- 1Institute of Human Genetics, Heidelberg University, Heidelberg, Germany
- 2Department of Zoology, University of Hohenheim, Stuttgart, Germany
- 3National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, United States
- 4Institute of Medical Genetics and Applied Genomics, University of Tübingen, Tübingen, Germany
- 5Division of Pediatric Endocrinology, Department of Pediatrics, Willem-Alexander Children’s Hospital, Leiden University Medical Center, Leiden, Netherlands
Very tall people attract much attention and represent a clinically and genetically heterogenous group of individuals. Identifying the genetic etiology can provide important insights into the molecular mechanisms regulating linear growth. We studied a three-generation pedigree with five isolated (non-syndromic) tall members and one individual with normal stature by whole exome sequencing; the tallest man had a height of 211 cm. Six heterozygous gene variants predicted as damaging were shared among the four genetically related tall individuals and not present in a family member with normal height. To gain insight into the putative role of these candidate genes in bone growth, we assessed the transcriptome of murine growth plate by microarray and RNA Seq. Two (Ift140, Nav2) of the six genes were well-expressed in the growth plate. Nav2 (p-value 1.91E-62) as well as Ift140 (p-value of 2.98E-06) showed significant downregulation of gene expression between the proliferative and hypertrophic zone, suggesting that these genes may be involved in the regulation of chondrocyte proliferation and/or hypertrophic differentiation. IFT140, NAV2 and SCAF11 have also significantly associated with height in GWAS studies. Pathway and network analysis indicated functional connections between IFT140, NAV2 and SCAF11 and previously associated (tall) stature genes. Knockout of the all-trans retinoic acid responsive gene, neuron navigator 2 NAV2, in Xenopus supports its functional role as a growth promotor. Collectively, our data expand the spectrum of genes with a putative role in tall stature phenotypes and, among other genes, highlight NAV2 as an interesting gene to this phenotype.
Introduction
Tall stature, by definition, comprises the upper 2.3 percent of a population (1). Pathological causes of tall stature are rare and include endocrine diseases (such as GH producing tumours) and various genetic conditions, such as chromosomal abnormalities (e.g., Klinefelter syndrome), monogenic conditions (Marfan (MIM #154700), Sotos (MIM #117550) or Weaver (MIM #277590) syndromes) or imprinting disorders (Beckwith-Wiedemann syndrome) (1, 2). In most cases, isolated (non-dysmorphic) tall stature runs in the family and gets the diagnostic label of familial or constitutional tall stature. Since no cause can be established in such cases, we suggested to use the term “idiopathic tall stature”, with two subgroups (familial and non-familial) (1).
Individuals with non-syndromic (idiopathic) tall stature that seek advice in a clinic, often remain undiagnosed. When genetic evaluations are being performed in these individuals, monogenic causes cannot be found in the great majority of cases, suggesting that oligogenic effects may underlie this phenotype. Isolated tall stature therefore remains a challenge for clinicians and geneticists alike. Several thousands of genes have been associated with height in Genome-wide association studies (GWAS) (3), but only a small number of those are considered of definitive diagnostic value for tall stature phenotypes. Examples of genes with diagnostic value comprise duplications of the SHOX and IGF1R genes and cases carrying an activating NPR2 mutation (4–8).
Here, we studied a three-generation family with extreme tall stature, and aimed to identify genetic variants shared by the tall members of this family. For this purpose, sequence variants in the tall individuals were analysed by whole exome sequencing and compared with data from a control cohort. Expression analysis in the growth plate and genome-wide association studies (GWAS) furthermore supported a causative role for three particular genes in tall stature. The strongest candidate gene was analysed in the amphibian model Xenopus.
Materials and methods
Clinical height data
Our index case, individual II.1, was extremely tall in adolescence and treated with high dose testosterone esters. At 56 years of age, we measured his height at 208.6 cm, which would translate to 211 cm (4.0 SDS) at 21 years (due to shrinking with age) (9). Yet, this may still be an underestimation in view of the treatment with testosterone esters in adolescence. Sitting height was 104 cm (sitting height/height -1.0 SDS) (10) and head circumference 60 cm (1.3 SDS) (11). He was married to a tall woman (II.2; 3.2 SDS) and had a tall son (III.1; 2.8 SDS). His sister (II.4; 3.2 SDS) married a man with normal height (II.3; 1.7 SDS) and had a daughter of normal height (III.2; -0.7 SDS). Height data were adjusted for secular trend and shrinking (9). Subjects II.1, II.4 and III.1 were examined in childhood and adolescence by experienced paediatric endocrinologists, who documented a stable height SDS and an absence of any signs of growth overproduction or dysmorphic syndrome. Further information on subjects and growth measurements, and on the calculation of adjusted height and shrinking, is given in the Supplementary Information.
Candidate genes identified in this study were also checked in all tall family members that we previously described in Weiss et al, 2021 (12).
Exome sequencing and filtering
Exome sequencing was conducted on genomic DNA from peripheral blood leucocytes. In brief, coding genomic regions were enriched using the SureSelect XT Human All Exon Kit V7 (Agilent Technologies, Santa Clara, CA, USA) for subsequent sequencing as 2x125 or 2x100 bp paired-end reads on a HiSeq2500 or NovaSeq6000 system (Illumina, San Diego, CA, USA). Generated sequences were analysed using the megSAP pipeline (https://github.com/imgag/megSAP). Clinical variant prioritization included different filtering steps and was conducted either A) in a single-mode (MAF [minor allele frequency] in 1000g, ExAC, gnomAD or in an in-house database set to <0.1% [autosomal-dominant] or <1% [autosomal-recessive]) or B) in a multi-mode (prioritizing only those variants shared by family members with tall stature), with three different sub-types: multi-sample dominant, multi-sample compound-heterozygous [both with a MAF cut-off of <0.1%] and multi-sample recessive [MAF cut-off: <1%]).
Sanger sequencing and analysis of variants
Variants of interest were confirmed by Sanger sequencing. Polymerase chain reaction (PCR) with primers indicated in Supplementary Table 1 was performed with HotStar Taq Polymerase and products analysed on agarose gels and subsequently sequenced using an ABI machine (Azenta Life Sciences).
We also checked for duplications including SHOX and IGF1R duplications and XYY in all the exome data sets and can exclude complete duplications. Partial duplications of these genes are also unlikely, but we cannot totally exclude them due to methodological limitations. We also checked for IGF-1R variants in the exome data set and re-checked the three known NPR2 mutations from the literature that are associated with tall stature and found no such variants in this family.
Variants were also checked in the “normal Non-Finnish control cohort” from gnomAD, which does not specify the height of the control individuals. Therefore, it is possible that single gene variants can be present in controls at low allele frequencies.
Network and pathway analysis
QIAGEN Ingenuity Pathway Analysis (IPA; https://digitalinsights.qiagen.com/) was applied to predict functional connections. IPA integrates selected omics data sets with mining techniques. Interpretation was in the context of protein networks that comprise protein-protein interactions and related biological functions and canonical signalling pathways.
Databases
TGP (https://browser.1000genomes.org); GnomAD (https://gnomad.broadinstitute.org/); CADD score (https://cadd.gs.washington.edu); dbSNP (https://www.ncbi.nlm.nih.gov); GTEx database (www.gtexportal.org); PROVEAN/SIFT (http://provean.jcvi.org/index.php); Polyphen2 (http://genetics.bwh.harvard.edu/pph2/); GWAS (www.ebi.ac.uk); Disease knowledge portal (https://cvd.hugeamp.org); IPA Ingenuity Systems (https://digitalinsights.qiagen.com/).
