- 1Department of Zoology, Hazara University, Mansehra, Pakistan
- 2Department of Biotechnology, University of Agriculture, Dera Ismail Khan, Pakistan
- 3Faculty of Veterinary and Animal Science, University of Agriculture, Dera Ismail Khan, Pakistan
- 4Department of Biotechnology and Genetic Engineering, Hazara University, Mansehra, Pakistan
- 5Department of Biochemistry, Faculty of Science, King Abdul-Aziz University, Jeddah, Saudi Arabia
- 6Department of Molecular Medicine, Institute of Basic Medical Sciences, University of Oslo, Oslo, Norway
Introduction: Polycystic Ovarian Syndrome (PCOS) is a globally prevalent condition that leads to infertility in women. While environmental factors contribute to PCOS, maternal genetics also play a significant role. Currently, there is no definitive test for identifying predisposition to PCOS. Hence, our objective is to discover novel maternal genetic risk factors for PCOS by investigating the genomes of patients from Pakistan.
Methods: We utilized Next-Generation Sequencing (NGS) to sequence the complete mitochondrial DNA of three PCOS patients. Subsequently, we employed MitoTIP (Mitochondrial tRNA Informatics Predictor) and PON-mt-tRNA tools to identify variations in the mitochondrial DNA. Our analysis focused on the genes MT-RNR1, MT-RNR2, MT-ATP6, MT-TL2, and MT-CYTB, which displayed common variations in all three genomes. Additionally, we observed individual variations. The D-loop region exhibited the highest frequency of mutations, followed by the non-coding regions of RNR1 and RNR2 genes. Moreover, we detected frameshift mutations in the mitochondrially encoded NADH Dehydrogenase 2 (MT-ND2) and mitochondrially encoded NADH Dehydrogenase 5 (ND5) genes within individual genomes.
Results: Our analysis unveiled six regions with common variations in the mitochondrial DNA of all three PCOS patients. Notably, the MT-RNR1, MT-RNR2, MT-ATP6, MT-TL2, and MT-CYTB genes exhibited these variations. Additionally, we identified individual variations in the mitochondrial DNA. The D-loop region displayed the highest mutation frequency, followed by the non-coding regions of RNR1 and RNR2 genes. Furthermore, frameshift mutations were detected in the MT-ND2 and ND5 genes within individual genomes.
Conclusion: Through our study, we have identified variations in mitochondrial DNA that may be associated with the development of PCOS and have the potential to serve as predisposition tests. Our findings highlight the presence of novel mutations in the MT-RNR1, MT-RNR2, MT-ATP6, MT-TL2, and MT-CYTB genes, as well as frameshift mutations in the MT-ND2 and ND5 genes. Pathogenicity analysis indicated that most variants were likely to result in benign cysts. However, the frameshift mutations in the ND2 gene were associated with a high risk of complications and pathogenicity in PCOS. This is the first report identifying these mutations and their association with PCOS, contributing to our understanding of the genetic factors underlying the condition.
Introduction
Polycystic ovarian syndrome (PCOS) is a complex endocrine disorder that affects up to 8%–13% of women in their reproductive age (1). PCOS is a growing concern worldwide, with increasing incidence rates reported (2). The disorder is characterized by the presence of numerous ovarian cysts visible through ultrasound inspection (3). PCOS is associated with various symptoms such as menstrual irregularities, hormonal dysfunction, dermatological issues, psychological problems, high cancer risk, and metabolic disorders (4). Various gynecological issues are associated with PCOS, including anovulatory infertility (5), variations in oocyte competency (OC), which can lead to subfertility (6, 7), endometrial dysfunction, and abnormal trophoblast invasion and placentation (8), which can increase the risk of miscarriage and pregnancy complications in women with PCOS (5). PCOS women who have hyperandrogenic conditions are also at a higher risk of developing pervasive developmental disorders (PDDs) (9). Hormonal dysfunction is considered a key feature of PCOS (8, 10), while insulin resistance and hyperandrogenism have also been reported, leading to decreased folliculogenesis and an increased risk of comorbidities and androgenic alopecia (11). The most prominent impact of PCOS on women’s lives is menstrual irregularities and ovarian cancer. PCOS is a multifactorial disorder, with several risk factors contributing to its etiology, including obesity, neuroendocrine status, environment or lifestyle, and genetic makeup (3). Variations in mitochondrial DNA (mtDNA) are increasingly recognized as a genetic cause (12, 13). Both nuclear and mitochondrial genetic variations have been associated with PCOS pathogenesis (14). Nuclear genes, including calpain 10 (CAPN10), cytochrome family P450, insulin (INS) gene, androgen receptor (AR), fat mass obesity (FTO) gene, and follicle-stimulating hormone receptor (FSHR) gene, have been shown to be associated with PCOS (15).
The mitochondrial genome is considered more vulnerable to oxidative damage and has a high mutation rate due to the lack of protective histones, inefficient DNA repair mechanisms, and its proximity to the electron transport chain (ETC), where oxygen-derived free radicals are frequently generated (16). Hence, the mitochondrial genome is considered a Pandora’s box of pathogenic mutations (16). This study was designed to screen the whole mitochondrial DNA (WMTDNA), comprising 37 mitochondrial genes using next-generation sequencing and to predict in silico the resultant common variations for pathogenicity. In silico analysis was used to evaluate the pathogenicity of mutations and their impact on subjects.
Materials and methods
Ethical statement
The experimental procedures were approved by the Ethical Committee of the Institution and Board of Advanced Studies and Research at Hazara University, Mansehra (21300), and Pakistan under notification number F.No.73/HU/ORIC/IBC/2017/400.
Consent, recruitment of patients, and families
The Rotterdam criteria have been used for diagnosing PCOS patients (10). According to these criteria, a patient must have two of the following three symptoms: hyperandrogenism (biochemical or clinical), oligo- or anovulation, and polycystic ovary morphology (PCOM), as determined by ultrasound inspection.
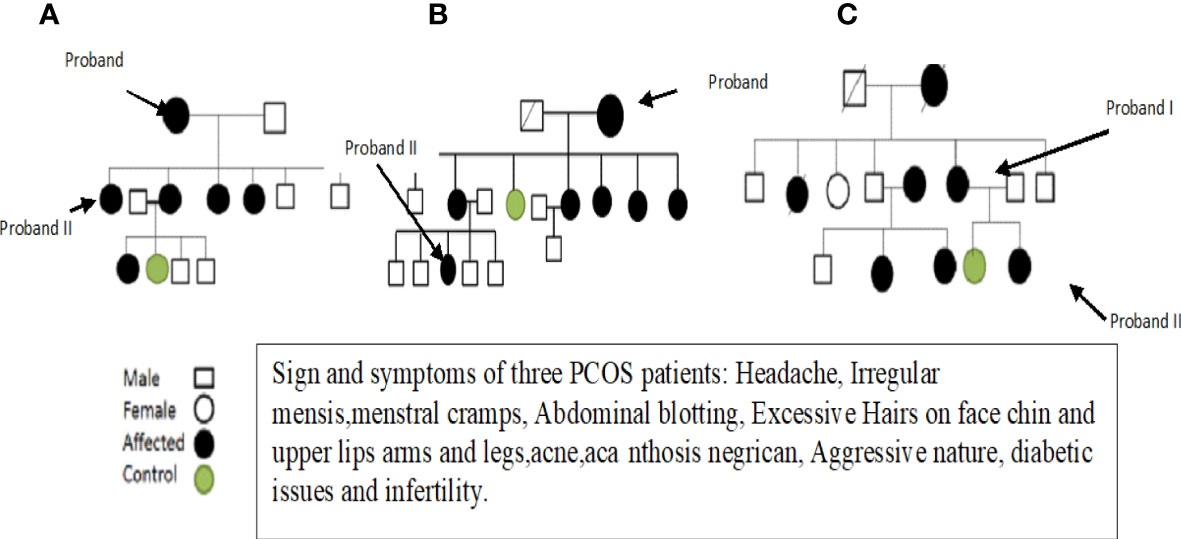
After obtaining informed consent and a physical examination by a gynecologist, selected patients were interviewed about their family history and other details, and pedigrees were constructed to trace the maternal inheritance pattern of their disorder. Saliva samples were then collected from each patient. Patients with hyperprolactinemia, thyroid and adrenal diseases, 21-hydroxylase deficiency, and androgen-secreting tumors were excluded because these disorders mimic the symptoms of PCOS. Finally, three fully expressed syndromic patients were selected and subjected to whole mitochondrial genome sequence (WMGS) analysis to draw the genetic portrait of mitochondrial mutational hotspots associated with maternally inherited PCOS. Variant calling and identification of homoplasmic and heteroplasmic mutations were carried out. After obtaining detailed family histories and information about deceased members, the pedigrees of the three families were constructed (see Figure 1). The mutations identified in WMGS were assessed in other family members of the probands through Sanger sequencing.
