- 1Novo Nordisk Foundation Center for Basic Metabolic Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
- 2The Wallenberg Laboratory, Department of Molecular and Clinical Medicine, Institute of Medicine, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden
Metagenomics studies have shown that type 2 diabetes (T2D) is associated with an altered gut microbiota. Whereas different microbiota patterns have been observed in independent human cohorts, reduction of butyrate-producing bacteria has consistently been found in individuals with T2D, as well as in those with prediabetes. Butyrate is produced in the large intestine by microbial fermentations, particularly of dietary fiber, and serves as primary fuel for colonocytes. It also acts as histone deacetylase inhibitor and ligand to G-protein coupled receptors, affecting cellular signaling in target cells, such as enteroendocrine cells. Therefore, butyrate has become an attractive drug target for T2D, and treatment strategies have been devised to increase its intestinal levels, for example by supplementation of butyrate-producing bacteria and dietary fiber, or through fecal microbiota transplant (FMT). In this review, we provide an overview of current literature indicating that these strategies have yielded encouraging results and short-term benefits in humans, but long-term improvements of glycemic control have not been reported so far. Further studies are required to find effective approaches to restore butyrate-producing bacteria and butyrate levels in the human gut, and to investigate their impact on glucose regulation in T2D.
Introduction
Type 2 diabetes (T2D) is a global concern and is projected to affect 700 million people by 2045 (1). Although lifestyle interventions (including diet, exercise and weight loss) are the first option for managing T2D, patients are often prescribed medications. A variety of drugs are already available, but side effects (such as pancreatitis and gastrointestinal complications) (2), and lifelong dependency on drugs entail a significant burden on the patients and on the healthcare system globally. Therefore, novel individualized therapies are being developed, focused on safety and personalized management of T2D.
‘Gut microbiota’ is a term used to describe the trillions of microbes that live in the gastrointestinal tract. The gut microbiota has been identified as a virtual organ interacting locally with the gut and systemically with other organs in the host to facilitate multiple physiological processes (3). The interest in understanding the composition and function of the gut microbiota has increased exponentially over the last two decades. From the initial studies addressing the possibility to culture and profile gut microbiota communities (4–7), the field has developed to describe the core human gut microbiota (8–12), its tremendous genetic potential (100 times larger than the human genome) (10, 13) and variations according to geographical location (14, 15), ethnicity (16, 17) and disease status (18), including T2D (19, 20). How an altered gut microbiota can impact metabolic health is debated, but metabolomics investigations have shown that the gut microbiota contribute to the variation of blood metabolites in humans (21), with important implications for metabolic regulation (22). Trimethylamine oxide (23), imidazole propionate (24) and indolepropionic acid (25) are examples of microbial metabolites associated with development or protection against metabolic diseases, and their specific roles in metabolic diseases as well as signaling mechanisms are currently under investigation.
Butyrate is one of the short chain fatty acids (SCFAs) produced as end-products of intestinal microbial fermentations (26, 27). Butyrate is absorbed rapidly in the gut and acts as signaling molecule in receptor-mediated signaling in numerous cell types (28). Microbial butyrate production in the human gut has been known for decades (29) before the large sequencing efforts of the gut microbiota started, but it was only in the last decade that metagenomics surveys consistently revealed in multiple independent human cohorts a decrease of butyrate-producing bacteria in individuals with T2D (19, 20). As restoration of butyrate-producing bacteria and butyrate levels might provide new treatment options for T2D, here we review recent literature on the association of butyrate and butyrate-producing bacteria with T2D, and discuss the therapeutic potential for management and treatment of this disease (Figure 1).
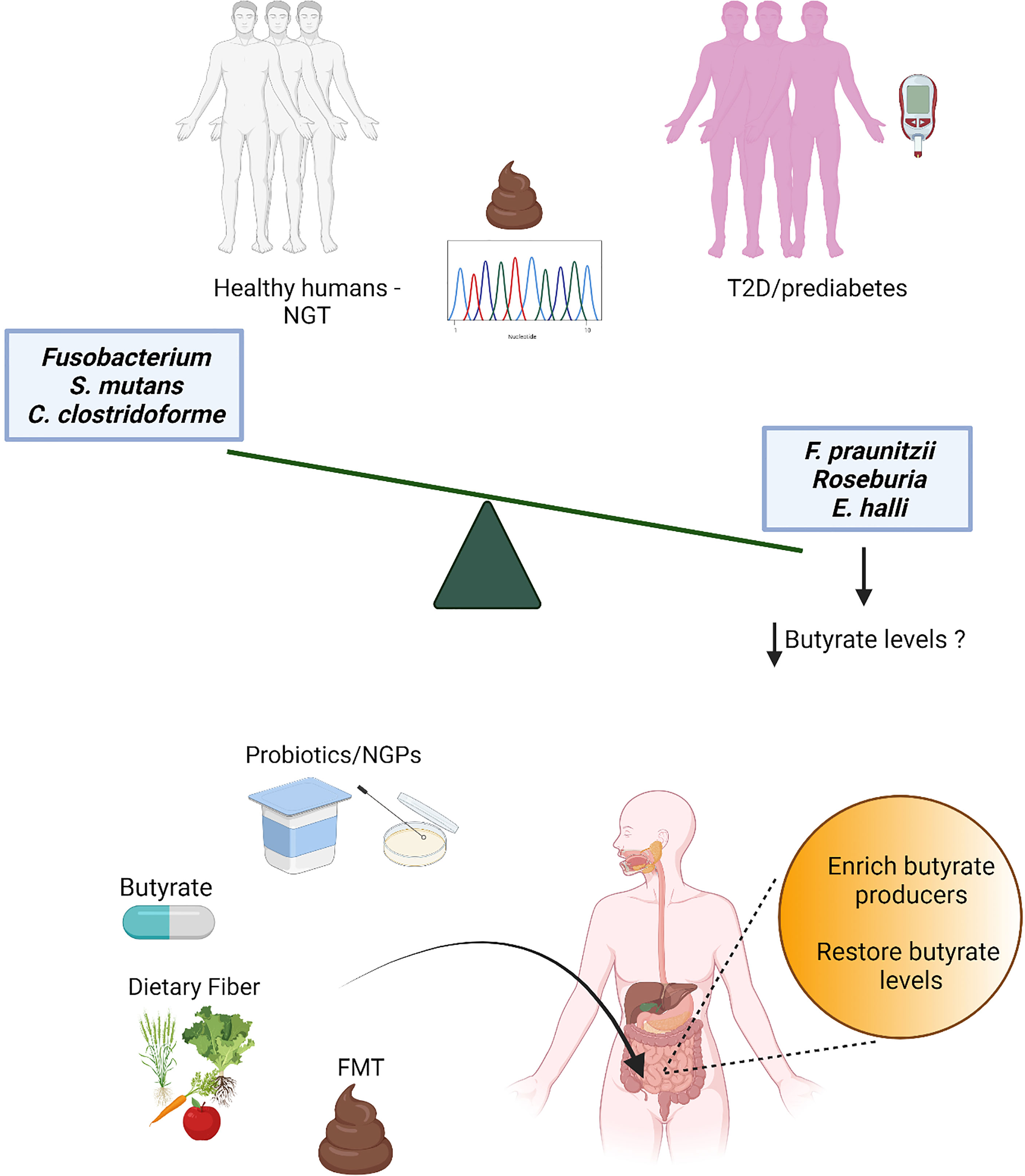
Figure 1 Several independent metagenomics studies have detected a decrease of butyrate producers in stools from individuals with prediabetes and type 2 diabetes (T2D) compared to individuals with normal glucose tolerance (NGT). While the loss of butyrate producers is robust and associated also with obesity and other cardiometabolic comorbidities (18), consistent patterns for increased microbial features have not been found, possibly due to the redundancy of the gut microbiota and stochasticity in gut microbiota alterations (30); only few gut microbiota species (mostly opportunistic pathogens) have been observed as increased in a limited number of studies. Thus, butyrate and butyrate producers have been selected as potential targets for the development of novel therapeutic strategies for T2D, such as direct butyrate administration, administration of butyrate producers and/or bacteria able to promote intestinal butyrate production [probiotics and next-generation probiotics (NGPs)], interventions with dietary fibers and fecal microbiota transplant (FMT).
