- 1Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals, Guangdong Research Center on Reproductive Control and Breeding Technology of Indigenous Valuable Fish Species, Fisheries College, Guangdong Ocean University, Zhanjiang, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), Zhanjiang, China
- 3Guangdong Havwii Agriculture Group Co., LTD, Zhanjiang, China
- 4Hainan Chenhai Aquatic Co., LTD, Sanya, China
- 5Key Laboratory of Utilization and Conservation for Tropical Marine Bioresources of Ministry of Education, Hainan Key Laboratory for Conservation and Utilization of Tropical Marine Fishery Resources, Yazhou Bay Innovation Institute, Hainan Tropical Ocean University, Sanya, China
Insulin-like growth factor 3 (IGF3) induces ovarian maturation in teleosts; however, research on its molecular regulatory mechanism remains deficient. Circular RNAs (circRNAs) and microRNAs (miRNAs) are involved in various biological processes, including reproduction. In this study, circRNAs and miRNAs involved in IGF3-induced ovarian maturation were evaluated in spotted scat (Scatophagus argus). In ovarian tissues, we identified 176 differentially expressed (DE) circRNAs and 52 DE miRNAs between IGF3 treatment and control groups. Gene Ontology (GO) enrichment analyses showed that host genes of DE circRNAs and target genes of DE miRNAs were enriched for various processes with a high degree of overlap, including cellular process, reproduction, reproductive process, biological adhesion, growth, extracellular region, cell junction, catalytic activity, and transcription factor activity. Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways included cell adhesion molecules, ECM–receptor interaction, regulation of actin cytoskeleton, focal adhesion, cell cycle, Hedgehog signaling pathway, phosphatidylinositol signaling system, PI3K-Akt signaling pathway, Apelin signaling pathway, Notch signaling pathway, insulin signaling pathway, and Rap1 signaling pathway. A circRNA–miRNA–mRNA regulatory network was constructed, including DE genes involved in reproduction (e.g., oocyte maturation, oocyte meiosis, and ECM remodeling), such as ccnd2, hecw2, dnm2, irs1, adam12, and cdh13. According to the regulatory network and tissue distribution, we identified one circRNA (Lachesis_group5:6245955|6270787) and three miRNAs (novel_miR_622, novel_miR_980, and novel_miR_64) that may exert regulatory effects in IGF3-induced ovarian maturation in S. argus. Taken together, this study provides a novel insight into the molecular mechanisms by which IGF3 functions in ovaries and highlights the effects of circRNAs and miRNAs in reproduction in S. argus.
Introduction
MicroRNAs (miRNAs) are endogenous non-coding RNAs (ncRNAs) with lengths of about 22 nucleotides. They exert post-transcriptional regulatory functions in plants and animals (1). They pair with the 3′ untranslated region (UTR) of the target genes, resulting in mRNA cleavage or translational repression (2; 1). The relationships between miRNAs and mRNAs are complex, e.g., a single miRNA may target several genes, while a single gene may be regulated by multiple miRNAs (3). MiRNAs are critical in the regulation of multiple biological processes, such as cell proliferation, differentiation, apoptosis, and gametogenesis (4–6). There is evidence for the roles of miRNAs in reproduction. For instance, miR-143 is involved in endocrine system regulation in mammalian gonads and functions in the maintenance of pregnancies (7, 8). In Oreochromis niloticus, miR-143 is highly expressed in mature gonads, indicating its functions in fish reproduction (9). In Danio rerio, the injection of miR430a-sox9a increases the proportion of spermatogonia, and the coinjection of miR430a-sox9a and miR218a-sox9b stimulates the renewal of ovarian follicles (10).
Circular RNAs (circRNAs) are another type of ncRNA. Because their 3′- and 5′-termini are joined together, circRNAs are characterized by covalent closed-loop structures (11). Studies have shown that circRNAs exert regulatory effects on multiple physiological processes (12). Some circRNAs involved in reproductive processes have been identified (13–15). CircRNAs can function as competing endogenous RNAs to regulate expression levels of target genes (16). For instance, circHIPK3 sponges miR-124a to regulate growth in human cells (17), and the circRNA chi_circ_0008219 also acts as a sponge for multiple miRNAs in pre-ovulatory ovarian follicles of goats (18).
The insulin-like growth factor (IGF) system has vital roles in various cellular processes, such as survival, growth, proliferation, and differentiation (19). IGF1 and IGF2 are regarded as typical ligands of the IGF family and are involved in follicle growth, meiotic resumption, and oocyte maturation (20–22). IGF3 is a newly discovered member of the IGF family exclusively found in teleosts (23). It can stimulate ovarian development and maturation in teleosts (24–27). Furthermore, the knockdown of igf3 in Cyprinus carpio via RNA interference significantly altered ncRNA expression profiles (28), indicating that ncRNAs are involved in IGF3-induced oocyte maturation; however, the molecular regulatory mechanism has not been determined.
Spotted scat (Scatophagus argus) is an economically important aquaculture species in south China (29–31) owing to its high nutritional value and colorful appearance (32–34). However, the development of artificial breeding methods for S. argus is limited by the inability of the ovaries of cultured S. argus to spontaneously mature (35, 36). Considering that IGF3 stimulates ovarian development and maturation in fish and these effects are clearly mediated by ncRNAs, studies of the molecular mechanism underlying IGF3-induced oocyte maturation in S. argus are warranted. The purpose of this study was to gain insight into circRNA–miRNA–mRNA interactions involved in the effects of IGF3 on oocyte maturation in S. argus.
Materials and methods
Experimental treatment and ethics statement
The experimental methods are described in a previous study (25). In brief, the open reading frame of igf3 in S. argus was cloned. The mature peptide of IGF3 was predicted, and the corresponding igf3 cDNA fragment was amplified. After purification, the igf3 cDNA fragment was ligated into the pMBP vector and was transfected into the Escherichia coli rosetta2 strain to produce recombinant IGF3 protein. After the recombinant IGF3 protein was obtained, in vitro experiments were conducted. Three adult female fishes (body weight ranging from 300 to 350 g) were sacrificed, and their ovaries (stage III) were dissected for further experiment. Ovary fragments were washed with Leibovitz’s L-15 medium (Gibco, American), then were incubated in a 24-well culture plate with L-15 medium containing penicillin and streptomycin (100 U/ml, Life Company, Shanghai, China). After pre-incubation (5% CO2, 27°C, 2 h), the medium was replaced with fresh medium containing 5 nM recombinant IGF3 protein, and the control group was not supplemented with recombinant IGF3. After incubation for 6 h, ovary fragments were collected and frozen in liquid nitrogen immediately. Then, samples were stored at −80°C for subsequent RNA extraction. The three treated ovary samples and three control ovary samples were used for RNA-seq and smallRNA-seq. In addition, 12 tissues of S. argus, including the brain, pituitary, heart, gill, liver, spleen, kidney, stomach, intestines, muscle, ovary, and testis, were collected in triplicate and stored at −80°C for subsequent RNA extraction. The 12 tissue samples were used for tissue distribution analysis of candidate RNAs.
All experimental procedures were in compliance with the Animal Research and Ethics Committees of Fisheries College of Guangdong Ocean University, China.
RNA library construction and sequencing
RNA extraction was performed with TRIzol (Invitrogen, USA) following the manufacturer’s protocol. The quality and concentration detection of RNA samples were evaluated using the NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies, USA). The purity and integrity of RNA samples were verified by Qubit 2.0 (Thermo Fisher Scientific, USA), Agilent 2100 (Agilent Technologies, USA), and electrophoresis methods. The total RNA with RNA integrity number (RIN) score > 7 was used for sequencing.
After verification of the RNA samples, circRNA libraries were constructed using Ribo-off rRNA Depletion Kit, and miRNA libraries were constructed using VAHTSTM Small RNA Library Prep Kit for Illumina. First rRNA of the samples were removed using the Epicenter RiboZero Kit. For circRNA libraries, linear RNAs were digested using RNase R, and RNA was randomly fragmented using a fragmentation buffer. After synthesis and purification, the cDNA was subjected to end repair, A-tailing, and sequencing adaptor ligation. For miRNA libraries, the small RNAs were ligated to 5′ and 3′ RNA/DNA chimeric oligonucleotide adaptors (Illumina); then, the ligation products were purified, and cDNA was synthesized. After purification and quality control, the cDNA libraries were sequenced on the Illumina NovaSeq 6000 platform (San Diego, USA) using NovaSeq 6000 S4 Reagent Kit by BioMarker Technologies company (Beijing, China). Raw data were uploaded to the National Center for Biotechnology Information (NCBI) (accession number PRJNA849653).
Sequencing quality control and analysis
To obtain clean data, raw sequencing data were filtered, including the removal of adapters, low-quality reads (q-value ≤ 20), or reads containing more than 5% unknown nucleotides (N) (37). Then, clean data were aligned to the genome of S. argus (38). The clean data of circRNA were aligned to the genome using HISAT2 with parameter –rna-strandness RF, and the clean data of miRNA were aligned to the genome using Bowtie with parameter -v 0. The mapped data were used for subsequent analyses.
Prediction and identification of circRNAs and miRNAs
The mapped reads from circRNA libraries were used for circRNA prediction using find_circ with default parameter (39). The mapped reads from miRNA libraries were aligned to the sequences of the mature miRNA in miRBase (v22) to identity the known miRNAs. Then, the novel miRNA were predicted using miRDeep2 with parameter -g -1 -b 0 (39).
Differential expression analysis
Differentially expressed (DE) circRNAs and miRNAs were detected between the treated group and the control group using DESeq2 with default parameter. The criteria for DE circRNA were fold change (FC) ≥ 2 and p < 0.05. The criteria for DE miRNA were fold change (FC) ≥ 1.5 and p ≤ 0.01. To avoid false positives, the Benjamini–Hochberg correction method was used, and the FDR was used as the key indicator for the DE miRNAs.
Functional enrichment analysis
A functional enrichment analysis of host genes of DE circRNAs was conducted with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using DAVID and KOBAS after non-redundant protein sequence database (NR) annotation. Target genes of miRNAs were predicted using miRanda and TargetScan and were also evaluated by GO and KEGG pathway enrichment analyses after NR annotation. The significance of enrichment analysis was indicated by p < 0.05.
circRNA–miRNA–mRNA network construction
To construct a preliminary network of circRNAs, miRNAs, and mRNAs, we focused on the DE circRNAs and DE miRNAs that related to DE genes with known roles in reproduction and ovarian maturation. According to the predicted circRNA–miRNA and miRNA–mRNA pairs, a circRNA–miRNA–mRNA coexpression network was constructed. The network was visualized using Cytoscape (http://www.cytoscape.org/).
