
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 08 July 2021
Sec. Reproduction
Volume 12 - 2021 | https://doi.org/10.3389/fendo.2021.675581
Background: Polycystic ovary syndrome (PCOS) is a common endocrine disorder worldwide. We aimed to examine the associations of two mitochondrial DNA (mtDNA) biomarkers in the peripheral blood, mtDNA copy number (CN), and mtDNA4977 deletion rate (DR), with PCOS in a clinical setting.
Methods: We performed a study involving 263 women with PCOS and 326 age-matched controls between June 2015 and June 2019. The mtDNA CN and mtDNA4977 DR were measured using multiplex probe-based qPCR. The associations of the mtDNA CN and mtDNA4977 DR with the risk of PCOS were estimated using logistic regression.
Results: Analysis of the associations between mtDNA biomarkers and PCOS indicate that the mtDNA CN (P = 0.003) and mtDNA4977 DR (P < 0.001) in PCOS patients were significantly higher than those in the controls. After adjusting for the body mass index, luteinizing hormone/follicle-stimulating hormone ratio, and testosterone level, only higher mtDNA4977 DR was associated with PCOS (odds ratio 1.053, 95% confidence interval 1.024 to 1.083; P < 0.001). The linear dose-response trends of the mtDNA4977 DR were also supported by the quartile analysis.
Conclusion: Multivariable models suggest that mtDNA4977 DR levels are strongly associated with PCOS and represent an independent risk factor for PCOS. Further investigation of the utility of mtDNA as a biomarker for PCOS is warranted.
Polycystic ovary syndrome (PCOS), characterized by polycystic ovarian morphology, hyperandrogenism, and/or ovulatory dysfunction, is a common endocrine disorder worldwide. According to the 2003 Rotterdam criteria, the prevalence of PCOS ranges from 4 to 19.9% (1, 2). Women with PCOS are frequently accompanied by infertility, chronic inflammation, and metabolic dysfunction. Consequently, the risk of type 2 diabetes, cardiovascular events, and pregnancy complications (e.g., gestational diabetes, premature birth, and eclampsia) increases in individuals with PCOS (3–5). Over the last few years, increased oxidative stress (OS) has been shown to be involved in the pathophysiology of PCOS (6–8). Previous studies have shown that the levels of circulating markers of OS in women with PCOS were abnormal, independent of weight status (9, 10). In addition, OS has been closely associated with PCOS-related complications, such as insulin resistance, obesity, low-level inflammation, and hyperandrogenism (11–13).
Mitochondria, the powerhouses of cells, regulate cellular ATP production and reactive oxygen species. It has been universally acknowledged that mitochondria are critically involved in the regulation of OS. Mitochondrial DNA (mtDNA) is a circular double-stranded genome, encoding 13 essential proteins involved in oxidative phosphorylation, 2 ribosomal RNAs, and 22 transfer RNAs (14). In contrast to the nuclear genome, mtDNA is present in multiple copies (10–10,000) per cell depending on the cell type. The mtDNA copy number (CN) is a relative measure of mitochondrial content, and its variation has been reported to be associated with some chronic diseases (15). Recently, some studies have shown abnormal mtDNA CN in PCOS patients, suggesting that mtDNA CN variation may be involved in the pathophysiology of PCOS, although the results are conflicting (16–20). In addition to mtDNA CN, deletions in mtDNA are associated with diverse human pathologies (21). The mtDNA 4977 bp (mtDNA4977) deletion, which sacrifices approximately one‐third of mtDNA, is the most common deletion in the mitochondrial genome and has been considered as potential biomarker that can reflect the relative integrity and damage of mtDNA (22). Considering the relationship between mtDNA4977 deletion and OS (23, 24), we hypothesized that the mtDNA4977 deletion rate (DR) is related to PCOS and serves as another potential biomarker for PCOS.
To date, the association between the mtDNA4977 DR and PCOS is unknown and the association between the mtDNA CN and PCOS remains controversial. The purpose of this study was to examine the associations between two mtDNA biomarkers in peripheral blood, mtDNA CN and mtDNA4977 DR, and PCOS in a large clinical setting.
