
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol., 15 June 2022
Sec. Experimental Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.883608
This article is part of the Research TopicNeuroendocrine Regulation of Feeding and Reproduction in FishView all 14 articles
 Chunyan Zhao1,2,3
Chunyan Zhao1,2,3 Bin Wang4
Bin Wang4 Yifan Liu2,3,5
Yifan Liu2,3,5 Chengcheng Feng2,3
Chengcheng Feng2,3 Shihong Xu2,3
Shihong Xu2,3 Wenqi Wang1
Wenqi Wang1 Qinghua Liu2,3
Qinghua Liu2,3 Jun Li2,3,6*
Jun Li2,3,6*Seasonal reproduction is generally controlled by the hypothalamus-pituitary-gonadal (HPG) axis in fish. Previous studies have demonstrated that the kisspeptin (Kiss)/kisspeptin receptor (Kissr) system, a positive regulator of the HPG axis, mediates the responses to environmental cues. Turbot (Scophthalmus maximus), a representative species of Pleuronectiformes, is one of the most commercially important fish species cultured in Europe and North China. However, the mechanisms by which the Kiss/Kissr system regulates the reproductive axis of turbot according to seasonal changes, especially photoperiod, have not been clearly characterized. In the current study, the cDNA sequences of kiss2/kissr2, along with kiss1/kissr3 which was thought to be lost in flatfish species, were cloned and functionally characterized. The kiss1, kiss2, and kissr3 transcripts were highly detected in the brain and gonad, while kissr2 mRNA was only abundantly expressed in the brain. Moreover, kiss/kissr mRNAs were further examined in various brain areas of both sexes. The kiss1, kissr2, kissr3 mRNAs were highly expressed in the mesencephalon, while a substantial degree of kiss2 transcripts were observed in the hypothalamus. During annual reproductive cycle, both kiss and kissr transcript levels declined significantly from the immature to mature stages and increased at the degeneration stage in the brains of both sexes, especially in the mesencephalon and hypothalamus. The ovarian kiss1, kiss2, and kissr2 mRNA levels were highest at the vitellogenic stage (mature stage), while expression of kissr3 was highest at the immature stage. The testicular kiss and kissr transcripts were highest in the immature and degeneration stages, and lowest at the mature stage. In addition, intraperitoneal injection of Kiss1-10 and Kiss2-10 significantly stimulated mRNA levels of pituitary lhβ, fhsβ, and gthα. In summary, two Kiss/Kissr systems were firstly proven in a flatfish species of turbot, and it has a positive involvement in controlling the reproduction of the Kiss/Kissr system in turbot. The results will provide preliminary information regarding how the Kiss/Kissr system controls seasonal reproduction in turbot broodstock.
In vertebrates, seasonal reproduction, as the most important biological rhythm, is classically controlled by the hypothalamus-pituitary-gonadal (HPG) axis. Hormones secreted from the HPG axis are regulated by seasonally changing patterns, such as photoperiod, which is an environmental signal that can trigger organism to sexually mature. The discovery of the kisspeptin/GPR54 signaling system has greatly improved our understanding of reproductive endocrinology (1–3). There is an abundance of evidence from mammals demonstrating that the kisspeptin/GPR54 system has a critical role in reproduction, especially in mediating the responses to environmental cues (4, 5). Previous studies in mammals have demonstrated that kisspeptin/GPR54 signaling, as a regulator of the HPG axis, regulates the secretion of gonadotropin-releasing hormone (GnRH) (2, 6). The influence of kisspeptin/GPR54 signaling on reproductive endocrine function has attracted increasing attention from reproductive physiologists.
The kisspeptin/GPR54 system is composed of the ligand, kisspeptin (Kiss), and its receptor, G protein-coupled receptor 54 (GPR54), now renamed kisspeptin receptor (Kissr) (7). The first kisspeptin transcript was isolated from human malignant melanoma cells and its cDNA was designated as Kiss1 (1). Subsequently, the Kiss1 cognate receptor gene (kissr1) was characterized in rats (8). Further studies on hypogonadism showed that mutation of kissr1 with the idiopathic hypothalamic hypogonadism (IHH) syndrome could cause sterile in mice (6, 9). In mammals, the role of the Kiss/Kissr system in the reproductive system was investigated, and has led to significant breakthroughs. It has been demonstrated that the Kiss/Kissr system controls the HPG axis by directly influencing GnRH neurons and regulating the secretion of the GnRH, resulting in the secretion of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) (10, 11).
Following the investigations in mammals, the Kiss/Kissr system has also been characterized in fish species. Parhar and coworkers first reported that another kisspeptin receptor kissr2 was co-expressed in GnRH neurons in Nile tilapia (Oreochromis niloticus), providing valuable insights into the kisspeptin system during normal sexual and reproductive development (12). Subsequently, up to two kiss genes (kiss1, kiss2) and three kissr genes (kissr1, kissr2, kissr3) have been identified in various teleosts, including zebrafish (Danio rerio) (13–15), medaka (Oryzias latipes) (16, 17), goldfish (Carassius auratus) (18, 19), European sea bass (Dicentrarchus labrax) (20, 21), orange-spotted grouper (Epinephelus coioides) (22), chub mackerel (Scomber japonicas) (23, 24), yellowtail kingfish (Seriola lalandi) (25, 26), European eel (Anguilla anguilla) (27, 28), pejerrey (Odontesthes bonariensis) (29, 30), lined seahorse (Hippocampus erectus) (31), yellowtail clownfish (Amphiprion clarkia) (32) as well as four flatfish species, Senegalese sole (Solea senegalensis) (33), Atlantic halibut (Hippoglossus hippoglossus) (34), Japanese flounder (Paralichthys olivaceus) (35), and half-smooth tongue sole (Cynoglossus semilaevis) (36, 37).
Kisspeptin has been proven as a key activator of the reproductive axis during the seasonal gonadal cycle in some fish species. For example, changes in expression of kiss/kissr mRNAs in golden mahseer (Tor putitora) and rohu (Labeo rohita) indicated that they have a role in gonadal development and annual reproductive cycle (38, 39). Increased transcription levels of the Kiss/Kissr system showed that it is related to the differentiation of the HPG axis during male development in pejerrey (30). Furthermore, kisspeptin neurons in the mediobasal hypothalamus indicated it is a site that is sensitive to feedback from the action of sex steroids in European sea bass (40). The administration of exogenous kisspeptin peptides has been shown to promote plasma levels of FSH and LH in several fish species (18, 31, 41, 42). However, Kiss knockout in zebrafish and medaka, the puberty onset and sexual differentiation are not affected (43–45). Kiss2 administration in recrudescent fish do not change the blood LH levels (43). It might be species-specific in the control of reproduction in teleosts. Of note, kisspeptin expression revealed specific patterns that coincided with seasonal environmental changes, more specifically photoperiod, and induced sexual development. Intracerebroventricular infusion of Kiss1 could override the inhibitory photoperiod and reactivate sexual activity in hamsters which are sexually active in long photoperiod and quiescent in short photoperiod (43, 46). Kiss1 in mice showed circadian patterns that peaked coincident with LH (46). Similarly, Kiss controlled of cyclic reproductive activity in teleost. Oocyte maturation, responding to day length, relied on the enhanced transcription of kiss and gnrh3 in the brain of zebrafish (47). The rhythms of kiss and gnrh/fshβ/lhβ mRNA expression further suggested that the natural photoperiod is involved in activating the reproductive axis during maturation in Atlantic cod (Gadus morhua), European sea bass and grass puffer (Takifugu niphobles) (48–50).
Turbot (Scophthalmus maximus) is classified into the Pleuronectiformes, and is one of the most commercially important fish species cultured in Europe and North China. During turbot aquaculture, photoperiod manipulation is used to induce egg and sperm production on a year-round basis (51–53). Previous studies have shown that exposure to continuous light reduced the proportion of females that mature and egg production by as much as 90% (54). Similarly, the administration of a long-term photoperiod around the spring equinox significantly decreased and delayed maturation in adult turbot (51, 55). Based on the previous studies, we hypothesized that there has a Kiss/Kissr system in turbot involved in the regulation of the entire reproductive axis in response to seasonal changes. Therefore, in this study, we aimed to identify the Kiss/Kissr system in turbot and investigate the expression profiles of kisspeptin genes in the brain and gonad of turbot within an annual reproductive axis under culture conditions exposed to artificial light. Moreover, the physiological role of Kiss1 and Kiss2 in regulating pituitary hormone gene expression was also examined. These data provide crucial information for controlling seasonal reproduction and breeding of turbot broodstock.
The turbot individuals used in this study were obtained from a fish farm in Shandong Province, China. All of the broodstock were over 2 years old and had previously undergone gonadal maturation. The fish were cultured under a simulated natural photoperiod (12L:12D) and ambient temperature of 17 ± 0.8°C before the reproductive season. During the breeding season, from November 2018 to April 2019, the lighting regime progressed through three stages: an inhibitory photoperiod (8L:16D) for about 1.5 months, a natural photoperiod (12L:12D) for about 0.5 months, and a prolonged photoperiod (16L:8D) for about 4 months. The fish were fed with enhanced nutrition (small yellow croaker Larimichthys polyactis and Pacific sand lance Ammodytes personatus). Three males and three females were collected for sampling every month throughout the reproductive season. Before sampling, turbots were anesthetized with a 0.05% MS222 (Sigma-Aldrich). The gonad from each turbot was divided into two parts, one part was fixed in Bouin’s fluid for histological analysis and the other was frozen in liquid nitrogen and then stored at -80°C. Besides gonads, brain and pituitary gland were also collected for qPCR analysis. In addition, various tissues, including brain, eye, skin, muscle, gill, liver, heart, spleen, kidney, stomach, intestine, liver, and gonad from immature male turbot were collected and stored at -80°C to examine the tissue distribution of the Kiss/Kissr system.
The fixed gonadal tissue samples were dehydrated through a graded series of alcohol concentrations and embedded in paraffin. Serial sections of 5 μm were sliced using a rotary microtome (Leica, Germany) and treated with HE (Haematoxylin and Eosin) stain. Stained sections were examined using a microscope Axio Scope A1 (Leica, Germany). The developmental stages of gonads were confirmed based on ovarian and testicular histology. All of the collected turbots were classified into II, III, IV, V, and VI stages according to the dominant gamete type and characteristics (56, 57).
Total RNA was extracted from the brains of immature turbot using the RNA Isolation Kit (Catalog No. 220011) (Fastagen Biotech, China). Quality and concentration of RNA were measured using 1.0% agarose gels electrophoresis and assessed by spectrophotometry (ND-2000, Nanodrop, USA). The first-strand cDNA was synthesized using a TransScript First-Strand cDNA synthesis kit (AT301-02) (Transgen, China) following the manufacturer’s instructions. Subsequently, cDNA fragments of turbot kiss1, kiss2, kissr2, and kissr3 were amplified with primers (Table 1). Finally, 5’-RACE and 3’-RACE were performed for full-length cDNAs using the SMARTer RACE cDNA Amplification Kit (Catalog No.634923) (Clontech, USA) with gene-specific primers (GSPs) (Table 1). The diluted first-round PCR products were used as templates for nested PCR with corresponding primers of GSP-nest (Table 1). All PCR programs were performed using a PTC-100 thermal cycle (Bio-Rad, USA). PCR was performed in a 25 μl reaction volume containing 1 μl of diluted cDNA, 1 μl of each primer (10 μM), 12.5 μl of Premix buffer (with dNTPs), and 9.5 μl ddH2O, with amplification procedure of denaturation at 94°C for 5minutes; 35 cycles of amplification at 94°C for 30 seconds, annealing with specific temperature of each primer (Table 1) for 45 seconds, elongation with 72°C for 30 seconds, an additional elongation at 72°C for 10 minutes.
