- 1Center for Reproductive Medicine, Department of Obstetrics and Gynecology, Peking University Third Hospital, Beijing, China
- 2National Clinical Research Center for Obstetrics and Gynecology (Peking University Third Hospital), Beijing, China
- 3Key Laboratory of Assisted Reproduction (Peking University), Ministry of Education, Beijing, China
- 4 Beijing Key Laboratory of Reproductive Endocrinology and Assisted Reproductive Technology, Beijing, China
- 5Research Units of Comprehensive Diagnosis and Treatment of Oocyte Maturation Arrest, Chinese Academy of Medical Sciences, Beijing, China
The mean age of women seeking infertility treatment has gradually increased over recent years. This has coincided with the emergence of in vitro maturation (IVM), a method used in assisted reproductive technology for patients with special requirements. However, when compared with conventional in vitro fertilization, IVM is associated with poor embryonic development potential and low live birth rates, thus limiting the widespread application of this technique. In this study, we performed RNA-sequencing transcriptomic assays and identified a total of 2,627 significant differentially expressed genes (DEGs) between IVM oocytes and in vivo matured oocytes from mice of advanced reproductive age. Next, Kyoto Encyclopedia of Genes and Genomes pathway analysis was used to identify the potential functions of the DEGs. The most significantly enriched pathway was oxidative phosphorylation (OXPHOS). In addition, we constructed a protein-protein interaction network to identify key genes and determined that most of the hub genes were mtDNA-encoded subunits of respiratory chain complex I. Antioxidant supplementation lead to an increase in ATP production and reduced the gene expression profile of the OXPHOS pathway in the IVM group. Moreover, alternative splicing (AS) events were identified during in vivo or in vitro oocyte maturation; data showed that skipped exons were the most frequent type of AS event. A number of genes associated with the OXPHOS pathway exhibited alterations in AS events, including Ndufa7, Ndufs7, Cox6a2, Ndufs5, Ndufb1, and Uqcrh. Furthermore, the process of IVO promoted the skipping of exon 2 in Ndufa7 and exon 3 in Ndufs7 compared with the IVM oocytes, as determined by semi−quantitative RT−PCR. Collectively, these findings provide potential new therapeutic targets for improving IVM of aged women who undergo infertility treatment.
Introduction
Global fertility has declined significantly over recent years and this has led to infertility becoming the third most prevalent disease after tumors and cardiovascular disease (1). The decline in fertility caused by advanced maternal age (≥ 35 years) has led to a significant increase in the incidence of infertility. Assisted reproductive technology (ART) offers a significant hope of pregnancy to women who are suffering from infertility. Notably, the mean age of women undergoing ART treatment has gradually increased (2, 3). In vitro maturation (IVM) of oocytes has emerged as an important ART procedure involving the in vitro culture and offers an alternative option for women with polycystic ovary syndrome (4). IVM can also prevent ovarian hyperstimulation syndrome during ovarian stimulation (5), can be of benefit to poor responders (6), and can help preserve fertility in patients being treated for cancer (7). However, previous studies have reported that maternal aging is associated with reduced IVM success rates and adverse events (8, 9). IVM is an alternative option for ovarian tissue cryopreservation (OTC); however, the success rates for IVM can be very low when used to mature oocytes retrieved from females of advanced age during OTC (10). Therefore, there is an urgent need to identify the molecular mechanisms underlying IVM so that this technique can be become a driving force for the implementation of ART technology.
The process of mitochondrial oxidative phosphorylation (OXPHOS) is a major metabolic pathway associated with ATP generation and involves five major membrane complexes (11). The OXPHOS system is the major source of reactive oxygen species (ROS) in mitochondria (12). Previous studies have revealed that the production of ROS via mitochondrial metabolism was higher in IVM oocytes than in vivo matured (IVO) oocytes (13, 14). This difference may be attributed to a number of environmental factors, including light exposure and the presence of antioxidants in the follicular fluid (15–17). In a previous study, Sanfins et al. reported that the fertilization rate was lower, and embryo development was delayed, in IVM oocytes than in IVO oocytes; these authors also reported a difference between these two types of oocytes with regards to cytoskeleton organization and nuclear lamina integrity (18, 19). Previous research identified several differentially expressed genes (DEGs) between IVM and IVO oocytes and demonstrated that the GATA-1/CREB1/WNT pathway is associated with the quality of human oocytes (20) and detected the cellular metabolism and genetic alterations in mouse oocytes (21). Furthermore, in our previous study, we investigated dynamic changes of gene expression between human IVM and IVO oocytes using single-cell transcriptome sequencing analysis. We found, for the first time, that metabolic function was impaired in human oocytes maintained in the in vitro environment (22). Our present study demonstrates, for the first time, the existence of altered alternative splicing (AS) events in the mitochondrial OXPHOS pathway between IVM and IVO oocytes from mice of advanced reproductive age.
Precursor mRNA refers to the process in which exons and introns are either excluded or included in gene expression, thus generating multiple different mature mRNAs and resulting in increased protein diversity (23, 24). A previous study revealed that approximately 95% of multi-exonic genes undergo AS regulation in humans, as determined by high-throughput sequencing technology (25). This regulatory system can be commonly categorized into five canonical patterns including skipped exon (SE), mutually exclusive exon (MXE), retained intron (RI), alternative 3´ and 5´ splice site (A3SS and A5SS) that are associated with the regulation of cell lineage differentiation, subcellular localization, tumor progression, and germ cell development (26–29). Furthermore, numerous reports have indicated that the aberrant pre-mRNA splicing of functional genes is involved in abnormal spermatogenesis and male infertility (30, 31). In mammalian oocytes, conserved stage-specific transcript variants indicate that alternative pre-mRNA splicing plays a crucial role in the accurate control of the maternal transcriptome (32, 33). For example, ESRP1 is involved in the AS programming of oocyte mRNA processing and the ESRP1 deletion-induced pre-mRNA splicing changes in spindle organization-related genes that lead to female infertility (34). Depletion of the nuclear m6A reader YTHDC1 causes extensive alternative polyadenylation and the 3′-UTR length is altered in oocytes, thus resulting in female sterility (35). Previous work in our group demonstrated that AS events are important regulatory mechanisms associated with the failure of maturation in human oocytes (36). However, our understanding of the differential expression of pre-mRNA splicing during the maturation of mammalian oocytes remains largely obscure.
In the present study, we used mice of advanced reproductive age as oocyte donors and identified differences in gene regulation during oocyte maturation when comparing IVO and IVM oocytes. Next, we investigated the molecular mechanisms underlying these changes at the level of the transcriptome. We performed KEGG pathway enrichment analysis and found that mitochondrial OXPHOS was significantly enriched. We also constructed an interaction network for the DEGs to investigate interacting partners and identified differential AS events among the DEGs that implied the potential physiological regulatory mechanisms may account for the inadequacies of oocytes derived by IVM. This research provides new insights into the outcomes of ART for women of advanced age with deficits in oocyte quality. Furthermore, we identified potential therapeutic biomarkers for clinical diagnosis that can be applied during the development of IVM oocytes.
Materials and Methods
Mice
All animal procedures were approved by the Institutional Animal Care and Use Committee of Peking University. Female Institute of Cancer Research (ICR) mice at the age of 42-45 weeks (reproductively old mice) were purchased from Charles River Laboratories (Beijing, China) and were maintained in accordance with the National Institutes of Health Guidelines for the Use of Laboratory Animals. ICR mice were housed in ideal conditions at 20-23°C under a 12-h light/dark cycle with free access to water and food.
Oocyte Collection and Culture
Germinal vesicle (GV)-intact oocytes were obtained as described previously (37). In brief, female ICR mice were intraperitoneally injected with 5 IU of pregnant mare’s serum gonadotropin (PMSG). Forty-six hours later, the animals were euthanized by cervical dislocation and the ovaries were harvested and transferred to a cell-culture dish containing pre-warmed M2 medium (Sigma, M7167). Cumulus-oocyte complexes were obtained from antral ovarian follicles by manual rupture. Then, denuded GV oocytes were obtained by removing cumulus cells with a narrow-bore glass pipette and gentle washing. For IVM, oocytes were cultured in M16 medium (Sigma, M7292) under paraffin oil for 12 h at 37°C in a 5% CO2 incubator. For melatonin treatment, oocytes were cultured in M16 medium at a final concentration of 1 μM melatonin (Sigma, M5250) in accordance with previous studies (38, 39). For IVO, mice injected with PMSG were further injected with 5 IU of human chorionic gonadotropin (hCG) 46 h later. Ovulated MII oocytes were collected from the oviduct ampullae of mice 14 h after hCG injection and cumulus cells were dispersed by brief exposure to M2 medium containing 1 mg/ml of hyaluronidase (Sigma, H3506).