DNA extraction, NGS analysis, and identification of variants
DNA was extracted from saliva samples using the phenol-chloroform method (17). Nanodrop quantification and gel electrophoresis were performed to determine the quantity and quality of the isolated DNA. Samples were then carefully labeled and stored at −20°C. The labeled samples were sent to a commercial company for DNA sequence analysis. Online DNA analysis tools like National Center for Biotechnology Information (NCBI) Blast, Universal Protein Resource (Uniprot), and UGENE were used to conduct further alignment and investigations. The resulting nucleotide sequences were compared to the revised Cambridge Reference Sequence (rCRS). We performed Sanger sequencing for validation of the identified common variations for D-loop, ATP6, MT-TL2, and CYTB.
In the UGENE (http://ugene.net/) editor, two nucleotide sequences were aligned, and variations were checked. Multiple sequence alignment was performed using integrated multiple sequence comparison by log expectations (MUSCLE) on UGENE.
Score-based evaluation of tRNA variants for pathogenicity and validation by in silico predictive tools
Mitochondrial tRNA Informatics Predictor (MitoTIP) and PON-mt-tRNA were utilized to determine the pathogenicity of mitochondrial variations. MitoTIP was employed to assess the pathogenicity status of genetic variations, while the PON-mt-tRNA, a multifactorial probability-based prediction approach, was used to classify the studied and identified mitochondrial variants. Some variants were reported to be deleterious, some benign, one novel frameshift, and one pathogenic based on the results from both MitoTIP and PON-mt-tRNA. The difference in the degree of predicted variants is attributed to the fact that both tools operate on distinct algorithms/principles and consider diverse factors (18–20).
Results
The current study focused on the mitochondrial genome of patients with maternally inherited PCOS. To achieve this, three patients were selected for the study, each from a different family with a history of maternally inherited PCOS. Furthermore, the experiment focused on analyzing the WMTDNA of these patients to identify any mutations or genetic variations that may be associated with the development of PCOS.
Clinical evaluations
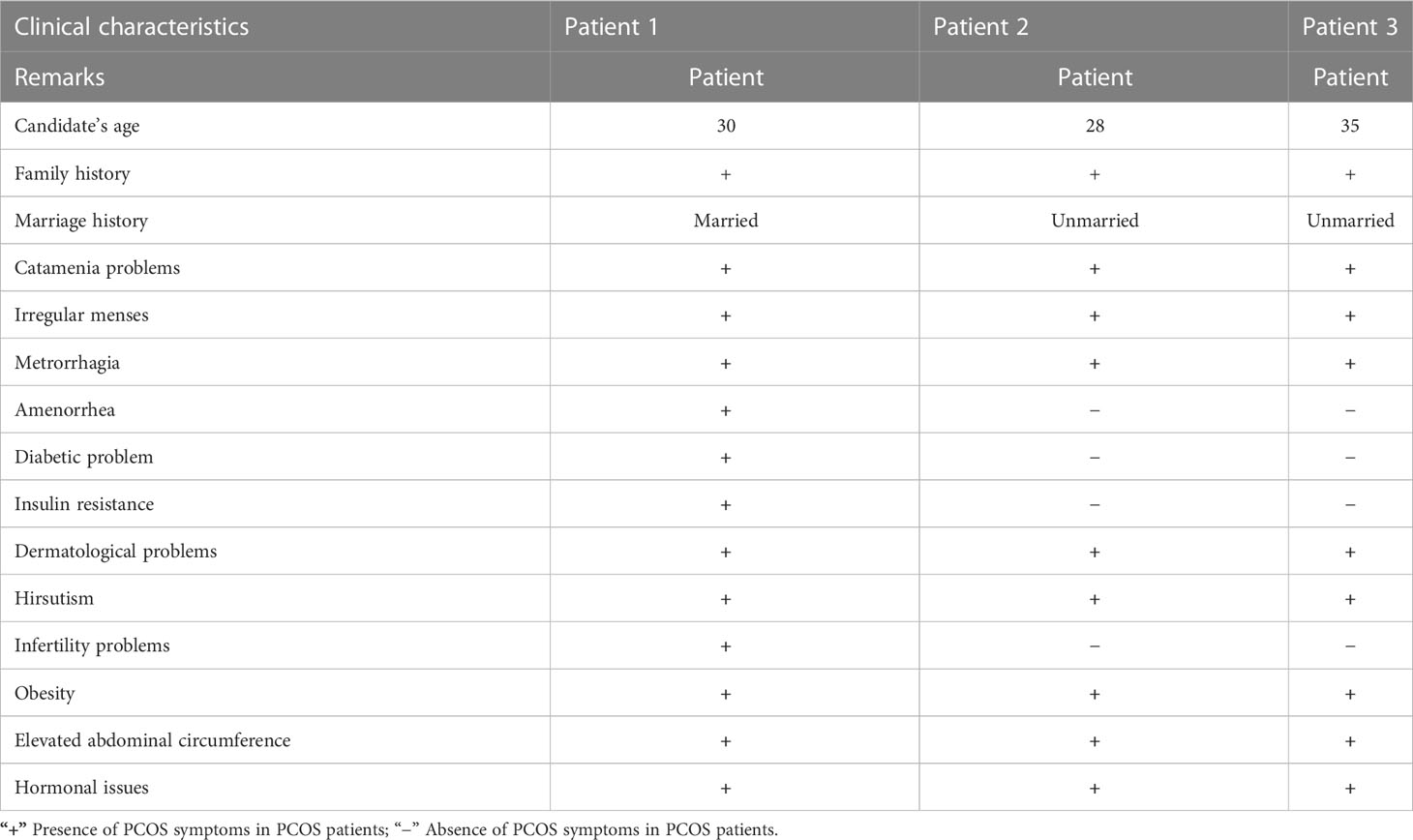
At the onset of the condition, the average age range was 30–35 years. Among these patients, two were unmarried and one was married but having infertility issues. One PCOS patient was suffering from diabetes due to insulin resistance, and the other two had catamenia, dermatological, metabolic, and hormonal issues. Other family members of these three patients also suffered from PCOS. Most of the symptoms, such as obesity, dermatological problems, catamenia, hirsutisms, and hormonal issues, are common in the three PCOS patients. The detailed clinical data of the PCOS patients are given in Table 1.
