
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 20 October 2022
Sec. Systems Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.989305
This article is part of the Research Topic Genetic Variants and Metabolic Diseases View all 10 articles
Histone deacetylase 11 (HDAC11) is the only member of the class IV HDAC, and the latest member identified. It is highly expressed in brain, heart, kidney and some other organs, and located in mitochondria, cytoplasm and nuclei, depending on the tissue and cell types. Although studies in HDAC11 total knockout mice suggest its dispensable features for tissue development and life, it participates in diverse pathophysiological processes, such as DNA replication, tumor growth, immune regulation, oxidant stress injury and neurological function of cocaine. Recent studies have shown that HDAC11 is also critically involved in the pathogenesis of some metabolic diseases, including obesity, diabetes and complications of diabetes. In this review, we summarize the recent progress on the role and mechanism of HDAC11 in the regulation of metabolic disorders, with the focus on its regulation on adipogenesis, lipid metabolism, metabolic inflammation, glucose tolerance, immune responses and energy consumption. We also discuss the property and selectivity of HDAC11 inhibitors and their applications in a variety of in vitro and in vivo models of metabolic disorders. Given that pharmacological and genetic inhibition of HDAC11 exerts a beneficial effect on various metabolic disorders, HDAC11 may be a potential therapeutic target to treat chronic metabolic diseases.
The removal of acetyl groups from e-lysine residues in proteins (1) connected to condensed chromatin structures that inhibit gene transcription (2) is catalyzed by a class of enzymes called histone deacetylases (HDACs). Mammals currently contain 18 HDACs that are classified into two families: the Zn2+- dependent or classical HDACs, and the nicotinamide adenine dinucleotide (NAD+)-dependent HDACs or sirtuins (SIRT). According to the homology of their catalytic domains, classical HDACs are further split into three classes: class I, class II, and class IV HDACs. Class I HDACs include HDAC1, HDAC2, HDAC3, and HDAC8, whereas class II HDACs include HDAC4, HDAC5, HDAC6, HDAC8, HDAC9, and HDAC10, and class IV HDACs include HDAC11 (1).
HDAC11, the solitary member of class IV HDAC, contains an open reading frame encoding a 347-residue protein and shares sequence homology with both class I and class II HDAC proteins in the catalytic core regions. HDAC11 is highly conserved, even in invertebrates and plants as the most recently identified (3–5) and combines with other HDACs to form functional complexes (6–8). Although HDAC11 structure has still not been discovered, it has been effectively modeled from HDAC8 structure (4, 9). HDAC11 can be degraded by the proteasome system and has an unstable half-life at around four hours (10). While most class I-III HDACs are involved in deacetylating their substrates (reviewed in (11)), HDAC11 has defattyacylase activity in addition to its deacetylase activity. In fact, as the only HDAC member that has a clear predilection for the removal of long-acyl instead of acetyl groups (12, 13), HDAC11 is the family’s most effective fatty deacetylase (9). It has been reported that the efficiency of HDAC11 defattyacylase activity is greater than 10,000 times its deacetylase activity (13). The activation of HDAC11 can be triggered by physiologic levels of free fatty acids and their metabolites (9).
HDAC11 is expressed in multiple organs and distributed in diverse organelles. It is primarily expressed in heart, kidney, smooth muscle (3), skeletal muscle (14–16), brain (3, 15, 17–20), testis (14, 21) and gall bladder (22). At cellular level, HDAC11 is abundant in the mitochondria of skeletal muscle cells, brain synapses (15), and the centrosomes of neurons from the dentate gyrus (19). But it locates predominantly in the cytoplasm of embryonic astrocyte precursors and mature cells (23), and the nucleus of activated astrocytes and oligodendrocytes (24). HDAC11 can be found both in the cytoplasm and the nucleus of newly isolated and unstimulated Treg cells (24), immature astrocytes and oligodendrocytes (23), retinal ganglion cells (25) and preadipocytes (26). In addition, HDAC11 is highly expressed in the rat brain, and pancreatic β cells (27).
Emerging evidence has indicated that HDAC11 is critically involved in physiological and pathological processes. HDAC11 has a variety of physiological functions, including immunomodulation (24, 28–36), genomic stability (21, 37–39), cell cycle progression (21, 40, 41), and nervous system development (42). Pathologically, HDAC11 plays a role in epithelial barrier dysfunction (43–45) and ischemic injury (46–48) and requried for the growth of several tumors (49–56), such as hepatic carcinoma (57–61), and lung cancer (62, 63). Moreover, it contributes to the development of some other diseases (56, 64), including hepatitis B (65–67) and age-related macular degeneration (68).
In the past two decades, HDACs have been revealed to be implicated in the regulation of multiple metabolic processes and pathogenesis of some metabolic disorders. For example, most class I HDAC members are associated with insulin resistance, energy metabolism and glucose homeostasis, and contribute to the pathogenesis of diabetes and its associated complications (69, 70), and obesity (71). Class II HDACs are required for regulating the transcription of genes associated with glucose homeostasis and hepatic gluconeogenesis (72). Moreover, HDACs are involved in the regulation of several events related to the pathogenesis of diabetes (i.e. oxidative stress, inflammation and fibrosis) and its associated complications (70, 73). Very recently, HDAC11 has been linked to the pathogenesis of obesity (74), diabetes, and diabetic complications (64, 75). Given that global deletion of HDAC11 in mice does not affect their development and health (24), pharmacological inhibition of HDAC11 could be a potential therapeutic approach for the treatment of metabolic disorders. In this review article, we summarize the role and possible mechanisms of HDAC11 in metabolic disorders, including obesity, metabolic inflammation, and diabetes and its complications, and provide detailed information about HDAC11 inhibitors developed so far.
Obesity is an excessive fat gain due to unbalanced energy intake and consumption (76), and its prevalence rises yearly in children and adults (77). HDAC11 is related to obesity in multiple ways.
HDAC11 participates in the regulation of adipogenesis. The differentiation of adipocytes is strictly controlled. Mature adipocytes are differentiated from mesenchymal precursor cells. Several essential adipose transcription factors, such as peroxisome proliferator‐activated receptor γ (PPARγ), CCAAT‐enhancer-binding protein β, and sterol regulatory element‐binding proteins regulate this process (78–80). It has also been reported that various HDACs, in particular, HDAC11 are critically involved in adipogenic differentiation (81–83). Silencing the HDAC11 gene by small interfering RNA results in reduced perilipin, adipoq, and PPARγ2 expression, and decreased formation of intracellular lipid droplets (84). By the use of HDAC11-KO mice and adipocytes from WT and HDAC11 KO mice exposed to FT895, it was also found that HDAC11 binds to a nearby gravin-α region and demyristoylates those spots. Gravin-α lysine myristoylation in brown and white adipocytes is necessary for the signal through β2- and β3-adrenergic receptors (β-ARs). Gravin-α lysine myristoylation induces the expression of protective thermogenic genes by directing β-ARs to lipid raft membrane microdomains and stimulating activation of PKA and its downstream signaling. These results establish reversible lysine myristoylation as a pattern of GPCR signaling regulation and emphasize the importance of HDAC11in regulating adipocyte phenotypes (85).
HDAC11 is essential for regulating the balance of brown adipose tissue (BAT) and white adipose tissue (WAT) (86). The WAT is the body’s greatest energy storage tissue, and can secretes cytokines and adipokines as part of its endocrine function; BAT is imperative in maintaining body temperature in newborns’ nonshivering thermogenesis (87–90). A role for HDAC11 in regulating adipose tissue and thermogenic capability has been suggested by the fact that HDAC11 is more expressed in WAT than BAT and that deletion of HDAC11 in mice enhances the development of BAT and “browning” of WAT (26). These are essential changes as WAT contributes to obesity by storing extra energy as fat in the body, while BAT is capable of turning fat into energy (90). Meanwhile, In HDAC11‐knockout (KO) mice, the histological study of BAT reveals a compacted tissue size with noticeably smaller lipid droplets (75). Mouse hepatic cell line AML12 with HDAC11 knockdown exhibits enhanced metabolic activity and oxygen consumption due to improved lipid oxidation capability (75). which is consistent with previous observations in skeletal muscle tissue (14).
