- 1Department of Neurology, Institute of Neuroscience, Key Laboratory of Neurogenetics and Channelopathies of Guangdong Province and the Ministry of Education of China, The Second Affiliated Hospital, Guangzhou Medical University, Guangzhou, China
- 2The First Clinical Medicine School of Guangzhou Medical University, Guangzhou, China
- 3The Second Clinical Medicine School of Guangzhou Medical University, Guangzhou, China
- 4Department of Obstetrics and Gynecology, The Sixth Affiliated Hospital, Sun Yat-sen University, Guangzhou, China
- 5Department of Obstetrics and Gynecology, Center for Reproductive Medicine, Guangdong Provincial Key Laboratory of Major Obstetric Diseases, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
- 6Key Laboratory for Reproductive Medicine of Guangdong Province, The Third Affiliated Hospital of Guangzhou Medical University, Guangzhou, China
Introduction: It has been established that UBR4 encodes E3 ubiquitin ligase, which determines the specificity of substrate binding during protein ubiquitination and has been associated with various functions of the nervous system but not the reproductive system. Herein, we explored the role of UBR4 on fertility with a Drosophila model.
Methods: Different Ubr4 knockdown flies were established using the UAS/GAL4 activating sequence system. Fertility, hatchability, and testis morphology were studied, and bioinformatics analyses were conducted. Our results indicated that UBR4 deficiency could induce male sterility and influent egg hatchability in Drosophila.
Results: We found that Ubr4 deficiency affected the testis during morphological analysis. Proteomics analysis indicated 188 upregulated proteins and 175 downregulated proteins in the testis of Ubr4 knockdown flies. Gene Ontology analysis revealed significant upregulation of CG11598 and Sfp65A, and downregulation of Pelota in Ubr4 knockdown flies. These proteins were involved in the biometabolic or reproductive process in Drosophila. These regulated proteins are important in testis generation and sperm storage promotion. Bioinformatics analysis verified that UBR4 was low expressed in cryptorchidism patients, which further supported the important role of UBR4 in male fertility.
Discussion: Overall, our findings suggest that UBR4 deficiency could promote male infertility and may be involved in the protein modification of UBR4 by upregulating Sfp65A and CG11598, whereas downregulating Pelota protein expression.
1 Introduction
Current evidence suggests that maintaining cellular protein homeostasis (proteostasis) largely relies on proteolysis (1). As a cellular mechanism that triggers proteasomal degradation, ubiquitin conjugation, based on ubiquitin and ubiquitin-like proteins (UBLs), plays an important role in controlling cell division, signal transduction, embryonic development, endocytic trafficking, and immune response (2).
UBR4 encodes the Ubiquitin Protein Ligase E3 Component N-Recognin 4; the homology of Drosophila is poe. It is an E3 ubiquitin ligase (E3 enzyme) that regulates protein degradation, removing damaged or misfolded proteins from the cellular environment, thereby regulating cellular homeostasis. The body utilizes the coordinated action of a series of enzymes in the ubiquitin–proteasome system (UPS), including ubiquitin-activating enzymes (E1 enzymes), ubiquitin-conjugating enzymes (E2 enzymes), and E3 ubiquitin ligases (E3 enzymes) (1–4). E3 enzymes are reportedly important in the ubiquitination process and determine substrate specificity (3). Protein error correction plays an extremely important role in physiological processes such as growth and development, reproductive inheritance, immunity, and endocrine of Drosophila and the human body.
Up to now, research on UBR4 has mainly focused on the nervous system (5–7). Interestingly, the number of Drosophila pupae decreased after Ubr4 gene knockdown in our preliminary experiments, suggesting that Ubr4 may be related to reproductive capacity. This phenomenon may be attributed to erroneous protein synthesis in the genitals, which may cause physiological and behavioral changes, including reduced egg production and feeding, decreased willingness to mate and storage/utilization of sperm, and altered gene expression (8, 9). Spermatogenesis abnormality is also another important reason for reproductive problems. In Drosophila, spermatogenesis starts at the apical tip of the testis, which hosts two stem cell populations, including the germline stem cells (GSCs) and somatic cyst stem cells (CySCs) (10–12). Zygotes produced by GSCs undergo four rounds of mitosis and then differentiate into 16 spermatogonia. The spermatogonia then differentiates to spermatocytes, round spermatozoa, long spermatozoa, and mature spermatozoa and complete an entire spermatogenesis process. The cyst cells differentiated from CySCs encase germ cells and support germ cell growth (13). The hub cells mainly maintain the self-renewal and differentiation of GSCs and CySCs (14, 15).
Herein, loss-of-function Drosophila models were applied as research vectors to detect the effect of the Ubr4 gene on Drosophila development and reproduction.
2 Materials and methods
2.1 Fly culture and qPCR
Drosophila was maintained on standard cornmeal and incubated under controlled conditions (temperature 25°C, humidity 60%–70%, and 12:12-h light/dark cycle). UAS-Ubr4-RNAi (THU1137, CG14472) was donated by Tsing Hua Fly Center (Tsinghua University, Beijing, China). The knockdown efficiency of RNAi was checked by qPCR (16). The primer sequencies were
Ubr4/Poe-F1: CCACCGTCACACACTTCAAT
Ubr4/Poe-R1: GGCAGCAGTCCATTACATCT
GAPDH-F1: CGTCAACGATCCCTTCATCGATGTC
GAPDH-R1: CAGCACTGGCCCAGTTGATGTTG.
2.2 Fertility and hatchability assays
The quantification of fertility and hatchability: Fertility refers to the ability of Drosophila to produce offspring, and our study mainly focused on the number of eggs it laid. Hatchability is the rate of eggs hatching into pupae and pupae hatching into adults. The details of analyses were shown below.
2.2.1 Ubr4 knockdown male fertility and hatchability assay
UAS-Ubr4-RNAi males were mated with tub-GAL4 females to generate Ubr4 knockdown flies (tub-GAL4>UAS-Ubr4-RNAi). UAS-Ubr4-RNAi males were used as their genetically matched controls. Subsequently, 3-day-old virgin knockdown and control males, isolated within 24 h of their eclosion, were mated with 3–5-day-old wild-type (Canton-S) virgin female flies for different assays, as described below.
Three males were mated with Canton-S virgin females in pairs. Then, males were discarded after 48 h. Mated females were allowed to lay eggs for 10 days with the change to fresh food vials at 48-h intervals for the fertility assay. The number of eggs laid, egg-to-pupa hatchability rate, and pupa-to-adult hatchability rate was calculated. Mated females were discarded after 10 days, and the resultant progenies were counted to determine fertility. For each group, the experiments were repeated three times with five to six replicates each.
2.2.2 Ubr4 knockdown female fertility and hatchability assay
Three-day-old Ubr4 knockdown (tub-GAL4>UAS-Ubr4-RNAi) and control (UAS-Ubr4-RNAi) virgin females, isolated within 24 h of their eclosion, were mated with 3–5-day-old Canton-S males for fertility assays.
After three males were mated with Canton-S virgin females in pairs, the males were discarded. We allowed mated females to lay eggs for 10 days, with fresh food vials changed every 48 h for the fertility assay. A fertility test was carried out by counting the progeny of females discarded after 10 days of mating. Hatchability is defined the same as above.
2.3 Larva development experiment
Newly Ubr4 knockdown (tub-GAL4>UAS-Ubr4-RNAi) and control (UAS-Ubr4-RNAi>Canton-S) instar larvae cultured in standard medium for 24, 48, 72, 96, and 120 h after egg laying (AEL) were heated with 40°C–50°C water for 15 min to foster the larvae to come out from the medium. Afterward, the collected larvae were washed with phosphate-buffered saline (PBS), and their length was measured under the microscope. Next, the number of larvae developing into pupae was counted at 6-h intervals each day until all larvae had hatched. Subsequently, the number of adults hatched from pupae at 6-h intervals was counted. At the same time, the time of hatching from larvae to pupae and pupae to adults were counted independently.
