
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 01 July 2022
Sec. Neuroendocrine Science
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.892758
This article is part of the Research Topic Genomic and Transcriptomic Insights into Neuroendocrine Evolution and Function View all 8 articles
 Bethany A. Crooks†
Bethany A. Crooks† Darrin Mckenzie†
Darrin Mckenzie† Luke C. Cadd
Luke C. Cadd Ciaran J. McCoy
Ciaran J. McCoy Paul McVeigh
Paul McVeigh Nikki J. Marks
Nikki J. Marks Aaron G. Maule
Aaron G. Maule Angela Mousley
Angela Mousley Louise E. Atkinson*
Louise E. Atkinson*The endocannabinoid signalling (ECS) system is a complex lipid signalling pathway that modulates diverse physiological processes in both vertebrate and invertebrate systems. In nematodes, knowledge of endocannabinoid (EC) biology is derived primarily from the free-living model species Caenorhabditis elegans, where ECS has been linked to key aspects of nematode biology. The conservation and complexity of nematode ECS beyond C. elegans is largely uncharacterised, undermining the understanding of ECS biology in nematodes including species with key importance to human, veterinary and plant health. In this study we exploited publicly available omics datasets, in silico bioinformatics and phylogenetic analyses to examine the presence, conservation and life stage expression profiles of EC-effectors across phylum Nematoda. Our data demonstrate that: (i) ECS is broadly conserved across phylum Nematoda, including in therapeutically and agriculturally relevant species; (ii) EC-effectors appear to display clade and lifestyle-specific conservation patterns; (iii) filarial species possess a reduced EC-effector complement; (iv) there are key differences between nematode and vertebrate EC-effectors; (v) life stage-, tissue- and sex-specific EC-effector expression profiles suggest a role for ECS in therapeutically relevant parasitic nematodes. To our knowledge, this study represents the most comprehensive characterisation of ECS pathways in phylum Nematoda and inform our understanding of nematode ECS complexity. Fundamental knowledge of nematode ECS systems will seed follow-on functional studies in key nematode parasites to underpin novel drug target discovery efforts.
Parasitic nematodes inflict a pervasive burden on human, animal and plant health (1). The rapid escalation of anthelmintic resistance, and an over reliance on a limited number of frontline anthelmintics, threatens the global sustainability of parasite control. The need for identification and validation of novel control strategies and chemotherapies for nematode parasites is urgent and requires a robust understanding of unexploited aspects of nematode biology that may offer a source of novel drug target candidates.
Neuromuscular signalling is the primary target for frontline anthelmintics because of its importance to nematode biology. Despite this, many facets of nematode neurobiology, including endocannabinoid signalling (ECS), remain uncharacterised and unexploited for parasite control. The ECS system is a complex lipid signalling pathway involved in the regulation of synaptic transmission via retrograde signalling (2, 3), and has been associated with a broad range of immunological, psychological, developmental, neuronal and metabolic physiologies in humans where it has significant therapeutic appeal (4). While mammalian (5–7) and invertebrate (8–13) ECS pathways have been studied extensively, our knowledge of the presence, structure and function of ECS in nematodes is limited (14–19 20).
In vertebrates, endocannabinoids (ECs) primarily activate the canonical cannabinoid G-protein coupled receptors (GPCRs) CB1 and CB2 (21–23), in addition to several other cannabinoid-associated receptors including, for example, transient receptor potential channels (TRP) (24, 25). In contrast, nematodes do not appear to possess homologs of the mammalian-like EC-GPCRs (CB1 and CB2). Instead, the nematode-specific GPCR NPR-19, has been functionally linked to ECS in C. elegans (Ce-NPR-19) (17, 19, 26, 27). Ce-NPR-19 displays only 23% sequence similarity with mammalian CB1 but possesses 50% of the key amino acids required for EC ligand (N-arachidonoylethanolamine; anandamide; AEA) binding (12, 27, 28). In addition, C. elegans NPR-32 is also activated in response to AEA (17), indicating that additional EC-GPCRs could also be present in nematodes.
In vertebrates the primary EC ligands 2-arachidonoylglycerol (2-AG) and AEA are enzymatically biosynthesised on demand, and subsequently metabolised post receptor activation (see Figure 1) (21, 29, 30). 2-AG synthesis occurs via the hydrolysis of diacylglycerol by diacylglycerol lipase (DAGL), while degradation can involve several enzymes including monoacylglycerol lipase (MAGL), lysophosphatidylserine lipase alpha/beta-hydrolase domain containing-12 (ABHD-12) , and alpha/beta-hydrolase domain containing 6 (ABHD-6) (see Figure 1A) (31–33). AEA is predominantly synthesised via the hydrolysis of N-arachidonyl phosphatidyl ethanol (NAPE) by N-arachidonyl phosphatidyl ethanol-phospholipase D (NAPE-PLD) and degraded by fatty acid amide hydrolase (FAAH) (see Figure 1B) (34–36). There is also evidence to suggest the presence of multiple alternative pathways for EC-ligand biosynthesis and degradation in mammals involving several alternative enzymes including alpha/beta-hydrolase domain containing-4, N-acyl phospholipase B (ABHD-4), lysophospholipase D (Lyso-PLD) and phospholipase A2 (PLA-2) (36–38).
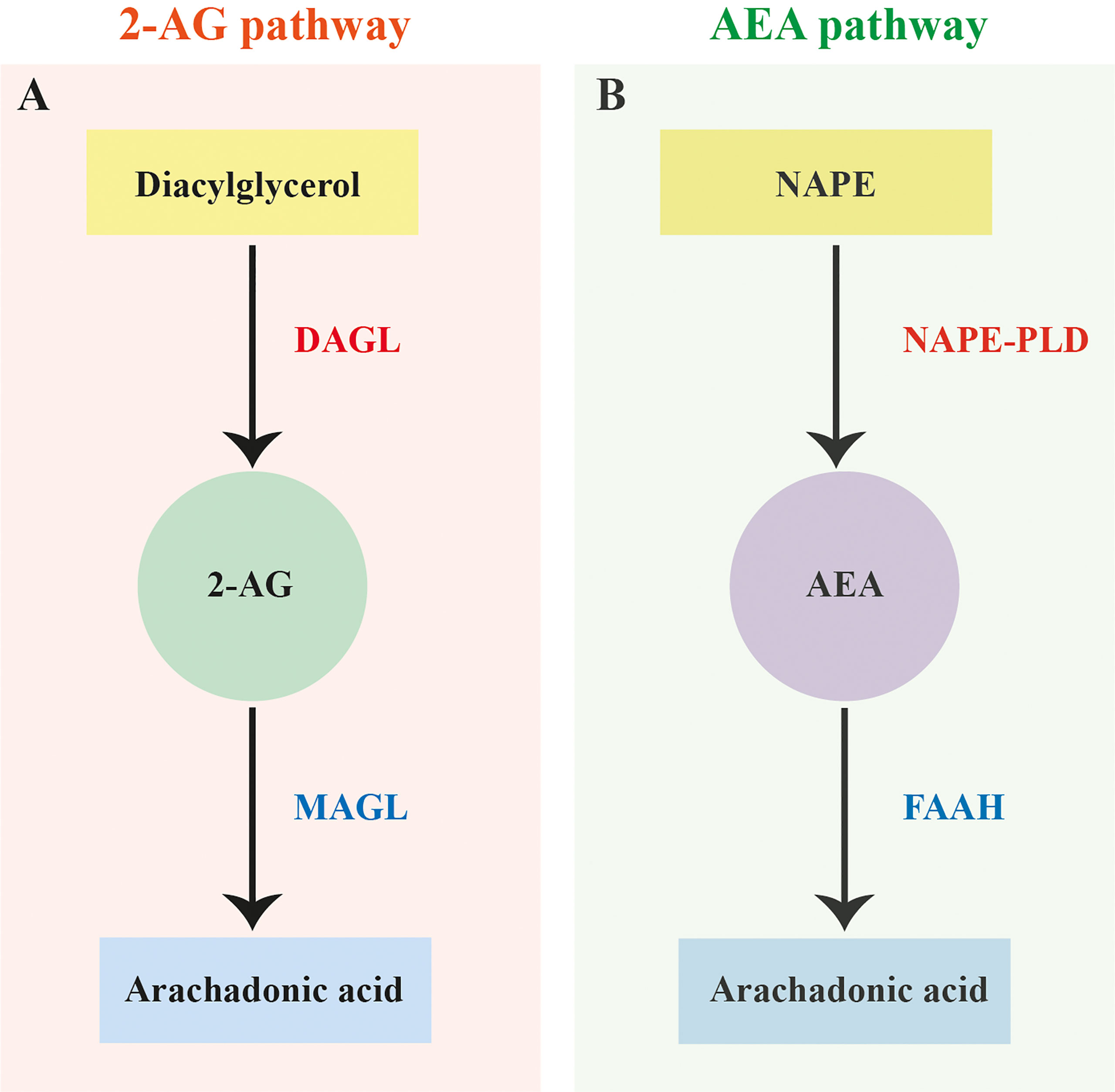
Figure 1 Canonical 2-arachidonoylglycerol and anandamide biosynthesis and degradation pathways. (A) Canonical 2-arachidonoylglycerol (2-AG) biosynthesis and degradation pathway based on vertebrates showing the hydrolysis of diacylglycerol by diacylglycerol lipase (DAGL) and degradation of 2-AG by monoacylglycerol lipase (MAGL). (B) Canonical anandamide (AEA) biosynthesis and degradation pathway based on vertebrates showing the hydrolysis of N-arachidonyl phosphatidyl ethanol (NAPE) by N-arachidonyl phosphatidyl ethanol-phospholipase D (NAPE-PLD) and degradation of AEA by fatty acid amide hydrolase (FAAH).
In nematodes 2-AG and AEA have been identified via mass-spectrometry in C. elegans, Pelodera strongyloides, Caenorhabditis briggsae and the rodent gastrointestinal nematode Nippostrongylus brasiliensis (18, 39). In addition, in C. elegans several of the hydrolytic enzymes linked to EC degradation (MAGL and FAAH) have been identified in silico (40, 41) and functionally characterised (42). Caenorhabditis elegans ECS has also been associated with a raft of important biological roles (15–17, 19, 27, 42–44).
Information on the presence and function of ECS in parasitic nematodes is limited to a single study that identified ECS enzymes and the putative EC-GPCR NPR-19 via bioinformatics in N. brasiliensis, Ancylostoma duodenale, Ancylostoma celanicum, Necator americanus, Steinernema carpocapsae, Ascaris lumbricoides, Strongyloides ratti, Strongyloides stercoralis, Wuchereria bancrofti and Toxocara canis (18). This work also demonstrated that EC’s may modulate the host immune response during parasite infection and that N. brasiliensis produces ECs throughout its lifecycle, most notably in the infective larval stage (18). This strongly supports the hypothesis that parasitic nematodes possess a functional ECS pathway. However, as these observations represent a small subset (6.7%) of available nematode genomes there remains an opportunity to exploit the recent expansion in nematode omics data to characterise the breadth and complexity of the ECS system across phylum Nematoda.
Here, we employed a bioinformatics driven in silico pipeline and phylogenetic analyses to identify the presence, and interrogate the conservation and expression profiles of ECS pathway effectors (EC-effectors) in all publicly available nematodes genomes and life stage and tissue-specific transcriptomes. Our data demonstrate that: (i) ECS is broadly conserved across phylum Nematoda, including in therapeutically and agriculturally relevant species; (ii) EC-effectors appear to display clade and lifestyle-specific conservation patterns; (iii) filarial species possess a reduced EC-effector complement; (iv) sequence analyses reveal key differences between nematode and vertebrate EC-effectors; (v) life stage-, tissue- and sex-specific EC-effector expression profiles suggest a role for ECS in therapeutically relevant parasitic nematodes. To our knowledge this study represents the most comprehensive, pan-phylum, analysis of the nematode ECS system, including in species with global therapeutic and agricultural significance. These data will facilitate basic research focused on of the role of EC’s in key aspects of nematode biology e.g. motility, sensory function, feeding, development, and will seed functional genomics studies in tractable parasitic nematodes to inform future novel anthelmintic target discovery pipelines.
