
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol., 12 October 2022
Sec. Diabetes: Molecular Mechanisms
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.953631
This article is part of the Research TopicProgression to Diabetes: Molecular and Cellular MechanismsView all 10 articles
 Meng Li1†
Meng Li1† Siqian Gong1†
Siqian Gong1† Xueyao Han1‡
Xueyao Han1‡ Lingli Zhou1
Lingli Zhou1 Simin Zhang1
Simin Zhang1 Qian Ren1
Qian Ren1 Xiaoling Cai1
Xiaoling Cai1 Yingying Luo1
Yingying Luo1 Wei Liu1
Wei Liu1 Yu Zhu1
Yu Zhu1 Xianghai Zhou1
Xianghai Zhou1 Yufeng Li2
Yufeng Li2 Linong Ji1*‡
Linong Ji1*‡Objectives: Mitochondrial DNA (mtDNA) plays an important role in the pathogenesis of diabetes. Variants in mtDNA have been reported in diabetes, but studies on the whole mtDNA variants were limited. Our study aims to explore the association of whole mtDNA variants with diabetes and diabetic kidney disease (DKD).
Methods: The whole mitochondrial genome was screened by next-generation sequencing in cohort 1 consisting of 50 early-onset diabetes (EOD) patients with a maternally inherited diabetes (MID) family history. A total of 42 variants possibly associated with mitochondrial diseases were identified according to the filtering strategy. These variants were sequenced in cohort 2 consisting of 90 EOD patients with MID. The association between the clinical phenotype and these variants was analyzed. Then, these variants were genotyped in cohort 3 consisting of 1,571 type 2 diabetes mellitus patients and 496 subjects with normal glucose tolerance (NGT) to analyze the association between variants with diabetes and DKD.
Results: Patients with variants in the non-coding region had a higher percentage of obesity and levels of fasting insulin (62.1% vs. 24.6%, P = 0.001; 80.0% vs. 26.5% P < 0.001). The patients with the variants in rRNA had a higher prevalence of obesity (71.4% vs. 30.3%, P = 0.007), and the patients with the variants in mitochondrial complex I had a higher percentage of the upper tertile of FINS (64.3% vs. 34.3%, P = 0.049). Among 20 homogeneous variants successfully captured, two known variants (m.A3943G, m.A10005G) associated with other mitochondrial diseases were only in the diabetic group, but not in the NGT group, which perhaps indicated its possible association with diabetes. The prevalence of DKD was significantly higher in the group with the 20 variants than those without these variants (18.7% vs. 14.6%, P = 0.049) in the participants with diabetes of cohort 3.
Conclusion: MtDNA variants are associated with MID and DKD, and our findings advance our understanding of mtDNA in diabetes and DKD. It will have important implications for the individual therapy of mitochondrial diabetes.
Mitochondrial diseases are a clinically heterogeneous cluster of inherited metabolic disease that could affect the function of the mitochondrial respiratory chain and oxidative phosphorylation system (1). Diabetes is the most well-described form of endocrine dysfunction in mitochondrial diseases. The mitochondrial genome is a maternally inherited 16,569-bp circular double-stranded DNA molecule that contains two ribosomal RNA genes, 22 transfer RNA (tRNA) genes, and 13 genes for the proteins of the oxidative phosphorylation system (2). Mitochondria are important for the synthesis of ATP and synthesis of the hormone. The mitochondrial DNA (mtDNA) variants possibly lead to the lack of ATP and impair hormone production when mitochondrial dysfunction occurs. In the process of insulin synthesis, mitochondria not only provide the necessary ATP for insulin exocytosis but also have a core function in glucose sensing and the induction of triggering and amplifying signals that adjust insulin secretion to glycaemia. Variants in the mitochondrial genomes are related to a cluster of metabolic diseases, including mitochondrial diabetes, insulin resistance (IR), non-alcoholic fatty liver disease (NAFLD), and metabolic syndrome (MetS) (3–6). Single mtDNA variants are associated with type 2 diabetes mellitus (T2DM). An mtDNA genome-wide association analysis indicated a potential role for mtDNA in diabetic kidney disease (DKD). As we know, the m.A3243G variant is the most common heteroplasmic mtDNA variant associated with mitochondrial diabetes. The m.A3243G variant reduced the synthesis of mtDNA-encoded proteins, which resulted in an imbalance within the mitochondrion between nuclear and mitochondrial encoded oxidative phosphorylation subunits (7, 8). Multisystemic disease manifestations can precede the onset of diabetes in patients with mitochondrial diseases (9). For example, sensorineural deafness and a family history of maternal inheritance should raise suspicion that the patient might have a mitochondrial disease. Other mtDNA variants associated with diabetes include point variants in the tRNA genes MT−TL1, MT−TK, MT−TS2, and MT−TE, as well as the variants in MT−ND6, which encodes the NADH-ubiquinone oxidoreductase chain 6 (10–12). Furthermore, IR also plays a key role in the development of T2DM and DKD. There is a clear evidence of an association between skeletal muscle IR and abnormalities in mitochondrial oxidative metabolism (13). However, most studies just focused on the association among single mtDNA loci and diabetes and DKD, and studies on the whole mtDNA gene with diabetes and DKD were limited. In addition, high-throughput sequencing could enhance the ability to identify mitochondrial respiratory chain complex deficiencies (14). Therefore, the study was designed as follows: to screen the whole mitochondrial gene variants in early-onset diabetes with the maternal family history using next-generation sequencing (NGS) and then to explore the mtDNA variants with diabetes and DKD. The study aims to help us elucidate novel insights into mitochondrial genetic etiology.
This study was conducted in compliance with the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of Peking University People’s Hospital. Written informed consent was obtained from all the study participants. A flowchart of the study is shown in Figure 1. The study was divided into three groups (1): The whole mitochondrial genome was screened by NGS in cohort 1 consisting of 50 early-onset diabetes (EOD) patients. The variants possibly associated with mitochondrial diseases were identified according to the filtering strategy. (2) The above variants were sequenced in cohort 2 with 90 EOD patients, and the clinical phenotypic features of patients with potential pathogenic variants were analyzed. (3) Cohort 3 was a case–control study that included 1,571 patients with T2DM and 496 healthy individuals with normal glucose tolerance (NGT) to analyze the different features of the patients with variants and without these variants.
Cohort 2 with 50 unrelated Chinese patients with EOD who had a maternally inherited diabetes (MID) family history were included from the inpatient and outpatient populations of the Department of Endocrinology and Metabolism at Peking University People’s Hospital from January 2013 to December 2017. Then, 40 unrelated EOD patients with MID were recruited to identify the potential disease-associated variants. Cohort 2 included the above 90 patients. All the patients were of northern Han Chinese ancestry and residents of Beijing. They have been diagnosed with diabetes according to the 1999 World Health Organization criteria (15). The inclusion criteria for the EOD patients were as follows: (1) diagnosed with T2DM ≤40 years, (2) patients with the typical clinical features of type 1 diabetes or other specific forms of diabetes (e.g., chronic pancreatitis) and those who were positive for anti-glutamic acid decarboxylase antibodies, anti-islet cell antibodies were excluded, and (3) without the m. A3243G variant. The clinical information of 90 EOD patients was collected, including demographics, initial presentation, the treatment of diabetes, physical examination results, and laboratory test results, which are summarized at the time of enrollment in Table 1. The patients were diagnosed as being overweight or obese, based on the criteria: overweight was defined as a body mass index (BMI) ≥24 kg/m2 and <28 kg/m2, and obesity was defined as a BMI ≥28 kg/m2. Coronary heart disease (CHD) was determined as a history of the disease. Cerebrovascular disease was determined as a history of transient ischemic attack or ischemic or hemorrhagic stroke. Hypertension was determined as a systolic blood pressure ≥140 mmHg and/or diastolic blood pressure ≥90 mmHg or the current treatment of hypertension. Dyslipidemia was defined as total cholesterol ≥5.18 mmol/L, triglycerides ≥1.70 mmol/L, LDL-c ≥3.37 mmol/L, HDL-c1.3 mmol/L in women or HDL-c<1.04 mmol/L in men, or treatment with antihyperlipidemic agents (16). Diabetic kidney disease (DKD) was defined as having a urinary albumin-to-creatinine ratio ≥30 mg/g or an estimated glomerular filtration rate (eGFR) <60 ml/min/1.73 m2. Patients diagnosed as having a urinary tract infection, other glomerular diseases, or gross hematuria were excluded. Homeostasis model assessment (HOMA) was used to evaluate β-cell function and insulin sensitivity. IR index (HOMA-IR) = [fasting insulin (Fins, mIU/L) × fasting plasma glucose (FPG, mmol/L)]/22.5, and β-cell function (HOMA-β) = [Fins (mIU/L) × 20]/[FPG (mmol/L) — 3.5] (17). When assessing the levels of HOMA-IR and HOMA-β, we excluded patients who were using insulin. MetS was assessed based on the International Diabetes Federation (IDF 2005) criteria: the diagnosis of central obesity (WC ≥ 90 cm in men or ≥ 80 cm in women), plus any two of the following four factors: (1) TG levels ≥ 1.7 mmol/L, (2) HDL-C < 1.03 mmol/L in men or < 1.29 mmol/L in women, (3) SBP ≥ 130 mmHg or DBP ≥ 85 mmHg or previous diagnosis with hypertension, (4) FBG ≥ 5.6 mmol/L, or previous diagnosis with type 2 diabetes mellitus (18).
DNA was extracted from peripheral blood using standard procedures. The whole mitochondrial DNA of 50 patients in cohort 1 was used with NGS. For them, the Agilent Sureselect XT2 capture chip was used for the mitochondrial gene (accession number: NC_012920.1). A HiSeq 2500 Sequencing System (Illumina, San Diego, CA, USA) was adopted for sequencing. Sequencing data were obtained with the target region coverage (~100%) and the sequencing depth on target > 2,000. All rare variants of mtDNA were validated by Sanger sequencing.
