- 1Yale College, New Haven, CT, United States
- 2Department of Molecular Biophysics and Biochemistry, New Haven, CT, United States
- 3Department of Pharmacology, New Haven, CT, United States
- 4Yale Cancer Center, Yale University, New Haven, CT, United States
The Src homology 2 (SH2) domain has a special role as one of the cornerstone examples of a “modular” domain. The interactions of this domain are very well-conserved, and have long been described as a bidentate, or “two-pronged plug” interaction between the domain and a phosphotyrosine (pTyr) peptide. Recent work has, however, highlighted unusual features of the SH2 domain that illustrate a greater diversity than was previously appreciated. In this review we discuss some of the novel and unusual characteristics across the SH2 family, including unusual peptide binding pockets, multiple pTyr recognition sites, recognition sites for unphosphorylated peptides, and recently identified variability in the conserved FLVR motif.
Introduction
In 1986, a new domain was identified within v-src, the transforming gene of the Rous sarcoma virus (1–3). This region had high sequence similarity across the then-known cytoplasmic protein-tyrosine kinases, and a conserved location N-terminal to the kinase domain (the first homology region identified in Src). The domain was termed the Src homology 2 (SH2) domain, and its identification heralded a new era in the understanding of molecular interactions and cellular signaling (2, 4). SH2 domains have been critical for development of key concepts such as the dependency of cytosolic signaling on post-translational modification-regulated protein interactions (5–8), and the modularity of protein domains (3, 9, 10). Over the years, extremely well conserved molecular mechanisms have been revealed which are used by SH2 domains to mediate their effects (11–13). These canonical features are well-documented, but unusual features also occur and increase the diversity of the fold. In this review, we discuss these unusual features and how they exhibit divergence from canonical SH2 domain architecture.
Global Features of The Canonical SH2 Domain
The primary molecular role of the SH2 domain is to directly bind phosphotyrosine (pTyr) residues (14). This is central to propagation of signaling by receptor and non-receptor tyrosine kinases such as the insulin receptor and the JAK kinases, so SH2 domains are critical to a range of fields including endocrinology (5, 15–18). The SH2-pTyr interaction is broadly independent of folding of the pTyr-ligand, and can be observed for denatured Tyr peptides (19–22), but is distinct from recognition of pTyr by for example the phosphotyrosine binding (PTB) domains (5). Thus, the binding of SH2 domains to short linear peptide motifs can be predictive for the interactome of specific SH2 domains (20–23). SH2 interaction selectivity has yielded extensive knowledge of their binding partner preferences and signaling networks (2, 5).
The mechanisms for SH2-ligand interaction are well-defined, with the first cohort of structures for this ~100 amino acid fold determined in 1992 and 1993 (24–28). These structures showed that the SH2 domain consists of a central β-sheet flanked by two α-helices. They revealed that the phosphorylated peptide binds perpendicularly to the β-sheet and docks into two abutting recognition sites formed by the β-sheet with each of the α-helices. This bidentate, or “two-pronged plug” (26), interaction provides both a deep basic pTyr binding site, and a specificity pocket that usually recognizes an amino acid three residues C-terminal to the pTyr (termed the +3 position), a mode of interaction that is consistent in vitro and in cells (3, 21, 22, 29) (Figure 1A). The nomenclature for the fold defines the antiparallel β-strands as βA-βG and the helices as αA and αB, with loops named by the flanking secondary structure (Figure 1B) (28). Thus, the pTyr pocket is canonically defined by residues of αA, βB, βC, βD, and by the BC “phosphate binding loop” (Figure 1C); and the specificity pocket by residues of αB, βG, and the BG and EF loops.
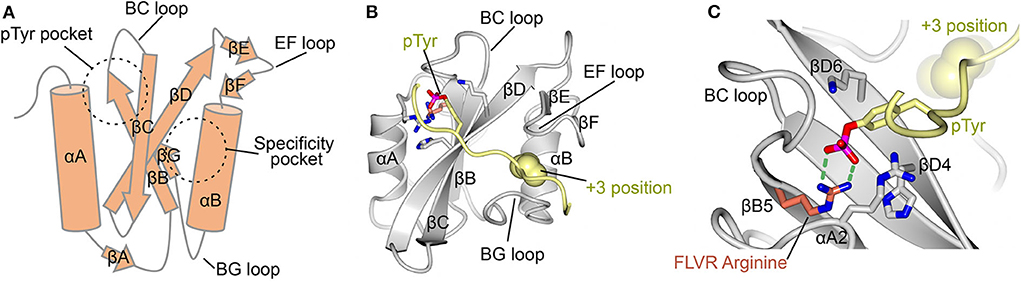
Figure 1. Canonical SH2 domain binding to phosphotyrosine peptide. (A) Schematic diagram of the SH2 domain. Important secondary structure elements are indicated. Locations of the pTyr binding pocket and the specificity pocket are shown. (B) Crystal structure of the LCK SH2 domain in complex with a pTyr peptide (PDB ID: 1LCJ) (28). SH2 domain shown in gray and the pTyr peptide in yellow. pTyr and important residues for its binding are shown in stick format. Ile three residues C-terminal to pTyr (+3 position) is shown in space filling format. (C) Close up of the pTyr binding pocket of LCK showing the FLVR arginine at position βB5 in orange and illustrating its H-bonding to pTyr (green dashed lines). Other important residues for pTyr binding include those at positions αA2, βD4, and βD6.
