
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 30 June 2022
Sec. Cancer Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.875442
This article is part of the Research Topic Targeting the Tumor Microenvironment for Effective Treatment of Gastrointestinal Cancers View all 5 articles
 JinHua He1†
JinHua He1† NaiTe Xi2†
NaiTe Xi2† ZePing Han1†
ZePing Han1† WenFeng Luo1
WenFeng Luo1 Jian Shen1
Jian Shen1 ShengBo Wang3
ShengBo Wang3 JianHao Li4*
JianHao Li4* ZhongHui Guo1*
ZhongHui Guo1* HanWei Cheng1*
HanWei Cheng1*Colorectal cancer (CRC) is one of the most common malignant tumors of the digestive tract worldwide and is a serious threat to human life and health. CRC occurs and develops in a multi-step, multi-stage, and multi-gene process, in which abnormal gene expression plays an important role. CRC is currently diagnosed via endoscopy combined with tissue biopsy. Compared with tissue biopsy, liquid biopsy technology has received increasingly more attention and applications in the field of molecular detection due to its non-invasive, safe, comprehensive, and real-time dynamic nature. This review article discusses the application and limitations of current liquid biopsy analytes in the diagnosis, treatment, and prognosis of CRC, as well as directions for their future development.
Colorectal cancer (CRC) is one of the most common cancers worldwide. Due to changes in living environments, eating habits, and lifestyles, the incidence of CRC is increasing yearly in both developed and developing countries. There are approximately 1.8 million new cases worldwide each year, and approximately 900,000 people die from CRC every year (1, 2). Population growth and the aging of society may lead to a further increase in the number of deaths in many countries and regions, with the number of deaths from CRC predicted to double by 2035 (3). The long treatment process and lower quality of life of CRC not only impose a high financial burden on most patients’ families but also cause tremendous psychological pressure on patients (4).
To date, the diagnosis of CRC usually relies on the examination of serum biomarker levels, tissue biopsy, and imaging. However, the diagnostic accuracy and sensitivity of pathological and imaging methods are still limited, and the specificity of the commonly used serum markers is poor.
The main treatment for CRC is surgical resection. Pathological stage at the time of diagnosis can provide an accurate prediction for those who are at high risk of recurrence. Postoperative chemotherapy can be added to reduce this risk. Despite this, 17%–40% of CRC patients undergo relapse (5, 6). A study in the United States from 2001 to 2007 showed that the prognosis of patients diagnosed with local tumors, regional tumor spread, and distant tumor spread are significantly different with 5-year survival rates of 90.1%, 69.2%, and 11.7%, respectively (7). Tumor, node, metastasis (TNM) staging of the tumor is important and is also the basis for adjuvant chemotherapy. Therefore, clinical scientists have been looking for reliable biomarkers to provide guidance for the early diagnosis, treatment, and prognosis of CRC.
Traditional serum tumor markers such as carcinoembryonic antigen, carbohydrate antigen 19-9, and carbohydrate antigen 72-4 have, to a certain extent, guiding significance for CRC. Studies have shown that the combined quantification of these three markers is more sensitive than individually, but specificity is reduced; compared with patients with normal levels of these markers, the overall survival time of patients with elevated levels of all three markers is significantly shorter and the recurrence rate is higher (8).
In the past 30 years, some progress has been made in the use of genetic biomarkers such as plasmacytoma variant translocation 1, long-chain non-coding RNAs, and KRAS gene. The levels of these markers may be increased in patients with CRC and they have certain guiding significance for diagnosis (9, 10). However, traditional serum tumor markers or gene-level biomarkers are not sufficiently specific or sensitive, and there are large individual differences in their levels. Currently, there is a lack of biomarkers that can guide early diagnosis, targeted therapy, prognosis, and monitoring of patients with CRC.
Liquid biopsy technology, as a branch of in vitro diagnosis, refers to a non-invasive blood test that can be used to monitor circulating tumor cells (CTCs), circulating tumor DNA (ctDNA) fragments, metabolites, and so on released into the blood by tumors or metastases. It is a breakthrough technology for the use of adjuvant treatment in cancer. Its main advantages are as follows: it can address the problems associated with precision medicine; it reduces harm from biopsies through the use of non-invasive sampling, effectively prolonging the survival period of patients; and (iii) it is cost effective (11–16). Liquid biopsy analytes include mainly CTCs, ctDNA, circulating tumor RNAs, circulating tumor microRNAs (ctmiRNAs), circulating free DNA (cfDNA), exosomes, extracellular vesicles, and metabolites. In this review, we discuss the role of CTCs, ctDNA, exosomes, ctmiRNAs, and metabolites and outline their use as potential biomarkers in the diagnosis, treatment, and prognosis of CRC.
CTCs are tumor cells that separate from tumor tissue and enter the circulatory system. The production of CTCs is a necessary condition for patients to develop a distant metastasis (17). CTCs detection technology mainly includes CTCs separation and enrichment technology and CTCs identification technology. The ideal separation, enrichment and detection technique of CTCs should meet the requirements of high sensitivity and specificity. CTCs must not be contaminated, and the collected CTCs can be identified and analyzed genetically (Figure 1). CTCs are not only an important prognostic indicator, they are also considered a very effective real-time biopsy tool and a window into the molecular mechanisms for pathogenicity and drug resistance in tumor cells (18). However, most CTCs die from shear stress, oxidative stress, and attack from the immune system. In the early stage of CRC, due to limitations of the detection methods, very few CTCs can be captured in the peripheral blood of CRC patients, and it is almost impossible to identify CTCs in the blood of early CRC patients, which limits the possibility of using CTCs for clinical analysis. After decades of development, tremendous breakthroughs have been made in the technology for detecting CTCs, including nucleic acid identification based on PCR, and enrichment methods using the physical and biological characteristics of CTCs (19, 20). Recently, Cheng et al. developed Hydro-Seq, a new type of microfluidic chip used to capture CTCs that can accurately separate ultra-high purity CTCs from patient blood samples, independent of white blood cells and red blood cells. By using Hydro-Seq, comprehensive analysis of CTCs can be performed in a high-throughput manner, which can effectively provide patients with treatment programs in clinical practice (21).
Based on the progress of CTC detection methods, researchers have performed in-depth studies on CTCs in CRC. In terms of early screening, in 620 patients (including patients with stage I–IV CRC and precancerous lesions), the quantitative detection of CTCs is generally accurate in judging the stage of all colorectal diseases including precancerous lesions with an accuracy of 88% (22). In terms of the clinical stage of the disease, the number of CTCs reflects the degree of tumor mutation in patients, to a certain extent. In patients with advanced CRC, the detection rate of CTCs increases as the tumor stage increases. Further research on the TNM staging system has shown that the number of detectable CTCs is positively correlated with the size and depth of invasion of the primary tumor, lymph node invasion, and distant metastasis, suggesting that CTCs can be used to judge lymph node invasion and distant metastasis (23–26).
