- Suntory Foundation for Life Sciences, Bioorganic Research Institute, Kyoto, Japan
Cnidarians are early-branching animals in the eukaryotic tree of life. The phylum Cnidaria are divided into five classes: Scyphozoa (true jellyfish), Cubozoa (box jellyfish), Hydrozoa (species, Hydra and Hydractinia), Anthozoa (sea anemone, corals, and sea pen), and Staurozoa (stalked jellyfish). Peptides play important roles as signaling molecules in development and differentiation in cnidaria. For example, cnidaria use peptides for cell-to cell communication. Recent discoveries show that Hydra neuropeptides control several biological processes including muscle contraction, neuron differentiation, and metamorphosis. Here, I describe the structure and functions of neuropeptides in Hydra and other cnidarian species. I also discuss that so-called primitive nervous system of Hydra is in more complex than generally believed. I also discuss how cnidaria use peptides for communication among cells rather than in higher animals.
Introduction
Molecular phylogenetic studies show that Cnidaria are the sister group of Bilateria. Ancestral Cnidarians diverged over 500 million years ago in animal evolution. Despite the long course of evolution, the nervous systems of cnidarians are differentiated (1). Cnidarian species are also mainly classified into two groups according to the unique life cycle, the anthozoans and medusozoans (1). Anthozoa lives exclusively as polyps. Among medusozoans, Cubozoa and Scyphozoa predominantly live as medusae. On the other hand, Hydrozoa usually follows a life cycle where the species alternate between these two forms except for Hydra and Hydractinia. Staurozoa lives exclusively as polyps.
Cnidaria such as Hydra are composed of multiple cell types that represent the fundamental architecture of multicellular organisms. Hydra exhibits a simple body plan with a head and tentacles on one end and a foot on the opposite end of a hypostome. The gastric region is located between the head and foot. The body is composed of two layers, ectoderm and endoderm, which are separated by an extracellular matrix, the mesoglea. The cells of both epithelial layers also function as muscle cells. Hydra also have multipotent interstitial stem cells, which differentiate into nerve cells (2), nematocytes (2), gland cells (3), and germ cells (4). Hydra as a member of cnidaria represents an attractive model to understand axial pattern formation into head- and foot-specific tissues.
The nervous system of Hydra is simple and is composed of a nerve net that extends throughout the animal. The cnidarian nervous system is mainly peptidergic (5). Classical molecules such as acetylcholine also contribute to the Hydra nervous system (6).
Peptides play important roles as hormones and neurotransmitters and they are involved in the maintenance of a variety of developmental stages. However, little is known about whether they are involved in differentiation and development. In Hydra, theoretical model suggests that small molecules such as peptides are transported to establish morphogenetic gradients that regulate patterning processes. To systematically identify and characterize peptide signaling molecules, we started the Hydra Peptide Project (7). By using the strategy illustrated in Figure 1, many peptides were extracted and purified with successive steps of high performance liquid chromatography (HPLC). Signaling peptides were identified by their effect on the gene expression profile of Hydra by using differential display (DD)-PCR. Positive peptides were chemically synthesized, the synthetic peptides were used for biological assays including behavioral (muscle contraction), neuron differentiation, and others. Furthermore, introduction of the Hydra Expressed Sequence Tag (EST) Project has enabled us to identify transcripts for novel peptides even more efficiently (Figure 1) (8).
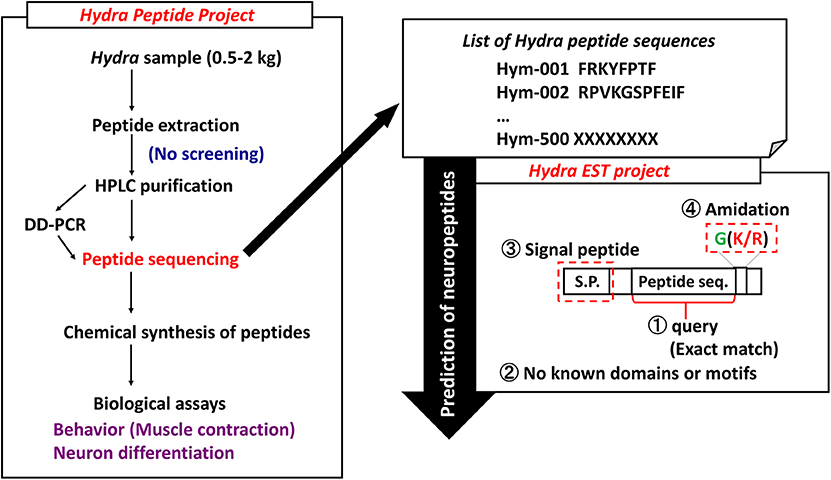
Figure 1. Strategy to identify Hydra neuropeptides. DD-PCR, differential display PCR; HPLC, high performance liquid chromatography.
The primary aim of the present review is to describe the structures and functions of peptide signaling molecules such as neuropeptides in cnidarians, especially in Hydra.
Cnidarian Neuropeptides
FMRFamide-Like Peptides (FLPs)
The peptide FMRFamide was originally purified from the cerebral ganglion of the clam Macrocallista nimbosa (9, 10). Other mollusks and members of most other phyla express peptides with a similar sequence. FMRFamides are categorized into two groups depending on the structural similarity with FMRFamide. The first category consists of FMRFamide-related peptides (FaRPs), which include encode for multiple peptides with the C-terminal FMRFamide or FLRFamide (11). The second category of FMRFamides includes FLPs, which are peptides that have only the RFamide sequence at C-termini (12). Therefore, FaRPs and all other RFamide peptides are considered FLPs. Krajniak (13) excellently reviewed FaRPs in invertebrates. This overview primarily focuses on cnidarian FLPs.
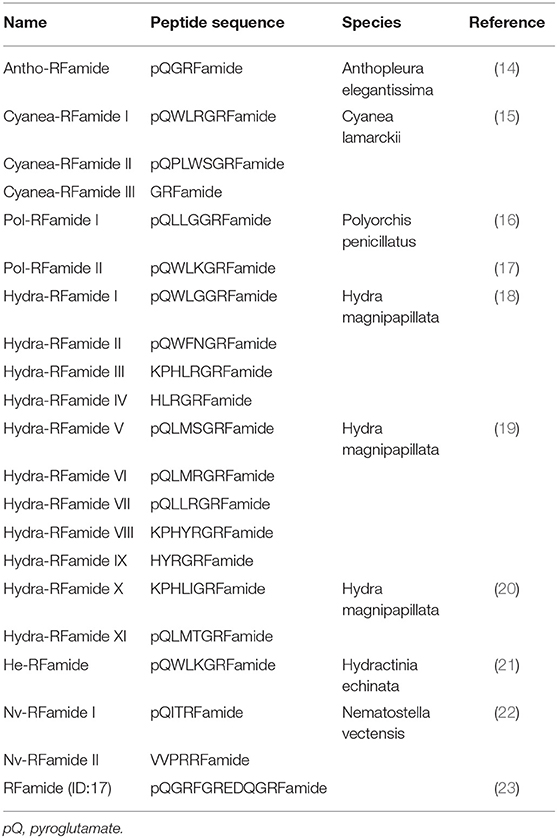
A variety of FLPs are expressed in the evolutionarily ancient nervous system of cnidarians (Table 1). Peptides with GRFamide at the C-terminus have been found in a scyphozoan (the jellyfish Cyanea lamarckii) (15), three hydrozoans (Hydra magnipapillata, the hydromedusa Polyorchis penicillatus, and Hydractinia echinata) (16–21), and an anthozoan (the sea anemone Anthopleura elegantissima) (14), whereas peptides with TRFamide and/or RRFamide at the C-terminus have been described in another anthozoan (the sea anemone Nematostella vectensis) (22). All mature neuropeptides are controlled by highly regulated secretion pathways. Usually, a precursor of a neuropeptide is incorporated as a preprohormone in the endoplasmic reticulum, where it is converted into a prohormone. Next, prohormones move to the Golgi apparatus for endoproteolysis and/or amidation at the C-terminus, which results in the final active peptide. FLPs have been identified in numerous cnidarians. A Calliactis parasitica cDNA includes 19 copies of Antho-RFamide (Table 1), two copies of FQGRFamide, and one copy of YVPGRYamide (24). Two cDNAs have been isolated from Anthopleura elegantissima; one cDNA includes 13 copies of Antho-RFamide (Table 1) and nine other FLPs; the second cDNA includes 14 copies of Antho-RFamide and eight other FLPs (25). Renilla koellikeri has 36 copies of Antho-RFamide (26). A Polyorchis penicillatus cDNA includes one copy of Pol-RFamide I (Table 1) and 11 copies of Pol-RFamide II (Table 1), in addition to another predicted FLP (27). In Hydra, RFamides are spliced from three different preprohormones called A, B, and C (Figure 2A). Preprohormone-A includes six Hydra-RFamides (Hydra-RFamide I-VI) (Figure 2A) (Table 1) (19). Preprohormone-B has one copy of Hydra-RFamide I and Hydra-RFamide II and three probable Hydra-RFamides (Hydra-RFamide V, VII, and VIII) (Figure 2A) (19). Preprohormone-C has one copy of Hydra-RFamide I and seven copies of additional neuropeptide sequences (one copy of pQWFSGRFamide and six copies of pQWLSGRFamide) (Figure 2A) (19). In Hydractinia echinata, one copy of He-RFamide is present (Table 1) (21). In Nematostella vectensis, three FLPs [Nv-RFamide I and II and RFamide (ID:17)] are present (Table 1) (22, 23). Collectively, precursor-encoding cnidarian FLP cDNAs yield many neuropeptides with great structural diversity, indicating that they have great functional diversity as well.
