n=4, 65-236 neurons per animal). This discrepancy between the RNAscope and dual IHC has numerous potential explanations, as detailed in the discussion.
Conditional deletion of VAX1 in VipCre neurons shortens SCN circadian period in females and males
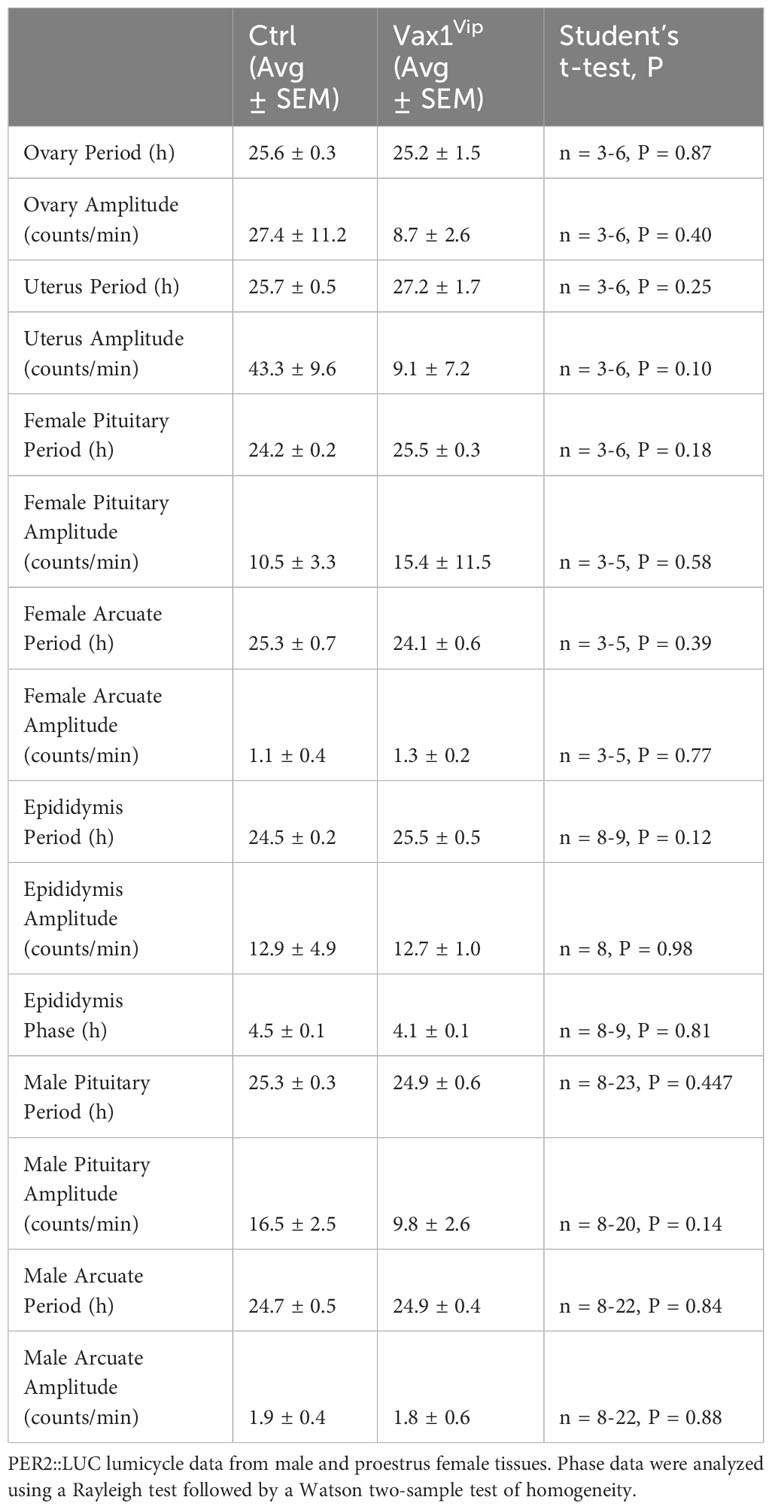
To determine the role of VAX1 in VIP neuron circadian output, we evaluated the wheel running behavior of Ctrl and Vax1flox/flox:VipCre (Vax1Vip) mice in LD and constant darkness (DD). Wheel running patterns in LD of Ctrl and Vax1Vip females and males were comparable (Figures 3A–I, LD). Light is a strong entraining signal of the SCN, and activity rhythms in LD can mask weakened SCN function (59). Following the initial LD period, mice were placed in DD for 28d to assess the endogenous free-running period. Both male and female Vax1Vip mice showed a significantly shortened free-running period (Tau) compared to Ctrls (Figure 3E, two-way ANOVA, male p = 0.0004, female p = <0.0001) with no differences in Chi2 amplitude (Qp). There were no changes in the number of wheel revolutions per day or activity duration (alpha, Figures 3F–I). To determine if the shortening of the free-running period during DD resulted from a change in the endogenous SCN circadian period, we generated triple transgenic mice crossing Vax1Vip mice with the PER2::LUC reporter mouse (60). In agreement with the significantly shortened behavioral period of Vax1Vip males and females on DD, we found that the SCN of Vax1Vip:PER2::LUC mice showed a significant shortening in period as compared to Ctrl males [Figure 4A, t(27) = 2.936, p = 0.0067] and females [Figure 4B, t(24) = 2.125, p = 0.0440]. No differences were found in the amplitude or phase relationships of PER2::LUC in the SCN of Vax1Vip males or females as compared to Ctrl, indicating that the rhythms in the SCN are not misaligned or significantly weakened (Figures 4C–F). Together these data show that loss of VAX1 in VIP neurons shortens SCN circadian output in both males and females.
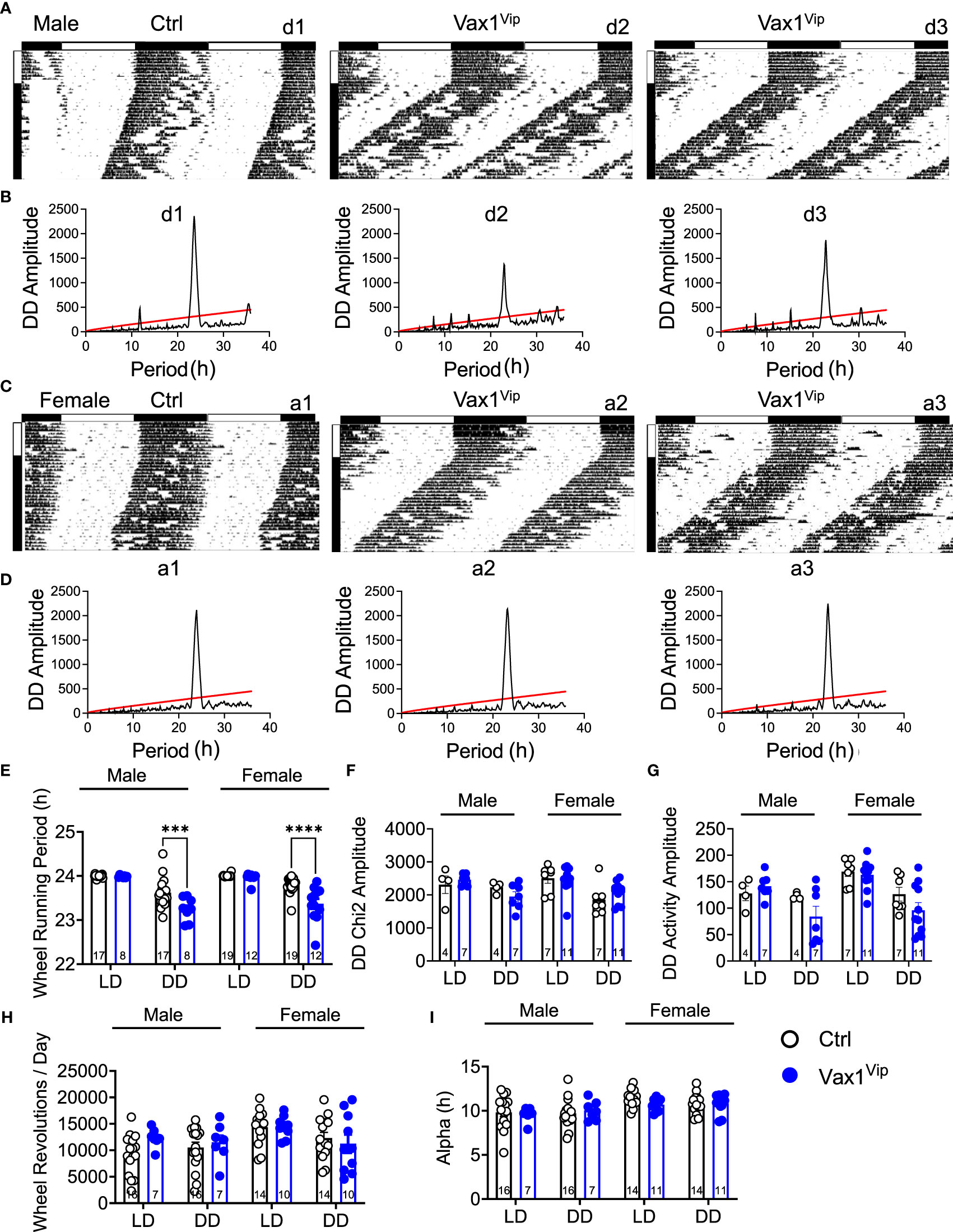
Figure 3 Vax1 deletion within VIP neurons shortens behavioral free-running period in males and females. (A, B) Male and (C, D) female Ctrl and Vax1Vip mice were single housed with running wheels. (A, C) Data show double plotted actogram activity with 14 days in LD12:12 (LD) followed by 28 days in constant darkness (DD). Data are presented in ClockLab normalized format. Horizontal bar above the actograms indicates lights on (white) and lights off (black) during the LD12:12 cycle. (B, D) Chi2 periodograms during 2 weeks in DD. Matching codes (a1, a2, etc.) on the upper right corner of each actogram and chi2 periodogram indicate data from a particular mouse, with variable scaling indicated in the upper left. 14-day average wheel-running data were used for indicated analysis parameters in LD and DD. Average histogram data for (E) Wheel-running period, (F) Chi2 amplitude, (G) activity amplitude, (H) wheel revolutions, and (I) activity duration (Alpha). Number within the bar indicates number of animals in each group. 3-way ANOVA, ***, p < 0.001; ****, p < 0.0001.
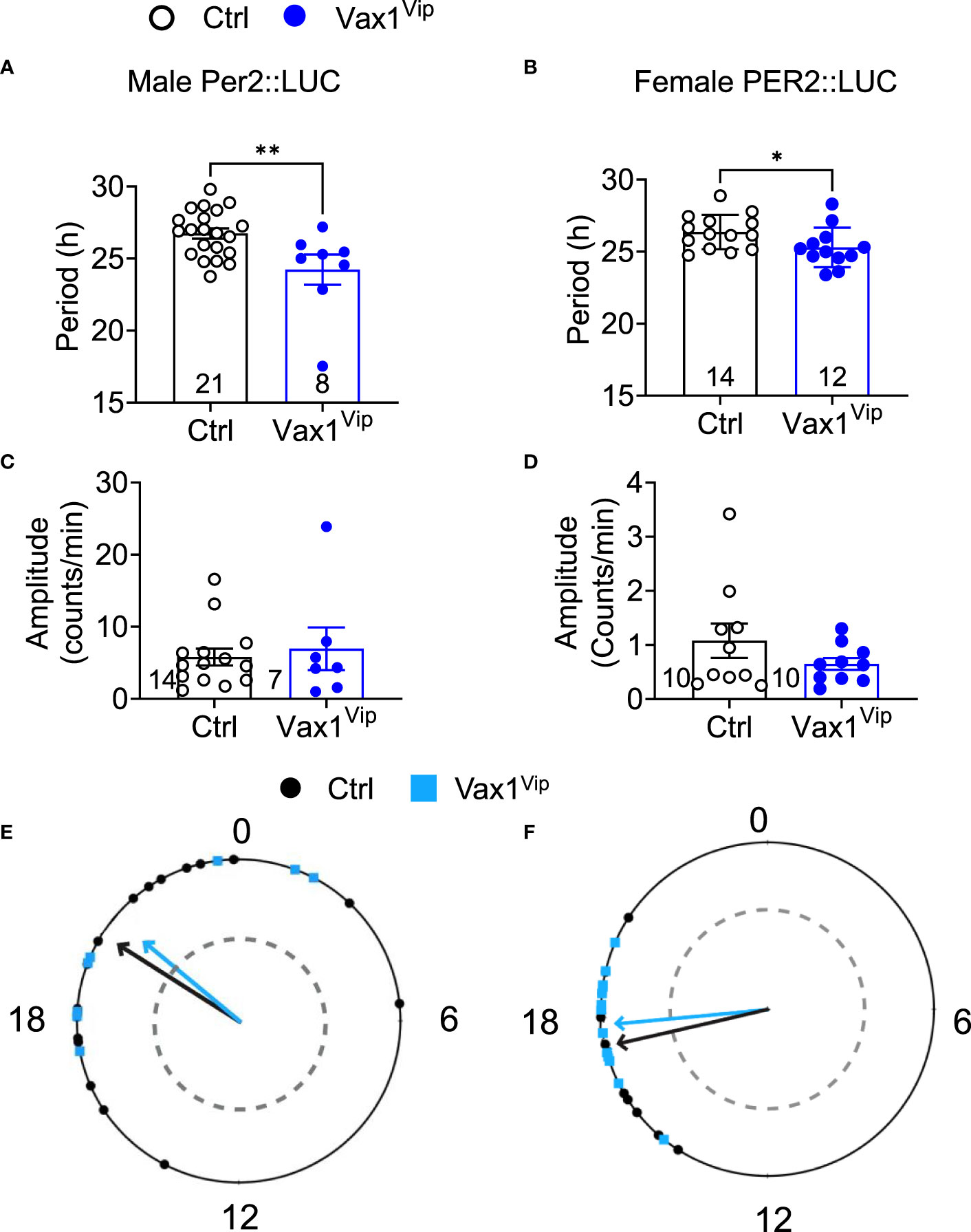
Figure 4 Vax1Vip SCN has a shortened PER2::LUC period in ex vivo culture. Histogram of PER2::LUC SCN (A, B) circadian period and (C, D) amplitude from control and Vax1Vip:PER2::LUC females (combined proestrus and diestrus) and males. Statistical analysis by Student’s t-test, *, p<0.05 **, p < 0.01, n = 7-16. (E) PER2::LUC phase (time of first peak) in the SCN of control and Vax1Vip:PER2::LUC males and (F) females, n = 7-16. Mean times of first peak are indicated by vector lines, and symbols indicate individual data points. Data were analyzed via the Rayleigh Test of Uniformity, where crossing the dotted gray line indicates significant clustering (p < 0.05), and the Watson’s Two-Sample Test of Homogeneity. No significant differences were found in females between estrous stages.
