
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 03 August 2022
Sec. Neuroendocrine Science
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.902737
This article is part of the Research Topic Neuroendocrine Regulation of Feeding and Reproduction in Fish View all 14 articles
Kisspeptin plays a vital role in mediating the stress-induced reproductive regulation. Cortisol, known as a stress-related hormone, is involved in gonadal development and sexual differentiation by binding with glucocorticoid receptor (GR) to regulate the expression of kiss gene. In the present study, cortisol treatment in yellowtail clownfish (Amphiprion clarkii) showed that the expression of kiss (kiss1 and kiss2) and gr (gr1 and gr2) genes were increased significantly. We demonstrated that the yellowtail clownfish Kiss neurons co-express the glucocorticoid receptors in the telencephalon, mesencephalon, cerebellum, and hypothalamus. We further cloned the promoter of kiss2 gene in yellowtail clownfish and identified the presence of putative binding sites for glucocorticoid receptors, estrogen receptors, androgen receptors, progesterone receptors, AP1, and C/EBP. Applying transient transfection in HEK293T cells of the yellowtail clownfish kiss2 promoter, cortisol (dexamethasone) treatment was shown to enhance the promoter activities of the yellowtail clownfish kiss2 gene in the presence of GRs. Deletion analysis of kiss2 promoter indicated that cortisol-induced promoter activities were located between position −660 and −433 with GR1, and −912 and −775 with GR2, respectively. Finally, point mutation studies on the kiss2 promoter showed that cortisol-stimulated promoter activity was mediated by one GRE site located at position −573 in the presence of GR1 and by each GRE site located at position −883, −860, −851, and −843 in the presence of GR2. Results of the present study provide novel evidence that cortisol could regulate the transcription of kiss2 gene in the yellowtail clownfish via GRE-dependent GR pathway.
Kisspeptin is regarded as the key factor of reproduction and plays a role in the hypothalamus–pituitary–gonad axis (HPG axis) in vertebrates (1, 2). Kisspeptin binds to its receptor (G protein-coupled receptor 54, GPR54) and releases gonadotropin-releasing hormone (GnRH) from the hypothalamus, thereby stimulating the secretion of the gonadotropic hormone (GtH). Knockout of Kiss1/Gpr54 in mice has been shown to prevent sexual maturity, cause gonad hypoplasia, hypogonadotropic hypogonadism, and infertility (3, 4). Moreover, kisspeptin has been reported to be involved in a variety of physiological activities, such as glucose homeostasis and light signal regulation (5, 6). Especially, kisspeptin is supposed to be involved regulation of various hormones on the hypothalamus–pituitary–adrenal axis (HPA axis) and the HPG axis under stress induction (7, 8).
Stress can lead to the dysfunction of the HPG axis and reproductive behavior through the HPA axis in vertebrates (9). There is a close relationship between stress and reproductive disorders (10). In patients with depression under stress, an excessive corticotropin-releasing hormone (CRH) level leads to the inhibition of the HPG axis, and increased cortisol level further inhibits the action of GnRH neurons, luteinizing hormone (LH) amplitude, follicle-stimulating hormone (FSH) levels, and LH pulse frequency (11–13). In mice, both psychosocial stress and unpredictable chronic stress reduce the expression of hypothalamic kiss1 and the activity of kisspeptin neuron (14, 15). Moreover, corticotropin-releasing hormone or corticosterone treatment suppress kiss1 expression and kisspeptin neuron activity in the brain of female rats and mice (16, 17). Glucocorticoid, a steroid regulated by stress, can bind with glucocorticoid receptor (GR) to regulate gene expression by associating with specific genomic glucocorticoid response elements (GREs) (18). In female rats, GR protein is detected in the kisspeptin neurons of periventricular nucleus continuum (AVPV/PeN) and arcuate nucleus (ARC), demonstrating that kisspeptin neurons can be modulated directly by glucocorticoid via GR (19). Additionally, GRE is found in the promoter regions of kiss genes in goldfish (Carassius auratus) (20). However, the molecular mechanism of glucocorticoid-regulated kiss genes in vertebrate species is still unknown.
Kisspeptin could be encoded by multiple genes in non-mammalian, and two paralogous kiss genes, known as kiss1 and kiss2, are found in some teleosts, such as zebrafish and medaka (2). The yellowtail clownfish (Amphiprion clarkii) is a protandrous teleost whose sex is associated with social status within a group, including a male–female breeding pair and some non-breeders (21, 22). In one social unit, females occupy the first dominant status and inhibited sexual development of subdominant male and non-breeders, and subdominant male could undergo sexual development to female after female disappeared or the largest non-breeder change sex to male after the disappearance of male (22, 23). The level of cortisol, the main component of glucocorticoid in teleosts, depends on their social status in the population. In the protogynous orange-spotted grouper (Epinephelus coioides), the female treated with cortisol will change to male (24). In our previous research, the higher levels of hypothalamic kiss2/gr2 expression and gonadal hormone were found in the subordinate of yellowtail clownfish (25, 26). Moreover, there is the sexually dimorphic distribution of kiss1 and kiss2 in the brain of yellowtail clownfish, especially in dorsal habenular nucleus (NHd) and dorsal part of the nucleus of the lateral recess (NRLd), which are involved in the regulation of reproductive function and environmental cues (27).
In order to better understand how cortisol exerts its action on the expression of kiss gene via the GR and thereafter regulates the reproduction in yellowtail clownfish, the expression of kiss and gr genes were examined after cortisol treatment. Moreover, the co-localization of gr1, gr2, and kiss1/kiss2 mRNA were also studied in the brain by RNAscope. Then, the promoter region of kiss2 was cloned by genome walking and predicted with the online tool for potential GR binding sites. In addition, after cortisol treatment, the kisspeptin promoter activities were detected in HEK-293T cells expressing yellowtail clownfish GR1 or GR2. Finally, the regulatory regions and binding sites of GR were identified by deletion analysis and site-directed mutagenesis analysis.
Sexually mature and 3-month-old immature yellowtail clownfish were purchased from a local aquarium market (Dongfang city, Hainan, China) in June of 2021. The fish, nine per group, were reared in glass tank (length, 45 cm; width, 35 cm; and height, 60 cm) with continuously flowing aerated seawater at 27 ± 1°C. The photoperiod was a 12:12-h light–dark cycle, with lights turn on at 07:00 and off at 19:00. The fish were fed with commercial feed twice a day (09:00 and 18:00) and reared for a period of 1 week before experiment.
All animals used in this study were conducted in accordance with the guidelines of the animal welfare of the National Committee and approval of the Institutional Animal Care and Use Committee of Hainan University (HNUAUCC-2021-00014).
The cortisol (hydrocortisone 21-hemisuccinate; MCE, NJ, USA) was dissolved in dimethyl sulfoxide (DMSO) and then diluted in the ratio of 10% cortisol, 40% polyethylene glycol 300 (MCE, NJ, USA), and 50% saline (0.9%). The immature fish (length, 4 ± 0.5 cm; weight, 2 ± 0.5 g) were anesthetized with MS-222 (Sigma, MO, USA) and then given an intraperitoneal injection with cortisol (10 or 50 mg/g body mass) at a volume of 10 μl/g body mass. The control group was injected the same liquid but without cortisol. We collected the whole brain at 6, 12, 24, and 48 h after injection, respectively.
The gonads of sexually mature yellowtail clownfish (length, 11 ± 1 cm; weight, 21 ± 5 g) were isolated and fixed in Bouin’s solution (Sigma, MO, USA) after anesthesia. The gonadal tissues were embedded in paraffin and cut into 5-μm paraffin sections for indentation of gonadal development. The brain of fish was fixed in 4% paraformaldehyde fix solution (Sigma, MO, USA) for in situ hybridization.
