
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Endocrinol. , 10 March 2021
Sec. Cellular Endocrinology
Volume 12 - 2021 | https://doi.org/10.3389/fendo.2021.634191
This article is part of the Research Topic Anti-obesity: Targeting Brown and Beige Adipocytes View all 14 articles
 Zhiyin Zhang1,2†
Zhiyin Zhang1,2† Qihan Wu1,2†
Qihan Wu1,2† Yang He1,2†
Yang He1,2† Peng Lu1,2
Peng Lu1,2 Danjie Li1,2
Danjie Li1,2 Minglan Yang1,2
Minglan Yang1,2 Weiqiong Gu1,2
Weiqiong Gu1,2 Ruixin Liu1,2*
Ruixin Liu1,2* Jie Hong1,2*
Jie Hong1,2* Jiqiu Wang1,2*
Jiqiu Wang1,2*Objective: The Iroquois homeobox 3 (IRX3) gene was recently reported to be a functional downstream target of a common polymorphism in the FTO gene, which encodes an obesity-associated protein; however, the role of IRX3 in energy expenditure remains unclear. Studies have revealed that the overexpression of a dominant–negative form of IRX3 in the mouse hypothalamus and adipose tissue promoted energy expenditure by enhancing brown/browning activities. Meanwhile, we and others recently demonstrated that IRX3 knockdown impaired the browning program of primary preadipocytes in vitro. In this study, we aimed to further clarify the effects of overexpressing human IRX3 (hIRX3) on brown/beige adipose tissues in vivo.
Methods: Brown/beige adipocyte-specific hIRX3-overexpressing mice were generated and the browning program of white adipose tissues was induced by both chronic cold stimulation and CL316,243 injection. Body weight, fat mass, lean mass, and energy expenditure were measured, while morphological changes and the expression of thermogenesis-related genes in adipose tissue were analyzed. Moreover, the browning capacity of primary preadipocytes derived from hIRX3-overexpressing mice was assessed. RNA sequencing was also employed to investigate the effect of hIRX3 on the expression of thermogenesis-related genes.
Results: hIRX3 overexpression in embryonic brown/beige adipose tissues (Rosa26hIRX3;Ucp1-Cre) led to increased energy expenditure, decreased fat mass, and a lean body phenotype. After acute cold exposure or CL316,243 stimulation, brown/beige tissue hIRX3-overexpressing mice showed an increase in Ucp1 expression. Consistent with this, induced hIRX3 overexpression in adult mice (Rosa26hIRX3;Ucp1-CreERT2) also promoted a moderate increase in Ucp1 expression. Ex vitro experiments further revealed that hIRX3 overexpression induced by Ucp1-driven Cre recombinase activity upregulated brown/beige adipocytes Ucp1 expression and oxygen consumption rate (OCR). RNA sequencing analyses indicated that hIRX3 overexpression in brown adipocytes enhanced brown fat cell differentiation, glycolysis, and gluconeogenesis.
Conclusion: Consistent with the in vitro findings, brown/beige adipocyte-specific overexpression of hIRX3 promoted Ucp1 expression and thermogenesis, while reducing fat mass.
Iroquois homeobox gene 3 (IRX3) encodes a transcription factor of the Iroquois family of homeodomain-containing proteins (1). IRX3 is initially expressed during embryogenesis, and is involved in the development and patterning of multiple tissues, including the nervous system, heart, and skeleton (2). Recent studies have indicated that IRX3, together with its homolog IRX5, may also have a role in energy balance and adiposity by regulating thermogenesis in brown adipose tissue (BAT) and the browning program in white adipose tissue (WAT). WAT stores energy in the form of triglycerides under excess caloric intake, whereas BAT dissipates energy through the activity of the inner mitochondrial membrane-localized uncoupled protein 1 (UCP1) to maintain body temperature hemostasis (3–5). Evidence for several genome-wide association studies (GWAS) has indicated that several common single nucleotide polymorphism (SNP) variants, such as rs1421085 and rs9930506, located in the first intron of fat mass and obesity associated (FTO) gene, are strongly associated with an increased risk of obesity (6–8), and the functional loci and biological targets associated with these variants are largely unknown.
Recently, IRX3 and IRX5, but not FTO, were proposed to function as targets of the rs1421085 variant in thermogenesis (9). However, whether IRX3 acts as an activator or repressor of thermogenesis, where the functional target(s) are located, and whether other candidate genes including FTO, IRX5, and Rpgrip1l were involved in thermogenesis regulation of rs1421085 (10, 11), remain unclear. Another study reported that the hypothalamic overexpression of a dominant–negative form of IRX3 (EnR-Irx3) (12), which can increase instead of suppress the transcriptional activities of the IRX3 protein in certain contexts (13), induced a lean body phenotype accompanied by an enhanced WAT browning capacity and activation of BAT (12). Additionally, the overexpression of this “dominant–negative” form of IRX3 (EnR-Irx3;aP2-Cre) in adipose tissue also induced a lean body phenotype with marked browning changes in WATs (14). In contrast, a recent study demonstrated that partial (approximately 50%) inhibition of endogenous hypothalamic IRX3 expression reduced thermogenesis in peripheral BAT and increased diet-induced body mass gain, thereby exacerbating obesity (15). We have previously also provided evidence that Irx3 knockdown can impair the thermogenic capacities of induced brown and beige adipocytes derived from preadipocytes of mouse inguinal WAT (iWAT) and BAT, and human subcutaneous WAT, respectively; and that missense mutations in IRX3 identified in humans markedly reduced the transcription of UCP1 in vitro (16). Consistent with these observations, another group recently showed that Irx3 knockout in mouse preadipocytes impaired both the early and late stages of adipogenic differentiation to beige adipocytes and preadipocyte mitochondrial respiration (17). These contradictory data highlight the importance of identifying the precise roles of human IRX3 (wild type) in brown/beige adipocytes in vivo.
To this end, we generated two brown/beige adipocyte-specific hIRX3-overexpressing mouse models by crossing Rosa26hIRX3 knock-in mice with Ucp1-Cre mice, resulting in the continuous induced expression of hIRX3 from the embryonic stage, as well as with Ucp1-CreERT2 mice, which expressed hIRX3 from adulthood following tamoxifen (TMX) injection. In Ucp1-Cre mice, hIRX3 overexpression in BAT led to increased energy expenditure, decreased fat mass, and a lean body phenotype, while hIRX3 overexpression in adulthood induced a subtle increase in thermogenesis after stimulation with a β3-AR agonist. Furthermore, hIRX3 overexpression significantly enhanced beige adipocyte differentiation concomitant with increased Ucp1 expression. Together, our results revealed that hIRX3 overexpression can promote thermogenesis in brown/beige adipose tissue in vivo, and provide a more comprehensive understanding of the role of IRX3 in energy balance and obesity.
To generate Rosa26hIRX3 (hIRX3 knock-in) mice, a human IRX3 cDNA-polyA cassette (GenBank accession number: NM_024336.2; Ensembl: ENSG00000177508) was cloned into intron 1 of the Rosa26 locus, and CAG-loxP-stop-loxP was inserted upstream of the cassette. Ucp1-Cre mice were obtained from the Jackson Laboratory (Jax no. 024670). To generate Ucp1-CreERT2 mice, a CreERT2-IRES-EGFP-PA cassette was knocked-in downstream of the ATG start codon of the mUcp1 gene such that the expression of CreERT2 and EGFP were under the control of mUcp1 regulatory sequences. Mouse genomic fragments were amplified with high-fidelity Taq DNA polymerase and were assembled into a targeting vector, together with recombination sites and selection markers, as indicated in the vector map in Supplementary Figure 1A. The final sequence of the targeting vector is shown in Supplementary Figure 1B. The constitutive hIRX3 knock-in allele was obtained after Flp-mediated recombination (Supplementary Figure 1C). C57BL/6 embryonic stem cells were used for gene targeting. Genotypes were verified by PCR (Supplementary Figure 1D). Maximal Cre recombinase mRNA expression was seen in BAT (Supplementary Figures 1E, F). Ucp1-Cre;Rosa26hIRX3 (U-IRX3ov) and Ucp1-CreERT2;Rosa26hIRX3 (iU-IRX3ov) mice were generated using the Cre/loxP system. Rosa26hIRX3 was used as a control with U-IRX3ov mice. As Ucp1-CreERT2 showed an approximately 50% decrease in Ucp1 protein expression compared with that of endogenous Ucp1 (Supplementary Figure 1G), we used Rosa26wild type(WT);Ucp1-CreERT2 as the control for iU-IRX3ov. iU-IRX3ov mice and control littermates were treated intraperitoneally with TMX at a dose of 100 mg/(kg·day−1) to induce hIRX3 expression (Supplementary Figure 1H-K). All animal procedures were approved by the Animal Care Committee of Shanghai Jiao Tong University School of Medicine and followed the guide for the care and use of laboratory animals.
For cold exposure, mice were placed individually in a room with the temperature set at 4°C for 7 days. The animals had free access to food and water during this period. Their core body temperature was measured using a rectal probe (Physitemp Instruments Inc., USA). CL316,243 (Sigma-Aldrich, USA) was injected interperitoneally at a dose of 1 mg/(kg·day−1), and the injection protocol was described in the results part for details.