VAX1 regulates molecular clock gene expression in VIP neurons through a direct mechanism
A shortened SCN period can be driven by both changes in SCN peptide expression and changes in molecular clock gene expression. We have previously shown that VAX1 promotes Per2-luciferase plasmid expression using transient transfection assays (25). However, we have not yet demonstrated whether this action is direct or indirect. To assess if VAX1 directly binds with our top candidate ATTA site of the mouse Per2 DNA regulatory region (25), we used EMSA. We found that VAX1 directly binds the ATTA site at +1774/1770bp from the transcriptional start site of the mouse Per2 gene (Figure 5A, Super shift is indicated by *). In addition to ATTA sites in the Per2 regulatory region, the Bmal1 regulatory region also contains numerous ATTA sites. Using transient transfections, we found that VAX1 promotes Bmal1-luciferase expression (Figure 5B). Site-directed mutagenesis of ATTA-like sites in the Bmal1 regulatory region (Table 1) showed a modest increase in fold-change of VAX1-driven Bmal1-luciferase expression in transfected cells (Figure 5B). Interestingly, VAX1 can directly bind to all the identified ATTA sites tested by EMSA (EMSA, Figure 5C, supershift indicated by *). This identifies for the first time that VAX1 can directly bind to the regulatory regions of Per2 and Bmal1 and provides a mechanism by which changes in VAX1 expression can directly impact molecular clock function. Next, to determine if loss of VAX1 in VIP neurons would significantly impact Bmal1 expression in Vip neurons, we performed RNAscope® for Bmal1, Vip, and Avp in the adult SCN of Ctrl and Vax1Vip females (Figure 6). Note these experiments were completed in proestrus at ZT16, with the goal of having the most hormonally challenging environment in the female body present at the time of sample collection. We found that in Ctrl and Vax1Vip females almost 100% of Vip and Avp neurons co-expressed Bmal1 (Figures 6A, B). Despite a trend in a reduction in Vip in the SCN of Vax1Vip females (Figure 6C), as well as a trend in the reduction in cells co-expressing Vip and Bmal1 (Figure 6D), no significant difference in any of the studied transcripts, or colocalization of transcripts were identified (Figures 6C, D).
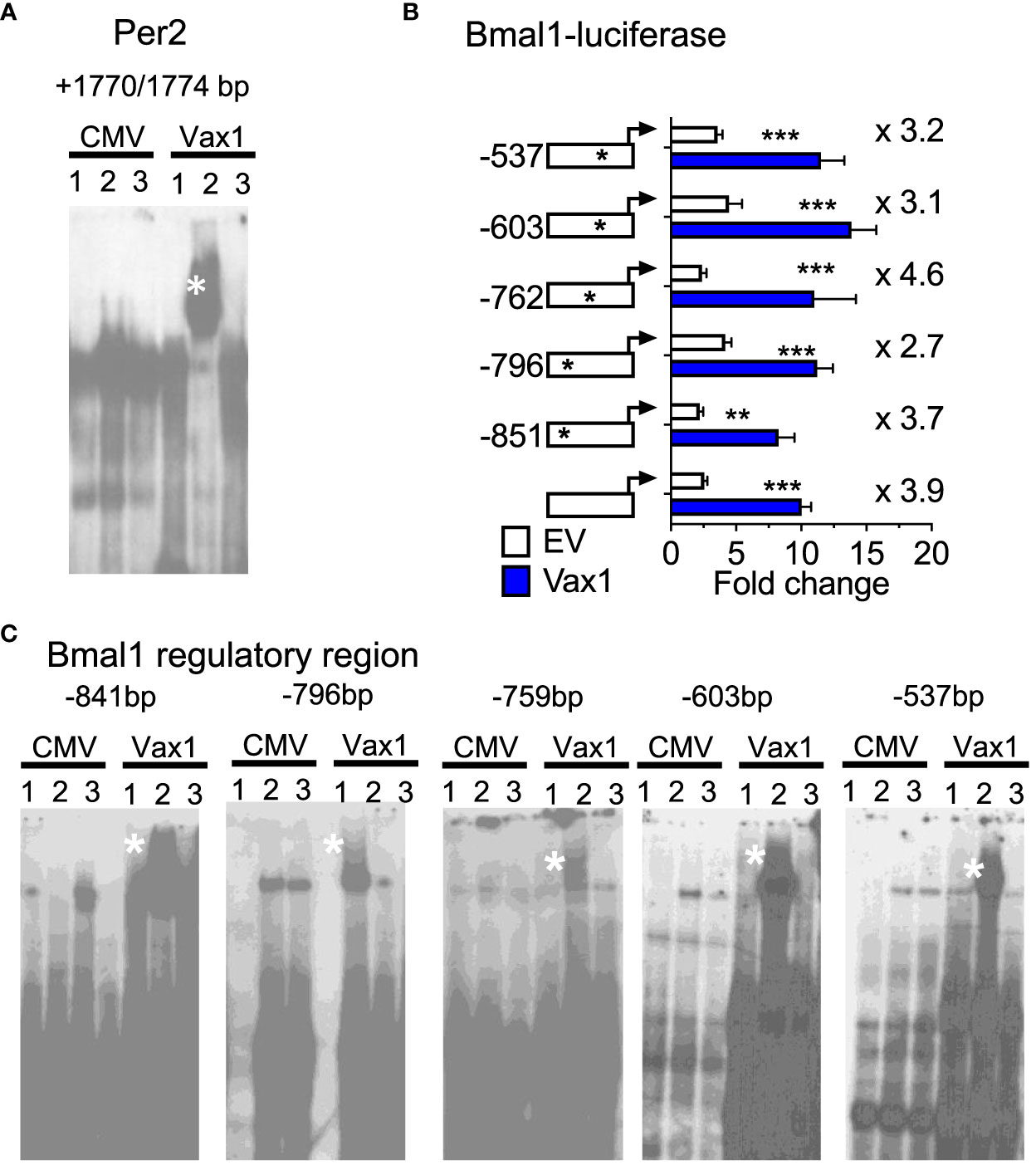
Figure 5 VAX1 binds directly to regulatory regions of molecular clock genes Per2 and Bmal1. (A, C) EMSA assay of COS-1 cells, represents in Lane 1: pCMV-Flag (CMV, empty vector), Lane 2: Vax1-Flag plasmid and Lane 3: Vax1-Flag plasmid + anti Flag antibody. White star indicates super shift. Example gels of n = 3. (B) Transient transfections of NIH3T3 cells with the mouse Bmal1 regulatory region driving luciferase (Bmal1-luciferase) with and without Vax1 overexpression vector (20 ng) or its empty vector (EV, pCMV6, 20 ng). Numbers indicated with the stars on the regulatory regions refer to ATTA sites that have been mutated (see Table 1). Statistical analysis by Two-way ANOVA mixed effect model, *, p < 0.05; **, p < 0.01; ***, p < 0.001, n = 4-6 in duplicate or triplicate.
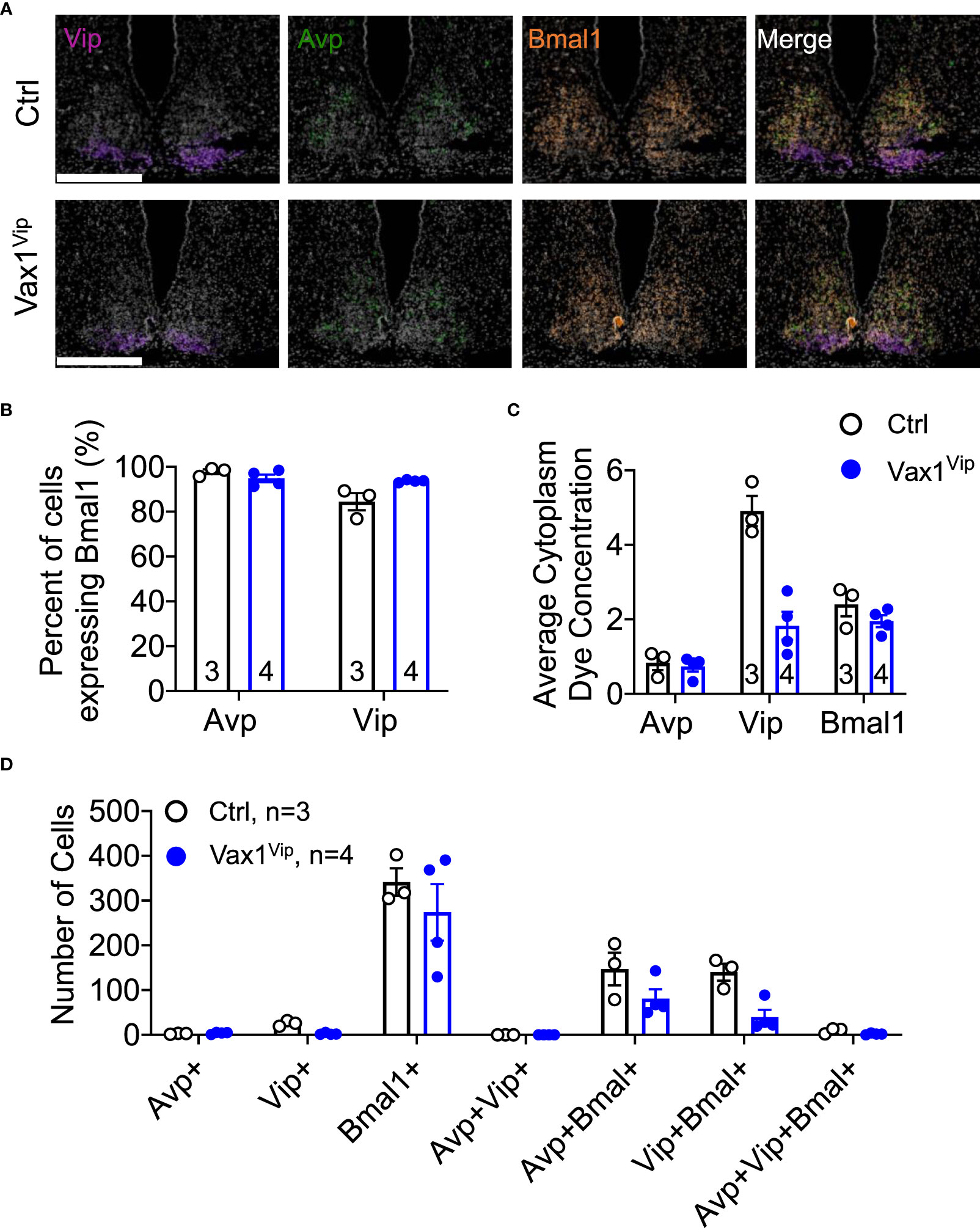
Figure 6 Loss of Vax1 in VIP neurons does not reduce Vip and Bmal1 expression in the SCN. RNAscope® assay at ZT16 in the SCN of proestrus Ctrl and Vax1Vip females. (A) Example images of RNAscope® assay for Vip (blue), Avp (red) and Bmal1 (green). N = 3-4 per group. Scale bar 300 µm. (B) Percentage of cells that co-express Bmal1 with Avp or Vip. Mann-Whitney, p > 0.05. (C) Average cytoplasm dye concentration reflecting mRNA transcripts for Avp, Vip, and Bmal1 Mann-Whitney, p > 0.05. (D) Number of cells expressing indicated combinations of Avp, Vip, and Bmal1. Mann-Whitney, p > 0.05.
