
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol., 26 August 2022
Sec. Bone Research
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.964103
This article is part of the Research TopicNew Progress in Understanding and Treatment of OsteoporosisView all 28 articles
Osteoporosis is a systemic degenerative bone disease characterized by low bone mass and damage to bone microarchitecture, which increases bone fragility and susceptibility to fracture. The risk of osteoporosis increases with age; with the aging of the global population, osteoporosis is becoming more prevalent, adding to the societal healthcare burden. Histone modifications such as methylation, acetylation, ubiquitination, and ADP-ribosylation are closely related to the occurrence and development of osteoporosis. This article reviews recent studies on the role of histone modifications in osteoporosis. The existing evidence indicates that therapeutic targeting of these modifications to promote osteogenic differentiation and bone formation may be an effective treatment for this disease.
Osteoporosis is a common skeletal disease characterized by a decrease in bone mass, changes in bone microarchitecture, and increased bone fragility and risk of fracture. Pain, fractures, and other complications of osteoporosis are associated with high rates of death and disability. Bone homeostasis (1), which is maintained under physiologic conditions by a balance between bone formation and resorption during bone remodeling, is critical for ensuring the long-term stability of bone morphology and strength (2). Although the pathogenesis of osteoporosis is not fully understood, an imbalance in bone homeostasis during bone reconstruction whereby bone resorption exceeds bone formation is a major cause (1). The function of osteoblasts and osteoclasts is regulated and influenced by many factors (3–5), and epigenetic studies have provided evidence for the role of histone modifications in the development of osteoporosis (6–8).
Histones H1, H2A, H2B, H3, and H4 are small proteins enriched in positively charged basic amino acids (arginine [R] and lysine [K]) that interact with the negatively charged phosphate groups in DNA and are enveloped by DNA to form nucleosomes, the basic structural unit of chromatin. Histone N-terminal R and K undergo covalent post-transcriptional modifications such as methylation, acetylation, ubiquitination, and ADP-ribosylation that affect histone binding to DNA and alter the structure and state (open vs. closed) of chromatin (9), and also affect the binding of transcription factors at gene promoters to influence gene regulation (10, 11). Histone modifications occur at every stage of development, growth, and aging and are a key aspect of epigenetic regulation that has been linked to the development and progression of multiple diseases (12).
There is increasing evidence that dysregulation of histone modification (methylation, acetylation, ubiquitination, and ADP-ribosylation) and impaired function of related enzymes contribute to the development of osteoporosis. However, molecular-level details of the relationship between these modifications and disease pathogenesis are lacking, and the full clinical significance of histone modifications in osteoporosis remains to be determined (13, 14). Nonetheless, histone modifications may be important for the diagnosis, treatment, and prognosis of osteoporosis and are potential therapeutic targets (15).
In this review, we summarize the current state of knowledge on histone modifications in osteoporosis. Many studies have demonstrated that regulators of histone modifications and their targets function in a complex regulatory network in cells. We discuss the evidence for targeting the regulation of histone modifications as a treatment for osteoporosis, as well as the potential utility of these modifications as disease markers.
Histone methylation usually occurs at R and K residues at the N terminus of histones. K residues can be mono-, di- or trimethylated and R residues can be mono- or dimethylated. Histone methylation positively and negatively regulates gene expression: H3K4me1, H3K4me3, H3K36me3, and H3K79me2 are associated with the activation of gene transcription whereas H3K27me3 and H3K9me3 are associated with transcriptional repression (16). Histone methylation is regulated by histone methyltransferases (HMTs) and histone demethylases (HMDs) (17). Thus, the expression of genes related to bone homeostasis and osteoporosis can be regulated by altering the level of histone methylation (Table 1).
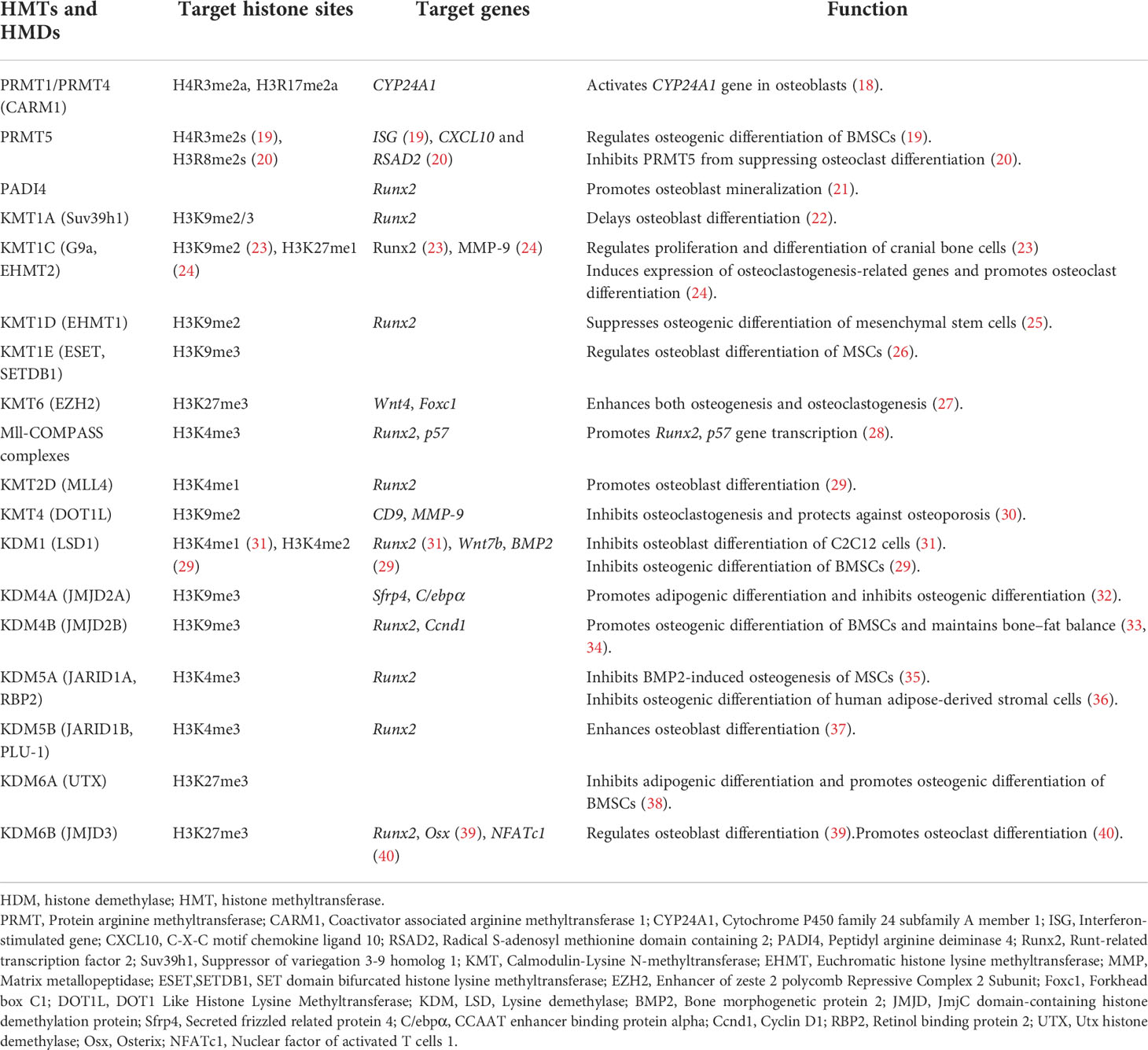
Table 1 Histone methyltransferases, histone demethylases, target histone sites, and their roles in the occurrence and development of osteoporosis.