Validation by real-time quantitative PCR
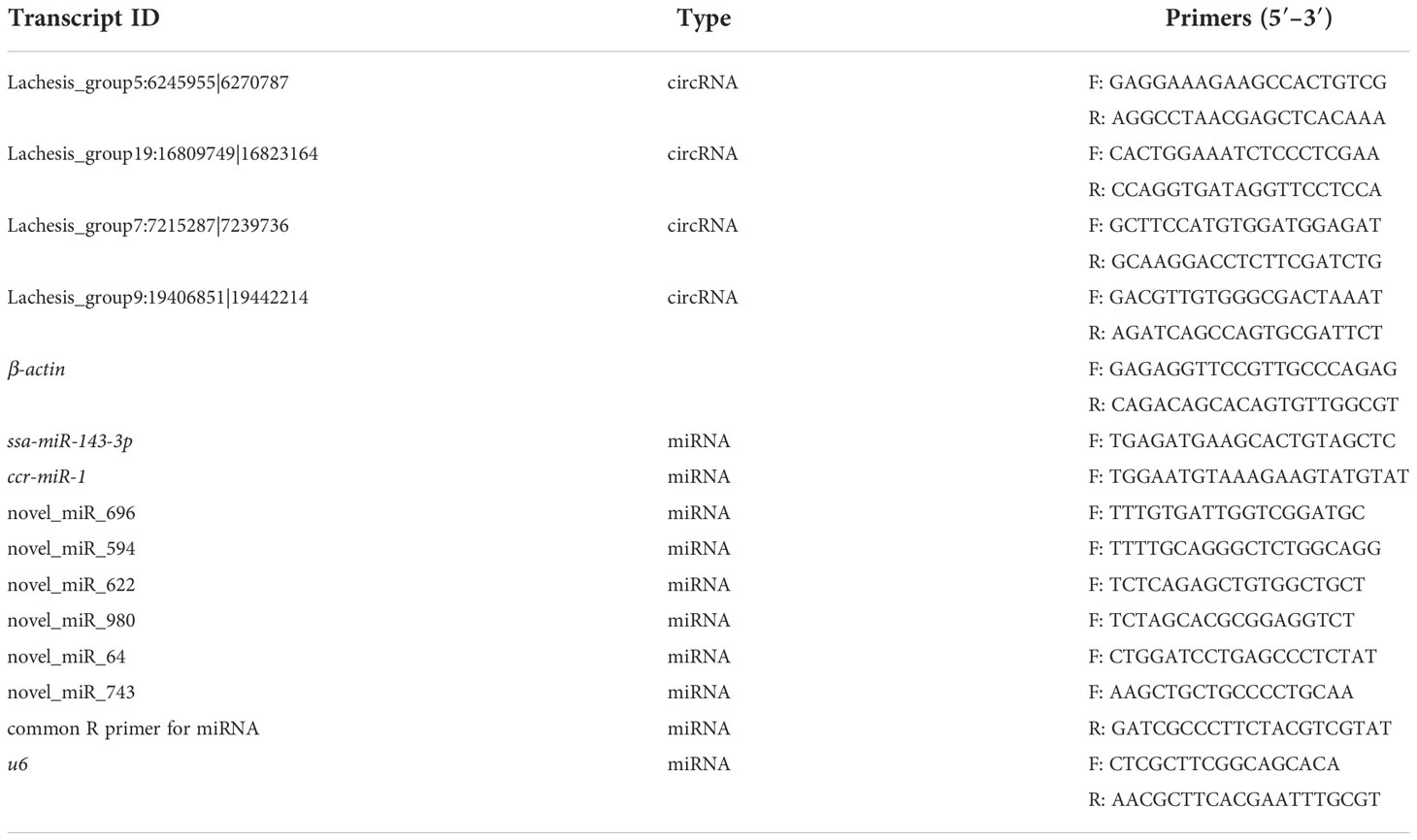
The cDNA was synthesized using TransScript® Uni All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (One-Step gDNA Removal) (TransGen Biotech, China) The cDNA was then used for RT-qPCR with PerfectStart® Green qPCR SuperMix (TransGen Biotech, China). The conditions were initial denaturation at 95°C for 2 min, followed by 45 cycles of 95°C for 20 s, 55°C for 30 s, and 72°C for 30 s. Relative expressions were calculated by the 2−ΔΔCt method. β-Actin was chosen as a reference gene for circRNAs, and u6 was the reference for miRNAs. All primer sequences are shown in Table 1.
Tissue distribution of candidate circRNA and miRNA by RT-qPCR
One circRNA (Lachesis_group5:6245955|6270787) and three miRNAs (novel_miR_622, novel_miR_980, and novel_miR_64) were selected for analyses of expression levels in 12 tissues (brain, pituitary, heart, gill, liver, spleen, kidney, stomach, intestines, muscle, ovary, and testis); these RNAs were associated with the DE genes involved in oocyte maturation. Triplicate total RNA samples from different tissues of adult S. argus individuals were prepared and reverse transcribed into cDNA. The methods for RNA isolation, reverse transcription, and RT-qPCR were the same as described above. The primers for circRNAs and miRNAs are listed in Table 1.
Results
Overview of RNA-Seq
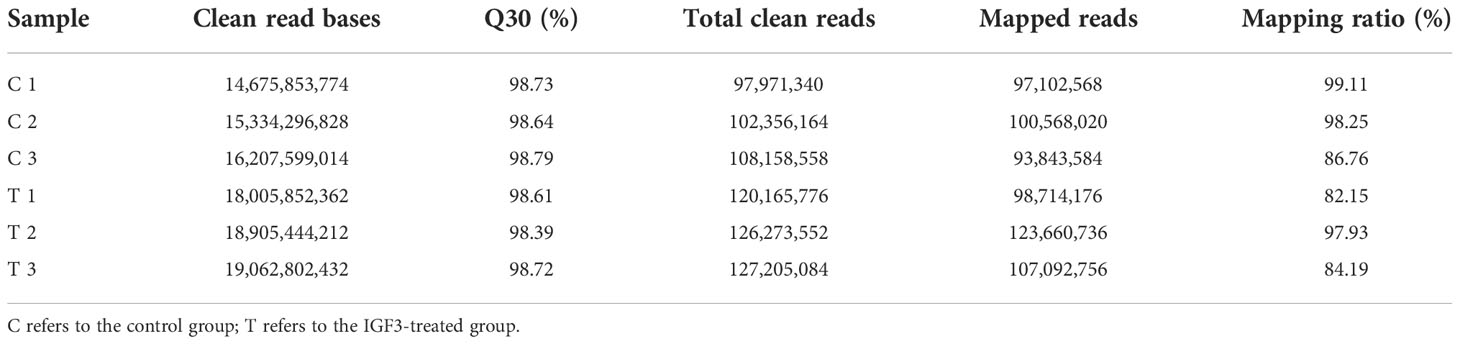
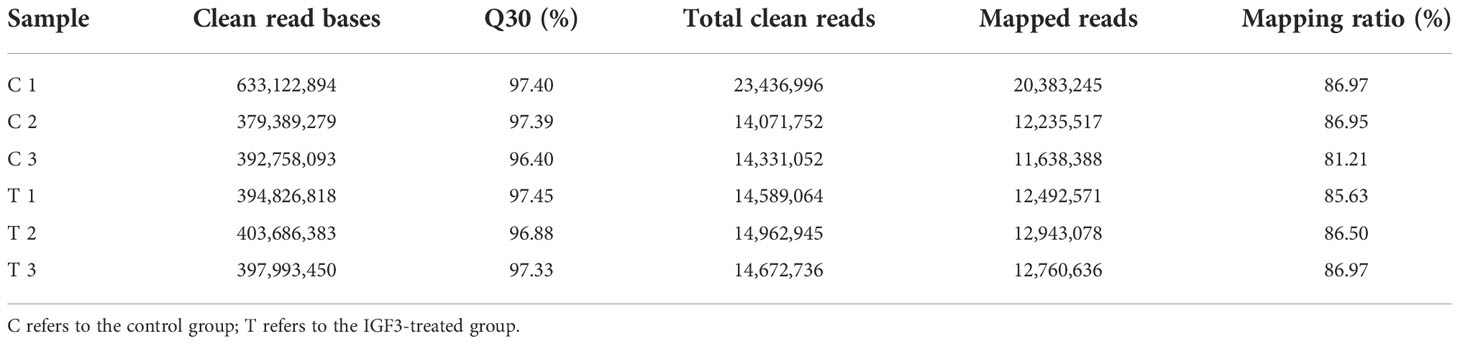
Six cDNA circRNA libraries and six cDNA small RNA libraries were constructed and sequenced. After quality control, 102.19 Gb of clean data for circRNA libraries was obtained with an average Q30 above 98.39%, and 2.6 Gb of clean data for small RNA libraries was obtained with an average Q30 above 96.40%. Then, clean reads were aligned to the S. argus reference genome. Percentages of clean circRNA reads aligned to the genome ranged from 82.15% to 99.11% (Table 2), and percentages of clean small RNA reads aligned to the genome ranged from 81.21% to 86.97% (Table 3). The mapped reads were used for further analyses. In total, 8,919 circRNAs and 2,073 miRNAs were identified (Figure 1). Among the identified miRNAs, there were 1,040 known miRNAs and 1,033 novel miRNAs.