Vax1Vip females have lengthened estrous cycles and deregulated sex steroids, whereas males have increased sperm count
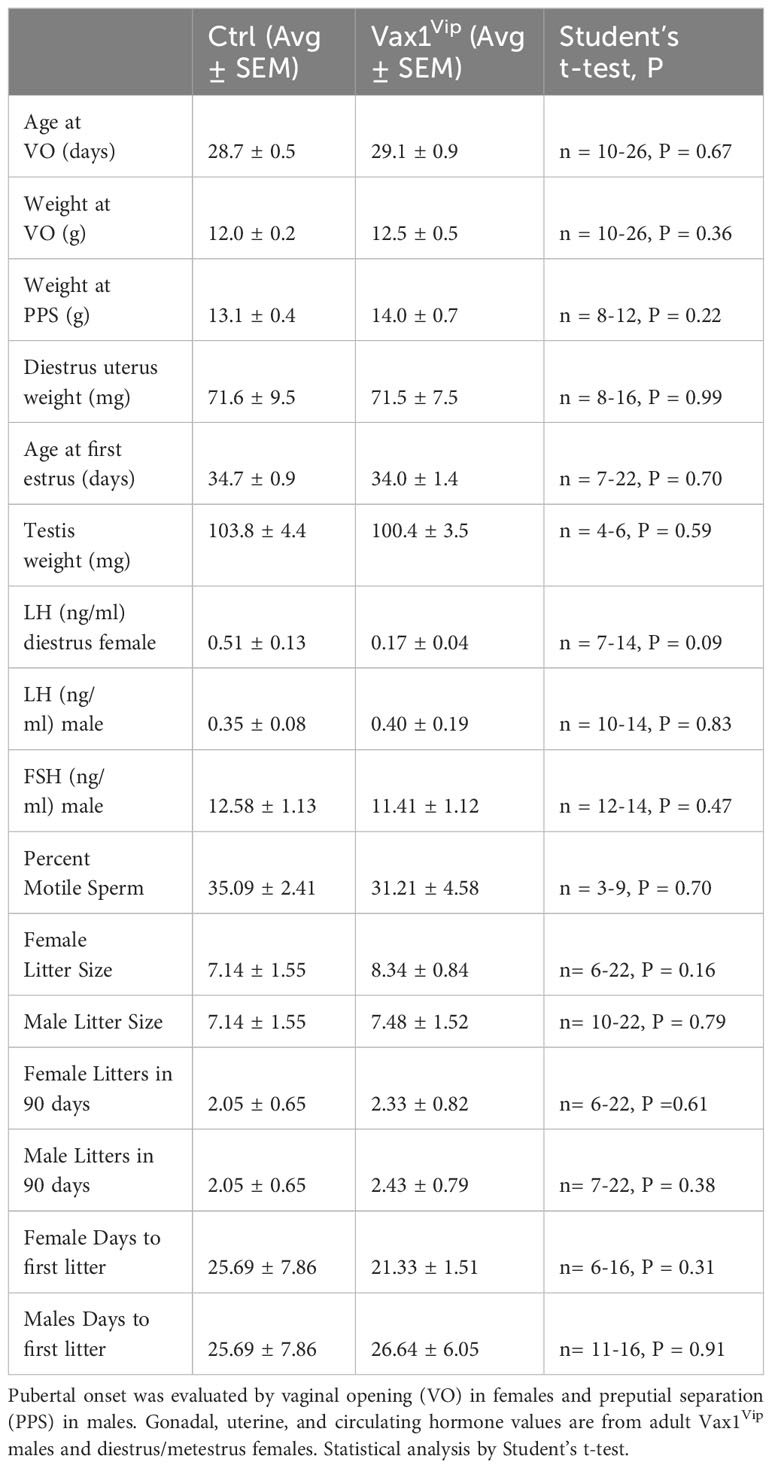
The SCN provides daily neuronal and hormonal signals aligning circadian timekeeping in peripheral reproductive tissues allowing coordination between hormone release and increased tissue sensitivity improving reproductive function (25, 27, 35, 57). To determine if Vax1Vip mice have misaligned circadian phase of their reproductive tissues, we recorded PER2::LUC expression in the pituitary, ovary (female), uterus (female), and epididymis (male) of triple transgenic mice. No tissues were found to have significant differences in period (Table 2), amplitude (Table 2), or time of first PER2::LUC peak (Table 2, phase). These data indicate that the weakened SCN output of Vax1Vip mice does not significantly impact circadian timekeeping in the studied peripheral tissues but does not preclude disruptions to fertility. To determine if reproductive function is impacted in Vax1Vip mice, we first evaluated pubertal onset. At pubertal onset, body weight was comparable between Ctrls and Vax1Vip in both sexes (Table 3). Male pubertal onset, as evaluated by preputial separation (PPS) was slightly delayed [Figure 7A, t(18) = 2.211, p = 0.040], whereas female pubertal onset, as assessed through vaginal opening (VO) and first estrous, were comparable between Ctrls and Vax1Vip females (Table 3). There was no impact on male reproductive function (Table 3) apart from significantly increased total sperm count [Figure 7B, t(12) = 3.101, p = 0.009]. Vax1Vip males had normal testis size and percent motile sperm (Table 3). The increase in total sperm pool was not associated with changes in basal LH and FSH levels in males (Table 3). Both Vax1Vip males and females were comparable to Ctrls for the number of litters generated in 90 days, days to first litter, and litter sizes (Table 3). Despite the normal fertility in females, Vax1Vip females had a significant lengthening of the estrous cycle [Figure 7C, t(19) = 2.307, p = 0.033] with a similar amount of time spent in each cycle stage as compared to Ctrls [Two-way ANOVA, F (1, 10) = 2.500, P=0.1449]. This lengthening in estrous cycles was associated with a reduction in FSH, estrogen, and ovarian weight in Vax1Vip females (Figures 7D–F), but did not impact basal LH or uterine weight in diestrus (Table 3).
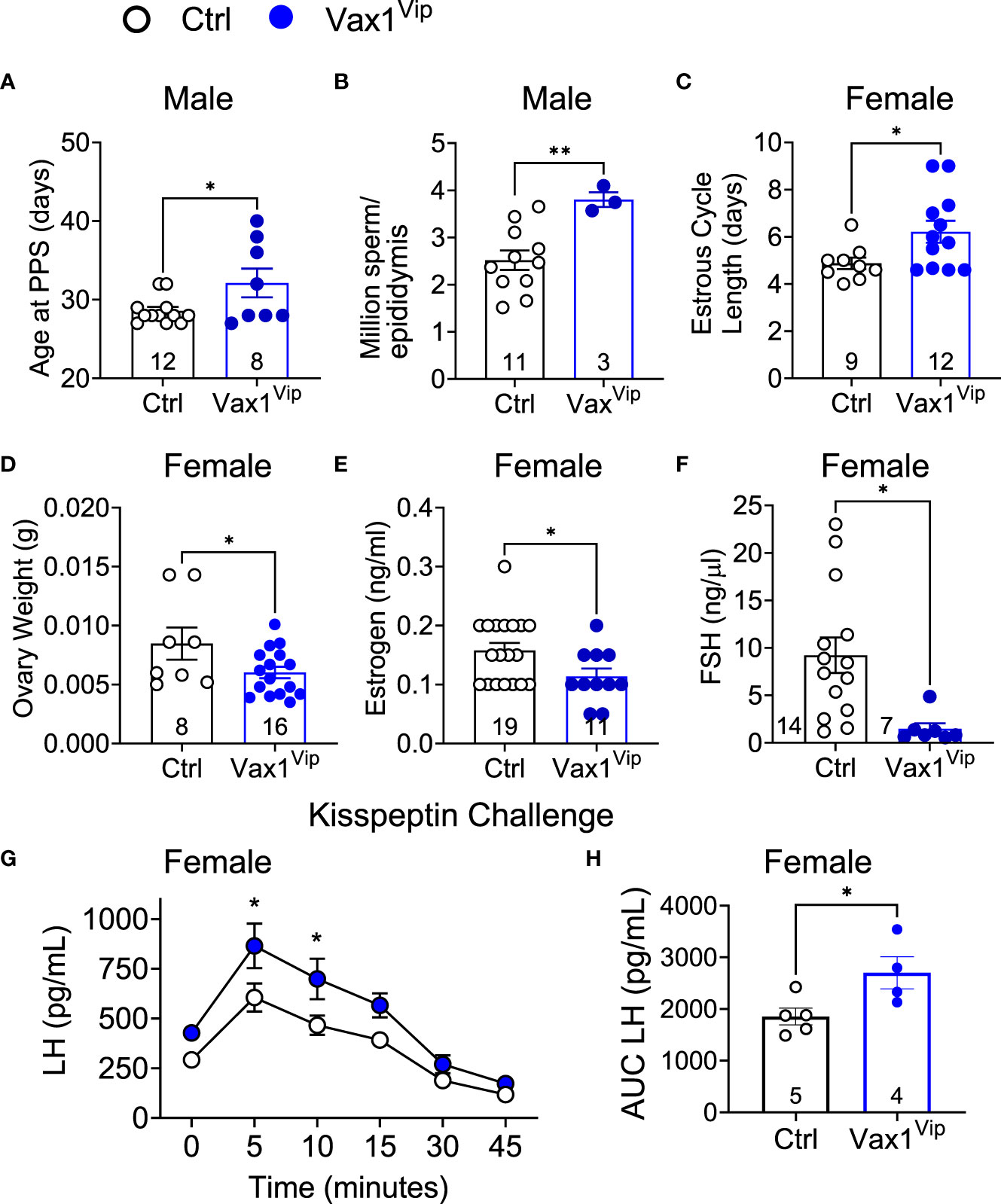
Figure 7 Vax1Vip females have a reduction in ovary weight, estrogen, and FSH, as well as an increased sensitivity to kisspeptin. (A) Age at preputial separation (PPS) and (B) million sperm per epididymis in Ctrl and Vax1Vip males, n indicated in graphs, Student’s t-test, *, p<0.05; **, p <0.01. (C) Estrous cycles were evaluated in females, and average estrous cycle length was established. Student’s t-test, *, p < 0.05. (D) Ovary weight, (E) circulating estrogen and (F) circulating FSH of diestrus females. n indicated in graphs, Student’s t-test, *, p < 0.05. (G) Circulating LH levels in diestrus females evaluated over a 45-minute time period in response to an i.p. kisspeptin injection. Mixed Effects analysis, *, p < 0.05. and (H) the resulting area under the curve. Student’s t-test, *, p < 0.05.
Reduced VIP in the SCN of Vax1Vip mice is associated with increased GnRH neuron sensitivity to kisspeptin in females, but not in males
VIP neurons from the SCN project directly to GnRH neurons and indirectly through AVP to kisspeptin neurons. The anterior pituitary releases LH into the circulation upon GnRH release at the median eminence, allowing an indirect approach to study GnRH neuron function. To determine if the reduction in VIP in the Vax1Vip SCN impacted GnRH neuron response to kisspeptin, we performed hormone challenges in mice. We first confirmed that the pituitary responded to GnRH by increasing LH release through an i.p. injection of GnRH. As expected, the fold change in LH in response to a GnRH challenge was comparable between Ctrl (9.07 ± 1.97, n = 4) and Vax1Vip males [12.09 ± 2.43, n = 6, t(8) = 0.885, p= 0.401], and between Ctrl (10.50 ± 3.37, n=8), and Vax1Vip females [6.95 ± 3.84, n = 4, t(10) = 0.641, p= 0.535]. To assess if the GnRH neuron response to kisspeptin was impacted in Vax1Vip males and females, we next performed an i.p. kisspeptin challenge. There were no differences between LH release in Vax1Vip (5798 pg/mL ± 583, n = 4) males and Ctrls [5325 pg/mL ± 670, n = 5, t(7) = 1.11, p = 0.303]. In contrast, Vax1Vip females had an increased release of LH in response to kisspeptin at 5 and 10 minutes as compared to Ctrls (Figure 7G, mixed-effects analysis, 5 minutes p = 0.0102, 10 minutes p = 0.0257), as well as an overall increase in LH release [Figure 7H, t(7) = 2.560, p = 0.0376]. Such alteration in the neuroendocrine network regulating LH release would be expected to impact female estrous cyclicity, which relies on precisely timed hormone release and sex-steroid-feedback.
