
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol., 16 December 2022
Sec. Cellular Endocrinology
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.1074911
This article is part of the Research TopicA Year in Review: Discussions in Cellular EndocrinologyView all 6 articles
Peroxisome proliferator-activated receptor α (PPARα) is a ligand-activated transcription factor that is involved in lipid metabolism of various tissues. Different metabolites of fatty acids and agonists like fibrates activate PPARα for its transactivative or repressive function. PPARα is known to affect diverse human diseases, and we focus on advanced studies of its transcriptional regulation in these diseases. In MAFLD, PPARα shows a protective function with its upregulation of lipid oxidation and mitochondrial biogenesis and transcriptional repression of inflammatory genes, which is similar in Alzheimer’s disease and cardiovascular disease. Activation of PPARα also prevents the progress of diabetes complications; however, its role in diabetes and cancers remains uncertain. Some PPARα-specific agonists, such as Wy14643 and fenofibrate, have been applied in metabolic syndrome treatment, which might own potential in wider application. Future studies may further explore the functions and interventions of PPARα in cancer, diabetes, immunological diseases, and neurodegenerative disease.
Peroxisome proliferator-activated receptor α (PPARα) is a ligand-activated transcription factor belonging to the NR1C nuclear receptor subfamily. Together with PPARγ and PPARβ/δ, all PPARs are responsible for the metabolism of lipid and glucose, as well as cell proliferation and differentiation, inflammation, vascular biology, and cancer (1). The expression of the three PPARs is quite different between organs, indicating their distinct physiological roles (2). PPARα is highly expressed in hepatocytes, cardiomyocytes, proximal renal tubular cells, and brown adipocytes. PPARβ/δ is more ubiquitous but mainly found in skeletal muscle, skin, adipose tissue, heart, liver, and inflammatory cells, whereas the localization of PPARγ is wider.
As the first identified member of the family in 1990, PPARα was found activated by a diverse class of rodent hepatocarcinogens that causes proliferation of peroxisomes (3). Subsequently, the other two members were identified and this family was verified as transcriptional factors (4, 5). PPARs are activated by various ligands, including different metabolites of fatty acids. However, most of the ligands and agonists seem not very selective, partly resulting from the highly similar structure of this family (2).
PPARα, which was firstly found as a member of the steroid hormone receptor superfamily of ligand-activated transcription factors, is mainly involved in regulation of lipid oxidation. Because lipid and even energy homeostasis maintains the function of the whole body, PPARα is logically associated with various diseases. Meanwhile, the decrease of PPARα is also found in various diseases including MAFLD, diabetes, Alzheimer’s disease, and cardiovascular disease (6–9), emphasizing the key role of PPARα in human diseases. This review summarized the recent findings of PPARα in metabolic syndrome, Alzheimer’s disease, and cardiovascular disease and discussed unsolved questions of the role of PPARα in cancers.
The PPARα protein possesses five main functional domains embodied in a modular canonical structure, namely, the activation function-1 (AF-1), the DNA-binding domain (DBD), the hinge region, the ligand-binding domain (LBD), and activation function 2 (AF-2) (5). The N-amino terminal end harbors AF-1 that operates autonomously in a ligand-independent manner. DBD consists of two highly conserved zinc finger-like motifs that promote the receptor’s binding to the PPRE sequence of the target genes, localizing in gene regulatory regions and organized as direct repeats of two hexamer core sequences AGG(A/T)CA. The hinge region that bridges the DBD to the LBD acts as a docking site for cofactors. In the C-terminal region, the LBD is responsible for ligand specificity and contains AF-2, where the ligand-containing LDB stabilizes and facilitates the interface of AF-2 so that PPARα can recruit co-activators (10).
It is worthy to notice that the PPAR family, including PPARα/β/γ, binds PPREs uniquely as heterodimers with the retinoid X receptor (RXR) (11). PPRE contains two core sequences separated by one nucleotide (DR-1), which provides a polarization signal to the PPAR/RXR heterodimer, whereas PPARs interact with 5′-extended hexamers and RXR binds to the downstream motif (12). Ligand-activated PPARα also recruits numerous co-activator proteins to form the transcriptionally active PPARα-interacting cofactor complex, depending on the AF-2 domain of PPARα (13). The complex contains members of the CBP/p300 and SRC/p160 family exhibiting HAT activity and the large complex of PBP/MED1 for transcription (14).
PPARα recognizes PPRE to activate transcription; however, it is interesting that almost half of the PPARα-binding regions in human hepatocytes are located within introns, whereas only 26% was in the promoter region (<2.5 kb). In addition, overlap chromatin binding regions of LXR–RXR and PPARα–RXR and co-enrichment of PPARα-binding regions in C/EBPα and TBP motifs together suggest that PPARα may influence gene expression through the formation of complexes (15).
PPARα mainly regulates the expression of genes involved in fatty acid transport and oxidation to control lipid homeostasis. Hepatic PPARα activity controls the expression of apolipoprotein, as functional PPRE has been identified in the promoters of the LPL, APOA5, APOA1, and APOA2 genes (16), whereas PPARα regulates ABCA1 in macrophage and intestine. Fatty acid oxidation in the liver and brown adipose tissue is also affected by PPARα-mediated transactivation. The expressions of Acox1, Cpt1, and Ehhadh, three important genes in mitochondrial fatty acid β-oxidation, are directly enhanced by PPARα (15). Moreover, Fgf21, a secretory hepatic factor participating in regulation of energy balance, is also a target of PPARα (17).
The models of PPARα transcriptional repression include PPRE-dependent or independent patterns. The independent manner of negative regulation is via protein–protein interactions, where a well-known example is that PPARα represses pro-inflammatory signaling pathways in acute inflammation. There are direct physical interactions between PPARα, the p65 Rel homology domain, and the N-terminus JNK-responsive part of cJun, which reduces IL-6 gene expression through the AP-1 and NF-κB signaling pathways (18). Moreover, ligand activation of GR and PPARα leads to the enhanced repression of IL-6 transcriptional activity, by the mechanism that stems from a direct GR–PPARα physical interaction (12). Another mechanism of PPRE-independent transcriptional repression occurs in the ERR-driven mitochondrial respiration due to PPARα–SIRT1 complex competitive binding to the hexameric ERRE motif with ERRs (19).
A novel model of PPRE-dependent transcriptional regulation has also been proposed on the repression of IL-6 expression. Through the physical interaction between PPRE and p65, PPARα abolishes p65 binding to the upstream NF-κB response element on the complement C3 promoter (20). These studies support the complex pattern of PPARα-mediated translational regulation.
The expression of PPARα, as an important nuclear factor in metabolic regulation, is reported under the effect of metabolites. Exposure of β cells to elevated glucose rapidly decreases PPARα gene expression (21), which is in requirement of phosphorylation of the sugar. A further study shows that AMPK, the energy sensor, activates the expression of PPARα, which is implicated in high glucose conditions (22). Despite that glucose and polyunsaturated fatty acid improve PPARα expression in different organs which have been reported (23, 24), the pattern is complex. Fatty acids show specific alterations on PPARα gene expression, whereas its mRNA expression was upregulated by SFA, MUFA, ALA, ARA, and DHA and downregulated by LNA and EPA (25). Recently, a human APP-dependent expression of PPARα in brains has also been found in AD patients, although the regulatory pattern is still unknown (8). Posttranscriptional regulation is also reported to play a role in PPARα expression, where miR20b suppresses PPARα expression by directly targeting its mRNA (26).
It has been reported since 1996 that PPARα could be phosphorylated (27), and following research has shown that phosphorylation of PPARα activates its transcriptional function. Phosphorylation of two serine sites, S12/S21, correlates with increased transactivation of PPARα in hepatocytes and cardiac myocytes, potentially via decreased co-repressor interaction with NCoR or increased interaction with a certain co-activator, PGC1α (28). These two sites are both targeted by mitogen-activated protein kinases (MAPKs) and cyclin-dependent kinase 7 (CDK7), which is associated with reduced adipose mass and increased energy expenditure (29, 30). Two sites in the hinge region, Ser 179 and 230, are reported to be phosphorylated in the PKC-dependent pathway, also participating in PPARα transcriptional activity (31). Moreover, some studies also show that phosphorylation of PPARα is associated with protein stability (32). S73 phosphorylation, an important event mediated by glycogen synthase kinase β (GSKβ), leads to the degradation of PPARα (33). Gilbert’s syndrome, a mouse model that shows the protective effect against hepatic steatosis, might be mediated by increased PPARα protein levels due to the reduction of S73 phosphorylation (34).
Poly-ubiquitination and the proteasome pathway also mediate the degradation of PPARα. Early findings implicated the E3 ligase MDM2 in the regulation of PPARα protein stability (35). Recently, the E3 ubiquitin ligase HUWE1 has been reported to affect PPARα stability to control hepatic fatty acid oxidation (36). Two members of the progestin and adipoQ receptor (PAQR) family, PAQR3 and PAQR9, display the regulatory function on HUWE1 combination with PPARα (37). PAQR3 pulled PPARα to Golgi apparently bound by HUWE1, whereas PAQR9 competitively combined with HUWE1 to avoid PPARα degradation. Except the regulation of protein stability, another research finds that the muscle-specific ubiquitin ligase MuRF1 can modify PPARα with mono-ubiquitination, leading to the decreased activity of PPARα due to its export from the nucleus (38).
Two lysine residues of PPARα, K185 and K358, have been reported to be subjected to SUMOylation (28). The modification of both residues increases the repressive ability of PPARα through enhanced co-repressor recruitment; however, K358 SUMOylation only occurs in female livers, suggesting a role in sexual dimorphism (39). Moreover, methylation of PPARα has recently been reported in neurons, which might affect protein stability (40).
