
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Endocrinol., 09 January 2023
Sec. Bone Research
Volume 13 - 2022 | https://doi.org/10.3389/fendo.2022.1044673
This article is part of the Research TopicGut Microbiota and Gut-Associated Metabolites (GAMs) in Bone HealthView all 5 articles
Rheumatoid arthritis (RA) is a chronic destructive autoimmune disease of the joints which causes significant pain, functional disability, and mortality. Although aberrant immune cell activation induced by the imbalance between T helper Th1/Th17 and Treg cells is implicated in the RA development, its etiopathogenesis remains unclear. The presence of mucosal inflammation and systemic IgA-isotype-autoantibodies (anti-citrullinated peptide antibodies and rheumatoid factor) in pre-clinical RA supports the mucosal origin hypothesis involving altered microbiota in disease development. The gut microbiota comprises diverse bacteria, fungal and viral components, which are critical in developing host immunity. Alterations in microbial abundance are known to exacerbate or attenuate immune responses in the gut microenvironment subsequently affecting the joints. Further, these changes can provide biomarkers for disease activity and outcome in RA. Most of the research till date has been focused on describing gut bacterial components in RA. Studies on gut mycobiome and virome components in RA are relatively new and burgeoning field. Given the paucity of mycobiome or virome specific studies in RA, this review, discusses the recent findings on alterations in gut bacterial, fungal, and viral components as well as their role in regulating the spectrum of immune-pathogenic events occurring in RA which might be explored in future as a potential therapeutic target. Further, we provide an overview on inter-kingdom interactions between bacteria, fungi, and viruses in RA. The current understanding on gut microbiota modulation for managing RA is also summarised.
Rheumatoid arthritis (RA) is an autoimmune disease characterized by synovial inflammation caused by leukocyte infiltration in the joints. It affects approximately 1% of the population worldwide (1). Although the pathogenesis of RA is not well understood, it involves complex interactions between immunological, genetic, and environmental factors (2, 3). Polymorphisms in genes such as HLA-DRB1, PTPN22, CTLA-4, and PADI4 are known to be associated with a genetic predisposition for RA pathogenesis. Environmental factors such as silica exposure, smoking, and infections also contribute to pathogenesis by inducing breakdown of immune tolerance to post-translationally modified proteins. These altered proteins are presented by dendritic cells (DCs) to T cells, which in turn activate B cells to undergo plasma cell differentiation leading to secretion of autoantibodies such as anti-citrullinated protein antibodies (ACPA) and rheumatoid factor (RF) (4, 5). These auto antibodies form immune complexes, which activate immune cells; these in turn recruit various other inflammatory cells to the joints contributing to localized destructive mechanisms. Moreover, DCs promote T-helper 17 (Th17) differentiation and inhibit regulatory T (Treg) cell differentiation, thus shifting the T cell balance toward inflammation. The activated T cells then stimulate effector cells such as macrophages, fibroblasts, osteoclasts, and chondrocytes, as well as their effector molecules, leading to cartilage and bone destruction (2–4).
Although the mechanisms underlying the RA pathogenesis remains elusive, altered gut microbiota composition, namely dysbiosis, influences the autoimmune responses and disease outcomes in RA (6–11). Most studies suggest that microbial colonization of gut begins during or after birth, whereas others suggest in utero acquisition probably through contact with placental microbiota, which originates from mother’s oral and gut microbiome (12–14). However, the evidence supporting in utero colonization are weak, further supporting its acquisition during and after birth (15). Gut microbiota is less diverse at the time of birth, however, their composition, abundance, diversity is significantly influenced by diet, the feeding mode (breast-fed or formula-fed) and during infant transition from milk to solid food within the first three years of life (16–18).
Among the different microbes in the gut microbiota, bacteria are the most prevalent. The gut is also colonized by other microorganisms, such as viruses, fungi, and archaea, which play pivotal roles in modulating the mucosal barrier and immune responses (19).
In this review, we discuss gut dysbiosis with an emphasis on the bacteriome, mycobiome, and virome in RA. Further, we explain how these three components of the gut microbiome interact to influence the disease pathophysiology. Further, we overview the mechanisms underlying the activation of innate and adaptive immune responses and link intestinal dysbiosis with the development and perpetuation of RA. Finally, we propose a few strategies on modulating the gut microbiota as potential treatment approaches in RA.
Google and PubMed searches were performed using the terms ‘microbiota in RA,’ ‘gut-joint axis in RA,’ ‘microbiome,’ ‘bacteriome in RA,’ ‘gut fungi in RA/autoimmune diseases,’ ‘virome in RA/autoimmune diseases,’ ‘inter-kingdom interactions’, ‘diet,’ and ‘probiotics.’
Accumulating evidence has shown the association of an altered gut microbiota with RA pathogenesis, which triggers arthritis based on genetic susceptibility and host-related factors (10, 20).
Studies comparing the gut microbiota in patients with RA and in healthy controls have revealed enrichment of gram-positive bacteria and reduction of gram-negative bacteria. The enriched gram-positive bacteria predominantly comprise Eggerthella lenta, Clostridium asparagiforme, Lachnospiraceae bacterium, and Gordonibacter pamelaeae, which can be normalized after treatment (8). Among bacteria, Prevotellaceae is the major bacterial family associated with dysbiosis. Various studies observed the overexpansion of Prevotella copri in fecal samples from patients with RA (21–23). Further, a peptide derived from a 27 kDa protein from Prevotella copri (Pc-p27) was found to be presented on HLA-DR and was shown to stimulate a Th1 cell response in patients with new onset RA (21, 22). Along with Prevotella, other rare microorganisms have also been described in RA. Chen et al. demonstrated the abundance of Collinsella and Eggerthella in patients with RA, along with an overall reduction in intestinal diversity compared to that in the first-degree relatives (FDR) of RA patients and healthy controls (24). A study by David et al. showed increased abundance of Clostridiaceae and Epsilonproteobacteria in RA (25). Further, enrichment of Lactobacillus salivarius and depletion of Haemophilus species has been reported in the gut of Chinese patients with RA (8). Other recent studies indicated an increased abundance of Klebsiella, Escherichia-shigella, Eisenbergiella, Lactobacillus, Streptococcus, Akkermansia and Flavobacterium and depletion of Bacteriodes fraglis, Faecalibacterium, Fusicatenibacter, Megamon, Bifidobacterium, Clostridium and Enterococcus genera in RA (26–31). These studies suggest that bacteriome have a role in RA pathogenesis.