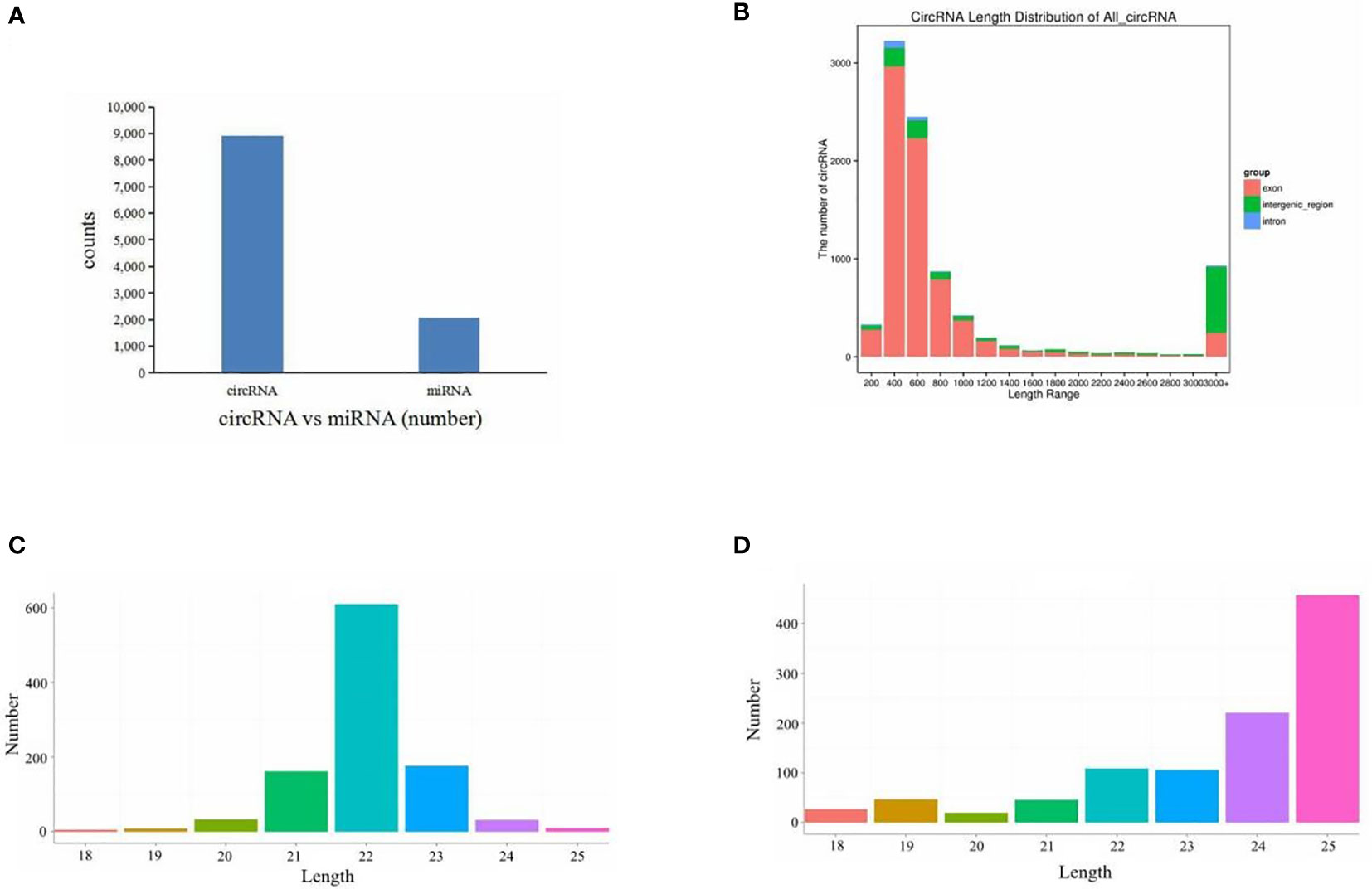
Figure 1 Overview of RNA sequencing. (A) Numbers of circRNAs and miRNAs. (B) All circRNA length distribution. (C) Known miRNA length distribution. (D) Novel miRNA length distribution.
Identification of differentially expressed circRNAs and miRNAs
DE RNAs were discovered by comparisons with the control group. After IGF3 protein treatment, 176 circRNAs and 52 miRNAs were DE. These included 49 upregulated circRNAs and 127 downregulated circRNAs in the treatment group (p < 0.05, fold change ≥ 2) and 39 upregulated miRNAs and 13 downregulated miRNAs (p < 0.01, fold change ≥ 1.5) (Supplementary Table S1). The DE circRNAs and miRNAs are summarized in a volcano plot exhibiting their upregulation or downregulation (Figures 2A, B). Results of a systematic clustering analysis of DE circRNAs and miRNAs are shown in heatmaps (Figures 2C, D).
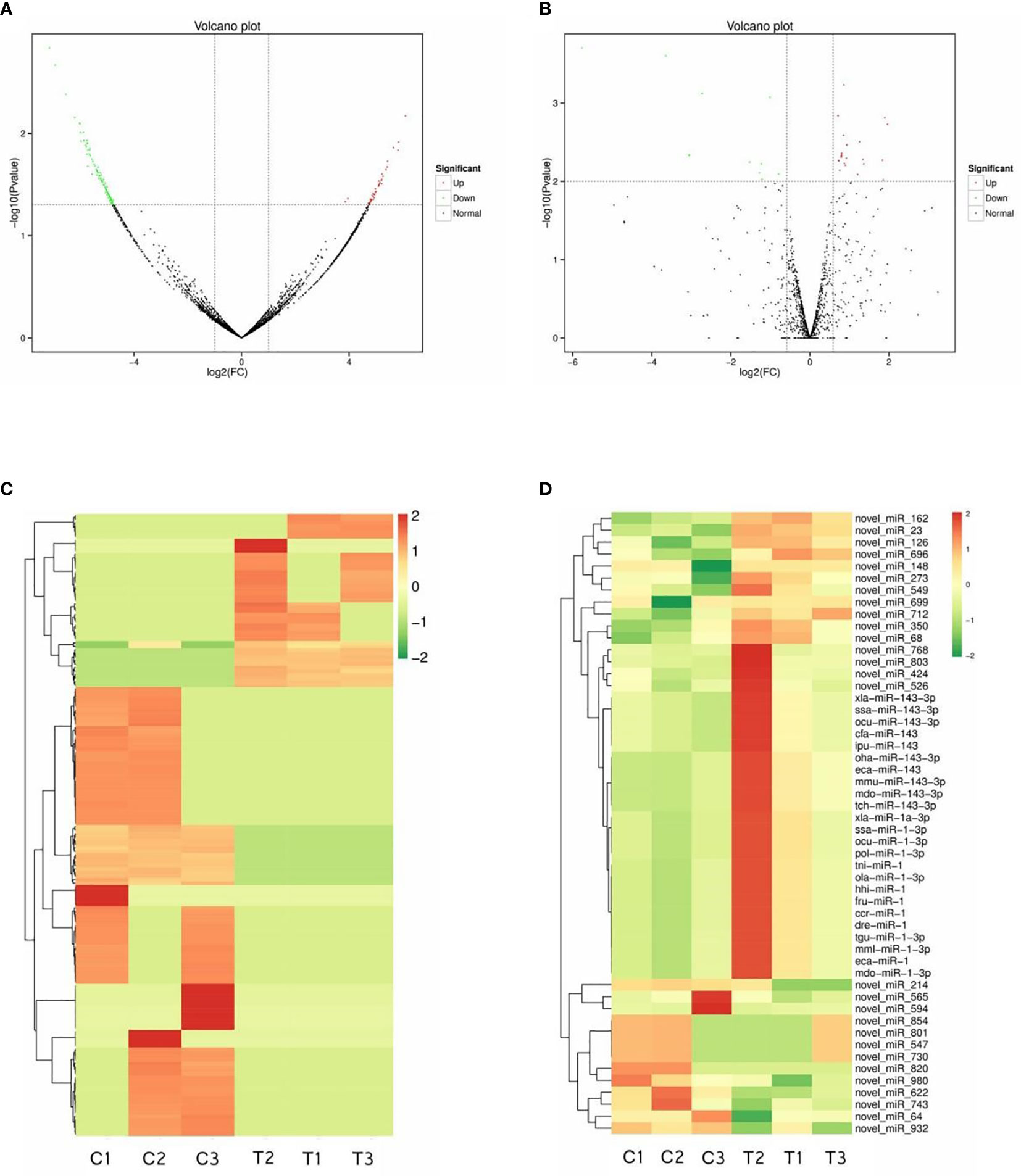
Figure 2 Differentially expressed circRNAs and miRNAs. Volcano plot figures of identified circRNAs (A) and miRNAs (B). Heatmaps of differentially expressed circRNAs (C) and miRNAs (D).
GO and KEGG pathway enrichment analysis
Host genes of all DE circRNAs and target genes of all DE miRNAs were predicted and annotated (Supplementary Table S2). The top significantly enriched GO terms in three main categories, namely, biological processes, cellular components, and molecular functions, of DE circRNAs and DE miRNAs were determined (Figures 3A, B). The enriched GO term for DE circRNAs and DE miRNAs overlapped substantially. In the biological process category, DE circRNAs and DE miRNAs were related to the following terms: cellular process, reproduction, reproductive process, biological adhesion, and growth. In the cellular component category, the terms membrane, extracellular region, cell junction, and extracellular region part were obtained. In the molecular function category, enrichment for binding, catalytic activity, molecular function regulator, nucleic acid binding transcription factor activity, transcription factor activity, and protein binding was detected (Supplementary Table S3).