In a study of 183 CRC patients, blood samples were collected at different time points during the pre-follow-up and follow-up periods. The results showed that the presence of CTCs before surgery was associated with a significant reduction in survival, and patients with a high risk of recurrence could be identified (27). Other studies have also shown that the presence of CTCs is an indicator for the poor prognosis of CRC patients (28, 29). The total number of CTCs and the number of CTCs with a mesenchymal phenotype are reported to be significantly related to advanced disease stage and the occurrence of metastasis (30). The total number of mesenchymal-type CTCs can be used as a prognostic marker of disease progression and metastasis in CRC patients. For example, 149 CRC patients were tested for CTCs according to tumor stage (TNM); as tumor stage increased, the number of CTCs also increased. Survival analysis showed that with the increase in T stage, the number of CTCs increased and the survival and risk curves changed faster (31). In addition, Nicolazzo et al. compared the characteristics of CTCs in the blood of 84 metastatic colon cancer patients with left colon cancer (LCC) and right colon cancer (RCC) and found that the CTCs present in both types of colon cancer showed phenotypic heterogeneity (20). Compared with proximal primary tumors, the CTCs found in patients with distal primary tumors are mainly mesenchymal. The CTCs of RCC patients are mostly apoptotic, whereas those of LCC patients are mainly mesenchymal. This may indicate that the biology of proximal and distal cancers differs substantially, which might be related to the different modes of tumor cell spread. The poor prognosis of RCC is not determined by the hematological dissemination of tumor cells, which is manifested mainly by the passive shedding of inactive cells. In contrast, subtypes of LCC with a poor prognosis can be identified reliably by the presence of mesenchyme-type CTCs (18).
cfDNA usually refers to degraded DNA fragments that are released into the plasma. cfDNA exists in various human body fluids. Under physiological conditions, cfDNA in the blood comes mainly from the necrosis and apoptosis of white blood cells. In some diseases and under special conditions, other cells also release cfDNA into the blood, including ctDNA derived from tumor cells. ctDNA fragments are derived mainly from necrotic tumor cells, apoptotic tumor cells, circulating tumor cells, and exosomes secreted by tumor cells (32). At present,cfDNA detection methods mainly include real-time fluorescence quantitative PCR technology, digital PCR (dPCR) technology, high-throughput sequencing (NGS) technology. Flight mass spectrometry. Detecting changes in the number of types of cfDNA in specific sequences can be used to identify cancer, and related diseases. Measuring the size of cfDNA can reveal which cell death mechanism it is produced by (Figure 2).
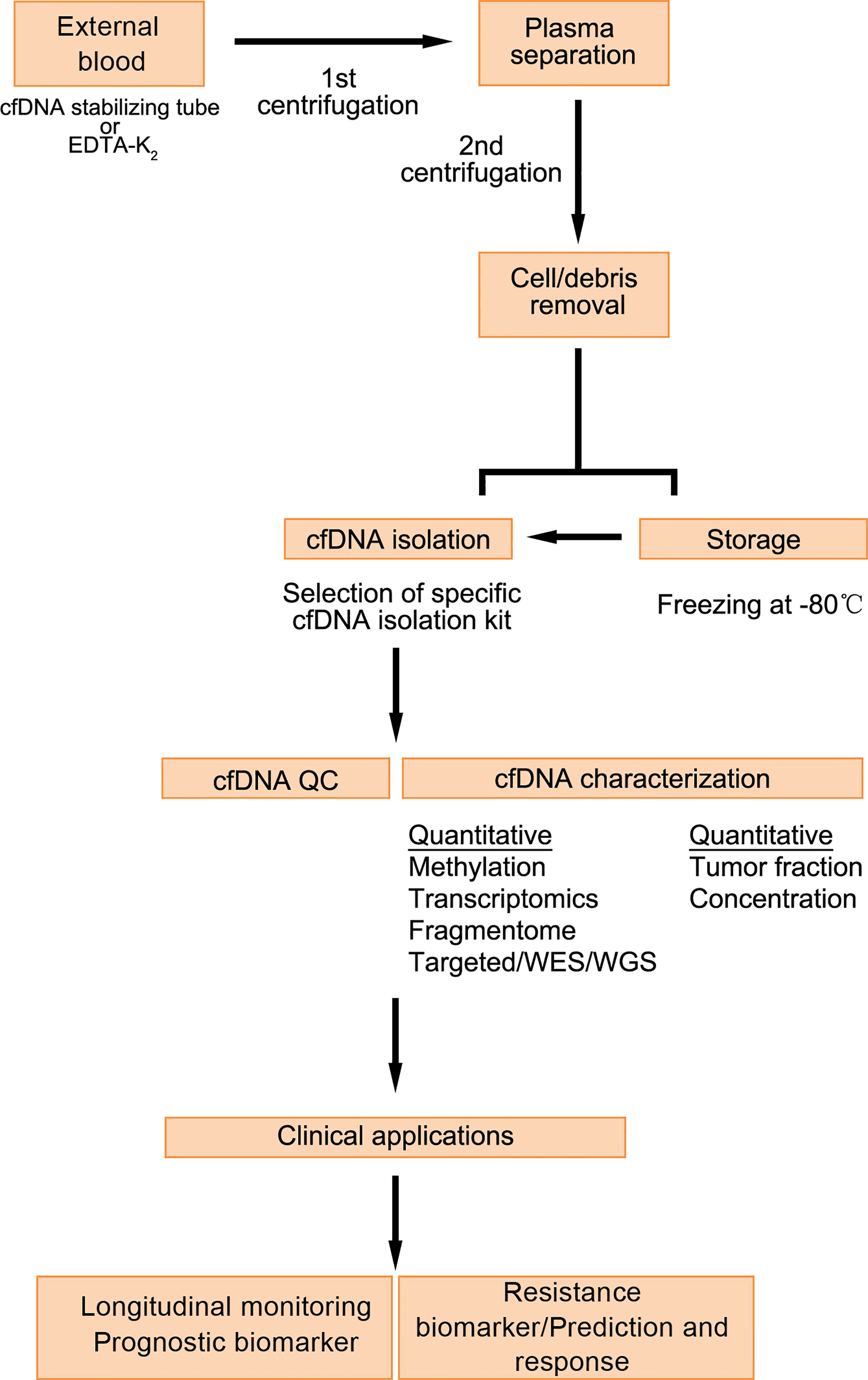
Figure 2 Detection process and application of cfDNA (33).
Under normal circumstances, cfDNA is produced mainly as small and uniform 185-200-bp fragments during cell apoptosis. Tumor cells undergo unique biological processes, including necrosis, apoptosis, and extracellular transport, resulting in large cfDNA fragments of different sizes greater than 200 bp (34). Interestingly, the proportion of cfDNA fragments smaller than 100 bp is higher in CRC patients than in healthy individuals, and the proportion of cfDNA fragments larger than 300 bp is lower in CRC patients than in healthy individuals (35). Although the consistency between cfDNA and genomic DNA in tumor biopsies is still controversial, there is evidence that there is a high degree of consistency between the two (34).Kang et al. performed ultra-deep sequencing of 10 genes (38 kb) with repeated mutations from plasma cfDNA, peripheral blood mononuclear cell genomic DNA, and available genomic DNA from matching tumor tissues from 54 patients with metastatic CRC. Analysis showed that the two types of samples generally had a high degree of coincidence, with a coincidence rate of 93% (34). Studies have pointed out that the level of cfDNA is significantly increased in the blood of patients with metastatic CRC (35). In comparison with other colorectal-related diseases, the cfDNA level of patients with primary CRC is significantly higher than that of patients with intestinal polyps (36).