Homology searches of the deduced turbot Kiss1, Kiss2, Kissr2, and Kissr3 sequences were performed using the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). The putative signal peptides were predicted by SignalP4.1. The putative transmembrane domain was predicted using the TMHMM server V2.0. The percentages of similarity and identity were calculated using LALIGN (http://www.ch.embnet.org/software/LALIGN-form.html). Multiple alignments of predicted amino acid sequences were conducted using BioEdit with the ClustalW alignment tool. A phylogenetic tree was constructed in the Mega7 software by the neighbor-joining method using bootstrapping with over 1,000 iterations.
The syntenic analysis of kiss1 and kiss2 genes among turbot, zebrafish, medaka and tilapia was conducted basing on the comparison the neighboring genes. Briefly, the protein sequences of neighbor genes of Kiss were predicted from the turbot chromosome sequences by FGENESH program. The identified neighboring protein sequences were annotated against NCBI by BLASTP. The conserved syntenic pattern of Kiss genes in other species were determined in Ensembl database (http://asia.ensembl.org/index.html) and Genomicus (https://www.genomicus.bio.ens.psl.eu/genomicus-83.01/cgi-bin/search.pl).
According to the sequence analysis, turbot Kiss1-10 (YNLNSFGLRY-NH2) and Kiss2-10 (FNFNPFGLRF-NH2) peptides were synthetized by ChinaPeptides Co., Ltd. (Shanghai, China) with purities of 98% and 97%, respectively, as determined by HPLC. For in vivo experiments, juvenile turbot (body weight (BW) = 9.575 ± 0.47 g) were used, at a rearing temperature of 16 ± 0.5°C with dissolved oxygen > 7.0 mg/L and a 12L:12D photoperiod.
The doses of Kiss peptides referred the administration experiments in some fish species, including goldfish and cinnamon clownfish (18, 31, 42). In these articles, it demonstrated that intraperitoneal injection of 0.01-1ug/g in 6h could stimulate expression of lhβ and fshβ. Therefore, doses of 100ng/g BW and 1000 ng/g BW with intraperitoneal injection were chosen. The synthetized peptides were dissolved in phosphate-buffered saline (PBS) and injected intraperitoneally into turbot anesthetized with 0.05% MS222. PBS alone acted as the negative control. At 3 and 6 h post-injection, sample fish were collected (n = 6) and their pituitary glands quickly dissected, frozen in liquid nitrogen, and stored at -80°C for qPCR analysis.
Quantitative real-time PCR was performed using the CFX96 Real-Time PCR Detection System (Bio-Rad, USA) with a SYBR Premix Ex Taq Kit (Takara, Japan) using the standard curve method with β-actin as the reference gene. Specific primers used for each target gene were designed by the Primer Premier 5.0 software (Table 2). The 20 μl reaction system contained 10 μl SYBR Premix Ex Taq, 0.2 μl forward primer (10 μM), 0.2 μl reverse primer (10 μM), 7.6 μl ddH2O, and 2 μl cDNA template. The PCR procedure was programmed according to the manufacturer’s protocol: 30 s at 95°C, 5 s at 95°C, and 30 s at 60°C for 40 cycles. A dissociation curve was performed at the end of each program to determine the amplification specificity. The relative gene expression levels were analyzed by the 2- ΔΔCT method.
Statistical analysis was carried out using SPSS version 21.0. All results are presented as means ± SEM. Gene expression during gonadal stages were analyzed using one-way analysis of variance (ANOVA) followed by Duncan’s multiple range tests. Differences were considered to be significant at P < 0.05. All assays were carried out independently in triplicate.
The full-length cDNA of turbot kiss1 (GenBank accession no. MW057929) was 580 bp in size, with an open reading frame (ORF) of 312 bp encoding a preprohormone of 104 amino acids. The full-length cDNA of turbot kiss2 (GenBank accession no. MW057930) was 622 bp, with an ORF of 369 bp encoding a precusor of 123 amino acids. Amino acid sequence analysis demonstrated that an obvious Kiss1 domain-YNLNSFGLRY (Kiss1-10) and Kiss2 domain-FNFNPFGLRF (Kiss2-10) were involved in turbot kisspeptin system (Supplementary Figure 1). However, the Kiss1-10 differed from Kiss2-10 by 4 amino acids in turbot. The sequence alignments of the deduced amino acid sequences of turbot Kiss1 and Kiss2 with other vertebrate species are presented in Supplementary Figure 2.
Notably, although the Kiss-10 regions are highly conserved across vertebrates, there were still 2 positions in Kiss1-10 and 3 positions in Kiss2-10 with different amino acids. For Kiss1-10, at the 3rd position from the N-terminus, most teleosts exhibit a Leucine and some exhibit phenylalanine, while mammals and amphibians exhibit a Tryptophan. For turbot Kiss2-10, the 3rd position from the N-terminus was identical to medaka, zebrafish, longtooth grouper (Epinephelus bruneus), rare minnow (Gobiocypris rarus), and Senegalese sole. However, turbot had Phenylalanine and Glycine at positions 6 and 7 from the N-terminus, which differed from the Leucine and Threonine in Japanese flounder.
Phylogenetic analysis revealed that the Kiss1 and Kiss2 sequences clustered in two separate clades (Figure 1). The synteny analysis of kiss showed that the kiss1 gene was usually positioned in the genomic regions, including plekha6, cntn2, psma5, foxp4, mapkapk2, and pik3c2b, and kiss2 gene was usually positioned including gys2, spx, golt1b, ldhba, slc25a3a, and strap (Figure 2).
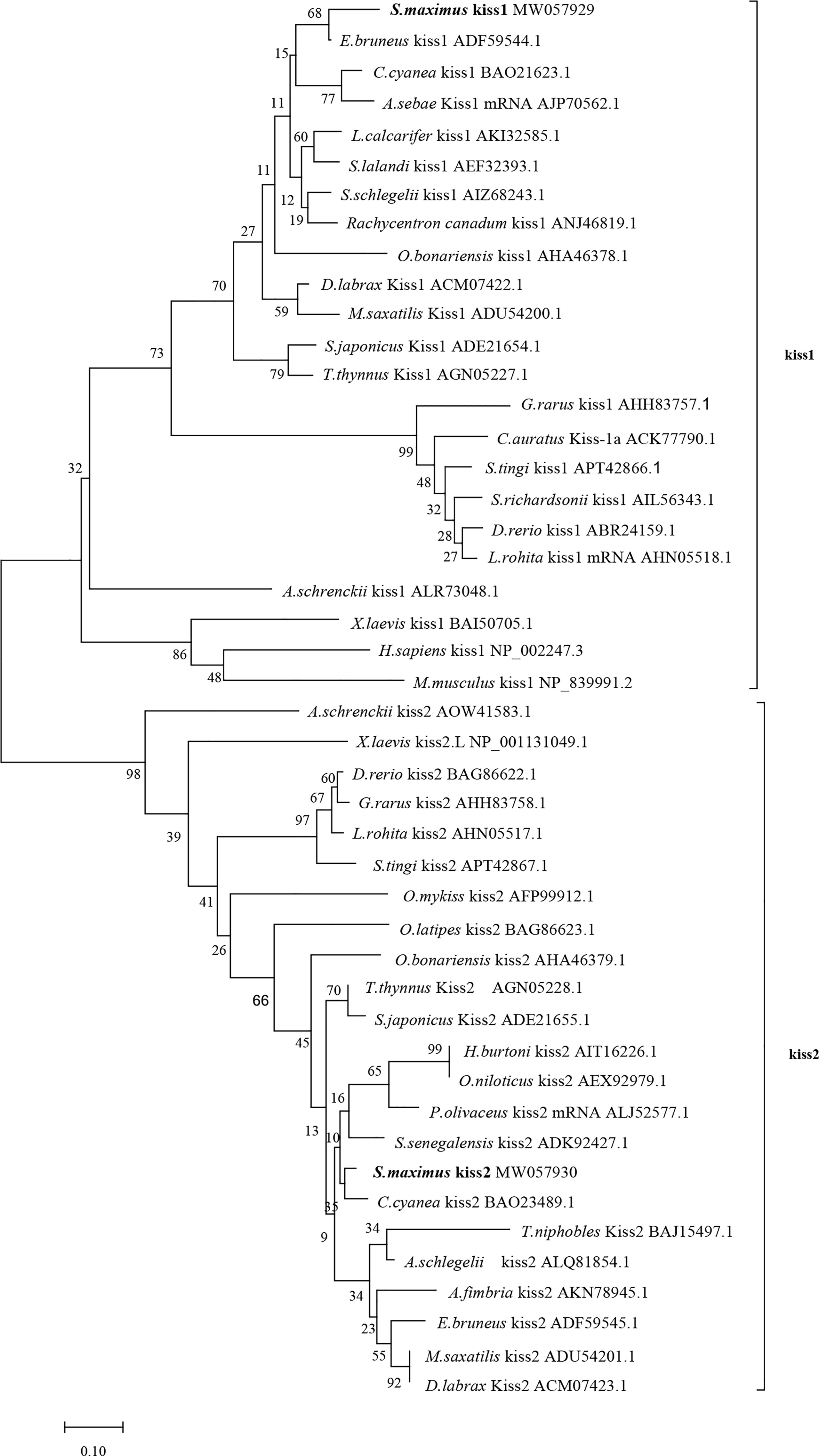
Figure 1 Phylogenetic tree of deduced amino acid sequences of Kiss1 and Kiss2 from turbot and other vertebrates was constructed by Mega7 with neighbor-joining method. The bootstrap values at nodes indicate bootstrap percentage value for 1000 replicates (>80%). Relative branch lengths indicate the evolutionary rates of the lineages. The GenBank accession numbers of the sequences were presented after the species name.
The full-length cDNA of turbot kissr3 (GenBank accession no. MT319113) was 2329 bp, with an ORF of 1092 bp encoding a peptide with 363 amino acids. The full-length cDNA of turbot kissr2 (GenBank accession no. MW057931) was 1578 bp, with an ORF of 1128 bp encoding a peptide with 375 amino acids. The deduced amino acids of Kissr3 and Kissr2 all contained a seven transmembrane domain (Supplementary Figure 3). The sequence alignment of the deduced amino acid sequences of turbot Kissr2 and Kissr3 to those of other vertebrate species is presented in Supplementary Figure 4. In the phylogenetic analysis, the two turbot Kissr clustered into the Kissr2 and Kissr3 clades, respectively (Figure 3).
