
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol., 10 May 2022
Sec. Experimental Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.863360
The giant wrasse Cheilinus undulatus is a protogynous socially hermaphroditic fish. However, the physiological basis of its sex reversal remains largely unknown. cyp19 is a key gender-related gene encoding P450 aromatase, which converts androgens to estrogens. cyp19 transcription regulation is currently unknown in socially sexually reversible fish. We identified NR5A1 by encoding SF-1, and FOXL2 from giant wrasse cDNA and cyp19a1a and cyp19a1b promoter regions were cloned from genomic DNA to determine the function of both genes in cyp19a1 regulation. Structural analysis showed that SF-1 contained a conserved DNA-binding domain (DBD) and a C-terminal ligand-binding domain (LBD). FOXL2 was comprised of an evolutionarily conserved Forkhead domain. In vitro transfection assays showed that SF-1 could upregulate cyp19a1 promoter activities, but FOXL2 could only enhance cyp19a1b promoter transcriptional activity in the HEK293T cell line. Furthermore, HEK293T and COS-7 cell lines showed that co-transfecting the two transcription factors significantly increased cyp19a1 promoter activity. The −120 to −112 bp (5′-CAAGGGCAC-3′) and −890 to −872 bp (5′-AGAGGAGAACAAGGGGAG-3′) regions of the cyp19a1a promoter were the core regulatory elements for SF-1 and FOXL2, respectively, to regulate cyp19a1b promoter transcriptional activity. Collectively, these results suggest that both FOXL2 and SF-1 are involved in giant wrasse sex reversal.
In many teleost species, sex fate is not an irreversible deterministic process. Instead, it is actively regulated via the suppression or activation of opposing genetic networks, creating the potential for flexibility in sexual phenotype in adulthood (1). Under natural conditions, many fish exhibit sequential hermaphroditism, including female-to-male (protogynous), male-to-female (protandrous), or bidirectional (serial) sex changes. Sex changes involve complex coordinated transformations across multiple biological systems, including behavioral, anatomical, neuroendocrine, and molecular axes. P450 aromatase is a key enzyme in the hormonal steroidogenic pathway (2), playing a switch role in converting androgen to estrogen and is the key enzyme in sex determination and differentiation (3). Estrogens and cyp19a1a are critical in controlling ovarian and testicular differentiation in gonochoristic and hermaphroditic fish species (4). Teleosts exhibit intense aromatase activity because of the strong expression of one of the two aromatase genes (aromatase A or cyp19a1a and aromatase B or cyp19a1b) that arise from a gene duplication event. Interestingly, only radial glial cells (RGC) express aromatase B in adult fish (5).
Fushi-tarazu factor-1 (FTZ-F1) is a member of the orphan nuclear receptor family discovered in Drosophila. According to function and expression type, FTZ-F1 gene is divided into two subgroups, and the liver receptor hormone-1 gene (LRH-1) constitutes the nuclear receptor subfamily 5 group A member 2 (NR5A2) subgroup, mainly expressed in the liver. Steroidogenic factor-1 (SF-1/adrenal 4 binding protein (Ad4BP)) is part of the NR5A1 subgroup and is mainly expressed in tissues related to steroid synthesis (6). Therefore, SF-1 is used as a steroid-producing factor to regulate gonadal development and sex determination (7). SF-1 potentially plays a role in transcriptional regulation of P450 aromatase gene (cyp19a1) in medaka ovarian follicles (8). SF-1 directly binds to the cyp19a1 promoter on a conserved binding site to activate cyp19a1 mRNA transcription in chicken ovaries (9). In mammals, SF-1 knockout male mice showed abnormal and underdeveloped testis structure. Additionally, the expression of cyp11a and the steroidogenic acute regulatory protein (STAR), two Leydig cell function markers, was impaired, indicating a defect in androgen biosynthesis (10). Collectively, SF-1 could be crucial in sex reversal in giant wrasses by regulating cyp19 gene transcription.
On the other hand, Forkhead (FH) box (FOX) L2 (FOXL2), another important transcription factor discovered in dermatolysis palpebrarum, is a member of the FOX transcription factor family. In recent decades, FOXL2 has been recognized as a key gene in ovarian differentiation and egg formation in vertebrates (especially mammals) (11). Many FOXL2 target genes have been discovered, including genes involved in steroid production (such as STAR, cyp17, and aromatase), inflammation (such as nuclear factor of activated T cells (NFAT), prostaglandin-endoperoxide synthase (PTGS2), and code dystrophy 2 (COD2)), and apoptosis/detoxification (such as manganese superoxide dismutase (MnSOD)) (12, 13). FOXL2 also participates in aromatase transcriptional regulation (14). In Nile tilapia, FOXL2 directly binds to the promoter region of cyp19a1 through its FH domain to activate cyp19a1 transcription. FOXL2 can also interact with the ligand-binding domain (LBD) of Ad4BP/SF-1 to enhance Ad4BP/SF-1-mediated cyp19a1 transcription (15). Recent studies using catfish have shown that FOXL2 mainly interacts with the cytochrome P450 protein family (16). Similarly, transient transfection of Japanese flounder showed that FOXL2 and cAMP analogs could activate cyp19a1 transcription in vitro (17). These results indicate that FOXL2 may be crucial in ovarian differentiation in bony fish by regulating aromatase expression and steroid production pathways. Recent studies have shown that FOXL2 can directly bind to SF-1 or act as a nuclear receptor co-modulator to regulate ovarian steroidogenesis and follicular development (15). Therefore, in this study, we presumed that FOXL2 and SF-1 were candidate factors that regulate cyp19a1 expression in giant wrasses.
As an imminent danger species, the fishing industry has sharply decreased the giant wrasse population (18). However, artificially reproducing this species is limited by the small number of male fish. As a social animal, a colony of giant wrasses allows only one male to be present (19). Indeed, research on sex reversal in giant wrasses is urgent. In the present study, we identified and cloned SF-1 and FOXL2 cDNA sequences from a giant wrasse and tested both factors in cyp19a1 transcriptional regulation by utilizing two cell lines (HEK293T and COS-7). Examining cyp19a1 promoter deletions and mutations confirmed the SF-1 and FOXL2 binding sites in the cyp19a1 promoter.
A 2-year-old wild and healthy giant wrasse was purchased from the Hainan Qionghai Fishing Port Terminal and Guangzhou Huangsha Fishery Market. The fish was anesthetized in ice, and samples from the brain and gonads were taken and stored in liquid nitrogen for future use. All animal handling procedures were conducted under the Institutional Animal Care and Use Committee of Hainan University.
RNA was extracted using TRIzol reagent, according to the manufacturer’s instructions. The Prime Script™ RT Reagent Kit with a gDNA Eraser (Perfect Real Time) was used to synthesize cDNA templates. The reverse transcription products were stored at −20°C for later use. cDNA fragments were cloned into PGEM-Teasy plasmids using the SF-1 F1, SF-1 R1, FOXL2 F1, and FOXL2 R1 primers to amplify the target fragments. For this study, SF-1 open reading frame (ORF) F1, SF-1 ORF R1, FOXL2 ORF F1, and FOXL2 ORF R1 were used to amplify HindIII, XhoI, and XhoI and EcoRI restriction sites. According to the restriction endonuclease site existing in pCDNA3.1, a suitable enzyme that was the same as the fragment multi-band restriction site was selected to double-digest the pCDNA3.1 vector. The SF-1 and FOXL2 ORF sequences were ligated into the pCDNA3.1 plasmid, and then the recombinant plasmid was transferred into the DH5α competent cell. Finally, the recombinant plasmid containing the target fragment was screened and identified, and the promoter sequence was obtained using the GenomeWalker method. The cloning method was performed according to the Universal GenomeWalker™ 2.0 User Manual. Table 1 lists all the primers used for cloning and vector construction.
Pomoter regions of the cyp19a1a and cyp19a1b were isolated by genome walking. The online software ALGGEN-PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) was used to predict the cyp19a1a and cyp19a1b transcription binding sites of SF-1 and FOXL2. The giant wrasse’s NR5A1 and FOXL2 gene sequences were spliced using DNAMAN software. The ORF was identified using the ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). The National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/) was used to identify the coding regions and amino acid sequences. The online software Signal P4.1 was used to predict the signal peptides of NR5A1 and FOXL2 genes in the giant wrasse. In addition, an adjacency (neighbor joining (NJ)) phylogenetic tree was constructed based on amino acid sequence alignment using MEGA 5.0. GeneDoc software was used for multiple sequence alignments.
In this study, the cyp19a1a and cyp19a1b promoters were analyzed using online software, and the SF-1 and FOXL2 binding positions in their sequences were predicted. The promoters were deleted from the analysis. Primers were designed with restriction sites AAGCTT and GGTACC of restriction endonuclease (HindIII and KpnI). Progressive deletion fragments were constructed using a pGL-4.10 fluorescent expression vector. Based on the effect of transcription factors on the progressive deletion of the promoter, the scope of key transcription binding sites was narrowed, and specific primers were designed to clone the full-length sequence of site-directed mutations by fusion PCR. The ORFs of NR5A1 and FOXL2 were ligated using gene-specific primers, restriction endonucleases, and the pCDNA3.1 vector to construct a site-mutant expression vector.
