- Bioorganic Research Institute, Suntory Foundation for Life Sciences, Kyoto, Japan
Omics studies contribute to the elucidation of genomes and profiles of gene expression. In the ascidian Ciona intestinalis Type A (Ciona robusta), mass spectrometry (MS)-based peptidomic studies have detected numerous Ciona-specific (nonhomologous) neuropeptides as well as Ciona homologs of typical vertebrate neuropeptides and hypothalamic peptide hormones. Candidates for cognate G protein-coupled receptors (GPCRs) for these peptides have been found in the Ciona transcriptome by two ways. First, Ciona homologous GPCRs of vertebrate counterparts have been detected by sequence homology searches of cognate transcriptomes. Second, the transcriptome-derived GPCR candidates have been used for machine learning-based systematic prediction of interactions not only between Ciona homologous peptides and GPCRs but also between novel Ciona peptides and GPCRs. These data have ultimately led to experimental evidence for various Ciona peptide-GPCR interactions. Comparative transcriptomics between the wildtype and Ciona vasopressin (CiVP) gene-edited Ciona provide clues to the biological functions of CiVP in ovarian follicular development and whole body growth. Furthermore, the transcriptomes of follicles treated with peptides, such as Ciona tachykinin and cionin (a Ciona cholecystokinin homolog), have revealed key regulatory genes for Ciona follicle growth, maturation, and ovulation, eventually leading to the verification of essential and novel molecular mechanisms underlying these biological events. These findings indicate that omics studies, combined with artificial intelligence and single-cell technologies, pave the way for investigating in greater details the nervous, neuroendocrine, and endocrine systems of ascidians and the molecular and functional evolution and diversity of peptidergic regulatory networks throughout chordates.
Introduction
Ascidians are aquatic organisms that live all over the world and belong to the phylum Urochordata and superphylum Chordata; they are one of the closest relatives of vertebrates (1–3). They have been used in developmental biology as model organisms for more than 50 years, and the genome of Ciona intestinalis Type A (Ciona robusta) was sequenced in 2002 (4). At the dawn of the omics era, expressed sequence tags (ESTs) and gene model analyses identified approximately 16,000 protein-coding genes and various vertebrate orthologs in the C. robusta genome (4, 5). Furthermore, the gene expression profiles during embryonic development have been determined by in situ hybridization and transcriptomics (6–9), and similarities between the transcriptional network of C. robusta and vertebrates have been clarified (10–13). These genomic and transcriptomic studies have highlighted the importance of C. robusta in comparative and evolutionary developmental and genome biology (13–16). In contrast, less attention was paid to the reproductive mechanisms of Ciona.
In vertebrates, ovarian follicular development is accurately regulated by the hypothalamic-pituitary-gonadal axis (HPG axis). A hypothalamic neuropeptide, gonadotropin-releasing hormone (GnRH), stimulates the synthesis and secretion of gonadotropins in the pituitary, which induces follicle growth, maturation, ovulation, and the production of sex steroid hormones for negative feedback regulation in the ovary (17–20). Recent studies have further revealed that more neuropeptides, including kisspeptin, neurokinin B, dynorphin A, and gonadotropin-inhibitory hormone, play important roles in the regulation of the HPG axis (21, 22). Thus, neuropeptides, as major signaling factors, are responsible for the regulation of the HPG axis in vertebrates. In contrast, C. robusta is not endowed with the HPG axis, leading to the presumption that neuropeptides produced in the neuroendocrine system directly regulate follicle development via neural network transport instead of regulation by the HPG axis. Combined with the crucial phylogenetic position of ascidians as a sister group of the vertebrates, elucidation of the mechanisms of reproduction regulated by ascidian neuropeptides will provide insight into the molecular and functional evolution of the endocrine and neuroendocrine systems in chordates.
As stated above, gene model analyses of C. robusta have found approximately 16,000 protein-coding genes. However, few neuropeptide genes were annotated on the original version, given that homology-based searching methods are frequently useless for the detection of small peptides or their genes due to far less sequence homology information, compared with that of proteins. Over the last two decades, peptidomic analyses have advanced due to mass spectrometry (MS)-based technology, leading to the sensitive detection of small amounts of peptides. An MS-based peptidomic analysis has characterized 33 peptides, including 26 novel peptides, from the Ciona neural complex followed by the detection of their genes by referencing the resultant peptide sequences to the genome/EST database and by localization of their gene expression (Figure 1A) (23). These ascidian neuropeptides are largely classified into two groups: (i) homologs of vertebrate peptides, and (ii) Ciona-specific peptides. The former group includes typical vertebrate neuropeptide homologs, including a vasopressin homolog (CiVP) (24), a tachykinin homolog (CiTK) (25), GnRH homologs (tGnRHs) (26), and a cholecystokinin/gastrin homolog (cionin) (27, 28), while the latter group contains CiLFs, CiYFVs, and CiNTLPs including partial neurotensin-like sequences (23) (Supplementary Table 1). The identification of these neuropeptides has paved the way to understanding the ascidian neuroendocrine and nervous systems involved in reproductive functions.
