
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 11 January 2023
Sec. Reproduction
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.1099270
This article is part of the Research Topic Novel Insights into Sperm Function and Selection: from Basic Research to Clinical Application View all 14 articles
 Haowei Bai1†
Haowei Bai1† Yanwei Sha2†
Yanwei Sha2† Yueqiu Tan3†
Yueqiu Tan3† Peng Li1
Peng Li1 Yuxiang Zhang1
Yuxiang Zhang1 Junwei Xu1
Junwei Xu1 Shuai Xu1
Shuai Xu1 Zhiyong Ji1
Zhiyong Ji1 Xiaobo Wang1
Xiaobo Wang1 Wei Chen1
Wei Chen1 Jianxiong Zhang1
Jianxiong Zhang1 Chencheng Yao1*
Chencheng Yao1* Zheng Li1*
Zheng Li1* Erlei Zhi1*
Erlei Zhi1*Introduction: Oligoasthenoteratozoospermia (OAT) is a major cause of infertility in males. Only a few pathogenic genes of OAT have been clearly identified till now. A large number of OAT-affected cases remain largely unknown.
Methods: Here, Whole-exome sequencing (WES) in 725 idiopathic OAT patients was performed. Ejaculated spermatozoa by OAT patients were microinjected into mouse oocytes to estimate fertilization potential. Diff-quick staining and transmission electron microscopy were performed to evaluate sperm morphology and ultrastructure. The protein expression level and localization In vitro were detected by Western Blotting and Immunocytochemistry.
Results: We identified four X-linked hemizygous deleterious variants of TAF7L—namely, c.1301_1302del;(p.V434Afs*5), c.699G>T;(p.R233S), c.508delA; (p. T170fs), c.719dupA;(p.K240fs) —in five probands. Intracytoplasmic sperm injection (ICSI) were carried out in M1, M2-1and M3 patient's wife. However only M1 patient’s wife became pregnant after embryo transfer. In vitro study demonstrated significantly reduced fertilization ability in patient with TAF7L mutation. The TAF7L mutation let to abnormal sperm head and impaired histone-to protamine exchange. Variant 719dupA (p. K240fs) resulted in producing a truncated TAF7L protein and localized massively within the nucleus. In addition, TAF7L expression were not able to be detected due to variants c.1301_1302del (p. V434Afs*5) and c.508delA (p. T170fs) In vitro.
Conclusion: Our findings support that TAF7L is one of pathogenic genes of OAT and deleterious mutations in TAF7L may cause impaired histone-to-protamine affected the chromatin compaction of sperm head.
Infertility affects about 15% of couples worldwide and a male infertility associated factor could be found in approximately half of all the couples (1, 2). Clinical manifestations of male infertility are mainly as follows, oligozoospermia, asthenozoospermia, teratozospermia, astheno-teratozospemia and oligoasthenoteratozoospermia(OAT) (3, 4). Varicocele, poor drug delivery, Y chromosomal microdeletion, and genetic abnormalities all contribute to male infertility (5).
Genetic research of male infertility related defect spermiogenesis has achieved some progress in recent years. Based on the above researches, different pathogenic genes have been classified as two specific phenotypes, including sperm head malformations (AURKC, SPATA16, DPY19L2, PICK1, IQCN), and multiple morphological abnormalities of the sperm flagella (DNAH1, CFAP65, CFAP47, CFAP44, CFAP43, CFAP251, DZIP1, DNAH8, DNAH10) (3, 6–14). However, the discovery of different pathogenic genes indicates that there is great genetic heterogeneity in male infertility, and abnormal sperm with the same morphology may also be caused by different genes. Therefore, the discovery of more pathogenic genes has an important role in the genetic analysis of male infertility.
Sex chromosomes not only determine the sex of a baby but also play a vital part in fertility. According to previous research, most of the genes that are expressed predominantly in testes were identified on sex chromosomes (15). Deletion mutations in certain genes can cause male infertility (16). It is well known that microdeletions in the Y chromosome play important role in the infertility of males, however, only a few genes present on the X chromosome are known to cause male infertility. For instance, it has been found that the hemizygous TEX11 variant causes meiosis arrest, giving rise to non-obstructive azoospermia (NOA) (17, 18). Also, hemizygous ADGRG2 variants were identified in obstructive azoospermia (OA)-affected patients (19). Hemizygous CFAP47 variants induced OAT and primary male infertility (12). However, many other X-linked genes responsible for male infertility related defect spermatogenesis are yet to be identified.
Another X-linked gene, TAF7L (also known as CT40), was linked to OAT and male infertility in the current study, and four X-linked hemizygous deleterious variations of TAF7L were detected in probands, but with significantly reduced in vitro fertilization. The results of this study will help in genetic counselling and treatments of infertility.
In the current study, 725 idiopathic OAT patients were recruited. According to the WHO Laboratory Manual for the Examination and Processing of Human Semen (5th edition), OAT was defined as sperm concentration <15×106/mL, progressive motility <32% and normal sperm morphology <4% with two times of semen analyses. 126 infertile men were recruited in the Department of Andrology, Urologic Medical Center, Shanghai General Hospital, Shanghai Jiao Tong University (Shanghai, China). 479 male infertile individuals were recruited in the Reproductive and Genetic Hospital of CITIC-Xiangya (Changsha, Hunan, China) and 120 in the Department of Reproductive Medicine, Xiamen Maternity and Child Care Hospital, Xiamen. (Fujian, Xiamen, China).
Patients with congenital diseases, deletions in the AZF gene, or factors linked to OAT, testicular cancer, cryptorchidism, or orchitis were excluded from the study. Patients who received chemotherapy or radiation were also excluded. The family histories of the patients were collected. For the control group, 20 healthy and fertility men from the Han Chinese ethnic group with a similar background of genetic makeup were included. Subjects of the control group exhibited normal semen analysis and they fathered one or more healthy babies.
Prior to participation, each participant signed a written informed consent form. The research was approved by the corresponding ethics committees of the Shanghai General Hospital, Shanghai Jiao Tong University(2021-SQ-112), the Reproductive and Genetic Hospital of CITIC Xiangya (LL-SC-2017-025 and LL-SC-2019-034), and the Xiamen Maternity and Child Care Hospital, Xiamen (Permit Number: 2020SQ199, KY-2019-060).
The manufacturer’s protocol was followed to extract whole DNA from blood samples of patients using TIANamp Blood DNA (TIANGEN Biotech, Beijing, China). Covaris focused ultrasonication was used for DNA fragmentation. xGen Exome Research Panel (Integrated DNA Technologies, Coralville, IA, USA) was used for capturing known exons and boundaries of the exon-intron followed by preparation of DNA sequences libraries according to the manufacturer’s protocol.
DNA was sequenced on a HiSeq X10 platform (Illumina, San Diego, CA, USA). Burrows-Wheeler Aliigner (BWA) was employed for the alignment of the sequencing reads with the human genome (GRCh37/hg19). GATK, VarSCan, Platypus, LoFreq, SNVer, Freebayes, VarDict and SAM tools were used for calling SNVs as well as indels within the intervals of captured coding exons. ANNOVAR software was used to filter and annotate variants. Those genetic variants were excluded which had higher than 1% allelic frequencies as per 1000 Gnomes Project and ExAC Browser while removing upstream, downstream and intronic variants. Frameshift, Nonsense, essential splice-site and missense variants with deleterious potential (SIFT, Mutation Taster and Polyphen-2) were retained for later analysis. Additionally, candidate genes were compared with pathogenic genes in mice (http://www.informatics.jax.org/mgihome/homepages/) and testis enriched genes in the database (http://www.proteinatlas.org/). The bioinformatic analysis and sequencing were performed with the collaboration of the Nuprobe company.