HEK 293T and COS7 cell lines were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM of L-glutamine, 100 U/ml of penicillin, and 0.1 mg/ml of streptomycin in 5% CO2 and cultured at 37°C. The confluent cells were seeded in a 24-well plate. The transfection rate was maintained at ~ 90% at the time of transfection. The specific experimental components were as follows: 1) 250 ng of complete 5′ upstream or deletion constructs (normal and mutated) of cyp19a1a and cyp19a1b promoters cloned into the pGL-4.10 (firefly luciferase) vector and 2) 200 ng of pcDNA3.1 expression plasmid (Invitrogen, Carlsbad, CA, USA) containing ORF cDNAs encoding SF-1 and FOXL2. A 5 ng/well of Renilla luciferase (CMV) vector (Promega, Madison, WI, USA) was used as an internal control. Luciferase assays were performed 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega). Renilla luciferase activity was used to normalize firefly luciferase activity. Luminescence was measured using a GloMax®96 microplate luminometer (Promega). The plasmids used in the transfection experiments were prepared using an Endo Free Plasmid Isolation Kit.
The experimental cell data were set up in four parallels per group. A one-way ANOVA was performed, and the results are expressed as the mean ± SD. Statistical significance was set at p < 0.05. * represents a significant difference between the data. SPSS and DPS software was used to perform the statistical analyses and constructed graphs using GraphPad Prism version 5.
The SF-1 and FOXL2 cDNA sequences were cloned and sequenced. The gene encoding SF-1 had a 1,467-bp ORF encoding 488 amino acid residues (Figure 1A). The predicted protein (SF-1) had a molecular weight of 55 kDa and a theoretical isoelectric point (pI) of 7.06. FOXL2 had a 921-bp ORF encoding 306 amino acid residues (Figure 1B). The predicted FOXL2 protein had a molecular weight of 34.59 kDa and a theoretical pI of 9.21.

Figure 1 Nucletide and amino acid sequences of C. undulates SF-1 and FOXL2. (A) Nucletide and amino acid sequences of C. undulates SF-1. The black box marks the start codon ATG and the stop codon TGA. The black typeface is the nucleotide sequence, the red typeface is the amino acid sequence. (B) Nucletide and amino acid sequences of C. undulates FOXL2. The black box marks the start codon ATG and the stop codon TAA.
Multiple sequence alignment was conducted to compare the amino acid sequences of SF-1 with those of other vertebrates. It showed that SF-1 was conserved in different aquatic animals (Figure 2A). Conserved domain analysis indicated that the SF-1 amino acid sequence contained two domains: a conserved DNA-binding domain (DBD) and a C-terminal LBD. FOXL2 consisted of an evolutionarily conserved FH domain (Figure 2B). In addition, a phylogenetic tree was constructed based on the SF-1 amino acid sequences from giant wrasse and 13 other species. As Figure 3A shows, SF-1 was most closely related to Labrus bergylta SF-1, clustered with Larimichthys crocea, and formed a subgroup. SF-1 also showed high similarity to other teleosts and had a relatively long homology with mammalian, avian, and amphibian species. The FOXL2 homology comparison results were similar to those of SF-1. FOXL2 showed the closest relationship with L. bergylta FOXL2. FOXL2 also showed relatively distant homology with mammalian, avian, and amphibian species (Figure 3B). In summary, the molecular evolution of SF-1 and FOXL2 was consistent with species evolution.
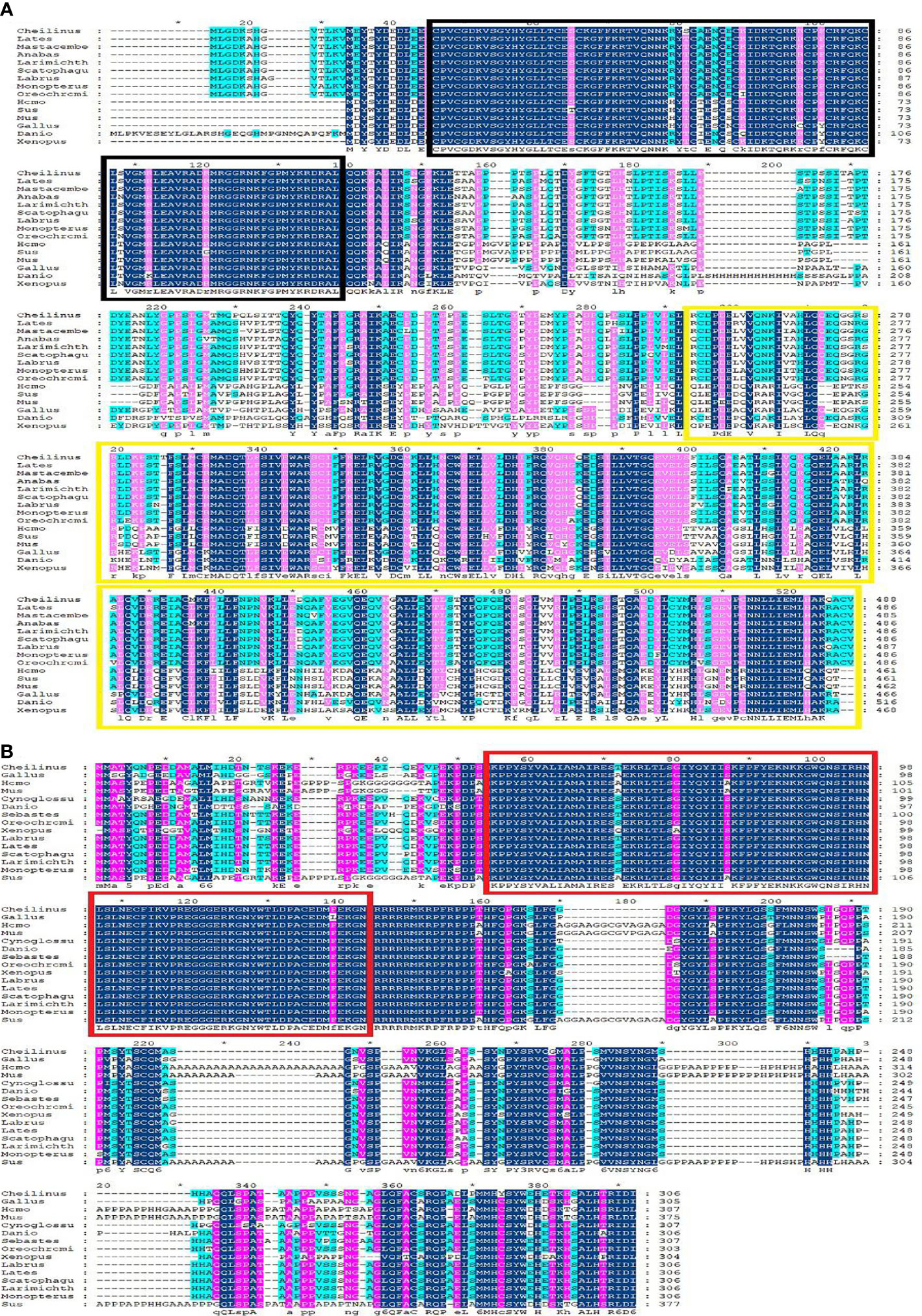
Figure 2 Amino acids sequence alignment of C. undulates SF-1 and FOXL2. (A) Amino acids sequence alignment of C. undulates SF-1. The black box represents the conserved DNA binding domain (DBD). The yellow box represents the C-terminal ligand binding domain (LBD). (B) Amino acids sequence alignment of C. undulates FOXL2. The red box represents the conserved Forkhead (FH) domain.
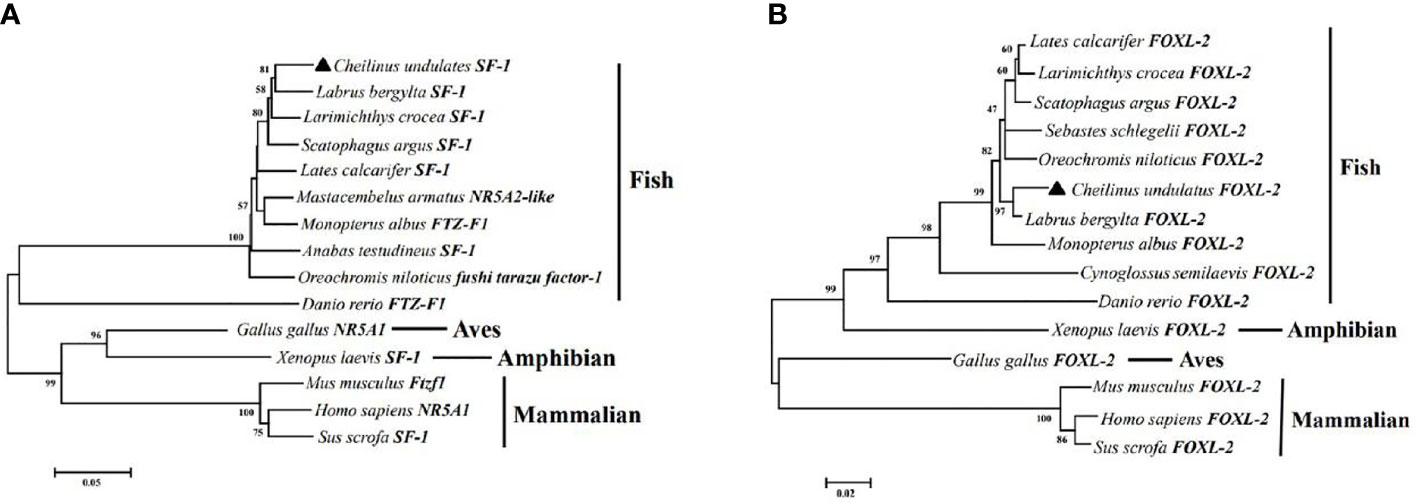
Figure 3 Phylogenetic analysis of C. undulates with other vertebrate SF-1 (A) and FOXL-2 (B). The pylogenetic tree is constructed using the Mega 5.0 adjacency method. Data were resampled with 1,000 bootstrap replicates.