The fat mass and lean mass of each mouse was measured using an EchoMRI-100H (EchoMRI, USA). The mice were placed in a Comprehensive Laboratory Animal Monitoring System (CLAMS, Columbus Instruments, USA) for the evaluation of whole-body energy metabolism. Oxygen and carbon dioxide consumption, as well as activity, was continuously measured for two days. The respiratory exchange ratio (RER) and energy expenditure were calculated based on the oxygen and carbon dioxide data and were normalized to body weight (18).
BATs and WATs were isolated, fixed in 4% paraformaldehyde, embedded in paraffin, and sliced into 5-μm (iWAT, eWAT) or 3-μm (BAT) sections for hematoxylin and eosin (H&E) staining. Images were captured under a microscope (Olympus, Japan). Pictures were scanned by Digital Pathology Slide Scanner (KF-PRO-120).
The stromal vascular fractions (SVFs) were isolated from the BAT and iWAT of five-week-old U-IRX3ov and control mice and then induced to fully differentiate into brown/beige adipocytes as previously described (5). In brief, the fat pads were isolated, cut into pieces, and digested with type II collagenase (Sigma) at 37°C for 30 min followed by quenching with DMEM/F12 supplemented with 10% FBS. The suspended samples were filtered using a 40-μm strainer (BD, USA) and then plated on culture dishes. The SVFs were first grown to 100% confluence in DMEM/F12 supplemented with 10% FBS (plus 1% penicillin/streptomycin and 1 mM L-glutamine), and then these primary preadipocytes were differentiated into brown/beige adipocytes in a cocktail containing 5 μg/ml insulin (Eli Lilly, USA), 1 μM dexamethasone, 1 μM rosiglitazone, 1 μM triiodothyronine (T3), and 0.5 mM IBMX (all Sigma–Aldrich, USA) for two days, and subsequently in medium with insulin, rosiglitazone, and T3 for another six days.
After eight days of induction, mature adipocytes were stained with Oil Red O. In brief, the cells were fixed in 4% paraformaldehyde for 30 min, rinsed, air-dried, and incubated with Oil Red O (Nanjing Jiancheng Bioengineering Institute, China) for 30 min. Images were captured under a microscope (Olympus).
SVFs were seeded in an XF24 V28 microplate (Agilent Technologies, USA) coated with poly-L-lysine. The induction protocol was as described in section 2.5. The oxygen consumption rate (OCR) was measured at induction day 4 using an XF24 analyzer (Agilent Technologies) following the manufacturer’s instructions. Briefly, the induced brown/beige adipocytes were washed with Seahorse assay medium, consisting of XF DMEM supplemented with 10 mM XF glucose, 1 mM XF pyruvate, and 2 mM XF L-glutamine, followed by incubation with 525 μl of assay medium at 37°C in an incubator without CO2 (Agilent Technologies) for 1 h. Respiratory inhibitors (75 μl) were loaded into the injection port to final concentrations of 1 mg/ml oligomycin, 2 mM FCCP, 0.5 mM antimycin A, and 0.5 mg/ml rotenone to detect uncoupled respiration, maximal respiration, and nonmitochondrial respiration, respectively. The final OCR results were standardized to the total protein content. The results are representative of at least three independent experiments.
Total RNA was extracted from cultured cells or frozen adipose tissue using the Eastep Super Total RNA Extraction Kit (Promega (Beijing) Biotech Co., China). The absorbance ratio at 260/280 nm and the RNA concentration of each sample were detected using a NanoDrop ND2000 spectrophotometer (Thermo Scientific). Reverse transcription was performed using the PrimeScript Reverse Transcript Master Mix (TaKaRa, Japan). qPCR was performed using a QuantStudio Dx Real-Time PCR Instrument (Applied Biosystems). The comparative ΔΔCt method was used to evaluate the relative mRNA levels; 36B4 served as the reference gene (Supplementary Table 1).
RNA sequencing was performed by NovelBio, Shanghai, China. The RNA quality was assessed using an Agilent 2200 and the RNA was stored at −80°C. RNA with an RNA integrity number (RIN) >7 was considered acceptable for cDNA library construction. cDNA libraries were constructed for each RNA sample using the TruSeq Stranded mRNA Library Prep Kit (Illumina) according to the manufacturer’s instructions. The libraries were quality controlled with Agilent 2200 and sequenced by HiSeq X (Illumina) as 150-bp paired-end reads. For the analysis of differentially expressed genes, P-value and false discovery rate (FDR) analysis were subjected to the following criteria: i) Fold change (FC) >2 or <0.5; ii) P-value <0.05, FDR <0.05. Fisher’s exact test was applied to identify significant GO categories and KEGG pathways (P-value <0.05). The approach for gene set enrichment analysis (GSEA) was in accordance with that previously reported (19). Genes were considered to be significantly differentially expressed when the FDR was less than 0.05 and the log2FC was more than 1.
Total protein was isolated using RIPA lysis buffer (Biocolors, China) with a protease inhibitor cocktail (Sigma). Western blotting was performed as previously described (5). The following antibodies were used: anti-IRX3 (ab174307, Abcam), anti-UCP1 (ab10983, Abcam), anti-Hsp90 (Cell Signaling Technology, 4877s), and anti-PGC-1α (Abcam, ab54481). The results are representative of at least three independent experiments.
Data are shown as means ± S.E.M, and the results were compared by two-tailed t-tests. A P-value <0.05 was considered to be significantly different. Spearman’s correlation analysis was performed to examine the associations between the expression of hIRX3 and mUcp1. For molecular experiments, data were generated from three independent experiments. Analyses were undertaken with GraphPad Prism version 8.2.1 (279) (GraphPad Software, Inc., La Jolla, CA, USA).
To clarify the physiological and biological roles of hIRX3 in thermogenesis in brown/browning adipose tissues, we generated a Ucp1-Cre-driven hIRX3 overexpression mouse model (Ucp1-Cre; Rosa26hIRX3, referred to as U-IRX3ov) (Figure 1A). When fed a normal chow diet, both male and female U-IRX3ov mice gained substantially less body weight than the controls (Rosa26hIRX3) at 10 weeks of age (Figure 1B and Supplementary Figures 2A–C). Body composition analysis revealed a lower fat mass percentage in U-IRX3ov mice when compared with controls; however, there was no significant difference in lean mass percentage between the two genotypes (Figures 1C, D and Supplementary Figures 2D, E). To test for potential alterations in energy balance, we undertook a comprehensive evaluation of the food intake, physical activities, and energy expenditure of the mice. We found that, with comparable daily food intake (Figure 1E and Supplementary Figure 2F), U-IRX3ov mice had greater O2 consumption, increased CO2 production, and greater total energy expenditure, especially at night, compared with controls (Figures 1F–I). U-IRX3ov mice showed a slight increase in physical activities in few hours of a day (X-and Y-axis), although the total increase was not statistically significant (Supplementary Figures 2G–I). Importantly, U-IRX3ov mice displayed higher average energy expenditure per hour compared with controls (Figure 1J). These findings suggested that U-IRX3ov mice gained less fat mass, which was likely due to increased energy expenditure.
