
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol., 23 June 2023
Sec. Experimental Endocrinology
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.1190834
This article is part of the Research TopicNeuropeptide Actions in Arthropod BiologyView all 7 articles
Background: Eicosanoids are a group of the oxygenated C20 polyunsaturated fatty acids and play crucial roles in mediating various insect physiological processes. Catalytic activity of phospholipase A2 (PLA2) provides an initial substrate, arachidonic acid (AA), for subsequent eicosanoid biosynthesis.
Results: This study identified four different secretory PLA2 (As-PLA2A–As-PLA2D) genes encoded in the Asian onion moth, Acrolepiopsis sapporensis. A phylogenetic analysis indicated that As-PLA2A and As-PLA2D are clustered with Group III PLA2s while As-PLA2B and As-PLA2C are clustered with Group XII and Group X PLA2s, respectively. Expression levels of these PLA2 genes increased along with larval development, especially in the fat body. A bacterial immune challenge upregulated the basal expression levels of the four PLA2 genes, which resulted in significant increases of the PLA2 enzyme activity. The enzyme activity was susceptible to a calcium chelator or reducing agent, suggesting Ca2+ dependency and disulfide linkage required for the catalytic activities of the secretory type of PLA2s. In addition, the PLA2 activity was also susceptible to bromophenacyl bromide (BPB), a specific inhibitor to sPLA2, but not to intracellular PLA2 inhibitors. An addition of BPB to the immune challenge significantly prevented hemocyte-spreading behavior of A. sapporensis. BPB treatment also suppressed a cellular immune response measured by hemocyte nodule formation. However, the immunosuppression was significantly rescued by the AA addition. To determine the PLA2(s) responsible for the immunity, individual RNA interference (RNAi) treatments specific to each of the four PLA2s were performed. Injection of gene-specific double-stranded RNAs caused significant reductions in the transcript level in all four PLA2s. In all four PLA2s, the RNAi treatments prevented the cellular immune response even after the immune challenge.
Conclusion: This study reports four secretory PLA2s encoded in A. sapporensis and their function in mediating cellular immunity.
Polyunsaturated fatty acids (PUFAs) are preferentially associated with phospholipid (PL) components of biological membranes. PUFAs are associated with biomembrane properties of fluidity and thickness (1), while they occur in much lower proportions in neutral and energy-storage lipids such as triacylglycerols (2, 3). Phospholipase A2 (PLA2) catalyzes the hydrolysis of PUFAs from PLs (4, 5). PLA2s exert a wide range of biological actions, such as dietary PL digestion, remodeling of lipid bilayer of cellular and subcellular membranes, signal transduction, and immunity (6). These diverse functions are supported by at least 16 groups (I-XVI) of PLA2s among biological systems (7). The six main types of PLA2 enzymes are the secreted (sPLA2 including I–III, V, IX–XIV, and XVI groups), cytosolic (cPLA2, IV), calcium-independent (iPLA2, VI), platelet-activating factor acetyl hydrolase (PAF-AH, VII, VIII), also known as lipoprotein-associated PLA2 (Lp-PLA2, VIIA), lysosomal PLA2 (LPLA2, XV), and adipose PLA2 (AdPLA) (8).
PLA2 acts as the first biochemical step in eicosanoid signaling by releasing certain PUFAs from biomembrane PLs, specifically C18 or C20 PUFAs that can serve as precursors to eicosanoid signaling responsible for defending insects from invading pathogens (9, 10). Park and Kim (11) first revealed the broad biological significance of PLA2 actions in insect immunity with their discovery that eicosanoid treatments rescued beet armyworms, Spodoptera exigua, from lethal bacterial, Xenorhabdus nematophila, infections. They also document the bacterial action of inhibiting PLA2 activity required to release PUFAs from PLs for eicosanoid biosynthesis. Park et al. (12) reported that an organic extract of the bacterial broth used to grow X. nematophila inhibits PLA2s from insect, prokaryote, and vertebrate sources. Later, nine secondary metabolites containing phenyl propylene skeletons including benzylideneacetone were chemically identified from the bacterial culture broth. These compounds are competitive inhibitors of PLA2 (13).
Due to the absence or near-absence of arachidonic acid (AA) in insect PLs, it was not immediately clear that eicosanoids signal insect immunity. Mammals maintain substantial proportions (~13.1%) of AA in PLs, from which it can be hydrolyzed for eicosanoid biosynthesis (14). Insects tend to maintain very low proportions of AA (often no more than trace amounts) and high proportions of linoleic acid (LA; 18:2n-6) in cellular PLs, which may help reduce oxidative damage to cellular PLs (15). Hasan et al. (10) demonstrated AA biosynthesis from LA in S. exigua following immune challenge via sequential elongase and desaturase activities. Indeed, the bacterial infections led to increased proportions of AA from undetectable in naïve S. exigua larvae to 0.20% after bacterial infections (10, 16). The increased AA was converted into various eicosanoids, such as prostaglandins (PGs) and epoxyeicosatrienoic acids that mediate cellular and humoral immune responses (17–19). The LA metabolites including C18 lipoxins signal immune reactions to insect pathogens such as malarial parasites in mosquitoes (20) and bacterial pathogens in S. exigua (21). Although little information is available about the role of eicosanoids in insect nervous systems, eicosanoids act as intracellular signals by stimulating neuropeptides such as calcitonin gene–related peptide release in mammalian trigeminal ganglion neurons, which are cranial nerves responsible for sensation and motor functions in the face (22). These findings suggest that PLA2 is an essential step in coordinating immune and nervous signalings in insects.
The Asiatic onion moth, Acrolepiopsis sapporensis, is native to Asia, including China, Mongolia, Russia, Japan, and Korea, and its larvae infest a high-value crop, the Welsh onion (23). Despite serious concerns of massive economic losses due to A. sapporensis infestation, very little research into identifying and developing molecular targets for development of new, effective management technologies has been reported for this costly pest insect. Insect immunity is a powerful and effective defense against a wide range of entomopathogens, which may be an effective target for novel A. sapporensis control measures. Because PLA2 is the first biochemical step in eicosanoid signaling targeting the gene encoding this enzyme, it would be a workable strategy to develop novel control agent(s) against A. sapporensis. Here, we suggest that application of immunosuppressants by targeting PLA2 will enhance the virulence of the entomopathogens as biological control agents, as demonstrated in lepidopteran (24), coleopteran (25), and dipteran (26) pests. In this regard, PLA2 would be reframed as a molecular target for development of novel insecticides to synergize the microbial insecticides in A. sapporensis (27). Here, we report on identification of four previously unknown secretory PLA2s of A. sapporensis and their physiological functions in mediating cellular immune responses.
A field population of A. sapporensis, used in all experiments reported in this paper, was collected from a Welsh onion (Allium fistulasum L.) field (Suanbo, Korea) and reared under laboratory conditions at 27 ± 2°C, photoperiod 16:8 h (L:D) with 65 ± 5% relative humidity. Adults were kept in an acrylic cage and fed ad libitum with 10% sucrose solution for their diet. Fresh onion leaves were placed in the cage for egg laying and replaced daily. The egg-laid leaves were placed in a breeding dish (90 mm diameter and 15 mm height) for larval hatching. Larvae underwent five instars (L1–L5) over 16 ± 2 days in the laboratory conditions. Resulting pupae were kept at 85% relative humidity for emergence.
Arachidonic acid (AA: 5,8,11,14-eicosatetraenoic acid), bromoenol lactone (BEL), and methyl arachidonyl fluorophosphate (MAFP) were purchased from Sigma-Aldrich Korea (Seoul, Korea) and dissolved in dimethyl sulfoxide (DMSO). Bromophenacyl bromide (BPB) was purchased from Tokyo Chemical (Tokyo, Japan). sPLA2 enzyme assay kits were purchased from Cayman Chemical (Ann Arbor, MI, USA). For RNA preparation, 1 ml of diethyl pyrocarbonate (DEPC) was combined with 1 L of deionized distilled water to prepare DEPC water and incubated for 12 h at 37°C. The water was then autoclaved twice and held at room temperature (RTP) until use. Phosphate-buffered saline (PBS) was prepared with 100 mM phosphate with 0.7% NaCl and adjusted to pH 7.4 with 1 N NaOH.
