
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol., 10 May 2023
Sec. Reproduction
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.1134868
 Mohamed Aboul Ezz1,2†‡
Mohamed Aboul Ezz1,2†‡ Alireza Mansouri1‡
Alireza Mansouri1‡ Ihshan Akthar1
Ihshan Akthar1 Mohamed Samy Yousef1,3
Mohamed Samy Yousef1,3 Rasoul Kowsar4
Rasoul Kowsar4 Akio Miyamoto1*
Akio Miyamoto1*Recently, we reported that sperm induce cluster of differentiation 44 (CD44) expression and Toll-like receptor 2 (TLR2)-mediated inflammatory response in bovine uterus. In the present study, we hypothesized that the interaction between CD44 of bovine endometrial epithelial cells (BEECs) and hyaluronan (HA) affects sperm attachment and thereby enhancing TLR2-mediated inflammation. To test our hypothesis, at first, in-silico approaches were employed to define the binding affinity of HA for CD44 and TLR2. Further, an in-vitro experiment using the sperm-BEECs co-culture model was applied to investigate the effect of HA on sperm attachment and inflammatory response. Here, low molecular weight (LMW) HA at different concentrations (0, 0.1, 1, or 10 µg/mL) was incubated with BEECs for 2 h followed by the co-culture without- or with non-capacitated washed sperm (106/ml) for additional 3 h was performed. The present in-silico model clarified that CD44 is a high-affinity receptor for HA. Moreover, TLR2 interactions with HA oligomer (4- and 8-mers) target a different subdomain (h-bonds) compared to TLR2-agonist (PAM3) which targets a central hydrophobic pocket. However, the interaction of LMW HA (32-mers) with TLR2 revealed no stability of HA at any pocket of TLR2. Notably, the immunofluorescence analysis revealed the HA localization in both endometrial stroma and epithelia of ex-vivo endometrial explant. Moreover, ELISA showed significant levels of HA in BEECs culture media. Importantly, BEECs pretreatment with HA prior to sperm exposure increased the number of attached sperm to BEECs, and upregulated the transcriptional levels of pro-inflammatory genes (TNFA, IL-1B, IL-8, and PGES) in BEECs in response to sperm. However, BEECs treated with HA only (no sperm exposure) did not show any significant effect on the transcript abundance of pro-inflammatory genes when compared to the non-treated BEECs. Altogether, our findings strongly suggest a possible cross-talk between sperm and endometrial epithelial cells via HA and HA binding receptors (CD44 and TLR2) to induce a pro-inflammatory response in bovine uterus.
Upon mating or artificial insemination (AI), semen provokes a physiological inflammatory reaction in the female genital tract of humans and animals (1–6). A such inflammatory reaction is characterized by the rapid and transient influx of immune cells, mainly polymorphonuclear cells (PMNs), into the uterine lumen which is critical for not only the removal of dead/excess sperm with associated contaminants (7), but also for preventing the activation of the acquired immune response towards male gametes (sperm) (8).
Bovine uterus has a well-regulated immune response to eliminate bacterial contamination after parturition, tolerate the allogenic sperm and accept semi-allogenic embryos (9). Pattern recognition receptors (PRRs), innate immune cell receptors expressed by the endometrium, have the ability to distinguish “infectious non-self” from “non-infectious self” (9–12). Among PRRs families, Toll-like receptors (TLRs) are involved in the initiation of inflammatory responses to either infections or sterile tissue injuries through recognition of highly conserved molecules called pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), respectively (13). Recently, our research group has demonstrated, via employing a series of in vitro- (14) and ex vivo (15) investigations, that active sperm attach to bovine endometrial epithelial cells (BEECs) and thereby stimulate a pro-inflammatory response through activation of TLR2 signaling pathway. However, which molecule(s) linking sperm to TLR2 pathway in BEECs is still unclear.
Different exogenous or endogenous molecules have been reported as ligands and/or regulator for TLR2 signaling (16); one of these molecules is hyaluronan (HA) (17). HA, a non-sulphated glycosaminoglycan, is normally present in most of mammalian tissues and fluids including those of reproductive system (18). Definitely, HA is secreted in the seminal- (19), oviductal- (20), uterine- (21), and cervical fluids (22) of different species including humans. As well, HA is localized to both granulosa- and cumulus cell layers of the ovarian follicles (23–25). So far, HA has gained a special relevance in the field of reproductive biology due to its participation in numerous physiological events such as ovulation (26), fertilization (27), and cervical ripening prior to labor induction (28). In the last few decades, several reports have linked HA to TLRs activation with a consequent initiation of a pro-inflammatory cascade (29) in a variety of cell types including endothelial cells, epithelial cells, fibroblasts, dendritic cells, as well as macrophages (30–34). In regard to sperm physiology, it has been shown that HA fragments, produced by sperm-released hyaluronidase, activate TLR2/4 signaling pathway with subsequent cytokine/chemokine production in the cumulus cells of cumulus-oocyte complexes (COCs) which is essential for accomplishment of fertilization process (27).
Cluster of differentiation 44 (CD44), a transmembrane protein with multiple isoforms due to its frequent alternative splicing and post-translational modifications, is a major HA receptor (35). CD44 proteins, a class of cell surface adhesion molecules, are ubiquitously expressed throughout the body (35, 36). Other than its well-known function in facilitating cell adhesion and migration, CD44 can mediate numerous cellular pathways via recruitment and assembly of signaling proteins (36). Further, CD44 with its ligand HA are involved, either via cell-cell interaction or cell-matrix interaction, in promotion of inflammatory process (36, 37). Ligation of CD44 to HA is crucial for leukocytic infiltration (38, 39), T-cell- proliferation and activation (40, 41), as well as cytokines and chemokines production (30, 42–44).
Lately, we have shown that sperm upregulate the gene and protein expression of CD44 adhesion molecule, in both BEECs- and uterine explant models, in the course of sperm-induced uterine inflammation in bovine (45). Altogether, HA could be a good candidate to act as a bridging ligand between the sperm cells from one side and CD44 of BEECs from the other side for regulating sperm attachment, and TLR2-mediated inflammation induced by the sperm. To test the above hypothesis, we first performed in-silico approaches to detect the binding affinity of HA molecules for CD44 and TLR2. Then, we determined the existence of HA in the uterine environment via immunofluorescence and enzyme-linked immunosorbent assay (ELISA). Further, sperm-BEECs in-vitro co-culture model was applied to investigate the impact of BEECs pretreatment with exogenous HA on sperm attachment and subsequent immune response.
For conducting the in-silico investigations, human HA-binding proteins [i.e., CD44 (PDB ID: 1UUH) and TLR2 glycoprotein (PDB ID: 2Z7X)] were applied; the sequence identity between human- and bovine HA binding proteins is 92.5 and 72.27 for CD44 antigen and TLR2 extracellular domain, respectively. Additionally, template-based modeling (46) showed that both proteins in human and bovine species have similar folding, as the template modelling (TM) scores of 0.8 and 0.86 were obtained for TLR2 and CD44, respectively (Supplementary Figure 1).
The three-dimensional (3-D) structure of HA with 4-, 8- and 32-mers were generated as previously described (47, 48). For optimizing of all the above molecules, molecular dynamics (MD) simulations were operated (49–52).
The optimized HA4, HA8 and HA32 structures, obtained from phase I, were used for docking studies by AutoDock VINA (v.1.2.0) (53). In this study, HA structures without any routable bond were considered to be ligands whereas the binding proteins as rigid entities were considered to be receptors. For docking calculation, the main binding sites of both proteins were selected as the grid box. To compare the interaction of HA molecules to the main binding site of TLR2, the complex of TLR2/1-Pam3CSK4 (PDB ID: 2Z7X) was used to perform MD simulation for 150 ns. PamCSK4 (PAM3) is a synthetic triacylated lipopeptide (LP) and specific agonist for TLR2 which mimics the acylated amino terminus of bacterial LPs. The ligand orientation of HA to the main binding site of receptor with the lowest binding energy were selected for further analysis and MD simulation.
MD simulation (during 150 ns) and molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) methods were used to calculate and predict the binding free energy (BFE) (Supplementary Data) (54). Radial distribution function (RDF) and center of mass (COM) were calculated to describe the atomic interaction between HA and hyaluronan binding proteins (55, 56).
The uterine samples used for immunostaining as well as isolation and culture of BEECs were brought from a local slaughterhouse (Hokkaido Livestock, Doto Plant Tokachi Factory; Obihiro, Hokkaido, Japan). Simply, the bovine uterine horns (contra-lateral to mature follicle) were carefully opened and grossly examined to be free from any abnormalities. Only healthy uterine horns (ipsi-lateral to mature follicle) from the pre-ovulatory phase (Days 19-22) were collected, immersed in physiological saline with antibiotics [1% penicillin-streptomycin (Gibco, Grand Island, NY, USA) and 1% amphotericin B (Gibco)], and then transported to the laboratory within 1-1.5 h on ice. Of note, the phase of estrous cycle was determined on the basis of corpus luteum appearance, size, and color as well as the follicular diameter (57).