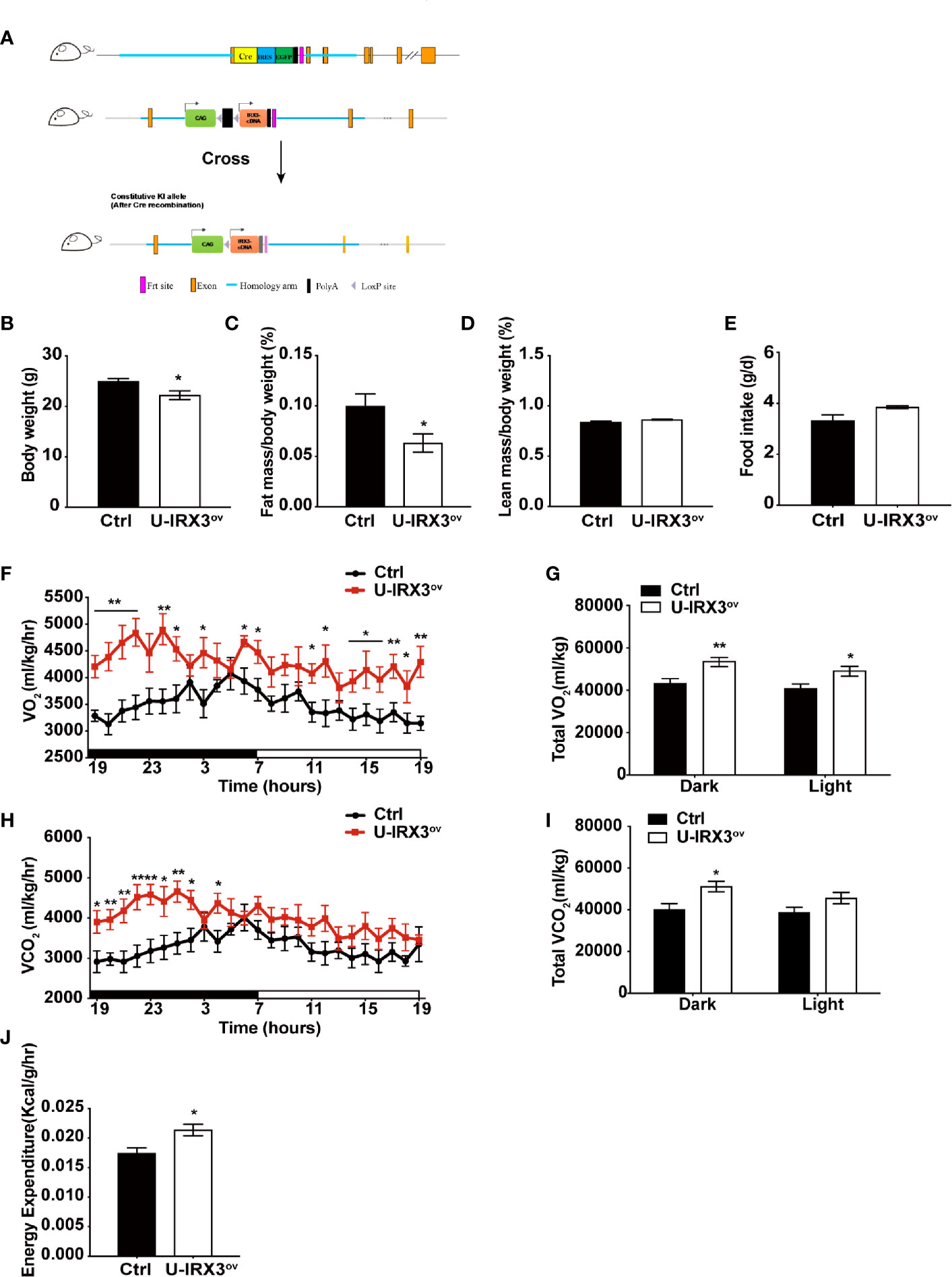
Figure 1 Brown/beige adipocyte-specific hIRX3 overexpression increases energy expenditure and induces a lean body phenotype in male mice. (A) Schematic diagram of the generation of U-IRX3ov (Rosa26-loxP-stop-loxP-hIRX3;Ucp1-Cre) mice. (B–D) Body weight (B), fat mass percentage (C), and lean mass percentage (D) of male U-IRX3ov and control mice at 10 weeks of age (n = 6~8). (E) Average daily food intake of male U-IRX3ov and control mice at 10 weeks of age (average of 3 individual measurements). (F–J) Whole-body oxygen (O2) consumption per hour (F) and per 12 h (G), carbon dioxide (CO2) production per hour (H) and per 12 h (I), and energy expenditure (J) of 10-week-old U-IRX3ov and control mice during a 24-h period (n = 6~8). Statistics were standardized by body weight. Data are shown as means ± SEM. *P < 0.05, **P < 0.01.
Next, to test the response of the mice to acute and chronic cold, we subjected the two groups of mice to cold treatment at 4°C. No significant differences in body temperature were observed during the first 6 h (Supplementary Figure 2J). However, after seven days of cold stimulation, the mice showed a subtle, but statistically insignificant, decrease in eWAT mass percentage (P = 0.08) (Figure 2A). With moderate hIRX3 overexpression in inguinal WAT (iWAT) and BAT (Figure 2B and Supplementary Figure 2K), U-IRX3ov mice showed marked morphological changes in iWAT, characterized by a more condensed texture with markedly increased number of smaller lipid droplets, as well as a significantly reduced droplet content in BAT, and an increase in the percentage of smaller adipocytes in eWAT compared with those in control mice (Figures 2C, D). We next examined the expression levels of thermogenesis-related genes in three adipose tissues, and found that the mRNA levels of Pgc-1α and Dio2 were increased in the BAT of U-IRX3ov mice (Figure 2E). Although the mRNA levels of Ucp1 were unchanged, the BAT of U-IRX3ov mice exhibited a small but substantial increase in UCP1 protein levels (Figures 2F, G). For iWAT, a marked increase in the mRNA expression levels of Ucp1, Cidea, Dio2, Cox7a1, and Cox8b was observed in U-IRX3ov mice (Figure 2H). Consistent with our previous in vitro findings, hIRX3 mRNA expression in vivo was also positively correlated with that of Ucp1 (Figure 2I). The protein levels of UCP1 and PGC-1α in iWAT were also increased in the iWAT of U-IRX3ov mice (Figure 2J). However, only the mRNA levels of Ucp1 and Cox8b, and the protein levels of PGC-1α, showed increased expression in the eWAT of U-IRX3ov mice (Figures 2K, L).
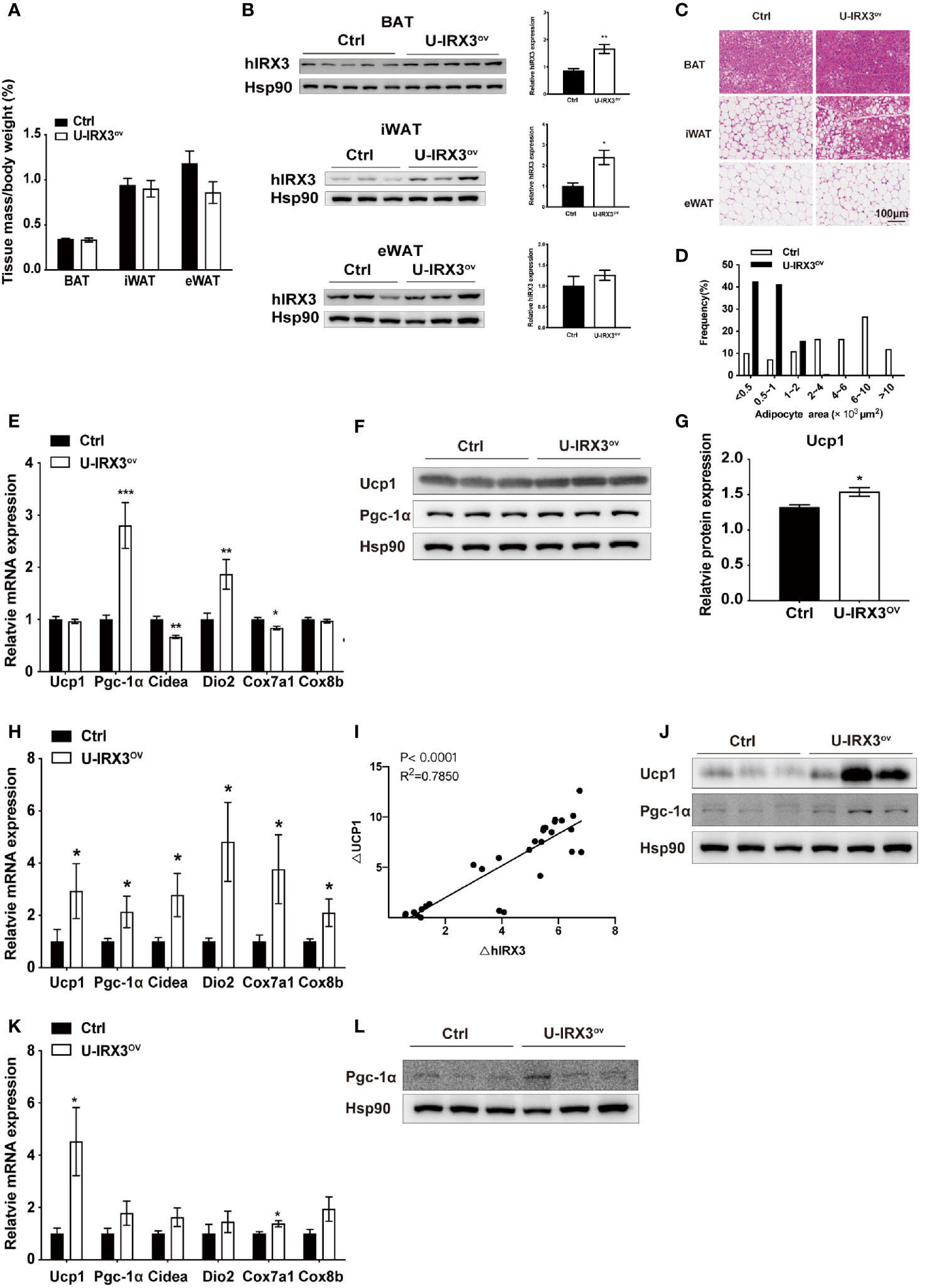
Figure 2 The overexpression of hIRX3 from the embryonic stage enhances cold-induced thermogenesis. (A–L) Ten-week-old U-IRX3ov and control mice were placed in a cold room at 4°C for 7 days. Tissue mass percentage (A) and protein levels of overexpressed hIRX3 in BAT, iWAT, and eWAT of the two groups of mice (n = 3~5). (B) Representative images of hematoxylin and eosin (H&E) staining of BAT (top), iWAT (middle), and eWAT (bottom) for the two groups of mice (C). (D) Adipocyte size distribution of the eWAT in (C). Scale bars, 100 μm. (E) The mRNA expression levels of thermogenesis-related genes in the BAT of the two groups of mice (n = 6~8). (F) The protein levels of UCP1 and PGC-1α in the BAT of the two groups of mice (n = 3). (G) The UCP1 protein expression level relative to that of Hsp90 in (F). (H) The mRNA expression levels of thermogenesis-related genes in the iWAT of the two groups of mice (n = 6~8). (I) Linear regression analyses of hIRX3 and Ucp1 RNA expression level of male (n=13) and female(n=16) U-IRX3ov and control mice (J) The protein levels of UCP1 and PGC-1α in the iWAT of the two groups of mice (n = 3). (K) The mRNA expression levels of thermogenesis-related genes in the eWAT of the two groups of mice (n = 6~8). (L) The protein levels PGC-1α in the eWAT of the two groups of mice (n = 3). Data are shown as means ± SEM. *P < 0.05.