This study was conducted at the International Peace Maternal and Child Health Hospital (IPMCH) of Shanghai Jiao Tong University School of Medicine (Shanghai, China). The study population included patients who visited the Reproductive Medicine Centre in the hospital from June 2015 to June 2019. The cases represented approximately 90% of the patients with PCOS who first attended the clinic during the study period. PCOS was diagnosed according to the 2003 Rotterdam criteria, which requires the presence of at least two of the following conditions: (1) ultrasonographic polycystic ovarian morphology, (2) clinical or biochemical hyperandrogenism, (3) Oligo- and/or anovulation (25). Women with oligomenorrhea or hyperandrogenism of other causes, including congenital adrenal hyperplasia, 21-hydroxylase-deficient non-classic adrenal hyperplasia, hyperprolactinemia, Cushing’s syndrome, androgen-secreting tumors, or androgenic/anabolic drug use or abuse, were excluded from this study. During the same period, women who attended the clinic due to male infertility or tubal factor infertility were recruited as controls and matched by age (in 5 y bands) with cases. All the control participants had regular menstrual cycles, normal androgen levels, and no hirsutism, diabetes, galactorrhea, or any endocrine or systemic disease that could affect reproductive physiology. Clinical information, including age and body mass index (BMI), and biochemical parameters, including follicle-stimulating hormone (FSH), luteinizing hormone (LH), estradiol (E2), progesterone (P), and testosterone (T) levels, were recorded. Overall, 263 PCOS cases and 326 age-matched controls were enrolled in this study. Peripheral blood was collected when they first came to the Reproductive Medicine Centre, to avoid the effect of drugs or treatment on the biochemical parameters, mtDNA CN, or mtDNA4977 DR.
Peripheral blood samples of the participants were collected, processed, aliquoted, and frozen at −80°C according to standardized procedures. Total DNA was extracted using a DNeasy Blood & Tissue Kit (Qiagen, cat. # 69504), following the manufacturer’s instructions. DNA was quantified, and DNA integrity was examined.
The mtDNA CN and mtDNA4977 DR were measured by real-time quantitative polymerase chain reaction (qPCR) using a QuantStudio 7 Flex real-time PCR machine. A multiplex qPCR method, which has been validated by several scientific studies, was used to measure the mtDNA CN and mtDNA4977 DR. Briefly, two segments, one in the major arc (mtMajArc) and the other in the minor arc (mtMinArc) of the mitochondrial genome, were used to assess the mtDNA4977 DR and mtDNA CN, respectively. TaqMan assays of mtMajArc and mtMinArc are detailed in Supplementary Table S1. RNase P (Thermo Fisher, cat. # 4403326) was used as the genomic DNA reference. Real-time PCR was performed under the same cycling conditions as previously described (26). Three technical replicates were performed. The mtDNA CN and mtDNA4977 DR were calculated using the following formulae: mtDNA CN = 2△CT(mtDNA CN), where △CT(mtDNA CN) = CTRNase P − CTmtMinArc; mtDNA4977 DR (%) = 2△CT(mtDNA4977 DR) ×100, where △CT(mtDNA4977 DR) = CTmtMinArc − CTmtMajArc.
The study was designed to include all adults with PCOS and age-matched controls who fulfilled the inclusion criteria at a ratio of 1:1.2 during the study period; hence, no formal calculation of the sample size was performed. Data were shown as the median (interquartile range, IQR) for non-normal distributions or the mean ± standard deviation (SD) for normal distributions. The mtDNA biomarkers were transformed using logarithmic conversion. Comparisons of normally distributed variables between groups were performed using unpaired t test, whereas comparisons of non-normally distributed ones were conducted using the Mann–Whitney U test. When more than two groups were compared, the Kruskal–Wallis test, followed by Bonferroni’s post hoc test, was used. The mtDNA CN and mtDNA4977 DR were analyzed as continuous variables and quartiles, with the first quartile as the reference group. Binary logistic regression was performed to assess the associations of the mtDNA CN and mtDNA4977 DR with PCOS. Odds ratios (ORs) and 95% confidence intervals (CIs) were reported for each analysis. All statistical analyses were performed using R statistical software, version 3.6.2. A P-value of 0.05 or less (two-sided) was considered significantly different.