Query sequences for 70 genes encoding a total of 14 C. elegans, C. briggsae, Caenorhabditis brenneri, Caenorhabditis japonica and Caenorhabditis remanei EC pathway effectors [seven EC receptors (ECRs) including receptors that have been closely linked to ECS, and seven endocannabinoid enzymes (ECEs)] were obtained from WormBase ParaSite v14 (WBP; https://parasite.wormbase.org) (48) (see File SI 1) (18, 27, 43, 45, 46). Note that in the nematode literature, NAPE-PLD orthologs are commonly referred to as NAPE (42, 43, 47) [WormBase Gene IDs; nape-1 WBGene00021371, nape-2 WBGene00021370 (48)], consequently we have continued to refer to nematode NAPE-PLD as NAPE in this study for consistency.
A Hidden Markov Model (HMM)-based approach has previously been reported for the identification of flatworm GPCRs (49). Predicted protein datasets were downloaded from WBP v14 for all nematodes with publicly available genome data (134 genomes; see File SI 2). Predicted protein datasets were concatenated for use as a predicted protein database for HMMERv3.3 HMM-searches. Profile HMMs for nematodes were constructed using predicted protein alignments of all candidate EC-effector protein homologs in C. elegans, C. briggsae, C. brenneri, C. japonica and C. remanei (see File SI 1). Multiple Sequence Alignments (MSAs) were generated using EMBL-EBI Clustal-Omega [https://www.ebi.ac.uk/Tools/msa/clustalo; (50)]. The hmmsearch function was employed to identify putative EC proteins within the nematode predicted protein datasets (see File SI 2) using default settings. Due to the volume of genomic data generated, the highest confidence hits were selected for each protein based on an inclusion threshold of E-value ≤0.01 and/or a score of ≥150.
Putative EC protein sequences identified via hmmsearch were then used as queries in reciprocal BLASTp searches of WBP (https://parasite.wormbase.org/Multi/Tools/Blast; default settings) and NCBI non-redundant (https://blast.ncbi.nlm.nih.gov; default settings) databases. Queries that failed to return a putative ECR or ECE hit within the top 4 BLAST results were excluded from further downstream analyses. All BLASTp searches returning negative hits (hits outside of the outlined inclusion criteria and those that returned no hits) were confirmed negative via tBLASTn searches; this approach also mitigated the impact of poor genome quality (false negatives) on our analyses where relevant.
Key EC ligand binding residues and functional motifs for mammalian and invertebrate ECRs and ECEs were identified from the published literature (see File SI 3) and all positive hits were examined visually for the presence of any key residues/motifs via multiple sequence alignments using EMBL-EBI Clustal-Omega [https://www.ebi.ac.uk/Tools/msa/clustalo (50)]. The presence of known protein family or structural domains in ECE BLAST hits was analysed using InterProScan [https:www.ebi.ac.uk/interpro/search/sequence-search, (51)]. Putative ECR hits were analysed for the presence of GPCR transmembrane (TM) domains using EMB TMpred server [https://embnet.vital-it.ch/software/TMPRED_form.html (52)]. Any putative ECE hits which lacked the required family/protein domains for designation as an ECE, or putative ECR sequences which possessed ≤ 3 TM domains, were excluded from further analysis. ECRs and ECEs were analysed for the presence of conservative substitutions of key residues or within binding motifs using WebLogo3 (53, 54) (see Supplementary Table 2).
MUSCLE was used to create multiple sequence alignments (MSAs) of protein sequences for all positive EC protein hits using MEGA X (55). For ECRs, alignments were manually edited to include only TM domains, for ECEs only functional domains were included in analysis. Functional domains for ECEs and TM domains for ECRs were identified via the NCBI Conserved Domains Database [CDD; https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (56)]. Maximum likelihood (ML) phylogenetic trees were constructed using PhyML [http://www.phylogeny.fr (57)] from the domain only MUSCLE MSAs with default parameters and branch support assessment using the approximate likelihood ratio test (aLRT) with “SH-like” parameters. Trees were exported from PhyML in Newick format and were drawn and annotated using the Interactive Tree of Life [iTOL; https://itol.embl.de (58).
180 publicly available transcriptome datasets (145 life stage- and 35 tissue-specific datasets) representing 32 nematode species were analysed in this study. One hundred and fifty publicly available life stage and tissue specific transcriptome datasets representing 27 nematode species were collated from WBP v14 Gene Expression database (48) and published literature (see File SI 2). WBP datasets (see File SI 2) consisted of metadata, raw counts, transcripts per million (TPM) and DESeq2 differential expression data (in log2foldchange and adjusted p value formats). Data for an additional 34 datasets, representing five species [Haemonchus contortus, Toxocara canis, Globodera pallida, Strongyloides venezuelensis and Strongylodies papillosus; see File SI 2] were identified from published literature, and metadata and raw counts were accessed from NCBI Sequence Read Archive [SRA; www.ncbi.nlm.nih.gov/sra] (59) for analysis. TPM and median TPM data for H. contortus, T. canis and G. pallida were downloaded using the European Bioinformatics Institute RNAseq-er Application Program Interface (60) (File SI 2).
S. venezuelensis and S. papillosus (raw counts and TPM) data were analysed using an established RNA-Seq pipeline (61). Briefly, raw sequences reads were processed into forward and reverse fastq files using the NCBI SRA Toolkit (62). Reads were then trimmed using Trimmomatic (v0.36; parameters: LEADING:5 TRAILING:5 SLIDINGWINDOW:3:15 MINLEN:34) (63) and sequences below this established inclusion threshold were removed. Corresponding genome assemblies [BioProject accessions; PRJEB530 and PRJEB525, respectively] (64) were downloaded from WormBase ParaSite v14 (48) and reads were mapped to the relevant genome using HISAT2 v2.1.0 (65). Raw gene counts were assigned via SubRead v 2.0.1 featureCounts (66). Raw counts of orthologous genes were transformed to TPM and subsequently median TPMs were calculated to represent raw gene expression in the life stages with RNA-Seq data available. An inclusion threshold for expression of 1.5 TPM was applied [TPM thresholds are typically set between 1-2 TPM (67, 68)], any transcripts which failed to meet the threshold for expression were excluded from downstream analysis.
Differential expression data for H. contortus, T. canis, G. pallida, S. venezuelensis and S. papillosus RNA-Seq data were generated using DESeq2 in the format of log2foldchange and adjusted p values (69, 70). Datasets were then mined for pathway protein gene IDs identified in the HMM searches. Heatmaps displaying log2AverageTPM were generated using the Heatmapper Expression protocol, with an average linkage clustering method and Pearson’s distance measurement method (71).
In this study we mined 133 nematode genomes (representing 109 species, 7 clades and 3 lifestyles) for 13 putative ECS pathway effectors, expanding upon previous studies (18). Our HMM-based in silico approach returned a total of 1289 putative ECS effector homologs (ECEs and ECRs; ECRs included some receptors linked to ECS function) (see Figure 2 and File SI 3). The data demonstrate that: (i) ECEs and ECRs display pan-phylum conservation; (ii) representatives of Clades 8 and 12 exhibit the lowest level of EC-effector conservation [clade 8: 68% and 79% of all possible ECEs and ECRs conserved, respectively; clade 12: 83% and 72% of all possible ECEs and ECRs conserved, respectively] and, (iii) free-living and parasitic nematodes display a comparable level of EC-effector conservation [free-living: 91% and 96% of all possible ECEs and ECRs conserved, respectively; parasitic: 83% and 92% of all possible ECEs and ECRs conserved, respectively] (see Figure 2). To our knowledge this is the first pan-phylum examination of nematode ECS profiles and represents a comprehensive analysis of ECS pathway conservation in parasitic species that impact human, animal and plant health. Several important points emerge from these data (see below).

Figure 2 Nematodes possess homologs of canonical endocannabinoid signalling pathway effectors that display broad pan-phylum conservation. (A) Pan-phylum conservation of ECS proteins in all nematodes that possess genome data. Black boxes represent the presence of a homolog. Core Eukaryotic Genes Mapping Approach (CEGMA)/Benchmarking Universal Single-Copy Orthologs (BUSCO) scores represent genome quality [data derived from WormbaseParasite, each circle represents 10% increase in genome quality, colours represent scale (red represents lower percentage genome quality, green represents higher percentage genome quality)]. Asterix denotes multiple genomes (*). Nematode genome references listed in File SI 2. All homolog gene IDs identified listed in File SI 3. Clades based on Holterman and Blaxter classifications where roman numerals represent Blaxter classification and numbers represent Holterman classification (i.e. 2/I denotes Holterman clade 2 and Blaxter classification clade I) (72, 73). (B) Bar chart displaying the percentage of nematode species that possess each ECR/ECE.
Nematode EC-effectors (ECEs responsible for synthesis and degradation of EC ligands, and putative ECRs) appear to be broadly conserved across phylum Nematoda (Figure 2A). ECEs display greater conservation than putative ECRs across all nematodes, with the exception of the ECE nape-1/2 which appears to be absent from 37% of nematodes, many of which are representatives of clades 8 and 9 (Figures 2A, B). The more conserved profile of ECEs versus ECRs is consistent with the requirement for specific ECEs in the biosynthesis and degradation of EC-ligands and the potential for redundancy among ECRs, which has been documented in other systems (74, 75).
Genes encoding the key ECEs responsible for 2-AG synthesis and metabolism, DAGL-2 and ABHD-12 respectively, are co-conserved in 87% of nematodes (see Figure 2), indicating that a significant proportion of nematode species are likely to possess a functional, canonical, 2-AG synthesis and degradation pathway. Most (95%) nematode species examined possess an gene ortholog of the 2-AG biosynthesis enzyme DAGL-2, while the gene encoding ABHD-12, responsible for catalysing 2-AG metabolism is present in 88% of species. Parascaris equorum and Globodera pallida do not appear to possess either a dagl-2 or abhd-12 homolog, however their genome quality is lower as indicated by CEGMA/BUSCO scores [see Figure 2; (76, 77)].
Genes encoding NAPE and FAAH, the primary enzymes responsible for the synthesis and metabolism of AEA, are co-conserved in 55% of nematode species, suggesting that a significant proportion nematodes have the ability to metabolise AEA (Figure 2). This is corroborated by studies that have identified AEA in several nematodes, including C. elegans and N. brasiliensis, via mass-spectrometry (18, 39). While 88% of species encode at least one FAAH homolog, the NAPE-encoding gene is conserved in only 63% of nematodes (either nape-1 or -2). Notably, many of the species that lack a nape homolog are filarial nematodes, Strongyloides species, or members of the Diplogasteroidea superfamily (Figure 2). Whilst the absence of nape in some species may be explained by genome quality, in other species with robust genome data an alternative AEA synthesis pathway may exist (see below). Previous studies have demonstrated the presence of two, functionally divergent, nape orthologs in C. elegans that occupy adjacent genomic positions (nape-1, IV:3739520.3740880; nape-2, IV:3735470.3738925; [WormBase; (78)], share 73% sequence identity, and display complete conservation of the NAPE-PLD signature sequence (43). Our pan-phylum analysis confirms that C. elegans is the only nematode species that possesses two distinct NAPE-encoding genes. In all other species that encode NAPE the same gene ID was returned for both the nape-1 and nape-2 BLASTp searches (one positive nape hit was considered a positive return for both nape-1 and nape-2 and was designated nape-1/2; see Figure 2 and File SI 3). The presence of a single nape-1/2 in all of the nematodes examined, including other Caenorhabditis species, suggests that nape-1 and -2 may have arisen as a result of a relatively recent gene duplication event in C. elegans (43).