We constructed a luciferase reporter system for cyp19a1a and cyp19a1b promoters and co-transfected the eukaryotic expression vectors of SF-1 and FOXL2 into HEK293T and COS7 cell lines to understand how SF-1 and FOXL2 influence the transcriptional activities of promoters. We used different processing times for different cell lines because the expression time of dual luciferase was unknown after transfection.
In the HEK293T cell line, we found that SF-1 significantly upregulated the transcriptional activity of cyp19a1 promoters 24 and 48 h after transfection, and FOXL2 significantly increased cyp19a1b promoter transcriptional activity only after 48 h of transfection (Figure 4, p < 0.05). At the same time, after SF-1 was transfected into COS-7 cells for 48 h, we observed that the transcriptional activities of cyp19a1a and cyp19a1b promoters were significantly increased. However, FOXL2 did not increase the transcriptional activity of the cyp19a1 promoters. Conversely, SF-1 and FOXL2 co-transfection significantly upregulated cyp19a1 promoter activity (Figure 5, p < 0.05). The results of the two cell lines showed that co-transfection of the two transcription factors significantly increased cyp19a1 promoter activity.
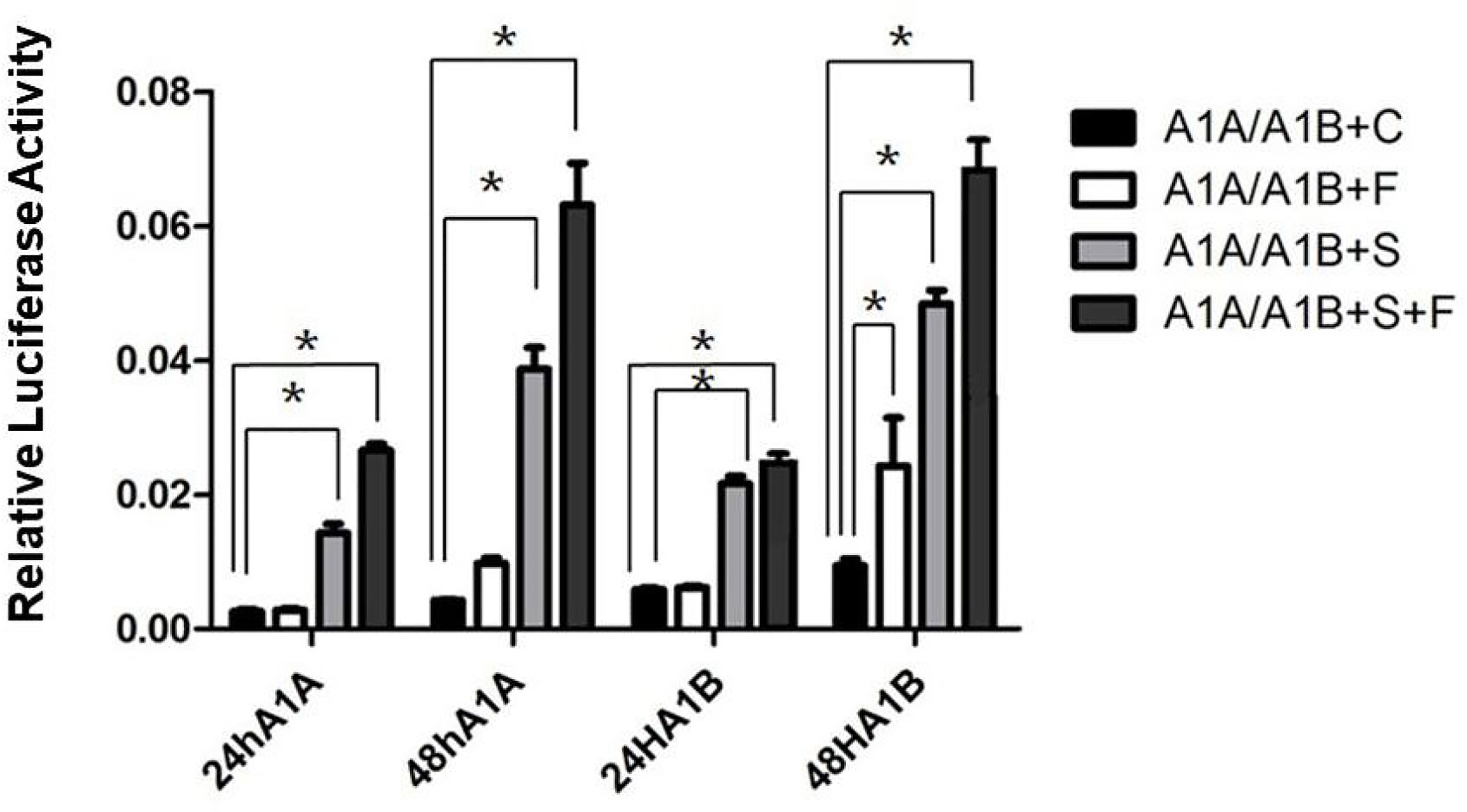
Figure 4 Transcriptional activities of cyp19a1a and cyp19a1b promoters in HEK293T after 24h and 48h transfection. S meaned that only transcription factor SF-1 was added, F meaned that only transcription factor FOXL2 was added. S+F meaned that transcription factors SF-1 and FOXL2 were added at the same time. One-way ANOVA was performed on the data and expressed as mean ± standard deviation (n = 4). *There is a significant difference between the data.
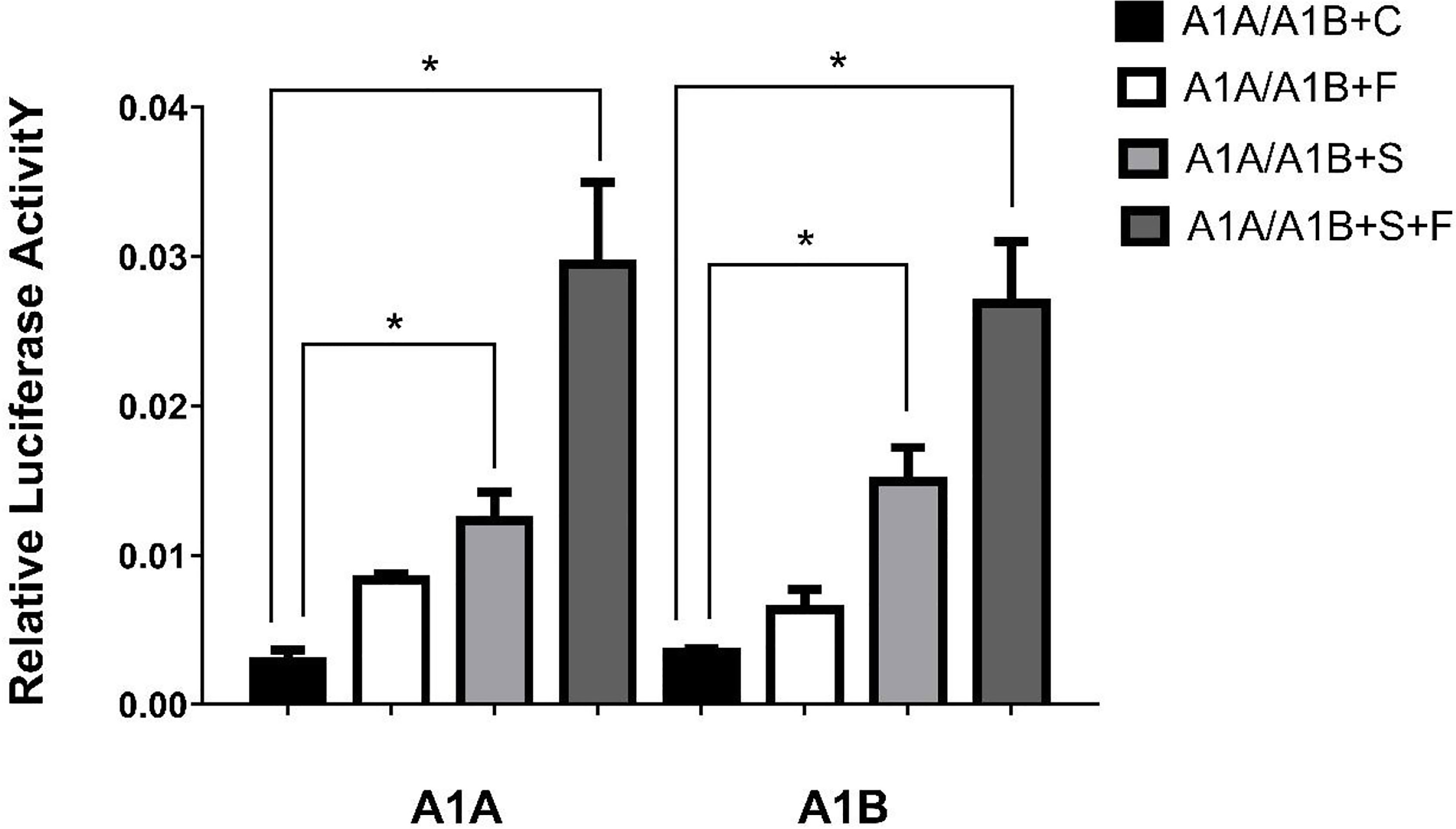
Figure 5 Transcriptional activities of cyp19a1a and cyp19a1b promoters in COS-7 after 48h transfection. S meaned that only transcription factor SF-1 was added, F meaned that only transcription factor FOXL2 was added. S+F meaned that transcription factors SF-1 and FOXL2 were added at the same time. One-way ANOVA was performed on the data and expressed as mean ± standard deviation (n = 4). *There is a significant difference between the data.