Evolutionarily, the SH2 domains appear early in the eukaryotic phylogenetic tree and are thought to have co-evolved with tyrosine kinases to the complex array of pTyr-responsive signaling is found in humans (30, 31). Indeed, an ancestral SH2 domain appears to have been identified in SPT6, a transcription elongation factor universally present from yeast to humans. As discussed below, SPT6 maintains the overall SH2 fold but binds to phospho-serine and phospho-threonine thus providing a stepping stone to pTyr binding (31, 32). Unusual SH2 domains have also been acquired and evolved by some bacteria, presumably for invasive purposes, and below we discuss some of those contained in the Legionella genome (33, 34).
Arginine βB5 (The FLVR of an SH2 Domain) and Other Conserved Features
Within the canonically defined pTyr pocket there are conserved residue motifs (21). The most critical motif for pTyr binding includes an arginine at the fifth position of βB, βB5, as part of a highly conserved “FLVR” or “FLVRES” amino acid motif (12, 25) (Figure 1C). As much as half of the free energy of binding is lost on point mutation of this residue (35, 36) resulting in a 1,000-fold reduction in binding affinity (25, 26), and it provides a floor at the base of the deep pTyr pocket which allows specificity toward pTyr over pSer/pThr (4, 8). The FLVR arginine residue is considered key to pTyr recognition, and is conserved in all but 3 of the 120+ human SH2 domains (31).
Other conserved residues which often work in concert with βB5 to coordinate pTyr have been identified. The most prominent are basic residues (arginine or lysine) at positions αA2 and βD6 (Figure 1C). Coordination of pTyr by both αA2 and βD6 is rarely observed in the same SH2 domain and this observation has allowed the definition of two major classes, the Src-like (with a basic residue at αA2), and the SAP-like (with a basic residue at βD6) SH2 domains, referencing two of the most well studied members of the family (33). Experimentally, however, Arg βB5 is the residue most often targeted by point mutagenesis to interrupt SH2-pTyr binding (37, 38).
Outside of the canonical binding site, which provides recognition of the pTyr and +3 positions, interfaces have been found to contribute to binding at a range extending to the −6 and +6 positions (7, 21). Larger interaction and alternative surfaces have also been observed, for example to achieve high FGFR1 selectivity the N-terminal SH2 domain of PLCγ1 uses an extended surface, but its C-terminal SH2 domain does not and is consequently a weaker binder (39). An alternate surface is also used by the SAP SH2 domain which interacts with the SH3 domain of Fyn using a region distal to its pTyr binding site (40). Despite these findings, however, the foundational conserved mode of binding for most SH2 domains to pTyr ligands is centered on the interactions of pTyr and the +3 position. Nonetheless, over the course of study of SH2 domains exceptions to these general rules have been observed.
Unusual SH2 Domains
The exceptions to the canonical “two-pronged plug” binding observed thus far create a diversity in the recognition patterns by which SH2 domains can bind their partners. These exceptions include unusual binding pockets, unique specificities, and dependency of oligomeric state for binding. Below, we highlight some of the mechanisms SH2 domains use to select binding partners and discuss diversity within the pTyr binding site, starting with ancestral SH2 domains before proceeding to eukaryotic SH2 domains.
Ancestral and Borrowed SH2 Domains
Probably the most ancient SH2 domain discovered to date is found in SPT6, an essential transcription elongation protein. This protein contains tandem SH2 domains which are the only two SH2 domains in yeast. They pack against one another and recognize extended phosphorylated serine and threonine peptides of RNA polymerase II (41). The C-terminal SH2 domain lacks a canonical phospho-binding site (32, 42), but instead has a pocket on its back side which binds a pSer in its binding partner. In contrast, the N-terminal SH2 domain has a near canonical phospho-binding pocket which recognizes pThr, and its recent structure-guided analysis showed that the N-SH2 pocket preferentially binds pThr followed by Tyr (41). This pT-X-Y motif makes use of the FLVR arginine to coordinate the pThr's phosphate, but the Tyr is also oriented into this pocket in a manner similar to the aromatic region of a canonical pTyr-SH2 interaction. The coordination of both pThr and Tyr by SPT6 therefore resembles a canonical pTyr-SH2 interaction, making this potentially the evolutionary stepping-stone to SH2-mediated pTyr recognition (41) (Figure 2A).
