
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 17 January 2023
Sec. Clinical Diabetes
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.1050200
This article is part of the Research Topic Clinical and Genetic Determinants of Diabetes and Complications View all 31 articles
Purpose: Diabetes mellitus is a systemic metabolic disorder which may target the lungs and lead to interstitial lung disease. The clinical characteristics and mechanisms of type 2 diabetes mellitus (T2DM) complicated with interstitial lung disease (ILD) have been studied. However, little work has been done to assess genetic contributions to the development of T2DM complicated with ILD.
Method: A pedigree of T2DM complicated with ILD was investigated, and the whole genome re-sequencing was performed to identify the genetic variations in the pedigree. According to the literature, the most valuable genetic contributors to the pathogenesis of T2DM complicated with ILD were screened out, and the related cellular functional experiments were also performed.
Results: A large number of SNPs, InDels, SVs and CNVs were identified in eight subjects including two diabetic patients with ILD, two diabetic patients without ILD, and four healthy subjects from the pedigree. After data analysis according to the literature, MUC5B SNP rs2943512 (A > C) was considered to be an important potentially pathogenic gene mutation associated with the pathogenesis of ILD in T2DM patients. In vitro experiments showed that the expression of MUC5B in BEAS-2B cells was significantly up-regulated by high glucose stimulation, accompanied by the activation of ERK1/2 and the increase of IL-1β and IL-6. When silencing MUC5B by RNA interference, the levels of p-ERK1/2 as well as IL-1β and IL-6 in BEAS-2B cells were all significantly decreased.
Conclusion: The identification of these genetic variants in the pedigree enriches our understanding of the potential genetic contributions to T2DM complicated with ILD. MUC5B SNP rs2943512 (A > C) or the up-regulated MUC5B in bronchial epithelial cells may be an important factor in promoting ILD inT2DM patients, laying a foundation for future exploration about the pathogenesis of T2DM complicated with ILD.
Diabetes mellitus (DM) is a systemic metabolic disorder characterized by chronic hyperglycemia due to insulin deficiency or resistance (1). The long-term effects of DM include neurological, micro-vascular and macro-vascular complications. The lungs are particularly susceptible targets of diabetic micro-vascular damage and non-enzymatic glycation as a result of their large alveolar-capillary network and abundant connective tissue. Diabetic patients frequently report respiratory symptoms, and diabetes related lung injuries have been observed in several studies (2, 3). Recently, epidemiological studies have suggested that type 2 diabetes (T2DM) is an independent risk factor for interstitial lung disease (ILD) (4–6).
Pulmonary function tests of patients with T2DM show restrictive ventilatory dysfunction and decreased diffusion capacity, including a reduction in forced expiratory volume in one second. High-resolution computed tomography (HRCT) images of the lungs from patients with T2DM are prone to show fibrotic pathological changes, such as a usual interstitial pneumonia pattern (7). However, there is still not enough HRCT data in patients with T2DM to generalize these results. Pathologically, nodular deposition of collagen in the middle of the alveolar walls, and increased thickness of the alveolar epithelial and endothelial capillary basal lamina have been reported to be the features of lung tissue in diabetic patients and animal diabetic models (8–10). A previous study reported significant increases in the cytokines in the bronchoalveolar lavage fluid of patients with T2DM compared to the controls (11), indicating that T2DM could induce inflammation which might promote pulmonary interstitial changes in the lungs. Altogether, clinical and experimental data from numerous studies have revealed that hyperglycemia or T2DM could promote the development of ILD. However, the pathogenic mechanisms involved in the association between T2DM and ILD haven’t been well understood. Previous studies considered unbalanced oxidative stress (12–14), overproduction of advanced glycation end-products (AGEs) and their receptors (15–18), epithelial to mesenchymal transition (EMT) (7, 19, 20), endoplasmic reticulum (ER) stress (21–24), and defects in bronchiolar surfactant layer (25–27) as mechanisms underlying T2DM complicated with ILD. In genetics, abundant variants associated with DM have been identified by genome-wide association studies (GWAS) (28, 29). Additionally, the genetic contributors to the development of ILD have also been identified, especially in cases of familial interstitial pneumonia (FIP) (30). However, it has not yet been determined about the genetic variations accounting for the development of ILD in T2DM.
In this study, a pedigree with T2DM complicated with ILD was investigated, and the whole genome re-sequencing of eight subjects from the pedigree was performed respectively to identify the genetic variations. After the analysis of genetic data based on literature, we proposed a non-synonymous single nucleotide variant (A>C; rs2943512) in MUC5B gene or the over-expressed MUC5B in bronchial epithelial cells might be an important factor in promoting ILD in T2DM patients. To our knowledge, this study is the first one presenting potential pathogenic genetic variants in a pedigree of T2DM complicated with ILD.
The subjects in this study were recruited from a pedigree with T2DM and interstitial lung disease in the Jilin Province of China (Figure 1). T2DM was diagnosed with the following criteria established by the American Diabetes Association: fasting plasma glucose concentration ≥ 126 mg/dl (7.0 mM), 2-hour post-load plasma glucose ≥200mg/dl (11.1 mM) after the oral glucose tolerance test, history of T2DM and/or on prescribed medication for diabetes. In addition, the tests of antibodies for T1DM were negative in each subject. Interstitial lung disease was collectively diagnosed by two radiologists and three respiratory physicians. Inclusion criteria for the subjects were as follows: 1) absence of systemic and metabolic disease other than obesity and T2DM; 2) absence of malignancy, infection, hepatic diseases, renal diseases, neurological diseases, cardiovascular events and endocrine dysfunction; and 3) absence of history of drug or alcohol abuse, defined as >80 g/day in men and >40 g/day in women. Recruited subjects are tagged in Figure 1. All subjects were informed of the purpose of the study and signed the consent. This study was approved by the Ethics Committee of the Second Hospital of Jilin University. Plasma of these subjects was withdrawn and stored at −80°C until analysis.
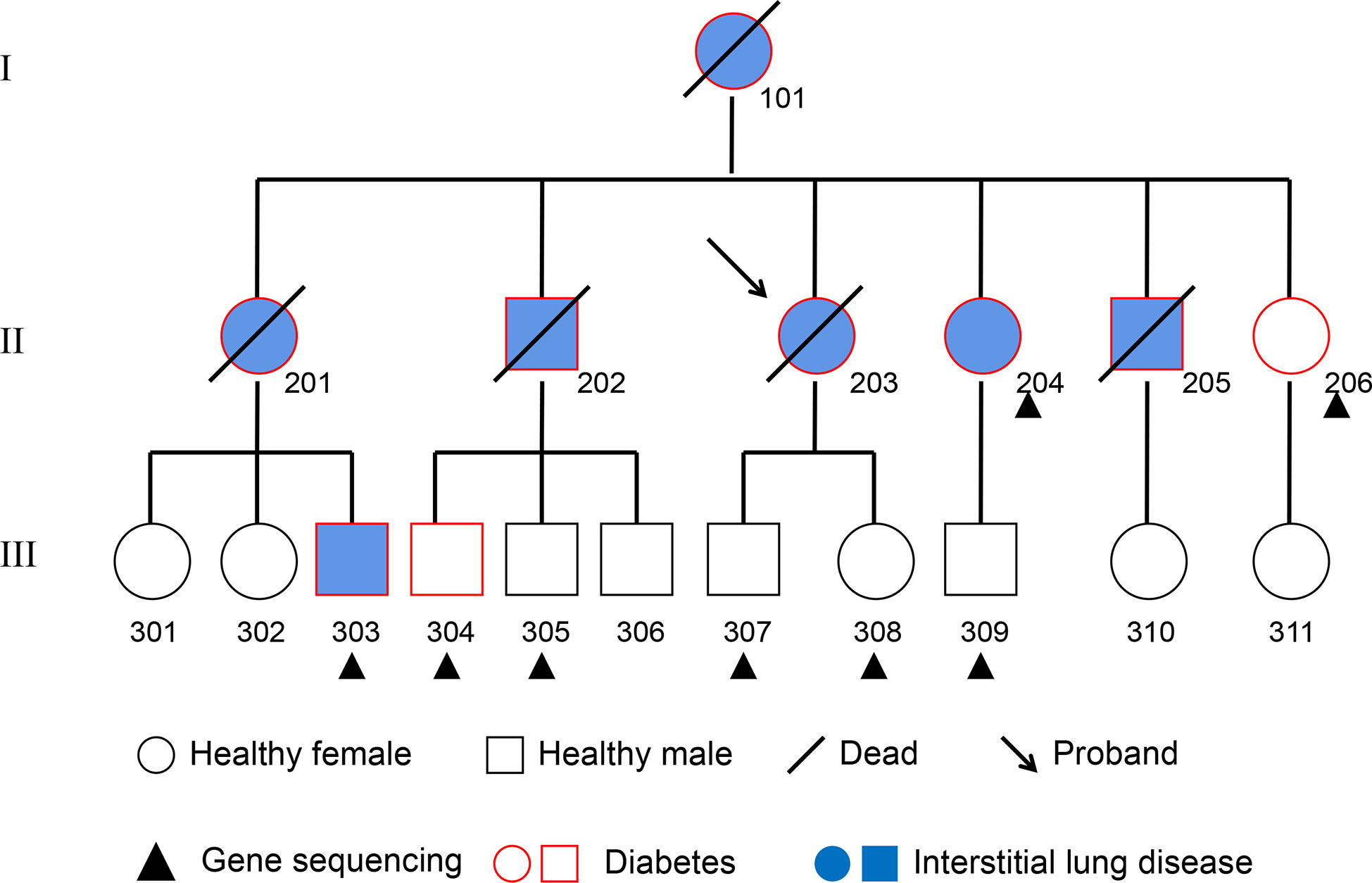
Figure 1 Pedigree Chart. The 8 subjects from the second and third generation who received whole-genome re-sequencing are marked with a black triangle. Squares represent males, circles represent females. The subjects affected by pulmonary fibrosis are painted blue, and the subjects affected by T2DM are indicated with a red border. Deceased individuals are labeled with a black slash.