Several genes are regulated by HMTs or HMDs during the differentiation and maturation of osteoblasts and osteoclasts (6, 14, 15). Suv39h1 is an H3K9 methyltransferase that can modify H3K9 with two or three methyl groups. H3K9me2 and H3K9me3 bind to the promoter of Runx2 (22)—a key gene involved in osteoblast differentiation and maturation (41)—and suppress gene transcription, thereby delaying osteoblast differentiation, which may be relevant to the pathogenesis of osteoporosis. EZH2 is a trimethyltransferase of H3K27; H3K27me3 activates transcription of the Wnt4 gene in osteoblasts to promote osteogenic differentiation (27). H3K27me3 also activates the transcription of Foxc1 in osteoclasts and promotes osteoclast differentiation (27). Thus, histone methylation plays opposing regulatory roles in the pathogenesis of osteoporosis. However, some of the evidence regarding the function of histone methylation in bone homeostasis is controversial. For instance, JMJD2A and JMJD2B are H3K9me3 demethylases; in one study, demethylation of H3K9me3 by JMJD2A promoted adipogenic differentiation and inhibited osteogenic differentiation (32), whereas another study found that H3K9me3 demethylation by JMJD2B promoted osteogenic differentiation of bone marrow-derived stem cells (BMSCs) and maintained bone–fat balance (33).
Acetylation was one of the first histone modifications found to affect transcriptional regulation and is therefore the most widely studied. Acetylation causes K residues in the N-terminal histone tails protruding from nucleosomes to become negatively charged, which repels negatively charged DNA and leads to relaxation of the chromatin structure. The open chromatin conformation allows transcription factors to bind more easily, resulting in an increase in gene expression (42, 43). Thus, histone acetylation is mainly associated with gene activation; it is known to be involved in cell cycle regulation, cell proliferation, and apoptosis, cellular differentiation, DNA replication and repair, nuclear import, and neuronal inhibition (44, 45), whereas dysregulation of histone acetylation has been implicated in osteoporosis progression (46–48).
Histone acetylation is regulated by HATs and HDACs (Table 2). The HAT family includes GNAT (HAT1, GCN5, PCAF) and MYST (Tip60, MOF, MOZ, MORF, HBO1) as well as CREB-binding protein (CBP)/p300, which share very high sequence similarity in the bromodomain, cysteine-histidine-rich region, and HAT structural domain and specifically bind phosphorylated CREB to enhance its transcription of cAMP-responsive genes.

Table 2 Histone acetyltransferases, histone deacetylases, and their roles in the occurrence and development of osteoporosis.
CBP/p300 has a regulatory role in bone formation, targeting transcription factors such as Runx2 during osteoblast differentiation. During parathyroid hormone-induced osteoblast differentiation, phosphorylated HDAC4 dissociates from Runx2, which interacts with CBP/P300 (67). Transforming growth factor beta-1(TGF-β1) and BMP2 stimulate ERK-mediated phosphorylation of Runx2 to promote its interaction with CBP/p300 (68). BMP2 activates SMAD1/5, leading to CBP/p300-mediated acetylation of Runx2, which enhances the expression of osteogenic genes such as alkaline phosphatase(ALP) and collagen type I (COL-I) (69, 70).
Runx2 (41), Sp7 (71), and FoxO1 (72) are important transcription factors for osteoblast differentiation and maturation that induce the transcription of downstream osteogenesis-related genes such as ALP, osteocalcin(OCN), osteopontin(OPN), and COL-1 and promote the maturation and mineralization of osteoblasts. As Runx2 transcription is initiated, PCAF and CBP/p300 acetylate histone H3 to promote osteoblast differentiation and maturation (52, 55). However, the transcription of Runx2, SP7, and FoxO1 was shown to be inhibited after HDAC reduced the histone acetylation level, leading to suppression of osteoblast differentiation and maturation (48, 57, 58, 61, 65) (Table 2). The differentiation and maturation of osteoclasts are controlled by the transcription factors NFATc1 (73) and NF-κB (74), among others. PCAF and CBP/p300 acetylate histone H3 to promote osteoclast differentiation, whereas differentiation is inhibited by HDAC-mediated H3 deacetylation (52, 55) (Table 2). These findings suggest that the balance between the activities of HATs and HDACs is critical for the regulation of transcription factors involved in osteoblast and osteoclast differentiation.
SIRTs are highly conserved HATs that transfer the acetyl group of a substrate to the ADP-ribosyl moiety of nicotinamide adenine dinucleotide (NAD+), NAD+ dependent protein deacetylation consumes NAD+, transfers the acetyl group from the lysine to ADP-ribose to form 2’-O-acetyl-ADPR, nicotinamide, and a deacetylated lysine. The SIRT family comprises SIRT1–7, of which SIRT1, SIRT6, and SIRT7 are mainly localized in the nucleus (75, 76). It should be noted that, most of the discussed Sirtuin effects do not exhibit their function by direct deacetylation of histones, but by deacetylation of other targets. SIRT1 deacetylates H3K9 and regulates of a variety of physiologic processes including metabolism, immune response, and aging (77), and has been linked to osteoporosis (78). The Wnt/β-catenin signaling pathway plays a central role in the differentiation of BMSCs into osteoblasts (79–82); SIRT1 deacetylates K49R or K345R of β-catenin, promoting its entry into the nucleus where it induces the transcription of osteogenic differentiation-related genes such as Cyclin D1 and C-myc and promotes the expression of Runx2 (83). SIRT1 also directly deacetylates Runx2, which in turn induces the transcription of genes that promote osteogenic differentiation of BMSCs (84). peroxisome proliferators-activated receptors gamma(PPARγ) is an important transcription factor for the adipogenic differentiation of BMSCs (85), which inhibits osteogenic differentiation and disrupts bone–fat balance; this is restored by reduction of PPARγ acetylation level by SIRT1 and consequent suppression of the adipogenic differentiation of BMSCs (86, 87) (Figure 1). In pre-oisteoblasts, SIRT1 also down-regulates PPARγ to promote osteogenic differentiation (88). Excessive reactive oxygen species (ROS) production in BMSCs under oxidative stress affects osteogenic differentiation (72). SIRT1 reduces the acetylation level of FoxO3, a transcription factor involved in the cellular response to oxidative stress, leading to FoxO3 transcription and the expression of antioxidant enzymes such as heme oxygenase 1(HO-1) and superoxide dismutase 2(SOD2) (89, 90). The subsequent removal of excess ROS in BMSCs restores their osteogenic differentiation capacity. FoxO3 transcription also promotes the expression of β-catenin, resulting in osteogenic differentiation (90) (Figure 1). Bone resorption depends on ROS produced in osteoclasts; SIRT1 was shown to reduce FoxO acetylation level in bone marrow macrophages(BMMs) and promote the expression of antioxidant enzymes that clear ROS (91). At the same time, SIRT1 reduced TNF-α acetylation level, thereby increasing the expression of transient receptor potential cation channel subfamily V member 1(TRPV1), ROS scavenging, and inhibiting bone resorption (92) (Figure 1).