Total RNA from the whole brain were extracted by the TRIzol method and then was reversed transcribed into cDNA using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). Primer sequences and primer efficiency for quantitative PCR (qPCR) are listed in Supplementary Table S1. The quantitative real-time PCR was performed by Roche Light Cycler 96 real-time PCR System using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) according to the manufacturer’s protocol. The qPCR program was as follows: denaturation at 95°C for 30 s, then followed by 40 cycles at 95°C for 5 s and 55–58°C for 30 s and 72°C for 30 s. Each sample was used in triplicate. The relative mRNA levels of kiss1, kiss2, gr1, and gr2 were evaluated using comparative threshold cycle (Ct) method with β-actin as internal reference gene and then calculated with the formula 2−ΔΔCt (28).
Brain samples of female yellowtail clownfish were fixed in 4% paraformaldehyde at 4°C overnight. Samples successively were immersed in 10%, 20%, and 30% sucrose containing phosphate-buffered saline (PBS) for dehydration. Cross-sections (10 μm) were generated after being frozen on dry ice in optimal cutting temperature (OCT) compound. RNAscope in situ hybridization was performed following the manufacturer’s protocol from Advanced Cell Diagnostics (ACD). All steps demanding incubation at 40°C were achieved in the HybEZ Oven (ACD, Hayward USA). Binding of the specific probes against kiss1 (1044931), kiss2 (1044941), gr1 (1088191), and gr2 (1088201) were detected with RNAscope@ Multiplex Fluorescent Reagent Kit v2 (ACD, Hayward, USA). Probes actb2 (1045881) and dapB (310043) were used as positive and negative controls, respectively. Images were taken by Nikon ECLIPSE Ti2 (Nikon, NY, USA).
Genomic DNA was extracted from yellowtail clownfish muscle using phenol-chloroform methods. The 5′-flanking region of the kiss2 was isolated in the reference to Universal Genome Walker Kit (Takara, Tokyo, Japan). The gene-specific primers were designed in the exon 1 based on the sequences of yellowtail clownfish kiss2 (GenBank: MK368702.1) and are shown in Supplementary Table S1. Products of primary PCR were diluted 100 times, and then, secondary PCR was performed with the diluted products as the template. The secondary PCR products were purified using FastPure Plasmid Mini Kit (Vazyme, Nanjing, China) and were subcloned into the pMD 19-T Vector (Takara, Tokyo, Japan) for sequencing. The transcriptional start site (TSS) of kiss2 was determined by our previous result of 5′-rapid amplification of cDNA ends. Transcription factor binding sites were predicted using the online PROMO (http://alggen.lsi.upc.es/recerca/menu_recerca.html) and gene-regulation tool (http://gene-regulation.com/cgi-bin/pub/programs/alibaba2/webbaba2.cgi).
A 1,442-bp 5′-flanking region and 48-bp exon 1 of kiss2 was obtained from a pair of primers containing two different restriction enzyme sites, respectively, namely, KpnI and XhoI. PCR was performed with PrimeSTAR HS DNA Polymerase (Takara, Tokyo, Japan). The PCR products and pGL4.10 vector (Promega, WI, USA) were digested by KpnI and XhoI restriction endonucleases (NEB, MA, USA). After purification, the digested products were ligated using T4 DNA Ligase Kit (NEB, MA, USA). The construction of recombinant vector above, namely, pkiss2-1442, was used as template to construct a series of deletion vectors, namely, pkiss2-912, pkiss2-775, pkiss2-660, pkiss2-433, and pkiss2-335. All constructs were sequenced ensuring accuracy. The recombinant vectors were extracted with Omega Endo-Free plasmid DNA mini kit II (OMEGA, GA, USA). Primers used in here are presented in Supplementary Table S1.
HEK-293T cells (Bosterbio, CA, USA) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum at 37°C with 5% CO2 and passed at least two generations prior to transfection. The healthy cells were seeded into 48-well plates, and approximately 1.5 × 105 cells/well were cultured for 12 h. Cells were then co-transfected with 0.5 μg pkiss2-1442/pkiss2-912/pkiss2-775/pkiss2-660/pkiss2-433/pkiss2-335, 0.05 μg pcDNA3.1-GR1/pcDNA3.1-GR2 (yellowtail clownfish glucocorticoid receptor expression plasmid) and 0.025 μg pRL-CMV in 250 μl Opti-MEM using EZ Trans (Liji, Shanghai, China). After transfection of 6 h, cells were treated with 10−7 M dexamethasone sodium (DXMS, exogenous cortisol). Luciferase activities were detected 24 h later with Dual Luciferase Kit (Promega, WI, USA) in GloMax Discover (Promega, WI, USA).
Mutations of putative GRE sites in the kiss2 promoter were carried out using a series of specific primers (Supplementary Table S1) by two rounds of PCR amplification. Briefly, primers containing restriction enzyme site and mutation point were used to amplify the mutated fragments of upstream and downstream from the mutation site. The first-round PCR conditions were as follows: denaturation at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s and 67–69°C for 30 s and 72°C for 1 min, with the final extension at 72°C for 10 min. The two PCR products were purified and mixed together for the second-round PCR. The conditions were as follows: denaturation at 95°C for 30 s, then followed by 9 cycles at 95°C for 5 s and 67–69°C for 30 s and 72°C for 1 min and 30 s. Then, primers for full-length promoter amplification of kiss2-1442-F and kiss2-1442-R were added and continued for an additional 20 cycles at 95°C for 5 s and 69°C for 30 s and 72°C for 1 min 30 s, with the final extension at 72°C for 10 min. PCR products were digested by KpnI and XhoI restriction endonucleases and were subcloned into pGL4.10 vector. All constructs were sequenced ensuring accuracy, and plasmid DNAs were extracted with Omega Endo-Free plasmid DNA mini kit II.
All data are shown as mean ± standard error of the mean (SEM). Statistical analysis was performed using one-way ANOVA followed by Tukey’s multiple comparisons test in GraphPad Prism 7.0 (GraphPad Software, SD, USA). Results were considered significantly different when p-value was <0.05 (p < 0.05).
After cortisol injection, real-time PCR was performed to investigate the expression profiles of kiss1, kiss2, gr1, and gr2 in the brain of immature yellowtail clownfish. The highest kiss1 levels were detected at cortisol treatment with concentrations of 10 and 50 μg/g after 6 h (Figure 1A). For kiss2, transcripts were elevated 2-fold at 6 h and 3.5-fold at 12 h in the 50 μg/g cortisol-treated group relative to the other groups and were highest at 48 h in the 10 μg/g cortisol treatment (Figure 1B). The gr1 mRNA levels were significantly higher at 24 and 48 h cortisol treatment with a dose of 10 μg/g (Figure 1C), but the most abundant gr2 transcripts were at 6 and 12 h cortisol treatment with 50 μg/g (Figure 1D). These results indicated that cortisol treatment enhanced the transcription of kiss1 and kiss2 and gr1 and gr2 in the brain of yellowtail clownfish.