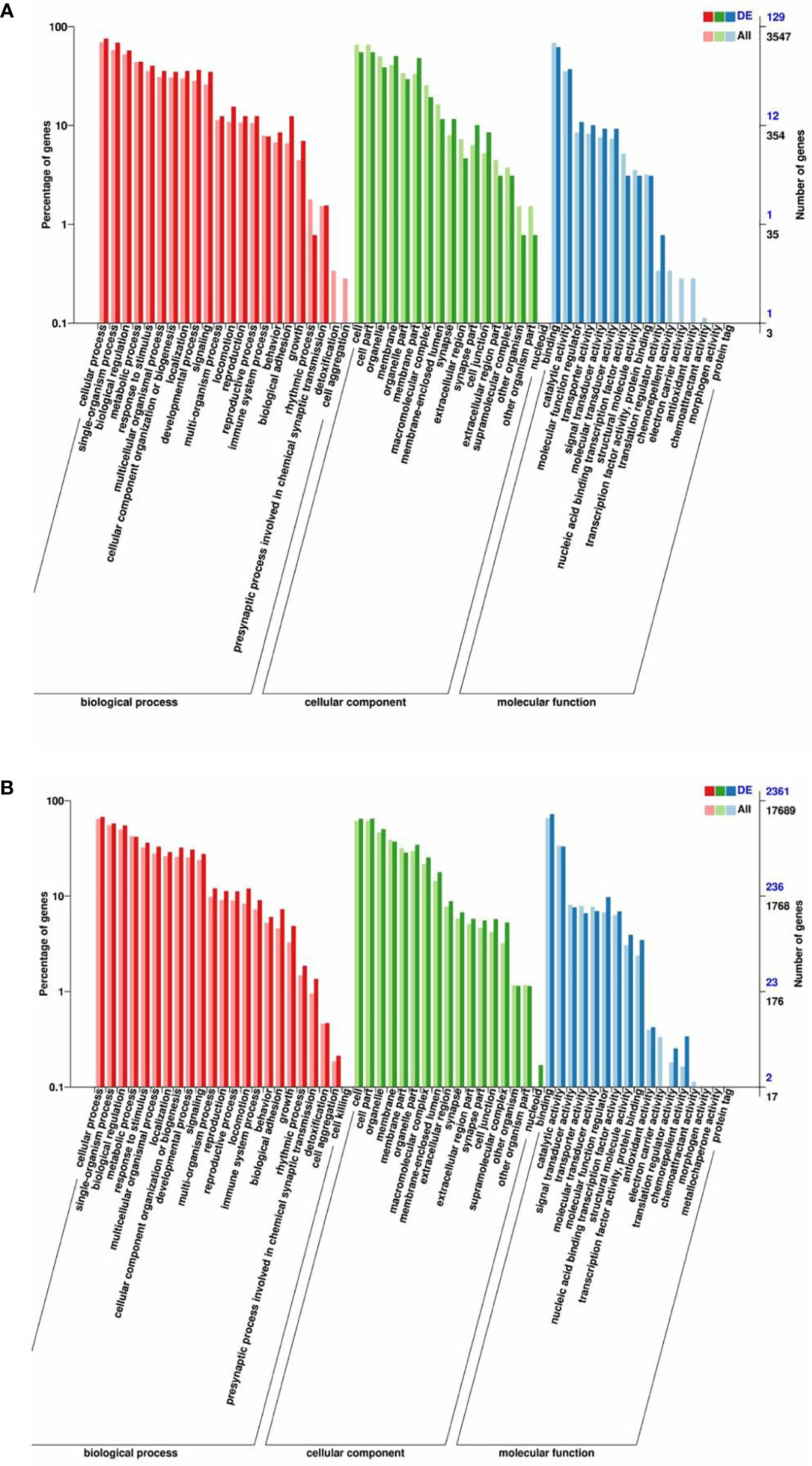
Figure 3 GO enrichment analysis of the host genes of DE circRNAs and the target genes of DE miRNAs. GO enrichment of the host genes of DE circRNAs (A) and the target genes of DE miRNAs (B). The terms representing biological process, cellular component, and molecular function are in red, green, and blue, respectively.
A KEGG pathway enrichment analysis was also carried out to uncover the functional pathways defined by DE circRNAs and DE miRNAs (Supplementary Table S4). Enriched KEGG pathways involving host genes of DE circRNAs included the Hedgehog signaling pathway, phosphatidylinositol signaling system, PI3K-Akt signaling pathway, cell adhesion molecules, ECM–receptor interaction, and regulation of actin cytoskeleton (Figure 4A). Enriched KEGG pathways for target genes of DE miRNAs included ECM–receptor interaction, ABC transporters, Hedgehog signaling pathway, Apelin signaling pathway, tight junction, focal adhesion, Notch signaling pathway, regulation of actin cytoskeleton, cell cycle, insulin signaling pathway, and Rap1 signaling pathway (Figure 4B).
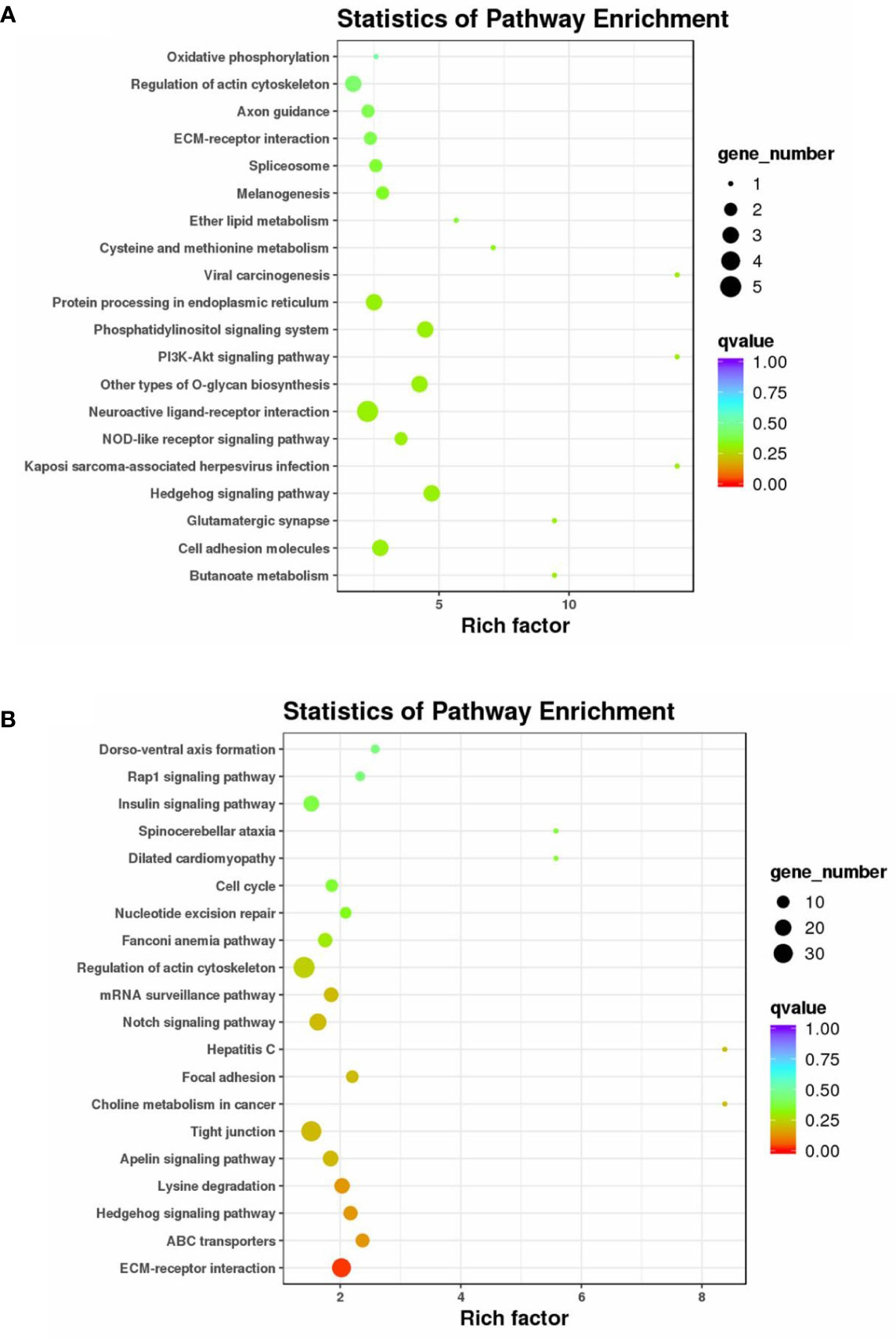
Figure 4 KEGG pathway enrichment analysis of the host genes of DE circRNAs and the target genes of DE miRNAs. KEGG enrichment of the host genes of DE circRNA (A) and the target genes of DE miRNAs (B). The color of the dot represents the q-value for each pathway. The size of the dot represents the number of genes enriched in each pathway.
Construction of a circRNA–miRNA–mRNA regulatory network
To further reveal the potential regulatory relationship between circRNAs, miRNAs, and mRNAs in IGF3-induced ovarian maturation, the DE mRNAs related to reproduction and ovarian maturation, DE miRNAs targeting these DE mRNAs, and DE circRNAs targeted by these DE miRNAs or related to these DE mRNAs were selected for network construction (Figure 5). The network included 18 DE circRNAs, 6 DE miRNAs, and 15 DE mRNAs. In this network, we found three miRNAs (novel_miR_622, novel_miR_980, and novel_miR_64) with multiple connections to DE mRNAs and one circRNA (Lachesis_group5:6245955|6270787) with multiple connections to DE miRNAs.
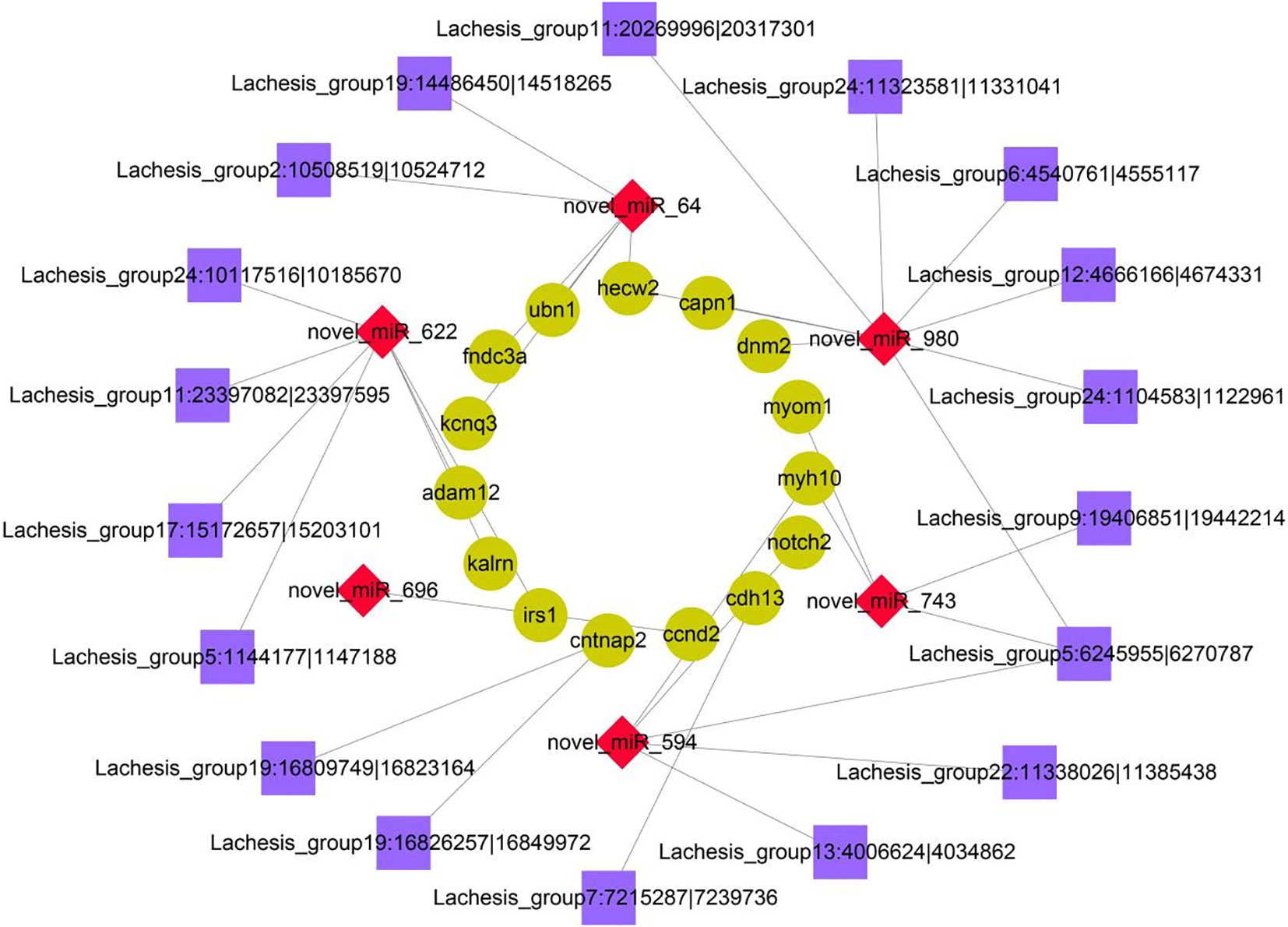
Figure 5 Coexpression network of DE circRNAs, miRNAs, and mRNAs. The cyan circular nodes represent mRNAs, the red diamond nodes represent miRNAs, and the purple square nodes represent circRNAs.