Fungi represent only 0.1–1.0% of the intestinal microbiota and are referred as mycobiota (32). Although mycobiota compositions have not been very well examined, yeast has been shown to be most dominant species of the human mycobiome, particularly Saccharomyces (S. cerevisiae), Malassezia (M. restricta), and Candida (C. albicans) (33). At the phylum level, increased abundance of Ascomycota and decreased abundance of Basidiomycota has been reported in the synovial fluid of patients with RA (34). Among the predominant fungi, C. albicans is the most frequently detected, and has been associated with many autoimmune disorders (35–37). Further, fecal samples of Chinese patients with RA showed increased abundance of Candida and Wallemia species and decreased abundance of Pholiota, Scedosporium, and Trichosporon species (38). Furthermore, a lower frequency of fungal genera such as Dendroclathra, Phacidium, and Septobasidium was found in fecal samples from older patients than younger patients with RA (38). Previous in vivo studies have implicated mycotoxins in RA pathogenesis (39). In addition, Th17 cells mediate immune responses against fungi, also play critical role in the pathogenesis of RA, further suggests that the differences in the fungal microbiome might be related to inflammatory responses in RA (36, 37, 40).
Gut viruses, collectively called the virome, are a largely understudied component of the human gut microbiome (41). A case-control study reported that crAss-like phages are significantly reduced in RA and that the hosts of these crAss-like phages are Bacteriodes vulgates and Firmicutes (42). Yutin et al. reported Prevotella copri as a host for crAss-like phages (43). A recent study characterizing the fecal virome of the RA-FDR revealed increased Bacteroidaceae-infecting phages. On stratifying the participants based on serology, the samples from ACPA-positive FDRs were found to be more enriched in Streptococcaceae- and Lachnospiraceae-infecting phages than those from ACPA-negative FDRs (44). Ruochun et al. reported enrichment of Lactococcus phages in the dental plaques of treated patients with RA as compared to untreated RA. However, minor changes in the gut virome were observed along with a significantly decreased abundance of the family Phycodnaviridae in treated patients with RA (45). As the role of the bacteriome in autoimmunity development has been well-established (46), the changes of bacteriophage abundance in at-risk individuals suggests that they might be involved in RA pathogenesis, perhaps by modulating bacterial taxa or by directly interacting with the host immune system.
The alterations in bacteriome, mycobiome and virome components of gut are listed in Table 1.
The presence of intestinal bacteria limits the fungal colonization or viral invasion of the intestine and vice versa (47–49). Interactions between bacterial, fungal, and viral components in the gut have the potential to enhance pathogenesis. Although reports on inter-kingdom bacteria-fungi interactions in RA are limited, studies on patients with inflammatory bowel disease (IBD) indicate that bacterial and fungal interactions influence gut inflammation (50, 51). Hoarau et al. demonstrated positive inter-kingdom correlations between E. coli, S. marcescens, and C. tropicalis in patients with Crohn’s disease (CD) and that C. tropicalis cooperates with E. coli, and S. marcescens to form biofilms consisting of fungal hyphae and other species-specific interactions, which have pathogenic potential to damage host tissues (50). Sokol et al. also observed a positive correlation for inter-kingdom bacteria-fungi interactions of Saccharomyces and Malassezia with several bacterial taxa in patients with IBD (35). Further, a study using two model yeasts, C. albicans and S. boulardi, showed that the presence of Enterobacteriaceae is required for the effect of fungi on gut inflammation in a dextran-sulphate sodium induced colitis model (51). These findings suggest that bacteria-fungal interactions might influence the immune system in RA.
Analysis of the virus-bacteria interactions in the gut has reported a correlation between the abundance of crAss-like phages and Bacteroides intestinalis in autoimmune diseases, especially RA and SLE. Further, a positive correlation between two bacterial clades i.e., Faecalibacterium spp. and Faecalibacterium cf. prausnitzii with Podoviridae abundance in patients with SLE was reported (42). Faecalibacterium is a bacterial genus which shows anti-inflammatory activity by producing short-chain fatty acids (52). Thus, the symbiotic relationship of Faecalibacterium and Podoviridae could be important to maintain immune homeostasis. Another study has reported that the bacteria-virus interaction network is disrupted in treated patients with RA compared to untreated patients and healthy controls (45). Moreover, phages can affect gut bacteria pathogenicity by altering their adhesion, invasion, colonization, and toxin production (53). Overall, these findings suggest that inter-kingdom interactions impact the host immune responses in RA.
Despite available evidence, the exact pathway by which dysbiosis specifically induces synovial inflammation remains poorly understood. The mechanisms by which bacterial dysbiosis favors RA initiation or progression include altered intestinal permeability, autoantigen modification, molecular mimicry, and immune/inflammatory system activation (54).