2.4 Testes morphology
Drosophila testes from 2-day-old young adults were dissected in 1× PBS, fixed in 1× PBS/4% PFA for 30 min at room temperature. After being washed three times with PBS, the testes were transferred to glass slides. A mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Abcam, USA) was dripped and reacted with the testes for 10 min before being covered with a coverslip. Testes morphology was observed with an inverted phase-contrast microscope (SP8; Zeiss, Jena, Germany). The number of spermatogenic cells was counted by ImageJ software (National Institutes of Health, Bethesda, MD, USA), and data were analyzed by GraphPad Prism 7.0.
2.5 Mass spectrometry and bioinformatics analyses
2.5.1 Extraction of protein and enzymatic hydrolysis of peptides
Testes were collected and frozen in liquid nitrogen before protein extraction. Tissue protein was extracted by SDT (4% (w/v) SDS, 100 mm Tris/HCl pH 7.6, 0.1 m DTT), then protein was quantified by BCA. AFign appropriate amount of protein was obtained from each sample, and enzymatic hydrolysis of trypsin was performed by filter-aided proteome preparation (FASP). Next, the peptide was desalted by the C18 Cartridge. After freeze-drying, the peptide was lyophilized and redissolved in 40 μl of 0.1% formic acid solution (OD280).
2.5.2 Protein identification and quantitative analysis
The MaxQuant software (version number 1.6.14) (17) was used for database identification and quantitative analysis. Mass spectrometry was carried out by Shanghai Applied Protein Technology Co., Ltd.
2.5.3 Bioinformatics analyses
The CTD (Comparative Toxicogenomics Database), which can be used for the prediction of correlations between genes, diseases, and chemicals (18), indicates that changes in UBR4 may be associated with cryptorchidism (http://ctdbase.org/). As one of the reasons for male infertility disorders, cryptorchidism, teratospermia, and azoospermia may be risk factors for the disease (19). To validate the findings, we downloaded the cryptorchidism dataset (GSE16191), teratospermia (GSE6872), and azoospermatism (GSE108886) from the GEO (Gene Expression Omnibus) database. With the screening criteria that (1) there were clear experimental and control groups, (2) the experimental group did not have any treatment other than cryptorchidism, (3) the samples were all from humans, and (4) the dataset had been borrowed from other articles, we finally chose the cryptorchidism dataset GSE16191 (20) and completed a bioinformatics analysis using R software (R.4.2.2), containing 16 diseased samples and 4 healthy control samples with tissue taken from whole testes (http://www.ncbi.nlm.nih.gov/geo/). The expression of three key genes, UBR4, PELO, and LIPA, was compared in groups to validate their up- and downregulation, and ROC curve analysis of key genes was performed to predict their diagnostic efficacy for cryptorchidism. A single-gene GSEA was performed to identify downstream genes in the functional pathway of the key genes. GO and KEGG analyses of downstream genes and key genes were also performed to search for related biological processes and enrichment pathways mediated by key genes.The quantitative information of the target protein set was normalized (normalized to (-1,1) interval). Then, the ComplexHeatmap R package (R Version 3.4) was used to classify the two dimensions (distance algorithm: Euclid, connection mode: average linkage) of the expression of samples and protein simultaneously. Finally, a hierarchical clustering heat map was generated.
GO annotation of the target protein set by Blast2GO can be summarized into four steps: sequence comparison (Blast), GO item mapping, GO annotation, and InterProScan annotation. The ProScan software package, which runs a scanning algorithm from the InterPro database in an integrated way to characterize the sequence, was used to obtain the domain annotation information of the target protein sequence in the Pfam database.
2.6 Statistical analysis
All quantitative data were expressed as mean ± SD. The Student’s t-test was used to compare two independent or paired samples. Multiple samples were compared using one-way ANOVA, and differences between the two groups were evaluated using Tukey’s post-hoc test. The data were analyzed using GraphPad Prism 7.00 and SPSS 20. A P-value < 0.05 was statistically significant.
3 Results
3.1 Ubr4 deficiency influences larval development
Flies crossing strategy in larval development assay was showed in Figure 1A. The Knockdown efficiency of RNAi is about 20% (Figure 1B). Compared with the control group, the length of tub-GAL4>UAS-Ubr4-RNAi larvae was lower than that of UAS-Ubr4-RNAi> Canton-S, especially for 48-h AEL and 72-h AEL. The length of 48h AEL tub-GAL4>UAS-Ubr4-RNAi larvae was significantly lower than that of UAS-Ubr4-RNAi> Canton-S (1.694 ± 0.544 mm [n = 28] vs. 2.159 ± 0.388 mm [n = 20]; **P = 0.002, Figure 1E). At the same time, the 72-h AEL tub-GAL4>UAS-Ubr4-RNAi larvae had a lower length than UAS-Ubr4-RNAi>Canton-S (2.847 ± 0.687 mm [n = 25] vs. 3.608 ± 0.744 mm [n = 27]; ***P = 0.0004, Figure 1F). However, when the larvae developed to 24-h AEL, 96-h AEL and 120-h AEL, there was no significant difference in larval length between the tub-GAL4>UAS-Ubr4-RNAi and UAS-Ubr4-RNAi> Canton-S flies (24-h AEL: 1.007 ± 0.292 m24-h AEL: 1.007 ± 0.292 mm [n = 21] vs. 1.112 ± 0.211 mm [n = 25]; P >0.05 Figure24-h AEL: 1.007 ± 0.292 mm [n=21] vs. 1.112 ± 0.211 mm [n=25]; P>0.05, Figure 1D. 96-h AEL: 4.125 ± 0.276 mm [n = 26] vs. 4.157 ± 0.340 mm [n = 21]; P > 0.05, Figure 1G. 120-h AEL: 4.253 ± 0.417 mm [n = 23] vs. 4.258 ± 0.588 mm [n = 25]; P > 0.05, Figure 1H). These results suggested that the heterogeneity in developmental speed accounted for the difference in larval length observed in our study.

Figure 1 Comparison of larval development between Ubr4 knockdown and wild type. (A) Experimental procedure. (B) Relative mRNA expression level in WT and Ubr4 knockdown flies. (C) The overall length variation (24–120 h) of larvae. (D) Comparison of the length of 24-h-old larvae. (E) Comparison of the length of 48-h-old larvae. (F) Comparison of the length of 72-h-old larvae. (G) Comparison of the length of 96-h-old larvae. (H) Comparison of the length of 120-h-old larvae. a. tub-Gal4>UAS-Ubr4-RNAi, b. UAS-Ubr4-RNAi. (I) Larval hatchability at different time points. (J) Pupae hatchability at different time points. ns, P>0.05, *P<0.05, **P<0.01, ***P<0.005.
Further experiments showed that there was no significant difference in the development time from larva stage to pupa and from pupa stage to adult between the tub-GAL4>UAS-Ubr4-RNAi and UAS-Ubr4-RNAi> Canton-S groups (Figures 1I, J).
