
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 19 November 2019
Sec. Reproduction
Volume 10 - 2019 | https://doi.org/10.3389/fendo.2019.00748
This article is part of the Research Topic Regulation of Dynamic Changes and Remodeling Events During the Formation, Rescue and Regression of the Corpus Luteum View all 11 articles
 Aamir S. Teeli1
Aamir S. Teeli1 Paweł Leszczyński1
Paweł Leszczyński1 Narayanan Krishnaswamy2
Narayanan Krishnaswamy2 Hidesato Ogawa3
Hidesato Ogawa3 Megumi Tsuchiya3
Megumi Tsuchiya3 Magdalena Śmiech1
Magdalena Śmiech1 Dariusz Skarzynski4
Dariusz Skarzynski4 Hiroaki Taniguchi1*
Hiroaki Taniguchi1*The corpus luteum (CL) is an important tissue of the female reproductive process which is established through ovulation of the mature follicle. Pulsatile release of prostaglandin F2α from the uterus leads to the regression of luteal cells and restarts the estrous cycle in most non-primate species. The rapid functional regression of the CL, which coincides with decrease of progesterone production, is followed by its structural regression. Although we now have a better understanding of how the CL is triggered to undergo programmed cell death, the precise mechanisms governing CL protein degradation in a very short period of luteolysis remains unknown. In this context, activation of ubiquitin-proteasome pathway (UPP), unfolded protein response (UPR) and autophagy are potential subcellular mechanisms involved. The ubiquitin-proteasome pathway (UPP) maintains tissue homeostasis in the face of both internal and external stressors. The UPP also controls physiological processes in many gonadal cells. Emerging evidence suggests that UPP dysfunction is involved in male and female reproductive tract dysfunction. Autophagy is activated when cells are exposed to different types of stressors such as hypoxia, starvation, and oxidative stress. While emerging evidence points to an important role for the UPP and autophagy in the CL, the key underlying transcriptional mechanisms have not been well-documented. In this review, we propose how CL regression may be governed by the ubiquitin-proteasome and autophagy pathways. We will further consider potential transcription factors which may regulate these events in the CL.
Corpus luteum (CL) formation, an integral part of the female reproductive process, is accomplished through ovulation of the mature follicle. The CL is a transient organ composed of various cells types. These include endothelial cells, immune cells, and luteal cells, which differentiate from follicular cells (granulosa and thecal cells) following ovulation. The CL development is classified as early, mid, late and regression stages in terms of its growth rate, neovascularisation, and rate of progesterone (P4) production. In the absence of pregnancy, luteolysis occurs with a decrease in P4 synthesis and secretion. Conversely, P4 produced by the CL maintains pregnancy in several species. To produce P4 in the CL, free cholesterol is transferred to the inner mitochondrial membrane by carrier proteins including steroidogenic acute regulatory protein (STAR). This process involving the P450 cholesterol side-chain cleavage enzyme (p450scc/CYP11A1) converts cholesterol to pregnenolone, the C-21 steroid precursor (1). STAR is a protein that governs the rate-limiting step of gonadal steroidogenesis.
In a non-fertile estrous or menstrual cycle, the CL undergoes luteolysis. In ruminants, pulsatile release of prostaglandin F2α (PGF2α) by the uterus leads to regression of luteal cells and renewal of the estrous cycle. The rapid functional regression of the CL, which is characterized by decreased P4 production, is followed by structural regression. During the structural regression, luteal cells undergo apoptosis (2–5). Failure of this mechanism is associated with dysfunction of the reproductive cycle and infertility. While we understand the process by which the CL undergoes programmed cell death, the mechanisms governing the degradation of a large quantity of CL proteins over a very short period of time is yet to be elucidated.
The ubiquitin-proteasome pathway (UPP) plays an important role in the degradation of unnecessary proteins. During this process, target proteins are first bound to small ubiquitin proteins and degraded. The UPP is a regulatory mechanism that maintains tissue homeostasis in response to various stressors including oxidative stress. This pathway acts through the endoplasmic reticulum (ER) (6). The UPP governs physiological processes in a variety of gonadal cells (7, 8). Emerging evidence suggests that UPP dysfunction leads to pathology within reproductive system (9). Additionally, the unfolded protein response (UPR) signaling pathway, a cellular stress response associated with the ER, is involved in the development, maintenance, and regression of the bovine CL (10). Female mice lacking Beclin1 (Becn1), a regulator of autophagy and the UPR system, display a preterm labor phenotype associated with P4 production dysfunction in ovarian granulosa cells (GCs) (11). Interestingly, GRP78, an ER chaperone protein essential for UPR, plays an integral role in the initiation of steroidogenesis through STAR activation at the mitochondrial membrane (12). Moreover, increased oxidative stress in the ER reduces testosterone production in Leydig cells (13). As such, the UPP and UPR are crucial to degrade unnecessary proteins and maintain cellular homeostasis. Unfortunately, the molecular mechanisms governing UPR function in the CL remain poorly understood.
Similar to the UPP system, autophagy degrades unnecessary proteins through the autophagosome. This process is involved in the metabolism of cellular components associated with the UPP under normal and pathologic conditions. The UPP and autophagy systems are closely related mechanisms that remove unnecessary cellar components. They act cooperatively to maintain cellular homeostasis (14). ER stress is an important trigger of the UPP and autophagy. Recent studies have demonstrated that ER stress and autophagy play important roles in structural regression of CL (15, 16). Together, these results suggest that ER stress-mediated autophagy may play an important role in luteolysis.
In view of these findings, we hypothesize that the UPP and autophagy may play important roles in the functional regulation of the CL and luteolysis. In this review, we will explore the molecular mechanisms governing luteal function and regression as well as its interplay with the proteasome-autophagy system.
Progesterone, the major hormone of CL is elaborated by small and large luteal cells, that are derived from the follicular theca interna and granulosa cells, respectively. The non-steroidogenic component of CL comprises of endothelial cells, pericytes, fibroblasts and immune cells. Small luteal cells are stimulated by luteinizing hormone (LH) which is secreted by the pituitary gland. P4 secreted by large luteal cells represents the basal P4 level. The P4 level derived from small luteal cells is known as LH-induced P4. The LH receptor is a G protein-coupled receptor with seven membrane spanning domains (17). Once LH binds to its receptor, the second messenger cyclic AMP is released and protein kinase A (PKA) is subsequently activated. PKA phosphorylates various proteins and in turn modulates their function (18). Thereafter, transcription factors induce the expression of steroidogenic enzymes including STAR, Cyp11a, and 3β-hydroxysteroid dehydrogenase (3β-HSD). STAR plays a role in transporting cholesterol to the mitochondria. P450scc converts cholesterol into pregnenolone in the mitochondria. Pregnenolone is finally converted to P4 by 3β-HSD in the luteal cells [(19), Figure 1].
