
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol., 07 April 2021
Sec. Translational and Clinical Endocrinology
Volume 12 - 2021 | https://doi.org/10.3389/fendo.2021.632335
This article is part of the Research TopicEmerging Roles of the Gut Microbiota in the Pathogenesis of Metabolic DisordersView all 11 articles
Mounting evidence suggested that the gut microbiota has a significant role in the metabolism and disease status of the host. In particular, Type 2 Diabetes (T2D), which has a complex etiology that includes obesity and chronic low-grade inflammation, is modulated by the gut microbiota and microbial metabolites. Current literature supports that unbalanced gut microbial composition (dysbiosis) is a risk factor for T2D. In this review, we critically summarize the recent findings regarding the role of gut microbiota in T2D. Beyond these associative studies, we focus on the causal relationship between microbiota and T2D established using fecal microbiota transplantation (FMT) or probiotic supplementation, and the potential underlying mechanisms such as byproducts of microbial metabolism. These microbial metabolites are small molecules that establish communication between microbiota and host cells. We critically summarize the associations between T2D and microbial metabolites such as short-chain fatty acids (SCFAs) and trimethylamine N-Oxide (TMAO). Additionally, we comment on how host genetic architecture and the epigenome influence the microbial composition and thus how the gut microbiota may explain part of the missing heritability of T2D found by GWAS analysis. We also discuss future directions in this field and how approaches such as FMT, prebiotics, and probiotics supplementation are being considered as potential therapeutics for T2D.
Diabetes is a metabolic disorder characterized by elevated blood glucose levels. The incidence of diabetes is widespread and the International Diabetes Federation (IDF) reports that 463 million people in the world are suffering from diabetes, which is estimated to reach 700 million by the year 2045 (1). In the USA, 13% of adults aged 18 or older have diabetes (2). Diabetes has been categorized into three classes (type 1, type 2, and gestational diabetes) depending on the underlying pathophysiology. Among them, type 2 diabetes (T2D) accounts for more than 90% of all diabetes (1, 2). Though genetic susceptibility is a critical determinant of T2D, non-genetic factors, including diet, physical activity, also play a significant role in the development and severity of T2D. The recent understanding that microbiota is a critical determinant of human health has opened a new avenue of basic and clinical research for T2D.
The microbiota refers to an assemblage of living microorganisms including bacteria, archaea, and fungi present in a defined environment (3). The microbiota can reside within or on the host and can modulate nutritional status, health, and diseases of the host (4). The most widely used technique to assay microbiota is to quantify the variable region(s) of the 16S rRNA gene (5). 16S rRNA gene analysis is economical and straightforward, however, it provides a limited regulation of taxonomic information, up to the genus level reliably. Alternatively, metagenomics, a whole-genome shotgun sequencing approach of all the DNA present in the sample, provides much better taxonomic resolution down to species or strain level. Additionally, metagenomics encompasses the collective functional genomes of all microorganisms, thus provides an opportunity for functional profiling of the metabolic pathways present in a community (6). The diversity and composition of the gut bacteria have been intensely studied, as well as their impact on health and diseases (7), including obesity (8), inflammation (9), and T2D (10). A better understanding of the link between the gut microbiota and metabolic disorders, especially T2D, may lead to advances in current treatment approaches, accurate disease monitoring, and development of novel therapeutics.
In addition to sex and age, both diet and the immune system contribute to the composition of the microbiota (11). The underlying architecture of the host’s genetics may also shape the community structure of the gut microbiota (12). Several genetic variants are associated with T2D susceptibility (13) and it is speculated that a part of “missing heritability” described in genome-wide association study (GWAS) studies (14) may be explained by gut microbiota. Moreover, growing evidence suggests that gut microbial metabolites regulate gene expression through a variety of classic signaling pathways (14) and more recently epigenetics (15). Thus, understanding the complex interactions between microbiome, microbial metabolome, and host genome will assist the development of novel therapeutics.
In this review, we critically summarize the recent developments describing the role of microbiota on T2D susceptibility, development, and severity. In particular, we focus on the underlying biochemical mechanisms by which gut microbiota may affect T2D. A number of these mechanisms may be mediated by the host genetics, and epigenetics thus may be viable targets for precision medicine. The potential effects of prebiotics, probiotics, medication, and intermittent fasting on the microbiota and T2D are extensively discussed. Finally, we comment on the future direction in this field.
A growing number of studies suggest that gut microbiota influences T2D susceptibility, development, severity, and progression. Dysbiosis, an alteration of a healthy microbiota, is associated with obesity, low-grade inflammation, insulin resistance, and T2D which potentially reflects a causal role linking these pathologies (16). Along with animal studies, numerous human cohorts also have reported specific gut bacteria enriched or depleted in T2D patients compared to healthy controls. A summary of the recent reports of altered microbiota found in T2D patients is depicted in Table 1, and the interactions between environmental factors, genetics, microbiota, microbial metabolites, obesity, inflammation, and T2D are shown in Figure 1.
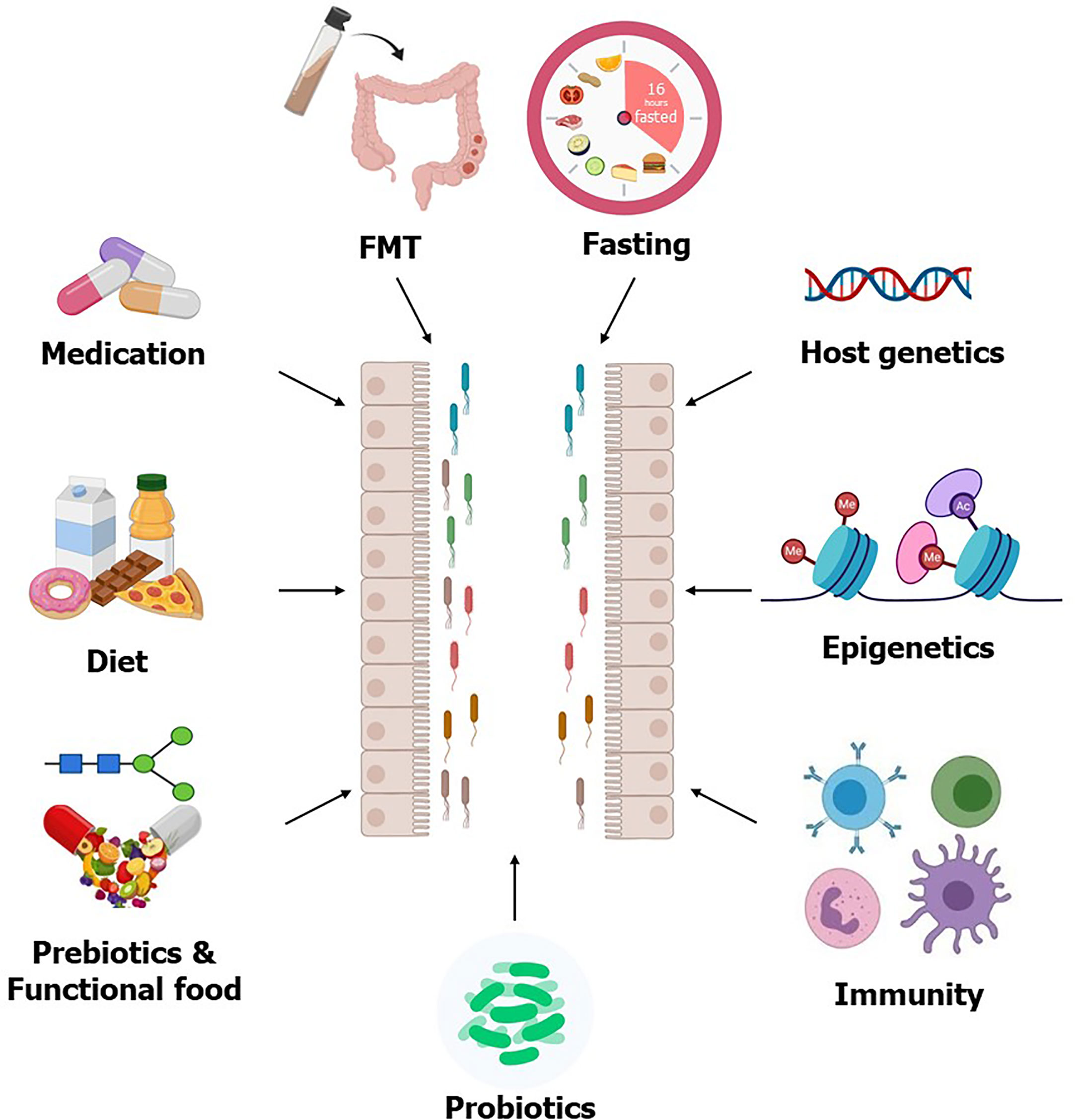
Figure 1 Factors affecting gut microbiota. The gut microbial composition can be modulated by different interventions such as prebiotics, probiotics, FMT, and intermittent fasting, all of which are considering as potential therapeutics for T2D. Host genetics, epigenetics, and immunity also modulate gut microbiota. Some T2D medication improves circulating glucose levels partly through modulating gut microbiota, which further supports the usability of the gut microbiota as therapeutics for T2D.
Landmark studies in the 2000s (27–29) demonstrated that the microbiota contributes to digestion, carbohydrate metabolism, obesity, and plasma glucose levels. Additionally, those studies established a causal relationship by showing that the susceptibility to obesity could be transferred between mice when the fecal microbiota of obese mice was transplanted into non-obese animals. Consistent with these findings, several other studies have reported enrichment or depletion of specific obesity-related gut bacteria and indicated a connection between gut microbiota, adiposity, and T2D. For example, increased abundance of Prevotella and decreased abundance of Bacteroides were associated with a higher risk of obesity with metabolic syndrome, while body mass index and body fat percentage were negatively correlated with Coprococcus abundance (30). More recent data suggests that the abundance of a bacteria in the Bacilli family was positively associated with fat mass, and negatively associated with lean mass and plasma glucose level (31). Additionally, Peptostreptococcaceae, Blautia, and a bacterium related to the Clostridiaceae family were positively associated with plasma glucose levels (31). In a recent randomized, double-blind, placebo-controlled clinical trial with overweight or obese insulin-resistant subjects, pasteurized Akkermansia muciniphila supplementation was associated with weight loss, improved insulin sensitivity, and reduced insulinemia (32). A potential mechanism of these positive effects is an interaction between temperature stable outer membrane protein Amuc 1100 found in pasteurized Akkermansia muciniphila and Toll-like receptor 2 (33).