From June 2015 to June 2019, 263 women with PCOS and 326 controls were included in this study. Demographic characteristics and biochemical parameters are summarized in Table 1. Significant differences were found between PCOS cases and controls with respect to BMI, FSH, LH, LH/FSH, T, and P (P < 0.001 for all comparisons), but not E2 (P = 0.07).
The mtDNA CN and mtDNA4977 DR of two groups were compared using the Mann–Whitney U test and Student’s t test, respectively. As shown in Figure 1, the mean log-transformed mtDNA CN in women with PCOS was significantly higher than that in the controls (2.00 versus 1.95, P = 0.002). Additionally, the mtDNA4977 DR was significantly higher in the PCOS patients than that of controls (1.52 versus 1.48, P < 0.001).
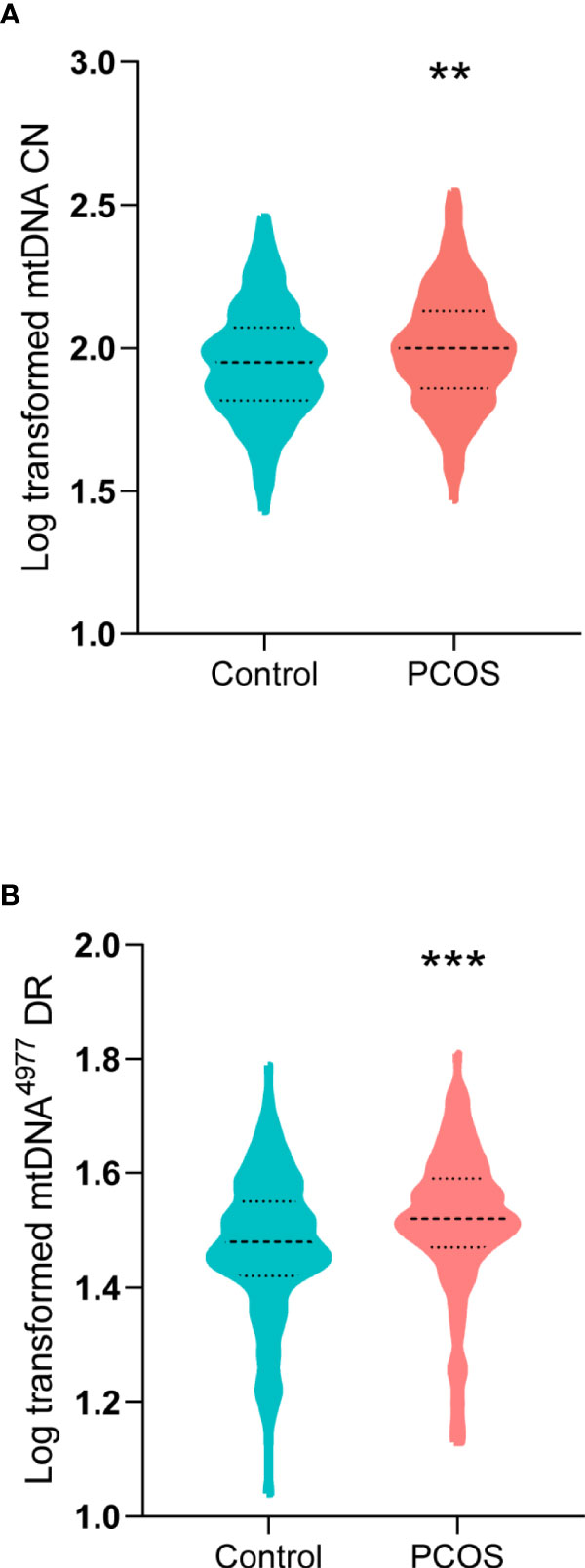
Figure 1 Mitochondrial DNA (mtDNA) copy number (CN) and mtDNA 4977 bp (mtDNA4977) deletion rate (DR) in women with PCOS and controls. (A) Log-transformed mtDNA CN in women with PCOS and controls (**P = 0.002). (B) Log-transformed mtDNA4977 DR in women with PCOS and controls (***P < 0.001).