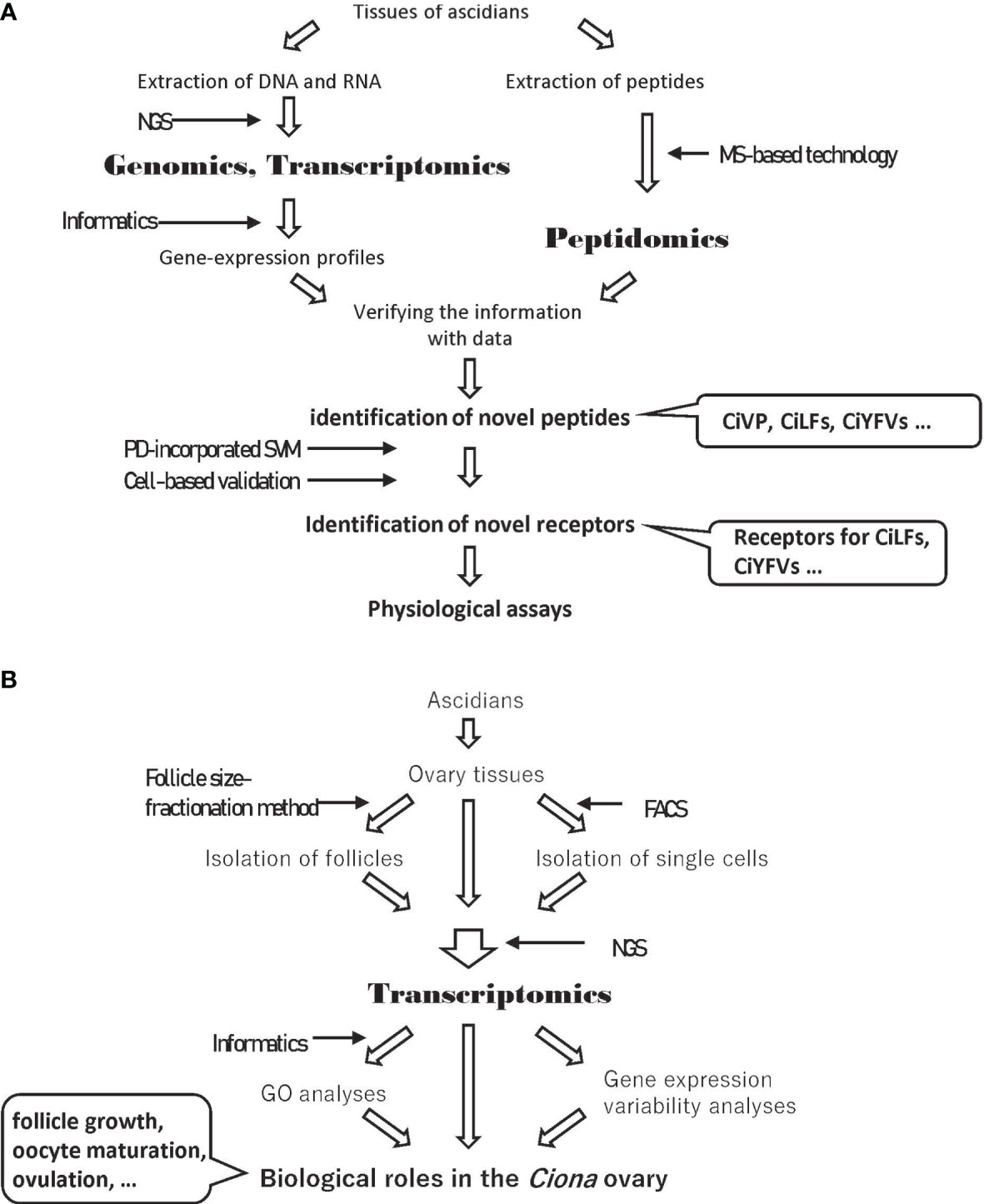
Figure 1 Experimental strategies including omics analyses. (A) Experimental strategy to identify novel ascidian peptides and their receptors. (B) Experimental strategy to elucidate the biology of the Ciona ovary using transcriptome data. NGS, Next Generation Sequencing; MS, Mass Spectrometry; PD, Peptide Descriptor; SVM, Support Vector Machine; FACS, Fluorescence-Activated Cell Sorting; GO, Gene Ontology.
Omics analyses are also powerful tools to identify comprehensive gene expression profiles. Gene ontology (GO) analyses using comprehensive gene expression profiles provide clues to the elucidation of the biological features and functions of target tissues or cells (Figure 1B). Comparisons of gene expression profiles between tissues treated or untreated with a target peptide or between target gene-knockdown organisms and wildtype organisms can identify key molecules induced by a target peptide (Figure 1B). In this review, we provide an overview of essential neuropeptidergic regulatory mechanisms underlying Ciona follicle growth, oocyte maturation, and ovulation, which have been elucidated by a combination of omics and physiological analyses.
Nervous System and Ovary in CIONA
Recent morphological studies using prohormone convertase 2 promoter-Kaede transgenic Ciona showed that the peptidergic nervous system directly innervates to the ovary (29, 30). The innervations of nerves to the ovary were also found in CiVP promoter-Venus transgenic Ciona, suggesting that CiVP acts as an endogenous factor in the ovary (31). In addition to these morphological characteristics of the peptidergic nervous system, expression of the receptors for neuropeptides, including CiTK, CiVP, and cionin, was detected in the ovary (24, 25, 32). These findings indicate that the ovary is a major target of the neuropeptides produced in the neural complex of Ciona (Figure 2A).
