
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 01 March 2022
Sec. Diabetes: Molecular Mechanisms
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.821091
This article is part of the Research Topic Monogenic Diabetes: from Genetics and Cell Biology to Clinical Practice View all 14 articles
 Yanwu Yang1*‡
Yanwu Yang1*‡ Michael D. Glidden2,3,4‡
Michael D. Glidden2,3,4‡ Balamurugan Dhayalan1‡
Balamurugan Dhayalan1‡ Alexander N. Zaykov5†
Alexander N. Zaykov5† Yen-Shan Chen1
Yen-Shan Chen1 Nalinda P. Wickramasinghe2
Nalinda P. Wickramasinghe2 Richard D. DiMarchi5
Richard D. DiMarchi5 Michael A. Weiss1*
Michael A. Weiss1*Toxic misfolding of proinsulin variants in β-cells defines a monogenic diabetes syndrome, designated mutant INS-gene induced diabetes of the young (MIDY). In our first study (previous article in this issue), we described a one-disulfide peptide model of a proinsulin folding intermediate and its use to study such variants. The mutations (LeuB15→Pro, LeuA16→Pro, and PheB24→Ser) probe residues conserved among vertebrate insulins. In this companion study, we describe 1H and 1H-13C NMR studies of the peptides; key NMR resonance assignments were verified by synthetic 13C-labeling. Parent spectra retain nativelike features in the neighborhood of the single disulfide bridge (cystine B19-A20), including secondary NMR chemical shifts and nonlocal nuclear Overhauser effects. This partial fold engages wild-type side chains LeuB15, LeuA16 and PheB24 at the nexus of nativelike α-helices α1 and α3 (as defined in native proinsulin) and flanking β-strand (residues B24-B26). The variant peptides exhibit successive structural perturbations in order: parent (most organized) > SerB24 >> ProA16 > ProB15 (least organized). The same order pertains to (a) overall α-helix content as probed by circular dichroism, (b) synthetic yields of corresponding three-disulfide insulin analogs, and (c) ER stress induced in cell culture by corresponding mutant proinsulins. These findings suggest that this and related peptide models will provide a general platform for classification of MIDY mutations based on molecular mechanisms by which nascent disulfide pairing is impaired. We propose that the syndrome’s variable phenotypic spectrum—onsets ranging from the neonatal period to later in childhood or adolescence—reflects structural features of respective folding intermediates.
The mutant proinsulin syndrome (MPS) is a prototypical disease of toxic protein misfolding. Unlike toxic extracellular aggregation as observed among neurodegenerative diseases (1) and diverse amyloid disorders (2), in the MPS a dominant mutation impairs proinsulin folding efficiency in a critical intracellular organelle: the endoplasmic reticulum (ER). Impaired foldability of the variant protein of pancreatic β-cells leads to aberrant aggregation and in turn induces chronic ER stress (3–5). Although the unfolded protein response (UPR) evolved as an adaptive pathway [broadly conserved among eukaryotic cells (6, 7)], its chronic activation in β-cells impairs glucose-stimulated insulin secretion and β-cell viability [for review, see (8, 9)]. Also designated mutant INS-gene induced diabetes of the young (MIDY) (4), MPS encompasses a range of patient phenotypes, representing subtypes of permanent neonatal diabetes mellitus (PNDM) to maturity-onset diabetes of the young (MODY) (10–13).
Among monogenic endocrine syndromes in general [such as complete or partial androgen insensitivity (14)], a given mutation may be associated with a range of phenotypes, even in the same kindred (15). A given mutation in the androgen receptor, for example, may be associated with male development, somatic female development with Mullerian regression, or ambiguous genitalia (16). The complexity of genotype-phenotype relationships (GPR) in such syndromes presumably reflects the influence of modifier genes in multigenic regulatory pathways (17). Modifier genes have also been inferred in the genetics of Type 1 diabetes mellitus (DM). GPRs in MPS may be more straightforward: the extent of β-cell dysfunction and velocity of β-cell loss (together determining age of diabetes onset in a particular patient) may reflect mutation-specific molecular properties, i.e., whether a given amino-acid substitution is associated with a severe (PNDM) or mild (MODY) perturbation to foldability. In our initial study [preceding article in this issue (18)] we designed a 49-residue peptide model of an early on-pathway proinsulin folding intermediate and its application to three representative MIDY mutations. This single-chain model contains only one disulfide bridge and is thus designated 1SS. This bridge (cystine B19-A20) is the first to accumulate among populated partial folds in the in vitro folding pathway of proinsulin and homologous factors (19, 20). The present study provides a detailed two-dimensional NMR study of the parent 1SS peptide and representative MIDY-related variants.
Native insulin contains two chains, B (30 residues) and A (21 residues) (21). Its native structure contains three α-helices stabilized by two inter-chain disulfide bridges (cystines B7-A7 and B19-A20) and one intrachain bridge (A6-A11; Figure 1A) (23). Whereas chain combination between the isolated chains is inefficient (24), cellular biosynthesis exploits a single-chain precursor, proinsulin, wherein disulfide pairing is intramolecular (25). Human proinsulin contains a disordered 35-residue connecting domain (C domain) between ThrB30 and GlyA1. Efficiency of disulfide pairing in single-chain precursors can be augmented by shortening the C domain or deleting it entirely (26). The present peptide model contains a peptide bond between residues B28 and A1 (red bars in Figure 1B); the native ProB28 is substituted by Lys to permit convenient enzymatic cleavage to a two-chain hormone (red in sequence N in Figure 1B) (27). The 49-residue synthetic precursor (designated “DesDi”) exhibits remarkable folding efficiency, enabling preparation of certain insulin analogs refractory to classical chain combination (27). In the 1SS model cystine B7-A7 (solvent exposed in native insulin) is pairwise substituted by Ser whereas cystine A6-A11 (buried in the core of native insulin) is pairwise substituted by Ala (18). Segmental α-helical propensity and solubility were augmented by acidic surface substitutions HisB10→Asp (28, 29) and ThrA8→Glu (30). The seven amino-acid substitutions in the parent 1SS peptide (positions B7, B10, B28, A6-A8 and A11) are highlighted in red in Figure 1B (entry 1SS-WT). Relative to an homologous two-chain peptide model of the corresponding 1SS IGF-I folding intermediate (Figure 1C) (22), we anticipated that the 1SS DesDi-based single-chain model would exhibit a conformational equilibrium biased toward a collapsed state (Figure 1D).
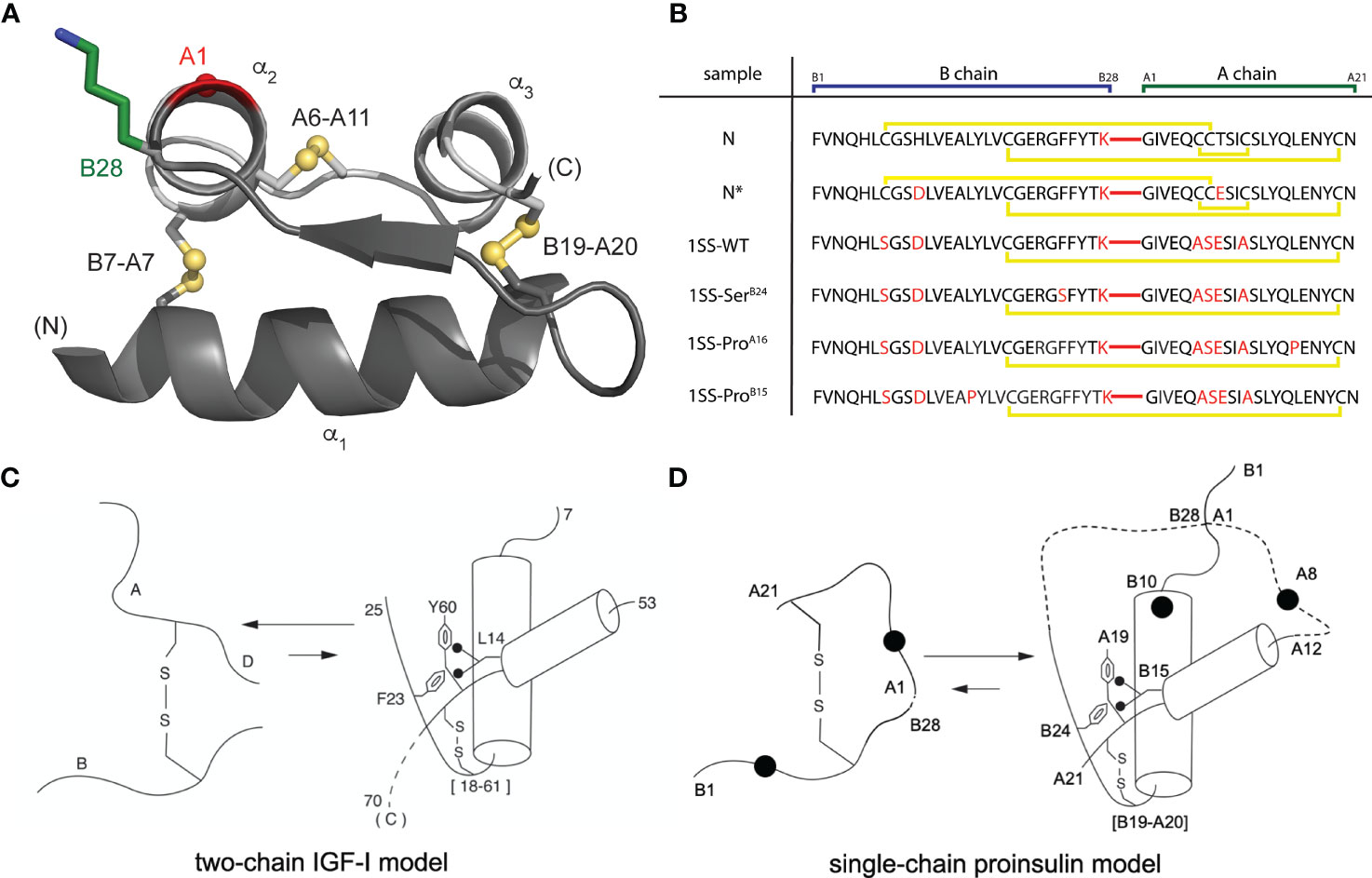
Figure 1 Single-chain DesDi as a proinsulin model. (A) Cartoon model of single-chain DesDi molecule that connects A- and B- chains through a linkage between LysB28 and GlyA1 (PDB entry 7JP3). Color legend: B chain, dark gray; A chain, light gray; LysB28 side chain, green sticks; GlyA1-Cα, red sphere; sulfur atoms in disulfide bonds are shown as spheres. All the spheres are set to one third Van der walls radii. N- and C- terminals are indicated. Insulin’s three characteristic helices are labeled as α1, α2, and α3. (B) DesDi protein sequences. Sample names at left report the presence of either one ([B19-A20]) or three (N and N*) disulfide bonds; yellow lines show disulfide linkages. A red line connecting the C-terminal B-domain and N-terminal A-domain sequences signifies the presence of a peptide bond between residues B28 and A1. All [B19-A20]-1SS samples and N* have additional mutations GluA8 and AspB10 to enhance solubility at neutral pH and the high protein concentrations needed for NMR spectroscopy. (C) General folding nucleus of insulin-related superfamily is shown with IGF-I as a model. (D) Folding nucleus of 1SS-DesDi single chain visualized as a proinsulin model. Initial pairing of B19-A20 disulfide in combination with the formation of two-helices (B9-B19 and A12-A20) are considered to be central events in the formation of folding nucleus. Dotted lines represent disordered regions. Larger circles represent the AspB10 and GluA8 substitutions. Key residues are highlighted by their sequence position [panel (C, D) modified from Hua et al. (22)].
To connect our model to patient phenotypes, three MIDY mutations were introduced into the 1SS peptide model (Figure 1B). Two (ProB15 and ProA16) are associated with neonatal-onset DM (31, 32); the third (SerB24) is associated with onset in early adulthood (33). The structural environments of these conserved side chains are shown in Figure 2A. Whereas the side chains of LeuB15 and LeuA16 are buried in the hydrophobic core (Figures 2B, C, E, F), the aromatic side chain of PheB24 packs within a crevice overlying internal cystine B19-A20 such that one side of the aromatic ring is exposed to solvent (Figures 2B–D). Initial characterization of these peptides was described in our companion article (18). Whereas the Pro substitutions introduced marked perturbations in folding efficiency in a mini-proinsulin [“DesDi”, (27)], SerB24 was well tolerated. Synthetic yields mirrored residual α-helix contents (as inferred from far-UV circular dichroism; CD) in the corresponding 1SS peptides (18). A further correlation was observed between these properties and a pertinent pathogenetic process: extent of ER stress induced in a human cell line (HEK 293T) on transient expression of the corresponding mutant proinsulins. The coherence of these correlations (18) motivated this companion study wherein NMR spectroscopy provides a residue-specific view.