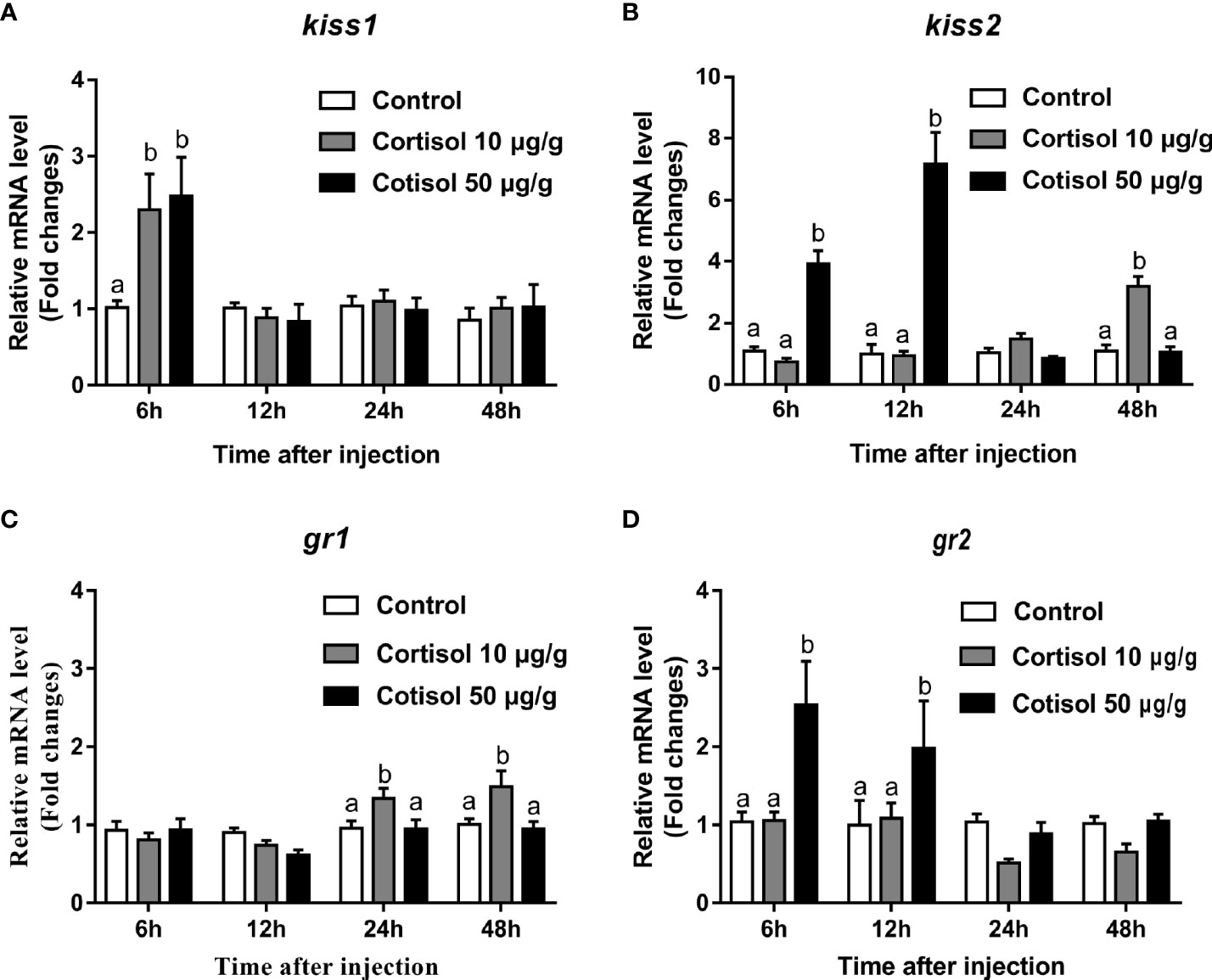
Figure 1 Effects of cortisol on kiss1, kiss2, gr1, and gr2 mRNA levels in the brain of yellowtail clownfish. Expression of kiss1 (A), kiss2 (B), gr1 (C), and gr2 (D) after injection of cortisol with both concentrations at 10 and 50 μg/g during 48 h. The data are expressed as mean ± SEM (n = 5–8). Bars with different letters indicate significant differences between treatments at the same sampling time (p < 0.05).
According to the distribution of kiss1 and kiss2 genes in the brain of yellowtail clownfish (unpublished), RNAscope in situ hybridization for gr1 and gr2 and kiss1/kiss2 genes were performed in areas of the telencephalon (Te), mesencephalon (Me), cerebellum (Ce), and hypothalamus (Hy). The co-expression of gr1, gr2, and kiss1 was found in the in the dorsal habenular nucleus (NHd), subdivision 2 of the medial dorsal telencephalic area (Dm2), subdivision 3 of the medial dorsal telencephalic area (Dm3), lateral posterior part of the dorsal telencephalic area (DIP), posterior portion of the dorsal telencephalon (DP), corpus of the cerebellum (CCe), and lateral part of the diffuse nucleus of the inferior lobe (NDLIl) (Figures 2A–E, F–K). The gr2 signal was more abundantly distributed than gr1 in NHd, Dm2, and CCe (Figures 2A, F, J) and weakly expressed in the DIP and DP (Figures 2H, I). The gr1, gr2, and kiss2 were simultaneously detected in the dorsal part of the nucleus of the lateral recess (NRLd), Dm2, Dm3, DIP, DP, CCe, optic tectum (OT), NDLIl, posterior part of glomerular nucleus (NGP), tegmentum (TEG), and periventricular nucleus of the posterior tuberculum (TPp) (Figures 3A–E, F–O). Compared with gr1, the stronger gr2 signals were detected in the NRLd, Dm2, Dm3, OT, NGP, and TPp (Figures 3A, F, G, K, M, O), but the weaker signaling molecules were examined in the DIP and NDLIl (Figures 3H, L).
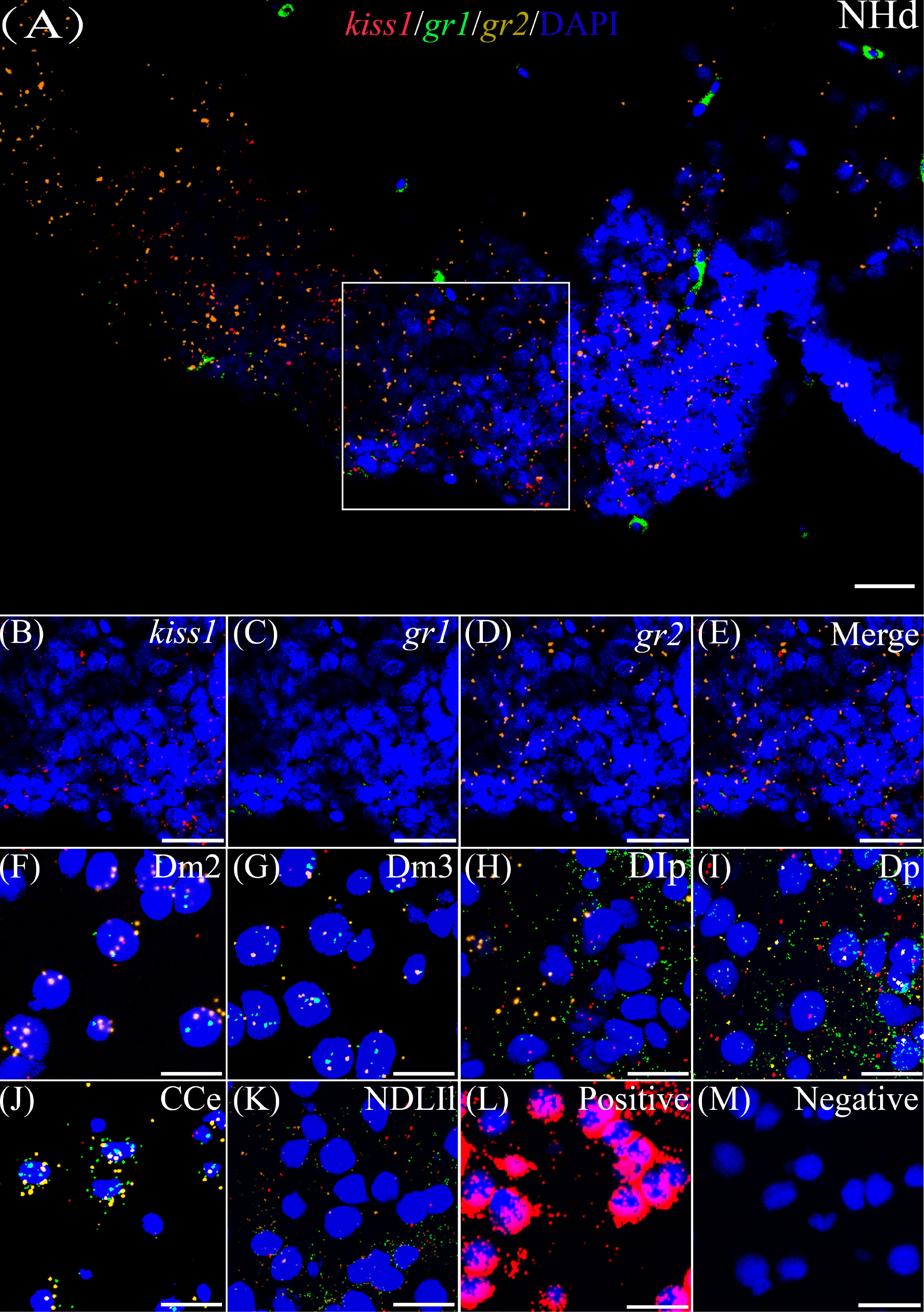
Figure 2 Co-expression of gr1, gr2, and kiss1. The view of NHd (A) shows kiss1 (red), gr1 (green), and gr2 (yellow) with DAPI cell nuclear staining (blue). (B–E) The view of boxed region in panel (A), showing kiss1 (red), gr1 (green), and gr2 (yellow) with DAPI cell nuclear staining (blue) in panels (B–D), respectively, and the “merge” in panel (E) shows kiss1 (red), gr1 (green), and gr2 (yellow) with DAPI cell nuclear staining (blue). Representative images display gr1 (green) and gr2 (yellow) co-expression of kiss1 (red) with DAPI cell nuclear staining (blue) in Dm2 (F), Dm3 (G), DIp (H), Dp (I), CCe (J), and NDLIl (K). Positive and negative controls are shown in panels (L, M), respectively. NHd, dorsal habenular nucleus; Dm2, subdivision 2 of the medial dorsal telencephalic area; Dm3, subdivision 3 of the medial dorsal telencephalic area; DIp, lateral posterior part of the dorsal telencephalic area; Dp, posterior portion of the dorsal telencephalon; CCe, corpus of the cerebellum; NDLIl, lateral part of the diffuse nucleus of the inferior lobe. Bars = 20 μm.