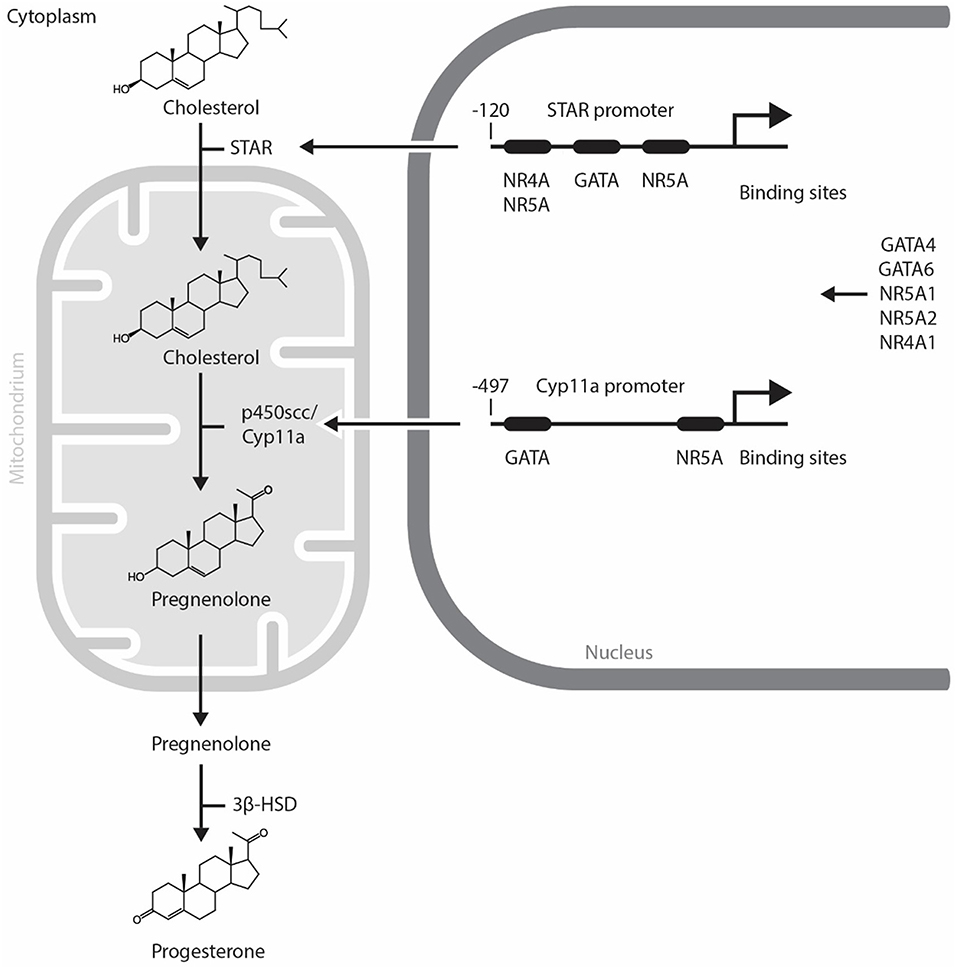
Figure 1. Mechanism of progesterone synthesis and its molecular regulation by GATA and NR5A transcription factors in steroidogenic cells. STAR plays a role in transporting cholesterol to the mitochondria. p450scc converts cholesterol into pregnenolone in the mitochondria. Pregnenolone is finally converted to progesterone by 3β-HSD in steroidogenic cells. STAR and Cyp11a promoters contain evolutionally conserved GATA and NR5A/4A binding sites. GATA, NR4A1, and NR5A transcription factors control the expression of steroidogenic enzymes including STAR, p450scc/Cyp11a. NR5A1, Nuclear Receptor Subfamily 5 Group A Member 1; NR5A2, Nuclear Receptor Subfamily 5 Group A Member 2; NR4A1, Nuclear Receptor Subfamily 4 Group A Member 1; p450scc, cytochrome p450 side-chain cleavage enzyme; STAR, Steroidogenic Acute Regulatory Protein; 3β-HSD, 3β-hydroxysteroid dehydrogenase.
The regulation of steroidogenesis in the CL involves the temporal expression of genes coding for a variety of steroidogenic enzymes. As the rapid upregulation of steroidogenic enzyme gene expression is required, it is likely that the acute regulation of steroidogenesis in the CL is regulated by transcription factors (Figure 1) and change in the expression and activity of these transcription factors trigger functional CL regression. In this regard, NR5A1 (Nuclear Receptor Subfamily 5 Group A Member 1, also known also as adrenal 4 binding protein/steroidogenic factor 1: Ad4BP/SF-1), a regulator of multiple P450 hydroxylases and other components of the steroidogenic program, was first isolated from the adrenal gland (20, 21). Since then, several researchers have identified other transcription factors that regulate the promoter activity of Star, CYP11A1, HSD3B2, and CYP17 [reviewed in (22)]. NR5A1 is abundant in the CL during the midluteal phase and binds to the Star promoter in vivo (23). Moreover, NR5A1 regulates Star and Cyp11a gene expressions in luteal cells and the CL of many species (23–26). A luteal cell-specific Nr5a1 knockout (KO) has not been reported. The ovaries of a newborn mice lacking Nr5a1 in GCs are comparable with wild type; however, Nr5a1-deficient adult females lack CL and suffer from sterility (27).
NR5A2 (also known as Fetoprotein transcription factor: FTF, liver receptor homolog 1: LRH-1), another NR5A family member, is also present in the ovary (28). Interestingly, NR5A2 recognizes the same consensus binding sequence as NR5A1 and may regulate similar steroidogenic enzyme target genes. NR5A2 is the most prominent NR5A factor in the CL (29). Similar to NR5A1, NR5A2 is a potent regulator of steroidogenic gene expression in the CL (23, 29). Luteal specific KO of Nr5a2 is linked with luteal insufficiency, which suggests that this factor plays a crucial role in luteal formation and function (30). Additionally, Nr5a1 and Nr5a2 KO mice individually exhibit luteal disruption with downregulated steroidogenic enzyme gene expression (functional luteal disruption) and severe structural damage (27, 29, 31). Both these factors, therefore, play prominent roles in luteal function, development, and regression.
While the role of NR4A1 (Nuclear Receptor Subfamily 4 Group A Member 1) in steroidogenesis in the CL is yet to be elucidated, its expression is upregulated by cAMP, a second messenger of several pituitary hormones including LH. NR4A1 is present in the theca cells, GCs, and luteal cells in the human ovary (32, 33). Moreover, NR4A1 is known to regulate StAR gene expression activity in mouse Leydig cells (34, 35). On the other hand, NR4A1 levels are upregulated by PGF2α in pseudopregnant rats (36). Further studies are needed to elucidate the control of NR4A1 levels and its role in luteal steroidogenesis.
The gonads also express several GATA factors that are known to regulate steroidogenic gene expression [reviewed in (37)]. In the CL of Gata4 and Gata6 conditional double knockdown mice, a reduction in P4 production is observed along with an acute inhibition of expression of genes in the steroidogenic pathway including Star, Hsd3b1, and Cyp11a [(38), Figure 2]. Moreover, GATA4 and GATA6 mRNA and protein were identified in bovine CL and it is suggested that GATA6 may be involved in the regulation of STAR expression in this species (23). As GATA4 and GATA6 are proposed to physically and functionally interact with NR5A1 and NR5A2 to upregulate steroidogenic enzyme gene transcription via the HSD3B2 and Cyp19a promoter (39), one can expect that they also play important roles in steroidogenesis in the CL. While we have good insights into how steroidogenesis is likely turned on by transcriptional factors in the CL, the mechanism governing its inhibition during functional luteolysis remains obscure. NR0B1 inhibits transcriptional cooperation between GATA4 and NR5A1 in testicular cells, suggesting that it might possibly mediate the inhibitory effect of PGF2α on P4 in the CL (40, 41).
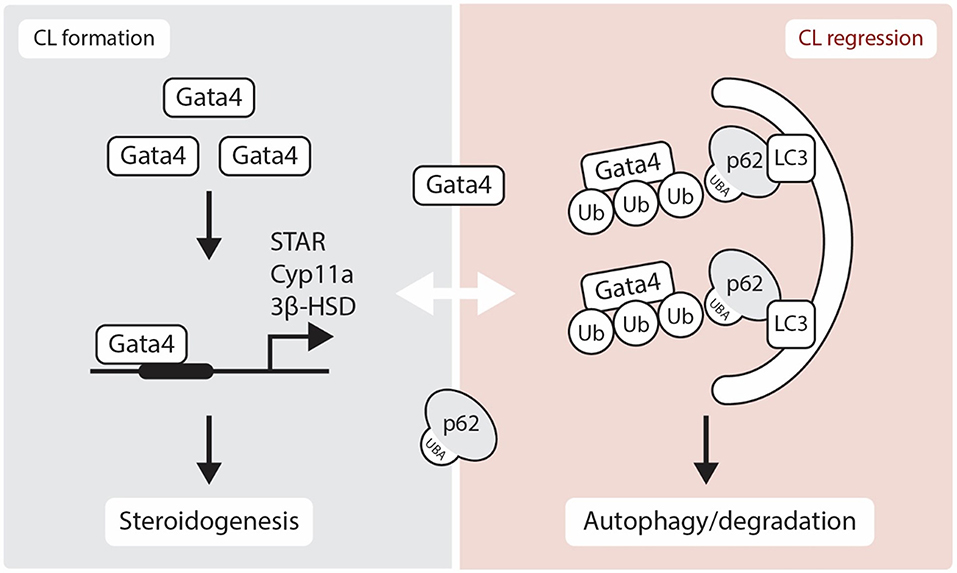
Figure 2. Schematic representation showing the role of GATA4 in luteogenesis and luteolysis. Gata4 is located in the nucleus and regulates several steroidogenic enzymes gene expression. Although p62 is thought to be localized in the cytoplasm, recent studies have suggested that p62 physically interacts with nuclear proteins. Here, we show a hypothetical regulation of GATA4 transcription factor through an interaction with p62 in the luteolytic CL. STAR, Steroidogenic Acute Regulatory Protein; 3β-HSD, 3β-hydroxysteroid dehydrogenase; CYP11A, Cytochrome P450 Family 11 Subfamily A Member; Ub, Ubiquitin; LC3, Microtubule-associated protein 1A/1B-light chain 3; UBA, Ubiquitin-Associated domain.
