
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol., 30 September 2021
Sec. Neuroendocrine Science
Volume 12 - 2021 | https://doi.org/10.3389/fendo.2021.718363
This article is part of the Research TopicNeuropeptide GPCRs in Neuroendocrinology, Volume IIView all 28 articles
 Louise E. Atkinson1†
Louise E. Atkinson1† Ciaran J. McCoy1†
Ciaran J. McCoy1† Bethany A. Crooks1
Bethany A. Crooks1 Fiona M. McKay1
Fiona M. McKay1 Paul McVeigh1
Paul McVeigh1 Darrin McKenzie1
Darrin McKenzie1 Allister Irvine1
Allister Irvine1 John Harrington2
John Harrington2 Bruce A. Rosa3,4
Bruce A. Rosa3,4 Makedonka Mitreva3,4
Makedonka Mitreva3,4 Nikki J. Marks1
Nikki J. Marks1 Aaron G. Maule1
Aaron G. Maule1 Angela Mousley1*
Angela Mousley1*Nematode parasites undermine human health and global food security. The frontline anthelmintic portfolio used to treat parasitic nematodes is threatened by the escalation of anthelmintic resistance, resulting in a demand for new drug targets for parasite control. Nematode neuropeptide signalling pathways represent an attractive source of novel drug targets which currently remain unexploited. The complexity of the nematode neuropeptidergic system challenges the discovery of new targets for parasite control, however recent advances in parasite ‘omics’ offers an opportunity for the in silico identification and prioritization of targets to seed anthelmintic discovery pipelines. In this study we employed Hidden Markov Model-based searches to identify ~1059 Caenorhabditis elegans neuropeptide G-protein coupled receptor (Ce-NP-GPCR) encoding gene homologs in the predicted protein datasets of 10 key parasitic nematodes that span several phylogenetic clades and lifestyles. We show that, whilst parasitic nematodes possess a reduced complement of Ce-NP-GPCRs, several receptors are broadly conserved across nematode species. To prioritize the most appealing parasitic nematode NP-GPCR anthelmintic targets, we developed a novel in silico nematode parasite drug target prioritization pipeline that incorporates pan-phylum NP-GPCR conservation, C. elegans-derived reverse genetics phenotype, and parasite life-stage specific expression datasets. Several NP-GPCRs emerge as the most attractive anthelmintic targets for broad spectrum nematode parasite control. Our analyses have also identified the most appropriate targets for species- and life stage- directed chemotherapies; in this context we have identified several NP-GPCRs with macrofilaricidal potential. These data focus functional validation efforts towards the most appealing NP-GPCR targets and, in addition, the prioritization strategy employed here provides a blueprint for parasitic nematode target selection beyond NP-GPCRs.
Nematode parasites continue to have a global impact on human health and agricultural productivity such that novel mode-of-action anthelmintics are critical for sustained parasite control, especially in light of the escalation in anthelmintic resistance (1–3). Whilst the nematode neuromuscular system is a proven drug target, it remains underexploited (4, 5). Indeed, the majority of frontline anthelmintics only target aspects of neuromuscular signaling controlled by ion channels, however the neuropeptide signaling system is also critical to normal nematode neuromuscular function (6, 7).
Within the neuromuscular signalling system, neuropeptide GPCRs (NP-GPCRs) have been identified as highly ‘druggable’ targets (8). Indeed, an estimated 34% of human drugs act on GPCRs (9, 10). Despite this, NP-GPCRs have yet to be exploited for chemotherapeutic control of nematode parasites. In part, this is due to limited knowledge of NP-GPCR profiles in key parasitic nematode species which would enable NP-GPCR target prioritization.
Recent advances in ‘omics’ technologies have driven a significant expansion of in silico data for parasitic nematodes (11), providing an opportunity for the identification of novel putative anthelmintic targets. However, the volume and complexity of the available datasets presents a significant challenge to target prioritization. In silico prioritization approaches are essential given the lack of tractable and scalable reverse genetics tools for parasitic nematode systems (12, 13).
Analysis of the Caenorhabditis elegans genome suggests the presence of 152 putative NP-GPCRs (14), several of which are likely to represent attractive and exploitable anthelmintic targets. Indeed, functional studies indicate that some Ce-NP-GPCR knockdown/knockout worms display aberrant phenotypes that include paralysis and death [see WormBase; Harris et al. (15)]. Despite this, we have limited knowledge of NP-GPCR encoding gene conservation and life-stage expression in therapeutically relevant parasitic nematodes. These data are essential to drive the prioritization of parasite NP-GPCR drug targets for functional validation and chemotherapeutic exploitation.
In this study we employed in silico approaches to: (i) characterise the NP-GPCR complements of 10 key parasitic nematode species; (ii) develop a novel nematode drug target prioritization pipeline that incorporates NP-GPCR conservation, expression and functional data, and (iii) identify NP-GPCRs that represent putative, novel, broad spectrum parasitic nematode control targets. Integration of these multi-omics-derived datasets provides a springboard for functional biology that will improve our understanding of fundamental nematode neurosignalling and support future anthelmintic discovery efforts.
Putative nematode NP-GPCRs were identified via multiple sequence alignment derived Hidden Markov Models (HMMs), using methods based on those previously described (16). Briefly, HMMs were constructed using predicted protein alignments of all putative C. elegans neuropeptide receptors (14). Alignments were generated using MEGA 7 with default MUSCLE settings (17). Distinct models were constructed with rhodopsin and secretin NP-GPCR family members (14) using default hmmbuild parameters [HMMER v3; Mistry et al. (18)]. hmmsearch (HMMER v3) was employed to identify potential NP-GPCRs within the predicted protein datasets of 10 phylogenetically dispersed nematode parasites (Trichuris muris, Trichinella spiralis, Romanomermis culicivorax, Ascaris suum, Brugia malayi, Dirofilaria immitis, Necator americanus, Haemonchus contortus, Bursaphelenchus xylophilus, Globodera pallida; see (Supplementary Table 1), using default settings. The putative NP-GPCR sequences identified via hmmsearch were then used as queries in BLASTp searches in the NCBI non-redundant database (https://blast.ncbi.nlm.nih.gov; default settings) to identify the most similar sequences in C. elegans. Queries that failed to return a putative NP-GPCR as the highest scoring pair/top hit were excluded from downstream analyses. Putative NP-GPCR sequences were then filtered based on the number of transmembrane (TM) domains, as predicted by hmmtop (19). Returns containing 4 or more TM regions were excluded from downstream phylogenetic analyses (see Supplementary Figure 1 for a species-specific summary of the TM domain composition of all returns present in the putative NP-GPCR datasets), but still included in the drug target prioritization pipeline (see Figure 1).
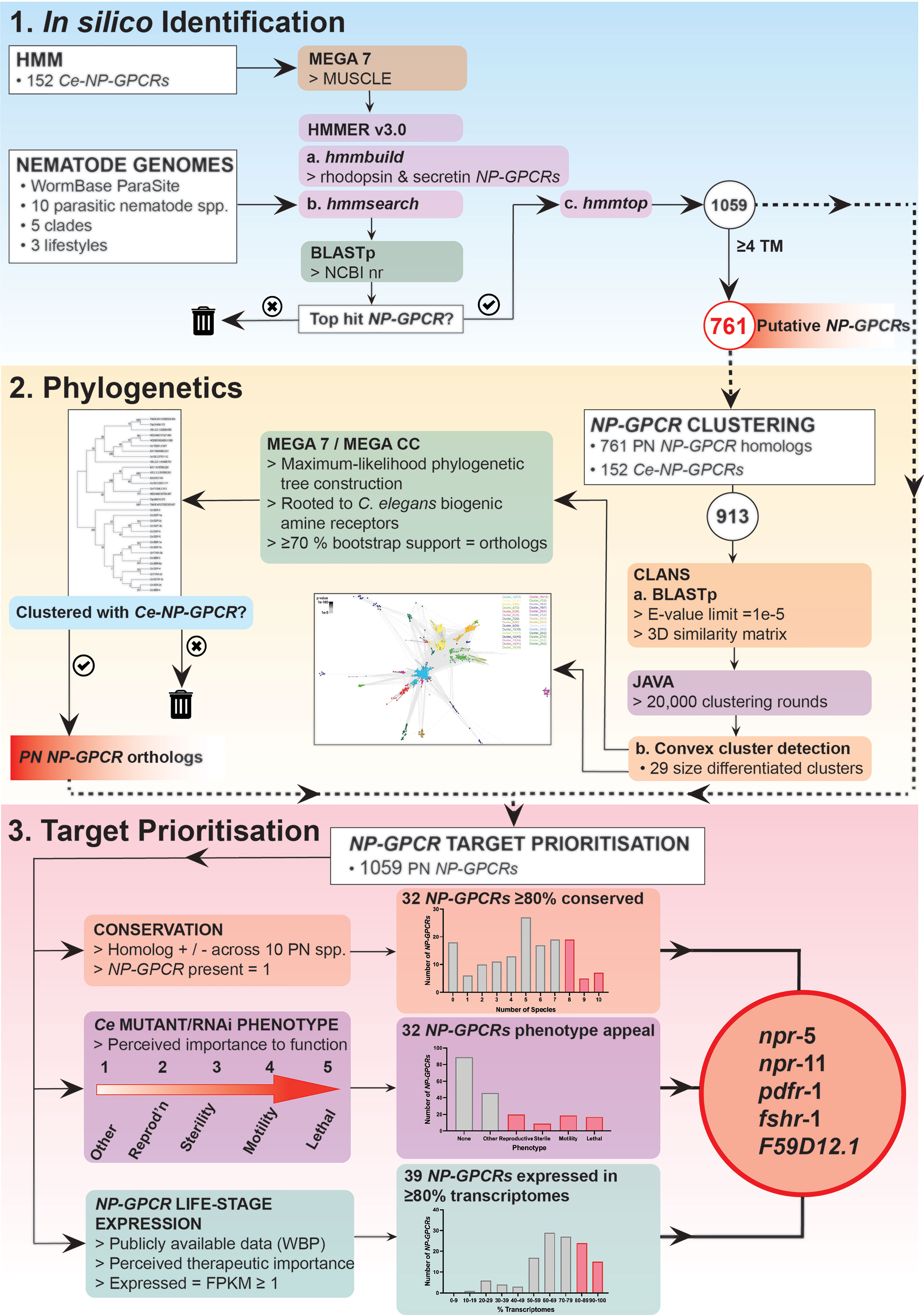
Figure 1 NP-GPCR drug target prioritization pipeline demonstrating NP-GPCR identification, phylogenetics and target prioritization workflow and a summary of the data generated at key stages in the pipeline. Five parasitic nematode NP-GPCRs are prioritised for validation in parasites: npr-5, -11, pdfr-1, fshr-1 and F59D12.1. HMM, Hidden Markov Models; TM, transmembrane domains; PN, parasitic nematode; CLANS, Cluster Analysis of Sequences; Ce, Caenorhabditis elegans; Reprod’n, reproduction; EXP, expression; WBP, WormBase Parasite; KO, knockout; RNAi, RNA interference.
