
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol., 20 September 2022
Sec. Cancer Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.971687
This article is part of the Research TopicNew Drugs for treating COVID-19 Cancer PatientsView all 9 articles
The variant virus-based 2019 coronavirus disease (COVID-19) pandemic has reportedly impacted almost all populations globally, characterized by a huge number of infected individuals. Clinical evidence proves that patients with cancer are more easily infected with severe acute respiratory disease coronavirus 2 (SARS-CoV-2) because of immunologic deficiency. Thus, there is an urgent need to develop candidate medications to treat patients with cancer plus COVID-19, including those with osteosarcoma (OS). Ferulic acid, a latent theriacal compound that has anti-tumor and antivirus activities, is discovered to have potential pharmacological use. Thus, in this study, we aimed to screen and determine the potential therapeutic targets of ferulic acid in treating patients with OS plus COVID-19 as well as the pharmacological mechanisms. We applied a well-established integrated methodology, including network pharmacology and molecular docking technique, to detail target prediction, network construction, gene ontology, and pathway enrichment in core targets. The network pharmacology results show that all candidate genes, by targeting autophagy, were the core targets of ferulic acid in treating OS and COVID-19. Through molecular docking analysis, the signal transducer and activator of transcription 3 (STAT3), mitogen-activated protein kinase 1 (MAPK1), and phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1) were identified as the pharmacological targets of ferulic acid in treating OS. These preclinical findings from bioinformatics analysis altogether effectively determined the pharmacological molecules and mechanisms via targeting autophagy, demonstrating the therapeutic effectiveness of ferulic acid against COVID-19 and OS.
The 2019 coronavirus disease (COVID-19), reportedly caused by severe acute respiratory disease coronavirus 2 (SARS-CoV-2), has developed into a cosmopolitical pandemic crisis, causing an imponderable death toll (1). The pathogenesis following SARS-CoV-2 infection may comprise acute respiratory distress syndrome and deadly pneumonia, also related to the cytokine storm that induces secondary tissue impairment (2). Despite the availability of COVID-19 vaccines, clinical evidence shows that emerging mutations result in unexpected escape from vaccine effect (3). The protection of existing vaccines may be affected over time against the highly infectious Omicron variant (4). Currently, few specific antiviral medications have been approved for treating COVID-19. From clinical observations, patients with cancer who become infected with SARS-CoV-2 may have a higher risk for in-hospital mortality than other infections (5). In addition, patients with cancer are believed to have a higher risk for severe COVID-19 due to their advanced age and pre-existing disorders (6). Osteosarcoma (OS) is one of the most malignant cancers in orthopedics, occurring in adolescents and the elderly aged >65 years (7). Recent data have reported that the 5-year survival rate of patients with OS has not meliorated in the past 30 years and has approached around 55–75% (8). Children with cancer such as metastatic OS are likely to have a higher risk of developing life-threatening COVID-19 (9). Patients with OS and COVID-19 are medically hard to treat as current clinical treatment is not yet available, and specific functions of the pathogenetic genes involved in OS and COVID-19 are still unreported. In China, traditional Chinese medicine (TCM) is commonly used for chronic clinical disorders, including malignant cancers (10). In anti-COVID-19 practice, the use of TCM for COVID-19 treatment has been recommended in most Chinese provinces, which is characterized by enhanced cure rate (11). Preclinical reports show that some new compounds isolated from TCM have potent antiviral action, including glycyrrhizic acid (12). Ferulic acid, termed 3-methoxy-4-hydroxycinnamic acid, has broad pharmacological properties, such as antioxidant function as well as antibacterial and antiviral actions (13). In addition, the anti-neoplastic effect of ferulic acid has been experimentally validated (14). Furthermore, ferulic acid may mediate potential anti-OS action in vitro by promoting the apoptotic pathway (15). Based on the current antiviral and anti-cancer characteristics of ferulic acid, this study was designed to investigate the potential efficacy and mechanisms of ferulic acid in treating OS and COVID-19 by using network pharmacology and molecular docking approaches. The preclinical findings may provide a new direction for the promising treatment of OS and COVID-19 in future clinical practice.
Expression profiling by array for OS was retrieved from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database (16), including GSE28424 [modulation of the OS expression phenotype by miRNAs (Illumina)], in which the dataset was based on the platform file GPL13376. Then, the GEO2R online tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was applied for gene differential analysis. The screening conditions were set as the difference fold |log2FC| >0.5, and the false discovery rate (FDR) was <0.05 to obtain the significantly differentially expressed genes of human OS and to draw the volcano plot associated with differential genes. Using the Genecards database (17) and the Online Mendelian Inheritance in Man (OMIM) database (18), relevant targets of COVID-19 were screened out accordingly.
The action targets of ferulic acid were obtained using the comparative toxicogenomics database, Swiss Target Prediction database, SuperPred database, and Pharmmapper database. The aforementioned targets were corrected using the reviewed (Swiss-Prot) and Human in the UniprotKB database (19). The OMIM database, GeneCards database, and National Center for Biotechnology Information gene function module were used to identify the autophagy-related targets. Finally, a Venn diagram analysis (20) was performed between the ferulic acid/autophagy-associated targets and OS/COVID-19-related targets to acquire the co-targets of ferulic acid-anti-OS/COVID-19 effect through autophagy.
The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses and visualization of core targets were performed using R-packages such as “ClusterProfiler”, “org.Hs.eg.Db”, and “ggplot2” in the R language. The gene annotation information was obtained from “org.Hs.eg.Db”, the adjusted p-value cutoff was 0.05, and the q-value cutoff was 0.05 during enrichment, resulting in an output corresponding to a bubble chart and a circle chart. In addition, Cytoscape_v3.8.2 was used to create a drug-target–GO–CC–MF-pathway disease visualization graph from the results of the biological process and pathway enrichment of ferulic acid against OS and COVID-19 targets through autophagy (21).