Common mitochondrial DNA mutations were identified in the coding and noncoding region of the three PCOS patients in the present study
The analysis of the whole mitochondrial genome has enabled us to identify a set of mutations present in three familial subjects with PCOS. The mutations that were identified in all three genomes are presented in Table 2. Specifically, eight mutations at eight different positions in six genes were identified as common among the three PCOS patients. Of these, two mutations were located in the D-loop region, one in RNR1, one in RNR2, one in ATP6, one in MT-TL2, and one in CYTB, while four were missense variants, two were identified in intergenic regions, and one was a noncoding transcript. To further validate these findings, the mutations were revalidated in other PCOS patients through Sanger sequencing of the individual positions, as depicted in Figure 2.
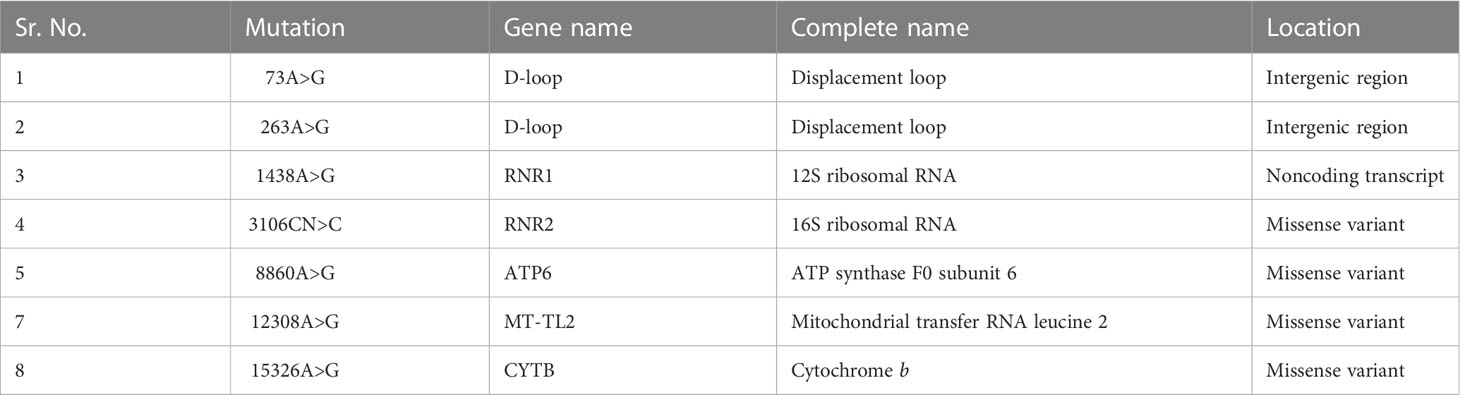
Table 2 Common mitochondrial DNA mutations identified in the coding and noncoding regions of three PCOS patients in the present study.
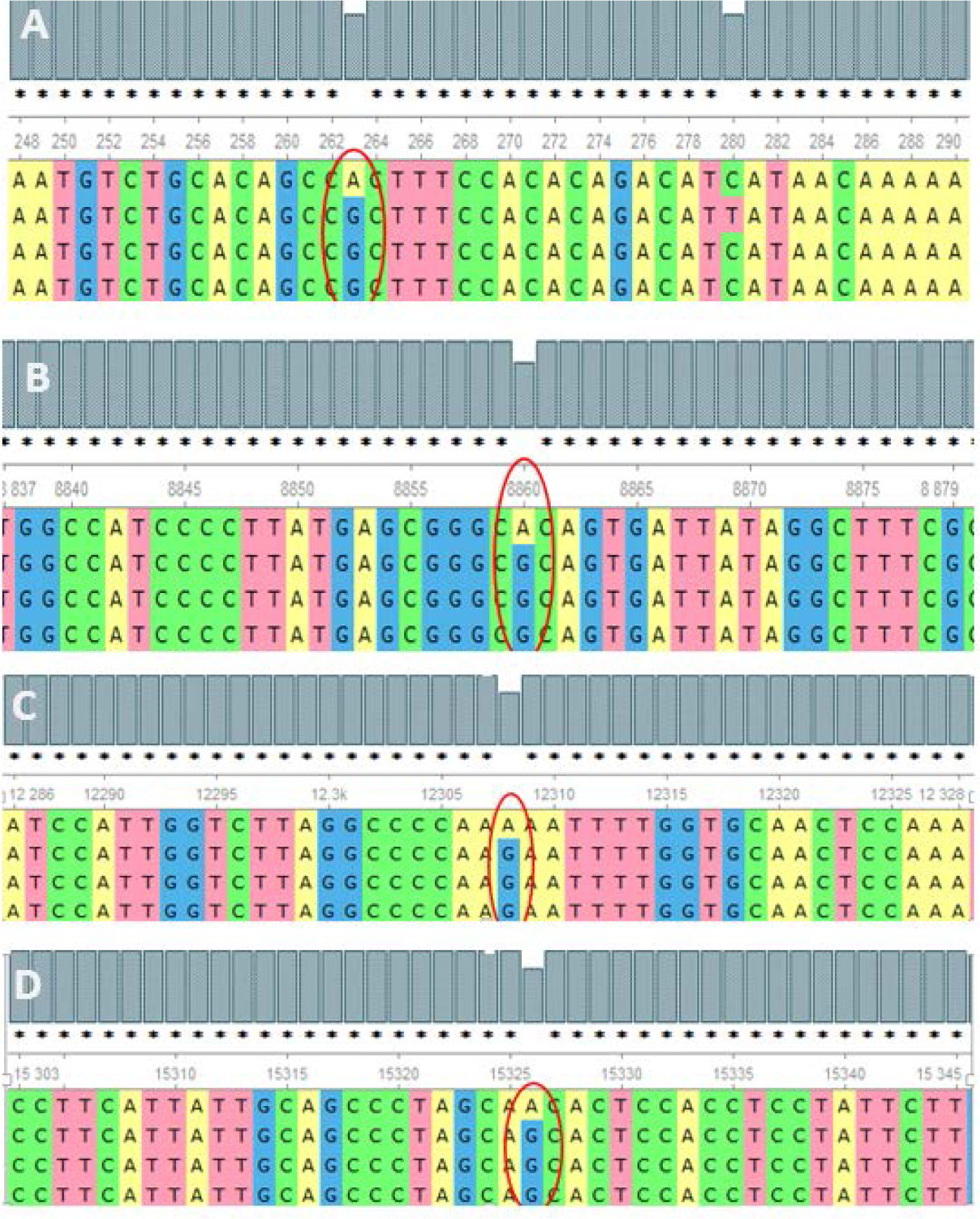
Figure 2 Alignment of the sequence resulted in nucleotides from maternally inherited PCOS-positive patients with rCRS Accession No. NC-012920.1 exhibiting mutations (encircled) at positions (A) 263A>G (D-loop), (B) 8860A>G (MT-ATP6), (C) 12308A>G (MT-TL2), and (D) 15326A>G MT-CYTB.
Mutations were identified in the whole mitochondrial genome sequence of the three PCOS patients in the present study
The nucleotide sequence analysis has identified 36 mutations in PCOS patient 1, including 11 variants in the D-loop region, seven variants in the RNR1 and RNR2 genes, nine synonymous mutations, and nine missense mutations (Figure 3).