As reviewed above that the LBD domain recognizes and binds ligands, the AF-2 helix is tightly packed against the LBD core for PPARα activation. Crystallography identifies tyrosine 314 as the main determinant of isotype ligand specificity (41), which affects the interaction with cofactors for transcriptional regulation. The PPARα ligands are fatty acid derivatives formed during various metabolic pathways including lipolysis, lipogenesis, and FA catabolism. Liver-specific knockout of fatty acid synthase (FAS), an enzyme catalyzing the synthesis of FA, results in NASH which could be reversed by PPARα agonists, identifying products of FAS as PPARα activators (42), which further reported phospholipid as a FAS-dependent lipid intermediate PPARα ligand. Because disruption of ACOX1 results in elevated PPARα target gene expression, substrates of ACOX-1 are likely PPARα endogenous agonists (43). Moreover, ATGL-dependent hydrolysis of TG also yields lipid PPARα ligands (44). All these studies support lipids as the endogenous ligand of PPARα, which suggests the balance regulatory function of PPARα in lipid metabolism.
Several chemical agonists have also been developed, at least 30 kinds of PPARα agonists or antagonists according to MCE (https://www.medchemexpress.cn/). Fibrates, including gemfibrozil, fenofibrate, and ciprofibrate, are clinically used in the treatment of primary hypertriglyceridemia. However, it is noted that fibrates are weak PPARα agonists and their selectivity should be concerned, especially fibrates which might also activate PPARγ and δ and even other proteins like NRF2. In some studies, fenofibrate is found to interact with over 80 proteins (45). Moreover, the potency of synthetic PPARα agonists may differ between human and mouse receptors, such as EC50 = 18,000 nM of fenofibrate in mouse but 30,000 in human (46). Wy14643 is another typical agonist of PPARα reversing insulin resistance and hepatic steatosis (47), although it also has the disadvantage of fibrates. Nevertheless, some dual-PPAR agonists like fenofibrate and saroglitazar have also been shown effective in clinical treatment (48). Some potent and selective PPARα modulators (SPPARMs), such as K-877, GW9578, and elafibranor, are currently under development for the treatment of NAFLD and diabetes (49), respectively.
NAFLD is the liver manifestation of the metabolic syndrome and includes the spectrum of liver steatosis (known as non-alcoholic fatty liver, NAFL) and steatohepatitis (known as non-alcoholic steatohepatitis, NASH) (50). With an increasing epidemic of obesity worldwide, the estimated global prevalence of NAFLD is over 25% (51). NAFLD is a consequence of caloric overload, which is commonly referred to as the hepatic manifestation of the metabolic syndrome. On the other side, NAFLD is strongly associated with several core components of metabolic syndrome including obesity, insulin resistance or T2DM, and dyslipidemia (52). Considering the pathogenesis and multiplicate clinical indications, metabolic dysfunction-associated fatty liver disease (MAFLD) has recently been suggested as a more appropriate overarching term (53, 54). For a more accurate description, in the following part we would use the term “MAFLD.”
As PPARα regulates lipid metabolism and the main ligands for PPARα are fatty acids, the function of this transcriptional factor in lipid accumulation was noticed early. In the fasting state, increased fatty acid oxidation produces acetyl-CoA and promotes ketone body biogenesis, which is upregulated by PPARα (37). PPARα-deficient mice display impaired fatty acid oxidation, lipid accumulation in the liver, and an inability to augment ketone body synthesis during fasting, which indicates that PPARα is critically involved in the fasting state. Furthermore, transcriptional analysis shows that PPARα in the liver regulates fatty acid transport, peroxisomal and mitochondrial β-oxidation, and lipolysis and influences the production of apolipoproteins (55), which suggests its key role in MAFLD (Figure 1). Clinical data showed that liver PPARα expression inversely correlates with NASH severity, and importantly, histological improvement is associated with an increase in expression of PPARα and its target genes (6). Mouse models are in line with these experimental findings, indicating that whole-body or hepatocyte-specific deletion of PPARα promotes MAFLD in the context of obesity (56). Moreover, in preclinical models, pharmacological activation of PPARα has preventive and curative effects on NASH due to activation of hepatic transport, oxidation, and metabolism of lipids (16, 57). Mitochondrial function is also impaired in the livers of patients with NASH, who have increased hepatic oxidative stress (58). PPARα protects the liver from ROS overload via hydrogen peroxide detoxification and decreases hepatic ROS pools by upregulating catalase expression (16). Activation of PPARα expression by KLF16 could improve steatohepatitis and insulin resistance through ROS reduction (59). Moreover, some compounds for MAFLD, such as geniposide and fenofibrates, have recently been reported to elevate ROS levels through activating PPARα expression (60). Some studies investigate that the expression of PPARα affects fibrosis by the collagen-associated pathway. PPARα regulates NASH-related fibrogenesis through dermatopontin, which is a protein involved in fibrogenesis and collagen deposition, and its expression is lowered by PPARα activation.
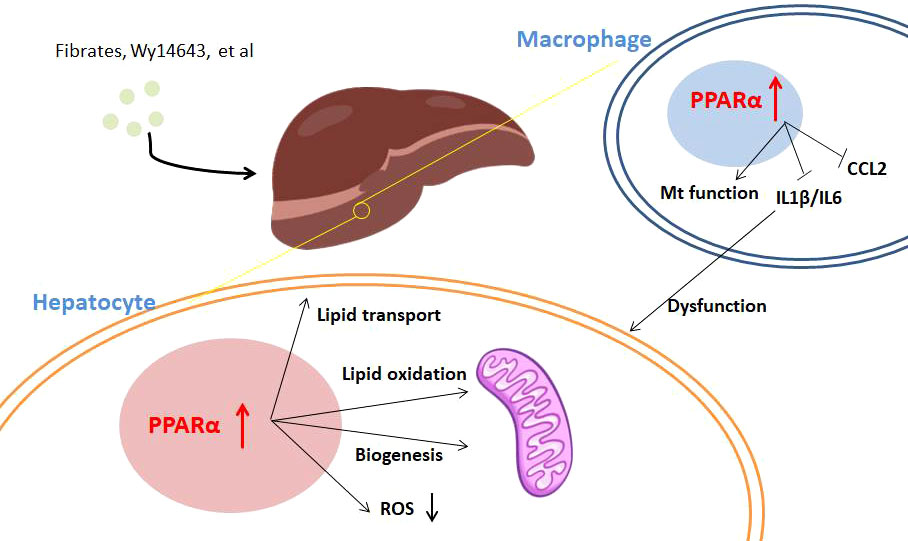
Figure 1 The regulation of PPARα in MAFLD. Activation of PPARα with agonists including fibrates and Wy14643 influences hepatocytes and immune cells in the liver. In hepatocytes, PPARα agonism promotes lipid transport and oxidation for lipid clearance; moreover, PPARα activation leads to ROS reduction. PPARα agonists inhibit proinflammatory cytokine production of immune cells in the liver, which protects against dysfunction of hepatocytes.
Despite hepatocytes, immune cells take part in MAFLD progress especially fibrosis. On the one hand, immune activation affects the PPARα signaling pathway. Activation of the JNK pathway, which drives HFD-induced insulin resistance, increases the expression of Ncor1 and Nrip1 co-repressors to negatively regulate PPARα target expression (61). In accordance with this theory, hepatic JNK deficiency in HFD-fed mice leads to increased expression of Fgf21 for improving systemic metabolism. On the other hand, different from transactivation of lipid metabolism in hepatocytes, PPARα shows repressive function in the expression of immune genes, as discussed above in PPARα transcriptional repression. PPARα agonism resulted in reduced numbers of activated macrophages, decreased levels of IL-1β and IL-6, and improved histological evidence of liver dysfunction and endothelial function (13, 62). Pan-PPAR agonists may counteract inflammation and NASH disease progression potently (63). Moreover, PPARα agonist Wy14643 treatment could alleviate steatosis and injury of the liver and decrease the level of chemokine CCL2 (64). However, clinical assessment of the effect of PPARα selective agonists on NASH and fibrosis is still lacking.
Interestingly, as we discussed above that PPARα agonists might activate other targets, some investigators think that these targets are also beneficial to MAFLD treatment. Transcriptional factors like PPARγ and SREBP-1c are also found to be modulated by fenofibrate (65). In some ways, combining different targets, like PPAR-γ and PPAR-α combined agonist therapy, is thought to be effective in controlling fructose-induced NASH (66) and pan-PPAR agonists are found to improve in MAFLD treatment (48, 63). However, the selective and pan agonists show different functions in some research and their availability and security need further control study.
The mechanism that PPARα decreases as NASH progresses remains to lead to different theories. Although the JNK pathway has been shown to decrease PPARα expression, the regulation in the early stage of NASH might be complicated. Epigenetic mechanisms such as H3K9me3 and H3K4me3 signatures being altered in the mouse hepatic PPARα promoter might be involved in this downregulation in the model of NASH (67). Posttranscriptional silencing of PPARα is also reported to occur in hepatocytes, via miR10b or miR21, whose expression is enhanced during NASH (16).
PPARα has been known to affect progress of type 2 diabetes. Some polymorphisms such as L162V and A268V were focused in diabetic patients, whereas L162V was early found to be associated with diabetes (68, 69). However, following genetic research showed that this mutation is associated with body mass index in patients with non-insulin-dependent diabetes mellitus (70) and fasting serum cholesterol concentrations (71), not directly affecting diabetes. Nevertheless, PPARα agonists show potential in diabetes treatment. PPARα agonists, mainly fenofibrate and Wy14643, improve glucose homeostasis by enhancing insulin sensitivity in adipose tissue and muscle (62, 72), which is probably the result of decreased lipid content in tissues by improving fatty acid β-oxidation. In addition, PPARα agonists can preserve pancreatic β-cell function, indicating that PPARα influences glucose homeostasis in part via effects on pancreas function (73). However, PPARα knockout also shows protective function on insulin resistance whereas fibrates do not seem to improve glucose homeostasis in humans (74, 75), making it confusing what role PPARα plays in glucose homeostasis.