Four PLA2 genes were obtained from an annotated A. sapporensis transcriptome (GenBank accession number of PRJNA834156). They were retrieved with accession numbers OQ625511 (As-PLA2A), OQ625512 (As-PLA2B), OQ625513 (As-PLA2C), and OQ625514 (As-PLA2D). MEGA6 (28) was used to construct phylogenetic trees using the neighbor-joining method and the Poisson correction model. Bootstrap values at each branch were calculated with 1,000 repeats. InterPro (http://www.ebi.ac.uk/interpro/) was used to predict the protein domain, and the SigmalP 5.0 server (http://services.healthtech.dtu.dk/service.SignalP-5.0) was performed to detect the N-terminal signal peptide. Sequence alignment was performed using the MegAlign program (DNAStar 7.0, Lasergene, Madison, WI, USA) to get consensus residues among different PLA2 genes.
Total RNAs were extracted from developmental stages and larval tissues indicated in Results using a Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. RT-premix (Intron Biotechnology, Seoul, Korea) with the oligo-dT primer was used to synthesize complementary DNA (cDNA) from the extracted RNA (1 µg per sample) and quantified using a spectrophotometer (NanoDrop, Thermo Fisher Scientific, Wilmington, DE, USA). RT-PCR was performed using DNA Taq polymerase (GeneAll, Seoul, Korea) with an initial heat treatment at 95°C for 2 min, followed by 35 cycles of DNA denaturation at 95°C for 30 s, annealing for 1 min at varied temperatures using different gene-specific primers (Table S1), and chain extension at 72°C for 1 min. PCR reaction samples (25 µl) consisted of 2.5 µl of 10× Taq buffer, 2.5 µl of 10× dNTP, 1 µl of Taq DNA polymerase, each 1 µl of forward and reverse primers (10 pmol), 1 µl of template (cDNA), and 16 µl of deionized distilled water. The PCR result was validated using 1% agarose gel electrophoresis. RT-qPCR was performed using a real-time PCR system (Step One Plus Real-Time PCR System, Applied Biosystems, Singapore) in accordance with the instructions provided by (29). The reaction mixture (20 µl) included 7 µl of deionized distilled water, 10 µl of Power SYBR Green PCR Master Mix, 1 µl of cDNA template (70 ng), and each 1 µl of forward and reverse primers (Table S1). After activating Hot-start Taq DNA polymerase at 94°C for 5 min, the reaction was amplified with 40 cycles of denaturation at 94°C for 30 s, annealing at different temperatures (Table S1) for 1 min, and extension at 72°C for 30 s. A ribosomal gene, RL32 was used to validate the cDNA integrity. Each treatment was replicated three times by preparing individual sample preparation. A comparative CT method was used to calculate the relative expression levels (30).
Escherichia coli Top10 was purchased from Invitrogen and grown overnight at 37°C in Luria–Bertani (LB) medium (BD, Franklin Lakes, NJ, USA). Then bacteria were heat-killed at 95°C for 10 min prior to immune challenge. For the immune challenge, E. coli (1.64 × 106 cells/larva) was injected with a micro-capillary using a shutter CO2-based pico-pump injector (PV830, World Precision Instrument, Sarasota, FL, USA) under a stereomicroscope (SZX-ILLK200, Olympus, Tokyo, Japan). To determine inhibitory effects, selected chemicals or prepared dsRNA was injected along with E. coli. To rescue the immunosuppression, AA was coinjected with E. coli.
A T7 promoter sequence was added to four gene-specific PLA2 primers at the 5’ end. Using these primers, the four As-PLA2 genes were amplified as just described. After amplifying all genes, the PCR products were used to prepare double-stranded RNA (dsRNA) by the Megascript RNAi Kit (Ambion, Austin, TX, USA) according to their instructions. A green fluorescence protein (GFP) was used as control dsRNA (dsCON). Before injection, dsRNA was combined with a transfection reagent (Metafectene Pro, Biontex, Planegg, Germany) in a 1:1 ratio. After that, 1 µg of dsRNA was injected into L5 larva as described above. RNAi efficiency was determined by RT-qPCR at time points indicated in Results. Each treatment was replicated three times with independent RNA preparations.
L5 larvae were immune challenged as described. Sterilized PBS was injected as a control treatment, represented as a naïve treatment. After removing the intestine, the remaining whole body was collected from naïve or challenged insects at time points indicated in Results into 1X PBS containing a protease inhibitor cocktail (Sigma-Aldrich Korea) and phenylmethylsulfonyl fluoride (Thermo Fisher Scientific Korea, Seoul, Korea). The samples were crushed completely and centrifuged at 500 × g for 5 min at 4°C. After centrifugation, the supernatants were transferred to new tubes. The collected supernatants were mixed with 4× denatured sample buffer [300 mM Tris-HCL, 600 mM dithiothreitol, 12% sodium dodecyl sulfate (SDS), 0.6% bromophenol blue, and 60% glycerol]. After a 5 min heat treatment at 95°C, the extracted proteins were separated using 10% SDS-PAGE at a constant 100 V. The nitrocellulose membrane (BioRad, Hercules, CA, USA) was used to transfer the separated proteins from the gel with a transfer buffer (25 mM Tris, 190 mM glycine, 20% methanol, pH 8.5) at 100 V for 50 min at 4°C. After the membrane was washed with PBS, the background in the membrane was blocked with 5% skim milk in PBS for 1 h at RTP. After discarding the blocking solution, the membrane was again incubated with a primary antibody (diluted by 1:5,000 with 5% skim milk) raised against sPLA2 of S. exigua (Se-sPLA2) (31) for 3 h at RTP. After washing three times with PBS, the membrane was incubated with anti-rabbit Immunoglobulin G (IgG)–alkaline phosphatase secondary antibody (Sigma-Aldrich Korea) (dilution by 1:30,000 with 5% skim milk) for 1 h at RTP. The membrane was washed three times with PBS. Finally, nitrocellulose membrane blots were incubated for approximately 2 min with nitro blue tetrazolium/5-bromo-4-chloro-3’-indolyphosphate p-toluidine salt as a substrate to reveal alkaline phosphatase activity.
A commercial PLA2 kit (Cayman Chemical, Ann Arbor, MI, USA) was used to determine the enzyme activities as detailed by Vatanparast and Kim (32). The samples were collected as just mentioned. Reaction mixtures (225 µl) were composed of 10 µl of each sample, 10 µl of Ellman’s reagent, 5 µl of assay buffer, and 200 µl of substrate (sPLA2). The same volume of reaction mixture consisting of 10 µl 5,5’-dithiobis 2-nitrobenzoic (DTNB), 15 µl assay buffer, and 200 µl substrate was used as a negative control. The amount of enzyme activity was measured using a spectrofluorometer (VICTOR multilabel plate reader, PerkinElmer, Waltham, MA, USA). Changes in absorbance at 405 nm of the reaction product were measured and plotted to obtain the slope of a linear portion of the curve. Enzyme activity was calculated with an extinction coefficient of DTNB (14,150 M-1cm-1). Specific enzyme activity was calculated by normalizing the spectrophotometer readings to the total amount of the protein in the samples (33). Each treatment was replicated thrice with individual sample preparations.
L5 larvae were used for assessing the hemocyte-spreading assay. The larvae were immune-challenged as just described. To inhibit the immune response, BPB (1 µg per larva) was coinjected. At 2 and 8 h post-infection (pi), 10 µl of hemolymph from each of 15 insects was collected on a glass slide, directly fixed with 4% paraformaldehyde dissolved in PBS and incubated for 10 min at RTP under darkness. After washing three times with 1X PBS, cells were permeabilized with 0.2% Triton-X in PBS for 2 min. Again, the cells were rinsed three times with PBS and incubated with 5% skim milk for 10 min at RTP. After washing with PBS, hemocytes were incubated with 1.2% fluorescein isothiocyanate (FITC)–tagged phalloidin for 1 h at RTP. After washing three times, the cells were incubated with 4´,6-diamidino-2-phenylindole (DAPI, 1 mg/ml). After 10 min incubation, cells were washed three times with PBS and observed under a fluorescence microscope (DM2500, Leica, Wetzlar, Germany) at ×200 magnification. Each treatment was replicated three times.
Overnight-cultured E. coli was used in the nodulation formation assay. Before injection, the live bacteria were killed by incubating at 95°C for 5 min and the bacterial cell number was counted using a hemocytometer (Neubauer improved bright line, Superior Marienfeld, Lauda-Konigshofen, Germany) under a phase contrast microscope (BX41, Olympus, Tokyo, Japan). L5 larvae were injected with 1 µl of E. coli (1.64 × 106 cells/larva) along with 1 µg of BPB or dsRNA using micro-capillaries. For the control treatment, the larvae were injected with E. coli without adding any inhibitors or with dsCON. Then, the injected larvae were reared under laboratory conditions for 8 h. Finally, the nodules were counted by dissecting the larvae under a stereomicroscope (Stemi SV 11, Zeiss, Jena, Germany) at ×50 magnification.