3.2 UBR4 is essential for male fertility but not female fertility
Wild-type females (Canton-S) mated to Ubr4 knockdown males (tub-Gal4>UAS-Ubr4-RNAi) produced significantly fewer fertilized eggs than those mated with wild-type (UAS-Ubr4-RNAi) males. The reduction in fertility observed in Drosophila could be due to several reasons, including fewer eggs being laid by mated females and/or reduced hatchability/increased mortality of these laid eggs during development (21). Therefore, to determine the cause of decreased fertility of Ubr4 knockdown males, we counted the number of eggs laid (fertility) on day 1, day 3, day 5, day 7, and day 9. Flies crossing strategy was shown in Figure 2A. Figure 2B showed the overview of eggs numbers of Ubr4 knockdown group and control group. Over 10 days, the number of eggs produced by the females mated with tub-GAL4>UAS-Ubr4-RNAi males was less than in those mated with Canton-S>UAS-Ubr4-RNAi males (53.83 ± 18.90 [n = 6] vs. 88.00 ± 25.10 [n = 4]; *P = 0.0388, Figure 2C), on day 5 (32.50 ± 16.53 [n = 6] vs. 65.75 ± 12.18 [n = 4]; **P = 0.0090, Figure 2F) and day 9 (55.00 ± 20.26 [n = 6] vs. 88.00 ± 13.90 [n = 4]; *P = 0.05, Figure 2H). No significant difference was shown on day 1, day 3, day 7 between the tub-Gal4>UAS-Ubr4-RNAi and UAS-Ubr4-RNAi> Canton-S flies (P>0.05, Figures 2B, 2D, 2G).
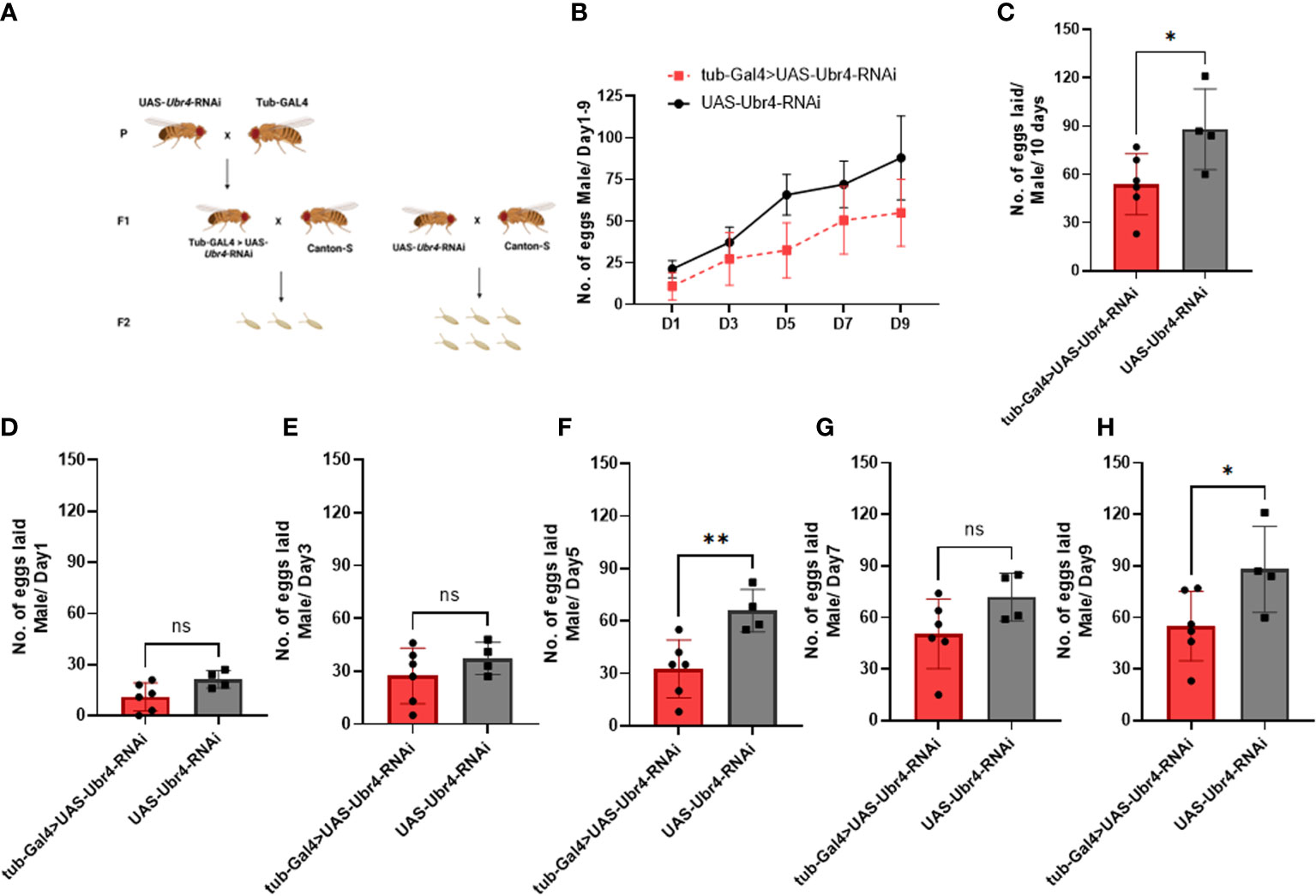
Figure 2 Reproductive performance of females mated to Ubr4 knockdown males. (A) Experimental procedure. (B) The overview of eggs laid from days 1 to 9. (C) The overall fertility (total no. of eggs laid/10 days) of mated females. (D) Day 1 fertility (no. of eggs laid/day from day 1) of mated females. (E) Day 3 fertility (no. of eggs laid/day from day 3) of mated females. (F) Day 5 fertility (no. of eggs laid/day from day 5) of mated females. (G) Day 7 fertility (no. of eggs laid/day from day 7) of mated females. (H) Day 9 fertility (no. of eggs laid/day from day 9) of mated females. ns, P>0.05, *P<0.05, **P<0.01.
However, after wild-type males (Canton-S) were mated to Ubr4-deficient females (tub-Gal4>UAS-Ubr4-RNAi), no significant difference was observed compared with the control (Canton-S males mated with UAS-Ubr4-RNAi) in the overall number of progenies produced over 10 days (P > 0.05, Figures 3B–H). Therefore, the Ubr4 gene has a more important influence on male fertility than on female fertility.