To further validate the promotive effect of hIRX3 on thermogenesis in BAT, we intraperitoneally injected CL316,243, an agonist of the β3-adrenergic receptor (β3-AR), into female U-IRX3ov and littermate control mice to activate BAT and induce the browning process. Male mice showed no significant change in mass under cold treatment; however, female U-IRX3ov mice displayed a reduction in BAT, iWAT, and gWAT (gonadal WAT) content (Supplementary Figure 3A). Histomorphological analysis revealed that the adipocytes were smaller and more condensed in all these adipose tissues (Supplementary Figures 3B–D). Both the mRNA and protein expression levels of Ucp1 and Pgc-1α were increased in the BAT of female U-IRX3ov mice (Supplementary Figures 3E, F). The expression of thermogenesis-related genes, such as Ucp1, Pgc-1α, Cidea, and Dio2, was enhanced in the iWAT of U-IRX3ov mice (Supplementary Figure 3G). Similarly, UCP1 protein levels showed an increasing trend in the iWAT of U-IRX3ov mice (Supplementary Figure 3H). Collectively, these results indicated that the overexpression of hIRX3 increased both cold treatment- and CL316,243-induced thermogenesis in brown/beige adipose tissues in vivo.
The protein expression of UCP1 in BAT first appears in late gestation and then rapidly increases at birth (20), allowing Ucp1 promoter-driven Cre recombinase to excise loxP-flanked (floxed) sequences (STOP in this study) and theoretically induce hIRX3 overexpression prepartum. A recent study demonstrated that Irx3 ablation in mouse preadipocytes attenuated the proliferation and early differentiation of beige adipocytes in vitro (17). To avoid the nonspecific consequences of overexpressing hIRX3 in brown preadipocytes, and to investigate the effects of transient hIRX3 overexpression in mature beige adipocytes in adult mice, we generated a TMX-inducible hIRX3 overexpressing mouse line (Ucp1-CreERT2; Rosa26hIRX3, iU-IRX3ov) by crossing Rosa26hIRX3 mice with Ucp1-CreERT2 mice (Figure 3A). Then, we treated eight-week-old male iU-IRX3ov and control mice with TMX (i.p. once/day) for 5 days during the 10-days CL316,243 injection interval (Figure 3B). The overexpression of hIRX3 in adipose tissue was validated by qPCR and Western-blot (Supplementary Figures 1H–K). No differences in body weight, fat mass percentage, lean mass percentage, or adipose tissue weight were observed between the two groups (Figures 3C–F). In the iWAT of iU-IRX3ov mice, there was a small but significant increase in Ucp1 mRNA and protein levels (p<0.05), while the expression of other thermogenesis-related genes showed an increasing, but insignificant, trend (Figures 3G–I). Interestingly, the mRNA expression of Ucp1, Pgc-1α, and Cidea was significantly increased in the eWAT of iU-IRX3ov mice when compared with controls (Figure 3J), which was accompanied by an increase in UCP1 protein levels (Figures 3K, L). Combined, these results indicated that the temporary overexpression of hIRX3 in WAT of adult mice, and especially in eWAT, can enhance thermogenesis, but with a relatively subtle effect when compared with that seen in U-IRX3ov mice.
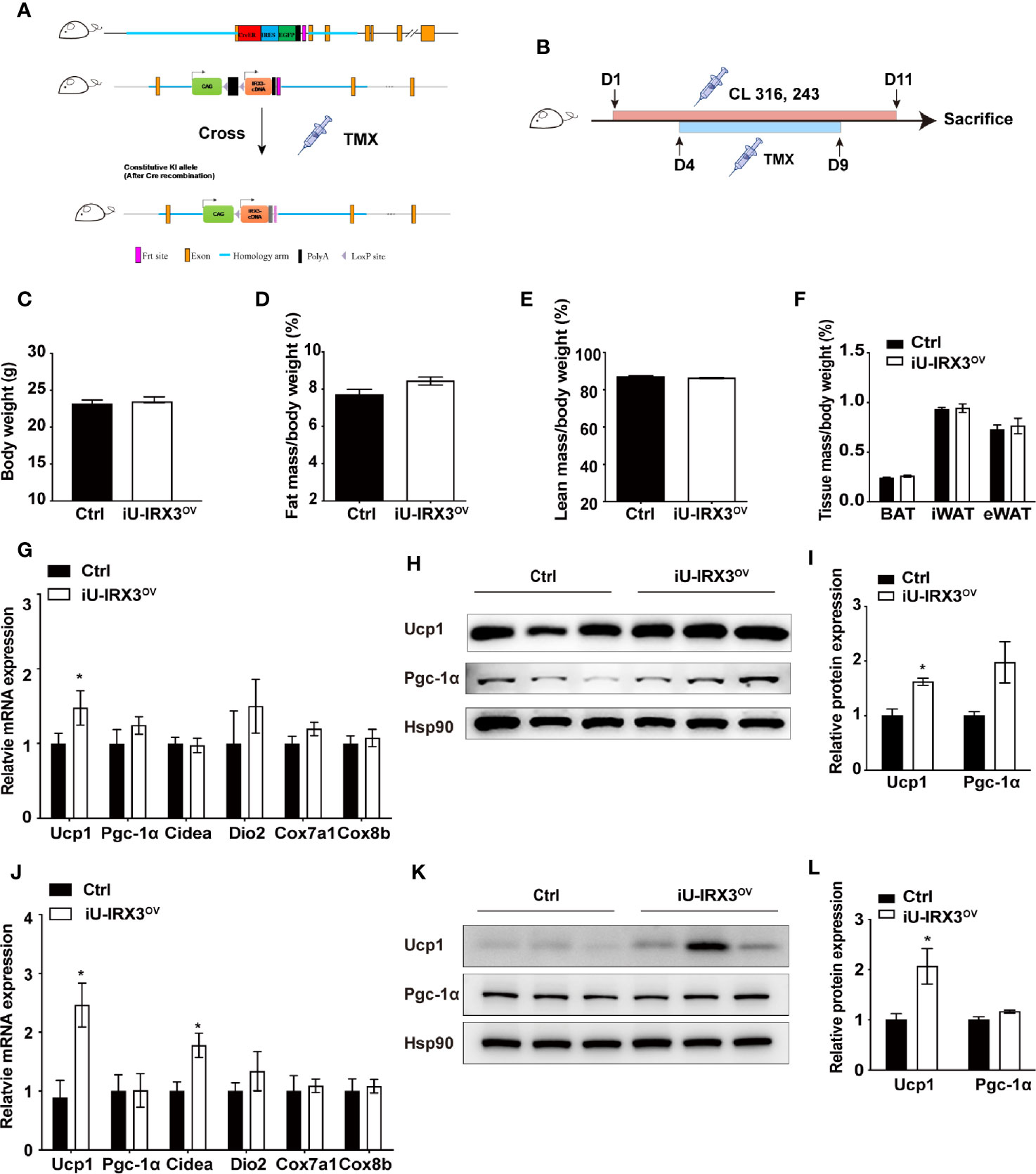
Figure 3 Overexpressing hIRX3 after adulthood increased the expression of thermogenesis-related genes under β3-AR agonist (CL316,243) stimulation. (A) Schematic representation of the strategy to generate iU-IRX3ov mice. (B) Timeline of CL316,243 and tamoxifen (TMX) injection in eight-week-old male iU-IRX3ov and control mice (n = 9~10). (C–F) Body weight (C), fat mass percentage (D), lean mass percentage (E), and tissue mass percentage of BAT, iWAT, and eWAT (F) in male iU-IRX3ov and control mice. (G) The relative mRNA expression levels of Ucp1 and other thermogenesis-related genes in the iWAT of iU-IRX3ov mice(n = 9~10). (H, I) Images and quantitative values of UCP1 and PGC-1α protein levels in iWAT (n = 3). (J) The relative mRNA expression levels of Ucp1 and other thermogenesis-related genes in eWAT (n = 9~10). (K, L) Images and quantitative values of UCP1 and PGC-1α protein levels in eWAT (n = 3). Data are shown as means ± SEM. *P < 0.05.