Further, all subjects were divided into four groups according to the quartiles of the LH/FSH ratio. A significant difference in mtDNA CN as well as mtDNA4977 DR was observed among the four groups (Supplementary Table S2). After Bonferroni’s correction, a significant difference was confirmed between the lowest and highest quartiles (P = 0.011) for the mtDNA4977 DR. However, no differences were found among the four groups with respect to the mtDNA CN.
The associations between two potential mtDNA biomarkers and PCOS were assessed using different logistic regression models (Table 2). The mtDNA CN was significantly associated with PCOS. However, after adjustment for BMI, LH/FSH ratio, and T level, the mtDNA CN was not associated with PCOS (OR = 1.004, 95% CI: 0.999, 1.008) as a linear continuous variable. This was consistent with the quartile analysis results, which showed no statistically significant associations between the first quartile and higher quartiles (Figure 2).
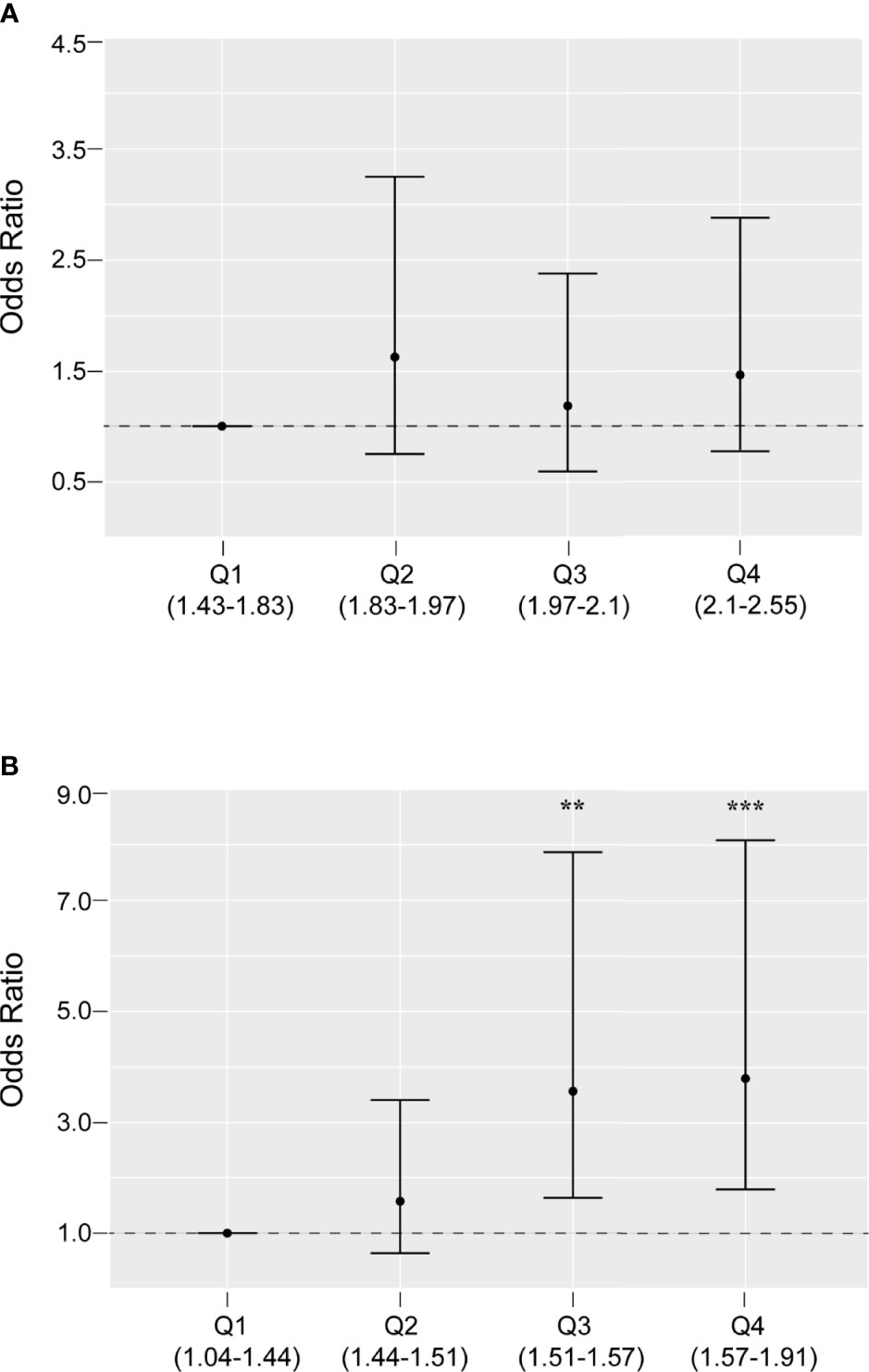
Figure 2 Odds ratios and 95% confidence intervals for PCOS by quartiles of mitochondrial DNA copy number (A) and 4977 bp deletion rate (B), adjusted for body mass index, luteinizing hormone/follicle-stimulating hormone ratio, and testosterone level. **P < 0.01, ***P < 0.001
Similarly, the mtDNA4977 DR was significantly associated with PCOS (P < 0.001). In addition, the linear dose-response trends were still apparent in the adjusted model. As shown in Table 2, the adjusted OR (95% CI) for PCOS by the mtDNA4977 DR was 1.053 (1.024, 1.083). This relationship was also supported by the quartile analysis results (Figure 2). Compared with the first quartile, the higher quartiles of the mtDNA4977 DR were associated with a higher risk of PCOS, especially the highest quartile (OR = 3.867, 95% CI: 1.847, 8.096). These results imply that the association between the mtDNA4977 DR and PCOS is independent of metabolic parameters.
