- 1Bioorganic Research Institute, Suntory Foundation for Life Sciences, Kyoto, Japan
- 2Shimoda Marine Research Center, University of Tsukuba, Shimoda, Japan
Oxytocin (OT) and vasopressin (VP) superfamily neuropeptides are distributed in not only vertebrates but also diverse invertebrates. However, no VPergic innervation of invertebrates has ever been documented. In the ascidian, Ciona intestinalis Type A (Ciona robusta), an OT/VP superfamily peptide was identified, and the Ciona vasopressin (CiVP) induces oocyte maturation and ovulation. In the present study, we characterize the innervation and phenotypes of genetically modified Ciona: CiVP promoter-Venus transgenic and CiVP mutants. CiVP promoter-Venus transgenic Ciona demonstrated that CiVP gene was highly expressed in the cerebral ganglion and several nerves. Fluorescence was also detected in the ovary of young CiVP promoter-Venus transgenic ascidians, suggesting that the CiVP gene is also expressed temporarily in the ovary of young ascidians. Furthermore, a marked decrease of post-vitellogenic (stage III) follicles was observed in the ovary of CiVP mutants, whereas pre-vitellogenic (stage I) and vitellogenic (stage II) follicles were increased in the mutant ovary, compared with that of wildtype Ciona. Gene expression profiles showed that the expression of various genes, including genes related to ovarian follicle growth, was altered in the ovary of CiVP mutants. Altogether, these results indicated that CiVP, mainly as a neuropeptide, plays pivotal roles in diverse biological functions, including growth of early-stage ovarian follicles via regulation of the expression of a wide variety of genes. This is the first report describing a VP gene promoter-transgenic and VP gene-edited invertebrate and also on its gene expression profiles and phenotypes.
Introduction
Oxytocin (OT) and vasopressin (VP) superfamily peptides are one of the most widely distributed neuropeptides and/or neurohypophysial hormones in both vertebrates and invertebrates (1–10). These peptides are typically comprised of 9 to 14 amino acids, and share Cys1 and Cys6, which form an intramolecular disulfide bridge essential for biological activities. Moreover, the vertebrate OT family peptides share a neutral aliphatic amino acid (Leu, Ile, Val or Thr), whereas the vertebrate VP family peptides contain a basic one (Lys or Arg) at position 8. These superfamily peptides play pleiotropic roles in reproductive behaviors, osmoregulation, food intake, learning, and social behavior and are implicated with pathological processes such as anxiety and autism (1, 2, 11–13). Molecular phylogenetic trees demonstrated that the OT and VP superfamily have been diverged via duplication of a common ancestral gene during the process of agnathan to gnathostome evolution (1, 2, 4, 9–12).
OT/VP superfamily peptides have been identified in most invertebrates including nematodes, mollusks, annelids, insects, echinoderms, amphioxus, and ascidians (1–10, 14–16). Invertebrate OT/VP peptides also manifest some unique biological effects: induction of egg-laying behavior in earthworm (2, 17, 18); induction of unusual extra-oral feeding behavior in starfish (19); indirect induction of diuretic activity in beetles via stimulation of Malpighian tubules (5); modulation of gustatory associative learning in nematodes via effects of salt chemotaxis (8); modulation of parental-offspring social behavior via food-leaving in nematodes (20).
Recently, we identified unique biological roles of the VP/OT peptide of the cosmopolitan species of ascidians, Ciona intestinalis Type A (Ciona robusta). The Ciona VP peptide, CiVP (3) triggers oocyte maturation via germinal vesicle breakdown and ovulation (21). However, the CiVPergic neuroanatomy and developmental gene expression profiles of the CiVP gene have yet to be investigated. In this paper, we present the expression profiles of CiVP promoter-Venus-Ciona transgenic larvae, young adults, and adults, and comparative transcriptomes between the wildtype and the genome-edited CiVP-knockout Ciona.
Materials and Methods
Animals
Wildtype Ciona were cultivated and collected at Onagawa Bay, Maizuru Bay, Sagami Bay, Tosa Bay, and Mukaishima, Ascidians were cultured using an inland system described by Joly et al. (22).
PCR Primers
All PCR primers were ordered from Thermo Fisher Scientific Japan (Yokohama, Japan) or SIGMA ALDRICH JAPAN (Tokyo, Japan)
Plasmid Construction for Generation of Transgenic Ascidians
5’-untranslated region (UTR) sequence of CiVP gene was obtained from Ghost Database (http://ghost.zool.kyoto-u.ac.jp/blast_kh.html). Ciona genomic was prepared from sperms using a Wizard genomic DNA purification kit (Promega, Madison, WI, USA). An approximately 5-kb segment of CiVP gene 5’-UTR was amplified using the Ciona genomic DNA and gene-specific primers with a Sal I site (gattaGTCGACttaatgagaattcgcgctc, gattaGTCGACatgattgaaaactgtctgtacat), and a thermal cycler GeneAmp PCR system 9700 (Thermo Fisher Scientific, Waltham, MA, USA). The sequence of the 5-kb segment was shown in Supplemental Figure 1. The 5-kb segment was inserted into a Sal I site of pSP-Venus vector, followed by two steps of amplification of the segment encoding CiVP 5’-UTR and Venus coding region. The first PCR was performed using the pSP-CiVP 5’-UTR-Venus plasmid with specific primers containing partial sequences of attB (AAAAAGCAGGCTTAATGAGAATTCGCGCTCCTT, AGAAAGCTGGGTTTACTTGTACAGCTCGTCCATG). The PCR program was 98°C for 30 sec, 10 cycles of 94°C for 15 sec, 60°C for 30 sec, and final extension at 68°C for 12 min. The second PCR was performed with the first PCR products and attB adaptor primers (GGGGACAAGTTTGTACAAAAAAGCAGGCT, GGGGACCACTTTGTACAAGAAAGCTGGGT). The PCR program was 98°C for 30 sec, 5 cycles of 94°C for 15 sec, 45°C for 30 sec, and 68°C for 12 min, 20 cycles of 94°C for 15 sec, 60°C for 30 sec, and final extension at 68°C for 12 min. The amplified attB-PCR segment was inserted into pDONR221 vector by BP Clonase (Thermo Fisher Scientific). Furthermore, the CiVP 5’ UTR-Venus segment in the entry clone was recombined by LR Clonase (Thermo Fisher Scientific) into the destination vector, pMiDestF, which encodes Minos inverted repeats [(23), https://marinebio.nbrp.jp/ciona/]. Schematic depiction of the CiVP transgenic vector is shown in Supplemental Figure 2A.
Generation of Transgenic Ascidians
Transgenic ascidian was generated as described previously (24, 25). In brief, Minos transposase mRNA was synthesized using Megascript T3 kit, poly (A) tailing kit (Ambion, Carlsbad, CA) and Cap structure analog (New England Biolabs, Ipswich, MA). Both Minos mRNA and pMiDestF-CiVP 5’- UTR-Venus vector were electroporated into dechorionated and fertilized Ciona eggs (26). Subsequently, the electroporated eggs were cultured on agar-coated petri dishes with Millipore-filtered sea water. We raised the founder ascidian emitting Venus fluorescence, and its sperms were mated with wildtype ascidian eggs to obtain F1 progeny. Gametes of grown F1 progeny were then mated to generate transgenic F2 progeny.
RT-PCR Analysis of CiVP mRNA
Total RNA extracted from young adult ascidian ovaries was reverse-transcribed to the template cDNA using Superscript III (Thermo Fisher Scientific). RT-PCR was performed using CiVP gene specific primers (TGCTCTAACATGGATTGG, GCACTTGTTGTAAGACAC), GAPDH gene specific primers (GACGAATTGGACGCTT, GATGGTTGTGAAAACTCC), ExTaq polymerase (TAKARA BIO INC, Kyoto, Japan) and a thermal cycler (model GeneAmp PCR system 9700; Thermo Fisher Scientific). The PCR program was 94°C for 3 min, 35 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, and 72°C for 3 min. The PCR solutions were electrophoresed with 1.5% agarose gel.
Generation of CiVP-Deleted Mutants by TALEN
TALE Targeting sites of CiVP gene were determined using TAL Effector Nucleotide Targeter 2.0 (https://tale-nt.cac.cornell.edu/) (27). Left targeting site (TTTGGACGCGTGTTTT) contains sequences encoding the N-terminal Cys-Phe of CiVP (amino acid sequence: CFFRDCSNMDWYR), while Right targeting site (GCGGTACCAATCCATGT) corresponds to the sequence encoding C-terminal Asn-Met-Asp-Trp-Tyr-Arg in CiVP. Plasmids encoding TALE binding to these CiVP sequences were constructed onto the TALEN backbone vector containing the ubiquitous promoter from Ciona elongation factor (EF1α) using Platinum Gate TALEN kit [addgene, Cambridge, MA, USA; (28–30)]. NBRP web site (https://marinebio.nbrp.jp/ciona/) shows sequence of the backbone vector named as pBSCiEF1aSanTAL-NG-2A-mcherry-ver181108. Schematic depiction of the CiVP TALEN construction verifying activity is shown in Supplemental Figure 2B. The EF1a>CiVP TALEN vectors were electroporated into Ciona embryos. At the tailbud stage, the electroporated animals having mCherry expression in the whole body were collected, and genomic DNA was extracted in bulk. The DNA region containing the TALEN target site was PCR amplified, and the PCR fragments were electrophoresed in 15% acrylamide gel. The PCR bands exhibiting mutation signatures were subcloned into a conventional vector, and their sequences were determined.