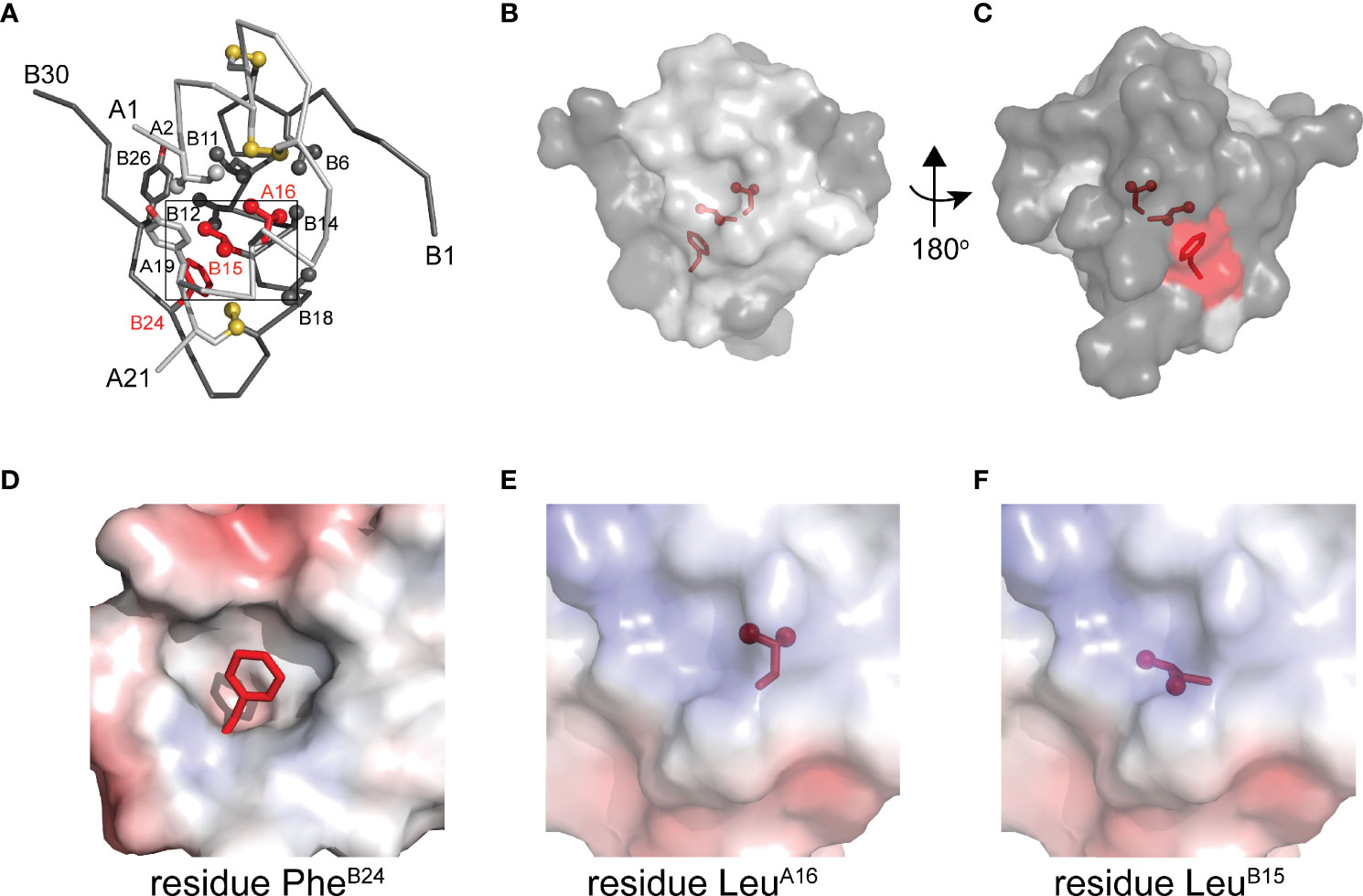
Figure 2 Native structure of insulin and sites of present MIDY mutations. (A) ribbon diagram of insulin monomer (PDB entry 4INS). Side chains of key residues are shown as sticks with methyl groups and sulfur atoms in disulfide bonds are shown as spheres (one third van der Waals radii). Clinical mutations under the study is shown in red and all other side chains follow the color code: A chain (light gray), B chain (dark gray). (B) Surface representation of insulin monomer (transparency set to 40%) showing the side chains of PheB24, LeuB15 and LeuA16 (sticks shown as in panel A) within the core of the molecule. The surface of these residues is shown in red. (C) A view after rotating the molecule in panel B by 180° through y-axis. (D–F) Respective close-up views of the environments of PheB24 (crevice), LeuA16 (core) and LeuB15 (core). In these panels the highlighted side chain is shown in red. These electrostatic surfaces were calculated in absence of indicated side chain.
In this companion study we employ two-dimensional 1H- and [1H, 13C]-NMR methods to interrogate the 1SS peptide models in relation to the native insulin. Analysis of main-chain 1H and 13C chemical shifts in the parent peptide (34–36) provided evidence for nativelike nascent α-helices in the B domain (residues B9-B19) and A domain (A12-A20), together in accordance with CD-defined α-helix content (18). Although chemical-shift dispersion in this and the variant partial folds is more limited than in the spectrum of a native insulin monomer (37), key side-chain resonance assignments were verified by site-specific 13C (and 13C, 15N) labeling, using selective labeled amino-acid precursors in chemical peptide synthesis. Analysis of signature chemical shifts and framework nuclear Overhauser effects (NOEs) provides evidence for a nativelike folding nucleus in the parent 1SS peptide that is dependent on maintenance of the B19-A20 disulfide bridge. The clinical mutations perturb this nascent structure in the order SerB24 (least perturbed) >> ProA16 > ProB15 (most perturbed). Together, these findings suggest that the DesDi-based 1SS model will provide a general platform for comparative biophysical studies of that subset of MIDY mutations that perturb initial closure of cystine B19-A20 in proinsulin biosynthesis in pancreatic β-cells.
Peptides were synthesized either with an ABI 433A Peptide Synthesizer (Applied Biosystems) or Tribute 2-Channel peptide synthesizer (Gyros Protein technologies) using a preprogrammed solid-phase fluorenylmethyloxycarbonyl chloride (Fmoc) protocol designed for standard 0.1mmol scale syntheses. ABI protocols consist of the following modules: cycle-1 [d], cycle-2 [aibde], cycle-3 with number of repetitions equal #aa-1 [afgbde], cycle-“#aa+2” [ffbdc], where #aa is a number of amino acids in the sequence. Automated couplings utilized diisopropylcarbodiimide (DIC)/6-Cl-hydroxybenzotriazole (6-Cl-HOBt) in N-methyl pyrrolidone (NMP) whereas Fmoc deprotections used 20% piperidine in NMP. α-Carboxyl-protected Asp was used in place of Asn in all syntheses of DesDi analogs to accommodate the use of ChemMatrix® Rink-Amide resin (loading = 0.46mmol/g). The Tribute peptide synthesizer used heating protocols: coupling was done at 6 min at 60°C except for Cys/His (2 min at 25°C, then 5 min at 60 °C) and Arg (20 min at 25 °C, then 5 min at 60°C); deprotection was done twice (30 sec at 50°C, then 3 min at 50°C). Reagent conditions were otherwise similar to ABI protocols except that DMF was used as solvent and choice of the resin was H-Asn(Trt)-HMPB-ChemMatrix® resin. Peptides were cleaved with a trifluoroacetic acid (TFA) cocktail (2.5% vol/vol of each: β-mercaptoethanol, triisopropylsilane, anisole, and water) followed by ether precipitation.
Crude peptides from ether precipitation were dissolved in glycine buffer (20mM glycine and 2mM cysteine hydrochloride, pH 10.5) to a final peptide concentration of 0.1mM. The pH of this solution was readjusted to 10.5 to account for traces of residual TFA present in lyophilized peptides. This solution was then stirred while open to air at 4°C until reaction completion (usually overnight). The pH of the solution was then lowered to ~2.0 with 5N HCl to neutralize the folding reaction. Folded peptide was then purified by preparative rp-HPLC with a 60 min 20→40% acetonitrile elution gradient (10-15mL/min flow rate) run by a Waters rp-HPLC (Milford, MA) equipped with a Waters Model 600 Controller, Waters 2487 Dual Wavelength Detector, ProStar Model 701 Fraction Collector, Kipp & Zonen BD41 chart recorder, and a Luna 10μm C8(2) 100A AXIA (250 × 21.2mm) column. Fractions containing clean peptide were pooled and lyophilized. Purity of the materials was confirmed by LCQ Advantage Ion Trap Mass Spectrometer System coupled to an Agilent 1100 Series HPLC system utilizing a TARGA C8 5-μm (250 mm x 4.6 mm) column and a 35 min 25→50% acetonitrile elution gradient (1mL/min flow rate). See Table S1 for LCMS retention times and mass verification.
Isotopically labeled 1SS and control peptides were prepared on 0.05 mmol scales. N* was assembled in entirety as a single batch whereas the 1SS peptides were assembled as a single batch (0.1mmol) through residue B25. At that point, half of the 1SS resin was used to complete assembly of the 1SS-SerB24 analog. The remaining resin was used to complete the 1SS synthesis. Coupling of isotopically labeled amino acids (Cambridge Isotopes Inc., Tewksbury, MA) was performed manually using a single 0.3mmol mixture consisting of equivalent amounts of labeled Fmoc amino acid, 1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU), and N,N-diisopropylethylamine (DIEA) reacted with each individual batch of resin (2.25X overall excess). The same automated synthesis protocol used for unlabeled peptide syntheses was then used for the addition of subsequent unlabeled amino acids to the labeled peptide assembly. The following labeled amino acids were used (residual positions given in blue in Scheme S1): Fmoc-Gly[13C2, 15N]-OH, Fmoc-Leu[13C6, 15N]-OH, Fmoc-Ile[13C6, 15N]-OH, Fmoc-Val[13C5, 15N]-OH, and Fmoc-Tyr[13C9, 15N]-OH. The peptides were cleaved from the resin by treatment with TFA cocktail as described above. The folding and purification of these isotope labeled 1SS peptides followed essentially same protocol as described in the companion article (18). Purity of the materials was confirmed by LC-MS with an Agilent 1260 Infinity/6120 Quadrupole instrument utilizing a Kinetex C8 2.6-μm 100A (75 mm x 2.1 mm) column and a 10 min 10-80% acetonitrile elution gradient (1mL/min flow rate) (See Figures S1-S3).
Wild-type insulin and insulin lispro were purified from U-100 pharmaceutical formulations of Humulin® and Humalog® (Eli Lilly and Co.), respectively, using preparative RP-HPLC (C4 10μm 250×20mm Proto 300 Column; Higgins Analytical, Inc.) utilizing buffer A (0.1% TFA in H2O) and a 10-min elution gradient of 20→70% buffer B (0.1% TFA in acetonitrile). Following lyophilization of the collected protein fraction, purity was verified using analytical HPLC (TARGA C8 5-μm [250 mm x 4.6 mm]; Higgins Analytical, Inc.) with a 35-min elution gradient of 25→50% buffer B; molar mass was verified with an Applied Biosystems 4700 Proteomics Analyzer utilizing MALDI-TOF in reflector mode. chromatographic retention times and mass measurements for these clinical analogs are given in Table S1.
Plasmids expressing full-length human proinsulin or variants were constructed by polymerase chain reaction (PCR). Mutations in proinsulin were introduced using QuikChange™ (Stratagene). Constructions were verified by DNA sequencing.
1H NMR spectra were acquired at a proton frequency of 700 MHz at pD 7.4 (direct meter reading) at 35°C. 1H-13C heteronuclear single-quantum coherence (HSQC) spectra were acquired at natural abundance as described. The spectra were obtained at 13C frequency of 176 MHz at a constant temperature of 308 K using the “hsqcetgp” Bruker pulse sequence as described by the manufacturer. Aliphatic 1H-13C HSQCs were acquired with FID size 2048 x 128, 800 scans, 1.0 sec relaxation delay, sweep widths 11 ppm (1H) and 70 ppm (13C) with offset 4.7 and 40 ppm for the 1H and 13C dimension, respectively. Similar parameters were used to acquire aromatic 1H-13C HSQC, except with sweep widths of 40 ppm and 125 ppm offset in 13C dimension. Data were processed with Topspin 4.0.6 (Bruker Biospin) and analyzed with Sparky software (38) using a 90° shifted-sine window function to a total of 2048 × 1024 data points (F2 × F1), followed by automated baseline- and phase correction. All NMR data were acquired using a BRUKER 700 MHz spectrometer equipped with quadruple [1H, 19F, 13C, 15N]-resonance liquid-helium-cooled cryoprobe.