Reduced P4 secretion begins during the late luteal phase and leads to CL structural regression. This structural regression occurs through apoptosis which involves nuclear fragmentation (3, 4) as well as caspase 3 and p53 activation (42–44). Detailed information of apoptosis-mediated CL regression is well-reviewed in (45). Our group has identified that apoptosis in bovine CL is induced by the interaction between cytokines and Fas/FasL (46). It is well-established that PGF2α triggers apoptosis during luteolysis. Immune cells and cytokines play important roles in structural luteolysis as evidenced by increased T-lymphocyte and macrophage influx during CL regression (47). In bovine luteal cells, Fas-FasL mediated cell death plays a crucial role in luteal cell apoptosis. This process is induced by interferon gamma (IFNγ) and tumor necrosis factor alpha (TNFα) plays a stimulatory role. Treatment of luteal cells with Fas ligand, in presence of IFNγ and TNFα, leads to the formation of apoptotic bodies, which supports the notion that these cytokines are implicated in luteal regression (46). Moreover, IFNγ and TNFα induce mouse luteal cell apoptosis (48). Macrophages degrade extracellular matrix (ECM) and phagocytize degenerated luteal cells leading to the release of cytokines including TNFα, interleukin-1β (IL-1), and IFNγ (49). The intraluteal TNFα level increases significantly during both spontaneous and induced in vivo luteolysis in microdialyzed CL (50). It is, therefore, likely that TNFα stimulates synthesis of luteal PGF2α. This modulation of TNFα levels also leads to increased nitrates/nitrites, and stabilization of nitric oxide metabolites (51). TNFα acts in concert with IFNγ to induce luteolysis (46, 52). Hojo et al. (53) have demonstrated that necroptosis is involved in structural regression of CL due to receptor-interacting serine/threonine-protein kinase (RIPK)1 and 3 induction following the treatment of the luteal cells with the inflammatory cytokines IFNγ and TNFα in bovine CL (53). Increased RIPK1 and 3 protein expression is also found in PGF2α-induced CL regression, suggesting necroptosis is involved in CL regression. Administration of PGF2α in livestock with functional CL induces luteal regression.
Following functional and structural regression of the CL, proteins in the CL are degraded and removed by regulatory mechanisms. We describe hereafter the proteasome-ubiquitin system and the autophagy mechanisms involved in protein degradation and removal of unnecessary tissue structures.
The UPP plays major roles in the degradation of unnecessary proteins. The targeted proteins are bound by small ubiquitin proteins (Figure 3). The role of ubiquitin is tightly regulated by several enzymes namely ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) (54). On the other hand, the ubiquitinated proteins are degraded by a huge protein complex called the 26S proteasome. The 26S proteasome complex consists of two subclass complexes: the 19S and 20S particles. It is known that the UPP plays important role in the reproductive system (7, 8). Emerging evidence also suggests that dysfunction of the UPS leads to multiple diseases, including the dysfunction of the male and female reproductive tracts (9, 55).
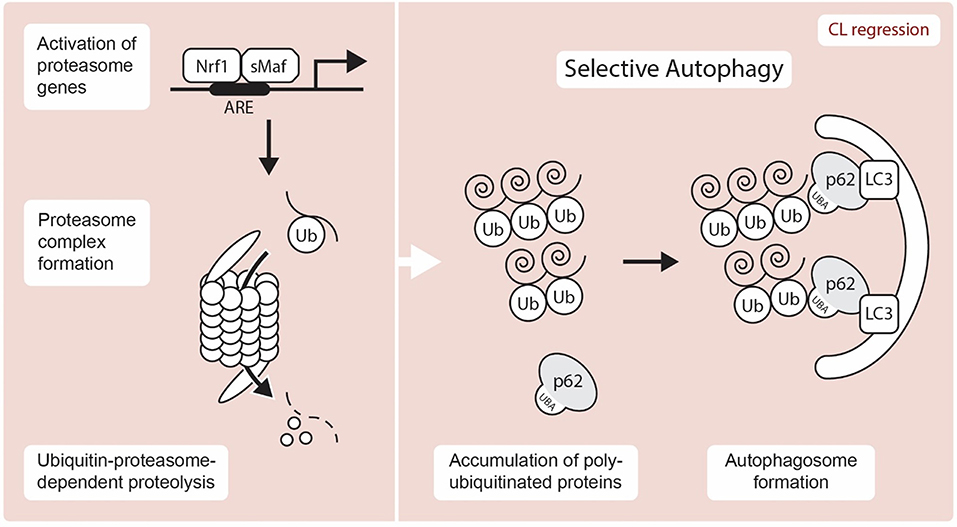
Figure 3. Schematic representation showing UPP and autophagy mediated proteolysis. A transcription factor, Nrf1 is liberated from the ER after stress and translocates to the nucleus, where it induces the expression of proteasome subunit genes through the ARE (antioxidant response element) by hetero-dimerizing with a small Maf (sMaf) protein. Selective autophagy is known to exert proteolysis through the recognition of unnecessary target protein via p62-LC3-autophagosome. Nrf1, NF-E2-related factor 1; sMAF, small musculoaponeurotic fibrosarcoma proteins; ARE, antioxidant response element; Ub, Ubiquitin; LC3, Microtubule-associated protein 1A/1B-light chain 3; UBA, Ubiquitin-Associated domain.
Surprisingly, the UPP system in the CL is not well-characterized and only a very limited number of proteasome genes have been identified in the CL. Nonetheless, the proteasome plays a central role in the degradation of unnecessary proteins, which are labeled with ubiquitin proteins. Since the mammalian CL is renewed after each infertile estrous/menstrual cycle, understanding how its proteins are degraded is an important question that remains unanswered. The proteasome inhibitors MG115 and MG132 reduced both mRNA and protein expression of StAR in the rat adrenal cortex (56). On the other hand, the stability of breast cancer susceptibility gene 1 (BRCA1) and its partner BRCA1-associated RING domain protein 1 (BARD1) is regulated by proteasome degradation in human ovarian GCs (57). This regulatory process is also associated with both cAMP-dependent and cAMP-independent steroidogenic processes. Forskolin-induced Cyp19a expression is blocked by MG132 treatment, which suggests that proteasome inhibition downregulates steroidogenesis. It appears that the proteasome-ubiquitin system plays an important role in steroidogenic gene expression and therefore further functional studies about the roles of UPP in luteal steroidogenesis are required. At the molecular level, NF-E2-related factor 1 (NRF1), a transcription factor involved in the stress response, has garnered significant attention due to its implication in protein clearance via the UPP (Figure 3). NRF1 is a member of the cap “n” collar (CNC)-bZip transcription factor family and is localized at the ER until an external stressor is present. Upon activation by an external stressor, NRF1 is liberated from the ER and translocates to the nucleus, where it stimulates the expression of proteasome subunit genes via the antioxidant response element (ARE) by hetero-dimerizing with a sMAF (small musculoaponeurotic fibrosarcoma) proteins and to maintain protein homeostasis [(58, 59); Figure 3]. Proteasome and deubiquitination enzyme gene expressions are regulated by NRF1 (60, 61). A recent study has demonstrated that deletion of Nrf1 causes downregulation of several proteasome genes with the accumulation of ubiquitinated proteins and p62/SQSTM1, an autophagy marker (61). Much like ubiquitination, deubiquitination also contributes to proteostasis by regulating cellular levels of free monomeric ubiquitin. However, while Proteasome subunit beta type (PSMB) 8 and 9 gene expression, has been identified in the bovine CL (62), the roles of the UPP and its regulation by NRF1 require further study.