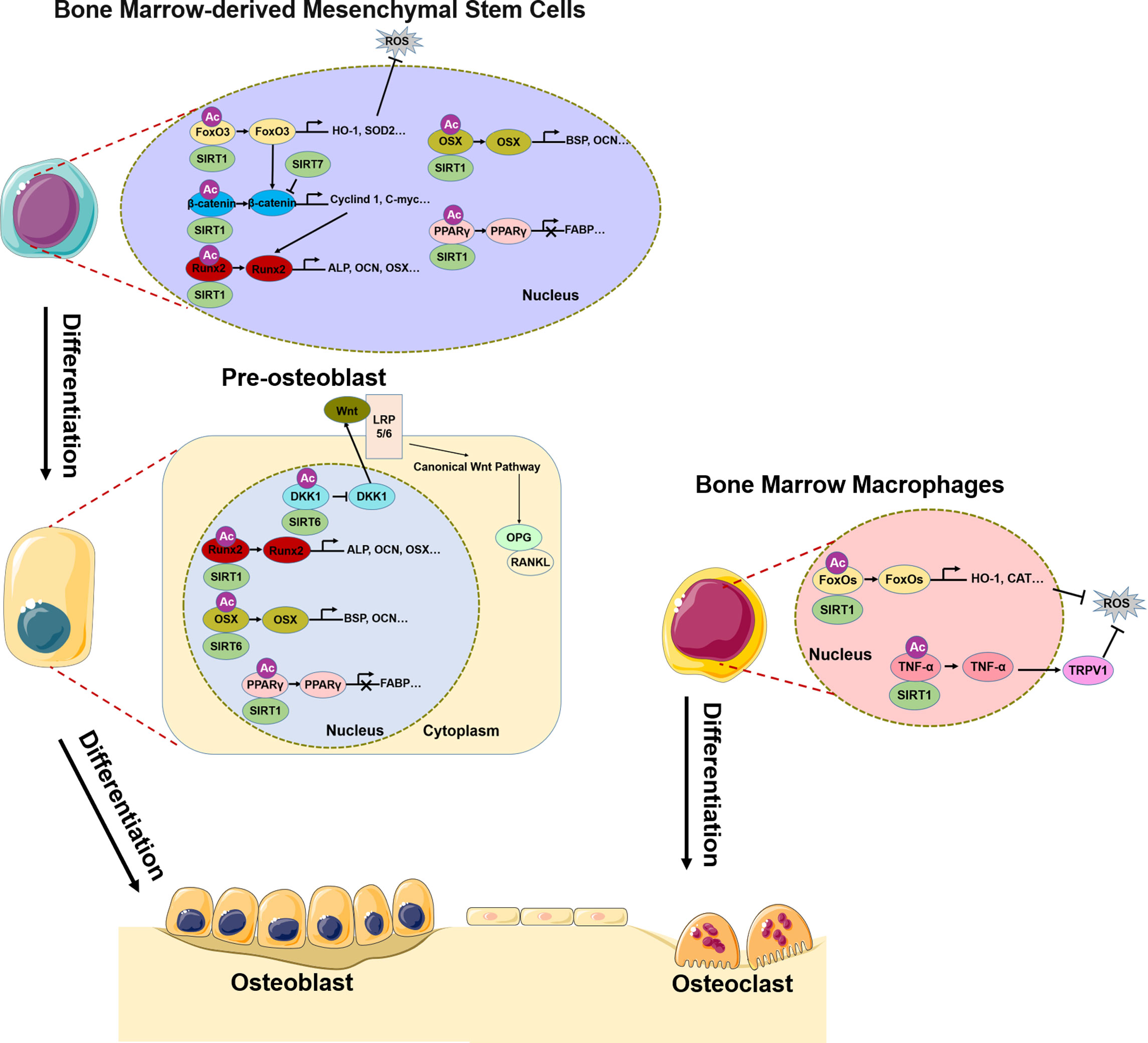
Figure 1 Role of SIRT1/6/7 in bone remodeling. In BMSCs, SIRT1, SIRT6, and SIRT7 promote osteogenic differentiation by regulating transcription factors FoxO3, β-catenin, Runx2, Osx, and PPARγ. In pre-osteoblasts, SIRT6 not only regulates Runx2 and Osx transcription, but also inhibits DKK1 transcription, activates canonical Wnt signaling, and promotes osteogenic differentiation. In BMMs, SIRT1 inhibits osteoclast differentiation by regulating FoxO and TNF-α transcription, which results in ROS scavenging.
SIRT6 and SIRT7 are involved in chromatin regulation and play multiple roles in metabolism, aging, and disease. In osteoblasts, SIRT6 reduces H3K9 acetylation level and promotes the transcription of osteogenic transcription factors Runx2 and Osx and the expression of osteogenic genes (93). Additionally, SIRT6 negatively regulates the expression of dickkopf1 (DKK1) (93), a secreted protein that binds to the Wnt receptor LRP5/6; this induces rapid endocytosis and reduces LRP5/6 in the cell membrane (94), thereby blocking the canonical Wnt signaling cascade. Blocking the Wnt pathway leads to a reduction in the synthesis of osteoprotegerin (OPG), which competitively binds to RANKL to inhibit osteoblast differentiation (95). Thus, reducing DKK1 expression facilitates the nuclear entry of β-catenin, which initiates the transcription of target genes that promote osteoblast differentiation while inhibiting those involved in osteoblast differentiation. Additionally, SIRT7 has the effect of reducing β-catenin level in BMSCs (96), although the significance of this observation in the context of bone homeostasis and osteoporosis remains unclear.
All histones can be ubiquitinated, with H2A and H2B being the most frequent targets. Histone ubiquitination plays a central role in the DNA damage response. Monoubiquitination of H2A, H2B, and H2AX has been observed at DNA double-strand break sites; the most common forms are monoubiquitination of K119 on H2A and K123 (yeast)/K120 (vertebrate) on H2B. H2A and H2B monoubiquitination was shown to be associated with gene silencing and transcriptional activation, respectively (97, 98).