The TALEN repeats were digested with SalI and BamHI, and were subcloned into pTnI>TALEN-NG::2A::mCherry with the In-fusion Cloning kit (TAKARA BIO INC). CiVP mutant line was generated by the electroporation-mediated germ cell regeneration method (31) using pTnI>CiVP-TALEN-L::2A::mCherry and pTnI>CiVP-TALEN-R::2A::mCherry. Schematic depiction of the CiVP knockout line generation is shown in Supplemental Figure 2C. Genomic DNA was extracted from the sperm of Ciona adults using a Wizard genomic DNA purification kit (Promega). The CiVP region was amplified using ExTaq (TAKARA BIO INC), CiVP gene-specific primers (AGATACAGACTGTATAGATTTC, AACCTGAAATGGTCTGTTATCC), and a thermal cycler (GeneAmp PCR system 9700) by the PCR program: 94°C for 3 min, 50 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec, and final extension at 72°C for 7 min. The PCR products were sequenced after subcloning in TOPO cloning vector (Thermo Fisher Scientific) and/or analyzed by heteroduplex mobility shift assay (32) to screen CiVP heterozygous mutants. Sperm of the heterozygous mutants were mated with wildtype ascidian eggs to generate the F1 progeny. Moreover, CiVP homozygous mutants were generated using gametes of F1 ascidians. The homogeneity of CiVP-TALEN mutants was confirmed by electrophoresis of PCR products amplified using the aforementioned CiVP gene-specific primers and Bsa I-digests. Sequencing analysis demonstrated that the CGGGACTGCTCT sequence in the CiVP-coding region of the genomic DNA was substituted with CGGTCTCT in the CiVP-TALEN mutants.
Microscopic Observation of Ciona Ovaries
Ciona ovaries were dissected from wild ascidians and mutant ascidians, and fixed in 4% paraformaldehyde/PBS at 4°C overnight. The fixed ovaries were dehydrated in ethanol and xylene, embedded in paraffin, cut into 10-μm thick sections, and were attached to Frontier coated glass slides (Matsunami Glass, Osaka, Japan). Paraffin was removed from the slides by xylene and ethanol. The deparaffinized sections were incubated in hematoxylin for 1 min, washed with water for 15 min, and incubated in eosin for 3 min. The stained sections were dehydrated in ethanol and xylene, and were observed under a microscope, ECLIPSE 600 (Nikon, Tokyo, Japan). The area of each ovary section was measured using Image J software and oocytes within the stained sections were counted to calculate oocyte occupancy.
RNA-seq of Ciona Ovaries
Total RNA was extracted from the ovaries of a CiVP homozygous mutant (CiVP mutant) and a wildtype ascidian using RNeasy mini kit (QIAGEN, Venlo, Netherlands). The quality of the RNA samples was evaluated using BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA) with an RNA6000 Pico Chip. A 0.2-μg aliquot of total RNA from each sample was used to construct cDNA libraries using TruSeq Stranded mRNA Sample preparation kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. The resulting cDNA library was validated using BioAnalyzer with a DNA1000 Chip and quantified using Cycleave PCR Quantification Kit (TAKARA BIO INC). Single end sequencing over 101 cycles was performed using HiSeq1500 (Illumina) in the rapid mode. Total reads were extracted with CASAVA v1.8.2 (Illumina). Then, PCR duplicates, adaptor sequences, and low-quality reads were removed from the extracted reads as follows. Briefly, if the first 10 bases of the two reads were identical and the entire reads exhibited > 90% similarity, the reads were defined as PCR duplicates. Remaining reads were then aligned with Bowtie version 2.2.3 to the C. intestinalis genes (KH, ver.2013), which was downloaded from Ghost Database (http://ghost.zool.kyoto-u.ac.jp/download_kh.html). We defined fragments per kilobase of transcript per million mapped reads (FPKM) value > 5 as expression genes in the ascidian ovary. Furthermore, we defined genes satisfying the condition of “FPKM value of CiVP mutant ovary/FPKM value of wildtype ovary” > 2 as genes upregulated in the ovary of CiVP mutant. Likewise, genes satisfying the condition of “FPKM value of wildtype ovary/FPKM value of CiVP mutant ovary” > 2 were defined as upregulated in the ovary of wildtype ascidian.
GO Analysis
Putative genes for the transcripts were annotated based on a homology search of the Swissprot database under the condition of e-values < 0.0001, using Blast2GO software (version 4.1) with default parameters (33). Moreover, these ascidian genes were mapped and annotated using Blast2GO software with default parameters. The putative genes were annotated to Gene Ontology (GO) terms (biological process, molecular function, and cellular component). Subsequently, the annotated biological information was compared using GO ‘is_a’ graphs as described previously (34, 35). The graphs were drawn with Cytoscape (http://www.cytoscape.org/). To quantify the enrichment of GO terms, we calculated enrichment scores as follows: where NX (GO) indicates the frequency of each GO term for CiVP mutant ovary (CiVP mutant << wildtype)- or wildtype ovary (wildtype << CiVP mutant)-specific genes (i.e., X), and Ntotal (X) indicates the frequency of Ci-VP mutant ovary (CiVP mutant << wildtype)- or wildtype ovary (wildtype << CiVP mutant)-specific genes (i.e., X) mapped to each GO term in the Blast2GO results. For collection, a pseudo-count was set as a fixed value.
Quantitative PCR
Total RNA was isolated from the ovary of wild ascidian and CiVP mutant. Each RNA was reverse-transcribed to the template cDNA at 50 C for 50 min. using the oligo (dT) anchor primer and Superscript III First Strand Synthesis Supermix (Thermo Fisher Scientific). Quantitative PCR was performed using Power SYBR Green PCR Master Mix (Bio-rad, Hercules, California, USA) and CFX96 Real-Time System (Bio-Rad) with the ΔΔCt method as previously reported (35). In brief, Ct values for Ci-GAPDH gene expression were used as standard values, and ΔΔCt values were calculated for each gene according to the manufacturer’s instruction. Moreover, relative expression values were calculated from ΔΔCt values. Primer sequences are shown in Supplemental Table 1.
Results and Discussion
Distribution of CiVPergic Neurons and Non-Neuronal Cells
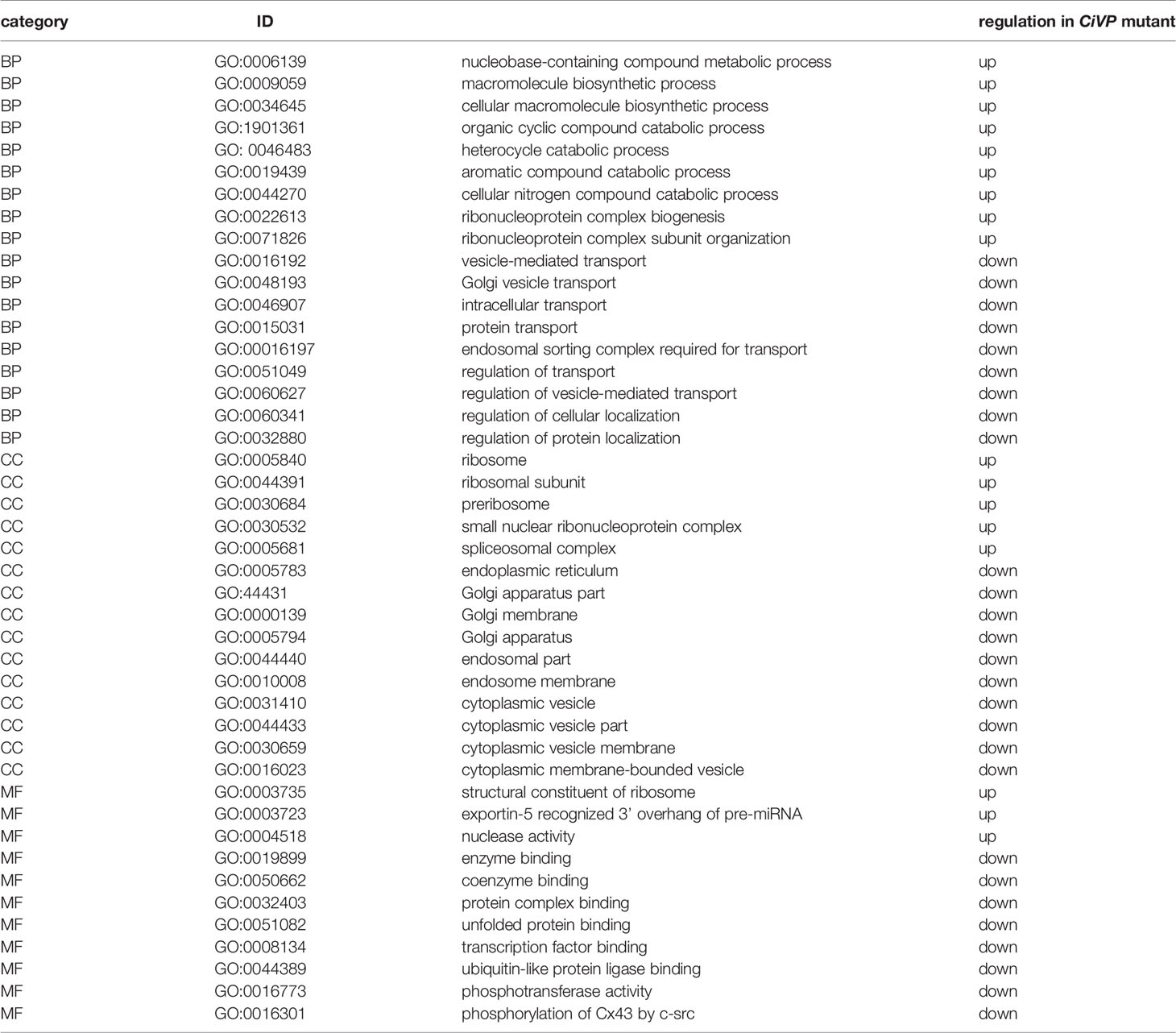
To examine CiVP gene expression in detail, we constructed transgenic ascidians introduced by the 5-kb CiVP promoter region-conjugated with the ORF of a fluorescent protein, Venus. Fluorescence was detected in the cerebral ganglion of CiVP promoter-Venus-transgenic larvae (Figure 1A). This fluorescent signal was in good agreement with the localization of the CiVP mRNA in wildtype Ciona larva but not in any earlier developmental stages as observed in our previous study (36), suggesting that the fluorescence derived from CiVP promoter-Venus construct indicates the expression of the CiVP gene in the transgenic Ciona.
