
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 17 February 2022
Sec. Experimental Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.820896
This article is part of the Research Topic Glucosensing Impact on Glucose Metabolism: From Fish to Mammals View all 5 articles
 Xiaozhu Wang1†
Xiaozhu Wang1† Song Xue1†
Song Xue1† Xiaowei Lei1†
Xiaowei Lei1† Wenqi Song1
Wenqi Song1 Lei Li2
Lei Li2 Xuan Li1
Xuan Li1 Yanbin Fu1
Yanbin Fu1 Cong Zhang3
Cong Zhang3 Hailin Zhang1,3
Hailin Zhang1,3 Yao Luo1
Yao Luo1 Meng Wang2*
Meng Wang2* Gufa Lin1,3*
Gufa Lin1,3* Chao Zhang1*
Chao Zhang1* Jing Guo1*
Jing Guo1*The Melanocortin-3 receptor (MC3R) and Melanocortin-4 receptor (MC4R), two members of the key hypothalamic neuropeptide signaling, function as complex mediators to control the central appetitive and energy homeostasis. The melanocortin 2 receptor accessory protein 2 (MRAP2) is well-known for its modulation on the trafficking and signaling of MC3R and MC4R in mammals. In this study, we cloned and elucidated the pharmacological profiles of MRAP2 on the regulation of central melanocortin signaling in a relatively primitive poikilotherm amphibian species, the Mexican axolotl (Ambystoma mexicanum). Our results showed the higher conservation of axolotl mc3r and mc4r across species than mrap2, especially the transmembrane regions in these proteins. Phylogenetic analysis indicated that the axolotl MC3R/MC4R clustered closer to their counterparts in the clawed frog, whereas MRAP2 fell in between the reptile and amphibian clade. We also identified a clear co-expression of mc3r, mc4r, and mrap2 along with pomc and agrp in the axolotl brain tissue. In the presence of MRAP2, the pharmacological stimulation of MC3R by α-MSH or ACTH significantly decreased. MRAP2 significantly decreased the cell surface expression of MC4R in a dose dependent manner. The co-localization and formation of the functional complex of axolotl MC3R/MC4R and MRAP2 on the plasma membrane were further confirmed in vitro. Dramatic changes of the expression levels of mc3r, mrap2, pomc, and agrp in the fasting axolotl hypothalamus indicated their critical roles in the metabolic regulation of feeding behavior and energy homeostasis in the poikilotherm aquatic amphibian.
As a key neuropeptide hormone family, the melanocortins and their receptors have been reported to participate in numerous physiological functions such as energy homeostasis, natriuresis, inflammation, and exocrine secretion (1, 2). The melanocortins consist of four members, namely, α-, β- and γ-melanocyte-stimulating hormone (MSH) and adrenocorticotropin (ACTH). All of these melanocortins were derived from the polypeptide precursor preprohormone proopiomelanocortin (POMC) through post-translational cleavage in a tissue-specific manner (3, 4). Given the fact that all melanocortins are derived from the same precursor, a tetrapeptide sequence (His-Phe-Arg-Trp) was conserved among them, which involves the melanogenic effects of melanocortins (2). POMC is enriched in the hypothalamus, specifically the arcuate nucleus, where it is processed to ACTH and α-MSH (5–7). To sustain the biological function of the melanocortins, they need to bind with the melanocortin receptors (MCRs), which belong to the seven-pass transmembrane guanine nucleotide-protein coupled receptor (GPCR) family (8).
Five melanocortin receptor subtypes (MC1R–MC5R) have been identified to be distributed in different tissues with segregated biological functions (2, 8, 9). The first MCR family member (MC1R) was cloned in 1992 (10, 11). As a key control element for melanogenesis, MC1R had an abundant expression in melanocytes and melanoma cells (12–15). MC2R, first cloned in the adrenal gland in 1993 (16), could extensively regulate the development, steroidogenesis, glucocorticoid synthesis, and neonatal gluconeogenesis of adrenal gland (17). Shortly after the identification of MC2R, MC3R (18) and MC4R (19) were cloned in the same year. Both MC3R and MC4R were critical for central control of appetite and energy homeostasis, which could be targeted as drug candidates for obesity or anorexia (20–22). MC3R, but not MC4R, was reported to highly express in dopaminergic neurons of the ventral tegmental area (VTA) and regulate the mesolimbic dopaminergic system and reward in a sex-dimorphism manner (23). MC4R widely expressed in the central nervous system (CNS), and its mutations could lead to monogenic obesity in human (24–26). The last MCR family member to be identified was MC5R in 1994 (27). MC5R generally expressed in multiple tissues, involving in exocrine secretion (28), immunoregulation (29) and other physiological functions (30).
In addition to the four melanocortins (α-, β- and γ-MSH and ACTH) and five MCRs (MC1R–MC5R), the melanocortin system also includes two endogenous antagonists of the MCRs: agouti (ASP) (31) and agouti-related protein (AgRP) (32). The binding affinities of five MCRs to their agonists (α-, β- and γ-MSH and ACTH) and antagonists (ASP and AGRP) vary from each other. Human MC1R has the highest binding affinity with α-MSH, almost equivalent affinity with ACTH, but less affinity with β- and γ-MSH (33). MC2R is the ACTH receptor, as ACTH can upregulate MC2R expression and increase the receptor numbers (34). There is no difference in the binding affinity of MC3R for any of the melanocortins, even γ-MSH can activate MC3R at the same extent (35). MC4R has a higher affinity for α-MSH and ACTH, a lower affinity for β-MSH, and the lowest affinity for γ-MSH (2). Surprisingly, MC4R can be antagonized by both ASP and AgRP, which distincts it from MC3R (2). The binding affinity of MC5R with α-MSH and ACTH is higher than β- and γ-MSH (2, 31).
A pair of GPCR single-transmembrane accessory proteins: melanocortin 2 receptor accessory protein (MRAP) and melanocortin 2 receptor accessory protein 2 (MRAP2) also function in the complex melanocortin regulatory system (36). MRAP2 can homodimerize and heterodimerize with MRAP in CHO cells (37). As a specific molecular chaperone for MC2R, MRAP was first reported to actively be involved in the trafficking of MC2R from the endoplasmic reticulum (ER) to the cell surface, suggesting its role as an accessory protein for MC2R (38). Subsequent study further extended the interaction of MRAP and MRAP2 from MC2R to each of the MCRs by Co-IP assay (37). The interaction of MC3R and MC4R with MRAP2 can also alter the MC3R/MC4R cell surface expression. For instance, the co-expression of human MC4R with MRAP or MRAP2 can reduce its cell surface expression, but the cell surface expression of human MC3R was found not to be altered (37, 39).
Several studies have been conducted to evaluate the pharmacological profiles of MC3R/MC4R in the presence of MRAPs in various vertebrates, such as human (40, 41), mouse (42), chicken (43), the clawed frog (44–46), zebrafish (47, 48), snakehead (49), orange-spotted grouper (50), channel catfish (51), topmouth culter (52, 53), and sea lamprey (54). For instance, human MRAP2 was found to promote the maximal activity of MC3R/MC4R stimulated by α-MSH, whereas the mutated MRAP2 variants failed to show the similar effect (40). Moreover, consistent observations were reported for human, chicken, and zebrafish that interaction with MRAP2 made MC4R becomes an ACTH receptor (41, 47, 55).
The melanocortin system, especially MC3R, MC4R, their POMC-derived agonists and endogenous regulator MRAPs, represents a fundamental component to regulate the food intake and energy balance. Multiple studies identified significant increases of adiposity in Agrp transgenic or Pomc, Mc3r/Mc4r deficient rodents, suggesting their essential role in body weight regulation and energy metabolism (56–60). For instance, Mc3r-deficient mice reveal a normal appetite but increased adiposity (61), whereas Mc4r-deficient mice was observed to have an increase in food intake, body weight, body length, and adiposity (62). However, almost all studies regarding melanocortin signaling are conducted in rodents or humans. Only few studies have been conducted to examine the melanocortin signaling in other vertebrates. For instance, MRAPs were identified to improve the MC4R ligand sensitivity to ACTH in chicken (43). Similarly, MRAPs were reported to mediate the pharmacological activities of the neuronal MC3R and MC4R signaling in clawed frog (44–46) and sea lamprey (54). Therefore, the interaction and regulation of the melanocortin signaling in other vertebrates remain unclear and intriguing.
The Mexican axolotl (Ambystoma mexicanum) serves as a fascinating tetrapod model for the developmental, evolutionary, and regeneration studies. One of the striking features for the axolotl is its enormous genome (~32 Gb), which consists of 14 chromosomes with large number of repetitive elements. The high percentage of repetitive sequences in the axolotl genome makes the genome assembly and gene annotation extremely difficult. Therefore, tremendous efforts have been made to construct a high-quality reference genome for the axolotl (63–65). The available reference genome allows us to conduct a comprehensive evaluation of the melanocortin system in the axolotl. In this study, we first identified the full-length sequence of mc3r, mc4r, and mrap2 in axolotl genome and examined the homology of these genes among species by phylogenetic and synteny analysis. We also revealed the tissue distribution of these genes in adult axolotl and their expression alterations in the hypothalamus after 3-week fasting, and finally elucidated a comprehensively pharmacological evaluation of MRAP2 on melanocortin signaling in axolotl model.
Animal procedures performed in this study were in accordance with the Association for Assessment and Accreditation of Laboratory Animal Care International (http://www.aaalac.org/index.cfm), and the protocols were approved by the Institutional Animal Care and Use Committees (IACUC) of Tongji University.
All the fragments were ligated into pcDNA3.1(+) plasmid, and the constructed plasmids were then verified by DNA sequencing. The pCre-luc (Santa Cruz, CA, USA) was kindly gifted by Xin Xie’s Lab of Tongji University. Human α-melanocyte stimulating hormone (α-MSH) and human adrenocorticotropin ACTH (1–24) was synthesized by Genescript (Nanjing, China). We purchased the paraformaldehyde (4%), phosphate buffered saline (PBS), bovine serum albumin, non-fat milk powder and β-mercaptoethanol from Sangon Biotech (Shanghai, China). TRNzol Universal Reagent and FastQuant RT Kit (with gDNase) were obtained from Tiangen Biotech (Beijing, China). Tetramethylbenzidine (TMB) chromogen solution was purchased from Beyotime® Biotechnology (Shanghai, China). Hydrochloric acid and sulfuric acid were obtained from Sinopharm Chemical Reagent Co., Ltd (Beijing, China). The antibodies used in this study included Rabbit anti-Flag (Cell Signaling Technology, USA), Mouse anti-HA (Sigma Aldrich, MO, USA), Mouse anti-Flag (Abcam, Cambridge, UK), Goat anti-Mouse IgG (horseradish peroxidase (HRP)-conjugated) (ABclonal Biotech Co., Ltd, Wuhan, China), and Goat Anti-Rabbit Alexa-Fluor 594 (Abcam). All primers used for full-length gene amplification, reverse transcription PCR (RT-PCR), and real-time quantitative PCR are listed in Supplemental Table 1 and synthesized by GENEWIZ (Suzhou, China).
To identify the axolotl mc3r, mc4r, and mrap2 gene sequences, we searched the axolotl RNA-Seq dataset with amino acid sequences of the corresponding protein sequences from human (Homo sapiens), mouse (Mus musculus), cattle (Bos taurus), chicken (Gallus gallus), common lizard (Zootoca vivipara), painted turtle (Chrysemys picta bellii), clawed frog (Xenopus tropicalis), and zebrafish (Danio rerio) as queries using TBLASTN. We then eliminated the duplicates from the initial sequence pool by using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). The resulting unique set of sequences was used for primer design to clone the full-length sequences of these genes. The mc3r and mc4r were amplified from genomic DNA extracted from axolotl hypothalamus. The mrap2 was amplified from cDNA of the axolotl brain tissue. The functional domain characteristics and transmembrane helices of mc3r, mc4r, and mrap2 were predicted by NCBI Conserved Domain Search (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
The protein sequences of MC3Rs, MC4Rs, and MRAP2s of various different species, namely, Human, Macaque, Sheep, Cattle, Mouse, Chicken, Snake, Common lizard, Painted turtle, Clawed Frog, Rainbow trout, Zebrafish, and Elephant shark were obtained from ENSEMBL genome browser (http://www.ensembl.org/) or NCBI database (http://www.ncbi.nlm.nih.gov/). The details of all used species and sequence information are listed in Supplemental Table 2. The multiple alignments of these selected protein sequences were aligned with MAFFT (66). To identify the consensus phylogenetic tree, the best-fit protein evolution model was searched using modeltest NG (67). The final ML phylogenetic tree was inferred with the multiple alignments of amino acid sequences using RAxML v8.2 (68) with the PROTGAMMAJTT protein model (best fit model identified by modeltest NG) and 1,000 rapid bootstrap replicates. Synteny analysis was performed by manually examining adjacent genes of mc3r, mc4r, and mrap2 among various species.