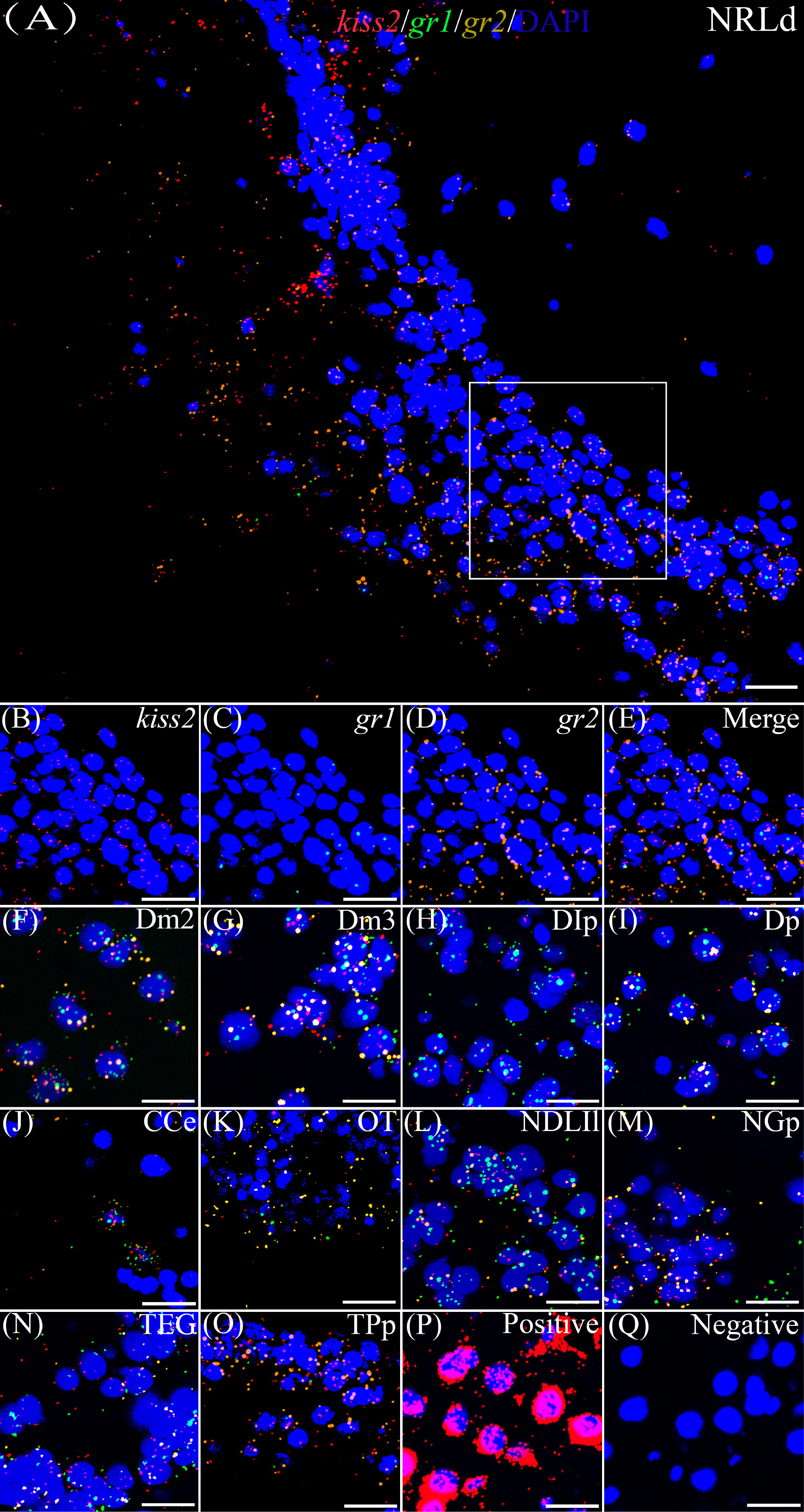
Figure 3 Co-expression of gr1, gr2, and kiss2. The view of NRLd (A) shows kiss2 (red), gr1 (green), and gr2 (yellow) with DAPI cell nuclear staining (blue). (B–E) The view of boxed region in panel (A), showing kiss2 (red), gr1 (green), and gr2 (yellow) with DAPI cell nuclear staining (blue) in panels (B–D), respectively; and the “merge” in panel (E) shows kiss2 (red), gr1 (green), and gr2 (yellow) with DAPI cell nuclear staining (blue). Representative images display gr1 (green) and gr2 (yellow) co-expression of kiss2 (red) with DAPI cell nuclear staining (blue) in Dm2 (F), Dm3 (G), DIp (H), Dp (I), CCe (J), OT (K), NDLIl (L), NGp (M), TEG (N), and TPp (O). Positive and negative controls are shown in panels (P, Q), respectively. NRLd, dorsal part of the nucleus of the lateral recess; Dm2, subdivision 2 of the medial dorsal telencephalic area; Dm3, subdivision 3 of the medial dorsal telencephalic area; DIp, lateral posterior part of the dorsal telencephalic area; Dp, posterior portion of the dorsal telencephalon; CCe, corpus of the cerebellum; OT, optic tectum; NDLIl, lateral part of the diffuse nucleus of the inferior lobe; NGp, posterior part of glomerular nucleus; TEG, tegmentum; TPp, periventricular nucleus of the posterior tuberculum. Bars = 20 μm.
To further investigate the transcriptional regulatory mechanism of kiss2 by cortisol in yellowtail clownfish, we isolated the 5′-flanking region of kiss2 gene. The putative kiss2 promoter sequence includes a 1,442-bp upstream of the transcription start site and a 48-bp first exon fragment (Figure 4). In silico analysis revealed that kiss2 promoter sequence possessed 13 potential glucocorticoid response elements (GREs). In addition, several motifs for other steroid receptors were identified on the kiss2 promoter, such as five androgen response elements, three estrogen response elements, and two progesterone response elements (Figure 4).