Obesity and dysbiosis may cause low-grade inflammation (Figure 2) which also contributes to insulin resistance and the development of T2D. Several studies have demonstrated associations between gut microbiota, or microbial components, and low-grade inflammation in T2D (34). An array of bacterial components such as lipopolysaccharides (LPS) (35), flagellin (36), and peptidoglycan (37) can elicit an inflammatory response. LPS binds to immune cell receptors such as Toll-like receptors and Nucleotide Oligomerization Domain (NOD)-like receptors and triggers the expression of proinflammatory mediators that fuel chronic inflammation, promoting metabolic dysregulation and development of T2D (38). The interaction of specific microbes in the gut with the immune system is complex. Some gut bacteria and microbial components promote low-grade inflammation, while others stimulate anti-inflammatory cytokines and chemokines. For example, induction of interleukin (IL)-10 and IL-22 by species of Roseburia, Bacteroides, Akkermansia, and Lactobacillus (33, 39–43) may contribute to restoring insulin sensitivity and improving glucose metabolism (43, 44). Similarly, Bacteroides thetaiotaomicron, Roseburia intestinalis, Clostridium clusters IV, and XIVa induce Treg cells (45, 46), which are tolerogenic immune cells and are important for maintaining a balance between pro and anti-inflammatory immune responses (47). Additionally, butyrate produced by the gut microbiota enhances colonic Treg differentiation through epigenetic modification of histone deacetylase inhibition (48, 49) and is discussed in detail below along with other short-chain fatty acids (SCFAs). Inhibition of pro-inflammatory cytokines and chemokines is another pathway that beneficial microbes use to prevent low-grade inflammation. Various species of Lactobacillus, Bacteroides, Roseburia, and Akkermansia can decrease pro-inflammatory cytokines such as IL-1β, IL-6, IL-8, IL-17, and tumor necrosis factor (TNF)-α (40, 50–52). Conversely, Fusobacterium nucleatum and Ruminococcus gnavus can increase inflammatory cytokine production. Therefore, depending on the composition, the gut microbiota may contribute to increased or decreased low-grade inflammation, impacting insulin sensitivity and T2D.
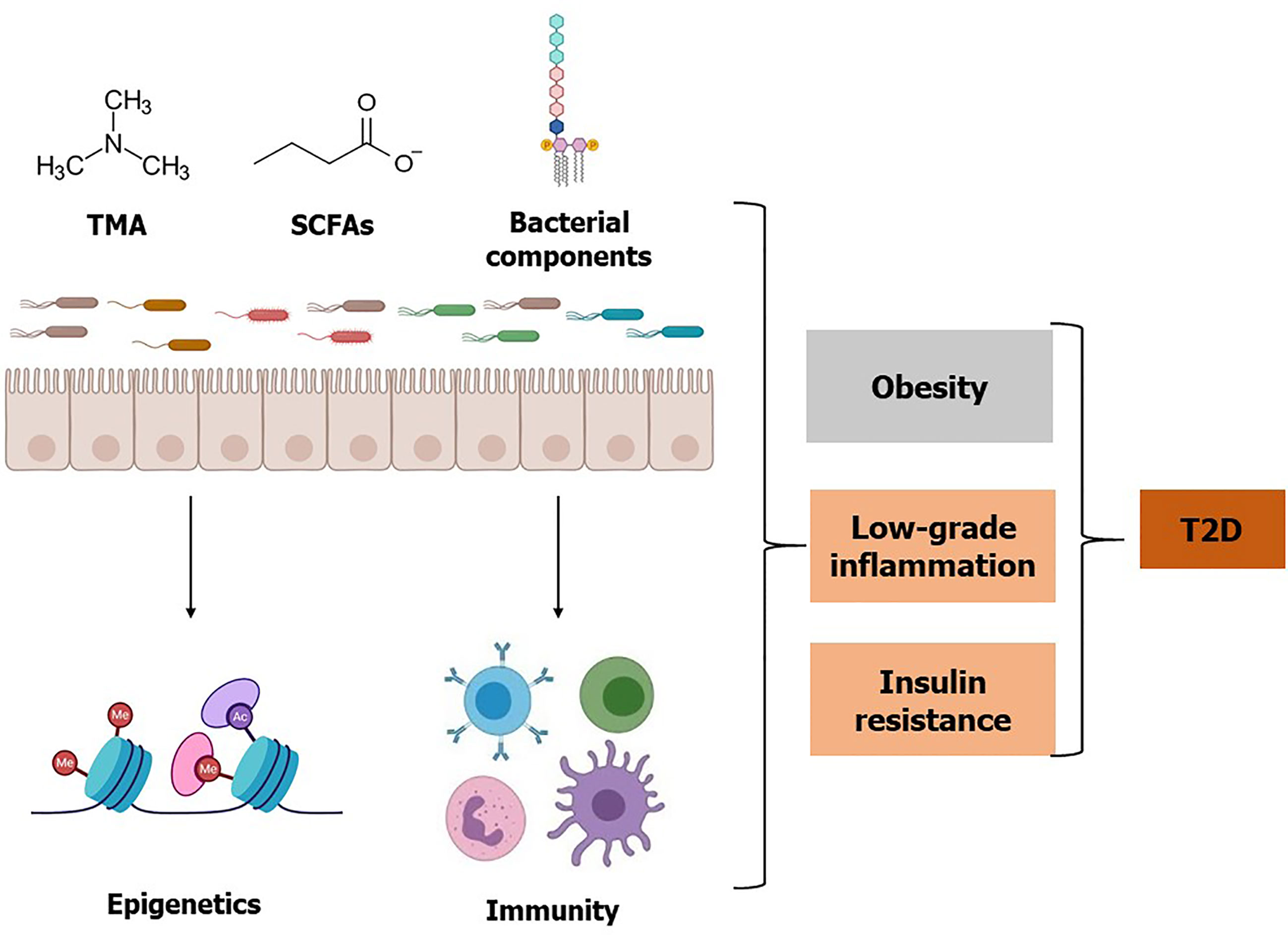
Figure 2 Effects of gut microbiota, microbial metabolites, and bacterial components on T2D. Gut microbiota and specific bacterial taxa are associated with a risk of obesity, low-grade inflammation, and insulin resistance. Microbial metabolite TMA is converted to TMAO by the host enzyme and elevated TMAO is associated with insulin resistance. Whereas some bacterial metabolites such as SCFAs may improve glucose homeostasis. Additionally, SCFAs influence epigenetic programming by inhibiting histone deacetylase enzyme activity, which may improve insulin resistance and T2D. Besides live bacteria, bacterial components such as LPS, flagellin, and peptidoglycan can elicit an inflammatory response and may contribute to the increased risk of T2D. Conversely, some bacterial components such as Amuc 1100 can improve T2D. Referred studies can be found on the main body of this review.
Gut microbiota profiling performed on large cohorts of T2D patients has found the abundance of several bacteria enriched or depleted in T2D subjects compared to controls (Table 1). In general, T2D patients commonly have a decreased abundance of SCFA producing bacteria (Eubacterium rectale, Faecalibacterium prausnitzii, Roseburia intestinalis, Roseburia inulinivorans, Akkermansia, and Bifidobacterium) and tryptophan metabolite producing bacteria (Lactobacillus, Bacteroides, Bifidobacterium, Peptostreptococcus, Ruminococcus, Ruminiclostridium, and Clostridium), and an increased abundance of opportunistic pathogens (Bacteroides caccae and Clostridium hathewayi), branch chain amino acid synthesizing bacteria (Bacteroides vulgatus and Prevotella copri), and sulfate-metabolizing bacteria (Desulfovibrio, Lactobacillus gasseri, and Lactobacillus reuteum) compared to healthy controls (17–22, 26, 53). However, we note that not all the data derived from observational studies have been consistent. For example, one study comparing significant differences in gut microbiota diversity between T2D patients and healthy individuals was conducted on fecal samples from 18 men (24). In this study, decreased Clostridia and increased Bacteroidetes and Proteobacteria were observed, while overall diversity of the gut microbiota was positively correlated with plasma glucose levels in T2D patients. However, these results have not been identified in three large-scale metagenomics analyses performed in Europe and China (17, 18). A decrease in Prevotella was observed in 50 Japanese T2D patients compared to the healthy subjects (53), but in studies of 291 Nigerians and 171 Chinese, increased Prevotella abundance was associated with T2D (23, 25). The reason for the discrepancy between studies may be due to a number of confounding variables such as diet, genetics, medication use, and sequencing techniques. Utilizing alternative approaches and developing new technologies are of critical need to determine which of the associations between the microbiota and T2D are causal and which of the microbial differences are responsive to T2D.
Beyond the direct effects of bacteria in the microbiota or their components, such as LPS, we now appreciate that the microbiota is a metabolically active “organ” that produces small biomolecules. In the following section, we highlight two important bacteria-derived metabolites TMAO and SFCAs, and briefly comment on other metabolites important for T2D. Gut microbiota is involved in the digestion of food ingredients and produces metabolites supporting physiological function in the human body (54). Microbial metabolites assist our interpretation of the underlying mechanisms by which gut bacterial taxa influence host health and disease (Figure 2 and Table 2).
Trimethylamine (TMA) is produced by intestinal microbial metabolism of dietary choline and carnitine and is transported to the liver via the portal vein. In the liver, TMA is converted to TMAO by the activity of flavin-containing monooxygenase 3 (FMO3) (70, 71). In vivo studies have identified several candidate microbial taxa associated with TMA/TMAO production including Anaerococcus hydrogenalis, Clostridium asparagiforme, C. hathewayi, C. sporogenes, Desulfovibrio desulfuricans, Edwardsiella tarda, Escherichia fergusonii, Proteus penneri, and Providencia rettgeri (55). TMAO concentrations are elevated in T2D patients, suggesting that this pathway is associated with T2D (72, 73). However, it is not yet clear if the elevated TMAO has a direct causal effect on T2D development or if it is a consequence of T2D. Animal studies have shown that TMAO consumption impairs glucose tolerance by mediating the insulin signaling pathway in the liver and upregulates the expression of pro-inflammatory mediators in adipose tissue (56). Reduction of plasma TMAO by FMO3 knockdown also decreases plasma glucose and insulin levels, whereas FMO3 overexpression increases plasma glucose level and induces insulin resistance (74). A similar relationship between TMAO and T2D may exist in humans as circulating TMAO concentration was found to be significantly higher in T2D patients compared to control subjects observed in a meta-analysis (75). In contrast, a recent Mendelian randomization analysis suggests that elevated circulating TMAO is a consequence of T2D not causal (76). Additional studies on the relationship between TMAO and T2D are needed to clarify these results.