Multiple fresh tissues (brain, pituitary, hypothalamus, eye, heart, liver, kidney, spleen, stomach, intestine, ovary, gill, ventral skin, dorsal skin, muscle, fat) from a female adult axolotl were collected, and their total RNA were extracted with TRNzol Universal reagent. The reverse transcription was conducted with 0.5 or 1 μg total RNA from each tissue using FastQuant RT Kit (with gDNase). The expression levels of mc3r, mc4r, mrap2, agrp, and pomc in each axolotl tissue were determined by RT-PCR, and products were separated on 1.5% agarose gel. To evaluate the expression patterns of mc3r, mc4r, and mrap2 in more axolotl tissues, we obtained the public available RNA-Seq data from Axolotl Transcriptomics website (https://portals.broadinstitute.org/axolotlomics/) from a axolotl cross-tissue transcriptome study (69).
Human Embryonic Kidney HEK293T cells were cultured in Dulbecco’s modified Eagles medium (DMEM) medium with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin–streptomycin. Cells were grown in a humidified 5% CO2 incubator at 37°C. Transfection was carried out with Polyethyleneimine (PEI) Transfection Reagent (Polysciences, Warrington, PA, USA) according to the manufacturer’s protocol. Empty pcDNA3.1(+) vector were added to maintain the constant total plasmid amounts for all transfections groups.
HEK293T cells at a density of 1 × 105 cells/ml were plated into 24-well plates before transfection. The plasmids of axolotl MC3R or MC4R and MRAP2 (at different ratios 1:0, 1:1, 1:3, and 1:6) were transiently co-transfected into HEK293T cells along with constant amounts of pCRE-Luc (firefly luciferase, Santa Cruz, CA, USA) to monitor the cAMP production, and pRL-TK reporter vectors (renilla luciferase, Promega, WI, USA) to normalize the transfection efficiency. Empty vectors were added to each group for maintaining an identical total transfection amount. After 24 h, cell culture medium was replaced with human α-MSH/ACTH (1–24) including indicated concentration in DMEM with 0.1% BSA and incubated for 4 h at 37°C/5% CO2, while negative controls were applied by using zero peptide concentration of α-MSH/ACTH (1–24). Luminescent assay was conducted with Dual-Glo Luciferase Assay System (Promega, WI, USA) according to the manufacturer’s protocol. Evaluation of firefly and Renilla luminescence were performed with Spectramax ID3 multimode microplate reader (Molecular Devices, CA, USA). The Firefly luciferase activities of HEK293T cells expressing axolotl MC3R/MC4R were further normalized to Renilla luciferase activities.
To examine the effect of axolotl MRAP2 on the cell surface expression of MC3R and MC4R, surface epitope detection by ELISA was conducted in HEK293T cells. In brief, HEK293T cells seeded in a 24-well plate coated with 0.01% poly-D-lysine were transiently transfected with N-terminally 3×HA receptors plasmids with or without different ratios of 2×Flag-MRAP2 plasmids (plasmid ratios of 1:0, 1:1, 1:3, and 1:6). After 24 h transfection, cells were fixed with 4% paraformaldehyde and then washed three times with D-PBS for 5 min each time. Next, cells were incubated for 45 min with blocking buffer (5% milk in D-PBS), followed by incubation with mouse anti-HA antibodies (1:2,000) for 2 h. After incubation, cells were washed again and incubated for 2 h with HRP-conjugated Goat anti Mouse IgG antibodies (1:2,000). Finally, after washing these cells with D-PBS for three times and incubating with TMB for 30 min, the reaction was terminated by sulfuric acid (2 mol/L). We measured the absorbance value at 450 nm using Spectramax ID3 multimode microplate reader.
To detect the interaction between axolotl MC3R/MC4R with MRAP2, HEK293T cells were grown on 0.01% poly-D-lysine coated cover glasses and 2Flag-MC3R/MC4R-VF1 (Venus Fragment 1) and 2Flag-MRAP2-VF2 (Venus Fragment 2) were transfected. 2Flag-MRAP2-VF1 and 2Flag-MRAP2-VF2 were transfected simultaneously as controls. After 24 h of transfection, we removed the culture medium, washed the cells with PBS, and fixed them with 4% paraformaldehyde solution for 15 min at room temperature. Cells washed by PBS were permeabilized, and non-specific binding sites were blocked in PBS containing 5% (w/v) milk for 1 h at room temperature. Permeabilized cells were then incubated overnight with 1:2,000 rabbit anti Flag monoclonal antibodies in PBS with 5% (w/v) milk at room temperature for 2 h and then the cells were washed by PBST (PBS supplement with 0.1% tween-20) 3 times for 5 min each. Then Goat anti-rabbit Alexa-Fluor 594 secondary antibodies (1:1,000) were added and incubated for 2 h in the dark. Finally, the coverslips were mounted with antifade reagent with DAPI (Cell Signaling Technology, USA) on microscope slides. Confocal microscopy imaging was conducted with a ×63 oil objective on a Zeiss confocal microscope, and the micrographs displayed in each group were exposed and identically processed.
To explore the protein interaction between the axolotl MC3R/MC4R and MRAP2, the N-terminally Flag tagged MRAP2 (2xFlag-MRAP2) and N-terminally 3×HA tagged receptors expression plasmids were constructed by cloning the coding sequence into the pcDNA3.1(+) vector. The tagged receptors and MRAP2 expression plasmids were then co-transfected into HEK293T cells and incubated for 36 h. The transfected cells were then lysed by lysis buffer with 0.75% TritonX-100 [150 mM NaCl,50 mM Tris–HCl (pH 7.9) and proteinase inhibitor cocktail (Roche, Penzberg, Germany)]. The co-IP experiment was conducted with supernatants centrifuged from the cell lysates by overnight incubating the samples with rabbit anti HA antibody at 4°C. Protein A/G Agarose beads (Beyotime, Shanghai, China) were added to the cell lysates right after the overnight incubation and rotated at 4°C for 3–4 h. Beads were then washed three times in lysis buffer and centrifuged, re-suspended in SDS loading buffer. After boiling for 15 min, proteins separated on sodium dodecyl sulfate-polyacrylamide gel (SDS/PAGE, 12%) electrophoresis along with the mouse anti-HA antibodies for receptor proteins and mouse anti-Flag antibodies for MRAP2 proteins were used to perform western blot.
To evaluate the metabolic expression changes of mc3r, mc4r, mrap2, pomc, and agrp after three-week fasting in the axolotl, 6 juvenile axolotls were used for the food deprivation experiment with 3 as a control group and 3 in the experimental group. All juvenile axolotls from both groups were kept at the room temperature with sufficient oxygen supplements, and water in each tank was changed every day. The animals in the control group were fed twice a day for 3 weeks, whereas the animals in the experimental group were fasted for 3 weeks without any food. The body weight for each axolotl was measured before and post the experiment. The hypothalamic tissues from both control and fasting group were dissected and stored in TRNzol. The total RNA extraction and first-strand reverse transcription were conducted with TRNzol Universal reagent and FastQuant RT Kit (with gDNase) following the standard protocols. We then diluted the cDNA reaction to 1:10 as a template for qPCR. The qRT-PCR was performed using SuperReal PreMix Plus (SYBR Green) with the PCR setting as following: preincubation at 95°C for 600 s; 45 cycles of 95°C 10 s, 60°C for 10 s, and 72°C for 10 s; melting at 95°C for 10 s, 65°C for 60 s and 97°C for 1 s. All reactions were run in three biological replicates for both control and fasting groups and repeated three times.
To validate the results of all experiments, we separately repeated these experiments at least three times. The dose–response curves for cAMP luminescent assay were analyzed with the nonlinear regression models, and the evaluation of the corresponding half-maximal effective concentration (EC50) values was calculated using GraphPad Prism 6 (GraphPad Software, Inc., CA, USA). The receptor surface expression was also analyzed using GraphPad Prism 6. We applied one-way ANOVA with Tukey post-test with a nominal significant level of 0.05 (*p <0.05). The final results were represented as mean ± SEM.
Using multiple blast tools from the NCBI database, we identified the full-length gene sequences of the melanocortin receptors (mc3r, mc4r) and melanocortin accessory protein (mrap2) in the axolotl genome (63–65). The axolotl mc3r contains 963 coding nucleotides for 320 amino acids and are located in Chr3P; mc4r contains 1,008 coding nucleotides for 324 amino acids and are located in Chr5P; and mrap2 contains 651 coding nucleotides for 216 amino acids and are located in Chr4P. Both axolotl MC3R and MC4R contain seven transmembrane (TM) regions like other GPCRs, while MRAP2 includes one TM domain like MRAPs in other species.
The multiple alignments of MC3R, MC4R, and MRAP2 protein sequences were carried out for representative vertebrates selected from each class, namely, primate (Human, Macaque), ruminant (Cattle, Sheep), rodent (Mouse), avian (Chicken), reptile (Turtle, Snake, Common lizard), amphibia (Clawed frog, Axolotl), teleost (Zebrafish, Rainbow trout), and cartilaginous fish (Elephant shark), as shown in Figures 1A–C, respectively (Supplemental Table 2). The axolotl MC3R and its orthologs across species have an average sequence identity 72%, sharing the highest identity with chicken (80%), and the lowest identity with rainbow trout (66%) (Figure 1A). In comparison with the axolotl MC3R, MC4R shares higher sequence identities (average identity of 80%) with its orthologs across species, namely, the lowest identity of 71% with zebrafish and the highest identity of 85% with turtle (Figure 1B). Notably, sequences in the 7 TMs for both MC3R and MC4R are more conserved across various species than that in the C-terminal and N-terminal. The axolotl MRAP2 and its orthologs across species has a relatively low sequence identity (average identity of 59%), whereas a striking homology was found in the transmembrane domain, suggesting its importance for maintaining conserved function of MRAP2.
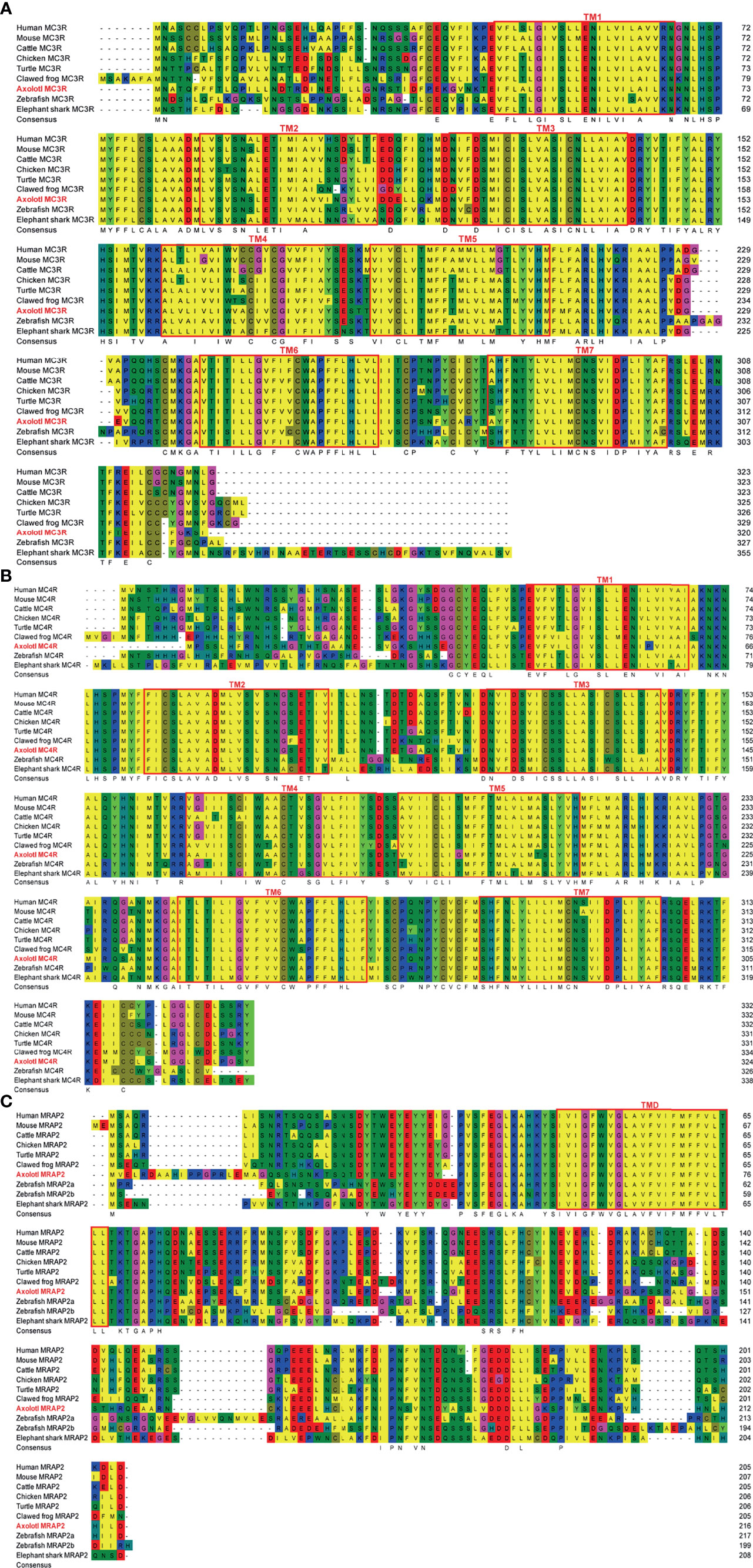
Figure 1 Sequence alignments of multiple MC3R, MC4R, and MRAP2 proteins. (A) Sequence alignment of multiple MC3R proteins. (B) Sequence alignment of multiple MC4R proteins. (C) Sequence alignment of multiple MRAP2 proteins. The transmembrane (TM) regions in the amMC3R and amMC4R are represented by red box and are numbered 1–7. The red box marks the transmembrane domains (TM) in the MRAP2.