All experiments in this study were performed with three replicates, and a Sigma plot was used to plot the results by mean and standard deviation. Means and variances of treatments were compared by a least squared difference (LSD) test of one-way analysis of variance (ANOVA) using PROC GLM of the SAS (34) program and discriminated at Type I error = 0.05.
Transcriptome analysis annotated four PLA2 genes: As-PLA2A, As-PLA2B, As-PLA2C, and As-PLA2D (Figure 1). A phylogenetic analysis with 11 known PLA2s groups (Figure 1A) included four PLA2 types: sPLA2, iPLA2, cPLA2, and PAF-AH. In this classification, the four A. sapporensis PLA2s were clustered with other sPLA2s. Specifically, As-PLA2A and As-PLA2D were assigned into Group III, As-PLA2B into Group XII and As-PLA2C into Group X. These four PLA2s were then compared in their functional domains using a proteomics bioinformatics tool (www.expasy.com). Each PLA2 has a separate signal peptide representing secretory proteins (Figure 1B). They also share calcium binding sites with a conserved glycine residue (Figure S1) indicating their dependence on Ca2+. In the catalytic domains, a histidine-aspartic acid (‘HD’, Figure S1) dyad occurred in all four PLA2s. Finally, more than 12 cysteine residues were encoded in the four PLA2s.
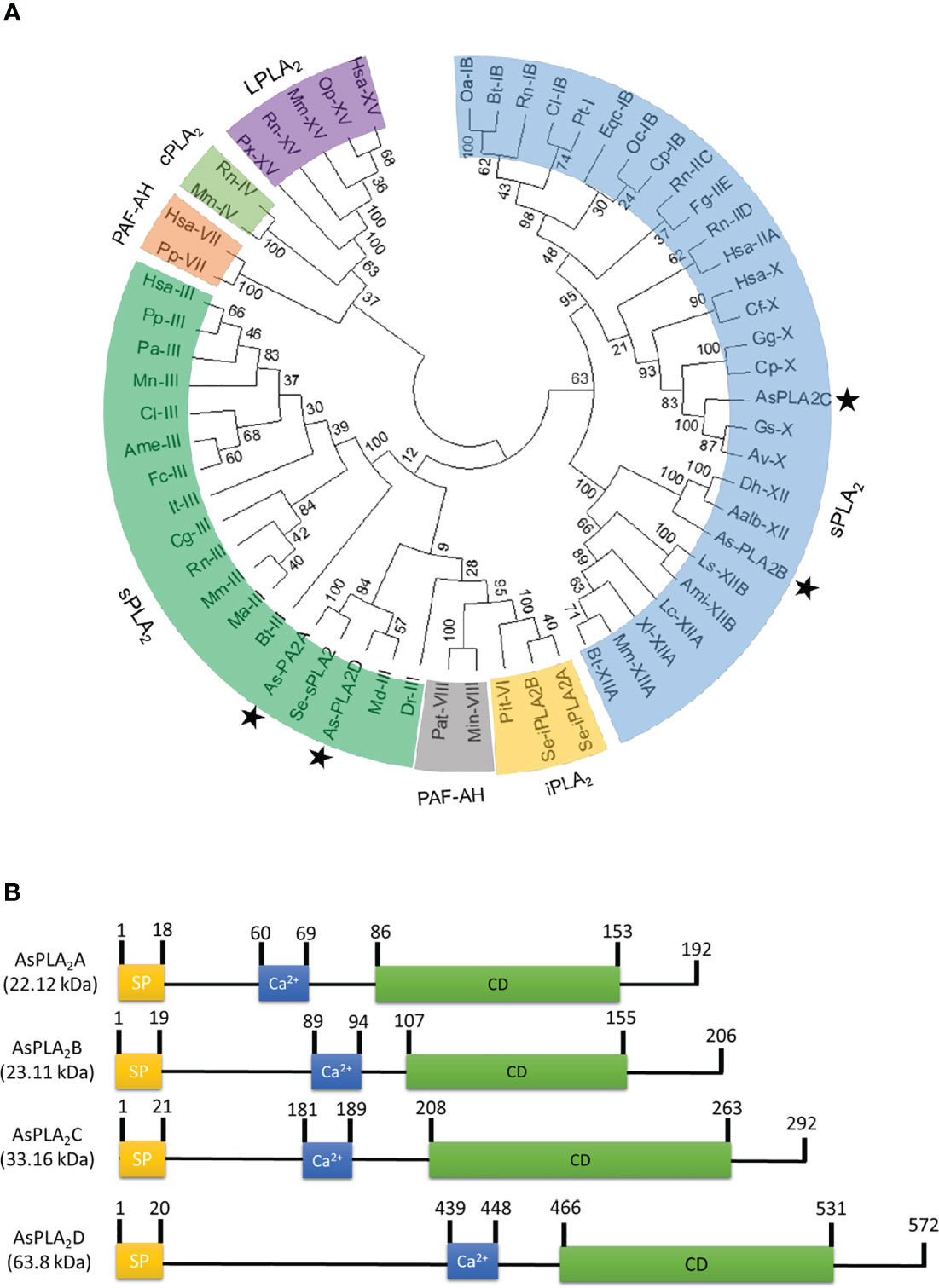
Figure 1 Identification of four phospholipase A2 (PLA2) genes (asterisks) from a transcriptome of A. sapporensis. (A) Phylogenetic tree of different PLA2s including four As-PLA2s using the MEGA6 program. Bootstrap values were obtained with 1,000 repetitions to support branching and clustering. Different types of PLA2s include secretory phospholipase A2 (sPLA2), calcium-independent phospholipase A2 (iPLA2), cytosolic phospholipase A2 (cPLA2), lysosomal phospholipase A2 (LPLA2), and platelet-activating factor acetyl hydrolase (PAF-AH). (B) Comparison of functional domains of four A. sapporensis sPLA2 (As-sPLA2) genes. ‘SP’, ‘Ca2+’, and ‘CD’ stand for signal peptide, calcium-binding site, and catalytic domain. InterPro (http://www.ebi.ac.uk/interpro/), Expasy (www.expasy.com) and SigmalP 5.0 server (http://services.healthtech.dtu.dk/service.SignalP-5.0) were used to predict domain and signal peptides. Molecular weights of the four As-PLA2s were predicted using the EditSeq program of DNAStar 7.0. The numbers above domains indicate the locations of amino acids.
All four PLA2s were expressed through development from larva to adult stages (Figure 2A). Their expression levels increased with larval development and showed the highest expressions at the final larval instar for As-PLA2A and As-PLA2B and at the penultimate instar for As-PLA2C and As-PLA2D. In the larval stage, the four PLA2s were expressed in all the tested tissues (hemocyte, fat body, and gut) with high expression at the fat body (Figure 2B). Among the four PLA2s, As-PLA2A was highly expressed in all tissues.