Another pathway that has been implicated in CL regression is ER stress pathway (10, 15, 63). The ER stress is a cellular phenomenon induced by diverse stimuli disturbing the protein folding in the ER (64, 65). In response to ER stress, UPR pathway is activated to restore the ER homeostasis. The UPR pathway involves the actions of three signaling proteins: protein kinase RNA-like ER kinase (PERK), inositol-requiring enzyme-1/X-box-binding protein (IRE1/XBP-1), and activating transcription factor 6 (ATF6) (66, 67). The PERK and ATF6 are normally in inactive form due to their association with BiP (Binding immunoglobulin Protein; also known as Glucose-regulated protein-Grp78), an ER resident chaperone (Figure 4). The main role of the CL is to secrete P4, which is essential for maintaining pregnancy. While the steroidogenic process diverges into several separate pathways which lead to the synthesis of different steroid products, StAR is significantly involved in this process and regulates the rate-limiting step in P4 production in the CL. Reduced expression of GRP78, a ER chaperone protein critical for UPR, cause inhibition of StAR protein expression and activity in steroidogenic cells (12). Moreover, female mice lacking Becn1, a regulator of autophagy and the UPR system, have a defect in P4 production in the ovarian GCs and display a preterm labor phenotype (11). These results suggest that the UPR system may regulate P4 production through various mechanisms. On the other hand, Skn1, an ortholog of Nrf1-3 in C. elegans, regulates UPR signaling and transcription factor genes. SKN-1 contributes to the expression of core UPR factors, pek-1 and atf-6 (68). Loss of Skn1 inhibits upregulation of Xbp1 gene expression by ER-stress. Chromatin immunoprecipitation studies indicate that endogenous SKN-1 accumulates at the xbp-1 site of transcription in the presence of ER stress (Figure 4). In the steroidogenic cells, intracellular cholesterol is stored in an esterified form. In response to trophic hormone signals coming from the pituitary (e.g., LH, ACTH), cholesterol esterase hydrolyzes cholesterol ester into free cholesterol in target steroidogenic cells. In luteal cells, cholesterol is converted into different types of steroids including P4 by several steroidogenic enzymes. The gene expression of these enzymes is tightly regulated by several transcription factors. Interestingly, a recent study has revealed that Nrf1 controls cholesterol homeostasis by binding to cholesterol (69). Thus, it is important to determine whether Nrf1 controls steroidogenesis via UPP and UPR modulation in the CL.
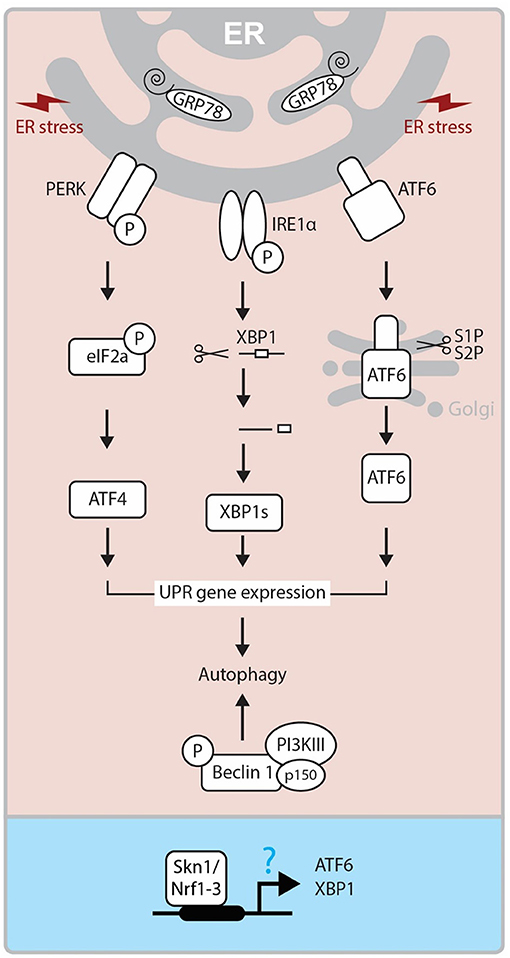
Figure 4. Molecular mechanism of UPR. ER Stress-activated three pathways (IRE, PERK, and ATF6) control the activity of XBP1s, ATF4, ATF6 transcription factors and regulate UPR related gene expressions, which triggers autophagy. On the other hand, Beclin1 forms a complex with PI3KIII and p150, and this also triggers autophagy mechanism. In Caenorhabditis elegans, SKN-1, the ortholog of human Nrf1-3, has been reported to regulate ATF6 and XBP1 expressions at the promoter level. ER, endoplasmic reticulum; GRP78, Glucose-regulated protein; PERK, PKR-like eukaryotic initiation factor 2α (eIF2α) kinase; IRE1, inositol-requiring transmembrane kinase/endoribonuclease 1; ATF6, activating transcription factor-6; XBP1, X-box-binding protein 1; XBP1s, spliced XBP1 protein; S1P and S2P, site-1 and site-2 proteases; PI3KIII, Class III PI 3-kinase; Skn1, Protein skinhead-1.
As an ER transmembrane protein, IRE1 is essential for UPR and is only activated through binding to unfolded proteins (70). Under the conditions of severe and chronic ER stress, the UPR pathway is not able to cope and cellular dysfunction or death ensues through the activation of both the extrinsic and intrinsic apoptosis pathways (71). Apoptosis is initiated by PERK/eIF2α mediated induction of proapoptotic TF CHOP (Transcriptional factor C/EBP homologous protein), and IRE1 dependent activation of TRAF2 (TNF receptor-associated factor 2). This process stimulates the c-Jun NH2-terminal kinase (JNK) pathway (64, 72). The role of ER stress-mediated apoptosis is well-established in many reproductive events, including follicular atresia (73) and CL regression (10, 15, 63). Yang et al. (15) demonstrated that ER stress markers like Grp78, CHOP, ATF6α and caspase 12 are significantly expressed at the mRNA and protein levels in the late luteal stage during spontaneous and PGF2α induced luteolysis (15). These findings were further supported by decreased expression of ER stress markers and apoptosis in luteal cells treated with tauroursodeoxycholic acid, which functions as a chemical chaperone and reduces ER stress (15, 63). Thus, it is plausible that ER stress-mediated UPR controls both functional and structural luteal regression although further studies are needed to elucidate its molecular mechanism.
Redundant cellular components are recognized and transferred to the lysosome by a mechanism known as autophagy. In autophagy, mitophagy selectively degrades mitochondria and pexophagy selectively degrade peroxisomes. Selective autophagy also plays major roles in the degradation of cellular components such as aggregated proteins (74, 75). To recognize target components and properly bring them to the lysosome, an adapter protein involved in autophagy requires at least two domains. One region binds to the target protein and the other domain is necessary for transporting the target to an autophagy mechanism. Autophagy receptor p62 (also known as sequestosome-1) is an adaptor protein which mediates the interaction between selected proteins and autophagosomes. The receptor p62 possesses a LC3-Interacting Region (LIR) motif that interacts with LC3. This interaction allows the receptor p62 to bind to the autophagosomal membrane (Figure 3). p62 co-aggregates with the target at the phagophore due to its homopolymerisation property mediated by its PB1(Phox/Bem1p) domain. Interaction of p62 with Atg8/LC3 on the autophagosomal membrane is extremely important for transport. An increase in the expression of LC3-II relative to the expression of LC3-I occurs during autophagy. Although studies on autophagy in the mammalian CL are limited, the link between autophagy and apoptosis is well-established (76, 77). In fact, expression of various autophagy related factors is increased in the late luteal and regression stages of the CL. Increased expression of beclin protein during the late luteal stage in the sow suggests a role in the removal of unwanted proteins during luteolysis (78). LC3-II, an autophagy marker is expressed more in the late than middle stage CL in cattle (42). Similar results were obtained in rat CL as well (16). These findings suggest that autophagy is highly involved in luteal regression in several species. Lipid droplets are unique organelles in the luteal cells enriched with cholesterol esters, that serve as precursor for steroidogenesis. Inhibition of autophagy in luteal cells via deletion of Becn1 causes failure of lipid droplet formation, and leads to reduced P4 secretion, demonstrating the critical role of Becn1/ autophagy in luteal steroidogenesis (11). Therefore, the relationship between steroid synthesis and autophagy in the CL should be studied. The p62 receptor exerts its physiological functions including signal transduction regulation, intracellular protein localization (trafficking), and selective autophagy of ubiquitinated proteins through its interaction with various proteins (79). The p62 receptor binds to ubiquitin containing aberrantly aggregated proteins. Since p62 has a ubiquitin associated (UBA) domain and binds to ubiquitinated proteins, p62 plays an important role in selective autophagy. Presently, LC3 and p62 are widely used as autophagy markers. Interestingly, recent studies have revealed that the p62 receptor, a primarily cytoplasmic protein, plays an important role in the nucleus by interacting with several nuclear factors (described below, Table 1) that are expressed in the CL (23, 36, 38).