To reveal the phylogenetic relationship of these melanocortin genes, the ML phylogenetic trees were constructed with the protein sequences of the representative vertebrates. The consensus phylogenetic trees of MC3R, MC4R, and MRAP2 are shown in Figure 2. The axolotl MC3R was grouped into the amphibian clade, and mostly related to the clawed frog MC3R with a bootstrap value of 72 (Figure 2A and Supplemental Table 2). Similarly, the axolotl MC4R was grouped into the amphibian clade, and mostly related to the clawed frog MC4R with a bootstrap value of 72 (Figure 2B and Supplemental Table 2). Unlike the axolotl MC3R/MC4R, MRAP2 was not grouped with the clawed frog MRAP2, but instead the axolotl MRAP2 fell in between the reptile and amphibian clade (Figure 2C and Supplemental Table 2). The phylogenetic relationships of MC3R and MC4R for other species were generally in accordance with their evolutionary clades, except for the elephant shark MC3R and MRAP2 which were grouped more closely with other non-teleost fish species instead of zebrafish and rainbow trout.
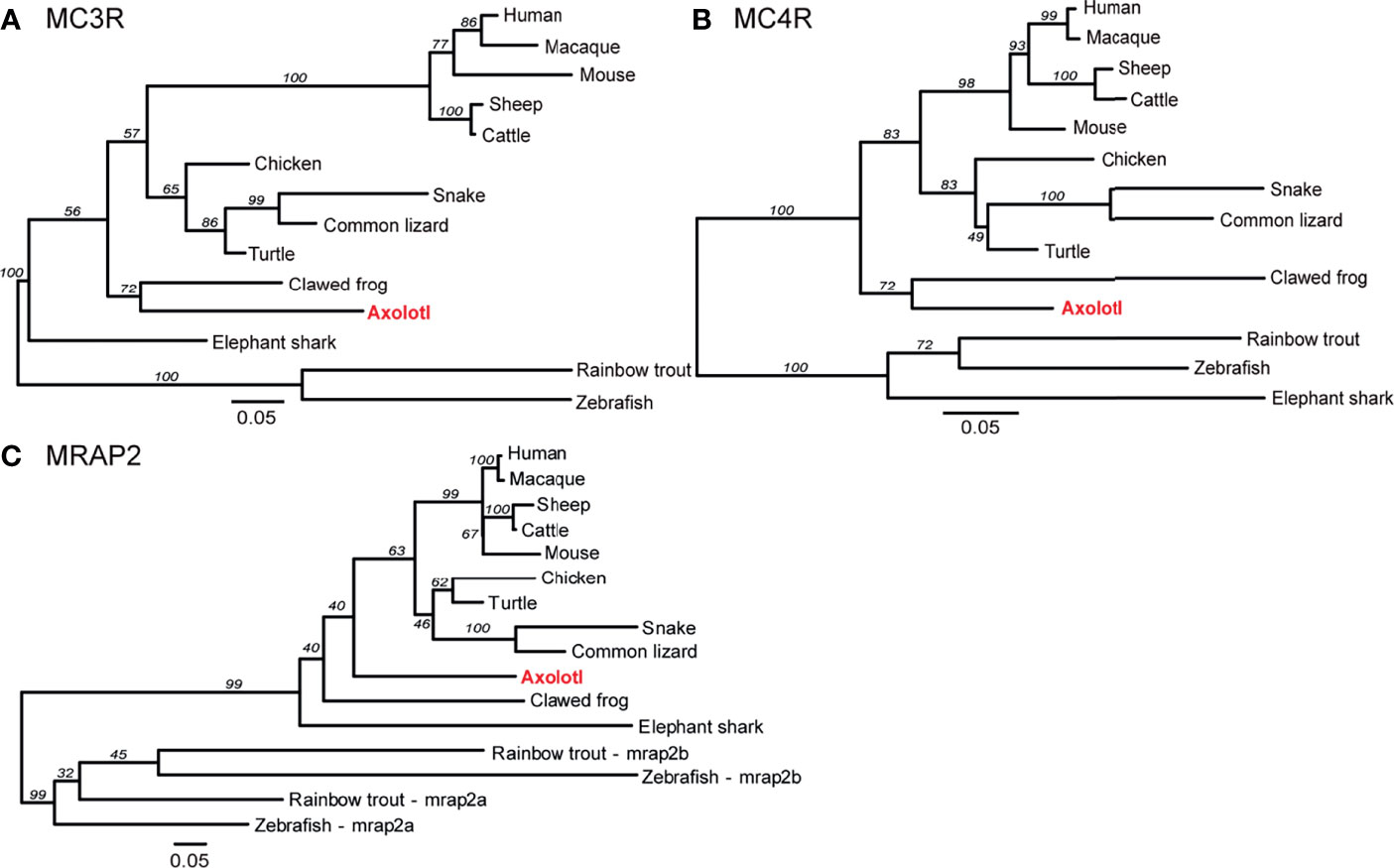
Figure 2 Phylogenetic analysis of MC3R, MC4R, and MRAP2. Phylogenetic trees of (A) MC3R, (B) MC4R, and (C) MRAP2. The ML phylogenetic tree was inferred based on multiple alignments of protein sequences using RAxML v8.2 with the PROTGAMMAJTT protein model and 1,000 rapid bootstrap replicates.
Synteny analysis was performed to provide additional aspects for the homology of mc3r, mc4r, and mrap2 genes across species, including human, mouse, chicken, turtle, clawed frog, zebrafish, and axolotl. We found that all flanking genes of axolotl MC3R shared flanking genes in other species except for diaph1 and bmp7 (as shown in blue, Figure 3A). The syntenic blocks of mc3r in axolotl were generally conserved with turtle, chicken, mouse, and human, while the flanking genes of mc3r in clawed frog and zebrafish varied from other species. In contrast, 5 out of 14 mc4r flanking genes in axolotl were conserved with the flanking genes in most species (as shown in blue, Figure 3B), suggesting that the syntenic blocks of axolotl mc4r were much less conserved with other species than that in axolotl mc3r. Moreover, we noticed that the flanking genes in other species, especially human, mouse, chicken, and turtle, were much more conserved with each other than with axolotl (shared flanking genes among other species without axolotl are shown in green, Figure 3B). The syntenic blocks of axolotl mrap2 were generally conserved with other species, especially between axolotl and turtle (Figure 3C). Similar with axolotl mc3r, 12 out of 14 mrap2 flanking genes in axolotl shared homologous in other species except for both copies (mrap2a and mrap2b) in zebrafish. The adjacent genes of two mrap2 copies in zebrafish were hardly overlapped with other species, suggesting a unique evolutionary pattern in teleost.
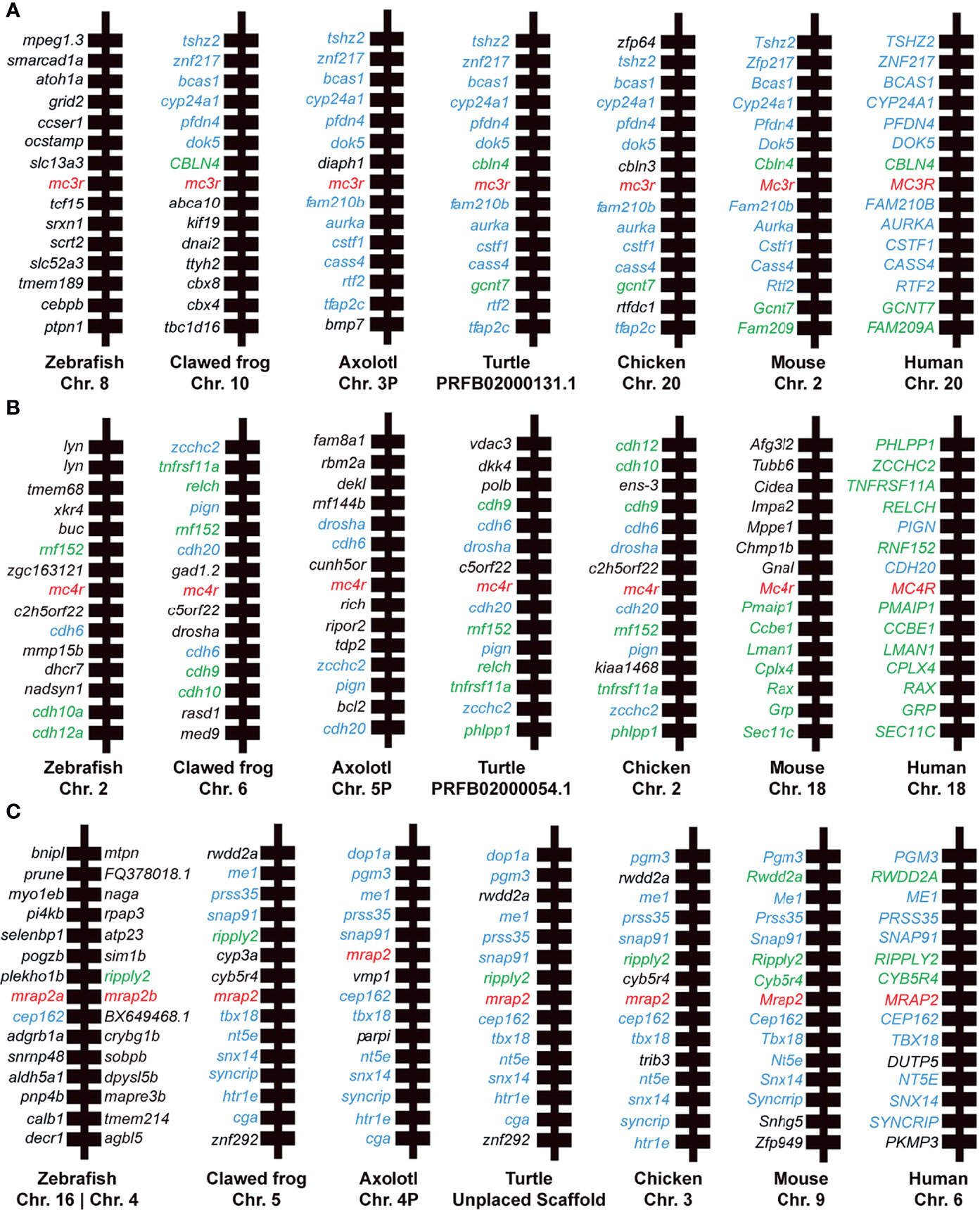
Figure 3 Synteny analysis of mc3r, mc4r, and mrap2. Chromosomal location and adjacent genes of (A) mc3r, (B) mc4r, and (C) mrap2 in following species: primate (Human, Macaque), ruminant (Cattle, Sheep), rodent (Mouse), avian (Chicken), reptile (Turtle, Snake, Common lizard), amphibia (Clawed frog, Axolotl), teleost (Zebrafish, Rainbow trout), and cartilaginous fish (Elephant shark). Genes showing conserved synteny with axolotl are shown in blue. Genes showing conserved synteny among at least two species but not with axolotl are shown in green.