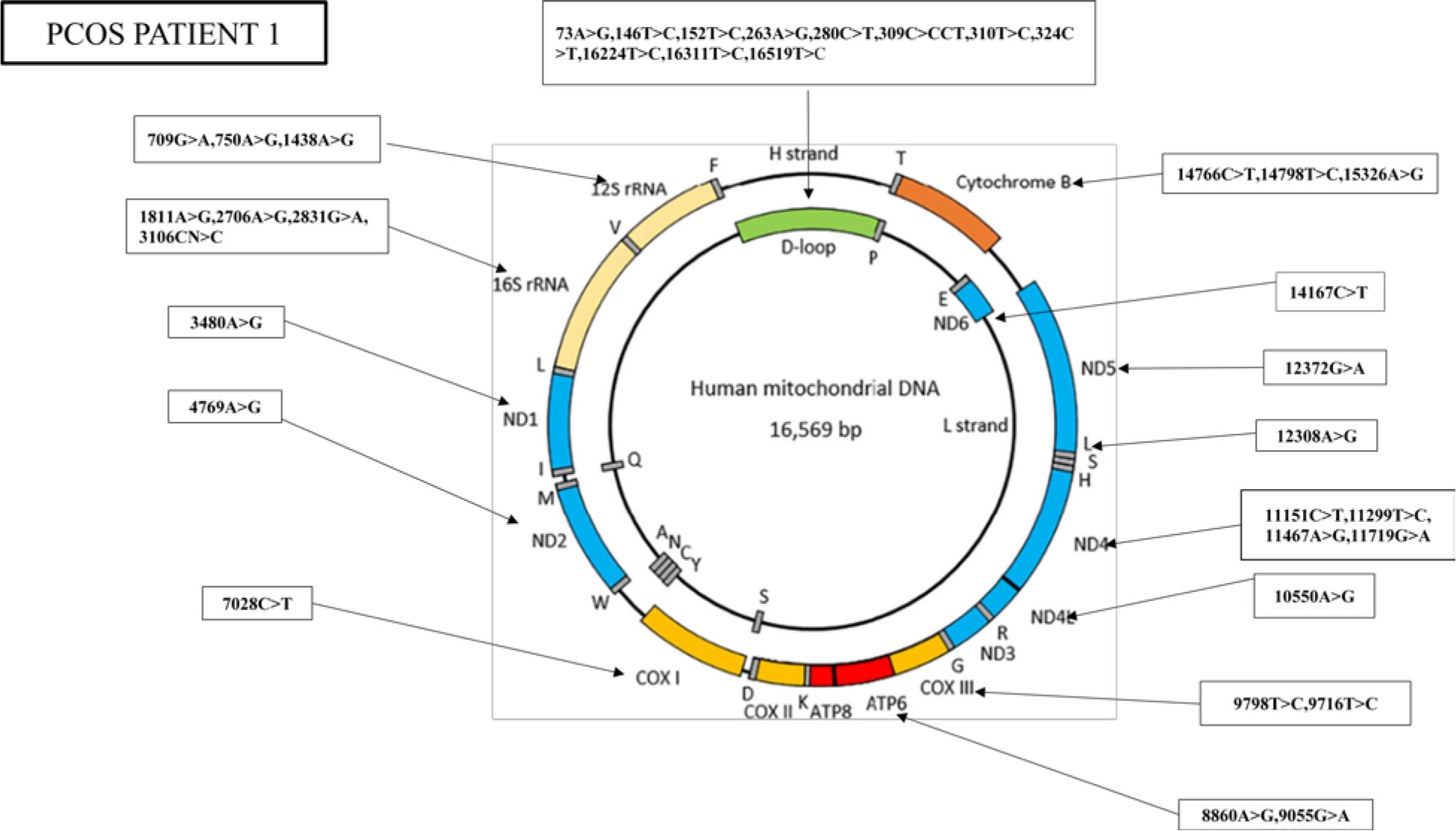
Figure 3 Whole mitogenome variants identified in PCOS patient 1. Mitochondria consist of two strands; an outer heavy strand and an inner light strand. Overall, 36 mutations were identified in the whole mitochondrial genome: 11 in D-loop, three in RNR1, four in RNR2, one in ND1, one in ND2, one in COX1, two in ATP6, two in −COX3, one in ND4L, four in ND4, one in ND5, one in ND6, and three in CYTB.
Similarly, the nucleotide sequence analysis has identified 38 mutations in PCOS patient 2, including 14 variants in the D-loop region, five variants in the RNR1 and RNR2 genes, nine synonymous mutations and eight missense mutations, and one stop-loss and one frameshift (Figure 4).
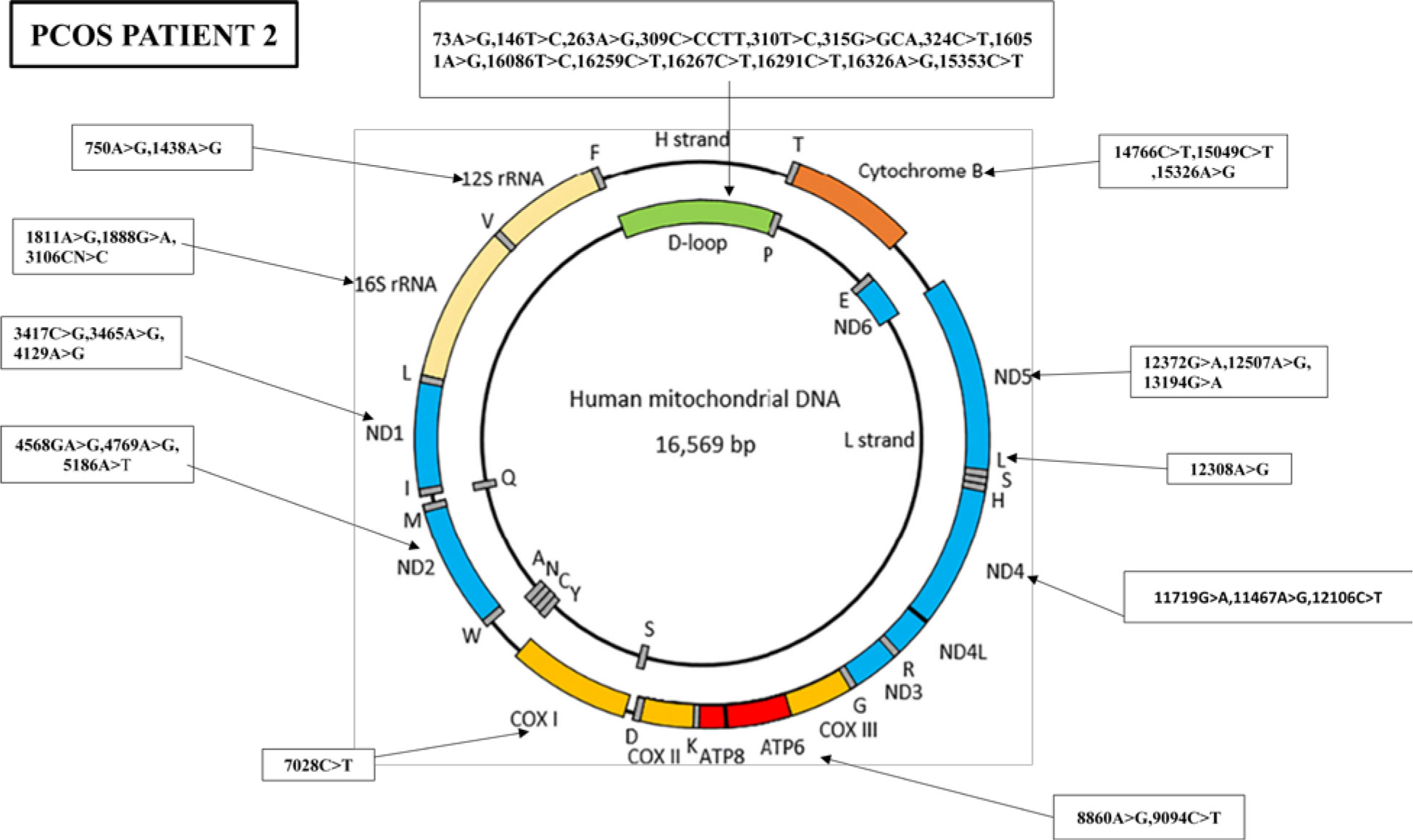
Figure 4 Mutations identified in the whole mitochondrial DNA sequence of PCOS-positive patient 2. Overall, 38 mutations were identified in the whole mitochondrial genome of PCOS patient 2: 14 mutations were found in D-loop, two in RNR1, three in RNR2, three in ND1, three in ND2, one in COX1, two in ATP6, three in ND4, three in ND5, one in MT-TL2, and three in CYTB.