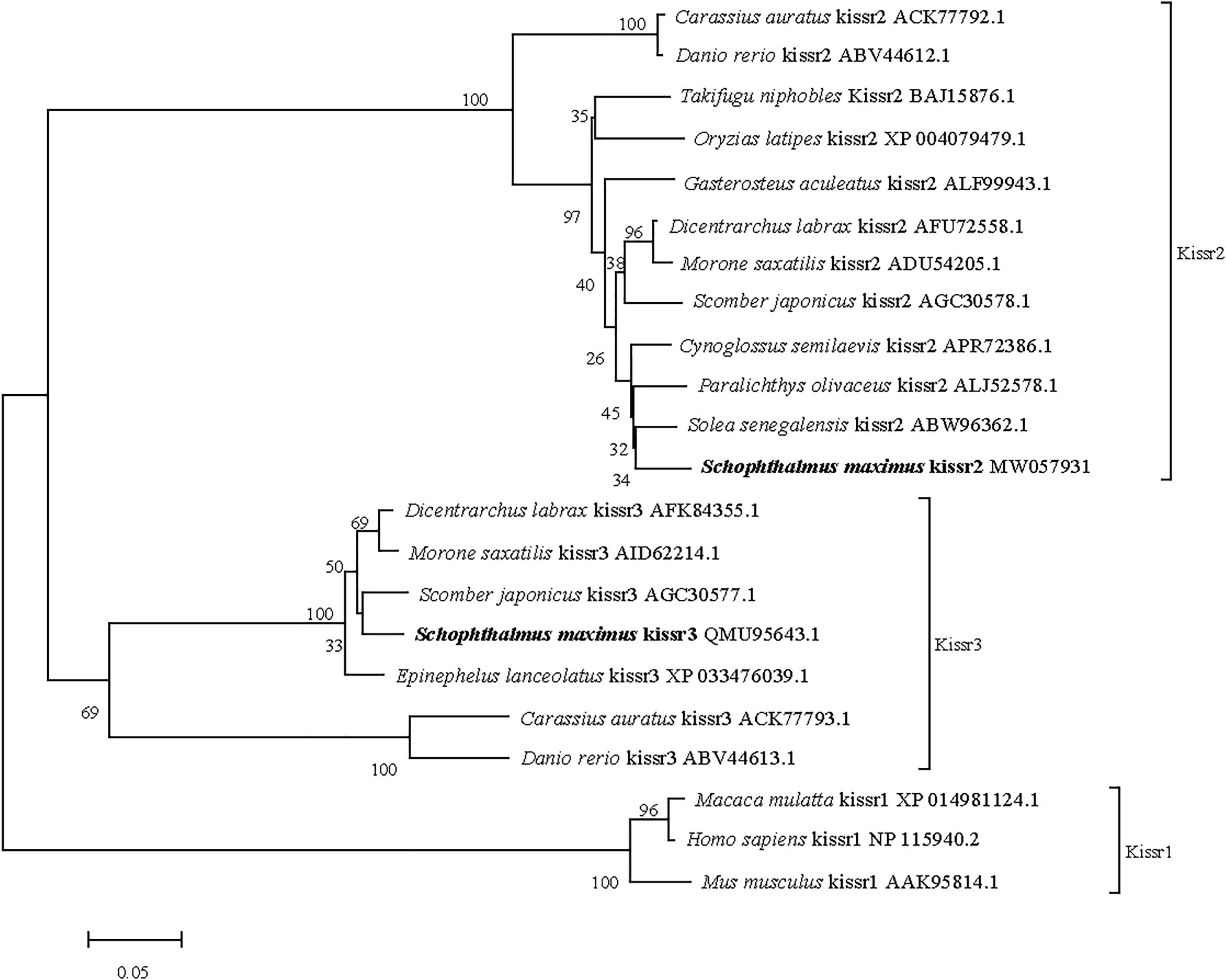
Figure 3 Phylogenetic tree of deduced amino acid sequences of Kissr3 and Kissr2 from turbot and other vertebrates was constructed by Mega7 with neighbor-joining method. The bootstrap values at nodes indicate bootstrap percentage value for 1000 replicates (>80%). Relative branch lengths indicate the evolutionary rates of the lineages. The GenBank accession numbers of the sequences were presented after the species name.
Expression patterns of turbot kiss (kiss1, kiss2) and kissr (kissr2, kissr3) genes in various tissues and different brain regions were detected by qPCR. Firstly, expression of tissues of brain, pituitary, gonad, eye, skin, muscle, gill, heart, spleen, kidney, stomach, intestine, and liver from adult male were shown in Figure 4. The kiss1 mRNA was highly expressed in the brain and gonad, but had low expression in the skin, pituitary, and intestine. The kiss2 mRNA was particularly highly expressed in brain and gonad, and barely expressed in other tissues. The kissr3 mRNA was significantly expressed in brain, gonad, and eye. While the kissr2 mRNA was abundantly expressed in brain and eye, but barely in gonad.
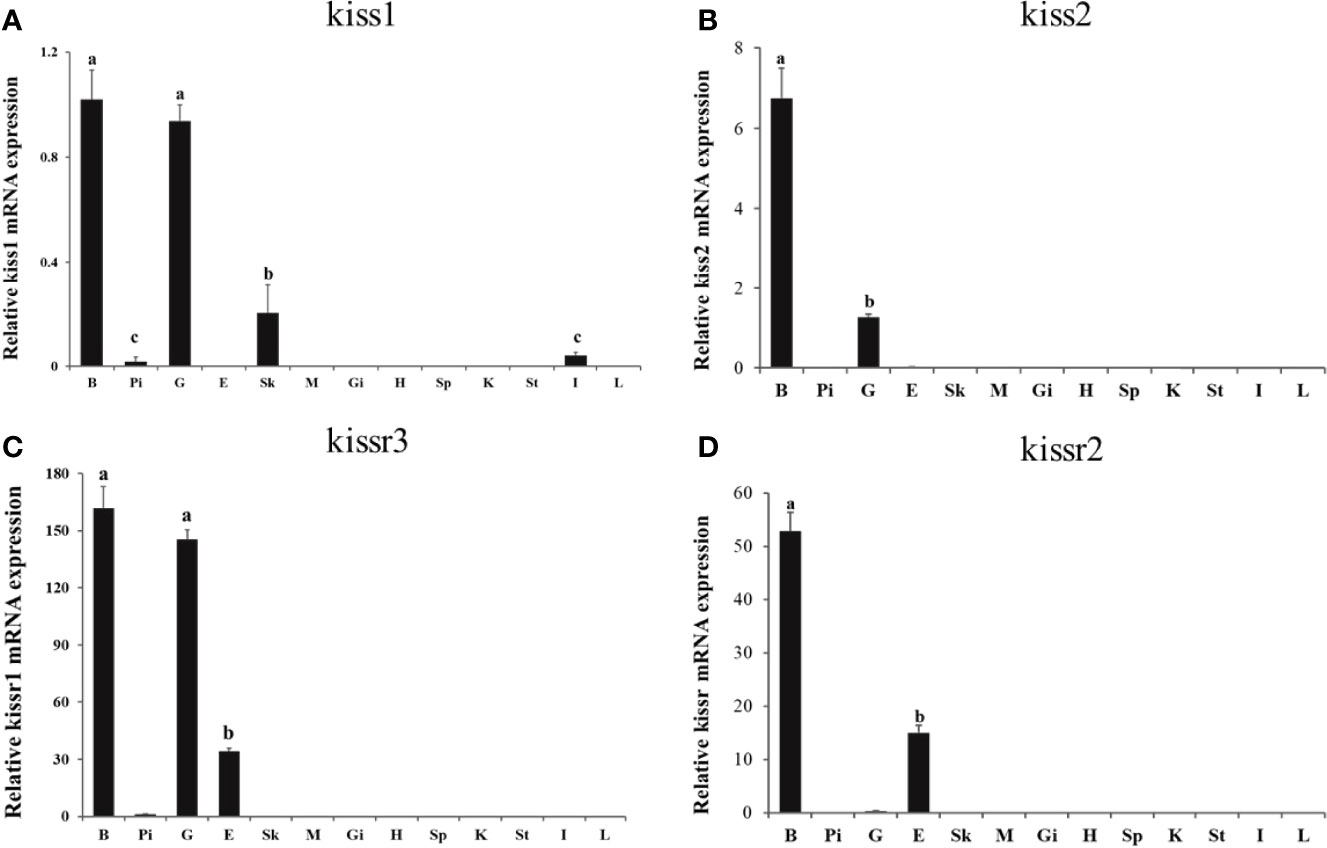
Figure 4 Tissue distribution of kiss/kissr genes in turbot. (A) kiss1, (B) kiss2, (C) kissr3, (D) kissr2. The relative abundance of kiss/kissr genes were expressed as mean ± SEM (n=3). Tissue abbreviations: B, brain; Pi, pituitary; G, gonad; E, eye; Sk, skin; M, muscle; Gi, gill; H, heart; Sp, spleen; K, kidney; St, stomach; I, intestine; L, liver. Different letters above bars represent statistical significance (p < 0.05) between two different tissues.
The expression patterns of turbot kiss/kissr mRNAs in central brain areas, including olfactory bulbs (Ob), telencephalon (Te) with preoptic area (POA), mesencephalon (Me) with thalamus, hypothalamus (Hy) with saccus vasculosus, cerebellum (Ce), medulla oblongata (Mo), and pituitary gland (Pi) were also analyzed (Figure 5). The expression in the eye was also investigated alongside the central brain areas. The kiss/kissr mRNAs were expressed in all central brain areas and eyes. The kiss1 mRNA expression was high in Me, but low in Mo and Hy. However, the kiss2 mRNA was highly detected in the Hy. The expressions of kissr3 and kissr2 mRNA were high in the Me and Hy. In addition, the expression patterns of kiss/kissr mRNAs were similar in the female and male turbot brains, however, expression levels were higher in the central brain areas of females than males.
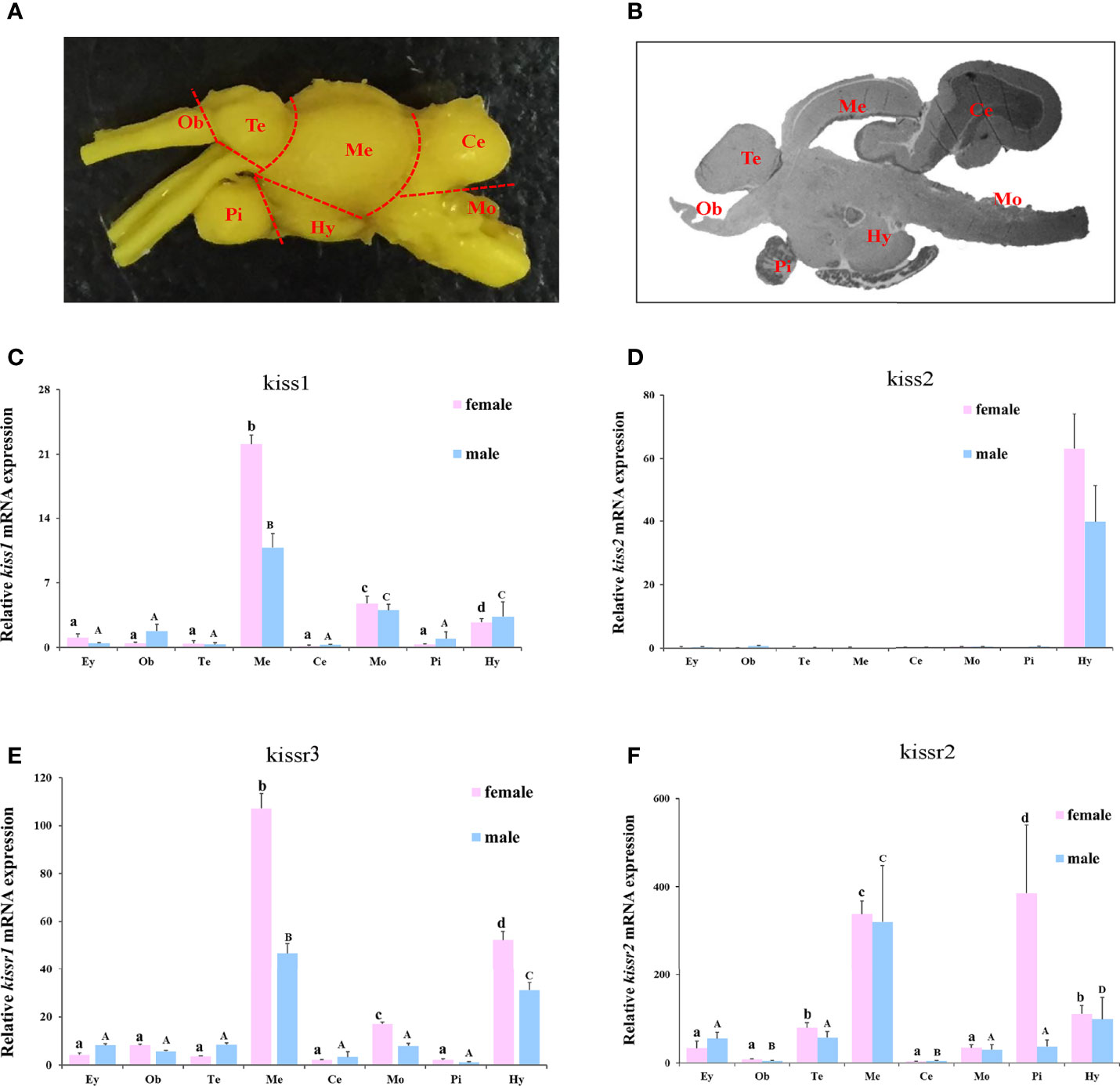
Figure 5 The turbot brain areas and expression of kiss/kissr genes in different brain areas and eyes. (A) morphology of turbot brain areas. (B) histology of turbot brain areas. (C) kiss1. (D) kiss2. (E) kissr3. (F) kissr2. The relative abundance of kiss/kissr genes were expressed as mean ± SEM (n = 3). Ey, eye; Ob, olfactory bulbs; Te, telencephalon; Me, mesencephalon; Ce, cerebellum; Hy, hypothalamus; Pi, pituitary gland; Mo, medulla oblongata. The different uppercase letters in the figure represent statistical significance (p < 0.05) in male. The different lowercase letters in the figure represent statistical significance (p < 0.05) in female.