Semen samples were collected from patients by asking them for masturbation, 3-7 days post-sexual intercourse. Analysis of the samples was performed in a laboratory as per 5th WHO guidelines. The morphology of sperm was analyzed with examination using Diff-Quick staining. Ejaculated sperm of one patient (family 1, M1: II-1) without enough spermatozoa were recovered by testicular biopsy on the day of oocyte retrieval. H&E staining was performed on testicular tissue to assess spermatogenesis.
Viable sperm for ICSI were selected by laser, the tips of immotile sperm were targeted with a laser beam of approximately 200 μJ with an irradiation time of about 2 ms. Those spermatozoa which presented with curling of the tails after the laser shot were regarded as viable, while others which did not curling of the tails were considered to be non-viable. And assisted oocyte activation (AOA) has been done to activate oocytes. After ICSI treatment, outcomes were obtained from three different hospitals. Results of the following parameters were included: the number of injected oocytes, fertilized oocytes, ICSI cycles, rate of embryos cleavage, 8 cells, blastocysts, transfer cycles, per cycle transferred embryos, rate of implantation, pregnancies and miscarriage.
Spermatozoa were fixed routinely as previously (5). After being embedded in Epon, ultrathin sections of sperm were stained with uranyl acetate, then photographed by TEM. At least 50 flagella with cross sections and several longitudinal sections were counted.
To further prove the different capacity of ejaculated spermatozoa between patients with TAF7L variants and healthy men, microinjection of spermatozoa into the mice oocytes were performed using a previously described procedure (20). For superovulation, mice were injected with 5 IU of equine chorionic gonadotropin. Mice were then given 5 IU of human chorionic gonadotropin (HCG) after 48 hours. After 16 hours of HCG injection, cumulus-oocyte complexes were harvested from oviducts and transferred to HEPES Chatot-Ziomek-Bavister (CZB) medium. To disperse cells of cumulus, they were treated with hyaluronidase (0.1%). Oocytes were rinsed two times and then transferred to fresh CZB drop. They were given at least 15 min incubation in plain CZB media. To artificially activate the oocytes, they were kept for 1 hour in CZB free of Ca2+ and supplemented with SrCL2 (10mM) followed by washing two times again. For resuming incubation, they were again transferred to a CZB medium. 100 sperm from each group were microinjected into the eggs with piezoelectric elements. To examine the formation of pronucleus and in vitro development, the oocytes in the CZB medium were kept for incubation at 37 °C and 5% CO2.
The spermatozoa of the proband (family 1, M1: II-1) and normal control were stained by immunofluorescent. The samples were fixed with 4% paraformaldehyde for 20 min. After 10 min washes with PBS, heat-induced antigen retrieval was carried out by boiling the slides in 10 mM citrate buffer (pH 6.0) with a microwave oven for 10 min. After three 10 min washes with 0.1% Triton X-100 in PBS, the slides were blocked with 5% BSA diluted in PBST for 1h and then the slides were incubated with anti-histone H3 (1:100; CST), anti-PRM2(1:100; Atlas) in PBST overnight, samples were incubated with fluorescein isothiocyanate-conjugated anti-rabbit IgG (1:200; Invitrogen), and peanut agglutinin (PNA) (1:300; Invitrogen) for 30 min at room temperature. The samples were washed three times in PBS and incubated 10-min with Hoechst33342(1:200; Invitrogen), imaged by fluorescence microscopy.
According to the manufacturer’s instruction, testicular RNA of patient (Family 1, M1: II-1) was extracted using the kit (Qiagen, USA). The purity was assessed by the A260/A280 ratio. Then we carried out the reverse transcription under the following conditions: 37 °C for 15 min, followed by 85 °C for 5 s. The products were performed under the following conditions: 95 °C for 30s, 40 cycles of 95 °C for 10 s, 60 °C for 30 s, and 95 °C 15s, 60°C for 60s, 95°C for 15s. GAPDH was used as an internal control and primers for real-time quantitative PCR are listed in Table S1.
Full-length cDNA encoding human TAF7L was amplified and cloned into pCMV7.1 vector with N-terminal FLAG tag. Site-directed mutagenesis was accomplished using a ClonExpress Ultra One Step Cloning Kit (C115-01, Vazyme, China). The wild-type and mutant clones were confirmed by Sanger sequencing. HEK293T cells were cultured in DMEM/high glucose (SH30243.FS, Cytiva) with 10% FBS (10099141C, Gibco) and 1% penicillin/streptomycin (15140122, Gibco) at 37°C in 5% CO2. TAF7L wild-type and mutant plasmids were transfected into HEK293T cells using the lipofectamine 3000 (L3000015, Invitrogen) according to the manufacturer’s protocol.
48 h after transfection, the cells were lysed by RIPA lysis buffer (89901, Thermo Fisher Scientific) on ice 20 min, followed by centrifugation at 14,000 × g for 15 min at 4°C to isolate the protein, which was quantified by BCA assay (23225, Thermo Fisher Scientific) and separated by 10% sodium dodecyl sulfate polyacrylamide gels and were transferred to a 0.22 µm PVDF membrane (ISEQ00010, Millipore). The membranes were blocked with 5% not fat milk in TBST buffer 1h. After washing three times with TBST for 5 min, then incubated at 4°C overnight with anti-FLAG rabbit monoclonal antibodies (dilution: 1:5,000; catalogue number: F1804, Sigma) or β-actin (dilution: 1:10,000; 66009-1-Ig, Proteintech). The membranes were washed with TBST three times and incubated with anti-Mouse IgG HRP (dilution: 1:5,000; sc-516102, Santa Cruz Biotechnology) for 1 h at room temperature followed by washing again with TBST three times. Finally, the blots were imaged on AI600 imager (GE healthcare)
In this study, WES of 725 idiopathic OAT patients was performed. Four X-linked hemizygous deleterious variants of TAF7L were identified in five probands (0.68%,5/725).
Patients were examined for physical parameters and they exhibited normal epididymis, scrotum, prostate, penis and testes development. Semen analysis revealed OAT phenotype as per the 5th edition of WHO guidelines. All patients had normal sex hormones, sex chromosomes and autosomal chromosomes. No Y chromosome microdeletions and no history of cancer, cryptorchidism, hypogonadism, alcoholism, or smoking were found.
To identify candidate genes that could be potentially associated with OAT, WES was performed. The workflow of data processing and analyses was described in Supplementary Figure 1. After excluding the polymorphisms, which were reported in previous studies. We found four deleterious variants X-Linked TAF7L variants in five Chinese OAT patients, the hemizygous frameshift [c.1301_1302del;(p.V434Afs*5)] in M1(Figure 1A; Table 1), a hemizygous missense mutation [c.699G>T;(p.R233S)] in M2-1 and M2-2, which was inherited from their parents (Figure 1B; Table 1). We also found the hemizygous frameshift [c.508delA; (p. T170fs)] in M3, the hemizygous frameshift [c.719dupA;(p.K240fs)] in M4 (Figures 1C, D; Table 1).