Library Preparation and Illumina NovaSeq 6000 Sequencing
Total RNA was extracted from 20 MII oocytes after IVM and IVO from three mice of advanced reproductive age reproductively (these were regarded as one group). Three biological replicates obtained via both IVM and IVO were used to construct a eukaryotic micro-library. Specifically, we used a SMART-Seq v4 Ultra Low Input RNA Kit (Takara Bio) to obtain full-length cDNA in accordance with the manufacturer’s instructions. cDNA synthesis and amplification were validated by an Agilent 2100 Bioanalyzer (Agilent Technologies) and a High Sensitivity DNA Kit (Agilent Technologies) in accordance with the manufacturer’s instructions. Then, the Nextera XT DNA Library Preparation Kit (Illumina) was used to construct a library that was suitable for sequencing. Finally, a paired-end RNA-seq library was performed on the Illumina NovaSeq 6000 system (2 × 150bp read length).
DEG Identification and Functional Enrichment Analysis
SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) software packages were used to filter the raw paired-end reads with default parameters. The clean reads were then aligned to the mouse reference genome (GRCm39) using the Hisat2 mapping tool (40). The fragments per kilobase per million reads (FPKM) method was then used to calculate gene expression levels; gene abundance was determined by utilizing the RSEM software workflow (41). The analysis of DEGs between the IVM and IVO groups was performed with the R statistical package DESeq2 (42) using |log2FC| ≥ 1 and adjusted P-value < 0.05 as thresholds for significance. KEGG pathway enrichment analyses of the identified DEGs were carried out using KOBAS software (43) which incorporates information from the KEGG database (https://www.genome.jp/kegg/).
The Identification of Alternative Splice Events
All alternative splice (AS) events in the IVM and IVO groups were detected by the replicate multivariate analysis of transcript splicing (rMATS) statistical method (44). Only isoforms that were similar to the reference or consisted of novel splice junctions were considered, and splicing differences were detected in five major forms (exon inclusion, exclusion, alternative 5′, 3′, and intron retention events).
RNA Extraction, qRT-PCR, and RT-PCR Analysis
Total RNA was extracted from 100 matured oocytes from twelve mice of advanced reproductive age using an RNeasy Micro Kit (Qiagen, 74034) in each group. cDNA was synthesized using the PrimeScript RT Master Mix Kit (Takara, RR036A) and qRT-PCR was performed using the PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, A25742) and the QuantStudio 3 Real-Time PCR System (Applied Biosystems, USA). Relative mRNA levels were normalized to Gapdh as a control standard and calculated using the 2-△△CT method. Semi-quantitative RT-PCR was performed using Premix Taq (Takara, RR901A) and specific primers were designed to flank the constitutively expressed exons of differentially spliced genes. PSI values were calculated using the ratio of the band intensity of the exon inclusion divided by the sum of the exon-included and exon-excluded bands, as described previously (45, 46). Primer sequences are given in Table S1.
Detection of Intracellular ROS Levels
For intracellular ROS detection, oocytes were incubated in M2 medium supplemented with 5 μM CM-H2DCFDA (Invitrogen, C6827) at 37°C for 30 min in a 5% CO2 atmosphere and a dark environment. Then, the oocytes were washed three times with pre-warmed M2 medium. Then, oocytes from the IVM and IVO groups were placed on glass-bottomed culture dishes and examined with a confocal laser scanning microscope (Carl Zeiss 710, Germany).
Determination of Adenosine 5′-Triphosphate Content
Adenosine 5′-triphosphate (ATP) content in a pool of 20 oocytes was measured with a bioluminescent somatic cell assay kit (Sigma, USA) as described previously (47, 48). A five-point standard curve (0, 0.1, 0.5, 1.0, 10, and 50 pmol of ATP) was generated in each assay, and ATP levels were calculated by using a formula derived from the linear regression of the standard curve.
Statistical Analysis
Data are expressed as mean ± standard error of the mean (SEM) from three independent experiments and statistical analyses were performed with GraphPad Prism 5 software (San Diego, CA, USA) and the Student’s t-test between the IVM and IVO groups. P < 0.05 was considered as statistically significant (*P < 0.05, **P < 0.01, and ***P < 0.001).
Results
Sequencing Data Summary
As shown in Figure 1A, we used ICR mice at the age of 42-45 weeks (a reproductive advanced age) that were near to the end of their reproductive lifespan and exhibited a decline in oocyte quality, as described in previous studies (49). These mice are commonly regarded as a natural aging model and were used as donors for IVM and IVO oocytes so that we could carry out RNA sequencing. First, we used cDNA to generate libraries for IVM and IVO oocytes, which were then sequenced by the Illumina NovaSeq 6000 platform (three biological replicates per group). Gene expression distribution statistics showed that the six groups of IVM and IVO oocytes from mice of advanced reproductive age presented with similar gene expression distributions (Figure 1B). A heatmap for the Pearson’s correlation coefficient indicated that the experiment was reliable, as determined by the fragments per kilobase million (FPKM) value calculated for each sample (Figure 1C). A total of 10,777 expressed genes were identified across all six samples, as shown on the UpSet plot (Figure 1D). When comparing the IVM and IVO groups, we identified 2,627 DEGs based on specific criteria (|log2FC| ≥ 1 and an adjusted P-value < 0.05). Of these, 2,254 genes were down-regulated, and 373 genes were up-regulated in IVO oocytes from mice of advanced reproductive age when compared with IVM oocytes (Figure 1E). A list of the DEGs is given in Supplementary Excel S1. Furthermore, principal components analysis (PCA) revealed that the DEG clusters of oocytes obtained from mice of advanced reproductive age in the IVO and IVM groups were separated (Figure 1F), thus implying that the different culture procedures had a substantial effect on transcriptomic profiles. In addition, we generated a heatmap of DEGs for the IVO and IVM groups using cluster analysis and the FPKM values (Figure 1G).
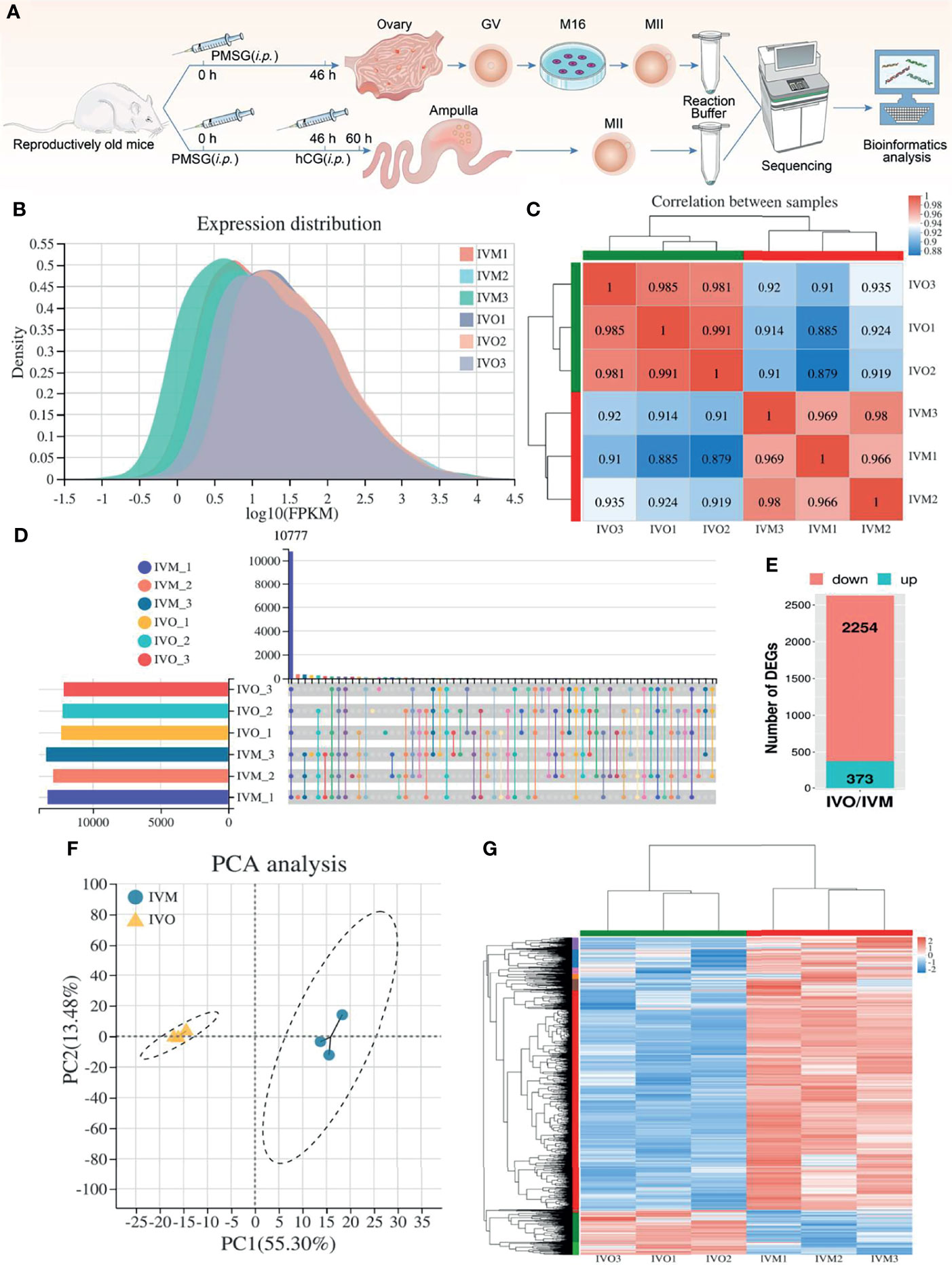
Figure 1 DEG analysis between IVO and IVM groups. (A) A schematic showing experimental design and time points. (B) Gene expression density distribution in the IVO and IVM groups. (C) Correlation analysis between the IVM and IVO groups. The degree of correlation is represented by different colors. (D) An UpSet plot summarizing the genes detected in the six groups. (E) A stacked column showing the numbers of DEGs in the IVO group compared with the IVM group. (F) PCA of gene expression profiles in oocytes between the IVM group (blue dots) and the IVO group (yellow triangles). (G) Heatmap showing DEGs; the red rows represent high expression levels and blue rows represent low expression levels. Each column represents an independent sample.