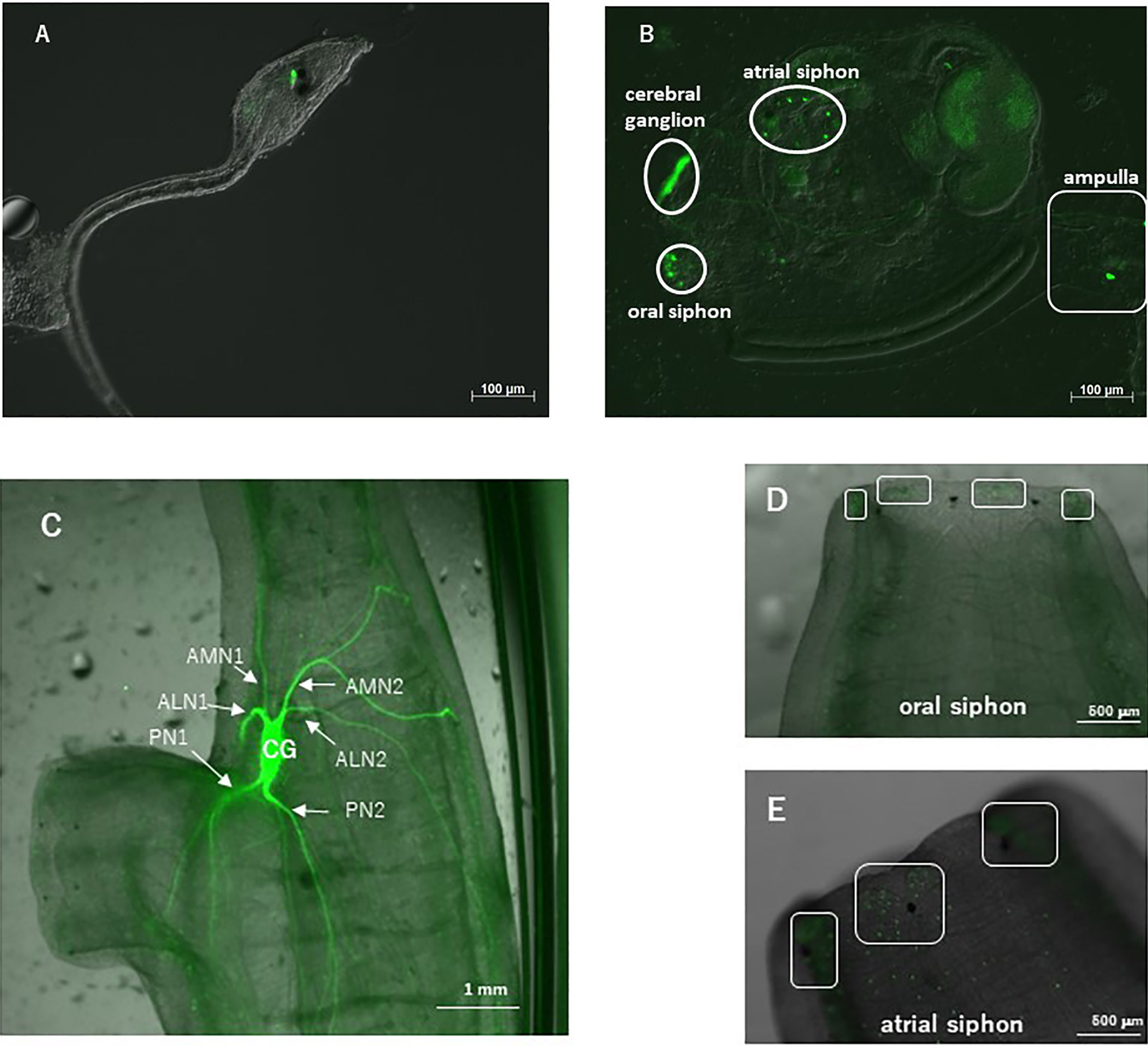
Figure 1 Observation of a CiVP promoter–Venus transgenic ascidian. (A) Swimming larva; (B) juvenile; (C) neural ganglion of adult; (D) oral siphon; and (E) atrial siphon. In (C), CG, cerebral ganglion; PN, posterior nerve; ALN, anterior lateral nerve; AMN, anterior medial nerve. In (D, E), boxed regions show numerous fluorescent cells.
Observation of the transgenic ascidians showed that Venus-derived fluorescence disappeared during the body rotation period, and gradually increased during the protostigmata period. Venus-derived fluorescence was detected not only in the cerebral ganglion with the highest intensity but also in the oral siphon, atrial siphon, and ampulla in the transgenic juveniles (Figure 1B). In the transgenic adults, intense Venus fluorescence was detected in the cerebral ganglion (Figure 1C), consistent with the previous study demonstrating that the CiVP gene was expressed exclusively in neurons in the cerebral ganglion of wild-type adult ascidians (3). Moreover, fluorescence was detected in several nerves including, posterior nerve 1, posterior nerve 2, anterior lateral nerve 1, anterior lateral nerve 2, anterior medial nerve 1, and anterior medial nerve 2. Particularly, the posterior nerve 2 connects to visceral nerves (37), suggesting that CiVP regulates functions of the stomach and intestine. In addition, weak fluorescence was detected in the oral siphon and atrial siphon of adult ascidians (Figures 1D, E). The anterior lateral and anterior medial nerves project to the oral siphon, and posterior nerves project to the atrial siphon (37), suggesting that CiVP regulates functions in the siphons. Fluorescence was also detected in the ovary of the transgenic young adult ascidian (Figure 2A). Since fluorescence is observed in some cells in the ovary, CiVP is likely to act as an endogenous factor in the ovary. Moreover, fluorescent nerve fibers projected to several organs including the stomach, intestines, and ovary (Figure 2A). These results suggested that CiVP originating from the neural tissue is involved in regulation of functions of the digestive tracts and ovary in the young adult ascidian. In our previous study, RT-PCR indicated that the CiVP gene was not expressed in the ovary of adult Ciona (3). To confirm the CiVP gene expression in the ovary of young adult Ciona, we performed RT-PCR for CiVP mRNA using three different wildtype young adult ascidian ovaries. RT-PCR for CiVP mRNA proved that the CiVP gene was expressed specifically in the ovaries of young adult ascidians (Figure 2B), although the elucidation of the biological significance of the gene expression awaits further study.