The CLANS algorithm (https://toolkit.tuebingen.mpg.de/#/tools/clans) was used to identify convex clusters within the NP-GPCR datasets (20). Parasite NP-GPCR hits (761 putative parasite NP-GPCR sequences with ≥4 TM domains) were analysed alongside all putative NP-GPCRs from C. elegans (14). NP-GPCR sequences were uploaded to the CLANS website; BLAST high scoring pairs were extracted up to an E-value limit of 1e-5, all other parameters remained at default. CLANS performed a series of all-against-all BLASTp comparisons between every sequence submitted, generating a 3D similarity matrix constructed from the e-values of each individual search. The CLANS file output was visualized and coloured after 20,000 clustering rounds using the Java-based desktop software. CLANS convex cluster detection algorithm was used to delineate clusters of sequences under default settings. Clusters were numbered according to size (Cluster 1 being the largest). Individual clusters were used in Maximum-likelihood phylogenetic tree construction using MEGA 7 or MEGA CC (17), depending on the computing requirements of individual trees. Note that, where CLANS delineated clusters within the previously defined NP-GPCR families (14), these clusters were amalgamated prior to further phylogenetic analyses. Similarly, satellite singleton (non-clustered) sequences and small clusters that lacked any putative C. elegans homolog were grouped with their nearest-neighbour cluster prior to tree construction. Sequences extracted from each cluster were aligned via default MUSCLE settings in MEGA 7. Alignments were analyzed using the ‘find best DNA/Protein Models (ML)’ option to determine the most appropriate model of evolution for tree construction. All trees were constructed using: the bootstrap method (500 replicates); the LG model of evolution (G+I) with 5 discrete Gamma categories; a partial deletion of gaps (80% site coverage cut-off); and the nearest-neighbour interchange algorithm with no branch swap filter. Trees were rooted using a selection of C. elegans biogenic amine receptor sequences (see Supplementary Figures 2–12). Returns that clustered with a specific Ce-NP-GPCR with ≥70% bootstrap support were considered orthologs. Where returns failed to cluster with a specific Ce-NP-GPCR, but clustered with multiple C. elegans paralogs within the same NP-GPCR family, they were assigned based on top BLAST hit.
A drug target prioritization pipeline based on: (i) NP-GPCR-encoding gene conservation (generated in this study); (ii) C. elegans derived functional data (15), and (iii) publicly available RNASeq data (see Supplementary Table 1), were collated and curated as outlined in Figure 1. Briefly, NP-GPCR conservation profiles across the nine key parasite species in this study were analysed using the phylogenetics approach described above. To enable the inclusion of all putative NP-GPCR hits in the prioritization pipeline, and to circumvent prioritization bias by losing those that possess partial sequence availability (<4 TM domains; not suitable for phylogenetic analyses), predicted proteins with <4 TM regions were included as homologs of the highest scoring C. elegans BLAST hit.
Phenotype data associated with C. elegans mutant/RNAi experiments for the 152 known NP-GPCRs were collated from observed phenotypes reported on WormBase [version WS280; Harris et al. (15)]. Each Ce-NP-GPCR encoding gene was scored based on phenotype significance (with relevance to anthelmintic target discovery), where no recorded phenotype scored 0, reproductive scored 2, sterility scored 3, motility scored 4, and lethality scored 5. Any other recorded phenotype scored 1. Many Ce-NP-GPCR encoding genes had multiple phenotypes recorded; in this scenario, phenotype scores were combined to provide an overall total phenotype score for each NP-GPCR. Where multiple phenotypes within the same category were recorded, the category was only scored once.
RNASeq data were accessed from published life-stage specific transcriptome datasets [untreated/wildtype: T. muris (21); A. suum (22, 23); B. malayi (24, 25); D. immitis (26); H. contortus (27), and G. pallida (28)]. An FPKM value of 1 was used as the threshold for transcript expression (where FPKM ≥1 was deemed to be expressed). RNASeq data for H. contortus and T. muris (raw counts and FPKM) were generated following an established RNASeq pipeline. Raw sequences reads [PRJEB1360 (27); PRJEB1054 (21)] were downloaded and split into forward and reverse fastq files using NCBI SRA Toolkit (29). Reads were trimmed using Trimmomatic [v0.36; parameter: LEADING:5 TRAILING:5 SLIDINGWINDOW:3:15 MINLEN:34 (30)]. Corresponding genome assemblies for H. contortus (27) and T. muris (21) respectively, were downloaded from WormBase ParaSite (WBP) FTP server (31) and reads were mapped to these genomes using HISAT2 [v2.1.0 (32)]. Following genome mapping, raw gene counts were assigned through use of SubRead v 2.0.1 featureCounts (33). Raw counts of orthologous genes in samples were transformed to FPKM using countToFPKM (34) and median FPKMs were calculated in order to represent raw gene expressions of various developmental stages in these nematodes.
In this study we identified 1059 putative Ce-NP-GPCR homologs in the predicted protein datasets of 10 phylogenetically dispersed nematode parasites (see Supplementary Table 2 and Figure 1). To our knowledge this is the most comprehensive analysis of NP-GPCR profiles in parasitic nematodes to date, spanning five phylogenetic clades and a range of parasitic lifestyles [human parasitic nematode (HPN), animal parasitic nematode (APN), plant parasitic nematode (PPN), entomopathogenic nematodes (EPN)]. Several key points emerge from this study:
All 10 parasitic nematodes examined in this study exhibited restricted profiles of the 152 Ce-NP-GPCRs [21-78% Ce-NP-GPCR profile (average 49.2%); see Supplementary Table 2, Supplementary Data Sheet 1 and Figure 2] this trend is similar to that noted previously (35, 36). Ascaris suum boasts the largest Ce-NP-GPCR complement of all parasites examined (78%; Figure 2A), including in comparison to the clade 9 species H. contortus and N. americanus which are more closely related to C. elegans. This suggest that A. suum has lost fewer NP-GPCR encoding genes than both H. contortus and N. americanus despite being more distantly related to C. elegans. The lowest complement of Ce-NP-GPCRs was identified in the clade 2 species T. muris (21%) and T. spiralis (22%; Figure 2A). The phylum spanning profile of NP-GPCR encoding gene complements reported here closely aligns with the parasitic nematode neuropeptide [FMRF-amide-like peptide (flp), and neuropeptide like protein (nlp)] profiles characterised previously (36, 37), where A. suum and the clade 2 species (T. spiralis and T. muris) also display the largest and smallest complements of parasite neuropeptide encoding genes respectively.