Hypothalamic imbalance between Vip and Avp in the Vax1Vip hypothalamus is associated with increased basal corticosterone and depressive-like symptoms in females
AVP is expressed outside the SCN and is highly expressed in the paraventricular nucleus (PVN), a direct target of SCN neurons. The PVN is a well-established relay site playing a significant role in several autonomic functions, including stress (61–63). To determine how Avp levels were impacted in the hypothalamus of Vax1Vip females, we analyzed Avp using RNAscope® assay. Interestingly, we found that Vax1Vip females, which have comparable Avp mRNA in the SCN to Ctrl (Figures 6C, D), display a significant increase in Avp transcript and cell numbers in the PVN (Figures 8A–C). This increase of Avp in the PVN of Vax1Vip females provides a potential link to the activation of the stress axis. In agreement with this, basal corticosterone levels were overall increased at ZT3 in Vax1Vip mice [Figure 8D, two-way ANOVA, F(1,12) = 5.868, p = 0.032]. Finally, to determine if these known risk factors for depression would reflect an increase in depressive-like behavior in Vax1Vip mice, we tested males and females in the Porsolt forced swim test. We found that Vax1Vip females (metestrus, ZT13) exhibited increased depressive-like behaviors as shown by an increase in the percentage of time floating (Figure 8E) as compared to Ctrl females [t(17) = 2.121, p = 0.0489], while Vax1Vip males were comparable to Ctrls [t(11) = 0.5889, p = 0.5678]. The increased time floating in the Porsolt forced swim tests of the VaxVip females correlated with increased circulating corticosterone 1 h after the swim test [Figure 8F, t(11) = 3.595, p = 0.0042].
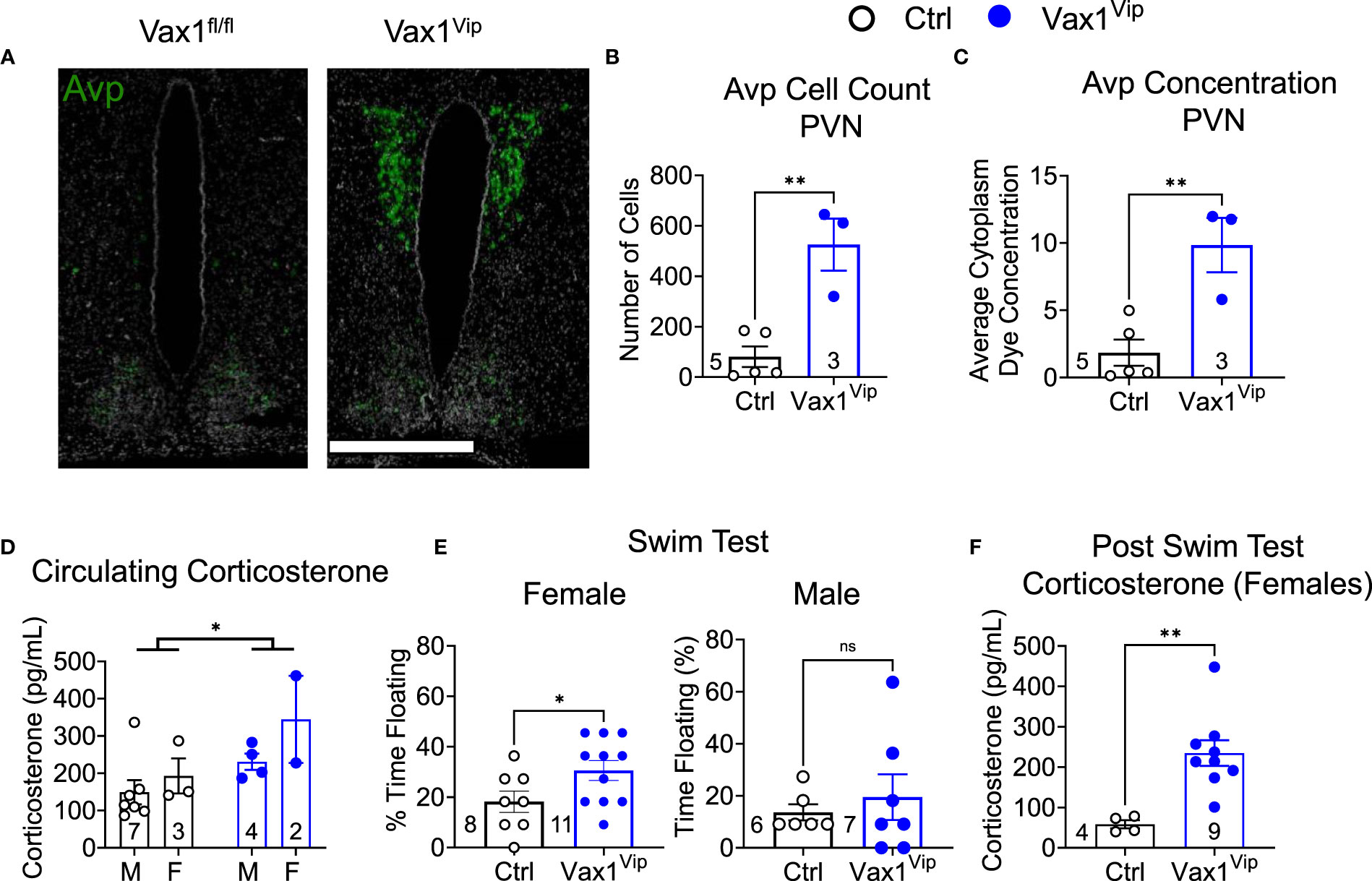
Figure 8 Vax1Vip females have increased Avp in the PVN, increased corticosterone, and increased depressive-like behaviors. (A) Example images of RNAscope® detection for Avp in the SCN and paraventricular nucleus (PVN) of proestrus females at ZT16. Scale bar is 600 µm, n=3-5. Quantification of RNAscope® assay by (B) cell counts and (C) dye concentration of Avp in the PVN, n=3-5, Student’s t-test, **, p <0.01. (D) Corticosterone was measured at ZT3-5 in males (M) and females (F), Two-way ANOVA, *, p<0.05. (E) To test depressive-like behavior, metestrus female or male mice were tested at ZT13 by a Porsolt forced swim test, and the % time floating assessed. N indicated in graph, Student’s t-test, *, p<0.05. (F) Corticosterone levels in circulating blood 1 h following the Porsult forced swim test. n = 4-9, Student’s t-test, **, p<0.01.
Discussion
This work explores VIP neurons within the SCN as a neurological point of overlap between circadian disruption, reproduction, and depressive-like behaviors. Here, we leverage the highly localized co-expression between the homeodomain transcription factor VAX1 in VIP neurons of the SCN to develop a conditional knockout mouse model that exhibits abnormal circadian timekeeping, reproductive axis function, and mood. Excitingly, this novel mouse model suggests that VIP neurons might provide a shared neurological underpinning between reproductive and mood disorders.
Vax1 is co-expressed in Vip, Nms, and Avp neurons from development to adulthood and regulates molecular clock gene expression
The SCN retains a high expression of numerous homeodomain transcription factors after development (26, 46, 64–66), including Vax1. Vax1 is critical in brain and neuronal development (25, 46, 47, 53, 67) and highly expressed in the developing mouse brain before it becomes refined to the adult hypothalamus, primarily in the SCN, during the early postnatal period (25, 58). We have previously shown that conditional deletion of Vax1 in late neuronal development using the SynapsinCre allele reduced VIP expression in the adult SCN (25), but this previous study did not address how many VIP neurons co-expressed Vax1. Here we find that ~100% of Vip-expressing neurons co-express Vax1 at P2 and maintain a close to 100% co-expression at P10 and P60. In addition, we show that Vax1 is also co-expressed by ~100% of Nms and Avp neurons at P2, P10, and P60. This shows for the first time that Vax1 is highly expressed in three primary SCN neuron populations from the early postnatal period into adulthood, suggesting a role of VAX1 in regulating the function of these neuronal populations. The close to 100% co-expression of Vax1 and Vip strengthens the value of our conditional knock-out model as a novel tool to specifically study SCN VIP neurons. This high level of overlap, coupled with the primary restriction of Vax1 expression within VIP neurons in adulthood to the SCN (25, 58), allows for targeted impairment of VIP neurons within the SCN alone. This approach allows us to build upon previous work that identified the importance of VIP in circadian and reproductive function using VIP knockout mice (36), while avoiding some pitfalls of other methods of selectively targeting VIP neurons within the SCN, such as damage to other neurons or brain nuclei via surgical methods due to the ventral location of the SCN (9, 68). One concern with conditional knockout mice is that off-target recombination may impact other brain or peripheral functions or have a negative impact on development. In our model, we found VipCre-mediated recombination outside of the SCN was restricted to the olfactory bulb, which also expresses Vax1. However, as a functioning olfactory bulb is pivotal to male mating (69) and our male mice bred normally, it is likely that the olfactory bulb is not significantly affected in this model. We also generated VipCre : Td mice to identify neurons that were targeted by the VipCre allele prior to tissue collection to validate that the VipCre allele specifically targeted VAX1 expressing neurons of the SCN. We confirmed that SCN tdTomato+ neurons colocalized with VAX1, and VAX1 neurons colocalized with tdTomato, although these percentages were lower than predicted by RNAScope® ISH assay. The differences in co-localization between mRNA and protein results could reflect different sensitivity limits between the visualization approaches, or alternatively may provide evidence that not all Vax1 mRNA is being translated into protein. Another possibility is that VAX1 expression could be circadian at the mRNA and/or protein level, leading to differences in expression that are time-of-day dependent, although future studies will be needed to determine this.
As a transcription factor that binds to ATTA and ATTA-like sites, a common sequence in the DNA, VAX1 has a high number of genes it can potentially regulate. Here, we build upon our previous work that determined the ability of VAX1 to promote Per2-luciferase expression (25) by providing evidence that this occurs via direct binding of VAX1 to regulatory regions of Per2. In addition to Per2, another core component of the molecular clock, Bmal1 also contains numerous ATTA-like sites in its regulatory region. We found here that VAX1 can directly bind to all the identified Bmal1 ATTA sites tested by EMSA, in addition to promoting Bmal1-luciferase expression. This work provides a mechanism by which changes in VAX1 expression can directly impact molecular clock function by VAX1 binding to the regulatory regions of Per2 and Bmal1. Excitingly, Vax1Vip mice presented with a shortening in SCN PER2::LUC period in ex vivo recordings, as well a shortened free-running period, together supporting a role of VAX1 as a novel regulator of both molecular clock expression and function. Future work will be required to determine if a loss of VAX1 in VIP neurons causes a circadian phase shift in VIP neurons, and/or if a loss of VAX1 leads to a reduction in molecular clock transcript expression, which could impact clock-controlled gene expression and phase.
Circadian timekeeping is impaired in Vax1Vip mice, where differences in SCN peptide transcript levels may underlie sex-specific vulnerability to circadian disruption
VIP neurons within the SCN are an important coordinator of the circadian timekeeping system (37). Given this, it is no surprise that the Vax1Vip mouse model demonstrates altered SCN output, as indicated by the shortened Vax1Vip free-running period in both sexes. This finding is consistent with work done by others indicating that a decrease in VIP within the SCN results in a shortened free-running period (36). However, VIP rarely acts alone in the regulation of circadian behaviors, and other peptides and components of the molecular clock exert strong influences on locomotor period (70). A shortened free-running period can be caused by an increase in SCN AVP (71–73) or reductions in Bmal1 expression (74). Others have found that Bmal1 is expressed rhythmically in Avp- and Vip-expressing neurons (75) and that deletion of Bmal1 from Avp-expressing neurons can lengthen free-running periods in mice (76). As Vax1Vip females do not show significant changes in Avp in the SCN, and only trended towards decreased Vip expression, in addition to a non-significant reduction in neurons co-expressing Bmal1 and Vip, the cause of the shortened period in Vax1Vip mice remains unknown. Future work will aim at determining if these trending reductions in Vip, combined with a trend in reduced Bmal1 expression in Vip neurons together might contribute to the shortened SCN period of Vax1Vip mice.