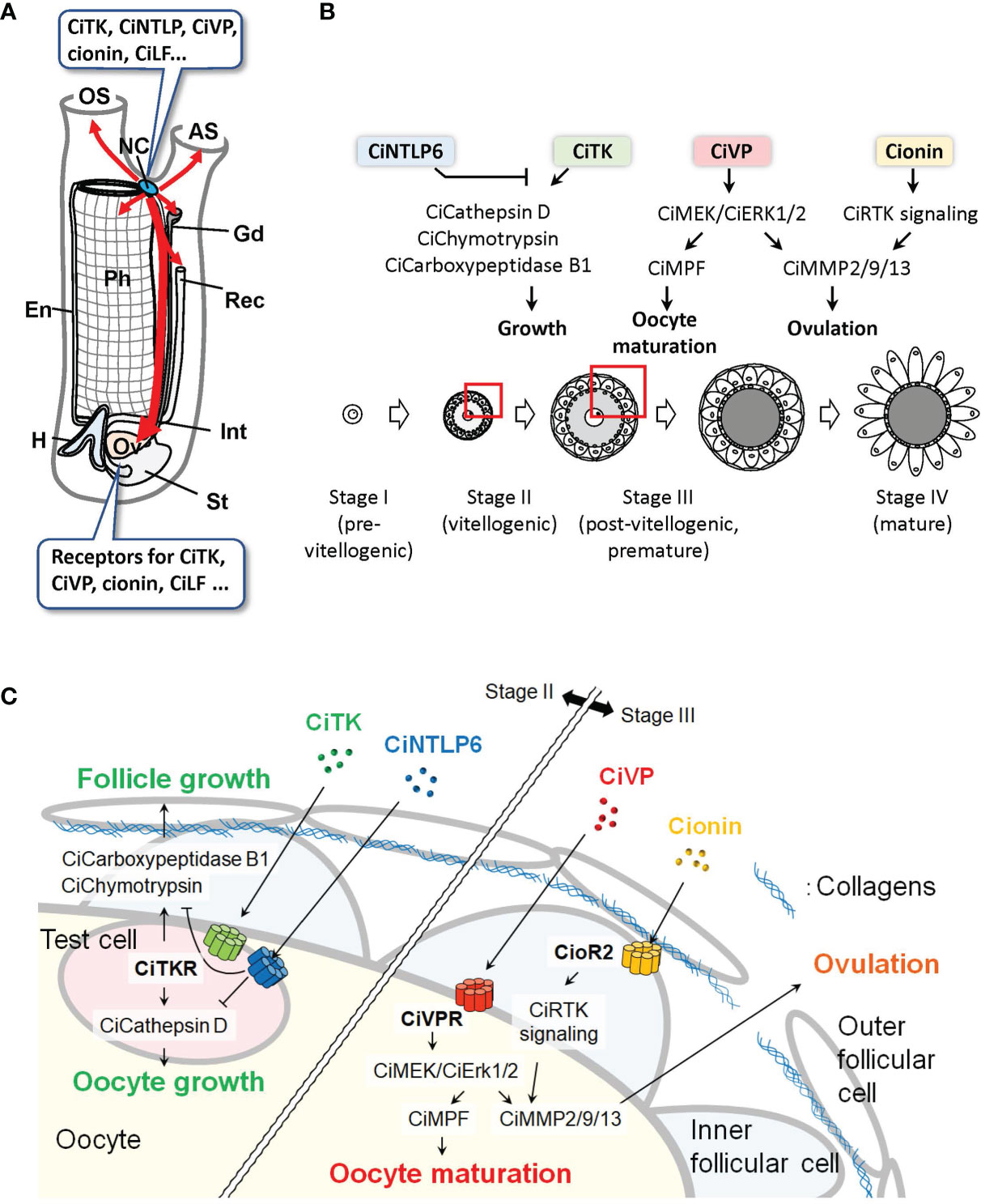
Figure 2 Peptidergic systems in Ciona. (A) Various neuropeptides are expressed in the neural complex. Peptidergic nerves directly innervate from the neural complex to the peripheral organs (red arrows). The ovary, which expresses various peptide receptors, is a major target of the neuropeptides. AS, atrial siphon; En, endostyle; Gd, gonoduct; H, heart; Int, intestine; NC, neural complex; OS, oral siphon; Ov, ovary; Ph, pharynx; Rec, rectum; St, stomach. (B, C) Peptidergic regulation of ovarian follicular growth, oocyte maturation, and ovulation in Ciona. Overview (B) and the enlarged structure of a follicle (C) as indicated by red boxes in (B) are shown. In vitellogenic (stage II) follicles, CiTK induces the expression of CiCathepsin D in test cells and CiChymotrypsin and CiCarboxypeptidase B1 in inner follicular cells and promotes follicle growth from stage II to stage III. CiNTLP6 suppresses CiTK-induced gene expression and subsequent follicle growth. In postvitellogenic (stage III) follicles, CiVP activates CiMEK/CiErk1/2 in oocytes and promotes oocyte maturation and ovulation via activation of CiMPF and CiMMP2/9/13, respectively. Cionin also induces CiMMP2/9/13 expression via CioR2 in inner follicular cells and RTK signaling and subsequent ovulation in stage III follicles.
Ciona ovarian follicles are classified into four major developmental stages: stage I (pre-vitellogenic), stage II (vitellogenic), stage III (post-vitellogenic and pre-mature), and stage IV (mature) oocytes (33). This classification is based on criteria including size, pigmentation of the oocytes, and changes in the cellular layer surrounding the oocytes (33). Recently, an efficient follicle size-fractionation method using stainless steel sieves of various particle sizes was developed (34, 35). This method enables physiological assays as well as transcriptome analyses of the respective stages of follicles. Furthermore, an in vitro evaluation of follicle growth, maturation, and ovulation of Ciona follicles has been established (34–37). These novel experimental procedures can lead to the investigation of the specific biological functions of neuropeptides on follicles at individual stages.
