- 1Division of Endocrinology and Diabetes, Children’s Hospital of Philadelphia, University of Pennsylvania, Philadelphia, PA, United States
- 2Department of Biomedical and Health Informatics, Children’s Hospital of Philadelphia, Philadelphia, PA, United States
- 3Division of Oncology, Children’s Hospital of Philadelphia, University of Pennsylvania, Philadelphia, PA, United States
- 4Cancer Predisposition Program, Children’s Hospital of Philadelphia, University of Pennsylvania, Philadelphia, PA, United States
- 5Abramson Cancer Center, University of Pennsylvania, Philadelphia, PA, United States
- 6Division of Cancer Epidemiology and Genetics, National Cancer Institute, Rockville, MD, United States
- 7Department of Pathology and Laboratory Medicine, Children’s Hospital of Philadelphia, University of Pennsylvania, Philadelphia, PA, United States
- 8Division of Endocrinology, The Hospital for Sick Children, Toronto, ON, Canada
DICER1 is a highly conserved RNase III endoribonuclease essential for the biogenesis of single-stranded mature microRNAs (miRNAs) from stem-loop precursor miRNAs. Somatic mutations in the RNase IIIb domain of DICER1 impair its ability to generate mature 5p miRNAs and are believed to drive tumorigenesis in DICER1 syndrome-associated and sporadic thyroid tumors. However, the DICER1-driven specific changes in miRNAs and resulting changes in gene expression are poorly understood in thyroid tissue. In this study, we profiled the miRNA (n=2,083) and mRNA (n=2,559) transcriptomes of 20 non-neoplastic, 8 adenomatous and 60 pediatric thyroid cancers (13 follicular thyroid cancers [FTC] and 47 papillary thyroid cancers [PTC]) of which 8 had DICER1 RNase IIIb mutations. All DICER1-mutant differentiated thyroid cancers (DTC) were follicular patterned (six follicular variant PTC and two FTC), none had lymph node metastasis. We demonstrate that DICER1 pathogenic somatic mutations were associated with a global reduction of 5p-derived miRNAs, including those particularly abundant in the non-neoplastic thyroid tissue such as let-7 and mir-30 families, known for their tumor suppressor function. There was also an unexpected increase of 3p miRNAs, possibly associated with DICER1 mRNA expression increase in tumors harboring RNase IIIb mutations. These abnormally expressed 3p miRNAs, which are otherwise low or absent in DICER1-wt DTC and non-neoplastic thyroid tissues, make up exceptional markers for malignant thyroid tumors harboring DICER1 RNase IIIb mutations. The extensive disarray in the miRNA transcriptome results in gene expression changes, which were indicative of positive regulation of cell-cycle. Moreover, differentially expressed genes point to increased MAPK signaling output and loss of thyroid differentiation comparable to the RAS-like subgroup of PTC (as coined by The Cancer Genome Atlas), which is reflective of the more indolent clinical behavior of these tumors.
Introduction
DICER1 is a highly conserved RNase III endoribonuclease essential for the biogenesis of single-stranded mature microRNAs (miRNAs) from stem-loop precursor miRNAs (pre-miRNAs) (1). It possesses two tandem endonuclease domains, RNase IIIa and RNase IIIb, which selectively cleaves 3p miRNA and 5p miRNA from the 3′ and 5′ pre-miRNA, respectively (2, 3). Therefore, each pre-miRNA will generate two mature miRNA strands (5p and 3p) that contain different mRNA-targeting sequences. The produced mature miRNAs are loaded into the RNA-induced silencing complex (RISC) and suppress gene expression by binding to the 3′ untranslated region (3′-UTR) of the target mRNAs (4). By targeting more than half of protein-coding genes, miRNAs are involved in virtually all developmental and pathological processes in animals, including malignant neoplasms (5, 6).
Pathogenic and likely pathogenic germline DICER1 variants are associated with DICER1-related tumor predisposition, a syndrome characterized by an increased risk for pleuropulmonary blastoma (PPB), pulmonary cysts, thyroid gland neoplasia (multinodular goiter, adenomas, and/or thyroid cancer), ovarian tumors (Sertoli-Leydig cell tumor, gynandroblastoma, and sarcoma), cystic nephroma, and Wilms tumor (7). DICER1-associated tumors typically harbor a germline loss-of-function (LOF) variant (nonsense, frame-shift, rarely missense; copy number changes are rarely observed) and a tumor-specific missense mutation in the DICER1 RNase IIIb domain affecting one of the five hotspot codons that encode key amino acids in the metal biding catalytic cleft of the nuclease domain: E1705, D1709, G1809, D1810, and E1813 (8–11). DICER1-related tumor predisposition therefore represents an unusual form of Knudson’s two-hit hypothesis, since the somatic mutation affects the ability of DICER1 RNase IIIb domain to process 5p miRNAs, while keeping its capacity to generate 3p miRNAs by the preserved RNase IIIa domain (2, 3, 12). This change in 5p:3p strand ratio alters the miRNA repertoire of tumor cells and is predicted to drastically alter gene expression.
Disruption of Dicer1 in mice leads to lethality early in embryonic development (13). Thyroid-specific inactivation of Dicer1 generates mice with severe hypothyroidism and marked loss of differentiation markers (Tg, Nis, Tshr and Tpo) (14), further emphasizing its role in thyroid pathogenesis. Indeed, DICER1 hotspot mutations have been reported in human thyroid neoplasms of follicular cell origin, including adenomas, papillary thyroid carcinoma (PTC), follicular thyroid carcinoma (FTC), and poorly differentiated thyroid cancers (PDTC) (15–22). The effects of DICER1 somatic mutations on miRNA biogenesis and the associated gene expression changes that are relevant for thyroid tumorigenesis are poorly understood. Experimental data suggest that the pathogenicity of somatic mutations in DICER1 is dependent on the cellular context and controlled by tissue-specific miRNAs and mRNAs (23, 24). Therefore, genome-wide miRNA/mRNA expression studies will be fundamental to determine tissue-specific miRNA signatures associated with DICER1-driven thyroid tumors and identify oncogenic pathways activated by RNase IIIb hotspot mutations. In this study, we aimed to evaluate the effects of DICER1 RNase IIIb mutations on miRNA and mRNA transcriptomes of differentiated thyroid cancers. We propose molecular mechanisms (gene networks and signaling pathways) underlying the tumorigenesis of follicular thyroid cells harboring DICER1 somatic mutations. We also uncover potential miRNA biomarkers for DICER1-driven malignant tumors that may be valuable in the clinical setting.
Material and methods
Patient samples
Tumor samples were obtained from the Children’s Hospital of Philadelphia (CHOP) and Hospital for Sick Children, Toronto, Ontario, Canada. This study included archived Formalin-Fixed Paraffin-Embedded (FFPE) samples from 20 non-neoplastic thyroids, 8 adenomatous nodules and 60 sporadic well-differentiated follicular derived thyroid cancers (DTC): 47 PTCs and 13 FTCs. Demographic information including age and sex was collected from each patient in addition to histopathologic results and tumor staging. Ethics approvals for collection and use of the patient samples were obtained from the CHOP Institutional Review Board as part of the Child and Adolescent Thyroid Consortium (CATC) Biorepository study (IRB# 20-018240).
miRNA and mRNA targeted next-generation sequencing (NGS) analysis
Tissue was cut into 5µm serial sections and mounted on glass microscope slides. Diagnosis was confirmed by a board-certified pathologist specialized in thyroid disease and regions of interest representing tumor foci on an H&E slide were selected and circled. Samples were sent to HTG Molecular Diagnostics in Tucson, AZ, and regions of interest were microdissected under a Leica LMD6500 laser capture microscope (LCM). Each sample was immediately suspended in EdgeSeq lysis buffer (HTG Molecular Diagnostics, Tucson, AZ) and miRNA/mRNA profiling was carried out using one FFPE tissue slide from each sample for each assay as previously described (25). miRNA and mRNA expression were profiled using HTG Edgeseq miRNA whole transcriptome assay and HTG EdgeSeq Oncology Biomarker Panel (OBP), respectively. HTG EdgeSeq system is a NGS application that measures gene expression without the need for extracting RNA. HTG Edgeseq miRNA whole transcriptome assay measures the expression of 2,083 human miRNAs described in the miRBase v20 database. HTG EdgeSeq OBP measures the expression of 2,559 genes associated with tumor biology, including 15 housekeeping genes. All miRNAs and mRNAs screened in this study are listed in Supplemental Tables 1, 2, respectively. Briefly, the workflow entailed automated, extraction-free sample preparation, quantitative nuclease protection using the EdgeSeq processor, and library generation and sequencing. The HTG EdgeSeq Parser (HTG Molecular, Tucson, AZ, USA) was used to align the FASTQ files to the probe list to collate the data. Data were provided as data tables of raw, quality control (QC) raw, counts per million (CPM), and median normalized counts.
