
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 31 March 2023
Sec. Cancer Endocrinology
Volume 14 - 2023 | https://doi.org/10.3389/fendo.2023.1093042
This article is part of the Research Topic Epitranscriptomics: at the Intersection of Metabolism and Aging View all 5 articles
 Wenbo Zou1
Wenbo Zou1 Hao Wang2
Hao Wang2 Dingguo Wu1
Dingguo Wu1 Yunyang Wu1
Yunyang Wu1 Kuiping Zhou1
Kuiping Zhou1 Yuanshu Lian1
Yuanshu Lian1 Gengyun Chang1
Gengyun Chang1 Yuze Feng1
Yuze Feng1 Jifeng Liang1
Jifeng Liang1 Gao Huang1*
Gao Huang1*Introduction: Malignant pancreatic cancer has poor long-term survival. Increasing evidence shows that FAM83A (family with sequence similarity 83 member A) plays a vital role in tumorigenesis and malignant progression in some human cancer types. The present study explored the potential mechanism of FAM83A in improving the prognosis of pancreatic cancer patients.
Methods: Transcriptomic and clinical data from patients were obtained from The Cancer Genome Atlas while FAM83A expression was measured in tumorous pancreatic tissue compared with normal controls by quantitative real-time PCR and immunohistochemistry.
Results: FAM83A is a vital prognostic indicator and potential oncogene in pancreatic cancer via pan-cancer analysis. In silico analysis revealed that AL049555.1/hsa-miR-129-5p axis was the pivotal upstream ncRNA- mediated pathway of FAM83A in pancreatic cancer. Furthermore, FAM83A expression was related to immune cell infiltration through vital immune-related genes including programmed cell death 1 (PDCD1), and tumorigenesis through common mutation genes including KRAS protooncogene GTPase (KRAS), and SMAD family member 4 (SMAD4). In summary, ncRNA-mediated upregulation of FAM83A is associated with poor long-term survival and immune cell infiltration in pancreatic cancer.
Discussion: FAM83A may be used as a novel survival-related and immune-related biomarker. This information suggests that FAM83A may be a novel therapeutic target for combined or individual treatment for patients with pancreatic cancer.
Pancreatic cancer (PC) has the highest mortality rates of all the cancer types, with an average 5-year survival rate of approximately 10% in the United States (1). A lack of obvious early symptoms and a difficult diagnosis led to poor treatment efficacy and prognosis, thus increasing mortality. Radical surgery is the only curative treatment, and adjuvant chemotherapy is currently the main treatment for advanced PC. The increasing development of various comprehensive treatments has gradually increased the overall survival (OS) of patients (1, 2); however, most patients lose the opportunity for surgical treatment because of distant metastasis or local invasion, and resistance to chemotherapy is inevitable (3, 4). Immunotherapy has recently revolutionized oncotherapy since it can mobilize the patient’s immune system to enhance its antitumor abilities (5). New human cancer-immune phenotypes were proposed: immune-inflamed, immune-excluded, and immune-desert tumors (6) highlighting that immune subtypes affect anticancer responses. Thus, exploring the biomarkers that participate in immune-subtype transition is essential to better understand the immunologic mechanisms and assist the immunotherapy of patients with PC. Our previous studies showed that several key genes play a vital role in immune subtype transition via plentiful bioinformatic analyses (7). According to the previous study, we explored the differential expressed markers among subtypes, and found the FAM83A play an essential role in the tumorigenesis and immune mechanism of pancreatic cancer. It may exert an effectiveness on the immune subtype transition. However, its specific prognostic value and impact of immune microenvironment in various cancers involved PC need to be further revealed and validated.
Family with sequence similarity 83 member A (FAM83A) is located on chromosome 8q24 and is a potential biomarker in human cancer (8). The conserved DUF1669 domain at the N-terminus participates in tumorigenesis and malignant progression of several human cancers (9–11). FAM83A is upregulated in various malignancies such as lung, breast, and cervical cancers (12–15) indicating that it is a biomarker of poor prognosis and suggests that FAM83A is a key oncogene (16). FAM83A depletion play an important role in breast cancer, FAM83A can reverse the malignant biological behaviors, and rendered the sensitive to EGFR-tyrosine kinase inhibitors (17). FAM83A upregulation promotes the progression of non-small cell lung cancer (NSCLC) by inhibiting the mitogen-activated protein kinase (MAPK) signal transduction pathway (12), and promotes tumorigenicity in NSCLC via the signal-regulated kinase (ERK) pathway and phosphatidylinositol-3-kinase/mammalian target of rapamycin (PI3K/Akt/mTOR) pathways (18). In addition, FAM83A/PI3K/AKT/c-Jun formed a feedback structure that induced malignant progression in hepatocellular carcinoma (19). FAM83A was also studied in PC: Upregulated FAM83A promotes cancer stem cell-like traits and enhances chemoresistance in PC showing that FAM83A plays a vital oncogenic role in the malignant progression of PC (20). Notably, FAM83A upregulates tetraspanin 1 (TSPAN1) through the WNT-CTNNB1 signaling pathway and affects macroautophagy/autophagy, suggesting that FAM83A and its target axis may predict prognosis and become a novel therapeutic target for patients with PC (21). Although several studies have revealed the mechanism of action of FAM83A in PC, but lack of some researches explore the upstream regulators of FAM83A in PC. Thus, more research is needed to explore the possible regulated pathways of FAM83A for assisting treatment in patients with this disease.
In the present study, the prognostic value of FAM83A in pan-cancer was also revealed using expression and survival analyses. The candidate noncoding RNAs (ncRNAs); microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) regulating FAM83A were detected in PC. Clinical correlation analyses of FAM83A expression identified the relationship of FAM83A expression with clinicopathologic characteristics and immune cell infiltration in PC. Finally, the correlation between FAM83A expression and key immune-related biomarkers and common mutation genes was investigated. In conclusion, ncRNA-mediated upregulation of FAM83A was associated with poor prognosis and immune status transition in PC patients.
The mRNA matrixes of 33 cancer types were downloaded from The Cancer Genome Atlas (TCGA) database (https://genome-cancer.ucsc.edu/) (22) and normalized for subsequent analyses. FAM83A differential expression analysis was performed in pan-cancer using “limma” package (23) with the threshold P value set to < 0.05. The GEPIA database (http://gepia.cancer-pku.cn/) was used to verify the differential expression of FAM83A in human cancer types (24).
Tumor tissues with pathological diagnosis of PC and paracancerous normal tissues were prospectively collected from the No.924 Hospital of PLA Joint Logistic Support Force, some of them were preserved in 10% neutral formalin-fixed for immunohistochemical (IHC) staining, and the other parts were preserved at -80°C refrigerator for quantitative real time-polymerase chain reaction (qRT-PCR). This study was approved by the ethics committee of the No.924 Hospital of PLA Joint Logistic Support Force. The patients/participants provided their written informed consent to participate in this study.
The predictive power of FAM83A in OS was produced using a Kaplan-Meier survival curve with “survival” and “survminer” R packages. Univariate Cox regression analyses evaluated the significance of FAM83A in predicting OS, disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) in screened cancer types. In addition, the correlation between FAM83A expression and the American Joint Committee on Cancer (AJCC) stage was analyzed in PC.