To identify the tissue distribution patterns of mc3r, mc4r, mrap2, agrp, and pomc in axolotl, quantitative RT-PCR analysis was performed with the cDNA libraries from 16 tissues of a female adult axolotl (Figure 4A). Moderate but clear expression of mc3r, mc4r, mrap2, agrp, and pomc were detected in the axolotl brain, hypothalamus and pituitary. In addition, axolotl mc3r expression was detected in eye, heart, liver, spleen, ovary, dorsal skin and fat, whereas axolotl mc4r showed a broad expression pattern across multiple tissues in axolotl. Similar with mc3r, axolotl mrap2 also expressed in several peripheral tissues including eye, heart, liver, stomach, ovary, gill and ventral skin. The axolotl agrp and pomc showed restricted expression profiles in brain, with very low level of pomc in liver, ovary, and dorsal skin.
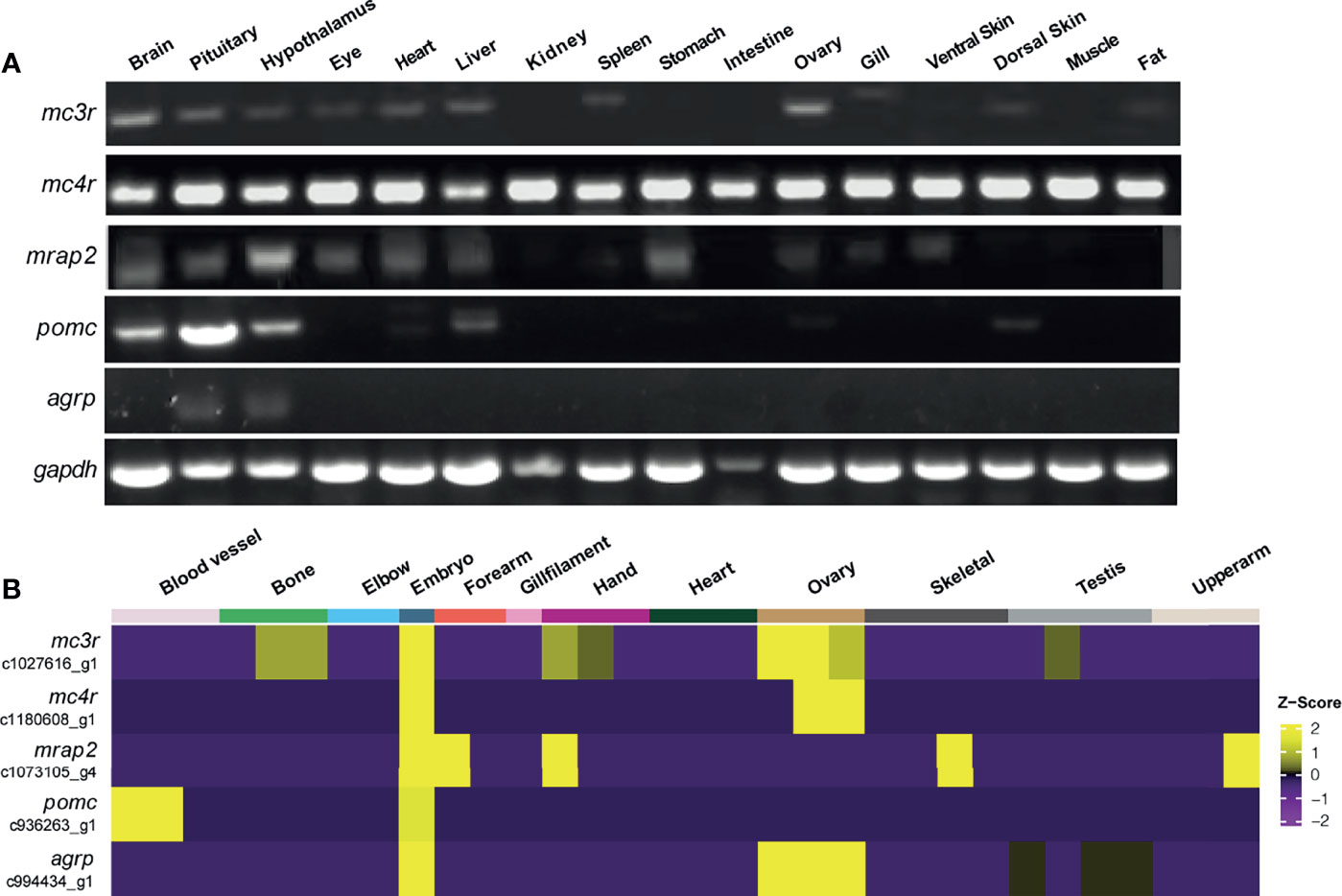
Figure 4 Tissue distribution of mc3r, mc4r, mrap2, agrp, and pomc mRNA in axolotl. (A) Tissue expression of mc3r, mc4r, mrap2, agrp, and pomc mRNA was analyzed by semi‐quantitative reverse transcription PCR using a housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (gapdh) as the internal control. (B) Heatmap showing the corresponding transcripts of mc3r, mc4r, mrap2, agrp, and pomc enriched in specific tissue types.
To reveal the expression profiles in more axolotl tissues, we obtained the published bulk RNA-seq data of axolotl tissues and annotated the corresponding transcripts for mc3r, mc4r, mrap2, agrp, and pomc to illustrate the expression patterns in various tissues (Figure 4B) (69). Interestingly, all 5 genes were highly expressed in axolotl embryo, suggesting their potential functional involvements for the embryonic development.
Next, we performed cAMP luminescent monitoring assay in HEK293 cells upon treatment by α-MSH or ACTH in the presence of different concentrations of axolotl MRAP2 (1:0, 1:1, 1:3, and 1:6) to characterize the pharmacological profiles of axolotl MC3R and MC4R in vitro. As shown in Figures 5A, B, we found that the luciferase signal of MC3R could be potentiated by α-MSH and ACTH without MRAP2 (logEC50 of α-MSH −9.95 ± 0.35 M and logEC50 of ACTH −9.51 ± 0.36 M, Table 1). Similar with MC3R, we also verified the stimulation of MC4R by α-MSH with logEC50 value of −9.84 ± 0.31 M and ACTH with logEC50 value of −9.62 ± 0.30 M in a dose-dependent manner (Figures 5C, D and Table 1). Strikingly, axolotl MC3R and MC4R both exhibited noticeable constitutive activities in the presence of MRAP2 proteins.
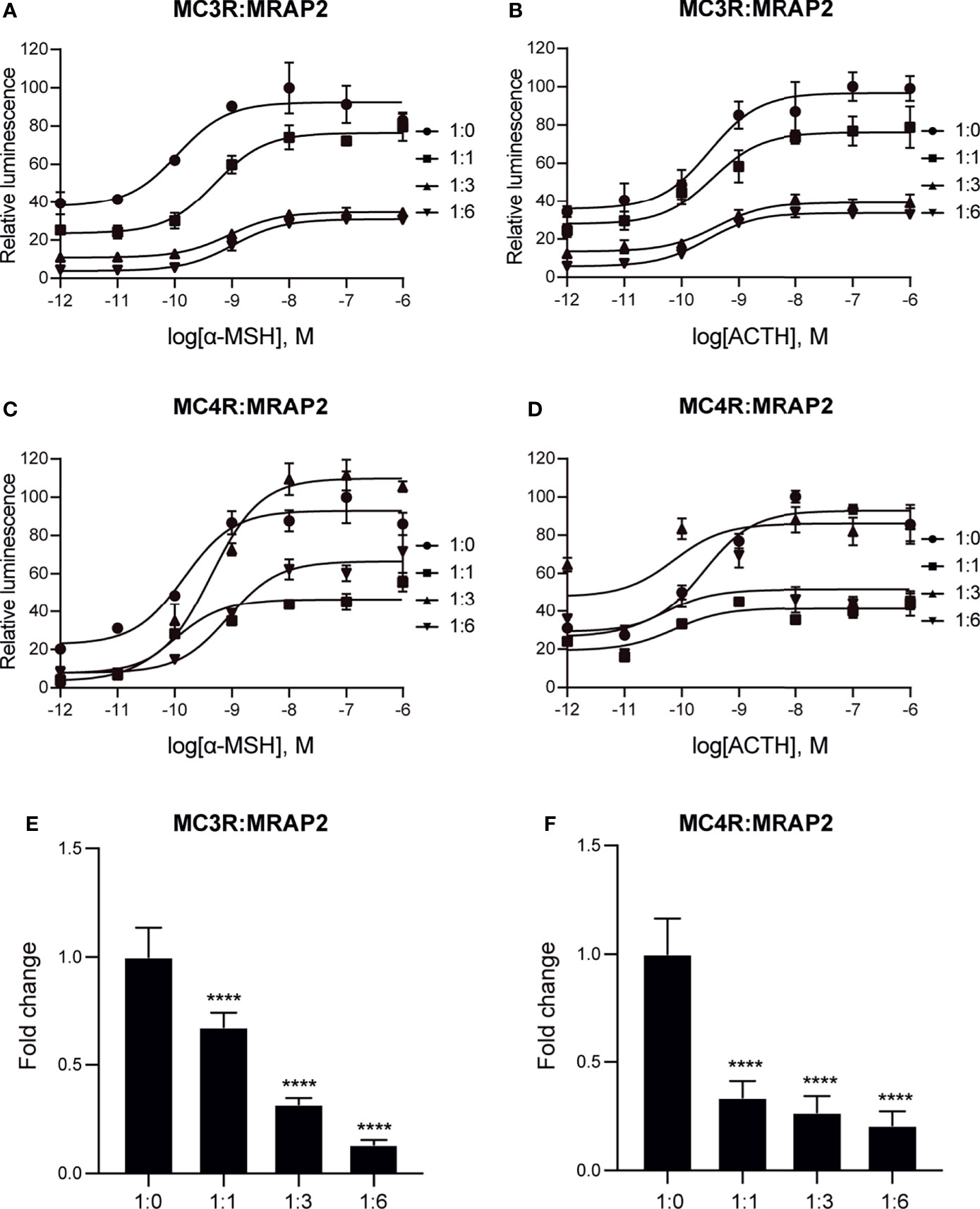
Figure 5 Pharmacological modulation of axolotl MC3R and MC4R by α-MSH and ACTH. (A) Effect of different doses (10−12–10−6 M) of α-MSH in activating axolotl MC3R in the presence of axolotl MRAP2 in HEK293T cells measured with Dual-glo luciferase assay. (B) Effect of different doses (10−12–10−6 M) of ACTH in activating axolotl MC3R in the presence of axolotl MRAP2 in HEK293T cells measured with Dual-glo luciferase assay. (C) Effect of different doses (10−12–10−6 M) of α-MSH in activating axolotl MC4R in the presence of axolotl MRAP2 in HEK293T cells measured with Dual-glo luciferase assay. (D) Effect of different doses (10−12–10−6 M) of ACTH in activating axolotl MC4R in the presence of axolotl MRAP2 in HEK293T cells measured with Dual-glo luciferase assay. (E, F) The constitutive activity of MC3R/MC4R in the presence of different amounts of MRAP2. ****p < 0.0001.
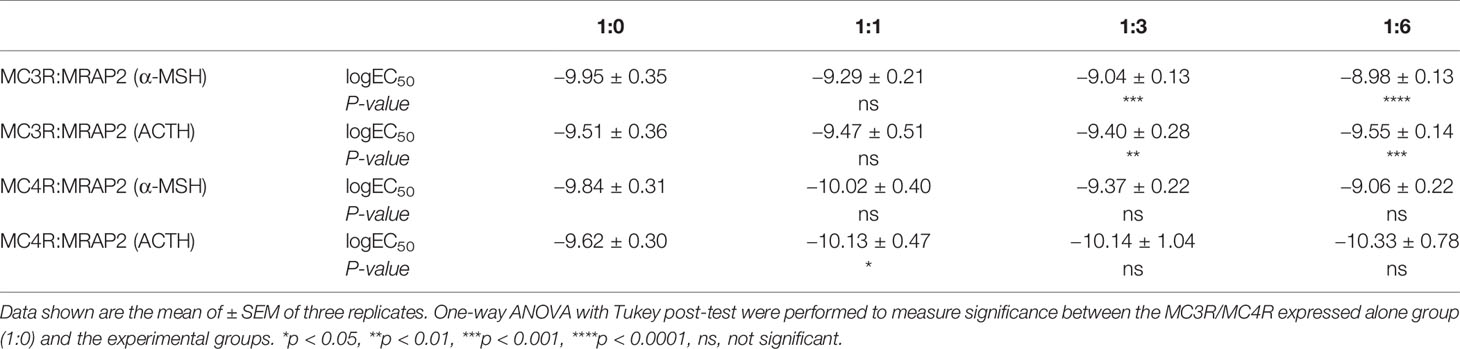
Table 1 EC50 values of α-MSH and ACTH in activating axolotl MC3R/MC4R in presence of different amounts of axolotl MRAP2.