SCFAs are microbial metabolites produced in the colon and are known to have a wide range of biochemical effects on the host (77). Anaerostipes, Blautia, Coprococcus, Eubacterium, Faecalibacterium, Marvinbryantia, Megasphaera, Roseburia, and Ruminococcus are among the primary gut microbes that produce SCFAs. The SCFAs acetate and butyrate improve glucose homeostasis by inducing intestinal production of glucagon-like peptide-1 (GLP-1) and peptide YY (PYY). These peptides in turn stimulate insulin secretion, suppress appetite, and slow gastric emptying (78–80). GLP-1 is released from colonic enteroendocrine L cells that are distributed along the length of the intestinal epithelium and are in direct contact with the gut microbiota (81). Additionally, SCFAs can regulate the intestinal immune system through G protein-coupled receptors (GPCRs) such as GPR41 and GPR43 (82). Reduced abundance of SCFA producing bacteria was observed in T2D patients suggesting that this pathway is altered in T2D (17, 83). A clinical trial demonstrated that selective enrichment of SCFA producing bacteria, achieved by dietary fiber supplementation, was associated with lower hemoglobin A1c (HbA1c) levels and improved glucose metabolism (84). Stool samples collected prior to and following the intervention were then mechanistically tested via adoptive transfer experiments using mice to establish causality (84). Overall, SCFAs are involved in glucose and lipid metabolism via activation of SCFA receptors (85). Therefore, SCFA could be an intermediate phenotype by which microbiota provides a beneficial effect of T2D prevention.
Besides TMAO and SCFAs there are several other microbial metabolites such as tryptophan catabolites: tryptamine, indole, indolelactic acid (ILA), indolepropionic acid (IPA), indoleacetic acid (IAA), indoleacrylic acid (IA), indolealdehyde (IAld), and 3-methylindole (skatole) [reviewed in (62)]; bile acids: deoxycholic acid (DCA) and lithocholic acid (LCA) [reviewed at (65, 86)], histidine catabolite: imidazole propionate (Imp) (87), branched-chain amino acids [reviewed at (88)], and hydrogen sulfide (84) have been investigated in the context of T2D (89–92). These microbial metabolites are involved in the regulation of host metabolism, immunity, gene expression, and intestinal integrity, creating an important link between the gut microbiota, and insulin resistance and T2D development. Individually, these results are intriguing but cumulatively the results are complex and heterogeneous. Alterations in the experimental system, environment, diet, or even circadian rhythms (93) all have been implicated as sources of variation contributing to the heterogeneity in the literature. Of particular interest to this review, the overall variation in the microbiota composition was better captured by a 17-day diet history (94). Therefore, information from longitudinal sampling and metabolomics is required to identify precise and dynamic interactions between diet, microbiota, and host.
In addition to environmental factors including gut microbiota and microbial metabolites, host genetic architecture is associated with T2D (13). Many studies demonstrate that host genetics influences the community structure of gut microbiota in humans. This opens several interesting hypotheses regarding host-microbe symbiosis and perhaps the microbiota as a mediating variable contributing to the missing heritability in GWAS. More specifically, the contribution of genetic polymorphisms associated with T2D may be partly mediated through gut microbiota. One could wonder if intervention in the gut microbiota may improve T2D in susceptible individuals. Here we provide some recent evidence implicating interactions between host genetics and the microbiome that affect T2D.
Human studies of monozygotic and dizygotic twins have demonstrated that host genetics contribute to the composition of the microbiota (12), by tolerating or rejecting several microbial taxa. For example, the abundance of Bifidobacterium, an important commensal bacterium for T2D, is associated with host genotype at the lactase gene locus (LCT, rs4988235, and rs1446585) (95). Individuals with the GG genotype have reduced lactase activity and harbor higher levels of Bifidobacterium in their gut. Mechanistically these individuals provide more lactose to the bifidobacteria for utilizing as an energy source, which enriches bifidobacteria in their gut. Establishing if an increase in Bifidobacterium due to LCT genotype affects T2D remains to be determined.
In addition to the LCT locus and Bifidobacterium, genetic studies are beginning to identify some bacteria associated with specific genetic loci. For example, a recent GWAS found an association between Ruminococcus and rs150018970 near the gene RAPGEF1 (96). RAPGEF1 is a signaling protein that transduces signals from GPCRs, which are involved in the regulation of gastrointestinal tract physiology, such as metabolism, immune cell differentiation, and tissue repair (97). Similarly, another study (98) found a quantitative trait locus for Butyricicoccus at the locus of SLC5A11 (rs72770483), which encodes a sodium-dependent myo-inositol/glucose co-transporter protein (99). These studies underscore the influence of host genetics on gut microbial colonization. However, further studies will be needed to determine to what extent host genetics affects the gut microbiota and T2D.
In addition to interactions between the host genotype and microbiota composition, we are now beginning to appreciate that microbial metabolites can influence host gene expression through epigenetic mechanisms (15). Thus, there seem to be bi-directional interactions with effects on both the host and specific bacteria of the microbiome. For example, in various tissues, including proximal colon, liver and white adipose tissue, microbial metabolites such as SCFAs influence epigenetic programming by inhibiting histone deacetylase (HDAC) enzyme activity, (100), which promotes de-condensation and relaxation of chromatin and increases chromatin accessibility to transcription factors (101). In particular, Faecalibacterium prausnitzii is one of the most abundant anaerobic bacteria in the healthy human gut that produces butyrate. Butyrate targeted inhibition of HDAC1 may have anti-inflammatory effects and ultimately improve insulin sensitivity by downregulating the IL-6/STAT3/IL-17 pathway (102). Butyrate may also influence differentiation of Th17 and Treg cells through enhanced Forkhead Box P3 (Foxp3) expression (102). In human adipose tissue, the epigenetic regulation of the expression of genes involved in glucose and energy homeostasis, such as insulin-like growth factor 2 mRNA binding protein 2(IGF2BP2), is associated with gut bacterial populations (103). These data support the idea that the gut microbiota could act as an epigenetic regulator in T2D (104). A genome-wide DNA methylation analysis of isolated human pancreatic islet cells harvested from donors with and without T2D revealed 853 unique differential DNA methylation genes, including 17 genes previously identified in GWAS such as TCF7L2, THADA, KCNQ1, FTO, and IRS1 associated with the risk of T2D (105). This reinforces the idea that genetic and epigenetic mechanisms may interact to affect pancreatic β-cell function, development of insulin resistance, and T2D. Understanding if the microbiota specifically aids the host epigenetic changes associated with T2D could be important in the development of novel therapies T2D or comorbidities such as obesity.
The associations between gut microbiota, microbial metabolites, and T2D, opened a new perspective for potential novel therapeutics for T2D. Several gut microbiota targeted therapeutics including fecal microbiota transplantation (FMT), medication, and dietary choices could be useful therapeutic strategies to manage T2D (Figure 1). Several clinical trials to evaluate the impact of these potential therapeutic agents on T2D are currently completed or in progress (Table 3).
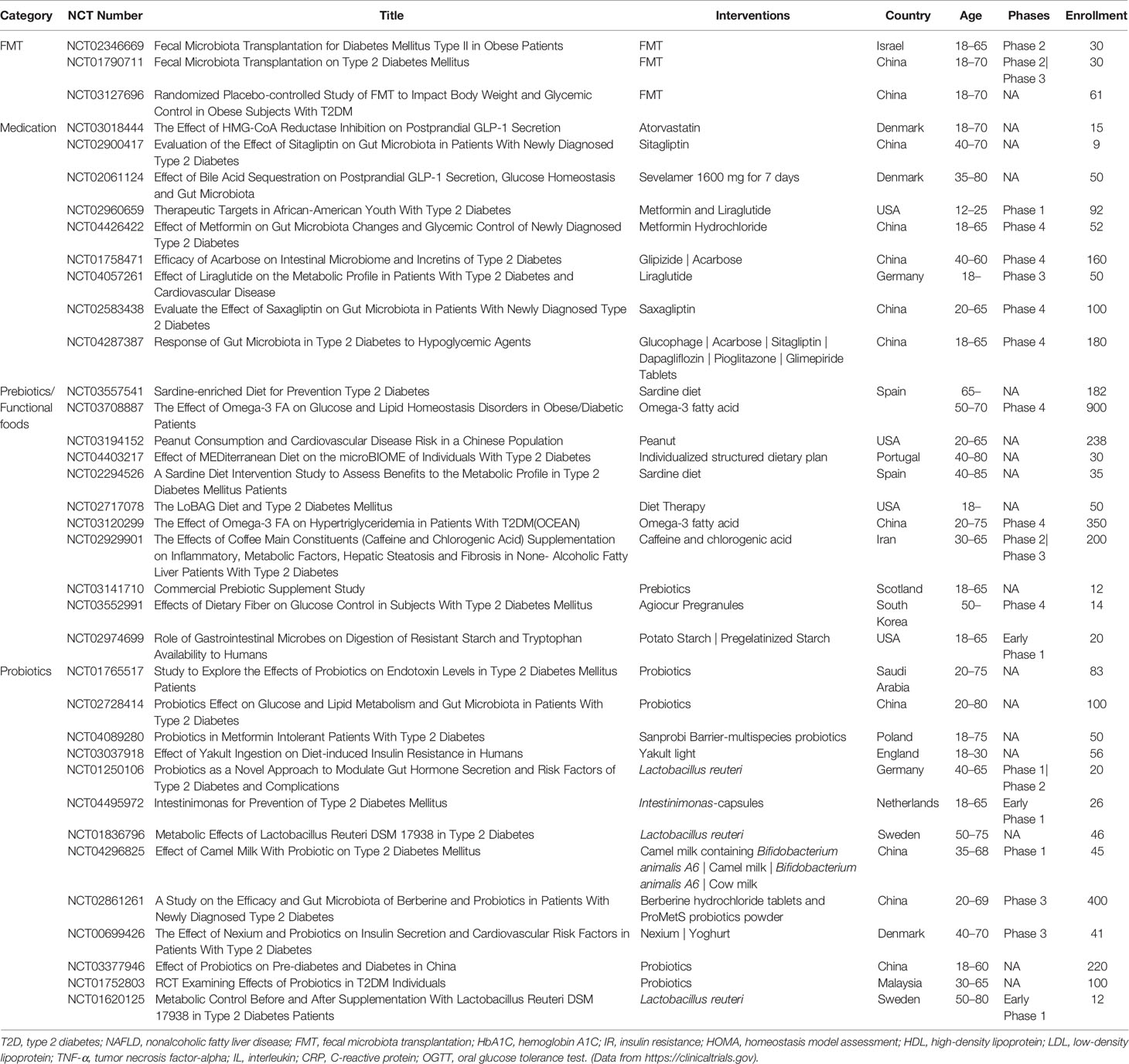
Table 3 Ongoing or completed clinical trials on T2D with FMT, medication, prebiotics/functional foods, or probiotics.