Alteration of intestinal permeability by the microbiota involves release of Zonulin, which decreases intestinal barrier function by displacement of the tight junction-forming proteins, zonula occludens 1 and occludins, from the junction complex, thereby increasing the penetrance of microbes or their products in the submucosa (55). Antigen presenting cells (APC) such as dendritic cells or macrophages respond to these microbes and their products and polarize T cells into Th1 and Th17 cells, which mainly produce IFN-γ and IL-17A, respectively (20). Group 3 innate lymphoid cells (ILC3) are pivotal in bridging the intestinal bacteriome and systemic immune responses (56). The penetrance of microbes and their products (ATP, free fatty acid receptor-2 agonists, aryl hydrocarbon ligands and retinoids) into the submucosa directly activates ILC3 and induces IL-22 secretion (57, 58). Other innate cells such as invariant natural killer T cells, mucosa-associated invariant T cells, macrophages, and monocytes also sense these bacteria and their products in the submucosa through various receptors such as Toll-like receptors (TLR), NOD-like receptors (NLR), c-type lectin receptors (CLR), and RIG-like receptors, thereby triggering the inflammatory cascade in the intestine and ultimately causing T cell activation and differentiation (20).
Another mechanism by which bacterial imbalance contributes to RA etiopathogenesis includes posttranslational modification of self-proteins such as hypercitrullination, which promotes ACPA production by inducing loss of T and B cell tolerance against citrullinated neoantigens (59–61). Some specific mucosa-associated bacteria have been implicated in promoting protein citrullination via activation of peptidyl-arginine deiminases after their release during cell damage (60). Autoantibodies promote RA pathogenesis by forming immune complexes with antigens, which activate innate immune cells by binding with Fc receptors as well as induce bone degradation by promoting osteoclastogenesis (62, 63).
In addition to autoantigen modification, mucosal and joint immune responses are considered to be linked by the molecular mimicry between certain bacterial antigens (epitopes from Prevotella) and autoantigens (N-acetylglucosamine-6-sulfatase). In the gut mucosa, Prevotella-derived epitopes are speculated to activate T cells, which then migrate to joints and cross-react with autoantigens to mount aberrant immune responses (64).
Another mechanism in RA immunopathogenesis involves the induction of inflammatory immune responses. Certain bacteria, such as segmented filamentous bacteria (SFB), Prevotella copri, and Lactobacillus species increase Th17 and Th1 cell responses in the gut mucosa (22, 65–67). In addition to the bacteriome, minor components of gut microbiota such as fungi and viruses also contribute to immune response activation, which may lead to RA development. The interactions between the gut microbiota and immune system are depicted in Figure 1.
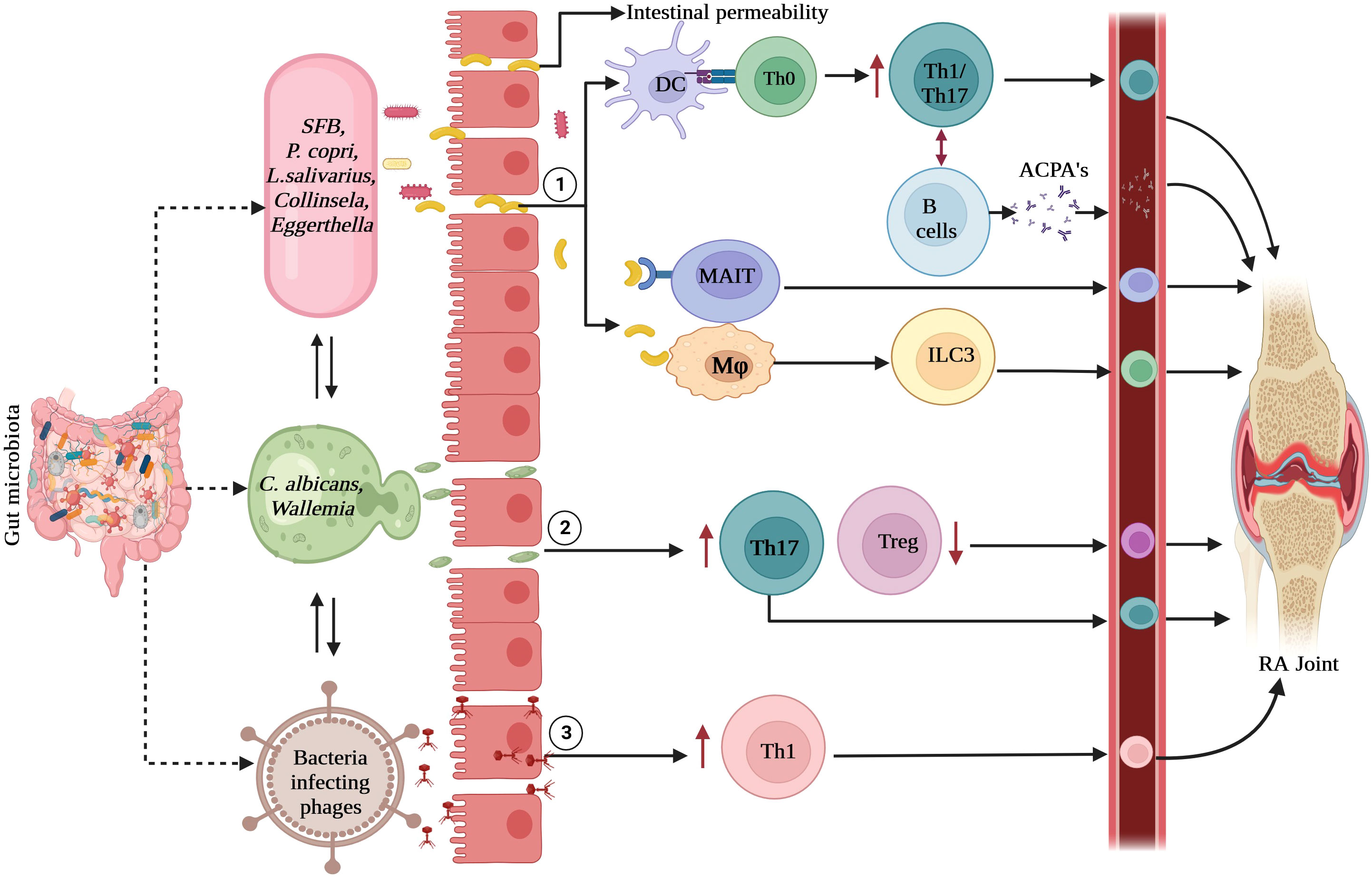
Figure 1 Interactions between gut microbiota and immune system: (1). Bacteria and its derived products activate gut epithelial and immune cells to enhance gut permeability which causes the migration of microbes into submucosa. These microbes are presented by dendritic cells to T cells which polarize into Th1 and Th17 cells. Gut colonization by SFB causes the induction of Th17 cells and P. copri and L. salivarius enhances the Th1 responses. These antigen-specific Th cells activate B cells to produce IgA-ACPA. Bacteria and its products directly activate macrophages and other innate immune cells such as ILC3 or MAIT cells which lead to gut inflammation locally and further activation of T cells. (2). The interactions between fungi mainly C. albicans and phagocytes leads to induction of increased Th17 responses and decreased Treg cells which contribute to local and systemic inflammation. (3). Viral dysbiosis causes induction of Th1 responses. Upon dysbiosis in RA, these innate and adaptive immune cells migrate to joints, exaggerating inflammation. RA, rheumatoid arthritis; Th, T-helper cells, ILC3, group 3 innate lymphoid cells; MAIT, mucosa associated invariant T cells; ACPA, anti-citrullinated peptide antibodies; SFB, segmented filamentous bacteria, P.copri, Prevotella copri; L. salivarius, Lactobacillus salivarius; C.albicans, Candida albicans.