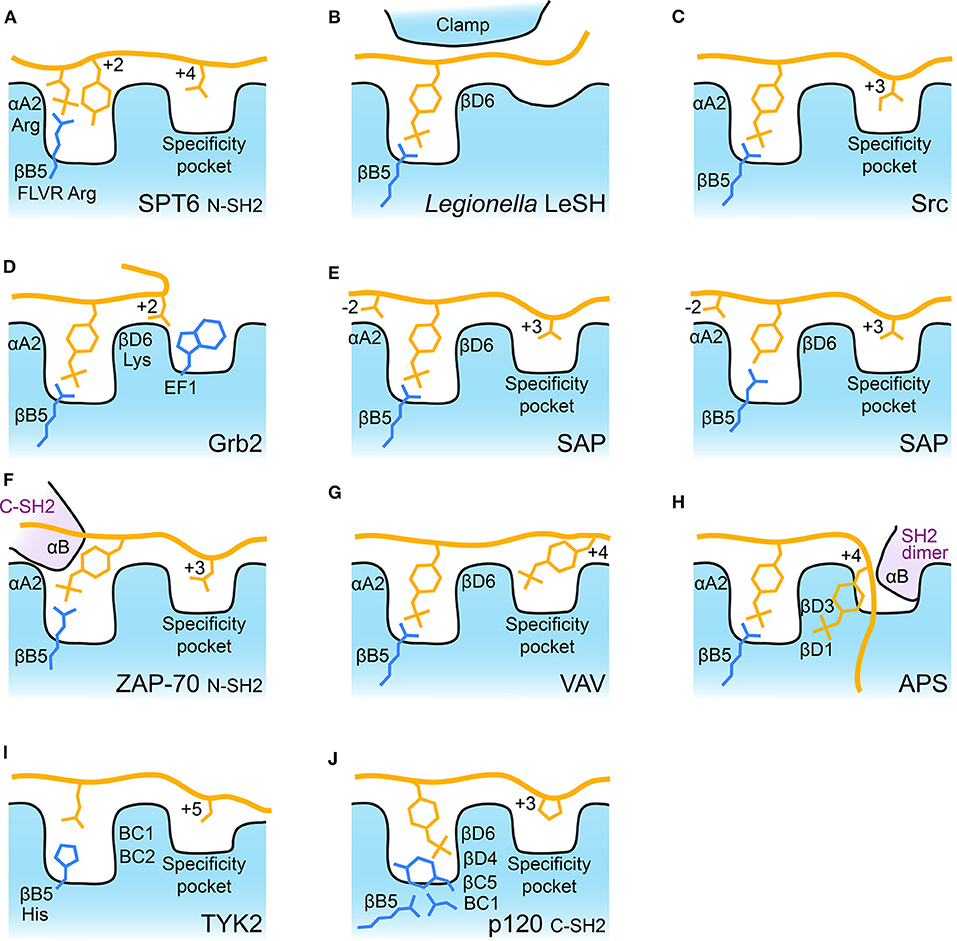
Figure 2. Schematic of SH2 binding modes. Schematic illustration of some of the major SH2 binding features discussed. For each panel the FLVR arginine residue, βB5, is indicated and depicted. Bound peptide is shown with some residues important for selectivity indicated. The FLVR Arg is shown in each schematic. Position αA2 is often Arg and βD6 is often Lys. (A) SPT6 bound to pThr peptide. (B) Legionella LeSH clamping pTyr peptide. βD6 is Arg in LeSH. (C) Src bound to pTyr peptide. (D) Grb2 illustrating Trp blockage of the canonical specificity pocket. (E) SAP bound to phosphorylated and unphosphorylated peptides. (F) ZAP-70 N-SH2 domain illustrating the contribution of residues from its C-SH2 αB helix to pTyr binding. (G) VAV bound to doubly phosphorylated peptide. (H) APS bound to doubly phosphorylated peptide with αB helix from its dimer partner indicated, βD1 and βD3 are both Lys residues. (I) TYK2 bound to cytokine receptor tail. The FLVR residue is a His (indicated), BC1 is Ser and BC2 is Thr. (J) p120RasGAP (p120) C-SH2 bound to phosphorylated peptide. The BC1 residue is an Asp which makes a salt bridge to the FLVR Arg, βB5. βC5 is a Tyr which blocks pTyr interaction with Arg, βB5. βD4 is Arg and βD6 is Lys, both of which coordinate pTyr.
In Gram-negative bacteria, a large number of SH2 domains have been discovered in the Legionella pneumophila genome (33). These are probably the result of horizontal gene transfer but expand the SH2 group to prokaryotes. The Legionella SH2 domains bind using a conserved FLVR arginine and coordinate pTyr in the canonical fashion, but show minimal selectivity for residues in the +3 region due to the lack of a specificity pocket. The structure of the LeSH SH2 domain demonstrates this novel architecture, whereby a large insert (located in the same region as the EF loop) is thought to undergo a “clamping” conformational reorganization which grasps the pTyr peptide and facilitates high affinity binding with low sequence selectivity (33) (Figure 2B). This is a completely divergent mechanism of achieving tight SH2-pTyr binding compared to all other SH2 domains, so it is interesting to speculate on the potential role of these newly identified SH2 domains in hijacking host cell signaling cascades.
Drivers of Substrate Selectivity
Among eukaryotes there is also great diversity in SH2 selectivity. This is most often driven by the selectivity pocket, and much work has been conducted to delineate the ability of individual SH2 domains to recognize specific sequences particularly at the pTyr +3 position (21, 29, 43), and to understand the structural and molecular basis for these selectivities (26, 28, 44) (Figure 2C). These studies have found that for some SH2 domains the mode of peptide binding can be variable. For example, in PLCγ1 a number of pTyr peptides have been shown to bind the FLVR arginine residue but to display significant differences in peptide binding (45). In contrast, the adaptor protein Grb2 and its homolog Gads demonstrate high selectivity by using canonical FLVR arginine interactions to recognize pTyr but an altered selectivity pocket, where a Trp residue contributed by the EF loop forces the bound peptide to turn, to achieve a strong preference for Asn at the +2 position (46–48) (Figure 2D). Furthermore, post-translational modifications can alter peptide selectivity, and this has been observed for multiple members of the Src-family kinase SH2 domains where phosphorylation (for example in Lck at residue Tyr192) alters the selectivity preference at the +2/+3 position (49–51).
Not all SH2 domains are restricted to binding pTyr. For example, the SH2 domain of SAP is mutated in X-linked lymphoproliferative syndrome (XLP) and can bind both phosphorylated and non-phosphorylated tyrosine peptides of the lymphocyte receptor SLAM (52, 53). This ability to bind unphosphorylated Tyr peptides is highly unusual for SH2 domains and is achieved by combining FLVR arginine interactions with either pTyr or Tyr and an extended “three-pronged” interaction (Figure 2E). SAP uses the pTyr and the +3 hydrophobic pocket to bind SLAM, but also utilizes a pocket which recognizes positions N-terminal to the pTyr (52). The extended interaction allows SAP to bind phosphorylated as well as unphosphorylated peptides, albeit with ~2–5 fold lower affinity than binding to the phosphorylated peptide (52, 53).