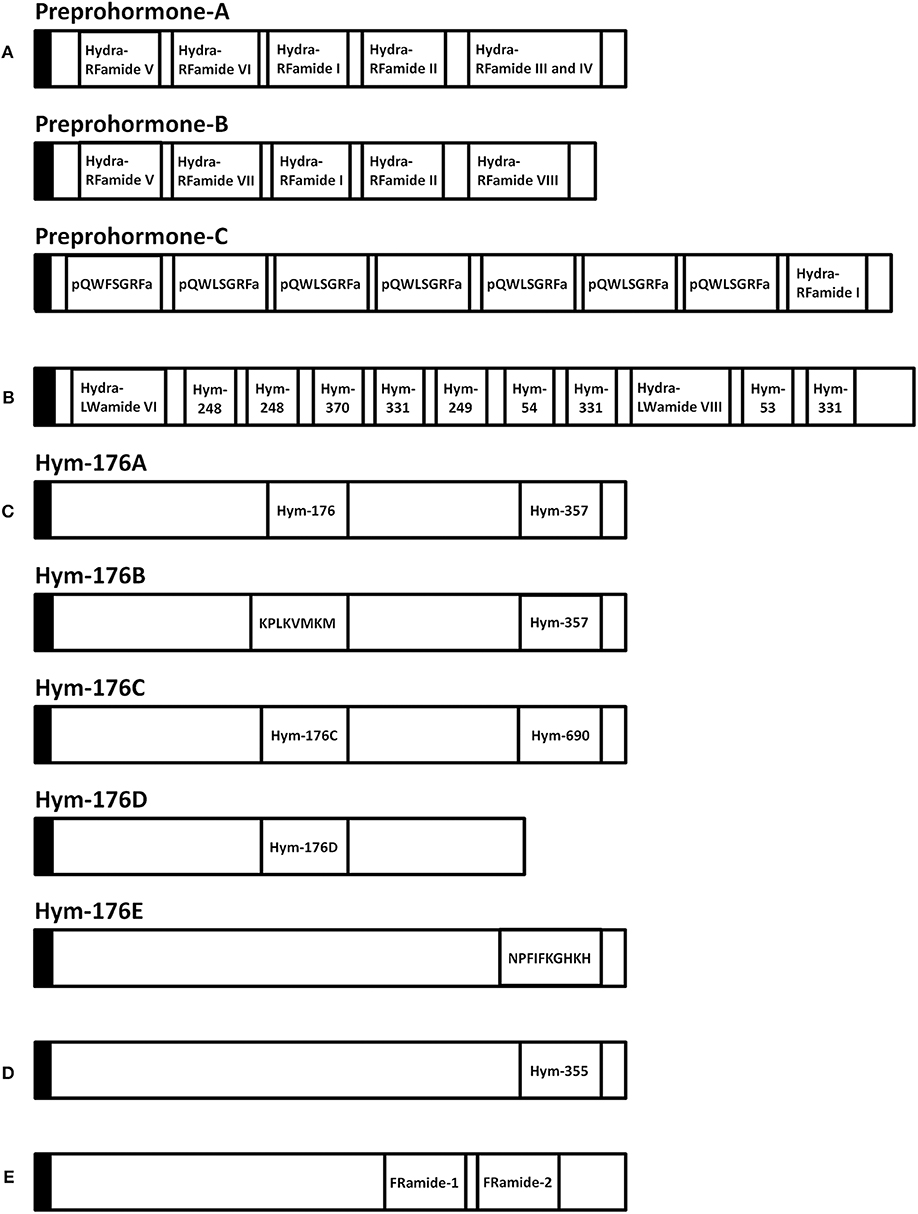
Figure 2. Schematic representation of eleven preprohormones in Hydra. (A) Preprohormone-A contains unprocessed Hydra-RFamide I, II, III, IV, V, and VI. Preprohormone-B contains unprocessed Hydra-RFamide I, II, V, VII, and VIII. Preprohormone-C contains Hydra-RFamide I and two putative neuropeptide sequences (pQWFSGRFa and pQWLSGRFa). (B) The GLWamide precursor contains unprocessed GLWamides (Hym-53, 54, 248, 249, 331, and 370) and two putative neuropeptide sequences (Hydra-LWamide VI and VIII). (C) The Hym-176 precursor (Hym-176A) contains one copy of unprocessed Hym-176 and Hym-357. Hym-176B contains Hym-357 and one putative neuropeptide (KPLKVMKM). Hym-176C and Hym-176D contain one copy of Hm-176-homologous peptide (Hym-176C and Hym-176D), respectively. Hym-176C also contains one unprocessed Hym-690. Hym-176E contains one putative neuropeptide sequence (NPFIFKGHKH). (D) The Hym-355 precursor contains one unprocessed Hym-355. (E) The FRamide precursor contains unprocessed FRamide-1 and−2. Black box: signal sequence, a: amide.
Cnidarian FLPs control several functions, such as muscle contraction, feeding, sensation, reproduction, metamorphosis, and movement of larvae. Treatment of the sea anemone Calliactis parasitica with Antho-RFamide increases muscle tone, contraction amplitude, and contraction of slow muscles (28). In individual autozooid polyps of Renilla koellikeri, Antho-RFamide also leads to tonic contractions in the rachis and peduncle (29). In Hydra, Hydra-RFamide III mediates pumping of the peduncle in a dose-dependent manner (30).
FMRFamide activates a Na+ channel identified in snails (31, 32). Three cation channel subunits of the degenerin (DEG)/epithelial Na+ channel (ENaC) gene family were cloned from the freshwater polyp Hydra magnipapillata and designated Hydra Na+ channel (HyNaC)2–4 (33). Subsequently, a novel subunit, designated HyNaC5, was cloned, and expression of the gene was shown to be co-localized with HyNaC2 and HyNaC3 at the base of the tentacles (34). Co-injection of HyNaC5 with HyNaC2 and HyNaC3 genes in Xenopus oocytes strongly enhances the current amplitude after peptide application and increases the affinity of the channel for Hydra-RFamide I and II (34). HyNaC2/3/5 is assembled into a functional heterotrimeric channel that is activated by Hydra-RFamide I with high affinity. The experimental data of HyNaCs suggested that secretion of Hydra-RFamide I and/or II induces tentacle contraction, perhaps during feeding (33, 34). Seven additional HyNaC subunits, HyNaC6-HyNaC12, were cloned, and all belong to the DEG/ENaC gene family (35). These subunits and the four originally identified subunits self-assemble in Xenopus oocytes to create 13 different ion channels that show high-affinity binding of Hydra-RFamide I and II. The HyNaC inhibitor, diminazene, slows tentacle movement in Hydra. Because Hydra express multiple peptide-gated ion channels with a restricted number of FLPs as ligands (35), FLPs may be important for fast transmission at neuro-muscular junction in cnidarians. The function of Hydra-RFamide IV in Hydra is unknown.