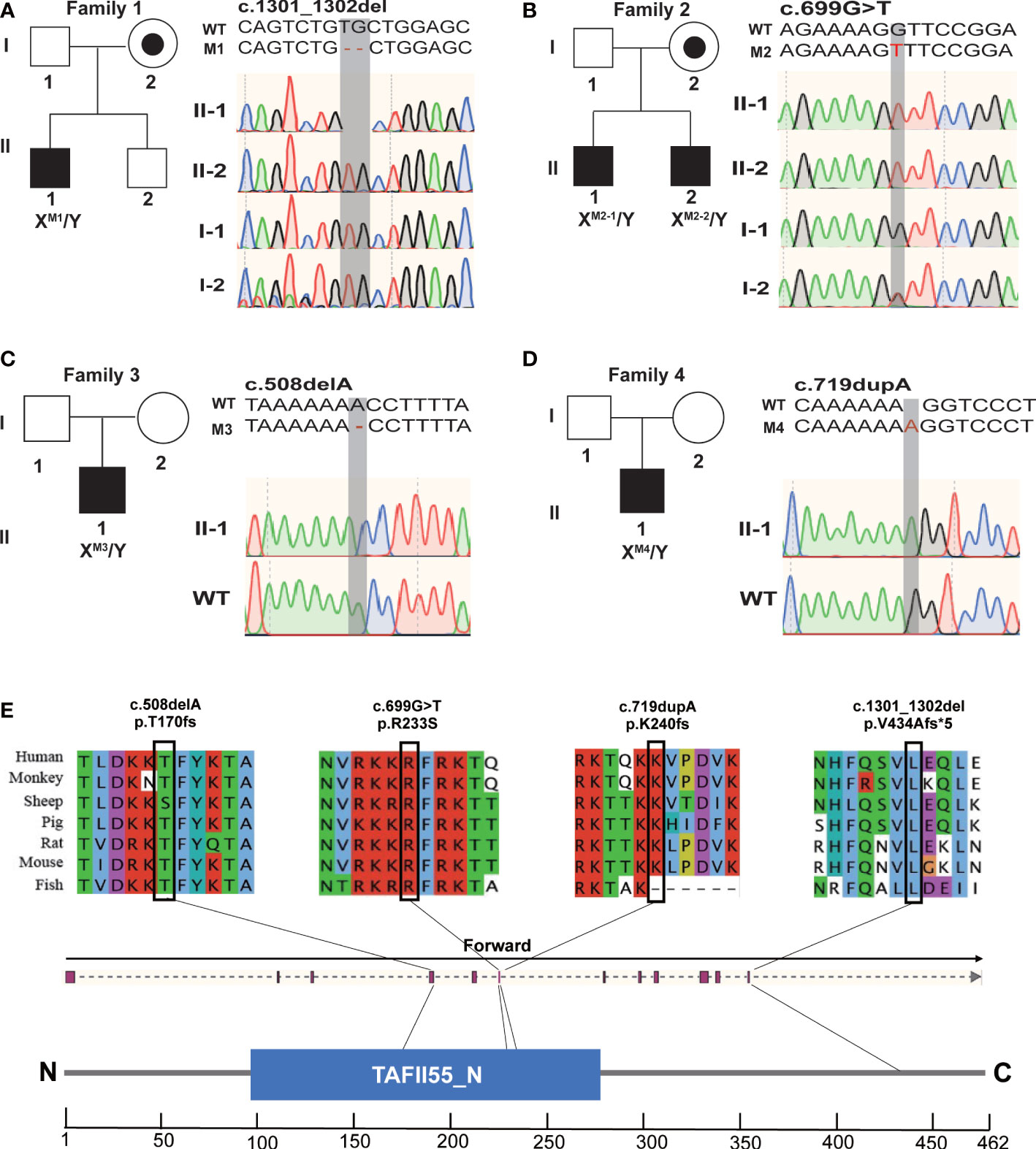
Figure 1 Hemizygous mutations in TAF7L identified in patients with OAT. (A–D) Pedigree analysis of the four patients affected by X-linked TAF7L Hemizygous variants that were identified by WES. Black filled squares indicate infertile men in these families. Sanger sequencing results are shown to the right of the pedigrees. The mutated positions are indicated by red words and black shadow boxes. (E) The genomic structure of TAF7L, with variants mapped to isoform 1 (GenBank accession number, NM_024885) The positions of the novel TAF7L variants identified in this study are indicated by black lines. Locations of the mutated sites in the exon structure of TAF7L(upper); Locations of the affected amino acids in the protein domain map of TAF7L (bottom). Sequence alignment shows conservation of the mutated residues among different species. Blue squares stand for TAFII55 protein conserved region according to the NCBI.
These hemizygous TAF7L variants were not detected in human population genome databases, including gnomAD and the 1000 Genomes Project (Table 1). In silico analysis of all these variants predicted them to be deleterious (SIFT, PolyPhen-2, M-CAP and CADD tools) (Table 1). Hence, these variants were classified as pathogenic following the American College of Medical Genetics and Genomics criteria (ACMG). All corresponding variant residues are highly conserved in many organisms (Figure 1E).
It is suggested that ICSI treatment can be effective in overcoming the physical problems faced by sperm from patients. The couples underwent ICSI by using spermatozoa of the subjects carrying hemizygous TAF7L mutation. Hence, the patients with TAF7L variants got fewer two-cell embryos and blastocysts than the control. These five patients with TAF7L variants didn’t get clinical pregnancy using ejaculated sperm, two of them chose donor’s sperm for ICSI treatment.
One patient (Family 1, M1: II-1), who had previously undergone seven ICSI cycles without conceiving with ejaculated sperm. In this time, ejaculated sperm of patient without enough spermatozoa were recovered by testicular biopsy on the day of oocyte retrieval. The couple got the successful clinical pregnancy with sperm of testes at the time of writing (Table 2).
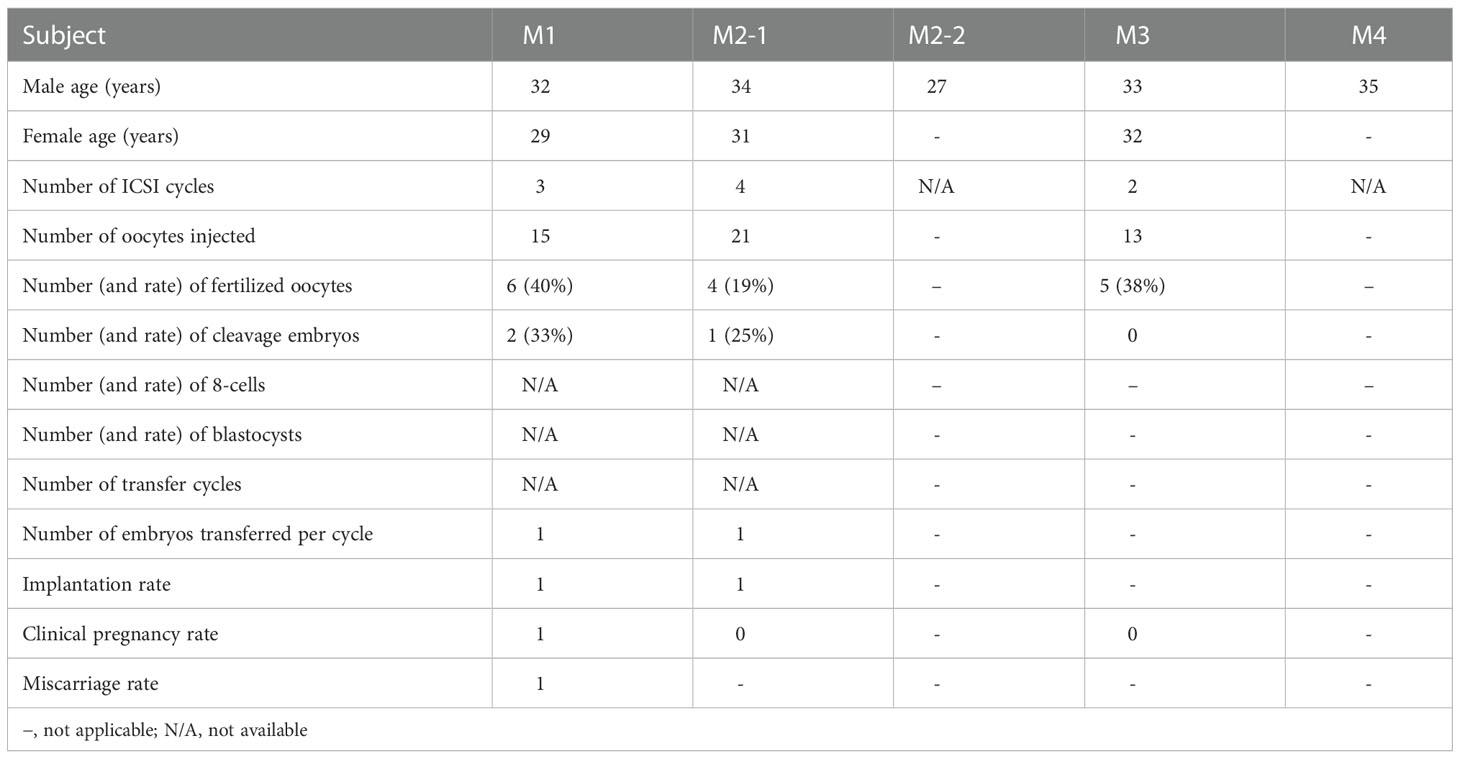
Table 2 Clinical outcomes of ICSI cycles using the spermatozoa from men harboring hemizygous TAF7L variants.