Bioinformatic analysis
The DESeq2 data normalization, analyses, and statistical comparisons between benign thyroid tissue and DICER1-mutant thyroid tumors were performed using the HTG EdgeSeq Reveal software version: 4.0.1. DESeq2 normalized data were logarithmically scaled for data visualization. Plots were created using GraphPad prism 9 (GraphPad software Inc., La Jolla, CA). TCGA RNA-Seq expression data for adult PTCs and non-neoplastic thyroid tissues were obtained from http://tcga-data.nci.nih.gov and http://gdac.broadinstitute.org/in September 2022. Functional enrichment analysis was performed using g:Profiler to search for genes significantly over-represented in the list of DEG, as compared to all background genes included in the HTG OBP panel (26). Predicted targets for different miRNAs were obtained from TargetScan Human release 8.0 (https://www.targetscan.org/vert_80/). Only predicted targets with a total context score <−0.2 were included in the analysis.
Biostatistical analysis
Differential expression was calculated using DESeq2 and only differentially expressed mRNAs and miRNAs with a false-discovery rate (FDR) < 0.05 were included. T-test analysis and One-Way ANOVA were performed with GraphPad prism 9. A two-sided p value < 0.05 was considered statistically significant.
Results
Pediatric thyroid tumors with DICER1 RNase IIIb mutations are associated with less invasive phenotype
In thyroid, DICER1-driven PTCs are infrequent in adult (0-0.4%) but comparatively more prevalent in pediatric PTCs and FTCs (5-10%) (15, 17, 20, 27, 28). Despite its overall low prevalence, we identified 12 cases of DTC with DICER1 hotspot mutations as part of the Child and Adolescent Thyroid Consortium (CATC) multi-institutional collaborative research program (nine from CHOP and three from Sick Kids). Detailed histopathologic features and molecular alterations of these 12 cases are summarized in Table 1. Four of these 12 have been previously reported and published: cases ID# 6, 7, 11, and 12 (20, 29). The 12 cancer cases with mutations in the RNase IIIb domain of DICER1 included nine females (75%) with mean age of 13.7 years (SD = 2.0) at surgery. DTC harboring DICER1 mutations were all of the follicular type: four FTCs and six follicular variant PTCs (fvPTC). None had extrathyroidal extension, lymph node or distant metastasis. Loss of function defects were found in six cases (three loss of heterozygosity (LOH) and three inactivating variants). To determine whether DICER1 mutations were somatic or germline, DNA from non-neoplastic thyroid tissue from the same cases was analyzed by conventional Sanger sequencing and somatic alterations are highlighted in bold in Table 1. All the RNase IIIb mutations tested and the three inactivating variants reported here (S125*, L777fs, G661Vfs*24) were found to be somatic by Sanger sequencing (Supplemental Figure 1). In 3 cases, DICER1 RNase IIIb alteration was revealed due to the miRNA signature (further discussed in the next section), but no tissue was available to confirm the mutational status. In one case (case 9), DICER1 positivity was detected by a commercially available somatic thyroid oncogene panel but the specific codon was not reported.
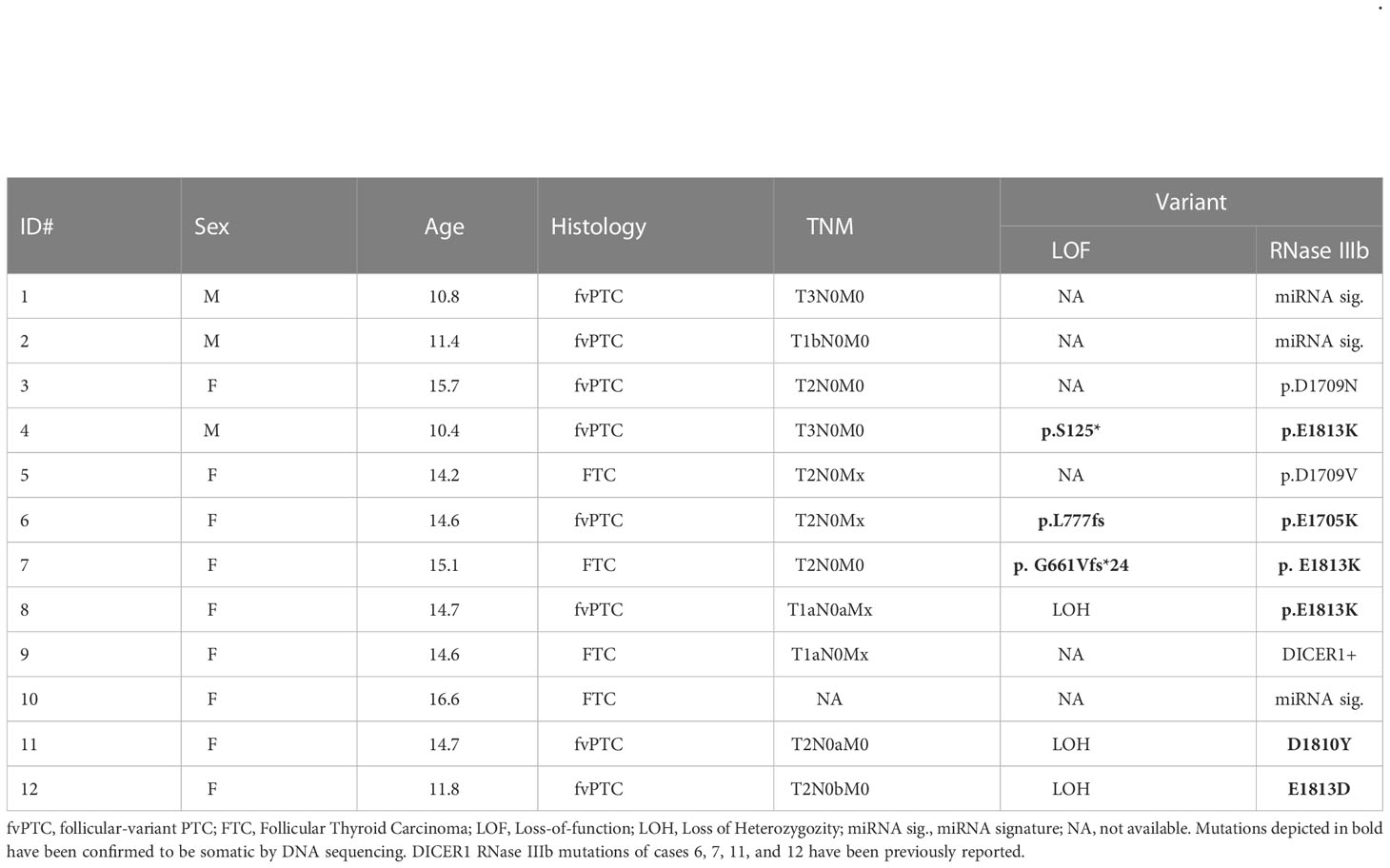
Table 1 Demographic and histopathological features of pediatric thyroid cancers with DICER1 RNase IIIb domain alterations.
DTC with DICER1 RNase IIIb mutations is associated with widespread miRNA dysregulation
FFPE tissue specimens were available for eight of 12 cases with DTCs harboring DICER1 RNase IIIb domain mutations. To determine the impact of DICER1 hotspot mutations on miRNA synthesis, we analyzed the differential miRNA expression in DICER1-mut DTC (six fvPTC and two FTC) vs non-neoplastic/adenomatous thyroids (n=26) using the HTG Edgeseq miRNA whole transcriptome assay which evaluates the expression of 2,083 human mature miRNAs. The DICER1 hotspot mutations in the RNase IIIb domain (E1705, D1709, D1810, and E1813) have been shown to impair the ability of DICER1 to generate mature 5p miRNAs as illustrated in Figure 1A (8). On par with its role in miRNA biogenesis, we show that tumors harboring these mutations demonstrate clear reduction of 5p miRNAs (regardless of the specific mutation site of the RNase IIIb domain), including those abundantly expressed in the non-neoplastic thyroid tissue and associated with tumor suppressor function in thyroid cancer such as let-7 and mir-30 families and mir-125b (30–32)(Figures 1B, C). Previous mechanistic studies have shown that RNase IIIa and IIIb activities are distinct and can be uncoupled (2, 33). We found that levels of 3p miRNAs were markedly increased when compared to non-neoplastic thyroid tissue and benign hyperplastic lesions (Figure 1B). This striking decrease in 5p:3p strand ratios caused by the RNase IIIb hotspot mutations is further illustrated by the differential processing of miR-20a pre-miRNA in DICER1-mut tumors (Figure 1D). While benign thyroids and DICER1-wt DTC show predominance of miR-20a-5p, DICER1-mut DTC have a prominent reduction in 5p:3p strand ratios and express mostly the 3p strand (miR-20a-3p). Moreover, we report a patient with bilateral FTC, with the carcinoma on the right lobe harboring biallelic DICER1 alterations (p.G661Vfs*24, p.E1813K) while the carcinoma on the left lobe was DICER1-wt (Figure 1E). Predictably, the DICER1-mutant FTC showed reduced levels of 5p miRNAs and increased levels of 3p miRNAs compared to the DICER1-wt FTC (Figure 1F). miR-451a is the only known miRNA for which maturation has been shown to be DICER1-independent (34). Indeed, expression levels of miR-451a were similar between the two tumors, on par with the non-requirement for DICER1 for its processing, and emphasizing the causal effect of the DICER1 on the downstream miRNA changes.