Functional Analysis of the DEGs
We mapped the DEGs from mice oocytes of an advanced reproductive age in the IVO and IVM groups to the KEGG pathways database terms in order to interpret the biological consequences of differential expression. KEGG enrichment analysis revealed significant enrichment in terms of OXPHOS pathways (map00190; 59 DEGs; P-adjusted = 2.03 × 10-6), non-alcoholic fatty liver disease (NAFLD) (map04932; 49 DEGs; P-adjusted = 3.11 × 10-5), Huntington’s disease (HD) (map05016; 76 DEGs; P-adjusted = 2.34 × 10-4), and Parkinson disease (PD) (map05012; 54 DEGs; P-adjusted = 6.48 × 10-4) (Figure 2A and Supplementary Excel S2). A KEGG enrichment chord plot, featuring the top four terms (the OXPHOS, NAFLD, HD, and PD pathways) suggested that several target genes did not uniquely exist in one single enriched pathway; rather, they were annotated in multiple terms (Supplementary Figure S1). A network diagram was constructed to identify significant interaction in the enriched KEGG pathways, which revealed that the OXPHOS pathway was critical (Figure 2B). Venn diagram analysis further showed that some of the DEGs were shared among different group combinations. For example, two up-regulated and 41 down-regulated overlapped DEGs were represented in the four KEGG pathways of mitochondrial OXPHOS and diseases related to mitochondria dysfunction (NAFLD, HD, and PD) (Figure 2C). In addition, the most significant enrichment pathway diagram (OXPHOS) featured many down-regulated genes in the IVO groups when compared with the IVM group (Figure 2D), thus implying that oocytes matured in vitro suffer from changes associated with oxidative stress and metabolic activity.
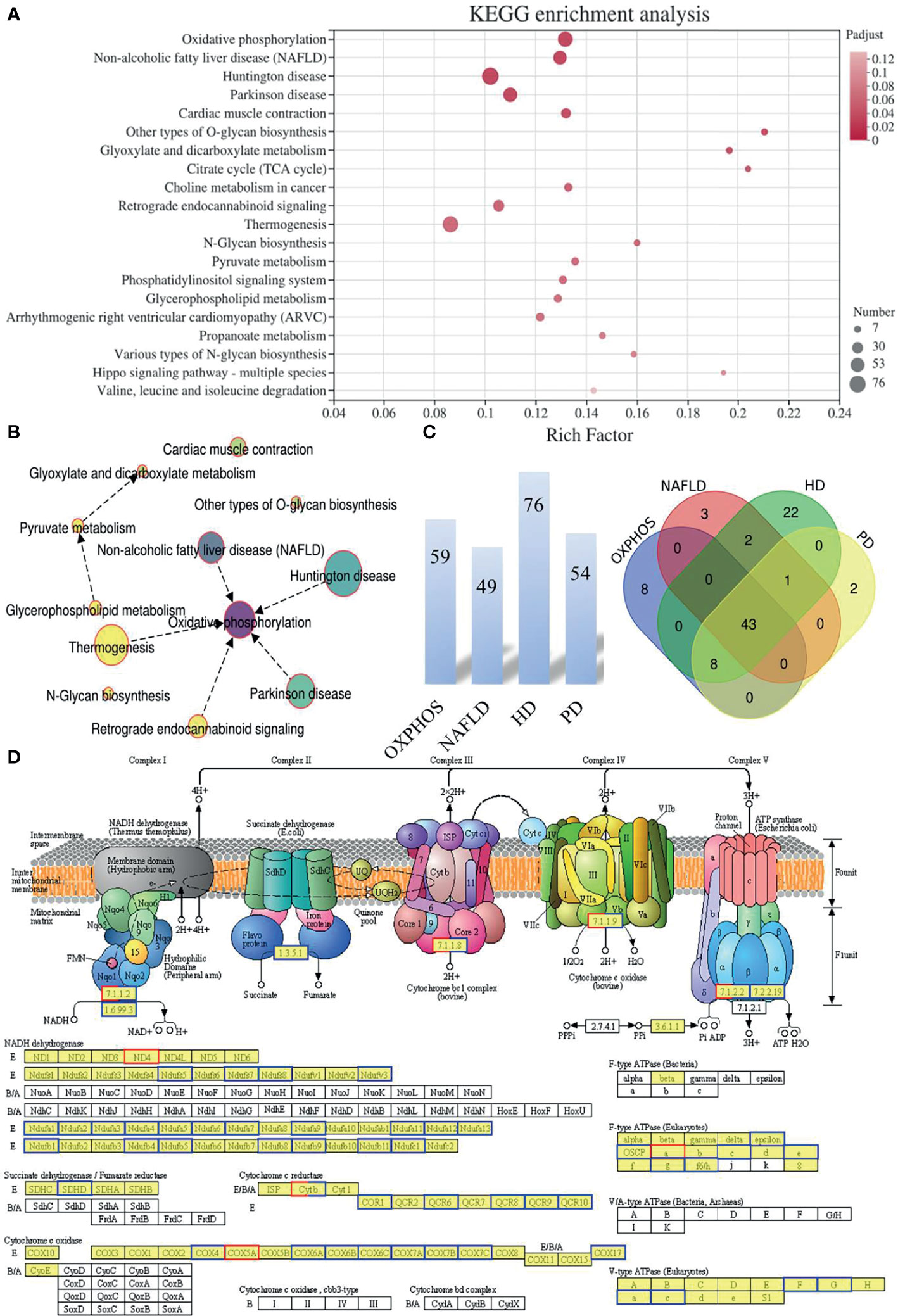
Figure 2 The functional pattern of gene expression between IVO and IVM groups. (A) The top 20 enriched KEGG pathways for the DEGs identified between the IVO and IVM groups. The X-axis represents the Rich Factor while the Y-axis represents the names of the pathways. The higher the Rich factor, the higher the degree of enrichment. (B) Network chart showing the significantly enriched KEGG pathways. (C) The number above each bar indicates the enriched DEGs of the top four terms (the OXPHOS, NAFLD, HD, and PD pathways) and the Venn diagram shows the overlapping DEGs between the top four enriched KEGG pathways. (D) KEGG pathway diagram for oxidative phosphorylation. Boxes with red borders represent up-regulated genes while boxes with blue borders represent down-regulated genes when the IVO group was compared with the IVM group.
qRT-PCR Validation of DEGs That Were Enriched in KEGG Pathways
A heatmap of overlapping DEGs from the top four enriched KEGG pathways showed that 43 protein-encoding genes, including NADH-ubiquinone oxidoreductase (complex I) subunits, cytochrome c oxidase subunits, and ubiquinol-cytochrome c reductase subunits, were significantly altered (Figure 3A). Volcano plots were also used to visualize overlapping DEGs, including two up-regulated and 41 down-regulated genes (Figure 3B) that met our selection criteria [P-adjusted < 0.05 and |log2FC| ≥ 1, as determined by statistical DESeq2 (42)]. To identify relationships between these DEGs, we constructed a protein-protein interaction (PPI) network using the STRING online database (50); this consisted of 48 nodes and 100 edges and featured 2254 down-regulated genes when the IVO groups were compared with the IVM groups (Figure 3C). Next, to validate the RNA sequencing data, the relative mRNA levels of DEGs were verified by qRT-PCR and were then classified by function into the NADH-ubiquinone oxidoreductase (complex I) subunits, ribosomal protein family, ubiquinol-cytochrome c reductase subunits, and cytochrome c oxidase subunits (Figures 3D, E). Results indicated that data were consistent with those derived from the transcriptomic analysis.