As contrast normal spermatogenesis, histological analysis of testis revealed that this patient (Family 1, M1: II-1) showed post meiotic arrest at the first stage of spermiogenesis, which revealed dramatically decreased elongated spermatids (Figure 2).
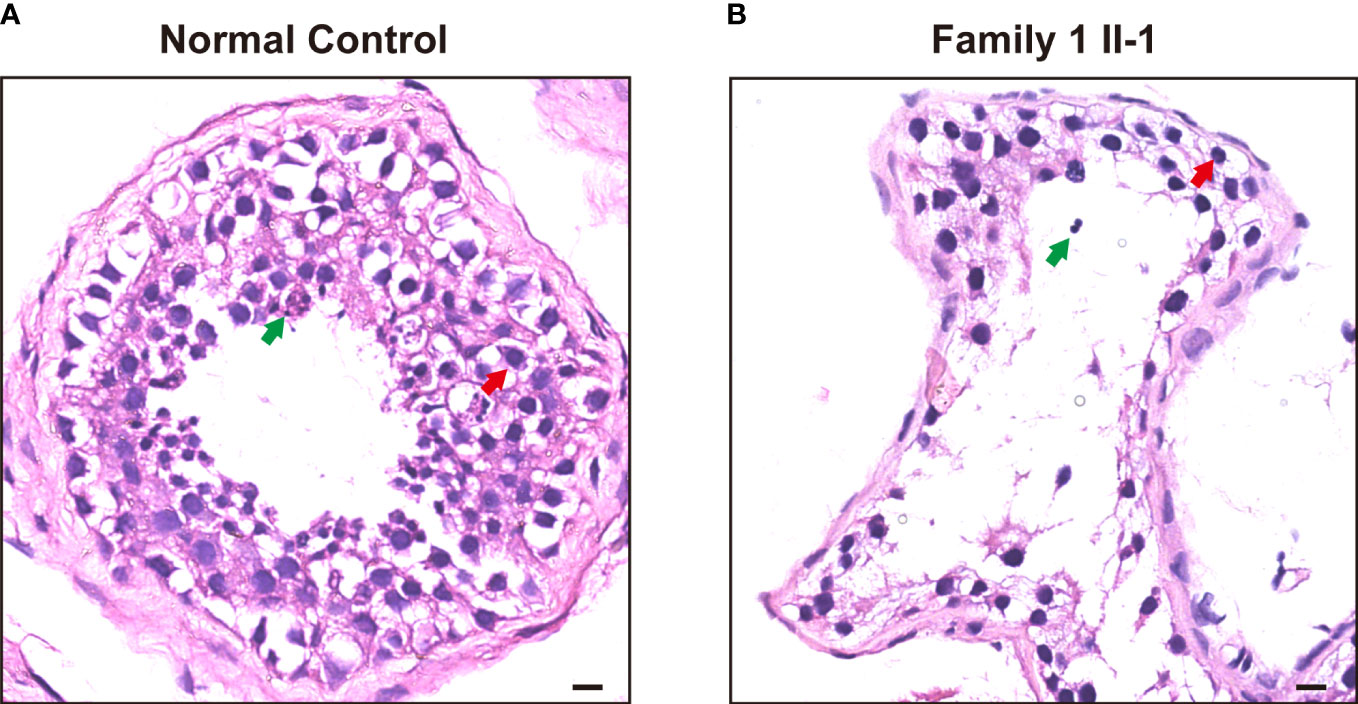
Figure 2 H&E staining of cross-sections in the subjects carrying hemizygous mutations in TAF7L and a patient with OA. (A) H&E staining of cross-sections of testicular biopsy from a patient with OA as the positive control and (B) the Family 1 II-1patient. Red arrows mark the spermatocyte. Green arrows mark the spermatozoa. Scale bars: 10μm.
To ascertain the fertilization capacity of sperm from patients with TAF7L variants, microinjection of spermatozoa into the mice oocytes were performed to demonstrate the capacity of spermatozoa derived from OAT-affected patients with TAF7L variants. Notably, the embryos from both TAF7L variants and control could develop into the two-cell stage. However, the two-cell rate was significantly reduced compared with the control (10% vs 64%) (Figure 3). Collectively, it is illustrated that the sperm from OAT affected patients with TAF7L variants showed reduced fertilization capacity compared to the normal sperm.
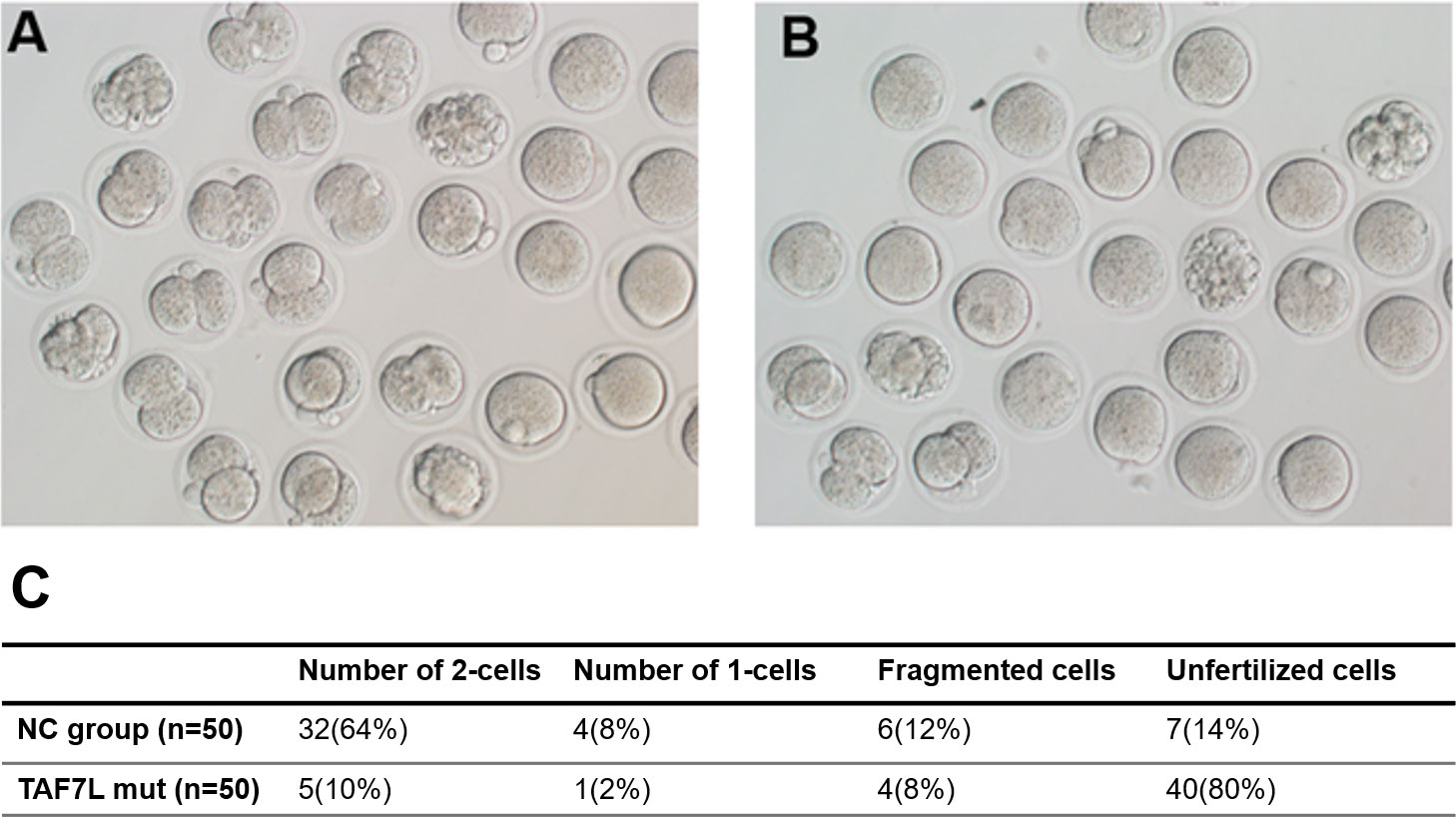
Figure 3 Embryos developmental potentials from oocytes of mouse fertilized with sperm carrying hemizygous mutations in TAF7L and the healthy man. (A) Two cells embryo formation from the ejaculated of healthy man. (B) Two cells embryo formation from ejaculated of the patient (Family 1 II-1) presented with OAT phenotype. (C) Statistics of pregnancy outcome from the healthy man and patient. NC: normal control.