To further evaluate how axolotl MRAP2 proteins alter the stimulation of axolotl MC3R and MC4R by α-MSH or ACTH in vitro, we examined different transient transfection ratios of MCRs to MRAP2 (1:0, 1:1, 1:3, 1:6). Noticeably, the stimulation of MC3R was dramatically suppressed by MRAP2 (Figures 5A, B and Table 1). The presence of MRAP2 could dose‐dependently decrease both the constitutive activity and maximal responsive plateau of MC3R by either α-MSH or ACTH (Figures 5A, B, E and Table 1). Similarly, the co-expression of MRAP2 revealed a mild alteration on the MC4R stimulation by two agonists (Figures 5C, D and Table 1). With different concentrations of MRAP2, the stimulation of MC4R by α-MSH and ACTH generally showed no significant change (p >0.05). The constitutive activity of MC4R significantly decreased in a dose-dependent manner with various proportions of MC4R and MRAP2 plasmids (Figure 5F).
To investigate the role of axolotl MRAP2 in the trafficking of axolotl MC3R/MC4R to the cell membrane, we conducted ELISA assay to measure the cell surface expression level of MC3R/MC4R in the presence of various ratios of MRAP2 plasmids. As shown in Figure 6A, no significant change was found in the cell surface expression level of MC3R in the presence of any amounts of MRAP2, suggesting that MRAP2 could not significantly affect the trafficking of MC3R to the cell membrane. Unlike MC3R, the cell surface expression of MC4R significantly decreased when co-expressed with MRAP2 in the ratios of 1:3 and 1:6 (Figures 6B). Moreover, the presence of MRAP2 decreased MC4R cell surface expression in a dose-dependent manner because the lower amount of MRAP2 (1:1) failed to significantly decrease the cell surface expression level of MC4R (Figure 6B). Taken together, the distinct effects of MRAP2 on the cell surface expression of MC3R and MC4R in axolotl suggested a complex mechanism of MRAP2 for modulating trafficking and post-translational cell surface translocation of MCRs.
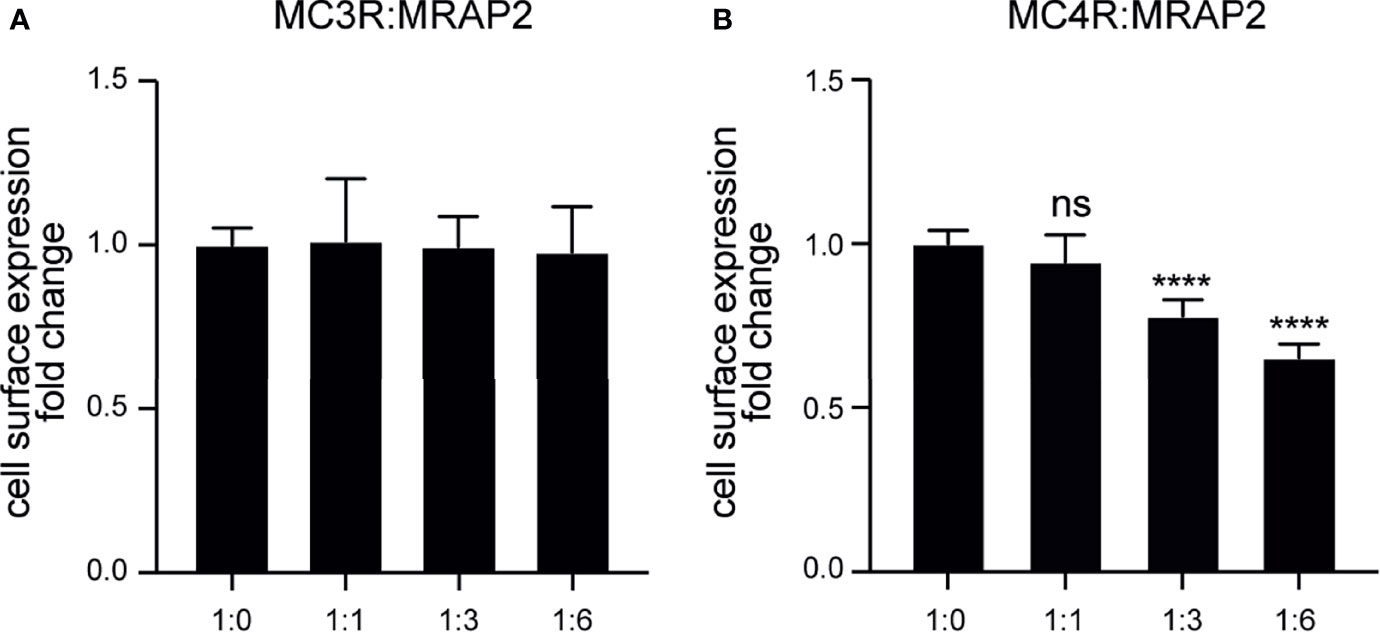
Figure 6 Modulation of the cell surface expression of axolotl MC3R and MC4R by MRAP2. (A) Cell surface expression of axolotl MC3R in presence of different amounts of MRAP2 (1:0, 1:1, 1:3, 1:6). (B) Cell surface expression of axolotl MC4R in presence of different amounts of MRAP2 (1:0, 1:1, 1:3, 1:6). The cell surface expression level of each receptor was expressed as the fold change of the measured surface epitopes level when the receptor was expressed alone (at the plasmid ratio of 1:0). Each data point represents mean ± SEM of three replicates (N = 3). One-way ANOVA with Tukey post-test were performed for all groups of data (****p < 0.0001, ns, not significant).
Given the fact that axolotl MRAP2 could regulate the trafficking and signaling of MC3R and MC4R, we further monitored the protein complex formation and co-localization in vitro by co-IP and BiFC assays in HEK293T cells. We found that both axolotl MC3R and MC4R could interact with MRAP2 and the coimmunoprecitated bands were observed on the SDS-PAGE gel (Figures 7A, B). To examine the co-localization of axolotl MC3R/MC4R with MRAP2, we conducted the BiFC assays using two complementary fragments of Venus fluorescent protein: Venus-F1/VF1 fused to MC3R/MC4R and Venus-F2/VF2 fused to MRAP2. The MRAP2 control experiment indicated a clear co-localization of MRAP2 with two different tags (Figure 7C). As shown in Figure 7D, we detected a stable fluorescence on the plasma membrane of MRAP2 and MC3R, suggesting their co-localization on the cell surface. Similarly, a steadily detectable fluorescence of MRAP2 and MC4R was also identified on the plasma membrane, confirming the co-localization and functional protein complex of MRAP2 and MC4R (Figure 7E).
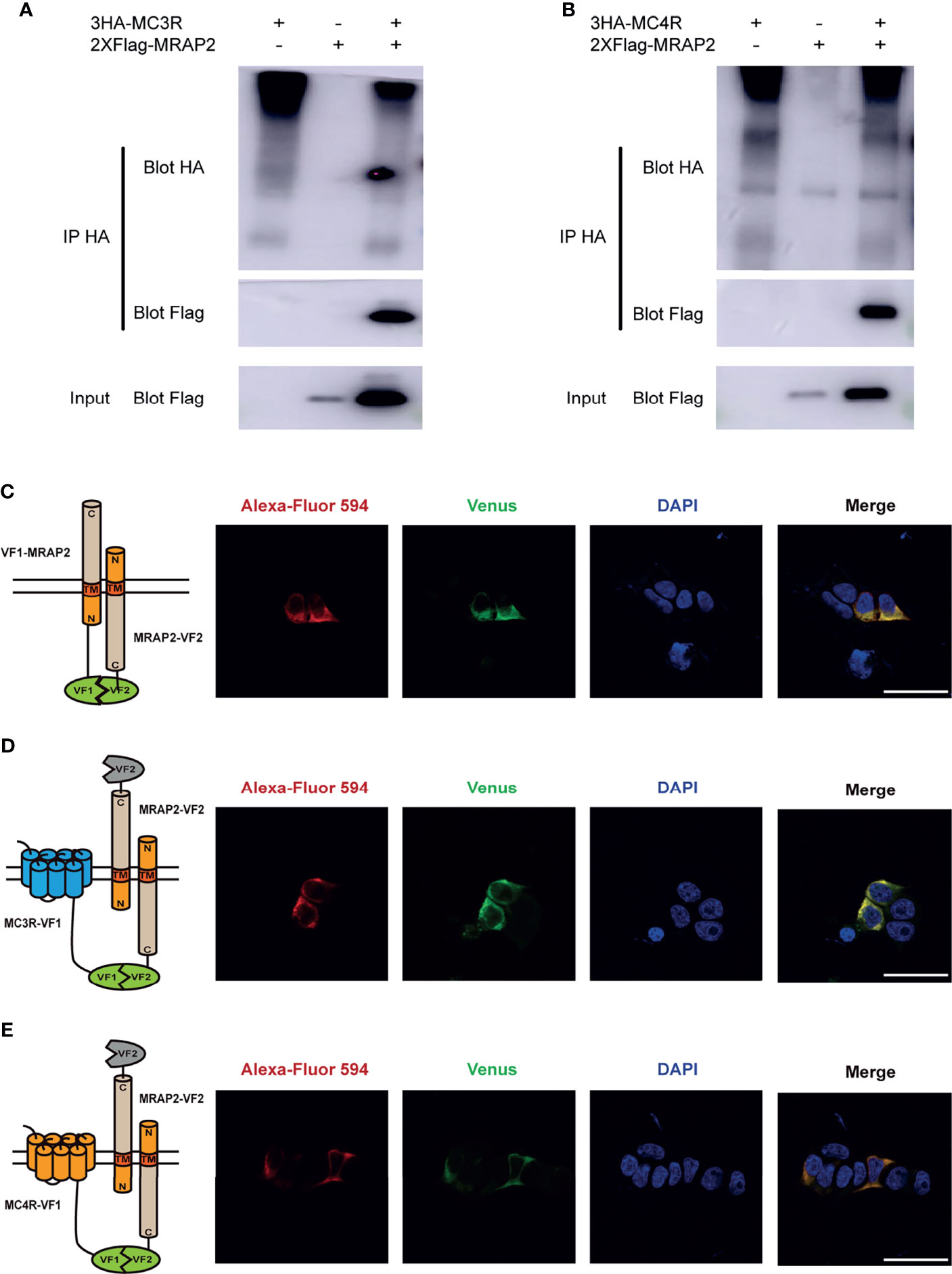
Figure 7 Interaction and co-localization of axolotl MC3R/MC4R and MRAP2 in vitro. (A) Coimmunoprecipitation of 3HA‐MC3R with 2xFlag‐MRAP2 in HEK293T cells. (B) Coimmunoprecipitation of 3HA‐MC4R with 2xFlag‐MRAP2 in HEK293T cells. (C) Confocal microscopy of the co-localization of MRAP2-VF1 and MRAP2-VF2 on the plasma membrane. (D) Confocal microscopy of the co-localization of MC3R and MRAP2 on the plasma membrane. (E) Confocal microscopy of the co-localization of MC4R and MRAP2 on the plasma membrane. Nuclei stained with DAPI are shown in blue and Venus fluorescence in green. Surface expression of MC3R/MC4R, and MRAP2 is shown in red, detected by anti‐Flag antibody and secondary anti‐rabbit Alexa Fluro 594 (Abcam). Scale bar = 50 µm.
To evaluate the metabolic expression dynamics of mc3r, mc4r, mrap2, agrp, and pomc in axolotl hypothalamus, we conducted a 3-week food deprivation experiment, and carried out qRT-PCR to compare the hypothalamic expression levels between the normal feeding and the 3-week fasting group. The axolotl mc3r was significantly upregulated in the fasting hypothalamus (Figure 8A), while mc4r had a mild but not significant upregulation upon fasting (Figure 8B). After 3-week fasting, the expression level of mrap2 significantly was downregulated with a P-value of 0.0007 (Figure 8C). Similar to previous fasting studies, pomc was significantly downregulated in hypothalamus after fasting, whereas agrp revealed an opposite pattern with significantly upregulation in the fasting hypothalamus (Figures 8D, E).