Of the seven putative ECRs included in this study NPR-19 and NPR-32 have been most closely linked to ECS in nematodes (16, 17, 27). Our in silico analysis reveals that 79% of the nematode species investigated in this study possess NPR-19 and 89% possess NPR-32, underscoring their putative importance to nematode biology. In addition to NPR-19 and -32, OCR-2 an ortholog of the human transient receptor potential vanilloid channel (TRPV) (79), has also been closely linked to ECS (25). OCR-2 regulates signal transmission and thus modulates several C. elegans behaviours (19, 80). It is interesting to note that 100% of nematode species examined in this study possess a gene encoding OCR-2, also suggesting a significant role in nematode biology. NPR-9 has been implicated in locomotion, regulation of innate immune responses, roaming and foraging behaviours in C. elegans (81–83), while GPR-55, the human ortholog of nematode NPR-9, is known to interact with human CB1 and CB2 to form functionally important heteromers (84, 85). The lower level of npr-9 conservation across phylum Nematoda revealed here (npr-9 conserved in 60% of species examined; Figure 2) may indicate a less conserved functional role for this putative ECR in nematodes.
Our data demonstrate distinct conservation patterns of EC-effectors across nematode clades. Whilst clade 9 and 10 nematodes exhibit the highest degree of EC-effector conservation, clade 8 species display the most reduced complements (Figure 2). This could be, in part, explained by lower CEGMA/BUSCO scores for clade 8 genomes, such that the profiles presented here may not be a true representation of EC-effector complements in this clade. Variable genome quality is an inherent caveat to in silico approaches such as those employed in this study however, the use of tBLASTn for all negative BLASTp returns can help to mitigate this in addition to continued improvements in genome quality (86).
For other clades there appear to be ECR specific trends. For example, npr-32 is broadly conserved across all clades, pointing towards a more conserved function for this receptor pan-phylum. Other ECRs display more clade-specific, restricted profiles, including nhr-49 which appears to be entirely absent from clade 2 (Figure 2A). Clade 2 nematode genome assemblies are high quality [as indicated by CEGMA/BUSCO scores; (48)] and likely provide a true reflection of the nhr-49 profile. Notably, npr-19 is also entirely absent from clade 12 species, indicating that alternative ECRs may contribute to the ECS pathway in these nematodes. Indeed, clade 12 nematodes exhibit broad conservation of other putative ECRs (e.g. npr-9, -32, nhr-49, octr-1, ser-4, ocr-2; see Figure 2A).
Whilst there appear to be limited differences between free-living and parasitic nematode EC-effector profiles in general [96% ECR conservation in free-living nematodes (FL) vs 92% in parasitic species; 91% ECE conservation in free-living nematodes vs 83% in parasitic species], filarial nematodes display distinct EC-effector gaps. For example, filarial species completely lack genes encoding NPR-9 and NAPE-1/2, and have a significantly reduced FAAH-2 encoding gene profile (Figure 2A). The absence of NAPE-1/2 points towards the presence of an alternative AEA synthesis pathway in filarids, which may also be the case for other species e.g. Pristionchus spp. and Strongyloides spp. that likewise lack NAPE-1/2 encoding genes (see below; Figure 2A).
Our data indicate that 41 nematode species lack genes encoding the nematode NAPE-PLD ortholog NAPE-1/2, the enzyme primarily responsible for AEA synthesis (Figure 2A) (34). In vertebrates, two additional pathways have been implicated in the synthesis of AEA: (i) hydrolysis of NAPE by ABHD-4 forming the intermediate glycerophosphoanandamide (Glycero-p-AEA) which, following further hydrolysis by glycerophosphodiester phosphodiesterase 1 (GDE-1), results in the formation of AEA (see Figure 3A ii); (ii) hydrolysis of NAPE by phospholipase A2 (PLA-2) to form the intermediate N-acyl-1-acyl-lyso-PE (lyso-NAPE) and subsequently, following hydrolysis by lysophospholipase D (Lyso-PLD), results in the formation of AEA (see Figure 3A iii) (87–90). To determine if alternative AEA synthesis pathways exist in nematodes that appear to lack the classical NAPE-1/2 AEA biosynthesis pathway, we mined all available genome data for abhd-4, gde-1, pla-2 and lyso-PLD.

Figure 3 Nematode species lacking the anandamide (AEA) synthesis enzyme N-arachidonyl phosphatidyl ethanol (NAPE) possess putative alternative AEA synthesis enzymes. (A) Diagram showing canonical (i) AEA synthesis pathway alongside two alternative synthesis pathways: (ii) hydrolysis of NAPE by Abhydrolase Domain Containing 4 (ABHD-4) forming the intermediate glycerophosphoanandamide (Glycero-p-AEA) and hydrolysis by glycerophosphodiester phosphodiesterase 1 (GDE-1) to synthesise AEA, and (iii) hydrolysis of NAPE by phospholipase A2 (PLA-2) to form the intermediate N-acyl-1-acyl-lyso-PE (lyso-NAPE) and hydrolysis by lysophospholipase D (Lyso-PLD) to synthesise AEA. (B) Conservation of genes encoding the alternative AEA synthesis enzymes ABHD-4, GDE-1, PLA-2, Lyso-PLD is shown in all nematodes that lack NAPE. Black boxes represent the presence of a homolog. Core Eukaryotic Genes Mapping Approach (CEGMA)/Benchmarking Universal Single-Copy Orthologs (BUSCO) scores represent genome quality [data derived from WormbaseParasite, each circle represents 10% increase in genome quality, colours represent scale (red represents lower percentage genome quality, green represents higher percentage genome quality)]. Asterix denotes multiple genomes (*). Nematode genome references listed in File SI 2. All homolog gene IDs identified listed in File SI 3. Clades based on Holterman and Blaxter classifications where roman numerals represent Blaxter classification and numbers represent Holterman classification (i.e. 2/I denotes Holterman clade 2 and Blaxter classification clade I) (72, 73).
Our data reveal that 39 of the 41 species that lack NAPE-1/2 possess at least one putative alternative AEA synthesis pathway (see Figure 3B, File SI 3). 95% of these species possess both abhd-4 and gde-1 (alternative pathway shown in Figure 3A ii), while 56% encode both PLA-2 and Lyso-PLD (alternative pathway shown in Figure 3A iii); 56% of nematodes encode the enzymes for both alternative AEA synthesis pathways. Therefore these data suggest that nematodes which lack the classical NAPE-1/2 biosynthesis pathway predominantly synthesise AEA via ABHD-4 and GDE-1. However, within clades 8, 9 and 10 there are examples of species which may have the ability to synthesise AEA via either alternative pathway e.g. Strongyloides species and several filarial nematodes (Figure 3B).
Interestingly in mammals, in addition to PLA-2, other enzymes have been linked to the synthesis of lyso-NAPE (38), for example, ABHD-4 can remove an acyl group from NAPE resulting in the creation of lyso-NAPE (89). Therefore the presence of abhd-4 in 100% of the nematodes investigated in this study suggests that some nematodes that possess lyso-PLD but lack pla-2 could compensate by employing ABHD-4 in the synthesis of lyso-NAPE.
Interestingly, it appears that a subset of the species that do encode NAPE-1/2 (20 key species selected for their relevance to human, veterinary or plant health and broad clade representation), also encode all of the alternative AEA synthesis enzymes (abhd-4, gde-1, pla-2 and lyso-PLD), with the exception of Trichuris muris which appears to lack pla-2 (see File SI 4). It is unclear whether NAPE-1/2 is indeed the predominant AEA synthesis pathway in these species or, if the alternative pathways also contribute to AEA production.
In silico evidence for the presence of putative, alternative, AEA biosynthesis enzymes in a range of therapeutically relevant nematodes strongly suggests that NAPE-1/2 independent pathways may contribute to AEA synthesis in these species. Further analysis, including mass spectrometry to isolate AEA, will begin to unravel the importance of alternative AEA biosynthesis pathways in these nematodes.
The mammalian AEA hydrolysis enzymes FAAH-1 and FAAH-2 possess four key residues required for catabolic activity (FAAH-1: K142, M191, S217, S241; FAAH-2: K131, C180, S206, S230) (42, 91, 92). Analysis of nematode FAAH-1 homologs identified in this study (see Figure 2) revealed that >90% possess the key mammalian binding site residues K142, S217 and S241, while 83% of identified nematode FAAH-2 homologs possess K131, S206 and S230 (see Figures 4A–C). However, 87% of nematode FAAH-1 homologs display a conservative substitution (methionine for leucine) at position 191 while 80% of nematode FAAH-2 homologs substitute cysteine for leucine at position 180 (Figures 4A–C). M191 has been implicated in mammalian FAAH-1 EC-derivative binding, with studies demonstrating lipophilic interactions between this residue and partial cannabinoid receptor agonists, linking it to AEA binding (93), however the significance of C180 (FAAH-2) is less clear (94–96).
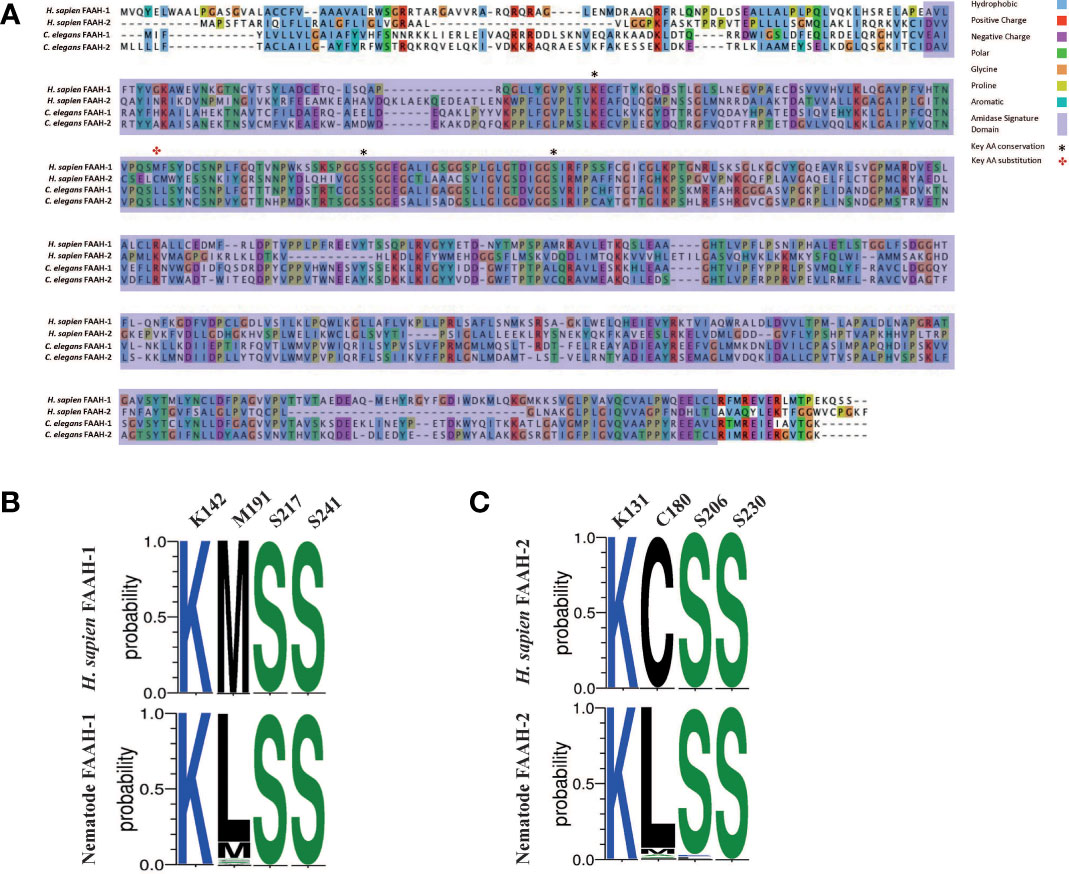
Figure 4 Nematode FAAH homologs conserve key functional domains and motifs, but display conservative substitutions in a key AEA binding site. (A) Protein sequence alignment of Homo sapiens fatty acid amide hydrolase (FAAH-1), H. sapiens FAAH-2, Caenorhabditis elegans FAAH-1 and C. elegans FAAH-2. Amino acids are highlighted in the same colour if > 60% of residues are conserved. Legend denotes conserved amino residues, key ligand binding residues in FAAH-1 and FAAH-2, and the amidase signature domain. AA denotes amino acid; H. sapiens denotes Homo sapiens, C. elegans denotes Caenorhabditis elegans. (B) Amino acid sequence-logo representing sequence diversity between key residues in nematode FAAH-1 homologs (consensus) vs H. sapiens FAAH-1 [O00519, FAAH1_HUMAN] and, (C) Amino acid sequence-logo representing sequence diversity between key residues in nematode FAAH-2 homologs vs H. sapiens FAAH-2. Key EC binding site residues were derived from H. sapiens FAAH-1 and -2 and are detailed in the top column of each table. Colours indicate hydrophobicity of amino acid residues (hydrophilic residues are blue, neutral residues are green and hydrophobic residues are black).