Mechanistically, uncoupling protein 1 (UCP1), a mitochondrial long-chain fatty acid/H+ symporter, and PGC1-α, a primary regulator of mitochondrial biogenesis, are both downregulated by HDAC11 to inhibit the BAT transcriptional program (26). HDAC11 deletion increases metabolic pool clearance, thermogenic capability, UCP1 expression in BAT, and energy expenditure. Through its physical interaction with BRD2(an enhancer regulating Ucp1 gene) (26), HDAC11 inhibits the thermogenic gene program. HDAC11 inhibition increases oxygen consumption and boosts adiponectin, a hormone that controls fatty acid oxidation, blood glucose levels, and stimulates lipid metabolism by activating the adiponectin-AdipoR-AMPK pathway (75).
Recent studies have also shown that HDAC11 is a critical regulator of the body’s overall metabolism. HDAC11 KO mice exhibit higher body temperatures than wild type (WT) controls both at room temperature (22°C) and during a 24-hour cold challenge (4°C), which is correlated with higher metabolic rate and oxygen consumption (26, 75). Importantly, HDAC11-deficient mice show alleviated hypercholesterolemia, hepatic steatosis and liver damage (26, 75).
Altogether, these results suggests that HDAC11 is a new metabolic regulator, lowering its levels might improve cells’ ability to adapt to an elevated energy requirement under stressful circumstances. Furthermore, as a result of the considerable rise in metabolic rate and oxygen consumption caused by HDAC11 inhibition, there is an increase in lipid oxidation and energy expenditure. Therefore, HDAC11 would be a prospective therapeutic target for obesity and the related metabolic effects.
Diabetes is a metabolic, chronic, multisystem disease and chronic exposure to hyperglycemia eventually leads to multiple complications, such as diabetic nephropathy, cardiovascular disease, retinopathy and neuropathy with considerable impact on the quality of life and overall life expectancy.
HDAC11 is essential for preserving insulin sensitivity and glucose homeostasis. In mice fed with high fat diet (HFD), HDAC11 deletion significantly decreases blood insulin levels, stabilizes blood glucose, and greatly reduces blood glucose levels after insulin challenge, thereby enhancing glucose tolerance and ameliorating diabetes (75). In addition, adiponectin significantly increases in HDAC11 KO mice (91). By uisng adiponectin-knockout mice fed on a HFD or either regular chow, it has been demonstrated that adipoR2-peroxisome proliferator-activated receptor α (PPARα) and adipoR1-AMP-activated protein kinase (AMPK) pathways play a major role in adiponectin signaling in the liver (91). The vital energy sensor AMPK has been linked to the control of the hepatic metabolic processes, such as gluconeogenesis. Increased energy expenditure, improved glucose tolerance, and lower plasma cholesterol levels all result from AdipoR2 KO. Lysophospholipids are one of adiponectin’s targets, and they are upregulated by a high-fat diet (HFD) and tend to cause hypertriglyceridemia, decreased glucose tolerance, and insulin resistance (91).
Diabetic nephropathy (DN) is a serious complication of diabetes. It presents as localized kidney inflammation and fibrosis that lead to structural remodeling (92–94).
Although there is no report about the role of HDAC11 in DN so far, HDAC11 is vital in the response to renal inflammation and fibrosis. Plasminogen agonist inhibitor type 1, a physiological inhibitor of fibrinolysis (PAI‐1), is evelvated in DN (95). Excess PAI‐1 lead to the accumulation of extracellular matrix proteins, whereas PAI‐1 deficiency protected the kidney from injury-induced fibrosis (96). In a murine model of renal ischemia/reperfusion (I/R), increased testosterone can decrease the ability of HDAC11 to bind to PAI-1 promoter, leading to increased histone 3 acetylation and PAI-1 expression and accelerated I/R-induced renal injury (46, 47). Moreover, HDAC11 expression are increased in the kidneys in animal models of renal fibrosis induced by unilateral ureteral obstruction and angiotensin II by suppressing Kruppel-like factor 15, an anti-fibrogenic factor (97). Since renal inflammation and fibrosis contribute to the pathogenesis of DN, it is speculated that HDAC11 would also play a role in the development of DN. Further studies are needed to address this issue.
Type 2 diabetes and cardiovascular diseases are predisposed to by obesity (98). Increased body weight can, in fact, cause metabolic changes in cardiomyocytes that switch them from processing fatty acids to sugar, which adds to lipid storage in the pericardium and, as a severe consequence of type 2 diabetes, causes myocardial infarction (99). Interestingly, inhibition of HDAC11 activity could prevent or ameliorate diabetic cardiomyopathy. In apoE mice fed with HFD, atherosclerosis and blood lipid levels have recently been shown to be alleviated by HDAC11-AS1. HDAC11-AS1 improves lipoprotein lipase (LPL), a crucial rate-limiting enzyme involved in triglyceride (TG) hydrolysis, via controlling adropin histone deacetylation both in vitro and in vivo (100). Another study shows that suppression of HDAC11 enhances the prevention of pyroptosis in human umbilical vein endothelial cells (HUVECs) triggered by TNF-α, indicating that vascular endothelial pyroptosis might be prevented through downregulation of HDAC11 related signaling pathways in atherosclerosis (AS) (101). In addition, a fructose injury-induced mouse model of diabetic heart failure that lacks HDAC11 had lower levels of apoptosis, dyslipidemia, inflammation, and oxidative stress (102). HDAC11 has also been suggested to be an essential regulator in heart failure (103). Therefore, HDAC11 contributes to the pathogenesis of diabetic Cardiopathy.
Metabolic disorders are closely associated with chronic mild inflammation (104–106). Most obese people develop inflammation in their adipose tissue, like chronically damaged tissue, along with immune cell remodeling and infiltration. During the early phases of adipose swelling and the progression of chronic obesity, inflammation is induced, and the immune system is irreversibly changed into a proinflammatory phenotype (107). Changes in adipose tissue function are related to obesity, and the loss of adipocytes also contributes to chronic mild inflammation (104). The regulating function of HDAC11 in metabolic inflammation is crucial.
HDAC11 regulates metabolic inflammation primarily through the control of the IL-10 released by antigen-presenting cells (APCs) (28). Inhibition of HDAC11 causes macrophages to express more IL-10, whereas overexpression of HDAC11 reduces IL-10 expression (108, 109). In addtion, HDAC11 overexpression in APCs is efficient in reactivating tolerant T cell responses and CD4+ T cells specific for antigens. And APC had the reverse result when HDAC11 expression was absent (33). Conversely, suppression of HDAC11 resulted in impaired antigen-specific expression, increased IL-10 expression, downregulated IL-12 expression and immune cell expression (such as myeloid-derived suppressor cells, neutrophils, and T cells), leading to immune tolerance (110, 111). In addition, muted HDAC11 transcripts boosted the synethesis of IL-17 and TNF in the supernatants of HL cells (112). Moreover, liver immune tolerance is regulated by HDAC11 through TNF‐α, interferon‐γ, IL‐2, and IL‐4 (80, 90, 113–120).
Most HDACi are pan-HDACi that target multiple HDACs with different nanomolar potency. Zinc-dependent catalytic processes are shared by Classes I, II, and IV HDACs. Many pan-HDACi have been synthesized, including Aes-135 (121), AR-42, belinostat (PXD101,PX105684), fimepinostat (CUDC-907) (9), FT895 (122), M344(D237, MS344), Panobinostat (LBH589, NVP-LBH589), pracinostat (SB939), dacinostat, quisinostat (JNJ-26481585), trichostatin A (TSA), vorinostat (34) (SAHA, MK0683), mocetinostat (123)(MGCD0103), tucidinostat (Chimade, HBI-8000, CS055), trapoxin A (124)(TpxA, C34H42N4O6), garcinol (125), romidepsin (126). Recently, Compound 8, a newly designed novel HDAC6 selective inhibitor with 2-mercaptoquinazolinone as the Cap Moiety, has displayed stronger inhibition activity against HDAC11 than Belinostat (127). The toxicity caused by general inhibition of HDACs restricts their potential utility. Among the pan-HDAC inhibitors, garcinol shows more HDAC11 selectivity and efficiency than other HDACs (125). The deacetylase and demyristoylase activities of HDAC11 are also suggested to be effectively inhibited by Fimepinostat (9). At concentrations of 0.02, 0.2, and 2μM, respectively, Suberoylanilide hydroxamic acid (SAHA) could suppress 10, 50, and 90% of HDAC11 activity (34). Additionally, it has been noted that trichostatin A (TSA) and romidepsin have a nanomolar potency toward HDAC11 (126). However, pracinostat, dacinostat, mocetinostat, quisinostat, trapoxin A, and trichostatin A have been found not as efficient in inhibiting HDAC11 deacetylation activity as reported before (9). Unexpectedly, butyrate, valproate, SAHA, and TSA could trigger myeloid cells to express HDAC11 (128). And low doses of MS275 have been found to show agonistic actions (129).