Expression analysis in growth plates
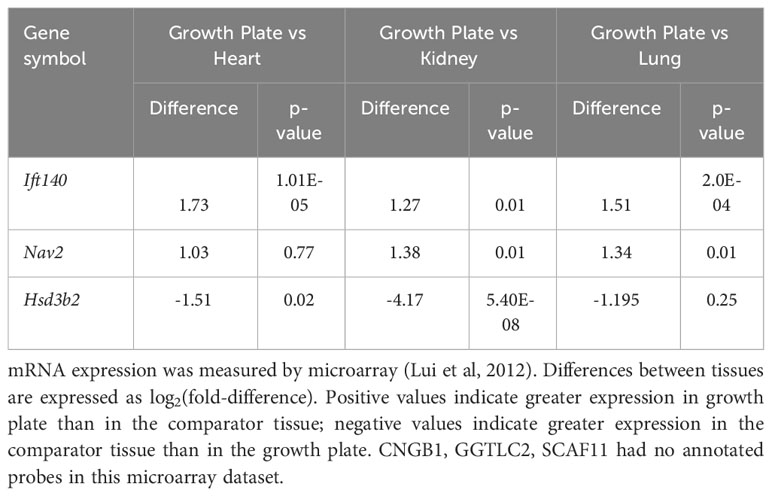
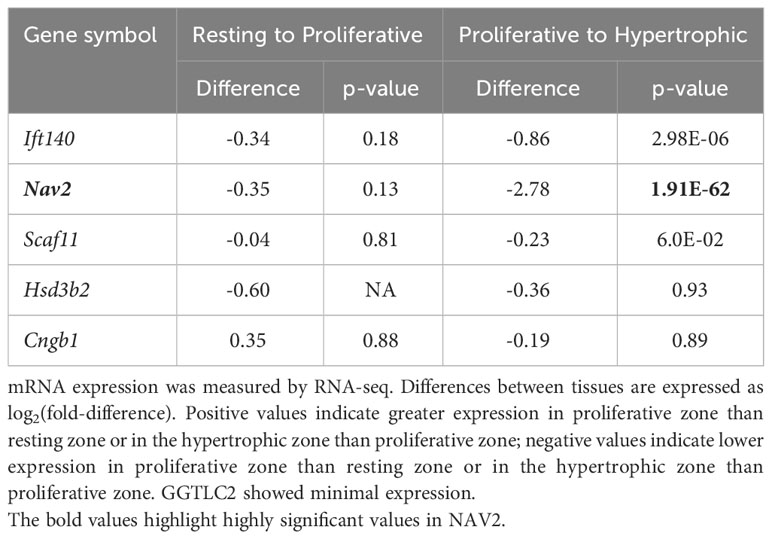
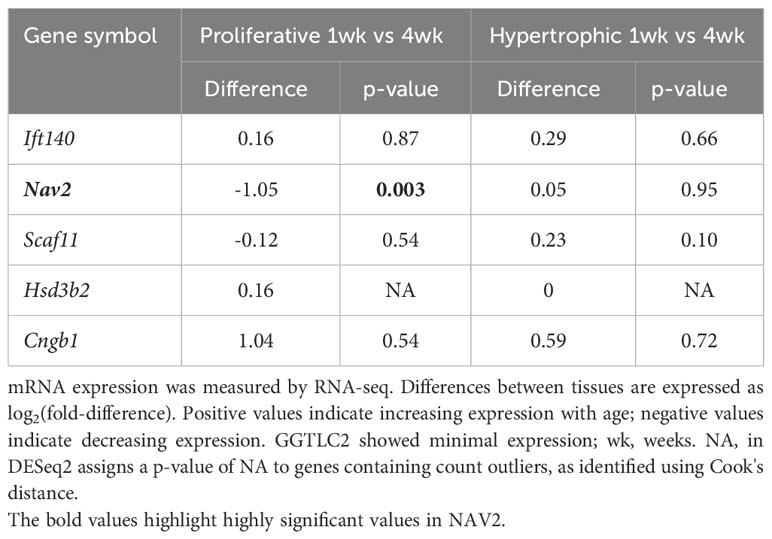
Gene expression analysis in mouse growth plate was carried out to analyse (1) growth plate specificity: comparing mRNA expression in growth plate of 1-week-old mice with expression in three different soft tissues (lung, kidney and heart) using microarray (13) (2); spatial regulation: comparing mRNA expression in resting, proliferative and hypertrophic zones in the 1-week-old mouse growth plate by RNA-Seq; and (3) temporal regulation: comparing mRNA expression at 1 and 4 weeks of age in proliferative and hypertrophic zones of mouse growth plate by RNA-Seq (14). For spatial comparison, chondrocytes from different zones of 1-week-old mouse proximal tibia growth plate were isolated by laser capture microdissection as previously described (15). The spatial analysis of gene expression in the growth plate has previously been validated by demonstrating that it correctly identifies the spatial expression pattern of known growth plate zone markers (14). Sequencing libraries were prepared using a TruSeq Stranded mRNA Prep Kit (Illumina). Sequencing was performed via a paired end 75 cycle on Illumina HiSeq 2500. RNA-Seq reads were trimmed with cutadapt (-AAGATCGGAAGAGCACACGTCTGAACTCCAGTCAAAGATCGGAAGAGCGTCGTGTAGGGAAAGA GTGT -overlap 6 -q 20 -minimum-length 25) and aligned using STAR (2 pass alignment) to mouse mm10 reference genome sequences. Log2 fold changes and p-values (adjusted for multiple comparison using the Benjamini Hochberg correction) were generated by DESeq2.
Functional studies in Xenopus
Xenopus laevis care and maintenance
Xenopus laevis was purchased from Nasco (901 Janesville Avenue, P.O. Box 901, Fort Atkinson, WI, USA). Handling, care and experimental manipulations of animals was approved by the Regional Government Stuttgart, Germany (V349/18 ZO) according to German regulations and laws (§6, article 1, sentence 2, nr. 4 of the animal protection act). Animals were kept at the appropriate conditions (pH=7.7, 20°C) in the animal facility. To induce ovulation, female frogs were injected subcutaneously with 300-700 units of human chorionic gonadotropin (Sigma). Individual embryos from one batch were randomly picked and used either as control specimens or for injections.
CRISPR/Cas9 sgRNA design and microinjections
Two sgRNA were designed using the publicly available ‘CRISPRscan’ software (16), each targeting a distinct exon in the orthologous Xenopus laevis nav2 gene (with each sgRNA targeting the L- and S-forms simultaneously). sgRNA1 was designed to mutate the conserved exon corresponding to the human exon where the R2142C missense mutation is located, potentially inducing a truncated version of the Nav2 protein (5’- GCGCCAGTACCTCTCCCACG -3’). sgRNA2 was designed to target a large 5’-located exon that is conserved in all transcript variants (corresponding to the human amino acid sequence AIPQPGA) (5’- GCAGTTGGGCTGGGGGATGG -3’). sgRNAs were transcribed with the MEGAshortscript T7 Kit from synthetic DNA oligomers and purified with the MEGAclear Transcription Clean-Up Kit (both ThermoFisher). Oligos for synthesis were:
sgRNA1_GCAGCTAATACGACTCACTATAGGGCCAGTACCTCTCCCACGGTTTTAGAGCTAGAAATAGCAAG.
sgRNA2_GCAGCTAATACGACTCACTATAGGAGTTGGGCTGGGGGATGGGTTTTAGAGCTAGAAATAGCAAG.
Embryos were injected with 1 ng Cas9 protein (PNA Bio) and 300 pg sgRNA at 1-cell stage, embryos were then cultivated at 18°C until stage 45 and fixed for length analyses. DNA was isolated from 10 mutant or 5 control embryos. Un-injected littermates served as controls. PCR-based amplification for DNA samples was according to the manufacturer’s protocol, adapted for taq polymerase conditions. PCR oligos were the following: sgRNA1(L)-F_5’-AACGGGTTTCCGGCTAACTG-3’ and R_5’- GAGGTGGCTTGGTTCATGGT-3’, sgRNA1(S)-F_5’- TTCTTAGAAGCTGGTGGTGGT-3’ and R_5’- GGGGCTGGAATAAAGACAGGA-3’, sgRNA2(L)-F_5’-TTTTCCTGGTGCGTGTGAGT-3’ and R_5’- ACCCTTCTAGGGGACTTCCG-3’, sgRNA2(S)-F_5’-CACACAAGGCAGGGAATGTG-3’ and R_5’- GGAGTCCAACCTCTCACAGC-3’. For verification of mutagenesis, amplified target sequences were Sanger sequenced using the respective PCR oligos. Sequence results were analyzed for mutagenesis rates (percentage of Indels, i.e. insertion or deletion of bases) in the targeted exons using Synthego’s ICE software [https://ice.synthego.com, (17)] and/or TIDE (18). Indel percentages for sgRNA1 were 38% (L-form) and 32% (S-form), for sgRNA2 93% (L-form) and 75% (S-form).