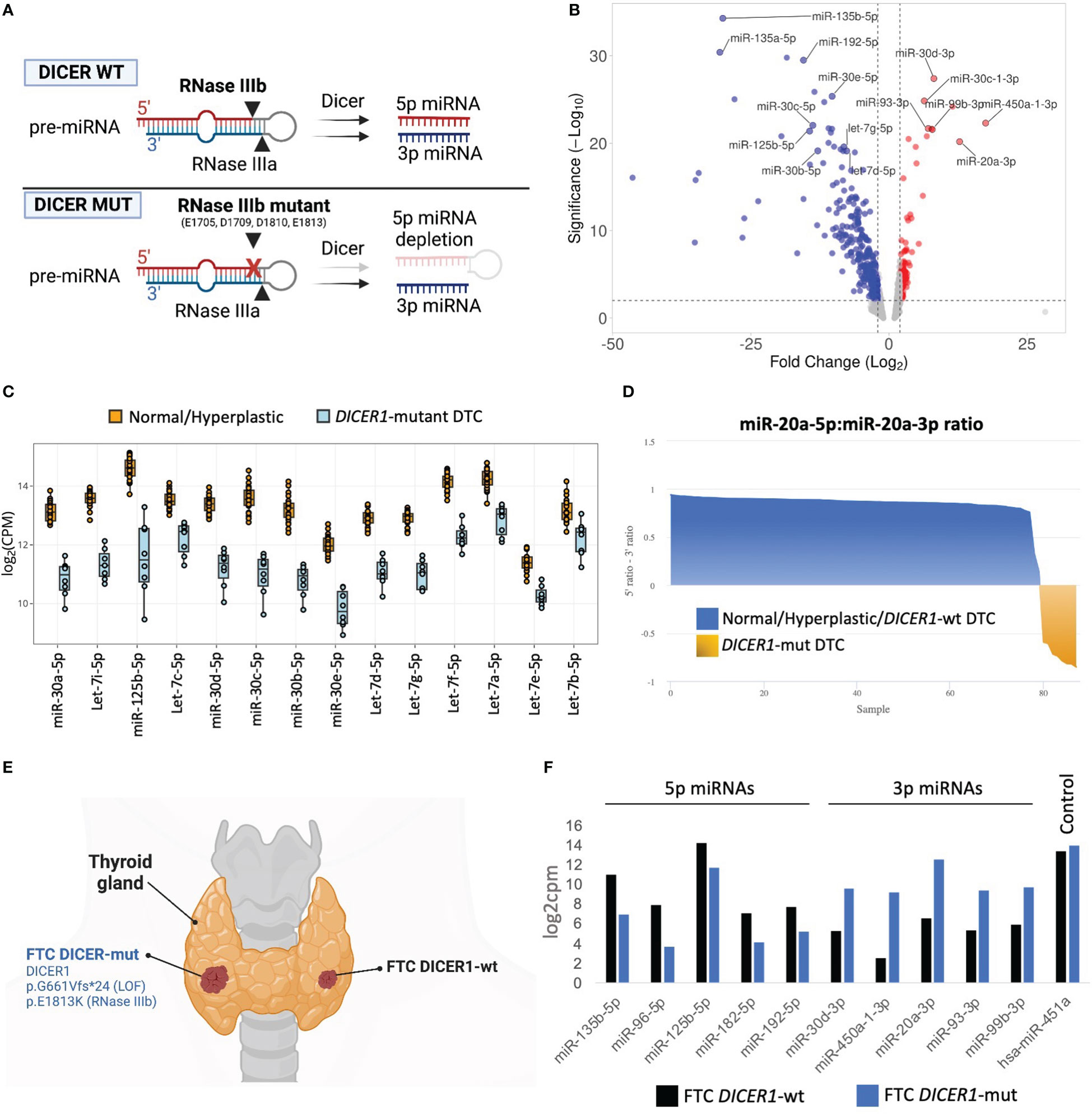
Figure 1 miRNA dysregulation in thyroid cancers harboring hotspot mutations in DICER1 RNase IIIb domain. (A) Scheme illustrating the mechanism by which the two RNase III domains of DICER1 cleave a pre-miRNA hairpin, and for specific loss of 5p miRNAs in RNase IIIb hotspot mutants. (B) Volcano plot highlighting miRNAs differentially expressed between 8 differentiated thyroid cancers harboring RNase IIIb mutations and 26 non-neoplastic/hyperplastic cases. Analysis shows general downregulation of 5p miRNAs and upregulation of 3p miRNAs. (C) Differential expression of selected miRNAs well-known for their tumor suppressor function in cancer, including let-7 and miR-30 families and miR-125b-5p. (D) Analysis of miR-20a pre-miRNA strand processing (5p:3p ratio) across normal/hyperplastic, DICER1-wt tumors and DICER1-mut tumors. The tumors with hotspot mutations of DICER1 (shown in yellow) exhibit selective defects in processing 5p miRNA strands (but not 3p), leading to overall decrease in 5p:3p strand ratios. (E) Scheme of patient diagnosed with bilateral follicular thyroid cancer (FTC), one with DICER1 RNase IIIb and one, DICER1-wt. Created with BioRender.com. (F) The 2 FTCs from the same patient show remarkable differences in miRNAs levels. DICER1-mut FTC (right lobe) shows reduction in 5p miRNAs and upregulation of 3p miRNAs when compared to DICER1-wt FTC (left lobe).
DICER1-mutant DTC exhibit unique miRNA signature
We next sought to determine whether this unique miRNA signature of tumors with RNase IIIb mutations could be used to identify thyroid cancers harboring DICER1 hotspot mutations. Principal component analysis (PCA) revealed that a classifier with as few as four 3p-miRNA markers (miR-20a-3p, miR-30d-3p, miR-99b-3p, and miR-450a-1-3p) could efficiently distinguish DICER1-mut DTC (regardless of histology: PTC or FTC) from non-neoplastic/benign hyperplastic and DICER1-wt PTCs/FTCs (Figure 2A). Two cases of benign hyperplastic nodules (adenomatous follicular hyperplasia) harboring DICER1 variants, one case with biallelic DICER1 alterations (p.E1813G, p.T847delinsFHKHS) and one with a variant of unknown significance (p.Q7R) showed an interesting pattern of miRNA expression. Both cases showed decreased expression of 5p miRNAs, comparable to that observed in DICER1-mut DTCs (Supplemental Figure 2). However, these DICER1-mut benign nodules had no increase of 3p miRNAs as observed for DICER1-mut DTCs (Figure 2B), indicating these 3p miRNAs are specifically upregulated in malignant cases with RNase IIIb mutation. One case of DTC harboring the DICER1 p.E1420del (VUS) mutation had no significant differences in miRNA levels. Therefore, 3p miRNAs are not only valuable to classify RNase IIIb mutants as benign or malignant but could also be causally involved in DICER1-driven malignant progression. The validity of these four miRNAs biomarkers was subsequently tested using the TCGA cohort comprising 496 adult PTC patients for which we know there are two cases with DICER1 hotspot mutations: p.E1813G and p.D1810H (17). The expression of these four miRNAs is also dramatically increased in two adult PTCs harboring DICER1 RNase IIIb mutation when compared to 59 non-neoplastic and 496 DICER1-wt PTCs (Figure 2C). One PTC case driven by NRAS p.Q61R and with a co-occurring DICER1 p.R1906S mutation (outside of the RNase IIIb domain) had no increase in the expression of these markers. We next sought to determine whether DICER1 mRNA expression was altered in any of the tumors. In both cohorts (CHOP and TCGA), we found DICER1 mRNA expression to be downregulated in DICER1-wt DTC compared to non-neoplastic cases. On the other hand, DTC with DICER1 RNase IIIb mutations were associated with increased expression of DICER1 mRNA and could explain, at least in part, the increased levels of 3p miRNAs observed in these cases (Figure 2D). The differential expression of selected 5p and 3p miRNAs (let-7i-5p, miR-30a-5p, miR-20a-3p and miR-99b-3p) was further validated by quantitative PCR in four DICER1-mut DTC and matched normals – cases ID# 5, 6, 7 and 8 from Table 1 (Supplemental Figure 3).