The morphology and histology of ovaries and testes during breeding season of turbot were examined. Five ovarian and five testicular developmental stages were identified from stage II to VI (Figure 6). First, the five ovarian stages are shown in Figure 6A1–5. At stage II, the ovary was present with light fleshy red color and the blood vessels on the ovarian membrane were not obvious. The germinal epithelium of the ovary was filled with oocytes (primary growth oocytes and perinucleolar oocytes) in stage II. At stage III, the appearance of the ovary was flesh-colored, and eggs could be seen in the ovary. The histological results showed the oocytes of stage III had developed into the previtellogenic stage and vitellogenesis had begun. At stage IV, the ovary exhibited significant growth and the oocytes were characterized as late vitellogenic. At stage V, matured oocytes were obvious in the ovary and the proportion of oocytes in the post vitellogenic stage increased. Stage VI was characterized by degenerated ovaries and there were only primary growth oocytes left.
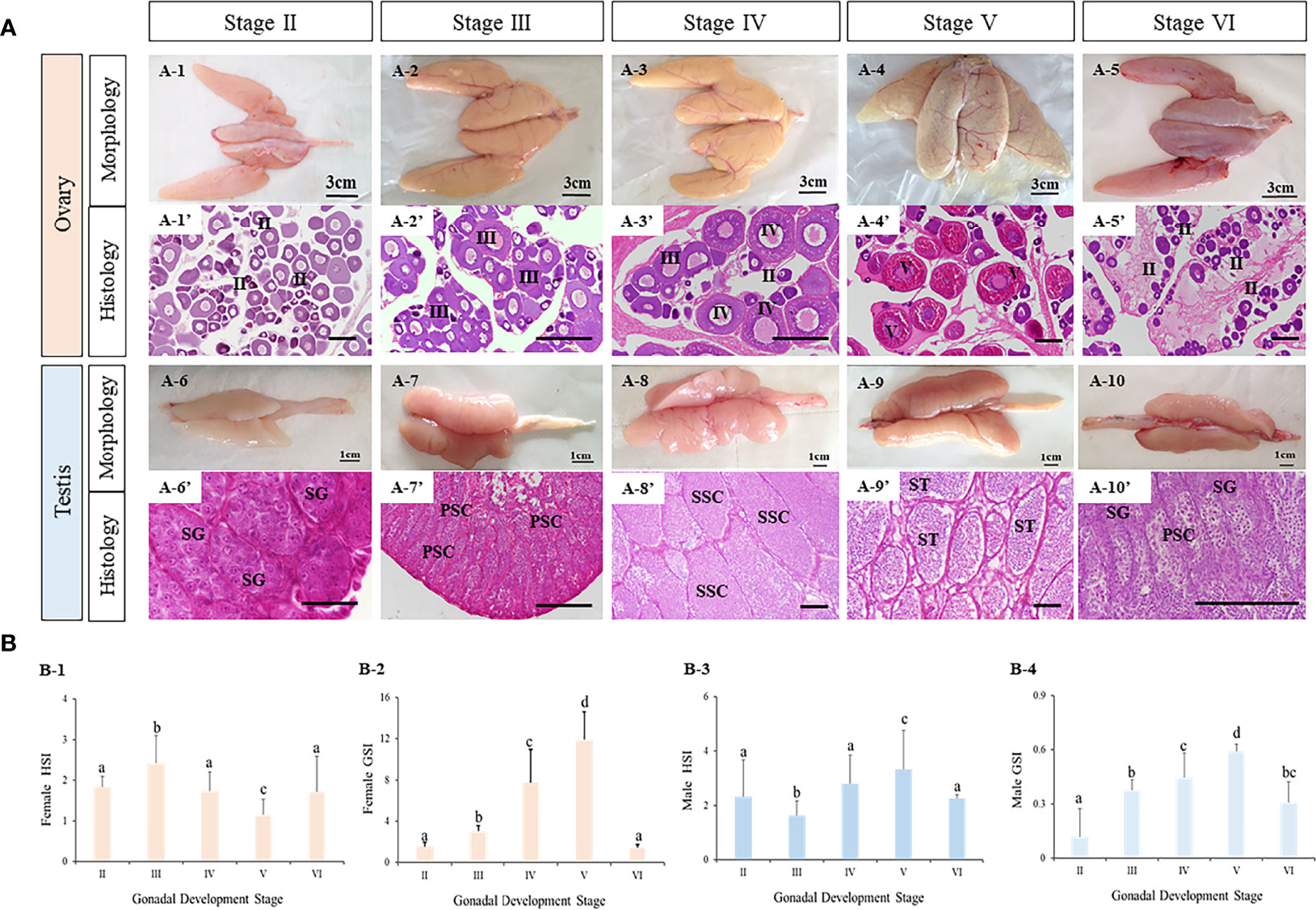
Figure 6 The characteristics of tourbot testes and ovaries during reproductive cycle. (A) morphology and histology of turbot testes and ovaries. A1-5, morphology of turbot ovaries. A1’-5’, histology of turbot ovaries. A6-10, morphology of turbot testes. A6’-10’, histology of turbot testes. (B) GSI and HSI of turbot testes and ovaries. B1-2, GSI and HSI of female turbot. B3-4, GSI and HSI of male turbot. (A) PSC, primary spermatocytes; SG, spermatogonia; SSC, secondary spermatocytes; ST, spermatids; SZ, spermatozoa; (B) I, previtellogenic oocytes. II, primary vitellogenic oocytes; III-IV, large growth of vitellogenic oocytes. Scale: A1-5, 3 cm; A6-10, 1 cm; A1’-10’, 50 μm. Different letters above bars represent statistical significance (p < 0.05) at different reproductive stages in (B).
Similarly, the appearance and major cell types in the five developmental stages of testis were observed (Figure 6A6–10). With development, the testis became bigger and wrinkles and blood vessels became clear. In addition, spermatogenesis was observed. At stage II, the testis contained a large number of spermatogonia. At stage III, spermatogenesis had begun and most germ cells had become primary spermatocytes. At stage IV, secondary spermatocytes appeared. At stage V, the seminiferous tubules were filled with spermatozoa with tails, indicating that spermatogenesis was nearly complete. With the release of spermatozoa, the testis were classified as stage VI.
Meanwhile, the gonadosomatic index (GSI, gonad weight/body weight) and hepatosomatic index (HSI, liver mass/body weight) were calculated (Figure 6B1–4). In both male and female turbot, the GSI of ovaries and testes all increased from stage II to V, reaching their peak values at stage V, before decreasing at stage VI. The HSI of ovaries at stage V was lower than all other stages. While in testes, the HSI was highest at stage V.
In females, kiss1 mRNA expression was very high during stage II, then gradually decreased from stage II to V, followed by a significant increase at stage VI (Figure 7A). The kiss2 expression level declined significantly from stage II to III, and did not change significantly from stage III to VI (Figure 7C). Meanwhile, the kissr3 and kissr2 mRNA expression profiles showed a similar pattern to kiss1 mRNA expression (Figures 7E, G). In males, the expression patterns of kiss1 and kiss2 mRNA were similar during the reproductive stages (Figures 7B, D). The highest expression levels appeared at stages II and VI. Their expression levels did not change significantly from stage III to V, but were significantly lower than stages II and VI. In addition, the expression patterns of kissr3 and kissr2 mRNA in males were similar to those in female turbot (Figures 7F, H).

Figure 7 Expression profiles of kiss/kissr genes throughout the reproductive cycle in the brains of female (A, C, E, G) and male (B, D, F, H). Error bars are presented as the mean±SEM. Different letters above bars represent statistical significance (p < 0.05) between two reproductive stages.
Furthermore, to explore expression patterns in different brain regions, the expression levels of kiss and kissr transcripts in the Me and Hy of both sexes during three gonadal development stages, immature stage (II), mature stage (IV-V), and degeneration stage (VI), were analyzed (Figure 8). The results showed that the expression levels of kiss and kissr transcripts declined significantly from the immature to mature stages, and increased at the degeneration stage. However, kiss2 expression in the Me of females was lowest at the degeneration stage. In males, kiss2 expression in the Me showed no significant differences among the different gonad developmental stages.