We found that there may be three binding sites for the SF-1 transcription factor and no binding sites for FOXL2 using online software to predict the region of the gonadal aromatase cyp19a1a promoter (Figure 6). We identified five SF-1 and three FOXL2 transcription binding sites in the brain aromatase cyp19a1b promoter (Figure 7).

Figure 6 Predicted binding sites of SF-1 on cyp19a1a promoter sequence. The underlined location is the binding site of the predicted transcription binding factor on the initiation. Numbers indicate the distance between the rightmost base in the row and the base of the transcription initiation codo. TSS: Initial site, arrow direction: transcriptional direction.

Figure 7 Predicted binding sites of SF-1 and FOXL2 on cyp19a1b promoter sequence. The underlined location is the binding site of the predicted transcription binding factor on the initiation. Numbers indicate the distance between the rightmost base in the row and the base of the transcription initiation codon. TSS: Initial site, arrow direction: transcriptional direction.
We adopted the technology of constructing a progressive deletion of the promoter, fusion PCR, and site-directed mutagenesis to determine the transcription binding sites of the two transcription factors in the aromatase promoters and identify the key elements in transcription regulation. Finally, we identified the key SF-1 and FOXL2 transcription binding sites in the aromatase cyp19a1 promoters.
With the use of the full length of the cyp19a1 promoter as a reference comparison, deleting −1,291 to −111 bp revealed that the transcriptional activity of the cyp19a1a promoter was significantly downregulated (Figure 8, p < 0.05). We conducted site-directed mutations from −1,265 to −1,256 bp and −119 to −112 bp to determine whether −120 to −112 bp was the key binding site of SF-1 on cyp19a1a. We found that mutating −120 to −112 bp significantly reduced cyp19a1a promoter activity (Figure 9, p < 0.05).
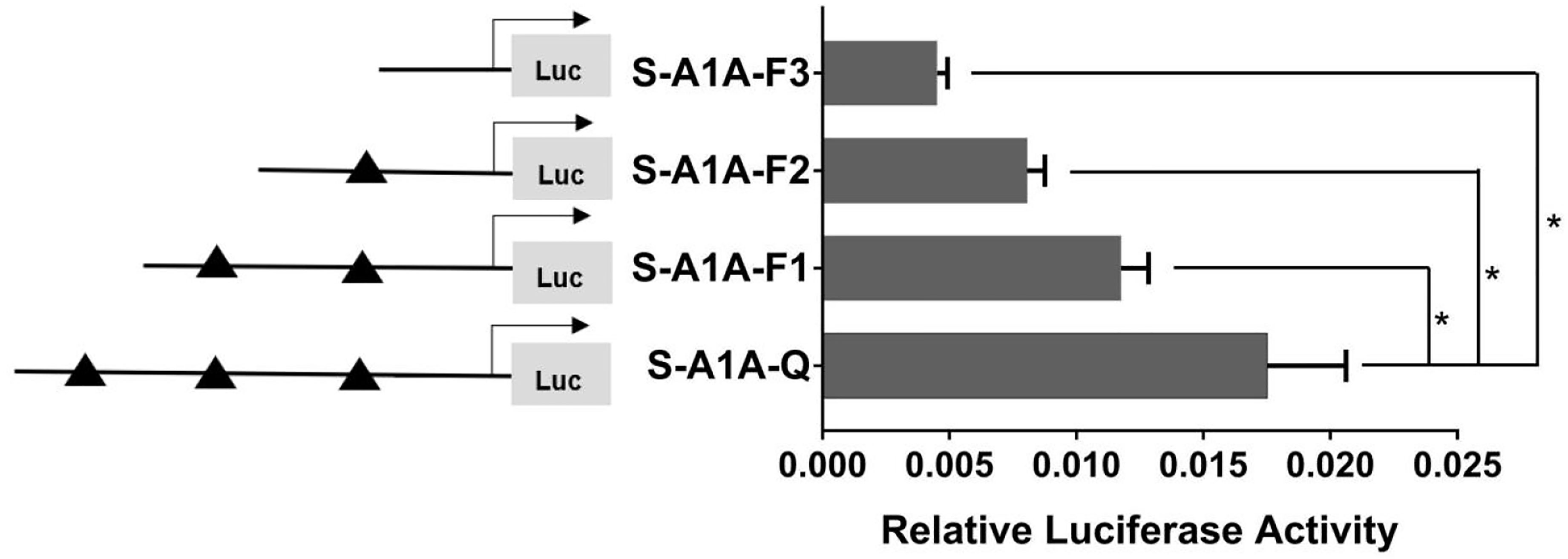
Figure 8 Effect of SF-1 on the transcriptional activity of the cyp19a1a promoter fragment of the progressive deletion site. On the left is a schematic diagram of the promoter deletion and the triangle represents the binding site. The right side is the expression of luciferase in the promoter recombinant vector. S-A1A-F1 (-1291 bp to -1255 bp deletion), S-A1A-F2 (-1291 bp to -111 bp deletion), S-A1A-F3 (-1291 bp to -21 bp deletion). The black triangle represents the SF-1 binding site. One-way ANOVA was performed on the data and expressed as mean ± standard deviation (n = 4). *There is a significant difference between the data.
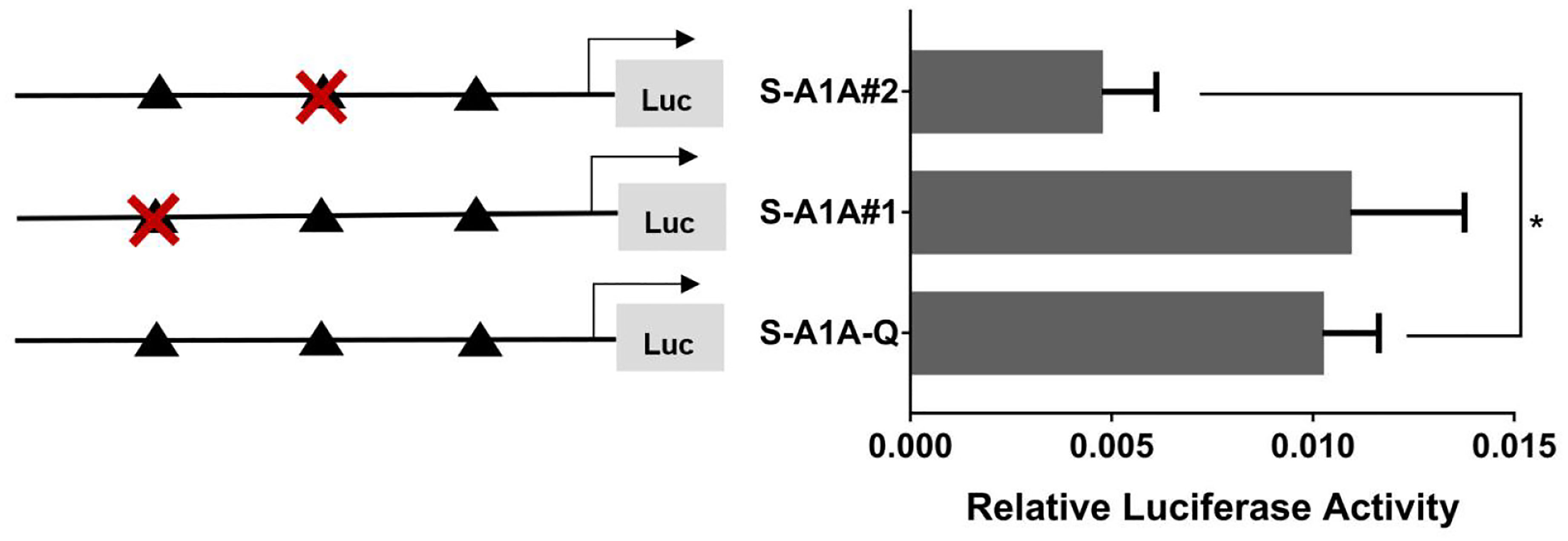
Figure 9 Effect of SF-1 on the transcriptional activity of the cyp19a1a promoter fragment mutated at the transcriptional site. On the left is a schematic diagram of the mutation of the promoter site, and the triangle represents the binding site. The right side is the expression of luciferase in the promoter recombinant vector. The black triangle with a red cross represents the SF-1 mutation site. One-way ANOVA was performed on the data and expressed as mean ± standard deviation (n = 4). *There is a significant difference between the data.