Tumor tissues with pathological diagnosis of PC and normal tissues were prospective collected from the No.924 Hospital of PLA Joint Logistic Support Force. TRIzol reagent(Ambion) was used to extracted total RNA; NanoPhotometer® C40 Touch (IMPLEN) was used to assess the RNA purity based on the ratio of OD260/280 and 260/230; Eppendorf Mastercycler® was used to perform reverse transcription of qualified RNA to single-stranded complementary DNA according to the manufacturer’s instructions; StepOnePlus Real-Time PCR system was used to implement real-time quantification; 18S rDNA was used as an internal reference, the cycle threshold (Ct) was recorded, and relative expression was calculated using the 2−ΔΔCt method. FAM83A, AL049555.1, hsa-miR-129-5p and 18S rDNA primer sequences are shown in Table S1.
Next, the PC, paracancerous and normal tissues preserved in in 10% neutral formalin-fixed were undergone IHC. The IHC staining of the collected tissue was performed according to the manufacturer’s protocol; the primary antibody is stated in Table S2. All images were obtained with an Olympus DP72 camera using Olympus image analysis software.
Univariate and multivariate Cox regression analyses were performed on the FAM83A expression level and clinical data including age, sex, race, and AJCC stage: T stage, N stage, and M stage. A nomogram was developed by combining the independent prognostic factors to predict the 1-, 2-, and 3-year survival of patients with PC, with calibration curves and receiver operating characteristic (ROC) curves generated to evaluate the performance of the nomogram. Analyses were performed using the “rms” and “foreign” R packages.
Candidate upstream binding miRNAs of FAM83A were predicted based on the seven prediction programs: PITA, RNA22, miRmap, microT, miRanda, PicTar, and TargetScan in starBase. miRNAs predicted by two or more programs were included in subsequent analyses. Expression correlation analysis of target miRNAs with FAM83A in PC was conducted using starBase (http://starbase.sysu.edu.cn/) (25) according to the competing endogenous RNA (ceRNA) hypothesis (26). Final regulatory networks were elucidated using Cytoscape version 3.7.2. Potential binding of lncRNAs to the pivotal miRNAs was also predicted by starBase.
GEPIA was used to evaluate the differential expression and prognostic value of candidate lncRNAs in PC by integrating transcriptomes from cancer patients and normal patients from the TCGA and Genotype-Tissue Expression (GTEx) portals (24).
FAM83A may participate in immune-subtype variation since it is significantly differentially expressed among the immune subtypes (7). The CIBERSORT algorithm was utilized to assess ICI in PC and estimate its distribution in 22 immune cell types by gene expression analysis (27). Spearman’s correlation analysis was used to evaluate the degree of correlation between FAM83A expression and ICI.
The correlation of FAM83A copy number variation with immune cell infiltration in PC was analyzed by TIMER (https://cistrome.shinyapps.io/timer/) (28) using a threshold of P < 0.05.
Immune-related genes including immunostimulatory factors, immunoinhibitory factors, chemokines, and chemokine receptors were extracted from the Tumor and Immune System Interaction Database (TISIDB) (http://cis.hku.hk/TISIDB/) and used to determine correlation coefficients using R software (29). In addition, common gene mutations were also used in the analysis.
R software (version 4.0.2), GraphPad Prism (version 8.0.1), and relevant website tools were used for all statistical analyses and graphics. Survival curves were generated using the Kaplan-Meier method and compared using the log-rank test. Statistical significance was set at P < 0.05 and all tests were two-tailed.
FAM83A was significantly differentially expressed in 15/33 types of human cancer analyzed from the TGCA database. Significantly high expression was observed in BLCA (bladder urothelial cancer), BRCA (breast invasive cancer), CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma), COAD (colon adenocarcinoma), ESCA (esophageal carcinoma), GBM (glioblastoma multiforme), HNSC (head and neck squamous cell carcinoma), LUAD (lung adenocarcinoma), LUSC (lung squamous cell carcinoma), PAAD (pancreatic adenocarcinoma), READ (rectum adenocarcinoma), STAD (stomach adenocarcinoma), and UCEC (uterine corpus endometrial carcinoma) cancer tissues, while significantly low expression was found in PRAD (prostate adenocarcinoma) and KICH (kidney chromophobe) compared with the normal tissue (Figure 1A).
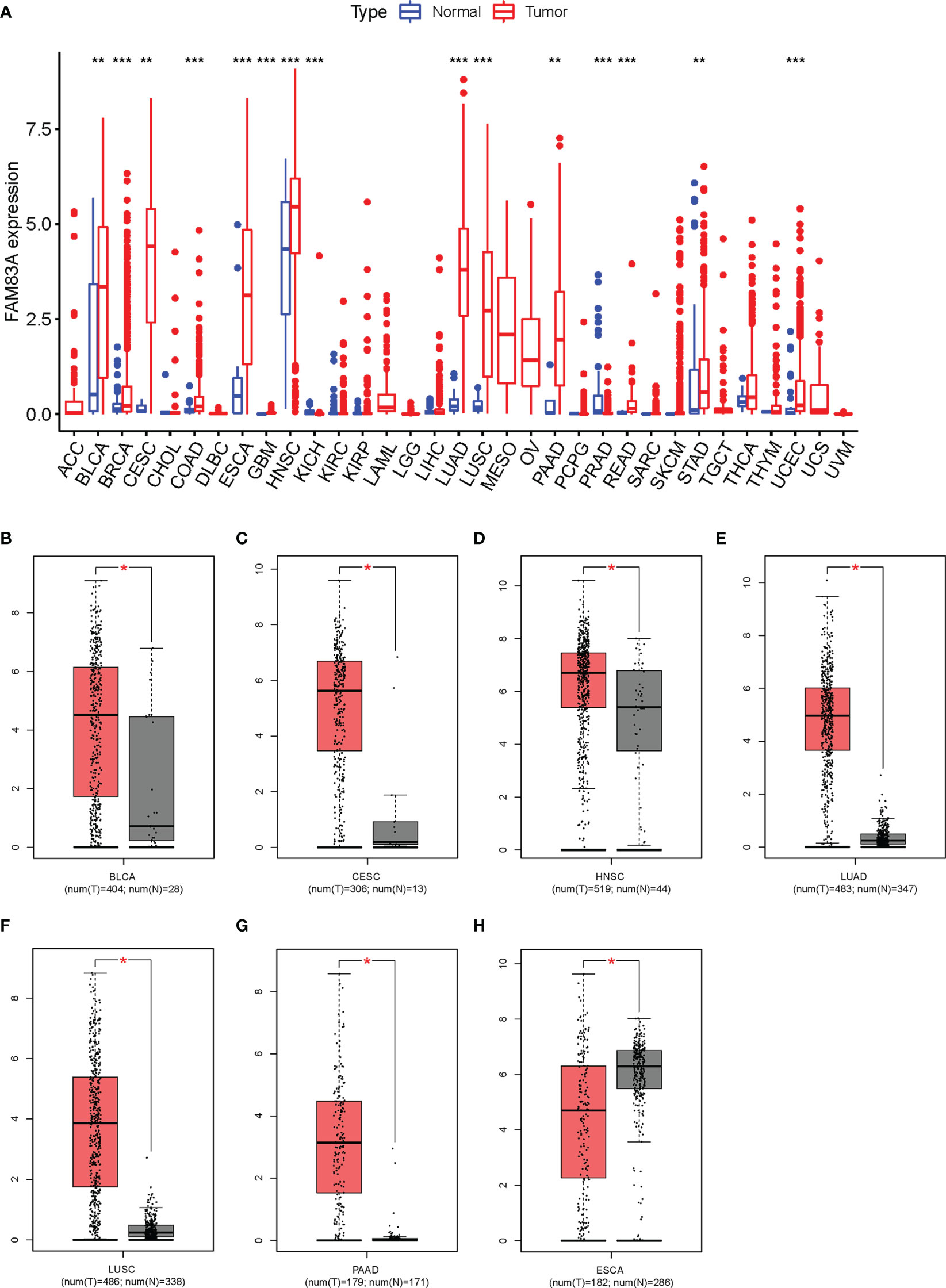
Figure 1 Expression analysis for FAM83A in multiple cancers: (A) The expression of FAM83A in human cancers based on TCGA database. (B–H) The differential expression of FAM83A between tumor and normal tissues in TCGA and GTEx: (B) BLCA (C), CESC (D), HNSC (E), LUAD (F), LUSC (G), PAAD (H), ESCA. *p value < 0.05; **p value < 0.01; ***p value < 0.001.
