
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 22 July 2022
Sec. Neuroendocrine Science
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.937218
This article is part of the Research Topic Genomic and Transcriptomic Insights into Neuroendocrine Evolution and Function View all 8 articles
Corticotropin-releasing hormone (CRH) was discovered for its role as a brain neurohormone controlling the corticotropic axis in vertebrates. An additional crh gene, crh2, paralog of crh (crh1), and likely resulting from the second round (2R) of vertebrate whole genome duplication (WGD), was identified in a holocephalan chondrichthyan, in basal mammals, various sauropsids and a non-teleost actinopterygian holostean. It was suggested that crh2 has been recurrently lost in some vertebrate groups including teleosts. We further investigated the fate of crh1 and crh2 in vertebrates with a special focus on teleosts. Phylogenetic and synteny analyses showed the presence of duplicated crh1 paralogs, crh1a and crh1b, in most teleosts, resulting from the teleost-specific WGD (3R). Crh1b is conserved in all teleosts studied, while crh1a has been lost independently in some species. Additional crh1 paralogs are present in carps and salmonids, resulting from specific WGD in these lineages. We identified crh2 gene in additional vertebrate groups such as chondrichthyan elasmobranchs, sarcopterygians including dipnoans and amphibians, and basal actinoperygians, Polypteridae and Chondrostei. We also revealed the presence of crh2 in teleosts, including elopomorphs, osteoglossomorphs, clupeiforms, and ostariophysians, while it would have been lost in Euteleostei along with some other groups. To get some insights on the functional evolution of the crh paralogs, we compared their primary and 3D structure, and by qPCR their tissue distribution, in two representative species, the European eel, which possesses three crh paralogs (crh1a, crh1b, crh2), and the Atlantic salmon, which possesses four crh paralogs of the crh1-type. All peptides conserved the structural characteristics of human CRH. Eel crh1b and both salmon crh1b genes were mainly expressed in the brain, supporting the major role of crh1b paralogs in controlling the corticotropic axis in teleosts. In contrast, crh1a paralogs were mainly expressed in peripheral tissues such as muscle and heart, in eel and salmon, reflecting a striking subfunctionalization between crh1a and b paralogs. Eel crh2 was weakly expressed in the brain and peripheral tissues. These results revisit the repertoire of crh in teleosts and highlight functional divergences that may have contributed to the differential conservation of various crh paralogs in teleosts.
Corticotropin-releasing hormone (CRH), first named CRF (F for factor) until its characterization, is the hypothalamic neurohormone of the corticotropic axis involved in the control of stress response. CRH control the production by the pituitary of corticotropin (ACTH), which itself controls the production of corticosteroids by the adrenals in mammals, birds and reptiles, and by the interrenals in amphibians and teleosts. Being able to activate other neuroendocrine axes, such as the thyrotropic and somatotropic axes, under environmental and internal stimuli, CRH has been proposed as the central coordinator of endocrine activation during life transitions such as metamorphosis in amphibians and teleosts, and egg hatching in birds and reptiles [for reviews: (1, 2)].
Initial evidence of the existence of CRH was reported in 1955 (3, 4), and over 25 years were needed to isolate CRH peptide in sheep (5). Subsequently, the ovine CRH precursor, named also prepro-CRH, was cloned and its primary structure described (6). Soon after, the characterization of CRH in rat (7, 8) and human (9) revealed the strong structural and molecular conservation of the CRH peptide that consists of an alpha helical peptide of 41 amino-acids (aa) which is C-terminally amidated and that is produced from the cleavage of a larger precursor ranging from 160 to 210 aa depending on the species [for review (10)]. In mammals, CRH-expressing neurons were found in paraventricular nucleus (PVN) with the majority of neurons projecting to the median eminence (11).
Other peptides showing similarities with CRH were characterized: sauvagine from the skin of neotropical frog Phyllomedusa sauvagei (12), urotensin-I from the urophysis of the white sucker Catostomus commersoni (13) and urocortin from rat midbrain (14), all now considered as “urocortin”, encoded by ucn (named ucn1, 15), a paralogous gene of crh [for review (16)]. Two other CRH-related peptides, named urocortin 2 (17) and urocortin 3 (18) were later found thanks to the completion of the human genome project. According to current evolutionary scenarios, ucn1 would be in fact more closely related to crh than to ucn2 and ucn3 [for review (16); (15, 19)]. Only one crh gene (crh) had been identified in vertebrates, until the discovery in 2011 of a second crh in the elephant shark Callorhinchus milii (20). This second crh gene (named crh2) was then identified in most vertebrate groups (21), a result confirmed by other studies (15, 19). Phylogenetic and syntenic analyses led to the hypothesis that the paralogs, the “classical” crh (also named crh1 for clarification, 15) and crh2, as well as ucn1 likely arose during the two rounds (1R/2R) of whole genome duplication (WGD) that occurred early in the vertebrate lineage (15, 21). In addition, it had been proposed that crh2 would have been lost repeatedly during vertebrate radiation, in teleosts, in amphibians and in placental mammals (15, 21, 22). Duplicated crh (crh1a and crh1b) were found in teleosts likely resulting from the teleost-specific WGD (3R) (15, 22). A further duplication of crh1a and crh1b resulting from salmonid-specific WGD (4R) led to four crh1 paralogs in salmonids (22, 23). Conservation of paralogs after duplication may be related to evolutionary selection for either amplification of initial function, sharing of initial pleiotropic functions (subfunctionalization) or emergence of a new function (neofunctionalization) [for review (24)].
In teleosts, as in other vertebrates, CRH conserved its role as a major stimulator of pituitary ACTH release in response to stress [for reviews (25, 26)]. CRH is also involved in metabolism, food intake [for review (27)], immunity [for review (28)] and locomotor activity [for review (29)]. In teleosts, CRH not only controls the corticotropic axis but is also a potent activator of the thyrotropic axis as in amphibians and birds. This suggests that CRH is able to control several physiological functions in teleost fish such as stress, osmoregulation, metabolism and may play an important role as central coordinator of the activation of endocrine axes for developmental transitions and adaptation to environmental changes [for reviews (1, 2)].
In the present study, we investigated crh paralogs in vertebrates and specially the fate of crh2 which was previously assumed to be lost in teleosts. In addition, to get some insights into the functional evolution of the crh paralogs, we analyzed their sequences, 3D structures and tissue distributions in two representative teleosts: a basal teleost, the European eel (Elopomorph), in which we revealed the presence of three crh paralogs (crh1a, crh1b and crh2), and the Atlantic salmon (Salmoniformes) which possesses four crh paralogs, all of the crh1-type. Our data show that teleost crh paralogs evolved distinct expression patterns and likely diverse functions.
Corticotropin-releasing hormone (crh) genes were sought in representative vertebrates with a special focus in the actinopterygian lineage. Sequences from the most closely related paralog to crh, ucn1, were also included. We screened the genomes of 70 vertebrate species. Gene sequences were retrieved from genome assemblies, either using GenBank gene prediction, or by an exhaustive Blast search against GenBank and UCSC genomic databases, and GenBank, UCSC and PhyloFish transcriptomic databases (30). We used genome assemblies and RNA libraries to search for non-annotated crh-like genes or to confirm gene loss. Coding sequence (CDS) of the prepropeptide were manually annotated by comparison with orthologous genes using CLC Main Workbench (QIAGEN). Sequences references and annotations are provided in Table S1. Phylogenetic analysis, supported by synteny analysis, allowed us to identify orthologs/paralogs and name or rename crh genes, accordingly. For gene nomenclature of WGD paralogs in teleosts, we used in the present study the letters “a/b” for teleost-specific 3R-duplicated genes, according to Zfin nomenclature conventions, and the symbols “α/β” for salmonid-specific 4R-duplicated genes according to Robertson and colleagues (31).
The signal peptide was predicted using SignalP 4.0 browser (32) and peptide cleavage site using both Modpred and NeuroPred webbrowser (33, 34).
Phylogenetic analyses were performed using 166 CRH and UCN1 sequences from 48 vertebrates. CRH/UCN1 mature peptides are highly conserved peptides, which makes it difficult to extract information from their amino-acid sequences, so we used the prepropeptide amino-acid sequences to infer the phylogenetic tree. Multiple sequence alignments of the CRH/UCN1 protein family were performed using the slow algorithm available on CLC workbench and further manually edited based on conserved amino-acid sequences.
The tree topology was inferred with a maximum likelihood analysis using PhyML 3.0 on the web browser of ATGC Montpellier bioinformatics platform and Seaview (35–37). The WAG substitution matrix (38) was chosen to infer the trees of CRH family. Strength of branch nodes was evaluated by both aLRT and bootstrap using 100 replicates.
To further resolve crh gene evolution in teleosts, the synteny of crh and ucn1 genes was investigated in actinopterygians, using a basal non-teleost actinopterygian, a Polyperidae, the reedfish (Erpetoichthys calabaricus) as template. The four chromosomes corresponding to the crh1/crh2/ucn1 tetraparalogon were identified in reedfish by comparing with the crh1/crh2/ucn1 paralogous chromosomes of spotted gar (15). Conserved neighboring gene families of crh1/crh2/ucn1 were identified by comparing manually NCBI predicted genes lists on the crh1/crh2/ucn1 chromosomes of reedfish (Figure S1). The following teleost species were studied: representatives of two basal groups, an elopomorph, European eel (Anguilla anguilla) and an osteoglossomorph, arowana (Scleropages formosus); a Clupeidae, herring (Denticeps clupeoides), a Cyprinidae, zebrafish (Danio rerio), an Esocidae, pike (Esox lucius) and a Salmonidae, Atlantic salmon (Salmo salar). Neighboring gene families with members inherited from 1 and 2R vertebrate WGD, 3R teleost WGD and 4R salmonid WGD were selected to illustrate the crh-gene paralogon evolution. Additional genes were used to further discriminate 3R- and 4R-paralogons. Neighboring gene identity was confirmed by examining close gene neighborhood using the genome browser, Genomicus (39). Non-predicted genes were sought by extensive blast against genome assemblies to confirm the gene loss. References of the gene neighborhood of the crh/ucn1 family is provided in Table S2. The neighboring genes of crh2 in arowana were reexamined together with those of another osteoglossomorph, Paramormyrops kingsleyae and are given in Table S2.
The structure of human, eel and salmon CRH peptides was predicted using iTASSER browser (40, 41). Protein model quality was assessed using MoldFold (42). All the five resulting models predicted by iTASSER, showed high quality score. For each peptide, the top model ranked was rendered using ChimeraX v1.3 (43). Logo sequences of the CRH precursors were generated from the alignment generated for the phylogenetic analysis using CLC Main Workbench (QIAGEN).
Female European eels (A. anguilla) were at the prepubertal “silver” stage, corresponding to the end of the continental stage of the eel life cycle, previous to migration to the ocean for reproduction. They were purchased from Gebr. Dil import-export BV (Akersloot, The Netherlands) and transferred to MNHN, France. Animals were anesthetized by cold and then killed by decapitation under the supervision of authorized person (KR; No. R-75UPMC-F1-08) according to the protocol approved by Cuvier Ethic Committee France (No. 68–027). The tissues were dissected, incubated in RNA later overnight at 4°C and stored at -20°C until RNA extraction.
For Atlantic salmon (S. salar), we reused the tissue samples collected for a previous study (44). These tissues had been collected from mixed sex (5 males and 5 females) juvenile fish of the Loire-Allier population raised indoor under natural water, temperature, and photoperiod conditions, at the Conservatoire National du Saumon Sauvage (CNSS) (44).
The following tissues, dissected according to (44, 45), were analyzed in both species: whole brain, pituitary, gill, heart, liver, spleen, kidney, intestine, fat, muscle, skin and gonad. For detailed brain distribution the following tissues were also analyzed: olfactory bulbs, telencephalon, mesencephalon, diencephalon, optic tectum, cerebellum, medulla oblongata as well as saccus vasculosus, epiphysis, pituitary and retina.
Total tissue RNA was extracted using Trizol according to the manufacturer recommendations. Tissues were homogenized in Trizol with steel beads, twice at 30Hz for 2-5 min, using a TissueLyzer II (Qiagen). Total RNA concentration was measured using a nanodrop spectrophotometer (Thermo Fisher Scientific) and treated with DNase I (Roche) according to the manufacturer’s instructions at 37°C for 20 min. DNase I was inactivated and removed by phenol extraction.
Complementary DNA was generated from 750 ng of denatured total RNA (at 65°C for 5 min) and 75 ng random primers using the superscript III (Invitrogen) under the following conditions: a primer hybridization step at 25°C for 10 min, followed by an extension step at 50°C for 60 min and an inactivation step at 70°C for 15 min. Two no-reverse transcriptase controls for potential DNA contamination were performed using total RNA from either brain or muscle in the same reaction conditions but without superscript III: no product or non-specific products were amplified by qPCR.
Many primer sets for each crh gene of eel and salmon were designed using primer 3 browser (Table S3). The design of primer sets for each gene were confined to the sequence parts showing nucleotide divergence between paralogs, which was especially critical for the pairs of salmon crh1 4R-paralogs that showed strong identity between each other. Each primer set was first tested on pools of tissues expressing the crh paralogs (brain and muscle). These tissues were used to produce qPCR standards: the standards were serial dilutions of cDNA from pooled samples of eel brain for eel crh1b and crh2, of eel muscle for eel crh1a, of salmon brain for salmon crh1bα and crh1bβ and of salmon muscle for salmon crh1aα and crh1aβ. Standard dilutions were run in duplicate to draw a calibration curve and measure amplification efficiency of the different primer sets. Primer sets were chosen according to the two following conditions: 1) they showed a good amplification efficiency (Table S3), 2) gene amplification efficiency was confirmed with two different primer sets positioned at different gene regions (same cycle quantification (Cq) for a same standard dilution) and 3) the Tm of the amplicon could be discriminated from the one resulting from primer-dimer amplification.
Quantitative PCR assays were performed using the LightCycler (Roche) with the LightCycler FastStart Master plus SYBR Green I kit (Roche) as recommended by the manufacturer and 500 nM of each primer. Each sample was run in duplicate. The PCR conditions were 95°C for 10 min followed by 50 cycles at 95°C for 5 sec, 60°C for 10 sec and 72°C for 5 sec. The specificity of amplified qPCR products was checked by performing melting curve analyses and by sequencing the amplicon. Transcript quantity was calculated using the LightCycler software from quantification cycle (Cq) determined by the automatic second derivative method, according to the calibration curve method. Samples were considered at the limit of detection when amplification was obtained after 42 Cq and a minimal value was assigned which was the lowest detectable dose of respective standard. Data are expressed as arbitrary units of gene transcript level/total RNA level. Results (means ± SEM) are presented as percentage per tissue for the tissue comparison, according to (46, 47).
The presence of crh/ucn1 genes was investigated in 70 vertebrates including 7 chondrichthyans, 14 sarcopterygians and 48 actinopterygians, and 252 sequences were retrieved from the genome assemblies of representative vertebrates (Table S1). A phylogenetic tree was inferred by maximum likelihood analysis from prepro-CRH/UCN1 amino-acid sequences (Figure 1); a second phylogenetic analysis was performed with a special focus on a larger number of actinopterygian species (Figure S2). Vertebrate CRH/UCN1 sequences clustered into three groups, including sequences of CRH1, CRH2 and UCN1 respectively (Figure 1). Three genes were present in a basal vertebrate (a cyclostome, the lamprey, Petromyzon marinus) as previously reported (19) and their amino-acid sequences branched at the basal position of each of the gnathostome CRH1, CRH2 and UCN1 clusters in agreement with vertebrate phylogeny (Figure 1).