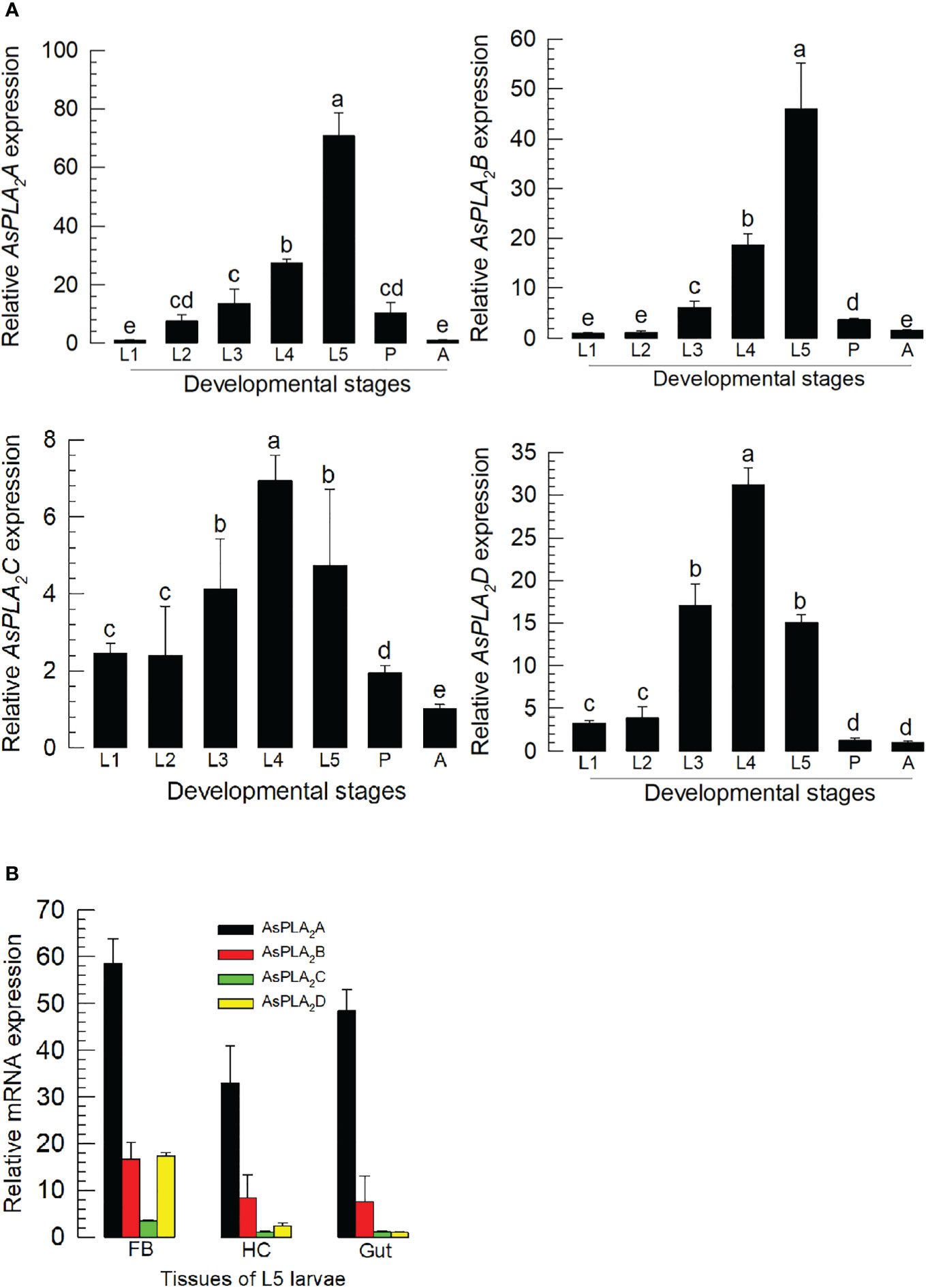
Figure 2 Expression profile of PLA2s in A. sapporensis. (A) Expression pattern of As-PLA2A, As-PLA2B, As-PLA2C, and As-PLA2D in whole body samples of different developmental stages: first to fifth instar larva (‘L1–L5’), pupa (‘P’), and adult (‘A’). Different letters above standard deviation bars indicate significant differences among means at Type I error = 0.05 [least squared difference (LSD) test]. (B) PLA2 expression analysis in different tissues of L5 larvae including the fat body (‘FB’), hemocyte (‘HC’), and gut. A ribosomal gene, RL32, was used as a reference gene. Each treatment was replicated three times with independent sample preparations.
PLA2 activity was monitored across developmental stages (Figure 3A). As expected, larval stage showed higher specific enzyme activity compared to pupal and adult stages. The larval fat body also expressed the highest enzyme activity compared to hemocytes, gut, and epidermis.
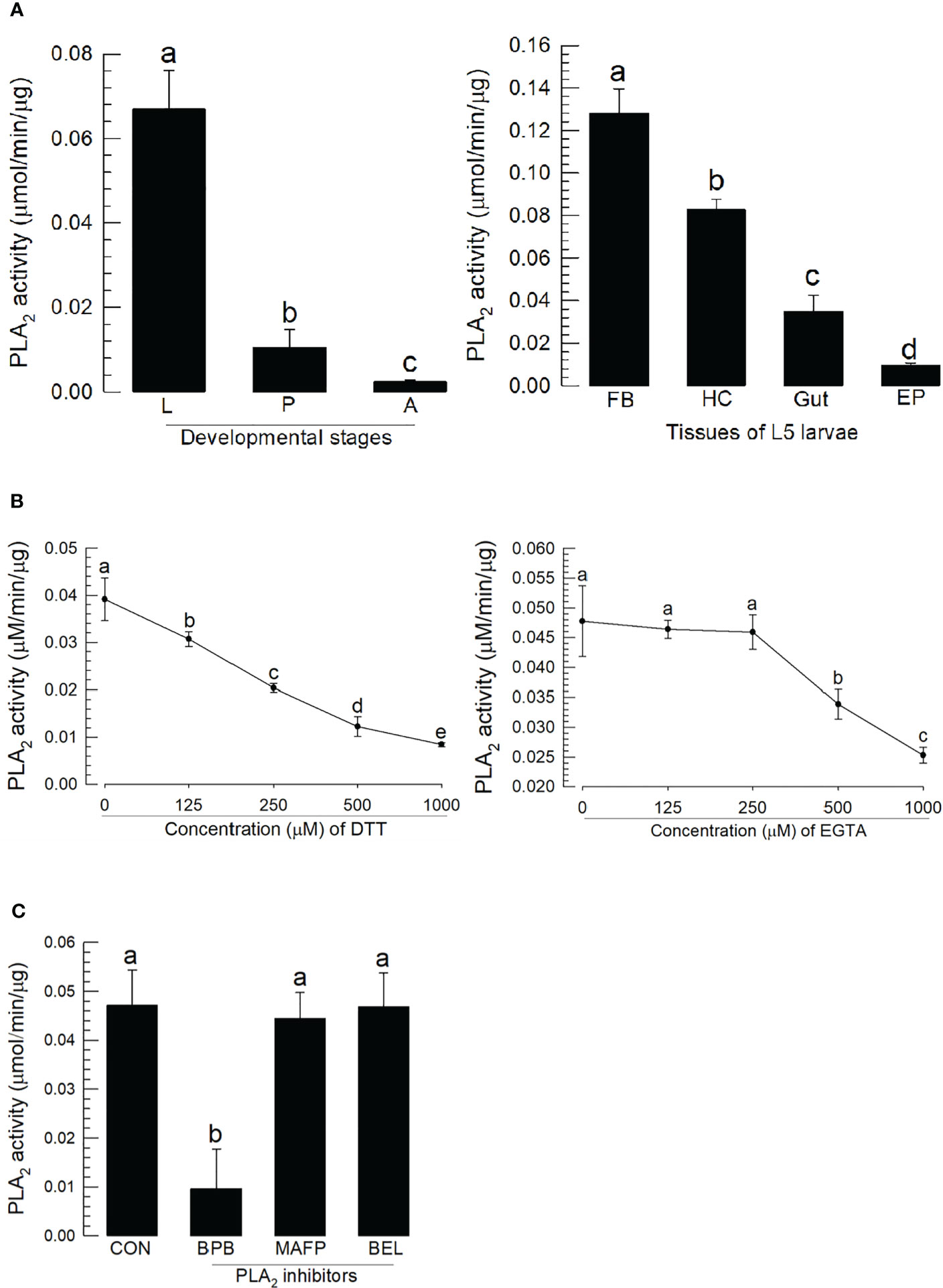
Figure 3 Characterization of A. sapporensis PLA2 enzyme activity. (A) Differential specific enzyme activities in developmental stages and larval tissues: larva (‘L’), pupa (‘P’), adult (‘A’), fat body (‘FB’), hemocyte (‘HC’), gut, and epidermis (‘EP’). (B) Inhibitory effects of dithiothreitol (DTT) and ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) on the PLA2 enzyme activity. The enzyme was extracted from the whole body of L5 larvae after removing the gut. (C) Influence of different PLA2 inhibitors [bromophenacyl bromide (BPB), bromoenol lactone (BEL), and methyl arachidonyl fluorophosphate] on the PLA2 enzyme activity. Different letters above standard deviation bars indicate significant differences among means at Type I error = 0.05 (LSD test).
We monitored the disulfide linkage and its influence on the catalytic activity, by running enzyme assays in the presence of dithiothreitol, which inhibited the enzyme activity in a dose-dependent manner (Figure 3B). We monitored Ca2+ dependence, by running assays in the presence of ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), which led to a dose-dependent inhibition of the activity. Three inhibitors specific to different types of PLA2s were applied to the reaction mixture (Figure 3C). BPB, a sPLA2-specific inhibitor, inhibited the PLA2 activity as expected; however, reactions in the presence of BEL (a iPLA2-specific inhibitor) or MAFP (a cPLA2-specific inhibitor) did not influence enzyme activity.