Highly specialized mechanoreceptor cells, called stinging cells or nematocytes, that are important for capturing prey and defense are present in cnidarians (36). Two- and three-cell synaptic pathways, including synapses between nematocytes and nearby nerve cells, are present in the epidermis of the sea anemone tentacles (37, 38). Cnidarian sensory function is probably mediated by FLPs, as evidenced by anti-FMRFamide and anti-RFamide antibody staining in the tentacles of four classes of cnidaria. Thus, FLPs probably mediate chemosensory regulation of cnidocyte discharge (39). The epidermal sensory cells of the spot ocellus in Aurelia are also positive for FMRFamide (40), which may inhibit spontaneous firing of nematocytes.
FLPs also play a key role in cnidarian reproduction, larval movement, and metamorphosis. Reproduction of colonial octocorals such as Renilla koellikeri occurs via spawning and exfoliation. Intact gamete follicles are released into the water during spawning. These follicles rupture during exfoliation, releasing the gametes. Antho-RFamide is present in ciliated neurons in the epithelium of follicles of Renilla koellikeri and induces exfoliation of the epithelium and subsequent release of the gametes into water (41). Light enhances the potency of Antho-RFamide (41).
The colony-forming marine hydroid, Hydractinia echinata, is closely related to freshwater Hydra. Fertilized eggs of this species undergo rapid cleavage divisions for about 1 day and develop into spindle-shaped planula larvae in about 3 days (42). Planula larvae are capable of migrating toward light (43), and they metamorphose into adult polyps when they receive appropriate environmental stimuli (44, 45). Hydra-RFamide I inhibits the migration of planula larvae, thus modulating phototaxis by inhibiting myomodulation (43). Metamorphosis is also inhibited by this peptide, leading to the suggestion that the function of endogenous FLPs is to stabilize the larval stage (46). Thus, FLPs may play a role in regulating the movement of planula larvae prior to metamorphosis, possibly linking movement to chemotactic or phototactic processes (47). Sensory neurons that express FLPs are present in planula larvae, suggesting that migration and metamorphosis of these animals may be mediated by secretion of endogenous neuropeptides in response to environmental stimuli.
GLWamides
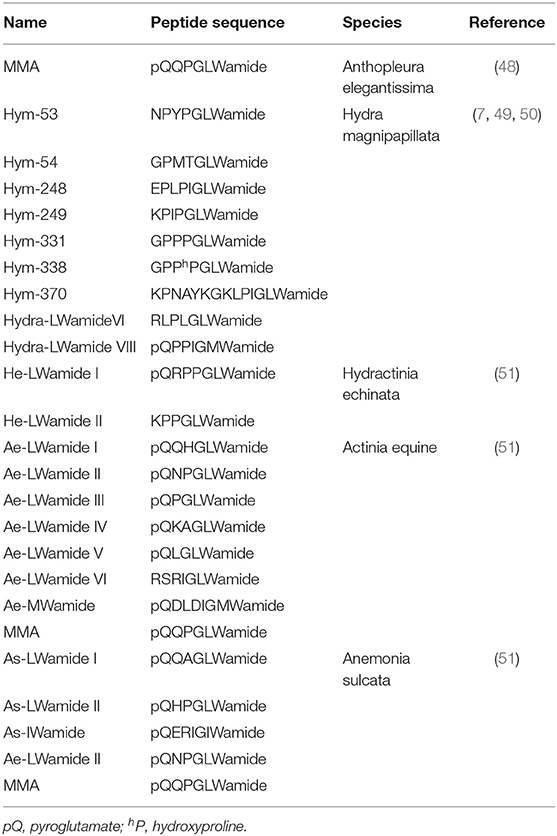
GLWamides are characterized by certain features at their N- and C-termini. Most GLWamides have a GLWamide motif at the C-terminus (Table 2). Seven GLWamide peptides are found in Hydra, and they include X-Pro or X-Pro-Pro at their N-termini (Table 2) (7, 49). In the anthozoan Anthopleura elegantissima, Metamorphosin A (MMA) that is a member of the GLWamide family has an N-terminal pyroglutamine (Table 2) (48). Both N-terminal modifications produce resistance to aminopeptidase (52).
GLWamide cDNAs are found in other cnidarians as well. A cDNA encoding a preprohormone with 11 immature peptide sequences, nine of which are unique, was cloned from Hydra magnipapillata (Figure 2B) (50). The corresponding gene includes one copy of Hym-53 (NPYPGLWamide), Hym-54 (GPMTGLWamide), Hym-249 (KPIPGLWamide), and Hym-370 (KPNAYKGKLPIGLWamide); two copies of Hym-248 (EPLPIGLWamide); and t copies of Hym-331 (GPPPGLWamide), as well as two additional putative GLWamides (Hydra-LWamide VI and VIII) (Table 2). Hydra-LWamide VIII is predicted from this cDNA and probably includes GMWamide at the C-terminus (50). A cDNA encoding GLWamides has been cloned from Hydractinia echinata (51) and includes one copy of He-LWamide I and 17 copies of He-LWamide II (Table 2). Two unique cDNAs have been cloned from the anthozoans Actinia equine and Anemonia sulcata (51). The Actinia gene includes one copy of MMA, Ae-LWamide IV, Ae-LWamide V, Ae-LWamide VI, and Ae-MWamide; two copies of Ae-LWamide I and Ae-LWamide III; and four copies of Ae-LWamide II (Table 2). In contrast, the Anemonia gene has one copy of MMA, Ae-LWamide II, and As-IWamide; two copies of As-LWamide II; and four copies of As-LWamide I (Table 2) (51). The preprohormones of anthozoans but not hydrozoans include MMA. The peptide is probably a prototype of the family (53). Two other peptides that are possibly generated from the preprohormones of Actinia and Anemonia are likely processed into -GMWamide (Ae-MWamide) and -GIWamide (As-IWamide) at their C-terminus (Table 2). Whether these two peptides and Hydra-LWamide VIII belong to the GLWamide family is uncertain, as substitution of the Leu residue in GLWamide with Met or Ile results in deactivation of contractile activity in the retractor muscle of the anthozoan Anthopleura fuscoviridis (54).
The various species of Hydractinia generally live on hermit crab shells. The Hydractinia life cycle includes a planula larval stage but no medusa stage. After attaching to snail shells, planula larvae undergo MMA-induced metamorphosis and become polyps after about 1 week (48, 55). MMA thus works as a neurohormone to mediate development in addition to its roles as a neurotransmitter and neuromodulator. In Hydractinia serrata, Hydra GLWamides also cause polyp development from planula larvae (7, 49). A common GLWamide sequence is required to induce metamorphosis in Hydractinia, and the GLWamide terminus and amidation are essential and specific for inducing metamorphosis (56). Substitution of Gly in GLWamide with another common amino acid (except Cys) decreases or completely inhibits potency of the peptide, and substitution of Leu or Trp in GLWamide with another common amino acid (except Cys) partially or completely blocks its potency for muscle contraction in Anthopleura fuscoviridis (54). The precise mechanism of how these peptides induce metamorphosis remains to be determined. Bacteria in the environment produce a chemical that can induce larvae to undergo metamorphosis (48). This chemical signal probably affects sensory neurons in the planula larvae that secrete endogenous GLWamides to induce a phenotypic change in the surrounding epithelial cells. Hydra lack a larval stage and develop directly into adults from embryos, and thus, how GLWamide peptides function during early development in Hydra is unclear.