Under homeostatic conditions, immune responses to yeast and filamentous fungi involve their recognition by CLR such as Dectin-1, Dectin-2, and Mincle, expressed by gut mononuclear phagocytic (MNPs) cells such as CX3CR1+ MNPs, which induce fungi-specific Th17 responses that orchestrate protective immunity in intestinal tissues (68, 69). However, dysregulated Th17 responses contribute to local and systemic autoimmune conditions, such as IBD and RA (70, 71), therefore connecting the altered gut mycobiota with inflammatory diseases, including RA.
Although viruses represent only a minor component of the gut microbiome, they are one of the key regulators of host immune responses. Several studies have reported a correlation between virome fluctuations and RA; however, the mechanistic understanding regarding how these viruses contribute to RA pathogenesis remains limited (42, 44). The contribution of phages in exacerbating gut inflammation in IBD involves induction of Th1 immune responses via TLR9 activation on APCs in response to viral DNA (72). Overall, phages play a critical role in the pathogenesis of inflammatory diseases like IBD and RA, and could be considered a potential therapeutic target.
The advent and rapid evolution of disease modifying therapies has dramatically changed the concept of treatment in patients with RA. Additionally, these gut microbiome-modifying therapies raise the possibility that modulating the gut microbiome can be a promising therapeutic or adjunct strategy for treating RA. The microbiome can be manipulated by various interventions such as diet, vitamins, plant extracts, prebiotics, probiotics, and fecal microbiota transplantation (FMT) (73, 74).
Numerous meta-analyses indicate that oral supplementation of omega-3 poly unsaturated fatty acids significantly reduces the levels of inflammation-related markers (75, 76). Dietary fiber consumption helps prevent the gut microbiome from eroding the gut mucosal barrier, thereby reducing pathogen infection and enhancing the quality of life (77, 78). By managing metabolic imbalance and regulating the gut microbiota, an appropriated diet may progressively alter the physiological state as well as aberrant immune responses.
The potential of the gut microflora to influence immune responses has sparked significant interest in the use of probiotic microorganisms for both preventive and therapeutic purposes (79, 80). As defined by the World Health Organization, probiotics are “live bacteria” that impart a health benefit to the host (81, 82). Lactobacillus spp. and Bifidobacterium spp. are widely used probiotics. For instance, L. casei has been shown to reduce the induction and disease progression of adjuvant-induced arthritis by restoring microbiome dysbiosis in the gut (83). Further, the osteoprotective role of Lactobacillus rhamnosus via immunomodulation has been demonstrated in the management of bone-related diseases (81). P. histicola MCI 001, a therapeutic bacterial strain, has been shown to treat RA by increasing the synthesis of short chain fatty acids, which modulate the gut immune response (84).
Apart from bacteria, immune system-boosting fungal extracts, such as those from Cordyceps militaris, have the potential to be used as adjuvant therapy in treating immunological diseases (85). Further, consumption of E. coli-targeting phages leads to an increase in the butyrate-producing bacterial genus Eubacterium and a decrease in Clostridium perfringens, thereby indicating the potential of bacteriophages in treating immune-mediated diseases (86).
A number of organic substances or plant extracts, including clematis, berberine triterpenoid saponins, and Paederia scandens extract (PSE), have also been reported to influence the gut microbiota (73). PSE can effectively reduce the serum levels of TNF-α, IL-1β, IL-6, IL-7, and IL-23 in a mouse model of RA, suggesting that these plant extracts could be used as a potential treatment modality for RA (87).
Success of Fecal microbiota transplantation using healthy donor microbiota, is another method for treating gut dysbiosis (88, 89); however, the mechanisms by which FMT and FVT components exert beneficial effects need further investigation.