FMT has gained attention over the past few years as a research method demonstrating the contribution of gut microbiota to a disease state. Most clinical trials with FMT have been performed in patients with Clostridium difficile infections (106, 107) and these studies have been successful. As an extension of these studies, several additional diseases such as T2D have been suggested responsive to microbiota transplantation (108). In rodent models, insulin sensitivity significantly improved after transferring microbiota in MyD88 deficient NOD mice (109). Similar studies where human microbiota from healthy Chinese subjects are transplanted into diabetic db/db mice remarkably lowers fasting blood glucose concentrations (110). Likewise, transplantation of fecal samples of patients treated with metformin into germ-free mice improves glucose tolerance (111). A limited number of studies have begun to suggest that FMT from lean subjects into patients improves insulin sensitivity which could be in part due to increased butyrate-producing bacteria (108). One study examined the effects of lean donor versus self-FMT on metabolic syndrome patients and found that insulin sensitivity improves significantly at 6-weeks after FMT in male recipients with the metabolic syndrome (112). However, FMT treatment sometimes failed to improve targeted clinical phenotypes. For example, one study failed to show reduced TMAO levels in the recipient of FMT from a vegan donor (112), who have altered intestinal microbiota compared to omnivores (114) and low production of TMAO (115). In addition to inconsistent results, the long-term effects of FMT have not been adequately examined. Thus, further studies are needed to evaluate the long-term effectiveness and potential side-effect of FMT in humans.
Metformin is a widely known common treatment for T2D but the exact mechanisms underlying the hypoglycemic effect are not yet fully understood. Metformin has been shown to have an inhibitory effect on T2D by activating AMP-activated protein kinase (AMPK) or inhibiting mitochondrial respiration and glycerophosphate dehydrogenase (116–118). Recently, evidence has been reported suggesting that the composition of the gut microbiota mediates the efficacy of metformin to lower blood glucose levels. The fact that intravenous injection of metformin, unlike oral administration, does not lower hyperglycemia, suggests that gut microbiota is an important part of metformin action (119). Indeed, metformin shifts the composition of gut microbiota in both mice and humans, making them more similar to the microbiota of a healthy host (111, 120, 121). Some of these gut microbiota changes have also been seen in healthy people who have not responded to glycemic control to metformin treatment, thus suggesting shifts in the gut microbiota induced by metformin itself, rather than simply reflecting lowered blood glucose level. Metformin influences the abundance of several microbial taxa, including increased abundance of A. muciniphila, Bifidobacterium bifidum, Bilophila wadsworthia, Escherichia, Lactobacillus, Shigella spp. as well as a reduced abundance of Clostridium spp. and Intestinibacter spp. (111, 122, 123). Regarding the effects of these changes on blood glucose, metagenomic analysis of microbial composition demonstrates changes in various functional pathways affecting the production of propionate and butyrate (124, 125). Metformin stimulates the activity of endocrine cells by regulating bile acid conversion, improving intestinal permeability, reducing endotoxin levels, and enhancing the release of GLP-1 and PYY peptides (126). Metformin also decreases the TMA level and the growth of bacteria that produce it in the gut, and thus the circulating TMAO level in mice (127). The fact that transferring the microbiota from metformin-treated mice improves metabolic traits in aged mice indicates that the shifts in the gut microbiota by metformin treatment are beneficial (111). The effect of the microbiota on the efficacy of metformin remains unclear as a recent study found that metformin’s ability to improve T2D in mice was not affected by the elimination of gut microbiota using gnotobiotic mice or antibiotics (128). Although previous studies did not directly demonstrate the role of gut microbiota in improving glycemic control by metformin, it is suggested that the anti-inflammatory activity of metformin could potentially play a role in eliciting some beneficial effects regardless of the gut microbiota.
Another anti-diabetic drug with a link to the microbiota is Acarbose, an α-glucosidase inhibitor. Acarbose suppresses the conversion of oligosaccharides to monosaccharides and disaccharides, delays the absorption of glucose in the intestine, and lowers blood glucose levels after a meal. Due to its effects on carbohydrate metabolism, Acarbose has been hypothesized to affect microbiota composition. In T2D patients Acarbose treatment alters the gut microbiota. The abundance of Dialister, B. longum, Faecalibacterium, and Lactobacillus increases, while the abundance of Butyricoccus, Phascolarctobacterium, and Ruminococcus is reduced. These changes in composition may improve gut health as evidenced by the decrease in circulating LPS levels (129–131). This alteration in the gut microbiota composition after Acarbose treatment suggests that the therapeutic effect of Acarbose may be partially mediated through microbiota. Whether these changes in microbial composition contribute to acarbose’s effect on lowering blood glucose has not been extensively studied. Similarly, liraglutide, a GLP-1 receptor agonist, stimulates satiety, slows gastric emptying, inhibits glucagon, and promotes insulin secretion. In animal studies, liraglutide increased the abundance of A. muciniphila, Allobaculum, Anaerostipes, Blautia, Butyricimonas, Desulfovibrio, Lactobacillus, Turicibacter, and SCFAs producing bacteria and decreased the abundance of Bacteroidales, Clostridiales, Proteobacteria (132, 133). These data suggest that the beneficial effect on hyperglycemia these drugs have, may in part be through the gut microbiota, although further clinical studies are needed.
Probiotics are live microorganisms that have a beneficial effect on human health (134). Various beneficial effects of taking probiotics have been reported, including improving gut health, alleviating symptoms of lactose intolerance, inhibiting the growth of pathogenic bacteria, producing SCFAs, balancing pH, and stimulating the immune system (135). The use of probiotics to manage T2D is of interest, but a limited number of studies have evaluated the effects in clinical settings. Preliminary studies indicated that alteration of the gut microbial composition by probiotics supplementation might improve T2D by reducing pro-inflammatory cytokines, intestinal permeability, and oxidative stress [reviewed at (136)]. Several bacterial species are used in commercial probiotics supplement products, including Bifidobacterium longum subsp. infantis, Lactobacillus, Streptococcus, Pediococcus, and Lactococcus species (137). L. gasseri, Lactobacillus helveticus, Lactobacillus casei, and Bifidobacterium bifidum probiotic reduce fasting blood glucose levels with HbA1c (138–140). Mechanistically, these probiotics have been shown to have antioxidant and immunomodulatory effects by reducing oxidative stress (140), reducing inflammatory molecules, and inhibiting effector functions of CD4+ T-cells (142), which may influence on the reducing blood glucose levels and T2D risk. A randomized, double-blind, placebo-controlled trial of administration of A. muciniphila in overweight/obesity insulin-resistant volunteers improved insulin sensitivity and reduced insulinemia, plasma total cholesterol, body fat mass, hip circumference, and level of blood markers associated with liver dysfunction and inflammation (32). Recent meta-analysis studies showed that the probiotic supplementation improved the fasting blood glucose, HbA1c, and homeostatic model assessment for insulin resistance (HOMA-IR) in T2D patients and thus can be recommended as complementary advice alongside medicine and lifestyle modifications for T2D treatment (143, 144).
Prebiotics are the non-digestible food ingredients that beneficially affect the host by selectively stimulating the growth and (or) the activity of one or a limited number of bacterial species already resident in the colon (145). Inulin, a linear β-2,1 fructosyl-fructose polydisperse carbohydrate material with or without a α-D-glucose moiety (146), is one of the most studied prebiotics. Inulin-type fructooligosaccharide (ITF) improved glycemia by increasing the production and release of the active forms of GLP-1 from the cecum and proximal colon and reducing plasma ghrelin concentration in the rat (147). The direct effect of inulin supplementation on the T2D is not conclusive in human clinical trials. One study reported that dietary inulin reduced fasting blood glucose, body weights, glycated hemoglobin, plasma LPS, IL-6, TNF-α and IL-17A in T2D patients (148). A recent placebo-controlled crossover clinical trial (149) found enrichment of Bifidobacterium and Bacteroides with a significantly higher fecal SCFAs concentration due to ITF consumption compared to placebo (150). Additionally, the relative abundance of Cyanobacteria and Bacteroides is increased, and a reduction in the relative abundance of Ruminiclostridium, Deferribacteres, and Tenericutes is observed due to inulin supplementation, indicating that the dietary inulin alleviates T2D via suppressing inflammation and modulating gut microbiota (148). A recent systemic review (151) has summarized clinical trials conducted to evaluate the effect of dietary inulin on Akkermansia muciniphila, which are usually present at a higher abundance in healthy individuals compared to T2D patients and found an increased abundance in the treatment group compared to controls. However, others found no effect largely due to interindividual variation at the baseline T2D phenotypes (152).
It should be noted that a symbiotic mixture of prebiotics and probiotics (134), supplementation could provide a better beneficial effect compared to prebiotic or probiotic alone (153). For example, Lactobacillus acidophilus DSM20079 induces 14·5-times more butyrate in the presence of inulin or pectin than glucose (154). Berberine, a natural plant alkaloid extracted from Berberis aristata and Coptis chinensis, is reported to be an effective remedy for T2D (155). A recent randomized, double-blind, placebo-controlled trial conducted in China demonstrates that administration of berberine with probiotics improves HbA1C levels compared to the group treated with berberine alone (156). A meta-analysis of randomized controlled trials reported that diets supplemented with either prebiotics or symbiotics improved fasting blood glucose and HbA1C in patients with T2D (157). Therefore, symbiotic products that selectively stimulate and (or) activate metabolism of probiotics could be recommended to effectively lower the risk of T2D.
Intermittent fasting (IF) is defined as a periodic dietary restriction, which has been shown to increase lifespan, and to reduce the risk of developing various age-related pathologies including T2D (158). Animal studies of IF have reported an improvement in body composition, glucose and lipid metabolism, decreased inflammation, and autophagy (159) and gut microbiota might play a pivotal role in this process (160, 161). Though most of the human IF studies show a beneficial effect, the results are not completely conclusive. Two recent reviews summarize the recent literature on the effect of IF on T2D (162, 163). In this portion of the review, we will critically evaluate the microbial aspect of the IF on T2D. A recent study (164) using diabetic mice reported that a 28-day IF intervention re-structured the gut microbiota by increasing the abundance of Aerococcus, Corynebacterium, Odoribacter, and Lactobacillus and decreasing the abundance of Streptococcus, Rummeliibacillus, and Candidatusarthromitu, which reduced plasma glucose and insulin levels, and improved energy metabolism. The changes in bacterial abundances due to IF are correlated with plasma secondary BAs concentration, increased villi length and reduced gut leakage accompanied by decreased plasma LPS levels (164), indicating improved low-grade inflammation (165). More importantly, the effect of IF on the T2D was suppressed by antibiotics treatment (164), suggesting that the microbiota is a causative agent of improvement in T2D by IF. An alternative to IF is a fasting-mimicking diet (FMD), which contains very low calories and low protein (166). Intermittent administration of FMD led to the reconstruction of gut microbiota by increasing the genera of Parabacteroides and Blautia while reducing Prevotellaceae, Alistipes, and Ruminococcaceae, along with normalized blood glucose levels, improved insulin sensitivity and β cell function in hyperglycemic db/db mice. This study further underscores that the loss of pancreatic islets and β cells can be prevented by the FMD-mediated altered gut microbiota (167), indicating that FMD improved T2D through pancreatic β cells function. Overall, IF may modulate gut microbiota and improve T2D. However, these findings need to be validated in human cohorts using longitudinal studies to establish the long-term effectiveness of IF in health outcomes including T2D.