Elevenostat (JB3-22) (21, 24), SIS7, SIS17 (130), and FT895 (122) are selective HDAC11 inhibitors. Nevertheless, the inhibitory capacity of elevenostat (JB3-22) on myristoylated and acetylated peptidic derivatives is extremely poor (9). To date, the HDAC11 inhibitors that are considered to be the most potent and selective are SIS17 and FT895. SIS17 is better than FT895 and SIS7 in terms of its cell permeability and metabolic stability (60), while FT895, SIS17, SIS7 can all inhibit HDAC11’s demyristoylase activity (130).
Though several HDAC11 inhibitors have been developed, only FT895 (85, 122), romidepsin (131), and quisinostat (97, 131, 132) have been reported to be utilized in animal studies. To explore the pharmacokinetic properties of FT895, it was injected to male Balb/c nude mice via i.v. at 1mg/kg) or i.p. at 5 mg/kg. After i.v. dosing, with a t1/2 of 9.4 hours, FT895 exhibits a high volume of distribution and a moderate clearance (42 mL/min/kg). In comparison, FT895 dosed intravenously has enhanced exposure, a similar half-life (10.2 h), a bioavailability of 81%, and sustains free drug levels above the cellular half-maximal inhibitory concentration (IC50) for up to 4 hours (122). Quisinostat (10 mg/kg Monday, Wednesday, and Friday) and romidepsin (0.3 mg/kg, 1 mg/kg, or 3 mg/kg Monday, Friday) were administered intraperitoneally (i.p.) for one week to tumor-bearing athymic NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. Romidepsin has unacceptable toxicity at 3 mg/kg; anemia and aspartate aminotransferase elevations are a result of 1 mg/kg dosing; Without causing considerable weight loss (>20%) or neurotoxicity, both 0.3 mg/kg and 1 mg/kg are tolerated. Treatment with quisinostat (10 mg/kg Monday, Wednesday, Friday) shows no systemic toxicity (131). Similarly, in BALB/c nude mice and NOD/SCID mice, quisinostat (3 and 10 mg/kg/day, i.p) has been used to treat tongue and esophageal squamous cell carcinoma (132, 133) and malaria (134). Romidepsin is also used in C57BL/6 (0.03mg/kg twice a week) (135, 136) and BALB/C(1mg/kg/2days) (137) mice for cancer treatment. Therefore, FT895, quisinostat and romidepsin are tolerable and safe in vivo.
In addition, quisinostat and romidepsin have been tested in clinical trials. The maximum tolerable dose of quisinostat for the treatment of cancer in patients is 10 mg administered orally three times per week along with bortezomib and dexamethasone, with median progression-free survival (PFS) 8.2 months and median duration of response 9.4 months (138). Combined with 5-azacytidine (AZA), romidepsin (14 mg/m2, day 8,15,22, per 35 days, IV) is used to treat peripheral T-cell lymphomas (PTCL) with the overall survival not met (at a median follow-up of 13.5 months), and the median progression-free survival (PFS) 8.0 months, duration of response 20.3 months (139). Romidepsin has also been reported to treat HIV-1-infected patients with a 5 mg/m2 dosage as a 4 hour infusion (140).
Thus, taking effectiveness, selectivity, toxicity, half-life, tolerance and survivability in vivo into consideration, FT895 exhibits pharmacokinetic properties that are reasonable in vivo research, and the most significant potential to advance into clinical trials.
The incidence of metabolic disorders is increasing worldwide, ranging from obesity to type 2 diabetes, leading to complications in the heart, kidney, retina, bone, skin and foot. HDAC11 participates in many aspects of metabolic diseases. HDAC11 mediates obesity and metabolic syndrome by regulating adipogenesis, increasing energy consumption and promoting lipid metabolism. It also contributes to adipose tissue inflammation by regulating immune responses and insulin resistance. HDAC11 was shown to have inhibitory roles in the development of diabetic cardiovascular disease. (Figure 1) In addition, a recent study shows that HDAC11 contributes to osteoporosis susceptibility and reduced peak bone mass through a mechanism of 11β-HSD2’s low-functional programming. This is triggered by corticosterone through GR/HDAC11 signaling, which amplifies the effect of corticosterone on inhibiting the function of BMSCs in osteogenesis (141, 142). However, studies on diabetic osteoporosis, lipoid nephrosis, fatty liver disease and obesity cardiomyopathy are still lacking. As such, further research on the effect of HDAC11 on metabolic diseases is required.
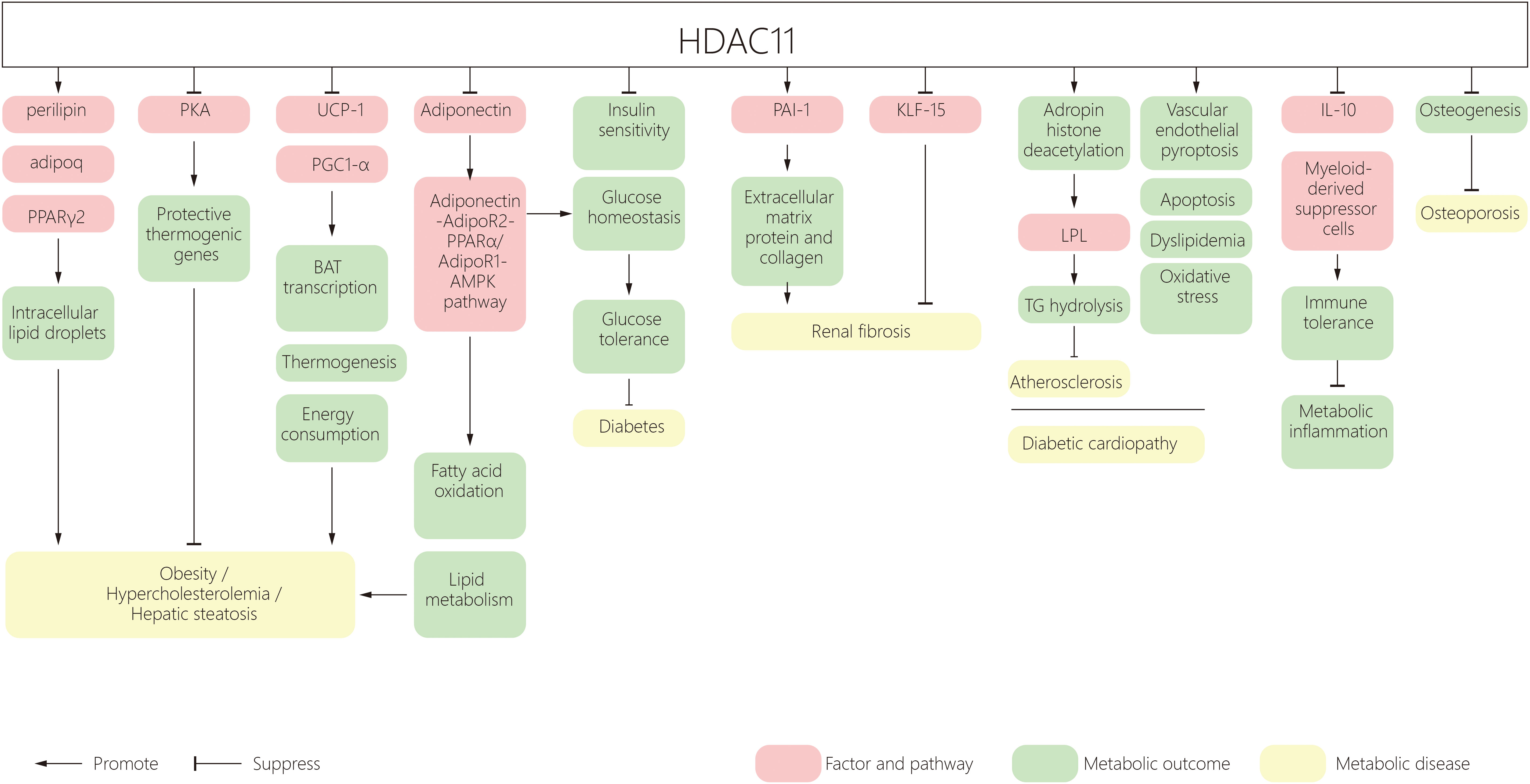
Figure 1 A schematic diagram of HDAC11 and its effect on metabolic disorders. HDAC11, Histone deacetylase 11; PPAR, peroxisome proliferator‐activated receptor; BAT, brown adipose tissue; UCP1, uncoupling protein 1; AMPK, AMP-activated protein kinase; PAI‐1, Plasminogen agonist inhibitor type 1; AP-2α, activator protein 2α; KLF15, Kruppel-like factor 15; LPL, Lipoprotein lipase; TG, triglyceride.