Whole-genome re-sequencing was performed in the HiSeq X ten PE150 NovaSeq 6000 system in Shanghai Genechem Company Limited on genomic DNA to identify copy number variations (CNVs), single nucleotide polymorphisms (SNPs), insertion-deletion (InDels), and chromosomal structural variations (SVs) in eight subjects from this pedigree. All data were processed using FastQC software developed by Babraham Bioinformatics. Sequence alignment was performed using Burrows-Wheeler Aligner (BWA) software (31). HaplotypeCaller software was applied for mutation detection (32). Variants were identified through SNV calling, SV calling and SNP calling. Annovar software was used to annotate any detected mutations (19).
Genes affected by CNVs, SVs, SNPs or InDels were selected for annotation by comparing with the reference genome. Genes or the corresponding proteins were uploaded to FunRich (Version 3.1.4) for GO classification, including cell component (CC), biological process (BP), molecular function (MF) and biological pathway enrichment analysis. The go-CC, GO-BP, GO-MF and biological pathway with P < 0.05 were identified as significant. Meanwhile, proteins were mapped using the online Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/version11.5) to construct the PPI network and identify possible relationships between proteins. The PPI network was constructed by setting the minimum required interaction score to medium confidence (0.4). The active interaction sources included were “texming”, “experiments”, “database”, “co-expressing”, “neighborhood”, “genefusion” and “co-occurence”.
Human bronchial epithelial cells(BEAS-2B) were from National Collection of Authenticated Cell Cultures (Shanghai, China). The cells were cultured in RPMI 1640 medium (ThermoFisher Scientific, USA) supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, and 10% fetal bovine serum (FBS; Hyclone Laboratories, USA). The cells were grown at 37°C in 5% CO2 fully humidified air and were subcultured twice weekly. The cells were seeded in wells of a 6-well plate at 1×105 cells/well. When growth was confluent, the cells were incubated in RPMI 1640 medium containing certain concentrations of D-Glucose (Sigma-Aldrich, USA) for the indicated times. The cell proliferation and viability of BEAS-2B cells was quantified by CCK-8 Kit (Beyotime Biotechnology, China).
BEAS-2B cells were seeded into a 96-well plate at 1x104 cells/well with 100 µl of 10% FBS RPMI1640 medium. After overnight incubation, the complete medium containing different concentrations of glucose (15 mM, 20 mM, 25 mM, 30 mM) replaced the original medium of each group for 12h, 24h, 48h and 72h. Then, 10 µl of CCK-8 solution was added to the medium of each group. After the cells were incubated in the dark at 37°C for an additional 1 h, the absorbance at a 450nm wavelength was detected. Then cell viability of each group was calculated.
Isolation of total RNA from the cultured cells was performed according to the manufacturer’s instructions of cell total RNA isolation kit (ForeGene, China). Each sample was reverse transcribed into cDNA using the Prime Script RT Regent Kit (Takara, Japan). The primer sequences used in the PCR were 5´- GCCCACATCTCCACC TATGAT-3´ (sense) and 5´-GCAGTTCTCGTTGTCCGTCA-3´ (antisense) for MUC5B. Real-time PCR was performed with the SYBR Green Realtime PCR Master Mix Kit (Solarbio, China). Data were normalized versus GAPDH. According to the Ct values, the expression of MUC5B relative to GAPDH was calculated by using 2-ΔΔCt formula.
The protein levels of MUC5B, IL-1β and IL-6 were determined by enzyme-linked immunosorbent assay (ELISA). The standard substance and samples of cell supernatants were prepared, and incubated at 37°C in the 96-well plate for 2 hours. The standard substance and samples were discarded, and then the plate was blocked with biotin-labeled antibody for 1 hour at 37°C. Wells were then washed three times with the washing buffer, and horseradish peroxidase(HRP)-conjugated secondary antibody was added to wells. 1 hour later at 37°C, wells were washed three times with the washing buffer, and the substrate solution was added to wells, followed by the incubation at 37°C for 20min from light. Color was developed using stopping solution. Optical densities were measured using an ELISA reader (BioTek Instruments, USA) at 450 nm and 570nm. According to the standard curve, the corresponding concentration of each sample was calculated.
Human BEAS-2B bronchial epithelial cells were seeded in a 6-well plate and treated with glucose for the indicated times and concentrations. The cells were then washed with cold PBS, and then exposed to the cold lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 150 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail, and bromophenol blue). The proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto a nitrocellulose membrane. The memebrane was then blocked with 5% nonfat dry milk in 25 mM Tris-HCl, 150 mM NaCl, and 0.2% Tween-20, and then incubated with the indicated primary antibody of p-JNK (#4668, Cell Signaling Technologies, USA), p-ERK1/2 (#4370, Cell Signaling Technologies, USA), ERK1/2 (#4695, Cell Signaling Technologies, USA), p-p38 (#4511, Cell Signaling Technologies, USA), p-IκBα (#2859, Cell Signaling Technologies, USA), and β-actin (#4970, Cell Signaling Technologies, USA), JNK (#9252, Cell Signaling Technologies, USA), P38 (#9212, Cell Signaling Technologies, USA), IκBα (#4812, Cell Signaling Technologies, USA) overnight at 4°C. Subsequently, the membrane was washed and incubated for 2 hour with secondary antibody conjugated to HRP (#7074,Cell Signaling Technologies, USA). Finally, the membrane was developed using an chemiluminescence reagent kit (Bio-Rad, USA) and exposed to the imager.
The MUC5B shRNA knockout plasmid (Psuper-MUC5B-SH) and the negative control shRNA-NC were designed and synthesized by Jiangsu Cencefe Co., Ltd. ShRNA sequence was designed according to the target gene sequence (5’-GGGAAGTCATCT ACAATAAGACC-3’) as follows: Top-Bgl II(60bp) 5’-gatccccGAAGTCATCTACAATAAGATTCAAGAGATCTTATTGTAGATGACTTCTTTTTa-3’; Bottom-Xho I(60bp), 5’-tcgat AAAAAGAAGTCATCTACAATAAGATCTCTTGAATCTTATTGTAGATGACTTCggg-3’.Plasmid transfection was carried out using the transfection reagent Lipofectamine 2000 (Invitrogen, USA), and the procedure was as follows according to the kit instruction (Invitrogen). Briefly, BEAS-2B cells were seeded in wells of a 6-well plate at 2×10 5 cells/well and incubated in RPMI 1640 medium. When the cells were confluent to 80%, MUC5B shRNA and Lipofectamine 2000 were incubated together in RPMI 1640 medium without serum to form a MUC5B shRNA-Lipofectamine complex. After the cells were washed with PBS, the complex-containing medium was then added to each well. After 48 hours of transfection with MUC5B shRNA, the cells were harvested for RT-PCR analysis of MUC5B mRNA. The same procedure was performed with control shRNA.
The results were expressed by mean ± standard deviation, analyzed and plotted by GraphPad Prism 6. The data were normally distributed by Pearson test. Comparisons were made using the Student t-test between two groups, one-way ANOVA test between multiple groups. Student-Newman-Keuls post hoc test was applied. For all tests, P-value less than 0.05 was considered statistically significant.
The introduction of this pedigree:
NO.203, the proband of the pedigree, female, 61 years old, homemaker. In December 2015, the patient was first admitted to the outpatient department of our hospital for dyspnea, and diagnosed as “ILD” (Figure 2A) and “T2DM”. However, the patient did not follow the doctor’s advice for treatment. Since then, her dyspnea had been progressively aggravated. In May 2016, the patient was admitted to the inpatient department for severe dyspnea. Past medical history: coronary heart disease for 20 years, T2DM for 30 years. She denied the history of long-term smoking, and the exposure to special drugs, dust and poison. Her deceased mother also suffered from T2DM and ILD. Physical examination on admission: tachypnea, cyanosis, right jugular vein swelling, crackles rale (Velcro) in both lower lungs by auscultation. Arterial blood gas analysis: pH 7.35, PCO2 35mmHg, PO2 57mmHg, SaO2 86%. Sinus tachycardia, pulmonary P wave and right ventricular high voltage could be detected in her electrocardiogram. Right ventricular hypertrophy, severe pulmonary hypertension, and left ventricular ejection fraction 70% were shown in her color doppler echocardiography. Chest CT images were shown in Figure 2B. Laboratory tests: WBC 11.0x109/L, neutrophil percentage 80%, hemoglobin 170g/L, platelet was normal in the blood routine test; the urine routine, liver and kidney function tests, D-dimer, procalcitonin, and fungal-D-glucan were all at normal level; brain natriuretic peptide (BNP) 126pg/mL; the antinuclear antibody spectrum, anti-neutrophil cytoplasmic antibodies, serum complements, rheumatoid factor, cyclic citrullinated peptide, immuno-globulins and anti-cardiolipin antibody were all negative or normal. Besides, no abnormalities were found in the tumor markers. The patient was given antibiotics and intravenous glucocorticoid, however, her condition improved slightly. Unfortunately, the patient died at home in late 2016.