Analysis of tadpole body length
Embryos were mildly fixed in 4% formaldehyde in MEM buffer (MOPS/EGTA/MgSO4) for 2hrs. For tadpole body length analyses, group pictures of all embryos were taken of each experiment at stage 44-45, using the same setup and magnification. Length was quantified by measuring distances from the mouth opening/cement gland (the most anterior head structures) to the tip of the tail (excluding the fin tissue) using the segmented line tool in ImageJ (19). Pixel to millimeter conversion was calibrated using a photographed millimeter scale. To ensure consistency of the measurements, a single observer performed these measurements in both controls and treated animals.
Statistical analysis
To be able to compile the data of the three different clutches, we normalized body lengths (in mm) of each single experiment to the median of its controls and then combined relative values of all three experiments. Using this approach, we found a statistically significant overall effect with Wilcoxon rank sum test (controls were normally distributed but not the treated animals for the dataset; p-value = 0.023).
Results
Identification of novel gene variants associated with tall stature
Exome sequencing was carried out in the index case (individual II.1) and three additional tall members of a three-generation family (individuals I.1, II.4, III.1; Figures 1A, B). All individuals were healthy and without organomegaly nor dysmorphic features. Filtered variants predicted as damaging (based on Provean, SIFT and PPH2 scores) and not rated as polymorphisms (by gnomAD) and with high CADD scores, as well as shared among the related tall individuals, were first validated by Sanger sequencing and then checked for absence in a family member with normal stature (individual III.2, Figure 1A). Missense variants in the following six genes were classified as likely pathogenic: CNGB1, GGTLC2, HSD3B2, IFT140, NAV2 and SCAF11 (Table 1). Comparison to the normal Non-Finnish controls from gnomAD revealed allele frequencies of the individual variants between 10-3 to 10-5. The likelihood that a control individual carries several of these variants at the same time is minimal.
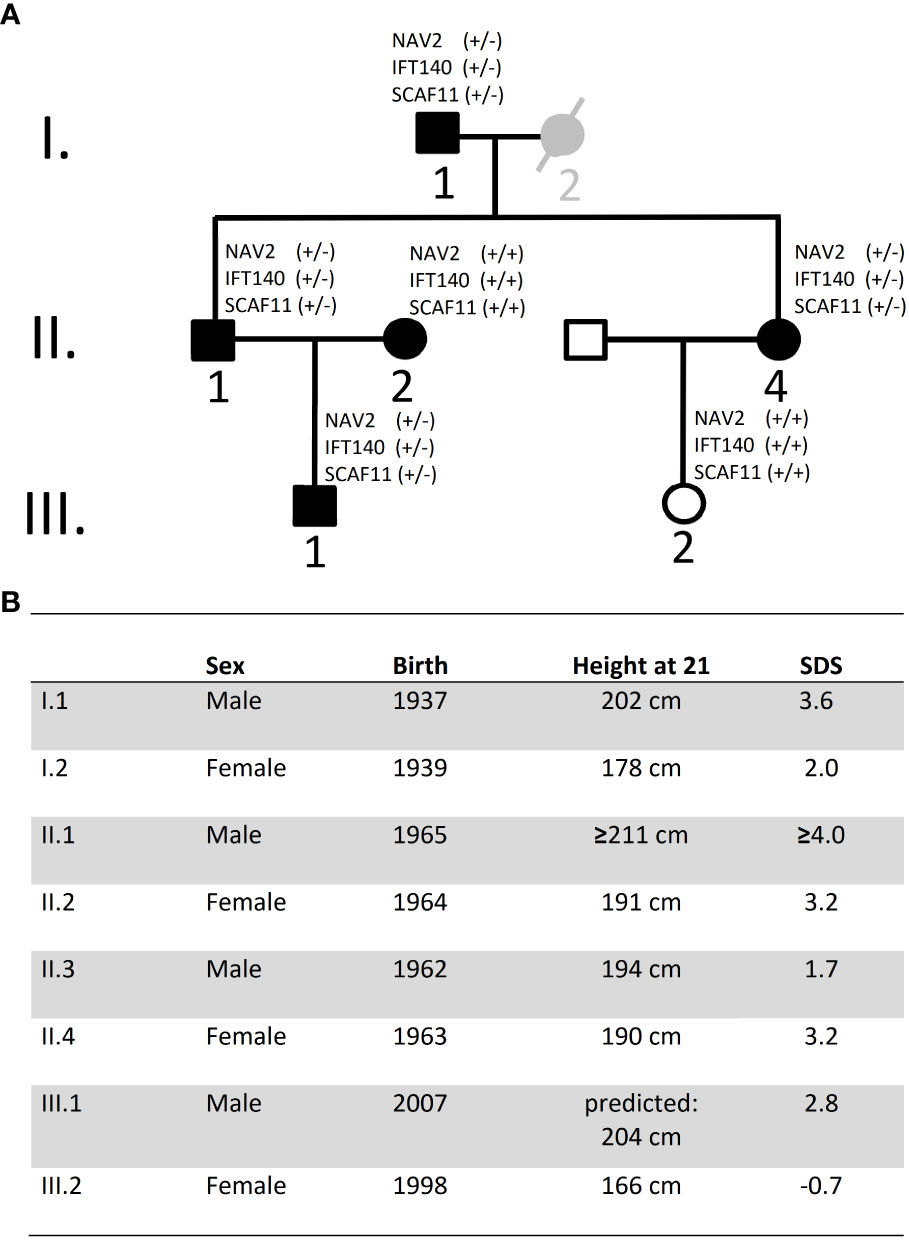
Figure 1 Pedigree and height data. (A) Pedigree of family: Black symbols indicate individuals with tall stature. Individual I.2 is considered as borderline tall stature (grey symbol). White symbols indicate normal stature. Exome sequencing was carried out on DNA from individuals I.1, II.1, II.4 and III.1. DNA was also available from I.2, II.2 and III.2 but not for exome sequencing; no DNA was available from individual II.3 (declined). (B) Data on height and year of birth of Family A Height data were adjusted for secular trend and shrinking. SDS, Standard deviation score.
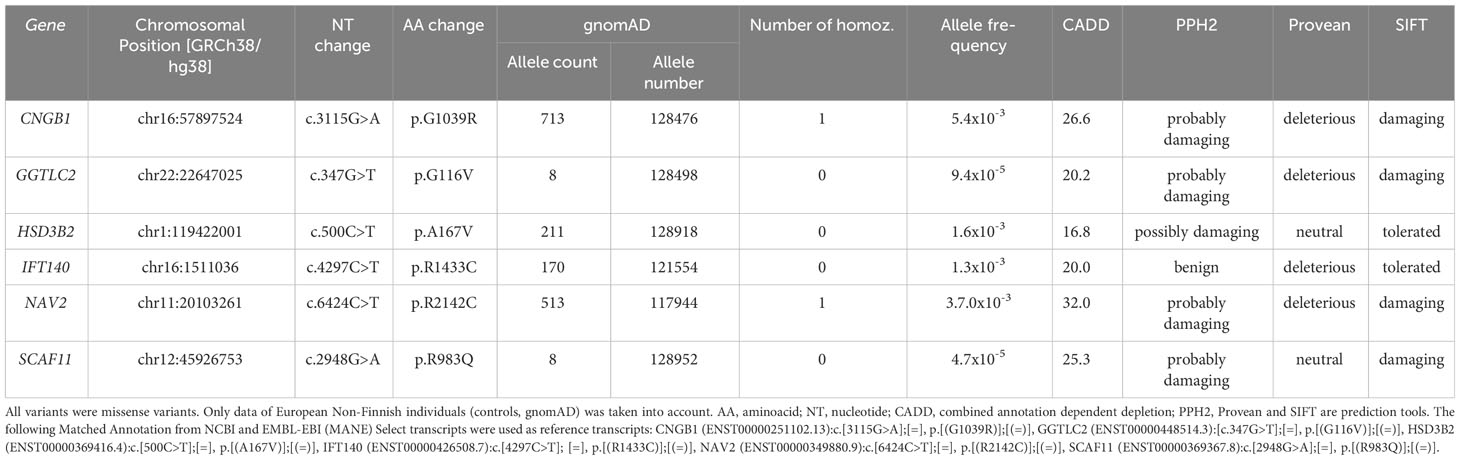
Table 1 Variants shared by tall individuals (I.1, II.1, III.1 and II.4) and not present in III.2 with normal height, considered damaging by at least one of the three prediction tools, and a CADD score above 15.