Motile planula larvae play a role in sexual reproduction in reef-building corals. These larvae undergo complex metamorphosis after adhering to a substrate, and a juvenile coral colony results. In Acropora, Hym-248 induces dose-dependent metamorphosis of nearly 100% of planula larvae into polyps (57). However, the effect of Hym-248 on metamorphosis is species-specific (57, 58). A Hym-248-specific receptor appears to exist in Acropora. The receptor may serve as a barrier to ensure specification in corals. In Hydractinia, the peptide for their receptors is loose. The possible receptors may share certain common sequences and binding sites. Hym-248-related peptide(s) are expected to be identified in Acropora.
In Hydra, all GLWamide peptides serve as myoactive peptides to activate sphincter muscle contraction and bud detachment (7). The sphincter muscle is involved in bud detachment. To test myoactivity in Hydra, nerve-free tissue of epithelial hydra is typically used (59, 60). When normal Hydra that contains nerve cells is treated with the peptides, they exhibit the same effect as epithelial Hydra. GLWamides are synthesized and expressed in nerve cells (49) and thus function as neurotransmitters or neuromodulators at the neuro-muscular junction. Hym-248, which is a Hydra GLWamide, induces both bud detachment and body elongation (49). Muscle tissue in Hydra runs perpendicular to the ectodermal and endodermal epithelial cells. Hym-248 may bind to two different types of receptors, one that binds all types of GLWamides and one that specifically binds to Hym-248. Substance P (SP) is a highly conserved member of the tachykinin peptide family that is widely expressed throughout the animal kingdom (61). It binds to tachykinin receptors [neurokinin-1, 2, and 3 receptor (NK1R, NK2R, and NK3R)] that belong to G-protein-coupled receptors (GPCRs). SP preferentially activates NK1R. This difference of specificity against other tachykinin peptides can be accounted for the conformational flexibility of the short and linear peptides and ligand binding affinity for the receptors (62). Probably, the features of both receptors for Hym-248 may depend on the ligand structure and binding affinity for receptors.
All GLWamide family peptides enhance retractor muscle contraction of Anthopleura (49). Nerve cells in the sea anemone retractor muscle stain strongly with a GLWamide motif-specific antibody, similar to the nervous system of Hydra (49).
In Hydractinia echinata, GLWamide and RFamide neuropeptides modulate planula larva migration. He-LWamide II, which is a GLWamide, induces migration by extending the active period (43). GLWamides and FLPs antagonize one another to modulate migration of Hydractinia echinata planula larvae.
In hydrozoan jellyfish, maturation of oocytes and spawning are initiated by light-dark cycles in natural conditions within 1 second (63). Exposure to Hym-53 for < 2 min is sufficient for oocyte maturation and spawning (64). Thus, neuropeptides function as hormones that modulate the first step that determines whether oocytes undergo irreversible meiosis after light exposure.
Hym-176 (APFIFPGPKVamide)
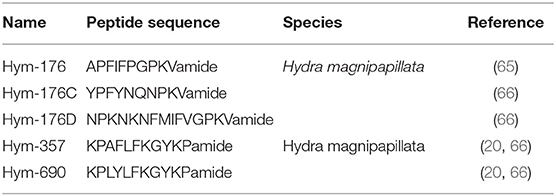
Hym-176 was a newly identified as a neuropeptide (Table 3) (7, 65). The gene that encodes Hym-176 is strongly expressed in the neurons of the lower peduncle and weakly expressed in the gastric region (67). This peptide induces contraction of the ectodermal muscle in Hydra (65). This region-specific neuron subset correlates with the myoactivity of the peptide. Hym-176 has no effects on muscle contraction in Anhtopleura, metamorphosis in Hydractinia, and oocyte maturation and spawning in Cytaeis. And also, the gene encoding the peptide (Hym-176A) is just isolated from Hydra (Figure 2C) (66, 67). Thus, the peptide is species-specific.
The gene that encodes Hym-176 also encodes a second peptide, Hym-357 (KPAFLFKGYKPamide) (Figure 2C) (Table 3). This neuropeptide was identified in a screen for myoactive peptides (20). Detailed observations suggest that Hym-357 neurons activate other neurons to release neurotransmitters for induction of muscle contraction.
To identify the homologous gene that encodes Hym-176, Noro and coworkers found four candidate genes in the freshwater polyp Hydra magnipapillata (66). No authentic Hym-176 is present in the four paralogues (Figure 2C) (66). The cDNAs, Hym-176C and Hym-176D, encode one copy of a Hym-176-homologous peptide (Figure 2C) (Table 3). Hym-357 is encoded in both the gene that encodes Hym-176 and the gene that encodes Hym-176B (Figure 2C) (66). Hym-176C encodes Hym-690 (KPLYLFKGYKPamide), which is closely related to Hym-357 (Figure 2C) (Table 3) (20). Hym-176E appears not to have Hym-176- and Hym-357-related peptides (Figure 2C). The function of Hym-176C and D and Hym-690 has not yet been characterized in Hydra.
Hym-355 (FPQSFLPRGamide)
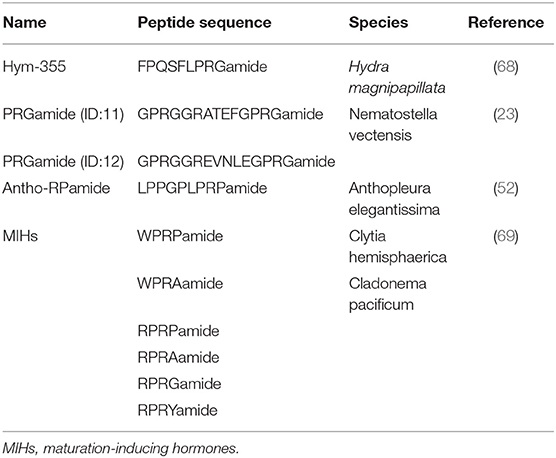
Hym-355 is a member of the PRXamide family of peptides that have PRXamide at their C-terminal region (Figure 2D) (Table 4) (68) and are subdivided into three groups in invertebrates: (a) neuropeptides that induce pheromone biosynthesis (70) and similar molecules, (b) small cardioactive peptides (71–73), and (c) antho-RPamide (52) and similar molecules. Antho-RPamide (LPPGPLPRPamide) is located in neurons of sea anemones and induces tentacle contraction. Thus, the peptide is involved in neurotransmission. PRXamide peptides have been identified in many invertebrates. Hym-355 is homologous to members of sub-group (c), including LPPGPLPRPamide (Anthopleura elegantissima) (Table 4), AAPLPRLamide (Urechis unicinctus) (74), QPPLPRYamide (Helix pomatia), and pQPPLPRYamide (Helix pomatia) (75). GPRGGRATEFGPRGamide and GPRGGREVNLEGPRGamide both have PRGamide at their C-termini and are expressed in the sea anemone Nematostella vectensis (Table 4) (23). The gene encoding the PRGamides is expressed in neurons (23), indicating that the PRGamides are neuropeptides.
Oxytocin-vasopressin superfamily peptides are neuropeptides synthesized in the hypothalamus and secreted from the posterior pituitary gland in mammals. Whether cnidarians express oxytocin/vasopressin superfamily peptides remains an open question in the field of comparative physiology of nervous systems. Immunohistochemical staining suggests that oxytocin/vasopressin superfamily peptides exist in the Hydra nervous system (76, 77). Morishita and coworkers (78) purified two peptides, Hym-355 and SFLPRGamide, from Hydra magnipapillata using HPLC fractionation and immunologic assays. They demonstrated that the antigen for vasopressin-like immunoreactivity is Hym-355 in the Hydra nervous system. The C-terminal region of Hym-355 (PRGamide) is identical to that of vasopressin. Neither antibody against the two peptides discriminates one peptide from the other. Thus, Koizumi et al. (79) performed immunohistochemistry with an anti–Hym-355 antibody and demonstrated immunoreactivity in the nerve rings of Cladonema radiatum and Turritopsis nutricula. However, whether Hym-355 functions as a neurohypophysial hormone is not well-understood.
The tissue of Hydra undergoes continuous renewal (Figure 3A). The number of neurons remains constant. Two groups of peptides, Hym-355 and PW family peptides, regulate this state (7, 68, 80). PW family peptides share the same sequence of Pro-Trp and are identified as epitheliopeptides (81).