Studies listed in Table 1 have shown the well-tolerated HDAC11 global deletion in mice, suggesting that its inhibition or depletion is without apparent side effects. Currently, toxicity and safe doses of HDAC11 inhibitors are far from clear and none of the inhibitors have been used in patients with metabolic disorders. Thus, more studies on the safe dosage and toxicity of HDAC11 inhibitors in animal models are needed before advancing them to human clinical trials. In addition, development of more effective HDAC11 inhibitors with enhanced selectivity is worth investigating.
HC drafted the article. The manuscript was edited by SZ, CX, and QC. After reviewing the manuscript, all authors gave their approval for publication.
This study is supported by US National Institutes of Health (2R01DK08506505A1 to SZ), the National Natural Science Foundation of China grants (81670623 and 81830021 to SZ), and National key R&D Program of China (2018YFA0108802 to SZ).
We are grateful to Dr. George Bayliss from Brown University for editing this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
HDAC11, Histone deacetylase 11; HDACs, Histone deacetylases; NAD+, nicotinamide adenine dinucleotide; SIRT, sirtuins; AMD, age-related macular degeneration; MS, multiple sclerosis; PPAR, peroxisome proliferator‐activated receptor; β-ARs, β-adrenergic receptors; WAT, white adipose tissue; BAT, brown adipose tissue; UCP1, uncoupling protein 1; WT, wild type; KO, knockout; AMPK, AMP-activated protein kinase; HFD, high fat diet; DN, Diabetic nephropathy; PAI‐1, Plasminogen agonist inhibitor type 1; AP-2α, activator protein 2α; KLF15, Kruppel-like factor 15; LPL, Lipoprotein lipase; TG, triglyceride; HUVECs, human umbilical vein endothelial cells; AS, atherosclerosis; APCs, antigen‐presenting cells; TSA, trichostatin A; TpxA, trapoxin A; SAHA, Suberoylanilide hydroxamic acid; i.p., intraperitoneal; IC50, half maximal inhibitory concentration; PFS, progression-free survival; AZA, 5-azacytidine; PTCL, Peripheral T-cell lymphomas; PFS, progression-free survival.
1. Seto E, Yoshida M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harbor Perspect Biol (2014) 6(4):a018713. doi: 10.1101/cshperspect.a018713
2. Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem (2007) 76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114
3. Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of Hdac11, a novel member of the human histone deacetylase family. J Biol Chem (2002) 277(28):25748–55. doi: 10.1074/jbc.M111871200
4. Thangapandian S, John S, Lee Y, Arulalapperumal V, Lee KW. Molecular modeling study on tunnel behavior in different histone deacetylase isoforms. PLos One (2012) 7(11):e49327. doi: 10.1371/journal.pone.0049327
5. Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat Rev Mol Cell Biol (2008) 9(3):206–18. doi: 10.1038/nrm2346
6. Bae J, Hideshima T, Tai YT, Song Y, Richardson P, Raje N, et al. Histone deacetylase (Hdac) inhibitor Acy241 enhances anti-tumor activities of antigen-specific central memory cytotoxic T lymphocytes against multiple myeloma and solid tumors. Leukemia (2018) 32(9):1932–47. doi: 10.1038/s41375-018-0062-8
7. Cheng F, Lienlaf M, Perez-Villarroel P, Wang HW, Lee C, Woan K, et al. Divergent roles of histone deacetylase 6 (Hdac6) and histone deacetylase 11 (Hdac11) on the transcriptional regulation of Il10 in antigen presenting cells. Mol Immunol (2014) 60(1):44–53. doi: 10.1016/j.molimm.2014.02.019
8. Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, et al. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol (2013) 9:672. doi: 10.1038/msb.2013.26
9. Kutil Z, Novakova Z, Meleshin M, Mikesova J, Schutkowski M, Barinka C. Histone deacetylase 11 is a fatty-acid deacylase. ACS Chem Biol (2018) 13(3):685–93. doi: 10.1021/acschembio.7b00942
10. Long C, Lai Y, Li T, Nyunoya T, Zou C. Cigarette smoke extract modulates pseudomonas aeruginosa bacterial load Via Usp25/Hdac11 axis in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol (2020) 318(2):L252–l63. doi: 10.1152/ajplung.00142.2019
11. Barnes CE, English DM, Cowley SM. Acetylation & Co: An expanding repertoire of histone acylations regulates chromatin and transcription. Essays Biochem (2019) 63(1):97–107. doi: 10.1042/ebc20180061
12. Moreno-Yruela C, Galleano I, Madsen AS, Olsen CA. Histone deacetylase 11 is an Ε-N-Myristoyllysine hydrolase. Cell Chem Biol (2018) 25(7):849–56.e8. doi: 10.1016/j.chembiol.2018.04.007
13. Cao J, Sun L, Aramsangtienchai P, Spiegelman NA, Zhang X, Huang W, et al. Hdac11 regulates type I interferon signaling through defatty-acylation of Shmt2. Proc Natl Acad Sci USA (2019) 116(12):5487–92. doi: 10.1073/pnas.1815365116
14. Hurtado E, Núñez-Álvarez Y, Muñoz M, Gutiérrez-Caballero C, Casas J, Pendás AM, et al. Hdac11 is a novel regulator of fatty acid oxidative metabolism in skeletal muscle. FEBS J (2021) 288(3):902–19. doi: 10.1111/febs.15456
15. Bryant DT, Landles C, Papadopoulou AS, Benjamin AC, Duckworth JK, Rosahl T, et al. Disruption to schizophrenia-associated gene Fez1 in the hippocampus of Hdac11 knockout mice. Sci Rep (2017) 7(1):11900. doi: 10.1038/s41598-017-11630-1
16. Sun L, Telles E, Karl M, Cheng F, Luetteke N, Sotomayor EM, et al. Loss of Hdac11 ameliorates clinical symptoms in a multiple sclerosis mouse model. Life Sci alliance (2018) 1(5):e201800039. doi: 10.26508/lsa.201800039
17. Liu H, Hu Q, D'Ercole AJ, Ye P. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in ol-1 oligodendroglia cells. Glia (2009) 57(1):1–12. doi: 10.1002/glia.20729
18. Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1-11 in the rat brain. J Mol Neuroscience: MN (2007) 31(1):47–58. doi: 10.1007/bf02686117
19. Watanabe Y, Khodosevich K, Monyer H. Dendrite development regulated by the schizophrenia-associated gene Fez1 involves the ubiquitin proteasome system. Cell Rep (2014) 7(2):552–64. doi: 10.1016/j.celrep.2014.03.022
20. Liu H, Hu Q, Kaufman A, D'Ercole AJ, Ye P. Developmental expression of histone deacetylase 11 in the murine brain. J Neurosci Res (2008) 86(3):537–43. doi: 10.1002/jnr.21521
21. Sui L, Zhang S, Huang R, Li Z. Hdac11 promotes meiotic apparatus assembly during mouse oocyte maturation Via decreasing H4k16 and Α-tubulin acetylation. Cell Cycle (Georgetown Tex) (2020) 19(3):354–62. doi: 10.1080/15384101.2019.1711315
22. Boltz TA, Khuri S, Wuchty S. Promoter conservation in hdacs points to functional implications. BMC Genomics (2019) 20(1):613. doi: 10.1186/s12864-019-5973-x
23. Tiwari S, Dharmarajan S, Shivanna M, Otteson DC, Belecky-Adams TL. Histone deacetylase expression patterns in developing murine optic nerve. BMC Dev Biol (2014) 14:30. doi: 10.1186/1471-213x-14-30
24. Huang J, Wang L, Dahiya S, Beier UH, Han R, Samanta A, et al. Histone/Protein deacetylase 11 targeting promotes Foxp3+ treg function. Sci Rep (2017) 7(1):8626. doi: 10.1038/s41598-017-09211-3
25. Schlüter A, Aksan B, Fioravanti R, Valente S, Mai A, Mauceri D. Histone deacetylases contribute to excitotoxicity-triggered degeneration of retinal ganglion cells in vivo. Mol Neurobiol (2019) 56(12):8018–34. doi: 10.1007/s12035-019-01658-x
26. Bagchi RA, Ferguson BS, Stratton MS, Hu T, Cavasin MA, Sun L, et al. Hdac11 suppresses the thermogenic program of adipose tissue Via Brd2. JCI Insight (2018) 3(15):120159. doi: 10.1172/jci.insight.120159