Type 2 Diabetes and Butyrate-Producing Bacteria
T2D has been associated with compositional and functional shifts in the gut microbiota. One of the striking features that was consistently observed in multiple cohorts across diverse geographical locations is the reduction of butyrate-producing bacteria in individuals with T2D. The first observations came from shotgun metagenomics studies of fecal communities in Chinese (20) and Swedish (19) individuals, and showed decreased abundance of several butyrate producers such as Roseburia and Faecalibacterium prausnitzii; these findings were later extended to individuals with T2D in Indian (31) and African (32) populations. Lower abundance of F. prausnitizii has also been found in the mesenchymal adipose tissue of obese individuals with T2D compared to normoglycemic controls matched for body mass index (33), reflecting the lower intestinal abundance of this bacterium in T2D independent of obesity, but linked to the metabolic status. The positive association between microbial potential for butyrate production and normoglycemia is supported by the results of a Dutch study that combined fecal microbiota metagenomics and human genome sequencing, and showed that higher potential for butyrate production driven by host genetics was linked to improved insulin response to an oral glucose load in normoglycemic individuals (34). Furthermore, one recent study has shown that also the diurnal oscillation of the gut microbiota is altered in T2D, and in particular that the abundance of several bacteria, among which Roseburia and F. prausnitzii, lost rhythmicity, and that the arrhythmicity might be important for risk classification and prediction of T2D (35). However, it is important to mention that other gut commensals (e.g. Akkermansia, Bifidobacterium and Bacteroides) are also decreased in T2D and might play additional roles for the modulation of gut barrier function, inflammation and metabolism, as reviewed elsewhere (36, 37).
The initial studies on the associations between gut microbiota and T2D did not account for medications used for T2D treatment; however, it is now established that numerous non-antibiotic drugs can influence the gut microbiota (38). Metformin is the first line of treatment in newly discovered T2D patients and was first demonstrated to influence gut microbiota composition in diet induced obese mouse models, where it resulted in enrichment of Akkermansia muciniphila (39, 40), a gut commensal showing beneficial effects on metabolism (41). Similar results were later observed also in large human cohorts (including both treatment naïve and metformin-treated T2D patients), in which shotgun sequencing of the fecal microbiota showed enrichment of butyrate producers, and in some cases also of A. muciniphila (42–45). By using germ-free (GF) mice, it was also shown that part of the antidiabetic effects of metformin can be explained by the microbiota shifts. GF mice are devoid of any microbiota, and are an efficient model to understand the functional significance of compositional changes in the gut microbiota: fecal microbiota transplant from metformin-treated individuals to GF mice were able to transfer the improved glucose tolerance phenotype to the recipients (45). In addition, gut microbiota has also been shown to interact with metformin and the diet to promote microbial synthesis of agmatine, specifically in individuals with T2D, an effector molecule able to regulate host lipid metabolism (46). Overall, these results indicate a possible beneficial role of the microbial shifts induced by metformin for the improvement of the glycemic and metabolic status in T2D. However, these studies also indicate that potential confounders [e.g., medications, dietary supplements and obesity (47)] should be considered in gut microbiota analyses for the identification of possible microbial signatures of T2D.
T2D is a chronic metabolic disorder that can remain undetected for a number of years. Prediabetes precedes T2D, and often presents with intermediate hyperglycemia, such as impaired fasting glucose (IFG), impaired glucose tolerance (IGT) or combined glucose intolerance (CGI) (48). Microbiota profiling in a Danish cohort diagnosed with prediabetes revealed lower abundance of butyrate-producing bacteria compared with age- and sex-matched individuals with normal glucose regulation (49). In addition, a metagenomic study in a T2D-treatment-naïve Swedish cohort has shown that microbiota composition is altered in IGT and CGI, and is characterized by a reduction in the abundance of butyrate-producing bacteria and the terminal genes for butyrate synthesis (43). These studies indicate that the butyrate-producing potential of the gut microbiome is depleted already in the prediabetes state, and suggest that replenishment of butyrate producers or butyrate levels might be important to delay or prevent progression to T2D. In contrast with the consistent finding of reduced abundance of butyrate-producing bacteria in T2D, different studies report extensive variation for the levels of butyrate in feces and/or blood of individuals with T2D compared to controls. In the large Dutch cohort, fecal butyrate levels did not correlate with either butyrate production potential or the selected anthropometric and glycemic traits, thus suggesting that fecal butyrate levels might not be representative of butyrate production and absorption (34). However, results from two smaller cohorts have showed decreased levels of SCFAs (including butyrate) in individuals with T2D as well as significant correlation with metabolic parameters (50, 51). Therefore, as butyrate is volatile and quickly absorbed and consumed by the colonic epithelium, static measurements in fecal and blood samples might not be sufficient to reveal an actual reduction of butyrate levels in T2D. Radioactive tracers might offer a better solution to trace the origin and absorption of butyrate, but would not be applicable to large-scale human cohort studies.
Butyrate – Production, Absorption and Physiological Roles
The SCFAs acetate, propionate and butyrate are produced by microbial fermentations in the gut (52). Most of the butyrate producers are distributed within multiple clusters of Clostridia, in the phylum Firmicutes (53). Butyrate is primarily a product of carbohydrate fermentation produced by condensation of two acetyl CoA molecules. However, lactate and by-products of amino acids metabolism are also cross-fed to generate butyrate (54). Two key enzymes have been identified for butyrate production by the gut microbiota from carbohydrates: the butyryl CoA:acetate CoA transferase (but) is the primary enzyme, whereas the butyrate kinase (buk) is present in a few strains (55). The terminal enzymes for butyrate production from amino acids are also known [i.e., butyryl CoA:4-hydroxybutyrate CoA transferase (4-hbt) and butyryl CoA:acetoacetate CoA transferase (ato)] (56), but their abundance in the human gut is lower compared to but and buk.
In humans, the major site of SCFAs and butyrate production is the colon, from where total SCFAs are drained into the portal blood with much higher concentration (375µmol/l) than in peripheral blood (79µmol/l) (29). However, approximately 95% of the butyrate produced in the gut lumen is rapidly absorbed by colonocytes and fuels cellular metabolism through mitochondrial β-oxidation (57). Colonic delivery of 13C labelled acetate, propionate and butyrate in healthy subjects revealed that only 36% of the acetate, 9% of the propionate and 2% of the butyrate could be recovered in blood samples collected over different time points during the day (58). Interestingly, 24% of acetate was bio-converted to butyrate, but the authors did not find significant correlation between the percentage of interconversion and the gene copies of but or buk in the fecal samples (58). These results confirm the observations from the large Dutch cohort (34), and thus indicate that the abundance of the genes does not fully reflect the activity, as different intestinal variables (e.g., pH, redox potential, lactate and acetate levels) can influence the rates of butyrate production (52).
In the GF mouse model, colonocytes are in a state of nutrient deficiency that causes cellular autophagy in the colonic epithelium (59) and slower intestinal transit (60). Supplementation of butyrate reverses these phenotypes (i.e. prevents autophagy and decreases intestinal transit time), indicating that microbially-produced butyrate is an important source of energy for colonic epithelial cells and its deficiency results in reversible adaptive mechanisms to cope with nutrient deficiency. In addition, mouse studies have shown that butyrate contributes to maintain the colonic environment anaerobic through the activation of the peroxisome proliferator-activated receptor-gamma in colonocytes, which results in the induction of β-oxidation (57). This process has been shown to consume oxygen and prevent colonic invasion by pathogenic Salmonella and Escherichia species (57). Because of its effects on colonocytes, butyrate levels in the gut might be important not only for signaling to the host (as described below) but also for the composition of the gut microbiota. Therefore, butyrate might be a particularly interesting therapeutic target.