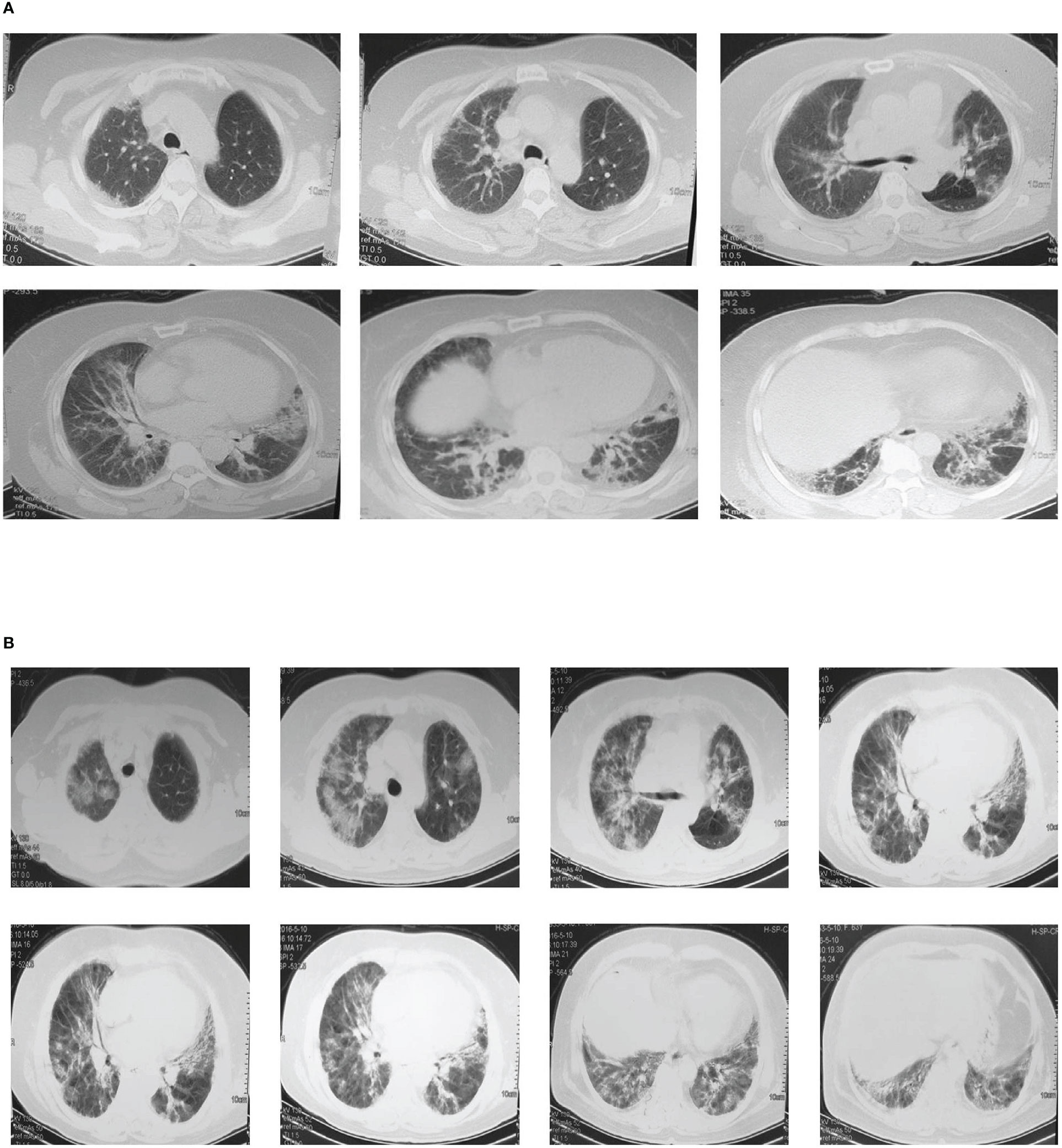
Figure 2 Chest CT images of the proband. (A) Multiple ground-glass and grid shadows distributing around the lungs and under the pleura were found in both lungs In December 2015. (B) Multiple patchy shadows, grid shadows and cable shadows were diffusely distributed in both lungs, and some of the shadows fused into large areas in May 2016.
NO.101, who had been deceased when the pedigree was investigated, suffered from T2DM and ILD.
NO.201, who had been deceased at the age of 38, suffered from T2DM and ILD.
NO.202, who had been deceased at the age of 40 when the pedigree was investigated, suffered from T2DM and ILD.
NO.204, who was still alive when the pedigree was investigated, had a history of T2DM for 8 years and ILD for 2 years. The patients once received more than 1 year of oral glucocorticoid treatment, which had been stopped when the pedigree was investigated. The insulin was still regularly used to treat her diabetes.
NO.205, who had been deceased at the age of 53 when the pedigree was investigated, suffered from T2DM and ILD.
NO.206, who had a history of T2DM for 5 years and denied any other diseases, was receiving insulin treatment when the pedigree was investigated. No abnormal changes showed in her chest HRCT images.
NO.303, who had a history of T2DM for 5 years, ILD for 1 year, was receiving regular oral glucocorticoid and insulin treatment when the pedigree was investigated.
NO.304, who had a history of T2DM for 6 years and denied any other diseases, was receiving regular insulin treatment when the pedigree was investigated. No abnormal changes showed in his chest HRCT images.
NO.307, 308, 301, 302, 311, 305, 306, 309 and 310: They didn’t have diabetes, cough, dyspnea and other respiratory symptoms. No abnormal changes showed in their chest HRCT images.
According to the inclusion criteria and informed consent, NO.204 (the proband’s younger sister), NO.206 (the proband’s youngest sister), NO.303 (the son of the proband’s eldest sister), NO.304 (the eldest son of the proband’s eldest brother), NO.305 (the second son of the proband’s eldest brother), NO.307 (son of the proband), NO.308 (daughter of the proband) and NO.309 (the son of the proband’s younger sister) of the pedigree map were included in this study. DNA was extracted from the blood of each subject, and the eight samples were re-sequenced to obtain the data of CNVs, SVs, SNPs and InDels from each sample.
We compared the sequencing data of each DNA sample with that of a control sample, and detected CNVs through the different distribution of reads. In this study, 23 genes existing in at least 7 subjects (> 80%) that might be affected by CNVs were selected out as shown in Table 1.
GO analysis and biological pathway analysis about the 23 genes were carried out. About cellular components, genes related to the nucleus accounted for the largest proportion. The molecular functions of 21.1% of the genes were unknown, while the rest genes were related to transcription factor activity, receptor activity, transferase activity and DNA binding. About biological processes, genes related to the metabolism of base, nucleoside, nucleotide and nucleic acid were the most abundant, followed by genes related to signal transduction and cell communication. The 23 genes were involved in many signaling pathways, such as ARF6 signaling pathway, PI3K signaling pathway, mTOR signaling pathway, ErbB signaling pathway, S1P1 signaling pathway, etc. Further analysis in STRING database didn’t detect significant enrichment.
According to the literature, the relevant studies on the 17 genes affected by CNV, including SH3RF3, UGT2B15, TRIM31, MICB, TRIM40, C6orf10, PGBD2, CHAMP1, PDPR, ZSCAN18, ERICH2, SIRPB1, APOBEC3B, RPL23AP82, CYB561D2, EPHA6 and HCG17, were not found in T2DM or ILD. The other 6 genes, including CREM, GCSH, KALRN, ECE2, HCG18 and GPSM1, had a certain role in the pathogenesis of T2DM and its complications. However, their roles in the development of ILD had not been studied yet.
SVs were detected by comparing the sequencing data of each sample with that of a control sample. In this study, 190 genes existing in at least 7 subjects (> 80%) that might be affected by SVs were selected out as shown in Table 2.
The 190 genes existing in at least 7 subjects which were affected by SVs all distributed on autosomes. About cell components, genes related to extracellular components accounted for the highest proportion. About cell function, the percentages of genes related to cell adhesion molecule activity, extracellular matrix composition and growth factor activity ranked among the top three. According to the biological process, the proportion of genes related to cell signal transduction and cell communication ranked among the top two. By biological pathway analysis, we found that the main pathways in which these genes were involved included neuronal system biological processes, EMT, chemical synaptic and postsynaptic transmission signaling. Further, the PPI network (Figure 3) of the proteins corresponding to the genes affected by SVs was constructed in the STRING database. The PPI network revealed that 15 proteins were associated with extracellular matrix components, 42 were involved in cell signal transduction, 54 belonged to glycoproteins, and 28 belonged to secretory proteins. The hub nodes and the four-color nodes in the PPI (the four-color nodes represented the proteins belonging to secretory protein, glycoprotein and extracellular matrix component at the same time, and also participating signal transduction) were analyzed, including PLG, MATN1, ANGPT1, MEPE, COL4A2, NID2, IMPG2, GDF10, SFRP1, ABI3BP, SBSPON, CDH2, SPP1, PTPRD and NRCAM. Whether each node played a role in the development of T2DM or ILD was still unclear. In addition, whether the genes corresponding to these nodes would form fusion genes, affect gene expression or change their phenotypes after being affected by SV had not been studied yet.