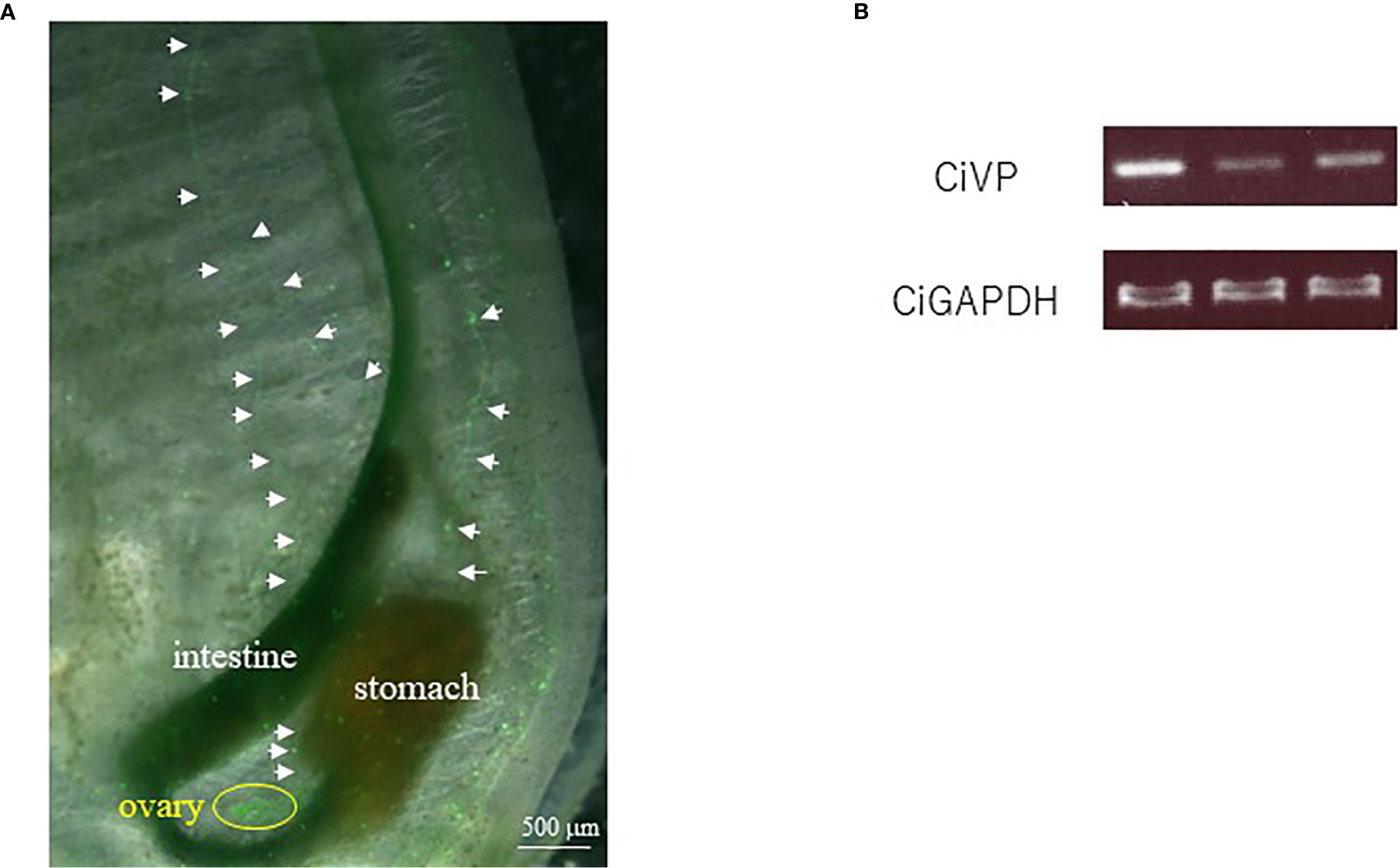
Figure 2 (A) Observation of the ovary of a CiVP promoter–Venus transgenic young adult ascidian. White arrows indicate fluorescent nerve fibers. (B) RT-PCR analysis of CiVP mRNA using three different wildtype young adult Ciona ovaries.
CiVP Mutants
To investigate biological functions of CiVP in vivo, we generated mutants of CiVP gene using the TALEN method. As a deletion site of the CiVP gene, we targeted the genome sequence encoding the N-terminal region of CiVP, since VP/OT family peptides including CiVP conserve Cys1 and Cys6 that form a disulfide bridge, and are responsible for activities of the family peptides (3, 7, 21). Thus, deletion of either of the cysteines is expected to leads to a loss of CiVP functions.
We selected the heterozygous mutants in which the CiVP gene has lost four nucleotides corresponding to Asp5 and Cys6 (Figure 3). CiVP mutant sperms were mated with wildtype ascidian eggs to generate F1 progeniy containing CiVP heterozygous mutation. F2 progeny harboring CiVP homozygous mutants were generated using gametes of the CiVP heterozygous mutants. The homozygous mutants were designated CiVP mutants. At the larval stage, CiVP homozygous mutants are indistinguishable from wildtype siblings, and the mutants metamorphosed to juveniles with normal morphology. Likewise, CiVP mutants exhibited growth similar to wildtype ascidians until reaching 2-cm body length. In contrast, growth of CiVP mutants gradually delayed compared with that of wildtype ascidians after 2-cm body length, and the growth of CiVP mutants stopped earlier than that of wildtype ascidians. As shown in Figure 4, the average body length of CiVP mutants and wildtype ascidians are approximately 3.5 cm and 6.2 cm, respectively. These results suggest that CiVP is involved in growth after ascidians reach a length of 2 cm. This is the first demonstration of a negative effect of deletion of an OT/VP gene deletion on total body growth of an animal.

Figure 3 CiVP amino acid sequence and CiVP gene sequence. Upper and lower sequences are for the wildtype ascidian and CiVP mutant, respectively. Cysteine residues that form a disulfide bridge are indicated in bold font. Hyphens in the nucleotide sequence of the CiVP mutant indicate nucleic acids deleted from the CiVP gene sequence.
Figure 4 Comparison of body length of wildtype ascidians and CiVP mutants (n = 4). WT and CiVP mut indicate wildtype ascidians and CiVP mutants, respectively. Data represent the mean ± S.E.M. *P < 0.05, compared with the length of wildtype ascidians.
Previously, the cognate CiVP receptor gene was shown to be expressed in the ovary (3). Furthermore, we substantiated that CiVP regulates oocyte maturation and ovulation (21). To examine the effect of CiVP on the whole ovary, we observed the ovaries of CiVP mutant and wildtype Ciona at the reproductive stage. Ciona follicles are classified into four growth stages (stage I – IV) (38). Pre-vitellogenic follicles, vitellogenic follicles, and post-vitellogenic follicles are classified into stages I, II, and III, respectively. Size of stage I, II, and III follicles are approximately 50, 70, and 100 μm in diameter, respectively. Stage I - III follicles are infertile, while stage IV follicles are fertile and ovulated (21, 38). We counted all hematoxylin/eosin-stained follicles in ovarian sections of CiVP mutant and wildtype ascidians. A total of 476 follicles were present in the section of CiVP mutant ovary, while 354 follicles were present in the section of wildtype Ciona ovary. In contrast, only 7 post-vitellogenic (stage III) follicles were detected in the section of CiVP mutant ovary, although 35 stage III follicles were observed in the section of wildtype Ciona ovary. In addition, we calculated the number of stage III follicles in a given area (mm2) of ovarian section of CiVP mutant and wildtype ascidians, revealing that the occupancy of stage III follicles in the mutant ovary section was 1.63 oocytes/mm2, which was markedly lower than 6.34 follicles/mm2 in the wildtype ovary section (Figure 5 and Table 1). Moreover, stage III follicles constituted 1.4% of all follicles in the CiVP mutant ovary, and 9.9% of all follicles in the section of the wildtype ascidian ovary (Figure 5 and Table 1). Since pre-vitellogenic follicles (stage I) and vitellogenic follicles (stage II) are more abundant in the CiVP mutant ovary, the number of all follicles per mm2 in the mutant section was approximately 1.7-fold greater than that of all follicles per mm2 in the wildtype ovarian section. Oocyte maturation is induced by CiVP in stage III follicles, leading to ovulation of fertile follicles (stage IV) (21, 38). Combined with these findings, the present results are consistent with the biological roles of CiVP, and also suggests that CiVP is involved in the stage I to III follicle growth. In other words, in the CiVP mutants, suppression of growth of stage II to -III may lead to the accumulation of stage I and stage II follicles, given that they are much smaller than stage III follicles, and thus a greater number of stage I and II follicles can occupy the ovary, compared with stage III follicles. This view is supported by the present results demonstrating that the CiVP mutant ovary harbors more follicles but fewer stage III follicles than the wildtype ovary (Figure 5 and Table 1).