The criteria for selecting the mtDNA variants possibly associated with mitochondrial diseases from the NGS data were as follows: (1) variant frequency in the cohort 1 was <20%; (2) sequencing depth >400 ×; (3) we searched the variants in the MITOMAP database (https://www.mitomap.org/MITOMAP) and selected the mtDNA variants that were possibly disease related based on the database. (4) The potential disease-related variants were validated by Sanger sequencing.
The potential disease-related variants screened and identified from cohort 1 were verified in cohort 2 by Sanger sequencing. PCR amplification was performed using PCR Enzyme Mix. The 24 pairs of mitochondrial primers are shown in Supplementary Table 1. The conditions for the PCR were as follows: initial denaturation at 94°C for 1 min, followed by 35 cycles of denaturation at 94°C for 30 s, primer annealing for 45 s at 61°C, and primer extension at 72°C for 2 min. A final extension was at 72°C for 5 min. The direct sequencing of PCR products was purified using a gel extraction kit (Axygen, California, USA) and performed with the ABI 3730xl DNA sequencer (Applied Biosystems) (19).
The sequencing data from cohort 2 were analyzed. The pathogenicity of these variants in the coding area and tRNA were evaluated by HmtVar (https://www.hmtvar.uniba.it/). The pathogenicity of non-synonymous CDS variants were classified into four categories: (1) polymorphic: disease score (DS) < 0.43, allele frequency (AF) > 0.003264; (2) likely polymorphic: DS < 0.43, AF ≤ 0.003264; (3) likely pathogenic: DS ≥ 0.43, AF > 0.003264; and (4) pathogenic: DS ≥ 0.43, AF ≤ 0.003264. The pathogenicity of tRNA variants was classified into four categories: (1) polymorphic: DS < 0.35, AF > 0.005020; (2) likely polymorphic: DS < 0.35, AF ≤ 0.005020; (3) likely pathogenic: DS ≥ 0.35, AF > 0.005020; (4) pathogenic: DS ≥ 0.35, AF ≤ 0.005020. The DS was evaluated according to the software of MutPred, Polyphen2, Panther, PhD SNP, and SNPs & GO.
A total of 1,571 T2DM patients and 496 healthy individuals with NGT were also enrolled in the case–control study. The inclusion criteria for the control samples were as follows: normal glucose tolerance confirmed via a 75 g oral glucose tolerance test according to the 1999 World Health Organization criteria (15); at the age of over 40; and hemoglobin A1c (HbA1c) <6%. Their clinical features are also summarized in Table 2.
The above selected mtDNA variants were screened in the case–control cohort (cohort 3) using high-throughput genotyping on a MassARRAY platform (Sequenom) to genotype the mtDNA variants. Duplicate DNA samples and wild genotypes confirmed by sequencing were simultaneously genotyped as quality controls.
Statistical tests were performed with the SPSS software 23.0 (Chicago, IL, USA). Continuous variables are presented as the means and standard deviations (means ± standard deviations) for normally distributed data or as medians (interquartile range) for non-normally distributed data. Categorical variables are presented as numbers and percentages. Student’s t-tests, Fisher’s exact tests, and chi-square tests were performed to compare the clinical difference. The Mann–Whitney rank-sum test was used to make comparisons for the non-normally distributed variable. When we analyzed the metabolic features between patients with 42 variants and without those variants in cohort 2 and cohort 3, the levels of FINS, HOMA-IR, and HOMA-β were grouped according to the tertiles. An ordered-classification chi-square test was used to compare the differences between groups. Logistic regression analysis was conducted to address the relation between variables. P < 0.05 was considered statistically significant.
The clinical characteristics of the enrolled participants are shown in Table 1. The whole mitochondrial genome was screened by NGS in cohort 1. In addition, the cohort 2 including 90 EOD patients was sequenced to analyze the variants possibly associated with metabolic diseases. The 90 patients had an MID family history, of which 59 (65.6%) were men and the average age at diagnosis was 30.2 ± 6.0 years. The mean BMI was 26.9 ± 4.2 kg/m2, and the average HbA1c was 8.5 ± 2.1%. The clinical characteristic features of cohort 3 are shown in Table 2.
Through the whole mitochondrial genome sequencing of 50 patients with MID, the mean coverage of the target genes was above 95.0%, and the mean depth was above 400 ×. In total, 474 variants on mtDNA were identified, including 225 variants in the coding region and 249 variants in the non-coding region. Among the coding region, 150 synonymous variants, one non-sense variant, and 74 non-synonymous variants were identified. According to the filtering strategy in the method, 42 mtDNA variants are identified in Table 3. Then, these 42 mtDNA variants were screened in cohort 2, and the frequency of these variants in cohort 2 are displayed in Table 3. Among them, seven variants were located in the D-loop, four variants in MT-ND1, two variants in MT-ND2, one in MT-CO2, two in MT-ATP6, two variants in MT-ND3, three variants in MT-ND4, five variants in MT-ND5, and one variant in MT-ND6. In addition, three variants were in MT-CYB, and six variants in tRNA, and six variants were in rRNA. The above 42 variants were located in complex I, complex III, complex IV, complex V and a non-coding area (including ND6 and D-loop), tRNA, and rRNA.
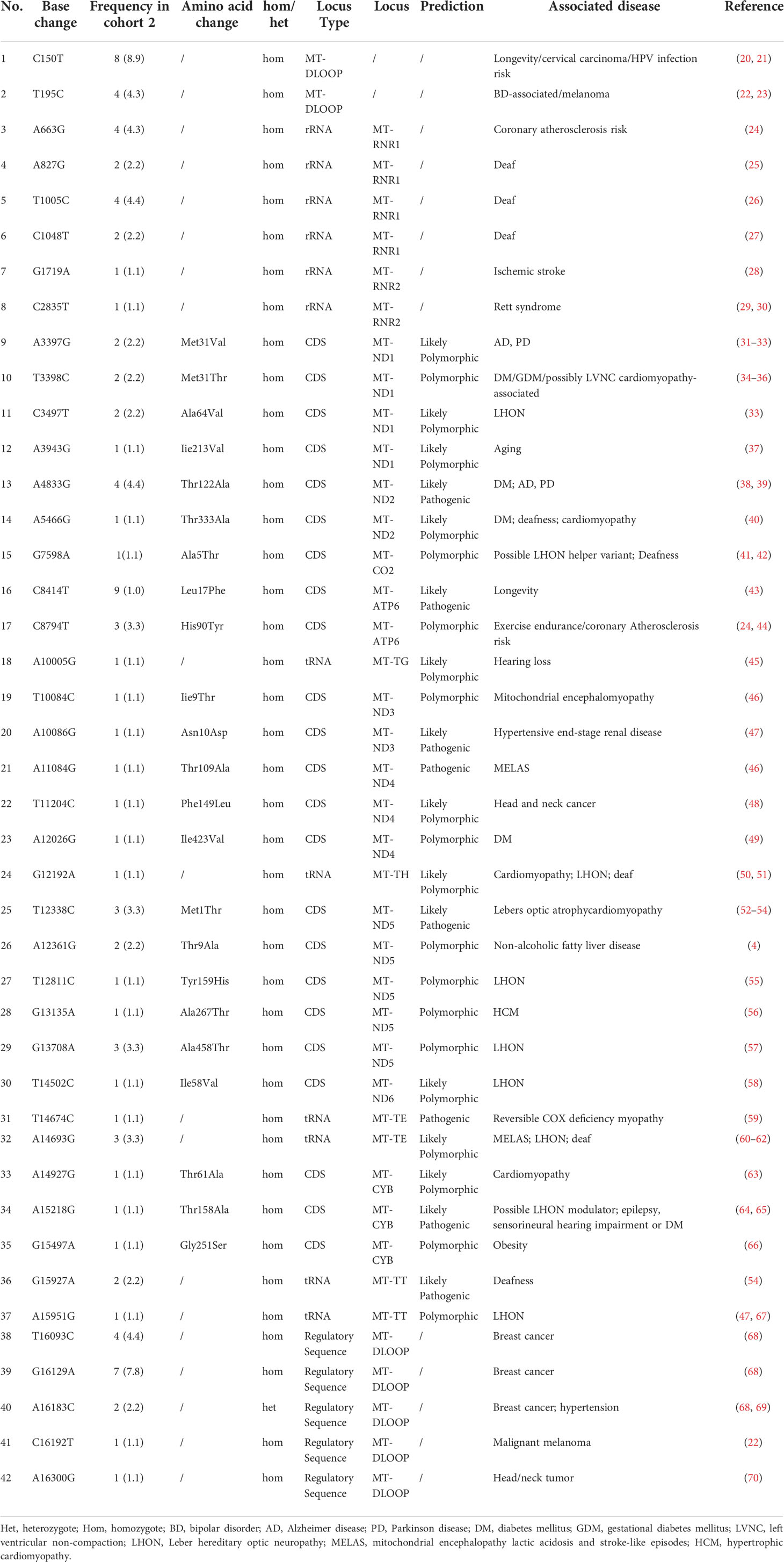
Table 3 The rare mitochondrial DNA (mtDNA) variants from cohort 2 possibly associated with mitochondrial disease (n=42).
Among the 42 mtDNA variants, five variants (m.T3398C, m.A4833G, m.A5466G, m.A12026G, m.A15218G) have been reported to be possibly associated with diabetes (Figure 2).