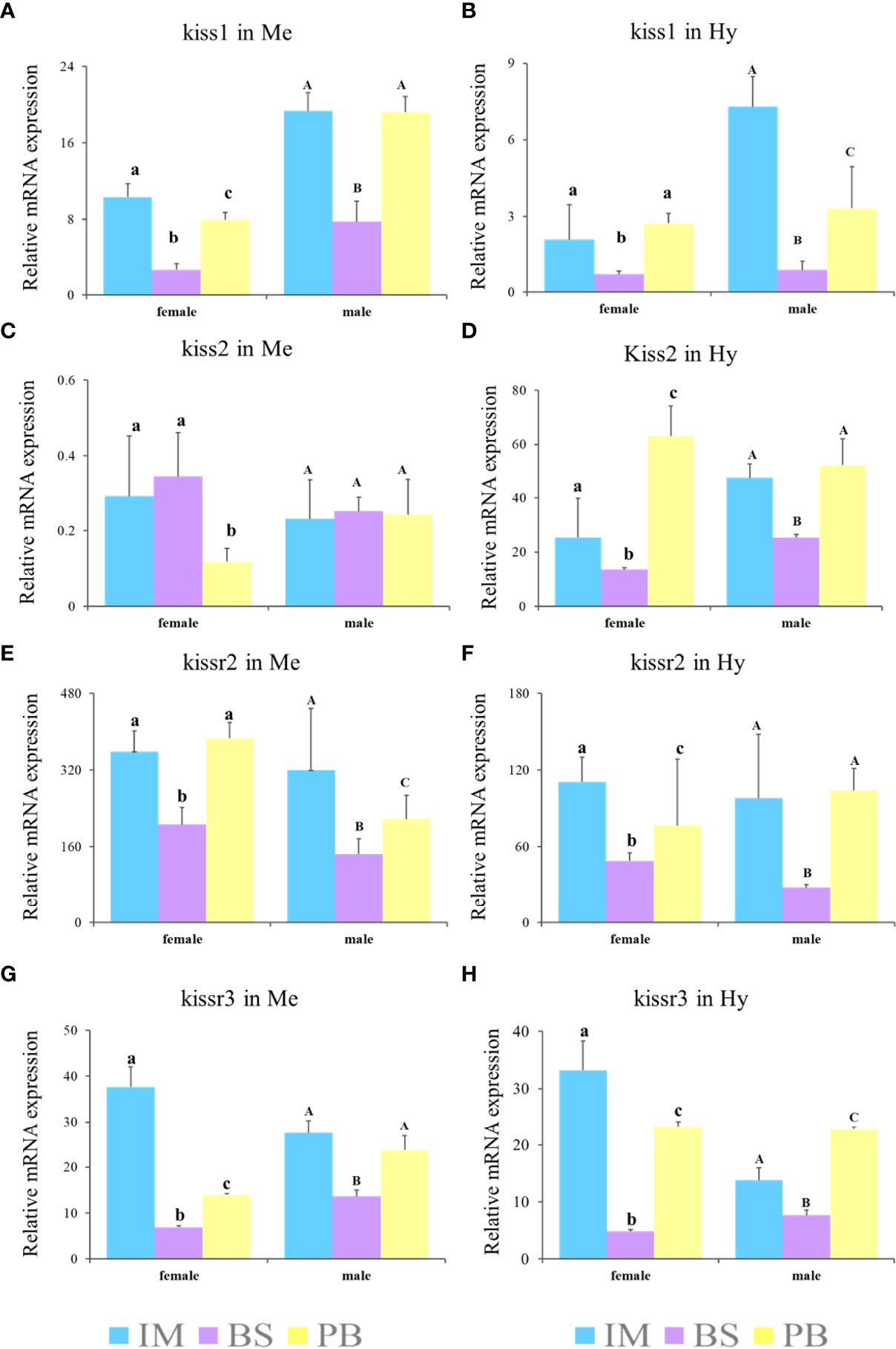
Figure 8 Expression profiles of kiss/kissr genes in Me (A, C, E, G) and Hy (B, D, F, H) at immature stage (IM), the breeding season (BS) and post-breeding season (PB). Error bars are presented as the mean±SEM. The different lowercase letters in the figure represent statistical significance (p < 0.05) between two stages in female. The different uppercase letters in the figure represent statistical significance (p < 0.05) between two stages in male.
In females, there was no a distinct trend of expression of kiss/kissr system. There was no difference of expression of kiss1 in stage V and IV. However, the expression of kiss1 in stage V was higher than that in stage II, III, and VI (Figure 9A). There was high expression of kiss2 and kissr2 mRNA at stage IV (Figures 9C, G). For kiss2, the lowest expression happened at stages III and VI. The lowest expression of kissr2 happened at stage VI. The highest kissr3 mRNA expression level appeared at the immature stage (II) (Figure 9E). In males, the highest kiss and kissr mRNA expression levels appeared at stages II and VI, and lowest at the mature stage (V) (Figures 9B, D, F, H).
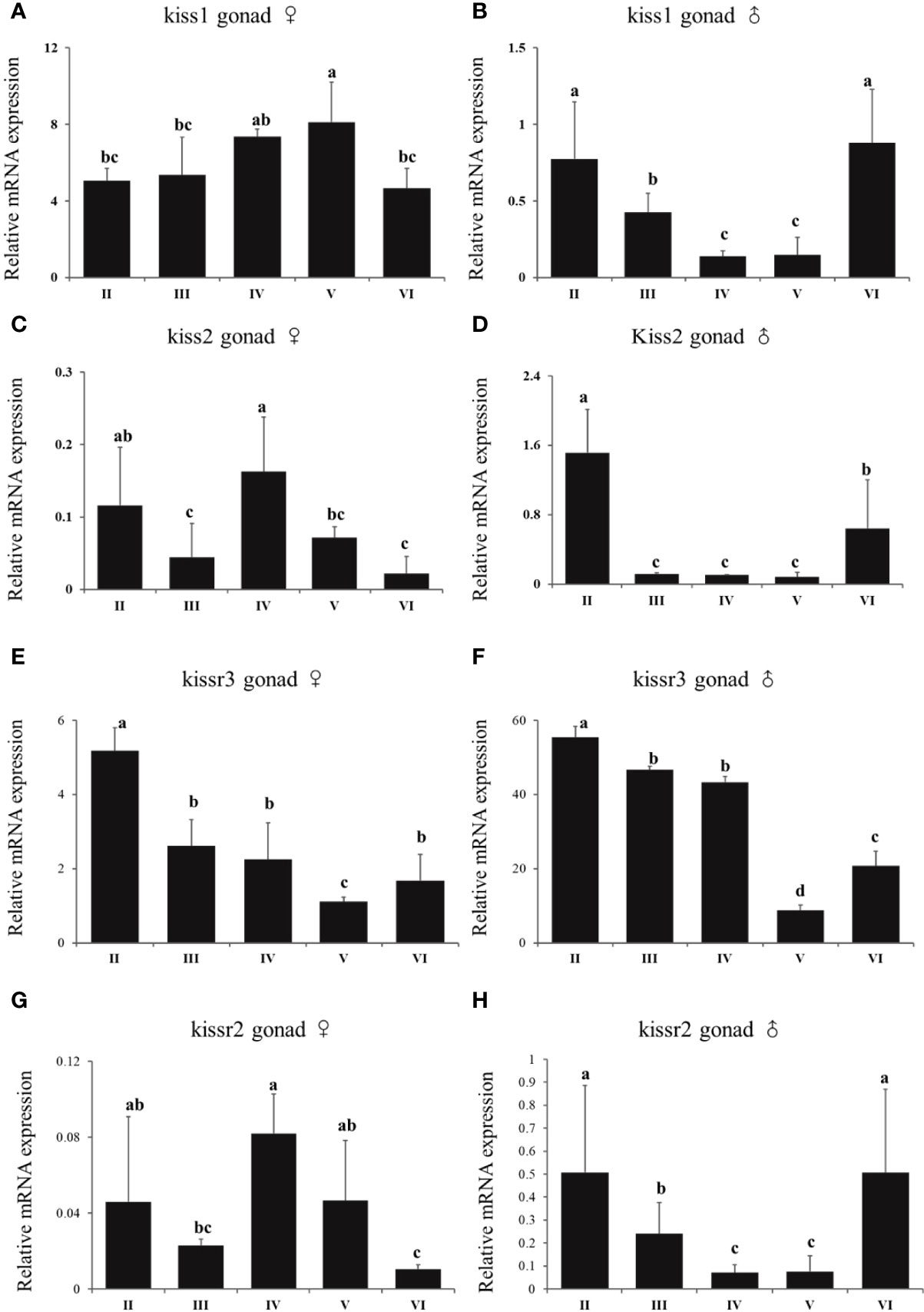
Figure 9 Expression profiles of kiss/kissr genes throughout the reproductive cycle in the gonads of female (A, C, E, G) and male (B, D, F, H). Error bars are presented as the mean±SEM. Different letters above bars represent statistical significance (p < 0.05) between two reproductive stages.
The changes of fshβ, lhβ, and gthα transcription levels were analyzed to investigate the physiological effects of Kiss1-10 and Kiss2-10 on pituitary function. The fshβ (Figure 10A), lhβ (Figure 10B), and gthα (Figure 10C) transcription levels increased significantly at 3 h and 6 h post-injection of 100 ng/g and1000 ng/g Kiss1-10 and Kiss2-10. The transcription levels of fshβ peaked at 3 h under 100 ng/g of Kiss1-10 and Kiss2-10, and 1000 ng/g of Kiss2-10. The lhβ transcription reached the highest level at 6 h under 1000 ng/g of Kiss1-10 and Kiss2-10. Gthα transcription increased significantly from 3 h to 6 h under the administration of both doses. It was note that, after 3h injection, the fshβ and Gthα mRNA had a lower expression with 1000ng/g than that with 100ng/g.
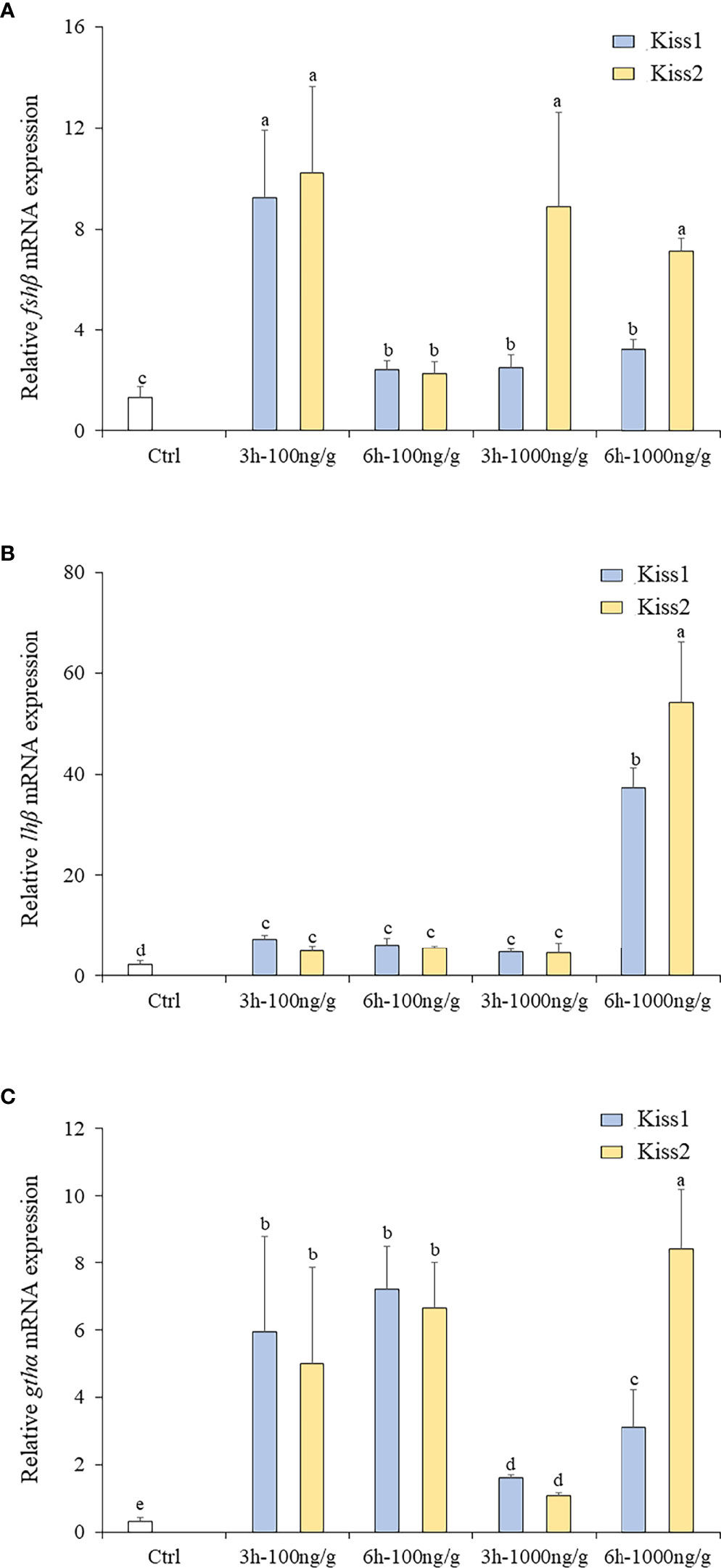
Figure 10 Expression profiles of fhsβ (A), lhβ (B) and gthα (C) in the pituitary of turbot by administration of Kiss1-10 and Kiss2-10 at 3 and 6 h post-injection. Error bars are presented as the mean ± SEM. Different letters above bars represent statistical significance (p < 0.05).