In this study, the elevated mtDNA CN and mtDNA4977 DR in the peripheral blood of PCOS patients compared with those in control subjects were observed. However, after adjusting for confounding factors (BMI, LH/FSH ratio, and T level), only the mtDNA4977 DR was independently associated with PCOS.
The mtDNA4977 deletion, a large deletion of almost one-third of the length of mtDNA, is the most common large deletion in the mitochondrial genome (22). The deleted region contains five tRNA genes and seven genes encoding four complex I subunits, one complex IV subunit, and two complex V subunits. Recently, the mtDNA4977 deletion has been shown to be a pathogenic mutation in humans, as it can cause a failure in ATP production and mitochondrial dysfunction (22). Increased OS has been reported to trigger the accumulation of deletions in mtDNA and cause mitochondrial dysfunction (22, 27). Consistent with evidence linking OS with mitochondrial destruction (10, 27), our study indicated that the mtDNA4977 DR is a risk factor for PCOS independent of metabolic parameters. Considering the evidence showing that PCOS patients exhibit elevated OS and reduced antioxidant capacity (28, 29), it is plausible to hypothesize that the high level of OS in PCOS may be a potential trigger of mtDNA4977 deletion. When the deletion rate reaches a threshold, it causes mitochondrial dysfunction and is involved in the pathogenesis of PCOS. Additionally, it is important to note that the mtDNA4977 DR in women with the highest LH/FSH ratio increased significantly. Since an increased LH/FSH ratio has been widely accepted as a specific endocrine profile parameter for many PCOS patients (30), such results could help confirm that a high mtDNA4977 DR in peripheral blood is associated with PCOS and that the mtDNA4977 DR could serve as a convincing and promising biomarker.