FAM83A expression levels in these 15 cancer types were reanalyzed using the GEPIA database with significantly higher expression observed in BLCA, CESC, HNSC, LUAD, LUSC, and PAAD cancer tissues and low expression in ESCA (Figures 1B–H). No significantly differentially expressed in other 8/15 types of human cancers (Figure S1). ESCA was omitted from further analysis since FAM83A expression was inconsistent between these two methods. These results suggested that upregulated FAM83A may play a crucial role in tumorigenesis.
Kaplan-Meier OS survival analysis for FAM83A in BLCA, CESC, HNSC, LUAD, LUSC, and PAAD showed that FAM83A was a positive risk factor for LUAD, LUSC, and PAAD patients (Figures 2A–C). Univariate Cox regression OS analyses suggested that FAM83A is a risk factor for patients with LUAD, LUSC, and PAAD in these six human cancer types (Figure 2D), while DSS, DFI, and PFI analyses revealed that FAM83A was a risk factor for patients with LUAD and PAAD (Figures 2E–G).
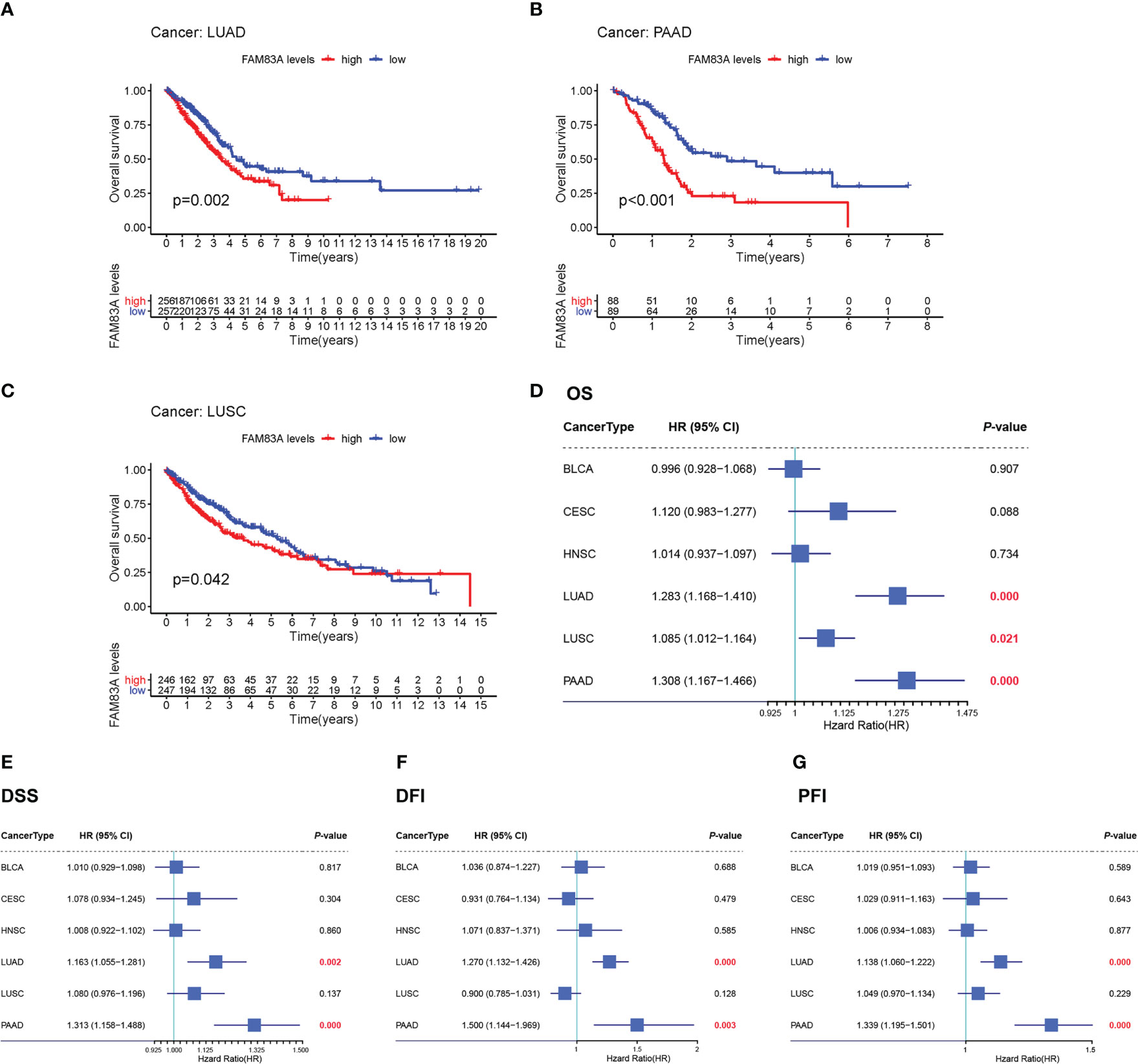
Figure 2 The overall survival (OS) and univariate Cox regression analysis for FAM83A in 6 human cancers based on TCGA: (A–C) The OS plot of FAM83A in LUAD (A), PAAD (B), LUSC (C). (D) the OS forest plot. (E) the DSS forest plot. (F) the DFI forest plot. (G) the PFI forest plot.
FAM83A was significantly highly expressed in tumorous PC tissues obtained from patients compared with that in normal controls (Figure 3A). The IHC results showed that the FAM83A located in cytoplasm, while FAM83A was also positively expressed in PC compared with paracancerous and normal tissues (Figures 3B–D).