Ring finger protein 40 (RNF40), an E3 ubiquitin ligase that monoubiquitinates H2B (99), was found to regulate the transcription of the osteogenic genes bone gamma-carboxyglutamate protein (BGLAP), ALP, and glucose-6-phosphate dehydrogenase (G6PD) to induce osteogenic differentiation of human BMSCs (100). The Tnfsf11 gene (encoding RANKL) is a target gene of H2Bub1 (101). It was reported that H2Bub1, whose expression was induced by RNF40, was required in the early stages of osteoblast differentiation and modulated osteoblast function by regulating VDR-induced Tnfsf11 expression in crosstalk between osteoblasts. The long noncoding RNA ODIR1 was significantly downregulated during osteogenic differentiation of human umbilical cord mesenchymal stem cells (hUC-MSCs); the interaction of ODIR1 with F-box protein 25 (FBXO25) increased monoubiquitination of H2BK120 (H2BK120ub), promoted the trimethylation of H3K4 (H3K4me3), and induced the expression of the transcription factor Osx, thereby enhancing the expression of the osteoblast markers OCN, OPN, and ALP. Thus, ODIR1 negatively regulates the osteogenic differentiation of hUC-MSCs via the FBXO25/H2BK120ub/H3K4me3/Osx axis (102).
Myb like, SWIRM and MPN domains 1(Mysm1) is an H2A deubiquitinating enzyme that regulates osteoblast differentiation and maturation by promoting Runx2 expression in osteoblasts. Mysm1−/− mice exhibit significant skeletal deformation and osteoporosis; however, osteogenic differentiation capacity was not significantly affected in MSCs lacking Mysm1. In p53−/−Mysm1−/− double knockout mice, p53 deletion rescued the skeletal defects and bone loss caused by Mysm1 deficiency. On the other hand, loss of p53 did not restore Runx2 expression in Mysm1−/− osteoblasts although MSCs proliferation and osteogenic differentiation was enhanced (103).
Poly (ADP-Ribose) polymerases (PARPs) (also known as ARTDs), are a family of 17 proteins, some of PARPs are mono-ADP-ribosyl transferases and some poly. PARPs uses NAD+ as substrate to transfer single or straight or branched ADP-ribose to itself or other target proteins, thus regulating various cellular responses. The most widely studied member of the PARPs family is PARP1, which is thought to play a role in DNA repair (104). PARP1 binds to DNA damage sites and catalyzes its own ADP-ribosylation reactions and the trans-modification of local substrate proteins, including DNA repair proteins, histones and other chromatin-associated proteins, to promote the repair of DNA lesions, influence chromatin structure and gene transcription (105, 106).
Induction of NFATc1 by macrophage colony-stimulating factor (M-CSF) and RANKL is essential for macrophage differentiation into osteoblasts (73). ADP-ribosylation of H2B at serine 7 by PARP1 reduced the occupancy of this histone at the NFATc1 promoter, reducing NFATc1 expression and osteoclast formation (107). Moreover, PARP1 inhibited the expression of osteoclast-promoting genes via regulation of histone ADP-ribosylation at the IL-1β promoter, which increased IL-1β expression (108). M-CSF induced PARP1 self–ADP-ribosylation in macrophages, resulting in PARP1 cleavage at D214 and its subsequent degradation; this stimulated RANKL-induced osteoclast differentiation and osteoclast maturation, whereas osteoclastogenesis was inhibited by expression of the cleavage-resistant D214N mutant form of PARP1 (109). The PARP inhibitor olaparib decreased ALP activity in preosteoblastic MC3T3-E1 cells and inhibited the formation of the mineralized nodules that characterize osteoblasts (110). In contrast, inhibiting the enzymatic activity of poly(ADP-Ribose) glycohydrolase (PARG) using the inhibitor PDD00017273 enhanced Runx2 PARylation (ribosylation) and osteoblast formation while having no effect on PARG mRNA expression.
Osteoporosis is a complex disease whose pathogenesis remains unclear, although an imbalance in bone homeostasis during bone reconstruction is thought to contribute. Accumulating evidence indicates that histone modifications play an essential role in various diseases including osteoporosis. Specifically, changes in the levels of histone modification and related enzymes can lead to altered expression of genes involved in bone formation or bone resorption, resulting in an imbalance in bone homeostasis and abnormal bone reconstruction.
According to this review, it can be proved that histone modifications play important role in osteoporosis, and related research results have been applied to the diagnosis and treatment of osteoporosis. Identifying specific biomarkers associated with osteoporosis will significantly improve the clinical diagnosis and treatment of this disease. In recent years, drugs targeting the activity of histone-modifying enzymes have been evaluated in studies focused on osteoporosis treatment. As an example, SOST—which negatively regulates bone formation—is regulated by a class I HDAC (111); meanwhile, the class I HDAC inhibitor MS-275 was shown to promote bone formation (112).
There have been a limited number of studies on the effects of histone modification on bone homeostasis, leaving many open questions. For example, the relationship between abnormal regulation of histone modifications in genes related to bone homeostasis and the development of osteoporosis requires clarification. It is unclear whether histone modifications are a primary cause of osteoporosis or a secondary effect, although it is clear that the effects of histone modifications are associated with the pathogenesis of osteoporosis. More in-depth mechanistic studies are expected to provide a better understanding of the effect of histone modification on individual bone cell type by using tissue-specific deletion and transgenic animal models. Further studies in this area can provide insight not only into disease pathogenesis, but also novel diagnostic biomarkers and therapeutic targets.
PS and TH performed the literature review and wrote the manuscript. CH consulted and summarized the literature. YW provided supervision and advisory during the writing process. DT reviewed the literature and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported in part by the National Natural Science Foundation of China (81730107, 81973883), the Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (ZYYCXTD-C-202202), the National Key R&D Program of China (2018YFC1704300), the Shanghai Scientific Research Project (19ZR1458000), the Shanghai Traditional Chinese Medicine Medical Center of Chronic Disease (2017ZZ01010), and the Innovation Team of the Ministry of Education (IRT1270).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kenkre JS, Bassett J. The bone remodelling cycle. Ann Clin Biochem (2018) 55(3):308–27. doi: 10.1177/0004563218759371
2. Intemann J, De Gorter DJJ, Naylor AJ, Dankbar B, Wehmeyer C. Importance of osteocyte-mediated regulation of bone remodelling in inflammatory bone disease. Swiss Med Weekly (2020) 150:w20187. doi: 10.4414/smw.2020.20187
3. Fang H, Deng Z, Liu J, Chen S, Deng Z, Li W. The mechanism of bone remodeling after bone aging. Clin Interv Aging (2022) 17:405–15.