To reveal the interaction between the ferulic acid targets and OS/COVID-19 targets, a protein–protein interaction (PPI) network was established through the Search Tool for the Retrieval of Interacting Genes (STRING) database (22). All core targets were transferred to this STRING database. Their size is in direct proportion to the enrichment method. The PPI network was assayed through the network analyzer from Cytoscape v3.7.1 to identify the core genes based on the betweenness, closeness, and degree scores.
Molecular docking analysis was performed to illustrate and validate the binding energy of the OS/COVID-19-associated proteins with ferulic acid. The three-dimensional structures of ferulic acid were collected from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) (23). The crystal structures of the screened proteins, including the signal transducer and activator of transcription 3 (STAT3), mitogen-activated protein kinase 1 (MAPK1), and phosphoinositide-3-kinase regulatory subunit 1 (PIK3R1), were identified from the Uniprot database (https://www.uniprot.org/) and transferred in Protein Data Bank (https://www.pdbus.org/) format. In addition, the Autodock (version 1.5.6) software was used to increase polar hydrogen and set the grid box, and the autodock vina function was further used to conduct the molecular docking assay. The binding energy of ferulic acid with these identified proteins was established, and then the Pymol software was used to visualize the optimal docking conformations (24).
We compared 19 normal group samples to four OS tumor group samples through the GEO database by retrieving the GSE28424 dataset. After screening, a total of 3,011 differential genes were obtained, including 1,500 upregulated genes and 1,511 downregulated genes, as shown in Figure 1. After searching databases such as Genecards, a total of 2,199 relevant COVID-19 target genes were obtained accordingly.
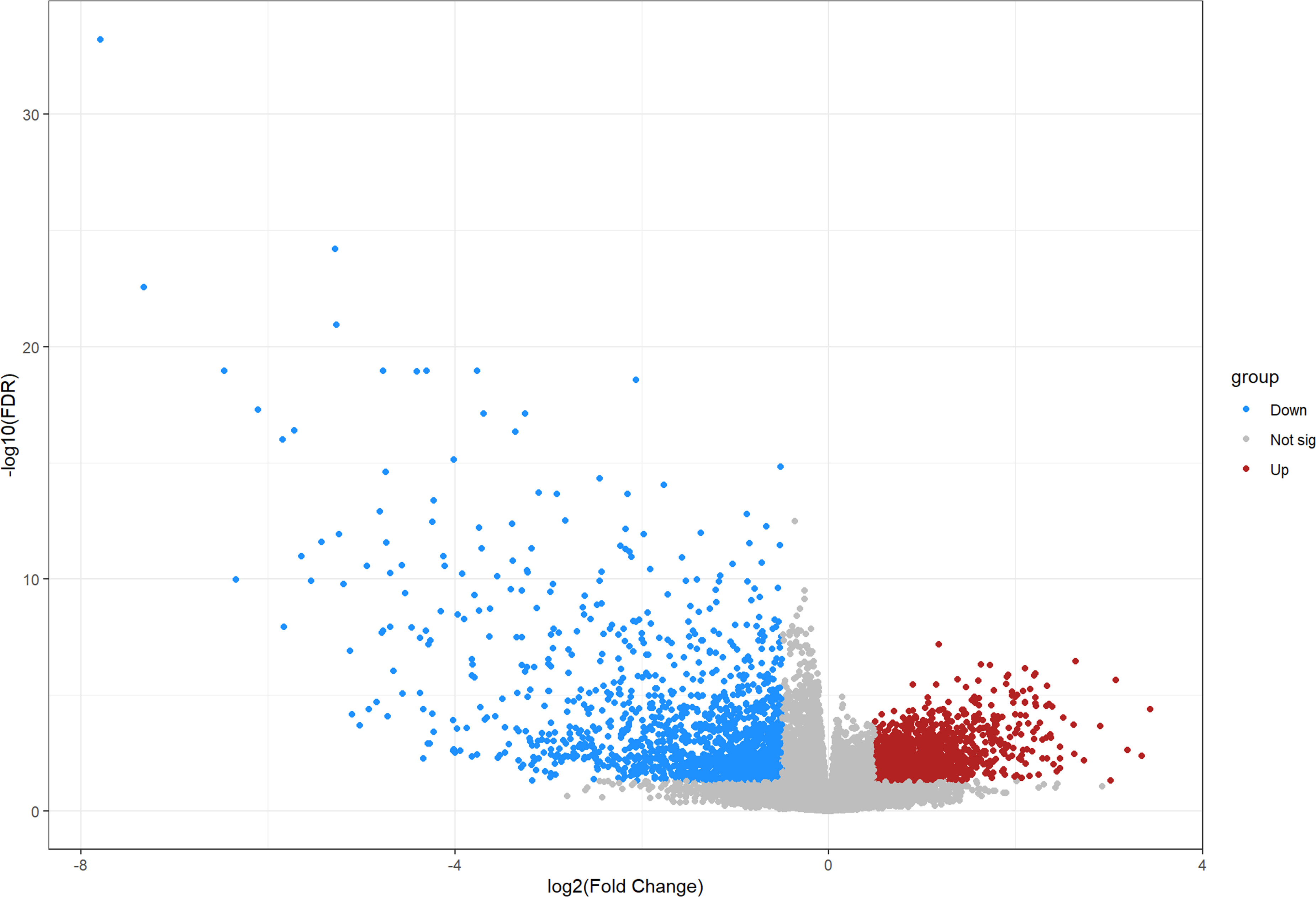
Figure 1 Volcano map illustrating all differentially expressed genes between osteosarcoma and non-osteosarcoma samples.
Using databases such as TCD to search for drug targets and correcting through the UniprotKB database, a total of 360 ferulic acid targets were obtained. The OMIM and other databases were used to obtain 870 autophagy-related targets. The obtained targets were mapped using a Venn diagram, and a total of five intersection targets were obtained, including MAPK1, TLR4, PIK3R1, STAT3, and PARP1 (Figure 2).