Binding Dual pTyrs by Tandem SH2 Domains
Some SH2 domains can bind peptides which contain multiple pTyr residues. These include the non-receptor tyrosine kinase ZAP-70, and its homolog, Syk that contain tandem SH2 domains (54, 55) which form a tight module and are linked by a small interdomain coiled coil domain (56). Their role is two-fold; they are critical for autoregulation of catalytic activity by orienting the interdomain coiled coil to pack against the kinase domain, and they recruit ZAP-70 to doubly-tyrosine phosphorylated ITAM receptors. To bind to ITAM cytoplasmic tails, the C-terminal ZAP-70 SH2 domain uses its conserved FLVR arginine and a canonical “two-pronged plug” type interaction (56). In contrast, the N-terminal SH2 domain uses a modified canonical interaction, where the FLVR arginine coordinates pTyr, but the binding site lies at the interface of the two domains and is completed by residues of the C-terminal SH2 domain so that recognition of the second pTyr requires both SH2 domains (Figure 2F) (56). This intercalated pTyr binding site is functionally important for ZAP-70 because its engagement in the context of ITAM binding induces a significant conformational change which releases autoinhibition and is an important step toward full kinase activity (54, 55, 57).
Binding Dual pTyrs by The Same SH2 Domain
Syk is itself phosphorylated, and two of these phosphosites are closely spaced (Y342 and Y346). Recognition of this tandem phosphorylated region is achieved by the SH2 domains of VAV and PLCγ1 (58, 59) in a phospho-state dependent fashion (60). The primary pTyr binding site is canonical and uses the FLVR arginine of VAV/PLCγ1, however, a second pTyr binding site is formed by basic residues of the βD strand and BG loop (Figure 2G). Interestingly, binding of the dual phosphopeptide by PLCγ1 SH2 induces significant conformational changes in the domain compared to when it is bound to mono-phosphorylated peptides (59). Src has also been shown to be able to bind to dual phosphorylated peptides, and structural and thermodynamic analysis of its binding to a doubly phosphorylated PDGF receptor peptide illustrate the coordination of a second pTyr by a basic residue of the βD strand as a potentially more common mechanism for SH2 domains (61).
Binding Quadruple pTyrs by an SH2 Module
Dual phosphotyrosine binding facilitated by βD residues is further expanded on by the adaptor protein APS, a substrate of insulin receptor tyrosine kinase which binds directly to the phosphorylated “activation loop” of the kinase (62). The SH2 domain of this adaptor protein exists as a dimer mediated primarily by its long αB helix (62). In the dimer, both extended αB helices reciprocally interrupt the canonical specificity pocket site of the other SH2 domain, so APS has reduced ability to bind to canonical extended backbone pTyr substrates in the normal fashion (62, 63). Instead, APS binds to two pTyr residues of insulin receptor with a two-pronged site that uses the canonical FLVR arginine pocket and a basic patch created by Lys residues on strand βD (Figure 2H). This is a dual pTyr recognition by both copies of the APS dimer that creates a quadruple phosphotyrosine binding module. From a signaling point of view, the quadruple phosphotyrosine binding does not interrupt insulin receptor's kinase activity, instead it facilitates phosphorylation of the APS C-terminal tail and recruitment of further signaling molecules to the insulin receptor (15).
A Diversity of FLVR
In the above examples, selectivity is mostly driven by alterations in the selectivity pocket or by unusual selectivity sites. These interactions are, however, extremely similar in their utilization of the FLVR arginine to coordinate pTyr (or pThr in SPT6 and Tyr in SAP). In contrast, there are very few SH2 domains which are not conserved at the FLVR arginine and consequently cannot bind pTyr via this residue. In humans these are RIN2, TYK2, and SH2D5; the FLVR arginine is replaced by histidine in RIN2 and TYK2, and a tryptophan in SH2D5. The consequences of this difference have been shown structurally for TYK2, a non-receptor tyrosine kinase of the Janus kinase (JAK) family (64). For all four JAKs (JAK1, JAK2, JAK3, and TYK2), the SH2 domain is a subsidiary component of a larger 4-sub-domain receptor binding module where the SH2 domains mediate a portion, but not the entirety, of the JAK-cytokine receptor interaction (64–67). For receptor binding, the SH2 domains use their canonical peptide substrate binding cleft but do not maintain the requirement to bind pTyr. They often bind an acidic Glu residue instead of pTyr and use an extended specificity pocket to further recognize the receptor tail (68). Evolutionarily, this allowed the FLVR arginine to diverge, and in TYK2 it is a His residue (Figure 2I). The Janus kinases therefore represent a class of SH2 domain that mediates protein-protein interactions without requiring FLVR arginine-pTyr recognition.