The parameters for semen analysis of TAF7L variants were obtained from laboratories according to the WHO5th guidelines. Semen analysis indicated severe reduced sperm concentration in all of five men harboring hemizygous TAF7L variants (Table 3). Furthermore, Diff-Quik staining displayed sperm head deformity (Figure 4A). Moreover, their nuclei were more efficiently stained with acidic aniline (Figure 4A), implying less condensed chromatin in these sperm head. TEM verified that the heads of sperm were less condensed in comparison with the controls (Figure 4B). These observations indicated of defective chromatin compaction in the heads of sperm.
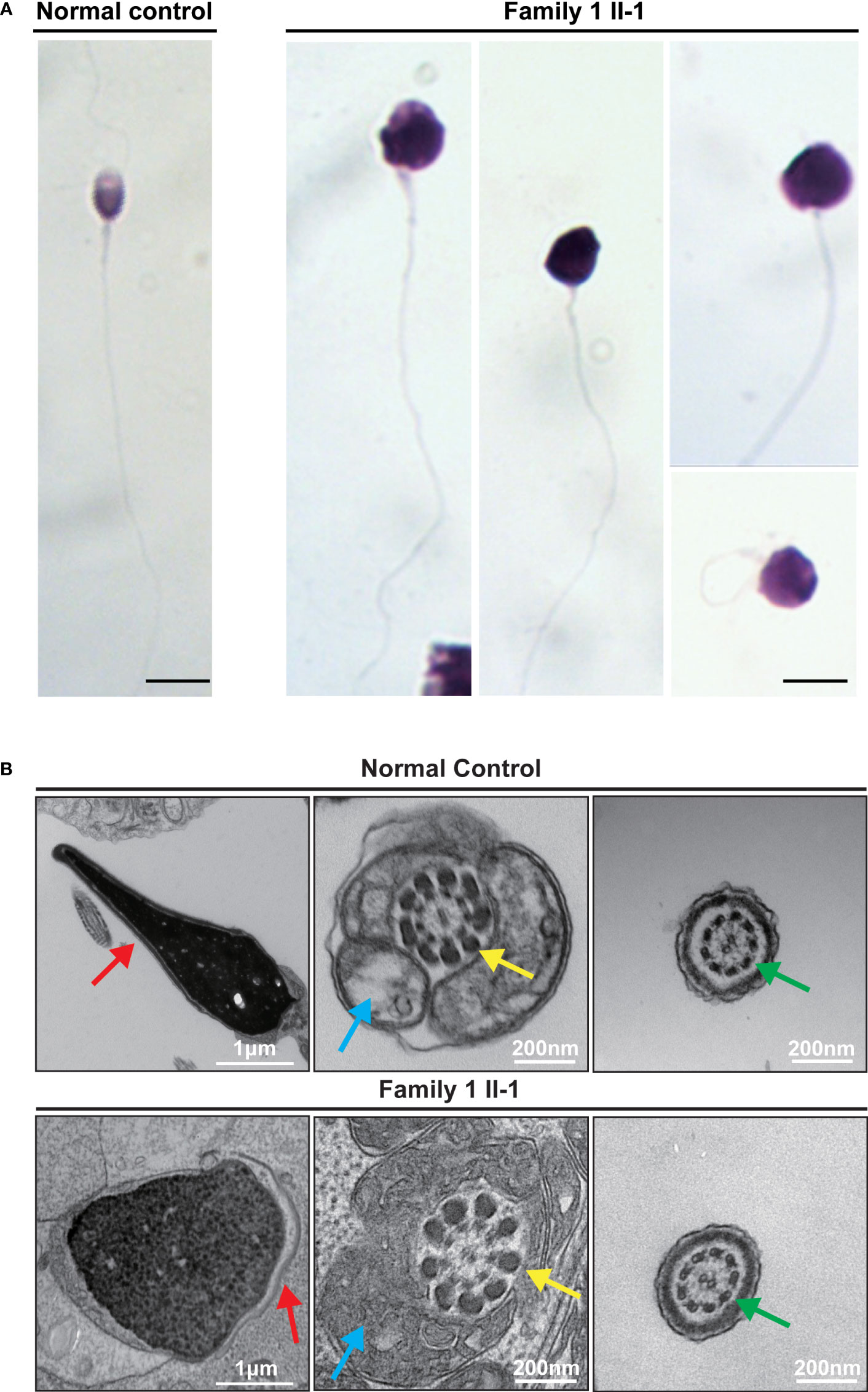
Figure 4 Sperm morphology and Ultrastructure Analysesin for the subjects carrying hemizygous mutations in TAF7L. (A) The morphology of the spermatozoa from a fertile control individual and subject Family 1 II-1 under light microscopy. Multiple images were taken, and typical features of abnormal spermatozoa are exemplified, such as abnormal head. Scale bars: 10 μm. (B) The longitudinal sections of sperm flagellar of a control individual. The normal sperm had a symmetrical mid-piece with smooth axoneme surrounding with a regularly arranged mitochondrial sheath. Family 1 II-1 sperm showed the less condensed head. Red arrows mark the acrosome. Blue arrows mark the mitochondrial sheath. Green arrows mark the axoneme. Yellow arrows mark the outer dense fibers.
To figure out the potential causes underlying defective chromatin compaction in the heads of sperm. Immunofluorescence staining verified abnormal retention of histone H3, and conversely, drastic reduced levels of protamine PRM2 in the heads of sperm (Family 1, M1: II-1) contrast to control (Figure 5).