Several studies have suggested that changes in the mtDNA CN are associated with the occurrence and development of PCOS (16, 17, 19). Lee et al. provided the first evidence of decreased peripheral mtDNA CN in PCOS patients (19). Recently, similar results were observed in two other studies, showing that the mtDNA CN in the peripheral blood of PCOS patients was significantly lower than that in healthy controls matched by age and BMI (16, 17). However, the result of our study revealed an elevated mtDNA CN in the peripheral blood of PCOS patients. Consistent with our study, Min et al. detected increased mtDNA CN and increased expression levels of mtDNA replication-related factors in PCOS-patient-derived induced pluripotent stem cells (31). This inconsistency may be attributed to participant preferences or the different sample size of the studied population. In addition, the extent of mtDNA CN alteration was found to be associated with the severity of PCOS (19), indicating that the composition of patients with different severities in different studies might also have influenced the association between the mtDNA CN and PCOS. Considering that this association is not independent of metabolic factors, we speculated that increased mtDNA CN is a compensatory mechanism for mitochondrial dysfunction caused by mtDNA damage, such as mtDNA4977 deletion. Further research is warranted to investigate the possible biological mechanisms of abnormal mtDNA CN in PCOS.
To our knowledge, this study is the first to evaluate the association between the mtDNA4977 DR and PCOS. The mtDNA4977 DR and mtDNA CN were measured simultaneously using an efficient and effective multiplex real-time PCR assay, making our results reliable (26, 32). Additionally, the sample size in this study was larger than that reported in previous similar studies. However, it should be noted that this study was a single-center study based on cross-sectional data in a clinical setting. Multicenter studies are required to validate these findings.
In summary, the present study demonstrates, for the first time, the contribution of the mtDNA4977 DR to PCOS, independent of metabolic parameters. Further study is warranted to elucidate the molecular mechanisms underlying this association.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by Ethics Committee of International Peace Maternal and Child Health Hospital (no. GKLW2018-12). The patients/participants provided their written informed consent to participate in this study.
All authors contributed to the article and approved the submitted version. The recruitment of participants and data collection were performed by SL, MY, and BH. The measurement of mtDNA biomarkers and data analysis were performed by BH, WS, and FG. The draft of the manuscript was written by MY and BH. The design of the study and writing review and editing were performed by CX and SL.
This work was supported by the National Natural Science Foundation of China (No. 81871136, 81501231, 81971344, and 81771638), the National Key Research and Development Program of China (No. 2016YFC0905103), the International Peace Maternity and Child Health Hospital Clinical Research Project (No. GFY5817 and GFY5818), the Shanghai Municipal Key Clinical Specialty and the Youth Science and Technology Innovation Studio, Shanghai Jiao Tong University School of Medicine.
Author BH was employed by company Shanghai WeHealth BioMedical Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the family members who participated in this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.675581/full#supplementary-material
1. Azziz R, Carmina E, Chen Z, Dunaif A, Laven JS, Legro RS, et al. Polycystic Ovary Syndrome. Nat Rev Dis Primers (2016) 2:16057. doi: 10.1038/nrdp.2016.57
2. Lizneva D, Suturina L, Walker W, Brakta S, Gavrilova-Jordan L, Azziz R. Criteria, Prevalence, and Phenotypes of Polycystic Ovary Syndrome. Fertil Steril (2016) 106:6–15. doi: 10.1016/j.fertnstert.2016.05.003
3. Rubin KH, Glintborg D, Nybo M, Abrahamsen B, Andersen M. Development and Risk Factors of Type 2 Diabetes in a Nationwide Population of Women With Polycystic Ovary Syndrome. J Clin Endocrinol Metab (2017) 102:3848–57. doi: 10.1210/jc.2017-01354
4. Torchen LC. Cardiometabolic Risk in PCOS: More Than a Reproductive Disorder. Curr Diabetes Rep (2017) 17:137. doi: 10.1007/s11892-017-0956-2
5. Duleba AJ, Dokras A. Is PCOS an Inflammatory Process? Fertil Steril (2012) 97:7–12. doi: 10.1016/j.fertnstert.2011.11.023
6. Ding Y, Xia B-H, Zhang C-J, Zhuo G-C. Mutations in Mitochondrial tRNA Genes May Be Related to Insulin Resistance in Women With Polycystic Ovary Syndrome. Am J Trans Res (2017) 9:2984–96.