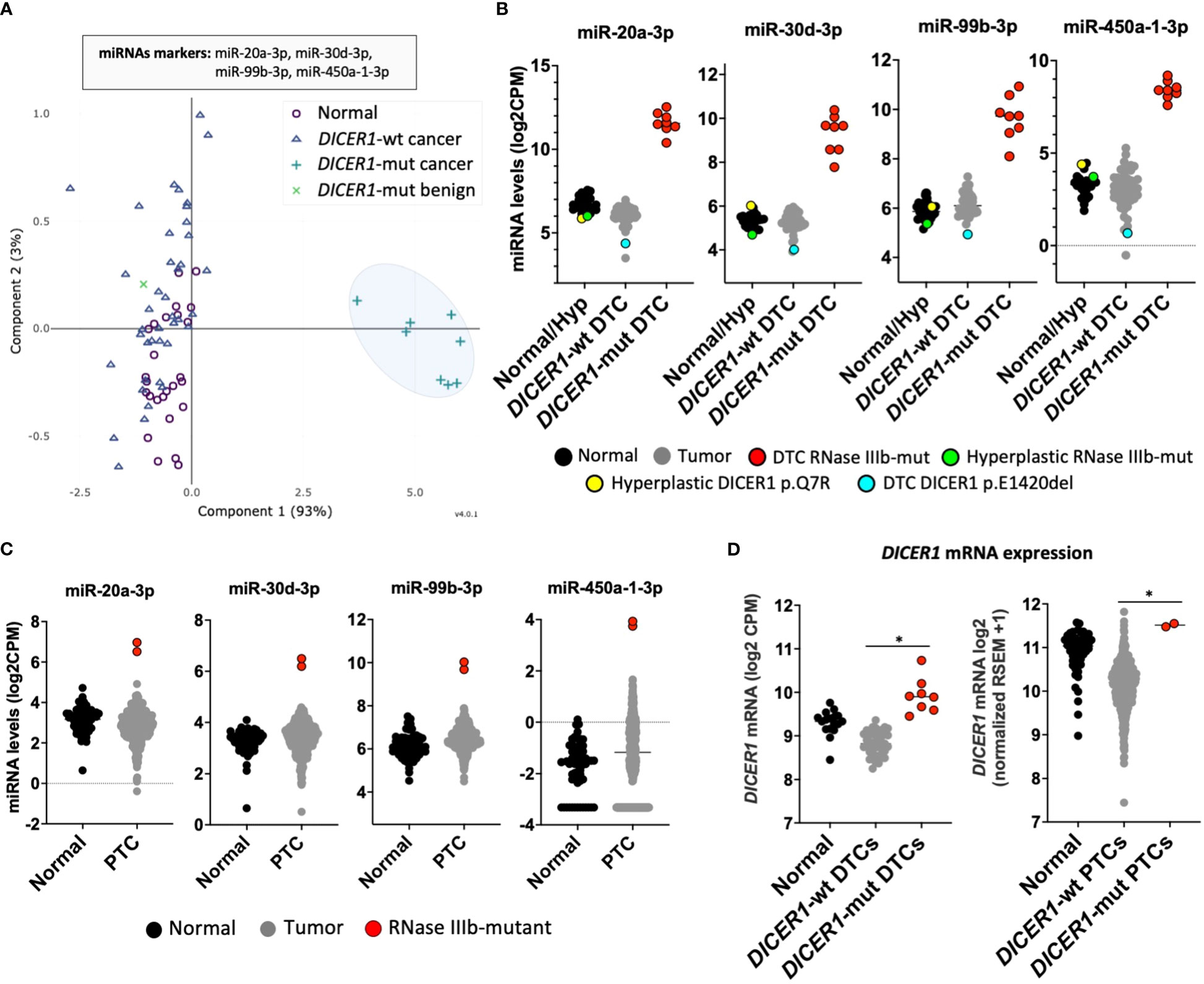
Figure 2 DICER1-mutant thyroid cancers exhibit unique miRNA signature. (A) Principal component analysis (PCA) of miRNA expression profiles from differentiated thyroid cancer (DTC) and non-neoplastic/hyperplastic shows that a set of four miRNAs (mir-20a-3p, mir-30d-3p, mir-99b-3p, and miR-450a-1-3p) can identify DTC with RNase IIIb mutation. (B) Expression of 3p miRNA markers: mir-20a-3p, mir-30d-3p, mir-99b-3p, and miR-450a-1-3p in 28 non-neoplastic/hyperplastic thyroids, 52 DICER1-wt DTC and 8 DICER1-mutant DTC. (C) Expression of miRNA markers: mir-20a-3p, mir-30d-3p, mir-99b-3p, and miR-450a-1-3p across 59 non-neoplastic thyroids and 506 papillary thyroid cancers (PTC) from the Thyroid Cancer The Cancer Genome Atlas (TCGA). Only the 2 PTC cases in the TCGA with DICER1 RNase IIIb domain mutations show overexpression of the 3p miRNA markers. (D) Expression of DICER1 mRNA in non-neoplastic/hyperplastic thyroid, DICER1-wt tumors and DICER1-mut tumors from our cohort (left) and Thyroid Cancer TCGA cohort (right). Asterisk represent p < 0.05.
DICER1 hotspot mutations are associated with increased MAPK output and loss of thyroid differentiation
In thyroid cancer, genetic alterations activating the MAPK pathway are highly prevalent and mutually exclusive to each other, indicating that harboring more than one of these mutations confers no clonal advantage (17, 29, 35). In this study, all cases with DICER1 RNase IIIb mutations lacked genetic alterations activating the MAPK pathway. A literature review of 42 thyroid cancers (including cases from this study) harboring DICER1 RNase IIIb mutations reinforced the mutually exclusive nature of these alterations in thyroid cancer (Figure 3A). The DICER1 mutations reviewed here - all in the RNase IIIb domain - are asymmetrically distributed between codons E1705, D1709, D1810, and E1813. E1813 is the most common alteration (20/42; 48%) overall and in each thyroid cancer type (PTC, FTC, PDTC) (Figures 3A, B). Even in more advanced forms of the disease such as PDTC, there was no overlap between DICER1 hotspot mutations and genetic alterations activating the MAPK pathway (18, 27). This supports the hypothesis that these alterations are associated with MAPK signaling activation. Indeed, mRNA expression of established MAPK output markers HMGA2, PPAT, EGR1 and CCND1 (from TCGA ERK-score (17)) is increased in DICER1-mut tumors when compared to non-neoplastic/hyperplastic thyroids (Figure 3C). Although it is not clear how DICER1 hotspot mutations activate MAPK, we found several positive regulators of the pathway to be upregulated in DICER1-mut tumors: NRG1, RTKs (FGFR3, ERBB2/3), BRAF and RAF1. We also found decreased expression of TPO, TFF3, LRP2 and increased expression of FN1, genes positively and negatively correlated with differentiation, respectively (Figure 3D). Although patients with DICER1 hotspot mutations have well differentiated thyroid tumors of follicular cell origin and are at low risk for lymph node metastasis, a fraction of cases are associated with high-risk PTC or PDTC (16). Our literature review of DICER-mutant cases shows that the one case of PTC with N1b disease harbored a TP53 truncating mutation (p.E343Afs*3) (Figure 3A). Moreover, five out of 12 (41%) DICER1-driven PDTC also had a TP53 mutation (4/12, 33%) or a TERT promoter mutation (1/12, 8%), alterations associated with progression of thyroid cancer (Figure 3A).