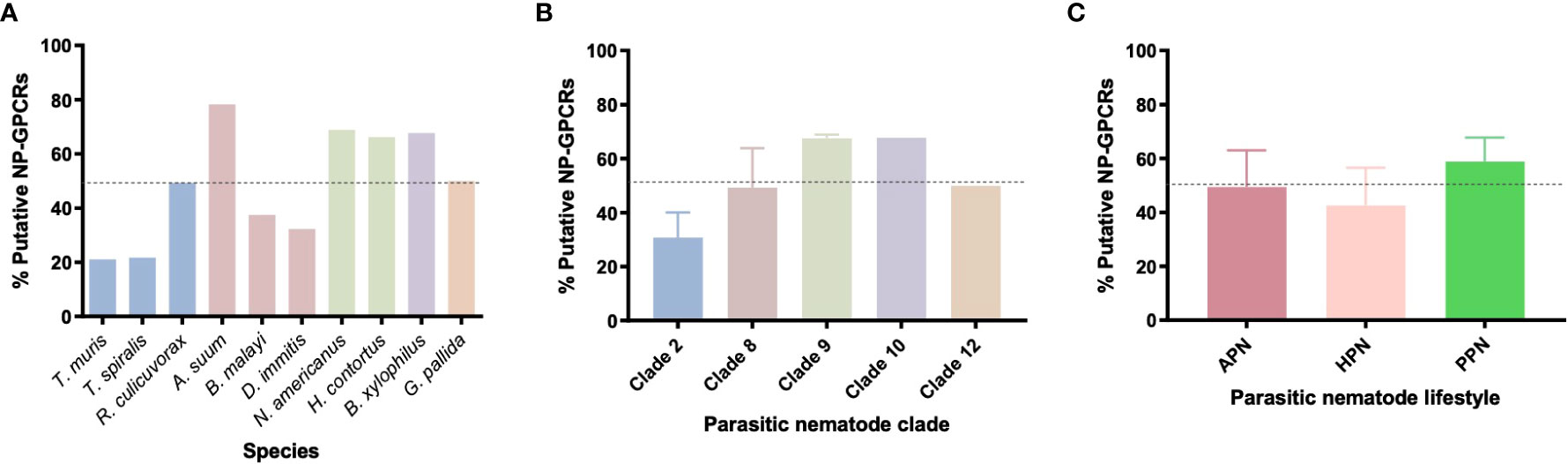
Figure 2 Nematode parasites have reduced and variable NP-GPCR complements across (A) 10 nematode species (B) phylogenetic clades and (C) parasitic lifestyles, expressed as a % of the predicted 152 Caenorhabditis elegans NP-GPCRs. Dotted line represents the average % (49.3%) of Ce-NP-GPCRs across all 10 species. APN, animal parasitic nematode; HPN, human parasitic nematode; PPN, plant parasitic nematode.
The HMM based approach employed also identified several biogenic amine GPCRs in addition to the NP-GPCRs reported here (data not shown); this provides confidence that all putative NP-GPCRs were identified in the available parasitic nematode datasets. Although a small number of divergent NP-GPCR sequences without an obvious C. elegans ortholog were identified in specific parasite species (see Supplementary Data Sheet 1 and Supplementary Figures 2–12), these were not broadly conserved across the parasite species examined.
The NP-GPCR encoding gene profiles of parasitic nematodes representing five nematode clades [2, 8, 9, 10, 12; Holterman et al. (38)] were examined in this study. Whilst only a small number of species from each clade were examined here, all clades exhibited a reduced complement of Ce-NP-GPCR homologs (see Figure 2B); clade 9 and 10 nematodes displayed the highest complement (both 68%) of Ce-NP-GPCRs and clade 2 displayed the most reduced (21%).
Within clades, variation in NP-GPCR complement was evident; for example, whilst the clade 2 species, T. muris and T. spiralis, displayed a highly similar, reduced, NP-GPCR complement (21%), an additional clade 2 species, R. culicivorax (entomopathogenic nematode), possessed 49% of Ce-NP-GPCR homologs (Figures 2A, B and Supplementary Table 2). Similarly, the clade 8 filarids, B. malayi and D. immitis, displayed reduced NP-GPCR complements relative to A. suum (clade 8). These data suggest multiple distinct gene loss events in the lineages that led to present day Trichuris/Trichinella and filarid spp. It is also likely that some of the 152 Ce-NP-GPCRs arose from gene duplication events that occurred in the lineages that led to the crown clades (clade 8-12; Holterman et al. ()), and so the NP-GPCRs absent from clade 2 species may not have been present in the last common ancestor of all nematodes. In contrast, both of the plant parasitic nematodes examined, B. xylophilus (clade 10) and G. pallida (clade 12), display relatively similar NP-GPCR profiles despite their distinct clade designations (Supplementary Table 2). The number of NP-GPCRs present appears to be consistent across nematode lifestyles (as defined here; Figure 2C), however the gene profiles are different since species have distinct gene repertoires.
The nematode parasite NP-GPCR profiles include representatives from the 17 rhodopsin and secretin receptor sub-families described in C. elegans (14). It is interesting to note that whilst there is broad representation across the majority of receptor sub-families (see Figure 3), there are also significant gaps in NP-GPCR complements especially within the Drosophila Dromyosuppressin (dmsr-10, 12-16) and Drosophila FMRF-amide (frpr-11-13) GPCR families (see Supplementary Table 2). Also evident are significant gaps in the otherwise broad NP-GPCR family profiles of the filarids including an absence of members of the Ghrelin-obstatin/neuromedin U, Galinin and Sex Peptide receptor families and a significant reduction in Neurokinin/neuropeptide FF/orexin receptor family members (see Supplementary Table 2).

Figure 3 CLANS analysis identifies NP-GPCR sub-families. Similarity matrix derived from all-against-all BLASTp comparisons between all identified nematode NP-GPCR sequences (E-value limit = 1e-5). NPF/Y, neuropeptide F/Y receptor family; SOMA, somatostatin receptor family; GAL, galanin receptor family; FRPR, FMRFamide Peptide Receptor family; aFRPR, another FMRFamide Peptide Receptor family; DMSR, Drosophila myosuppressin receptor family; GO/NU, Ghrelin-obstatin/neuromedin U receptor family; NK/NPFF/Orexin, Neurokinin/neuropeptide FF/orexin receptor family; GnRH, Gonadotropin-releasing hormone receptor family; GC, Gastrin-cholecystokinin receptor family; VP, Related Vasopressin receptor family; SexP, Related to Sex peptide receptor family; DR, related to fly ortholog (TF315326), plus related family (TF315359); TAG-89-like, Related family with no specific orthologs (TF318526); AEX-2-like, Related family with no specific orthologs (TF316587); TF317595, Related family with fly ortholog; TF315508, Related family with no specific orthologs; TF316160, Related family with no specific orthologs; FSHR, follicle-stimulating hormone receptor; DmDopEcR, Drosophila Dopamine/Ecdysteroid receptor; SEC, secretin-type receptors.
The majority of NP-GPCRs are broadly expressed across the lifecycle stages of key species in this study (those with available life-stage specific RNASeq data; FPKM ≥1) indicating their general importance to nematode biology (see Supplementary Table 3). NP-GPCRs display differential expression patterns across life-stages in all parasitic nematodes examined (T. muris, A. suum, B. malayi, D. immitis, H. contortus, G. pallida; see Supplementary Table 3. Whilst it is interesting to note that the majority of NP-GPCRs are expressed in all life stages, including in adult nematodes, there appears to be a general upregulation of NP-GPCR expression in the larval stages of a number of species including B. malayi (L3), T. muris (L2), G. pallida (J2), D. immitis (microfilariae). This indicates that, whilst NP-GPCRs have an important role across the nematode lifecycle, there may be an enhancement of NP-GPCR signaling in the larval stages that could reflect a significant need for movement/migration and development at this stage. Whether these patterns of NP-GPCR expression can be directly tied to variation in gene function between lifecycle stages remains to be investigated.
The volume and complexity of the NP-GPCR profiles outlined above challenge the ability to prioritise the most attractive NP-GPCRs for validation as novel drug targets. In silico approaches offer a novel route to exploit available datasets and integrate information to direct drug target selection (39–42). Here we present a novel in silico nematode parasite NP-GPCR drug target prioritization pipeline that incorporates pan-phylum NP-GPCR conservation (generated in this study), parasite life-stage specific expression, and C. elegans-derived phenotype data to assess the target appeal of NP-GPCRs for nematode control (Figure 1).
Seven of the 152 Ce-NP-GPCRs are conserved across all 10 parasitic nematodes examined (gnrr-1, ckr-2, frpr-19, C01F1.4, F59D12.1, pdfr-1 and seb-3; see Supplementary Table 2 and Figure 4). An additional six NP-GPCRs are conserved in nine of the 10 key species examined (npr-4, daf-38, dmsr-2, dmsr-8, T11F9.1, H09F14.1; see Supplementary Table 2 and Figure 4) and a further 18 are conserved in eight of the 10 parasites in this study (npr-5, npr-11, npr-35, npr-16, npr-32, ntr-2, sprr-1, frpr-5, frpr-7, frpr-9, frpr-18, dmsr-1, dmsr-6, dmsr-7, F40A3.7, aexr-1, fshr-1, F13H6.5; see Supplementary Table 2 and Figure 4). Eighteen NP-GPCRs were not identified in any parasite species (see Supplementary Table 2), and six NP-GPCRs (dmsr-11, frpr-16, gnrr-7, npr-33, D1014.2, ZK863.1; present in only one species) show highly restricted patterns of conservation.