Mammalian FAAH-1 and -2 also possess a highly conserved 130 bp amidase signature domain that enables enzyme characterisation (97, 98); this domain is conserved in 94% of the nematode species examined in this study (Figure 4A).
While these data demonstrate that nematodes possess homologs for FAAH enzymes that display broad conservation with vertebrates, the presence of a distinct substitution in a key AEA binding site across many nematodes may highlight the potential for drug target selectivity towards parasitic nematode species.
In mammals, MAGL is primarily responsible for 2-AG degradation and thus the termination of EC signalling (31, 99). Additional 2-AG degradation enzymes, ABHD-6 and -12, have also been reported (33, 100) however, ABHD-6 is thought to play a less significant role in 2-AG degradation (31, 99).
The nematode literature presents conflicting data on the identity of the 2-AG degradation enzyme; indeed prior to this study it was unclear whether nematodes possess a true ortholog of MAGL (19, 27) or, if the nematode 2-AG degradation enzyme is actually an ABHD-12 ortholog (11, 18). It is interesting to note that previous work indicates that magl homologs are absent in some nematode species (18), which has been confirmed here using our pan-phylum in silico approach (Figure 2A). Indeed, in our analyses only two nematode species (of the 109 in this study) returned an magl-like sequence within the top 5 BLASTp hits (Strongylus vulgaris, SVUK_0001964001; Steinernema scapterisci, L892_g30127.t1), both of which failed to meet E-value inclusion criteria. The absence of MAGL in nematodes indicates the presence of an alternative 2-AG degradation enzyme.
In light of the limited role for ABHD-6 in mammalian 2-AG degradation and the absence of MAGL across nematodes (as reported here and in previous studies), we focused our attention on ABHD-12 as a putative alternative to MAGL in nematodes. Pan-phylum analysis of nematode genomes identified orthologs for ABHD-12 in 88% of nematode species examined. Significantly, phylogenetic analyses of these putative abhd-12 homologs demonstrated that 99% of nematode BLAST returns for abhd-12 cluster more strongly with human abdh-12 than human magl (Figure 5 and Supplementary Figure 1) suggesting that the nematode 2-AG hydroxylase enzymes identified here, and originally designated as magl in C. elegans [Y97E10AL.2; (19)], are orthologous with abhd-12.
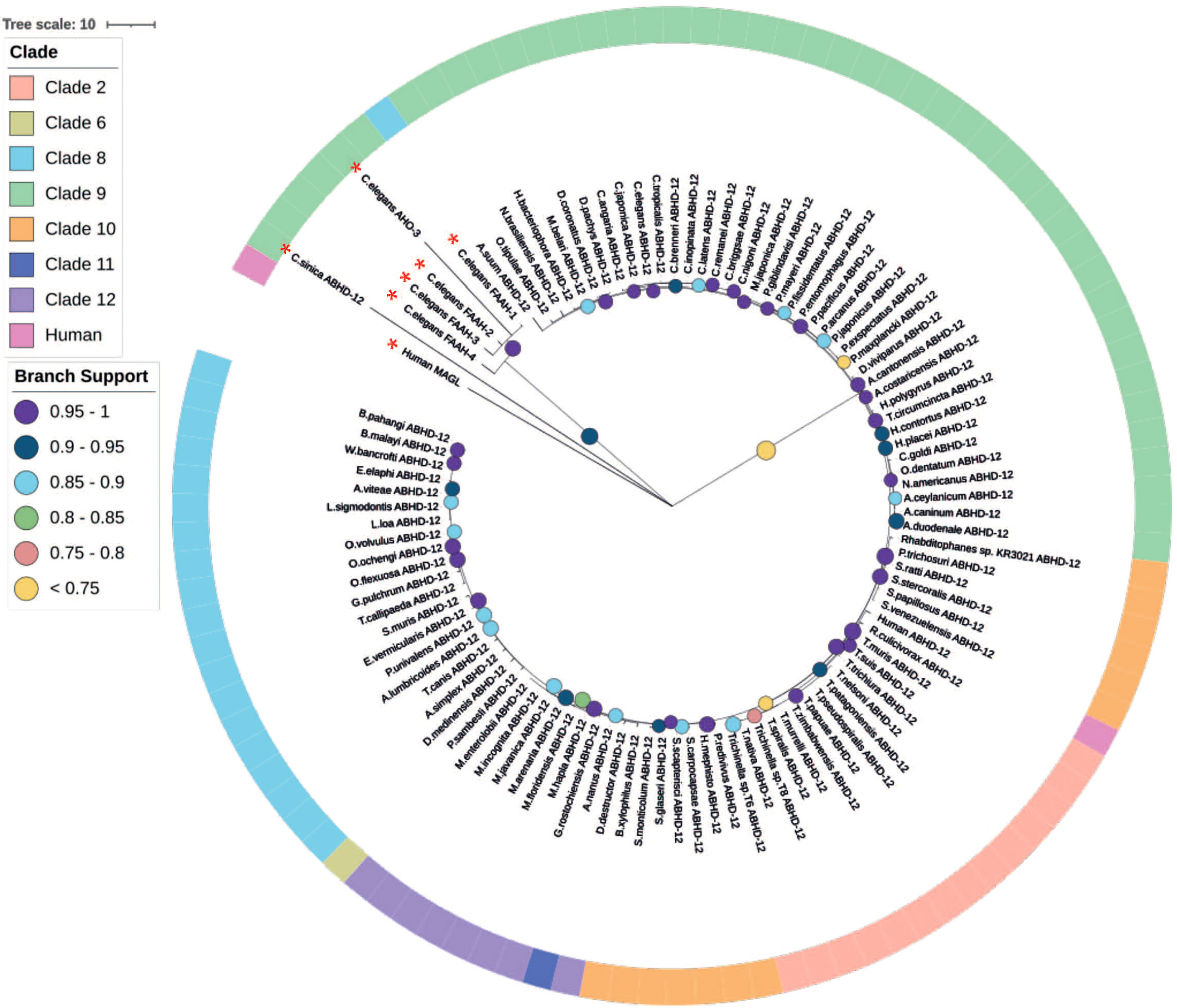
Figure 5 Maximum likelihood phylogeny of nematode ABHD-12 homologs. 98 nematode lysophosphatidylserine lipase alpha/beta-hydrolase domain containing-12 (ABHD-12) homologs are shown in addition to Homo sapiens ABHD-12 [Q8N2K0 (ABD12_HUMAN)] H. sapiens monoacylglycerol lipase (MAGL) [Q99685 (MGLL_HUMAN)], Caenorhabditis elegans fatty acid amide hydrolase (FAAH-1-4) [WBGene00015047, WBGene00015048, WBGene00019068, WBGene00013232] and C. elegans AHO-3 [WBGene00045192; alpha/beta hydrolase containing protein]. Non-ABHD-12 homologs are marked with a red asterisk (*). Outer ring denotes nematode clade and coloured circles represent branch support values. Tree was generated from an alignment trimmed to include protein functional domains. Branch supports indicate statistical support from approximate likelihood ratio test (aLRT).
NPR-19 has been identified as a putative EC-GPCR in C. elegans and appears to modulate key aspects of nematode biology including axon regeneration, locomotion, modulation of noiceception and feeding, and therefore may represent a promising anthelmintic target (27).
78% of the nematodes examined in this study possess an npr-19 ortholog (see Figure 2 and Figure 6A). Phylogenetic analysis demonstrates that nematode npr-19 orthologs failed to cluster with human CB1 and CB2, confirming that NPR-19 is not the direct ortholog of the human EC-GPCRs receptors (CB1 and CB2) (Figure 6A and Figure SI 1). Further analysis of the nematode npr-19 orthologs revealed that whilst the nematode NPR-19 consensus sequence has only 23% similarity to human CB1, several of the known human CB1 EC binding residues (N46, D88, F189, L193, F379, S383) (101, 102) are conserved (Figure 6B). This indicates that nematode NPR-19 orthologs are likely to possess EC ligand binding capacity and aligns with previous work in C. elegans (27). When examined at the clade level, sequence analysis showed that two conservative substitutions are present at position K192 (82% of clade 2 species substitute K192 for N192), while species in clades 8, 9, 10 and 12 substitute K192 for D192. K192 forms a hydrogen bond with the amide oxygen of AEA, implicating this residue in ligand binding (101) (see Figure 6B).
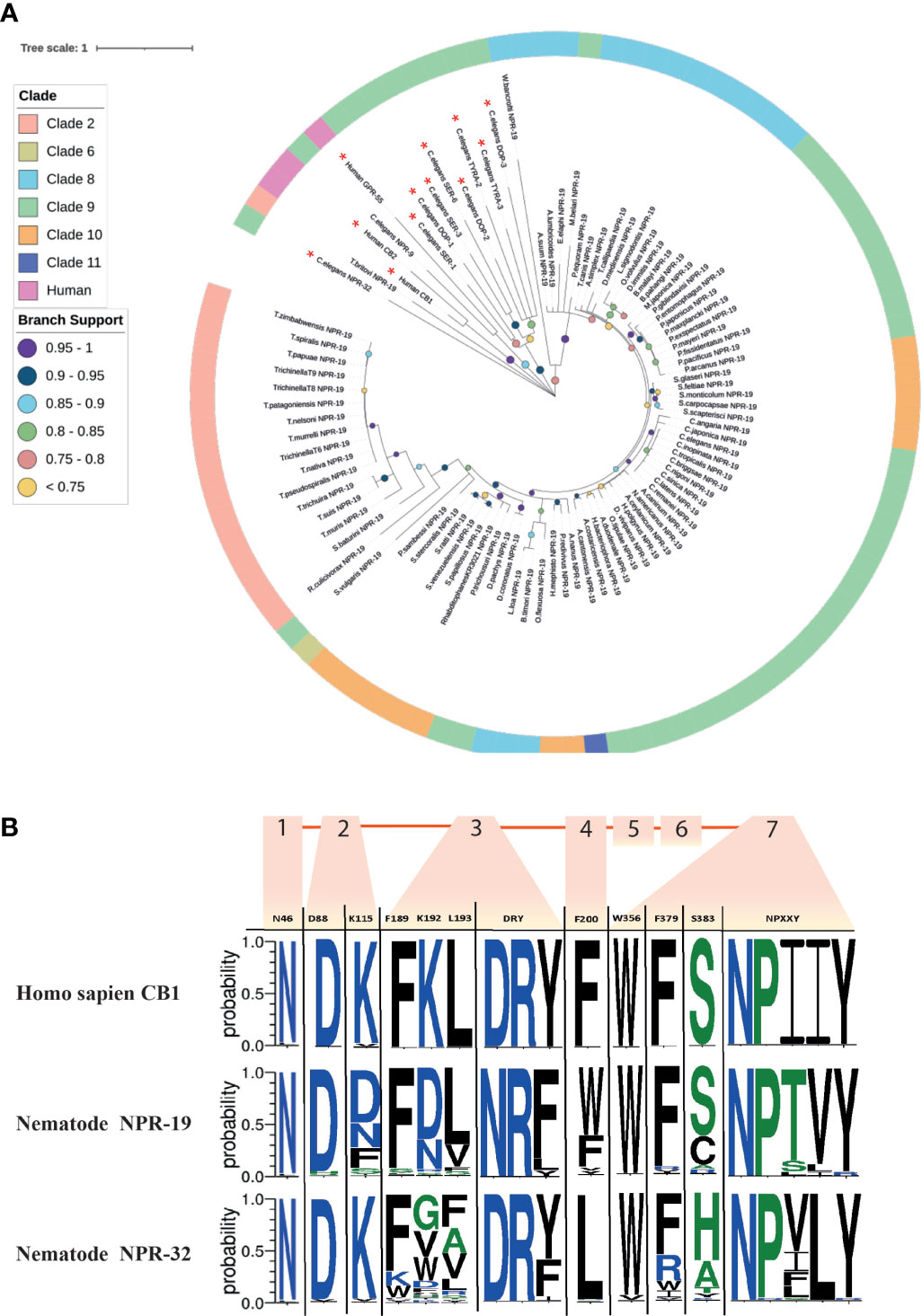
Figure 6 Maximum likelihood phylogeny of nematode npr-19 homologs. (A) 85 nematode NPR-19 homologs are shown in addition to Homo sapiens CB1 and CB2 [P21554 (CNR1_HUMAN), P34972 (CNR2_HUMAN)], H. sapiens GPR-55 [Q9Y2T6 (GPR55_HUMAN)] and several Caenorhabditis elegans biogenic amine receptors (serotonin [SER-1,-3,-6; WBGene00004776, WBGene00004778, WBGene00021897] dopamine [DOP-1-3; WBGene00001052, WBGene00001053, WBGene00020506] and tyramine [TYRA-2 and -3; WBGene00017157, WBGene00006475]). Non-NPR-19 homologs are marked with a red asterisk (*). Outer ring denotes nematode clade and coloured circles represent branch support values. Tree was generated from an alignment trimmed to include functional domains. Branch supports indicate statistical support from approximate likelihood ratio test (aLRT). (B) Amino acid sequence-logo demonstrating sequence diversity between nematode NPR-19 and NPR-32 orthologs (consensus) and H. sapiens CB1 [P21554 (CNR1_HUMAN)]. Known vertebrate endocannabinoid binding and GPCR motifs/residues are indicated in the top row of the sequence logo table, transmembrane regions 1-7 are indicated by orange boxes and numbers, amino acid colours indicate hydrophobicity of amino acid residues (hydrophilic residues are blue, neutral residues are green and hydrophobic residues are black).