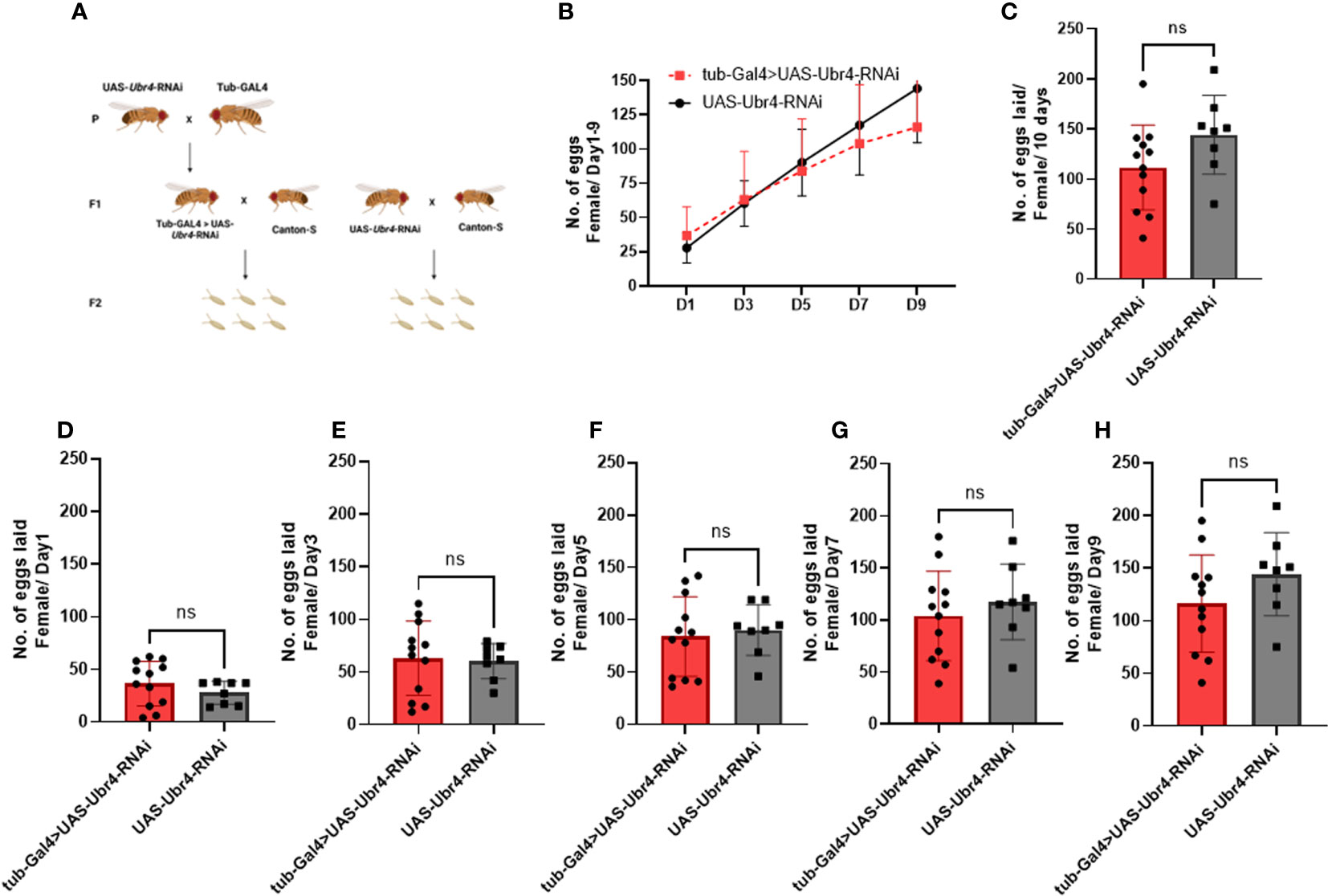
Figure 3 Reproductive performance of Ubr4 knockdown females mated to wild males. (A) Experimental procedure. (B) The overview of eggs laid from days 1 to 9. (C) The overall fertility (total no. of eggs laid/10 days) of mated females. (D) Day 1 fertility (no. of eggs laid/day from day 1) of mated females. (E) Day 3 fertility (no. of eggs laid/day from day 3) of mated females. (F) Day 5 fertility (no. of eggs laid/day from day 5) of mated females. (G) Day 7 fertility (no. of eggs laid/day from day 7) of mated females. (H) Day 9 fertility (no. of eggs laid/day from day 9) of mated females. ns, P>0.05.
To determine hatchability, we focused on the number of eggs that grew into pupae (egg hatchability) and the pupae that developed into adults (pupae hatchability). Analysis of the hatchability showed no significant difference in pupa-hatching rates between the pupae from Ubr4 knockdown parents and the pupae from wild-type parents (Figure 4B). In the egg hatchability, the hatching rate of the eggs reproduced from wild-type mothers with Ubr4 knockdown fathers is lower than that from wild-type parents (**P = 0.0090, Figure 4A), whereas the hatching rate of eggs reproduced form Ubr4 knockdown mothers with wild-type fathers is higher than that from wild-type parents (*P = 0.0175, Figure 4A).
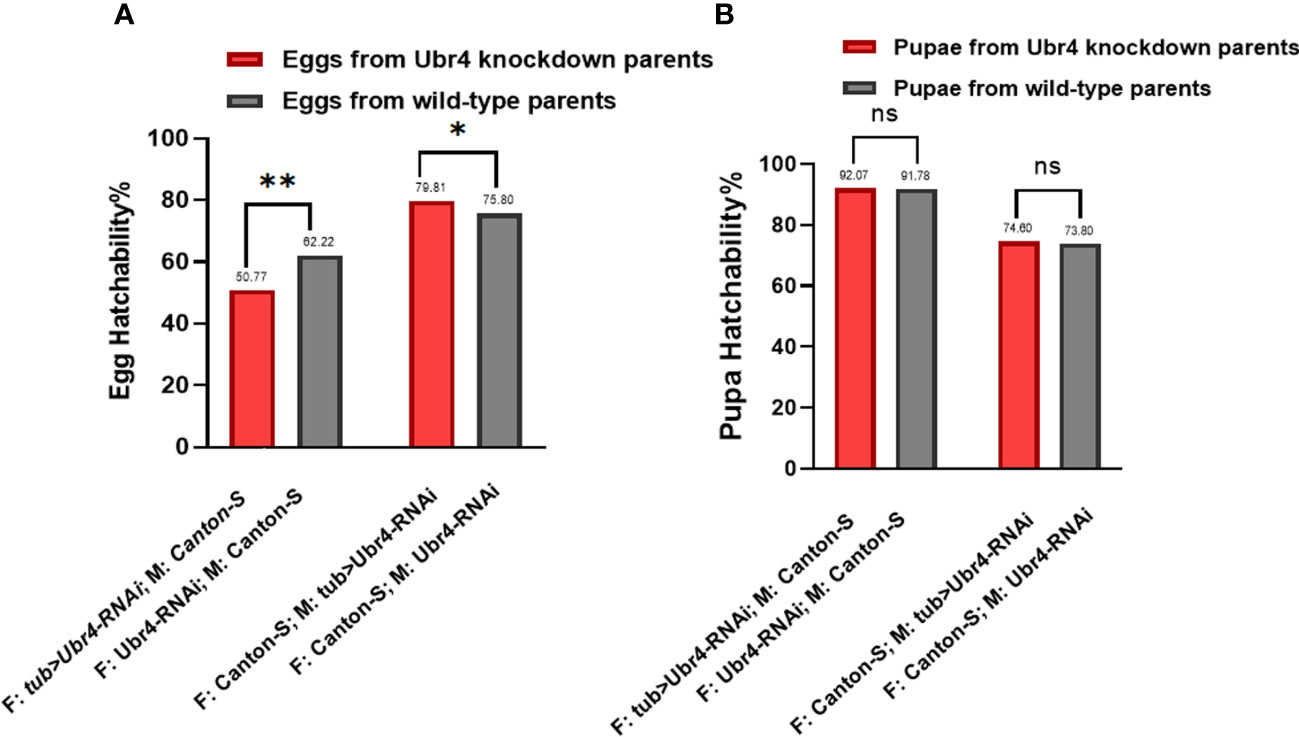
Figure 4 Egg hatchability and pupa hatchability. The hatchability rate was analyzed in two aspects, egg hatching rate (A) and pupa-to-adult rate (B). Two gender groups were included: one was Ubr4 knockdown males mated to wild females, and the other was Ubr4 knockdown females mated to wild males. F, father; M, mother. ns, P>0.05, *P<0.05, **P<0.01.
3.3 Ubr4 deficiency affected testis morphology
Morphological changes in Ubr4 knockdown flies were studied by confocal imaging. DAPI, a well-established blue-emitting fluorescent compound, was used to visualize the cell nucleus. The testis cells of the Ubr4 knockdown flies appear slightly obscure compared with wild-type flies, which required stronger laser power to be visualized (550 vs. 480). In general, the Hub cells, CySCs, GSCs, and Sp labeled by DAPI are easier to be observed whereas the spermatocytes and cyst cells are not easy to be observed. Thus, knockdown of Ubr4 may alter the ratio of cell types in the testis. However, further experiments with a cell-type-specific antibody should be used for studying which stage of germ cells or what kind of somatic cells are affected.
Furthermore, we tried to identify the number of spermatogenic cells in the germinal center of each group. The whole testes can be observed under a 20× confocal microscope, and spermatogonia at various stages were visible with 40× magnification (Figures 5A, B). The testes in the Ubr4 knockdown group had a collapsed morphology and a disorganized internal structure compared with the control group. The number of spermatogenic cells was analyzed by ImageJ. Statistical results showed that there was a significant difference in the number of spermatogenic cells near the head of the testis (59.42 ± 10.62 [n = 12] vs. 96.62 ± 25.15 [n = 13]; ****P < 0.0001, Figure 5C).