Ovarian Follicular Development
Consistent with transcriptomic data showing high expression of the Citk receptor gene during follicle development (34), our in vitro incubation system and morphological evaluation of isolated follicles demonstrated that CiTK stimulates follicle growth from stage II to stage III (36). Microarray and biochemical analyses of CiTK-treated and -untreated follicles revealed that CiTK induces the expression and enzymatic activities of chymotrypsin, carboxy-peptidase B1, and cathepsin D (36). Moreover, CiTK was shown to directly induce the gene expression and resulting enzymatic activity of cathepsin D in test cells (oocyte-accessory cells) expressing the CiTK receptor, and to secondarily activate the gene expression and resulting enzymatic activity of chymotrypsin and carboxypeptidase B1 in follicle cells (37). These proteases are all responsible for CiTK-induced follicle growth, given that inhibition of any one of these proteases resulted in the arrest of follicle growth (36). Also of interest is that CiNTLP6 treatment for stage II follicles suppresses CiTK-induced gene expression of these proteases and the subsequent follicle development to stage III (23). Consequently, CiTK and CiNTLP6 regulate follicle development from stage II to stage III via activation of the gene expression and enzymatic activities of cathepsin D in test cells and chymotrypsin and carboxypeptidase B1 in follicle cells (Figures 2B, C) (23, 36, 37). These findings provide evidence that CiTK is essential for the development of premature follicles.
The expression of the CiVP receptor also suggests important roles of CiVP in the ovary (24). In a TALEN-based CiVP mutant of Ciona, the number of stage III follicles is markedly decreased and the numbers of stage I and stage II follicles are increased in the ovaries (31). In the ovary of CiVP-mutant Ciona, transcriptome analysis and subsequent gene ontology (GO) analysis showed that the downregulated genes were categorized as the genes related to “Binding”, “Phosphorylation”, and “Organelles”, whereas upregulated genes were categorized as the genes related to “Nucleus” or “Ribosomes” (31). Interestingly, the ceramide synthase homologous gene that participates in the removal of low-quality early follicles as a lipid mediator in mammals is downregulated in CiVP-mutants (31). Protein kinase C β, which is an important factor for vesicle transport during the secretion of signaling molecules as well as organelle formation, is also downregulated in CiVP-mutants (31). In contrast, the MutL-homolog 3 gene, which is known to repair DNA mismatch during DNA replication and meiosis, is upregulated in CiVP-mutants (31). These findings suggest that CiVP plays multiple important roles to maintain the normal growth of early-stage follicles via the regulation of diverse genes.
Oocyte Maturation and Ovulation
In addition to the in vivo analysis of CiVP (31), in vitro assays and transcriptomic analyses following the aforementioned ovarian follicle fractionation have helped clarify the roles of neuropeptides in Ciona oocyte maturation and ovulation (34, 35). In line with receptor expression in the ovary (24, 32), CiVP and cionin have been demonstrated to promote follicle maturation and ovulation in vitro (34, 38). Transcriptomic analyses of fractionated follicles have contributed a great deal to our understanding of the essential molecular mechanisms underlying CiVP- and cionin-induced oocyte maturation and ovulation; the expressions of Civp receptor and the Ciona-extracellular signal-related kinase homolog, CiErk1/2, were elevated toward oocyte maturation and ovulation. Gene expression of a matrix metalloproteinase, CiMmp2/9/13, was suppressed in MEK (MAPK kinase)-inhibited follicles (i.e., CiErk1/2-inhibited immature, unovulated), compared with those that are uninhibited (mature, ovulated) (34). The expressions of the receptor tyrosine kinase (RTK) signaling genes and CiMmp2/9/13 were upregulated in cionin-treated follicles, compared with untreated ones (38). Physiological assays and qRT-PCR validation following transcriptomic analyses have verified that CiVP activates CiErk1/2 and promotes oocyte maturation via activation of the maturation promoting factor and ovulation via induction of CiMmp2/9/13 expression (34), and that cionin also induces CiMmp2/9/13 expression and ovulation via upregulation of the RTK signaling genes (38). Combined with visualizing the entire projection of peptidergic neurons from the neural complex to the peripheral tissues (29, 30) and in vivo analysis of CiVP (31), in vitro physiological analyses have revealed that neuropeptides such as CiVP and cionin are produced in the neural complex (23, 24, 32) and regulate follicular development, oocyte maturation, and ovulation directly through the neuroendocrine system. Consequently, a combination of the classical bioassays and transcriptomic analyses has revealed the major regulatory pathways of Ciona oocyte maturation and ovulation (Figures 2B, C).