We next investigated the effects of hIRX3 overexpression on the differentiation and thermogenic capacities of induced brown and beige adipocytes obtained from primary preadipocyte SVFs. Over expression of hIRX3 was proved by qPCR and Western-blot (Figures 4E, K and Supplementary Figure 3I). BAT SVF derived from U-IRX3ov mice showed markedly enhanced adipogenic differentiation capacity compared with controls, as evidenced by the increased number of multilocular lipid droplets in Oil Red O staining (Figure 4A). We also assessed the thermogenic capacity of induced brown adipocytes, represented by the mitochondrial OCR. Compared with controls, there was a significant increase in basal respiration and proton leak, as well as an increasing trend for the maximal respiration and ATP production capacity of the induced brown adipocytes derived from U-IRX3ov mice. This suggested that hIRX3 overexpression led to an increase in thermogenesis (Figures 4B, C). Meanwhile, the mRNA levels of thermogenesis-related genes, such as Ucp1, Pgc-1α, Cidea, Cox7a1, and Cox8b, were significantly increased in induced brown adipocytes derived from U-IRX3ov mice (Figure 4D). Consistent with these results, the protein levels of UCP1 and PGC-1α were also enhanced by hIRX3 overexpression (Figures 4E, F). Additionally, when the SVFs obtained from the iWAT of U-IRX3ov and control mice were induced to beige adipocytes in vitro, we found that adipogenesis, thermogenic capacity, and expression of thermogenesis-related genes were all increased in the U-IRX3ov group (Figures 4G–L). Together, these results indicated that the thermogenic capacity can be improved in brown and beige adipocytes following hIRX3 overexpression mediated by Ucp1 promoter-driven Cre recombinase activity in vitro.
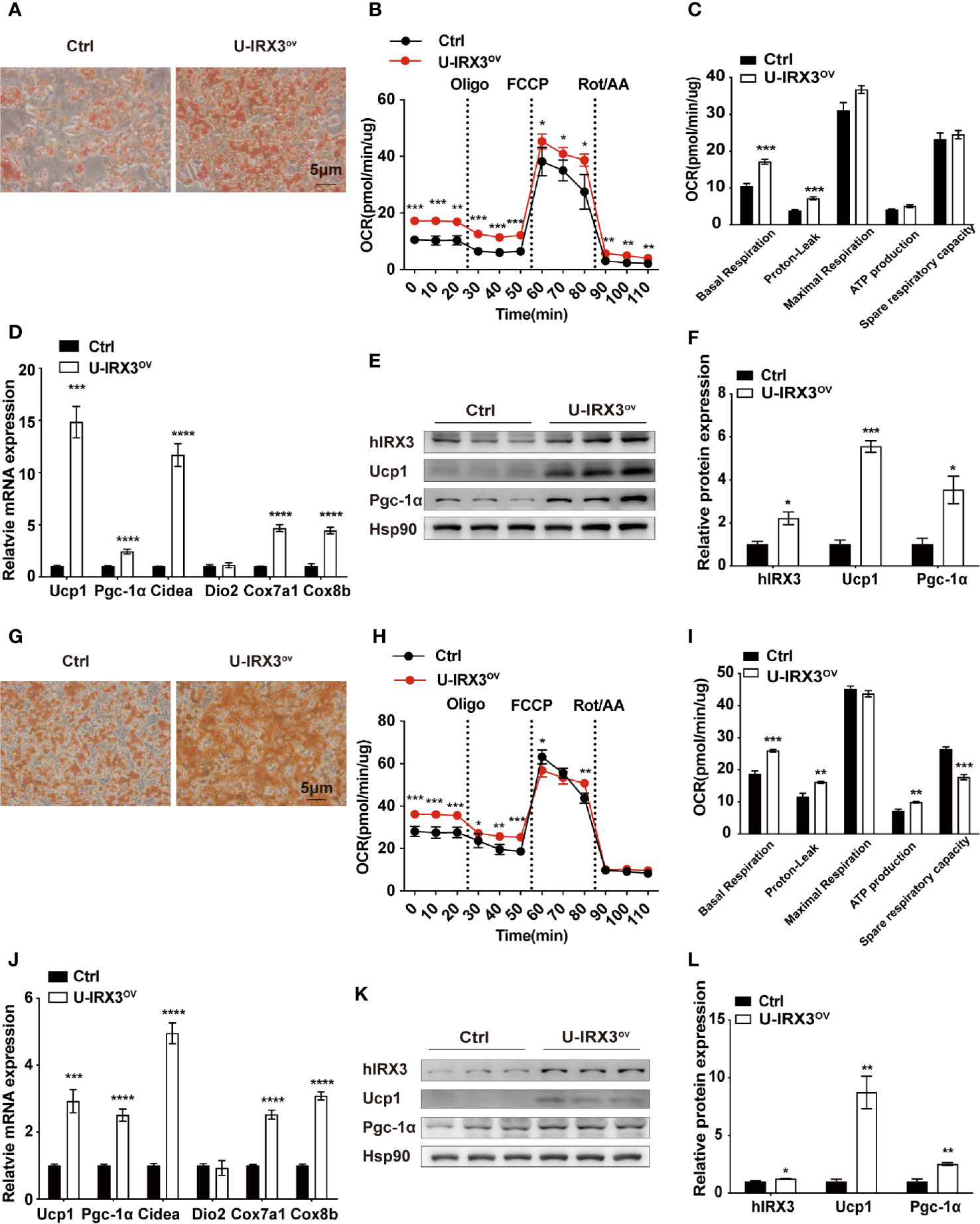
Figure 4 The overexpression of hIRX3 increases thermogenesis in brown/beige adipocytes in vitro. (A–F) The overexpression of hIRX3 in stromal vascular fractions (SVFs) derived from BAT of U-IRX3ov and control mice. Oil Red O staining (A), oxygen consumption rate (OCR) (B, C), mRNA expression levels of thermogenesis-related genes (D), and the protein expression (E) and their quantitative values (F), including hIRX3, PGC-1a, and UCP1, in induced brown adipocytes after five days of differentiation. (G–L) The overexpression of hIRX3 in SVFs derived from the iWAT of U-IRX3ov and control mice. Oil Red O staining of beige adipocytes after eight days of differentiation (G); the OCR of beige adipocytes after eight days of differentiation (H, I); the mRNA expression levels of thermogenesis-related genes (J), and the protein expression (K) and their quantitative values (L), including hIRX3, PGC-1a, and UCP1, in induced beige adipocytes after eight days of differentiation. Data are shown as means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To further elucidate the effects of hIRX3 overexpression on thermogenesis, we performed RNA-seq analysis on induced brown adipocytes derived from the SVFs of U-IRX3ov and control mice. In total, we identified 665 differentially expressed genes (FC <2, FDR <0.05) between the U-IRX3ov and control groups, 248 of which were upregulated and 417 downregulated (Supplementary Tables 2 and 3). Notably, and consistent with the qPCR findings, several genes annotated as being positively related to the browning program, such as Ucp1, Cidea, Pgc-1α, Cox7a1, and Cox8b, were markedly upregulated in the U-IRX3ov group (Figure 5A). Gene Ontology (GO) analysis revealed that the upregulated genes were enriched in biological processes involved in brown fat cell differentiation, glycolysis, gluconeogenesis, and other metabolic process associated with energy expenditure; meanwhile, the downregulated genes were mainly associated with immune system process, inflammatory response, and cell adhesion (Figure 5B), processes that are usually suppressed during brown adipogenesis or thermogenesis (21–23). Gene set enrichment analysis (GSEA) further indicated a marked overlap between enriched genes and the gene signature activated during brown adipogenesis (Figure 5C). We then explored protein–protein connection of upregulated genes, and clustered connected genes which classified into the same KEGG pathway. The results showed the upregulated genes were enriched in pathways related to thermogenesis, oxidative phosphorylation, glycolysis/gluconeogenesis, and PPAR signaling (Figure 5D). Collectively, these data suggested that hIRX3 overexpression in induced brown adipocytes can promote Ucp1 expression, brown cell adipocyte differentiation, and thermogenesis.