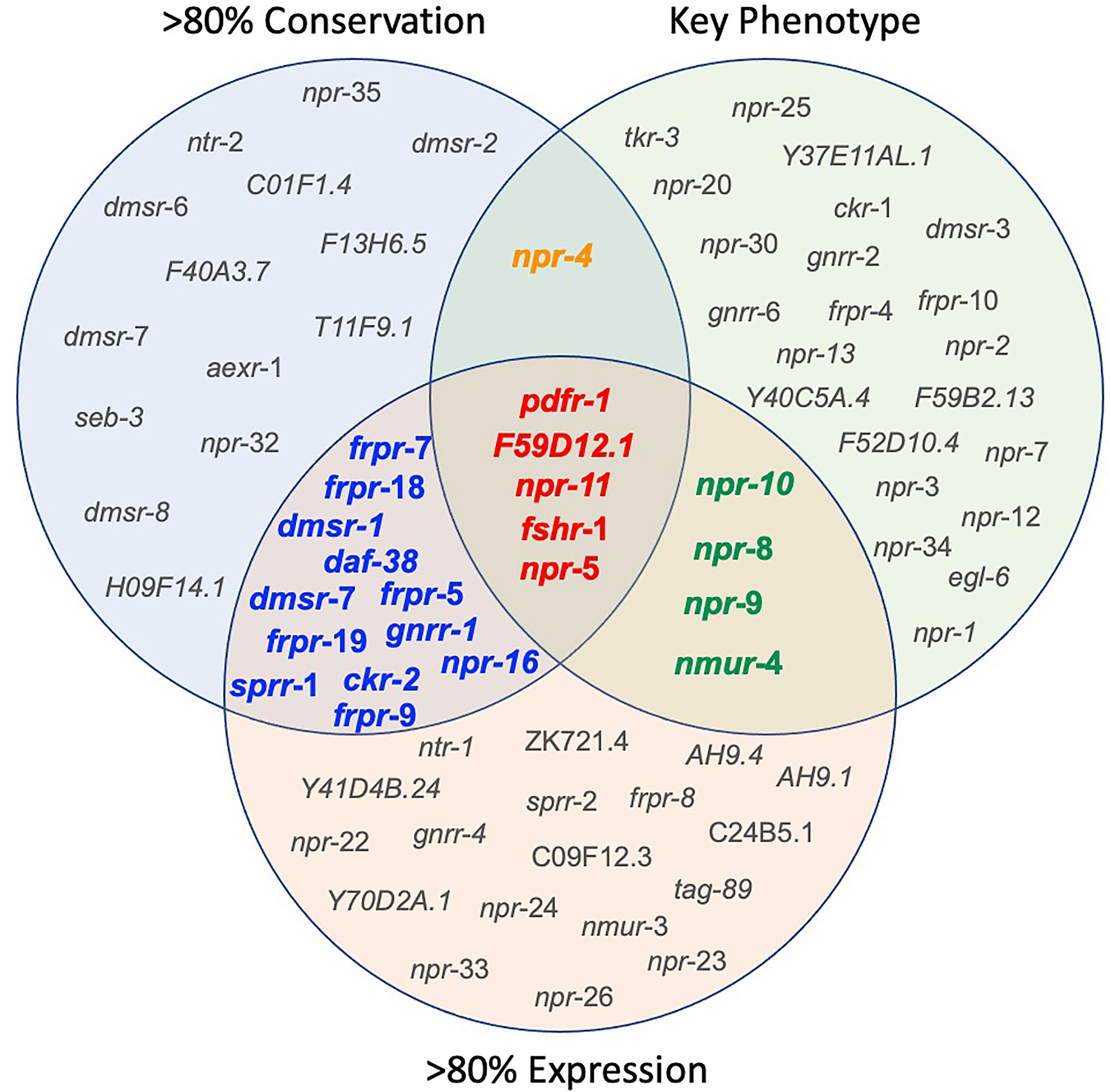
Figure 4 Integration of nematode ‘omics data informs NP-GPCR target prioritization. Venn Diagram illustrating the NP-GPCRs emerging from the prioritization pipeline following consideration of NP-GPCR conservation, NP-GPCR expression in key therapeutically relevant parasitic nematode lifecycle stages, and Ce-NP-GPCR null mutant/RNAi phenotype. Based on currently available data, the most appealing broad-spectrum NP-GPCR targets are highlighted in red. Receptors highlighted in orange represents those that share broad spectrum and key phenotype (lethality and locomotory) appeal; those highlighted in blue share broad spectrum conservation and expression attributes, and those highlighted in green share key phenotype and broad spectrum expression. >80% expression = NP-GPCRs that are expressed (FPKM>1) in more than 80% of the therapeutically relevant lifecycle stages analysed.
Caenorhabditis elegans functional data may inform NP-GPCR target appeal through the collation and consideration of phenotype information. In this study we collated phenotype data from C. elegans null mutant/RNAi experiments for the 152 NP-GPCRs [see WormBase (15); see Supplementary Table 4]. Each NP-GPCR was scored based on perceived phenotype significance to nematode biology and/or established anthelmintic endpoints, and therefore potential drug target appeal (see Materials and Methods). Often, multiple phenotypes were attributed to individual NP-GPCRs, therefore scores were added to yield an overall phenotype score for each receptor (see Supplementary Table 4). Several key points emerge from these datasets: (i) 89 of the 152 putative Ce-NP-GPCRs had no associated null mutant/RNAi phenotype(s) which may reflect a combination of: (a) lack of functional analyses data for Ce-NP-GPCRs, (b) use of an unsuitable C. elegans post-functional genomics bioassay and/or, (c) functional redundancy in nematode neuropeptidergic signalling systems; (ii) 16 Ce-NP-GPCRs had a lethal phenotype reported in at least one study (npr-5, nmur-4, npr-20, tkr-3, npr-30, gnrr-2, gnrr-6, ckr-1, frpr-4, frpr-10, dmsr-3, Y37E11AL.1, Y40C5A.4, F59B2.13, F52D10.4, fshr-1; see Supplementary Table 4); (iii) npr-3, -4, -7, -11, -12, -34, egl-6 and pdfr-1 also scored highly as these NP-GPCRs are associated with atypical locomotion, sterility or reproductive phenotypes in at least one study. Although the scoring system adopted here elevates the scores of NP-GPCRs that fall into multiple phenotype categories, the appeal of mutant/RNAi phenotypes associated with, for example, only locomotion should not be ignored; the NP-GPCRs associated with locomotion (in at least one study) include npr-1, -2, -8, -9, -10, -13, -25, and F59D12.1.
It is interesting to note that of the 16 NP-GPCRs that are associated with lethal phenotypes in C. elegans three (npr-5, ckr-1 and Y40C5A.4) are present in the most important APN/HPN species in this study (A. suum, B. malayi, D. immitis, N. americanus, H. contortus; see Supplementary Table 2). 16 additional NP-GPCRs emerged from the available phenotype data with appealing locomotory, reproductive and/or sterility phenotypes. Of these, five (npr-1, -11, -13, pdfr-1, F59D12.1) are present in the most important APN/HPN species in this study (see Supplementary Table 2). Finally, of the seven NP-GPCRs completely conserved in the parasitic species examined in this study, F59D12.1 and pdfr-1 have been linked to deleterious C. elegans phenotype post RNAi/knockout (see Supplementary Table 4).
In the context of this study, there are several important caveats to the extrapolation of the WormBase derived C. elegans phenotype data for drug target prioritisation including: (i) the reported differences between phenotypes recorded for multiple distinct mutations associated with the same gene, as well as RNAi animals (typically performed in RNAi hypersensitive mutant strains), (ii) the variable and often specific nature of the phenotype screens employed, and (iii) the bias in the volume/quality of functional data for specific NP-GPCRs or GPCR families. Whilst we have attempted to incorporate all of the observed C. elegans phenotypes recorded on WormBase regardless of experimental approach (how the mutant was generated, phenotype screens employed), the major caveats outlined above somewhat limit the utility of these data, and emphasise the need for functional analysis of all highly conserved and highly expressed NP-GPCRs in parasitic nematodes. Despite this, the approach offers a route to prioritising drug target candidates for functional validation in low throughput parasite platforms.
The format of our prioritisation pipeline allows for the distinct prioritisation of NP-GPCRs based on conservation, expression and/or C. elegans phenotype. This enables for the segregation or integration of prioritisation criteria as required and for the addition of phenotype data as they become available.