Sex differences are well-documented in SCN morphology and cellular function, previously reviewed in several studies (77, 78). There is strong evidence that VIP is sexually dimorphic, with increased VIP expression in human males (79, 80), as well as increased Vip transcript in male nocturnal laboratory rats (81) and diurnal Nile Grass rats (82). Our data support and extend these findings, where Ctrl male mice also exhibit increased Vip transcript and VIP compared to females. A potential mechanism guiding this sex difference could rely on the influence of the gonadal hormones estradiol and testosterone, which modulate VIP expression in the SCN (81–84). Our data, and others, suggest that sex differences in both Vip transcript and peptide levels (78) occur post-puberty. Recent work has demonstrated detailed spatial patterning of the onset and development of Avp and Vip transcription (85). Our data support the conclusion that SCN Vip neuron development is not complete at P10; however, we find decreased Vip transcript and protein in the adult female SCN compared to males, while increased VIP-TdT+ cell numbers were found in a lateral cluster of the SCN (85). There are several potential rationales for these differences, including delays between transcription onset and VipCre-driven TdTomato expression that may lead to different results from our mRNA transcript measure. Nevertheless, these neuropeptide sex differences highlight the need for continued investigation in both males and females to further our understanding of how SCN function drives circadian output in both sexes and the role of sex steroids therein.
Vax1Vip males exhibit normal fertility, while females display modest dysregulation of the reproductive axis
Circadian timekeeping is essential for coordinating hormone release and increased tissue sensitivity within reproductive tissues (25, 27, 35). The SCN modulates the timing of hormone release through direct and indirect projections to GnRH neurons, which in turn regulate the release of LH and FSH (86–88). These hormones are required for reproductive health through the production of testosterone and spermiogenesis in males and ovulation, embryo implantation, and follicular health in females (89–91). Though more frequently studied in females, current studies suggest that severe circadian disruption, through changes either in light exposure or via direct disruption to the SCN, may have a mild influence on male fertility (92). Interestingly, we found that Vax1Vip males had delayed pubertal onset. Given the importance of both SCN Avp and Vip in LH release (32, 93, 94), a required hormone for pubertal onset (95), it will be of interest for future studies to examine SCN neuropeptide expression in Vax1Vip males undergoing puberty. Interestingly, aside from delayed pubertal onset, there were no significant deficits in Vax1Vip male reproductive function, and Vax1Vip males displayed an increase in total sperm through an unknown mechanism. These data support a theory that a less stringent circadian control may favor male fertility. Together our data, coupled with evidence from genetic knockout and light-disruption studies (92, 96, 97), indicate that male fertility is resilient when faced with circadian challenges.
In contrast to males, circadian disruption and impaired SCN function are known to have a strong impact on female reproduction (27, 98–100). VIP, in particular, has an important role in the female reproductive axis, where VIP knockout females have lengthened estrous cycles and are sub-fertile, resulting in fewer liters of smaller sizes (19). It is likely that some of the sub-fertility in full body VIP knockout females is driven by VIP neurons within the SCN, as our Vax1Vip females also displayed lengthened estrous cycles. Our data are further supported by work in progress, showing that surgically ablated VIP neurons within the SCN also lengthened estrous cycles (101). Although Vax1Vip estrous cycles were lengthened, we did not see changes in litter sizes or number of litters that are associated with full VIP knockout females. Taken together, these data indicate estrous cycle length is influenced by Vip-expressing neurons within the SCN.
The estrous cycle is regulated by hormonal feedback throughout the reproductive axis. Within the hypothalamus, VIP neurons directly project onto GnRH neurons to regulate the frequency of GnRH release to the pituitary (31) and indirectly through projections onto AVP neurons in the anteroventral paraventricular nucleus to regulate kisspeptin neurons that modulate the surge release of GnRH needed for ovulation (102, 103). Although only a single dose of kisspeptin was tested, we found that Vax1Vip females had a greater release of LH in response to a kisspeptin challenge than Ctrls, a difference we did not observe in the males. The normal pituitary response to GnRH and normal circadian rhythms in ex vivo pituitary explants of Vax1Vip females, combined with the absence of Vax1 in gonadotropes as shown by qPCR in isolated gonadotrope cells from female mice (53, 104) and single cell RNAseq [personal communication, (104, 105)], suggest it is unlikely that the increased LH release in response to kisspeptin is associated with abnormal gonadotrope function. One possibility for the sex difference in kisspeptin-induced changes in LH release in Vax1Vip mice might be linked to differences in the neuronal circuit encompassing the sexually dimorphic anteroventral periventricular nucleus kisspeptin neurons (106). This neuronal population is larger in females than in males and plays a central role in the LH surge (107, 108). Interestingly, a comparable increased sensitivity to a kisspeptin challenge in females was also observed in full body Bmal1 knockout mice (8), where the mechanism for this increase remains unknown. Future work to determine how neuronal network changes, with or without intact molecular clock function, impact GnRH neuron sensitivity to kisspeptin will be of interest. Although the kisspeptin challenge elicited a greater LH release in the Vax1Vip females than Ctrls, basal LH levels were comparable to Ctrls, and Vax1Vip females exhibited a decrease in circulating FSH. LH and FSH production and release are controlled in a great part by the pulsatile pattern of GnRH release (87, 109, 110). Repeated blood sampling of Vax1Vip mice would have been an ideal approach to assess for changes in pulsatile hormone release, however, as Vax1Vip mice present with normal litter sizes, time to first litter, ovarian phase, which indicate overall normal ovarian function, and an increased activation of the stress axis, which is a suppressor of GnRH release (111), we decided against completing a LH and FSH pulse analysis due to the confounding effect of corticosterone on these data. In the future, we hope to better elucidate the relationship between stress and pulsatile hormones in this mouse model.
VAX1 in postnatal VIP neurons regulates female depressive-like behavior
Changes in the neuroendocrine regulation of FSH release from the pituitary in Vax1Vip mice is a potential pathway causing the reduction in estrogen of these mice. FSH is a limiting factor in the conversion of testosterone into estrogen in the granulosa cells of the ovary (112, 113), from where estrogen enters the general circulation. It is important to note that both FSH and estrogen are circadian (114, 115), thus a limit of this study, with blood sampling at a single time point, is our inability to assess if the reduction in these hormones might be due to a phase shift in hormone release. While primarily associated with its role in reproduction, estrogen has a multitude of functions, including altering the sensitivity of neuronal circuits (116, 117), regulating the activity of the stress axis (118), and displaying strong correlations with mood (119–121). Low estrogen has been correlated with depressive-like behaviors in women (122). In rodents, increasing estrogen has been shown to exert an anti-depressant-like effect during the Porsolt forced swim test, a test that is thought to reflect depressive-like behavior in rodents (123, 124). Additionally, an imbalance of progesterone and estrogen is associated with a higher incidence of mood disorders, including premenstrual dysphoric disorder (PMDD), a depressive disorder that presents with severe physical and physiological symptoms during the luteal phase of the menstrual cycle (125–128). In addition to recapitulating the low estradiol (or imbalance of progesterone and estrogen) as a risk factor for mood disorders, Vax1Vip females also display another hallmark of PMDD, weakened SCN output (129, 130). Excitingly, Vax1Vip females (but not males) have increased depressive-like behavior. Taken together, these data point to a novel role of VAX1 in regulating VIP neuron modulation of mood and the reproductive axis and raise the potential of Vax1Vip females to serve as a new model for mood disorders that are tied to reproductive cycles, such as PMDD. Furthermore, VIP-and AVP-expressing neurons contribute to the regulation of the stress axis (131–134). Stressful situations result in an increase in signal from the hypothalamus, which translates to higher levels of corticosterone via activation of the hypothalamic-pituitary-adrenal (stress) axis. Notably, the hypothalamus is comprised of several nuclei, including the SCN (135) and the PVN (136, 137), with varying roles and contributions to the stress axis. Within the SCN, reductions in Vip have been correlated with increased stress (138), whereas the VIP neuron target, the PVN, is a central relay station of the stress axis (63, 139). Thus, the reduction of Vip in the SCN of Vax1Vip females is a likely contributing factor in the increased corticosterone levels found in Vax1Vip females, both at baseline and in response to a stressor. Specifically within the PVN, AVP is known to stimulate the stress axis (140, 141), and AVP expression in the PVN is comparable between control and VIP knockout males (142). This suggests that the reduction in VIP in the SCN of Vax1Vip mice may not be driving the changes in Avp mRNA within the PVN, but more likely is the result of other VAX1 targets that impact VIP neuron communication with PVN neurons, such as GABA (143, 144).
Conclusion and summary
Due to the abundance of VIP throughout the brain and body, it is difficult to study subsets of neurons expressing VIP without invasive surgery, which can lead to damaged brain tissue and a variety of other complications. In this study, we leveraged the close to 100% overlap of Vax1 expression specifically within SCN Vip neurons, to generate a conditional knockout mouse model to study this subset of VIP neurons. We found that deletion of Vax1 from SCN VIP neurons results in mice with altered circadian rhythms. Excitingly, Vax1Vip females had disrupted reproductive axis function, low estrogen, and high corticosterone, as well as an increase in depressive-like behaviors. Together, these data provide us with an exciting new model to study the genetic and neurological overlap between circadian disruption, female reproductive health, and depressive-like behaviors.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal studies were approved by Michigan State University Institutional Animal Care & Use Committee and the University of California, San Diego Institutional Animal Care and Use Committee. The studies were conducted in accordance with the local legislation and institutional requirements.
Author contributions
BV: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. AY: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing, Supervision, Validation. JB: Data curation, Investigation, Writing – review & editing. BJ: Data curation, Writing – review & editing. DN: Data curation, Writing – review & editing, Investigation, Methodology. KJ: Data curation, Investigation, Writing – review & editing, Formal analysis. FR: Data curation, Investigation, Writing – review & editing, Conceptualization, Funding acquisition. EH: Investigation, Writing – review & editing. LC: Investigation, Writing – review & editing. DG: Investigation, Writing – review & editing. LS: Investigation, Writing – review & editing, Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Software, Supervision. MG: Investigation, Writing – review & editing, Conceptualization, Funding acquisition, Methodology, Software. KT: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Writing – review & editing, Visualization. PM: Conceptualization, Funding acquisition, Methodology, Software, Writing – review & editing, Formal analysis, Supervision, Validation. HH: Conceptualization, Formal analysis, Funding acquisition, Methodology, Software, Supervision, Validation, Writing – review & editing, Data curation, Investigation, Project administration, Resources, Visualization, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Institutes of Health (NIH) Grants R01 HD072754, R01 HD100580, and R01 HD082567 (to PM). It was also supported by NIH/Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) P50 HD012303 as part of the National Centers for Translational Research in Reproduction and Infertility (PM). PM was also partially supported by P30 DK063491, P30 CA023100, and P42 ES010337. KT was partially supported by K99/R00 NS119291, T32 HD007203, P42 ES010337, and F32 HD090837. HH was partially supported by K99/R00 HD084759 and the United States Department of Agriculture National Institute of Food and Agriculture Hatch project MICL1018024. AY and BL were partially supported by T32 HD087166. AY was partially supported by F32 HD107852. EH was supported by T32 GM008666 and T32 GM007198. Work in the MG laboratory was supported by Office of Naval Research N00014-13-1-0285. JB and LC were partially supported by the Frontiers of Innovation Scholars Program, UC San Diego. LC was also supported by The Endocrine Society Summer Research Fellowship Award and the Ledell Family Research Undergraduate Research Scholarship Award for Science and Engineering. The University of Virginia, Center for Research in Reproduction, Ligand Assay and Analysis Core, is supported by the NIH/NICHD Grant P50 HD028934. FR was supported by the ENDURE/Bridge to the PhD in Neuroscience Program R25-NS090989.