Validation by RT-qPCR
The RNA-seq results were validated by an RT-qPCR analysis of randomly selected DE circRNAs and miRNAs. The expression trends for all four DE circRNAs and eight DE miRNAs were consistent with the RNA-Seq results (Figure 6). The circRNAs (Lachesis_group5:6245955|6270787, Lachesis_group19:16809749|16823164, and Lachesis_group7:7215287|7239736) were upregulated, and the circRNA (Lachesis_group9:19406851|19442214) was downregulated. The miRNAs (ssa-miR-143-3p, ccr-miR-1, and novel_miR_696) were upregulated, and the miRNAs (novel_miR_594, novel_miR_622, novel_miR_980, novel_miR_64, and novel_miR_743) were downregulated. The expression trends for DE mRNAs were also consistent with RT-qPCR results from a previous study (40). Generally, the results of the qPCR analysis validated the RNA-seq results, confirming the accuracy and reliability of the results.
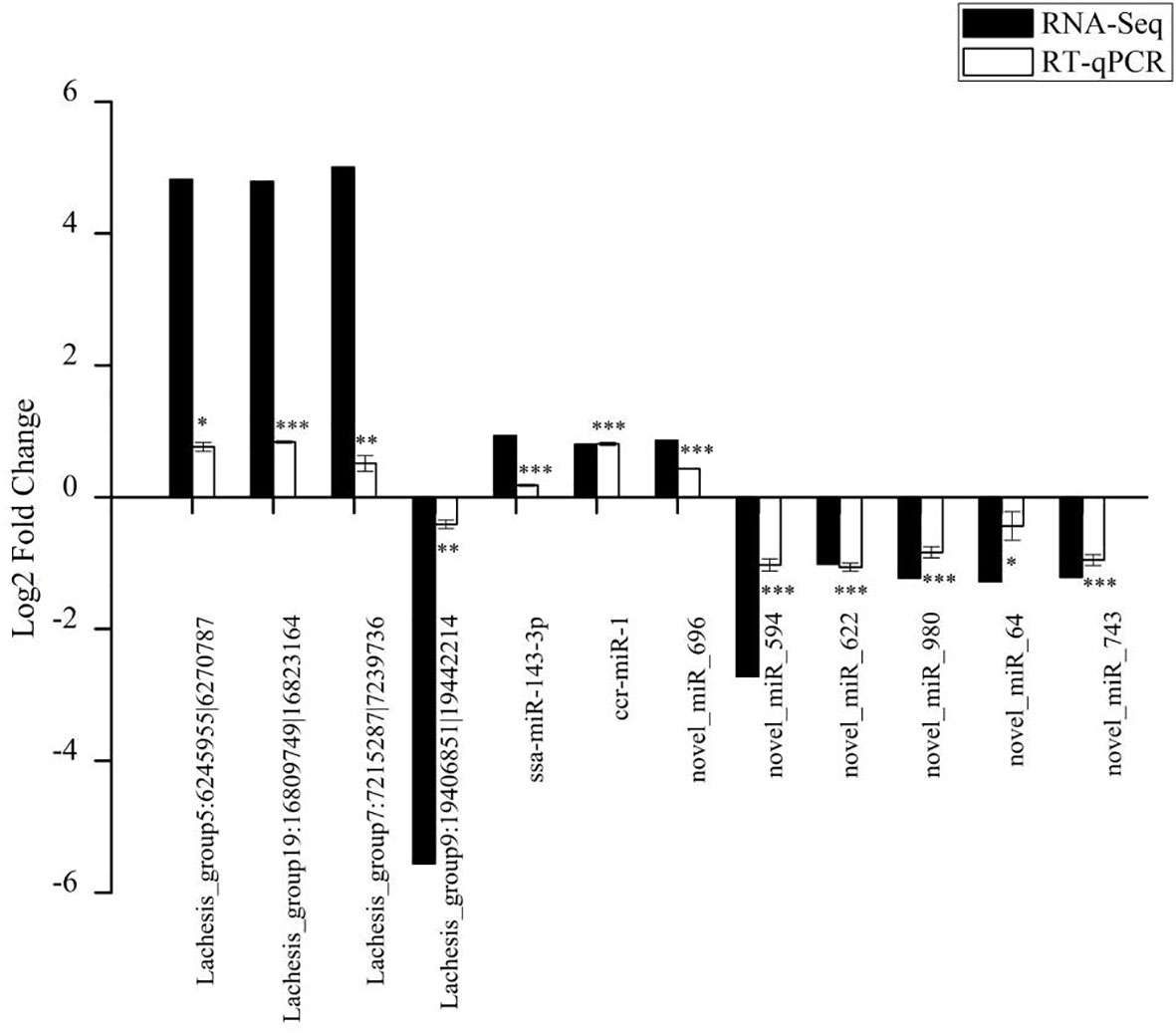
Figure 6 Validation of randomly selected four DE circRNAs and eight DE miRNAs of high-throughput sequencing results using qRT-PCR analysis. Data of qRT-PCR are expressed as the mean value ± SEM (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001.
Tissue distribution of four candidate RNAs
From the circRNA–miRNA–mRNA regulatory network, we found one circRNA (Lachesis_group5:6245955|6270787) and three miRNAs (novel_miR_622, novel_miR_980, and novel_miR_64) with multiple connections. These RNAs were selected for an analysis of expression distribution in 12 tissues of S. argus by RT-qPCR. The four candidate RNAs were all widely distributed across the 12 tissues (Figure 7). The circRNA (Lachesis_group5: 6245955|6270787) was expressed differentially in the gonads of different sexes, with low expression in the ovary but high expression in the testis (Figure 7A). All three miRNAs were expressed at low levels in gonads, and the expression levels of novel_miR_622 and novel_miR_64 were lowest in the ovary (Figures 7B–D).
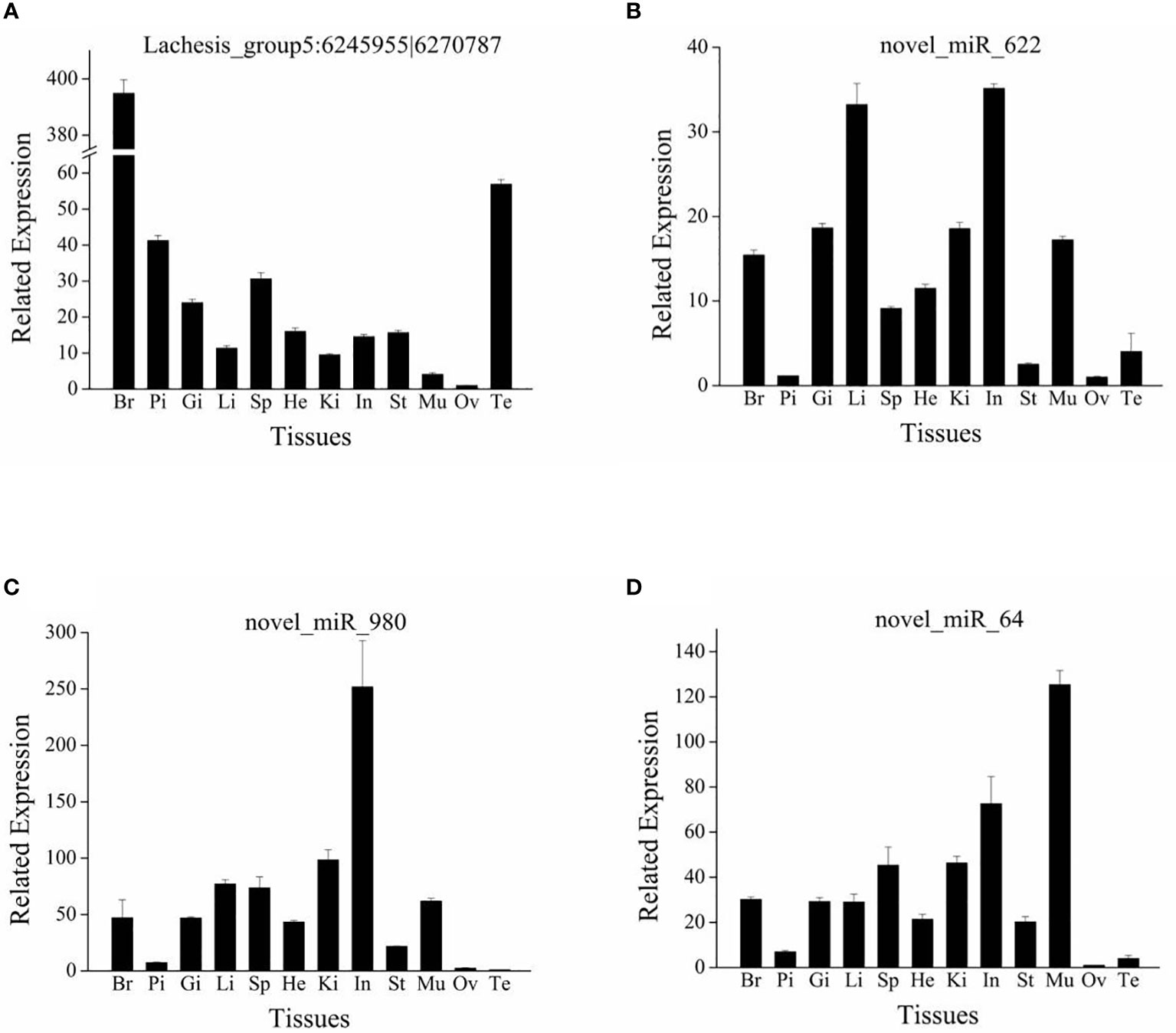
Figure 7 Tissue distribution of circRNA Lachesis_group5:6245955|6270787 (A) and three miRNAs novel_miR_622 (B), novel_miR_980 (C), and novel_miR_64 (D). Br, brain; Pi, pituitary; Gi, gill; Li, liver; Sp, spleen; He, heart; Ki, kidney; In, intestine; St, stomach; Mu, muscle; Ov, ovary; Te, testis. Data are expressed as the mean value ± SEM (n = 3).