Gut microbiome plays a vital role in maintaining host immune homeostasis. Recent data from various studies indicate that dysbiosis occurs in pre-clinical phase of disease and influences the development of RA. Previous narrative reviews on gut microbiota are mainly focussed on bacteriome dysbiosis in RA. There is limited data on fungal and virome alterations in patients with RA. Further, inter-kingdom associations between bacteriome, fungal and viral components of gut have not been comprehensively evaluated till late. Therefore, this review discusses the available published literature on alterations in the gut biomes specially mycobiome and virome along with bacteriome in RA. Further, we are also narrating the role of gut microbial components in influencing immune system.
Studies showing the effect of dysbiosis on aberrant immune responses have opened new avenues of research and have already been exploited for novel treatment opportunities such as maintaining the gut barrier and inhibition of immune cell migration from the gut to joints. However, future studies are needed to define the causes of dysbiosis and to determine exactly how and when gut dysbiosis influences RA development.
As discussed in this review, most studies have demonstrated an association of the disease with altered microbial composition. However, a deeper understanding of this relationship and the mechanistic pathways influencing disease development is required for obtaining effective diagnostic, prognostic, and therapeutic targets. These findings provide guidance for manipulating the microbiome as a preventive, adjunctive, or therapeutic treatment strategy in RA. Over the past decade, several studies have shown that restoration of gut microbial homeostasis can be achieved via nutritional changes, administration of probiotics, and FMT. Therefore, clinical trials of these approaches are needed to determine the potential of gut microbiota modification as a component of therapeutic toolbox for RA.
SD, JS and AS participated in the literature search and writing the review. YK, SC, RM provided valuable inputs in preparing and editing the manuscript. SD and AS created the illustrations. LR contributed to the conceptualization, design, writing, and editing of the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Van der Woude D, van der Helm-van Mil AHM. Update on the epidemiology, risk factors, and disease outcomes of rheumatoid arthritis. Best Pract Res Clin Rheumatol (2018) 32:174–87. doi: 10.1016/j.berh.2018.10.005
2. Firestein GS, McInnes IB. Immunopathogenesis of rheumatoid arthritis. Immunity (2017) 46:183–96. doi: 10.1016/j.immuni.2017.02.006
3. Scherer HU, Häupl T, Burmester GR. The etiology of rheumatoid arthritis. J Autoimmun (2020) 110:102400. doi: 10.1016/j.jaut.2019.102400
4. McInnes IB, Schett G. The pathogenesis of rheumatoid.arthritis. N Engl J Med (2011) 365:2205–19. doi: 10.1056/NEJMra1004965
5. Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature (2014) 506:376–81. doi: 10.1038/nature12873
6. Xu H, Zhao H, Fan D, Liu M, Cao J, Xia Y, et al. Interactions between gut microbiota and immunomodulatory cells in rheumatoid arthritis. Mediators Inflamm (2020) 2020:1430605. doi: 10.1155/2020/1430605
7. Horta-Baas G, Romero-Figueroa MDS, Montiel-Jarquín AJ, Pizano-Zárate ML, García-Mena J, Ramírez-Durán N. Intestinal dysbiosis and rheumatoid arthritis: A link between gut microbiota and the pathogenesis of rheumatoid arthritis. J Immunol Res (2017) 2017:4835189. doi: 10.1155/2017/4835189
8. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med (2015) 21:895–905. doi: 10.1038/nm.3914
9. Gupta VK, Cunningham KY, Hur B, Bakshi U, Huang H, Warrington KJ, et al. Gut microbial determinants of clinically important improvement in patients with rheumatoid arthritis. Genome Med (2021) 13:149. doi: 10.1186/s13073-021-00957-0
10. Kang Y, Cai Y, Zhang X, Kong X, Su J. Altered gut microbiota in RA: Implications for treatment. Z Rheumatol (2017) 76:451–7. doi: 10.1007/s00393-016-0237-5
11. Mei L, Yang Z, Zhang X, Liu Z, Wang M, Wu X, et al. Sustained drug treatment alters the gut microbiota in rheumatoid arthritis. Front Immunol (2021) 12. doi: 10.3389/fimmu.2021
12. Wassenaar TM, Panigrahi P. Is a foetus developing in a sterile environment? Lett Appl Microbiol (2014) 59:572–9. doi: 10.1111/lam.12334
13. Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PloS One (2014) 9:e90784. doi: 10.1371/journal.pone.0090784
14. Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep (2016) 6:23129. doi: 10.1038/srep23129
15. Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA (2010) 107:11971–5. doi: 10.1073/pnas.1002601107
16. Gioia C, Lucchino B, Tarsitano MG, Iannuccelli C, Di Franco M. Dietary habits and nutrition in rheumatoid arthritis: Can diet influence disease development and clinical manifestations? Nutrients (2020) 12:1456. doi: 10.3390/nu12051456
17. Ma J, Li Z, Zhang W, Zhang C, Zhang Y, Mei H, et al. Comparison of gut microbiota in exclusively breast-fed and formula-fed babies: A study of 91 term infants. Sci Rep (2020) 10:15792. doi: 10.1038/s41598-020
18. Niu J, Xu L, Qian Y, Sun Z, Yu D, Huang J, et al. Evolution of the gut microbiome in early childhood: A cross-sectional study of Chinese children. Front Microbiol (2020) 11:439. doi: 10.3389/fmicb.2020.00439
19. Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature (2007) 449:804–10. doi: 10.1038/nature06244
20. Zaiss MM, Joyce Wu H-J, Mauro D, Schett G, Ciccia F. The gut-joint axis in rheumatoid arthritis. Nat Rev Rheumatol (2021) 17:224–37. doi: 10.1038/s41584-021-00585-3
21. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal prevotella copri correlates with enhanced susceptibility to arthritis. Elife (2013) 2:e01202. doi: 10.7554/eLife.01202
22. Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE, et al. Evidence of the immune relevance of prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol (Hoboken NJ) (2017) 69:964–75. doi: 10.1002/art.40003
23. Alpizar-Rodriguez D, Lesker TR, Gronow A, Gilbert B, Raemy E, Lamacchia C, et al. Prevotella copri in individuals at risk for rheumatoid arthritis. Ann Rheum Dis (2019) 78:590–3. doi: 10.1136/annrheumdis-2018-214514
24. Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med (2016) 8:43. doi: 10.1186/s13073-016-0299-7
25. Muñiz Pedrogo DA, Chen J, Hillmann B, Jeraldo P, Al-Ghalith G, Taneja V, et al. An increased abundance of clostridiaceae characterizes arthritis in inflammatory bowel disease and rheumatoid arthritis: A cross-sectional study. Inflammation Bowel Dis (2019) 25:902–13. doi: 10.1093/ibd/izy318
26. Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol (2008) 35:1500–5.