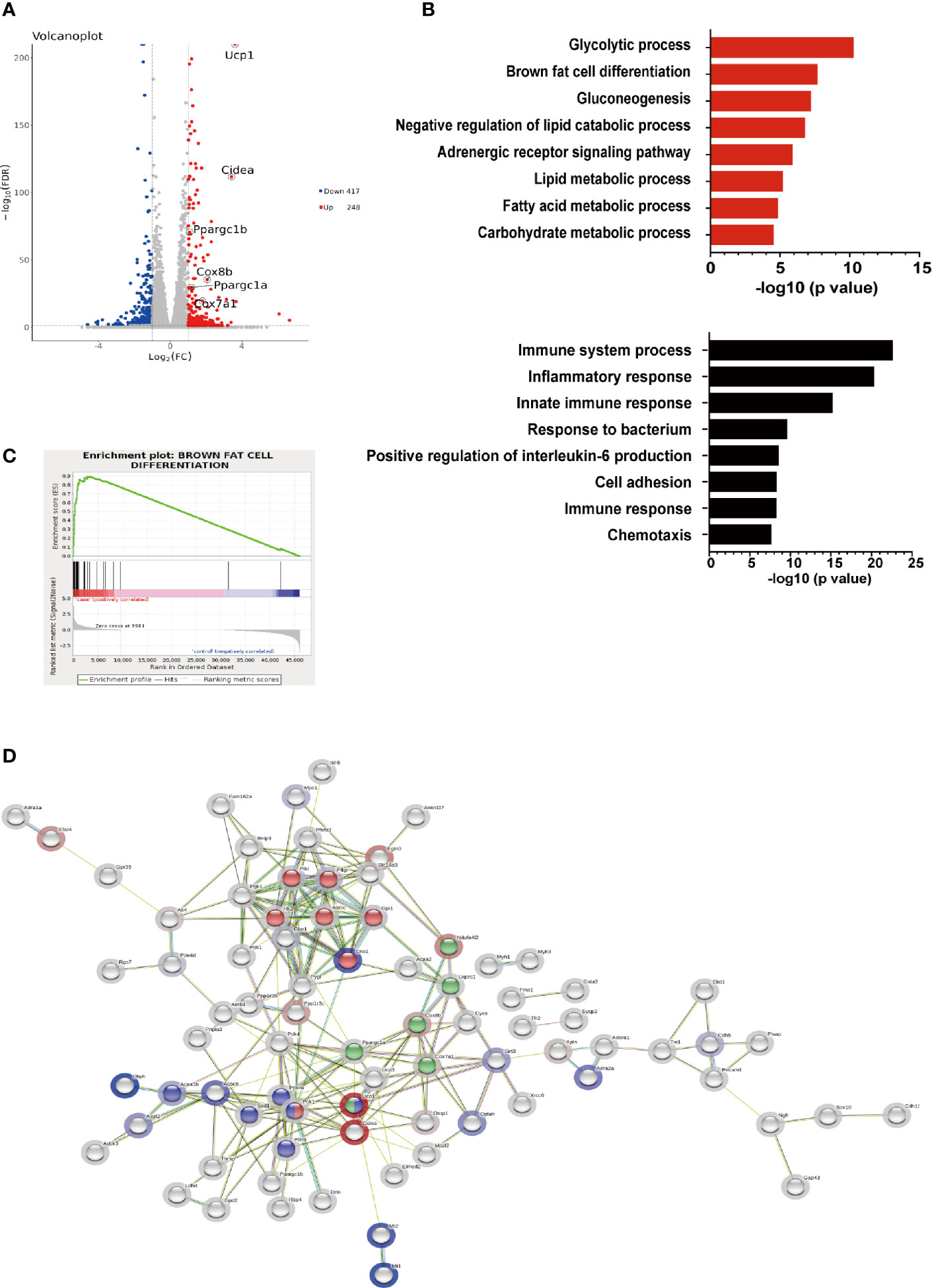
Figure 5 The gene expression profile of hIRX3-overexpressing brown adipocytes. (A–D) After eight days of differentiation, RNA-seq analysis was performed on induced brown adipocytes derived from BAT of U-IRX3ov and littermate control mice (n = 3). (A) Volcano plot of the differentially expressed genes; red: upregulated, blue: downregulated. The x-axis represents the log-fold change, and the y-axis represents the −log10 of the false discovery rate (FDR). Several key thermogenesis-related markers are circled. (B) The top gene ontology (GO) biological process terms enriched (P < 0.05, Fisher’s test) among genes that show significantly higher (top 8, red) or lower (top 8, black) expression (P < 0.05, DESeq) in the U-IRX3ov group relative to controls. (C) Gene set enrichment analysis (GSEA) of overlap between genes upregulated following hIRX3 overexpression and the brown fat cell differentiation gene signature in the GO analysis. NES, normalized enrichment score; p, empirical p-value. (D) A protein–protein interaction network of the upregulated genes (P < 0.05, log2FC ≥1, group average count ≥50, n = 129) was constructed using the STRING database (https://string-db.org) applying an interaction score >0.4. The halo color represents the log2FC value from low (blue) to high (red). Gene clusters for the main KEGG pathways are shown based on node color (red for glycolysis/gluconeogenesis, blue for PPAR signaling pathway, green for thermogenesis).
In this study, we generated two genetically modified mouse models to clarify the effects of hIRX3 overexpression in vivo, and present evidence that hIRX3 overexpression in mouse brown/beige adipocytes leads to an enhancement of thermogenesis. Using the U-IRX3ov line, in which hIRX3 is continuously expressed from an early stage of life, we found that the functional abilities of BAT and the browning program of WAT were both enhanced, which resulted in the increased expression of thermogenesis-related genes (including Ucp1), increased energy expenditure, smaller lipid droplets, and lower fat mass percentage. Using the iU-IRX3ov mouse line, in which the expression of hIRX3 is transiently induced in adulthood, we identified a weak but similar increase in the expression of thermogenesis-related genes in iWAT and eWAT.
Several GWAS studies have demonstrated that genetic variations (SNPs) within the first and second introns of the FTO gene are positively associated with an increased risk of obesity (6–8, 24–26). Among these SNPs, one variant—rs1421085—was recently identified as an underlying cause of adiposity through increasing IRX3 expression via long-range chromatin interaction (9, 12). However, whether IRX3 augments or attenuates the thermogenic capacities of brown/beige adipocytes remains unclear (27–29). In contrast to the findings that IRX3 is an inhibitor of the browning progress, we previously reported that IRX3 expression is elevated in human and mouse brown/beige adipocytes, and that the browning program of adipocytes is repressed when IRX3 is knockdown in vitro (16, 17). Importantly, we found that IRX3 could upregulate the transcription of Ucp1 by specific binding to the ACATGTGT motif (−3470 to −3463 bp) upstream of the transcription starting site of the mouse Ucp1 gene. In this study, we aimed to further clarify the role of the human IRX3 in thermogenesis and its effect on Ucp1 expression, especially in Ucp1-expressing brown/beige adipocytes. We found that hIRX3 enhanced thermogenesis in both hIRX3-overexpression mouse models (noninducible and TMX-inducible). Of note, thermogenesis was more prominent in U-IRX3ov (noninducible Ucp1-Cre) mice than in iU-IRX3ov (inducible Ucp1-CreERT2) mice. This difference could be attributed to the temporal, spatial, and dosage differences in hIRX3 expression between the two models. First, hIRX3 was expected to be expressed in brown adipocytes of U-IRX3ov mice shortly after birth, similar to that observed for normal Ucp1 expression (30), whereas hIRX3 expression was only relatively weakly induced in brown adipocytes of adult iU-IRX3ov mice after TMX injection. Second, there may be heterogeneity in the spatial and quantitative expression of Cre recombinase: the ectogenic Ucp1-Cre construct was inserted spontaneously, thus Cre expression would have been extensively induced (might not restricted to brown and beige adipose tissue), whereas the endogenous expression of Ucp1 would not be affected (31). On the other hand, in Ucp1-CreERT2, the Cre sequence was directly inserted into exon 1 of Ucp1, which means the Cre only expressed where endogenous Ucp1 appeared, but at the same time impaired endogenous Ucp1 expression and led to Ucp1 protein reduced by half. Thus the Cre recombinase in the Ucp1-CreERT2 model likely showed weak gene-editing activity. Notably, the effect of IRX3 on thermogenesis may change according to the stage of adipocyte development or differentiation status. By isolating preadipocytes from U-IRX3ov mice and inducing brown/beige adipogenesis in vitro, we found that hIRX3 overexpression following the initiation of Ucp1 mRNA expression could effectively enhance the thermogenic capacity and Ucp1 expression of mature brown/beige adipocytes. Nevertheless, the RNA-seq results showed that genes that were upregulated following hIRX3 overexpression were primarily enriched in processes such as brown cell differentiation and oxidative phosphorylation, further supporting a promotive role for hIRX3 in energy expenditure.
The overexpression of hIRX3 either from an early stage of life or only in adulthood promoted the thermogenic potential of brown/beige adipocytes, which was consistent with our previous findings in vitro (16). However, other studies have indicated that IRX3 may be a negative regulator of thermogenesis through central or peripheral regulation (9, 12). Nobrega et al. previously demonstrated that obesity-associated FTO variants were positively associated with IRX3 expression in the brain (12), and observed a 25%–30% decrease in body weight in Irx3 global knockout mice fed a normocaloric diet (NCD) compared with wild-type controls, which was attributable to a significant increase in brown/beige adipocyte function. Additionally, the hypothalamic overexpression of a dominant–negative form of mouse Irx3 (Ins2-Cre;Irx3DN) using EnR-Irx3, in which the EnR element was employed to inactivate Irx3 expression, also resulted in a prominent increase in the thermogenic capacities of brown/beige adipocytes. A different group subsequently identified that rs1421085, a leading FTO gene variant, disrupted a conserved binding motif for the ARID5B repressor, leading to the specific disinhibition of IRX3 expression in preadipocytes (not in mature white adipocytes) and, eventually, to the repression of preadipocyte thermogenic capacity or the impairment of adipocyte development at a very early stage of induction (day 2) in vitro (9). To support this, the authors generated adipose-specific Irx3 dominant–negative (aP2-Cre;Irx3DN) mice and found phenotypes similar to those of Ins2-Cre;Irx3DN mice. Both studies supposed a negative role for Irx3 in thermogenesis.