CNGB1 (cyclic nucleotide gated channel subunit beta 1) encodes an ion channel protein; GGTLC2 (gamma-glutamyltransferase light chain 2) is involved in RNA binding and spliceosomal complex assembly; HSD3B2 (3-beta-hydroxysteroid dehydrogenase) is essential for synthesis of multiple steroid hormones; IFT140 (Intraflagellar transport 140 homolog) is indispensable for intraflagellar transport in primary cilia; NAV2 (Neuron navigator 2) is an all-trans retinoic acid responsive gene; and SCAF11 (SR-related CTD associated factor 11) encodes an RNA binding protein and is a regulator of spliceosome assembly.
From all affected amino acids of the six genes, species conservation was highest for NAV2 (conserved between human and Drosophila) and lowest for HSD3B2 (Table 2). GTEx expression analysis indicated that CNGB1 is expressed in a brain-specific way, GGTL2 in a testis-specific and HSDB2 in an adrenal gland-specific manner, whereas expression of IFT140, NAV2 and SCAF11 is considered ubiquitous (Supplementary Table 2).
Association with height by GWAS
We next determined whether the identified six genes were associated with height in GWAS studies using the Disease knowledge portal (https://cvd.hugeamp.org). We found IFT140, NAV2 and SCAF11 to be associated with height with genome-wide significance (P-values between 10-11 and 10-14), while no associations were reported for CNGB1, GGTLC2 and HSD3B2 (Supplementary Table 3). Because it is unknown whether common genetic variants (SNPs) associated with height are concentrated around the same genes where rare variants are identified, we decided that further evidence was needed.
Network analysis
To further address the biological relevance of the identified gene variants, network analysis by Ingenuity Pathway Analysis (IPA) was carried out. IPA was used on the six novel candidate genes for tall stature together with 86 genes previously associated with tall stature by GWAS and literature data (Supplementary Table 4) (12, 20, 21). The newly identified six candidate genes were analysed to assess whether functional connections exist with known protein networks (Figure 2; Supplementary Figure 1). Our network analysis indicated direct protein-protein interactions between GGTLC2 and TLX3 (22), between IFT140 and CRK (23), and between SCAF11 and YWHAE (24) (Figure 2). Protein-protein interactions also exist between SCAF11 and the estrogen receptor ESR1 and the transcription factor TCF7 (25), and between HSD3B2 and ESR1 (26) (Figure 2). The all-trans retinoic acid-responsive NAV2 protein interacts with YWHAE, KRAS, HRAS and the multiprotein complex Vitamin D receptor-interacting protein mediator/TRAP (for thyroid hormone receptor-associated protein), which is highly associated with height (3.79E-28) (27–29). TRAP interacts with various transcription factors and acts as a transcriptional co-activator. CNGB1 did not interact with any of the known tall stature- associated genes.
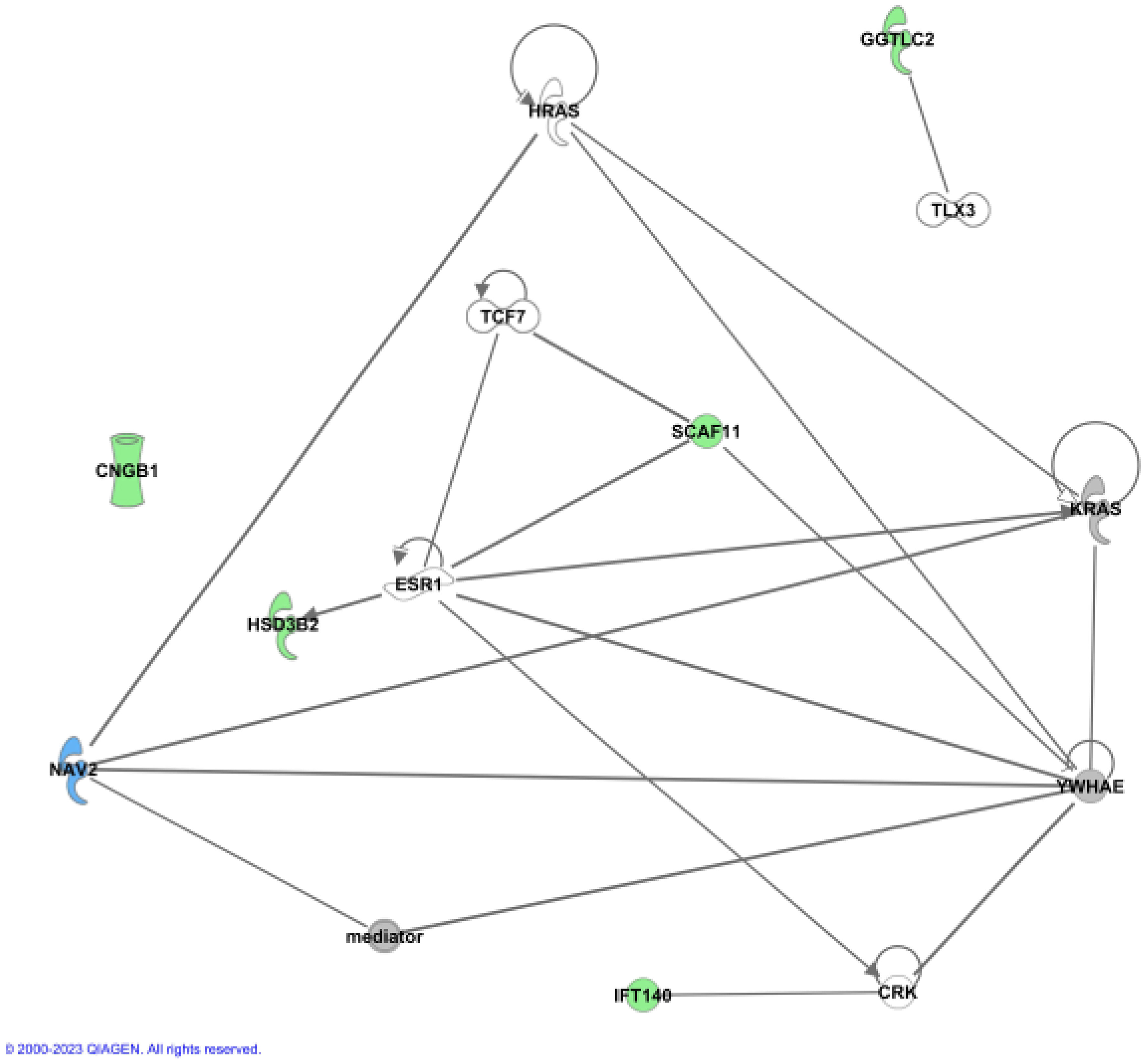
Figure 2 Ingenuity (IPA) Network analysis. The newly identified genes were used to show functional connections in context of known protein networks, based on data from the literature. No connections to known proteins could be established for CNGB1.
Together, these data provide experimentally-based evidence that five of the six candidate genes are involved in protein-protein interactions with other known genes previously associated with height.