NPR-32 is implicated in C. elegans axon regeneration (17) and, in addition to NPR-19, is believed to be a putative EC-GPCR (27). In this study we identified 97 NPR-32 homologs (Figure 2 and Supplementary Figure 1) which share (consensus sequence) only 21% identity with the human EC-GPCR CB1, but conserve several key residues (N46, D88, K115, F189; Figure 6B) that are believed to be important for ECS function (17).
In addition, CB1 possesses a “toggle switch” (residue W356), a putative molecular hinge that interacts with F200 to change the form and state of the receptor which in turn aids EC ligand binding (103, 104). In nematodes the “toggle switch” W356 is conserved in >85% of NPR-19 and -32 orthologs, whereas NPR-19 F200 is substituted for W200 in 59% of species and NPR-32 F200 is substituted for L200 in 90% of species, (see Figure 6B). The significance of these observations will be revealed through molecular docking studies, crystal structure analysis, and functional genomics in relevant parasite species, and will inform the role and importance of these receptors in nematode ECS biology.
EC-effector expression has previously been examined in several parasites demonstrating differential expression across life stages (18). Here we further profiled EC-effector expression in 32 nematode species representing several distinct lifestyles (see File SI 2).
Our data demonstrate that several of the putative ECRs examined in this study are upregulated in third-stage larvae (L3) of several parasites including Ancylostoma ceylanicum, Teladorsagia circumcincta, Dictyocaulus viviparus, H. contortus, S. ratti, S. stercoralis and Onchocerca volvulus (see Supplementary Figure 2). Parasitic nematode L3 larvae are analogous to the dauer life stage of C. elegans; both C. elegans dauer and parasitic nematode L3 stages display similar physiology, are in arrested development, are non-feeding and are highly resistant to their environment (105–107). L3 parasites of species such as those outlined above transition from arrested (dauer-like) L3 larvae to infective L3 (iL3) larvae either constitutively or via the influence of host and environmental factors (108). The ECS pathway has been implicated in antagonization of dauer formation and abolishing dauer larval arrest via stimulation of cholesterol in C. elegans and, in turn, promotion of nematode growth and development (15). Thus, the upregulation of putative ECRs in L3 stages of parasitic nematodes could suggest an analogous role for EC signalling in parasite growth and development at a critical stage in the parasitic lifecycle.
Upregulation of putative ECRs, and a key ECE (dagl-2) associated with EC ligand biosynthesis, is evident in Strongyloides iL3s (see Supplementary Figure 2). In contrast, the ECEs responsible for EC ligand degradation (abhd-12 and faah-1-4) are downregulated at the iL3 stage (see Supplementary Figure 2). The opposite expression profile is noted in the adult life stage (free-living and parasitic females) of Strongyloides spp. where EC-degradation enzymes are upregulated and putative ECRs and dagl-2 are downregulated (see Supplementary Figure 2). These data suggest that higher levels of EC ligands may exist in the iL3 stage of S. ratti and S. stercoralis and is consistent with the elevated production of EC-ligands by N. brasiliensis iL3s (18). Together these data indicate that the ECS system may be involved in processes linked to host infection. Parasitic nematodes exploit numerous sensory cues and mechanisms in order to find their host (109), thus the upregulation of EC-effectors in iL3s may also implicate EC signalling in sensory perception, host-seeking, and the establishment of host infection. These data will direct future functional genomics studies around the role of EC signalling in host finding and infection in tractable parasitic nematodes.
Expression profiling of EC-effectors in sex-specific transcriptome data reveal differential expression patterns in male and female nematodes of several species (see Supplementary Figure 2). In T. circumcincta EC-ligand degradation enzymes are broadly downregulated in adult males, and upregulated in adult females (Supplementary Figure 2). Conversely, O. volvulus exhibits upregulation of all pathway components in adult males, and downregulation in adult females (Supplementary Figure 2). Sex-specific expression of EC-effectors is common in mammalian species, where they exhibit alternative actions on neuropsychiatric processes and reproductive events (110–112). In addition the ECS pathway has been implicated in mammalian fertility regulation (113–117) and in invertebrate reproduction (118, 119). Interrogation of expression at the tissue level is challenging in nematodes where data sets are limited to species which are readily amenable to dissection, for example Ascaris suum and Dirofilaria immitis. Whilst it would appear that EC-effectors are upregulated in reproductive tissues (e.g. in A. suum), further analysis across more tissue types and species is required before meaningful comparisons can be made (data not shown). While the role of EC signalling in the regulation of vertebrate and invertebrate reproduction has been documented, ECS system function in nematode reproduction is yet to be determined. Indeed, enhancing the ability to generate life- and tissue-specific data from key parasitic nematodes will inform functional biology.
In silico approaches and the proliferation of nematode omics resources provide a valuable opportunity to identify putative novel anthelmintic drug targets for the control of parasite disease. This study focuses on the characterisation of the nematode ECS pathway, driven by its putative biological importance and therapeutic appeal (16, 27, 28, 39, 42). Here we: (i) provide a comprehensive pan-phylum overview of EC-effector complements in nematodes, that represent divergent clades and lifestyles; (ii) unravel the complexity of the nematode ECS to identify putative species- and lifestyle-specific EC-pathways and drug target selectivity, and (iii) reveal life stage-, and sex-specific EC-effector expression patterns in relevant parasite species. These data will direct the selection of novel ECS pathway targets for functional validation efforts in parasitic nematodes to inform biology and anthelmintic drug discovery pipelines.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
LA, AM, AGM, and NM designed the research. BC, DM and LC, performed the research. BC, DM and LC, analysed the data with assistance from CM and PM. LA, AM, BC, AGM and NM wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by: the Academy of Medical Sciences Springboard Award (SBF004\1018 to LA); the Biotechnology and Biological Sciences Research Council (BB/H019472/1 to AM); the Biotechnology and Biological Sciences Research Council/Boehringer Ingelheim (BB/T016396/1 to AM, NM, AGM, and LA); the Department of Education and Learning for Northern Ireland (studentships awarded to BC and LC); the Department of Agriculture, Environment and Rural Affairs for Northern Ireland (studentship awarded to DM).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank WormBase ParaSite for helpful assistance with transcriptome resources.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.892758/full#supplementary-material
File SI 1 | Caenorhabditis spp. EC-effector gene IDs. List of EC-effector gene IDs from Caenorhabditis spp. that were used as query sequences in this study.
File SI 2 | Nematode genome and transcriptome accession numbers and citations. List of accession numbers for all genome and transcriptome datasets used in this study along with original citations.
File SI 3 | Nematode HMM/BLAST hit gene IDs. List of all BLAST hit gene IDs generated in this study.
File SI 4 | Presence of alternative AEA synthesis pathways in NAPE encoding species. Presence/absence list of alternative AEA synthesis pathway enzymes in 20 key species that do possess NAPE orthologs (Tab 1) and BLAST hit gene IDs (Tab 2).
Supplementary Table 1 | Table of EC-effectors included in study.
Supplementary Table 2 | Table of EC-effector motifs and relevant citations.
Supplementary Figure 1 | Maximum likelihood phylogeny of: (A) 98 nematode ABHD-12 homologs. Homo sapiens ABHD-12 [Q8N2K0 (ABD12_HUMAN)] H. sapiens MAGL [Q99685(MGLL_HUMAN)], Caenorhabditis elegans FAAH-1-4 [WBGene00015047, WBGene00015048, WBGene00019068, WBGene00013232] and C. elegans AHO-3 [WBGene00045192; alpha/beta hydrolase containing protein] also included; (B) 85 nematode NPR-19 homologs. Homo sapiens CB1 and CB2 [P21554 (CNR1_HUMAN), P34972 (CNR2_HUMAN)], H. sapiens GPR-55 [Q9Y2T6 (GPR55_HUMAN)] and several C. elegans biogenic amine receptors (serotonin [SER-1,-3,-6; WBGene00004776, WBGene00004778, WBGene00021897] dopamine [DOP-1-3; WBGene00001052, WBGene00001053, WBGene00020506] and tyramine [TYRA-2 and -3; WBGene00017157, WBGene00006475]) also included. (C) 89 nematode NPR-9 homologs. Homo sapiens CB1 and CB2 [P21554 (CNR1_HUMAN), P34972 (CNR2_HUMAN)], H. sapiens GPR-55 [Q9Y2T6 (GPR55_HUMAN)] and several C. elegans biogenic amine receptors (serotonin [SER-1,-3,-6; WBGene00004776, WBGene00004778, WBGene00021897] dopamine [DOP-1-3; WBGene00001052, WBGene00001053, WBGene00020506] and tyramine [TYRA-2 and -3; WBGene00017157, WBGene00006475]) also included. (D) 97 nematode NPR-32 homologs. Homo sapiens CB1 and CB2 [P21554 (CNR1_HUMAN), P34972 (CNR2_HUMAN)], H. sapiens GPR-55 [Q9Y2T6 (GPR55_HUMAN)] and several C. elegans biogenic amine receptors (serotonin [SER-1,-3,-6; WBGene00004776, WBGene00004778, WBGene00021897] dopamine [DOP-1-3; WBGene00001052, WBGene00001053, WBGene00020506] and tyramine [TYRA-2 and -3; WBGene00017157, WBGene00006475]) also included. (E) 78 nematode NHR-49 homologs. Homo sapiens PPARG [P37231 (PPARG_HUMAN)], H. sapiens PPARD [Q03181 (PPARD_HUMAN)], H. sapiens PPARA [Q07869 (PPARA_HUMAN)], C. elegans NHR-88 [WBGene00003678], C. elegans NHR-64 [WBGene00003654] and C. elegans NHR-35 [WBGene00003628] also included. (F) 107 nematode OCR-2 homologs. Homo sapiens TRPV1-3 [Q8NER1 (TRPV1_HUMAN), Q9Y5S1 (TRPV2_HUMAN), Q8NET8 (TRPV3_HUMAN)], alongside C. elegans OSM-9 [WBGene00003889], C. elegans OCR-1 [WBGene00003838], C. elegans OCR-3 [WBGene00003840] and C. elegans UNC-44 [WBGene00006780] also shown. (G) 99 nematode OCTR-1 homologs. Homo sapiens CB1 and CB2 [P21554 (CNR1_HUMAN), P34972 (CNR2_HUMAN)], H. sapiens GPR-55 [Q9Y2T6 (GPR55_HUMAN)], H. sapiens ADA2A-C [P08913 (ADA2A_HUMAN). P18089 (ADA2B_HUMAN), P18825 (ADA2C_HUMAN)] and several C. elegans biogenic amine receptors (serotonin [SER-1,-3,-6; WBGene00004776, WBGene00004778, WBGene00021897] dopamine [DOP-1-3; WBGene00001052, WBGene00001053, WBGene00020506] and tyramine [TYRA-2 and -3; WBGene00017157, WBGene00006475]) also shown. (H) 100 nematode SER-4 homologs. Homo sapiens CB1 and CB2 [P21554 (CNR1_HUMAN), P34972 (CNR2_HUMAN)], H. sapiens GPR-55 [Q9Y2T6 (GPR55_HUMAN) and several C. elegans biogenic amine receptors (serotonin [SER-1,-3,-6; WBGene00004776, WBGene00004778, WBGene00021897] dopamine [DOP-1-3; WBGene00001052, WBGene00001053, WBGene00020506] and tyramine [TYRA-2 and -3; WBGene00017157, WBGene00006475]) also shown. Outer colours represent nematode clade and circles represent branch support values. Tree generated from an alignment trimmed to include functional domains. Branch supports indicate statistical support from approximate likelihood ratio test (aLRT).