Interestingly, both groups crossed Rosa26-loxP-stop-loxP-EnR-Irx3DN (a theoretical dominant–negative form of Irx3) with corresponding Cre tool mice to produce the tissue-specific “knockout” models (9, 12), which could be intriguing and tricky. First, mice from both the aP2-Cre;Irx3DN and Ins2-Cre;Irx3DN lines appeared to be markedly smaller compared with controls in early life (even at three weeks), and showed a large difference (approximately 10 g) in body weight at eight weeks of age (9, 12). This indicated that these mice likely had growth impairment or developmental defects, and the increased thermogenic capacities of the evaluated adipose tissues might have been due to impaired development (32). Furthermore, the blotting of hypothalamic proteins showed concomitant similar expression levels of endogenous wild-type Irx3 and exogenous EnR-Irx3 in Ins2-Cre;EnR-Irx3 mice. Indeed, the phenotypes of the Ins2-Cre;EnR-Irx3 mice were similar to those of U-IRX3ov mice, in which we employed Rosa26-loxP-stop-loxP-hIRX3 using a similar strategy to that used by the Nobrega group, and where the mice displayed a reduced less body weight and increased thermogenesis in adipose tissue. However, no short stature was observed among either U-IRX3ov or iU-IRX3ov mice. Notably, in a different study, the authors infected isolated neonatal ventricular myocytes (NVMS) with adenovirus encoding wild-type Irx3, a dominant Irx3 activator (VP16-Irx3), or a dominant Irx3 repressor (EnR-Irx3), and found that Cx40/Gja5, an IRX3 target gene, was significantly upregulated following the overexpression of wild-type Irx3 and EnR-Irx3, but not VP16-Irx3 (13). These results raised the possibility that, under certain conditions, EnR-Irx3 may exert a wild-type Irx3-like function. To address this, adipocyte-specific Irx3 knockout models (without growth defects) using floxed Irx3 would be beneficial for elucidating the roles of endogenous Irx3 in thermogenesis.
Mellgren et al., who first identified a role for rs1421085 in the regulation of IRX3 expression (9), further showed that the constitutive and complete absence of endogenous Irx3 in embryonic fibroblasts leads to the loss of adipogenic differentiation capacity (17). ME3 cells lacking Irx3 cannot initiate differentiation, and show profoundly inhibited mitochondrial respiration and a significant decrease in Ucp1 and Pgc-1α levels when treating with brown adipocyte induction protocol (17). Accordingly, shRNA-induced Irx3 knockdown in preadipocytes from both iWAT and BAT significantly repressed the thermogenic capacity and Ucp1 expression in induced mature brown/beige adipocytes (at differentiation days 6–8) (16). However, the same group previously reported that silencing Irx3 in wild-type preadipocytes derived from noncarriers (of rs1421085) did not inhibit Ucp1 expression and thermogenic capacity at differentiation day 2, at which stage Ucp1 expression and lipid droplet formation were almost undetectable (14). The thermogenesis phenotypes of preadipocytes on different induction day could be in different condition, which would be much essential and valuable, the detailed mechanism underlying these discrepancies was needed to be studied discreetly. Additionally, differences in species, cell lines, induction cocktail composition, IRX3 dosage, and genetic background used by different groups are likely to lead to different outcomes.
In addition, another group presented evidence showing that partial inhibition of hypothalamic Irx3 by lentiviral knockdown led to diet-induced adiposity, possibly through increasing caloric intake and reducing energy expenditure (15), in contrast to the metabolic phenotypes observed in Ins2-Cre;EnR-Irx3 mice. Further research using other neuron-specific Irx3 knockout models is required to clarify the central regulatory roles of Irx3 in peripheral brown/beige adipocyte thermogenesis.
We previously reported that IRX3 directly binds to the Ucp1 promoter and enhances its transcription in vitro. Consistent with this result, in this study, we further showed that Ucp1 expression was increased n hIRX3-overexpressing brown adipocytes in vivo, as was the expression of several other thermogenesis-related genes, including Pgc-1α, Cidea, and Dio2. Whether IRX3 can regulate other genes, including the above genes that form part of the transcription complex that binds to the enhancer region of Ucp1 to increase thermogenesis, remains to be clarified. Moreover, RNA-seq analysis revealed a marked upregulation in the expression of mitochondria-related (such as Cox7a1, Cox8b, and Uqcrb) and lipid metabolism-related (such as Scd1 and Acaa2) genes under the condition of hIRX3 overexpression.
In summary, our research revealed that hIRX3 exerts a regulatory role in energy homeostasis by promoting thermogenesis in brown/beige adipose tissues. Adult humans have active depots of BATs, while the size and activity of BAT depots in obese individuals are largely reduced (33–36). Whether FTO variants (including rs1421085) contribute to thermogenesis, and the exact role of IRX3 in this process, merit further and urgent investigation.
The data presented in the study are deposited in online repositories, accession number can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Animal Care Committee of Shanghai Jiao Tong University School of Medicine.
JW, JH, and RL designed the experiments and supervised the study. ZZ, QW, YH, PL, DL, and MY carried out the animal and molecular experiments. ZZ analyzed the data. ZZ and JW wrote the manuscript. WG and JH contributed to text revision and discussion. All authors contributed to the article and approved the submitted version.
This work was supported by grants from National Key Research and Development Program of China (2018YFC1313802), the National Natural Science Foundation of China (91957124 and 81822009), and the Outstanding Academic Leader Project of Shanghai Municipal Health Commission (2018BR01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Chi-Chuang Hui (from the Hospital for Sick Children, and Department of Molecular Genetics, University of Toronto) for his comments and suggestions to this project.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.634191/full#supplementary-material
Supplementary Figure 1 | Generation of Ucp1-CreERT2; Rosa26hIRX3(iU-IRX3ov) mouse model. (A) Final Targeting Vector. Linearization site: NotI. (B) The sequence of the final targeting vector. (C) Overview of the targeting strategy. (D) DNA PCR screen of Ucp1-CreERT2 mice. Recombinant allele target band: 413bp; WT allele target band: 139bp. (E) Cre mRNA expression in varies tissue (relative to 36B4, normalized to control mice, n=4~6). (F) IHC for BAT UCP1 expression in Rosa26wild type(WT);Ucp1-CreERT2 (control) male mice after 7-days cold exposure. GFP antibody (CST 2956s, 1:200). Photos were taken by LSM710 (Zeiss, 40×). (G) Ucp1 protein expression in BAT of control and Ucp1-CreERT2 mice (Het in 3rd lane, HO in 4th lane). (H) The mRNA expression levels of hIRX3(top) and mIRX3 (below) in BAT, iWAT, eWAT of male iU-IRX3ov and control mice after CL316,243 and TMX injection (n = 9~10).(I-K) The protein expression levels of hIRX3 in BAT (I), iWAT (J) and eWAT (K) of male iU-IRX3ov and control mice after CL316,243 and TMX injection (n = 3~5).
Supplementary Figure 2 | Growth curve, body weight, physical activities and cold tolerance test in U-IRX3ov mice. (A, B) Growth curve of NCD fed U-IRX3ov and control from 4 weeks to 10 weeks. (A) Male body weight (n=6~8). (B) Female body weigth (n=9~11). (C-F) Body weight (C), fat mass percentage (D), lean mass percentage (E) and average food intake (F) of female U-IRX3ov and Ctrl (n = 9~11, average of 3 individual measurements). (G-I) 24-hour (left) and total (right) physical activities on X-axis (G), Y-axis (H) and Z-axis (I) of male U-IRX3ov and control mice. (J) Body temperature changes of male U-IRX3ov and control mice (n=9~10) for the first 6 h in 4°C cold room. (K) The mRNA expression levels of hIRX3 (left) and mIRX3 (right) in BAT, iWAT and eWAT of male U-IRX3ov and control mice after cold stimulation (n = 9~10).
Supplementary Figure 3 | Overexpression of hIRX3 from the embryonic stage enhances β3-AR agonist-induced thermogenesis in female U-IRX3ov mice (A-H). Tissue mass percentage (A), representative images of HE staining of BAT (top), iWAT (middle) and gWAT (bottom) (B) and adipocyte size distribution of iWAT(C), gWAT (D) of 10-week-old female U-IRX3ov and control mice after 1-h CL 316,243 (1.5 mg/kg body weight) injection, the mRNA expression levels of thermogenesis-related genes (n = 9~11) (E), the protein levels of Ucp1 and Pgc-1α in iWAT (n = 3) (F), the mRNA expression levels of thermogenesis-related genes (n = 6~8) (G), and the protein levels of Ucp1 and Pgc-1α in gWAT (n = 3) (H). Expression level of hIRX3 mRNA (I) and mIRX3 mRNA (J) in SVF of male U-IRX3ov mice BAT(n=6) and iWAT n=(4). Scale bars, 100 μm. Data are shown as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Table 1 | Mouse and human qPCR primers sequences.
Supplementary Table 2 | All down regulated genes in RNA-sequencing analysis in Figure 5.
Supplementary Table 3 | All up regulated genes in RNA-sequencing analysis Figure 5.