Expression in the growth plate
To gain insight into the functional role of these candidate genes in bone growth, we assessed mRNA expression in murine growth plates. Growth plates are the cartilaginous structures residing near the ends of long bones, responsible for bone elongation and therefore linear growth. We evaluated microarray and RNA-Seq data (13, 14) in mouse growth plates (and other tissues) focusing on three comparisons. First, we compared 1-week old whole growth plate tissues with three soft tissues (lung, kidney, and heart) to compare expression of each gene of interest in the growth plate to expression in other tissues (13). Second, we compared expression in three different zones: the resting, proliferative, and hypertrophic zone in 1-week old mice by RNA-seq. We asked whether a gene showed a spatial gene expression pattern suggesting functional importance for stem cell maintenance (resting zone), cell division (proliferative zone), or terminal differentiation (hypertrophic zone). Third, we compared 1- versus 4-week old proliferative and hypertrophic zones using RNAseq (14), asking whether a gene showed a temporal decline or increase in gene expression that would suggest regulation of proliferation or differentiation, as growth slows with age.
Our analysis showed that Ift140 and Nav2 were well expressed in the growth plate, showing much higher levels compared to soft tissues analysed (Table 3A). Ift140 and Nav2, and to a lesser extent Scaf11, also showed significant downregulation of gene expression from proliferative to hypertrophic zone at 1 week. Ift140 with fold change of -0.86 and a p-value of 2.98E-06 as well as in particular, Nav2 with a fold change of -2.78 and a p-value of 1.91E-62 showed high changes between proliferative compared to hypertrophic zone, suggesting that they might be involved in chondrocyte proliferation and/or hypertrophic differentiation (Table 3B). Interestingly, Nav2 expression decreases in the proliferative zone from 1- to 4-week, further suggesting that Nav2 may act as an important regulator of chondrocyte proliferation (Table 3C).
In summary, our expression data are consistent with a functional role of both Ift140 and Nav2 in the growth plate, with Nav2 being the strongest of the two.
Knockout of the NAV2 ortholog in Xenopus supports a developmental role for body size
To functionally analyse NAV2 in an animal model, we used a CRISPR/Cas9-based knockout approach in the African clawed frog Xenopus laevis. Due to restrictions by animal protection, we could only raise tadpoles until stage 45, i.e. about 5 days after fertilization. Thus, we could test for a developmental function of the Xenopus nav2 gene during early body growth. To target the highly conserved exon harbouring the human arginine to cysteine missense mutation (R2142C), a single-guide RNA (sgRNA1) was designed. Injection of this sgRNA1 should generate a nonsense mutation or frameshift, leading to a truncated protein lacking the last six exons of the frog Nav2 protein, or alternatively, result in nonsense-mediated decay of the mRNA. We performed three injection experiments, injecting one-cell stage embryos and raised them until tadpole stage to quantify body sizes. Successful gene editing of injected specimens was evaluated by target sequence amplification and Sanger sequencing, demonstrating mutagenesis in about 35% of gene copies (for details, see Method section). Total tadpole body length of injected embryos showed increased body sizes in all three experiments (Exp1: +0.97%, Exp2: +3.36%, Exp3: +4.23%) (Figure 3). As the expected size differences were small and wild type body sizes differ naturally from batch to batch due to minor age differences, we could only compare between mutant and the corresponding wildtype. To be able to combine the data of the three different clutches, we normalized body lengths of each experiment to its controls. Statistical analysis of the combination of relative body sizes showed a significant increase after nav2 gene editing.
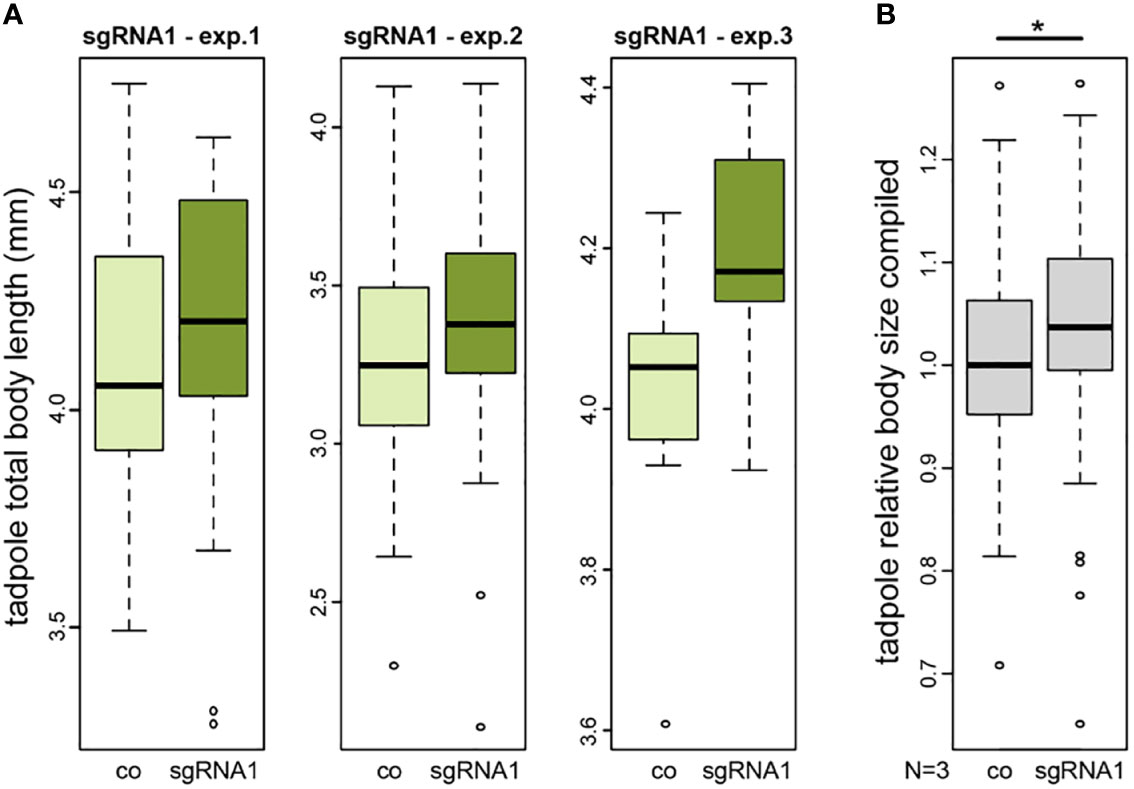
Figure 3 CRISPR/Cas-mediated knockout of nav2 causes enhanced embryonic growth. (A, B) Tadpole total body length measurements of three independent experiments. All experiments revealed a slightly increased average body size in nav2 sgRNA-injected specimens as compared to control littermates. (A) Three single experiments with sgRNA1 that specifically targets an exon orthologous to the exon harboring the human NAV2 missense mutation p.R2201C (exp.1, n=38 animals, stage 45; exp.2, n=102 animals, stage 44; exp.3, n=18 animals, stage 45); (B) Combined data of the three experiments in (A) after normalization to the mean of their controls revealed a significant increase of relative body sizes after sgRNA1 injections (p-value=0.023). Boxplot whiskers (error bars) are 1.5 * IQR +Q3 (upper), or 1.5 * IQR -Q1 (lower). co, control; exp., experiment; sgRNA, single-guide RNA; mm, millimeter.
We also designed a second sgRNA2 targeting a large 5’ exon, to cause a complete nav2 gene knockout (see Methods, for details). Sequencing confirmed a mutagenesis rate of 75-93%. Again, no early phenotype was observed in two different experiments, but when raised to tadpole stage 45, about 50% of injected specimens started to develop organ defects and edemas from stage 42 onwards (Supplementary Figures 2A–C). The massive body defects resulted in general body size reduction, potentially overruling any increase in length (Supplementary Figure 2D). Yet, when we compared the other 50%, crispants without gross defects, we also found mildly but not statistically significant increase in body sizes, as previously in the knockout approach with sgRNA1 (Exp4: +0.47%, Exp5: +1.49%) (Supplementary Figure 2E).
Taken together, our data on nav2 mutants provide independent functional verification of mildly increased body size of 1-4% in five different experiments already at early stages of tadpole development.
Two unrelated tall individuals with NAV2 variants
After we identified the candidate genes in this family, we re-examined exome sequencing data from a previously reported Dutch family with tall stature (12) and identified a rare NAV2 variant in two family members, but not in the other candidate genes of the current study. This NAV2 variant [c.G6112A, p.(E2038K)] was predicted as probably damaging by PPH2, damaging by Provean, deleterious by Sift and disease-causing by Mutation taster, with a CADD score of 26.8. The affected variant was conserved between human, mouse, chicken and Xenopus.