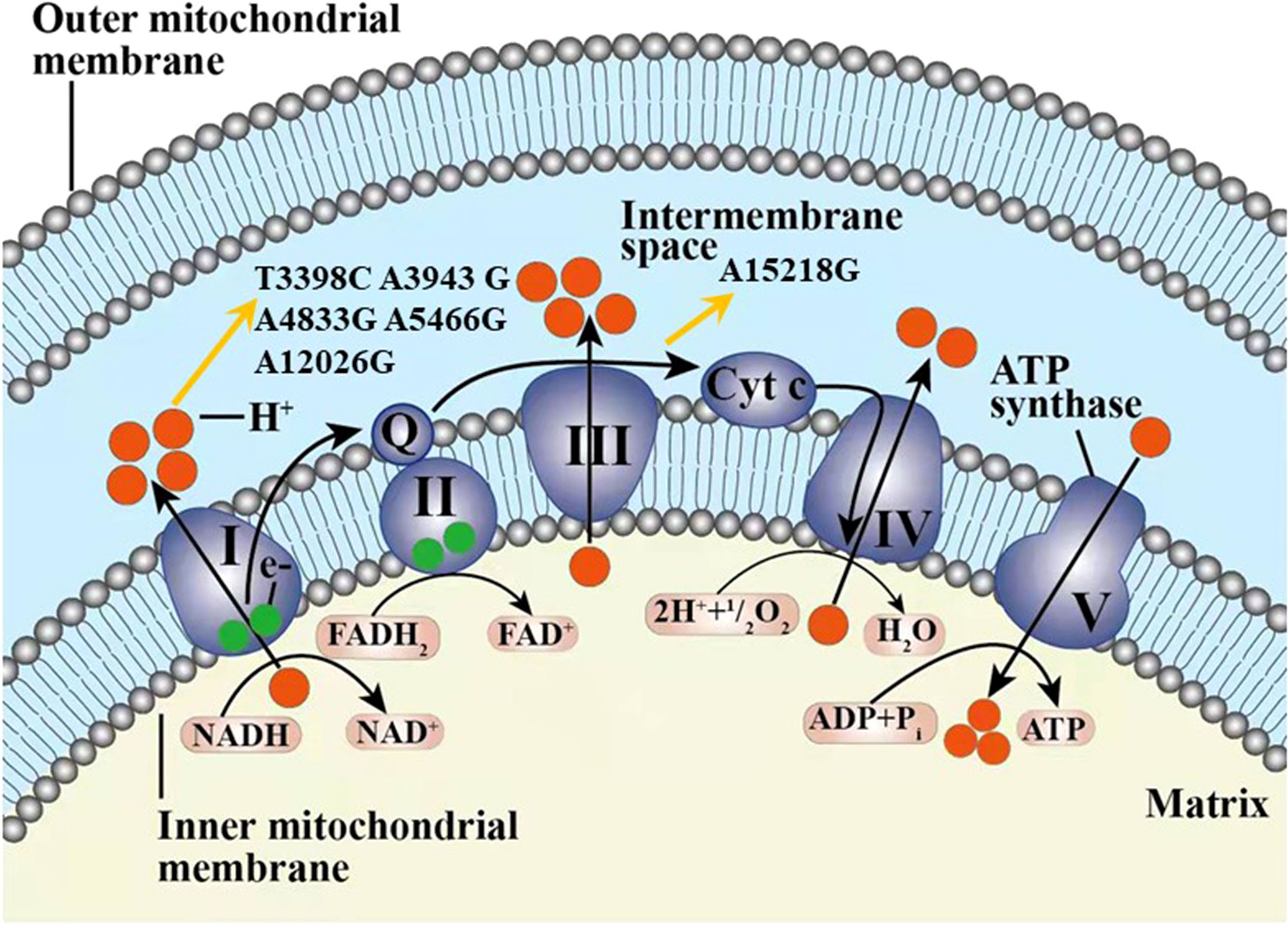
Figure 2 The six variants from cohort 2 and one variant from cohort 3 found in our study possibly associated with diabetes in our study displayed in the electron transport chain of mitochondria, including T3398C, A3943G, A4833G, A5466G, A12026G, and A15218G. The complexes I-V marked as I-V were included in the respiratory chain and oxidative phosphorylation system. Q, coenzyme Q10; Cyt c, cytochrome c; e-, electron.
Compared with the patients without 42 variants, the patients with these variants had higher levels of FINS and HOMA-IR with a significant difference [14.6 (10.0, 26.7) vs. 8.1 (5.8, 13.4), P = 0.006; 6.5 (3.2, 10.2) vs. 3.3 (1.8, 4.3), P = 0.002]. There was no difference between the two groups of BMI, waist circumference, and HbA1c. In addition, the prevalence of DKD was significantly higher in the group with the variants than those without the variants (48.4% vs. 20.0%, P = 0.001) in Table 4.
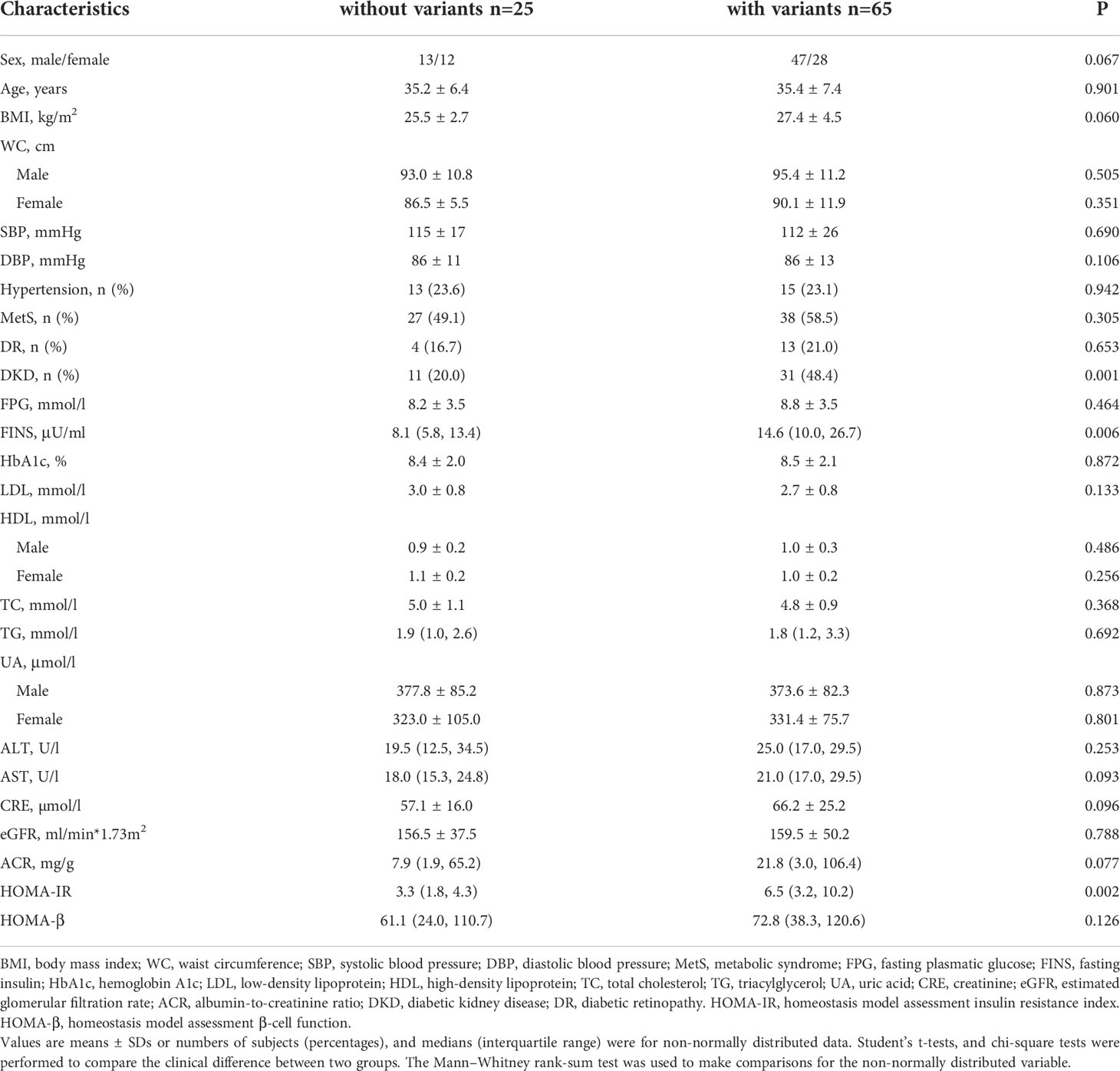
Table 4 Comparison of subjects with the 42 variants of mtDNA with the subjects without those variants in cohort 2.
A total of 42 mtDNA variants were in different domains of mitochondria including complex I, complex III, complex IV, complex V, non-coding region, rRNA, and tRNA. Patients with variants in the non-coding region had a higher percentage of obesity and levels of FINS than those without these variants with significant difference (62.1% vs. 24.6%, P = 0.001; 80.0% vs. 26.5% P < 0.001). The patients with the variants in rRNA had a higher prevalence of obesity (71.4% vs. 30.3%, P = 0.007), and the patients with the variants in complex I had a higher level of FINS (64.3% vs. 34.3%, P = 0.049). There was no significant difference on the frequency of different BMIs and levels of FINS, HOMA-IR, and HOMA-β of the patients with variants in other domains of mitochondria in Tables 5, 6.