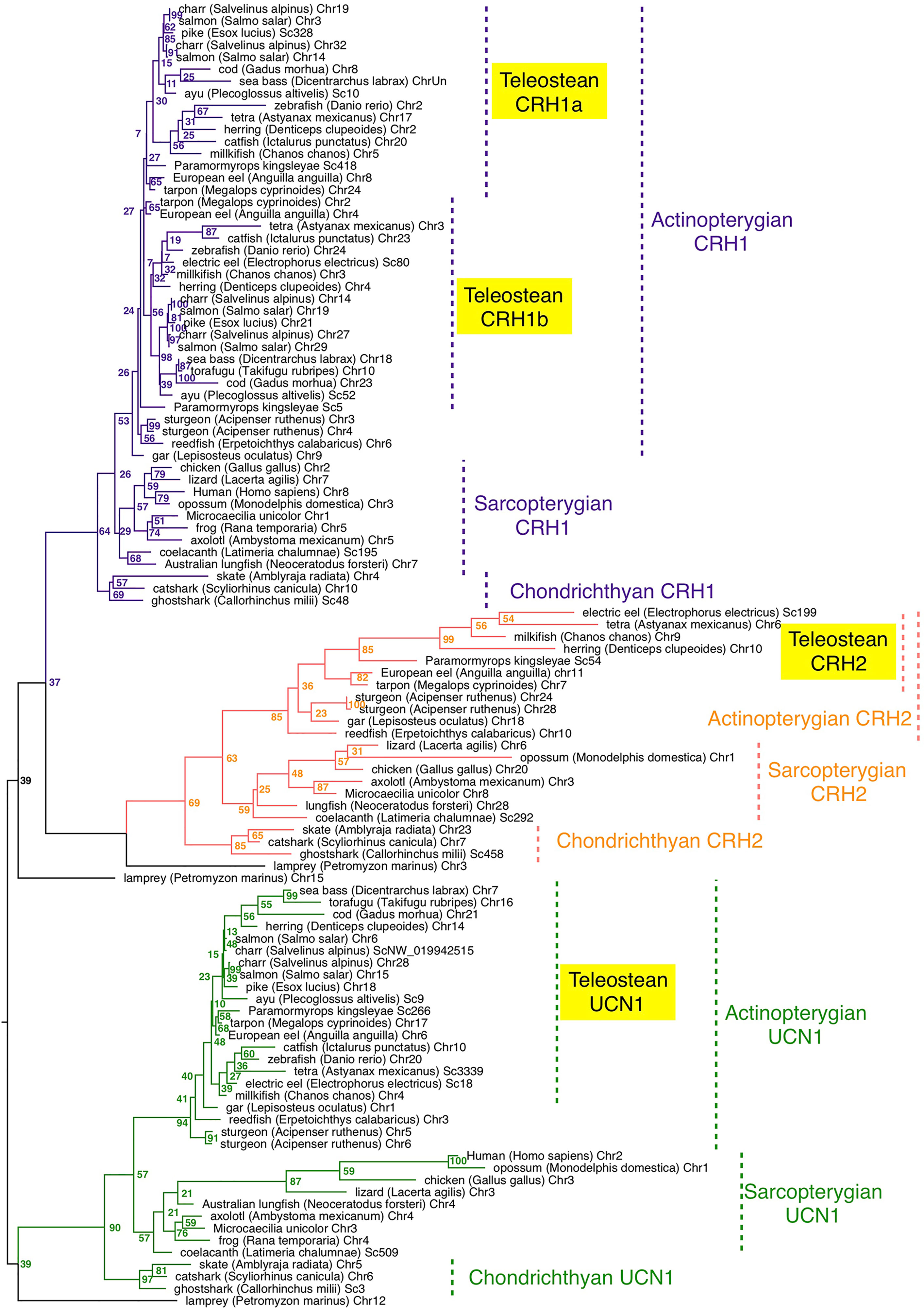
Figure 1 Maximum-likelihood phylogenetic tree of CRH prepropeptide amino acid sequences of vertebrate representatives. Phylogenetic relationships of CRH was inferred using the PhyML algorithm with the WAG substitution matrix and the best nearest neighbour interchange (NNI) and Subtree Pruning and Regrafting (SPR) improvement algorithm. Numbers at the node indicate the confidence percent of 100 bootstrap replications. The three gnathostome monophyletic clades are indicated with different branch colors and the corresponding CRH clade names are indicated using the same colour. Taxonomic group names are indicated at the right of the tree. The list of corresponding gene references is provided in Table S1.
In chondrichthyans, three genes, crh1, crh2 and ucn1, were present in a holocephalan (elephant shark, C. milii) as previously described, and were also retrieved in the present study in representatives of elasmobranchs, including selachians such as catshark, Scyliorhinus canicula and Batoidae such as the thorny skate, Amblyraja radiata (Table S1). Chondrichthyan amino-acid sequences branched at the basis of the osteichthyan CRH1, CRH2 and UCN1 clusters, respectively, also in accordance with vertebrate phylogeny (Figure 1).
In basal sarcopterygians, we found three genes (crh1, crh2 and ucn1) in actinistians (coelacanth, Latimeria chalumae) as previously reported, branching at the basis of the sarcopterygian sequences. We predicted two to three genes in the current status of the lungfish (Dipnoi) genome assembly (Table S1). The genome of the Australian lungfish (Neoceratodus forsteri) encoded indeed two full-length genes, corresponding to crh2 and ucn1 according to our phylogenetic analysis (Figure 1), as well as one crh1 pseudogene showing a frameshift due to indels at two different positions on the CDS. In the West African lungfish (Protopterus annectens) only two genes (crh1 and ucn1) could be retrieved (Figure S2; Table S1). In amphibians, we retrieved only two genes (crh1 and ucn1) in various anuran species (such as Rana temporaria) as previously reported for Xenopus. In contrast we identified a third gene, corresponding to crh2 according to our phylogenetic analysis, in representative species of other amphibian groups, Gymnophiona (Microceacilla unicolor), and Caudata (axolotl, Ambystoma mexicanum) (Figure 1). The three genes (crh1, crh2 and ucn1) were present in representative species of the various groups of sauropsids including birds, as recently reported (48), as well as in protherian and metatherian mammals, in agreement with previous studies (21) (Figure 1; Table S1).
In basal actinopterygians, we retrieved three genes, corresponding to crh1, crh2 and ucn1, in a representative of cladistian Polypteridae (reedfish, Erpetoichthys calabricus), as well as in two representative species of another non-teleost group, holosteans (spotted gar, Lepisosteus oculatus, as previously reported; and bowfin, Amia calva) (Figures 1, S2). In chondrostean species, including Polyodontinae (paddlefish, Polyodon spathula), and Acipenserinae (sterlet sturgeon, Acipenser ruthenus), we found six genes corresponding to duplicated genes for crh1, crh2 and ucn1 (Figures 1, S2) likely resulting from the polyploidization of the genome in these species.
In basal teleosts, four genes were predicted in elopomorphs, Anguillidae (European, American and Japanese eels, Anguilla sp), and Megalopydae (tarpon, Megalops cyprinoides) (Table S1), corresponding to duplicated crh1 (crh1a and crh1b), a single crh2 and a single ucn1, according to our phylogenetic analyses (Figures 1; S2). Crh1b paralog was present in all teleost species investigated in the present study, while crh1a could not be found in the fugu (Takifugu rubripes) in agreement with previous studies (22). Among clupeiforms, full-length crh1a was found in Denticipitidae (dendicle herring), and in Clupeidae (Atlantic herring, Clupea harengus and American shad, Alosa sapidissima), while a crh1a pseudogene was retrieved in an Engraulidae (anchovy, Coilia nasus) (Figure S2; Table S1). No crh1a sequence could be found in another Clupeidae (pilchard, Sardina pilchardus) nor in a gymnotiform (electric eel, Electrophorus electricus), but this may be due to the current status of their genome assemblies. We identified crh2, previously assumed to be lost in teleosts, as a single gene not only in elopomorphs as mentioned above but also in osteoglossomorphs, including Mormyridae (elephant fish, P. kingsleyae), Gymnarchidae (aba, Gymnarchus niloticus), and Osteoglossidae (arapaima, Arapaima gigas), and partial sequence in arowana, as well as in Clupeiformes (denticle and Atlantic herrings, American shad; Alice shad, (Alosa alosa), pilchard, anchovy) and in several ostariophysian groups including Gonorynchiformes (milkfish Chanos chanos), Characiformes (Mexican tetra, Astyanax mexicanus) and Gymnotiformes (electric eel) (Figure 1 and S2; Table S1). In contrast, we could not retrieve any crh2 gene in some other ostariophysian groups, Silurifomes and Cypriniformes. Crh2 gene could not be found either in any euteleostean species investigated, including esociforms, salmoniforms, galaxiiforms, osmeriforms, gadiforms, nor in any of the acanthopterygian groups. A single ucn1 gene could be retrieved in all teleost genomes investigated, and was duplicated in salmonids and some polyploid cyprinids, in relation to the further genome duplication in these lineages (Figures 1 and S2; Table S1). We also retrieved two ucn1 genes in denticle herring, one on the chromosome 14 and the other one on an unplaced scaffold (Tables S1, S3). These two genes showed 99.98% nucleotide identity indicating they might result either from recent sequential duplication or from a genome assembly artefact. In the present study, only the UCN1 carried by the chromosome 14 was further considered.
The gene family members xkr4/5/6/7/9, nkain2/3/4, bhlhe22/23, dnajc5/dnajc5g, trim54/55/101, sulf1/2, eya1/2/4, rock1/2, dlgap1/2/4, lpin1/2/3, and emilin1/2/3 were found to surround the crh1/crh2/ucn1 family genes on the tetraparalogous chromosomic regions of the reedfish (Figures 2–4 and S1), in agreement with their 1R and 2R origin. Additional neighboring gene families were used to further analyze the impact of 3R and 4R in teleosts, such as cspp1 on crh1 paralogon, nol4l, commd7 and uckl1 on crh2 paralogon, pomc and mpv7 on ucn1 paralogon. The synteny comparison among actinopterygians showed that the crh1, crh2 and ucn1 paralogons were duplicated by the teleost 3R WGD and further duplicated by salmonid 4R WGD (SS4R) (Figures 2–4).
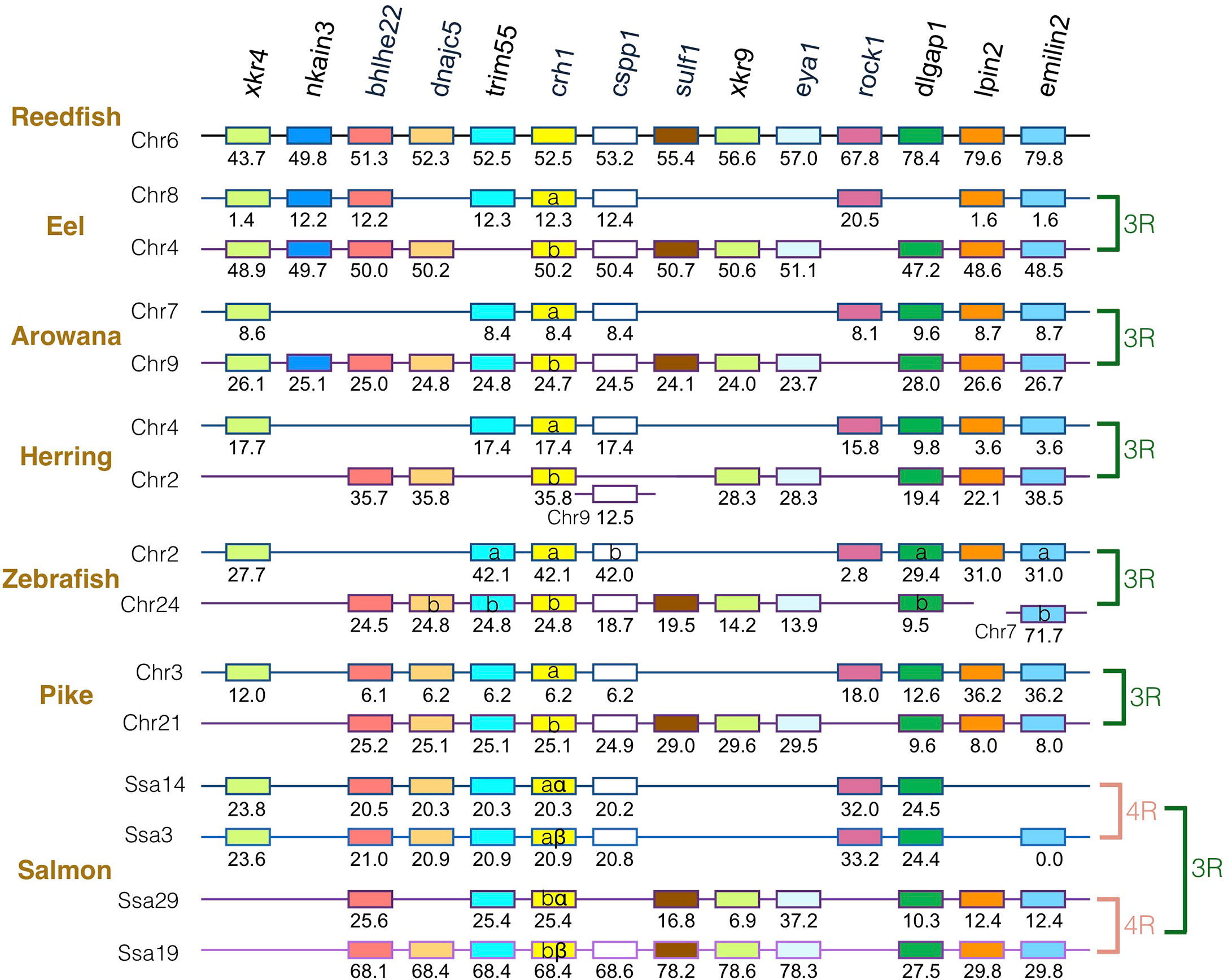
Figure 2 Synteny of crh1 genomic region in actinopterygians. Crh1 gene neighborhood is compared between teleost representatives, eel, arowana, herring, zebrafish, pike and salmon. The polypteriform actinopterygian, reedfish, was used as model of the gene arrangement prior the teleost genome duplication 3R. Chromosomes are indicated by Chr or Ssa for salmon. Gene position are indicated under the genes. Loss of crh1 gene is indicated with a red cross. Neighboring genes non-represented are considered as lost. Gene references are presented in Table S3. Letters a and b indicate the 3R-teleost duplicated paralogs and the symbols α and β the 4R-salmonid duplicated paralogs. Members of gene families conserved on the vertebrate tetraparalogon (issued from 1R/2R WGD) are with the same color in Figures 3–4.
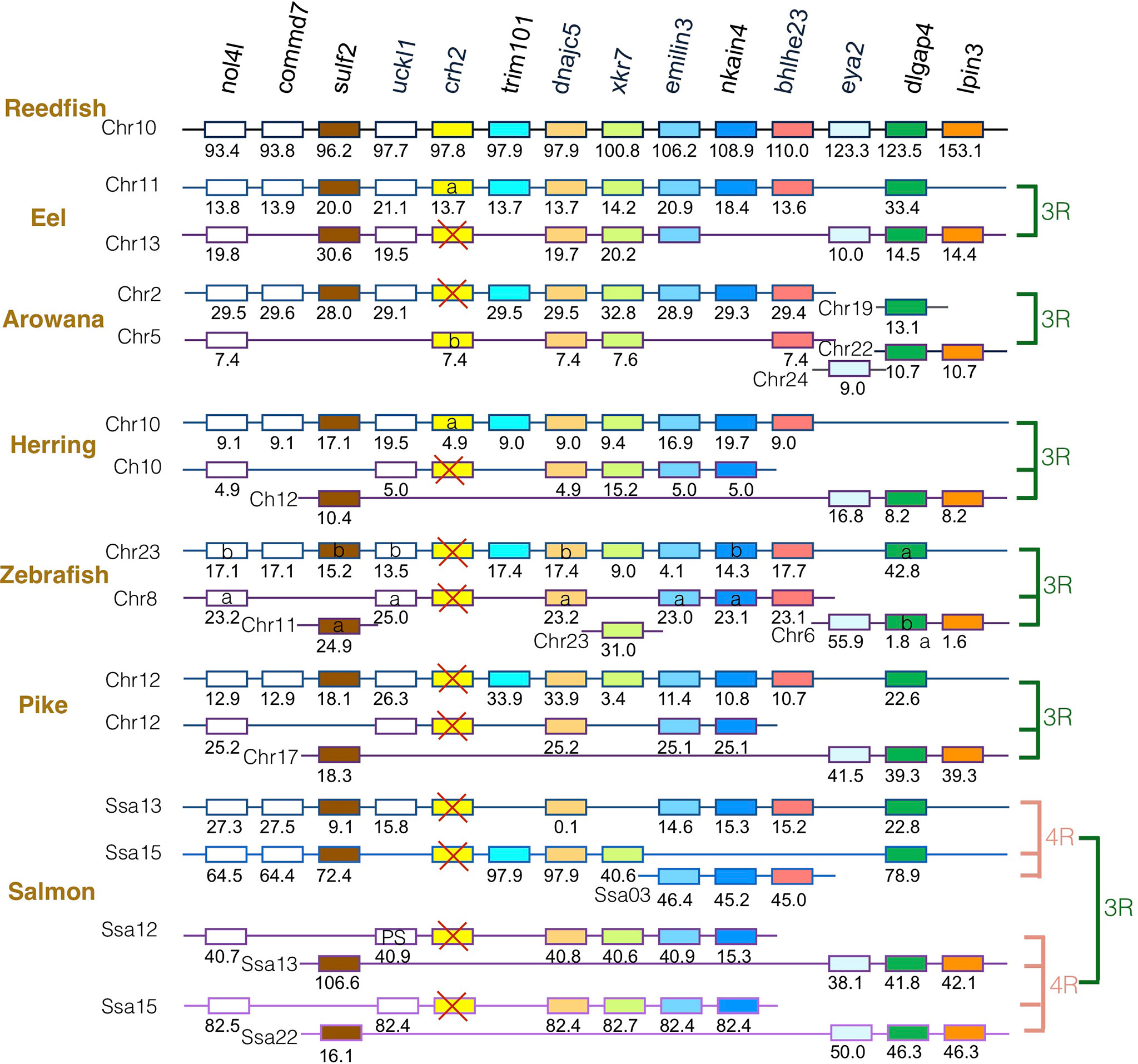
Figure 3 Synteny of crh2 genomic region in actinopterygians. Crh2 gene neighborhood is compared between teleost representatives, eel, arowana, herring, zebrafish, pike and salmon. The polypteriform actinopterygian, reedfish, was used as model of the gene arrangement prior the teleost genome duplication 3R. Chromosomes are indicated by Chr or Ssa for salmon. Gene position are indicated under the genes. Loss of crh2 gene is indicated with a red cross. Neighboring genes non-represented are considered as lost. Gene references are presented in Table S3. Letters a and b indicate the 3R-teleost duplicated paralogs and the symbols α and β the 4R-salmonid duplicated paralogs. Members of gene families conserved on the vertebrate tetraparalogon (issued from 1R/2R WGD) are with the same color in Figures 3, 4.