Our recent discovery has further illustrated the diversity of SH2 domains. One of the first SH2 proteins to be identified was p120RasGAP (RasGAP, RASA1) (69–71), and for 30 years it was thought to contain two canonical SH2 domains (termed the N- and C-terminals SH2s) (72–74). We investigated p120RasGAP, and our crystal structure and mutagenic analysis of the N-terminal SH2 domain showed this to be correct for the N-terminal domain (75), however, we found that the C-terminal SH2 domain assumes a completely unexpected mode of pTyr recognition (76). Instead of contacting the conserved phosphotyrosine as observed in every other SH2-pTyr interaction, the FLVR arginine at residue position 377 makes a salt bridge to an aspartic acid at position 380. This salt bridge is unprecedented in SH2 domains and requires that pTyr binding is achieved by a unique mechanism that uses multi-dentate recognition by basic residues at βD4 and βD6, and by residues in the BC loop (Figure 2J). The reason for this unusual binding mode is currently unknown, but it is highly conserved over evolution, only diverging in extremely ancient examples of p120RasGAP (76). Despite being one of the first identified and best studied SH2 domain proteins, the C-terminal SH2 domain of p120RasGAP has revealed another mechanism by which SH2 domains can achieve their purpose as protein interaction domains.
Conclusions
The SH2 domain is one of the best studied and understood protein-interaction folds. It has a wide range of functions, and its interactions with partner proteins are generally well-understood. Although the key findings made in the early 1990's describing the molecular basis of SH2-pTyr interactions have been shown to be remarkably robust across the fold and across evolution, there are exceptions to these findings. These include unusual peptide binding pockets, extra pTyr interaction sites, recognition and binding of unphosphorylated peptides, and unusual pTyr recognition sites. The recent discovery of a unique pTyr binding site in p120RasGAP, one of the earliest identified SH2 domains, illustrate the continued discoveries of novelty and diversity among this important group, and the surprises it continues to yield.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This research was partially funded by Mr. Morton and Mrs. Maggie Rosenfeld, the Rosenfeld Science Scholarship and the Yale College Dean's Office to RJ, and by NIH Grants R01GM102262 and R01NS117609, and American Heart Association Grant 19IPLOI34740007 to TB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Amy Stiegler Wyler for helpful comments and discussions.
Abbreviations
APS, Adapter protein with pleckstrin homology and Src homology 2 domains; FGFR1, Fibroblast growth factor receptor 1; Gads, Grb2-related adaptor downstream of Shc; Grb2, Growth factor receptor-bound protein 2; ITAM, Immunoreceptor tyrosine-based activation motif; JAK, Janus kinase; LeSH, Legionella SH2; PDGF, Platelet-derived growth factor; PLCγ1, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1; PTB, phosphotyrosine binding domain; pTyr, Phosphotyrosine; RIN2, Ras and Rab interactor 2; SAP, SLAM-associated protein; SH2, Src homology 2; SH2D5, SH2 domain-containing protein 5; SLAM, Signaling lymphocytic activation molecule; SPT6, Transcription elongation factor SPT6; Syk, Spleen tyrosine kinase; TYK2, Non-receptor tyrosine-protein kinase TYK2; XLP, X-linked lymphoproliferative syndrome; ZAP-70, ζ-chain associated protein of 70kDa.
References
1. Sadowski I, Stone JC, Pawson T. A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol Cell Biol. (1986) 6:4396–408. doi: 10.1128/MCB.6.12.4396
2. Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. (2004) 116:191–203. doi: 10.1016/S0092-8674(03)01077-8
4. Pawson T, Gish G. SH2 and SH3 domains: from structure to function. Cell. (1992) 71:359–62. doi: 10.1016/0092-8674(92)90504-6
5. Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE. (2003) 2003:RE12. doi: 10.1126/stke.2003.191.re12
6. Liu BA, Jablonowski K, Raina M, Arce M, Pawson T, Nash PD. The human and mouse complement of SH2 domain proteins-establishing the boundaries of phosphotyrosine signaling. Mol Cell. (2006) 22:851–68. doi: 10.1016/j.molcel.2006.06.001
7. Liu BA, Engelmann BW, Nash PD. The language of SH2 domain interactions defines phosphotyrosine-mediated signal transduction. FEBS Lett. (2012) 586:2597–605. doi: 10.1016/j.febslet.2012.04.054
8. Kaneko T, Huang H, Cao X, Li X, Li C, Voss C, et al. Superbinder SH2 domains act as antagonists of cell signaling. Sci Signal. (2012) 5:ra68. doi: 10.1126/scisignal.2003021
9. Shah NH, Amacher JF, Nocka LM, Kuriyan J. The src module: an ancient scaffold in the evolution of cytoplasmic tyrosine kinases. Crit Rev Biochem Mol Biol. (2018) 53:535–63. doi: 10.1080/10409238.2018.1495173