Butyrate also serves as histone deacetylase (HDAC) inhibitor to regulate the expression of genes by epigenetic mechanisms. Supernatants from cultures of butyrate-producing bacteria from the human gut microbiota express HDAC inhibitory activity to class I and II HDACs (61). T2D is associated with epigenetic changes in multiple organs (62) and it seems plausible that microbially-produced butyrate could be one of the contributing factors. In a non-obese diabetic mouse model, it has been observed that the reduction of intestinal butyrate associated with T2D caused an increase in colonic HDAC activity resulting in production of reactive oxygen species and alteration of colonic permeability (63). Furthermore, supplementation of sodium butyrate has been shown to ameliorate palmitate-induced insulin resistance in muscle cells by promoting hyperacetylation of insulin receptor substrate-1 in an in vitro study (64). Consistent with this study, supplementation of sodium butyrate to mice modulated mitochondrial chromatin structure (65) and lowered HDAC activity in skeletal muscle resulting in improved insulin sensitivity (66). Finally, sodium butyrate treatment has been shown to suppress HDAC activity also in mouse liver, leading to reduced gluconeogenesis and improved glucose homeostasis (67). These animal studies suggest that reduction of butyrate in T2D may alter gene expression by epigenetic mechanisms leading to insulin resistance, which can be reversed by butyrate supplementation. However, this observation awaits validation in human cohorts.
Butyrate (as well as the other SCFAs) also acts as a signaling molecule and is identified as a ligand for G-protein coupled receptors (GPCRs), such as FFAR3, FFAR2 and GPR109A. Human orthologs of FFAR2 have similar affinity for acetate and propionate followed by butyrate, while FFAR3 has higher affinity for propionate and butyrate compared to acetate (68). Niacin is identified as the most potent ligand of GPR109A, though butyrate also shows weak binding (69). These receptors are located in distal regions of the intestine and in adipocytes (70). In the intestinal lumen, the concentrations of SCFAs are supramaximal and therefore it is thought that these receptors are localized on the basolateral side of the intestinal epithelium (70). FFAR3 and FFAR2 are present on hormone-producing enteroendocrine cells (EECs): exposure to SCFAs stimulates EECs differentiation (71), and binding of SCFAs to FFAR3 and FFAR2 results in altered gene expression and secretion of gut hormones, such as peptide YY (PYY) and glucagon like peptide-1 (GLP-1) (72, 73). GLP-1 is one of the gut hormones that profoundly affects glucose regulation by promoting post-prandial insulin secretion, and GLP-1 based drugs are approved for the treatment of T2D (74). Supplementation of butyrate along with inulin have been shown to increase GLP-1 levels in individuals with T2D with significant improvement in the glycemic status (75). However, it is important to note that the in vitro studies mentioned here report different effects after stimulation with a mix of SCFAs or with individual SCFAs, indicating that the effects on EECs are not exclusively mediated by butyrate. Nevertheless, in human cell lines, butyrate selectively stimulates PYY secretion through mechanisms largely driven by HDAC inhibition (76).
In addition to signaling in the gut, activation of SCFAs-binding receptors might be important also in the adipose tissue. Selective chemical agonism of GPR109A in individuals with T2D decreased fasting glucose, but not Hb1Ac, through inhibition of lipolysis in adipocytes as demonstrated by the decreased circulating levels of non-esterified fatty acids in the patients (77, 78). Consistent with these results, overexpression of FFAR2 in adipose tissue protected mice from gut microbiota-dependent diet-induced obesity (79). Additionally, a role for butyrate in the stimulation of thermogenesis in brown and white adipose tissue has been demonstrated in rodents (66), with potential relevance for the regulation of glycaemia. However, oral supplementation of butyrate in individuals with T2D did not alter brown adipose tissue activity (80).
Finally, butyrate signaling might also be important for islets function. In vitro experiments have showed that addition of butyrate in culture media reduced streptozotocin-induced islet cell death (81). Moreover, supplementing HFD with 5% butyrate in a T2D mouse model prevented β-cell expansion and fat accumulation in the pancreas (82). In contrast, an oral dose of 4g butyrate to individuals with type 1 diabetes for a month did not improve β-cell function or islet autoimmunity (83).
Therefore, SCFAs (including butyrate) may play important roles in metabolic control, particularly via regulation of EECs and adipocyte lipolysis. However, specific targeting of the GPCRs in humans might be difficult due to their complex chemistry and diverse functions in different tissues (84). Additionally, applications might be limited by the lack of concordance between mouse and human studies that can possibly be explained by differences in dose, route and duration of administration, discrepancy between experimental models for diabetes and human diabetes, and specific intestinal environments in the different hosts. Nevertheless, the animal studies suggest that butyrate influences the regulation of glucose metabolism through multiple pathways that, if further characterized and validated in humans, can possibly be harnessed for the development of therapeutic strategies (Figure 2).
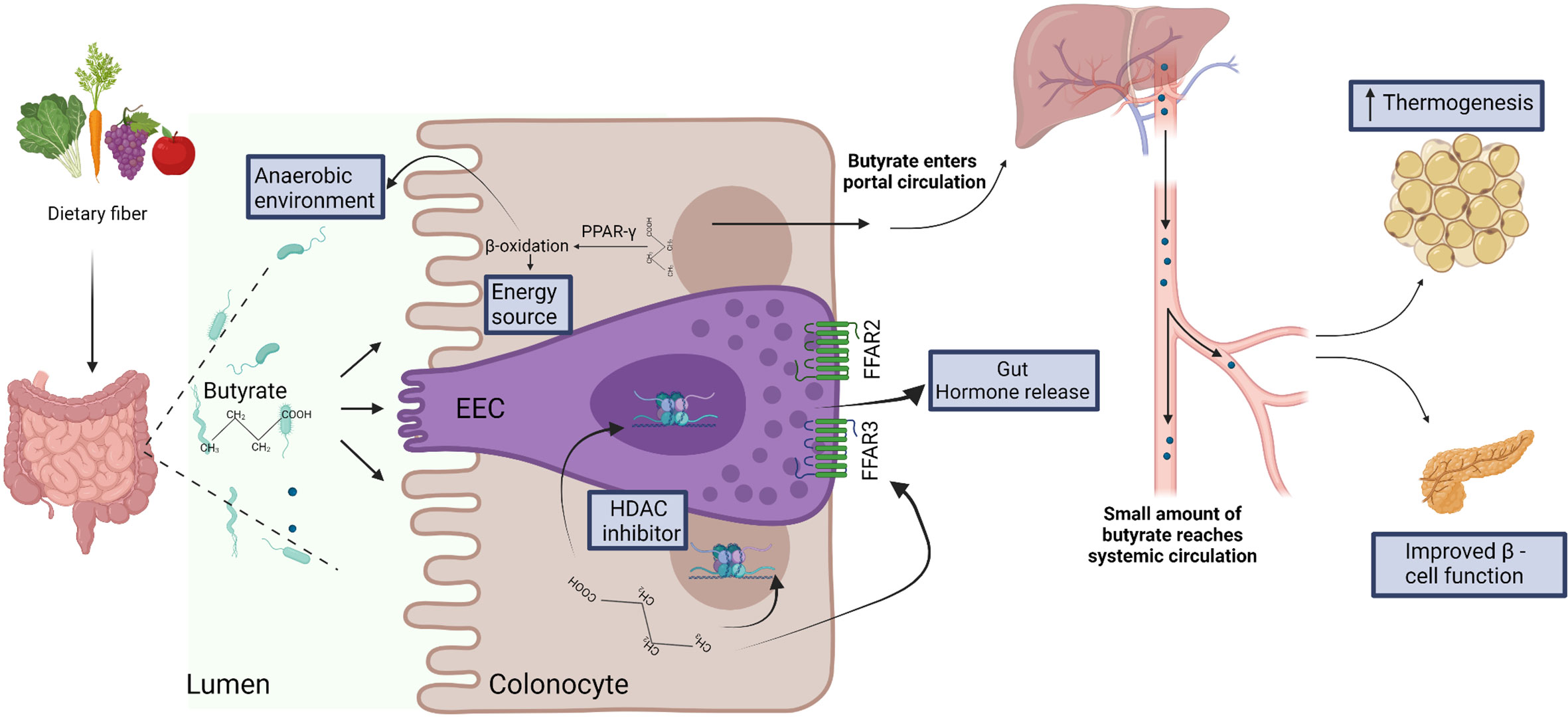
Figure 2 Dietary fiber is fermented by the gut microbiota to produce short chain fatty acids, including butyrate. Butyrate is efficiently absorbed by colonocytes and is utilized as energy source. Butyrate-mediated activation of the peroxisome proliferator-activated receptor-gamma (PPAR-γ) induces β-oxidation and consumption of oxygen, thus facilitating the establishment of anaerobic conditions that are required for the growth and function of several anaerobic gut commensals (57). In the specialized enteroendocrine cell (EEC), butyrate binds free fatty acid receptors (FFAR) FFAR2 and FFAR3 and regulates gut hormone release (73). Butyrate also acts as histone deacetylase (HDAC) inhibitor to regulate gene expression in EEC (76) and enterocytes (63). After absorption and utilization by colonocytes, the residual butyrate is first drained into the portal circulation, and then into the peripheral systemic circulation (29). In the systemic circulation, butyrate may regulate thermogenesis in brown adipose tissue (66) and β-cell function in pancreas (82).