4. Seely KD, Kotelko CA, Douglas H, Bealer B, Brooks AE. The human gut microbiota: A key mediator of osteoporosis and osteogenesis. Int J Mol Sci (2021) 22(17):9452. doi: 10.3390/ijms22179452
5. Song S, Guo Y, Yang Y, Fu D. Advances in pathogenesis and therapeutic strategies for osteoporosis. Pharmacol Ther (2022) 237:108168. doi: 10.1016/j.pharmthera.2022.108168
6. Xu F, Li W, Yang X, Na L, Chen L, Liu G. The roles of epigenetics regulation in bone metabolism and osteoporosis. Front Cell Dev Biol (2020) 8:619301. doi: 10.3389/fcell.2020.619301
7. Avilkina V, Chauveau C, Ghali Mhenni O. Sirtuin function and metabolism: Role in pancreas, liver, and adipose tissue and their crosstalk impacting bone homeostasis. Bone (2022) 154:116232. doi: 10.1016/j.bone.2021.116232
8. Zhao L, Yin B, Ma Q, Shi Y, Wang C, Ye L. The involvement of histone methylation in the osteogenic differentiation of mesenchymal stem cells. Curr Stem Cell Res Ther (2021) 16(8):915–23. doi: 10.2174/1574888X16666210203110354
9. Liu CF, Abnousi A, Bazeley P, Ni Y, Morley M, Moravec CS, et al. Global analysis of histone modifications and long-range chromatin interactions revealed the differential cistrome changes and novel transcriptional players in human dilated cardiomyopathy. J Mol Cell Cardiol (2020) 145:30–42. doi: 10.1016/j.yjmcc.2020.06.001
10. Morgan MAJ, Shilatifard A. Reevaluating the roles of histone-modifying enzymes and their associated chromatin modifications in transcriptional regulation. Nat Genet (2020) 52(12):1271–81. doi: 10.1038/s41588-020-00736-4
11. Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol (2015) 16(3):178–89. doi: 10.1002/wdev.109
12. Saul D, Kosinsky RL. Epigenetics of aging and aging-associated diseases. Int J Mol Sci (2021) 22(1):401. doi: 10.3390/ijms22010401
13. Ghayor C, Weber FE. Epigenetic regulation of bone remodeling and its impacts in osteoporosis. Int J Mol Sci (2016) 17(9):1446. doi: 10.3390/ijms17091446
14. Vrtacnik P, Marc J, Ostanek B. Epigenetic mechanisms in bone. Clin Chem Lab Med (2014) 52(5):589–608. doi: 10.1515/cclm-2013-0770
15. Holroyd C, Harvey N, Dennison E, Cooper C. Epigenetic influences in the developmental origins of osteoporosis. Osteoporos Int (2012) 23(2):401–10. doi: 10.1007/s00198-011-1671-5
16. Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol (2007) 8(12):983–94. doi: 10.1038/nrm2298
17. Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet (2012) 13(5):343–57. doi: 10.1038/nrg3173
18. Moena D, Merino P, Lian JB, Stein GS, Stein JL, Montecino M. Switches in histone modifications epigenetically control vitamin D3-dependent transcriptional upregulation of the CYP24A1 gene in osteoblastic cells. J Cell Physiol (2020) 235(6):5328–39. doi: 10.1002/jcp.29420
19. Kota SK, Roening C, Patel N, Kota SB, Baron R. PRMT5 inhibition promotes osteogenic differentiation of mesenchymal stromal cells and represses basal interferon stimulated gene expression. Bone (2018) 117:37–46. doi: 10.1016/j.bone.2018.08.025
20. Dong Y, Song C, Wang Y, Lei Z, Xu F, Guan H, et al. Inhibition of PRMT5 suppresses osteoclast differentiation and partially protects against ovariectomy-induced bone loss through downregulation of CXCL10 and RSAD2. Cell Signal (2017) 34:55–65. doi: 10.1016/j.cellsig.2017.03.004
21. Zhai Q, Zhao Y, Wang L, Dai Y, Zhao P, Xiang X, et al. CircRNA hsa_circ_0008500 acts as a miR-1301-3p sponge to promote osteoblast mineralization by upregulating PADI4. Front Cell Dev Biol (2020) 8:602731. doi: 10.3389/fcell.2020.602731
22. Kim HJ, Park JW, Lee KH, Yoon H, Shin DH, Ju UI, et al. Plant homeodomain finger protein 2 promotes bone formation by demethylating and activating Runx2 for osteoblast differentiation. Cell Res (2014) 24(10):1231–49. doi: 10.1038/cr.2014.127
23. Kim K, Shin Y, Kim J, Ulmer TS, An W. H3K27me1 is essential for MMP-9-dependent H3N-terminal tail proteolysis during osteoclastogenesis. Epigenet Chromatin (2018) 11(1):23. doi: 10.1186/s13072-018-0193-1
24. Ideno H, Nakashima K, Komatsu K, Araki R, Abe M, Arai Y, et al. G9a is involved in the regulation of cranial bone formation through activation of Runx2 function during development. Bone (2020) 137:115332. doi: 10.1016/j.bone.2020.115332
25. Huang H, Dou L, Song J, Luo J. CBFA2T2 is required for BMP-2-induced osteogenic differentiation of mesenchymal stem cells. Biochem Biophys Res Commun (2018) 496(4):1095–101. doi: 10.1016/j.bbrc.2018.01.144
26. Lawson KA, Teteak CJ, Gao J, Li N, Hacquebord J, Ghatan A, et al. ESET histone methyltransferase regulates osteoblastic differentiation of mesenchymal stem cells during postnatal bone development. FEBS Lett (2013) 587(24):3961–7. doi: 10.1016/j.febslet.2013.10.028
27. Cao X, He W, Rong K, Xu S, Chen Z, Liang Y, et al. DZNep promotes mouse bone defect healing via enhancing both osteogenesis and osteoclastogenesis. Stem Cell Res Ther (2021) 12(1):605. doi: 10.1186/s13287-021-02670-6
28. Rojas A, Sepulveda H, Henriquez B, Aguilar R, Opazo T, Nardocci G, et al. Mll-COMPASS complexes mediate H3K4me3 enrichment and transcription of the osteoblast master gene Runx2/p57 in osteoblasts. J Cell Physiol (2019) 234(5):6244–53. doi: 10.1002/jcp.27355
29. Munehira Y, Yang Z, Gozani O. Systematic analysis of known and candidate lysine demethylases in the regulation of myoblast differentiation. J Mol Biol (2017) 429(13):2055–65. doi: 10.1016/j.jmb.2016.10.004
30. Gao Y, Ge W. The histone methyltransferase DOT1L inhibits osteoclastogenesis and protects against osteoporosis. Cell Death Dis (2018) 9(2):33. doi: 10.1038/s41419-017-0040-5
31. Sun J, Ermann J, Niu N, Yan G, Yang Y, Shi Y, et al. Histone demethylase LSD1 regulates bone mass by controlling WNT7B and BMP2 signaling in osteoblasts. Bone Res (2018) 6:14. doi: 10.1038/s41413-018-0015-x