It is worth noting that browning of white adipocytes is thought to own the capability to counteract diabetes, which promotes lipid oxidation and glucose metabolism of the whole body (76, 77). Some early research showed that agonists for PPARα and PPARγ both affect brown adipocyte function, whereas 16 shared PPARα/γ target genes, like Ctsz, were found to regulate brown adipocyte thermogenesis (78). Activation of PPARα is reported to increase energy expenditure and insulin sensitivity in obese mice (79); meanwhile, fenofibrate is also responsible for countering brown adipose tissue whitening (80). In this aspect, PPARα activation is beneficial to glucose homeostasis through control of obesity (Figure 2).
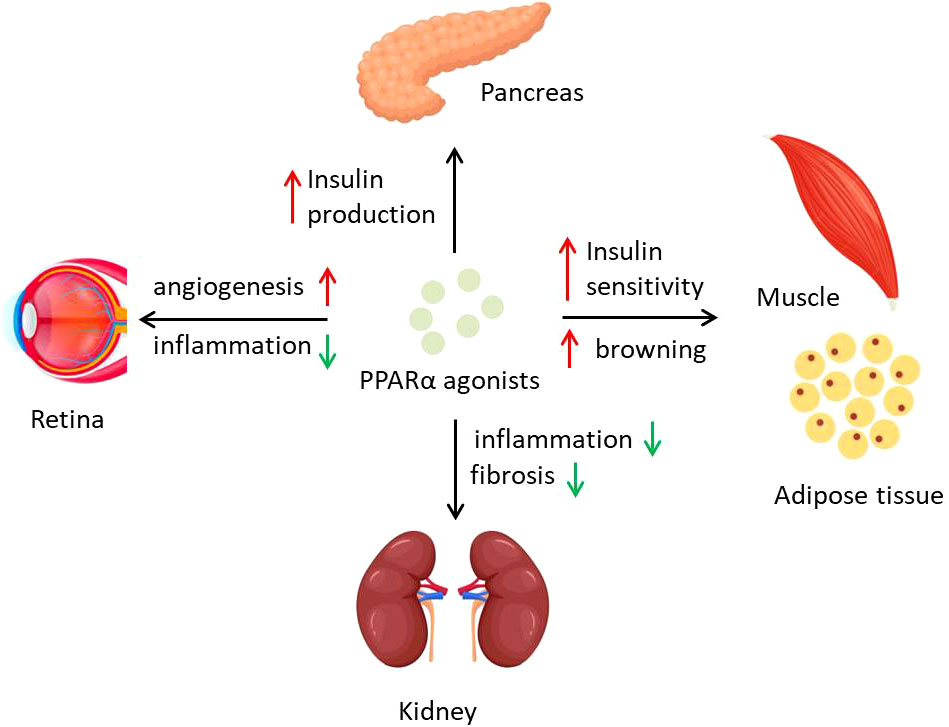
Figure 2 PPARα activation regulates diabetes and diabetic complications. PPARα agonists affect different organs to counter diabetes and its complications. PPARα activation preserves islets function of insulin production and promotes energy expenditure of adipose tissue and insulin sensitivity of muscle. Inflammation in kidney and retina could be inhibited by PPARα agonists, which prevent the process of DN and DR.
Complex diabetes complications lead to poor prognosis of diabetic patients and even high risk of death. Some complications that resulted from hyperglycemia, including atherosclerosis, retinopathy, and diabetic nephropathy, are associated with PPARα function. Some studies point that PPARα expression is downregulated during diabetes, mediated by not only transcriptional regulation but also posttranslational modification (9, 40). Early research reported that naturally occurring variations of PPARα function influenced plasma lipid concentrations in type II diabetic patients but not healthy people, demonstrating that PPARα is a link between diabetes and dyslipidemia (68). Activation of PPARα by berberine is thought to attenuate diabetic atherosclerosis in ApoE-/- mice (81). Fenofibrate as a PPARα agonist is used in various diabetic complications. A clinical research, field study showed that treatment with fenofibrate in individuals with type 2 diabetes mellitus reduces the need for laser treatment for diabetic retinopathy (DR) by 37%, although its mechanism might not be related to lipid oxidation but to angiogenesis (82). Following research showed that endothelial colony-forming cells (ECFCs) from PPARα−/− mice displayed impaired proliferation and migration, whereas activation of PPARα by fenofibrate normalized retinal vascular degeneration (9). Another study on type 1 diabetes indicated that intraocular injection of fenofibrate ameliorates retinal inflammation in OIR rats, and these therapeutic effects on DR are PPARα dependent, suggesting PPARα as a potential target of DR cure (83). The mechanisms on why PPARα activation could reverse the DR might be partly explained by promoter methylation and protein degradation in high glucose conditions (40). PPARα has also been recently found to play roles in progress of diabetic nephropathy (DN). Lipid accumulation and metabolism are tightly associated with DN progress (84), when PPARα deficiency appears to aggravate the severity of DN through an increase in extracellular matrix formation and inflammation (85). PPARα alleviating DN seems to be mostly through alterations of inflammation, like adiponectin exerting renoprotective effects against DN by activating AMPK-PPARα (86). In addition, Annexin A1 in diabetic mice regulates the AMPK-PPARα-CPT1 pathway to attenuate inflammation in the pathogenesis of DN (87), whereas fenofibrate could also attenuate renal fibrosis through blocking the canonic Wnt signaling and activating the antioxidant effects (88), together indicating the important role of PPARα in DN.
Recently, there has been growing concern for the function of PPARα in the brain. PPARα protein was observed to localize in different regions of the hippocampus including CA1, CA2, CA3, and dentate gyrus (89), suggesting its role in neurodegenerative disorders. Alzheimer’s disease (AD) is one of progressive neurodegenerative diseases with classic memory impairment and cognitive disorder, where genomic locus-encoding proteins for lipid metabolism showed involvement in disease regulation (90). Some early research reported an association of the PPARα L162V polymorphism with AD risk (91), whereas recently the expression and transcriptional activity of PPARα have been found to correlate with the expression of hAPP (8), which is thought to be one of the main causes of AD. On the other side, diverse activation of PPARα has been reported to weaken AD progress. A combination of low-dose gemfibrozil and retinoic acid could induce lysosomal biogenesis through the PPARα pathway and enhance the uptake of Aβ in astrocytes to alleviate AD (92). Amyloid pathology, memory deficits, and anxiety were reversed in the mouse model of AD treated with either gemfibrozil or Wy14643, mediated by a PPARα-dependent enhancement of autophagosome biogenesis (93). Fenofibrate-mediated PPARα activation also reduces amyloidogenic processing of APP in APP/PS1 transgenic mice (94).
Diverse mechanisms have been brought to address PPARα’s function in AD (Figure 3). Several studies report that PPARα plays an essential role in maintaining brain energy supply by modulating ketogenesis (90), due to ketone bodies’ function of protecting hippocampal neurons from Aβ toxicity (95, 96). Because mitochondrial disturbances play a crucial role both in aging and in neurodegenerative disorders, the regulation of PPARα in AD is also thought to be associated with mitochondrial function. In this area, some authors show that PPARα promotes glutamate transporter-1 endocytosis in astrocytes (97) and PARP1–PPARα–PGC1α regulates mitochondrial biogenesis and oxidative stress in neurons (98). Regulation of amyloid metabolism is thought to be another main function of PPARα as PPARα activity is associated with APP expression. The GW6471 PPARα antagonist inhibits APP knockdown-induced increases of synaptic activity in cortical cultures, whereas Wy14643 shows a reverse function, through the regulation of synaptic Aβ activity (8). Gemfibrozil or Wy14643 enhances autophagy in the APP-PSEN1ΔE9 mouse model to clear Aβ (93), and lysosome-mediated Aβ clearance regulated by PPARα also supports this pathway (92). Another point showed that activation of PPARα stimulates ADAM10-mediated proteolysis of Aβ in hippocampal culture (99), together indicating the regulation of Aβ metabolism by PPARα. Other pathways, like enhancing the level of brain-derived neurotrophic factor (BDNF) in the hippocampus, might partly take part in this therapy (100).
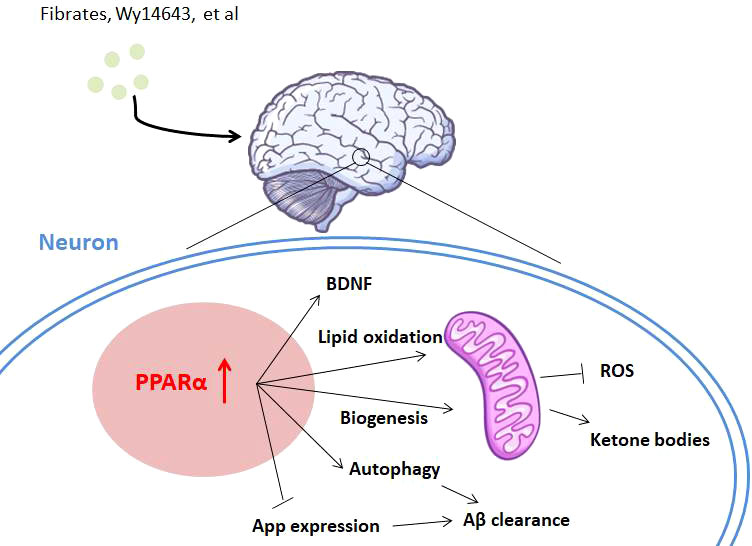
Figure 3 The role of PPARα in Alzheimer’s disease. PPARα is involved in Alzheimer’s disease with diverse mechanisms. Activation of PPARα promotes lipid oxidation and ketogenesis through mitochondrial biogenesis. PPARα activation also leads to Aβ clearance by App repression and autophagy of neurons. Neuroprotective factors like BDNF might also be activated by PPARα agonists for AD treatment.