The available nematode RNASeq data suggest that parasite NP-GPCRs are broadly expressed across the species examined in this study (see Supplementary Table 3; for example, of the 13 genes that were conserved in at least nine of the 10 parasite species examined (see NP-GPCRs Have Conservation Profiles That Highlight Their Appeal as Broad Spectrum Drug Targets), the majority are also expressed in therapeutically relevant lifecycle stages (including: adult H. contortus; microfilariae and adult B. malayi and D. immitis; adult A. suum; adult T. muris; see Supplementary Table 3) underpinning the appeal of NP-GPCRs as therapeutic targets. In this context, the NP-GPCR expression data on their own do not discriminate sufficiently to prioritise a reduced cohort of broad spectrum drug targets however, in the scenario where a species focused/narrow spectrum target is desirable, ranking candidate drug targets based on expression data is more informative. For example, 21 NP-GPCRs are expressed in all therapeutically relevant stages of A. suum (npr-1, -16, -23, -33, gnrr-2, daf-38, ckr-2, frpr-5, -7, -9, -18, -19, sprr-1, dmsr-1, -4, C17H11.1, C24B5.1, tag-89, fshr-1, F59D12.1, pdfr-1); of these, npr-1, fshr-1, F59D12.1 and pdfr-1 also display defective phenotypes in C. elegans (see NP-GPCRs Are Associated With C. elegans Phenotypes That May Have Drug Target Appeal).
The data presented here identify 17 NP-GPCRs as the most appealing broad spectrum drug target candidates (>80% conservation and expression across key parasitic nematodes; see Figures 1 and 4). Parasitic nematode reverse genetics platforms are low throughput necessitating a focus on a smaller subset of NP-GPCRs. With this in mind, npr-5, npr-11, pdfr-1, fshr-1 and F59D12.1 step forward as initial candidates for functional validation (Figures 1 and 4).
Two of the NP-GPCRs that emerge from our pipeline as appealing targets (npr-5 and -11) have been linked to several peptides. NPR-11 has been functionally linked to NLP-1 and FLP-34 and, heterologously matched with FLP-21, -18, -34, -15 and -27 (43–47). NPR-5 is also functionally linked to FLP-18 and heterologously linked to FLP-18 and -21 (46, 48–50). Interestingly FLP-18 signalling has been shown to be important to nematode biology and is associated with a raft of biological processes including chemosensation, heat avoidance, reversal length, foraging behaviour, metabolism, locomotion quiescence during lethargus, and dauer formation (49–55); these data enhance the appeal of NPR-5 and -11 as putative novel drug targets. Significantly, npr-5 is conserved in 84% of 134 nematode genomes, representing 109 species, 7 clades and 3 distinct lifestyles (56), highlighting the importance of NPR-5 across phylum Nematoda.
PDFR-1 is related to arthropod Pigment Dispersing Factor Receptor and, more distantly, to vertebrate Calcitonin and Vasoactive Intestinal Peptide receptors (57). These receptors function in the control of circadian rhythms and arousal (51, 57, 58). PDFR-1 in C. elegans has been deorphanised heterologously and functionally to PDF-1 and PDF-2 (NLP-37) peptides, which modulate locomotion (57, 59). PDFR-1 signalling has also been implicated in inducing extended roaming states, arousal of locomotory behaviour following lethargus, and in the promotion of male mate searching behaviour in C. elegans (51, 60, 61). Notably, pdf-1 and pdfr-1 were present together in 96% of 134 nematode genomes (unpublished observations).
The remaining prioritised receptors are orphan NP-GPCRs that have not yet been linked to a cognate ligand. F59D12.1, also known as PCDR-1 (Pathogen Clearance Defective Receptor), has been associated with locomotion via RNAi experiments which resulted in slow and paralyzed worms (62). PCDR-1 also plays a key role in pathogen clearance of Microbacterium nematophilum infection in C. elegans (63). The functional data available for fshr-1 indicate that mutant C. elegans (tm3954) and RNAi worms display lethal phenotypes (15, 64).
Whilst several additional NP-GPCRs (gnrr-1, ckr-2, frpr-19, C01F1.4, seb-3) were conserved in all species examined in this study, the limited functional information available for these receptors has precluded their prioritisation as the most appealing targets at this point (see Figure 4). This reflects a major gap in the NP-GPCR null mutant/RNAi phenotype data and is a caveat to drug prioritisation in this context. In addition, the scale and scope of the post-functional genomics phenotype screens performed in Ce-NP-GPCR null mutant/RNAi experiments are: (i) highly variable and often gene dependent and, (ii) focus almost exclusively on loss of function screens (lack of over-expression data), such that this results in a degree of bias within the NP-GPCR prioritisation pipeline whereby highly conserved receptors that simply lack phenotype data are not emerging among the prioritised subset. Indeed, this is supported by a lack of correlation between parasitic nematode NP-GPCR conservation and the C. elegans derived phenotype data reported here (Spearman’s rho; Figure 5). It is also interesting to note that some of the NP-GPCRs highlighted above, that are broadly conserved but were not prioritised due to lack of phenotype data, are also broadly expressed across nematode lifecycle stages (e.g. gnrr-1, ckr-2, frpr-19 expressed in 83%, 97% and 100% of the transcriptomes examined respectively); this suggests potential functional importance and should form the focus of future functional analyses in parasitic nematodes.
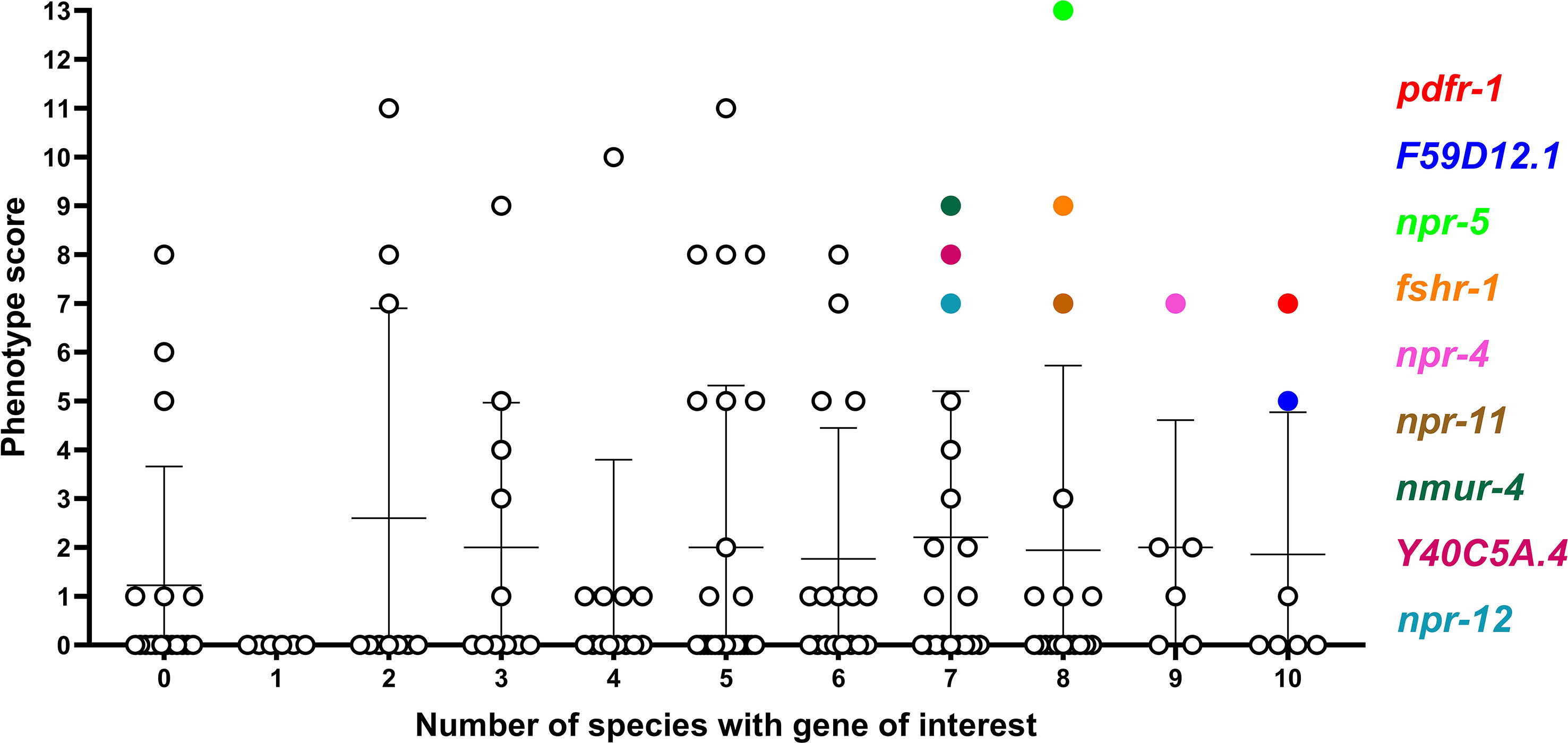
Figure 5 Comparison of phenotype score and NP-GPCR conservation. Phenotype score (sum of all phenotype categories associated with each gene) was plotted against the number of species which possess an NP-GPCR of interest. There is a no statistically significant correlation (Spearman rho) between phenotype score and NP-GPCR encoding gene conservation. Only genes displaying both a high level of conservation and phenotype score are highlighted.