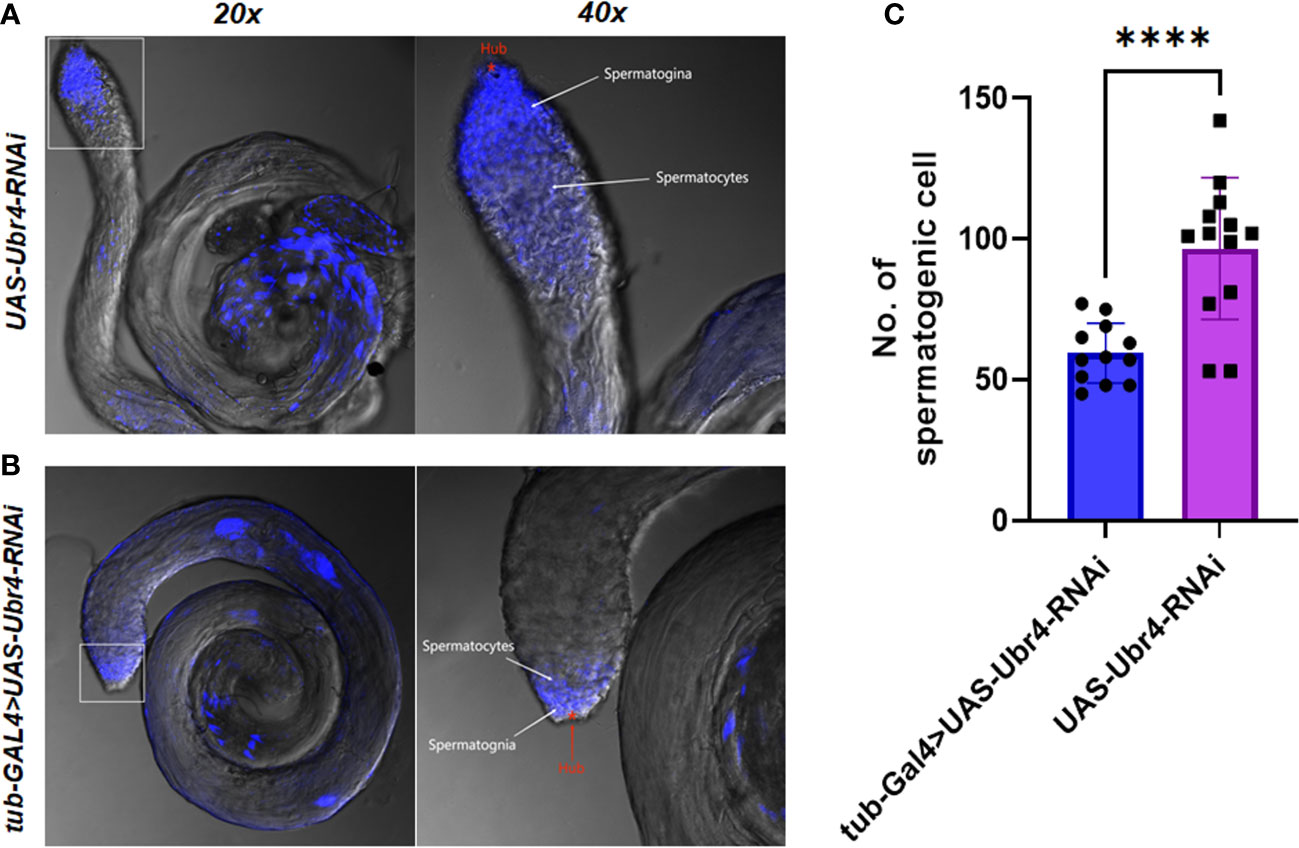
Figure 5 Testis morphology and spermatogenic cells To assess the effect of Ubr4 deficiency, testes were analyzed at the ultrastructural level. Images showing the structure of WT Drosophila male testis (A) and Ubr4 knockdown Drosophila (B). (C) Quantity of spermatogenic cells in testis. ****P<0.001.
3.4 Pelota was downregulated, whereas CG11598 and Sfp65A were upregulated in the testis of Ubr4-deficient flies
To better understand the differential protein expression patterns between WT and Ubr4-knockdown testis, the mechanisms underlying Ubr4-knockdown-induced lower reproduction capacities in male Drosophila were explored by label-free proteomics. There were 363 differentially expressed proteins between WT and Ubr4 knockdown Drosophila (Figure 6A). In comparison with WT control Drosophila, we found 188 upregulated proteins and 175 downregulated proteins in Ubr4-knockdown Drosophila (P < 0.05, Figure 6B). Heatmaps showed the alterations in the expression of 363 proteins (Figure 6C). Protein nodes that were upregulated (above 2.0-fold expression, red color) or downregulated (below 0.5-fold expression, blue color) were displayed using a gradient coloring scheme. Among these 363 proteins, CG11598 (orthologous to human LIPA) was the most upregulated and Sfp65A was the second, whereas Pelota (orthologous to human PELO) exhibited a significant downregulation after Ubr4 knockdown (Figure 6C). Next, we conducted GO annotation of the 363 differential proteins affected by the knockdown of Ubr4 to determine their function. It was suggested that the upregulation of Sfp65A and downregulation of Pelota were associated with GO terms Reproduction and Multicellular organism reproduction (Figure 6D), related to the reproductive process in Drosophila. CG11598 was related to GO terms Metabolic process, Primary metabolic process, and Hydrolase activity, which suggested that upregulation of CG11598 may reduce male fertility through biometabolic processes. The number of differential proteins was counted at the GO secondary functional annotation level, including Biologic Process (BP), Cellular Component (CC), and Molecular Function (MF). The following are related to the GOs most affected by the loss of Ubr4. The delta DNA polymerase complex and DNA polymerase complex were mainly enriched in CC. Regulation of endosome size, negative regulation of ERBB signaling pathway, and negative regulation of the mRNA metabolic process were significantly enriched in BP by the mutant of Ubr4. Glycine binding, neurotransmitter binding, glucosidase activity, and lipase activity enriched in MF also showed an important interaction after knockdown of Ubr4 (Figure 6E).

Figure 6 Mass spectrometry and bioinformatics analyses (A) Volcano plot showing the differentially expressed proteins from the WT control and the Ubr4 knockdown group in the testis of Drosophila. The vertical lines mark the rate of the Ubr4-knockdown group compared to the control group, and the horizontal lines mark the P value. The upper right quadrants show the proteins upregulated compared to the control group, and the upper left quadrants contain downregulated compared to the control group. (B) Proteomics data show significant differences between WT and Ubr4-knockdown Drosophila testis in terms of upregulated and downregulated proteins. (C) The heatmap displays the difference in protein expression between Ubr4-knockdown and WT controls (P < 0.05). (D) GO enrichment analysis of the above differentially expressed proteins highlighted that the upregulation of Sfp65A and downregulation of Pelota were involved in the reproductive process. (E) GO enrichment analysis of key targets. It included Biologic Process (BP), Cellular Component (CC), and Molecular Function (MF).
3.5 The function of UBR4 tested and verified with bioinformatics analyses
CG11598 is orthologous to human LIPA, and Pelota is orthologous to human PELO. Sfp65A has no homologous gene in human. Therefore, this study focuses on the expression of UBR4, LIPA, and PELO in the cryptorchidism dataset.