The preferential activation of PPARα also affects other neurodegenerative disorders. Fenofibrate reduces neuroinflammation and blocks neurodegeneration in a mouse model of ALS, whose mRNA analysis indicated a significant effect of this drug on transcription of anti-inflammatory and antioxidative genes (101). In another experimental animal model of Parkinson’s disease (PD), a neuroprotective effect of fenofibrate was also observed (102). Studies showing that PPARα directly participates in these diseases are still not sufficient. However, it is clear that mitochondrial metabolism, which is mediated by PPARα, plays a central role in the disorders. Whether PPARα agonists are effective for diverse neuron disorders deserves further exploration.
Cardiovascular disease (CVD) still remains the leading cause of death globally, accounting for 17.9 million deaths per year according to WHO (https://www.who.int/en/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)). Both obesity and diabetes have been implicated as major risk factors for CVD (7), suggesting the importance of lipid and glucose metabolic homeostasis in CVD. As we have discussed above on the function of PPARα in metabolic syndrome, PPARα is probably involved in the regulation of CVD. Moreover, PPARα is expressed in the vasculature and its expression is detected in ECs, vascular smooth muscle cells (VSMCs), and monocytes/macrophages, which are all associated with the progress of CVD (7).
Early studies have shown that PPARα protects the health of the cardiovascular system. Fenofibrate-induced PPARα activation protects against endothelin-induced cardiac hypertrophy and failure through negative regulation of AP-1 binding activity (103). PPARα agonists decrease macrophage-laden atherosclerotic lesions in a non-diabetic mouse model (104). PPARα agonist GW7647 treatment of LDL receptor-null mice is shown to inhibit both atherosclerosis and the formation of macrophage foam cells in the peritoneal cavity (105). Fibrate therapy results in an increase in apoA transcription and a subsequent increase in HDL levels (106); in addition, fibrates influence reverse cholesterol transport via an upregulation of the ATP-binding cassette transporter (ABCA1) (107) and by an increase in the hepatic uptake of HDL (108), together alleviating the progress of atherosclerosis, the main inducement of CVD.
PPARα influences CVD, which might be mainly through metabolic regulation (Figure 4). Cardiac-specific overexpression of PPARα results in hypertrophy and failure in association with intracellular accumulation of neutral lipids (109). Treatment of human macrophages with PPARα agonists increases the expression of cholesterol efflux proteins such as ABCA1 and SR-B1 (107). In atherosclerosis, PPARα plays important roles in lipid homeostasis in different tissues as discussed above. The mitochondrial states regulated by PPARα also influence health of the heart. Mitochondrial fatty acid oxidation causes alterations such as heart failure, ischemic heart disease, and diabetic cardiomyopathy, when the expression of PPARα is decreased (110). ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPARα and PGC1, where PPARα agonists completely reverse the mitochondrial defects and restore normal heart function (111). Moreover, overexpression of PPARα ameliorates doxorubicin-induced cardiotoxicity by reducing mitochondria-dependent apoptosis (112). PPARα has also been implicated in the regulation of redox responses in the endothelium, for example inducing the expression of SOD1 and also attenuating the induction of NOX in primary ECs (113, 114).
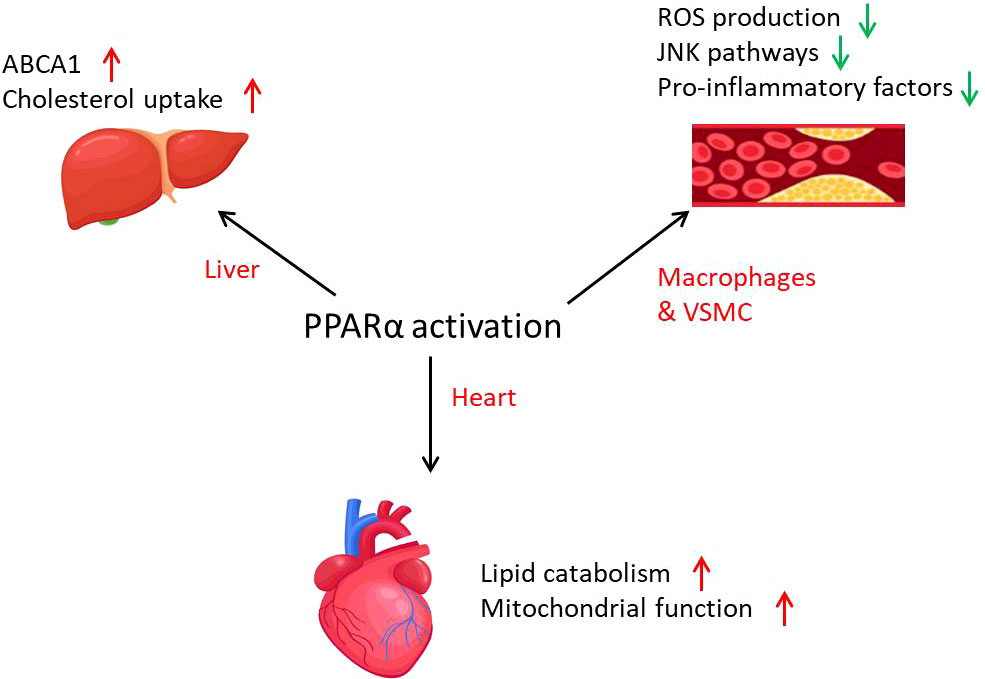
Figure 4 The role of PPARα in cardiovascular diseases. PPARα is involved in cardiovascular diseases through lipid metabolism activation and inflammation inhibition. PPARα activation promotes mitochondrial function and lipid catabolism in the heart and cholesterol uptake in the liver. In atherosclerosis, PPARα activation leads to the decrease of inflammation in macrophages and VSMCs.
In humans, PPARα trans-repression occurs not only in the liver but also in isolated vascular endothelial cells, linking PPARα to systemic inflammation and atherosclerosis. In LDLR-deficient mice, macrophage-specific overexpression of PPARα is reported to reduce atherosclerosis (115). An in vitro study suggested that PPARα activation protects against cardiac hypertrophy and failure partly via inhibition of the JNK pathway (103). Several PPARα agonists could inhibit the synthesis of proinflammatory mediators such as IL-1-mediated activation of IL-6 and prostaglandin along with cyclooxygenase-2 through suppression of NF-κB signaling in VSMCs (116, 117). Ligand activation of PPAR-α in macrophages inhibits the activation of inducible NOS and production of TNF-α and MMP9 (7). These results investigate that PPARα regulates CVD by its metabolic and immune regulatory function, where PPARα agonists show potential in CVD treatment.
The complex roles of PPARα in cancers have recently been focused by scientists Figure 5. TCGA data suggest that tumor microenvironment characteristics were correlated with the expression level of PPARα in pan-cancer (118). As a highly expressed transcriptional factor for lipid metabolism in the liver, PPARα is essential for MAFLD progress. However, some studies suggest that long-term activation of PPARα induced hepatocellular carcinoma in mice and was essential for the development of hepatic steatosis (119). Administration of fibrates and Wy14643 promoted hepatocyte proliferation and resulted in significant hepatomegaly in vivo (120). A recent study suggests that PPARα activation promotes hepatocyte proliferation through UHRF1–CDH1-mediated epigenetic modulation (121), which might partly explain the phenotype. However, it still remains a question that PPARα is negatively correlated with HCC in human, which seems different from data in mice (122). In breast cancer, the role of PPARα also seems bidirectional. Six PPARα polymorphisms are evaluated in association with incident breast cancer, from which rs4253760 is found associated with a nearly 100% relative increase in the risk of postmenopausal breast cancer (123). Fenofibrate induces apoptosis of triple-negative breast cancer cells via activation of the NF-κB pathway (124), and Wy14643 shows toxicity to breast cancer cells via PPARα–CYP1B1 expression (125), suggesting the therapy potential of PPARα in breast cancer. However, fibrates’ influence on proliferation of breast cancer might be dose-dependent, whereas low doses of fibrates stimulate proliferation of MCF-7 cells but high doses suppress it (126). Moreover, PPARα-selective antagonist GW6471 inhibits cell growth by inducing energy imbalance and metabolic stress (127). Studies on PPARα in colorectal cancer are relatively lacking compared with breast and liver cancers. Intestinal PPARα shows protective function against colon carcinogenesis via regulation of methyltransferases DNMT1 and PRMT6 (128), whereas its target HMGCS2 promotes cancer proliferation in another research (129). In a word, although PPARα is associated with the progress of pan-cancer according to omics analysis, the role of PPARα in cancer remains uncertain.
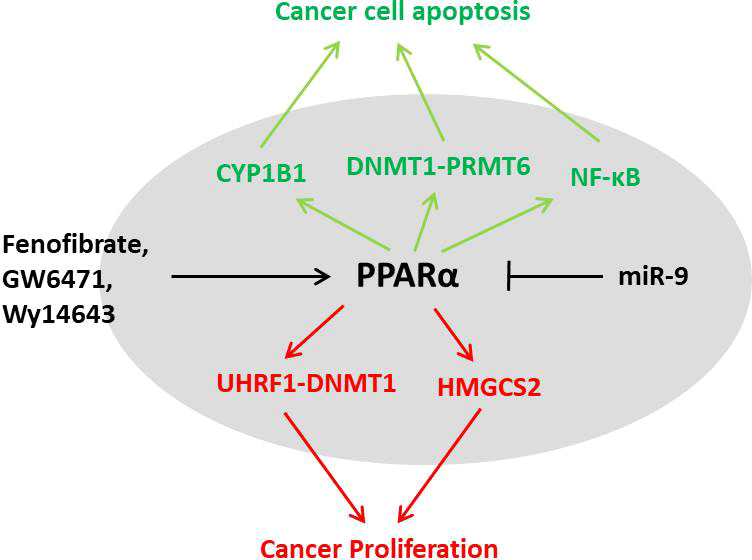
Figure 5 The dual characters of PPARα in cancer. PPARα activation could result in proliferation of cancer cells through the UHRF1–DNMT1 pathway and HMGCS2; however, the agonists could also lead to cell apoptosis by CYP1B, PRMT6, or NF-κB pathways. The role of PPARα in different cancers needs further investigation.