When we deleted the transcription binding sites that predicted SF-1 in the cyp19a1b promoter (Figure 10, p < 0.05), the mutations at these sites did not significantly reduce cyp19a1b promoter activity (Figure 11, p < 0.05). Therefore, we determined that these sites were not the core regulatory sites of SF-1 in the cyp19a1b promoter.
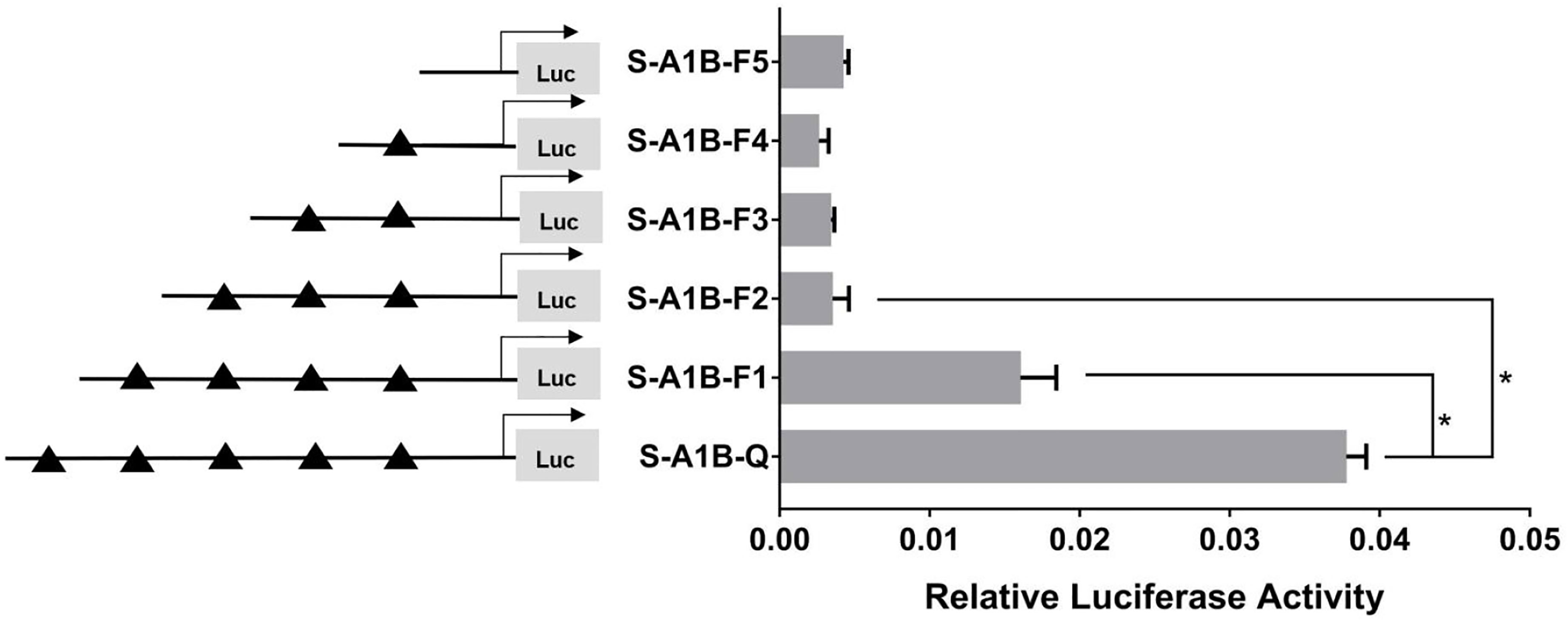
Figure 10 Effect of SF-1 on the transcriptional activity of the cyp19a1b promoter fragment of the progressive deletion site. On the left is a schematic diagram of the promoter deletion and the triangle represents the binding site. The right side is the expression of luciferase in the promoter recombinant vector. S-A1B-F1 (-2379 bp to -1795 bp deletion), S-A1B-F2 (-2379 bp to -1534 bp deletion), S-A1B-F3 (-2379 bp to -1501 bp deletion), S-A1B-F4 (-2379 bp to -864 bp delettion), S-A1B-F5 (2379 bp to -504 bp deletion). The black triangle represents the SF-1 binding site. One-way ANOVA was performed on the data and expressed as mean ± standard deviation (n = 4). *There is a significant difference between the data.
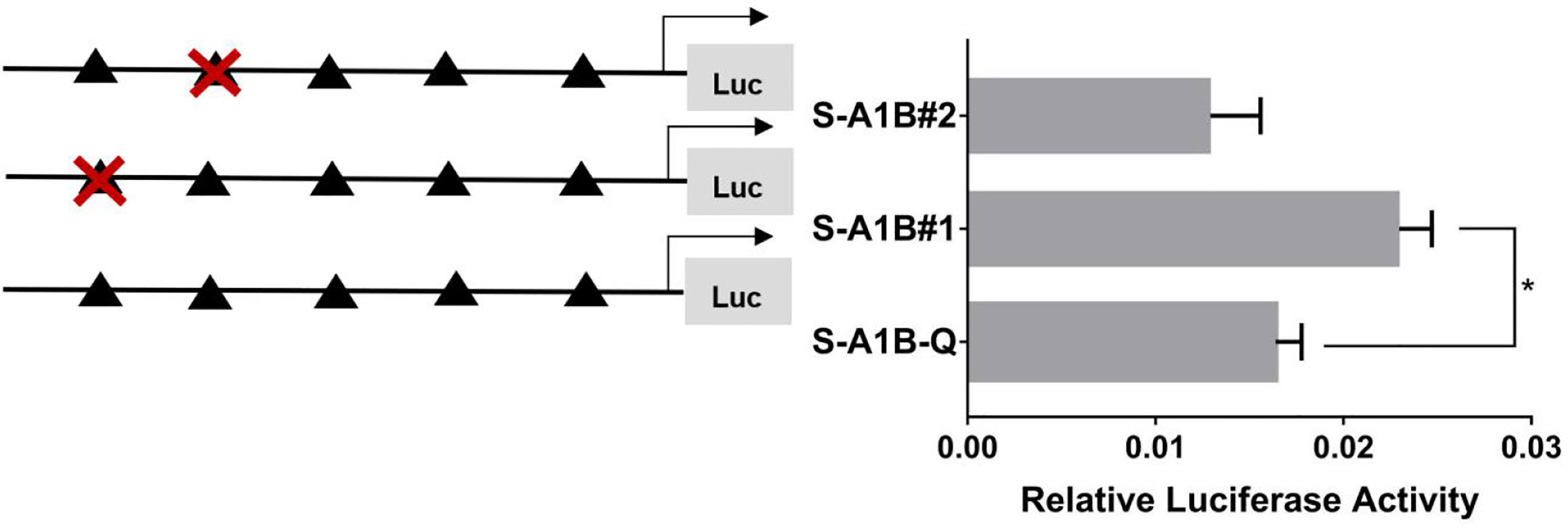
Figure 11 Effect of SF-1 on the transcriptional activity of the cyp19a1b promoter fragment mutated at the transcriptional site. On the left is a schematic diagram of the mutation of the promoter site, and the triangle represents the binding site. The right side is the expression of luciferase in the promoter recombinant vector. The black triangle with a red cross represents the SF-1 mutation site. One-way ANOVA was performed on the data expressed as mean ± standard deviation (n = 4). *There is a significant difference between the data.
In summary, we concluded that −120 to −112 bp (5′-CAAGGGCAC-3′) was the core regulatory element for SF-1 to regulate the transcriptional activity of the cyp19a1a promoter.
As Figure 13 shows, we found that after deleting −2,379 to −1,097, −2,379 to −1,026, and −2,379 to −871 bp, the transcription activities decreased significantly from the full length of the promoters (Figure 12, p < 0.05). Based on the predicted results from −2,379 to −871 bp, we performed site-directed mutations at the following three transcription sites: −1,116 to −1,098, −1,035 to −1,027, and −890 to −872 bp. After mutating −890 to −872 bp, cyp19a1b promoter activity decreased significantly (Figure 13, p < 0.05).