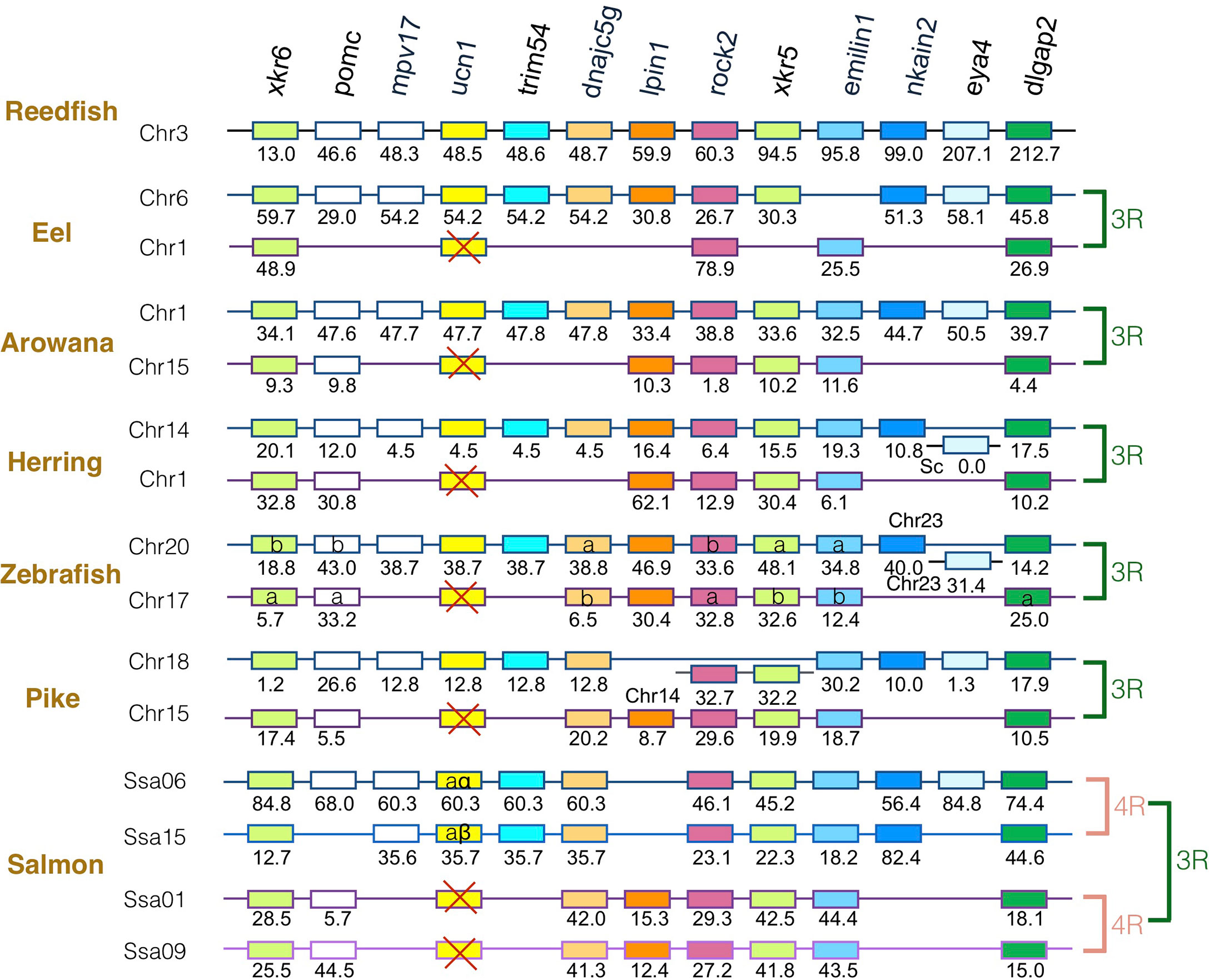
Figure 4 Synteny of ucn1 genomic region in actinopterygians. Ucn1 gene neighborhood is compared between teleost representatives, eel, arowana, herring, zebrafish, pike and salmon. The polypteriform actinopterygian, reedfish, was used as model of the gene arrangement prior the teleost genome duplication 3R. Chromosomes are indicated by Chr or Ssa for salmon. Gene position are indicated under the genes. Loss of ucn1 gene is indicated with a red cross. Neighboring genes non-represented are considered as lost. Gene references are presented in Table S3. Letters a and b indicate the 3R-teleost duplicated paralogs and the symbols α and β the 4R-salmonid duplicated paralogs. Members of gene families conserved on the vertebrate tetraparalogon (issued from 1R/2R WGD) are with the same color in Figures 2–3.
Concerning crh1 paralogon (Figure 2), 3R-duplicated crh1 paralogs (crh1a and crh1b) were conserved in eel, arowana, zebrafish, pike, as well as 3R-duplicated neighboring gene paralogs such as cspp1 and dlgpa1. The salmonid 4R WGD led to quadruplicated crh1 paralogs (crh1aα, crh1aβ, crh1bα, crh1bβ) as well as quadruplicated neighboring genes bhlhe22, trim55 and dlgap1.
Concerning crh2 paralogon (Figure 3), some translocation events occurred in this genomic region. It could still be inferred that some neighboring genes, such as nol4l, sulf2, dnajc5, nkain4 and dlgap4, were conserved as duplicates after 3R and as quadruplicate after 4R. In contrast, a single crh2 gene was present in eel, arowana and herring, and we could not find any crh2 gene in the genome assemblies of zebrafish, pike and salmon. Comparison of neighboring genes conserved as single paralogs allowed to infer that a different crh2 paralog was conserved in eel and herring versus in arowana. The crh2 paralog named crh2a conserved in eel and herring was located in the paralogon bearing the single conserved paralog of commd7, trim101, nkain4, while the crh2 paralog named crh2b conserved in arowana was located on the other paralogon. Since the crh2 gene retrieved from the arowana genome assembly is fragmented, we also examined the crh2 gene neighborhood in another osteoglossomorph species belonging to the Mormyridae, the P. kingsleyae for which crh2 gene encodes a putative functional Crh2 (Table S2). This confirmed the presence of a crh2 (crh2b) gene in the other 3R- ohnologous region than the one carrying eel and herring crh2 (crh2a) gene. The lack of crh2 in pike and salmon suggests that a loss of crh2 already occurred in their common ancestor and therefore no impact of 4R. We observed a chromosome fusion between the two crh2 genomic regions in herring, pike and salmon that may have occurred prior the radiation of Clupeocephala. Such genome reorganization between paralogous regions have already been described for other gene duplicated paralogs such as the melatonin receptors (47). These events were related to intense rate of interchromosomal rearrangements that occurred in some lineages during the rediploidization process after the 3R (49).
Concerning ucn1 paralagon, some neighboring genes such as xkr6, rock2, emilin1, dlgap2 were conserved as duplicates in eel, arowana, herring, zebrafish, and pike and as quadruplicates in salmon, reflecting the impact of 3R and 4R on this genomic region. In contrast, ucn1 was found as a single gene in eel, arowana, herring, zebrafish and pike. This single ucn1 was located in all these species on the paralogon bearing the single conserved paralog of mpv7, trim54, nkain2 and eye4 neighboring genes, indicating that the same ucn1 paralog was conserved in the different teleost species. Synteny analysis indicated that this single ucn1 gene was inherited by salmonids and duplicated by the 4R.
Both European eel crh1a and crh1b paralogous genes, located on the chromosome 8 and 4 respectively, code for 164 amino acid (aa) precursors, prepro-Crh1a and prepro-Crh1b respectively, including a 24 aa signal peptide required for the hormone release, a 97 aa cryptic region with at its C-terminus a proteolytic cleavage site at the position R121, and the 41 aa C-terminal region coding for CRH peptide with a C-terminal Gly-Lys amidation site. At the difference of the prepro-Crh1b, the prepro-Crh1a showed putative additional cleavage sites at the positions R83 and R103. Japanese eel crh (BAP90537), previously characterized (50), is the ortholog to European eel crh1b. The eel crh2 gene located on the chromosome 11 encodes a 149 aa prepropeptide including a 23 aa signal peptide, a cryptic region of 81 aa at the N-terminus and a proteolytic cleavage site at the position R104, and the C-terminal region encoding a 43 aa mature Crh2 with a C-terminal amidation site. The prepro-Crh2 shows a putative additional cleavage site at the position R33. The eel ucn1 gene located on chromosome 6 codes for a 168 aa prepropeptide, including a 24 aa signal peptide, a 101 aa cryptic region, the C-terminal region coding for a 41 aa Ucn1 peptide with a C-terminal amidation site.
The two Atlantic salmon crh1aα and crh1aβ 4R-paralogous genes (located on the chromosomes 14 and 3, respectively) encode 171 and 170 aa prepropeptides, including a 24 aa signal peptide, a 100 and 101 aa cryptic region, respectively, and a C-terminal sequence encoding a 44 aa Crh1 mature peptide with a C-terminal amidation site. The two salmon crh1bα and crh1bβ 4R-paralogous genes (located on chromosomes 29 and 19) both encode for a 167 aa prepropeptide, including a 24 aa signal peptide, and a C-terminal sequence encoding for a 41 aa Crh1 mature peptide with a C-terminal amidation site. The two salmon ucn1α and ucn1β 4R-paralogous genes (located on chromosomes 6 and 15) code for 165 and 162 aa prepropeptides respectively, including a 24 aa signal peptide, a cryptic region of 98 aa and 95 aa respectively, and a 41 aa Ucn1 peptide with a C-terminal amidation site (Figures 5 and S3).
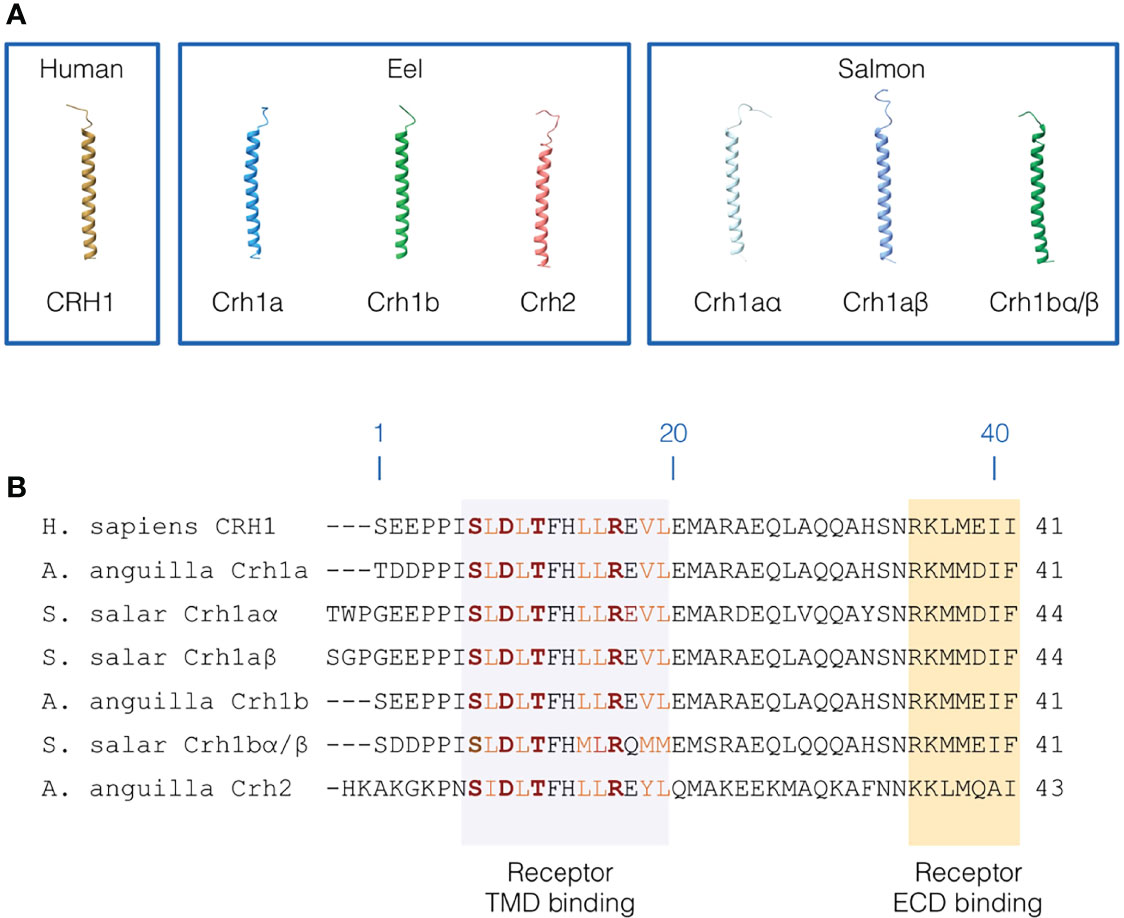
Figure 5 Primary and 3D structure of CRH peptides. (A) 3D structure of human CRH1 peptide, and the three CRH peptides (3R-Crh1a and b, Crh2) in European eel and four CRH peptides (4R-Crh1aα and aβ, 4R-Crh1bα/bβ which have the same sequence) in Atlantic salmon. CRH peptide structures were modelled using iTASSER. (B) Sequence alignment of human CRH1, eel 3R-Crh1a and b and Crh2, and Atlantic salmon 4R-Crh1aα, aβ, and 4R-Crh1bα/bβ. Residue numbering are based on human CRH1. The region of CRH peptides interacting with the CRH receptor transmembrane domain are highlighted in light purple and the region of CRH peptides interacting with the CRH receptor extracellular domain are highlighted in orange. Conserved hydrophobic residues involved in the receptor binding are indicated in orange and the residues involved in electrostatic interaction with the receptor are indicated in red and bold.
Comparison with salmonid cDNA sequences from previous studies indicates that: - Atlantic salmon “crh” (DY733166) (22, 51, 52), Atlantic salmon “crf1b1” (23), Atlantic salmon “crf-b1” (53), and trout, Oncorhynchus mykiss, crh (NM_001124286) (46) correspond to salmon crh1bα in our study; - Atlantic salmon “crf1b2” (23), Atlantic salmon “crf-b2” (53), and trout “crh2” (AY156929 and NM_001124627) (46, 54) correspond to salmon crh1bβ.
The aa sequences of European eel 3R-paralogs Crh1a and Crh1b shared 65 and 90% identity for the prepropeptides and the peptides, respectively. Eel prepro-Crh1a and prepro-Crh1b shared 38 and 42% identity with human prepro-CRH1, and 19 and 16% identity with human prepro-UCN1. Eel Crh1a and Crh1b peptides shared 86 and 95% identity with human CRH peptide, and 49 and 44% identity with human UCN1 peptide. When comparing eel Crh1a and b with eel Crh2 the percentage of identity fell down to 18 and 19% for the prepropeptides and 49% for the peptides. Eel Crh2 peptide shared higher identity with other Crh2 peptides: 88% for the tarpon, 39-67% for the other teleosts, 72-81% for the non-teleost actinopterygians, 41-67% for the sarcopterygians, 56-60% for the chondrichthyans. Eel prepro-Crh2 shared 15 and 11% identity with human prepro-CRH1 and prepro-UCN1, respectively, and eel Crh2 peptide 54 and 36% identity with the human CRH1 and UCN1 peptides. Eel Ucn1 shows 50 and 21% identity with human prepro-UCN1 and prepro-CRH1, respectively, and 60 and 56% identity for the corresponding peptides.
Atlantic salmon 4R-Crh1aα and Crh1aβ paralogs shared 86 and 89% identity between the prepropeptides and the peptides, respectively and the 4R-Crh1bα and Crh1bβ paralogs showed 93 and 100% identity between the prepropeptides and the peptides, respectively. When Crh1aα/β and Crh1bα/β pairs of paralogs were compared, the identity fell down to the range of 56 to 58% between prepropeptides, and to the range of 66 to 68% between peptides. The salmon Crh1aα and Crh1aβ prepropeptides showed 36 and 35% identity with the human prepro-CRH1, respectively, and the Crh1aα and Crh1aβ peptides shared 77 and 82% identity with human CRH1 peptide. The salmon Crh1bα and Crh1bβ prepropeptide showed 38 and 37% identity with the human prepro-CRH1 respectively and Crh1bα/bβ peptides showed 76% identity with human CRH1 peptide, respectively. The percentage of identity fell down when compared with human prepro-UCN1 (18-19%) and UCN1 peptide (40-43%). The two salmon 4R-Ucn1 showed higher identity with human UCN1 (21 and 22% for the prepro-UCN1 respectively, and both 64% for UCN1 peptide) than with human CRH1 (18 and 17% for the prepropeptides, respectively, and both 56% for the peptides).
Using a sequence logo, the overall comparison of CRH1, CRH2 and UCN1 prepropeptides sequences from various vertebrates used in the phylogeny (Figure 1; Table S1), revealed a high sequence conservation of CRH1, CRH2 and UCN1 peptides and of the signal peptides. The conservation of some parts of the cryptic regions was also observed for CRH1 prepropeptide, to lesser extent for UCN1 prepropeptide, but not for CRH2 prepropeptide (Figure S3).
In order to get some insights on potential differences in structure-function relationships between CRH paralogous peptides conserved in eel and in salmon, we further examined their aa sequences and 3D structures.