Supplementary Figure 2 | Life stage and sex-specific expression profiles of EC-effectors. (A) Ancylostoma ceylanicum, (B) Teladorsagia circumcincta, (C) Dictyocaulus viviparus, (D) Haemonchus contortus, (E) Strongyloides ratti, (F) Strongyloides stercoralis, (G) Trichuris muris and (H) Onchocerca volvulus. Expression heatmaps generated from log2TPM values of all EC-effector transcripts. Average Clustering Method & Pearson’s Distance Measurement Method employed. Life stage and sex-specific data are arranged in columns, rows indicate individual EC-effector transcripts as denoted by effector abbreviation/gene ID. Coloured circles represent EC-receptors (orange), EC biosynthesis enzymes (green) and EC degradation enzymes (purple). [Life stages include; L1, L2, activated L3 (L3A), not activated L3 (L3NA), untreated L3 (L3UT), adult female (AF), adult male (AM) hypobiotic larvae (Lhyp), mixed L1 & L2 (L1+L2), pre-adult L5 female (L5AF), pre-adult L5 male (L5AM), pre-adult L5 mixed gender (L5 Mixed), Adult L5 (L5A), infective larvae (iL3), free-living female (FL Female), tissue migrating L3 (L3+), parasitic female (P Female), post-free living L1 (PFLL1), post-parasitic L1 (PPL1), post-parasitic L3 (PPL3)].
2-AG, 2-arachidonoylglycerol; ABHD-4, Abhydrolase Domain Containing 4, N-Acyl Phospholipase B; ABHD-5, Abhydrolase Domain Containing 5; ABHD-6, Abhydrolase Domain Containing 6; ABHD-12, Abhydrolase Domain Containing 12, Lysophospholipase; AEA, Anandamide/N-arachidonoylethanolamine; BUSCO, Benchmarking Universal Single-Copy Orthologs; CB1, Cannabinoid Receptor 1; CB2, Cannabinoid Receptor 2; CEGMA, Core Eukaryotic Genes Mapping Approach; DAGL, Diacylglycerol lipase; EC, Endocannabinoid; EC-GPCR, Endocannabinoid G-protein coupled receptor; ECEs, Endocannabinoid Enzymes; ECRs, Endocannabinoid Receptors; ECS, Endocannabinoid signalling; FAAH-1, Fatty Acid Amide Hydrolase 1; FAAH-2, Fatty Acid Amide Hydrolase 2; GDE-1,Glycerophosphodiester phosphodiesterase 1; Glycero-p-AEA, Glycerophosphoanandamide; GPR-55, G-protein coupled receptor 55; HMM,Hidden Markov Model; Lyso-NAPE, N-acyl-1-acyl-lyso-PE; Lyso-PLD, Lysophospholipase D; MAGL, Monoacylglycerol-lipase; MSA, Multiple Sequence Alignment; NAPE, N-arachidonyl phosphatidyl ethanol-phospholipase D; NAPE-1, N-acyl phosphatidylethanolamine-specific phospholipase-1; NAPE-2, N-acyl phosphatidylethanolamine-specific phospholipase-2; NAPE-PLD, N-Acyl Phosphatidyl Ethanolamine specific phospholipase D; NHR-49, Nuclear Hormone Receptor-49; NPR-9, Neuropeptide Receptor-9; NPR-19, Neuropeptide Receptor-19; NPR-32, Neuropeptide Receptor-32; OCR-2, Osin-9 and Capsacin receptor-related 2; OCTR-1, Octopamine Receptor-1; PLA-2, Phospholipase A2; SER-4, Serotonin/Octopamine receptor family-4; TRPV, Transient receptor potential vanilloid channel.
1. Robertson LJ, Torgerson PR, Van Der Giessen J. Foodborne Parasitic Diseases in Europe: Social Cost-Benefit Analyses of Interventions. Trends Parasitol (2018) 34:919–23. doi: 10.1016/j.pt.2018.05.007
2. Castillo PE, Younts TJ, Chávez AE, Hashimotodani Y. Endocannabinoid signaling and synaptic function. Neuron (2012) 76:70–81. doi: 10.1016/j.neuron.2012.09.020
3. Zou S, Kumar U. Cannabinoid Receptors and the Endocannabinoid System: Signaling and Function in the Central Nervous System. Int J Mol Sci (2018) 19:833. doi: 10.3390/ijms19030833
4. Kaur R, Ambwani SR, Singh S. Endocannabinoid System: A Multi-Facet Therapeutic Target. Curr Clin Pharmacol (2016) 11:110–7. doi: 10.2174/1574884711666160418105339
5. De Azua IR, Lutz B. Multiple Endocannabinoid-Mediated Mechanisms in the Regulation of Energy Homeostasis in Brain and Peripheral Tissues. Cell Mol Life Sci (2019) 76:1341–63. doi: 10.1007/s00018-018-2994-6
6. Mouslech Z, Valla V. Endocannabinoid System: An Overview of its Potential in Current Medical Practice. Neuroendocrinol Lett (2009) 30:153–79.
7. Fucich EA, Mayeux JP, Mcginn MA, Gilpin NW, Edwards S, Molina PE. A Novel Role for the Endocannabinoid System in Ameliorating Motivation for Alcohol Drinking and Negative Behavioral Affect After Traumatic Brain Injury in Rats. J Neurotrauma (2019) 36:1847–55. doi: 10.1089/neu.2018.5854
8. Salzet M, Stefano GB. Evidence for an Invertebrate Neuroendocrine System: Neuropeptide Proccessing in Leech-Host Communication. Trends Comp Biochem Physiol (1998) 5:85–98.
9. Salzet M, Stefano G. The Endocannabinoid System in Invertebrates. Prostaglandins Leukotrienes Essential Fatty Acids (plefa) (2002) 66:353–61. doi: 10.1054/plef.2001.0347
10. Elphick MR. The Evolution and Comparative Neurobiology of Endocannabinoid Signalling. Philos Trans r Soc Lond b Biol Sci (2012) 367:3201–15. doi: 10.1098/rstb.2011.0394
11. Clarke TL, Johnson RL, Simone JJ, Carlone RL. The Endocannabinoid System and Invertebrate Neurodevelopment and Regeneration. Int J Mol Sci (2021) 22:2103. doi: 10.3390/ijms22042103
12. Mosca F, Zarivi O, Battista N, Maccarrone M, Tiscar PG. The Endocannabinoid System in the Mediterranean Mussel Mytilus galloprovincialis: Possible Mediators of the Immune Activity? Int J Mol Sci (2021) 22:4954. doi: 10.3390/ijms22094954
13. Di Marzo V, De Petrocellis L, Bisogno T, Melck D. Metabolism of Anandamide And 2-Arachidonoylglycerol: An Historical Overview and Some Recent Developments. Lipids (1999) 34Suppl:S319–25. doi: 10.1007/BF02562332
14. Aarnio V, Lehtonen M, Storvik M, Callaway JC, Lakso M, Wong G. Caenorhabditis Elegans Mutants Predict Regulation of Fatty Acids and Endocannabinoids by the Cyp-35a Gene Family. Front Pharmacol (2011) 2:12. doi: 10.3389/fphar.2011.00012
15. Galles C, Prez GM, Penkov S, Boland S, Porta EO, Altabe SG, et al. Endocannabinoids in Caenorhabditis Elegans are Essential for the Mobilization of Cholesterol From Internal Reserves. Sci Rep (2018) 8:6398. doi: 10.1038/s41598-018-24925-8
16. Pastuhov SI, Fujiki K, Nix P, Kanao S, Bastiani M, Matsumoto K, et al. Endocannabinoid-Goα Signalling Inhibits Axon Regeneration in Caenorhabditis Elegans by Antagonizing Gqα-Pkc-Jnk Signalling. Nat Commun (2012) 3:1–9. doi: 10.1038/ncomms2136
17. Pastuhov SI, Matsumoto K, Hisamoto N. Endocannabinoid Signaling Regulates Regenerative Axon Navigation in Caenorhabditis Elegans via the Gpcrs Npr-19 and Npr-32. Genes to Cells (2016) 21:696–705. doi: 10.1111/gtc.12377
18. Batugedara HM, Argueta D, Jang JC, Lu D, Macchietto M, Kaur J, et al. Host-And Helminth-Derived Endocannabinoids That Have Effects on Host Immunity are Generated During Infection. Infect Immun (2018) 86:e00441–18. doi: 10.1128/IAI.00441-18
19. Oakes M, Law WJ, Komuniecki R. Cannabinoids Stimulate the Trp Channel-Dependent Release of Both Serotonin and Dopamine to Modulate Behavior in C. Elegans. J Neurosci (2019) 39:4142–52. doi: 10.1523/JNEUROSCI.2371-18.2019
20. Estrada-Valencia R, de Lima ME, Colonnello A, Rangel-Lopez E, Saraiva NR, de Avila DS, et al. The Endocannabinoid System in Caenorhabditis Elegans. Rev Physiol Biochem Pharmaco. (2021). doi: 10.1007/112_2021_64
21. Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, et al. Isolation and Structure of a Brain Constituent That Binds to the Cannabinoid Receptor. Science (1992) 258:1946–9. doi: 10.1126/science.1470919
22. Mackie K. Cannabinoid Receptors: Where They are and What They do. J Neuroendocrinol (2008) 20:10–4. doi: 10.1111/j.1365-2826.2008.01671.x
23. Sun L, Tai L, Qiu Q, Mitchell R, Fleetwood-Walker S, Joosten EA, et al. Endocannabinoid Activation of Cb1 Receptors Contributes to Long-Lasting Reversal of Neuropathic Pain by Repetitive Spinal Cord Stimulation. Eur J Pain (2017) 21:804–14. doi: 10.1002/ejp.983
24. Morales P, Reggio PH. An Update on non-Cb1, non-Cb2 Cannabinoid Related G-Protein-Coupled Receptors. Cannabis Cannabinoid Res (2017) 2:265–73. doi: 10.1089/can.2017.0036
25. Muller C, Morales P, Reggio PH. Cannabinoid Ligands Targeting Trp Channels. Front Mol Neurosci (2018) 11:487. doi: 10.3389/fnmol.2018.00487
26. Oakes MD. Uncovering Cannabinoid Signaling in C. Elegans: A New Platform to Study the Effects of Medicinal Cannabis. (Ohio LINK Electronic Theses and Dissertation centre:University of toledo) (2018).