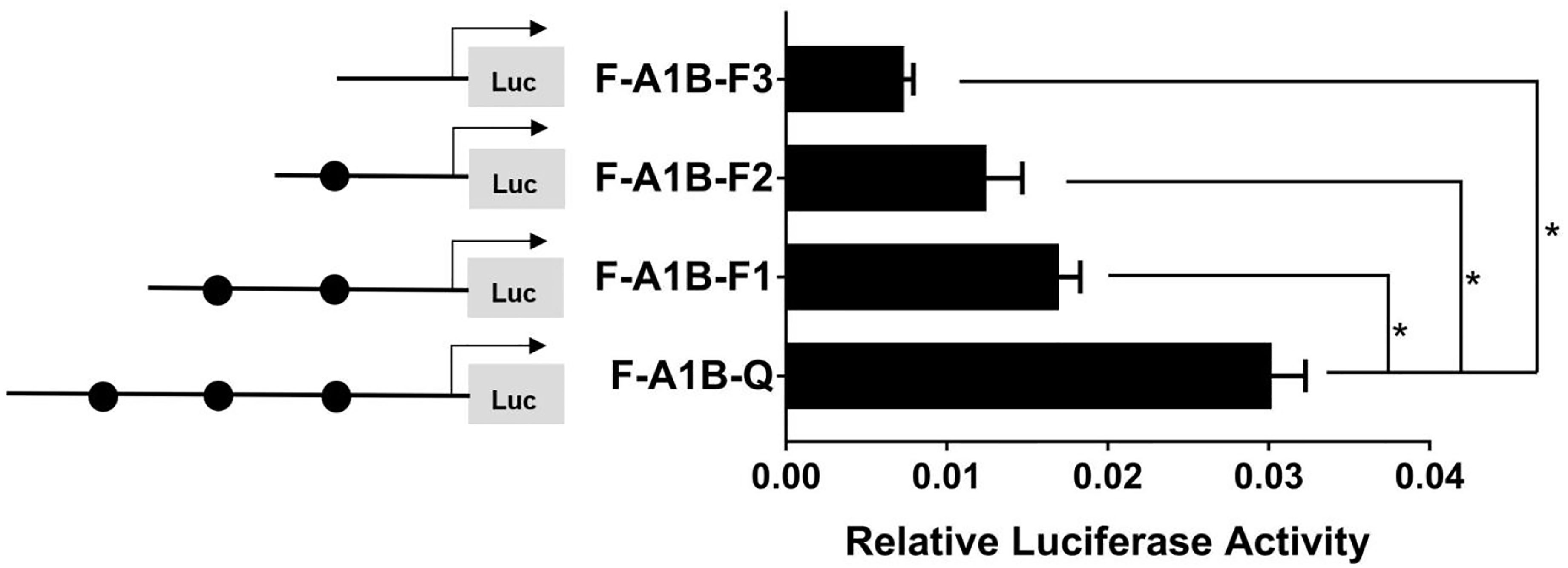
Figure 12 Effect of FOXL2 on the transcriptional activity of the cyp19a1b promoter fragment at the progressive deletion site. On the left is a schematic diagram of the promoter deletion and the circle represents the binding site. The right side is the expression of luciferase in the promoter recombinant vector. F-A1B-F1 (-2379 bp to -1097 bp deletion), F-A1B-F2 (-2379 bp to -1026 bp deletion), F-A1B-F3 (-2379 bp to -871 bp deletion). The black circle represents the FOXL2 binding site. One-way ANOVA was performed on the data and expressed as mean ± standard deviation (n = 4). *There is a significant difference between the data.
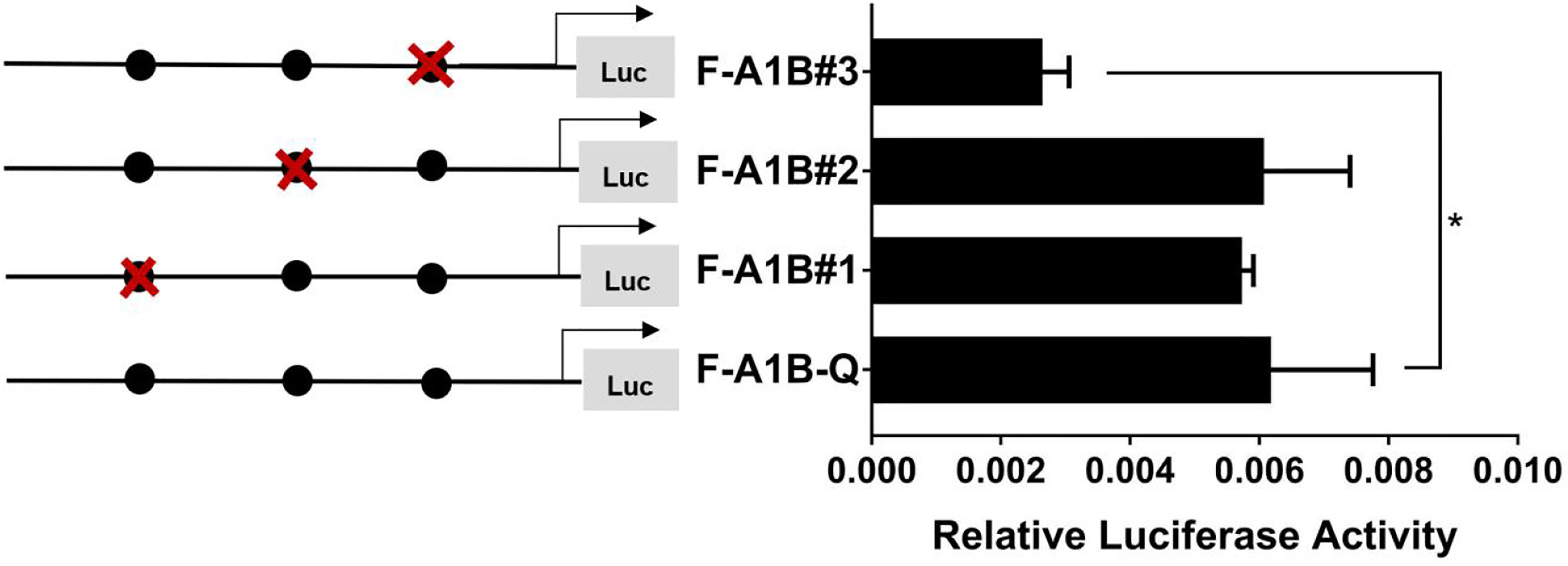
Figure 13 Effect of FOXL2 on transcriptional activity of the cyp19a1b promoter fragment mutated at the transcriptional site. On the left is a schematic diagram of the mutation of the promoter site, and the circle represents the binding site. The right side is the expression of luciferase in the promoter recombinant vector. The black circle with a red cross represents the FOXL2 mutation site. One-way ANOVA was performed on the data and expressed as mean ± standard deviation (n = 4). *There is a significant difference between the data.
Therefore, we determined that −890 to −872 bp (5′-AGAGGAGAACAAGGGGAG-3′) was the core regulatory element for FOXL2 to regulate the transcriptional activity of the cyp19a1b promoter.
cyp19a1 is the switch enabling androgen to estrogen conversion, affecting gonadal differentiation and sex ratios by regulating the biosynthesis of gonadal steroid hormones (20). We cloned two genes encoding the SF-1 and FOXL2 transcription factors and constructed a phylogenetic tree and conserved domains. Phylogenetic trees showed that the giant wrasse’s SF-1 and FOXL2 sequences clustered into the teleost group. Similar to a study on Cyprinus carpio SF-1, our conserved domain analysis found that the SF-1 amino acid sequence contained two domains: a conserved DBD and a C-terminal LBD (21). Similarly, FOXL2 consisted of an evolutionarily conserved FH domain. This result was the same as that of the conserved domain in the human FOXL2 gene (22).
Evidence supports that NR5A members (FTZ-F1, Ad4BP/SF-1, or LRH-1) and FOXL2 play a role in the brain. Previous studies have potentiated that NR5A members and FOXL2 are involved in cyp19a1a regulation in some fish and mammals (23–26). Furthermore, the combination of FOXL2 and SF-1 can significantly enhance aromatase cyp19a1 transcription (27). Similarly, our results showed that co-transfection of SF-1 and FOXL2 can enhance the transcriptional activity of the cyp19a1 promoters, which indicated a synergistic effect between SF-1 and FOXL2. The synergistic effects of SF-1 and FOXL2 have also been verified in other teleosts. For example, in catfish, FTZ-F1/SF-1 and FOXL2 bind to the promoter and upregulate transcription of the aromatase gene in the brain (28). In tilapia, FOXL2 binds to the promoter and interacts with Ad4BP/SF-1 to upregulate aromatase gene transcription in a female-specific manner (15). Similar results were also obtained in ricefield eels, where NR5A1A (one of the genes encoding NR5A homologs) cooperated with FOXL2 to upregulate cyp19a1a promoter activity (29). However, the synergistic mechanism by which SF-1 and FOXL2 act remains unclear. Studies have shown that SF-1-induced anti-Mullerian hormone (AMH) regulation via functional FOXL2 occurs through protein–protein interactions between FOXL2 and SF-1 in human granulosa cells (30). In addition, Nagahama et al. reported that FOXL2 could interact with the LBD of NR5A1 through the FH domain to form a heterodimer in tilapia (15). Researchers in Quebec, Canada, provided a more detailed explanation regarding this synergistic mechanism and found that the zinc finger region of GATA binding protein 4 (GATA-4) mediated GATA-4/SF-1 synergy. This synergy is the result of direct protein–protein interactions (31). These results collectively indicate that FOXL2 and SF-1 may affect cyp19a1 transcriptional regulation through a specific connection or interaction.