The variant [c.G6112A, p.(E2038K)] was identified in the two tallest family members, the index case (height 3.5 SDS) and his father (height 3.2. SDS). The variant was not carried by the sister of the index case (height 2.6 SDS) nor in the paternal uncle (height 2.9 SDS). All tall family members, however, shared damaging heterozygous variants in five different genes. This provides further evidence that damaging NAV2 variants, in conjunction with other genes, can be associated with tall stature phenotypes.
Discussion
In contrast to the success at genomic variant detection, the ability to distinguish pathogenic from benign missense variants by computational and experimental approaches is still challenging. Assessments of such variants, in particular missense variants, relies mostly on computational algorithms based on conservation. We have used a combinatorial approach to identify variants in new candidate genes for stature and classified the pathogenicity of these variants. This analysis enabled the identification of six candidate gene variants with a spectrum of likelihood of causality that were shared between all the genetically related family members with tall stature. The variants were not shared with the genetically unrelated spouse who married into this family, providing further evidence of a genetically heterogenous phenotype. Further evaluation of these six genes provided additional support on three of those genes (IFT140, NAV2 and SCAF11) with the highest likelihood of having an effect for IFT140 and in particular NAV2 by expression analysis, GWAS studies on height, network analysis and growth plate expression analysis. Damaging NAV2 variants (c.G6112A, p.E2038K) were also identified in two further unrelated tall individuals reported recently (12). Phenotype analysis of nav2 mutants in Xenopus provided further functional support.
SCAF11 is expressed in many tissues, and diseases associated with SCAF11 include Corneal Endothelial Dystrophy as well as various types of cancer. Among its related pathways is the TNFR1 Pathway. Protein-protein interactions exist between SCAF11 and the transcription factor TCF7 and between SCAF11 and YWHAE, involved in signal transduction (24, 26). SCAF11 also interacts with the estrogen receptor ESR1 which itself is very highly associated with height (5.58E-87) (25). It is therefore tempting to speculate that the detected variant p.R983Q interferes with normal interaction of SCAF11 and ESR1.
Missense variants in IFT140 have been reported to lead to “short-rib thoracic dysplasia 9 with or without polydactyly” (SRTD9, MIM #266920), also known as Mainzer-Saldino syndrome or retinitis pigmentosa 80 (RP80, MIM #617781) (30). Common signs and symptoms include a small chest and short ribs which restrict the growth and thorax expansion. The missense variant causing R1433C that we identified is monoallelic (biallelic in SRTD9) and resides in a different region at the 3´end of the IFT140 gene. In N-ethyl-N-nitrosourea (ENU) induced mutagenesis, Ift140 mutations have been shown to be associated with skeletal abnormality in mice, including craniofacial malformation, hindlimb polydactyly and forelimb poly- and oligodactyly (31). Molecular analysis of Ift140 cauli/cauli limb buds revealed altered spatial patterning and regulation of multiple growth factors, predominantly those in the cilia-dependent hedgehog signalling pathways. Indian hedgehog is an important factor in regulation within the growth plate, essential in the conversion from prehypertrophic to hypertrophic chondrocytes in the growth plate. Another interesting finding is that cilia play an important role in hedgehog signalling. Evidence that ciliary gene variants contribute to familial tall stature has been previously highlighted by our group (12). The ciliary protein IFT140 also interacts with the ciliary protein NEK2, and it is interesting that its homolog, NEK1, was previously identified as a gene underlying tall stature (12). Provided that the IFT140 variant would have a gain of function, it could increase growth by slowing down the maturation of the epiphyseal chondrocytes. We therefore consider IGF140 a reasonably strong candidate gene, but the evidence for IFT140 is not as strong as for NAV2. Two or three knockout strategies at the same time are technical difficult and possibly even lethal in Xenopus, therefore we chose the best candidate, NAV2, for the functional studies.
Like SCAF11 and IFT140, NAV2 is expressed ubiquitously. It encodes a member of a gene family involved in cellular growth and cytoskeletal dynamics. Multiple transcript variants encoding different isoforms have been found for this gene. The affected amino acid in NAV2 is conserved to Drosophila, supporting strong biological significance. Nav2 function has been addressed in mammalian nervous system development, and shown to be required for normal cranial nerve development and also blood pressure regulation. Biallelic truncating variants in NAV2 have been associated with a neurodevelopmental phenotype (32). NAV2 is a vitamin A metabolite responsive gene and numerous cellular functions, including bone cell functions, are mediated by vitamin A (33). Vitamin A (retinol, a precursor of retinoic acid) cannot only stimulate the formation of bone-resorbing osteoclasts but also inhibit their formation (34). It has been shown that retinoic acid negatively regulates longitudinal bone growth by inhibiting growth plate chondrocyte proliferation, chondrocyte hypertrophy, and matrix synthesis (35). NAV2 positively modulates inflammatory response of fibroblast-like synoviocytes through activating Wnt/β-catenin signalling pathway in rheumatoid arthritis, which is also an important pathway in chondrocyte biology (36). NAV2 also interacts directly with several protooncogenes, like KRAS and HRAS, and the multiprotein complex “mediator”, also termed TRAP, which is highly associated with height (1.8E-26) (28). Nav2 expression showed high changes between proliferative compared to hypertrophic zone, suggesting Nav2 may act as an important regulator of chondrocyte proliferation.
Collectively, our analysis revealed several novel high-confidence candidate genes associated with isolated tall stature. A growing number of genes with both a gain of function and loss of function mutations associated with opposing effects on the phenotype are known in the literature (20). Opposing skeletal phenotypes (tall and short stature) resulting from pathogenic heterozygous variants in the same gene are highly suggestive of shared pathophysiology. Evidence has been provided, for example in IGF1R, NPPC, NPR2, FGFR3, as well as FBN1 and FBN2 for a bidirectional effect on height [summarized in (20)]. While FGFR3 gain of function mutations lead to short stature (achondroplasia, ACH, MIM #100800 and hypochondroplasia, HCH, MIM #146000), FGFR3 loss of function mutations cause camptodactyly, tall stature, and hearing loss syndrome (CATSHLS, MIM #610474) (37, 38). Loss of function mutations in the natriuretic peptide receptor B (NPR2) have been reported in causing acromesomelic dysplasia 1 (AMD1, MIM #602875) (39), while gain of function mutations in the same gene result in overgrowth disorder (5, 7). Gain of function effects of large translocations and duplications have been also described for NPPC, IGF1R and SHOX (20, 40).
Gain of function mutations, however, are hard to identify from in silico predictions and require functional validation. To develop this work further, we have assessed the effect of our strongest candidate by carrying out a Nav2 knockdown in the animal model Xenopus laevis. Our data show a small (1-4%) but consistent increase in body size already at early stages of tadpole development (stage 45) in all experiments, suggesting that our observation is the first sign of the beginning process of enhanced growth during post-metaphoric stages (41). The mild increase in body size was not sufficient to obtain statistical significance in the three single experiments, but sgRNA1 data combination after normalization of body sizes resulted in a significant average increase (Figure 3). Stage 45 in tadpoles corresponds approximately to the time around the date of birth of mouse or human. As an amphibian species, this animal model has some limitations which relates to the later larval development that takes more time and incorporates a striking change of anatomy when going through metamorphosis. Its unique advantages are, however, the well characterized external embryonic development, the high numbers of embryos available from one female and the large size of its oocytes which allows for manipulations in one- to 64-cell embryos (e.g. CRISPR/Cas9-mediated loss-of-function). On top of these benefits, as a basal tetrapod species, Xenopus genetics and physiology are still close enough to humans (42, 43).
Body height in particular is conferred by gene variants acting together, supporting an oligogenic inheritance. For these reasons we anticipate and cannot rule out that further gene variants that did not quite meet our selection criteria may also contribute to the phenotype by an accumulation of small or tiny effects, as rare gene variants with stronger effect and risk factors with minor effect likely work together.