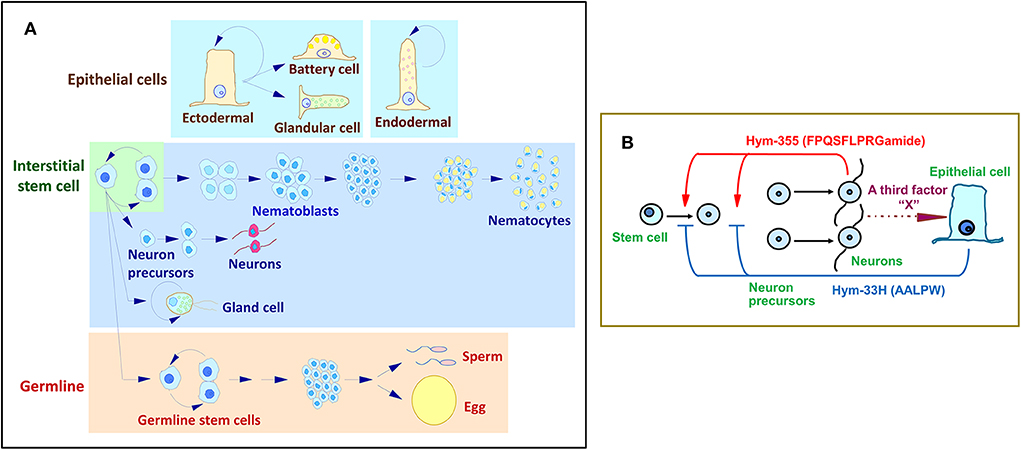
Figure 3. Hydra stem cell system. (A) Cell differentiation in Hydra. (B) A feedback model for the control of neuron differentiation that involves the antagonistic action of Hym-355 and the PW peptide, Hym-33H.
Hym-355 increases early neuron differentiation, and Hym-33H (AALPW) blocks neuron differentiation (68, 80). Simultaneous treatment with Hym-355 and Hym-33H results in a normal level of neuron differentiation. Taken together, the observations are consistent with a feedback model that modulates the homeostasis of neuronal differentiation in Hydra (Figure 3B) (68). This model suggests that Hym-355, which is synthesized by neurons, enhances early neuronal differentiation. To balance differentiation, epithelial cells produce PW peptides. A third factor termed as X in Figure 3B may control synthesis and secretion of PW family peptides. Hym-355, PW peptides, and the putative third factor may work together to maintain a constant neuronal density in Hydra. Hym-355 induces interstitial stem cells to undergo neuron differentiation and also induces retractor muscle contraction in the sea anemone Anthopleura fuscoviridis (68).
A member of the GLWamide family, Hym-53 (NPYPGLWamide) (Table 2), and Hym-355 induce oocyte maturation and spawning, but the effect of Hym-53 is stronger than that of Hym-355. Hym-355-like immunoreactivity is observed in neurons in Cytaeis (63). Possibly, neurons expressing Hym-53- and Hym-355-like peptides contribute downstream of light receptors in oocyte maturation and spawning in Cytaeis. Takeda and coworkers demonstrated that endogenous peptides including W/RPRPamide peptides are involved in oocyte maturation (Table 4) (69). RPRYamide, RPRGamide, WPRAamide, and RPRAamide may act as maturation-inducing hormones (MIHs) (Table 4) (69). Takeda et al. (69) also demonstrated that MIH peptides are synthesized by neurons in the gonad, and probably act on the oocyte surface. They propose that hydrozoan MIHs and neuropeptides are evolutionally linked to regulate reproduction upstream of MIHs in bilaterian species (69).
FRamide Family
During research aimed at systematic identification of peptide signaling molecules in Hydra (7), two novel neuropeptides, FRamide-1 (IPTGTLIFRamide) and FRamide-2 (APGSLLFRamide), were identified (Table 5) (82). Among Hydra EST and genome databases (8), we can rapidly identify peptide transcripts and their genes. The two peptides and the single gene encoding both peptides were identified using this exact approach (Figure 2E).
FRamide-1 (IPTGTLIFRamide) and FRamide-2 (APGSLLFRamide) exhibit opposing effects even though they are encoded by the same gene. The former peptide evokes body column elongation due to endodermal muscle contraction, whereas the latter peptide evokes body column contraction due to ectodermal muscle contraction (82). Two explanations for these seemingly contradictory observations are possible. One possibility is that the release of each peptide is differentially regulated (83, 84), and the other possibility is that each peptide is processed in a different type of neuron (85). Additionally, the opposing effects of FRamide family peptides may be ligand binding affinity for one receptor (62). In higher animals, most neuropeptides bind to GPCRs that are localized at the target cell. To understand the opposite effects, identification of FRamide-specific receptors on the target cells is important.
Conclusion
Neuropeptides released from nerve cells in response to a variety of stimuli are mandatory for fine-tuned regulation of behavior, reproduction, metamorphosis, and tissue maintenance (Figure 4). Here, I described 57 types of neuropeptides so far identified in cnidarians. However, the study of neuropeptides is still in its infancy. Additional novel peptides will likely be found (86), including neuropeptides, thus enabling elucidation of the mechanisms that regulate the physiology and development of cnidarians and increasing our understanding of peptide function in other species.
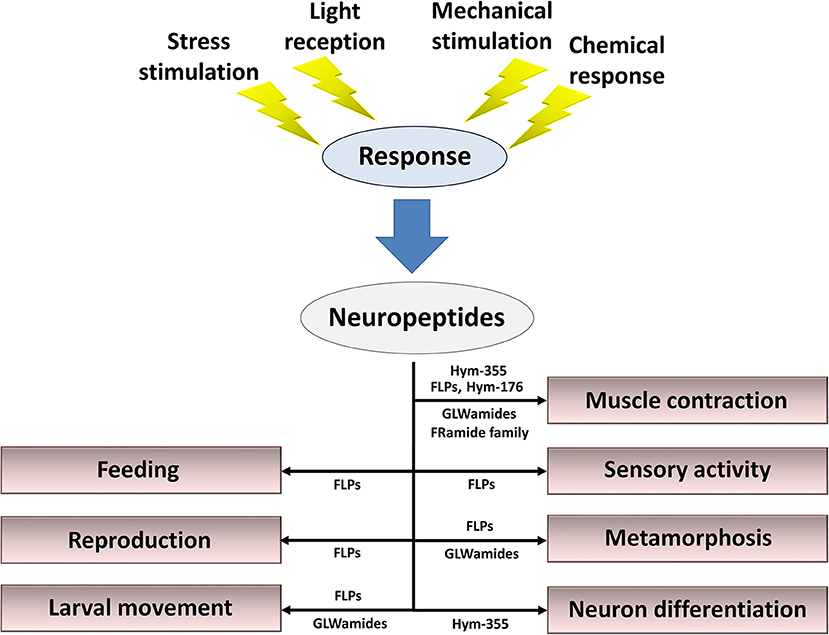
Figure 4. Summary of roles for neuropeptides in the control of behavior, reproduction, metamorphosis, and tissue maintenance. Cnidarian peptide signaling molecules function together and/or separately to maintain the organism's lifestyle in response to stress stimulation, light reception, mechanical stimulation, and chemical stimulation.
It is important to elucidate functional interaction between neuropeptides and receptors for the verification of their biological roles and evolutionary processes. However, no receptors for the neuropeptides remain to be identified in Hydra and cnidarians. Recently, Shiraishi and coworkers developed the machine-learning-assisted strategy for the identification of novel peptide–receptor pairs (87). As they indicate the multiplicity of use of the strategy, it is worth to use the strategy for increasing the receptor (especially GPCR) repertoire as many as possible on Hydra and cnidarians. When neuropeptide-GPCR pairs are efficiently and systematically elucidated in a phylogenetically critical Hydrozoa Hydra magnipapillata, Hydra provides cnidarian perspectives into evolution of GPCRs.