27. Lee JY, Mannaa M, Kim Y, Kim J, Kim GT, Seo YS. Comparative analysis of fecal microbiota composition between rheumatoid arthritis and osteoarthritis patients. Genes (2019) 10:748. doi: 10.3390/genes10100748
28. Sun Y, Chen Q, Lin P, Xu R, He D, Ji W, et al. Characteristics of gut microbiota in patients with rheumatoid arthritis in shanghai, China. Front Cell Infect Microbiol (2019) 9:369. doi: 10.3389/fcimb.2019.00369
29. Rodrigues GSP, Cayres LCF, Gonçalves FP, Takaoka NNC, Lengert AH, Tansini A, et al. Detection of increased relative expression units of Bacteroides and Prevotella, and decreased Clostridium leptum in stool samples from Brazilian rheumatoid arthritis patients: A pilot study. Microorganisms (2019) 7:413. doi: 10.3390/microorganisms7100413
30. Chen Y, Ma C, Liu L, He J, Zhu C, Zheng F, et al. Analysis of gut microbiota and metabolites in patients with rheumatoid arthritis and identification of potential biomarkers. Aging (2021) 13:23689–701. doi: 10.18632/aging.203641
31. Yu D, Du J, Pu X, Zheng L, Chen S, Wang N, et al. The gut microbiome and metabolites are altered and interrelated in patients with rheumatoid arthritis. Front Cell Infect Microbiol (2021) 11:763507. doi: 10.3389/fcimb.2021.763507
32. Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, et al. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere (2018) 3:e00092–18. doi: 10.1128/mSphere.00092-18
33. Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, et al. The gut mycobiome of the human microbiome project healthy cohort. Microbiome (2017) 5:153. doi: 10.1186/s40168-017-0373-4
34. Hammad DBM, Liyanapathirana V, Tonge DP. Molecular characterisation of the synovial fluid microbiome in rheumatoid arthritis patients and healthy control subjects. PloS One (2019) 14:e0225110. doi: 10.1371/journal.pone.0225110
35. Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, et al. Fungal microbiota dysbiosis in IBD. Gut (2017) 66:1039–48. doi: 10.1136/gutjnl-2015-310746
36. Iliev ID, Cadwell K. Effects of intestinal fungi and viruses on immune responses and inflammatory bowel diseases. Gastroenterology (2021) 160:1050–66. doi: 10.1053/j.gastro.2020.06.100
37. Patel DD, Kuchroo VK. Th17 cell pathway in human immunity: Lessons from genetics and therapeutic interventions. Immunity (2015) 43:1040–51. doi: 10.1016/j.immuni.2015.12.003
38. Sun X, Wang Y, Li X, Wang M, Dong J, Tang W, et al. Alterations of gut fungal microbiota in patients with rheumatoid arthritis. PeerJ (2022) 10:e13037. doi: 10.7717/peerj.13037
39. Jahreis S, Kuhn S, Madaj AM, Bauer M, Polte T. Mold metabolites drive rheumatoid arthritis in mice via promotion of IFN-gamma- and IL-17-producing T cells. Food Chem Toxicol (2017) 109:405–13. doi: 10.1016/j.fct.2017.09.027
40. Van Hamburg JP, Tas SW. Molecular mechanisms underpinning T helper 17 cell heterogeneity and functions in rheumatoid arthritis. J Autoimmun (2018) 87:69–81. doi: 10.1016/j.jaut.2017.12.006P
41. Carding SR, Davis N, Hoyles L. Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther (2017) 46:800–15. doi: 10.1111/apt.14280
42. Tomofuji Y, Kishikawa T, Maeda Y, Ogawa K, Nii T, Okuno T, et al. Whole gut virome analysis of 476 Japanese revealed a link between phage and autoimmune disease. Ann Rheum Dis (2022) 81:278–88. doi: 10.1136/annrheumdis-2021-221267
43. Yutin N, Benler S, Shmakov SA, Wolf YI, Tolstoy I, Rayko M, et al. Analysis of metagenome-assembled viral genomes from the human gut reveals diverse putative CrAss-like phages with unique genomic features. Nat Commun (2021) 12:1044. doi: 10.1038/s41467-021-21350-w
44. Mangalea MR, Paez-Espino D, Kieft K, Chatterjee A, Chriswell ME, Seifert JA, et al. Individuals at risk for rheumatoid arthritis harbor differential intestinal bacteriophage communities with distinct metabolic potential. Cell Host Microbe (2021) 29:726–39.e5. doi: 10.1016/j.chom.2021.03.020
45. Guo R, Li S, Zhang Y, Zhang Y, Wang G, Ullah H, et al. Dysbiotic oral and gut viromes in untreated and treated rheumatoid arthritis patients. Microbiol Spectr (2022) 10:e0034822. doi: 10.1128/spectrum.00348-22
46. Krych Ł, Nielsen DS, Hansen AK, Hansen CHF. Gut microbial markers are associated with diabetes onset, regulatory imbalance, and IFN-γ level in NOD mice. Gut Microbes (2015) 6:101–9. doi: 10.1080/19490976.2015.1011876
47. Samonis G, Gikas A, Anaissie EJ, Vrenzos G, Maraki S, Tselentis Y, et al. Prospective evaluation of effects of broad-spectrum antibiotics on gastrointestinal yeast colonization of humans. Antimicrob Agents Chemother (1993) 37:51–3. doi: 10.1128/AAC.37.1.51