Restoration of Butyrate in Type 2 Diabetes
Restoration of the intestinal levels of butyrate might be a novel strategy for the treatment of T2D, that could also be added to conventional therapy with lifestyle management and glucose-lowering drugs. In recent years, a number of studies have attempted to replenish butyrate levels and butyrate-producers in the gut using different approaches as discussed below: direct supplementation of butyrate or butyrate-producing bacteria, dietary supplementation of fibers to feed microbial butyrate production, and fecal microbiota transplantation.
Supplementation of Butyrate
Butyrate can be supplemented as sodium conjugate or as tributyrin (a triglyceride in which glycerol is esterified with three butyrate molecules). In mice with diet-induced obesity, oral supplementation of 400mg/kg of butyrate improved glucose tolerance and increased the expression of phosphorylated adenosine monophosphate kinase (AMPK) as well as glucose transporter-4 in the adipose tissue, and reversed some of the gut microbiota alterations caused by the high-fat diet (HFD) (85). In another mouse study, addition of 5% butyrate to HFD increased energy expenditure, improved insulin sensitivity, and induced adaptive thermogenesis in BAT followed by increased AMPK activity and mitochondrial biogenesis in muscle cells (66). As an alternative, tributyrin supplementation in diet-induced obese mice was also shown to improve glucose tolerance and inflammatory status (86), indicating that direct supplementation of butyrate might have beneficial effects on both metabolic and inflammatory parameters relevant for the pathophysiology of T2D.
With convincing results in mouse studies, butyrate supplementation was also tested in individuals with and without metabolic syndrome who were given 4g sodium butyrate in capsules for a period of 4 weeks (80). In this study, butyrate supplementation did not increase butyrate levels either in feces or plasma, but it improved both peripheral and hepatic insulin sensitivity in individuals without metabolic syndrome. In another study, oral butyrate supplementation at the same dose improved the inflammatory status in individuals with metabolic syndrome, but no effect on insulin sensitivity was measured in this study (87). Additional studies might be required to determine effective doses of butyrate in humans, or other methods of administration and delivery of butyrate to the colonic epithelium that mimics the production by the gut microbiota.
Butyrate-Producing Bacteria
Live bacteria that provide health benefits when consumed are generally called probiotics, and traditional Lactobacillus probiotics have demonstrated some efficacy for hyperglycemia and insulin sensitivity in human cohorts (88–91). In two independent studies, supplementation of Lactobacillus paracasei or Bifidobacterium bifidum to healthy individuals increased fecal butyrate levels (92, 93), indicating that traditional probiotics may modulate the activity of butyrate producers. However, the intestinal microbes that have been found as decreased in T2D in metagenomics studies are not traditional probiotics, and are being explored to produce next-generation probiotics (NGPs) (94, 95). For the butyrate producers, oral administration of Clostridium butyricum to mice lacking the leptin receptor, or to mice on HFD injected with streptozotocin to induce diabetes, was shown to improve oral glucose tolerance and insulin levels, and to increase the abundance of butyrate producers and fecal butyrate levels (96). In another study, oral administration of Eubacterium hallii to mice lacking the leptin receptor improved insulin sensitivity and increased energy expenditure (97). However, administration of E. hallii strain L2-7 (now reclassified as Anaerobutyricum soehngenii) to individuals with insulin resistance improved insulin sensitivity only in individuals with a specific gut microbiota at baseline (98), reflecting both the resilience of the human gut microbiota and the ecological interactions of commensal microbes in the communities that might be species-specific. To produce effective NGPs, advanced data-driven metagenomics approaches (99) and specific isolation efforts might be required to develop synthetic microbial communities targeted to produce butyrate.
Dietary Fiber
Since butyrate-producing bacteria feed upon dietary fiber, dietary supplementation with fiber may provide a feasible option to increase the levels and the activity of the bacteria, and increase the intestinal levels of butyrate. In a randomized clinical study by Zhao et al., supplementation of a mix of dietary fibers to individuals with T2D improved glycemic parameters, accompanied by increased abundance of acetate- and butyrate-producing bacteria and increased fecal levels of acetate and butyrate (100). In another study, combining a mix of butyrate-producing species (E. hallii, Clostridium beijerinckii and C. butyricum), with other gut bacteria (A. muciniphila and Bifidobacterium infantis) and inulin as fermentable fiber modestly increased butyrate levels and improved oral glucose tolerance and glycated hemoglobin levels in individuals with T2D (101). Finally, dietary supplementation of inulin along with sodium butyrate in capsules for 45 days improved fasting glucose and waist-to-hip ratio in individuals with T2D (75). These studies clearly indicate that dietary fiber itself or in combination with NGPs or butyrate can improve glucose control in T2D. However, strategies to maintain patient compliance and investigations of long-term effects of these supplements are still warranted. Additionally, as it is now evident that the baseline gut microbiota is a strong predictor of success for dietary interventions (102, 103), probiotic administrations (89) and microbiota transplantations (104), stratification of individuals with T2D based on their microbiota may help to achieve better metabolic outcomes.
Microbiota Transplantation
Microbiota transplants from mouse models (105) and humans (45, 106) into GF mice have successfully demonstrated the transmissibility of donor’s phenotypes. Therefore, human-to-human fecal microbiota transplants (FMT) [that have shown unprecedented success for the treatment of Clostridium difficile infections (107)] have recently been attempted for the treatment of T2D. When insulin resistant individuals were administered with duodenal infusion their own fecal microbiota (autologous) or fecal microbiota from a healthy lean donor (allogenic), the allogenic group displayed an improved insulin sensitivity (104, 108). The improved phenotype was observed 6 weeks after transplant in both studies, but not 18 weeks post-FMT (104). Change in butyrate producers (such as Roseburia, Eubacterium and Butyrivibiro) in feces and in the small intestine was observed in both studies, but increase in butyrate levels were observed only in one (108). In another study, daily cellulose supplementation after a single-dose oral FMT in individuals with metabolic syndrome improved insulin sensitivity 6 weeks after treatment compared to baseline. The authors found that this outcome was linked to higher GLP-1 secretion and better engraftment of the donor microbiota; however, they did not report significant changes for either the proportions of butyrate producers or fecal butyrate levels (109). Overall, the studies on FMT indicate that, while this procedure may improve insulin sensitivity in the short-term, the host gut microbiota is resilient enough to drift away the foreign microbial community in the long-term. In addition, while FMT is generally associated with mild side-effects, major adverse events have also been reported (110). These observations question the feasibility and applicability of FMT as a way of treatment for T2D.