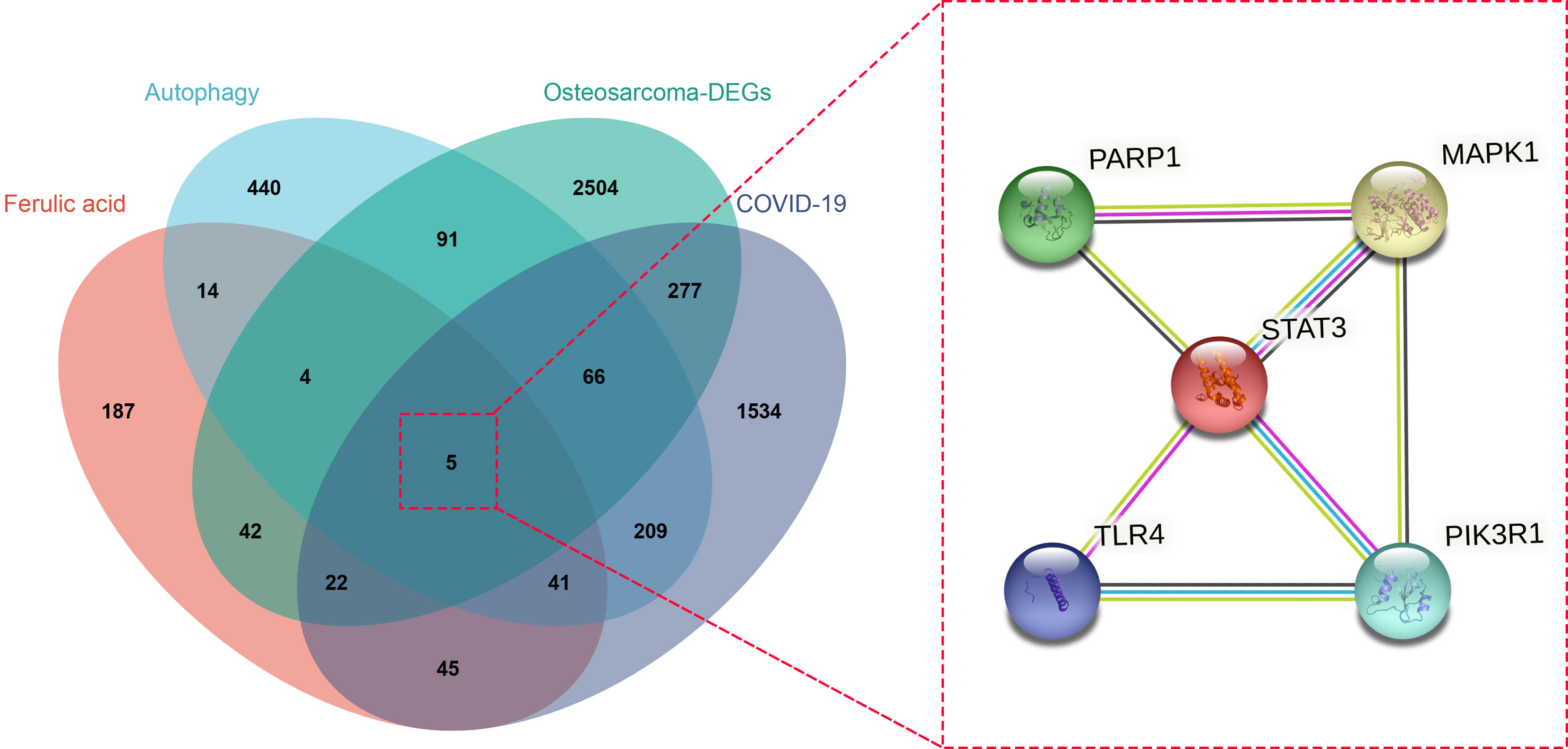
Figure 2 Venn diagram of ferulic acid, autophagy, and osteosarcoma/COVID-19-targeted genes is displayed, and a protein–protein interaction network was established from this diagram.
To better indicate the biological characteristics of the identified intersection targets, the R language software was used to implement GO and KEGG enrichment assays. Using parametric analysis, the GO-based biological process (BP) terms (Figure 3) mainly comprised protein localization to the nucleus, cellular response to peptide, astrocyte differentiation, myeloid cell differentiation, positive regulation of protein localization to the nucleus, positive regulation of tumor necrosis factor production, positive regulation of tumor necrosis factor superfamily cytokine production, regulation of protein localization to the nucleus, protein import into the nucleus, and import into the nucleus. Furthermore, the key cellular component (CC) terms (Figure 4) suggested the functions of postsynaptic density, asymmetric synapse, postsynaptic specialization, neuron to neuron synapse, early endosome, transcription regulator complex, pseudopodium, perinuclear endoplasmic reticulum, phagocytic cup, and phosphatidylinositol 3-kinase complex. Other molecular function (MF) terms (Figure 5) indicated phosphatase binding, phosphotyrosine residue binding, protein phosphorylated amino acid binding, signaling adaptor activity, phosphoprotein binding, nuclear receptor binding, protein phosphatase binding, cytokine receptor binding, protein–macromolecule adaptor activity, and RNA polymerase II-specific DNA-binding transcription factor binding. The KEGG enrichment assay was executed to cluster the pharmacological signaling pathways exerted by ferulic acid. A total of 42 molecular pathways with FDR <0.05 were identified (Figure 6). These enrichment data indicated the KEGG pathways of ferulic acid, including the HIF-1 signaling pathway, EGFR tyrosine kinase inhibitor resistance, Toll-like receptor signaling pathway, FoxO signaling pathway, apoptosis, signaling pathways regulating the pluripotency of stem cells, necroptosis, neutrophil extracellular trap formation, chemokine signaling pathway, and VEGF signaling pathway. Detailed enrichment terms were visualized in ferulic acid–OS/COVID-19–BP–CC–MF clusters through Cytoscape_v3.8.2 software analysis (Figure 7).