The endometrial sections used for immunostaining were prepared as previously described (15). In brief, the endometrial tissue explants were dissected from the glandular (intercaruncular) endometrial regions. Then, the explants were fixed in formalin, embedded in paraffin, and 4 µm thick endometrial sections were prepared. HA localization to the bovine endometrium was determined by immunofluorescence staining using biotinylated HA-binding protein (bHABP) according to (58, 59) protocols with minor modifications. Briefly, the endometrial sections were incubated with 1% BSA for 30 min at room temperature (RT) to block the non-specific binding sites. Afterwards, the sections were incubated with bHABP (5 µg/mL, #385911, Calibiochem) overnight at 4 °C followed by labeling with Streptavidin-Alexa Fluor conjugate (2 µg/ml, #S11223, Thermo Fisher Scientific) for 1h at RT. The sections were then mounted using VECTASHIELD mounting medium with DAPI (H-1200, Vector Laboratories). Negative controls were performed by pre-digesting sections with hyaluronidase (#H3506, Sigma Aldrich) to ensure the specificity of the reaction. The fluorescence signal was captured using an all-in-one fluorescence microscope (Keyence, BZ-X800) using BZ-X GFP (OP-87763)-, and BZ-X DAPI (OP-87762) filters set for the green- and blue wavelengths, respectively. Exposure time was constant for all sections including negative control. The experiment was repeated three times using endometrial explants from three different uteri.
Following the previously described protocols (60, 61) with minor modification, BEECs were isolated (from the obtained uterine horns) and then cultured. In brief, the epithelial cells were detached and suspended in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F12 (DMEM/F12) (Gibco) supplemented with 22 mM NaHCO3 (Sigma-Aldrich, St. Louis, MO, USA), 0.1% gentamicin (Sigma-Aldrich), 1% amphotericin B (Gibco), and 10% heat-inactivated fetal calf serum (FCS) (Bio Whittaker, Walkersville, MD, USA). Then, cells were seeded in 25 cm2 culture flasks (Nalge Nunc International, Roskilde, Denmark), and cultured at 38.5˚C in a humidified atmosphere of 5% CO2 in air. Upon reaching 70-80% (sub-confluence), the cells were passaged, trypsinized, and re-seeded (at 1 x 105 cells/ml) in 1.5 ml/well culture medium (DMEM/F12, 22 mM NaHCO3, 0.1% gentamicin, 1% amphotericin, and 5% FCS) in 12 well plates (Nalge Nunc International) until sub-confluence. These plated cells were exposed to estradiol-17β (E2; 50 pg/ml) and progesterone (P4; 1 ng/ml) throughout the whole culture period to simulate the pre-ovulatory phase in situ (62, 63). The purity of epithelial cells in our model (>98%) was ensured via immunofluorescence labelling with a monoclonal antibody against cytokeratin (anti-cytokeratin 8 + 18; ab53280, Abcam, Tokyo, Japan) (64).
For getting sperm, frozen 0.5 ml semen straws were obtained from three highly fertile Holstein bulls belonging to Genetics Hokkaido Association, Hokkaido, Japan. Frozen semen straws from three bulls were thawed in a water bath at 38.5°C for 30 sec, pooled, and washed 3 times in a Tyrode’s albumin, lactate, and pyruvate medium (Sp-TALP) (65). The progressive motility of the recovered sperm, as assessed by visual examination using a light microscope equipped with a stage warmer, was around 50%.
Sub-confluent BEECs monolayers in 12-well plates were incubated in 1 ml/well culture medium (DMEM/F12, 22 mM NaHCO3, 0.1% gentamicin, 1% amphotericin, 0.1% FCS as well as E2 and P4 at the above-mentioned concentrations) supplemented with LMW HA (Select-HATM Hyaluronan; mol wt 25-75 kDa, #S0326, Sigma) at different concentrations (0, 0.1, 1, or 10 µg/mL) for 2 h followed by the co-culture without- or with non-capacitated washed sperm (106/ml) for additional 3 h (64). This experiment was repeated five times using epithelial cells from five different uteri.
At the end of co-culture period, BEECs-conditioned media were collected, centrifuged twice at 1000 g for 10 min at 4 °C and kept at -80 °C for HA quantification. Commercially available HA ELISA kit (DHYALO, R&D Systems, Minneapolis, MN) was used for determination of HA concentrations in BEECs-conditioned media. A 50 µL aliquot of each sample was analyzed according to the manufacturer’s instructions. All samples were run in duplicate. Optical density (OD) readings were performed at 450 nm. Control group was run to determine the baseline concentration of HA in DMEM culture media. This experiment was repeated four times using epithelial cells from four different uteri.
To determine the number of attached sperm, BEECs monolayers in 12-well plates (1 ml/well culture medium without- or with HA) were exposed to washed sperm (106/ml) for 30 min followed by video capturing using a light microscope (at 200 x magnification) equipped with a stage warmer and digital camera connected to EOS utility software® (Canon U.S.A., Inc.); the focus was adjusted during video capturing to visualize all attached sperm. Five random fields were captured per each group. Of note, videos of the different groups within the same experiments were captured under the same field area and video setting (15).
On the other hand, to determine the number of sperm remained attached, BEECs monolayers in 12-well plates (2 ml/well culture medium without- or with HA) were exposed to 106 sperm/well for 3 h. Afterwards, the upper 1.5 ml media were very gently aspirated, centrifuged at 1000 x g, and the pellet was then re-suspended and counted independently by two investigators using a hemocytometer. To calculate the number of sperm remained attached to BEECs, we subtracted the number of detached sperm (dead and/or floating) from the total number of sperm (106/well) (64).
At the end of sperm-BEECs co-culture, RNA was extracted from BEECs using Trizol reagent (Invitrogen, Carlsbad, CA, USA), quantified by means of a NanoDrop Spectrophotometer 2000c (Thermo Scientific, Waltham, MA, USA), and then pure RNA samples (i.e., A260/A280 ratio were between 1.8 and 2.0) were kept in RNA storage solution (Ambion, Austin, TX, USA) at -80°C till cDNA synthesis (66).
The synthesis of cDNA was performed as previously decribed (67) with minor modifications. First, the extracted RNA was subjected to a DNase treatment step using RQ1 RNase-Free DNase kit (Promega, Madison, WI, USA) to remove residual genomic DNA as well as other contaminants. At such step, 1 μg of extracted RNA was incubated with the first mixure [1 μl of RQ1 RNase-free DNase 10X Reaction Buffer, 2 μl of RQ1 RNase-free DNase (1 unit/μl), and Nuclease-free water (Invitrogen, Carlsbad, CA, USA) to a total volume of 10 μl] for 30 min at 37°C in a thermal cycler (Eppendorf, Hamburg, Germany), then 1 μl of the RQ1 DNase Stop solution was added for 10 min at 65°C to stop this reaction. Afterwards, the first-strand cDNA was produced via SuperScript II Reverse Transcriptase kit (Invitrogen) according to the manufacturer instructions. In brief, the DNase-treated RNA was incubated with the second mixture [1.5 μl of 3 μg/μl random primer, 1.5 μl of 10 mM PCR Nucleotide Mix (dNTP) (Roche Diagnostics, Indianapolis, IN, USA) and Nuclease-free water to a total volume of 18 μl] at 65°C for 5 min. Then, the third mixture [6 μl of 5X First-Strand Buffer, 3 μl of 0.1M dithiothreitol and 1.5 μl of 40 units/μl Ribonuclease Inhibitor Recombinant (Toyobo, Osaka, Japan)] was added per each tube and incubated at 42°C for 2 min. Finally, 0.2 μl of 200 units/μl SuperScript II Reverse Transcriptase was added and the thermal cycler was programmed at 25°C for 10 min, 42°C for 50 min and then 70°C for 15 min. The synthesized cDNA was stored at -30°C.
The transcriptional levels of TLR2, Tumor necrosis factor (TNF) alpha (TNFA), Interleukin (IL)-1 beta (IL-1B), IL-8, and Prostaglandin E synthase (PGES) were detected via a quantitative real-time polymerase chain reaction (PCR) by means of an iQ5 real-time PCR detection system (Bio-Rad Laboratories, Tokyo, Japan). To clarify, a total 10 μl reaction mix [i.e., 2 μl/sample synthesized cDNA, 5 μl of QuantiTect SYBR Green PCR Master Mix (QIAGEN, Hilden, Germany), 0.2 μl of the targeted primer pairs (listed in Supplementary Table 1), and 2.8 μl nuclease-free water (Invitrogen)] was run in amplification program with an initial denaturation step at 95°C for 15 min, followed by 40 cycles of denaturation at 95°C for 15 sec, annealing at 51°C for 30 sec, extension at 72°C for 20 sec. A negative control (reactions containing nuclease-free water or non-reverse transcribed RNA) were involved in each run. It should be emphasized that we here used Primer Express Software v3.0.1 (Thermo Scientific) to design the used primers’ pairs. The calculated cycle threshold (Ct) values were normalized against ACTB (β-actin); no significant variances were detected in β-actin mRNA expression among the different treatments. The delta-delta Ct (2–ΔΔCt) method was applied to estimate the fold change between the different samples (64, 67, 68).
Each experiment was repeated at least three times using epithelial cells from 3–4 different uteri. In each uterus, 3 replicates were performed (3 wells per treatment per experiment) and data are presented as mean ± standard error of the mean (SEM). Student’s t-test was applied to compare the data between two groups, while one-way ANOVA followed by Tukey’s multiple comparisons test was used for more than two groups. The results were considered statistically significant at P< 0.05 and P< 0.0001.
The determination of initial structure of HA-receptors complex for MD simulations and energy analysis have been performed using docking simulations. The top three binding orientations of HA with main binding site of proteins with estimated BFEs through Autodock VINA were detected (Supplementary Figure 2). HA showed a potential interaction with CD44 through crystallographic mode (residues presented in the main binding site like Ile 96, Tyr 79, Ser 109, etc.). HA was not able to interact with the internal pocket of TLR2 and the residues involved in TLR2-HA interaction (mainly hydrogen bonds) were different form TLR2-PAM3 interaction (hydrophobic bonds) through Ligplot analysis (69) (Supplementary Figure 3).