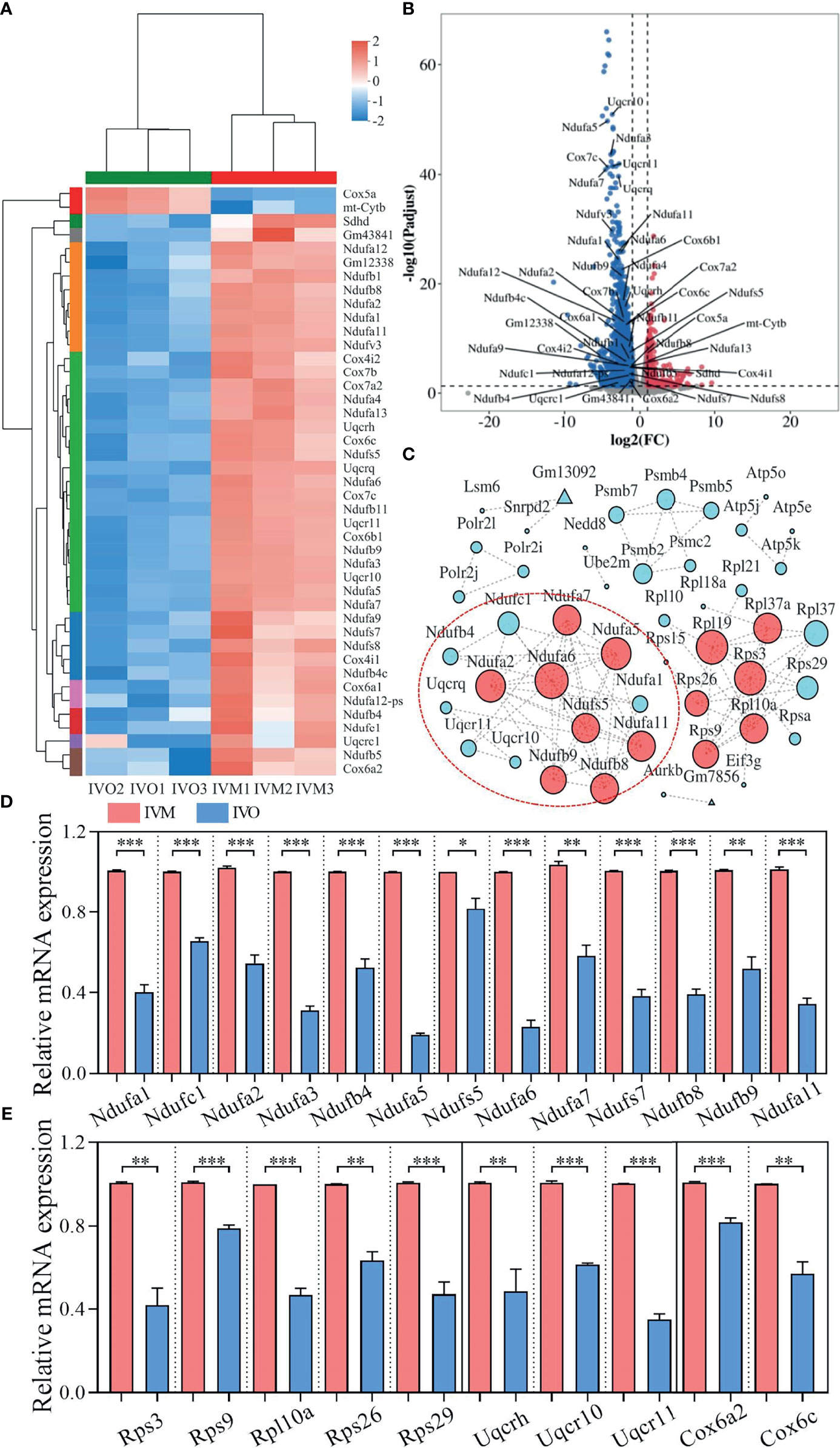
Figure 3 DEG analysis and network patterns for the top four enriched KEGG pathways. (A) Heatmap diagram showing the top four enriched KEGG pathways with overlapping DEGs (P-adjusted < 0.05 and |log2FC| ≥ 1). (B) Volcano plots showing overlapping DEGs in the top four KEGG enrichment analyses. Blue dots indicate down-regulated genes while red dots indicate up-regulated genes. (C) A PPI network for the down-regulated genes was constructed using the STRING database to compare the IVO and IVM groups. The node size represents the number of linked lines and connectivity. (D, E) The relative mRNA levels of DEGs including NADH-ubiquinone oxidoreductase (complex I) subunits, the ribosomal protein family, ubiquinol-cytochrome c reductase subunits, and cytochrome c oxidase subunits between the IVM and IVO groups, as determined by RNA-seq and confirmed by qRT-PCR assay. Data represent the mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementation With Antioxidant Melatonin Changed mRNA Levels of the DEGs
Next, we classified DEGs between the IVO and IVM groups in the OXPHOS system into five respiratory chain complexes within the mitochondria. Analysis showed that respiratory chain complex I had the most DEGs, as shown by a gene list and heatmaps (Figures 4A, B). We also measured the ROS levels in IVM and IVO oocytes from mice of advanced reproductive age; the fluorescence intensity of ROS was significantly higher in IVM oocytes compared with IVO oocytes (11.20 ± 1.36; n = 38 vs. 4.82 ± 0.60; n = 35, P < 0.05; Figure 4C), thus showing that changes in the expression of OXPHOS pathway genes have an effect on ROS level. The synthesis of ATP through oxidative metabolism was associated with oocyte maturation and the reduced ATP production and increased ROS accumulation are linked to poor oocyte quality according to a previous study (51). These results revealed that the ATP content was lower in IVM oocytes than in IVO oocytes, whereas with melatonin supplementation (1 μM), the ATP generation was restored in the melatonin treated IVM group, as shown in Figure 4D. This suggested that IVM-induced mitochondrial dysfunction in reproductively old mice oocytes could be rescued by melatonin treatment. In addition, to ascertain if antioxidant supplementation could change the gene expression profile observed in the IVM group, oocytes were cultured in melatonin supplemented M16 medium; the mRNA expression of genes regulating oxidative phosphorylation were shown to be reduced in oocytes when compared to those without melatonin (Figure 4E).
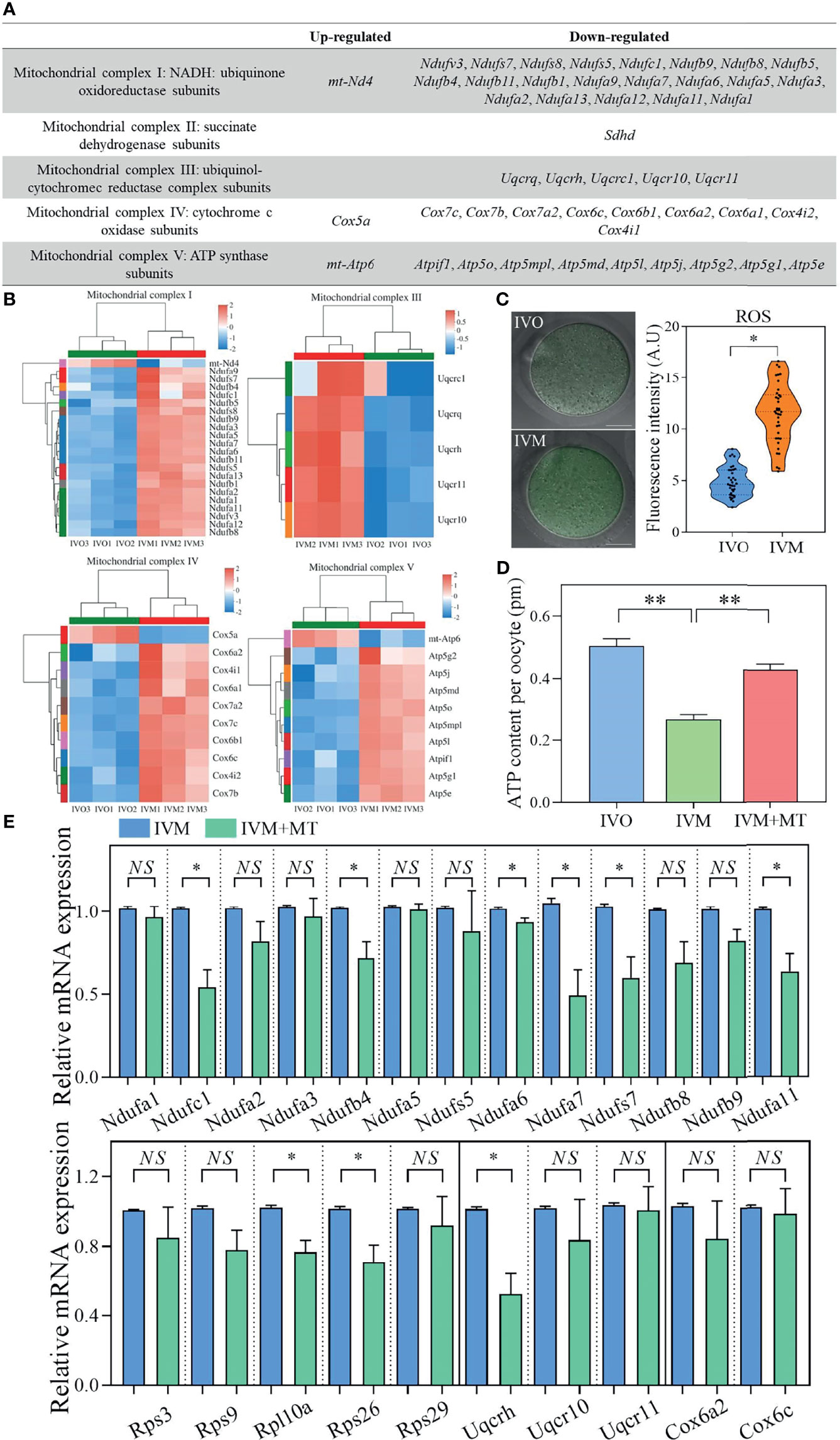
Figure 4 (A) List of up- and down-regulated genes in the OXPHOS system of five respiratory chain complexes in the mitochondria when compared between IVO and IVM groups. (B) Heatmaps corresponding to the identified DEGs. The red rows represent high expression levels and the blue rows represent low expression levels. Each column represents an independent sample. (C) Representative images of ROS signals in IVO and IVM groups. Scale bars = 20 μm. The fluorescence intensity of ROS in IVO and IVM groups. (D) Graphs showing the ATP levels in IVO, IVM, and melatonin-treated IVM oocytes (n = 20 for each group). (E) The relative mRNA levels of target genes including NADH-ubiquinone oxidoreductase (complex I) subunits, the ribosomal protein family, ubiquinol-cytochrome c reductase subunits, and cytochrome c oxidase subunits between the IVM and melatonin-treated IVM oocytes. Data represent the mean ± SEM of three independent experiments. NS, not significant, *P < 0.05, and **P < 0.01.