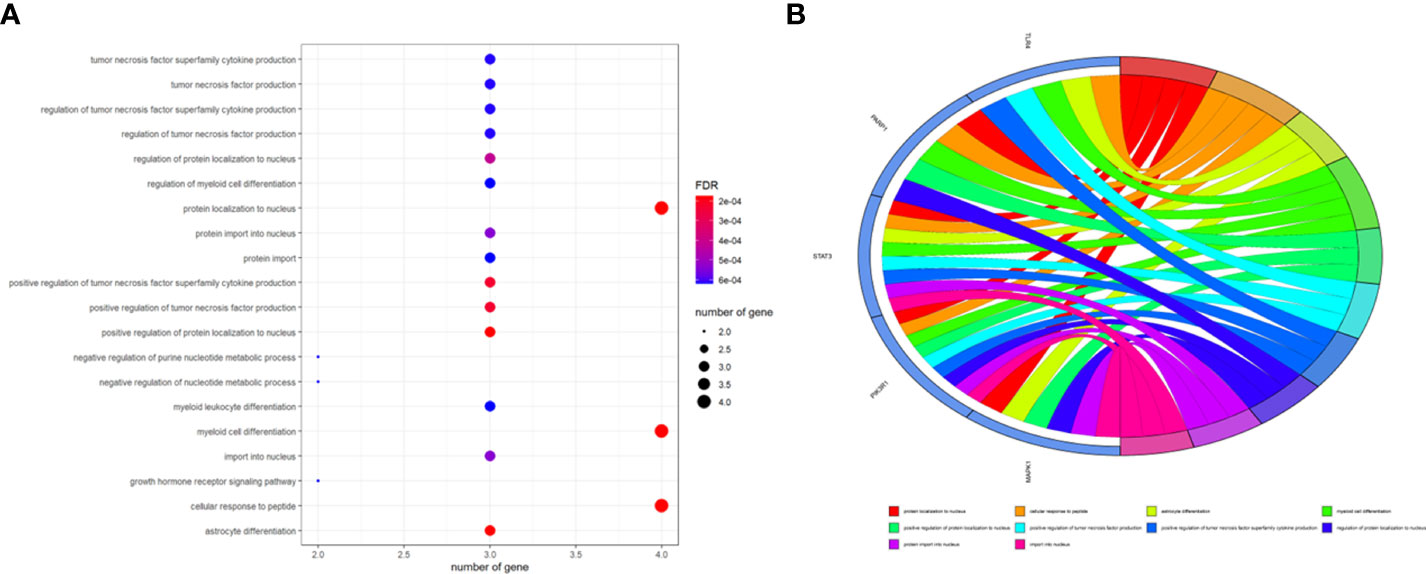
Figure 3 The main terms for biological process, cellular component, and molecular function are presented using bar plots (A). The top terms of the functional enrichment analysis are displayed through the bubble chart (B).
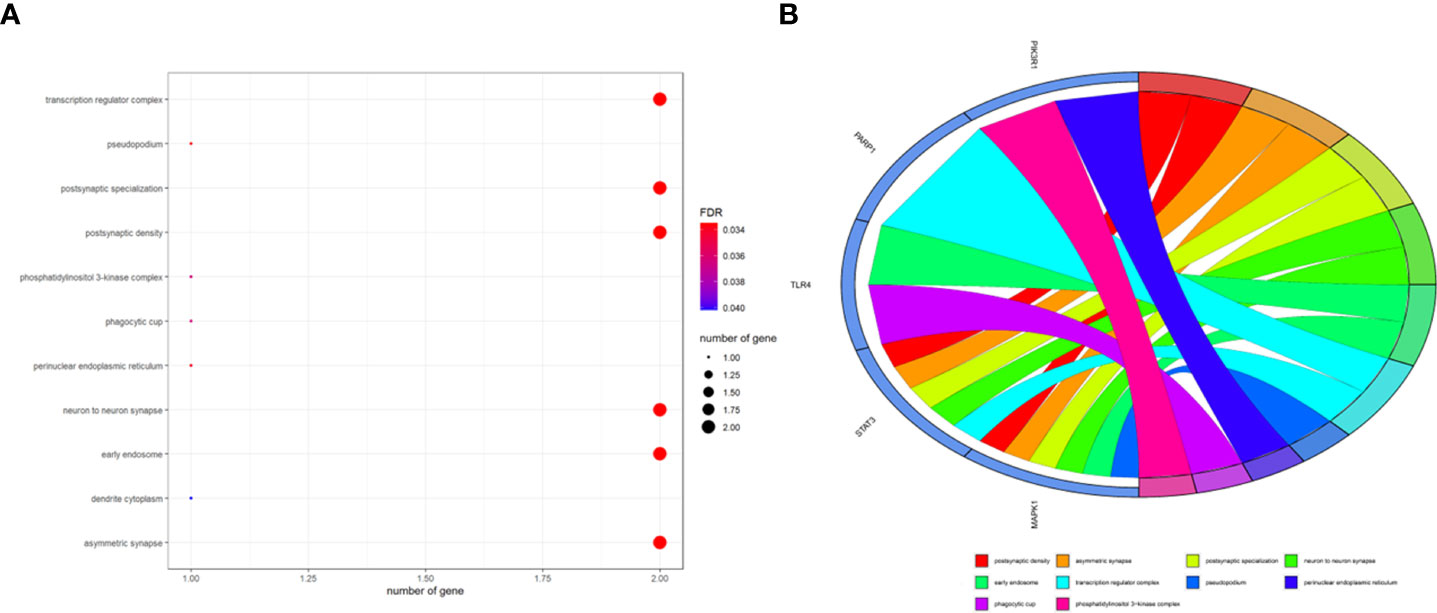
Figure 4 The main terms for biological process, cellular component, and molecular function are presented using bar plots (A). The top terms of the functional enrichment analysis are displayed through the bubble chart (B).