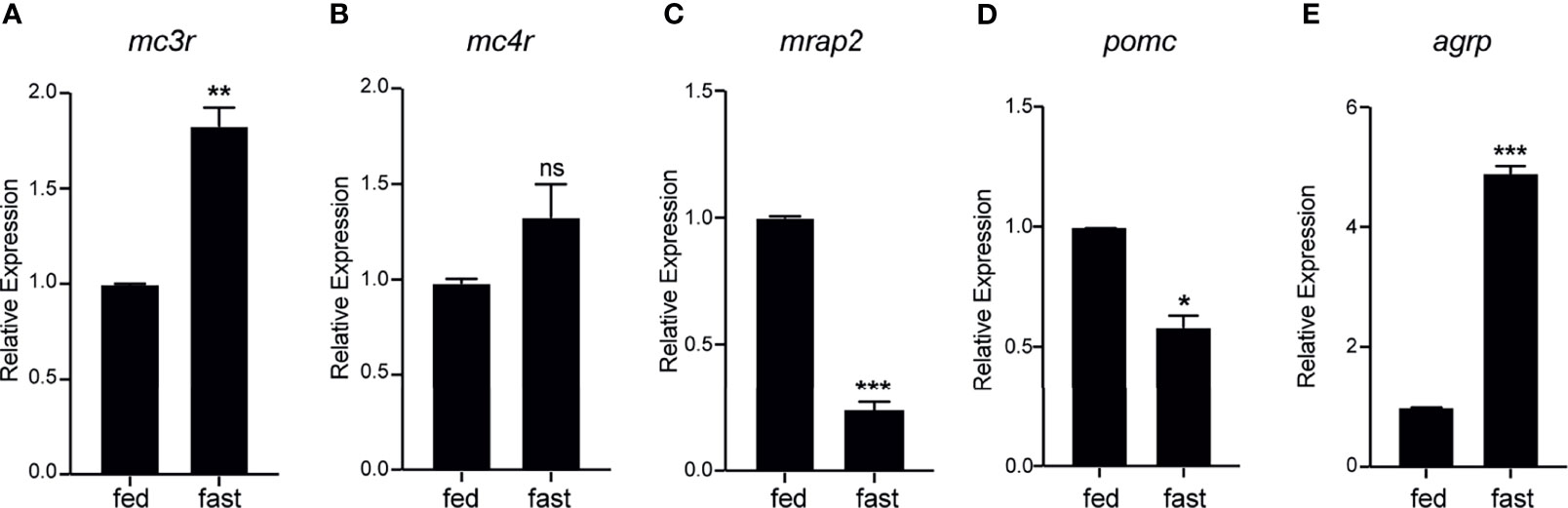
Figure 8 Fasting induced expression dynamics of mc3r, mc4r, mrap2, agrp, and pomc in the axolotl hypothalamus. Juvenile axolotls were fasted for 3 weeks and the expression levels of (A) mc3r, (B) mc4r, (C) mrap2, (D) pomc, and (E) agrp in the hypothalamus were analyzed by qRT-PCR. Data are analyzed by two-tailed t-test compared with the normal feeding group, graphs are shown as mean ± SEM. *P < 0.05, **P < 0.01; ***P < 0.001; ns, not significant.
The vital physiological roles of MC3R/MC4R and MRAP2 on the regulation of food intake and energy expenditure have been well studied in mammals, especially in human and mouse. As a phylogenetically important tetrapod species, the axolotl shares a similar vertebrate body shape with human but owns a remarkable capability to regenerate limbs and other tissues upon severe damage. We have previously shown that proper activation of MC4R signaling was essential for the limb regeneration in the Xenopus model (55). However, the pharmacological profiles of melanocortin receptor and the accessory protein system remain unknown in the axolotl. Here in this study, with multiple experimental approaches, we investigated the expression and function of axolotl neural melanocortin signaling. We cloned the full-length coding sequences of axolotl mc3r, mc4r, and mrap2 and performed the phylogenetic and synteny analysis of these genes among multiple species. We also examined the tissue expression patterns of these genes in axolotl and evaluated the pharmacological profiles of axolotl MC3R/MC4R in the presence of MRAP2. Finally, we determined the expressional dynamics of mc3r, mc4r, mrap2, pomc, and agrp in the fasting axolotl hypothalamus. This is the first comprehensive pharmacological evaluation of MRAP2 on axolotl neural melanocortin signaling.
In this study, obtaining the full-length sequences of axolotl mc3r, mc4r, and mrap2 required extra efforts due to the lack of complete gene annotation of the axolotl genome. Similar to other species, axolotl mc3r and mc4r are also intronless. So we cloned the full-length sequences from the hypothalamus derived genomic DNA. The full length of axolotl MC3R/MC4R protein is 320 and 324 amino acids, respectively, which is similar to the corresponding homology of other species (high identity >65%). The axolotl MRAP2 consists of 216 amino acids but is less conserved comparing to the corresponding genes of other species (average identity of 59%). The phylogenetic analysis indicates that both axolotl MC3R and MC4R are closely grouped with the corresponding counterparts in the clawed frog, suggesting the potential functional similarity of these MCRs among these species. We next compared the flanking genes of mc3r and mc4r across 7 different species and found the flanking region of mc3r was more conserved among these species than that of mc4r. Syntenic blocks of mc3r and mc4r in the zebrafish varied evidently from other species, indicating an early evolutionary separation of these genes in teleosts. The phylogenetic analysis of MRAP2 revealed a less conserved relationship among species, and axolotl MRAP2 fell in between the reptile and amphibian clade. A unique feature of two copies of mrap2 existent in the zebrafish and rainbow trout genome, suggests that there could be a teleost-specific duplication of mrap2 genes in the teleosts. The synteny analysis of mrap2 revealed that the flanking region of mrap2 was generally conserved among species, except for that in the zebrafish.
The tissue distribution of mc3r, mc4r, and mrap2 are identified in many species. In human, MC3R is enriched in the brain region and has a very low abundance in other peripheral tissues such as testis and adrenal gland. Human MC4R exhibits high expression level in the brain, fallopian tube, and retina, moderate abundance in thalamus, olfactory bulb, and spinal cord, but low abundance in salivary gland, adrenal gland, spleen, pituitary, stomach, testis, and ovary. In addition to high expression level of human brain, MRAP2 shows a broader distribution among other human tissues than MC3R/MC4R, indicating that its function might not limit to the energy homeostasis. In the clawed frog, mc3r and mc4r are expressed in brain region with low abundance in some peripheral tissues, whereas mrap2 distributes widely among multiple tissues including brain (45, 46). Moreover, in the zebrafish only mrap2a expressed in larval zebrafish, while mrap2b expression emerged until the adult stage, suggesting that these two genes could participate in the metabolic regulation at different life stages (48). In our study, a clear co-expression of mc3r, mc4r, mrap2, pomc, and agrp was observed in the axolotl brain region, indicating the potential interaction of these 5 genes for regulating the feeding behavior and energy metabolism in the central nervous system of axolotl.
MRAP2 could interact with MCRs and alter their sensitivity to α-MSH and ACTH in a bi-direction manner among different species. Human MRAP2 was reported to interact with all MCRs (37), and the co-expression of MRAP2 could change MC4R to an ACTH receptor (41). However, mouse MRAP2 could enhance the receptor sensitivity of MC4R to the α-MSH (42). In the clawed frog, MC3R and MC4R could respond to both α-MSH and ACTH, but α-MSH or ACTH seems to be more efficient for activating MC3R than MC4R (45). In chicken, MC3R and MC4R responded to α-MSH and ACTH equivalently, however, MC4R was found to become ACTH-preferred receptor in the presence of MRAP2 (43). Similarly, zebrafish MRAP2a facilitated the pharmacological profile of MC4R by preferably increasing its response to ACTH (47). In tilapia, MRAP2 was reported to reduce the MC4R signaling in response to α-MSH (70), whereas the feline MRAP2 was shown to enhance MC4R signaling stimulated by α-MSH (71). Here, we conducted cAMP luminescent assay to evaluate the pharmacological profiles of axolotl MC3R and MC4R in response to α-MSH and ACTH in vitro. We identified a clear stimulation of MC3R and MC4R by α-MSH in a dose-dependent manner (Figures 5A, B). These findings strongly suggested a conserved physiological function of axolotl MC3R and MC4R in response to the natural agonist α-MSH.
The constitutive activities of MCRs can affect the regulation of their physiological functions. We found that axolotl MC3R exhibited a high constitutive activity, that was significantly decreased by MRAP2 (Figure 5E). MC3R was also reported to have high constitutive activity in other species, such as zebrafish (72), channel catfish (51), and topmouth culter (53). In contrast to our result, the constitutive activity of MC3R was reported to be enhanced by both MRAP and MRAP2 in the clawed frog (45). MC4R exhibits strong constitutive activity in mammals (73–75), chicken (43), and amphibians (44, 45). Moreover, many teleost species have been reported to detect high constitutive activities of MC4R, namely, zebrafish (48), spotted scat (76), grass carp (77), swamp eel (78), Orange-spotted grouper (50), spotted sea bass (79), Nile tilapia (70), topmouth culter (52), and snakehead (49). Several MC4R mutants identified from obese patients have been reported to show a decreased constitutive activity but no other functional changes compared to normal MC4R (80). Constitutive activity defect of MC4R leads to obesity suggesting the basal component activity of MC4R acts as a tonic satiety signal, which may be necessary to maintain long-term energy homeostasis in human. In our study, we demonstrated the native basal cAMP activity of axolotl MC3R/MC4R signaling. MRAP2 reduced the constitutive activities of MC3R and MC4R indicating that MRAP2 could negatively influence the endogenous signaling cascades of axolotls.
Next our ELISA assay demonstrated that axolotl MRAP2 significantly decreased the cell surface expression of MC4R in a dose-dependent manner but did not affected the trafficking of MC3R. In other species, MRAP2 showed varied effect in the modulation of MC3R/MC4R translocation to the cell surface. For instance, the surface expression of clawed frog MC3R was suppressed by MRAP2 (45), whereas in our study, axolotl MRAP2 did not affect the cell surface expression of MC3R. MRAP2 showed no effect on cell surface expression of MC4R in both chicken (43) and snakehead (49). However, MRAP2 could significantly decrease the surface level of MC4R in tilapia (70). We further confirmed the direct interaction and co-localization of both MC3R and MC4R with MRAP2 proteins on the plasma membrane by co-IP and BiFC assays (Figure 7). Our results were consistent with some previous studies in other vertebrates such as the clawed frog (45) and sea lamprey (54). Taken together, multiple studies from different species demonstrated that MRAP2 acted as a bi-functional player in modulating the MCRs trafficking to the plasma membrane, suggesting its species-specific role in modulating the cell surface expression of MC3R/MC4R.
The food deprivation experiment further confirmed the vital role of melanocortin signaling in regulating the appetite and energy homeostasis. Our results showed that axolotl mc3r and agrp significantly upregulated in the fasting hypothalamus, whereas the expression levels of axolotl mrap2 and pomc significantly reduced in hypothalamus upon 3-week fasting (Figure 8). As a key regulator of central appetite, multiple studies had reported the significant expressional dynamics of agrp and pomc expression in the central nervous system upon fasting treatment. For example, the elevation of Agrp (81) and the inhibition of Pomc expression (82) were reported in mouse hypothalamus after fasting. Studies in teleost fish had shown consistent results, for instance, agrp was significantly upregulated whereas pomc was not altered by fasting in zebrafish adult brain (83). A significant hypothalamic upregulation of agrp1 and downregulation of pomca2 were identified after fasting in Atlantic Salmon (84).
In conclusion, our study comprehensively examined and elucidated the pharmacological and physiological regulation of central melanocortin signaling in the axolotl. We found a relatively conserved protein structure of axolotl MC3R, MC4R, and MRAP2 in comparison with other species, especially the TM regions. A co-expression profile of mc3r, mc4r, and mrap2 along with pomc and agrp in the axolotl brain regions and the significant elevation of mc3r and agrp and reduction of mrap2 and pomc in the hypothalamus upon fasting further confirmed the vital role of melanocortin signaling and their physiological interaction in regulating central feeding behavior and energy balance. The pharmacological profiles of axolotl MC3R and MC4R also demonstrated that both receptors could be activated by α-MSH and ACTH. The MRAP2 showed no effect on the cell surface expression level of MC3R, but significantly negative effect on the cell surface translocation of MC4R in a dose-dependent manner. The co-localization and functional complex formation of axolotl MC3R/MC4R with MRAP2 were confirmed by co-IP and BiFC assays in vitro. Taken together, pharmacological evaluation of the central melanocortin signaling of a primitive poikilotherm axolotl elucidated the vital physiological role in the regulation of appetite and energy balance in an amphibian species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by the Tongji University.