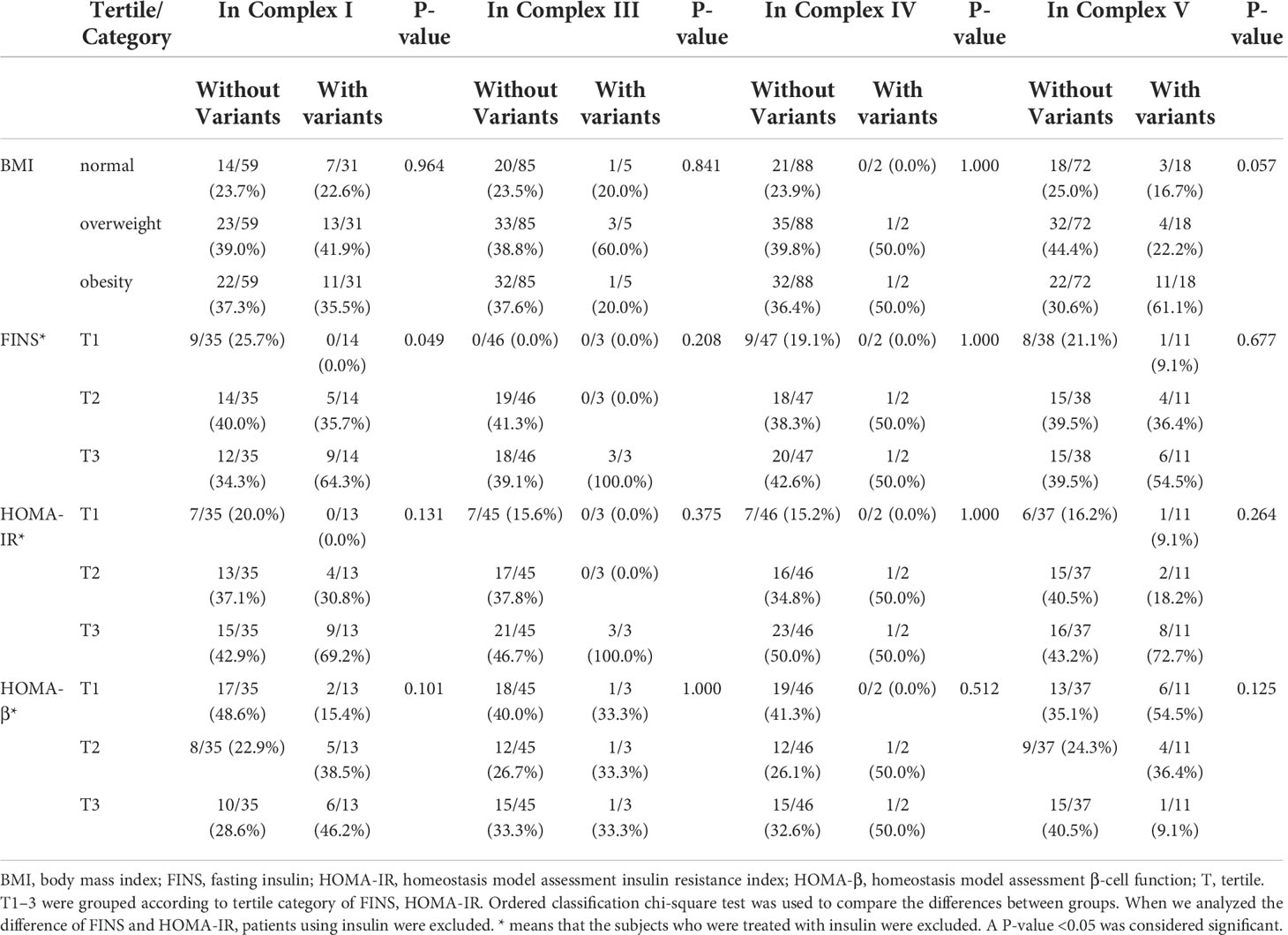
Table 5 Comparison of body mass index (BMI) and other traits among the patients with variants and without these variants in the complex I, III, IV, and V domains of mitochondria.
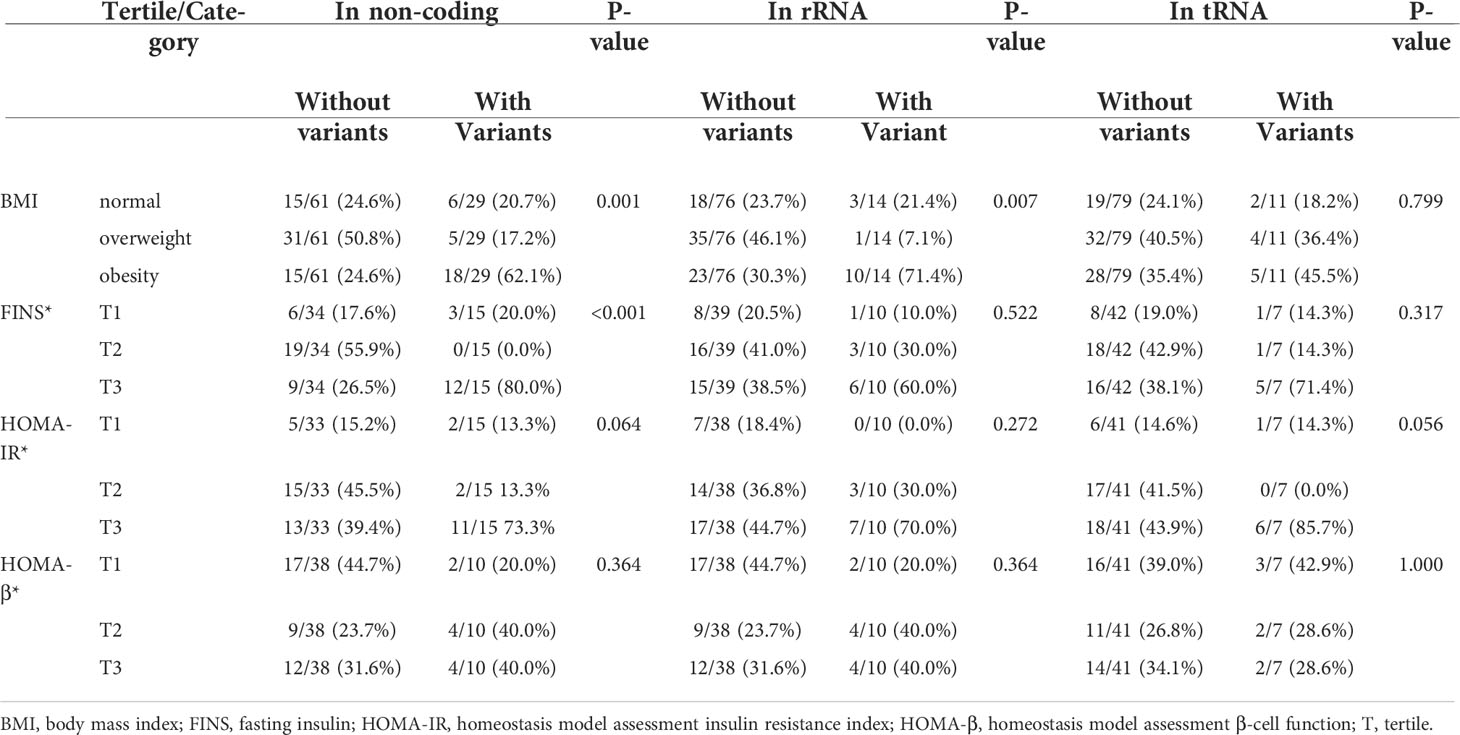
Table 6 Comparison of BMI and other traits among the patients with variants and without these variants in the non-coding region, transfer RNA, and rRNA of mitochondria.
In cohort 3, 42 mtDNA variants were screened using high-throughput genotyping, to investigate the phenotype and genotype of mtDNA variants. After the sequencing, 20 homogeneous variants associated with mitochondrial diseases were successfully captured and met the criteria of quality control in Table 7 and Figure 3. Excluding the variant of m.C3497T, there was no significant difference on the frequency of the mtDNA variants in the diabetic group and NGT group. We further enlarged 200 NGT subjects to screen the m.C3497T variant and found that there was no significant difference between the two groups.
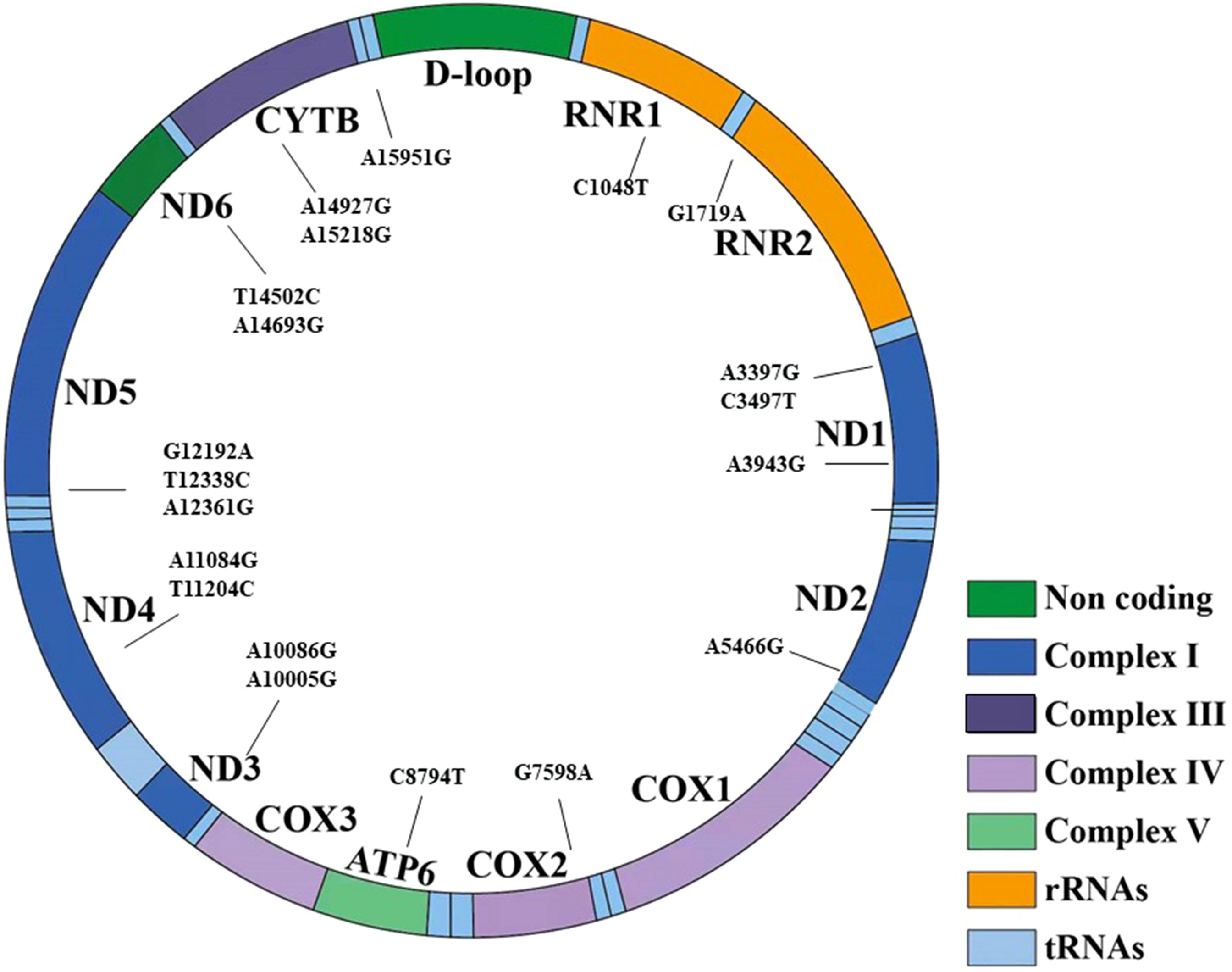
Figure 3 The 20 variants have been shown in the structure of the mitochondrial genes and different domains. It included seven regions: non-coding region, complex I, complex III, complex IV, complex V, transfer RNA, and rRNA.