Butyrate as Therapy – Conclusions
Butyrate has long been known as a microbial fermentation product of dietary fibers in the gut, and references of butyrate-producing bacteria isolated from dietary sources emerged already in late 1940 (111). The recent association of T2D with reduction of butyrate-producing bacteria has spurred interest to explore the therapeutic potential of butyrate for the treatment of T2D but, while the results of experimental studies overall look promising, human interventions have only shown positive outcomes in the short term, and might have important limitations. In particular, current studies based on the metagenomic profiling of DNA are not able to determine the activity of butyrate producers in the human gut. Probiotics, NGPs and fiber supplementations might be successful strategies to increase butyrate-producing bacteria and improve hyperglycemia and insulin resistance, but their effects might be dependent on the individualized gut microbiota at baseline (responders vs. non-responders) and/or mediated by multiple undefined mechanisms besides butyrate production. FMT seems promising for the restoration of the gut microbiota and to improve insulin sensitivity, but it is impractical to perform such a highly invasive procedure in humans for short-term benefits. Future studies are required to gain a better understanding of the intestinal conditions that might influence butyrate production in individuals with T2D, in relation both to the diet and the individualized gut microbiota; for example, ingestible electronic capsules able to monitor microbial fermentations directly in the gut (112) could be used to characterize intestinal conditions, responses to fibers and microbiota profiles linked to homeostatic butyrate production. Furthermore, robust methods for the measurement of butyrate, tracer studies and live-detection of butyrate-producing bacteria [for example, by flow cytometry (113)] might help to strengthen the association of butyrate with T2D and identify new potential NGPs or synthetic microbial communities for butyrate-based management of T2D.
Author Contributions
TA conceptualized the manuscript. TA and VT wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Novo Nordisk Foundation (Grant no. NNF15OC0016798).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Figures created using Biorender.com.
References
1. Cho NH, Shaw JE, Karuranga S, Huang Y, da Rocha Fernandes JD, Ohlrogge AW, et al. IDF Diabetes Atlas: Global Estimates of Diabetes Prevalence for 2017 and Projections for 2045. Diabetes Res Clin Pract (2018) 138:271–81. doi: 10.1016/j.diabres.2018.02.023
2. Engler C, Leo M, Pfeifer B, Juchum M, Chen-Koenig D, Poelzl K, et al. Long-Term Trends in the Prescription of Antidiabetic Drugs: Real-World Evidence From the Diabetes Registry Tyrol 2012-2018. BMJ Open Diabetes Res Care (2020) 8:e001279. doi: 10.1136/bmjdrc-2020-001279
3. O'Hara AM, Shanahan F. The Gut Flora as a Forgotten Organ. EMBO Rep (2006) 7:688–93. doi: 10.1038/sj.embor.7400731
4. Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic Analysis of the Human Distal Gut Microbiome. Science (2006) 312:1355–9. doi: 10.1126/science.1124234
5. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the Human Intestinal Microbial Flora. Science (2005) 308:1635–8. doi: 10.1126/science.1110591
6. Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct Analysis of Genes Encoding 16S rRNA From Complex Communities Reveals Many Novel Molecular Species Within the Human Gut. Appl Environ Microbiol (1999) 65:4799–807. doi: 10.1128/AEM.65.11.4799-4807.1999
7. Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, et al. Extensive Personal Human Gut Microbiota Culture Collections Characterized and Manipulated in Gnotobiotic Mice. Proc Natl Acad Sci USA (2011) 108:6252–7. doi: 10.1073/pnas.1102938108
8. Salonen A, Salojarvi J, Lahti L, de Vos WM. The Adult Intestinal Core Microbiota Is Determined by Analysis Depth and Health Status. Clin Microbiol Infect (2012) 18(Suppl 4):16–20. doi: 10.1111/j.1469-0691.2012.03855.x
9. Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A Core Gut Microbiome in Obese and Lean Twins. Nature (2009) 457:480–4. doi: 10.1038/nature07540
10. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature (2010) 464:59–65. doi: 10.1038/nature08821
11. Human Microbiome Project C. Structure, Function and Diversity of the Healthy Human Microbiome. Nature (2012) 486:207–14. doi: 10.1038/nature11234
12. Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-Level Analysis of Gut Microbiome Variation. Science (2016) 352:560–4. doi: 10.1126/science.aad3503
13. Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An Integrated Catalog of Reference Genes in the Human Gut Microbiome. Nat Biotechnol (2014) 32:834–41. doi: 10.1038/nbt.2942
14. Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human Gut Microbiome Viewed Across Age and Geography. Nature (2012) 486:222–7. doi: 10.1038/nature11053
15. Rampelli S, Schnorr SL, Consolandi C, Turroni S, Severgnini M, Peano C, et al. Metagenome Sequencing of the Hadza Hunter-Gatherer Gut Microbiota. Curr Biol (2015) 25:1682–93. doi: 10.1016/j.cub.2015.04.055
16. Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, et al. Depicting the Composition of Gut Microbiota in a Population With Varied Ethnic Origins But Shared Geography. Nat Med (2018) 24:1526–31. doi: 10.1038/s41591-018-0160-1
17. Brooks AW, Priya S, Blekhman R, Bordenstein SR. Gut Microbiota Diversity Across Ethnicities in the United States. PLoS Biol (2018) 16:e2006842. doi: 10.1371/journal.pbio.2006842
18. Duvallet C, Gibbons SM, Gurry T, Irizarry RA, Alm EJ. Meta-Analysis of Gut Microbiome Studies Identifies Disease-Specific and Shared Responses. Nat Commun (2017) 8:1784. doi: 10.1038/s41467-017-01973-8
19. Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut Metagenome in European Women With Normal, Impaired and Diabetic Glucose Control. Nature (2013) 498:99–103. doi: 10.1038/nature12198
20. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A Metagenome-Wide Association Study of Gut Microbiota in Type 2 Diabetes. Nature (2012) 490:55–60. doi: 10.1038/nature11450
21. Bar N, Korem T, Weissbrod O, Zeevi D, Rothschild D, Leviatan S, et al. A Reference Map of Potential Determinants for the Human Serum Metabolome. Nature (2020) 588:135–40. doi: 10.1038/s41586-020-2896-2
22. Krautkramer KA, Fan J, Backhed F. Gut Microbial Metabolites as Multi-Kingdom Intermediates. Nat Rev Microbiol (2021) 19:77–94. doi: 10.1038/s41579-020-0438-4
23. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal Microbiota Metabolism of L-Carnitine, A Nutrient in Red Meat, Promotes Atherosclerosis. Nat Med (2013) 19:576–85. doi: 10.1038/nm.3145
24. Koh A, Molinaro A, Stahlman M, Khan MT, Schmidt C, Manneras-Holm L, et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling Through Mtorc1. Cell (2018) 175:947–61.e17. doi: 10.1016/j.cell.2018.09.055