The nuclear transcription factor GATA4 binds to p62 and is degraded through autophagy [(90), Figure 2]. The lysosomal autophagic pathway regulates GATA4 during ionizing radiation-induced and progerin-induced senescence in human mesenchymal stem cells (hMSCs) and fibroblasts, respectively (80, 81). In both fibroblast and hMSCs senescent cells, GATA4 accumulates as a consequence of the loss of physical interaction between p62 and GATA4 (80, 81). While the various functions of GATA factors in luteal P4 regulation are well-characterized, their role in autophagy is yet to be elucidated (Figure 2). Accordingly, SKN-1/Nrf1-3 and ELT-2/GATA transcription factors may regulate the expression of proteasome subunit genes as well as oxidative and heat-stress response genes (91). As NRF family members play prominent roles in the proteasome-autophagy system (61, 92), the relationship between GATA factors and NRF family members in the CL merits further investigation.
Androgen receptor-interacting protein 4 (ARIP4) interacts with sumoylated nuclear receptors such as NR5A1, NR5A2, NR3C1 (GR: glucocorticoid receptor) and NR3C4 (AR: androgen receptor) (93). Multiple studies have shown that steroidogenic genes are regulated by NR5A1. This occurs through direct interaction with transcription factors such as GATA4 (39, 94). The UBA domain of p62 interacts with a novel domain in ARIP4, named SQSTM1/p62 interaction Domain (SID). This domain possesses binding properties similar to ubiquitin. The p62 receptor negatively regulates ARIP4 levels under starvation induced autophagy. This indicates that the interaction between ARIP4 and p62 is involved in the regulation of ARIP4 protein levels during autophagy (82). Considering the dual role of ARIP4 in steroidogenic gene regulation via NR5A1 and autophagy via p62, it will be interesting to further investigate its role during active steroidogenesis and autophagy-mediated CL regression (95, 96).
NR4A, a member of the nuclear receptor superfamily, plays an important role in a variety of cellular processes (97). NR4A1 functions as a nuclear transcriptional factor and activates steroidogenic gene expression in gonadal cells (32, 98). Hu et al. (89) first showed that NR4A1 physically interacts with p62 and accumulates in the mitochondria when mouse embryonic fibroblasts (MEFs) are treated with TNFα and celastrol (89). This suggests that TNFα-mediated apoptosis in the CL may be controlled by NR4A1-dependent regulation of mitochondrial autophagy. Further studies are needed to investigate autophagic regulation of nuclear receptors by p62 in the CL.
TP53 INP1 (tumor protein 53-inducible nuclear protein 1) is a tumor suppressor whose expression is reduced in various cancers. TP53 INP1-LC3 binding occurs via its functional LC3 interaction region (LIR). When TP53 INP1 is highly expressed, TP53 INP1-LC3 interaction is stronger than the p62-LC3 interaction. This inhibits the binding of LC3 to p62 and in turn enhances p62-mediated protein degradation (83). TP53 INP1 also induces autophagy-dependent cell death (83). Since P53 protein is an apoptotic factor in the CL (43), functional analysis of TP53 INP1 in the CL is needed.
PARP-1 [Poly (ADP-ribose) polymerase 1], a key factor in DNA repair, is a partner protein of p62 (85). PARP-1 also binds to LC3 and phosphorylated Unc-51 like autophagy activating kinase 1(ULK1), which are both key factors in autophagy. Rad51 plays a central role in DNA double strand break (DSB) repair through homologous recombination (HR) and also interacts with p62 (84). As autophagy occurs primarily in the cytoplasm, elucidating the role of crosstalk between nuclear localized proteins and autophagy signaling is crucial.
It is worth noting that p62 regulates the binding between Nrf2 and Keap1 (Kelch ECH associating protein 1) although they interact in the nucleus (99). The Nrf2-Keap1 interaction is known to be one of the cellular mechanisms to protect the cells from oxidative stresses. To regulate this mechanism, Nrf2 transcription factor is continuously degraded when its partner protein Keap1 binds. In canonical pathway, the binding of Keap1 controls Nrf2 transcriptional activity (100). However, in non-canonical pathway, p62 binds to Keap1, and this interaction induces a selective autophagy pathway and prevent the interaction between Keap1 and Nrf2 (99). Consequently, Nrf2 transcriptional activity is enhanced and this acts as stress sensor mechanism. To our surprise, many factors identified as nuclear partners of p62 are transcription factors involved in gene regulation of steroidogenic enzymes. Thus, in the near future, the relationship between luteal functional regulation and autophagy-controlled luteal regression will be clarified and we will have better understanding of how luteal function (P4 production) and regression (apoptosis/autophagy) are orchestrated.
Understanding the intracellular homeostatic mechanisms during the maintenance and lysis of CL will pave way for addressing infertility due to luteal dysfunction. Previous research has suggested that dysfunction of the ubiquitin-proteasome and autophagy systems leads to many disorders, including diseases of the male and female reproductive system. However, its molecular basis is not well-studied. We hope that transcriptional regulation of proteasome and autophagy systems during luteolysis will be unraveled in the near future. Elucidation of the transcription factor-proteasome/autophagy axis also could enable efficient recovery from stress situations which will lead to a significant advancement in the field of animal reproduction.
AT and HT conceived, coordinated, wrote and edited the manuscript. AT, NK, HO, and MT wrote, interpreted data, and revised the manuscript. PL, MŚ, and DS edited and revised the manuscript.
This study was supported by a Polish grant from the KNOW (Leading National Research Centre) Scientific Consortium Healthy Animal - Safe Food, decision of Ministry of Science and Higher Education No. 05-1/KNOW2/2015. This study was also supported by the Polish National Science Center MINIATURA1 (2017/01/X/NZ9/01263) for HT.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Dr. Tobias Hohenauer for help with illustrations.
1. Lavoie HA, King SR. Transcriptional regulation of steroidogenic genes: STARD1, CYP11A1 and HSD3B. Exp Biol Med. (2009) 234:880–907. doi: 10.3181/0903-MR-97
2. Bacci ML, Barazzoni AM, Forni M, Costerbosa GL. In situ detection of apoptosis in regressing corpus luteum of pregnant sow: evidence of an early presence of DNA fragmentation. Domest Anim Endocrinol. (1996) 13:361–72. doi: 10.1016/0739-7240(96)00049-5
3. Juengel JL, Garverick HA, Johnson AL, Youngquist RS, Smith MF. Apoptosis during luteal regression in cattle. Endocrinology. (1993) 132:249–54. doi: 10.1210/endo.132.1.8419126
4. Rueda BR, Tilly KI, Botros IW, Jolly PD, Hansen TR, Hoyer PB, et al. Increased bax and interleukin-1beta-converting enzyme messenger ribonucleic acid levels coincide with apoptosis in the bovine corpus luteum during structural regression. Biol Reprod. (1997) 56:186–93. doi: 10.1095/biolreprod56.1.186
5. Rueda BR, Wegner JA, Marion SL, Wahlen DD, Hoyer PB. Internucleosomal DNA fragmentation in ovine luteal tissue associated with luteolysis: in vivo and in vitro analyses. Biol Reprod. (1995) 52:305–12. doi: 10.1095/biolreprod52.2.305
6. Reichmann D, Voth W, Jakob U. Maintaining a healthy proteome during oxidative stress. Mol Cell. (2018) 69:203–13. doi: 10.1016/j.molcel.2017.12.021
7. Bose R, Manku G, Culty M, Wing SS. Ubiquitin-proteasome system in spermatogenesis. Adv Exp Med Biol. (2014) 759:181–213. doi: 10.1007/978-1-4939-0817-2_9
8. Yi YJ, Nagyova E, Manandhar G, Prochazka R, Sutovsky M, Park CS, et al. Proteolytic activity of the 26S proteasome is required for the meiotic resumption, germinal vesicle breakdown, and cumulus expansion of porcine cumulus-oocyte complexes matured in vitro. Biol Reprod. (2008) 78:115–26. doi: 10.1095/biolreprod.107.061366
9. Hou CC, Yang WX. New insights to the ubiquitin-proteasome pathway (UPP) mechanism during spermatogenesis. Mol Biol Rep. (2013) 40:3213–30. doi: 10.1007/s11033-012-2397-y
10. Park HJ, Park SJ, Koo DB, Kong IK, Kim MK, Kim JM, et al. Unfolding protein response signaling is involved in development, maintenance, and regression of the corpus luteum during the bovine estrous cycle. Biochem Biophys Res Commun. (2013) 441:344–50. doi: 10.1016/j.bbrc.2013.10.056
11. Gawriluk TR, Ko C, Hong X, Christenson LK, Rucker EB III. Beclin-1 deficiency in the murine ovary results in the reduction of progesterone production to promote preterm labor. Proc Natl Acad Sci USA. (2014) 111:E4194–203. doi: 10.1073/pnas.1409323111
12. Prasad M, Pawlak KJ, Burak WE, Perry EE, Marshall B, Whittal RM, et al. Mitochondrial metabolic regulation by GRP78. Sci Adv. (2017) 3:e1602038. doi: 10.1126/sciadv.1602038
13. Chen H, Jin S, Guo J, Kombairaju P, Biswal S, Zirkin BR. Knockout of the transcription factor Nrf2: effects on testosterone production by aging mouse Leydig cells. Mol Cell Endocrinol. (2015) 409:113–20. doi: 10.1016/j.mce.2015.03.013