In the present study, we cloned and characterized the Kiss/Kissr system (kiss1, kiss2, kissr2, and kissr3) in turbot, and provided preliminary information on its roles throughout the seasonal reproductive period. To date, numerous studies have reported on the Kiss/Kissr system in vertebrates. In most mammals, there is only one kisspeptin gene, kiss1, except in the platypus which contains two forms, kiss1 and kiss2 (58). However, two forms of kiss genes have been identified in several fish species, such as zebrafish, medaka, goldfish, European seabass, among others (59). However, in almost 20 fish species, only kiss2 can be found, such as the three-spined stickleback (Gasterosteus aculeatus), Nile tilapia (Oreochromis niloticus), and orange-spotted grouper (Epinephelus coioides) (20, 22, 60, 61).
Until now, four kiss and four kissr genes have been identified, indicating the Kiss/Kissr system has a high diversity in vertebrates (62). In this study, two kiss genes (kiss1 and kiss2) and two kissr genes (kissr3 and kissr2) were isolated in turbot. However, previous studies in other Pleuronectiformes only found kiss2 and kissr2, including Japanese flounder (35), half-smooth tongue sole (36, 37), and Senegalese sole (63). Thus, a hypothesis that kiss1 was probably lost in Pleuronectiformes was put forward. Many researchers have tried to explain the lack of kiss1 and establish a clearer picture of their molecular diversity in evolutionary terms of whole genome duplication (35, 62). While teleost-specific whole genome duplication (also known as the third round,3R) should have generated additional Kiss paralogs, there is a loss of Kiss1 in some teleosts (62). The reason might be a massive loss of 3R-Kiss paralogs shortly after the 3R event. However, according to synteny analysis and cognate neighboring genes, it is confirmed that turbot possesses kiss1 gene. It’s worth noting that Pleuronectiformes was split into ‘real flatfish Pleuronectoidei’ (RFP) and ‘flatfish-like Psettodoidei’ (FLP) lineages by genome analyses, and it was confirmed a polyphyletic origin for these two lineages in recent research (64). Meanwhile, there were morphological and genetic difference between the two lineages. Though it is clustered into RFP, the turbot seemed to be closer to FLP which forms one clade with Perciformes species according to the phylogenetic analysis. Therefore, it is supposed that turbot might manage to maintain kiss1 as an exceptional case within Pleuronectiforms.
Generally, the kiss gene encodes a polypeptide precursor, which comprises the mature peptide kisspeptin. Matured kisspeptin peptides encompass a C-terminal 10- decapeptide, namely Kiss-10 (20, 58). Positions 1 and 10 correspond to aromatic amino acids that are fully conserved among vertebrates, and consists of the “Y-Y type” and “F-F type” forms (35). In this study, turbot Kiss1-10 was YNLNSFGLRY, belonging to the “Y-Y type”, and Kiss2-10 was FNFNPFGLRF, belonging to the “F-F type”. Positions 3 and 5 of Kiss1-10 and positions 3, 7, and 8 of Kiss2-10 always exhibited variations in this study. This feature may help in generating specific antibodies against distinct kisspeptins for functional investigations (61).
The receptors of Kiss ligands were GPR54, now known as Kissr, and were first discovered in the rat (8). Previous studies showed that Kissr have been highly conserved throughout evolutionary history. The amino acid sequence comprised seven transmembrane helices (TM1 to TM7) and has a high shared identity among different vertebrate species (58). To date, four Kissr paralogous have been described. In teleosts, all species investigated so far have possessed the kissr2 gene, indicating the existence of one ancestral Kiss receptor (7). In this paper, phylogenetic analyses have revealed that turbot has two kissr paralogous, kissr2 and kissr3, as in some fish species, including zebrafish, medaka, and chub mackerel (14, 58, 65). In addition, three different kissr were found in the European eel (Anguilla anguilla) (28), which suggested that teleost species also possess a diversity of kissr genes.
Tissue expression profiles of kiss and kissr genes have been reported in many fish species (9, 35, 59). Generally, the highest expression of kiss/kissr genes has been found in the brain and gonads of investigated species, as in turbot, demonstrating its conserved expression in the CNS along with its putative role in fish reproduction. It is worth mentioning that, in the present study, kiss1 was more widely expressed in different tissues than kiss2. The kissr mRNA was also expressed in the eye, and kiss1 mRNA was expressed in the intestine, which was in line with previous studies, suggesting that the Kiss/Kissr system might have additional roles in turbot beyond reproduction (14, 20). In addition, differences in expression levels of kiss/kissr among different brain regions were found in turbot. Expression levels of kiss1 were highest in the Me and Hy, and kiss2 was highest in the Hy, which was consistent with their expression patterns in adults of many other fish, including medaka, European sea bass, striped bass (Morone saxatilis), and Nile tilapia, among others (16, 60, 66). While turbot kissr2 and kissr3 had wide distributions, and kissr3 had wider distribution than kissr2. A similarly wide distribution was observed in the medaka brain (17). However, the opposite was true in zebrafish and European seabass, with kissr2 showing a wider distribution than kissr3 (61, 67). These data suggest that the Kiss/Kissr system has diverse functions in the brain.
Numerous previous studies have used different approaches to help clarify the reproductive function of the Kiss/Kissr system, which has been established as an important regulator in the HPG axis of mammals and teleosts (10, 11, 68). The relative expression profiles of multiple genes in the Kiss/Kissr system in the brain and gonad were analyzed in different gonad developmental stages in the present study. First, the expression profiles of the four kiss/kissr genes were similar in the brains of both sexes, where the expression of the kiss/kissr genes gradually decreased from the immature stage to the mature stage, and increased again during the postspawning period, with the same result in the Me and Hy. The kiss/kissr genes can be different during the gonadal stages between males and females in different fish species (24, 30, 31, 68). For example, kiss1 levels continuously declined from the immature to postspawning stages in male chub mackerel, but its level in females showed no significant difference during ovarian development (24). The expression of kiss2 in the brain of seahorses significantly declined at the pregnancy stage. In fathead minnow (Pimephales promelas) the expression of kissr2 mRNA was low in the advanced gonadal stages in males (69). In addition, the expression profiles of four kiss/kissr genes in the testis and ovaries of turbot showed different expression profiles. Studies in the Senegalese sole revealed the genes of the Kiss/Kissr system were expressed in all germ cell types during spermatogenesis (63). Moreover, in sea bass, expressions of kissr2 and kissr3 peaked at the initiation and completion of the spermiation period (21), which was consistent with the kissr expression patterns in male turbot. These results in the brain and gonad of both male and female turbot suggest the positive involvement of the Kiss/Kissr system in turbot reproduction cycle, as in other fishes and mammals. Moreover, the Kiss/Kissr system is essential in other reproductive activity. Kiss2 is involved in sex differentiation in chub mackerel and pejerrey (30, 65). Kiss1 and Kiss2 is essential for male spermiation in the striped bass (66).
It is well known that seasonal fish reproduction is modulated by a wide variety of environmental factors. Photoperiod is generally thought to synchronize sexual maturation and determine spawning time through the activation of neuroendocrine pathways in temperate regions (48, 49). The discovery of the Kiss/Kissr system has provided important insights into the relationship between seasonal reproduction and photoperiod. In mammals, studies have revealed that the Kiss/Kissr system is the most potent activator of GnRH neurons, and the kisspeptin neurons project directly to GnRH neurons and kissr is located at the key site of GnRH neurons (59, 67, 70). Evidence in sheep has suggested that kiss neurons in the arcuate nucleus could directly innervate GnRH neuron somata and dendrites to mediate the induction of lhβ and fhsβ gene expression (71). The in vivo action of Kiss also investigated in some fish species. In lined seahorse, the injection of Kiss2-10 significantly increased fhsβ and lhβ at 6h post-injection and it was higher than 3h in the expression of fhsβ (31). Significant increases in fhsβ and lhβ mRNA levels were seen in the pituitary of zebrafish injected with Kiss2 at 12h after injection (15, 47). Treatment with 0.1 and 0.5 μg/g of Kiss significantly increased the GTH mRNA levels in the pituitary of female and male cinnamon clownfish at 2, 4 and 6 weeks after injection (42). Moreover, in goldfish, compared to 0.01μg/g, the high doses of 0.1 and 1.0 μg/g significantly increased serum LH levels after 6 h, indicating a dose-dependent manner (19). Therefore, it is concluded that the high dose increased relative mRNA levels (fhsβ, lhβ and GTH) at detection time. In the present study, Kiss1-10 and Kiss2-10 stimulated the expression of lhβ, fhsβ, and gthα in the pituitary of turbot, indicating that the Kiss/Kissr system could elevate GTH release in turbot, which is conserved among vertebrates (31). Different from previous studies was that the expression of fhsβ and gthα mRNA at high dose of 1000ng/g after 3h injection was lower than 100ng/g after 3h injection. It was proposed that the Kiss directly increased the expression of lhβ, fhsβ, and gthα in pituitary cells, but the effects depended on the time course and dose. And the investigation of an extended period in the Kiss to genes in pituitary cells would be done in future.
On the other hand, GnRH neurons projecting to the pineal gland, which is a key gland in transducing light signals in the circadian production of melatonin, provide support for the role of GnRH in the transduction of seasonal photoperiod changes (49). Taken together, these observations show the importance of the Kiss/Kissr system in the mechanisms regulating seasonal reproduction. Furthermore, recent studies in mammals and in seasonal breeders of fish have demonstrated that reproduction can be controlled via the Kiss/Kissr system. In European sea bass, manipulating the photoperiod affected the expression of kiss1 and gnrh2 in the forebrain-midbrain to activate the reproductive axis (49). Increased kiss2 and kissr2 gene expression in Atlantic cod also showed the potential role of the Kiss/Kissr system in the entrainment of reproduction (49, 62). Similarly, changing the photoperiod in the present study directly regulated the seasonal gonadal development and modulated the expression of the kiss/kissr genes. However, further investigation is needed to better characterize the mechanism by which the Kiss/Kissr system affects reproduction in turbot.
In conclusion, the present study investigated the Kiss/Kissr system and the changes in the expression of kiss and kissr genes during the reproductive cycle in turbot. All four highly conserved kisspeptin genes (kiss1, kiss2, kissr2, and kissr3) of the Kiss/Kissr system were present in this species, which was different from other Pleuronectiformes species. The expression levels of kiss and kissr in HPG during the reproductive stages and in the pituitary gland after administration of synthetic Kiss1-10 and Kiss2-10 suggest the positive involvement of the Kiss/Kissr system in controlling the seasonal reproductive cycle in turbot.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by Animal Research and Ethics Committees of Qingdao Agricultural University.