Furthermore, the WMTDNA nucleotide sequence analysis has identified 19 mutations in PCOS-positive patient 3, including five variants in the D-loop region, four variants in the RNR1 and RNR2 gene, five synonymous mutations and three missense mutations, and one stop-loss and one frameshift (Figure 5).
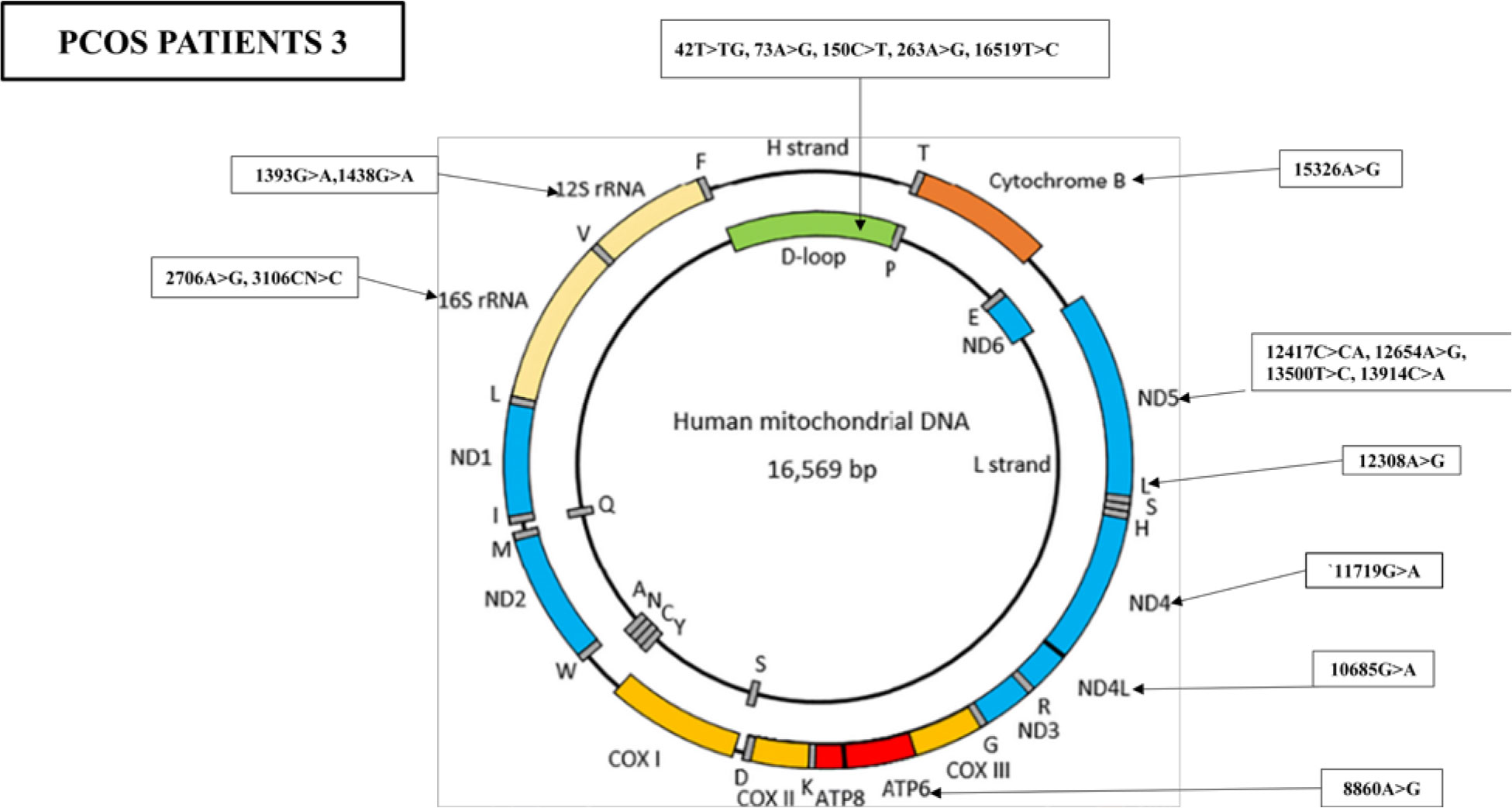
Figure 5 Mutations identified in the whole mitochondrial DNA sequence of PCOS patient 3. Overall, 19 mutations were identified in the whole mitochondrial genome: five mutations were found in D-loop, two variants in RNR1, two variants in RNR2, one in ATP6, one in ND4, one in ND4L, one in TL2, four in ND5, and one in CYTB.
Pathogenicity status of the studied mutations
The pathogenicity of the identified variants was evaluated using different tools. Results revealed that among all the common mutations identified in the three whole mitogenomes of PCOS patients after NGS analysis, mutations reported in RNR1 at positions 709A>G, 750A>G, 1438A>G, and 1393 G>A were found to be resulting in translation defects that were pathogenic. MT-12S rRNA variations are possibly associated with disruption of mitochondrial function. The mutations found in RNR2 at positions 2706A>G, 2831G>A, 3106CN>C, and 1888G>A. These mutations are present at the 530 loops of the ribosome, and they affect the codon–anticodon interaction at the A site (acceptor site). These mutations lead to the improper movement of the small subunit (SSU) head during translocation and finally the translocation (Table 3).
ML probability of pathogenicity: average probability of pathogenicity predicted by 20 machine learning (random forests) predictors
In addition, a mutation in the MT-ATP6 gene at position 8860A>G with a score of 0.1 was declared as tolerated with a score of 0.003. A mutation in mitochondrial transfer RNA leucine 2 at position 12308A>G with a score of 11.8509 was declared likely neutral with a score of 0.41. A mutation at position 11719G>A in the ND4 gene is a synonymous variant and pose no impact on the amino acid type.
Discussion
Overall, the present study aimed to investigate the mitochondrial genome of patients with maternally inherited PCOS. Three patients, each from a different family with a history of maternally inherited PCOS, were selected for the study. The study focused on analyzing the WMTDNA of these patients to identify any mutations or genetic variations that could be associated with the development of PCOS. By analyzing the mitochondrial genome of these patients, we aimed to gain insights into the potential genetic factors that contribute to the development of this disorder, specifically those that are passed down maternally.