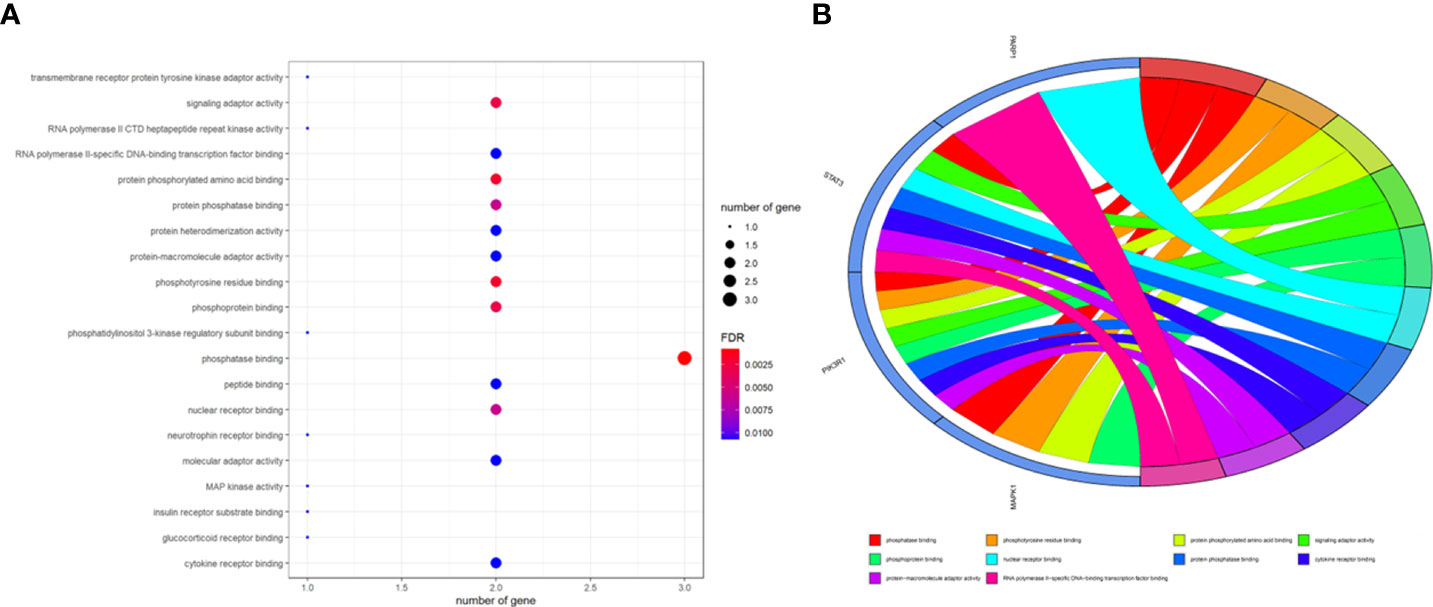
Figure 5 The main terms for biological process, cellular component, and molecular function are presented using bar plots (A). The top terms of the functional enrichment analysis are displayed through the bubble chart (B).
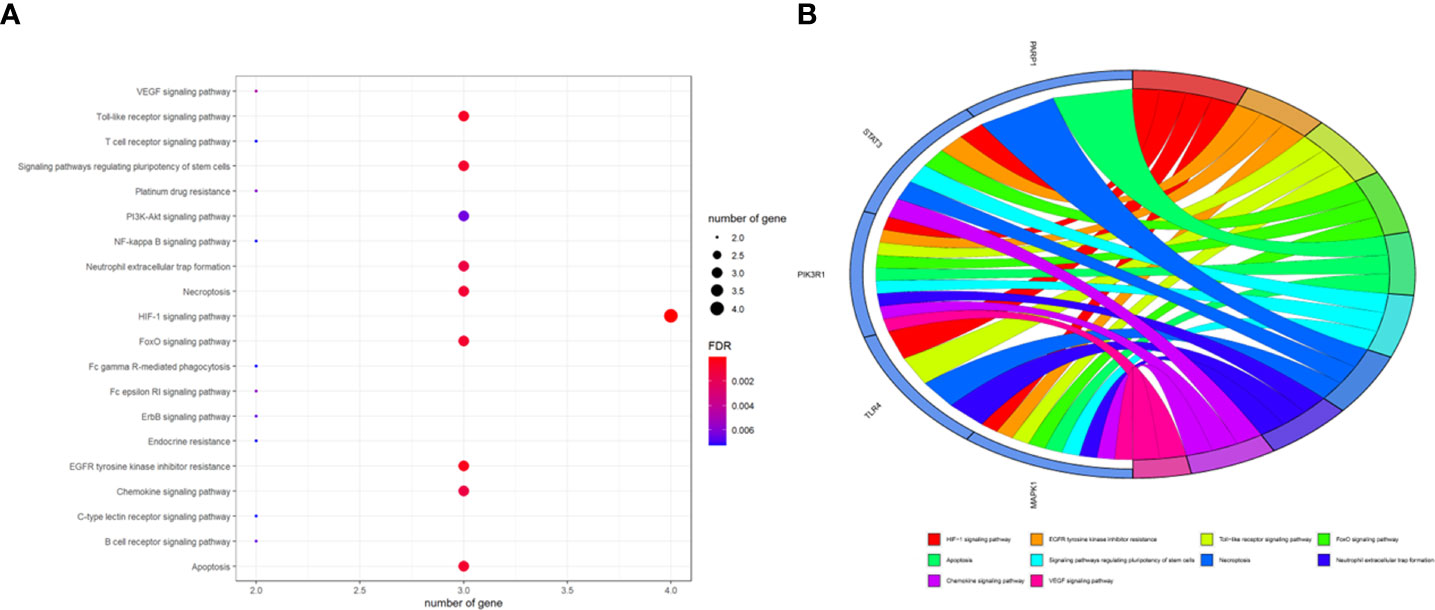
Figure 6 Summary of the Kyoto Encyclopedia of Genes and Genomes enrichment analysis characterized by signaling mechanisms, as shown in bubble chart (A) and circle diagram (B).
To further ascertain the core targets, topological parameters and target degrees of freedom were measured through Cytoscape_v3.8.2, wherein the filter condition range was set to three to four. As a result, all top three targets, including STAT3, MAPK1, and PIK3R1, were identified as the core genes before further in silico imitation analysis.