In other invertebrates, species-specific neuropeptides have been identified as maturation-inducing hormones (MIHs) such as W/RPRPamide for jellyfish in the phylum Cnidaria (39), relaxin-like gonad stimulating peptide for starfish (40), and cubifrin (NGIWYamide) for sea cucumber (41) in the phylum Echinodermata. In contrast to the variable MIHs, some molecular mechanisms underlying oocyte maturation and ovulation have been conserved among phyla. For example, the MOS (MAPK kinase kinase)/MAPK-maturation promoting factor (MPF, a complex of Cdk1 and Cyclin B) pathway in oocyte maturation is conserved in jellyfish and vertebrates (42). Moreover, degradation of extracellular matrix by matrix metalloproteinases (MMPs) was also found to be important for ovulation in fish (Vertebrata) (43, 44) and in Drosophila (Arthropod) (45). Furthermore, vasotocin, a non-mammalian vertebrate VP family peptide, has been reported to induce oocyte maturation and ovulation in catfish (Vertebrata) (46, 47), and RTK signaling was also reported to be involved in ovulation in mouse (Vertebrata) (48). Collectively, these studies, combined with the aforementioned Ciona (Urochordata) oocyte maturation and ovulation processes, shed light on species-specificity of triggering peptides in invertebrates and on the conserved mechanisms of oocyte maturation and ovulation among phyla. Further studies on oocyte maturation and ovulation in various animal species will pave the way for understanding the evolutionary processes of these biological events.
Conclusion and Perspectives
Omics studies are powerful tools to reveal the molecular mechanisms of biological events. In studies on Ciona, peptidomics and transcriptomics and subsequent molecular and physiological analyses have revealed that neuropeptides, including CiTK, CiNTLP6, CiVP, and cionin, regulate follicle growth, oocyte maturation, and ovulation (Figures 1, 2B, C). In addition to omics, newly developed techniques including follicle fractionation and gene-knockout have also contributed to the analysis of signaling networks induced by neuropeptides (Figure 1B). Moreover, a comparison of gene expression profiles between the tissues treated or untreated with a target peptide or between target gene-knockdown organisms and wildtype organisms will lead to the elucidation of reliable signaling networks induced by the target peptide (Figure 1B). Furthermore, comparisons of transcriptomic profiles of target peptide receptor homolog-expressing cells in closely related species will elucidate the evolution and diversification of signaling networks induced by the target peptide.
To develop omics-based biology to the next stage, single-cell transcriptomes, whose expression profiles exhibit more relevant signaling networks than whole-tissue transcriptomes, will play important roles. In C. robusta, Horie et al. analyzed a single-cell transcriptomic profile of the ascidian embryo at the mid-tailbud stage and determined the regulatory mechanism for dopaminergic neurons on the basis of marker gene homolog expression (49). The transcriptome profile showed that the Pancreas associated transcription factor 1a (Ptf1a) homolog gene is specifically expressed in ascidian dopaminergic neuron cells, and knock-down and misexpression of the Ptf1a homolog gene suggest that it functions as a crucial factor in dopaminergic neuronal differentiation (49). Thus, the profiling of single-cell transcriptomic data is a useful method to find crucial factors for biological functions. Moreover, fluorescence-activated cell sorting (FACS) is also a useful technique that can collect single cells expressing a target protein, including receptors of neuropeptides on the plasma membrane. FACS sorting and subsequent transcriptomic analyses of single cells expressing a target receptor provide clues that clarify the effects of the target peptide, uncovering the mechanism of signal transduction via peptide-receptor interactions in target cells (Figure 1B).
C. robusta possesses neuropeptides that are categorized as homologs of mammalian peptides or Ciona-specific peptides. Homology-based searches have contributed to the identification of ascidian peptides and receptors of mammalian homologs. However, homology-based identification of receptors for Ciona-specific peptides is virtually impossible due to the low sequence identity of Ciona-specific peptides or their receptors to known ones. In mammals, ligand-screening of orphan receptors has been widely employed by high-throughput reverse-pharmacological assays based on evaluating typical signal transductions. Due to the time, cost, and labor required, only a limited number of ligand-receptor pairs have been identified recently (50). During the past decade, artificial intelligence (AI) technologies have been developed and utilized to search for ligand-receptor pairs within enormous amounts of omics data (51–53). In a machine-learning system, chemical, physicochemical, and biochemical properties and sequence information of “known” ligand-receptor pairs are converted to numerical vectors (descriptors) and used to predict “novel” ligand-receptor pairs. Recently, the peptide descriptor (PD)-incorporated support vector machine (SVM) was developed to predict novel neuropeptide-receptor interactions, and receptors for Ciona-specific peptides were predicted (54). The predicted receptors for Ciona-specific peptides, CiLF1, CiLF2, CiLF5, CiLF6, CiLF7, CiLF8, CiYFV1, CiYFV3, and CiNTLP2, were validated by intracellular calcium mobilization assays (Figure 1A) (54). Thus, AI-based prediction of ligand-receptor pairs without sequence homology and molecular phylogenetic relatedness is contributing to the deorphanization of receptors. In other words, these studies obviously demonstrate the usefulness of omics data in “post-omics” research strategies, such as the machine learning-based prediction of novel peptide-receptor pairs. CiLF receptors are expressed in the ovaries (54), suggesting that at least several CiLFs participate in ovarian events, including follicle growth, oocyte maturation, or ovulation. In addition, transcriptomic profiles of Ciona follicles suggest that several orphan receptors are expressed in the follicles (34). The determination of novel peptide-receptor pairs using PD-incorporated SVM and cell-based validations will surely lead to the verification of the biological events in the ascidian ovary.
Taken together, the integration of omics analyses, physiological analyses, single-cell technologies, and AI technologies will provide novel insight into the mechanisms of the endocrine system and signaling networks in Ciona.
Author Contributions
TK, TO, SM, and HS contributed to the conception and design of the manuscript. TK, TO, SM, TY and HS wrote sections of the manuscript. TK, TO, SM, TS, AS, and HS designed the figures. All authors approved the submitted version.