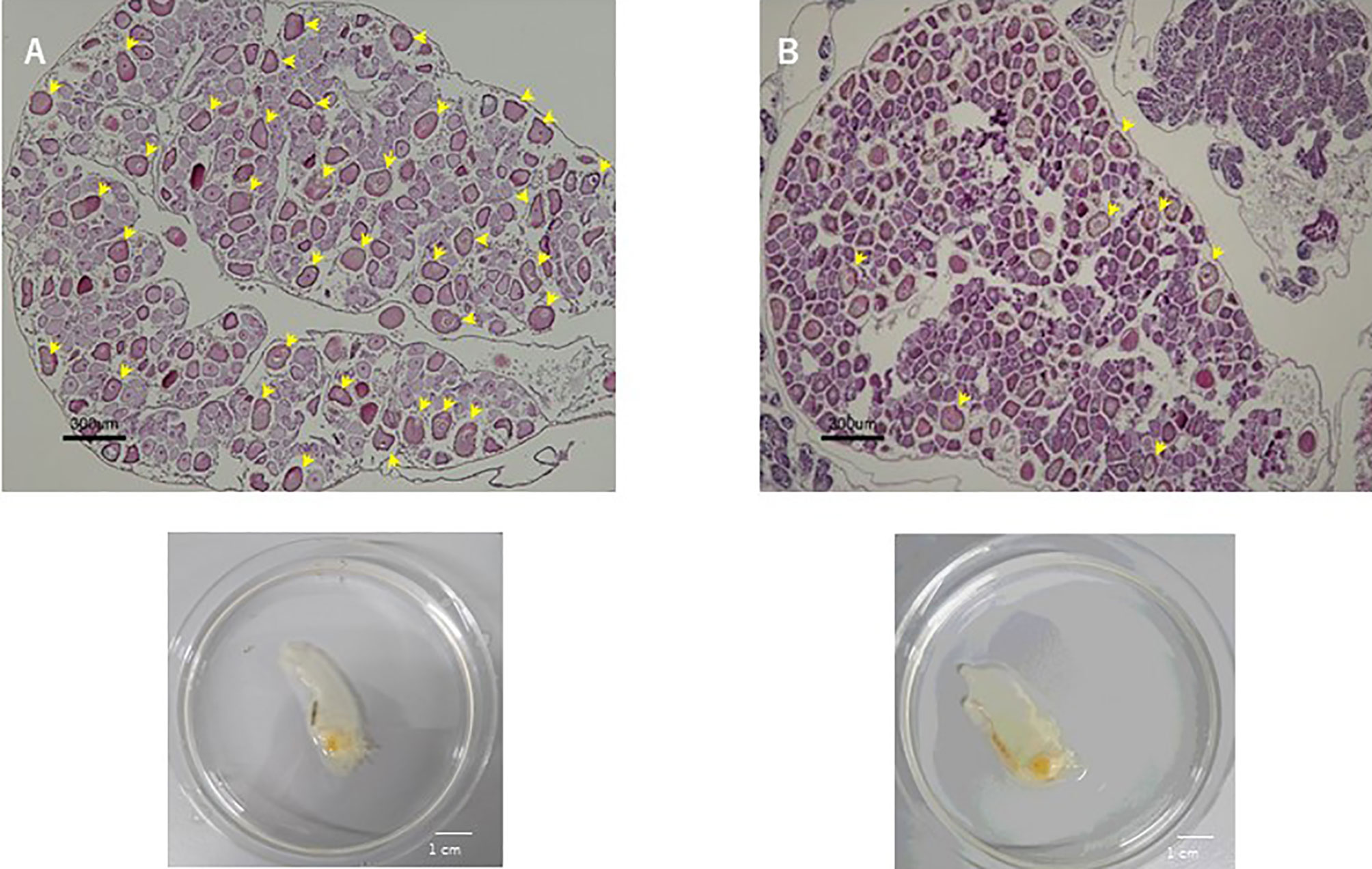
Figure 5 Hematoxylin/eosin staining of ovary sections of wildtype ascidian (A) and CiVP mutant (B). Arrows indicate stage III follicles.
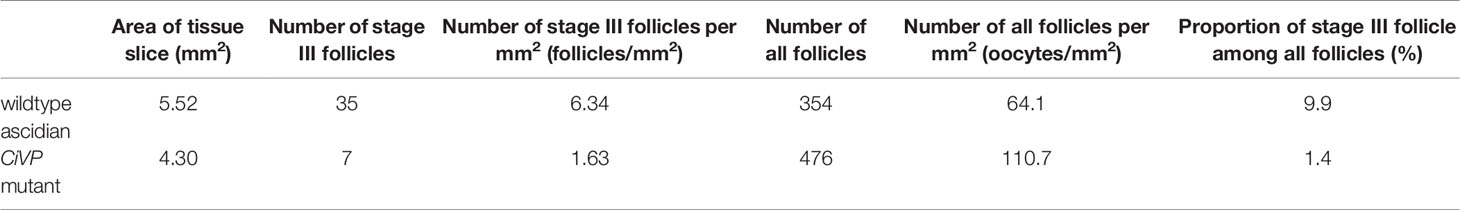
Table 1 The number and ratio of stage III follicles in a section of the ovaries of wildtype Ciona and CiVP mutant (n=2).
OT/VP gene- or their receptor gene-knockout mice exhibited no obvious deficiencies in fertility, follicle maturation, or reproductive behavior, although OT gene-knockout mice cannot release milk and OT receptor gene-knockout mice fail to lactate and exhibited a decrease in maternal behaviors [(39, 40), and Mouse Genome Informatics; http://www.informatics.jax.org]. These findings confirm that follicles normally grow in VP/OT signaling-deficient mutants. Likewise, analyses of nematodes, C. elegans indicated that the null mutants of the OT/VP superfamily peptide gene are as viable and fertile as wildtype nematodes, and exhibited normal egg-laying behavior and number of progeny but male nematodes exhibited low success in mating behavior (14). In addition, injection of the cognate OT/VP peptides resulted in induction of egg-laying behavior of annelids (2, 7, 18, 19). In the present study, stage III follicles were reduced in the ovary of CiVP mutants (Figure 5 and Table 1), demonstrating that CiVP is responsible for follicle growth in addition to oocyte maturation and ovulation (21). In combination, the present results suggest species-specific biological roles of OT/VP superfamily peptides in reproductive functions such as follicle growth. Investigation of the underlying molecular mechanisms is underway.
GO Analysis of the Ovary of CiVP Mutants
RNA-seq for ovaries of the CiVP mutant and wildtype ascidian using Hiseq 1500 yielded 40 million and 44 million reads, respectively for 101 paired-end reads. The resultant fastq files were deposited in SRA database (accession no. SRR12791386 and SRR12791387). Reads originating from each ovary were mapped against the C. intestinalis gene database (http://ghost.zool.kyoto-u.ac.jp/cgi-bin/gb2/gbrowse/kh/) to a total of 53,083 and 52,637 genes, respectively. The gene expression levels were assessed based on FPKM values were used. A total of 18,377 and 21,933 non-overlapping genes exhibiting FPKM values >5 were eventually identified in the CiVP mutant and wildtype ovaries, respectively. Figure 6 shows a scatter plot of FPKM values of these genes.
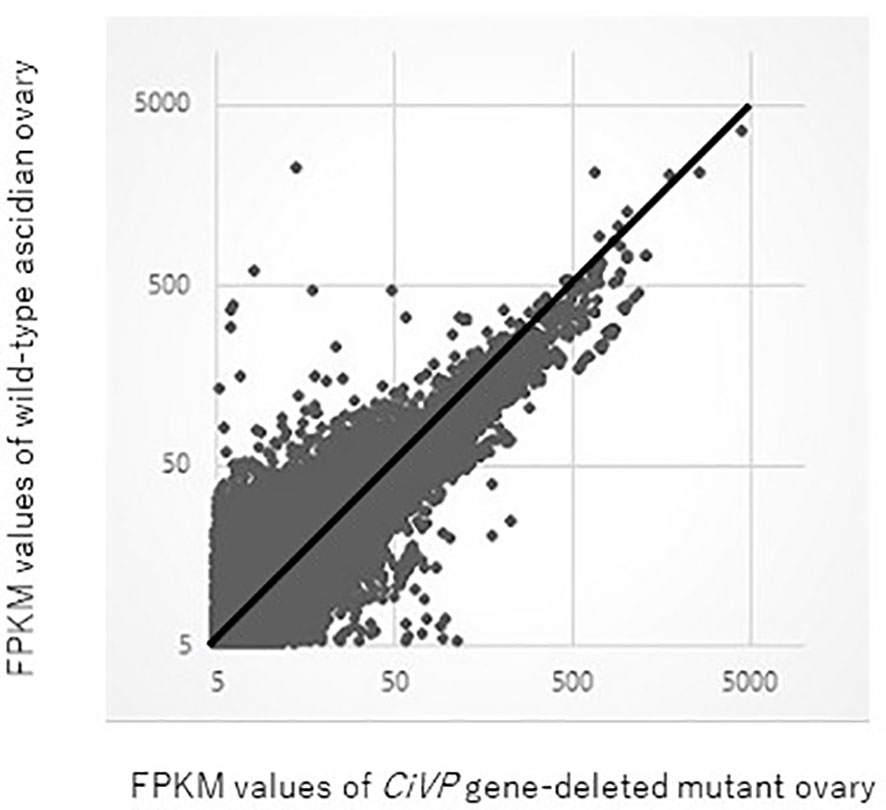
Figure 6 Comparative scatter plot analysis of gene expression levels in the ovary of CiVP mutant and wildtype Ciona. Horizontal axis shows FPKM values of the CiVP mutant, whereas the vertical axis shows FPKM values for wildtype Ciona.