27. Lundh M, Christensen DP, Rasmussen DN, Mascagni P, Dinarello CA, Billestrup N, et al. Lysine deacetylases are produced in pancreatic beta cells and are differentially regulated by proinflammatory cytokines. Diabetologia (2010) 53(12):2569–78. doi: 10.1007/s00125-010-1892-8
28. Yanginlar C, Logie C. Hdac11 is a regulator of diverse immune functions. Biochim Biophys Acta Gene Regul Mech (2018) 1861(1):54–9. doi: 10.1016/j.bbagrm.2017.12.002
29. Sahakian E, Chen J, Powers JJ, Chen X, Maharaj K, Deng SL, et al. Essential role for histone deacetylase 11 (Hdac11) in neutrophil biology. J leukoc Biol (2017) 102(2):475–86. doi: 10.1189/jlb.1A0415-176RRR
30. Zhang HP, Wang L, Fu JJ, Fan T, Wang ZL, Wang G. Association between histone hyperacetylation status in memory T lymphocytes and allergen-induced eosinophilic airway inflammation. Respirology (Carlton Vic) (2016) 21(5):850–7. doi: 10.1111/resp.12774
31. Yang H, Yang J, Cheng H, Cao H, Tang S, Wang Q, et al. Probiotics ingestion prevents Hdac11-induced Dec205+ dendritic cell dysfunction in night shift nurses. Sci Rep (2019) 9(1):18002. doi: 10.1038/s41598-019-54558-4
32. Yang H, Yang LT, Liu J, Tang S, Zhao X, Wang Q, et al. Circadian protein clk suppresses transforming growth factor-Β expression in peripheral b cells of nurses with day-night shift rotation. Am J Trans Res (2018) 10(12):4331–7.
33. Villagra A, Cheng F, Wang HW, Suarez I, Glozak M, Maurin M, et al. The histone deacetylase Hdac11 regulates the expression of interleukin 10 and immune tolerance. Nat Immunol (2009) 10(1):92–100. doi: 10.1038/ni.1673
34. Woods DM, Woan KV, Cheng F, Sodré AL, Wang D, Wu Y, et al. T Cells lacking Hdac11 have increased effector functions and mediate enhanced alloreactivity in a murine model. Blood (2017) 130(2):146–55. doi: 10.1182/blood-2016-08-731505
35. Chen J, Cheng F, Sahakian E, Powers J, Wang Z, Tao J, et al. Hdac11 regulates expression of C/Ebpβ and immunosuppressive molecules in myeloid-derived suppressor cells. J leukoc Biol (2021) 109(5):891–900. doi: 10.1002/jlb.1a1119-606rrr
36. Heim CE, Bosch ME, Yamada KJ, Aldrich AL, Chaudhari SS, Klinkebiel D, et al. Lactate production by staphylococcus aureus biofilm inhibits Hdac11 to reprogramme the host immune response during persistent infection. Nat Microbiol (2020) 5(10):1271–84. doi: 10.1038/s41564-020-0756-3
37. Glozak MA, Seto E. Acetylation/Deacetylation modulates the stability of DNA replication licensing factor Cdt1. J Biol Chem (2009) 284(17):11446–53. doi: 10.1074/jbc.M809394200
38. Toropainen S, Väisänen S, Heikkinen S, Carlberg C. The down-regulation of the human myc gene by the nuclear hormone 1alpha,25-dihydroxyvitamin D3 is associated with cycling of corepressors and histone deacetylases. J Mol Biol (2010) 400(3):284–94. doi: 10.1016/j.jmb.2010.05.031
39. Bagui TK, Sharma SS, Ma L, Pledger WJ. Proliferative status regulates Hdac11 mrna abundance in nontransformed fibroblasts. Cell Cycle (Georgetown Tex) (2013) 12(21):3433–41. doi: 10.4161/cc.26433
40. Byun SK, An TH, Son MJ, Lee DS, Kang HS, Lee EW, et al. Hdac11 inhibits myoblast differentiation through repression of myod-dependent transcription. Molecules Cells (2017) 40(9):667–76. doi: 10.14348/molcells.2017.0116
41. Blixt NC, Faulkner BK, Astleford K, Lelich R, Schering J, Spencer E, et al. Class ii and iv hdacs function as inhibitors of osteoclast differentiation. PLos One (2017) 12(9):e0185441. doi: 10.1371/journal.pone.0185441
42. Yue L, Sharma V, Horvat NP, Akuffo AA, Beatty MS, Murdun C, et al. Hdac11 deficiency disrupts oncogene-induced hematopoiesis in myeloproliferative neoplasms. Blood (2020) 135(3):191–207. doi: 10.1182/blood.2019895326
43. Zhang R, Ge J. Proteinase-activated receptor-2 modulates ve-cadherin expression to affect human vascular endothelial barrier function. J Cell Biochem (2017) 118(12):4587–93. doi: 10.1002/jcb.26123
44. Zhou B, Zeng S, Li N, Yu L, Yang G, Yang Y, et al. Angiogenic factor with G patch and fha domains 1 is a novel regulator of vascular injury. Arteriosclerosis thrombosis Vasc Biol (2017) 37(4):675–84. doi: 10.1161/atvbaha.117.308992
45. Liu FH, Li SS, Li XX, Wang S, Li MG, Guan L, et al. Vitamin D3 induces vitamin d receptor and Hdac11 binding to relieve the promoter of the tight junction proteins. Oncotarget (2017) 8(35):58781–9. doi: 10.18632/oncotarget.17692
46. Mrug M, Sanders PW. Beware the low Hdac11: Males at risk for ischemic kidney injury. Am J Physiol Renal Physiol (2013) 305(7):F973–4. doi: 10.1152/ajprenal.00308.2013
47. Kim JI, Jung KJ, Jang HS, Park KM. Gender-specific role of Hdac11 in kidney ischemia- and reperfusion-induced pai-1 expression and injury. Am J Physiol Renal Physiol (2013) 305(1):F61–70. doi: 10.1152/ajprenal.00015.2013
48. Sillesen M, Bambakidis T, Dekker SE, Fabricius R, Svenningsen P, Bruhn PJ, et al. Histone deactylase gene expression profiles are associated with outcomes in blunt trauma patients. J Trauma acute Care Surg (2016) 80(1):26–32. doi: 10.1097/ta.0000000000000896
49. Klieser E, Urbas R, Stättner S, Primavesi F, Jäger T, Dinnewitzer A, et al. Comprehensive immunohistochemical analysis of histone deacetylases in pancreatic neuroendocrine tumors: Hdac5 as a predictor of poor clinical outcome. Hum Pathol (2017) 65:41–52. doi: 10.1016/j.humpath.2017.02.009
50. Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, et al. Ythdf1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun (2019) 10(1):4892. doi: 10.1038/s41467-019-12801-6
51. Leslie PL, Chao YL, Tsai YH, Ghosh SK, Porrello A, Van Swearingen AED, et al. Histone deacetylase 11 inhibition promotes breast cancer metastasis from lymph nodes. Nat Commun (2019) 10(1):4192. doi: 10.1038/s41467-019-12222-5
52. Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-the road to new treatment strategies. Lancet (London England) (2017) 389(10087):2430–42. doi: 10.1016/s0140-6736(16)32454-0
53. van Schaijik B, Davis PF, Wickremesekera AC, Tan ST, Itinteang T. Subcellular localisation of the stem cell markers Oct4, Sox2, nanog, Klf4 and c-myc in cancer: A review. J Clin Pathol (2018) 71(1):88–91. doi: 10.1136/jclinpath-2017-204815
54. Bora-Singhal N, Mohankumar D, Saha B, Colin CM, Lee JY, Martin MW, et al. Novel Hdac11 inhibitors suppress lung adenocarcinoma stem cell self-renewal and overcome drug resistance by suppressing Sox2. Sci Rep (2020) 10(1):4722. doi: 10.1038/s41598-020-61295-6
55. Dallavalle S, Musso L, Cincinelli R, Darwiche N, Gervasoni S, Vistoli G, et al. Antitumor activity of novel Pola1-Hdac11 dual inhibitors. Eur J medicinal Chem (2022) 228:113971. doi: 10.1016/j.ejmech.2021.113971
56. Liu SS, Wu F, Jin YM, Chang WQ, Xu TM. Hdac11: A rising star in epigenetics. Biomedicine pharmacother (2020) 131:110607. doi: 10.1016/j.biopha.2020.110607
57. Freese K, Seitz T, Dietrich P, Lee SML, Thasler WE, Bosserhoff A, et al. Histone deacetylase expressions in hepatocellular carcinoma and functional effects of histone deacetylase inhibitors on liver cancer cells in vitro. Cancers (2019) 11(10):1587. doi: 10.3390/cancers11101587
58. Gong D, Zeng Z, Yi F, Wu J. Inhibition of histone deacetylase 11 promotes human liver cancer cell apoptosis. Am J Trans Res (2019) 11(2):983–90.