Among the 20 homogeneous variants, two variants of m.A3943G (Figure 3) and m.A10005G were just found in the diabetic group and did not exist in the NGT group. The detailed clinical features are shown in Supplementary Table 2.
In the diabetic group, the levels of FINS, BMI, HOMA-IR, and HOMA-β were similar between the group with the 20 variants and those without the variants. The prevalence of MetS was similar in the two groups (56.8% vs. 57.2%, P = 0.886). The prevalence of DKD was significantly higher in the group with the variants than those without the variants (18.7% vs. 14.6%, P = 0.049) in Table 8.
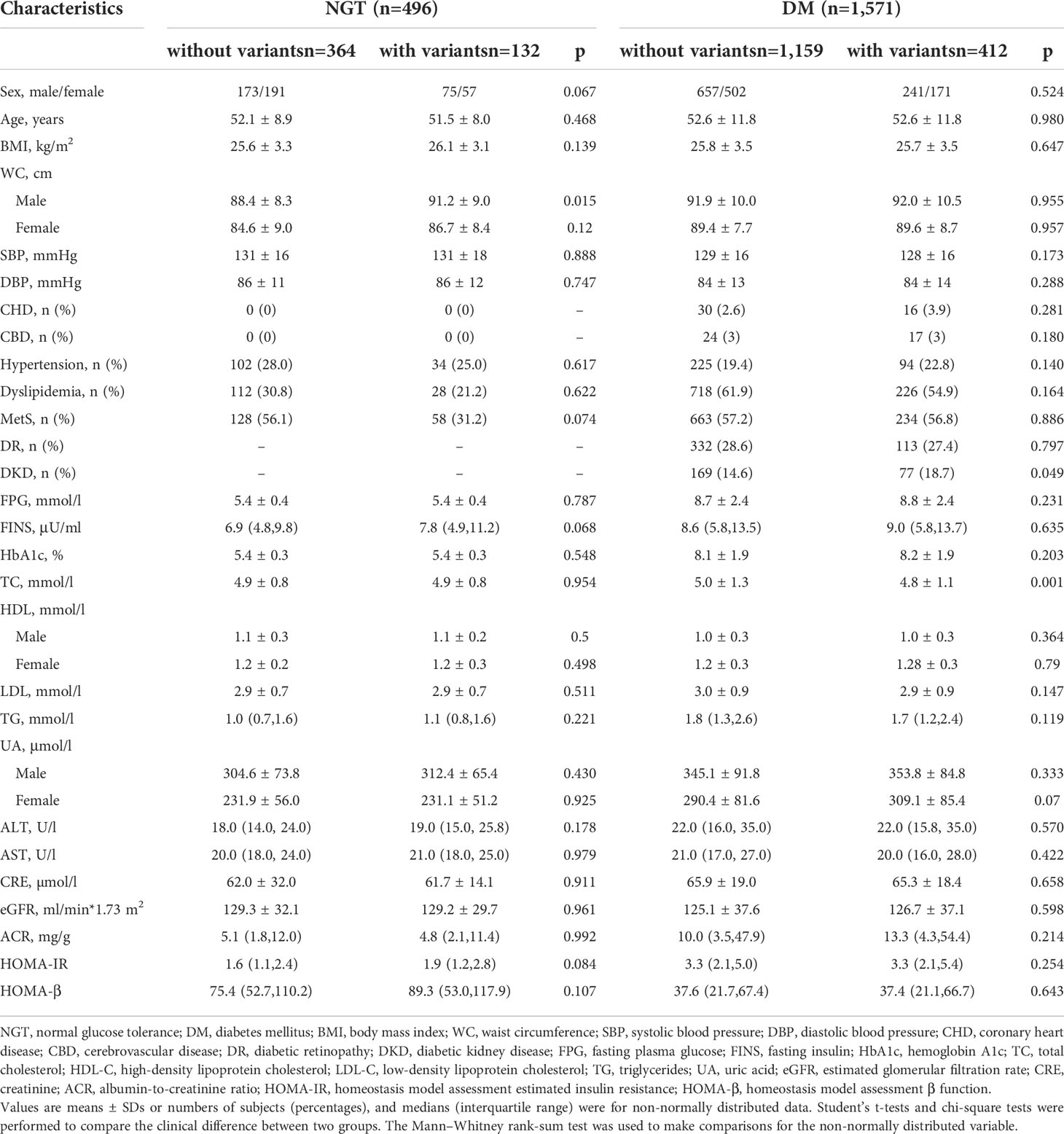
Table 8 The comparison of the subjects without the 20 variants and with the 20 variants in normal glucose tolerance and diabetes mellitus groups from cohort 3.
We divided the patients into two groups: (1) without variants and (2) with one-to-three variants. In the logistic regression analysis, compared with the group without the variants, the OR of DKD for the group with one-to-three variants was 1.422 (95% CI: 1.055, 1.915, P = 0.021). After adjustments for age, the OR was 1.434 (95% CI: 1.061, 1.937, P = 0.019). In the adjusted model 3 that was further fitted with age and sex, the OR of DKD for the group with one-to-three variants was 1.433 (95% CI: 1.060, 1.936, P = 0.019).
The 20 mtDNA variants were in six domains of mitochondria including complex I, complex III, complex V, non-coding, rRNA, and rRNA. There was no significant difference of the prevalence of patients with normal BMI/overweight/obesity, FINS tertiles, HOMA-IR tertiles, and HOMA-β tertiles between patients with variants in these domains of mitochondria and without these variants in Supplementary Table 3 and Table 4.
In our study, we analyzed whole mitochondrial DNA sequence variants in EOD with MID. We identified that seven variants, of which five variants were in complex I of mitochondrial, one variant in complex III, and one in tRNA from cohort 2 and cohort 3, were possibly diabetes associated. Patients with variants in the non-coding region had a higher percentage of obesity and levels of FINS. The patients with 20 homozygous variants had a significantly higher prevalence of DKD than those without these variants in diabetic patients. As we know, it was the first study to investigate the whole mtDNA sequence variants with diabetes and DKD in the Chinese population, and our study has provided a new insight into the mitochondrial genetic role in diabetes and its complication via.
Mitochondria are essential in energy metabolism and cellular survival and play a central role in numerous metabolic processes. IR plays a key role in the development of T2DM and DKD. Several abnormalities in mitochondrial oxidative metabolism, including the reduced content and size of mitochondria (11), impaired mitochondrial respiration, and reduced electron transport chain activity (11, 12, 17), as well as the transcriptional downregulation of genes involved in mitochondrial oxidative metabolism (13, 14, 19), have been associated with obesity, T2DM, and aging in human skeletal muscle. These alterations are collectively termed mitochondrial dysfunction and are often associated with an impaired insulin-mediated increase in transcript levels and ATP synthesis in the mitochondria of skeletal muscle in insulin-resistant individuals (9, 38). Skeletal muscle is essential for metabolism because of its role in glucose uptake and its importance in exercise. It plays an important role in peripheral IR (71). In our study of cohort 2, we found that 42 rare variants were associated with IR. In addition, patients with variants in the non-coding region had a higher percentage of obesity and higher FINS. These variants were in the D-loop region, and previously, variants in the D-loop have been shown to be associated with breast cancer and other tumors (22, 68–70). The D-loop is the main regulation region of mitochondrial transcription (72). In addition, variants in the 12S rRNA gene were found to have a higher percentage of obesity. These variants in the 12S rRNA gene have been reported to be associated with a coronary atherosclerosis risk, deafness, and ischemic stroke (24–28). These data strongly indicate that the D-loop region and 12S rRNA may be associated with obesity. After enlarging the sample size, we found that there was an association of mitochondrial variants with DKD. The levels of FINS and HOMA-IR had an upward trend in the group with 20 variants, although there was no significant statistical difference. This may be related to the heteroplasmy of mitochondria and the loss of information by high-throughput sequencing.