Substantial evidence suggests the gut microbiota, and the metabolites it produces, are critical to the etiology of T2D. A strategy including FMT, medication, prebiotics, probiotics, functional food, and intermittent fasting has been suggested as strategies to reduce T2D. However, most studies have focused on the characterization of gut microbiota rather than functional validation of specific microbial taxa affecting T2D risk. Identifying specific causally related microbial taxa or microbial metabolites responsible for the pathogenesis of T2D could provide interesting new opportunities for the diagnosis, treatment, and prevention of T2D.
Recently several novel approaches have been taken to directly modify the gut microbiota. For example, one study (168) reported a novel approach to remodeling the gut microbiota using cyclic d,l-α-peptides. Alternatively, the FXR agonist fexaramine, which was not absorbed by the intestine, binds the FXR receptor on intestinal cells and induces enteric fibroblast growth factor 15 that leads to alterations in bile acid composition, reduces diet-induced weight gain, body-wide inflammation, and hepatic glucose production (169). These studies suggest that the development of therapeutics targeting the microbiome instead of the host is a viable strategy for T2D.
As discussed above, probiotic supplementation and FMT studies have established a causal relationship between gut microbiota and T2D. However, studies (170, 171) using FMT have demonstrated that the relationship between gut microbiota and disease phenotype is more complex than usual thought. For example, FMT may not always be able to transfer the beneficial clinical phenotype, instead sometimes can be resulted in a detrimental opposite effect. A recent FMT study (31) in which the gut microbiota of C57BL/6J mice ablated using antibiotics was reconstituted with either C57BL/6J or WSB/EiJ fecal microbiota. C57BL/6J mice are more susceptible to obesity, diabetes, and atherosclerosis compared to WSB/EiJ mice (170). Paradoxically, mice reconstituted with WSB/EiJ microbiota had significantly higher fat mass compared to the mice reconstituted with C57BL/6J microbiota. Moreover, among the members of gut microbiota, only the bacterial community is being studied extensively. To date, enteric virus, fungal, or archaea communities are still underappreciated mostly because of the assay difficulties and lack of standard reference databases and thus their contribution to T2D remains largely unknown. Therefore, incorporating these members in analysis may potentially lead to the development of novel therapeutics for T2D.
Computational approaches such as machine learning facilitate the analysis of large “-omics” datasets through the development of algorithms and mathematical models designed to predict outcomes. It remains to be determined how these novel computational approaches can be harnessed to further our understanding of the microbiota’s role in T2D but initial studies are promising. Recently two studies used machine learning tools to explore the role of the microbiome in precision nutrition (172) and to predict cirrhosis based on gut-microbiota features (173). Thus, use of these novel computational approaches may further our understanding of the metabolic consequences of how alterations in dietary habits, microbiota, metabolomics, genetics, and epigenetics, interact to alter metabolism. A better understanding of the interactions between microbiota, lifestyle, and host factors such as genetics and epigenetics might lead to a novel therapeutic approach for T2D.
BB supervised all portions of the review process, interpreted the results, and mentored manuscript writing. MH and MK conducted the literature search, extracting the information, and drafting the manuscript. MH and MK also addressed co-authors’ comments and concerns. BB, MH, and MK critically revised the manuscript. BB had primary responsibility for the final content. All authors contributed to the article and approved the submitted version.
This research was supported in part by NIH grant 5R01HL128572 (BB), NIFA grant 2019-07731 (BB) and USDA project 2032-51530-025-00D (BB). The USDA is an equal-opportunity employer.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The figures were created by using tools available at BioRender.com.
1. IDF. IDF Diabetes Atlas. 9th Vol. 2019. Brussels, Belgium: International Diabetes Federation (2019).
2. Control CfD, Prevention. National diabetes statistics report, 2020. Atlanta, GA: Centers for Disease Control and Prevention, US Department of Health and Human Services (2020).
3. Berg G, Rybakova D, Fischer D, Cernava T, Vergès M-CC, Charles T, et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome (2020) 8(1):103. doi: 10.1186/s40168-020-00875-0
4. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol (1977) 31(1):107–33. doi: 10.1146/annurev.mi.31.100177.000543
5. Ju F, Zhang T. 16S rRNA gene high-throughput sequencing data mining of microbial diversity and interactions. Appl Microbiol Biotechnol (2015) 99(10):4119–29. doi: 10.1007/s00253-015-6536-y
6. Thomas T, Gilbert J, Meyer F. Metagenomics-a guide from sampling to data analysis. Microbial Inf Experiment (2012) 2(1):3. doi: 10.1186/2042-5783-2-3
7. Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev (2010) 90(3):859–904. doi: 10.1152/physrev.00045.2009
8. Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature (2006) 444(7122):1022–3. doi: 10.1038/4441022a
9. Tilg H, Zmora N, Adolph TE, Elinav E. The intestinal microbiota fuelling metabolic inflammation. Nat Rev Immunol (2020) 20(1):40–54. doi: 10.1038/s41577-019-0198-4
10. Gurung M, Li Z, You H, Rodrigues R, Jump DB, Morgun A, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine (2020) 51:102590. doi: 10.1016/j.ebiom.2019.11.051
11. Gülden E. Lifestyle factors affecting the gut microbiota’s relationship with type 1 diabetes. Curr Diabetes Rep (2018) 18(11):111. doi: 10.1007/s11892-018-1098-x
12. Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human genetics shape the gut microbiome. Cell (2014) 159(4):789–99. doi: 10.1016/j.cell.2014.09.053
13. Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, et al. The genetic architecture of type 2 diabetes. Nature (2016) 536(7614):41–7. doi: 10.1038/nature18642
14. Maher B. The case of the missing heritability: when scientists opened up the human genome, they expected to find the genetic components of common traits and diseases. But they were nowhere to be seen. Brendan Maher shines a light on six places where the missing loot could be stashed away. Nature (2008) 456(7218):18–22. doi: 10.1038/456018a
15. Qin Y, Wade PA. Crosstalk between the microbiome and epigenome: messages from bugs. J Biochem (2018) 163(2):105–12. doi: 10.1093/jb/mvx080
16. Sircana A, Framarin L, Leone N, Berrutti M, Castellino F, Parente R, et al. Altered Gut Microbiota in Type 2 Diabetes: Just a Coincidence? Curr Diabetes Rep (2018) 18(10):98. doi: 10.1007/s11892-018-1057-6
17. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature (2012) 490(7418):55–60. doi: 10.1038/nature11450
18. Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature (2013) 498(7452):99–103. doi: 10.1038/nature12198
19. Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature (2016) 535(7612):376–81. doi: 10.1038/nature18646
20. Wu H, Tremaroli V, Schmidt C, Lundqvist A, Olsson LM, Kramer M, et al. The Gut Microbiota in Prediabetes and Diabetes: A Population-Based Cross-Sectional Study. Cell Metab (2020) 32(3):379–90 e3. doi: 10.1016/j.cmet.2020.06.011
21. Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PloS One (2013) 8(8):e71108. doi: 10.1371/journal.pone.0071108
22. Li Q, Chang Y, Zhang K, Chen H, Tao S, Zhang Z. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci Rep (2020) 10(1):5450. doi: 10.1038/s41598-020-62224-3
23. Doumatey AP, Adeyemo A, Zhou J, Lei L, Adebamowo SN, Adebamowo C, et al. Gut Microbiome Profiles Are Associated With Type 2 Diabetes in Urban Africans. Front Cell Infect Microbiol (2020) 10:63:63. doi: 10.3389/fcimb.2020.00063
24. Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PloS One (2010) 5(2):e9085. doi: 10.1371/journal.pone.0009085
25. Wang J, Li W, Wang C, Wang L, He T, Hu H, et al. Enterotype Bacteroides Is Associated with a High Risk in Patients with Diabetes: A Pilot Study. J Diabetes Res (2020) 2020:6047145. doi: 10.1155/2020/6047145
26. Salamon D, Sroka-Oleksiak A, Kapusta P, Szopa M, Mrozińska S, Ludwig-Słomczyńska AH, et al. Characteristics of gut microbiota in adult patient … tes based on next−generation sequencing of the 16S rRNA gene fragment. Pol Arch Intern Med (2018) 128(6):336–43. doi: 10.20452/pamw.4246
27. Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci (2005) 102(31):11070–5. doi: 10.1073/pnas.0504978102
28. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature (2006) 444(7122):1027–31. doi: 10.1038/nature05414
29. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci (2004) 101(44):15718–23. doi: 10.1073/pnas.0407076101
30. Naderpoor N, Mousa A, Gomez-Arango LF, Barrett HL, Dekker Nitert M, de Courten B. Faecal Microbiota Are Related to Insulin Sensitivity and Secretion in Overweight or Obese Adults. J Clin Med (2019) 8(4):452. doi: 10.3390/jcm8040452
31. Huda MN, Winnike JH, Crowell JM, O’Connor A, Bennett BJ. Microbial modulation of host body composition and plasma metabolic profile. Sci Rep (2020) 10(1):1–13. doi: 10.1038/s41598-020-63214-1
32. Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med (2019) 25(7):1096–103. doi: 10.1038/s41591-019-0495-2
33. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med (2017) 23(1):107–13. doi: 10.1038/nm.4236
34. Furet J-P, Kong L-C, Tap J, Poitou C, Basdevant A, Bouillot J-L, et al. Differential adaptation of human gut microbiota to bariatric surgery–induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes (2010) 59(12):3049–57. doi: 10.2337/db10-0253
35. Rathinam VA, Zhao Y, Shao F. Innate immunity to intracellular LPS. Nat Immunol (2019) 20(5):527–33. doi: 10.1038/s41590-019-0368-3
36. Vijayan A, Rumbo M, Carnoy C, Sirard J-C. Compartmentalized antimicrobial defenses in response to flagellin. Trends Microbiol (2018) 26(5):423–35. doi: 10.1016/j.tim.2017.10.008
37. Wolf AJ, Underhill DM. Peptidoglycan recognition by the innate immune system. Nat Rev Immunol (2018) 18(4):243. doi: 10.1038/nri.2017.136
38. Lee YS, Wollam J, Olefsky JM. An integrated view of immunometabolism. Cell (2018) 172(1-2):22–40. doi: 10.1016/j.cell.2017.12.025