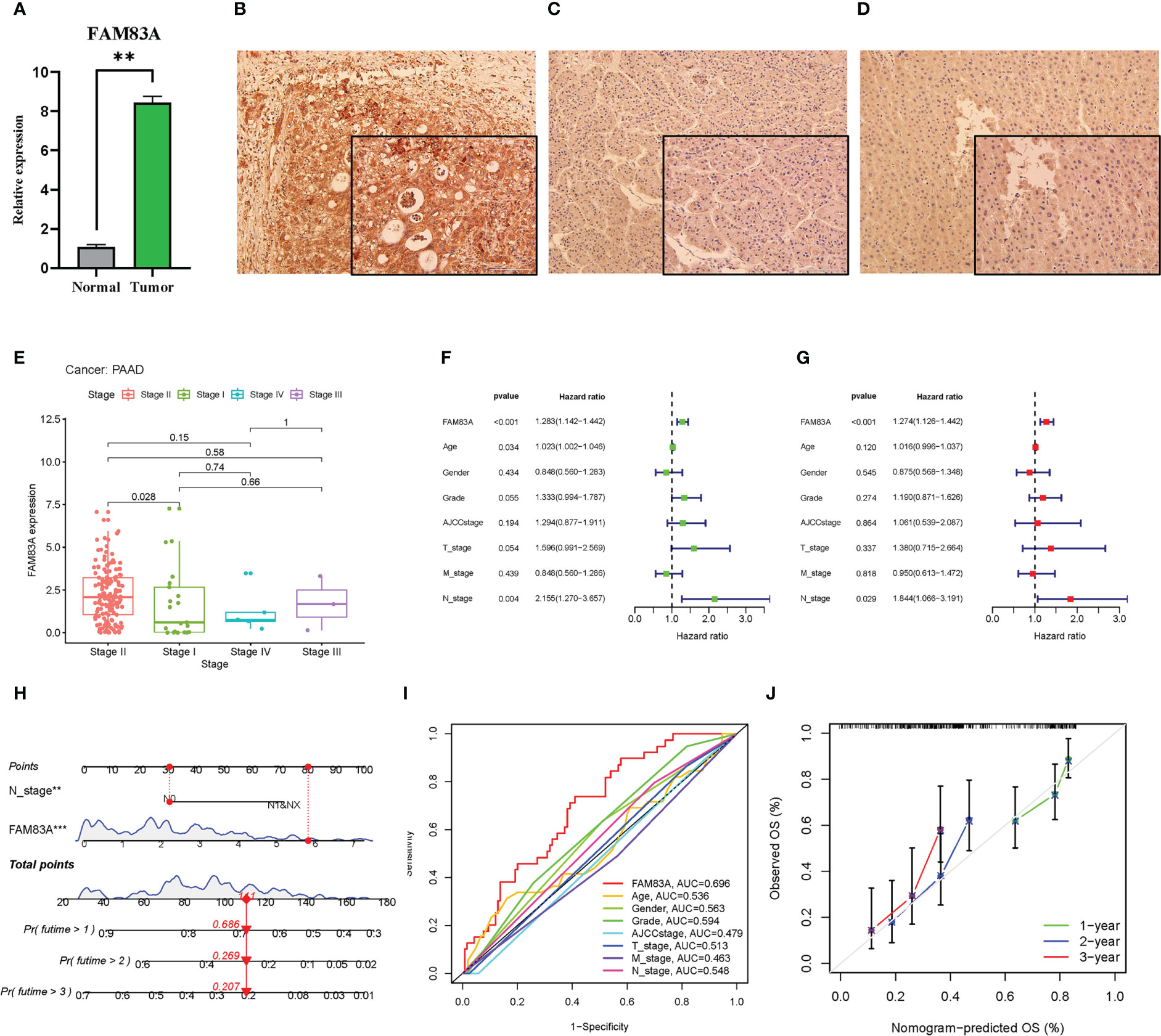
Figure 3 The laboratory experiment and clinical significance of FAM83A in PAAD: (A–D) The laboratory experiment. (A) The qRT-PCR result of FAM83A between the tumor and normal tissues. (B) The Immunohistochemistry (IHC) staining plot in PC. (C) The IHC staining plot in para-cancerous tissue. (D) The IHC staining plot in normal tissue. (E-J) clinical significance of FAM83A. (E) The correlation analysis of FAM83A with AJCC stage. (F) Univariate Cox regression analysis. (G) Multivariate Cox regression analysis. (H) The nomogram based on FAM83A expression and N stage. (I) The multivariate ROC curves. (J) The calibration curves at 1-, 2-, 3- years. **p value < 0.01; ***p value < 0.001.
Increased FAM83A expression was found in late-stage PAAD after clinical correlation analysis of different AJCC stages in these six cancer types (Figure 3E). This indicated that FAM83A may be considered a detrimental prognostic biomarker in PC patients. Incorporation of clinicopathologic characteristics into the prognosis analysis showed that FAM83A and the N stage significantly correlated with OS in both univariate and multivariate Cox regression analyses (Figures 3F, G: P < 0.001 and P = 0.029, respectively). Integration of FAM83A and N stages into a risk model for predicting 1-, 2-, and 3-year survival rates, and FAM83A was the strongest prognostic indicator compared with other variates with an 0.696 area under curve (AUC) value of multivariate ROC (Figures 3H, I). The calibration curves confirmed that the nomogram adequately predicted the OS probability at 1-, 2-, and 3- years (Figure 3J) indicating that FAM83A is a good predictive indicator for patients with PC.
ncRNA-related pathways play pivotal roles in gene expression with miRNAs inhibiting target gene expression (30). It was predicted that eleven upstream miRNAs bind to FAM83A (Table S3), and Cytoscape established the miRNA-FAM83A regulatory network (Figure 4). FAM83A significantly negatively correlated with hsa-miR-129-5p in PC by expression correlation analysis (Table 1, r = -0.322, P < 0.001). There was a statistically significant correlation between hsa-miR-367-3p and FAM83A, however, the correlation coefficient was weak.
The starBase database identified a total of 118 potential upstream lncRNAs of hsa-miR-129-5p (Table S4) and a lncRNA-hsa-miR-129-5p regulatory network was established using Cytoscape (Figure S2). Differential expression and prognostic values of these lncRNAs in PC analyzed by the GEPIA database showed that only AL049555.1 (RP3-523K23.2, ENSG00000261116) was significantly upregulated in PC compared with normal controls (Figure 5A). Increased AL049555.1 expression indicated poorer OS and relapse-free survival in PC, even though P > 0.05 in the latter (Figures 5B–C). AL049555.1 was strongly negatively correlated with hsa-miR-129-5p (r = -0.393, P value < 0.001) and strongly positively correlated with FAM83A (r = 0.518, P value < 0.001) (Figures 5D, E). Meanwhile, hsa-miR-129-5p was strongly negatively correlated with FAM83A (r = -0.322, P value < 0.001) (Figure 5F). This survival and correlation analysis suggested that the novel lncRNA, AL049555.1, may be the upstream lncRNA of the hsa-miR-129-5p/FAM83A axis in PC. Finally, in the qRT-PCR assay, hsa-miR-129-5p was significantly lowly expressed in tumorous PC tissues compared to that in normal controls, and AL049555.1 was significantly highly expressed in tumorous PC tissues obtained from patients compared with that in normal controls (Figure S3), which shows the ncRNAs (hsa-miR-129-5p and AL049555.1) play the pivotal roles in tumorigenesis and progression of PC, and may be considered to have abilities to regulate the expression of FAM83A.