Funding
This work was supported by grants from the Japan Society for the Promotion of Science (http://www.jsps.go.jp/english/index.html) to TK (JP20K06752), SM (JP19K16182), and HS (JP19K06752).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.858885/full#supplementary-material
References
1. Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and Not Cephalochordates Are the Closest Living Relatives of Vertebrates. Nature (2006) 439:965–8. doi: 10.1038/nature04336
2. Denoeud F, Henriet S, Mungpakdee S, Aury JM, Da Silva C, Brinkmann H, et al. Plasticity of Animal Genome Architecture Unmasked by Rapid Evolution of a Pelagic Tunicate. Science (2010) 330:1381–5. doi: 10.1126/science.1194167
3. Satoh N, Rokhsar D, Nishikawa T. Chordate Evolution and the Three-Phylum System. Proc Biol Sci (2014) 281:20141729. doi: 10.1098/rspb.2014.1729
4. Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, et al. The Draft Genome of Ciona Intestinalis: Insights Into Chordate and Vertebrate Origins. Science (2002) 298:2157–67. doi: 10.1126/science.1080049
5. Satou Y, Kawashima T, Shoguchi E, Nakayama A, Satoh N. An Integrated Database of the Ascidian, Ciona Intestinalis: Towards Functional Genomics. Zoolog Sci (2005) 22:837–43. doi: 10.2108/zsj.22.837
6. Imai K, Hino K, Yagi K, Satoh N, Satou Y. Gene Expression Profiles of Transcription Factors and Signaling Molecules in the Ascidian Embryo: Towards a Comprehensive Understanding of Gene Networks. Development (2004) 131:4047–58. doi: 10.1242/dev.01270
7. Imai KS, Levine M, Satoh N, Satou Y. Regulatory Blueprint for a Chordate Embryo. Science (2006) 312:1183–7. doi: 10.1126/science.1123404
8. Imai KS, Daido Y, Kusakabe TG, Satou. Cis-Acting Transcriptional Repression Establishes a Sharp Boundary in Chordate Embryos. Science (2012) 337:964–7. doi: 10.1126/science.1222488
9. Azumi K, Sabau SV, Fujie M, Usami T, Koyanagi R, Kawashima T, et al. Gene Expression Profile During the Life Cycle of the Urochordate Ciona Intestinalis. Dev Biol (2007) 308:572–82. doi: 10.1016/j.ydbio.2007.05.022
10. Liu B, Satou Y. The Genetic Program to Specify Ectodermal Cells in Ascidian Embryos. Dev Growth Differ (2020) 62:301–10. doi: 10.1111/dgd.12660
11. Lemaire P. Evolutionary Crossroads in Developmental Biology: The Tunicates. Development (2011) 138:2143–52. doi: 10.1242/dev.048975
12. Satoh N. The Ascidian Tadpole Larva: Comparative Molecular Development and Genomics. Nat Rev Genet (2003) 4:285–95. doi: 10.1038/nrg1042
13. Satou Y, Nakamura R, Yu D, Yoshida R, Hamada M, Fujie M, et al. A Nearly Complete Genome of Ciona Intestinalis Type A (C. Robusta) Reveals the Contribution of Inversion to Chromosomal Evolution in the Genus Ciona. Genome Biol Evol (2019) 11:3144–57. doi: 10.1093/gbe/evz228
14. Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, et al. Improved Genome Assembly and Evidence-Based Global Gene Model Set for the Chordate Ciona Intestinalis: New Insight Into Intron and Operon Populations. Genome Biol (2008) 9:R152. doi: 10.1186/gb-2008-9-10-r152
15. Cao C, Lemaire LA, Wang W, Yoon PH, Choi YA, Parsons LR, et al. Comprehensive Single-Cell Transcriptome Lineages of a Proto-Vertebrate. Nature (2019) 571:349–54. doi: 10.1038/s41586-019-1385-y
16. Horie R, Hazbun A, Chen K, Cao C, Levine M, Horie T. Shared Evolutionary Origin of Vertebrate Neural Crest and Cranial Placodes. Nature (2018) 560:228–32. doi: 10.1038/s41586-018-0385-7
17. Edson MA, Nagaraja AK, Matzuk MM. The Mammalian Ovary From Genesis to Revelation. Endocr Rev (2009) 30:624–712. doi: 10.1210/er.2009-0012
18. Richards JS, Pangas SA. The Ovary: Basic Biology and Clinical Implications. J Clin Invest (2010) 120:963–72. doi: 10.1172/JCI41350
19. Von Stetina JR, Orr-Weaver TL. Developmental Control of Oocyte Maturation and Egg Activation in Metazoan Models. Cold Spring Harb Perspect Biol (2011) 3:a005553. doi: 10.1101/cshperspect.a005553
20. Richards JS, Ascoli M. Endocrine, Paracrine, and Autocrine Signaling Pathways That Regulate Ovulation. Trends Endocrinol Metab (2018) 29:313–25. doi: 10.1016/j.tem.2018.02.012
21. Tsutsui K, Ubuka T, Son YL, Bentley GE, Kriegsfeld LJ. Contribution of GnIH Research to the Progress of Reproductive Neuroendocrinology. Front Endocrinol (Lausanne) (2015) 6:179. doi: 10.3389/fendo.2015.00179