7. Ding Y, Jiang Z, Xia B, Zhang L, Zhang C, Leng J. Mitochondria-Targeted Antioxidant Therapy for an Animal Model of PCOS-IR. Int J Mol Med (2019) 43:316–24. doi: 10.3892/ijmm.2018.3977
8. Liu J, Zhang D. The Role of Oxidative Stress in the Pathogenesis of Polycystic Ovary Syndrome. Sichuan Da Xue Xue Bao Yi Xue Ban (2012) 43:187–90. doi: 10.1007/s11783-011-0280-z
9. Murri M, Luque-Ramírez M, Insenser M, Ojeda-Ojeda M, Escobar-Morreale HF. Circulating Markers of Oxidative Stress and Polycystic Ovary Syndrome (PCOS): A Systematic Review and Meta-Analysis. Hum Reprod Update (2013) 19:268–88. doi: 10.1093/humupd/dms059
10. Chen L, Xu WM, Zhang D. Association of Abdominal Obesity, Insulin Resistance, and Oxidative Stress in Adipose Tissue in Women With Polycystic Ovary Syndrome. Fertil Steril (2014) 102:1167–74.e4. doi: 10.1016/j.fertnstert.2014.06.027
11. Zuo T, Zhu M, Xu W. Roles of Oxidative Stress in Polycystic Ovary Syndrome and Cancers. Oxid Med Cell Longev (2016) 2016:8589318. doi: 10.1155/2016/8589318
12. Savic-Radojevic A, Bozic Antic I, Coric V, Bjekic-Macut J, Radic T, Zarkovic M, et al. Effect of Hyperglycemia and Hyperinsulinemia on Glutathione Peroxidase Activity in Non-Obese Women With Polycystic Ovary Syndrome. Hormones (Athens Greece) (2015) 14:101–8. doi: 10.14310/horm.2002.1525
13. González F, Nair KS, Daniels JK, Basal E, Schimke JM. Hyperandrogenism Sensitizes Mononuclear Cells to Promote Glucose-Induced Inflammation in Lean Reproductive-Age Women. Am J Physiol Endocrinol Metab (2012) 302:E297–306. doi: 10.1152/ajpendo.00416.2011
14. Benkhalifa M, Ferreira YJ, Chahine H, Louanjli N, Miron P, Merviel P, et al. Mitochondria: Participation to Infertility as Source of Energy and Cause of Senescence. Int J Biochem Cell Biol (2014) 55:60–4. doi: 10.1016/j.biocel.2014.08.011
15. Castellani CA, Longchamps RJ, Sun J, Guallar E, Arking DE. Thinking Outside the Nucleus: Mitochondrial DNA Copy Number in Health and Disease. Mitochondrion (2020) 53:214–23. doi: 10.1016/j.mito.2020.06.004
16. Reddy TV, Govatati S, Deenadayal M, Sisinthy S, Bhanoori M. Impact of Mitochondrial DNA Copy Number and Displacement Loop Alterations on Polycystic Ovary Syndrome Risk in South Indian Women. Mitochondrion (2019) 44:35–40. doi: 10.1016/j.mito.2017.12.010
17. Saeed NA, Hamzah IH, Al-Gharrawi SA. Polycystic Ovary Syndrome Dependency on mtDNA Mutation; Copy Number and its Association With Insulin Resistance. BMC Res Notes (2019) 12:455. doi: 10.1186/s13104-019-4453-3
18. Ding Y, Xia B-H, Zhang C-J, Zhuo G-C. Mitochondrial tRNALeu(UUR) C3275T, tRNAGln T4363C and tRNALys A8343G Mutations may be Associated With PCOS and Metabolic Syndrome. Gene (2018) 642:299–306. doi: 10.1016/j.gene.2017.11.049
19. Lee S-H, Chung D-J, Lee H-S, Kim T-J, Kim M-H, Jeong HJ, et al. Mitochondrial DNA Copy Number in Peripheral Blood in Polycystic Ovary Syndrome. Metab: Clin Exp (2011) 60:1677–82. doi: 10.1016/j.metabol.2011.04.010
20. Ding Y, Zhuo G, Zhang C, Leng J. Point Mutation in Mitochondrial tRNA Gene is Associated With Polycystic Ovary Syndrome and Insulin Resistance. Mol Med Rep (2016) 13:3169–72. doi: 10.3892/mmr.2016.4916