27. Oakes MD, Law WJ, Clark T, Bamber BA, Komuniecki R. Cannabinoids Activate Monoaminergic Signaling to Modulate Key C. Elegans Behaviors. J Neurosci (2017) 37:2859–69. doi: 10.1523/JNEUROSCI.3151-16.2017
28. McPartland JM, Glass M. Functional Mapping of Cannabinoid Receptor Homologs in Mammals, Other Vertebrates, and Invertebrates. Gene (2003) 312:297–303. doi: 10.1016/S0378-1119(03)00638-3
29. Ligumsky M, Kaminiski NE, Schatz AR, Compton DR, Pertwee RG, Griffin G, et al. Identification of an Endogenous 2-Monoglyceride, Present in Canine Gut, That Binds to Cannabinoid Receptors. Biochem Pharmacol (1995) 50:83–90. doi: 10.1016/0006-2952(95)00109-D
30. Mechoulam R, Fride E. The Unpaved Road to the Endogenous Brain Cannabinoid Ligands, the Anandamides. London: Academic Press (1995).
31. Murataeva N, Straiker A, Mackie K. Parsing the Players: 2-Arachidonoylglycerol Synthesis and Degradation in the Cns. Br J Pharmacol (2014) 171:1379–91. doi: 10.1111/bph.12411
32. Clapper JR, Henry CL, Niphakis MJ, Knize AM, Coppola AR, Simon GM, et al. Monoacylglycerol Lipase Inhibition in Human and Rodent Systems Supports Clinical Evaluation of Endocannabinoid Modulators. J Pharmacol Exp Ther (2018) 367:494–508. doi: 10.1124/jpet.118.252296
33. Blankman JL, Simon GM, Cravatt BF. A Comprehensive Profile of Brain Enzymes That Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem Biol (2007) 14:1347–56. doi: 10.1016/j.chembiol.2007.11.006
34. Di Marzo V. Biosynthesis and Inactivation of Endocannabinoids: Relevance to Their Proposed Role as Neuromodulators. Life Sci (1999) 65:645–55. doi: 10.1016/S0024-3205(99)00287-8
35. Biringer RG. The Rise and Fall of Anandamide: Processes That Control Synthesis, Degradation, and Storage. Mol Cell Biochem (2021) 476:1–23. doi: 10.1007/s11010-021-04121-5
36. Liu J, Wang L, Harvey-White J, Osei-Hyiaman D, Razdan R, Gong Q, et al. A Biosynthetic Pathway for Anandamide. Proc Natl Acad Sci (2006) 103:13345–50. doi: 10.1073/pnas.0601832103
37. Basavarajappa BS. Critical Enzymes Involved in Endocannabinoid Metabolism. Protein Pept Lett (2007) 14:237–46. doi: 10.2174/092986607780090829
38. Sun Y-X, Tsuboi K, Okamoto Y, Tonai T, Murakami M, Kudo I, et al. Biosynthesis of Anandamide and N-Palmitoylethanolamine by Sequential Actions of Phospholipase A2 and Lysophospholipase D. Biochem J (2004) 380:749–56. doi: 10.1042/bj20040031
39. Lehtonen M, Reisner K, Auriola S, Wong G, Callaway JC. Mass-Spectrometric Identification of Anandamide and 2-Arachidonoylglycerol in Nematodes. Chem Biodiversity (2008) 5:2431–41. doi: 10.1002/cbdv.200890208
40. Piomelli D, Tarzia G, Duranti A, Tontini A, Mor M, Compton TR, et al. Pharmacological Profile of the Selective Faah Inhibitor Kds-4103 (Urb597). CNS Drug Rev (2006) 12:21–38. doi: 10.1111/j.1527-3458.2006.00021.x
41. Long JZ, Nomura DK, Cravatt BF. Characterization of Monoacylglycerol Lipase Inhibition Reveals Differences in Central and Peripheral Endocannabinoid Metabolism. Chem Biol (2009) 16:744–53. doi: 10.1016/j.chembiol.2009.05.009
42. Lucanic M, Held JM, Vantipalli MC, Klang IM, Graham JB, Gibson BW, et al. N-Acylethanolamine Signalling Mediates the Effect of Diet on Lifespan in Caenorhabditis Elegans. Nature (2011) 473:226. doi: 10.1038/nature10007
43. Harrison N, Lone MA, Kaul TK, Rodrigues PR, Ogungbe IV, Gill MS. Characterization of N-Acyl Phosphatidylethanolamine-Specific Phospholipase-D Isoforms in the Nematode Caenorhabditis Elegans. PloS One (2014) 9. doi: 10.1371/journal.pone.0113007
44. Lin YH, Chen YC, Kao TY, Lin YC, Hsu TE, Wu YC, et al. Diacylglycerol Lipase Regulates Lifespan and Oxidative Stress Response by Inversely Modulating TOR Signaling in Drosophila and C. Elegans. Aging Cell. (2014) 13Suppl:755–64. doi: 10.1111/acel.12232
45. Van Gilst MR, Hadjivassiliou H, Jolly A, Yamamoto KR. Nuclear Hormone Receptor Nhr-49 Controls Fat Consumption and Fatty Acid Composition in C. Elegans. PloS Biol (2005) 3:301–12. doi: 10.1371/journal.pbio.0030053
46. Jose AM, Bany IA, Chase DL, koelle MR. A Specific Subset of Transient Receptor Potential Vanilloid-Type Channel Subunits in Caenorhabditis Elegans Endocrine Cells Function as Mixed Heteromers to Promote Neurotransmitter Release. Genetics (2007) 175:93–105. doi: 10.1534/genetics.106.065516
47. McPartland JM, Matias I, Di Marzo V, Glass M. Evolutionary Origins of the Endocannabinoid System. Gene (2006) 370:64–74. doi: 10.1016/j.gene.2005.11.004
48. Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M. Wormbase Parasite– a Comprehensive Resource for Helminth Genomics. Mol Biochem Parasitol (2017) 215:2–10. doi: 10.1016/j.molbiopara.2016.11.005
49. McVeigh P, Mccammick E, Mccusker P, Wells D, Hodgkinson J, Paterson S, et al. Profiling G Protein-Coupled Receptors of Fasciola Hepatica Identifies Orphan Rhodopsins Unique to Phylum Platyhelminthes. Int J Parasitol: Drugs Drug Resistance (2018) 8:87–103. doi: 10.1016/j.ijpddr.2018.01.001
50. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol Syst Biol (2011) 7. doi: 10.1038/msb.2011.75
51. Jones P, Binns D, Chang H-Y, Fraser M, Li W, Mcanulla C, et al. Interproscan 5: Genome-Scale Protein Function Classification. Bioinformatics (2014) 30:1236–40. doi: 10.1093/bioinformatics/btu031
52. Hofmann K. Tmbase-A Database of Membrane Spanning Proteins Segments. Biol Chem Hoppe-seyler (1993) 374:166.
53. Schneider TD, Stephens RM. Sequence Logos: A New Way to Display Consensus Sequences. Nucleic Acids Res (1990) 18:6097–100. doi: 10.1093/nar/18.20.6097
54. Crooks GE, Hon G, Chandonia J-M, Brenner SE. Weblogo: A Sequence Logo Generator. Genome Res (2004) 14:1188–90. doi: 10.1101/gr.849004
55. Stecher G, Tamura K, Kumar S. Molecular Evolutionary Genetics Analysis (Mega) for Macos. Mol Biol Evolution (2020) 37:1237–9. doi: 10.1093/molbev/msz312
56. Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. Cdd/sparcle: The Conserved Domain Database in 2020. Nucleic Acids Res (2020) 48:d265–8. doi: 10.1093/nar/gkz991
57. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny. Fr: Robust Phylogenetic Analysis for the non-Specialist. Nucleic Acids Res (2008) 36:w465–9. doi: 10.1093/nar/gkn180
58. Letunic I, Bork P. Interactive Tree of Life (Itol) V5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res (2021) 49:w293–6. doi: 10.1093/nar/gkab301
59. National Centre for Biotechnology Information. Sequence Read Archive (Sra). Bethesda (md: National Library of Medicine (US (2009).
60. Petryszak R, Fonseca NA, Füllgrabe A, Huerta L, Keays M, Tang YA, et al. The Rnaseq-Er Api—a Gateway to Systematically Updated Analysis of Public Rna-Seq Data. Bioinformatics (2017) 33:2218–20. doi: 10.1093/bioinformatics/btx143
61. Lu MR, Lai C-K, Liao B-Y, Tsai IJ. Comparative Transcriptomics Across Nematode Life Cycles Reveal Gene Expression Conservation and Correlated Evolution in Adjacent Developmental Stages. Genome Biol Evol (2020) 12:1019–30. doi: 10.1093/gbe/evaa110
62. Sra Toolkit Development Team. Sra Toolkit (2020). Available at: http://ncbi.github.io/sra-tools/ (Accessed 2020).
63. Bolger AM, Lohse M, Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
64. Hunt VL, Tsai IJ, Coghlan A, Reid AJ, Holroyd N, Foth BJ, et al. The Genomic Basis of Parasitism in the Strongyloides Clade of Nematodes. Nat Genet (2016) 48:299–307. doi: 10.1038/ng.3495
65. Kim D, Langmead B, Salzberg SL. Hisat: A Fast Spliced Aligner With Low Memory Requirements. Nat Methods (2015) 12:357–60. doi: 10.1038/nmeth.3317
66. Liao Y, Smyth GK, Shi W. Featurecounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics (2014) 30:923–30. doi: 10.1093/bioinformatics/btt656
67. Wagner GP, Kin K, Lynch VJ. A Model Based Criterion for Gene Expression Calls Using Rna-Seq Data. Theory Biosci (2013) 132:159–64. doi: 10.1007/s12064-013-0178-3
68. Soneson C, Robinson M D. Bias, Robustness and Scalability in Single-Cell Differential Expression Analysis. Nat Methods (2018) 15:255. doi: 10.1038/nmeth.4612
69. Love MI, Huber W, Anders S. Moderated Estimation of Fold Change and Dispersion for Rna-Seq Data With Deseq2. Genome Biol (2014) 15:550. doi: 10.1186/s13059-014-0550-8