PCOS is a common endocrine disorder that affects reproductive-age women worldwide and is the leading cause of ovulatory dysfunction and infertility (21). PCOS is characterized by hyperandrogenism, anovulation, and polycystic ovaries, along with insulin resistance, obesity, and metabolic disorders (1). Several predisposing risk factors, including genetic, neuroendocrine, lifestyle/environmental, and obesity, have been linked to PCOS development (3). Moreover, mtDNA mutations are also considered to contribute to the pathogenesis of PCOS. MI mutations that were potentially associated with PCOS-IR were as follows: mt-tRNALeu(UUR) A3302G and C3275A mutations; mt-tRNAGln T4363C and T4395C mutations; mt-tRNASer(UCN) C7492T mutation; mt-tRNAAsp A7543G mutation, mt-tRNALys A8343G mutation, mt-tRNAArg T10454C mutation, and mt-tRNAGlu A14693G mutation (22). They utilized a whole mitochondrial genome sequencing analysis for three maternally inherited PCOS familial subjects. Their findings revealed several variations in mtDNA. Patient 1 had 11 variants in the d-loop region, including 73A>G, 146T>C, 152T>C, 263A>G, 280C>T, 309C>CCT, 310T>C, 324C>T, 16224T>C, 16311T>C, and 16519T>C. The variant 73A>G, 146T>C, and 152T>C have been previously reported to be associated with gastric colon and oral cancer, PCOS, and breast cancer (23–25). Women with PCOS may be more susceptible to some cancers due to their abnormal metabolic and hormonal conditions.
Prolonged hormone stimulation is known to be associated with the development of endometrial, ovarian, and breast cancers in women (24, 26). In this study, we identified several mtDNA variants in the three maternally inherited PCOS familial subjects that have been previously reported to be associated with various types of cancer and other diseases. Variant 263A>G was found to be linked with perilesional skin and skin tumors and PCOS (27). Variants 310T>C and 324C>T have been associated with epithelial ovarian cancer (28), while 16224T>C and 16311T>C have been linked to perilesional skin and skin tumors (27). Variant 309C>CCT has been associated with the etiology of malignant melanoma (29). One variant in the COX1 gene, which has been reported to be associated with PCOS, esophageal cancer, congenital contract, and obesity, was found in patient 1 (23, 30, 31). Similarly, variant 11719G>A has been linked to breast cancer, obesity, and PCOS (24, 31). Moreover, one variant reported in transfer RNA leucine 2 at nucleotide position 12308A>G has been associated with breast cancer, colorectal cancer, prostate and kidney cancer, Alzheimer’s disease, and cardiomyopathy (24, 25, 32, 33).
Our investigation identified three variants in the CYTB gene, including 14766C>T and 14798T>C, which have not been previously reported in association with PCOS (27). Women with a predisposition to PCOS may experience metabolic and hormonal issues such as insulin resistance and hyperandrogenism, which can lead to weight gain and eventually obesity. Obesity, in turn, can exacerbate the symptoms of PCOS, resulting in further metabolic complications and reproductive abnormalities. Our study also identified two mitochondrial variants in the ATP6 gene, namely 8860A>G and 9055G>A. These variants have been previously associated with hypertrophic cardiomyopathy (25) and nonsyndromic hearing loss (28). The vascular characteristics of arterial walls involved in the atherogenic process may be directly influenced by androgen excess in PCOS, as reported by Wu et al. (34) in 2020. Furthermore, mutations were discovered in both the coding and noncoding regions of mitochondria in selected patients who suffered from various issues such as skin problems, hormonal imbalances, diabetes, menstrual problems, and infertility. Other family members were also identified as having this disorder (Table 1).
Fourteen variants were identified in the D-loop of patient 2, including 73A>G, previously found to be associated with gastric colon and oral cancer (24), 146T>C with oral cancer (23) and PCOS, 263A>G in association with PCOS (24), 310T>C with malignant melanoma (26), and 513G>GCA with nodular sclerosing Hodgkin lymphoma (Mitomap). Variants not previously reported in the literature with any disease but identified in our study that may be associated with PCOS are 16051A>G, 16086T>C, 16259C>A, 16267C>T, 16291C>T, 16326A>G, and 16353C>T.
In PCOS patient 2, two variants were identified in RNR1, including 750A>G, which has been reported to be associated with brain tumors (28), obesity (31), and PCOS (24), while 1438A>G is associated with obesity (31), type 2 diabetes, Parkinson’s disease, and PCOS. In RNR2, three variants were reported, including 1811A>G, which has been associated with congenital cataracts (30) and PCOS (24). However, 1888G>A and 3106CN>C have not been reported in previous literature with any disease. Three variants were identified in ND1, including 3417C>G, 3465A>G, and 4129A>G, with no previous reports in association with any disorder, making them novel findings in this investigation. Similarly, three variants were reported in ND2, including the 4769A>G (frameshift) variant previously associated with esophageal cancer and PCOS (24), while 4569GA>G and 5186A>T (stop loss) variants have not been reported in previous studies with any disease. In the ATP6 gene, two variants were identified at position 8860A>G, reported to be associated with hypertrophic cardiomyopathy and PCOS, while 9094C>T is associated with primary ovarian insufficiency (35). In PCOS patient 2, a single variant was found in transfer RNA leucine 2, previously associated with breast cancer (24), and colorectal and kidney cancer (33). Three variants were identified in the cytochrome b gene, including 15326A>G, previously associated with PCOS (27), while 14766C>T and 15049C>T mutations have not been reported in any previous studies.
Five variants were identified in the mitochondrial D-loop region of PCOS patient 3, including 73A>G, 150C>T, 263A>G, and 16519T>C, previously associated with PCOS and gastric, colon, and oral cancers. In addition, 42T>TG was also identified. Two variants were identified in RNR1, including 1393G>A with no previous reports and 1438A>G with a reported association with obesity (31), type 2 diabetes, Parkinson’s disease, and PCOS (27). However, 3106CN>C has not been reported in previous literature with any disease.
One variant was identified in ND4L at position 10685G>A, and four variants were identified in ND5 at positions 12417C>CA, 12654A>G, 13500T>C, and 13914C>A. One variant was identified in ND6 at position 14305G>A, and one variant was identified at position 15326A>G, both associated with PCOS (27). We identified mutations in both the coding and noncoding regions of mitochondria, which may have phenotypic effects on patients. PCOS patient 3 also suffered from skin issues such as acne and hirsutism, hormonal problems, diabetes, menstrual problems, frequent urination, and some psychological issues. Other family members were also found to be suffering from this disorder (refer to Table 1). The identified mutations and their biological effects in our current study and in other previous studies have been summarized in Table 4.