Expression grouping box plots showed good between-group differences for UBR4 (*P = 0.02) and LIPA (*P = 0.049) but confirmed that the between-group differences for PELO were not statistically significant (P = 0.06), and the up- and downregulation were consistent with the experimental results (Figure 7A). ROC curve analysis (Figure 7B) showed relatively high diagnostic efficacy for all three genes (AUC > 0.8). Single-gene GSEA predicted key genes downstream, and six results were screened (MAPK14, NFKB1, MAPK13, MAPK11, MAPK9, and PIK3CD) (Figure 7C). GO analysis showed that BP was mainly enriched in the regulation of interleukin-12 production, stress-activated protein kinase signaling cascade, and stress-activated MAPK cascade. MF is mainly enriched in protein serine/threonine/tyrosine kinase activity and MAP kinase activity. CC is predominantly enriched in the secretory granule lumen and is enriched in ficolin-1-rich granule and specific granule. KEGG was mainly enriched in Chagas disease, AGE-RAGE signaling pathway in diabetic complications, and prolactin signaling pathway (Figure 7D).

Figure 7 Bioinformatics analyses of UBR4 and its related genes. (A) Box plot for expression control of UBR4, LIPA, and PELO. (B) Two hub genes were applied for the ROC (Receiver Operating Characteristic) evaluation in GSE16191. (C) Six downstream genes of the hub genes (UBR4, LIPA, and PELO) were selected using a Venn plot to visualize the single-gene GSEA (gene set enrichment analysis). (D) The GO\KEGG analysis of the hub genes and the downstream genes. ROC, receiver operating characteristic; GSEA, gene set enrichment analysis; GO, Genetic Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes. ns, P>0.05, *P<0.05.
4 Discussion
Infertility is characterized by failure to establish a clinical pregnancy after 12 months of regular and unprotected sexual intercourse and is estimated to affect between 8% and 12% of reproductive-aged couples worldwide. Males are responsible for 20%–30% of infertility cases but contribute to 50% of cases overall (22, 23). Therefore, it is important to understand the mechanisms underlying fertility to improve our management of this patient population. In the present study, animal experiments indicated that UBR4 represented a potential infertility gene that could induce infertility and abnormal testis structure in male Drosophila.
It is well-established that UBR4 plays a role in the nervous system (24). To our knowledge, little study has hitherto assessed the role of Ubr4 in the reproductive capability of Drosophila (25). Herein, we used bioinformatics and fly models to further confirm its important role in fertility. We provided compelling evidence that Ubr4 deficiency could induce a decline in male reproductive capacity and abnormal testis structure. Intriguingly, no significant difference was observed in egg production of Ubr4-knockdown female Drosophila. As expected, similar hatchability rates were observed in both gender groups. The egg-hatching rate was significantly different between Ubr4 knockdown and WT. The reasons might be the reduced fertility of the parents and the restricted development of the offspring. Firstly, knocking down the Ubr4 males mated with normal females may lead to a decrease in the quality and quantity of eggs produced, which may be manifested by a decrease in egg survival and an increase in the rate of pseudo-eggs. Secondly, after Ubr4 was knocked down, the genotypes of both offspring in the two gender groups were divided into four types, namely, (1) tub-Gal4/+>UAS-Ubr4-RNAi/+; (2) tub-Gal4/+>+/+; (3) +/+>UAS-Ubr4-RNAi/+; and (4) +/+>+/+, whereas the genotype of control groups was total +/+>UAS-Ubr4-RNAi/+. The egg-to-pupa hatchability can reflect the capacity of eggs to grow into pupae. Without Ubr4 gene knockdown in the offspring of the control group, it can be explained why there was a significant difference in its egg hatchability. Indeed, differences may be observed between the Ubr4 knockdown and the WT group if the quality of eggs cannot be ensured during production. Results showed that Ubr4 knockdown resulted in greater inhibition of egg hatchability compared with the hatching rate of pupa-to-adult. The reasons behind this may be that the previous stages eliminated the quality of some of the poor eggs from developing into pupae. The remaining pupae, however, have sufficient capacity to hatch into adults. As previously mentioned, the offspring gene remained consistent irrespective of the parent genotype accounting for the lack of difference between both groups. It has been reported that UBR4 encodes an E3 ubiquitination ligase (E3 enzyme), which regulates protein degradation and removes damaged or misfolded proteins from the cellular environment, thereby regulating cellular homeostasis (26). Protein error correction plays an important role in the development, reproductive inheritance, immunity, and endocrine and other important physiological processes of Drosophila (27–29).
It is now understood that UBR4, as an E3 enzyme, regulates the degradation of damaged or misfolded proteins (26). It is highly conceivable that UBR4 gene knockdown disrupts the mechanisms that degrade erroneous proteins. We examined the protein expression in the UBR4-knockdown group and the WT control group to explore the mechanism behind the phenomenon. Among the GO terms that were most affected after knocking down UBR4, Delta DNA polymerase complex, DNA polymerase complex, and Lipase activity may be associated with the mechanism of male sterility after UBR4 was knockdown. DNA polymerase complex has been reported to contribute to various disorders, including cancer and/or developmental defects (30). Lipase activity was associated with disorders of lipid metabolism, which has been reported to reduce semen quality and disrupted the blood–testis barrier integrity (31, 32). Downregulated expression of Pelo protein and upregulated Sfp65A protein was observed in the testis of UBR4-knockdown Drosophila. GO annotation indicated that Sfp65A and Pelo were associated with the reproductive process whereas CG11598 was related to biometabolic processes. Since the protein regulation ability of E3 ubiquitin ligase decreased following UBR4 knockdown in Drosophila, Sfp65A protein exhibited a significant increase.
The increased proteins were likely damaged or misfolded and escaped from the regulatory process. LIPA encodes lysosomal acid lipase (LAL), which catalyzes the breakdown of LDL to produce free fatty acids and cholesterol (33). The study by JF-SHI showed that disruption of cholesterol homeostasis in semen affects semen quality and disrupts the blood–testis barrier, leading to reduced male fertility (31, 34).
Sfp is a non-sperm component of the ejaculate, which can increase male fitness in various aspects, encompassing sperm storage promotion, temporarily improving the female egg production rate, and decreasing female sexual receptivity (35) to ensure the quality and quantity of progeny production and reduced sperm competition (36–38). Therefore, the increase in damaged or erroneous Sfp65A proteins is associated with decreased ability to protect the normal reproductive ability of male Drosophila.
The Pelo protein has been documented in the mRNA surveillance pathway. Li et al. showed that Pelo forms complexes with Hbs1 to regulate multiple processes during spermatogenesis with the help of an mRNA surveillance pathway (39). Yang et al. indicated that Pelo played a significant role in silencing transposable elements (TEs) at the translation level, which was critical for genome integrity and primarily depended on Piwi proteins and associated RNAs (40). Both researchers concluded that the Pelo protein was important in stabilizing the quality of sperm and the structure of the genitals. On the other hand, the Sfp65A protein, which was significantly upregulated by inhibiting UBR4 in Drosophila, has been associated with sperm development mediated by the Spliceosome pathway (41).
In the human cryptorchidism dataset (20), UBR4, PELO, and LIPA corresponded to the up- and downregulation of homologous genes in Drosophila in this experiment. This confirms that the effects of UBR4 on male reproduction are likely to be reflected in human cryptorchidism. In terms of diagnostic efficacy, the AUC values for all three key genes were in the range of 0.810–0.880, indicating that these genes have moderate accuracy in diagnostic tests (42) and may be promising targets for the diagnosis of cryptorchidism.
