
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Endocrinol. , 28 October 2020
Sec. Neuroendocrine Science
Volume 11 - 2020 | https://doi.org/10.3389/fendo.2020.610274
This article is a correction to:
New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts
A Corrigendum on
New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts
By Maugars G, Nourizadeh-Lillabadi R and Weltzien F-A (2020). Front. Endocrinol. 11:538196. doi: 10.3389/fendo.2020.538196
In the original article, there was a mistake in the Figure 9 as published. In the Figure, we have represented the mtnr1ba gene in the herring while it has been lost in this lineage. The corrected Figure 9 appears below.
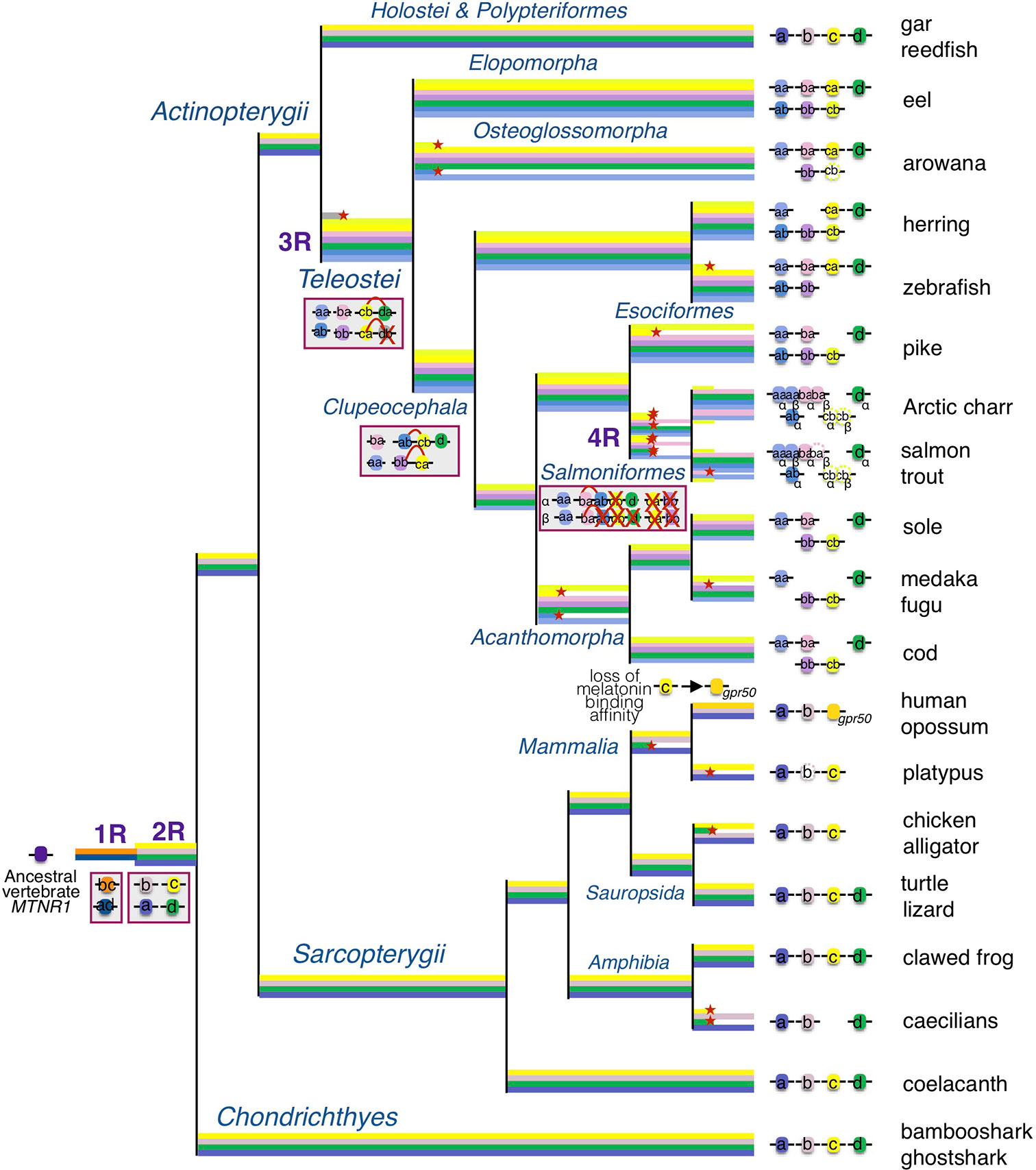
Figure 9 Evolutionary scenario of the melatonin receptors in vertebrates. This evolutionary scenario was developed on the basis of the phylogeny and synteny analyses presented in the original article. The four receptor genes are derived from duplication of an ancient mtnr1 gene through vertebrate tetraploidization (1R and 2R). The teleost 3R event generated duplicates of the four mtnr subtypes. Multiple and selective gene losses occurred, leading to mtnr gene repertoires differing between the main gnathostome lineages. Genome tetraploidization events (1R, 2R, 3R, and 4R) are indicated in purple. Major gene gain and loss events as well as chromosome rearrangements are indicated in red boxes. Genes located on the same linkage group are represented by clusters on the genomic DNA line. Red cross indicates gene loss. The red arc indicates genomic region fusion events. Colored tree branches represent mtnr gene lineages. Red star * on a tree branch indicates gene loss. The receptor identities are indicated in or beside the boxes. Paralogs originating from teleost 3R are designated by a and b suffixes, and salmonid 4R paralogs are designated by α and β suffixes.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Keywords: melatonin receptors, gene duplication, vertebrates, teleosts, medaka, phylogeny, synteny, functional evolution
Citation: Maugars G, Nourizadeh-Lillabadi R and Weltzien F-A (2020) Corrigendum: New Insights Into the Evolutionary History of Melatonin Receptors in Vertebrates, With Particular Focus on Teleosts. Front. Endocrinol. 11:610274. doi: 10.3389/fendo.2020.610274
Received: 25 September 2020; Accepted: 02 October 2020;
Published: 28 October 2020.
Edited and reviewed by: Dan Larhammar, Uppsala University, Sweden
Copyright © 2020 Maugars, Nourizadeh-Lillabadi and Weltzien. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gersende Maugars, Z2Vyc2VuZGUubWF1Z2Fyc0BubWJ1Lm5v; Finn-Arne Weltzien, Zmlubi1hcm5lLndlbHR6aWVuQG5tYnUubm8=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.