AS Events Associated With DEGs Detected by RNA-Seq Analysis
The mRNA precursors produced by most eukaryotic gene transcription only correspond to a translated protein. However, AS is a process that improves biodiversity by producing mRNA precursors for genes that produce different mRNA splicing isoforms by five different splicing mechanisms under certain conditions: SE, A5SS, A3SS, MXE, and RI (52, 53). We determined the distribution of AS events in the IVM and IVO groups and generated a stacked bar chart that identified SE as the most frequent type of AS event during both in vivo and in vitro oocyte maturation (Figure 5A). In addition, we generated pie charts that illustrated the proportion of each AS event according to the total number of events; SE events represented 81.07% and 79.61% of all AS events in the IVM and IVO groups, respectively (Figure 5B). Furthermore, we determined the differential AS events that occurred in genes that were up-regulated in the IVO group. Specifically, there were 35 (81.4%) SE events, five (11.6%) MXE events, one (2.3%) A3SS event, one (2.3%) A5SS event, and one (2.3%) RI event. Then, we determined the differential AS events for the down-regulated genes in the IVO groups. Specifically, there were 163 (75.5%) SE events, 20 (9.3%) MXE events, 16 (7.4%) A3SS events, ten (4.6%) A5SS events, and seven (3.2%) RI events (Figure 5B). The different types of AS events corresponding to DEGs including Ndufa7, Ndufs7, Cox6a2, Ndufs5, Ndufb1, and Uqcrh were listed in each enriched KEGG term (Figure 5C). These AS events included the SE, A3SS, and A5SS types, thus indicating splicing diversity during oocyte maturation. Next, Sashimi plots were used to visualize the differential splicing patterns with RNA-seq reads coverage across each part of the splice junction between two exons for Ndufa7 and Ndufs7 (Figures 5D, E). Experimental semi-quantitative RT-PCR validation was performed with specific primers and exon 2 of Ndufa7 and exon 3 of Ndufs7 was skipped in the IVO oocytes from mice of advanced reproductive age (Figures 5F, G).
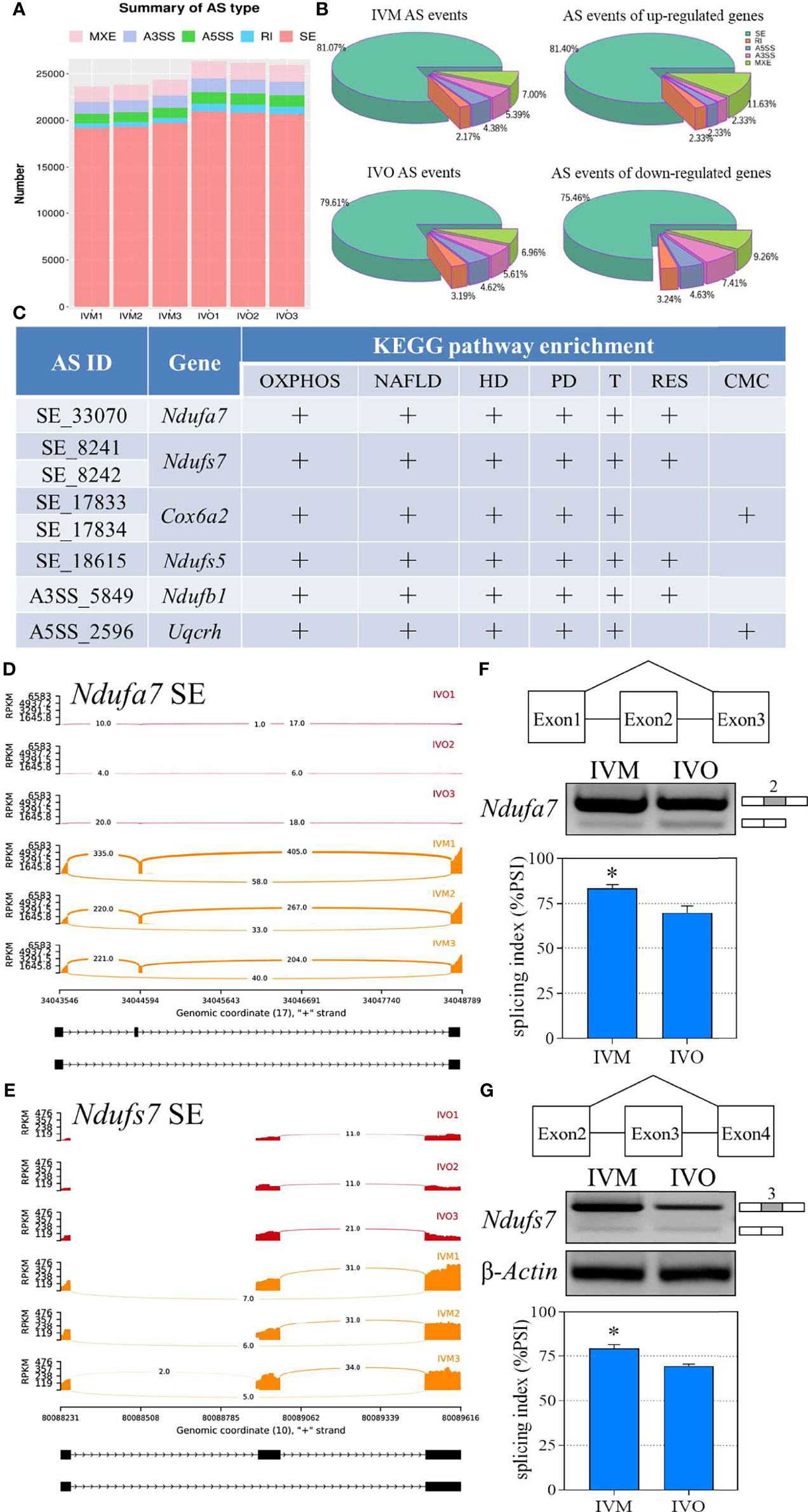
Figure 5 Statistical analysis of the AS events for KEGG enrichment terms. (A) A comparison of the number of AS events between the IVO and IVM groups. (B) The proportion of AS events of the IVM group, IVO group, the up-regulated and down-regulated genes. (C) List of significantly different AS events for the top four KEGG enriched overlapping DEGs (OXPHOS, oxidative phosphorylation; NAFLD, non-alcoholic fatty liver disease; HD, Huntington disease; PD, Parkinson disease; T, Thermogenesis; RES, retrograde endocannabinoid signaling; CMC, cardiac muscle contraction. Sashimi plots depicting the visualize specific splice sites of splicing events for exon skipping of the Ndufa7 (D) and Ndufs7 (E) genes. The red plots represent the IVO group while the orange plots represent the IVM group. The distribution of the number of junction reads is shown for each sample. The schematic in the upper panels represent the different splicing pattern. Representative agarose gels of semi-quantitative RT-PCR for Ndufa7 exon 2 (F) and Ndufs7 exon 3 (G) are shown in the middle panels. β-Actin served as an internal control. The histogram in the lower panel shows the average PSI that the IVO oocytes present exon inclusion suppressed compared with the IVM oocytes. Data represent the mean ± SEM of three independent experiments. *P < 0.05.
Discussion
The mean age of women at the birth of their first child has increased gradually over the past few decades (54, 55), which has led to a decline in both oocyte quantity and quality. The application of IVM as an ART procedure provides patients with an alternative treatment option that does not involve gonadotrophin stimulation, thus preventing the risk of OHSS and reducing medication costs (56). However, several clinical outcomes, including implantation rates, pregnancy rates, and live birth rates, are known to be lower for IVM than in vitro fertilization (57). In the present study, we applied transcriptomic analysis of oocytes derived from mice of advanced reproductive age and identified several DEGs in MII oocytes when comparing between an IVM group and an IVO group. Next, we performed KEGG enrichment analyses to investigate the potential functions of the DEGs. We found that mitochondrial OXPHOS was the most significantly enriched pathway including 59 target genes, followed by NAFLD, HD, and PD. In addition, the OXPHOS is the pathway with the most interactions with other pathways, as visualized by a PPI network.