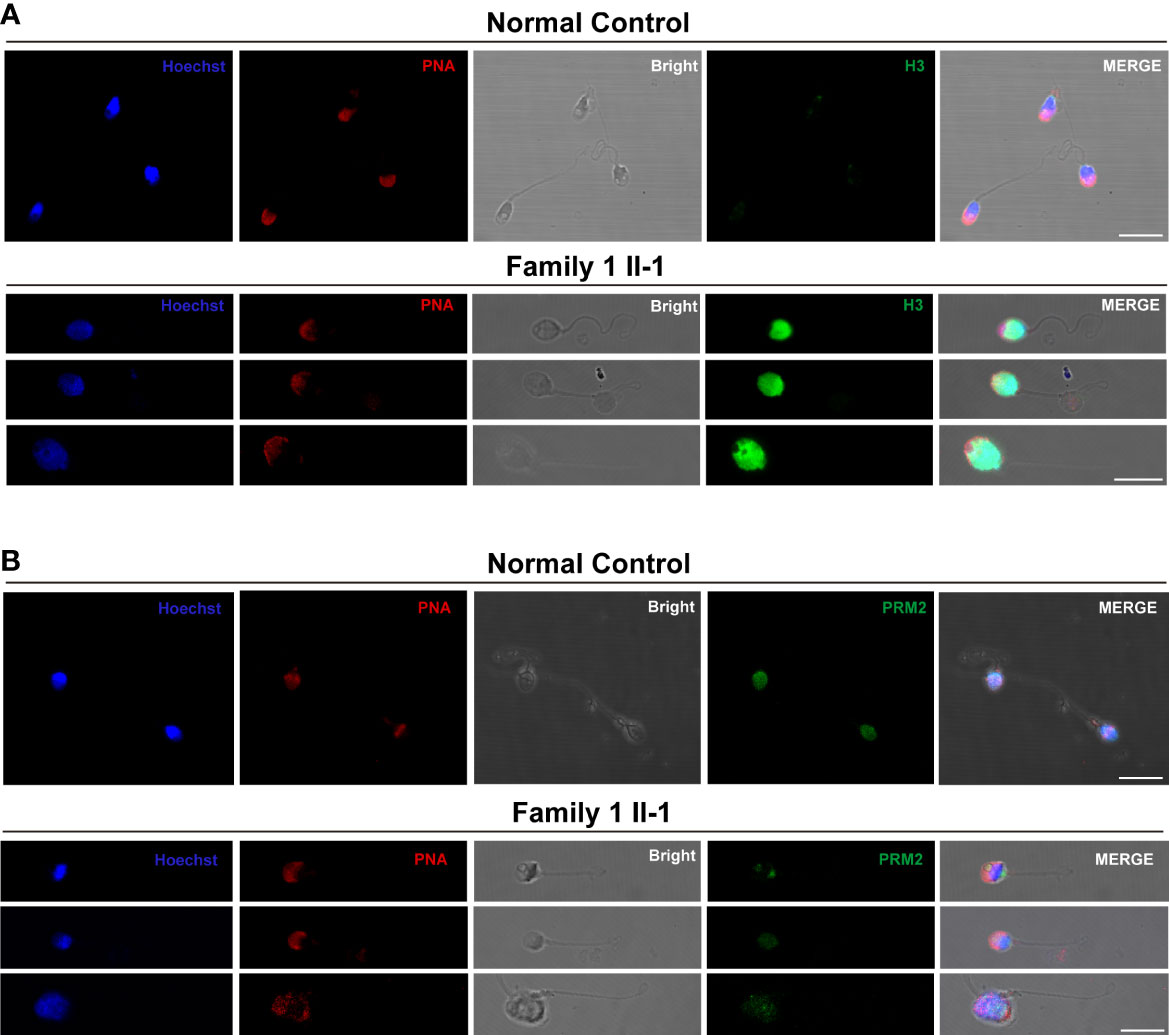
Figure 5 The Histone-to-Protamine Exchange Is Impaired during Spermiogenesis in the subjects carrying hemizygous mutations in TAF7L. (A) Representative confocal images of immunostaining with Hoechst (blue), PNA (red), Bright, and anti-H3 antibody (green) on sperm from TAF7L variants -affected patients and normal man. Scale bars: 10μm. (B) Representative confocal images of immunostaining with Hoechst (blue), PNA (red), Bright, and anti-PRM2 antibody (green) on sperm from TAF7L variants -affected patients and normal man. Scale bars: 10μm.
Subsequently, As shown in Figure 6, RT-PCR analysis revealed that the testicular tissue of patient (Family 1, M1: II-1) had significantly reduced expression of protamine PRM2 mRNA (p < 0.001). And these experiments indicated impaired histone-to-protamine exchange during spermiogenesis.

Figure 6 Differential abundance of the PRM1, PRM2, H2A and H3 transcripts in the subject carrying hemizygous mutations in TAF7L and normal men. (A–D) The mRNA content of PRM1 and PRM2 is significantly lower in TAF7L variants -affected patient compared with normal man. H2A and H3 mRNA ratio in TAF7L variants -affected patient samples are close to normal man. NS, not significant; **** indicates p <0.0001.
To determine the functional phenotypes of missense and LoF variants of human TAF7L variants in vitro. 293T cells were transiently transfected with the TAF7L wild-type and four mutant TAF7L plasmids with 3×FLAG tagged. Compared to wild-type, Immunofluorescence analysis showed that variant c.1301_1302del (p. V434Afs*5) and c.508delA (p. T170fs) had no immunofluorescence signal was detected above the background (Figures 7A, B, D), c. 719dupA (p. K240fs) signal was localized massively within the nucleus (Figure 7E). But variant c.699G>T;(p. R233S) location were no different compared to WT (Figure 7C). The WB data were similar with the IF results. The c. 1301_1302del (p. V434Afs*5), c. 508delA (p. T170fs) and c.719dupA (p. K240fs) mutations yielded a truncated protein, c. 1301_1302del (p. V434Afs*5) and c.508delA (p. T170fs) showed nearly undetectable expression protein level and that the c.719dupA (p. K240fs) mutation generated a normal expression truncated protein. There was no change in protein level for the c. 699G>T (p. R233S) mutation (Figure 7F).
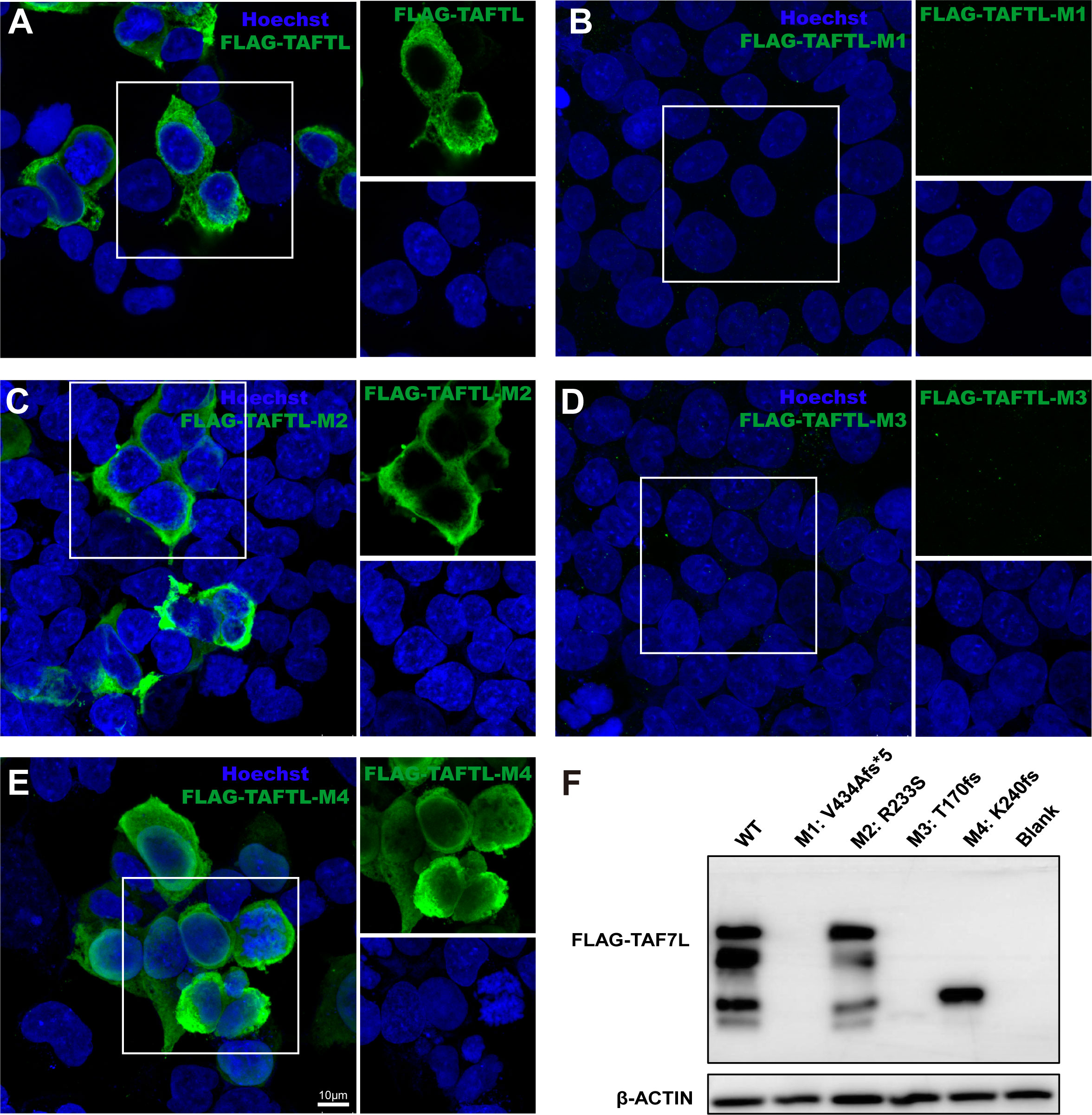
Figure 7 Effects of the TAF7L variants in in vitro. (A–E) Immunofluorescence analysis of TAF7L expression in HEK293T cell transfected with FLAG-tagged wild-type and mutant plasmids. (F) Western blot analysis of TAF7L-3×FLAG protein level in HEK293T transfected with wild-type and mutant plasmids.