32. Qi Q, Wang Y, Wang X, Yang J, Xie Y, Zhou J, et al. Histone demethylase KDM4A regulates adipogenic and osteogenic differentiation via epigenetic regulation of C/EBPalpha and canonical wnt signaling. Cell Mol Life Sci (2020) 77(12):2407–21. doi: 10.1007/s00018-019-03289-w
33. Deng P, Yuan Q, Cheng Y, Li J, Liu Z, Liu Y, et al. Loss of KDM4B exacerbates bone-fat imbalance and mesenchymal stromal cell exhaustion in skeletal aging. Cell Stem Cell (2021) 28(6):1057–73 e7. doi: 10.1016/j.stem.2021.01.010
34. Ye L, Fan Z, Yu B, Chang J, Al Hezaimi K, Zhou X, et al. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell (2012) 11(1):50–61. doi: 10.1016/j.stem.2012.04.009
35. Wang C, Wang J, Li J, Hu G, Shan S, Li Q, et al. KDM5A controls bone morphogenic protein 2-induced osteogenic differentiation of bone mesenchymal stem cells during osteoporosis. Cell Death Dis (2016) 7(8):e2335. doi: 10.1038/cddis.2016.238
36. Ge W, Shi L, Zhou Y, Liu Y, Ma GE, Jiang Y, et al. Inhibition of osteogenic differentiation of human adipose-derived stromal cells by retinoblastoma binding protein 2 repression of RUNX2-activated transcription. Stem Cells (2011) 29(7):1112–25. doi: 10.1002/stem.663
37. Rojas A, Aguilar R, Henriquez B, Lian JB, Stein JL, Stein GS, et al. Epigenetic control of the bone-master Runx2 gene during osteoblast-lineage commitment by the histone demethylase JARID1B/KDM5B. J Biol Chem (2015) 290(47):28329–42. doi: 10.1074/jbc.M115.657825
38. Hemming S, Cakouros D, Isenmann S, Cooper L, Menicanin D, Zannettino A, et al. EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells (2014) 32(3):802–15. doi: 10.1002/stem.1573
39. Yang D, Okamura H, Nakashima Y, Haneji T. Histone demethylase Jmjd3 regulates osteoblast differentiation via transcription factors Runx2 and osterix. J Biol Chem (2013) 288(47):33530–41. doi: 10.1074/jbc.M113.497040
40. Yasui T, Hirose J, Tsutsumi S, Nakamura K, Aburatani H, Tanaka S. Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1. J Bone Miner Res (2011) 26(11):2665–71. doi: 10.1002/jbmr.464
41. Komori T. Whole aspect of Runx2 functions in skeletal development. Int J Mol Sci (2022) 23(10):5776. doi: 10.3390/ijms23105776
42. Verdone L, Agricola E, Caserta M, Di Mauro E. Histone acetylation in gene regulation. Brief Funct Genomic Proteomic (2006) 5(3):209–21. doi: 10.1093/bfgp/ell028
43. Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol (2014) 15(11):703–8. doi: 10.1038/nrm3890
44. Peleg S, Feller C, Ladurner AG, Imhof A. The metabolic impact on histone acetylation and transcription in ageing. Trends Biochem Sci (2016) 41(8):700–11. doi: 10.1016/j.tibs.2016.05.008
45. Arora I, Sharma M, Sun LY, Tollefsbol TO. The epigenetic link between polyphenols, aging and age-related diseases. Genes (2020) 11(9):1094. doi: 10.3390/genes11091094
46. Guo Q, Kang H, Wang J, Dong Y, Peng R, Zhao H, et al. Inhibition of ACLY leads to suppression of osteoclast differentiation and function Via regulation of histone acetylation. J Bone Miner Res (2021) 36(10):2065–80. doi: 10.1002/jbmr.4399
47. Hu M, Xing L, Zhang L, Liu F, Wang S, Xie Y, et al. NAP1L2 drives mesenchymal stem cell senescence and suppresses osteogenic differentiation. Aging Cell (2022) 21(2):e13551. doi: 10.1111/acel.13551
48. Ma C, Gao J, Liang J, Dai W, Wang Z, Xia M, et al. HDAC6 inactivates Runx2 promoter to block osteogenesis of bone marrow stromal cells in age-related bone loss of mice. Stem Cell Res Ther (2021) 12(1):484. doi: 10.1186/s13287-021-02545-w
49. Jing H, Su X, Gao B, Shuai Y, Chen J, Deng Z, et al. Epigenetic inhibition of wnt pathway suppresses osteogenic differentiation of BMSCs during osteoporosis. Cell Death Dis (2018) 9(2):176. doi: 10.1038/s41419-017-0231-0
50. Liu Y, Cheng W, Zhao Y, Gao L, Chang Y, Tong Z, et al. Cyclic mechanical strain regulates osteoblastic differentiation of mesenchymal stem cells on TiO2 nanotubes through GCN5 and wnt/beta-catenin. Front Bioeng Biotechnol (2021) 9:735949. doi: 10.3389/fbioe.2021.735949
51. Zhang P, Liu Y, Jin C, Zhang M, Tang F, Zhou Y. Histone acetyltransferase GCN5 regulates osteogenic differentiation of mesenchymal stem cells by inhibiting NF-kappaB. J Bone Miner Res (2016) 31(2):391–402. doi: 10.1002/jbmr.2704
52. Zhang P, Liu Y, Jin C, Zhang M, Lv L, Zhang X, et al. Histone H3K9 acetyltransferase PCAF is essential for osteogenic differentiation through bone morphogenetic protein signaling and may be involved in osteoporosis. Stem Cells (2016) 34(9):2332–41. doi: 10.1002/stem.2424
53. Lian WS, Ko JY, Chen YS, Ke HJ, Hsieh CK, Kuo CW, et al. MicroRNA-29a represses osteoclast formation and protects against osteoporosis by regulating PCAF-mediated RANKL and CXCL12. Cell Death Dis (2019) 10(10):705. doi: 10.1038/s41419-019-1942-1
54. Kim JH, Kim K, Youn BU, Jin HM, Kim JY, Moon JB, et al. RANKL induces NFATc1 acetylation and stability via histone acetyltransferases during osteoclast differentiation. Biochem J (2011) 436(2):253–62. doi: 10.1042/BJ20110062
55. Krishnan RH, Sadu L, Das UR, Satishkumar S, Pranav Adithya S, Saranya I, et al. Role of p300, a histone acetyltransferase enzyme, in osteoblast differentiation. Differentiation (2022) 124:43–51. doi: 10.1016/j.diff.2022.02.002
56. Bachagol D, Joseph GS, Ellur G, Patel K, Aruna P, Mittal M, et al. Stimulation of liver IGF-1 expression promotes peak bone mass achievement in growing rats: a study with pomegranate seed oil. J Nutr Biochem (2018) 52:18–26. doi: 10.1016/j.jnutbio.2017.09.023
57. Zhu X, Yu J, Du J, Zhong G, Qiao L, Lin J. LncRNA HOXA-AS2 positively regulates osteogenesis of mesenchymal stem cells through inactivating NF-kappaB signalling. J Cell Mol Med (2019) 23(2):1325–32. doi: 10.1111/jcmm.14034
58. Dou C, Li N, Ding N, Liu C, Yang X, Kang F, et al. HDAC2 regulates FoxO1 during RANKL-induced osteoclastogenesis. Am J Physiol Cell Physiol (2016) 310(10):C780–C7. doi: 10.1152/ajpcell.00351.2015