Acknowledgments
We wish to thank the staff of the Nikon Imaging Center at UC San Diego for microscopy training. We would like to thank MSU Precision Health Program (PHP) Tissue Analysis Core for technical assistance. We thank Kierra Jursch for scoring the male behavior videos.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1269672/full#supplementary-material
References
1. Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet (2006) 15 Spec No(suppl_2):R271–7. doi: 10.1093/hmg/ddl207
2. Pfeffer M, Müller CM, Mordel J, Meissl H, Ansari N, Deller T, et al. The mammalian molecular clockwork controls rhythmic expression of its own input pathway components. J neuroscience : Off J Soc Neurosci (2009) 29(19):6114–23. doi: 10.1523/JNEUROSCI.0275-09.2009
3. Kudo T, Block GD, Colwell CS. The circadian clock gene period1 connects the molecular clock to neural activity in the suprachiasmatic nucleus. ASN Neuro (2015) 7(6):175909141561076. doi: 10.1177/1759091415610761
4. Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell (2000) 103(7):1009–17. doi: 10.1016/S0092-8674(00)00205-1
5. Lowrey PL, Takahashi JS. MAMMALIAN CIRCADIAN BIOLOGY: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet (2004) 5(1):407–41. doi: 10.1146/annurev.genom.5.061903.175925
6. Boden MJ, Varcoe TJ, Voultsios A, Kennaway DJ. Reproductive biology of female Bmal1 null mice. Reprod (Cambridge England) (2010) 139(6):1077–90. doi: 10.1530/REP-09-0523
7. Schoeller EL, Clark DD, Dey S, Cao NV, Semaan SJ, Chao LW, et al. Bmal1 is required for normal reproductive behaviors in male mice. Endocrinol Oxford Acad (2016) 157(12):4914–29. doi: 10.1210/EN.2016-1620
8. Tonsfeldt KJ, Schoeller EL, Brusman LE, Cui LJ, Lee J, Mellon PL. The contribution of the circadian gene bmal1 to female fertility and the generation of the preovulatory luteinizing hormone surge. J Endocr Soc (2019) 3(4):716–33. doi: 10.1210/js.2018-00228
9. Mazuski C, Chen SP, Herzog ED. Different roles for VIP neurons in the neonatal and adult suprachiasmatic nucleus. J Biol rhythms (2020) 35(5):465–75. doi: 10.1177/0748730420932073
10. Maywood ES, Reddy AB, Wong GKY, O’Neill JS, O’Brien JA, McMahon DG, et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr biol : CB (2006) 16(6):599–605. doi: 10.1016/j.cub.2006.02.023
11. Kuljis DA, Loh DH, Truong D, Vosko AM, Ong ML, McClusky R, et al. Gonadal- and sex-chromosome-dependent sex differences in the circadian system. Endocrinol United States (2013) 154(4):1501–12. doi: 10.1210/en.2012-1921
12. Evans JA, Gorman MR. In synch but not in step: Circadian clock circuits regulating plasticity in daily rhythms. Neuroscience (2016) 320:259–80. doi: 10.1016/j.neuroscience.2016.01.072
13. Patton AP, Hastings MH. The suprachiasmatic nucleus. Curr Biol (2018) 28(15):R816–22. doi: 10.1016/j.cub.2018.06.052
14. LeSauter J, Silver R. Localization of a suprachiasmatic nucleus subregion regulating locomotor rhythmicity. J Neurosci Soc Neurosci (1999) 19(13):5574–85. doi: 10.1523/JNEUROSCI.19-13-05574.1999
15. Kriegsfeld LJ, LeSauter J, Silver R. Targeted microlesions reveal novel organization of the hamster suprachiasmatic nucleus. J neuroscience : Off J Soc Neurosci (2004) 24(10):2449–57. doi: 10.1523/JNEUROSCI.5323-03.2004
16. Nielsen HS, Hannibal J, Fahrenkrug J. Vasoactive intestinal polypeptide induces per1 and per2 gene expression in the rat suprachiasmatic nucleus late at night. Eur J Neurosci (2002) 15(3):570–4. doi: 10.1046/j.0953-816x.2001.01882.x
17. Jung H, Choe Y, Kim H, Park N, Son GH, Khang I, et al. Involvement of CLOCK : BMAL1 heterodimer in serum-responsive mPer1 induction. NeuroReport (2003) 14(1):15–9. doi: 10.1097/00001756-200301200-00003
18. Alvarez JD, Hansen A, Ord T, Bebas P, Chappell PE, Giebultowicz JM, et al. The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J Biol Rhythms (2008) 23(1):26–36. doi: 10.1177/0748730407311254
19. Loh DH, Kuljis DA, Azuma L, Wu Y, Truong D, Wang HB, et al. Disrupted reproduction, estrous cycle, and circadian rhythms in female mice deficient in vasoactive intestinal peptide. J Biol Rhythms (2014) 29(5):355–69. doi: 10.1177/0748730414549767
20. Li M, Arimura A. Neuropeptides of the pituitary adenylate cyclase-activating polypeptide/vasoactive intestinal polypeptide/growth hormone-releasing hormone/secretin family in testis. Endocrine (2003) 20(3):201–14. doi: 10.1385/ENDO:20:3:201
21. Lacombe A, Lelievre V, Roselli CE, Muller J-M, Waschek JA, Vilain E. Lack of vasoactive intestinal peptide reduces testosterone levels and reproductive aging in mouse testis. J Endocrinol (2007) 194(1):153–60. doi: 10.1677/JOE-07-0102
22. Dolatshad H, Campbell EA, O’Hara L, Maywood ES, Hastings MH, Johnson MH. Developmental and reproductive performance in circadian mutant mice. Hum Reprod (Oxford England) (2006) 21(1):68–79. doi: 10.1093/humrep/dei313
23. Li C, Xiao S, Hao J, Liao X, Li G. Cry1 deficiency leads to testicular dysfunction and altered expression of genes involved in cell communication, chromatin reorganization, spermatogenesis, and immune response in mouse testis. Mol Reprod Dev (2018) 85(4):325–35. doi: 10.1002/mrd.22968
24. Hoffmann HM, Pandolfi EC, Larder R, Mellon PL. Haploinsufficiency of homeodomain proteins six3, vax1, and otx2 causes subfertility in mice via distinct mechanisms. Neuroendocrinology (2019) 109(3):200–7. doi: 10.1159/000494086
25. Hoffmann HM, Meadows JD, Breuer JA, Yaw AM, Nguyen D, Tonsfeldt KJ, et al. The transcription factors SIX3 and VAX1 are required for suprachiasmatic nucleus circadian output and fertility in female mice. J Neurosci Res (2021) 99(10):2625–45. doi: 10.1002/jnr.24864
26. Meadows JD, Breuer JA, Lavalle SN, Hirschenberger MR, Patel MM, Nguyen D, et al. Deletion of Six3 in post-proliferative neurons produces weakened SCN circadian output, improved metabolic function, and dwarfism in male mice. Mol Metab (2022) 57:101431. doi: 10.1016/j.molmet.2021.101431
27. Sen A, Hoffmann HM. Role of core circadian clock genes in hormone release and target tissue sensitivity in the reproductive axis. Mol Cell Endocrinol (2020) 501:110655. doi: 10.1016/j.mce.2019.110655
28. Yaw AM, DeVries BM, Hoffmann HM. Disrupted circadian rhythms and neuroendocrine function in fertility’. In: Biological implications of circadian disruption. Cambridge University Press (2023). p. 206–22. doi: 10.1017/9781009057646.010
29. Van Der Beek EM, Horvath TL, Wiegant VM, Van Den Hurk R, Buijs RM, et al. Evidence for a direct neuronal pathway from the suprachiasmatic nucleus to the gonadotropin-releasing hormone system: Combined tracing and light and electron microscopic immunocytochemical studies. J Comp Neurol (1997) 384(4):569–79. doi: 10.1002/(SICI)1096-9861(19970811)384:4<569::AID-CNE6>3.0.CO;2-0
30. Williams WP, Jarjisian SG, Mikkelsen JD, Kriegsfeld LJ. Circadian control of kisspeptin and a gated gnRH response mediate the preovulatory luteinizing hormone surge. Endocrinology (2011) 152(2):595–606. doi: 10.1210/en.2010-0943
31. Piet R, Dunckley H, Lee K, Herbison AE. Vasoactive intestinal peptide excites gnRH neurons in male and female mice. Endocrinology (2016) 157(9):3621–30. doi: 10.1210/en.2016-1399
32. Schafer D, Kane G, Colledge WH, Piet R, Herbison AE. Sex- and sub region-dependent modulation of arcuate kisspeptin neurones by vasopressin and vasoactive intestinal peptide. J Neuroendocrinol (2018) 30(12):e12660. doi: 10.1111/jne.12660
33. Garcia JE, Seegar Jones G, Wright GL. Prediction of the time of ovulation. Fertility Sterility (1981) 36(3):308–15. doi: 10.1016/S0015-0282(16)45730-4
34. Shoham Z, Schacter M, Loumaye E, Weissman A, MacNamee M, Insler V. The luteinizing hormone surge–the final stage in ovulation induction: modern aspects of ovulation triggering. Fertility sterility (1995) 64(2):237–51. doi: 10.1016/s0015-0282(16)57717-6
35. Mereness AL, Murphy ZC, Forrestel AC, Butler S, Ko C, Richards JS, et al. Conditional deletion of bmal1 in ovarian theca cells disrupts ovulation in female mice. Endocrinology (2016) 157(2):913–27. doi: 10.1210/en.2015-1645
36. Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, et al. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiology-Regulatory Integr Comp Physiol (2003) 285(5):R939–49. doi: 10.1152/ajpregu.00200.2003
37. Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci (2005) 8(4):476–83. doi: 10.1038/nn1419
38. Joye DAM, Rohr KE, Keller D, Inda T, Telega A, Pancholi H, et al. Reduced VIP expression affects circadian clock function in VIP-IRES-CRE mice (JAX 010908). J Biol Rhythms (2020) 35(4):340–52. doi: 10.1177/0748730420925573
39. Rabinovici J. The differential effects of FSH and LH on the human ovary. Bailliere’s Clin obstetrics gynaecol (1993) 7(2):263–81. doi: 10.1016/s0950-3552(05)80130-0
40. Howles CM. Role of LH and FSH in ovarian function. Mol Cell Endocrinol Elsevier (2000) 161(1–2):25–30. doi: 10.1016/S0303-7207(99)00219-1
41. Eriksson O, Bäckström T, Stridsberg M, Hammarlund-Udenaes M, Naessén T. Differential response to estrogen challenge test in women with and without premenstrual dysphoria. Psychoneuroendocrinology (2006) 31(4):415–27. doi: 10.1016/j.psyneuen.2005.10.004
42. Huhtaniemi IT, Themmen APN. Mutations in human gonadotropin and gonadotropin-receptor genes. Endocrine (2005) 26(3):207–18. doi: 10.1385/ENDO:26:3:207
43. Studd J, Nappi RE. Reproductive depression. Gynecological Endocrinol (2012) 28(sup1):42–5. doi: 10.3109/09513590.2012.651932
44. Joffe H, de Wit A, Coborn J, Crawford S, Freeman M, Wiley A, et al. Impact of estradiol variability and progesterone on mood in perimenopausal women with depressive symptoms. J Clin Endocrinol Metab (2020) 105(3):e642–50. doi: 10.1210/clinem/dgz181
45. Soria V, Martínez-Amorós È, Escaramís G, Valero J, Pérez-Egea R, García C, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology (2010) 35(6):1279–89. doi: 10.1038/npp.2009.230
46. Pandolfi EC, Breuer JA, Nguyen Huu VA, Talluri T, Nguyen D, Lee JS, et al. The homeodomain transcription factors vax1 and six6 are required for SCN development and function. Mol Neurobiol (2020) 57(2):1217–32. doi: 10.1007/s12035-019-01781-9
47. Tonsfeldt KJ, Mellon PL, Hoffmann HM. Circadian rhythms in the neuronal network timing the luteinizing hormone surge. Endocrinology (2022) 163(2):bqab268. doi: 10.1210/endocr/bqab268
48. Council NR. Guide for the care and use of laboratory animals. Guide for the care and use of laboratory animals (National Academies Press). (2011). doi: 10.17226/12910
49. Lucas RJ, Allan A, Brainard G, Brown T, Dauchy RT, Didikoglu A, et al. In the eye of the beholder: measuring and standardising light for laboratory mammals. Preprints (2023). p. 2023091766. doi: 10.20944/preprints202309.1766.v1
50. McDowell RJ, Didikoglu A, Woelders T, Gatt MJ, Hut RA, Brown TM, et al. ‘Beyond lux: methods for species and photoreceptor-specific quantification of ambient light for mammals. bioRxiv (2023). doi: 10.1101/2023.08.25.554794