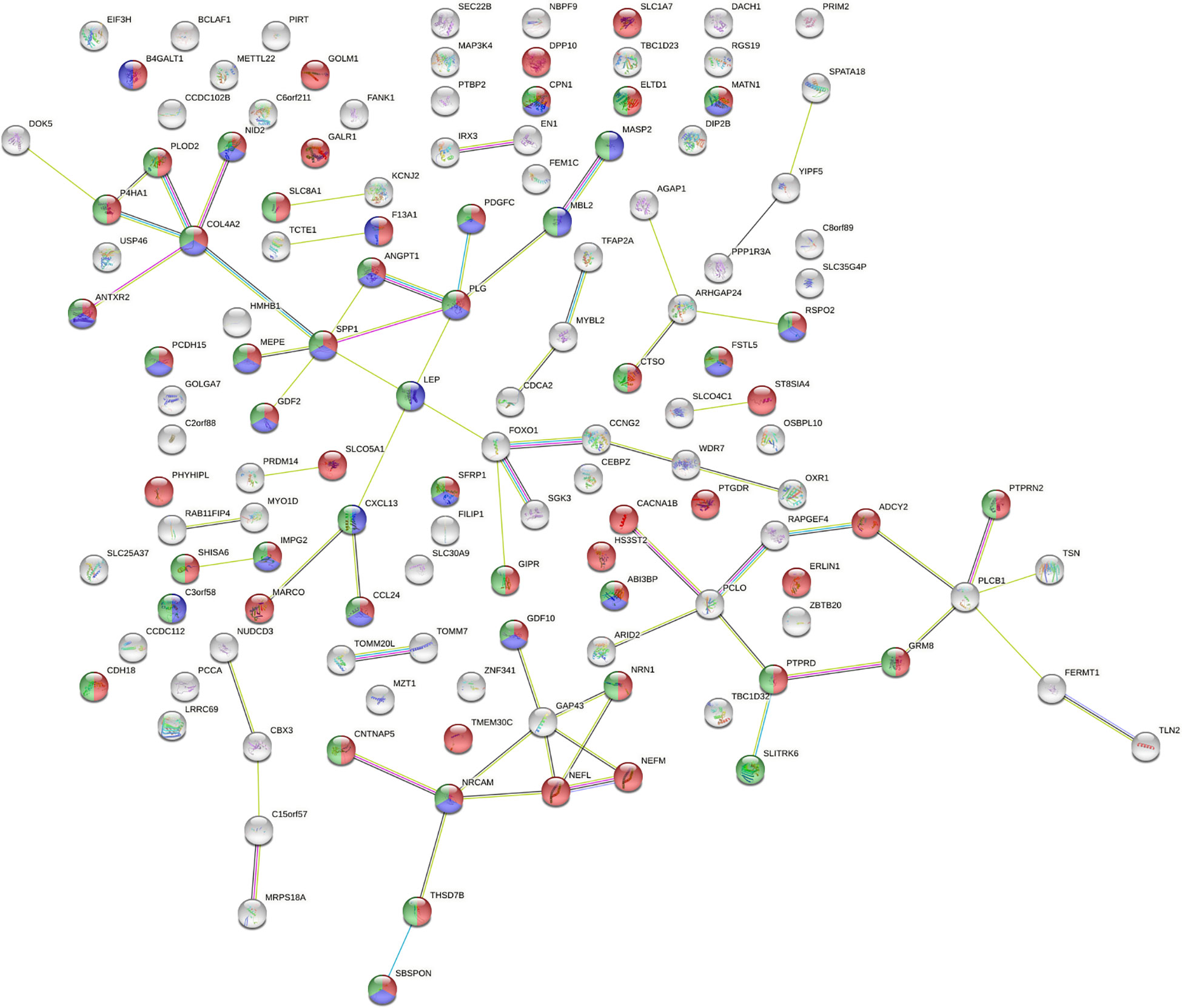
Figure 3 Analysis of protein-protein interaction network (PPI) affected by SVs. The number of nodes was 130, with each node representing a protein, 53 red nodes representing glycoproteins, 41 green nodes representing cell signal transduction, and 28 blue nodes representing secretory proteins. There were a total of 68 lines between nodes, each line representing the interaction between two proteins. Red line – gene fusion, green line – gene neighborhood, blue line – gene co-occurrence, purple line – experimentally determined, yellow line – textming, light blue line – from curated databases, black line – co-expression. PPI enrichment P = 1.57x10-4.
20263-20507 SNPs in 3’-UTR, 5’-UTR and exonic regions were genotyped in each subject, including synonymous, non-synonymous, stop-gain and stop-loss. 510-587 InDels were genotyped in each subject, including non-frameshift deletion, non- frameshift insertion, frameshift deletion, frameshift insertion, stop-gain, and stop-loss.
70 common InDels distributed in the coding region on the autosomes and X chromosomes in at least 7 subjects, including 40 frameshift deletions and frameshift insertions. The genes with frameshift deletions are shown in Table 3, and the genes with frameshift insertions are shown in Table 4.
GO classification and biological pathway analysis were performed about the 40 genes with frameshift deletion. About the cell components, the genes related to cell membrane components were in the majority. About the molecular functions, the function of 26.5% of the genes were unknown, while the rest were related to G protein coupled receptor activity and cellular structural molecular activity. In the biological process, the biological process of 26.5% genes was unknown, while the rest were mostly related to cell signaling transduction and cell communication. The signaling pathways in which these genes were involved are numerous, including ARF6 signaling pathway, PI3K signaling pathway, mTOR signaling pathway, ErbB signaling pathway, S1P1 signaling pathway, and IGF1 signaling pathway, etc. Further analysis in STRING database did not detect significant enrichment of the corresponding proteins. According to the literature, only the variants of SLC22A1, TBP, ORAI1, SARM1 and COL18A1 were found to play a certain role in the development of T2DM or be the risk factors of T2DM. However, the remaining 35 genes were rarely studied in the field of T2DM or ILD.
GO classification and biological pathway analysis were carried out about the 30 genes with frameshift insertion. About cellular component, genes related to cytoplasm and nucleus accounted for a large proportion. The molecular function of 45.8% of the genes were unknown, while the rest were related to immune protein activity, transcription factor activity, and G-protein-coupled receptor activity. In biological process, the biological process of 33.3% of the genes were unknown, while the remaining related to cell signalling transduction and cell communication accounted for a large proportion. The 30 genes were involved in numerous signaling pathways, such as IL-3-mediated signaling pathway, IL-5-mediated signaling pathway, PEGFR signaling pathway, GMCSF-mediated signaling pathway, ErbB signaling pathway, S1P1 signaling pathway, IGF1 signaling pathway, etc. Further analysis in STRING database did not detect significant enrichment of the corresponding proteins. 5 mutated genes including GIGYF2, ATG3, SRA1, WNK1 and CLECL1 were found to be involved in the development of T2DM and its complications, or be the risk factors for T2DM, according to the very few related studies. The remaining 25 genes had not yet been studied in T2DM or ILD.
The number of SNPs detected by each subjects was about 20,000. In order to simplify the search scope and effectively query the SNPs that may be relevant to the T2DM complicated with ILD, we searched according to the mutated genes which had been identified to be associated with the development of ILD (including familial pulmonary fibrosis) in previous studies, including AKAP13, ATP11A, CDKN1A, DPP9, DSP, ELMOD2, FAM13A, HLA-DRB1, IL1RN, IL8, MAPT, MDGA2, MUC2, MUC5B, OBFC1, SPPL2C, TERC, TERT, TGFB1, TLR3, TOLLIP and TP53 (30). 38 SNPs within the specific genes in at least 7 subjects were summarized in Table 5.
The 38 SNPs were found in 10 genes including AKAP13, ATP11A, DSP, FAM13A, IL1RN, MAPT, TP53, MUC2, MUC5B, OBFC1 and SPPL2C. According to the literature, the roles of AKAP13 SNPs rs8110, rs13225, rs3169121, rs2542604 and rs1808339, ATP11A SNPs rs7985702 and rs1046790, IL-1RN SNP RS315951, TP53 SNP rs2909430, MUC2 SNPs rs41411848, rs41345745 and rs57737240, MAPT SNP rs2258689, OBFC1 SNPs rs10786775, rs2487999, rs4917405 and rs911547, as well as SPPL2c SNPs rs242944 and rs171443 in the pathogenesis of T2DM or ILD were unclear; OBFC1 SNP rs4387287 might be closely related to the susceptibility of T2DM, but its role in ILD remained unclear; DSP rs2076295 and FAM13A rs2609255 had been confirmed to be associated with some types of ILD, but their roles in ILD remained unclear; MUC5B SNP rs2943512 had been identified to be significantly associated with the susceptibility of T2DM, and the over-expressed MUC5B in the distal airway and alveolar cavity had been confirmed to be closely related to the development of ILD.
To sum up, the roles of most variants in the pathogenesis of T2DM or ILD were unclear. Nevertheless, MUC5B SNP rs2943512 (A > C) was considered to be a potentially pathogenic mutation associated with T2DM complicated with ILD. Next, the function experiment of MUC5B in bronchial epithelial cells was carried out, laying a foundation for the mechanism exploration of T2DM complicated with ILD.
The viability of BEAS-2B cells stimulated by high glucose was detected by CCK-8 assay. The results demonstrated that high glucose 25mM (48h), 30mM (48h), 20mM (72h), 25mM (72h) and 30mM (72h) could inhibited cell growth significantly (***P<0.001) (Figure 4).
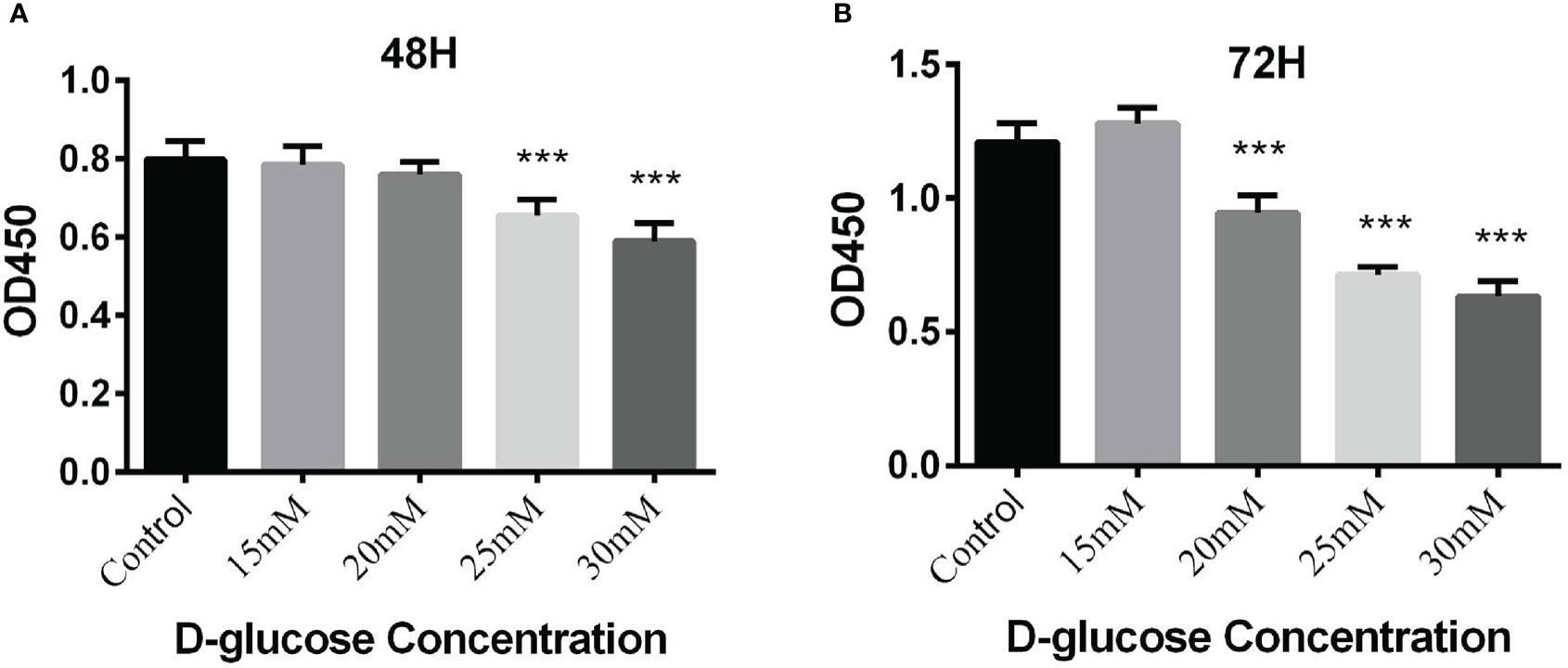
Figure 4 (A) Proliferation of BEAS-2B cells cultured in the medium containing different concentrations of glucose for 48 hours by CCK-8 assay; (B) Proliferation of BEAS-2B cells cultured in the medium containing different concentrations of glucose for 72 hours by CCK-8 assay (***P < 0.001).