The cells of Hydra are well-characterized and belong to the epithelial cell lineage and the interstitial stem cell lineage (Figure 3A). However, knowledge of the molecules and biochemical mechanisms of the cells remains limited. The single-cell RNA sequencing technique sheds light on the complete molecular diversity of the cells in Hydra. Siebert and coworkers (88) applied this approach to the homeostatic adult Hydra. They drew a molecular map of the Hydra nervous system and unlocked the door toward understanding the molecular basis of morphogenesis and regeneration in Hydra.
Author Contributions
TT wrote the original review manuscript draft.
Funding
This work was supported by a Grant-in-Aid for Scientific Research (C) to TT (Grant Number 17K07495).
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author acknowledges a grant from the JSPS KAKENHI to TT (Grant Number 17K07495).
References
1. Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, Chera S. Origins of neurogenesis, a cnidarian view. Dev. Biol. (2009) 332:2–24. doi: 10.1016/j.ydbio.2009.05.563
2. David CN, Gierer A. A cell cycle kinetics and development of Hydra attenuate. III. Nerve and nematocyte differentiation. J Cell Sci. (1974) 16:3375.
3. Schmidt T, David CN. Gland cells in Hydra: Cell cycle kinetics and development. J Cell Sci. (1986) 85:197–215.
4. Bosch TCG, David CN. Stem cells of Hydra magnipapillata can differentiate intosomatic cells and germ line cells. Dev Biol. (1987) 121:182–91. doi: 10.1016/0012-1606(87)90151-5
5. Grimmelikhuijzen CJP, Leviev I, Carstensen K. Peptides in the nervous system of cnidrians: structure, function and biosynthesis. Int Rev Cytol. (1996) 167:37–89. doi: 10.1016/S0074-7696(08)61345-5
6. Kass-Simon G, Pierobon P. Cnidarian chemical neurotransmission, an updated overview. Comp Biochem Physiol A Mol Integr Physiol. (2007) 146:9–25. doi: 10.1016/j.cbpa.2006.09.008
7. Takahashi T, Muneoka Y, Lohmann Y, deHaro MSL, Solleder G, Bosch TCG, et al. Systematic isolation of peptide signaling molecues regulating development in hydra: LWamide and PW families. Proc Natl Acad Sci USA. (1997) 94:1241–6. doi: 10.1073/pnas.94.4.1241
8. Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, et al. The dynamic genome of Hydra. Nature. (2010) 464:592–6. doi: 10.1038/nature08830
9. Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science. (1977) 97:670–1. doi: 10.1126/science.877582
10. Price DA, Greenberg MJ. Purification and characterization of a cadioexcitatory neuropeptide from the central ganglia of a bivalve mollusk. Prep Biochem. (1977) 7:261–81. doi: 10.1080/00327487708061643
11. Price DA, Greenberg MJ. The hunting of the FaRPs: The distribution of FMRFamide-related peptides. Biol Bull. (1989) 177:198–205. doi: 10.2307/1541933
12. Espinoza E, Carrigan M, Thomas SG, Shaw G, Edison AS. A statistical view of FMRFamide neuropeptide diversity. Mol Neurobiol. (2000) 21:35–56. doi: 10.1385/MN:21:1-2:035
13. Krajniak KG. Invertebrate FMRFamide related peptides Protein Pept Lett. (2013) 20:647–70. doi: 10.2174/0929866511320060005
14. Grimmelikhuijzen CJP, Graff D. Isolation of pyroGlu-Gly-Arg-Phe-NH2 (Antho-RFamide), a neuropeptide from sea anemones. Proc Natl Acad Sci USA. (1986) 83:9817–21. doi: 10.1073/pnas.83.24.9817
15. Moosler A, Rhinehart KL, Grimmelikhuijzen CJP. Isolation of three novel peptides, the Cyanea-RFamides I-III, from scyphomedusae. Biochem Biophys Res Commun. (1997) 236:743–9. doi: 10.1006/bbrc.1997.7022
16. Grimmelikhuijzen CJP, Hahn M, Rhinehart KL, Spencer AN. Isolation of pyroGlu-Leu-Leu-Gly-Gly-Arg-Phe-NH2 (Pol-RFamide), a novel neuropeptide from hydromedusae. Brain Res. (1988) 475:198–203. doi: 10.1016/0006-8993(88)90219-3
17. Grimmelikhuijzen CJP, Rhinehart KL, Spencer AN. Isolation of the neuropeptide less than Glu-Trp-Leu-Lys-Gly-Arg-Phe-NH2 (Pol-RFamide II) from the hydromedusa Polyorchis penicillatus. Biochem Biophys Res Commun. (1992) 183:375–82. doi: 10.1016/0006-291X(92)90491-3
18. Moosler A, Rhinehart KL, Grimmelikhuijzen CJP. Isolation of four novel neuropeptides, the Hydra-RFamides I-IV, from Hydra magnipapillata. Biochem Biophys Res Commun. (1996) 229:596–602. doi: 10.1006/bbrc.1996.1849
19. Darmer D, Hauser F, Nothacker HP, Bosch TCG, Williamson M, Grimmelikhuijzen CJP. Three different prohormones yield a variety of Hydra-RFamide (Arg-Phe-NH2) neuropeptide in Hydra magnipapillata. Biochem J. (1998) 332:403–12. doi: 10.1042/bj3320403
20. Fujisawa T. Hydra peptide project 1993–2007. Dev Growth Differ. (2008) 50:S257–68. doi: 10.1111/j.1440-169X.2008.00997.x
21. Gajewski M, Schmutzler C, Plickert G. Structure of neuropeptide precursors in cnidarian. Ann N Y Acad Sci. (1998) 839:311–5. doi: 10.1111/j.1749-6632.1998.tb10782.x
22. Anctil M. Chemical transmission in the sea anemone Nematostella vectensis: a genomic perspective. Comp Biochem Physiol D. (2009) 4:268–89. doi: 10.1016/j.cbd.2009.07.001
23. Hayakawa E, Watanabe H, Menschaert G, Holstein TW, Baggerman G, Schoofs L. A combined strategy of neuropeptide prediction and tandem mass spectrometry identifies evolutionarily conserved ancient neuropeptides in the sea anemone Nematostella vectensis. PLoS ONE. (2019) 14:e0215185. doi: 10.1371/journal.pone.0215185
24. Darmer D, Schmutzler C, Diekhoff D, Grimmelikhuijzen CJP. Primary structure of the precursor for the sea anemone neuropeptide Antho-RFamide (< Glu-Gly-Arg-Phe-NH2). Proc Natl Acad Sci USA. (1991) 88:2555–9. doi: 10.1073/pnas.88.6.2555
25. Schmutzler C, Darmer D, Diekhoff D, Grimmelikhuijzen CJP. Identification of a novel type of processing sites in the precursor for the sea anemone neuropeptide Antho-RFamide (< Glu-Gly-Arg-Phe-NH2) from Anthopleura elegantissima. J Biol Chem. (1992) 267:22534–41.
26. Reinscheid RK, Grimmelikhuijzen CJP. Primary structure of the precursor for the anthozoan neuropeptide Antho-RFamide from Renilla kollikeri: evidence for unusual processing enzymes. J Neurochem. (1994) 62:1214–22. doi: 10.1046/j.1471-4159.1994.62031214.x
27. Schmutzler C, Diekhoff D, Grimmelikhuijzen CJP. The primary structure of the Pol-RFamide neuropeptide precursor protein from the hydromedusa Polyorchis penicillatus indicates a novel processing proteinase activity. Biochem J. (1994) 299:431–6. doi: 10.1042/bj2990431
28. McFarlane ID, Graff D, Grimmelikhuijzen CJP. Excitatory actions of Antho-RFamide, an anthozoan neuropeptide, on muscles and conducting systems in the sea anemone. Calliactis parasitica. J Exp Biol. (1987) 133:157–68.