Narrow spectrum anthelmintics have proven utility in nematode parasite control strategies (65). The drug target prioritisation pipeline presented here also enables the prioritisation of narrow spectrum drug targets that are relevant to the control of specific parasite species or life-stages. For example, no macrofilaricidal anthelmintics currently exist (66). Several NP-GPCRs (npr-5, -19, -23, -29, gnrr-1, -4, ckr-1, -2, frpr-8, -19, Y40C5A.4, aexr-1, fshr-1 and F59D12.1) emerge from our pipeline as NP-GPCRs that are expressed in both adult male and female B. malayi and D. immitis (see Supplementary Table 3). Some of these NP-GPCRs have already been prioritised as broad-spectrum targets (see Several Parasitic Nematode NP-GPCRs Emerge as the Most Appealing Broad Spectrum Drug Targets; npr-5, fshr-1 and F59D12.1) however, an additional two NP-GPCRs (ckr-1 and Y40C5A.4) emerge that are also appealing as microfilaricides; this underscores the utility of the NP-GPCR prioritisation pipeline in teasing out species specific therapeutic targets (see Supplementary Tables 2–4).
Recently improved parasite ‘omics’ data have driven a paradigm-shift towards mechanism-directed drug target screening approaches, providing an opportunity to identify the most attractive nematode parasite targets. Our focus on NP-GPCRs as therapeutic targets is driven by their importance to nematode biology (8), however the number and diversity of nematode NP-GPCRs is currently a hinderance to functional validation and successful exploitation. Here we present data on NP-GPCR conservation and the application of a drug target prioritisation pipeline that highlights the most attractive parasitic nematode NP-GPCRs for parasite control at this time. These data: (i) provide a comprehensive library of NP-GPCRs in key nematode parasites; (ii) enable the selection of both broad and narrow spectrum control targets; (iii) inform future validation efforts for NP-GPCRs in key parasitic nematode systems which are currently significantly lacking and, (iv) will expedite the anthelmintic development pipeline via informed target selection.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
AM, LA, CM, AGM, and NM designed the research. LA, CM, BC, PM, DM, AI, FM, and BAR performed the research. LA, CM, and BC analysed the data with assistance from BAR and MM. AM, LA, CM, AGM, NM, and JH wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by: the Biotechnology and Biological Sciences Research Council (BB/H019472/1 to AM); the Biotechnology and Biological Sciences Research Council/Merial Animal Health grant (BB/M010392/1 to AM, NM, and AGM); the Biotechnology and Biological Sciences Research Council/Boehringer Ingelheim (BB/T016396/1 to AM, NM, AGM, and LA); the Department of Education and Learning for Northern Ireland (studentships awarded to FM and BC); the Department of Agriculture, Environment and Rural Affairs for Northern Ireland (studentships awarded to DM and AI); the National Institutes of Health-National Institute of Allergy and Infectious Diseases grant (AI159450 to MM).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank Dr James Cotton, Dr Shelly Michalski, and Dr Elodie Ghedin for providing resources and advice.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.718363/full#supplementary-material
Supplementary Figure 1 | Nematode parasite NP-GPCR predicted transmembrane domains. Sequences with < 4 transmembrane (TM) domains were excluded from further phylogenetic analysis. Full length sequences are expected to contain 7 TM domains. Proportion of GPCRs presenting 7 TM domains can be used as a crude but relevant proxy for NP-GPCR annotation/dataset quality in this instance.
Supplementary Figure 2–12 | Maximum likelihood phylogenies constructed using the LG model of evolution (+G +I; with 5 discrete Gamma categories), 500 bootstrap replicates, partial deletion of gaps (80% site coverage cut-off) and the nearest-neighbour interchange algorithm with no branch swap filter. For rhodopsin-type GPCR phylogenies, trees were rooted using a selection of C. elegans biogenic amine receptors.
Supplementary Data Sheet 1 | HMM search output and post-phylogenetic analysis summary highlighting the specific nematode parasite NP-GPCR encoding gene IDs identified here.
Supplementary Table 1 | Parasitic nematode species examined in this study. APN, animal parasitic nematode; HPN, human parasitic nematode; PPN, plant parasitic nematode; EPN, entomopathogenic nematode. Genomes and transcriptomes used: Trichuris muris (21), Trichinella spiralis (67), Romanomermis culicivorax (68), Ascaris suum (22, 23), Brugia malayi (24, 25, 69), Dirofilaria immitis (26, 70), Necator americanus (71), Haemonchus contortus (27), Bursaphelenchus xylophilus (72), Globodera pallida (28).
Supplementary Table 2 | Ce-NP-GPCR complements of 10 key nematode species. A black box indicates the presence of a homologue identified via BLAST search. A grey box indicates the absence of a gene. GO/NU, Ghrelin-obstatin/neuromedin U receptor family; NK/NPFF/Orexin, Neurokinin/neuropeptide FF/orexin receptor family; GnRH, Gonadotropin-releasing hormone receptor family; GC, Gastrin-cholecystokinin receptor family; VP, Related Vasopressin receptor family; Sex P, Related to Sex peptide receptor family; Drosophila related, related to fly ortholog (CG33639); FSHR, follicle-stimulating hormone receptor; DmDopEcR, Drosophila Dopamine/Ecdysteroid receptor.
Supplementary Table 3 | Ce-NP-GPCR binary expression patterns in parasitic nematodes. A black box indicates the presence of a NP-GPCR homologue that is not expressed in a specific life cycle stage (FPKM <1). A red box indicates the presence of a NP-GPCR homologue expressed in a specific life cycle stage (FPKM > 1). A grey box indicates the absence of a gene. GO/NU, Ghrelin-obstatin/neuromedin U receptor family; NK/NPFF/Orexin, Neurokinin/neuropeptide FF/orexin receptor family; GnRH, Gonadotropin-releasing hormone receptor family; GC, Gastrin-cholecystokinin receptor family; VP, Related Vasopressin receptor family; SexP, Related to Sex peptide receptor family; Drosophila related, related to fly ortholog (CG33639); FSHR, follicle-stimulating hormone receptor; DmDopEcR, Drosophila Dopamine/Ecdysteroid receptor.
Supplementary Table 4 | Ce-NP-GPCR knock out/RNAi phenotype summary. A coloured box indicates phenotype linked to each Ce-NP-GPCR encoding gene. A grey box indicates the absence of a phenotype category. GO/NU, Ghrelin-obstatin/neuromedin U receptor family; NK/NPFF/Orexin, Neurokinin/neuropeptide FF/orexin receptor family; GnRH, Gonadotropin-releasing hormone receptor family; GC, Gastrin-cholecystokinin receptor family; VP, Related Vasopressin receptor family; SexP, Related to Sex peptide receptor family; Drosophila related, related to fly ortholog (CG33639); FSHR, follicle-stimulating hormone receptor; DmDopEcR, Drosophila Dopamine/Ecdysteroid receptor.
1. Jones JT, Haegeman A, Danchin EG, Gaur HS, Helder J, Jones MG, et al. Top 10 Plant-Parasitic Nematodes in Molecular Plant Pathology. Mol Plant Pathol (2013) 14:946–61. doi: 10.1111/mpp.12057
2. Hotez PJ, Lo NC. Neglected Tropical Diseases: Public Health Control Programs and Mass Drug Administration. In: Hunter’s Tropical Medicine and Emerging Infectious Diseases. Elsevier (2020). p. 209–13.