The OXPHOS pathway is the major source of ATP production in mitochondria and involves five multi-subunit protein complexes that are associated with oocyte maturation (14), fertilization (58), and embryonic development (59). Furthermore, studies have shown that mitochondria are the major source of ROS and, with increasing age, the amount of ROS increases, thus causing cellular damage and a decline in oocyte quality (51). The OXPHOS system contains five respiratory chain complexes within the mitochondria and by analyzing the gene list and a heatmap, we found that respiratory chain complex I had the most DEGs, results of which were similar to a previous study that showed that complexes I and III of the mitochondrial respiratory chain are key contributors to the production of ROS (60, 61). Moreover, several extracellular factors are known to play vital roles in ROS defense during oocyte development in vivo. For example, follicular fluid has the ability to prevent the harmful effects of ROS due to the involvement of redox reactions (62, 63), and the enclosed cumulus cells act as scavengers by protecting oocytes against ROS toxicity (63, 64). In addition, the influence of physical factors, such as exposure to cool white or warm white fluorescent lights, can promote the abnormal accumulation of ROS in the development of mammalian zygotes (65). This explains why the levels of ROS are increased in IVM-derived oocytes due to lack of in vivo milieu (66); this concurs with the findings of the present study. Furthermore, several previous publications have shown that antioxidant supplementation of the IVM medium significantly reduced the levels of intracellular ROS, thus enhancing oocyte maturation rate and increasing the quality of oocytes from mice of advanced reproductive age (67, 68). In our study, we demonstrate that antioxidant melatonin supplementation may reduce the gene expression profile of the OXPHOS pathway and resolve the problem of insufficient ATP production in the IVM group.
We next constructed a PPI network for down-regulated genes and identified several genes that exhibited strong associations with ribosomal protein family genes, including Rpl10a, Rpl19, Rps3, Rpl37a, Rps9, and Rps26, that presented strong associations with hub nodes. A previous study showed that Rps26 acts as a key regulator during follicular development and oocyte growth by controlling histone methylation and mRNA synthesis activity; these are key mechanisms for female fertility (69). Recently, Peng et al. reported that RpS3 deficiency leads to a lower blastocyst formation rate and causes early embryonic arrest (70). Collectively, the identification of these related genes and enriched pathways has highlighted the potential mechanisms underlying the poor clinical outcomes and embryonic development associated with the IVM procedure and identify new therapeutic molecular targets.
AS is a complex process that enables genome complexity and biological diversity among eukaryotes; these events occur in approximately 95% of multi-exon genes in humans (25). We investigated the functionality of AS events in IVM and IVO MII oocytes from mice of advanced reproductive age using RNA-seq analysis and found that SE was the predominant form of AS event. Previous studies have revealed that AS events are associated with meiosis in mouse oocytes and that the depletion of SRSF3 results in meiotic resumption defects due to the dysregulation of Brd8 and Pdlim7 exon inclusion (33). The knockdown of Esrp1 induces female infertility and oocyte meiotic arrest by affecting pre-mRNA splicing alterations involving Lsm14b, Trb53bp1, and Rac1 (34). In our study, the DEGs between the IVO and IVM groups included Ndufa7, Ndufs7, Cox6a2, Ndufs5, Ndufb1, and Uqcrh, all of which were enriched in the mitochondrial OXPHOS pathway involving SE, A3SS, or A5SS splicing events, and overlapped with non-alcoholic fatty liver disease, Huntington disease, Parkinson disease, and thermogenesis-enriched pathways. Recent research has shown that the knockout of UQCRH reinforces the Warburg effect (WE) (71) and the WE plays a critical role in embryogenesis by supporting rapid cell proliferation (72). Previously, our group reported that calcium signaling was inhibited in IVM oocytes but not IVO oocytes in humans (22) and calcium treatment leads to a decreased content of NDUFS7 in the mitochondria (73); this might explain the fact that our transcriptomic analysis found a reduction of Ndufs7 in the IVO group. Future research needs to investigate AS events in these DEGs by either knockdown or exogenous overexpression experiments. In the present study, we also found that the Zfp982 gene undergoes the most frequent AS events, including SE, MXE, A3SS, and RI events (Supplementary Figure S2). Dehghanian et al. reported that Zfp982 acts as a transcription factor by interacting with YAP1 to maintain stemness in mouse embryonic stem cells (74). Furthermore, Obox1 as an oocyte-specific factor can promote induced pluripotent stem cells induction by regulating mesenchymal-to-epithelial transition during reprogramming by inhibiting cell-cycle-related gene expression (75). Little is known about AS events in oocyte maturation; consequently, the alternative RNA splicing genes identified in this study deserve further to be investigated, particularly with regards to their mechanisms of action.
In the present study, we systematically compared the expression profiles of DEGs between groups of IVM and IVO oocytes from mice of advanced reproductive age. We identified the potential functions of these DEGs and used KEGG enrichment and PPI network analysis to determine that OXPHOS was the most significantly enriched and interactive pathway. Heatmaps of the five respiratory chain complexes, particularly complex I, indicated the potential involvement of specific antioxidants. Notably, the AS events detected in the present study demonstrated the diversity of post-transcriptional gene regulation in MII oocytes when cultured in different processes. Therefore, the identified key genes may represent potential therapeutic targets for the treatment of infertility in women of advanced reproductive age.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE182711.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Peking University.
Author Contributions
JQ designed and supervised the study. HQ performed the experimental work and data analysis with help from YQ. HQ drafted the manuscript with support from YQ, RL, and JQ. JQ, HQ, and YQ provided the funding. All authors contributed to the article and approved the submitted version.
Funding
This project is funded by the China Postdoctoral Science Foundation (nos. 2019TQ0009 and 2020M670060), the National Key Research and Development Program of China (no. 2019YFA0801400), the National Natural Science Foundation of China (nos. 81521002, 81730038, and 31801244), and CAMS Innovation Fund for Medical Sciences (no. 2019-I2M-5-001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.816606/full#supplementary-material
Supplementary Figure 1 | The chord diagram represents DEGs associated with the top four enriched KEGG pathways (the OXPHOS, NAFLD, HD, and PD) that are shown on the right. The color links merge genes with the log fold change are located on the left.
Supplementary Figure 2 | (A) List of genes undergoing three or more AS events, including Zfp982, Pbdc1, Obox1, Phf20, Tnfsf13b, Platr30, Scml2, and Trmt11. (B) List of genes undergoing six or more AS events. (C) The top 20 enriched KEGG pathways for differential AS events of the DEGs between the IVO and IVM groups.
References
1. Qiao J, Feng HL. Assisted Reproductive Technology in China: Compliance and Non-Compliance. Transl Pediatr (2014) 3:91–7. doi: 10.3978/j.issn.2224-4336.2014.01.06
2. Bryant J, Sullivan E, Dean J. Assisted Reproductive Technology in Australia and New Zealand 2002. AIHW Cat. No. PER 26. Sydney: Australian Institute of Health and Welfare National Perinatal Statistics Unit (Assisted Reproductive Technology Series No. 8). Sydney: Australian Institute Health and Welfare National Perinatal Statistics Unit (2004). Available at: https://www.aihw.gov.au/getmedia/ede07613-7ab9-4a4f-8ee9-2e99dd6a9c41/artanz02.pdf.aspx?inline=true.
3. Newman JE, Paul RC, Chambers GM. Assisted Reproductive Technology in Australia and New Zealand 2018. Sydney: National Perinatal Epidemiology and Statistics Unit, The University of New South Wales, Sydney (2020).
4. Julania S, Walls ML, Hart R. The Place of In Vitro Maturation in PCO/PCOS. Int J Endocrinol (2018) 2018:5750298. doi: 10.1155/2018/5750298
5. Namavar Jahromi BM, Parsanezhad MM, Shomali ZM, Bakhshai PM, Alborzi MM, Moin Vaziri NMDP, et al. Ovarian Hyperstimulation Syndrome: A Narrative Review of Its Pathophysiology, Risk Factors, Prevention, Classification, and Management. Iran J Med Sci (2018) 43:248–60.
6. Liu J, Lim J-H, Chian R-C. IVM as an Alternative for Poor Responders. In-Vitro Maturation of Human Oocytes. In: Basic Science to Clinical Applications. London: Informa Healthcare (2007). p. 333–44.
7. Shalom-Paz E, Almog B, Shehata F, Huang J, Holzer H, Chian R-C, et al. Fertility Preservation for Breast-Cancer Patients Using IVM Followed by Oocyte or Embryo Vitrification. Reprod Biomed Online (2010) 21:566–71. doi: 10.1016/j.rbmo.2010.05.003
8. Aflatoonian A, Eftekhar M, Mohammadian F, Yousefnejad F. Outcome of Assisted Reproductive Technology in Women Aged 40 Years and Older. Iran J Reprod Med (2011) 9:281–4.