25. de Mello VD, Paananen J, Lindstrom J, Lankinen MA, Shi L, Kuusisto J, et al. Indolepropionic Acid and Novel Lipid Metabolites Are Associated With a Lower Risk of Type 2 Diabetes in the Finnish Diabetes Prevention Study. Sci Rep (2017) 7:46337. doi: 10.1038/srep46337
26. Fleming SE, Rodriguez MA. Influence of Dietary Fiber on Fecal Excretion of Volatile Fatty Acids by Human Adults. J Nutr (1983) 113:1613–25. doi: 10.1093/jn/113.8.1613
27. Flint HJ, Scott KP, Louis P, Duncan SH. The Role of the Gut Microbiota in Nutrition and Health. Nat Rev Gastroenterol Hepatol (2012) 9:577–89. doi: 10.1038/nrgastro.2012.156
28. Zhang L, Liu C, Jiang Q, Yin Y. Butyrate in Energy Metabolism: There Is Still More to Learn. Trends Endocrinol Metab (2021) 32:159–69. doi: 10.1016/j.tem.2020.12.003
29. Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short Chain Fatty Acids in Human Large Intestine, Portal, Hepatic and Venous Blood. Gut (1987) 28:1221–7. doi: 10.1136/gut.28.10.1221
30. Zaneveld JR, McMinds R, Vega Thurber R. Stress and Stability: Applying the Anna Karenina Principle to Animal Microbiomes. Nat Microbiol (2017) 2:17121. doi: 10.1038/nmicrobiol.2017.121
31. Bhute SS, Suryavanshi MV, Joshi SM, Yajnik CS, Shouche YS, Ghaskadbi SS, et al. Gut Microbial Diversity Assessment of Indian Type-2-Diabetics Reveals Alterations in Eubacteria, Archaea, and Eukaryotes. Front Microbiol (2017) 8:214. doi: 10.3389/fmicb.2017.00214
32. Doumatey AP, Adeyemo A, Zhou J, Lei L, Adebamowo SN, Adebamowo C, et al. Gut Microbiome Profiles Are Associated With Type 2 Diabetes in Urban Africans. Front Cell Infect Microbiol (2020) 10:63. doi: 10.3389/fcimb.2020.00063
33. Anhe FF, Jensen BAH, Varin TV, Servant F, Van Blerk S, Richard D, et al. Type 2 Diabetes Influences Bacterial Tissue Compartmentalisation in Human Obesity. Nat Metab (2020) 2:233–42. doi: 10.1038/s42255-020-0178-9
34. Sanna S, van Zuydam NR, Mahajan A, Kurilshikov A, Vich Vila A, Vosa U, et al. Causal Relationships Among the Gut Microbiome, Short-Chain Fatty Acids and Metabolic Diseases. Nat Genet (2019) 51:600–5. doi: 10.1038/s41588-019-0350-x
35. Reitmeier S, Kiessling S, Clavel T, List M, Almeida EL, Ghosh TS, et al. Arrhythmic Gut Microbiome Signatures Predict Risk of Type 2 Diabetes. Cell Host Microbe (2020) 28:258–72.e6. doi: 10.1016/j.chom.2020.06.004
36. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of Gut Microbiota in Type 2 Diabetes Pathophysiology. EBioMedicine (2020) 51:102590. doi: 10.1016/j.ebiom.2019.11.051
37. Cunningham AL, Stephens JW, Harris DA. Gut Microbiota Influence in Type 2 Diabetes Mellitus (T2DM). Gut Pathog (2021) 13:50. doi: 10.1186/s13099-021-00446-0
38. Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive Impact of Non-Antibiotic Drugs on Human Gut Bacteria. Nature (2018) 555:623–8. doi: 10.1038/nature25979
39. Lee H, Lee Y, Kim J, An J, Lee S, Kong H, et al. Modulation of the Gut Microbiota by Metformin Improves Metabolic Profiles in Aged Obese Mice. Gut Microbes (2018) 9:155–65. doi: 10.1080/19490976.2017.1405209
40. Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An Increase in the Akkermansia Spp. Population Induced by Metformin Treatment Improves Glucose Homeostasis in Diet-Induced Obese Mice. Gut (2014) 63:727–35. doi: 10.1136/gutjnl-2012-303839
41. Cani PD, de Vos WM. Next-Generation Beneficial Microbes: The Case of Akkermansia Muciniphila. Front Microbiol (2017) 8:1765. doi: 10.3389/fmicb.2017.01765
42. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling Type 2 Diabetes and Metformin Treatment Signatures in the Human Gut Microbiota. Nature (2015) 528:262–6. doi: 10.1038/nature15766
43. Wu H, Tremaroli V, Schmidt C, Lundqvist A, Olsson LM, Kramer M, et al. The Gut Microbiota in Prediabetes and Diabetes: A Population-Based Cross-Sectional Study. Cell Metab (2020) 32:379–90.e3. doi: 10.1016/j.cmet.2020.06.011
44. de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, et al. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia Muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care (2017) 40:54–62. doi: 10.2337/dc16-1324
45. Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin Alters the Gut Microbiome of Individuals With Treatment-Naive Type 2 Diabetes, Contributing to the Therapeutic Effects of the Drug. Nat Med (2017) 23:850–8. doi: 10.1038/nm.4345
46. Pryor R, Norvaisas P, Marinos G, Best L, Thingholm LB, Quintaneiro LM, et al. Host-Microbe-Drug-Nutrient Screen Identifies Bacterial Effectors of Metformin Therapy. Cell (2019) 178:1299–312.e29. doi: 10.1016/j.cell.2019.08.003
47. Thingholm LB, Ruhlemann MC, Koch M, Fuqua B, Laucke G, Boehm R, et al. Obese Individuals With and Without Type 2 Diabetes Show Different Gut Microbial Functional Capacity and Composition. Cell Host Microbe (2019) 26:252–64.e10. doi: 10.1016/j.chom.2019.07.004
48. Tabak AG, Herder C, Rathmann W, Brunner EJ, Kivimaki M. Prediabetes: A High-Risk State for Diabetes Development. Lancet (2012) 379:2279–90. doi: 10.1016/S0140-6736(12)60283-9
49. Allin KH, Tremaroli V, Caesar R, Jensen BAH, Damgaard MTF, Bahl MI, et al. Aberrant Intestinal Microbiota in Individuals With Prediabetes. Diabetologia (2018) 61:810–20. doi: 10.1007/s00125-018-4550-1
50. Adachi K, Sugiyama T, Yamaguchi Y, Tamura Y, Izawa S, Hijikata Y, et al. Gut Microbiota Disorders Cause Type 2 Diabetes Mellitus and Homeostatic Disturbances in Gut-Related Metabolism in Japanese Subjects. J Clin Biochem Nutr (2019) 64:231–8. doi: 10.3164/jcbn.18-101
51. Zhao L, Lou H, Peng Y, Chen S, Zhang Y, Li X. Comprehensive Relationships Between Gut Microbiome and Faecal Metabolome in Individuals With Type 2 Diabetes and Its Complications. Endocrine (2019) 66:526–37. doi: 10.1007/s12020-019-02103-8
52. Louis P, Flint HJ. Formation of Propionate and Butyrate by the Human Colonic Microbiota. Environ Microbiol (2017) 19:29–41. doi: 10.1111/1462-2920.13589
53. Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The Microbiology of Butyrate Formation in the Human Colon. FEMS Microbiol Lett (2002) 217:133–9. doi: 10.1111/j.1574-6968.2002.tb11467.x
54. Vital M, Karch A, Pieper DH. Colonic Butyrate-Producing Communities in Humans: An Overview Using Omics Data. mSystems (2017) 2:e00130–17. doi: 10.1128/mSystems.00130-17
55. Louis P, Young P, Holtrop G, Flint HJ. Diversity of Human Colonic Butyrate-Producing Bacteria Revealed by Analysis of the Butyryl-CoA:acetate CoA-Transferase Gene. Environ Microbiol (2010) 12:304–14. doi: 10.1111/j.1462-2920.2009.02066.x
56. Vital M, Howe AC, Tiedje JM. Revealing the Bacterial Butyrate Synthesis Pathways by Analyzing (Meta)Genomic Data. mBio (2014) 5:e00889. doi: 10.1128/mBio.00889-14
57. Byndloss MX, Olsan EE, Rivera-Chavez F, Tiffany CR, Cevallos SA, Lokken KL, et al. Microbiota-Activated PPAR-Gamma Signaling Inhibits Dysbiotic Enterobacteriaceae Expansion. Science (2017) 357:570–5. doi: 10.1126/science.aam9949