Challenge by injecting a non-pathogenic bacterium, E. coli, led to more than twofold increases in expression of the PLA2 genes in fat body, hemocyte, and gut (Figure 4A). The gene encoding As-PLA2C was the most highly induced in fat body by almost 13-folds while other PLA2s showed approximately 5-fold induction. This induction of the gene expressions was further supported by the increase of the enzyme activity, in which there was an approximate sixfold increase in PLA2 activity over the following 8~10 h (Figure 4B). As early as 1 h postinjection (pi), PLA2 activity was highly induced and kept the increase mode up to 8 h pi. The induced enzyme activity was decreased after 10 h pi. To visualize the PLA2s of A. sapporensis, a Western blot analysis was conducted using a polyclonal antibody raised against Se-sPLA2 of another lepidopteran. S. exigua (Figure 4C). As-PLA2A and As-PLA2D share sequence homologies (38.4%–53.6%) with that of Se-sPLA2, while As-PLA2B and As-PLA2C exhibit relatively low homologies (9.8%–11.0%) (Figure S2). The Western analysis showed four bands at approximately 25 kDa (‘P1’), 38 kDa (‘P2’), 55 kDa (‘P3’), and 90 kDa (‘P4’). After the immune challenge, an additional band (‘P5’) were detected at approximately 65 kDa.
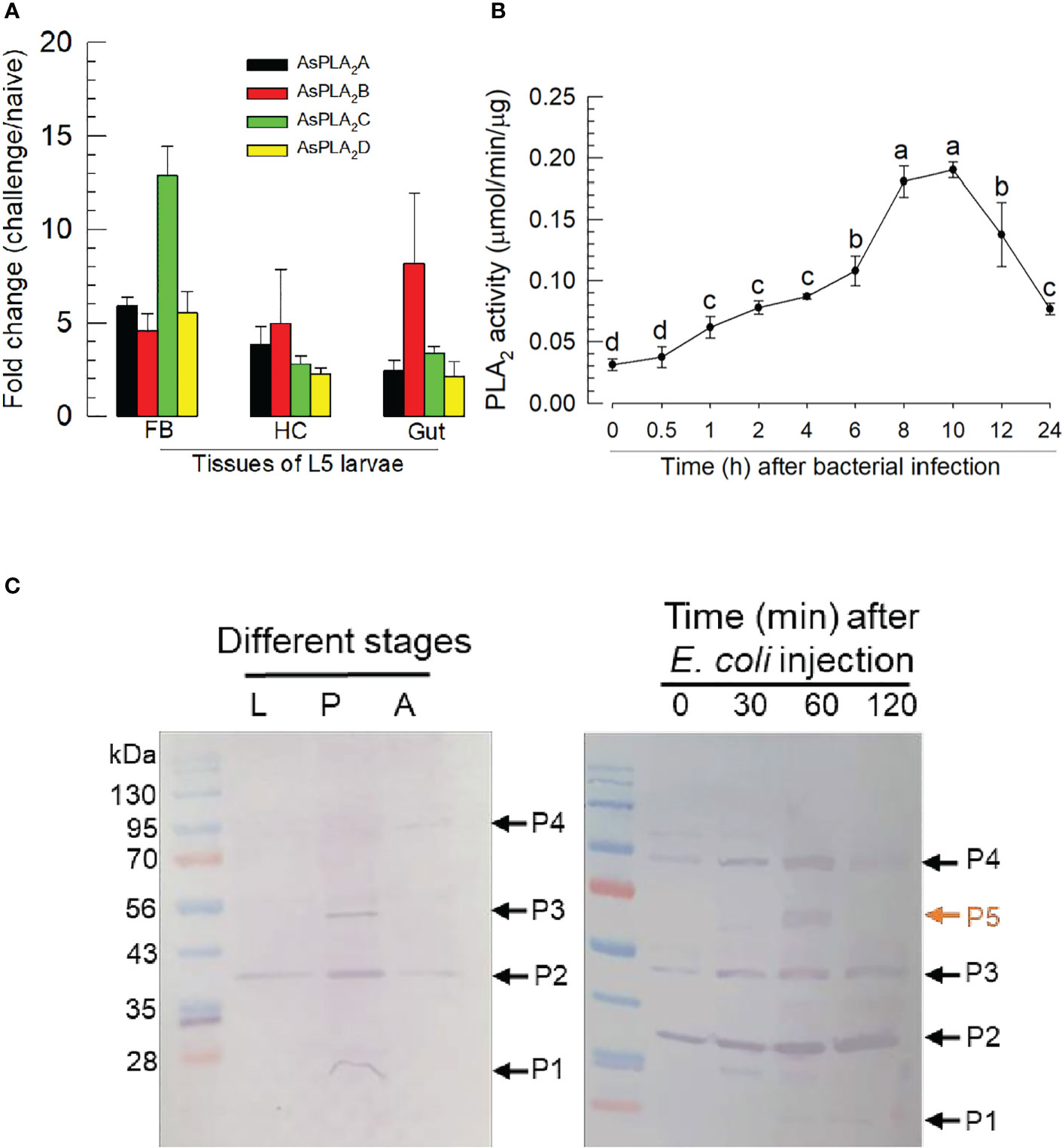
Figure 4 Inducible PLA2 enzyme activity in A. sapporensis larvae after immune challenge. (A) Inducible expressions of four As-PLA2 genes after the immune challenge in different larval tissues: fat body (‘FB’), hemocyte (‘HC’), and gut. (B) Induced PLA2 activities after the immune challenge. Enzyme was extracted from the whole body of L5 larvae after removing the gut. (C) Western analysis of As-PLA2s in naïve and challenged larvae. Three developmental stages (left panel) used naïve insects. After the immune challenge (right panel), the gut-removed whole body to avoid any other PLA2s in the gut lumen was used for the immunoblotting analysis with a polyclonal antibody raised against S. exigua sPLA2. Here, ‘0 min’ sample represents just before the immune challenge. Each lane contained 50 μg proteins. ‘P1–P5’ indicate five protein bands detected by the Western blot analysis. Each qPCR or enzyme assay was replicated three times with independent sample preparations. Different letters above standard deviation bars indicate significant differences among means at Type I error = 0.05 (LSD test).
The upregulations of PLA2 gene expression and enzyme activity upon the bacterial infection, suggesting its physiological role in immune responses. Hemocytes were spread after the immune challenge as early as 2 h pi and kept the spread behavior 8 h pi (Figure 5A). In contrast, BPB injection along with the immune challenge suppressed the hemocyte behavior. As this hemocyte behavior is required for cellular immune responses, we assessed nodule formation of the hemocytes against the immune challenge. In control larvae, approximately 25 nodules were formed after the bacterial injection at 8 h pi (Figure 5B). However, the BPB addition significantly prevented this nodule formation in a dose-dependent manner. Interestingly, the addition of arachidonic acid (a catalytic product of PLA2) significantly rescued the inhibitory activity of BPB.