9. Wiser A, Son WY, Shalom-Paz E, Reinblatt SL, Tulandi T, Holzer H. How Old Is Too Old for In Vitro Maturation (IVM) Treatment? Eur J Obstet Gynecol Reprod Biol (2011) 159:381–3. doi: 10.1016/j.ejogrb.2011.09.009
10. Karavani G, Wasserzug-Pash P, Mordechai-Daniel T, Bauman D, Klutstein M, Imbar T. Age-Dependent In Vitro Maturation Efficacy of Human Oocytes - Is There an Optimal Age? Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.667682
11. Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP. Structure of a Mitochondrial Supercomplex Formed by Respiratory-Chain Complexes I and III. Proc Natl Acad Sci USA (2005) 102:3225–9. doi: 10.1073/pnas.0408870102
12. Roth Z. Symposium Review: Reduction in Oocyte Developmental Competence by Stress Is Associated With Alterations in Mitochondrial Function. J Dairy Sci (2018) 101:3642–54. doi: 10.3168/jds.2017-13389
13. Combelles CM, Gupta S, Agarwal A. Could Oxidative Stress Influence the In-Vitro Maturation of Oocytes? Reprod BioMed Online (2009) 18:864–80. doi: 10.1016/S1472-6483(10)60038-7
14. Sugimura S, Matoba S, Hashiyada Y, Aikawa Y, Ohtake M, Matsuda H, et al. Oxidative Phosphorylation-Linked Respiration in Individual Bovine Oocytes. J Reprod Dev (2012) 58:636–41. doi: 10.1262/jrd.2012-082
15. Goto Y, Noda Y, Mori T, Nakano M. Increased Generation of Reactive Oxygen Species in Embryos Cultured In Vitro. Free Radic Biol Med (1993) 15:69–75. doi: 10.1016/0891-5849(93)90126-F
16. Guérin P, El Mouatassim S, Ménézo Y. Oxidative Stress and Protection Against Reactive Oxygen Species in the Pre-Implantation Embryo and Its Surroundings. Hum Reprod Update (2001) 7:175–89. doi: 10.1093/humupd/7.2.175
17. Lord T, Aitken RJ. Oxidative Stress and Ageing of the Post-Ovulatory Oocyte. Reproduction (2013) 146:R217–227. doi: 10.1530/REP-13-0111
18. Sanfins A, Plancha CE, Overstrom EW, Albertini DF. Meiotic Spindle Morphogenesis in In Vivo and In Vitro Matured Mouse Oocytes: Insights Into the Relationship Between Nuclear and Cytoplasmic Quality. Hum Reprod (2004) 19:2889–99. doi: 10.1093/humrep/deh528
19. Sanfins A, Plancha CE, Albertini DF. Pre-Implantation Developmental Potential From In Vivo and In Vitro Matured Mouse Oocytes: A Cytoskeletal Perspective on Oocyte Quality. J Assist Reprod Genet (2015) 32:127–36. doi: 10.1007/s10815-014-0363-4
20. Lee AWT, Ng JKW, Liao J, Luk AC, Suen AHC, Chan TTH, et al. Single-Cell RNA Sequencing Identifies Molecular Targets Associated With Poor In Vitro Maturation Performance of Oocytes Collected From Ovarian Stimulation. Hum Reprod (2021) 36:1907–21. doi: 10.1093/humrep/deab100
21. Zhang H-L, Xu Y, Ju J-Q, Pan Z-N, Liu J-C, Sun S-C. Increased Environment-Related Metabolism and Genetic Expression in the In Vitro Matured Mouse Oocytes by Transcriptome Analysis. Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.642010
22. Zhao H, Li T, Zhao Y, Tan T, Liu C, Liu Y, et al. Single-Cell Transcriptomics of Human Oocytes: Environment-Driven Metabolic Competition and Compensatory Mechanisms During Oocyte Maturation. Antioxid Redox Signal (2019) 30:542–59. doi: 10.1089/ars.2017.7151
23. Black DL. Mechanisms of Alternative Pre-Messenger RNA Splicing. Annu Rev Biochem (2003) 72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720
24. Chen M, Manley JL. Mechanisms of Alternative Splicing Regulation: Insights From Molecular and Genomics Approaches. Nat Rev Mol Cell Biol (2009) 10:741–54. doi: 10.1038/nrm2777
25. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep Surveying of Alternative Splicing Complexity in the Human Transcriptome by High-Throughput Sequencing. Nat Genet (2008) 40:1413–5. doi: 10.1038/ng.259
26. Link S, Grund SE, Diederichs S. Alternative Splicing Affects the Subcellular Localization of Drosha. Nucleic Acids Res (2016) 44:5330–43. doi: 10.1093/nar/gkw400
27. Fiszbein A, Kornblihtt AR. Alternative Splicing Switches: Important Players in Cell Differentiation. Bioessays (2017) 39:1–11. doi: 10.1002/bies.201600157
28. Sun C, Jin K, Zuo Q, Sun H, Song J, Zhang Y, et al. Characterization of Alternative Splicing (AS) Events During Chicken (Gallus Gallus) Male Germ-Line Stem Cell Differentiation With Single-Cell RNA-Seq. Animals (Basel) (2021) 11:1469–80. doi: 10.3390/ani11051469
29. Zhang Y, Qian J, Gu C, Yang Y. Alternative Splicing and Cancer: A Systematic Review. Signal Transduct Targeted Ther (2021) 6:78. doi: 10.1038/s41392-021-00486-7
30. Bao J, Tang C, Li J, Zhang Y, Bhetwal BP, Zheng H, et al. RAN-Binding Protein 9 Is Involved in Alternative Splicing and Is Critical for Male Germ Cell Development and Male Fertility. PloS Genet (2014) 10:e1004825. doi: 10.1371/journal.pgen.1004825
31. Iwamori N, Tominaga K, Sato T, Riehle K, Iwamori T, Ohkawa Y, et al. MRG15 Is Required for Pre-mRNA Splicing and Spermatogenesis. Proc Natl Acad Sci USA (2016) 113:E5408–15. doi: 10.1073/pnas.1611995113
32. Salisbury J, Hutchison KW, Wigglesworth K, Eppig JJ, Graber JH. Probe-Level Analysis of Expression Microarrays Characterizes Isoform-Specific Degradation During Mouse Oocyte Maturation. PloS One (2009) 4:e7479. doi: 10.1371/journal.pone.0007479
33. Do DV, Strauss B, Cukuroglu E, Macaulay I, Wee KB, Hu TX, et al. SRSF3 Maintains Transcriptome Integrity in Oocytes by Regulation of Alternative Splicing and Transposable Elements. Cell Discovery (2018) 4:33. doi: 10.1038/s41421-018-0032-3
34. Yu L, Zhang H, Guan X, Qin D, Zhou J, Wu X. Loss of ESRP1 Blocks Mouse Oocyte Development and Leads to Female Infertility. Development (2021) 148:196931–44. doi: 10.1242/dev.196931
35. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD, et al. Nuclear M6a Reader YTHDC1 Regulates Alternative Polyadenylation and Splicing During Mouse Oocyte Development. PloS Genet (2018) 14:e1007412. doi: 10.1371/journal.pgen.1007412
36. Li J, Lu M, Zhang P, Hou E, Li T, Liu X, et al. Aberrant Spliceosome Expression and Altered Alternative Splicing Events Correlate With Maturation Deficiency in Human Oocytes. Cell Cycle (2020) 19:2182–94. doi: 10.1080/15384101.2020.1799295
37. Qin H, Qu Y, Yuan Y-F, Li Y-Y, Qiao J. RBM14 Modulates Tubulin Acetylation and Regulates Spindle Morphology During Meiotic Maturation in Mouse Oocytes. Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.635728
38. Guo Y, Sun J, Bu S, Li B, Zhang Q, Wang Q, et al. Melatonin Protects Against Chronic Stress-Induced Oxidative Meiotic Defects in Mice MII Oocytes by Regulating SIRT1. Cell Cycle (2020) 19:1677–95. doi: 10.1080/15384101.2020.1767403
39. Wang J, Zhuo Z, Ma X, Liu Y, Xu J, He C, et al. Melatonin Alleviates the Suppressive Effect of Hypoxanthine on Oocyte Nuclear Maturation and Restores Meiosis via the Melatonin Receptor 1 (MT1)-Mediated Pathway. Front Cell Dev Biol (2021) 9. doi: 10.3389/fcell.2021.648148
40. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-Based Genome Alignment and Genotyping With HISAT2 and HISAT-Genotype. Nat Biotechnol (2019) 37:907–15. doi: 10.1038/s41587-019-0201-4
41. Li B, Dewey CN. RSEM: Accurate Transcript Quantification From RNA-Seq Data With or Without a Reference Genome. BMC Bioinf (2011) 12:323. doi: 10.1186/1471-2105-12-323
42. Love MI, Huber W, Anders S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data With Deseq2. Genome Biol (2014) 15:550. doi: 10.1186/s13059-014-0550-8
43. Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: A Web Server for Annotation and Identification of Enriched Pathways and Diseases. Nucleic Acids Res (2011) 39:W316–22. doi: 10.1093/nar/gkr483
44. Shen S, Park JW, Lu Z-X, Lin L, Henry MD, Wu YN, et al. rMATS: Robust and Flexible Detection of Differential Alternative Splicing From Replicate RNA-Seq Data. Proc Natl Acad Sci (2014) 111:E5593–601. doi: 10.1073/pnas.1419161111
45. Gueroussov S, Weatheritt RJ, O'hanlon D, Lin ZY, Narula A, Gingras AC, et al. Regulatory Expansion in Mammals of Multivalent hnRNP Assemblies That Globally Control Alternative Splicing. Cell (2017) 170:324–339.e323. doi: 10.1016/j.cell.2017.06.037
46. Li J, Zhao H, Xing Y, Zhao T, Cai L, Yan Z. A Genome-Wide Analysis of the Gene Expression and Alternative Splicing Events in a Whole-Body Hypoxic Preconditioning Mouse Model. Neurochem Res (2021) 46:1101–11. doi: 10.1007/s11064-021-03241-0
47. Zeng J, Jiang M, Wu X, Diao F, Qiu D, Hou X, et al. SIRT4 Is Essential for Metabolic Control and Meiotic Structure During Mouse Oocyte Maturation. Aging Cell (2018) 17:e12789. doi: 10.1111/acel.12789
48. Miao Y, Cui Z, Gao Q, Rui R, Xiong B. Nicotinamide Mononucleotide Supplementation Reverses the Declining Quality of Maternally Aged Oocytes. Cell Rep (2020) 32:107987. doi: 10.1016/j.celrep.2020.107987
49. Li C, He X, Huang Z, Han L, Wu X, Li L, et al. Melatonin Ameliorates the Advanced Maternal Age-Associated Meiotic Defects in Oocytes Through the SIRT2-Dependent H4K16 Deacetylation Pathway. Aging (Albany NY) (2020) 12:1610–23. doi: 10.18632/aging.102703
50. Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING V11: Protein-Protein Association Networks With Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res (2019) 47:D607–d613. doi: 10.1093/nar/gky1131
51. Sasaki H, Hamatani T, Kamijo S, Iwai M, Kobanawa M, Ogawa S, et al. Impact of Oxidative Stress on Age-Associated Decline in Oocyte Developmental Competence. Front Endocrinol (2019) 10. doi: 10.3389/fendo.2019.00811
53. Blencowe BJ. Alternative Splicing: New Insights From Global Analyses. Cell (2006) 126:37–47. doi: 10.1016/j.cell.2006.06.023
54. Mathews TJ, Hamilton BE. Mean Age of Mothers Is on the Rise: United States, 2000–2014. NCHS Data Brief. Hyattsville, MD: National Center for Health Statistics (2016).