Human CRH1 was predicted to adopt a variable N-terminal loop conformation (low structure confidence) and a long α-helical conformation for the residues 7 to 39 (Figure 5A). We found a similar 3D structure for eel and salmon Crh peptides. In the eel, the α-helix was predicted for aa 8-39 for both Crh1a and Crh1b and for aa 9-41 for Crh2. In salmon, Crh1aα and Crh1aβ have both a α-helical structure for the residues 10-42 and 9-43, respectively, and Crh1bα/Crh1bβ (which have exactly the same sequence) have a α-helical structure for 8-39 residues. Like in human CRH1, the N-terminal structure adopts a variable loop conformation in eel and salmon Crh peptides (Figure 5A).
The Crh1 or Crh2 peptides in eel and salmon conserved most of the first 7-18 residues involved in human in receptor binding including, the hydrophobic residues (L8, L10, L14, L15, V18 and L19) shaping hydrophobic interactions and the residues (S7, D9, T11 and R16) involved in electrostatic interactions stabilizing the bond peptide conformation within the peptide binding pocket (Figure 5B) (55–57).
In order to get more insight into the fate and functional evolution of the 3R- and the 4R-crh paralogs, we compared crh paralog tissue expression in both eel and salmon.
In eel, the three crh paralogs (crh1a, crh1b and crh2) were not only expressed in the brain but also in other tissues, with differential tissue distribution profiles (Figure 6A). All crh paralog transcripts were found in the brain, but comparison of their mean cycle quantification (Cq) suggested the following range of expression: crh1b (Cq 24.5) > crh1a (Cq 26.0) > crh2 (Cq 29.5). Crh1a gene showed the highest expression in the muscle (Cq 23.3), followed by gonads, heart and various other tissues including the brain; its expression in muscle was 15.5-fold than in the brain. In contrast, crh1b gene was mainly expressed in the brain while its transcript was also detected at a low level in the eye and at the limit of detection in the other tissues (Figure 6A). Crh2 gene was expressed at a low level in the gonad (Cq 27.3), intestine and spleen, and at lower levels or at the limit of detection in the other tissues. Regarding the distribution of the crh paralogs in the brain regions, the two crh1a and b paralogs showed a similar expression profiles but with much higher levels for crh1b than crh1a as mentioned above. For both paralogs, higher transcript levels were found in the frontal brain including olfactory bulbs, telencephalon, mesencephalon and diencephalon, and in medulla oblongata as compared to lower levels in optic tectum, cerebellum epiphysis, retina, saccus vasculosus and cerebellum (Figure 6B). In the pituitary, crh1a was moderately expressed, while crh1b levels were at the limit of the detection. Crh2 transcript was detected in the different brain regions but at very low levels.
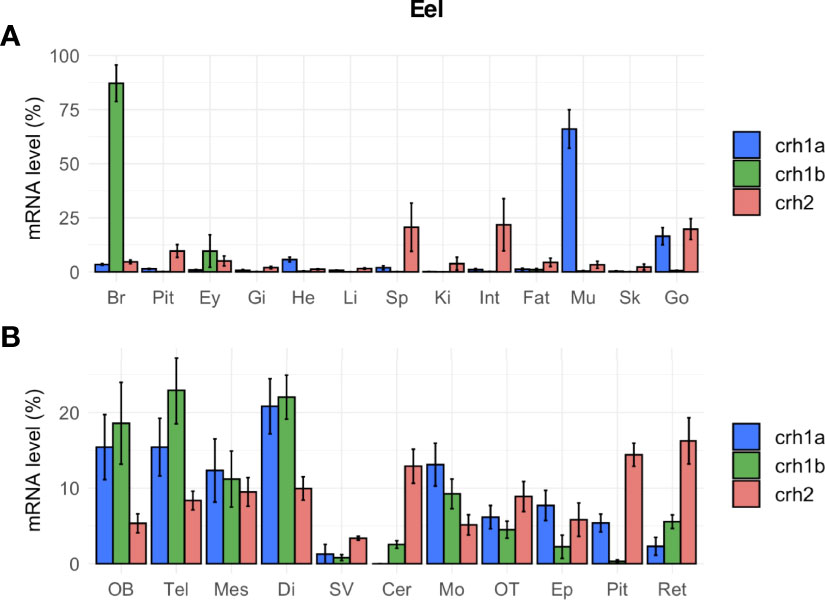
Figure 6 Tissue distribution of crh paralogs in silver female European eels as measured by qPCR. (A) crh1a, crh1b and crh2 expression in the brain (Br), pituitary (Pit), eyes (Ey), and various peripheral organs, gills (Gi), heart (He), liver (Li), spleen (Sp), kidney (Ki), intestine (Int), fat (Fat), muscle (Mu), skin (Sk) and gonad (Go). (B) crh1a, crh1b and crh2 expression in various brain regions: olfactory bulb (OB), telencephalon (Tel), mesencephalon (Mes), diencephalon (Di), saccus vasculosus (SV), cerebellum (Cer), medulla oblongata (Mo), optic tectum (OT), epiphysis (Ep), pituitary (Pit) and retina (Ret). Results (means ± SEM; n=9-10) are presented as percentage per tissue.
In salmon, the four crh1 paralogous genes (crh1aα, crh1aβ, crh1bα, crh1bβ) were expressed in the brain (Figure 7A). Comparison of their mean Cq suggested the following range of expression in the brain: crh1bβ (Cq 25.9) > crh1bα, (Cq 28.3) > crh1aα, (Cq 29.0) > crh1aβ (Cq 29.4). The pair of 4R-crh1aα and crh1aβ paralogs showed a similar tissue distribution profile, with highest expression in muscle [crh1aα (Cq 23.8) and crh1aβ (Cq 26.7)], followed by heart, then skin, retina, brain and pituitary (Figure 7A). The paralogs crh1aα and crh1aβ showed respectively 13- and 3-fold higher transcript levels in the muscle than in the brain. In contrast, the crh1b 4R-pair of paralogs, crh1bα and crh1bβ, are mainly expressed in the brain with very low or no detectable expression in the other tissues. The paralogs, crh1bα and crh1bβ showed, respectively 403- and 50-fold higher transcript levels in the brain than in the muscle. When comparing the expression of 4R-pairs of paralogs in various brain regions, some differences were observed between the two crh1a paralogs, with the crh1aβ mostly expressed in the retina while the crh1aα expression more widely distributed (Figure 7B).
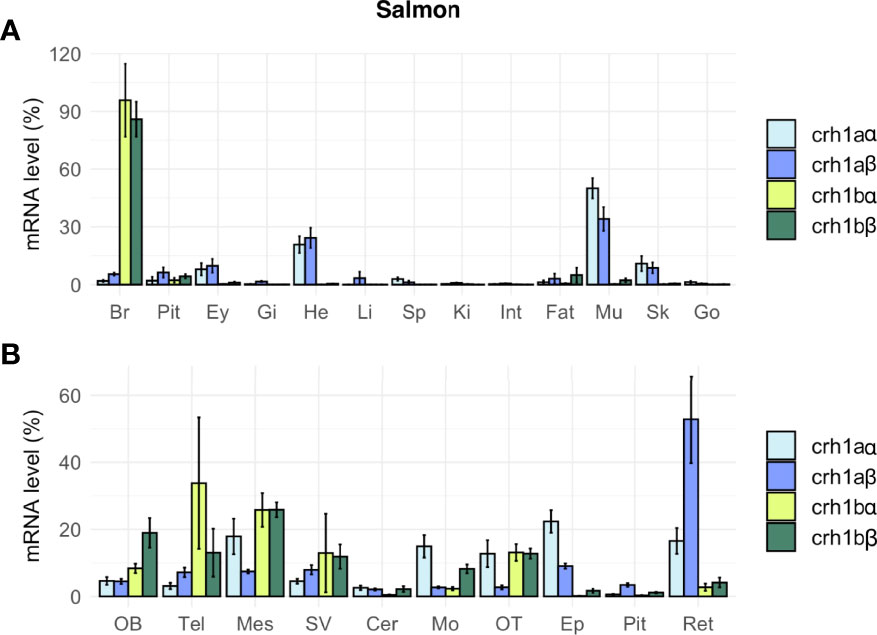
Figure 7 Tissue distribution of crh paralogs in juvenile Atlantic salmon as measured by qPCR. (A) crh1aα, crh1aβ, crh1bα and crh1bβ expression in the brain (Br), pituitary (Pit), eyes (Ey), aand various peripheral organs, gills (Gi), heart (He), liver (Li), spleen (Sp), kidney (Ki), intestine (Int), fat (Fat), muscle (Mu), skin (Sk) and gonad (Go). (B) crh1aα, crh1aβ, crh1bα and crh1bβ expression in various brain regions: olfactory bulb (OB), telencephalon (Tel), mesencephalon (Mes), diencephalon (Di), saccus vasculosus (SV), cerebellum (Cer), medulla oblongata (Mo), optic tectum (OT), epiphysis (Ep), pituitary (Pit) and retina (Ret). Results (means ± SEM; n=8-10) are presented as percentage per tissue.
The evolution of the CRH neuropeptide family has been previously investigated by other authors, unveiling the existence of five crh paralogs in vertebrates, crh (crh1), crh2, ucn (ucn1), ucn2 and ucn3, while a single related peptide gene would be present in invertebrate genomes, named in arthropods, diuretic hormone 44 (DH44). The timing of the crh/ucn family diversification has been under debate (15, 19, 58). It was first presumed that the whole crh/ucn gene family expanded from a chordate crh/ucn single ancestor gene, through the two vertebrate WGD (58). However, the comparison of gene chromosome location showed that the crh/ucn family evolved in two distinct gene environments, suggesting that the crh/ucn family was generated by a first local duplication of a crh/ucn ancestor gene, followed by the two successive rounds of vertebrate WGD along with the secondary losses of some paralogs during the rediploidization events (15, 19, 59). Contrary to what the “crh” versus “ucn” gene names suggest, sequence similarities, together with the location of crh1/crh2/ucn1 and ucn2/ucn3 in two distinct gene environments, point out a closer relationship of ucn (ucn1) with crh1 and crh2 than with ucn2 and ucn3. After the local duplication of the common ancestral crh/ucn gene, the two vertebrate WGD would have given rise on the one hand to the triplet crh, crh2 and ucn1, and on the other hand to ucn2 and ucn3 genes (15, 19, 21). Therefore, based on their phylogenetical relationships, and according to the nomenclature recommended for 1R/2R duplicate genes, ucn (ucn1) could be named “crh3”.
In the present study, we further investigated the evolutionary history of the two crh (crh1 and crh2) and their closest paralog ucn1 throughout vertebrate radiation. Taking advantage of the increasing number of genomes being sequenced, we investigated additional species and key group representatives. In our phylogenetical analysis of the prepropeptide amino acid sequences, the three sequences present in the lamprey, a representative of the most basal group of vertebrates (cyclostomes), clustered at the basis of each vertebrate CRH1, CRH2 and UCN1 clades, respectively, supporting the early origin in vertebrates of the triplet crh1/crh2/ucn1 (19). We also further assessed the presence of the triplet crh1, crh2 and ucn1 in the various gnathostome lineages, in agreement with previous studies (15, 21). Among chondrichthyans, in addition to holocephalan (elephant shark) as previously described, we also showed the presence of crh1, crh2 and ucn1 in representatives of the two subclasses of elasmobranchs, selacians (catshark), and Batoidae (thorny skate), allowing to generalize the presence of the triplet to all chondrichthyan lineages. Various studies suggest the conservation of CRH role as regulator of the stress axis in chondrichthyans as in other vertebrates [for review (60)]. Regarding CRH2, although it has been first discovered in the holocephan elephant shark, knowledge of its role and mode of action remained to be studied.
In basal sarcopterygians, we took advantage of the recent release of two lungfish giant genomes (61, 62) to search for crh/ucn sequences. Thus, in addition to actinistians (coelacanth), where the three genes have been previously reported (15, 21), we also identified the triplet in dipnoans (lungfish). In Australian lungfish we identified full-length crh2 and ucn1, but revealed a pseudogene for crh1, while in West African lungfish we were able to retrieve full-length crh1 and ucn1 but no crh2 gene. If these findings do not result from genome assembly artefacts, this suggests species-specific variations in crh1 and crh2 conservation or loss among lungfish, with possible compensation of function between crh1 and crh2.
In amphibians, crh1 and ucn1 genes, as well as the sauvagine gene, were previously identified while crh2 gene was reported to have been lost in this lineage (15, 21). Isolated from the skin in the leaf frogs P. sauvagii and Pachymedusa dacnicolor, sauvagine is considered as a Ucn1 peptide coded by a ucn1 paralog gene that may have recently highly diverged [for review (63); (64)]. Likely present only in the Phyllomedusinae family, we could not identify any “sauvagine” gene in the available amphibian genomes corresponding to other families. We retrieved only two genes (crh1 and ucn1) in various anuran species (such as Rana temporaria) as previously reported for Xenopus. However, in opposite to the previous assumption of the loss of crh2 in amphibians, we revealed the presence of a crh2 gene in representatives of caudatan and gymnophionan (such as the axolotl) amphibian lineages. The loss of crh2 gene would thus be specific to the anuran lineage. In sauropsids, our study further assessed the conservation of the three crh genes, crh1, crh2 and ucn1 in various lineages, including birds, as recently reported (48). In mammals, we found a crh2 gene only in monotremes and marsupials, confirming its loss in the eutherian lineage (15, 21). These data indicate repeated independent losses of crh2 in tetrapods, such as in anuran amphibians and eutherian mammals. This opens the way to new investigation on the role and mode of action of crh2 in order to understand how its function has evolved through the tetrapod radiation.
In actinopterygians, the crh gene triplet was previously reported in the spotted gar (21, 22), a representative of holosteans, the sister group of teleosts, and belonging with teleosts to the neopterygians. Our study also showed the conservation of crh1, crh2 and ucn1 genes in another holostean species, the bowfin. We also identified the triplet in the reedfish, a representative of the basal actinopterygian group of, Polypteriformes. When we looked at chondrosteans, number of copies was doubled for crh1, crh2 and ucn1 genes in an Acipenseridae, the sterlet and in a Polyondontidae, the paddlefish. Chondrosteans, both Acipenseridae and Polyondotidae, are known to have experienced lineage-specific WGD (65, 66). In our phylogeny analysis, crh2 duplicated paralogs clustered together on one side the sterlet paralogs, and on the other side the paddlefish paralogs (Figure S2). The crh2 pairing suggests independent gene doubling events for the crh2 paralogs in sterlet and paddlefish lineages. Similar relationships have been recently observed for the oxytocin and vasotocin receptors (67) and is in agreement with the independent lineage specific WGD in the Acipenseridae and Polyondotidae (65, 67, 68). Surprisingly, crh1 and ucn1 sterlet duplicated paralogs clustered with one of the respective paddlefish paralogs, with well supported nodes, suggesting the duplication events that produced the crh1 and ucn1 doubling in chondrosteans may have preceded the split between paddlefish and acipenser lineages. Further investigation may clarify the pattern of the impact of lineage-specific WGD on the fate of crh1/crh2/ucn1 triplet on chondrosteans, including also investigation of other sturgeon species which displayed additional WGD events.
We investigated, by phylogeny and synteny analyses, the impact of the teleost-specific WGD (3R) that occurred early in the teleost lineage, and of the additional WGD (4R) that occurred in some specific groups, as well as of gene losses, on crh1 repertoire in various extant teleost species. We found that crh1 duplicated paralogs (crh1a and crh1b) are present in basal teleosts, elopomorphs (eel and tarpon), osteoglossomorphs (arowana) as well as in many clupeocephalan representatives. Synteny analysis supported the 3R WGD origin of crh1a and crh1b paralogs in all teleosts. After 3R, both crh1a and crh1b paralogs have been conserved in most teleost species studied, with a few exceptions where crh1a was missing, such as in a tetraodontiform, the fugu, in agreement with a previous report (22). Our observations further suggest that crh1a may have also been lost independently in some other teleost species, such as in some clupeiforms, with crh1a being present in some Denticipitidae (dendicle herring) and Clupeidae (Atlantic herring, American shad) but detected as a pseudogene in an Engraulidae (anchovy) and missing in some other Clupeidae (pilchard). Crh1a was also missing in a gymnotiform (electric eel). Further studies are needed to assess if the absence crh1a in these species is related to a loss of function or just a genome sequencing artefact. The conservation of crh1b paralog in all teleosts compared to the loss of the crh1a paralog in some species, suggests that the crh1b paralog is subjected to a higher evolutionary pressure than the crh1a duplicate across teleost radiation.
The crh1a and crh1b paralogs were further duplicated in the polyploid carp (Cyprinus carpio) and barbel (Sinocyclocheilus grahami) while the other ostariophysians had only a single copy of each paralog. The doubling pairs of paralogs of carp and barbel showed orthologous relationships (Figure S2) suggesting duplicated pairs arose from the specific cyprinid WGD allotetraploidization event (CC4R) that occurred after 23 Mya and before the split between Sinocyclocheilus and Cyprinus lineages (9.7 Mya) (69). Our phylogenetic and syntenic analyses indicated that the four Atlantic salmon crh1 paralogs paired with the four of other salmonid species, were located on ohnologous genomic regions, in agreement with the 4R-origin of salmonid-specific crh1a and crh1b paralog doubling (22, 23). In the present study, we named the four salmonid paralogs crh1aα, crh1aβ, crh1bα and crh1bβ, respectively, in agreement with the nomenclature for salmonid 4R duplicated genes (31).