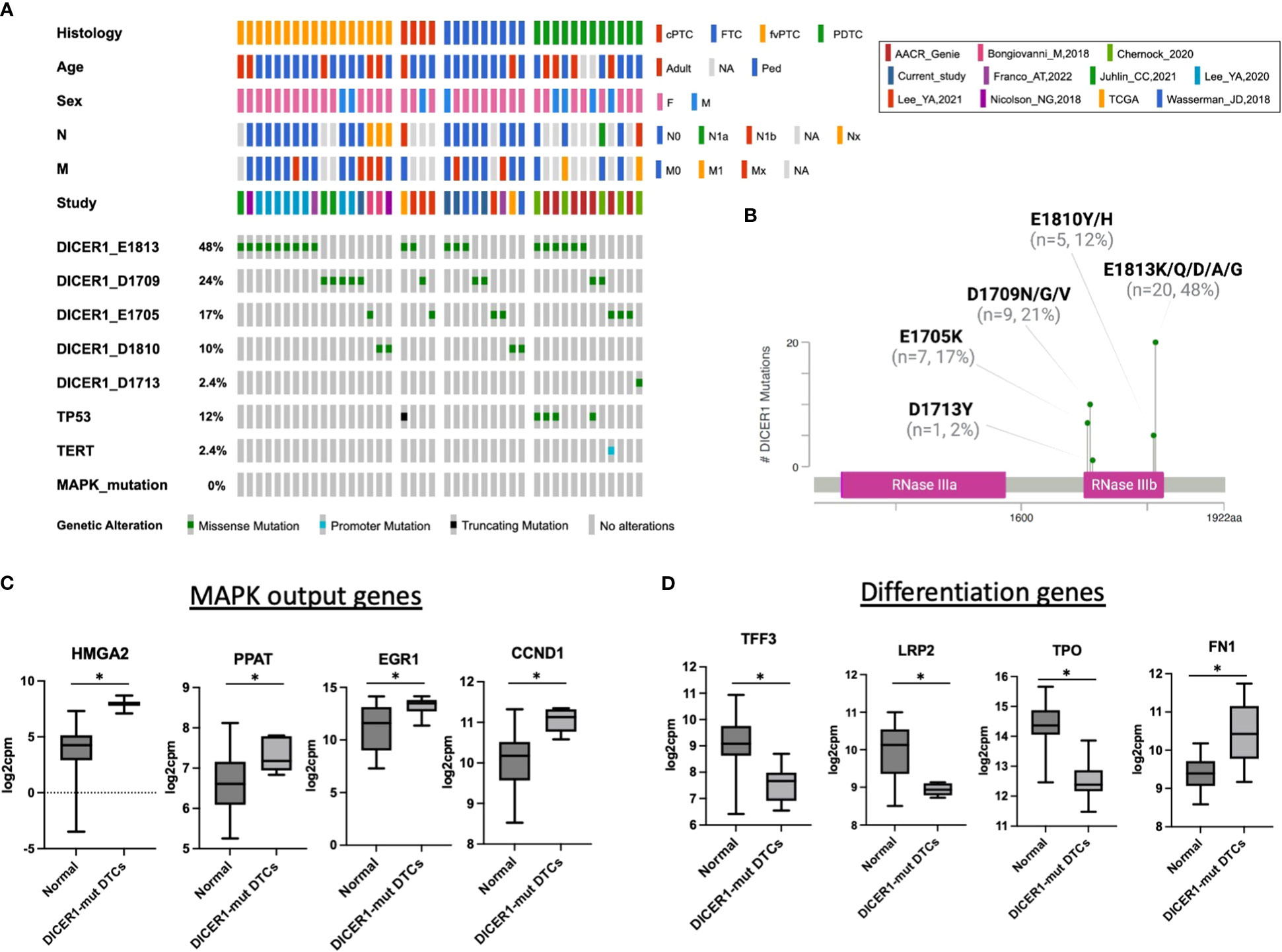
Figure 3 DICER1 RNase IIIb mutations are associated with increased MAPK output and loss of differentiation. (A) Oncoprint plot displaying 42 cases of DICER1-mut thyroid cancers including the cases from current study and a review of the literature. Only mutations in the RNase IIIb domain and from malignant tumors were included. Clinicopathologic characteristics include age, sex, histology, N status and M status. The different studies included in the design of this plot are shown. DICER1 RNase IIIb mutations were categorized according to mutated codon. cPTC, classic papillary thyroid cancer; FTC, follicular thyroid cancer; fvPTC, follicular variant papillary thyroid cancer; PDTC, poorly differentiated thyroid cancer; N, lymph node metastasis status; M, distant metastasis status. (B) DICER1 mutation lollipop plot depicting RNase IIIb mutations from 42 patients with thyroid cancer. (C) Expression of selected MAPK output genes present in the HTG Oncology Biomarker Panel in non-neoplastic/hyperplastic vs DICER1-mutant cases. (D) Expression of genes related to thyroid differentiation in non-neoplastic/hyperplastic vs DICER1-mutant cases. Asterisk represent p < 0.05.
DICER1-mutation impact the expression of genes related to cell-cycle
To understand the molecular mechanisms (gene networks and signaling pathways) underlying tumorigenesis of follicular cells harboring DICER1 mutations, we analyzed the mRNA expression profiles of the same pediatric thyroid tumors using the HTG Edgeseq OBP panel, which evaluates the mRNA expression of 2,559 genes related to cancer (Supplemental Table 2). We performed a differential gene expression analysis between eight DICER1-mut tumors and 26 non-neoplastic/hyperplastic thyroids and observed 569 differentially expressed genes (DEG) between the two groups (247 up and 322 down, FDR<0.05) (Figure 4A). To evaluate the pathways enriched among the DEGs from the attained dataset, we used g:Profiler web-based tool (v11.5) (26). Functional enrichment analysis of these 247 upregulated genes exposed a significant enrichment for cell-cycle genes in four different databases: Gene Ontology (GO), Reactome, KEGG and WikiPathways (Figure 4B). Among the upregulated genes in DICER1-mutant compared to non-neoplastic and hyperplastic lesions (FC≥2, and FDR≤.05), were genes related to cell-cycle, such as the transcription factors E2F1 and E2F5, different cyclins (CCNB1, CCND1, CCND2, CCNE2, CCNF), SKP2, TOP2A, MCM2, and the established clinical marker of cell proliferation, MKI67 (Figure 4C). E2F transcription factor binding sites were also found to be significantly overrepresented in the collection of upregulated genes based on TRANSFAC analysis, pointing to these transcription factors as important hubs in the cell-cycle regulation of DICER1-mut thyroid tumors (Figure 4B). While E2F1 was upregulated in all tumors (DICER1-wt and DICER1-mut), E2F5 was specifically upregulated in DICER1-mut tumors. Remarkably, we found that let-7, mir-17-5p, mir-96-5p and mir-181-5p, all of which were found to be downregulated in DICER1-mut tumors, have predicted binding sites that are conserved among vertebrates in the 3’ UTR of E2F5 (Figures 4D, E). Indeed, the regulation of E2F5 mRNA expression by some of these miRNAs has already been previously validated in other cancer types (36–38). The differential expression of selected mRNAs associated with cell-cycle (E2F5, SKP2), MAPK signaling output (HMGA2, CCND1) and thyroid differentiation (TPO) was further validated by quantitative PCR in four DICER1-mut DTC and matched normals – cases ID# 5, 6, 7 and 8 from Table 1 (Supplemental Figure 4).