Figure 4 Sequence analysis of 5′-flanking region for kiss2. The numbering of the sequence is relative to the transcription start site marked on # and designated as +1. Putative binding sites for transcription factors are underlined and labeled. Transcription factor binding sites were predicted using the online PROMO and gene-regulation tool.
GR-negative HEK293T cells were transfected with the recombinant vector for kiss2 promoter (pkiss2-1442), in combination with expression plasmids for yellowtail clownfish glucocorticoid receptor (GR1 or GR2), to analyze the transcriptional regulation of kiss2 by cortisol. Basal promoter activity was examined for the kiss2 promoter, indicating that functional promoter activity existed in the 5′-flanking region (Figure 5A). DXMS significantly increased kiss2 promoter activity in the presence of GR, being more efficient in GR1 than GR2 (Figure 5B). The kiss2 promoter activities were detected after treatment with different DXMS concentrations and significantly upregulated at 10−7 M DXMS in the presence of GR1 or GR2 (Figures 6A, B). Therefore, this concentration of DXMS was chosen for the subsequent investigation.
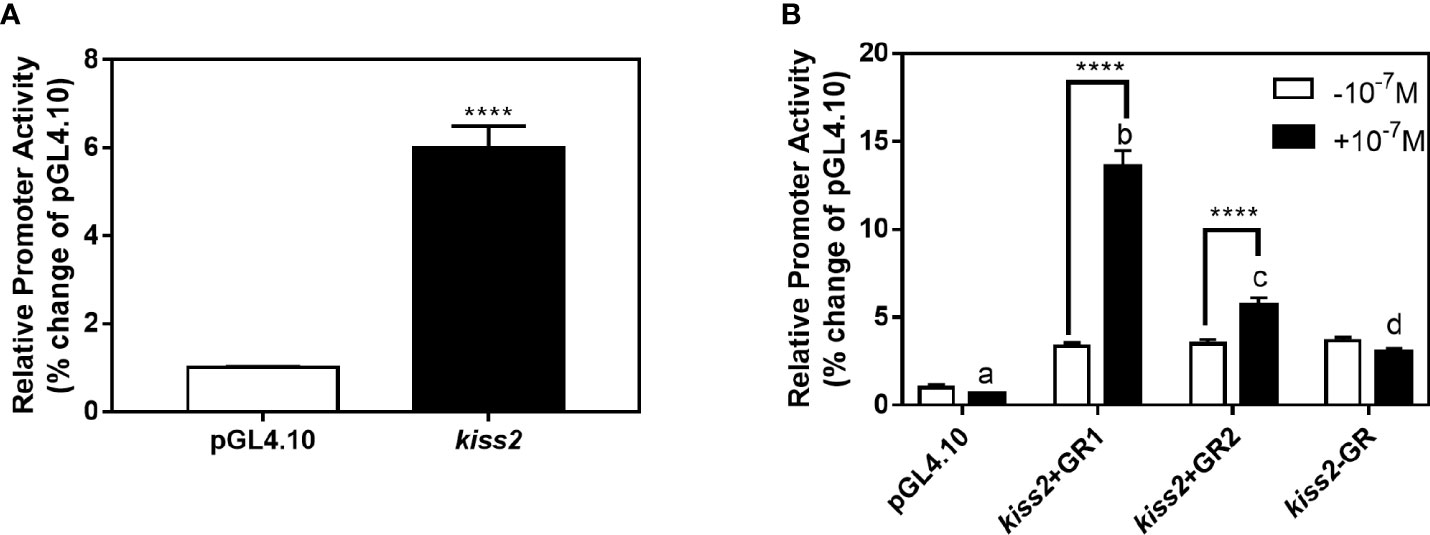
Figure 5 Effects of cortisol on kiss2 promoter activity. (A) Basic activity of kiss2 gene promoter in HEK-293T cell lines. The cells were transfected with 0.5 μg pkiss2-1442 and 0.025 μg pRL-CMV, or 0.5 μg pGL4.10 and 0.025 μg pRL-CMV as the control. Luciferase activity was measured after 24 h. Relative promoter activities are expressed as percentage of pGL4.10. (B) The activities of kiss2 promoter in the presence of cortisol in HEK293T cell lines. Cells were co-transfected with 0.5 μg pkiss2-1442, and 0.025 μg pRL-CMV with or without 0.05 μg yellowtail clownfish glucocorticoid receptor (GR1 or GR2) expression plasmid. The transfected cells were treated with or without 10−7 M cortisol. The luciferase activity was measured 24 h later. Relative promoter activities are expressed as percentage of pGL4.10 in the absence of cortisol. Data are represented as mean ± SEM (n = 4). **** (p < 0.0001) indicates that significant difference compared with the corresponding control. The different letters mean significant differences between groups with cortisol treatment (p < 0.05).
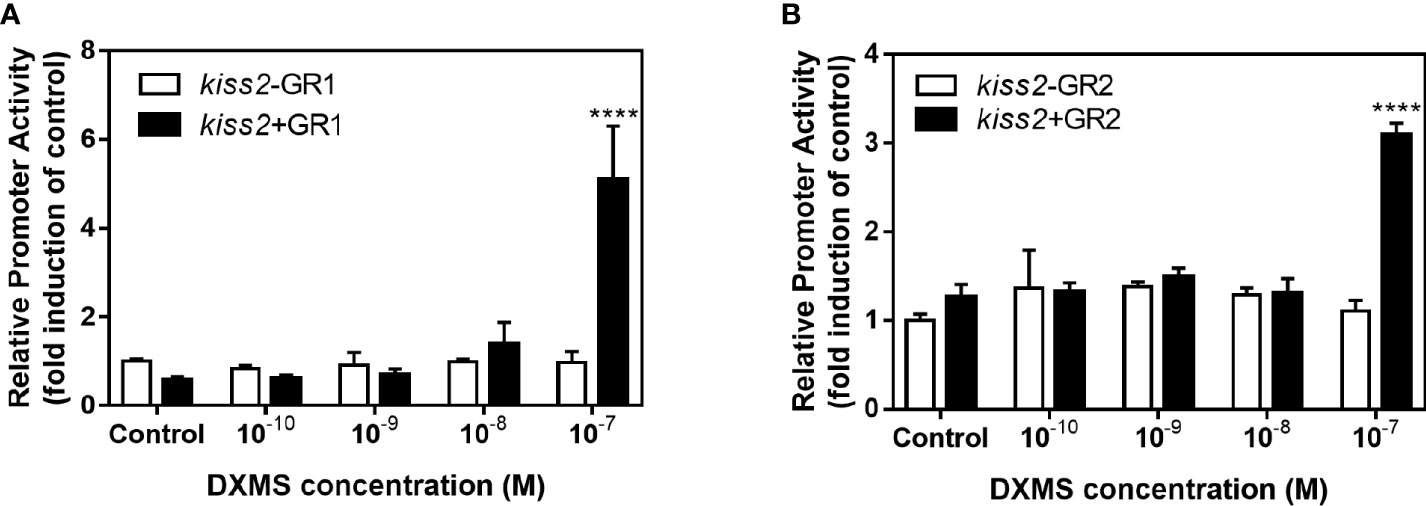
Figure 6 Effects of cortisol with different concentrations on the activities of kiss2 gene promoter. HEK-293T cells were transfected with 0.5 μg pkiss2-1442, 0.05 μg GR1 (A) or GR2 (B) and 0.025 μg pRL-CMV; 0.5 μg pGL4.10 co-transfected with 0.025 μg pRL-CMV as the control. Cells were treated with or without cortisol. Luciferase activity was detected after 24 h. Data are represented as mean ± SEM (n = 4). **** (p < 0.0001) indicates significant differences between groups with cortisol treatment.