XW, GL, JG, and ChZ participated in the design of the study. XW, SX, and XiL performed the experiments. LL, CoZ, WS, XL, YF, YL and HZ contributed to data collection and data analysis. XW, GL, and ChZ contributed to the manuscript writing. All authors contributed to the article and approved the submitted version.
The work was supported by grants from the National Key Research and Development Program of China (Grant Nos. 2019YFA0111400 and 2017YFA0103902); the National Natural Science Foundation of China (Grant Nos. 31771283, 31771608, and 31801226); the Innovative Research Team of High-level Local Universities in Shanghai (Grant No. SSMU-ZDCX20180700) and a Key Laboratory Program of the Education Commission of Shanghai Municipality (Grant No. ZDSYS14005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.820896/full#supplementary-material
1. Catania A, Gatti S, Colombo G, Lipton JM. Targeting Melanocortin Receptors as a Novel Strategy to Control Inflammation. Pharmacol Rev (2004) 56(1):1–29. doi: 10.1124/pr.56.1.1
2. Cone RD. Studies on the Physiological Functions of the Melanocortin System. Endocr Rev (2006) 27(7):736–49. doi: 10.1210/er.2006-0034
3. Millington GW. Proopiomelanocortin (POMC): The Cutaneous Roles of Its Melanocortin Products and Receptors. Clin Exp Dermatol (2006) 31(3):407–12. doi: 10.1111/j.1365-2230.2006.02128.x
4. Cawley NX, Li Z, Loh YP. 60 YEARS OF POMC: Biosynthesis, Trafficking, and Secretion of Pro-Opiomelanocortin-Derived Peptides. J Mol Endocrinol (2016) 56(4):T77–97. doi: 10.1530/JME-15-0323
5. Turner JD, Keith AB, Smith AI, McDermott JR, Biggins JA, Edwardson JA. Studies on the Characterisation of Alpha-MSH-Like Immunoreactivity in Rat Hypothalamus. Regul Pept (1983) 5(3):283–93. doi: 10.1016/0167-0115(83)90259-8
6. Reudelhuber TL. Salt-Sensitive Hypertension: If Only it Were as Simple as Rocket Science. J Clin Invest (2003) 111(8):1115–6. doi: 10.1172/JCI18397
7. Bicknell AB. The Tissue-Specific Processing of Pro-Opiomelanocortin. J Neuroendocrinol (2008) 20(6):692–9. doi: 10.1111/j.1365-2826.2008.01709.x
8. Dores RM, Londraville RL, Prokop J, Davis P, Dewey N, Lesinski N. Molecular Evolution of GPCRs: Melanocortin/melanocortin Receptors. J Mol Endocrinol (2014) 52(3):T29–42. doi: 10.1530/JME-14-0050
9. Yang Y. Structure, Function and Regulation of the Melanocortin Receptors. Eur J Pharmacol (2011) 660(1):125–30. doi: 10.1016/j.ejphar.2010.12.020
10. Chhajlani V, Wikberg JE. Molecular Cloning and Expression of the Human Melanocyte Stimulating Hormone Receptor cDNA. FEBS Lett (1992) 309(3):417–20. doi: 10.1016/0014-5793(92)80820-7
11. Mountjoy KG, Robbins LS, Mortrud MT, Cone RD. The Cloning of a Family of Genes That Encode the Melanocortin Receptors. Science (1992) 257(5074):1248–51. doi: 10.1126/science.1325670
12. Cone RD, Lu D, Koppula S, Vage DI, Klungland H, Boston B, et al. The Melanocortin Receptors: Agonists, Antagonists, and the Hormonal Control of Pigmentation. Recent Prog Horm Res (1996) 51:287–317; discussion 318.
13. Rees JL. The Melanocortin 1 Receptor (MC1R): More Than Just Red Hair. Pigment Cell Res (2000) 13(3):135–40. doi: 10.1034/j.1600-0749.2000.130303.x
14. Schaffer JV, Bolognia JL. The Melanocortin-1 Receptor: Red Hair and Beyond. Arch Dermatol (2001) 137(11):1477–85. doi: 10.1001/archderm.137.11.1477
15. Park J, Jeong D, Jang B, Oh ES. The Melanocortin-1 Receptor Reversely Regulates the Melanin Synthesis and Migration of Melanoma Cells via Dimerization-Induced Conformational Changes. Biochem Biophys Res Commun (2019) 518(4):739–45. doi: 10.1016/j.bbrc.2019.08.123
16. Chhajlani V, Muceniece R, Wikberg JE. Molecular Cloning of a Novel Human Melanocortin Receptor. Biochem Biophys Res Commun (1993) 195(2):866–73. doi: 10.1006/bbrc.1993.2125
17. Chida D, Nakagawa S, Nagai S, Sagara H, Katsumata H, Imaki T, et al. Melanocortin 2 Receptor is Required for Adrenal Gland Development, Steroidogenesis, and Neonatal Gluconeogenesis. Proc Natl Acad Sci USA (2007) 104(46):18205–10. doi: 10.1073/pnas.0706953104
18. Gantz I, Konda Y, Tashiro T, Shimoto Y, Miwa H, Munzert G, et al. Molecular Cloning of a Novel Melanocortin Receptor. J Biol Chem (1993) 268(11):8246–50. doi: 10.1016/S0021-9258(18)53088-X
19. Gantz I, Miwa H, Konda Y, Shimoto Y, Tashiro T, Watson SJ, et al. Molecular Cloning, Expression, and Gene Localization of a Fourth Melanocortin Receptor. J Biol Chem (1993) 268(20):15174–9. doi: 10.1016/S0021-9258(18)82452-8
20. Tao YX. The Melanocortin-4 Receptor: Physiology, Pharmacology, and Pathophysiology. Endocr Rev (2010) 31(4):506–43. doi: 10.1210/er.2009-0037
21. Demidowich AP, Jun JY, Yanovski JA. Polymorphisms and Mutations in the Melanocortin-3 Receptor and Their Relation to Human Obesity. Biochim Biophys Acta Mol Basis Dis (2017) 1863(10 Pt A):2468–76. doi: 10.1016/j.bbadis.2017.03.018
22. Lotta LA, Mokrosinski J, Mendes de Oliveira E, Li C, Sharp SJ, Luan J, et al. Human Gain-Of-Function MC4R Variants Show Signaling Bias and Protect Against Obesity. Cell (2019) 177(3):597–607 e599. doi: 10.1016/j.cell.2019.03.044
23. Lippert RN, Ellacott KL, Cone RD. Gender-Specific Roles for the Melanocortin-3 Receptor in the Regulation of the Mesolimbic Dopamine System in Mice. Endocrinology (2014) 155(5):1718–27. doi: 10.1210/en.2013-2049
24. Vaisse C, Clement K, Guy-Grand B, Froguel P. A Frameshift Mutation in Human MC4R is Associated With a Dominant Form of Obesity. Nat Genet (1998) 20(2):113–4. doi: 10.1038/2407
25. Tao YX. Mutations in Melanocortin-4 Receptor and Human Obesity. Prog Mol Biol Transl Sci (2009) 88:173–204. doi: 10.1016/S1877-1173(09)88006-X
26. Liu T, Ji RL, Tao YX. Naturally Occurring Mutations in G Protein-Coupled Receptors Associated With Obesity and Type 2 Diabetes Mellitus. Pharmacol Ther (2021):108044. doi: 10.1016/j.pharmthera.2021.108044
27. Gantz I, Shimoto Y, Konda Y, Miwa H, Dickinson CJ, Yamada T. Molecular Cloning, Expression, and Characterization of a Fifth Melanocortin Receptor. Biochem Biophys Res Commun (1994) 200(3):1214–20. doi: 10.1006/bbrc.1994.1580
28. Chen W, Kelly MA, Opitz-Araya X, Thomas RE, Low MJ, Cone RD. Exocrine Gland Dysfunction in MC5-R-Deficient Mice: Evidence for Coordinated Regulation of Exocrine Gland Function by Melanocortin Peptides. Cell (1997) 91(6):789–98. doi: 10.1016/s0092-8674(00)80467-5
29. Taylor AW, Kitaichi N, Biros D. Melanocortin 5 Receptor and Ocular Immunity. Cell Mol Biol (Noisy-le-grand) (2006) 52(2):53–9. doi: 10.1170/T708
30. Xu Y, Guan X, Zhou R, Gong R. Melanocortin 5 Receptor Signaling Pathway in Health and Disease. Cell Mol Life Sci (2020) 77(19):3831–40. doi: 10.1007/s00018-020-03511-0
31. Lu D, Willard D, Patel IR, Kadwell S, Overton L, Kost T, et al. Agouti Protein is an Antagonist of the Melanocyte-Stimulating-Hormone Receptor. Nature (1994) 371(6500):799–802. doi: 10.1038/371799a0
32. Yang Y, Harmon CM. Molecular Signatures of Human Melanocortin Receptors for Ligand Binding and Signaling. Biochim Biophys Acta Mol Basis Dis (2017) 1863(10 Pt A):2436–47. doi: 10.1016/j.bbadis.2017.04.025
33. Suzuki I, Cone R, Im S, Nordlund J, Abdel-Malek Z. Binding Capacity and Activation of the MC1 Receptors by Melanotropic Hormones Correlate Directly With Their Mitogenic and Melanogenic Effects on Human Melanocytes. Endocrinology (1996) 137:1627–33. doi: 10.1210/endo.137.5.8612494
34. Mountjoy KG, Bird IM, Rainey WE, Cone RD. ACTH Induces Up-Regulation of ACTH Receptor mRNA in Mouse and Human Adrenocortical Cell Lines. Mol Cell Endocrinol (1994) 99(1):R17–20. doi: 10.1016/0303-7207(94)90160-0
35. Roselli-Rehfuss L, Mountjoy KG, Robbins LS, Mortrud MT, Low MJ, Tatro JB, et al. Identification of a Receptor for Gamma Melanotropin and Other Proopiomelanocortin Peptides in the Hypothalamus and Limbic System. Proc Natl Acad Sci USA (1993) 90(19):8856–60. doi: 10.1073/pnas.90.19.8856
36. Tao YX, Conn PM. Chaperoning G Protein-Coupled Receptors: From Cell Biology to Therapeutics. Endocr Rev (2014) 35(4):602–47. doi: 10.1210/er.2013-1121
37. Chan LF, Webb TR, Chung TT, Meimaridou E, Cooray SN, Guasti L, et al. MRAP and MRAP2 Are Bidirectional Regulators of the Melanocortin Receptor Family. Proc Natl Acad Sci USA (2009) 106(15):6146–51. doi: 10.1073/pnas.0809918106
38. Metherell LA, Chapple JP, Cooray S, David A, Becker C, Ruschendorf F, et al. Mutations in MRAP, Encoding a New Interacting Partner of the ACTH Receptor, Cause Familial Glucocorticoid Deficiency Type 2. Nat Genet (2005) 37(2):166–70. doi: 10.1038/ng1501
39. Webb TR, Clark AJ. Minireview: The Melanocortin 2 Receptor Accessory Proteins. Mol Endocrinol (2010) 24(3):475–84. doi: 10.1210/me.2009-0283
40. Liang J, Li L, Jin X, Xu B, Pi L, Liu S, et al. Pharmacological Effect of Human Melanocortin-2 Receptor Accessory Protein 2 Variants on Hypothalamic Melanocortin Receptors. Endocrine (2018) 61(1):94–104. doi: 10.1007/s12020-018-1596-2
41. Soletto L, Hernandez-Balfago S, Rocha A, Scheerer P, Kleinau G, Cerda-Reverter JM. Melanocortin Receptor Accessory Protein 2-Induced Adrenocorticotropic Hormone Response of Human Melanocortin 4 Receptor. J Endocr Soc (2019) 3(2):314–23. doi: 10.1210/js.2018-00370
42. Asai M, Ramachandrappa S, Joachim M, Shen Y, Zhang R, Nuthalapati N, et al. Loss of Function of the Melanocortin 2 Receptor Accessory Protein 2 is Associated With Mammalian Obesity. Science (2013) 341(6143):275–8. doi: 10.1126/science.1233000
43. Zhang J, Li X, Zhou Y, Cui L, Li J, Wu C, et al. The Interaction of MC3R and MC4R With MRAP2, ACTH, Alpha-MSH and AgRP in Chickens. J Endocrinol (2017) 234(2):155–74. doi: 10.1530/JOE-17-0131