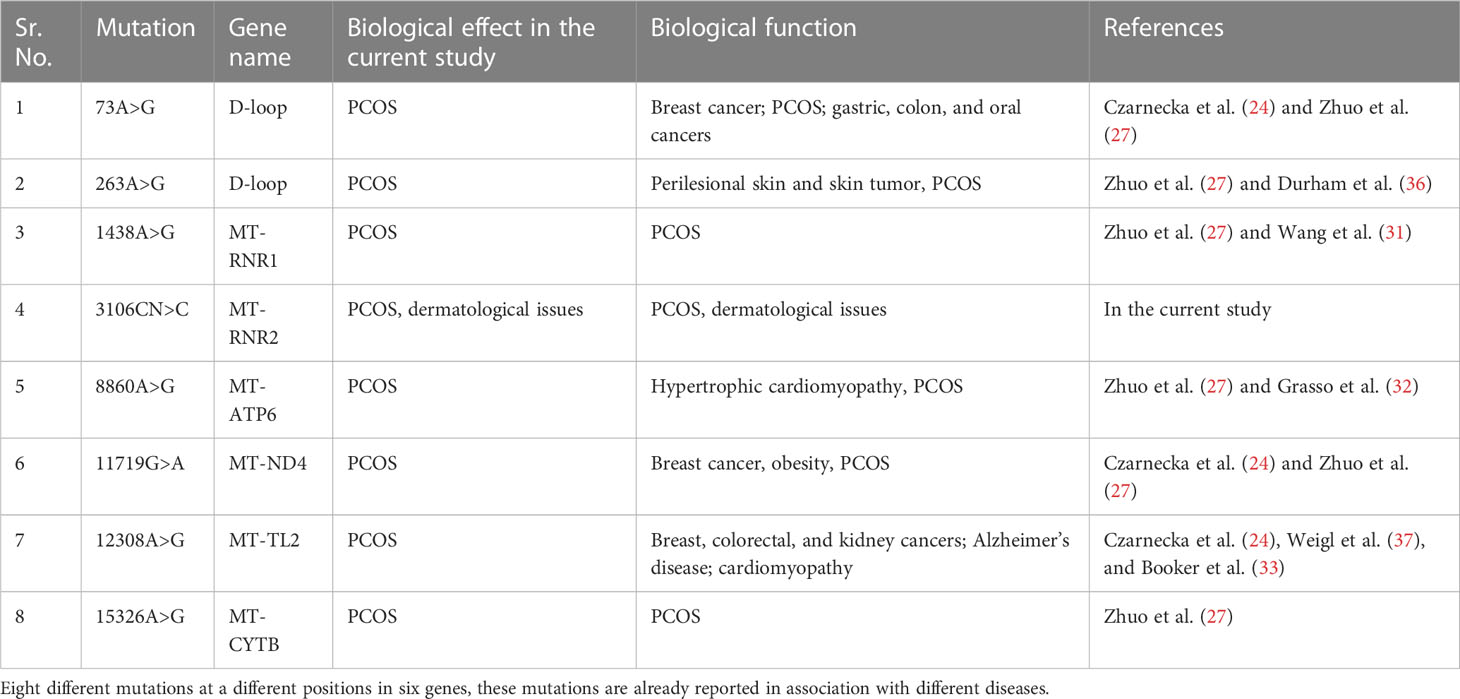
Table 4 Mitochondrial DNA mutations identified in our stud of the WMTDNA of PCOS patients and their association with other diseases.
Conclusions
In conclusion, studying the mutation spectrum of the entire mitochondrial genome is a valuable tool for investigating various maternally inherited genetic disorders in humans, including PCOS. Mitochondrial dynamics, including the mutational spectrum, can be explored to elucidate the etiology of such genetic diseases, given that mitochondria have all the mechanisms for energy transduction in many cells and organs. Homoplasmic variations can have catastrophic consequences, while heteroplasmic mutations may have a lesser impact. This study identified mutations in the D-loop, MT-RNR1, MT-RNR2, MT-ATP6, MT-TL2, and MT-CYTB genes that are most likely associated with the etiology of maternally inherited PCOS in Pakistan. To help patients receive appropriate treatment, genetic testing for the condition and public awareness efforts should be implemented. To validate the association of these mutations with PCOS, it is recommended to predict the defects and replicate the sequence analysis with a larger sample size in other parts of the world. Furthermore, the paternal inheritance pattern should be studied in PCOS patients from Pakistani families. Overall, this study highlights the potential of mitochondrial genetic variations as a novel biomarker for PCOS diagnosis and management. Further research is needed to establish a causal relationship between these mutations and the development of PCOS.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by Ethical Committee of the Institution and Board of Advanced Studies and Research at Hazara University, Mansehra (21300), Pakistan under notification numberF.No.73/HU/ORIC/IBC/2017/400. The patients/participants provided their written informed consent to participate in this study.
Author contributions
SB, MK, GA, ST, and MZ designed the study and wrote the manuscript. ST and MZ supervised the manuscript. SB, TN, QU, MK, AU, MF, SG, MN, and GA helped in the collection of data resources and editing of the final version of the manuscript. Moreover, data analysis and collection were completed by SB, TN, and MF. All authors contributed to the article and approved the submitted version.
Funding
The authors acknowledge HEC for funding the project under NRPU10076.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zeng X, Xie YJ, Liu YT, Long SL, Mo ZC. Polycystic ovarian syndrome: correlation between hyperandrogenism, insulin resistance and obesity. Clinica chimica Acta (2020) 502:214–21. doi: 10.1016/j.cca.2019.11.003
2. Sharma M, Khapre M, Saxena V, Kaushal P. Polycystic ovary syndrome among Indian adolescent girls–a systematic review and metanalysis. Nepal J Epidemiol (2021) 11(3):1063. doi: 10.3126/nje.v11i3.38460
3. Bulsara J, Patel P, Soni A, Acharya S. A review: brief insight into polycystic ovarian syndrome. Endocrine Metab Sci (2021) 3:100085. doi: 10.1016/j.endmts.2021.100085
4. Setji TL, Brown AJ. Polycystic ovary syndrome: diagnosis and treatment. Am J Med (2007) 120(2):128–32. doi: 10.1016/j.amjmed.2006.06.029
5. Palomba S. Is fertility reduced in ovulatory women with polycystic ovary syndrome? an opinion paper. Hum Reprod (2021) 36(9):2421–8. doi: 10.1093/humrep/deab181
6. Palomba S, Daolio J, La Sala GB. Oocyte competence in women with polycystic ovary syndrome. Trends Endocrinol Metab (2017) 28(3):186–98. doi: 10.1016/j.tem.2016.11.008
7. Harris HR, Titus LJ, Cramer DW, Terry KL. Long and irregular menstrual cycles, polycystic ovary syndrome, and ovarian cancer risk in a population-based case-control study. Int J Cancer (2017) 140(2):285–91. doi: 10.1002/ijc.30441
8. Palomba S, Piltonen TT, Giudice LC. Endometrial function in women with polycystic ovary syndrome: a comprehensive review. Hum Reprod Update (2021) 27(3):584–618. doi: 10.1093/humupd/dmaa051
9. Palomba S, Marotta R, Di Cello A, Russo T, Falbo A, Orio F, et al. Pervasive developmental disorders in children of hyperandrogenic women with polycystic ovary syndrome: a longitudinal case–control study. Clin Endocrinol (2012) 77(6):898–904. doi: 10.1111/j.1365-2265.2012.04443.x
10. Azziz R. Diagnosis of polycystic ovarian syndrome: the Rotterdam criteria are premature. J Clin Endocrinol Metab (2006) 91(3):781–5. doi: 10.1210/jc.2005-2153
11. Rojas J, Chávez M, Olivar L, Rojas M, Morillo J, Mejías J, et al. Polycystic ovary syndrome, insulin resistance, and obesity: navigating the pathophysiologic labyrinth. Int J Reprod Med (2014) 2014, 1–17. doi: 10.1155/2014/719050
12. Shukla P, Mukherjee S, Patil A. Identification of variants in mitochondrial d-loop and oriL region and analysis of mitochondrial DNA copy number in women with polycystic ovary syndrome. DNA Cell Biol (2020) 39(8):1458–66. doi: 10.1089/dna.2019.5323
13. Finsterer J. Mitochondrial dysfunction in polycystic ovary syndrome. Reprod Sci (2022) 30(5):1435–1442. doi: 10.1007/s43032-022-01100-z
14. Dabravolski SA, Nikiforov NG, Eid AH, Nedosugova LV, Starodubova AV, Popkova TV, et al. Mitochondrial dysfunction and chronic inflammation in polycystic ovary syndrome. Int J Mol Sci (2021) 22(8):3923. doi: 10.3390/ijms22083923
15. Scarfò G, Daniele S, Fusi J, Gesi M, Martini C, Franzoni F, et al. Metabolic and molecular mechanisms of diet and physical exercise in the management of polycystic ovarian syndrome. Biomedicines (2022) 10(6):1305. doi: 10.3390/biomedicines10061305
16. Hahn A, Zuryn S. Mitochondrial genome (mtDNA) mutations that generate reactive oxygen species. Antioxidants (2019) 8(9):392. doi: 10.3390/antiox8090392
17. Aidar M, Line SRP. A simple and cost-effective protocol for DNA isolation from buccal epithelial cells. Braz Dental J (2007) 18:148–52. doi: 10.1590/S0103-64402007000200012
18. Niroula A, Vihinen M. PON-mt-tRNA: a multifactorial probability-based method for classification of mitochondrial tRNA variations. Nucleic Acids Res (2016) 44(5):2020–7. doi: 10.1093/nar/gkw046
19. Sonney S, Leipzig J, Lott MT, Zhang S, Procaccio V, Wallace DC, et al. Predicting the pathogenicity of novel variants in mitochondrial tRNA with MitoTIP. PloS Comput Biol (2017) 13(12):e1005867. doi: 10.1371/journal.pcbi.1005867
20. Yarham JW, Al-Dosary M, Blakely EL, Alston CL, Taylor RW, Elson JL, et al. A comparative analysis approach to determining the pathogenicity of mitochondrial tRNA mutations. Hum Mutat (2011) 32(11):1319–25. doi: 10.1002/humu.21575
21. Barbosa G, de Sá LBPC, Rocha DRTW, Arbex AK. Polycystic ovary syndrome (PCOS) and fertility. Open J Endocrine Metab Dis (2016) 6(1):58–65. doi: 10.4236/ojemd.2016.61008
22. Ding Y, Xia BH, Zhang CJ, Zhuo GC. Mutations in mitochondrial tRNA genes may be related to insulin resistance in women with polycystic ovary syndrome. Am J Trans Res (2017) 9(6):2984.