Using the full length of the kiss2 promoter vector as a template, a series of deletion constructs were established as shown in the left panel of Figures 7, 8. In the presence of GR1, the promoter activity of kiss2 was significantly higher after 10−7 M DXMS treatment (Figure 7A). Deletions of kiss2 promoter to position −433 (pkiss2-433) abolished cortisol-induced promoter activity, indicating that the region from −660 to −433 is relevant with cortisol-induced promoter activity by GR1 (Figure 7A). Site-directed mutagenesis was conducted to determine whether the GRE at −573 (−573 TGTAC−569) was the key regulatory site on kiss2 promoter. Mutation of a GRE at −573 eradicated cortisol-induced promoter activity (Figure 8A). However, we found that mutation in other GRE sites at −1,236 (−1,236AGTTCT−1,231) or −1,188 (−1,188AGGAT−1,184) did not change cortisol-induced promoter activity (Figure 8B). Thus, the GRE at −573 is critical for cortisol/GR1-induced kiss2 promoter activity.

Figure 7 Deletion analysis of the kiss2 promoter. Schematic of the putative GRE sites and deletion constructs of kiss2 promoter is shown on the left. HEK-293T cells were transfected with deletion constructs and pRL-CMV with GR1 (A) or GR2 (B) expression plasmid. Cells were incubated with or without 10−7 M cortisol treatment. Relative promoter activities are expressed as percentage of pGL4.10 in the absence of cortisol. Data are represented as mean ± SEM (n = 4). **(p < 0.01) and **** (p < 0.0001) indicate the significant differences compared with the corresponding control.
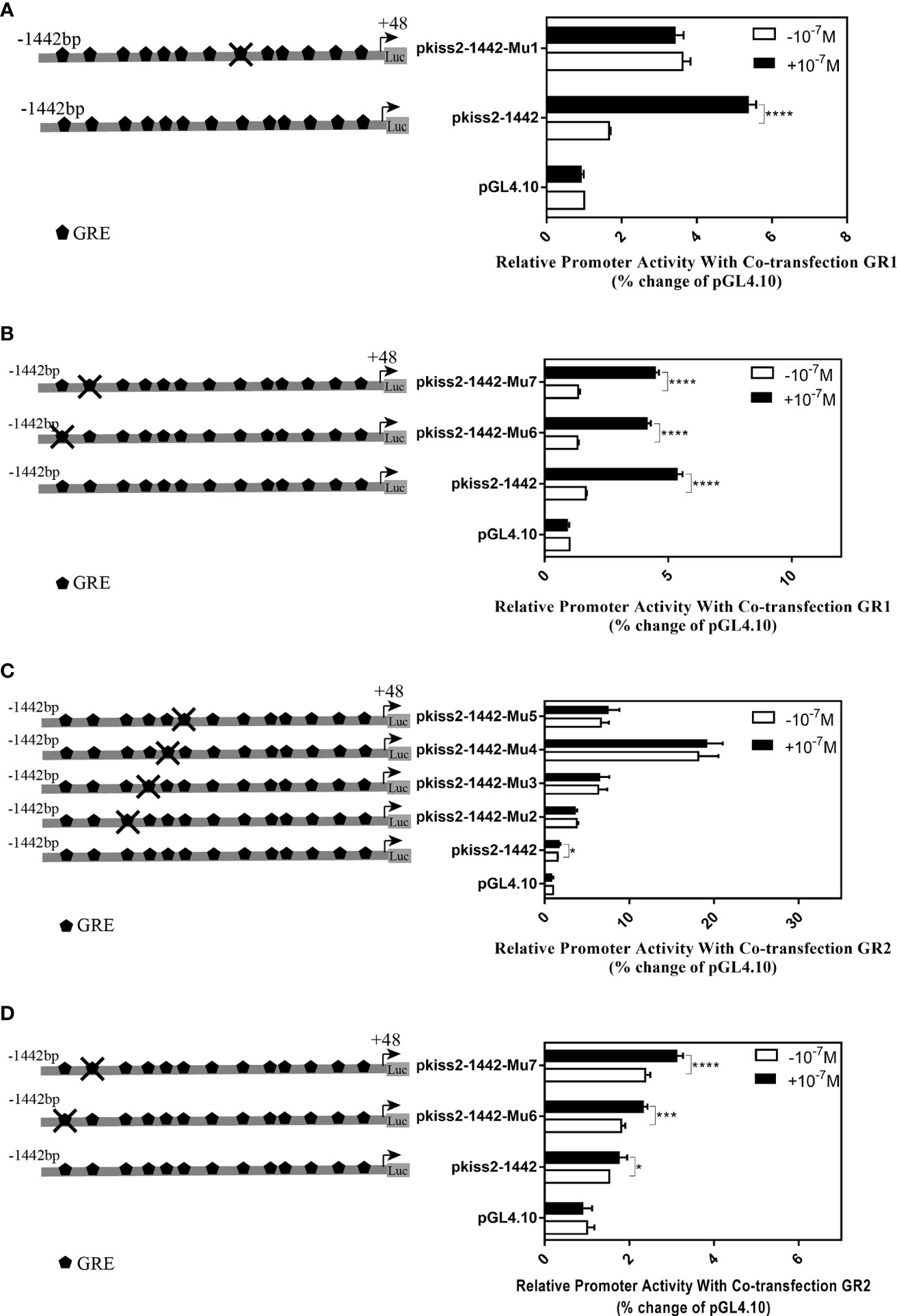
Figure 8 Mutations of the putative glucocorticoid receptor binding sites in kiss2 promoter. Schematic of mutated GRE sites is shown on the left. HEK-293T cells were transfected with GRE-mutated promoter constructs and pRL-CMV with GR1 (A, B) or GR2 (C, D) expression plasmid. Cells were incubated with or without 10−7 M cortisol treatment. Relative promoter activities are expressed as percentage of pGL4.10 in the absence of cortisol. Data are represented as mean ± SEM (n = 4). * (p < 0.05), *** (p < 0.001), and **** (p < 0.0001) indicate significant differences compared with the corresponding control.
In the presence of GR2, 10−7 M DXMS treatment significantly upregulated the wild-type kiss2 promoter activity (Figure 7B).Truncation of the kiss2 promoter to −775 bp abolished by cortisol-induced promoter activity (Figure 7B). Mutations of two GRE at −1,236 or −1,188 of the kiss2 promoter still responded to the cortisol treatment (Figure 8D). However, cortisol-induced promoter activities of kiss2 were removed in mutations of the following four GRE at −883 (−883AGGAT−879), −860 (−860TGTAC−856), −851 (−851AGTTCT−846), and −843 (−843ATCCT−839), indicating that these GRE binding sites contribute to cortisol-/GR2-induced kiss2 promoter activity (Figure 8C).
Previous studies in vertebrates have demonstrated that kisspeptin plays a vital role in mediating the stress-induced reproductive regulation (7). Cortisol, the main steroid hormone for stress response, is involved in gonadal development and sexual differentiation by regulating kiss genes (26). This study aims to observe the molecular mechanism of glucocorticoid regulation of the kiss genes in teleosts. In rodents, the hypothalamic kiss1 mRNA level and kisspeptin neuron activity are reduced after the administration of cortisol (16, 17). There is only one kiss gene in rodents, but there are two kiss genes (kiss1 and kiss2) in several teleost fish participating in the reproductive regulation (2). Cortisol treatment showed that the expression of kiss (kiss1 and kiss2) and gr (gr1 and gr2) genes were increased significantly in the brain of yellowtail clownfish. In addition, elevation of two kiss genes expression by cortisol has been reported in zebrafish (29). Moreover, our previous study revealed that kiss2, E2, and testosterone (T) levels are higher in the subordinate than in the dominant yellowtail clownfish (25). A study in protandrous false clown anemonefish (Amphiprion ocellaris) showed that the social rank reflects the blood cortisol value (30).