44. Li L, Xu Y, Zheng J, Kuang Z, Zhang C, Li N, et al. Pharmacological Modulation of Dual Melanocortin-4 Receptor Signaling by Melanocortin Receptor Accessory Proteins in the Xenopus Laevis. J Cell Physiol (2021) 236(8):5980–93. doi: 10.1002/jcp.30280
45. Tai X, Xue S, Zhang C, Liu Y, Chen J, Han Y, et al. Pharmacological Evaluation of MRAP Proteins on Xenopus Neural Melanocortin Signaling. J Cell Physiol (2021) 236(9):6344–61. doi: 10.1002/jcp.30306
46. Xu Y, Li L, Zheng J, Wang M, Jiang B, Zhai Y, et al. Pharmacological Modulation of the cAMP Signaling of Two Isoforms of Melanocortin-3 Receptor by Melanocortin Receptor Accessory Proteins in the Tetrapod Xenopus Laevis. Endocr Connect (2021) 10(11):1477–88. doi: 10.1530/EC-21-0179
47. Josep Agulleiro M, Cortes R, Fernandez-Duran B, Navarro S, Guillot R, Meimaridou E, et al. Melanocortin 4 Receptor Becomes an ACTH Receptor by Coexpression of Melanocortin Receptor Accessory Protein 2. Mol Endocrinol (2013) 27(11):1934–45. doi: 10.1210/me.2013-1099
48. Sebag JA, Zhang C, Hinkle PM, Bradshaw AM, Cone RD. Developmental Control of the Melanocortin-4 Receptor by MRAP2 Proteins in Zebrafish. Science (2013) 341(6143):278–81. doi: 10.1126/science.1232995
49. Wen ZY, Liu T, Qin CJ, Zou YC, Wang J, Li R, et al. MRAP2 Interaction With Melanocortin-4 Receptor in SnakeHead (Channa Argus). Biomolecules (2021) 11(3):481. doi: 10.3390/biom11030481
50. Rao YZ, Chen R, Zhang Y, Tao YX. Orange-Spotted Grouper Melanocortin-4 Receptor: Modulation of Signaling by MRAP2. Gen Comp Endocrinol (2019) 284:113234. doi: 10.1016/j.ygcen.2019.113234
51. Yang LK, Zhang ZR, Wen HS, Tao YX. Characterization of Channel Catfish (Ictalurus Punctatus) Melanocortin-3 Receptor Reveals a Potential Network in Regulation of Energy Homeostasis. Gen Comp Endocrinol (2019) 277:90–103. doi: 10.1016/j.ygcen.2019.03.011
52. Tao M, Ji RL, Huang L, Fan SY, Liu T, Liu SJ, et al. Regulation of Melanocortin-4 Receptor Pharmacology by Two Isoforms of Melanocortin Receptor Accessory Protein 2 in Topmouth Culter (Culter Alburnus). Front Endocrinol (Lausanne) (2020) 11:538. doi: 10.3389/fendo.2020.00538
53. Ji RL, Huang L, Wang Y, Liu T, Fan SY, Tao M, et al. Topmouth Culter Melanocortin-3 Receptor: Regulation by Two Isoforms of Melanocortin-2 Receptor Accessory Protein 2. Endocr Connect (2021) 10(11):1489–501. doi: 10.1530/EC-21-0459
54. Zhu M, Xu B, Wang M, Liu S, Zhang Y, Zhang C. Pharmacological Modulation of MRAP2 Protein on Melanocortin Receptors in the Sea Lamprey. Endocr Connect (2019) 8(4):378–88. doi: 10.1530/EC-19-0019
55. Zhang M, Chen Y, Xu H, Yang L, Yuan F, Li L, et al. Melanocortin Receptor 4 Signaling Regulates Vertebrate Limb Regeneration. Dev Cell (2018) 46(4):397–409.e395. doi: 10.1016/j.devcel.2018.07.021
56. Graham M, Shutter JR, Sarmiento U, Sarosi I, Stark KL. Overexpression of Agrt Leads to Obesity in Transgenic Mice. Nat Genet (1997) 17(3):273–4. doi: 10.1038/ng1197-273
57. Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted Disruption of the Melanocortin-4 Receptor Results in Obesity in Mice. Cell (1997) 88(1):131–41. doi: 10.1016/s0092-8674(00)81865-6
58. Butler AA, Kesterson RA, Khong K, Cullen MJ, Pelleymounter MA, Dekoning J, et al. A Unique Metabolic Syndrome Causes Obesity in the Melanocortin-3 Receptor-Deficient Mouse. Endocrinology (2000) 141(9):3518–21. doi: 10.1210/endo.141.9.7791
59. Chen AS, Metzger JM, Trumbauer ME, Guan XM, Yu H, Frazier EG, et al. Role of the Melanocortin-4 Receptor in Metabolic Rate and Food Intake in Mice. Transgenic Res (2000) 9(2):145–54. doi: 10.1023/a:1008983615045
60. Challis BG, Coll AP, Yeo GS, Pinnock SB, Dickson SL, Thresher RR, et al. Mice Lacking Pro-Opiomelanocortin Are Sensitive to High-Fat Feeding But Respond Normally to the Acute Anorectic Effects of Peptide-YY(3-36). Proc Natl Acad Sci USA (2004) 101(13):4695–700. doi: 10.1073/pnas.0306931101
61. Chen AS, Marsh DJ, Trumbauer ME, Frazier EG, Guan XM, Yu H, et al. Inactivation of the Mouse Melanocortin-3 Receptor Results in Increased Fat Mass and Reduced Lean Body Mass. Nat Genet (2000) 26(1):97–102. doi: 10.1038/79254
62. Butler AA, Marks DL, Fan W, Kuhn CM, Bartolome M, Cone RD. Melanocortin-4 Receptor is Required for Acute Homeostatic Responses to Increased Dietary Fat. Nat Neurosci (2001) 4(6):605–11. doi: 10.1038/88423
63. Nowoshilow S, Schloissnig S, Fei JF, Dahl A, Pang AWC, Pippel M, et al. The Axolotl Genome and the Evolution of Key Tissue Formation Regulators. Nature (2018) 554(7690):50–5. doi: 10.1038/nature25458
64. Smith JJ, Timoshevskaya N, Timoshevskiy VA, Keinath MC, Hardy D, Voss SR. A Chromosome-Scale Assembly of the Axolotl Genome. Genome Res (2019) 29(2):317–24. doi: 10.1101/gr.241901.118
65. Schloissnig S, Kawaguchi A, Nowoshilow S, Falcon F, Otsuki L, Tardivo P, et al. The Giant Axolotl Genome Uncovers the Evolution, Scaling, and Transcriptional Control of Complex Gene Loci. Proc Natl Acad Sci USA (2021) 118(15):e2017176118. doi: 10.1073/pnas.2017176118
66. Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol (2013) 30(4):772–80.
67. Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol Biol Evol (2020) 37(1):291–4. doi: 10.1093/molbev/msz189
68. Stamatakis A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics (2014) 30(9):1312–3. doi: 10.1093/bioinformatics/btu033
69. Bryant DM, Johnson K, DiTommaso T, Tickle T, Couger MB, Payzin-Dogru D, et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep (2017) 18(3):762–76. doi: 10.1016/j.celrep.2016.12.063
70. Wang M, Chen Y, Zhu M, Xu B, Guo W, Lyu Y, et al. Pharmacological Modulation of Melanocortin-4 Receptor by Melanocortin Receptor Accessory Protein 2 in Nile Tilapia. Gen Comp Endocrinol (2019) 282:113219. doi: 10.1016/j.ygcen.2019.113219
71. Habara M, Mori N, Okada Y, Kawasumi K, Nakao N, Tanaka Y, et al. Molecular Characterization of Feline Melanocortin 4 Receptor and Melanocortin 2 Receptor Accessory Protein 2. Gen Comp Endocrinol (2018) 261:31–9. doi: 10.1016/j.ygcen.2018.01.020
72. Renquist BJ, Zhang C, Williams SY, Cone RD. Development of an Assay for High-Throughput Energy Expenditure Monitoring in the Zebrafish. Zebrafish (2013) 10(3):343–52. doi: 10.1089/zeb.2012.0841
73. Tao YX. Constitutive Activity in Melanocortin-4 Receptor: Biased Signaling of Inverse Agonists. Adv Pharmacol (2014) 70:135–54. doi: 10.1016/B978-0-12-417197-8.00005-5
74. Zhao L, Zhou X, Rokas A, Cone RD. Functional Variants of the Melanocortin-4 Receptor Associated With the Odontoceti and Mysticeti Suborders of Cetaceans. Sci Rep (2017) 7(1):5684. doi: 10.1038/s41598-017-05962-1
75. Zhang J, Li J, Wu C, Hu Z, An L, Wan Y, et al. The Asp298Asn Polymorphism of Melanocortin-4 Receptor (MC4R) in Pigs: Evidence for its Potential Effects on MC4R Constitutive Activity and Cell Surface Expression. Anim Genet (2020) 51(5):694–706. doi: 10.1111/age.12986
76. Li JT, Yang Z, Chen HP, Zhu CH, Deng SP, Li GL, et al. Molecular Cloning, Tissue Distribution, and Pharmacological Characterization of Melanocortin-4 Receptor in Spotted Scat, Scatophagus Argus. Gen Comp Endocrinol (2016) 230–231:143–152. doi: 10.1016/j.ygcen.2016.04.010
77. Li L, Yang Z, Zhang YP, He S, Liang XF, Tao YX. Molecular Cloning, Tissue Distribution, and Pharmacological Characterization of Melanocortin-4 Receptor in Grass Carp (Ctenopharyngodon Idella). Domest Anim Endocrinol (2017) 59:140–51. doi: 10.1016/j.domaniend.2016.11.004
78. Yi TL, Yang LK, Ruan GL, Yang DQ, Tao YX. Melanocortin-4 Receptor in Swamp Eel (Monopterus Albus): Cloning, Tissue Distribution, and Pharmacology. Gene (2018) 678:79–89. doi: 10.1016/j.gene.2018.07.056
79. Zhang KQ, Hou ZS, Wen HS, Li Y, Qi X, Li WJ, et al. Melanocortin-4 Receptor in Spotted Sea Bass, Lateolabrax Maculatus: Cloning, Tissue Distribution, Physiology, and Pharmacology. Front Endocrinol (Lausanne) (2019) 10:705. doi: 10.3389/fendo.2019.00705
80. Srinivasan S, Lubrano-Berthelier C, Govaerts C, Picard F, Santiago P, Conklin BR, et al. Constitutive Activity of the Melanocortin-4 Receptor is Maintained by its N-Terminal Domain and Plays a Role in Energy Homeostasis in Humans. J Clin Invest (2004) 114(8):1158–64. doi: 10.1172/JCI21927
81. Hahn TM, Breininger JF, Baskin DG, Schwartz MW. Coexpression of Agrp and NPY in Fasting-Activated Hypothalamic Neurons. Nat Neurosci (1998) 1(4):271–2. doi: 10.1038/1082
82. Mizuno TM, Kleopoulos SP, Bergen HT, Roberts JL, Priest CA, Mobbs CV. Hypothalamic Pro-Opiomelanocortin mRNA is Reduced by Fasting and in Ob/Ob and Db/Db Mice, But is Stimulated by Leptin. Diabetes (1998) 47(2):294–7. doi: 10.2337/diab.47.2.294
83. Song Y, Golling G, Thacker TL, Cone RD. Agouti-Related Protein (AGRP) is Conserved and Regulated by Metabolic State in the Zebrafish, Danio Rerio. Endocrine (2003) 22(3):257–65. doi: 10.1385/ENDO:22:3:257
Keywords: Ambystoma mexicanum, MRAP2, MC3R, MC4R, metabolism
Citation: Wang X, Xue S, Lei X, Song W, Li L, Li X, Fu Y, Zhang C, Zhang H, Luo Y, Wang M, Lin G, Zhang C and Guo J (2022) Pharmacological Evaluation of Melanocortin 2 Receptor Accessory Protein 2 on Axolotl Neural Melanocortin Signaling. Front. Endocrinol. 13:820896. doi: 10.3389/fendo.2022.820896
Received: 23 November 2021; Accepted: 05 January 2022;
Published: 17 February 2022.
Edited by:
Ya-Xiong Tao, Auburn University, United StatesReviewed by:
Renlei Ji, Auburn University, United StatesCopyright © 2022 Wang, Xue, Lei, Song, Li, Li, Fu, Zhang, Zhang, Luo, Wang, Lin, Zhang and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meng Wang, d20xOTkyMTI3NjUyOUAxNjMuY29t; Gufa Lin, bGluZ3VmYUB0b25namkuZWR1LmNu; Chao Zhang, emhhbmdjaGFvQHRvbmdqaS5lZHUuY24=; Jing Guo, Z2hhcHB5akAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.