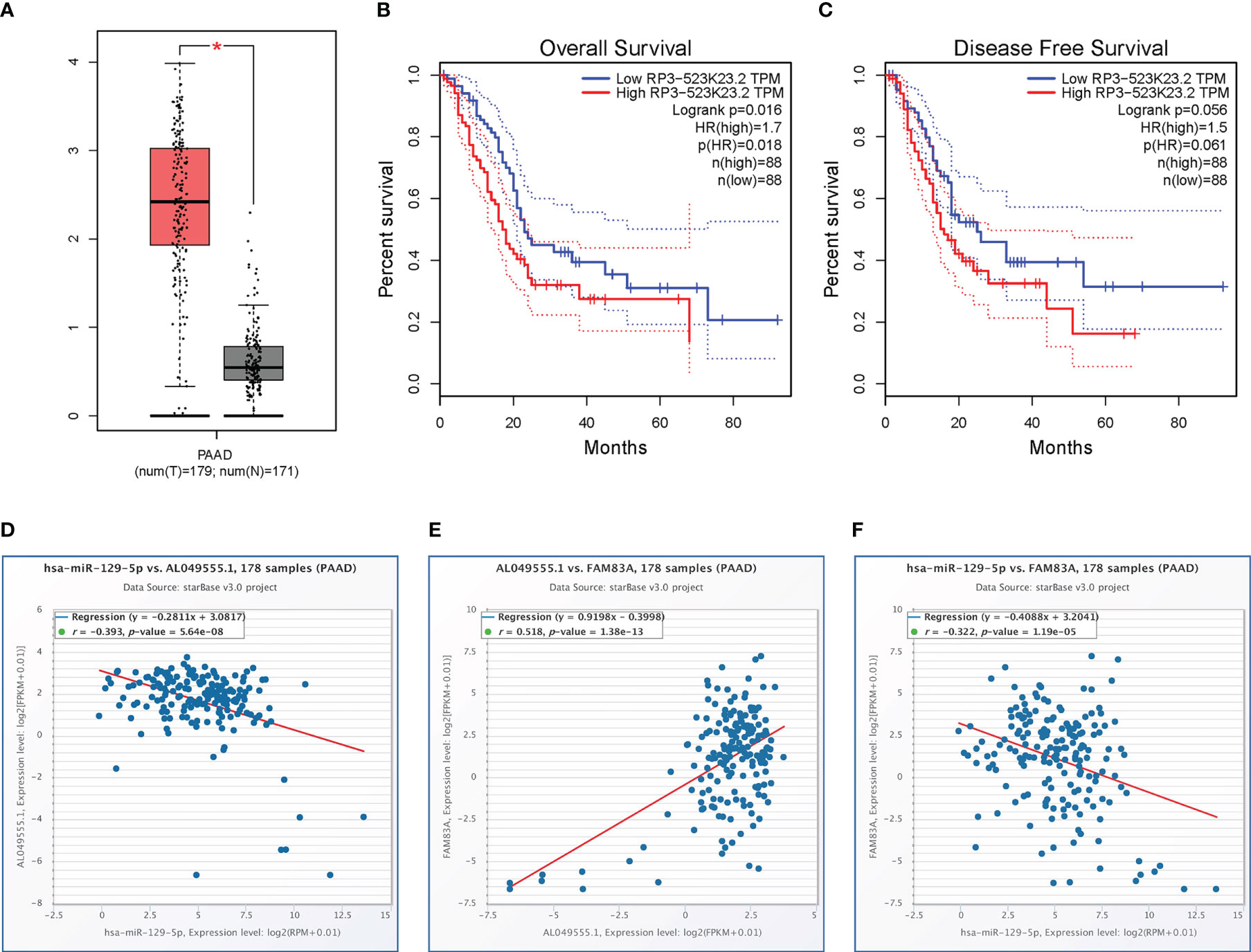
Figure 5 Expression analysis and survival analysis for AL049555.1, and relationship among AL049555.1/hsa−miR−129−5p/FAM83A axis: (A) The expression of AL049555.1 based on TCGA and GTEx. (B-C) survival analysis of OS and RFS for AL049555.1. (D) The AL049555.1 was strong negatively corelated with hsa-miR-129-5p. (E) The AL049555.1 was strong positively corelated with FAM83A. (F) hsa-miR-129-5p was strong negatively corelated with FAM83A. *p value < 0.05.
FAM83A expression was significantly positively correlated with memory B cells and M0 macrophage infiltration in PC, with significant changes in B cells, CD4+ T cells, and CD8+ T cell infiltration levels under various FAM83A copy numbers in PC (Figures 6A–C). This suggested that FAM83A variation may influence ICI in the tumor microenvironment of PC.