As a transcription factor with fatty acids to be the natural ligands, PPARα is a key TF in lipid metabolism. Its transactivation of lipid oxidation and trans-repression of inflammation, together with abundant modification and agonists, suggests that PPARα is involved in the regulation of diverse human diseases. PPARα shows protective effects in metabolic syndromes including MAFLD and diabetes, as well as its benefit for cardiovascular health. Selective agonists such as fenofibrate and Wy14643 show great potential in treatment of these diseases. However, the function of PPARα in cancer remains a puzzle.
In perspective, there are still lots of questions that should be answered in this area. Our knowledge on modification of PPARα is deficiency, which is obviously associated with metabolic conditions of the cell. How PPARα affects diabetes particularly insulin resistance needs more evidence. Whether and how PPARα directly regulates neuroinflammatory diseases lacks focus. The role of PPARα in different cancers still needs further exploration; its contradicting function in some research indicates that the function of this factor might be tightly associated with metabolic states, or even the regulation of PPARα is only a consequence of therapies. Moreover, clinical researchers should also pay attention to the targeted tissues of PPARα agonists, which might lead to a systemic influence on adipose, liver, muscle, heart, etc.
YL conceived the study and accomplished the major part of writing, YW and P-FL provided perspectives and carried out the modification and perfection. All the authors contributed to the finalization of this manuscript.
This work was supported by the Fujian Science and Technology Innovation Platform (No.2020Y2016 to Xiamen Cardiovascular Hospital).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Mirza AZ, Althagafi II, Shamshad H. Role of PPAR receptor in different diseases and their ligands: Physiological importance and clinical implications. Eur J Med Chem (2019) 166:502–13. doi: 10.1016/j.ejmech.2019.01.067
2. Wang Y, Nakajima T, Gonzalez FJ, Tanaka N. PPARs as metabolic regulators in the liver: Lessons from liver-specific PPAR-null mice. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21062061
3. Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature (1990) 347:645–50. doi: 10.1038/347645a0
4. Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci U.S.A. (1994) 91:7355–9. doi: 10.1073/pnas.91.15.7355
5. Tahri-Joutey M, Andreoletti P, Surapureddi S, Nasser B, Cherkaoui-Malki M, Latruffe N. Mechanisms mediating the regulation of peroxisomal fatty acid beta-oxidation by PPARalpha. Int J Mol Sci (2021) 22. doi: 10.3390/ijms22168969
6. Francque S, Verrijken A, Caron S, Prawitt J, Paumelle R, Derudas B, et al. PPARalpha gene expression correlates with severity and histological treatment response in patients with non-alcoholic steatohepatitis. J Hepatol (2015) 63:164–73. doi: 10.1016/j.jhep.2015.02.019
7. Han L, Shen WJ, Bittner S, Kraemer FB, Azhar S. PPARs: regulators of metabolism and as therapeutic targets in cardiovascular disease. part I: PPAR-alpha. Future Cardiol (2017) 13:259–78. doi: 10.2217/fca-2016-0059
8. Saez-Orellana F, Leroy T, Ribeiro F, Kreis A, Leroy K, Lalloyer F, et al. Regulation of PPARalpha by APP in Alzheimer disease affects the pharmacological modulation of synaptic activity. JCI Insight (2021) 6. doi: 10.1172/jci.insight.150099
9. Shao Y, Chen J, Dong LJ, He X, Cheng R, Zhou K, et al. A protective effect of PPARalpha in endothelial progenitor cells through regulating metabolism. Diabetes (2019) 68:2131–42. doi: 10.2337/db18-1278
10. Kawasaki M, Kambe A, Yamamoto Y, Arulmozhiraja S, Ito S, Nakagawa Y, et al. Elucidation of molecular mechanism of a selective PPARalpha modulator, pemafibrate, through combinational approaches of X-ray crystallography, thermodynamic analysis, and first-principle calculations. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21010361
11. Gearing KL, Gottlicher M, Teboul M, Widmark E, Gustafsson JA. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc Natl Acad Sci U.S.A. (1993) 90:1440–4.doi: 10.1073/pnas.90.4.1440
12. Pawlak M, Lefebvre P, Staels B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J Hepatol (2015) 62:720–33. doi: 10.1016/j.jhep.2014.10.039
13. Pawlak M, Bauge E, Bourguet W, De Bosscher K, Lalloyer F, Tailleux A, et al. The transrepressive activity of peroxisome proliferator-activated receptor alpha is necessary and sufficient to prevent liver fibrosis in mice. Hepatology (2014) 60:1593–606. doi: 10.1002/hep.27297
14. Surapureddi S, Yu S, Bu H, Hashimoto T, Yeldandi AV, Kashireddy P, et al. Identification of a transcriptionally active peroxisome proliferator-activated receptor alpha -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proc Natl Acad Sci U.S.A. (2002) 99:11836–41. doi: 10.1073/pnas.182426699
15. van der Meer DL, Degenhardt T, Vaisanen S, de Groot PJ, Heinaniemi M, de Vries SC, et al. Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res (2010) 38:2839–50. doi: 10.1093/nar/gkq012
16. Gross B, Pawlak M, Lefebvre P, Staels B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat Rev Endocrinol (2017) 13:36–49. doi: 10.1038/nrendo.2016.135
17. Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab (2007) 5:415–25. doi: 10.1016/j.cmet.2007.05.003
18. Delerive P, De Bosscher K, Besnard S, Vanden Berghe W, Peters JM, Gonzalez FJ, et al. Peroxisome proliferator-activated receptor alpha negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-kappaB and AP-1. J Biol Chem (1999) 274:32048–54. doi: 10.1074/jbc.274.45.32048
19. Oka S, Alcendor R, Zhai P, Park JY, Shao D, Cho J, et al. PPARalpha-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab (2011) 14:598–611. doi: 10.1016/j.cmet.2011.10.001
20. Mogilenko DA, Kudriavtsev IV, Shavva VS, Dizhe EB, Vilenskaya EG, Efremov AM, et al. Peroxisome proliferator-activated receptor alpha positively regulates complement C3 expression but inhibits tumor necrosis factor alpha-mediated activation of C3 gene in mammalian hepatic-derived cells. J Biol Chem (2013) 288:1726–38. doi: 10.1074/jbc.M112.437525
21. Roduit R, Morin J, Masse F, Segall L, Roche E, Newgard CB, et al. Glucose down-regulates the expression of the peroxisome proliferator-activated receptor-alpha gene in the pancreatic beta -cell. J Biol Chem (2000) 275:35799–806. doi: 10.1074/jbc.M006001200
22. Joly E, Roduit R, Peyot ML, Habinowski SA, Ruderman NB, Witters LA, et al. ). glucose represses PPARalpha gene expression via AMP-activated protein kinase but not via p38 mitogen-activated protein kinase in the pancreatic beta-cell. J Diabetes (2009) 1:263–72. doi: 10.1111/j.1753-0407.2009.00043.x
23. Hajjar T, Meng GY, Rajion MA, Vidyadaran S, Othman F, Farjam AS, et al. Omega 3 polyunsaturated fatty acid improves spatial learning and hippocampal peroxisome proliferator activated receptors (PPARalpha and PPARgamma) gene expression in rats. BMC Neurosci (2012) 13:109. doi: 10.1186/1471-2202-13-109
24. Wan CW, Wong CN, Pin WK, Wong MH, Kwok CY, Chan RY, et al. Chlorogenic acid exhibits cholesterol lowering and fatty liver attenuating properties by up-regulating the gene expression of PPAR-alpha in hypercholesterolemic rats induced with a high-cholesterol diet. Phytother Res (2013) 27:545–51. doi: 10.1002/ptr.4751
25. Coccia E, Varricchio E, Vito P, Turchini GM, Francis DS, Paolucci M. Fatty acid-specific alterations in leptin, PPARalpha, and CPT-1 gene expression in the rainbow trout. Lipids (2014) 49:1033–46. doi: 10.1007/s11745-014-3939-y
26. Lee YH, Jang HJ, Kim S, Choi SS, Khim KW, Eom HJ, et al. Hepatic MIR20B promotes nonalcoholic fatty liver disease by suppressing PPARA. Elife (2021) 10. doi: 10.7554/eLife.70472
27. Shalev A, Siegrist-Kaiser CA, Yen PM, Wahli W, Burger AG, Chin WW, et al. The peroxisome proliferator-activated receptor alpha is a phosphoprotein: regulation by insulin. Endocrinology (1996) 137:4499–502. doi: 10.1210/endo.137.10.8828512
28. Brunmeir R, Xu F. Functional regulation of PPARs through post-translational modifications. Int J Mol Sci (2018) 19. doi: 10.3390/ijms19061738
29. Compe E, Drane P, Laurent C, Diderich K, Braun C, Hoeijmakers JH, et al. Dysregulation of the peroxisome proliferator-activated receptor target genes by XPD mutations. Mol Cell Biol (2005) 25:6065–76. doi: 10.1128/MCB.25.14.6065-6076.2005
30. Juge-Aubry CE, Hammar E, Siegrist-Kaiser C, Pernin A, Takeshita A, Chin WW, et al. Regulation of the transcriptional activity of the peroxisome proliferator-activated receptor alpha by phosphorylation of a ligand-independent trans-activating domain. J Biol Chem (1999) 274:10505–10. doi: 10.1074/jbc.274.15.10505
31. Blanquart C, Mansouri R, Paumelle R, Fruchart JC, Staels B, Glineur C. ). the protein kinase c signaling pathway regulates a molecular switch between transactivation and transrepression activity of the peroxisome proliferator-activated receptor alpha. Mol Endocrinol (2004) 18:1906–18. doi: 10.1210/me.2003-0327