All HA polymers (HA4, HA8 and HA32) could not stay in the vicinity of the main binding site of TLR2 during MD simulation (Figure 1). However, CD44 has the main binding domain for all HAs. Moreover, the BFE of HAs-CD44 proposed the high affinity of HA to this receptor (-723.4, -1272.5 and -1680.6 kJ/mol for HA4-CD44, HA8-CD44 and HA32-CD44, receptively). Additionally, HA with higher number of monomer (HA32) showed stronger interaction to the main binding site of CD44 comparing to HA4 or HA8, since more residues in the binding pocket involved in the h-bond interaction (Supplementary Figure 4). RDF (calculated during the last 10 ns simulation time) of the complex of HA in the main binding site of receptors was showed in Figure 2A. RDF calculated for PAM3 and HAs induced individual peaks for PAM3 at TLR2 main binding distance of ∼ 5Å. As for HA4 and HA8 in HA-TLR2 complexes, the peaks of RDF were appeared at distance of ~ 30 Å. Regarding CD44 crystallographic mode, the RDF of HA4, HA8 and HA32 has peaks at ~ 7Å which indicating a high tendency of HA to CD44.
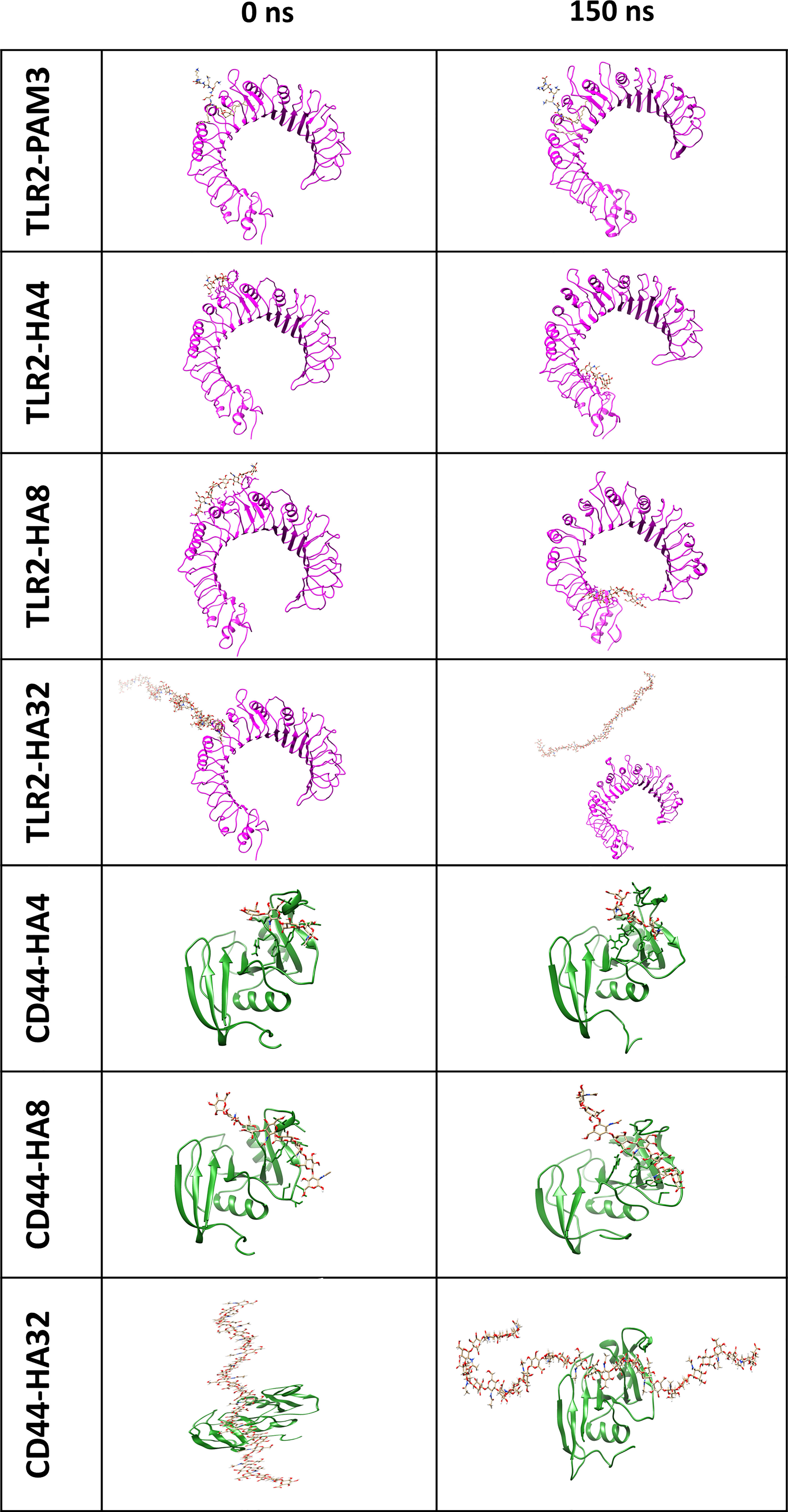
Figure 1 The first and final snapshots of HA/TLR2 and HA/CD44 complexes (with representation of magenta and green ribbon for TLR2 and CD44, respectively and stick for HA) after 0 and 150 ns simulation time. Note that, HA could not stay and bind to the main binding site of TLR2, as PAM3 during 150 MD simulation time, however, HA oligomers (HA4 and HA8) moved and stayed in peripheral domain of TLR2. Concerning CD44, HA indicated a strong affinity since the location of HA did not change the after applying 150 ns simulation time.
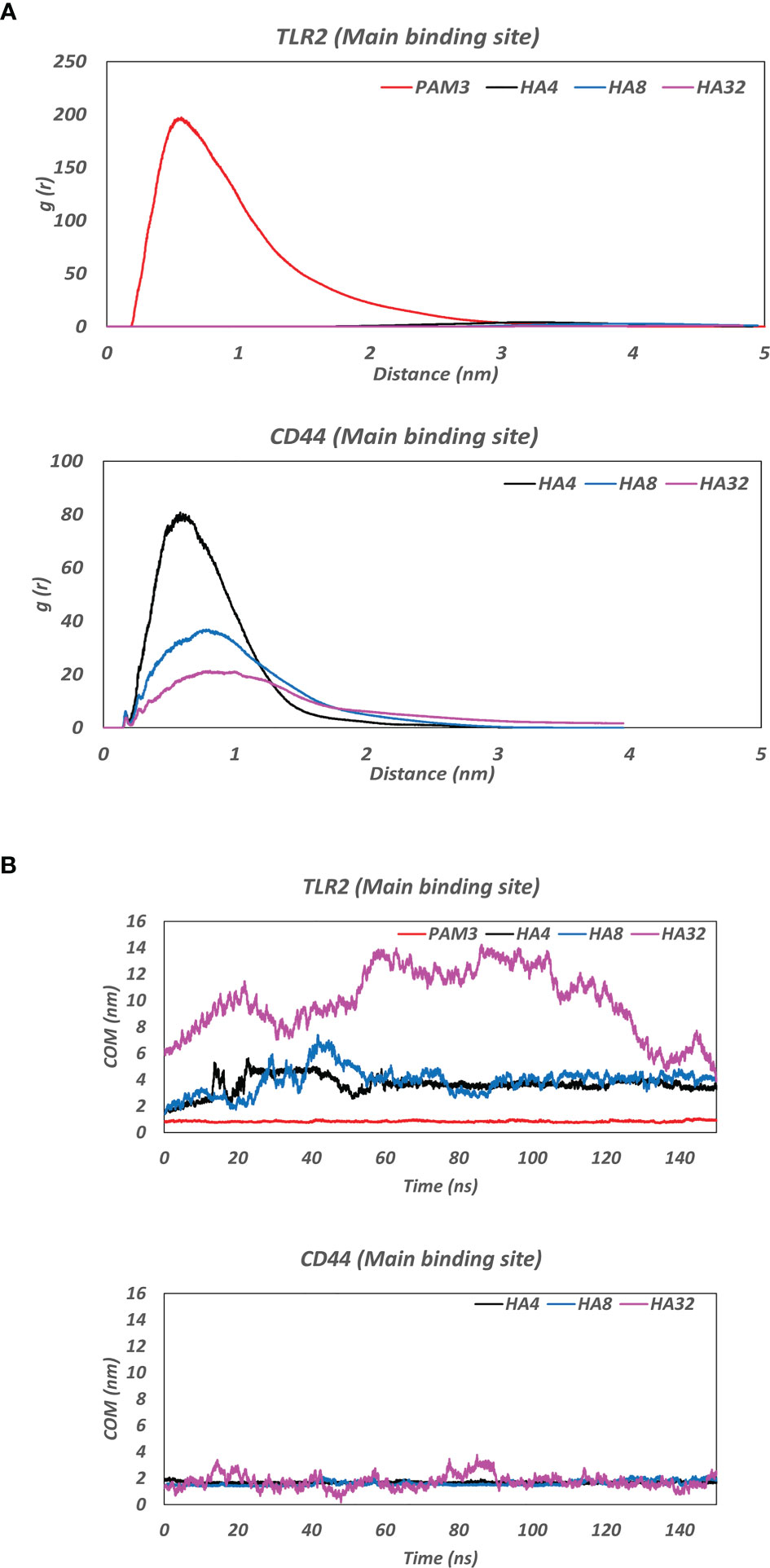
Figure 2 (A) Radial distribution function (RDF) of the complex of HA and the main binding site of receptors (TLR2 and CD44) during the last 10 ns MD simulation. Note the clear peak for PAM3-TLR2 at ∼ 5Å and for HAs-CD44 at ~ 7Å which indicating a high affinity of ligands (PAM3 and HA) to their receptors (TLR2 and CD44, respectively). (B) The average of center of mass (COM) distance between HA and the main binding site of receptors (TLR2 and CD44) during 150ns MD simulation. Note the fluctuations between TLR2 before 60 ns MD. COM distance between both HAs and CD44 was nearly constant around 2nm throughout the 150 ns MD.