As common CRC mutation genes, KRAS and BRAF have a high detection value. cfDNA analysis can be used to detect KRAS and BRAF mutations. In the case of KRAS mutations, cfDNA analysis has diagnostic value for patients with a poor prognosis (37, 38). In addition, analysis of BRAF mutations in cfDNA can be used as a prognostic indicator for CRC. In patients with high levels of BRAF mutations, overall survival is usually low (39). Similarly, the detection of BRAF mutations in cfDNA plays an important role in the selection of treatment options for CRC patients (40). Siravegna et al. used digital PCR to quantitatively analyze the mutation levels of undetermined genes in the ctDNA of metastatic CRC patients, revealing the dynamic process of tumor evolution in the process of drug therapy, and also providing a basis for metastatic CRC-targeted therapy strategies (41). These studies have shown that the detection of cfDNA has guiding significance for the diagnosis, prognosis, and postoperative chemotherapy of CRC patients.
Because genetic and epigenetic changes affect the development of CRC, the use of DNA methylation as a biomarker to improve current diagnosis, screening, prognosis, and treatment has great potential (42). Abnormal methylation patterns can be detected in cfDNA. For example, the hypermethylation of ALX4, FBN2, HLTF, P16, TMEFF1, and VIM in cfDNA is associated with poor prognosis in patients with CRC. At the same time, the hypermethylation of APC, NEUROG1, RASSF1A, RASSF2A, SDC2, SEPT9, TAC1, and THBD in cfDNA can be detected in patients with early CRC, while the hypermethylation of P16 and TFPI2 in cfDNA is related to the recurrence of CRC (43). The methylation status of the SEPT9 gene promoter in cfDNA can be used as a biomarker for detecting early CRC with a sensitivity of 30%–75% and specificity of approximately 90%. The large difference in sensitivity and specificity may be related to the relatively small number of patients examined and the technology used to detect SEPT9 methylation (44–48).
miRNAs are a group of non-coding RNAs, 18–25 nucleotides in length, whose main function is to affect the expression of post-translational genes. They work by binding to the 3′-untranslated region of their target gene mRNA (49–51). The expression of miRNAs is related to tumor tissue type, degree of differentiation, aggressiveness, treatment prognosis, and other clinical biological characteristics (52). Tumor cells release miRNAs specific to their metabolism into the peripheral blood, which are called ctmiRNAs, and they are carried by exosomes, which can protect them from RNase degradation (53, 54). The detection of ctmiRNAs was verified by fluorescence probe q-PCR. Specific miRNA primers and joint primers were used for rapid amplification of miRNA during the detection process. At the same time, fluorescence probe was used to combine the detection process to ensure high specificity and sensitivity of the amplification process. miRNA molecules can be specifically detected and miRNA molecules with high similarity can be distinguished (Figure 3). Therefore, the detection of ctmiRNAs in the serum or plasma of CRC patients has great potential for diagnosis, prognosis, and treatment guidance.
ctmiRNAs can be used not only to screen patients with CRC but also to distinguish CRC from other gastrointestinal tumors. As early as 2008, Hunter et al. found that a subset of 69 ctmiRNAs were highly expressed in the serum of CRC patients but were almost undetectable in the serum of healthy volunteers (55) Kosaka et al. performed ctmiRNA expression profile analysis using tissue and plasma samples from CRC patients and healthy volunteers. They found that the levels of miR-17-3p and miR-92 were significantly increased in the serum of CRC patients; however, after surgical removal of the tumor, their expression was significantly decreased, suggesting that the expression of miR-17-3p and miR-92 in serum was specific for screening CRC (56). miR-21 is a secreted miRNA and one of the most intensively studied ctmiRNAs. It is usually released by cancer cells and is abundant in plasma and serum (57). miR-21 participates in the metabolic process of CRC by targeting the expression of PTEN, PDCD, and DKK2 and activating the Wnt/β-catenin signaling pathway (58–60). Some studies have pointed out that the expression level of miR-21 in the serum of CRC patients is positively correlated with its expression level in tumor tissues, and miR-21 expression in the serum is significantly higher in CRC patients than in patients with colorectal adenoma and healthy individuals. Therefore, serum miR-21 may become a promising biomarker for screening CRC patients (61–64).
Exosomes are small vesicles secreted by cells that contain proteins, DNA, mRNAs, and some non-coding RNAs (65). At present, exosomes are mostly isolated by ultrafast centrifugation, magnetic bead immunocapture, precipitation or filtration. The size and morphology were analyzed by electron microscopy, cell surface markers were analyzed by flow cytometry, proteins were analyzed by Western blot and ELISA, or RNA was analyzed by qPCR and next-generation sequencing (Figure 4).
They are means of communication between cells. Studies have found that exosomes are related to the occurrence and development of tumors, and there is a correlation between metastasis and drug resistance (66–68). Tumor cells use exosomes as carriers to help them escape immune system surveillance. On the one hand, exosomes guide the direction of tumor cell metastasis, and on the other hand, they also create a microenvironment suitable for tumor growth. The information carried by exosomes is diverse (69, 70). The proteins and nucleic acids they carry can be used for the early diagnosis of cancer, recurrence monitoring, drug resistance monitoring, and other related analyses (71, 72). Moreover, exosomes are more abundant than CTCs, which makes them easier to be enriched. Exosomes effectively protect the nucleic acids they carry from degradation, which overcomes the problem that ctDNA is easily degraded in blood, and have broad prospects in clinical applications.