In many bony fish, SF-1 binding sites exist in the promoter region of the gonadal aromatase cyp19a1 (32–35). In Oncorhynchus mykiss, it was found that only −150 to −142 bp and −118 to −110 bp were the core transcription binding sites in the cyp19a1a promoter (36). In the orange-spotted grouper, deletion of the region (−246 to −112 bp) containing two conserved FTZ-F1 sites significantly reduced the transcriptional activity of the cyp19a1a promoter (37). We hypothesized that −120 to −112 bp were the key transcriptional binding sites of SF-1 in the cyp19a1a promoter. Collectively, these results indicated that SF-1 family proteins are common transcriptional regulators of gonad-type cyp19a1a genes in fish and provided evidence that they regulate cyp19a1 expression in vertebrate ovaries. Interestingly, we did not find the core regulatory site of SF-1 in the cyp19a1b promoter. From fish to mammals, SF-1 appears to be the common transcription factor of gonad-type cyp19a1a but not brain-type cyp19a1b. Other studies on aromatase promoters have found that in some fish, including ricefield eel (38), Epinephelus akaara (37), zebrafish (34), and Sparus macrocephalus (39), only the cyp19a1a promoter contains the SF-1 binding site, while the brain-type cyp19a1b promoter does not. Therefore, we speculated that Cheilinus undulatus may be the same as these fish. Thus, this explanation is reasonable. For example, the SF-1 binding site was found in the mouse brain-type cyp19a promoter (40). A previous study found that NR5A1 knockout mice still contained aromatase in some of their brain cells (41), suggesting that SF-1 may not be necessary for cyp19a1b expression.
Previous studies have shown that FOXL2 is involved in regulating cyp19a1 promoter transcription in vertebrates. FOXL2 and SF-1 co-regulate aromatase transcription (15, 27, 28). However, some researchers believe that FOXL2 directly binds to some DNA sites through its FH. Other studies have shown that FOXL2 binds to the half-site sequence of nuclear receptors, namely, the SF-1 response element (SFRE). The SFRE acts on the aromatase promoter (42). Our experiment found that deleting or mutating −890 to −872 bp significantly reduced cyp19a1b promoter activity. Therefore, we speculated that −890 to −872 bp is the core element for FOXL2 to regulate cyp19a1b transcriptional activity. Our results are consistent with those reported for tilapia. A mutation in the FOXL2 binding site (−545 to −538 bp) in the cyp19a1b promoter of tilapia significantly reduced the activity of the promoter (15). Similarly, when a FOXL2 binding site in the promoter of catfish cyp19a1b was mutated from −977 to −954 bp, cyp19a1b transcriptional activity was significantly downregulated, indicating that this region was the core element of FOXL2 to regulate cyp19a1b transcriptional activity (28). These studies all point to the possible involvement of FOXL2 in regulating cyp19a1b promoter transcription.
In conclusion, this study found that SF-1 and FOXL2 can regulate cyp19a1a and cyp19a1b promoters, respectively. SF-1 and FOXL2 contain key binding elements in the cyp19a1a and cyp19a1b promoters, respectively. The combination of SF-1 and FOXL2 enhanced aromatase promoter activity in giant wrasse. This study lays a theoretical foundation for identifying the aromatase gene transcriptional regulatory pathway in giant wrasse.
The raw data supporting the conclusions of this article will be made available by the authors on reasonable request.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Hainan University. Written informed consent was obtained from the owners for the participation of their animals in this study.
YZ, YW and XJ was the main performer of the experiment. XJ and YZ completed the manuscript, the revision of the article and result graphing. SB analyzed the data and submited the article. XW, FS provided suggestions for writing articles. JL designed all the experiments, provided the required facilities and guidance. All authors read the final article and approved its submission.
This work was supported by the National Natural Science Foundation (Grant No. 3166130111).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Gemmell NJ, Todd EV, Goikoetxea A, Ortega-Recalde O, Hore TA. Natural Sex Change in Fish. Curr Top Dev Biol (2019) 134:71–117. doi: 10.1016/bs.ctdb.2018.12.014
2. Piferrer F, Blázquez M, Navarro L, González A. Genetic, Endocrine, and Environmental Components of Sex Determination and Differentiation in the European Sea Bass (Dicentrarchus Labrax L.). Gen Comp Endocrinol (2005) 142(1-2):102–10. doi: 10.1016/j.ygcen.2005.02.011
3. Tao W, Yuan J, Zhou L, Sun L, Sun Y, Yang S, et al. Characterization of Gonadal Transcriptomes From Nile Tilapia (Oreochromis Niloticus) Reveals Differentially Expressed Genes. PloS One (2013) 8(5):e63604. doi: 10.1371/journal.pone.0063604
4. Guiguen Y, Fostier A, Piferrer F, Chang CF. Ovarianaromatase and Estrogens: A Pivotal Role for Gonadal Sex Differentiation and Sex Change in Fish. Gen Comp Endocrinol (2010) 165(3):352–66. doi: 10.1016/j.ygcen.2009.03.002
5. Diotel N, Le Page Y, Mouriec K, Tong SK, Pellegrini E, Vaillant C, et al. Aromatase in the Brain of Teleost Fish: Expression, Regulation and Putative Functions. Front Neuroendocrinol (2010) 31(2):172–92. doi: 10.1016/j.yfrne.2010.01.003
6. Cao J, Chen J, Jiang Z, Luo Y, Gan XI. Cloning and Expression Characteristics of FTZ-F1 Gene in Jifu Tilapia. China Fisheries Society. In: Aquatic Biotechnology Professional Committee of China Fisheries Society and Fish Sex Determination Mechanism Proceedings of the Symposium on and Regulation Technology, vol. 27. (2011). Available at: https://doc.paperpass.com/conference/20110002zho_23201.html?utm_source=doc_share.
7. Hoivik EA, Lewis AE, Aumo L, Bakke M. Molecular Aspects of Steroidogenic Factor 1 (SF-1). Mol Cell Endocrinol (2010) 315(1):27–39. doi: 10.1016/j.mce.2009.07.003
8. Watanabe M, Tanaka M, Kobayashi D, Yoshiura Y, Oba Y, Nagahama Y. Medaka (Oryzias Latipes) FTZ-F1 Potentially Regulates the Transcription of P-450 Aromatase in Ovarian Follicles: cDNA Cloning and Functional Characterization. Mol Cell Endocrinol (1999) 149(1-2):221–8. doi: 10.1016/s0303-7207(99)00006-4
9. Wang J, Gong Y. Transcription of CYP19A1 is Directly Regulated by SF-1 in the Theca Cells of Ovary Follicles in Chicken. Gen Comp Endocrinol (2017) 247:1–7. doi: 10.1016/j.ygcen.2017.03.013
10. Xie Q. Steroidogenic Factor 1 (SF-1) is Essential for Steroidogenesis, Gonadal Development and Sex Differentiation in Nile Tilapia. Chongqing: Southwest University (2016) p. 17–8. Available at: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=1016908992.nh&DbName=CDFD2017.
11. Matsumoto Y, Crews D. Molecular Mechanisms of Temperature-Dependent Sex Determination in the Context of Ecological Developmental Biology. Mol Cell Endocrinol (2012) 354(1-2):103–10. doi: 10.1016/j.mce.2011.10.012
12. Escudero JM, Haller JL, Clay CM, Escudero KW. Microarray Analysis of Foxl2 Mediated Gene Regulation in the Mouse Ovary Derived KK1 Granulosa Cell Line: Over-Expression of Foxl2 Leads to Activation of the Gonadotropin Releasing Hormone Receptor Gene Promoter. J Ovarian Res (2010) 3(1):4–4. doi: 10.1186/1757-2215-3-4
13. Frank B, Daniel V, Jean D, Veitia RA. Potential Targets of FOXL2, a Transcription Factor Involved in Craniofacial and Follicular Development, Identified by Transcriptomics. Proc Natl Acad Sci USA (2007) 104(9):3330–5. doi: 10.1073/pnas.0611326104
14. Govoroun MS, Pannetier M, Pailhoux E, Cocquet J, Brillard JP, Couty I, et al. Isolation of Chicken Homolog of the FOXL2 Gene and Comparison of its Expression Patterns With Those of Aromatase During Ovarian Development. Dev Dyn (2004) 231(4):859–70. doi: 10.1002/dvdy.20189
15. Wang DS, Kobayashi T, Zhou LY, Paul-Prasanth B, Ijiri S, Sakai F, et al. Foxl2 Up-Regulates Aromatase Gene Transcription in a Female-Specific Manner by Binding to the Promoter as Well as Interacting With Ad4 Binding Protein/Steroidogenic Factor 1. Mol Endocrinol (2007) 21(3):712–25. doi: 10.1210/me.2006-0248
16. Bhat IA, Rather MA, Dar JY, Sharma R. Molecular Cloning, Computational Analysis and Expression Pattern of Forkhead Box L2 (Foxl2) Gene in Catfish. Comput Biol Chem (2016) 64:9–18. doi: 10.1016/j.compbiolchem.2016.05.001
17. Yamaguchi T, Yamaguchi S, Hirai T, Kitano T. Follicle-Stimulating Hormone Signaling and Foxl2 are Involved in Transcriptional Regulation of Aromatase Gene During Gonadal Sex Differentiation in Japanese Flounder, Paralichthys Olivaceus. Biochem Biophys Res Commun (2007) 359(4):935–40. doi: 10.1016/j.bbrc.2007.05.208
18. Chen M, Luo J, Chen G, Wen X, Guangcan WU, Wenshen LI, et al. Embryonic Development and Morphologic Observations of Newly-Hatched Cheilinus Undulatus Larvae. Prog Fishery Sci (2015) 36(5):38–9. Available at: http://journal.yykxjz.cn/yykxjz/ch/reader/view_abstract.aspx?file_no=20150506&flag=1.