3. Charlier J, Rinaldi L, Musella V, Ploeger HW, Chartier C, Vineer HR, et al. Initial Assessment of the Economic Burden of Major Parasitic Helminth Infections to the Ruminant Livestock Industry in Europe. Prev Vet Med (2020) 182:105103. doi: 10.1016/j.prevetmed.2020.105103
4. Wolstenholme AJ. Ion Channels and Receptor as Targets for the Control of Parasitic Nematodes. Int J Parasitol: Drugs Drug Resist (2011) 1:2–13. doi: 10.1016/j.ijpddr.2011.09.003
5. Martin RJ, Robertson AP. Control of Nematode Parasites With Agents Acting on Neuro-Musculature Systems: Lessons for Neuropeptide Ligand Discovery. Neuropeptide Syst as Targets Parasite Pest Control (2010) 692:138–54. doi: 10.1007/978-1-4419-6902-6_7
6. McVeigh P, Geary TG, Marks NJ, Maule AG. The FLP-Side of Nematodes. Trends Parasitol (2006) 22:385–96. doi: 10.1016/j.pt.2006.06.010
7. Frooninckx L, Van Rompay L, Temmerman L, Van Sinay E, Beets I, Janssen T, et al. Neuropeptide GPCRs in C. Elegans. Front Endocrinol (2012) 3:167. doi: 10.3389/fendo.2012.00167
8. McVeigh P, Atkinson L, Marks NJ, Mousley A, Dalzell JJ, Sluder A, et al. Parasite Neuropeptide Biology: Seeding Rational Drug Target Selection? Int J Parasitol Drugs Drug Resist (2012) 2:76–91. doi: 10.1016/j.ijpddr.2011.10.004
9. Santos R, Ursu O, Gaulton A, Bento AP, Donadi RS, Bologa CG, et al. A Comprehensive Map of Molecular Drug Targets. Nat Rev Drug Discov (2017) 16:19–34. doi: 10.1038/nrd.2016.230
10. Hauser AS, Attwood MM, Rask-Andersen M, Schiöth HB, Gloriam DE. Trends in GPCR Drug Discovery: New Agents, Targets and Indications. Nat Rev Drug Discov (2017) 16:829–42. doi: 10.1038/nrd.2017.178
11. Bolt BJ, Rodgers FH, Shafie M, Kersey PJ, Berriman M, Howe KL. Using WormBase ParaSite: An Integrated Platform for Exploring Helminth Genomic Data. Methods Mol Biol (2018) 1757:471–91. doi: 10.1007/978-1-4939-7737-6_15
12. McCoy CJ, Atkinson LE, Robb E, Marks NJ, Maule AG, Mousley A. Tool-Driven Advances in Neuropeptide Research From a Nematode Parasite Perspective. Trends Parasitol (2017) 33:986–1002. doi: 10.1016/j.pt.2017.08.009
13. Ward JD. Rendering the Intractable More Tractable: Tools From Caenorhabditis Elegans Ripe for Import Into Parasitic Nematodes. Genetics (2015) 201:1279–94. doi: 10.1534/genetics.115.182717
14. Hobert O. The Neuronal Genome of Caenorhabditis Elegans. WormBook (2013) 13:1–106. doi: 10.1895/wormbook.1.161.1
15. Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Cho J, et al. WormBase: A Modern Model Organism Information Resource. Nucleic Acids Res (2020) 48:D762–7. doi: 10.1093/nar/gkz920
16. McVeigh P, McCammick E, McCusker P, Wells D, Hodgkinson J, Paterson S, et al. Profiling G Protein-Coupled Receptors of Fasciola Hepatica Identifies Orphan Rhodopsins Unique to Phylum Platyhelminthes. Int J Parasitol Drugs Drug Resist (2018) 8:87–103. doi: 10.1016/j.ijpddr.2018.01.001
17. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol (2016) 33:1870–4. doi: 10.1093/molbev/msw054
18. Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res (2013) 41:e121. doi: 10.1093/nar/gkt263
19. Tusnády GE, Simon I. The HMMTOP Transmembrane Topology Prediction Server. Bioinformatics (2001) 17:849–50. doi: 10.1093/bioinformatics/17.9.849
20. Frickey T, Lupas A. CLANS: A Java Application for Visualizing Protein Families Based on Pairwise Similarity. Bioinformatics (2004) 20:3702–4. doi: 10.1093/bioinformatics/bth444
21. Foth BJ, Tsai IJ, Reid AJ, Bancroft AJ, Nichol S, Tracey A, et al. Whipworm Genome and Dual-Species Transcriptome Analyses Provide Molecular Insights Into an Intimate Host-Parasite Interaction. Nat Genet (2014) 46:693–700. doi: 10.1038/ng.3010
22. Jex AR, Liu S, Li B, Young ND, Hall RS, Li Y, et al. Ascaris Suum Draft Genome. Nature (2011) 479:529–33. doi: 10.1038/nature10553
23. Rosa BA, Jasmer DP, Mitreva M. Genome-Wide Tissue-Specific Gene Expression, Co-Expression and Regulation of Co-Expressed Genes in Adult Nematode Ascaris Suum. PloS Negl Trop Dis (2014) 8:e2678. doi: 10.1371/journal.pntd.0002678
24. Choi Y-J, Ghedin E, Berriman M, McQuillan J, Holroyd N, Mayhew GF, et al. A Deep Sequencing Approach to Comparatively Analyze the Transcriptome of Lifecycle Stages of the Filarial Worm, Brugia Malayi. PloS negl Trop Dis (2011) 5:e1409–9. doi: 10.1371/journal.pntd.0001409
25. Ballesteros C, Tritten L, O’Neill M, Burkman E, Zaky WI, Xia J, et al. The Effect of In Vitro Cultivation on the Transcriptome of Adult Brugia Malayi. PloS Negl Trop Dis (2016) 10:e0004311. doi: 10.1371/journal.pntd.0004311
26. Luck AN, Evans CC, Riggs MD, Foster JM, Moorhead AR, Slatko BE, et al. Concurrent Transcriptional Profiling of Dirofilaria Immitis and Its Wolbachia Endosymbiont Throughout the Nematode Life Cycle Reveals Coordinated Gene Expression. BMC Genomics (2014) 15:1041. doi: 10.1186/1471-2164-15-1041
27. Laing R, Kikuchi T, Martinelli A, Tsai IJ, Beech RN, Redman E, et al. The Genome and Transcriptome of Haemonchus Contortus, a Key Model Parasite for Drug and Vaccine Discovery. Genome Biol (2013) 14:R88. doi: 10.1186/gb-2013-14-8-r88
28. Cotton JA, Lilley CJ, Jones LM, Kikuchi T, Reid AJ, Thorpe P, et al. The Genome and Life-Stage Specific Transcriptomes of Globodera Pallida Elucidate Key Aspects of Plant Parasitism by a Cyst Nematode. Genome Biol (2014) 15:R43. doi: 10.1186/gb-2014-15-3-r43
29. Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration. The Sequence Read Archive. Nucleic Acids Res (2011) 39:D19–21.
30. Bolger AM, Lohse M, Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
31. Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M. WormBase ParaSite– a Comprehensive Resource for Helminth Genomics. Mol Biochem Parasitol (2017) 215:2–10. doi: 10.1016/j.molbiopara.2016.11.005
32. Kim D, Langmead B, Salzberg SL. HISAT: A Fast Spliced Aligner With Low Memory Requirements. Nat Methods (2015) 12:357–60. doi: 10.1038/nmeth.3317
33. Liao Y, Smyth GK, Shi W. Featurecounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics (2014) 30:923–30. doi: 10.1093/bioinformatics/btt656
34. Alhendi ASN. countToFPKM: Convert Counts to Fragments per Kilobase of Transcript per Million (FPKM). R package version 1.0.0 (2019). Available at: https://CRAN.R-project.org/package=countToFPKM
35. Cardoso JC, Félix RC, Fonseca VG, Power DM. Feeding and the Rhodopsin Family G-Protein Coupled Receptors in Nematodes and Arthropods. Front Endocrinol (Lausanne) (2012) 3:157. doi: 10.3389/fendo.2012.00157
36. McCoy CJ, Atkinson LE, Zamanian M, McVeigh P, Day TA, Kimber MJ, et al. New Insights Into the FLPergic Complements of Parasitic Nematodes: Informing Deorphanisation Approaches. EuPA Open Proteom (2014) 3:262–72. doi: 10.1016/j.euprot.2014.04.002
37. McKay FM, McCoy CJ, Marks NJ, Maule AG, Atkinson LE, Mousley A. In Silico Analyses of Neuropeptide-Like Protein (NLP) Profiles in Parasitic Nematodes. bioRxiv (2021). doi: 10.1101/2021.03.03.433794
38. Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, et al. Phylum-Wide Analysis of SSU rDNA Reveals Deep Phylogenetic Relationships Among Nematodes and Accelerated Evolution Toward Crown Clades. Mol Biol Evol (2006) 23:1792–800. doi: 10.1093/molbev/msl044
39. Abubucker S, Martin J, Taylor CM, Mitreva M. HelmCoP: An Online Resource for Helminth Functional Genomics and Drug and Vaccine Targets Prioritization. PloS One (2011) 6:e21832. doi: 10.1371/journal.pone.0021832
40. Taylor CM, Fischer K, Abubucker S, Wang Z, Martin J, Jiang D, et al. Targeting Protein-Protein Interactions for Parasite Control. PloS One (2011) 6:e18381. doi: 10.1371/journal.pone.0018381
41. Taylor CM, Martin J, Rao RU, Powell K, Abubucker S, Mitreva M. Using Existing Drugs as Leads for Broad Spectrum Anthelmintics Targeting Protein Kinases. PloS Pathog (2013) 9:e1003149. doi: 10.1371/journal.ppat.1003149
42. Doyle MA, Gasser RB, Woodcroft BJ, Hall RS, Ralph SA. Drug Target Prediction and Prioritization: Using Orthology to Predict Essentiality in Parasite Genomes. BMC Genomics (2010) 11:1–14. doi: 10.1186/1471-2164-11-222
43. Lowery DE, Geary TG, Kubiak TM, Larsen MJ. G Protein-Coupled Receptor-Like Receptors and Modulators Thereof, United States, Patent 6,632,621. Pharmacia & Upjohn Company (2003).