The BEAS-2B cells in normal RPMI1640 medium (D-glucose concentration 11.11mM) was regarded as control, while the cells in medium containing 20mM, 25mM and 30mM glucose for 72h were experimental groups. MUC5B mRNA and MUC5B protein in the supernatant in these groups were detected, respectively. The results showed that compared with the control group, MUC5B mRNA in 20mM, 25mM and 30mM high-glucose groups were statistically increased at 72h (***P < 0.001) (Figure 5A), and MUC5B in the supernatant of 25mM and 30mM high-glucose groups were statistically increased at 72h (***P < 0.001) (Figure 5B). Finally, 30mM glucose stimulation for 72h was chosen as the subsequent experimental conditions.
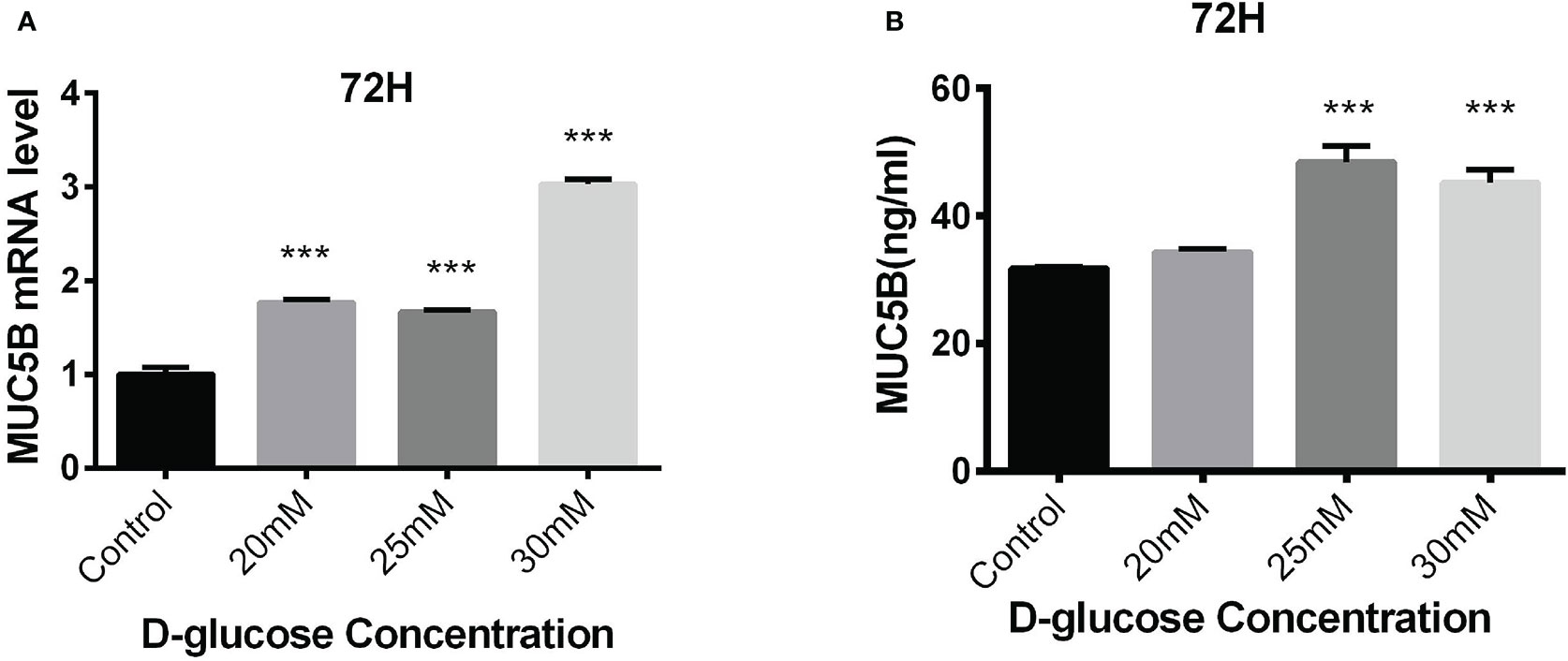
Figure 5 Effects of high glucose on the expression of MUC5B in BEAS-2B cells. (A) Compared with the control group, MUC5B mRNA in BEAS-2B cells were significantly increased after 20mM, 25mM and 30mM high-glucose stimulation for 72 hours (***P < 0.001). (B) Compared with the control group,MUC5B in the supernatant were significantly increased after 25mM and 30mM high-glucose stimulation for 72 hours (***P < 0.001).
To identify the effects of MUC5B on the production of cytokines in BEAS-2B cells, the cells were cultured in medium containing 30mM glucose for 72 h following the transfection of MUC5B shRNA into the cells (Figure 6A). Subsequently, IL-1 β and IL-6 in the supernatant were detected by ELISA, respectively. (Figures 6B, C). The results showed that the concentrate ions of IL-1β and IL-6 in high glucose group were both significantly increased compared to the control (***P < 0.001). While, compared to the high glucose group, the concentrations of IL-1β and IL-6 were both significantly decreased when MUC5B was knockdown even stimulated by high glucose (**P <0.01, ***P < 0.001).
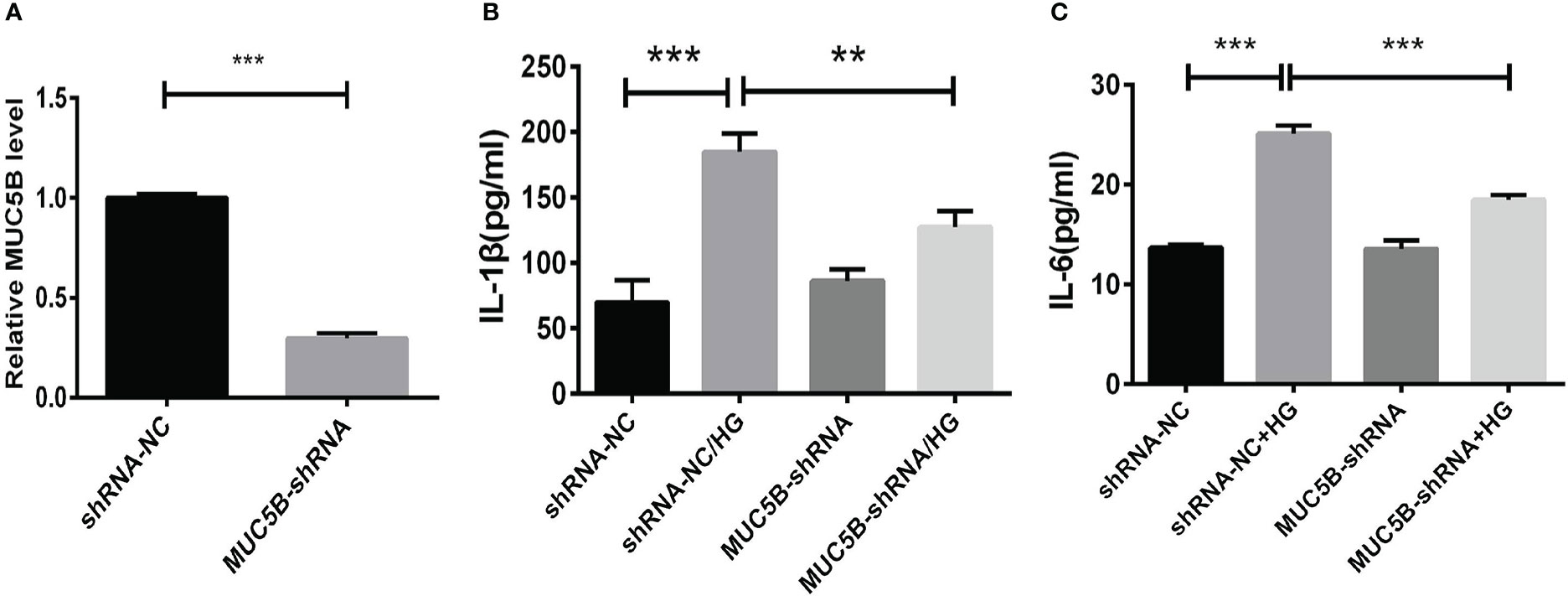
Figure 6 Effects of MUC5B on cytokine production in BEAS-2B cells stimulated by high glucose. (A) The efficiency of MUC5B-ShRNA transfection into BEAS-2B cells was detected by RT-PCR. After MUC5B knockdown, BEAS-2B cells were stimulated by 30mM glucose for 72 hours. (B) IL-1 β and (C) IL-6 in the supernatant were both significantly increased compared to the control (**P < 0.01, ***P < 0.001).
After culturing BEAS-2B cells in the medium containing 30mM glucose for 15min, 30min, 1h, 3h and 6h, p-ERK1/2, p-P38, p-JNK, and p-IκB in each group were detected. The results showed that compared with the control group, p-ERK1/2 was up-regulated when stimulated by the high glucose for 15min and 30min (Figure 7A,*P <0.05, ***P < 0.001). However, no change of p-P38, p-JNK or p-IκB was detected at each point from 15min to 6h (Figure 7B).