14. Lilienbaum A. Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol. (2013) 4:1–26.
15. Yang Y, Sun M, Shan Y, Zheng X, Ma H, Ma W, et al. Endoplasmic reticulum stress-mediated apoptotic pathway is involved in corpus luteum regression in rats. Reprod Sci. (2015) 22:572–84. doi: 10.1177/1933719114553445
16. Choi J, Jo M, Lee E, Choi D. The role of autophagy in corpus luteum regression in the rat. Biol Reprod. (2011) 85:465–72. doi: 10.1095/biolreprod.111.091314
17. Gilchrist RL, Ryu KS, Ji I, Ji TH. The luteinizing hormone/chorionic gonadotropin receptor has distinct transmembrane conductors for cAMP and inositol phosphate signals. J Biol Chem. (1996) 271:19283–7. doi: 10.1074/jbc.271.32.19283
18. Kallen CB, Arakane F, Christenson LK, Watari H, Devoto L, Strauss JF III. Unveiling the mechanism of action and regulation of the steroidogenic acute regulatory protein. Mol Cell Endocrinol. (1998) 145:39–45. doi: 10.1016/S0303-7207(98)00167-1
19. Sasano H, Suzuki T. Localization of steroidogenesis and steroid receptors in human corpus luteum. Classification of human corpus luteum (CL) into estrogen-producing degenerating CL, and nonsteroid-producing degenerating CL. Semin Reprod Endocrinol. (1997) 15:345–51. doi: 10.1055/s-2008-1068372
20. Morohashi K, Hatano O, Nomura M, Takayama K, Hara M, Yoshii H, et al. Function and distribution of a steroidogenic cell-specific transcription factor, Ad4BP. J Steroid Biochem Mol Biol. (1995) 53:81–8. doi: 10.1016/0960-0760(95)00041-W
21. Sadovsky Y, Crawford PA, Woodson KG, Polish JA, Clements MA, Tourtellotte LM, et al. Mice deficient in the orphan receptor steroidogenic factor 1 lack adrenal glands and gonads but express P450 side-chain-cleavage enzyme in the placenta and have normal embryonic serum levels of corticosteroids. Proc Natl Acad Sci USA. (1995) 92:10939–43. doi: 10.1073/pnas.92.24.10939
22. King SR, LaVoie HA. Gonadal transactivation of STARD1, CYP11A1 and HSD3B. Front Biosci. (2012) 17:824–46. doi: 10.2741/3959
23. Taniguchi H, Komiyama J, Viger RS, Okuda K. The expression of the nuclear receptors NR5A1 and NR5A2 and transcription factor GATA6 correlates with steroidogenic gene expression in the bovine corpus luteum. Mol Reprod Dev. (2009) 76:873–80. doi: 10.1002/mrd.21054
24. Liu Z, Simpson ER. Steroidogenic factor 1 (SF-1) and SP1 are required for regulation of bovine CYP11A gene expression in bovine luteal cells and adrenal Y1 cells. Mol Endocrinol. (1997) 11:127–37. doi: 10.1210/me.11.2.127
25. Mamluk R, Greber Y, Meidan R. Hormonal regulation of messenger ribonucleic acid expression for steroidogenic factor-1, steroidogenic acute regulatory protein, and cytochrome P450 side-chain cleavage in bovine luteal cells. Biol Reprod. (1999) 60:628–34. doi: 10.1095/biolreprod60.3.628
26. Sugawara T, Kiriakidou M, McAllister JM, Kallen CB, Strauss JF III. Multiple steroidogenic factor 1 binding elements in the human steroidogenic acute regulatory protein gene 5'-flanking region are required for maximal promoter activity and cyclic AMP responsiveness. Biochemistry. (1997) 36:7249–55. doi: 10.1021/bi9628984
27. Jeyasuria P, Ikeda Y, Jamin SP, Zhao L, De Rooij DG, Themmen AP, et al. Cell-specific knockout of steroidogenic factor 1 reveals its essential roles in gonadal function. Mol Endocrinol. (2004) 18:1610–9. doi: 10.1210/me.2003-0404
28. Duggavathi R, Volle DH, Mataki C, Antal MC, Messaddeq N, Auwerx J, et al. Liver receptor homolog 1 is essential for ovulation. Genes Dev. (2008) 22:1871–6. doi: 10.1101/gad.472008
29. Zhang C, Large MJ, Duggavathi R, DeMayo FJ, Lydon JP, Schoonjans K, et al. Liver receptor homolog-1 is essential for pregnancy. Nat Med. (2013) 19:1061–6. doi: 10.1038/nm.3192
30. Bertolin K, Gossen J, Schoonjans K, Murphy BD. The orphan nuclear receptor Nr5a2 is essential for luteinization in the female mouse ovary. Endocrinology. (2014) 155:1931–43. doi: 10.1210/en.2013-1765
31. Meinsohn MC, Morin F, Bertolin K, Duggavathi R, Schoonjans K, Murphy BD. The orphan nuclear receptor liver homolog receptor-1 (Nr5a2) regulates ovarian granulosa cell proliferation. J Endocr Soc. (2017) 2:24–41. doi: 10.1210/js.2017-00329
32. Li M, Xue K, Ling J, Diao FY, Cui YG, Liu JY. The orphan nuclear receptor NR4A1 regulates transcription of key steroidogenic enzymes in ovarian theca cells. Mol Cell Endocrinol. (2010) 319:39–46. doi: 10.1016/j.mce.2010.01.014
33. Park JI, Park HJ, Choi HS, Lee K, Lee WK, Chun SY. Gonadotropin regulation of NGFI-B messenger ribonucleic acid expression during ovarian follicle development in the rat. Endocrinology. (2001) 142:3051–9. doi: 10.1210/en.142.7.3051
34. Martin LJ, Boucher N, Brousseau C, Tremblay JJ. The orphan nuclear receptor NUR77 regulates hormone-induced StAR transcription in Leydig cells through cooperation with Ca2+/calmodulin-dependent protein kinase I. Mol Endocrinol. (2008) 22:2021–37. doi: 10.1210/me.2007-0370
35. Martin LJ, Tremblay JJ. The nuclear receptors NUR77 and SF1 play additive roles with c-JUN through distinct elements on the mouse Star promoter. J Mol Endocrinol. (2009) 42:119–29. doi: 10.1677/JME-08-0095
36. Qi L, Guo N, Wei Q, Jin P, Wang W, Mao D. The involvement of NR4A1 and NR4A2 in the regulation of the luteal function in rats. Acta Histochem. (2018) 120:713–19. doi: 10.1016/j.acthis.2018.07.007
37. Viger RS, Guittot SM, Anttonen M, Wilson DB, Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Mol Endocrinol. (2008) 22:781–98. doi: 10.1210/me.2007-0513
38. Convissar SM, Bennett J, Baumgarten SC, Lydon JP, DeMayo FJ, Stocco C. GATA4 and GATA6 knockdown during luteinization inhibits progesterone production and gonadotropin responsiveness in the corpus luteum of female mice. Biol Reprod. (2015) 93:1–10. doi: 10.1095/biolreprod.115.132969
39. Martin LJ, Taniguchi H, Robert NM, Simard J, Tremblay JJ, Viger RS. GATA factors and the nuclear receptors, steroidogenic factor 1/liver receptor homolog 1, are key mutual partners in the regulation of the human 3beta-hydroxysteroid dehydrogenase type 2 promoter. Mol Endocrinol. (2005) 19:2358–70. doi: 10.1210/me.2004-0257
40. Sandhoff TW, McLean MP. Repression of the rat steroidogenic acute regulatory (StAR) protein gene by PGF2alpha is modulated by the negative transcription factor DAX-1. Endocrine. (1999) 10:83–91. doi: 10.1385/ENDO:10:1:83