10. Pawson T. Protein modules and signalling networks. Nature. (1995) 373:573–80. doi: 10.1038/373573a0
11. Schaffhausen B. SH2 domain structure and function. Biochim Biophys Acta. (1996) 1242:61–75. doi: 10.1016/0304-419X(95)00004-Y
12. Bradshaw JM, Waksman G. Molecular recognition by SH2 domains. Adv Protein Chem. (2002) 61:161–210. doi: 10.1016/S0065-3233(02)61005-8
13. Waksman G, Kuriyan J. Structure and specificity of the SH2 domain. Cell. (2004) 116 (2 Suppl.):S45–8. doi: 10.1016/S0092-8674(04)00043-1
14. Matsuda M, Mayer BJ, Fukui Y, Hanafusa H. Binding of transforming protein, P47gag-crk, to a broad range of phosphotyrosine-containing proteins. Science. (1990) 248:1537–9. doi: 10.1126/science.1694307
15. Hubbard SR. The insulin receptor: both a prototypical and atypical receptor tyrosine kinase. Cold Spring Harb Perspect Biol. (2013) 5:a008946. doi: 10.1101/cshperspect.a008946
16. Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. (2010) 141:1117–34. doi: 10.1016/j.cell.2010.06.011
17. Schindler C, Levy DE, Decker T. JAK-STAT signaling: from interferons to cytokines. J Biol Chem. (2007) 282:20059–63. doi: 10.1074/jbc.R700016200
18. Anderson D, Koch CA, Grey L, Ellis C, Moran MF, Pawson T. Binding of SH2 domains of phospholipase C gamma 1, GAP, and Src to activated growth factor receptors. Science. (1990) 250:979–82. doi: 10.1126/science.2173144
19. Matsuda M, Mayer BJ, Hanafusa H. Identification of domains of the v-crk oncogene product sufficient for association with phosphotyrosine-containing proteins. Mol Cell Biol. (1991) 11:1607–13. doi: 10.1128/MCB.11.3.1607
20. Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. (1993) 72:767–78. doi: 10.1016/0092-8674(93)90404-E
21. Huang H, Li L, Wu C, Schibli D, Colwill K, Ma S, et al. Defining the specificity space of the human SRC homology 2 domain. Mol Cell Proteomics. (2008) 7:768–84. doi: 10.1074/mcp.M700312-MCP200
22. Songyang Z, Shoelson SE, McGlade J, Olivier P, Pawson T, Bustelo XR, et al. Specific motifs recognized by the SH2 domains of Csk 3BP2, fps/fes, Grb-2, HCP, SHC, Syk and Vav. Mol Cell Biol. (1994) 14:2777–85. doi: 10.1128/MCB.14.4.2777
23. Hidaka M, Homma Y, Takenawa T. Highly conserved eight amino acid sequence in SH2 is important for recognition of phosphotyrosine site. Biochem Biophys Res Commun. (1991) 180:1490–7. doi: 10.1016/S0006-291X(05)81364-6
24. Booker GW, Breeze AL, Downing AK, Panayotou G, Gout I, Waterfield MD, et al. Structure of SH2 domain of the p85a subunit of phosphatidylinositol-3-OH kinase. Nature. (1992) 358:684–7. doi: 10.1038/358684a0
25. Waksman G, Kominos D, Robertson SC, Pant N, Baltimore D, Birge RB, et al. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature. (1992) 358:646–53. doi: 10.1038/358646a0
26. Waksman G, Shoelson SE, Pant N, Cowburn D, Kuriyan J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell. (1993) 72:779–90. doi: 10.1016/0092-8674(93)90405-F
27. Overduin M, Rios CB, Mayer BJ, Baltimore D, Cowburn D. Three-dimensional solution structure of the src homology 2 domain of c-abl. Cell. (1992) 70:697–704. doi: 10.1016/0092-8674(92)90437-H
28. Eck MJ, Shoelson SE, Harrison SC. Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck1. Nature. (1993) 362:87–91. doi: 10.1038/362087a0
29. Kaneko T, Huang H, Zhao B, Li L, Liu H, Voss CK, et al. Loops govern SH2 domain specificity by controlling access to binding pockets. Sci Signal. (2010) 3:ra34. doi: 10.1126/scisignal.2000796
30. Liu BA, Shah E, Jablonowski K, Stergachis A, Engelmann B, Nash PD. The SH2 domain-containing proteins in 21 species establish the provenance and scope of phosphotyrosine signaling in eukaryotes. Sci Signal. (2011) 4:ra83. doi: 10.1126/scisignal.2002105
31. Liu BA, Nash PD. Evolution of SH2 domains and phosphotyrosine signalling networks. Philos Trans R Soc Lond B Biol Sci. (2012) 367:2556–73. doi: 10.1098/rstb.2012.0107
32. Sun M, Lariviere L, Dengl S, Mayer A, Cramer P. A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II C-terminal repeat domain (CTD). J Biol Chem. (2010) 285:41597–603. doi: 10.1074/jbc.M110.144568
33. Kaneko T, Stogios PJ, Ruan X, Voss C, Evdokimova E, Skarina T, et al. Identification and characterization of a large family of superbinding bacterial SH2 domains. Nat Commun. (2018) 9:4549. doi: 10.1038/s41467-018-06943-2
34. Gomez-Valero L, Rusniok C, Cazalet C, Buchrieser C. Comparative and functional genomics of legionella identified eukaryotic like proteins as key players in host-pathogen interactions. Front Microbiol. (2011) 2:208. doi: 10.3389/fmicb.2011.00208
35. Waksman G, Kumaran S, Lubman O. SH2 domains: role, structure and implications for molecular medicine. Expert Rev Mol Med. (2004) 6:1–18. doi: 10.1017/S1462399404007331