In contrast to the conservation of 3R-duplicated crh1a and crh1b paralogs in most teleost species, we found a single ucn1 paralog type in all teleosts investigated, in agreement with previous reports (15, 21, 70). Synteny analysis showed that the same ucn1 paralog was conserved in the different teleost species, supporting an early loss after 3R of the other ucn1 paralog. Two ucn1 paralogs were found in polyploid cyprinids, and in salmonids. Synteny analysis further supported that the single ucn1 gene was inherited by salmonid ancestor and duplicated by the 4R.
The absence of crh2 gene in teleosts was assumed to result from the loss of crh2 gene in a teleost ancestor prior the 3R WGD (15, 19, 21, 22). In order to trace the evolutionary trajectory of the crh2 gene in teleosts, we investigated a large number of species from various teleost groups. Strikingly, our gene search and phylogenetic analysis revealed the presence of a crh2 gene in representative species of basal teleost taxa, elopomorphs and osteoglossomorphs, as well as of some basal clupeocephalan orders such as Clupeiformes, Gonorynchiformes, Characiformes and Gymnotiformes. In contrast, crh2 gene was missing in Siluriformes and Cypriniformes as well as in all euteleosts investigated.
In teleost species where a crh2 was present, a single gene was identified. The presence of only a single crh2 gathering in one clade for teleost species in the phylogenetic analysis, may have suggested an early loss of one of the 3R-duplicated paralogs before the teleost radiation. Synteny analysis confirmed that crh2 gene environment has been duplicated in accordance with the 3R WGD event. However, gene neighborhood exploration revealed that the crh2 gene was not located on the same paralogon in osteoglossopmorphs, as compared to elopomorphs and clupeocephalans, and corresponded to different 3R-paralogs. This raises the question about the timing of crh2 3R-duplicate gene loss. According to our synteny study, both 3R-crh2 paralogs would have been retained after the divergence of the teleosts, with elopomorphs and clupeocephalans having conserved one 3R crh2 paralog, that we named crh2a and lost the crh2b paralog, while osteoglossomorphs having conserved the crh2b paralog and lost the crh2a paralog. The lack of crh2 gene in cypriniforms and siluriforms indicates in addition to the loss of crh1b in a clupocephalan ancestor, crh2a paralog was lost independently twice in the ostariophysian lineage, one time before the radiation of cypriniforms and the other time before the radiation of siluriforms. We could not find any crh2 genes either in any representatives of basal euteleostean taxa, including esociforms, salmoniforms, galaxiiforms and osmeroiforms, supporting the loss of crh2b paralog before the radiation of clupeocepalans, followed by the loss of crh2a paralog before the radiation of euteleosts.
The present data allow us to refute the previous assumption of the absence of crh2 in teleosts and to infer the evolutionary scenario of crh2 in teleosts. Crh2, present in basal actinopterygian taxa, polypterids, chondrosteans, and holosteans, would have been inherited by teleost ancestor and duplicated via the teleost-specific 3R WGD into crh2a and crh2b paralogs. The crh2a paralog would have been lost in osteoglossomorphs while it would have been conserved in elopomorphs and clupeocephalans. Conversely, crh2b would have been conserved in osteoglossomorphs but lost independently in elopomorphs and clupeocephalans. While crh2a has been conserved in various basal clupeocephalan taxa (Clupeiformes, Gonorynchiformes, Characiformes and Gymnotiformes), it would have been lost independently in some other basal clupeocephalan taxa (Siluriformes, Cypriniformes), as well in the euteleostean lineage leading to the lack of any crh2 in the majority of extant teleost species.
The vertebrate CRH1 and CRH2 precursors have all the same overall structure, including a signal peptide, a cryptic region with at its C-terminus a proteolytic cleavage site and the C-terminal region coding for the CRH peptide with a C-terminal Gly-Lys amidation site. The different branch sizes of CRH prepropeptides in our inferred phylogenetic tree indicates different selective constraints between the prepro-CRH. Together with the sequence alignment it showed a higher variability of CRH2 than CRH1 prepropeptide sequences among vertebrates in agreement with previous observations (15, 21). This suggests a higher evolutionary constraint on CRH1 than CRH2, which is also supported by the conservation of crh1 gene in all vertebrates, while crh2 gene had been repeatedly lost in various lineages, such as in anuran amphibians, eutherian mammals, and some teleost taxa. The strong conservation of CRH1 has been associated to the evolutionary pressure related to its central role as coordinator of the corticotropic axis in all vertebrates, while the potential physiological roles of CRH2 have still to be investigated.
When comparing the sequences of Crh1a, Crh1b and Crh2 peptides themselves, a striking sequence conservation is observed, as for example seen in eel and salmon. In salmon, the 4R-duplicated Crh1bα and bβ peptides even have exactly the same sequence. 3D structure prediction revealed, as for human CRH1, the conservation of the alpha helical structure in eel and salmon Crh peptides including eel Crh2. The action of the neuropeptides of CRH family, including CRH1 and CRH2, UCN1 as well as UCN2 and UCN3, is mediated by receptors belonging to the class B of G protein-coupled receptors (GPCR) of the secretin-like receptor superfamily [ (71, 72); for review (73)]. Two CRH receptors, CRHR1 and CRHR2 arose from the vertebrate WGD and have been first characterized in mammals (71, 74–78). The recent single-particle cryoelectron microscopy (cryo-EM) resolution of CRH1 and UCN1 complexes to the CRH receptors revealed a conserved mode of ligand binding and receptor activation for the CRH family peptides and their receptors (55, 56). CRH binding to the receptor induces the rearrangement of the receptor transmembrane domains driving the receptor activation and the coupling with G proteins (55). The C-terminal helical segment of CRH peptide interacts with the receptor extracellular loops promoting recognition specificity and the N-terminal helix segment that bind to the receptor transmembrane domain core pocket stimulates the receptor activation and G protein coupling (55, 57). In addition to the helical structure conservation, Crh peptides in both salmon and eel conserved the residues reported in human CRH as the key residues for the receptor recognition and activation. These data suggest that all these peptides conserved CRH biological ability to stimulate CRH receptors. CRH role has been investigated in teleosts using heterologous CRH1 peptides in vitro and in vivo. In eel, bovine CRH is active and able to stimulate in vitro release of growth hormone (GH) by pituitary cells (79). In salmon, ovine CRH stimulates thyroid-stimulating hormone (TSH) release (80), as well as transcription of gh and tshβ subunit paralogs (81) in vitro. Central administration of ovine or of rat/human CRH increases locomotor activity in Chinook salmon (Oncorhynchus tshawytscha) (82) and downstream migration in smolt coho salmon (Oncorhynchus kisutch) (83).
While CRH1 peptide in mammals is known to exert mainly its actions through the activation of the CRHR1 receptor for which it shows higher affinity, little is known about CRH2 receptor preference. The functional characterization of chicken (Gallus gallus) CRH2 using recombinant chicken CRHR1 and CRHR2 expressed in CHO cells revealed CRH2 is more selective for CRHR2 than CRHR1 while CRH1 activates both receptors with similar potency (48).
Gene coding for CRH receptors (crhr1 and crhr2) underwent duplication during the teleost 3R WGD, but one of the 3R-crhr2 paralog would have been early lost in the teleost lineage. Some species have conserved the two 3R-paralogs of crhr1 such as the tilapia (Oreochromis niloticus) (77). Japanese medaka (Oryzias latipes), belongs to the species that have conserved the pair of 3R-crh1 paralogs, but a single paralog of Crha and Crhb (70). Both medaka Crh1a and Crh1b peptides were reported to be able to stimulate the two CRH receptor types (Crhr1 and Crhr2) transiently expressed in HEK293T cells and subsequently cAMP production with an EC50 for Crh1b peptide slightly lower for both receptors (70). Eels have conserved 3 genes coding for CRHR, two crhr1 and a single crhr2 (LOC118216809, LOC118218028 and LOC118234689). Atlantic salmon has two pair for both crhr1 (LOC106600876 and LOC105023366) and crhr2 (LOC106569328 and LOC106599903), each duplicate arising likely from the duplication of a single 3R-paralog crhr1 and crhr2 during the salmonid tetraploidization (4R). Crhr1 and Crhr2 in chum salmon (Oncorhynchus keta) show highly conserved amino acid sequences with human orthologs (84). Functional study of these receptors showed that rat/human CRH can bind and activate both receptor type with similar potency (84). Further functional experiments using homologous CRH peptides and CRHR will contribute to deeper understanding of the functional evolution of the multiple CRH-CRHR systems in fish.
Conservation of paralogous gene after duplication is under selective pressure likely related to either amplification of function, functional partitioning (subfunctionalization), or emergence of new function (neofunctionalization). The strong conservation of CRH1 and CRH2 peptide sequences and 3D structure as discussed above suggests that conservation of multiple crh genes is not related to major differences in the abilities of the peptides to bind and activate CRH receptors. Selection may have rather targeted differential crh gene tissue expression and regulation. In the present study we got some insights on the potential differential functions of crh paralogous genes by comparing their tissue expression in two key models, a basal teleost, an elopomorph, the European eel, which has conserved both 3R-crh1a and crh1b paralogs, as well as a single crh2 gene, and a salmonid, the Atlantic salmon, which possesses four crh1 paralogs resulting from 4R duplication of crh1a and crh1b, and no crh2 gene. Furthermore, in addition to the relevance of their phylogenetical position and crh repertoire among teleosts, eel and salmon are also of wide interest in biology, ecology, conservation or aquaculture. It should be noted that our study was performed on juvenile fish and that the relative tissue expression of crh paralogs may vary according to maturation status and other physiological conditions, such as stress.
All crh paralog transcripts could be detected by qPCR in the brain, in the eel and salmon, including in the brain region (mesencephalon/diencephalon) where hypophysiotropic neurons are located. Early immunocytochemical studies in mammals located CRH neurons in the paraventricular nucleus of the hypothalamus (11). Since then, this principal center of CRH expression, with neurons projecting to the median eminence, has been confirmed in other tetrapods [birds: (85, 86); reptiles: (87); amphibians: (88–90). In teleosts, due to the absence of hypophyseal portal system and the direct innervation of the pituitary by hypophysiotropic neurons, the situation is slightly different, with CRH neurons in the preoptic area projecting up to the pituitary [Carassius auratus and C. carpio: (91); white sucker C. commersoni: (92, 93); Anguilla species: (94); Salmo, Oncorhynchus and Anguilla species, Mugil ramada and Myoxocephalus octodecimspinosus: (95); rainbow trout: (96); Chinook salmon: (97); tilapia Oreochromis mossambicus: (98)].
In our study, the detection in eel and salmon of the expression of each crh paralog in the brain hypophysiotropic region, suggests that all crh paralogs may potentially be involved in the major role of CRH as brain regulator of the corticotropic axis. However, comparison of Cq values indicates that crh1b paralog is expressed at a higher levels than crh1a in the brain in the eel, and a similar situation is observed in the salmon for the 4R-pair of crh1b paralogs as compared to the 4R-pair of crh1a paralogs. Furthermore, comparison of the distribution of each paralog in the brain and various peripheral tissues showed that crh1b paralogs (crh1b in the eel and the pair of 4R-crh1b in salmon) are mostly expressed in the brain, while crh1a paralogs (crh1a in eel and the pair of 4R-crh1a in salmon) are mostly expressed in the muscle. Our results are in accordance with the distribution of the 3R-crh1 paralogs in other teleosts (22; 70). In medaka crh1a is more expressed in the muscle, heart and gonad than in the brain. In the brain, crh1b is broadly expressed including in the hypophysiotropic neurons in zebrafish, in Astatolipia burtoni and medaka while crh1a paralogs are found in different location through the brain including the ventral hypothalamus but not expressed in the hypophysiotropic neurones (22; 70).
This suggests an early functional partitioning between crh1 paralogs issued from the 3R before teleost radiation, with crh1b playing a major role in the brain and crh1a in the muscle. The potential major role of crh1b paralog in the neuroendocrine control of the corticotropic action, may represent the evolutionary constraint that led to the conservation of crh1b paralog in all extant teleost species while crh1a paralog has been lost in some taxa/species such as in tetraodontiforms (fugu) and some clupeiforms and gymnotiforms.
Concerning 4R-pairs of crh1a and crh1b paralogs in salmon, their general central and peripheral tissue distribution is quite similar within each pair, reflecting some low differentiation of 4R-issued paralogs. Some differences could still be observed when comparing the expression of salmon 4R-paralogs between various brain regions, as also previously reported (23). The authors showed that crh1bα (in their article crf1b1) was the most abundant in the post-smolt brain but that the four paralogs may be involved in the response to various stress exposures (hypoxia, chasing, combination of both and confinement) (23). Crh1bα (in their article crf-b1) was also suggested to be involved in the activation of the pituitary interrenal axis leading to the elevation of cortisol at smoltification (53).
Apart from the hypothalamus, a widespread distribution of CRH peptides or transcripts in the brain has been reported in other vertebrates [mammal: (99); bird: (100); reptile: (87); amphibian: (89)]. In our study, the different paralogs characterized in the European eel and the Atlantic salmon are expressed in the various brain regions. This supports that in addition to be the activator of neuroendocrine axes, CRH1 is involved in the control of many other brain physiological functions including various behaviors [for review: (10)] such as food intake and feeding behavior [mammals: (101); teleosts: (29, 102)], sensory processing, locomotion and migration [vertebrates: (103); teleosts: (29)].
CRH1, originally isolated from the hypothalamus for its neuroendocrine role on the pituitary, is also secreted locally in various peripheral tissues, where it can exert autocrine or paracrine effects, as its receptors are widely distributed [for review: (10)].
Strikingly, the highest level of expression for eel crh1a and salmon 4R-crh1a pair was observed in the skeletal muscle, as compared to other tissues including the brain and various peripheral tissues. A relatively high expression of crh1a was also seen in the heart in salmon and eel. In contrast, crh1b paralog transcripts were at low levels in the skeletal muscle in the eel and undetectable in the salmon, and crh1b paralogs were not detected in the heart in both species. Mirroring the potential major role of crh1b paralog in the brain, this suggests a major potential role of crh1a paralog in the control of muscle and heart functions in teleosts, further supporting the functional partitioning between 3R-crh1 paralogs.
The expression of crh1 in skeletal and cardiac muscles has already been reported in other teleosts. In A. burtoni, crh1b expression was detected using RT-PCR in skeletal muscle and heart (104), but these results were not confirmed for heart and not tested for muscle by qPCR (59). Crh1b was also detected by qPCR in muscle and heart of Schizothorax prenanti at low and middle levels, respectively (105), and of Schizothorax davidi at low levels (106). In the medaka, high levels of crh1a transcripts have been reported in muscle and heart (70). In zebrafish, both crh1a and crh1b mRNAs were detected in the heart (107). These data suggest possible species-specific variations in the functional partitioning and the relative roles of crh1a and crh1b paralogs in skeletal muscle and heart, according to teleost species.
Expression of crh1 has also been reported in the heart of Xenopus laevis, but was absent in the muscle (108). No matter which paralogs are involved, the expression of crh1 in the heart in teleosts allows to raise the hypothesis that CRH1 may have a protective role against stressors in the heart, as shown for CRH-related peptides in mammals [for review: (109)]. In zebrafish, it has recently been reported that hypoxia-reperfusion exposure increased cardiac crh1b expression, and rat/human CRH was protective against hypoxia/reoxygenation-induced apoptosis in vitro in this tissue (107). These data suggest a potential direct action of CRH-like peptides on cardiac myocytes without the involvement of nervous system. CRH in teleosts may also modulate glucose uptake and insulin sensitivity, as urocortin 2 does in mouse skeletal muscle (110). Finally, it can be hypothesized that CRH1 modulation of locomotor and migratory activities in teleosts, as shown in salmon species [Chinook salmon: (82, 111); chum salmon: (83); coho salmon: (112)], may be mediated not only via central actions on behavior and neuroendocrine axes, but also via direct peripheral actions on heart and muscle function. Injection of CRH1 in hypophysectomized rats increases locomotor activity demonstrating that CRH1 can produce behavioral activation independently of its effect on the corticotropic axis (113). In salmon and eel, CRH1 effects in the brain would be mostly ensured by crh1b paralogs while the effects on skeletal muscle and heart by crh1a paralogs.
In the eel, after muscle and heart, crh1a paralog was found to be expressed in the gonads (ovarian tissue from prepubertal silver eels). Low expression of crh1a 4R-paralogs could also be detected in ovarian tissues in immature smolt salmons. In contrast, crh1b paralog transcripts were not detectable in the gonads of both species. Expression of crh1 has been previously reported in the ovary or testis of some other teleosts, such as common carp (114), A. burtoni [crh1b: (104)], fathead minnow (Pimephales promelas) (115). A recent study in zebrafish showed that both crhα and crhβ paralogs (corresponding to crh1a and crh1b) are expressed in the ovary, with a differential regulation of their expression according to vitellogenic stages (116). Furthermore, the authors demonstrated an inhibitory effect of CRH1 on estradiol production by zebrafish follicular cells in vitro. In mammals, CRH1 immunoreactivity or expression was reported in gonads, testis [rat: (117–119)] and ovary [rat: (120); human: (121, 122)]. In the testis, CRH1, produced by the Leydig cells of the testis, exerts autocrine inhibition of testosterone biosynthesis (118, 123). Similarly, in the ovary, CRH1 inhibits steroid biosynthesis as shown in vitro [rat and human granulosa cells: (124); human granulosa-lutein cells: (125); human thecal cells: (126); mouse preantral follicles: (127)]. Altogether these data suggest a conserved direct inhibitory role of CRH1 on gonadal steroidogenesis in vertebrates, which may participate, together with interactions between neuroendocrine axes, in the well-known stress-related impairment of reproduction, as observed in many species [for review (128)]. In teleosts, this role of CRH1 on the gonads may likely be fulfilled by either one or the other crh1 paralog according to species.