41. Tremblay JJ, Viger RS. Nuclear receptor Dax-1 represses the transcriptional cooperation between GATA-4 and SF-1 in Sertoli cells. Biol Reprod. (2001) 64:1191–9. doi: 10.1095/biolreprod64.4.1191
42. Aboelenain M, Kawahara M, Balboula AZ, Montasser A, Zaabel SM, Okuda K, et al. Status of autophagy, lysosome activity and apoptosis during corpus luteum regression in cattle. J Reprod Dev. (2015) 61:229–36. doi: 10.1262/jrd.2014-135
43. Nakamura T, Sakamoto K. Reactive oxygen species up-regulates cyclooxygenase-2, p53, and Bax mRNA expression in bovine luteal cells. Biochem Biophys Res Commun. (2001) 284:203–10. doi: 10.1006/bbrc.2001.4927
44. Trott EA, Plouffe L Jr, Hansen K, McDonough PG, George P, Khan I. The role of p53 tumor suppressor gene and bcl-2 protooncogene in rat corpus luteum death. Am J Obstet Gynecol. (1997) 177:327–31. discussion 331–2. doi: 10.1016/S0002-9378(97)70194-7
45. Rolaki A, Drakakis P, Millingos S, Loutradis D, Makrigiannakis A. Novel trends in follicular development, atresia and corpus luteum regression: a role for apoptosis. Reprod Biomed Online. (2005) 11:93–103. doi: 10.1016/S1472-6483(10)61304-1
46. Taniguchi H, Yokomizo Y, Okuda K. Fas-Fas ligand system mediates luteal cell death in bovine corpus luteum. Biol Reprod. (2002) 66:754–9. doi: 10.1095/biolreprod66.3.754
47. Penny LA, Armstrong D, Bramley TA, Webb R, Collins RA, Watson ED. Immune cells and cytokine production in the bovine corpus luteum throughout the oestrous cycle and after induced luteolysis. J Reprod Fertil. (1999) 115:87–96. doi: 10.1530/jrf.0.1150087
48. Jo T, Tomiyama T, Ohashi K, Saji F, Tanizawa O, Ozaki M, et al. Apoptosis of cultured mouse luteal cells induced by tumor necrosis factor-alpha and interferon-gamma. Anat Rec. (1995) 241:70–6. doi: 10.1002/ar.1092410110
49. Niswender GD, Juengel JL, Silva PJ, Rollyson MK, McIntush EW. Mechanisms controlling the function and life span of the corpus luteum. Physiol Rev. (2000) 80:1–29. doi: 10.1152/physrev.2000.80.1.1
50. Shaw DW, Britt JH. Concentrations of tumor necrosis factor alpha and progesterone within the bovine corpus luteum sampled by continuous-flow microdialysis during luteolysis in vivo. Biol Reprod. (1995) 53:847–54. doi: 10.1095/biolreprod53.4.847
51. Skarzynski DJ, Bah MM, Deptula KM, Woclawek-Potocka I, Korzekwa A, Shibaya M, et al. Roles of tumor necrosis factor-α of the estrous cycle in cattle: an in vivo study. Biol Reprod. (2003) 69:1907–13. doi: 10.1095/biolreprod.103.016212
52. Petroff MG, Petroff BK, Pate JL. Mechanisms of cytokine-induced death of cultured bovine luteal cells. Reproduction. (2001) 121:753–60. doi: 10.1530/rep.0.1210753
53. Hojo T, Siemieniuch MJ, Lukasik K, Piotrowska-Tomala KK, Jonczyk AW, Okuda K, et al. Programmed necrosis - a new mechanism of steroidogenic luteal cell death and elimination during luteolysis in cows. Sci Rep. (2016) 6:38211. doi: 10.1038/srep38211
54. Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. (2009) 85:12–36. doi: 10.2183/pjab.85.12
55. Hu X, Roberts JR, Apopa PL, Kan YW, Ma Q. Accelerated ovarian failure induced by 4-vinyl cyclohexene diepoxide in Nrf2 null mice. Mol Cell Biol. (2006) 26:940–54. doi: 10.1128/MCB.26.3.940-954.2006
56. Rucinski M, Tortorella C, Ziolkowska A, Nowak M, Nussdorfer GG, Malendowicz LK. Steroidogenic acute regulatory protein gene expression, steroid-hormone secretion and proliferative activity of adrenocortical cells in the presence of proteasome inhibitors: in vivo studies on the regenerating rat adrenal cortex. Int J Mol Med. (2008) 21:593–7. doi: 10.3892/ijmm.21.5.593
57. Lu Y, Amleh A, Sun J, Jin X, McCullough SD, Baer R, et al. Ubiquitination and proteasome-mediated degradation of BRCA1 and BARD1 during steroidogenesis in human ovarian granulosa cells. Mol Endocrinol. (2007) 21:651–63. doi: 10.1210/me.2006-0188
58. Katsuoka F, Yamamoto M. Small Maf proteins (MafF, MafG, MafK): history, structure and function. Gene. (2016) 586:197–205. doi: 10.1016/j.gene.2016.03.058
59. Kim HM, Han JW, Chan JY. Nuclear factor erythroid-2 like 1 (NFE2L1): structure, function and regulation. Gene. (2016) 584:17–25. doi: 10.1016/j.gene.2016.03.002
60. Taniguchi H, Okamuro S, Koji M, Waku T, Kubo K, Hatanaka A, et al. Possible roles of the transcription factor Nrf1 (NFE2L1) in neural homeostasis by regulating the gene expression of deubiquitinating enzymes. Biochem Biophys Res Commun. (2017) 484:176–83. doi: 10.1016/j.bbrc.2017.01.038
61. Tsuchiya Y, Taniguchi H, Ito Y, Morita T, Karim MR, Ohtake N, et al. The casein kinase 2-nrf1 axis controls the clearance of ubiquitinated proteins by regulating proteasome gene expression. Mol Cell Biol. (2013) 33:3461–72. doi: 10.1128/MCB.01271-12
62. Cannon MJ, Pate JL. Expression and regulation of interferon gamma-inducible proteasomal subunits LMP7 and LMP10 in the bovine corpus luteum. Biol Reprod. (2003) 68:1447–54. doi: 10.1095/biolreprod.102.010249
63. Park HJ, Park SJ, Koo DB, Lee SR, Kong IK, Ryoo JW, et al. Progesterone production is affected by unfolded protein response (UPR) signaling during the luteal phase in mice. Life Sci. (2014) 113:60–7. doi: 10.1016/j.lfs.2014.07.033
64. Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. (2013) 1833:3460–70. doi: 10.1016/j.bbamcr.2013.06.028
65. Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci USA. (2005) 102:18773–84. doi: 10.1073/pnas.0509487102
66. Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. (2011) 333:1891–4. doi: 10.1126/science.1209126
67. Olzmann JA, Kopito RR, Christianson JC. The mammalian endoplasmic reticulum-associated degradation system. Cold Spring Harb Perspect Biol. (2013) 5:10. doi: 10.1101/cshperspect.a013185
68. Glover-Cutter KM, Lin S, Blackwell TK. Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. (2013) 9:e1003701. doi: 10.1371/journal.pgen.1003701
69. Widenmaier SB, Snyder NA, Nguyen TB, Arduini A, Lee GY, Arruda AP, et al. NRF1 is an ER membrane sensor that is central to cholesterol homeostasis. Cell. (2017) 171:1094–109.e15. doi: 10.1016/j.cell.2017.10.003
70. Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol Rev. (2011) 91:1219–43. doi: 10.1152/physrev.00001.2011
71. Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, et al. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. (2010) 8:e1000415. doi: 10.1371/journal.pbio.1000415
72. Rasheva VI, Domingos PM. Cellular responses to endoplasmic reticulum stress and apoptosis. Apoptosis. (2009) 14:996–1007. doi: 10.1007/s10495-009-0341-y