70. Rs Team. Rstudio: Integrated Development for R. In: Rstudio, vol. 639. . boston, ma: Inc. (2015). p. 640.
71. Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, et al. Heatmapper: Web-Enabled Heat Mapping for All. Nucleic Acids Res (2016) 44:w147–53. doi: 10.1093/nar/gkw419
72. Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, et al. A Molecular Evolutionary Framework for the Phylum Nematoda. Nature (1998) 392:71–5. doi: 10.1038/32160
73. Holterman M, Van Der Wurff A, Van Den Elsen S, Van Megen H, Bongers T, Holovachov O, et al. Phylum-Wide Analysis of Ssu Rdna Reveals Deep Phylogenetic Relationships Among Nematodes and Accelerated Evolution Toward Crown Clades. Mol Biol Evol (2006) 23:1792–800. doi: 10.1093/molbev/msl044
74. Paulsen RT, Burrell BD. Comparative Studies of Endocannabinoid Modulation of Pain. Philos Trans R Soc b (2019) 374:20190279. doi: 10.1098/rstb.2019.0279
75. Lu H-C, Mackie K. Review of the Endocannabinoid System. Biol Psychiatry: Cogn Neurosci Neuroimaging (2021) 6:607–15. doi: 10.1016/j.bpsc.2020.07.016
76. Parra G, Bradnam K, Ning Z, Keane T, Korf I. Assessing the Gene Space in Draft Genomes. Nucleic Acids Res (2009) 37:289–97. doi: 10.1093/nar/gkn916
77. Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. Busco: Assessing Genome Assembly and Annotation Completeness With Single-Copy Orthologs. Bioinformatics (2015) 31:3210–2. doi: 10.1093/bioinformatics/btv351
78. Davis P, Zarowiecki M, Arnaboldi V, Becerra A, Cain S, Chan J, et al. Wormbase in 2022—Data, Processes, and Tools for Analyzing Caenorhabditis Elegans. Genetics (2022) 220. doi: 10.1093/genetics/iyac003
79. De Bono M, Tobin DM, Davis MW, Avery L, Bargmann CI. Social Feeding in Caenorhabditis Elegans is Induced by Neurons That Detect Aversive Stimuli. Nature (2002) 419:899–903. doi: 10.1038/nature01169
80. Lee BH, Ashrafi K. A Trpv Channel Modulates C. Elegans Neurosecretion, Larval Starvation Survival, and Adult Lifespan. PloS Genet (2008) 4:e1000213. doi: 10.1371/journal.pgen.1000213
81. Bendena WG, Boudreau JR, Papanicolaou T, Maltby M, Tobe SS, Chin-Sang ID. A Caenorhabditis Elegans Allatostatin/Galanin-Like Receptor Npr-9 Inhibits Local Search Behavior in Response to Feeding Cues. Proc Natl Acad Sci (2008) 105:1339–42. doi: 10.1073/pnas.0709492105
82. Campbell JC, Polan-Couillard LF, Chin-Sang ID, Bendena WG. Npr-9, a Galanin-Like G-Protein Coupled Receptor, and Glr-1 Regulate Interneuronal Circuitry Underlying Multisensory Integration of Environmental Cues in Caenorhabditis Elegans. PloS Genet (2016) 12. doi: 10.1371/journal.pgen.1006050
83. Yu Y, Zhi L, Wu Q, Jing L, Wang D. Npr-9 Regulates the Innate Immune Response in Caenorhabditis Elegans by Antagonizing the Activity of Aib Interneurons. Cell Mol Immunol (2018) 15:27–37. doi: 10.1038/cmi.2016.8
84. Balenga N, Martínez-Pinilla E, Kargl J, Schröder R, Peinhaupt M, Platzer W, et al. Heteromerization of Gpr 55 and Cannabinoid Cb 2 Receptors Modulates Signalling. Br J Pharmacol (2014) 171:5387–406. doi: 10.1111/bph.12850
85. Kargl J, Balenga N, Parzmair GP, Brown AJ, Heinemann A, Waldhoer M. The Cannabinoid Receptor Cb1 Modulates the Signaling Properties of the Lysophosphatidylinositol Receptor Gpr55. J Biol Chem (2012) 287:44234–48. doi: 10.1074/jbc.M112.364109
86. Doyle SR, Cotton JA. Genome-Wide Approaches to Investigate Anthelmintic Resistance. Trends Parasitol (2019) 35:1237–301. doi: 10.1016/j.pt.2019.01.004
87. Wang J, Ueda N. Biology of Endocannabinoid Synthesis System. Prostaglandins Other Lipid Mediators (2009) 89:112–9. doi: 10.1016/j.prostaglandins.2008.12.002
88. Muccioli GG. Endocannabinoid Biosynthesis and Inactivation, From Simple to Complex. Drug Discovery Today (2010) 15:474–83. doi: 10.1016/j.drudis.2010.03.007
89. Simon GM, Cravatt BF. Endocannabinoid Biosynthesis Proceeding Through Glycerophospho-N-Acyl Ethanolamine and a Role for α/β-Hydrolase 4 in This Pathway. J Biol Chem (2006) 281:26465–72. doi: 10.1074/jbc.M604660200
90. Astarita G, Piomelli D. Lipidomic Analysis of Endocannabinoid Metabolism in Biological Samples. J Chromatogr b (2009) 877:2755–67. doi: 10.1016/j.jchromb.2009.01.008
91. Sirrs S, Van Karnebeek CD, Peng X, Shyr C, Tarailo-Graovac M, Mandal R, et al. Defects in Fatty Acid Amide Hydrolase 2 in a Male With Neurologic and Psychiatric Symptoms. Orphanet J Rare Dis (2015) 10:1–10. doi: 10.1186/s13023-015-0248-3
92. Haq I, Kilaru A. An Endocannabinoid Catabolic Enzyme Faah and its Paralogs in an Early Land Plant Reveal Evolutionary and Functional Relationship With Eukaryotic Orthologs. Sci Rep (2020) 10:1–14. doi: 10.1038/s41598-020-59948-7
93. Chicca A, Arena C, Bertini S, Gado F, Ciaglia E, Abate M, et al. Polypharmacological Profile of 1, 2-Dihydro-2-Oxo-Pyridine-3-Carboxamides in the Endocannabinoid System. Eur J Medicinal Chem (2018) 154:155–71. doi: 10.1016/j.ejmech.2018.05.019
94. Janowitz T, Kneifel H, Piotrowski M. Identification and Characterization of Plant Agmatine Iminohydrolase, the Last Missing Link in Polyamine Biosynthesis of Plants. FEBS Lett (2003) 544:258–61. doi: 10.1016/S0014-5793(03)00515-5
95. Rossignoli G, Grottesi A, Bisello G, Montioli R, Borri Voltattorni C, Paiardini A, et al. Cysteine 180 is a Redox Sensor Modulating the Activity of Human Pyridoxal 5′-Phosphate Histidine Decarboxylase. Biochemistry (2018) 57:6336–48. doi: 10.1021/acs.biochem.8b00625
96. Facchiano A, Marabotti A. Analysis of Galactosemia-Linked Mutations of Galt Enzyme Using a Computational Biology Approach. Protein Eng Design Selection (2010) 23:103–13. doi: 10.1093/protein/gzp076
97. Ahn K, Johnson DS, Cravatt BF. Fatty Acid Amide Hydrolase as a Potential Therapeutic Target for the Treatment of Pain and Cns Disorders. Expert Opin Drug Discovery (2009) 4:763–84. doi: 10.1517/17460440903018857
98. Chebrou H, Bigey F, Arnaud A, Galzy P. Study of the Amidase Signature Group. Biochim Biophys Acta (bba)-protein Structure Mol Enzymology (1996) 1298:285–93. doi: 10.1016/S0167-4838(96)00145-8
99. Kano M, Ohno-Shosaku T, Hashimotodani Y, Uchigashima M, Watanabe M. Endocannabinoid-Mediated Control of Synaptic Transmission. Physiol Rev (2009) 89:309–80. doi: 10.1152/physrev.00019.2008
100. Savinainen J, Saario S, Laitinen J. The Serine Hydrolases Magl, Abhd6 and Abhd12 as Guardians of 2-Arachidonoylglycerol Signalling Through Cannabinoid Receptors. Acta Physiologica (2012) 204:267–76. doi: 10.1111/j.1748-1716.2011.02280.x
101. McAllister SD, Rizvi G, Anavi-Goffer S, Hurst DP, Barnett-Norris J, Lynch DL, et al. An Aromatic Microdomain at the Cannabinoid Cb1 Receptor Constitutes an Agonist/Inverse Agonist Binding Region. J Medicinal Chem (2003) 46:5139–52. doi: 10.1021/jm0302647
102. Reggio PH. Endocannabinoid Binding to the Cannabinoid Receptors: What is Known and What Remains Unknown. Curr Medicinal Chem (2010) 17:1468–86. doi: 10.2174/092986710790980005
103. Pei Y, Mercier RW, Anday JK, Thakur GA, Zvonok AM, Hurst D, et al. Ligand-Binding Architecture of Human Cb2 Cannabinoid Receptor: Evidence for Receptor Subtype-Specific Binding Motif and Modeling Gpcr Activation. Chem Biol (2008) 15:1207–19. doi: 10.1016/j.chembiol.2008.10.011
104. Al-Zoubi R, Morales P, Reggio PH. Structural Insights Into Cb1 Receptor Biased Signaling. Int J Mol Sci (2019) 20:1837. doi: 10.3390/ijms20081837
105. Hotez P, Hawdon J, Schad G. Hookworm Larval Infectivity, Arrest and Amphiparatenesis: The Caenorhabditis Elegans Daf-C Paradigm. Parasitol Today (1993) 9:23–6. doi: 10.1016/0169-4758(93)90159-D
106. Viney ME, Thompson F, Crook M. Tgf-β and the Evolution of Nematode Parasitism. Int J Parasitol (2005) 35:1473–5. doi: 10.1016/j.ijpara.2005.07.006
107. Crook M. The Dauer Hypothesis and the Evolution of Parasitism: 20 Years on and Still Going Strong. Int J Parasitol (2014) 44:1–8. doi: 10.1016/j.ijpara.2013.08.004
108. Vlaar L.E., Bertran A., Rahimi M., Dong D., Kammenga J.E, Helder J., et al. On the Role of Dauer in the Adaptation of Nematodes to a Parasitic Lifestyle. Parasit Vectors (2021) 14, 554. doi: 10.1186/s13071-021-04953-6
109. Gang SS, Castelletto ML, Yang E, Ruiz F, Brown TM, Bryant AS, et al. Chemosensory Mechanisms of Host Seeking and Infectivity in Skin-Penetrating Nematodes. Proc Natl Acad Sci (2020) 117:17913–23. doi: 10.1073/pnas.1909710117
110. Karasu T, Marczylo T, Maccarrone M, Konje J. The Role of Sex Steroid Hormones, Cytokines and the Endocannabinoid System in Female Fertility. Hum Reprod Update (2011) 17:347–61. doi: 10.1093/humupd/dmq058
111. Huang GZ, Woolley CS. Estradiol Acutely Suppresses Inhibition in the Hippocampus Through a Sex-Specific Endocannabinoid and Mglur-Dependent Mechanism. Neuron (2012) 74:801–8. doi: 10.1016/j.neuron.2012.03.035
112. Viveros M, Llorente R, Suarez J, Llorente-Berzal A, Lopez-Gallardo M, Rodriguez De Fonseca F. The Endocannabinoid System in Critical Neurodevelopmental Periods: Sex Differences and Neuropsychiatric Implications. J Psychopharmacol (2012) 26:164–76. doi: 10.1177/0269881111408956
113. Kolodny RC, Masters WH, Kolodner RM, Toro G. Depression of Plasma Testosterone Levels After Chronic Intensive Marihuana Use. New Engl J Med (1974) 290:872–4. doi: 10.1056/NEJM197404182901602
114. Nielsen JE, Rolland AD, Rajpert-De Meyts E, Janfelt C, Jørgensen A, Winge SB, et al. Characterisation and Localisation of the Endocannabinoid System Components in the Adult Human Testis. Sci Rep (2019) 9:1–14. doi: 10.1038/s41598-019-49177-y
115. Schuel H. Tuning the Oviduct to the Anandamide Tone. J Clin Invest (2006) 116:2087–90. doi: 10.1172/JCI29424
116. El-Talatini MR, Taylor AH, Elson JC, Brown L, Davidson AC, Konje JC. Localisation and Function of the Endocannabinoid System in the Human Ovary. PloS One (2009) 4. doi: 10.1371/journal.pone.0004579
117. Maccarrone M. Endocannabinoids: Friends and Foes of Reproduction. Prog Lipid Res (2009) 48:344–54. doi: 10.1016/j.plipres.2009.07.001
118. Schuel H, Berkery D, Schuel R, Chang MC, Zimmerman AM, Zimmerman S. Reduction of the Fertilizing Capacity of Sea Urchin Sperm by Cannabinoids Derived From Marihuana. I. Inhibition of the Acrosome Reaction Induced by Egg Jelly. Mol Reprod Dev (1991) 29:51–9. doi: 10.1002/mrd.1080290109
Keywords: endocannabinoid, endocannabinoid signalling, nematode, parasite, genome, transcriptome, drug target, endocannabinoid receptor
Citation: Crooks BA, Mckenzie D, Cadd LC, McCoy CJ, McVeigh P, Marks NJ, Maule AG, Mousley A and Atkinson LE (2022) Pan-phylum In Silico Analyses of Nematode Endocannabinoid Signalling Systems Highlight Novel Opportunities for Parasite Drug Target Discovery. Front. Endocrinol. 13:892758. doi: 10.3389/fendo.2022.892758
Received: 09 March 2022; Accepted: 20 April 2022;
Published: 01 July 2022.
Edited by:
Dan Larhammar, Uppsala University, SwedenReviewed by:
Brian Burrell, University of South Dakota, United StatesCopyright © 2022 Crooks, Mckenzie, Cadd, McCoy, McVeigh, Marks, Maule, Mousley and Atkinson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Louise E. Atkinson, bC5hdGtpbnNvbkBxdWIuYWMudWs=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.