Figure 2B illustrates the distance averages of COM between HAs and main binding sites of receptors. Regarding the main binding site of TLR2, there were fluctuations between 3 and 4 nm for HA4 and HA8 and became stable after 60 ns, as they moved and stabilized into another subdomain in TLR2. However, as for HA32, the ligand failed to interact with any pockets of TLR2. Looking at CD44, the fluctuation of COM distance was not changed during 150 simulation time (the amplitude of the oscillations is between around 1.5 and 2 nm for all HA). HA tried to preserve the initial distance (positions obtained from docking simulation) during 150 ns MD simulation time.
To ensure the localization of HA to bovine endometrium, the endometrial sections were subjected to immunostaining using bHABP. Indeed, HA was localized to bovine endometrium. Although a higher expression was observed in the endometrial stroma, HA was also expressed by luminal- and glandular epithelia of bovine endometrium (Figure 3). Negative control sections treated with hyaluronidase showed no staining, demonstrating the specificity of the staining.
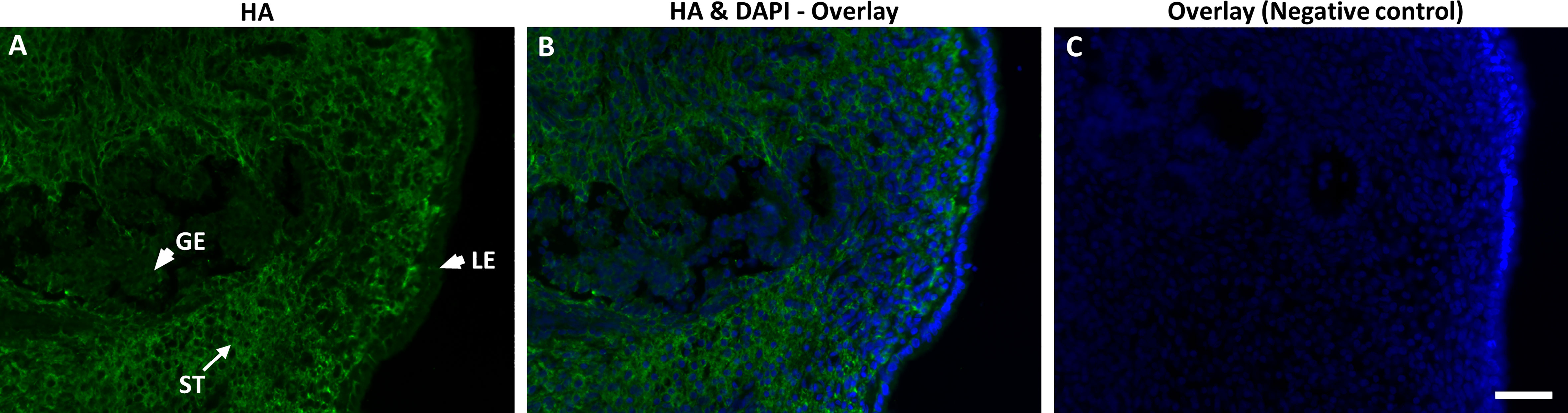
Figure 3 Immunofluorescence localization of hyaluronan (HA) using biotinylated HA-binding protein (bHABP) in bovine endometrium at pre-ovulatory stage visualized using streptavidin-Alexa Fluor conjugate. (A, B) HA shown as green staining while (B, C) nuclei stained blue with DAPI. Arrows indicate strong expression of HA in stroma (ST), while arrowheads indicate weak expression in luminal (LE) and glandular epithelium (GE). (C) The negative control sections pre-incubated with hyaluronidase shows no staining. Bar = 50µm.
Concurrently, the conditioned media in-vitro produced via incubating the culture media with BEECs monolayers showed detectable levels of HA with an average of 16.05 ± 2.33 ng/ml.
Accordingly, it was essential to elucidate the possible role(s) of HA in sperm-BEECs crosstalk. BEECs monolayers were enriched with different concentrations of HA (0, 0.1, 1, or 10 µg/mL) for 2 h prior to the co-culture with sperm for further 3 h. At a glance, we observed that the number of attached sperm was significantly higher in BEECs treated with HA (at either 1 or 10 µg/mL) than in the non-treated BEECs (Figure 4A). To confirm such observation, we quantified the number of sperm remained attached to BEECs at the end of co-culture period (3 h). Likewise, BEECs pretreatment with HA dose-dependently increased the number of remained attached sperm (Figure 4B).
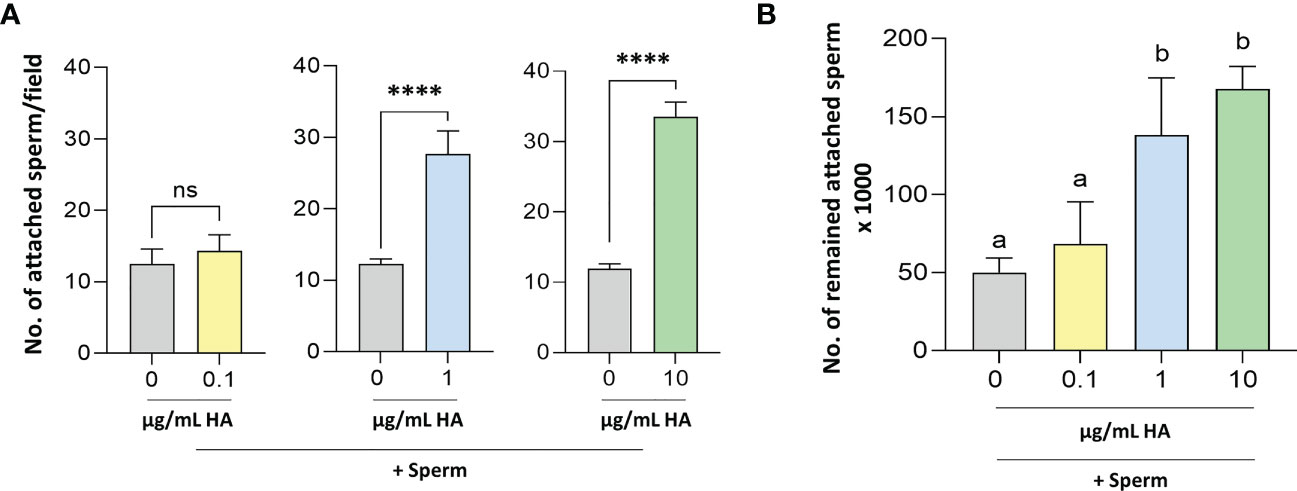
Figure 4 Determination of the number of (A) attached and (B) remained attached sperm to BEECs pretreated without- with HA. Sperm cells at 106/ml Sp-TALP were exposed to HA (at 0, 0.1, 1, or 10 µg/mL) for 0, 30, 60, and 120 min. Data are presented as mean ± SEM of 3 independent experiments using epithelial cells from 3 different uteri (3 wells per treatment per experiment). Different letters denote a significant variance (P<0.05) between the different groups when compared with One-way ANOVA followed by Tukey’s multiple comparisons test, while asterisks denote a significant variance (**** means P<0.0001) between the defined groups when compared using Student’s t-test.
To exclude the possibility that HA could adversely affect sperm dynamics, sperm cells at 106/ml were exposed to HA at the above concentrations for 2 h. Then, the progressive motility of recovered sperm was assessed at 0-, 30-, 60-, and 120-min post-exposure. Importantly, HA did not affect sperm progressive motility over the exposure period (Supplementary Figure 5).
Afterwards, it was crucial to investigate the effect of BEECs pretreatment with HA on sperm-induced inflammation. Therefore, BEECs monolayers were co-incubated with HA at either 0, 0.1, 1, or 10 µg/mL for 2 h followed by the co-culture with sperm for additional 3 h. Then, mRNA expressions of TLR2, pro-inflammatory- cytokines (TNFA and IL-1B) and chemokines (IL-8) as well as PGES were quantified in BEECs via a real-time PCR. Our data showed that the pre-incubation of BEECs with HA at lower concentration (0.1 µg/mL) has no effect on TLR2, TNFA, IL-1B, IL-8, and PGES mRNA expressions in BEECs triggered with sperm (Figure 5A), while HA at medium concentration (1 µg/mL) increases the stimulatory effect of sperm on the abundances of TLR2, TNFA, IL-1B, IL-8, and PGES transcripts in BEECs (Figure 5B). However, we recorded lower transcriptional levels of the aforementioned genes upon sperm co-culture with BEECs treated with HA at higher concentration (10 µg/mL) when compared to the non-treated BEECs (Figure 5C).