Protein secondary structure was inferred from selected 1H and 13C secondary chemical shifts (1HN, 1Hα, 13Cα and 13Cβ) as described (35, 39, 40). In such algorithms 13Cα and 1Hα chemical shifts distinguish α-helix from β-strand or random coil (41) whereas 13Cβ secondary shifts are more sensitive to β-strand. These chemical shifts in the parent 1SS model were assigned on the basis of 2D 1H-1H NOESY, TOCSY, DQF-COSY in D2O and H2O (10% D2O) and natural abundance 1H-13C HSQC spectra. Corresponding secondary shifts were extracted from observed chemical shifts (Δ = δobs - δcoil). Secondary structural elements were predicted by TALOSplus (34–36).
Structural ensembles were calculated by simulated annealing using XPLOR-NIH (42–44). The models of the one-disulfide proinsulin and one-disulfide DesDi intermediates (containing cystine B19-A20) were generated using distance restraints pertaining to residues A16-A21 and B15-B26 as observed in an engineered proinsulin (45) or an engineered insulin monomer (46). To allow for protein flexibility in these partial folds, upper bounds on long-range distance restraints were increased by 3 Å relative to NMR-derived upper bounds obtained in prior studies of insulin and proinsulin (45, 46).
One-dimensional 1H-NMR spectra of the 1SS peptide model and its variants were presented in our companion study (18) in relation to spectra of the native state (provided as Figure S4 for convenience of the reader). Molecular properties of these peptides are summarized in Table S1. Although the 1D spectra were in overall accordance with trends in synthetic yield, CD deconvolution and redox stability (18), interpretation was limited by the small number of resolved features. To circumvent this limitation, 2D homonuclear and 2D 1H-13C HSQC NMR spectra were obtained at natural abundance. Analysis was undertaken in reference to baseline HSQC spectra of native DesDi (as the single-chain precursor and as cleaved two-chain hormone analog; Figures 3A, B). Near-complete assignment of 1HN, 1Hα, 13Cα and 13Cβ resonances in the parent 1SS peptide model enabled mapping of secondary structure based on pattern of secondary chemical shifts (Figure 4) (34–36). α-Helical segments comprise residues B9-B18 and A13-A19 (Figure 4E), a subset of native secondary structure. Chemical shifts (referenced below) and estimates of chemical-shift dispersion are tabulated in Tables S2–S9.
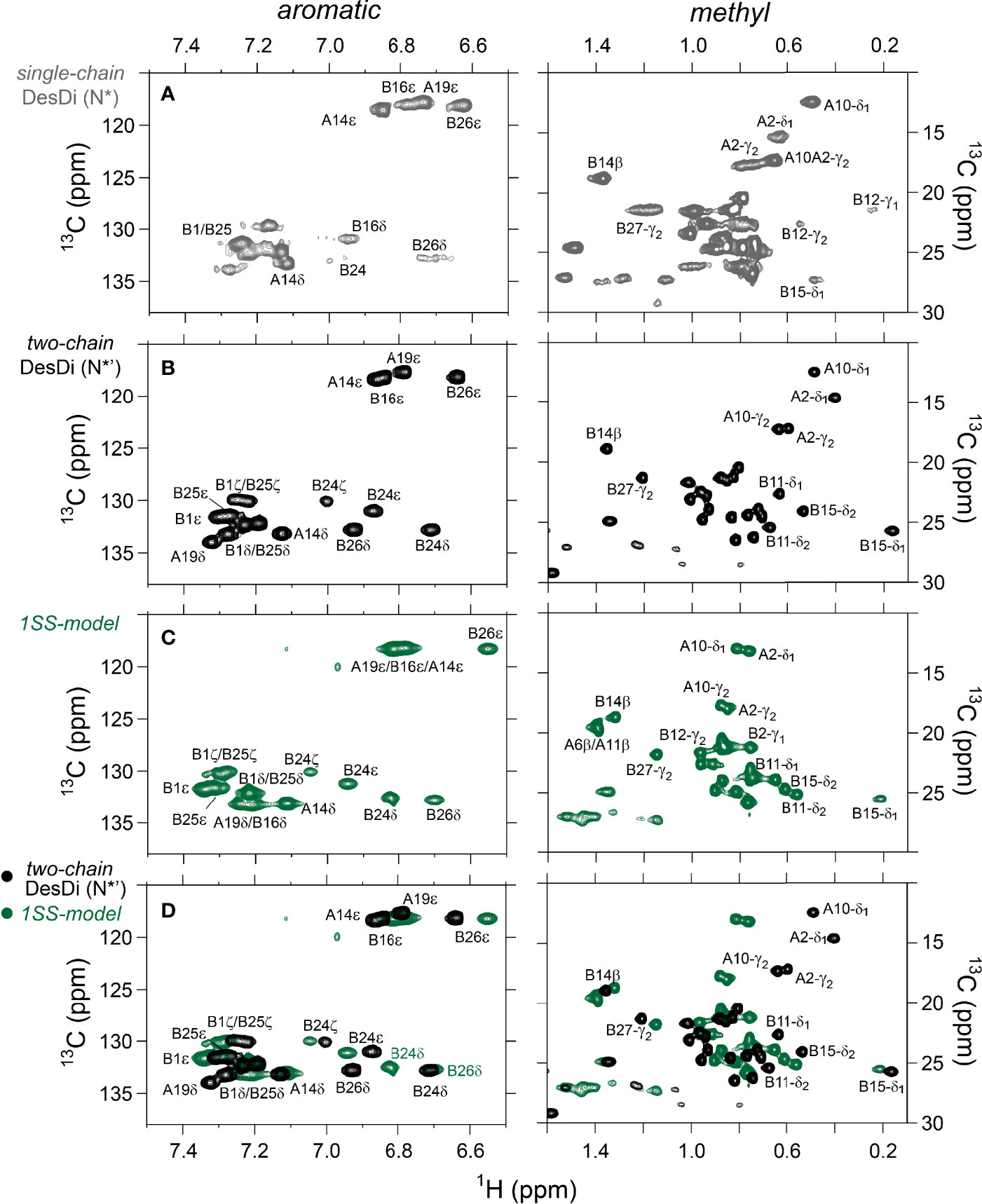
Figure 3 Nature abundance 1H,13C HSQC NMR spectra of DesDi analogs in aromatic (left panel) and methyl region (right panel). (A) single-chain DesDi (N*); (B) two-chain DesDi (N*’); (C) single-chain 1SS-DesDi model (green) and (D) the 1H-13C HSQC spectral overlay of two-chain DesDi (N*’, black) and 1SS-model (green). Spectra were acquired at a 1H frequency of 700 MHz at pD 7.4 (direct meter reading) and at 35°C in D2O. Selected resonance assignments are as indicated.
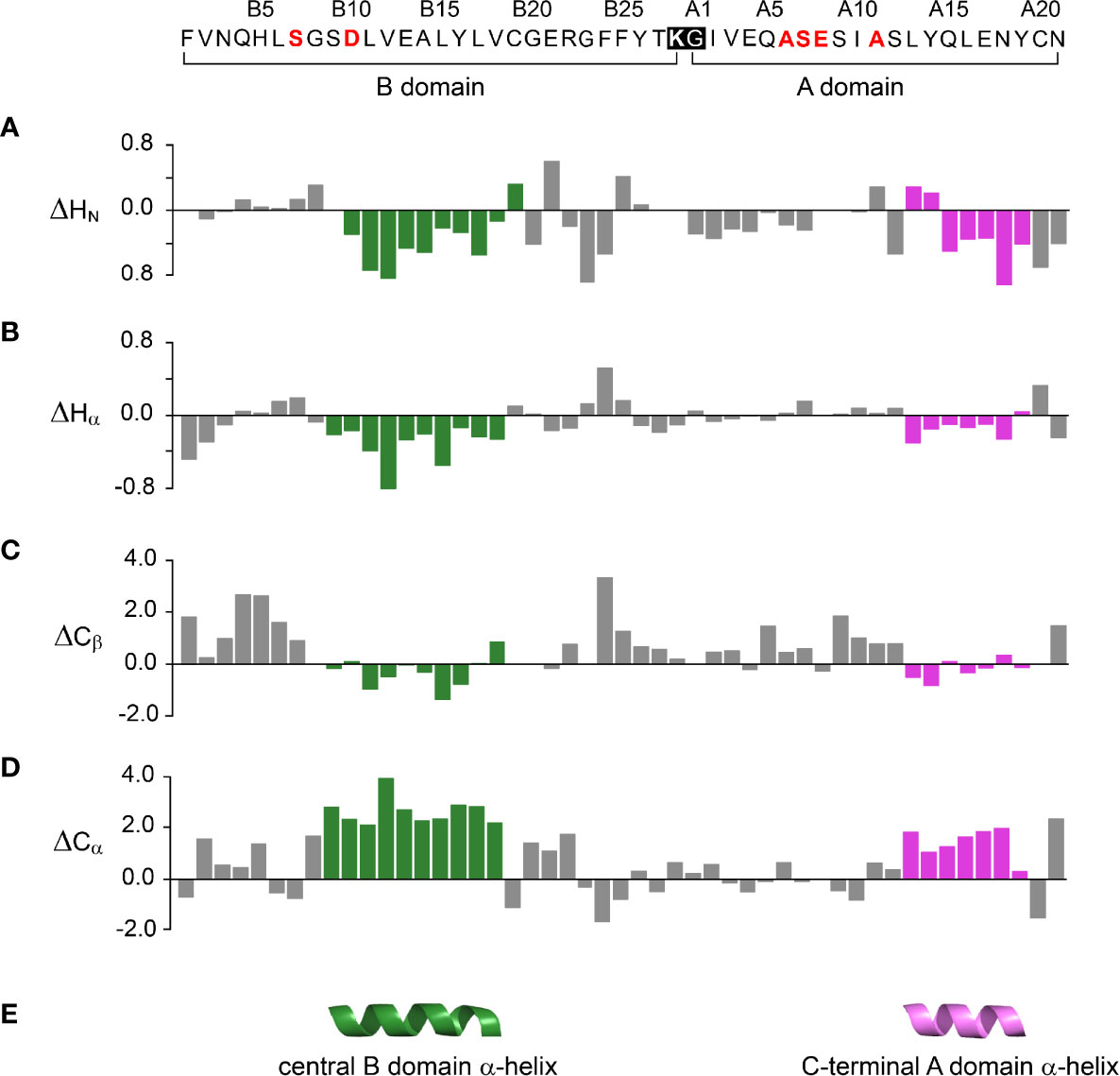
Figure 4 Chemical shift index (CSI) and inferred secondary structure of parent peptide model. Top, 49-residue polypeptide sequence with six amino-acid substitutions in red. DesDi’s engineered junction between LysB28 (bold) and GlyA1 (27) is highlighted within black box. The secondary shift was defined as the difference of observed chemical shift and random coil shift (Δ = δobs - δcoil). Positive (negative) values indicate that the observed chemical shift is downfield (upfield) of expected random-coil frequency (34, 35). (A–D) CSI-related secondary chemical shifts by residue: (A) amide protons (ΔHN); (B) α-protons (ΔHα); (C) β-carbons (ΔCβ); (D) α-carbons (ΔCα). (E) Inferred elements of secondary structure as predicted by TALOSplus (34–36): central B-domain α-helix (residues B9-B18) and C-terminal A-domain α-helix (residues A13-A19). Corresponding residue-specific secondary shifts are given in Table S9.
1H-13C HSQC spectra provide correlations between a 13C atom and an attached proton (1H) via a one-bond J-coupling (47, 48). Initial spectra were obtained at 35 °C. Key resonance assignments in the parent 1SS peptide and 1SS-SerB24 variant were verified by site-specific 13C labeling of residues ValB12, LeuB15, GlyB23, PheB24, TyrB26 and IleA2 (Scheme S1 and Figure S5). In each figure aromatic 2D spectra are shown at left, and aliphatic spectra at right. The spectrum of the native state is better illustrated by two-chain [AspB10, GluA8]-DesDi-insulin (Figure 3B; black) than its single-chain precursor (Figure 3A; gray) due to selected resonance broadening in the latter spectrum; such broadening may reflect partial dimerization (stronger in single-chain analogs) and/or conformational exchange intermediate on the timescale of NMR chemical shifts. The baseline 2D 1H-13C HSQC spectrum of two-chain [AspB10, GluA8]-DesDi-insulin is notable for its resolution of many individual spin systems; selected resonance assignments are provided in Figures 3A, B. Of particular interest are the aromatic resonance of PheB24 and TyrB26, which pack against the central B-chain α-helix and influence the chemical shifts (via their aromatic ring currents) of the side chains of LeuB11 and LeuB15. These respective upfield aromatic and upfield methyl resonances provide markers of the native B-chain supersecondary structure (49, 50). The upfield chemical shifts of IleA2 by contrast reflects A-chain supersecondary structure (in particular the aromatic ring current of TyrA19); that of IleA10 reflects inter-chain packing of HisB5 (51, 52). The latter structural features (and their NMR signatures) are reinforced by cystines A6-A11 and B7-A7 in native insulin.