Exosomes are involved in the occurrence and development of a variety of malignant tumors and have been considered by many scholars as a reliable biomarker that can be used to assist cancer prediction and diagnosis and guide treatment (73, 74)Exosomes involved in the occurrence and progression of malignant tumors are called tumor-derived exosomes (TDEs). TDEs are widely involved in tumor cell proliferation and metastasis and mediate the transmission of information between tumor cells and the tumor microenvironment, thereby regulating related signaling pathways, transformation of recipient cells, and expression of tumor genes. The tumor-associated miRNAs carried by TDEs interact with stromal cells in the tumor microenvironment to regulate tumor progression, angiogenesis, and immune escape, thereby causing changes in cell metabolism (75). In a recent study, Shang et al. identified a new type of CRC-derived exosomal circular RNA, called circPACRGL, which is a circulating exosomal RNA secreted by CRC cells (76) circPACRGL acts as a “sponge” to adsorb miR-142-3p/miR-506-3p and promote the proliferation, migration, and invasion of CRC cells through the transforming growth factor-β1 axis (75). However, TDEs do not only affect the gene expression of malignant tumors, they also participate in the distant metastasis of malignant tumors. For example, CXCL13 induces the formation of a pre-metastatic niche, which in turn promotes liver metastasis of CRC (76). Zhao et al. found that CRC cell-derived exosomes mediate miR-934 to induce the polarization of M2 macrophages by down-regulating the expression of PTEN and activating the PI3K/AKT signaling pathway. Polarized M2 macrophages can secrete chemokines (77). Liu et al. found that exosome-mediated miR-106b-3p promotes distant metastasis of CRC by down-regulating the expression of DLC-1 (78). Zou et al. reported that serum exosome-mediated miR-150-5p expression levels are significantly reduced in CRC patients and closely related to poor differentiation, positive lymph node metastasis, and advanced TNM staging (79). Similarly, Liu et al. found that circulating exosome-mediated miR-27a/miR-130a are highly expressed in the plasma of CRC patients, and their plasma concentration is negatively correlated with prognosis (80). Ren et al. found that miR-196b-5p is highly enriched in serum exosomes of CRC patients and related to poor prognosis (81). Some exosome-mediated miRNAs can also be used to predict treatment response, for example, miR-21-5p, miR-1246, miR-1229-5p, and miR-96-5p in 5-FU/oxaliplatin therapy. Their expression is significantly higher in oxaliplatin-resistant patients than in a sensitive control group, which shows that they have great value in identifying 5-FU/oxaliplatin-resistant/sensitive patients (82). Exosome-mediated miRNAs can also be used as biomarkers for recurrence in CRC patients. For example, the expression level of miR-17-92a mediated by exosomes in serum is related to the recurrence of CRC, and patients with high serum miR-17-92a levels often have a poor prognosis (83)
Although there have been a large number of studies, research assessing the feasibility of exosomes as biomarkers in malignant tumors is only in its infancy. The main disadvantage of using exosomes is the lack of a technical consensus, which leads to differences in the results presented by different technologies. Therefore, before exosomes become clinical biomarkers, efforts must be made to standardize every procedure for their detection.
Metabolomics is an important branch of systems biology as well as a supplement to genomics, transcriptomics, and proteomics. Metabolomics integrates information modeling and systems mainly through high-throughput detection and data processing (84). Metabolomics can identify metabolites with a molecular weight of less than 1 kDa, such as sugars, lipids, and vitamins. The changes in their metabolic levels can be used to determine pathophysiological state at a certain period of time. Small molecular metabolites in this state can become markers for disease screening and early diagnosis (85). The research process for metabolomics involves sample preparation, detection and analysis of the metabolites in the sample, and statistical processing of the data obtained from metabolite analysis of the sample (Figure 5). Samples for detection and analysis are often biological fluids (urine and blood), cells, or tissues that can be obtained easily in a non-invasive manner (86).
Serum has always been the sample of choice for the identification of metabolites because it is easy to obtain and reflects a patient’s metabolic profile at the time of collection. Qiu et al. analyzed the serum metabolomics characteristics of 64 CRC patients and 65 healthy individuals using two methods, i.e., gas chromatography high-throughput time-of-flight (TOF) mass spectrometry (MS) and ultra-high pressure liquid chromatography quadruple-TOF-MS, and identified 33 different metabolites, among which the abnormal metabolism of glycolysis, arginine, proline, and fatty acid oleamide may be related to the pathogenesis of CRC (87). Uchiyama et al. used capillary electrophoresis-TOF-MS to analyze samples from 56 patients with CRC, 59 patients with colon adenoma, and 60 healthy individuals, and identified 139 metabolites. Among them, benzoic acid may be a diagnostic marker for CRC (88).
The analysis of metabolites in plasma is not only limited to CRC and healthy individuals but can also be used for precancerous lesions related to CRC. Gumpenberger et al. compared the plasma metabolites of 88 patients with CRC and 400 patients with colorectal adenoma. Compared with colorectal adenoma patients, the plasma levels of 1-methylnicotinamide and carnitine were higher, the concentrations of bilirubin, lysolecithin, choline, and polyunsaturated fatty acids were decreased, while taurine, caffeine-related metabolites, and hypoxanthine were increased in patients with CRC (89).
In patients with CRC at different stages, the analysis of metabolites in plasma also has a certain guiding role. For example, in serum metabolite analysis of 744 patients with stage I–IV CRC, compared with stage I patients, there was no significant difference in plasma metabolite concentrations in stage II patients; sphingomyelin C26:0 concentration was significantly reduced and sphingomyelin C18:0 and phosphatidylcholine C32:0 plasma concentrations were increased in stage III patients; citrulline, histidine, phosphatidylcholine (diacyl) C34:4, and phospholipids were significantly reduced and acylcholine (acyl-alkyl) C40:1 and lysophosphatidylcholine (acyl) C16:0 and C17:0 were significantly reduced in stage IV patients (90).
In addition, metabolite analysis has guiding significance for the differentiation of LCC and RCC. Deng et al. used ultra-high performance liquid chromatography quadruple-TOF-MS to analyze the plasma metabolites of 147 patients with LCC and 105 patients with RCC. Compared with LCC patients, anserine, L-arginine, gamma-glutamyl gamma-aminobutyraldehyde, and pyridoxal 5′-phosphate levels were reduced in RCC patients. In contrast, compared with LCC patients, trimethylamine oxide and indoxyl sulfate levels were increased in RCC patients (91).
In summary, liquid biopsies have great significance for the individualized and precise treatment of CRC. For liquid biopsy analytes, ctDNA, CTCs, ctmiRNAs, exosomes, and metabolites have their own advantages and disadvantages (Table 1). In essence, they all obtain tumor genetic material and metabolites for molecular diagnosis. They should not exclude or oppose each other but should be complementary, reflecting the characteristics of tumors from different times, areas, and perspectives. Obviously, the more accurate and comprehensive our understanding of the molecular characteristics of tumors more targeted and precise treatment strategies can be developed. However, the constantly changing characteristics of tumor heterogeneity and the fact that tissue specimens are not easy to obtain have made the role and value of liquid biopsies increasingly important. We believe that with the continued improvement and development of liquid biopsy-related technologies, liquid biopsies will play an increasingly important role in individualized precision medicine for CRC.
JHH, NTX and ZPH wrote the original draft, JS, SBW, ZHG and WFL reviewed and edited the manuscript. HWC and JHL supervised the writing. All authors have contributed the manuscript and approved the submitted version.