22. Uenoyama Y, Inoue N, Nakamura S, Tsukamura H. Central Mechanism Controlling Pubertal Onset in Mammals: A Triggering Role of Kisspeptin. Front Endocrinol (Lausanne) (2019) 10:312. doi: 10.3389/fendo.2019.00312
23. Kawada T, Ogasawara M, Sekiguchi T, Aoyama M, Hotta K, Oka K, et al. Peptidomic Analysis of the Central Nervous System of the Protochordate, Ciona Intestinalis: Homologs and Prototypes of Vertebrate Peptides and Novel Peptides. Endocrinology (2011) 152:2416–27. doi: 10.1210/en.2010-1348
24. Kawada T, Sekiguchi T, Itoh Y, Ogasawara M, Satake H. Characterization of a Novel Vasopressin/Oxytocin Superfamily Peptide and its Receptor From an Ascidian Ciona Intestinalis. Peptides (2008) 29:1672–8. doi: 10.1016/j.peptides.2008.05.030
25. Satake H, Ogasawara M, Kawada T, Masuda K, Aoyama M, Minakata H, et al. Tachykinin and Tachykinin Receptor of an Ascidian, Ciona Intestinalis: Evolutionary Origin of the Vertebrate Tachykinin Family. J Biol Chem (2004) 279:53798–805. doi: 10.1074/jbc.M408161200
26. Adams BA, Tello JA, Erchegyi J, Warby C, Hong DJ, Akinsanya KO, et al. Six Novel Gonadotropin-Releasing Hormones are Encoded as Triplets on Each of Two Genes in the Protochordate, Ciona Intestinalis. Endocrinology (2003) 144:1907–19. doi: 10.1210/en.2002-0216
27. Johnsen AH, Rehfeld JF. Cionin: A Disulfotyrosyl Hybrid of Cholecystokinin and Gastrin From the Neural Ganglion of the Protochordate Ciona Intestinalis. J Biol Chem (1990) 265:3054–58. doi: 10.1016/S0021-9258(19)39732-7
28. Schjoldager B, Park J, Johnsen AH, Yamada T, Rehfeld JF. Cionin, a Protochordean Hybrid of Cholecystokinin and Gastrin: Biological Activity in Mammalian Systems. Am J Physiol (1991) 260(6 Pt 1):G976–82. doi: 10.1152/ajpgi.1991.260.6.G977
29. Osugi T, Sasakura Y, Satake H. The Nervous System of the Adult Ascidian Ciona Intestinalis Type A (Ciona Robusta): Insights From Transgenic Animal Models. PloS One (2017) 12:e0180227. doi: 10.1371/journal.pone.0180227
30. Osugi T, Sasakura Y, Satake H. The Ventral Peptidergic System of the Adult Ascidian Ciona Robusta (Ciona Intestinalis Type A) Insights From a Transgenic Animal Model. Sci Rep (2020) 10:1892. doi: 10.1038/s41598-020-58884-w
31. Kawada T, Shiraishi A, Matsubara S, Hozumi A, Horie T, Sasakura Y, et al. Vasopressin Promoter Transgenic and Vasopressin Gene-Edited Ascidian, Ciona Intestinalis Type A (Ciona Robusta): Innervation, Gene Expression Profiles, and Phenotypes. Front Endocrinol (Lausanne) (2021) 12:668564. doi: 10.3389/fendo.2021.668564
32. Sekiguchi T, Ogasawara M, Satake H. Molecular and Functional Characterization of Cionin Receptors in the Ascidian, Ciona Intestinalis: The Evolutionary Origin of the Vertebrate Cholecystokinin/Gastrin Family. J Endocrinol (2012) 213:99–106. doi: 10.1530/JOE-11-0410
33. Prodon F, Chenevert J, Sardet C. Establishment of Animal-Vegetal Polarity During Maturation in Ascidian Oocytes. Dev Biol (2006) 290:297–311. doi: 10.1016/j.ydbio.2005.11.025
34. Matsubara M, Shiraishi A, Osugi T, Kawada T, Satake H. The Regulation of Oocyte Maturation and Ovulation in the Closest Sister Group of Vertebrates. Elife (2019) 8:e49062. doi: 10.7554/eLife.49062
35. Matsubara S, Shiraishi A, Osugi T, Kawada T, Satake H. Fractionation of Ovarian Follicles and In Vitro Oocyte Maturation and Ovulation Assay of Ciona Intestinalis Type A. Bio Protoc (2020) 10:e3577. doi: 10.21769/BioProtoc.3577
36. Aoyama M, Kawada T, Fujie M, Hotta K, Sakai T, Sekiguchi T, et al. A Novel Biological Role of Tachykinins as an Up-Regulator of Oocyte Growth: Identification of an Evolutionary Origin of Tachykininergic Functions in the Ovary of the Ascidian, Ciona Intestinalis. Endocrinology (2008) 149:4346–56. doi: 10.1210/en.2008-0323
37. Aoyama M, Kawada T, Satake H. Localization and Enzymatic Activity Profiles of the Proteases Responsible for Tachykinin-Directed Oocyte Growth in the Protochordate, Ciona Intestinalis. Peptides (2012) 34:186–92. doi: 10.1016/j.peptides.2011.07.019
38. Osugi T, Miyasaka N, Shiraishi A, Matsubara S, Satake H. Cionin, a Vertebrate Cholecystokinin/Gastrin Homolog, Induces Ovulation in the Ascidian Ciona Intestinalis Type A. Sci Rep (2021) 11:10911. doi: 10.1038/s41598-021-90295-3