The 2D 1H-13C HSQC spectrum of the parent 1SS model (Figure 3C; green) is remarkable for preservation of some upfield shifts (PheB24, TyrB26, LeuB11 and LeuB15) but not others (IleA2 and IleA10). The former subset of nativelike chemical shifts indicates maintenance of B-chain secondary structure flanking CysB19 (and therefore the B19-A20 disulfide bridge) whereas the latter attenuation of structure-dependent secondary shifts indicates destabilization of long-range structure involved the A2-A10 segment in accordance with removal of the B7-A7 and A6-A11 disulfide bridges (50, 53, 54). This combination of preserved and attenuated chemical shifts in the spectrum of the parent 1SS model is illustrated by overlay of its spectrum (green) and that of two-chain [AspB10, GluA8]-DesDi-insulin (black) in Figure 3D. The spectrum of the parent 1SS peptide in turn provides a baseline for comparative analysis of the MIDY-derived variants (Figure 5). Chemical-shift dispersion is markedly attenuated in each of the variants (Figures 5B–D), most markedly in 1SS-ProB15 (Figure 5D). In the case of 1SS-SerB24 (Figure 5B) such attenuation may reflect both loss of ordered structure and removal of the PheB24 aromatic ring current (52), potentially a confounding issue (the same issue pertains to SerB24-insulin analogs with native pairing; Figure S6). A subtle trend is observed in the 1Hϵ chemical shift of TyrB26 (arrows in Figures 5B–D) in which slight upfield shifts are observed in two case (1SS-SerB24 > 1SS-ProA16), but not observed in 1SS-ProB15. This trend is shown in an enlargement of spectra in Figure S7.
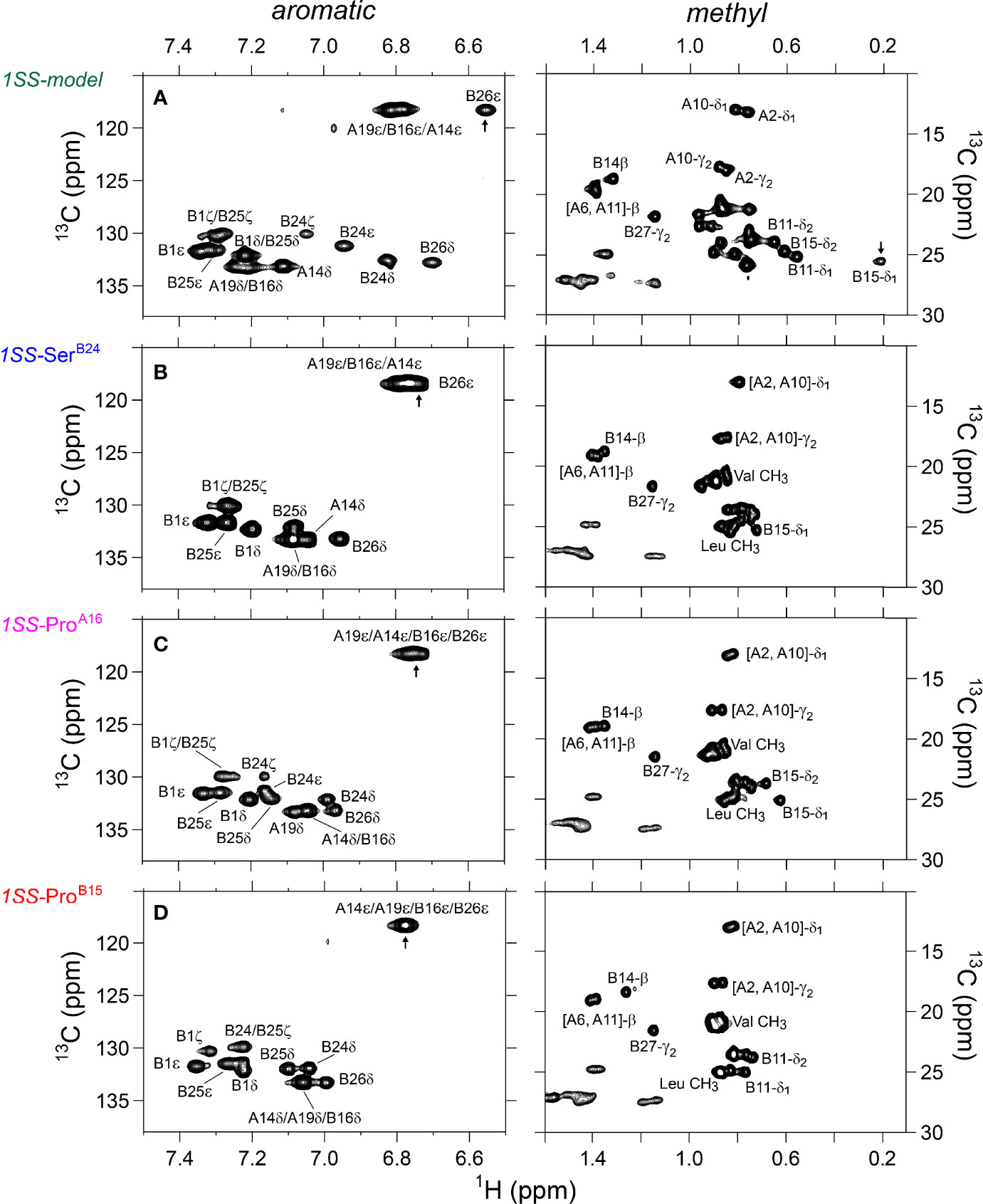
Figure 5 Nature abundance 1H-13C HSQC NMR spectra of single-chain 1SS-DesDi analogs in aromatic (left panel) and methyl region (right panel). (A) single-chain 1SS model; (B) single-chain 1SS-SerB24 analog. (C) single-chain 1SS-ProA16 analog and (D) single-chain 1SS-ProB15 analog. Spectra were acquired at a 1H NMR frequency of 700 MHz at pD 7.4 (direct meter reading) and 35°C in D2O. Selected resonance assignments are as indicated.
Analysis of 1H-13C chemical shifts was extended by 2D 1H-1H NOE spectroscopy (NOESY). Of particular interest are inter-proton NOEs (reflecting distances < 5 Å) between aromatic and aliphatic side chains. Such NOEs are prominent in the spectrum of native insulin (upper panel of Figures 6A, B), shown in relation to the corresponding TOCSY (total correlation spectroscopy) spectra of aromatic spin systems (lower panel). These inter-residue NOEs in part retained in the NOESY spectrum of the parent 1SS peptide (upper panel in Figure 6C). Particularly notable is the retention of close contacts between PheB24 and TyrB26 and the methyl groups of LeuB15, resolved due to native-like chemical-shift dispersion. A subtle feature is observed in the aromatic TOCSY spectra: the spin systems of TyrB16 and TyrA19, downfield of the mobile and solvent-exposed side chain of TyrA14 in native insulin (lower panels of Figures 6A, B), is retained in attenuated form in the TOCSY spectrum of 1SS peptide (lower panel of Figure 6C). NOEs between aromatic and aliphatic protons are observed in the spectra of the variants, but with decreased dispersion (inset boxes in Figures 6D–F); in the case of 1SS-ProB15, the overall integrated cross-peak envelope intensity is reduced (Figure 6F). Although as expected the aromatic spin system of PheB24 is absent in the TOCSY spectrum of 1SS-SerB24 (lower panel of Figure 6D), subtle upfield shifts of PheB24 are retained in 1SS-ProA16 and 1SS-ProB15. These trends are shown in expanded form in Figure S8. We imagine that the latter conformational ensembles contain a minor fraction of compact substates with long-range contacts, which nonetheless are less populated than in the parent 1SS peptide. This interpretation is supported by more detailed examination of these NOESY regions (Figures 7C–F) in relation to the structural relationships in native insulin (Figures 7A, B). Aromatic-methyl NOEs in the parent 1SS model are shown in expanded form in Figure 7C; long-range contacts are prominent in the neighborhood of cystine B19-A20, proposed to constitute the specific folding nucleus of proinsulin (green side chains in Figure 7A) (22). Also observed are long-range NOEs from TyrB26 to the methyl groups of IleA2 and ValA3, presumably reinforced by the B28-A1 peptide bond in the DesDi framework and foreshadowing subsequent steps in A-domain segmental folding associated with pairing of the remaining two cystines. This subset of these nativelike long-range NOEs can be resolved in the variants despite their attenuated chemical-shift dispersion (Figures 7D–F).
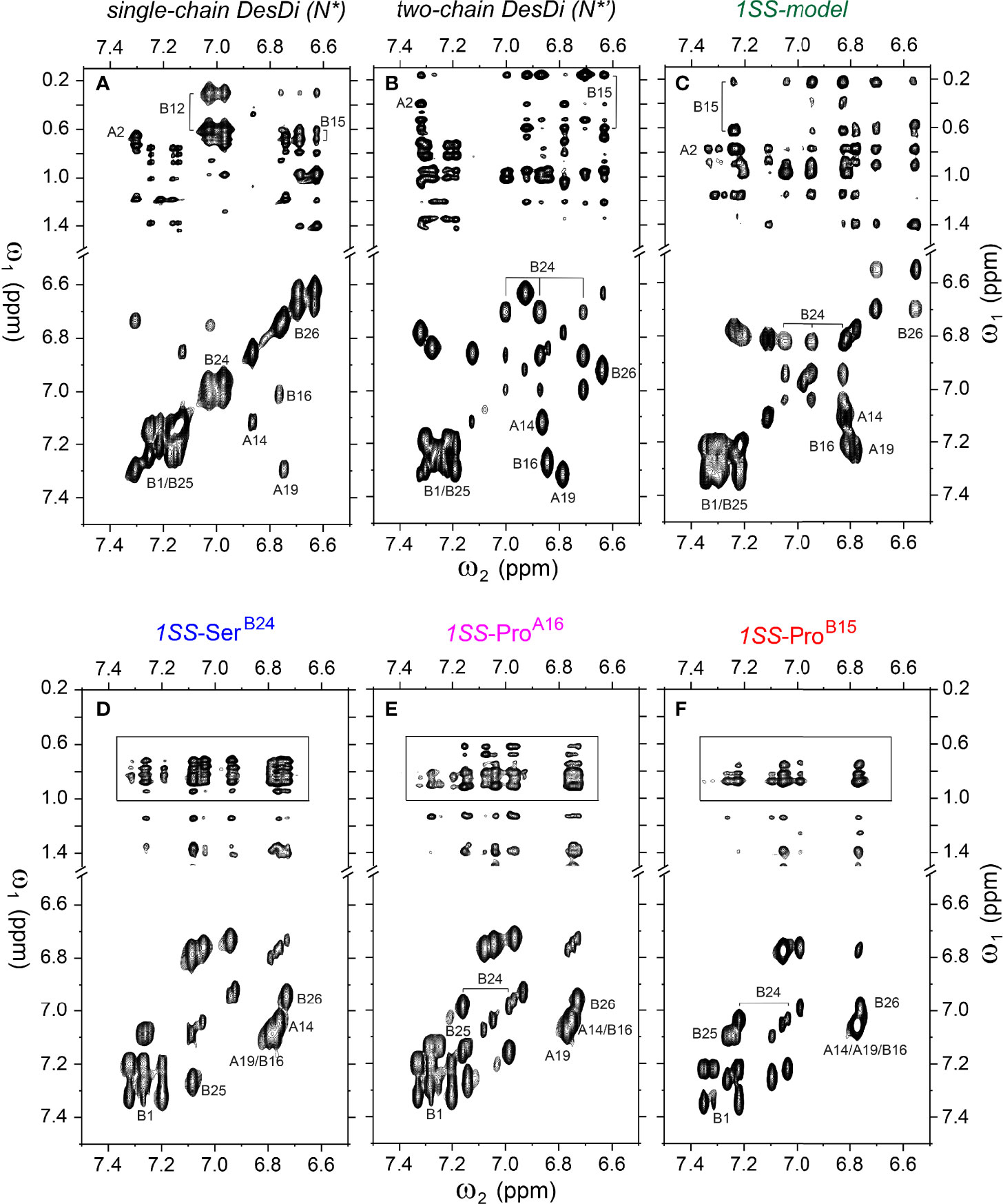
Figure 6 Homonuclear 2D-NMR spectra: NOESY spectra (mixing time 150 ms) showing NOEs from aromatic protons to methyl protons (top panel) and TOCSY spectra (mixing time 55 ms) showing aromatic resonance correlation (bottom panel). (A) single-chain DesDi (N*); (B) two-chain DesDi (N*’); (C) single-chain DesDi 1SS model; (D) single-chain 1SS-SerB24 analog; (E) single-chain 1SS-ProA16 analog and (F) single-chain 1SS-ProB15 analog. The boxes in panel D-F indicate spectral expansion for detailed analysis. Spectra were acquired at pD 7.4 (direct meter reading) and 35°C in D2O.