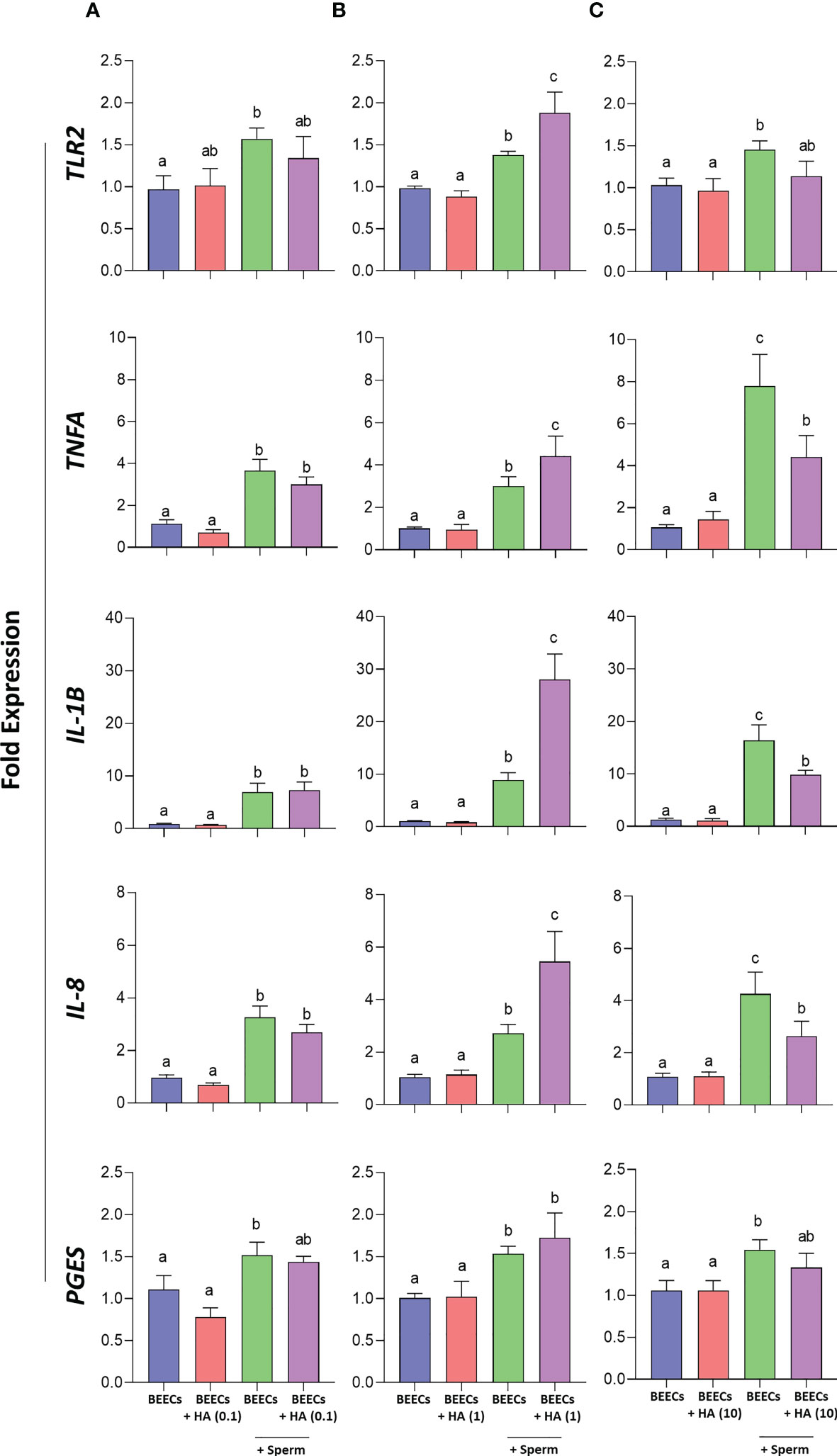
Figure 5 Effect of BEECs pretreatment with HA on sperm- induced inflammation. (A–C) BEECs monolayers were incubated without- or with HA (at 0.1, 1, or 10 µg/mL) for 2 h prior to the co-culture with 106 sperm/mL for 3 h. Then, mRNA expressions of TLR2, TNFA, IL-1B, IL-8, and PGES were quantified in BEECs via a real-time PCR. Data are presented as mean ± SEM of 3 independent experiments using epithelial cells from 3 different uteri (3 wells per treatment per experiment). Different letters denote a significant variance (P<0.05) between the different groups when compared with One-way ANOVA followed by Tukey’s multiple comparisons test.
In support of our previous investigations concerning sperm-maternal immunological crosstalk (14, 15, 45, 64, 67, 70, 71), the present study using in-silico analyses combined with in-vitro co-culture model of sperm with endometrial epithelial cells provides several lines of evidence that HA, primarily through CD44 interaction, has the capacity to facilitate sperm attachment to the endometrial epithelia and thereby triggering sperm-induced inflammatory response in bovine uterus via TLR2 signaling transduction.
HA, the most abundant glycosaminoglycans in female reproductive organs (18), interact with CD44 but could not interact with the main binding site of TLR2. The present study showed that HA has no affinity to the main binding site of TLR2, compared with PAM3 during MD simulation time. Notably, the main binding site of TLR2 is the internal hydrophobic channel, which highly attract lipoprotein, such as PAM3 (72).
According to the Ligplot result, the main bond types for HA interaction were hydrogen bonds (h-bonds). For instance, HA create h-bonds with residues of CD44 (Arg 41, Tyr 42 etc.) to interact strongly. TLR2 presented h-bonds (such as Arg 508, Arg 447 and Arg 486) in a peripheral subdomain, away from the main pocket, which involved in the HA-TLR2 interaction (for 4- and 8-mers HA). It is obvious that polar and electricity charged amino acids play an important role in HA binding affinity. CD44 has three topographically main binding for HA, however HA has the high potential affinity only to crystallographic mode (47, 73). In harmony to previous literatures (47, 73), our computational analysis confirmed the strong interaction between crystallographic binding mode of CD44 with all HAs (BFE: -723.4, -1272.5 and -1680.6 kJ/mol for HA4, HA8 and HA32 respectively). Hence, our in-silico investigations strongly suggest that HA has higher binding affinity to the main binding site of CD44 than TLR2.
Based on the above computational analyses, it was fundamental to confirm the existence of HA in bovine uterus. Importantly, the immunohistochemical detection of HA in the pre-ovulatory uterus showed its localization to both endometrial stroma and epithelia. The staining revealed that HA were strongly localized within the stroma and weakly expressed by luminal and glandular epithelium of the endometrium. Similar observations (i.e., intense expression in stroma) were reported in other species such as in ovine (74) and mouse (21) endometrium. In ovine species, HA was expressed strongly in the follicular stage endometrium compared to luteal stage (74). In bovine oviducts, HA was strongly localized to the stroma of the oviductal villi and no expression was observed in the luminal epithelium (59). The observed lower expression of HA in endometrial epithelium may be due to the release of luminal and glandular epithelial HA into the uterine cavity. It is likely that, the lower level of epithelial HA would favor a diffusion from stroma to epithelium which could lead to a dynamic releasing of HA from stroma to the uterine lumen. In support of this, the present ELISA result demonstrated that BEECs have the ability to release detectable levels of HA into the culture media over the co-incubation period.
Accordingly, we next aimed to investigate the effect of BEECs enrichment with HA prior to the co-culture with sperm on the subsequent sperm-BEECs interaction. Interestingly, BEECs pre-enrichment with HA dose-dependently increased the number of sperm attached to BEECs; such phenomenon could be referred to HA-CD44 interaction. In light of our in-silico investigations, HA has a much stronger binding affinity to CD44 than TLR2. As well, we have previously reported that CD44 adhesion molecule plays a principal role in sperm attachment to the endometrial epithelia in bovine uterus; the addition of anti-CD44 neutralizing antibody negatively impacted sperm-BEECs interaction (45). Besides, it has been reported that sperm cells could express CD44 (75). Altogether, we speculate that the exogenous HA added to BEECs culture media prior to sperm exposure could act as bridging ligand between sperm from one side and CD44 of BEECs from the other side. Such model has been extensively studied in leukocyte trafficking. Namely, leukocytic infiltration from the bloodstream into inflamed tissues or organs requires binding interactions between adhesion molecules on leukocytes from one side and endothelial cells from the other side; HA-CD44 interaction has been implicated in regulation of rolling-, adhesion- as well as invasion processes ending with leukocytic infiltration (36).
Since sperm attachment to the endometrial epithelia is a prerequisite for promotion of a pro-inflammatory response in bovine uterus via activation of TLR2 signaling pathway (14, 45, 64, 70), it was necessary to determine whether enhancing sperm attachment by the aid of BEECs pretreatment with HA could stimulate a much stronger inflammatory response in BEECs. Strikingly, our PCR data showed that BEECs pretreatment with medium HA concentration (i.e., 1 µg/mL) upregulates mRNA expressions TLR2, pro-inflammatory- cytokines (TNFA and IL-1B) and chemokines (IL-8) as well as prostaglandins E synthesis (PGES). However, higher exogenous HA concentrations (i.e., 10 µg/mL) added to BEECs prior to exposure to sperm weakened sperm-triggered inflammation in BEECs.
In the past few decades, HA was considered as inert constituent of extracellular matrix, but it is currently categorized as “dynamic” molecule with a continuous turnover to HA molecules of various sizes: high molecular weight (HMW) HA, low molecular weight (LMW) HA as well as oligosaccharides. Under the physiological conditions, HMW HA primarily contributes to tissue integrity (76, 77). Upon tissue injury, HMW HA molecules are rapidly degraded into LMW HA molecules, referred as HA fragments. Via binding with HA receptors, mainly CD44 adhesion molecule (37), these HA fragments have the capacity to activate a pro-inflammatory response with a subsequent transcription of pro-inflammatory genes (29) such as cytokines (TNFA, IL-8, and IL-12), chemokines [macrophage inflammatory protein (MIP), keratinocyte chemoattractant (KC), macrophage chemoattractant protein-1 (MCP-1), and IFN induced protein-10], matrix-modifying enzymes (MMEs), inducible nitric oxide synthase (iNOS), as well as plasminogen activator inhibitor (30, 42, 44, 78, 79). Concurrent with their capability to induce inflammation through CD44 interaction, HA fragments can signal through a TLR-mediated pathway. Using various murine models, it has been established that HA fragments stimulate cytokine and chemokine production through activation of TLR2/4 signaling pathway in dendritic cells (80), macrophages (81), as well as COCs (27).