21. Fontana GA, Gahlon HL. Mechanisms of Replication and Repair in Mitochondrial DNA Deletion Formation. Nucleic Acids Res (2020) 48:11244–58. doi: 10.1093/nar/gkaa804
22. Yusoff AA, Abdullah WS, Khair SZ, Radzak SM. A Comprehensive Overview of Mitochondrial DNA 4977-Bp Deletion in Cancer Studies. Oncol Rev (2019) 13:409. doi: 10.4081/oncol.2019.409
23. Jou M-J, Peng T-I, Wu H-Y, Wei Y-H. Enhanced Generation of Mitochondrial Reactive Oxygen Species in Cybrids Containing 4977-Bp Mitochondrial DNA Deletion. Ann N Y Acad Sci (2005) 1042:221–8. doi: 10.1196/annals.1338.024
24. Berneburg M, Gattermann N, Stege H, Grewe M, Vogelsang K, Ruzicka T, et al. Chronically Ultraviolet-Exposed Human Skin Shows a Higher Mutation Frequency of Mitochondrial DNA as Compared to Unexposed Skin and the Hematopoietic System. Photochem Photobiol (1997) 66:271–5. doi: 10.1111/j.1751-1097.1997.tb08654.x
25. Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 Consensus on Diagnostic Criteria and Long-Term Health Risks Related to Polycystic Ovary Syndrome. Fertil Steril (2004) 81:19–25. doi: 10.1016/j.fertnstert.2003.10.004
26. Wu H, Whitcomb BW, Huffman A, Brandon N, Labrie S, Tougias E, et al. Associations of Sperm Mitochondrial DNA Copy Number and Deletion Rate With Fertilization and Embryo Development in a Clinical Setting. Hum Reprod (Oxford England) (2019) 34:163–70. doi: 10.1093/humrep/dey330
27. Blajszczak C, Bonini MG. Mitochondria Targeting by Environmental Stressors: Implications for Redox Cellular Signaling. Toxicology (2017) 391:84–9. doi: 10.1016/j.tox.2017.07.013
28. Sulaiman MA, Al-Farsi YM, Al-Khaduri MM, Saleh J, Waly MI. Polycystic Ovarian Syndrome Is Linked to Increased Oxidative Stress in Omani Women. Int J Womens Health (2018) 10:763–71. doi: 10.2147/IJWH.S166461
29. Moti M, Amini L, Mirhoseini Ardakani SS, Kamalzadeh S, Masoomikarimi M, Jafarisani M. Oxidative Stress and Anti-Oxidant Defense System in Iranian Women With Polycystic Ovary Syndrome. Iran J Reprod Med (2015) 13:373–8.
30. Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 Consensus on Diagnostic Criteria and Long-Term Health Risks Related to Polycystic Ovary Syndrome (PCOS). Hum Reprod (Oxford England) (2004) 19:41–7. doi: 10.1093/humrep/deh098
31. Min Z, Gao Q, Zhen X, Fan Y, Tan T, Li R, et al. New Insights Into the Genic and Metabolic Characteristics of Induced Pluripotent Stem Cells From Polycystic Ovary Syndrome Women. Stem Cell Res Ther (2018) 9:210. doi: 10.1186/s13287-018-0950-x
Keywords: polycystic ovary syndrome, mitochondrial DNA deletion, mitochondrial DNA copy number, mitochondria, infertility
Citation: Ye M, Hu B, Shi W, Guo F, Xu C and Li S (2021) Mitochondrial DNA 4977 bp Deletion in Peripheral Blood Is Associated With Polycystic Ovary Syndrome. Front. Endocrinol. 12:675581. doi: 10.3389/fendo.2021.675581
Received: 03 March 2021; Accepted: 17 June 2021;
Published: 08 July 2021.
Edited by:
Ravinder Anand-Ivell, University of Nottingham, United KingdomReviewed by:
Li Li, Guangzhou Medical University, ChinaCopyright © 2021 Ye, Hu, Shi, Guo, Xu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuyuan Li, c2h1eXVhbmxpODE2QDE2My5jb20=; Chenming Xu, Y2hlbm1pbmdfeHUyMDA2QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.