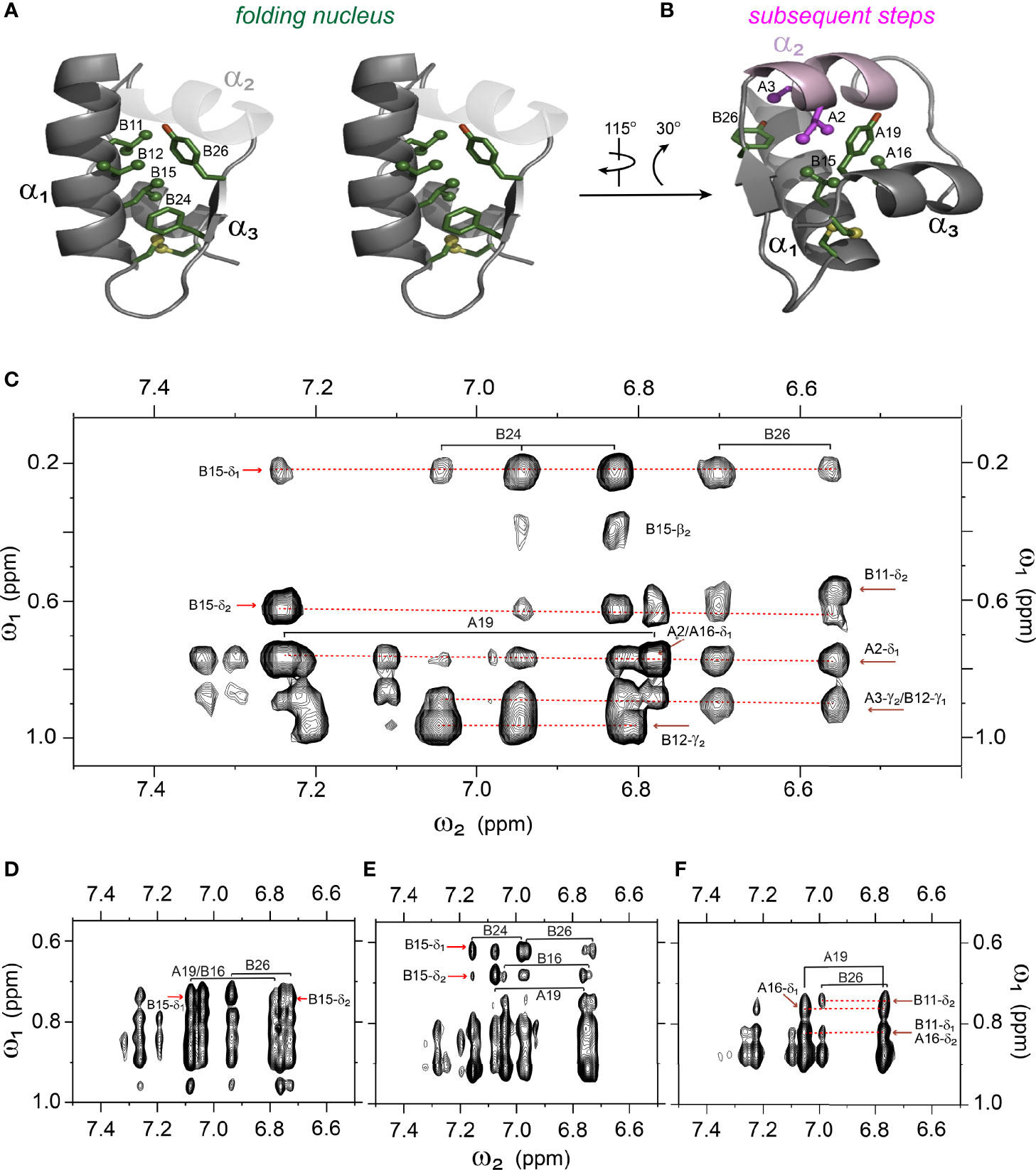
Figure 7 Spectral expansion of 2D-NOESY revealing signature long-range NOEs in the variants of single-chain 1SS-DesDi. (A) stereo view of the ribbon structure of single-chain DesDi (PDB entry 7JP3). The side-chains of indicated residues assembled a hydrophobic cluster that functions as a folding nucleus. (B) the ribbon structure of single-chain DesDi showing subsequent steps of insulin folding. (C) NOESY expansion of single-chain DesDi 1SS model. (D) single-chain 1SS-SerB24 analog. (E) single-chain 1SS-ProA16 analog and (F) single-chain 1SS-ProB15 analog. All spectra were acquired at pD 7.4 (direct meter reading) and at 35°C in D2O.
The above degree of organization in the nascent structure of the parent 1SS model is dependent on pairing of CysB19 and CysA20. Whereas overlay of the 1H-13C HSQC spectra of the parent 1SS peptide and two-chain [AspB10, GluA8]-DesDi-insulin reveals similar limits of dispersion (green versus black in Figure 8A; inset vertical and horizontal arrows), reduction of the 1SS disulfide bridge by deuterated dithiothreitol (to yield a linear peptide) led to loss of 1H-13C chemical-shift dispersion (green versus brown in Figure 8B). The reduced 1SS peptide exhibits limited dispersion, with pattern that is similar in detail to that of 1SS-ProB15 (brown versus red in Figure 8C). Possible transient or nascent long-range interactions in the linear 1SS peptide have not been investigated. 1H-13C HSQC spectra of the four 1SS peptides are overlaid in Figure 8D; relative main-chain dispersions exhibit the qualitative trend: parent > SerB24 > ProA16 > ProB15 in accordance with the findings above. This trend was made quantitative by detailed analysis of respective secondary shifts [in reference to tabulated random-coil values (55)] as shown in the four histograms in Figure S11. Reliance of 1Hα/13Cα chemical shifts circumvents the confounding absence of the B24 ring current in 1SS-SerB24 as these resonances are less influenced by aromatic ring currents (52, 56). The greater main-chain chemical-shift dispersion in 1SS-SerB24 and 1SS-ProA16 relative to 1SS-ProB15 was accentuated by lowering the temperature from 35 to 10°C. Stacked plots of 1D 1H-NMR spectra are shown for each 1SS peptide as a function of temperature in the range 5-35°C (in steps of 5°C) in Figure S9. At lower temperatures spectra of the parent 1SS peptide, 1SS-SerB24 and 1SS-ProA16 exhibit conformational broadening of upfield aromatic and aliphatic features, suggesting slowing of conformational fluctuations into the millisecond regime characteristic of intermediate exchange on the time scale of 1H-NMR chemical shifts. This phenomenon is not observed in the spectrum of 1SS-ProB15. Comparison of 1Hα-13Cα cross peaks in respective HSQC spectra likewise highlights the anomalous temperature-independence of the 1SS-ProB15 spectrum (Figure S10). This trend extends to the aromatic and methyl regions of the HSQC spectra (Figure S12). The chemical shift of TyrB26 Hϵ and a resolved methyl resonance in the reduced 1SS peptide are likewise independent of temperature (Figures S12, S13). We speculate that the anomalous NMR properties of 1SS-ProB15, indicating loss of nascent structure relative to the other 1SS peptides, rationalizes this mutation’s essentially complete block to the folding of ProB15-DesDi, even in the presence of stabilizing substitutions AspB10 and GluA8 [preceding article in this issue (18)].
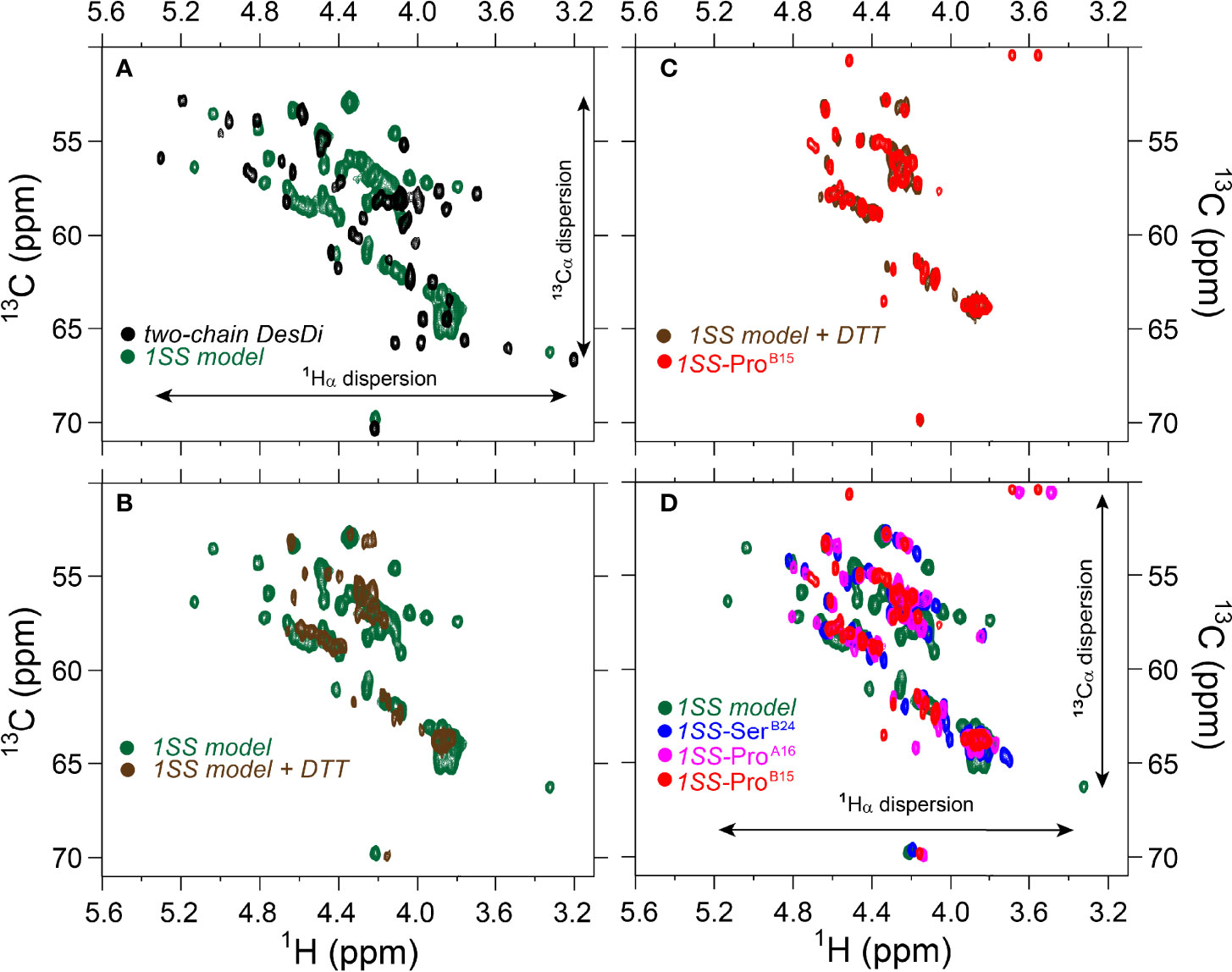
Figure 8 Comparison of nature abundance 1H-13C HSQC spectra of single-chain DesDi analogs in 1Hα/13Cα region. (A) 1H-13C HSQC spectral overlay of two-chain DesDi (N*’, black) and single-chain DesDi 1SS model (green); (B) 1H-13C HSQC spectral overlay of single-chain DesDi 1SS model (green) and 1SS model in 70 mM deuterated dithiothreitol (DTT) (brown); (C) spectral overlay of single-chain 1SS-ProB15 analog (red) and 1SS-model in 70 mM deuterated dithiothreitol (brown); (D) spectral overlay of single-chain DesDi 1SS model (green), single-chain 1SS-SerB24 analog (blue), single-chain 1SS-ProA16 analog (magenta) and single-chain 1SS-ProB15 analog (red). Spectra were acquired at a 1H frequency of 700 MHz at pD 7.4 (direct meter reading) and at 35°C in D2O.
The discovery of proinsulin by Steiner and colleagues in 1967 [(25, 57); for review, see (58)] solved a problem encountered in the chemical synthesis of insulin: inefficient specific disulfide pairing encountered in chain combination (24). Although the isolated A- and B chains of insulin contain sufficient information to specify the native structure (59), yield is reduced by competing off-pathway reactions, including formation of cyclic peptides and amyloid. Proinsulin is nonetheless itself difficult to fold. The majority of human cell lines in routine laboratory use do not efficiency fold proinsulin (4), leading to detectable disulfide isomers (3). The specialized folding environment of the β-cell ER is adapted to the biochemical demands of proinsulin biosynthesis, and yet even so physiological overexpression of the INS gene—as a compensatory response to peripheral insulin resistance (60)—can induce chronic ER stress and contribute to the progression of prediabetes and Type 2 DM (8). The growing collection of MIDY mutations in proinsulin associated with toxic misfolding leading to β-cell dysfunction (9) has motivated the hypothesis that insulin sequences, well conserved among vertebrates (23, 61), are entrenched at the edge of foldability (46). The marginal stability of insulin and proinsulin, relative to such classical model proteins as bovine pancreatic trypsin inhibitor and hen egg white lysozyme, is associated with qualitative differences in their respective refolding properties (62–67) (see also Supplemental Discussion).