To confirm the potential targets of ferulic acid, these STAT3, MAPK1, and PIK3R1 proteins were computationally docked with ferulic acid based on the binding energy scores, characterized by 6SM8, 3SA0, and 6OCO binding active sites. The amino acid residue GLY-1020 (3.4 Å) formed a functional hydrogen bond between ferulic acid and 6SM8 (STAT3), and the free docking energy was -6.3 kcal/mol (Figure 8A). Moreover, the amino acid residues ASP-111 (2.3 Å), MET-108 (2.3 Å), and LYS-114 (2.4 Å) formed a functional hydrogen bond between ferulic acid and 3SA0 (MAPK1), with a free docking energy of -5.5 kcal/mol (Figure 8B). Finally, the amino acid residues SER-831 (3.0 Å) and VAL-828 (2.6 Å) formed a functional hydrogen bond between ferulic acid and 6OCO (PIK3R1), and the free docking energy was -5.7 kcal/mol (Figure 8C).
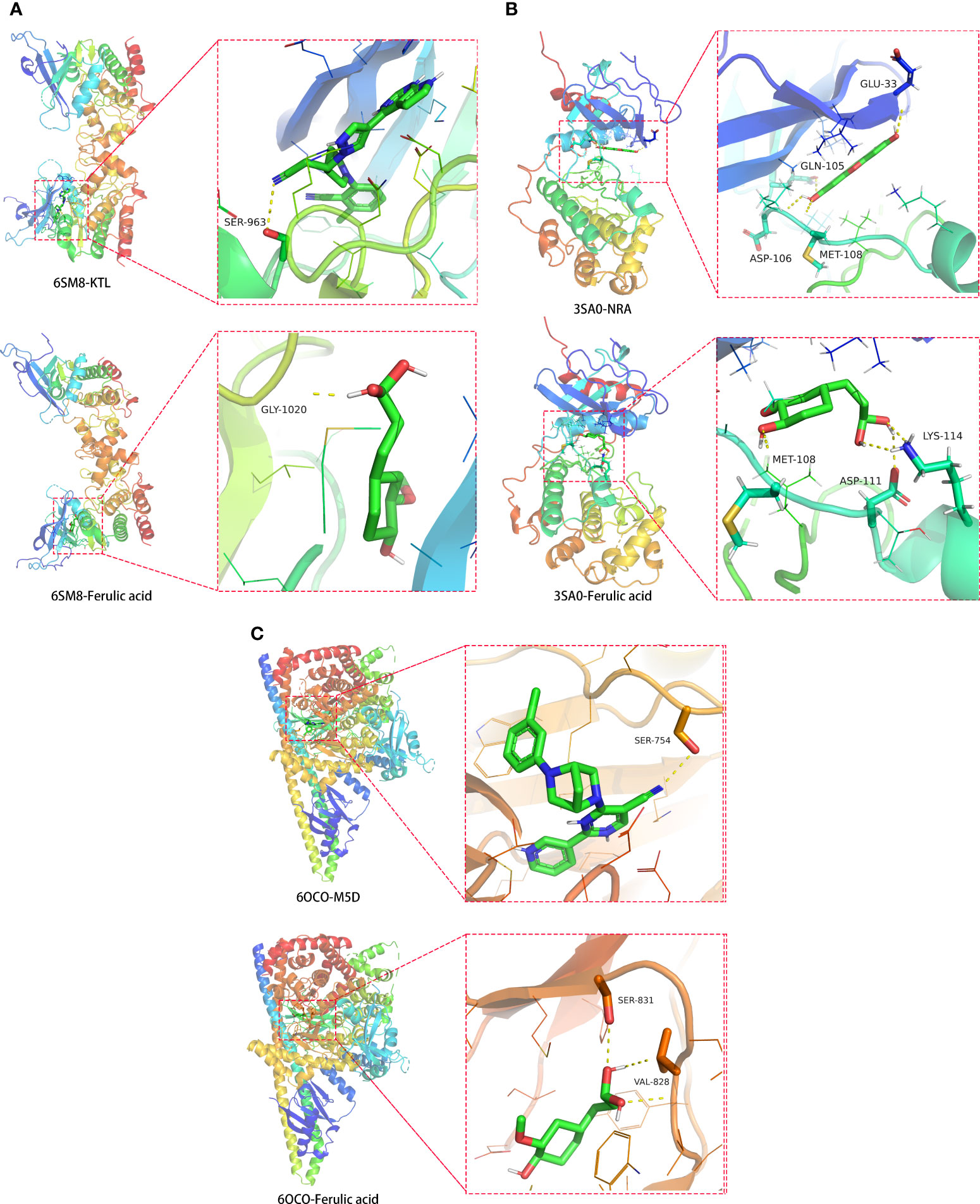
Figure 8 Molecular docking models of ferulic acid binding to the identified STAT3, MAPK1, and PIK3R1 target proteins with respect to (A) 6SM8, (B) 3SA0, and (C) 6OCO.