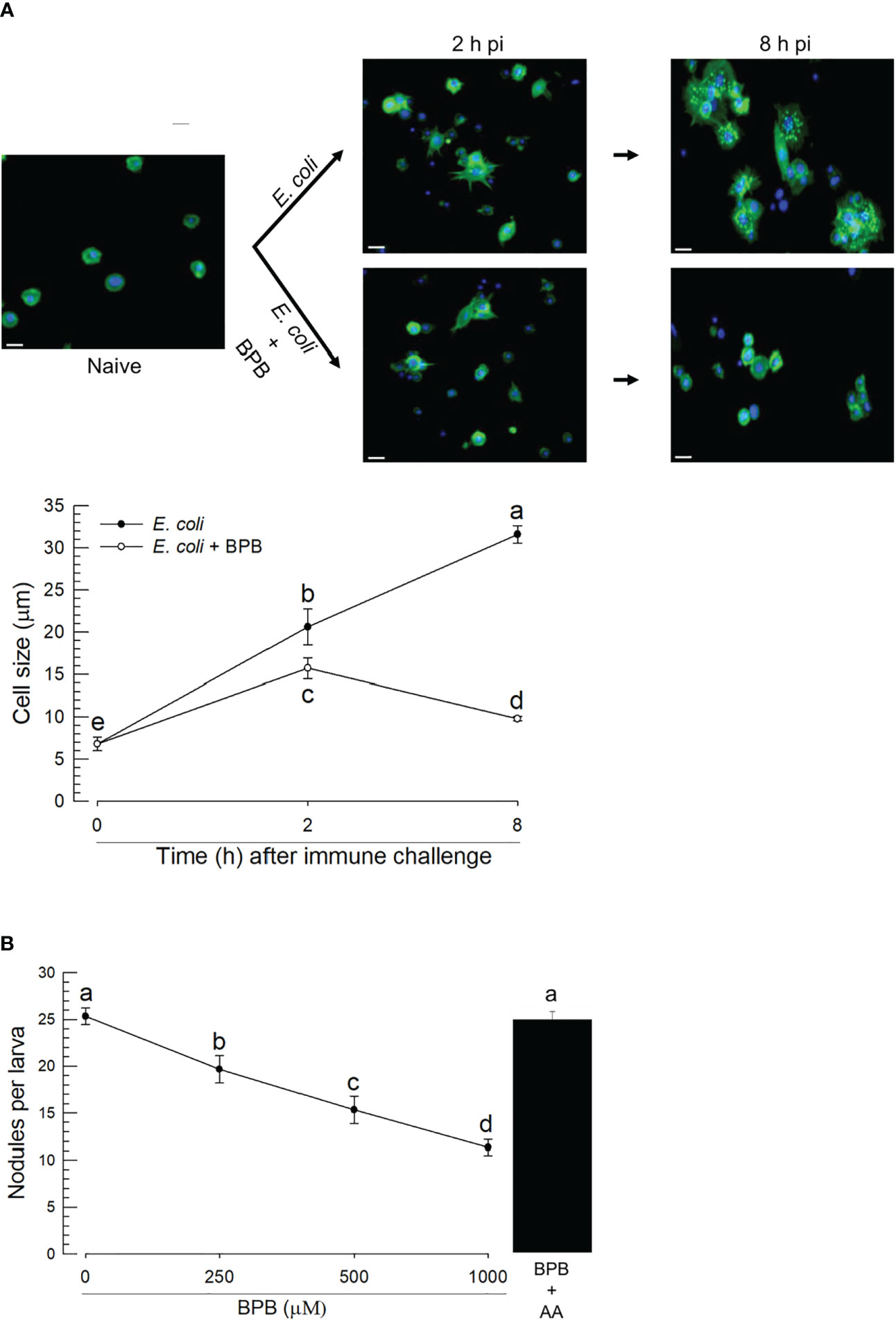
Figure 5 Functional association of PLA2 activity with cellular immune responses in A. sapporensis. To inhibit PLA2 activity, BPB (1 μg per larva) was injected to the hemocoel of L5 larvae. (A) Influence of BPB on hemocyte-spreading behavior at 2 and 8 h after the immune challenge. The white scale bars represent 10 μm. Hemocyte cell size represents the longest cell diameter on the image. Each measurement used randomly chosen 30 hemocytes and was replicated three times with independently prepared hemocyte samples. (B) Inhibitory effect of BPB on nodule formation after the immune challenge. After 8 h pi with immune challenge along with BPB, the larvae were dissected to evaluate the number of nodules. Only E. coli was injected for a control treatment. AA (arachidonic acid, 1 μg per larva) was injected along with BPB. Different letters above the error show significant differences among means at Type I error = 0.05 (LSD test).
To determine which sPLA2(s) of A. sapporensis might be associated with the cellular immune responses, individual RNAi treatments were applied to each of four As-sPLA2 genes (Figure 6). Injection of dsRNAs specific to each of As-PLA2 genes significantly suppressed their target genes for at least 48 h pi (Figure 6A). Under these individual RNAi conditions, PLA2 enzyme activities were monitored after immune challenge (Figure 6B). As expected, the immune challenge significantly induced the enzyme activity. However, all the four individual RNAi treatments significantly suppressed the induction of the enzyme activity after the immune challenge. The four individual RNAi treatments then inhibited the nodule formation after the immune challenge (Figure 6C).
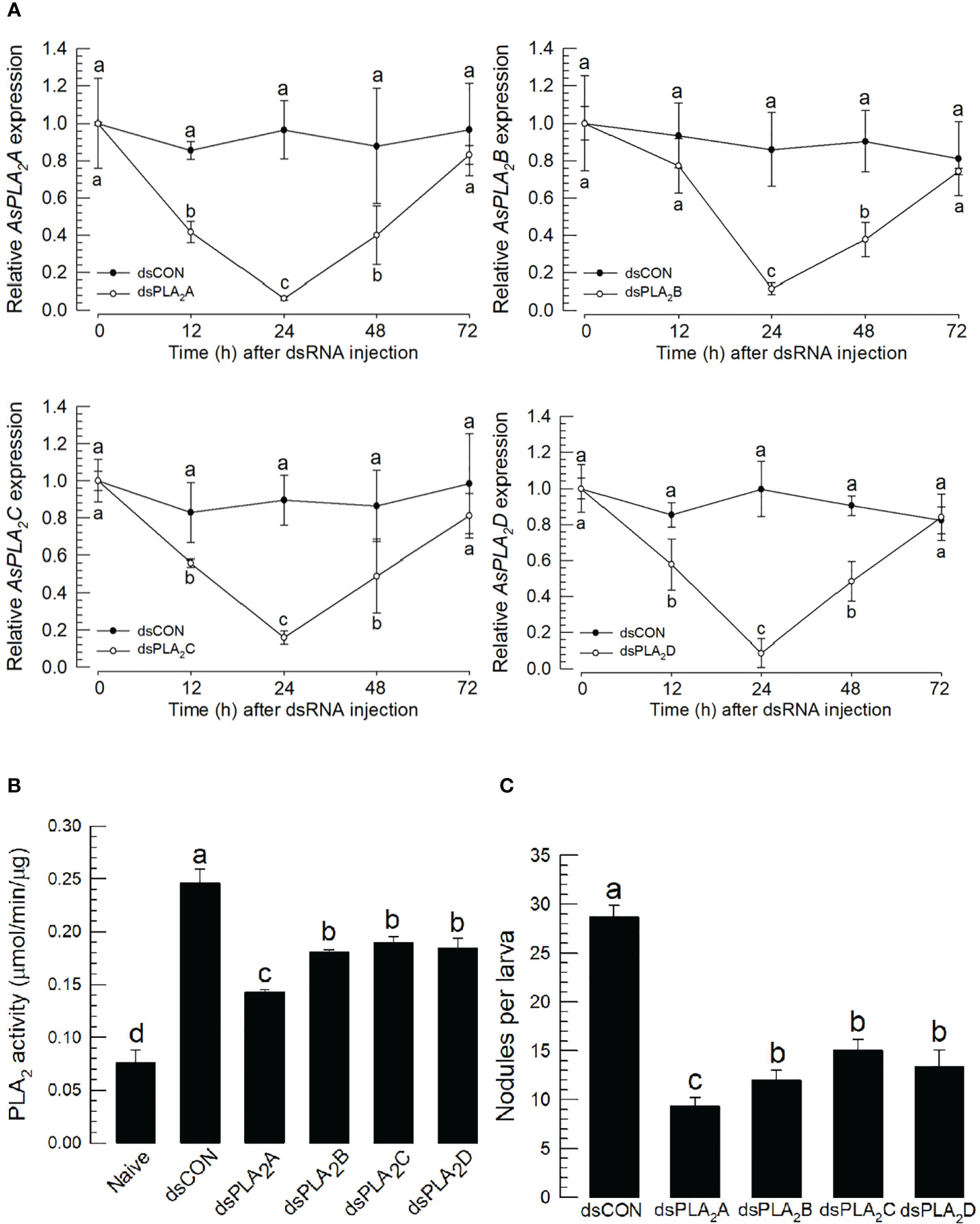
Figure 6 Functional assay of each As-sPLA2 in mediating cellular immune response in A. sapporensis. Individual RNA interference (RNAi) treatments were applied by injecting 1 μg of dsRNA (‘dsPLA2A–dsPLA2D’) to L5 larva. A green fluorescence protein was used as a control dsRNA treatment (‘dsCON’). (A) Reduction of specific As-sPLA2 genes by individual RNAi treatments. (B) Suppression of PLA2 activity induced by the immune challenge by the individual RNAi treatments. (B) Influence of the individual RNAi treatments on nodule formation. Each treatment was replicated thrice with independent sample preparations. Different letters above the standard deviation bars show significant differences among means at Type I error = 0.05 (LSD test).