In this and our companion study (18) we have introduced a single-chain peptide model of an early proinsulin folding intermediate. A framework (“DesDi”) was provided by an innovative mini-proinsulin containing a peptide bond between residues B28 and A1, with ProB28 substituted by Lys to enable facile enzymatic cleavage to liberate an active insulin analog (27). The B28-A1 peptide bond enables successful oxidative folding of the 49-residue synthetic precursor even in the presence of mutations (such as ValA16) that otherwise block classical chain combination (68). The B28-A1 inter-chain tether in DesDi presumably favors a productive orientation between A- and B-domain folding determinants and limits off-pathway events. We further stabilized DesDi by enhancing the α-helical propensity of the central B-domain segment [HisB10→Asp (29)] and N-terminal A-domain segment [ThrA8 →Glu (30)]. Increasing the net negative charge through these acidic substitutions would also be expected to enhance solubility and retard competing formation of amyloid (69). A one-disulfide model of an initial proinsulin folding intermediate was thus obtained by pairwise substitution of exposed cystine B7-A7 by Ser and internal cystine A6-A11 by Ala (18).
The present study builds on our foundational characterization of the 1SS peptide model to interrogate nascent structure by two-dimensional 1H and 1H-13C NMR spectroscopy. In accordance with prior NMR studies of a two-chain peptide model of a one-disulfide IGF-I folding intermediate (22), the parent 1SS model contains a subset of native secondary structure: central B-domain α-helix (residues B9-B19), C-terminal A-domain α-helix (A12-A20) and nascent β-strand (B24-B26). Molecular models of the parent model and the corresponding proinsulin intermediate are shown in Figure 9A in relation to the solution structure of an engineered proinsulin monomer (45). In these models cystine B19-A20 is integral to the hydrophobic mini-core formed at the confluence of the nascent elements of secondary structure. We envision that this nativelike subdomain represents the first organized nucleus in a series of successive folding landscapes (Figure 9B). Although disulfide chemistry in polypeptides can exhibit (especially at basic pH) complex patterns of native and non-native disulfide exchange and rearrangement, this structural perspective offers a simplified view of the predominant proinsulin folding scheme at neutral pH (Figure 9C). This scheme in principle provides a framework for interpreting clinical mutations that impair folding efficiency.
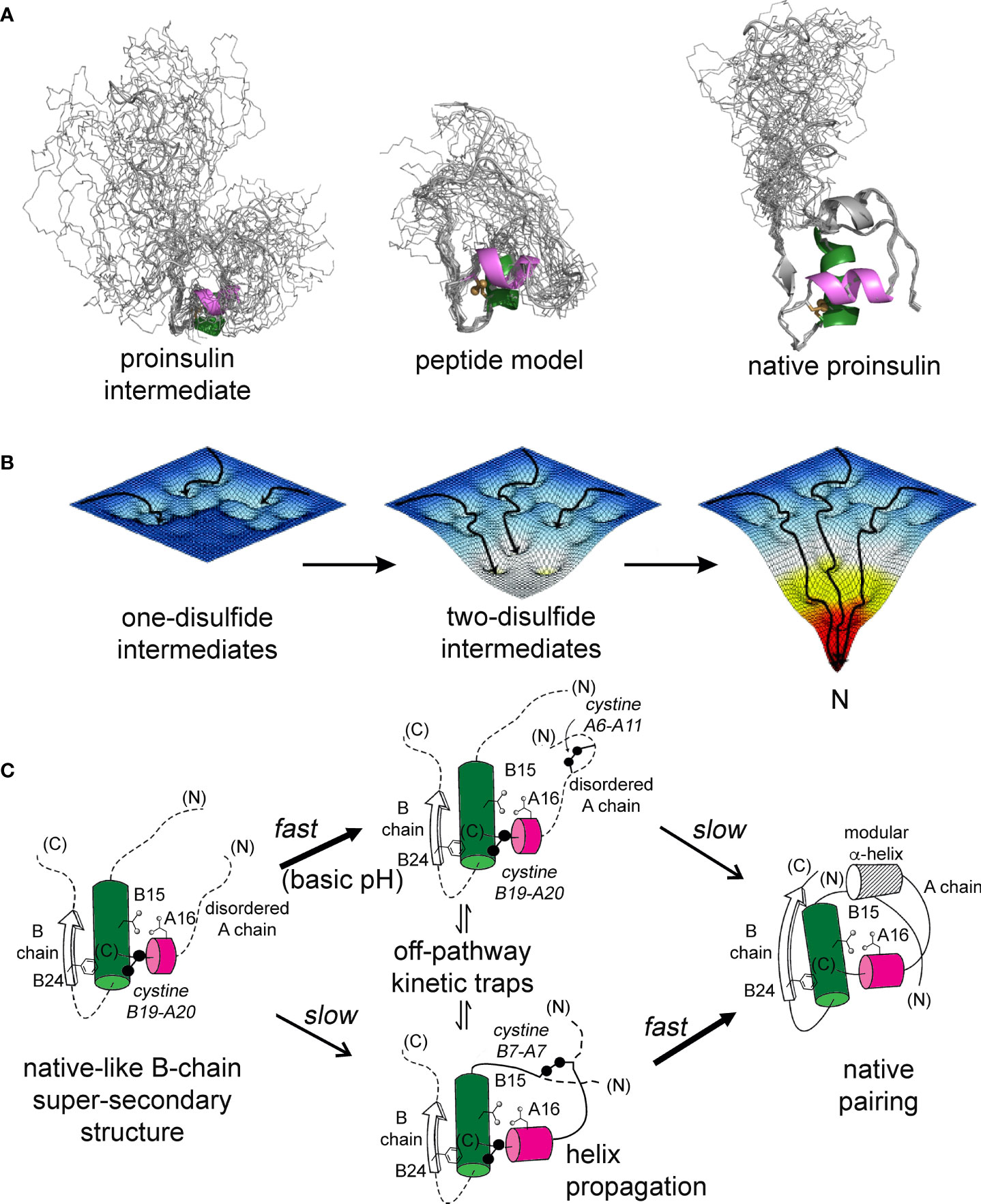
Figure 9 Proinsulin peptide models and hierarchical disulfide pathway. (A) Predicted structural ensembles for one-disulfide 86-residue proinsulin intermediate containing cystine B19-A20 (left) and corresponding 49-residue one-disulfide model (middle) relative to the solution structure of an engineered proinsulin monomer [PDB entry 2KQP; (45)] (right). In each ensemble α-helices are shown in green (central B domain) and magenta (C-terminal A domain). The simulated ensembles were generated by restrained molecular dynamics using XPLOR-NIH software (43, 44) and visualized by PyMol (https://sourceforge.net/projects/pymol/). (B) Free-energy landscapes as envisioned to govern proinsulin folding: disulfide pairing follows a sequence of folding trajectories on successively steeper landscapes. (C) Preferred pathway of disulfide pairing begins with cystine B19-A20 (left), directed by a nascent hydrophobic core formed by the central B-domain α-helix (residues B9–B19), part of the C-terminal B-chain β-strand (B24-B26) and part of the C-terminal A-domain α-helix (A16–A20). Alternative pathways mediate formation of successive disulfide bridges (middle panel) en route to the native state (right panel). This pathway is perturbed by diverse MIDY mutations. Nascent α-helices are color-coded as in (A). Panel (B) is adapted from an image kindly provided by J. Williamson; Panel (C) is adapted from reference (70).
Comparative NMR studies of the variant 1SS peptides suggest structural mechanisms of impaired foldability. In particular, patterns of chemical shifts and NOEs provide evidence of native-like tertiary structure in the neighborhood of cystine B19-A20 and its destabilization in the variant peptides in rank order SerB24 >> ProA16 > ProB15 (least organized). Their respective ensembles of partial folds each exhibit a subset of nativelike long-range NOEs—presumably reflecting fractional occupancies of analogous molten-globule states that foreshadow native structural relationships—but with progressively more complete averaging of chemical shifts in this series (Figure S14). Among these 1SS peptides and native insulin, striking correlations are observed between CD-defined α-helix contents [in the same rank order (18)] and NMR parameters: mean 1Hα chemical-shift dispersion (Figure 10A) and average 1Hα/13Cα main-chain secondary shifts (Figure 10B). The biological importance of these CD- and NMR-derived biophysical parameters is demonstrated by their further correlation with levels of ER stress induced by expression of the corresponding mutant proinsulin (Figures 10C, D) (18). Although each MIDY mutation alters an invariant framework residue—conserved among both vertebrate insulins and vertebrate insulin-like growth factors (23, 61, 71) —the less severe biophysical consequences of PheB24→Ser in consistent with the delayed onset of DM in patients with this mutation (4, 33). Although ProB15 is more profoundly perturbing than is ProA16, each is associated with neonatal-onset DM (31, 32) and so must surpass the threshold for post-natal β-cell ER stress leading to the rapid progression of β-cell dysfunction and death (8).
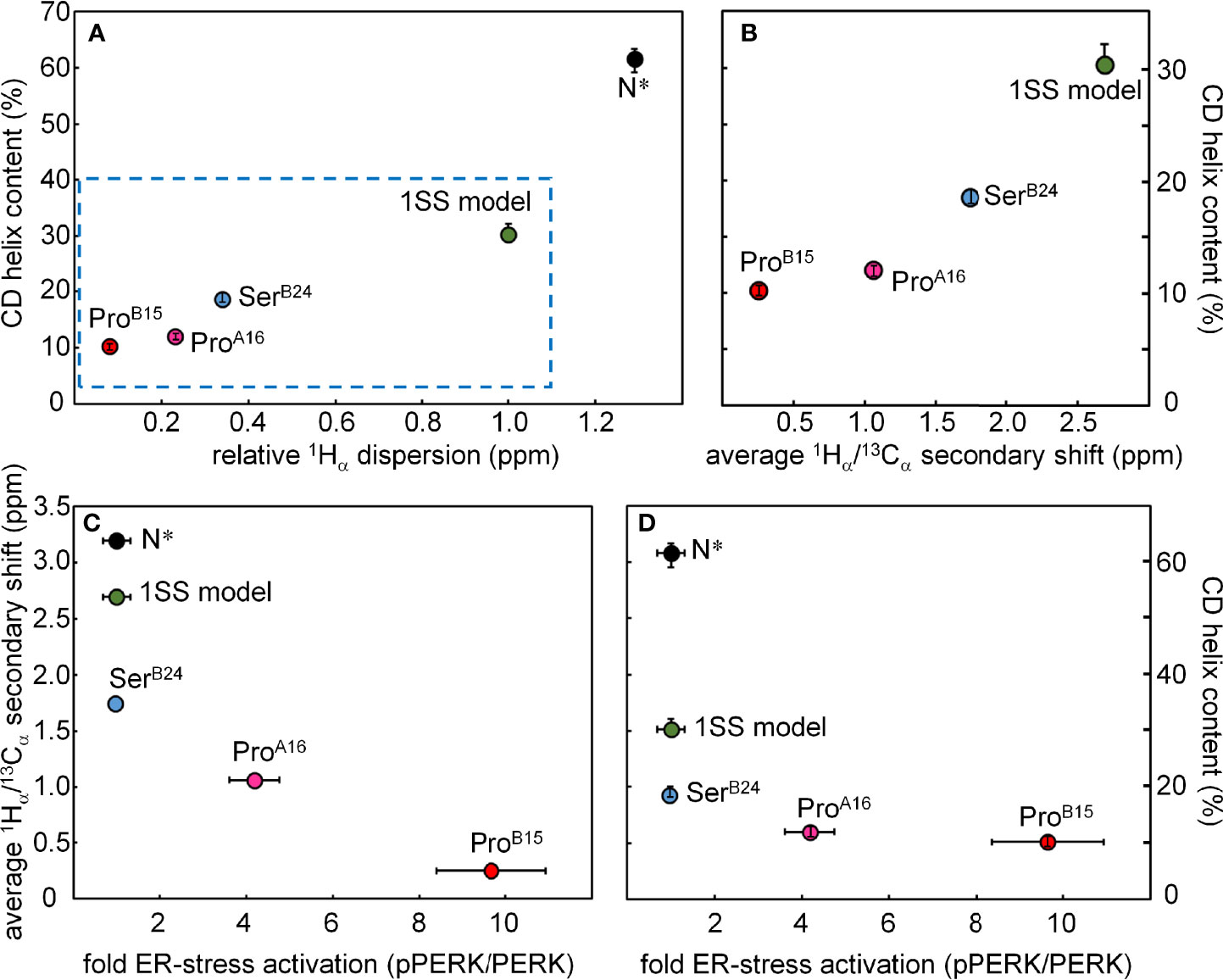
Figure 10 Plots showing correlations among biophysical features, NMR parameters and cellular ER-stress activation. (A) The plot shows the relationship between the relative 1Hα dispersion (X axis; ppm) CD-derived helix contents of DesDi variants (Y axis; %). N* analogs provide control data pertaining to the single-chain ground state stabilized by the three native disulfide bridges; sequences contain the same favorable substitutions at B10 and A8 as in the 1SS analogs. Relative 1Hα dispersion was defined by the difference of 1Hα chemical-shift dispersion of DesDi analogs and base line control (1SS model in 70 mM deuterated dithiothreitol). (B) Correlation between CD-derived helix contents (Y axis) with the average 1Hα/13Cα secondary shift (X-axis). Average 1Hα/13Cα secondary shift was calculated using equation ΔB12, ΔB14 and ΔB18 are 1Hα/13Cα secondary shift of residues ValB12, AlaB14 and ValB18, respectively. (C, D) Relationship between ER stress in cell-based assay and NMR/biophysical parameters. (C) Relationship between fold-change in ER stress (X axis; pPERK/PERK ratio) and 1Hα/13Cα secondary shift (Y axis; ppm) among DesDi variants. (D) Relationship between ER stress and α-helix content of related DesDi variants (Y-axis; %).