59. Molstad DHH, Mattson AM, Begun DL, Westendorf JJ, Bradley EW. Hdac3 regulates bone modeling by suppressing osteoclast responsiveness to RANKL. J Biol Chem (2020) 295(51):17713–23. doi: 10.1074/jbc.RA120.013573
60. Nakatani T, Partridge NC. MEF2C interacts with c-FOS in PTH-stimulated Mmp13 gene expression in osteoblastic cells. Endocrinology (2017) 158(11):3778–91. doi: 10.1210/en.2017-00159
61. Li J, Liu C, Li Y, Zheng Q, Xu Y, Liu B, et al. TMCO1-mediated Ca(2+) leak underlies osteoblast functions via CaMKII signaling. Nat Commun (2019) 10(1):1589. doi: 10.1038/s41467-019-09653-5
62. Wein MN, Spatz J, Nishimori S, Doench J, Root D, Babij P, et al. HDAC5 controls MEF2C-driven sclerostin expression in osteocytes. J Bone Miner Res (2015) 30(3):400–11. doi: 10.1002/jbmr.2381
63. Pham L, Kaiser B, Romsa A, Schwarz T, Gopalakrishnan R, Jensen ED, et al. HDAC3 and HDAC7 have opposite effects on osteoclast differentiation. J Biol Chem (2011) 286(14):12056–65. doi: 10.1074/jbc.M110.216853
64. Jensen ED, Schroeder TM, Bailey J, Gopalakrishnan R, Westendorf JJ. Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. J Bone Miner Res (2008) 23(3):361–72. doi: 10.1359/jbmr.071104
65. Fu Y, Zhang P, Ge J, Cheng J, Dong W, Yuan H, et al. Histone deacetylase 8 suppresses osteogenic differentiation of bone marrow stromal cells by inhibiting histone H3K9 acetylation and RUNX2 activity. Int J Biochem Cell Biol (2014) 54:68–77. doi: 10.1016/j.biocel.2014.07.003
66. Xiao H, Wu Z, Li B, Shangguan Y, Stoltz JF, Magdalou J, et al. The low-expression programming of 11beta-HSD2 mediates osteoporosis susceptibility induced by prenatal caffeine exposure in male offspring rats. Br J Pharmacol (2020) 177(20):4683–700. doi: 10.1111/bph.15225
67. Shimizu E, Nakatani T, He Z, Partridge NC. Parathyroid hormone regulates histone deacetylase (HDAC) 4 through protein kinase a-mediated phosphorylation and dephosphorylation in osteoblastic cells. J Biol Chem (2014) 289(31):21340–50. doi: 10.1074/jbc.M114.550699
68. Gomathi K, Rohini M, Partridge NC, Selvamurugan N. Regulation of transforming growth factor-beta1-stimulation of Runx2 acetylation for matrix metalloproteinase 13 expression in osteoblastic cells. Biol Chem (2022) 403(3):305–15. doi: 10.1515/hsz-2021-0292
69. Jeon EJ, Lee KY, Choi NS, Lee MH, Kim HN, Jin YH, et al. Bone morphogenetic protein-2 stimulates Runx2 acetylation. J Biol Chem (2006) 281(24):16502–11. doi: 10.1074/jbc.M512494200
70. Jun JH, Yoon WJ, Seo SB, Woo KM, Kim GS, Ryoo HM, et al. BMP2-activated Erk/MAP kinase stabilizes Runx2 by increasing p300 levels and histone acetyltransferase activity. J Biol Chem (2010) 285(47):36410–9. doi: 10.1074/jbc.M110.142307
71. Hojo H, Ohba S. Sp7 action in the skeleton: Its mode of action, functions, and relevance to skeletal diseases. Int J Mol Sci (2022) 23(10):5647. doi: 10.3390/ijms23105647
72. Ma X, Su P, Yin C, Lin X, Wang X, Gao Y, et al. The roles of FoxO transcription factors in regulation of bone cells function. Int J Mol Sci (2020) 21(3):692. doi: 10.3390/ijms21030692
73. Kang JY, Kang N, Yang Y-M, Hong JH, Shin DM. The role of Ca2+-NFATc1 signaling and its modulation on osteoclastogenesis. Int J Mol Sci (2020) 21(10):3646. doi: 10.3390/ijms21103646
74. Yao Z, Getting SJ, Locke IC. Regulation of TNF-induced osteoclast differentiation. Cells (2021) 11(1):132. doi: 10.3390/cells11010132
75. Wang Y, He J, Liao M, Hu M, Li W, Ouyang H, et al. An overview of sirtuins as potential therapeutic target: Structure, function and modulators. Eur J Med Chem (2019) 161:48–77. doi: 10.3390/cells11010132
76. Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun (2016) 7:12235. doi: 10.1038/ncomms12235
77. Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol (2010) 5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250
78. Li Q, Cheng JC, Jiang Q, Lee WY. Role of sirtuins in bone biology: Potential implications for novel therapeutic strategies for osteoporosis. Aging Cell (2021) 20(2):e13301. doi: 10.1111/acel.13301
79. Matsushita Y, Nagata M, Kozloff KM, Welch JD, Mizuhashi K, Tokavanich N, et al. A wnt-mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration. Nat Commun (2020) 11(1):332. doi: 10.1038/s41467-019-14029-w
80. Yu S, Zhu K, Lai Y, Zhao Z, Fan J, Im HJ, et al. atf4 promotes beta-catenin expression and osteoblastic differentiation of bone marrow mesenchymal stem cells. Int J Biol Sci (2013) 9(3):256–66. doi: 10.7150/ijbs.5898
81. Zhou T, Gao B, Fan Y, Liu Y, Feng S, Cong Q, et al. Piezo1/2 mediate mechanotransduction essential for bone formation through concerted activation of NFAT-YAP1-ß-catenin. eLife (2020) 9:e52779. doi: 10.7554/eLife.52779
82. Zhu Y, Wang Y, Jia Y, Xu J, Chai Y. Catalpol promotes the osteogenic differentiation of bone marrow mesenchymal stem cells via the wnt/β-catenin pathway. Stem Cell Res Ther (2019) 10(1):37. doi: 10.1186/s13287-019-1143-y
83. Simic P, Zainabadi K, Bell E, Sykes DB, Saez B, Lotinun S, et al. SIRT1 regulates differentiation of mesenchymal stem cells by deacetylating beta-catenin. EMBO Mol Med (2013) 5(3):430–40. doi: 10.1002/emmm.201201606
84. Shakibaei M, Shayan P, Busch F, Aldinger C, Buhrmann C, Lueders C, et al. Resveratrol mediated modulation of sirt-1/Runx2 promotes osteogenic differentiation of mesenchymal stem cells: potential role of Runx2 deacetylation. PloS One (2012) 7(4):e35712. doi: 10.1371/journal.pone.0035712
85. Evseeva MN, Balashova MS, Kulebyakin KY, Rubtsov YP. Adipocyte biology from the perspective of in vivo research: Review of key transcription factors. Int J Mol Sci (2021) 23(1):322. doi: 10.3390/ijms23010322
86. Han L, Zhou R, Niu J, McNutt MA, Wang P, Tong T. SIRT1 is regulated by a PPARγ–SIRT1 negative feedback loop associated with senescence. Nucleic Acids Res (2010) 38(21):7458–71. doi: 10.1093/nar/gkq609
87. Bäckesjö C-M, Li Y, Lindgren U, Haldosén L-A. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Mineral Res (2006) 21(7):993–1002. doi: 10.1359/jbmr.060415