On the other hand, it has been shown that CD44 plays a role in stimulating TLR2 downstream targets with a subsequent progression of osteoarthritis as evidenced by the upregulation of NFκB-, IL-1B- as well as TNFA gene expression in human macrophages. This study demonstrated that a reduction in CD44 levels in macrophages, via using CD44- specific antibody or knockdown, prior to TLR2 activation downregulates NF-κB transcription and thereby lowers proinflammatory cytokines’ (IL-1B and TNFA) production. In addition, they reported that pretreatment of human macrophages with higher doses of HA (100, 250, or 500 µg/mL) dose-dependently inhibits their pro-inflammatory response upon TLR2 ligation (82). Based on the latter finding, we can conclude that BEECs treatment with a higher dose of HA (10 µg/mL) prior to sperm exposure could reduce CD44-meditated TLR2 activation and thereby diminish sperm-induced inflammation in BEECs.
Importantly, it has been shown that both TLR2 and CD44 molecules can function as coreceptors after stimulation of immune cells with TLR2 agonist (Zymosan) (83). Moreover, HA can form a triple complex with CD44 and TLR2 to promote the invasiveness and pro-inflammatory environment in cancer cells (84). Therefore, it seems that sperm-uterine immune-crosstalk could be regulated by CD44 and TLR2 under the effect of HA. However further investigations are necessary to clarify this phenomenon. Moreover, additional studies are required to identify the post-translational modification (PTMs) and how polymorphism affect TLR2 response toward different ligands in bovine species.
In conclusion, the present in-silico and in-vitro investigations proposed a possible cross-talk between sperm and uterine epithelial cells via hyaluronan and hyaluronan binding receptors (CD44 and TLR2), to induce pro-inflammatory response in bovine uterus (Figure 6). The physiological molecule HA increase the sperm attachment to BEECs via CD44 which in turn activate TLR2 signaling pathway ending with transcription of the pro-inflammatory genes in response to sperm. Further investigations are required to define the specific molecule(s) from the sperm side which regulate the TLR2 to induce inflammation after sperm attachment.
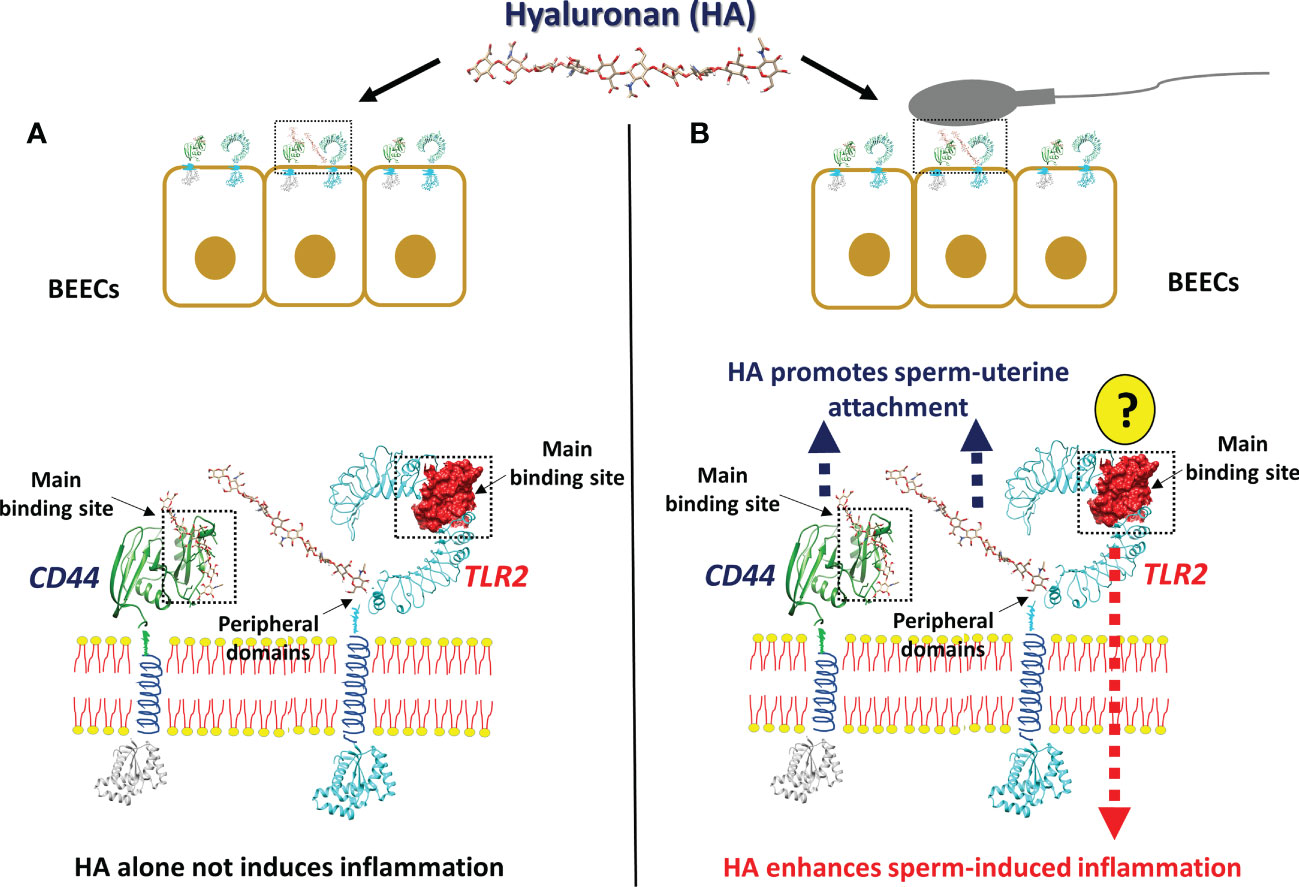
Figure 6 The working hypothesis of the impact of Hyaluronan (HA) on sperm-induced inflammation in BEECs. The anticipated model indicating that, stimulation of BEECs with the most abundant endogenous ligands and molecules in bovine uterine lumen HA, prior to sperm co-culture, accelerate sperm attachment and subsequent inflammation, due to the strong interaction between HA and its receptors. (A) HA alone is unable to induce inflammation, since it is unable to interact with the main binding site of TLR2 which is responsible for the triggering of inflammation. (B) In the presence of sperm, HA could have an interaction with the main binding site of CD44 and with the peripheral domains of TLR2, which is involved in sperm attachment. After the sperm attachment, the unknown molecules (?) from sperm side should trigger the main binding site of TLR2 to induce inflammatory cascade.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by committee on the ethics of animal experiments of the Obihiro University of Agriculture and Veterinary Medicine, Japan (Permit number 27-74).
ME, AlM, IA, MY, RK, and AkM conceived and designed the experiments. ME, AlM, IA, and MY performed the experiments. ME, AlM, IA, and MY analyzed the data. AlM, RK, and AkM provided reagents/materials/analysis tools. ME, AlM, IA, MY, RK, and AM wrote the manuscript. All authors contributed to the article and approved the submitted version.
The present research work was funded by a Grant-in-Aid for Scientific Research (20H03122 and 23H02356) of Japan Society for the Promotion of Science (JSPS), and in part by Livestock Promotional Funds of Japan Racing Association (JRA).