39. Shen Z, Zhu C, Quan Y, Yang J, Yuan W, Yang Z, et al. Insights into Roseburia intestinalis which alleviates experimental colitis pathology by inducing anti-inflammatory responses. J Gastroenterol Hepatol (2018) 33(10):1751–60. doi: 10.1111/jgh.14144
40. Chang Y-C, Ching Y-H, Chiu C-C, Liu J-Y, Hung S-W, Huang W-C, et al. TLR2 and interleukin-10 are involved in Bacteroides fragilis-mediated prevention of DSS-induced colitis in gnotobiotic mice. PloS One (2017) 12(7):e0180025. doi: 10.1371/journal.pone.0180025
41. Li X, Wang N, Yin B, Fang D, Jiang T, Fang S, et al. Effects of Lactobacillus plantarum CCFM0236 on hyperglycaemia and insulin resistance in high-fat and streptozotocin-induced type 2 diabetic mice. J Appl Microbiol (2016) 121(6):1727–36. doi: 10.1111/jam.13276
42. Chen P, Zhang Q, Dang H, Liu X, Tian F, Zhao J, et al. Antidiabetic effect of Lactobacillus casei CCFM0412 on mice with type 2 diabetes induced by a high-fat diet and streptozotocin. Nutrition (2014) 30(9):1061–8. doi: 10.1016/j.nut.2014.03.022
43. Wang X, Ota N, Manzanillo P, Kates L, Zavala-Solorio J, Eidenschenk C, et al. Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature (2014) 514(7521):237–41. doi: 10.1038/nature13564
44. Dagdeviren S, Young Jung D, Friedline RH, Noh HL, Kim JH, Patel PR, et al. IL-10 prevents aging-associated inflammation and insulin resistance in skeletal muscle. FASEB J (2017) 31(2):701–10. doi: 10.1096/fj.201600832R
45. Hoffmann TW, Pham H-P, Bridonneau C, Aubry C, Lamas B, Martin-Gallausiaux C, et al. Microorganisms linked to inflammatory bowel disease-associated dysbiosis differentially impact host physiology in gnotobiotic mice. ISME J (2016) 10(2):460–77. doi: 10.1038/ismej.2015.127
46. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature (2013) 500(7461):232–6. doi: 10.1038/nature12331
47. Shi H, Chi H. Metabolic Control of Treg Cell Stability, Plasticity, and Tissue-Specific Heterogeneity. Front Immunol (2019) 10:2716. doi: 10.3389/fimmu.2019.02716
48. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature (2013) 504(7480):446–50. doi: 10.1038/nature12721
49. Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr (2003) 133(7):2485S–93S. doi: 10.1093/jn/133.7.2485S
50. Tian P, Li B, He C, Song W, Hou A, Tian S, et al. Antidiabetic (type 2) effects of Lactobacillus G15 and Q14 in rats through regulation of intestinal permeability and microbiota. Food Funct (2016) 7(9):3789–97. doi: 10.1039/C6FO00831C
51. Zhang L, Qin Q, Liu M, Zhang X, He F, Wang G. Akkermansia muciniphila can reduce the damage of gluco/lipotoxicity, oxidative stress and inflammation, and normalize intestine microbiota in streptozotocin-induced diabetic rats. Pathog Dis (2018) 76(4):fty028. doi: 10.1093/femspd/fty028
52. Liu W-C, Yang M-C, Wu Y-Y, Chen P-H, Hsu C-M, Chen L-W. Lactobacillus plantarum reverse diabetes-induced Fmo3 and ICAM expression in mice through enteric dysbiosis-related c-Jun NH2-terminal kinase pathways. PloS One (2018) 13(5):e0196511. doi: 10.1371/journal.pone.0196511
53. Sato J, Kanazawa A, Ikeda F, Yoshihara T, Goto H, Abe H, et al. Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes Care (2014) 37(8):2343–50. doi: 10.2337/dc13-2817
54. Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res (2013) 69(1):52–60. doi: 10.1016/j.phrs.2012.10.020
55. Qi J, You T, Li J, Pan T, Xiang L, Han Y, et al. Circulating trimethylamine N-oxide and the risk of cardiovascular diseases: a systematic review and meta-analysis of 11 prospective cohort studies. J Cell Mol Med (2018) 22(1):185–94. doi: 10.1111/jcmm.13307
56. Gao X, Liu X, Xu J, Xue C, Xue Y, Wang Y. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J Biosci Bioeng (2014) 118(4):476–81. doi: 10.1016/j.jbiosc.2014.03.001
57. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes (2016) 7(3):189–200. doi: 10.1080/19490976.2015.1134082
58. Hirasawa A, Hara T, Katsuma S, Adachi T, Tsujimoto G. Free fatty acid receptors and drug discovery. Biol Pharm Bull (2008) 31(10):1847–51. doi: 10.1248/bpb.31.1847
59. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther (2008) 27(2):104–19. doi: 10.1111/j.1365-2036.2007.03562.x
61. Koh A, Mannerås-Holm L, Yunn NO, Nilsson PM, Ryu SH, Molinaro A, et al. Microbial Imidazole Propionate Affects Responses to Metformin through p38γ-Dependent Inhibitory AMPK Phosphorylation. Cell Metab (2020) 32(4):643–53. doi: 10.1016/j.cmet.2020.07.012
62. Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun (2018) 9(1):1–10. doi: 10.1038/s41467-018-05470-4
63. Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature (2017) 551(7682):648–52. doi: 10.1038/nature24661
64. Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity (2014) 41(2):296–310. doi: 10.1016/j.immuni.2014.06.014
65. Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol (2018) 15(2):111–28. doi: 10.1038/nrgastro.2017.119
66. Chiang JY. Bile acids: regulation of synthesis. J Lipid Res (2009) 50(10):1955–66. doi: 10.1194/jlr.R900010-JLR200
67. Zhou H, Hylemon PB. Bile acids are nutrient signaling hormones. Steroids (2014) 86:62–8. doi: 10.1016/j.steroids.2014.04.016
68. Chen X, Yang W. Branched-chain amino acids and the association with type 2 diabetes. J Diabetes Investig (2015) 6(4):369–70. doi: 10.1111/jdi.12345
69. Mutaguchi Y, Kasuga K, Kojima I. Production of d-Branched-Chain Amino Acids by Lactic Acid Bacteria Carrying Homologs to Isoleucine 2-Epimerase of Lactobacillus buchneri. Front Microbiol (2018) 9:1540. doi: 10.3389/fmicb.2018.01540
70. Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature (2011) 472(7341):57–63. doi: 10.1038/nature09922
71. Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab (2013) 17(1):49–60. doi: 10.1016/j.cmet.2012.12.011
72. Tang WH, Wang Z, Li XS, Fan Y, Li DS, Wu Y, et al. Increased Trimethylamine N-Oxide Portends High Mortality Risk Independent of Glycemic Control in Patients with Type 2 Diabetes Mellitus. Clin Chem (2017) 63(1):297–306. doi: 10.1373/clinchem.2016.263640
73. Dambrova M, Latkovskis G, Kuka J, Strele I, Konrade I, Grinberga S, et al. Diabetes is Associated with Higher Trimethylamine N-oxide Plasma Levels. Exp Clin Endocrinol Diabetes (2016) 124(4):251–6. doi: 10.1055/s-0035-1569330
74. Shih DM, Wang Z, Lee R, Meng Y, Che N, Charugundla S, et al. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J Lipid Res (2015) 56(1):22–37. doi: 10.1194/jlr.M051680
75. Zhuang R, Ge X, Han L, Yu P, Gong X, Meng Q, et al. Gut microbe-generated metabolite trimethylamine N-oxide and the risk of diabetes: A systematic review and dose-response meta-analysis. Obes Rev (2019) 20(6):883–94. doi: 10.1111/obr.12843
76. Jia J, Dou P, Gao M, Kong X, Li C, Liu Z, et al. Assessment of Causal Direction Between Gut Microbiota-Dependent Metabolites and Cardiometabolic Health: A Bidirectional Mendelian Randomization Analysis. Diabetes (2019) 68(9):1747–55. doi: 10.2337/db19-0153
77. Torres LE, Melian K, Moreno A, Alonso J, Sabatier CA, Hernandez M, et al. Prevalence of vacA, cagA and babA2 genes in Cuban Helicobacter pylori isolates. World J Gastroenterol (2009) 15(2):204–10. doi: 10.3748/wjg.15.204
78. Holst JJ. The physiology of glucagon-like peptide 1. Physiol Rev (2007) 87(4):1409–39. doi: 10.1152/physrev.00034.2006
79. Everard A, Cani PD. Gut microbiota and GLP-1. Rev Endocrine Metab Disord (2014) 15(3):189–96. doi: 10.1007/s11154-014-9288-6
80. Parnell JA, Reimer RA. Weight loss during oligofructose supplementation is associated with decreased ghrelin and increased peptide YY in overweight and obese adults. Am J Clin Nutr (2009) 89(6):1751–9. doi: 10.3945/ajcn.2009.27465
81. Chimerel C, Emery E, Summers DK, Keyser U, Gribble FM, Reimann F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep (2014) 9(4):1202–8. doi: 10.1016/j.celrep.2014.10.032
82. Macia L, Thorburn AN, Binge LC, Marino E, Rogers KE, Maslowski KM, et al. Microbial influences on epithelial integrity and immune function as a basis for inflammatory diseases. Immunol Rev (2012) 245(1):164–76. doi: 10.1111/j.1600-065X.2011.01080.x
83. McCreight LJ, Bailey CJ, Pearson ER. Metformin and the gastrointestinal tract. Diabetologia (2016) 59(3):426–35. doi: 10.1007/s00125-015-3844-9
84. Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science (2018) 359(6380):1151–6. doi: 10.1126/science.aao5774
85. Mandaliya DK, Seshadri S. Short Chain Fatty Acids, pancreatic dysfunction and type 2 diabetes. Pancreatology (2019) 19(2):280–4. doi: 10.1016/j.pan.2019.01.021
86. Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens (2014) 3(1):14–24. doi: 10.3390/pathogens3010014
87. Koh A, Molinaro A, Stahlman M, Khan MT, Schmidt C, Manneras-Holm L, et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell (2018) 175(4):947–61.e17. doi: 10.1016/j.cell.2018.09.055
88. Bloomgarden Z. Diabetes and branched-chain amino acids: What is the link? J Diabetes (2018) 10(5):350–2. doi: 10.1111/1753-0407.12645
89. Katsuma S, Hirasawa A, Tsujimoto G. Bile acids promote glucagon-like peptide-1 secretion through TGR5 in a murine enteroendocrine cell line STC-1. Biochem Biophys Res Commun (2005) 329(1):386–90. doi: 10.1016/j.bbrc.2005.01.139
90. Chen M-l, Yi L, Zhang Y, Zhou X, Ran L, Yang J, et al. Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. MBio (2016) 7(2):e02210–15. doi: 10.1128/mBio.02210-15
91. Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab (2009) 9(4):311–26. doi: 10.1016/j.cmet.2009.02.002
92. Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, et al. Metabolite profiles and the risk of developing diabetes. Nat Med (2011) 17(4):448–53. doi: 10.1038/nm.2307
93. Thaiss CA, Itav S, Rothschild D, Meijer MT, Levy M, Moresi C, et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature (2016) 540(7634):544–51. doi: 10.1038/nature20796
94. Johnson AJ, Vangay P, Al-Ghalith GA, Hillmann BM, Ward TL, Shields-Cutler RR, et al. Daily Sampling Reveals Personalized Diet-Microbiome Associations in Humans. Cell Host Microbe (2019) 25(6):789–802. doi: 10.1016/j.chom.2019.05.005
95. Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe (2016) 19(5):731–43. doi: 10.1016/j.chom.2016.04.017
96. Hughes DA, Bacigalupe R, Wang J, Rühlemann MC, Tito RY, Falony G, et al. Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nat Microbiol (2020) 5(9):1079–87. doi: 10.1038/s41564-020-0743-8
97. Cohen LJ, Esterhazy D, Kim S-H, Lemetre C, Aguilar RR, Gordon EA, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature (2017) 549(7670):48–53. doi: 10.1038/nature23874