GR signals are detected in the Kiss neurons of periventricular nucleus continuum (AVPV/PeN) and arcuate nucleus (ARC) by double-labeling immunohistochemistry in female rats (19). In the present study, we have demonstrated that Kiss neurons co-expressed the glucocorticoid receptors in the Te, Me, Ce, and Hy, suggesting that cortisol could directly affect kisspeptin neurons via GR in yellowtail clownfish. Habenular kiss1 and serotonin-related genes are downregulated after exposure to alarm substance (AS), and Kiss1 antagonist injection can reduce AS-evoked fish fear response, indicating that habenular kisspeptin modulates fear in zebrafish (31). The yellowtail clownfish non-breeders are always attacked by both female and male, and hypothalamic gr2 levels of non-breeders are significantly higher than that of the others in one social group (21, 26). From our results showing gr genes co-expression with kiss1 in NHd, we raise the possibility that habenular kiss1 is involved in the regulation of fear response in the yellowtail clownfish. Moreover, kiss2 levels in the NRLd are decreased under testosterone (T) treatment in sea bass (32). In yellowtail clownfish, gr1 and gr2 were found to be co-expressed with kiss2 in the NRLd, indicating that this region may participate in stress-induced reproductive functions via kiss2.
Multiple GR binding sites were found in kiss2 promoter in the yellowtail clownfish by silicon analysis, implying that kiss2 could be regulated by cortisol via GR through binding with GRE. In the present study, cortisol injection also enhanced the gr1 and gr2 mRNA levels in the brain. Other binding sites such as AP1, Sp1, and C/EBP were also predicted in the kiss2 promoter region. AP1 could interact with GR for GR-regulated transcription and recruitment to co-occupied AP1 binding site by DNaseI accessibility and chromatin immunoprecipitation with high-throughput sequencing (33). In addition, a series of binding sites for sex steroid receptors existed in the kiss2 promoter of yellowtail clownfish, which is similar to the results of goldfish and zebrafish (20, 34), indicating that the potential ability of kiss2 is involved in the regulation of reproduction. Our previous study found that hypothalamic kiss2 had higher expression in non-breeders than females and males, which may contribute to the regulation of gonad development under social stress in the yellowtail clownfish (25).
The cortisol treatment could enhance yellowtail clownfish kiss2 promoter activities in HEK293T cells in the presence of GR, whereby GR1 was more effective than GR2. In yellowtail clownfish, GR1 contains conserved nine amino acids, which are present in the most known teleostean GR1 proteins but absent in other vertebrates (26, 35, 36). A previous study revealed that the additional nine amino acids made GR1a to better bind with single GRE than GR1b in rainbow trout (Oncorhynchus mykiss) (37). Thus, we speculate that GR1 has a better binding affinity for GRE than GR2 in the yellowtail clownfish. Altogether, GRs play a vital role in the mediation of cortisol effect on kiss2 promoter.
GR can activate or repress gene expression by binding with GRE directly or interacting with other transcription factors (38). Using a series of deletion constructs, we have demonstrated that cortisol-induced promoter activities of kiss2 gene were located between position −660 and −433 with GR1, and −912 and −775 with GR2, respectively. Point mutations in the kiss2 promoter were generated by site-directed mutagenesis. Our results showed that in the presence of GR1, cortisol-stimulated promoter activity was only mediated by one GRE site located at the position of −573, whereas in the presence of GR2, promoter activity could be modulated by all four GRE sites located at positions −883, −860, −851, and −843. Therefore, the kiss2 gene is regulated by cortisol through the GRE-dependent mechanism in yellowtail clownfish. It has also been reported that there is a synergistic action between the enhancer binding protein (C/EBP) and GR in the regulation of thymidine kinase promoter activity (39).
In conclusion, the present study demonstrated for the first time the molecular mechanism of glucocorticoid regulation of the kiss genes in teleosts. It was found that cortisol treatment could upregulate the expression levels of kiss and gr genes in the yellowtail clownfish. The Kiss neurons coexpressed the glucocorticoid receptors in Te, Me, Ce, and Hy. Cortisol could enhance kiss2 promoter activity in the presence of GRs and was more effective with GR1 than GR2. Moreover, cortisol was shown to regulate kiss2 promoter activity by one GRE site through GR1 and four GRE sites via GR2. Our findings demonstrate that cortisol could directly regulate the expression of kiss2 gene via the GRE-dependent GR pathway in the yellowtail clownfish.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Committee of Hainan University (HNUAUCC-2021-00014).
S-YB planned and wrote the manuscript, participated in experiments and composed the figures. Y-YZ planned, edited, and drafted the manuscript. XZ, T-XL, D-CZ, and Z-XH participated in experiments. QW planned, edited, supervised, and reviewed the manuscript. All authors read the final article and approved its submission.
This work was supported by National Natural Science Foundation of China (NSFC Grant No. 31760759).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.902737/full#supplementary-material
1. Castellano J, Roa J, Luque R, Dieguez C, Aguilar E, Pinilla L, et al. KiSS-1/kisspeptins and the metabolic control of reproduction: physiologic roles and putative physiopathological implications. Peptides (2009) 30:139–45. doi: 10.1016/j.peptides.2008.06.007
2. Ogawa S, Parhar IS. Anatomy of the kisspeptin systems in teleosts. Gen Comp Endocrinol (2013) 181:169–74. doi: 10.1016/j.ygcen.2012.08.023
3. De Roux N, Genin E, Carel J-C, Matsuda F, Chaussain J-L, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci (2003) 100:10972–6. doi: 10.1073/pnas.1834399100
4. Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr, Shagoury JK, et al. The GPR54 gene as a regulator of puberty. Obstetrical Gynecol Survey (2004) 59:351–3. doi: 10.1056/NEJMoa035322
5. Hussain MA, Song W-J, Wolfe A. There is kisspeptin–and then there is kisspeptin. Trends Endocrinol Metab (2015) 26:564–72. doi: 10.1016/j.tem.2015.07.008
6. Espigares F, Rocha A, Gómez A, Carrillo M, Zanuy S. Photoperiod modulates the reproductive axis of European sea bass through regulation of kiss1 and gnrh2 neuronal expression. Gen Comp Endocrinol (2017) 240:35–45. doi: 10.1016/j.ygcen.2016.09.007
7. Iwasa T, Matsuzaki T, Yano K, Mayila Y, Irahara M. The roles of kisspeptin and gonadotropin inhibitory hormone in stress-induced reproductive disorders. Endocrine J (2018) 65:133–40. doi: 10.1507/endocrj.EJ18-0026
8. Wang B-Q, Chen Y-Y, Lan X-X, Zhou Z-Y, Xu X-X, Wu X-Q. The effect of neonatal immune challenge on reproduction by altering intraovarian kisspeptin/GPR54 system in the rat. Reprod Toxicol (2017) 74:40–7. doi: 10.1016/j.reprotox.2017.08.021