TCM is found to have anti-inflammatory, antiviral, and immunoregulatory activities and is also characterized by a candidate curative effect against COVID-19 (25). It is of interest that bioinformatics analysis through the network pharmacology methodology has demonstrated that niacin may be used to potentially treat colorectal cancer and COVID-19 (21). By using comprehensive bioinformatics approaches, we aimed to reveal the potential molecular mechanisms and therapeutic targets of ferulic acid, a functional molecule, in the treatment of OS and COVID-19. A previous study indicates that ferulic acid has proven antiviral action against PPV infection through effective regulation of the caspase-dependent apoptotic pathway (26). Another in vitro experiment suggests that ferulic acid restrains NLRP3 inflammasome activities and lowers the synthesis of inflammatory cytokines through the autophagy channel (27). Thus, ferulic acid may mediate a potential action against COVID-19 by inhibiting virus infection and inflammatory reaction. Furthermore, in the cell culture model of 143B and MG63 OS cells, ferulic acid plays anti-OS effects by inactivating the PI3K/Akt pathway (28). Taking all the present information together, we surmised that ferulic acid may exert a potential action against OS and COVID-19. In our bioinformatics analysis, the clinicopathological features showed 3,011 differentially expressed genes, 1,500 and 1,511 of which were up- and downregulated, respectively. These differentially expressed genes indicate potential diagnostic biomarkers for human OS. In addition, we identified 2,199 genes that are mutual for OS and COVID-19, 360 ferulic acid target genes, and 870 autophagy target genes. Using Venn diagram mapping, we further identified five core ferulic acid targets against OS and COVID-19 via autophagy, including MAPK1, TLR4, PIK3R1, STAT3, and PARP1. Among these core target genes, TLR4, STAT3, and PARP1 are associated with the development of COVID-19 (29–31). Moreover, the GO and KEGG enrichment assays revealed that biological processes were distinctly enriched in the positive regulation of tumor necrosis factor production, particularly the positive regulation of tumor necrosis factor superfamily cytokine production. More data have suggested that ferulic acid might inhibit autophagy-dependent cell proliferation in cancers (32) as well as inflammation-related SARS-CoV-2 infection (33). Following the use of the molecular docking analysis, we ultimately identified the core ferulic acid target proteins against OS and COVID-19, namely, STAT3, MAPK1, and PIK3R1. Among these, STAT3 could be developed as a promising therapeutic target for patients with cancer who contract COVID-19 (34). Furthermore, other preclinical evidence shows that both MAPK1 and PIK3R1 could be potential pharmacological targets against human cancers (35, 36). The molecular docking data of these core target proteins suggest that ferulic acid may function in multiple disease stages of patients with cancer plus a SARS-CoV-2 infection. However, our current study still has certain limitations. Experimental validation in vitro and in vivo was not performed as models of SARS-CoV-2 infection are currently unavailable commercially. Further clinical trials using ferulic acid to treat OS and COVID-19 are still unconducted.
This study systematically revealed the therapeutic targets and mechanisms of ferulic acid for the potential treatment of OS and COVID-19 through network pharmacology and molecular docking validation. In addition, our results indicate that ferulic acid could regulate multiple anti-cancer and antiviral signaling pathways to exert anti-OS and anti-COVID-19 properties. Our findings conclude that ferulic acid may be a promising therapeutic compound for potentially treating OS and COVID-19. As potential limitation in the current bioinformatics report, we still need further experimental confirmation of our current bioinformatics findings.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
LJ conceived and designed the study. GP, TY, and HL performed the data analysis and data interpretation. GP, TY, and HL conducted the bioinformatics and statistical analyses. LJ prepared and revised the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by the Key Laboratory of Tumor Immunology and Pathology Army Medical University (grant no. 2018jsz104) and Guangxi Natural Science Foundation of China (grant no. 2020JJA140172).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.971687/full#supplementary-material
1. Merad M, Blish CA, Sallusto F, Iwasaki A. The immunology and immunopathology of COVID-19. Science. (2022) 375(6585):1122–7. doi: 10.1126/science.abm8108
2. Marc F, Moldovan CM, Hoza A, Magheru S, Ciavoi G, Farcas DM, et al. Comparative study of cytokine storm treatment in patients with COVID-19 pneumonia using immunomodulators. J Clin Med (2022) 11(10):2945. doi: 10.3390/jcm11102945
3. Kang YF, Sun C, Sun J, Xie C, Zhuang Z, Xu HQ, et al. Quadrivalent mosaic HexaPro-bearing nanoparticle vaccine protects against infection of SARS-CoV-2 variants. Nat Commun (2022) 13(1):2674. doi: 10.1038/s41467-022-30222-w
4. Garcia-Valtanen P, Hope CM, Masavuli MG, Yeow AEL, Balachandran H, Mekonnen ZA, et al. SARS-CoV-2 omicron variant escapes neutralizing antibodies and T cell responses more efficiently than other variants in mild COVID-19 convalescents. Cell Rep Med (2022) 3(6):100651. doi: 10.1016/j.xcrm.2022.100651
5. Kodde C, Bonsignore M, Schöndube D, Bauer T, Hohenstein S, Bollmann A, et al. Mortality in cancer patients with SARS-CoV-2 or seasonal influenza: an observational cohort study from a German-wide hospital network. Infection (2022), 1–9. doi: 10.1007/s15010-022-01852-5
6. Rüthrich MM, Giessen-Jung C, Borgmann S, Classen AY, Dolff S, Grüner B, et al. COVID-19 in cancer patients: clinical characteristics and outcome-an analysis of the LEOSS registry. Ann Hematol (2021) 100(2):383–93. doi: 10.1007/s00277-020-04328-4
7. Kim HJ, Chalmers PN, Morris CD. Pediatric osteogenic sarcoma. Curr Opin Pediatr (2010) 22(1):61–6. doi: 10.1097/MOP.0b013e328334581f