44. Chalasani SH, Kato S, Albrecht DR, Nakagawa T, Abbott L, Bargmann CI. Neuropeptide Feedback Modifies Odor-Evoked Dynamics in Caenorhabditis Elegans Olfactory Neurons. Nat Neurosci (2010) 13:615. doi: 10.1038/nn.2526
45. Hapiak V, Summers P, Ortega A, Law WJ, Stein A, Komuniecki R. Neuropeptides Amplify and Focus the Monoaminergic Inhibition of Nociception in Caenorhabditis Elegans. J Neurosci (2013) 33:14107–16. doi: 10.1523/JNEUROSCI.1324-13.2013
46. Gershkovich MM, Groß VE, Kaiser A, Prömel S. Pharmacological and Functional Similarities of the Human Neuropeptide Y System in C. Elegans Challenges Phylogenetic Views on the FLP/NPR System. Cell Commun Signal (2019) 17:123. doi: 10.1186/s12964-019-0436-1
47. Fadda M, De Fruyt N, Borghgraef C, Watteyne J, Peymen K, Vandewyer E, et al. NPY/NPF-Related Neuropeptide FLP-34 Signals From Serotonergic Neurons to Modulate Aversive Olfactory Learning in Caenorhabditis Elegans. J Neurosci (2020) 40:6018–34. doi: 10.1523/JNEUROSCI.2674-19.2020
48. Kubiak TM, Larsen MJ, Bowman JW, Geary TG, Lowery DE. FMRFamide-Like Peptides Encoded on the Flp-18 Precursor Gene Activate Two Isoforms of the Orphan Caenorhabditis Elegans G-Protein-Coupled Receptor Y58G8A.4 Heterologously Expressed in Mammalian Cells. Biopolymers (2008) 90:339–48. doi: 10.1002/bip.20850
49. Cohen M, Reale V, Olofsson B, Knights A, Evans P, de Bono M. Coordinated Regulation of Foraging and Metabolism in C. Elegans by RFamide Neuropeptide Signaling. Cell Metab (2009) 9:375–85. doi: 10.1016/j.cmet.2009.02.003
50. Stawicki TM, Takayanagi-Kiya S, Zhou K, Jin Y. Neuropeptides Function in a Homeostatic Manner to Modulate Excitation-Inhibition Imbalance in C. elegans. PloS Genet (2013) 9:e1003472. doi: 10.1371/journal.pgen.1003472
51. Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, Kaplan JM. Analysis of NPR-1 Reveals a Circuit Mechanism for Behavioral Quiescence in C. elegans. Neuron (2013) 78:869–80. doi: 10.1016/j.neuron.2013.04.002
52. Luo J, Xu Z, Tan Z, Zhang Z, Ma L. Neuropeptide Receptors NPR-1 and NPR-2 Regulate Caenorhabditis Elegans Avoidance Response to the Plant Stress Hormone Methyl Salicylate. Genetics (2015) 199:523–31. doi: 10.1534/genetics.114.172239
53. Bhardwaj A, Thapliyal S, Dahiya Y, Babu K. FLP-18 Functions Through the G-Protein-Coupled Receptors NPR-1 and NPR-4 to Modulate Reversal Length in Caenorhabditis Elegans. J Neurosci (2018) 38:4641–54. doi: 10.1523/JNEUROSCI.1955-17.2018
54. Nkambeu B, Salem JB, Leonelli S, Marashi FA, Beaudry F. EGL-3 and EGL-21 are Required to Trigger Nocifensive Response of Caenorhabditis Elegans to Noxious Heat. Neuropeptides (2019) 73:41–8. doi: 10.1016/j.npep.2018.11.002
55. Park J, Choi W, Dar AR, Butcher RA, Kim K. Neuropeptide Signaling Regulates Pheromone-Mediated Gene Expression of a Chemoreceptor Gene in C. elegans. Molecules Cells (2019) 42:28. doi: 10.14348/molcells.2018.0380
56. Atkinson LE, Liu Y, McKay F, Vandewyer E, Viau C, Irvine A, et al. Ascaris suum Informs Extrasynaptic Volume Transmission in Nematodes. ACS Chem Neurosci (2021). doi: 10.1021/acschemneuro.1c00281
57. Janssen T, Husson SJ, Lindemans M, Mertens I, Rademakers S, Ver Donck K, et al. Functional Characterization of Three G Protein-Coupled Receptors for Pigment Dispersing Factors in Caenorhabditis Elegans. J Biol Chem (2008) 283:15241–9. doi: 10.1074/jbc.M709060200
58. Janssen T, Husson SJ, Meelkop E, Temmerman L, Lindemans M, Verstraelen K, et al. Discovery and Characterization of a Conserved Pigment Dispersing Factor-Like Neuropeptide Pathway in Caenorhabditis Elegans. J Neurochem (2009) 111:228–41. doi: 10.1111/j.1471-4159.2009.06323.x
59. Meelkop E, Temmerman L, Janssen T, Suetens N, Beets I, Van Rompay L, et al. PDF Receptor Signaling in Caenorhabditis Elegans Modulates Locomotion and Egg-Laying. Mol Cell Endocrinol (2012) 361:232–40. doi: 10.1016/j.mce.2012.05.001
60. Barrios A, Ghosh R, Fang C, Emmons SW, Barr MM. 1 Neuropeptide Signaling Modulates a Neural Circuit for Mate-Searching Behavior in C. elegans. Nat Neurosci (2012) 15:1675–82. doi: 10.1038/nn.3253
61. Flavell SW, Pokala N, Macosko EZ, Albrecht DR, Larsch J, Bargmann CI. Serotonin and the Neuropeptide PDF Initiate and Extend Opposing Behavioral States in C. elegans. Cell (2013) 154:1023–35. doi: 10.1016/j.cell.2013.08.001
62. Keating CD, Kriek N, Daniels M, Ashcroft NR, Hopper NA, Siney EJ, et al. Whole-genome analysis of 60 G protein-coupled receptors in Caenorhabditis elegans by gene knockout with RNAi. Curr Biol (2003) 13(19):1715–20. doi: 10.1016/j.cub.2003.09.003
63. Anderson A, Chew YL, Schafer W, McMullan R. Identification of a Conserved, Orphan G Protein-Coupled Receptor Required for Efficient Pathogen Clearance in Caenorhabditis Elegans. Infect Immun (2019) 87:e00034–19. doi: 10.1128/IAI.00034-19
64. Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, et al. Genome-Wide RNAi of C. Elegans Using the Hypersensitive Rrf-3 Strain Reveals Novel Gene Functions. PloS Biol (2003) 1:E12. doi: 10.1371/journal.pbio.0000012
65. Hansen TV, Cirera S, Neveu C, Courtot E, Charvet CL, Calloe K, et al. The Narrow-Spectrum Anthelmintic Oxantel Is a Potent Agonist of a Novel Acetylcholine Receptor Subtype in Whipworms. PloS Pathog (2021) 17:e1008982. doi: 10.1371/journal.ppat.1008982
66. Geary TG, Maule A. Neuropeptide Systems as Targets for Parasite and Pest Control. Landes Bioscience (2010). doi: 10.1007/978-1-4419-6902-6
67. Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, Martin J, et al. The Draft Genome of the Parasitic Nematode Trichinella Spiralis. Nat Genet (2011) 43:228–35. doi: 10.1038/ng.769
68. Schiffer PH, Kroiher M, Kraus C, Koutsovoulos GD, Kumar S, Camps JI, et al. The Genome of Romanomermis Culicivorax: Revealing Fundamental Changes in the Core Developmental Genetic Toolkit in Nematoda. BMC Genomics (2013) 14:1–16. doi: 10.1186/1471-2164-14-923
69. Tracey A, Foster JM, Paulini M, Grote A, Mattick J, Tsai YC, et al. Nearly Complete Genome Sequence of Brugia Malayi Strain Fr3. Microbiol Resour Announc (2020) 9(24):e00154-20. doi: 10.1128/MRA.00154-20
70. Godel C, Kumar S, Koutsovoulos G, Ludin P, Nilsson D, Comandatore F, et al. The Genome of the Heartworm, Dirofilaria Immitis, Reveals Drug and Vaccine Targets. FASEB J (2012) 26:4650–61. doi: 10.1096/fj.12-205096
71. Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, et al. Genome of the Human Hookworm Necator Americanus. Nat Genet (2014) 46:261–9. doi: 10.1038/ng.2875
Keywords: neuropeptide, G-protein coupled receptor, FMRF-amide like peptide, drug target, nematode parasite
Citation: Atkinson LE, McCoy CJ, Crooks BA, McKay FM, McVeigh P, McKenzie D, Irvine A, Harrington J, Rosa BA, Mitreva M, Marks NJ, Maule AG and Mousley A (2021) Phylum-Spanning Neuropeptide GPCR Identification and Prioritization: Shaping Drug Target Discovery Pipelines for Nematode Parasite Control. Front. Endocrinol. 12:718363. doi: 10.3389/fendo.2021.718363
Received: 31 May 2021; Accepted: 20 July 2021;
Published: 30 September 2021.
Edited by:
Liliane Schoofs, KU Leuven, BelgiumReviewed by:
Joao Carlos dos Reis Cardoso, University of Algarve, PortugalCopyright © 2021 Atkinson, McCoy, Crooks, McKay, McVeigh, McKenzie, Irvine, Harrington, Rosa, Mitreva, Marks, Maule and Mousley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angela Mousley, YS5tb3VzbGV5QHF1Yi5hYy51aw==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.