39. Takeda N, Kon Y, Quiroga Artigas G, Lapébie P, Barreau C, Koizumi O, et al. Identification of Jellyfish Neuropeptides That Act Directly as Oocyte Maturation-Inducing Hormones. Development (2018) 145:dev156786. doi: 10.1242/dev.156786
40. Mita M, Yoshikuni M, Ohno K, Shibata Y, Paul-Prasanth B, Pitchayawasin S, et al. A Relaxin-Like Peptide Purified From Radial Nerves Induces Oocyte Maturation and Ovulation in the Starfish, Asterina Pectinifera. Proc Natl Acad Sci USA (2009) 106:9507–12. doi: 10.1073/pnas.0900243106
41. Kato S, Tsurumaru S, Taga M, Yamane T, Shibata Y, Ohno K, et al. Neuronal Peptides Induce Oocyte Maturation and Gamete Spawning of Sea Cucumber, Apostichopus Japonicus. Dev Biol (2009) 326:169–76. doi: 10.1016/j.ydbio.2008.11.003
42. Jessus C, Munro C, Houliston E. Managing the Oocyte Meiotic Arrest-Lessons From Frogs and Jellyfish. Cells (2020) 9:1150. doi: 10.3390/cells9051150
43. Takahashi T, Hagiwara A, Ogiwara K. Follicle Rupture During Ovulation With an Emphasis on Recent Progress in Fish Models. Reproduction (2019) 157:R1–R13. doi: 10.1530/REP-18-0251
44. Zhu Y. Metalloproteases in Gonad Formation and Ovulation. Gen Comp Endocrino (2021) 314:113924. doi: 10.1016/j.ygcen.2021.113924
45. Deady LD, Shen W, Mosure SA, Spradling AC, Sun J. Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in Drosophila. PloS Genet (2015) 11:e1004989. doi: 10.1371/journal.pgen.1004989
46. Singh V, Chaube R, Joy KP. Vasotocin Stimulates Maturation-Inducing Hormone, Oocyte Maturation and Ovulation in the Catfish Heteropneustes Fossilis: Evidence for a Preferential Calcium Involvement. Theriogenology (2021) 167:51–60. doi: 10.1016/j.theriogenology.2021.03.001
47. Joy KP, Chaube R. Vasotocin–A New Player in the Control of Oocyte Maturation and Ovulation in Fish. Gen Comp Endocrinol (2015) 221:54–63. doi: 10.1016/j.ygcen.2015.02.013
48. Bernard V, Bouilly J, Kramer P, Carré N, Schlumberger M, Visser JA, et al. The Tyrosine Kinase Inhibitor Sunitinib Affects Ovulation But Not Ovarian Reserve in Mouse: A Preclinical Study. PloS One (2016) 11:e0152872. doi: 10.1371/journal.pone.0152872
49. Horie T, Horie R, Chen K, Cao C, Nakagawa M, Kusakabe TG, et al. Regulatory Cocktail for Dopaminergic Neurons in a Protovertebrate Identified by Whole-Embryo Single-Cell Transcriptomics. Genes Dev (2018) 32:1297–302. doi: 10.1101/gad.317669.118
50. Laschet C, Dupuis N, Hanson J. The G Protein-Coupled Receptors Deorphanization Landscape. Biochem Pharmacol (2018) 153:62–74. doi: 10.1016/j.bcp.2018.02.016
51. Yabuuchi H, Niijima S, Takematsu H, Ida T, Hirokawa T, Hara T, et al. Analysis of Multiple Compound-Protein Interactions Reveals Novel Bioactive Molecules. Mol Syst Biol (2011) 7:390–400:472. doi: 10.1038/msb.2011.5
52. Bushdid C, de March CA, Fiorucci S, Matsunami H, Golebiowski J. Agonists of G-Protein-Coupled Odorant Receptors Are Predicted From Chemical Features. J Phys Chem Lett (2018) 9:2235–40. doi: 10.1021/acs.jpclett.8b00633
53. Raschka S, Kaufman B. Machine Learning and AI-Based Approaches for Bioactive Ligand Discovery and GPCR-Ligand Recognition. Methods (2020) 180:89–110. doi: 10.1016/j.ymeth.2020.06.016
Keywords: ascidian, Ciona intestinalis, peptidomics, transcriptome, peptide, receptor
Citation: Kawada T, Osugi T, Matsubara S, Sakai T, Shiraishi A, Yamamoto T and Satake H (2022) Omics Studies for the Identification of Ascidian Peptides, Cognate Receptors, and Their Relevant Roles in Ovarian Follicular Development. Front. Endocrinol. 13:858885. doi: 10.3389/fendo.2022.858885
Received: 20 January 2022; Accepted: 10 February 2022;
Published: 07 March 2022.
Edited by:
Maurice Richard Elphick, Queen Mary University of London, United KingdomReviewed by:
Daniel Thiel, University of Exeter, United KingdomKazue Nagasawa, Tohoku University, Japan
Copyright © 2022 Kawada, Osugi, Matsubara, Sakai, Shiraishi, Yamamoto and Satake. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honoo Satake, c2F0YWtlQHN1bmJvci5vci5qcA==
†These authors have contributed equally to this work
 Tsuyoshi Kawada
Tsuyoshi Kawada Tomohiro Osugi
Tomohiro Osugi Shin Matsubara
Shin Matsubara Tsubasa Sakai
Tsubasa Sakai Akira Shiraishi
Akira Shiraishi Tatsuya Yamamoto
Tatsuya Yamamoto Honoo Satake
Honoo Satake