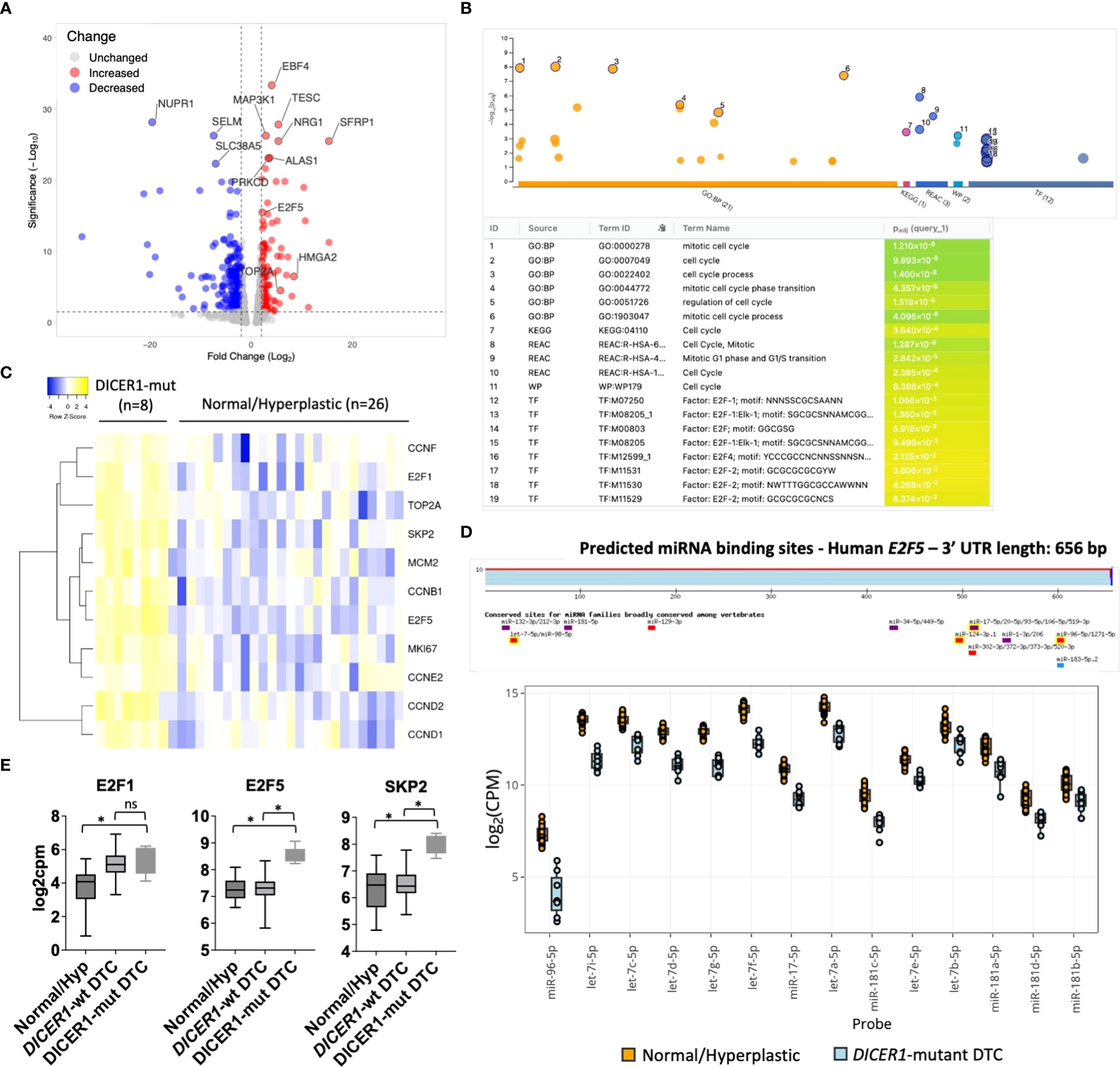
Figure 4 DICER1 RNase IIIb mutations positively regulate mitotic cell-cycle genes. (A).Volcano plot highlighting differentially expressed mRNAs between DICER1-mut differentiated thyroid cancer (DTC) (n=8) vs. non-neoplastic/benign hyperplastic group (n=26). The blue dots denote downregulated gene expression and the red dots denote upregulated gene expression. Grey dots denote gene expression without significant differences (FC>2 and FDR≤.05). (B) Functional enrichment analysis using g:Profiler version e106_eg53_p16_65fcd97. Analysis of 247 upregulated genes reveals enrichment for genes related to mitotic cell-cycle in different databases (GO, KEGG, Reactome, Wikipathways) and over-representation of E2F transcription factor binding sites according to TRANSFAC database. (C) Heatmap displaying increased expression of E2F1 and E2F5 transcription factors and selected genes related to cell-cycle in DICER1-mutant tumors vs. non-neoplastic/benign hyperplastic tissues. Yellow: upregulated genes; Blue: downregulated genes. (D) Predicted miRNA binding sites in the 3’ untranslated region (UTR) of E2F5 using TargetscanHuman 8.0 (Top) and downregulation of miRNAs predicted to target E2F5 3’ UTR in DICER1-mut vs non-neoplastic/hyperplastic tissues (bottom). (E) Increased expression of E2F1, E2F5 and SKP2 mRNA in DICER1-mut DTC vs. DICER1-wt DTC and non-neoplastic/hyperplastic tissues. Asterisk represent p < 0.05; ns: not significant.
Discussion
DICER1 is a highly conserved RNase III endoribonuclease which plays a critical role in the biogenesis of miRNAs. Several studies have reported biallelic DICER1 variants in syndrome-related tumors as well as DICER1-driven sporadic tumors (20, 22, 39), however the mechanisms by which the mutation affects miRNA and mRNA transcriptomes remain to be fully elucidated. Pediatric thyroid tumors are a particularly interesting model to dissect the effects of DICER1 mutations on miRNA and mRNA changes. These tumors have a relative quiet genome, with low mutational burden (40). Moreover, DICER1-mut DTC typically lack other tumor-initiating driver mutations, suggesting that DICER1 mutations are the sole drivers of these tumors. The lack of additional genomic changes in these tumors affords a clear panorama of the miRNA and mRNA changes triggered by DICER1 mutations. In fact, our analysis uncovered a clinically relevant miRNA signature in DICER1-mut malignant nodules that could help guide thyroid nodule diagnosis and management. The downstream changes in gene expression caused by DICER1 mutations and miRNA dysregulation revealed an important role of the RNase IIIb mutation in the activation of the MAPK pathway, loss of differentiation and positive regulation of cell-cycle.
DICER1 RNase IIIb mutations are found in both benign and malignant follicular derived thyroid tumors, predominantly in follicular-patterned lesions. Even when malignant, DICER1-mut tumors are associated with less invasive disease and generally have an excellent prognosis, clinically resembling RAS-mutant thyroid tumors. However, DICER1-mut DTC can progress to PDTC, similar to RAS-mutant tumors (16, 27). Therefore, it is critical to add molecular layers of information to improve the risk assessment of these thyroid neoplasms. The set of 3p miRNAs found to be specifically upregulated in DICER1-driven malignant tumors may be effective markers to discriminate the malignant status of nodules harboring the RNase IIIb mutation.
As we expand our multi-institutional collaborations within the CATC, it will be critical to evaluate a greater number of benign and malignant tumors with RNase IIIb mutations to validate these findings. The detection of 3p miRNAs associated with DICER1 mutations provides a promising path to enhance the diagnostic risk of malignancy in thyroid nodules. It is worth noting that commercially available thyroid-specific miRNA panels should be used with caution in pediatric thyroid cancers where DICER1 mutations are more frequent (41). Some miRNAs included in these panels, such as mir-146b-5p and mir-31-5p, are consistently increased in malignant thyroid disease but found to be markedly reduced in DICER1-mut DTCs based on data from this study as well as the Thyroid Cancer TCGA (17). Relying on these panels to assess the risk of malignancy could increase the possibility of a benign call for a DICER1-mut malignant nodule.
DICER1 mRNA itself has a long 3’ UTR (~4,000 nucleotides) which allows multiple miRNAs to bind and repress its expression. Some miRNAs that have been reported to target the DICER1 3’ UTR include let-7 (42), miR-9 (43), and miR-146b-5p (44), all of which are 5p miRNAs downregulated in DICER1-mut tumors. These findings point to a negative feedback mechanism in the regulation of DICER1 expression that could explain the increase in DICER1, and consequently, the increase in 3p miRNAs observed in DICER1-mut cases.
In thyroid cancer, DICER1 hotspot mutations are mutually exclusive with MAPK mutations, suggesting their ability to activate this pathway. Indeed, we found increased expression of positive modulators of the pathway as well as MAPK signaling output markers. Because the mRNA expression analysis utilized in this study was limited to the ~2,500 genes included in the HTG OBP panel, we were unable to assess the ERK score (52-gene list signature) and the Thyroid Differentiation Score (TDS, 16-gene list signature) generated by the Thyroid Cancer TCGA to evaluate MAPK signaling output and thyroid differentiation, respectively (17). However, two previous reports of integrated mutational and gene expression analysis with two DICER1-mut cases reported increased MAPK output and decreased TDS for DICER1-mut tumors (17, 45). In both studies, MAPK output and TDS scores were intermediate between non-neoplastic and BRAF/fusion-positive tumors, similar to the RAS-like subgroup of tumors from the thyroid cancer TCGA (well-differentiated tumors with favorable prognosis), strongly suggesting that RNase IIIb mutations have the ability to activate the pathway. Interestingly, this appears to be exclusive to thyroid cancers as in other tumor types such as endometrial cancers, pleuropulmonary blastomas, sarcomas, Sertoli-Leydig cell tumor, colorectal cancer, and cutaneous melanoma, DICER1 hotspot mutations frequently co-occur with MAPK alterations and/or other driver alterations (10, 46).