59. Bi L, Ren Y, Feng M, Meng P, Wang Q, Chen W, et al. Hdac11 regulates glycolysis through the Lkb1/Ampk signaling pathway to maintain hepatocellular carcinoma stemness. Cancer Res (2021) 81(8):2015–28. doi: 10.1158/0008-5472.Can-20-3044
60. Wang W, Ding B, Lou W, Lin S. Promoter hypomethylation and mir-145-5p downregulation- mediated Hdac11 overexpression promotes sorafenib resistance and metastasis of hepatocellular carcinoma cells. Front Cell Dev Biol (2020) 8:724. doi: 10.3389/fcell.2020.00724
61. Fei Q, Song F, Jiang X, Hong H, Xu X, Jin Z, et al. Lncrna St8sia6-As1 promotes hepatocellular carcinoma cell proliferation and resistance to apoptosis by targeting mir-4656/Hdac11 axis. Cancer Cell Int (2020) 20:232. doi: 10.1186/s12935-020-01325-5
62. Kirchner M, Kluck K, Brandt R, Volckmar AL, Penzel R, Kazdal D, et al. The immune microenvironment in egfr- and Erbb2-mutated lung adenocarcinoma. ESMO Open (2021) 6(5):100253. doi: 10.1016/j.esmoop.2021.100253
63. Gimenez-Xavier P, Pros E, Bonastre E, Moran S, Aza A, Graña O, et al. Genomic and molecular screenings identify different mechanisms for acquired resistance to met inhibitors in lung cancer cells. Mol Cancer Ther (2017) 16(7):1366–76. doi: 10.1158/1535-7163.Mct-17-0104
64. Núñez-Álvarez Y, Suelves M. Hdac11: A multifaceted histone deacetylase with proficient fatty deacylase activity and its roles in physiological processes. FEBS J (2022) 289(10):2771–92. doi: 10.1111/febs.15895
65. Yuan Y, Zhao K, Yao Y, Liu C, Chen Y, Li J, et al. Hdac11 restricts hbv replication through epigenetic repression of cccdna transcription. Antiviral Res (2019) 172:104619. doi: 10.1016/j.antiviral.2019.104619
66. Cheng Y, Shi W, Cui X, Sun L, Nan Y, Yao H, et al. Long noncoding rna Tfap2a-As1 suppressed hepatitis b virus replication by modulating mir-933/Hdac11. Dis Markers (2022) 2022:7733390. doi: 10.1155/2022/7733390
67. Wu J, Niu Q, Yuan J, Xu X, Cao L. Lncrna-Cd160 decreases the immunity of Cd8 (+) T cells through epigenetic mechanisms in hepatitis b virus infection. Oncol Lett (2020) 20(1):235–47. doi: 10.3892/ol.2020.11534
68. Wang J, Zibetti C, Shang P, Sripathi SR, Zhang P, Cano M, et al. Atac-seq analysis reveals a widespread decrease of chromatin accessibility in age-related macular degeneration. Nat Commun (2018) 9(1):1364. doi: 10.1038/s41467-018-03856-y
69. Makkar R, Behl T, Arora S. Role of hdac inhibitors in diabetes mellitus. Curr Res Trans Med (2020) 68(2):45–50. doi: 10.1016/j.retram.2019.08.001
70. Dewanjee S, Vallamkondu J, Kalra RS, Chakraborty P, Gangopadhyay M, Sahu R, et al. The emerging role of hdacs: Pathology and therapeutic targets in diabetes mellitus. Cells (2021) 10(6):1340. doi: 10.3390/cells10061340
71. Bagchi RA, Weeks KL. Histone deacetylases in cardiovascular and metabolic diseases. J Mol Cell Cardiol (2019) 130:151–9. doi: 10.1016/j.yjmcc.2019.04.003
72. Zeng Z, Liao R, Yao Z, Zhou W, Ye P, Zheng X, et al. Three single nucleotide variants of the hdac gene are associated with type 2 diabetes mellitus in a Chinese population: A community-based case-control study. Gene (2014) 533(1):427–33. doi: 10.1016/j.gene.2013.09.123
73. Wang J, Li J, Zhang X, Zhang M, Hu X, Yin H. Molecular mechanisms of histone deacetylases and inhibitors in renal fibrosis progression. Front Mol Biosci (2022) 9:986405. doi: 10.3389/fmolb.2022.986405
74. Bhaskara S. Histone deacetylase 11 as a key regulator of metabolism and obesity. EBioMedicine (2018) 35:27–8. doi: 10.1016/j.ebiom.2018.08.008
75. Sun L, Marin de Evsikova C, Bian K, Achille A, Telles E, Pei H, et al. Programming and regulation of metabolic homeostasis by Hdac11. EBioMedicine (2018) 33:157–68. doi: 10.1016/j.ebiom.2018.06.025
76. Zhou YT, Wang ZW, Higa M, Newgard CB, Unger RH. (1999). Reversing adipocyte differentiation: Implications for treatment of obesity, in: Proceedings of the National Academy of Sciences of the United States of America 96:2391–5. doi: 10.1073/pnas.96.5.2391
77. Hales CM, Carroll MD, Fryar CD, Ogden CL. Prevalence of obesity among adults and youth: United states, 2015-2016. NCHS Data Brief (2017) 288):1–8.