19. Dorenbosch M, Grol MGG, Nagelkerken I, Velde VD. eagrass Beds and Mangroves as Potential Nurseries for the Threatened Indo-Pacific Humphead Wrasse, Cheilinus Undulatus and Caribbean Rainbow Parrotfish,Scarus Guacamaia. Biol Conserv (2006) 129:277–82. doi: 10.1016/j.biocon.2005.10.032
20. Fan Z, Zou Y, Jiao S, Tan X, Wu Z, Liang D, et al. Significant Association of Cyp19a Promoter Methylation With Environmental Factors and Gonadal Differentiation in Olive Flounder Paralichthys Olivaceus. Comp Biochem Physiol Part A: Mol Integr Physiol (2017) 208:70–9. doi: 10.1016/j.cbpa.2017.02.017
21. Zhao W. Structure, Expression Pattern and its Relationship With miRNA Targets Analysis of Sf-1 Gene in Yellow River Camp. Henan: the Graduate School of Henan Normal University (2019) p. 3–4. doi: 10.27118/d.cnki.ghesu.2019.000080
22. Pelletier GJ, Brody SL, Liapis H, White RA, Hackett BP. A Human Forkhead/Winged-Helix Transcription Factor Expressed in Developing Pulmonary and Renal. Am J Physiol (1998) 274(3):L351–9. doi: 10.1152/ajplung.1998.274.3.L351
23. Pannetier M, Fabre S, Batista F, Kocer A, Renault L, Jolivet G, et al. FOXL2 Activates P450 Aromatase Gene Transcription: Towards a Better Characterization of the Early Steps of Mammalian Ovarian Development. J Mol Endocrinol (2006) 36(3):399–413. doi: 10.1677/jme.1.01947
24. Vincenzo P, Rosa S, Adele C, Maggiolini M, Bourguiba S, Delalande C, et al. Differential Expression of Steroidogenic Factor-1/Adrenal 4 Binding Protein and Liver Receptor Homolog-1 (LRH-1)/Fetoprotein Transcription Factor in the Rat Testis: LRH-1 as a Potential Regulator of Testicular Aromatase Expression. Endocrinology (2004) 145(5):2186. doi: 10.1210/en.2003-1366
25. Fan Z, Zou Y, Liang D, Tan X, You F. Roles of Forkhead Box Protein L2 (Foxl2) During Gonad Differentiation and Maintenance in a Fish, the Olive Flounder (Paralichthys Olivaceus). Reproduction Fertil Dev (2019) 31(11):1742–52. doi: 10.1071/RD18233
26. Yoshiura Y, Senthilkumaran B, Watanabe M, Oba Y, Kobayashi T, Nagahama Y. Synergistic Expression of Ad4bp/Sf-1 and Cytochrome P-450 Aromatase (Ovarian Type) in the Ovary of Nile Tilapia, Oreochromis Niloticus, During Vitellogenesis Suggests Transcriptional Interaction. Biol Reprod (2003) 68(5):1545–53. doi: 10.1095/biolreprod.102.010843
27. Fleming NI, Knower KC, Lazarus KA, Fuller PJ, Simpson ER, Clyne CD. Aromatase is a Direct Target of FOXL2: C134W in Granulosa Cell Tumors via a Single Highly Conserved Binding Site in the Ovarian Specific Promoter. PloS One (2010) 5(12):e14389. doi: 10.1371/journal.pone.0014389
28. Sridevi P, Chaitanya RK, Dutta-Gupta A, Senthilkumaran B. FTZ-F1 and FOXL2 Up-Regulate Catfish Brain Aromatase Gene Transcription by Specific Binding to the Promoter Motifs. Biochim Biophys Acta (2012) 1819(1):57–66. doi: 10.1016/j.bbagrm.2011.10.003
29. Yan T, Lu H, Sun C, Peng Y, Meng F, Gan R, et al. Nr5a Homologues in the Ricefield Eel Monopterus Albus: Alternative Splicing, Tissue-Specific Expression, and Differential Roles on the Activation of Cyp19a1a Promoter In Vitro. Gen Comp Endocrinol (2021) 312:113871. doi: 10.1016/j.ygcen.2021.113871. ISSN 0016-6480.
30. Jin H, Won M, Park SE, Lee S, Park M, Bae J. FOXL2 Is an Essential Activator of SF-1-Induced Transcriptional Regulation of Anti-Müllerian Hormone in Human Granulosa Cells. PloS One (2016) 11(7):e0159112. doi: 10.1371/journal.pone.0159112
31. Tremblay JJ, Viger RS. Transcription Factor GATA-4 Enhances Müllerian Inhibiting Substance Gene Transcription Through a Direct Interaction With the Nuclear Receptor SF-1. Mol Endocrinol (1999) 13(8):1388–401. doi: 10.1210/mend.13.8.0330
32. Callard GV. Differential Tissue Distribution, Developmental Programming, Estrogen Regulation and Promoter Characteristics of Cyp19 Genes in Teleost Fish. J Steroid Biochem Mol Biol (2001) 1-5(79):305–14. doi: 10.1016/s0960-0760(01)00147-9
33. Kazeto Y, Ijiri S, Place AR, Zohar Y, Trant JM. The 5’-Flanking Regions of CYP19A1 and CYP19A2 in Zebrafish. Biochem Biophys Res Commun (2001) 288(3):503. doi: 10.1006/bbrc.2001.5796
34. Tanaka M. Structure and Promoter Analysis of the Cytochrome P-450 Aromatase Gene of the Teleost Fish, Medaka (Oryzias Latipes). J Biochem (1995) 4(117):719–25. doi: 10.1093/oxfordjournals.jbchem.a124768
35. Xiaotian C. Two Types of Aromatase With Different Encoding Genes, Tissue Distribution and Developmental Expression in Nile Tilapia (Oreochromis Niloticus). Gen Comp Endocrinol (2005) 141(2):101–15. doi: 10.1093/oxfordjournals.jbchem.a124768
36. Kanda H, Okubo T, Omori N, Niihara H, Matsumoto N, Yamada K, et al. Transcriptional Regulation of the Rainbow Trout CYP19a Gene by FTZ-F1 Homologue. J Steroid Biochem Mol Biol (2006) 99(2):85–92. doi: 10.1016/j.jsbmb.2005.12.005
37. Zhang W, Lu H, Jiang H, Li M, Zhang S, Liu Q, et al. Isolation and Characterization of Cyp19a1a and Cyp19a1b Promoters in the Protogynous Hermaphrodite Orange-Spotted Grouper (Epinephelus Coioides). Gen Comp Endocrinol (2012) 175(3):473–87. doi: 10.1016/j.ygcen.2011.12.005
38. Zhang W, Yang Y, Peng Y, Zhang S, Zhang Y, Wu C, et al. Differential Synergism of Ftz-F1 Homologues and Foxl2 on the Activation of Cyp19a1a Gene From Rice Field Eel Monopterus Albus, a Protogynous Hermaphroditic Teleost. Biol Reprod (2010) 82(1):134–5. doi: 10.1095/biolreprod.109.076729
39. Ten-Tsao W, Shigeho I, Yonathan Z. Molecular Biology of Ovarian Aromatase in Sex Reversal: Complementary DNA and 5’-Flanking Region Isolation and Differential Expression of Ovarian Aromatase in the Gilthead Seabream (Sparus Aurata). Biol Reprod (2006) 74(5):857. doi: 10.1095/biolreprod.105.045351
40. Honda SI, Harada N, Abe-Dohmae S, Takagi Y. Identification of Cis-Acting Elements in the Proximal Promoter Region for Brain-Specific Exon 1 of the Mouse Aromatase Gene. Mol Brain Res (1999) 66(1–2):122–32. doi: 10.1016/s0169-328x(99)00017-0
41. Shinoda K, Lei H, Yoshii H, Nomura M, Nagano M, Shiba H, et al. Developmental Defects of the Ventromedial Hypothalamic Nucleus and Pituitary Gonadotroph in the Ftz-F1 Disrupted Mice. Dev Dynamics (2010) 204(1):22–9. doi: 10.1002/aja.1002040104
Keywords: promoter, SF-1, FOXL2, cyp19a1a, cyp19a1b, transcriptional regulation
Citation: Ji X, Bu S, Zhu Y, Wang Y, Wen X, Song F and Luo J (2022) Identification of SF-1 and FOXL2 and Their Effect on Activating P450 Aromatase Transcription via Specific Binding to the Promoter Motifs in Sex Reversing Cheilinus undulatus. Front. Endocrinol. 13:863360. doi: 10.3389/fendo.2022.863360
Received: 31 January 2022; Accepted: 08 March 2022;
Published: 10 May 2022.
Edited by:
Gustavo M. Somoza, Instituto Tecnológico de Chascomús (CONICET-UNSAM), ArgentinaCopyright © 2022 Ji, Bu, Zhu, Wang, Wen, Song and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Luo, bHVvamlhbkBoYWluYW51LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.