73. Lin P, Yang Y, Li X, Chen F, Cui C, Hu L, et al. Endoplasmic reticulum stress is involved in granulosa cell apoptosis during follicular atresia in goat ovaries. Mol Reprod Dev. (2012) 79:423–32. doi: 10.1002/mrd.22045
74. Svenning S, Johansen T. Selective autophagy. Essays Biochem. (2013) 55:79–92. doi: 10.1042/bse0550079
75. Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol. (2016) 428(9 Pt A):1714–24. doi: 10.1016/j.jmb.2016.02.004
76. Patra S, Panigrahi DP, Praharaj PP, Bhol CS, Mahapatra KK, Mishra SR, et al. Dysregulation of histone deacetylases in carcinogenesis and tumor progression: a possible link to apoptosis and autophagy. Cell Mol Life Sci. (2019) 76:3263. doi: 10.1007/s00018-019-03098-1
77. Zhou J, Peng X, Mei S. Autophagy in ovarian follicular development and Atresia. Int J Biol Sci. (2019) 15:726–37. doi: 10.7150/ijbs.30369
78. Grzesiak M, Michalik A, Rak A, Knapczyk-Stwora K, Pieczonka A. The expression of autophagy-related proteins within the corpus luteum lifespan in pigs. Domest Anim Endocrinol. (2018) 64:9–16. doi: 10.1016/j.domaniend.2018.03.004
79. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. (2011) 147:728–41. doi: 10.1016/j.cell.2011.10.026
80. Kang C, Xu Q, Martin TD, Li MZ, Demaria M, Aron L, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. (2015) 349:aaa5612. doi: 10.1126/science.aaa5612
81. Lee JY, Yu KR, Lee BC, Kang I, Kim JJ, Jung EJ, et al. GATA4-dependent regulation of the secretory phenotype via MCP-1 underlies lamin A-mediated human mesenchymal stem cell aging. Exp Mol Med. (2018) 50:63. doi: 10.1038/s12276-018-0092-3
82. Tsuchiya M, Isogai S, Taniguchi H, Tochio H, Shirakawa M, Morohashi K, et al. Selective autophagic receptor p62 regulates the abundance of transcriptional coregulator ARIP4 during nutrient starvation. Sci Rep. (2015) 5:14498. doi: 10.1038/srep14498
83. Seillier M, Peuget S, Gayet O, Gauthier C, N'Guessan P, Monte M, et al. TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell Death Differ. (2012) 19:1525–35. doi: 10.1038/cdd.2012.30
84. Hewitt G, Carroll B, Sarallah R, Correia-Melo C, Ogrodnik M, Nelson G, et al. SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy. (2016) 12:1917–30. doi: 10.1080/15548627.2016.1210368
85. Yan S, Liu L, Ren F, Gao Q, Xu S, Hou B, et al. Sunitinib induces genomic instability of renal carcinoma cells through affecting the interaction of LC3-II and PARP-1. Cell Death Dis. (2017) 8:e2988. doi: 10.1038/cddis.2017.387
86. Wild P, McEwan DG, Dikic I. The LC3 interactome at a glance. J Cell Sci. (2014) 127(Pt 1):3–9. doi: 10.1242/jcs.140426
87. Mitani T, Minami M, Harada N, Ashida H, Yamaji R. Autophagic degradation of the androgen receptor mediated by increased phosphorylation of p62 suppresses apoptosis in hypoxia. Cell Signal. (2015) 27:1994–2001. doi: 10.1016/j.cellsig.2015.07.009
88. Diradourian C, Le May C, Cauzac M, Girard J, Burnol AF, Pegorier JP. Involvement of ZIP/p62 in the regulation of PPARalpha transcriptional activity by p38-MAPK. Biochim Biophys Acta. (2008) 1781:239–44. doi: 10.1016/j.bbalip.2008.02.002
89. Hu M, Luo Q, Alitongbieke G, Chong S, Xu C, Xie L, et al. Celastrol-induced Nur77 interaction with TRAF2 alleviates inflammation by promoting mitochondrial ubiquitination and autophagy. Mol Cell. (2017) 66:141–53.e6. doi: 10.1016/j.molcel.2017.03.008
90. Kobayashi S, Volden P, Timm D, Mao K, Xu X, Liang Q. Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J Biol Chem. (2010) 285:793–804. doi: 10.1074/jbc.M109.070037
91. Keith SA, Maddux SK, Zhong Y, Chinchankar MN, Ferguson AA, Ghazi A, Fisher AL. Graded proteasome dysfunction in caenorhabditis elegans activates an adaptive response involving the conserved SKN-1 and ELT-2 transcription factors and the autophagy-lysosome pathway. PLoS Genet. (2016) 12:e1005823. doi: 10.1371/journal.pgen.1005823
92. Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. (2003) 23:8786–94. doi: 10.1128/MCB.23.23.8786-8794.2003
93. Ogawa H, Komatsu T, Hiraoka Y, Morohashi K. Transcriptional suppression by transient recruitment of ARIP4 to sumoylated nuclear receptor Ad4BP/SF-1. Mol Biol Cell. (2009) 20:4235–45. doi: 10.1091/mbc.e08-12-1247
94. Bouchard MF, Taniguchi H, Viger RS. The effect of human GATA4 gene mutations on the activity of target gonadal promoters. J Mol Endocrinol. (2009) 42:149–60. doi: 10.1677/JME-08-0089
95. Baba T, Otake H, Inoue M, Sato T, Ishihara Y, Moon JY, et al. Ad4BP/SF-1 regulates cholesterol synthesis to boost the production of steroids. Commun Biol. (2018) 1:18. doi: 10.1038/s42003-018-0020-z
96. Syu JS, Baba T, Huang JY, Ogawa H, Hsieh CH, Hu JX, et al. Lysosomal activity maintains glycolysis and cyclin E1 expression by mediating Ad4BP/SF-1 stability for proper steroidogenic cell growth. Sci Rep. (2017) 7:240. doi: 10.1038/s41598-017-00393-4
97. Beard JA, Tenga A, Chen T. The interplay of NR4A receptors and the oncogene-tumor suppressor networks in cancer. Cell Signal. (2015) 27:257–66. doi: 10.1016/j.cellsig.2014.11.009
98. Martin LJ, Tremblay JJ. The human 3beta-hydroxysteroid dehydrogenase/Delta5-Delta4 isomerase type 2 promoter is a novel target for the immediate early orphan nuclear receptor Nur77 in steroidogenic cells. Endocrinology. (2005) 146:861–9. doi: 10.1210/en.2004-0859
99. Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. (2010) 12:213–23. doi: 10.1038/ncb2021
Keywords: ubiquitin-proteasome, autophagy, steroidogenesis, corpus luteum, transcription factors
Citation: Teeli AS, Leszczyński P, Krishnaswamy N, Ogawa H, Tsuchiya M, Śmiech M, Skarzynski D and Taniguchi H (2019) Possible Mechanisms for Maintenance and Regression of Corpus Luteum Through the Ubiquitin-Proteasome and Autophagy System Regulated by Transcriptional Factors. Front. Endocrinol. 10:748. doi: 10.3389/fendo.2019.00748
Received: 29 April 2019; Accepted: 16 October 2019;
Published: 19 November 2019.
Edited by:
Jens Vanselow, Leibniz Institute for Farm Animal Biology, GermanyReviewed by:
Andrea S. Cupp, University of Nebraska-Lincoln, United StatesCopyright © 2019 Teeli, Leszczyński, Krishnaswamy, Ogawa, Tsuchiya, Śmiech, Skarzynski and Taniguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroaki Taniguchi, aC50YW5pZ3VjaGlAaWdoei5wbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.