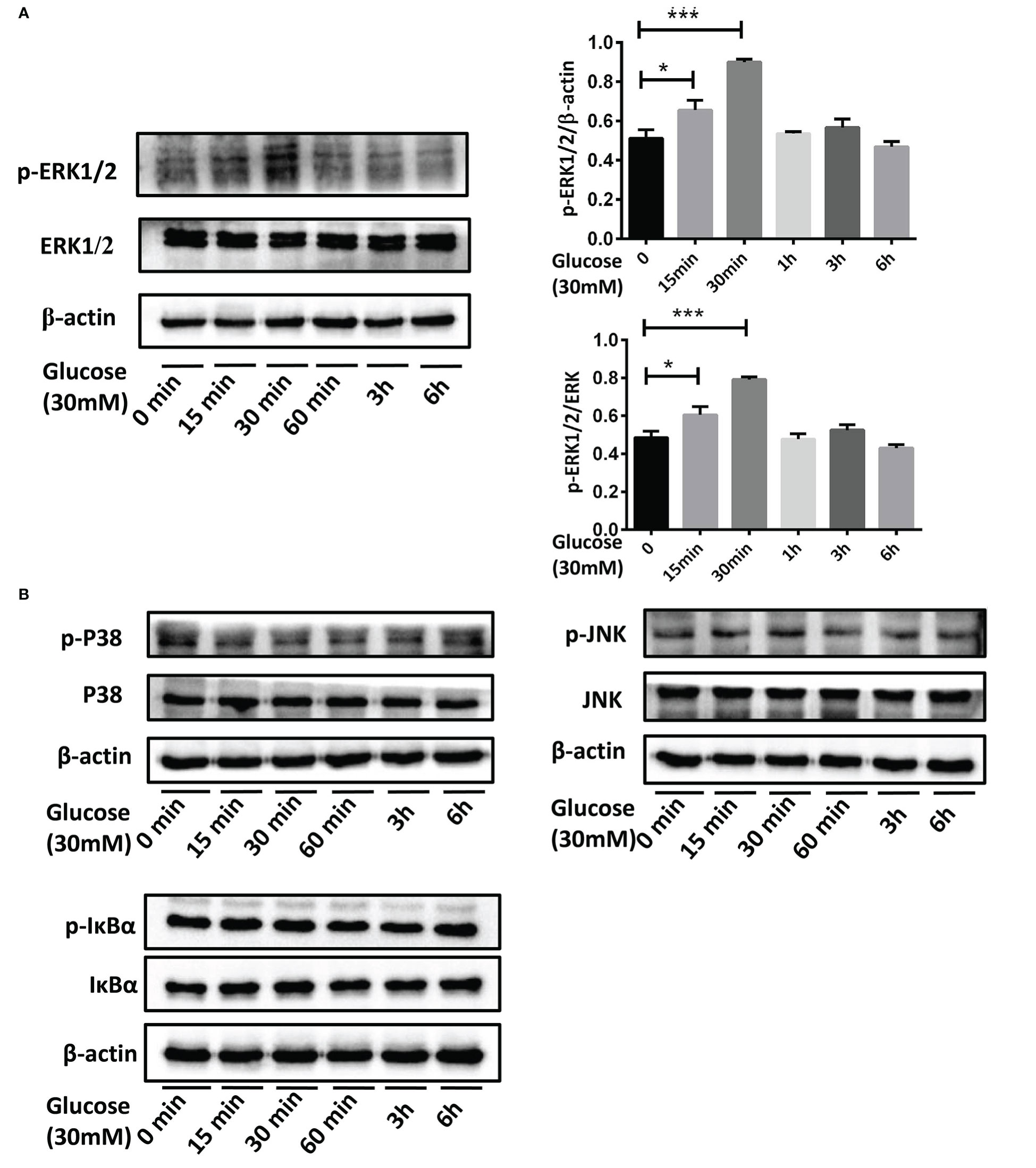
Figure 7 The activation of MAPK and NF-κB pathways in BEAS-2B cells stimulated by high glucose. (A) Compared with the control group, p-ERK1/2 in BEAS-2B cells was up-regulated when stimulated by 30mM glucose for 15 minutes and 30 minutes. (B) P38, JNK or IκB didn’t activate at each point from 15 minutes to 6 hours when stimulated by 30mM glucose. The blots and densitometry analysis data are representative of three independent experiments. *P <0.05, and ***P < 0.001.
To confirm our suppose that over-expressed MUC5B could promote the synthesis of IL-1β and IL-6 by activating ERK1/2, BEAS-2B cells were stimulated by 30mM glucose for 30min following the silencing of MUC5B gene. Subsequently, p-ERK1/2 in each group was detected by western blot. The results showed that compared with the high glucose group, p-ERK1/2 in the MUC5B knockdown cells stimulated by high glucose was significantly down-regulated (Figure 8, **P <0.01), suggesting over-expressed MUC5B could promote ERK1/2 activation in BEAS-2B cells when stimulated by high glucose.
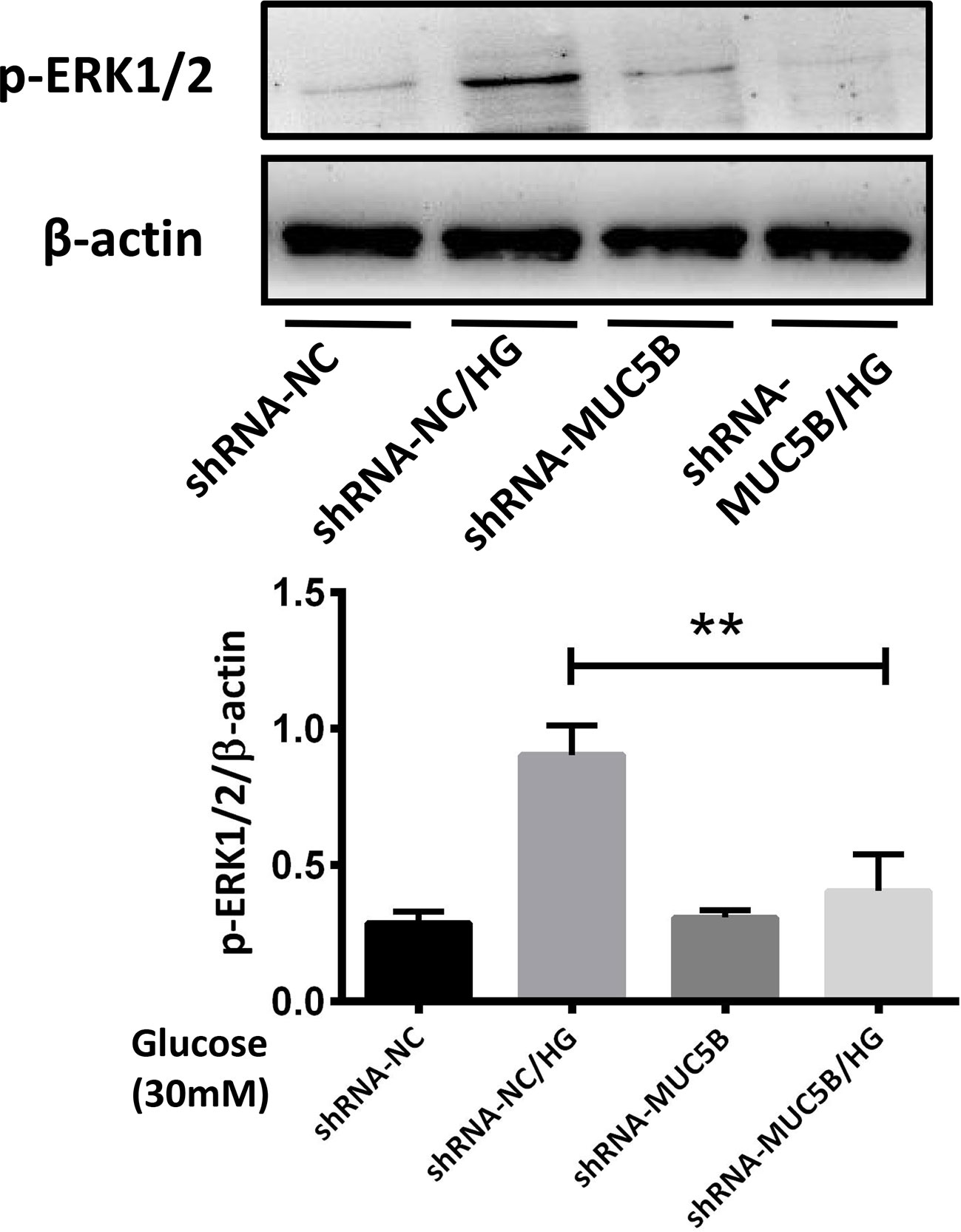
Figure 8 MUC5B promoted ERK1/2 activation in BEAS-2B cells stimulated by high glucose. Compared with high glucose group, p-ERK1/2 in the MUC5B knockdown cells stimulated by 30mM glucose for 30min was significantly down-regulated (**P <0.01). The blots and densitometry analysis data are representative of three independent experiments. **P <0.01.
T2DM and ILD both belong to complex diseases. In recent years, genetic studies about the both diseases have been gradually extensive, and mutations related to the risks or pathogenesis of the two diseases have been constantly revealed. With further studies, it has been recognized that ILD could be a complication of T2DM. However, the relevant studies about genetic variations promoting ILD in T2DM patients are still rare.
In this study, a pedigree with T2DM complicated with ILD including three generations was selected. Familial T2DM is not rare in clinical practice, however, pedigree with both T2DM and ILD are indeed rare. In this pedigree, almost all the first and second generation members had both T2DM and ILD. 8 living members of the pedigree were included as subjects. By conducting the whole-genome re-sequencing of each subject’s blood DNA sample, it was found that the healthy subjects from the third generation also had the same potential pathogenic genetic variants as the patients from the second and third generation. It suggested that T2DM was relevant to the development of ILD, and T2DM complicated with ILD might be heredofamilial.