32. Blanquart C, Mansouri R, Fruchart JC, Staels B, Glineur C. Different ways to regulate the PPARalpha stability. Biochem Biophys Res Commun (2004) 319:663–70. doi: 10.1016/j.bbrc.2004.05.035
33. Hinds TD Jr., Burns KA, Hosick PA, McBeth L, Nestor-Kalinoski A, Drummond HA, et al. Biliverdin reductase a attenuates hepatic steatosis by inhibition of glycogen synthase kinase (GSK) 3beta phosphorylation of serine 73 of peroxisome proliferator-activated receptor (PPAR) alpha. J Biol Chem (2016) 291:25179–91. doi: 10.1074/jbc.M116.731703
34. Hinds TD Jr., Hosick PA, Chen S, Tukey RH, Hankins MW, Nestor-Kalinoski A, et al. Mice with hyperbilirubinemia due to gilbert's syndrome polymorphism are resistant to hepatic steatosis by decreased serine 73 phosphorylation of PPARalpha. Am J Physiol Endocrinol Metab (2017) 312:E244–52. doi: 10.1152/ajpendo.00396.2016
35. Gopinathan L, Hannon DB, Peters JM, Vanden Heuvel JP. Regulation of peroxisome proliferator-activated receptor-alpha by MDM2. Toxicol Sci (2009) 108:48–58. doi: 10.1093/toxsci/kfn260
36. Zhao Z, Xu D, Wang Z, Wang L, Han R, Wang Z, et al. Hepatic PPARalpha function is controlled by polyubiquitination and proteasome-mediated degradation through the coordinated actions of PAQR3 and HUWE1. Hepatology (2018) 68:289–303. doi: 10.1002/hep.29786
37. Lin Y, Chen L, You X, Li Z, Li C, Chen Y. PAQR9 regulates hepatic ketogenesis and fatty acid oxidation during fasting by modulating protein stability of PPARalpha. Mol Metab (2021) 53:101331. doi: 10.1016/j.molmet.2021.101331
38. Rodriguez JE, Liao JY, He J, Schisler JC, Newgard CB, Drujan D, et al. The ubiquitin ligase MuRF1 regulates PPARalpha activity in the heart by enhancing nuclear export via monoubiquitination. Mol Cell Endocrinol (2015) 413:36–48. doi: 10.1016/j.mce.2015.06.008
39. Leuenberger N, Pradervand S, Wahli W. Sumoylated PPARalpha mediates sex-specific gene repression and protects the liver from estrogen-induced toxicity in mice. J Clin Invest (2009) 119:3138–48. doi: 10.1172/JCI39019
40. Zhu Y, Wang X, Zhou X, Ding L, Liu D, Xu H. DNMT1-mediated PPARalpha methylation aggravates damage of retinal tissues in diabetic retinopathy mice. Biol Res (2021) 54:25. doi: 10.1186/s40659-021-00347-1
41. Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, et al. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U.S.A. (2001) 98:13919–24. doi: 10.1073/pnas.241410198
42. Chakravarthy MV, Pan Z, Zhu Y, Tordjman K, Schneider JG, Coleman T, et al. "New" hepatic fat activates PPARalpha to maintain glucose, lipid, and cholesterol homeostasis. Cell Metab (2005) 1:309–22. doi: 10.1016/j.cmet.2005.04.002
43. Fan CY, Pan J, Usuda N, Yeldandi AV, Rao MS, Reddy JK. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. implications for peroxisome proliferator-activated receptor alpha natural ligand metabolism. J Biol Chem (1998) 273:15639–45. doi: 10.1074/jbc.273.25.15639
44. Sapiro JM, Mashek MT, Greenberg AS, Mashek DG. Hepatic triacylglycerol hydrolysis regulates peroxisome proliferator-activated receptor alpha activity. J Lipid Res (2009) 50:1621–9. doi: 10.1194/jlr.M800614-JLR200
45. Mahmoudi A, Butler AE, Jamialahmadi T, Sahebkar A. Target deconvolution of fenofibrate in nonalcoholic fatty liver disease using bioinformatics analysis. BioMed Res Int (2021) 2021:3654660. doi: 10.1155/2021/3654660
46. Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem (2000) 43:527–50. doi: 10.1021/jm990554g
47. Chou CJ, Haluzik M, Gregory C, Dietz KR, Vinson C, Gavrilova O, et al. WY14,643, a peroxisome proliferator-activated receptor alpha (PPARalpha ) agonist, improves hepatic and muscle steatosis and reverses insulin resistance in lipoatrophic a-ZIP/F-1 mice. J Biol Chem (2002) 277:24484–9. doi: 10.1074/jbc.M202449200
48. Kaul U, Parmar D, Manjunath K, Shah M, Parmar K, Patil KP, et al. New dual peroxisome proliferator activated receptor agonist-saroglitazar in diabetic dyslipidemia and non-alcoholic fatty liver disease: integrated analysis of the real world evidence. Cardiovasc Diabetol (2019) 18:80. doi: 10.1186/s12933-019-0884-3
49. Fruchart JC. Selective peroxisome proliferator-activated receptor alpha modulators (SPPARMalpha): the next generation of peroxisome proliferator-activated receptor alpha-agonists. Cardiovasc Diabetol (2013) 12:82. doi: 10.1186/1475-2840-12-82
50. Francque S, Szabo G, Abdelmalek MF, Byrne CD, Cusi K, Dufour JF, et al. Nonalcoholic steatohepatitis: the role of peroxisome proliferator-activated receptors. Nat Rev Gastroenterol Hepatol (2021) 18:24–39. doi: 10.1038/s41575-020-00366-5
51. Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology (2016) 64:73–84. doi: 10.1002/hep.28431
52. Yki-Järvinen H. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol (2014) 2:901–10. doi: 10.1016/S2213-8587(14)70032-4
53. Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero-Gomez M, et al. A new definition for metabolic dysfunction-associated fatty liver disease: An international expert consensus statement. J Hepatol (2020) 73:202–9. doi: 10.1016/j.jhep.2020.03.039
54. Eslam M, Sanyal AJ, George J, International Consensus P. MAFLD: A consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology (2020) 158:1999–2014 e1991. doi: 10.1053/j.gastro.2019.11.312
55. Bougarne N, Weyers B, Desmet SJ, Deckers J, Ray DW, Staels B, et al. Molecular actions of PPARalpha in lipid metabolism and inflammation. Endocr Rev (2018) 39:760–802. doi: 10.1210/er.2018-00064
56. Regnier M, Polizzi A, Smati S, Lukowicz C, Fougerat A, Lippi Y, et al. Hepatocyte-specific deletion of pparalpha promotes NAFLD in the context of obesity. Sci Rep (2020) 10:6489. doi: 10.1038/s41598-020-63579-3
57. Ip E, Farrell GC, Robertson G, Hall P, Kirsch R, Leclercq I. Central role of PPARalpha-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology (2003) 38:123–32. doi: 10.1053/jhep.2003.50307
58. Koliaki C, Szendroedi J, Kaul K, Jelenik T, Nowotny P, Jankowiak F, et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metab (2015) 21:739–46. doi: 10.1016/j.cmet.2015.04.004
59. Sun N, Shen C, Zhang L, Wu X, Yu Y, Yang X, et al. Hepatic kruppel-like factor 16 (KLF16) targets PPARalpha to improve steatohepatitis and insulin resistance. Gut (2021) 70:2183–95. doi: 10.1136/gutjnl-2020-321774
60. Shen B, Feng H, Cheng J, Li Z, Jin M, Zhao L, et al. Geniposide alleviates non-alcohol fatty liver disease via regulating Nrf2/AMPK/mTOR signalling pathways. J Cell Mol Med (2020) 24:5097–108. doi: 10.1111/jcmm.15139
61. Vernia S, Cavanagh-Kyros J, Garcia-Haro L, Sabio G, Barrett T, Jung DY, et al. The PPARalpha-FGF21 hormone axis contributes to metabolic regulation by the hepatic JNK signaling pathway. Cell Metab (2014) 20:512–25. doi: 10.1016/j.cmet.2014.06.010
62. Rodriguez-Vilarrupla A, Lavina B, Garcia-Caldero H, Russo L, Rosado E, Roglans N, et al. PPARalpha activation improves endothelial dysfunction and reduces fibrosis and portal pressure in cirrhotic rats. J Hepatol (2012) 56:1033–9. doi: 10.1016/j.jhep.2011.12.008
63. Lefere S, Puengel T, Hundertmark J, Penners C, Frank AK, Guillot A, et al. Differential effects of selective- and pan-PPAR agonists on experimental steatohepatitis and hepatic macrophages(). J Hepatol (2020) 73:757–70. doi: 10.1016/j.jhep.2020.04.025
64. Larter CZ, Yeh MM, Van Rooyen DM, Brooling J, Ghatora K, Farrell GC. Peroxisome proliferator-activated receptor-alpha agonist, wy 14,643, improves metabolic indices, steatosis and ballooning in diabetic mice with non-alcoholic steatohepatitis. J Gastroenterol Hepatol (2012) 27:341–50. doi: 10.1111/j.1440-1746.2011.06939.x
65. Zhang Y, Jia XB, Liu YC, Yu WQ, Si YH, Guo SD. Fenofibrate enhances lipid deposition via modulating PPARgamma, SREBP-1c, and gut microbiota in ob/ob mice fed a high-fat diet. Front Nutr (2022) 9:971581. doi: 10.3389/fnut.2022.971581
66. Abd El-Haleim EA, Bahgat AK, Saleh S. )Effects of combined PPAR-gamma and PPAR-alpha agonist therapy on fructose induced NASH in rats: Modulation of gene expression. Eur J Pharmacol (2016) 773:59–70. doi: 10.1016/j.ejphar.2016.01.011
67. Jun HJ, Kim J, Hoang MH, Lee SJ. Hepatic lipid accumulation alters global histone h3 lysine 9 and 4 trimethylation in the peroxisome proliferator-activated receptor alpha network. PloS One (2012) 7:e44345. doi: 10.1371/journal.pone.0044345
68. Flavell DM, Pineda Torra I, Jamshidi Y, Evans D, Diamond JR, Elkeles RS, et al. Variation in the PPARalpha gene is associated with altered function in vitro and plasma lipid concentrations in type II diabetic subjects. Diabetologia (2000) 43:673–80. doi: 10.1007/s001250051357
69. Lacquemant C, Lepretre F, Pineda Torra I, Manraj M, Charpentier G, Ruiz J, et al. Mutation screening of the PPARalpha gene in type 2 diabetes associated with coronary heart disease. Diabetes Metab (2000) 26:393–401.