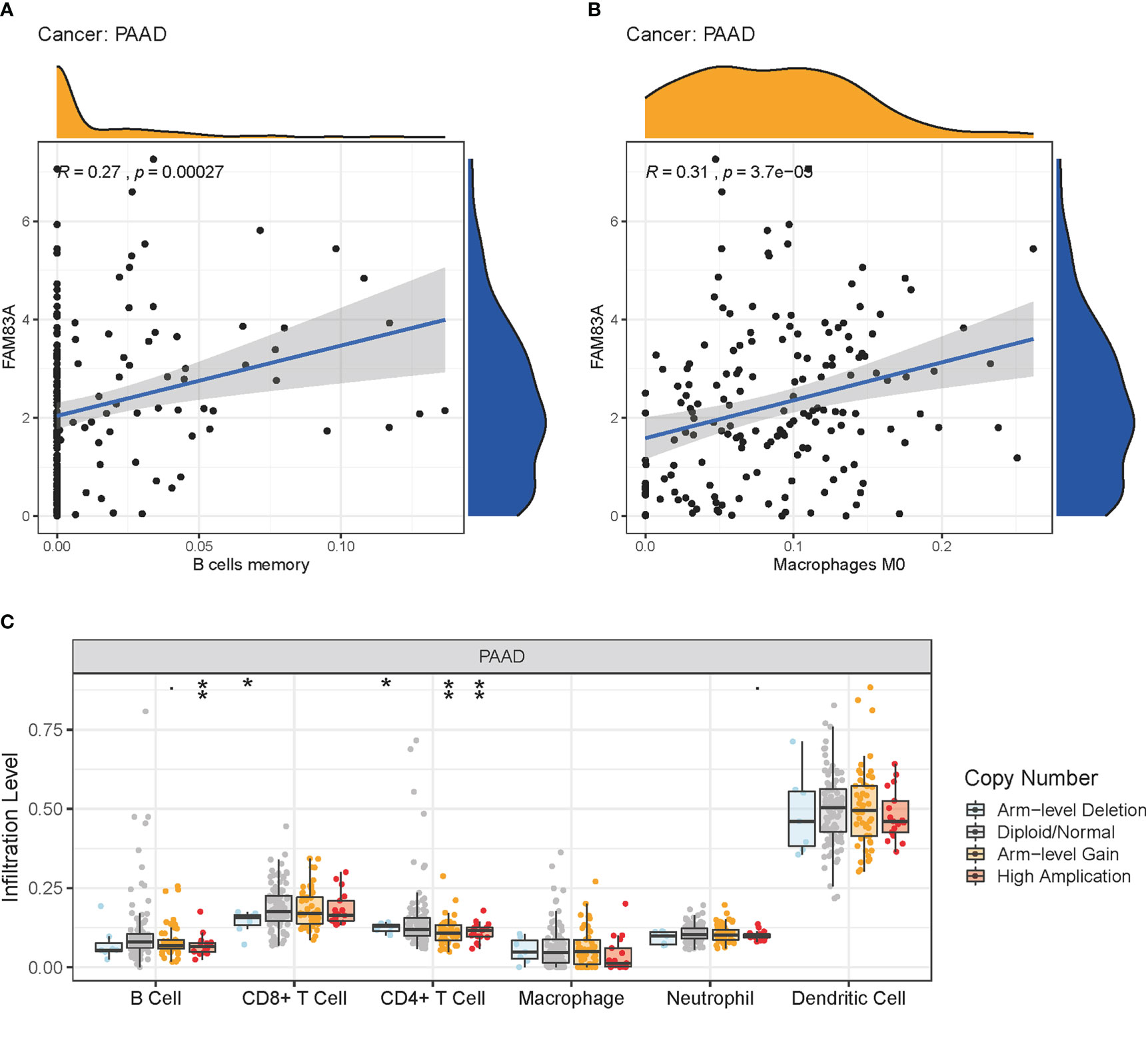
Figure 6 Immune cell infiltration (ICI) analysis: (A-B) The relationship of FAM83A expression with ICI in PC. (A) Memory B cells. (B) Macrophages M0. (C) ICI level under copy numbers variation of FAM83A in PC. *p value < 0.05; **p value < 0.01.
FAM83A expression correlated with a series of immune-related genes in PC such as CD160 and interleukin 10 (IL10), and a vital immune checkpoint protein, programmed cell death 1 (PDCD1); Chemokines, C-C motif chemokine ligand 4 (CCL4), CCL7, CCL13, and their receptors: CCR3, CCR4, and CCR6; and common mutation genes including: KRAS proto-oncogene, GTPase (KRAS), SMAD family member 4 (SMAD4) (Figures 7A–E). The positive interaction between the FAM83A axis expression, the immune landscape, and gene mutations will assist future immunotherapy for patients with PC.
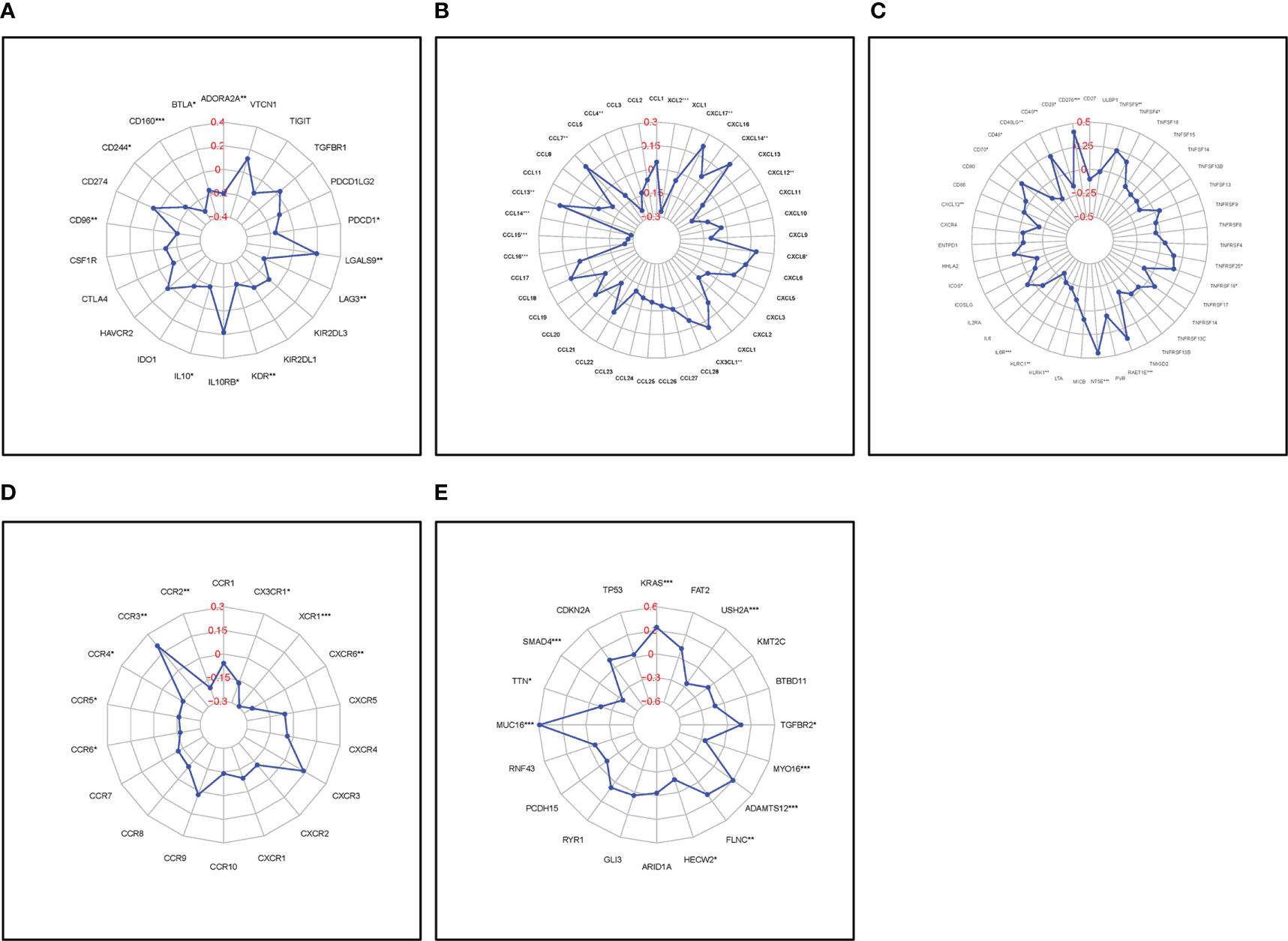
Figure 7 Correlation of FAM83A expression with immune-related biomarkers and common mutation genes: (A) Immunostimulatory factors. (B) Immunoinhibitory factors. (C) Chemokines. (D) Chemokine receptors. (E) Mutation genes. Red numbers are considered correlation coefficients.