GO analysis using Blast 2 GO indicated that frequent GO terms were similar between the CiVP mutant and wildtype ovaries. Subsequently, we calculated the ratio of the FPKM values for CiVP mutants to the FPKM values for wildtype ascidians to detect genes upregulated in the premature or mature ovaries, indicating that 1,976 genes and 5,644 genes were upregulated in the CiVP mutant and wildtype ovaries with FPKM CiVP mutant value/FPKM wildtype ratios of > 2 or < 0.5, respectively. Homology searching of these genes using NCBI blastp against the Swissprot database under the condition of e-value <0.0001 indicated that 86.2% (n = 1,704) and 96.8% (n = 5,463) of genes upregulated in the ovary of CiVP mutants and wildtype ascidians, respectively, were homologous to known proteins. Furthermore, GO data enrichment scores for these genes were calculated to examine the features of the ovary of the CiVP mutants. Annotated biological information was compared between CiVP mutant and wildtype ovaries using a heat map in graph view (Figure 7). Red indicates high enrichment score for the CiVP mutant, and blue shows low enrichment score. For simple interpretation, a GO term with a GO level <2 or a GO term with an enrichment score between −1.2 and 1.2 would be represented as a zero-sized node. Figure 7 and Table 2 show data for three enrichment analyses for the representative GO categories biological process, cellular component, and molecular function. Enrichment analysis of biological process indicates that genes related to biosynthetic processes were upregulated in the ovary of CiVP mutants (Figure 7A and Table 2). Furthermore, genes related to ribonucleoprotein were also upregulated in the mutant ovary (Figure 7A and Table 2). On the other hand, genes related to transport and localization were downregulated (Figure 7A and Table 2). Enrichment analyses of cell components showed that genes related to ribosome were upregulated in the CiVP mutant ovary (Figure 7B and Table 2). In contrast, enrichment analyses of cell components indicated that genes related to various organelles were downregulated in the ovary of a CiVP mutant (Figure 7B and Table 2). Expression profiles of genes related to the biological process and cellular component categories suggested that formation of organelles in ovary cells is suppressed by deletion of CiVP, whereas the synthesis of biomolecules appears to be activated (Table 2), ultimately disrupting cellular homeostasis or normal cell division. Enrichment analysis of the molecular function category indicated that genes related to binding and phosphorylation were downregulated in the ovary of CiVP mutants, whereas genes related to the nucleus or ribosomes were upregulated (Figure 7C and Table 2). The expression of genes related to nucleotide and protein synthesis was upregulated in the ovary of CiVP mutants, whereas the expression of genes related to organelles was downregulated (Figure 7C and Table 2). Thus, formation and activation of organelles are likely to be suppressed in the ovary of CiVP mutants, although nucleotides and proteins are synthesized. In Ciona ovarian follicles, accessory cells surrounding the oocytes, such as test cells, are produced from stage I to II, and are actively proliferated from stage II to III (38). Particularly, the number of accessory cells markedly increases during follicle growth from stage II to III (38). Consequently, it is presumed that formation and activation of organelles are upregulated according to the proliferation of accessory cells in the ovary of wildtype ascidians, whereas genes related to organelles are downregulated in the ovary of CiVP mutants. These findings are compatible with the upregulated expression of genes related to nuclei and ribosome in the ovary of CiVP mutant (Figure 7 and Table 2).
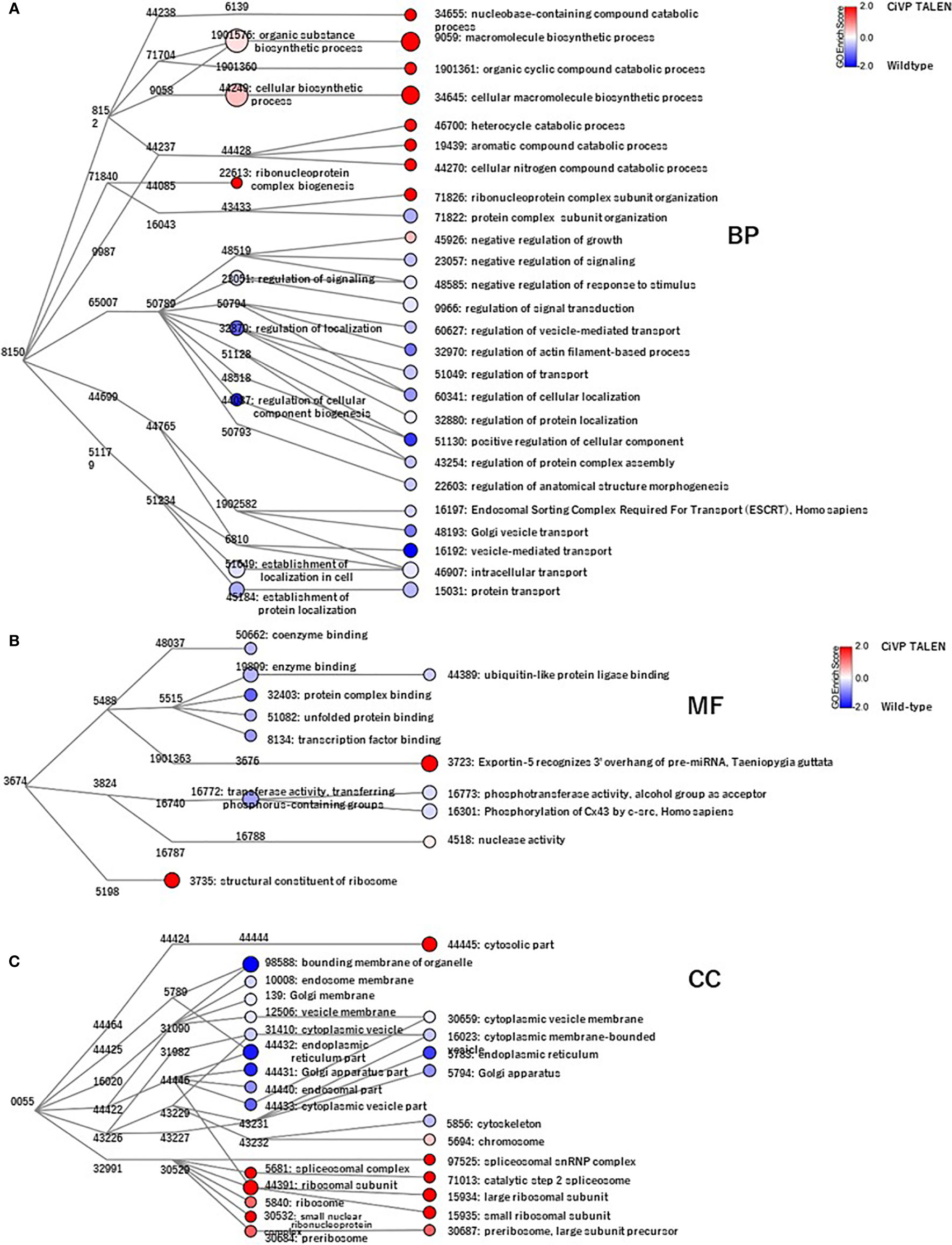
Figure 7 Transcriptomic analyses of CiVP mutant and wildtype Ciona ovaries. GO enrichment analysis of 343 differentially expressed genes in premature and mature ovaries. GO terms were categorized as 344 “(A), Biological Process” (BP), “(B), Molecular Function” (MF), and “(C), Cell Component” (CC). Each node represents a GO term. Branches of GO hierarchical trees not exhibiting significantly enriched GO terms are not shown. The size of each node circle indicates the number of genes with that GO term. The color of each node indicates the enrichment score. Red and blue indicate high enrichment scores of genes expressed in the ovary of CiVP mutant and wildtype ascidians, respectively. Edges indicate ‘is_a’ connections between GO terms.
Quantitative PCR for genes detected by RNA-seq and GO analysis between wildtype and CiVP mutant revealed 1.5 to 2-fold differential expression of various genes. For example, ceramide synthase homologous gene showed 1.5-fold lower expression in the ovary of CiVP mutant than that of wildtype ascidian (Figure 8). Ceramide is known to participate in the removal of low-quality early follicles as a lipid mediator in mammals (41, 42). As shown in Figure 5, the ovary of CiVP mutant possesses far fewer stage III (post-vitellogenic) follicles than that of wildtype, demonstrating that normal growth from stage II to III is inhibited by occupation of early follicles that cannot grow. Altogether, the present study suggests that CiVP is involved in quality control of follicle maturation via upregulation of ceramide synthase gene expression.