Mitochondrial respiratory Complex I plays an important role in the process of ATP synthesis, which is the rate-limiting enzyme in electron transport chain (ETC) (73, 74). Current evidence suggests that the defects in complex I may be associated with metabolic diseases, including diabetes, IR, and NAFLD (3–5). In addition, complex I is also a major source of reactive oxygen species (ROS) in mitochondria (75), which has been implicated in the pathology of Parkinson disease (76) and ageing (77). Previous studies have demonstrated that the pivotal role of the ND6 epigenetic network could regulate mitochondrial function and affect IR (5). It indicated that the function of complex I may be associated with IR. Five of the above seven mtDNA variants were found in complex I in our study, which were previously reported in patients with diabetes (34, 38, 40, 78). It may be due to the influence on the structure and the function of complex I, leading to IR and defecting the function of islets; thereby, these were considered to be possibly diabetes and DKD associated.
m.A10005G was previously reported to be possibly associated with hearing loss (45). m.A10005G is located in the D-loop and had a crucial role in tRNA recognition by cognate aminoacyl-tRNA synthetases (79). Another variant was the m.A15218G variant in MT-CYB. Previous research associated m.A15218G with epilepsy, diabetes, and Leber hereditary optic neuropathy (LHON) (78, 80). This variant led to the substitution of a conserved hydrophilic threonine by a hydrophobic alanine at position 159 of the cytochrome b subunit of complex III. In addition, the position was highly conservative (81). The cytochrome b subunit of complex III formed the catalytic core (82). The evolutionary conservation of the amino acid position, results from the prediction algorithms, and the homoplasic nature of the variant suggest that m.A15218G is mildly deleterious (64).
Current studies have shown that increased ROS was associated with diabetic kidney injury (83). Hyperglycemia would stimulate an increase in aerobic glycolysis with the resultant increase in substrate delivery to the mitochondria and a subsequent increase in mitochondrial activity (84). Increased tricarboxylic acid cycle (TCA) activity would produce more NADH and FADH2, which would then be used in the ETC in the mitochondria. The above evidence may partly explain the reason why the mtDNA variation could lead to DKD. The variety of mtDNA variants could alternate the structure of mitochondria and affect different aspects of mitochondrial function, thereby causing diabetes and DKD.
Diabetes due to mtDNA variants requires individualized treatment. Metformin is not recommended in the case of mitochondrial diabetes because it was shown to cause lactic acidosis. SGLT-2i could lead to lower blood sugar and body weight; reduced cardiovascular events risks; and the improvement of the inflammation status, liver steatosis, and uric acid concentrations (85). GLP-1 Ras also had the protective effect on the cardiovascular system. Since mtDNA variants are known to elevate the level of ROS, they are especially dangerous for the cardiovascular system, where they can cause cardiomyopathy, atherosclerosis, and hypertension. Diabetic patients with identified mtDNA variants are considered a high-risk group for cardiovascular diseases. Therefore, the cardioprotective effects of SGLT-2i, GLP-1 RA, and related substances have significant benefits in comparison to alternative treatments (86). In addition, we also need to pay attention to new strategies aimed at restoring complex I activity, reducing oxidative stress in alleviating IR, and the treatment of diabetes.
There are also three limitations in our study. First, the software we used was aimed to evaluate the coding variants and tRNA, and the variants in rRNA were unavailable. The pathogenicity of these variants is needed to be further investigated. Secondly, the sample size to evaluate the potential disease-associated variants was limited; thus, a larger sample size of population is needed to further investigate the variants in the mitochondrial diabetes and DKD for future studies. Thirdly, the exact mechanism of the potential disease-associated variants that possibly cause disease is further needed for the research in vivo and in vitro studies.
In our study, we found that seven variants in mitochondrial complex I, D-loop, and complex I were possibly associated with diabetes and 20 mtDNA variants may be associated with a higher risk of DKD. Although it has been proposed that the variants might impair the function of mitochondrial complex I and elevate the levels of ROS, the molecular mechanism linking diabetes and DKD is further needed to be investigated. Prospective and larger-scale studies are needed to establish the clinical relevance of this association.
The datasets presented in this article are not readily available due to ethical restrictions. Requests to access the datasets should be directed to the corresponding author.
The studies involving human participants were approved by the Ethics Committee of Peking University People’s Hospital. The patients/participants provided their written informed consent to participate in this study.
ML conducted this study, collected and explored the data, and completed the original manuscript. LJ contributed to the study design, critically revised the manuscript, and obtained the funding. XH also contributed to the study design and critically revised the manuscript. SG contributed to the sample collection and analysis of the data and reviewed the manuscript. LZ, SZ, QR, XC, YYL, WL, YZ, XZ, and YFL contributed to the data and sample collection and reviewed the manuscript. All authors read the final manuscript and approved final submission.
This research was supported by grants from National Key Research and Development Program of China (2016YFC1304901), Beijing Municipal Science and Technology Committee Funding (Z141100007414002 and D131100005313008), National High-Technology Research and Development Program of China (2012AA02A509), Bethune Merck Research Fund for young and middle-aged doctors (2119000314), and Major Chronic Non-communicable Disease Prevention and Control Research(2016YFC1305600).
We would like to express our gratitude to the subjects for their contributions to the study, as well as to all the physicians and nurses for their work in this procedure of the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.953631/full#supplementary-material
mtDNA, mitochondrial DNA; NGS, next-generation sequencing; EOD, early-onset diabetes; MID, maternally inherited diabetes; DKD, diabetic kidney disease; tRNA, transfer RNA; IR, insulin resistance; NAFLD, non-alcoholic fatty liver disease; MetS, metabolic syndrome; FINS, fasting insulin; FPG, fasting plasma glucose; HOMA, homeostasis model assessment; HbA1c, hemoglobin A1c; LHON, Leber hereditary optic neuropathy.
1. Chinnery PF. Primary mitochondrial disorders overview. In: GeneReviews. Seattle: University of Washington Press (1993).
2. Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res (2016) 44:D1251–1257. doi: 10.1093/nar/gkv1003
3. Sharma V, Sharma I, Singh VP, Verma S, Pandita A, Singh V, et al. mtDNA G10398A variation provides risk to type 2 diabetes in population group from the jammu region of India. Meta Gene (2014) 2:269–73. doi: 10.1016/j.mgene.2014.02.003
4. Lu MY, Huang JF, Liao YC, Bai RK, Trieu RB, Chuang WL, et al. Mitochondrial polymorphism 12361A>G is associated with nonalcoholic fatty liver disease. Transl Res (2012) 159:58–9. doi: 10.1016/j.trsl.2011.10.011
5. Cao K, Lv W, Wang X, Dong S, Liu X, Yang T, et al. Hypermethylation of hepatic mitochondrial ND6 provokes systemic insulin resistance. Adv Sci (Weinh) (2021) 8:2004507. doi: 10.1002/advs.202004507
6. Palmieri VO, De Rasmo D, Signorile A, Sardanelli AM, Grattagliano I, Minerva F, et al. T16189C mitochondrial DNA variant is associated with metabolic syndrome in Caucasian subjects. Nutrition (2011) 27:773–7. doi: 10.1016/j.nut.2010.08.016
7. Janssen GM, Maassen JA, van Den Ouweland JM. The diabetes-associated 3243 mutation in the mitochondrial tRNA(Leu(UUR)) gene causes severe mitochondrial dysfunction without a strong decrease in protein synthesis rate. J Biol Chem (1999) 274:29744–8. doi: 10.1074/jbc.274.42.29744
8. Maassen JA, t Hart LM, Janssen GM, Reiling E, Romijn JA, Lemkes HH. Mitochondrial diabetes and its lessons for common type 2 diabetes. Biochem Soc Trans (2006) 34:819–23. doi: 10.1042/bst0340819
9. Guillausseau PJ, Dubois-Laforgue D, Massin P, Laloi-Michelin M, Bellanné-Chantelot C, Gin H, et al. Heterogeneity of diabetes phenotype in patients with 3243 bp mutation of mitochondrial DNA (Maternally inherited diabetes and deafness or MIDD). Diabetes Metab (2004) 30:181–6. doi: 10.1016/s1262-3636(07)70105-2