A surprising aspect of the present NMR studies is extent to which the parent 1SS peptide retains native-like spectroscopic features. Such structure is lost on reduction of cystine B19-A20. We ascribe the nascent organization of the parent 1SS peptide to (a) the B28-A1 peptide bond, which orients flanking B- and A-domain segments, and (b) stabilizing α-helical substitutions AspB10 and GluA8. Although such extensive nascent structure would not be expected in a one-disulfide analog of proinsulin (86 residues), the richness of the 1SS NMR spectrum suggests that it would be of future interest to develop a biosynthetic expression system, so that uniform 13C and 15N isotopic labeling would enable application of powerful 3D/4D heteronuclear NMR methods (72, 73), including residual dipolar couplings (74). We foresee that such high-resolution analysis would enable comparison between the nascent structure and dynamics of the 1SS peptide and the classic globular structure of native insulin (23). Further, such complete 1H, 13C and 15N characterization would provide a rigorous platform for comparative studies of those MIDY mutations that perturb pairing of CysB19 and CysA20.
The spectrum of diabetes-associated mutations in the insulin gene implicates diverse genotype-phenotype relationships (9, 75). These include not only mutations toxic misfolding of proinsulin with the ER (the present focus), but also those affecting upstream translocation of nascent preproinsulin (76, 77) and downstream trafficking, prohormone processing, and receptor binding (75). Each class of clinical mutations promises an opportunity to dissect respective molecular mechanisms critical to wild-type hormone biosynthesis and function. Because mutations may introduce mild, intermediate or severe biochemical perturbations, their comparative study may reveal quantitative thresholds of dysfunction associated with clinical features, such as age of diabetes onset or degree of genetic penetrance. Adult-onset SerB24 represents a mild perturbation of folding efficiency whereas both neonatal-onset ProA16 and ProB15 mutations—albeit distinct in location and degree of structural perturbation—must be below the threshold of foldability required for β-cell viability. Extending the present approach to additional MIDY mutations may define molecular determinants of this threshold. The mutant proinsulin syndrome thus promises to provide an intriguing model to relate chemistry to biology in a prototypical disease of intracellular protein misfolding.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Chemical peptide syntheses were performed by BD, AZ, and RD. NMR studies were performed and interpreted by YY, MG, NW, and MW. Figures were prepared by YY, BD, MG, AZ, Y-SC and MW. The Supplementary Material was prepared by YY, BD, AZ and MW. All authors contributed to editing the manuscript with first draft prepared by MG and MW. Overall experimental design and oversight were provided by MW. All authors contributed to the article and approved the submitted version.
This work was supported in part by grants to MW from the National Institutes of Health (R01 DK040949 and R01 DK069764). AZ was supported in part by Callibrium, LLC; the authors declare that this funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. MG was a Pre-doctoral Fellow of the National Institutes of Health (Medical Scientist Training Program 5T32GM007250-38 and Fellowship 1F30DK104618-01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Prof. Peter Arvan (University of Michigan), G.I. Bell (University of Chicago) and L. Philipson (University of Chicago) for helpful discussion and communication of genetic results prior to publication; Drs. M. Jarosinski (IUSM) and J.P. Mayer (Indiana University) for advice on peptide synthesis; and Prof. P. Arvan, M. Liu (Tianjin Research Institute of Endocrinology, Tianjin, China) and R. Wek (IUSM) for advice regarding ER stress assays. We dedicate this article to the memory of the late Prof. C.M. Dobson (Cambridge University, UK), to whom the senior author (MW) is grateful for ever-kind encouragement since his years as a graduate student.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.821091/full#supplementary-material

1. Dobson CM. Protein Folding and Misfolding. Nature (2003) 426(6968):884–90. doi: 10.1038/nature02261
2. Chiti F, Dobson CM. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu Rev Biochem (2017) 86:27–68. doi: 10.1146/annurev-biochem-061516-045115
3. Liu M, Hodish I, Rhodes CJ, Arvan P. Proinsulin Maturation, Misfolding, and Proteotoxicity. Proc Natl Acad Sci USA (2007) 104(40):15841–6. doi: 10.1073/pnas.0702697104
4. Liu M, Haataja L, Wright J, Wickramasinghe NP, Hua Q-X, Phillips NF, et al. Mutant INS-Gene Induced Diabetes of Youth: Proinsulin Cysteine Residues Impose Dominant-Negative Inhibition on Wild-Type Proinsulin Transport. PloS One (2010) 5(10):e13333. doi: 10.1371/journal.pone.0013333
5. Sun J, Cui J, He Q, Chen Z, Arvan P, Liu M. Proinsulin Misfolding and Endoplasmic Reticulum Stress During the Development and Progression of Diabetes. Mol Aspects Med (2015) 42:105–18. doi: 10.1016/j.mam.2015.01.001
6. Walter P, Ron D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science (2011) 334(6059):1081–6. doi: 10.1126/science.1209038
7. Wek RC, Cavener DR. Translational Control and the Unfolded Protein Response. Antioxid Redox Signal (2007) 9(12):2357–72. doi: 10.1089/ars.2007.1764
8. Liu M, Weiss MA, Arunagiri A, Yong J, Rege N, Sun J, et al. Biosynthesis, Structure, and Folding of the Insulin Precursor Protein. Diabetes Obes Metab (2018) 20:28–50. doi: 10.1111/dom.13378
9. Støy J, De Franco E, Ye H, Park S-Y, Bell GI, Hattersley AT. In Celebration of a Century With Insulin–Update of Insulin Gene Mutations in Diabetes. Mol Metab (2021) 52:101280. doi: 10.1016/j.molmet.2021.101280
10. Støy J, Edghill EL, Flanagan SE, Ye H, Paz VP, Pluzhnikov A, et al. Insulin Gene Mutations as a Cause of Permanent Neonatal Diabetes. Proc Natl Acad Sci USA (2007) 104(38):15040–4. doi: 10.1073/pnas.0707291104
11. Colombo C, Porzio O, Liu M, Massa O, Vasta M, Salardi S, et al. Seven Mutations in the Human Insulin Gene Linked to Permanent Neonatal/Infancy-Onset Diabetes Mellitus. J Clin Investig (2008) 118(6):2148–56. doi: 10.1172/JCI33777
12. Aguilar-Bryan L, Bryan J. Neonatal Diabetes Mellitus. Endocr Rev (2008) 29(3):265–91. doi: 10.1210/er.2007-0029
13. Edghill EL, Flanagan SE, Patch A-M, Boustred C, Parrish A, Shields B, et al. Insulin Mutation Screening in 1,044 Patients With Diabetes: Mutations in the INS Gene Are a Common Cause of Neonatal Diabetes But a Rare Cause of Diabetes Diagnosed in Childhood or Adulthood. Diabetes (2008) 57(4):1034–42. doi: 10.2337/db07-1405
14. Hughes IA, Davies JD, Bunch TI, Pasterski V, Mastroyannopoulou K, MacDougall J. Androgen Insensitivity Syndrome. Lancet (2012) 380(9851):1419–28. doi: 10.1016/S0140-6736(12)60071-3
15. Boehmer AL, Br̈ggenwirth H, van Assendelft C, Otten BJ, Verleun-Mooijman MC, Niermeijer MF, et al. Genotype Versus Phenotype in Families With Androgen Insensitivity Syndrome. J Clin Endocrinol Metab (2001) 86(9):4151–60. doi: 10.1210/jcem.86.9.7825
16. Galani A, Kitsiou-Tzeli S, Sofokleous C, Kanavakis E, Kalpini-Mavrou A. Androgen Insensitivity Syndrome: Clinical Features and Molecular Defects. Hormones (2008) 7(3):217–29. doi: 10.14310/horm.2002.1201
17. Chakravorty S, Hegde M. Inferring the Effect of Genomic Variation in the New Era of Genomics. Hum Mutat (2018) 39(6):756–73. doi: 10.1002/humu.23427
18. Dhayalan B, Glidden MD, Zaykov A, Chen Y-S, Yang Y, Phillips NB, et al. Peptide Model of the Mutant Proinsulin Syndrome. I. Design and Clinical Correlation. (2022). Front Endocrinol (2022) 13:821069. doi: 10.3389/fendo.2022.821069
19. Miller JA, Narhi LO, Hua QX, Rosenfeld R, Arakawa T, Rohde M, et al. Oxidative Refolding of Insulin-Like Growth Factor 1 Yields Two Products of Similar Thermodynamic Stability: A Bifurcating Protein-Folding Pathway. Biochemistry (1993) 32:5203–13. doi: 10.1021/bi00070a032
20. Qiao Z-S, Min C-Y, Hua Q-X, Weiss MA, Feng Y-M. In Vitro Refolding of Human Proinsulin Kinetic Intermediates, Putative Disulfide-Forming Pathway, Folding Initiation Site, and Potential Role of C-Peptide in Folding Process. J Biol Chem (2003) 278(20):17800–9. doi: 10.1074/jbc.M300906200
21. Ryle A, Sanger F, Smith L, Kitai R. The Disulphide Bonds of Insulin. Biochem J (1955) 60(4):541–56. doi: 10.1042/bj0600541
22. Hua QX, Mayer J, Jia W, Zhang J, Weiss MA. The Folding Nucleus of the Insulin Superfamily: A Flexible Peptide Model Foreshadows the Native State. J Biol Chem (2006) 281:28131–42. doi: 10.1074/jbc.M602616200
23. Baker EN, Blundell TL, Cutfield JF, Dodson EJ, Dodson GG, Hodgkin DMC, et al. The Structure of 2Zn Pig Insulin Crystals at 1.5 Å Resolution. Philos Trans R Soc Lond B Biol Sci (1988) 319(1195):369–456. doi: 10.1098/rstb.1988.0058
24. Katsoyannis P. Synthesis of Insulin. Science (1966) 154:1509–14. doi: 10.1126/science.154.3756.1509
25. Steiner DF, Cunningham D, Spigelman L, Aten B. Insulin Biosynthesis: Evidence for a Precursor. Science (1967) 157(3789):697–700. doi: 10.1126/science.157.3789.697
26. Markussen J. Comparative Reduction/Oxidation Studies With Single Chain Des-(B30) Insulin and Porcine Proinsulin. Int J Pept Protein Res (1985) 25:431–4. doi: 10.1111/j.1399-3011.1985.tb02197.x
27. Zaykov AN, Mayer JP, Gelfanov VM, DiMarchi RD. Chemical Synthesis of Insulin Analogs Through a Novel Precursor. ACS Chem Biol (2014) 9(3):683–91. doi: 10.1021/cb400792s
28. Schwartz GP, Burke GT, Katsoyannis PG. A Superactive Insulin: [B10-Aspartic Acid]Insulin(Human). Proc Natl Acad Sci USA (1987) 84:6408–11. doi: 10.1073/pnas.84.18.6408
29. Kaarsholm NC, Norris K, Jorgensen RJ, Mikkelsen J, Ludvigsen S, Olsen OH, et al. Engineering Stability of the Insulin Monomer Fold With Application to Structure-Activity Relationships. Biochemistry (1993) 32:10773–8. doi: 10.1021/bi00091a031
30. Weiss MA, Hua Q-X, Jia W, Nakagawa SH, Chu Y-C, Hu S-Q, et al. Activities of Monomeric Insulin Analogs at Position A8 Are Uncorrelated With Their Thermodynamic Stabilities. J Biol Chem (2001) 276(43):40018–24. doi: 10.1074/jbc.M104634200
31. Ortolani F, Piccinno E, Grasso V, Papadia F, Panzeca R, Cortese C, et al. Diabetes Associated With Dominant Insulin Gene Mutations: Outcome of 24-Month, Sensor-Augmented Insulin Pump Treatment. Acta Diabetol (2016) 53(3):499–501. doi: 10.1007/s00592-015-0793-1