9. Ralph C, Lehman M, Goodman R, Tilbrook A. Impact of psychosocial stress on gonadotrophins and sexual behaviour in females: role for cortisol. Reproduction (2016) 152:R1–R14. doi: 10.1530/REP-15-0604
10. Padda J, Khalid K, Hitawala G, Batra N, Pokhriyal S, Mohan A, et al. Depression and its effect on the menstrual cycle. Cureus (2021) 13. doi: 10.7759/cureus.16532
11. Claes S. Corticotropin-releasing hormone (CRH) in psychiatry: from stress to psychopathology. Ann Med (2004) 36:50–61. doi: 10.1080/07853890310017044
12. Joseph DN, Whirledge S. Stress and the HPA axis: balancing homeostasis and fertility. Int J Mol Sci (2017) 18:2224. doi: 10.3390/ijms18102224
13. Raftogianni A, Roth LC, García-González D, Bus T, Kühne C, Monyer H. Deciphering the contributions of CRH receptors in the brain and pituitary to stress-induced inhibition of the reproductive axis. Front Mol Neurosci (2018) 11:305. doi: 10.3389/fnmol.2018.00305
14. Hirano T, Kobayashi Y, Omotehara T, Tatsumi A, Hashimoto R, Umemura Y, et al. Unpredictable chronic stress-induced reproductive suppression associated with the decrease of kisspeptin immunoreactivity in male mice. J Vet Med Sci (2014) 76, 1201–8. doi: 10.1292/jvms.14-0177
15. Yang JA, Song CI, Hughes JK, Kreisman MJ, Parra RA, Haisenleder DJ. Acute psychosocial stress inhibits LH pulsatility and Kiss1 neuronal activation in female mice. Endocrinology (2017) 158:3716–23. doi: 10.1210/en.2017-00301
16. Kinsey-Jones J, Li X, Knox A, Wilkinson E, Zhu X, Chaudhary A. Down-regulation of hypothalamic kisspeptin and its receptor, Kiss1r, mRNA expression is associated with stress-induced suppression of luteinising hormone secretion in the female rat. J Neuroendocrinol (2009) 21:20–9. doi: 10.1111/j.1365-2826.2008.01807.x
17. Luo E, Stephens SB, Chaing S, Munaganuru N, Kauffman AS, Breen KM. Corticosterone blocks ovarian cyclicity and the LH surge via decreased kisspeptin neuron activation in female mice. Endocrinology (2016) 157:1187–99. doi: 10.1210/en.2015-1711
18. Yamamoto K, Darimont B, Wagner R, Iniguez-Lluhi J. Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harbor Symp quant Biol (1998), 63, 587–98. doi: 10.1101/sqb.1998.63.587
19. Takumi K, Iijima N, Higo S, Ozawa H. Immunohistochemical analysis of the colocalization of corticotropin-releasing hormone receptor and glucocorticoid receptor in kisspeptin neurons in the hypothalamus of female rats. Neurosci Lett (2012) 531:40–5. doi: 10.1016/j.neulet.2012.10.010
20. Wang Q, Sham KW, Ogawa S, Li S, Parhar IS, Cheng CH. Regulation of the two kiss promoters in goldfish (Carassius auratus) by estrogen via different ERα pathways. Mol Cell Endocrinol (2013) 375:130–9. doi: 10.1016/j.mce.2013.04.023
21. Fricke H, Fricke S. Monogamy and sex change by aggressive dominance in coral reef fish. Nature (1977) 266:830–2. doi: 10.1038/266830a0
22. Fricke HW. Social control of sex: field experiments with the anemonefish amphiprion bicinctus. Z für Tierpsychologie (1983) 61:71–7. doi: 10.1111/j.1439-0310.1983.tb01327.x
23. Hattori A, Yanagisawa Y. Life-history pathways in relation to gonadal sex differentiation in the anemonefish, amphiprion clarkii, in temperate waters of Japan. Environ Biol Fishes (1991) 31:139–55. doi: 10.1007/BF00001015
24. Chen J, Peng C, Yu Z, Xiao L, Yu Q, Li S. The administration of cortisol induces female-to-male sex change in the protogynous orange-spotted grouper, epinephelus coioides. Front Endocrinol (2020) 11:12. doi: 10.3389/fendo.2020.00012
25. Zhang H, Zhang Y, Guo Y, Zhang X, Wang Q, Liu X. Kiss2 but not kiss1 is involved in the regulation of social stress on the gonad development in yellowtail clownfish, amphiprion clarkii. Gen Comp Endocrinol (2020) 298:12. doi: 10.3389/fendo.2020.00012
26. Zhang Y, Zhang H, Wang J, Zhang X, Bu S, Liu X. Molecular characterization and expression patterns of glucocorticoid receptor (GR) genes in protandrous hermaphroditic yellowtail clownfish, amphiprion clarkii. Gene (2020) 745:144651. doi: 10.1016/j.gene.2020.144651
27. Zhang Y, Zhang X, Bu S, Zhang W, Li T, Zheng D, et al. Sexually dimorphic distribution of kiss1 and kiss2 in the brain of yellowtail clownfish, Amphiprion clarkii. Endocr Connect (2022) in press. doi: 10.1530/EC-22-0136
28. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
29. Khor YM, Soga T, Parhar IS. Early-life stress changes expression of GnRH and kisspeptin genes and DNA methylation of GnRH3 promoter in the adult zebrafish brain. Gen Comp Endocrinol (2016) 227:84–93. doi: 10.1016/j.ygcen.2015.12.004
30. Iwata E, Mikami K, Manbo J, Moriya-Ito K, Sasaki H. Social interaction influences blood cortisol values and brain aromatase genes in the protandrous false clown anemonefish, amphiprion ocellaris. Zoological Sci (2012) 29:849–55. doi: 10.2108/zsj.29.849
31. Ogawa S, Nathan FM, Parhar IS. Habenular kisspeptin modulates fear in the zebrafish. Proc Natl Acad Sci (2014) 111:3841–6. doi: 10.1073/pnas.1314184111
32. Alvarado M, Servili A, Molés G, Gueguen M-M, Carrillo M, Kah O. Actions of sex steroids on kisspeptin expression and other reproduction-related genes in the brain of the teleost fish European sea bass. J Exp Biol (2016) 219:3353–65. doi: 10.1242/jeb.137364
33. Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell (2011) 43:145–55. doi: 10.1016/j.molcel.2011.06.016
34. Nocillado JN, Mechaly AS, Elizur A. In silico analysis of the regulatory region of the yellowtail kingfish and zebrafish kiss and kiss receptor genes. Fish Physiol Biochem (2013) 39:59–63. doi: 10.1007/s10695-012-9642-0
35. Ducouret B, Tujague M, Ashraf J, Mouchel N, Servel N, Valotaire Y. Cloning of a teleost fish glucocorticoid receptor shows that it contains a deoxyribonucleic acid-binding domain different from that of mammals. Endocrinology (1995) 136:3774–83. doi: 10.1210/en.136.9.3774
36. Stolte EH, van Kemenade BLV, Savelkoul HF, Flik G. Evolution of glucocorticoid receptors with different glucocorticoid sensitivity. J Endocrinol (2006) 190:17–28. doi: 10.1677/joe.1.06703
37. Lethimonier C, Tujague M, Kern L, Ducouret B. Peptide insertion in the DNA-binding domain of fish glucocorticoid receptor is encoded by an additional exon and confers particular functional properties. Mol Cell Endocrinol (2002) 194:107–16. doi: 10.1016/S0303-7207(02)00181-8
38. Bamberger CM, Schulte HM, Chrousos GP. Molecular determinants of glucocorticoid receptor function and tissue sensitivity to glucocorticoids. Endocrine Rev (1996) 17:245–61. doi: 10.1210/edrv-17-3-245
Keywords: cortisol, stress, kiss2 promoter, glucocorticoid receptor, Amphiprion clarkii
Citation: Bu S-Y, Zhang Y-Y, Zhang X, Li T-X, Zheng D-C, Huang Z-X and Wang Q (2022) Regulation of the kiss2 promoter in yellowtail clownfish (Amphiprion clarkii) by cortisol via GRE-dependent GR pathway. Front. Endocrinol. 13:902737. doi: 10.3389/fendo.2022.902737
Received: 23 March 2022; Accepted: 04 July 2022;
Published: 03 August 2022.
Edited by:
Bin Wang, Chinese Academy of Fishery Sciences (CAFS), ChinaReviewed by:
Alejandro S. Mechaly, CONICET Instituto de Investigaciones en Biodiversidad y Biotecnología (INBIOTEC), ArgentinaCopyright © 2022 Bu, Zhang, Zhang, Li, Zheng, Huang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Wang, d2FuZ3FpYW4xNzBAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.