36. Bradshaw JM, Mitaxov V, Waksman G. Investigation of phosphotyrosine recognition by the SH2 domain of the Src kinase. J Mol Biol. (1999) 293:971–85. doi: 10.1006/jmbi.1999.3190
37. Bibbins KB, Boeuf H, Varmus HE. Binding of the Src SH2 domain to phosphopeptides is determined by residues in both the SH2 domain and the phosphopeptides. Mol Cell Biol. (1993) 13:7278–87. doi: 10.1128/MCB.13.12.7278
38. Mayer BJ, Jackson PK, Van ER, Baltimore D. Point mutations in the abl SH2 domain coordinately impair phosphotyrosine binding in vitro and transforming activity in vivo. Mol Cell Biol. (1992) 12:609–18. doi: 10.1128/MCB.12.2.609
39. Bae JH, Lew ED, Yuzawa S, Tome F, Lax I, Schlessinger J. The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell. (2009) 138:514–24. doi: 10.1016/j.cell.2009.05.028
40. Chan B, Lanyi A, Song HK, Griesbach J, Simarro-Grande M, Poy F, et al. SAP couples fyn to SLAM immune receptors. Nat Cell Biol. (2003) 5:155–60. doi: 10.1038/ncb920
41. Sdano MA, Fulcher JM, Palani S, Chandrasekharan MB, Parnell TJ, Whitby FG, et al. A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription. Elife. (2017) 6:e28723. doi: 10.7554/eLife.28723
42. Diebold ML, Loeliger E, Koch M, Winston F, Cavarelli J, Romier C. Noncanonical tandem SH2 enables interaction of elongation factor Spt6 with RNA polymerase II. J Biol Chem. (2010) 285:38389–98. doi: 10.1074/jbc.M110.146696
43. Liu H, Huang H, Voss C, Kaneko T, Qin WT, Sidhu S, et al. Surface loops in a single SH2 domain are capable of encoding the spectrum of specificity of the SH2 Family. Mol Cell Proteomics. (2019) 18:372–82. doi: 10.1074/mcp.RA118.001123
44. Bradshaw JM, Grucza RA, Ladbury JE, Waksman G. Probing the “two-pronged plug two-holed socket” model for the mechanism of binding of the Src SH2 domain to phosphotyrosyl peptides: a thermodynamic study. Biochemistry. (1998) 37:9083–90. doi: 10.1021/bi973147k
45. McKercher MA, Guan X, Tan Z, Wuttke DS. Multimodal recognition of diverse peptides by the C-terminal SH2 domain of phospholipase c-gamma1 protein. Biochemistry. (2017) 56:2225–37. doi: 10.1021/acs.biochem.7b00023
46. Inaba S, Numoto N, Ogawa S, Morii H, Ikura T, Abe R, et al. Crystal structures and thermodynamic analysis reveal distinct mechanisms of CD28 phosphopeptide binding to the src homology 2 (SH2) domains of three adaptor proteins. J Biol Chem. (2017) 292:1052–60. doi: 10.1074/jbc.M116.755173
47. Ogura K, Tsuchiya S, Terasawa H, Yuzawa S, Hatanaka H, Mandiyan V, et al. Solution structure of the SH2 domain of Grb2 complexed with the Shc-derived phosphotyrosine-containing peptide. J Mol Biol. (1999) 289:439–45. doi: 10.1006/jmbi.1999.2792
48. Rahuel J, Gay B, Erdmann D, Strauss A, Garcia-Echeverria C, Furet P, et al. Structural basis for specificity of Grb2-SH2 revealed by a novel ligand binding mode. Nat Struct Biol. (1996) 3:586–9. doi: 10.1038/nsb0796-586
49. Granum S, Sundvold-Gjerstad V, Gopalakrishnan RP, Berge T, Koll L, Abrahamsen G, et al. The kinase Itk and the adaptor TSAd change the specificity of the kinase Lck in T cells by promoting the phosphorylation of Tyr192. Sci Signal. (2014) 7:ra118. doi: 10.1126/scisignal.2005384
50. Courtney AH, Amacher JF, Kadlecek TA, Mollenauer MN, Au-Yeung BB, Kuriyan J, et al. A phosphosite within the SH2 domain of lck regulates its activation by CD45. Mol Cell. (2017) 67:498–511.e6. doi: 10.1016/j.molcel.2017.06.024
51. Jin LL, Wybenga-Groot LE, Tong J, Taylor P, Minden MD, Trudel S, et al. Tyrosine phosphorylation of the Lyn Src homology 2 (SH2) domain modulates its binding affinity and specificity. Mol Cell Proteomics. (2015) 14:695–706. doi: 10.1074/mcp.M114.044404
52. Poy F, Yaffe MB, Sayos J, Saxena K, Morra M, Sumegi J, et al. Crystal structures of the XLP protein SAP reveal a class of SH2 domains with extended, phosphotyrosine-independent sequence recognition. Mol Cell. (1999) 4:555–61. doi: 10.1016/S1097-2765(00)80206-3
53. Hwang PM, Li C, Morra M, Lillywhite J, Muhandiram DR, Gertler F, et al. A “three-pronged” binding mechanism for the SAP/SH2D1A SH2 domain: structural basis and relevance to the XLP syndrome. Embo J. (2002) 21:314–23. doi: 10.1093/emboj/21.3.314
54. Au-Yeung BB, Deindl S, Hsu LY, Palacios EH, Levin SE, Kuriyan J, et al. The structure, regulation, and function of ZAP-70. Immunol Rev. (2009) 228:41–57. doi: 10.1111/j.1600-065X.2008.00753.x
55. Au-Yeung BB, Shah NH, Shen L, Weiss A. ZAP-70 in signaling, biology, and disease. Annu Rev Immunol. (2018) 36:127–56. doi: 10.1146/annurev-immunol-042617-053335
56. Hatada MH, Lu X, Laird ER, Green J, Morgenstern JP, Lou M, et al. Molecular basis for the interaction of the protein tyrosine kinase ZAP-70 with the T-cell receptor. Nature. (1995) 377:32–8. doi: 10.1038/377032a0