To analyze the downstream genes predicting key genes, we screened six results (MAPK14, NFKB1, MAPK13, MAPK11, MAPK9, and PIK3CD) by single-gene GSEA and showed genes to be members of the IκB family, the JNK subfamily, and the MAPK family, a group of proteins that bind directly to the transcription factor NF-κB and regulate NF-κB activity, a key regulator of NF-κB activation (43, 44), and NF-κB has previously been shown to be associated with male sterility disorders (45). In GO and KEGG analyses, transitional secretion of interleukin-12 may cause abortion in women and abnormal sperm function in men (46, 47). The stress-activated protein kinase signaling cascade in BP annotation, and the stress-activated MAPK cascade and the MAP kinase activity in MF annotation, were all relative to the MAPK signaling pathway. By linking the results with the GSEA, the results suggest that activation of the MAPK signaling pathway has an important role in key genes such as UBR4, affecting male sterility. According to the current study, the MAPK-JNK signaling pathway regulates a variety of important physiopathological effects such as cell proliferation, differentiation, stress, and inflammatory responses (48, 49). Importantly, MAPK signaling regulates male fertility and plays multiple roles in various biological processes in germ cells and is closely linked to processes such as germ cell apoptosis and epididymal maturation (50). More specifically, the MAP4K4-JNK signaling pathway stimulates the proliferation of human spermatogonial stem cells and inhibits apoptosis, which is closely associated with male infertility (51). Thus, the MAPK pathway consisting of these downstream genes may be a key part of the mechanism linking the action of genes such as UBR4. In future studies, we will further explore the specific mechanism of action of UBR4 in the MAPK signaling pathway.
In conclusion, we provide preliminary evidence that after UBR4 knockdown, male Drosophila fertility declines due to abnormal testis structure and low egg hatchability. Nonetheless, further studies are warranted to explore the underlying mechanisms.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
S-MX, J-XL, X-CC, J-DQ, and Y-LM designed the study and wrote the manuscript. C-QL, X-XZ and Y-ML performed the larva development, fertility and hatchability experiments. S-MX and J-XL performed testis dissection and analyzed the overall data. Q-WL and D-YH performed Bioinformatics analyses Q-WL and D-YH performed Bioinformatics analyses Q-WL and D-YH performed Q-WL and D-YH performed Bioinformatics analyses. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Tsinghua Fly Center for donating transgenic RNAi lines. This study was funded by Guangzhou Medical University (Funding No. 2021A021 to S-MX), Guangdong Basic and Applied Basic Research Foundation (Funding No. 2022A1515111094 to X-CC, Funding No. 2022A1515111123 to J-DQ), and Guangzhou Basic and Applied Basic Research Project (Funding No. 2023A04J0579 to Y-LM). The Guangzhou Medical University Student Innovation Ability Enhancement Plan (Funding No. 02-408-2203-2060).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Ella H, Reiss Y, Ravid T. The hunt for degrons of the 26S proteasome. Biomolecules (2019) 9(6). doi: 10.3390/biom9060230
2. Huang DT, Walden H, Duda D, Schulman BA. Ubiquitin-like protein activation. Oncogene (2004) 23(11):1958–71. doi: 10.1038/sj.onc.1207393
3. Stewart MD, Ritterhoff T, Klevit RE, Brzovic PS. E2 enzymes: more than just middle men. Cell Res (2016) 26(4):423–40. doi: 10.1038/cr.2016.35
4. Garcia-Barcena C, Osinalde N, Ramirez J, Mayor U. How to inactivate human ubiquitin E3 ligases by mutation. Front Cell Dev Biol (2020) 8:39. doi: 10.3389/fcell.2020.00039
5. Kim ST, Lee YJ, Tasaki T, Hwang J, Kang MJ, Yi EC, et al. The n-recognin UBR4 of the n-end rule pathway is required for neurogenesis and homeostasis of cell surface proteins. PloS One (2018) 13(8):e0202260. doi: 10.1371/journal.pone.0202260
6. Hegazi S, Cheng AH, Krupp JJ, Tasaki T, Liu J, Szulc DA, et al. UBR4/POE facilitates secretory trafficking to maintain circadian clock synchrony. Nat Commun (2022) 13(1):1594. doi: 10.1038/s41467-022-29244-1
7. Ling HH, Beaule C, Chiang CK, Tian R, Figeys D, Cheng HY. Time-of-day- and light-dependent expression of ubiquitin protein ligase E3 component n-recognin 4 (UBR4) in the suprachiasmatic nucleus circadian clock. PloS One (2014) 9(8):e103103. doi: 10.1371/journal.pone.0103103
8. Avila FW, Sirot LK, LaFlamme BA, Rubinstein CD, Wolfner MF. Insect seminal fluid proteins: identification and function. Annu Rev Entomol (2011) 56:21–40. doi: 10.1146/annurev-ento-120709-144823
9. Sirot LK, LaFlamme BA, Sitnik JL, Rubinstein CD, Avila FW, Chow CY, et al. Molecular social interactions: drosophila melanogaster seminal fluid proteins as a case study. Adv Genet (2009) 68:23–56. doi: 10.1016/S0065-2660(09)68002-0
10. Amoyel M, Anderson J, Suisse A, Glasner J, Bach EA. Socs36E controls niche competition by repressing MAPK signaling in the drosophila testis. PloS Genet (2016) 12(1):e1005815. doi: 10.1371/journal.pgen.1005815
11. Yu J, Lan X, Chen X, Yu C, Xu Y, Liu Y, et al. Protein synthesis and degradation are essential to regulate germline stem cell homeostasis in drosophila testes. Development (2016) 143(16):2930–45. doi: 10.1242/dev.134247
12. Davies EL, Fuller MT. Regulation of self-renewal and differentiation in adult stem cell lineages: lessons from the drosophila male germ line. Cold Spring Harb Symp Quant Biol (2008) 73:137–45. doi: 10.1101/sqb.2008.73.063
13. Spradling A, Fuller MT, Braun RE, Yoshida S. Germline stem cells. Cold Spring Harb Perspect Biol (2011) 3(11):a002642. doi: 10.1101/cshperspect.a002642
14. Senos Demarco R, Uyemura BS, D'Alterio C, Jones DL. Mitochondrial fusion regulates lipid homeostasis and stem cell maintenance in the drosophila testis. Nat Cell Biol (2019) 21(6):710–20. doi: 10.1038/s41556-019-0332-3
15. Chang YC, Tu H, Chen JY, Chang CC, Yang SY, Pi H. Reproduction disrupts stem cell homeostasis in testes of aged male drosophila via an induced microenvironment. PloS Genet (2019) 15(7):e1008062. doi: 10.1371/journal.pgen.1008062
16. Liu CQ, Qu XC, He MF, Liang DH, Xie SM, Zhan XX, et al. Efficient strategies based on behavioral and electrophysiological methods for epilepsy-related gene screening in the drosophila model. Front Mol Neurosci (2023) 16:1121877. doi: 10.3389/fnmol.2023.1121877
17. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol (2008) 26(12):1367–72. doi: 10.1038/nbt.1511
18. Davis AP, Wiegers TC, Johnson RJ, Sciaky D, Wiegers J, Mattingly CJ. Comparative toxicogenomics database (CTD): update 2023. Nucleic Acids Res (2023) 51(D1):D1257–62. doi: 10.1093/nar/gkac833
19. Foresta C, Zuccarello D, Garolla A, Ferlin A. Role of hormones, genes, and environment in human cryptorchidism. Endocr Rev (2008) 29(5):560–80. doi: 10.1210/er.2007-0042