Data availability statement
The data presented in the study are deposited in the LOVD repository, variant numbers 0000921484, 0000921485, 0000921486, 0000921487, 0000921488, 0000921489, 0000921490.
Ethics statement
The studies involving humans were approved by the research protocol (P06.118) “Genetic diagnostics of very tall stature”, Leiden University Medical Center. The study was also part of the Research Program “Whole exome genetic analyses in rare hereditary diseases” at Heidelberg University. The study was conducted in accordance with the guidelines of the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article. Handling, care and experimental manipulations of animals was approved by the Regional Government Stuttgart, Germany (V349/18 ZO) according to German regulations and laws (§6, article 1, sentence 2, nr. 4 of the animal protection act).
Author contributions
BW: Methodology, Writing – review & editing, Data curation, Formal analysis, Investigation. TO: Writing – review & editing, Data curation, Formal analysis, Investigation. PV: Data curation, Formal analysis, Conceptualization, Methodology, Writing – original draft. JL: Data curation, Formal analysis, Methodology, Writing – review & editing. RR: Data curation, Writing – review & editing. SV: Data curation, Writing – review & editing. SW: Data curation, Writing – review & editing, Formal analysis, Methodology. SH: Formal analysis, Writing – review & editing. JB: Formal analysis, Writing – review & editing. JMW: Writing – review & editing, Conceptualization, Writing – original draft. GAR: Conceptualization, Writing – original draft, Writing – review & editing, Funding acquisition, Methodology, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by the Faculty of Medicine at Heidelberg University and the Intramural Program of the National Institute of Child Health and Human Development, National Institutes of Health. For the publication fee we acknowledge financial support by the Deutsche Forschungsgemeinschaft within the programme Open Access, as well as by Heidelberg University.
Acknowledgments
We thank Beate Kootz, Karin Hammann, Olga Seibel-Kelemen and Nicolas Casadei (Tübingen) for excellent diagnostic, technical and project management support. We thank Gerdine A. Kamp and Hester Havers for assistance in the collection of auxology data from the cases. We are grateful to the staff of the Leiden laboratory for diagnostic genome analysis (LDGA), supervised by Monique Losekoot, for logistic support in purifying and shipping DNA to Heidelberg. We are most grateful to the members of this tall family for their voluntary contribution to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1258313/full#supplementary-material
Supplementary Table 1 | Primer.
Supplementary Table 2 | GTEX data.
Supplementary Table 3 | GWAS on height.
Supplementary Table 4 | List of associated tall stature genes.
Supplementary Figure 1 | Ingenuity (IPA) Network analysis. Known genes from the literature (n=86; in green colour) underlying tall stature were analysed using the IPA network analysis. The newly identified genes were added to predict functional connections in the context of known protein networks.
Supplementary Figure 2 | nav2 sgRNA2 causes wild type appearance in about half of the animals and organ defects and reduced embryonic growth in the other half. (A–C) Overall appearance of representative stage 45 control specimen (A, A’), or nav2 sgRNA2-injected specimen (B, B’), illustrating organ malformations with edema formation in about 50% of nav2 sgRNA2 injections. Tadpoles are shown in lateral (A, B) and ventral view (A’, B’). (C, D) Quantification of total body length of tadpoles with organ defects (grey fraction in C), showing reduced body length when compared to control specimens (A, C). (E) Quantification of total body length of tadpoles without organ defects (dark blue fraction in C) of the two individual experiments with sgRNA2 (exp.1, n=53 animals, stage 45; exp.2, n=31 animals, stage 45) demonstrating a mild but not statistically significant increase in body size. co, control; sgRNA, single-guide RNA; mm, millimeter; OD, organ defects; st., stage; wta, wild type appearance.
References
1. Lauffer P, Kamp GA, Menke LA, Wit JM, Oostdijk W, on behalf of the Dutch Working Group on T, et al. Towards a rational and efficient diagnostic approach in children referred for tall stature and/or accelerated growth to the general paediatrician. Horm Res Paediatr (2019) 91(5):293–310. doi: 10.1159/000500810
2. Corredor B, Dattani M, Gertosio C, Bozzola M. Tall stature: A challenge for clinicians. Curr Pediatr Rev (2019) 15(1):10–21. doi: 10.2174/1573396314666181105092917
3. Yengo L, Vedantam S, Marouli E, Sidorenko J, Bartell E, Sakaue S, et al. A saturated map of common genetic variants associated with human height. Nature (2022) 610(7933):704–12. doi: 10.1038/s41586-022-05275-y
4. Benito-Sanz S, Barroso E, Heine-Suner D, Hisado-Oliva A, Romanelli V, Rosell J, et al. Clinical and molecular evaluation of SHOX/PAR1 duplications in Leri-Weill dyschondrosteosis (LWD) and idiopathic short stature (ISS). J Clin Endocrinol Metab (2011) 96(2):E404–12. doi: 10.1210/jc.2010-1689
5. Hannema SE, van Duyvenvoorde HA, Premsler T, Yang RB, Mueller TD, Gassner B, et al. An activating mutation in the kinase homology domain of the natriuretic peptide receptor-2 causes extremely tall stature without skeletal deformities. J Clin Endocrinol Metab (2013) 98(12):E1988–98. doi: 10.1210/jc.2013-2358
6. Kant SG, Kriek M, Walenkamp MJ, Hansson KB, van Rhijn A, Clayton-Smith J, et al. Tall stature and duplication of the insulin-like growth factor I receptor gene. Eur J Med Genet (2007) 50(1):1–10. doi: 10.1016/j.ejmg.2006.03.005
7. Miura K, Namba N, Fujiwara M, Ohata Y, Ishida H, Kitaoka T, et al. An overgrowth disorder associated with excessive production of cGMP due to a gain-of-function mutation of the natriuretic peptide receptor 2 gene. PloS One (2012) 7(8):e42180. doi: 10.1371/journal.pone.0042180
8. Thomas NS, Harvey JF, Bunyan DJ, Rankin J, Grigelioniene G, Bruno DL, et al. Clinical and molecular characterization of duplications encompassing the human SHOX gene reveal a variable effect on stature. Am J Med Genet A (2009) 149A(7):1407–14. doi: 10.1002/ajmg.a.32914
9. Niewenweg R, Smit ML, Walenkamp MJ, Wit JM. Adult height corrected for shrinking and secular trend. Ann Hum Biol (2003) 30(5):563–9. doi: 10.1080/0301446032000112661
10. Fredriks AM, van Buuren S, van Heel WJ, Dijkman-Neerincx RH, Verloove-Vanhorick SP, Wit JM. Nationwide age references for sitting height, leg length, and sitting height/height ratio, and their diagnostic value for disproportionate growth disorders. Arch Dis Child (2005) 90(8):807–12. doi: 10.1136/adc.2004.050799
11. Schonbeck Y, Talma H, van Dommelen P, Bakker B, Buitendijk SE, HiraSing RA, et al. The world's tallest nation has stopped growing taller: the height of Dutch children from 1955 to 2009. Pediatr Res (2013) 73(3):371–7. doi: 10.1038/pr.2012.189
12. Weiss B, Eberle B, Roeth R, de Bruin C, Lui JC, Paramasivam N, et al. Evidence that non-syndromic familial tall stature has an oligogenic origin including ciliary genes. Front Endocrinol (Lausanne) (2021) 12:660731. doi: 10.3389/fendo.2021.660731