This work was supported by Science and Technology Project of Panyu District, Guangzhou (2019-Z04-02; 2020-Z04-026; 2021-Z04-053), Guangzhou Health and Family Planning Commission Program (No. 20181A011118; 20192A011027; 20191A011119; 20201A010085; 20212A010025), Guangzhou Science and Technology Plan Project (No. 202002030032 ), Medical Science and Technology Research Foundation of Guangdong Province (No. A2020304; A2022524), Scientific Research Fund project of Guangzhou Panyu Central Hospital (No. 2021Y002; 2021Y004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CRC, Colorectal Cancer; TNM, T, Primary Tumor; N, Regional Lymph Nodes, M: Distant Metastasis; CTC, Circulating Tumor Cell; CtDNA, Circulating Tumor DNA; CtmiRNA, Circulating Tumor miRNA; CfDNA, Circulating Free DNA; LCC, Left colon cancer; RCC, Right colon cancer; PBMC, Peripheral blood mononuclear cell; OS, Overall survival; PCR, Polymerase Chain Reaction; mCRC, Metastatic colon cancer; ALX4, Aristaless-like Homeobox 4; FBN2, fibrillin-2; HLTF, Helicase-like transcription factor; TMEFF1, transmembrane protein with EGF-like and two follistatinlike domains 1; VIM, vimentin; APC, Adenomatous Polyposis Colon; NEUROG1, Neurogenin 1; P16, Cyclin-dependent kinase inhibitor 2a; RASSF1A, Ras association domain family member 1; RASSF2A, Ras association domain family member 2; SDC2, Syndecan 2; SEPT9, Septin 9; TAC1, tachykinin precursor 1; THBD, Thrombomodulin; TFPI, Tissue factor pathway inhibitor 2; CRCR, Colorectal cancer recurrence; PTEN, phosphatase and tensin homolog deleted on chromosome ten; PDCD, Programmed cell death; DKK2, Dickkopf 2; CRA, colorectal adenoma; TDE, Tumor-derived exosomes; M2, Macrophages 2; CXCL13, chemokine (C-X-C motif) ligand 13; DLC-1, Deleted in Liver Cancer-1; GC-TOF-MS, Gas Chromatography High Throughput Time of Flight Mass Spectrometry; UPLC-QTOF-MS, Ultra high pressure liquid chromatography time-of-flight mass spectrometer; LysoPC, Lysophosphatidylcholine; EV, Extracellular vesicle foam; ELISA, Enzyme-Linked Immunosorbnent Assay.
1. GBD 2017 Colorectal Cancer Collaborators. The Global, Regional,and National Burden of Colorectal Cancer and Its Attributable Risk Factors in 195 Countries and Territories, 1990-2017:a Systematic Analysis for the Global Burden of Disease Study. Lancet Gastroenterol Hepatol (2019) 4(12):913–33. doi: 10.1016/S2468-1253(19)30345-0
2. Bray F, Ferlay J, Soerjomataram I, Siegel R, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2018) 68(6):94–424. doi: 10.3322/caac.21492
3. Araghi M, Soerjomataram I, Jenkins M, Brierley J, Morris E, Bray F, et al. Global Trends in Colorectal Cancer Mortality: Projections to the Year 2035. Int J Cancer (2019) 144(12):2992–3000. doi: 10.1002/ijc.32055
4. Sharp L, O'Leary E, O'Ceilleachair A, Skally M, Hanly P. Financial Impact of Colorectal Cancer and Its Consequences: Associations Between Cancer-Related Financial Stress and Strain and Health-Related Quality of Life. Dis Colon Rectum (2018) 61(1):27–35. doi: 10.1097/DCR.0000000000000923
5. Duineveld LA, van Asselt KM, Bemelman WA, Smits AB, Tanis PJ, van Weert HC, et al. Symptomatic and Asymptomatic Colon Cancer Recurrence: A Multicenter Cohort Study. Ann Fam Med (2016) 14(3):215–20. doi: 10.1370/afm.1919
6. Zare-Bandamiri M, Fararouei M, Zohourinia S, Daneshi N, Dianatinasab M. Risk Factors Predicting Colorectal Cancer Recurrence Following Initial Treatment: A 5-Year Cohort Study. Asian Pac J Cancer Prev (2017) 18(9):2465–70. doi: 10.22034/APJCP.2017.18.9.2465
7. Brenner H, Kloor M, Pox CP. Colorectal Cancer. Lancet (2014) 383(9927):1490–502. doi: 10.1016/S0140-6736(13)61649-9
8. Wu T, Mo Y, Wu C. Prognostic Values of CEA, CA19-9, and CA72-4 in Patients With Stages I-III Colorectal Cancer. Int J Clin Exp Pathol (2020) 13(7):1608–14.
9. Ogunwobi OO, Mahmood F, Akingboye A. Biomarkers in Colorectal Cancer: Current Research and Future Prospects. Int J Mol Sci (2020) 21(15):5311. doi: 10.3390/ijms21155311
10. Li AJ, Li HG, Tang EJ, Wu W, Chen Y, Jiang HH, et al. PIK3CA and TP53 Mutations Predict Overall Survival of Stage II/III Colorectal Cancer Patients. World J Gastroentero (2018) 24(5):631–40. doi: 10.3748/wjg.v24.i5.631
11. Lu T, Li J. Clinical Applications of Urinary Cell-Free DNA in Cancer: Current Insights and Promising Future. Am J Cancer Res (2017) 7(11):2318–32.
12. Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, et al. Identification of Ras Oncogene Mutations in the Stool of Patients With Curable Colorectal Tumors. Science (1992) 256(5053):102–5. doi: 10.1126/science.1566048
13. De Mattos-Arruda L, Mayor R, Ng CKY, Weigelt B, Martínez-Ricarte F, Torrejon D, et al. Cerebrospinal Fluid-Derived Circulating Tumour DNA Better Represents the Genomic Alterations of Brain Tumours Than Plasma. Nat Commun (2015) 6:8839. doi: 10.1038/ncomms9839
14. Stefancu A, Badarinza M, Moisoiu V, Iancu SD, Serban O, Leopold N, et al. SERS-Based Liquid Biopsy of Saliva and Serum From Patients With Sjögren's Syndrome. Anal Bioanal Chem (2019) 411(22):5877–83. doi: 10.1007/s00216-019-01969-x
15. Song Z, Cai Z, Yan J, Shao YW, Zhang Y. Liquid Biopsies Using Pleural Effusion-Derived Exosomal DNA in Advanced Lung Adenocarcinoma. Transl Lung Cancer Res (2019) 8(4):392–400. doi: 10.21037/tlcr.2019.08.14
16. Peterson VM, Castro CM, Chung J, Miller NC, Ullal AV, Castano MD, et al. Ascites Analysis by a Microfluidic Chip Allows Tumor-Cell Profiling. Proc Natl Acad Sci USA (2013) 110(51):E4978–4986. doi: 10.1073/pnas.1315370110
17. Meng S, Tripathy D, Frenkel EP, Shete S, Naftalis EZ, Huth JF, et al. Circulating Tumor Cells in Patients With Breast Cancer Dormancy. Clin Cancer Res (2004) 10(24):8152–62. doi: 10.1158/1078-0432.CCR-04-1110
18. Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Prognostic Significance of Circulating Tumor Cells in Patients With Metastatic Colorectal Cancer. Ann Oncol (2009) 20(7):1223–9. doi: 10.1093/annonc/mdn786
19. Marcuello M, Vymetalkova V, Neves RPL, Duran-Sanchon S, Vedeld HM, Tham E, et al. Circulating Biomarkers for Early Detection and Clinical Management of Colorectal Cancer. Mol Aspects Med (2019) 69:107–22. doi: 10.1016/j.mam.2019.06.002
20. Nicolazzo C, Raimondi C, Gradilone A, Emiliani A, Zeuner A, Francescangeli F, et al. Circulating Tumor Cells in Right- and Left-Sided Colorectal Cancer. Cancers (Basel) (2019) 11(8):1042. doi: 10.3390/cancers11081042
21. Cheng YH, Chen YC, Lin E, Brien R, Jung S, Chen YT, et al. Hydro-Seq Enables Contamination-Free High-Throughput Single-Cell RNA-Sequencing for Circulating Tumor Cells. Nat Commun (2019) 10(1):2163. doi: 10.1038/s41467-019-10122-2
22. Tsai WS, Nimgaonkar A, Segurado O, Chang Y, Hsieh B, Shao HJ, et al. Prospective Clinical Study of Circulating Tumor Cells for Colorectal Cancer Screening. J Clin Oncol (2018) 36(4):556. doi: 10.1200/JCO.2018.36.4_suppl.556
23. Romiti A, Raffa S, Di Rocco R, Roberto M, Milano A, Zullo A, et al. Circulating Tumor Cells Count Predicts Survival in Colorectal Cancer Patients. J Gastrointestin Liver Dis (2014) 23(3):279–84. doi: 10.15403/jgld.2014.1121.233.arom1
24. Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J, et al. Neutrophils Escort Circulating Tumour Cells to Enable Cell Cycle Progression. Nature (2019) 566(7745):553–7. doi: 10.1038/s41586-019-0915-y
25. Mohme M, Riethdorf S, Pantel K. Circulating and Disseminated Tumour Cells - Mechanisms of Immune Surveillance and Escape. Nat Rev Clin Oncol (2017) 14(3):155–67. doi: 10.1038/nrclinonc.2016.144
26. Rahbari NN, Bork U, Schölch S, Reissfelder C, Thorlund K, Betzler A, et al. Metastatic Spread Emerging From Liver Metastases of Colorectal Cancer: Does the Seed Leave the Soil Again? Ann Surg (2016) 263(2):345–52. doi: 10.1097/SLA.0000000000001341
27. van Dalum G, Stam GJ, Scholten LF, Mastboom WJ, Vermes I, Tibbe AG, et al. Importance of Circulating Tumor Cells in Newly Diagnosed Colorectal Cancer. Int J Oncol (2015) 46(3):1361–8. doi: 10.3892/ijo.2015.2824
28. Tan Y, Wu H. The Significant Prognostic Value of Circulating Tumor Cells in Colorectal Cancer: A Systematic Review and Meta-Analysis. Curr Probl Cancer (2018) 42(1):95–106. doi: 10.1016/j.currproblcancer.2017.11.002
29. Bork U, Rahbari NN, Schölch S, Reissfelder C, Kahlert C, Büchler MW, et al. Circulating Tumour Cells and Outcome in Non-Metastatic Colorectal Cancer: A Prospective Study. Br J Cancer (2015) 112(8):1306–13. doi: 10.1038/bjc.2015.88
30. Wang W, Wan L, Wu S, Yang J, Zhou Y, Liu F, et al. Mesenchymal Marker and LGR5 Expression Levels in Circulating Tumor Cells Correlate With Colorectal Cancer Prognosis. Cell Oncol (Dordr) (2018) 41(5):495–504. doi: 10.1007/s13402-018-0386-4
31. Pan RJ, Hong HJ, Sun J, Yu CR, Liu HS, Li PY, et al. Detection and Clinical Value of Circulating Tumor Cells as an Assisted Prognostic Marker in Colorectal Cancer Patients. Cancer Manag Res (2021) 13:4567–78. doi: 10.2147/CMAR.S300554
32. Schwarzenbach H, Hoon DS, Pantel K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat Rev Cancer (2011) 11(6):426–37. doi: 10.1038/nrc3066
33. Casanova-Salas I, Athie A, Boutros PC, Del Re M, Miyamoto DT, Pienta KJ, et al. Quantitative and Qualitative Analysis of Blood-Based Liquid Biopsies to Inform Clinical Decision-Making in Prostate Cancer. Eur Urol (2021) 79(6):762–71. doi: 10.1016/j.eururo.2020.12.037
34. Kang JK, Heo S, Kim HP, Song SH, Yun H, Han SW, et al. Liquid Biopsy-Based Tumor Profiling for Metastatic Colorectal Cancer Patients With Ultra-Deep Targeted Sequencin. PloS One (2020) 15(5):e0232754. doi: 10.1371/journal.pone.0232754
35. Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-Free DNA Comprises an In Vivo Nucleosome Footprint That Informs Its Tissues-Of-Origin. Cell (2016) 164(1-2):57–68. doi: 10.1016/j.cell.2015.11.050
36. Mouliere F, El Messaoudi S, Pang D, Dritschilo A, Thierry AR. Multi-Marker Analysis of Circulating Cell-Free DNA Toward Personalized Medicine for Colorectal Cancer. Mol Oncol (2014) 8(5):927–41. doi: 10.1016/j.molonc.2014.02.005
37. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res (1977) 37(3):646–50.