In the pituitary, we detected a low expression of 3R-paralog crh1a in eel and of 4R-paralog crh1aβ in salmon. This is in agreement with the recent report of the expression of “crf1a2” (corresponding to crh1aβ in our study) in the salmon pituitary (23). In other teleosts, the expression of crh in the pituitary has been mostly studied for crh1b, the firstly identified “classical” crh, and absence [goldfish, Northern-blot: (129); European flounder (Platichthys flesus), RT-PCR: (130); A. burtoni RT-PCR: (104)] or low expression [S. prenanti, qPCR: (105); grass carp (Ctenopharyngodon idellus), RNAseq: (131); RT-PCR: (59)] were reported. When crh1a was studied, no expression could be detected in the pituitary [medaka, ISH: (70); grass carp, RNAseq: (131); A. burtoni RT-PCR: (59)]. Together with ours, these data suggest species-specific variation in the expression of 3R-crh1a or b paralog in the pituitary across teleost species. In other vertebrates, the amphibian Xenopus laevis (108) and various mammals [rat: (132); baboon: (133)], CRH expression (transcript or peptide) have been reported in the pituitary. Altogether, this indicates that CRH may exert a paracrine/autocrine role in the pituitary in addition to its neuroendocrine major role, in teleosts as in other vertebrates.
In the retina, we detected the expression of crh1a in the eel while the crh1b was at the limit of the detection. In the salmon, the 4R-crh1aβ was much more expressed than the other 4R-crh1 paralogs. The relatively higher expression of a crh1a paralog in both eel and salmon as compare to crh1b reflects a functional differentiation of the 3R paralogs in these species. The low retinal expression levels of 4R-crh1aα as compared to 4R-crh1aβ indicates that functional differentiation also occurred between the 4R-crh1a paralogs. Early immunocytochemical studies reported the presence of CRH in the retina in teleosts [goldfish: (134)] as well as in birds [chicken: (135)], reptiles [turtle: (136)] and mammals [rat: (137, 138)]. Among teleosts, when retina was tested, it was shown to be a site of expression of crh1 [crh1a in medaka: (70); crh1b in Shizothorax species: (105, 106); crh1a and crh1b in A. burtoni: (59, 104)]. Using ISH, 22 observed some differences in the retinal expression of crh1 a and b paralogs between two teleosts, A. burtoni and zebrafish (22). Crh1a expression was either absent (zebrafish) or present (A. burtoni) in the retina, while crh1b expression was present but in distinct cells: amacrine and ganglion cells in zebrafish; amacrine and bipolar cells in A. burtoni. As for other organs, this suggests species-specific variations among teleosts in the respective roles of crh1a and b paralogs in the retina. Whatever the crh1 paralog involved, the expression of CRH in the retina supports possible local autocrine/paracrine actions of CRH1 in the neuromodulation of retinal function in teleosts (22).
This study is the first one to demonstrate the presence of crh2 gene in teleosts and thus to investigate the tissue distribution of its expression in a teleost species. Measure of crh2 transcripts by qPCR in the eel revealed a tissue expression profile distinct from those of crh1a and crh1b. Crh2 expression was low and widely distributed in central and peripheral tissues, with no striking major site of expression. The general tissue distribution shows that, differently from crh1a and b, the main sites of expression of crh2 are the gonads, the intestine and the spleen. However, even in these tissues, crh2 transcripts were at lower (gonads, spleen) or similar (intestine) levels than those of crh1a, as suggested by Cq comparison, while crh1b transcripts were at the limit of detection. CRH activation of the corticotropic axis, in response to stress, is known to stimulate digestive tract motility as for instance in human (139). The expression of crh2 and crh1a in the eel intestine suggests an additional autocrine/paracrine action of CRH in addition to its central effect. The expression of crh2 and crh1a in the spleen in the eel also suggests a direct action of CRH on immune function. Chronic stress is recognized to impair immunity and notably humoral response in mammals, an effect involving not only CRH-activated corticotropic axis but also a direct effect of CRH on immune cells as indicated by the expression of CRH receptors by splenic B cells (140). Crh2 expression in the digestive tract and spleen of chicken has also been reported (48). When looking at the detailed central distribution, crh2 expression was very low, with slightly higher transcript levels in the retina, pituitary and cerebellum than in the other brain parts, a distribution profile also different from those of crh1a and b.
The low and wide expression of crh2 in the eel does not support any major specific function for crh2, as compared to crh1a and crh1b which in contrast may play out predominant roles in the muscle and in the brain, respectively. To the best of our knowledge only one other study investigated the central and peripheral tissue distribution of crh2 in vertebrates: in the chicken, qPCR analysis showed that crh2 is widely expressed in various brain regions as well in multiple peripheral tissues (48), similarly to our finding in the eel. Differently, a very limited distribution of crh2 was observed in the spotted gar, but using ISH on the brain, with crh2 expression restricted to a specific nucleus of the hindbrain and in particular no expression detected in the retina. Further studies, including representatives of other actinopterygians as well as of other vertebrate taxa having conserved crh2, are clearly needed to evaluate common and divergent patterns of crh2 expression across vertebrate radiation.
The low expression and wide distribution pattern of crh2 in a basal teleost, the eel, suggests a low specific evolutionary constraint, paving the way to the repeated losses of crh2 through teleost radiation, such as shown in our study for siluriforms, cypriniforms and the euteleostean lineage, leading to the absence of crh2 in the majority of extant teleost species. A similar situation may have occurred in tetrapods with the losses of crh2 in anuran amphibians and eutherian mammals.
The pituitary gland and its control by brain neurohormones is a major anatomical and functional innovation of vertebrates. Families of neuropeptides and receptors, pre-existing in metazoans before the emergence of vertebrates, have been diversified via the two vertebrate WGD and recruited for the control of the pituitary (for review: 24). This is the case for CRH, the neurohormone responsible for the control of the vertebrate corticotropic axis, which represents a typical case of neofunctionalization in the CRH/UCN family, as compared to the ancestral role in non-vertebrate metazoans. The present study further assesses the role of vertebrate crh1 paralog in the neuroendocrine control of the corticotropic axis, a strong evolutionary constrain that led to its conservation in all extant vertebrates. It highlights the subfunctionalization between the duplicated crh1 paralogs issued from teleost-specific WGD (3R), with crh1b assuming the neuroendocrine control of the pituitary and being conserved in all extant teleost species, while crh1a being mainly involved in local autocrine/paracrine functions, with a lower selective pressure leading to species-specific variations in crh1a expression and functions, up to the loss of crh1a paralog in some teleost species. Concerning the more recently identified vertebrate crh2 paralog, this study reveals some wider conservation across vertebrates than previously assumed, with its presence in additional vertebrate groups including elasmobranchs, dipnoans, caudatan and gymnophionan amphibians, actinopterygian polypterids, chondrosteans and teleosts. The low and wide tissue expression of crh2 as observed in a basal teleost, the eel, suggests various local autocrine/paracrine functions. Future studies should aim at investigating the regulation of crh2 expression in relation to development, maturation and stress challenges in teleost species, such as the eel, that have retained this paralog. These results revisit the repertoire of crh in teleosts and highlight functional divergences that may have contributed to the conservation of various crh paralogs in teleosts. This study also supports that no major specific function of crh2 paralog would have led to low evolutionary constraint and repeated losses of crh2 across vertebrate radiation, such as in some amphibians, in eutherian mammals and in various teleosts including the large group of euteleosts.
All relevant data is contained within the article: The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Cuvier Ethic Committee France (No. 68–027).
Design of the study: GM, PM, KR, SD. Experiments: GM, XM. Analysis and interpretation of data: GM, SA, KR, SD. Writing of the manuscript: GM, KR, SD. All the authors reviewed and approved the submitted version of the manuscript.
This work was supported by grants from Alliance Sorbonne University DESYNCHRO SU-14-R-CDV-07-1 and from the French National Research Agency SALTEMP No ANR-12-ADAP-0021 and NEMO No. ANR- 14-CE02-0020.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank E. Ryckelynck and his team from Nodaiwa (Paris, France) and the team of CNSS, as well as Dr. A-G Lafont for their kind cooperation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.937218/full#supplementary-material
Supplementary Table 1 | References of crh neuropeptide sequences used for phylogenetic analysis. The table provides the species names, gene prediction/annotation, names, identifiers, and locus references.
Supplementary Table 2 | References and location of the genes used for the crh neuropeptide synteny analysis. The table lists the neighboring genes of the crh genes in reedfish, eel, arowana, herring, zebrafish, pike, salmon. The data includes gene identifiers, and location.
Supplementary Table 3 | Primer sets used for quantitative PCR. The table provides the name and 5’-3’ nucleotide sequences of the primer sets used for qPCR of the crh1 and crh2 paralog genes and gene references in European eel and Atlantic salmon.
Supplementary Figure 1 | Venn diagram showing the distribution of the gene family members on the crh-bearing chromosomes in the reedfish genomes.
Supplementary Figure 2 | Maximum-likelihood phylogenetic tree of CRH precursor amino acid sequences with a focus in actinopterygian representatives. Maximum-likelihood phylogenetic tree of CRH precursor amino acid sequences with a focus in actinopterygian representatives. Phylogenetic relationships of the prepro-CRH was inferred using the PhyML algorithm with the WAG substitution matrix and the best nearest neighbour interchange (NNI) and Subtree Pruning and Regrafting (SPR) improvement algorithm. Numbers at the node indicate the confidence percent of 100 bootstrap replication. The three gnathostome monophyletic clades are indicated with different branch colors, purple for the CRH1, pink for the CRH2 and green for the UCN1. Taxonomic group names are indicated at the right of the tree, with colors and the corresponding animal silhouettes (http://www.phylopic.org).
Supplementary Figure 3 | Sequence logo of CRH1, CRH2 and UCN1 prepropeptides. The logo sequence was built for each CRH prepropeptides from the sequence alignment of the phylogenetic tree (Figure 1) using CLC Main Workbench (QIAGEN). Consensus position of different domains in the CRH prepropeptides are underlined: the signal peptide, the cryptic region, the CRH peptides cleavage site (indicated by an arrow) and the CRH peptides with its amidation site. It shows a high conservation of CRH1, CRH2 and UCN1 peptide sequences and a good conservation of the peptide signal. Conserved motifs are observed within the cryptic of CRH1 and UCN1 precursors but not for CRH2 precursors that show high sequence divergences within the cryptic regions. CRH1 precursor sequences of mammals and sauropsids are longer than other CRH1 sequences (around 30 aa longer), the additional residues are distributed between the conserved motifs of cryptic region.
1. Rousseau K, Dufour S, Sachs LM. Interdependence of Thyroid and Corticosteroid Signaling in Vertebrate Developmental Transitions. Front Ecol Evol (2021) 9:735487. doi: 10.3389/fevo.2021.735487
2. Watanabe Y, Grommen SVH, De Groef B. Corticotropin-Releasing Hormone: Mediator of Vertebrate Life Stage Transitions? Gen Comp Endocrinol (2016) 228:60–8. doi: 10.1016/j.ygcen.2016.02.012
3. Guillemin R, Rosenberg B. Humoral Hypothalamic Control of Anterior Pituitary : A Study With Combined Tissue Cultures. Endocrinology (1955) 57:599–607. doi: 10.1210/endo-57-5-599
4. Saffran M, Schally AV, Benfey BG. Stimulation of the Release of Corticotropin From the Adenohypophysis by a Neurohypophysial Factor. Endocrinology (1955) 57:439–44. doi: 10.1210/endo-57-4-439
5. Vale W, Spiess J, Rivier C, Rivier J. Characterization of a 41-Residue Ovine Hypothalamic Peptide That Stimulates Secretion of Corticotropin and β-Endorphin. Science (1981) 213:1394–7. doi: 10.1097/00006254-198205000-00013
6. Furutani Y, Morimoto Y, Shibahara S, Noda M, Takahashi H, Hirose T, et al. Cloning and Sequence Analysis of cDNA for Ovine Corticotropin-Releasing Factor Precursor. Nature (1983) 301:537–40. doi: 10.1038/301537a0
7. Jingami H, Mizuno N, Takahashi H, Shibahara S, Furutani Y, Imura H, et al. Cloning and Sequence Analysis of cDNA for Rat Corticotropin-Releasing Factor Precursor. FEBS Lett (1985) 191:63–6. doi: 10.1093/nar/16.19.9338
8. Rivier J, Spiess J, Vale W. Characterization of Rat Hypothalamic Corticotropin-Releasing Factor. Proc Natl Acad Sci U S A (1983) 80:4851–5. doi: 10.1073/pnas.80.15.4851
9. Shibahara S, Morimoto Y, Furutani Y, Notake M, Takahashi H, Shimizu S, et al. Isolation and Sequence Analysis of the Human Corticotropin-Releasing Factor Precursor Gene. EMBO J (1983) 2:775–9. doi: 10.1002/j.1460-2075.1983.tb01499.x
10. Lovejoy DA, Balment RJ. Evolution and Physiology of the Corticotropin-Releasing Factor (CRF) Family of Neuropeptides in Vertebrates. Gen Comp Endocrinol (1999) 115:1–22. doi: 10.1006/gcen.1999.7298
11. Bloom FE, Battenberg ELF, Rivier J, Vale W. Corticotropin Releasing Factor (CRF): Immunoreactive Neurones and Fibers in Rat Hypothalamus. Regul Pept (1982) 4:43–8. doi: 10.1016/0167-0115(82)90107-0
12. Montecucchi P, Anastasi A, de Castaglione R, Erspamer V. Isolation and Amino Ai-Cid Composition of Sauvagine. Int J Pept Protein Res (1980) 16:191–9. doi: 10.1111/j.1399-3011.1980.tb02952.x
13. Lederis K, Letter A, McMaster D, Moore G, Schlesinger D. Complete Amino Acid Sequence of Urotensin I, a Hypotensive and Corticotropin-Releasing Neuropeptide From Catostomus. Science (1982) 218:162–5. doi: 10.1126/science.6981844