29. Anctil M, Grimmelikhuijzen CJP. Excitatory action of the native neuropeptide Antho-RFamide in muscles in the pennatulid Renilla kollikeri. Gen Pharmacol. (1989) 20:381–4. doi: 10.1016/0306-3623(89)90277-2
30. Shimizu H, Fujisawa T. Peduncle of Hydra and the heart of higher organisms share a common ancestral origin. Genesis. (2003) 36:182–6. doi: 10.1002/gene.10213
31. Cottrell GA, Green KA, Davis NW. The neuropeptide Phe-Met-Arg-Phe-NH2(FMRFamide) can activate a ligand-gated ion channel in Helix neurons. Pflugers Arch. (1990) 416:612–4. doi: 10.1007/BF00382698
32. Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. (1995) 378:730–3. doi: 10.1038/378730a0
33. Golubovic A, Kuhn A, Williamson M, Kalbacher H, Holstein TW, Grimmelikhuijzen CJP, et al. A peptide-gated ion channel from the freshwater polyp Hydra. J Biol Chem. (2007) 282:35098–103. doi: 10.1074/jbc.M706849200
34. Dürrnagel S, Kuhn A, Tsiairis CD, Williamson M, Kalbacher H, Grimmelikhuijzen CJP, et al. Three homologous subunits form a high affinity peptide-gated ion channel in Hydra. J Biol Chem. (2010) 285:11958–65. doi: 10.1074/jbc.M109.059998
35. Assmann M, Kuhn A, Dürrnagel S, Holstein TW, Gründer S. The comprehensive analysis of DEG/ENaC subunits in Hydra reveals a large variety of peptide-gated channels, potentially involved in neuromuscular transmission. BMC Biol. (2014) 12:84. doi: 10.1186/s12915-014-0084-2
36. Tardent P. The cnidarian cnidocyte, a high tech cellular weaponry. BioEssays. (1995) 17:351–62. doi: 10.1002/bies.950170411
37. Holtmann M, Thurm U. Mono- and oligo-vesicular synapses and their connectivity in a Cnidarian sensory epithelium (Coryne tubulosa). J Comp Neurol. (2001) 432:537–49. doi: 10.1002/cne.1118
38. Westfall JA, Elliott CF, Carlin RW. Ultrastructural evidence for two-cell and three-cell neural pathways in the tentacle epidermis of the sea anemone Aiptasia pallida. J Morphol. (2002) 251:83–92. doi: 10.1002/jmor.1075
39. Anderson PA, Thompson LF, Moneypenny CG. Evidence for a common pattern of peptidergic innavations of cnidocytes. Biol Bull. (2004) 207:141–6. doi: 10.2307/1543588
40. Nakanishi N, Hartenstein V, Jacobs DK. Development of the rhopalial nervous system in Aurelia sp. 1 (Cnidaria, Scyphozoa). Dev Genes Evol. (2009) 219:301–17. doi: 10.1007/s00427-009-0291-y
41. Tremblay ME, Henry J, Anctil M. Spawning and gamete follicle rupture in the cnidarian Renilla Koellikeri: effects of putative neurohormones. Gen Comp Endocrinol. (2004) 137:9–18. doi: 10.1016/j.ygcen.2004.02.009
42. Plickert G, Kroiher M, Munck A. Cell proliferation and early differentiation during embryonic development and metamorphosis of Hydractinia echinata. Development. (1988) 103:795–803.
43. Katsukura Y, Ando H, David CN, Grimmelikhuijzen CJP, Sugiyama T. Control of planula migration by LWamide and RFamide neuropeptides in Hydractinia echinata. J Exp Biol. (2004) 207:1803–10. doi: 10.1242/jeb.00974
44. Leitz T. Metamorphosin A and related compounds: a novel family of neuropeptides with morphogenetic activity. Ann N Y Acad Sci. (1998) 839:105–10. doi: 10.1111/j.1749-6632.1998.tb10740.x
45. Leitz T. Induction of metamorphosis of the marine hydrozoan Hydractinia echnata Fleming, 1828. Biofouling. (1998) 12:173–87. doi: 10.1080/08927019809378353
46. Katsukura Y, David CN, Grimmelikhuijzen CJP, Sugiyama T. Inhibition of metamorphosis by RFamide neuropeptides in planula larvae of Hydractinia echinata. Dev Genes Evol. (2003) 213:579–86. doi: 10.1007/s00427-003-0361-5
47. Seipp S, Schmich J, Will B, Schetter Plickert G, Leitz T. Neuronal cell death during metamorphosis of Hydractinia echinata. (Cnidaria, Hydrozoa). Invertebr Neurosci. (2010) 10:77–91. doi: 10.1007/s10158-010-0109-7
48. Leitz T, Morand K, Mann M. Metamorphosin A: a novel peptide controlling development of the lower metazoan Hydractinia echinata (Coelenterata, Hydrozoa). Dev Biol. (1994) 163:440–6. doi: 10.1006/dbio.1994.1160
49. Takahashi T, Kobayakawa Y, Muneoka Y, Fujisawa Y, Mohri S, Hatta M, et al. Identification of a new member of the GLWamide peptide family: physiological activity and cellular localization in cnidarian polyps. Comp Biochem Physiol Part B. (2003) 135:309–24. doi: 10.1016/S1096-4959(03)00088-5
50. Leviev I, Williamson M, Grimmelikhuijzen CJP. Molecular cloning of a preprohormone from Hydra magnipapillata containing multiple copies of Hydra-LWamide (Leu-Trp-NH2) neuropeptides: evidence for processing at Ser and Asn residues. J Neurochem. (1997) 68:1319–25. doi: 10.1046/j.1471-4159.1997.68031319.x
51. Gajewski M, Leitz T, Schlosherr J, Plickert G. LWamides from cnidaria constitute a novel family of neuropeptides with morphogenetic activity. Roux's Arch Dev Biol. (1996) 205:232–42. doi: 10.1007/BF00365801
52. Carstensen K, Rinehart KL, McFarlane ID, Graff D, Grimmelikhuijzen CJP. Isolation of Leu-Pro-Pro-Gly-Pro-Leu-Pro-Arg-Pro-NH2 (Antho-RPamide), and N-terminally protected, biologically active neuropeptide from sea anemones. Peptides. (1992) 13:851–7. doi: 10.1016/0196-9781(92)90040-A
53. Leitz T, Lay M. Metamorphosin A is a neuropeptide. Roux's Arch Dev Biol. (1995) 204:276–9. doi: 10.1007/BF00208495
54. Takahashi T, Ohtani M, Muneoka Y, Aimoto S, Hatta M, Shimizu H, et al. Structure-activity relation of LWamide peptides synthesized with a multipeptide synthesizer. In: Kitada C, editor. Peptide Chemistry. Osaka: Protein Research Foundation (1997). p. 193–6.
55. Takahashi T, Hatta M. The importance of GLWamide neuropeptides in Cnidarian development and physiology. J Amino Acids. (2011) 2011:1–8. doi: 10.4061/2011/424501
56. Schmich J, Trepel S, Leitz T. The role of GLWamides in metamorphosis of Hydractinia echinata. Dev Genes Evol. (1998) 208:267–73. doi: 10.1007/s004270050181
57. Iwao K, Fujisawa T, Hatta M. A cnidarian neuropeptide of the GLWamide family indices metamorphosis of reef-building corals in the genus Acropora. Coral Reefs. (2002) 21:127–9. doi: 10.1007/s00338-002-0219-8
58. Erwin PM, Szmant AM. Settlement induction of Acropora palmate planulae by a GLW-amide neuropeptide. Coral Reefs. (2010) 29:929–39. doi: 10.1007/s00338-010-0634-1