8. Saraf AJ, Fenger JM, Roberts RD. Osteosarcoma: Accelerating progress makes for a hopeful future. Front Oncol (2018) 8:4. doi: 10.3389/fonc.2018.00004
9. Offenbacher R, Fabish L, Baker A, Chou AJ, Loeb DM. Respiratory failure in a child with pulmonary metastatic osteosarcoma and COVID-19. J Pediatr Hematol Oncol (2021) 43(6):859–60. doi: 10.1097/MPH.0000000000001897
10. Oyenihi OR, Oyenihi AB, Erhabor JO, Matsabisa MG, Oguntibeju OO. Unravelling the anticancer mechanisms of traditional herbal medicines with metabolomics. Molecules. (2021) 26(21):6541. doi: 10.3390/molecules26216541
11. Zhang DH, Zhang X, Peng B, Deng SQ, Wang YF, Yang L, et al. Network pharmacology suggests biochemical rationale for treating COVID-19 symptoms with a traditional Chinese medicine. Commun Biol (2020) 3(1):466. doi: 10.1038/s42003-020-01190-y
12. Li R, Wu K, Li Y, Liang X, Lai KP, Chen J. Integrative pharmacological mechanism of vitamin c combined with glycyrrhizic acid against COVID-19: findings of bioinformatics analyses. Brief Bioinform (2021) 22(2):1161–74. doi: 10.1093/bib/bbaa141
13. Antonopoulou I, Sapountzaki E, Rova U, Christakopoulos P. Ferulic acid from plant biomass: A phytochemical with promising antiviral properties. Front Nutr (2022) 8:777576. doi: 10.3389/fnut.2021.777576
14. Gupta A, Singh AK, Loka M, Pandey AK, Bishayee A. Ferulic acid-mediated modulation of apoptotic signaling pathways in cancer. Adv Protein Chem Struct Biol (2021) 125:215–57. doi: 10.1016/bs.apcsb.2020.12.005
15. Zhang XD, Wu Q, Yang SH. Ferulic acid promoting apoptosis in human osteosarcoma cell lines. Pak J Med Sci (2017) 33(1):127–31. doi: 10.12669/pjms.331.12066
16. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Res (2013) 41:991–5. doi: 10.1093/nar/gks1193
17. Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinf (2016) 54:1.30.1–1.30.33. doi: 10.1002/cpbi.5
18. Hamosh A, Scott AF, Amberger J, Bocchini C, Valle D, McKusick VA. Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res (2002) 30(1):52–5. doi: 10.1093/nar/30.1.52
19. UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res (2021) 49:480–9. doi: 10.1093/nar/gkaa1100
20. Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. Jvenn: an interactive Venn diagram viewer. BMC Bioinf (2014) 15(1):293. doi: 10.1186/1471-2105-15-293
21. Li R, Li Y, Liang X, Yang L, Su M, Lai KP. Network pharmacology and bioinformatics analyses identify intersection genes of niacin and COVID-19 as potential therapeutic targets. Brief Bioinform (2021) 22:1279–90. doi: 10.1093/bib/bbaa300
22. Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res (2021) 49:605–12. doi: 10.1093/nar/gkaa1074
23. Wang Y, Bryant SH, Cheng T, Wang J, Gindulyte A, Shoemaker BA, et al. PubChem BioAssay: 2017 update. Nucleic Acids Res (2017) 45:955–63. doi: 10.1093/nar/gkw1118
24. Qin X, Huang C, Wu K, Li Y, Liang X, Su M, et al. Anti-coronavirus disease 2019 (COVID-19) targets and mechanisms of puerarin. J Cell Mol Med (2021) 25:677–85. doi: 10.1111/jcmm.16117
25. Hu Z. COVID-19 patients' views and experiences of traditional Chinese medicine treatment in south Africa. Altern Ther Health Med (2022), AT7248.
26. Ma X, Guo Z, Zhang Z, Li X, Wang X, Liu Y, et al. Ferulic acid isolated from propolis inhibits porcine parvovirus replication potentially through bid-mediate apoptosis. Int Immunopharmacol (2020) 83:106379. doi: 10.1016/j.intimp.2020.106379
27. Liu Y, Shi L, Qiu W, Shi Y. Ferulic acid exhibits anti-inflammatory effects by inducing autophagy and blocking NLRP3 inflammasome activation. Mol Cell Toxicol (2022), 1–11. doi: 10.1007/s13273-021-00219-5
28. Wang T, Gong X, Jiang R, Li H, Du W, Kuang G. Ferulic acid inhibits proliferation and promotes apoptosis via blockage of PI3K/Akt pathway in osteosarcoma cell. Am J Transl Res (2016) 8(2):968–80.
29. Atalis A, Keenum MC, Pandey B, Beach A, Pradhan P, Vantucci C, et al. Nanoparticle-delivered TLR4 and RIG-I agonists enhance immune response to SARS-CoV-2 subunit vaccine. J Control Release (2022) 347:476–88. doi: 10.1016/j.jconrel.2022.05.023
30. Luo W, Ding R, Guo X, Zhan T, Tang T, Fan R, et al. Clinical data mining reveals gancao-banxia as a potential herbal pair against moderate COVID-19 by dual binding to IL-6/STAT3. Comput Biol Med (2022) 145:105457. doi: 10.1016/j.compbiomed.2022.105457
31. Lampropoulou DI, Bala VM, Zerva E, Pliakou E, Filippou D, Gazouli M, et al. The potential role of the combined PARP-1 and VEGF inhibition in severe SARS-CoV-2 (COVID-19) infection. J Infect Dev Ctries (2022) 16(1):101–11. doi: 10.3855/jidc.15386
32. Gao J, Yu H, Guo W, Kong Y, Gu L, Li Q, et al. The anticancer effects of ferulic acid is associated with induction of cell cycle arrest and autophagy in cervical cancer cells. Cancer Cell Int (2018) 18:102. doi: 10.1186/s12935-018-0595-y
33. DiNicolantonio JJ, McCarty MF, Assanga SI, Lujan LL, O'Keefe JH. Ferulic acid and berberine, via Sirt1 and AMPK, may act as cell cleansing promoters of healthy longevity. Open Heart (2022) 9(1):e001801. doi: 10.1136/openhrt-2021-001801
34. Bosch-Barrera J, Roqué A, Teixidor E, Carmona-Garcia MC, Arbusà A, Brunet J, et al. Clinical management of COVID-19 in cancer patients with the STAT3 inhibitor silibinin. Pharm (Basel) (2021) 15(1):19. doi: 10.3390/ph15010019
35. Liu Y, Wang D, Li Z, Li X, Jin M, Jia N, et al. Pan-cancer analysis on the role of PIK3R1 and PIK3R2 in human tumors. Sci Rep (2022) 12(1):5924. doi: 10.1038/s41598-022-09889-0
Keywords: osteosarcoma, COVID-19, ferulic acid, autophagy, therapeutic targets
Citation: Pang G, Yi T, Luo H and Jiang L (2022) Preclinical findings: The pharmacological targets and molecular mechanisms of ferulic acid treatment for COVID-19 and osteosarcoma via targeting autophagy. Front. Endocrinol. 13:971687. doi: 10.3389/fendo.2022.971687
Received: 17 June 2022; Accepted: 08 August 2022;
Published: 20 September 2022.
Edited by:
Ka Wu, Nanning Second People’s Hospital, ChinaCopyright © 2022 Pang, Yi, Luo and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihe Jiang, amlhbmdsaWhlQHltdW4uZWR1LmNu; amlhbmdsaWhlQGd4dS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.