48. Maraki S, Hajiioannou I, Anatoliotakis N, Plataki M, Chatzinikolaou I, Zoras O, et al. Ceftriaxone and dexamethasone affecting yeast gut flora in experimental mice. J Chemother (1999) 11:363–6. doi: 10.1179/joc.1999.11.5.363
49. Lv Z, Xiong D, Shi J, Long M, Chen Z. The interaction between viruses and intestinal microbiota: A review. Curr Microbiol (2021) 78:3597–608. doi: 10.1007/s00284-021-02623-5
50. Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, et al. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial crohn’s disease. MBio (2016) 7:e01250–16. doi: 10.1128/mBio.01250-16
51. Sovran B, Planchais J, Jegou S, Straube M, Lamas B, Natividad JM, et al. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome (2018) 6:152. doi: 10.1186/s40168-018-0538-9
52. Ferreira-Halder CV, Faria AV de S, Andrade SS. Action and function of faecalibacterium prausnitzii in health and disease. Best Pract Res Clin Gastroenterol (2017) 31:643–8. doi: 10.1016/j.bpg.2017.09.011
53. Popescu M, Van Belleghem JD, Khosravi A, Bollyky PL. Bacteriophages and the immune system. Annu Rev Virol (2021) 8:415–35. doi: 10.1146/annurev-virology-091919-074551
54. Reyes-Castillo Z, Valdés-Miramontes E, Llamas-Covarrubias M, Muñoz-Valle JF. Troublesome friends within us: The role of gut microbiota on rheumatoid arthritis etiopathogenesis and its clinical and therapeutic relevance. Clin Exp Med (2021) 21:1–13. doi: 10.1007/s10238-020-00647-y
55. Sturgeon C, Fasano A. Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue barriers (2016) 4:e1251384. doi: 10.1080/21688370.2016.1251384
56. Mauro D, Macaluso F, Fasano S, Alessandro R, Ciccia F. ILC3 in axial spondyloarthritis: the gut angle. Curr Rheumatol Rep (2019) 21:37. doi: 10.1007/s11926-019-0834-9
57. Chun E, Lavoie S, Fonseca-Pereira D, Bae S, Michaud M, Hoveyda HR, et al. Metabolite-sensing receptor Ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity (2019) 51:871–84.e6. doi: 10.1016/j.immuni.2019.09.014
58. Li S, Bostick JW, Zhou L. Regulation of innate lymphoid cells by aryl hydrocarbon receptor. Front Immunol (2018) 8:1909. doi: 10.3389/fimmu.2017.01909
59. Holers VM, Demoruelle MK, Kuhn KA, Buckner JH, Robinson WH, Okamoto Y, et al. Rheumatoid arthritis and the mucosal origins hypothesis: Protection turns to destruction. Nat Rev Rheumatol (2018) 14:542–57. doi: 10.1038/s41584-018-0070-0
60. Darrah E, Andrade F. Rheumatoid arthritis and citrullination. Curr Opin Rheumatol (2018) 30:72–8. doi: 10.1097/BOR.0000000000000452
61. Volkov M, van Schie KA, van der Woude D. Autoantibodies and b cells: The ABC of rheumatoid arthritis pathophysiology. Immunol Rev (2020) 294:148–63. doi: 10.1111/imr.12829
62. Clavel C, Nogueira L, Laurent L, Iobagiu C, Vincent C, Sebbag M, et al. Induction of macrophage secretion of tumor necrosis factor alpha through fcgamma receptor IIa engagement by rheumatoid arthritis-specific autoantibodies to citrullinated proteins complexed with fibrinogen. Arthritis Rheum (2008) 58:678–88. doi: 10.1002/art.23284
63. Harre U, Georgess D, Bang H, Bozec A, Axmann R, Ossipova E, et al. Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest (2012) 122:1791–802. doi: 10.1172/JCI60975
64. Pianta A, Arvikar SL, Strle K, Drouin EE, Wang Q, Costello CE, et al. Two rheumatoid arthritis-specific autoantigens correlate microbial immunity with autoimmune responses in joints. J Clin Invest (2017) 127:2946–56. doi: 10.1172/JCI93450
65. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity (2010) 32:815–27. doi: 10.1016/j.immuni.2010.06.001
66. Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol (2016) 68:2646–61. doi: 10.1002/art.39783
67. Brisbin JT, Gong J, Parvizi P, Sharif S. Effects of lactobacilli on cytokine expression by chicken spleen and cecal tonsil cells. Clin Vaccine Immunol (2010) 17:1337–43. doi: 10.1128/CVI.00143-10
68. Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, et al. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science (2018) 359:232–6. doi: 10.1126/science.aao1503
69. Speakman EA, Dambuza IM, Salazar F, Brown GD. T Cell antifungal immunity and the role of c-type lectin receptors. Trends Immunol (2020) 41:61–76. doi: 10.1016/j.it.2019.11.007
70. Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the c-type lectin receptor dectin-1 influence colitis. Science (2012) 336:1314–7. doi: 10.1126/science.1221789
71. Wang T, Pan D, Zhou Z, You Y, Jiang C, Zhao X, et al. Dectin-3 deficiency promotes colitis development due to impaired antifungal innate immune responses in the gut. PloS Pathog (2016) 12:e1005662. doi: 10.1371/journal.ppat.1005662