57. Yan Q, Barros T, Visperas PR, Deindl S, Kadlecek TA, Weiss A, et al. Structural basis for activation of ZAP-70 by phosphorylation of the SH2-kinase linker. Mol Cell Biol. (2013) 33:2188–201. doi: 10.1128/MCB.01637-12
58. Chen CH, Martin VA, Gorenstein NM, Geahlen RL, Post CB. Two closely spaced tyrosines regulate NFAT signaling in B cells via Syk association with Vav. Mol Cell Biol. (2011) 31:2984–96. doi: 10.1128/MCB.05043-11
59. Groesch TD, Zhou F, Mattila S, Geahlen RL, Post CB. Structural basis for the requirement of two phosphotyrosine residues in signaling mediated by Syk tyrosine kinase. J Mol Biol. (2006) 356:1222–36. doi: 10.1016/j.jmb.2005.11.095
60. Chen CH, Piraner D, Gorenstein NM, Geahlen RL C. Beth post: differential recognition of syk-binding sites by each of the two phosphotyrosine-binding pockets of the Vav SH2 domain. Biopolymers. (2013) 99:897–907. doi: 10.1002/bip.22371
61. Lubman OY, Waksman G. Structural and thermodynamic basis for the interaction of the Src SH2 domain with the activated form of the PDGF beta-receptor. J Mol Biol. (2003) 328:655–68. doi: 10.1016/S0022-2836(03)00344-9
62. Hu J, Liu J, Ghirlando R, Saltiel AR, Hubbard SR. Structural basis for recruitment of the adaptor protein APS to the activated insulin receptor. Mol Cell. (2003) 12:1379–89. doi: 10.1016/S1097-2765(03)00487-8
63. Hu J, Hubbard SR. Structural basis for phosphotyrosine recognition by the Src homology-2 domains of the adapter proteins SH2-B and APS. J Mol Biol. (2006) 361:69–79. doi: 10.1016/j.jmb.2006.05.070
64. Wallweber HJ, Tam C, Franke Y, Starovasnik MA, Lupardus PJ. Structural basis of recognition of interferon-alpha receptor by tyrosine kinase 2. Nat Struct Mol Biol. (2014) 21:443–8. doi: 10.1038/nsmb.2807
65. Zhang D, Wlodawer A, Lubkowski J. Crystal structure of a complex of the intracellular domain of interferon lambda receptor 1 (IFNLR1) and the FERM/SH2 domains of human JAK1. J Mol Biol. (2016) 428:4651–68. doi: 10.1016/j.jmb.2016.10.005
66. Ferrao R, Wallweber HJ, Ho H, Tam C, Franke Y, Quinn J, et al. The structural basis for class ii cytokine receptor recognition by JAK1. Structure. (2016) 24:897–905. doi: 10.1016/j.str.2016.03.023
67. McNally R, Toms AV, Eck MJ. Crystal structure of the FERM-SH2 module of human Jak2. PLoS ONE. (2016) 11:e0156218. doi: 10.1371/journal.pone.0156218
68. Ferrao R, Lupardus PJ. The Janus Kinase (JAK) FERM and SH2 Domains: bringing specificity to JAK-Receptor interactions. Front Endocrinol. (2017) 8:71. doi: 10.3389/fendo.2017.00071
69. Trahey M, McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. (1987) 238:542–5. doi: 10.1126/science.2821624
70. Vogel US, Dixon RA, Schaber MD, Diehl RE, Marshall MS, Scolnick EM, et al. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. (1988) 335:90–3. doi: 10.1038/335090a0
71. Trahey M, Wong G, Halenbeck R, Rubinfeld B, Martin GA, Ladner M, et al. Molecular cloning of two types of GAP complementary DNA from human placenta. Science. (1988) 242:1697–700. doi: 10.1126/science.3201259
72. Pamonsinlapatham P, Hadj-Slimane R, Lepelletier Y, Allain B, Toccafondi M, Garbay C, et al. p120-Ras GTPase activating protein (RasGAP): a multi-interacting protein in downstream signaling. Biochimie. (2009) 91:320–8. doi: 10.1016/j.biochi.2008.10.010
73. McCormick F, Adari H, Trahey M, Halenbeck R, Koths K, Martin GA, et al. Interaction of ras p21 proteins with GTPase activating protein. Cold Spring Harb Symp Quant Biol. (1988) 53:849–54. doi: 10.1101/SQB.1988.053.01.097
74. Adari H, Lowy DR, Willumsen BM, Der CJ, McCormick F. Guanosine triphosphatase activating protein (GAP) interacts with the p21 ras effector binding domain. Science. (1988) 240:518–21. doi: 10.1126/science.2833817
75. Jaber Chehayeb R, Stiegler AL, Boggon TJ. Crystal structures of p120RasGAP N-terminal SH2 domain in its apo form and in complex with a p190RhoGAP phosphotyrosine peptide. PLoS ONE. (2019) 14:e0226113. doi: 10.1371/journal.pone.0226113
Keywords: Src-homology 2, phosphotyrosine, FLVR motif, protein-protein interaction, signal transduction, protein domains
Citation: Jaber Chehayeb R and Boggon TJ (2020) SH2 Domain Binding: Diverse FLVRs of Partnership. Front. Endocrinol. 11:575220. doi: 10.3389/fendo.2020.575220
Received: 22 June 2020; Accepted: 12 August 2020;
Published: 18 September 2020.
Edited by:
Ichiro Maruyama, Okinawa Institute of Science and Technology Graduate University, JapanReviewed by:
Anne Spurkland, University of Oslo, NorwayOliver Hantschel, École Polytechnique Fédérale de Lausanne, Switzerland
Copyright © 2020 Jaber Chehayeb and Boggon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Titus J. Boggon, dGl0dXMuYm9nZ29uQHlhbGUuZWR1
 Rachel Jaber Chehayeb1,2
Rachel Jaber Chehayeb1,2 Titus J. Boggon
Titus J. Boggon