Malignant pancreatic tumors are increasingly threatening human life and health and imposing a serious disease burden on society worldwide (1, 5). The very high mortality rate in PC places importance on exploring novel clinical biomarkers and staging systems to provide personalized treatments to improve patients’ long-term survival (1). Several studies showed that FAM83A plays a pivotal role in the tumorigenesis of various malignancies (12, 14, 21), however, its role in PC is poorly understood. In this study, we revealed FAM83A is associated with poor long-term survival and immune cell infiltration in pancreatic cancer. FAM83A may be used as a novel survival-related and immune-related biomarker. It suggests that FAM83A may be a novel therapeutic target for combined or individual treatment for patients with pancreatic cancer.
Many recent studies have demonstrated that ncRNAs (lncRNA, miRNA, and circular RNA) involved in tumorigenesis affect the expression level of target genes via the ceRNA action mechanism (31–33). Eleven candidate upstream miRNA binding targets of FAM83A were determined using seven prediction programs in the Starbase database. Correlation analysis between the candidate miRNAs and FAM83A identified hsa-miR-129-5p as the most adaptive upstream miRNA of FAM83A which suppresses its expression. Previous studies showed that hsa−miR−129−5p plays a vital regulatory role in glioma (34) and is involved in chemoresistance in ovarian cancer cells (35). However, few studies have focused on the potential mechanism of hsa−miR−129−5p in regulating target genes and tumorigenesis of PC. Our findings are the first to show that hsa−miR−129−5p is a candidate miRNA that negatively regulates FAM83A expression and participates in PC suppression.
ceRNA hypothesis states that lncRNAs are positively correlated with mRNA expression since they negatively regulate miRNA expression. Thus, lncRNAs of hsa−miR−129−5p/FAM83A axis is potentially positively correlated with carcinogenesis in PC. One hundred and eighteen potential lncRNAs of hsa−miR−129−5p/FAM83A axis was predicted, with only AL049555.1 detected as the candidate lncRNA following correlation, survival, and differential expression analysis. AL049555.1 is a novel lncRNA so its mechanism of action in human cancers has not been studied. This is the first study showing that AL049555.1/hsa−miR−129−5p/FAM83A axis is a vital ceRNA regulatory network involved in promoting PC development.
Various studies demonstrated that immune cells play a pivotal role in the tumor immune microenvironment which may affect the response to immunotherapy (36, 37) and the rate of ICI directly affects patient prognosis (38, 39). This study showed that FAM83A variation significantly influenced the ICI landscape of PC through significantly positive correlations with memory B cells and M0 macrophage infiltrations, with significant changes in B cells, CD4+ T cells, and CD8+ T cell infiltration levels.
Several immune-related genes including the immune checkpoint are closely related to the development of malignant tumors and affect the response to immunotherapy in patients with PC. For instance, targeted inhibition of programmed cell death 1 (PD-1) or programmed cell death ligand 1 (PD-L1) successfully treated various tumors (40, 41). This study showed that FAM83A expression correlated with various immune-related biomarkers such as PDCD1 which suggests that targeting FAM83A may influence the response to immunotherapy in PC.
High mutation rates of KRAS and tumor protein p53 (TP53) are highly related to the immune status of tumors suggesting that they are predictors of responses to PD1/PDL1 immunotherapy (42, 43). This study showed that upregulated FAM83A positively correlated with KRAS and mucin 16, cell surface associated (MUC16), and negatively correlated with SMAD4 and titin (TTN) indicating that targeting FAM83A may affect the key gene mutation proteins and further influence immunotherapy responses in PC.
In present study, four endpoints (OS, DSS, DFI, and PFI) survival analyses of several cancer types indicated that PC patients with high FAM83A expression had shorter long-term survival. The clinical prognostic values of FAM83A in PC carried out using a correlation analysis between FAM83A expression and clinicopathological characteristics showed increased FAM83A expression at later AJCC stages in PC. A nomogram was constructed and validated, providing novel predictive tools for clinicians showing that FAM83A expression and the N stage are independent risk factors using univariate and multivariate Cox regression.
Despite these significant findings, some limitations must be noted. First, all the obtained transcriptome data were analyzed retrospectively based on a multi database with intrinsic bias. Although, in vitro experiment validated FAM83A expression in tumorigenic PC tissues and in silico analysis propose a novel AL049555.1/hsa−miR−129−5p/FAM83A axis in tumorigenesis and reveal the axis-related immune microenvironment status in patients with PC, our results will need to be validated in a large independent cohort. Therefore, further abundant basic laboratory experiments and clinical trials are required to validate this work.
A novel PC AL049555.1/hsa−miR−129−5p/FAM83A axis was constructed (Figure 8). FAM83A accelerated tumorigenesis and development and executed its carcinogenic role by influencing ICI, immune-related biomarker expression, and expression of commonly mutated genes.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the ethics committee of the No.924 Hospital of PLA Joint Logistic Support Force. The patients/participants provided their written informed consent to participate in this study.
Study concept and design: GH, WZ. Drafting of the manuscript: GH, WZ, HW. Acquisition of data, analysis, and interpretation of data: HW, DW, YW, KZ, YL, GC. Critical revision of the manuscript: GH. Statistical analysis: HW, YF, JL. Study supervision: GH. All authors contributed to the article and approved the submitted version.