As discussed above, we found hemizygous mutations of TAF7L among five non-related OAT patients (0.68%,5/725). The samples were collected from three different hospitals. The WES results showed that the subjects did not carry any other known pathogenic variant related to OAT genes except the hemizygous TAF7L mutation. Remarkably, all the TAF7L mutations were either completely missing in the public databases (1000 Genomes and gnomAD) or they showed low allelic frequencies. They were indicated to be potentially pathogenic by in-silico study. Therefore, it is needed to explore the OAT phenotypes in such cases by hemizygous deleterious variants in TAF7L.
TAF7L (also known as CT40) is a paralogue of TAF7 which is a subunit of TFIID and is specific to germ cells. TAF7L encodes a protein associated with transcription factors and is mainly expressed in testis (21). Previously it was found that mice deficient with TAF7L exhibited low fertility, abnormal morphology of sperm and reduced motility (22).Previous study mutation frequencies in Taf7l gene were likely to be polymorphisms in human. Only one study previously reported a deleterious mutation (D136G) in the X-linked TAF7L gene as a potential cause of oligozoospermia in men, but the morphology of sperm was not described in this study. And the corresponding D144G substitution in the mouse Taf7l gene does not affect male fertility (23).Therefore, this is a great clinical heterogeneity in man with Taf7l variants.
Herein, we identified four deleterious variants TAF7L in human. Instead of causing oligozoospermia, the mutation causes OAT, including a hemizygous frameshift mutation of TAF7L [c. 1301_1302del;(p. V434Afs*5)] in M1(Figure 1A), one hemizygous missense mutation [c. 699G>T;(p. R233S)] in M2-1 and M2-2, it was confirmed that this variant was inherited from their parents (Figure 1B). We also found the hemizygous frameshift mutation [c. 508delA;(p. T170fs), c. 719dupA;(p. K240fs)] in M3 and M4, respectively, (Figures 1C, D; Table 1). These four TAF7L variants were not found in the control group of human population genome databases and in silico analysis predicted as deleterious. In vitro tests confirmed the protein-truncating mutation c. 1301_1302del;(p. V434Afs*5), c. 508delA;(p. T170fs), c. 719dupA;(p. K240fs) causes abnormal expression of TAF7L.
No medicine is available for the treatment of OAT patients which can improve the quality of the ejaculated spermatozoa, which renders ICSI as the only solution for conception. However, studies have found that different mutations in various genes cause OAT with various types of clinical outcomes. For instance, biallelic mutations of CEP135 (MIM: 301057) resulted in pregnancy failure, while oligozoospermia due to CFAP47 (MIM:301057) mutation showed good clinical results (12). In this study, the outcomes of the ICSI treatment from three different hospitals showed that the number of blastocysts and two cells embryo formation by TAF7L variants was lower compared to the control.
In the current study, these three patients with TAF7L variants didn’t get clinical pregnancy using ejaculated sperm, two of them chose donor’s sperm for ICSI treatment. Ejaculated sperm of patient (Family 1, M1: II-1) without enough spermatozoa were recovered by testicular biopsy on the day of oocyte retrieval. The couple got successful clinical pregnancy at the time of writing (Table 2). Based on the current clinical evaluation of female fertility, we believe that their fertility were normal. Hence, this study supports the idea that male infertility could be because of hemizygous TAF7L variants due to the low fertilization capacity of sperm.
To further prove this hypothesis that the capacity of low spermatozoa derived from OAT patients with TAF7L variants, microinjection of spermatozoa into the mice oocytes was performed. The two-cell rate was significantly reduced than the control (10% vs 64%). This indicated the sperm from OAT patients with TAF7L variants showed reduced in-vitro fertilization capacity through ICSI treatment.
Why the low capacity of spermatozoa derived from OAT patients with TAF7L variants? In this study, we confirmed that the defective chromatin compaction in sperm head of proband (Family 1, M1: II-1). Previous study indicated that Taf7l and Trf2 coregulate the expression of protamine 1/2 (Prm1/2) genes, and ChIP-qPCR analysis confirmed dramatically diminished in Taf7l-null testes at target protamine 1/2 (Prm1/2) promoters (24). In this study, we found that abnormal retention of histone H3 and reduced levels of protamine PRM2 were in the heads of sperm. RT-PCR analysis revealed that the testicular tissue of patient (Family 1, M1: II-1) had significantly reduced expression of protamine PRM2 mRNA, So we speculated that the abnormal of histone-to-protamine exchange may induce the defective chromatin compaction of sperm head. Because the histone-to-protamine exchange is crucial for producing functional sperm, these results implied that a defect histone-to-protamine exchange may underlie the capacity deficiency of TAF7L variants sperm.
The datasets for this article are not publicly available due to concerns regarding participant/patient anonymity. Requests to access the datasets should be directed to the corresponding authors.
The research was approved by the corresponding ethics committees of the Shanghai General Hospital, Shanghai Jiao Tong University(2021-SQ-112), the Reproductive and Genetic Hospital of CITIC Xiangya (LL-SC-2017-025 and LL-SC-2019-034), and the Xiamen Maternity and Child Care Hospital, Xiamen (Permit Number: 2020SQ199, KY-2019-060)
CY, ZL and EZ designed the study, directed and supervised the research. HB, YS and YT collected the data. YZ, JX, SX, performed experiments. PL and ZJ have drafted the work. XW, WC, JZ enrolled the patients and collected clinical information. HB wrote the manuscript, with input from others. CY, ZL and EZ substantively revised it. All authors contributed to the article and approved the submitted version.
This study was supported by National Natural Science Foundation of China (Grant Nos. 82171586, 82071697, 32271163 and 81871200), the Medical Innovation Project of Fujian Province (Grant No. 2020CXB051).