51. Hoffmann HM, Trang C, Gong P, Kimura I, Pandolfi EC, Mellon PL. Deletion of vax1 from gonadotropin-releasing hormone (GnRH) neurons abolishes gnRH expression and leads to hypogonadism and infertility. J neuroscience : Off J Soc Neurosci (2016) 36(12):3506–18. doi: 10.1523/JNEUROSCI.2723-15.2016
52. Porsolt RD, Le Pichon M, Jalfre M. Depression: a new animal model sensitive to antidepressant treatments. Nature (1977) 266(5604):730–2. doi: 10.1038/266730a0
53. Hoffmann HM, Tamrazian A, Xie H, Pérez-Millán MI, Kauffman AS, Mellon PL. Heterozygous deletion of ventral anterior homeobox (Vax1) causes subfertility in mice. Endocrinology (2014) 155(10):4043–53. doi: 10.1210/en.2014-1277
54. Hoffmann HM, Gong P, Tamrazian A, Mellon PL. Transcriptional interaction between cFOS and the homeodomain-binding transcription factor VAX1 on the GnRH promoter controls Gnrh1 expression levels in a GnRH neuron maturation specific manner. Mol Cell Endocrinol (2018) 461:143–54. doi: 10.1016/j.mce.2017.09.004
55. Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep (2017) 7(1):16878. doi: 10.1038/s41598-017-17204-5
56. Sempere LF, Zaluzec E, Kenyon E, Kiupel M, Moore A. ‘Automated five-color multiplex co-detection of microRNA and protein expression in fixed tissue specimens’. Methods Mol Biol (2020) 2148:257–76. doi: 10.1007/978-1-0716-0623-0_17
57. Yaw AM, Duong TV, Nguyen D, Hoffmann HM. Circadian rhythms in the mouse reproductive axis during the estrous cycle and pregnancy. J Neurosci Res (2021) 99(1):294–308. doi: 10.1002/jnr.24606
58. Hoffmann HM, Larder R, Lee JS, Hu RJ, Trang C, Devries BM, et al. Differential CRE expression in lhrh-cre and gnRH-cre alleles and the impact on fertility in otx2-flox mice. Neuroendocrinology (2019) 108(4):328–42. doi: 10.1159/000497791
59. Redlin U. Neural basis and biological function of masking by light in mammals: suppression of melatonin and locomotor activity. Chronobiol Int (2001) 18(5):737–58. doi: 10.1081/cbi-100107511
60. Yoo S-H, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci (2004) 101(15):5339–46. doi: 10.1073/pnas.0308709101
61. Buijs RM, Van Eden CG. The integration of stress by the hypothalamus, amygdala and prefrontal cortex: balance between the autonomic nervous system and the neuroendocrine system. Prog Brain Res (2000) pp:117–32. doi: 10.1016/S0079-6123(00)26011-1
62. Ulrich-Lai YM, Herman JP. Neural regulation of endocrine and autonomic stress responses. Nat Rev Neurosci Nat Publishing Group (2009) 10(6):397–409. doi: 10.1038/nrn2647
63. Busnardo C, Tavares RF, Resstel LBM, Elias LLK, Correa FMA. Paraventricular nucleus modulates autonomic and neuroendocrine responses to acute restraint stress in rats. Autonomic Neurosci (2010) 158(1–2):51–7. doi: 10.1016/j.autneu.2010.06.003
64. Conte I, Morcillo J, Bovolenta P. Comparative analysis of Six3 and Six6 distribution in the developing and adult mouse brain. Dev Dynamics (2005) 234(3):718–25. doi: 10.1002/dvdy.20463
65. Hatori M, Gill S, Mure LS, Goulding M, O’Leary DDM, Panda S. Lhx1 maintains synchrony among circadian oscillator neurons of the SCN. eLife (2014) 3(July2014):e03357. doi: 10.7554/eLife.03357
66. Bedont JL, Blackshaw S. Constructing the suprachiasmatic nucleus: a watchmaker’s perspective on the central clockworks. Front Syst Neurosci (2015) 9:74(MAY). doi: 10.3389/fnsys.2015.00074
67. Hallonet M, Hollemann T, Pieler T, Gruss P. Vax1, a novel homeobox-containing gene, directs development of the basal forebrain and visual system. Genes Dev (1999) 13(23):3106–14. doi: 10.1101/gad.13.23.3106
68. Shimizu K, Fukada Y. Stereotaxic surgery for suprachiasmatic nucleus lesions in mice. Bio-protocol (2017) 7(12):e2346. doi: 10.21769/BioProtoc.2346
69. Murphy MR, Schneider GE. Olfactory bulb removal eliminates mating behavior in the male golden hamster. Science (1970) 167(3916):302–4. doi: 10.1126/science.167.3916.302
70. Ono D, Honma K, Honma S. Roles of neuropeptides, VIP and AVP, in the mammalian central circadian clock. Front Neurosci (2021) 15:650154. doi: 10.3389/fnins.2021.650154
71. Mieda M, Okamoto H, Sakurai T. Manipulating the cellular circadian period of arginine vasopressin neurons alters the behavioral circadian period. Curr biol : CB (2016) 26(18):2535–42. doi: 10.1016/j.cub.2016.07.022
72. Rohr KE, Telega A, Savaglio A, Evans JA. Vasopressin regulates daily rhythms and circadian clock circuits in a manner influenced by sex. Hormones Behav (2021) 127:104888. doi: 10.1016/j.yhbeh.2020.104888
73. Tsuno Y, Peng Y, Horike S, Wang M, Matsui A, Yamagata K, et al. In vivo recording of suprachiasmatic nucleus dynamics reveals a dominant role of arginine vasopressin neurons in circadian pacesetting. PloS Biol (2023) 21(8):e3002281. doi: 10.1371/journal.pbio.3002281
74. Haque SN, Booreddy SR, Welsh DK. Effects of BMAL1 manipulation on the brain’s master circadian clock and behavior. Yale J Biol Med (2019) 92(2):251–8.
75. Wen S, Ma D, Zhao M, Xie L, Wu Q, Gou L, et al. Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat Neurosci (2020) 23(3):456–67. doi: 10.1038/s41593-020-0586-x
76. Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, et al. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron (2015) 85(5):1103–16. doi: 10.1016/j.neuron.2015.02.005
77. Bailey M, Silver R. Sex differences in circadian timing systems: Implications for disease. Front Neuroendocrinol (2014) 35(1):111–39. doi: 10.1016/j.yfrne.2013.11.003
78. Joye DAM, Evans JA. Sex differences in daily timekeeping and circadian clock circuits. Semin Cell Dev Biol (2022) 126:45–55. doi: 10.1016/j.semcdb.2021.04.026
79. Zhou J. VIP neurons in the human SCN in relation to sex, age, and Alzheimer’s disease. Neurobiol Aging. Elsevier (1995) 16(4):571–6. doi: 10.1016/0197-4580(95)00043-E
80. Hofman MA, Zhou J-N, Swaab DF. Suprachiasmatic nucleus of the human brain: An immunocytochemical and morphometric analysis. Anatomical Rec (1996) 244(4):552–62. doi: 10.1002/(SICI)1097-0185(199604)244:4<552::AID-AR13>3.0.CO;2-O
81. Krajnak K, Kashon ML, Rosewell KL, Wise PM. Aging Alters the Rhythmic Expression of Vasoactive Intestinal Polypeptide mRNA But Not Arginine Vasopressin mRNA in the Suprachiasmatic Nuclei of Female Rats. J Neurosci (1998) 18(12):4767–74. doi: 10.1523/JNEUROSCI.18-12-04767.1998
82. Mahoney MM, Ramanathan C, Hagenauer MH, Thompson RC, Smale L, Lee T. Daily rhythms and sex differences in vasoactive intestinal polypeptide, VIPR2 receptor and arginine vasopressin mRNA in the suprachiasmatic nucleus of a diurnal rodent, Arvicanthis niloticus. Eur J Neurosci (2009) 30(8):1537–43. doi: 10.1111/j.1460-9568.2009.06936.x
83. Gozes I, Werner H, Fawzi M, Abdelatty A, Shani Y, Fridkin M, et al. Estrogen regulation of vasoactive intestinal peptide mRNA in rat hypothalamus. J Mol Neurosci (1989) 1(1):55–61. doi: 10.1007/BF02896857
84. Watanobe H, Takebe K. A comparative study of the effects of neonatal androgenization and estrogenization on vasoactive intestinal peptide levels in the anterior pituitary and the hypothalamus of adult female rats. Neuroendocrinology (1992) 56(5):653–9. doi: 10.1159/000126289
85. Carmona-Alcocer V, Brown LS, Anchan A, Rohr KE, Evans JA. Developmental patterning of peptide transcription in the central circadian clock in both sexes. Front Neurosci (2023) 17:1177458(May). doi: 10.3389/fnins.2023.1177458
86. Bruni JF, Huang H-H, Marshall S, Meites J. Effects of single and multiple injections of synthetic gnRH on serum LH, FSH and testosterone in young and old male rats. Biol Reprod (1977) 17(3):309–12. doi: 10.1095/biolreprod17.3.309
87. Hall JE, Brodie TD, Badger TM, Rivier J, Vale W, Conn PM, et al. Evidence of differential control of FSH and LH secretion by gonadotropin-releasing hormone (GnRH) from the use of a GnRH antagonist. J Clin Endocrinol Metab (1988) 67(3):524–31. doi: 10.1210/jcem-67-3-524
88. Vizcarra JA, Wettemann RP, Braden TD, Turzillo AM, Nett TM. Effect of gonadotropin-releasing hormone (GnRH) pulse frequency on serum and pituitary concentrations of luteinizing hormone and follicle-stimulating hormone, GnRH receptors, and messenger ribonucleic acid for gonadotropin subunits in cows. Endocrinol Oxford Acad (1997) 138(2):594–601. doi: 10.1210/endo.138.2.4938
89. Hillier SG. Current concepts of the roles of follicle stimulating hormone and luteinizing hormone in folliculogenesis. Hum Reprod (1994) 9(2):188–91. doi: 10.1093/oxfordjournals.humrep.a138480
90. Christensen A, Bentley G, Cabrera R, Ortega H, Perfito N, Wu T, et al. Hormonal regulation of female reproduction. Hormone Metab Res (2012) 44(08):587–91. doi: 10.1055/s-0032-1306301
91. Dutta S, Sengupta P, Muhamad S. Male reproductive hormones and semen quality. Asian Pacific J Reprod (2019) 8(5):189. doi: 10.4103/2305-0500.268132
92. Li T, Bai Y, Jiang Y, Jiang K, Tian Y, Gu J, et al. The potential impacts of circadian rhythm disturbances on male fertility. Front Endocrinol (2022) 13:1001316. doi: 10.3389/fendo.2022.1001316
93. Harney JP, Scarbrough K, Rosewell KL, Wise PM. In vivo antisense antagonism of vasoactive intestinal peptide in the suprachiasmatic nuclei causes aging-like changes in the estradiol-induced luteinizing hormone and prolactin surges. Endocrinology (1996) 137(9):3696–701. doi: 10.1210/endo.137.9.8756535
94. Russo KA, La JL, Stephens SBZ, Poling MC, Padgaonkar NA, Jennings KJ, et al. Circadian control of the female reproductive axis through gated responsiveness of the RFRP-3 system to VIP signaling. Endocrinology (2015) 156(7):2608–18. doi: 10.1210/en.2014-1762
95. Michael SD, Kaplan SB, Macmillan BT. Peripheral plasma concentrations of LH, FSH, prolactin and GH from birth to puberty in male and female mice. Reproduction (1980) 59(1):217–22. doi: 10.1530/jrf.0.0590217
96. Kennaway DJ, Boden MJ, Varcoe TJ. Circadian rhythms and fertility. Mol Cell Endocrinol (2012) 349(1):56–61. doi: 10.1016/j.mce.2011.08.013
97. Peterlin A, Kunej T, Peterlin B. The role of circadian rhythm in male reproduction. Curr Opin Endocrinol Diabetes Obes (2019) 26(6):313–6. doi: 10.1097/MED.0000000000000512
98. Williams W. P.III, Kriegsfeld LJ. Circadian control of neuroendocrine circuits regulating female reproductive function’. Front Endocrinol (2012) 3:60(May). doi: 10.3389/fendo.2012.00060
99. Miller BH, Takahashi JS. Central circadian control of female reproductive function. Front Endocrinol (2013) 4:195(January). doi: 10.3389/fendo.2013.00195
100. Yaw A, McLane-Svoboda A, Hoffmann H. ‘Shiftwork and light at night negatively impact molecular and endocrine timekeeping in the female reproductive axis in humans and rodents’. Int J Mol Sci (2020) 22(1):324. doi: 10.3390/ijms22010324
101. Kahan A, Coughlin GM, Borsos M, Brunton BW, Gradinaru V. Dysregulated mammalian estrus cycle rescued by timed activation of VIP neurons in the circadian pacemaker and late afternoon light exposure. bioRxiv (2023). doi: 10.1101/2023.01.14.524075