20. Hadziselimovic F, Hadziselimovic NO, Demougin P, Krey G, Hoecht B, Oakeley EJ. EGR4 is a master gene responsible for fertility in cryptorchidism. Sex Dev (2009) 3(5):253–63. doi: 10.1159/000249147
21. Ram KR, Wolfner MF. Sustained post-mating response in drosophila melanogaster requires multiple seminal fluid proteins. PloS Genet (2007) 3(12):e238. doi: 10.1371/journal.pgen.0030238
22. Vander Borght M, Wyns C. Fertility and infertility: definition and epidemiology. Clin Biochem (2018) 62:2–10. doi: 10.1016/j.clinbiochem.2018.03.012
23. Inhorn MC, Patrizio P. Infertility around the globe: new thinking on gender, reproductive technologies and global movements in the 21st century. Hum Reprod Update (2015) 21(4):411–26. doi: 10.1093/humupd/dmv016
24. Haddadi M, Ataei R. Wde, calpA, if, dap160, and poe genes knock down drosophila models exhibit neurofunctional deficit. Gene (2022) 829:146499. doi: 10.1016/j.gene.2022.146499
25. Richards S, Hillman T, Stern M. Mutations in the drosophila pushover gene confer increased neuronal excitability and spontaneous synaptic vesicle fusion. Genetics (1996) 142(4):1215–23. doi: 10.1093/genetics/142.4.1215
26. Kim JG, Shin HC, Seo T, Nawale L, Han G, Kim BY, et al. Signaling pathways regulated by UBR box-containing E3 ligases. Int J Mol Sci (2021) 22(15). doi: 10.3390/ijms22158323
27. Phillips BP, Miller EA. Membrane protein folding and quality control. Curr Opin Struct Biol (2021) 69:50–4. doi: 10.1016/j.sbi.2021.03.003
28. Stephani M, Picchianti L, Gajic A, Beveridge R, Skarwan E, Sanchez de Medina Hernandez V, et al. A cross-kingdom conserved ER-phagy receptor maintains endoplasmic reticulum homeostasis during stress. Elife (2020) 9. doi: 10.7554/eLife.58396
29. Carr PA, Park JS, Lee YJ, Yu T, Zhang S, Jacobson JM. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res (2004) 32(20):e162. doi: 10.1093/nar/gnh160
30. Barbari SR, Shcherbakova PV. Replicative DNA polymerase defects in human cancers: consequences, mechanisms, and implications for therapy. DNA Repair (Amst) (2017) 56:16–25. doi: 10.1016/j.dnarep.2017.06.003
31. Lu JC, Jing J, Chen L, Ge YF, Feng RX, Liang YJ, et al. Analysis of human sperm DNA fragmentation index (DFI) related factors: a report of 1010 subfertile men in China. Reprod Biol Endocrinol (2018) 16(1):23. doi: 10.1186/s12958-018-0345-y
32. Wang S, Qian Z, Ge X, Li C, Xue M, Liang K, et al. LncRNA Tug1 maintains blood-testis barrier integrity by modulating Ccl2 expression in high-fat diet mice. Cell Mol Life Sci (2022) 79(2):114. doi: 10.1007/s00018-022-04142-3
33. Li F, Zhang H. Lysosomal acid lipase in lipid metabolism and beyond. Arterioscler Thromb Vasc Biol (2019) 39(5):850–6. doi: 10.1161/ATVBAHA.119.312136
34. Shi JF, Li YK, Ren K, Xie YJ, Yin WD, Mo ZC. Characterization of cholesterol metabolism in sertoli cells and spermatogenesis (Review). Mol Med Rep (2018) 17(1):705–13. doi: 10.3892/mmr.2017.8000
35. Gillott C. Male Accessory gland secretions: modulators of female reproductive physiology and behavior. Annu Rev Entomol (2003) 48:163–84. doi: 10.1146/annurev.ento.48.091801.112657
36. Fricke C, Wigby S, Hobbs R, Chapman T. The benefits of male ejaculate sex peptide transfer in drosophila melanogaster. J Evol Biol (2009) 22(2):275–86. doi: 10.1111/j.1420-9101.2008.01638.x
37. Wigby S, Sirot LK, Linklater JR, Buehner N, Calboli FC, Bretman A, et al. Seminal fluid protein allocation and male reproductive success. Curr Biol (2009) 19(9):751–7. doi: 10.1016/j.cub.2009.03.036
38. Findlay GD, Yi X, Maccoss MJ, Swanson WJ. Proteomics reveals novel drosophila seminal fluid proteins transferred at mating. PloS Biol (2008) 6(7):e178. doi: 10.1371/journal.pbio.0060178
39. Li Z, Yang F, Xuan Y, Xi R, Zhao R. Pelota-interacting G protein Hbs1 is required for spermatogenesis in drosophila. Sci Rep (2019) 9(1):3226. doi: 10.1038/s41598-019-39530-6
40. Yang F, Zhao R, Fang X, Huang H, Xuan Y, Ma Y, et al. The RNA surveillance complex pelo-Hbs1 is required for transposon silencing in the drosophila germline. EMBO Rep (2015) 16(8):965–74. doi: 10.15252/embr.201540084
41. Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Lührmann R. Conservation of the protein composition and electron microscopy structure of drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol (2009) 29(1):281–301. doi: 10.1128/MCB.01415-08
42. Akobeng AK. Understanding diagnostic tests 1: sensitivity, specificity and predictive values. Acta Paediatr (2007) 96(3):338–41. doi: 10.1111/j.1651-2227.2006.00180.x
43. Mulero MC, Huxford T, Ghosh G. NF-kappaB, IkappaB, and IKK: integral components of immune system signaling. Adv Exp Med Biol (2019) 1172:207–26. doi: 10.1007/978-981-13-9367-9_10
44. Yamamoto Y, Gaynor RB. IkappaB kinases: key regulators of the NF-kappaB pathway. Trends Biochem Sci (2004) 29(2):72–9. doi: 10.1016/j.tibs.2003.12.003
45. Chen H, Ruan YC, Xu WM, Chen J, Chan HC. Regulation of male fertility by CFTR and implications in male infertility. Hum Reprod Update (2012) 18(6):703–13. doi: 10.1093/humupd/dms027
46. Ledee-Bataille N, Dubanchet S, Coulomb-L'hermine A, Durand-Gasselin I, Frydman R, Chaouat G. A new role for natural killer cells, interleukin (IL)-12, and IL-18 in repeated implantation failure after in vitro fertilization. Fertil Steril (2004) 81(1):59–65. doi: 10.1016/j.fertnstert.2003.06.007
47. Seshadri S, Bates M, Vince G, Jones DI. The role of cytokine expression in different subgroups of subfertile men. Am J Reprod Immunol (2009) 62(5):275–82. doi: 10.1111/j.1600-0897.2009.00736.x
48. Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell (2000) 103(2):239–52. doi: 10.1016/S0092-8674(00)00116-1
49. Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer (2009) 9(8):537–49. doi: 10.1038/nrc2694
50. Luo D, He Z, Yu C, Guan Q. Role of p38 MAPK signalling in testis development and Male fertility. Oxid Med Cell Longev (2022) 2022:6891897. doi: 10.1155/2022/6891897
Keywords: male fertility, hatchability, drosophila, testes, UBR4
Citation: Xie S-M, Lai J-X, Liu C-Q, Zhang X-X, Lin Y-M, Lan Q-W, Hong D-Y, Chen X-C, Qiao J-D and Mao Y-L (2023) UBR4 deficiency causes male sterility and testis abnormal in Drosophila. Front. Endocrinol. 14:1165825. doi: 10.3389/fendo.2023.1165825
Received: 14 February 2023; Accepted: 05 June 2023;
Published: 10 July 2023.
Edited by:
Shun Bai, University of Science and Technology of China, ChinaReviewed by:
Javier Morante, Spanish National Research Council (CSIC), SpainDi Che, Guangzhou Medical University, China
Bo Zheng, Nanjing Medical University, China
Copyright © 2023 Xie, Lai, Liu, Zhang, Lin, Lan, Hong, Chen, Qiao and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing-Da Qiao, Sm9hcXVpbnFqZEAxNjMuY29t; Yu-Ling Mao, bWFveXVsaW5nMDEyMUAxNjMuY29t
†These authors have contributed equally to this work
‡ORCID: Jing-Da Qiao, orcid.org/0000-0002-4693-8390
Yu-Ling Mao, orcid.org/0000-0002-9931-6579
 Shi-Ming Xie1,2†
Shi-Ming Xie1,2† Chu-Qiao Liu
Chu-Qiao Liu Yong-Miao Lin
Yong-Miao Lin Jing-Da Qiao
Jing-Da Qiao Yu-Ling Mao
Yu-Ling Mao