Discussion
Oocyte development and maturation, essential processes for successful reproduction, play critical roles in artificial breeding in the aquaculture industry (41). Research has revealed that IGF3 can stimulate oocyte development and maturation in teleosts (24–27). The molecular regulatory mechanism underlying IGF3-induced oocyte maturation remains unclear; accordingly, a RNA-seq analysis was performed in this study. Considering the key roles of miRNAs and circRNAs in reproduction (9, 10, 14), detailed RNA profiles of IGF3-induced oocyte maturation in S. argus were obtained. Finally, we identified 176 DE circRNAs and 52 DE miRNAs between the control group and IGF3-treated group.
A KEGG enrichment analysis of DE circRNAs and miRNAs revealed some overlapping terms, such as ECM–receptor interaction, and regulation of actin cytoskeleton. The extracellular matrix (ECM) has contributed to ovarian remodeling during the reproductive cycle and is essential for follicular development and ovulation (42). Additionally, the cytoskeleton plays vital roles in oocyte meiotic maturation by regulating spindle assembly, spindle length, and chromosome segregation (43). Some of the pathways identified in the KEGG enrichment analysis have been the focus of recent research, such as the Hedgehog, PI3K-Akt, Apelin, Notch, insulin, and Rap1 signaling pathways. The hedgehog signaling pathway participates in steroidogenesis in gonad tissues (44, 45), and IGF3 stimulates steroidogenic enzymes in the ovaries of O. niloticus (46). In D. rerio, IGF3 can induce the maturation of oocytes by activating the PI3K-Akt signaling pathway (23). Apelin13 significantly upregulates the phosphorylation levels of both Akt and AMPK, resulting in the proliferation and migration of cells (47). The Notch signaling pathway is involved in the regulation of female germ cell meiosis progression and early oogenesis (48). Furthermore, this pathway is required for ovarian follicle formation and follicular growth (49). The insulin signaling pathway plays an important role via downstream dietary and neuronal signaling in the regulation of various metabolic outputs, including reproductive capacity (50, 51). Furthermore, defects in insulin/IGF1 signaling can cause infertility (52, 53). Rap1b mediates the mitogenic signal of cAMP leading to an increase in G1/S phase entry (54). The activation of Rap1 can activate B-Raf/MEK/Erk and Akt signaling pathways to stimulate cell proliferation and survival and can induce integrin-mediated cell adhesion (55).
In the circRNA–miRNA–mRNA network, several DE genes related to reproduction and ovarian maturation were included. For instance, various DE genes, such as ccnd2, hecw2, and dnm2, were related to oocyte meiotic division, which is a prerequisite for oocyte maturation (56, 57). D-Type cyclins (CCNDs) are cell cycle proteins; CCND2 plays an essential role in oocyte meiosis during the reproductive cycle (58, 59). Furthermore, CCND2 is associated with the proliferation of granulosa cells and ovulation (60). E3 ubiquitin-protein ligase (HECW2) enhances the activation of transcription and is involved in the regulation of the mitotic metaphase/anaphase transition (61, 62). Dynamin 2 (DNM2) binds to and hydrolyzes GTP to produce microtubule bundles and is involved in cytokinesis (63).
During the reproductive cycle, the ECM plays an essential role in ovarian remodeling in follicular development and ovulation (42). Research has revealed that adhesion proteins, such as fibronectin and laminin, are components of the ECM in follicular cells (64, 65). In the network, several cell-adhesion-related genes were detected, such as adam12 and cdh13. The gene adam12 encodes a member of a family of proteins that are structurally associated with snake venom disintegrins and has been implicated in cell–cell and cell–matrix interactions, such as fertilization (66, 67). Cadherin 13, encoded by cdh13, is a member of calcium-dependent cell adhesion proteins and is localized to the surface of the cell membrane (68).
Insulin exerts various effects by binding to the insulin receptor (IR). After ligand binding, the IR undergoes conformational changes to activate various cellular substrates, including the insulin receptor substrate (IRS) (69). Four IRSs have been identified in various mammals (69). All four IRSs are able to interact with all the regulatory subunits of PI3K and to activate the catalytic subunit (70). In the present study, irs1 was upregulated after IGF3 treatment and was involved in the circRNA–miRNA–mRNA network. IRS1 is reported to be involved in insulin responses, notably mitogenesis (71). A lack of IRS1 in mice leads to defective reproductive function (72).
Among the DE miRNAs, most of the known miRNAs belonged to the miR-143 and miR-1 families. Research has revealed that miR-143 plays an important role in reproduction. At early time points, miR-143 exerts regulatory functions in the formation of primordial follicles, and its regulatory effects continue throughout ovarian development (73). Additionally, miR-143 is highly expressed in mature gonads of O. niloticus and Trachinotus ovatus, indicating that it regulates fish reproduction during the maturation period (9, 74). Furthermore, miR-143 has been regarded as a critical regulator of the endocrine system in gonadal functions and pregnancy maintenance in mammal (7, 8). It can promote progesterone production in ovarian granulosa cells by interacting with FSH (75). Furthermore, miR-143 is a transcriptional target of the Notch signaling pathway (76) and modulates cytoskeletal dynamics (77). In contrast, miR-1 is involved in cell junctions (78) and has regulatory functions on cell proliferation and cell cycle progression (79, 80).
Among the DE novel miRNAs, we found three miRNAs (novel_miR_622, novel_miR_980, and novel_miR_64) targeting the DE genes that were related to reproduction and ovarian maturation, such as irs1, adam12, hecw2, and dnm2 (61, 63, 67, 71). The tissue distribution results revealed that expression levels of these three novel miRNAs were low in gonads. These results were consistent with the observation that IGF3 downregulated the three miRNAs. IGF3 may induce maturation via these three miRNAs. However, the expression of the candidate circRNA (Lachesis_group5:6245955|6270787) differed between the ovary and testis (i.e., expression levels were low in the ovary and high in the testis). This circRNA may play a role in the reproduction of S. argus; however, it may bias towards male sex. The present results demonstrated that the ncRNAs may be involved in IGF3-induced ovarian maturation in S. argus. These results are expected to provide useful information about fish reproduction.
Conclusions
In this study, circRNA and miRNA expression profiles involved in IGF3-induced ovarian maturation in S. argus were identified. An enrichment analysis of host genes of DE circRNAs and target genes of DE miRNAs revealed the roles of several pathways related to reproductive functions and oocyte maturation, such as the Hedgehog, PI3K-Akt, Apelin, Notch, insulin, and Rap1 signaling pathways. A circRNA–miRNA–mRNA regulatory network was constructed, from which DE genes involved in reproduction were identified, such as ccnd2, hecw2, dnm2, irs1, adam12, and cdh13. These DE genes are involved in oocyte maturation, such as oocyte meiosis and ECM remodeling. Based on the regulatory network and tissue distribution, we identified one circRNA (Lachesis_group5:6245955|6270787) and three miRNAs (novel_miR_622, novel_miR_980, and novel_miR_64) that may exert regulatory functions in IGF3-induced ovarian maturation in S. argus. Taken together, this study provided a novel insight into the molecular mechanisms underlying the effects of IGF3 in the ovary and highlighted the roles of circRNAs and miRNAs in S. argus reproduction.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA849653.
Ethics statement
The animal study was reviewed and approved by Animal Research and Ethics Committees of Fisheries College of Guangdong Ocean University.
Author contributions
ZL: investigation, data curation, formal analysis, and writing—original draft. YG: investigation and methodology. CN: data curation and formal analysis. HDC: data curation and formal analysis. CH: methodology and resources. GZ: methodology. HH: data curation, and writing—review and editing. GL: methodology and resources. HPC: data curation, funding acquisition, methodology, resources, supervision, writing—original draft, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This article was supported by National Natural Science Foundation of China (Nos. 32273131 and 32172971), South China Aquaculture Breeding Funding (2022RHDKFKT01), the Key Research and Development Program of Guangdong (2021B202020002), the talent team tender grant of Zhanjiang Marine Equipment and Biology (2021E05035), the Guangdong Basic and Applied Basic Research Foundation (2019A1515010958), and the Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang) (ZJW-2019-06).