88. Qu B, Ma Y, Yan M, Gong K, Liang F, Deng S, et al. Sirtuin1 promotes osteogenic differentiation through downregulation of peroxisome proliferator-activated receptor gamma in MC3T3-E1 cells. Biochem Biophys Res Commun (2016) 478(1):439–45. doi: 10.1016/j.bbrc.2016.06.154
89. Sun W, Qiao W, Zhou B, Hu Z, Yan Q, Wu J, et al. Overexpression of Sirt1 in mesenchymal stem cells protects against bone loss in mice by FOXO3a deacetylation and oxidative stress inhibition. Metabolism (2018) 88:61–71. doi: 10.1016/j.metabol.2018.06.006
90. Iyer S, Han L, Bartell SM, Kim HN, Gubrij I, de Cabo R, et al. Sirtuin1 (Sirt1) promotes cortical bone formation by preventing beta-catenin sequestration by FoxO transcription factors in osteoblast progenitors. J Biol Chem (2014) 289(35):24069–78. doi: 10.1074/jbc.M114.561803
91. Kim H-N, Han L, Iyer S, de Cabo R, Zhao H, O'Brien CA, et al. Sirtuin1 suppresses osteoclastogenesis by deacetylating FoxOs. Mol Endocrinol (2015) 29(10):1498–509. doi: 10.1210/me.2015-1133
92. Yan S, Miao L, Lu Y, Wang L. Sirtuin 1 inhibits TNF-alpha-mediated osteoclastogenesis of bone marrow-derived macrophages through both ROS generation and TRPV1 activation. Mol Cell Biochem (2019) 455(1-2):135–45. doi: 10.1007/s11010-018-3477-7
93. Sugatani T, Agapova O, Malluche HH, Hruska KA. SIRT6 deficiency culminates in low-turnover osteopenia. Bone (2015) 81:168–77. doi: 10.1016/j.bone.2015.07.018
94. Glantschnig H, Scott K, Hampton R, Wei N, McCracken P, Nantermet P, et al. A rate-limiting role for dickkopf-1 in bone formation and the remediation of bone loss in mouse and primate models of postmenopausal osteoporosis by an experimental therapeutic antibody. J Pharmacol Exp Ther (2011) 338(2):568–78. doi: 10.1124/jpet.111.181404
95. Diarra D, Stolina M, Polzer K, Zwerina J, Ominsky MS, Dwyer D, et al. Dickkopf-1 is a master regulator of joint remodeling. Nat Med (2007) 13(2):156–63. doi: 10.1038/nm1538
96. Chen EEM, Zhang W, Ye CCY, Gao X, Jiang LLJ, Zhao TTF, et al. Knockdown of SIRT7 enhances the osteogenic differentiation of human bone marrow mesenchymal stem cells partly via activation of the wnt/beta-catenin signaling pathway. Cell Death Dis (2017) 8(9):e3042. doi: 10.1038/cddis.2017.429
97. Chen Z, Djekidel MN, Zhang Y. Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos. Nat Genet (2021) 53(4):551–63. doi: 10.1038/s41588-021-00821-2
98. Wang Y, Yang L, Zhang X, Cui W, Liu Y, Sun QR, et al. Epigenetic regulation of ferroptosis by H2B monoubiquitination and p53. EMBO Rep (2019) 20(7):e47563. doi: 10.15252/embr.201847563
99. Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biochem Sci (2012) 37(2):58–65. doi: 10.1016/j.tibs.2011.11.001
100. Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, Konig A, et al. The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol Cell (2012) 46(5):705–13. doi: 10.1016/j.molcel.2012.05.022
101. Najafova Z, Liu P, Wegwitz F, Ahmad M, Tamon L, Kosinsky RL, et al. RNF40 exerts stage-dependent functions in differentiating osteoblasts and is essential for bone cell crosstalk. Cell Death Differ (2021) 28(2):700–14. doi: 10.1038/s41418-020-00614-w
102. He S, Yang S, Zhang Y, Li X, Gao D, Zhong Y, et al. LncRNA ODIR1 inhibits osteogenic differentiation of hUC-MSCs through the FBXO25/H2BK120ub/H3K4me3/OSX axis. Cell Death Dis (2019) 10(12):947. doi: 10.1038/s41419-019-2148-2
103. Haffner-Luntzer M, Kovtun A, Fischer V, Prystaz K, Hainzl A, Kroeger CM, et al. Loss of p53 compensates osteopenia in murine Mysm1 deficiency. FASEB J (2018) 32(4):1957–68. doi: 10.1096/fj.201700871R
104. Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res (2006) 34(21):6170–82. doi: 10.1093/nar/gkl840
105. Langelier MF, Zandarashvili L, Aguiar PM, Black BE, Pascal JM. NAD(+) analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat Commun (2018) 9(1):844. doi: 10.1038/s41467-018-03234-8
106. Kraus WL, Lis JT. PARP goes transcription. Cell (2003) 113(6):677–83. doi: 10.1016/s0092-8674(03)00433-1
107. Wang C, Xiao J, Nowak K, Gunasekera K, Alippe Y, Speckman S, et al. PARP1 hinders histone H2B occupancy at the NFATc1 promoter to restrain osteoclast differentiation. J Bone Miner Res (2020) 35(4):776–88. doi: 10.1002/jbmr.3927
108. Robaszkiewicz A, Qu C, Wisnik E, Ploszaj T, Mirsaidi A, Kunze FA, et al. ARTD1 regulates osteoclastogenesis and bone homeostasis by dampening NF-κB-dependent transcription of IL-1β. Sci Rep (2016) 6(1):21131. doi: 10.1038/srep21131
109. Wang C, Qu C, Alippe Y, Bonar SL, Civitelli R, Abu-Amer Y, et al. Poly-ADP-ribosylation-mediated degradation of ARTD1 by the NLRP3 inflammasome is a prerequisite for osteoclast maturation. Cell Death Dis (2016) 7:e2153. doi: 10.1038/cddis.2016.58
110. Sasaki Y, Nakatsuka R, Inouchi T, Masutani M, Nozaki T. Inhibition of poly (ADP-ribose) glycohydrolase accelerates osteoblast differentiation in preosteoblastic MC3T3-E1 cells. Int J Mol Sci (2022) 23(9):5041. doi: 10.3390/ijms23095041
111. Baertschi S, Baur N, Lueders-Lefevre V, Voshol J, Keller H. Class I and IIa histone deacetylases have opposite effects on sclerostin gene regulation. J Biol Chem (2014) 289(36):24995–5009. doi: 10.1074/jbc.M114.564997
Keywords: osteoporosis, histone modification, osteoblast, osteoclast, differentiation
Citation: Sun P, Huang T, Huang C, Wang Y and Tang D (2022) Role of histone modification in the occurrence and development of osteoporosis. Front. Endocrinol. 13:964103. doi: 10.3389/fendo.2022.964103
Received: 08 June 2022; Accepted: 09 August 2022;
Published: 26 August 2022.
Edited by:
Gordon L. Klein, University of Texas Medical Branch at Galveston, United StatesReviewed by:
Baicheng He, Chongqing Medical University, ChinaCopyright © 2022 Sun, Huang, Huang, Wang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongjun Wang, eWp3YW5nODg4OEAxMjYuY29t; Dezhi Tang, ZHp0YW5nNzAyQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.