59. Marcum BA, Campbell RD. Development of hydra. lacking nerve and intestinal cells. J Cell Sci. (1978) 29:17–33.
60. Campbell RD. Elimination of Hydra. intestinal and nerve cells by means of colchicines. J Cell Sci. (1976) 21:1–13.
61. Zieglgänsberger W. Substance P and pain chronicity. Cell Tissue Res. (2019) 375:227–41. doi: 10.1007/s00441-018-2922-y
62. Ganjiwale A, Cowsik SM. Molecular recognition of tachykinin receptor selective agonists: insights from structural studies. Mini Rev Med Chem. (2013) 13:2036–46. doi: 10.2174/13895575113139990079
63. Takeda N, Kyozuka K, Deguchi R. Increase in intracellular cAMP is a prerequisite signal for initiation of physiological oocyte meiosis maturation in the hydrozoan Cytaeis uchidae. Dev Biol. (2006) 298:248–58. doi: 10.1016/j.ydbio.2006.06.034
64. Takeda N, Nakajima Y, Koizumi O, Fujisawa T, Takahashi T, Matsumoto M, et al. Neuropeptides trigger oocyte maturation and subsequent spawning in the hydrozoan jellyfish Cytaeis uhcidae. Mol Reprod Dev. (2013) 80:223–232. doi: 10.1002/mrd.22154
65. Yum S, Takahashi T, Koizumi O, Ariura Y, Kobayakawa Y, Mohri S, et al. A novel neuropeptide, Hym-176, induces contraction of the ectodermal muscle in Hydra magnipapillata. Biochem Biophys Res Commun. (1998) 248:584–90. doi: 10.1006/bbrc.1998.8831
66. Noro Y, Yum S, Fujisawa-Nishimiya C, Busse C, Shimizu H, Mineta K, et al. Regionalized nervous system in Hydra and the mechanism of its development. Gene Expr Patterns. (2019) 31:42–59. doi: 10.1016/j.gep.2019.01.003
67. Yum S, Takahashi T, Hatta M, Fujisawa T. The structure and expression of a preprohormone of a neuropeptide, Hym-176 in Hydra magnipapillata. FEBS Lett. (1998) 439:31–4. doi: 10.1016/S0014-5793(98)01314-3
68. Takahashi T, Koizumi O, Ariura Y, Romanovitch A, Bosch TCG, Kobayakawa Y, et al. A novel neuropeptide, Hym-355, positively regulates neuron differentiation in Hydra. Development. (2000) 127:997–1005. doi: 10.1016/S1095-6433(99)90367-7
69. Takeda N, Kon Y, Artigas GQ, Lapebie P, Barreau C, Koizumi O, et al. Identification of jellyfish neuropeptides that act directly as oocyte maturation-inducing hormones. Development. (2018) 145:dev156786. doi: 10.1242/dev.156786
70. Raina AK, Jaffe H, Kempe TG, Keim P, Blacher RW, Fales HM, et al. Identification of a neuropeptide hormone that regulates sex pheromone production in female moths. Science. (1989) 244:796–8. doi: 10.1126/science.244.4906.796
71. Muneoka Y, Takahashi T, Kobayashi M, Ikeda T, Minakata H, Nomoto K. Phylogenetic aspects of structure and action of molluscan neuropeptides. In: Davey KG, Peter RE, Tobe SS, editors. Perspectives in Comparative Endocrinology. Toronto, ON: National Research Council of Canada (1994). p. 109–18.
72. Morris HR, Panico M, Karplus A, Lloyd PE, Piniker B. Identification by FAB-MS of the structure of a new cardioactive peptide from Aplysia. Nature. (1982) 300:643–5. doi: 10.1038/300643a0
73. Lloyd PE, Kupfermann I, Weiss KR. Sequence of small cardioactive peptide A: a second member of a class of neuropeptides in Aplysia. Peptides. (1987) 8:179–83. doi: 10.1016/0196-9781(87)90184-7
74. Ikeda T, Kubota I, Miki W, Nose T, Takao T, Shimonishi Y, et al. Structures and actions of 20 novel neuropeptides isolated from the ventral nerve cords of an echiuroid worm, Urechis unicinctus. In: Yanaihara N, editor. Peptide Chemistry 1992. Leiden: Protein Research Foundation (1993). p. 583–5. doi: 10.1007/978-94-011-1474-5_168
75. Minakata H, Ikeda T, Fujita T, Kiss T, Hiripi L, Muneoka Y, et al. Neuropeptides isolated from Helix pomatia. Part 2. FMRFamide-related peptides, S-Iamide peptides, FR peptides and others. In: Yanaihara N, editor. Peptide Chemistry 1992. Leiden: Protein Research Foundation (1993) 579–82. doi: 10.1007/978-94-011-1474-5_167
76. Grimmelikhuijzen CJP, Dierickx K, Boer GJ. Oxytocin/vasopressin-like immunoreactivity is present in the nervous system of hydra. Neuroscience. (1982) 7:3191–9. doi: 10.1016/0306-4522(82)90241-X
77. Koizumi O, Bode HR. Plasticity in the nervous system of adult hydra. III. Conversion of neurons to expression of a vasopressin-like immunoreactivity depends on axial location. J Neurosci. (1991) 11:2011–20. doi: 10.1523/JNEUROSCI.11-07-02011.1991
78. Morishita F, Nitagai Y, Furukawa Y, Matsushima O, Takahashi T, Hatta M, et al. Identification of a vasopressin-like immunoreactive substance in hydra. Peptides. (2003)24:17–26. doi: 10.1016/S0196-9781(02)00272-3
79. Koizumi O, Hamada S, Minobe S, Hamaguchi-Hamada K, Kurumata-Shigeto M, Nakamura M. The nerve ring in cnidarians: its presence and structure in hydrozoan medusa. Zoology. (2015) 118:79–88. doi: 10.1016/j.zool.2014.10.001
80. Takahashi T, Koizumi O, Hayakawa E, Minobe S, Suetsugu R, Kobayakawa Y, et al. Further characterization of the PW peptide family that inhibits neuron differentiation in Hydra. Dev Genes Evol. (2009) 219:119–29. doi: 10.1007/s00427-009-0272-1
81. Takahashi T. Neuropeptides and epitheliopeptides: structural and functional diversity in an ancestral metazoan Hydra. Protein Pept Lett. (2013) 20:671–80. doi: 10.2174/0929866511320060006
82. Hayakawa E, Takahashi T, Nishimiya-Fujisawa C, Fujisawa T. A novel neuropeptide (FRamide) family identified by a peptidomic approach in Hydra magnipapillata. FEBS J. (2007) 274:5438–48. doi: 10.1111/j.1742-4658.2007.06071.x
83. Eipper B, Mains RE. Structure and biosynthesis of proadrenocorticotropin/endorphin and released peptides. Endocr Rev. (1980) 1:1–27. doi: 10.1210/edrv-1-1-1
84. Rosa PA, Policastro P, Herbert E. A cellular basis for the differences in regulation of synthesis and secretion of ACTH/endorphin peptides in anterior and intermediate lobes of the pituitary. J Exp Biol. (1980) 89:215–37.
85. Klumperman J, Spijker S, van Minnen J, Sharp-Baker H, Smit AB, Geraerts WPM. Cell type-specific sorting of neuropeptides: a mechanism to modulate peptide composition of large-dense-core vesicles. J Neurosci. (1996) 16:7930–40. doi: 10.1523/JNEUROSCI.16-24-07930.1996
86. Koch TL, Grimmelikhuijzen CJP. Global neuropeptide annotations from the genomes and transcriptomes of Cubozoa, Scyphozoa, Staurozoa (Cnidaria: Medusozoa), and Octocorallia (Chidaria: Anthozoa). Front. Endocrinol. (2019) 10:831. doi: 10.3389/fendo.2019.00831
87. Shiraishi A, Okuda T, Miyasaka N, Osugi T, Okuno Y, Inoue J, et al. Repertoires of G protein-coupled receptors for Ciona-specific neuropeptides. Proc. Natl. Acad. Sci. USA. (2019) 116:7847–56. doi: 10.1073/pnas.1816640116
Keywords: Hydra, Cnidaria, neuropeptide, metamorphosis, myoactivity, interstitial stem cell, neuron differentiation
Citation: Takahashi T (2020) Comparative Aspects of Structure and Function of Cnidarian Neuropeptides. Front. Endocrinol. 11:339. doi: 10.3389/fendo.2020.00339
Received: 03 March 2020; Accepted: 30 April 2020;
Published: 27 May 2020.
Edited by:
James A. Carr, Texas Tech University, United StatesReviewed by:
Hervé Tostivint, Muséum National d'Histoire Naturelle, FranceJoao Carlos dos Reis Cardoso, University of Algarve, Portugal
Copyright © 2020 Takahashi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toshio Takahashi, dGFrYWhhc2hpQHN1bmJvci5vci5qcA==
 Toshio Takahashi
Toshio Takahashi