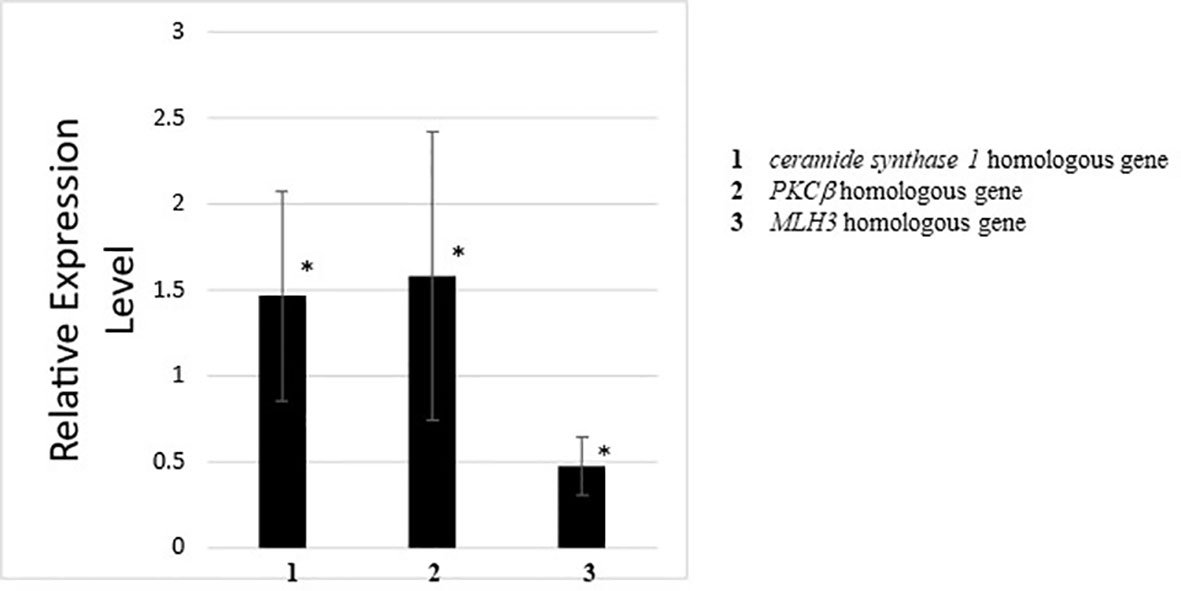
Figure 8 Quantitative PCR analysis of the expression of ceramide synthase 1 homologous gene, PKCβ homologous gene, and MLH3 homologous gene in the ovary of wildtype ascidians versus CiVP knockout ascidians. The relative expression of each gene regulated in ovaries treated with CiVP was calculated using ΔΔCt values. Data represent the mean ± S.E.M. for three independent experiments. *P < 0.05, compared with the relative expression value for the wildtype ascidian ovary.
Expression of the PKCβ homologous gene was downregulated 1.6-fold in the ovary of CiVP mutants versus wildtype ascidians (Figure 8). PKCβ has multi-functions including construction of organelle in mammals. For example, PKCβ-knockout rat mast cells exhibited reduced vesicle fusion (43), indicating that PKCβ is an important factor for vesicle transport during the secretion of signal molecules as well as organelle formation. This function of PKCβ coincides with the result of GO analysis demonstrating that genes related to organelles were downregulated in the ovary of CiVP mutants (Figure 7 and Table 2). PKCβ thus appears to be related to impaired follicle growth in Ciona at least in part via a disruption of vesicle transport, although the investigation of the detailed molecular mechanisms awaits further study.
Expression of the MLH3 gene was upregulated 2.1-fold in the ovary of CiVP mutants versus wildtype ascidians (Figure 8). MLH3 is known to repair DNA mismatch during DNA replication and meiosis (44). It is noteworthy that ascidian meiotic competence is acquired in stage III follicles (45) which are far fewer in the ovary of CiVP mutants (Figure 5). In combination, these results suggest that MLH3 plays a major role in DNA replication in the Ciona ovary. Furthermore, as stated above, Ciona follicle accessory cells—such as test cells—are actively proliferated at stage I and II (38). Consequently, the present study, combined with these findings, supports the view that higher expression of MLH3 is functionally implicated with more abundant immature follicles (stage I and II) in the ovary of CiVP mutants, compared with those of the wildtype. Taken together, these gene expression profiles suggest that CiVP plays a variety of roles in the control of the growth of early-stage follicles via regulation of diverse genes, although verification of the precise molecular mechanisms awaits further study.
In conclusion, we have presented the CiVPergic innervation and phenotypes and gene expression profiles of CiVP mutant Ciona. This is the first demonstration of both neuropeptide promoter-transgenic and neuropeptide gene-deleted Ciona adults, in particular, insufficient growth of the whole body and ovarian follicles in Ciona adults. Consequently, the present study will pave the way for investigations of not only various biological roles of CiVP but also the functional evolution of VP/OT family peptides.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
TK and HS conceived and designed the study. AH, TH, and YS constructed the transgenic and gene-deletion ascidians. AS, SM, and TK conducted RNA-seq experiments and analyzed the data. TK performed quantitative PCR analyses. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the Japan Society for the Promotion of Science to TK (JP23570100, JP26440172).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the National BioResource Project (Ciona intestinalis) for cultivating and providing wildtype, transgenic, and gene-edited ascidians.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.668564/full#supplementary-material
References
1. Hoyle CH. Neuropeptide families: evolutionary perspectives. Regul Pept (1998) 73:1–33. doi: 10.1016/s0006-8993(99)01975-7
2. Satake H, Takuwa K, Minakata H, Matsushima O. Evidence for Conservation of the vasopressin/oxytocin Superfamily in Annelida. J Biol Chem (1999) 274:5605–11. doi: 10.1074/jbc.274.9.5605
3. Kawada T, Sekiguchi T, Itoh Y, Ogasawara M, Satake H. Characterization of a novel vasopressin/oxytocin superfamily peptide and its receptor from an ascidian, Ciona intestinalis. Peptides (2008) 29:1672–8. doi: 10.1016/j.peptides.2008.05.030
4. Stafflinger E, Hansen KK, Hauser F, Schneider M, Cazzamali G, Williamson M, et al. Cloning and identification of an oxytocin/vasopressin-like receptor and its ligand from insects. Proc Natl Acad Sci USA (2008) 105:3262–7. doi: 10.1073/pnas.0710897105
5. Aikins MJ, Schooley DA, Begum K, Detheux M, Beeman RW, Park Y. Vasopressin-like peptide and its receptor function in an indirect diuretic signaling pathway in the red flour beetle. Insect Biochem Mol Biol (2008) 38:740–8. doi: 10.1016/j.ibmb.2008.04.006
6. Elphick MR, Rowe ML. NGFFFamide and echinotocin: structurally unrelated myoactive neuropeptides derived from neurophysin-containing precursors in sea urchins. J Exp Biol (2009) 212:1067–77. doi: 10.1242/jeb.027599
7. Kawada T, Sekiguchi T, Sugase K, Satake H. Evolutionary aspects of molecular forms and biological functions of oxytocin family peptides. In: Jastrow H, Feuerbach D, editors. Handbook of Oxytocin Research: Synthesis, Storage and Release, Actions and Drug Forms. New York, USA: Nova Science Publishers Inc (2009), ISBN: 978-1-60876-023-7p. 59–85.