CZ: Performed experiments, analyzed the data, wrote, and approved the manuscript; BW: Performed experiments, analyzed the data, edited and approved the manuscript; YL, CF, SX, WW, and QL: Performed experiments and reviewed the manuscript; JL: Designed research, edited and approved the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Natural Science Foundation of China (31802319, 32072949 and 31702321), Advanced Talents Foundation of QAU (6631119055), Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0402), China Agriculture Research System (CARS-47).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.883608/full#supplementary-material
1. Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE, et al. Kiss-1, a Novel Human Malignant Melanoma Metastasis-Suppressor Gene. J Natl Cancer Institute (1996) 88(23):1731–7. doi: 10.1093/jnci/88.23.1731
2. De Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic Hypogonadism Due to Loss of Function of the Kiss1-Derived Peptide Receptor Gpr54. Proc Natl Acad Sci USA (2003) 100(19):10972–6. doi: 10.1073/pnas.1834399100
3. Elizur A. The Kiss1/Gpr54 System in Fish. Peptides (2009) 30(1):164–70. doi: 10.1016/j.peptides.2008.08.018
4. Kauffman AS, Clifton DK, Steiner RA. Emerging Ideas About Kisspeptin– Gpr54 Signaling in the Neuroendocrine Regulation of Reproduction. Trends Neurosci (2007) 30(10):504–11. doi: 10.1016/j.tins.2007.08.001
5. Roa J, Tena-Sempere M. Kiss-1 System and Reproduction: Comparative Aspects and Roles in the Control of Female Gonadotropic Axis in Mammals. Gen Comp Endocr (2007) 153(1):132–40. doi: 10.1016/j.ygcen.2007.01.026
6. Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr., Shagoury JK, et al. The Gpr54 Gene as a Regulator of Puberty. New Engl J Med (2003) 349(17):1614–27. doi: 10.1056/NEJMoa035322
7. Pasquier J, Kamech N, Lafont A-G, Vaudry H, Rousseau K, Dufour S. Kisspeptin/Kisspeptin Receptors. J Mol Endocrinol (2014) 52(3):T101–T17. doi: 10.1530/jme-13-0224
8. Lee DK, Nguyen T, O’Neill GP, Cheng R, Liu Y, Howard AD, et al. Discovery of a Receptor Related to the Galanin Receptors. FEBS Lett (1999) 446(1):103–7. doi: 10.1016/S0014-5793(99)00009-5
9. Funes S, Hedrick JA, Vassileva G, Markowitz L, Abbondanzo S, Golovko A, et al. The Kiss-1 Receptor Gpr54 Is Essential for the Development of the Murine Reproductive System. Biochem Bioph Res Co (2003) 312(4):1357–63. doi: 10.1016/j.bbrc.2003.11.066
10. Navarro VM, Castellano JM, Fernández-Fernández R, Tovar S, Roa J, Mayen A, et al. Effects of Kiss-1 Peptide, the Natural Ligand of Gpr54, on Follicle-Stimulating Hormone Secretion in the Rat. Endocrinology (2005) 146(4):1689–97. doi: 10.1210/en.2004-1353
11. Roseweir AK, Millar RP. The Role of Kisspeptin in the Control of Gonadotrophin Secretion. Hum Reprod Update (2008) 15(2):203–12. doi: 10.1093/humupd/dmn058
12. Parhar IS, Ogawa S, Sakuma Y. Laser-Captured Single Digoxigenin-Labeled Neurons of Gonadotropin-Releasing Hormone Types Reveal a Novel G Protein-Coupled Receptor (Gpr54) During Maturation in Cichlid Fish. Endocrinology (2004) 145(8):3613–8. doi: 10.1210/en.2004-0395
13. van Aerle R, Kille P, Lange A, Tyler CR. Evidence for the Existence of a Functional Kiss1/Kiss1 Receptor Pathway in Fish. Peptides (2008) 29(1):57–64. doi: 10.1016/j.peptides.2007.10.018
14. Biran J, Ben-Dor S, Levavi-Sivan B. Molecular Identification and Functional Characterization of the Kisspeptin/Kisspeptin Receptor System in Lower Vertebrates. Biol Reprod (2008) 79(4):776–86. doi: 10.1095/biolreprod.107.066266
15. Kitahashi T, Ogawa S, Parhar IS. Cloning and Expression of Kiss2 in the Zebrafish and Medaka. Endocrinology (2009) 150(2):821–31. doi: 10.1210/en.2008-0940
16. Kanda S, Akazome Y, Matsunaga T, Yamamoto N, Yamada S, Tsukamura H, et al. Identification of Kiss-1 Product Kisspeptin and Steroid-Sensitive Sexually Dimorphic Kisspeptin Neurons in Medaka (Oryzias Latipes). Endocrinology (2008) 149(5):2467–76. doi: 10.1210/en.2007-1503
17. Mitani Y, Kanda S, Akazome Y, Zempo B, Oka Y. Hypothalamic Kiss1 But Not Kiss2 Neurons Are Involved in Estrogen Feedback in Medaka (Oryzias Latipes). Endocrinology (2010) 151(4):1751–9. doi: 10.1210/en.2009-1174
18. Li S, Zhang Y, Liu Y, Huang X, Huang W, Lu D, et al. Structural and Functional Multiplicity of the Kisspeptin/Gpr54 System in Goldfish (Carassius Auratus). J Endocrinol (2009) 201(3):407–18. doi: 10.1677/joe-09-0016
19. Yang B, Jiang Q, Chan T, Ko WK, Wong AO. Goldfish Kisspeptin: Molecular Cloning, Tissue Distribution of Transcript Expression, and Stimulatory Effects on Prolactin, Growth Hormone and Luteinizing Hormone Secretion and Gene Expression Via Direct Actions at the Pituitary Level. Gen Comp Endocrinol (2010) 165(1):60–71. doi: 10.1016/j.ygcen.2009.06.001
20. Felip A, Zanuy S, Pineda R, Pinilla L, Carrillo M, Tena-Sempere M, et al. Evidence for Two Distinct Kiss Genes in Non-Placental Vertebrates That Encode Kisspeptins With Different Gonadotropin-Releasing Activities in Fish and Mammals. Mol Cell Endocrinol (2009) 312(1-2):61–71. doi: 10.1016/j.mce.2008.11.017
21. Felip A, Espigares F, Zanuy S, Gomez A. Differential Activation of Kiss Receptors by Kiss1 and Kiss2 Peptides in the Sea Bass. Reproduction (2015) 150(3):227–43. doi: 10.1530/REP-15-0204
22. Shi Y, Zhang Y, Li S, Liu Q, Lu D, Liu M, et al. Molecular Identification of the Kiss2/Kiss1ra System and Its Potential Function During 17alpha-Methyltestosterone-Induced Sex Reversal in the Orange-Spotted Grouper. Epinephelus Coioides. Biol Reprod (2010) 83(1):63–74. doi: 10.1095/biolreprod.109.080044
23. Selvaraj S, Ohga H, Nyuji M, Kitano H, Nagano N, Yamaguchi A, et al. Subcutaneous Administration of Kiss1 Pentadecapeptide Accelerates Spermatogenesis in Prepubertal Male Chub Mackerel (Scomber Japonicus). Comp Biochem Phys A (2013) 166(2):228–36. doi: 10.1016/j.cbpa.2013.06.007
24. Selvaraj S, Kitano H, Fujinaga Y, Ohga H, Yoneda M, Yamaguchi A, et al. Molecular Characterization, Tissue Distribution, and Mrna Expression Profiles of Two Kiss Genes in the Adult Male and Female Chub Mackerel (Scomber Japonicus) During Different Gonadal Stages. Gen Comp Endocrinol (2010) 169(1):28–38. doi: 10.1016/j.ygcen.2010.07.011
25. Nocillado JN, Zohar Y, Biran J, Levavi-Sivan B, Elizur A. Chronic Kisspeptin Administration Stimulated Gonadal Development in Pre-Pubertal Male Yellowtail Kingfish (Seriola Lalandi; Perciformes) During the Breeding and Non-Breeding Season. Gen Comp Endocrinol (2013) 191:168–76. doi: 10.1016/j.ygcen.2013.06.005
26. Nocillado JN, Biran J, Lee YY, Levavi-Sivan B, Mechaly AS, Zohar Y, et al. The Kiss2 Receptor (Kiss2r) Gene in Southern Bluefin Tuna, Thunnus Maccoyii and in Yellowtail Kingfish, Seriola Lalandi - Functional Analysis and Isolation of Transcript Variants. Mol Cell Endocrinol (2012) 362(1-2):211–20. doi: 10.1016/j.mce.2012.06.024
27. Pasquier J, Lafont AG, Denis F, Lefranc B, Dubessy C, Moreno-Herrera A, et al. Eel Kisspeptins: Identification, Functional Activity, and Inhibition on Both Pituitary Lh and Gnrh Receptor Expression. Front Endocrinol (Lausanne) (2018) 8:3389/fendo.2017.00353. doi: 10.3389/fendo.2017.00353
28. Pasquier J, Lafont A-G, Jeng S-R, Morini M, Dirks R, van den Thillart G, et al. Multiple Kisspeptin Receptors in Early Osteichthyans Provide New Insights Into the Evolution of This Receptor Family. PLoS One (2012) 7(11):e48931. doi: 10.1371/journal.pone.0048931
29. Mechaly AS, Tovar Bohorquez MO, Mechaly AE, Suku E, Perez MR, Giorgetti A, et al. Evidence of Alternative Splicing as a Regulatory Mechanism for Kissr2 in Pejerrey Fish. Front Endocrinol (Lausanne) (2018) 9:3389/fendo.2018.00604. doi: 10.3389/fendo.2018.00604
30. Tovar Bohorquez MO, Mechaly AS, Hughes LC, Campanella D, Orti G, Canosa LF, et al. Kisspeptin System in Pejerrey Fish (Odontesthes Bonariensis). Characterization and Gene Expression Pattern During Early Developmental Stages. Comp Biochem Physiol A Mol Integr Physiol (2017) 204:146–56. doi: 10.1016/j.cbpa.2016.11.014
31. Zhang H, Zhang B, Qin G, Li S, Lin Q. The Roles of the Kisspeptin System in the Reproductive Physiology of the Lined Seahorse (Hippocampus Erectus), an Ovoviviparous Fish With Male Pregnancy. Front Neurosci (2018) 12:3389/fnins.2018.00940. doi: 10.3389/fnins.2018.00940
32. Zhang H, Zhang YY, Guo Y, Zhang X, Wang Q, Liu XC, et al. Kiss2 But Not Kiss1 Is Involved in the Regulation of Social Stress on the Gonad Development in Yellowtail Clownfish, Amphiprion Clarkii. Gen Comp Endocr (2020) 298:113551. doi: 10.1016/j.ygcen.2020.113551
33. Mechaly AS, Vinas J, Piferrer F. Identification of Two Isoforms of the Kisspeptin-1 Receptor (Kiss1r) Generated by Alternative Splicing in a Modern Teleost, the Senegalese Sole (Solea Senegalensis). Biol Reprod (2009) 80(1):60–9. doi: 10.1095/biolreprod.108.072173
34. Mechaly AS, Vinas J, Murphy C, Reith M, Piferrer F. Gene Structure of the Kiss1 Receptor-2 (Kiss1r-2) in the Atlantic Halibut: Insights Into the Evolution and Regulation of Kiss1r Genes. Mol Cell Endocrinol (2010) 317(1-2):78–89. doi: 10.1016/j.mce.2009.11.005
35. Song H, Wang M, Wang Z, Yu H, Wang Z, Zhang Q. Identification and Characterization of Kiss2 and Kissr2 Homologs in Paralichthys Olivaceus. Fish Physiol Biochem (2016) 42(4):1073–92. doi: 10.1007/s10695-016-0199-1
36. Wang B, Liu Q, Liu X, Xu Y, Shi B. Molecular Characterization of Kiss2 Receptor and In Vitro Effects of Kiss2 on Reproduction-Related Gene Expression in the Hypothalamus of Half-Smooth Tongue Sole (Cynoglossus Semilaevis). Gen Comp Endocr (2017) 249:55–63. doi: 10.1016/j.ygcen.2017.04.006