38. Hao TB, Shi W, Shen XJ, Qi J, Wu XH, Wu Y, et al. Circulating Cell-Free DNA in Serum as a Biomarker for Diagnosis and Prognostic Prediction of Colorectal Cancer. Br J Cancer (2014) 111(8):1482–9. doi: 10.1038/bjc.2014.470
39. Mohan S, Ayub M, Rothwell DG, Gulati S, Kilerci B, Hollebecque A, et al. Analysis of Circulating Cell-Free DNA Identifies KRAS Copy Number Gain and Mutation as a Novel Prognostic Marker in Pancreatic Cancer. Sci Rep (2019) 9(1):11610. doi: 10.1038/s41598-019-47489-7
40. Janku F, Huang HJ, Claes B, Falchook GS, Fu S, Hong D, et al. BRAF Mutation Testing in Cell-Free DNA From the Plasma of Patients With Advanced Cancers Using a Rapid, Automated Molecular Diagnostics System. Mol Cancer Ther (2016) 15(6):1397–404. doi: 10.1158/1535-7163.MCT-15-0712
41. Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal Evolution and Resistance to EGFR Blockade in the Blood of Colorectal Cancer Patients. Nat Med (2015) 21(7):827. doi: 10.1038/nm0715-827b
42. Hong DS, Morris VK, El Osta B, Sorokin AV, Janku F, Fu S, et al. Phase Ib Study of Vemurafenib in Combination With Irinotecan and Cetuximab in Patients With BRAF-Mutated Metastatic Colorectal Cancer and Advanced Cancers. J Clin Oncol (2015) 33(Suppl 15):3511–1. doi: 10.1158/2159-8290.CD-16-0050
43. Lam K, Pan K, Linnekamp JF, Medema JP, Kandimalla R. DNA Methylation Based Biomarkers in Colorectal Cancer: A Systematic Review. Biochim Biophys Acta (BBA) - Rev Cancer (2016) 1866(1):106–20. doi: 10.1016/j.bbcan.2016.07.001
44. Rasmussen SL, Krarup HB, Sunesen KG, Pedersen IS, Madsen PH, Thorlacius-Ussing O, et al. Hypermethylated DNA as a Biomarker for Colorectal Cancer: A Systematic Review. Colorectal Dis (2016) 18(6):549–61. doi: 10.1111/codi.13336
45. Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, et al. DNA Methylation Biomarkers for Blood-Based Colorectal Cancer Screening. Clin Chem (2008) 54(2):414–23. doi: 10.1373/clinchem.2007.095992
46. Grützmann R, Molnar B, Pilarsky C, Habermann JK, Schlag PM, Saeger HD, et al. Sensitive Detection of Colorectal Cancer in Peripheral Blood by Septin 9 DNA Methylation Assay. PloS One (2008) 3(11):e3759. doi: 10.1371/journal.pone.0003759
47. deVos T, Tetzner R, Model F, Weiss G, Schuster M, Distler J, et al. Circulating Methylated SEPT9 DNA in Plasma Is a Biomarker for Colorectal Cancer. Clin Chem (2009) 55(7):1337–46. doi: 10.1373/clinchem.2008.115808
48. Lee HS, Hwang SM, Kim TS, Kim DW, Park DJ, Kang SB, et al. Circulating Methylated Septin 9 Nucleic Acid in the Plasma of Patients With Gastrointestinal Cancer in the Stomach and Colon. Transl Oncol (2013) 6(3):290–6. doi: 10.1593/tlo.13118
49. Masuda T, Hayashi N, Kuroda Y, Ito S, Eguchi H, Mimori K.. MicroRNAs as Biomarkers in Colorectal Cancer. Cancers (2017) 9(9):124. doi: 10.3390/cancers9090124
50. Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: Synthesis, Mechanism, Function, and Recent Clinical Trials. Biochim Biophys Acta (2010) 1803(11):1231–43. doi: 10.1016/j.bbamcr.2010.06.013
51. Mansoori B, Mohammadi A, Shirjang S, Baradaran B. Micro-RNAs: The New Potential Biomarkers in Cancer Diagnosis, Prognosis and Cancer Therapy. Cell Mol Biol (Noisy-le-grand) (2015) 61(5):1–10.
52. Mansoori B, Mohammadi A, Shir Jang S, Baradaran B. Mechanisms of Immune System Activation in Mammalians by Small Interfering RNA (siRNA). Artif Cells Nanomed Biotechnol (2016) 44(7):1589–96. doi: 10.3109/21691401.2015.1102738
53. Brase JC, Wuttig D, Kuner R, Sültmann H. Serum microRNAs as Non-Invasive Biomarkers for Cancer. Mol Cancer (2010) 9:306. doi: 10.1186/1476-4598-9-306
54. Montano M. MicroRNAs: miRRORS of Health and Disease. Transl Res (2011) 157(4):157–62. doi: 10.1016/j.trsl.2011.02.001
55. Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA Expression in Human Peripheral Blood Microvesicles. PloS One (2008) 3(11):e3694. doi: 10.1371/journal.pone.0003694
56. Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in Body Fluid: A New Potential Biomarker for Cancer Diagnosis and Prognosis. Cancer Sci (2010) 101(10):2087–92. doi: 10.1111/j.1349-7006.2010.01650.x
57. Kawakita A, Yanamoto S, Yamada S, Naruse T, Takahashi H, Kawasaki G, et al. MicroRNA-21 Promotes Oral Cancer Invasion via the Wnt/β-Catenin Pathway by Targeting DKK2. Pathol Oncol Res (2014) 20(2):253–61. doi: 10.1007/s12253-013-9689-y
58. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in Serum: A Novel Class of Biomarkers for Diagnosis of Cancer and Other Diseases. Cell Res (2008) 18(10):997–1006. doi: 10.1038/cr.2008.282
59. Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, et al. Differential Expression of microRNAs in Plasma of Patients With Colorectal Cancer: A Potential Marker for Colorectal Cancer Screening. Gut (2009) 58(10):1375–81. doi: 10.1136/gut.2008.167817
60. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 Regulates Expression of the PTEN Tumor Suppressor Gene in Human Hepatocellular Cancer. Gastroenterology (2007) 133(2):647–58. doi: 10.1053/j.gastro.2007.05.022
61. Talotta F, Cimmino A, Matarazzo MR, Casalino L, De Vita G, D'Esposito M, et al. An Autoregulatory Loop Mediated by miR-21 and PDCD4 Controls the AP-1 Activity in RAS Transformation. Oncogene (2009) 28(1):73–84. doi: 10.1038/onc.2008.370
62. Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, et al. Serum miR-21 as a Diagnostic and Prognostic Biomarker in Colorectal Cancer. J Natl Cancer Inst (2013) 105(12):849–59. doi: 10.1093/jnci/djt101
63. Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, et al. Plasma miR-21: A Potential Diagnostic Marker of Colorectal Cancer. Ann Surg (2012) 256(3):544–51. doi: 10.1097/SLA.0b013e318265bd6f
64. Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, et al. Serum miR-21 and miR-92a as Biomarkers in the Diagnosis and Prognosis of Colorectal Cancer. Tumour Biol (2013) 34(4):2175–81. doi: 10.1007/s13277-013-0753-8
65. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: Composition, Biogenesis, and Mechanisms in Cancer Metastasis and Drug Resistance. Mol Cancer (2019) 18(1):75. doi: 10.1186/s12943-019-0991-5
66. Guo W, Li Y, Pang W, Shen H. Exosomes: A Potential Therapeutic Tool Targeting Communications Between Tumor Cells and Macrophages. Mol Ther (2020) 28(9):1953–64. doi: 10.1016/j.ymthe.2020.06.003