102. d’Anglemont de Tassigny X, Fagg LA, Carlton MBL, Colledge WH. Kisspeptin can stimulate gonadotropin-releasing hormone (GnRH) release by a direct action at GnRH nerve terminals. Endocrinology (2008) 149(8):3926–32. doi: 10.1210/en.2007-1487
103. da Silva Mansano N, da S, Paradela RS, Bohlen TM, Zanardi IM, Chaves FM, Silveira MA, et al. Vasoactive intestinal peptide exerts an excitatory effect on hypothalamic kisspeptin neurons during estrogen negative feedback. Mol Cell Endocrinol (2022) 542:111532. doi: 10.1016/j.mce.2021.111532
104. Lavalle SN, Chou T, Hernandez J, Naing NCP, Tonsfeldt KJ, Hoffmann HM, et al. Kiss1 is differentially regulated in male and female mice by the homeodomain transcription factor VAX1. Mol Cell Endocrinol (2021) 534:111358. doi: 10.1016/j.mce.2021.111358
105. Ho Y, Hu P, Peel MT, Chen S, Camara PG, Epstein DJ, et al. Single-cell transcriptomic analysis of adult mouse pituitary reveals sexual dimorphism and physiologic demand-induced cellular plasticity. Protein Cell (2020) 11(8):565–83. doi: 10.1007/s13238-020-00705-x
106. Jamieson BB, Bouwer GT, Campbell RE, Piet R. Estrous cycle plasticity in the central clock output to kisspeptin neurons: implications for the preovulatory surge. Endocrinol Oxford Acad (2021) 162(6):1–20. doi: 10.1210/endocr/bqab071
107. Roa J, Castellano JM, Navarro VM, Handelsman DJ, Pinilla L, Tena-Sempere M. Kisspeptins and the control of gonadotropin secretion in male and female rodents. Peptides (2009) 30(1):57–66. doi: 10.1016/j.peptides.2008.08.009
108. Kauffman AS. Coming of age in the Kisspeptin Era: Sex differences, development, and puberty. Mol Cell Endocrinol (2010) 324(1–2):51–63. doi: 10.1016/j.mce.2010.01.017
109. Thompson IR, Kaiser UB. GnRH pulse frequency-dependent differential regulation of LH and FSH gene expression. Mol Cell endocrinol Elsevier (2014) 385(1–2):28–35. doi: 10.1016/j.mce.2013.09.012
110. Stamatiades GA, Carroll RS, Kaiser UB. GnRH—A key regulator of FSH. Endocrinol Oxford Acad (2019) 160(1):57–67. doi: 10.1210/en.2018-00889
111. Luo E, Stephens SBZ, Chaing S, Munaganuru N, Kauffman AS, Breen KM. Corticosterone blocks ovarian cyclicity and the LH surge via decreased kisspeptin neuron activation in female mice. Endocrinol Oxford Acad (2016) 157(3):1187–99. doi: 10.1210/en.2015-1711
112. Layman LC, McDonough PG. Mutations of follicle stimulating hormone-beta and its receptor in human and mouse: genotype/phenotype. Mol Cell endocrinol Elsevier (2000) 161(1–2):9–17. doi: 10.1016/s0303-7207(99)00217-8
113. Siegel ET, Kim H-G, Nishimoto HK, Layman LC. The molecular basis of impaired follicle-stimulating hormone action: evidence from human mutations and mouse models. Reprod Sci (Thousand Oaks Calif.) (2013) 20(3):211–33. doi: 10.1177/1933719112461184
114. Jean-Faucher C, Berger M, de Turckheim M, Veyssière G, Jean C. Circadian variations in plasma LH and FSH in juvenile and adult male mice. Hormone Res (1986) 23(3):185–92. doi: 10.1159/000180321
115. Bao A-M, Ji Y-F, Van Someren EJW, Hofman MA, Liu R-Y, Zhou J-N. Diurnal rhythms of free estradiol and cortisol during the normal menstrual cycle in women with major depression. Hormones Behav (2004) 45(2):93–102. doi: 10.1016/j.yhbeh.2003.09.004
116. Nunemaker CS, DeFazio RA, Moenter SM. Estradiol-sensitive afferents modulate long-term episodic firing patterns of GnRH neurons. Endocrinology (2002) 143(6):2284–92. doi: 10.1210/endo.143.6.8869
117. Piet R, Fraissenon A, Boehm U, Herbison AE. Estrogen permits vasopressin signaling in preoptic kisspeptin neurons in the female mouse. J neuroscience : Off J Soc Neurosci (2015) 35(17):6881–92. doi: 10.1523/JNEUROSCI.4587-14.2015
118. Goel N, Workman JL, Lee TT, Innala L, Viau V. Sex differences in the HPA axis. Compr Physiol (2014) 4(3):1121–55. doi: 10.1002/cphy.c130054
119. Fink G, Sumner BEH, Rosie R, Grace O, Quinn JP. Estrogen control of central neurotransmission: Effect on mood, mental state, and memory. Cell Mol Neurobiol (1996) 16(3):325–44. doi: 10.1007/BF02088099
120. Payne JL. The role of estrogen in mood disorders in women. Int Rev Psychiatry (Abingdon England) (2003) 15(3):280–90. doi: 10.1080/0954026031000136893
121. Wharton W, Gleason CE, Sandra O, Carlsson CM, Asthana S. Neurobiological underpinnings of the estrogen - mood relationship. Curr Psychiatry Rev Bentham Sci Publishers (2012) 8(3):247–56. doi: 10.2174/157340012800792957
122. Walf AA, Frye CA. A review and update of mechanisms of estrogen in the hippocampus and amygdala for anxiety and depression behavior. Neuropsychopharmacology (2006) 31(6):1097–111. doi: 10.1038/sj.npp.1301067
123. Rachman IM, Unnerstall JR, Pfaff DW, Cohen RS. Estrogen alters behavior and forebrain c-fos expression in ovariectomized rats subjected to the forced swim test. Proc Natl Acad Sci United States America (1998) 95(23):13941–6. doi: 10.1073/pnas.95.23.13941
124. Rocha BA, Fleischer R, Schaeffer JM, Rohrer SP, Hickey GJ. 17 Beta-estradiol-induced antidepressant-like effect in the forced swim test is absent in estrogen receptor-beta knockout (BERKO) mice. Psychopharmacology (2005) 179(3):637–43. doi: 10.1007/s00213-004-2078-1
125. Toffoletto S, Lanzenberger R, Gingnell M, Sundström-Poromaa I, Comasco E. Emotional and cognitive functional imaging of estrogen and progesterone effects in the female human brain: A systematic review. Psychoneuroendocrinol Psychoneuroendocrinol (2014) 50:28–52. doi: 10.1016/j.psyneuen.2014.07.025
126. Yonkers KA, Simoni MK. Premenstrual disorders. Am J Obstetrics Gynecology (2018) 218(1):68–74. doi: 10.1016/j.ajog.2017.05.045
127. Sundström-Poromaa I, Comasco E, Sumner R, Luders E. Progesterone – friend or foe? Front Neuroendocrinol (2020) 59:100856. doi: 10.1016/j.yfrne.2020.100856
128. Kundakovic M, Rocks D. Sex hormone fluctuation and increased female risk for depression and anxiety disorders: From clinical evidence to molecular mechanisms. Front Neuroendocrinol (2022) 66:101010. doi: 10.1016/j.yfrne.2022.101010
129. Parry BL, Berga SL, Mostofi N, Klauber MR, Resnick A. Plasma melatonin circadian rhythms during the menstrual cycle and after light therapy in premenstrual dysphoric disorder and normal control subjects. J Biol rhythms (1997) 12(1):47–64. doi: 10.1177/074873049701200107
130. Shechter A, Boivin DB. Sleep, hormones, and circadian rhythms throughout the menstrual cycle in healthy women and women with premenstrual dysphoric disorder. Int J Endocrinol (2010) 2010:1–17. doi: 10.1155/2010/259345
131. Nowak M, Markowska A, Nussdorfer GG, Tortorella C, Malendowicz LK. Evidence that endogenous vasoactive intestinal peptide (VIP) is involved in the regulation of rat pituitary-adrenocortical function: in vivo studies with a VIP antagonist. Neuropeptides Churchill Livingstone (1994) 27(5):297–303. doi: 10.1016/0143-4179(94)90111-2
132. Alexander SL, Irvine CHG, Donald RA. Dynamics of the regulation of the hypothalamo–pituitary–adrenal (HPA) axis determined using a nonsurgical method for collecting pituitary venous blood from horses. Front Neuroendocrinol (1996) 17(1):1–50. doi: 10.1006/frne.1996.0001
133. Nussdorfer GG, Malendowicz LK. Role of VIP, PACAP, and related peptides in the regulation of the hypothalamo-pituitary-adrenal axis. Peptides (1998) 19(8):1443–67. doi: 10.1016/s0196-9781(98)00102-8
134. Scott LV, Dinan TG. Vasopressin and the regulation of hypothalamic-pituitary-adrenal axis function: Implications for the pathophysiology of depression. Life Sci Pergamon (1998) 62(22):1985–98. doi: 10.1016/S0024-3205(98)00027-7
135. Weaver DR. The suprachiasmatic nucleus: A 25-year retrospective. J Biol Rhythms (1998) 13(2):100–12. doi: 10.1177/074873098128999952
136. Benarroch EE. Paraventricular nucleus, stress response, and cardiovascular disease. Clin Autonomic Res Springer (2005) 15(4):254–63. doi: 10.1007/s10286-005-0290-7
137. Ferguson AV, Latchford KJ, Samson WK. The paraventricular nucleus of the hypothalamus – a potential target for integrative treatment of autonomic dysfunction. Expert Opin Ther Targets (2008) 12(6):717–27. doi: 10.1517/14728222.12.6.717
138. Handa RJ, Zoeller RT, McGivern RF. Changes in vasoactive intestinal peptide and arginine vasopressin expression in the suprachiasmatic nucleus of the rat brain following footshock stress. Neurosci Lett (2007) 425(2):99–104. doi: 10.1016/j.neulet.2007.08.044
139. Herman JP, Cullinan WE, Ziegler DR, Tasker JG. Role of the paraventricular nucleus microenvironment in stress integration. Eur J Neurosci (2002) 16(3):381–5. doi: 10.1046/j.1460-9568.2002.02133.x
140. Wotjak CT, Kubota M, Liebsch G, Montkowski A, Holsboer F, Neumann I, et al. Release of vasopressin within the rat paraventricular nucleus in response to emotional stress: A novel mechanism of regulating adrenocorticotropic hormone secretion? J Neurosci Soc Neurosci (1996) 16(23):7725–32. doi: 10.1523/JNEUROSCI.16-23-07725.1996
141. Itoi K, Jiang Y-Q, Iwasaki Y, Watson SJ. Regulatory mechanisms of corticotropin-releasing hormone and vasopressin gene expression in the hypothalamus. J Neuroendocrinol (2004) 16(4):348–55. doi: 10.1111/j.0953-8194.2004.01172.x
142. Varadarajan S, Tajiri M, Jain R, Holt R, Ahmed Q, LeSauter J, et al. Connectome of the suprachiasmatic nucleus: new evidence of the core-shell relationship’. Soc Neurosci (2018) 5(5). doi: 10.1523/ENEURO.0205-18.2018
143. Gonchar Y. Multiple distinct subtypes of GABAergic neurons in mouse visual cortex identified by triple immunostaining. Front Neuroanat (2008) 1:3.2007(MAR). doi: 10.3389/neuro.05.003.2007
Keywords: ventral anterior homeobox 1, vasoactive intestinal peptide, arginine vasopressin, suprachiasmatic nucleus, reproduction, premenstrual disorders, circadian rhythm, cortisol
Citation: Van Loh BM, Yaw AM, Breuer JA, Jackson B, Nguyen D, Jang K, Ramos F, Ho EV, Cui LJ, Gillette DLM, Sempere LF, Gorman MR, Tonsfeldt KJ, Mellon PL and Hoffmann HM (2023) The transcription factor VAX1 in VIP neurons of the suprachiasmatic nucleus impacts circadian rhythm generation, depressive-like behavior, and the reproductive axis in a sex-specific manner in mice. Front. Endocrinol. 14:1269672. doi: 10.3389/fendo.2023.1269672
Received: 30 July 2023; Accepted: 28 November 2023;
Published: 22 December 2023.
Edited by:
Natalia Schlabritz-Lutsevich, Advanced Fertility Centers, LLC, United StatesReviewed by:
Donají Chi-Castañeda, Universidad Veracruzana, MexicoEric L Bittman, University of Massachusetts Amherst, United States
Anat Kahan, Hebrew University of Jerusalem, Israel
Copyright © 2023 Van Loh, Yaw, Breuer, Jackson, Nguyen, Jang, Ramos, Ho, Cui, Gillette, Sempere, Gorman, Tonsfeldt, Mellon and Hoffmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanne M. Hoffmann, aGFubmVAbXN1LmVkdQ==
†These authors have contributed equally to this work
 Brooke M. Van Loh
Brooke M. Van Loh Alexandra M. Yaw
Alexandra M. Yaw Joseph A. Breuer2
Joseph A. Breuer2 Krystal Jang
Krystal Jang Fabiola Ramos
Fabiola Ramos Lorenzo F. Sempere
Lorenzo F. Sempere Michael R. Gorman
Michael R. Gorman Karen J. Tonsfeldt
Karen J. Tonsfeldt Hanne M. Hoffmann
Hanne M. Hoffmann