Here, we report identification of four newly discovered genes encoding PLA2s in A. sapporensis and lay out an argument that these genes can be developed into targets that will effectively cripple insect immune reactions to infections. First, based on a field study conducted in Turkish fields surrounding Kahramanmaraş, Tunaz and Stanley (35) reported that 98% of all collected insect specimens either were or had been infected by naturally occurring microbes, indicated by the presence of melanized nodules in their hemocoels. These data document the need and deployment of effective innate immunity in insect biology. Second, phylogenic and domain analyses of the genes encoding PLA2s document identification of the genes. Third, qPCR analysis shows the genes are most highly expressed in larvae and, within larvae, the genes are expressed in the fat body, hemocytes, and alimentary canal. Fourth, our data show that the enzymes encoded by the four genes are sensitive to the expected inhibitory conditions for PLA2s. Fifth, bacterial infection led to increased expressions of genes encoding the four PLA2s and in PLA2 enzyme activity. Sixth, treating larvae with inhibitors of PLA2 activity led to reductions in two specific immune actions: hemocyte spreading and nodulation. While these points are not a comprehensive summary, they amount to a strong argument that PLA2s play crucial roles in mediating immune responses in A. sapporensis and possibly other pest insect species
The PLA2 activity reported here can be inhibited by a sPLA2-specific inhibitor, but not by iPLA2- or cPLA2-inhibitors, indicating that is it a sPLA2. All four PLA2s are secretory enzymes assigned to three sPLA2 groups (III, X, and XII). PLA2s are classified into 16 groups based on amino acid sequences; they include sPLA2, iPLA2, and cPLA2 (7). In this classification, Group III sPLA2s consists of PLA2s derived from insects and divided into two subgroups (venomous and non-venomous) (36). The lepidopteran, S. exigua, expresses a single sPLA2, Se-sPLA2, classified into Group III. This enzyme mediates immune responses (31). As-PLA2A and As-PLA2D are closely clustered with Se-sPLA2 in our phylogenetic analysis, from which we suggest that they act in mediating immune responses. As-PLA2B is assigned into Group XII. Another Group XII PLA2 identified from a hemipteran Rhodnius prolixus produces eicosanoids via releasing PUFAs from cellular PLs (37). In a mosquito, Aedes albopictus, four PLA2s including Group XII are identified and speculated to be associated with biosynthesis of PGE2 for oogenesis (38). PGE2 also acts in mediating insect immune responses (15). This also suggested that As-PLA2B acts in mediating immune responses. We assigned As-PLA2C into Group X in our phylogenetic analysis, the first insect PLA2s classified into this group.
The expression profiles of these As-sPLA2 genes indicates changing PLA2 enzyme activities during immature development. Gene expressions increased during larval development, with highest expression and enzyme activity during late larval instars. Insect sPLA2 also act in lipid digestion by producing lysophospholipids, which are presumed to act like mammalian bile because insects do not produce bile (39). The increase of the PLA2 activity reported here may be consistent with increased feeding activity during larval development. The correlation between gene expression and the enzyme activity was also recorded after the immune challenge. Expression of the As-sPLA2 genes was highly induced after immune challenge, which may have led to significant increases in PLA2 enzyme activities. Based on the high sequence similarities between As-sPLA2s and Se-sPLA2, we performed a western analysis to assess the change in PLA2 protein. Naïve larvae showed four bands (‘P1–P4’), in which the P1 protein band matched the predicted molecular weights of As-PLA2A or As-PLA2B while P2 appeared to match with the size of As-PLA2C. However, protein bands at 55 kDa (‘P3’) and 90 kDa (‘P4’) were not explained by current four PLA2s. In contrast, the protein samples extracted from immune-challenged larvae had an additional band (‘P5’) that matched the predicted molecular weight of As-PLA2D. Ongoing research will clarify these points.
PLA2 activity mediates A. sapporensis cellular immune responses. We recorded hemocyte spreading, a visible immune reaction to infections that was inhibited in larvae treated with an inhibitor of PLA2 activity. Hemocyte spreading is a phase of cellular immune actions including phagocytosis, nodulation, and encapsulation (40). The spreading involves cytoskeleton rearrangement and local volume change, which are mediated by eicosanoids (41). Influencing cytoskeletons may be a common PG function in animal cells. Green et al. (42) reported earlier that cytoskeleton components are also specific targets of PG signaling in Drosophila egg development. As already mentioned, PLA2 is the first step in eicosanoid biosynthesis and inhibiting this enzyme likely inhibits such subcellular actions generally.
Individual RNAi treatments suppressed their target mRNA levels for at least 48 h pi. Our immune assays were performed at 24 h pi, when the RNAi effects were maximal for all four PLA2s. The RNAi efficacies are supported by the decreases in PLA2 activities. Under these RNAi conditions, the cellular immune response, nodulation, was suppressed following the four RNAi treatments. Our interpretation is that each of the four PLA2s is associated with cellular immune responses. We suggest that each of these four enzymes is required for cellular immune responses because RNAi treatment against one of the four PLA2s significantly suppressed nodulation. Multiple PLA2s forms are expressed in specific mammalian tissues. Rat brain expresses six PLA2s including four sPLA2s, an iPLA2, and cPLA2 in different regions (43), which may be required for regulating synaptic plasticity. Here, we speculate that the four A. sapporensis PLA2s may be expressed in different hemocyte subtypes of hemocytes and they may act in hemocyte-specific immune responses. For example, two cell types, granulocytes and plasmatocytes, form multicell layers around pathogens and a third type, oenocytoids, releases prophenoloxidase during nodule formation (40, 44).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
MH: methodology, resources, software, formal analysis, investigation, writing—original draft, and visualization. JH: methodology, resources, software, formal analysis, and investigation. YK: conceptualization, methodology, project administration, writing—original draft, writing—review and editing, and funding acquisition. All authors contributed to the article and approved the submitted version.
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through the Exporting Promotion Technology Development Program funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (321100-3), Republic of Korea. This study was also funded by a research grant from Andong National University to YK.
We deeply express our gratitude to two guest editors, Qisheng Song and David Stanley, on a kind invitation to this special issue. Also, we appreciate an extraordinary effort from David Stanley to edit the original manuscript, which has improved the readability of the text for readers.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1190834/full#supplementary-material
1. Baccouch R, Shi Y, Vernay E, Mathelié-Guinlet M, Taib-Maamar N, Villette S, et al. The impact of lipid polyunsaturation on the physical and mechanical properties of lipid membranes. Biochim Biophys Acta Biomembr (2023) 1865:184084. doi: 10.1016/j.bbamem.2022.184084
2. Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J (2000) 79:328–39. doi: 10.1016/S0006-3495(00)76295-3
3. Rajamoorthi K, Petrache HI, McIntosh TJ, Brown MF. Packing and viscoelasticity of polyunsaturated ω-3 and ω-6 lipid bilayers as seen by 2H NMR and X-ray diffraction. J Am Chem Soc (2005) 127:1576–88. doi: 10.1021/ja046453b
4. Stanley D. Prostaglandins and other eicosanoids in insects: biological significance. Annu Rev Entomol (2006) 51:25–44. doi: 10.1146/annurev.ento.51.110104.151021