13. Lui JC, Nilsson O, Chan Y, Palmer CD, Andrade AC, Hirschhorn JN, et al. Synthesizing genome-wide association studies and expression microarray reveals novel genes that act in the human growth plate to modulate height. Hum Mol Genet (2012) 21(23):5193–201. doi: 10.1093/hmg/dds347
14. Lui JC, Jee YH, Garrison P, Iben JR, Yue S, Ad M, et al. Differential aging of growth plate cartilage underlies differences in bone length and thus helps determine skeletal proportions. PloS Biol (2018) 16(7):e2005263. doi: 10.1371/journal.pbio.2005263
15. Kikani B, Lui JC. Laser capture microdissection of mouse growth plate cartilage. Methods Mol Biol (2021) 2245:105–19. doi: 10.1007/978-1-0716-1119-7_8
16. Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, et al. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods (2015) 12(10):982–8. doi: 10.1038/nmeth.3543
17. Conant D, Hsiau T, Rossi N, Oki J, Maures T, Waite K, et al. Inference of CRISPR edits from sanger trace data. CRISPR J (2022) 5(1):123–30. doi: 10.1089/crispr.2021.0113
18. Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res (2014) 42(22):e168. doi: 10.1093/nar/gku936
19. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods (2012) 9(7):671–5. doi: 10.1038/nmeth.2089
20. Estrada K, Froelich S, Wuster A, Bauer CR, Sterling T, Clark WT, et al. Identifying therapeutic drug targets using bidirectional effect genes. Nat Commun (2021) 12(1):2224. doi: 10.1038/s41467-021-21843-8
21. Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) (2016) 2016. doi: 10.1093/database/baw100
22. Luck K, Kim DK, Lambourne L, Spirohn K, Begg BE, Bian W, et al. A reference map of the human binary protein interactome. Nature (2020) 580(7803):402–8. doi: 10.1038/s41586-020-2188-x
23. Grossmann A, Benlasfer N, Birth P, Hegele A, Wachsmuth F, Apelt L, et al. Phospho-tyrosine dependent protein-protein interaction network. Mol Syst Biol (2015) 11(3):794. doi: 10.15252/msb.20145968
24. Nassa G, Giurato G, Salvati A, Gigantino V, Pecoraro G, Lamberti J, et al. The RNA-mediated estrogen receptor alpha interactome of hormone-dependent human breast cancer cell nuclei. Sci Data (2019) 6(1):173. doi: 10.1038/s41597-019-0179-2
25. Chen YJ, Lee MT, Yao HC, Hsiao PW, Ke FC, Hwang JJ. Crucial role of estrogen receptor-alpha interaction with transcription coregulators in follicle-stimulating hormone and transforming growth factor beta1 up-regulation of steroidogenesis in rat ovarian granulosa cells. Endocrinology (2008) 149(9):4658–68. doi: 10.1210/en.2008-0063
26. Astori A, Tingvall-Gustafsson J, Kuruvilla J, Coyaud E, Laurent EMN, Sunnerhagen M, et al. ARID1a associates with lymphoid-restricted transcription factors and has an essential role in T cell development. J Immunol (2020) 205(5):1419–32. doi: 10.4049/jimmunol.1900959
27. Cho NH, Cheveralls KC, Brunner AD, Kim K, Michaelis AC, Raghavan P, et al. OpenCell: Endogenous tagging for the cartography of human cellular organization. Science (2022) 375(6585):eabi6983. doi: 10.1126/science.abi6983
28. Ebmeier CC, Taatjes DJ. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc Natl Acad Sci USA (2010) 107(25):11283–8. doi: 10.1073/pnas.0914215107
29. Kovalski JR, Bhaduri A, Zehnder AM, Neela PH, Che Y, Wozniak GG, et al. The functional proximal proteome of oncogenic ras includes mTORC2. Mol Cell (2019) 73(4):830–44 e12. doi: 10.1016/j.molcel.2018.12.001
30. Perrault I, Saunier S, Hanein S, Filhol E, Bizet AA, Collins F, et al. Mainzer-Saldino syndrome is a ciliopathy caused by IFT140 mutations. Am J Hum Genet (2012) 90(5):864–70. doi: 10.1016/j.ajhg.2012.03.006
31. Caruana G, Farlie PG, Hart AH, Bagheri-Fam S, Wallace MJ, Dobbie MS, et al. Genome-wide ENU mutagenesis in combination with high density SNP analysis and exome sequencing provides rapid identification of novel mouse models of developmental disease. PloS One (2013) 8(3):e55429. doi: 10.1371/journal.pone.0055429
32. Accogli A, Lu S, Musante I, Scudieri P, Rosenfeld JA, Severino M, et al. Loss of neuron navigator 2 impairs brain and cerebellar development. Cerebellum (2022) 22(2):206–22. doi: 10.1007/s12311-022-01379-3
33. Muley PD, McNeill EM, Marzinke MA, Knobel KM, Barr MM, Clagett-Dame M. The atRA-responsive gene neuron navigator 2 functions in neurite outgrowth and axonal elongation. Dev Neurobiol (2008) 68(13):1441–53. doi: 10.1002/dneu.20670
34. Conaway HH, Henning P, Lerner UH. Vitamin a metabolism, action, and role in skeletal homeostasis. Endocr Rev (2013) 34(6):766–97. doi: 10.1210/er.2012-1071
35. De Luca F, Uyeda JA, Mericq V, Mancilla EE, Yanovski JA, Barnes KM, et al. Retinoic acid is a potent regulator of growth plate chondrogenesis. Endocrinology (2000) 141(1):346–53. doi: 10.1210/endo.141.1.7283
36. Wang R, Li M, Wu W, Qiu Y, Hu W, Li Z, et al. NAV2 positively modulates inflammatory response of fibroblast-like synoviocytes through activating Wnt/beta-catenin signaling pathway in rheumatoid arthritis. Clin Transl Med (2021) 11(4):e376. doi: 10.1002/ctm2376
37. Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, et al. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature (1994) 371(6494):252–4. doi: 10.1038/371252a0
38. Toydemir RM, Brassington AE, Bayrak-Toydemir P, Krakowiak PA, Jorde LB, Whitby FG, et al. A novel mutation in FGFR3 causes camptodactyly, tall stature, and hearing loss (CATSHL) syndrome. Am J Hum Genet (2006) 79(5):935–41. doi: 10.1086/508433
39. Wang SR, Jacobsen CM, Carmichael H, Edmund AB, Robinson JW, Olney RC, et al. Heterozygous mutations in natriuretic peptide receptor-B (NPR2) gene as a cause of short stature. Hum Mutat (2015) 36(4):474–81. doi: 10.1002/humu.22773
40. Marchini A, Ogata T, Rappold GA. A track record on SHOX: from basic research to complex models and therapy. Endocr Rev (2016) 37(4):417–48. doi: 10.1210/er.2016-1036
41. Zahn N, James-Zorn C, Ponferrada VG, Adams DS, Grzymkowski J, Buchholz DR, et al. Normal Table of Xenopus development: a new graphical resource. Development (2022) 149(14):1–16. doi: 10.1242/dev.200356
42. Nenni MJ, Fisher ME, James-Zorn C, Pells TJ, Ponferrada V, Chu S, et al. Xenbase: facilitating the use of Xenopus to model human disease. Front Physiol (2019) 10:154. doi: 10.3389/fphys.2019.00154
Keywords: isolated tall stature, growth plate, NAV2, all-trans retinoic acid, oligogenic inheritance, IFT140, Xenopus
Citation: Weiss B, Ott T, Vick P, Lui JC, Roeth R, Vogel S, Waldmüller S, Hoffmann S, Baron J, Wit JM and Rappold GA (2023) Identification of novel genes including NAV2 associated with isolated tall stature. Front. Endocrinol. 14:1258313. doi: 10.3389/fendo.2023.1258313
Received: 13 July 2023; Accepted: 07 November 2023;
Published: 12 December 2023.
Edited by:
Pierre De Meyts, Université Catholique de Louvain, BelgiumReviewed by:
Justin Davies, University of Southampton, United KingdomJohn Pintar, Rutgers, The State University of New Jersey, United States
Copyright © 2023 Weiss, Ott, Vick, Lui, Roeth, Vogel, Waldmüller, Hoffmann, Baron, Wit and Rappold. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gudrun A. Rappold, Z3VkcnVuLnJhcHBvbGRAbWVkLnVuaS1oZWlkZWxiZXJnLmRl
†These authors share last authorship
 Birgit Weiss1
Birgit Weiss1 Tim Ott
Tim Ott Philipp Vick
Philipp Vick Julian C. Lui
Julian C. Lui Sandra Hoffmann
Sandra Hoffmann Jeffrey Baron
Jeffrey Baron Jan M. Wit
Jan M. Wit Gudrun A. Rappold
Gudrun A. Rappold