Conflict of interest
Author HDC is employed by Guangdong Havwii Agriculture Group Co., Ltd, Zhanjiang, China. CH and GZ are employed by Hainan Chenhai Aquatic Co., Ltd, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.998207/full#supplementary-material
References
1. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat Cell Biol (2009) 11(3):228–34. doi: 10.1038/NCB0309-228
2. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
3. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet (2010) 11(9):597–610. doi: 10.1038/nrg2843
4. Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in drosophila. Cell (2003) 113:25–36. doi: 10.1016/S0092-8674(03)00231-9
5. Nixon B, Stanger SJ, Mihalas BP, Reilly JN, Anderson AL, Tyagi S, et al. The MicroRNA signature of mouse spermatozoa is substantially modified during epididymal maturation. Biol Reprod (2015) 93(4):91, 1–20. doi: 10.1095/biolreprod.115.132209
6. Qi R, Huang J, Wang Q, Liu H, Wang R, Wang J, et al. MicroRNA-224-5p regulates adipocyte apoptosis induced by TNF J. MicroRN-κp Regulates Adipurnal Cell Physiol (2017) 233(2):1236–46. doi: 10.1002/jcp.25992
7. Hu S-J, Ren G, Liu J-L, Zhao Z-A, Yu Y-S, Su R-W, et al. MicroRNA expression and regulation in mouse uterus during embryo implantation. J Biol Chem (2008) 283(34):23473–84. doi: 10.1074/jbc.M800406200
8. Kim SY, Romero R, Tarca AL, Bhatti G, Lee J, Chaiworapongsa T, et al. miR-143 regulation of prostaglandin-endoperoxidase synthase 2 in the amnion: Implications for human parturition at term. PLos One (2011) 6(9):e24131. doi: 10.1371/journal.pone.0024131.t001
9. Xiao J, Zhong H, Zhou Y, Yu F, Gao Y, Luo Y, et al. Identification and characterization of MicroRNAs in ovary and testis of Nile tilapia (Oreochromis niloticus) by using solexa sequencing technology. PLos One (2014) 9(1):e86821. doi: 10.1371/journal.pone.0086821
10. Guo H, Du X, Zhang Y, Wu J, Wang C, Li M, et al. Specific miRNA-G protein-coupled receptor networks regulate Sox9a/Sox9b activities to promote gonadal rejuvenation in zebrafish. Stem Cells (2019) 37(9):1189–99. doi: 10.1002/stem.3040
11. Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett (2015) 365(2):141–8. doi: 10.1016/j.canlet.2015.06.003
12. Chen I, Chen C-Y, Chuang T-J. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Reviews: RNA (2015) 6(5):563–79. doi: 10.1002/wrna.1294
13. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
14. Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep (2014) 9(5):1966–80. doi: 10.1016/j.celrep.2014.10.062
15. Zhang C, Liu J, Lai M, Li J, Zhan J, Wen Q, et al. Circular RNA expression profiling of granulosa cells in women of reproductive age with polycystic ovary syndrome. Arch Gynecol Obstetrics (2019) 300(2):431–40. doi: 10.1007/s00404-019-05129-5
16. Yang J-J, Tao H, Deng Z-Y, Lu C, Li J. Non-coding RNA-mediated epigenetic regulation of liver fibrosis. Metabolism (2015) 64(11):1386–94. doi: 10.1016/j.metabol.2015.08.004
17. Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun (2016) 7:11215. doi: 10.1038/ncomms11215
18. Tao H, Xiong Q, Zhang F, Zhang N, Liu Y, Suo X, et al. Circular RNA profiling reveals chi_circ_0008219 function as microRNA sponges in pre-ovulatory ovarian follicles of goats (Capra hircus). Genomics (2018) 110(4):257–66. doi: 10.1016/j.ygeno.2017.10.005
19. Wood AW, Duan C, Howard AB. Insulin-like growth factor signaling in fish. Int Rev Cytology (2005) 243(243):215–85. doi: 10.1079/SUM2005312
20. Mukherjee D, Mukherjee D, Sen U, Paul S, Bhattacharyya SP. In vitro effects of insulin-like growth factors and insulin on oocyte maturation and maturation-inducing steroid production in ovarian follicles of common carp, cyprinus carpio. Comp Biochem Physiol Part A: Mol Integr Physiol (2006) 144(1):63–77. doi: 10.1016/j.cbpa.2006.01.012
21. Reinecke M. Insulin-like growth factors and fish reproduction. Biol Reprod (2010) 82(4):656–61. doi: 10.1095/biolreprod.109.080093
22. Reinecke M, Björnsson BT, Dickhoff WW, McCormick SD, Navarro I, Power DM, et al. Growth hormone and insulin-like growth factors in fish: Where we are and where to go. Gen Comp Endocrinol (2005) 142(1-2):20–4. doi: 10.1016/j.ygcen.2005.01.016
23. Li J, Liu Z, Kang T, Li M, Cheng C. Igf3 a novel player in fishreproduction. Biol Reprod (2021) 104(6):1194–204. doi: 10.1093/biolre/ioab042
24. Li J, Chu L, Sun X, Liu Y, Cheng CHK. IGFs mediate the action of LH on oocyte maturation in zebrafish. Mol Endocrinol (2015) 29(3):373–83. doi: 10.1210/me.2014-1218
25. Li Z, Ren X, Guo Y, Ru X, Tian C, Shi H, et al. Identification and ovarian developmental regulation of insulin-like growth factor 3 in spotted scat (Scatophagus argus). Aquaculture Rep (2021) 21:100866. doi: 10.1016/j.aqrep.2021.100866
26. Song F, Wang L, Zhu W, Fu J, Dong J, Dong Z. A novel igf3 gene in common carp (Cyprinus carpio): Evidence for its role in regulating gonadal development. PLos One (2016) 11(12):e0168874. doi: 10.1371/journal.pone.0168874
27. Yang H, Chen H, Zhao H, Liu L, Xie Z, Xiao L, et al. Molecular cloning of the insulin-like growth factor 3 and difference in the expression of igf genes in orange-spotted grouper (Epinephelus coioides). Comp Biochem Physiol Part B: Biochem Mol Biol (2015) 186:68–75. doi: 10.1016/j.cbpb.2015.04.005
28. Song F, Wang L, Zhu W, Dong Z. Long noncoding RNA and mRNA expression profiles following igf3 knockdown in common carp, cyprinus carpio. Sci Data (2019) 6:190024. doi: 10.1038/sdata.2019.24
29. Assan D, Wang Y, Mustapha UF, Ndandala CB, Li Z, Li G-L, et al. Neuropeptide y in spotted scat (Scatophagus Argus), characterization and functional analysis towards feed intake regulation. Fishes (2022) 7(3):111. doi: 10.3390/fishes7030111
30. Chen H, Li Z, Wang Y, Yang W, Shi H, Li S, et al. Relationship between body weight and morphological traits in female and Male spotted scat (Scatophagus argus). Pakistan J Zoology (2021) 54(4):1–9. doi: 10.17582/journal.pjz/20210305150335
31. Wang Y, Yang W, Ren X, Jiang D, Deng S, Chen H, et al. Distribution patterns of microsatellites and development of polymorphic markers from scatophagus argus genome. J Guangdong Ocean Univ (2020) 40(4):7–17. doi: 10.3969/j.issn.1673-9159.2020.04.002
32. Amarasinghe US, Amarasinghe MD, Nissanka C. Investigation of the negombo estuary (Sri Lanka) brush park fishery, with an emphasis on community-based management. Fisheries Manage Ecol (2002) 9(1):41–56. doi: 10.1046/j.1365-2400.2002.00250.x
33. Chen H, Jiang D, Li Z, Wang Y, Yang X, Li S, et al. Comparative physiological and transcriptomic profiling offers insight into the sexual dimorphism of hepatic metabolism in size-dimorphic spotted scat (Scatophagus argus). Life (2021) 11(6):589. doi: 10.3390/life11060589
34. Gupta S. An overview on morphology, biology, and culture of spotted scat scatophagus argus (Linnaeus 1766). Rev Fisheries Sci Aquaculture (2016) 24(2):203–12. doi: 10.1080/23308249.2015.1119800
35. Li ZY, Hong G, Wang YR, Li GL, Zhu CH, Chen HP, et al. cDNA cloning and mRNA expression analysis of Sox3 in scatophagus argus. Genomics Appl Biol (2020) 39(7):2980–8. doi: 10.13417/j.gab.039.002980
36. Shi H, Ru X, Liu Y, Zheng Y, Wu X, Huang Y, et al. Transcriptomic analysis of hypothalamus in female scatophagus argus after 17β-estradiol injection. J Guangdong Ocean Univ (2021) 41(2):76–85. doi: 10.3969/j.issn.1673-9159.2021.02.11
37. Li Z, Tian C, Huang Y, Lin X, Wang Y, Jiang D, et al. A first insight into a draft genome of silver sillago (Sillago sihama) via genome survey sequencing. Animals (2019) 9(10):756. doi: 10.3390/ani9100756
38. Huang Y, Mustapha UF, Huang Y, Tian C, Yang W, Chen H, et al. A chromosome–level genome assembly of the spotted scat (Scatophagus argus). Genome Biol Evol (2021) 13(6):evab092. doi: 10.1093/gbe/evab092
39. Liang Y, Zhang Y, Xu L, Zhou D, Jin Z, Zhou H, et al. CircRNA expression pattern and ceRNA and miRNA–mRNA networks involved in anther development in the CMS line of brassica campestris. Int J Mol Sci (2019) 20(19):4808. doi: 10.3390/ijms20194808
40. Wang Y, Li Z, Guo Y, Nong C, Ndandala CB, Yang H, et al. Analysis on lncRNA and mRNA expression profiles of IGF3-induced ovarian maturation in spotted scat (Scatophagus argus). Aquaculture Rep (2022) 27:101367. doi: 10.1016/j.aqrep.2022.101367
41. Das D, Khan PP, Maitra S. Endocrine and paracrine regulation of meiotic cell cycle progression in teleost oocytes: cAMP at the centre of complex intra-oocyte signalling events. Gen Comp Endocrinol (2017) 241:33–40. doi: 10.1016/j.ygcen.2016.01.005
42. Curry TE, Osteen KG. The matrix metalloproteinase system: Changes, regulation, and impact throughout the ovarian and uterine reproductive cycle. Endocrine Rev (2003) 24(4):428–65. doi: 10.1210/er.2002-0005
43. Ou X-H, Li S, Xu B-Z, Wang Z-B, Quan S, Li M, et al. p38α MAPK is a MTOC-associated protein regulating spindle assembly, spindle length and accurate chromosome segregation during mouse oocyte meiotic maturation. Cell Cycle (2014) 9(20):4130–43. doi: 10.4161/cc.9.20.13389