70. Evans D, Aberle J, Wendt D, Wolf A, Beisiegel U, Mann WA. A polymorphism, L162V, in the peroxisome proliferator-activated receptor alpha (PPARalpha) gene is associated with lower body mass index in patients with non-insulin-dependent diabetes mellitus. J Mol Med (Berl). (2001) 79:198–204. doi: 10.1007/s001090100189
71. Sparso T, Hussain MS, Andersen G, Hainerova I, Borch-Johnsen K, Jorgensen T, et al. Relationships between the functional PPARalpha Leu162Val polymorphism and obesity, type 2 diabetes, dyslipidaemia, and related quantitative traits in studies of 5799 middle-aged white people. Mol Genet Metab (2007) 90:205–9. doi: 10.1016/j.ymgme.2006.10.007
72. Kim H, Haluzik M, Asghar Z, Yau D, Joseph JW, Fernandez AM, et al. Peroxisome proliferator-activated receptor-alpha agonist treatment in a transgenic model of type 2 diabetes reverses the lipotoxic state and improves glucose homeostasis. Diabetes (2003) 52:1770–8. doi: 10.2337/diabetes.52.7.1770
73. Lalloyer F, Vandewalle B, Percevault F, Torpier G, Kerr-Conte J, Oosterveer M, et al. Peroxisome proliferator-activated receptor alpha improves pancreatic adaptation to insulin resistance in obese mice and reduces lipotoxicity in human islets. Diabetes (2006) 55:1605–13. doi: 10.2337/db06-0016
74. Black RN, Ennis CN, Young IS, Hunter SJ, Atkinson AB, Bell PM. The peroxisome proliferator-activated receptor alpha agonist fenofibrate has no effect on insulin sensitivity compared to atorvastatin in type 2 diabetes mellitus; a randomised, double-blind controlled trial. J Diabetes Complications. (2014) 28:323–7. doi: 10.1016/j.jdiacomp.2014.01.001
75. Guerre-Millo M, Rouault C, Poulain P, Andre J, Poitout V, Peters JM, et al. PPAR-alpha-null mice are protected from high-fat diet-induced insulin resistance. Diabetes (2001) 50:2809–14. doi: 10.2337/diabetes.50.12.2809
76. Cheng L, Wang J, Dai H, Duan Y, An Y, Shi L, et al. Brown and beige adipose tissue: a novel therapeutic strategy for obesity and type 2 diabetes mellitus. Adipocyte (2021) 10:48–65. doi: 10.1080/21623945.2020.1870060
77. Kaisanlahti A, Glumoff T. Browning of white fat: agents and implications for beige adipose tissue to type 2 diabetes. J Physiol Biochem (2019) 75:1–10. doi: 10.1007/s13105-018-0658-5
78. Shen Y, Su Y, Silva FJ, Weller AH, Sostre-Colon J, Titchenell PM, et al. Shared PPARalpha/gamma target genes regulate brown adipocyte thermogenic function. Cell Rep (2020) 30:3079–3091 e3075. doi: 10.1016/j.celrep.2020.02.032
79. Rachid TL, Silva-Veiga FM, Graus-Nunes F, Bringhenti I, Mandarim-de-Lacerda CA, Souza-Mello V. Differential actions of PPAR-alpha and PPAR-beta/delta on beige adipocyte formation: A study in the subcutaneous white adipose tissue of obese male mice. PloS One (2018) 13:e0191365. doi: 10.1371/journal.pone.0191365
80. Miranda CS, Silva-Veiga F, Martins FF, Rachid TL, Mandarim-De-Lacerda CA, Souza-Mello V. PPAR-alpha activation counters brown adipose tissue whitening: a comparative study between high-fat- and high-fructose-fed mice. Nutrition (2020) 78:110791. doi: 10.1016/j.nut.2020.110791
81. Man B, Hu C, Yang G, Xiang J, Yang S, Ma C. Berberine attenuates diabetic atherosclerosis via enhancing the interplay between KLF16 and PPARalpha in ApoE(-/-) mice. Biochem Biophys Res Commun (2022) 624:59–67. doi: 10.1016/j.bbrc.2022.07.072
82. Keech AC, Mitchell P, Summanen PA, O'Day J, Davis TM, Moffitt MS, et al. Effect of fenofibrate on the need for laser treatment for diabetic retinopathy (FIELD study): a randomised controlled trial. Lancet (2007) 370:1687–97. doi: 10.1016/S0140-6736(07)61607-9
83. Chen Y, Hu Y, Lin M, Jenkins AJ, Keech AC, Mott R, et al. Therapeutic effects of PPARalpha agonists on diabetic retinopathy in type 1 diabetes models. Diabetes (2013) 62:261–72. doi: 10.2337/db11-0413
84. Herman-Edelstein M, Scherzer P, Tobar A, Levi M, Gafter U. Altered renal lipid metabolism and renal lipid accumulation in human diabetic nephropathy. J Lipid Res (2014) 55:561–72. doi: 10.1194/jlr.P040501
85. Park CW, Kim HW, Ko SH, Chung HW, Lim SW, Yang CW, et al. Accelerated diabetic nephropathy in mice lacking the peroxisome proliferator-activated receptor alpha. Diabetes (2006) 55:885–93. doi: 10.2337/diabetes.55.04.06.db05-1329
86. Kim Y, Lim JH, Kim MY, Kim EN, Yoon HE, Shin SJ, et al. The adiponectin receptor agonist AdipoRon ameliorates diabetic nephropathy in a model of type 2 diabetes. J Am Soc Nephrol (2018) 29:1108–27. doi: 10.1681/ASN.2017060627
87. Wu L, Liu C, Chang DY, Zhan R, Zhao M, Man Lam S, et al. The attenuation of diabetic nephropathy by annexin A1 via regulation of lipid metabolism through the AMPK/PPARalpha/CPT1b pathway. Diabetes (2021) 70:2192–203. doi: 10.2337/db21-0050
88. Cheng R, Ding L, He X, Takahashi Y, Ma JX. Interaction of PPARalpha with the canonic wnt pathway in the regulation of renal fibrosis. Diabetes (2016) 65:3730–43. doi: 10.2337/db16-0426
89. Roy A, Jana M, Corbett GT, Ramaswamy S, Kordower JH, Gonzalez FJ, et al. Regulation of cyclic AMP response element binding and hippocampal plasticity-related genes by peroxisome proliferator-activated receptor alpha. Cell Rep (2013) 4:724–37. doi: 10.1016/j.celrep.2013.07.028
90. Saez-Orellana F, Octave JN, Pierrot N. Alzheimer's disease, a lipid story: Involvement of peroxisome proliferator-activated receptor alpha. Cells (2020) 9. doi: 10.3390/cells9051215
91. Brune S, Kolsch H, Ptok U, Majores M, Schulz A, Schlosser R, et al. Polymorphism in the peroxisome proliferator-activated receptor alpha gene influences the risk for alzheimer's disease. J Neural Transm. (Vienna). (2003) 110:1041–50. doi: 10.1093/humupd/dmz034
92. Raha S, Ghosh A, Dutta D, Patel DR, Pahan K. Activation of PPARalpha enhances astroglial uptake and degradation of beta-amyloid. Sci Signal (2021) 14:eabg4747. doi: 10.1126/scisignal.abg4747
93. Luo R, Su LY, Li G, Yang J, Liu Q, Yang LX, et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy (2020) 16:52–69. doi: 10.1080/15548627.2019.1596488
94. Zhang H, Gao Y, Qiao PF, Zhao FL, Yan Y. Fenofibrate reduces amyloidogenic processing of APP in APP/PS1 transgenic mice via PPAR-alpha/PI3-K pathway. Int J Dev Neurosci (2014) 38:223–31. doi: 10.1016/j.ijdevneu.2014.10.004
95. Cunnane SC, Courchesne-Loyer A, Vandenberghe C, St-Pierre V, Fortier M, Hennebelle M, et al. Can ketones help rescue brain fuel supply in later life? implications for cognitive health during aging and the treatment of alzheimer's disease. Front Mol Neurosci (2016) 9:53. doi: 10.3389/fnmol.2016.00053
96. Kashiwaya Y, Takeshima T, Mori N, Nakashima K, Clarke K, Veech RL. D-beta-hydroxybutyrate protects neurons in models of alzheimer's and parkinson's disease. Proc Natl Acad Sci U.S.A. (2000) 97:5440–4. doi: 10.1073/pnas.97.10.5440
97. Huang HT, Liao CK, Chiu WT, Tzeng SF. Ligands of peroxisome proliferator-activated receptor-alpha promote glutamate transporter-1 endocytosis in astrocytes. Int J Biochem Cell Biol (2017) 86:42–53. doi: 10.1016/j.biocel.2017.03.008
98. Wojtowicz S, Strosznajder AK, Jezyna M, Strosznajder JB. The novel role of PPAR alpha in the brain: Promising target in therapy of alzheimer's disease and other neurodegenerative disorders. Neurochem Res (2020) 45:972–88. doi: 10.1007/s11064-020-02993-5
99. Corbett GT, Gonzalez FJ, Pahan K. Activation of peroxisome proliferator-activated receptor alpha stimulates ADAM10-mediated proteolysis of APP. Proc Natl Acad Sci U.S.A. (2015) 112:8445–50. doi: 10.1073/pnas.1504890112
100. Jiang B, Wang YJ, Wang H, Song L, Huang C, Zhu Q, et al. Antidepressant-like effects of fenofibrate in mice via the hippocampal brain-derived neurotrophic factor signalling pathway. Br J Pharmacol (2017) 174:177–94. doi: 10.1111/bph.13668