We acknowledge TCGA, UCSC Xena, starBase, and TISIDB databases for providing their platforms and contributors for uploading their meaningful datasets, and thank all individuals who participated in this study and donated samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1093042/full#supplementary-material
1. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet (2020) 395:2008–20. doi: 10.1016/S0140-6736(20)30974-0
2. Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol (2018) 15:333–48. doi: 10.1038/s41575-018-0005-x
3. Li Q, Yang G, Feng M, Zheng S, Cao Z, Qiu J, et al. NF-κB in pancreatic cancer: Its key role in chemoresistance. Cancer Lett (2018) 421:127–34. doi: 10.1016/j.canlet.2018.02.011
4. Yu S, Zhang C, Xie KP. Therapeutic resistance of pancreatic cancer: Roadmap to its reversal. Biochim Biophys Acta Rev Cancer (2021) 1875:188461. doi: 10.1016/j.bbcan.2020.188461
5. Schizas D, Charalampakis N, Kole C, Economopoulou P, Koustas E, Gkotsis E, et al. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat Rev (2020) 86:102016. doi: 10.1016/j.ctrv.2020.102016
6. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature (2017) 541:321–30. doi: 10.1038/nature21349
7. Wang Z, Zou W, Wang F, Zhang G, Chen K, Hu M, et al. Identification of the immune cell infiltration landscape in pancreatic cancer to assist immunotherapy. Future Oncol (2021) 17(31):4131–43. doi: 10.2217/fon-2021-0495
8. Li Y, Dong X, Yin Y, Su Y, Xu Q, Zhang Y, et al. BJ-TSA-9, a novel human tumor-specific gene, has potential as a biomarker of lung cancer. Neoplasia (2005) 7:1073–80. doi: 10.1593/neo.05406
9. Fulcher LJ, Bozatzi P, Tachie-Menson T, Wu KZL, Cummins TD, Bufton JC, et al. The DUF1669 domain of FAM83 family proteins anchor casein kinase 1 isoforms. Sci Signal (2018) 11(531):eaao2341. doi: 10.1126/scisignal.aao2341
10. Bozatzi P, Sapkota GP. The FAM83 family of proteins: from pseudo-PLDs to anchors for CK1 isoforms. Biochem Soc Trans (2018) 46:761–71. doi: 10.1042/BST20160277
11. Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol Cancer Res (2014) 12:1156–65. doi: 10.1158/1541-7786.MCR-13-0289
12. Wang Y, Xu R, Zhang D, Lu T, Yu W, Wo Y, et al. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl Lung Cancer Res (2019) 8:862–75. doi: 10.21037/tlcr.2019.11.04
13. Liu C, Jiang Y, Han B. miR-613 suppresses chemoresistance and stemness in triple-negative breast cancer by targeting FAM83A. Cancer Manag Res (2020) 12:12623–33. doi: 10.2147/CMAR.S276316
14. Xu J, Lu W. FAM83A exerts tumor−suppressive roles in cervical cancer by regulating integrins. Int J Oncol (2020) 57:509–21. doi: 10.3892/ijo.2020.5078
15. Liu Z, Zhang Y, Dang Q, Wu K, Jiao D, Li Z, et al. Genomic alteration characterization in colorectal cancer identifies a prognostic and metastasis biomarker: FAM83A|IDO1. Front Oncol (2021) 11:632430. doi: 10.3389/fonc.2021.632430
16. Zhou C, Zhu X, Liu N, Dong X, Zhang X, Huang H, et al. B-lymphoid tyrosine kinase-mediated FAM83A phosphorylation elevates pancreatic tumorigenesis through interacting with β-catenin. Signal Transduct Target Ther (2023) 8:66. doi: 10.1038/s41392-022-01268-5
17. Lee SY, Meier R, Furuta S, Lenburg ME, Kenny PA, Xu R, et al. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest (2012) 122:3211–20. doi: 10.1172/JCI60498
18. Hu H, Wang F, Wang M, Liu Y, Wu H, Chen X, et al. FAM83A is amplified and promotes tumorigenicity in non-small cell lung cancer via ERK and PI3K/Akt/mTOR pathways. Int J Med Sci (2020) 17:807–14. doi: 10.7150/ijms.33992
19. Liu C, Peng X, Li Y, Liu S, Hou R, Zhang Y, et al. Positive feedback loop of FAM83A/PI3K/AKT/c-jun induces migration, invasion and metastasis in hepatocellular carcinoma. BioMed Pharmacother (2020) 123:109780. doi: 10.1016/j.biopha.2019.109780
20. Chen S, Huang J, Liu Z, Liang Q, Zhang N, Jin Y. FAM83A is amplified and promotes cancer stem cell-like traits and chemoresistance in pancreatic cancer. Oncogenesis (2017) 6:e300. doi: 10.1038/oncsis.2017.3
21. Zhou C, Liang Y, Zhou L, Yan Y, Liu N, Zhang R, et al. TSPAN1 promotes autophagy flux and mediates cooperation between WNT-CTNNB1 signaling and autophagy via the MIR454-FAM83A-TSPAN1 axis in pancreatic cancer. Autophagy (2021) 17(10):3175–95. doi: 10.1080/15548627.2020.1826689
22. Goldman MJ, Craft B, Hastie M, Repečka K, McDade F, Kamath A, et al. Visualizing and interpreting cancer genomics data via the xena platform. Nat Biotechnol (2020) 38:675–8. doi: 10.1038/s41587-020-0546-8
23. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res (2015) 43:e47. doi: 10.1093/nar/gkv007
24. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res (2017) 45:W98–w102. doi: 10.1093/nar/gkx247
25. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res (2014) 42(Database issue):D92–7. doi: 10.1093/nar/gkt1248
26. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
27. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods (2015) 12:453–7. doi: 10.1038/nmeth.3337
28. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res (2017) 77:e108–10. doi: 10.1158/1538-7445.AM2017-108
29. Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics (2019) 35:4200–2. doi: 10.1093/bioinformatics/btz210
30. Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA (2003) 100(17):9779–84. doi: 10.1073/pnas.1630797100
31. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature (2014) 505:344–52. doi: 10.1038/nature12986
32. Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet (2015) 52:710–8. doi: 10.1136/jmedgenet-2015-103334
33. Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discovery (2013) 3:1113–21. doi: 10.1158/2159-8290.CD-13-0202
34. Datta I, Noushmehr H, Brodie C, Poisson LM. Expression and regulatory roles of lncRNAs in G-CIMP-low vs G-CIMP-high glioma: an in-silico analysis. J Transl Med (2021) 19:182. doi: 10.1186/s12967-021-02844-z
35. Liu Q, Liu DW, Zheng MJ, Deng L, Wang HM, Jin S, et al. Human epididymis protein 4 promotes p−glycoprotein−mediated chemoresistance in ovarian cancer cells through interactions with annexin II. Mol Med Rep (2021) 24(1):496. doi: 10.3892/mmr.2021.12135
36. Torphy RJ, Schulick RD, Zhu Y. Understanding the immune landscape and tumor microenvironment of pancreatic cancer to improve immunotherapy. Mol Carcinog (2020) 59:775–82. doi: 10.1002/mc.23179
37. Upadhrasta S, Zheng L. Strategies in developing immunotherapy for pancreatic cancer: Recognizing and correcting multiple immune “Defects” in the tumor microenvironment. J Clin Med (2019) 8(9):1472. doi: 10.3390/jcm8091472
38. Ino Y, Yamazaki-Itoh R, Shimada K, Iwasaki M, Kosuge T, Kanai Y, et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer (2013) 108:914–23. doi: 10.1038/bjc.2013.32
39. Tahkola K, Mecklin JP, Wirta EV, Ahtiainen M, Helminen O, Böhm J, et al. High immune cell score predicts improved survival in pancreatic cancer. Virchows Arch (2018) 472:653–65. doi: 10.1007/s00428-018-2297-1
40. Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol (2008) 26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331
41. Yi M, Jiao D, Xu H, Liu Q, Zhao W, Han X, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol Cancer (2018) 17:129. doi: 10.1186/s12943-018-0864-3
42. Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res (2017) 23:3012–24. doi: 10.1158/1078-0432.CCR-16-2554
Keywords: FAM83A, pancreatic cancer, prognosis, immune cell infiltration, immunotherapy
Citation: Zou W, Wang H, Wu D, Wu Y, Zhou K, Lian Y, Chang G, Feng Y, Liang J and Huang G (2023) ncRNA-mediated upregulation of FAM83A is associated with poor prognosis and immune infiltration in pancreatic cancer. Front. Endocrinol. 14:1093042. doi: 10.3389/fendo.2023.1093042
Received: 08 November 2022; Accepted: 13 March 2023;
Published: 31 March 2023.
Edited by:
Dragana Nikitovic, University of Crete, GreeceCopyright © 2023 Zou, Wang, Wu, Wu, Zhou, Lian, Chang, Feng, Liang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gao Huang, aHVhbmdnYW8xOTgxQDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.