10. Murphy R, Turnbull DM, Walker M, Hattersley AT. Clinical features, diagnosis and management of maternally inherited diabetes and deafness (MIDD) associated with the 3243A>G mitochondrial point mutation. Diabetic Med J Br Diabetic Assoc (2008) 25:383–99. doi: 10.1111/j.1464-5491.2008.02359.x
11. Kishimoto M, Hashiramoto M, Araki S, Ishida Y, Kazumi T, Kanda E, et al. Diabetes mellitus carrying a mutation in the mitochondrial tRNA(Leu(UUR)) gene. Diabetologia (1995) 38:193–200. doi: 10.1007/bf00400094
12. Whittaker RG, Schaefer AM, McFarland R, Taylor RW, Walker M, Turnbull DM. Prevalence and progression of diabetes in mitochondrial disease. Diabetologia (2007) 50:2085–9. doi: 10.1007/s00125-007-0779-9
13. Peppa M, Koliaki C, Nikolopoulos P, Raptis SA. Skeletal muscle insulin resistance in endocrine disease. J BioMed Biotechnol (2010) 2010:527850. doi: 10.1155/2010/527850
14. Kohda M, Tokuzawa Y, Kishita Y, Nyuzuki H, Moriyama Y, Mizuno Y, et al. A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PloS Genet (2016) 12:e1005679. doi: 10.1371/journal.pgen.1005679
15. Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. part 1: Diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabetes Med (1998) 15:539–53. doi: 10.1002/(sici)1096-9136(199807)15:7<539::Aid-dia668>3.0.Co;2-s
16. National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III). Third report of the national cholesterol education program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult treatment panel III) final report. Circulation (2002) 106:3143–421. doi: 10.1161/circ.106.25.3143
17. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia (1985) 28:412–9. doi: 10.1007/bf00280883
18. Alberti KGMM, Zimmet P, Shaw J. Metabolic syndrome–a new world-wide definition. a consensus statement from the international diabetes federation. Diabetes Med (2006) 23:469–80. doi: 10.1111/j.1464-5491.2006.01858.x
19. Wang Q, Liu M, Xu C, Tang Z, Liao Y, Du R, et al. Novel CACNA1S mutation causes autosomal dominant hypokalemic periodic paralysis in a Chinese family. J Mol Med (Berl) (2005) 83:203–8. doi: 10.1007/s00109-005-0638-4
20. Niemi AK, Moilanen JS, Tanaka M, Hervonen A, Hurme M, Lehtimäki T, et al. A combination of three common inherited mitochondrial DNA polymorphisms promotes longevity in Finnish and Japanese subjects. Eur J Hum Genet (2005) 13:166–70. doi: 10.1038/sj.ejhg.5201308
21. Zhai K, Chang L, Zhang Q, Liu B, Wu Y. Mitochondrial C150T polymorphism increases the risk of cervical cancer and HPV infection. Mitochondrion (2011) 11:559–63. doi: 10.1016/j.mito.2011.02.005
22. Ebner S, Lang R, Mueller EE, Eder W, Oeller M, Moser A, et al. Mitochondrial haplogroups, control region polymorphisms and malignant melanoma: a study in middle European caucasians. PloS One (2011) 6:e27192. doi: 10.1371/journal.pone.0027192
23. Rollins B, Martin MV, Sequeira PA, Moon EA, Morgan LZ, Watson SJ, et al. Mitochondrial variants in schizophrenia, bipolar disorder, and major depressive disorder. PloS One (2009) 4:e4913. doi: 10.1371/journal.pone.0004913
24. Sawabe M, Tanaka M, Chida K, Arai T, Nishigaki Y, Fuku N, et al. Mitochondrial haplogroups a and M7a confer a genetic risk for coronary atherosclerosis in the Japanese elderly: an autopsy study of 1,536 patients. J Atheroscler Thromb (2011) 18:166–75. doi: 10.5551/jat.6742
25. Chaig MR, Zernotti ME, Soria NW, Romero OF, Romero MF, Gerez NM. A mutation in mitochondrial 12S rRNA, A827G, in argentinean family with hearing loss after aminoglycoside treatment. Biochem Biophys Res Commun (2008) 368:631–6. doi: 10.1016/j.bbrc.2008.01.143
26. Li Z, Li R, Chen J, Liao Z, Zhu Y, Qian Y, et al. Mutational analysis of the mitochondrial 12S rRNA gene in Chinese pediatric subjects with aminoglycoside-induced and non-syndromic hearing loss. Hum Genet (2005) 117:9–15. doi: 10.1007/s00439-005-1276-1
27. Abreu-Silva RS, Rincon D, Horimoto AR, Sguillar AP, Ricardo LA, Kimura L, et al. The search of a genetic basis for noise-induced hearing loss (NIHL). Ann Hum Biol (2011) 38:210–8. doi: 10.3109/03014460.2010.513774
28. Rosa A, Fonseca BV, Krug T, Manso H, Gouveia L, Albergaria I, et al. Mitochondrial haplogroup H1 is protective for ischemic stroke in Portuguese patients. BMC Med Genet (2008) 9:57. doi: 10.1186/1471-2350-9-57
29. Tang J, Qi Y, Bao XH, Wu XR. Mutational analysis of mitochondrial DNA of children with rett syndrome. Pediatr Neurol (1997) 17:327–30. doi: 10.1016/s0887-8994(97)00151-3
30. Cardaioli E, Dotti MT, Hayek G, Zappella M, Federico A. Studies on mitochondrial pathogenesis of rett syndrome: Ultrastructural data from skin and muscle biopsies and mutational analysis at mtDNA nucleotides 10463 and 2835. J Submicrosc Cytol Pathol (1999) 31:301–4.
31. Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MF, Mirra SS, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics (1993) 17:171–84. doi: 10.1006/geno.1993.1299
32. Cavelier L, Erikson I, Tammi M, Jalonen P, Lindholm E, Jazin E, et al. MtDNA mutations in maternally inherited diabetes: presence of the 3397 ND1 mutation previously associated with alzheimer's and parkinson's disease. Hereditas (2001) 135:65–70. doi: 10.1111/j.1601-5223.2001.00065.x
33. Mitchell AL, Elson JL, Howell N, Taylor RW, Turnbull DM. Sequence variation in mitochondrial complex I genes: Mutation or polymorphism? J Med Genet (2006) 43:175–9. doi: 10.1136/jmg.2005.032474
34. Jaksch M, Hofmann S, Kaufhold P, Obermaier-Kusser B, Zierz S, Gerbitz KD. A novel combination of mitochondrial tRNA and ND1 gene mutations in a syndrome with MELAS, cardiomyopathy, and diabetes mellitus. Hum Mutat (1996) 7:358–60. doi: 10.1002/(sici)1098-1004(1996)7:4<358::Aid-humu11>3.0.Co;2-1
35. Tang S, Batra A, Zhang Y, Ebenroth ES, Huang T. Left ventricular noncompaction is associated with mutations in the mitochondrial genome. Mitochondrion (2010) 10:350–7. doi: 10.1016/j.mito.2010.02.003
36. Finsterer J, Bittner R, Bodingbauer M, Eichberger H, Stöllberger C, Blazek G. Complex mitochondriopathy associated with 4 mtDNA transitions. Eur Neurol (2000) 44:37–41. doi: 10.1159/000008190
37. Tranah GJ, Nalls MA, Katzman SM, Yokoyama JS, Lam ET, Zhao Y, et al. Mitochondrial DNA sequence variation associated with dementia and cognitive function in the elderly. J Alzheimers Dis (2012) 32:357–72. doi: 10.3233/jad-2012-120466
38. Ohkubo E, Aida K, Chen J, Hayashi JI, Isobe K, Tawata M, et al. A patient with type 2 diabetes mellitus associated with mutations in calcium sensing receptor gene and mitochondrial DNA. Biochem Biophys Res Commun (2000) 278:808–13. doi: 10.1006/bbrc.2000.3867
39. Takasaki S. Mitochondrial SNPs associated with Japanese centenarians, alzheimer's patients, and parkinson's patients. Comput Biol Chem (2008) 32:332–7. doi: 10.1016/j.compbiolchem.2008.03.014
40. Sakaue S, Ohmuro J, Mishina T, Miyazaki H, Yamaguchi E, Nishimura M, et al. A case of diabetes, deafness, cardiomyopathy, and central sleep apnea: Novel mitochondrial DNA polymorphisms. Tohoku J Exp Med (2002) 196:203–11. doi: 10.1620/tjem.196.203
41. Chen T, Liu Q, Jiang L, Liu C, Ou Q. Mitochondrial COX2 G7598A mutation may have a modifying role in the phenotypic manifestation of aminoglycoside antibiotic-induced deafness associated with 12S rRNA A1555G mutation in a han Chinese pedigree. Genet Test Mol Biomarkers (2013) 17:122–30. doi: 10.1089/gtmb.2012.0251
42. Zhadanov SI, Atamanov VV, Zhadanov NI, Schurr TG. De novo COX2 mutation in a LHON family of Caucasian origin: implication for the role of mtDNA polymorphism in human pathology. J Hum Genet (2006) 51:161–70. doi: 10.1007/s10038-005-0340-y
43. Jiang C, Cui J, Liu F, Gao L, Luo Y, Li P, et al. Mitochondrial DNA 10609T promotes hypoxia-induced increase of intracellular ROS and is a risk factor of high altitude polycythemia. PloS One (2014) 9:e87775. doi: 10.1371/journal.pone.0087775
44. Tanaka M, Takeyasu T, Fuku N, Li-Jun G, Kurata M. Mitochondrial genome single nucleotide polymorphisms and their phenotypes in the Japanese. Ann N Y Acad Sci (2004) 1011:7–20. doi: 10.1007/978-3-662-41088-2_2
45. Zheng J, Bai X, Xiao Y, Ji Y, Meng F, Aishanjiang M, et al. Mitochondrial tRNA mutations in 887 Chinese subjects with hearing loss. Mitochondrion (2020) 52:163–72. doi: 10.1016/j.mito.2020.03.005
46. Lertrit P, Noer AS, Jean-Francois MJ, Kapsa R, Dennett X, Thyagarajan D, et al. A new disease-related mutation for mitochondrial encephalopathy lactic acidosis and strokelike episodes (MELAS) syndrome affects the ND4 subunit of the respiratory complex I. Am J Hum Genet (1992) 51:457–68.
47. Levin L, Zhidkov I, Gurman Y, Hawlena H, Mishmar D. Functional recurrent mutations in the human mitochondrial phylogeny: Dual roles in evolution and disease. Genome Biol Evol (2013) 5:876–90. doi: 10.1093/gbe/evt058
48. Allegra E, Garozzo A, Lombardo N, De Clemente M, Carey TE. Mutations and polymorphisms in mitochondrial DNA in head and neck cancer cell lines. Acta Otorhinolaryngol Ital (2006) 26:185–90.