72. Gogokhia L, Buhrke K, Bell R, Hoffman B, Brown DG, Hanke-Gogokhia C, et al. Expansion of bacteriophages is linked to aggravated intestinal inflammation and colitis. Cell Host Microbe (2019) 25:285–99.e8. doi: 10.1016/j.chom.2019.01.008
73. Horta-Baas G, Sandoval-Cabrera A, Romero-Figueroa M del S. Modification of gut microbiota in inflammatory arthritis: Highlights and future challenges. Curr Rheumatol Rep (2021) 23:67. doi: 10.1007/s11926-021-01031-9
74. Dourado E, Ferro M, Sousa Guerreiro C, Fonseca JE. Diet as a modulator of intestinal microbiota in rheumatoid arthritis. Nutrients (2020) 12:3504. doi: 10.3390/nu12113504
75. Zampelas A, Panagiotakos DB, Pitsavos C, Das UN, Chrysohoou C, Skoumas Y, et al. Fish consumption among healthy adults is associated with decreased levels of inflammatory markers related to cardiovascular disease: The ATTICA study. J Am Coll Cardiol (2005) 46:120–4. doi: 10.1016/j.jacc.2005.03.048
76. Gioxari A, Kaliora AC, Marantidou F, Panagiotakos DP. Intake of ω-3 polyunsaturated fatty acids in patients with rheumatoid arthritis: A systematic review and meta-analysis. Nutrition (2018) 45:114–24.e4. doi: 10.1016/j.nut.2017.06.023
77. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell (2016) 167:1339–53.e21. doi: 10.1016/j.cell.2016.10.043
78. Häger J, Bang H, Hagen M, Frech M, Träger P, Sokolova MV, et al. The role of dietary fiber in rheumatoid arthritis patients: A feasibility study. Nutrients (2019) 11:2392. doi: 10.3390/nu11102392
79. Zamani B, Golkar HR, Farshbaf S, Emadi-Baygi M, Tajabadi-Ebrahimi M, Jafari P, et al. Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis: A randomized, double-blind, placebo-controlled trial. Int J Rheum Dis (2016) 19:869–79. doi: 10.1111/1756-185X.12888
80. Vaghef-Mehrabany E, Alipour B, Homayouni-Rad A, Sharif S-K, Asghari-Jafarabadi M, Zavvari S. Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis. Nutrition (2014) 30:430–5. doi: 10.1016/j.nut.2013.09.007
81. Sapra L, Dar HY, Bhardwaj A, Pandey A, Kumari S, Azam Z, et al. Lactobacillus rhamnosus attenuates bone loss and maintains bone health by skewing treg-Th17 cell balance in ovx mice. Sci Rep (2021) 11:1807. doi: 10.1038/s41598-020-80536-2
82. Sapra L, Shokeen N, Porwal K, Saini C, Bhardwaj A, Mathew M, et al. Bifidobacterium longum ameliorates ovariectomy-induced bone loss via enhancing anti-osteoclastogenic and immunomodulatory potential of regulatory b cells (Bregs). Front Immunol (2022) 13:875788. doi: 10.3389/fimmu.2022.875788
83. Pan H, Guo R, Ju Y, Wang Q, Zhu J, Xie Y, et al. A single bacterium restores the microbiome dysbiosis to protect bones from destruction in a rat model of rheumatoid arthritis. Microbiome (2019) 7:107. doi: 10.1186/s40168-019-0719-1
84. Balakrishnan B, Luckey D, Bodhke R, Chen J, Marietta E, Jeraldo P, et al. Prevotella histicola protects from arthritis by expansion of allobaculum and augmenting butyrate production in humanized mice. Front Immunol (2021) 12:609644. doi: 10.3389/fimmu.2021.609644
85. Shin S, Kwon J, Lee S, Kong H, Lee S, Lee C-K, et al. Immunostimulatory effects of cordyceps militaris on macrophages through the enhanced production of cytokines via the activation of NF-κB. Immune Netw (2010) 10:55. doi: 10.4110/in.2010.10.2.55
86. Febvre H, Rao S, Gindin M, Goodwin N, Finer E, Vivanco J, et al. PHAGE study: Effects of supplemental bacteriophage intake on inflammation and gut microbiota in healthy adults. Nutrients (2019) 11:666. doi: 10.3390/nu11030666
87. Xiao M, Fu X, Ni Y, Chen J, Jian S, Wang L, et al. Protective effects of paederia scandens extract on rheumatoid arthritis mouse model by modulating gut microbiota. J Ethnopharmacol (2018) 226:97–104. doi: 10.1016/j.jep.2018.08.012
88. Zeng J, Peng L, Zheng W, Huang F, Zhang N, Wu D, et al. Fecal microbiota transplantation for rheumatoid arthritis: A case report. Clin Case Rep (2021) 9:906–9. doi: 10.1002/ccr3.3677
Keywords: gut microbiota, dysbiosis, rheumatoid arthritis, immune responses, adjuvant therapy
Citation: Dagar S, Singh J, Saini A, Kumar Y, Chhabra S, Minz RW and Rani L (2023) Gut bacteriome, mycobiome and virome alterations in rheumatoid arthritis. Front. Endocrinol. 13:1044673. doi: 10.3389/fendo.2022.1044673
Received: 14 September 2022; Accepted: 20 December 2022;
Published: 09 January 2023.
Edited by:
Rupesh K. Srivastava, All India Institute of Medical Sciences, IndiaReviewed by:
Valentina Caputi, University College Cork, IrelandCopyright © 2023 Dagar, Singh, Saini, Kumar, Chhabra, Minz and Rani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lekha Rani, bGVraGFwZ2lAeWFob28uY29t
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.