14. Vaughan J, Donaldson C, Bittencourt J, Perrin MH, Lewis K, Sutton S, et al. Urocortin, a Mammalian Neuropeptide Related to Fish Urotensin I and to Corticotropin-Releasing Factor. Nature (1995) 378:287–92. doi: 10.1038/378287a0
15. Cardoso JCR, Bergqvist CA, Félix RC, Larhammar D. Corticotropin-Releasing Hormone Family Evolution: Five Ancestral Genes Remain in Some Lineages. J Mol Endocrinol (2016) 57:73–86. doi: 10.1530/JME-16-0051
16. Lovejoy DA, de Lannoy L. Evolution and Phylogeny of the Corticotropin-Releasing Factor (CRF) Family of Peptides: Expansion and Specialization in the Vertebrates. J Chem Neuroanat (2013) 54:50–6. doi: 10.1016/j.jchemneu.2013.09.006
17. Reyes TM, Lewis K, Perrin MH, Kunitake KS, Vaughan J, Arias CA, et al. Urocortin II: A Member of the Corticotropin-Releasing Factor (CRF) Neuropeptide Family That is Selectively Bound by Type 2 CRF Receptors. Proc Natl Acad Sci U S A (2001) 98:2843–8. doi: 10.1073/pnas.051626398
18. Lewis K, Li C, Perrin MH, Blount A, Kunitake K, Donaldson C, et al. Identification of Urocortin III, an Additional Member of the Corticotropin-Releasing Factor (CRF) Family With High Affinity for the CRF2 Receptor. Proc Natl Acad Sci U S A (2001) 98:7570–5. doi: 10.1073/pnas.121165198
19. Cardoso JCR, Bergqvist CA, Larhammar D. Corticotropin-Releasing Hormone (CRH) Gene Family Duplications in Lampreys Correlate With Two Early Vertebrate Genome Doublings. Front Neurosci (2020) 14:672. doi: 10.3389/fnins.2020.00672
20. Nock TG, Chand D, Lovejoy DA. Identification of Members of the Gonadotropin-Releasing Hormone (GnRH), Corticotropin-Releasing Factor (CRF) Families in the Genome of the Holocephalan, Callorhinchus Milii (Elephant Shark). Gen Comp Endocrinol (2011) 171:237–44. doi: 10.1016/j.ygcen.2011.02.001
21. Grone BP, Maruska KP. A Second Corticotropin-Releasing Hormone Gene (CRH2) is Conserved Across Vertebrate Classes and Expressed in the Hindbrain of a Basal Neopterygian Fish, the Spotted Gar (Lepisosteus Oculatus). J Comp Neurol (2015) 523:1125–43. doi: 10.1002/cne.23729
22. Grone BP, Maruska KP. Divergent Evolution of Two Corticotropin-Releasing Hormone (CRH) Genes in Teleost Fishes. Front Neurosci (2015) 9:365. doi: 10.3389/fnins.2015.00365
23. Lai F, Royan MR, Gomes AS, Espe M, Aksnes A, Norberg B, et al. The Stress Response in Atlantic Salmon (Salmo Salar L.): Identification and Functional Characterization of the Corticotropin-Releasing Factor (Crf) Paralogs. Gen Comp Endocrinol (2021) 313:113894. doi: 10.1016/j.ygcen.2021.113894
24. Dufour S, Quérat B, Tostivint H, Pasqualini C, Vaudry H, Rousseau K. Origin and Evolution of the Neuroendocrine Control of Reproduction in Vertebrates, With Special Focus on Genome and Gene Duplications. Physiol Rev (2020) 100:869–943. doi: 10.1152/physrev.00009.2019
25. Flik G, Klaren PHM, Van Den Burg EH, Metz JR, Huising MO. CRF and Stress in Fish. Gen Comp Endocrinol (2006) 146:36–44. doi: 10.1016/j.ygcen.2005.11.005
26. Wendelaar Bonga SE. The Stress Response in Fish. Physiol Rev (1997) 77:591–625. doi: 10.1152/physrev.1997.77.3.591
27. Volkoff H. The Neuroendocrine Regulation of Food Intake in Fish: A Review of Current Knowledge. Front Neurosci (2016) 10:540. doi: 10.3389/fnins.2016.00540
28. Guo H, Dixon B. Understanding Acute Stress-Mediated Immunity in Teleost Fish. Fish Shellfish Immunol Rep (2021) 2:100010. doi: 10.1016/j.fsirep.2021.100010
29. Matsuda K. Regulation of Feeding Behavior and Psychomotor Activity by Corticotropin-Releasing Hormone (CRH) in Fish. Front Neurosci (2013) 7:91. doi: 10.3389/fnins.2013.00091
30. Pasquier J, Cabau C, Nguyen T, Jouanno E, Severac D, Braasch I, et al. Gene Evolution and Gene Expression After Whole Genome Duplication in Fish: The PhyloFish Database. BMC Genomics (2016) 17:1–10. doi: 10.1186/s12864-016-2709-z
31. Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, et al. Lineage-Specific Rediploidization is a Mechanism to Explain Time-Lags Between Genome Duplication and Evolutionary Diversification. Genome Biol (2017) 18:1–14. doi: 10.1186/s13059-017-1241-z
32. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating Signal Peptides From Transmembrane Regions. Nat Methods (2011) 8:785–6. doi: 10.1038/nmeth.1701
33. Pejaver V, Hsu WL, Xin F, Dunker AK, Uversky VN, Radivojac P. The Structural and Functional Signatures of Proteins That Undergo Multiple Events of Post-Translational Modification. Protein Sci (2014) 23:1077–93. doi: 10.1002/pro.2494
34. Southey BR, Amare A, Zimmerman TA, Rodriguez-Zas SL, Sweedler JV. NeuroPred: A Tool to Predict Cleavage Sites in Neuropeptide Precursors and Provide the Masses of the Resulting Peptides. Nucleic Acids Res (2006) 34:267–72. doi: 10.1093/nar/gkl161
35. Gouy M, Guindon S, Gascuel O. Sea View Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol Biol Evol (2010) 27:221–4. doi: 10.1093/molbev/msp259
36. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst Biol (2010) 59:307–21. doi: 10.1093/sysbio/syq010
37. Lefort V, Longueville JE, Gascuel O. SMS: Smart Model Selection in PhyML. Mol Biol Evol (2017) 34:2422–4. doi: 10.1093/molbev/msx149
38. Whelan S, Goldman N. A General Empirical Model of Protein Evolution Derived From Multiple Protein Families Using a Maximum-Likelihood Approach. Mol Biol Evol (2001) 18:691–9. doi: 10.1093/oxfordjournals.molbev.a003851
39. Louis A., Nguyen N. T. T., Muffato M., Roest Crollius H.. Genomicus update 2015: KaryoView and MatrixView Provide a Genome-Wide Perspective to Multispecies Comparative Genomics. Nucleic Acids Research (2015) 43(D1):D682–D689
40. Roy A, Kucukural A, Zhang Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat Protoc (2010) 5:725–38. doi: 10.1038/nprot.2010.5
41. Yang J, Zhang Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res (2015) 43:W174–81. doi: 10.1093/nar/gkv342
42. McGuffin LJ, Aldowsari FMF, Alharbi SMA, Adiyaman R. ModFOLD8: Accurate Global and Local Quality Estimates for 3D Protein Models. Nucleic Acids Res (2021) 49:W425–30. doi: 10.1093/nar/gkab321
43. Pettersen E, Goddard T, Huang C, Meng E, Couch G, Croll T, et al. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci (2020) 30:70–82. doi: 10.1002/pro.3943
44. Fleming MS, Maugars G, Lafont A-G, Rancon J, Fontaine R, Nourizadeh-Lillabadi R, et al. Functional Divergence of Thyrotropin Beta-Subunit Paralogs Gives New Insights Into Salmon Smoltification Metamorphosis. Sci Rep (2019) 9:4561. doi: 10.1038/s41598-019-40019-5
45. Maugars G, Dufour S, Cohen-Tannoudji JL, Quérat B. Multiple Thyrotropin B-Subunit and Thyrotropin Receptor-Related Genes Arose During Vertebrate Evolution. PloS One (2014) 9:e111361. doi: 10.1371/journal.pone.0111361
46. Doyon C, Gilmour KM, Trudeau VL, Moon TW. Corticotropin-Releasing Factor and Neuropeptide Y mRNA Levels are Elevated in the Preoptic Area of Socially Subordinate Rainbow Trout. Gen Comp Endocrinol (2003) 133:260–71. doi: 10.1016/S0016-6480(03)00195-3
47. Maugars G, Nourizadeh-Lillabadi R, Weltzien FA. New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts. Front Endocrinol (Lausanne) (2020) 11:538196. doi: 10.3389/fendo.2020.538196
48. Bu G, Fan J, Yang M, Lv C, Lin Y, Meng F, et al. Identification of a Novel Functional Corticotropin-Releasing Hormone (CRH2) in Chickens and its Roles in Stimulating Pituitary Tshβ Expression and ACTH Secretion. Front Endocrinol (Lausanne) (2019) 10:595. doi: 10.3389/fendo.2019.00595
49. Braasch I, Gehrke A, Smith J, Kawasaki K, Et A. The Spotted Gar Genome Illuminates Vertebrate Evolution and Facilitates Human-to-Teleost Comparisons. Nat Genet (2016) 48:427–37. doi: 10.1038/ng.3526
50. Amano M, Mizusawa N, Okubo K, Amiya N, Mizusawa K, Chiba H, et al. Cloning of Corticotropin-Releasing Hormone (CRH) Precursor cDNA and Immunohistochemical Detection of CRH Peptide in the Brain of the Japanese Eel, Paying Special Attention to Gonadotropin-Releasing Hormone. Cell Tissue Res (2014) 356:243–51. doi: 10.1007/s00441-013-1784-6
51. Koop BF, Von Schalburg KR, Leong J, Walker N, Lieph R, Cooper GA, et al. A Salmonid EST Genomic Study: Genes, Duplications, Phylogeny and Microarrays. BMC Genomics (2008) 9:545. doi: 10.1186/1471-2164-9-545
52. Pooley NJ, Tacchi L, Secombes CJ, Martin SAM. Inflammatory Responses in Primary Muscle Cell Cultures in Atlantic Salmon (Salmo Salar). BMC Genomics (2013) 14:747. doi: 10.1186/1471-2164-14-747
53. Culbert BM, Regish AM, Hall DJ, McCormick SD, Bernier NJ. Neuroendocrine Regulation of Plasma Cortisol Levels During Smoltification and Seawater Acclimation of Atlantic Salmon. Front Endocrinol (2022) 13:859817. doi: 10.3389/fendo.2022.859817
54. Pérez-Maceira JJ, Otero-Rodiño C, Mancebo MJ, Soengas JL, Aldegunde M. Food Intake Inhibition in Rainbow Trout Induced by Activation of Serotonin 5-HT2C Receptors is Associated With Increases in POMC, CART and CRF mRNA Abundance in Hypothalamus. J Comp Physiol B Biochem Syst Environ Physiol (2016) 186:313–21. doi: 10.1007/s00360-016-0961-9
55. Liang YL, Belousoff MJ, Zhao P, Koole C, Fletcher MM, Truong TT, et al. Toward a Structural Understanding of Class B GPCR Peptide Binding and Activation. Mol Cell (2020) 77:656–668.e5. doi: 10.1016/j.molcel.2020.01.012
56. Ma S, Shen Q, Zhao LH, Mao C, Zhou XE, Shen DD, et al. Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors. Mol Cell (2020) 77:669–680.e4. doi: 10.1016/j.molcel.2020.01.013
57. Seidel L, Zarzycka B, Zaidi SA, Katritch V, Coin I. Structural Insight Into the Activation of a Class B G-Protein-Coupled Receptor by Peptide Hormones in Live Human Cells. Elife (2017) 6:1–25. doi: 10.7554/eLife.27711
58. Endsin MJ, Michalec O, Manzon LA, Lovejoy DA, Manzon RG. CRH Peptide Evolution Occurred in Three Phases: Evidence From Characterizing Sea Lamprey CRH System Members. Gen Comp Endocrinol (2017) 240:162–73. doi: 10.1016/j.ygcen.2016.10.009
59. Grone BP, Butler JM, Wayne CR, Maruska KP. Expression Patterns and Evolution of Urocortin and Corticotropin-Releasing Hormone Genes in a Cichlid Fish. J Comp Neurol (2021) 529:2596–619. doi: 10.1002/cne.25113
60. Bouyoucos IA, Schoen AN, Wahl RC, Anderson WG. Ancient Fishes and the Functional Evolution of the Corticosteroid Stress Response in Vertebrates. Comp Biochem Physiol Part A Mol Integr Physiol (2021) 260:111024. doi: 10.1016/j.cbpa.2021.111024
61. Meyer A, Schloissnig S, Franchini P, Du K, Woltering JM, Irisarri I, et al. Giant Lungfish Genome Elucidates the Conquest of Land by Vertebrates. Nature (2021) 590:284–9. doi: 10.1038/s41586-021-03198-8
62. Wang K, Wang J, Zhu C, Yang L, Ren Y, Ruan J, et al. African Lungfish Genome Sheds Light on the Vertebrate Water-to-Land Transition. Cell (2021) 184:1362–1376.e18. doi: 10.1016/j.cell.2021.01.047
63. Denver RJ. Stress Hormones Mediate Environment-Genotype Interactions During Amphibian Development. Gen Comp Endocrinol (2009) 164:20–31. doi: 10.1016/j.ygcen.2009.04.016
64. Boorse GC, Crespi EJ, Dautzenberg FM, Denver RJ. Urocortins of the South African Clawed Frog, Xenopus Laevis: Conservation of Structure and Function in Tetrapod Evolution. Endocrinology (2005) 146:4851–60. doi: 10.1210/en.2005-0497
65. Cheng P, Huang Y, Du H, Li C, Lv Y, Ruan R, et al. Draft Genome and Complete Hox-Cluster Characterization of the Sterlet Sturgeon (Acipenser Ruthenus). Front Genet (2019) 10:776. doi: 10.3389/fgene.2019.00776
66. Du K, Stöck M, Kneitz S, Klopp C, Woltering JM, Adolfi MC, et al. The Sterlet Sturgeon Genome Sequence and the Mechanisms of Segmental Rediploidization. Nat Ecol Evol (2020) 4:841–52. doi: 10.1038/s41559-020-1166-x
67. Ocampo Daza D, Bergqvist CA, Larhammar D. The Evolution of Oxytocin and Vasotocin Receptor Genes in Jawed Vertebrates: A Clear Case for Gene Duplications Through Ancestral Whole-Genome Duplications. Front Endocrinol (Lausanne) (2022) 12:792644. doi: 10.3389/fendo.2021.792644
68. Cheng P, Huang Y, Lv Y, Du H, Ruan Z, Li C, et al. The American Paddlefish Genome Provides Novel Insights Into Chromosomal Evolution and Bone Mineralization in Early Vertebrates. Mol Biol Evol (2021) 38:1595–607. doi: 10.1093/molbev/msaa326
69. Xu P, Xu J, Liu G, Chen L, Zhou Z, Peng W, et al. The Allotetraploid Origin and Asymmetrical Genome Evolution of the Common Carp Cyprinus Carpio. Nat Commun (2019) 10:1–11. doi: 10.1038/s41467-019-12644-1
70. Hosono K, Kikuchi Y, Miyanishi H, Hiraki-Kajiyama T, Takeuchi A, Nakasone K, et al. Teleocortin: A Novel Member of the CRH Family in Teleost Fish. Endocrinology (United States) (2015) 156:2949–57. doi: 10.1210/en.2015-1042
71. Chen R, Lewis KA, Perrin MH, Vale WW. Expression Cloning of a Human Corticotropin-Releasing-Factor Receptor. Proc Natl Acad Sci U S A (1993) 90:8967–71. doi: 10.1073/pnas.90.19.8967
72. Lovenberg TW, Liaw CW, Grigoriadis DE, Clevenger W, Chalmers DT, De Souza EB, et al. Cloning and Characterization of a Functionally Distinct Corticotropin-Releasing Factor Receptor Subtype From Rat Brain. Proc Natl Acad Sci U S A (1995) 92:836–40. doi: 10.1073/pnas.92.3.836
73. Chalmers DT, Lovenberg TW, Grigoriadis DE, Behan DP, De Souza EB. Corticotrophin-Releasing Factor Receptors: From Molecular Biology to Drug Design. Trends Pharmacol Sci (1996) 17:166–72. doi: 10.1016/0165-6147(96)81594-X
74. Chang CP, Pearse RV, O’Connell S, Rosenfeld MG. Identification of a Seven Transmembrane Helix Receptor for Corticotropin-Releasing Factor and Sauvagine in Mammalian Brain. Neuron (1993) 11:1187–95. doi: 10.1016/0896-6273(93)90230-O
75. Perrin M, Donaldson C, Chen R, Blount A, Berggren T, Bilezikjian L, et al. Identification of a Second Corticotropin-Releasing Factor Receptor Gene and Characterization of a cDNA Expressed in Heart. Proc Natl Acad Sci U S A (1995) 92:2969–73. doi: 10.1073/pnas.92.7.2969
76. Perrin M, Donaldson C, Chen R, Lewis K, Vale W. Cloning and Functional Expression of a Rat Brain Corticotropin Releasing Factor (CRF) Receptor. Endocrinology (1993) 133:3058–61. doi: 10.1210/endo.133.6.8243338
77. Cardoso JCR, Félix RC, Bergqvist CA, Larhammar D. New Insights Into the Evolution of Vertebrate CRH (Corticotropin-Releasing Hormone) and Invertebrate DH44 (Diuretic Hormone 44) Receptors in Metazoans. Gen Comp Endocrinol (2014) 209:162–70. doi: 10.1016/j.ygcen.2014.09.004
78. Hwang JI, Moon MJ, Park S, Kim DK, Cho EB, Ha N, et al. Expansion of Secretin-Like G Protein-Coupled Receptors and Their Peptide Ligands via Local Duplications Before and After Two Rounds of Whole-Genome Duplication. Mol Biol Evol (2013) 30:1119–30. doi: 10.1093/molbev/mst031
79. Rousseau K, Le Belle N, Marchelidon J, Dufour S. Evidence That Corticotropin-Releasing Hormone Acts as a Growth Hormone-Releasing Factor in a Primitive Teleost, the European Eel (Anguilla Anguilla). J Neuroendocrinol (1999) 11:385–92. doi: 10.1046/j.1365-2826.1999.00334.x
80. Larsen DA, Swanson P, Dickey JT, Rivier J, Dickhoff WW. In Vitro Thyrotropin-Releasing Activity of Corticotropin-Releasing Hormone-Family Peptides in Coho Salmon, Oncorhynchus Kisutch. Gen Comp Endocrinol (1998) 109:276–85. doi: 10.1006/gcen.1997.7031