78. James AW. Review of signaling pathways governing msc osteogenic and adipogenic differentiation. Scientifica (2013) 2013:684736. doi: 10.1155/2013/684736
79. Moseti D, Regassa A, Kim WK. Molecular regulation of adipogenesis and potential anti-adipogenic bioactive molecules. Int J Mol Sci (2016) 17(1):124. doi: 10.3390/ijms17010124
80. Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol (2006) 7(12):885–96. doi: 10.1038/nrm2066
81. Haberland M, Carrer M, Mokalled MH, Montgomery RL, Olson EN. Redundant control of adipogenesis by histone deacetylases 1 and 2. J Biol Chem (2010) 285(19):14663–70. doi: 10.1074/jbc.M109.081679
82. Yoo EJ, Chung JJ, Choe SS, Kim KH, Kim JB. Down-regulation of histone deacetylases stimulates adipocyte differentiation. J Biol Chem (2006) 281(10):6608–15. doi: 10.1074/jbc.M508982200
83. Zhou Y, Peng J, Jiang S. Role of histone acetyltransferases and histone deacetylases in adipocyte differentiation and adipogenesis. Eur J Cell Biol (2014) 93(4):170–7. doi: 10.1016/j.ejcb.2014.03.001
84. Yang H, Chen L, Sun Q, Yao F, Muhammad S, Sun C. The role of Hdac11 in obesity-related metabolic disorders: A critical review. J Cell Physiol (2021) 236(8):5582–91. doi: 10.1002/jcp.30286
85. Bagchi RA, Robinson EL, Hu T, Cao J, Hong JY, Tharp CA, et al. (2022). Reversible lysine fatty acylation of an anchoring protein mediates adipocyte adrenergic signaling, in: Proceedings of the National Academy of Sciences of the United States of America 119. doi: 10.1073/pnas.2119678119
86. Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: Tracking obesity to its source. Cell (2007) 131(2):242–56. doi: 10.1016/j.cell.2007.10.004
87. Harms M, Seale P. Brown and beige fat: Development, function and therapeutic potential. Nat Med (2013) 19(10):1252–63. doi: 10.1038/nm.3361
88. Kuhn E, Binart N, Lombès M. [Brown, white, beige: The color of fat and new therapeutic perspectives for obesity]. Annales d'endocrinologie (2012) 73 Suppl 1:S2–8. doi: 10.1016/s0003-4266(12)70009-4
89. Pfeifer A, Hoffmann LS. Brown, beige, and white: The new color code of fat and its pharmacological implications. Annu Rev Pharmacol Toxicol (2015) 55:207–27. doi: 10.1146/annurev-pharmtox-010814-124346
90. Sidossis L, Kajimura S. Brown and beige fat in humans: Thermogenic adipocytes that control energy and glucose homeostasis. J Clin Invest (2015) 125(2):478–86. doi: 10.1172/jci78362
91. Liu Y, Sen S, Wannaiampikul S, Palanivel R, Hoo RL, Isserlin R, et al. Metabolomic profiling in liver of adiponectin-knockout mice uncovers lysophospholipid metabolism as an important target of adiponectin action. Biochem J (2015) 469(1):71–82. doi: 10.1042/bj20141455
92. Magee C, Grieve DJ, Watson CJ, Brazil DP. Diabetic nephropathy: A tangled web to unweave. Cardiovasc Drugs Ther (2017) 31(5-6):579–92. doi: 10.1007/s10557-017-6755-9
93. Moreno JA, Gomez-Guerrero C, Mas S, Sanz AB, Lorenzo O, Ruiz-Ortega M, et al. Targeting inflammation in diabetic nephropathy: A tale of hope. Expert Opin investigational Drugs (2018) 27(11):917–30. doi: 10.1080/13543784.2018.1538352
94. Zheng Z, Zheng F. Immune cells and inflammation in diabetic nephropathy. J Diabetes Res (2016) 2016:1841690. doi: 10.1155/2016/1841690
95. Yu FN, Hu ML, Wang XF, Li XP, Zhang BH, Lu XQ, et al. Effects of microrna-370 on mesangial cell proliferation and extracellular matrix accumulation by binding to canopy 1 in a rat model of diabetic nephropathy. J Cell Physiol (2019) 234(5):6898–907. doi: 10.1002/jcp.27448
96. Paueksakon P, Revelo MP, Ma LJ, Marcantoni C, Fogo AB. Microangiopathic injury and augmented pai-1 in human diabetic nephropathy. Kidney Int (2002) 61(6):2142–8. doi: 10.1046/j.1523-1755.2002.00384.x
97. Mao L, Liu L, Zhang T, Qin H, Wu X, Xu Y. Histone deacetylase 11 contributes to renal fibrosis by repressing Klf15 transcription. Front Cell Dev Biol (2020) 8:235. doi: 10.3389/fcell.2020.00235
98. Williams R, Karuranga S, Malanda B, Saeedi P, Basit A, Besançon S, et al. Global and regional estimates and projections of diabetes-related health expenditure: Results from the international diabetes federation diabetes atlas, 9th edition. Diabetes Res Clin Pract (2020) 162:108072. doi: 10.1016/j.diabres.2020.108072
99. Park WY, Yiannakou I, Petersen JM, Hoffmann U, Ma J, Long MT. Sugar-sweetened beverage, diet soda, and nonalcoholic fatty liver disease over 6 years: The framingham heart study. Clin Gastroenterol Hepatol (2021). doi: 10.1016/j.cgh.2021.11.001
100. Li L, Xie W. Lncrna Hdac11-As1 suppresses atherosclerosis by inhibiting Hdac11-mediated adropin histone deacetylation. J Cardiovasc Trans Res (2022). doi: 10.1007/s12265-022-10248-7
101. Yao F, Jin Z, Lv X, Zheng Z, Gao H, Deng Y, et al. Hydroxytyrosol acetate inhibits vascular endothelial cell pyroptosis Via the Hdac11 signaling pathway in atherosclerosis. Front Pharmacol (2021) 12:656272. doi: 10.3389/fphar.2021.656272
102. Fan XD, Wan LL, Duan M, Lu S. Hdac11 deletion reduces fructose-induced cardiac dyslipidemia, apoptosis and inflammation by attenuating oxidative stress injury. Biochem Biophys Res Commun (2018) 503(2):444–51. doi: 10.1016/j.bbrc.2018.04.090
103. Lin Z, Chang J, Li X, Wang J, Wu X, Liu X, et al. Association of DNA methylation and transcriptome reveals epigenetic etiology of heart failure. Funct Integr Genomics (2022) 22(1):89–112. doi: 10.1007/s10142-021-00813-9
104. Iyengar NM, Gucalp A, Dannenberg AJ, Hudis CA. Obesity and cancer mechanisms: Tumor microenvironment and inflammation. J Clin Oncol (2016) 34(35):4270–6. doi: 10.1200/jco.2016.67.4283
105. Lee YM, Yoon Y, Yoon H, Park HM, Song S, Yeum KJ. Dietary anthocyanins against obesity and inflammation. Nutrients (2017) 9(10):1089. doi: 10.3390/nu9101089
106. Wu H, Ballantyne CM. Skeletal muscle inflammation and insulin resistance in obesity. J Clin Invest (2017) 127(1):43–54. doi: 10.1172/jci88880
107. Saltiel AR, Olefsky JM. Inflammatory mechanisms linking obesity and metabolic disease. J Clin Invest (2017) 127(1):1–4. doi: 10.1172/jci92035
108. Georgopoulos K. From immunity to tolerance through hdac. Nat Immunol (2009) 10(1):13–4. doi: 10.1038/ni0109-13
109. Wang X, Wu Y, Jiao J, Huang Q. Mycobacterium tuberculosis infection induces il-10 gene expression by disturbing histone deacetylase 6 and histonedeacetylase 11 equilibrium in macrophages. Tuberculosis (Edinburgh Scotland) (2018) 108:118–23. doi: 10.1016/j.tube.2017.11.008
110. Wang L, Tao R, Hancock WW. Using histone deacetylase inhibitors to enhance Foxp3 (+) regulatory T-cell function and induce allograft tolerance. Immunol Cell Biol (2009) 87(3):195–202. doi: 10.1038/icb.2008.106
111. Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med (2007) 13(11):1299–307. doi: 10.1038/nm1652
112. Buglio D, Khaskhely NM, Voo KS, Martinez-Valdez H, Liu YJ, Younes A. Hdac11 plays an essential role in regulating Ox40 ligand expression in Hodgkin lymphoma. Blood (2011) 117(10):2910–7. doi: 10.1182/blood-2010-08-303701