1. Scarlett K, Pattabiraman V, Barnett P, Liu D, Anderson LM. The proangiogenic effect of iroquois homeobox transcription factor Irx3 in human microvascular endothelial cells. J Biol Chem (2015) 290:6303–15. doi: 10.1074/jbc.M114.601146
2. Holmquist Mengelbier L, Lindell-Munther S, Yasui H, Jansson C, Esfandyari J, Karlsson J, et al. The Iroquois homeobox proteins IRX3 and IRX5 have distinct roles in Wilms tumour development and human nephrogenesis. J Pathol (2019) 247:86–98. doi: 10.1002/path.5171
3. Shabalina IG, Petrovic N, deJong JMA, Kalinovich AV, Cannon B, Nedergaard J. UCP1 in Brite/Beige adipose tissue mitochondria is functionally thermogenic. Cell Rep (2013) 5:1196–203. doi: 10.1016/j.celrep.2013.10.044
4. Wang W, Seale P. Control of brown and beige fat development. Nat Rev Mol Cell Biol (2016) 17:691–702. doi: 10.1038/nrm.2016.96
5. Wang J, Liu R, Wang F, Hong J, Li X, Chen M, et al. Ablation of LGR4 Promotes Energy Expenditure by Driving White-To-Brown Fat Switch. Nat Cell Biol (2013) 15:1455–63. doi: 10.1038/ncb2867
6. Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PloS Genet (2007) 3:1200–10. doi: 10.1371/journal.pgen.0030115
7. Liu C, Mou S, Cai Y. FTO gene variant and risk of overweight and obesity among children and adolescents: a systematic review and meta-analysis. PloS One (2013) 8:e82133. doi: 10.1371/journal.pone.0082133
8. Dina C, Meyre D, Gallina S, Durand E, Körner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet (2007) 39:724–6. doi: 10.1038/ng2048
9. Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med (2015) 373:895–907. doi: 10.1056/NEJMoa1502214
10. Stratigopoulos G, Burnett LC, Rausch R, Gill R, Penn DB, Skowronski AA, et al. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J Clin Invest (2016) 126:1897–910. doi: 10.1172/JCI85526
11. Hunt LE, Noyvert B, Bhaw-Rosun L, Sesay AK, Paternoster L, Nohr EA, et al. Complete re-sequencing of a 2Mb topological domain encompassing the FTO/IRXB genes identifies a novel obesity-associated region upstream of IRX5. Genome Med (2015) 7:1–14. doi: 10.1186/s13073-015-0250-3
12. Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gómez-Marín C, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature (2014) 507:371–5. doi: 10.1038/nature13138
13. Zhang SS, Kim KH, Rosen A, Smyth JW, Sakuma R, Delgado-Olguiń P, et al. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc Natl Acad Sci USA (2011) 108:13576–81. doi: 10.1073/pnas.1106911108
14. Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med (2015) 373:895–907. doi: 10.1056/NEJMoa1502214
15. de Araujo TM, Razolli DS, Correa-da-Silva F, de Lima-Junior JC, Gaspar RS, Sidarta-Oliveira D, et al. The partial inhibition of hypothalamic IRX3 exacerbates obesity. EBioMedicine (2019) 39:448–60. doi: 10.1016/j.ebiom.2018.11.048
16. Zou Y, Lu P, Shi J, Liu W, Yang M, Zhao S, et al. IRX3 Promotes the Browning of White Adipocytes and Its Rare Variants are Associated with Human Obesity Risk. EBioMedicine (2017) 24:64–75. doi: 10.1016/j.ebiom.2017.09.010
17. Bjune JI, Dyer L, Røsland GV, Tronstad KJ, Njølstad PR, Sagen JV, et al. The homeobox factor Irx3 maintains adipogenic identity. Metabolism (2020) 103:154014. doi: 10.1016/j.metabol.2019.154014
18. Tschöp MH, Speakman JR, Arch JRS, Auwerx J, Brüning JC, Chan L, et al. A guide to analysis of mouse energy metabolism. Nat Methods (2012) 9:57–63. doi: 10.1038/nmeth.1806
19. Subramaniana A, Tamayoa P, Moothaa VK, Mukherjeed S, Eberta BL, Gillettea MA, et al. Mesirova k aBroad. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U.S.A. (2005) 102:15545–50. doi: 10.3969/j.issn.0372-2112.2018.08.016
20. Symonds ME, Mostyn A, Pearce S, Budge H, Stephenson T. Endocrine and nutritional regulation of fetal adipose tissue development. J Endocrinol (2003) 179:293–9. doi: 10.1677/joe.0.1790293
21. Sun Y, Wang R, Zhao S, Li W, Liu W, Tang L, et al. FGF9 inhibits browning program of white adipocytes and associates with human obesity. J Mol Endocrinol (2019) 62:79–90. doi: 10.1530/JME-18-0151
22. Keipert S, Kutschke M, Ost M, Schwarzmayr T, van Schothorst EM, Lamp D, et al. Long-Term Cold Adaptation Does Not Require FGF21 or UCP1. Cell Metab (2017) 26:437–46. doi: 10.1016/j.cmet.2017.07.016
23. Zhao Y, Pan J, Wang Y, Zou Y, Guo L, Qian S, et al. Stimulation of histamine H4 receptor participates in cold-induced browning of subcutaneous white adipose tissue. Am J Physiol Metab (2019) 317(6):E1158–71. doi: 10.1152/ajpendo.00131.2019
24. Babenko V, Babenko R, Gamieldien J, Markel A. FTO haplotyping underlines high obesity risk for European populations. BMC Med Genomics (2019) 12:109–15. doi: 10.1186/s12920-019-0491-x
25. Peters U, North KE, Sethupathy P, Buyske S, Haessler J, Jiao S, et al. A systematic mapping approach of 16q12.2/FTO and BMI in more than 20,000 African Americans narrows in on the underlying functional variation: results from the Population Architecture using Genomics and Epidemiology (PAGE) study. PloS Genet (2013) 9:e1003171. doi: 10.1371/journal.pgen.1003171
26. Couto Alves A, De Silva NMG, Karhunen V, Sovio U, Das S, Taal HR, et al. GWAS on longitudinal growth traits reveals different genetic factors influencing infant, child, and adult BMI. Sci Adv (2019) 5:eaaw3095. doi: 10.1126/sciadv.aaw3095
27. Inagaki T. Regulations of Adipocyte Phenotype and Obesity by IRX3. Positive or Negative? EBioMedicine (2017) 24:7–8. doi: 10.1016/j.ebiom.2017.09.032
28. Tung YCL, Yeo GSH, O’Rahilly S, Coll AP. Obesity and FTO: Changing focus at a complex locus. Cell Metab (2014) 20:710–8. doi: 10.1016/j.cmet.2014.09.010
29. Cedernaes J, Benedict C. Human obesity: FTO, IRX3, or both? Mol Metab (2014) 3:505–6. doi: 10.1016/j.molmet.2014.05.003
30. Xue B, Rim JS, Hogan JC, Coulter AA, Koza RA, Kozak LP. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. J Lipid Res (2007) 48:41–51. doi: 10.1194/jlr.M600287-JLR200
31. Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, et al. Ribosomal Profiling Provides Evidence for a Smooth Muscle- Like Origin of Beige Adipocytes. Cell Metab (2014) 19:810–20. doi: 10.1016/j.cmet.2014.03.025.Ribosomal
32. Koizumi A, Sasano T, Kimura W, Miyamoto Y, Aiba T, Ishikawa T, et al. Genetic defects in a His-Purkinje system transcription factor, IRX3, cause lethal cardiac arrhythmias. Eur Heart J (2016) 37:1469–75. doi: 10.1093/eurheartj/ehv449
33. Somm E, Henry H, Bruce SJ, Aeby S, Rosikiewicz M, Sykiotis GP, et al. β-Klotho deficiency protects against obesity through a crosstalk between liver, microbiota, and brown adipose tissue. JCI Insight (2017) 2. doi: 10.1172/jci.insight.91809
34. Riis-Vestergaard MJ, Richelsen B, Bruun JM, Li W, Hansen JB, Pedersen SB. Beta-1 and not Beta-3 adrenergic receptors may be the primary regulator of human brown adipocyte metabolism. J Clin Endocrinol Metab (2020) 105:E994–E1005. doi: 10.1210/clinem/dgz298
35. Chevalier C, Stojanović O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell (2015) 163:1360–74. doi: 10.1016/j.cell.2015.11.004
Keywords: IRX3, Ucp1, thermogenesis, obesity, overexpression
Citation: Zhang Z, Wu Q, He Y, Lu P, Li D, Yang M, Gu W, Liu R, Hong J and Wang J (2021) IRX3 Overexpression Enhances Ucp1 Expression In Vivo. Front. Endocrinol. 12:634191. doi: 10.3389/fendo.2021.634191
Received: 27 November 2020; Accepted: 04 February 2021;
Published: 10 March 2021.
Edited by:
Jae B. Kim, Seoul National University, South KoreaReviewed by:
Lei Sun, National University of Singapore, SingaporeCopyright © 2021 Zhang, Wu, He, Lu, Li, Yang, Gu, Liu, Hong and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ruixin Liu, eGluZXIxOTgyODdAMTYzLmNvbQ==; Jie Hong, aG9uZ2ppZUBtZWRtYWlsLmNvbS5jbg==; Jiqiu Wang, d2FuZ2pxQHNoc211LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.