The common genetic variants of at least 7 subjects were further screened out after gene sequencing, involving numerous SVs, CNVs, SNPs and InDels. Previous studies considered that the susceptibility of many diseases was basically attributed to SNPs, however, SNP can only explain a part of the heritability of diseases. Our study also provided genes which were affected by InDels, SVs and CNVs, so as to provide more genetic information for the mechanism research of ILD in diabetic patients. Up to now, the roles of most variants in T2DM or ILD remained unclear, and only a small part might be involved in the development of T2DM or ILD. While, MUC5B variation was the specific one found to be related to both T2DM and ILD in the study, and MUC5B SNP rs2943512 (A > C) was considered to be a potentially pathogenic mutation associated with T2DM complicated with ILD. Meanwhile, the over-expressed MUC5B protein in the distal airway and alveolar cavity had also been considered to be closely related to the development of idiopathic pulmonary fibrosis (IPF) (33, 34). Therefore, it was of great significance to investigate the role of MUC5B SNP rs2943512 (A > C) or abnormal MUC5B protein in the development of T2DM complicated with ILD.
MUC5B encodes mucin 5B protein, which is a glycosylated macromolecular component of mucus and produced by mucinous cells in bronchial submucosal glands and type II alveolar epithelial (ATII) cells. Normally, MUC5B plays the physiological roles of maintaining airway homeostasis (35), involving capturing inhaled particles and bacteria which are transported out of the airway by cilia oscillating or coughing. In addition, MUC5B could also helps remove endogenous debris, including dead epithelial cells and white blood cells. When MUC5B is over-expressed by stimulus, the capacity of mucociliary clearance will be impaired, resulting in excessive retention of inhaled particles, microorganisms or endogenous inflammatory debris, and mediating the reactive fibrosis in the bronchoalveolar region, promoting the interstitial lesions (33, 34). Although the mechanisms of MUC5B mediating the interstitial fibrosis are still unknown, it is speculated that it may be related to the injury of ATII cells caused by the excessive MUC5B.
Both the endogenous factors (such as genetics and aging) and environmental factors have been implicated in ATII damages. Infection, drugs, poisons, etc., all cause a certain degree of damages to ATII in ILD with known etiology. In T2DM, the accompanied inflammation is not only closely related to the development of diabetic complications (36), but also could cause the impairment of lung function due to significantly elevated cytokines such as TNF-α, IL-1β and IL-6 (37). However, whether high glucose promotes the development of ILD by causing inflammatory injury of ATII cells needs to be further studied.
In our study, we identified MUC5B rs2943512 (A>C) in the pedigree. However, the effects of this SNP on the expression of MUC5B in ATII cells is unclear. The previous study showed that the expression of MUC5B was significantly elevated in pancreatic tissue of patients with T2DM compared to those without T2DM (38), therefore, we speculated that MUC5B rs2943512 (A>C) might cause the over-expression of MUC5B in lung tissue of T2DM patients. Even beyond the potential influence of the genetic factor of MUC5B SNP rs2943512, whether high glucose itself could cause the over-expression of MUC5B to mediate ATII cells injury and thus trigger pulmonary fibrosis is unclear. Therefore, in order to clarify the relationship between high glucose, MUC5B and ATII injury, this study selected human bronchial epithelial cells BEAS-2B to be stimulated by high glucose in this experiments, to simulate ATII cells (39) in diabetic patients. The results showed that the transcriptional and translational levels of MUC5B in BEAS-2B cells were significantly up-regulated, suggesting that MUC5B existed in the distal airways and ATII cells in the lung tissues of T2DM patients. While this hypothesis needs to be confirmed by in vivo experiments in the future.
To explore the potential mechanisms that over-expressed MUC5B promoting fibrosis in alveolar region, we started the experiment from the perspective of inflammatory injury of ATII cells, and detected the cytokines in BEAS-2B cells under high glucose stimulation. The results showed the increased IL-1β and IL-6 was accompanied by the over-expressed MUC5B. Numerous studies have confirmed that IL-1β could cause apoptosis in different types of cell including ATII. Moreover, IL-1β could also play a pro-fibrotic role in certain pathological conditions (40–43). In vivo experiments have shown that IL-1β induces progressive pulmonary fibrosis through long-term activation of TGF-β signaling (40). In vitro, IL-1β could promote EMT by activating TGF-β in bronchial epithelial cells (44, 45). In addition, IL-1β also stimulates the release of IL-6, which not only play the pro-inflammatory role, but also aggravates the pulmonary fibrosis by activating STAT3 pathway (46).
To clarify the interaction between MUC5B and the cytokines, RNA interference was applied to silence MUC5B before BEAS-2B cells were stimulated by high glucose. It was found that IL-1β and IL-6 were significantly decreased in MUC5B knockdown BEAS-2B cells. This finding had rarely been reported in previous studies, and suggested that MUC5B could promote the production of IL-1β and IL-6 in bronchial epithelial cells when stimulated by high glucose, and the over-expressed MUC5B causing inflammatory damage to ATII cells might initiating the pulmonary interstitial fibrosis.
This study also explored the mechanisms of over-expressed MUC5B promoting the synthesis of IL-1β and IL-6 in BEAS-2B cells. It was found that high glucose could cause the activation of ERK1/2 in BEAS-2B cells, while the activation was significantly decreased after MUC5B silencing, suggesting that the changes in MUC5B transcriptional or translational level might affect ERK1/2 activation, which at least partly explained the changes in IL-1β and IL-6. However, whether MUC5B also affect the synthesis of IL-1β and IL-6 through other pathways remains to be explored. In addition, the molecular mechanisms by which high glucose promotes the over- expression of MUC5B as well as MUC5B promotes the activation of ERK1/2 are both unclear at present, which are needed to be studied in the future.In addition,the lack of further experiments to validate the implications of MUC5B site-specific mutation for the functions of BEAS-2B cells and for the aggravation of T2DM and ILD is also one of the limitations of this study. Therefore, we will also focus on the cell and animal experiments related to MUC5B site-specific mutation in the next step.
In this study, a pedigree with T2DM complicated with ILD was selected, and 8 members of the pedigree were included as subjects. Whole-genome resequencing of each subject’s blood was conducted, and the common genetic variants of at least 7 subjects were further screened out, involving numerous SVs, CNVs, SNPs and InDels. The identification of these genetic variants in the pedigree enriches our understanding of the potential genetic contributions to T2DM complicated with ILD. MUC5B SNP rs2943512 (A > C) or the up-regulated MUC5B in bronchial epithelial cells may be an important factor in promoting ILD in T2DM patients, making MUC5B a potential biological marker for the development of ILD in diabetic patients, and laying a foundation for future exploration about the pathogenesis of T2DM complicated with ILD.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA919498.
The studies involving human participants were reviewed and approved by the Ethics Committee of The Second Hospital of Jilin University, China. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
QZ and YW collected, analyzed and interpreted the data, drafted and revised the manuscript. CT conducted the cellular experiments, and helped revise the manuscript. JYu and YL helped process and analyze the data. JYa conceived, designed and supervised the study, and guided the revision of the manuscript. All authors read and approved the final manuscript.
This study was supported by grants from the Science and Technology Department of Jilin Province (20190201279JC), and the Finance Department of Jilin Province (20106145).
We thank the patients and their families for their participation in this study. We thank the Science and Technology Department of Jilin Province and the Finance Department of Jilin Province for their funding.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. A. American Diabetes. 2. classification and diagnosis of diabetes: Standards of medical care in diabetes-2018. Diabetes Care (2018) 41:S13–27. doi: 10.2337/dc18-S002
2. De Santi F, Zoppini G, Locatelli F, Finocchio E, Cappa V, Dauriz M, et al. Type 2 diabetes is associated with an increased prevalence of respiratory symptoms as compared to the general population. BMC Pulm Med (2017) 17:101. doi: 10.1186/s12890-017-0443-1
3. Ehrlich SF, Quesenberry CP Jr., Van Den Eeden SK, Shan J, Ferrara A. Patients diagnosed with diabetes are at increased risk for asthma, chronic obstructive pulmonary disease, pulmonary fibrosis, and pneumonia but not lung cancer. Diabetes Care (2010) 33:55–60. doi: 10.2337/dc09-0880
4. Hu Y, Ma Z, Guo Z, Zhao F, Wang Y, Cai L, et al. Type 1 diabetes mellitus is an independent risk factor for pulmonary fibrosis. Cell Biochem Biophys (2014) 70:1385–91. doi: 10.1007/s12013-014-0068-4
6. Klein OL, Kalhan R, Williams MV, Tipping M, Lee J, Peng J, et al. Lung spirometry parameters and diffusion capacity are decreased in patients with type 2 diabetes. Diabetes Med (2012) 29:212–9. doi: 10.1111/j.1464-5491.2011.03394.x
7. Talakatta G, Sarikhani M, Muhamed J, Dhanya K, Somashekar BS, Mahesh PA, et al. Diabetes induces fibrotic changes in the lung through the activation of TGF-beta signaling pathways. Sci Rep (2018) 8:11920. doi: 10.1038/s41598-018-30449-y
8. Popov D, Simionescu M. Structural and transport property alterations of the lung capillary endothelium in diabetes. Ital J Anat Embryol (2001) 106:405–12.