8. Beets I, Janssen T, Meelkop E, Temmerman L, Suetens N, Rademakers S, et al. Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science (2012) 338:543–5. doi: 10.1126/science.1226860
9. Satake H, Matsubara S, Shiraishi A, Yamamoto T, Osugi T, Sakai T, et al. Neuropeptides, Peptide Hormones, and Their Receptors of a Tunicate, Ciona Intestinalis. Results Probl Cell Differ (2019) 68:107–25. doi: 10.1007/978-3-030-23459-1_5
10. Odekunle EA, Elphick MR. Comparative and Evolutionary Physiology of Vasopressin/ Oxytocin-Type Neuropeptide Signaling in Invertebrates. Front Endocrinol (Lausanne) (2020) 11:225. doi: 10.3389/fendo.2020.00225
11. Gimpl G, Fahrenholz F. The Oxytocin Receptor System: Structure, Function, and Regulation. Physiol Rev (2001) 81:629–83. doi: 10.1152/physrev.2001.81.2.629
12. Frank E, Landgraf R. The vasopressin system–from antidiuresis to psychopathology. Eur J Pharmacol (2008) 583:226–42. doi: 10.1016/j.ejphar.2007.11.063
13. Stoop R. Neuromodulation by Oxytocin and Vasopressin. Neuron (2012) 76:142–59. doi: 10.1016/j.neuron.2012.09.025
14. Garrison JL, Macosko EZ, Bernstein S, Pokala N, Albrecht DR, Bargmann CI. Oxytocin/vasopressin-related peptides have an ancient role in reproductive behavior. Science (2012) 338:540–3. doi: 10.1126/science.1226201
15. van Kesteren RE, Smit AB, Dirks RW, With ND, Geraerts WP, Joosse J. Evolution of the vasopressin/oxytocin superfamily: characterization of a cDNA encoding a vasopressin-related precursor, preproconopressin, from the mollusc Lymnaea stagnalis. Proc Natl Acad Sci USA (1992) 89:4593–7. doi: 10.1073/pnas.89.10.4593
16. Oumi T, Ukena K, Matsushima O, Ikeda T, Fujita T, Minakata H, et al. Annetocin: an oxytocin-related peptide isolated from the earthworm, Eisenia foetida. Biochem Biophys Res Commun (1994) 198:393–9. doi: 10.1006/bbrc.1994.1055
17. Ukena K, Iwakoshi-Ukena E, Hikosaka A. Unique form and osmoregulatory function of a neurohypophysial hormone in a urochordate. Endocrinology (2008) 149:5254–61. doi: 10.1210/en.2008-0607
18. Kawada T, Kanda A, Minakata H, Matsushima O, Satake H. Identification of a novel receptor for an invertebrate oxytocin/vasopressin superfamily peptide: molecular and functional evolution of the oxytocin/vasopressin superfamily. Biochem J (2004) 382:231–7. doi: 10.1042/BJ20040555
19. Odekunle EA, Semmens DC, Martynyuk N, Tinoco AB, Garewal AK, Patel RR, et al. Ancient role of vasopressin/oxytocin-type neuropeptides as regulators of feeding revealed in an echinoderm. BMC Biol (2019) 17:60. doi: 10.1186/s12915-019-0680-2
20. Scott E, Hudson A, Feist E, Calahorro F, Dillon J, de Freitas R, et al. An oxytocin-dependent social interaction between larvae and adult C. elegans. Sci Rep (2017) 7:10122. doi: 10.1038/s41598-017-09350-7
21. Matsubara S, Shiraishi A, Osugi T, Kawada T, Satake H. The regulation of oocyte maturation and ovulation in the closest sister group of vertebrates. elife (2019) 8:e49062. doi: 10.7554/eLife.49062
22. Joly JS, Kano S, Matsuoka T, Auger H, Hirayama K, Satoh N, et al. Culture of Ciona intestinalis in closed systems. Dev Dyn (2007) 236:1832–40. doi: 10.1002/dvdy.21124
23. Sasakura Y, Suzuki M, Hozumi A, Inaba K, Satoh N. Maternal factor-mediated epigenetic gene silencing in the ascidian Ciona intestinalis. Mol Genet Genomics (2010) 283:99–110. doi: 10.1007/s00438-009-0500-4
24. Sasakura Y, Awazu S, Chiba S, Satoh N. Germ-line transgenesis of the Tc1/mariner superfamily transposon Minos in Ciona intestinalis. Proc Natl Acad Sci USA (2003) 100:7726–30. doi: 10.1073/pnas.1230736100
25. Hozumi A, Mita K, Miskey C, Mates L, Izsvak Z, Ivics Z, et al. Germline transgenesis of the chordate Ciona intestinalis with hyperactive variants of sleeping beauty transposable element. Dev Dyn (2013) 242:30–43. doi: 10.1002/dvdy.23891
26. Hikosaka A, Kusakabe T, Satoh N, Makabe KW. Introduction and Expression of Recombinant Genes in Ascidian Embryos. Dev Growth Differ (1992) 34:627–34. doi: 10.1111/j.1440-169X.1992.tb00031.x
27. Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res (2012) 40:W117–22. doi: 10.1093/nar/gks608
28. Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res (2011) 39:e82. doi: 10.1093/nar/gkr218
29. Sakuma T, Ochiai H, Kaneko T, Mashimo T, Tokumasu D, Sakane Y, et al. Repeating pattern of non-RVD variations in DNA-binding modules enhances TALEN activity. Sci Rep (2013) 3:3379. doi: 10.1038/srep03379
30. Treen N, Yoshida K, Sakuma T, Sasaki H, Kawai N, Yamamoto T, et al. Tissue-specific and ubiquitous gene knockouts by TALEN electroporation provide new approaches to investigating gene function in Ciona. Development (2014) 141:481–7. doi: 10.1242/dev.099572
31. Yoshida K, Nakahata A, Treen N, Sakuma T, Yamamoto T, Sasakura Y. Hox-mediated endodermal identity patterns pharyngeal muscle formation in the chordate pharynx. Development (2017) 144:1629–34. doi: 10.1242/dev.144436
32. Ota S, Hisano Y, Muraki M, Hoshijima K, Dahlem TJ, Grunwald DJ, et al. Efficient identification of TALEN-mediated genome modifications using heteroduplex mobility assays. Genes Cells (2013) 18:450–8. doi: 10.1111/gtc.12050
33. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics (2005) 21:3674–6. doi: 10.1093/bioinformatics/bti610
34. Shiraishi A, Murata J, Matsumoto E, Matsubara S, Ono E, Satake H. De Novo Transcriptomes of Forsythia koreana Using a Novel Assembly Method: Insight into Tissue- and Species-Specific Expression of Lignan Biosynthesis-Related Gene. PloS One (2016) 11:e0164805. doi: 10.1371/journal.pone.0164805
35. Kawada T, Shiraishi A, Aoyama M, Satake H. Transcriptomes of the Premature and Mature Ovaries of an Ascidian, Ciona intestinalis. Front Endocrinol (2017) 8:88. doi: 10.3389/fendo.2017.00088
36. Hamada M, Shimozono N, Ohta N, Satou Y, Horie T, Kawada T, et al. Expression of neuropeptide- and hormone-encoding genes in the Ciona intestinalis larval brain. Dev Biol (2011) 352:202–14. doi: 10.1016/j.ydbio.2011.01.006
37. Osugi T, Sasakura Y, Satake H. The nervous system of the adult ascidian Ciona intestinalis Type A (Ciona robusta): Insights from transgenic animal models. PloS One (2017) 12:e0180227. doi: 10.1371/journal.pone.0180227
38. Prodon F, Chenevert J, Sardet C. Establishment of animal-vegetal polarity during maturation in ascidian oocytes. Dev Biol (2006) 290:297–311. doi: 10.1016/j.ydbio.2005.11.025
39. Nishimori K, Young LJ, Guo Q, Wang Z, Insel TR, Matzuk MM. Oxytocin is required for nursing but is not essential for parturition or reproductive behavior. Proc Natl Acad Sci USA (1996) 93:11699–704. doi: 10.1073/pnas.93.21.11699
40. Takayanagi Y, Yoshida M, Bielsky IF, Ross HE, Kawamata M, Onaka T, et al. Pervasive social deficits, but normal parturition, in oxytocin receptor-deficient mice. Proc Natl Acad Sci USA (2005) 102:16096–101. doi: 10.1073/pnas.0505312102
41. Santana P, Lanes L, Hernandez I, Gonzalez-Robayna I, Tabraue C, Gonzalez-Reyes J, et al. Interleukin-1 beta stimulates sphingomyelin hydrolysis in cultured granulosa cells: evidence for a regulatory role of ceramide on progesterone and prostaglandin biosynthesis. Endocrinology (1996) 137:2480–9. doi: 10.1210/endo.137.6.8641202
42. Itami N, Shirasuna K, Kuwayama T, Iwata H. Palmitic acid induces ceramide accumulation, mitochondrial protein hyperacetylation, and mitochondrial dysfunction in porcine oocytes. Biol Reprod (2018) 98:644–53. doi: 10.1093/biolre/ioy023
43. Malmersjö S, Di Palma S, Diao J, Lai Y, Pfuetzner RA, Wang AL, et al. Phosphorylation of residues inside the SNARE complex suppresses secretory vesicle fusion. EMBO J (2016) 35:1810–21. doi: 10.15252/embj.201694071
44. Brown MS, Lim E, Chen C, Nishant KT, Alani E. Genetic analysis of mlh3 mutations reveals interactions between crossover promoting factors during meiosis in baker’s yeast. G3 (Bethesda) (2013) 3:9–22. doi: 10.1534/g3.112.004622
Keywords: ascidian, Ciona intestinalis, oxytocin, vasopressin, ovary, transgenic, gene edition
Citation: Kawada T, Shiraishi A, Matsubara S, Hozumi A, Horie T, Sasakura Y and Satake H (2021) Vasopressin Promoter Transgenic and Vasopressin Gene-Edited Ascidian, Ciona intestinalis Type A (Ciona robusta): Innervation, Gene Expression Profiles, and Phenotypes. Front. Endocrinol. 12:668564. doi: 10.3389/fendo.2021.668564
Received: 16 February 2021; Accepted: 25 March 2021;
Published: 06 May 2021.
Edited by:
Takayoshi Ubuka, Cancer Medical Service, JapanReviewed by:
Takehiro Kusakabe, Konan University, JapanKazuyoshi Ukena, Hiroshima University, Japan
Toshio Sekiguchi, Kanazawa University, Japan
Copyright © 2021 Kawada, Shiraishi, Matsubara, Hozumi, Horie, Sasakura and Satake. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Honoo Satake, c2F0YWtlQHN1bmJvci5vci5qcA==
 Tsuyoshi Kawada
Tsuyoshi Kawada Akira Shiraishi
Akira Shiraishi Shin Matsubara
Shin Matsubara Akiko Hozumi2
Akiko Hozumi2 Takeo Horie
Takeo Horie Yasunori Sasakura
Yasunori Sasakura Honoo Satake
Honoo Satake