113. Lian ZR, Xu YF, Wang XB, Gong JP, Liu ZJ. Suppression of histone deacetylase 11 promotes expression of il-10 in kupffer cells and induces tolerance following orthotopic liver transplantation in rats. J Surg Res (2012) 174(2):359–68. doi: 10.1016/j.jss.2010.12.035
114. Luo XQ, Shao JB, Xie RD, Zeng L, Li XX, Qiu SQ, et al. Micro rna-19a interferes with il-10 expression in peripheral dendritic cells of patients with nasal polyposis. Oncotarget (2017) 8(30):48915–21. doi: 10.18632/oncotarget.16555
115. Ramos-Nino ME. The role of chronic inflammation in obesity-associated cancers. ISRN Oncol (2013) 2013:697521. doi: 10.1155/2013/697521
116. Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell (2014) 156(1-2):20–44. doi: 10.1016/j.cell.2013.12.012
117. Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J (2012) 279(15):2610–23. doi: 10.1111/j.1742-4658.2012.08644.x
118. Scott I. Regulation of cellular homoeostasis by reversible lysine acetylation. Essays Biochem (2012) 52:13–22. doi: 10.1042/bse0520013
119. Selvi RB, Kundu TK. Reversible acetylation of chromatin: Implication in regulation of gene expression, disease and therapeutics. Biotechnol J (2009) 4(3):375–90. doi: 10.1002/biot.200900032
120. Shukla V, Vaissière T, Herceg Z. Histone acetylation and chromatin signature in stem cell identity and cancer. Mutat Res (2008) 637(1-2):1–15. doi: 10.1016/j.mrfmmm.2007.07.012
121. Shouksmith AE, Shah F, Grimard ML, Gawel JM, Raouf YS, Geletu M, et al. Identification and characterization of aes-135, a hydroxamic acid-based hdac inhibitor that prolongs survival in an orthotopic mouse model of pancreatic cancer. J medicinal Chem (2019) 62(5):2651–65. doi: 10.1021/acs.jmedchem.8b01957
122. Martin MW, Lee JY, Lancia DR Jr., Ng PY, Han B, Thomason JR, et al. Discovery of novel n-Hydroxy-2-Arylisoindoline-4-Carboxamides as potent and selective inhibitors of Hdac11. Bioorganic medicinal Chem Lett (2018) 28(12):2143–7. doi: 10.1016/j.bmcl.2018.05.021
123. Fournel M, Bonfils C, Hou Y, Yan PT, Trachy-Bourget MC, Kalita A, et al. Mgcd0103, a novel isotype-selective histone deacetylase inhibitor, has broad spectrum antitumor activity in vitro and in vivo. Mol Cancer Ther (2008) 7(4):759–68. doi: 10.1158/1535-7163.Mct-07-2026
124. Wang P, Wang Z, Liu J. Correction to: Role of hdacs in normal and malignant hematopoiesis. Mol Cancer (2020) 19(1):55. doi: 10.1186/s12943-020-01182-w
125. Son SI, Su D, Ho TT, Lin H. Garcinol is an Hdac11 inhibitor. ACS Chem Biol (2020) 15(11):2866–71. doi: 10.1021/acschembio.0c00719
126. Kutil Z, Mikešová J, Zessin M, Meleshin M, Nováková Z, Alquicer G, et al. Continuous activity assay for Hdac11 enabling reevaluation of hdac inhibitors. ACS omega (2019) 4(22):19895–904. doi: 10.1021/acsomega.9b02808
127. Bui HTB, Nguyen PH, Pham QM, Tran HP, Tran Q, Jung H, et al. Target design of novel histone deacetylase 6 selective inhibitors with 2-mercaptoquinazolinone as the cap moiety. Molecules (Basel Switzerland) (2022) 27(7):2204. doi: 10.3390/molecules27072204
128. Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia (2005) 19(10):1751–9. doi: 10.1038/sj.leu.2403910
129. Tian Y, Lv W, Li X, Wang C, Wang D, Wang PG, et al. Stabilizing Hdac11 with saha to assay slow-binding benzamide inhibitors. Bioorganic medicinal Chem Lett (2017) 27(13):2943–5. doi: 10.1016/j.bmcl.2017.05.004
130. Son SI, Cao J, Zhu CL, Miller SP, Lin H. Activity-guided design of Hdac11-specific inhibitors. ACS Chem Biol (2019) 14(7):1393–7. doi: 10.1021/acschembio.9b00292
131. Vitanza NA, Biery MC, Myers C, Ferguson E, Zheng Y, Girard EJ, et al. Optimal therapeutic targeting by hdac inhibition in biopsy-derived treatment-naïve diffuse midline glioma models. Neuro-oncology (2021) 23(3):376–86. doi: 10.1093/neuonc/noaa249
132. Wang X, Liu K, Gong H, Li D, Chu W, Zhao D, et al. Death by histone deacetylase inhibitor quisinostat in tongue squamous cell carcinoma Via apoptosis, pyroptosis, and ferroptosis. Toxicol Appl Pharmacol (2021) 410:115363. doi: 10.1016/j.taap.2020.115363
133. Zhong L, Zhou S, Tong R, Shi J, Bai L, Zhu Y, et al. Preclinical assessment of histone deacetylase inhibitor quisinostat as a therapeutic agent against esophageal squamous cell carcinoma. Investigational New Drugs (2019) 37(4):616–24. doi: 10.1007/s10637-018-0651-4
134. Li R, Ling D, Tang T, Huang Z, Wang M, Ding Y, et al. Discovery of novel plasmodium falciparum Hdac1 inhibitors with dual-stage antimalarial potency and improved safety based on the clinical anticancer drug candidate quisinostat. J medicinal Chem (2021) 64(4):2254–71. doi: 10.1021/acs.jmedchem.0c02104
135. Afaloniati H, Poutahidis T, Giakoustidis A, Gargavanis A, Giakoustidis D, Angelopoulou K. Romidepsin hepatocellular carcinoma suppression in mice is associated with deregulated gene expression of bone morphogenetic protein and notch signaling pathway components. Mol Biol Rep (2021) 48(1):551–62. doi: 10.1007/s11033-020-06089-9
136. Afaloniati H, Angelopoulou K, Giakoustidis A, Hardas A, Pseftogas A, Makedou K, et al. Hdac1/2 inhibitor romidepsin suppresses den-induced hepatocellular carcinogenesis in mice. OncoTar Ther (2020) 13:5575–88. doi: 10.2147/ott.S250233
137. Shi Y, Fu Y, Zhang X, Zhao G, Yao Y, Guo Y, et al. Romidepsin (Fk228) regulates the expression of the immune checkpoint ligand pd-L1 and suppresses cellular immune functions in colon cancer. Cancer immunol immunother (2021) 70(1):61–73. doi: 10.1007/s00262-020-02653-1
138. Moreau P, Facon T, Touzeau C, Benboubker L, Delain M, Badamo-Dotzis J, et al. Quisinostat, bortezomib, and dexamethasone combination therapy for relapsed multiple myeloma. Leukemia lymphoma (2016) 57(7):1546–59. doi: 10.3109/10428194.2015.1117611
139. Falchi L, Ma H, Klein S, Lue JK, Montanari F, Marchi E, et al. Combined oral 5-azacytidine and romidepsin are highly effective in patients with ptcl: A multicenter phase 2 study. Blood (2021) 137(16):2161–70. doi: 10.1182/blood.2020009004
140. Moltó J, Rosás-Umbert M, Miranda C, Manzardo C, Puertas MC, Ruiz-Riol M, et al. Pharmacokinetic/Pharmacodynamic analysis of romidepsin used as an hiv latency reversing agent. J antimicrobial chemother (2021) 76(4):1032–40. doi: 10.1093/jac/dkaa523
141. Xiao H, Wu Z, Li B, Shangguan Y, Stoltz JF, Magdalou J, et al. The low-expression programming of 11β-Hsd2 mediates osteoporosis susceptibility induced by prenatal caffeine exposure in Male offspring rats. Br J Pharmacol (2020) 177(20):4683–700. doi: 10.1111/bph.15225
Keywords: HDAC11, metabolic disorders, obesity, diabetic complications, diabetes
Citation: Chen H, Xie C, Chen Q and Zhuang S (2022) HDAC11, an emerging therapeutic target for metabolic disorders. Front. Endocrinol. 13:989305. doi: 10.3389/fendo.2022.989305
Received: 08 July 2022; Accepted: 10 October 2022;
Published: 20 October 2022.
Edited by:
Tarunveer Singh Ahluwalia, Steno Diabetes Center Copenhagen (SDCC), DenmarkReviewed by:
SMITHA GEORGE, Van Andel Institute, United StatesCopyright © 2022 Chen, Xie, Chen and Zhuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shougang Zhuang, c3podWFuZ0BsaWZlc3Bhbi5vcmc=; Z2FuZ3podWFuZ0Bob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.