67. Tutanov O, Orlova E, Proskura K, Grigor’eva A, Yunusova N, Tsentalovich Y, et al. Proteomic Profiling of Plasma and Total Blood Exosomes in Breast Cancer: A Potential Role in Tumor Progression, Diagnosis, and Prognosis. Front Oncol (2020) 10(580891):1–15. doi: 10.3390/biom10040495
68. Masaoutis C, Korkolopoulou P, Theocharis S. Exosomes in Sarcomas: Tiny Messengers With Broad Implications in Diagnosis, Surveillance, Prognosis and Treatment. Cancer Lett (2019) 449:172–7. doi: 10.1016/j.canlet.2019.02.025
69. Li L, Cao B, Liang X, Lu S, Luo H, Wang Z, et al. Microenvironmental Oxygen Pressure Orchestrates an Anti- and Pro-Tumoral γδ T Cell Equilibrium via Tumor-Derived Exosomes. Oncogene (2019) 38(15):2830–43. doi: 10.1038/s41388-018-0627-z
70. Lukic A, Wahlund CJE, Gómez C, Brodin D, Samuelsson B, Wheelock CE, et al. Exosomes and Cells From Lung Cancer Pleural Exudates Transform LTC4 to LTD4, Promoting Cell Migration and Survival via Cyslt1. Cancer Lett (2019) 444:1–8. doi: 10.1016/j.ymthe.2019.07.003
71. Jiang K, Yang J, Guo S, Zhao G, Wu H, Deng G. Peripheral Circulating Exosome-Mediated Delivery of miR-155 as a Novel Mechanism for Acute Lung Inflammation. Mol Ther (2019) 27(10):1758–71. doi: 10.1016/j.ymthe.2019.07.003
72. Jabbari N, Akbariazar E, Feqhhi M, Rahbarghazi R, Rezaie J. Breast Cancer-Derived Exosomes: Tumor Progression and Therapeutic Agents. J Cell Physiol (2020) 235(10):6345–56. doi: 10.1002/jcp.29668
73. Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, et al. Isolation and Characterization of Exosomes for Cancer Research. J Hematol Oncol (2020) 13(1):152. doi: 10.1186/s13045-020-00987-y
74. Kok VC, Yu CC. Cancer-Derived Exosomes: Their Role in Cancer Biology and Biomarker Development. Int J Nanomed (2020) 15:8019–36. doi: 10.2147/IJN.S272378
75. Tomasetti M, Lee W, Santarelli L, Neuzil J. Exosome-Derived microRNAs in Cancer Metabolism: Possible Implications in Cancer Diagnostics and Therapy. Exp Mol Med (2017) 49(1):e285. doi: 10.1038/emm.2016.153
76. Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL Promotes Progression of Colorectal Cancer via the miR-142-3p/miR-506-3p- TGF-β1 Axis. Mol Cancer (2020) 19(1):117. doi: 10.1186/s12943-020-01235-0
77. Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, et al. Tumor-Derived Exosomal miR-934 Induces Macrophage M2 Polarization to Promote Liver Metastasis of Colorectal Cancer. J Hematol Oncol (2020) 13(1):156. doi: 10.1186/s13045-020-00991-2
78. Liu H, Liu Y, Sun P, Leng K, Xu Y, Mei L, et al. Colorectal Cancer-Derived Exosomal miR-106b-3p Promotes Metastasis by Down-Regulating DLC-1 Expression. Clin Sci (Lond) (2020) 134(4):419–34. doi: 10.1042/CS20191087
79. Zou SL, Chen YL, Ge ZZ, Qu YY, Cao Y, Kang ZX. Downregulation of Serum Exosomal miR-150-5p Is Associated With Poor Prognosis in Patients With Colorectal Cancer. Cancer biomark (2019) 26(1):69–77. doi: 10.3233/CBM-190156
80. Liu X, Pan B, Sun L, Chen X, Zeng K, Hu X, et al. Circulating Exosomal miR-27a and miR-130a Act as Novel Diagnostic and Prognostic Biomarkers of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev (2018) 27(7):746–54. doi: 10.1158/1055-9965.EPI-18-0067
81. Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y, et al. Maintenance of Cancer Stemness by miR-196b-5p Contributes to Chemoresistance of Colorectal Cancer Cells via Activating STAT3 Signaling Pathway. Oncotarget (2017) 8(30):49807–23. doi: 10.18632/oncotarget.17971
82. Jin G, Liu Y, Zhang J, Bian Z, Yao S, Fei B, et al. A Panel of Serum Exosomal microRNAs as Predictive Markers for Chemoresistance in Advanced Colorectal Cancer. Cancer Chemother Pharmacol (2019) 84(2):315–25. doi: 10.1007/s00280-019-03867-6
83. Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, et al. Exosomal microRNA in Serum Is a Novel Biomarker of Recurrence in Human Colorectal Cance. Br J Cancer (2015) 113(2):275–81. doi: 10.1038/bjc.2015.201
84. Nicholson JK, Lindon JC, Holmes E. 'Metabonomics': Understanding the Metabolic Responses of Living Systems to Pathophysiological Stimuli via Multivariate Statistical Analysis of Biological NMR Spectroscopic Data. Xenobiotica (1999) 29(11):1181–9. doi: 10.1080/004982599238047
85. Fiehn O. Combining Genomics, Metabolome Analysis, and Biochemical Modelling to Understand Metabolic Networks. Comp Funct Genomics (2001) 2(3):155–68. doi: 10.1002/cfg.82
86. Kuehnbaum NL, Britz-McKibbin P. New Advances in Separation Science for Metabolomics: Resolving Chemical Diversity in a Post-Genomic Era. Chem Rev (2013) 113(4):2437–68. doi: 10.1021/cr300484s
87. Qiu Y, Cai G, Su M, Chen T, Zheng X, Xu Y, et al. Serum Metabolite Profiling of Human Colorectal Cancer Using GC-TOFMS and UPLC-QTOFMS. J Proteome Res (2009) 8(10):4844–50. doi: 10.1021/pr9004162
88. Uchiyama K, Yagi N, Mizushima K, Higashimura Y, Hirai Y, Okayama T, et al. Serum Metabolomics Analysis for Early Detection of Colorectal Cancer. J Gastroenterol (2017) 52(6):677–94. doi: 10.1007/s00535-016-1261-6
89. Gumpenberger T, Brezina S, Keski-Rahkonen P, Baierl A, Robinot N, Leeb G, et al. Untargeted Metabolomics Reveals Major Differences in the Plasma Metabolome Between Colorectal Cancer and Colorectal Adenomas. Metabolites (2021) 11(2):119. doi: 10.3390/metabo11020119
90. Geijsen AJMR, van Roekel EH, van Duijnhoven FJB, Achaintre D, Bachleitner-Hofmann T, Baierl A, et al. Plasma Metabolites Associated With Colorectal Cancer Stage: Findings From an International Consortium. Int J Cancer (2020) 146(12):3256–66. doi: 10.1002/ijc.32666
Keywords: colorectal cancer, circulating tumor cell, circulating tumor DNA, circulating tumor miRNA, exosomes
Citation: He JH, Xi NT, Han ZP, Luo WF, Shen J, Wang SB, Li JH, Guo ZH and Cheng HW (2022) The Role of Liquid Biopsy Analytes in Diagnosis, Treatment and Prognosis of Colorectal Cancer. Front. Endocrinol. 13:875442. doi: 10.3389/fendo.2022.875442
Received: 14 February 2022; Accepted: 26 April 2022;
Published: 30 June 2022.
Edited by:
Simone Di Franco, University of Palermo, ItalyReviewed by:
Miriam Gaggianesi, University of Palermo, ItalyCopyright © 2022 He, Xi, Han, Luo, Shen, Wang, Li, Guo and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: HanWei Cheng, ZG9jdGVyd2VpQHNpbmEuY29t; ZhongHui Guo, Nzc1NDk5OTI5QHFxLmNvbQ==; JianHao Li, MTg5MjIyMzgwMzJAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.