49. Tawata M, Ohtaka M, Iwase E, Ikegishi Y, Aida K, Onaya T. New mitochondrial DNA homoplasmic mutations associated with Japanese patients with type 2 diabetes. Diabetes (1998) 47:276–7. doi: 10.2337/diab.47.2.276
50. Shin WS, Tanaka M, Suzuki J, Hemmi C, Toyo-oka T. A novel homoplasmic mutation in mtDNA with a single evolutionary origin as a risk factor for cardiomyopathy. Am J Hum Genet (2000) 67:1617–20. doi: 10.1086/316896
51. Mimaki M, Ikota A, Sato A, Komaki H, Akanuma J, Nonaka I, et al. A double mutation (G11778A and G12192A) in mitochondrial DNA associated with leber's hereditary optic neuropathy and cardiomyopathy. J Hum Genet (2003) 48:47–50. doi: 10.1007/s100380300005
52. Liu Z, Song Y, Gu S, He X, Zhu X, Shen Y, et al. Mitochondrial ND5 12338T>C variant is associated with maternally inherited hypertrophic cardiomyopathy in a Chinese pedigree. Gene (2012) 506:339–43. doi: 10.1016/j.gene.2012.06.071
53. Zhang J, Ji Y, Lu Y, Fu R, Xu M, Liu X, et al. Leber's hereditary optic neuropathy (LHON)-associated ND5 12338T > C mutation altered the assembly and function of complex I, apoptosis and mitophagy. Hum Mol Genet (2018) 27:1999–2011. doi: 10.1093/hmg/ddy107
54. Chen B, Sun D, Yang L, Zhang C, Yang A, Zhu Y, et al. Mitochondrial ND5 T12338C, tRNA(Cys) T5802C, and tRNA(Thr) G15927A variants may have a modifying role in the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three han Chinese pedigrees. Am J Med Genet Part A (2008) 146a:1248–58. doi: 10.1002/ajmg.a.32285
55. Zhou HP, Ishikawa H, Yasumoto R, Sakurai K, Sawamura H. Leber hereditary optic neuropathy harboring a rare m.12811 T>C mitochondrial DNA mutation. Can J Ophthalmol (2021) 56:e82–4. doi: 10.1016/j.jcjo.2020.12.022
56. Wei YL, Yu CA, Yang P, Li AL, Wen JY, Zhao SM, et al. Novel mitochondrial DNA mutations associated with Chinese familial hypertrophic cardiomyopathy. Clin Exp Pharmacol Physiol (2009) 36:933–9. doi: 10.1111/j.1440-1681.2009.05183.x
57. Jha RK, Dawar C, Hasan Q, Pujar A, Gupta G, Vishnu VY, et al. Mitochondrial genetic heterogeneity in leber's hereditary optic neuropathy: Original study with meta-analysis. Genes (Basel) (2021) 12:1300. doi: 10.3390/genes12091300
58. Zhang J, Zhou X, Zhou J, Li C, Zhao F, Wang Y, et al. Mitochondrial ND6 T14502C variant may modulate the phenotypic expression of LHON-associated G11778A mutation in four Chinese families. Biochem Biophys Res Commun (2010) 399:647–53. doi: 10.1016/j.bbrc.2010.07.135
59. Horvath R, Kemp JP, Tuppen HA, Hudson G, Oldfors A, Marie SK, et al. Molecular basis of infantile reversible cytochrome c oxidase deficiency myopathy. Brain (2009) 132:3165–74. doi: 10.1093/brain/awp221
60. Young WY, Zhao L, Qian Y, Li R, Chen J, Yuan H, et al. Variants in mitochondrial tRNAGlu, tRNAArg, and tRNAThr may influence the phenotypic manifestation of deafness-associated 12S rRNA A1555G mutation in three han Chinese families with hearing loss. Am J Med Genet Part A (2006) 140:2188–97. doi: 10.1002/ajmg.a.31434
61. Tzen CY, Thajeb P, Wu TY, Chen SC. Melas with point mutations involving tRNALeu (A3243G) and tRNAGlu(A14693g). Muscle Nerve (2003) 28:575–81. doi: 10.1002/mus.10473
62. Tong Y, Mao Y, Zhou X, Yang L, Zhang J, Cai W, et al. The mitochondrial tRNA(Glu) A14693G mutation may influence the phenotypic manifestation of ND1 G3460A mutation in a Chinese family with leber's hereditary optic neuropathy. Biochem Biophys Res Commun (2007) 357:524–30. doi: 10.1016/j.bbrc.2007.03.189
63. Sun Y, Han X. Study progress in mitochondrial cardiomyopathy. J Appl Clin Pediatr (2011) 26:50–2. doi: 10.3969/j.issn.1003-515X.2011.01.019
64. Soini HK, Moilanen JS, Finnila S, Majamaa K. Mitochondrial DNA sequence variation in Finnish patients with matrilineal diabetes mellitus. BMC Res Notes (2012) 5:350. doi: 10.1186/1756-0500-5-350
65. Jancic J, Rovcanin B, Djuric V, Pepic A, Samardzic J, Nikolic B, et al. Analysis of secondary mtDNA mutations in families with leber's hereditary optic neuropathy: Four novel variants and their association with clinical presentation. Mitochondrion (2020) 50:132–8. doi: 10.1016/j.mito.2019.10.011
66. Liguori R, Mazzaccara C, Pasanisi F, Buono P, Oriani G, Finelli C, et al. The mtDNA 15497 G/A polymorphism in cytochrome b in severe obese subjects from southern Italy. Nutr Metab Cardiovasc Dis (2006) 16:466–70. doi: 10.1016/j.numecd.2005.06.009
67. Li R, Qu J, Zhou X, Tong Y, Hu Y, Qian Y, et al. The mitochondrial tRNA(Thr) A15951G mutation may influence the phenotypic expression of the LHON-associated ND4 G11778A mutation in a Chinese family. Gene (2006) 376:79–86. doi: 10.1016/j.gene.2006.02.014
68. Tommasi S, Favia P, Weigl S, Bianco A, Pilato B, Russo L, et al. Mitochondrial DNA variants and risk of familial breast cancer: An exploratory study. Int J Oncol (2014) 44:1691–8. doi: 10.3892/ijo.2014.2324
69. Buford TW, Manini TM, Kairalla JA, McDermott MM, Vaz Fragoso CA, Chen H, et al. Mitochondrial DNA sequence variants associated with blood pressure among 2 cohorts of older adults. J Am Heart Assoc (2018) 7:e010009. doi: 10.1161/jaha.118.010009
70. Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Sci (New York NY) (2000) 287:2017–9. doi: 10.1126/science.287.5460.2017
71. Merz KE, Thurmond DC. Role of skeletal muscle in insulin resistance and glucose uptake. Compr Physiol (2020) 10:785–809. doi: 10.1002/cphy.c190029
72. Suzuki M, Toyooka S, Miyajima K, Iizasa T, Fujisawa T, Bekele NB, et al. Alterations in the mitochondrial displacement loop in lung cancers. Clin Cancer Res (2003) 9:5636–41. doi: 10.1038/nrm3997
73. Sazanov LA. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol (2015) 16:375–88. doi: 10.1038/nrm3997
74. Lapuente-Brun E, Moreno-Loshuertos R, Acín-Pérez R, Latorre-Pellicer A, Colás C, Balsa E, et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Sci (New York NY) (2013) 340:1567–70. doi: 10.1126/science.1230381
75. Murphy MP. How mitochondria produce reactive oxygen species. Biochem J (2009) 417:1–13. doi: 10.1042/bj20081386
76. Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in parkinson's disease. Science (2003) 302:819–22. doi: 10.1126/science.1087753
77. Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell (2005) 120:483–95. doi: 10.1016/j.cell.2005.02.001
78. Soini HK, Moilanen JS, Vilmi-Kerälä T, Finnilä S, Majamaa K. Mitochondrial DNA variant m.15218A > G in Finnish epilepsy patients who have maternal relatives with epilepsy, sensorineural hearing impairment or diabetes mellitus. BMC Med Genet (2013) 14:73. doi: 10.1186/1471-2350-14-73
79. Asahara H, Himeno H, Tamura K, Hasegawa T, Watanabe K, Shimizu M. Recognition nucleotides of escherichia coli tRNA(Leu) and its elements facilitating discrimination from tRNASer and tRNA(Tyr). J Mol Biol (1993) 231:219–29. doi: 10.1006/jmbi.1993.1277
80. Hudson G, Mowbray C, Elson JL, Jacob A, Boggild M, Torroni A, et al. Does mitochondrial DNA predispose to neuromyelitis optica (Devic's disease)? Brain (2008) 131:e93. doi: 10.1093/brain/awm224
81. Tanaka M, Cabrera VM, González AM, Larruga JM, Takeyasu T, Fuku N, et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res (2004) 14:1832–50. doi: 10.1101/gr.2286304
82. Legros F, Chatzoglou E, Frachon P, Ogier De Baulny H, Laforêt P, Jardel C, et al. Functional characterization of novel mutations in the human cytochrome b gene. Eur J Hum Genet (2001) 9:510–8. doi: 10.1038/sj.ejhg.5200678
83. Forbes JM, Coughlan MT, Cooper ME. Oxidative stress as a major culprit in kidney disease in diabetes. Diabetes (2008) 57:1446–54. doi: 10.2337/db08-0057
84. Brownlee M. The pathobiology of diabetic complications: A unifying mechanism. Diabetes (2005) 54:1615–25. doi: 10.2337/diabetes.54.6.1615
85. Bonora BM, Avogaro A, Fadini GP. Extraglycemic effects of SGLT2 inhibitors: A review of the evidence. Diab Metab syndrome Obes Targets Ther (2020) 13:161–74. doi: 10.2147/dmso.S233538
86. Zheng SL, Roddick AJ, Aghar-Jaffar R, Shun-Shin MJ, Francis D, Oliver N, et al. Association between use of sodium-glucose cotransporter 2 inhibitors, glucagon-like peptide 1 agonists, and dipeptidyl peptidase 4 inhibitors with all-cause mortality in patients with type 2 diabetes: A systematic review and meta-analysis. JAMA (2018) 319:1580–91. doi: 10.1001/jama.2018.3024
Keywords: mitochondrial DNA, mitochondrial diabetes, diabetes, diabetic kidney disease, insulin resistance
Citation: Li M, Gong S, Han X, Zhou L, Zhang S, Ren Q, Cai X, Luo Y, Liu W, Zhu Y, Zhou X, Li Y and Ji L (2022) Contribution of mitochondrial gene variants in diabetes and diabetic kidney disease. Front. Endocrinol. 13:953631. doi: 10.3389/fendo.2022.953631
Received: 26 May 2022; Accepted: 04 August 2022;
Published: 12 October 2022.
Edited by:
Carlos Guillén, Complutense University, SpainReviewed by:
Andrei I. Tarasov, Ulster University, United Kingdom;Lili Ding, Shanghai University of Traditional Chinese Medicine, ChinaCopyright © 2022 Li, Gong, Han, Zhou, Zhang, Ren, Cai, Luo, Liu, Zhu, Zhou, Li and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linong Ji, amlsbkBiam11LmVkdS5jbg==
†These authors have contributed equally to this work
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.