55. Guzzo KB, Payne KK. Average Age at First Birth 1970 - 2017. Family Profiles, FP-18-25. Bowling Green, OH: National Center for Family & Marriage Research (2018). doi: 10.25035/ncfmr/fp-18-25
56. Vuong LN, Ho TM, Gilchrist RB, Smitz J. The Place of In Vitro Maturation in Assisted Reproductive Technology. Fertil Reprod (2019) 1:11–5. doi: 10.1142/S2661318219300022
57. Gremeau AS, Andreadis N, Fatum M, Craig J, Turner K, Mcveigh E, et al. In Vitro Maturation or In Vitro Fertilization for Women With Polycystic Ovaries? A Case-Control Study of 194 Treatment Cycles. Fertil Steril (2012) 98:355–60. doi: 10.1016/j.fertnstert.2012.04.046
58. Lopes AS, Lane M, Thompson JG. Oxygen Consumption and ROS Production Are Increased at the Time of Fertilization and Cell Cleavage in Bovine Zygotes. Hum Reprod (2010) 25:2762–73. doi: 10.1093/humrep/deq221
59. Alcolea MP, Colom B, Lladó I, García-Palmer FJ, Gianotti M. Mitochondrial Differentiation and Oxidative Phosphorylation System Capacity in Rat Embryo During Placentation Period. Reproduction (2007) 134:147–54. doi: 10.1530/REP-07-0012
60. Brand MD. Mitochondrial Generation of Superoxide and Hydrogen Peroxide as the Source of Mitochondrial Redox Signaling. Free Radic Biol Med (2016) 100:14–31. doi: 10.1016/j.freeradbiomed.2016.04.001
61. Mazat JP, Devin A, Ransac S. Modelling Mitochondrial ROS Production by the Respiratory Chain. Cell Mol Life Sci (2020) 77:455–65. doi: 10.1007/s00018-019-03381-1
62. Freitas C, Neto AC, Matos L, Silva E, Ribeiro Â, Silva-Carvalho JL, et al. Follicular Fluid Redox Involvement for Ovarian Follicle Growth. J Ovarian Res (2017) 10:44. doi: 10.1186/s13048-017-0342-3
63. Da Broi MG, Giorgi VSI, Wang F, Keefe DL, Albertini D, Navarro PA. Influence of Follicular Fluid and Cumulus Cells on Oocyte Quality: Clinical Implications. J Assist Reprod Genet (2018) 35:735–51. doi: 10.1007/s10815-018-1143-3
64. Shaeib F, Khan SN, Ali I, Thakur M, Saed MG, Dai J, et al. The Defensive Role of Cumulus Cells Against Reactive Oxygen Species Insult in Metaphase II Mouse Oocytes. Reprod Sci (2016) 23:498–507. doi: 10.1177/1933719115607993
65. Takenaka M, Horiuchi T, Yanagimachi R. Effects of Light on Development of Mammalian Zygotes. Proc Natl Acad Sci USA (2007) 104:14289–93. doi: 10.1073/pnas.0706687104
66. Li X, Mu Y, Elshewy N, Ding D, Zou H, Chen B, et al. Comparison of IVF and IVM Outcomes in the Same Patient Treated With a Modified IVM Protocol Along With an Oocytes-Maturing System Containing Melatonin: A Pilot Study. Life Sci (2021) 264:118706. doi: 10.1016/j.lfs.2020.118706
67. Ben-Meir A, Burstein E, Borrego-Alvarez A, Chong J, Wong E, Yavorska T, et al. Coenzyme Q10 Restores Oocyte Mitochondrial Function and Fertility During Reproductive Aging. Aging Cell (2015) 14:887–95. doi: 10.1111/acel.12368
68. Wu X, Hu F, Zeng J, Han L, Qiu D, Wang H, et al. NMNAT2-Mediated NAD(+) Generation Is Essential for Quality Control of Aged Oocytes. Aging Cell (2019) 18:e12955. doi: 10.1111/acel.12955
69. Liu XM, Yan MQ, Ji SY, Sha QQ, Huang T, Zhao H, et al. Loss of Oocyte Rps26 in Mice Arrests Oocyte Growth and Causes Premature Ovarian Failure. Cell Death Dis (2018) 9:1144. doi: 10.1038/s41419-018-1196-3
70. Peng H, Zhao Y, Chen J, Huo J, Zhang Y, Xiao T. Knockdown of Ribosomal Protein S3 Causes Preimplantation Developmental Arrest in Mice. Theriogenology (2019) 129:77–81. doi: 10.1016/j.theriogenology.2019.02.022
71. Luo Y, Medina Bengtsson L, Wang X, Huang T, Liu G, Murphy S, et al. UQCRH Downregulation Promotes Warburg Effect in Renal Cell Carcinoma Cells. Sci Rep (2020) 10:15021. doi: 10.1038/s41598-020-72107-2
72. Krisher RL, Prather RS. A Role for the Warburg Effect in Preimplantation Embryo Development: Metabolic Modification to Support Rapid Cell Proliferation. Mol Reprod Dev (2012) 79:311–20. doi: 10.1002/mrd.22037
73. Mohsin AA, Thompson J, Hu Y, Hollander J, Lesnefsky EJ, Chen Q. Endoplasmic Reticulum Stress-Induced Complex I Defect: Central Role of Calcium Overload. Arch Biochem Biophys (2020) 683:108299. doi: 10.1016/j.abb.2020.108299
74. Dehghanian F, Bovio PP, Hojati Z, Vogel T. ZFP982 Confers Mouse Embryonic Stem Cell Characteristics by Regulating Expression of Nanog, Zfp42 and Dppa3. bioRxiv (2020) 2020. 06.03.131847. doi: 10.1101/2020.06.03.131847
Keywords: oocyte, in vitro maturation, oxidative phosphorylation, transcriptome, differentially expressed genes, alternative splicing
Citation: Qin H, Qu Y, Li R and Qiao J (2022) In Vivo and In Vitro Matured Oocytes From Mice of Advanced Reproductive Age Exhibit Alternative Splicing Processes for Mitochondrial Oxidative Phosphorylation. Front. Endocrinol. 13:816606. doi: 10.3389/fendo.2022.816606
Received: 16 November 2021; Accepted: 03 January 2022;
Published: 26 January 2022.
Edited by:
Wenpei Xiang, Huazhong University of Science and Technology, ChinaReviewed by:
Mingtian Deng, Nanjing Agricultural University, ChinaXiuliang Dai, Changzhou Maternal and Child Health Care Hospital, China
Chun Yuan, Nanjing Medical University, China
Copyright © 2022 Qin, Qu, Li and Qiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Qiao, amllLnFpYW9AMjYzLm5ldA==
 Hao Qin
Hao Qin Yi Qu
Yi Qu Rong Li
Rong Li Jie Qiao
Jie Qiao