101. Esmaeili MA, Yadav S, Gupta RK, Waggoner GR, Deloach A, Calingasan NY, et al. Preferential PPAR-alpha activation reduces neuroinflammation, and blocks neurodegeneration in vivo. Hum Mol Genet (2016) 25:317–27. doi: 10.1093/hmg/ddv477
102. Uppalapati D, Das NR, Gangwal RP, Damre MV, Sangamwar AT, Sharma SS. Neuroprotective potential of peroxisome proliferator activated receptor- alpha agonist in cognitive impairment in parkinson's disease: Behavioral, biochemical, and PBPK profile. PPAR. Res (2014) 2014:753587. doi: 10.1155/2014/753587
103. Irukayama-Tomobe Y, Miyauchi T, Sakai S, Kasuya Y, Ogata T, Takanashi M, et al. Endothelin-1-induced cardiac hypertrophy is inhibited by activation of peroxisome proliferator-activated receptor-alpha partly via blockade of c-jun NH2-terminal kinase pathway. Circulation (2004) 109:904–10. doi: 10.1161/01.CIR.0000112596.06954.00
104. Hennuyer N, Tailleux A, Torpier G, Mezdour H, Fruchart JC, Staels B, et al. PPARalpha, but not PPARgamma, activators decrease macrophage-laden atherosclerotic lesions in a nondiabetic mouse model of mixed dyslipidemia. Arterioscler Thromb Vasc Biol (2005) 25:1897–902. doi: 10.1161/01.ATV.0000175756.56818.ee
105. Li AC, Binder CJ, Gutierrez A, Brown KK, Plotkin CR, Pattison JW, et al. Differential inhibition of macrophage foam-cell formation and atherosclerosis in mice by PPARalpha, beta/delta, and gamma. J Clin Invest (2004) 114:1564–76. doi: 10.1172/JCI18730
106. Vu-Dac N, Schoonjans K, Laine B, Fruchart JC, Auwerx J, Staels B. Negative regulation of the human apolipoprotein a-I promoter by fibrates can be attenuated by the interaction of the peroxisome proliferator-activated receptor with its response element. J Biol Chem (1994) 269:31012–8. doi: 10.1016/S0021-9258(18)47383-8
107. Chinetti G, Lestavel S, Bocher V, Remaley AT, Neve B, Torra IP, et al. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat Med (2001) 7:53–8. doi: 10.1038/83348
108. Chinetti G, Gbaguidi FG, Griglio S, Mallat Z, Antonucci M, Poulain P, et al. CLA-1/SR-BI is expressed in atherosclerotic lesion macrophages and regulated by activators of peroxisome proliferator-activated receptors. Circulation (2000) 101:2411–7. doi: 10.1161/01.CIR.101.20.2411
109. Finck BN, Lehman JJ, Leone TC, Welch MJ, Bennett MJ, Kovacs A, et al. The cardiac phenotype induced by PPARalpha overexpression mimics that caused by diabetes mellitus. J Clin Invest (2002) 109:121–30. doi: 10.1172/JCI0214080
110. Fillmore N, Mori J, Lopaschuk GD. Mitochondrial fatty acid oxidation alterations in heart failure, ischaemic heart disease and diabetic cardiomyopathy. Br J Pharmacol (2014) 171:2080–90. doi: 10.1111/bph.12475
111. Haemmerle G, Moustafa T, Woelkart G, Buttner S, Schmidt A, van de Weijer T, et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-alpha and PGC-1. Nat Med (2011) 17:1076–85. doi: 10.1038/nm.2439
112. Wang W, Fang Q, Zhang Z, Wang D, Wu L, Wang Y. PPARalpha ameliorates doxorubicin-induced cardiotoxicity by reducing mitochondria-dependent apoptosis via regulating MEOX1. Front Pharmacol (2020) 11:528267. doi: 10.3389/fphar.2020.528267
113. Ibarra-Lara L, Hong E, Soria-Castro E, Torres-Narvaez JC, Perez-Severiano F, Del Valle-Mondragon L, et al. Clofibrate PPARalpha activation reduces oxidative stress and improves ultrastructure and ventricular hemodynamics in no-flow myocardial ischemia. J Cardiovasc Pharmacol (2012) 60:323–34. doi: 10.1097/FJC.0b013e31826216ed
114. Salazar G. NADPH oxidases and mitochondria in vascular senescence. Int J Mol Sci (2018) 19. doi: 10.3390/ijms19051327
115. Babaev VR, Ishiguro H, Ding L, Yancey PG, Dove DE, Kovacs WJ, et al. Macrophage expression of peroxisome proliferator-activated receptor-alpha reduces atherosclerosis in low-density lipoprotein receptor-deficient mice. Circulation (2007) 116:1404–12. doi: 10.1161/CIRCULATIONAHA.106.684704
116. Duan SZ, Usher MG, Mortensen RM. PPARs: the vasculature, inflammation and hypertension. Curr Opin Nephrol Hypertens (2009) 18:128–33. doi: 10.1097/MNH.0b013e328325803b
117. Marx N, Duez H, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors and atherogenesis: regulators of gene expression in vascular cells. Circ Res (2004) 94:1168–78. doi: 10.1161/01.RES.0000127122.22685.0A
118. Huang R, Zhang J, Li M, Yan P, Yin H, Zhai S, et al. The role of peroxisome proliferator-activated receptors (PPARs) in pan-cancer. PPAR. Res (2020) 2020:6527564. doi: 10.1155/2020/6527564
119. Tanaka N, Moriya K, Kiyosawa K, Koike K, Gonzalez FJ, Aoyama T. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest (2008) 118:683–94. doi: 10.1172/JCI33594
120. Peters JM, Cattley RC, Gonzalez FJ. Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator wy-14,643. Carcinogenesis (1997) 18:2029–33. doi: 10.1093/carcin/18.11.2029
121. Aibara D, Takahashi S, Yagai T, Kim D, Brocker CN, Levi M, et al. Gene repression through epigenetic modulation by PPARA enhances hepatocellular proliferation. iScience (2022) 25:104196. doi: 10.1016/j.isci.2022.104196
122. Drakaki A, Hatziapostolou M, Polytarchou C, Vorvis C, Poultsides GA, Souglakos J, et al. Functional microRNA high throughput screening reveals miR-9 as a central regulator of liver oncogenesis by affecting the PPARA-CDH1 pathway. BMC Cancer (2015) 15:542. doi: 10.1186/s12885-015-1562-9
123. Golembesky AK, Gammon MD, North KE, Bensen JT, Schroeder JC, Teitelbaum SL, et al. Peroxisome proliferator-activated receptor-alpha (PPARA) genetic polymorphisms and breast cancer risk: a long island ancillary study. Carcinogenesis (2008) 29:1944–9. doi: 10.1093/carcin/bgn154
124. Li T, Zhang Q, Zhang J, Yang G, Shao Z, Luo J, et al. Fenofibrate induces apoptosis of triple-negative breast cancer cells via activation of NF-kappaB pathway. BMC Cancer (2014) 14:96. doi: 10.1186/1471-2407-14-96
125. Hwang YP, Won SS, Jin SW, Lee GH, Pham TH, Choi JH, et al. WY-14643 regulates CYP1B1 expression through peroxisome proliferator-activated receptor alpha-mediated signaling in human breast cancer cells. Int J Mol Sci (2019) 20. doi: 10.3390/ijms20235928
126. Tauber Z, Koleckova M, Cizkova K. Peroxisome proliferator-activated receptor a (PPARa)-cytochrome P450 epoxygenases-soluble epoxide hydrolase axis in ER + PR + HER2- breast cancer. Med Mol Morphol. (2020) 53:141–8. doi: 10.1007/s00795-019-00240-7
127. Castelli V, Catanesi M, Alfonsetti M, Laezza C, Lombardi F, Cinque B, et al. PPARalpha-selective antagonist GW6471 inhibits cell growth in breast cancer stem cells inducing energy imbalance and metabolic stress. Biomedicines (2021) 9. doi: 10.3390/biomedicines9020127
128. Luo Y, Xie C, Brocker CN, Fan J, Wu X, Feng L, et al. Intestinal PPARalpha protects against colon carcinogenesis via regulation of methyltransferases DNMT1 and PRMT6. Gastroenterology (2019) 157:744–759 e744. doi: 10.1053/j.gastro.2019.05.057
Keywords: PPARα (peroxisome proliferator-activated receptor alpha), transcription, MAFLD, diabetes, alzhaimer’s disease (AD), cardiovascular diseases, cancer
Citation: Lin Y, Wang Y and Li P-f (2022) PPARα: An emerging target of metabolic syndrome, neurodegenerative and cardiovascular diseases. Front. Endocrinol. 13:1074911. doi: 10.3389/fendo.2022.1074911
Received: 20 October 2022; Accepted: 23 November 2022;
Published: 16 December 2022.
Edited by:
Ashu Johri, Independent Researcher, New York, NY, United StatesReviewed by:
Makoto Makishima, Nihon University, JapanCopyright © 2022 Lin, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yijun Lin, bGlueWlqdW45NUAxMjYuY29t; Yan Wang, d3lAbWVkbWFpbC5jb20uY24=; Pei-feng Li, TGlweGlhQHllYWgubmV0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.