32. Flanagan SE, De Franco E, Allen HL, Zerah M, Abdul-Rasoul MM, Edge JA, et al. Analysis of Transcription Factors Key for Mouse Pancreatic Development Establishes NKX2-2 and MNX1 Mutations as Causes of Neonatal Diabetes in Man. Cell Metab (2014) 19(1):146–54. doi: 10.1016/j.cmet.2013.11.021
33. Shoelson S, Polonsky K, Zeidler A, Rubenstein A, Tager H. Human Insulin B24 (Phe→Ser). Secretion and Metabolic Clearance of the Abnormal Insulin in Man and in a Dog Model. J Clin Invest (1984) 73(5):1351–8. doi: 10.1172/JCI111338
34. Wishart DS, Sykes BD, Richards FM. The Chemical Shift Index: A Fast and Simple Method for the Assignment of Protein Secondary Structure Through NMR Spectroscopy. Biochemistry (1992) 31(6):1647–51. doi: 10.1021/bi00121a010
35. Wishart DS, Sykes BD. The 13C Chemical-Shift Index: A Simple Method for the Identification of Protein Secondary Structure Using 13C Chemical-Shift Data. J Biomol NMR (1994) 4(2):171–80. doi: 10.1007/BF00175245
36. Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A Hybrid Method for Predicting Protein Backbone Torsion Angles From NMR Chemical Shifts. J Biomol NMR (2009) 44(4):213–23. doi: 10.1007/s10858-009-9333-z
37. Hua Q-X, Jai W, Weiss MA. Conformational Dynamics of Insulin. Front Endocrinol (2011) 2:48. doi: 10.3389/fendo.2011.00048
38. Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: Enhanced Software for Biomolecular NMR Spectroscopy. Bioinformatics (2015) 31(8):1325–7. doi: 10.1093/bioinformatics/btu830
39. Szilágyi L, Jardetzky O. α-Proton Chemical Shifts and Secondary Structure in Proteins. J Magn Reson (1989) 83(3):441–9. doi: 10.1016/0022-2364(89)90341-7
40. Wüthrich K. NMR With Proteins and Nucleic Acids. Europhys News (1986) 17(1):11–3. doi: 10.1051/epn/19861701011
41. Wang Y, Jardetzky O. Probability-Based Protein Secondary Structure Identification Using Combined NMR Chemical-Shift Data. Protein Sci (2002) 11(4):852–61. doi: 10.1110/ps.3180102
42. Nilges M, Clore GM, Gronenborn AM. Determination of Three-Dimensional Structures of Proteins From Interproton Distance Data by Hybrid Distance Geometry-Dynamical Simulated Annealing Calculations. FEBS Lett (1988) 229(2):317–24. doi: 10.1016/0014-5793(88)81148-7
43. Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR Molecular Structure Determination Package. J Magn Reson (2003) 160(1):65–73. doi: 10.1016/S1090-7807(02)00014-9
44. Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor–NIH for NMR Molecular Structure Determination. Prog Nucl Magn Reson Spectrosc (2006) 48(1):47–62. doi: 10.1016/j.pnmrs.2005.10.001
45. Yang Y, Hua QX, Liu J, Shimizu EH, Choquette MH, Mackin RB, et al. Solution Structure of Proinsulin: Connecting Domain Flexibility and Prohormone Processing. J Biol Chem (2010) 285:7847–51. doi: 10.1074/jbc.C109.084921
46. Rege NK, Liu M, Yang Y, Dhayalan B, Wickramasinghe NP, Chen Y-S, et al. Evolution of Insulin at the Edge of Foldability and its Medical Implications. Proc Natl Acad Sci USA (2020) 117(47):29618–28. doi: 10.1073/pnas.2010908117
47. Bodenhausen G, Ruben DJ. Natural Abundance Nitrogen-15 NMR by Enhanced Heteronuclear Spectroscopy. Chem Phys Lett (1980) 69(1):185–9. doi: 10.1016/0009-2614(80)80041-8
48. Vuister GW, Bax A. Resolution Enhancement and Spectral Editing of Uniformly 13C-Enriched Proteins by Homonuclear Broadband 13C Decoupling. J Magn Reson (1992) 98(2):428–35. doi: 10.1016/0022-2364(92)90144-V
49. Hua QX, Shoelson SE, Weiss MA. Nonlocal Structural Perturbations in a Mutant Human Insulin: Sequential Resonance Assignment and 13C-Isotope-Aided 2D-NMR Studies of [PheB24→Gly]insulin With Implications for Receptor Recognition. Biochemistry (1992) 31(47):11940–51. doi: 10.1021/bi00162a037
50. Hua Q-X, Hu S-Q, Frank BH, Jia W, Chu Y-C, Wang S-H, et al. Mapping the Functional Surface of Insulin by Design: Structure and Function of a Novel A-Chain Analogue. J Mol Biol (1996) 264(2):390–403. doi: 10.1006/jmbi.1996.0648
51. Hua Q-X, Liu M, Hu S-Q, Jia W, Arvan P, Weiss MA. A Conserved Histidine in Insulin Is Required for the Foldability of Human Proinsulin: Structure and Function of an AlaB5 Analog. J Biol Chem (2006) 281(34):24889–99. doi: 10.1074/jbc.M602617200
52. Jacoby E, Hua QX, Stern AS, Frank BH, Weiss MA. Structure and Dynamics of a Protein Assembly. 1H-NMR Studies of the 36 kDa R6 Insulin Hexamer. J Mol Biol (1996) 258(1):136–57. doi: 10.1006/jmbi.1996.0239
53. Weiss MA, Hua QX, Jia W, Chu YC, Wang RY, Katsoyannis PG. Hierarchiacal Protein "Un-Design": Insulin's Intrachain Disulfide Bridge Tethers a Recognition α-Helix. Biochemistry (2000) 39:15429–40. doi: 10.1021/bi001905s
54. Hua QX, Nakagawa SH, Jia W, Hu SQ, Chu YC, Katsoyannis PG, et al. Hierarchical Protein Folding: Asymmetric Unfolding of an Insulin Analogue Lacking the A7-B7 Interchain Disulfide Bridge. Biochemistry (2001) 40:12299–311. doi: 10.1021/bi011021o
55. Schwarzinger S, Kroon GJ, Foss TR, Wright PE, Dyson HJ. Random Coil Chemical Shifts in Acidic 8 M Urea: Implementation of Random Coil Shift Data in NMRView. J Biomol NMR (2000) 18(1):43–8. doi: 10.1023/A:1008386816521
56. Hoch JC, Dobson CM, Karplus M. Fluctuations and Averaging of Proton Chemical Shifts in the Bovine Pancreatic Trypsin Inhibitor. Biochemistry (1982) 21(6):1118–25. doi: 10.1021/bi00535a002
57. Steiner DF, Oyer PE. The Biosynthesis of Insulin and a Probable Precursor of Insulin by a Human Islet Cell Adenoma. Proc Natl Acad Sci USA (1967) 57(2):473–80. doi: 10.1073/pnas.57.2.473
58. Steiner DF. On the Discovery of Precursor Processing. In: Mbikay M, Seidah N, editors. Proprotein Convertases. Methods Mol Biol (Methods and Protocols). Humana Press (2011). vol 768. doi: 10.1007/978-1-61779-204-5_1
59. Wei J, Xie L, Lin Y-Z, Tsou C-L. The Pairing of the Separated A and B Chains of Insulin and Its Derivatives, FTIR Studies. Biochim Biophys Acta (1992) 1120(1):69–74. doi: 10.1016/0167-4838(92)90425-D
60. Johnson JD. On the Causal Relationships Between Hyperinsulinaemia, Insulin Resistance, Obesity and Dysglycaemia in Type 2 Diabetes. Diabetologia (2021) 64(10):2138–46. doi: 10.1007/s00125-021-05505-4
61. Conlon JM. Molecular Evolution of Insulin in Non-Mammalian Vertebrates. Am Zool (2000) 40(2):200–12. doi: 10.1093/icb/40.2.200
62. van Mierlo CP, Darby NJ, Keeler J, Neuhaus D, Creighton TE. Partially Folded Conformation of the (30-51) Intermediate in the Disulphide Folding Pathway of Bovine Pancreatic Trypsin Inhibitor: 1H and 15N Resonance Assignments and Determination of Backbone Dynamics From 15N Relaxation Measurements. J Mol Biol (1993) 229(4):1125–46. doi: 10.1006/jmbi.1993.1108
63. Miranker A, Radford SE, Karplus M, Dobson CM. Demonstration by NMR of Folding Domains in Lysozyme. Nature (1991) 349(6310):633–6. doi: 10.1038/349633a0
64. Staley JP, Kim PS. Formation of a Native-Like Subdomain in a Partially Folded Intermediate of Bovine Pancreatic Trypsin Inhibitor. Protein Sci (1994) 3(10):1822–32. doi: 10.1002/pro.5560031021
65. Barbar E, Barany G, Woodward C. Dynamic Structure of a Highly Ordered. β-Sheet Molten Globule: Multiple Conformations With a Stable Core. Biochemistry (1995) 34(36):11423–34. doi: 10.1021/bi00036a015
66. Ittah V, Haas E. Nonlocal Interactions Stabilize Long Range Loops in the Initial Folding Intermediates of Reduced Bovine Pancreatic Trypsin Inhibitor. Biochemistry (1995) 34(13):4493–506. doi: 10.1021/bi00013a042
67. Barbar E, Hare M, Daragan V, Barany G, Woodward C. Dynamics of the Conformational Ensemble of Partially Folded Bovine Pancreatic Trypsin Inhibitor. Biochemistry (1998) 37(21):7822–33. doi: 10.1021/bi973102j
68. Liu M, Wan Z-l, Chu Y-C, Aladdin H, Klaproth B, Choquette M, et al. Crystal Structure of a “Nonfoldable” Insulin: Impaired Folding Efficiency Despite Native Activity. J Biol Chem (2009) 284(50):35259–72. doi: 10.1074/jbc.M109.046888
69. DuBay KF, Pawar AP, Chiti F, Zurdo J, Dobson CM, Vendruscolo M. Prediction of the Absolute Aggregation Rates of Amyloidogenic Polypeptide Chains. J Mol Biol (2004) 341(5):1317–26. doi: 10.1016/j.jmb.2004.06.043
70. Weiss MA. Diabetes Mellitus Due to the Toxic Misfolding of Proinsulin Variants. FEBS Lett (2013) 587(13):1942–50. doi: 10.1016/j.febslet.2013.04.044
71. Conlon JM. Evolution of the Insulin Molecule: Insights Into Structure-Activity and Phylogenetic Relationships. Peptides (2001) 22(7):1183–93. doi: 10.1016/S0196-9781(01)00423-5
72. Bax A. Multidimensional Nuclear Magnetic Resonance Methods for Protein Studies. Curr Opin Struct Biol (1994) 4(5):738–44. doi: 10.1016/S0959-440X(94)90173-2
73. Clore GM, Gronenborn AM. Applications of Three-and Four-Dimensional Heteronuclear NMR Spectroscopy to Protein Structure Determination. Prog Nucl Magn Reson Spectrosc (1991) 23(1):43–92. doi: 10.1016/0079-6565(91)80002-J
74. Bax A, Grishaev A. Weak Alignment NMR: A Hawk-Eyed View of Biomolecular Structure. Curr Opin Struct Biol (2005) 15(5):563–70. doi: 10.1016/j.sbi.2005.08.006
75. Steiner DF, Tager HS, Chan SJ, Nanjo K, Sanke T, Rubenstein AH. Lessons Learned From Molecular Biology of Insulin-Gene Mutations. Diabetes Care (1990) 13:600–9. doi: 10.2337/diacare.13.6.600
76. Liu M, Lara-Lemus R, Shan S-o, Wright J, Haataja L, Barbetti F, et al. Impaired Cleavage of Preproinsulin Signal Peptide Linked to Autosomal-Dominant Diabetes. Diabetes (2012) 61(4):828–37. doi: 10.2337/db11-0878
Keywords: monogenic diabetes, peptide chemistry, protein folding, folding nucleus, oxidative folding intermediate, NMR spectroscopy
Citation: Yang Y, Glidden MD, Dhayalan B, Zaykov AN, Chen Y-S, Wickramasinghe NP, DiMarchi RD and Weiss MA (2022) Peptide Model of the Mutant Proinsulin Syndrome. II. Nascent Structure and Biological Implications. Front. Endocrinol. 13:821091. doi: 10.3389/fendo.2022.821091
Received: 23 November 2021; Accepted: 21 January 2022;
Published: 01 March 2022.
Edited by:
Jiajun Zhao, Shandong Provincial Hospital, ChinaReviewed by:
Cheng Hu, Shanghai Jiao Tong University, ChinaCopyright © 2022 Yang, Glidden, Dhayalan, Zaykov, Chen, Wickramasinghe, DiMarchi and Weiss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanwu Yang, eWFuZ3lhbndAaXUuZWR1; Michael A. Weiss, d2Vpc3NtYUBpdS5lZHU=
†Present address: Alexander N. Zaykov, Novo-Nordisk Research Center Indianapolis, Indianapolis, IN, United States
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.