23. Datta S, Majumder M, Biswas NK, Sikdar N, Roy B. Increased risk of oral cancer in relation to common Indian mitochondrial polymorphisms and autosomal GSTP1 locus. Cancer (2007) 110(9):1991–9. doi: 10.1002/cncr.23016
24. Czarnecka AM, Krawczyk T, Plak K, Klemba A, Zdrozny M, Arnold RS, et al. Mitochondrial genotype and breast cancer predisposition. Oncol Rep (2010) 24(6):1521–34. doi: 10.3892/or_00001014
25. Mohammed FMA, Mosaieby E, Houshmand M. Mitochondrial A12308G alteration in tRNALeu (CUN) in colorectal cancer samples. Diagn Pathol (2015) 10(1):1–4. doi: 10.1186/s13000-015-0337-6
26. Li J, Liu L, Feng Z, Wang X, Huang Y, Dai H, et al. Tumor markers CA15-3, CA125, CEA and breast cancer survival by molecular subtype: a cohort study. Breast Cancer (2020) 27:621–30. doi: 10.1007/s12282-020-01058-3
27. Zhuo G, Feng G, Leng J, Yu L, Jiang Y. A 9-bp deletion homoplasmy in women with polycystic ovary syndrome revealed by mitochondrial genome-mutation screen. Biochem Genet (2010) 48:157–63. doi: 10.1007/s10528-009-9308-5
28. Mkaouar-Rebai E, Tlili A, Masmoudi S, Charfeddine I, Fakhfakh F. New polymorphic mtDNA restriction site in the 12S rRNA gene detected in Tunisian patients with non-syndromic hearing loss. Biochem Biophys Res Commun (2008) 369(3):849–52. doi: 10.1016/j.bbrc.2008.02.107
29. Ebner S, Lang R, Mueller EE, Eder W, Oeller M, Moser A, et al. Mitochondrial haplogroups, control region polymorphisms and malignant melanoma: a study in middle European caucasians. PloS One (2011) 6(12):e27192. doi: 10.1371/journal.pone.0027192
30. Roshan M, Kabekkodu SP, Vijaya PH, Manjunath K, Graw J, Gopinath PM, et al. Analysis of mitochondrial DNA variations in Indian patients with congenital cataract. Mol Vision (2012) 18:181.
31. Wang B, Qiao L, Wang Y, Zeng J, Chen D, Guo H, et al. Mitochondrial DNA D-loop lesions with the enhancement of DNA repair contribute to gastrointestinal cancer progression. Oncol Rep (2018) 40(6):3694–704. doi: 10.3892/or.2018.6724
32. Grasso M, Diegoli M, Brega A, Campana C, Tavazzi L, Arbustini E. The mitochondrial DNA mutation T12297C affects a highly conserved nucleotide of tRNALeu (CUN) and is associated with dilated cardiomyopathy. Eur J Hum Genet (2001) 9(4):311–5. doi: 10.1038/sj.ejhg.5200622
33. Booker LM, Habermacher GM, Jessie BC, Sun QC, Baumann AK, Amin M, et al. North American white mitochondrial haplogroups in prostate and renal cancer. J Urol (2006) 175(2):468–73. doi: 10.1016/S0022-5347(05)00163-1
34. Wu CH, Chiu LT, Chang YJ, Lee CI, Lee MS, Lee TH, et al. Hypertension risk in young women with polycystic ovary syndrome: a nationwide population-based cohort study. Front Med (2020) 7:574651. doi: 10.3389/fmed.2020.574651
35. Venkatesh S, Kumar M, Sharma A, Kriplani A, Ammini AC, Talwar P, et al. Oxidative stress and ATPase6 mutation is associated with primary ovarian insufficiency. Arch gynecology obstetrics (2010) 282:313–8. doi: 10.1007/s00404-010-1444-y
36. Durham SE, Krishnan KJ, Betts J, Birch-Machin MA. Mitochondrial DNA damage in non-melanoma skin cancer. Br J Cancer (2003) 88(1):90–5. doi: 10.1038/sj.bjc.6600773
Keywords: mitochondrial DNA, mutations, PCOS, genome, sequence analysis, pathogenicity
Citation: Bibi S, Abbas G, Khan MZ, Nawaz T, Ullah Q, Uddin A, Khan MF, Ghafoor SU, Nadeem MS, Tabassum S and Zahoor M (2023) The mutational analysis of mitochondrial DNA in maternal inheritance of polycystic ovarian syndrome. Front. Endocrinol. 14:1093353. doi: 10.3389/fendo.2023.1093353
Received: 08 November 2022; Accepted: 17 April 2023;
Published: 22 August 2023.
Edited by:
Stefano Palomba, Magna Græcia University, ItalyReviewed by:
Jason Mears, Case Western Reserve University, United StatesTsung-Hsien Lee, Chung Shan Medical University, Taiwan
Copyright © 2023 Bibi, Abbas, Khan, Nawaz, Ullah, Uddin, Khan, Ghafoor, Nadeem, Tabassum and Zahoor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sadia Tabassum, c2FhZGlhLnRhYmFzc3VtODFAaHUuZWR1LnBr; Muhammad Zahoor, bXVoYW1tYWQuemFob29yQG1lZGlzaW4udWlvLm5v
†These authors have contributed equally to this work
 Shaheen Bibi1†
Shaheen Bibi1† Ghulam Abbas
Ghulam Abbas Muhammad Zahoor Khan
Muhammad Zahoor Khan Qudrat Ullah
Qudrat Ullah Aziz Uddin
Aziz Uddin Sajid Ul Ghafoor
Sajid Ul Ghafoor Muhammad Shahid Nadeem
Muhammad Shahid Nadeem Sadia Tabassum
Sadia Tabassum Muhammad Zahoor
Muhammad Zahoor