98. Consortium G. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science (2015) 348(6235):648–60.
99. Coady MJ, Wallendorff B, Gagnon DG, Lapointe J-Y. Identification of a novel Na+/myo-inositol cotransporter. J Biol Chem (2002) 277(38):35219–24. doi: 10.1074/jbc.M204321200
100. Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell (2016) 64(5):982–92. doi: 10.1016/j.molcel.2016.10.025
101. Bolduc J-F, Hany L, Barat C, Ouellet M, Tremblay MJ. Epigenetic metabolite acetate inhibits class I/II histone deacetylases, promotes histone acetylation, and increases HIV-1 integration in CD4+ T cells. J Virol (2017) 91(16):e01943–16. doi: 10.1128/JVI.01943-16
102. Zhou L, Zhang M, Wang Y, Dorfman RG, Liu H, Yu T, et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm bowel Dis (2018) 24(9):1926–40. doi: 10.1093/ibd/izy182
103. Ramos-Molina B, Sanchez-Alcoholado L, Cabrera-Mulero A, Lopez-Dominguez R, Carmona-Saez P, Garcia-Fuentes E, et al. Gut Microbiota Composition Is Associated With the Global DNA Methylation Pattern in Obesity. Front Genet (2019) 10:613. doi: 10.3389/fgene.2019.00613
104. Watson MM, Søreide K. The Gut Microbiota Influence on Human Epigenetics, Health, and Disease. Handb Epigenet Elsevier (2017), 495–510. doi: 10.1016/B978-0-12-805388-1.00032-8
105. Dayeh T, Volkov P, Salö S, Hall E, Nilsson E, Olsson AH, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PloS Genet (2014) 10(3):e1004160. doi: 10.1371/journal.pgen.1004160
106. Kassam Z, Lee CH, Yuan Y, Hunt RH. Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol (2013) 108(4):500–8. doi: 10.1038/ajg.2013.59
107. Spinner JA, Bocchini CE, Luna RA, Thapa S, Balderas MA, Denfield SW, et al. Fecal microbiota transplantation in a toddler after heart transplant was a safe and effective treatment for recurrent Clostridiodes difficile infection: A case report. Pediatr Transplant (2020) 24(1):e13598. doi: 10.1111/petr.13598
108. Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat Rev Gastroenterol Hepatol (2016) 13(9):508–16. doi: 10.1038/nrgastro.2016.98
109. Peng J, Narasimhan S, Marchesi JR, Benson A, Wong FS, Wen L. Long term effect of gut microbiota transfer on diabetes development. J Autoimmun (2014) 53:85–94. doi: 10.1016/j.jaut.2014.03.005
110. Zhang PP, Li LL, Han X, Li QW, Zhang XH, Liu JJ, et al. Fecal microbiota transplantation improves metabolism and gut microbiome composition in db/db mice. Acta Pharmacol Sin (2020) 41(5):678–85. doi: 10.1038/s41401-019-0330-9
111. Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med (2017) 23(7):850–8. doi: 10.1038/nm.4345
112. Kootte RS, Levin E, Salojarvi J, Smits LP, Hartstra AV, Udayappan SD, et al. Improvement of Insulin Sensitivity after Lean Donor Feces in Metabolic Syndrome Is Driven by Baseline Intestinal Microbiota Composition. Cell Metab (2017) 26(4):611–9.e6. doi: 10.1016/j.cmet.2017.09.008
113. Smits LP, Kootte RS, Levin E, Prodan A, Fuentes S, Zoetendal EG, et al. Effect of vegan fecal microbiota transplantation on carnitine-and choline-derived trimethylamine-N-oxide production and vascular inflammation in patients with metabolic syndrome. J Am Heart Assoc (2018) 7(7):e008342. doi: 10.1161/JAHA.117.008342
114. Wu GD, Compher C, Chen EZ, Smith SA, Shah RD, Bittinger K, et al. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut (2016) 65(1):63–72. doi: 10.1136/gutjnl-2014-308209
115. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med (2013) 19(5):576–85. doi: 10.1038/nm.3145
116. Baur JA, Birnbaum MJ. Control of gluconeogenesis by metformin: does redox trump energy charge? Cell Metab (2014) 20(2):197–9. doi: 10.1016/j.cmet.2014.07.013
117. El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, Leverve X. Dimethylbiguanide Inhibits Cell Respiration via an Indirect Effect Targeted on the Respiratory Chain Complex I. J Biol Chem (2000) 275(1):223–8. doi: 10.1074/jbc.275.1.223
118. Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest (2001) 108(8):1167–74. doi: 10.1172/jci200113505
119. Bonora E, Cigolini M, Bosello O, Zancanaro C, Capretti L, Zavaroni I, et al. Lack of effect of intravenous metformin on plasma concentrations of glucose, insulin, C-peptide, glucagon and growth hormone in non-diabetic subjects. Curr Med Res Opin (1984) 9(1):47–51. doi: 10.1185/03007998409109558
120. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature (2015) 528(7581):262–6. doi: 10.1038/nature15766
121. Ejtahed HS, Tito RY, Siadat SD, Hasani-Ranjbar S, Hoseini-Tavassol Z, Rymenans L, et al. Metformin induces weight loss associated with gut microbiota alteration in non-diabetic obese women: a randomized double-blind clinical trial. Eur J Endocrinol (2019) 180(3):165–76. doi: 10.1530/EJE-18-0826
122. Rosario D, Benfeitas R, Bidkhori G, Zhang C, Uhlen M, Shoaie S, et al. Understanding the Representative Gut Microbiota Dysbiosis in Metformin-Treated Type 2 Diabetes Patients Using Genome-Scale Metabolic Modeling. Front Physiol (2018) 9:775. doi: 10.3389/fphys.2018.00775
123. Malik F, Mehdi SF, Ali H, Patel P, Basharat A, Kumar A, et al. Is metformin poised for a second career as an antimicrobial? Diabetes Metab Res Rev (2018) 34(4):e2975. doi: 10.1002/dmrr.2975
124. Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol (2014) 80(19):5935–43. doi: 10.1128/AEM.01357-14
125. de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, Velasquez-Mejia EP, Carmona JA, Abad JM, et al. Metformin Is Associated With Higher Relative Abundance of Mucin-Degrading Akkermansia muciniphila and Several Short-Chain Fatty Acid-Producing Microbiota in the Gut. Diabetes Care (2017) 40(1):54–62. doi: 10.2337/dc16-1324
126. Rodriguez J, Hiel S, Delzenne NM. Metformin: old friend, new ways of action-implication of the gut microbiome? Curr Opin Clin Nutr Metab Care (2018) 21(4):294–301. doi: 10.1097/MCO.0000000000000468
127. Kuka J, Videja M, Makrecka-Kuka M, Liepins J, Grinberga S, Sevostjanovs E, et al. Metformin decreases bacterial trimethylamine production and trimethylamine N-oxide levels in db/db mice. Sci Rep (2020) 10(1):14555. doi: 10.1038/s41598-020-71470-4
128. Adeshirlarijaney A, Zou J, Tran HQ, Chassaing B, Gewirtz AT. Amelioration of metabolic syndrome by metformin associates with reduced indices of low-grade inflammation independently of the gut microbiota. Am J Physiol Endocrinol Metab (2019) 317(6):E1121–E30. doi: 10.1152/ajpendo.00245.2019
129. Su B, Liu H, Li J, Sunli Y, Liu B, Liu D, et al. Acarbose treatment affects the serum levels of inflammatory cytokines and the gut content of bifidobacteria in Chinese patients with type 2 diabetes mellitus. J Diabetes (2015) 7(5):729–39. doi: 10.1111/1753-0407.12232
130. Gu Y, Wang X, Li J, Zhang Y, Zhong H, Liu R, et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun (2017) 8(1):1785. doi: 10.1038/s41467-017-01682-2
131. Zhang X, Fang Z, Zhang C, Xia H, Jie Z, Han X, et al. Effects of Acarbose on the Gut Microbiota of Prediabetic Patients: A Randomized, Double-blind, Controlled Crossover Trial. Diabetes Ther (2017) 8(2):293–307. doi: 10.1007/s13300-017-0226-y
132. Zhang Q, Xiao X, Zheng J, Li M, Yu M, Ping F, et al. Featured article: Structure moderation of gut microbiota in liraglutide-treated diabetic male rats. Exp Biol Med (Maywood) (2018) 243(1):34–44. doi: 10.1177/1535370217743765
133. Wang L, Li P, Tang Z, Yan X, Feng B. Structural modulation of the gut microbiota and the relationship with body weight: compared evaluation of liraglutide and saxagliptin treatment. Sci Rep (2016) 6:33251. doi: 10.1038/srep33251
134. Schrezenmeir J, de Vrese M. Probiotics, prebiotics, and synbiotics–approaching a definition. Am J Clin Nutr (2001) 73(2 Suppl):361S–4S. doi: 10.1093/ajcn/73.2.361s
135. Markowiak P. Śliżewska K. Effects of probiotics, prebiotics, and synbiotics on human health. Nutrients (2017) 9(9):1021. doi: 10.3390/nu9091021
136. Kim YA, Keogh JB, Clifton PM. Probiotics, prebiotics, synbiotics and insulin sensitivity. Nutr Res Rev (2018) 31(1):35–51. doi: 10.1017/S095442241700018X
137. Kechagia M, Basoulis D, Konstantopoulou S, Dimitriadi D, Gyftopoulou K, Skarmoutsou N, et al. Health benefits of probiotics: a review. Int Scholarly Res Notices (2013) 2013:1–7. doi: 10.5402/2013/481651