We thank the patients and their family members for their support during this research study, and all the individuals who participated in and supported this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.1099270/full#supplementary-material
OAT, Oligoasthenoteratozoospermia; WES, Whole-exome sequencing; NOA, non-obstructive azoospermia; HCG, human chorionic gonadotropin; CZB, Chatot-Ziomek-Bavister; PNA, peanut agglutinin; TEM, Transmission electron microscopy analysis; ICSI, Intracytoplasmic sperm injection.
1. Curi SM, Ariagno JI, Chenlo PH, Mendeluk GR, Pugliese MN, Sardi Segovia LM, et al. Asthenozoospermia: Analysis of a large population. Arch Androl (2003) 49(5):343–9. doi: 10.1080/01485010390219656
2. Coutton C, Escoffier J, Martinez G, Arnoult C, Ray PF. Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod Update (2015) 21(4):455–85. doi: 10.1093/humupd/dmv020
3. Tang S, Wang X, Li W, Yang X, Li Z, Liu W, et al. Biallelic mutations in CFAP43 and CFAP44 cause Male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet (2017) 100(6):854–64. doi: 10.1016/j.ajhg.2017.04.012
4. McLachlan RI. Approach to the patient with oligozoospermia. J Clin Endocrinol Metab (2013) 98(3):873–80. doi: 10.1210/jc.2012-3650
5. Liu C, He X, Liu W, Yang S, Wang L, Li W, et al. Bi-allelic mutations in TTC29 cause Male subfertility with asthenoteratospermia in humans and mice. Am J Hum Genet (2019) 105(6):1168–81. doi: 10.1016/j.ajhg.2019.10.010
6. Ghedir H, Gribaa M, Mamai O, Ben Charfeddine I, Braham A, Amara A, et al. Macrozoospermia: screening for the homozygous c.144delC mutation in AURKC gene in infertile men and estimation of its heterozygosity frequency in the Tunisian population. J Assist Reprod Genet (2015) 32(11):1651–8. doi: 10.1007/s10815-015-0565-4
7. ElInati E, Fossard C, Okutman O, Ghedir H, Ibala-Romdhane S, Ray PF, et al. A new mutation identified in SPATA16 in two globozoospermic patients. J Assist Reprod Genet (2016) 33(6):815–20. doi: 10.1007/s10815-016-0715-3
8. Shang YL, Zhu FX, Yan J, Chen L, Tang WH, Xiao S, et al. Novel DPY19L2 variants in globozoospermic patients and the overcoming this male infertility. Asian J Androl (2019) 21(2):183–9. doi: 10.4103/aja.aja_79_18
9. Ben Khelifa M, Coutton C, Zouari R, Karaouzene T, Rendu J, Bidart M, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet (2014) 94(1):95–104. doi: 10.1016/j.ajhg.2013.11.017
10. Sha Y, Chen Y, Wang X, Meng R, Yang X, Li Y, et al. Biallelic mutations in IQCN, encoding a novel acroplaxome protein, lead to fertilization failure and male infertility with defects in the acrosome and shaping of the spermatid head in humans and mice. Life Med (2022) lnac050. doi: 10.1093/lifemedi/lnac050
11. Liu C, Miyata H, Gao Y, Sha Y, Tang S, Xu Z, et al. Bi-allelic DNAH8 variants lead to multiple morphological abnormalities of the sperm flagella and primary Male infertility. Am J Hum Genet (2020) 107(2):330–41. doi: 10.1016/j.ajhg.2020.06.004
12. Liu C, Tu C, Wang L, Wu H, Houston BJ, Mastrorosa FK, et al. Deleterious variants in X-linked CFAP47 induce asthenoteratozoospermia and primary male infertility. Am J Hum Genet (2021) 108(2):309–23. doi: 10.1016/j.ajhg.2021.01.002
13. Lv M, Liu W, Chi W, Ni X, Wang J, Cheng H, et al. Homozygous mutations in DZIP1 can induce asthenoteratospermia with severe MMAF. J Med Genet (2020) 57(7):445–53. doi: 10.1136/jmedgenet-2019-106479
14. Tu C, Cong J, Zhang Q, He X, Zheng R, Yang X, et al. Bi-allelic mutations of DNAH10 cause primary male infertility with asthenoteratozoospermia in humans and mice. Am J Hum Genet (2021) 108(8):1466–77. doi: 10.1016/j.ajhg.2021.06.010
15. Vockel M, Riera-Escamilla A, Tuttelmann F, Krausz C. The X chromosome and male infertility. Hum Genet (2021) 140(1):203–15. doi: 10.1007/s00439-019-02101-w
16. Vogt PH, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, Kiesewetter F, et al. Human y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet (1996) 5(7):933–43. doi: 10.1093/hmg/5.7.933
17. Boroujeni PB, Sabbaghian M, Totonchi M, Sodeifi N, Sarkardeh H, Samadian A, et al. Expression analysis of genes encoding TEX11, TEX12, TEX14 and TEX15 in testis tissues of men with non-obstructive azoospermia. JBRA Assist Reprod (2018) 22(3):185–92. doi: 10.5935/1518-0557.20180030
18. Yatsenko AN, Georgiadis AP, Ropke A, Berman AJ, Jaffe T, Olszewska M, et al. X-Linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med (2015) 372(22):2097–107. doi: 10.1056/NEJMoa1406192
19. Zhang DL, Sun YJ, Ma ML, Wang YJ, Lin H, Li RR, et al. Gq activity- and beta-arrestin-1 scaffolding-mediated ADGRG2/CFTR coupling are required for male fertility. Elife (2018) 7:e33432. doi: 10.7554/eLife.33432
20. Yang S, Ping P, Ma M, Li P, Tian R, Yang H, et al. Generation of haploid spermatids with fertilization and development capacity from human spermatogonial stem cells of cryptorchid patients. Stem Cell Rep (2014) 3(4):663–75. doi: 10.1016/j.stemcr.2014.08.004
21. Akinloye O, Gromoll J, Callies C, Nieschlag E, Simoni M. Mutation analysis of the X-chromosome linked, testis-specific TAF7L gene in spermatogenic failure. Andrologia (2007) 39(5):190–5. doi: 10.1111/j.1439-0272.2007.00789.x
22. Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, et al. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci (2003) 116(Pt 9):1847–58. doi: 10.1242/jcs.00391
23. Ling L, Li F, Yang P, Oates RD, Silber S, Kurischko C, et al. Genetic characterization of a missense mutation in the X-linked TAF7L gene identified in an oligozoospermic man. Biol Reprod (2022) 107(1):157–67. doi: 10.1093/biolre/ioac093
Keywords: TAF7L, hemizygous variant, oligoasthenoteratozoospermia, ICSI, male infertility
Citation: Bai H, Sha Y, Tan Y, Li P, Zhang Y, Xu J, Xu S, Ji Z, Wang X, Chen W, Zhang J, Yao C, Li Z and Zhi E (2023) Deleterious variants in TAF7L cause human oligoasthenoteratozoospermia and its impairing histone to protamine exchange inducing reduced in vitro fertilization. Front. Endocrinol. 13:1099270. doi: 10.3389/fendo.2022.1099270
Received: 15 November 2022; Accepted: 05 December 2022;
Published: 11 January 2023.
Edited by:
Tao Luo, Nanchang University, ChinaReviewed by:
Yang Xiaoyu, Nanjing Medical University, ChinaCopyright © 2023 Bai, Sha, Tan, Li, Zhang, Xu, Xu, Ji, Wang, Chen, Zhang, Yao, Li and Zhi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chencheng Yao, eWFvY2hlbmNoZW5nQDEyNi5jb20=; Zheng Li, bGl6aGVuZ2Jvc2hpQDE2My5jb20=; Erlei Zhi, emh6emVyMTk4NUBhbHVtbmkuc2p0dS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.