5. Burke JE, Dennis EA. Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res (2009) 50:237–42. doi: 10.1194/jlr.R800033-JLR200
6. Kim Y, Stanley D. Eicosanoid signaling in insect immunology: new genes and unresolved issues. Genes (2021) 12:211. doi: 10.3390/genes12020211
7. Dennis E, Cao J, Hsu Y-H, Magrioti V, Kokotos G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev (2011) 111:6130–85. doi: 10.1021/cr200085w
8. Murakami M, Taketomi Y, Miki Y, Sato H, Hirabayashi T, Yamamoto K. Recent progress in phospholipase a research: from cells to animals to humans. Prog Lipid Res (2011) 50:152–92. doi: 10.1016/j.plipres.2010.12.001
9. Kim Y, Ahmed S, Stanley D, An C. Eicosanoid-mediated immunity in insects. Dev Comp Immunol (2018) 83:130–43. doi: 10.1016/j.dci.2017.12.005
10. Hasan MA, Ahmed S, Kim Y. Biosynthetic pathway of arachidonic acid in Spodoptera exigua in response to bacterial challenge. Insect Biochem Mol Biol (2019) 111:103179. doi: 10.1016/j.ibmb.2019.103179
11. Park Y, Kim Y. Eicosanoids rescue Spodoptera exigua infected with Xenorhabdus nematophilus, the symbiotic bacteria to the entomopathogenic nematode Steinernema carpocapsae. J Insect Physiol (2000) 46:1469–76. doi: 10.1016/S0022-1910(00)00071-8
12. Park Y, Kim Y, Stanley DW. The bacterium Xenorhabdus nematophila inhibits phospholipases A2 from insect, prokaryote, and vertebrate sources. Naturwissenschaften (2004) 91:371–3. doi: 10.1007/s00114-004-0548-2
13. Seo S, Lee S, Hong Y, Kim Y. Phospholipase A2 inhibitors synthesized by two entomopathogenic bacteria, Xenorhabdus nematophila and Photorhabdus temperata subsp. temperata. Appl Environ Microbiol (2012) 78:3816–23. doi: 10.1128/AEM.00301-12
14. Borkman M, Storlien LH, Pan DA, Jenkins AB, Chisholm DJ, Campbell LV. The relation between insulin sensitivity and the fatty-acid composition of skeletal-muscle phospholipids. N Engl J Med (1993) 328:238–44. doi: 10.1056/NEJM199301283280404
15. Stanley D, Kim Y. Prostaglandins and other eicosanoids in insects: biosynthesis and biological actions. Front Physiol (2018) 9:1927. doi: 10.3389/fphys.2018.01927
16. Kim Y, Lee SH, Seo SH, Kim KW. Fatty acid composition of different tissues of Spodoptera exigua larvae and a role of cellular phospholipase A2. Korean J Appl Entomol (2016) 55:129–38. doi: 10.5656/KSAE.2016.04.0.011
17. Ahmed S, Stanley D, Kim Y. An insect prostaglandin E2 synthase acts in immunity and reproduction. Front Physiol (2018) 9:1231. doi: 10.3389/fphys.2018.01231
18. Sajjadian SM, Ahmed S, Al Baki MA, Kim Y. Prostaglandin D2 synthase and its functional association with immune and reproductive processes in a lepidopteran insect, Spodoptera exigua. Gen Comp Endocrinol (2020) 287:113352. doi: 10.1016/j.ygcen.2019.113352
19. Vatanparast M, Lee DH, Kim Y. Biosynthesis and immunity of epoxyeicosatrienoic acids in a lepidopteran insect, Spodoptera exigua. Dev Comp Immunol (2020) 107:103643. doi: 10.1016/j.dci.2020.103643
20. Xu J, Morisseau C, Hammock BD. Expression and characterization of an epoxide hydrolase from Anopheles gambiae with high activity on epoxy fatty acids. Insect Biochem Mol Biol (2014) 54:42–52. doi: 10.1016/j.ibmb.2014.08.004
21. Vatanparast M, Ahmed S, Lee DH, Hwang SH, Hammock B, Kim Y. EpOMEs act as immune suppressors in a lepidopteran insect, Spodoptera exigua. Sci Rep (2020) 10:20183. doi: 10.1038/s41598-020-77325-2
22. Iliff JJ, Fairbanks SL, Balkowiec A, Alkayed NJ. Epoxyeicosatrienoic acids are endogenous regulators of vasoactive neuropeptide release from trigeminal ganglion neurons. J Neurochem (2010) 115:1530–42. doi: 10.1111/j.1471-4159.2010.07059.x
23. Park HH, Kim KH, Park CG, Choi YS, Lee SG. Injury characteristics of allium leafminer, Acrolepiopsis sapporensis (Lepidoptera: acrolepiidae) in welsh onion and damage assessment according to larval density levels during summer. Korean J Appl Entomol (2012) 51:383–8. doi: 10.5656/KSAE.2012.09.0.054
24. Eom S, Park Y, Kim H, Kim Y. Development of a high efficient “Dual bt-plus” insecticide using a primary form of an entomopathogenic bacterium, Xenorhabdus nematophila. J Microbiol Biotechnol (2014) 24:507–21. doi: 10.4014/jmb.1310.10116
25. Kim E, Jeoung S, Park Y, Kim K, Kim Y. A novel formulation of Bacillus thuringiensis for the control of brassica leaf beetle, Phaedon brassicae (Coleoptera: chrysomelidae). J Econ Entomol (2015) 108:2556–65. doi: 10.1093/jee/tov245
26. Park Y, Jung J, Kim Y. A mixture of Bacillus thuringiensis subsp. israelensis with Xenorhabdus nematophila-cultured broth enhances toxicity against mosquitoes Aedes albopictus and Culex pipiens pallens. J Econ Entomol (2016) 109:1086–93. doi: 10.1093/jee/tow063
27. Jin G, Hrithik TH, Kim Y. Control efficacy of bacterial secondary metabolites of Xenorhabdus hominickii against a fungal pathogen, Alternaria alternata, infecting welsh onion. Korean J Pestic Sci (2023) 27:1–8. doi: 10.7585/kjps.2023.27.1.23
28. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol (2013) 30:2725–29. doi: 10.1093/molbev/mst197
29. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem (2009) 55:611–22. doi: 10.1373/clinchem.2008.112797
30. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2-ΔΔCT method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
31. Vatanparast M, Ahmed S, Sajjadian SM, Kim Y. A prophylactic role of a secretory PLA2 of Spodoptera exigua against entomopathogens. Dev Comp Immunol (2019) 95:108–17. doi: 10.1016/j.dci.2019.02.008
32. Vatanparast M, Kim Y. Functional interaction of bacterial virulence factors of Xenorhabdus nematophila with a calcium-independent cytosolic PLA2 of Spodoptera exigua. J Invertebr Pathol (2020) 169:107309. doi: 10.1016/j.jip.2019.107309
33. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye finding. Anal Biochem (1972) 72:248–54. doi: 10.1016/0003-2697(76)90527-3
35. Tunaz H, Stanley D. An immunological axis of biocontrol: infections in field-trapped insects. Naturwissenschaften (2009) 96:115–9. doi: 10.1007/s00114-009-0572-3
36. Stanley D, Kim Y. Why most insects have very low proportions of C20 polyunsaturated fatty acids: the oxidative damage hypothesis. Arch Insect Biochem Physiol (2020) 103:e21622. doi: 10.1002/arch.21622
37. Defferrari MS, Lee DH, Fernandes CL, Orchard I, Carlini CR. A phospholipase A2 gene is linked to jack bean urease toxicity in the chagas’ disease vector. Rhodnius prolixus. Biochim Biophys Acta (2014) 1840:396–405. doi: 10.1016/j.bbagen.2013.09.016
38. Choi D, Al Baki MA, Ahmed S, Kim Y. Aspirin inhibition of prostaglandin synthesis impairs mosquito egg development. Cells (2022) 11:4092. doi: 10.3390/cells11244092
39. Sajjadian SM, Vatanparast M, Stanley D, Kim Y. Secretion of secretory phospholipase A2 into Spodoptera exigua larval midgut lumen and its role in lipid digestion. Insect Mol Biol (2019) 28:773–84. doi: 10.1111/imb.12588
40. Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol (2002) 32:1295–309. doi: 10.1016/S0965-1748(02)00092-9
41. Ahmed S, Kim Y. PGE2 mediates hemocyte-spreading behavior by activating aquaporin via cAMP and rearranging actin cytoskeleton via Ca2+. Dev Comp Immunol (2021) 125:104230. doi: 10.1016/j.dci.2021.104230
42. Green CM, Spracklen AJ, Fagan TN, Tootle TL. Drosophila fascin is a novel downstream target of prostaglandin signaling during actin remodeling. Mol Biol Cell (2012) 23:4567–78. doi: 10.1091/mbc.e12-05-0417
43. Molloy GY, Rattray M, Williams RJ. Genes encoding multiple forms of phospholipase A2 are expressed in rat brain. Neurosci Lett (1998) 258:139–42. doi: 10.1016/S0304-3940(98)00838-6
Keywords: eicosanoid, PLA2, immunity, nodulation, RNAi, Acrolepiopsis sapporensis
Citation: Hrithik MTH, Hong J and Kim Y (2023) Identification of four secretory phospholipase A2s in a lepidopteran insect, Acrolepiopsis sapporensis, and their functional association with cellular immune responses. Front. Endocrinol. 14:1190834. doi: 10.3389/fendo.2023.1190834
Received: 21 March 2023; Accepted: 07 June 2023;
Published: 23 June 2023.
Edited by:
David Stanley, United States Department of Agriculture, United StatesReviewed by:
Bin Li, Nanjing Normal University, ChinaCopyright © 2023 Hrithik, Hong and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonggyun Kim, aG9zYW5uYUBhbnUuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.