58. Boets E, Gomand SV, Deroover L, Preston T, Vermeulen K, De Preter V, et al. Systemic Availability and Metabolism of Colonic-Derived Short-Chain Fatty Acids in Healthy Subjects: A Stable Isotope Study. J Physiol (2017) 595:541–55. doi: 10.1113/JP272613
59. Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, et al. The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon. Cell Metab (2011) 13:517–26. doi: 10.1016/j.cmet.2011.02.018
60. Wichmann A, Allahyar A, Greiner TU, Plovier H, Lunden GO, Larsson T, et al. Microbial Modulation of Energy Availability in the Colon Regulates Intestinal Transit. Cell Host Microbe (2013) 14:582–90. doi: 10.1016/j.chom.2013.09.012
61. Yuille S, Reichardt N, Panda S, Dunbar H, Mulder IE. Human Gut Bacteria as Potent Class I Histone Deacetylase Inhibitors In Vitro Through Production of Butyric Acid and Valeric Acid. PLoS One (2018) 13:e0201073. doi: 10.1371/journal.pone.0201073
62. Ling C, Ronn T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab (2019) 29:1028–44. doi: 10.1016/j.cmet.2019.03.009
63. Noureldein MH, Bitar S, Youssef N, Azar S, Eid AA. Butyrate Modulates Diabetes-Linked Gut Dysbiosis: Epigenetic and Mechanistic Modifications. J Mol Endocrinol (2020) 64:29–42. doi: 10.1530/JME-19-0132
64. Chriett S, Zerzaihi O, Vidal H, Pirola L. The Histone Deacetylase Inhibitor Sodium Butyrate Improves Insulin Signalling in Palmitate-Induced Insulin Resistance in L6 Rat Muscle Cells Through Epigenetically-Mediated Up-Regulation of Irs1. Mol Cell Endocrinol (2017) 439:224–32. doi: 10.1016/j.mce.2016.09.006
65. Henagan TM, Stefanska B, Fang Z, Navard AM, Ye J, Lenard NR, et al. Sodium Butyrate Epigenetically Modulates High-Fat Diet-Induced Skeletal Muscle Mitochondrial Adaptation, Obesity and Insulin Resistance Through Nucleosome Positioning. Br J Pharmacol (2015) 172:2782–98. doi: 10.1111/bph.13058
66. Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate Improves Insulin Sensitivity and Increases Energy Expenditure in Mice. Diabetes (2009) 58:1509–17. doi: 10.2337/db08-1637
67. Khan S, Jena G. Sodium Butyrate Reduces Insulin-Resistance, Fat Accumulation and Dyslipidemia in Type-2 Diabetic Rat: A Comparative Study With Metformin. Chem Biol Interact (2016) 254:124–34. doi: 10.1016/j.cbi.2016.06.007
68. Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, et al. The Orphan G Protein-Coupled Receptors GPR41 and GPR43 Are Activated by Propionate and Other Short Chain Carboxylic Acids. J Biol Chem (2003) 278:11312–9. doi: 10.1074/jbc.M211609200
69. Tunaru S, Kero J, Schaub A, Wufka C, Blaukat A, Pfeffer K, et al. PUMA-G and HM74 Are Receptors for Nicotinic Acid and Mediate Its Anti-Lipolytic Effect. Nat Med (2003) 9:352–5. doi: 10.1038/nm824
70. Husted AS, Trauelsen M, Rudenko O, Hjorth SA, Schwartz TW. GPCR-Mediated Signaling of Metabolites. Cell Metab (2017) 25:777–96. doi: 10.1016/j.cmet.2017.03.008
71. Petersen N, Reimann F, Bartfeld S, Farin HF, Ringnalda FC, Vries RG, et al. Generation of L Cells in Mouse and Human Small Intestine Organoids. Diabetes (2014) 63:410–20. doi: 10.2337/db13-0991
72. Arora T, Vanslette AM, Hjorth SA, Backhed F. Microbial Regulation of Enteroendocrine Cells. Med-Cambridge (2021) 2:553–70. doi: 10.1016/j.medj.2021.03.018
73. Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, et al. Short-Chain Fatty Acids Stimulate Glucagon-Like Peptide-1 Secretion via the G-Protein-Coupled Receptor FFAR2. Diabetes (2012) 61:364–71. doi: 10.2337/db11-1019
74. Aroda VR, Bain SC, Cariou B, Piletic M, Rose L, Axelsen M, et al. Efficacy and Safety of Once-Weekly Semaglutide Versus Once-Daily Insulin Glargine as Add-on to Metformin (With or Without Sulfonylureas) in Insulin-Naive Patients With Type 2 Diabetes (SUSTAIN 4): A Randomised, Open-Label, Parallel-Group, Multicentre, Multinational, Phase 3a Trial. Lancet Diabetes Endo (2017) 5:355–66. doi: 10.1016/S2213-8587(17)30085-2
75. Roshanravan N, Mahdavi R, Alizadeh E, Jafarabadi MA, Hedayati M, Ghavami A, et al. Effect of Butyrate and Inulin Supplementation on Glycemic Status, Lipid Profile and Glucagon-Like Peptide 1 Level in Patients With Type 2 Diabetes: A Randomized Double-Blind, Placebo-Controlled Trial. Horm Metab Res (2017) 49:886–91. doi: 10.1055/s-0043-119089
76. Larraufie P, Martin-Gallausiaux C, Lapaque N, Dore J, Gribble FM, Reimann F, et al. SCFAs Strongly Stimulate PYY Production in Human Enteroendocrine Cells. Sci Rep (2018) 8:74. doi: 10.1038/s41598-017-18259-0
77. Dobbins R, Byerly R, Gaddy R, Gao F, Mahar K, Napolitano A, et al. GSK256073 Acutely Regulates NEFA Levels via HCA2 Agonism But Does Not Achieve Durable Glycaemic Control in Type 2 Diabetes. A Randomised Trial. Eur J Pharmacol (2015) 755:95–101. doi: 10.1016/j.ejphar.2015.03.005
78. Dobbins RL, Shearn SP, Byerly RL, Gao FF, Mahar KM, Napolitano A, et al. GSK256073, a Selective Agonist of G-Protein Coupled Receptor 109A (GPR109A) Reduces Serum Glucose in Subjects With Type 2 Diabetes Mellitus. Diabetes Obes Metab (2013) 15:1013–21. doi: 10.1111/dom.12132
79. Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. The Gut Microbiota Suppresses Insulin-Mediated Fat Accumulation via the Short-Chain Fatty Acid Receptor GPR43. Nat Commun (2013) 4:1829. doi: 10.1038/ncomms2852
80. Bouter K, Bakker GJ, Levin E, Hartstra AV, Kootte RS, Udayappan SD, et al. Differential Metabolic Effects of Oral Butyrate Treatment in Lean Versus Metabolic Syndrome Subjects. Clin Transl Gastroenterol (2018) 9:155. doi: 10.1038/s41424-018-0025-4
81. Hu S, Kuwabara R, de Haan BJ, Smink AM, de Vos P. Acetate and Butyrate Improve Beta-Cell Metabolism and Mitochondrial Respiration Under Oxidative Stress. Int J Mol Sci (2020) 21:1542. doi: 10.3390/ijms21041542
82. Matheus VA, Monteiro L, Oliveira RB, Maschio DA, Collares-Buzato CB. Butyrate Reduces High-Fat Diet-Induced Metabolic Alterations, Hepatic Steatosis and Pancreatic Beta Cell and Intestinal Barrier Dysfunctions in Prediabetic Mice. Exp Biol Med (Maywood) (2017) 242:1214–26. doi: 10.1177/1535370217708188
83. de Groot PF, Nikolic T, Imangaliyev S, Bekkering S, Duinkerken G, Keij FM, et al. Oral Butyrate Does Not Affect Innate Immunity and Islet Autoimmunity in Individuals With Longstanding Type 1 Diabetes: A Randomised Controlled Trial. Diabetologia (2020) 63:597–610. doi: 10.1007/s00125-019-05073-8
84. Milligan G, Shimpukade B, Ulven T, Hudson BD. Complex Pharmacology of Free Fatty Acid Receptors. Chem Rev (2017) 117:67–110. doi: 10.1021/acs.chemrev.6b00056
85. Gao F, Lv YW, Long J, Chen JM, He JM, Ruan XZ, et al. Butyrate Improves the Metabolic Disorder and Gut Microbiome Dysbiosis in Mice Induced by a High-Fat Diet. Front Pharmacol (2019) 10:1040. doi: 10.3389/fphar.2019.01040
86. Sato FT, Yap YA, Crisma AR, Portovedo M, Murata GM, Hirabara SM, et al. Tributyrin Attenuates Metabolic and Inflammatory Changes Associated With Obesity Through a GPR109A-Dependent Mechanism. Cells (2020) 9. doi: 10.3390/cells9092007