81. Fleming MS, Maugars G, Martin P, Dufour S, Rousseau K. Differential Regulation of the Expression of the Two Thyrotropin Beta Subunit Paralogs by Salmon Pituitary Cells In Vitro. Front Endocrinol (Lausanne) (2020) 11:603538. doi: 10.3389/fendo.2020.603538
82. Clements S, Schreck CB, Larsen DA, Dickhoff WW. Central Administration of Corticotropin Releasing Hormone Stimulates Locomotor Activity in Juvenile Chinook Salmon (Oncorhynchus Tshawytscha). Gen Comp Endocrinol (2002) 125:319–27. doi: 10.1006/gcen.2001.7707
83. Ojima D, Iwata M. Central Administration of Growth Hormone-Releasing Hormone Triggers Downstream Movement and Schooling Behavior of Chum Salmon (Oncorhynchus Keta) Fry in an Artificial Stream. Comp Biochem Physiol A Mol Integr Physiol (2009) 152:293–8. doi: 10.1016/j.cbpa.2008.06.037
84. Pohl S, Darlison MG, Clarke WC, Lederis K, Richter D. Cloning and Functional Pharmacology of Two Corticotropin-Releasing Factor Receptors From a Teleost Fish. Eur J Pharmacol (2001) 430:193–202. doi: 10.1016/S0014-2999(01)01391-7
85. Péczely P, Antoni FA. Comparative Localization of Neurons Containing Ovine Corticotropin Releasing Factor (CRF)-Like and Neurophysin-Like Immunoreactivity in the Diencephalon of the Pigeon (Columba Livia Domestica). J Comp Neurol (1984) 228:69–80. doi: 10.1002/cne.902280108
86. Vandenborne K, De Groef B, Geelissen SME, Boorse GC, Denver RJ, Kühn ER, et al. Molecular Cloning and Developmental Expression of Corticotropin-Releasing Factor in the Chicken. Endocrinology (2005) 146:301–8. doi: 10.1210/en.2004-0608
87. Bugnon C, Fellmann D, Gouget A, Bresson J, Clavequin M, Hadjyiassemis M, et al. Corticoliberin Neurons: Cytophysiology, Phylogeny and Ontogeny. J Steroid Biochem (1984) 20:183–95. doi: 10.1016/0022-4731(84)90205-X
88. Bhargava S, Rao PDP. Distribution of Corticotropin-Releasing Factor Immunoreactive Neurons in the Brain of the Tigerfrog, Rana Tigrina. Neurosci Lett (1993) 154:27–30. doi: 10.1016/0304-3940(93)90163-F
89. Fasolo A, Andreone C, Vandesande F. Immunohistochemical Localization of Corticotropin-Releasing Factor (CRF)-Like Immunoreactivity in the Hypothalamus of the Newt, Triturus Cristatus. Neurosci Lett (1984) 49:135–42. doi: 10.1016/0304-3940(84)90149-6
90. Olivereau M, Vandesande F, Boucique E, Ollevier F, Olivereau J. Immunocytochemical Localization and Spatial Relation to the Adenohypophysis of a Somatostatin-Like and Corticotropin-Releasing Factor-Like Peptide in the Brain of Four Amphibian Species. Cell Tissue Res (1987) 247:317–24. doi: 10.1007/BF00218313
91. Olivereau M, Ollevier F, Vandesande F, Verdonck W. Immunocytochemical Identification of CRF-Like and SRIF-Like Peptides in the Brain and the Pituitary of Cyprinid Fish. Cell Tissue Res (1984) 237:379–82. doi: 10.1007/BF00217162
92. Yulis CR, Lederis K, Wong KL, Fisher AWF. Localization of Urotensin I- and Corticotropin-Releasing Factor-Like Immunoreactivity in the Central Nervous System of Catostomus Commersoni. Peptides (1986) 7:79–86. doi: 10.1016/0196-9781(86)90065-3
93. Yulis CR, Lederis K. Co-Localization of the Immunoreactivities of Corticotropin-Releasing Factor and Arginine Vasotocin in the Brain and Pituitary System of the Teleost Catostomus Commersoni. Cell Tissue Res (1987) 247:267–73. doi: 10.1007/BF00218308
94. Olivereau M, Moons L, Olivereau J, Vandesande F. Coexistence of Corticotropin-Releasing Factor-Like Immunoreactivity and Vasotocin in Perikarya of the Preoptic Nucleus in the Eel. Gen Comp Endocrinol (1988) 70:41–8. doi: 10.1016/0016-6480(88)90092-5
95. Olivereau M, Olivereau J. Localization of CRF-Like Immunoreactivity in the Brain and Pituitary of Teleost Fish. Peptides (1988) 9:13–21. doi: 10.1016/0196-9781(88)90004-6
96. Ando H, Hasegawa M, Ando J, Urano A. Expression of Salmon Corticotropin-Releasing Hormone Precursor Gene in the Preoptic Nucleus in Stressed Rainbow Trout. Gen Comp Endocrinol (1999) 113:87–95. doi: 10.1006/gcen.1998.7182
97. Matz SP, Hofeldt GT. Immunohistochemical Localization of Corticotropin-Releasing Factor in the Brain and Corticotropin-Releasing Factor and Thyrotropin-Stimulating Hormone in the Pituitary of Chinook Salmon (Oncorhynchus Tshawytscha). Gen Comp Endocrinol (1999) 114:151–60. doi: 10.1006/gcen.1999.7253
98. Pepels PPLM, Meek J, Wendelaar Bonga SE, Balm PHM. Distribution and Quantification of Corticotropin-Releasing Hormone (CRH) in the Brain of the Teleost Fish Oreochromis Mossambicus (Tilapia). J Comp Neurol (2002) 453:247–68. doi: 10.1002/cne.10377
99. Fischman AJ, Moldow RL. Distribution of CRF-Like Immunoreactivity in the Rabbit. Peptides (1982) 3:841–3. doi: 10.1016/0196-9781(82)90025-0
100. Józsa R, Scanes CG, Vigh S, Mess B. Functional Differentiation of the Embryonic Chicken Pituitary Gland Studied by Immunohistological Approach. Gen Comp Endocrinol (1979) 39:158–63. doi: 10.1016/0016-6480(79)90221-1
101. Richard D, Lin Q, Timofeeva E. The Corticotropin-Releasing Factor Family of Peptides and CRF Receptors: Their Roles in the Regulation of Energy Balance. Eur J Pharmacol (2002) 440:189–97. doi: 10.1016/S0014-2999(02)01428-0
102. Bernier NJ. The Corticotropin-Releasing Factor System as a Mediator of the Appetite-Suppressing Effects of Stress in Fish. Gen Comp Endocrinol (2006) 146:45–55. doi: 10.1016/j.ygcen.2005.11.016
103. Lowry CA, Moore FL. Regulation of Behavioral Responses by Corticotropin-Releasing Factor. Gen Comp Endocrinol (2006) 146:19–27. doi: 10.1016/j.ygcen.2005.12.006
104. Chen CC, Fernald RD. Sequences, Expression Patterns and Regulation of the Corticotropin-Releasing Factor System in a Teleost. Gen Comp Endocrinol (2008) 157:148–55. doi: 10.1016/j.ygcen.2008.04.003
105. Wang T, Zhou C, Yuan D, Lin F, Chen H, Wu H, et al. Schizothorax Prenanti Corticotropin-Releasing Hormone (CRH): Molecular Cloning, Tissue Expression, and the Function of Feeding Regulation. Fish Physiol Biochem (2014) 40:1407–15. doi: 10.1007/s10695-014-9935-6
106. Yuan D, Wang B, Tang T, Lei L, Zhou C, Li Z, et al. Characterization and Evaluation of the Tissue Distribution of CRH, Apelin, and GnRH2 Reveal Responses to Feeding States in Schizothorax Davidi. Fish Physiol Biochem (2021) 47:421–38. doi: 10.1007/s10695-020-00922-5
107. Williams TA, Bergstrome JC, Scott J, Bernier NJ. CRF and Urocortin 3 Protect the Heart From Hypoxia/Reoxygenation-Induced Apoptosis in Zebrafish. Am J Physiol Regul Integr Comp Physiol (2017) 313:R91–R100. doi: 10.1152/ajpregu.00045.2017
108. Boorse GC, Denver RJ. Widespread Tissue Distribution and Diverse Functions of Corticotropin-Releasing Factor and Related Peptides. Gen Comp Endocrinol (2006) 146:9–18. doi: 10.1016/j.ygcen.2005.11.014
109. Davidson SM, Rybka AE, Townsend PA. The Powerful Cardioprotective Effects of Urocortin and the Corticotropin Releasing Hormone (CRH) Family. Biochem Pharmacol (2009) 77:141–50. doi: 10.1016/j.bcp.2008.08.033
110. Chen A, Brar B, Choi CS, Rousso D, Vaughan J, Kuperman Y, et al. Urocortin 2 Modulates Glucose Utilization and Insulin Sensitivity in Skeletal Muscle. Proc Natl Acad Sci U S A (2006) 103:16580–5. doi: 10.1073/pnas.0607337103
111. Clements S, Schreck CB. Central Administration of Corticotropin-Releasing Hormone Alters Downstream Movement in an Artificial Stream in Juvenile Chinook Salmon (Oncorhynchus Tshawytscha). Gen Comp Endocrinol (2004) 137:1–8. doi: 10.1016/j.ygcen.2004.02.004
112. Ojima D, Iwata M. Central Administration of Growth Hormone-Releasing Hormone and Corticotropin-Releasing Hormone Stimulate Downstream Movement and Thyroxine Secretion in Fall-Smolting Coho Salmon (Oncorhynchus Kisutch). Gen Comp Endocrinol (2010) 168:82–7. doi: 10.1016/j.ygcen.2010.04.007
113. Eaves M, Thatcher-Britton K, Rivier J, Vale W, Koob GF. Effects of Corticotropin Releasing Factor on Locomotor Activity in Hypophysectomized Rats. Peptides (1985) 6:923–6. doi: 10.1016/0196-9781(85)90323-7
114. Huising MO, van der Aa LM, Metz JR, de Fátima Mazon A, van Kemenade BML, Flik G. Corticotropin-Releasing Factor (CRF) and CRF-Binding Protein Expression in and Release From the Head Kidney of Common Carp: Evolutionary Conservation of the Adrenal CRF System. J Endocrinol (2007) 193:349–57. doi: 10.1677/JOE-07-0070
115. Garcia-Reyero N, Villeneuve D, Kroll K, Liu L, Orlando E, Watanabe K, et al. Expression Signatures for a Model Androgen and Antiandrogen in the Fathead Minnow (Pimephales Promelas) Ovary. Environ Sci Technol (2009) 43:2614–9. doi: 10.1021/es8024484
116. Zhou H, Chen A, Lu W. Corticotropin-Releasing Hormone Reduces Basal Estradiol Production in Zebrafish Follicular Cells. Mol Cell Endocrinol (2021) 527:111222. doi: 10.1016/j.mce.2021.111222
117. Audhya T, Hollander CS, Schlesinger DH, Hutchinson B. Structural Characterization and Localization of Corticotropin-Releasing Factor in Testis. Biochim Biophys Acta (BBA)/Protein Struct Mol (1989) 995:10–6. doi: 10.1016/0167-4838(89)90226-4
118. Fabbri A, Tinajero J, Dufau M. Corticotropin-Releasing Factor is Produced by Rat Leydig Cells and has a Major Local Antireproductive Role in the Testis. Endocrinology (1990) 127:1541–3. doi: 10.1210/endo-127-3-1541
119. Yoon D, Sklar C, David R. Presence of Immunoreactive Corticotropin-Releasing Factor in Rat Testis. Endocrinology (1988) 122:759–61. doi: 10.1210/endo-122-2-759
120. Mastorakos G, Webster EL, Friedman TC, Chrousos GP. Immunoreactive Corticotropin-Releasing Hormone and its Binding Sites in the Rat Ovary. J Clin Invest (1993) 92:961–8. doi: 10.1172/JCI116672
121. Asakura H, Zwain IH, Yen SSC. Expression of Genes Encoding Corticotropin-Releasing Factor (CRF), Type 1 CRF Receptor, and CRF-Binding Protein and Localization of the Gene Products in the Human Ovary. J Clin Endocrinol Metab (1997) 82:2720–5. doi: 10.1210/jc.82.8.2720
122. Mastorakos G, Scopa CD, Vryonidou A, Friedman TC, Kattis D, Phenekos C, et al. Presence of Immunoreactive Corticotropin-Releasing Hormone in Normal and Polycystic Human Ovaries. J Clin Endocrinol Metab (1994) 79:1191–7. doi: 10.1210/jc.79.4.1191
123. Ulisse S, Fabbri A, Dufau ML. Corticotropin-Releasing Factor Receptors and Actions in Rat Leydig Cells. J Biol Chem (1989) 264:2156–63. doi: 10.1016/s0021-9258(18)94155-4
124. Calogero AE, Burrello N, Negri-Cesi P, Papale L, Palumbo MA, Cianci A, et al. Effects of Corticotropin-Releasing Hormone on Ovarian Estrogen Production In Vitro. Endocrinology (1996) 137:4161–6. doi: 10.1210/endo.137.10.8828472
125. Ghizzoni L, Mastorakos G, Vottero A, Barreca A, Furlini M, Cesarone A, et al. Corticotropin-Releasing Hormone (CRH) Inhibits Steroid Biosynthesis by Cultured Human Granulosa-Lutein Cells in a CRH and Interleukin-1 Receptor- Mediated Fashion. Endocrinology (1997) 138:4806–11. doi: 10.1210/endo.138.11.5474
126. Erden HF, Zwain IH, Asakura H, Yen SSC. Corticotropin-Releasing Factor Inhibits Luteinizing Hormone-Stimulated P450c17 Gene Expression and Androgen Production by Isolated Thecal Cells of Human Ovarian Follicles. J Clin Endocrinol Metab (1998) 83:448–52. doi: 10.1210/jc.83.2.448
127. Dinopoulou V, Partsinevelos GA, Mavrogianni D, Anagnostou E, Drakakis P, Makrigiannakis A, et al. The Effect of CRH and its Inhibitor, Antalarmin, on in Vitro Growth of Preantral Mouse Follicles, Early Embryo Development, and Steroidogenesis. Endocrinology (2013) 154:222–31. doi: 10.1210/en.2012-1838
128. Rousseau K, Prunet P, Dufour S. Special Features of Neuroendocrine Interactions Between Stress and Reproduction in Teleosts. Gen Comp Endocrinol (2021) 300:113634. doi: 10.1016/j.ygcen.2020.113634
129. Bernier NJ, Lin X, Peter RE. Differential Expression of Corticotropin-Releasing Factor (CRF) and Urotensin I Precursor Genes, and Evidence of CRF Gene Expression Regulated by Cortisol in Goldfish Brain. Gen Comp Endocrinol (1999) 116:461–77. doi: 10.1006/gcen.1999.7386
130. Lu E, Dow L, Gumusgoz S, Brierley MJ, Warne JM, McCrohan CR, et al. Coexpression of Corticotropin-Releasing Hormone and Urotensin I Precursor Genes in the Caudal Neurosecretory System of the Euryhaline Flounder (Platichthys Flesus): A Possible Shared Role in Peripheral Regulation. Endocrinology (2004) 145:5786–97. doi: 10.1210/en.2004-0144
131. Ye C, Xu S, Hu Q, Zhou L, Qin X, Jia J, et al. Global View of Neuropeptides and Their Receptors in the Brain and Pituitary of Grass Carp (Ctenopharyngodon Idellus). Aquaculture (2019) 512:734360. doi: 10.1016/j.aquaculture.2019.734360
132. Thompson R. C, Seasholtz A. F, Lebo R, Herbert E. Rat corticotropin-releasing hormone gene: sequence and tissue-specific expression. Molecular Endocrinology (1987) 1(5):363–70. doi: 10.1126/science.3629259
133. Dotzler SA, Digeronimo RJ, Yoder BA, Siler-Khodr TM. Distribution of Corticotropin Releasing Hormone in the Fetus, Newborn, Juvenile, and Adult Baboon. Pediatr Res (2004) 55:120–5. doi: 10.1203/01.PDR.0000100460.00639.F4
134. Sakanaka M, McMaster D, Chohan K, Shibasaki T, Stell WK, Lederis K. Urotensin I-Like Immunoreactivity in Amacrine Cells of the Goldfish Retina. Neurosci Lett (1987) 76:96–100. doi: 10.1016/0304-3940(87)90199-6
135. Kiyama H, Shiosaka S, Kuwayama Y, Shibasaki T, Ling N, Tohyama M. Corticotropin-Releasing Factor in the Amacrine Cells of the Chicken Retina. Brain Res (1984) 298:197–200. doi: 10.1016/0006-8993(84)91170-3
136. Williamson DE, Eldred WD. Amacrine and Ganglion Cells With Corticotropin-Releasing-Factor-Like Immunoreactivity in the Turtle Retina. J Comp Neurol (1989) 280:424–35. doi: 10.1002/cne.902800308
137. Skofitsch G, Jacobowitz DM. Corticotropin Releasing Factor-Like Immunoreactive Neurons in the Rat Retina. Brain Res Bull (1984) 12:539–42. doi: 10.1016/0361-9230(84)90169-2
138. Zhang D, Yeh HH. Histogenesis of Corticotropin Releasing Factor-Like Immunoreactive Amacrine Cells in the Rat Retina. Dev Brain Res (1990) 53:194–9. doi: 10.1016/0165-3806(90)90006-K
139. Fukudo S, Nomura T, Hongo M. Impact of Corticotropin-Releasing Hormone on Gastrointestinal Motility and Adrenocorticotropic Hormone in Normal Controls and Patients With Irritable Bowel Syndrome. Gut (1998) 42:845–9. doi: 10.1136/gut.42.6.845
Keywords: corticotropin-releasing hormone, phylogeny, synteny, tissue distribution, vertebrates, eel, salmon
Citation: Maugars G, Mauvois X, Martin P, Aroua S, Rousseau K and Dufour S (2022) New Insights Into the Evolution of Corticotropin-Releasing Hormone Family With a Special Focus on Teleosts. Front. Endocrinol. 13:937218. doi: 10.3389/fendo.2022.937218
Received: 05 May 2022; Accepted: 09 June 2022;
Published: 22 July 2022.
Edited by:
Elizabeth Amy Williams, University of Exeter, United KingdomReviewed by:
Nicholas J. Bernier, University of Guelph, CanadaCopyright © 2022 Maugars, Mauvois, Martin, Aroua, Rousseau and Dufour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gersende Maugars, Z2Vyc2VuZGUubWF1Z2Fyc0Bob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.