The authors thank Genetics Hokkaido Association, Shimizu-Cho, Hokkaido, Japan, for providing cryopreserved semen straws used in this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1134868/full#supplementary-material
1. Kaeoket K, Persson E, Dalin AM. Influence of pre-ovulatory insemination and early pregnancy on the infiltration by cells of the immune system in the sow endometrium. Anim Reprod Sci (2003) 75(1):55–71. doi: 10.1016/S0378-4320(02)00230-0
2. McMaster MT, Newton RC, Dey SK, Andrews GK. Activation and distribution of inflammatory cells in the mouse uterus during the preimplantation period. J Immunol (1992) 148(6):1699–705. doi: 10.4049/jimmunol.148.6.1699
3. O’Leary S, Jasper MJ, Warnes GM, Armstrong DT, Robertson SA. Seminal plasma regulates endometrial cytokine expression, leukocyte recruitment and embryo development in the pig. Reproduction (2004) 128(2):237–47. doi: 10.1530/rep.1.00160
4. Scott JL, Ketheesan N, Summers PM. Spermatozoa and seminal plasma induce a greater inflammatory response in the ovine uterus at oestrus than dioestrus. Reprod Fertil Dev (2009) 21(7):817–26. doi: 10.1071/rd09012
5. Thompson LA, Barratt CL, Bolton AE, Cooke ID. The leukocytic reaction of the human uterine cervix. Am J Reprod Immunol (1992) 28(2):85–9. doi: 10.1111/j.1600-0897.1992.tb00765.x
6. Canisso IF, Segabinazzi LGTM, Fedorka CE. Persistent breeding-induced endometritis in mares - a multifaceted challenge: from clinical aspects to immunopathogenesis and pathobiology. Int J Mol Sci (2020) 21(4):1432. doi: 10.3390/ijms21041432
7. Katila T. Post-mating inflammatory responses of the uterus. Reprod Domest Anim (2012) 47 Suppl 5:31–41. doi: 10.1111/j.1439-0531.2012.02120.x
8. Hansen PJ. The immunology of early pregnancy in farm animals. Reprod Domest Anim (2011) 46 Suppl 3:18–30. doi: 10.1111/j.1439-0531.2011.01850.x
9. Robertson SA. Control of the immunological environment of the uterus. Rev Reprod (2000) 5(3):164–74. doi: 10.1530/ror.0.0050164
10. Bazer FW, Wu G, Spencer TE, Johnson GA, Burghardt RC, Bayless K. Novel pathways for implantation and establishment and maintenance of pregnancy in mammals. Mol Hum Reprod (2010) 16(3):135–52. doi: 10.1093/molehr/gap095
11. Quayle AJ. The innate and early immune response to pathogen challenge in the female genital tract and the pivotal role of epithelial cells. J Reprod Immunol (2002) 57(1-2):61–79. doi: 10.1016/s0165-0378(02)00019-0
12. Janeway CA Jr., Medzhitov R. Innate immune recognition. Annu Rev Immunol (2002) 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359
13. Koga K, Mor G. Expression and function of toll-like receptors at the maternal-fetal interface. Reprod Sci (2008) 15(3):231–42. doi: 10.1177/1933719108316391
14. Ezz MA, Marey MA, Elweza AE, Kawai T, Heppelmann M, Pfarrer C, et al. TLR2/4 signaling pathway mediates sperm-induced inflammation in bovine endometrial epithelial cells in vitro. PloS One (2019) 14(4):e0214516. doi: 10.1371/journal.pone.0214516
15. Akthar I, Suarez SS, Morillo VA, Sasaki M, Ezz MA, K-i T, et al. Sperm enter glands of preovulatory bovine endometrial explants and initiate inflammation. Reproduction (2020) 159(2):181–92. doi: 10.1530/REP-19-0414
16. Erridge C. Endogenous ligands of TLR2 and TLR4: agonists or assistants? J Leukoc Biol (2010) 87(6):989–99. doi: 10.1189/jlb.1209775
17. Ebid R, Lichtnekert J, Anders HJ. Hyaluronan is not a ligand but a regulator of toll-like receptor signaling in mesangial cells: role of extracellular matrix in innate immunity. ISRN Nephrol (2014) 2014:714081. doi: 10.1155/2014/714081
18. Fouladi-Nashta AA, Raheem KA, Marei WF, Ghafari F, Hartshorne GM. Regulation and roles of the hyaluronan system in mammalian reproduction. Reproduction (2017) 153(2):R43–r58. doi: 10.1530/rep-16-0240
19. Kershaw-Young CM, Evans G, Maxwell WM. Glycosaminoglycans in the accessory sex glands, testes and seminal plasma of alpaca and ram. Reprod Fertil Dev (2012) 24(2):362–9. doi: 10.1071/rd11152
20. Tienthai P, Kjellén L, Pertoft H, Suzuki K, Rodriguez-Martinez H. Localization and quantitation of hyaluronan and sulfated glycosaminoglycans in the tissues and intraluminal fluid of the pig oviduct. Reprod Fertil Dev (2000) 12(3-4):173–82. doi: 10.1071/rd00034
21. Teixeira Gomes RC, Verna C, Nader HB, dos Santos Simões R, Dreyfuss JL, Martins JR, et al. Concentration and distribution of hyaluronic acid in mouse uterus throughout the estrous cycle. Fertil Steril (2009) 92(2):785–92. doi: 10.1016/j.fertnstert.2008.07.005
22. Perry K, Haresign W, Wathes DC, Khalid M. Hyaluronan (HA) content, the ratio of HA fragments and the expression of CD44 in the ovine cervix vary with the stage of the oestrous cycle. Reproduction (2010) 140(1):133–41. doi: 10.1530/rep-09-0424
23. Kimura N, Konno Y, Miyoshi K, Matsumoto H, Sato E. Expression of hyaluronan synthases and CD44 messenger RNAs in porcine cumulus-oocyte complexes during In vitro Maturation1. Biol Reprod (2002) 66(3):707–17. doi: 10.1095/biolreprod66.3.707
24. Schoenfelder M, Einspanier R. Expression of hyaluronan synthases and corresponding hyaluronan receptors is differentially regulated during oocyte maturation in Cattle1. Biol Reprod (2003) 69(1):269–77. doi: 10.1095/biolreprod.102.011577
25. Chavoshinejad R, Marei WF, Hartshorne GM, Fouladi-Nashta AA. Localisation and endocrine control of hyaluronan synthase (HAS) 2, HAS3 and CD44 expression in sheep granulosa cells. Reprod Fertil Dev (2016) 28(6):765–75. doi: 10.1071/rd14294
26. Salustri A, Yanagishita M, Hascall VC. Synthesis and accumulation of hyaluronic acid and proteoglycans in the mouse cumulus cell-oocyte complex during follicle-stimulating hormone-induced mucification. J Biol Chem (1989) 264(23):13840–7. doi: 10.1016/S0021-9258(18)80077-1
27. Shimada M, Yanai Y, Okazaki T, Noma N, Kawashima I, Mori T, et al. Hyaluronan fragments generated by sperm-secreted hyaluronidase stimulate cytokine/chemokine production via the TLR2 and TLR4 pathway in cumulus cells of ovulated COCs, which may enhance fertilization. Development (2008) 135(11):2001–11. doi: 10.1242/dev.020461
28. El Maradny E, Kanayama N, Kobayashi H, Hossain B, Khatun S, Liping S, et al. The role of hyaluronic acid as a mediator and regulator of cervical ripening. Hum Reprod (1997) 12(5):1080–8. doi: 10.1093/humrep/12.5.1080
29. Petrey AC, de la Motte CA. Hyaluronan, a crucial regulator of inflammation. Front Immunol (2014) 5:101. doi: 10.3389/fimmu.2014.00101
30. McKee CM, Penno MB, Cowman M, Burdick MD, Strieter RM, Bao C, et al. Hyaluronan (HA) fragments induce chemokine gene expression in alveolar macrophages. the role of HA size and CD44. J Clin Invest (1996) 98(10):2403–13. doi: 10.1172/jci119054
31. Termeer CC, Hennies J, Voith U, Ahrens T, Weiss JM, Prehm P, et al. Oligosaccharides of hyaluronan are potent activators of dendritic cells. J Immunol (2000) 165(4):1863–70. doi: 10.4049/jimmunol.165.4.1863
32. Beck-Schimmer B, Oertli B, Pasch T, Wüthrich RP. Hyaluronan induces monocyte chemoattractant protein-1 expression in renal tubular epithelial cells. J Am Soc Nephrol (1998) 9(12):2283–90. doi: 10.1681/asn.V9122283
33. Montesano R, Kumar S, Orci L, Pepper MS. Synergistic effect of hyaluronan oligosaccharides and vascular endothelial growth factor on angiogenesis. vitro. Lab Invest (1996) 75(2):249–62.
34. Mascarenhas MM, Day RM, Ochoa CD, Choi WI, Yu L, Ouyang B, et al. Low molecular weight hyaluronan from stretched lung enhances interleukin-8 expression. Am J Respir Cell Mol Biol (2004) 30(1):51–60. doi: 10.1165/rcmb.2002-0167OC
35. Thapa R, Wilson GD. The importance of CD44 as a stem cell biomarker and therapeutic target in cancer. Stem Cells Int (2016) 2016:2087204–. doi: 10.1155/2016/2087204
36. McDonald B, Kubes P. Interactions between CD44 and hyaluronan in leukocyte trafficking. Front Immunol (2015) 6:68. doi: 10.3389/fimmu.2015.00068
37. Misra S, Hascall VC, Markwald RR, Ghatak S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front Immunol (2015) 6:201. doi: 10.3389/fimmu.2015.00201
38. Springer TA. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell (1994) 76(2):301–14. doi: 10.1016/0092-8674(94)90337-9
39. DeGrendele HC, Estess P, Picker LJ, Siegelman MH. CD44 and its ligand hyaluronate mediate rolling under physiologic flow: a novel lymphocyte-endothelial cell primary adhesion pathway. J Exp Med (1996) 183(3):1119–30. doi: 10.1084/jem.183.3.1119
40. Sugiyama K, Komada Y, Deguchi T, Zhang XL, Azuma E, Ido M, et al. CD3-mediated T cell activation is inhibited by anti-CD44 monoclonal antibodies directed to the hyaluronan-binding region. Immunol Invest (1999) 28(2-3):185–200. doi: 10.3109/08820139909061147
41. Shimizu Y, Van Seventer GA, Siraganian R, Wahl L, Shaw S. Dual role of the CD44 molecule in T cell adhesion and activation. J Immunol (1989) 143(8):2457–63. doi: 10.4049/jimmunol.143.8.2457
42. Hodge-Dufour J, Noble PW, Horton MR, Bao C, Wysoka M, Burdick MD, et al. Induction of IL-12 and chemokines by hyaluronan requires adhesion-dependent priming of resident but not elicited macrophages. J Immunol (1997) 159(5):2492–500. doi: 10.4049/jimmunol.159.5.2492
43. Denning SM, Le PT, Singer KH, Haynes BF. Antibodies against the CD44 p80, lymphocyte homing receptor molecule augment human peripheral blood T cell activation. J Immunol (1990) 144(1):7–15. doi: 10.4049/jimmunol.144.1.7
44. Horton MR, Shapiro S, Bao C, Lowenstein CJ, Noble PW. Induction and regulation of macrophage metalloelastase by hyaluronan fragments in mouse macrophages. J Immunol (1999) 162(7):4171–6. doi: 10.4049/jimmunol.162.7.4171
45. Elesh IF, Marey MA, Zinnah MA, Akthar I, Kawai T, Naim F, et al. Peptidoglycan switches off the TLR2-mediated sperm recognition and triggers sperm localization in the bovine endometrium. Front Immunol (2020) 11:619408. doi: 10.3389/fimmu.2020.619408
46. Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res (2005) 33(7):2302–9. doi: 10.1093/nar/gki524
47. Vuorio J, Vattulainen I, Martinez-Seara H. Atomistic fingerprint of hyaluronan–CD44 binding. PloS Comput Biol (2017) 13(7):e1005663. doi: 10.1371/journal.pcbi.1005663
48. Paul D, Santra S, Jana M. Interation between CD44 and HA16: an investigation on multiple binding modes of the complex by using molecular dynamics simulation studies. J Indian Chem Soc (2019) 96:851–61.
49. Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun (1995) 91(1):43–56. doi: 10.1016/0010-4655(95)00042-E
50. Bjelkmar P, Larsson P, Cuendet MA, Hess B, Lindahl E. Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theory Comput (2010) 6(2):459–66. doi: 10.1021/ct900549r
51. Brooks BR, Brooks CL 3rd, Mackerell AD Jr., Nilsson L, Petrella RJ, Roux B, et al. CHARMM: the biomolecular simulation program. J Comput Chem (2009) 30(10):1545–614. doi: 10.1002/jcc.21287
52. Zoete V, Cuendet MA, Grosdidier A, Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem (2011) 32(11):2359–68. doi: 10.1002/jcc.21816
53. Trott O, Olson AJ. AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem (2010) 31(2):455–61. doi: 10.1002/jcc.21334
54. Kumari R, Kumar R, Lynn A. G_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model (2014) 54(7):1951–62. doi: 10.1021/ci500020m
55. Barzegar A, Mansouri A, Azamat J. Molecular dynamics simulation of non-covalent single-walled carbon nanotube functionalization with surfactant peptides. J Mol Graph Model (2016) 64:75–84. doi: 10.1016/j.jmgm.2016.01.003
56. Mansouri A, Mahnam K. Designing new surfactant peptides for binding to carbon nanotubes via computational approaches. J Mol Graph Model (2017) 74:61–72. doi: 10.1016/j.jmgm.2017.02.016
57. Wijayagunawardane MP, Miyamoto A, Cerbito WA, Acosta TJ, Takagi M, Sato K. Local distributions of oviductal estradiol, progesterone, prostaglandins, oxytocin and endothelin-1 in the cyclic cow. Theriogenology (1998) 49(3):607–18. doi: 10.1016/s0093-691x(98)00011-9
58. Tammi R, Rönkkö S, Agren UM, Tammi M. Distribution of hyaluronan in bull reproductive organs. J Histochem Cytochem (1994) 42(11):1479–86. doi: 10.1177/42.11.7523491
59. Ulbrich SE, Schoenfelder M, Thoene S, Einspanier R. Hyaluronan in the bovine oviduct–modulation of synthases and receptors during the estrous cycle. Mol Cell Endocrinol (2004) 214(1-2):9–18. doi: 10.1016/j.mce.2003.12.002
60. Skarzynski DJ, Miyamoto Y, Okuda K. Production of prostaglandin f(2alpha) by cultured bovine endometrial cells in response to tumor necrosis factor alpha: cell type specificity and intracellular mechanisms. Biol Reprod (2000) 62(5):1116–20. doi: 10.1095/biolreprod62.5.1116
61. Tanikawa M, Lee HY, Watanabe K, Majewska M, Skarzynski DJ, Park SB, et al. Regulation of prostaglandin biosynthesis by interleukin-1 in cultured bovine endometrial cells. J Endocrinol (2008) 199(3):425–34. doi: 10.1677/joe-08-0237
62. Ulbrich SE, Frohlich T, Schulke K, Englberger E, Waldschmitt N, Arnold GJ, et al. Evidence for estrogen-dependent uterine serpin (SERPINA14) expression during estrus in the bovine endometrial glandular epithelium and lumen. Biol Reprod (2009) 81(4):795–805. doi: 10.1095/biolreprod.108.075184
63. Cerbito WA, Miyamoto A, Balagapo CR Jr., Natural NG, Miyazawa K, Sato K. Prostaglandin E2 levels in uterine tissues and its relationship with uterine and luteal progesterone during the estrous cycle in dairy cows. Theriogenology (1994) 42(6):941–50. doi: 10.1016/0093-691x(94)90117-2
64. Elweza AE, Ezz MA, Acosta TJ, Talukder AK, Shimizu T, Hayakawa H, et al. A proinflammatory response of bovine endometrial epithelial cells to active sperm. vitro. Mol Reprod Dev (2018) 85(3):215–26. doi: 10.1002/mrd.22955
65. Parrish JJ, Susko-Parrish J, Winer MA, First NL. Capacitation of bovine sperm by heparin. Biol Reprod (1988) 38(5):1171–80. doi: 10.1095/biolreprod38.5.1171
66. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem (1987) 162(1):156–9. doi: 10.1006/abio.1987.9999
67. Yousef MS, Marey MA, Hambruch N, Hayakawa H, Shimizu T, Hussien HA, et al. Sperm binding to oviduct epithelial cells enhances TGFB1 and IL10 expressions in epithelial cells as well as neutrophils In vitro: prostaglandin E2 as a main regulator of anti-inflammatory response in the bovine oviduct. PloS One (2016) 11(9):e0162309. doi: 10.1371/journal.pone.0162309
68. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
69. Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng (1995) 8(2):127–34. doi: 10.1093/protein/8.2.127
70. Akthar I, Marey MA, Kim Y, Shimada M, Suarez SS, Miyamoto A. Sperm interaction with the uterine innate immune system: toll-like receptor 2 (TLR2) is a main sensor in cattle. Reprod Fertil Dev (2021) 34(2):139–48. doi: 10.1071/rd21265
71. Marey MA, Aboul Ezz M, Akthar I, Yousef MS, Imakawa K, Shimada M, et al. Sensing sperm via maternal immune system: a potential mechanism for controlling microenvironment for fertility in the cow. J Anim Sci (2020) 98(Suppl 1):S88–s95. doi: 10.1093/jas/skaa147
72. Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell (2007) 130(6):1071–82. doi: 10.1016/j.cell.2007.09.008
73. Lintuluoto M, Horioka Y, Hongo S, Lintuluoto JM, Fukunishi Y. Molecular dynamics simulation study on allosteric regulation of CD44-hyaluronan binding as a force sensing mechanism. ACS Omega (2021) 6(12):8045–55. doi: 10.1021/acsomega.0c05502
74. Raheem KA, Marei WF, Mifsud K, Khalid M, Wathes DC, Fouladi-Nashta AA. Regulation of the hyaluronan system in ovine endometrium by ovarian steroids. Reproduction (2013) 145(5):491–504. doi: 10.1530/rep-13-0001
75. Bains R, Adeghe J, Carson RJ. Human sperm cells express CD44. Fertil Steril (2002) 78(2):307–12. doi: 10.1016/s0015-0282(02)03230-2
76. Laurent TC, Laurent UB, Fraser JR. Functions of hyaluronan. Ann Rheum Dis (1995) 54(5):429–32. doi: 10.1136/ard.54.5.429
77. Fraser JR, Laurent TC, Laurent UB. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med (1997) 242(1):27–33. doi: 10.1046/j.1365-2796.1997.00170.x
78. Horton MR, McKee CM, Bao C, Liao F, Farber JM, Hodge-DuFour J, et al. Hyaluronan fragments synergize with interferon-gamma to induce the c-X-C chemokines mig and interferon-inducible protein-10 in mouse macrophages. J Biol Chem (1998) 273(52):35088–94. doi: 10.1074/jbc.273.52.35088
79. McKee CM, Lowenstein CJ, Horton MR, Wu J, Bao C, Chin BY, et al. Hyaluronan fragments induce nitric-oxide synthase in murine macrophages through a nuclear factor kappaB-dependent mechanism. J Biol Chem (1997) 272(12):8013–8. doi: 10.1074/jbc.272.12.8013
80. Tesar BM, Jiang D, Liang J, Palmer SM, Noble PW, Goldstein DR. The role of hyaluronan degradation products as innate alloimmune agonists. Am J Transplant (2006) 6(11):2622–35. doi: 10.1111/j.1600-6143.2006.01537.x
81. Scheibner KA, Lutz MA, Boodoo S, Fenton MJ, Powell JD, Horton MR. Hyaluronan fragments act as an endogenous danger signal by engaging TLR2. J Immunol (2006) 177(2):1272–81. doi: 10.4049/jimmunol.177.2.1272
82. Qadri M, Almadani S, Jay GD, Elsaid KA. Role of CD44 in regulating TLR2 activation of human macrophages and downstream expression of proinflammatory cytokines. J Immun (2018) 200(2):758–67. doi: 10.4049/jimmunol.1700713
83. Kawana H, Karaki H, Higashi M, Miyazaki M, Hilberg F, Kitagawa M, et al. CD44 suppresses TLR-mediated inflammation. J Immun (2008) 180(6):4235–45. doi: 10.4049/jimmunol.180.6.4235
Keywords: uterus, sperm, hyaluronan, CD44, TLR2
Citation: Ezz MA, Mansouri A, Akthar I, Yousef MS, Kowsar R and Miyamoto A (2023) Hyaluronan regulates sperm-induced inflammatory response by enhancing sperm attachment to bovine endometrial epithelial cells via CD44: in-silico and in-vitro approaches. Front. Endocrinol. 14:1134868. doi: 10.3389/fendo.2023.1134868
Received: 03 January 2023; Accepted: 18 April 2023;
Published: 10 May 2023.
Edited by:
Fred Sinowatz, Ludwig Maximilian University of Munich, GermanyReviewed by:
Ralf Einspanier, Freie Universität Berlin, GermanyCopyright © 2023 Ezz, Mansouri, Akthar, Yousef, Kowsar and Miyamoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akio Miyamoto, YWtpb21peWFAb2JpaGlyby5hYy5qcA==
†Present address: Mohamed Aboul Ezz, Division of Animal Sciences, University of Missouri, Columbia, MO, United States
‡These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.