87. Cleophas MCP, Ratter JM, Bekkering S, Quintin J, Schraa K, Stroes ES, et al. Effects of Oral Butyrate Supplementation on Inflammatory Potential of Circulating Peripheral Blood Mononuclear Cells in Healthy and Obese Males. Sci Rep (2019) 9:775. doi: 10.1038/s41598-018-37246-7
88. Andreasen AS, Larsen N, Pedersen-Skovsgaard T, Berg RM, Moller K, Svendsen KD, et al. Effects of Lactobacillus Acidophilus NCFM on Insulin Sensitivity and the Systemic Inflammatory Response in Human Subjects. Br J Nutr (2010) 104:1831–8. doi: 10.1017/S0007114510002874
89. Mobini R, Tremaroli V, Stahlman M, Karlsson F, Levin M, Ljungberg M, et al. Metabolic Effects of Lactobacillus Reuteri DSM 17938 in People With Type 2 Diabetes: A Randomized Controlled Trial. Diabetes Obes Metab (2017) 19:579–89. doi: 10.1111/dom.12861
90. Simon MC, Strassburger K, Nowotny B, Kolb H, Nowotny P, Burkart V, et al. Intake of Lactobacillus Reuteri Improves Incretin and Insulin Secretion in Glucose-Tolerant Humans: A Proof of Concept. Diabetes Care (2015) 38:1827–34. doi: 10.2337/dc14-2690
91. Sun J, Buys NJ. Glucose- and Glycaemic Factor-Lowering Effects of Probiotics on Diabetes: A Meta-Analysis of Randomised Placebo-Controlled Trials. Br J Nutr (2016) 115:1167–77. doi: 10.1017/S0007114516000076
92. Ferrario C, Taverniti V, Milani C, Fiore W, Laureati M, De Noni I, et al. Modulation of Fecal Clostridiales Bacteria and Butyrate by Probiotic Intervention With Lactobacillus Paracasei DG Varies Among Healthy Adults. J Nutr (2014) 144:1787–96. doi: 10.3945/jn.114.197723
93. Gargari G, Taverniti V, Balzaretti S, Ferrario C, Gardana C, Simonetti P, et al. Consumption of a Bifidobacterium Bifidum Strain for 4 Weeks Modulates Dominant Intestinal Bacterial Taxa and Fecal Butyrate in Healthy Adults. Appl Environ Microbiol (2016) 82:5850–9. doi: 10.1128/AEM.01753-16
94. Martin R, Langella P. Emerging Health Concepts in the Probiotics Field: Streamlining the Definitions. Front Microbiol (2019) 10:1047. doi: 10.3389/fmicb.2019.01047
95. Langella P, Guarner F, Martin R. Editorial: Next-Generation Probiotics: From Commensal Bacteria to Novel Drugs and Food Supplements. Front Microbiol (2019) 10:1973. doi: 10.3389/fmicb.2019.01973
96. Jia L, Li D, Feng N, Shamoon M, Sun Z, Ding L, et al. Anti-Diabetic Effects of Clostridium Butyricum CGMCC0313.1 Through Promoting the Growth of Gut Butyrate-Producing Bacteria in Type 2 Diabetic Mice. Sci Rep (2017) 7:7046. doi: 10.1038/s41598-017-07335-0
97. Udayappan S, Manneras-Holm L, Chaplin-Scott A, Belzer S, Herrema S, Dallinga-Thie S, et al. Oral Treatment With Eubacterium Hallii Improves Insulin Sensitivity in Db/Db Mice. NPJ Biofilms Microbiomes (2016) 2:16009. doi: 10.1038/npjbiofilms.2016.9
98. Gilijamse PW, Hartstra AV, Levin E, Wortelboer K, Serlie MJ, Ackermans MT, et al. Treatment With Anaerobutyricum Soehngenii: A Pilot Study of Safety and Dose-Response Effects on Glucose Metabolism in Human Subjects With Metabolic Syndrome. NPJ Biofilms Microbiomes (2020) 6:16. doi: 10.1038/s41522-020-0127-0
99. Clark RL, Connors BM, Stevenson DM, Hromada SE, Hamilton JJ, Amador-Noguez D, et al. Design of Synthetic Human Gut Microbiome Assembly and Butyrate Production. Nat Commun (2021) 12:3254. doi: 10.1038/s41467-021-22938-y
100. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut Bacteria Selectively Promoted by Dietary Fibers Alleviate Type 2 Diabetes. Science (2018) 359:1151–6. doi: 10.1126/science.aao5774
101. Perraudeau F, McMurdie P, Bullard J, Cheng A, Cutcliffe C, Deo A, et al. Improvements to Postprandial Glucose Control in Subjects With Type 2 Diabetes: A Multicenter, Double Blind, Randomized Placebo-Controlled Trial of a Novel Probiotic Formulation. BMJ Open Diabetes Res Care (2020) 8:e001319. doi: 10.1136/bmjdrc-2020-001319
102. Jie Z, Yu X, Liu Y, Sun L, Chen P, Ding Q, et al. The Baseline Gut Microbiota Directs Dieting-Induced Weight Loss Trajectories. Gastroenterology (2021) 160:2029–42.e16. doi: 10.1053/j.gastro.2021.01.029
103. Rodriguez J, Hiel S, Neyrinck AM, Le Roy T, Potgens SA, Leyrolle Q, et al. Discovery of the Gut Microbial Signature Driving the Efficacy of Prebiotic Intervention in Obese Patients. Gut (2020) 69:1975–87. doi: 10.1136/gutjnl-2019-319726
104. Kootte RS, Levin E, Salojarvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of Insulin Sensitivity After Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell Metab (2017) 26:611–9.e6.
105. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI, et al. An Obesity-Associated Gut Microbiome With Increased Capacity for Energy Harvest. Nature (2006) 444:1027–31. doi: 10.1038/nature05414
106. Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut Microbiota From Twins Discordant for Obesity Modulate Metabolism in Mice. Science (2013) 341:1241214. doi: 10.1126/science.1241214
107. van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium Difficile. N Engl J Med (2013) 368:407–15. doi: 10.1056/NEJMoa1205037
108. Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF, et al. Transfer of Intestinal Microbiota From Lean Donors Increases Insulin Sensitivity in Individuals With Metabolic Syndrome. Gastroenterology (2012) 143:913–6.e7. doi: 10.1053/j.gastro.2012.06.031
109. Mocanu V, Zhang Z, Deehan EC, Kao DH, Hotte N, Karmali S, et al. Fecal Microbial Transplantation and Fiber Supplementation in Patients With Severe Obesity and Metabolic Syndrome: A Randomized Double-Blind, Placebo-Controlled Phase 2 Trial. Nat Med (2021) 27:1272–9. doi: 10.1038/s41591-021-01399-2
110. Baxter M, Colville A. Adverse Events in Faecal Microbiota Transplant: A Review of the Literature. J Hosp Infect (2016) 92:117–27. doi: 10.1016/j.jhin.2015.10.024
111. Clark FM, Dehr A. A Study of Butyric Acid-Producing Anaerobes Isolated From Spoiled Canned Tomatoes. Food Res (1947) 12:122–8. doi: 10.1111/j.1365-2621.1947.tb16401.x
112. Kalantar-Zadeh K, Berean KJ, Burgell RE, Muir JG, Gibson PR. Intestinal Gases: Influence on Gut Disorders and the Role of Dietary Manipulations. Nat Rev Gastroenterol Hepatol (2019) 16:733–47. doi: 10.1038/s41575-019-0193-z
Keywords: type 2 diabetes (T2D), microbiota, butyrate, metabolic disease, short chain fatty acids (SCFAs)
Citation: Arora T and Tremaroli V (2021) Therapeutic Potential of Butyrate for Treatment of Type 2 Diabetes. Front. Endocrinol. 12:761834. doi: 10.3389/fendo.2021.761834
Received: 20 August 2021; Accepted: 23 September 2021;
Published: 19 October 2021.
Edited by:
Peiyuan Yin, Dalian Medical University, ChinaReviewed by:
Pierre Larraufie, INRA UMR1319 Microbiologie de l’Alimentation au Service de la Santé, FranceVan B. Lu, Western University, Canada
Raylene A. Reimer, University of Calgary, Canada
Copyright © 2021 Arora and Tremaroli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tulika Arora, YXJvcmFAc3VuZC5rdS5kaw==
 Tulika Arora
Tulika Arora Valentina Tremaroli
Valentina Tremaroli