9. Carlson EC, Audette JL, Veitenheimer NJ, Risan JA, Laturnus DI, Epstein PN. Ultrastructural morphometry of capillary basement membrane thickness in normal and transgenic diabetic mice. Anat Rec A Discovery Mol Cell Evol Biol (2003) 271:332–41. doi: 10.1002/ar.a.10038
10. Vracko R, Thorning D, Huang TW. Basal lamina of alveolar epithelium and capillaries: quantitative changes with aging and in diabetes mellitus. Am Rev Respir Dis (1979) 120:973–83. doi: 10.1164/arrd.1979.120.5.973
11. Kim YJ, Park JW, Kyung SY, Lee SP, Chung MP, Kim YH, et al. Clinical characteristics of idiopathic pulmonary fibrosis patients with diabetes mellitus: the national survey in Korea from 2003 to 2007. J Korean Med Sci (2012) 27:756–60. doi: 10.3346/jkms.2012.27.7.756
12. Stavniichuk R, Shevalye H, Lupachyk S, Obrosov A, Groves JT, Obrosova IG, et al. Peroxynitrite and protein nitration in the pathogenesis of diabetic peripheral neuropathy. Diabetes Metab Res Rev (2014) 30:669–78. doi: 10.1002/dmrr.2549
13. Richter K, Konzack A, Pihlajaniemi T, Heljasvaara R, Kietzmann T. Redox-fibrosis: Impact of TGFbeta1 on ROS generators, mediators and functional consequences. Redox Biol (2015) 6:344–52. doi: 10.1016/j.redox.2015.08.015
14. Toda N, Mukoyama M, Yanagita M, Yokoi H. CTGF in kidney fibrosis and glomerulonephritis. Inflammation Regener (2018) 38:14. doi: 10.1186/s41232-018-0070-0
15. Sanchez E, Lecube A, Betriu A, Hernandez C, Lopez-Cano C, Gutierrez-Carrasquilla L, et al. Subcutaneous advanced glycation end-products and lung function according to glucose abnormalities: The ILERVAS project. Diabetes Metab (2019) 45:595–8. doi: 10.1016/j.diabet.2018.04.002
16. Caspar-Bell G, Dhar I, Prasad K. Advanced glycation end products (AGEs) and its receptors in the pathogenesis of hyperthyroidism. Mol Cell Biochem (2016) 414:171–8. doi: 10.1007/s11010-016-2669-2
17. Prasad K, Mishra M, Stress AGE-RAGE. Stressors, and antistressors in health and disease. Int J Angiol (2018) 27:1–12. doi: 10.1055/s-0037-1613678
18. Machahua C, Montes-Worboys A, Llatjos R, Escobar I, Dorca J, Molina-Molina M, et al. Increased AGE-RAGE ratio in idiopathic pulmonary fibrosis. Respir Res (2016) 17:144. doi: 10.1186/s12931-016-0460-2
19. Zou XZ, Gong ZC, Liu T, He F, Zhu TT, Li D, et al. Involvement of epithelial-mesenchymal transition afforded by activation of LOX-1/ TGF-beta1/KLF6 signaling pathway in diabetic pulmonary fibrosis. Pulm Pharmacol Ther (2017) 44:70–7. doi: 10.1016/j.pupt.2017.03.012
20. Song JS, Kang CM, Park CK, Yoon HK, Lee SY, Ahn JH, et al. Inhibitory effect of receptor for advanced glycation end products (RAGE) on the TGF-beta-induced alveolar epithelial to mesenchymal transition. Exp Mol Med (2011) 43:517–24. doi: 10.3858/emm.2011.43.9.059
21. Ayaub EA, Kolb PS, Mohammed-Ali Z, Tat V, Murphy J, Bellaye PS, et al. GRP78 and CHOP modulate macrophage apoptosis and the development of bleomycin-induced pulmonary fibrosis. J Pathol (2016) 239:411–25. doi: 10.1002/path.4738
22. Engin F. ER stress and development of type 1 diabetes. J Investig Med (2016) 64:2–6. doi: 10.1097/JIM.0000000000000229
23. Maamoun H, Abdelsalam SS, Zeidan A, Korashy HM, Agouni A. Endoplasmic reticulum stress: A critical molecular driver of endothelial dysfunction and cardiovascular disturbances associated with diabetes. Int J Mol Sci (2019) 20(7):1658. doi: 10.3390/ijms20071658
24. Burgos-Moron E, Abad-Jimenez Z, Maranon AM, Iannantuoni F, Escribano-Lopez I, Lopez-Domenech S, et al. Relationship between oxidative stress, ER stress, and inflammation in type 2 diabetes: The battle continues. J Clin Med (2019) 8(9):1385. doi: 10.3390/jcm8091385
25. Chiba H, Otsuka M, Takahashi H. Significance of molecular biomarkers in idiopathic pulmonary fibrosis: A mini review. Respir Investig (2018) 56:384–91. doi: 10.1016/j.resinv.2018.06.001
26. Wang K, Ju Q, Cao J, Tang W, Zhang J. Impact of serum SP-a and SP-d levels on comparison and prognosis of idiopathic pulmonary fibrosis: A systematic review and meta-analysis. Med (Baltimore) (2017) 96:e7083. doi: 10.1097/MD.0000000000007083
27. Lopez-Cano C, Lecube A, Garcia-Ramirez M, Munoz X, Sanchez E, Seminario A, et al. Serum surfactant protein d as a biomarker for measuring lung involvement in obese patients with type 2 diabetes. J Clin Endocrinol Metab (2017) 102:4109–16. doi: 10.1210/jc.2017-00913
28. Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. Large-Scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet (2012) 44:981–90. doi: 10.1038/ng.2383
29. Roizen JD, Bradfield JP, Hakonarson H. Progress in understanding type 1 diabetes through its genetic overlap with other autoimmune diseases. Curr Diabetes Rep (2015) 15:102. doi: 10.1007/s11892-015-0668-4
30. Borie R, Le Guen P, Ghanem M, Taille C, Dupin C, Dieude P, et al. The genetics of interstitial lung diseases. Eur Respir Rev (2019). 28(153):190053. doi: 10.1183/16000617.0053-2019
31. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics (2009) 25:1754–60. doi: 10.1093/bioinformatics/btp324
32. Navarro J, Nevado B, Hernandez P, Vera G, Ramos-Onsins SE. Optimized next-generation sequencing genotype-haplotype calling for genome variability analysis. Evol Bioinform Online (2017) 13:1176934317723884. doi: 10.1177/1176934317723884
33. Boucher RC. Idiopathic pulmonary fibrosis–a sticky business. N Engl J Med (2011) 364:1560–1. doi: 10.1056/NEJMe1014191
34. Button B, Cai LH, Ehre C, Kesimer M, Hill DB, Sheehan JK, et al. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science (2012) 337:937–41. doi: 10.1126/science.1223012
35. Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, et al. Muc5b is required for airway defence. Nature (2014) 505:412–6. doi: 10.1038/nature12807
36. Lontchi-Yimagou E, Sobngwi E, Matsha TE, Kengne AP. Diabetes mellitus and inflammation. Curr Diabetes Rep (2013) 13:435–44. doi: 10.1007/s11892-013-0375-y
37. Dennis RJ, Maldonado D, Rojas MX, Aschner P, Rondon M, Charry L, et al. Inadequate glucose control in type 2 diabetes is associated with impaired lung function and systemic inflammation: a cross-sectional study. BMC Pulm Med (2010) 10:38. doi: 10.1186/1471-2466-10-38
38. Chen G, Zhang Z, Adebamowo SN, Liu G, Adeyemo A, Zhou Y, et al. Common and rare exonic MUC5B variants associated with type 2 diabetes in han Chinese. PloS One (2017) 12:e0173784. doi: 10.1371/journal.pone.0173784
39. Zhang C, Zhu X, Hua Y, Zhao Q, Wang K, Zhen L, et al. YY1 mediates TGF-beta1-induced EMT and pro-fibrogenesis in alveolar epithelial cells. Respir Res (2019) 20:249. doi: 10.1186/s12931-019-1223-7
40. Kolb M, Margetts PJ, Anthony DC, Pitossi F, Gauldie J. Transient expression of IL-1beta induces acute lung injury and chronic repair leading to pulmonary fibrosis. J Clin Invest (2001) 107:1529–36. doi: 10.1172/JCI12568
41. Pan LH, Ohtani H, Yamauchi K, Nagura H. Co-Expression of TNF alpha and IL-1 beta in human acute pulmonary fibrotic diseases: an immunohistochemical analysis. Pathol Int (1996) 46:91–9. doi: 10.1111/j.1440-1827.1996.tb03584.x
42. Mozaffarian A, Brewer AW, Trueblood ES, Luzina IG, Todd NW, Atamas SP, et al. Mechanisms of oncostatin m-induced pulmonary inflammation and fibrosis. J Immunol (2008) 181:7243–53. doi: 10.4049/jimmunol.181.10.7243
43. Wilson MS, Madala SK, Ramalingam TR, Gochuico BR, Rosas IO, Cheever AW, et al. Bleomycin and IL-1beta-mediated pulmonary fibrosis is IL-17A dependent. J Exp Med (2010) 207:535–52. doi: 10.1084/jem.20092121
44. Fan JM, Huang XR, Ng YY, Nikolic-Paterson DJ, Mu W, Atkins RC, et al. Interleukin-1 induces tubular epithelial-myofibroblast transdifferentiation through a transforming growth factor-beta1-dependent mechanism in vitro. Am J Kidney Dis (2001) 37:820–31. doi: 10.1016/S0272-6386(01)80132-3
45. Doerner AM, Zuraw BL. TGF-beta1 induced epithelial to mesenchymal transition (EMT) in human bronchial epithelial cells is enhanced by IL-1beta but not abrogated by corticosteroids. Respir Res (2009) 10:100. doi: 10.1186/1465-9921-10-100
Keywords: type 2 diabetes mellitus, interstitial lung disease, MUC5B, pedigree, whole genome re-sequencing
Citation: Zhang Q, Wang Y, Tian C, Yu J, Li Y and Yang J (2023) Clinical characteristics and genetic analysis of a Chinese pedigree of type 2 diabetes complicated with interstitial lung disease. Front. Endocrinol. 13:1050200. doi: 10.3389/fendo.2022.1050200
Received: 21 September 2022; Accepted: 28 December 2022;
Published: 17 January 2023.
Edited by:
Sen Li, Beijing University of Chinese Medicine, ChinaReviewed by:
Bijesh P, Council of Scientific and Industrial Research (CSIR), IndiaCopyright © 2023 Zhang, Wang, Tian, Yu, Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junling Yang, anVubGluZ0BqbHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.