138. Yun S, Park H, Kang J. Effect of Lactobacillus gasseri BNR17 on blood glucose levels and body weight in a mouse model of type 2 diabetes. J Appl Microbiol (2009) 107(5):1681–6. doi: 10.1111/j.1365-2672.2009.04350.x
139. Hove KD, Brons C, Faerch K, Lund SS, Rossing P, Vaag A. Effects of 12 weeks of treatment with fermented milk on blood pressure, glucose metabolism and markers of cardiovascular risk in patients with type 2 diabetes: a randomised double-blind placebo-controlled study. Eur J Endocrinol (2015) 172(1):11–20. doi: 10.1530/EJE-14-0554
140. Sharma P, Bhardwaj P, Singh R. Administration of Lactobacillus casei and Bifidobacterium bifidum Ameliorated Hyperglycemia, Dyslipidemia, and Oxidative Stress in Diabetic Rats. Int J Prev Med (2016) 7:102. doi: 10.4103/2008-7802.188870
141. Villarini M, Caldini G, Moretti M, Trotta F, Pasquini R, Cenci G. Modulatory activity of a Lactobacillus casei strain on 1,2-dimethylhydrazine-induced genotoxicity in rats. Environ Mol Mutagen (2008) 49(3):192–9. doi: 10.1002/em.20367
142. So JS, Kwon HK, Lee CG, Yi HJ, Park JA, Lim SY, et al. Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Mol Immunol (2008) 45(9):2690–9. doi: 10.1016/j.molimm.2007.12.010
143. Bock PM, Telo GH, Ramalho R, Sbaraini M, Leivas G, Martins AF, et al. The effect of probiotics, prebiotics or synbiotics on metabolic outcomes in individuals with diabetes: a systematic review and meta-analysis. Diabetologia (2020) 64(1):26–41. doi: 10.1007/s00125-020-05295-1
144. Kasinska MA, Drzewoski J. Effectiveness of probiotics in type 2 diabetes: a meta-analysis. Pol Arch Med Wewn (2015) 125(11):803–13. doi: 10.20452/pamw.3156
145. Gibson GR, Roberfroid MB. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr (1995) 125(6):1401–12. doi: 10.1093/jn/125.6.1401
146. Roberfroid MB. Introducing inulin-type fructans. Br J Nutr (2005) 93 Suppl 1:S13–25. doi: 10.1079/bjn20041350
147. Cani PD, Dewever C, Delzenne NM. Inulin-type fructans modulate gastrointestinal peptides involved in appetite regulation (glucagon-like peptide-1 and ghrelin) in rats. Br J Nutr (2004) 92(3):521–6. doi: 10.1079/bjn20041225
148. Li K, Zhang L, Xue J, Yang X, Dong X, Sha L, et al. Dietary inulin alleviates diverse stages of type 2 diabetes mellitus via anti-inflammation and modulating gut microbiota in db/db mice. Food Funct (2019) 10(4):1915–27. doi: 10.1039/C8FO02265H
149. Birkeland E, Gharagozlian S, Birkeland KI, Valeur J, Mage I, Rud I, et al. Prebiotic effect of inulin-type fructans on faecal microbiota and short-chain fatty acids in type 2 diabetes: a randomised controlled trial. Eur J Nutr (2020) 59(7):3325–38. doi: 10.1007/s00394-020-02282-5
150. Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia (2007) 50(11):2374–83. doi: 10.1007/s00125-007-0791-0
151. Verhoog S, Taneri PE, Roa Diaz ZM, Marques-Vidal P, Troup JP, Bally L, et al. Dietary Factors and Modulation of Bacteria Strains of Akkermansia muciniphila and Faecalibacterium prausnitzii: A Systematic Review. Nutrients (2019) 11(7):1565. doi: 10.3390/nu11071565
152. McRorie JW, Gibb RD, McKeown NM. Inulin-type fructans have no significant beneficial effects on lipid or glucose metabolism. Eur J Clin Nutr (2017) 71(5):677. doi: 10.1038/ejcn.2017.15
153. Morshedi M, Saghafi-Asl M, Hosseinifard ES. The potential therapeutic effects of the gut microbiome manipulation by synbiotic containing-Lactobacillus plantarum on neuropsychological performance of diabetic rats. J Transl Med (2020) 18(1):18. doi: 10.1186/s12967-019-02169-y
154. Nazzaro F, Fratianni F, Nicolaus B, Poli A, Orlando P. The prebiotic source influences the growth, biochemical features and survival under simulated gastrointestinal conditions of the probiotic Lactobacillus acidophilus. Anaerobe (2012) 18(3):280–5. doi: 10.1016/j.anaerobe.2012.03.002
155. Zhang Y, Li X, Zou D, Liu W, Yang J, Zhu N, et al. Treatment of type 2 diabetes and dyslipidemia with the natural plant alkaloid berberine. J Clin Endocrinol Metab (2008) 93(7):2559–65. doi: 10.1210/jc.2007-2404
156. Zhang Y, Gu Y, Ren H, Wang S, Zhong H, Zhao X, et al. Gut microbiome-related effects of berberine and probiotics on type 2 diabetes (the PREMOTE study). Nat Commun (2020) 11(1):5015. doi: 10.1038/s41467-020-18414-8
157. Mahboobi S, Rahimi F, Jafarnejad S. Effects of Prebiotic and Synbiotic Supplementation on Glycaemia and Lipid Profile in Type 2 Diabetes: A Meta-Analysis of Randomized Controlled Trials. Adv Pharm Bull (2018) 8(4):565–74. doi: 10.15171/apb.2018.065
158. Anson RM, Guo Z, de Cabo R, Iyun T, Rios M, Hagepanos A, et al. Intermittent fasting dissociates beneficial effects of dietary restriction on glucose metabolism and neuronal resistance to injury from calorie intake. Proc Natl Acad Sci (2003) 100(10):6216–20. doi: 10.1073/pnas.1035720100
159. Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab (2012) 15(6):848–60. doi: 10.1016/j.cmet.2012.04.019
160. Zhang C, Li S, Yang L, Huang P, Li W, Wang S, et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Commun (2013) 4(1):1–10. doi: 10.1038/ncomms3163
161. Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab (2014) 20(6):1006–17. doi: 10.1016/j.cmet.2014.11.008
162. de Cabo R, Mattson MP. Effects of Intermittent Fasting on Health, Aging, and Disease. N Engl J Med (2019) 381(26):2541–51. doi: 10.1056/NEJMra1905136
163. Zhu S, Surampudi P, Rosharavan B, Chondronikola M. Intermittent fasting as a nutrition approach against obesity and metabolic disease. Curr Opin Clin Nutr Metab Care (2020) 23(6):387–94. doi: 10.1097/MCO.0000000000000694
164. Liu Z, Dai X, Zhang H, Shi R, Hui Y, Jin X, et al. Gut microbiota mediates intermittent-fasting alleviation of diabetes-induced cognitive impairment. Nat Commun (2020) 11(1):855. doi: 10.1038/s41467-020-14676-4
165. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet–induced obesity and diabetes in mice. Diabetes (2008) 57(6):1470–81. doi: 10.2337/db07-1403
166. Brandhorst S, Choi IY, Wei M, Cheng CW, Sedrakyan S, Navarrete G, et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab (2015) 22(1):86–99. doi: 10.1016/j.cmet.2015.05.012
167. Wei S, Han R, Zhao J, Wang S, Huang M, Wang Y, et al. Intermittent administration of a fasting-mimicking diet intervenes in diabetes progression, restores β cells and reconstructs gut microbiota in mice. Nutr Metab (2018) 15(1):1–12. doi: 10.1186/s12986-018-0318-3
168. Chen PB, Black AS, Sobel AL, Zhao Y, Mukherjee P, Molparia B, et al. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat Biotechnol (2020) 38(11):1288–97. doi: 10.1038/s41587-020-0549-5
169. Fang S, Suh JM, Reilly SM, Yu E, Osborn O, Lackey D, et al. Intestinal FXR agonism promotes adipose tissue browning and reduces obesity and insulin resistance. Nat Med (2015) 21(2):159–65. doi: 10.1038/nm.3760
170. Lee KT, Karunakaran S, Ho MM, Clee SM. PWD/PhJ and WSB/EiJ mice are resistant to diet-induced obesity but have abnormal insulin secretion. Endocrinology (2011) 152(8):3005–17. doi: 10.1210/en.2011-0060
171. Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang Y, et al. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics (2015) 47(6):187–97. doi: 10.1152/physiolgenomics.00136.2014
172. Berry SE, Valdes AM, Drew DA, Asnicar F, Mazidi M, Wolf J, et al. Human postprandial responses to food and potential for precision nutrition. Nat Med (2020) 26(6):964–73. doi: 10.1038/s41591-020-0934-0
Keywords: microbiota (16S), type 2 diabetes (T2D), metabolites, probiotics, prebioitcs, intermittent fasting, genetics, epigenetics
Citation: Huda MN, Kim M and Bennett BJ (2021) Modulating the Microbiota as a Therapeutic Intervention for Type 2 Diabetes. Front. Endocrinol. 12:632335. doi: 10.3389/fendo.2021.632335
Received: 23 November 2020; Accepted: 04 February 2021;
Published: 07 April 2021.
Edited by:
Antonio Salgado Somoza, Independent Researcher, Neufchateau, BelgiumReviewed by:
Ger Rijkers, University College Roosevelt, NetherlandsCopyright © 2021 Huda, Kim and Bennett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian J. Bennett, YnJpYW4uYmVubmV0dEB1c2RhLmdvdg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.