44. Huang C-CJ, Yao HH-C. Diverse functions of hedgehog signaling in formation and physiology of steroidogenic organs. Mol Reprod Dev (2010) 77(6):489–96. doi: 10.1002/mrd.21174
45. Walterhouse DO, Lamm MLG, Villavicencio E, Iannaccone PM. Emerging roles for hedgehog-Patched-Gli signal transduction in Reproduction1. Biol Reprod (2003) 69(1):8–14. doi: 10.1095/biolreprod.103.015941
46. Li M, Wu F, Gu Y, Wang T, Wang H, Yang S, et al. Insulin-like growth factor 3 regulates expression of genes encoding steroidogenic enzymes and key transcription factors in the Nile tilapia Gonad1. Biol Reprod (2012) 86(5):1–10. doi: 10.1095/biolreprod.111.096248
47. Zhang P, Liu M-Y, Zhu W, Yang X-M, Wei M. Apelin promotes angiogenesis of myocardial micro-vascular endothelial cells via Akt/AMPK pathway. Int J Cardiovasc Dis (2013) 40(1):5. doi: 10.3969/j.issn.1673-6583.2013.01.015
48. Feng Y-M, Liang G-J, Pan B, Qin X, Zhang X-F, Chen C-L, et al. Notch pathway regulates female germ cell meiosis progression and early oogenesis events in fetal mouse. Cell Cycle (2014) 13(5):782–91. doi: 10.4161/cc.27708
49. Vanorny DA, Prasasya RD, Chalpe AJ, Kilen SM, Mayo KE. Notch signaling regulates ovarian follicle formation and coordinates follicular growth. Mol Endocrinol (2014) 28(4):499–511. doi: 10.1210/me.2013-1288
50. Badisco L, Van Wielendaele P, Vanden Broeck J. Eat to reproduce: a key role for the insulin signaling pathway in adult insects. Front Physiol (2013) 4:202. doi: 10.3389/fphys.2013.00202
51. Burks DJ, Mora J, Schubert M, Withers DJ, White MF. IRS-2 pathways integrate female reproduction and energy homeostasis. Nature (2000) 407(6802):377–82. doi: 10.1038/35030105
52. Garofalo RS. Genetic analysis of insulin signaling in drosophila. Trends Endocrinol Metab (2002) 13(4):156–62. doi: 10.1016/S1043-2760(01)00548-3
53. Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, et al. The fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in c. elegans. Nat (1997) 389(30):994–9. doi: 10.1038/40194
54. Lou L, Urbani J, Ribeiro-Neto F, Altschuler DL. cAMP inhibition of akt is mediated by activated and phosphorylated Rap1b. J Biol Chem (2002) 277(36):32799–806. doi: 10.1074/jbc.M201491200
55. Jin A, Kurosu T, Tsuji K, Mizuchi D, Arai A, Fujita H, et al. BCR/ABL and IL-3 activate Rap1 to stimulate the b-Raf/MEK/Erk and akt signaling pathways and to regulate proliferation, apoptosis, and adhesion. Oncogene (2006) 25(31):4332–40. doi: 10.1038/sj.onc.1209459
56. Nagahama Y, Yamashita M. Regulation of oocyte maturation in fish. Development Growth Differentiation (2008) 50:S195–219. doi: 10.1111/j.1440-169X.2008.01019.x
57. Zhao H, Ge J, Wei J, Liu J, Liu C, Ma C, et al. Effect of FSH on E2/GPR30-mediated mouse oocyte maturation in vitro. Cell Signalling (2020) 66:109464. doi: 10.1016/j.cellsig.2019.109464
58. Shi X, Xiong X, Lan D, Chen W, Hu J, Cai W, et al. Cloning and expression analysis of CCND2 gene in yak ovaries during different periods of estrus. Scientia Agricult. Sin (2017) 50(13):2604–13. doi: 10.3864/j.issn.0578-1752.2017.13.018
59. Zhang Q, Sakamoto K, Wagner K-U. D-type cyclins are important downstream effectors of cytokine signaling that regulate the proliferation of normal and neoplastic mammary epithelial cells. Mol Cell Endocrinol (2014) 382(1):583–92. doi: 10.1016/j.mce.2013.03.016
60. Hernandez-Gonzalez I, Gonzalez-Robayna I, Shimada M, Wayne CM, Ochsner SA, White L, et al. Gene expression profiles of cumulus cell oocyte complexes during ovulation reveal cumulus cells express neuronal and immune-related genes: Does this expand their role in the ovulation process? Mol Endocrinol (2006) 20(6):1300–21. doi: 10.1210/me.2005-0420
61. Lu L, Hu S, Wei R, Qiu X, Lu K, Fu Y, et al. The HECT type ubiquitin ligase NEDL2 is degraded by anaphase-promoting Complex/Cyclosome (APC/C)-Cdh1, and its tight regulation maintains the metaphase to anaphase transition. J Biol Chem (2013) 288(50):35637–50. doi: 10.1074/jbc.M113.472076
62. Miyazaki K, Ozaki T, Kato C, Hanamoto T, Fujita T, Irino S, et al. A novel HECT-type E3 ubiquitin ligase, NEDL2, stabilizes p73 and enhances its transcriptional activity. Biochem Biophys Res Commun (2003) 308(1):106–13. doi: 10.1016/s0006-291x(03)01347-0
63. Thompson HM, Skop AR, Euteneuer U, Meyer BJ, Mcniven MA. The Large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr Biol (2002) 12(24):2111–7. doi: 10.1016/S0960-9822(02)01390-8
64. Yasuda K, Hagiwara E, Takeuchi A, Mukai C, Matsui C, Sakai A. Changes in the distribution of tenascin and fibronectin in the mouse ovary during folliculogenesis, atresia, corpus luteum formation and luteolysis. Zoological Sci (2005) 22(2):237. doi: 10.2108/zsj.22.237
65. Zhao Y, Luck MR. Gene expression and protein distribution of collagen,fibronectin and laminin in bovine follicles and corpora lutea. J Reprod Fertil (1995) 104(1):115. doi: 10.1530/jrf.0.1040115
66. Iba K, Albrechtsen R, Gilpin B, Fröhlich C, Loechel F, Zolkiewska A, et al. The cysteine-rich domain of human Adam 12 supports cell adhesion through syndecans and triggers signaling events that lead to β1 integrin–dependent cell spreading. J Cell Biol (2000) 149(5):1143. doi: 10.1083/jcb.149.5.1143
67. Leyme A, Bourd-Boittin K, Bonnier D, Falconer A, Arlot-Bonnemains Y, Théret N, et al. Identification of ILK as a new partner of the ADAM12 disintegrin and metalloprotease in cell adhesion and survival. Mol Biol Cell (2012) 23(17):3461–72. doi: 10.1091/mbc.e11-11-0918
68. Widschwendter A, Ivarsson L, Blassnig A, Müller HM, Fiegl H, Wiedemair A, et al. CDH1 and CDH13 methylation in serum is an independent prognostic marker in cervical cancer patients. Int J Cancer (2004) 109(2):163–6. doi: 10.1002/ijc.11706
69. Dupont J, Scaramuzzi RJ. Insulin signalling and glucose transport in the ovary and ovarian function during the ovarian cycle. Biochem J (2016) 473:1483–501. doi: 10.1042/BCJ20160124
70. Wijesekara N, Konrad D, Eweida M, Jefferies C, Liadis N, Giacca A, et al. Muscle-specific pten deletion protects against insulin resistance and diabetes. Mol Cell Biol (2005) 25(3):1135–45. doi: 10.1128/mcb.25.3.1135-1145.2005
71. Smith LK, Vlahos CJ, Reddy KK, Falck JR, Garner CW. Wortmannin and LY294002 inhibit the insulin-induced down-regulation of IRS-1 in 3T3-L1 adipocytes. Mol Cell Endocrinol (1995) 113(1):73–81. doi: 10.1016/0303-7207(95)03622-e
72. Kadowaki T, Tamemoto H, Tobe K, Terauchi Y, Yazaki Y. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Diabetic Med (1996) 13(9 Suppl 6):S103–8. doi: 10.1002/dme.1996.13.s6.103
73. Zhang J, Ji X, Zhou D, Li Y, Lin J, Liu J, et al. miR-143 is critical for the formation of primordial follicles in mice. Front Biosci (2013) 18:588–97. doi: 10.2741/4122
74. He P, Wei P, Chen X, Lin Y, Peng J. Identification and characterization of microRNAs in the gonad of trachinotus ovatus using solexa sequencing. Comp Biochem Physiol Part D: Genomics Proteomics (2019) 30:312–20. doi: 10.1016/j.cbd.2019.03.010
75. Gao X-B, Yao N, Zheng P-P, Luo H-Y, Ma X, Lu C-L. Role of miR-143 in FSH mediated progesterone production in ovarian granulosa cells. J Reprod Med (2019) 28(6):684–90. doi: 10.3969/j.issn.1004-3845.2019.06.018
76. Boucher JM, Peterson SM, Urs S, Zhang C, Liaw L. The miR-143/145 cluster is a novel transcriptional target of jagged-1/Notch signaling in vascular smooth muscle cells. J Biol Chem (2011) 286(32):28312–21. doi: 10.1074/jbc.M111.221945
77. Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev (2009) 23(18):2166–78. doi: 10.1101/gad.1842409
78. Simon DJ, Madison JM, Conery AL, Thompson-Peer KL, Soskis M, Ruvkun GB, et al. The MicroRNA miR-1 regulates a MEF-2-Dependent retrograde signal at neuromuscular junctions. Cell (2008) 133(5):903–15. doi: 10.1016/j.cell.2008.04.035
79. Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol (2011) 11:34. doi: 10.1186/1471-213x-11-34
Keywords: IGF3, oocyte maturation, ovary, transcriptome, circRNA, miRNA, scatophagus argus
Citation: Li Z, Guo Y, Ndandala CB, Chen H, Huang C, Zhao G, Huang H, Li G and Chen H (2022) Analysis of circRNA and miRNA expression profiles in IGF3-induced ovarian maturation in spotted scat (Scatophagus argus). Front. Endocrinol. 13:998207. doi: 10.3389/fendo.2022.998207
Received: 19 July 2022; Accepted: 03 November 2022;
Published: 25 November 2022.
Edited by:
Sebastian Boltana, University of Concepcion, ChileReviewed by:
Kailasam Muniyandi, Central Institute of Brackishwater Aquaculture (ICAR), IndiaGustavo Núñez-Acuña, University of Concepcion, Chile
Copyright © 2022 Li, Guo, Ndandala, Chen, Huang, Zhao, Huang, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huapu Chen, Y2hwc3lzdUBob3RtYWlsLmNvbQ==; Hai Huang, SHVhbmdoYWk3NEAxMjYuY29t
 Zhiyuan Li
Zhiyuan Li Yuwen Guo1,2
Yuwen Guo1,2 Hai Huang
Hai Huang Huapu Chen
Huapu Chen