37. Wang B, Liu Q, Liu X, Xu Y, Song X, Shi B. Molecular Characterization of Kiss2 and Differential Regulation of Reproduction-Related Genes by Sex Steroids in the Hypothalamus of Half-Smooth Tongue Sole (Cynoglossus Semilaevis). Comp Biochem Physiol A Mol Integr Physiol (2017) 213:46–55. doi: 10.1016/j.cbpa.2017.08.003
38. Shahi N, Singh AK, Sahoo M, Mallik SK, Thakuria D. Molecular Cloning, Characterization and Expression Profile of Kisspeptin1 and Kisspeptin1 Receptor at Brain-Pituitary-Gonad (Bpg) Axis of Golden Mahseer, Tor Putitora (Hamilton, 1822) During Gonadal Development. Comp Biochem Phys B (2017) 205:13–29. doi: 10.1016/j.cbpb.2016.11.010
39. Saha A, Pradhan A, Sengupta S, Nayak M, Samanta M, Sahoo L, et al. Molecular Characterization of Two Kiss Genes and Their Expression in Rohu (Labeo Rohita) During Annual Reproductive Cycle. Comp Biochem Phys B (2016) 191:135–45. doi: 10.1016/j.cbpb.2015.10.008
40. Alvarado MV, Servili A, Moles G, Gueguen MM, Carrillo M, Kah O, et al. Actions of Sex Steroids on Kisspeptin Expression and Other Reproduction-Related Genes in the Brain of the Teleost Fish European Sea Bass. J Exp Biol (2016) 219(21):3353–65. doi: 10.1242/jeb.137364
41. Zmora N, Stubblefield JD, Wong T-T, Levavi-Sivan B, Millar RP, Zohar Y. Kisspeptin Antagonists Reveal Kisspeptin 1 and Kisspeptin 2 Differential Regulation of Reproduction in the Teleost, Morone Saxatilis. Biol Reprod (2015) 93(3):76. doi: 10.1095/biolreprod.115.131870
42. Kim NN, Shin HS, Choi YJ, Choi CY. Kisspeptin Regulates the Hypothalamus-Pituitary-Gonad Axis Gene Expression During Sexual Maturation in the Cinnamon Clownfish, Amphiprion Melanopus. Comp Biochem Phys B (2014) 168:19–32. doi: 10.1016/j.cbpb.2013.11.002
43. Simonneaux V, Ansel L, Revel FG, Klosen P, Pévet P, Mikkelsen JD. Kisspeptin and the Seasonal Control of Reproduction in Hamsters. Peptides (2009) 30(1):146–53. doi: 10.1016/j.peptides.2008.06.006
44. Tang H, Liu Y, Luo D, Ogawa S, Yin Y, Li S, et al. The Kiss/Kissr Systems Are Dispensable for Zebrafish Reproduction: Evidence From Gene Knockout Studies. Endocrinology (2015) 156(2):589–99. doi: 10.1210/en.2014-1204
45. Nakajo M, Kanda S, Karigo T, Takahashi A, Akazome Y, Uenoyama Y, et al. Evolutionally Conserved Function of Kisspeptin Neuronal System Is Nonreproductive Regulation as Revealed by Nonmammalian Study. Endocrinology (2018) 159(1):163–83. doi: 10.1210/en.2017-00808
46. Robertson JL, Clifton DK, de la Iglesia HO, Steiner RA, Kauffman AS. Circadian Regulation of Kiss1 Neurons: Implications for Timing the Preovulatory Gonadotropin-Releasing Hormone/Luteinizing Hormone Surge. Endocrinology (2009) 150(8):3664–71. doi: 10.1210/en.2009-0247
47. Carnevali O, Gioacchini G, Maradonna F, Olivotto I, Migliarini B. Melatonin Induces Follicle Maturation in Danio Rerio. PLoS One (2011) 6(5):e19978. doi: 10.1371/journal.pone.0019978
48. Cowan M, Davie A, Migaud H. Photoperiod Effects on the Expression of Kisspeptin and Gonadotropin Genes in Atlantic Cod, Gadus Morhua, During First Maturation. Comp Biochem Phys A (2012) 163(1):82–94. doi: 10.1016/j.cbpa.2012.05.191
49. Espigares F, Rocha A, Gómez A, Carrillo M, Zanuy S. Photoperiod Modulates the Reproductive Axis of European Sea Bass Through Regulation of Kiss1 and Gnrh2 Neuronal Expression. Gen Comp Endocr (2017) 240:35–45. doi: 10.1016/j.ygcen.2016.09.007
50. Ando H, Ogawa S, Shahjahan M, Ikegami T, Doi H, Hattori A, et al. Diurnal and Circadian Oscillations in Expression of Kisspeptin, Kisspeptin Receptor and Gonadotrophin-Releasing Hormone 2 Genes in the Grass Puffer, a Semilunar-Synchronised Spawner. J Neuroendocrinol (2014) 26(7):459–67. doi: 10.1111/jne.12165
51. Imsland AK, Gunnarsson S, Roth B, Foss A, Le Deuff S, Norberg B, et al. Long-Term Effect of Photoperiod Manipulation on Growth, Maturation and Flesh Quality in Turbot. Aquaculture (2013) 416-417:152–60. doi: 10.1016/j.aquaculture.2013.09.005
52. Imsland AK, Folkvord A, Stefansson SO. Growth, Oxygen Consumption and Activity of Juvenile Turbot (Scophthalmus Maximus L.) Reared Under Different Temperatures and Photoperiods. Netherlands J Sea Res (1995) 34(1):149–59. doi: 10.1016/0077-7579(95)90023-3
53. Foss A, Imsland AK, Roth B, Schram E, Stefansson SO. Effects of Chronic and Periodic Exposure to Ammonia on Growth and Blood Physiology in Juvenile Turbot (Scophthalmus Maximus). Aquaculture (2009) 296(1):45–50. doi: 10.1016/j.aquaculture.2009.07.013
54. Imsland AK, Dragsnes M, Stefansson SO. Exposure to Continuous Light Inhibits Maturation in Turbot (Scophthalmus Maximus). Aquaculture (2003) 219(1):911–9. doi: 10.1016/S0044-8486(03)00034-6
55. Imsland AK, Folkvord A, Jónsdóttir ÓDB, Stefansson SO. Effects of Exposure to Extended Photoperiods During the First Winter on Long-Term Growth and Age at First Maturity in Turbot (Scophthalmus Maximus). Aquaculture (1997) 159(1):125–41. doi: 10.1016/S0044-8486(97)00152-X
56. Xue R, Wang XY, Xu SH, Liu YF, Feng CC, Zhao CY, et al. Expression Profile and Localization of Vitellogenin Mrna and Protein During Ovarian Development in Turbot (Scophthalmus Maximus). Comp Biochem Phys B (2018) 226:53–63. doi: 10.1016/j.cbpb.2018.08.002
57. Zhao C, Xu S, Liu Y, Feng C, Xiao Y, Wang Y, et al. Changes of Melatonin and its Receptors in Synchronizing Turbot (Scophthalmus Maximus) Seasonal Reproduction and Maturation Rhythm. Acta Oceanol. Sin (2022) 41:84–98. doi: 10.1007/s13131-021-1923-y
58. Lee YR, Tsunekawa K, Moon MJ, Um HN, Hwang J-I, Osugi T, et al. Molecular Evolution of Multiple Forms of Kisspeptins and Gpr54 Receptors in Vertebrates. Endocrinology (2009) 150(6):2837–46. doi: 10.1210/en.2008-1679
59. Selvaraj S, Kitano H, Ohga H, Yamaguchi A, Matsuyama M. Expression Changes of Mrnas Encoding Kisspeptins and Their Receptors and Gonadotropin-Releasing Hormones During Early Development and Gonadal Sex Differentiation Periods in the Brain of Chub Mackerel (Scomber Japonicus). Gen Comp Endocr (2015) 222:20–32. doi: 10.1016/j.ygcen.2014.09.019
60. Ogawa S, Ng KW, Xue X, Ramadasan PN, Sivalingam M, Li S, et al. Thyroid Hormone Upregulates Hypothalamic Kiss2 Gene in the Male Nile Tilapia, Oreochromis Niloticus. Front Endocrinol (2013) 4:3389/fendo.2013.00184. doi: 10.3389/fendo.2013.00184
61. Escobar S, Felip A, Gueguen M-M, Zanuy S, Carrillo M, Kah O, et al. Expression of Kisspeptins in the Brain and Pituitary of the European Sea Bass (Dicentrarchus Labrax). J Comp Neurol (2013) 521(4):933–48. doi: 10.1002/cne.23211
62. Dufour S, Quérat B, Tostivint H, Pasqualini C, Vaudry H, Rousseau K. Origin and Evolution of the Neuroendocrine Control of Reproduction in Vertebrates, With Special Focus on Genome and Gene Duplications. Physiol Rev (2019) 100(2):869–943. doi: 10.1152/physrev.00009.2019
63. Mechaly AS, Viñas J, Piferrer F. Gene Structure Analysis of Kisspeptin-2 (Kiss2) in the Senegalese Sole (Solea Senegalensis): Characterization of Two Splice Variants of Kiss2, and Novel Evidence for Metabolic Regulation of Kisspeptin Signaling in Non-Mammalian Species. Mol Cell Endocrinol (2011) 339(1):14–24. doi: 10.1016/j.mce.2011.03.004
64. Lü Z, Gong L, Ren Y, Chen Y, Wang Z, Liu L, et al. Large-Scale Sequencing of Flatfish Genomes Provides Insights Into the Polyphyletic Origin of Their Specialized Body Plan. Nat Genet (2021) 53(5):742–51. doi: 10.1038/s41588-021-00836-9
65. Ohga H, Fujinaga Y, Selvaraj S, Kitano H, Nyuji M, Yamaguchi A, et al. Identification, Characterization, and Expression Profiles of Two Subtypes of Kisspeptin Receptors in a Scombroid Fish (Chub Mackerel). Gen Comp Endocr (2013) 193:130–40. doi: 10.1016/j.ygcen.2013.07.016
66. Zmora N, Stubblefield J, Zulperi Z, Biran J, Levavi-Sivan B, Muñoz-Cueto JA, et al. Differential and Gonad Stage-Dependent Roles of Kisspeptin1 and Kisspeptin2 in Reproduction in the Modern Teleosts, Morone Species. Biol Reprod (2012) 86(6):177. doi: 10.1095/biolreprod.111.097667
67. Ogawa S, Ng KW, Ramadasan PN, Nathan FM, Parhar IS. Habenular Kiss1 Neurons Modulate the Serotonergic System in the Brain of Zebrafish. Endocrinology (2012) 153(5):2398–407. doi: 10.1210/en.2012-1062
68. Ohga H, Selvaraj S, Matsuyama M. The Roles of Kisspeptin System in the Reproductive Physiology of Fish With Special Reference to Chub Mackerel Studies as Main Axis. Front Endocrinol (2018) 9:3389/fendo.2018.00147. doi: 10.3389/fendo.2018.00147
69. Filby AL, van Aerle R, Duitman J, Tyler CR. The Kisspeptin/Gonadotropin-Releasing Hormone Pathway and Molecular Signaling of Puberty in Fish. Biol Reprod (2008) 78(2):278–89. doi: 10.1095/biolreprod.107.063420
70. Piet R, de Croft S, Liu X, Herbison AE. Electrical Properties of Kisspeptin Neurons and Their Regulation of Gnrh Neurons. Front Neuroendocrinol (2015) 36:15–27. doi: 10.1016/j.yfrne.2014.05.006
Keywords: turbot, kisspeptin, kisspeptin receptor, reproduction, gonadal development
Citation: Zhao C, Wang B, Liu Y, Feng C, Xu S, Wang W, Liu Q and Li J (2022) New Evidence for the Existence of Two Kiss/Kissr Systems in a Flatfish Species, the Turbot (Scophthalmus maximus), and Stimulatory Effects on Gonadotropin Gene Expression. Front. Endocrinol. 13:883608. doi: 10.3389/fendo.2022.883608
Received: 25 February 2022; Accepted: 10 May 2022;
Published: 15 June 2022.
Edited by:
Yong Zhu, East Carolina University, United StatesReviewed by:
Agata Żmijewska, University of Warmia and Mazury in Olsztyn, PolandCopyright © 2022 Zhao, Wang, Liu, Feng, Xu, Wang, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Li, anVubGlAcWRpby5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.