The enrichment of cell-cycle genes found in DICER1-mutant tumors agrees with previous reports demonstrating that DICER1 RNase IIIb mutations seem to confer a proliferative advantage to affected cells (47, 48). Although sustaining proliferative signaling is a hallmark of cancer, the unique mechanism among these thyroid tumors indicates that E2F transcription factors, specially E2F5, may have an important role in this process. Moreover, the increased expression of the S-phase kinase protein 2 SKP2 (also specific to DICER1-mut tumors), a major ubiquitin ligase that controls abundance of cell-cycle regulatory proteins at the G1–S transition may be an important target for therapies of these tumors (49), particularly when progression to high-grade PTC or PDTC occurs. This study highlights a previously unreported mechanism by which DICER1 mutations may induce transformation, and suggest novel therapeutic approaches for treating these rare malignancies.
The rarity of tumors bearing DICER1 variants represents a significant challenge to studying the role of DICER1 mutations in oncogenesis. Most studies report no more than two or three cases harboring the RNase IIIb mutation, making it difficult to correlate these alterations with clinical features and molecular changes. The CATC is building a collaborative research community dedicated to studying thyroid cancers and other thyroid-related conditions, including the establishment of a biorepository, in pediatric patients. Through these initial efforts and networks within the CATC, we were able to collect eight cases. Future studies will seek to leverage the growing network of the CATC and to utilize the signature reported here to identify additional DICER1-mutant tumors (which are enriched in the pediatric population) to further elucidate the mechanisms of DICER1-induced transformation.
To our knowledge, there are no commercially available cancer cell lines with spontaneous hotspot mutation in the RNase IIIb domain of DICER1 (50). In addition, different mouse models and human cell lines have been established to study DICER1 effects in cancer by simply knocking out the gene. The generation of proper models inactivating the RNase IIIb domain of DICER1 while preserving function of the RNase IIIa will be important for proper mechanistic studies of DICER1-related tumors.
In conclusion, our integrated analysis of mutational data, miRNAs, and mRNA profiling suggests that DTC with DICER1 RNase IIIb hotspot mutation generate an unbalanced expression of 5p:3p miRNAs that could be valuable for diagnostic and prognostic purposes. These tumors are associated with an increase in MAPK output and decreased differentiation, resembling the RAS-like tumors subgroup from the adult thyroid cancer TCGA, which is correspondingly reflected in their clinically indolent behavior. Mechanistic studies will be necessary to further understand the pathways and biological networks associated with the DICER1-driven changes in miRNAs, especially those changes in 3p miRNAs, which seem to be relevant in malignant progression.
Data availability statement
The data presented in the study are deposited in the NCBI repository, Bioproject ID: PRJNA934932 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA934932).
Ethics statement
The study was approved by Children’s Hospital of Philadelphia Institutional Review Board (IRB# 20-018240). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
JR-F, AF, JW, and AB designed the research. JR-F, AF, JW, and AB conducted the research. JR-F and AF wrote the article and JR-F had primary responsibility for final content. Financial support: AF, AB, JW, and DS; Administrative support: AF, AI, AB, and MS; Provision of study materials or patients: AB, JW, AI, TP, and JB. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by a grant from NIH R01CA214511 (AF), The Children’s Hospital of Philadelphia Frontier Programs (AB, AF), Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, and National Institutes of Health (DRS) and the Garron Family Cancer Centre (JW).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1083382/full#supplementary-material
References
1. Foulkes WD, Priest JR, Duchaine TF. Dicer1: Mutations, micrornas and mechanisms. Nat Rev Cancer (2014) 14(10):662–72. doi: 10.1038/nrc3802
2. Gurtan AM, Lu V, Bhutkar A, Sharp PA. In vivo structure-function analysis of human dicer reveals directional processing of precursor mirnas. RNA (2012) 18(6):1116–22. doi: 10.1261/rna.032680.112
3. Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human dicer and bacterial rnase iii. Cell (2004) 118(1):57–68. doi: 10.1016/j.cell.2004.06.017
4. Ha M, Kim VN. Regulation of microrna biogenesis. Nat Rev Mol Cell Biol (2014) 15(8):509–24. doi: 10.1038/nrm3838
5. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mrnas are conserved targets of micrornas. Genome Res (2009) 19(1):92–105. doi: 10.1101/gr.082701.108
6. Di Leva G, Garofalo M, Croce CM. Micrornas in cancer. Annu Rev Pathol (2014) 9:287–314. doi: 10.1146/annurev-pathol-012513-104715
7. Schultz KAP, Stewart DR, Kamihara J, Bauer AJ, Merideth MA, Stratton P, et al. Dicer1 tumor predisposition. In: Adam MP, Everman DB, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, et al, editors. Genereviews((R)). (Seattle, WA: NCBI Bookshelf) (1993).
8. Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, et al. Recurrent somatic Dicer1 mutations in nonepithelial ovarian cancers. N Engl J Med (2012) 366(3):234–42. doi: 10.1056/NEJMoa1102903
9. Anglesio MS, Wang Y, Yang W, Senz J, Wan A, Heravi-Moussavi A, et al. Cancer-associated somatic Dicer1 hotspot mutations cause defective mirna processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J Pathol (2013) 229(3):400–9. doi: 10.1002/path.4135
10. Pugh TJ, Yu W, Yang J, Field AL, Ambrogio L, Carter SL, et al. Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of Tp53 and two hits in Dicer1 resulting in retention of 5p-derived mirna hairpin loop sequences. Oncogene (2014) 33(45):5295–302. doi: 10.1038/onc.2014.150
11. Witkowski L, Mattina J, Schonberger S, Murray MJ, Choong CS, Huntsman DG, et al. Dicer1 hotspot mutations in non-epithelial gonadal tumours. Br J Cancer (2013) 109(10):2744–50. doi: 10.1038/bjc.2013.637
12. Thunders M, Delahunt B. Gene of the month: Dicer1: Ruler and controller. J Clin Pathol (2021) 74(2):69–72. doi: 10.1136/jclinpath-2020-207203
13. Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet (2003) 35(3):215–7. doi: 10.1038/ng1253
14. Frezzetti D, Reale C, Cali G, Nitsch L, Fagman H, Nilsson O, et al. The microrna-processing enzyme dicer is essential for thyroid function. PloS One (2011) 6(11):e27648. doi: 10.1371/journal.pone.0027648
15. Lee YA, Im SW, Jung KC, Chung EJ, Shin CH, Kim JI, et al. Predominant Dicer1 pathogenic variants in pediatric follicular thyroid carcinomas. Thyroid (2020) 30(8):1120–31. doi: 10.1089/thy.2019.0233
16. Chernock RD, Rivera B, Borrelli N, Hill DA, Fahiminiya S, Shah T, et al. Poorly differentiated thyroid carcinoma of childhood and adolescence: A distinct entity characterized by Dicer1 mutations. Mod Pathol (2020) 33(7):1264–74. doi: 10.1038/s41379-020-0458-7
17. Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell (2014) 159(3):676–90. doi: 10.1016/j.cell.2014.09.050
18. Chong AS, Nikiforov YE, Condello V, Wald AI, Nikiforova MN, Foulkes WD, et al. Prevalence and spectrum of Dicer1 mutations in adult-onset thyroid nodules with indeterminate cytology. J Clin Endocrinol Metab (2021) 106(4):968–77. doi: 10.1210/clinem/dgab025
19. Juhlin CC, Stenman A, Zedenius J. Macrofollicular variant follicular thyroid tumors are Dicer1 mutated and exhibit distinct histological features. Histopathology (2021) 79(4):661–6. doi: 10.1111/his.14416
20. Wasserman JD, Sabbaghian N, Fahiminiya S, Chami R, Mete O, Acker M, et al. Dicer1 mutations are frequent in adolescent-onset papillary thyroid carcinoma. J Clin Endocrinol Metab (2018) 103(5):2009–15. doi: 10.1210/jc.2017-02698
21. Yoo SK, Lee S, Kim SJ, Jee HG, Kim BA, Cho H, et al. Comprehensive analysis of the transcriptional and mutational landscape of follicular and papillary thyroid cancers. PloS Genet (2016) 12(8):e1006239. doi: 10.1371/journal.pgen.1006239
22. Rutter MM, Jha P, Schultz KA, Sheil A, Harris AK, Bauer AJ, et al. Dicer1 mutations and differentiated thyroid carcinoma: Evidence of a direct association. J Clin Endocrinol Metab (2016) 101(1):1–5. doi: 10.1210/jc.2015-2169
23. Bahubeshi A, Tischkowitz M, Foulkes WD. Mirna processing and human cancer: Dicer1 cuts the mustard. Sci Transl Med (2011) 3(111):111ps46. doi: 10.1126/scitranslmed.3002493
24. Guo Z, Maki M, Ding R, Yang Y, Zhang B, Xiong L. Genome-wide survey of tissue-specific microrna and transcription factor regulatory networks in 12 tissues. Sci Rep (2014) 4:5150. doi: 10.1038/srep05150
25. Girard L, Rodriguez-Canales J, Behrens C, Thompson DM, Botros IW, Tang H, et al. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clin Cancer Res (2016) 22(19):4880–9. doi: 10.1158/1078-0432.CCR-15-2900
26. Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, et al. G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res (2019) 47(W1):W191–W8. doi: 10.1093/nar/gkz369
27. Ghossein CA, Dogan S, Farhat N, Landa I, Xu B. Expanding the spectrum of thyroid carcinoma with somatic Dicer1 mutation: A survey of 829 thyroid carcinomas using msk-impact next-generation sequencing platform. Virchows Arch (2022) 480(2):293–302. doi: 10.1007/s00428-021-03212-4
28. Lee YA, Im SW, Jung KC, Chung EJ, Shin CH, Kim JI, et al. Predominant dicer1 pathogenic variants in pediatric follicular thyroid carcinomas. Thyroid (2020) 30(8):1120–31. doi: 10.1089/thy.2019.0233
29. Franco AT, Ricarte-Filho JC, Isaza A, Jones Z, Jain N, Mostoufi-Moab S, et al. Fusion oncogenes are associated with increased metastatic capacity and persistent disease in pediatric thyroid cancers. J Clin Oncol (2022) 40(10):1081–90. doi: 10.1200/JCO.21.01861
30. Boufraqech M, Nilubol N, Zhang L, Gara SK, Sadowski SM, Mehta A, et al. Mir30a inhibits lox expression and anaplastic thyroid cancer progression. Cancer Res (2015) 75(2):367–77. doi: 10.1158/0008-5472.CAN-14-2304
31. Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, et al. Specific micrornas are downregulated in human thyroid anaplastic carcinomas. Oncogene (2007) 26(54):7590–5. doi: 10.1038/sj.onc.1210564
32. Ricarte-Filho JC, Fuziwara CS, Yamashita AS, Rezende E, da-Silva MJ, Kimura ET. Effects of let-7 microrna on cell growth and differentiation of papillary thyroid cancer. Transl Oncol (2009) 2(4):236–41. doi: 10.1593/tlo.09151
33. Vedanayagam J, Chatila WK, Aksoy BA, Majumdar S, Skanderup AJ, Demir E, et al. Cancer-associated mutations in Dicer1 rnase iiia and iiib domains exert similar effects on mirna biogenesis. Nat Commun (2019) 10(1):3682. doi: 10.1038/s41467-019-11610-1
34. Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent mirna biogenesis pathway that requires ago catalysis. Nature (2010) 465(7298):584–9. doi: 10.1038/nature09092
35. Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of braf mutations in thyroid cancer: Genetic evidence for constitutive activation of the Ret/Ptc-Ras-Braf signaling pathway in papillary thyroid carcinoma. Cancer Res (2003) 63(7):1454–7.
36. Lal A, Thomas MP, Altschuler G, Navarro F, O’Day E, Li XL, et al. Capture of microrna-bound mrnas identifies the tumor suppressor mir-34a as a regulator of growth factor signaling. PloS Genet (2011) 7(11):e1002363. doi: 10.1371/journal.pgen.1002363
37. Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, et al. Microrna directly enhances mitochondrial translation during muscle differentiation. Cell (2014) 158(3):607–19. doi: 10.1016/j.cell.2014.05.047
38. Gurtan AM, Ravi A, Rahl PB, Bosson AD, JnBaptiste CK, Bhutkar A, et al. Let-7 represses Nr6a1 and a mid-gestation developmental program in adult fibroblasts. Genes Dev (2013) 27(8):941–54. doi: 10.1101/gad.215376.113
39. Khan NE, Bauer AJ, Schultz KAP, Doros L, Decastro RM, Ling A, et al. Quantification of thyroid cancer and multinodular goiter risk in the Dicer1 syndrome: A family-based cohort study. J Clin Endocrinol Metab (2017) 102(5):1614–22. doi: 10.1210/jc.2016-2954
40. Pozdeyev N, Gay LM, Sokol ES, Hartmaier R, Deaver KE, Davis S, et al. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res (2018) 24(13):3059–68. doi: 10.1158/1078-0432.CCR-18-0373
41. Finkelstein SD, Sistrunk JW, Malchoff C, Thompson DV, Kumar G, Timmaraju VA, et al. A retrospective evaluation of the diagnostic performance of an interdependent pairwise microrna expression analysis with a mutation panel in indeterminate thyroid nodules. Thyroid (2022) 32:1362–71. doi: 10.1089/thy.2022.0124
42. Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. Let-7 regulates dicer expression and constitutes a negative feedback loop. Carcinogenesis (2008) 29(11):2073–7. doi: 10.1093/carcin/bgn187
43. Leucci E, Zriwil A, Gregersen LH, Jensen KT, Obad S, Bellan C, et al. Inhibition of mir-9 de-represses hur and Dicer1 and impairs Hodgkin lymphoma tumour outgrowth in vivo. Oncogene (2012) 31(49):5081–9. doi: 10.1038/onc.2012.15
44. Ramirez-Moya J, Wert-Lamas L, Riesco-Eizaguirre G, Santisteban P. Impaired microrna processing by Dicer1 downregulation endows thyroid cancer with increased aggressiveness. Oncogene (2019) 38(27):5486–99. doi: 10.1038/s41388-019-0804-8
45. Stosic A, Fuligni F, Anderson ND, Davidson S, de Borja R, Acker M, et al. Diverse oncogenic fusions and distinct gene expression patterns define the genomic landscape of pediatric papillary thyroid carcinoma. Cancer Res (2021) 81(22):5625–37. doi: 10.1158/0008-5472.CAN-21-0761
46. Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet (2013) 45(10):1113–20. doi: 10.1038/ng.2764
47. Chen J, Wang Y, McMonechy MK, Anglesio MS, Yang W, Senz J, et al. Recurrent Dicer1 hotspot mutations in endometrial tumours and their impact on microrna biogenesis. J Pathol (2015) 237(2):215–25. doi: 10.1002/path.4569
48. Wang Y, Chen J, Yang W, Mo F, Senz J, Yap D, et al. The oncogenic roles of Dicer1 rnase iiib domain mutations in ovarian sertoli-leydig cell tumors. Neoplasia (2015) 17(8):650–60. doi: 10.1016/j.neo.2015.08.003
49. Rodriguez S, Abundis C, Boccalatte F, Mehrotra P, Chiang MY, Yui MA, et al. Therapeutic targeting of the E3 ubiquitin ligase Skp2 in T-all. Leukemia (2020) 34(5):1241–52. doi: 10.1038/s41375-019-0653-z
Keywords: DICER1, mutation, RNAse IIIb, pediatric, thyroid, cancer, microRNA, MAPK
Citation: Ricarte-Filho JC, Casado-Medrano V, Reichenberger E, Spangler Z, Scheerer M, Isaza A, Baran J, Patel T, MacFarland SP, Brodeur GM, Stewart DR, Baloch Z, Bauer AJ, Wasserman JD and Franco AT (2023) DICER1 RNase IIIb domain mutations trigger widespread miRNA dysregulation and MAPK activation in pediatric thyroid cancer. Front. Endocrinol. 14:1083382. doi: 10.3389/fendo.2023.1083382
Received: 28 October 2022; Accepted: 01 February 2023;
Published: 21 February 2023.
Edited by:
Willem Staels, Vrije Universiteit Brussel (VUB), BelgiumReviewed by:
Muhammad Riaz Khan, Université de Sherbrooke, CanadaKristian Schafernak, Phoenix Children’s Hospital, United States
Copyright © 2023 Ricarte-Filho, Casado-Medrano, Reichenberger, Spangler, Scheerer, Isaza, Baran, Patel, MacFarland, Brodeur, Stewart, Baloch, Bauer, Wasserman and Franco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aime T. Franco, ZnJhbmNvYTFAY2hvcC5lZHU=; Julio C. Ricarte-Filho, cmljYXJ0ZWZpakBjaG9wLmVkdQ==
 Julio C. Ricarte-Filho
Julio C. Ricarte-Filho Victoria Casado-Medrano1
Victoria Casado-Medrano1 Garrett M. Brodeur
Garrett M. Brodeur Douglas R. Stewart
Douglas R. Stewart Zubair Baloch
Zubair Baloch Andrew J. Bauer
Andrew J. Bauer